
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 16 December 2020
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.608589
This article is part of the Research TopicSignaling Events in Regulating Leaf SenescenceView all 11 articles
Leaf senescence is regulated by a large number of internal and environmental factors. Here, we report that AtUSR1 (U-box Senescence Related 1) which encodes a plant Ring/U-box protein, is involved in age-dependent and dark-induced leaf senescence in Arabidopsis. Expression of AtUSR1 gene in leaves was up-regulated in darkness and during aging. Plants of usr1, an AtUSR1 gene knock-down mutant, showed a significant delay in age-dependent and dark-induced leaf senescence and the delayed senescence phenotype was rescued when the AtUSR1 gene was transferred back to the mutant plants. Meanwhile, overexpression of AtUSR1 caused accelerated leaf senescence. Furthermore, the role of AtUSR1 in regulating leaf senescence is related to MYC2-mediuated jasmonic acid (JA) signaling pathway. MeJA treatments promoted the accumulation of AtUSR1 transcripts and this expression activation was dependent on the function of MYC2, a key transcription factor in JA signaling. Dual-luciferase assay results indicated that MYC2 promoted the expression of AtUSR1. Overexpression of AtUSR1 in myc2 mutant plants showed precocious senescence, while myc2 mutation alone caused a delay in leaf senescence, suggesting that AtUSR1 functions downstream to MYC2 in the JA signaling pathway in promoting leaf senescence.
As a process of programmed cell death, leaf senescence is important in plants’ development and response to environmental stresses (Guo and Gan, 2005; Kim et al., 2018). Under harsh environmental conditions, in order to complete their life cycle, stressed plants often endeavor to reallocate nutrients to reproductive organs via the process of leaf senescence (Sedigheh et al., 2011). Leaf senescence can be induced by a large number of endogenous and environmental factors including age, plant hormones, light conditions, abiotic stresses, and pathogen infection. Once senescence is initiated, significant changes in gene expression, metabolic, and physiological activities take place and the execution of leaf senescence eventually leads to programmed cell death (Quirino et al., 2000; Khanna-Chopra, 2012; Guo, 2013). During senescence, visible leaf yellowing can be observed as the result of chloroplast degeneration which is associated with drastic metabolic reprogramming including degradation of macromolecules, enhanced reactive oxygen species (ROS), and occurrence of membrane ion leakage (Ansari et al., 2014; Gan, 2018). During senescence, the expression of genes related to photosynthesis is down-regulated, whereas genes involved in senescence execution generally have increased expression. Genes encoding components of protein degradation pathways, for example, comprise a significant proportion of Senescence Associated Genes (SAGs; Guo et al., 2004; Lim et al., 2007; Gregersen et al., 2008; Zhang et al., 2010). As an essential part of protein degradation, the ubiquitin/26S proteasome pathway is not only important in targeting and degrading proteins, but also involved in regulation of the senescence process. Mutation in several components in the ubiquitin/proteasome system led to altered leaf senescence (Woo et al., 2001; So et al., 2019; Shim et al., 2020), suggesting a regulatory role of the ubiquitin pathway in leaf senescence. As a critical component of the ubiquitin pathway, E3 ubiquitin ligase is involved in polyubiquitination of target proteins and participates in various plant development processes including senescence. In rice, RING-type ubiquitin ligase GW2 acts as a positive regulator of leaf senescence by affecting chlorophyll degradation (Shim et al., 2020). Senescence-associated E3 ubiquitin ligase1/Plant U-box44 (SAUL1/PUB44) regulates dark-induced leaf senescence in Arabidopsis by affecting stability of the chloroplast-localized Abscisic acid (ABA)-responsive factor Senescence-Associated Protein (CSAP; So et al., 2019). RING-type E3 ligases SEVEN IN ABSENTIA OF ARABIDOPSIS THALIANA1 (SINAT1), SINAT2, and SINAT6 control autophagosome formation and leaf senescence via controlling stability of the autophagy-related protein ATG13 in Arabidopsis (Qi et al., 2020). U-box E3 ubiquitin ligases PUB12 and PUB13 play multiple functions in immunity, flowering, and senescence. Loss-of-function mutations of PUB12 and PUB13 led to early senescence phenotypes in Arabidopsis under stress conditions (Zhou et al., 2015). BIG BROTHER (BB) also encodes an E3 ligase and is involved in organ size and leaf senescence. Arabidopsis plants overexpressing BB displayed early senescence, while bb mutants showed delayed senescence phenotypes (Vanhaeren et al., 2017). In rice, dark-induced senescence results in releasing of cytochrome f (Cyt f) from chloroplasts into the cytoplasm, where Cyt f functions to activate caspase-3-like activities by interacting with E3-ubiquitin ligases and RPN9b, the subunit of the ubiquitin proteasome system, ultimately resulting in programmed cell death (PCD) process (Wang et al., 2014).
Phytohormones play very important roles during leaf senescence. ABA, ethylene, jasmonic acid (JA), and salicylic acid (SA) promote senescence, while cytokinins and auxin inhibit this process (Zhang and Guo, 2018; Woo et al., 2019). As a positive regulator of leaf senescence, JA accumulation increases during senescence (He et al., 2002; Hu et al., 2017). Activation of the JA biosynthetic gene LIPOXYGENASE2 (LOX2) by transcription factor TEOSINTE BRANCHED/CYCLOIDEA/PCF4 (TCP4) resulted in early senescence (Suraneni et al., 2010). JA-biosynthetic enzyme 3-ketoacyl-CoA thiolase 2 (KAT2) participates in the catabolism associated with senescence, as well in the early events required for leaf senescence (Castillo and Leon, 2008). As a JA receptor, CORONATINE INSENSITIVE 1 (COI1) is an F-box protein forming complexes with JASMONATE ZIM-domain proteins (JAZ) to mediate their degradation via the 26S proteasome pathway (Xie et al., 1998; Thines et al., 2007; Shan et al., 2011; Kim et al., 2013). The JA-insensitive coi1-1 mutant displays delayed leaf senescence in Arabidopsis (Castillo and Leon, 2008). In the rice (Oryza sativa) genome, there are three COI homologs named OsCOI1a, OsCOI1b, and OsCOI2, respectively. oscoi1b-1 mutant plants displayed delayed leaf senescence under dark and natural conditions. 35S:OsCOI1a or 35S:OsCOI1b could rescue the delayed senescence phenotype of coi1-1 in Arabidopsis, indicating that both OsCOI1a and OsCOI1b play a role in promoting leaf senescence in rice (Lee et al., 2015). JAZs function as negative regulators of the JA signaling pathway via repressing transcriptional activation activities of downstream transcription factors (Chini et al., 2007; Song et al., 2011). JAZ proteins form complex with NOVEL INTERACTOR of JAZs (NINJA)/TOPLESS to restrain JA response by directly regulating various transcription factors (Pauwels et al., 2010; Causier et al., 2012; Kim et al., 2013). JAZ4 and JAZ8 were reported to inhibit JA–induced senescence through interacting with transcription factor WRKY57, which was proposed to play as a balance internode between JA and auxin signaling in regulating senescence (Jiang et al., 2014). JA also interacts with ethylene in regulating senescence in which JAZ proteins interact with ETHYLENE-INSENSITIVE3(EIN3) and ETHYLENE-INSENSITIVE3-LIKE 1(EIL1), two of the key components of the ethylene signaling pathway (Zhu et al., 2011). Furthermore, crosstalk between JA and SA in regulating leaf senescence has been reported. Senescence regulating TF WRKY53 was found to be antagonistically regulated by JA and SA signals (Miao and Zentgraf, 2007). Among the proteins inhibited by JAZ complexes, MYC2, MYC3, MYC4, and MYC5 displayed accelerated protein degradation under dark or shade conditions but were stabilized in light and after JA treatments (Zhu et al., 2015; Yu et al., 2016). These MYC transcription factors have been shown to be involved in multiple JA response processes via regulating expression of JA responsive genes (Figueroa and Browse, 2012; Zhu et al., 2015; Hu et al., 2017). Recently, It was reported that JA inducible gene DNA binding-with-one-finger 2.1(Dof2.1) functions in enhancing JA-induced leaf senescence via a MYC2–Dof2.1–MYC2 feed-forward loop (Zhuo et al., 2020).
Dark-induced leaf senescence has been widely studied, in which Ethylene, JA, and Nitric oxide (NO) all play regulatory roles (Fujiki et al., 2001). NO was shown to be involved in regulating dark-induced senescence via ETHYLENE INSENSITIVE 2(EIN2) of the ethylene signaling pathway (Niu and Guo, 2012; Liu and Guo, 2013). It has been shown that endogenous JA content and expression of JA biosynthetic genes increased during dark-induced senescence (Hu et al., 2017; Huang et al., 2017). JAZ7 was reported to regulate dark-induced senescence by interacting with COI1 and MYC2. In Arabidopsis, knock-out mutant of JAZ7 displayed accelerated senescence (Yu et al., 2016). Previous study has demonstrated that MYC2 regulated JA-induced leaf senescence in Arabidopsis by binding to the promoter and activating the expression of SENESCENCE-ASSOCIATED GENE 29(SAG29). On the other hand, the bHLH transcription factors (TF) including bHLH03, bHLH13, bHLH14, and bHLH17 repressed the senescence process via antagonistically binding to the promoter and repressing the expression of SAG29, resulting in fine-tuned control of JA-induced leaf senescence which assists plants in adapting various environmental changes (Qi et al., 2015). Additionally, ROS plays important roles in JA-induced leaf senescence. JA enhances the accumulation of H2O2 and reduced H2O2 content suppresses JA-induced leaf senescence. Results from a recent study revealed that MYC2 was involved in JA-induced H2O2 accumulation through down-regulating the expression of CATALASE 2 (CAT2) by directly binding to its promoter (Zhang Y. et al., 2020).
