
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 January 2021
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.607878
 Jinsen Cai1,2,3
Jinsen Cai1,2,3 Weiwei Cai1,2,3
Weiwei Cai1,2,3 Xueying Huang1,2,3
Xueying Huang1,2,3 Sheng Yang1,2,3
Sheng Yang1,2,3 Jiayu Wen1,2,3
Jiayu Wen1,2,3 Xiaoqin Xia1,2,3
Xiaoqin Xia1,2,3 Feng Yang1,2,3
Feng Yang1,2,3 Yuanyuan Shi1,2,3
Yuanyuan Shi1,2,3 Deyi Guan1,2,3
Deyi Guan1,2,3 Shuilin He1,2,3*
Shuilin He1,2,3*Low-phosphorus stress (LPS) and pathogen attack are two important stresses frequently experienced by plants in their natural habitats, but how plant respond to them coordinately remains under-investigated. Here, we demonstrate that CaWRKY58, a known negative regulator of the pepper (Capsicum annuum) response to attack by Ralstonia solanacearum, is upregulated by LPS. Virus-induced gene silencing (VIGS) and overexpression of CaWRKY58 in Nicotiana benthamiana plants in combination with chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSA) demonstrated that CaWRKY58 positively regulates the response of pepper to LPS by directly targeting and regulating genes related to phosphorus-deficiency tolerance, including PHOSPHATE STARVATION RESPONSE1 (PHR1). Yeast two-hybrid assays revealed that CaWRKY58 interacts with a 14-3-3 protein (Ca14-3-3); this interaction was confirmed by pull-down, bimolecular fluorescence complementation (BiFC), and microscale thermophoresis (MST) assays. The interaction between Ca14-3-3 and CaWRKY58 enhanced the activation of PHR1 expression by CaWRKY58, but did not affect the expression of the immunity-related genes CaNPR1 and CaDEF1, which are negatively regulated by CaWRKY58 in pepper upon Ralstonia solanacearum inoculation. Collectively, our data indicate that CaWRKY58 negatively regulates immunity against Ralstonia solanacearum, but positively regulates tolerance to LPS and that Ca14-3-3 transcriptionally activates CaWRKY58 in response to LPS.
Plants are confronted with fluctuating ecological environments and are often exposed to biotic or abiotic stresses. Therefore, they have evolved sophisticated defense mechanisms to perceive stress, and to initiate and translate signaling pathways into appropriate defense responses that involve massive transcriptional reprogramming via transcription factors (Moore et al., 2011). To maximize fitness, plant growth, development, and responses to environmental cues must be tightly and coordinately regulated, which requires extensive crosstalk among plant responses to stresses, growth, and development (Fujita et al., 2006; Xia et al., 2015; Li et al., 2020). A single transcription factor is often involved in regulating several apparently disparate processes (Rushton et al., 2010), but the molecular details underlying this coordination often remain poorly understood.
Low-phosphorus stress (LPS) is one of the most important abiotic stresses experienced by plants, due to the requirement of phosphorus (P) for plant growth and development, but its limited availability in natural soils (Shen et al., 2011). Under this selection pressure, plants have evolved strategies to maximize its availability and to adapt to LPS. Massive transcriptional reprogramming activated by LPS has frequently been observed in plant species (Lan et al., 2013; Oono et al., 2013; O’Rourke et al., 2013; Fan et al., 2014; Deng et al., 2018; Xue et al., 2018). Many genes are transcriptionally activated by LPS, leading to enhanced P acquisition and utilization; these include genes that encode proteins such as Pi transporters (Miao et al., 2009; Remy et al., 2012; Ayadi et al., 2015; Liu et al., 2018a; Zhang et al., 2019), H+-ATPase (Yuan et al., 2017), phosphate transporter PHT4;6 (Hassler et al., 2012), PHOSPHATE STARVATION RESPONSE 1 (PHR1; Motte and Beeckman, 2017; Wang et al., 2019), protein kinases (Lan et al., 2013; Lei et al., 2014), purple acid phosphatases (Lan et al., 2013; Liu et al., 2018c), SPX (Zhang et al., 2016; Liu et al., 2018b; Osorio et al., 2019), and PHOSPHATE1 (PHO1; Hamburger et al., 2002). The processes involved in LPS responses include alterations in root architecture (Jain et al., 2007; Peret et al., 2011, 2014; Lopez-Arredondo et al., 2014; Postma et al., 2014; Strock et al., 2018), modification of the soil chemistry surrounding roots (Lopez-Arredondo et al., 2014) and the activation of metabolism to efficiently use phosphorus (Plaxton and Tran, 2011; Lopez-Arredondo et al., 2014). The transcription of many transcription factor genes is altered in response to LPS; these transcription factors act as positive or negative modulators of the response and include members of the MYB (Khan et al., 2014; Zhou et al., 2017; Wang et al., 2019), WRKY (Wang et al., 2014; Su et al., 2015; Dai et al., 2016), JAZ (Jasmonate-ZIM domain) (Aparicio-Fabre et al., 2013), bHLH (Valdes-Lopez and Hernandez, 2008b), zinc-finger (Devaiah et al., 2007b; Ding et al., 2016), AP2/ERF (Ramaiah et al., 2014), CCAAT box-binding (NF-Y; Qu et al., 2015), and auxin response factor (ARF; Shen et al., 2013) families, as well as PHR1 (Motte and Beeckman, 2017; Wang et al., 2019). Some of these transcription factors might be modulated in a context-dependent manner by interacting with other regulators, such as SPX domain-containing proteins (Zhong et al., 2018) and WRKY proteins (Zhou et al., 2017). In addition, the response of plants to phosphorus deficiency is closely related to other biological processes, such as the response to NO3− (Medici et al., 2015), Fe starvation (Dai et al., 2016), cold stress (Dai et al., 2016), S, Fe, Zn (Briat et al., 2015), K (Wang et al., 2002) and in particular, to plant immunity (Hiruma et al., 2016; Luo et al., 2019). The silencing of TaPT29-6A, a Pi transporter that plays a major role in Pi uptake from soil by roots, significantly increased the susceptibility of wheat plants to biotrophic, hemi-biotrophic, and necrotrophic pathogens (Zhang et al., 2019). Furthermore, the levels of secondary metabolites involved in plant immune reactions, including benzoxazinoids and flavonoids, differed significantly in plants grown under Pi-deficient conditions (Luo et al., 2019), indicating extensive crosstalk between plant response to phosphorus-deficiency stress and pathogen attack. However, the underlying mechanism for this crosstalk remains largely uncharacterized.
The WRKY proteins constitute one of the largest transcription factor families in many species (Eulgem et al., 2000), and WRKY family members have been implicated in diverse biological processes and particularly in plant immunity, by specifically targeting the conserved cognate W-box within the promoter regions of their target genes (Eulgem, 2006; Eulgem and Somssich, 2007; Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012; Buscaill and Rivas, 2014; Birkenbihl et al., 2017). WRKY transcription factors, including AtWRKY6 (Bakshi et al., 2015; Decker et al., 2017; Ye et al., 2018), WRKY42 (Sunar et al., 2015), WRKY45 (Wang et al., 2014), and WRKY75 in Arabidopsis (Devaiah et al., 2007a; Encinas-Villarejo et al., 2009), GbWRKY1 in cotton (Zhang et al., 2012) and OsWRKY74 in rice (Dai et al., 2016) function in response to LPS. The importance of WRKY transcription factors in the coordination of biological processes might reflect their flexible and diverse regulatory mechanisms, which are achieved by interactions with an array of proteins such as VQ proteins, MAPKs, chromatin remodeling proteins, calmodulin proteins, 14-3-3 proteins, WRKY or other transcription factors (Chi et al., 2013). However, the function of the majority of WRKY transcription factors in the plant LPS response and other biological processes has not been extensively characterized.
The 14-3-3 proteins are highly conserved eukaryotic proteins characterized by a conserved central core flanked by divergent regions at the N and C termini. These regions can be subdivided into two distinct groups: the ε group, and the plant-specific non-ε group. The 14-3-3 proteins act as molecular scaffolds or chaperones by physically interacting with target proteins via phosphorylated motifs containing phosphoserine residues of (R/K)XX(pS/pT)XP, (R/K)XXX(pS/pT)XP, and pS/pT-X1-2-COOH, in which pS and pT denote a phosphoserine and a phosphothreonine, respectively (Ormancey et al., 2017). The target proteins include H(+)-ATPase (Xu et al., 2016), nitrate reductase (Xu et al., 2016), ubiquitin ligase (Lu et al., 2016), protein kinases such as CDPKs (Ito et al., 2014), and transcription factors such as AtWRI1 (Kong and Ma, 2018), PIF7 (Huang et al., 2018), and bZIP (Takeo and Ito, 2017; Luang et al., 2018). The interaction of 14-3-3 proteins with other proteins alters the activity, stability, subcellular localization, or composition of the protein complex and this regulates physiological processes in plants that include metabolism, transport, growth, development, and stress responses (Ormancey et al., 2017). The expression of several 14-3-3 proteins is affected in planta in response to phosphorus deprivation (Cao et al., 2007) and these proteins have been implicated in plant responses to phosphorus deficiency (Cao et al., 2007; Ding et al., 2012; Xu et al., 2012a,b; Li et al., 2017; Yuan et al., 2017; Zhang et al., 2018). They function by modulating H+ efflux through affecting Arabidopsis plasma membrane H+-ATPase2 (AHA2) or AHA7 (Li et al., 2017), regulating leaf carbon allocation, increasing phloem sucrose transport to promote root growth, or activating root plasma membrane H(+)-ATPases to release more protons under phosphorus deficiency (Xu et al., 2012a,b). However, knowledge concerning the role of 14-3-3 proteins and the molecular basis for the plant response to phosphorus deficiency remain elusive.
Pepper (Capsicum annuum L.) is a member of the solanaceae and is an agriculturally important vegetable crop. It is mainly distributed or planted in uplands that contain soil-borne pathogens, including Ralstonia solanacearum, and that confront plants with other stresses such as LPS, which frequently cause growth retardation and yield loss (Forster et al., 1998). The application of phosphite, which causes phosphorus deficiency and growth retardation in pepper plants, significantly reduces the incidence of Phytophthora crown rot (Forster et al., 1998), indicates potential crosstalk between responses to pathogen attack and LPS. Notably, the WRKY protein CaWRKY58 was previously shown to negatively regulate the response of pepper to Ralstonia solanacearum (Wang et al., 2013). Here, we report that CaWRKY58 is transcriptionally induced by LPS and positively regulates the response of pepper to LPS by directly targeting PHR1 and physically interacting with 14-3-3.
