
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 January 2021
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.603178
This article is part of the Research Topic Photosynthesis Under Fluctuating Light View all 15 articles
 Yun Zhao1,2
Yun Zhao1,2 Ting Min2
Ting Min2 Miaojin Chen3
Miaojin Chen3 Hongxun Wang4
Hongxun Wang4 Changqing Zhu1
Changqing Zhu1 Rong Jin1
Rong Jin1 Andrew C. Allan5,6
Andrew C. Allan5,6 Kui Lin-Wang5
Kui Lin-Wang5 Changjie Xu1*
Changjie Xu1*Red coloration contributes to fruit quality and is determined by anthocyanin content in peach (Prunus persica). Our previous study illustrated that anthocyanin accumulation is strongly regulated by light, and the effect of induction differs according to light quality. Here we showed that both ultraviolet-A (UVA) and ultraviolet-B (UVB) irradiation promoted anthocyanin biosynthesis in “Hujingmilu” peach fruit, and a combination of UVA and UVB had additional effects. The expression of anthocyanin biosynthesis and light signaling related genes, including transcription factor genes and light signaling elements, were induced following UV irradiation as early as 6 h post-treatment, earlier than apparent change in coloration which occurred at 72 h. To investigate the molecular mechanisms for UVA- and UVB-induced anthocyanin accumulation, the genes encoding ELONGATED HYPOCOTYL 5 (HY5), CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), Cryptochrome (CRY), and UV RESISTANCE LOCUS 8 (UVR8) in peach were isolated and characterized through functional complementation in corresponding Arabidopsis (Arabidopsis thaliana) mutants. PpHY5 and PpCOP1.1 restored hypocotyl length and anthocyanin content in Arabidopsis mutants under white light; while PpCRY1 and PpUVR8.1 restored AtHY5 expression in Arabidopsis mutants in response to UV irradiation. Arabidopsis PpHY5/hy5 transgenic lines accumulated higher amounts of anthocyanin under UV supplementation (compared with weak white light only), especially when UVA and UVB were applied together. These data indicated that PpHY5, acting as AtHY5 counterpart, was a vital regulator in UVA and UVB signaling pathway. In peach, the expression of PpHY5 was up-regulated by UVA and UVB, and PpHY5 positively regulated both its own transcription by interacting with an E-box in its own promoter, and the transcription of the downstream anthocyanin biosynthetic genes chalcone synthase 1 (PpCHS1), chalcone synthase 2 (PpCHS2), and dihydroflavonol 4-reductase (PpDFR1) as well as the transcription factor gene PpMYB10.1. In summary, functional evidence supports the role of PpHY5 in UVA and UVB light transduction pathway controlling anthocyanin biosynthesis. In peach this is via up-regulation of expression of genes encoding biosynthetic enzymes, as well as the transcription factor PpMYB10.1 and PpHY5 itself.
Anthocyanins, a group of secondary metabolites known as flavonoid compounds, are important water-soluble pigments widely distributed in plants where they perform a multitude of biological functions such as protecting against a variety of abiotic stresses [ultraviolet (UV) radiation, nutrient deficiency, low temperature, drought, etc.], attracting pollinators and seed dispersers, and defense against pathogens and herbivores (Schaefer et al., 2004; Gould et al., 2010). In addition, they are also recognized to have potential human health benefits in aspects such as prevention from cancer and heart disease (Tsuda, 2012; Zhang et al., 2015). Hence, anthocyanin is considered as an essential determinant for fruit quality.
The anthocyanin biosynthetic pathway has been extensively studied and found to be relatively conserved among plant species (Winkel-Shirley, 2001). Anthocyanins are biosynthesized via the phenylpropanoid pathway, which is catalyzed stepwise by a series of enzymes including phenylalanine ammonia lyase (PAL), cinnamic acid-4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), flavonoid 3’,5’-hydroxylase (F3’5’H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT; Jaakola, 2013; Zhang et al., 2014). The expression of the genes that encode these enzymes are transcriptionally regulated by MYB, basic Helix-Loop-Helix (bHLH), and WD40 repeat proteins, often in a form of trimeric protein complex of MYB-bHLH-WD40 (the MBW complex; Lin-Wang et al., 2010; BenSimhon et al., 2011; An et al., 2012; Chagné et al., 2013; Wang N. et al., 2018). In peach (Prunus persica), several relevant transcription factors (PpMYB10s, PpbHLH3, and PpWD40-1) have been functionally confirmed to control anthocyanin biosynthesis (Ravaglia et al., 2013; Rahim et al., 2014; Uematsu et al., 2014; Zhou et al., 2014, 2018; Tuan et al., 2015).
Anthocyanin biosynthesis is not only developmentally regulated but also influenced by various environmental and endogenous stimuli as well (Guo et al., 2008). As the primary energy source and an informational signal for regulating plant growth and development, light is one of the most critical environmental factors (Kami et al., 2010). Light profoundly influences complex signaling pathways such as photomorphogenesis, circadian rhythms and the regulation of secondary metabolism throughout the life of plants (Gelderen et al., 2018).
Previous research has revealed the significance of light on the induction of anthocyanin biosynthesis in fruits such as bilberry (Vaccinium myrtillus, Uleberg et al., 2012), Chinese bayberry (Myrica rubra, Niu et al., 2010), grapes (Vitis vinifera, Azuma et al., 2012), raspberry (Rubus idaeus, Wang et al., 2009) and cranberry (Vaccinium macrocarpon, Zhou and Singh, 2004), etc. The effects of light on anthocyanin accumulation vary with different light wavelengths, where UV light always presents a more profound effect on anthocyanin biosynthesis compared with white light (Zoratti et al., 2014). For instance, ultraviolet-B (UVB) irradiation significantly increased anthocyanin content in the peel of apple (Malus domestica) and pear (Pyrus pyrifolia) fruits (Hagen et al., 2007; Qian et al., 2013; Sun et al., 2014; Henry-Kirk et al., 2018). Tomato (Solanum lycopersicum) grown under ultraviolet-A (UVA) supplementation visibly accumulated more anthocyanin in a short time (Guo and Wang, 2010; Castagna et al., 2014). In strawberries (Fragaria ananassa), blue light and UVC irradiation stimulated anthocyanin accumulation and enhanced the antioxidant activities (Li et al., 2014; Xu et al., 2014). In peach, it has also been demonstrated that UV lights are stronger stimulants for anthocyanin accumulation (Zhao et al., 2017).
Higher plants have evolved extremely advanced systems to sense changes in ambient light conditions (Casal, 2013), by being equipped with specialized sensory photoreceptors for specific wavelengths of light, including red/far-red light receptor phytochromes (PHYA to PHYE) (Oh and Montgomery, 2017), blue/UVA light receptor cryptochromes (CRY1 and CRY2) or phototropins (PHOTs; Lin, 2002; Liu et al., 2011; Wang Q. et al., 2018), and UVB light receptor UV RESISTANCE LOCUS 8 (UVR8; Rizzini et al., 2011; Yin and Ulm, 2017; Liang et al., 2019). Photoreceptors are indispensable for light-induced anthocyanin biosynthesis (Yang et al., 2016), as in Arabidopsis (Arabidopsis thaliana), the photoreceptor deficient mutants uvr8 and cry1cry2 respectively have a weak response in UVB and blue light/UVA-induced photomorphogenesis like blocking the inhibition of hypocotyl elongation and the induction of anthocyanin biosynthesis (Wade et al., 2001; Morales et al., 2013; Wang Q. et al., 2018).
After perceiving light, this signal is transmitted from photoreceptors to downstream light signal transduction factors to regulate the process of light-induced anthocyanin biosynthesis both transcriptionally and post-translationally (Casal, 2013). ELONGATED HYPOCOTYL 5 (HY5), a basic leucine zipper (bZIP) transcription factor, is a master regulator in light signal transduction pathway that acts downstream of multiple photoreceptors to respond to photomorphogenesis (Holm et al., 2002; Gangappa and Botto, 2016). HY5 is considered as a positive regulator to participate in light-induced anthocyanin biosynthesis in Arabidopsis, apple, grape, and pear (Loyola et al., 2016; An et al., 2017; Nawkar et al., 2017; Tao et al., 2018). AtHY5 targets the anthocyanin biosynthetic genes (CHS, F3H) and MYB [PRODUCTION OF FLAVONOL GLYCO-SIDES 1 (PFG1)/MYB12, MYB75/PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1)] to activate their expression by direct binding to the promoters (Chattopadhyay et al., 1998; Lee et al., 2007; Stracke et al., 2010; Shin et al., 2013). In eggplant (Solanum melongena) under blue light, SmHY5 has also been revealed to accelerate anthocyanin accumulation by regulating the expression of downstream genes encoding enzymes of the anthocyanin biosynthetic pathway (Jiang et al., 2016). In apple, MdHY5 promoted the expression of MdMYB10 to regulate anthocyanin accumulation (An et al., 2017). CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), an E3 ubiquitin ligase, is a crucial repressor for photomorphogenesis and anthocyanin biosynthesis, targeting HY5 for proteasome-mediated degradation (Osterlund et al., 2000; Lau and Deng, 2012). In pear, PpyCOP1 indirectly interacted with PpyHY5 to mediate regulation of anthocyanin accumulation (Tao et al., 2018). Additionally, apple MdCOP1 was also found to be critical for the ubiquitination and degradation of MdMYB1, a positive regulator for anthocyanin biosynthesis, further leading to the negative regulation for the coloration in apple fruit (Li et al., 2012).
Anthocyanin accumulation is essential for the quality of peach fruit. Our previous study revealed that UV light enhances anthocyanin content in peach peel (Zhao et al., 2017). However, how photoreceptors and light signal transduction elements mediate UV light-induced anthocyanin accumulation has not been elucidated. In this study, the effect of UVA and UVB irradiation on the coloration of peach fruit was examined. We cloned and functionally characterized PpHY5 and provided molecular evidence that PpHY5 plays a vital role in anthocyanin biosynthesis in response to UVA and UVB. These results contributed to our understanding of the mechanism underlying UV light-induced anthocyanin biosynthesis in peach, as well as providing evidence for improving fruit pigmentation.
The peach cultivar (cv) “Hujingmilu” was used in this study. All fruit were bagged with commercial yellow paper bags at 42 days after full blossom and collected at just before turning stage. For UV irradiation treatment experiments, all collected fruit were transferred to a growth chamber at a constant 20°C and irradiated with continuous UVA (320–400 nm, 1,000 μw/cm2) or UVB (280–320 nm, 58 μw/cm2) or both together for 0, 6, 12, 24, 48, 72, and 144 h, respectively. The fruit kept in the dark served as the control. Peel tissue from 30 fruit of each treatment were separated simultaneously into three biological replicates (10 fruit for each replicate) at every sampling time, then immediately frozen in liquid nitrogen and stored at −80°C for subsequent experiments.
The Arabidopsis hy5 (SALK_056405), cop1 (SALK_022133), uvr8 (SALK_033468), and cry1 (SALK_069292) mutants with the Columbia genetic background were obtained from The Arabidopsis Information Resource (TAIR). For hypocotyl length and anthocyanin accumulation assays, seeds of wild type (WT), above-mentioned mutants as well as transgenic lines obtained in this study were sterilized and germinated on Murashige and Skoog plates supplemented with 0.6% agar and 1% sucrose, and subjected to a chilling treatment at 4°C for 2 days in the dark. Then these seedlings were transferred to a growth chamber at 24°C under the long-day condition (16 h photoperiod/8 h dark cycle) of white light in normal intensity (40 w/m2) for growth. For UV light treatment experiment, seedlings were grown under continuous weak white light (WWL, 40 μw/cm2) supplemented with UVA (40 μw/cm2) or UVB (10 μw/cm2) or both together at 24°C. And the seedlings kept under only WWL served as the control. After growth under different light conditions as indicated for 5 or 7 days, respectively, the hypocotyl length was measured and seedlings were sampled for subsequent experiments.
