
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 15 January 2021
Sec. Plant Membrane Traffic and Transport
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.599944
This article is part of the Research Topic Plant Membrane Transport and Ionic Homeostasis in Hostile Soils View all 8 articles
Ca2+ is a second messenger that mediates plant responses to abiotic stress; Ca2+ signals need to be decoded by Ca2+ sensors that translate the signal into physiological, metabolic, and molecular responses. Recent research regarding the Ca2+ sensor CALCINEURIN B-LIKE PROTEIN 10 (CBL10) has resulted in important advances in understanding the function of this signaling component during abiotic stress tolerance. Under saline conditions, CBL10 function was initially understood to be linked to regulation of Na+ homeostasis, protecting plant shoots from salt stress. During this process, CBL10 interacts with the CBL-interacting protein kinase 24 (CIPK24, SOS2), this interaction being localized at both the plasma and vacuolar (tonoplast) membranes. Interestingly, recent studies have exposed that CBL10 is a regulator not only of Na+ homeostasis but also of Ca2+ under salt stress, regulating Ca2+ fluxes in vacuoles, and also at the plasma membrane. This review summarizes new research regarding functions of CBL10 in plant stress tolerance, predominantly salt stress, as this is the most commonly studied abiotic stress associated with the function of this regulator. Special focus has been placed on some aspects that are still unclear. We also pay particular attention on the proven versatility of CBL10 to activate (in a CIPK-dependent manner) or repress (by direct interaction) downstream targets, in different subcellular locations. These in turn appear to be the link through which CBL10 could be a key master regulator of stress signaling in plants and also a crucial participant in fruit development and quality, as disruption of CBL10 results in inadequate Ca2+ partitioning in plants and fruit. New emerging roles associated with other abiotic stresses in addition to salt stress, such as drought, flooding, and K+ deficiency, are also addressed in this review. Finally, we provide an outline of recent advances in identification of potential targets of CBL10, as CBL10/CIPKs complexes and as CBL10 direct interactions. The aim is to showcase new research regarding this master regulator of abiotic stress tolerance that may be essential to the maintenance of crop productivity under abiotic stress. This is particularly pertinent when considering the scenario of a projected increase in extreme environmental conditions due to climate change.
Plants have evolved a complex system to interact with the environment during evolution, which is able to perceive, transduce, and trigger responses to stresses at molecular, cellular, and physiological levels. In this system, calcium ion (Ca2+) plays a key role as a second messenger in plant responses to a wide array of environmental stimuli, including abiotic and biotic stress. Advancing in the knowledge of Ca2+ -signaling processes is essential in breeding programs for developing crops with enhanced tolerance to adverse environmental conditions. A Ca2+ signal is generated when a controlled increase of cytosolic Ca2+ occurs. Rise of cytosolic Ca2+ is derived either from intracellular stores or from the apoplast. The nature of the stimulus has an influence on the frequency, amplitude, and shape of the Ca2+ signal (Dodd et al., 2010). The stimulus generates temporal variations in Ca2+ concentration in cytosol, in such a way that the cation encodes precise information, the so-called Ca2+ signature. This signature defines the nature and magnitude of the response (Allen et al., 2001; McAinsh and Pittman, 2009). The sources of this Ca2+ are the plasma membrane, vacuole, and endoplasmic reticulum (Peiter, 2011). In this picture, activating or deactivating the different components of Ca2+ homeostasis (ionic channels or transporters) is requested to generate such Ca2+ signal.
For the initiation of the signaling triggered by Ca2+ and the activation of cellular and physiological responses, appropriate Ca2+ sensors are needed to detect changes in these Ca2+ transients. These sensors are Ca2+ -binding proteins and are grouped in three major families: calmodulin (CaM) and CaM-like proteins, Ca2+ -dependent protein kinases, and calcineurin B–like (CBL) proteins. This last family is constituted by EF-hand Ca2+ sensors that present the highest similarity with the regulatory B subunit of phosphatase calcineurin (CNB) protein found in animals. CBLs act as “sensor relays” undergoing conformational changes upon Ca2+ binding, allowing for the binding and activation of a family of serine/threonine protein kinases designated as CBL-interacting protein kinases (CIPKs; recently reviewed in Manishankar et al., 2018; Saito and Uozumi, 2020; Tang et al., 2020). CBL/CIPK complexes have been extensively studied by means of the analysis of mutants in Arabidopsis, and it has been demonstrated that these complexes mediate Ca2+ signals elicited by different environmental stimuli such as cold, abscisic acid (ABA), salinity, osmotic stress, low K+ concentration, high pH, and pathogen infection, among others (Kim, 2013). As a result, different CBL/CIPK combinations may engender temporal and spatial specificity in Ca2+ signaling and are able to connect diverse stimuli to define cellular responses (Batistič and Kudla, 2012). The intracellular localization of the CBL/CIPK complexes (plasma membrane, nucleus, vacuolar membrane, endoplasmic reticulum, or cytoplasm) is determined by CBLs allowing for spatial specificity in target recognition (D’Angelo et al., 2006; Batistič and Kudla, 2009; Batistič et al., 2010). But binding CIPKs is not the only way of action of CBLs as it has been reported that they may interact in a direct way with target proteins, in a CIPK-independent manner, negatively regulating their activity (Ren et al., 2013; Cho et al., 2016). CALCINEURIN B-LIKE PROTEIN 10 (CBL10), also known as SOS3-LIKE Ca2+ BINDING PROTEIN8 (SCABP8), has been recently identified in Arabidopsis (Kim et al., 2007; Quan et al., 2007), and recent research shows its function is associated with regulation of ion homeostasis, protecting plant shoots from salt stress. In recent years, new roles in several biotic and abiotic stresses have been attributed to CBL10. In addition, in recent years, progress in establishing the mechanisms of action of CBL10 has been particularly fast, and it has provided a wealth of information on plant cell signaling, which is summarized below.
As already said, CBL10 was first identified as a crucial regulator of salt tolerance in Arabidopsis (Kim et al., 2007; Quan et al., 2007). The identification and characterization of the first two T-DNA cbl10 mutants (Atcbl10 mutants) by these research groups showed that disruption of the CBL10 gene caused salt hypersensitive phenotypes specifically in shoots, whereas root growth was significantly less affected. In a first phase, salinity reduced chlorophyll content of rosette leaves and inhibited stem elongation of adult mutant plants; later on, the collapsed stems wilted and dried, and finally mutant plants died. The expression profile of the CBL10 gene detected in Arabidopsis wild-type (WT) plants supported such specific shoot salt sensitivity observed in the mutant plants, as transcript amounts were much higher in shoots than in roots (Kim et al., 2007; Quan et al., 2007). Following these pioneering studies, CBL10 has been identified in other species as a crucial regulator in shoot response to salt toxicity. Two CBL10 homologs have been identified in poplar (Populus trichocarpa), PtCBL10A and PtCBL10B (Li et al., 2013; Tang et al., 2014); the first one was ubiquitously expressed at low levels, whereas the second was mainly expressed in the green-aerial tissues. Salt tolerance by maintaining ion homeostasis in shoot tissues is conferred by overexpression of either PtCBL10A or PtCBL10B, indicating a key role for the gene products in plant response to salt toxicity in shoot. Furthermore, expression of either one in Arabidopsis cbl10 mutants is able to rescue its salt-sensitive shoot phenotype. Other studies have shown that the heterologous overexpression of homolog CBL10s from different species such as tobacco (Dong et al., 2015) enhances the salt tolerance of Arabidopsis.
After identification of the Arabidopsis cbl10 mutants, the only other identified and characterized cbl10 mutant has been a tomato T-DNA mutant (Slcbl10 mutant), identified by our research group. We observed that in this tomato mutant CBL10 disruption induced severe alterations in the shoot apex at different developmental stages (Egea et al., 2018). Thus, in both in vitro and in vivo salt stress assays, the Slcbl10 mutant exhibited shoot growth inhibition, hypocotyl thickening, chlorosis at the edge of young leaves, and apical necrosis, and finally plants died because of apical collapse. In agreement with previous studies of Atcbl10 mutants (Kim et al., 2007; Quan et al., 2007), the SlCBL10 gene was mainly expressed in aerial tissues of tomato plants. Moreover, expression analysis revealed an upregulation of SlCBL10 in all vegetative tissues in plants subjected to salinity, especially in upper mature leaves (first fully developed leaves from the apex). Our study demonstrated that the transcriptional activity of SlCBL10 in this specific tissue is critically involved in the adaptive response of tomato plants to salt stress as it protects shoot apical meristems and growing tissues from physiological damage caused by salinity. In addition, molecular complementation assays proved that SlCBL10 is orthologous to the Arabidopsis CBL10 gene and that it rescues the shoot salt-sensitive phenotype of the Arabidopsis cbl10 mutant.
