
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 December 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.606463
The genera Arnebia and Lithospermum (Lithospermeae-Boraginaceae) comprise 25–30 and 50–60 species, respectively. Some of them are economically valuable, as their roots frequently contain a purple-red dye used in the cosmetic industry. Furthermore, dried roots of Arnebia euchroma, A. guttata, and Lithospermum erythrorhizon, which have been designated Lithospermi Radix, are used as traditional Korean herbal medicine. This study is the first report on the floral micromorphology and complete chloroplast (cp) genome sequences of A. guttata (including A. tibetana), A. euchroma, and L. erythrorhizon. We reveal great diversity in floral epidermal cell patterns, gynoecium, and structure of trichomes. The cp genomes were 149,361–150,465 bp in length, with conserved quadripartite structures. In total, 112 genes were identified, including 78 protein-coding regions, 30 tRNA genes, and four rRNA genes. Gene order, content, and orientation were highly conserved and were consistent with the general structure of angiosperm cp genomes. Comparison of the four cp genomes revealed locally divergent regions, mainly within intergenic spacer regions (atpH-atpI, petN-psbM, rbcL-psaI, ycf4-cemA, ndhF-rpl32, and ndhC-trnV-UAC). To facilitate species identification, we developed molecular markers psaA- ycf3 (PSY), trnI-CAU- ycf2 (TCY), and ndhC-trnV-UAC (NCTV) based on divergence hotspots. High-resolution phylogenetic analysis revealed clear clustering and a close relationship of Arnebia to its Lithospermum sister group, which was supported by strong bootstrap values and posterior probabilities. Overall, gynoecium characteristics and genetic distance of cp genomes suggest that A. tibetana, might be recognized as an independent species rather than a synonym of A. guttata. The present morphological and cp genomic results provide useful information for future studies, such as taxonomic, phylogenetic, and evolutionary analysis of Boraginaceae.
Arnebia Forssk., an economically and medicinally important plant group of the Boraginaceae Juss. (tribe Lithospermeae Dumort.) comprises 25–30 species, distributed across Southwest and Central Asia, the Himalayas, Northeast Africa, and (Southeast) Mediterranean (Zhu et al., 1995; Mabberley, 2008; Ambrish and Srivastava, 2014; Coppi et al., 2015). Plants in the Arnebia are distinguishable from other Boraginaceae based on an actinomorphic corolla devoid of appendages, di- or tetra-stigmatic flowers, a style divided in two or four parts, a fruiting calyx often hardened and enclosing the nutlets, and nutlets without stipe (Forsskål, 1775; Zhu et al., 1995; Coppi et al., 2015). The roots of certain Arnebia taxa, which frequently contain a purple-red dye, have been used as a colorant in food and cosmetic preparations. Moreover, anti-microbial, anti-inflammatory, anti-oxidant, wound healing, anti-tumor, cardiotonic, and antineoplastic properties have been reported for A. benthamii (Wall. ex G. Don) I.M. Johnst. (Katoch et al., 2016), A. densiflora Ledeb. (Akkol et al., 2009; Kosger et al., 2009), A. euchroma (Royle ex Benth.) I.M. Johnst. (Damianakos et al., 2012; Bo et al., 2019), A. hispidissima (Lehm.) DC. (Jain et al., 2000; Shukla et al., 2001), and A. nobilis Rech.f. (Khatoon et al., 2003; Garg et al., 2014). The roots of Lithospermum L., another genus in the tribe Lithospermeae, also contain a red dye and have been reported to possess various phytomedical activities (Ishida and Sakaguchi, 2007). The dried roots of A. euchroma, A. guttata Bunge, and Lithospermum erythrorhizon Siebold & Zucc. are used as an important herbal remedy, known as Lithospermi Radix or Arnebiae Radix, which is designated as a medicine in the Korean and Chinese Herbal Pharmacopeia (Song J. H. et al., 2019; Korea Institute of Oriental Medicine [KIOM], 2020). Because their morphological, micromorphological, and molecular characteristics have not been compared and described in detail yet, the different species of medicinal borages have been used as a single remedy. Thus, there is a need to conduct comprehensive comparisons of their biological properties. Moreover, revealing the taxonomic identity of A. tibetana Kurz is essential to determine its medicinal properties. The taxonomic status of this species has been controversial. Several authors place A. tibetana as a synonym of A. guttata (Kazmi, 1970; Ambrish and Srivastava, 2014; Urgamal et al., 2014), whereas others consider that the biennial habitat and absence of blackish-purple spots on corolla lobes of A. tibetana support its classification as a separate species, independent from A. guttata (Zhu et al., 1995; Lazkov and Sultanova, 2014).
Floral micromorphological characteristics observed using scanning electron microscopy (SEM) have proven to be useful for taxonomic delimitation and phylogenetic inference of various groups, including Adoxaceae (Konarska, 2017), Crassulaceae (Giuliani et al., 2018), Nymphaeaceae (Coiro and Barone Lumaga, 2018), Lentibulariaceae (Płachno et al., 2019), and Rosaceae (Song et al., 2020). Notably, taxonomic and systematic relevance of floral micromorphology, including that of the corolla, calyx, style, and stigma, has been confirmed within Boraginaceae (Bigazzi et al., 1999; Bacchetta et al., 2000; Bigazzi and Selvi, 2000; Akçin, 2009).
Chloroplasts are key organelles in plants, where they mediate photosynthesis, carbon fixation, and biosynthesis of starch, fatty acids, amino acids, and pigments (Jansen and Ruhlman, 2012; Daniell et al., 2016). Chloroplast (cp) genomes of angiosperms reach 72 to 217 kb in size (Sugiura, 1992, 1995) and exhibit a quadripartite structure consisting of a large single-copy (LSC) and small single-copy (SSC) of a large inverted repeat (IR) region. Most angiosperm cp genomes contain approximately 80 protein-coding genes, four ribosomal RNA (rRNA) genes, and 30 transfer RNA (tRNA) genes (Wicke et al., 2011). The angiosperm cp genome structure and gene content are highly conserved due to lack of recombination, maternal inheritance, and elevated copy numbers (Wolfe et al., 1987; Delannoy et al., 2011). Hence, cp genomes have facilitated species classification, construction of high-resolution phylogenetic trees, and identification of evolutionary relationships in Plantae (Jansen et al., 2007; Parks et al., 2009). Furthermore, comparative studies of cp genomes provide useful information on gene rearrangements, including expansion or contraction of IR regions, as well as gene content, gene duplication or loss, genetic variance caused by insertion/deletion (indel), single sequence repeats (SSR), and single nucleotide polymorphism. These genetic markers can be used for species identification and classification, population genetics, and evolutionary studies (Hamilton et al., 2003; Chen et al., 2013). The remarkable reproducibility of methods based on these markers has facilitated the authentication and quality control of herbal medicine (Park et al., 2018a,b). Several studies have compared cp genomes to develop indel markers aimed at identifying herbal medicinal plants (Kim et al., 2015; Park et al., 2017, 2018b) and promoting their conservation.
The family Boraginaceae comprises 90 genera and 1,600 to 1,700 species according to the Angiosperm Phylogeny Group (APG) IV classification system (Cohen, 2014). Previous studies based on internal transcribed spacer (ITS) regions and a few cp loci have clarified the evolutionary relationships of Boraginaceae. Cohen (2014) classified 224 species of Boraginaceae based on their morphological characteristics, whereas Chacón et al. (2016) reclassified the monophyletic relationship between previously recognized clades and the position of various unplaced genera using 170 Boraginaceae species. However, lack of genomic resources has prevented the resolution of existing controversies regarding the Arnebia genus (Coppi et al., 2015). At present, the cp genome has been fully sequenced solely in Borago officinalis (NC_046796.1), Onosma fuyunensis (NC_049569.1), and Tiquilia plicata (MG573056). Only additional cp genome data will reveal the phylogenetic relationship between Arnebia and other genera in the Boraginaceae family, as well as correct species classification.
Here, we conducted a microscopic analysis of the floral organs of A. guttata, A. tibetana, A. euchroma, and L. erythrorhizon, and sequenced the cp genomes of these species. Moreover, we employed this information to verify the taxonomic and phylogenetic identity of A. guttata and A. tibetana.
Plant material used in this study was collected from natural populations in Mongolia, Kyrgyzstan, and Korea during a three-year survey of the vascular flora of Central Asia (2018–2020). Protologues and other relevant taxonomic literature were searched and used for accurate identification of all studied species (Royle, 1839; Bunge, 1840; Kurz, 1874; Johnston, 1924; Fang and Staples, 1995; Song J. H. et al., 2019; Kim, 2019). To confirm the consistency of all studied characteristics, at least two populations were compared for each species: A. guttata (two individuals), A. tibetana (10 individuals), A. euchroma (ten individuals), and L. erythrorhizon (10 individuals).
The samples were assigned identification numbers, and voucher specimens were deposited in the Korean Herbarium of Standard Herbal Resources (Index Herbariorum code: KIOM) at the Korea Institute of Oriental Medicine, Naju, South Korea. Moreover, the same collections were also deposited in both Institute for Biology of National Academy of Science (FRU) and the National University of Mongolia (UBU). Information about samples used for morphological, micromorphological, and cp genome sequence analyses is listed in Supplementary Table 1.
For detailed floral micromorphological observations, fully mature reproductive organs were examined using a stereo microscope (SZX16; Olympus, Tokyo, Japan). For scanning electron microscopic observations, the dried samples from voucher specimens were rehydrated overnight in a wetting agent (Agepon: distilled water, 1:200; Agfa Gevaert, Leverkusen, Germany). The floral samples were dehydrated through a graded ethanol series (50, 70, 90, 95, and 100% ethanol) at room temperature for 1 h at each ethanol concentration. The dehydrated material was immersed in liquid CO2 for critical-point drying (SPI-13200JE-AB; SPI Supplies, West Chester, PA, United States) and subsequent mounting on aluminum stubs using a double-sided adhesive conductive carbon disk (05073-BA; SPI Supplies). All samples were coated with gold using an ion-sputtering device (208HR; Cressington Scientific Instruments Ltd., Watford, United Kingdom) and were observed using a low-voltage field emission scanning electron microscope (JSM-7600F; JEOL, Tokyo, Japan) at an accelerating voltage of 5–10 kV and working distance of 8–10 mm.
Total DNA of the four tested species was extracted using the modified CTAB method (Allen et al., 2006). Four libraries were prepared from total genomic DNA using the TruSeq DNA Nano kit following the manufacturer’s protocols and generated using the NextSeq500 platform (Illumina, San Diego, CA, United States), generating 1.6–2 Gb of paired-end (2 × 150 bp) reads. The generated reads were trimmed and their quality was checked using CLC quality trim implemented in the CLC Assembly Cell software (ver 4.2.1; CLC Inc., Aarhus, Denmark). Trimmed paired-end reads (Phred score ≥ 20) were assembled using the CLC genome assembler (ver. 4.2.1; CLC Inc.) with default parameters. SOAP de novo gap closer was used to fill gaps based on alignment of paired-end reads (Luo et al., 2012). Contigs were aligned against NCBI nrDB to detect cp contigs and were retrieved from total contigs using Nucmer (Delcher et al., 2003). Aligned contigs were ordered using the cp genome sequences of Abeliophyllum distichum (MK616470.1), Schrebera orientalis (NC_042266.1), and Forestiera ligustrina (MH817903.1) as references. The complete cp genomes were validated by PCR-based sequencing using sequence-specific primers (Supplementary Table 2). PCR products of the four junctions (LSC/IRa, IRa/SSC, SSC/IRb, and IRb/LSC) were compared to complete cp genome sequences (Supplementary Table 3). Finally, total trimmed paired-end reads were mapped onto complete genome sequences using BWA ver. 0.7.25 (Li and Durbin, 2010; Supplementary Figure 1).