Leaf senescence is regulated by a large number of different signals with complex cross-talks among different signaling pathways. Identification and characterization of key senescence regulators that modulate different signaling pathways are therefore of great significance in understanding the molecular mechanisms underlying leaf senescence. In this study, we identified a novel senescence regulator, Ring/U-box protein AtUSR1. Expression of AtUSR1 was induced by age, darkness, and several plant hormones including JA and ABA. Functional analyses revealed that AtUSR1 plays a positive role in regulating leaf senescence, potentially through regulating the JA signaling pathway. Further study demonstrated that MYC2 can regulate AtUSR1 expression and AtUSR1 likely functions downstream of the JA-MYC2 signaling pathway in regulating leaf senescence.
Arabidopsis thaliana ecotype Col-0 was used in this study. The usr1 (Salk_095353) and myc2 [Salk_017005, also referred as jin1-9 (Lorenzo et al., 2004; Jung et al., 2015)] mutant lines were obtained from the Arabidopsis Biological Resource Center (ABRC). Mutants homozygous were obtained by genotyping. Arabidopsis seeds were sterilized in 70% ethanol for 3 min, then washed 3 times by sterile water, and placed on 0.5 × Murashige and Skoog (MS) medium plates, stratified at 4°C for 3 day in the dark, then transferred into continuous light conditions for germination. Plants were grown in soil at 22–24°C under continuous light (100 μmol m–2 s–1) conditions in a growth room.
For dark treatments of detached leaves, the fifth or sixth leaves from 30-day-old plants were detached and treated in the dark for designated time on filter papers soaked with treatment buffer (3 mM MES, 0.5 × MS, pH 5.8). For dark-induced senescence of attached leaves, fully expanded non-senescence leaves were wrapped with aluminum foil for 6 days.
For hormone treatments, fully expanded non-senescence leaves of 4-week-old plants were detached and immersed in hormone treatment solutions (3 mM MES, 0.5 × MS, pH 5.8) with or without phytohormones (10 μM ABA, 50 μM MeJA, 50 μM IAA, or 10 μM 1-aminocyclopropane-1-carboxylic acid (ACC)) for designated time.
To generate the 35S::AtUSR1 construct, full-length coding sequence (CDS) of AtUSR1 was polymerase chain reaction (PCR)-amplified and cloned into the pEarleyGate202 (Earley et al., 2006) vector with via gateway cloning methods according to the manufacturer’s instructions (Invitrogen). To obtain the AtUSR1 complementation construct, AtUSR1 genomic DNA including the promoter region was PCR-amplified using primers proUSR1-gate-F and AtUSR1gateOER and subcloned into pEarleyGate302 (Earley et al., 2006). To obtain the proUSR1::GUS construct, the promoter of AtUSR1 was obtained using primers proUSR1-Glucuronidase (GUS)-F/R, then subcloned into pCAMBIA3301-GUS vector (Abdollahi et al., 2007)at the restriction enzymes digest sites HindIII and NcoI. To generate Dual-luciferase assay constructs, the promoter of AtUSR1 was amplified using primers proUSR1-pGreenHindIIIF and proUSR1-pGreenBamH1R, then subcloned into the pGreenII 0800-LUC vector at the HindIII and BamH1 sites. Transgenic plants were obtained via the floral-dip method (Clough and Bent, 1998). T3 generation of overexpression plants were used for phenotyping analysis. Primers used in this study are listed in Supplementary Table 1.
Histochemical staining was carried out as described previously (Jefferson et al., 1987). Briefly, the sixth leaves of 4-week-old transgenic plants harboring the proUSR1::GUS construct were incubated in the 5-bromo-4-chloro-3-indolyl-β-D-glucuronide solution (0.5 g/mL of 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 0.5 mM of potassium ferricyanide, 0.5 mM of potassium ferrocyanide, and 0.1 M of sodium phosphate, pH 7.4) at 37°C for 12 h. Subsequently, the leaves were decolorized in 100% (v/v) ethanol.
Nitro-blue tetrazolium (NBT) chloride staining was carried out as previously described (Zhang Z. et al., 2020). Briefly, the fifth leaves of 4-week-old plants were detached and incubated in the NBT staining buffer (0.5 mg/mL NBT in 10 mM potassium phosphate buffer, pH 7.6) overnight, then decolorized in the fixative solution (ethanol: acetic acid: glycerol, 3:1:1) and kept in the ethanol: glycerol (4:1) solution at 4°C. Quantitative H2O2 measurement was performed according to the methods described previously (Lee et al., 2012).
Chlorophyll was extracted from leaves at different growth stages using 95% ethanol, and chlorophyll content was determined by detecting the absorbance at 665 nm and 649 nm using a UV2400 UV/VIS spectrophotometer as previously described (Zhang and Guo, 2018). The photochemical efficiency of photosystem II (PSII; Fv/Fm) was measured using a Chlorophyll Fluorescence Imaging System (Technologica, United Kingdom). Measurements of relative electrolyte leakage were carried out using a bench-top conductivity meter (CON500, CLEAN Instruments). Detached leaves were collected and washed three times using deionized water and immersed in water. Initial conductivity data was collected using the conductor followed by a final conductivity data reading after boiling in 100°C for 10 min to maximum membrane disruption. Total membrane ion leakage was calculated as following: initial conductivity/final conductivity × 100%. Membrane leakage rate was described in the form of percentage of initial conductivity over final conductivity, with 100% ion leakage meaning complete disruption of the membrane system.
Total RNA was extracted using TRIzol following the manufacturer’s instructions (Invitrogen). The first-strand cDNA was obtained by the Transgene Kit (Transgene Company), which includes the elimination of contaminant genomic DNA. qRT-PCR was carried out using the SYBR Premix Ex Taq (Takara). Each reaction was designed with three technical replicates. Data analysis was done using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The Ct was calculated using the ACTIN2 gene as an internal control. Three biological replicates were performed for each genotype. Primers used are listed in Supplementary Table 1.
For dual-luciferase assay, the pGreenII 0800-LUC vector (Hellens et al., 2000) harboring a firefly luciferase (LUC) gene driven by the AtUSR1 promoter was generated. The Renilla luciferase (REN) gene driven by the CaMV35S promoter was used as an internal control. 35S::AtUSR1 was inserted into the effector plasmid. The reporter and effector plasmids were co-transformed into Arabidopsis mesophyll protoplasts isolated from young and senescence leaves of 4-week–old plants according to the methods described previously (Zhuo et al., 2020). Dual-luciferase assay was performed according to the manufacturer (Promega), Briefly, the luciferase extracted by Passive Lysis buffer (PLB), then value of firefly and REN was obtained in an muti-mode microplate reader (TECAN, Infinite M200 PRO).
The accession numbers for the genes mentioned in this article are listed as follows: AtUSR1(At1g14200), MYC2 (At1g32640), MYC3(At5g46760), MYC4(At4g17880), MYC5(At5g46830), WRKY57(At1g69310), JAZ7(At2g34600), SAG12(At5g45890), SAG13(At2g29350), RBCS3B(At5g38410), LOX2(At3g45140), and ACTIN2(At3g18780).
To identify new regulators of leaf senescence, the GENEVESTIGATOR database1 was analyzed and a RING/U-box gene named AtUSR1 (U-box Senescence Related 1, At1g14200), was identified to have enhanced expression in senescing leaves. Results of qRT-PCR analysis showed that AtUSR1 was significantly up-regulated in senescing leaves (Figure 1A). Within an individual leaf, significant increase of AtUSR1 expression was observed from the base to the tip where senescence has been initiated (Figure 1B). Expression of SENESCENCE-ASSOCIATED GENE 12 (SAG12) was used as an indicator of senescence progress in these analyses. Different from the expression of SAG12that was not detected until the last stage of leaf senescence, the expression peak of AtUSR1 appeared at the early senescence stage. Under darkness, the level of AtUSR1 transcripts in detached leaves increased gradually and reached a peak at 12 h after dark treatments (Figure 1C).