One of the latest studies on CBL10 has been performed in Eutrema (Eutrema salsugineum; formerly Thellungiella halophila), a halophytic relative of Arabidopsis, used as a model for understanding plant adaptation to soil salinity (Monihan et al., 2019). In this study, CBL10 gene (EsCBL10) was found to be duplicated (EsCBL10a and EsCBL10b), linking this duplication to the high salt tolerance of Eutrema, as this species maintains its growth in salt-affected soils where most crop plants are not able to grow. Downregulation, using amiRNAs lines, of either of the duplicated EsCBL10 genes individually reduced growth in salt stress conditions, hinting that both genes operate in response to salinity. If downregulation of both EsCBL10 genes is combined, an even greater decrease in growth occurred. This observation suggests that the two genes have either additive effects or different functions, so increasing the number and expanding the activities of this Ca2+ sensor via gene duplication confers an adaptive benefit for plant growth in saline soils. While similar levels of EsCBL10a transcripts were found throughout the plant (roots, leaves, and flowers), expression profile of EsCBL10b was more similar to that of AtCBL10 in Arabidopsis; both transcripts were detected in higher amounts in leaves than in roots. However, when transcript accumulation was compared between leaves and flowers, levels of EsCBL10b transcripts were similar in both organs, while transcripts of AtCBL10 were mainly detected in leaves and only weakly in flowers. Surprisingly, in tomato plants, the highest expression level of CBL10 was found in flowers (Egea et al., 2018). In fact, tomato Slcbl10 (Egea et al., 2018) and also Arabidopsis Atcbl10 (Monihan et al., 2016) displayed an altered flower development under salt stress conditions, which in the case of the tomato mutant led to a significant reduction in fruit yield and quality. Taken together, CBL10 is a key gene involved in protection of the shoot, of vegetative, and reproductive organs under saline conditions, and this function seems to be conserved among species. However, more studies are needed to elucidate the specific function of CBL10 in flower development as this has been attributed to an indirect effect of the altered function of CBL10 in vegetative tissues.
The main effect induced by salinity is the toxic effect; this is widely recognized in numerous species to be largely due to Na+, as the accumulation of this cation in cytoplasm interferes with multiple metabolic processes (Munns and Tester, 2008). Thus, plants with either less uptake or increased cellular efflux of Na+ are generally more tolerant to salinity (Hasegawa, 2013; Maathuis et al., 2014). However, one of the common characteristics of cbl10 mutants, both in tomato and Arabidopsis, is a significantly lower Na+ accumulation in shoots under salt stress conditions. Therefore, it is difficult to understand how the salt hypersensitive phenotypes of cbl10 mutants are associated with low shoot Na+ accumulation, when the opposite would be expected.
The mechanism of salinity tolerance in glycophytes is characterized by a favored Na+ accumulation in mature leaves and stem, preventing the cation to reach the shoot apex and other growing tissues (Cuartero et al., 2010). In order to explain how Slcbl10 plants accumulate less Na+ but exhibit a salt-hypersensitive phenotype, a precise investigation on Na+ partitioning in the tomato mutant was carried out, analyzing separately the young and mature parts of the shoot (Egea et al., 2018). One of the features of Slcbl10 is its inability to retain Na+ in mature tissues so it reaches the shoot apex. This fact matches with the damage observed in apex of the mutant and strongly supports the hypothesis that SlCBL10 gene is involved in shoot apex protection in salt stress conditions. Therefore, the fact that a low accumulation of Na+ causes a greater salt sensitivity in cbl10 mutants is due to an altered distribution of Na+ within the shoot.
There is controversy regarding the mechanism by which CBL10 is involved in the regulation of Na+ homeostasis. Whereas some authors place CBL10 at the plasma membrane as a member of the Salt Overly Sensitive (SOS) pathway, involved in Na+ extrusion to apoplast, others have localized CBL10 at the tonoplast, suggesting that it could be involved in the activation of a still unknown Na+ transporter, compartmentalizing excess of Na+ into the vacuole. Both transport processes are recognized as tolerance mechanisms evolved by plants to cope with Na+ toxicity by avoiding its accumulation in cytoplasm (Munns and Tester, 2008).
In the SOS pathway, a Ca2+ -binding protein SOS3 (also known as Calcineurin B-Like Protein 4, CBL4) senses the salt-elicited Ca2+ signal and then binds to and activates the SOS2 Ser/Thr protein kinase (also known as CBL-Interacting Protein Kinase 24, CIPK24). Later, the SOS2/SOS3 complex phosphorylates and activates the plasma membrane Na+/H+ antiporter SOS1, which extrudes excess of Na+ from the cytosol to extracellular medium (Qiu et al., 2002; Quintero et al., 2002). The first study involving CBL10 in the SOS pathway was carried out by Quan et al. (2007). These authors showed that AtCBL10 interacted with AtCIPK24 both in vitro and in vivo, and such interaction enhanced the kinase activity of AtSOS2. By transient expression of AtCBL10 fused to green fluorescent protein in onion epidermal cells, they also showed that AtCBL10 recruits AtCIPK24 to the plasma membrane, being the N-terminal hydrophobic domain of AtCBL10 protein crucial for such subcellular localization. Subsequently, this research group supported and complemented this hypothesis by determining that AtCIPK24 also phosphorylates AtCBL10 at Ser-237 and that such phosphorylation induced by salt stress occurs at the membrane level, stabilizing CBL10–CIPK24 interaction and enhancing AtSOS1 activity (Lin et al., 2009; Figure 1). Results from these studies demonstrate that although AtCBL10 exhibited a higher ability for similar functions than AtSOS3, both CBL proteins must be involved in different regulatory functions in the plant response to salt stress in Arabidopsis, as AtSOS3 is expressed only in roots, whereas AtCBL10 is mainly expressed in shoots. cbl10 and sos3 Arabidopsis mutants display shoot- and root-specific salt sensitivities, respectively, and these should arise, at least partially, from these different spatial expression profiles. The activation by CBL10 of Na+/H+ antiporter SOS1 at the plasma membrane when the former interacts with CIPK8 instead of CIPK24 has been recently demonstrated (Yin et al., 2019). Thus, it seems that CBL10–CIPK8 complex positively regulates SOS1 activity and that the CBL10–CIPK8-SOS1 pathway is able to efficiently extrude the excess Na+ in yeast cells subjected to salinity. In Arabidopsis plants exposed to increasing salinity conditions, activation of SOS1 by phosphorylation triggered by interacting CBL10–CIPK8 occurs at the same sites of the C-terminus domain (Ser-1136 and Ser-1138) as for the same reaction produced by CBL10–SOS2 (Figure 1).