Fruit surface color measurement was carried out using a reflectance spectrophotometer (Hunter Lab Mini Scan XE Plus colorimeter) at four evenly distributed equatorial positions of per fruit. The raw data as L∗ (lightness, from black to white), a∗ (the degree of green-red variation), b∗ (the degree of blue-yellow variation) values were recorded and the color index of red grapes (CIRG) was calculated according to the formula CIRG = (180 − H)/(L∗ + C), while H = arctan (b∗/a∗) and C = [(a∗)2+(b∗)2]0.5 (Carreño et al., 1995; Zhang et al., 2008).
Anthocyanin was extracted from peach fruit peel and Arabidopsis seedlings by using the method as described in our previous study (Zhao et al., 2017). High-performance liquid chromatography (HPLC) analysis for quantification of anthocyanin was performed using an Agilent 1260 liquid chromatograph system (Agilent Technologies, CA, United States) equipped with a ZORBAX SB-C18 column following a previously reported protocol (Cheng et al., 2014). The UV-visible light detector wavelength was set at 520 nm. Cyanidin 3-O-glucoside chloride was employed as the authentic standard.
Total RNA was isolated from peach tissue samples and Arabidopsis by using a modified cetyltrimethylammonium bromide (CTAB) method (Liu et al., 2015) and the TRIzol Reagent Kit (Ambion, Hopkinton, MA, United States), respectively. First-strand cDNA was synthesized with HiScript® II Q Select RT SuperMix (Vazyme) after removal of genomic DNA contamination by TURBO DNase (Ambion) following the manufacturer’s protocol. Reverse transcription quantitative PCR (RT-qPCR) was performed using the SsoFast EvaGreen Supermix kit (Bio-Rad, CA, United States) with the CFX96 instrument (Bio-Rad, CA, United States) according to the following program: 95°C for 3 min, followed by 45 cycles of 95°C for 10 s, 60°C for 30 s and then 95°C for 10 s followed by a continuous increase from 65°C to 95°C at the heating rate of 0.5°C/s for dissociation curve analysis (Zhao et al., 2017). The PpTEF2 (GenBank accession: No. JQ732180) and ATACT2 (GenBank accession: No. AT3G18780) genes were used as the internal reference genes to normalize expression values for peach and Arabidopsis, respectively (Tong et al., 2009; Zhou et al., 2013). The primer sequences for RT-qPCR were designed with Primer Premier 5 and listed in Supplementary Table 1.
RNA sequencing (RNA-Seq) of peach fruit samples (irradiated with different UV-light conditions for 0, 6, 12, 24, and 48 h) was conducted using the Xten platform by staff at Biomarker (Beijing, China) with three biological replicates for each sample. Data analysis was conducted as described in our previous work (Zhao et al., 2017). Transcript abundances were normalized by calculating FPKM (expected number of Fragments Per Kilobase of transcript sequence per Million base pairs sequenced).
The full-length coding sequence of PpHY5 (Prupe.1G478400), PpHYH (Prupe.1G208500), PpCOP1.1 (Prupe.5G031300), PpUVR8.1 (Prupe.4G277200), PpCRY1 (Prupe.1G517600) was isolated and recombined into the pGreenII 0029 62-SK vector (PpHY5-SK, PpHYH-SK, PpCOP1.1-SK, PpUVR8.1-SK, PpCRY1-SK). The promoter sequence (1500 bp upstream of the initiation codon) of structural genes (PpCHS1, PpCHS2, PpCHI, PpF3H, PpF3’H, PpDFR, PpANS, and PpUFGT) and transcription factors (PpMYB10.1/2/3) were constructed into the pGreen II 0800-LUC vector (PpCHS1-LUC, PpCHS2-LUC, PpCHI-LUC, PpF3H-LUC, PpF3’H-LUC, PpDFR-LUC, PpANS-LUC, PpUFGT-LUC, and PpMYB10.1/2/3-LUC). In experiments for verifying the function of E-box motifs in PpHY5 promoter, mutations on the PpHY5 promoter were conducted using the Fast Mutagenesis System Kit (Transgene) and inserted into the pGreenII 0800-LUC vector, named PpHY5m1-LUC, PpHY5m2-LUC, PpHY5m3-LUC, and PpHY5m4-LUC, respectively. For subcellular localization analysis, the coding sequence of each gene without stop codon was fused with green fluorescent protein (GFP) and cloned into the pCAMBIA super 1300-eGFP vector (PpHY5-GFP, PpCOP1.1-GFP). All constructs were transformed into Agrobacterium tumefaciens strain GV3101 (MP90) by electroporation with GenePulser XcellTM Electroporation Systems (Bio-Rad, Hercules, CA, United States) for subsequent experiments. Primers used for plasmid construction were described as listed in Supplementary Table 2.
The dual-luciferase assay was performed with Nicotiana benthamiana leaves by using a previously reported protocol (Zhao et al., 2020). Agrobacterium cultures containing recombinant pGreenII 0029 62-SK vectors harboring transcription factor genes and the cultures containing recombinant pGreen II 0800-LUC vector harboring target gene promoters were infiltrated into N. benthamiana leaves with a ratio of 10:1 (v/v). At three days following infiltration, the firefly luciferase (LUC) and renilla luciferase (REN) activities were measured using dual-Luciferase Reporter Assay System (Promega, MI, United States) with Modulus Luminometer. When the ratio of LUC/REN was over two and the difference was highly significant (P < 0.01) as compared with the control, the transcription factor was considered to be able to activate the promoter. For each transcription factor and promoter interaction, dual-luciferase assays were conducted with at least three independent experiments (six replicate reactions in each experiment).
The Arabidopsis transformation was performed by using the floral dip method (Zhang et al., 2006). SilwetL-77 (Real-Times, Beijing, China) was applied as a surfactant in Agrobacterium-based transformation of Arabidopsis. Transgenic seeds were sterilized and placed on Murashige and Skoog medium containing 50 mg/mL kanamycin for transgenic plant selection. For each target gene, at least three lines were characterized and three of them with highest expression levels of target gene were selected, and individuals of the T2 were used for further phenotype analysis. After growth for 5 or 7 days, the hypocotyl length measurement of at least 30 Arabidopsis seedlings was conducted for each replicate. All experiments were carried out with three independent biological replicates.
The Agrobacterium strain carrying the 35S::PpHY5-GFP or 35S::PpCOP1.1-GFP recombinant vector was infiltrated into transgenic N. benthamiana plant with a red fluorescent nuclear marker (Nucleus-RFP) according to a previous report (Zhao et al., 2020), while 35S::GFP was used as a negative control. The GFP florescence of the transiently expressed leave was observed under a Zeiss LSM710NLO confocal laser scanning microscope.
The predictions of conserved cis-regulatory elements in the promoter were analyzed using the PlantCARE database.1
The statistical significance of differences was analyzed using SPSS statistical software (SPSS 19.0, SPSS Inc., Chicago, IL, United States) with Student’s t-test. Graphs were plotted using Origin 8.0 (Origin Lab Corporation, Northampton, MA, United States).
To investigate the influence of different UV light treatments on anthocyanin biosynthesis in peach, the fruit were given either a continuous UVA or UVB or UVA + UVB treatment. The control fruit was kept in the dark and did not develop red color over time. Under UV irradiation, the peel color remained green within 24 h and a few red spots began to be slightly visible at 48 h. At 72 and 144 h following irradiation, the fruit turned deep red (Figure 1A). These observations matched well with the CIRG values (Figure 1B). Under dark condition, the concentration of anthocyanin in peel was very low and showed no significant increase over time (Figure 1B). In contrast, anthocyanin accumulated following irradiation with UVA or UVB or UVA + UVB, and the effects varied significantly among different UV lights conditions. The concentration of anthocyanin rose sharply to 3.12 mg/100gFW (15-fold higher than the control) under UVA exposure and 2.11 mg/100gFW (10-fold higher than the control) under UVB exposure at 144 h respectively. During the irradiation period, the highest anthocyanin content was detected in fruit under UVA + UVB treatment, reaching 3.47 mg/100gFW at 72 h (2–3 fold higher than UVA or UVB) and 11.59 mg/100gFW at 144 h (3-6 fold higher than UVA or UVB), respectively (Figure 1B), which was consistent with the visual appearance.
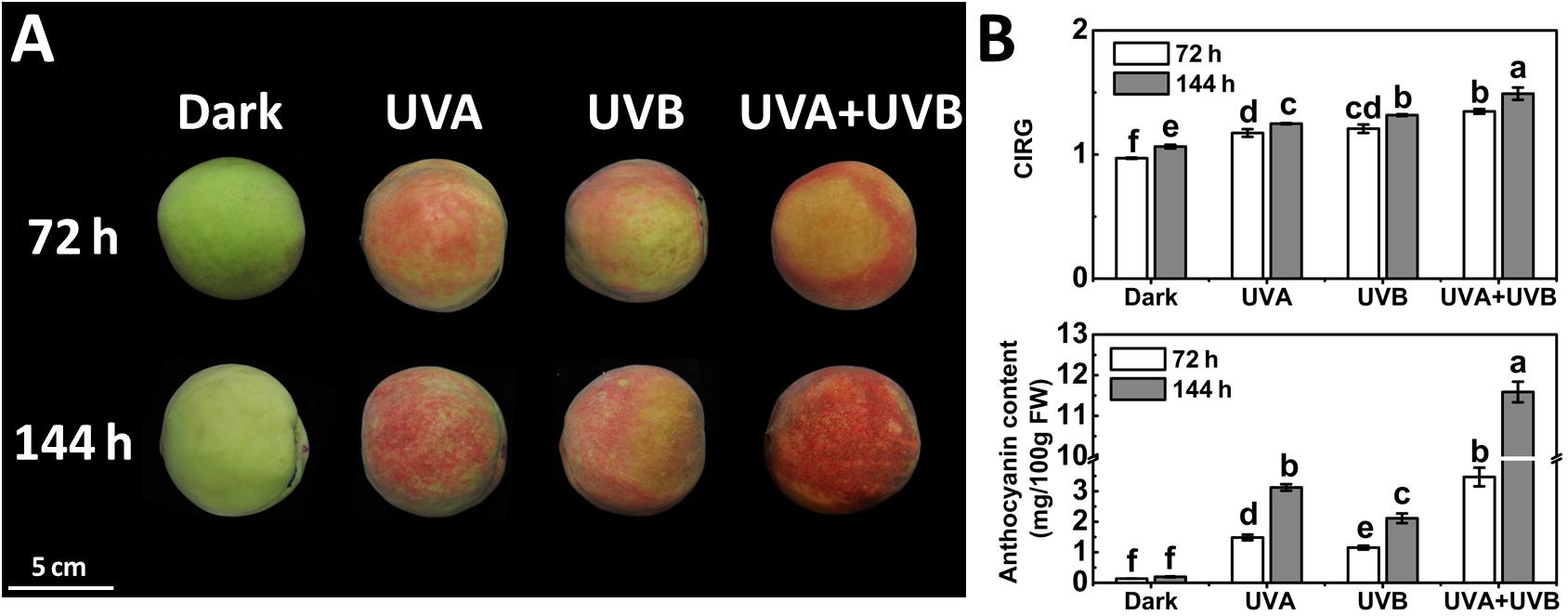
Figure 1. Coloration (A) and the color index of red grapes (CIRG) values and anthocyanin content (B) in the peach peel after continuous UV irradiation or kept in dark. UVA, 1,000 μw/cm2; UVB, 58 μw/cm2. Scale bar indicated 5 cm. Data were presented as means (±SE) from three independent biological replicates. Values labeled with different letters indicated a significant difference at P < 0.05.
