- Soybean Research Institute, National Center for Soybean Improvement, Key Laboratory of Biology and Genetic Improvement of Soybean (General, Ministry of Agriculture), State Key Laboratory of Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, College of Agriculture, Nanjing Agricultural University, Nanjing, China
High-temperature (HT) is one of the most important environmental factors that negatively impact the yield of some soybean cytoplasmic male sterility (CMS)-based hybrid (F1) combinations. The response of soybean to HT, especially at the male organ development stage, is poorly understood. To investigate the molecular mechanisms of the response from soybean CMS-based F1 male organ to HT, a detailed transcriptomics analysis was performed during flower bud development of soybean HT-tolerant and HT-sensitive CMS-based F1 combinations (NF1 and YF1) under normal-temperature and HT conditions. Obvious HT damage was observed by subjecting YF1 with HT, such as indehiscent anthers and decreased pollen fertility, whereas the male fertility of NF1 was normal. In total, 8,784 differentially expressed genes (DEGs) were found to respond to HT stress, which were mainly associated with anther/pollen wall development, carbohydrate metabolism and sugar transport, and auxin signaling. The quantitative real-time PCR (qRT-PCR) analysis and substance content detection also revealed that HT caused male fertility defects in YF1 by altering pectin metabolism, auxin, and sugar signaling pathways. Most importantly, the sugar signaling-PIF-auxin signaling pathway may underlie the instability of male fertility in YF1 under HT. Furthermore, HT induced the expression of heat shock factor (HSF) and heat shock protein (HSP) gene families. Overexpression of GmHSFA2 in Arabidopsis can promote the expression of HT protective genes (such as HSP20) by binding to the HSE motifs in their promoters, so as to improve the HT tolerance during flowering. Our results indicated that GmHSFA2 acted as a positive regulator, conferring HT tolerance improvement in soybean CMS-based F1. GmHSFA2 may be directly involved in the activation of male fertility protection mechanism in the soybean CMS-based F1 under HT stress.
Introduction
Temperature is an important ecological factor affecting physiological and biochemical processes in plants. The physiological damage caused by a high-temperature (HT) environment to plants is called HT stress (Puteh et al., 2013). For plants, even an increase of 1°C in the threshold level is considered as HT stress (Teixeira et al., 2013). The development of male organs in plants is extremely sensitive to temperature, and continuous HT stress will cause microspore abortion, anther indehiscence, filament shortening, and abnormal pollen viability or germination (Djanaguiraman et al., 2018; Begcy et al., 2019). In recent years, with the increase of global temperature, HT stress has become a serious factor affecting crop growth and development (Min et al., 2013; Li et al., 2018).
The “cytoplasmic male sterility (CMS)-based” breeding system is composed of the CMS line and its corresponding maintainer line and restorer line, which is one of the most widely used breeding systems in crop hybrid (F1) seed production (Chen and Liu, 2014). At present, the “CMS-based” matching system has been successfully applied in rice, maize, soybean, and other crops (Chen and Liu, 2014). In the process of hybridization, the sterility of the male sterile line can be restored by the fertility restorer gene of the male restorer line. Due to the genetic effects of cytoplasmic and nuclear interactions between the CMS line and its restorer line, CMS-based F1 is generally more sensitive to the external environment than conventional materials, especially for gametophyte sterile material, in which only about 50% of its CMS-based F1 pollen is fertile (Xie, 2008). Under the influence of certain conditions (including HT stress), the percentage of fertile pollen may be greatly reduced for CMS-based F1 and eventually fail to develop normal seeds (Xie, 2008).
It has been reported that HT is one of the main factors affecting the difference of CMS-based F1 fertility restoration in some plants, such as cotton and rice (Zhao et al., 2009; Zhang et al., 2019). Continuous HT stress resulted in insufficient anther dehiscence, decreased pollen survival rate, and finally decreased yield of CMS-based F1. In general, soybeans begin to bloom in late July. However, extreme HT frequently occurred in July and August in the Huanghuai region and South China, the main producing areas of summer-sown soybean in China. Similar to rice and cotton, the male fertility of soybean CMS-based F1 is also unstable under HT (Nie et al., 2017).
In recent years, it has been reported that fertility-enhancing genes and DNA methylation are involved in the fertility regulation of cotton CMS-based F1 (Wang, 2019; Zhang et al., 2019). Wang (2019) found that pollen fertility of cotton CMS-based F1 is related to the restorer gene and fertility-enhancing gene such as GST (Zhu, 2005). Under the same nuclear genetic background of the restorer gene, the restorer line with strong adaptability can be selected using different ecological environments (Wang, 2019). Zhang et al. (2019) found that HT-induced DNA methylation maintained the dynamic balance of ATP synthesis and ROS production by upregulating the expression of mitochondrial respiratory chain-related genes, so as to ensure the normal fertility recovery ability of the cotton CMS-D2 system under HT stress. However, no study has focused on the molecular mechanism of HT-induced male fertility instability in soybean CMS-based F1. In order to better understand the molecular mechanism of difference in male fertility restoration of soybean CMS-based F1 under HT stress, anther/pollen morphology observation, RNA sequencing (RNA-seq), physiological and biochemical determination, and gene functional verification were performed. Cytological observation showed that soybean HT-sensitive CMS-based F1 was mainly characterized by anther indehiscence and decreased pollen fertility under HT stress. Based on the analysis of differentially expressed genes (DEGs) and differential metabolites, we found that genes or substances related to anther/pollen wall development and auxin metabolism, carbohydrate metabolism, sugar transport, transcription factors (TFs), and heat shock proteins (HSP) may be involved in the fertility regulation of soybean CMS-based F1 under HT. Most importantly, it was found that GmHSFA2 can regulate HSP and galactinol synthase (GolS)-related genes to improve HT tolerance of plants.
Materials and Methods
Plant Materials and HT Treatment
Two soybean CMS-based F1 combinations of the CMS system with different degrees of HT stress tolerance in the field were used in this study, namely, NF1 and YF1, which are tolerant and sensitive to HT stress, respectively. The hybridization of the CMS line NJCMS1A and its restorer lines N4608 and YY6 was carried out in the field at Dangtu Experimental Station (National Center for Soybean Improvement, Nanjing Agricultural University, Dangtu, Anhui, China) in the summer of 2017. And the F1 seeds of (NJCMS1A × N4608) and (NJCMS1A × YY6) were harvested in the autumn, which were designated as NF1 and YF1, respectively, in this study. The plants were grown in illuminated incubators (RXZ-430D, Ningbo Jiangnan, Ningbo, China) at 26 ± 1/20 ± 1°C (day/night) with a 12-h light/12-h dark photoperiod during seedling. The flowering plants were grown in an illuminated incubator at 30°C/24°C (day/night) considered as a normal-temperature (NT) condition. For temperature gradient treatment, three individual flowering plants (R1 stage) of each combination were incubated at 38/32°C and 34/28°C (day/night) for 7 days. During HT treatment, the flowering plants (R1 stage) were incubated at 38/32°C in an illuminated incubator. Because it is very difficult to judge the precise development stage of pollen from the appearance of the flower buds in soybean as described previously (Ding et al., 2016), after HT treatment for 7 days, flower buds of different sizes were collected from NF1 and YF1 plants under NT and HT, respectively, and then immediately frozen in liquid nitrogen and stored at −80°C for RNA isolation. To analyze the expression patterns of GmHSFA2 (Glyma.14G096800) and GmHSP20a (Glyma.12G013100) genes, flowering plants from N4608 were initially exposed to 40°C for 7 days of HT treatment and then transferred to NT (30°C) for recovery. Flower buds of different sizes were sampled at time points of 0, 1st, 3rd, 5th, 7th day and 1 day after recovery. Flower buds of each genotype were collected from three individual plants as three independent biological replicates for NF1NT, YF1NT, NF1HT, YF1HT, and N4608.
The Arabidopsis thaliana Columbia (Col-0) ecotype was used as wild-type (WT) control. The 35S:GmHSFA2, pGmHSFA2:GUS, and 35S:GmHSP20a transgenic plants were all in the Col-0 background. The seeds were vernalized for 2 days at 4°C and then cultivated on a prefertilized soil mixture (nutritional soil, perlite, and vermiculite at a 3:1:1 ratio) at 23°C with long-day conditions (16 h light/8 h dark) in an illuminated incubator (RXZ-430D, Ningbo Jiangnan, Ningbo, China). To evaluate the HT damage on inflorescence and the expression levels of GmHSFA2 downstream regulatory genes under HT stress, three 35S:GmHSFA2 transgenic lines and WT were exposed to HT stress at 45/40°C (day/night) for 3 days. The HT treatment on male fertility was performed as Kim et al. (2001) described. The 35S:GmHSFA2 and 35S:GmHSP20a transgenic plants (two lines for each transgenic type) and WT were held in an illuminated incubator (RXZ-430D, Ningbo Jiangnan, Ningbo, China) at 42°C for 4 h and then transferred to normal growth conditions. All types were grown at 23°C as control.
RNA Isolation and cDNA Library Construction
Total RNA from the flower buds of NF1NT, YF1NT, NF1HT, and YF1HT (three independent biological replicates for each genotype) was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. In order to obtain mitochondrial and chloroplast-related genes, this study refers to the cDNA library construction of prokaryote, considering that the plant mitochondrial and chloroplast genomes are similar to its ring genome. So after total RNA was extracted, sample mRNA was enriched by removing rRNA by a Ribo-ZeroTM Magnetic Kit (Epicentre). Then the enriched mRNA was fragmented into short fragments using a fragmentation buffer and reverse transcribed into cDNA with random primers. A second-strand cDNA was synthesized by DNA polymerase I, RNase H, dNTP, and buffer. Then the cDNA fragments were purified with a QIAquick PCR extraction kit, end repaired, poly(A) added, and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeqTM 2500 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Data Analysis of RNA-Seq
Raw reads were filtered to obtain high-quality reads by removing reads containing adapters or more than 10% of unknown nucleotides (N) and more than 50% of low-quality (Q-value ≤ 20) bases. The rRNA mapped reads were removed by a short-reads alignment tool Bowtie 2 (Langmead and Salzberg, 2012). Clean reads (the rRNA removed reads) were subsequently aligned with the soybean Williams 82 reference genome (Wm82.a2.v1) using TopHat2 (version 2.0.3.12, Kim et al., 2013). Gene abundances were quantified by software RSEM (Li and Dewey, 2011), and the gene expression level was normalized by using the fragments per kilobase of transcript per million mapped reads (FPKM) method (Mortazavi et al., 2008).
Subsequent data were analyzed using repeated correlation analysis (RCA) and principal component analysis (PCA). The correlation coefficient between the two replicas was calculated to evaluate repeatability between samples. The closer the correlation coefficient gets to 1, the better the repeatability between two parallel experiments. The PCA was performed with R package models1; it is largely used to reveal the relationship of NF1NT, YF1NT, NF1HT, and YF1HT. To identify DEGs across samples or groups, the edge R package (see text footnote 1) was used. Only genes with | Log2FC (fold change)| ≥ 1 and false discovery rate (FDR) ≤ 0.05 were identified as significant DEGs. Gene Ontology (GO) enrichment analysis provides all GO terms that are significantly enriched in DEGs compared to genomic backgrounds and maps all DEGs to GO terms in the GO database.2 GO terms with FDR ≤ 0.05 were considered to be significantly enriched. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was performed in the KEGG database web server3 (Kanehisa et al., 2008). Pathways with FDR ≤ 0.05 were defined as significantly enriched pathways in DEGs.
Plant Transformation
Full-length CDS clones of the GmHSFA2 (Glyma.14G096800) and GmHSP20a (Glyma.12G013100) genes were obtained from SoyBase.4 Two overexpression constructs were generated by inserting the full-length GmHSFA2 and GmHSP20a CDS fragments into the binary vector pCAMBIA3301-26 after the CaMV 35S promoter, using a one-step cloning kit (Vazyme, Nanjing, China) and designated as 35S:GmHSFA2 and 35S:GmHSP20a, respectively. The promoter of GmHSFA2 (2,000 bp) was amplified by PCR using N4608 DNA and replaced the 35S promoter of pCAMBIA3301-GUS using HindIII and NcoI digestion, resulting in a plasmid of pGmHSFA2:GUS. All the above overexpression vectors were introduced into Agrobacterium tumefaciens strain EHA105 via the freeze–thaw method. Agrobacterium-mediated floral dip method was used for Arabidopsis transformation (Clough and Bent, 1998). The specific primers used for CDS and promoter cloning are given in Supplementary Table 1. Transgenic plants (T0, T1, T2, and T3) were screened by the Murashige and Skoog medium glufosinate (20 mg/L).
GUS Staining and Plant Trait Investigation
The inflorescence of pGmHSFA2:GUS plant materials was GUS stained following the protocol of Jefferson et al. (1987). The morphology of anthers from opened flowers of soybean and Arabidopsis was observed under an Olympus CX31 microscope (Japan). Pollen viability of soybean and Arabidopsis was analyzed by I2-KI staining (Nie et al., 2019) and Alexander’s staining (Ding et al., 2020), respectively. The stamen and pistil length of Arabidopsis was measured with the cellSens software (Olympus, Japan). Nine flower buds/flowers of each genotype/line were collected from three individual plants to measure the length of stamen/pistil and observe the fertility of pollen. Student’s t-test was performed to compare the trait differences between the experimental group and the control group.