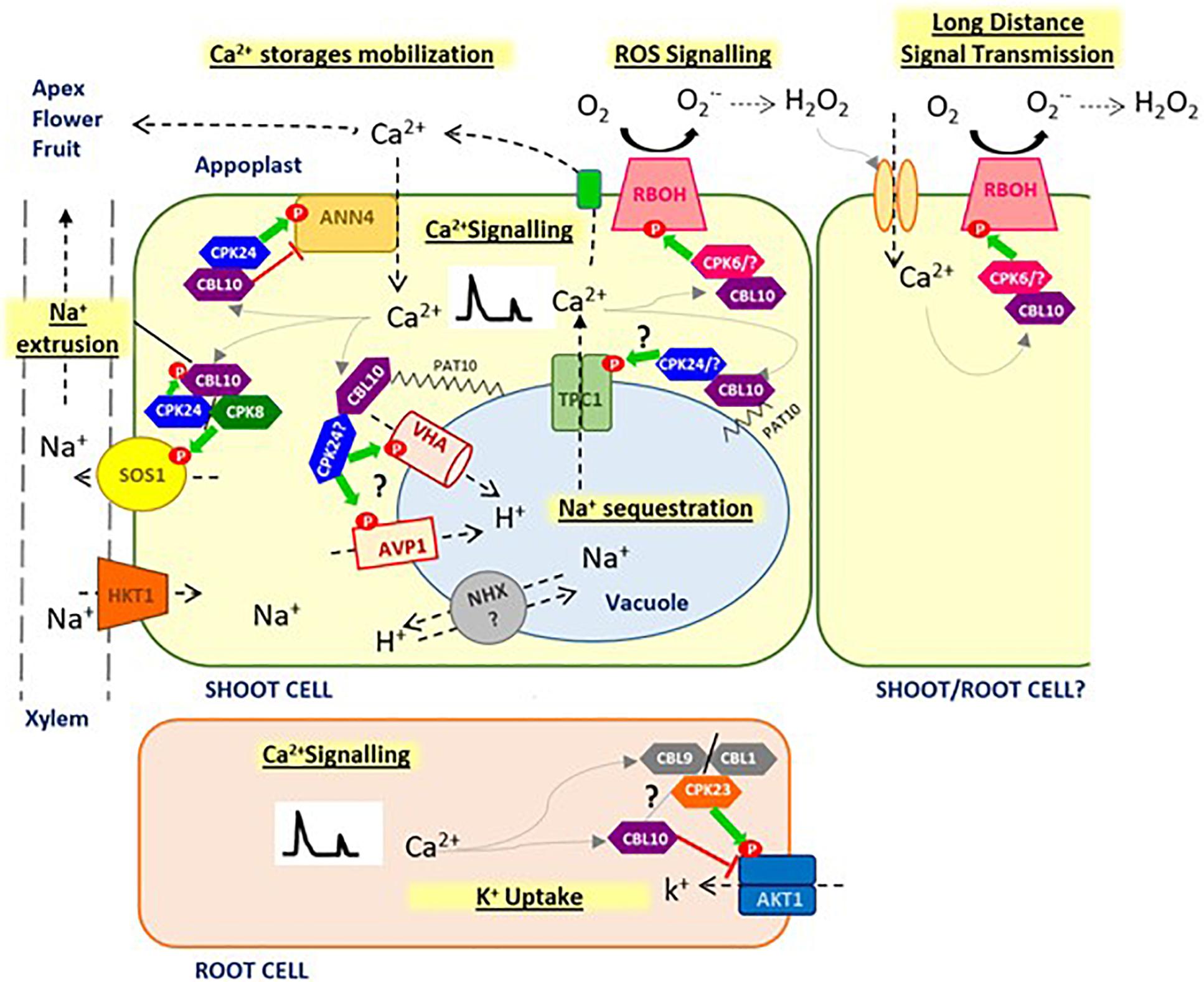
Figure 1. Proposed model for CBL10 function. CBL10 functions as a crucial regulator in Na+ homeostasis under salt stress conditions, interacting with CIPK24 (SOS2) and CIPK8, and activating SOS1 pathway at plasma membrane for Na+ extrusion to apoplast and may be interacting with vacuolar H+ pumps (AVP1, VHA) facilitating Na+ sequestration into the vacuole. CBL10 also plays a key role in Ca2+ homeostasis, which in turn hints at this Ca2+ -sensor protein as a key master regulator of stress signaling in plants, involved in generation and fine tuning of Ca2+ signatures in the cytoplasm, through the coordination of different Ca2+ channels at different cellular localizations, such as plasma membrane (AtANN4) and probably tonoplast (TPC1), as well as in the cross-talk between Ca2+ and ROS, through regulation of RBOHs activity, for a rapid long-distance transmission of signals. CBL10 seems also to be involved in mobilizing Ca2+ stored in vacuole toward demanding tissues to compensate for Ca2+ deficiency caused by salinity and thus allowing development of the meristem and reproductive organs. Finally, functions of CBL10 in root have been associated with K+ uptake by negative regulation of AKT1 channel. Green arrows indicate activation by phosphorylation; red line indicates repression by direct interaction; gray arrows indicate activation by Ca2+, and question mark indicates hypothesized signaling processes that have not been verified yet.
In Eutrema, CBL10 is also involved in the SOS pathway; the two identified duplicate EsCBL10 genes complemented the Atcbl10 mutant, which demonstrates the conservation of the function of CBL10 in these species. Nevertheless, a divergence for the activity of both Eutrema proteins was evidenced when assessing the different capacities of EsCBL10a and EsCBL10b to trigger SOS pathway (Monihan et al., 2019). Cross-species complementation assays have allowed to confirm that only EsCBL10b is able to enhance activation of the SOS pathway; furthermore, in yeast, the interaction of two-hybrid assays of this regulator was observed with AtSOS2 at a similar level as AtCBL10. EsCBL10a is able to activate the SOS pathway but only weakly and displays little interaction with CIPK24, hinting at a function for EsCBL10a not performed by AtCBL10 or EsCBL10b. Experiments with chimeric proteins revealed that changes in the N-terminal domain are responsible for the role of EsCBL10b in the SOS pathway and of EsCBL10a in an alternative pathway, and these changes may be involved in the different localizations of the proteins and even in different CIPK interactions.
Despite all these studies that involve CBL10 with the SOS pathway, the in vivo functional relationship of SOS and CBL10–regulated processes remains unclear. In fact, controversial results have been obtained when Na+ concentration and distribution were analyzed in plants showing impairment in the function of CBL10. Thus, while sos mutants accumulate more Na+ than WT plants, the opposite is observed in cbl10 mutants; i.e., there is less accumulation of Na+ in the plant (Kim et al., 2007; Quan et al., 2007; Egea et al., 2018). Furthermore, when any SOS mutation (sos3 or sos1) is combined with the cbl10 mutation, a reduced Na+ accumulation occurs compared with the simple sos1 mutation under salinity, despite the fact that the double mutants exhibited similar salt-sensitive phenotypes to sos1 at the whole plant level (Yang et al., 2019). If the function of CBL10 only resulted in the extrusion of Na+ out of the cell via the SOS pathway, a greater accumulation of Na+ in the plant would be expected, as occurs in any of the sos mutants, but not the opposite, suggesting that CBL10 regulates other transport processes besides that involved in the SOS pathway.
In addition to extruding Na+ out of the cell, plants have evolved other tolerance mechanisms to cope with salt stress such as compartmentalizing excess of Na+ into the vacuole (Munns and Tester, 2008). The first study involving CBL10 in this strategy suggested that the high salt sensitivity of the Atcbl10 mutant was due to alteration in Na+ vacuolar sequestration in leaves (Kim et al., 2007). Thus, when storage in vacuoles was lower, the amount of saline ions that would penetrate plant cells before these die of salt toxicity was lesser, which would explain why the Atcbl10 mutant contains less Na+ than the WT, although the former is much more salt-sensitive. SOS2-CBL10 was localized at the tonoplast instead of the plasma membrane in the study of Kim et al. (2007); moreover, Waadt et al. (2008) confirmed this subcellular localization by bimolecular fluorescence complementation. In other species such as Populus, the interaction of CBL10 and CIPK24 was also observed at the vacuolar level (Tang et al., 2014).
Post-translational modification studies on the CBL10 protein support such subcellular localization at the tonoplast: one cysteine residue of CBL10 at the N-terminus, Cys-38 (C38), was predicted to be an S-acylation site by CSS-Palm prediction software (Ren et al., 2008). Chai et al. (2019) proposed S-Acyl Transferase 10 (PAT10) protein as the enzyme responsible for CBL10 association with tonoplast through S-acylation of CBL10 at C38 (Figure 1). Thus, the point mutation of C38S in CBL10 determined an intracellular localization mainly situated in cytoplasm (Chai et al., 2019). However, the expression of the point-mutated version CBL10C38S was at least in part able to enhance growth of cbl10 mutant, suggesting that the role of CBL10 in salinity responses might be involved at subcellular locations other than tonoplast. Given that S-acylation is a reversible process, this modification could be involved in regulation of CBL10 partitioning among different cellular membranes-compartments (plasma membrane, tonoplast, chloroplast membrane, etc.). Nevertheless, a consensus model has not still been found in order to predict how S-acylation could modulate the traffic of a specific protein among diverse cell membranes, although some studies suggest that the degree of S-acylation could play a role in this modulation (Chamberlain and Shipston, 2015). Regarding this aspect, several N-terminal cysteine residues of CBL10 have been predicted to be S-acylation sites (Ren et al., 2008).
Identification of targets of CBL10 at the tonoplast for vacuolar Na+ sequestration still remains a challenge. Initial studies suggested that CBL10–CIPK24 complex may regulate vacuolar transporters Na+/H+ Exchangers (NHXs) for Na+ sequestration by phosphorylation. In support of this hypothesis in a study of salt tolerance in soybean (Glycine max), it was observed that the salt-tolerant variety S111-9 exhibited much higher Na+ accumulation in the vacuole than the salt-sensitive variety Melros, which was associated with a higher expression of the CBL10, CIPK24, and NHX genes (He et al., 2015). However, earlier studies have shown evidence that tonoplast-localized NHX is mainly involved in K+ storage, with little bearing on Na+ sequestration (Bassil et al., 2011; Barragán et al., 2012; Andres et al., 2014). In addition, recent genetic evidence in Arabidopsis implies that vacuole-localized antiporters NHX1-4 exhibit Na+-transport activities, but their contribution to vacuolar Na+ compartmentation is small, as the quadruple knockout mutant nhx1/2/3/4 does not show higher susceptibility to NaCl than WT. Furthermore, vacuoles isolated from the quadruple mutant still retain Na+ uptake capability, which hints at the existence in tonoplast of NHX-independent Na+ transporters (Bassil et al., 2019).
