- 1Guangdong Provincial Key Laboratory of Marine Biotechnology, STU-UNIVPM Joint Algal Research Center, College of Sciences, Institute of Marine Sciences, Shantou University, Shantou, China
- 2Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
- 3Faculty of Marine Sciences, Lasbela University of Agriculture, Water & Marine Sciences, Uthal, Pakistan
Different anthropogenic activities result in the continuous increase of metal lead (Pb) in the environment and adversely affect living organisms. Therefore, it is important to investigate the tolerance mechanism in a model organism. Chlamydomonas reinhardtii is an important green eukaryotic model microalga for studying different kinds of biological questions. In this study, the responses of C. reinhardtii were revealed via a comprehensive approach, including physiological, genomic, transcriptomic, glycomic, and bioinformatic techniques. Physiological results showed that the growth rate and soluble protein content were significantly reduced under the high lead stress. Also, the results obtained from the genomic and transcriptomic analyses presented that the endoplasmic reticulum-mediated protein quality control (ERQC) system and endoplasmic reticulum-associated degradation (ERAD) pathway were activated under the third day of high lead stress. The unique upregulated protein disulfide isomerase genes on the ERQC system were proposed to be important for the protein level and protein quality control. The accumulation of specific N-glycans indicated that specific N-glycosylation of proteins might alter the biological functions of proteins to alleviate the Pb stress in alga and/or lead to the degradation of incomplete/misfolded proteins. At the same time, it was observed that genes involved in each process of ERAD were upregulated, suggesting that the ERAD pathway was activated to assist the degradation of incomplete/misfolded proteins. Therefore, it is reasonable to speculate that the reduction of protein level under the high lead stress was related to the activated ERQC system and QRAD pathway. Our findings will provide a solid and reliable foundation and a proposed ERAD working model for further in-depth study of the ERQC system and ERAD pathway under the Pb stress and even other biotic and abiotic stresses.
Introduction
Metal pollution is one of the most serious global environmental problems threatening algae, plants, animals, and even human beings (Peharec Štefanić et al., 2012). Among all metals, lead (Pb) is one of the most toxic metals, with a long half-life. When Pb was released into the environment, algae and plants were physiologically affected, particularly in terms of the inhibition of growth and photosynthesis (Du et al., 2018), the increased production of reactive oxygen species (Szivák et al., 2009), and the changed phytochelatin production and kinetics, metabolism, and ultrastructure of cells (Sharma and Dubey, 2005), and the damage of lipid, protein, and nucleic acid (Hasan et al., 2017). In addition, gene expression and protein translation were also affected by Pb in algae and plants via transcriptome and proteome analysis (Wang et al., 2013; Li et al., 2016). Transcriptome results implied that differentially expressed genes involved in the cell wall and glutathione metabolism were upregulated, but genes related to carbohydrate metabolism were downregulated during Pb stress (Wang et al., 2013). Proteome analysis showed that proteins involved in signal transduction, redox, stress, and transport were upregulated, whereas proteins related to nucleotide, energy, protein, and carbohydrate metabolisms were downregulated under the lead stress (Li et al., 2016; Du et al., 2018). The inconsistency between transcript and protein levels suggested that both transcriptional and translational regulations played important roles in response to Pb contamination (Li et al., 2016).
Nascent translated proteins in response to stresses were largely dependent on co-/post-translational modifications (e.g., N-glycosylation) that altered the properties and biological functions of proteins, including activity, localization, and/or interaction with other proteins and consequently affected the phenotype of organisms (Peharec Štefanić et al., 2012). N-glycosylation happens in the endoplasmic reticulum, and the Golgi apparatus is a major co-/post-translational modification in the secretory pathway of proteins. Suitable complex N-glycan formation and processing regulate glycoproteins for a plant to deal with different stresses. For example, it was documented that mannosidase-mediated trimming of N-glycans was important for Arabidopsis tolerance to salt and osmotic stresses (Liu et al., 2018). Xylose residues on N-glycans created a regulatory role for seed germination, rice growth, and development under phytohormone treatments and other abiotic stresses, including temperature, drought, salinity, and oxygen-deficiency stresses (Takano et al., 2015). Additionally, missing core fucose residues but not xylose in N-glycans strengthened salt sensitivity of Arabidopsis hybrid glycosylation mutant (Kaulfürst-Soboll et al., 2011). On the contrary, some N-glycans were proved to be critical for mature N-glycosylation of proteins but not essential for plants’ defense under abiotic stresses. For example, 1,3-mannosyltransferase-dependent N-glycans did not affect Arabidopsis phenotype under normal and high temperature or sale/osmotic stress conditions (Kajiura et al., 2010). Therefore, the structure of N-glycans played an important role in protein folding and quality control and defense under different stresses. However, so far, a complete gap exists in the role of N-glycans formation and protein N-glycosylation in the responses of algae to biotic and abiotic stresses, especially Pb stress.
Nascent glycopeptides are initially folded to their correct conformation in the ER. This process is aided by a known endoplasmic reticulum (ER)-mediated protein quality control (ERQC) mechanism (Blanco-Herrera et al., 2015). Firstly, two glucose residues on the N-glycans of nascent glycopeptides are sequentially removed by glucosidases I and II (GI and GII), producing mono-glucosylated N-glycans. The trimmed N-glycan structures are further recognized by the ER lectin chaperones calnexin (CNX) and/or calreticulin (CRT). Meanwhile, some other ER chaperones and folding enzymes [e.g., protein disulfide isomerases (PDIs)] are also recruited by CNX/CRT for high-specificity high-affinity binding with the mono-glucosylated N-glycans (Nagashima et al., 2018). Subsequently, the remaining glucose residue is removed by GII, the nascent glycoprotein is released from CNX/CRT, and the folding process is finished. Finally, correctly folded glycoproteins will be exported to the Golgi apparatus for further modification and transported to final destinations. However, incomplete/misfolded glycoproteins are recognized and re-glucosylated by an ER-resident folding sensor UDP-glucose: glycoprotein glucosyltransferase (UGGT) for reassociation with CNX/CRT (Caramelo and Parodi, 2008). One or more CNX/CRT cycles are performed until incomplete/misfolded glycoproteins achieving their native conformations. If the glycoproteins that fail to fold its native structure after multiple CNX/CRT cycles are removed from the ERQC system and degraded by an ER-associated degradation pathway (ERAD) (Shin et al., 2018). ERAD mainly contains three steps: (1) the marking of terminally misfolded glycoproteins, (2) recruitment, retrotranslocation, and ubiquitination of misfolded glycoproteins, and (3) extraction, processing, and delivery of misfolded glycoproteins to the proteasome for final degradation. It was known that ERQC is an environmental sensor and responder in plants under different stresses. When the demands for protein folding exceed the capacity of ERQC under stress, the ER protein folding mechanism reaches its limit (Liu and Li, 2014). In this situation, incomplete/misfolded glycoproteins are largely accumulated in the ER, stimulating an unfolded protein response (UPR) (Liu and Howell, 2010). Subsequently, UPR will alleviate ER stress via upregulating the expression of genes involved in ERQC and/or ERAD. For example, the NtPDI gene involved in ERQC was upregulated under the Cd stress in Nicotiana tabacum (Xu et al., 2013). AtHRD1 (a gene associated with ERAD) mutant was more sensitive to Se stress in Arabidopsis (Van Hoewyk, 2013). Therefore, it is curious to study the ERQC and ERAD mechanisms under the lead stress and see how the ERQC and ERAD affect the protein level and quality control.
Chlamydomonas reinhardtii is a model green alga that is widely used to study flagellar biogenesis, circadian rhythms, photosynthesis, responses to nutrient deficiency, and so on (Merchant et al., 2007; Harris, 2013). So far, the responses of C. reinhardtii to Pb stress were also studied, such as the role of phytochelatin in Pb detoxification (Scheidegger et al., 2011), bioaccumulation and biosorption of Pb (Stewart et al., 2015), and its compartmentalization in vacuoles (Sánchez-Marín et al., 2014). Furthermore, it was documented that exposure to metals led to changes in the expression of proteins in C. reinhardtii (Gillet et al., 2006; Walliwalagedara et al., 2012). However, it is still unknown how the Pb stress changes protein folding and quality control and the effects of the ERQC system and ERAD in C. reinhardtii under the Pb stress.
In this study, a comprehensive approach, including physiological, genomic, transcriptomic, glycomic, and bioinformatic analyses, was carried out to investigate the physiological effects, especially the protein content, and the ERQC and ERAD occurring in C. reinhardtii under Pb stress. Results revealed that the growth was significantly inhibited; soluble protein content was extremely reduced after the third day of high lead stress. N-glycan structures of glycoproteins were changed, indicating their importance for the protein level and protein quality control. Additionally, it was speculated that the ERQC system and QRAD were activated to modify and/or remove incomplete/misfolded proteins to maintain ER homeostasis under the third day of high lead stress. Therefore, it was concluded that the reduction of protein content might be owing to the activated ERQC system and ERAD pathway. Our findings will provide an important opportunity to advance the understanding of the effects of the ERQC system and ERAD pathway on the protein level and quality control under the Pb stress.