Gene annotation of A. guttata, A. tibetana, A. euchroma, and L. erythrorhizon cp genomes was performed using GeSeq (Tillich et al., 2017), and annotation results were concatenated using an in-house script pipeline. Protein-coding sequences (CDS) were manually curated and confirmed using Artemis (Carver et al., 2008), and checked against the NCBI protein database. The tRNAs were confirmed with tRNAscan-SE 1.21 (Lowe and Eddy, 1997). IR region sequences were confirmed using IR finder and RepEx (Gurusaran et al., 2013). Circular maps of the three Arnebia and one Lithospermum cp genomes were obtained using OGDRAW (Greiner et al., 2019). GC content and relative synonymous codon usage (RSCU) of the four cp genomes were analyzed using MEGA7 software (Kumar et al., 2016). Codon usage distribution of six Boraginaceae cp genomes was visualized using the Heatmapper program, with average linkage of clustering and Euclidean distance measurement methods (Babicki et al., 2016). RSCU < 1.00 indicated a codon that was used less frequently than expected; whereas RSCU > 1.00 indicated a codon that was used more frequently than expected. MAUVE V2.3.1 was used to identify local collinear blocks of six Boraginaceae species (Arnebia guttata, A. tibetana, A. euchroma, Lithospermum erythrorhizon, Borago officinalis, and Tiquilia plicata; Darling et al., 2010). The mVISTA program in Shuffle-LAGAN mode was used to compare the cp genomes with A. guttata as reference (Frazer et al., 2004). DnaSP version 6 was used to calculate nucleotide variability (Pi) among cp genomes (Rozas et al., 2017). Substitution rates Ka and Ks were estimated with KaKs Calculator ver. 2.0 (Wang et al., 2010).
We used REPuter to find forward and reverse repeats with minimal length of 20 bp, identity of 90%, and a Hamming distance of 3 (Kurtz et al., 2001). SSRs were detected using MISA (Beier et al., 2017), with the minimum number of repeat parameters set to 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively. Tandem repeats ≥20 bp were identified using Tandem repeats finder (Benson, 1999) with minimum alignment score of 50 and maximum period size of 500; the identity of repeats was set to ≥90%.
Primers for indel markers were designed using NCBI Primer-BLAST. Specificity of indel markers was confirmed by PCR with 20 ng of genomic DNA extracted from eight samples of the four species. Reactions were carried out in a 20-μL PCR mixture with 10 pmol of PSY, TCY, and NCTV primers using a ProFlex PCR system (Applied Biosystems, Waltham, MA, United States) with the following amplification parameters: initial denaturation at 95°C for 2 min; 35 cycles at 95°C for 50 s, 60°C for 50 s, and 72°C for 50 s; final extension at 72°C for 5 min. PCR products were separated on a 2% agarose gel for 40 min at 150 V. The germplasms of the three Arnebia and one Lithospermum species are listed in Supplementary Table 4. PSY, TCY, and NCTV primer sequences are listed in Supplementary Table 5.
A total of eight cp genomes, namely seven from Boraginaceae, one from Oleaceae, were used for phylogenetic analysis, along with Forsythia suspensa (NC_036367) as outgroup. Of these, 4 chloroplast genome sequences were downloaded from the NCBI GenBank (Supplementary Table 6). We used two matrices, one composed of 72 conserved protein-coding genes (CDS) and one of the whole chloroplast genomes. Using MAFFT ver. 7.388 (Katoh and Standley, 2013) two data set were aligned, respectively. Each aligned gene was extracted using Geneious program1, and then the genes were arranged alphabetically. The concatenated gene dataset was created using Geneious program in CDS data set. The alignments data sets were filtered to remove ambiguously aligned regions using Gblocks ver. 5 (Castresana, 2000). The best-fitting model of nucleotide substitutions was determined using Akaike information criterion in jModelTest V2.1.10 (Darriba et al., 2012; Supplementary Table 7) and the GTR+I+G model was chosen for Maximum likelihood (ML) analysis and GTR+G model was selected for Bayesian Inference (BI) analysis. ML analysis was performed using RaxML v 8.0.5 (Stamatakis, 2014) with 1,000 bootstrap replicates. BI analysis was carried out using MrBayes 3.2.2 (Ronquist et al., 2012), with two independent runs of four simultaneous chains that were run for 5,000,000 generations using Markov chain Monte Carlo algorithm. Trees were sampled every 5,000 generations, with the first 25% discarded as burn-in. Trees were determined from 50% majority-rule consensus to estimate posterior probabilities. The reconstructed trees were visualized using Figtree V.1.4.2 (Rambaut, 2014).
Floral external and epidermal micromorphological characteristics (calyx, corolla, and gynoecium surfaces) are summarized in Table 1, while representative photo- and micrographs are shown in Figures 1–3. Although limited variation was observed between individuals of the same species, the four species could be clearly distinguished by their floral characteristics.
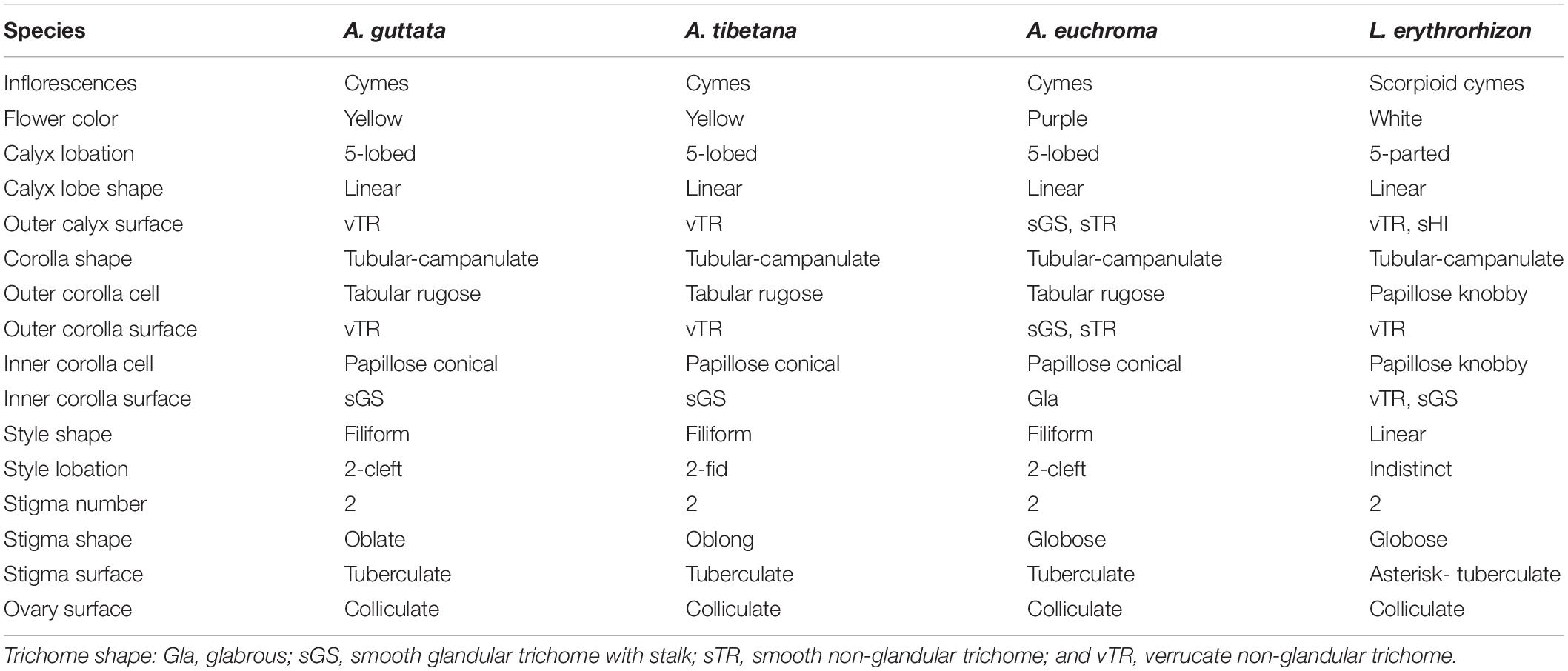
Table 1. Floral morphological and micromorphological characteristics of Arnebia and Lithospermum species.

Figure 1. Flowers of Arnebia and Lithospermum species. (A) Arnebia guttata, (B) Arnebia tibetana, (C) Arnebia euchroma, and (D) Lithospermum erythrorhizon.

Figure 2. Calyx and corolla micromorphological characteristics of Arnebia and Lithospermum species. (A,E,I,M) Arnebia guttata, (B,F,J,N) Arnebia tibetana, (C,G,K,O) Arnebia euchroma, and (D,H,L,P) Lithospermum erythrorhizon.
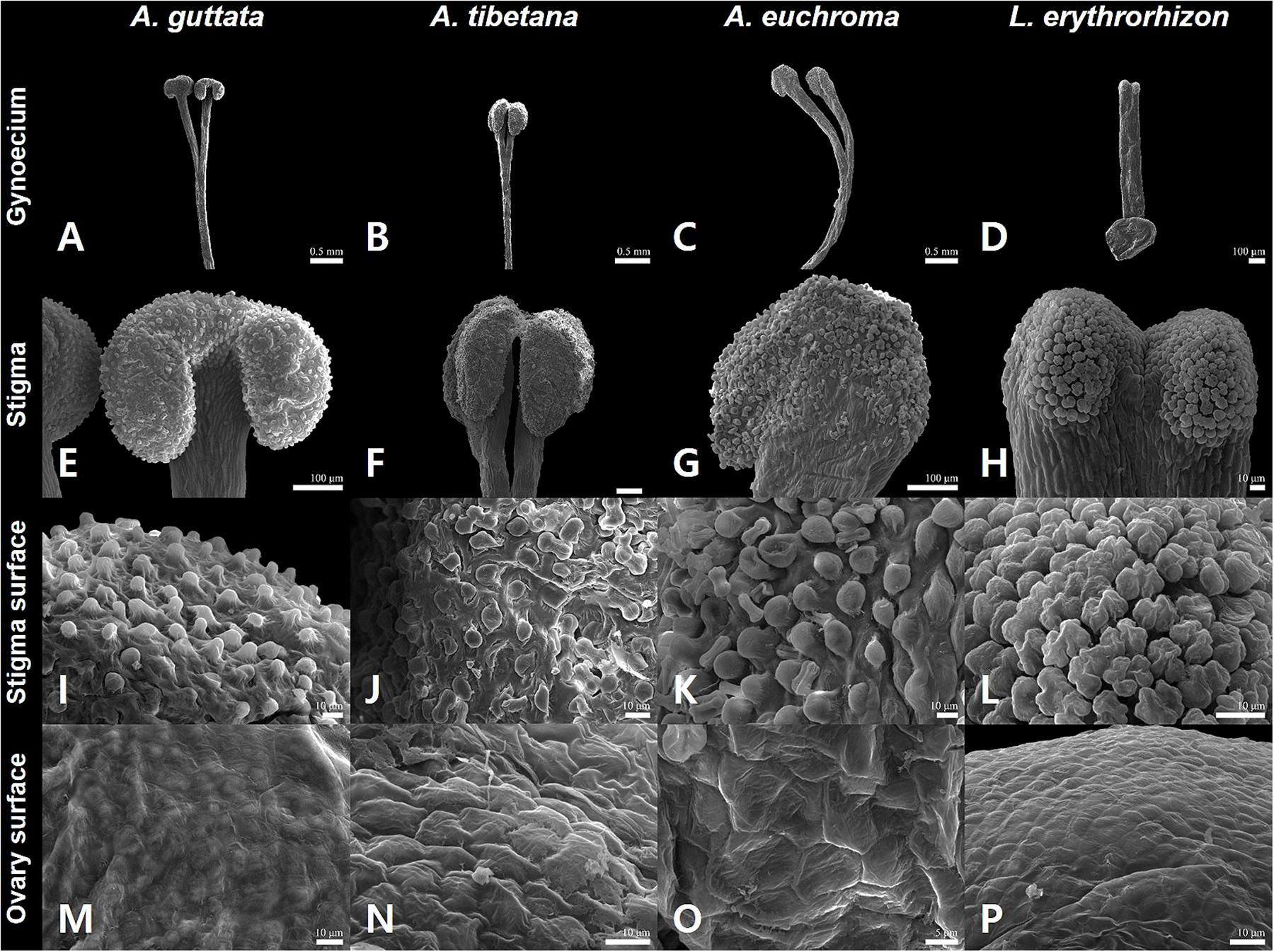
Figure 3. Gynoecium characteristics of Arnebia and Lithospermum species. (A,E,I,M) Arnebia guttata, (B,F,J,N) Arnebia tibetana, (C,G,K,O) Arnebia euchroma, and (D,H,L,P) Lithospermum erythrorhizon.