Quantitative Real-Time PCR Analysis
The quantitative real-time PCR (qRT-PCR) was used to validate the gene expression levels in soybean and Arabidopsis. All primers (Supplementary Table 1) were designed based on the mRNA sequences and synthesized commercially (General Biosystems, Chuzhou, China). Total RNA from the same soybean samples that constructed the cDNA library was used for the validation of RNA-seq. According to the procedures provided in the HiScript Q RT SuperMix for the qPCR kit (+gDNA wiper, Vazyme, Nanjing, China), 1 μg of total RNA was reverse-transcribed using an Oligo(dT) primer. The mRNA qRT-PCR analysis was carried out using AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) on a Bio-Rad CFX96 instrument (CFX96 Touch, Bio-Rad, United States). For Arabidopsis, all reactions were run with three independent biological replicates, each comprising three individual plants, and AtActin (accession number: NM_001338359.1) was used as internal control genes. For soybean, GmTubulin (accession number: NM_001252709.2) was used as internal control genes. The NF1 and WT under the NT condition were used as the control in qRT-PCR experiments on soybean and Arabidopsis, respectively. The relative expression levels of the genes were quantified using the 2–ΔΔCt method (Livak and Schmittgen, 2001). Student’s t-test was performed to compare mRNA expression differences between the experimental group and the control group.
Substance Content and Enzyme Activity Assays
Flower buds of different sizes were collected from NF1 and YF1 plants (three independent biological replicates for each genotype) under NT and HT for substance content and enzyme activity assays. The contents of Suc, Glc, starch, and IAA were determined on a UV–vis spectrophotometer (EU-2600D, Onlab, Shanghai, China) using a Suc assay kit (Jiancheng, Nanjing, China), Glc assay kit (Sinobestbio, Shanghai, China), starch assay kit (Sinobestbio, Shanghai, China), and IAA assay kit (Mallbio, Nanjing, China), respectively, by following the manufacturer’s protocol. The pectinase activity was measured at 540 nm on a microplate reader (SpectraMax iD5, Molecular Devices, United States) using the pectinase assay kit (Sinobestbio, Shanghai, China) by following the manufacturer’s protocol. Three independent biological replicates were assayed, and one-way ANOVA and Duncan’s test were performed for statistical analysis.
Subcellular Localization and Yeast One-Hybrid Assay
The open reading frame (ORF) (after removal of the stop codon) of GmHSFA2 was integrated into the 5′ end of the green fluorescent protein (GFP) coding region in the pCAMBIA3301-GFP vector using BglII digestion, resulting in a plasmid of GmHSFA2-GFP. Both GmHSFA2-GFP and pCAMBIA3301-GFP (control) constructs were transformed into tobacco (Nicotiana benthamiana) leaves according to the protocol of Sparkes et al. (2006). The treated seedlings of tobacco were grown at 23°C with long-day conditions (16 h light/8 h dark) in an illuminated incubator (RXZ-430D, Ningbo Jiangnan, Ningbo, China) for 3 days and then observed under a confocal laser scanning microsystem LSM780 (Carl Zeiss, Jena, Germany) with 488-nm excitation wavelengths.
The direct interaction between GmHSFA2 and the promoter of GmHSP20a was detected by the yeast one-hybrid (Y1H) assay system. Four tandem cis-acting HSE motifs present in the promoter region of GmHSP20a were amplified by PCR using N4608 DNA and integrated into the PAbAi vector, yielding pAbAi-pGmHSP20a as bait, while the full-length CDS of GmHSFA2 was amplified from the 35S:GmHSFA2 vector and inserted into a pGADT7 vector, yielding a pGADT7-GmHSFA2 construct as prey. The primers used are listed in Supplementary Table 1. The pGBKT7-pGmHSP20a was first introduced into the Y1H gold yeast (Clontech) and cultured on SD/–Ura and SD/–Ura/A medium for self-activating detection. After that, the PGADT7, negative control, and positive control vectors were introduced and cultured on SD/–Ura/A for spot assay.
Results
Characterization of Soybean CMS-Based F1 Male Fertility Under HT Stress
To explore the mechanism of male fertility instability under HT stress, two soybean CMS-based F1 combinations were used in this study, namely, NF1 (HT tolerant) and YF1 (HT sensitive) (Supplementary Figure 1). There was no difference in male fertility between the two combinations under NT (30°C) according to gradient temperature treatment (Supplementary Figures 2A,B and Figure 1). However, obvious HT damage was observed by subjecting YF1 to gradient temperatures (30, 34, and 38°C, such as worse anther dehiscence and gradually decreasing pollen fertility (Supplementary Figures 2A,B). When the temperature reached 38°C (HT treatment in this study), the male fertility of YF1 was significantly affected, and YF1 finally displayed forms of anther indehiscence and decreased pollen fertility, while NF1 performed normally (Figure 1).
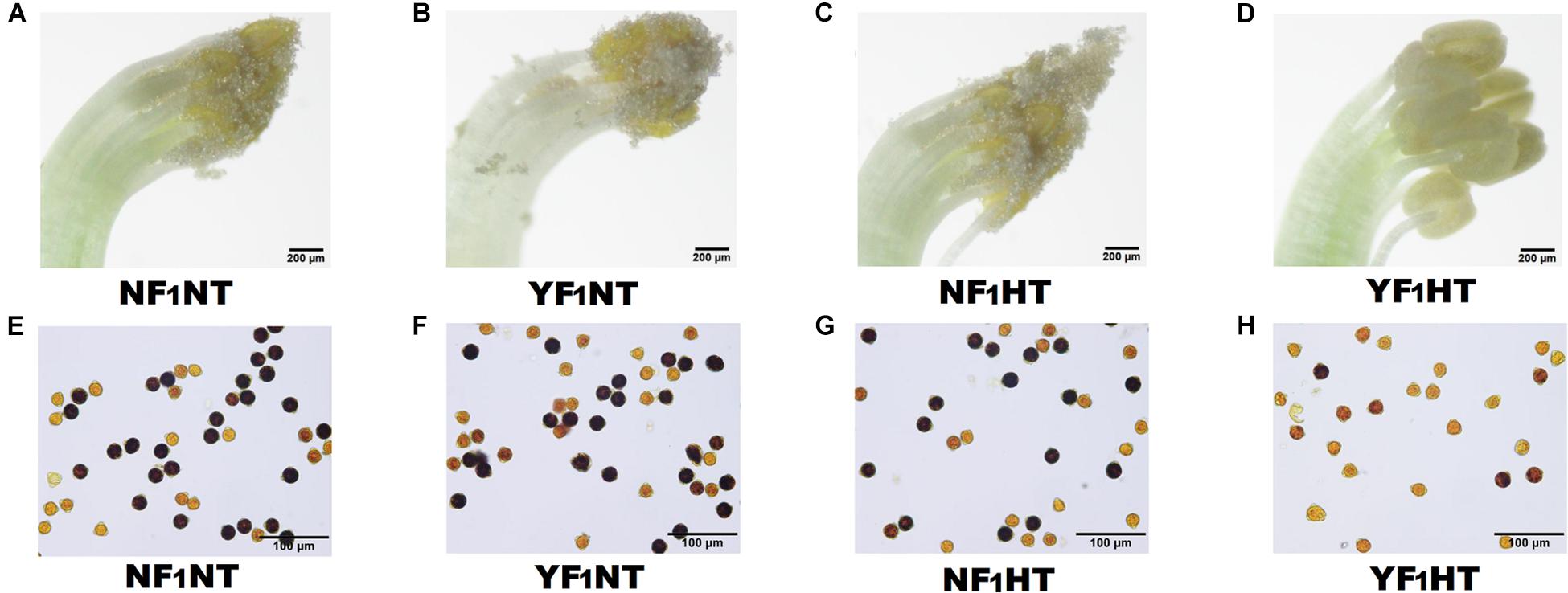
Figure 1. Phenotypic characteristics of soybean male fertility under NT and HT conditions. (A–D) Soybean anthers from NF1 and YF1 under NT and HT conditions. (E–H) Soybean pollens from NF1 and YF1 under NT and HT conditions. The sterile pollen was stained bright yellow by I2-KI solution.
Transcriptomics Analysis in Flower Buds of NF1 and YF1 Under HT Stress
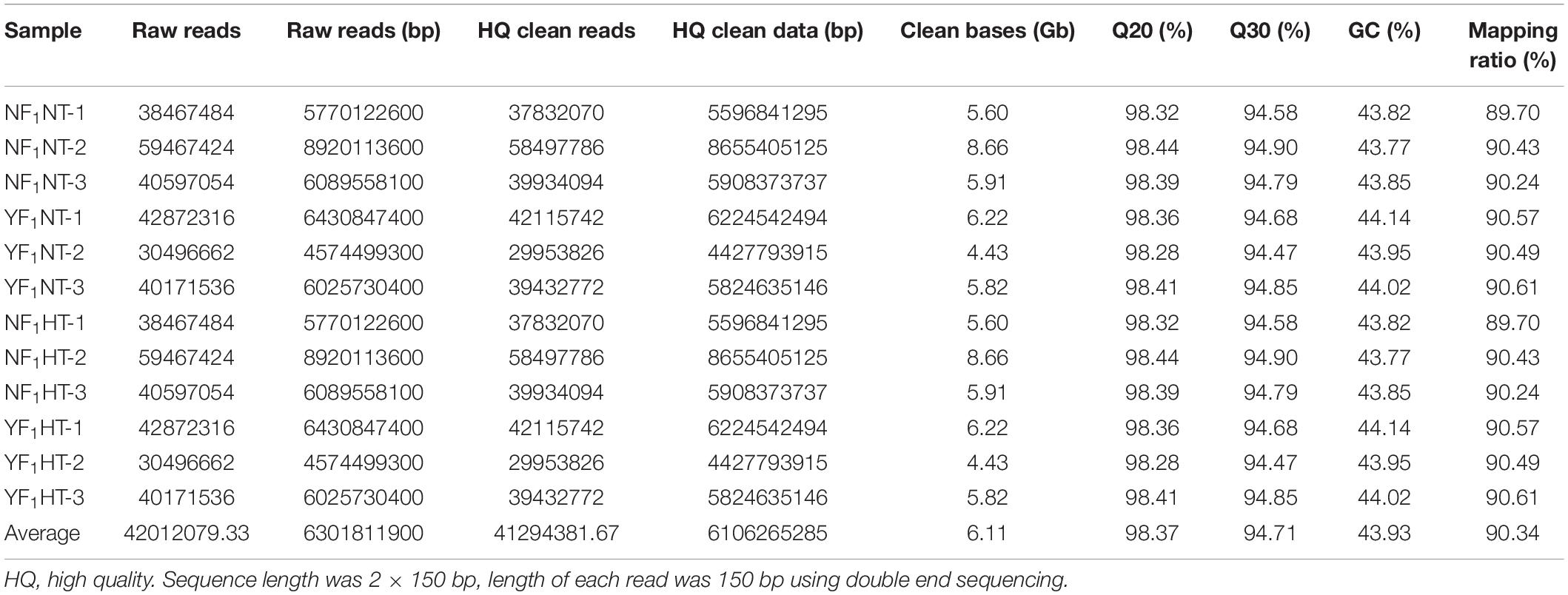
To gain insight into the molecular mechanism of male fertility reduction in soybean HT-sensitive CMS-based F1 under HT stress, RNA-seq was performed for both HT-tolerant and HT-sensitive F1 during flower bud development under NT and HT conditions. A total of 50.41 million raw reads were generated from 12 samples with an average read length of 150 bp (Table 1). After removal of reads containing adapters, poly(N) greater than 10%, and low-quality sequences, an average of 6.11-Gb clean data were obtained for each sample (Table 1). After removal of reads mapped on rRNA, 89.70–90.57% of clean reads were mapped to the soybean reference genome (Gmax_275_Wm82.a2.v1) (Table 1).
Principal component analysis was used to analyze the relationship between two genotypes under NT and HT conditions. The first principal component (PC1) accounted for 78.2% of the variance, and the second principal component (PC2) accounted for 12.0% of the variance (Supplementary Figure 3). With the exception of NF1HT, the three biological replicas in each group were clustered closely together. In general, NF1 and YF1 were significantly different under NT and HT conditions. As shown in Supplementary Figure 4, the correlation coefficients (R2) between the biological replicates of each group were greater than 0.96 and close to 1, indicating that each group had good repeatability.