Pepper (inbred line HN42) and N. benthamiana plants were cultivated as described previously (Cheng et al., 2017) before growth in hydroponic culture. The roots of plants at the seven-leaf stage were cleared and plants were transferred from soil to nutrient solution containing sufficient phosphorus (0.83 mM NH4+, 9 mM NO3−, 0.83 mM HPO32+, 6.0 mM K+, 1.5 mM Ca2+, 0.75 Mm Mg2+, 0.75 mM SO42−, 15.8 μM Fe2+, 10.3 μM Mn2+, 4.2 μM Zn2+, 43.5 μM B+, and 2.14 μM Cu2+). After 7 days, the plants were transferred to the above-mentioned hydroponic culture solution in which 0.83 mM HPO32+ was replaced with 0.08 mM HPO32+ to provide a LPS treatment. The plants were harvested at 7 dpt (days post treatment) and 30 dpt, to measure root system architecture (RSA) and root phosphorus content, respectively.
The full-length open reading frames (ORFs) of CaWRKY58 and Ca14-3-3 were cloned into the entry vector pDONR207 by BP reaction. After confirmation by sequencing, the ORFs were recombined into the destination vectors pEarleyGate103 [for C-terminal green fluorescent protein (GFP) fusions], pDEST-15 (for N-terminal GST fusions), pDEST17 (for N-terminal 6 × His fusions and expression in Escherichia coli) by LR reactions, using Gateway cloning (Invitrogen, Carlsbad, CA, United States). To construct vectors for VIGS-mediated gene silencing, a specific 300–400 bp fragment within the 3' UTRs of CaWRKY58 or CaPHR1 was amplified with specific primer pairs, using genomic DNA from pepper accession Zunla-1 as a template. The specificity of the amplicon was confirmed by BLAST search against the pepper genome.1 The PCR amplicon was cloned into pDONR207 by BP reaction and the identity of the clones was confirmed by sequencing before further subcloning into the PYL279 (pTRV2) vector, to generate pTRV2:CaWRKY58, or pTRV2:CaPHR1. All vectors were introduced into Agrobacterium tumefaciens strain GV3101.
Agrobacterium tumefaciens GV3101 cells containing pTRV1, pTRV2:00, pTRV2:Ca LPS, pTRV2:CaWRKY58, or pTRV2:CaPHR1 were grown overnight in LB medium supplemented with appropriate antibiotics and were then pelleted and resuspended to a final cell density of OD600 = 0.8 in infiltration medium (10 mM MES, 10 mM MgCl2, 200 μM acetosyringone, and pH5.4). GV3101 cells harboring pTRV1 were mixed with cells containing pTRV2:00, pTRV2:CaPDS, pTRV2:CaWRKY58 or pTRV2:CaPHR1 in a 1:1 ratio and infiltrated into the cotyledons of 2-week-old pepper seedlings. The plants were then placed in a growth chamber at 16°C in the dark for 56 h, and were then transferred to a growth room at 25°C and 60% humidity, with a light intensity of 60–70 μmol photons m−2 s−1 and a 16-h light/8-h dark photoperiod, until the plants infiltrated with pTRV:CaPDS exhibited a bleached phenotype, indicative of successful silencing of the PHYTOENE DESATURASE gene. Around 36 pepper plants for each gene every time, and the experiment was replicated three times.
Transgenic N. benthamiana plants were generated by leaf transformation as described previously (Dang et al., 2013), using the plant expression vector pEarleyGate103-CaWRKY58. Transgenic lines (T0) were selected with 0.04% BASTA and 20 BASTA-resistant T0 plants were further confirmed by PCR and RT-PCR using a CaWRKY58-specific primer pair (Supplementary Table S1). Seeds of T1 lines were harvested from BASTA-resistant and self-pollinated T0 plants; similarly, seeds of T2 lines were harvested from BASTA-resistant and self-pollinated plants of T1 lines. Plants of T2 lines were used for future analysis after confirmation by RT-PCR using CaWRKY58-specific primer pair and western blotting using an anti-GFP antibody.
Measurement of Pi concentration and plant total P content was performed as described previously (Nanamori et al., 2004; Zhou et al., 2008; Wu et al., 2011). The Pi concentration was normalized to fresh weight (FW) and the total P content was normalized to dry weight. Six biological replicates were performed in the experiment.
The plant expression vector pEarleyGate103-CaWRKY58 (Ca14-3-3) was transformed into GV3101 cells, which were infiltrated into leaves of pepper or N. benthamiana, following the method described previously (Guan et al., 2018). Leaves were harvested at indicated time points for further experiments, including subcellular protein localization and total RNA extraction.
To purify soluble CaWRKY58-6 × His and Ca14-3-3-GST fusion proteins, a pDEST17 plasmid harboring the full-length ORF of CaWRKY58 or the pDEST-15 vector containing the full-length ORF of Ca14-3-3 was introduced into E. coli strain BL21. Expression of the fusion proteins was induced by the addition of 1 mM (final concentration) isopropyl β-D-1-thiogalactopyranoside (IPTG) at 20°C for 12 h. The presence of the soluble proteins in the supernatant of the E. coli BL21 cell lysate was confirmed by SDS-PAGE gel electrophoresis.
Electrophoretic mobility shift assay (EMSA) was performed as described previously (Heravi and Altenbuchner, 2014; Shen et al., 2020), using CaWRKY58-6 × His purified from E. coli strain BL21 and the promoter fragments containing the wild-type or mutated W-box obtained from Cy5-labeled probes (CaPHR1-Pcy5, mCaPHR1-Pcy5) or Cy5-nonlabeled probes (CaPHR1-P; Supplementary Table S1). Before incubation with CaWRKY58-6 × His, CaPHR1-Pcy5, and CaPHR1-P were mixed in different ratios to assay their competitive interaction with CaWRKY58-6 × His. The EMSA blot image was generated with Odyssey CLX (LI-COR). The experiment was replicated three times.
The yeast two-hybrid assay was performed as described previously (Liu et al., 2015), the ORF of CaWRKY58 was cloned into the pDONR201 (satellite vector) and transferred into the pDEST32 (destination vector) for the yeast two-hybrid screen. The yeast two-hybrid library screening was performed according to the manufacturer’s instructions (Invitrogen).
The ORF of CaWRKY58 and Ca14-3-3 were cloned into the entry vector pDONR207 first, then the ORF of CaWRKY58 was recombined into destination vector pEYFP-N1 (CaWRKY58-YFPN) and the ORF of Ca14-3-3 was recombined into destination vector pEYFP-C1 (Ca14-3-3-YFPC). The constructs were transformed into the Agrobacterium tumefaciens strain GV3101, respectively. The GV3101 cells containing the two vectors were mixed in a 1:1 ratio and were infiltrated into N. benthamiana leaves. The cell layers of infiltrated leaves were visualized by microscopy (Leica, Germany) at 48 hours post infiltration (hpi). The experiment was replicated three times.
The interaction between CaWRKY58 and Ca14-3-3 was confirmed by microscale thermophoresis (MST; Zillner et al., 2012). For this, Ca14-3-3-GFP, or GFP isolated by immunoprecipitation with an anti-GFP antibody from pepper leaves transiently overexpressing Ca14-3-3-GFP or GFP were used as targets, and CaWRKY58-6 × His was used as a ligand. Protein-protein interactions between Ca14-3-3-GFP or GFP, and CaWRKY58-6 × His, were measured using 20 nM Ca14-3-3-GFP or GFP. The CaWRKY58-6 × His solution was diluted to a concentration range from 1.0E−10 to 1.0E−3 mM and the CaWRKY58-6 × His fusion protein was incubated with the labeled protein for 10 min in interaction buffer. The samples were then loaded into Monolith NT.115 Capillaries (Cat. MO-K002, NanoTemper Technologies, Germany) using 50% IR laser power and an LED excitation source, with λ = 470 nm at ambient temperature. The Kd values were calculated for interactions between Ca14-3-3-GFP and CaWRKY58-6 × His or GFP and CaWRKY58-6XHis using NanoTemper Analysis 1.2.20 software (Zillner et al., 2012; Papageorgiou et al., 2016). The experiment was replicated three times.
The physical interaction between CaWRKY58 and Ca14-3-3 was confirmed by a pull-down assay. Ca14-3-3-GST fusion protein isolated from E. coli was incubated with GSH magnetic beads (Beaver, Suzhou, China) for 1 h, and CaWRKY58-GFP isolated from pepper leaves that transiently overexpressed CaWRKY58-GFP was added to the mixture and incubated for 1 h. After washing and elution, an appropriate amount of 1 M tris-HCl (pH 8.0) solution was added to adjust the pH. The protein was examined by immunoblotting with anti-GFP and anti-GST antibodies (Guan et al., 2018). The experiment was replicated three times.
A BIO-RAD Real-time PCR system (Foster City, CA, United States) and SYBR Premix Ex Taq II system (TaKaRa, Dalian, China) were used to quantify gene expression. Total RNA extraction and real-time RT-PCR were carried out as described previously (Dang et al., 2013; Cai et al., 2015; Guan et al., 2018). The specific primer pairs used are listed in Supplementary Table S1. The relative transcript level of each sample was normalized to that of CaACTIN (GQ339766). Six biological replicates were performed in the experiment.
Chromatin immunoprecipitation (ChIP) was performed as described previously (Guan et al., 2018). Briefly, CaWRKY58-GFP was transiently overexpressed in pepper leaves via agroinfiltration, and the DNA-protein complexes were isolated, sheared into fragments of 300–500 bp in length and immunoprecipitated using an anti-GFP antibody. The DNA was purified and used as a template for PCR and qPCR. The specific primer pairs of the fragment containing W-box in the promoter of CaPHR1 and CaNPR1 are listed in Supplementary Table S1. The experiment was replicated three times.
The Ralstonia solanacearum (FJC100301) cultivation and inoculation was performed as described previously (Dang et al., 2013; Shen et al., 2020). The pepper root was harvested at 24 h post inoculation for transcript level detection of related genes.