All studied borages presented cyme inflorescences: L. erythrorhizon had scorpioid cymes with paired flowers, A. guttata and A. tibetana displayed crowded cymes with many flowers, and A. euchroma exhibited terminal cymes with numerous flowers. The corolla was yellow with or without dark spots on corolla lobes in A. guttata and A. tibetana (Figures 1A,B), dark purple to purple in A. euchroma (Figure 1C), and white in L. erythrorhizon (Figure 1D). The calyx of the three Arnebia species was 5-lobed; whereas that of L. erythrorhizon was 5-parted and more deeply lobed than in Arnebia specimens.
The outer calyx of A. guttata and A. tibetana was densely covered with non-glandular trichomes (Figures 2A,B); whereas that of A. euchroma was densely covered with both non-glandular trichomes and glandular trichomes with stalk (Figure 2C). Hirsute, thick, and long non-glandular trichomes were found in L. erythrorhizon (Figure 2D). Non-glandular trichome surfaces were generally verrucate (Figures 2E,F); whereas glandular trichomes were always smooth (Figure 2G). Only in L. erythrorhizon, non-glandular trichomes were both verrucate and smooth (Figure 2H). The shape of the corolla was tubular-campanulate in all species; however, cellular and trichome distribution patterns were quite different. All Arnebia species presented a tabular rugose cellular pattern on the outer corolla (Figures 2I–K); whereas L. erythrorhizon exhibited a papillose knobby pattern (Figure 2L). The verrucate, non-glandular trichomes were sparsely to moderately pubescent on the outer corolla surface of A. guttata, A. tibetana, and L. erythrorhizon. Smooth glandular trichomes with stalk and smooth non-glandular trichomes were only found in A. euchroma. The inner corolla cells of all Arnebia species were papillose conical and 10.70 ± 2.69 (S.D.) μm tall (Figures 2M–O), whereas those of L. erythrorhizon were papillose knobby and 36.74 ± 6.22 (S.D.) μm tall (Figure 2P). A. guttata and A. tibetana had smooth glandular trichomes with stalk on the inner corolla surface, A. euchroma was glabrous, and L. erythrorhizon had both smooth glandular trichomes with stalk and verrucate non-glandular trichomes.
The gynoecium varied between species (Figure 3). All Arnebia species had a filiform style and two stigmas (Figures 3A–C); whereas L. erythrorhizon had an indistinct broad linear style with two stigmas (Figure 3D). Stylar lobation in Arnebia was of two types: 2-cleft (divided almost through the middle) in A. guttata (Figure 3A) and A. euchroma (Figure 3C), and 2-fid (divided through the top) in A. tibetana (Figure 3B). The stigma of Arnebia species presented different shapes: oblate in A. guttata (Figure 3E), oblong in A. tibetana (Figure 3F), and globose in A. euchroma (Figure 3G). In contrast, cellular surface patterns were of the same tuberculate type (Figures 3I–K). L. erythrorhizon had a globose stigma with asterisk-tuberculate cellular surface pattern (Figure 3L). All studied species shared a colliculate ovary surface (Figures 3M–P).
We sequenced four Boraginaceae accessions: A. guttata, A. tibetana, A. euchroma, and L. erythrorhizon at approximately 601X, 324X, 447X, and 433X coverage, respectively. This generated 1.6–2.0 Gb raw paired-end reads and 1.3–1.7 Gb trimmed reads (Supplementary Table 8). The complete circular cp genome was 149,316–150,465 bp and matched the quadripartite structure of cp genomes, consisting of a pair of IRs (17,144–17,311 bp) separated by LSC (80,411–81,348 bp) and SSC (25,856–25,979 bp) regions (Figure 4, Table 2, and Supplementary Figure 2). The complete cp genomes were all of high quality (Supplementary Figure 1). Overall GC content was approximately 37%, and slightly higher in the IR (43%) than LSC (35%) or SSC (31%) regions. The four cp genomes comprised 112 genes, namely 78 protein-coding, four rRNA, and 30 tRNA genes. They had 18 intron-containing genes, 15 of which had a single intron and two of which (ycf3, clpP) had three introns with duplicate genes (ndhB, trnI-GAU, and trnA-UGC) in the IR regions (Supplementary Table 10). Analysis of codon usage and anticodon recognition patterns revealed 24,529–25,929 codons, among which leucine, isoleucine, and serine were the most abundant (Supplementary Figure 3). To identify codon patterns, we analyzed codon distribution in six cp genomes using the hierarchical clustering method (Supplementary Figure 4). Red and green indicate weak (RSCU > 1) and strong (RSCU < 1) codon bias, respectively. Codons with an A or T in the third position had a strong codon bias. Most Boraginaceae displayed a similar pattern, with high RSCU values for GGT (Guanine), AGG (Arginine), and GGG (Guanine); only B. officinalis presented particularly low RSCU values.
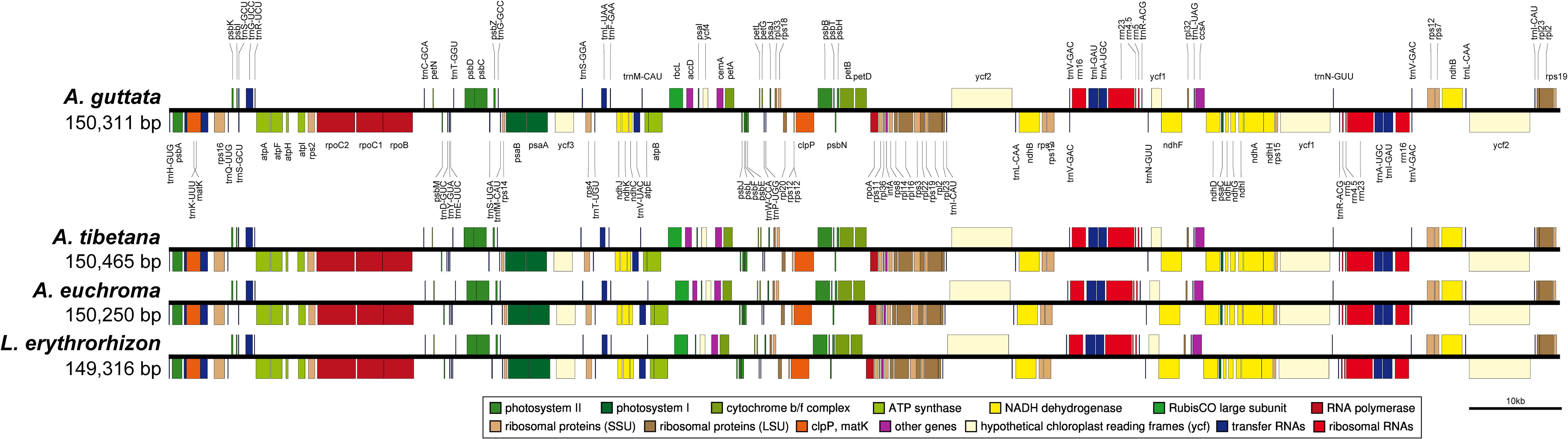
Figure 4. Linear gene map of chloroplast genomes from Arnebia and Lithospermum species. Genes are transcribed left to right. Genes above the line are positioned in the forward direction (left to right). Genes below the line are positioned in the reverse direction (right to left).
REPuter-based analysis of repetitive sequences, such as forward and reverse repeats, yielded 39–55 repeats (Figure 5A). In the three Arnebia cp genomes, they were located in the intragenic spacer (IGS) region, while in L. erythrorhizon they were particularly abundant in exons. MISA software (Beier et al., 2017) identified a total of 420–439 SSRs (Figure 5B), mostly in the LSC and particularly within the IGS region. The pentanucleotide motif was the most abundant in all accessions, followed by hexanucleotide motif repeats (Figure 5C). Tandem repeats finder (Benson, 1999) located most tandem repeats in the LSC (Figure 5D). They were generally 31–50 bp long and only 2–5 tandem repeats >71 bp in length were identified in all four accessions.
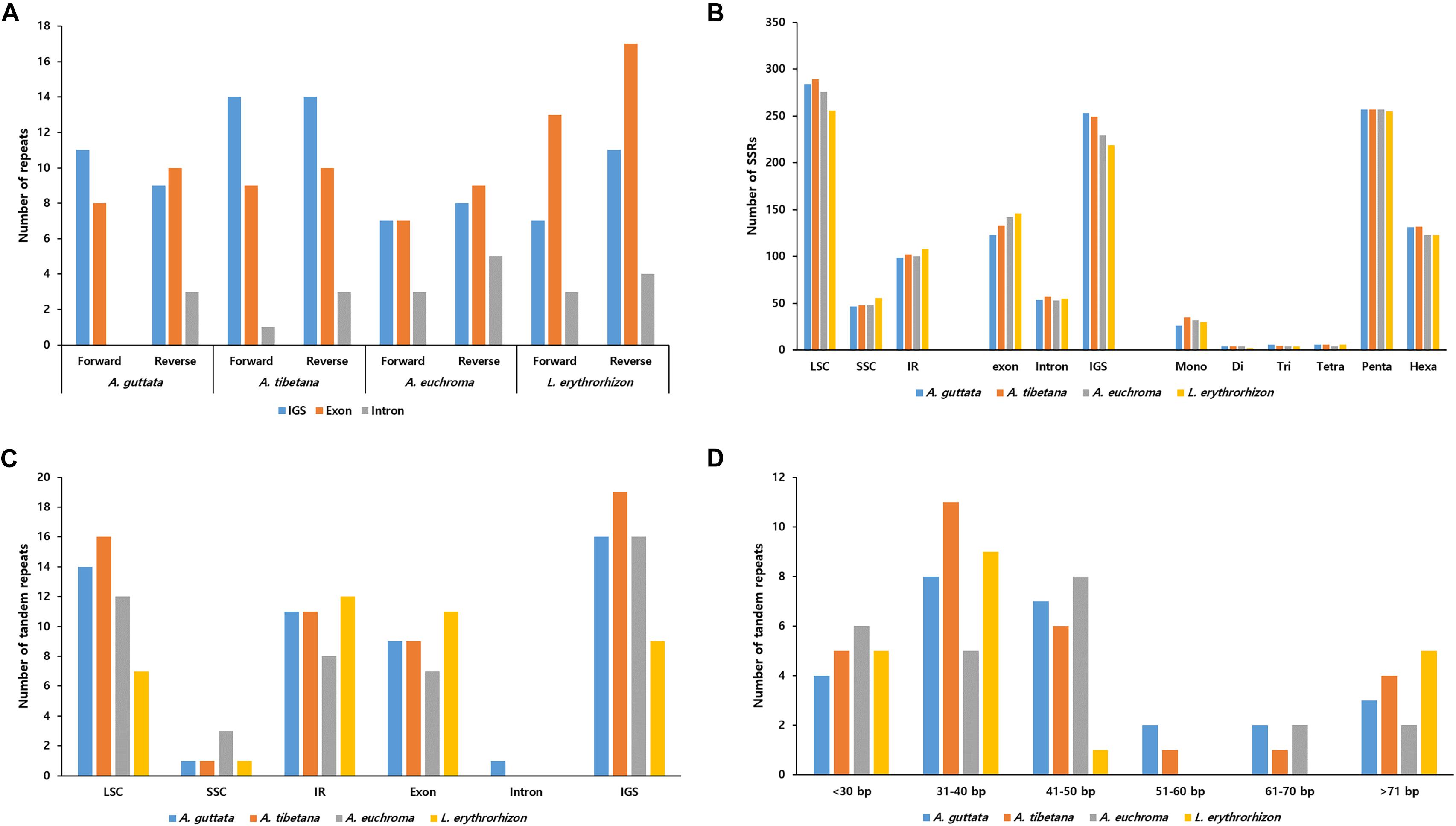
Figure 5. Distribution of repeat sequences in Arnebia and Lithospermum chloroplast genomes. (A) Number of forward and reverse repeats in genomic regions. (B) Distribution of SSRs in intergenic spacer (IGS), exon, and intron regions. (C) Distribution of tandem repeats in genomic regions. (D) Distribution of tandem repeats’ length.