Materials and Methods
Microalgal Strain and Culture Conditions
Chlamydomonas reinhardtii (CC-503 cw903 mt+) from Freshwater Algae Culture Collection (Institute of Hydrobiology, Wuhan, China) was used in this study. Algae were firstly preincubated in Tris-acetate-phosphate medium under the condition of 22°C, 12-h light:12-h darkness photoperiod, 0.08 mol photons m–2 s–1 light intensity in 100 rpm rotary shaker (Hutner, 1990). During the mid-exponential period of algal growth, cells were cultured in fresh modified Tris-acetate-phosphate medium (1.0 × 105 cells ml–1), where the KH2PO4 was substituted for Na2-glycerol-2-phosphate (Kopittke et al., 2008). pH was controlled to around 6 (Hassler et al., 2004). Ten millimolar 2-(N-morpholino) ethane sulfonate (MES, sodium salt, Sigma) medium was used as a pH buffer. Finally, algal mediums containing 0, 3, and 80 μmol L–1 Pb(NO3)2 were prepared. Nitrate was used as a counter ion, as its low possibility to form metal complexes. Nitric acid was applied to the adjustment of the pH value. All experiments were performed in triplicates.
Measurements of Cell Density and Soluble Protein Content
On 1/2, 1, 3, 5, and 7 days, cell density of C. reinhardtii was analyzed by an electronic particle counter (Orifice, 50 μm; Multisizer II; Beckham Coulter, Fullerton, CA, United States) (Zheng et al., 2013).
On the third and seventh days, 40-ml C. reinhardtii culture was centrifuged at 3,000 × g for 10 min. Concentrated C. reinhardtii cells were resuspended in 1 ml of 20-mM phosphate-buffered saline buffer (pH 7.4) and 10-μl 25 × protease inhibitor cocktail and sonicated on 3 s/off 5 s for 3 min, 90 W. After centrifugation at 12,000 × g for 20 min, 50-μl aliquot of the supernatant was used to determine the soluble protein concentration with Coomassie brilliant blue kit (Jiancheng Biotech Company, Nanjing, China) via UV-VIS spectrophotometry at 595 nm. To purify protein, the remaining supernatant was exhaustively dialyzed against 2 L of water at 4°C for 48 h. During the process of dialysis, water was refreshed every 8 h. All the steps for protein preparation were carried out at 4°C. The dialyzed samples were lyophilized for the next experiments.
Release of N-Glycans With PNGase F and in situ Derivatization With Fmoc
On the third day, 40-ml algal cells were harvested for the extraction of total glycoprotein. The release and derivatization of N-glycans from protein samples were mainly performed as described in previous article (Kamoda et al., 2005; Nakano et al., 2009). Briefly, 10 μg of each glycoprotein sample was completely dissolved in 100 μl of 20-mM phosphate buffer (pH 8.5). One microliter PNGase F (one unit) was added to the glycoprotein sample and incubated at 37°C for 2 h to release N-glycans. Subsequently, the mixture was added 300-μl deionized water for dilution and 200-μl fresh solution of Fmoc-Cl in acetone (50 mg ml–1) and incubated at 37°C for 2 h to derive the N-glycans. Afterward, to remove excess Fmoc reagent, 300-μl chloroform was used to wash N-glycans. The washing steps were repeated at least five times. Finally, the chloroform layer was removed, and the aqueous layer containing Fmoc-linked N-glycans of each sample was dried by an evaporator and dissolved in 20-μl double distilled water. Five microliters was used for the electrospray ionization mass spectrometry (ESI-MS) experiment.
Electrospray Ionization Mass Spectrometry Conditions
The acquisition range was m/z 1,200–1,900. The mass spectrometer was calibrated via a tune mix (Thermo Scientific). Mass spectra were viewed and analyzed via software Xcalibur V2.2. Additionally, Glycoworkbench software was used to annotate and visualize the structures of N-glycans as described in previous article (Pedersen et al., 2017; Lucas et al., 2018, 2019).
The relative abundance of the different N-glycans under lead stresses was based on the signal intensities of the corresponding Fmoc-labeled N-glycans obtained from ESI-MS analysis as previously described (Nakano et al., 2009; Mathieu-rivet et al., 2013). Briefly, each signal intensity of N-glycans was used to calculate their relative abundance compared with the total signal intensity of all identified N-glycans.
In silico Genome Analysis
On the third day, algal cells under the control and the lead stresses were used to do transcriptomic analysis. The transcriptomic data were referenced from our recent article (Zheng et al., 2020). The sequence reads had already been available by the National Center for Biotechnology Information sequence read archives (SRR10269729, SRR10269730, SRR10269727, SRR10269728, SRR10269731, and SRR10269732). Genes with | log2 fold change| ≥ 1.5 and FDR < 0.01 (adjusted P-value, determined by the Benjamini and Hochberg multiple-testing correction implemented in the “p.adjust” method of R) were defined as differentially expressed genes.
Protein sequences involved in the ERQC system and ERAD pathway of Arabidopsis thaliana were retrieved from Phytozome 121. The Hidden Markov model (HMM) files corresponding to different domains were downloaded from the Pfam protein family database2 against protein sequences from A. thaliana. HMMER 3.0 was used to search the genes involved in the ERQC system and ERAD pathway from the C. reinhardtii genome database. From the proteins obtained using the HMM, E-value < 1e-20 was set to isolate high-quality proteins. All candidate genes based on HMMER results were further manually analyzed by confirming the existence of the core sequences using the SMART program3 and National Center for Biotechnology Information Conserved Domains Database4.
The genome database of green algae C. reinhardtii, Ostreococcus lucimarinus, Chlorella variabilis, and plant A. thaliana were downloaded from Ensembl5. Full length and PDI domain sequences of all PDI proteins from algae C. reinhardtii, O. lucimarinus, C. variabilis, and plant A. thaliana were aligned using ClustalW2, and a phylogenetic tree was produced using MEGA 6.0 via the neighbor-joining method, with 1,000 replicates of bootstrap analysis. The phylogenetic tree of PDI domains was modified and improved by EvolView. Motifs of all PDI proteins were predicted by the motif elicitation program (MEME)6. InterPro database was used for the second identification of conserved motifs7. Transmembrane domains of proteins were compared among different servers, including genome database, TOPCONS8, TMHMM Server v. 2.09, ΔG predictor10, and SOSUI. Putative N-terminal signal peptide was predicted using SignalP 3.011 using default settings. PredictProtein was used to predict the secondary structure12.
Data Statistical Analysis
All the data from experiments were shown as mean ± standard deviation. Data were analyzed using SPSS 20.0 software with one-way ANOVA. The statistical significance was considered at ∗P < 0.05 for significant and ∗P < 0.01 for highly significant values. All figures were generated by Origin 8.0 and Adobe Illustrator CS6.
Results
Cell Density and Soluble Protein Content of C. reinhardtii Under the Lead Stress
The cell density of C. reinhardtii was measured under the lead stress (Figure 1A). It was shown that the cell density did not have a significant difference between control and low lead stress. However, the cell number was extremely dramatically decreased after 3 days of high lead stress (P < 0.01). Soluble protein content was shown in Figure 1B. Obviously, soluble protein content was significantly reduced on the third and seventh days under the high lead stress compared with that under the control and low lead stress (P < 0.01).
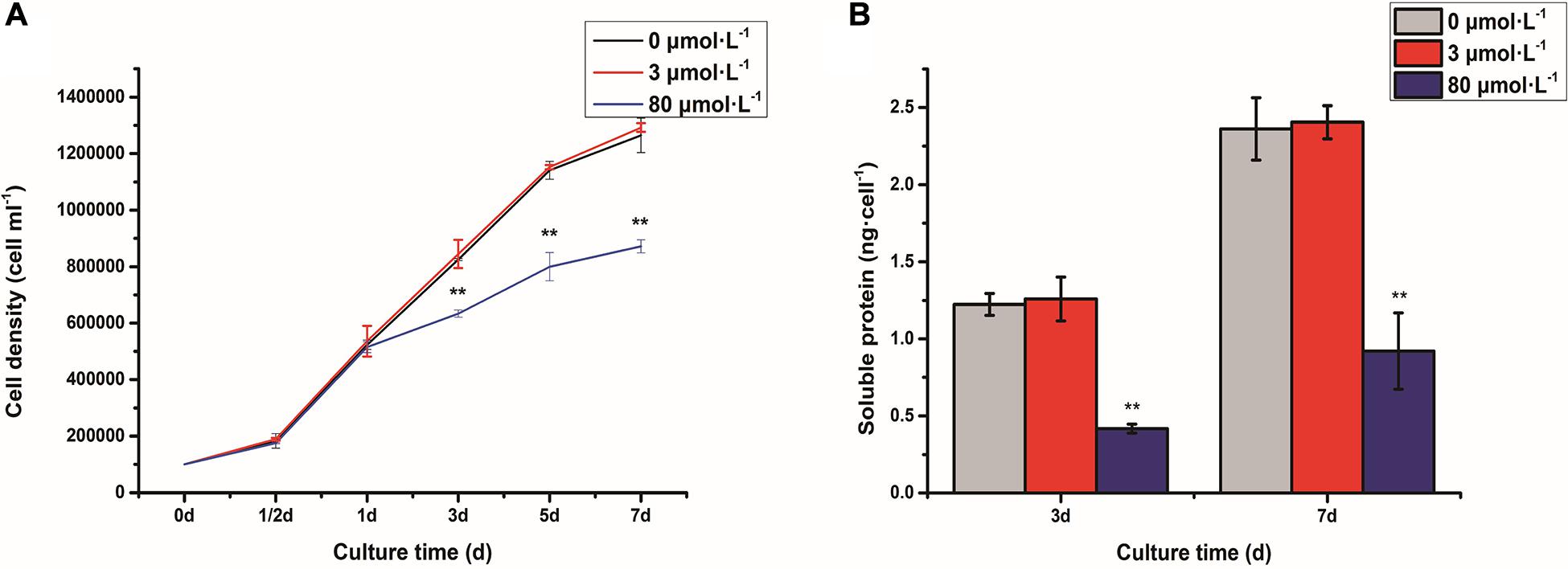
Figure 1. Cell density (A) and soluble protein content (B) of Chlamydomonas reinhardtii under the lead stress. **P < 0.01 representing extremely significant differences in mean value between different stresses. Error bars indicate SD for three biological replicates. Cell density was referenced from our recent article (Zheng et al., 2020).