Characterization of the Slcbl10 tomato mutant showed that, in response to salinity, the expression pattern of main genes responsible for Na+ distribution and compartmentation SlNHX4, SlSOS1, and SlSOS2, SlHKT1;2 (HIGH-AFFINITY K+ TRANSPORTER 2) was altered (Egea et al., 2018). In addition, the vacuolar H+ pumps SlAVP1 (H+-PYROPHOSPHATASE, AVP1) and SlVHA-A1 (VACUOLAR H+-ATPases, V-ATPase) gene expressions are altered in the upper leaves close to the apical tissues of the tomato mutant. A severe downregulation of both proton vacuolar pumps (SlAVP1 and SlVHA-A1) responsible for generating the required driving force for Na+ compartmentation into the vacuole, as well as moderated downregulation of the Na+/H+ antiporter NHX4, was observed in theSlcbl10 mutant, revealing that loss of function of SlCBL10 impaired the ability of tomato leaves to sequester Na+ into vacuoles. An upregulation of SlSOS1 and a severe downregulation of SlHKT1;2 were also observed in mutant leaves, indicating a higher Na+ extrusion from the cytoplasm to the apoplast and a lower Na+ influx into the cells, respectively, avoiding in such a way the excessive accumulation of Na+ in cytoplasm as a consequence of impaired vacuolar Na+ storage. Such an expression pattern resulted in a reduced upload of Na+ from the xylem, allowing toxic ions to reach the shoot apex, inducing its collapse and subsequent plant death (Egea et al., 2018). These findings reveal that SlCBL10 is required for the regulation of Na+ homeostasis through the activity of gene products involved in the compartmentalization of Na+ into vacuoles (Figure 1); this is in agreement with the first hypothesis postulated by Kim et al. (2007), and it is supported by several studies that have established the subcellular localization of CBL10–SOS2 at the tonoplast (Waadt et al., 2008; Tang et al., 2014). Although more studies are necessary to achieve a definitive conclusion, our results revealed two vacuolar H+ pumps, SlAVP1 and SlVHA-A1, as potential targets of SlCBL10 in tomato plants (Egea et al., 2018; Figure 1). In this sense, Batelli et al. (2007) indicated that SOS2 is able to interact and activate the vacuolar H+ pump V-ATPase. To drive the compartmentalization of Na+ in vacuole mediated by vacuolar Na+/H+ antiporters, an electrochemical gradient of H+ generated by the vacuolar H+ pumps AVP1 and V-ATPase is needed (Maeshima, 2000; Hasegawa, 2013), which could explain the controversy generated in previous studies around vacuolar Na+/H+ antiporters as targets for the CBL10–CIPK24 complex. However, a study in Arabidopsis has shown that the vacuolar proton pump V-ATPase (VHA-a2, VHA-a3) seems not to be involved, at least directly, in sequestration of Na+ in the vacuole. It was observed in the tested experimental conditions that H+ pump AVP1 activity is enough to carry on this process (Krebs et al., 2010). Conversely, the isoform localized at the trans-Golgi network-early endosome (VHA-a1) seems to play a key role in detoxification of Na+, pointing out an important role of the endosomal system in Na+ uptake (Krebs et al., 2010). In this regard, it would be interesting to verify if a subcellular localization of CBL10 at the endoplasmic reticulum and Golgi-endosomal occurs, as S-acylation also confers ability to attach to the membranes confining the ER and Golgi compartments (Chamberlain and Shipston, 2015).
In summary, CBL10 plays a key physiological role in protecting different tissues and organs against salt stress, which requires a proper distribution of Na+ across tissues and organs, and transport of Na+ from the cytosol to vacuoles. A possibility could be, at least for tomato, that participation of CBL10 in both Na+ transport processes could be tissue-dependent. Thus, in young growing tissues, which are little vacuolated, CBL10 could be mainly active at the plasma membrane, involving Na+ extrusion out of the cell, which would explain why the loss of function of CBL10 in tomato causes a greater accumulation of Na+ in these tissues. However, in cells of mature leaves, equipped with giant vacuoles, CBL10 could be mainly associated with the tonoplast, participating in sequestration of Na+ in the vacuole, and hence, the loss of its function causes a lower accumulation of Na+ (Egea et al., 2018). Nevertheless, in addition to both membranes, plasma membrane, and tonoplast, CBL10 has also been associated with other subcellular locations. Translocon of the Outer Membrane of the Chloroplasts 34 (TOC34) was identified as a novel interacting partner of CBL10 occurring at the outer membrane of chloroplasts (Cho et al., 2016), an evidence of the ability of CBL10 to relay Ca2+ signals in more diverse subcellular locations than it was currently known.
Initial studies aimed at the identification of the functional role of CBL10 during salt tolerance were focused on Na+ homeostasis, although changes in Ca2+ contents were also observed in plants where CBL10 was disrupted. First, in cbl10 mutants of Arabidopsis, a higher Na+ and lower Ca2+ accumulation in flowers was detected under salt stress conditions, and the research performed reached the conclusion that CBL10 is crucial for reproductive development (Monihan et al., 2016). These authors also determined that CBL10 function in reproductive development is independent of CIPK24 and SOS1, as loss of CIPK24 activity had no effect on seed development; furthermore, salt stress affected reproductive development in different ways in Arabidopsis sos1 and cbl10 mutants. Thus, it was proposed that CBL10 might interact with another CIPK in flowers to regulate Na+ levels during reproductive development, or alternatively, CBL10 could interact with a protein that does not belong to the CIPK family. However, no function in direct regulation of Ca2+ homeostasis in plants was attributed to CBL10 in the mentioned study (Monihan et al., 2016).
More recently, evidence from the research on Slcbl10 tomato mutant implicated CBL10 in cellular functions involved not only in Na+ but also in Ca2+ homeostasis (Egea et al., 2018). Taking into account that Ca2+ is normally compartmentalized within vacuoles, mainly in stems and upper leaves close to apical tissues, the mobilization of Ca2+ vacuolar reservoirs by plants subjected to salt stress is essential for growth continuance of sink organs such as juvenile tissues and fruits (Bonales-Alatorre et al., 2013; Zhai et al., 2015). Results showed that Ca2+ content in stems and upper mature leaves was reduced by salinity in WT but not in Slcbl10 mutant, which suggests that the mutant encountered difficulties in mobilizing Ca2+ reservoirs (Egea et al., 2018). Moreover, by means of reciprocal grafting assays between WT and Slcbl10, we demonstrated that it was retention of Ca2+ in mature tissues and not a higher transport of it from root to shoot the cause of the higher Ca2+ content found in Slcbl10 mutant organs under salinity. If that is the case, retention of Ca2+ in vacuolated upper mature leaves of Slcbl10 might prevent the ion reaching developing tissues and fruits in the required concentration. Blossom end rot (BER) is the most known symptom of Ca2+ deficiency disorders in tomato fruit (de Freitas et al., 2014; Zhai et al., 2015), and we observed a very high BER incidence in fruits of Slcbl10 mutants (Egea et al., 2018). Poor plant development, inhibited growth of the apical meristem, smaller leaves that showed evident chlorosis at their edges, and thickened petioles and stems, all of them phenotypical changes induced by Ca2+ deficiency, were observed during long-term assays in salt-treated tomato mutant plants. In conclusion, the retention of Ca2+ in Slcbl10 upper mature leaves causes an inefficient supply of Ca2+ to other demanding organs, as it has been observed in shoot apices and flowers, severely limiting their development under saline conditions. This fact, together with the higher Na+ accumulation observed in mutant shoot apex and flowers, indicates that SlCBL10 gene participates in the maintenance of an adequate low Na+/Ca2+ ratio in tomato apex and flowers under saline conditions. Certainly the function of CBL10 in controlling Na+ homeostasis has been previously demonstrated in Arabidopsis and other species, but the analysis of Slcbl10 tomato mutants has emphasized this role of CBL10 in salt tolerance. However, it is of even greater interest that this study presented evidence of CBL10 involvement in regulation of Ca2+ homeostasis (Egea et al., 2018).