Identification of DEGs in Response to Heat Stress
Significantly DEGs were screened between the different samples with the criteria of fold change ≥ 2 and FDR ≤ 0.05. To determine the genes that were differentially expressed between two genotypes under NT and HT conditions, four comparisons (NF1NT vs YF1NT, NF1NT vs NF1HT, YF1NT vs YF1HT, and NF1HT vs YF1HT) were performed. Under the NT condition, a total of 1,385 (294 upregulated and 1,091 downregulated) DEGs were identified for the comparison of NF1NT vs YF1NT (Figure 2A). After HT stress, 13,491 genes were differentially expressed in different comparisons. Among these, a total of 10,093 (2,199 upregulated and 7,894 downregulated) and 6,309 (2,162 upregulated and 4,147 downregulated) DEGs were identified for the comparisons of NF1NT vs NF1HT and YF1NT vs YF1HT, respectively (Figure 2A). We identified 4,187 (1,200 upregulated and 2,987 downregulated) DEGs that were in common among these two pairs. A total of 2,181 (1,247 upregulated and 934 downregulated) DEGs were identified for the pair of NF1HT vs YF1HT. The Venn diagram showed that the groups NF1NT vs YF1NT and NF1HT vs YF1HT had only 386 (107 upregulated and 279 downregulated) DEGs in common. However, the DEGs under HT stress accounted for 56.45% of the total DEGs (3,180 DEGs) of these two combinations. This indicated that most DEGs had differential expression changes in response to HT stress. Based on the Venn diagram, we found that 4,519 DEGs showed the same expression pattern between NF1 and YF1 and that the remaining 9,359 DEGs were upregulated or downregulated in different comparisons under HT stress (Figures 2B,C). Twelve DEGs were randomly selected for qRT-PCR verification, and the coincidence rate between qRT-PCR results and RNA-seq data was 100% (Supplementary Figure 5), supporting the reliability of expression patterns revealed by RNA-seq.
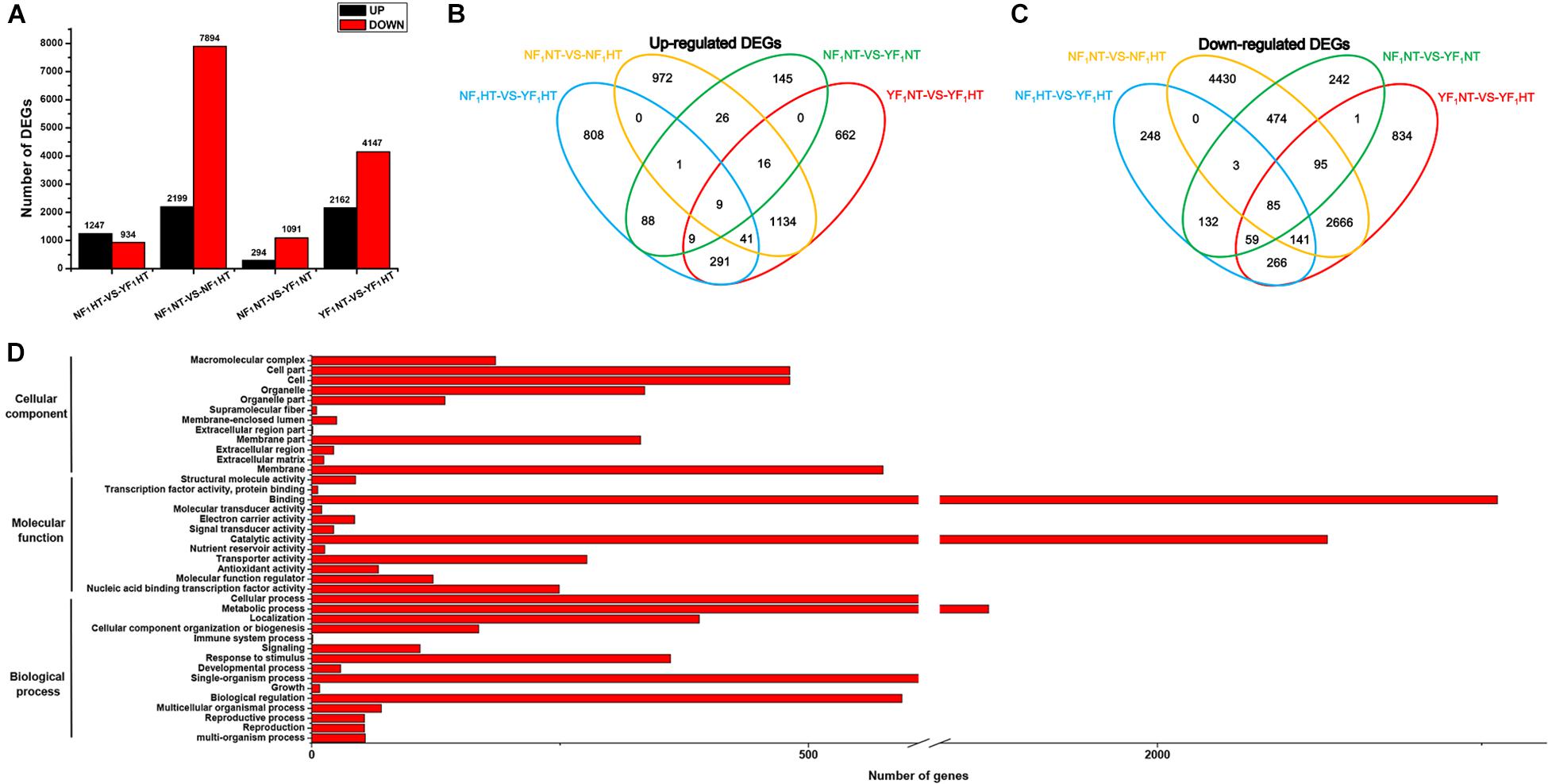
Figure 2. Analysis of DEGs between NF1 and YF1 under NT and HT conditions. (A) Number of DEGs that were up- and down-regulated under NT and HT conditions. (B,C) Venn diagram of common up- and down-regulated DEGs under NT and HT conditions. (D) GO classification of DEGs. The x-axis indicates the number of DEGs, and the y-axis indicates the GO groups.
Functional Classification of DEGs in Response to Heat Stress
Among the 9,359 DEGs between the NT and HT samples, 2,244 upregulated and 5,965 downregulated genes were identified, and the other 575 DEGs were upregulated or downregulated in different combinations at the same time. In order to understand the potential functions in the list of DEGs, all 8,784 DEGs were further analyzed for GO functional annotations. The results revealed that 4,482 DEGs could be classified into 39 GO terms: 3,024 DEGs participated in biological processes, 4,034 DEGs had molecular functions, and 1,018 DEGs had cellular components (Figure 2D and Supplementary Tables 2–4). At the biological process level, the DEGs are enriched into 23 biological processes (p.adjust ≤ 0.05), including pollination (GO:0009856), reproduction (GO:0000003), phosphorylation (GO:0016310), response to oxidative stress (GO:0006979), and oxidation-reduction process (GO:0055114). In particular, we also observed two DEGs in the GO terms response to heat (GO:0009408) and response to temperature stimulus (GO:0009266). Similarly, large numbers of DEGs were also enriched in the molecular function and cellular component, including pectinesterase activity (GO:0030599), peroxidase activity (GO:0004601), antioxidant activity (GO:0016209), cell wall (GO:0005618), and membrane (GO:0016020).
To explore the biological pathways on the reproductive development of soybean CMS-based F1 on which HT has an important influence, KEGG pathway analysis was further performed for these DEGs. A total of 13 significant KEGG pathways (Q-value ≤ 0.05) were enriched for 1,409 DEGs (Supplementary Table 5 and Figure 3A), including pentose and glucuronate interconversions, phenylpropanoid biosynthesis, and starch and sucrose metabolism (Figure 3B). Most importantly, HT stress-induced DEGs were mostly enriched in pentose and glucuronate interconversions, starch and sucrose metabolism, phenylpropanoid biosynthesis, flavonoid biosynthesis, and circadian rhythm–plant pathways, which belong to carbohydrate metabolism, biosynthesis of other secondary metabolites, and environmental adaptation classes, respectively, for the comparisons NF1HT vs YF1HT and YF1NT vs YF1HT (Supplementary Figures 6A,D). This is consistent with the male sterile phenotype of YF1 under HT.
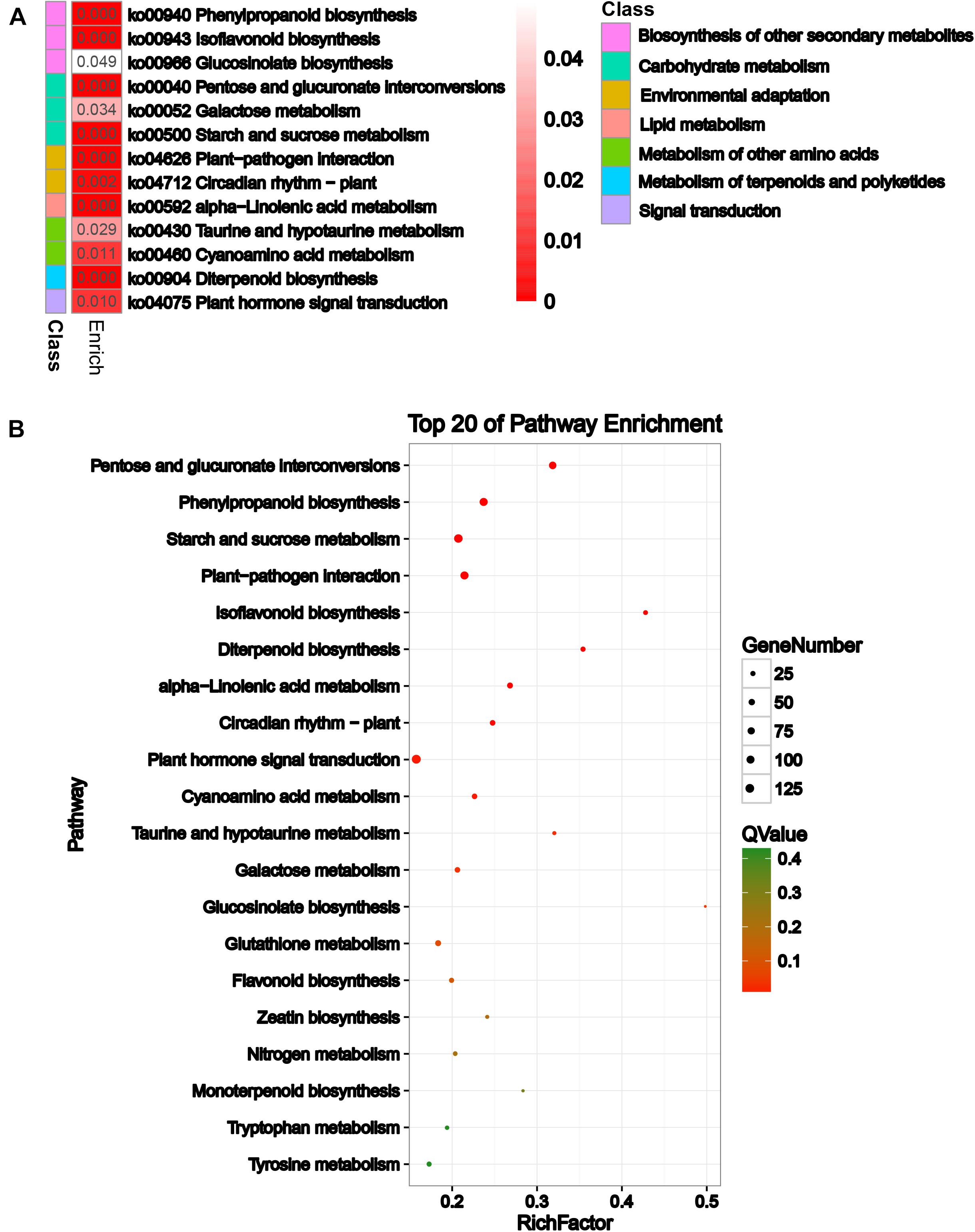
Figure 3. KEGG enrichment analysis of the DEGs. (A) KEGG analysis with heat map. (B) Top 20 of pathway enrichment. The x-axis indicates the rich factor corresponding to each pathway, and the y-axis indicates name of the KEGG pathway. The color of the point represents the P-values of the enrichment analysis. The size and color of bubbles represent the number and degree of enrichment of DEGs, respectively.
HT Caused Anther Defects by Altering Anther/Pollen Wall Development
Based on the expression level, some enzyme genes involved in pectin metabolism were identified, which included pectate lyase (PL), pectin methylesterase (PME, also named pectinesterase), polygalacturonase (PG), and exopolygalacturonase (Exo-PG). Eleven PLs, 21 PMEs, 17 PGs, and 4 Exo-PGs were downregulated in flower buds of YF1 compared with those of NF1 under the HT condition (Figure 4A and Supplementary Table 6). The results also showed lower expressions of PLs, PMEs, PGs, and Exo-PGs in flower buds of YF1 compared with those of NF1 under the NT condition (Figure 4A and Supplementary Figures 7A–D). Most importantly, RNA-seq data in Phytozome v12.0 showed that these four type genes were highly expressed in flowers of soybean (Figure 4B). This indicated that the pectinase activity of YF1 anthers was defective under the HT condition, which led to abnormal formation of the anther cell wall and finally affected anther dehiscence (Figure 1D). To further confirm this result, the pectinase activity was assessed under NT and HT conditions (Figure 4C). However, pectinase activity in the YF1 decreased only slightly compared with that in NF1 under HT, which may be due to the pectinase being composed of PG, PL, and PME (Li et al., 2019) that have different activities in the pollen-related tissues and need to be further studied.