In order to further explore the molecular mechanisms for anthocyanin accumulation in response to UV lights, we analyzed the transcript levels of genes encoding anthocyanin biosynthetic enzymes (PpPAL, Pp4CL, PpCHS1, PpCHS2, PpCHI, PpF3H, PpDFR1, PpDFR2, PpANS, and PpUFGT), transport genes PpGST1 and regulatory genes (PpMYB10.1, PpMYB10.2, and PpMYB10.3) in peel during the early irradiation period (0, 6, 12, 24, and 48 h) before the appearance of red coloration. Under dark condition, the assayed genes exhibited low expression levels and for some genes such as PpCHS1, PpDFR1, and PpUFGT, the transcript levels showed little change over time (Figure 2). In comparison to the low expression levels at the initial point (0 h), the expression profiles of the biosynthetic genes were up-regulated to various degrees in peel of fruit when exposed to the UV treatments. The transcripts of all assayed biosynthetic genes under different UV light conditions were elevated than control (Figure 2). After exposure to UV light for 6 h, the expression levels of all analyzed biosynthetic genes sharply increased (Figure 2). For example, the transcript levels of PpCHS1 and PpUFGT in the UV light-exposed fruit were at least 6-fold and 9-fold higher than the control. However, after exposure to UV light for 12 h, the transcript levels of PpCHI, PpF3H, PpANS, and PpGST1 decreased slightly. Pp4CL and PpCHS1 showed the highest expression after 24 h of UV light irradiation. Although there was a slight decline in the transcript abundance of some genes after irradiation for 48 h, the expression was consistently higher than the control, for instance, the transcript levels of PpDFR1 were at least 7-fold higher, 14-fold higher and 28-fold higher in the UV light-exposed fruit than that in the dark, respectively. In particular, the expression of PpGST1 continuously remained very low in the fruit under dark condition, while was strongly stimulated by UV light, showing a 43-fold, 155-fold, and 332-fold elevation at 48 h in UVA, UVB, and UVA + UVB light exposed fruit, respectively.
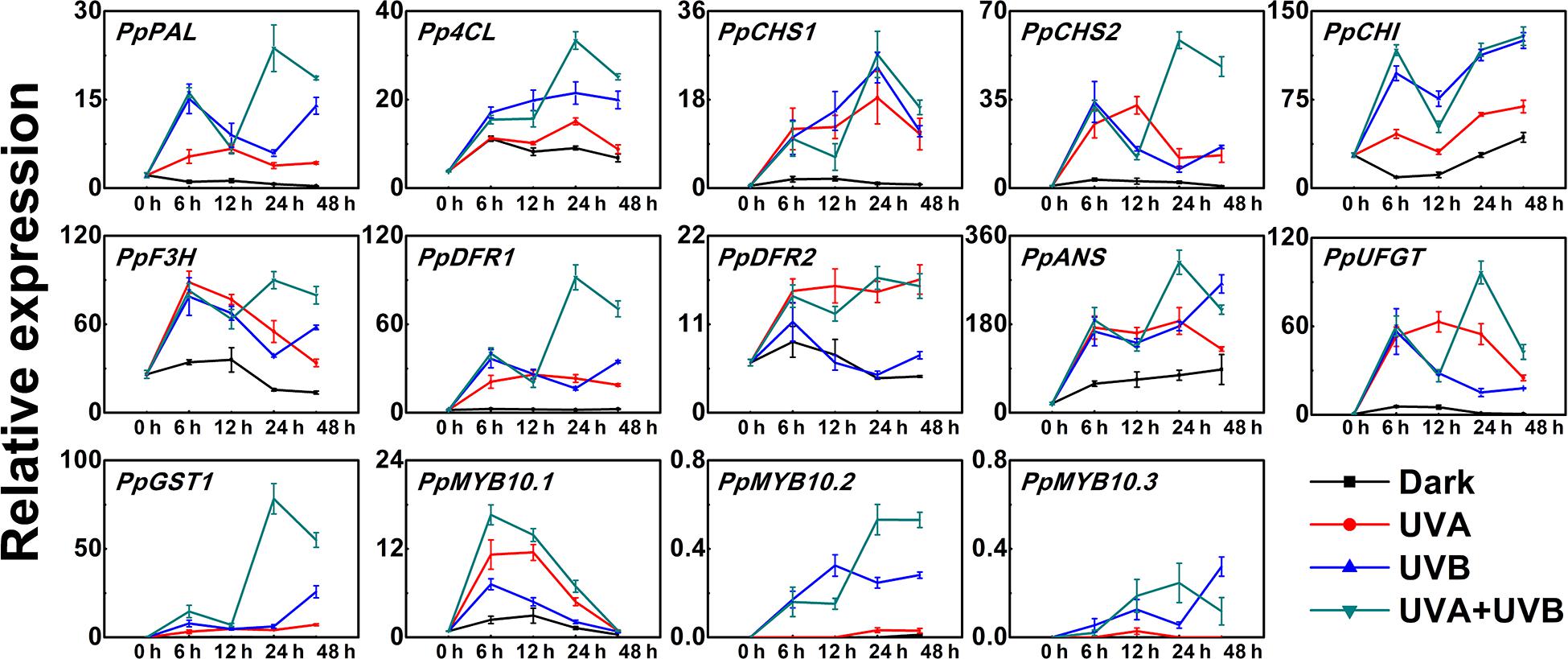
Figure 2. Time course of the transcript levels of anthocyanin biosynthesis related genes in peach peel in response to continuous UV lights. UVA, 1,000 μw/cm2; UVB, 58 μw/cm2. Data were presented as means (±SE) from three independent biological replicates.
The expression patterns of the regulatory transcription factor genes (PpMYB10.1, PpMYB10.2, and PpMYB10.3) showed elevation by UV, but with different profiles. The transcript level of PpMYB10.1 was strongly induced by UV light and then decreased after prolonged exposure. The expression of PpMYB10.2 and PpMYB10.3 presented little change under UVA irradiation and transcript abundance was lower compared with PpMYB10.1 (Figure 2). The transcript levels of most genes were generally highest in peel of fruit irradiated with UVA + UVB, followed by UVA and UVB, while the expression were almost undetectable or showed low levels in control. Even though there were slight declines in transcription level at some time points, most genes reached the maximum transcription levels when treated with UVA + UVB for 24 h, which ensured the high transcription abundance detected in fruit at later stages. The results suggested that the coloration of the peel was able to be induced by the irradiation of UVA or UVB, and there was a synergistic effect of UVA and UVB in stimulating anthocyanin biosynthesis.
In Arabidopsis, it has been demonstrated that HY5 and COP1 play vital roles in light signal transduction pathway (Lau and Deng, 2012; Gangappa and Botto, 2016). Previously, we identified potential homologous genes in peach (named PpHY5, PpHYH, PpCOP1.1, and PpCOP1.2) (Zhao et al., 2017). To clarify their functions in UV light induced anthocyanin biosynthesis in peach fruit, the expression levels following UV light exposure were monitored.
PpHY5 expression exhibited a strong response to UV light irradiation (Figure 3). The transcript level of PpHY5 was low at the beginning (0 h); with UV there was a sharp increase which peaked within the first 6 h, and showed the highest accumulation level under UVA + UVB (about 4-fold higher than the control); from this peak there was a gradual decrease but expression remained higher than control (Figure 3). In Arabidopsis, HY5 HOMOLOG (HYH) has multiple overlapping functions with HY5 (Holm et al., 2002). In our study, the transcript level of PpHYH was similar to those of PpHY5, which increased at the beginning of irradiation but eventually decreased with continuous UV light (Figure 3). By contrast, the expression of PpCOP1.1 decreased gradually following irradiation with UVA or UVB or UVA + UVB while slightly increased in the control (Figure 3). However, PpCOP1.2 remained at quite low levels of expression with no obvious variation during the entire irradiation period among all different treatments (Figure 3). Overall, expression of PpHY5 and PpHYH increased following UV treatment while that of PpCOP1.1 was reduced. The transcript levels of these genes also responded quickly, within 6 h after the start of the treatment.
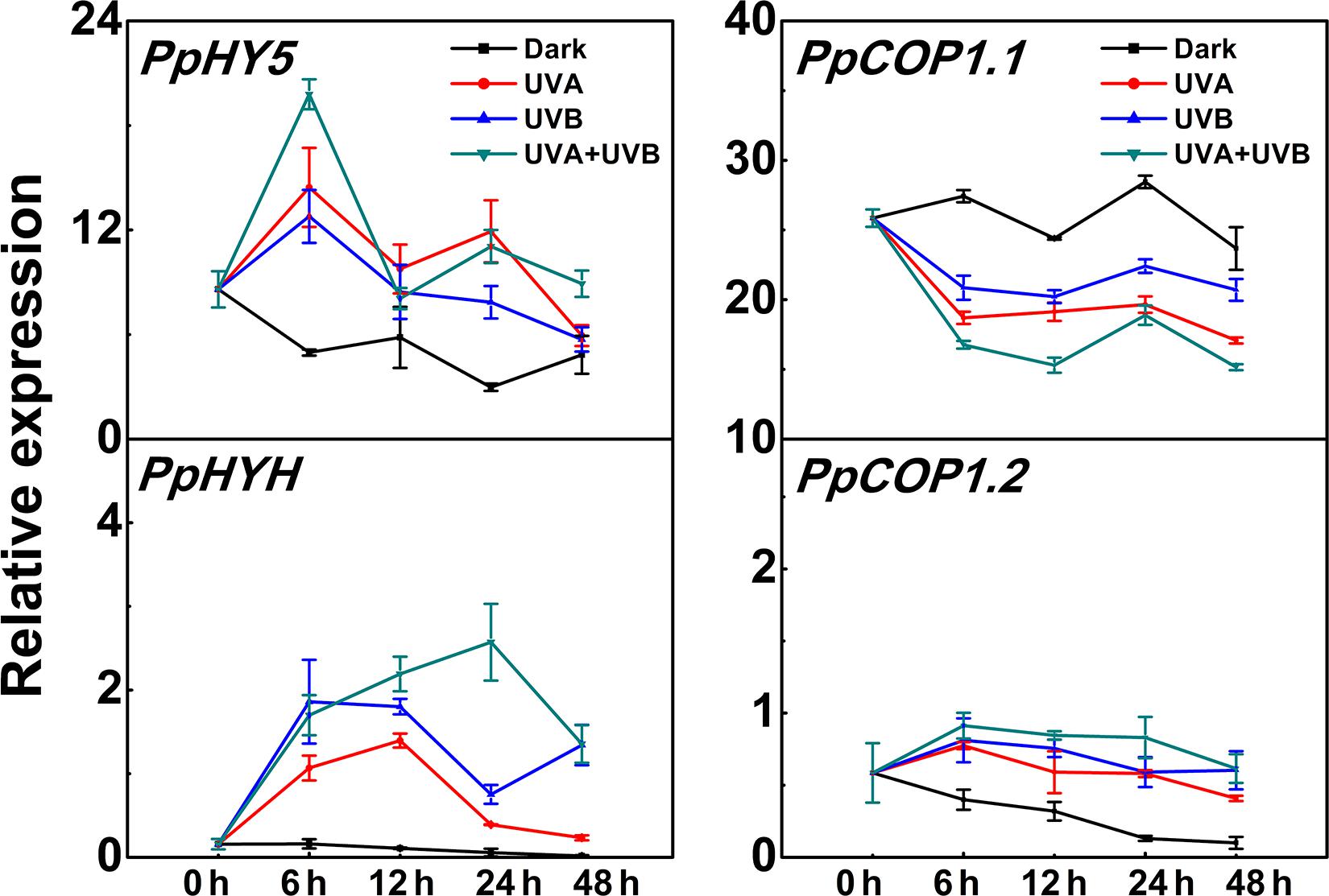
Figure 3. Time course of the transcript levels of PpHY5, PpHYH and PpCOP1.1, PpCOP1.2 in peach peel in response to continuous UV lights. UVA, 1,000 μw/cm2; UVB, 58 μw/cm2. Data were presented as means (±SE) from three independent biological replicates.