Endoplasmic Reticulum-Mediated Protein Quality Control System in C. reinhardtii Under Lead Stresses
Homology searches based on sequence similarities with ER-resident proteins in the N-glycosylation pathway were carried out in the genome database of C. reinhardtii. The results were presented in Table 1. The N-glycosylation pathway of protein in eukaryotes firstly takes place in the cytosolic side and lumen of the ER. Many glycosyltransferases and glucosidases are involved in the N-glycosylation pathway in the ER, such as ALG, RTF, CNX, CRT, RPN, and STT3. In this study, only four genes related to the N-glycosylation pathway (ALG14, RPN1, RPN2, and STT3B) were differentially expressed under the third day of high lead stress (Table 1). Among these four genes ALG14, RPN2, and STT3B were upregulated, RPN1 was downregulated under the high lead stress compared with that in the control and the low lead stress.
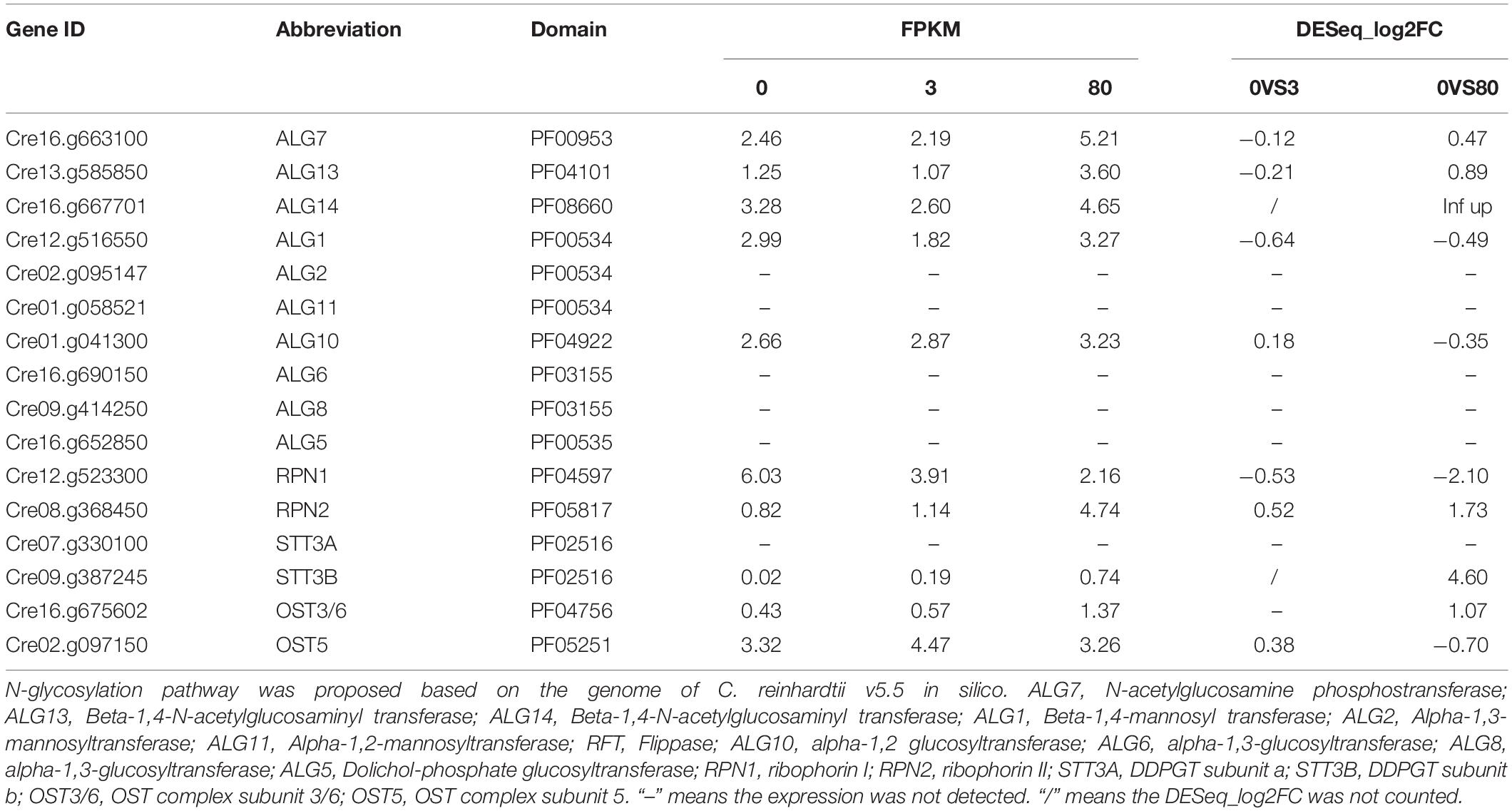
Table 1. Expression of putative genes involved in the ER protein N-glycosylation of Chlamydomonas reinhardtii under the third day of lead stress.
Although bioinformatic analyses gave rise to thorough insights into the N-glycosylation pathway in the ER of C. reinhardtii and the expression of related genes from transcriptome analysis, no information about N-glycans of glycoproteins that happened under the lead stress can be inferred. To investigate the impact of lead stress on the biosynthesis of glycans N-linked to proteins, N-glycans from control, low lead and high lead stressed cells were released from total intracellular protein by PNGase F. The released N-glycans were then labeled with Fmoc-Cl and analyzed by ESI-MS. The results are presented in Figure 2. Based on the m/z values of [M+Na]+ ions, ions were majorly assigned to Fmoc derivatives of structure from Man3GlcNAc2 to Man5GlcNAc2. Nevertheless, no obvious ion was assigned to high mannose structures, such as Man8–9GlcNAc2. Furthermore, a linear Man5GlcNAc2 oligosaccharide was observed from three groups.
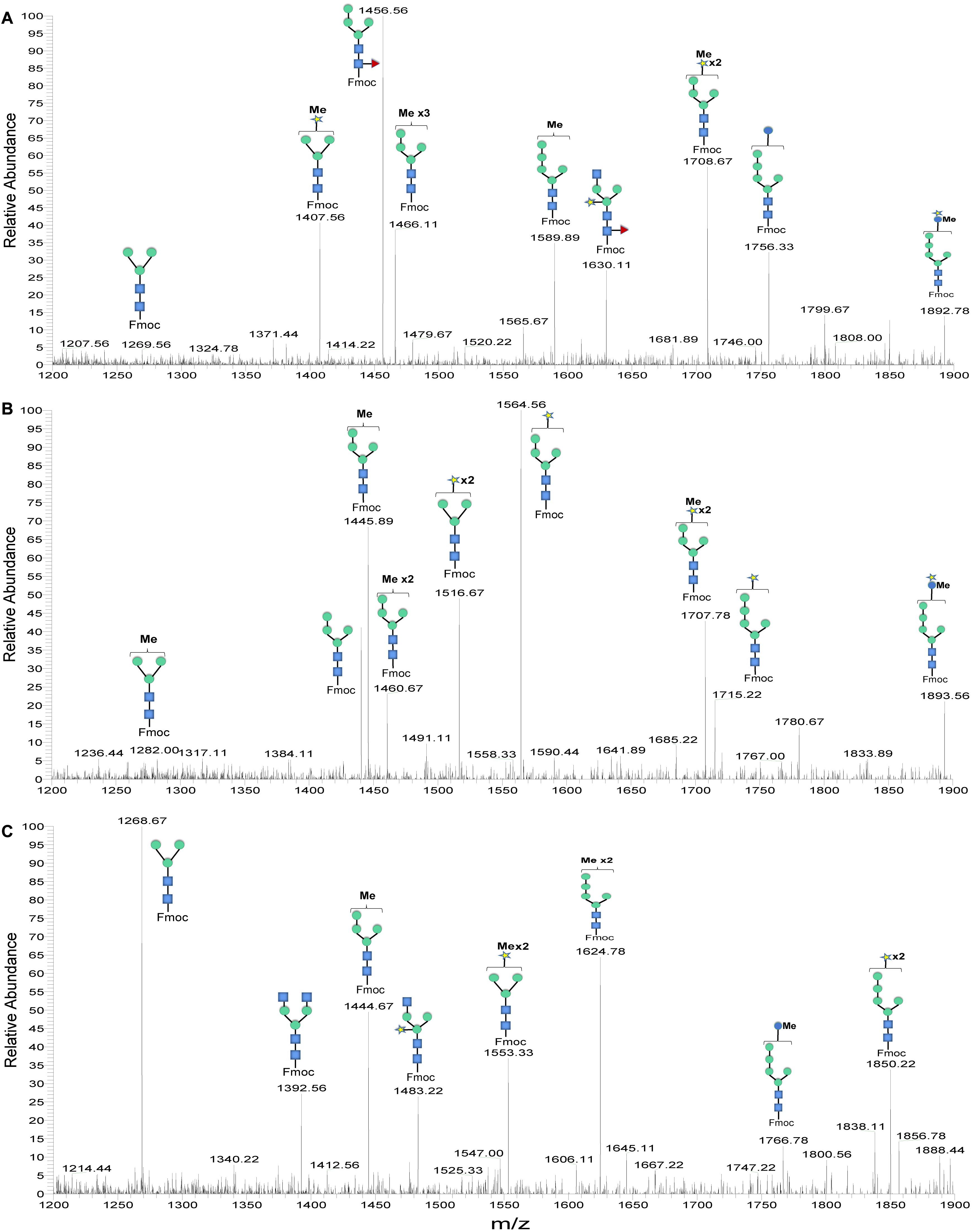
Figure 2. ESI mass spectra of Fmoc-labeled N-glycans released from Chlamydomonas reinhardtii soluble proteins using PNGase F. (A) Control group; (B) low lead stress; (C) high lead stress. Me, methylation; blue square, N-acetylglucosamine; green circle, mannose; white star, xylose; red triangle, fucose; blue circle, glucose; ×2, twice. Monosaccharides and Me depicted above the solid horizontal bracket can be bound to any subjacent residue. Consortium of Functional Glycomics nomenclature was used for the N-glycan structures (Varki et al., 2009).