The expression profiles of key genes involved in Ca2+ fluxes such as Cation Exhanger Transporter 1 (CAX1), AVP1, and VHA-A1, required for Ca2+ compartmentalization into the vacuole (Cheng et al., 2003; Pittman et al., 2009), and vacuolar ion channel Two Pore Channel1 (TPC1) involved in Ca2+ release from vacuoles (Hedrich and Marten, 2011), were comparatively analyzed in Slcbl10 mutants and WT tomato plants (Egea et al., 2018). Results hinted at the role that SlCBl10 could play in regulating TPC1 activity, suggesting that the CBL10–CIPK24 complex could directly regulate the TPC1 channel aperture through phosphorylation. In support of this hypothesis, the regulation of TPC1 channel has been proposed to be fulfilled by phosphorylation (Kintzer and Stroud, 2016). Nevertheless, CBL10 could also play an indirect role in regulating this TPC1 activity mediating the acidification of vacuoles through the regulation of vacuolar H+ pumps, as low pH is required for aperture of the channel (Kintzer and Stroud, 2016). In fact, both SlAVP1 and SlV-ATPase were also identified in this study as potential targets for SlCBL10 (Egea et al., 2018). Interestingly, the temporal expression profile induced by salt treatment of these four genes in tomato is basically similar, a result that supports the idea of a direct relationship among the functions of these gene products in response to salinity in tomato (Egea et al., 2018).
TPC1 channel, in addition to its function as a calcium-release channel, is also a permeable channel for K+ and Na+ ions (Hedrich et al., 2018). In addition, Ca2+ and Na+ ions seem to play opposite roles in the gates operation of TPC1 channel. Thus, when luminal Ca2+ concentration increases, activation of the channel is inhibited, whereas the presence of high Na+ concentration in lumen seems to alleviate the inhibitory effect of Ca2+ (Hedrich et al., 2018). Nevertheless, very little is known up to now regarding the regulation of this channel in salt stress conditions. If CBL10 is implicated in its regulation, this could explain the perturbations found both in Ca2+ and Na+ homeostasis in Slcbl10 tomato mutant. On the other hand, the correct activity of the vacuolar H+ pumps is also essential for both Ca2+ and Na+ vacuolar fluxes, providing the necessary driving force to their transport against their electrochemical gradient, which is in accordance with the observed alteration in the distribution of these ions in Slcbl10 mutants under salt stress (Figure 1). In support of these hypotheses, yeast two-hybrid assay has allowed to demonstrate that SOS2 directly interacted with V-ATPase regulatory subunits B1 and B2 in Arabidopsis (Batelli et al., 2007). The premise that Ca2+ is also a critical messenger in signal transduction coordinating numerous responses to environmental challenges (Kudla et al., 2018) remarks the fact that the role of CBL10 in plant stress signaling through its participation in vacuolar Ca2+ release to the cytoplasm remains an exciting perspective to be investigated.
As said, Ca2+ is implicated in the fine regulation of responses to developmental processes and environmental perturbations such as biotic and abiotic stresses acting as a key second messenger. Ca2+ accumulation events in the cytosol are featured by specific temporal and spatial profiles, and these diverge in amplitude, frequency, and duration (Steinhorst and Kudla, 2013). Spatially defined Ca2+ signals are produced because of the slow diffusion of Ca2+ in the cytoplasm, combined with a tight regulation of its release from and uptake into the apoplast or diverse intracellular stores. These Ca2+ signals encode information about specific stimuli, such as a salt stress signal that is presented to the cell as a particular Ca2+ signature. Among intracellular stores, the vacuole is the one by far able to accumulate more remobilizable Ca2+ in most plant cells. Total and free Ca2+ contents in the vacuole change depending on plant species, cell type, and environmental conditions, and these probably have an impact on vacuolar function and release of vacuolar Ca2+ (Peiter, 2011). The only vacuolar Ca2+ -permeable channel cloned to date is the slow vacuolar channel TPC1. The involvement of TPC1 in activating-expanding the Ca2+ wave in the cytoplasm has been reported, but the exact role of this channel in mediation of stress-induced Ca2+ changes and the identification of regulatory mechanisms acting to modulate TPC1 channel gating remains to be elucidated (Evans et al., 2016). Results from analysis of Slcbl10 tomato mutant suggest that CBL10 could be directly or indirectly linked to TPC1 channel regulation (Egea et al., 2018) and therefore with vacuolar Ca2+ release to the cytoplasm with a signaling function (Figure 1).
What is more, recent studies have demonstrated that plant systemic signaling is propped up by a ROS-assisted Ca2+ -induced Ca2+ -release mechanism involving ROS production by Respiratory Burst Oxidase Homolog proteins (RBOHs), a family of membrane-bound NADPH-oxidase enzymes, and Ca2+ release for this process depends on activity of the vacuolar channel TPC1 (Choi et al., 2017). This system established a direct connection between ROS production and Ca2+ signaling pathways, and it enables the cell-to-cell communication as a rapid long-distance transmission of signals in plants. In the tomato plant, CBL10, in addition to its possible direct or indirect role in regulating TPC1 channel, has been linked with activation of a plasma membrane RBOH involved in ROS production (Figure 1) in response to biotic stress (de la Torre et al., 2013). CBL10 interacts and recruits CIPK6 to the plasma membrane, where the complex CBL10–CIPK6 activates by phosphorylation of the RBOHB protein contributing to ROS generation during the effector-triggered immunity occurring in the interaction of Pseudomonas syringae pv. tomato DC3000 and Nicotiana benthamiana. In Arabidopsis, AtCBL10 has also been described as a signaling component for ROS production induced by brassinosteroids under salt stress conditions (Kang and Nam, 2016), but if this function is mediated in a CIPK-dependent manner, activating an RBOH, has not been determined. Further studies are needed to confirm if CBL10 could be a key link coordinating TPC1 and RBOH in the cross-talk between Ca2+ and ROS for stress signaling in plants.
Others studies hint at a relationship between CBL10 and the generation and fine-tuning of specific Ca2+ signatures in cytoplasm under stress conditions. Thus, in rice, it has been found that CBL10 is involved in Ca2+ cytoplasmic rise in response to hypoxia as a consequence of flooding (Ye et al., 2018). Analysis of OsCBL10 promoter sequences of eight cultivars of rice differing in their tolerance to flooding at the germination stage revealed variations that might contribute to their divergent tolerance. The flood-tolerant rice cultivars showed a steady low expression of OsCBL10 and lower Ca2+ influx to the cytoplasm, in comparison with flood-sensitive cultivars. In agreement with this observation, OsCBL10 overexpression lines, with enhanced cytoplasmic Ca2+ flux in comparison to WT, were more sensitive to flooding and hypoxia during rice germination. A study in Arabidopsis also determined that CBL10, in tandem with CIPK24, induced and mediated the fine-tuning of an AtANNEXIN4 (AtANN4)–dependent Ca2+ signature under salt stress (Ma et al., 2019). AtANNs constitute a family of Ca2+ -dependent membrane-binding proteins that mediate salt-induced influx of Ca2+ contributing to the increase of cytoplasmic Ca2+ with important effects on stress tolerance. AtANN4 mediates salt stress–induced Ca2+ transients in plants subjected to salinity, leading to a speedy increment of cytosolic Ca2+ levels and resulting in SOS pathway activation. CBL10 promotes phosphorylation of AtANN4 mediated by CIPK24 and this further enhances the interaction of AtANN4 and CBL10. Such direct interaction represses AtANN4-mediated Ca2+ transients that generate a salt-specific Ca2+ signal (Figure 1). In fact, higher increases in cytoplasmic Ca2+ in sos2 and cbl10 mutants than in WT plants have been determined, revealing that both CIPK24 and CBL10, when targeted at AtANN4, are involved in a negative feedback loop in order to maintain a specific amplitude and frequency of Ca2+ concentration in the cytosol. The final result is activation of the SOS pathway at the plasma membrane by a specific Ca2+ signature generated in the whole process. Given this demonstrated function of CBL10 in Arabidopsis, it would be necessary to prove if changes in the expression of TPC1 observed in Slcbl10 tomato mutant could be a consequence rather than the cause of its altered cytoplasmic Ca2+ (Egea et al., 2018).
Taken together, these results seem to place CBL10 as a key master regulator of stress signaling in plants. On the one hand, CBL10 is involved in the generation and fine tuning of Ca2+ signatures in the cytoplasm that activate different responses depending on the perceived stress, through the coordination of Ca2+ influxes from the apoplast and vacuole via regulation of different channels such as AtANN4 (at plasma membrane) and the potential vacuolar channel TPC1. On the other hand, CBL10 might also be crucial in the cross-talk between Ca2+ and ROS, through regulation of RBOHs activity, for rapid long-distance transmission of signals (Figure 1).