To confirm the roles of PpHY5 and PpCOP1.1 as light signal transduction related genes, we conducted the functional complementation assays with the Arabidopsis hy5 mutant and cop1 mutant transformed with PpHY5 and PpCOP1.1, respectively. At least three independent transgenic lines of each background were generated and confirmed by RT-qPCR. The transgenic lines (PpHY5/hy5, PpCOP1.1/cop1) with high expression of PpHY5 or PpCOP1.1 were chosen to compare with WT and hy5 or cop1 mutant by measuring hypocotyl length and anthocyanin content under all conditions in 5-day-old seedlings.
For plants grown under white light for five days, hy5 exhibited the longest hypocotyl phenotype (11.47 mm), while cop1 exhibited the shortest hypocotyl phenotype (3.55 mm) (Figure 4A). The constitutive overexpression of PpHY5 or PpCOP1.1 in corresponding mutant background restored the hypocotyl length under white light (Figure 4A). The concentrations of anthocyanin in all lines also showed difference to some degrees (Figure 4B). Compared to WT, cop1 displayed significantly higher accumulation of anthocyanin, and a lower level was observed in hy5. The anthocyanin levels were restored and enhanced in PpHY5/hy5 transgenic lines, reaching a level that was about 4.1-fold higher than that in hy5 mutant background (Figure 4B). In comparison, the anthocyanin accumulation was inhibited in the PpCOP1.1-complemented lines and decreased by 32% than those in cop1 mutant background (Figure 4B). Likewise, the genes encoding enzymes of the anthocyanin biosynthetic pathway (PAL, CHS, CHI, F3H, DFR, ANS, and UFGT) exhibited similar transcript accumulation trends consistent with anthocyanin content (Figure 4C). All these structural genes were generally lowly expressed in hy5 and highly expressed in cop1 relative to WT. For instance, the expression levels of ANS and UFGT decreased significantly in hy5 and were about 50 and 75% lower, whereas at least 5-fold and 4-fold higher in cop1 than that measured in WT. No significant differences in transcript levels of anthocyanin biosynthetic structural genes were found between PpHY5/hy5 or PpCOP1.1/cop1 transgenic lines and WT.
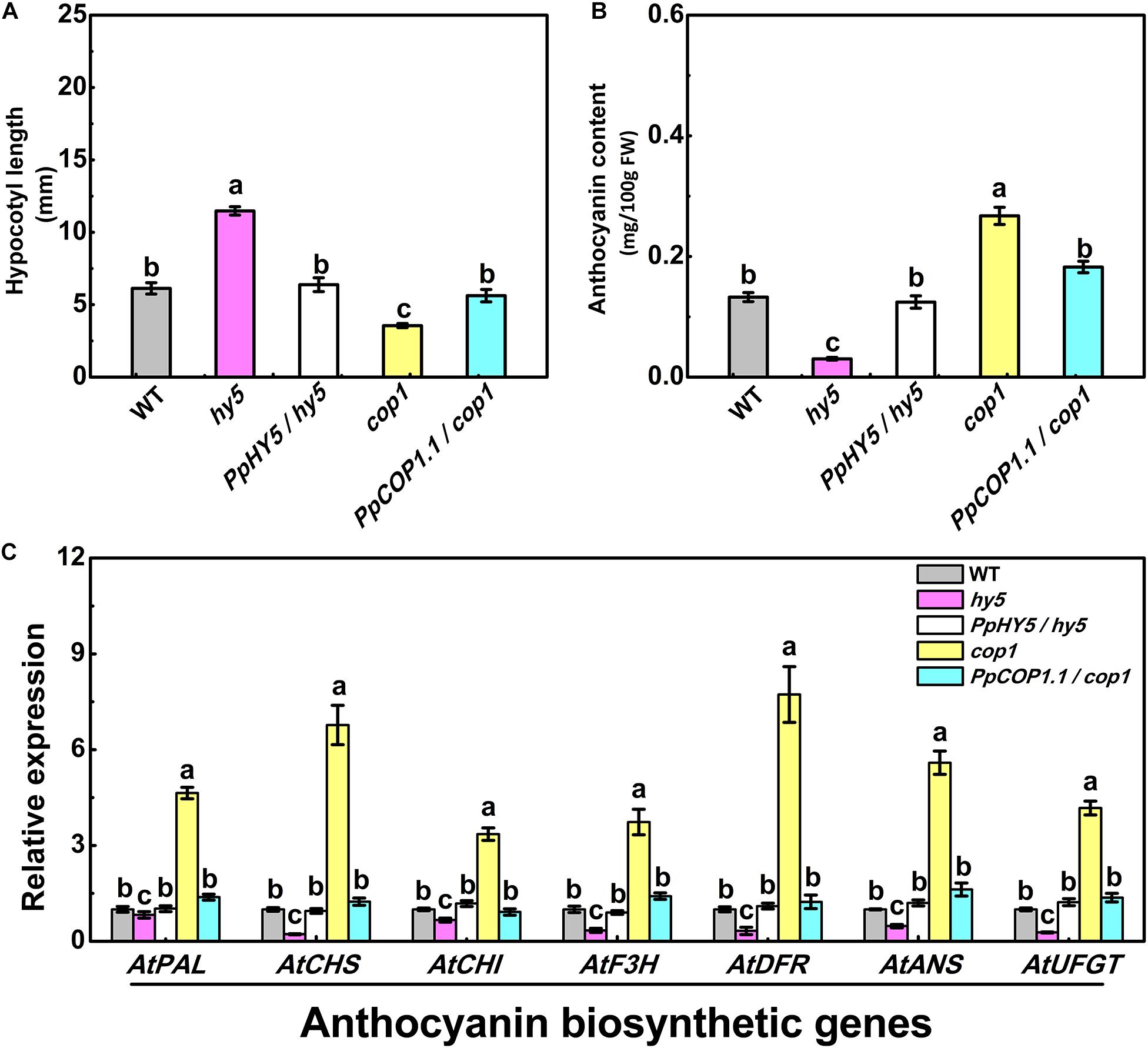
Figure 4. Hypocotyl length (A), anthocyanin content (B) and the relative expression levels of anthocyanin biosynthesis related genes (C) in 5-day-old seedlings of wild type (WT), mutant (hy5, cop1) and transgenic plants (PpHY5/hy5, PpCOP1.1/cop1) grown under long-day condition (16 h 40 w/m2 white light/8 h dark cycle). For Arabidopsis transgenic plants, three independent lines for each target gene were used. Data were presented as means (±SE) from three independent biological replicates with 30 or more Arabidopsis seedlings collected for each replicate. Values labeled with different letters indicated a significant difference at P < 0.05.
To visualize the subcellular localization of PpHY5 and PpCOP1.1, we performed the confocal microscopy assays of N. benthamiana leaves transiently transformed with PpHY5-GFP and PpCOP1.1-GFP. Our results showed that both PpHY5-GFP and PpCOP1.1-GFP displayed signal in the nucleus under white light (Supplementary Figure 1). This was in consistent with the subcellular localization of AtHY5 and AtCOP1 (Hardtke et al., 2000; Osterlund et al., 2000; Lau and Deng, 2012). These data suggested that PpHY5 and PpCOP1.1 were the functional orthologous of AtHY5 and AtCOP1 and were able to participate in white-light-related photomorphogenesis.
In Arabidopsis, AtHY5 is a key factor in photomorphogenic responses and acts as a downstream signaling intermediate of AtCRY1 and AtUVR8 (Binkert et al., 2014; Marzi et al., 2020). To determine whether the peach counterpart of these genes function in the same signaling cascade in Arabidopsis, we generated the PpHY5/hy5, PpCRY1/cry1, and PpUVR8.1/uvr8 transgenic lines. Five-day-old seedlings of WT, cry1 mutant and PpCRY1/cry1 transgenic lines were exposed to WWL + UVA for periods of 0–24 h and the expression of AtHY5 was examined. Upon WWL + UVA exposure, the transcript levels of AtHY5 were up-regulated and reached the maximum at 4 h (the upregulation ratio of PpCRY1/cry1 was 13-fold, for cry1 mutant it was 3-fold, for WT it was 16-fold), then a decreased occurred with the increment of irradiation time (Figure 5A). In comparison, AtHY5 in PpCRY1-overexpression seedling in cry1 background exhibited a similar transcript accumulation pattern as WT which was consistently higher than cry1 mutant. With complementation of the cry1 mutant, it can be concluded that PpCRY1 was a functional counterpart of AtCRY1 and could regulate AtHY5 expression in response to UVA irradiation.