N-glycan profiles were different among the three groups. To characterize the difference of N-glycan profiles under the third day of the lead stress, the relative abundance of N-glycan structures was compared, as shown in Table 2. Compared with N-linked glycans in the control group, fucose modifying N-glycans (i.e., GlcNAc1Man3Xyl1Fuc1GlcNAc2 and Man4Fuc1GlcNAc2) were not observed under the lead stress. In addition to these two fucose modifying N-glycans, some N-glycan structures were also not identified from the total glycoproteins under the lead stress, including Glc1Man5GlcNAc2, Me3Man4GlcNAc2, Me1Man5GlcNAc2, and Xyl1Me1Man3GlcNAc2. Differently, Me1Man4GlcNAc2 was the unique oligosaccharide structure identified under the low and high lead stresses but not in the control group. Diversely, complex oligosaccharide Me2Man5GlcNAc2, GlcNAc2Man3GlcNAc2, GlcNAc1Man3Xyl1GlcNAc2, Glc1Me1Man5GlcNAc2, Xyl2Man5 GlcNAc2, and Xyl1Me2Man3GlcNAc2 structures were identified under the high lead stress but not under the control and low lead stress. Also, it was shown that the relative abundance of Me2Man5GlcNAc2 oligosaccharide structure was a little bit higher than that of the other oligosaccharide structures. Furthermore, converse to oligosaccharides Xyl2Me1Man4GlcNAc2 and Glc1Xyl1Me1Man5GlcNAc2 in control and low lead stress groups, these two complex N-glycans were not remarkably observed in the high lead stress group. It was observed that oligosaccharides Man4Fuc1GlcNAc2 (28.64%), Xyl1Man4GlcNAc2 (26.83%), and Man3GlcNAc2 (28.45%) had the highest relative abundance under the control group, low lead stress, and high lead stress, respectively.
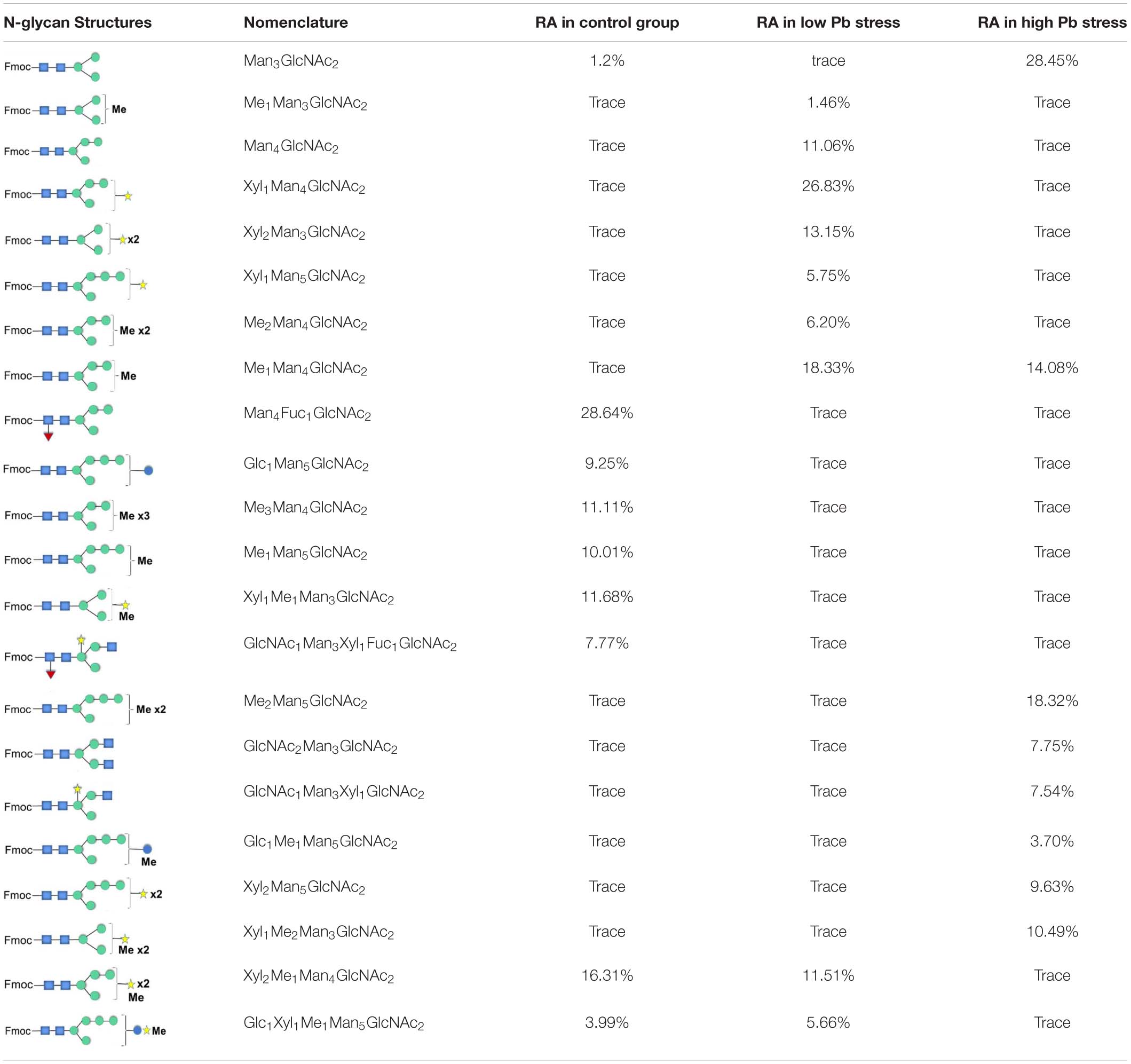
Table 2. Relative abundance (RA) of the N-glycans found on Chlamydomonas reinhardtii soluble proteins under the third day of lead stress.
Additionally, genes involved in the core region of the ERQC (CNX, CRT, GI, GIIA, GIIB, UGGT, and PDIs) systems were also analyzed from the transcriptome of C. reinhardtii under the lead stress. It was shown that only PDIs were differentially expressed and significantly upregulated under the third day of high lead stress (Table 3). In the genome of C. reinhardtii, eight PDI genes were annotated to PDI1-5, PDIA1, and PDIA6. Although the eighth PDI was only predicted to have a thioredoxin domain, therefore, it was speculated to be PDIx here. Among all these eight PDI genes, PDI2, PDIA1, PDIA6, and PDIx were upregulated under the high lead stress.