We compared IR contraction and expansion of the Boraginaceae accessions with those of Forsythia suspensa (Supplementary Figure 5). In spite of similar IR length, from 25,217 to 25,979 bp, differences in IR expansions and contractions were observed. The rps19 gene of five Boraginaceae (A. guttata, A. tibetana, A. euchroma, L. erythrorhizon, and B. officinalis) was located entirely in the LSC region, but was expanded beyond that in Tiquilia plicata. The ycf1 gene was located at the junction of IRb/SSC and SSC/IRa, with the border between IRa/SSC extending into ycf1. The IRb/SSC border was also located in the ycf1 gene, but it expanded by the same length also into the ψycf1 gene. Overall, IRb was found to be expanded in all cp genomes assessed. The trnH gene was located in the LSC region, 16–40 bp from the IRa/LSC boundary. The rpl2 gene of T. plicata was duplicated in the IR regions flanking border junctions. Generally, Arnebia and Lithospermum appeared to be highly conserved.
We analyzed sequence identity using MAUVE and mVISTA programs, with A. guttata as reference. Overall, Boraginaceae cp genome structure was well conserved (Figure 6 and Supplementary Figure 6), with genic regions being more conserved than IGS regions. Most genic regions were highly convergent; the only divergent regions were detected in matK, rpoC2, rpoC1, ycf1, ycf2, and ndhF genes. The highest divergence was observed in IGS regions, such as atpH-atpI, petN-psbM, rbcL-psaI, ycf4-cemA, ndhF-rpl32, and ndhC-trnV-UAC. The cp genomes of A. guttata and A. tibetana were particularly highly conserved. Mildly divergent regions were identified in trnS-GCU-trnG-UCC, trnC-GCA-petN, psaA-ycf3, trnI-CAU-ycf2, psbE-petL, clpP, and rps15-ycf1 (Figure 6). Nucleotide diversity (Pi) for the cp genome of the four species was calculated as 0.015 using the DNA SP program. IR regions (Pi = 0.007) were more variable than LSC (Pi = 0.0046) or SSC (Pi = 0.019) regions. Single-copy regions presented higher Pi values than IR regions. In particular, trnH-psbA (Pi = 0.078) and atpB-rbcL (Pi = 0.038) appeared highly diverse (Figure 7). In spite of the morphological and cp genome similarity between A. guttata and A. tibetana, the atpB-rbcL region was more diverse (Pi = 0.048) than the average for the four cp genomes.
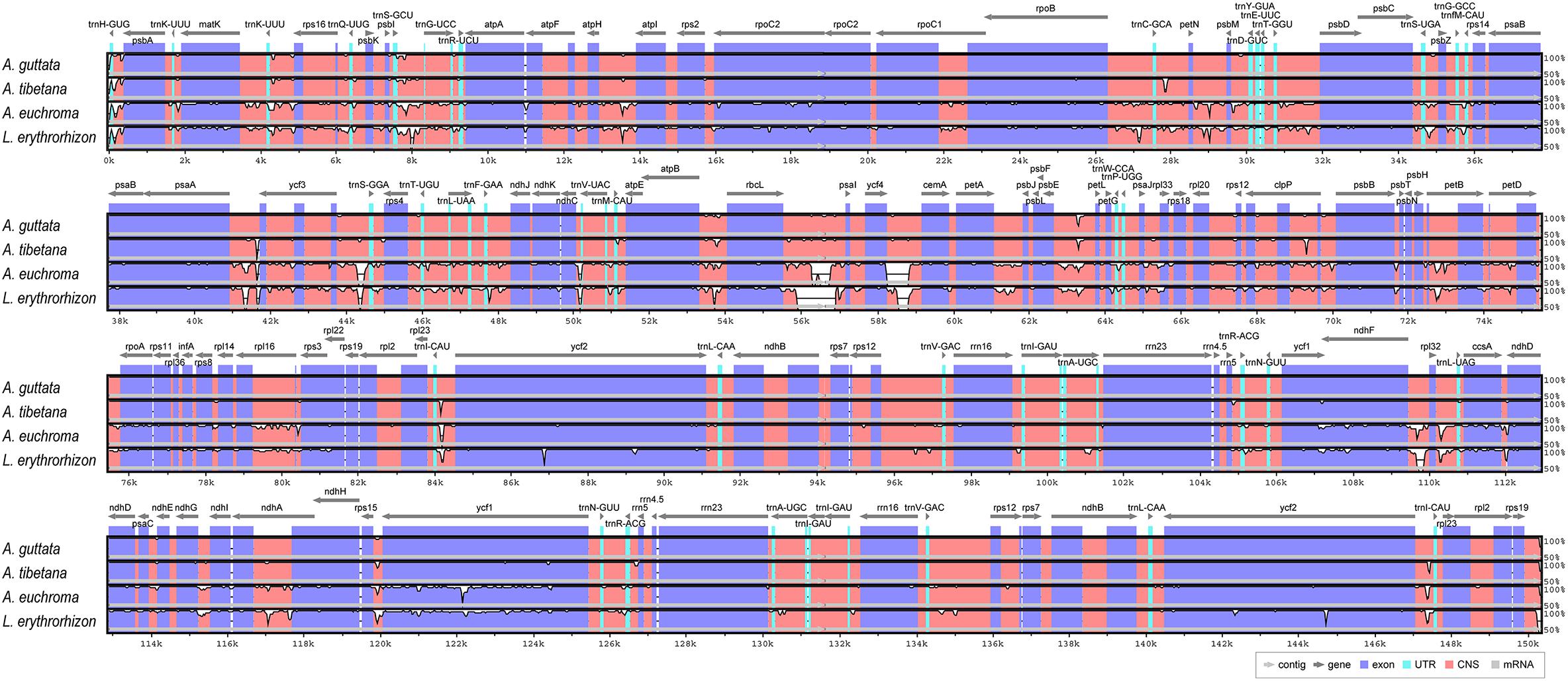
Figure 6. Comparison of Arnebia and Lithospermum chloroplast genomes using mVISTA. Complete cp genomes of five Trichosanthes were compared against A. guttata as reference. Blue block: conserved genes, sky-blue block: transfer RNA (tRNA) and ribosomal RNA (rRNA), and red block: conserved non-coding sequences (CNS). Regions with sequence variation among the five Trichosanthes are denoted in white.
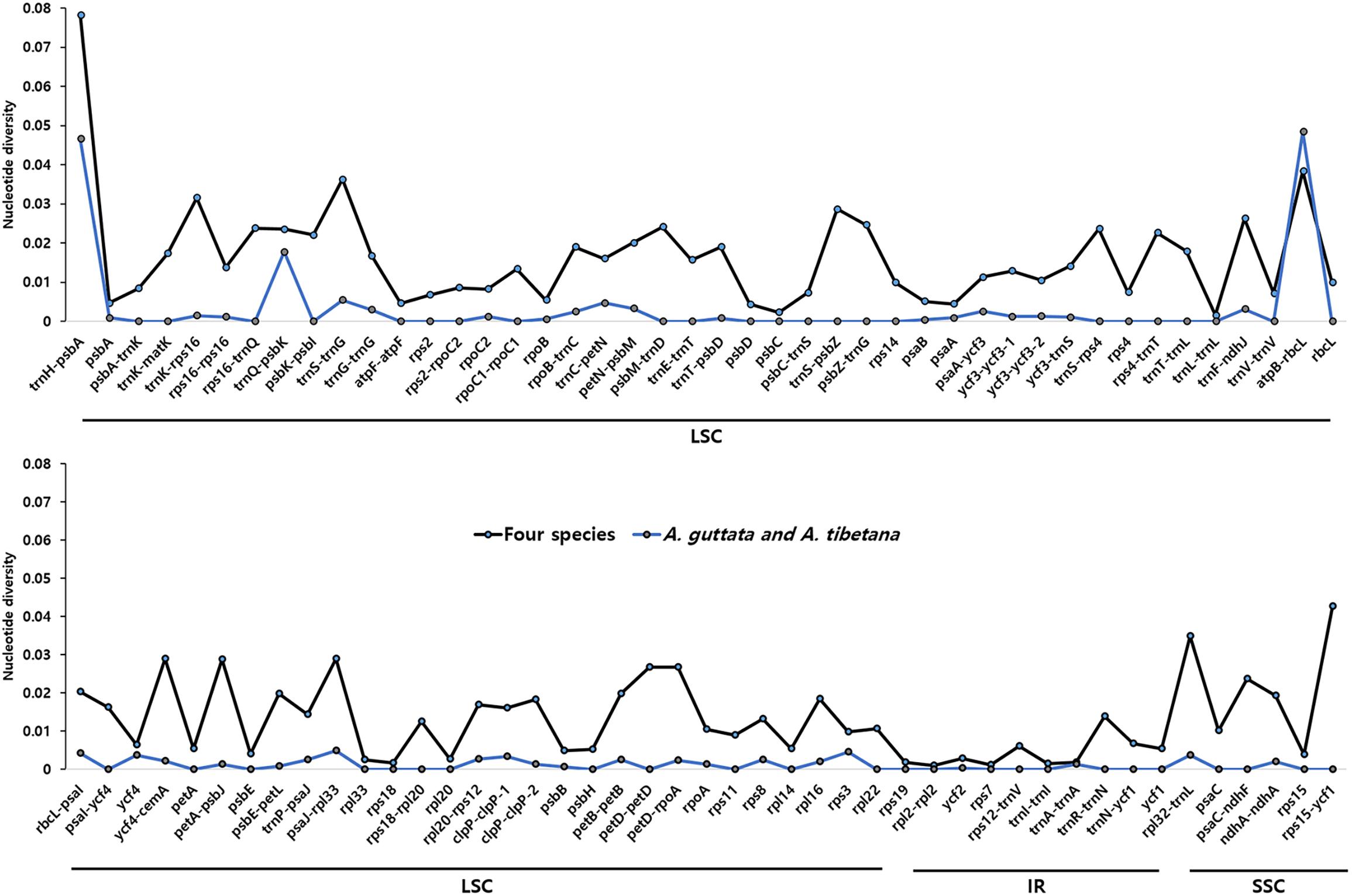
Figure 7. Comparison of nucleotide diversity (Pi) values among Arnebia and Lithospermum species. The dotted line indicates the average Pi value in Arnebia and Lithospermum chloroplast genomes and excludes regions with Pi = 0.
Using the non-synonymous substitution to synonymous substitution (Ka/Ks) ratio, a total of 77 genes were calculated to identify selective pressure for the three Arnebia species, L. erythrorhizon, and B. officinalis, with T. plicata as reference (Supplementary Figure 7). Most genes were conserved and exhibited a Ka/Ks ratio < 1. Only rbcL was positively selected in all five Boraginaceae cp genomes (Ka/Ks = 1.064–1.286).
Herbal medicinal plants represent important resources. To ensure efficient identification among the four tested species, we developed indel marker sets targeting three intergenic regions: psaA- ycf3 (PSY), trnI-CAU- ycf2 (TCY), and ndhC-trnV-UAC (NCTV; Figure 8). The PSY (psaA- ycf3) marker could discriminate clearly between the four species as it yielded fragments of 764 bp with A. guttata, 715 bp with A. tibetana, 701 bp with A. euchroma, and 629 bp with L. erythrorhizon. TCY (trnI-CAU- ycf2) was employed to distinguish A. tibetana, which yielded a fragment of 429 bp, from A. guttata (399 bp), A. euchroma (369 bp), and L. erythrorhizon (367 bp). Finally, NCTV (ndhC-trnV-UAC) could discriminate A. euchroma and L. erythrorhizon from A. guttata and A. tibetana.