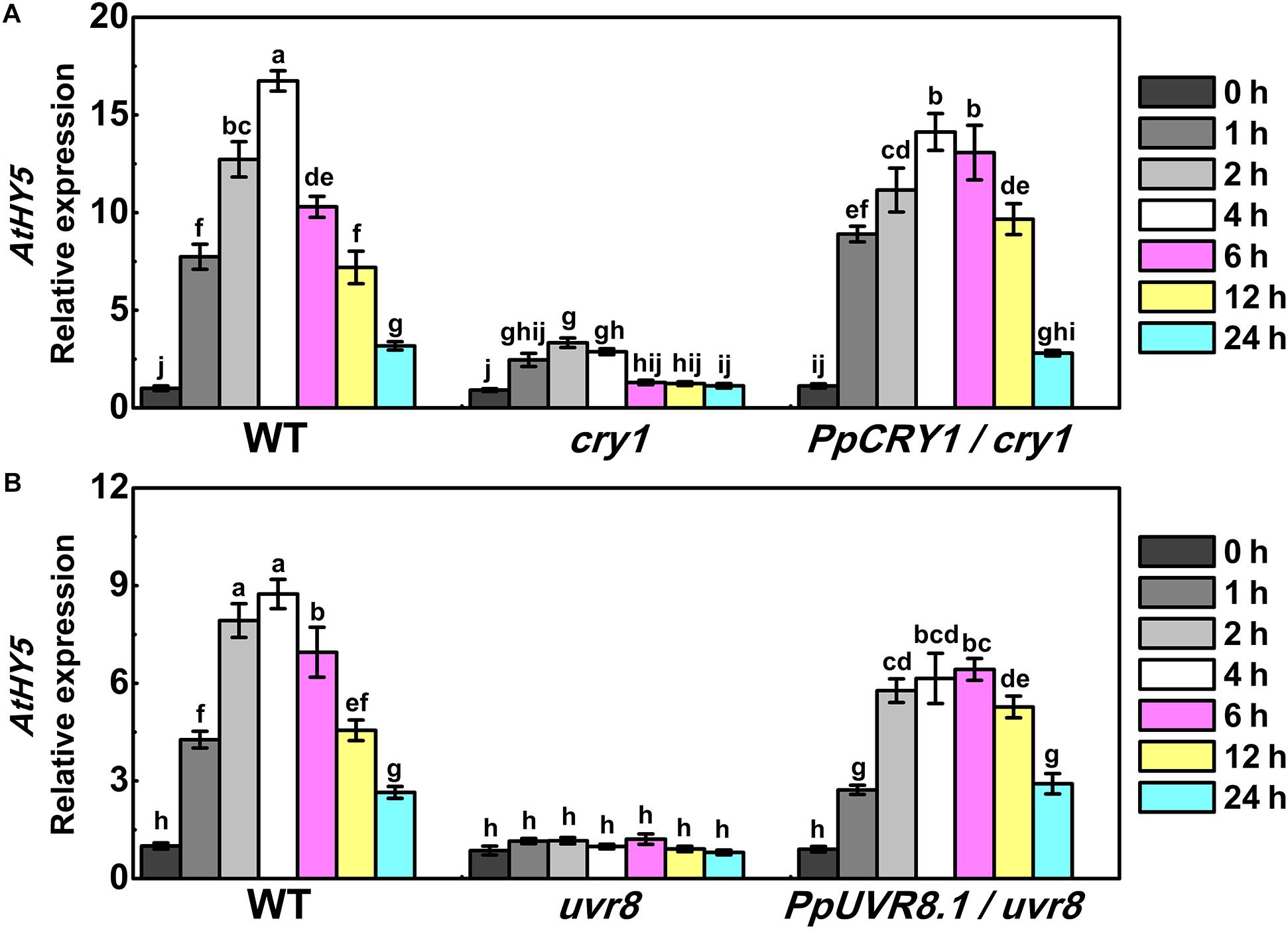
Figure 5. The expression of AtHY5 in the wild type (WT), cry1 mutant and PpCRY1/cry1 transgenic plant with different durations of continuous weak white light (WWL, 40 μw/cm2) + UVA irradiation (40 μw/cm2) (A) or in the WT, uvr8 mutant and PpUVR8.1/uvr8 transgenic plant with different durations of continuous WWL (40 μw/cm2) + UVB irradiation (10 μw/cm2) (B). For Arabidopsis transgenic plants, three independent lines for each target gene were used. 5-day-old seedlings grown under constant WWL were subjected to WWL + UVA or WWL + UVB irradiation for the indicated time span. Data were presented as means (±SE) from three independent biological replicates with 30 or more Arabidopsis seedlings collected for each replicate.
Similarly, after five days growth under white light, WT, uvr8 mutant and PpUVR8.1/uvr8 transgenic lines were transferred to WWL + UVB condition for periods of 0-24 h. Before WWL + UVB irradiation (0 h), AtHY5 displayed relatively low transcription levels in all lines (Figure 5B). After exposure to WWL + UVB, a UV-induced elevation in AtHY5 transcription occurred in WT, while no increase was observed in uvr8. However, the overexpression of PpUVR8.1 in uvr8 restored the ability of UVB to increase AtHY5 transcription. The transcript levels of AtHY5 showed a similar pattern in seedlings of PpUVR8.1/uvr8 transgenic lines and WT during the irradiation period, decreasing gradually after a rapid peaking within 4 h. This suggested that the PpUVR8.1 was a functional counterpart of AtUVR8 and could regulate the expression of AtHY5 in response to UVB irradiation.
As described above, AtHY5 participates in both UVA and UVB signaling pathways in the PpCRY1 and PpUVR8.1 dependent manner respectively, leading to regulation of hypocotyl elongation and anthocyanin biosynthesis. The PpHY5/hy5 transgenic line was used for measuring hypocotyl length and anthocyanin content under WWL, WWL + UVA, WWL + UVB or WWL + UVA + UVB condition. Among hy5, WT and PpHY5/hy5 transgenic line under different light conditions, hy5 consistently showed the longest hypocotyl length and the lowest anthocyanin content relative to other plants (Figure 6). When overexpressing PpHY5 in hy5, the phenotypes of inhibition of hypocotyl elongation and anthocyanin accumulation were restored to levels in WT. Upon UV light exposure, the hypocotyl length was suppressed and anthocyanin increased in PpHY5/hy5 transgenic line and WT as compared with those under WWL, but such phenotypes of photomorphogenesis were compromised in hy5 (Figure 6). A more profound effect on photomorphogenesis was observed when irradiated with both UVA and UVB (Figure 6). The observation was consistent with the result in peach fruit under UV light treatments, indicating that PpHY5 played a similar role to AtHY5, which directly participated in UVA- and UVB-induced photomorphogenic responses and acted as a central positive regulator downstream of PpCRY1 and PpUVR8.1 photoreceptors.
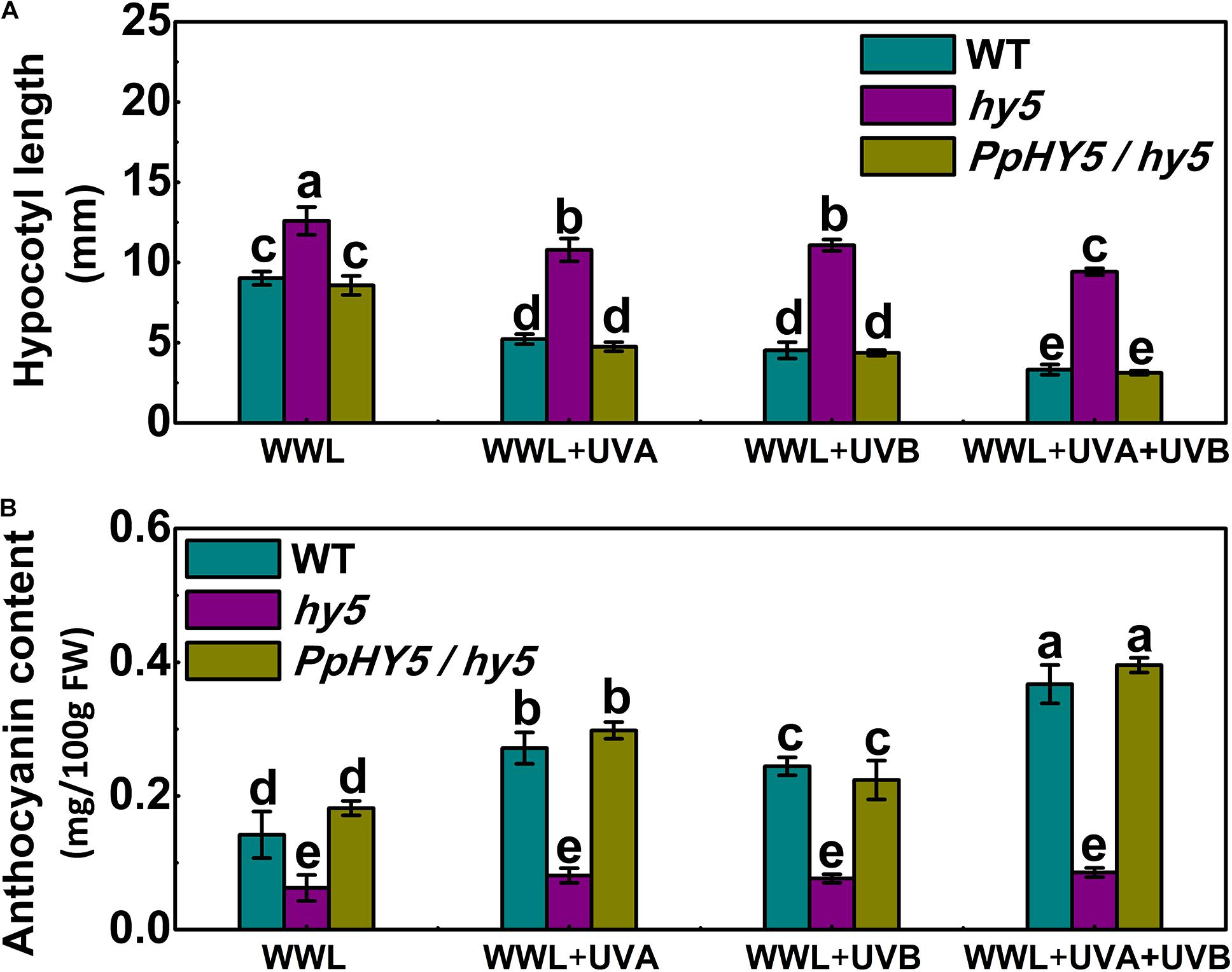
Figure 6. Effect of different light treatments on hypocotyl length (A) and anthocyanin content (B) of 7-day-old seedlings of wild type (WT), hy5 mutant and PpHY5/hy5 transgenic plant. Seedlings were grown under continuous weak white light (WWL, 40 μw/cm2) supplemented with/without UVA (40 μw/cm2) or UVB (10 μw/cm2) or both together for different UV light treatments. For Arabidopsis transgenic plant, three independent lines were used. Data were presented as means (±SE) from three independent biological replicates with 30 or more Arabidopsis seedlings collected for each replicate. Values labeled with different letters indicated a significant difference at P < 0.05.
Previous studies have demonstrated that Arabidopsis HY5 binds to the G-box (CACGTG) and ACE (ACGT containing element) motifs in the promoters of target genes in response to light (Lee et al., 2007). In Arabidopsis PpHY5/hy5 transgenic lines, it was found that PpHY5 promoted the expression of anthocyanin biosynthetic structural genes (Figure 4C). In order to elucidate the function of PpHY5 in the regulation of anthocyanin accumulation in peach, the promoter regions (1,500 bp length) of anthocyanin biosynthetic genes and PpMYB10s were isolated.
Analysis of promoter sequences revealed the presence of putative cis-regulatory elements related to light signaling. The HY5-binding motifs (G-box and/or ACE motifs) were distributed in the promoters of 11 genes (PpCHS1, PpCHS2, PpCHI, PpF3H, PpF3’H, PpDFR1, PpANS, PpUFGT, and PpMYB10.1/2/3), with the total number ranging from one to seven for each promoter (Figure 7A). For example, five G-box motifs were identified in the PpCHS1 promoter, four G-box motifs, and one ACE motif were identified in the PpMYB10.1 promoter, while only one G-box motif and one ACE motif were identified in the PpANS promoter (Figure 7A). To validate the effect of PpHY5 on the transcription of anthocyanin biosynthetic genes, the promoter sequences of 11 genes were isolated and the dual-luciferase assay was conducted. When co-infiltrated with PpHY5, the highest activity was observed in the promoter of PpCHS1 (5.3-fold) followed by PpMYB10.1 (4.9-fold), PpCHS2 (4.1-fold), and PpDFR1 (3.3-fold) (Figure 7B). However, no obvious enhancements in luciferase enzyme activities were detected for other genes. Similarly, infiltration of PpHYH was also able to activate the promoters of both PpCHS1 (2.9-fold) and PpDFR1 (2.6-fold), while no obvious change in activation was observed on the promoters of either PpCHS2 or PpMYB10.1 or other genes (Figure 7B). Compared with PpHY5, PpHYH showed a similar but weaker ability to activate the promoters of both PpCHS1 and PpDFR1. Taken together, these findings indicated that PpHY5 functioned as a positive transcriptional regulator for anthocyanin accumulation via activation of anthocyanin biosynthetic genes, possibly via binding directly to their promoters at G−box and ACE motifs.