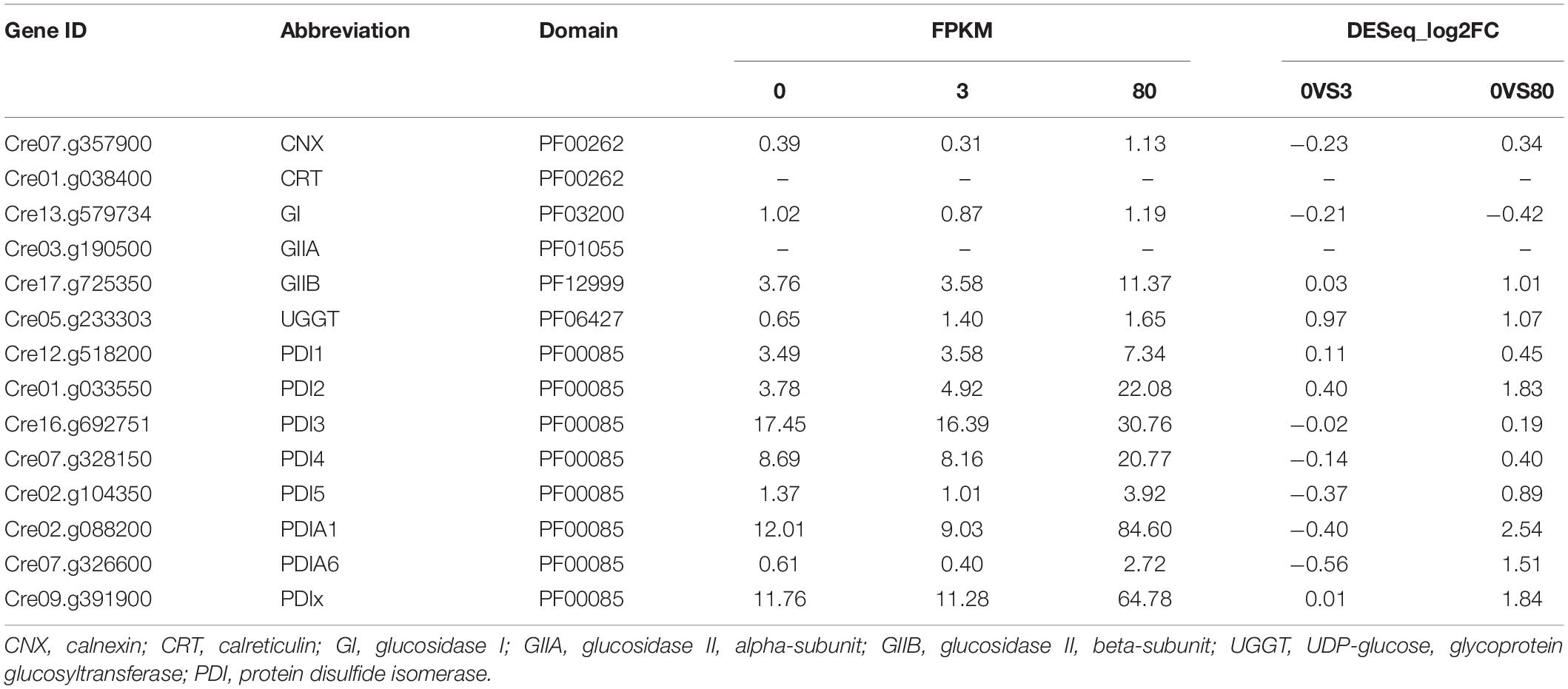
Table 3. Expression of genes involved in the core region of ERQC system in Chlamydomonas reinhardtii under the third day of lead stress.
To further study the PDI genes, bioinformatic analysis was carried out. The expression profiles of eight PDIs were shown in Figure 3A. Obviously, PDI2, PDIA1, PDIA6, and PDIx were all upregulated. In the genome of C. reinhardtii, 17 nuclear chromosomes were found. The positions of the identified PDI genes were drafted to chromosomes by using Mapchart software. Ultimately, eight PDI genes were located on six chromosomes (Figure 3B). Obviously, there is only one PDI gene on each of 1, 9, 12, and 16 chromosomes. They were PDI2 (Cre01g033550), PDIx (Cre09g391900), PDI1 (Cre12g518200), and PDI3 (Cre16g692751), respectively. Additionally, two PDI genes were distributed on chromosomes 2 and 7. They were PDIA1 (Cre02g088200) and PDI5 (Cre02g104350) on chromosome 2 and PDIA6 (Cre07g326600) and PDI4 (Cre07g328150) on chromosome 7. Contrary to PDIA1 and PDI5 genes anchored remotely on chromosome 2, whereas PDIA6 and PDI4 genes were clustered within a very short distance on chromosome 7, less than 0.2 Mb. It was also shown that the four upregulated genes (PDI2, PDIA1, PDIA6, and PDIx) were located on four different chromosomes.
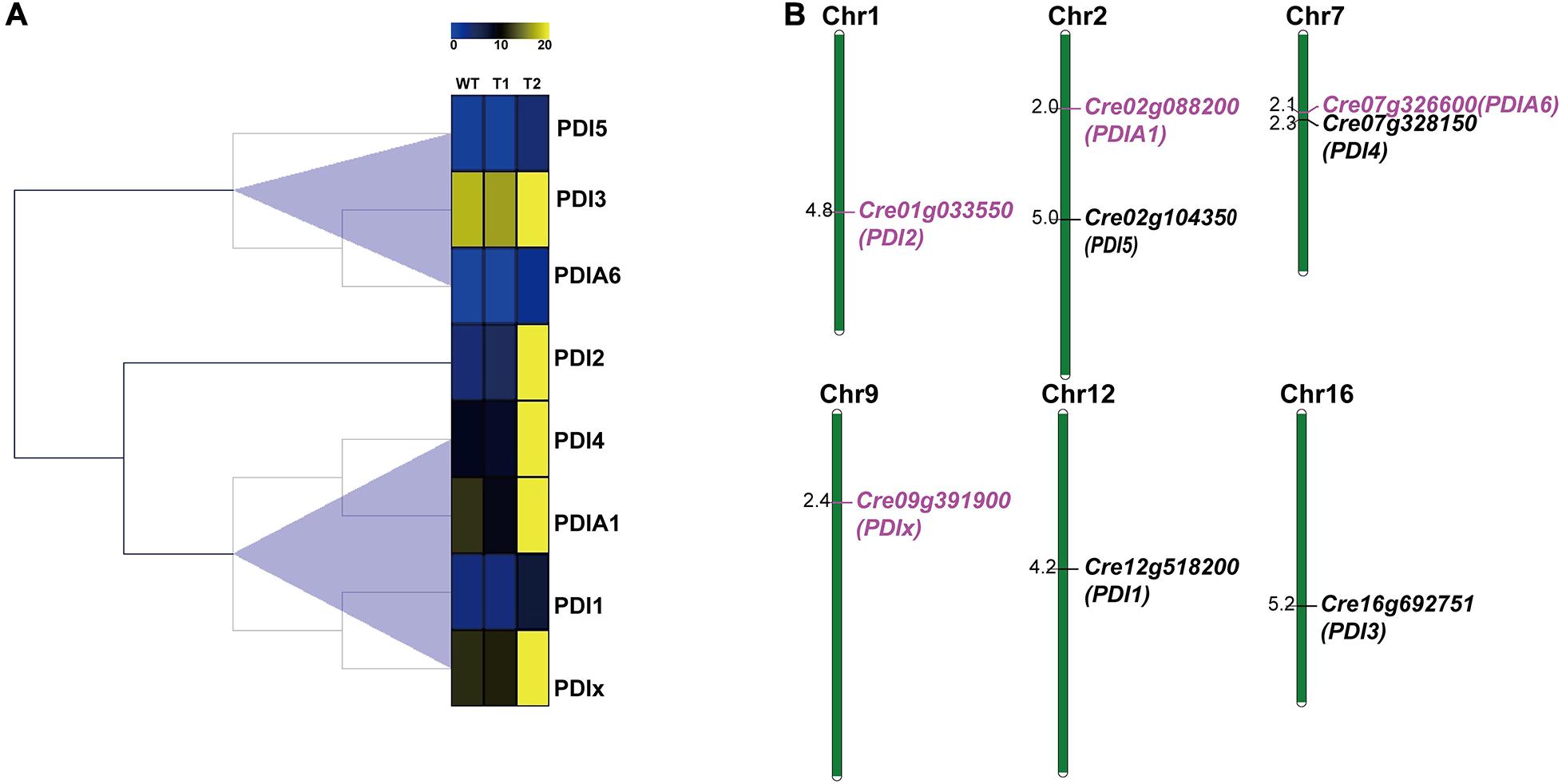
Figure 3. Expression profiles and chromosome distributions of eight PDI genes in Chlamydomonas reinhardtii. (A) Hierarchical clustering of expression of eight PDI genes in C. reinhardtii. (B) Eight PDI genes are distributed to six chromosomes. Number of chromosomes was presented at the top of each chromosome. Some other chromosomes without PDI gene location were not shown in this figure. Scale on the left of each chromosome in megabases (Mb). Genes in purple represent the upregulation expression under the high lead stress.
Fourteen PDI proteins were obtained from A. thaliana by using HMM and BLASTP. As annotated in the previous article, all these 14 PDIs were named AtPDI1-14 in A. thaliana (Yuen et al., 2016). Meanwhile, 8 and 10 PDI proteins were, respectively, achieved from the genomes of green algae O. lucimarinus and C. variabilis. All PDIs harbor one or more catalytic domains; the catalytic domains are homologous to the redox protein, thioredoxin. Therefore, the protein sequences of catalytic domains were obtained from all these PDI proteins and phylogenetically analyzed (Figure 4). Based on phylogenetic results, all catalytic domains were classified into three groups, namely, PDI domain I, PDI domain II, and PDI domain I/II groups. The PDI domain I group was the largest group and contained 28 catalytic domains. The PDI domain II group included nine PDI catalytic domains. Specifically, some PDI domains I and II were mixed in the third group, called PDI domain I/II group. It contained 14 domains, nine domain I and five domain II. Among four upregulated genes, PDIA1 (Cre02g088200) was predicted to have two thioredoxin domains; one domain was clustered into the PDI domain I group, and the other one was assembled with the PDI II domain group. In contrast, the other three PDIs only harbored one thioredoxin domain. As shown in Figure 4, the catalytic domain of PDIx (Cre09g391900) was grouped in PDI domain I. The catalytic domains of PDI2 (Cre01g033550) and PDIA6 (Cre07g326600) were clustered together within the PDI domain I/II group.