CBL10 exerts its role as repressor by direct interaction in other targets, aside from AtANN4. One of these other targets is the K+ channel AKT1 at the plasma membrane of root cells in Arabidopsis, involved in K+ uptake (Ren et al., 2013). AKT1 activity is induced by the complex that protein kinase CIPK23 forms with two CBL proteins, CBL1/CBL9 (Li et al., 2006; Xu et al., 2006). Ren et al. (2013) showed that CBL10 can directly interact with AKT1, impairing AKT1-mediated inward K+ currents. In fact, overexpression of AtCBL10 (CBL10–OE) caused a phenotype with a degree of sensitiveness as high as that of akt1 mutant under low K+ conditions, with lower K+ contents in both CBL10–OE lines and akt1 mutant plants than in WT. This study concluded that a competition exists between CBL10 and CIPK23 for binding AKT1, negatively modulating its activity in Arabidopsis (Figure 1). However, another study carried out in cotton has shown that the homolog CBL10 in this species is also able to interact with CIPK23 (Lu et al., 2017). Taking into account these results and the precedents in the case of regulation of ATANN4 activity, it remains to be verified whether CBL10 could participate in both processes, the activation of the K+ channel via its interaction with CIPK23 and the repression of it by direct interaction, modulating K+ flux in roots. In Eutrema, CBL10 was also related with regulation of K+ homeostasis in roots, in particular EsCBL10a isoform, suggesting this could interact with AtCIPK6 and/or AtCIPK16 proteins to regulate K+ levels in roots (Monihan et al., 2019).
Another direct interaction of CBL10 with a repressive function is the one described for TOC34 protein (Cho et al., 2016). TOC34 is a member of the TOC (translocon of the outer membrane of the chloroplasts) complex with GTPase activity, regulating protein importation into chloroplasts. The in vivo interaction of CBL10 and TOC34 that takes place at the outer membrane of chloroplasts has been observed. Additionally, the physical association of CBL10 with TOC34 has been determined in vitro, resulting in a significant diminution of the TOC34 GTPase activity. Taking into account the critical functions of chloroplasts in photosynthesis and other metabolic processes, the integration of this organelle within the Ca2+ -signaling network should be considered, and its regulation is certainly needed for adaptation to environmental changes.
Given the demonstrated ability of CBL10 to activate (through a CIPK-dependent manner) or repress (by direct interaction) the activity of an array of targets, it seems logical that loss of function of CBL10 may have positive or negative effects against different types of stress. Thus, the role of CBL10 as a positive regulator of salt stress is well-established (Kim et al., 2007; Quan et al., 2007; Lin et al., 2009; Tang et al., 2014; Kang and Nam, 2016; Egea et al., 2018; Ma et al., 2019; Monihan et al., 2019; Yang et al., 2019), and it appears to display a positive role in pathogen infection (de la Torre et al., 2013). However, opposite effects have been described for other stresses, such as flooding during rice germination in the mentioned research work of Ye et al. (2018), where a lower constitutive OsCBL10 gene expression was linked with a higher hypoxia tolerance, where it was observed that OsCBL10 overexpression lines exhibited significant susceptibility to flooding and hypoxia during their germination. Also, under K+ deficit, overexpression of AtCBL10 caused a more sensitive phenotype with lower K+ accumulation than in WT plants (Ren et al., 2013). Another stress described in the literature in which CBL10 seems to be implied as a negative regulator is drought. Thus, loss of function of CBL10 in Arabidopsis improved the resumed growth of plants after a period of dehydration compared with WT, whereas CBL10–OE plants did not recover such growth (Kang and Nam, 2016). The stomatal sensitivity of Atcbl10 mutants and CBL10–OE plants to ABA was similar to that of WT plants, and the analysis of the expression of drought marker genes (COR15A, RD29A, and DREB2A) did not clarify the drought tolerance mechanism observed in the mutant, hinting at the fact that the drought tolerant Atcbl10 phenotype is caused by specific but still unknown processes. Further studies in this direction should be carried out to clarify the relationship between CBL10 and drought stress as well as with other abiotic stressors that have not been studied to date, such as the high or cold temperatures that now commonly occur in crop plantations as a consequence of climate change.
In recent years, research on the Ca2+ sensor CBL10 has yielded important advances in the understanding of functions of this signaling component in different facets of plant development and stress tolerance. Altogether, results from these studies seem to point to CBL10 as a key master regulator of stress in plants. This hypothesis is supported by the demonstrated ability of CBL10 to interact with diverse CIPKs, downward activating specific targets, as well as through the demonstrated possibility of its direct interaction with targets repressing their activity, in such a way that opens the possibility of modulating the response to a particular stressor (Figure 2). The role of CBL10 has mainly been studied in the salt stress response, although recent research is expanding its role to examine other stresses, such as biotic (only studied in tomato up to date) and other abiotic ones such as drought, flooding, and low K+. To further advance in our understanding of the regulatory roles of CBL10 of different stressors, a crucial but challenging aim is to identify new target transporters activated by CBL10–CIPK complexes or repressed by the direct interaction with CBL10.
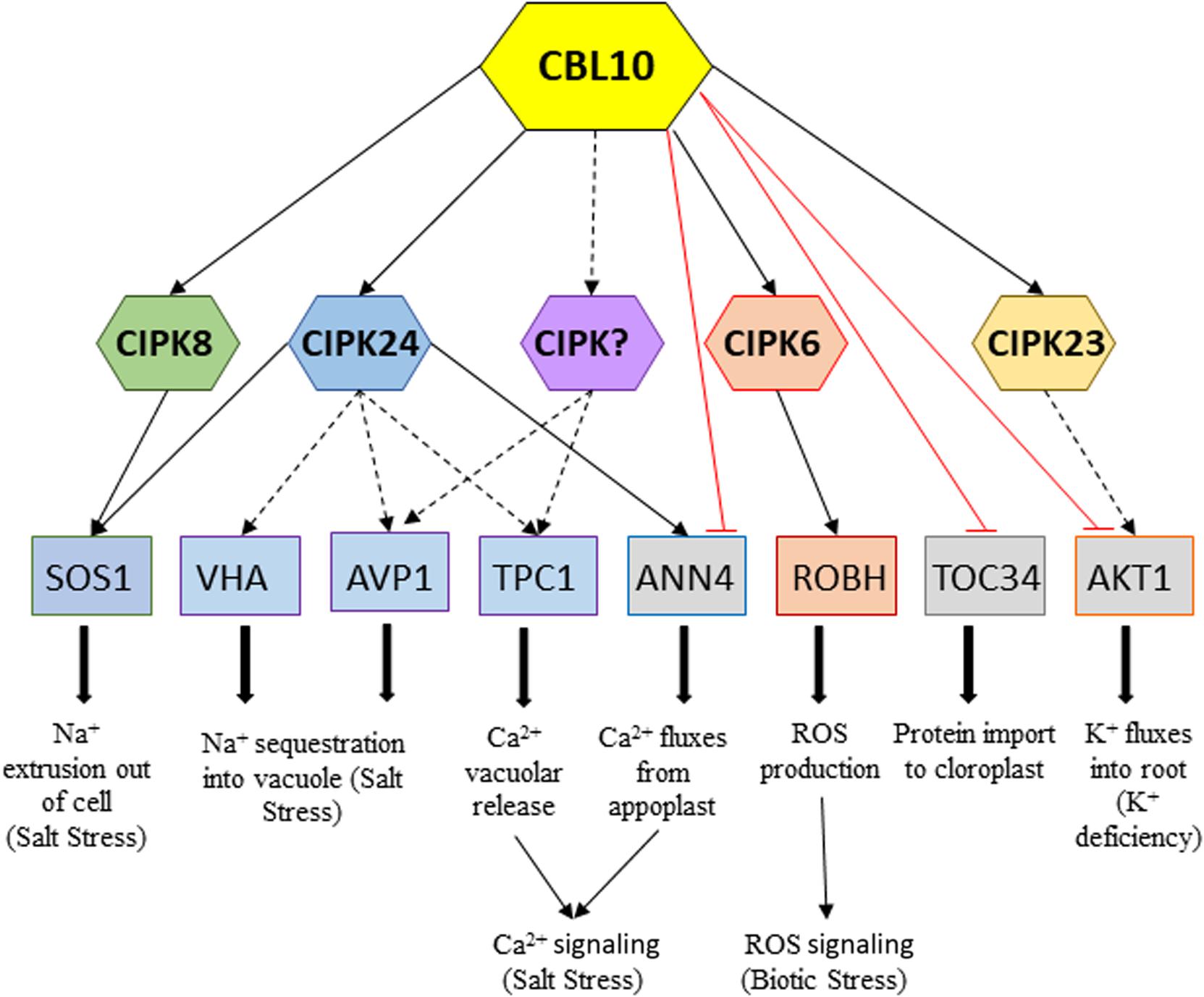
Figure 2. Network of downstream CBL10–CIPKs interactions and CBL10 direct interactions activating or repressing targets in response to different stresses, respectively. Continuous black arrows indicate proved interaction and/or activation by phosphorylation; dotted black arrows indicate interactions still remain to be demonstrated, and red lines indicate demonstrated repressive interactions.