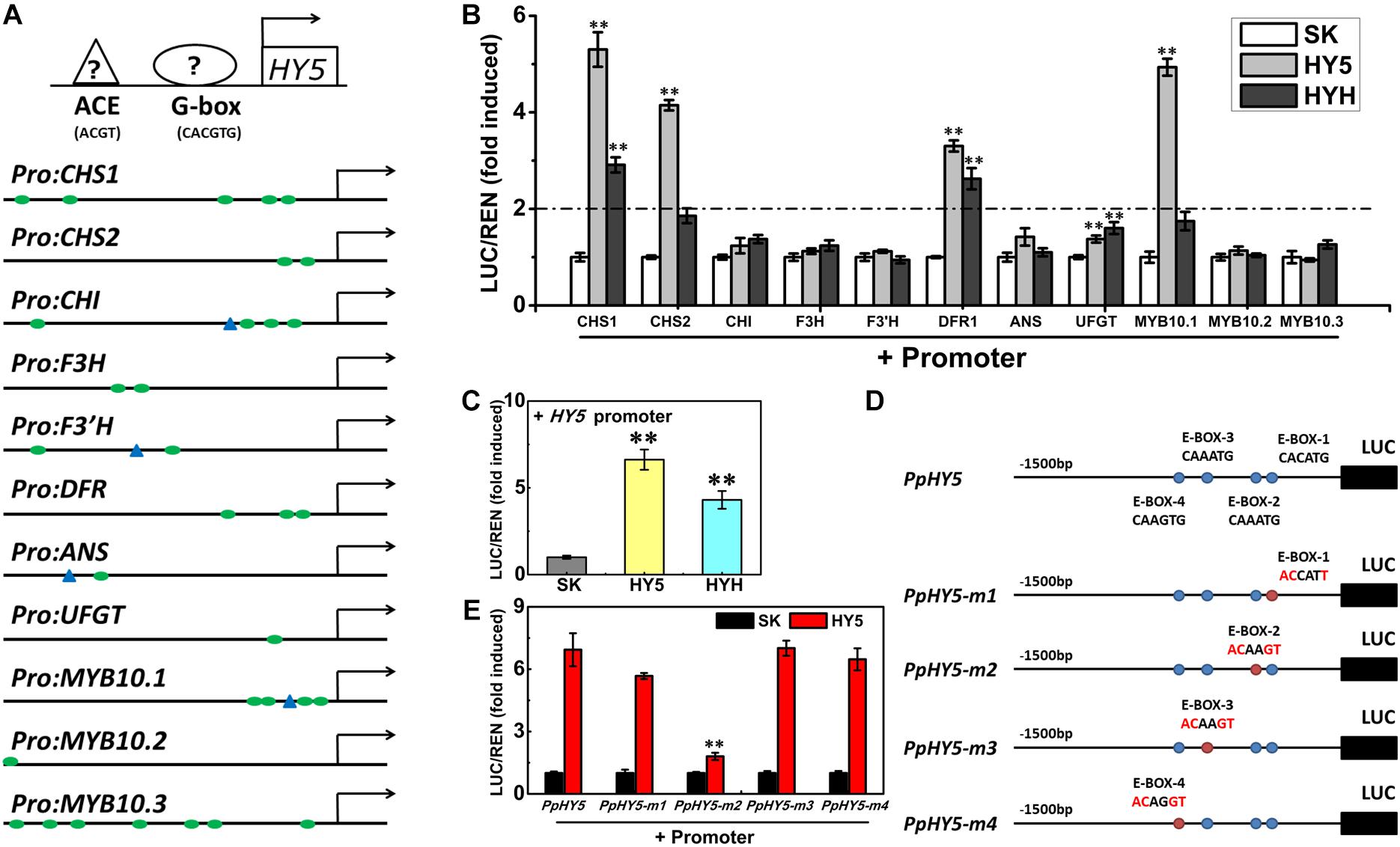
Figure 7. Activation/self-activation of anthocyanin biosynthesis related genes by PpHY5 or PpHYH. (A) Upstream sequences (1500 bp) of anthocyanin biosynthesis related genes with G-box and ACGT containing element (ACE) motifs presented as green circles and blue triangles in the promoters, respectively. (B) Effects of PpHY5 or PpHYH on the promoter activity of anthocyanin biosynthesis related genes in dual-luciferase assays. (C) Effects of PpHY5 or PpHYH on the promoter activity of PpHY5 in dual-luciferase assays. (D,E) Schematic diagrams of motif mutations for the PpHY5 promoter (D) and effects of PpHY5 on the activity of original and mutated promoters in dual-luciferase assays (E). The ratio of LUC/REN of the empty vector (SK) plus promoter was used as the calibrator (set as 1). LUC: firefly luciferase; REN: renilla luciferase; green circles: G-box; blue triangles: ACE motif; blue circles: E-box; red circles: mutated E-box. Data were presented as means (±SE) from six independent biological replicates and Student’s t-test was used for statistical analyses compared with corresponding control. Asterisks (∗∗) indicated a highly significant difference at P < 0.01.
We also analyzed the effects of PpHY5 and PpHYH on the promoter activity of PpHY5 itself. PpHY5 and PpHYH significantly stimulated PpHY5 promoter activity with the ratio of LUC to REN of 6.6 and 4.3 respectively (Figure 7C). To investigate the specificity of the interaction between PpHY5 and the PpHY5 promoter, we performed a combination of cis-regulatory element mutations and the dual-luciferase assays to compare the activation levels. Four predicted E-box motifs were found in the promoter of PpHY5 and designated as E-box-1, E-box-2, E-box-3 and E-box-4 respectively (Figure 7D). As shown in Figure 7E, when E-box-2 (CAAATG) within the PpHY5 promoter was mutated to ACAAGT, the trans-activation activity of PpHY5 on PpHY5-m2 promoter was attenuated by 74%. In contrast, PpHY5 still showed similar activation towards other mutated PpHY5 promoters (PpHY5-m1, PpHY5-m3 and PpHY5-m4) with the induction of 5.6-, 7-, and 6.4-fold, with no significant difference relative to the native PpHY5 promoter (Figure 7E). These results suggested that PpHY5 might recognize E-box-2 to specifically activate its own transcription.
Peach is an economically important fruit crop cultivated worldwide. Fruit coloration is one of the critical traits of exterior quality and commercial value for the consideration in consumer choice. Anthocyanin is the predominant component responsible for red pigmentation in peach fruit (Cheng et al., 2014). Light is an essential environmental factor influencing the production of anthocyanin and fruit coloration in peach, indicating the light signaling pathway is involved in the process (Zoratti et al., 2014). In this study, we conducted the comprehensive analysis of the effects of different UV lights on anthocyanin accumulation in peach. The results showed that peach fruit accumulated anthocyanin under UVA or UVB treatment and the induction was strengthened by the irradiation of UVA and UVB together (Figure 1). The induction was due to enhanced expression of genes encoding enzymes of the anthocyanin biosynthetic pathway.
The effects of UV irradiation on anthocyanin accumulation were not apparent until after continuous UV light treatment for 72 and 144 h. However, the transcript levels of multiple genes were induced in a short time (Figure 2). The results were similar with the significant elevation in the transcripts of AtCHS, AtF3H, and AtDFR in Arabidopsis under blue light treatment for 1 h (Cominelli et al., 2008). Maximum transcriptional response was observed following irradiation with of UVA + UVB, which correlated with anthocyanin accumulation. The molecular evidence of the synergistic effect of UVA + UVB on the regulation of anthocyanin biosynthesis needs further study.
HY5, a member of the bZIP family transcription factor, is an essential regulator in multifaceted developmental processes, such as photomorphogenesis, pigment accumulation, chloroplast development, nutrient assimilation, and carbon/nitrogen balance (Gangappa and Botto, 2016). HY5 has also been functionally characterized to be involved in the regulation of anthocyanin accumulation in response to light in a variety of plants (Shin et al., 2013; Jiang et al., 2016; An et al., 2017). After exposure to blue light, the transcript level of SlHY5 in tomato was induced sharply within 3 h, and the SlHY5 protein accumulated steadily and peaked at 48 h (Liu et al., 2018). In peach, PpHY5 had a strong response to both UVA and UVB at the transcription level, and the expression rapidly increased within 6 h. Expression of PpHYH, the closest homolog of PpHY5, was also induced by UVA and UVB treatments. Meanwhile, a decrease in the transcription level of PpCOP1.1 was observed.
In Arabidopsis under white light, the hy5 mutant exhibited skotomorphogenic phenotype with less anthocyanin content and longer hypocotyl length, conversely the cop1 mutant presented photomorphogenic phenotype with higher anthocyanin content and short hypocotyl length. Overexpression of PpHY5 in hy5 and PpCOP1.1 in cop1 restored wild type phenotypes under white light (Figure 4A). Moreover, the RT-qPCR analysis showed that anthocyanin biosynthetic structural genes, including AtPAL, AtCHS, AtCHI, AtF3H, AtDFR, AtANS, and AtUFGT, were significantly up-regulated in PpHY5/hy5 and down-regulated in PpCOP1.1/cop1 transgenic lines than those in the corresponding mutant (Figure 4C). The phenotypes in Arabidopsis transgenic lines and mutants implied that PpHY5 and PpCOP1.1 acted as counterparts. Under WWL, PpHY5 also directly participated in UVA- and UVB-induced anthocyanin accumulation which depended on PpCRY1 and PpUVR8.1 photoreceptors, indicating that PpHY5 exhibited a positive regulatory role in anthocyanin biosynthesis in response to various wavelengths of light environment.
We further investigated the role of PpHY5 in the regulation of anthocyanin accumulation using the dual-luciferase assay. PpHY5 positively activated the transcription of PpCHS1, PpDFR1, PpCHS2, and PpMYB10.1 (Figure 7B). This is similar to results seen in tomato, apple, and pear (An et al., 2017; Liu et al., 2018; Tao et al., 2018). We also found that PpHYH had a similar effect to PpHY5 in regulating anthocyanin accumulation (Figure 7B). In addition, PpHY5 could bind to the E-box to regulate its own transcription (Figure 7E). As hypothesized, UVA and UVB light signals for anthocyanin biosynthesis were via PpHY5 in peach (Figure 8). These findings suggested that PpHY5 had a conserved function across different species and various wavelengths of light in regulating anthocyanin accumulation.