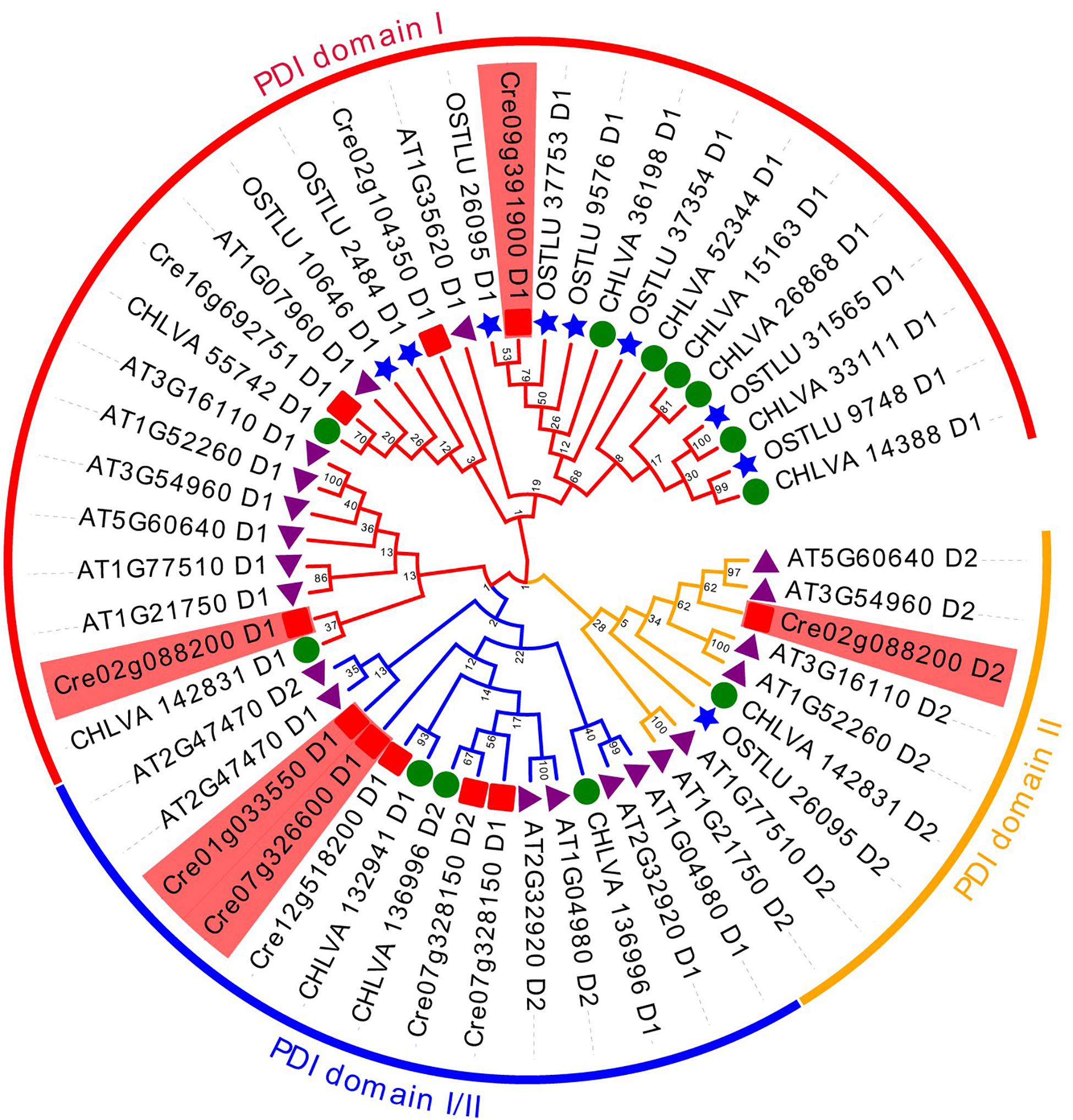
Figure 4. Unrooted phylogenetic analysis of PDI genes from different green algae and plants using the domain sequences. Neighbor-joining (NJ) tree with 1,000 bootstrap replicates was reconstructed using MEGA 6 software and online EvolView. Four green algal species are Chlamydomonas reinhardtii, Ostreococcus lucimarinus, Chlorella variabilis, and plant Arabidopsis thaliana. Different colored arcs/branches are different groups of PDI domains, including red arc for PDI domain I, orange arc for PDI domain II and blue arc for PDI domain I/II. Solid red rectangles represent PDI domains from C. reinhardtii. Solid blue stars represent PDI domains from O. lucimarinus. Solid green circles represent PDI domains from C. variabilis. Solid purple triangles represent PDI domains from plant A. thaliana. Five domains in red background are from four upregulated PDI genes in C. reinhardtii.
Phylogenetic analysis of PDI full-length sequences and their predicted motifs are illustrated in Figure 5. Based on the phylogenetic and motif analysis, all these PDI proteins were classified into night subfamilies, including PDI-A, PDI-B, PDI-C, PDI-D, PDI-E, PDI-F, PDI-M, PDI-S, and PDI-L subfamilies (Figures 5A,B). A search on the MEME program identified 10 motifs in all PDI proteins. The number of motifs in PDIs ranged from 2 to 10, and the length of motifs varied from 21 (motifs 2–3 and motif 7) to 100 (motif 4) amino acids (Figure 5B). All PDIs from the three green algae (C. reinhardtii, O. lucimarinus, and C. variabilis) were not clustered with PDI-A protein from terrestrial plant A. thaliana. The next two purple branches were named PDI-F with a motif 1 (a thioredoxin domain) and a C-terminal alpha helix. These PDI-F proteins were very short, ranging from 131 to 245 amino acids. Two PDI proteins, respectively, from C. reinhardtii (Cre12g518200_PDI1) and C. variabilis (CHLVA_132941) were composed of 540 and 466 amino acids. These two proteins displayed a central thioredoxin domain with a standard YAPWCGHC active sequence (Figures 5A–C). These two specific proteins were grouped into the PDI-D subfamily. Four proteins (Cre07g328150_PDI4, CHLVA_136996, AT2G32920_PDI9, and AT1G04980_PDI10) were divided into PDI-M subfamily. All these four PDI-M proteins possessed two thioredoxin domains with a strictly conserved WCGHC active site. A slightly modified ER retention sequences were observed at the C-terminus of proteins, [K/V/N] [E/D/G] [E/D] L. Considering the absence of motifs 9 and 10 in PDI-M compared with PDI-L, these four proteins (440–486 amino acids) were slightly shorter than that found in PDI-L (501–597 amino acids).