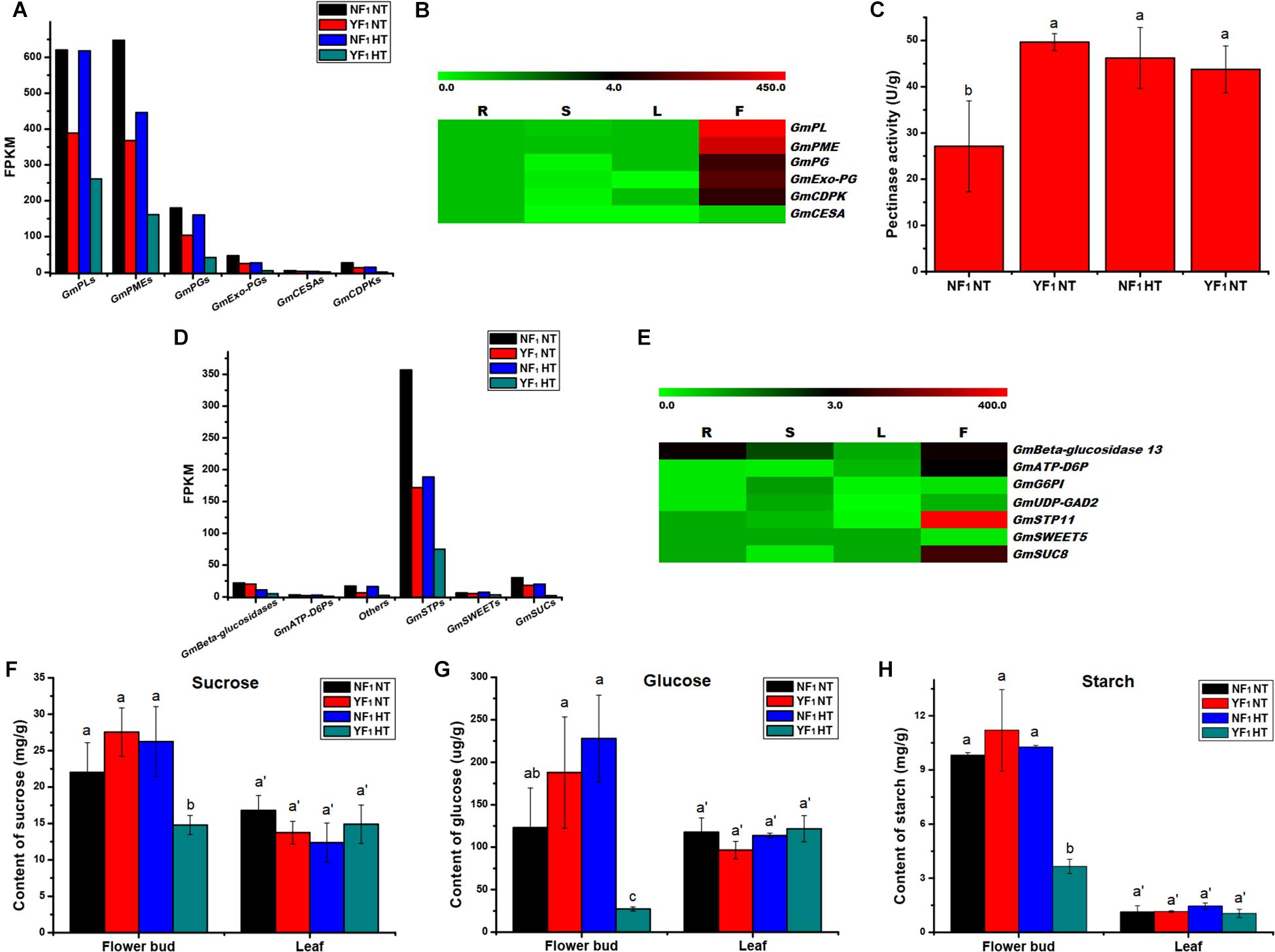
Figure 4. Effect of HT on anther/pollen wall development, carbohydrate metabolism and sugar transport. (A) DEGs related to anther/pollen wall development in RNA-seq data. (B) Heat map of the DEGs related to anther/pollen wall development in four different tissues. The color scale represents the relative transcript abundance of the DEGs in four soybean tissues. The heat map was conducted using MeV 4.9 software. The FPKM values were obtained from the RNA-seq data in Phytozome v12.0. R, root; S, stem; L, leaf; F, flower. (C) Pectinase activity in flower buds of NF1 and YF1 under NT and HT conditions (n = 3). Data are presented as means ± standard deviation (SD) from independent biological replicates. Values with different letter indicates statistical differences (one-way ANOVA, Duncan, P < 0.05). (D) DEGs related to carbohydrate metabolism and sugar transport in RNA-seq data. (E) Heat map of the DEGs related to carbohydrate metabolism and sugar transport in four different tissues. The color scale represents the relative transcript abundance of the DEGs in four soybean tissues. The heat map was conducted using MeV 4.9 software. The FPKM values were obtained from the RNA-seq data in Phytozome v12.0. R, root; S, stem; L, leaf; F, flower. (F–H) Suc, glc, and starch contents in flower buds and leaves of NF1 and YF1 under NT and HT conditions (n = 3). Data are presented as means ± standard deviation (SD) from independent biological replicates. Values with different letter indicates statistical differences (one-way ANOVA, Duncan, P < 0.05).
The RNA data and qRT-PCR also revealed that three cellulose synthase proteins (GmCESAs) were downregulated in flower buds of YF1 compared with those of NF1 under the HT condition (Supplementary Figure 7E), and they were also involved in the pollen wall development of plants (Wang et al., 2011). In addition, three pollen-specific protein (GmCDPK) DEGs were downregulated in flower buds of YF1 under both NT and HT conditions (Supplementary Figure 7F). These results indicated that pectinase, cellulose, and CDPK are associated with anther defects in YF1 under HT.
Carbohydrate Metabolism and Sugar Transport in Flower Buds of Soybean HT-Sensitive CMS-Based F1 Were Disrupted Under HT
A lot of DEGs involved in carbohydrate metabolism during soybean CMS-based F1 flower bud development under HT were found. Among them, there were 79, 117, and 31 DEGs that participated in pentose and glucuronate interconversions, starch and sucrose metabolism, and galactose metabolism pathways, respectively (Supplementary Table 5 and Figure 3). Further analysis indicated that many genes related to carbohydrate metabolism and sugar transport were downregulated, such as PLs, PMEs, PGs, Exo-PGs, beta-glucosidase 13, ATP-dependent 6-phosphofructokinase 7 (ATP-D6P7), UDP-glucuronic acid decarboxylase 2 (UDP-GAD2), sugar transport protein 11 (STP11), bidirectional sugar transporter SWEET5 (SWEET5), and sucrose transport protein SUC8-like (SUC8) (Supplementary Tables 6, 7). This result was also confirmed by qRT-PCR analysis (Supplementary Figures 7, 8, and Figures 4A,D). Most importantly, most of them were highly expressed in flowers of soybean (Figures 4B,E).
Carbohydrate analysis revealed that sucrose (Suc) and glucose (Glc) accumulation in flower buds of YF1 was reduced compared with NF1 under HT (Figures 4F,G). Interestingly, Suc and Glc content in either NF1 or YF1 leaves showed no difference under HT compared with NT (Figures 4F,G). Based on starch content detection, starch accumulation in YF1 flower buds was also lower than that in NF1 flower buds under HT (Figure 4H). All these results revealed that abnormal carbohydrate transport and accumulation affected pollen development, which was consistent with the results of KEGG analysis and reduction of pollen fertility in YF1HT.
HT Caused Instability of Male Fertility in YF1 by Altering Auxin Signaling
During soybean CMS-based F1 flower bud development under HT, many DEGs were found to be involved in plant hormone signal transduction (Supplementary Table 5). Among them, genes encoding proteins involved in auxin biosynthesis (YUCCA11 and GH3.1), auxin response protein (IAA29), and auxin-induced genes (except for AUX10A5 and AUXX15) were downregulated (Supplementary Figure 9). In addition, the expression of IAA regulator PHYTOCHROME-INTERACTING FACTOR genes (PIF1 and PIF4) were upregulated (Supplementary Table 8 and Figure 5A). Furthermore, the concentration of endogenous IAA in YF1 flower buds was lower than that of NF1 under HT (Figure 5B). These results suggested that the reduction in auxin concentration is caused by a decrease in auxin metabolism gene expression, which may lead to anther defects such as anther indehiscence in YF1 under HT afterward.
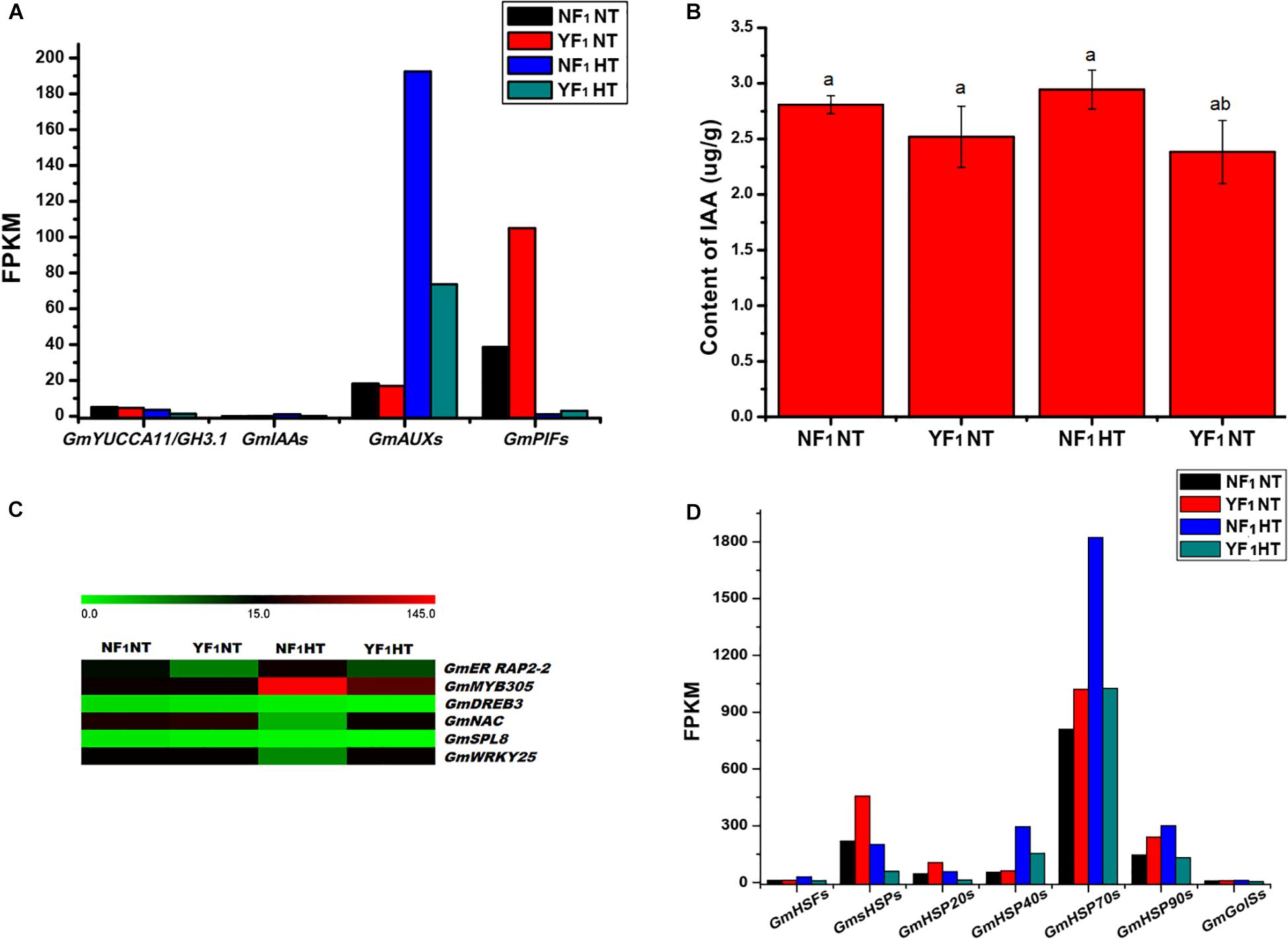
Figure 5. Effect of HT on auxin signaling, transcription factors, HSP and GolS. (A) DEGs related to auxin signaling in RNA-seq data. (B) IAA content in flower buds of NF1 and YF1 under NT and HT conditions (n = 3). Data are presented as means ± SD from independent biological replicates. Values with different letter indicates statistical differences (one-way ANOVA, Duncan, P < 0.05). (C) Heat map of the DEGs related to transcription factors in flower buds of NF1 and YF1 under NT and HT conditions. The color scale represents the relative transcript abundance of the DEGs in flower bud of soybean. The heat map was conducted using MeV 4.9 software. (D) DEGs related to HSF, HSP, and GolS in RNA-seq data.