The function of CBL10 seems to involve an organ-dependent component: in roots, its main function is related to the regulation of K+ homeostasis, whereas in shoots it is related to regulation of Na+ and Ca2+ homeostasis. Regarding its role in Na+ homeostasis, there is controversy with respect to the response mechanism triggered by CBL10. On the one hand, it has been demonstrated that CBL10 is able to activate the SOS pathway in the plasma membrane, for Na+ extrusion to the apoplast. On the other hand, CBL10 has also been localized in the tonoplast, supporting its role in Na+ sequestration in vacuoles. Taken together, these results and the patterns of Na+ distribution in plants where CBL10 has been blocked or inhibited, indicate its role in young tissues, where few vacuoles are present, would be Na+ extrusion to apoplast, via the SOS pathway, in order to prevent an excessive accumulation that would lead to cessation of plant growth and development. But there are elements of evidence supporting the hypothesis, which still needs to be verified, that CBL10 may participate in Na+ sequestration in vacuoles of cells from mature leaves, avoiding the risk that this toxic ion could be transported to and reach too high concentrations in young growing tissues. In this last scenario, identification of CBL10 targets in tonoplast for vacuolar Na+ sequestration is therefore a priority in future research.
Regarding Ca2+ homeostasis, CBL10 seems to be involved in mobilizing vacuolar Ca2+ toward demanding tissues, in order to compensate for Ca2+ deficiency caused by salinity and thus allowing the development of the meristem and reproductive organs. Furthermore, it could also be involved in Ca2+ flux from vacuoles to cytoplasm with signaling purposes. Within this framework, some tonoplast proteins have been identified as potential targets for CBL10, such as the H+ pumps AVP1 and V-ATPase (VHA), and TPC1 channel, although direct regulation by CBL10 in a CIPK-dependent or -independent manner remains to be explored. On the other hand, given the role of CBL10 in Ca2+ homeostasis, it would be very interesting to elucidate the possible roles of CBL10 in fruit development, production, and quality, as well as in the incidence of common fruit physiological disorders that have been linked to Ca2+ deficiency in fruit (e.g., BER in tomatoes). In fact, Ca2+ has an important role in membrane stabilization, water relations, and cell wall properties modification via cross-linking of de-esterified pectins, and these processes may have an important impact in fruit development, physical traits, and disease susceptibility (Hocking et al., 2016). It is even possible that CBL10 might have a role in the postharvest life of fruit. In support of this last hypothesis, a transcriptomic study of oranges stored at room temperature has shown results of an increase in the homolog CBL10 expression during fruit senescence (Tang et al., 2016). With regard to regulation of Ca2+ homeostasis, results from different studies seem to place CBL10 as a key master regulator of stress signaling in plants, involved in the generation and fine tuning of Ca2+ signatures in the cytoplasm, through coordination of different Ca2+ channels at different cellular locations, such as the plasma membrane (AtANN4) or the tonoplast (TPC1), as well as in the cross-talk between Ca2+ and ROS via regulation of RBOH activity, for rapid long-distance signal transmission. However, the direct interaction of CBL10 in the regulation of TPC1 needs to be verified, and identification of specific RBOHs regulated by CBL10 for long-distance signal transmission requires further research.
In conclusion, despite so important advances achieved in recent years, further and deeper studies are required to elucidate the complex roles of CBL10 in key functions during plant development under abiotic stress conditions.
FAP and YE wrote the original draft copy. FBF and AO-A revised and edit the manuscript contributing with new ideas. RL and IE conceived the idea of the review article, supervised the project, revised the manuscript, and finished the writing.
This work was supported by the grants PID2019-110833RB-C31, PID2019-110833RB-C33, and RYC2018-023956-I02019 from the Spanish Ministry of Science and Innovation (MICI/AEI/FEDER, UE). YE thanks the Spanish Ministry of Education for grant FPU17/02019 from the National Programme for Training of University Ph.D. Students.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to express my sincere gratitude to M. C. Bolarín, for her so valuable teaching and continuous support over the years, without which this manuscript, among many others, could not have been produced.
Allen, G. J., Chu, S. P., Harrington, C. L., Schumacher, K., Hoffmann, T., Tang, Y. Y., et al. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411, 1053–1057. doi: 10.1038/35082575
Andres, Z., Pérez-Hormaeche, J., Leidi, E. O., Schlücking, K., Steinhorst, L., McLachlan, D. H., et al. (2014). Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc. Natl. Acad. Sci. U.S.A. 111, E1806–E1814. doi: 10.1073/pnas.1320421111
Barragán, V., Leidi, E. O., Andrés, Z., Rubio, L., Luca, A., Fernández, J. A., et al. (2012). Ion Exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate Cell Turgor and Stomatal function in Arabidopsis. Plant Cell 24, 1127–1142. doi: 10.1105/tpc.111.095273
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M. A., Ushijima, K., Nakano, R., et al. (2011). The Arabidopsis Na+/H+ Antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Bassil, E., Zhang, S., Gong, H., Tajima, H., and Blumwald, E. (2019). Cation specificity of vacuolar NHX-Type Cation/H+ antiporters. Plant Physiol. 179, 616–629. doi: 10.1104/pp.18.01103
Batelli, G., Verslues, P. E., Agius, F., Qiu, Q., Fujii, H., Pan, S., et al. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27, 7781–7790. doi: 10.1128/MCB.00430-07
Batistič, O., and Kudla, J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793, 985–992. doi: 10.1016/j.bbamcr.2008.10.006
Batistič, O., and Kudla, J. (2012). Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 1820, 1283–1293. doi: 10.1016/j.bbagen.2011.10.012
Batistič, O., Waadt, R., Steinhorst, L., Held, K., and Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. doi: 10.1111/j.1365-313X.2009.04045.x
Bonales-Alatorre, E., Pottosin, I., Shabala, L., Chen, Z. H., Zeng, F., Jacobsen, S. E., et al. (2013). Differential activity of plasma and vacuolar membrane transporters contributes to genotypic differences in salinity tolerance in a halophyte species, Chenopodium quinoa. Int. J. Mol. Sci. 14, 9267–9285. doi: 10.3390/ijms14059267
Chai, S., Ge, F. R., Zhang, Y., and Li, S. (2019). S-acylation of CBL10/SCaBP8 by PAT10 is crucial for its tonoplast association and function in salt tolerance. J. Integr. Plant Biol. 62, 718–722. doi: 10.1111/jipb.12864
Chamberlain, L. H., and Shipston, M. J. (2015). The physiology of protein S-acylation. Physiol. Rev. 95, 341–376. doi: 10.1152/physrev.00032.2014
Cheng, N. H., Pittman, J. K., Barkla, B. J., Shigaki, T., and Hirschi, K. D. (2003). The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15, 347–364. doi: 10.1105/tpc.007385
Cho, J. H., Lee, J. H., Park, Y. K., Choi, M., and Kim, K. N. (2016). Calcineurin B-like protein CBL10 directly interacts with TOC34 (Translocon of the Outer Membrane of the Chloroplasts) and decreases its GTPase activity in Arabidopsis. Front. Plant Sci. 7:1911. doi: 10.3389/fpls.2016.01911
Choi, W. G., Miller, G., Wallace, I., Harper, J., Mittler, R., and Gilroy, S. (2017). Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J. 90, 698–707. doi: 10.1111/tpj.13492
Cuartero, J., Bolarin, M. C., Moreno, V., and Pineda, B. (2010). “Molecular tools for enhancing salinity tolerance in plants,” in Molecular Techniques in Crop Improvement, eds S. M. Jain and D. S. Brar (New York, NY: Springer), 373–405. doi: 10.1007/978-90-481-2967-6_16
D’Angelo, C., Weinl, S., Batistič, O., Pandey, G. K., Cheong, Y. H., Schültke, S., et al. (2006). Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48, 857–872. doi: 10.1111/j.1365-313X.2006.02921