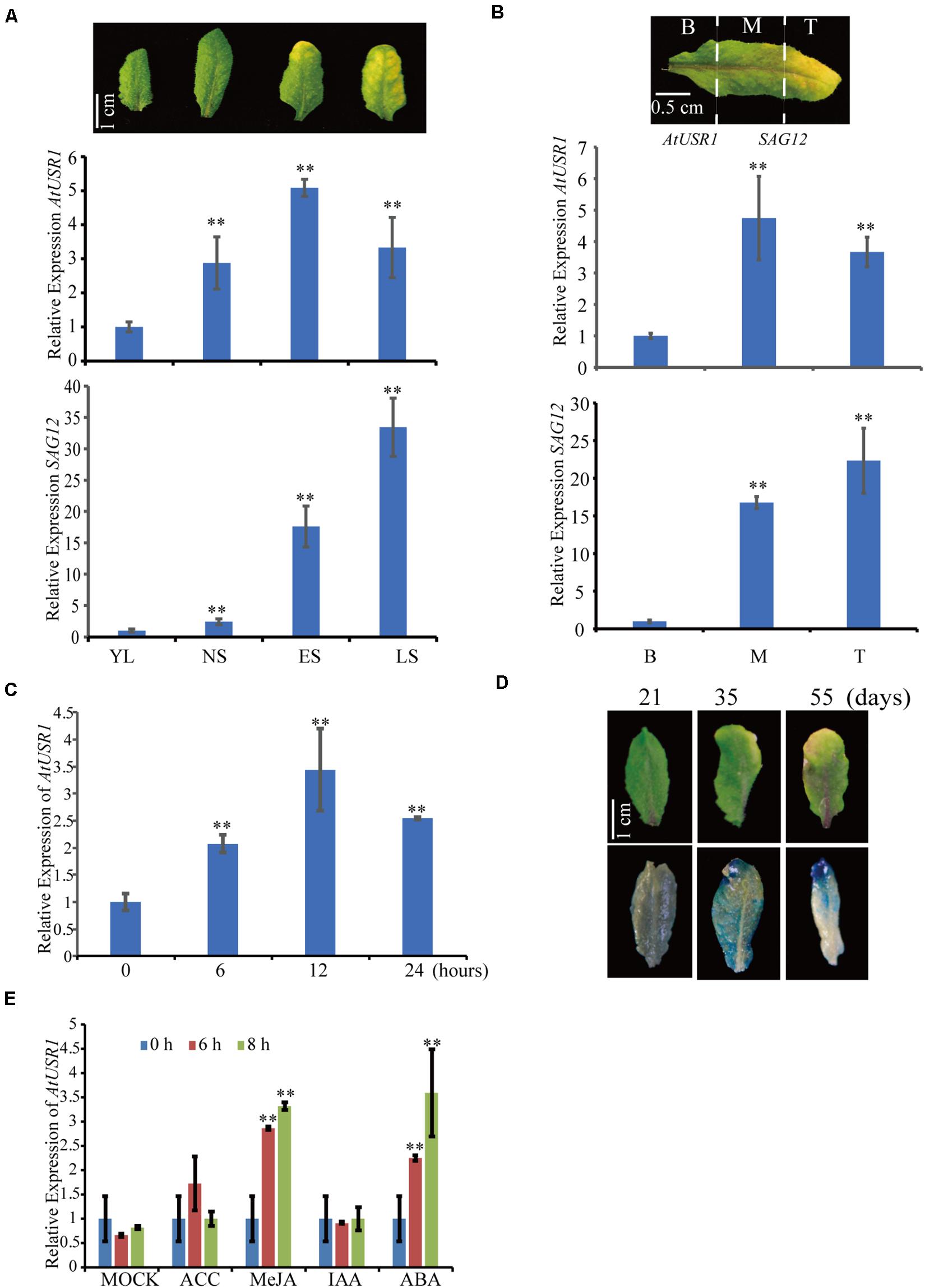
Figure 1. The expression pattern of AtUSR1. (A) The transcript level of AtUSR1 was enhanced in an age dependent manner and reached its peak at the ES stage. According to the description of Guo and Gan (2006), gene expression was detected at four different stages of leaf senescence in Arabidopsis. YL, young leaves; NS, fully expanded mature leaves without senescence symptoms; ES, early senescent leaves; and LS, late senescent leaves. The relative values of gene expression (AtUSR1 and SAG12) were calculated based on comparison with gene expression from YL, which was set as 1. (B) The expression pattern of AtUSR1 in indifferent parts of a yellowing leaf; B, Base; M, Middle; and T, Tip. (C) The expression pattern of AtUSR1 under dark conditions. The relative values of gene expression were calculated based on comparison with gene expression from B, which was set as 1. (D) Histochemical staining of GUS activity detection in leaves at different timepoints as indicated. (E) The expression of AtUSR1 was induced by MeJA and ABA treatments. * and ** indicate significant difference at 0.01 < P < 0.05 and P < 0.01 levels using student’s t-test. Data are shown as the mean ± SD from three independent experiments. Significance analysis was only performed on AtUSR1 expression.
At different leaf developmental stages, as indicated by GUS staining of proAtUSR1::GUS transgenic plants, higher promoter activities of AtUSR1 were detected in yellowing leaves with dark blue staining, whereas less AtUSR1 promoter activities were detected in younger leaves with light blue staining (Figure 1D).
We further analyzed the effect of phytohormones on the expression of AtUSR1. No obvious difference was detected when leaves were treated with ACC or IAA, while significant increase of AtUSR1 expression was observed when leaves were treated with JA or ABA (Figure 1E).
To investigate the function of AtUSR1 in leaf senescence, we obtained an AtUSR1 knock-down mutant (usr1) in which a T-DNA fragment is inserted in the 5′UTR region of this gene (Supplementary Figure 1A). The AtUSR1 expression level was significantly lower in usr1 than in WT Col-0 (Supplementary Figure 1B). A complementation assay was carried out in which the full length genomic DNA of AtUSR1 (proAtUSR1::AtUSR1) was used to transform the usr1 mutant to generate complementation plants. Under our growth conditions, older leaves from 6-week-old Col-0 plants started to exhibit visible yellowing, while the counterpart leaves from usr1 plants were still green. The complementation plants displayed similar senescence progress to Col-0 (Figures 2A,B), indicating that the proUSR1::AtUSR1 transgene was able to rescue the delayed senescence phenotype of usr1 plants.
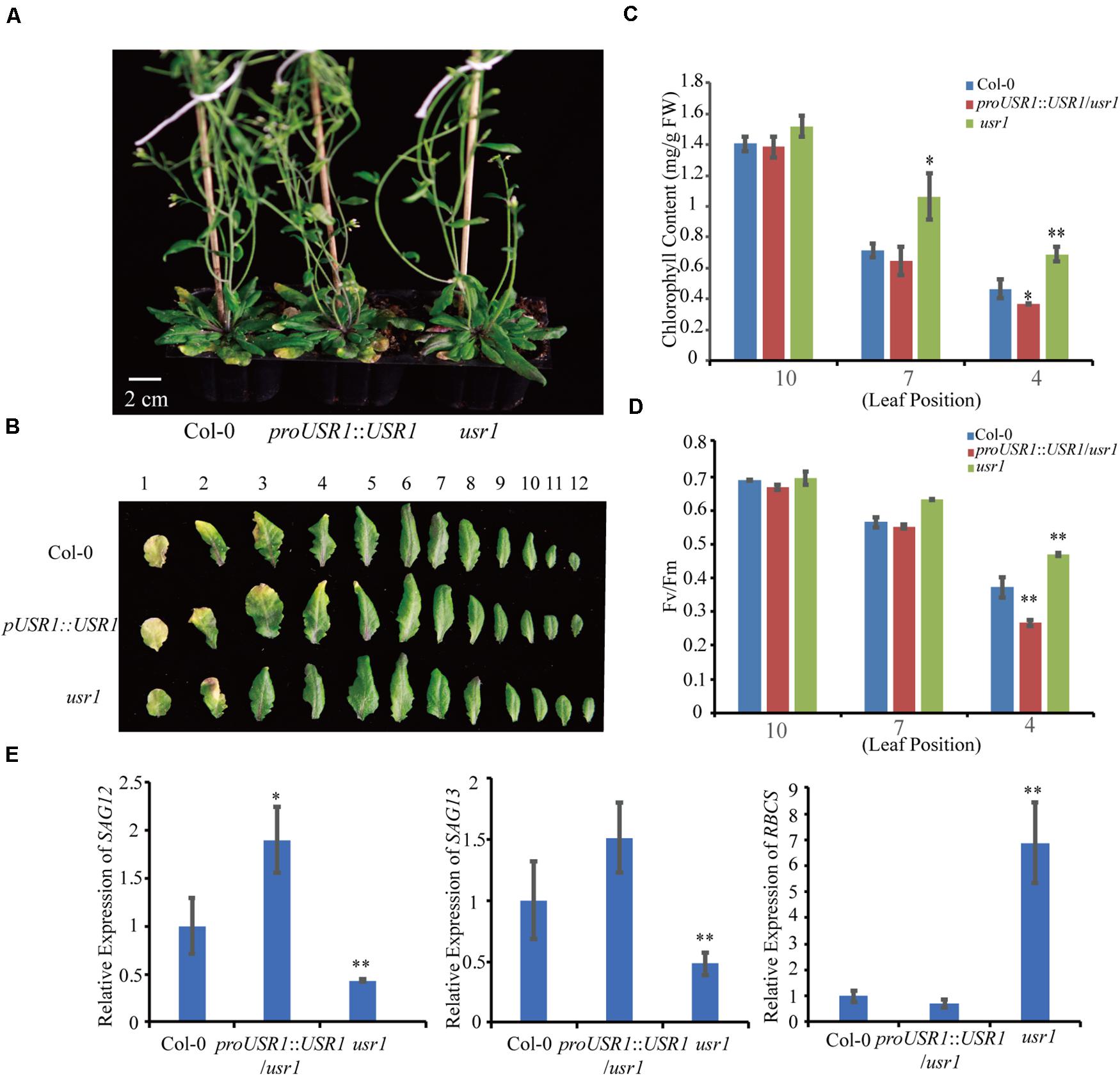
Figure 2. The usr1 mutant plants show delayed leaf senescence. (A) The senescence phenotype of plants with different genotype as indicated after grown under continuously light for 40 days. usr1 mutant shows delayed leaf senescence and complementation plants proUSR1::USR1/usr1 rescued the delayed leaf senescence phenotype of usr1 plants. (B) Senescence phenotypes of detached 1st–12th leaves of different genotypes as indicated. (C,D) Chlorophyll content and Fv/Fm detection of leaves from different leaf positions in Col-0, usr1, and proUSR1::USR1/usr1 plants as indicated. (E) Expression of SAGs genes including SAG12, SAG13, and RBCS in plants of different genotypes as indicated. Single and double asterisk indicate significant difference at 0.01 < P < 0.05 and P < 0.01 levels using student’s t-test. Data are shown as the mean ± SD from three independent experiments.