TFs and HSP May Participate in the Male Fertility Regulation of Soybean CMS-Based F1 Under HT
Our transcriptomics analysis indicated that numerous HT-responding genes encoding TF are involved in heat signal transduction, such as heat shock factor (HSF), ethylene-responsive TF RAP2-2 (ER RAP2-2), myb-related protein 305 (MYB305), dehydration-responsive element-binding protein 3 (DREB3), NAC, squamosa promoter-binding-like protein 8 (SPL8), and WRKY25 (Figure 5C). As shown in Supplementary Figure 10, both transcriptomics and qRT-PCR analyses indicated that ER RAP2-2, MYB305, DREB3, and SPL8 were downregulated in YF1HT compared with NF1HT, and NAC and WRKY25 were activated by HT in YF1.
Remarkably, the rapid response to heat triggered downregulation of a substantial number of HSF and HSP genes in YF1HT (Figure 5D). The results showed that 55 DEGs about HSFs and HSPs were identified in the NT and HT comparison (Supplementary Table 9 and Supplementary Figure 11). Five GmHSFs, namely, four HSFA and one HSFB genes, were induced in NF1 but repressed in YF1 under HT. In this study, a total of 50 GmHSP genes were identified to be upregulated in NF1HT, including 25 small GmHSP (GmsHSP), 6 GmHSP20, 8 GmHSP40 (DnaJ protein, Georgopoulos et al., 1980), 8 GmHSP70, and 3 GmHSP90 genes.
Both GmHSFA2 and Its Downregulated Gene GmHSP20a Overexpression Conferred Tolerance to HT Stress During Flowering in Arabidopsis
According to the RNA-seq and qRT-PCR analyses, GmHSFA2 (Glyma.14G096800) was induced and inhibited by HT in NF1 and YF1, respectively (Supplementary Table 9). Its role in HT response was further analyzed. Bioinformatics analysis showed that GmHSFA2 had high sequence identity with AtHSFA2 and SoHSFA2, which contained a 1,095-bp ORF and predicted to encode 364 amino acids (Figures 6A,B). The alignment revealed that the GmHSFA2 has the typical domains of HSFA2, including a conserved DNA binding domain (DBD), an oligomerization domain (OD) with two adjacent hydrophobic heptad repeats (HR-A/B), a nuclear localization signal (NLS), and an AHA motif (Figure 6B). Subcellular localization analysis showed that the 35S:GmHSFA2-GFP fusion protein was exclusively localized in the nucleus, which was consistent with the predicted NLS domain between the OD and AHA motif (Figures 6B,C). GUS staining of three pGmHSFA2:GUS-transformed Arabidopsis lines confirmed that GmHSFA2 was expressed only in early-stage anthers of inflorescence during flowering (Figure 6D). The expression patterns of GmHSFA2 under the HT condition (40°C) were evaluated by qRT-PCR using RNA samples extracted from flower buds of soybean, and the NT condition (30°C) was used as a control. The expression level of GmHSFA2 increased significantly with time and peaked at the seventh day and then decreased rapidly after recovery with NT for 1 day (Supplementary Figure 12). This implies that GmHSFA2 was extremely sensitive to HT during flowering in soybean reproductive tissues.
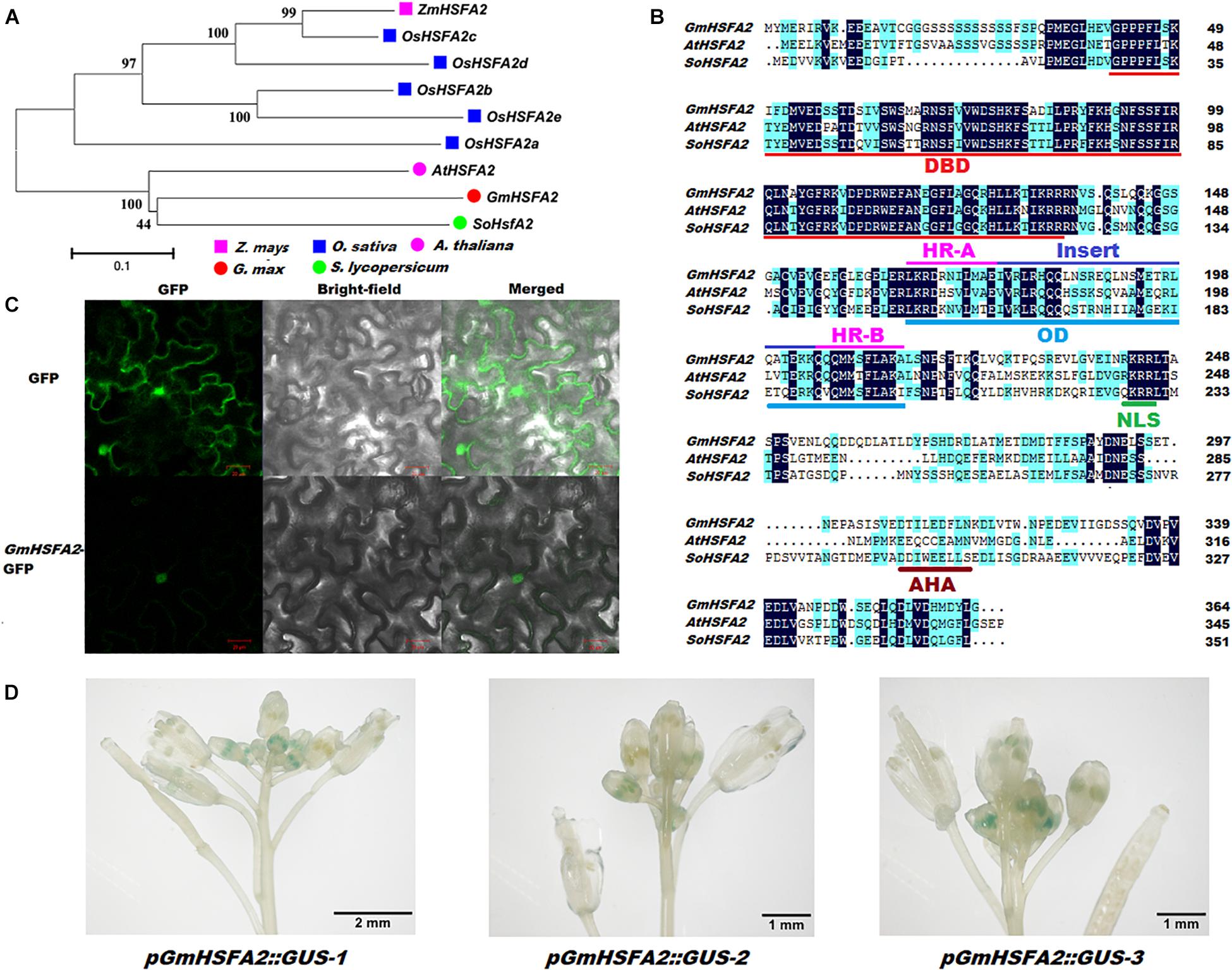
Figure 6. Identification of GmHSFA2. (A) Phylogenetic tree analysis of HSFA2 in five plant species. The phylogenetic tree was constructed using MEGA 5.02 based on the Neighbor-joining (NJ) method. Bootstrap values in percentage (1000 replicates) are labeled on the nodes. (B) Amino acids sequence comparison and conserved domains of GmHSFA2 and its closest orthologs AtHSFA2 and SoHSFA2 in Arabidopsis and tomato, respectively. Sequence alignment was performed with DNAMAN. Dark blue and light blue regions indicate identical and similar amino acids among the three sequences, respectively. DBD, DNA-binding domain; OD, oligomerization domain; HR, hydrophobic heptad repeats; NLS, nuclear localization signal; AHA motif, activator motif. (C) Subcellular localization of GmHSFA2. (D) pGmHSFA2:GUS expression pattern in inforescence with developing flower buds during flowering in Arabidopsis.
To further confirm the role of GmHSFA2 in HT tolerance during flowering, three lines of Arabidopsis overexpressing GmHSFA2 with different expression levels were selected for HT treatment (Figure 7A). In the HT tolerance assay (45°C for 3 days) during flowering, the top of the inflorescence of transgenic plants basically kept normal growth while that of the WT wilted (Supplementary Figure 13). Most importantly, HT treatment increased the stability of 35S:GmHSFA2 transgenic plants under HT stress (42°C for 4 h), which showed anther dehiscence and only a little pollen abortion after HT treatment for 2 and 6 days, respectively (Figures 7C,D), while the rate of stamen length/pistil length in both WT and transgenic lines decreased after 2 days of HT treatment (Figure 7E). However, the WT showed pollen shrinkage with anther indehiscence and male sterility (no pollen grains or most pollen abortion) after 2 and 6 days of HT treatment, respectively (Figures 7C,D,F).
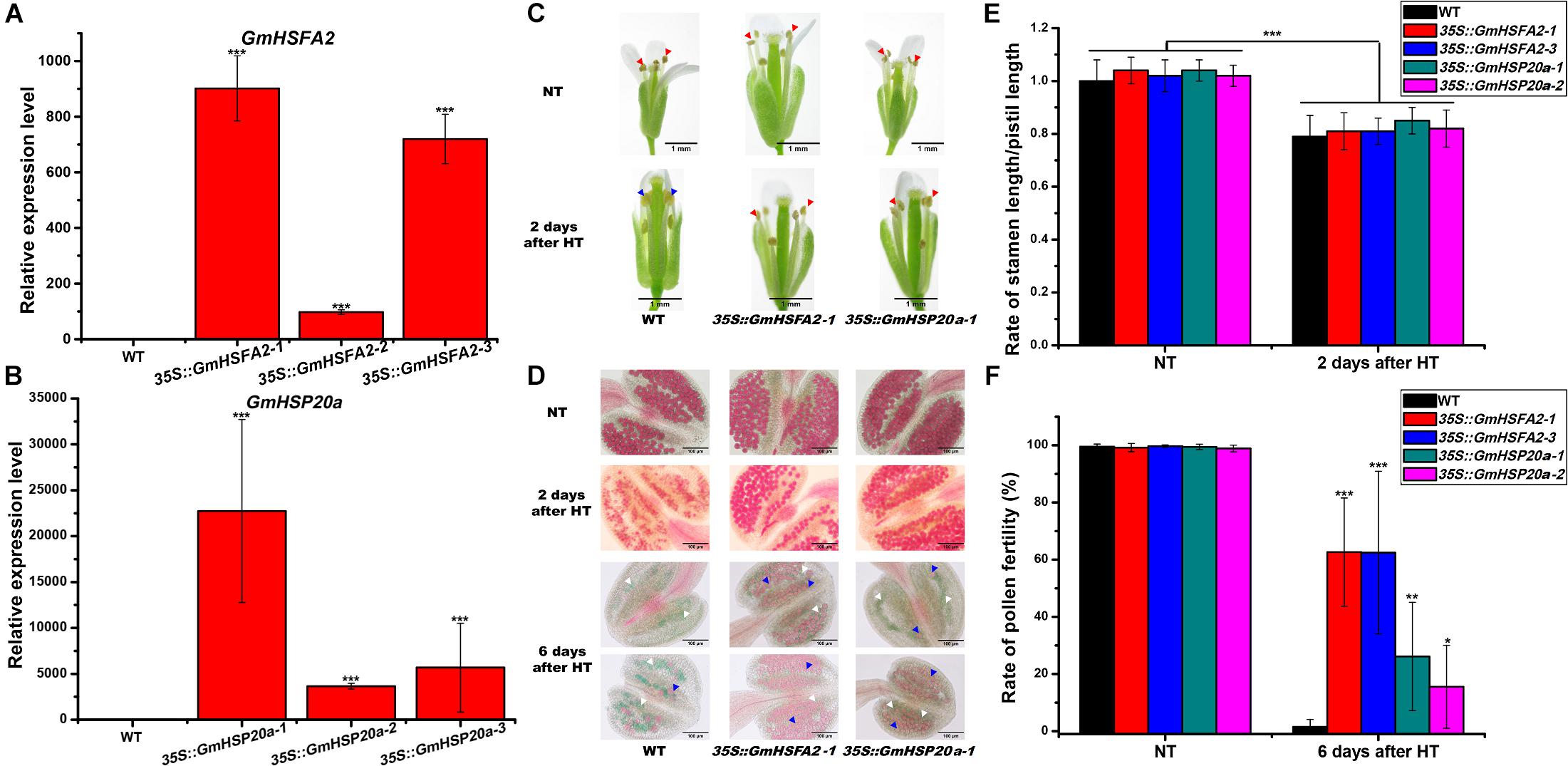
Figure 7. Effect of HT on male fertility in WT, 35S:GmHSFA2 and 35S:GmHSP20a lines. (A,B) GmHSFA2 and GmHSP20a expression levels in WT, 35S:GmHSFA2 and 35S:GmHSP20a lines. The WT under NT condition was used as the control and qRT-PCR data are expressed as the mean values ± SD of three independent biological replicates. (C) Phenotype of anthers in WT, 35S:GmHSFA2 and 35S:GmHSP20a lines under NT and HT conditions. The red and blue arrows indicate dehiscent and indehiscent anthers, respectively. (D) Phenotype of pollen in WT, 35S:GmHSFA2 and 35S:GmHSP20a lines under NT and HT conditions. The red pollens pointed by blue arrows and green pollens pointed by white arrows indicate fertile pollens and sterile pollens, respectively. (E,F) Calculation of rate of stamen length/pistil length and rate of pollen fertility in WT, 35S:GmHSFA2 and 35S:GmHSP20a lines under NT and HT conditions (n = 9). Asterisk indicates statistical differences, *P < 0.05; **P < 0.01; ***P < 0.001.