CaWRKY58 is constitutively expressed in pepper plants, but is downregulated by Ralstonia solanacearum infection (RSI) and negatively regulates the response to RSI (Wang et al., 2013). To test the potential role of CaWRKY58 in other plant biological processes, we quantified the CaWRKY58 transcript level in pepper plants challenged with several different stresses (salinity, heat stress, and drought), including LPS. Expression of CaWRKY58 was upregulated by LPS at 3 and 5 dpt (Figure 1A), indicating that it might be involved in the LPS response.
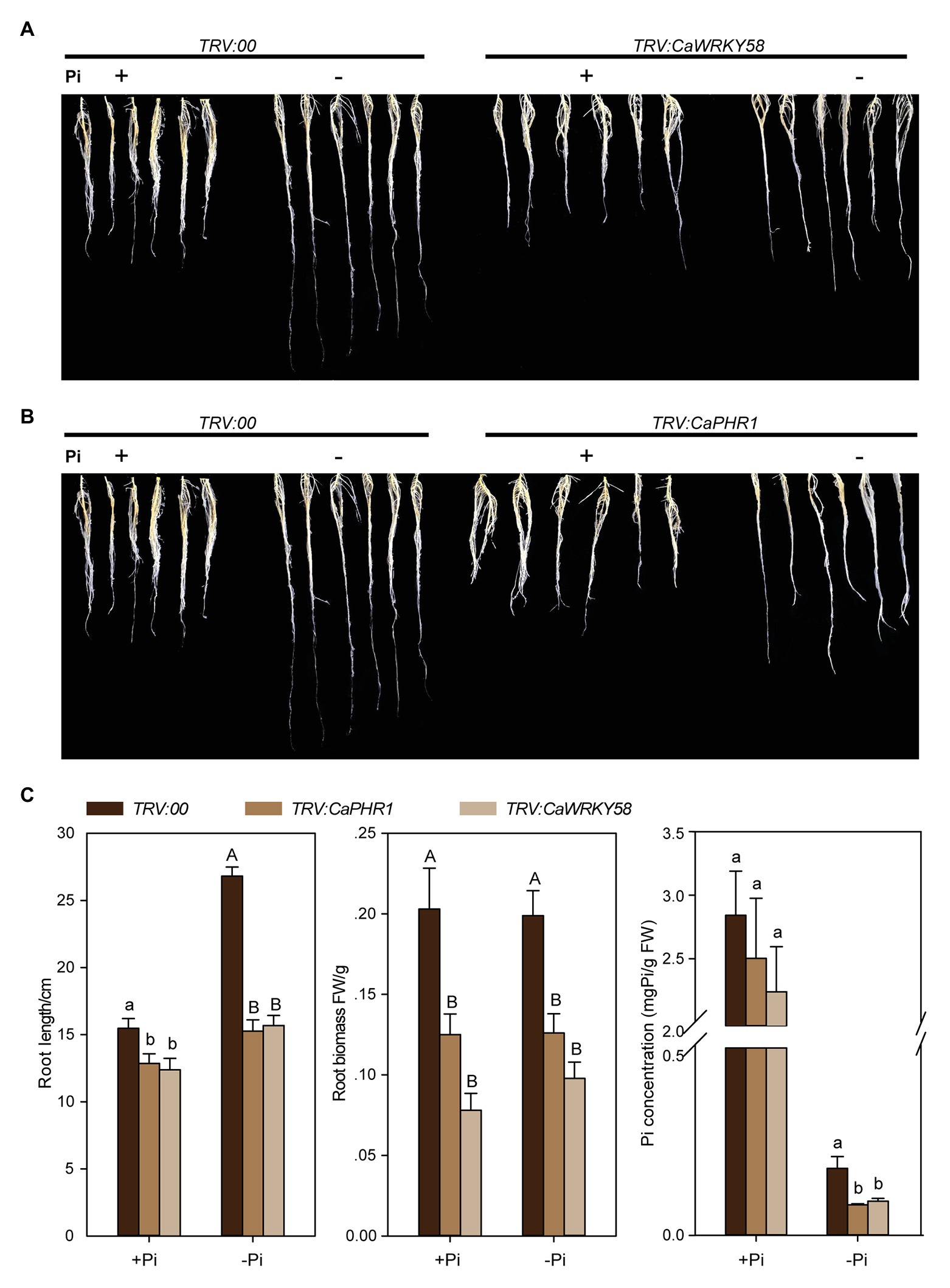
Figure 1. CaWRKY58 is induced by and plays a role in pepper response to low-phosphorus stress (LPS). (A), Relative transcript level of CaWRKY58 in roots of pepper plants challenged with low-phosphorus stress LPS measured by quantitative real-time RT-PCR (qRT-PCR) at 1, 3, and 5 dpt (days post treatment). (B), Root system architecture (RSA) of Nicotiana benthamiana plants overexpressing CaWRKY58 following exposure to LPS. Transgenic N. benthamiana plants at the 4–6 leaf stage were grown in nutrient solution with sufficient Pi and were transferred to Pi-deficient solution for 7 days. Control plants were cultured in nutrient solution with sufficient Pi, (bars = 5 cm). (C), Root length and weight of N. benthamiana plants overexpressing CaWRKY58 following exposure to LPS. FW: fresh weight, WT: wild type, CaWRKY58-OE: N. benthamiana plants overexpressing CaWRKY58, +Pi: with sufficient Pi, −Pi: with low Pi. In (A,C), the data represent the mean ± SD for three biological replicates. The uppercase and lowercase above the bars indicate significant differences (p < 0.01) and (p < 0.05), respectively, according to Fisher’s protected least significant difference (LSD) test.
To further explore the role of CaWRKY58 in the response to LPS in pepper, N. benthamiana plants were generated that transgenically overexpressed CaWRKY58. Twenty T0 transgenic plants were produced, and the corresponding T1 and T2 lines were produced. Three of the resulting lines with high levels of CaWRKY58 expression were selected for further study. Transgenic and control plants were subjected to LPS, and their RSA and root Pi content were quantified. The three transgenic N. benthamiana lines exhibited an enlarged RSA and enhanced root Pi content compared with control plants (Figures 1B,C, Supplementary Figure S1), suggesting that overexpression of CaWRKY58 enhanced the tolerance of N. benthamiana to LPS.
To test the potential role of CaWRKY58 in the response of pepper plants to LPS, we generated CaWRKY58-silenced pepper plants via virus-induced gene silencing (VIGS) using a vector described previously (Wang et al., 2013). The efficiency and specificity of CaWRKY58 silencing were assayed, and the transcript level of CaWRKY58 in TRV:CaWRKY58 pepper plants was approximately 20–30% that in control plants, the transcript level of CaWRKY65, whose sequence is more highly similar to that of CaWRKY58 than other WRKY members in pepper, was not significantly affected by CaWRKY58 silencing (Supplementary Figure S2), indicating that silencing was specific for CaWRKY58. The CaWRKY58-silenced pepper plants were subjected to LPS, and the RSA of TRV:CaWRKY58 and wild-type control plants was enlarged upon LPS treatment compared with that in non-limiting phosphorus conditions, and the RSA of TRV:CaWRKY58 plants under LPS was significantly reduced compared with that of control plants (Figures 2A,C). In parallel, the Pi content was analyzed and was significantly lower in TRV:CaWRKY58 plants than in wild-type plants (Figure 2C). Collectively, these data indicate that CaWRKY58 positively regulates the response of pepper plants to LPS treatment.
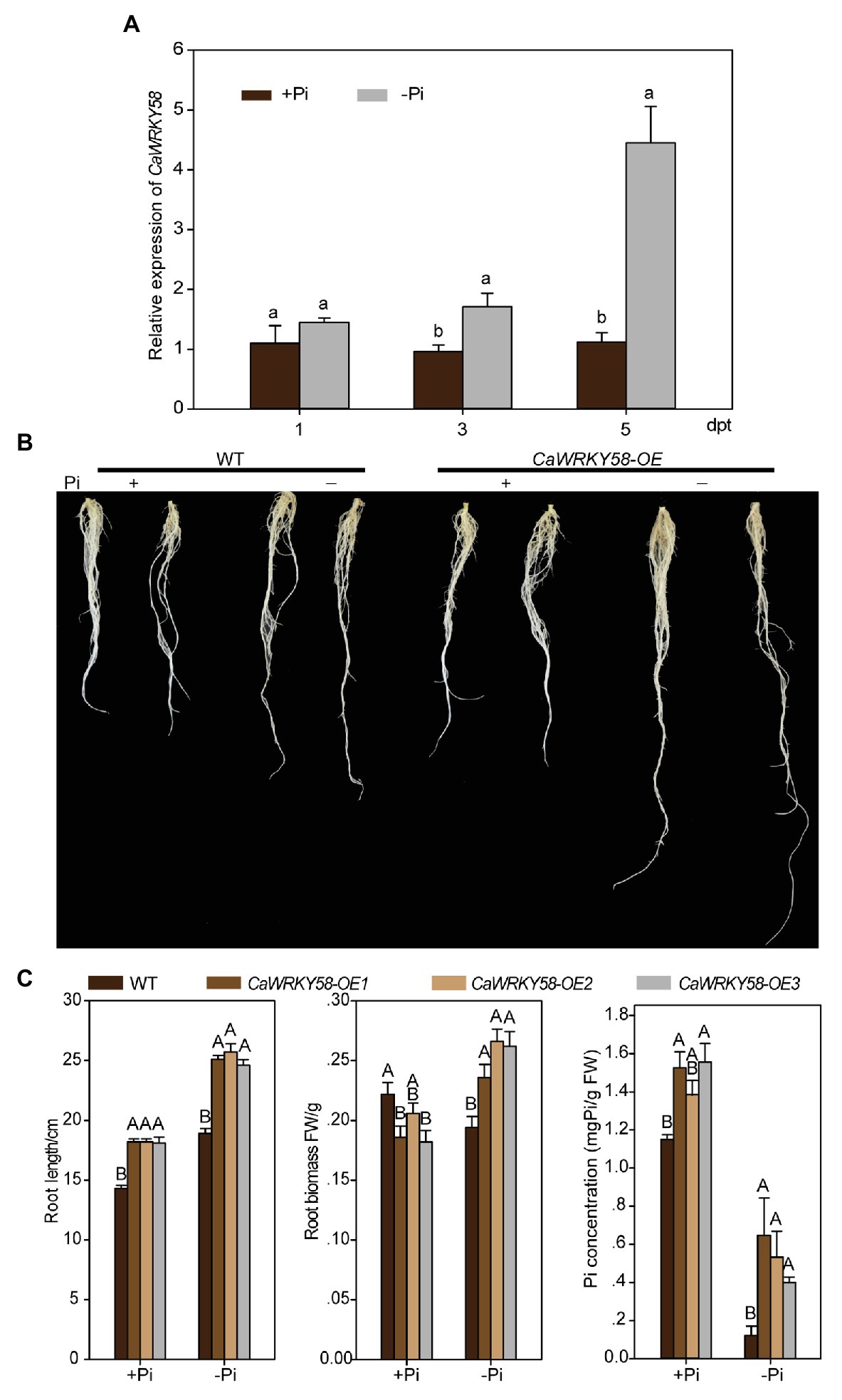
Figure 2. The effect of Pi deficiency on root growth of CaPHR1 and CaWRKY58-silenced pepper plants. (A), RSA of CaWRKY58-silenced pepper plants upon LPS. (B), RSA of CaPHR1-silenced pepper plants treated with LPS at 7 dpt; both CaWRKY58 and CaPHR1 were silenced by virus-induced gene silencing (VIGS; bars = 3 cm). (C), Root length and root weight of pepper plants subjected to LPS at 7 dpt. FW: fresh weight. In (C), the data represent the mean ± SD for three biological replicates. The uppercase and lowercase above the bars indicate significant differences (p < 0.01) and (p < 0.05), respectively, according to Fisher’s protected LSD test.