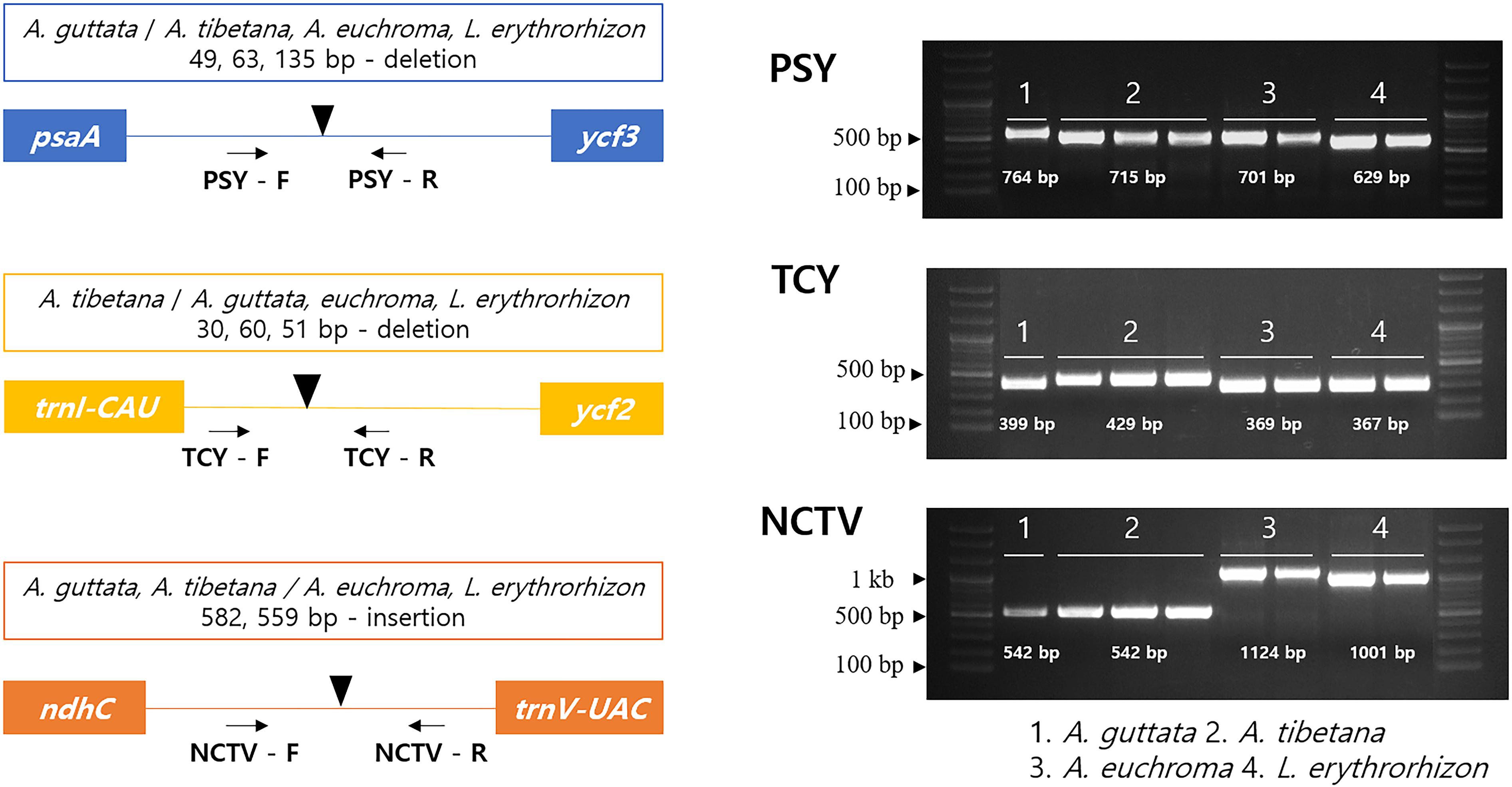
Figure 8. Schematic representation of indel markers for Arnebia and Lithospermum species. Primers for PSY (psaA- ycf3), TCY (trnI-CAU-ycf2), and NCTV (ndhC-trnV-UAC) were tested. Control samples were used in a marker test (Supplementary Table 4). 1: A. guttata; 2: A. tibetana; 3: A. euchroma; and 4: L. erythrorhizon.
To identify the phylogenic relationships between Arnebia and Lithospermum species within the Boraginaceae family, we aligned the whole chloroplast genomes and 71 conserved protein- CDS (65,155 bp) shared by the three Arnebia and one Lithospermum accessions examined in this study, with F. suspensa from the Oleaceae used as outgroup (Figure 9 and Supplementary Figure 8). ML and BI topologies were highly congruent for both the whole chloroplast genomes and CDS data. All phylogenetic trees except Lithospermum were strongly supported (ML = 100%, BI = 1.0). Seven species within the Boraginaceae clustered consistently with respect to the APG IV classification system. Overall, clustering revealed that A. guttata and A. tibetana shared the closest phylogenetic relationship, with A. euchroma as their sister group and Lithospermum clearly distinct from Arnebia.
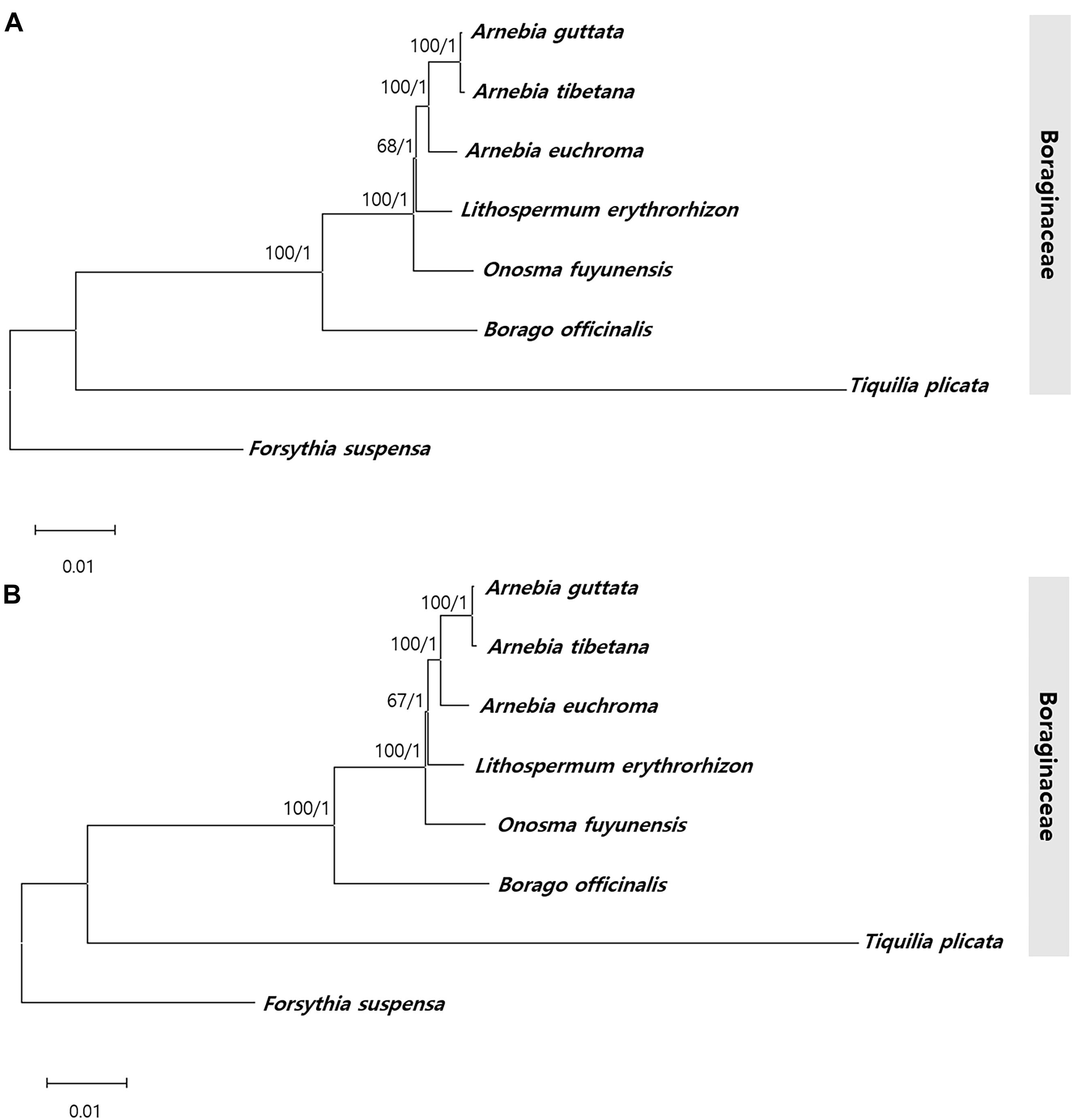
Figure 9. Phylogenetic tree from Arnebia and Lithospermum using maximum likelihood bootstraps and Bayesian posterior probabilities. (A) Whole chloroplast genome dataset topology, (B) 72 CDS dataset topology. Maximum likelihood topology is shown with bootstrap support values/Bayesian posterior probabilities given at each node.
Although all studied species shared the same calyx lobe or corolla shape and ovary surface cell patterns, Arnebia and Lithospermum exhibited substantial diversity with respect to floral epidermis cell patterns, gynoecium, and structure of trichomes.
Based on floral morphology, only L. erythrorhizon had scorpioid cymes and white flowers with faucal appendages (also referred to as “faucal scale”) on the corolla tube, deeply lobed calyx, and indistinct linear-shaped style (Table 1). Faucal appendages, which appear in certain species, represent a useful diagnostic tool in Boraginaceae (Zhu et al., 1995; Cohen, 2011; Weigend et al., 2016). Previous studies have suggested that these appendages could be modified through development of specialized epidermal cells to better attract pollinators (Kelley and Wilken, 1993; Cohen, 2011). Accordingly, L. erythrorhizon with faucal appendages and Arnebia species without faucal appendages may attract different pollinators. In Arnebia, A. euchroma could be easily distinguished from other species based on morphological traits, including corolla color, plant height, root length, and leaf shape (Song J. H. et al., 2019).
The observations of members of the tribe Boragineae (genera Gastrocotyle, Hormuzakia, and Phyllocara) have revealed that the floral trichome characteristics, such as trichome surface pattern, density, length, and cell number, have taxonomic implications in the context of their generic classification (Bigazzi et al., 1999). Our floral micromorphological data also confirmed the usefulness of trichome types for the identification of medicinal borage species. The morphology and the distribution of trichomes in A. guttata were similar to those of A. tibetana (verrucate, non-glandular trichome on both the outer calyx and corolla). In contrast, smooth, hirsute non-glandular trichomes on the outer calyx were exclusive to L. erythrorhizon; whereas non-glandular and glandular trichomes with stalk on both the outer calyx and corolla were unique to A. euchroma. Hirsute, rigid trichomes form physical barriers against insects as part of a “greasy pole strategy” (Kerner, 1876; Harley, 1991; Eigenbrode, 2004; Jeffree, 2006). Floral glands, which contain glucose or phenolic compounds, may provide additional chemical protection against herbivore attacks (McKey, 1979; Sudd and Sudd, 1985; Nicolson and Thornburg, 2007; Baker-Meio and Marquis, 2012; Guesdon et al., 2019). Thus, outer calyx trichomes and outer floral glands on the studied species are probably associated with attracting anti-herbivore protectors. Further morpho-anatomical and histochemical studies are required to confirm their detailed internal structures and functions.
A conical or papillose corolla epidermis is found on various angiosperm species (Christensen and Hansen, 1998; Whitney and Glover, 2007; Song et al., 2020), where its tactile cues may promote pollination by certain insects (Whitney et al., 2009, 2011). All studied species displayed a papillose cell pattern on the inner corolla. Only L. erythrorhizon had a unique papillose knobby pattern, which resulted in much higher and sharper pointed cells than a papillose conical type, (average papillose knobby cell height, 36.74 μm; papillose conical cell height, 10.70 μm), with non-glandular and glandular trichomes. As with the presence or absence of faucal appendages, inner corolla cellular patterns are also associated with pollinators. More comprehensive studies on differences in pollination position, visiting rate of pollinator, or pollen shape in Lithospermum and Arnebia will provide better understanding of their reproduction.