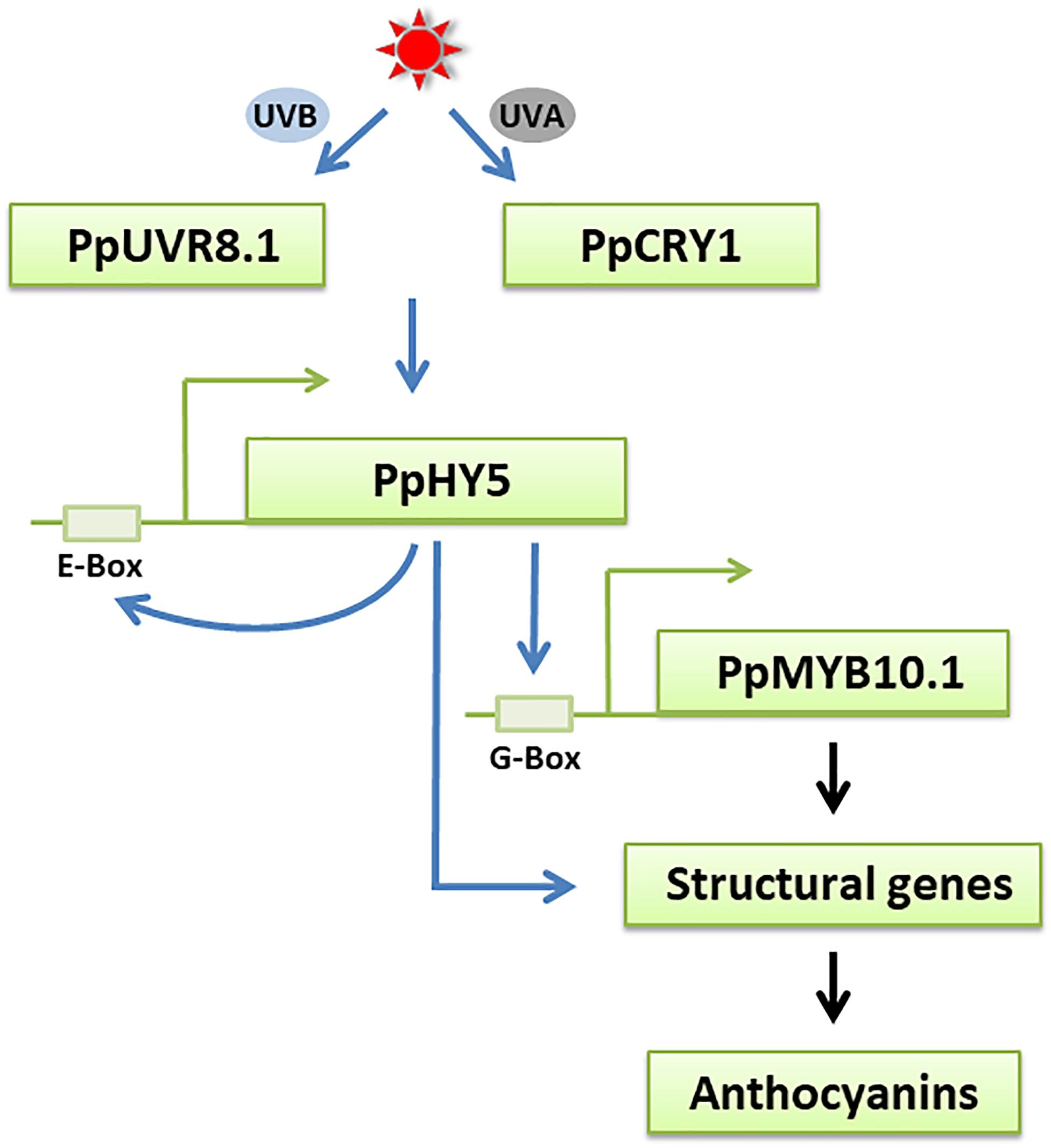
Figure 8. A possible model for UVA- and UVB-induced anthocyanin biosynthesis in peach fruit. The UV light receptors PpCRY1 and PpUVR8.1 respectively perceive UVA and UVB light signals and influences the expression of PpHY5, an essential regulator in UVA and UVB signaling pathway. PpHY5 positively induces its own transcription by interacting with an E-box in its own promoter and activates the expression of PpMYB10.1 to regulate anthocyanin accumulation. The transcription of anthocyanin biosynthetic structural genes is directly regulated by PpMYB10.1 and some of them were also regulated by PpHY5. The blue arrows indicate the pathways verified in the present work. The black arrows indicate the pathways that have been previously reported in peach.
Many studies have revealed that the photoreceptors are indispensable for anthocyanin biosynthesis. VviUVR1 and MpUVR8, the UVB receptors in grape and liverwort, played a predominant role in the UVB-induced flavonoid accumulation (Loyola et al., 2016; Clayton et al., 2018). The phenotypes of alleviated hypocotyl elongation inhibition and decreased anthocyanin accumulation were observed in the antisense Brassica napus CRY1 (BnCRY1) transgenic plant (Chatterjee et al., 2006). In apple, cryptochrome genes MdCRY1 and MdCRY2 have been identified to regulate anthocyanin accumulation and MdCRY2 also participated in the photoperiodic control of flowering (Li et al., 2013a,b). Overexpression of SlCRY1a led to higher anthocyanin accumulation in tomato (Liu et al., 2018) and SlUVR8 mediated the expression of SlCHS in response to the UVB environment (Li et al., 2018). We cloned the PpCRY1 and PpUVR8.1 from peach and characterized their functions as UVA and UVB receptors by complementation of Arabidopsis mutants. In our study, overexpressing PpUVR8.1 or PpCRY1 in the corresponding transgenic line promoted the expression of AtHY5 in response to UVA or UVB irradiation, respectively. The results provided evidences that PpCRY1 and PpUVR8.1 acted as the photoreceptors of UVA and UVB and were associated with eliciting the transcript accumulation of HY5.
In conclusion, our study showed that UVA and UVB accelerated anthocyanin biosynthesis in the peel of “Hujingmilu” peach. The functions of crucial genes and photoreceptors in UV light signaling transduction pathways from peach were clarified. The data suggested that PpHY5 was both involved in UVA- and UVB-induced anthocyanin accumulation via a relatively conserved signaling pathway in PpCRY1-dependent and PpUVR8.1-dependent manner, respectively (Figure 8). PpHY5 regulated anthocyanin accumulation by activating the expression of anthocyanin biosynthetic-related genes. However, the results were mainly based on transcriptional control, which still needs to be verified at the post-transcriptional level in future experiments. Additionally, it would be worthy to investigate whether a direct interaction or an indirect interaction containing other unknown factors exists between PpCRY1 or PpUVR8.1 and PpHY5.
The raw data were deposited in NCBI sequence read archive (SRA) under accession number PRJNA665861.
YZ carried out the experiment, analyzed the data, and drafted the manuscript. MC helped to provide the experiment materials. CZ and RJ helped to provide technical assistance. TM, HW, AA, KL-W, and CX were involved in the revision of the manuscript. CX initiated the project, designed the research framework, coordinated the study, and participated in writing the manuscript. All authors read and approved the final manuscript.
This study was supported by the National Natural Science Foundation in China (31572102), Zhejiang Provincial Science and Technology Project for the Belt and Road (2016C04001), and the Open Project Fund of Hubei Key Laboratory for Processing and Transformation of Agricultural Products (2019HBSQGDKFB05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.603178/full#supplementary-material
Supplementary Figure 1 | Subcellular localization of PpHY5 and PpCOP1.1 in Nicotiana benthamiana leaves under white light. eGFP, GFP channel; Nucleus-RFP, transgenic N. benthamiana plants with red florescence in the nucleus; Bright-field, light microscopy image; Merged, merged image of the GFP and Bright channels. Scale bars indicated 20 μm.
Supplementary Table 1 | Primers used for reverse transcription quantitative PCR analysis.
Supplementary Table 2 | Primers used for plasmid construction. The restriction sites are underlined.
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4:17023. doi: 10.1038/hortres.2017.23
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., and Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
Azuma, A., Yakushiji, H., Koshita, Y., and Kobayashi, S. (2012). Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 236, 1067–1080. doi: 10.1007/s00425-012-1650-x
BenSimhon, Z., Judeinstein, S., NadlerHassar, T., and Trainin, T. (2011). A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234, 865–861. doi: 10.1007/s00425-011-1438-4
Binkert, M., Kozma-Bognár, L., Terecskei, K., De, V. L., Nagy, F., and Ulm, R. (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26, 4200–4213. doi: 10.1105/tpc.114.130716
Carreño, J., Martínez, A., Almela, L., and Fernández-López, J. A. (1995). Proposal of an index for the objective evaluation of the colour of red table grapes. Food Res. Int. 28, 373–377. doi: 10.1016/0963-9969(95)00008-A
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Castagna, A., Dall’Asta, C., Chiavaro, E., Galaverna, G., and Ranieri, A. (2014). Effect of post-harvest UV-B irradiation on polyphenol profile and antioxidant activity in flesh and peel tomato fruits. Food Bioproc. Technol. 7, 2241–2250. doi: 10.1007/s11947-013-1214-5
Chagné, D., Lin-Wang, K., Espley, R. V., Volz, R. K., How, N. M., Rouse, S., et al. (2013). An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 161, 225–239. doi: 10.1104/pp.112.206771
Chatterjee, M., Sharma, P., and Khurana, J. P. (2006). Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 141, 61–74. doi: 10.1104/pp.105.076323
Chattopadhyay, S., Ang, L. H., Puente, P., Deng, X. W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. doi: 10.1105/tpc.10.5.673
Cheng, J., Wei, G., Zhou, H., Gu, C., Vimolmangkang, S., Liao, L., et al. (2014). Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 166, 1044–1058. doi: 10.1104/pp.114.246876
Clayton, W. A., Albert, N. W., Thrimawithana, A. H., McGhie, T. K., Deroles, S. C., Schwinn, K. E., et al. (2018). UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants. Plant J. 96, 503–517. doi: 10.1111/tpj.14044
Cominelli, E., Gusmaroli, G., Allegra, D., Galbiati, M., Wade, H. K., and Jenkins, G. I. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. Plant Physiol. 165, 886–894. doi: 10.1016/j.jplph.2007.06.010
Gangappa, S. N., and Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant. 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gelderen, K. V., Kang, C., and Pierik, R. (2018). Light signaling, root development, and plasticity. Plant Physiol. 176, 1049–1060. doi: 10.1104/pp.17.01079
Gould, K. S., Dudle, D. A., and Neufeld, H. S. (2010). Why some stems are red: cauline anthocyanins shield photosystem II against high light stress. J. Exp. Bot. 61, 2707–2717. doi: 10.1093/jxb/erq106
Guo, J., and Wang, M. (2010). Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (Solanum lycopersicum L.). Plant Growth Regul. 62, 1–8. doi: 10.1007/s10725-010-9472-y
Guo, J., Han, W., and Wang, M. H. (2008). Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: a review. Afr. J. Biotechnol. 7, 4966–4972. doi: 10.5897/AJB08.090
Hagen, S. F., Borge, G., Bengtsson, G. B., Bilger, W., Berge, A., and Haffner, K. (2007). Phenolic contents and other health and sensory related properties of apple fruit (Malus domestica Borkh., cv. Aroma): effect of postharvest UV-B irradiation. Postharvest Biol. Technol. 45, 1–10. doi: 10.1016/j.postharvbio.2007.02.002
Hardtke, C. S., Gohda, K., Osterlund, M. T., Oyama, T., Okada, K., and Deng, X. W. (2000). HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19, 4997–5006. doi: 10.1093/emboj/19.18.4997
Henry-Kirk, R. A., Plunkett, B., Hall, M., McGhie, T., Allan, A. C., Wargent, J. J., et al. (2018). Solar UV light regulates flavonoid metabolism in apple (Malus domestica). Plant Cell Environ. 41, 675–688. doi: 10.1111/pce.13125
Holm, M., Ma, L. G., Qu, L., and Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jiang, M., Ren, L., Lian, H., Liu, Y., and Chen, H. (2016). Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 249, 46–58. doi: 10.1016/j.plantsci.2016.04.001
Kami, C., Lorrain, S., Hornitschek, P., and Fankhauser, C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66. doi: 10.1016/S0070-2153(10)91002-8
Lau, O. S., and Deng, X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. doi: 10.1016/j.tplants.2012.05.004
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. doi: 10.1105/tpc.106.047688
Li, D., Luo, Z., Mou, W., Wang, Y., and Ying, T. (2014). ABA and UV-C effects on quality, antioxidant capacity and anthocyanin contents of strawberry fruit (Fragaria ananassa Duch.). Postharvest Biol. Technol. 90, 56–62. doi: 10.1016/j.postharvbio.2013.12.006
Li, H. R., Li, Y. X., Deng, H., Sun, X. C., Wang, A. Q., Tang, X. F., et al. (2018). Tomato UV-B receptor SlUVR8 mediates plant acclimation to UV-B radiation and enhances fruit chloroplast development via regulating SlGLK2. Sci Rep. 8:6097. doi: 10.1038/s41598-018-24309-y