Total chlorophyll contents of the 4th and 7th leaves of usr1 plants were significantly higher than that of Col-0 (Figure 2C). As an indicator of photosynthetic efficiency, the Fv/Fm ratio in usr1 leaves was significantly higher than Col-0 (Figure 2D). Meanwhile, the complementation plants displayed similar chlorophyll contents and Fv/Fm values to Col-0 (Figures 2C,D). Similarly, the usr1 mutant displayed lower transcripts of the senescence marker genes SAG12 and SAG13 but higher expression of the photosynthetic gene RBCS compared with Col-0 and the complementation plants (Figure 2E).
In this study, proUSR1::USR1/usr1 transgenic plants displayed an slightly early senescence phenotype compared with WT (Figure 2). One possibility is that multiple copies of the T-DNA insertion led to higher expression of AtUSR1 than WT (Supplementary Figure 1).
To further understand the role of AtUSR1 in leaf senescence, transgenic plants harboring 35S::AtUSR1 were generated and two representative lines (35S::AtUSR1#1, 35S::AtUSR1#6) were used for further characterization (Supplementary Figure 2). Under continuous light, 5-week-old plants of 35S::AtUSR1#1 and 35S::AtUSR1#6 displayed a premature leaf senescence phenotype compared with Col-0 (Figures 3A,B). Consistent with the visible phenotype, chlorophyll levels, and Fv/Fm ratios of both AtUSR1 overexpression lines were lower than Col-0 (Figures 3C,D). The expression of SAG12 and SAG13 was significantly increased while the expression of RBCS was reduced in AtUSR1 overexpression plants compared with Col-0 (Figure 3E). Together, these results suggested that AtUSR1 acted as a positive regulator of leaf senescence.
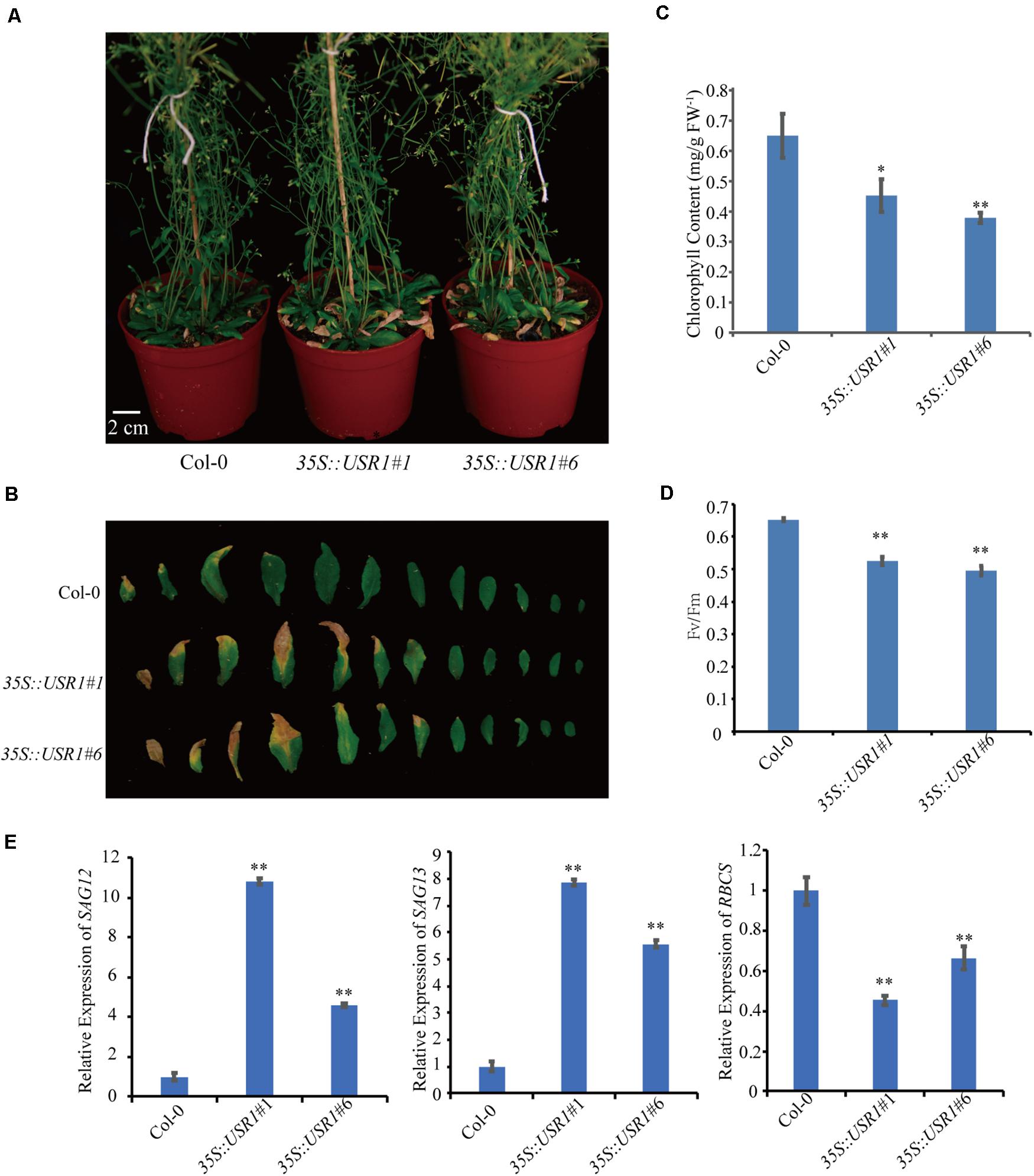
Figure 3. AtUSR1 overexpression accelerates leaf senescence. (A) The precocious senescence phenotype of AtUSR1 overexpression lines after grown under continuously light condition for 40 days. (B) The phenotype of 1st–12th leaves detached from plants with different genotypes. (C,D) Chlorophyll content and Fv/Fm detection at different leaf positions in Col-0, two independent AtUSR1 overexpression plants as indicated. (E) Expression pattern of SAG12, SAG13, and RBCS in plants overexpressing AtUSR1 and in Col-0. Single and double asterisk indicate significant difference at 0.01 < P < 0.05 and P < 0.01 levels using student’s t-test. Data are shown as the mean ± SD from three independent experiments.
Since AtUSR1 expression was induced by both senescence and darkness, we studied the role of AtUSR1 in dark-induced senescence. After incubated under dark conditions for 5 days, detached leaves of the usr1 mutant retained green color while the 35S::AtUSR1#1 and 35S::AtUSR1#6 leaves showed accelerated leaf yellowing compared to that of Col-0 (Figure 4A). Total chlorophyll levels and Fv/Fm ratios of the AtUSR1-overexpressing lines were significantly lower than the usr1 mutant after the dark treatment (Figures 4B,C).
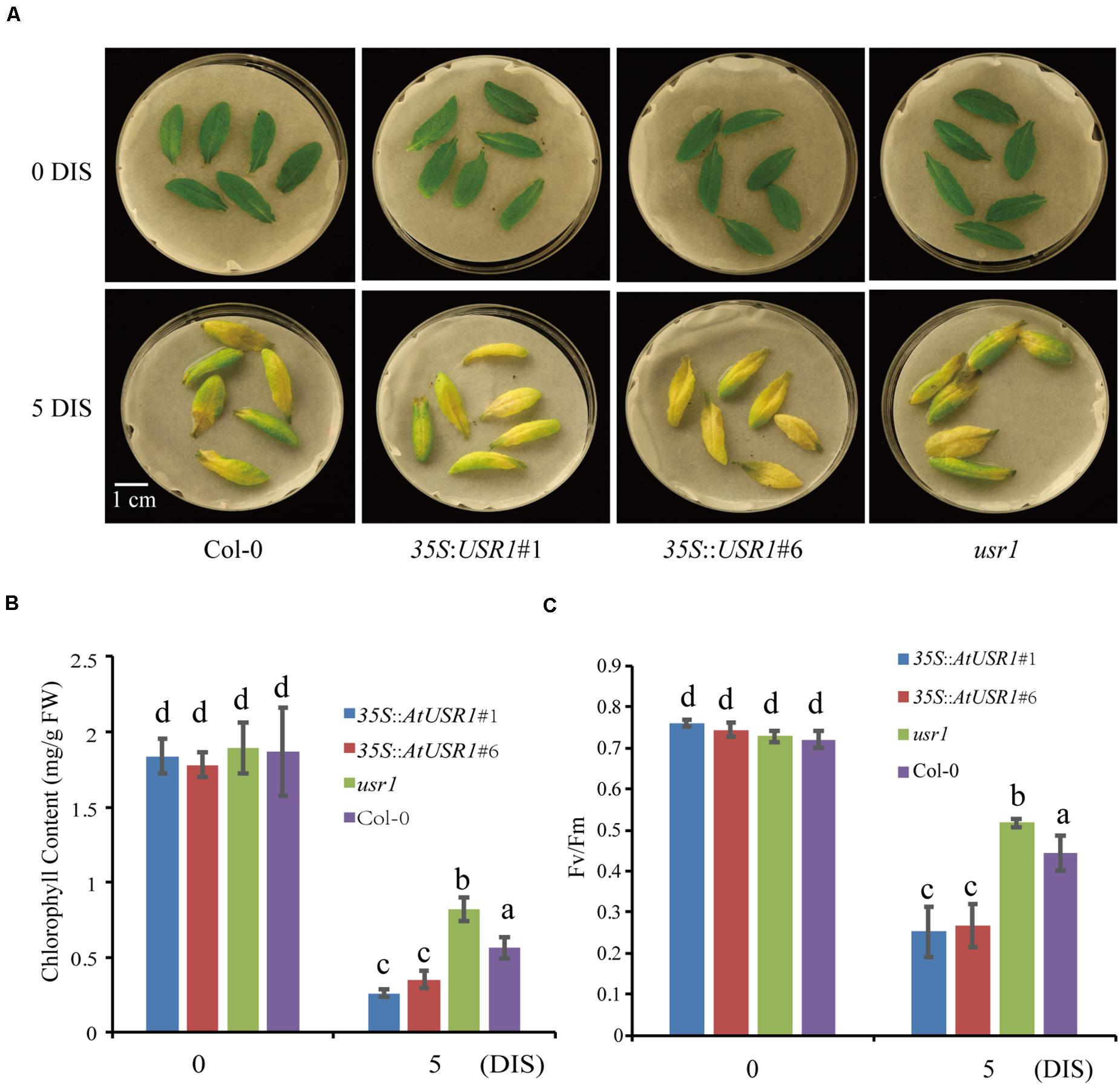
Figure 4. AtUSR1 is involved in dark-induced leaf senescence. (A) AtUSR1 Overexpression lines displayed precocious leaf senescence while the usr1 mutant showed delayed senescence under dark conditions. (B,C) Total chlorophyll content and Fv/Fm of different genotypes as indicated. The data in (B,C) different letters above columns indicate significant differences according to Duncan’s multiple range test (P < 0.05). Data are shown as the mean ± SD from three independent experiments.