Given that WRKY proteins function mainly by binding the conserved W-box in promoters of their target genes, and CaWRKY58 regulates the response of pepper plants to LPS, we hypothesized that CaWRKY58 positively regulates the response to LPS by binding to the W-box in the promoters of genes related to LPS tolerance. To identify the potential target genes of CaWRKY58 in response to LPS, the promoters of potential LPS-responsive genes were screened for the presence of a W-box, which was identified in the promoter of CaPHR1 (LOC107863907) in the pepper genome. CaPHR1 is the ortholog of PHR1, and PHR1 is a MYB transcription factor that positively regulates the response of different plant species to LPS (Nilsson et al., 2007; Valdes-Lopez et al., 2008a; Ren et al., 2012; Ruan et al., 2015; Pacak et al., 2016; Xue et al., 2017). To assess whether CaPHR1 is regulated by CaWRKY58 in response to LPS, the level of CaPHR1 expression was quantified in the leaves of pepper plants that transiently overexpressed CaWRKY58 and was significantly higher than that in the leaves of control plants. However, the expression of CaPHO2 (the ortholog of PHOSPHATE2 in pepper) was not significantly different to that in control plants (Figure 3A). By contrast, LPS significantly upregulated CaPHR1, whereas silencing CaWRKY58 by VIGS significantly blocked this upregulation in leaves (Figure 3B). To elucidate the role of CaPHR1 in the response to LPS, CaPHR1 was silenced by VIGS and the silenced plants were subjected to LPS. A reduced RSA was observed in TRV:CaPHR1 plants compared to in the wild-type plants at 7 dpt, which was similar to that in TRV:CaWRKY58 plants (Figures 2B,C). These results indicate that CaWRKY58 positively regulates the response to LPS by regulating the expression of CaPHR1.
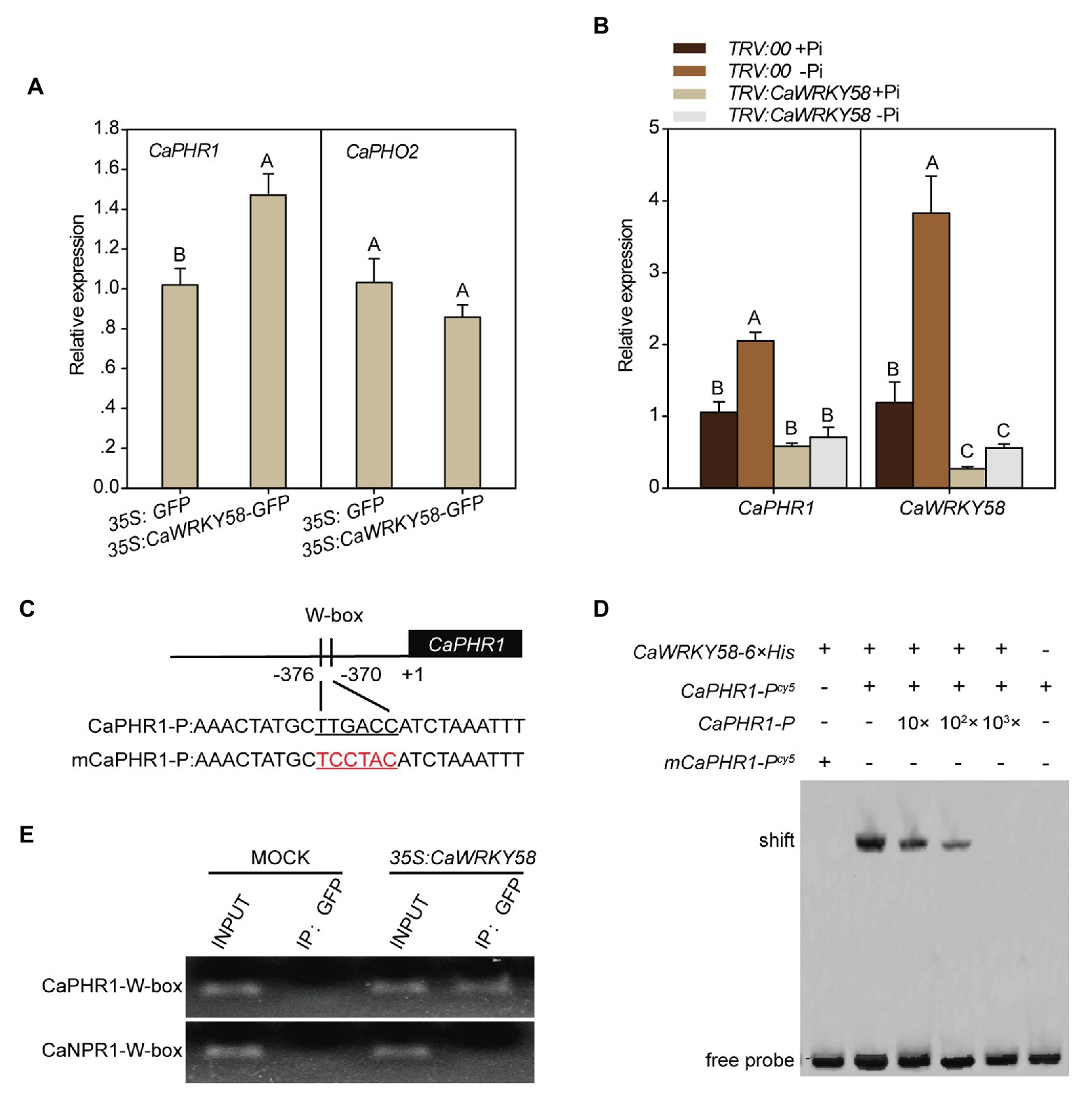
Figure 3. CaWRKY58 directly binds the promoter of CaPHR1 via the W-box and transcriptionally regulates its transcription. (A), Effect of transient overexpression of CaWRKY58-GFP on the transcription of CaPHR1 and CaPHO2. The relative transcript level of CaPHR1 to that in the leaves of plants transiently overexpressing GFP was detected by RT-qPCR at 48 hours post infiltration (hpi). (B), Effect of CaWRKY58 silencing on the transcription of CaPHR1 and CaPHO2 in plants with and without LPS at 5 dpt. (C), Location of the W-box in the promoter of CaPHR1 and the sequence of the mutated W-box. (D), Electrophoretic mobility shift assay (EMSA) for the binding of CaWRKY58-6 × His to the CaPHR1 promoter containing the W-box (CaPHR1-P) and to the promoter fragment of CaPHR1 containing a mutated W-box (mCaPHR1-P). (E), Chromatin immunoprecipitation (ChIP)-qPCR assay showing that CaWRKY58 binds the W-box within the promoter of CaPHR1. CaWRKY58-GFP was transiently overexpressed in leaves for 48 h and the chromatin was isolated, sheared into 300–500 bp fragments and immunoprecipitated using an anti-GFP antibody. The purified DNA fragments were used as a template for qPCR with a specific primer pair to the promoter fragment of CaNPR1 and CaPHR1 containing the W-box. In (A,B), the data represent the mean ± SD for three biological replicates. The uppercase above the bars indicates a significant difference (p < 0.01) according to Fisher’s protected LSD test.
To analyze whether CaPHR1 is directly targeted by CaWRKY58, the ability of CaWRKY58 to bind the CaPHR1 promoter was studied by EMSA using CaWRKY58-6 × His purified from E. coli and the promoter of CaPHR1 (CaPHR1-Pcy5), which contains a W-box. We detected a mobility shift when CaWRKY58-6 × His was incubated with excess CaPHR1-P (CaPHR1-Pcy5), but no mobility shift was detected when CaWRKY58-6 × His was incubated with excess CaPHR1-P containing a mutated W-box (mCaPHR1-Pcy5; Figures 3C,D). In parallel, a ChIP-PCR assay showed that CaWRKY58-GFP was enriched at the W-box containing CaPHR1 promoter but not at the W-box containing CaNPR1 promoter (mCaPHR1-P; Figure 3E). These results indicate that CaWRKY58 directly bind to the promoter of CaPHR1.