The expression levels of GmHSFA2 downstream regulatory genes (AtsHSP, AtHSP20, AtHSP40, AtHSP70, AtHSP90, AtGolS1, and AtGolS2) under HT stress during flowering in 35S:GmHSFA2 plants were compared by qRT-PCR analysis. The transcripts of most of them except AtHSP70 and AtHSP90 were all higher than those in the WT under the NT condition (Figure 8A). After HT treatment, all of these downstream regulator genes were upregulated in 35S:GmHSFA2 plants compared with WT (Figure 8A). Moreover, the fold changes in the expression levels of almost all genes (except AtHSP40) between the two materials under HT were higher than that under the NT condition (Figure 8A). Most importantly, both RNA-seq data and qRT-PCR analysis showed that their homologous genes in soybean NF1 were upregulated by HT induction (Figure 8B and Supplementary Figures 11D–J).
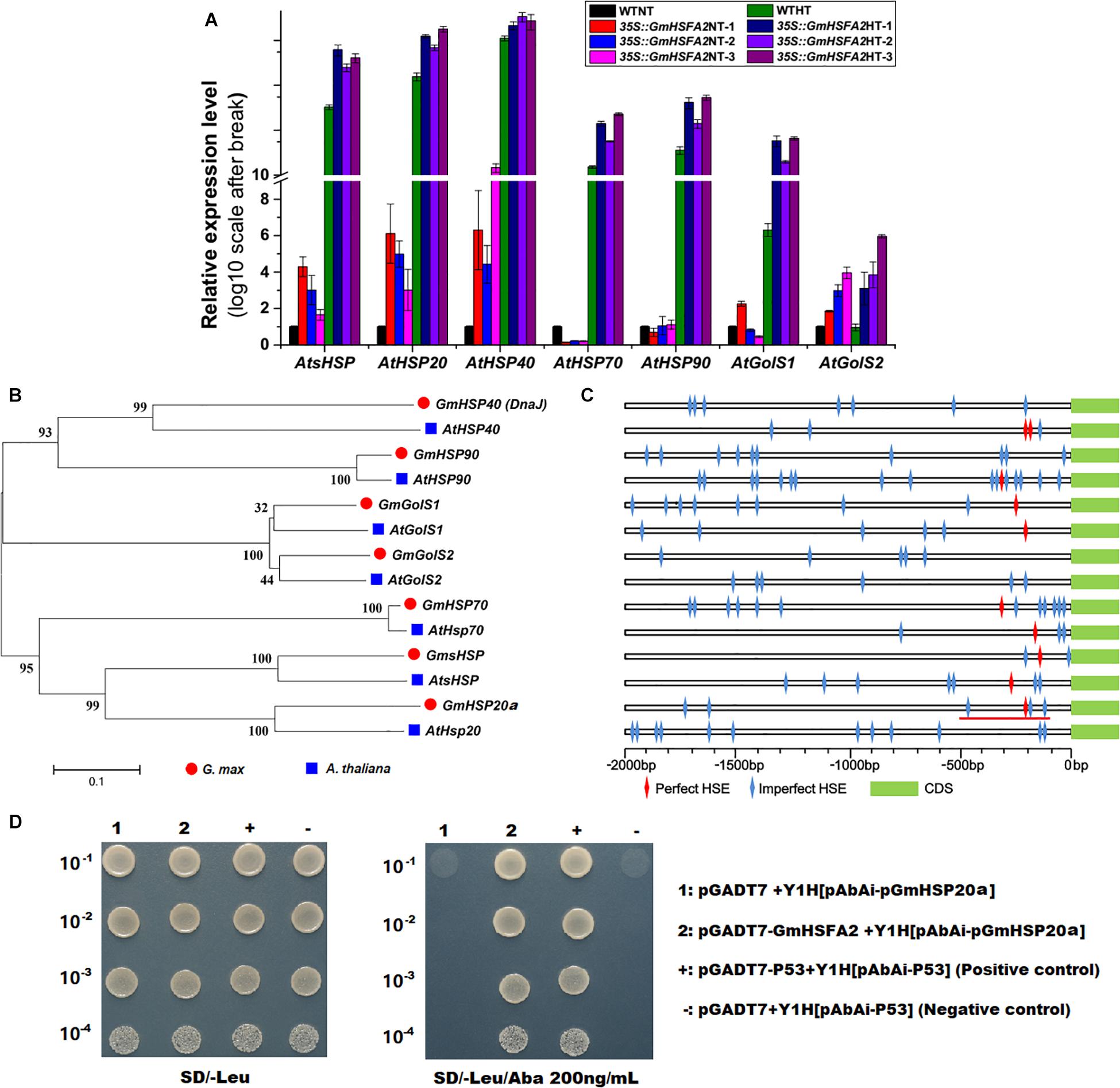
Figure 8. Effect of GmHSFA2 on its downstream regulatory genes. (A) Expression levels of HSP and GolS in 35S:GmHSFA2 Arabidopsis transgenic and WT plants under NT and HT conditions. The WT under NT condition was used as the control and qRT-PCR data are expressed as the mean values ± SD of three independent biological replicates. (B,C) Phylogenetic tree and schematic of HSP and GolS. The phylogenetic tree was constructed using MEGA 5.02 based on the NJ method. Bootstrap values in percentage (1000 replicates) are labeled on the nodes. Red line labeled cis-acting HSE motifs (–500 to –101 bp relative to the translational start codon of GmHSP20a) were used for cloning and Y1H assay. (D) The interaction between GmHSFA2 and the promoter of GmHSP20a in yeast. The relevant bacterial solution was diluted in gradient of 1:10, 1:100, 1:1000, and 1:10,000, and grown on medium.
Bioinformatics analysis showed that perfect and imperfect HSE motifs are distributed within promoter regions of selected HSPs and GolSs in both soybean and Arabidopsis (Figure 8C). Furthermore, we investigated a direct link between GmHSFA2 and the promoter of a selected GmHSP20a (Glyma.12G013100) by Y1H assay (Figure 8D). In addition, the expression trend of GmHSP20a was consistent with that of GmHSFA2 under HT stress (Supplementary Figure 12). Most importantly, the 35S:GmHSP20a Arabidopsis transgenic lines also improved HT tolerance during flowering (Supplementary Figure 13 and Figures 7B–D,F). All these results show that GmHSFA2 might improve the HT tolerance of soybean CMS-based F1 and transgenic Arabidopsis by regulating the expression changes of HSP and GolS.
Discussion
The CMS-based hybridization method has been widely used in plant hybrid breeding due to its effective way of hybrid seed production by use of the CMS line, maintainer line, and restorer line. However, increasing evidence has indicated that male fertility of CMS-based F1 is affected by climate conditions such as HT stress (Zhao et al., 2009; Zhang et al., 2019; Nie et al., 2017). In this study, two soybean CMS-based F1 combinations, NF1 and YF1, were employed, and it was found that the male fertility of YF1 was obviously damaged by HT, such as anther indehiscence and decreased pollen fertility, thereby decreasing soybean yield (Supplementary Figure 1). Furthermore, RNA-seq and functional study of GmHSFA2 were adopted to globally identify the DEGs and pathways participating in male fertility regulation of soybean CMS-based F1 under HT.
Abnormal Anther/Pollen Development Is Related to Male Fertility Instability of HT-Sensitive F1 Under HT
In our RNA-seq, many PL, PME, PG, and Exo-PG genes showed differential expression between NF1 and YF1 under the HT condition (Figures 4A–C). Among them, pectinase (PL, PME, and PG) is a key enzyme involved in the degradation of plant pectin and participates in the regulation of anther/pollen development (Micheli, 2001; Ogawa et al., 2009; Corral-Martínez et al., 2016; Li et al., 2019). It has been shown that pectinase activity was decreased in anthers of Qx-115 (anther indehiscent phenotype material of chrysanthemum) during anther development (Li et al., 2019). Pectinase has been extensively studied in many plants. Wei et al. (2019) found that PL, Exo-PG, and PME were related to the fertility restorer of the CMS line in pepper. In Brassica campestris, downregulation of BcPLL9 and BcPLL10 results in disorder of pectin metabolism in pollen and finally leads to male semi-sterility (Jiang et al., 2014a, b). Also, in B. campestris, Huang et al. (2009) found that a PG gene (BcMF2) was specifically expressed in the tapetum and pollen and that its inhibition led to pollen deformity with abnormal intine development. Except PLs and PGs, PMEs are also important for pollen development in plants. Recently, a CRISPR/Cas9 system-induced BcPME37c mutant has been characterized, and its mutation caused the abnormal thickening of the pollen intine in B. campestris (Xiong et al., 2019). The downregulation of pectinase genes in YF1HT may reduce the degradation of pectin, thus changing the maintenance of the anther wall, leading to anther indehiscence in YF1HT, and needs further research.
Carbohydrate Undersupply and Sugar Transport Blockage Are Two of the Main Causes for Male Fertility Instability in YF1HT
Our RNA-seq analysis found that compared with NF1HT, the expression of hundreds of DEGs related to carbohydrate metabolism in YF1HT was downregulated, including PLs, PMEs, and beta-glucosidase (Supplementary Table 7). In addition, many sugar transport-related DEGs, such as STP11, SWEET5, and SUC8, are also decreased in expression (Supplementary Table 7). Based on the determination of Suc, Glc, and starch contents, it is speculated that their reduction affected the male fertility of YF1 under HT stress (Figure 9). Moreover, similar results were found in tomato and cotton, where a decrease in sugar affected their male reproductive development under HT (Firon et al., 2006; Min et al., 2014).
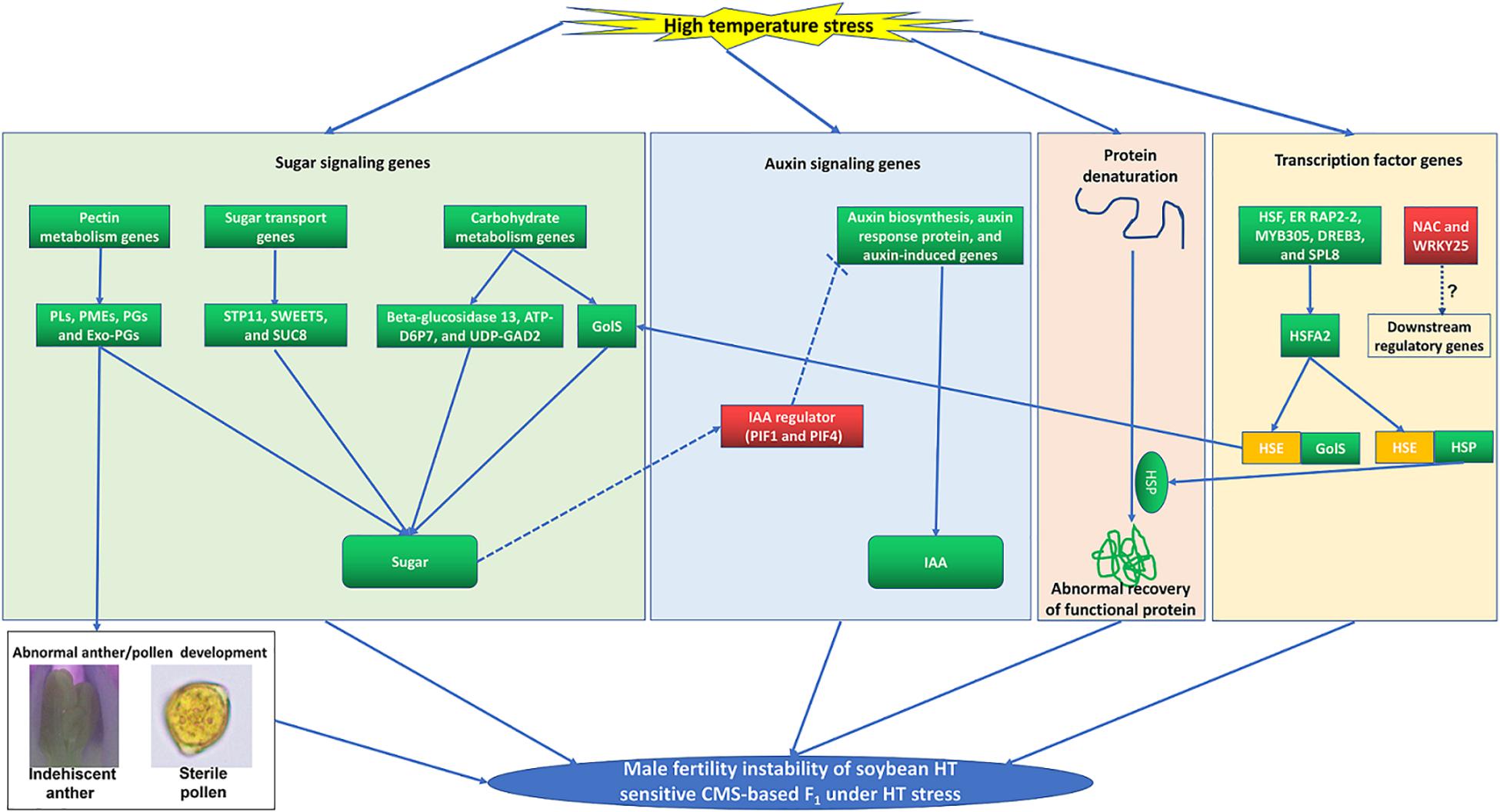
Figure 9. A proposed model incorporating the main processes that involved in the male fertility instability of soybean HT sensitive CMS-based F1 under HT stress. The up-regulated and down-regulated of gene or metabolite content are in red and green backgrounds, respectively.