Gynoecium characteristics, such as stylar lobation or stigma number and shape in Arnebia and related genera, are critical for distinguishing between groups and species (Zhu et al., 1995; Ambrish and Srivastava, 2014). In particular, the degree of lobation on the style is a key characteristic to discriminate the genera Arnebia and Huynhia, as well as Arnebia species (Coppi et al., 2015). Here, we detected differences in stylar lobation and stigma shape between the very similar A. guttata and A. tibetana species. All A. guttata had a consistently 2-fid style with oblong stigmas; whereas A. tibetana had a 2-cleft style with oblate stigmas. Notably, all Arnebia species shared the same tuberculate stigma cell pattern.
Based on the floral external and epidermal micromorphological characteristics of three Arnebia and one Lithospermum species, we provide the following identification key.
1. Flower color white; calyx 5-parted; outer and inner corolla surface with papillose knobby cells; style linear, indistinct; stigma asterisk-tuberculate …………………….. L. erythrorhizon
1’. Flower color purple or yellow; calyx 5-lobed; outer corolla surface with tabular rugose and inner corolla with papillose conical cells; style filiform, distinct; stigma tuberculate ….2
2. Flower color purple; both outer calyx and outer corolla surfaces pubescent with smooth glandular and non-glandular trichomes; inner corolla surface glabrous; stigma globose ………………………………………………………… A. euchroma
2’. Flower color yellow; both outer calyx and outer corolla surfaces pubescent with verrucate non-glandular trichomes; inner corolla surface pubescent with smooth glandular trichomes with stalk; stigma not-globose ………. 3
3. Style 2-cleft; stigma oblate ……………………………….. A. guttata
3’. Style 2-fid; stigma oblong ………………………………. A. tibetana
We determined the complete cp genomes of three Arnebia and one Lithospermum accessions. The organization of these four cp genomes follows that of other Boraginaceae (Chen and Zhang, 2019; Guo et al., 2020), and includes a typical quadripartite structure with LSC and SSC regions separated by two IRs. Genomes in the tested Arnebia and Lithospermum species possess 112 unique genes. Their gene order, GC content, genomic structure, and overall length (149,316–150,465 bp) conform to previously described angiosperm cp genomes (Millen et al., 2001). Codon usage is a key factor for correct expression of genetic information, and plays an important role in shaping cp genome evolution (Wang et al., 2016; Yi et al., 2018). Arnebia and Lithospermum accessions possess nearly identical codons, which are similar to those of other cp genomes (Wang et al., 2016; Raman et al., 2019). RSCU values indicated synonymous codon usage bias, with a disproportionate number of codons having an A or T in the third position. This phenomenon has been described in many angiosperm cp genomes and is consistent with the pattern in seed plants. Indeed, high RSCU values correspond to more highly conserved cp genes (Wang et al., 2016; Ivanova et al., 2017; Zuo et al., 2017). We surveyed the RSCU values for six Boraginaceae cp genomes downloaded from NCBI. Half of the codons had a weak codon bias (Supplementary Figure 4), denoted in green in the figure (RSCU > 1). This result is similar to other cp genomes, and most codons with high RSCU values had an A or T in the third position. The cp genome of B. officinalis displayed slightly different codon bias than other Boraginaceae species. In particular, GGT (Guanine), AGG (Arginine), and GGG (Guanine) codons had relatively low RSCU values; whereas TGT (Cysteine) and CGA (Arginine) had somewhat higher RSCU values.
The mVISTA results show that the cp genomes of Arnebia and Lithospermum possess relatively high diversity, with genic regions being more conserved than intergenic spacer regions, as is typical in angiosperm cp genomes (Shaw et al., 2007; Huo et al., 2019; Song J. H. et al., 2019). Even though the cp genomes of A. guttata and A. tibetana displayed closer proximity than other cp genomes, trnS-GCU-trnG-UCC, trnC-GCA-petN, psaA-ycf3, trnI-CAU-ycf2, psbE-petL, clpP, and rps15-ycf1 regions were found to be hotspots of genetic variation (Figure 4). Hotspot regions in plants indicate underlying evolution (Morton and Clegg, 1993; Maier et al., 1995; Liu H. et al., 2018; Liu H. et al., 2019) and can be used to distinguish species or genera. Molecular markers relative to these regions serve for the identification of valuable herbal medicinal plants (Cho et al., 2015; Hong et al., 2017; Park et al., 2018a,b). In terms of nucleotide diversity, most divergent regions were non-coding, which is again consistent with other cp genomes (Schroeder et al., 2016; Zhitao et al., 2017; Liu E. et al., 2019; Smidt et al., 2020). Other cp genomes were reported to have highly variable non-coding regions at trnS-GCU-trnG-UCC, trnC-GCA-petN, psaA-ycf3, trnI-CAU-ycf2, psbE-petL, and rps15-ycf1. We noticed higher diversity in the atpB-rbcL region between A. guttata and A. tibetana compared to the other four species. This divergence indicated a species-unique region, which could be clearly distinguished and used for identification.
Usually, angiosperm cp genomes are highly conserved in length, structure, gene number, and gene order (Jansen and Ruhlman, 2012). IR contraction and expansion cause the length of the genome to vary (Raubeson et al., 2007). Here, we compared six Boraginaceae cp genomes, including Arnebia and Lithospermum, against F. suspensa (family Oleaceae) as reference (Supplementary Figure 5). The six Boraginaceae cp genomes were well conserved in terms of gene order and positions. Moreover, except for T. plicata, they all indicated IR contraction. Specifically, rps2 was located in LSC/IRa and ycf1 was identified in the IRa/SSC junction, which overlapped with ndhF. These findings revealed the occurrence of IR contraction and expansion, as well as their effect on genome length.
Most genes in the Boraginaceae exhibited stable gene evolution (purifying selection). Positive selection was detected in rbcL, a gene encoding the large subunit of Rubisco, which is commonly observed in angiosperm cp genomes due to the elevated selective pressure for this protein (Kapralov and Filatov, 2007; Shi et al., 2019; Yao et al., 2019). Large-scale studies confirmed that positive selection of rbcL is widespread among land plants. Thus, Kapralov and Filatov (2007) evaluated positive selection in rbcL in over 3,000 species. and reported positive selection in most land plants but not in algae and cyanobacteria. A similar study supported the positive selection of rbcL in Ilex using 240 Ilex sequences (Yao et al., 2019). Positive selection in the chloroplast genome, including at rbcL, has been reported in numerous plant taxa (Bock et al., 2014). The results of our study corroborated the results of the previous studies. The positive selection suggests ongoing adaptation to the environment (Kimura, 1989; Raman and Park, 2016; Ivanova et al., 2017).
Taken together, we demonstrated that Boraginaceae chloroplast genomes are similar to other angiosperm genomes, whereas sequence divergence of the chloroplast genome was identified between A. guttata and A. tibetana (Figures 6, 7). Previously, species identification of A. guttata and A. tibetana based on only general morphological characterization was difficult, and their taxonomic classification was also unclear. However, we found the noticed evidence for distinguishing this group. The complete sequences of Arnebia chloroplast genomes enabled accurate comparative analysis and the identification of species-specific traits for accurate classification of Arnebia species.
Artaxa guttata, A. euchroma, and L. erythrorhizon are valuable medicinal plants in traditional Korean herbal medicine (Korea Institute of Oriental Medicine [KIOM], 2020). To ensure quality control, safety, and effectiveness of their ingredients, Herbal Pharmacopeia mandates that herbal medicinal materials should be kept in pure form. Adulterants may cause negative side effects and quality problems. The purity of herbal medicinal ingredients can be tested with species-specific molecular markers. Here, we developed indel marker sets to distinguish between four species of medicinal Boraginaceae. Although cp genomes are highly conserved within the same genus, they frequently possess insertion/deletion regions. We detected the conserved indel regions psaA- ycf3, trnI-CAU- ycf2, and ndhC-trnV-UAC. The corresponding marker sets psaA- ycf3 (PSY), trnI-CAU- ycf2 (TCY), and ndhC-trnV-UAC (NCTV) successfully discriminated between the four species, as well as between the closely related A. guttata and A. tibetana. The rapid and accurate species identification achieved by these marker sets will help distinguish Arnebia and Lithospermum without having to rely on morphological analysis, and will prevent incorrect classification of ambiguous samples from valuable Arnebia species.
Complete cp genomes provide sufficient information to study the phylogenetic relationships among plants, including at low taxonomy level and for unresolved taxa (Jansen et al., 2007; Park et al., 2019, 2020). ITS and cp loci have confirmed the correct classification of numerous Boraginaceae species and inference of their evolutionary relationships based on morphological characteristics (Cohen, 2014). However, the Arnebia genus maintains an ambiguous phylogenetic position with respect to other monophyletic species and genera, such as Onosma, Cerinthe, Moltkia, and Cystostemon. This phylogenetic relationship derives from the use of only two Arnebia species out of 224 Boraginaceae. The monophyletic relationship between previously recognized clades and the position of various unplaced genera in Boraginaceae have been reclassified (Chacón et al., 2016); however, infrageneric classification of Arnebia species has remained unresolved (Coppi et al., 2015). Although it comprises only 25–30 species, the genus Arnebia is valuable in traditional herbal medicine. Here, we attempted to determine phylogenetic relationships within Boraginaceae using cp genomes. ML and BI values based on the whole chloroplast genome and CDS dataset offered powerful evidence (Figure 9) of clear species clustering within families according to the APG IV classification system. In Boraginaceae, A. guttata and A. tibetana were found to group monophyletically with a sister group, A. euchroma, which is consistent with previous findings (Wu et al., 2017; Liu J. et al., 2020).
Since A. tibetana was firstly described by Kurz (1874), this species has been regarded as a synonym of A. guttata (Kazmi, 1970; Ambrish and Srivastava, 2014; Urgamal et al., 2014) or as an independent species (Zhu et al., 1995; Lazkov and Sultanova, 2014). In Flora of China, A. tibetana is placed as a subspecies owing to its narrow distribution in Xizang, Xinjiang in China, and adjacent Xinjiang in Kyrgyzstan and its absence of blackish purple spots on corolla lobes (Zhu et al., 1995). Moreover, Lazkov and Sultanova (2014) recognized and listed A. guttata and A. tibetana as independent species in the Flora of Kyrgyzstan on the basis of the different life cycles (A. guttata, perennial; A. tibetana, biennial). During our field survey, we found that the flowers with or without blackish purple spots on corolla lobes in both A. guttata and A. tibetana. The blackish purple spots appear during the flowering season, but disappear after pollination and at withering in A. guttata and A. tibetana. Thus, this traditional diagnostic character for distinguishing A. tibetana (Zhu et al., 1995) is not useful because they are the seasonal variation. However, based on our consistently observed floral morphological characteristics, such as style lobation, and stigma shape, and genetic distance of cp genomes reported in this study, A. tibetana is recognized as an independent species following Kurz (1874)’s taxonomic concept with newly found diagnostic characteristics, 2-fid style and oblong stigma.
Floral morphological and micromorphological characteristics, as well as chloroplast (cp) genome sequences of selected species in Arnebia and Lithospermum genera were analyzed. The floral characteristics reveal great diversity in floral epidermis cell patterns, gynoecium, and structure of trichomes. Chloroplast genomes of the four species were well conserved with respect to genome length, gene number, gene orientation, and local divergence. Most variable sequences were detected in non-coding regions. These hotspots were used to develop new indel molecular marker sets for efficient and rapid species identification. We successfully established phylogenetic relationships among Arnebia species in Boraginaceae using cp genome sequences. Accordingly, we suggest that A. tibetana be recognized as an independent species following Kurz (1874)’s taxonomic concept based on its narrow distribution, different life cycle, and newly found gynoecium characteristics and genetic distance of cp genomes in this study. Morphological characteristics, genomic data, and taxonomic concept presented in our study for medicinal Arnebia and Lithospermum species will be helpful in further research on Boraginaceae.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi. nlm.nih.gov/genbank/, MT975391; https://www.ncbi.nlm.nih.gov/genbank/, MT975392; https://www.ncbi.nlm.nih.gov/genbank/, MT975393; https://www.ncbi.nlm.nih.gov/genbank/, MT975394; https://datadryad.org/stash, https://doi.org/10.5061/dryad.gtht76hk7.