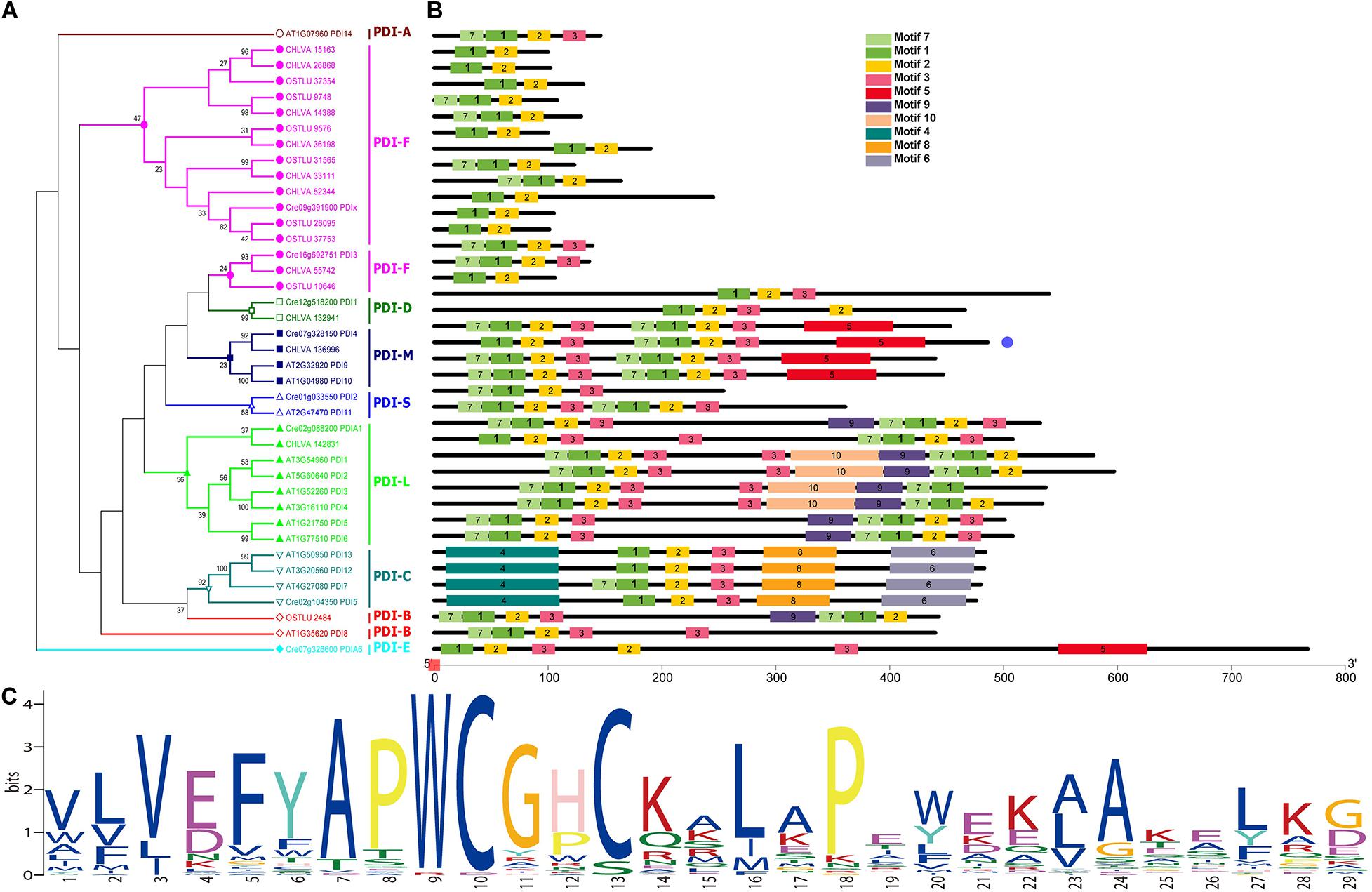
Figure 5. Phylogenetic relationships and conserved motifs of PDI genes from algae and plant. (A) Phylogenetic tree was constructed based on the full-length sequences of algal and plant PDI proteins using MEGA 6 software. Protein in dark red belongs to PDI-A subfamily (hollow circle). Proteins in purple belong to PDI-F subfamily (solid circle). Proteins in dark green belong to PDI-D subfamily (hollow box). Proteins in dark blue belong to PDI-M subfamily (solid box). Proteins in light blue belong to PDI-S subfamily (hollow triangle). Proteins in green belong to PDI-L subfamily (solid triangle). Proteins in cyan belong to PDI-C subfamily (inverted hollow triangle). Proteins in red belong to PDI-B subfamily (hollow rhombus). Proteins in aqua green belong to PDI-E subfamily (solid rhombus). (B) Motif structure of all PDI proteins. Motifs, numbers 1–10, are shown in different colored boxes. Detailed sequence information for each motif is presented in Supplementary Figure 1. Length of each PDI protein can be estimated using the scale at the bottom of the structure of conserved motifs. (C) The sequence logo for motif I (thioredoxin domain).
In this phylogenetic analysis, only two proteins (Cre01g033550_PDI2 and AT2G47470_PDI11) were grouped into the PDI-S subfamily, as shown in Figure 5A. It was observed that PDI11 from A. thaliana contained two thioredoxin domains, whereas Chlamydomonas PDI2 possessed only one thioredoxin domain. The prediction of protein secondary structure showed that these two PDI-S proteins contained a putative alpha-helical conformation and a predicted peptide/protein binding position on the C-terminus. Eight proteins belonging to the PDI-L subfamily were clustered into one branch. These PDI-L proteins were predicted to have two thioredoxin domains (motif I). It was shown that all the PDI-L proteins contained a classical C-terminal ER retention signal (KDEL). In addition to motif 1, all PDI-L proteins harbored conserved motif 2, 3, and 7. Some PDI-L proteins were predicted to have additional motifs, such as motif 9 and 10 in the PDI-L subfamily. In C. reinhardtii, PDIA1 was phylogenetically clustered into this PDI-L subfamily.
The next subfamily is PDI-C in a cyan mark. All these PDI-C proteins contained one thioredoxin domain and some other conserved motifs, including motifs 2, 3, 4, 6, and 8. Motif 4 was predicted to be the N-terminal of ER-Golgi intermediate compartment clusters from the pfam analysis. In contrast, motifs 6 and 8 were COPII coated ER vesicle transporter domains. Additionally, the search of protein sequences showed that all PDI-C proteins without N-terminal signal peptide contained two predicted transmembrane anchoring domains on the terminus. PDI5 harboring these characteristics was classified into this PDI-C subfamily in C. reinhardtii. PDIA6 from C. reinhardtii formed an individual clade in this phylogenetic tree. It was the largest PDI protein with 767 amino acids. Analysis of signal peptide, a transmembrane domain, and conserved PDI domain showed that PDIA6 included one putative thioredoxin domain and a predicted TMD. Compared with PDI-E in Arabidopsis (Selles et al., 2011), PDIA6 was primarily classified into the PDI-E subfamily in this study.
Endoplasmic Reticulum-Associated Degradation Pathway in C. reinhardtii Under Lead Stresses
Incomplete/misfolded glycoproteins will be removed from the ERQC system after multiple CNX/CRT cycles and degraded by an ERAD pathway. What happened in the ERAD pathway of C. reinhardtii under the lead stress? A total of 43 genes with detected expression were identified from the transcriptome of C. reinhardtii under the third day of lead stress (Table 4). All 43 ERAD-related genes were not differentially expressed under the low lead stress. However, 14 genes were significantly upregulated under high lead stress. These upregulated genes were subdivided into different groups, including Uba, Ubc, Ube, Ubx, Ufd, Ubp, and Otu groups.
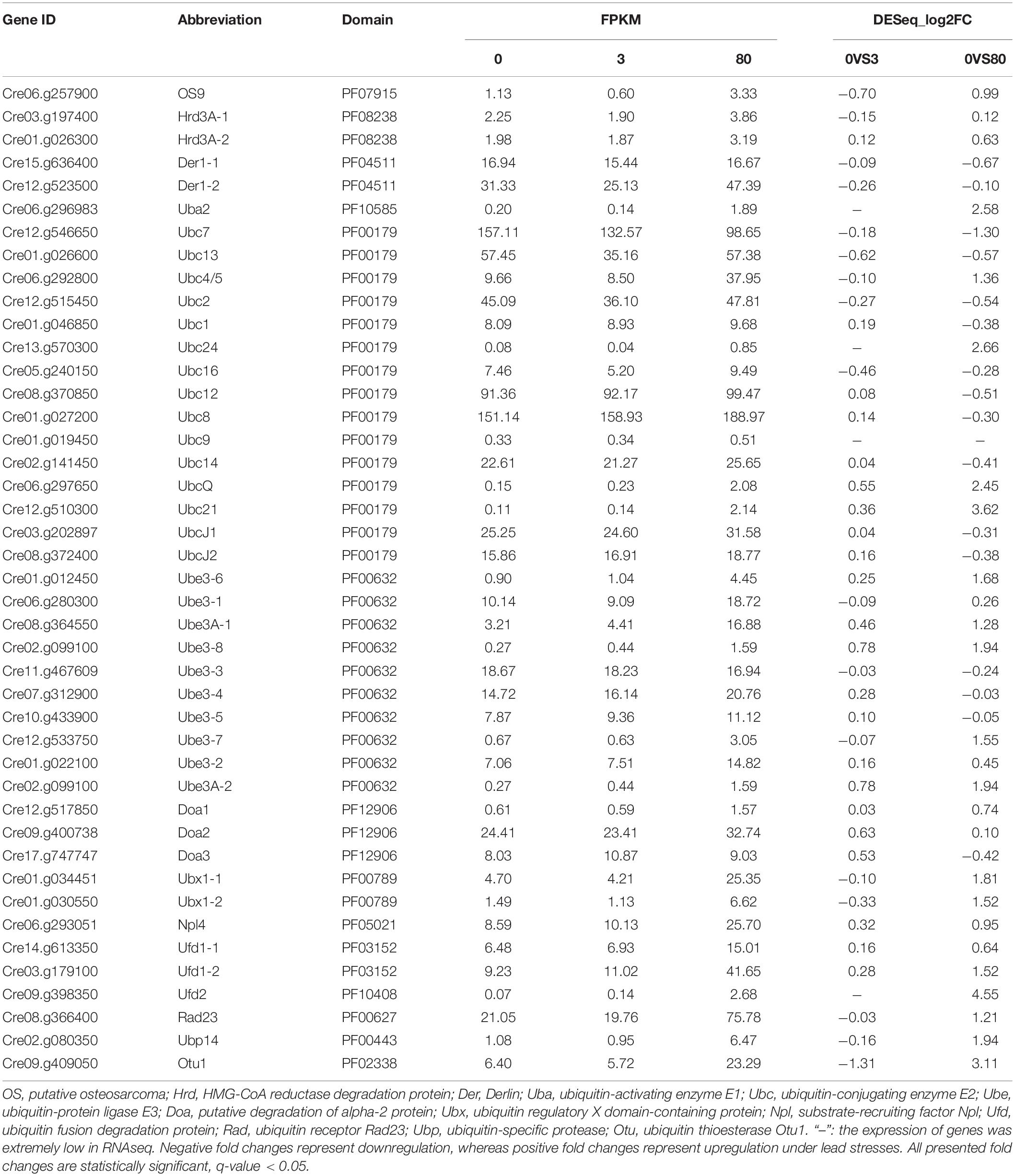
Table 4. Expression of putative genes involved in the ERAD pathway in Chlamydomonas reinhardtii under the third day of lead stress.