Li, Y. Y., Mao, K., Zhao, C., Zhang, R. F., Zhao, X. Y., Zhang, H. L., et al. (2013a). Molecular cloning of cryptochromes 1 from apple and its functional characterization in Arabidopsis. Plant Physiol. Biochem. 67, 169–177. doi: 10.1016/j.plaphy.2013.02.031
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, R. F., Zhang, H. L., et al. (2013b). Molecular cloning and functional analysis of a blue light receptor gene MdCRY2 from apple (Malus domestica). Plant Cell Rep. 32, 555–566. doi: 10.1007/s00299-013-1387-4
Liang, T., Yang, Y., and Liu, H. T. (2019). Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 221, 1247–1252. doi: 10.1111/nph.15469
Lin, C. T. (2002). Blue light receptors and signal transduction. Plant Cell 14, 207–225. doi: 10.1105/tpc.000646
Lin-Wang, K., Bolitho, K., Grafton, K., and Karunairetnam, S. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Liu, C. C., Chi, C., Jin, L. J., Zhu, J. H., Yu, J. Q., and Zhou, Y. H. (2018). The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 41, 1762–1775. doi: 10.1111/pce.13171
Liu, H. T., Liu, B., Zhao, C. X., Pepper, M., and Lin, C. T. (2011). The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691. doi: 10.1016/j.tplants.2011.09.002
Liu, T., Song, S., Yuan, Y. B., Wu, D. J., Chen, M. J., Sun, Q. N., et al. (2015). Improved peach peel color development by fruit bagging. Enhanced expression of anthocyanin biosynthetic and regulatory genes using white non-woven polypropylene as replacement for yellow paper. Sci. Hortic. 184, 142–148. doi: 10.1016/j.scienta.2015.01.003
Loyola, R., Herrera, D., Mas, A., Wong, D. C., Holl, J., Cavallini, E., et al. (2016). The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67, 5429–5445. doi: 10.1093/jxb/erw307
Marzi, D., Brunetti, P., Mele, G., Napoli, N., Calò, L., Spaziani, E., et al. (2020). Light controls stamen elongation via cryptochromes, phytochromes and COP1 through HY5 and HYH. Plant J. 103, 379–394. doi: 10.1111/tpj.14736
Morales, L. O., Brosche, M., Vainonen, J., Jenkins, G. I., Wargent, J. J., Sipari, N., et al. (2013). Multiple roles for UV RESISTANCE LOCUS8 in regulating gene expression and metabolite accumulation in Arabidopsis under solar ultraviolet radiation. Plant Physiol. 161, 744–759. doi: 10.1104/pp.112.211375
Nawkar, G. M., Kang, C. H., Maibam, P., Park, J. H., Jung, Y. J., Chae, H. B., et al. (2017). HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 2084–2089. doi: 10.1073/pnas.1609844114
Niu, S. S., Xu, C. J., Zhang, W. S., Zhang, B., Li, X., Lin-Wang, K., et al. (2010). Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 231, 887–899. doi: 10.1007/s00425-009-1095-z
Oh, S., and Montgomery, B. L. (2017). Phytochromes: Where to start. Cell 171, 1254–1256. doi: 10.1016/j.cell.2017.11.020
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Qian, M., Zhang, D., Yue, X., Wang, S., and Li, X. (2013). Analysis of different pigmentation patterns in “Mantianhong” (Pyrus pyrifolia Nakai) and “Cascade” (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci. Hortic. 151, 75–82. doi: 10.1016/j.scienta.2012.12.020
Rahim, M. A., Busatto, N., and Trainotti, L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240, 913–929. doi: 10.1007/s00425-014-2078-2
Ravaglia, D., Espley, R. V., Henry-Kirk, R. A., Andreotti, C., and Ziosi, V. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13:68. doi: 10.1186/1471-2229-13-68
Rizzini, L., Favory, J. J., Cloix, C., and Faggionato, D. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
Schaefer, H. M., Schaefer, V., and Levey, D. J. (2004). How plant-animal interactions signal new insights in communication. Trends Ecol. Evol. 19, 577–584. doi: 10.1016/j.tree.2004.08.003
Shin, D. H., Choi, M. G., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Stracke, R., Favory, J. J., Gruber, H., Bartelniewoehner, L., Bartels, S., Binkert, M., et al. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103. doi: 10.1111/j.1365-3040.2009.02061.x
Sun, Y. W., Qian, M. J., Wu, R. Y., Niu, Q. F., Teng, Y. W., and Zhang, D. (2014). Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia Nakai) in response to optimum light and temperature. Postharvest Biol. Technol. 91, 64–71. doi: 10.1016/j.postharvbio.2013.12.015
Tao, R. Y., Bai, S. L., Ni, J. B., Yang, Q. S., Zhao, Y., and Teng, Y. W. (2018). The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 248, 37–48. doi: 10.1007/s00425-018-2877-y
Tong, Z., Gao, Z., Wang, F., Zhou, J., and Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10:71. doi: 10.1186/1471-2199-10-71
Tsuda, T. (2012). Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 56, 159–170. doi: 10.1002/mnfr.201100526
Tuan, P. A., Bai, S. L., Yaegaki, H., Tamura, T., Hihara, S., Moriguchi, T., et al. (2015). The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 15:280. doi: 10.1186/s12870-015-0664-5
Uematsu, C., Katayama, H., and Makino, I. (2014). Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J. Exp. Bot. 65, 1081–1094. doi: 10.1093/jxb/ert456
Uleberg, E., Rohloff, J., Jaakola, L., Trost, K., Junttila, O., and Haggman, H. (2012). Effects of temperature and photoperiod on yield and chemical composition of northern and southern clones of bilberry (Vaccinium myrtillus L.). J. Agric. Food Chem. 60, 10406–10414. doi: 10.1021/jf302924m
Wade, H. K., Bibikova, T. N., and Valentine, W. J. (2001). Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J. 25, 675–685. doi: 10.1046/j.1365-313x.2001.01001.x
Wang, N., Qu, C. Z., Jiang, S. H., Chen, Z. J., Xu, H. F., Fang, H. C., et al. (2018). The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 96, 39–55. doi: 10.1111/tpj.14013
Wang, Q., Zuo, Z. C., Wang, X., Liu, Q., Gu, L. F., Oka, Y., et al. (2018). Beyond the photocycle-how cryptochromes regulate photoresponses in plants? Curr. Opin. Plant Biol. 45, 120–126. doi: 10.1016/j.pbi.2018.05.014
Wang, S. Y., Chen, C. T., and Wang, C. Y. (2009). The influence of light and maturity on fruit quality and flavonoid content of red raspberries. Food Chem. 112, 676–684. doi: 10.1016/j.foodchem.2008.06.032
Winkel-Shirley, B. (2001). Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Xu, F., Cao, S., Shi, L., Chen, W., and Su, X. (2014). Blue light irradiation affects anthocyanin content and enzyme activities involved in postharvest strawberry fruit. J. Agric. Food Chem. 62, 4778–4783. doi: 10.1021/jf501120u
Yang, D. Y., Seaton, D. D., Krahmer, J., and Halliday, K. J. (2016). Photoreceptor effects on plant biomass, resource allocation, and metabolic state. Proc. Natl. Acad. Sci. U. S. A. 113, 7667–7672. doi: 10.1073/pnas.1601309113
Yin, R., and Ulm, R. (2017). How plants cope with UV-B: from perception to response. Curr. Opin. Plant Biol. 37, 42–48. doi: 10.1016/j.pbi.2017.03.013
Zhang, W. S., Li, X., Zheng, J. T., Wang, G. Y., Sun, C. D., Ferguson, I. B., et al. (2008). Bioactive components and antioxidant capacity of Chinese bayberry (Myrica rubra Sieb. and Zucc.) fruit in relation to fruit maturity and postharvest storage. Eur. Food Res. Technol. 227, 1091–1097. doi: 10.1007/s00217-008-0824-z
Zhang, X. N., Huang, H. Z., Zhao, X. Y., Lv, Q., Sun, C. D., Li, X., et al. (2015). Effects of flavonoids-rich Chinese bayberry (Myrica rubra) pulp extracts on glucose consumption in human HepG2 cells. J. Funct. Foods. 14, 144–153. doi: 10.1016/j.jff.2015.01.030
Zhang, X., Henriques, R., Lin, S., Niu, Q., and Chua, N. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, Y., Butelli, E., and Martin, C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi: 10.1016/j.pbi.2014.05.011
Zhao, Y., Dong, W. Q., Wang, K., Zhang, B., Allan, A. C., Lin-Wang, K., et al. (2017). Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Front. Plant Sci. 8:1552. doi: 10.3389/fpls.2017.01552
Zhao, Y., Dong, W. Q., Zhu, Y. C., Allan, A. C., Lin-Wang, K., and Xu, C. J. (2020). PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 18, 1284–1295. doi: 10.1111/pbi.13291
Zhou, H., Lin-Wang, K., Wang, F. R., Andrew, A. C., and Han, Y. P. (2018). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 4, 1919–1934. doi: 10.1111/nph.15486
Zhou, Y., and Singh, B. R. (2004). Effect of light on anthocyanin levels in submerged, harvested cranberry fruit. Biomed. Biotechnol. 5, 259–263. doi: 10.1155/S1110724304403027
Zhou, Y., Guo, D., Li, J., Cheng, J., Zhou, H., Gu, C., et al. (2013). Coordinated regulation of anthocyanin biosynthesis through photorespiration and temperature in peach (Prunus persica). Tree Genet Genomes. 9, 265–278. doi: 10.1007/s11295-012-0552-1
Zhou, Y., Zhou, H., Lin-wang, K., Vimolmangkang, S., Espley, R. V., Wang, L., et al. (2014). Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 14:1–13. doi: 10.1186/s12870-014-0388-y
Keywords: peach, anthocyanin, UVA, UVB, HY5, photoreceptor, PpMYB10.1
Citation: Zhao Y, Min T, Chen M, Wang H, Zhu C, Jin R, Allan AC, Lin-Wang K and Xu C (2021) The Photomorphogenic Transcription Factor PpHY5 Regulates Anthocyanin Accumulation in Response to UVA and UVB Irradiation. Front. Plant Sci. 11:603178. doi: 10.3389/fpls.2020.603178
Received: 05 September 2020; Accepted: 28 December 2020;
Published: 18 January 2021.
Edited by:
Jeremy Harbinson, Wageningen Plant Research, Wageningen University and Research, NetherlandsReviewed by:
Weiqiang Li, RIKEN, JapanCopyright © 2021 Zhao, Min, Chen, Wang, Zhu, Jin, Allan, Lin-Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjie Xu, Y2hqeHVAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.