Many studies have shown that genes related to either carbohydrate metabolism or sugar transport are associated with male sterility in plants. For example, our previous study found that male sterility of the soybean CMS line is associated with alterations in carbohydrate metabolism (Li et al., 2015). In cucumber, the downregulation of sugar transporters CsHT1 and CsSUT1 inhibits pollen germination and causes male sterility, respectively (Cheng et al., 2015; Sun et al., 2019). At the same time, they also protect against HT stress during pollen development (Frank et al., 2009; Min et al., 2013, 2014). Min et al. (2014) found that HT disrupted anther carbohydrate metabolism in cotton, including starch and Suc metabolism pathways, leading to abnormal male fertility development in H05 (HT-sensitive line) under HT. Further investigation demonstrated that GhCKI caused pollen abortion and anther indehiscence in cotton via inactivating starch synthase (Min et al., 2013). In tomato, HT-induced expressions of carbohydrate metabolism and sugar transport genes, such as sucrose phosphate synthase and sorbitol transporter, were involved in the HT response during pollen development (Frank et al., 2009). Thus, we speculate that carbohydrate undersupply and sugar transport blockage are two of the main causes for male fertility instability in YF1 under HT and need to be verified in future studies.
Sugar Signaling-PIF-Auxin Signaling Pathway May Underlie Instability of Male Fertility in YF1 Under HT
Sugar plays a vital role as a protector defending against HT stress during male reproductive organ development (Min et al., 2013, 2014). And auxin is also closely related to instability of male fertility in plants under HT stress (Sakata et al., 2010; Higashitani, 2013; Min et al., 2014; Ding et al., 2017). In barley and Arabidopsis, HT can induce downregulation of genes related to auxin biosynthesis (YUC2, YUC6, and TAA1/TIR2), resulting in a sharp decrease in endogenous auxin level and eventually anther abortion (Sakata et al., 2010). Furthermore, exogenous auxin could completely reverse the male sterility of barley and Arabidopsis under HT stress (Sakata et al., 2010). Previous research has uncovered a pathway where sugar signaling is involved in plant growth by regulating auxin metabolism through the PIF protein (Stewart et al., 2011; Min et al., 2014). The PIF protein is an IAA regulator (Sun et al., 2012, 2019), and it is also involved in the HT response (Leivar and Quail, 2011). Furthermore, PIF expression was induced by low content of sugar, which altered auxin metabolism afterward and led to male sterility in cotton and cucumber eventually (Min et al., 2014; Sun et al., 2019). Similar to the cotton male sterility induced by HT stress, the sugar content and the expression levels of PIFs (GmPIF1 and GmPIF4) in flower buds of YF1 were also altered under HT (Supplementary Table 8). Meanwhile, downregulated auxin signaling genes and content were observed in flower buds of YF1 under HT, indicating that PIF might act as a negative regulator of IAA biosynthesis, which is consistent with the results in Arabidopsis and cucumber (Sairanen et al., 2012; Sun et al., 2019). However, Min et al. (2014) showed that PIF acts as a positive regulator of HT-induced IAA biosynthesis in cotton. It appears that the sugar signaling-PIF-auxin signaling pathway acts as a master switch role during the male organ development under HT stress in soybean CMS-based F1, which needs further study (Figure 9).
TFs Is Required for Enhanced Activation of HT Stress Response and Increased Thermotolerance in Soybean CMS-Based F1
Transcription factors are central regulators of gene expression affecting plant HT responses (Li et al., 2018). Many TF families, including ER, MYB, DREB, SPL, and HSF, are involved in HT stress response and enhanced tolerance in both model and crop plants (Hong et al., 2009; El-Kereamy et al., 2012; Wan et al., 2014; Chao et al., 2017; Li et al., 2018). In our study, some TF family members were upregulated in NF1HT related to YF1HT, including GmER RAR2-2, GmMYB305, GmDREB3, GmSPL8, and HSFA2, which may confer tolerance to NF1 under HT stress (Figure 5C and Supplementary Table 9). However, some TF family members may play as negative regulators, such as GmNAC and GmWRKY25, which were upregulated in YF1HT compared with NF1HT (Figure 5C). In Arabidopsis, a NAC-like gene (AtAIF) was found to be an inhibitor that controls anther dehiscence (Shih et al., 2014). Similarly, the overexpression of GhWRKY22, GmWRKY45, and AtWRKY27 in Arabidopsis displayed the male fertility defect with decreased pollen viability (Mukhtar et al., 2017; Wang et al., 2019; Li et al., 2020). Most importantly, Dang et al. (2018) found that the overexpression of CaWRKY27 in Arabidopsis inhibited the scavenging of H2O2 and played a negative regulator role in HT stress.
Although great progress has been made in deciphering the response of TFs such as HSF to HT stress in Arabidopsis, maize, tomato, tall fescue, and other plants (Charng et al., 2007; Giorno et al., 2010; Fragkostefanakis et al., 2016; Wang et al., 2017; Gu et al., 2019), few HSF genes have been elucidated in soybean, especially on the stability of male fertility. Four GmHSFA2 genes (Glyma.13G105700, Glyma.14G096800, Glyma.17G053700, and Glyma.17G227600) were found in this study (Supplementary Table 10), and one of them (Glyma.17G227600) was overexpressed in Arabidopsis, showing the characteristics of HT and drought resistance during seedling in previous studies (Li et al., 2014). In this study, only GmHSF-30 (Glyma.14G096800 and GmHSFA2 in this study) was induced by HT in soybean CMS-based F1 flower buds during flowering at the mRNA level (Supplementary Figure 11A). Fragkostefanakis et al. (2016) found that HSFA2 is an important coactivator of HSFA1a during HT to control pollen viability by regulated HSP101 and HSP17.7C-CI in tomato. In rice, HSF and HSP genes including HSFA2a and HSP17.9A are highly induced in HT-tolerant material rather in HT-sensitive varieties during anthesis under HT stress (González-Schain et al., 2016). In tomato, HT induced expressions of HSFA2, sHSP genes, HSP70, and HSP101 during pollen development. In this study, both HSFA2 and HSP (sHSP, HSP20, HSP40, HSP70, and HSP70) were induced by HT stress during flower bud development. Most importantly, a functional study found that HSFA2 was directly involved in HT stress response and that inhibition of HSFA2 reduces the viability and germination rate of tomato pollen under HT (Giorno et al., 2010; Fragkostefanakis et al., 2016). It has been reported that HT stress causes male sterility by affecting anther dehiscence and pollen production at a specific stage in Arabidopsis (Kim et al., 2001), and similar results were obtained in this study. Most importantly, ectopically expressing GmHSFA2 enhanced HT tolerance in Arabidopsis, suggesting that it positively regulated HT tolerance during flowering in plants.
Our results suggest that GmHSFA2 is a key regulator in response to HT stress. However, its regulatory molecular mechanism in soybean is still unknown. Many studies have shown that HSF promotes HT tolerance by binding to the HSE motifs in the promoter of HSP and GolS (Busch et al., 2005; Kotak et al., 2007; von Koskull-Doering et al., 2007; Fragkostefanakis et al., 2016; Wang et al., 2017; Gu et al., 2019). Frank et al. (2009) found that HT induced expressions of HSF and GolS during pollen development in tomato. However, the relationship among them during pollen development under HT stress is still unknown, especially in soybean. In our study, multiple HSP and GolS genes, including sHSP, HSP20, HSP40, HSP70, HSP90, GolS1, and GolS2, were upregulated by overexpression of GmHSFA2 in Arabidopsis compared with WT under HT (Figure 8A). Most importantly, their homologous genes in soybean were also upregulated in NF1HT related to YF1HT, according to the RNA-seq and qRT-PCR analyses (Figure 5D). Furthermore, multiple HSE motifs were found in their promoters, and the Y1H assay revealed that there was a direct link between GmHSFA2 and the promoter of GmHSP20a, indicating that GmHSFA2 could regulate these genes (Figure 9). And HSP and helper molecular chaperones can help inactivated proteins reassemble into active high-level structures and maintain normal cell functions (Scharf et al., 2012). Most importantly, overexpression of GmHSP20a in Arabidopsis also conferred plant HT tolerance during flowering (Figures 7C,D,F). However, its HT tolerance was lower than that of 35S:GmHSFA2 transgenic plants under HT stress, indicating that GmHSP20a was only one of the downstream regulator genes of GmHSFA2. All the above results revealed that a complex TF regulatory network exists in soybean CMS-based F1 (Figure 9). As a key regulator in response to HT stress, the regulation mechanism of GmHSFA2 in soybean needs to be explicated further.
Data Availability Statement
The datasets generated by this study can be found in the NCBI using accession number PRJNA677945.
Author Contributions
XD and SY conceived and designed the experiments. XD, QG, and QL performed the experiments. XD wrote the manuscript. SY and JG revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key R&D Program of China (2016YFD0101500 and 2016YFD0101504), the Fundamental Research Funds for the Central Universities (KYT201801), and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.600217/full#supplementary-material
Abbreviations
ATP-D6P7, ATP-dependent 6-phosphofructokinase 7; CMS, cytoplasmic male sterility; DEGs, differentially expressed genes; DREB, dehydration-responsive element binding; Exo-PG, exopolygalacturonase; Glc, glucose; GolS, galactinol synthase; ER, ethylene responsive; FPKM, fragments per kilobase of transcript per million mapped reads; GFP, green fluorescent protein; GO, Gene Ontology; GUS, β-glucuronidase; HT, high-temperature; HSF, heat shock factor; HSP, heat shock protein; KEGG, Kyoto Encyclopedia of Genes and Genomes; NJ, Neighbor joining; NT, Normal temperature; PCA, principal component analysis; PG, polygalacturonase; PIF, PHYTOCHROME-INTERACTING FACTOR; PL, pectate lyase; PME, pectin methylesterase; qRT-PCR, quantitative real-time PCR; RCA, repeated correlation analysis; SD, standard deviation; SPL, squamosa promoter-binding protein-like; Suc, sucrose; TFs, transcription factors; UDP-GAD, UDP-glucuronic acid decarboxylase; Y1H, yeast one-hybrid; WT, wild-type.