We also studied dark-induced senescence of attached leaves from Col-0, AtUSR1 overexpression, and usr1 plants. Fully expanded non-senescence leaves were wrapped with aluminum foil for dark treatments. 6 days after treatments, the wrapped leaves of usr1 plants stayed green while dark-treated leaves of the 35S::AtUSR1 lines displayed senescence symptoms compared with Col-0 (Supplementary Figure 2A). Compared to Col-0, chlorophyll content was higher in leaves of usr1 and lower in 35S::AtUSR1 leaves while membrane leakage rates were lower in usr1 leaves and higher in 35S::AtUSR1 leaves (Supplementary Figures 2B,C). The above described results suggested that AtUSR1 positively regulates dark-induced senescence as well.
Reactive oxygen species play important roles in senescence as well as stress responses (Sedigheh et al., 2011; Jajic et al., 2015). Cellular levels of ROS in fully expanded non-senescence leaves with different genotypes were measured by NBT staining and H2O2 quantification. The results showed that leaves of the 35S::AtUSR1 lines displayed stronger while the usr1 mutant showed weaker NBT staining compared with Col-0, suggesting that AtUSR1 rendered plant accumulating more ROS in leaves (Figure 5A). Leaves from 35S::AtUSR1#1 to 35S::AtUSR1#6 lines contained significantly higher amount of H2O2 while the usr1 mutant had lower levels of H2O2 accumulation compared with Col-0 (Figure 5B).
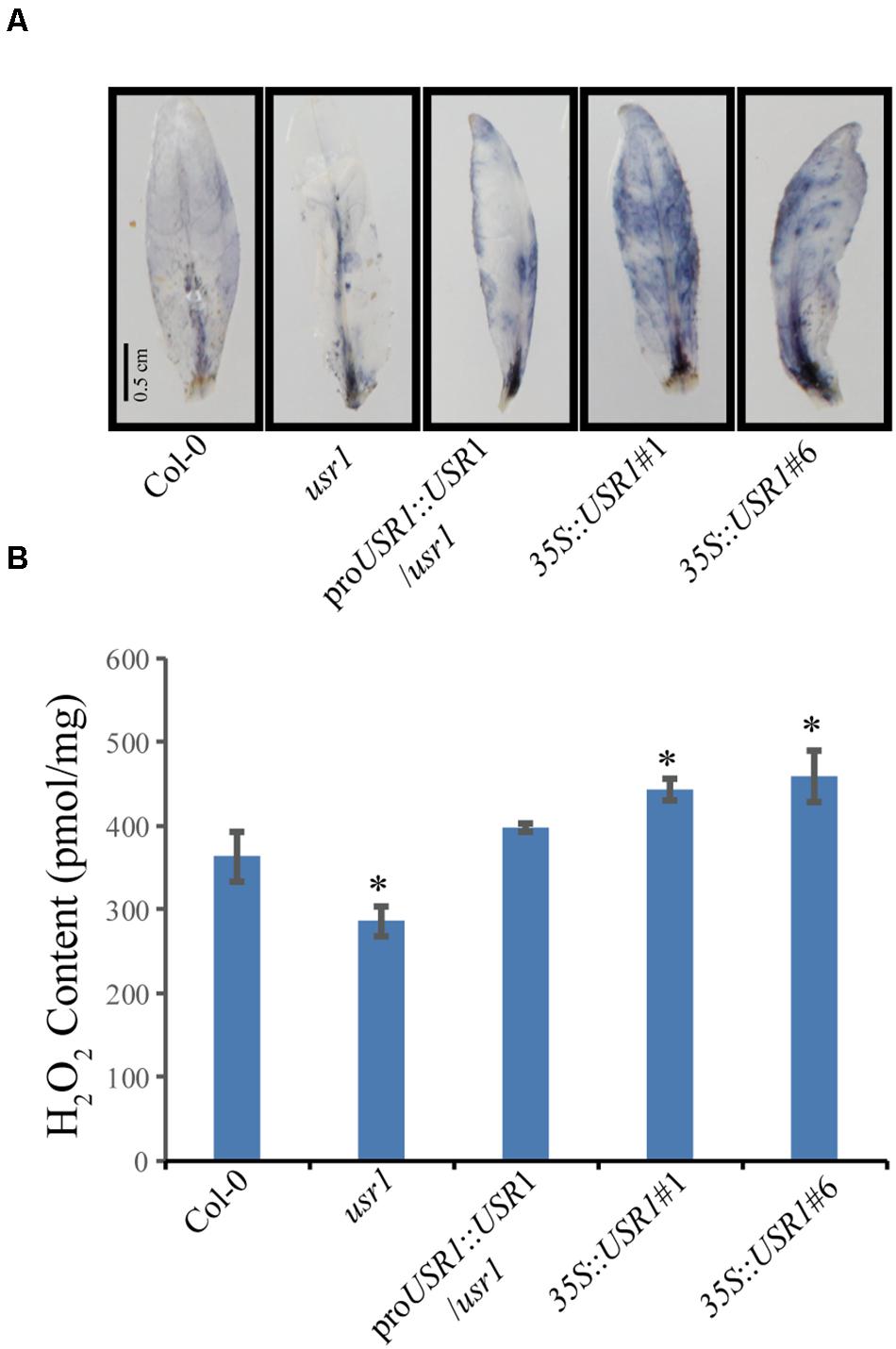
Figure 5. AtUSR1 affects ROS accumulation. (A) The NBT staining results of leaves from different genotypes. (B) Accumulation of H2O2 in detached leaves of different genotypes as indicated. *Indicate significant difference at 0.01 < P < 0.05 levels using student’s t-test. Data are shown as the mean ± SD from three independent experiments.
Jasmonic acid is a senescence-promoting signal in both age-dependent and dark-induced senescence (Yu et al., 2016; Hu et al., 2017). Our data indicated that AtUSR1 expression was up-regulated by JA treatments (Figure 1E). We therefore tested the role of AtUSR1 in JA-induced senescence. After treated with exogenous MeJA for 5 days, detached leaves from 35S::AtUSR1 plants were completely yellow, while usr1 leaves remained mostly green and leaves of Col-0 were in-between (Figure 6A). Chlorophyll content and Fv/Fm ratio data also indicated that the usr1 mutation delayed while AtUSR1 overexpression accelerated JA-induced senescence on detached leaves (Figures 6B,C).