de Freitas, S. T., McElrone, A. J., Shackel, K. A., and Mitcham, E. J. (2014). Calcium partitioning and allocation and blossom-end rot development in tomato plants in response to whole-plant and fruit-specific abscisic acid treatments. J. Exp. Bot. 6, 235–247. doi: 10.1093/jxb/ert364
de la Torre, F., Gutiérrez-Beltrán, E., Pareja-Jaime, Y., Chakravarthy, S., Martin, G. B., and del Pozo, O. (2013). The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25, 2748–2764. doi: 10.1105/tpc.113.113530
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Dong, L., Wang, Q., Manik, S. M. N., Song, Y., Shi, S., Su, Y., et al. (2015). Nicotiana sylvestris calcineurin B-like protein NsylCBL10 enhances salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 12, 2053–2063. doi: 10.1007/s00299-015-1851-4
Egea, I., Pineda, B., Ortíz-Atienza, A., Plasencia, F. A., Drevensek, S., García-Sogo, B., et al. (2018). The SlCBL10 Calcineurin B-Like protein ensures plant growth under salt stress by regulating Na+ andCa2+ homeostasis. Plant Physiol. 176, 1676–1693. doi: 10.1104/pp.17.01605
Evans, M. J., Choi, W. G., Gilroy, S., and Morris, R. J. (2016). A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171, 1771–1784. doi: 10.1104/pp.16.00215
Hasegawa, P. M. (2013). Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 92, 19–31. doi: 10.1016/j.envexpbot.2013.03.001
He, Y., Fu, J., Yu, C., Wang, X., Jiang, Q., Hong, J., et al. (2015). Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 66, 6877–6889. doi: 10.1093/jxb/erv392
Hedrich, R., and Marten, I. (2011). TPC1-SV channels gain shape. Mol. Plant 4, 428–441. doi: 10.1093/mp/ssr017
Hedrich, R., Mueller, T. D., Becker, D., and Marten, I. (2018). Structure and function of TPC1 vacuole SV channel gains shape. Mol. Plant 11, 764–775. doi: 10.1016/j.molp.2018.03.017
Hocking, B., Tyerman, S. D., Burton, R. A., and Gilliham, M. (2016). Fruit calcium: transport and physiology. Front. Plant Sci. 7:569. doi: 10.3389/fpls.2016.00569
Kang, H. K., and Nam, K. H. (2016). Reverse function of ROS-induced CBL10 during salt and drought stress responses. Plant Sci. 243, 49–55. doi: 10.1016/j.plantsci.2015.11.006
Kim, B. G., Waadt, R., Cheong, Y. H., Pandey, G. K., Dominguez-Solis, J. R., Schültke, S., et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52, 473–484. doi: 10.1111/j.1365-313X.2007.03249.x
Kim, K. N. (2013). Stress responses mediated by the CBL calcium sensors in plants. Plant Biotechnol. Rep. 7, 1–8. doi: 10.1007/s11816-012-0228-1
Kintzer, A. F., and Stroud, R. M. (2016). Structure, inhibition and regulation of two pore channel TPC1 from Arabidopsis thaliana. Nature 531, 258–262. doi: 10.1038/nature17194
Krebs, M., Beyhl, D., Görlich, E., Al-Rasheid, K. A. S., Marten, M., Stierhof, Y. D., et al. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 107, 3251–3256. doi: 10.1073/pnas.0913035107
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 218, 414–431. doi: 10.1111/nph.14966
Li, D. D., Xia, X. L., Yin, W. L., and Zhang, H. C. (2013). Two poplar calcineurin B-like proteins confer enhanced tolerance to abiotic stresses in transgenic Arabidopsis thaliana. Biol. Plant 57, 70–78. doi: 10.1007/s10535-012-0251-7
Li, L., Kim, B. G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca2+ signalling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 103, 12625–12630. doi: 10.1073/pnas.0605129103
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619. doi: 10.1105/tpc.109.066217
Lu, T., Zhang, G., Sun, L., Wang, J., and Hao, F. (2017). Genome-wide identification of CBL family and expression analysis of CBLs in response to potassium deficiency in cotton. PeerJ 5:e3653. doi: 10.7717/peerj.3653
Ma, L., Ye, J., Yang, Y., Lin, H., Yue, L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48, 697–709. doi: 10.1016/j.devcel.2019.02.010
Maathuis, F. J. M., Ahmad, A., and Patishtan, J. (2014). Regulation of Na+ fluxes in plants. Front. Plant Sci. 4:467. doi: 10.3389/fpls.2014.00467
Maeshima, M. (2000). Vacuolar H+-pyrophosphatase. Biochim. Biophys. Acta 1465, 37–51. doi: 10.1016/S0005-2736(00)00130-9
Manishankar, P., Wang, N., Köster, P., Alatar, A. A., and Kudla, J. (2018). Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 69, 4215–4226. doi: 10.1093/jxb/ery201
Monihan, S. M., Magness, C. A., Yadegari, R., Smith, S. E., and Schumaker, K. S. (2016). Arabidopsis CALCINEURIN B-LIKE10 functions independently of the SOS pathway during reproductive development in saline conditions. Plant Physiol. 171, 369–379. doi: 10.1104/pp.16.00334
Monihan, S. M., Ryu, C. H., Magness, C. A., and Schumaker, K. S. (2019). Linking duplication of a calcium sensor to salt tolerance in Eutrema salsugineum. Plant Physiol. 179, 1176–1192. doi: 10.1104/pp.18.01400
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Peiter, E. (2011). The plant vacuole: emitter and receiver of calcium signals. Cell Calcium 50, 120–128. doi: 10.1016/j.ceca.2011.02.002
Pittman, J. K., Edmond, C., Sunderland, P. A., and Bray, C. M. (2009). A cation regulated and proton gradient-dependent cation transporter from Chlamydomonas reinhardtii has a role in calcium and sodium homeostasis. J. Biol. Chem. 284, 525–533. doi: 10.1074/jbc.M807173200
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431. doi: 10.1105/tpc.106.042291
Quintero, F. J., Ohta, M., Shi, H., Zhu, J. K., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644. doi: 10.1093/protein/gzn039
Ren, X. L., Qi, G. N., Feng, H. Q., Zhao, S., Zhao, S. S., Wang, Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74, 258–266. doi: 10.1111/tpj.12123
Saito, S., and Uozumi, N. (2020). Calcium-regulated phosphorylation systems controlling uptake and balance of plant nutrients. Front. Plant Sci. 11:44. doi: 10.3389/fpls.2020.00044
Steinhorst, L., and Kudla, J. (2013). Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol. 163, 471–485. doi: 10.1104/pp.113.222950
Tang, N., Deng, W., Hu, N., Chen, N., and Li, Z. (2016). Metabolite and transcriptomic analysis reveals metabolic and regulatory features associated with Powell orange pulp deterioration during room temperature and cold storage. Postharvest Biol. Technol. 112, 75–86. doi: 10.1016/j.postharvbio.2015.10.008
Tang, R. J., Wang, C., Li, K., and Luan, S. (2020). The CBL–CIPK calcium signaling network: unified paradigm from 20 years of discoveries. Trends Plant Sci. 25, 604–617. doi: 10.1016/j.tplants.2020.01.009
Tang, R. J., Yang, Y., Yang, L., Liu, H., Wang, C. T., Yu, M. M., et al. (2014). Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ. 37, 573–588. doi: 10.1111/pce.12178
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R., and Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Yang, Y., Zhang, C., Tang, R. J., Xu, H. X., Lan, W. Z., Zhao, F., et al. (2019). Calcineurin B-like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int. J. Mol. Sci. 20:2421. doi: 10.3390/ijms20102421
Ye, N. H., Wang, F. Z., Shi, L., Chen, M. X., Cao, Y. Y., Zhu, F. Y., et al. (2018). Natural variation in the promoter of rice calcineurin B-likeprotein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J. 94, 612–625. doi: 10.1111/tpj.13881
Yin, X., Xia, Y., Xie, Q., Cao, Y., Wang, Z., Hao, G., et al. (2019). The protein kinase complex CBL10–CIPK8–SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 71, 1801–1814. doi: 10.1093/jxb/erz549
Keywords: Na+ homeostasis, Cacpsdummy2+ homeostasis, Cacpsdummy2+-mediated stress signaling, Cacpsdummy2+-ROS cross-talk, salinity, fruit development, CBL-CIPK
Citation: Plasencia FA, Estrada Y, Flores FB, Ortíz-Atienza A, Lozano R and Egea I (2021) The Ca2+ Sensor Calcineurin B–Like Protein 10 in Plants: Emerging New Crucial Roles for Plant Abiotic Stress Tolerance. Front. Plant Sci. 11:599944. doi: 10.3389/fpls.2020.599944
Received: 28 August 2020; Accepted: 09 December 2020;
Published: 15 January 2021.
Edited by:
Sergey Shabala, University of Tasmania, AustraliaReviewed by:
Jose M. Pardo, Institute of Plant Biochemistry and Photosynthesis (IBVF), SpainCopyright © 2021 Plasencia, Estrada, Flores, Ortíz-Atienza, Lozano and Egea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabel Egea, aWVnZWFAY2ViYXMuY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.