Footnotes
- ^ http://www.r-project.org/
- ^ http://www.geneontology.org/
- ^ http://www.kegg.jp/kegg/
- ^ https://www.soybase.org/
References
Begcy, K., Nosenko, T., Zhou, L. Z., Fragner, L., Weckwerth, W., and Dresselhaus, T. (2019). Male sterility in maize after transient heat stress during the tetrad stage of pollen development. Plant Physiol. 181, 683–700. doi: 10.1104/pp.19.00707
Busch, W., Wunderlich, M., and Schöffl, F. (2005). Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 41, 1–14. doi: 10.1111/j.1365-313X.2004.02272.x
Chao, L. M., Liu, Y. Q., Chen, D. Y., Xue, X. Y., Mao, Y. B., and Chen, X. Y. (2017). Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 10, 735–748. doi: 10.1016/j.molp.2017.03.010
Charng, Y. Y., Liu, H. C., Liu, N. Y., Chi, W. T., Wang, C. N., Chang, S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Chen, L. T., and Liu, Y. G. (2014). Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Cheng, J. T., Wang, Z. Y., Yao, F. Z., Gao, L. H., Ma, S., Sui, X. L., et al. (2015). Down-regulating CsHT1, a cucumber pollen-specific hexose transporter, inhibits pollen germination, tube growth, and seed development. Plant Physiol. 168, 635–647. doi: 10.1104/pp.15.00290
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Corral-Martínez, P., García-Fortea, E., Bernard, S., Driouich, A., and Seguí-Simarro, J. M. (2016). Ultrastructural immunolocalization of arabinogalactan protein, pectin and hemicellulose epitopes through anther development in Brassica napus. Plant Cell Physiol. 57, 2161–2174. doi: 10.1093/pcp/pcw133
Dang, F. F., Lin, J. H., Xue, B. P., Chen, Y. P., Guan, D. Y., Wang, Y. F., et al. (2018). CaWRKY27 negatively regulates H2O2-mediated thermotolerance in pepper (Capsicum annuum). Front. Plant Sci. 9:1633. doi: 10.3389/fpls.2018.01633
Ding, X. L., Li, J. J., Zhang, H., He, T. T., Han, S. H., Li, Y. W., et al. (2016). Identification of miRNAs and their targets by high-throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genomics 17:24. doi: 10.1186/s12864-015-2352-0
Ding, X. L., Ruan, H., Yu, L. F., Li, Q., Song, Q. J., Yang, S. P., et al. (2020). miR156b from soybean CMS line modulates floral organ development. J. Plant Biol. 63, 141–153. doi: 10.1007/s12374-020-09237-7
Ding, Y. H., Ma, Y. Z., Liu, N., Xu, J., Hu, Q., Li, Y. Y., et al. (2017). microRNAs involved in auxin signaling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 91, 977–994. doi: 10.1111/tpj.13620
Djanaguiraman, M., Perumal, R., Jagadish, S. V. K., Ciampitti, I. A., Welti, R., and Prasad, P. V. V. (2018). Sensitivity of sorghum pollen and pistil to high-temperature stress. Plant Cell Environ. 41, 1065–1082. doi: 10.1111/pce.13089
El-Kereamy, A., Bi, Y. M., Ranathunge, K., Beatty, P. H., Good, A. G., and Rothstein, S. J. (2012). The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One 7:e52030. doi: 10.1371/journal.pone.0052030
Firon, N., Shaked, R., Peet, M. M., Pharr, D. M., Zamski, E., Rosenfeld, K., et al. (2006). Pollen grains of heat tolerant tomato cultivars retain higher carbohydrate concentration under heat stress conditions. Sci. Hortic. 109, 212–217. doi: 10.1016/j.scienta.2006.03.007
Fragkostefanakis, S., Mesihovic, A., Simm, S., Paupière, M. J., Hu, Y., Paul, P., et al. (2016). HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol. 170, 2461–2477. doi: 10.1104/pp.15.01913
Frank, G., Pressman, E., Ophir, R., Althan, L., Shaked, R., Freedman, M., et al. (2009). Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot. 60, 3891–3908. doi: 10.1093/jxb/erp234
Georgopoulos, C. P., Heil, A. L., Yochem, J., and Feiss, M. (1980). Identification of the E. coli dnaJ gene product. Mol. Gen. Genet. 178, 583–588. doi: 10.1007/bf00337864
Giorno, F., Wolters-Arts, M., Grillo, S., Scharf, K. D., Vriezen, W. H., and Mariani, C. (2010). Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot. 61, 453–462. doi: 10.1093/jxb/erp316
González-Schain, N., Dreni, L., Lawas, L. M., Galbiati, M., Colombo, L., Heuer, S., et al. (2016). Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol. 57, 57–68. doi: 10.1093/pcp/pcv174
Gu, L., Jiang, T., Zhang, C. X., Li, X. D., Wang, C. M., Zhang, Y. M., et al. (2019). Maize HSFA2 and HSBP2 antagonistically modulate raffinose biosynthesis and heat tolerance in Arabidopsis. Plant J. 100, 128–142. doi: 10.1111/tpj.14434
Higashitani, A. (2013). High temperature injury and auxin biosynthesis in microsporogenesis. Front. Plant Sci. 4:47. doi: 10.3389/fpls.2013.00047
Hong, B., Ma, C., Yang, Y. J., Wang, T., Yamaguchi-Shinozaki, K., and Gao, J. P. (2009). Over-expression of AtDREB1A in chrysanthemum enhances tolerance to heat stress. Plant Mol. Biol. 70, 231–240. doi: 10.1007/s11103-009-9468-z
Huang, L., Cao, J. S., Zhang, A. H., Ye, Y. Q., Zhang, Y. C., and Liu, T. T. (2009). The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. J. Exp. Bot. 60, 301–313. doi: 10.1093/jxb/ern295
Jefferson, R., Kavanagh, T., Bevan, M. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jiang, J. J., Yao, L. N., Yu, Y. J., Liang, Y., Jiang, J. X., Ye, N. H., et al. (2014a). PECTATE LYASE-LIKE 9 from Brassica campestris is associated with intine formation. Plant Sci. 229, 66–75. doi: 10.1016/j.plantsci.2014.08.008
Jiang, J. J., Yao, L. N., Yu, Y. J., Lv, M. L., Miao, Y., and Cao, J. S. (2014b). PECTATE LYASE-LIKE10 is associated with pollen wall development in Brassica campestris. J. Integr. Plant Biol. 56, 1095–1105. doi: 10.1111/jipb.12209
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kim, S. Y., Hong, C. B., and Lee, I. (2001). Heat shock stress causes stage-specific male sterility in Arabidopsis thaliana. J. Plant Res. 114, 301–307. doi: 10.1007/PL00013991
Kotak, S., Larkindale, J., Lee, U., von Koskull-Döring, P., Vierling, E., and Scharf, K. D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. doi: 10.1016/j.pbi.2007.04.011
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, B. J., Gao, K., Ren, H. M., and Tang, W. Q. (2018). Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 60, 757–779. doi: 10.1111/jipb.12701
Li, C., Liu, X. Y., Ruan, H., Zhang, J. Y., Xie, F. B., Gai, J. Y., et al. (2020). GmWRKY45 enhances tolerance to phosphate starvation and salt stress, and changes fertility in transgenic Arabidopsis. Front. Plant Sci. 10:1714. doi: 10.3389/fpls.2019.01714
Li, J. J., Han, S. H., Ding, X. L., He, T. T., Dai, J. Y., Yang, S. P., et al. (2015). Comparative transcriptome analysis between the cytoplasmic male sterile line NJCMS1A and its maintainer NJCMS1B in soybean (Glycine max (L.) Merr.). PLoS One 10:e0126771. doi: 10.1371/journal.pone.0126771
Li, P. S., Yu, T. F., He, G. H., Chen, M., Zhou, Y. B., Chai, S. C., et al. (2014). Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics 15:1009. doi: 10.1186/1471-2164-15-1009
Li, Q., Wu, Z., Wu, H. J., Fang, W. M., Chen, F. D., and Teng, N. J. (2019). Transcriptome profiling unravels a vital role of pectin and pectinase in anther dehiscence in Chrysanthemum. Int. J. Mol. Sci. 20:5865. doi: 10.3390/ijms20235865
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Micheli, F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419. doi: 10.1016/S1360-1385(01)02045-3
Min, L., Li, Y. Y., Hu, Q., Zhu, L. F., Gao, W. H., Wu, Y. L., et al. (2014). Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 164, 1293–1308. doi: 10.1104/pp.113.232314
Min, L., Zhu, L. F., Tu, L. L., Deng, F. L., Yuan, D. J., and Zhang, X. L. (2013). Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J. 75, 823–835. doi: 10.1111/tpj.12245
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mukhtar, M. S., Liu, X. Y., and Somssich, I. E. (2017). Elucidating the role of WRKY27 in male sterility in Arabidopsis. Plant Signal. Behav. 12:e1363945. doi: 10.1080/15592324.2017.1363945
Nie, Z. X., Zhao, T. J., Liu, M. F., Dai, J. Y., He, T. T., Lyu, D., et al. (2019). Molecular mapping of a novel male-sterile gene msNJ in soybean [Glycine max (L.) Merr.]. Plant Reprod. 32, 371–380. doi: 10.1007/s00497-019-00377-6
Nie, Z. X., Zhao, T. J., Yang, S. P., and Gai, J. Y. (2017). Development of a cytoplasmic male-sterile line NJCMS4A for hybrid soybean production. Plant Breed. 136, 516–525. doi: 10.1111/pbr.12488
Ogawa, M., Kay, P., Wilson, S., and Swain, S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233. doi: 10.1105/tpc.108.063768
Puteh, A. B., Thuzar, M., Mohd Monjurul, A. M., Abdullah, N. A. P. B., and Mohd Ridzwan, A. H. (2013). Soybean [Glycine max (L.) Merrill] seed yield response to high temperature stress during reproductive growth stages. Aust. J. Crop Sci. 7, 1472–1479.
Sairanen, I., Novák, O., Pěnčík, A., Ikeda, Y., Jones, B., Sandberg, G., et al. (2012). Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24, 4907–4916. doi: 10.1105/tpc.112.104794
Sakata, T., Oshino, T., Miura, S., Tomabechi, M., Yuta, T., Higashitani, N., et al. (2010). Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. U.S.A. 107, 8569–8574. doi: 10.1073/pnas.1000869107
Scharf, K. D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Shih, C. F., Hsu, W. H., Peng, Y. J., and Yang, C. H. (2014). The NAC-like gene ANTHER INDEHISCENCE FACTOR acts as a repressor that controls anther dehiscence by regulating genes in the jasmonate biosynthesis pathway in Arabidopsis. J. Exp. Bot. 65, 621–639. doi: 10.1093/jxb/ert412
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Stewart, J. L., Maloof, J. N., and Nemhauser, J. L. (2011). PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS One 6:e19894. doi: 10.1371/journal.pone.0019894
Sun, J. Q., Qi, L. L., Li, Y. N., Chu, J. F., and Li, C. Y. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8:e1002594. doi: 10.1371/journal.pgen.1002594
Sun, L. L., Sui, X. L., Lucas, W. J., Li, Y. X., Feng, S., Ma, S., et al. (2019). Down-regulation of the sucrose transporter CsSUT1 causes male sterility by altering carbohydrate supply. Plant Physiol. 180, 986–997. doi: 10.1104/pp.19.00317
Teixeira, E. I., Fischer, G., van Velthuizen, H., Walter, C., and Ewert, F. (2013). Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 170, 206–215. doi: 10.1016/j.agrformet.2011.09.002
von Koskull-Doering, P., Scharf, K. D., and Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Wan, L. Y., Wu, Y. S., Huang, J. Q., Dai, X. F., Lei, Y., Yan, L. Y., et al. (2014). Identification of ERF genes in peanuts and functional analysis of AhERF008 and AhERF019 in abiotic stress response. Funct. Integr. Genomics 14, 467–477. doi: 10.1007/s10142-014-0381-4
Wang, W., Wang, L., Chen, C., Xiong, G. Y., Tan, X. Y., Yang, K. Z., et al. (2011). Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 62, 5161–5177. doi: 10.1093/jxb/err221
Wang, X. D. (2019). Overview of the study and application of cytoplasmic male sterility in cotton. Sci. Agric. Sin. 52, 1341–1354. doi: 10.3864/j.issn.0578-1752.2019.08.005
Wang, X. Y., Huang, W. L., Liu, J., Yang, Z. M., and Huang, B. R. (2017). Molecular regulation and physiological functions of a novel FaHsfA2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnol. J. 15, 237–248. doi: 10.1111/pbi.12609
Wang, Y., Li, Y., He, S. P., Gao, Y., Wang, N. N., Lu, R., et al. (2019). A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol. Biochem. 141, 231–239. doi: 10.1016/j.plaphy.2019.06.005
Wei, B. Q., Wang, L. L., Bosland, P. W., Zhang, G. Y., and Zhang, P. (2019). Comparative transcriptional analysis of Capsicum flower buds between a sterile flower pool and a restorer flower pool provides insight into the regulation of fertility restoration. BMC Genomics 20:837. doi: 10.1186/s12864-019-6210-3
Xie, P. D. (2008). Review of soybean male sterility and heterosis utilization. J. Shenyang Agric. Univ. 39, 131–136.
Xiong, X. P., Zhou, D., Xu, L. A., Liu, T. T., Yue, X. Y., Liu, W. M., et al. (2019). BcPME37c is involved in pollen intine formation in Brassica campestris. Biochem. Biophys. Res. Commun. 517, 63–68. doi: 10.1016/j.bbrc.2019.07.009
Zhang, M., Zhang, X. X., Guo, L. P., Qi, T. X., Liu, G. Y., Feng, J. J., et al. (2019). Single-base resolution methylomes of cotton CMS system reveal epigenomic changes in response to high-temperature stress during anther development. J. Exp. Bot. 71, 951–969. doi: 10.1093/jxb/erz470
Zhao, Z. W., Zeng, Z. H., Chen, X., Jiang, X. Y., Lei, K. R., and Yao, N. (2009). Influences of natural high-temperature stress on pollen fertility and main agronomic traits of hybrid Rice. Acta Agric. Jiangxi 21, 19–21. doi: 10.3969/j.issn.1001-8581.2009.08.008
Keywords: soybean, CMS-based F1, HT stress, RNA-seq, HSFA2
Citation: Ding X, Guo Q, Li Q, Gai J and Yang S (2020) Comparative Transcriptomics Analysis and Functional Study Reveal Important Role of High-Temperature Stress Response Gene GmHSFA2 During Flower Bud Development of CMS-Based F1 in Soybean. Front. Plant Sci. 11:600217. doi: 10.3389/fpls.2020.600217
Received: 29 August 2020; Accepted: 18 November 2020;
Published: 15 December 2020.
Edited by:
Rosa M. Rivero, Spanish National Research Council, SpainReviewed by:
BihO Cao, South China Agricultural University, ChinaMonika Dalal, Indian Council of Agricultural Research (ICAR), India
Ling Min, Huazhong Agricultural University, China
Copyright © 2020 Ding, Guo, Li, Gai and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouping Yang, c3B5dW5nQDEyNi5jb20=
 Xianlong Ding
Xianlong Ding Qingling Guo
Qingling Guo Junyi Gai
Junyi Gai Shouping Yang
Shouping Yang