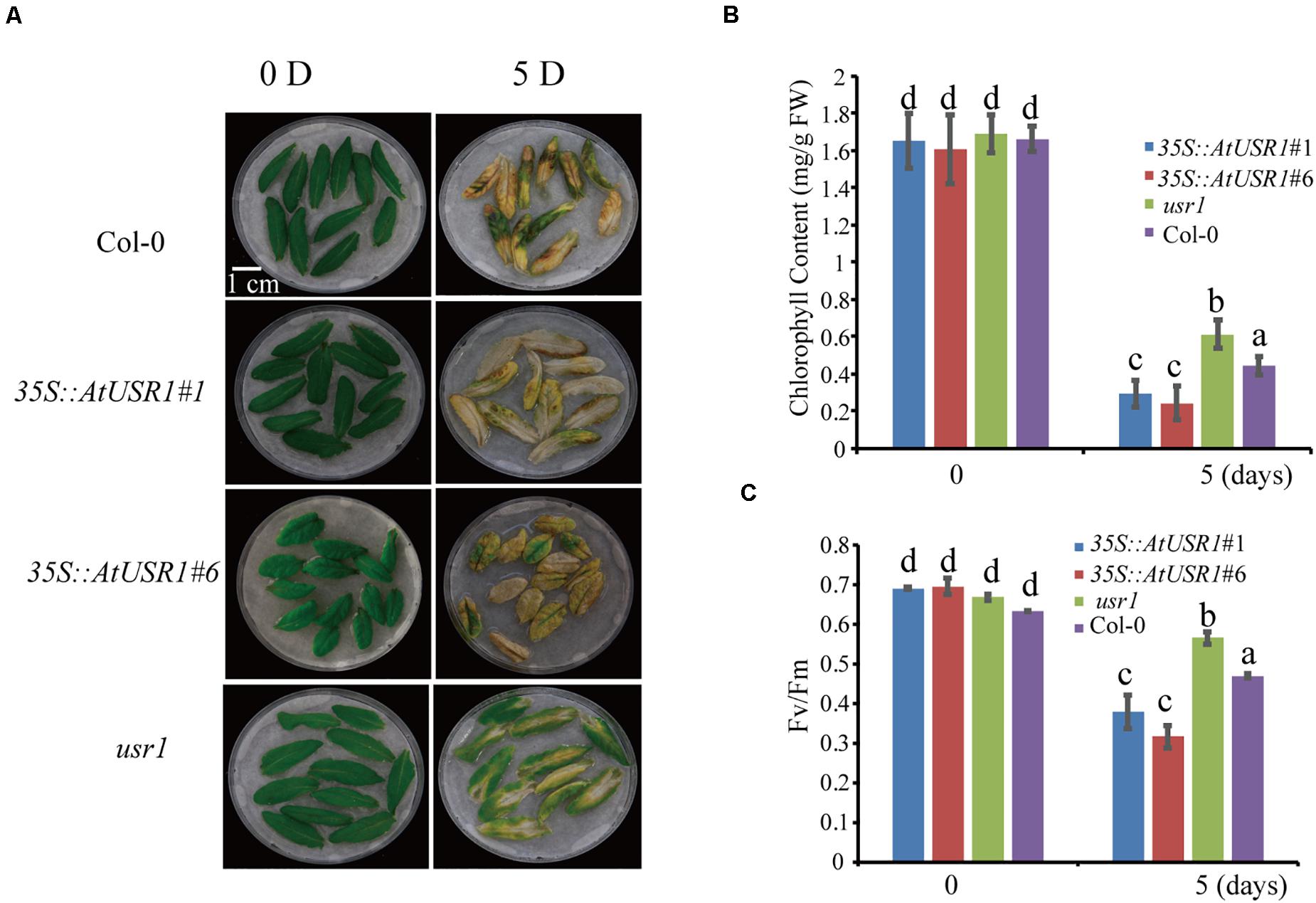
Figure 6. AtUSR1 plays as positive role in JA mediated leaf senescence. (A) AtUSR1 overexpression enhanced JA-induced leaf senescence while usr1 mutation delayed this process. Detached Sixth leaves of different genotypes were treated with 50 μM MeJA for 5 days. (B,C) Total chlorophyll content and Fv/Fm in the leaf samples of different genotypes as indicated. The data in (B,C) different letters above columns indicate significant differences according to Duncan’s multiple range test (P < 0.05). Data are shown as the mean ± SD from three independent experiments.
Previous studies have demonstrated that MYC2 plays an essential role in JA-induced senescence (Yu et al., 2016). Transcription factor MYC2 binds to the G-box motif (CACGTG) and its variants such as E-box (CANNTG), G/A box (CACGAG), and G/C box (CACGCG) to regulate expression of target genes (Dombrecht et al., 2007; Zhuo et al., 2020). Interestingly, using PLACE2, we have identified a E-box [CAN(A)N(T)TG] motif in the promoter region of AtUSR1 (−130 bp to −124 bp upstream of the translation start site).
We then examined the expression of AtUSR1 in the myc2 mutant under MeJA treatments. Detached fully expanded leaves were treated with MeJA for 8 h, after which AtUSR1 expression was observed to be significantly increased in Col-0 but this increase was significantly reduced in the myc2 mutant (Figure 7A), suggesting that the JA-induction of AtUSR1 expression is partially MYC2-dependent.
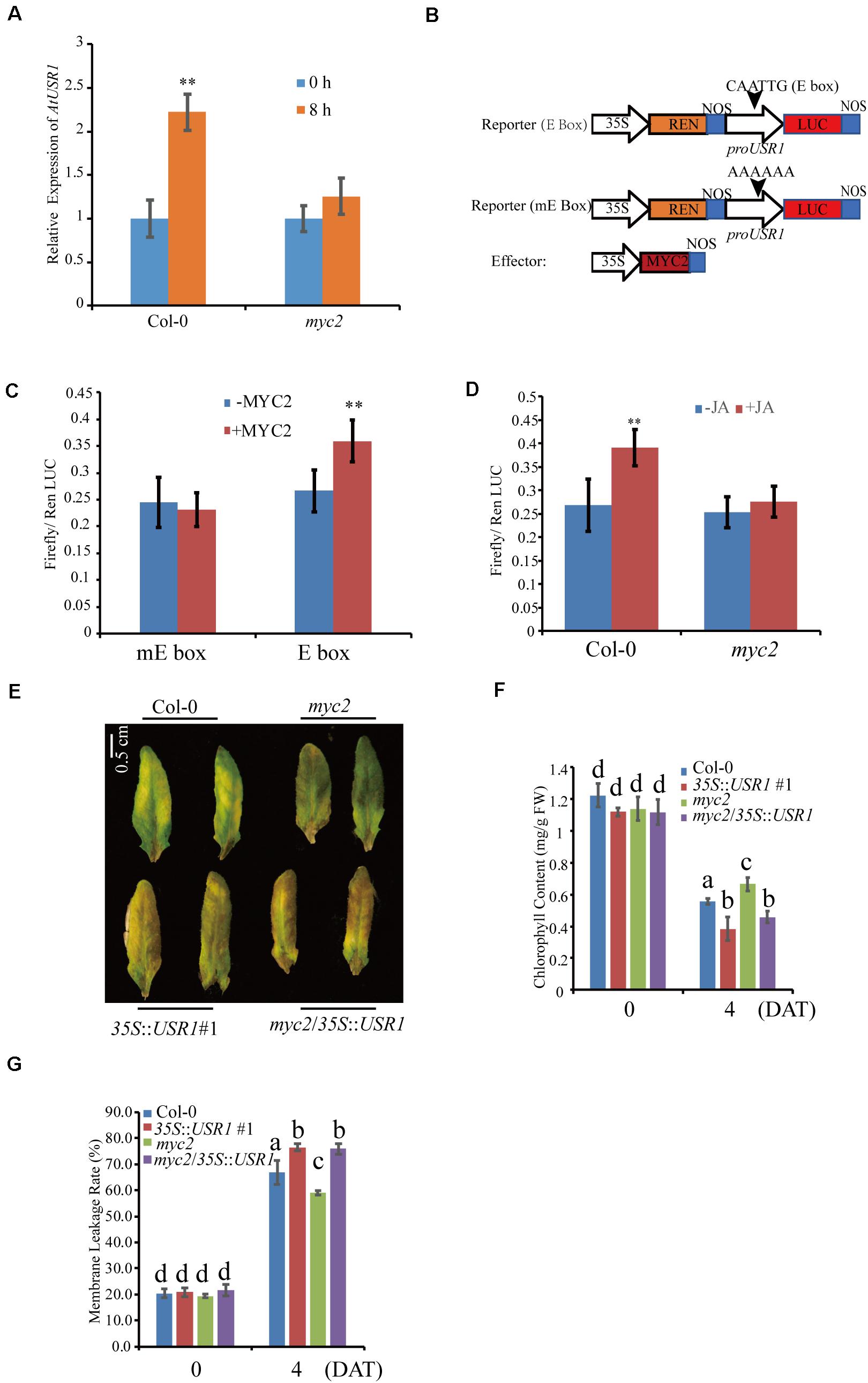
Figure 7. AtUSR1 functions downstream to MYC2. (A) The expression pattern of AtUSR1 in Col-0 and the myc2 mutant under 100 μM MeJA treatments. (B) Schematic diagram of the reporter and effector constructs used in the dual luciferase assay. (C) MYC2 increased the activities of the AtUSR1 promoter. The promoter of AtUSR1 containing an E-box (CAGCGT) or a mutant version of the E-box (AAAAAA) fused with LUC was co-transformed with an effecter construct with or without 35S::MYC2 into Col-0 protoplasts. Ren LUC activity acted as internal control. The experiments presented here were done only with Col-0 plants (labeled as –MYC2). (D) MeJA induced the expression of AtUSR1 and this induction was MYC2-dependent. The reporter construct (proAtUSR1::LUC) was transformed into protoplasts of Col-0 or myc2, respectively. Ren Luc acted as an internal control. (E) The cross line myc2/35S::USR1 rescued the delayed senescence phenotype of myc2 under MeJA treatments. (F) Chlorophyll contents in different genotypes as indicated. (G) Membrane leakage rates in different genotypes as indicated. **Indicate significant difference at P < 0.01 levels using student’s t-test. Data are shown as the mean ± SD from three independent experiments. The data in (F,G) different letters above columns indicate significant differences according to Duncan’s multiple range test (P < 0.05).
We further analyzed the MYC2 regulation of AtUSR1 expression via a dual-luciferase assay. In the reporter construct, the Firefly luciferase gene was driven by the AtUSR1 promoter containing the E-box sequence and the REN gene was used as an internal reference (Figure 7B). The reporter construct was used in co-transfecting of Col-0 protoplasts together with an effector construct in which MYC2 was under the control of the 35S promoter. The ratio of Firefly/Renilla activities was significantly higher when the effector construct harboring 35S::MYC2 was co-transformed with the reporter construct containing the AtUSR1 promoter. The increase in Firefly/Renilla ratio was not detected when the E-box on the AtUSR1 promoter was replaced by a mutated sequence (Figure 7C). These results suggest that MYC2 could regulate the expression of AtUSR1 and this regulation is dependent on the E-box on its promoter.