The function of WRKY transcription factors is often modulated by other regulatory proteins via protein-protein interactions (Chi et al., 2013). To identify the potential interacting partners of CaWRKY58, a yeast two-hybrid assay was performed using CaWRKY58 as a bait. Among the 25 positive clones, a 14-3-3 protein (LOC107867389) was identified. Because the deduced amino-acid sequence contained a conserved 14-3-3 domain and exhibited a high sequence identity to 14-3-3 proteins (Supplementary Figure S3), we named this protein Ca14-3-3. The expression level of Ca14-3-3 was significantly enhanced by LPS treatment (Figure 4A). The interaction between CaWRKY58 and Ca14-3-3 was confirmed by a pull-down assay by incubating Ca14-3-3-GST purified from E. coli with proteins isolated from pepper leaves that transiently overexpressed CaWRKY58-GFP. Following purification with GST magnetic beads, the protein complex that included Ca14-3-3-GST was immunoblotted with an anti-GFP antibody, which detected the presence of CaWRKY58-GFP. As expected, this demonstrated that Ca14-3-3 interacted with CaWRKY58 (Figure 4B). In parallel, the interaction between 14-3-3 and CaWRKY58 was further confirmed by bimolecular fluorescence complementation (BiFC) in N. benthamiana leaves. The leaves were analyzed at 48 hpi and YFP fluorescence was detected in the nucleus of epidermal cells of leaves co-expressing CaWRKY58 and Ca14-3-3 fusion proteins, but no fluorescence was observed in control leaves (Figure 4C). In addition, we purified CaWRKY58-6 × His from E. coli soluble extracts. Purified CaWRKY58-6 × His fusion protein was serially diluted and mixed with a constant amount of Ca14-3-3-GFP before performing an MST assay, which allowed the dissociation constant (Kd) of the CaWRKY58-6 × His-Ca14-3-3-GFP complex to be calculated. The CaWRKY58-Ca14-3-3 protein pair produced a clear binding curve, with a Kd of 2.0971E−7 M (Figure 4D). Collectively, these data indicate that CaWRKY58 and Ca14-3-3 physically interact with each other.
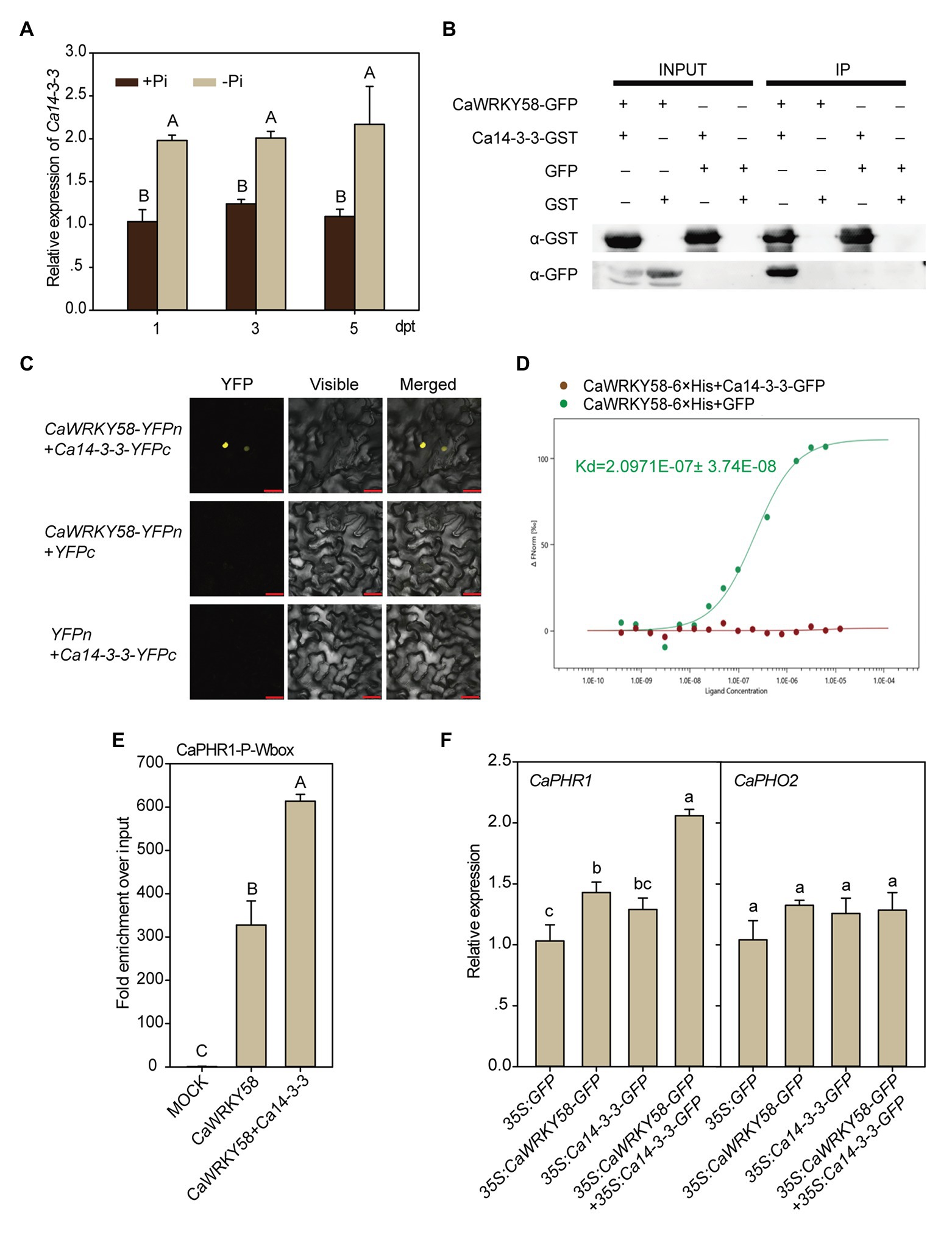
Figure 4. The transcriptional activation of CaPHR1 by CaWRKY58 is enhanced by interaction between CaWRKY58 and Ca14-3-3. (A), Transcription of Ca14-3-3 was induced by LPS in roots of pepper plants. (B), Pull-down assay to show the interaction between Ca14-3-3 and CaWRKY58. CaWRKY58-GFP was transiently overexpressed in leaves of pepper plants by agroinfiltration. Ca14-3-3-GST fusion protein purified from Escherichia coli (BL21) by GST magnetic beads was used as the bait protein, the protein complex was further purified using GST magnetic beads, and CaWRKY58 in the protein complex was detected by immunoblotting with an anti-GFP antibody. (C), Interaction between Ca14-3-3 and CaWRKY58 assayed by bimolecular fluorescence complementation (BiFC; bars = 25 μM). (D), Ca14-3-3/CaWRKY58 interaction was assayed by microscale thermophoresis (MST), CaWRKY58-6 × His (as the ligand) was purified from E. coli (BL21) and Ca14-3-3-GFP (as the target) was transiently overexpressed in leaves of pepper plants by agroinfiltration. (E), Effect of transient overexpression of Ca14-3-3 on the binding of CaWRKY58 to the CaPHR1 promoter by ChIP-qPCR assay. (F), Effect of transient overexpression of Ca14-3-3 on CaPHR1 and CaPHO2 transcription with or without CaWRKY58 transient co-overexpression. In (A,E,F), the data represent the mean ± SD for three biological replicates. The uppercase and lowercase above the bars indicate significant differences (p < 0.01) and (p < 0.05), respectively, according to Fisher’s protected LSD test.
Because CaWRKY58 interacts with Ca14-3-3 and directly transcriptionally upregulates CaPHR1 during the response of pepper to LPS, we hypothesized that Ca14-3-3 might affect the function of CaWRKY58 in this process. To test this hypothesis, we analyzed the effect of transient co-overexpression of CaWRKY58 and Ca14-3-3 in leaves of pepper plants on the transcription of CaPHR1, by qRT-PCR. The transcript level of CaPHR1 was promoted by transient overexpression of CaWRKY58 alone, and this promotion was enhanced by the additional transient overexpression of Ca14-3-3, although the overexpression of Ca14-3-3 alone did not significantly enhance the transcription of CaPHR1. By contrast, the expression of CaPHO2 was not affected by either transient overexpression of CaWRKY58 or its transient overexpression together with Ca14-3-3 (Figure 4F). To test whether binding of the CaPHR1 promoter by CaWRKY58 was enhanced by the interaction of Ca14-3-3 with CaWRKY58; ChIP-qPCR assay was performed on chromatin isolated from leaves of pepper plants that either transiently overexpressed CaWRKY58-GFP alone, or CaWRKY58-GFP and Ca14-3-3-GFP together. The chromatin was sheared into fragments 300–500 bp in length and was immunoprecipitated with an anti-GFP antibody, and DNA derived from the immunoprecipitated chromatin was used as template for ChIP-qPCR using a primer pair specific for the W-box within the CaPHR1 promoter. The W-box sequence was significantly enriched by transient co-overexpression of Ca14-3-3-FLAG and CaWRKY58-GFP (Figure 4E). Consistent with this, the transcript level of CaPHR1 in plants that transiently co-expressed CaWRKY58-GFP and Ca14-3-3-GFP was significantly higher than that in plants that expressed CaWRKY58-GFP or Ca14-3-3-GFP alone (Figure 4F). These data indicate that the transcriptional regulation of CaPHR1 by CaWRKY58 is enhanced by Ca14-3-3.
Previous study showed that CaWRKY58 negatively regulates the response of pepper to RSI by downregulating immunity-related genes, including CaDEF1 and CaNPR1 (Wang et al., 2013). Here, we demonstrate that CaWRKY58 positively regulates the response to LPS by upregulating CaPHR1. To address whether CaWRKY58 regulates its target genes in a context-dependent manner, the transcript levels of CaPHR1, CaNPR1, and CaDEF1 were quantified in pepper plants challenged with LPS and RSI. The expression of CaPHR1 was only upregulated by LPS, whereas that of CaDEF1 and CaNPR1 was upregulated by RSI, indicating that the expression of these genes is context dependent (Supplementary Figure S4). Furthermore, the transcript levels of CaDEF1, and CaNPR1 in CaWRKY58-silenced pepper plants were only enhanced in the presence of sufficient Pi, but not under LPS (Supplementary Figure S5). Furthermore, although the positive transcriptional regulation of CaPHR1 by CaWRKY58 was enhanced by transient co-overexpression of Ca14-3-3, the transcriptional regulation of CaDEF1 and CaNPR1 by CaWRKY58 was not affected by Ca14-3-3 (Supplementary Figure S6). These data indicate that the activation of CaPHR1 expression by CaWRKY58 occurs only in LPS but not in RSI; on the other hand, the negative regulation of CaDEF1 and CaNPR1 by CaWRKY58 does not occur under LPS.