IP and J-HS designed the experimental framework and drafted and revised the manuscript. J-HS, SY, and BM collected and identified plant materials. J-HS performed microscopic analysis, whereas IP performed molecular experiments and carried out genome analysis. IP, J-HS, and BM revised the manuscript. All authors contributed to the experiments and approved the final manuscript.
This research was funded by a grant from the Development of Sustainable Application for Standard Herbal Resources (KSN2012320) from the Korea Institute of Oriental Medicine (KIOM), South Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank directors of the following herbaria: FRU, UBU. Dr. G.A. Lazkov (National Academy of Sciences, Kyrgyzstan; FRU) and Dr. Prof. B. Oyuntsetseg (National University of Mongolia, Mongolia; UBU) for permitting the examination of specimens, as well as Dr. Prof. H.J. Choi (Changwon National University, South Korea), for constructive comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.606463/full#supplementary-material
Akçin, Ö. E. (2009). Micromorphological and anatomical studies on petals of 11 Turkish Onosma L. (Boraginaceae) taxa. Bangladesh J. Plant Taxon. 16, 157–164. doi: 10.3329/bjpt.v16i2.3928
Akkol, E. K., Koca, U., Peşin, I., Yılmazer, D., Toker, G., and Yeşilada, E. (2009). Exploring the wound healing activity of Arnebia densiflora (Nordm.) Ledeb. by in vivo models. J. Ethnopharmacol. 124, 137–141. doi: 10.1016/j.jep.2009.03.019
Allen, G., Flores-Vergara, M., Krasynanski, S., Kumar, S., and Thompson, W. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Ambrish, K., and Srivastava, S. K. (2014). Taxonomic studies on the genus Arnebia Forssk. (Boraginaceae) in India. Taiwania 59, 315–325.
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153.
Bacchetta, G., Brullo, S., and Selvi, F. (2000). Echium anchusoides (Boraginaceae), a new species from Sardinia (Italy). Nord. J. Bot. 20, 271–278. doi: 10.1111/j.1756-1051.2000.tb00743.x
Baker-Meio, B., and Marquis, R. J. (2012). Context-dependent benefits from ant–plant mutualism in three sympatric varieties of Chamaecrista desvauxii. ıJ. Ecol. 100, 242–252. doi: 10.1111/j.1365-2745.2011.01892.x
Beier, S., Thiel, T., Munch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bigazzi, M., and Selvi, F. (2000). Stigma form and surface in the tribe Boragineae (Boraginaceae): micromorphological diversity, relationships with pollen, and systematic relevance. Can. J. Bot. 78, 388–408. doi: 10.1139/b00-014
Bigazzi, M., Selvi, F., and Fiorini, G. (1999). A reappraisal of the generic status of Gastrocotyle, Hormuzakia and Phyllocara (Boraginaceae) in the light of micromorphological and karyological evidence. Edinb. J. Bot. 56, 229–251. doi: 10.1017/s0960428600001104
Bo, S., Dan, M., Li, W., and Zhang, P. (2019). Characterizations and immunostimulatory activities of a polysaccharide from Arnebia euchroma (Royle) Johnst. roots. Int. J. Biol. Macromol. 125, 791–799. doi: 10.1016/j.ijbiomac.2018.11.238
Bock, D. G., Andrew, R. L., and Rieseberg, L. H. (2014). On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 23, 4899–4911. doi: 10.1111/mec.12920
Bunge, A. A. (1840). Arnebia guttata Bunge, Index Seminum, Vol. 7. St. Petersburg: St Petersburg Press, 1840.
Carver, T., Berriman, M., Tivey, A., Patel, C., Bohme, U., Barrell, B. G., et al. (2008). Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676. doi: 10.1093/bioinformatics/btn529
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chacón, J., Luebert, F., Hilger, H. H., Ovchinnikova, S., Selvi, F., Cecchi, L., et al. (2016). The borage family (Boraginaceae s. str.): a revised infrafamilial classification based on new phylogenetic evidence, with emphasis on the placement of some enigmatic genera. Taxon 65, 523–546. doi: 10.12705/653.6
Chen, C. W., Huang, Y. M., Kuo, L. Y., Nguyen, Q. D., Luu, H. T., Callado, J. R., et al. (2013). trnL-F is a powerful marker for DNA identification of field vittarioid gametophytes (Pteridaceae). Ann. Bot. 111, 663–673. doi: 10.1093/aob/mct004
Chen, Q., and Zhang, D. (2019). The complete chloroplast genome sequence of Onosma paniculatum Bur. et Franch. (Boraginaceae), a medicinal plant in Yunnan and its adjacent regions. Mitochondrial DNA Part B Resour. 4, 3330–3332. doi: 10.1080/23802359.2019.1673230
Cho, K. S., Yun, B. K., Yoon, Y. H., Hong, S. Y., Mekapogu, M., Kim, K. H., et al. (2015). Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLoS One 10:e0125332. doi: 10.1371/journal.pone.0125332
Christensen, K. I., and Hansen, H. V. (1998). SEM studies of epidermal patterns of petals in the angiosperms. Opera Bot. 135, 5–91. doi: 10.5091/plecevo.2013.738
Cohen, J. I. (2011). A phylogenetic analysis of morphological and molecular characters of Lithospermum L. (Boraginaceae) and related taxa: evolutionary relationships and character evolution. Cladistics 27, 559–580. doi: 10.1111/j.1096-0031.2011.00352.x
Cohen, J. I. (2014). A phylogenetic analysis of morphological and molecular characters of Boraginaceae: evolutionary relationships, taxonomy, and patterns of character evolution. Cladistics 30, 139–169. doi: 10.1111/cla.12036
Coiro, M., and Barone Lumaga, M. R. (2018). Disentangling historical signal and pollinator selection on the micromorphology of flowers: an example from the floral epidermis of the Nymphaeaceae. Plant Biol. 20, 902–915. doi: 10.1111/plb.12850
Coppi, A., Cecchi, L., Nocentini, D., and Selvi, F. (2015). Arnebia purpurea: a new member of formerly monotypic genus Huynhia (Boraginaceae-Lithospermeae). Phytotaxa 204, 123–136. doi: 10.11646/phytotaxa.204.2.3
Damianakos, H., Kretschmer, N., Sykłowska-Baranek, K., Pietrosiuk, A., Bauer, R., and Chinou, I. (2012). Antimicrobial and cytotoxic isohexenylnaphthazarins from Arnebia euchroma (Royle) Jonst. (Boraginaceae) callus and cell suspension culture. Molecules 17, 14310–14322. doi: 10.3390/molecules171214310
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17:134.
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Delannoy, E., Fujii, S., Colas Des Francs-Small, C., Brundrett, M., and Small, I. (2011). Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28, 2077–2086. doi: 10.1093/molbev/msr028
Delcher, A. L., Salzberg, S. L., and Phillippy, A. M. (2003). Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinform. 10, 10.3.1–10.3.18.
Eigenbrode, S. D. (2004). The effects of plant epicuticular waxy blooms on attachment and effectiveness of predatory insects. Arthropod Struct. Dev. 33, 91–102. doi: 10.1016/j.asd.2003.11.004
Fang, R. C., and Staples, G. W. (1995). “Arnebia Forsskål,” in Flora of China 16: Gentianaceae through Boraginaceae, eds Z.-Y. Wu and P. H. Raven (St. Louis, MO: Missouri Botanical Garden), 344–346.
Forsskål, P. (1775). Flora Aegyptiaco-Arabica: Sive Descriptiones Plantarum Quas per Aegyptum Inferiorem et Arabiam Felicem. Hauniae: Ex Officina Mölleri.
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279.
Garg, S., Chandra, A., Mazumder, A., and Mazumder, R. (2014). Green synthesis of silver nanoparticles using Arnebia nobilis root extract and wound healing potential of its hydrogel. Asian J. Pharm. 8, 95–101. doi: 10.4103/0973-8398.134925
Giuliani, C., Foggi, B., and Mariotti Lippi, M. (2018). Floral morphology, micromorphology and palinology of selected Sedum s.l. species (Crassulaceae). Plant Biosyst. 152, 333–348. doi: 10.1080/11263504.2016.1271056
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64.
Guesdon, I. R., Amorim, A. M., and Meira, R. M. S. A. (2019). Functional role and evolutionary contributions of floral gland morphoanatomy in the Paleotropical genus Acridocarpus (Malpighiaceae). PLoS One 14:e0222561. doi: 10.1371/journal.pone.0222561
Guo, X., Wang, X., Wang, Q., Liu, C., Zhang, R., Cheng, A., et al. (2020). The complete chloroplast genome sequence of Borago officinalis Linn.(Boraginaceae) and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 5, 1461–1462. doi: 10.1080/23802359.2020.1741467
Gurusaran, M., Ravella, D., and Sekar, K. (2013). RepEx: repeat extractor for biological sequences. Genomics 102, 403–408. doi: 10.1016/j.ygeno.2013.07.005
Hamilton, M. B., Braverman, J. M., and Soria-Hernanz, D. F. (2003). Patterns and relative rates of nucleotide and insertion/deletion evolution at six chloroplast intergenic regions in new world species of the Lecythidaceae. Mol. Biol. Evol. 20, 1710–1721. doi: 10.1093/molbev/msg190
Harley, R. (1991). “The greasy pole syndrome,” in Ant-Plant Interactions, eds C. R. Huxley and D. F. Cutler (Oxford: Oxford University Press), 430–433.
Hong, S. Y., Cheon, K. S., Yoo, K. O., Lee, H. O., Cho, K. S., Suh, J. T., et al. (2017). Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front. Plant Sci. 8:1696. doi: 10.3389/fpls.2017.01696
Huo, Y., Gao, L., Liu, B., Yang, Y., Kong, S., Sun, Y., et al. (2019). Complete chloroplast genome sequences of four Allium species: comparative and phylogenetic analyses. Sci. Rep. 9:12250.
Ishida, T., and Sakaguchi, I. (2007). Protection of human keratinocytes from UVB-induced inflammation using root extract of Lithospermum erythrorhizon. Biol. Pharm. Bull. 30, 928–934. doi: 10.1248/bpb.30.928
Ivanova, Z., Sablok, G., Daskalova, E., Zahmanova, G., Apostolova, E., Yahubyan, G., et al. (2017). Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 8:204. doi: 10.3389/fpls.2017.00204
Jain, S. C., Singh, B., and Jain, R. (2000). Arnebins and antimicrobial activities of Arnebia hispidissima DC. Cell cultures. Phytomedicine 6, 474–476. doi: 10.1016/s0944-7113(00)80077-x
Jansen, R. K., Cai, Z., Raubeson, L. A., Daniell, H., Depamphilis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104, 19369–19374. doi: 10.1073/pnas.0709121104
Jansen, R. K., and Ruhlman, T. A. (2012). “Plastid genomes of seed plants,” in Genomics of Chloroplasts and Mitochondria, Vol. 35, eds R. Bock and V. Knoop (Dordrecht: Springer), 103–126. doi: 10.1007/978-94-007-2920-9_5
Jeffree, C. E. (2006). “The fine structure of the plant cuticle,” in Biology of the Plant Cuticle, eds M. Riederer and C. Müller (Oxford: Blackwell), 11–125. doi: 10.1002/9780470988718.ch2
Kapralov, M. V., and Filatov, D. A. (2007). Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol. Biol. 7:73. doi: 10.1186/1471-2148-7-73
Katoch, P., Rana, S., Kumar, D., Kumar, S., and Bhushan, S. (2016). Concurrent NP-HPTLC determination of shikonin and β, β-Dimethylacryl shikonin in Arnebia benthamii. J. Chromatogr. Sci. 54, 1421–1427. doi: 10.1093/chromsci/bmw096
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kazmi, S. M. A. (1970). A revision of the Boraginaceae of West Pakistan and Kashmir. J. Arnold Arbor. 51, 133–184.
Kelley, W. A., and Wilken, D. (1993). “Cryptantha,” in The Jepson Manual: Higher Plants of California, ed. J. C. Hickman (Berkeley, CA: University of California Press), 369–378.