Next, we studied the role of MYC2 in JA-regulated expression of AtUSR1 using the same dual luciferase strategy. The 35S::MYC2 effector construct was co-transfected with a reporter construct in which firefly LUC was driven by the AtUSR1 promoter. In the same reporter construct, Renilla LUC driven by the 35S promoter was used as an internal control. This combination was co-transfected into Col-0 protoplasts with or without JA treatments. The results indicated that Firefly/Renilla was significantly higher under JA treatments compared with the mock. The increase of Firefly/Renilla caused by JA treatments disappeared when myc2 mutant protoplasts were transfected, suggesting that the MeJA induction of AtUSR1 promoter activities was affected in the myc2 mutant (Figure 7D).
Mutation of MYC2 has been reported to delay JA-induced chlorophyll degradation (Zhu et al., 2015). To further study the relationship between AtUSR1 and MYC2 in JA-induced senescence, we obtained myc2 mutant plants harboring 35S::AtUSR1 (referred to as myc2/35S::AtUSR1) and senescence phenotypes of detached leaves were examined in presence of MeJA. Similar to 35S::AtUSR1 leaves that displayed early senescence phenotypes compared to Col-0, leaves of myc2/AtUSR1OE plants also showed precocious senescence, rescuing the delayed senescence phenotype of the myc2 mutant (Figure 7E). Moreover, the effects of myc2 mutation on chlorophyll content and Fv/Fm of MeJA-treated leaves were reduced by AtUSR1 overexpression (Figures 7F,G). The above-described results suggested that AtUSR1 acted downstream to MYC2 in mediating JA-induced senescence.
As the last stage of development, leaf senescence is a crucial process that influences photosynthetic capacity and reallocation of nutrients from senescing leaves into young leaves and reproductive organs (Ahmad and Guo, 2019; Krieger-Liszkay et al., 2019). Timely senescence is essential for plants’ reproductive success. Precocious or early senescence caused by harsh growth conditions, on the other hand, compromises crop yield in an agricultural setting (Wu et al., 2012; Bengoa Luoni et al., 2019). Initiation and progression of leaf senescence can be affected by a large number of endogenous and environmental factors including developmental stage, age, phytohormones, environmental cues such as temperature, darkness, and pathogens (Bresson et al., 2018). All these factors and related signaling pathways form a complex network in fine-tuning the initiation and progression of leaf senescence. Identifying senescence-regulating genes and clarifying their functions are of great importance in devising genetic strategies of manipulating senescence for improving crop yield and production traits (Jing and Nam, 2012).
A large number of genes undergo substantial changes in expression during senescence, including genes related to protein degradation, nutrient remobilization, chlorophyll metabolism, and transcription regulation (Sato et al., 2009; Goud and Kachole, 2011; Izumi and Ishida, 2011; Sakuraba et al., 2012; Tamary et al., 2019). Also included are regulators involved in signaling transduction in response to phytohormones such as ethylene, JA, ABA, auxin, and cytokinins (Jan et al., 2019).
Here we report the function of a ring/u-box protein AtUSR1 in promoting leaf senescence mediated by the JA-MYC2 pathway. Firstly, the expression of AtUSR1 was age-dependent and was induced by darkness, JA, and ABA treatments (Figure 1). Interestingly, different from the expression of SAG12 that was not detected until the last stage of leaf senescence, the expression peak of AtUSR1 appeared at the early senescence stage indicating that AtUSR1 plays potential roles in the initial of senescence stage. Leaves of usr1 plants showed an obvious delayed senescence phenotype and proAtUSR1::AtUSR1 was able to rescue the mutant phenotype (Figure 2). Overexpression of AtUSR1 caused early leaf senescence (Figure 3). In addition, results from dark treatments of usr1 and 35S::AtUSR1 plants indicated that AtUSR1 functioned in accelerating dark-induced senescence as well (Figure 4). Further study demonstrated that AtUSR1overexpression promoted JA-induced senescence and knocking-down of AtUSR1 delayed this process (Figure 6).
In this study, we demonstrated that AtUSR1 acts downstream of MYC2 in JA-induced leaf senescence. Firstly, MeJA treatments enhanced AtUSR1 expression in a MYC2-dependent manner (Figure 7A). Results of dual luciferase assays indicated that MYC2 could regulate AtUSR1 expression (Figures 7C,D). In the presence of JA, JAZ proteins undergo degradation via ubiquitin pathways mediated by the F-box protein COI1. MYC2 is thus released from the JAZ-MYC2 complex and function to activate the expression of downstream genes including AtUSR1, that functions in promoting leaf senescence (Figure 7). In the absence of JA, the JAZ-MYC2 complexes inhibit the activity of MYC2 and AtUSR1 transcription is compromised.
We also found that AtUSR1 affects ROS homeostasis. ROS over-accumulated in 35S::AtUSR1 leaves and the usr1 mutant had less ROS (Figure 5). ROS are known to be involved in multiple biological processes in plants such as leaf senescence, stress response, and hypersensitive response (Sedigheh et al., 2011; Jajic et al., 2015). Since ROS can interplay with other signaling molecules in regulating plant development and stress responses (Hoque et al., 2012; Li et al., 2018), it’s reasonable to hypothesize that AtUSR1 could also be involved in multiple signaling pathways. Significant overlap of gene expression changes exists between natural senescence, treatments of senescence-promoting phytohormones and stress conditions (Guo, 2013). AtUSR1 can potentially function as an internode factor that can be affected by multiple senescence-promoting signaling pathways induced by age, darkness, and JA.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YG conceived the project. ZZ and YG designed the research, performed data analysis, and wrote the manuscript. ZZ and MX performed experiments. All authors contributed to the article and approved the submitted version.
This work was supported by the Agricultural Science and Technology Innovation Program, Chinese Academy of Agricultural Sciences (ASTIP-TRI02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.608589/full#supplementary-material
Supplementary Figure 1 | Genotyping of the usr1 mutant. (A) The Gene structure of AtUSR1 and the usr1 (Salk_095353) T-DNA insertion in the 5′UTR region. (B) Transcript levels of AtUSR1 in Col-0, usr1 mutant, and complementation plants harboring proAtUSR1::USR1/usr1.
Supplementary Figure 2 | Expression of AtUSR1 in two independent overexpression lines.
Supplementary Figure 3 | Phenotype of attached leaves under dark conditions. (A) Fully expanded leaves were wrapped with aluminum foil for 6 days. (B) Chlorophyll contents of different genotypes as indicated. (C) Membrane leakage rates of different genotypes as indicated.
Supplementary Table 1 | Primers used in this study.
Abdollahi, M. R., Moieni, A., Mousavi, A., Salmanian, A. H., Jalali Javaran, M., and Majdi, M. (2007). Effect of integrated bombardment and Agrobacterium transformation system on transient GUS expression in hypocotyls of rapeseed (Brassica napus L. cv. PF704) microspore-derived embryos. Pak. J. Biol. Sci. 10, 3141–3145. doi: 10.3923/pjbs.2007.3141.3145
Ahmad, S., and Guo, Y. (2019). Signal transduction in leaf senescence: progress and perspective. Plants 8:405. doi: 10.3390/plants8100405
Ansari, M. I., Hasan, S., and Jalil, S. U. (2014). Leaf senescence and GABA shunt. Bioinformation 10, 734–736. doi: 10.6026/97320630010734
Bengoa Luoni, S., Astigueta, F. H., Nicosia, S., Moschen, S., Fernandez, P., and Heinz, R. (2019). Transcription factors associated with leaf senescence in crops. Plants 8:411. doi: 10.3390/plants8100411
Bresson, J., Bieker, S., Riester, L., Doll, J., and Zentgraf, U. (2018). A guideline for leaf senescence analyses: from quantification to physiological and molecular investigations. J. Exp. Bot. 69, 769–786. doi: 10.1093/jxb/erx246
Castillo, M. C., and Leon, J. (2008). Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 59, 2171–2179. doi: 10.1093/jxb/ern079
Causier, B., Ashworth, M., Guo, W., and Davies, B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438. doi: 10.1104/pp.111.186999
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Figueroa, P., and Browse, J. (2012). The Arabidopsis JAZ2 promoter contains a G-Box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 53, 330–343. doi: 10.1093/pcp/pcr178
Fujiki, Y., Yoshikawa, Y., Sato, T., Inada, N., Ito, M., Nishida, I., et al. (2001). Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111, 345–352. doi: 10.1034/j.1399-3054.2001.1110312.x
Gan, S. (2018). Concepts and types of senescence in plants. Methods Mol. Biol. 1744, 3–8. doi: 10.1007/978-1-4939-7672-0_1
Goud, P. B., and Kachole, M. S. (2011). Role of chloroplastidial proteases in leaf senescence. Plant Signal. Behav. 6, 1371–1376. doi: 10.4161/psb.6.9.16316
Gregersen, P. L., Holm, P. B., and Krupinska, K. (2008). Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 10(Suppl. 1), 37–49. doi: 10.1111/j.1438-8677.2008.00114.x
Guo, Y. (2013). Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 82, 519–528.
Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549.
Guo, Y., and Gan, S. (2005). Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 71, 83–112.
Guo, Y., and Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
He, Y., Fukushige, H., Hildebrand, D. F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/a:1006496308160
Hoque, T. S., Uraji, M., Ye, W., Hossain, M. A., Nakamura, Y., and Murata, Y. (2012). Methylglyoxal-induced stomatal closure accompanied by peroxidase-mediated ROS production in Arabidopsis. J. Plant Physiol. 169, 979–986. doi: 10.1016/j.jplph.2012.02.007
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Huang, H., Liu, B., Liu, L., and Song, S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68, 1349–1359. doi: 10.1093/jxb/erw495
Izumi, M., and Ishida, H. (2011). The changes of leaf carbohydrate contents as a regulator of autophagic degradation of chloroplasts via Rubisco-containing bodies during leaf senescence. Plant Signal. Behav. 6, 685–687. doi: 10.4161/psb.6.5.14949
Jajic, I., Sarna, T., and Strzalka, K. (2015). Senescence, stress, and reactive oxygen species. Plants 4, 393–411. doi: 10.3390/plants4030393
Jan, S., Abbas, N., Ashraf, M., and Ahmad, P. (2019). Roles of potential plant hormones and transcription factors in controlling leaf senescence and drought tolerance. Protoplasma 256, 313–329.