Although WKRY and 14-3-3 proteins have been previously been shown to participate in plant immunity and the LPS response, and some members of both families physically interact, their functions in crosstalk between plant immunity and response to LPS remain uninvestigated. In this study, we provide evidence that in addition to acting as a negative regulator of immunity against RSI in pepper, CaWRKY58 positively regulates tolerance to LPS, an effect that is mediated by interaction with Ca14-3-3.
The data from CaWRKY58 upregulation against LPS and the data from loss--of-function and gain-of-function assay indicate that CaWRKY58 positively regulates the response to LPS by regulating CaPHR1, which related to LPS tolerance (Motte and Beeckman, 2017; Wang et al., 2019), as well as by modulating RSA, a key parameter that is positively related to LPS tolerance (Jain et al., 2007; Mori et al., 2016). Primary root growth was strongly inhibited by LPS in Arabidopsis (Peret et al., 2011; Gutierrez-Alanis et al., 2018); however, we observed that the growth of primary and lateral roots in pepper was promoted by P deficiency, which was enhanced by CaWRKY58 overexpression (Figure 1B). This indicates that the regulation of RSA by LPS might differ among plant species. The data also indicate that CaWRKY58 positively regulates CaPHR1 expression directly, because the W-box within the CaPHR1 promoter was bound by CaWRKY58 according to ChIP-qPCR and EMSA (Figure 3D). PHR1 is a MYB-CC-type transcription factor that plays a key role in regulating the expression of Pi starvation-induced (PSI) genes, which leads to enhanced phosphate uptake (Nilsson et al., 2007). We conclude that CaWRKY58 positively regulates the LPS response in pepper by targeting PHR1.
In addition to regulation at the transcriptional level, the expression and function of transcription factors is frequently regulated post-translationally via interactions with other proteins (Schutze et al., 2008; Chi et al., 2013; Alves et al., 2014; Pireyre and Burow, 2015; Ohama et al., 2017). In particular, WRKY transcription factors interact with other WRKY proteins, VQ motif-containing proteins, MAPKs, chromatin remodeling proteins, calmodulin, and 14-3-3 proteins (Chi et al., 2013). 14-3-3 proteins regulate transcription by interacting with the transcription factors or activators RSG (Igarashi et al., 2001; Ishida et al., 2008), GmMYB176 (Li et al., 2012), MYBS2 (Chen et al., 2019), and CBF (Liu et al., 2017), and modifying their subcellular localization, stability, or transcription, leading to an appropriate transcriptional output. Moreover, 14-3-3 proteins have been implicated in the response to P deficiency (Wang et al., 2002; Xu et al., 2012b); however, it remains unknown whether 14-3-3 proteins function as transcription activators in these conditions. The data here indicate that the transcriptional activation of CaPHR1 by CaWRKY58 is enhanced by the interaction between CaWRKY58 and Ca14-3-3, suggesting that Ca14-3-3 might promote the transcriptional activation activity of CaWRKY58 during the P-deficiency response. Because the interaction between 14-3-3 proteins and their protein partners is either dependent on or related to phosphorylation (Li et al., 2012; Ito et al., 2014; Chen et al., 2019) mediated by kinases including CDPKs (Ishida et al., 2008; Liu et al., 2017), we speculate that the interaction between Ca14-3-3 and CaWRKY58 might be associated with phosphorylation mediated by unidentified kinases. The further identification of these kinases might provide insight into the mechanism that underlies the LPS response in pepper.
We demonstrated previously that CaWRKY58 negatively regulates the response to RSI in pepper (Wang et al., 2013); therefore, CaWRKY58 functions in the response of pepper to both RSI and LPS. These two stresses activate convergent signaling nodes, such as WRKY45 (Shimono et al., 2007; Wang et al., 2014), WRKY75 (Devaiah et al., 2007a; Encinas-Villarejo et al., 2009), and PR10a (Huang et al., 2016), suggesting that plant immunity might be closely related to phosphorus nutrition. On the one hand, application of sufficient phosphate fertilizer is generally essential to increase root ramification and strength, thereby conferring vitality and disease resistance to plants (Sharma et al., 2013). By contrast, phosphate starvation can result in the repression of plant immunity (Finkel et al., 2019), and in Arabidopsis, PHR1 also directly represses immune responses (Castrillo et al., 2017). On the other hand, pathogen infection might decrease phosphate availability (Petters et al., 2002). However, the significance of these shared regulatory proteins in plant adaptation to the environment and their contribution to the relationship between plant phosphorus nutrition and disease resistance remains to be elucidated. The data here showed that LPS tolerance and immunity-related genes were differentially activated in pepper plants upon RSI and LPS treatments, and immunity-related genes, including CaDEF1 and CaNPR1, were not negatively regulated by CaWARKY58 in plants exposed to LPS treatment, or following transient co-overexpression of CaWRKY58 and Ca14-3-3. This indicates that CaWRKY58 and Ca14-3-3 contribute to the positive regulation of CaWRKY58 in response to LPS, but do not contribute to its negative regulation in immunity against RSI.
Collectively, the data in this study show that CaWRKY58 acts as positive regulator of LPS in pepper plants by directly targeting and regulating CaPHR1 and Ca14-3-3 acts as a specific transcriptional activator of CaWRKY58 during the LPS response.
The original contributions presented in the study are included in the article/Supplementary Material and further inquiries can be directed to the corresponding author.
SH and JC conceived the research and designed the experiments. JC, XH, SY, WC, JW, XX, FY, and YS performed the experiments. JC, SY, and DG analyzed the data. SH wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (31572136, 31372061, and 31902032), Development Fund Project of Fujian Agriculture and Forestry University (CXZX2016158, CXZX2017548).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Mark D. Curtis for kindly providing the Gateway destination vectors and Dr. S. P. Dinesh-Kumar of Yale University for the pTRV1 and pTRV2 vectors.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.607878/full#supplementary-material
Alves, M. S., Dadalto, S. P., Goncalves, A. B., de Souza, G. B., Barros, V. A., and Fietto, L. G. (2014). Transcription factor functional protein-protein interactions in plant defense responses. Proteome 2, 85–106. doi: 10.3390/proteomes2010085
Aparicio-Fabre, R., Guillen, G., Loredo, M., Arellano, J., Valdes-Lopez, O., Ramirez, M., et al. (2013). Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol. 13:26. doi: 10.1186/1471-2229-13-26
Ayadi, A., David, P., Arrighi, J. F., Chiarenza, S., Thibaud, M. C., Nussaume, L., et al. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 167, 1511–1526. doi: 10.1104/pp.114.252338
Bakshi, M., Vahabi, K., Bhattacharya, S., Sherameti, I., Varma, A., Yeh, K. W., et al. (2015). WRKY6 restricts Piriformospora indica-stimulated and phosphate-induced root development in Arabidopsis. BMC Plant Biol. 15:305. doi: 10.1186/s12870-015-0673-4
Birkenbihl, R. P., Liu, S., and Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.2017.04.004
Briat, J. F., Rouached, H., Tissot, N., Gaymard, F., and Dubos, C. (2015). Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front. Plant Sci. 6:290. doi: 10.3389/fpls.2015.00290
Buscaill, P., and Rivas, S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. doi: 10.1016/j.pbi.2014.04.004
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Cao, A., Jain, A., Baldwin, J. C., and Raghothama, K. G. (2007). Phosphate differentially regulates 14-3-3 family members and GRF9 plays a role in pi-starvation induced responses. Planta 226, 1219–1230. doi: 10.1007/s00425-007-0569-0
Castrillo, G., Teixeira, P. J., Paredes, S. H., Law, T. F., de Lorenzo, L., Feltcher, M. E., et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. doi: 10.1038/nature21417
Chen, Y. S., Ho, T. D., Liu, L., Lee, D. H., Lee, C. H., Chen, Y. R., et al. (2019). Sugar starvation-regulated MYBS2 and 14-3-3 protein interactions enhance plant growth, stress tolerance, and grain weight in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 21925–21935. doi: 10.1073/pnas.1904818116
Cheng, W., Xiao, Z., Cai, H., Wang, C., Hu, Y., Xiao, Y., et al. (2017). A novel leucine-rich repeat protein, CaLRR51, acts as a positive regulator in the response of pepper to Ralstonia solanacearum infection. Mol. Plant Pathol. 18, 1089–1100. doi: 10.1111/mpp.12462
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J. Q., et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6, 287–300. doi: 10.1093/mp/sst026
Dai, X., Wang, Y., and Zhang, W. H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Decker, E. L., Alder, A., Hunn, S., Ferguson, J., Lehtonen, M. T., Scheler, B., et al. (2017). Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol. 216, 455–468. doi: 10.1111/nph.14506
Deng, Q. W., Luo, X. D., Chen, Y. L., Zhou, Y., Zhang, F. T., Hu, B. L., et al. (2018). Transcriptome analysis of phosphorus stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff). Biol. Res. 51:7. doi: 10.1186/s40659-018-0155-x
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007a). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971
Devaiah, B. N., Nagarajan, V. K., and Raghothama, K. G. (2007b). Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 145, 147–159. doi: 10.1104/pp.107.101691