Kerner, A. (1876). Die Schutzmittel der Blüthen Gegen Unberufene Gäste. Wien: Zoologisch-Botanische Gesellschaft.
Khatoon, S., Mehrotra, B. N., and Mehrotra, S. (2003). Pharmacognostic evaluation of ratanjot-Arnebia nobilis Rech. f. Nat. Prod. Sci. 9, 286–290.
Kim, K., Lee, S. C., Lee, J., Lee, H. O., Joh, H. J., Kim, N. H., et al. (2015). Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One 10:e0117159. doi: 10.1371/journal.pone.0117159
Kim, Y. D. (2019). “Boraginaceae,” in The Genera of Vascular Plants of Korea, ed. C. W. Park (Fort Lauderdale, FL: Academy Publishing Co), 1057–1070.
Kimura, M. (1989). The neutral theory of molecular evolution and the world view of the neutralists. Genome 31, 24–31. doi: 10.1139/g89-009
Konarska, A. (2017). Comparative micromorphology and anatomy of flowers and floral secretory structures in two Viburnum species. Protoplasma 254, 523–537. doi: 10.1007/s00709-016-0972-0
Korea Institute of Oriental Medicine [KIOM] (2020). Defining Dictionary for Medicinal Herbs. Available online at: http://boncho.kiom.re.kr/codex/ (accessed September 1, 2020).
Kosger, H. H., Ozturk, M., Sokmen, A., Bulut, E., and Ay, S. (2009). Wound healing effects of Arnebia densiflora root extracts on rat palatal mucosa. Eur. J. Dent. 3, 96–99. doi: 10.1055/s-0039-1697414
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Lazkov, G. A., and Sultanova, B. A. (2014). Checklist of Vascular Plants of Kyrgyzstan. Bishkek: Kyrgyz Academy of Sciences.
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Liu, E., Yang, C., Liu, J., Jin, S., Harijati, N., Hu, Z., et al. (2019). Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci. Rep. 9:809.
Liu, H., He, J., Ding, C., Lyu, R., Pei, L., Cheng, J., et al. (2018). Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe anemoneae (Ranunculaceae). Front. Plant Sci. 9:1097. doi: 10.3389/fpls.2018.01097
Liu, H., Su, Z., Yu, S., Liu, J., Yin, X., Zhang, G., et al. (2019). Genome comparison reveals mutation hotspots in the chloroplast genome and phylogenetic relationships of Ormosia Species. Biomed Res. Int. 2019:7265030.
Liu, J., Zhao, C., Guo, L., Ma, S., Zheng, J., and Wang, J. (2020). The plant original identification of Inner Mongolia Arnebia radix based on morphology and DNA barcoding. Res. Sq. PPR196478. doi: 10.21203/rs.3.rs-50697/v1
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.
Mabberley, D. J. (2008). Mabberley’s Plant Book: A Portable Dictionary of Plants, their Classification and Uses. Cambridge: Cambridge University Press.
Maier, R. M., Neckermann, K., Igloi, G. L., and Kossel, H. (1995). Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. doi: 10.1006/jmbi.1995.0460
McKey, D. (1979). “The distribution of secondary compounds within plants,” in Herbivores: Their Interactions with Secondary Plant Metabolites, eds G. A. Rosenthal and D. H. Janzen (New York, NY: Academic Press), 55–133.
Millen, R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. doi: 10.2307/3871412
Morton, B. R., and Clegg, M. T. (1993). A chloroplast DNA mutational hotspot and gene conversion in a noncoding region near rbcL in the grass family (Poaceae). Curr. Genet. 24, 357–365. doi: 10.1007/bf00336789
Nicolson, S. W., and Thornburg, R. W. (2007). “Nectar chemistry,” in Nectaries and Nectar, eds S. W. Nicolson, M. Nepi, and E. Pacini (Dordrecht: Springer), 215–263. doi: 10.1007/978-1-4020-5937-7_5
Park, I., Kim, W. J., Yang, S., Yeo, S. M., Li, H., and Moon, B. C. (2017). The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS One 12:e0184257. doi: 10.1371/journal.pone.0184257
Park, I., Song, J. H., Yang, S., and Moon, B. C. (2020). Comparative analysis of Actaea chloroplast genomes and molecular marker development for the identification of authentic Cimicifugae Rhizoma. Plants 9:157. doi: 10.3390/plants9020157
Park, I., Yang, S., Kim, W. J., Noh, P., Lee, H. O., and Moon, B. C. (2018a). Authentication of herbal medicines Dipsacus asper and Phlomoides umbrosa using DNA barcodes, chloroplast genome, and sequence characterized amplified region (SCAR) Marker. Molecules 23:1748. doi: 10.3390/molecules23071748
Park, I., Yang, S., Kim, W. J., Noh, P., Lee, H. O., and Moon, B. C. (2018b). The complete chloroplast genomes of six Ipomoea species and indel marker development for the discrimination of authentic Pharbitidis Semen (Seeds of I. nil or I. purpurea). Front. Plant Sci. 9:965. doi: 10.3389/fpls.2018.00965
Park, I., Yang, S., Kim, W. J., Song, J. H., Lee, H. S., Lee, H. O., et al. (2019). Sequencing and comparative analysis of the chloroplast genome of Angelica polymorpha and the development of a novel indel marker for species identification. Molecules 24:1038. doi: 10.3390/molecules24061038
Parks, M., Cronn, R., and Liston, A. (2009). Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84. doi: 10.1186/1741-7007-7-84
Płachno, B. J., Stpiczyńska, M., Świa̧tek, P., Lambers, H., Cawthray, G. R., Nge, F. J., et al. (2019). Floral micromorphology and nectar composition of the early evolutionary lineage Utricularia (subgenus Polypompholyx, Lentibulariaceae). Protoplasma 256, 1531–1543. doi: 10.1007/s00709-019-01401-2
Raman, G., and Park, S. (2016). The complete chloroplast genome sequence of Ampelopsis: gene organization, comparative analysis, and phylogenetic relationships to other angiosperms. Front. Plant Sci. 7:341. doi: 10.3389/fpls.2016.00341
Raman, G., Park, S., Lee, E. M., and Park, S. (2019). Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Sci. Rep. 9:5028.
Rambaut, A. (2014). FigTree v 1.4. 2 Molecular Evolution, Phylogenetics and Epidemiology. Edinburgh: The University of Edinburgh.
Raubeson, L. A., Peery, R., Chumley, T. W., Dziubek, C., Fourcade, H. M., Boore, J. L., et al. (2007). Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174. doi: 10.1186/1471-2164-8-174
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sanchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Schroeder, H., Cronn, R., Yanbaev, Y., Jennings, T., Mader, M., Degen, B., et al. (2016). Development of molecular markers for determining continental origin of wood from white oaks (Quercus L. sect. Quercus). PLoS One 11:e0158221. doi: 10.1371/journal.pone.0158221
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Shi, H., Yang, M., Mo, C., Xie, W., Liu, C., Wu, B., et al. (2019). Complete chloroplast genomes of two Siraitia Merrill species: comparative analysis, positive selection and novel molecular marker development. PLoS One 14:e0226865. doi: 10.1371/journal.pone.0226865
Shukla, Y. N., Srivastava, A., and Singh, S. C. (2001). New naphthoquinones from Arnebia hispidissima roots. Planta Med. 67, 575–577. doi: 10.1055/s-2001-16470
Smidt, E. C., Paez, M. Z., Vieira, L. D. N., Viruel, J., De Baura, V. A., Balsanelli, E., et al. (2020). Characterization of sequence variability hotspots in Cranichideae plastomes (Orchidaceae, Orchidoideae). PLoS One 15:e0227991. doi: 10.1371/journal.pone.0227991
Song, J. H., Choi, G., Yang, S., and Moon, B. C. (2019). Morphological characteristics, distribution and taxonomic consideration of the Arnebia euchroma and A. guttata. Korea J. Herbol. 34, 63–70.
Song, J. H., Roh, H. S., and Hong, S. P. (2020). Petal micromorphology and its systematic implications in Rosaceae tribe Spiraeeae. Brittonia 72, 111–122. doi: 10.1007/s12228-020-09609-w
Song, Y., Zhang, Y., Xu, J., Li, W., and Li, M. (2019). Characterization of the complete chloroplast genome sequence of Dalbergia species and its phylogenetic implications. Sci. Rep. 9:20401.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sudd, J. H., and Sudd, M. E. (1985). Seasonal changes in the response of wood-ants (Formica lugubris) to sucrose baits. Ecol. Entomol. 10, 89–97. doi: 10.1111/j.1365-2311.1985.tb00538.x
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11.
Urgamal, M., Oyuntsetseg, B., Nyambayar, D., and Dulamsuren, C. (2014). Conspectus of the Vascular Plants of Mongolia. Ulaanbaatar: Admon Printing Press.
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8, 77–80. doi: 10.1016/s1672-0229(10)60008-3
Wang, Y., Zhan, D. F., Jia, X., Mei, W. L., Dai, H. F., Chen, X. T., et al. (2016). Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Front. Plant Sci. 7:280. doi: 10.3389/fpls.2016.00280
Weigend, M., Selvi, F., Thomas, D. C., and Hilger, H. H. (2016). “Boraginaceae,” in Flowering Plants. Eudicots, the Families and Genera of Vascular Plants 14, eds J. W. Kadereit and V. Bittrich (Cham: Springer International Publishing), 41–102.
Whitney, H. M., Bennett, K. V., Dorling, M., Sandbach, L., Prince, D., Chittka, L., et al. (2011). Why do so many petals have conical epidermal cells? Ann. Bot. 108, 609–616. doi: 10.1093/aob/mcr065
Whitney, H. M., Chittka, L., Bruce, T. J., and Glover, B. J. (2009). Conical epidermal cells allow bees to grip flowers and increase foraging efficiency. Curr. Biol. 19, 948–953. doi: 10.1016/j.cub.2009.04.051
Whitney, H. M., and Glover, B. J. (2007). Morphology and development of floral features recognised by pollinators. Arthropod Plant Int. 1, 147–158. doi: 10.1007/s11829-007-9014-3
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Muller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, F.-Y., Tang, C.-Y., Guo, Y.-M., Bian, Z.-W., Fu, J.-Y., Lu, G.-H., et al. (2017). Transcriptome analysis explores genes related to shikonin biosynthesis in Lithospermeae plants and provides insights into Boraginales’ evolutionary history. Sci. Rep. 7:4477.
Yao, X., Tan, Y.-H., Yang, J.-B., Wang, Y., Corlett, R. T., and Manen, J.-F. (2019). Exceptionally high rates of positive selection on the rbcL gene in the genus Ilex (Aquifoliaceae). BMC Evol. Biol. 19:192. doi: 10.1186/s12862-019-1521-1
Yi, S., Li, Y., and Wang, W. (2018). Selection shapes the patterns of codon usage in three closely related species of genus Misgurnus. Genomics 110, 134–142. doi: 10.1016/j.ygeno.2017.09.004
Zhitao, N., Shuying, Z., Jiajia, P., Ludan, L., Jing, S., and Xiaoyu, D. (2017). Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 7:2073.
Zhu, G., Riedl, H., and Kamelin, R. V. (1995). “Boraginaceae,” in Flora of China 16: Gentianaceae through Boraginaceae, eds Z.-Y. Wu and P. H. Raven (St. Louis, MO: Missouri Botanical Garden), 329–427.
Keywords: Boraginaceae, micromorphology, plastid genome, phylogenetic analysis, molecular marker
Citation: Park I, Yang S, Song J-H and Moon BC (2020) Dissection for Floral Micromorphology and Plastid Genome of Valuable Medicinal Borages Arnebia and Lithospermum (Boraginaceae). Front. Plant Sci. 11:606463. doi: 10.3389/fpls.2020.606463
Received: 15 September 2020; Accepted: 16 November 2020;
Published: 04 December 2020.
Edited by:
Petr Koutecký, University of South Bohemia, CzechiaReviewed by:
James Cohen, Weber State University, United StatesCopyright © 2020 Park, Yang, Song and Moon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Ho Song, c29uZ2poQGtpb20ucmUua3I=; Byeong Cheol Moon, YmNtb29uQGtpb20ucmUua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.