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jiang, Y., Liang, G., Yang, S., and Yu, D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26, 230–245. doi: 10.1105/tpc.113.117838
Jing, H. C., and Nam, H. G. (2012). Leaf senescence in plants: from model plants to crops, still so many unknowns. J. Integr. Plant Biol. 54, 514–515. doi: 10.1111/j.1744-7909.2012.01148.x
Jung, C., Zhao, P., Seo, J. S., Mitsuda, N., Deng, S., and Chua, N. H. (2015). PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 27, 2016–2031. doi: 10.1105/tpc.15.00385
Khanna-Chopra, R. (2012). Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481. doi: 10.1007/s00709-011-0308-z
Kim, J., Dotson, B., Rey, C., Lindsey, J., Bleecker, A. B., Binder, B. M., et al. (2013). New clothes for the jasmonic acid receptor COI1: delayed abscission, meristem arrest and apical dominance. PLoS One 8:e60505. doi: 10.1371/journal.pone.0060505
Kim, J., Kim, J. H., Lyu, J. I., Woo, H. R., and Lim, P. O. (2018). New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 69, 787–799. doi: 10.1093/jxb/erx287
Krieger-Liszkay, A., Krupinska, K., and Shimakawa, G. (2019). The impact of photosynthesis on initiation of leaf senescence. Physiol. Plant. 166, 148–164. doi: 10.1111/ppl.12921
Lee, S., Seo, P. J., Lee, H. J., and Park, C. M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70, 831–844. doi: 10.1111/j.1365-313X.2012.04932.x
Lee, S. H., Sakuraba, Y., Lee, T., Kim, K. W., An, G., Lee, H. Y., et al. (2015). Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 57, 562–576. doi: 10.1111/jipb.12276
Li, Z., Wang, F., Zhao, Q., Liu, J., and Cheng, F. (2018). Involvement of NADPH oxidase isoforms in the production of O2- manipulated by ABA in the senescing leaves of early-senescence-leaf (esl) mutant rice (Oryza sativa). PLoS One 13:e0190161. doi: 10.1371/journal.pone.0190161
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Liu, F., and Guo, F. Q. (2013). Nitric oxide deficiency accelerates chlorophyll breakdown and stability loss of thylakoid membranes during dark-induced leaf senescence in Arabidopsis. PLoS One 8:e56345. doi: 10.1371/journal.pone.0056345
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorenzo, O., Chico, J. M., Sanchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Miao, Y., and Zentgraf, U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19, 819–830. doi: 10.1105/tpc.106.042705
Niu, Y. H., and Guo, F. Q. (2012). Nitric oxide regulates dark-induced leaf senescence through EIN2 in Arabidopsis. J. Integr. Plant Biol. 54, 516–525. doi: 10.1111/j.1744-7909.2012.01140.x
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Perez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Qi, H., Li, J., Xia, F. N., Chen, J. Y., Lei, X., Han, M. Q., et al. (2020). Arabidopsis SINAT proteins control autophagy by mediating ubiquitylation and degradation of ATG13. Plant Cell 32, 263–284. doi: 10.1105/tpc.19.00413
Qi, T., Wang, J., Huang, H., Liu, B., Gao, H., Liu, Y., et al. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27, 1634–1649. doi: 10.1105/tpc.15.00110
Quirino, B. F., Noh, Y. S., Himelblau, E., and Amasino, R. M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5, 278–282.
Sakuraba, Y., Schelbert, S., Park, S. Y., Han, S. H., Lee, B. D., Andres, C. B., et al. (2012). STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24, 507–518. doi: 10.1105/tpc.111.089474
Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A., and Kusaba, M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57, 120–131. doi: 10.1111/j.1365-313X.2008.03670.x
Sedigheh, H. G., Mortazavian, M., Norouzian, D., Atyabi, M., Akbarzadeh, A., Hasanpoor, K., et al. (2011). Oxidative stress and leaf senescence. BMC Res. Notes 4:477. doi: 10.1186/1756-0500-4-477
Shan, X., Li, C., Peng, W., and Gao, B. (2011). New perspective of jasmonate function in leaf senescence. Plant Signal. Behav. 6, 575–577. doi: 10.4161/psb.6.4.14899
Shim, K. C., Kim, S. H., Jeon, Y. A., Lee, H. S., Adeva, C., Kang, J. W., et al. (2020). A RING-type E3 ubiquitin ligase, OsGW2, controls chlorophyll content and dark-induced senescence in rice. Int. J. Mol. Sci. 21:1704. doi: 10.3390/ijms21051704
So, W. M., Kim, S. Y., Hyoung, S., and Shin, J. S. (2019). The novel protein CSAP accelerates leaf senescence and is negatively regulated by SAUL1 in the dark. Plant Cell Rep. 39, 325–334. doi: 10.1007/s00299-019-02493-z
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. doi: 10.1105/tpc.111.083089
Suraneni, M. V., Schneider-Broussard, R., Moore, J. R., Davis, T. C., Maldonado, C. J., Li, H., et al. (2010). Transgenic expression of 15-lipoxygenase 2 (15-LOX2) in mouse prostate leads to hyperplasia and cell senescence. Oncogene 29, 4261–4275. doi: 10.1038/onc.2010.197
Tamary, E., Nevo, R., Naveh, L., Levin-Zaidman, S., Kiss, V., Savidor, A., et al. (2019). Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct 3:e00127. doi: 10.1002/pld3.127
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Vanhaeren, H., Nam, Y. J., De Milde, L., Chae, E., Storme, V., Weigel, D., et al. (2017). Forever young: the role of ubiquitin receptor DA1 and E3 ligase BIG BROTHER in controlling leaf growth and development. Plant Physiol. 173, 1269–1282. doi: 10.1104/pp.16.01410
Wang, H., Zhu, X., Li, H., Cui, J., Liu, C., Chen, X., et al. (2014). Induction of caspase-3-like activity in rice following release of cytochrome-f from the chloroplast and subsequent interaction with the ubiquitin-proteasome system. Sci. Rep. 4:5989. doi: 10.1038/srep05989
Woo, H. R., Chung, K. M., Park, J. H., Oh, S. A., Ahn, T., Hong, S. H., et al. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. doi: 10.1105/tpc.010061
Woo, H. R., Kim, H. J., Lim, P. O., and Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376.
Wu, X. Y., Kuai, B. K., Jia, J. Z., and Jing, H. C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 54, 936–952. doi: 10.1111/jipb.12005
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yu, J., Zhang, Y., Di, C., Zhang, Q., Zhang, K., Wang, C., et al. (2016). JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 67, 751–762. doi: 10.1093/jxb/erv487
Zhang, A., Lu, Q., Yin, Y., Ding, S., Wen, X., and Lu, C. (2010). Comparative proteomic analysis provides new insights into the regulation of carbon metabolism during leaf senescence of rice grown under field conditions. J. Plant Physiol. 167, 1380–1389. doi: 10.1016/j.jplph.2010.05.011
Zhang, Y., Ji, T. T., Li, T. T., Tian, Y. Y., Wang, L. F., and Liu, W. C. (2020). Jasmonic acid promotes leaf senescence through MYC2-mediated repression of CATALASE2 expression in Arabidopsis. Plant Sci. 299:110604. doi: 10.1016/j.plantsci.2020.110604
Zhang, Z., and Guo, Y. (2018). Hormone treatments in studying leaf senescence. Methods Mol. Biol. 1744, 125–132. doi: 10.1007/978-1-4939-7672-0_11
Zhang, Z., Li, W., Gao, X., Xu, M., and Guo, Y. (2020). DEAR4, a member of DREB/CBF family, positively regulates leaf senescence and response to multiple stressors in Arabidopsis thaliana. Front. Plant Sci. 11:367. doi: 10.3389/fpls.2020.00367
Zhou, J., Lu, D., Xu, G., Finlayson, S. A., He, P., and Shan, L. (2015). The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J. Exp. Bot. 66, 3353–3366. doi: 10.1093/jxb/erv148
Zhu, X., Chen, J., Xie, Z., Gao, J., Ren, G., Gao, S., et al. (2015). Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84, 597–610. doi: 10.1111/tpj.13030
Zhu, Z., An, F., Feng, Y., Li, P., Xue, L., Mu, A., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 12539–12544. doi: 10.1073/pnas.1103959108
Keywords: MYC2, JA, leaf senescence, AtUSR1, ring/U-box
Citation: Zhang Z, Xu M and Guo Y (2020) Ring/U-Box Protein AtUSR1 Functions in Promoting Leaf Senescence Through JA Signaling Pathway in Arabidopsis. Front. Plant Sci. 11:608589. doi: 10.3389/fpls.2020.608589
Received: 21 September 2020; Accepted: 17 November 2020;
Published: 16 December 2020.
Edited by:
Alejandro Ferrando, Universitat Politècnica de València, SpainReviewed by:
Krzysztof Zienkiewicz, Nicolaus Copernicus University in Toruń, PolandCopyright © 2020 Zhang, Xu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfeng Guo, Z3VveW9uZ2ZlbmdAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.