Ding, Y., Ndamukong, I., Xu, Z., Lapko, H., Fromm, M., and Avramova, Z. (2012). ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 8:e1003111. doi: 10.1371/journal.pgen.1003111
Ding, W., Wang, Y., Fang, W., Gao, S., Li, X., and Xiao, K. (2016). TaZAT8, a C2H2-ZFP type transcription factor gene in wheat, plays critical roles in mediating tolerance to pi deprivation through regulating P acquisition, ROS homeostasis and root system establishment. Physiol. Plant. 158, 297–311. doi: 10.1111/ppl.12467
Encinas-Villarejo, S., Maldonado, A. M., Amil-Ruiz, F., de los Santos, B., Romero, F., Pliego-Alfaro, F., et al. (2009). Evidence for a positive regulatory role of strawberry (Fragaria x ananassa) Fa WRKY1 and Arabidopsis at WRKY75 proteins in resistance. J. Exp. Bot. 60, 3043–3065. doi: 10.1093/jxb/erp152
Eulgem, T. (2006). Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2:e126. doi: 10.1371/journal.ppat.0020126
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Fan, F., Cui, B., Zhang, T., Qiao, G., Ding, G., and Wen, X. (2014). The temporal transcriptomic response of Pinus massoniana seedlings to phosphorus deficiency. PLoS One 9:e105068. doi: 10.1371/journal.pone.0105068
Finkel, O. M., Salas-Gonzalez, I., Castrillo, G., Spaepen, S., Law, T. F., Teixeira, P., et al. (2019). The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 17:e3000534. doi: 10.1371/journal.pbio.3000534
Forster, H., Adaskaveg, J. E., Kim, D. H., and Stanghellini, M. E. (1998). Effect of phosphite on tomato and pepper plants and on susceptibility of pepper to phytophthora root and crown rot in hydroponic culture. Plant Dis. 82, 1165–1170. doi: 10.1094/PDIS.1998.82.10.1165
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Guan, D., Yang, F., Xia, X., Shi, Y., Yang, S., Cheng, W., et al. (2018). CaHSL1 acts as a positive regulator of pepper thermotolerance under high humidity and is transcriptionally modulated by CaWRKY40. Front. Plant Sci. 9:1802. doi: 10.3389/fpls.2018.01802
Gutierrez-Alanis, D., Ojeda-Rivera, J. O., Yong-Villalobos, L., Cardenas-Torres, L., and Herrera-Estrella, L. (2018). Adaptation to phosphate scarcity: tips from Arabidopsis roots. Trends Plant Sci. 23, 721–730. doi: 10.1016/j.tplants.2018.04.006
Hamburger, D., Rezzonico, E., MacDonald-Comber Petetot, J., Somerville, C., and Poirier, Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14, 889–902. doi: 10.1105/tpc.000745
Hassler, S., Lemke, L., Jung, B., Mohlmann, T., Kruger, F., Schumacher, K., et al. (2012). Lack of the Golgi phosphate transporter PHT4;6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation in Arabidopsis. Plant J. 72, 732–744. doi: 10.1111/j.1365-313X.2012.05106.x
Heravi, K. M., and Altenbuchner, J. (2014). Regulation of the Bacillus subtilis mannitol utilization genes: promoter structure and transcriptional activation by the wild-type regulator (MtlR) and its mutants. Microbiology 160, 91–101. doi: 10.1099/mic.0.071233-0
Hiruma, K., Gerlach, N., Sacristan, S., Nakano, R. T., Hacquard, S., Kracher, B., et al. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. doi: 10.1016/j.cell.2016.02.028
Huang, L. F., Lin, K. H., He, S. L., Chen, J. L., Jiang, J. Z., Chen, B. H., et al. (2016). Multiple patterns of regulation and overexpression of a ribonuclease-like pathogenesis-related protein gene, OsPR10a, conferring disease resistance in rice and Arabidopsis. PLoS One 11:e0156414. doi: 10.1371/journal.pone.0156414
Huang, X., Zhang, Q., Jiang, Y., Yang, C., Wang, Q., and Li, L. (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. elife 7:e31636. doi: 10.7554/eLife.31636
Igarashi, D., Ishida, S., Fukazawa, J., and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13, 2483–2497. doi: 10.1105/tpc.010188
Ishida, S., Yuasa, T., Nakata, M., and Takahashi, Y. (2008). A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20, 3273–3288. doi: 10.1105/tpc.107.057489
Ishihama, N., and Yoshioka, H. (2012). Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15, 431–437. doi: 10.1016/j.pbi.2012.02.003
Ito, T., Nakata, M., Fukazawa, J., Ishida, S., and Takahashi, Y. (2014). Scaffold function of Ca2+−DEPENDENT PROTEIN KINASE: tobacco Ca2+-DEPENDENT PROTEIN KINASE1 transfers 14-3-3 to the substrate REPRESSION OF SHOOT GROWTH after phosphorylation. Plant Physiol. 165, 1737–1750. doi: 10.1104/pp.114.236448
Jain, A., Poling, M. D., Karthikeyan, A. S., Blakeslee, J. J., Peer, W. A., Titapiwatanakun, B., et al. (2007). Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 144, 232–247. doi: 10.1104/pp.106.092130
Khan, G. A., Bouraine, S., Wege, S., Li, Y., de Carbonnel, M., Berthomieu, P., et al. (2014). Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 65, 871–884. doi: 10.1093/jxb/ert444
Kong, Q., and Ma, W. (2018). WRINKLED1 as a novel 14-3-3 client: function of 14-3-3 proteins in plant lipid metabolism. Plant Signal. Behav. 13:e1482176. doi: 10.1080/15592324.2018.1482176
Lan, P., Li, W., and Schmidt, W. (2013). Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genomics 14:210. doi: 10.1186/1471-2164-14-210
Lei, L., Li, Y., Wang, Q., Xu, J., Chen, Y., Yang, H., et al. (2014). Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol. 203, 1146–1160. doi: 10.1111/nph.12872
Li, X., Chen, L., and Dhaubhadel, S. (2012). 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 71, 239–250. doi: 10.1111/j.1365-313X.2012.04986.x
Li, L. Q., Huang, L. P., Pan, G., Liu, L., Wang, X. Y., and Lu, L. M. (2017). Identifying the genes regulated by AtWRKY6 using comparative transcript and proteomic analysis under phosphorus deficiency. Int. J. Mol. Sci. 18:1046. doi: 10.3390/ijms18051046
Li, B., Tang, M., Caseys, C., Nelson, A., Zhou, M., Zhou, X., et al. (2020). Epistatic transcription factor networks differentially modulate Arabidopsis growth and defense. Genetics 214, 529–541. doi: 10.1534/genetics.119.302996
Liu, P., Cai, Z., Chen, Z., Mo, X., Ding, X., Liang, C., et al. (2018c). A root-associated purple acid phosphatase, SgPAP23, mediates extracellular phytate-P utilization in Stylosanthes guianensis. Plant Cell Environ. 41, 2821–2834. doi: 10.1111/pce.13412
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128. doi: 10.1016/j.molcel.2017.02.016
Liu, Z. Q., Qiu, A. L., Shi, L. P., Cai, J. S., Huang, X. Y., Yang, S., et al. (2015). SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 66, 3683–3698. doi: 10.1093/jxb/erv161
Liu, N., Shang, W., Li, C., Jia, L., Wang, X., Xing, G., et al. (2018b). Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 8:170231. doi: 10.1098/rsob.170231
Liu, C., Su, J., Stephen, G. K., Wang, H., Song, A., Chen, F., et al. (2018a). Overexpression of phosphate transporter gene CmPht1;2 facilitated pi uptake and alternated the metabolic profiles of chrysanthemum under phosphate deficiency. Front. Plant Sci. 9:686. doi: 10.3389/fpls.2018.00686
Lopez-Arredondo, D. L., Leyva-Gonzalez, M. A., Gonzalez-Morales, S. I., Lopez-Bucio, J., and Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
Lu, Y., Yasuda, S., Li, X., Fukao, Y., Tohge, T., Fernie, A. R., et al. (2016). Characterization of ubiquitin ligase SlATL31 and proteomic analysis of 14-3-3 targets in tomato fruit tissue (Solanum lycopersicum L). J. Proteome 143, 254–264. doi: 10.1016/j.jprot.2016.04.016
Luang, S., Sornaraj, P., Bazanova, N., Jia, W., Eini, O., Hussain, S. S., et al. (2018). The wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase. Plant Mol. Biol. 96, 543–561. doi: 10.1007/s11103-018-0713-1
Luo, B., Ma, P., Nie, Z., Zhang, X., He, X., Ding, X., et al. (2019). Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J. 97, 947–969. doi: 10.1111/tpj.14160
Medici, A., Marshall-Colon, A., Ronzier, E., Szponarski, W., Wang, R., Gojon, A., et al. (2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6:6274. doi: 10.1038/ncomms7274
Miao, J., Sun, J., Liu, D., Li, B., Zhang, A., Li, Z., et al. (2009). Characterization of the promoter of phosphate transporter TaPHT1.2 differentially expressed in wheat varieties. J. Genet. Genom. 36, 455–466. doi: 10.1016/S1673-8527(08)60135-6
Moore, J. W., Loake, G. J., and Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23, 2809–2820. doi: 10.1105/tpc.111.087346
Mori, A., Fukuda, T., Vejchasarn, P., Nestler, J., Pariasca-Tanaka, J., and Wissuwa, M. (2016). The role of root size versus root efficiency in phosphorus acquisition in rice. J. Exp. Bot. 67, 1179–1189. doi: 10.1093/jxb/erv557
Motte, H., and Beeckman, T. (2017). PHR1 balances between nutrition and immunity in plants. Dev. Cell 41, 5–7. doi: 10.1016/j.devcel.2017.03.019
Nanamori, M., Shinano, T., Wasaki, J., Yamamura, T., Rao, I. M., and Osaki, M. (2004). Low phosphorus tolerance mechanisms: phosphorus recycling and photosynthate partitioning in the tropical forage grass, Brachiaria hybrid cultivar Mulato compared with rice. Plant Cell Physiol. 45, 460–469. doi: 10.1093/pcp/pch056
Nilsson, L., Muller, R., and Nielsen, T. H. (2007). Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30, 1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x
Ohama, N., Sato, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
Oono, Y., Kobayashi, F., Kawahara, Y., Yazawa, T., Handa, H., Itoh, T., et al. (2013). Characterisation of the wheat (Triticum aestivum L.) transcriptome by de novo assembly for the discovery of phosphate starvation-responsive genes: gene expression in pi-stressed wheat. BMC Genomics 14:77. doi: 10.1186/1471-2164-14-77
Ormancey, M., Thuleau, P., Mazars, C., and Cotelle, V. (2017). CDPKs and 14-3-3 proteins: emerging duo in signaling. Trends Plant Sci. 22, 263–272. doi: 10.1016/j.tplants.2016.11.007
O’Rourke, J. A., Yang, S. S., Miller, S. S., Bucciarelli, B., Liu, J., Rydeen, A., et al. (2013). An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 161, 705–724. doi: 10.1104/pp.112.209254
Osorio, M. B., Ng, S., Berkowitz, O., De Clercq, I., Mao, C., Shou, H., et al. (2019). SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 181, 332–352. doi: 10.1104/pp.18.00594
Pacak, A., Barciszewska-Pacak, M., Swida-Barteczka, A., Kruszka, K., Sega, P., Milanowska, K., et al. (2016). Heat stress affects pi-related genes expression and inorganic phosphate deposition/accumulation in barley. Front. Plant Sci. 7:926. doi: 10.3389/fpls.2016.00926
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Papageorgiou, A. C., Adam, P. S., Stavros, P., Nounesis, G., Meijers, R., Petratos, K., et al. (2016). HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses. Extremophiles 20, 695–709. doi: 10.1007/s00792-016-0859-1
Peret, B., Clement, M., Nussaume, L., and Desnos, T. (2011). Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 16, 442–450. doi: 10.1016/j.tplants.2011.05.006
Peret, B., Desnos, T., Jost, R., Kanno, S., Berkowitz, O., and Nussaume, L. (2014). Root architecture responses: in search of phosphate. Plant Physiol. 166, 1713–1723. doi: 10.1104/pp.114.244541
Petters, J., Gobel, C., Scheel, D., and Rosahl, S. (2002). A pathogen-responsive cDNA from potato encodes a protein with homology to a phosphate starvation-induced phosphatase. Plant Cell Physiol. 43, 1049–1053. doi: 10.1093/pcp/pcf117
Pireyre, M., and Burow, M. (2015). Regulation of MYB and bHLH transcription factors: a glance at the protein level. Mol. Plant 8, 378–388. doi: 10.1016/j.molp.2014.11.022
Plaxton, W. C., and Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156, 1006–1015. doi: 10.1104/pp.111.175281
Postma, J. A., Dathe, A., and Lynch, J. P. (2014). The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 166, 590–602. doi: 10.1104/pp.113.233916
Qu, B., He, X., Wang, J., Zhao, Y., Teng, W., Shao, A., et al. (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167, 411–423. doi: 10.1104/pp.114.246959
Ramaiah, M., Jain, A., and Raghothama, K. G. (2014). Ethylene response Factor070 regulates root development and phosphate starvation-mediated responses. Plant Physiol. 164, 1484–1498. doi: 10.1104/pp.113.231183
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sa-Correia, I., and Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Ren, F., Guo, Q. Q., Chang, L. L., Chen, L., Zhao, C. Z., Zhong, H., et al. (2012). Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS One 7:e44005. doi: 10.1371/journal.pone.0044005
Ruan, W., Guo, M., Cai, L., Hu, H., Li, C., Liu, Y., et al. (2015). Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol. Biol. 87, 429–440. doi: 10.1007/s11103-015-0289-y
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schutze, K., Harter, K., and Chaban, C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13, 247–255. doi: 10.1016/j.tplants.2008.03.002
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587. doi: 10.1186/2193-1801-2-587
Shen, C., Wang, S., Zhang, S., Xu, Y., Qian, Q., Qi, Y., et al. (2013). OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L). Plant Cell Environ. 36, 607–620. doi: 10.1111/pce.12001
Shen, L., Yang, S., Guan, D., and He, S. (2020). CaCML13 acts positively in pepper immunity against Ralstonia solanacearum infection forming feedback loop with CabZIP63. Int. J. Mol. Sci. 21:4186. doi: 10.3390/ijms21114186
Shen, J., Yuan, L., Zhang, J., Li, H., Bai, Z., Chen, X., et al. (2011). Phosphorus dynamics: from soil to plant. Plant Physiol. 156, 997–1005. doi: 10.1104/pp.111.175232
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Strock, C. F., Morrow de la Riva, L., and Lynch, J. P. (2018). Reduction in root secondary growth as a strategy for phosphorus acquisition. Plant Physiol. 176, 691–703. doi: 10.1104/pp.17.01583
Su, T., Xu, Q., Zhang, F. C., Chen, Y., Li, L. Q., Wu, W. H., et al. (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 167, 1579–1591. doi: 10.1104/pp.114.253799
Sunar, K., Dey, P., Chakraborty, U., and Chakraborty, B. (2015). Biocontrol efficacy and plant growth promoting activity of Bacillus altitudinis isolated from Darjeeling hills, India. J. Basic Microbiol. 55, 91–104. doi: 10.1002/jobm.201300227
Takeo, K., and Ito, T. (2017). Subcellular localization of VIP1 is regulated by phosphorylation and 14-3-3 proteins. FEBS Lett. 591, 1972–1981. doi: 10.1002/1873-3468.12686
Valdes-Lopez, O., Arenas-Huertero, C., Ramirez, M., Girard, L., Sanchez, F., Vance, C. P., et al. (2008a). Essential role of MYB transcription factor: PvPHR1 and microRNA: pvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 31, 1834–1843. doi: 10.1111/j.1365-3040.2008.01883.x
Valdes-Lopez, O., and Hernandez, G. (2008b). Transcriptional regulation and signaling in phosphorus starvation: what about legumes? J. Integr. Plant Biol. 50, 1213–1222. doi: 10.1111/j.1744-7909.2008.00758.x
Wang, Y., Dang, F., Liu, Z., Wang, X., Eulgem, T., Lai, Y., et al. (2013). CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14, 131–144. doi: 10.1111/j.1364-3703.2012.00836.x
Wang, Y. H., Garvin, D. F., and Kochian, L. V. (2002). Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 130, 1361–1370. doi: 10.1104/pp.008854
Wang, H., Xu, Q., Kong, Y. H., Chen, Y., Duan, J. Y., Wu, W. H., et al. (2014). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. doi: 10.1104/pp.113.235077
Wang, Y., Zhang, F., Cui, W., Chen, K., Zhao, R., and Zhang, Z. (2019). The FvPHR1 transcription factor control phosphate homeostasis by transcriptionally regulating miR399a in woodland strawberry. Plant Sci. 280, 258–268. doi: 10.1016/j.plantsci.2018.12.025
Wu, Z., Ren, H., McGrath, S. P., Wu, P., and Zhao, F. J. (2011). Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 157, 498–508. doi: 10.1104/pp.111.178921
Xia, X. J., Zhou, Y. H., Shi, K., Zhou, J., Foyer, C. H., and Yu, J. Q. (2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 66, 2839–2856. doi: 10.1093/jxb/erv089
Xu, W., Jia, L., Shi, W., Liang, J., and Zhang, J. (2012a). Smart role of plant 14-3-3 proteins in response to phosphate deficiency. Plant Signal. Behav. 7, 1047–1048. doi: 10.4161/psb.20997
Xu, W., Shi, W., Jia, L., Liang, J., and Zhang, J. (2012b). TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ. 35, 1393–1406. doi: 10.1111/j.1365-3040.2012.02497.x
Xu, H., Zhao, X., Guo, C., Chen, L., and Li, K. (2016). Spinach 14-3-3 protein interacts with the plasma membrane H(+)-ATPase and nitrate reductase in response to excess nitrate stress. Plant Physiol. Biochem. 106, 187–197. doi: 10.1016/j.plaphy.2016.04.043
Xue, Y. B., Xiao, B. X., Zhu, S. N., Mo, X. H., Liang, C. Y., Tian, J., et al. (2017). GmPHR25, a GmPHR member up-regulated by phosphate starvation, controls phosphate homeostasis in soybean. J. Exp. Bot. 68, 4951–4967. doi: 10.1093/jxb/erx292
Xue, Y., Zhuang, Q., Zhu, S., Xiao, B., Liang, C., Liao, H., et al. (2018). Genome wide transcriptome analysis reveals complex regulatory mechanisms underlying phosphate homeostasis in soybean nodules. Int. J. Mol. Sci. 19:2924. doi: 10.3390/ijms19102924
Ye, Q., Wang, H., Su, T., Wu, W. H., and Chen, Y. F. (2018). The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low-pi stress in Arabidopsis. Plant Cell 30, 1062–1076. doi: 10.1105/tpc.17.00845
Yuan, W., Zhang, D., Song, T., Xu, F., Lin, S., Xu, W., et al. (2017). Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 68, 1731–1741. doi: 10.1093/jxb/erx040
Zhang, Y., Hu, L., Yu, D., Xu, K., Zhang, J., Li, X., et al. (2019). Integrative analysis of the wheat PHT1 gene family reveals a novel member involved in arbuscular mycorrhizal phosphate transport and immunity. Cell 8:490. doi: 10.3390/cells8050490
Zhang, L., Li, G., Li, Y., Min, J., Kronzucker, H. J., and Shi, W. (2018). Tomato plants ectopically expressing Arabidopsis GRF9 show enhanced resistance to phosphate deficiency and improved fruit production in the field. J. Plant Physiol. 226, 31–39. doi: 10.1016/j.jplph.2018.04.005
Zhang, J., Xu, L., Wang, F., Deng, M., and Yi, K. (2012). Modulating the root elongation by phosphate/nitrogen starvation in an OsGLU3 dependant way in rice. Plant Signal. Behav. 7, 1144–1145. doi: 10.4161/psb.21334
Zhang, J., Zhou, X., Xu, Y., Yao, M., Xie, F., Gai, J., et al. (2016). Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 248, 82–91. doi: 10.1016/j.plantsci.2016.04.010
Zhong, Y., Wang, Y., Guo, J., Zhu, X., Shi, J., He, Q., et al. (2018). Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 219, 135–148. doi: 10.1111/nph.15155
Zhou, J., Jiao, F., Wu, Z., Li, Y., Wang, X., He, X., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. doi: 10.1104/pp.107.111443
Zhou, X., Zha, M., Huang, J., Li, L., Imran, M., and Zhang, C. (2017). StMYB44 negatively regulates phosphate transport by suppressing expression of PHOSPHATE1 in potato. J. Exp. Bot. 68, 1265–1281. doi: 10.1093/jxb/erx026
Keywords: Capsicum annuum, CaWRKY58, Ca14-3-3, CaPHR1, phosphorus deficiency
Citation: Cai J, Cai W, Huang X, Yang S, Wen J, Xia X, Yang F, Shi Y, Guan D and He S (2021) Ca14-3-3 Interacts With CaWRKY58 to Positively Modulate Pepper Response to Low-Phosphorus Starvation. Front. Plant Sci. 11:607878. doi: 10.3389/fpls.2020.607878
Received: 18 September 2020; Accepted: 16 December 2020;
Published: 14 January 2021.
Edited by:
Stephan Pollmann, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Byoung-Cheorl Kang, Seoul National University, South KoreaCopyright © 2021 Cai, Cai, Huang, Yang, Wen, Xia, Yang, Shi, Guan and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuilin He, c2hsaGVmYWZ1QDE2My5jb20=; c2hsaGUyMDEzMDRAYWxpeXVuLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.