Discussion
In algae, Pb can change the algal ultrastructure, effect the growth, photosynthesis, respiration, and the activity of related enzymes, and reduce polysaccharides and protein concentrations. In agreement with previous studies in Chlorella sorokiniana 211-8k, Chlorella sp. FleB1, and Scenedesmus YaA6, the growth of C. reinhardtii was also inhibited under the high lead stress here (Carfagna et al., 2013; Dao and Beardall, 2016). Meanwhile, the concentration of soluble protein was significantly reduced in C. reinhardtii under the high lead stress; this result was consistent with a decrease of soluble protein levels in green microalga C. sorokiniana (Carfagna et al., 2013). Additionally, many studies showed that protein levels could be affected by lead stress (Harrison et al., 1997; Shanab et al., 2012; Qiao et al., 2013; Dao and Beardall, 2016; Li et al., 2016). However, it is still unclear why the protein levels were changed under lead stress.
An efficiently and accurately ERQC system was important in the protein quality control and protein level, as it could differentiate terminally incomplete/misfolded proteins, stopped the futile CNX/CRT folding cycles of the proteins, and eliminated them via a multistep degradation process called ERAD (Smith et al., 2011). It was already known that more than half of secretory and membrane proteins were co-translationally N-glycosylated when transporting into ER (Lizak et al., 2011). Correctly N-linked structures of proteins played an important role in protein folding and quality control (Aebi et al., 2010). In silico analysis of the C. reinhardtii genome, ALG7, ALG13/14, ALG1, ALG2, ALG11, ALG5, ALG6, ALG8, and ALG10 were identified. Among them, ALG13/ALG14 catalyzes the addition of the second core GlcNAc residue to Dol-PP-GlcNAc (Lannoo and Van Damme, 2015). In this study, the ALG14 gene was a unique differentially expressed ALG gene under the high lead stress from the transcriptome analysis of C. reinhardtii. Putative C. reinhardtii ALG14 exhibited 40% of identity with ALG14 from A. thaliana (Mathieu-rivet et al., 2013). It was certified that ALG14 organized the formation of a multiglycosyltransferase complex during the initiation of N-glycan biosynthesis (Kellokumpu et al., 2016). Oligosaccharyltransferase (OST), gatekeeper to the secretory pathway, catalyzes the first step in the biosynthesis of N-glycoproteins (Dempski and Imperiali, 2002). Six subunits of the OST complex (Ribophorin I and II, STT3A, STT3B, OST3/6, and OST5) were predicted from the C. reinhardtii genome. STT3A and STT3B are catalytically active subunits of heteromeric membrane-bound protein OST complex. Ribophorin I was downregulated, and Ribophorin II and STT3B were upregulated in C. reinhardtii under the high lead stress. It was displayed that putative C. reinhardtii STT3A/STT3B exhibited 27% (At5g19960) and 57% (At1g34130) of identity with orthologs from A. thaliana (Mathieu-rivet et al., 2013). On the contrary to the STT3B-deficiency, STT3A-deficient plants displayed an obvious change in N-glycosylation efficiency and affected the biogenesis of heavily glycosylated proteins (e.g., the pattern recognition receptor) (Nekrasov et al., 2009; Häweker et al., 2010). However, stt3a and stt3b double knockout were gametophytic lethal in A. thaliana, suggesting that these two catalytic subunits are very important for the N-glycosylation of plant proteins (Koiwa, 2003). Therefore, it was speculated that the upregulated STT3B was important in the N-glycosylation of Chlamydomonas proteins under the lead stress.
Owing to the four differentially expressed genes (ALG14, RPN1, RPN2, and STT3B genes) in C. reinhardtii, N-glycan structures were also analyzed in this study. So far, N-glycosylation of proteins in C. reinhardtii has received very little attention. The first study in 2011 showed that C. reinhardtii harbored glycoproteins with mammalian-like sialylated N-linked glycans (Mamedov and Yusibov, 2011). Glycoproteins from C. reinhardtii were also found to carry oligomannosidic N-glycans (Man2–5GlcNAc2) containing 6-O-methylated mannoses and one or two xylose residues (Mathieu-rivet et al., 2013). Subsequently, branched Man5GlcNAc2 N-glycans were re-evaluated as being linear oligosaccharide chains based on the analysis of ESI-MSn (Vanier et al., 2017). Compared with these studies in C. reinhardtii, consistent N-glycan structures with Man5 were also identified in this study via the analysis of ESI-MS under the lead stress. Compared with control and low lead stress, the relative abundance of Man3GlcNAc2 was extremely high under the high lead stress. A similar result was reported by a previous study, where it showed that Man3GlcNAc2 glycan was accumulated, protein N-glycosylation was reduced, and the UPR was activated by osmotic stress and hormone treatment in ALG11 mutant, indicating that protein N-glycosylation was important for plant development and the response to abiotic stresses (Zhang et al., 2009). Six specific N-glycans were only observed under high lead stress, although the two N-glycans from glycoproteins were found under the control and low lead stress but not under the high lead stress. One possible explanation for the present or absent N-glycans under the high lead stress is that these modifications were important for the specific functions of glycoproteins to alleviate the Pb stress in the alga. Another possible explanation is that these specific N-glycosylated proteins were accumulated in the ER, stimulating a UPR and finally degraded via the ERAD pathway. The complex N-linked glycans were widely studied in plant stress tolerance, such as Man4GlcNAc2 and Man5GlcNAc2 (Frank et al., 2008; von Schaewen et al., 2008; Nagashima et al., 2018). To our knowledge, the effects of altered N-glycan structures in microalga under the abiotic stress was the first time to be reported here.
Interestingly, in the core region of ERQC, PDI genes were only significantly upregulated under the lead stress. PDI, a molecular chaperone, is involved in the formation, reduction, and isomerization of disulfide bonds for the correct folding of nascent proteins in ER and prevention of misfolding during stress (Strasser, 2020). PDI could be recruited by CNX/CRT for the high-specificity binding between GlcMan9GlcNAc2 and CNX/CRT during the ERQC system (Liu and Li, 2014). The Arabidopsis genome encodes 14 PDI-like proteins harboring one or two thioredoxin-like domains; they are classified into six PDI subfamilies (PDI-A, PDI-B, PDI-C, PDI-L, PDI-M, and PDI-S) (Strasser, 2020). Eight PDI-like proteins (CrPDI1-5, CrPDIA1, CrPDIA6, and CrPDIx) were annotated in C. reinhardtii genome. Among the eight PDI-like proteins, four upregulated PDI genes (CrPDI2, CrPDIA1, CrPDIA6, and CrPDIx) were found from the transcriptome of C. reinhardtii under high lead stress. In a previous article, CrPDI2 was certified to regulate circadian rhythm and interact with peroxiredoxin, especially during night in C. reinhardtii (Filonova et al., 2013). Although CrPDI2 contained one thioredoxin domain, it was clustered with AtPDI11 harboring two thioredoxin domains in the PDI-S subfamily of Arabidopsis. The function of AtPDI11 has remained unknown so far (Yuen et al., 2016). The thioredoxin domain of CrPDI2 (Cre02g033550 D1) and domains from AtPDI11 (AT2G47470 D1/D2) were together clustered into a monophyletic group in PDI domain I/II, indicating that the appearance of a second domain in A. thaliana might result from duplication events during chlorophyte evolution. A similar relationship was also observed in terrestrial plants, and it was supported by the fact that genome duplication and rearrangements were important in the evolution of plants (Selles et al., 2011). CrPDIA1 harboring two thioredoxin domains and an ER retention domain KEDL was divided into the PDI-L subfamily with six PDI from A. thaliana. The mechanism of ERQC was perturbed in the AtUGGT mutant and finally caused the upregulation of the PDI-L2-1 gene (Blanco-Herrera et al., 2015). However, CrPDIA6 containing one thioredoxin domain and long protein sequences was clustered in an independent clade PDI-E. The specific structure suggested that CrPDIA6 might evolve independently from other PDIs. CrPDIx formed a large clade with PDI-F subfamily members. It was known that PDI could aid the correct folding and assembly of nascent proteins in ER to prevent the aggregation of proteins under some unfavorable environmental conditions, such as heat and drought stresses (Branlard et al., 2020). The role of increased PDI protein was to enhance the salt tolerance of Medicago sativa L (Rahman et al., 2015). Therefore, it is reasonable to speculate that the upregulation of four CrPDI genes was important for the tolerance of C. reinhardtii under the lead stress. Together with the upregulation of CrPDI genes in this study, PDI proteins were also differentially expressed in response to other metal stresses, such as arsenic (As), cadmium (Cd), and copper (Cu). The exposure of A. thaliana to Cd showed the upregulation of PDI protein (Sarry et al., 2006). The expression of PDI protein was reduced by excessive As- or Cu-treatments in rice Oryza sativa (Ahsan et al., 2007a, 2008). However, the PDI content was increased by exposure to excessive Cd (Ahsan et al., 2007b). It was suggested that the expression of PDI protein was dependent on species and metals. Furthermore, a previous study indicated that PDI played a vital role in the first committed step of the ERAD pathway under metal stresses (Anelli et al., 2007; Liu et al., 2016). PDIs in plants and animals were verified to form a complex with incomplete/misfolded proteins for the following degradation under the stresses (Liu and Li, 2014; Strasser, 2020), whereas the mechanism of algal PDIs in retaining incomplete/misfolded proteins is still unknown.
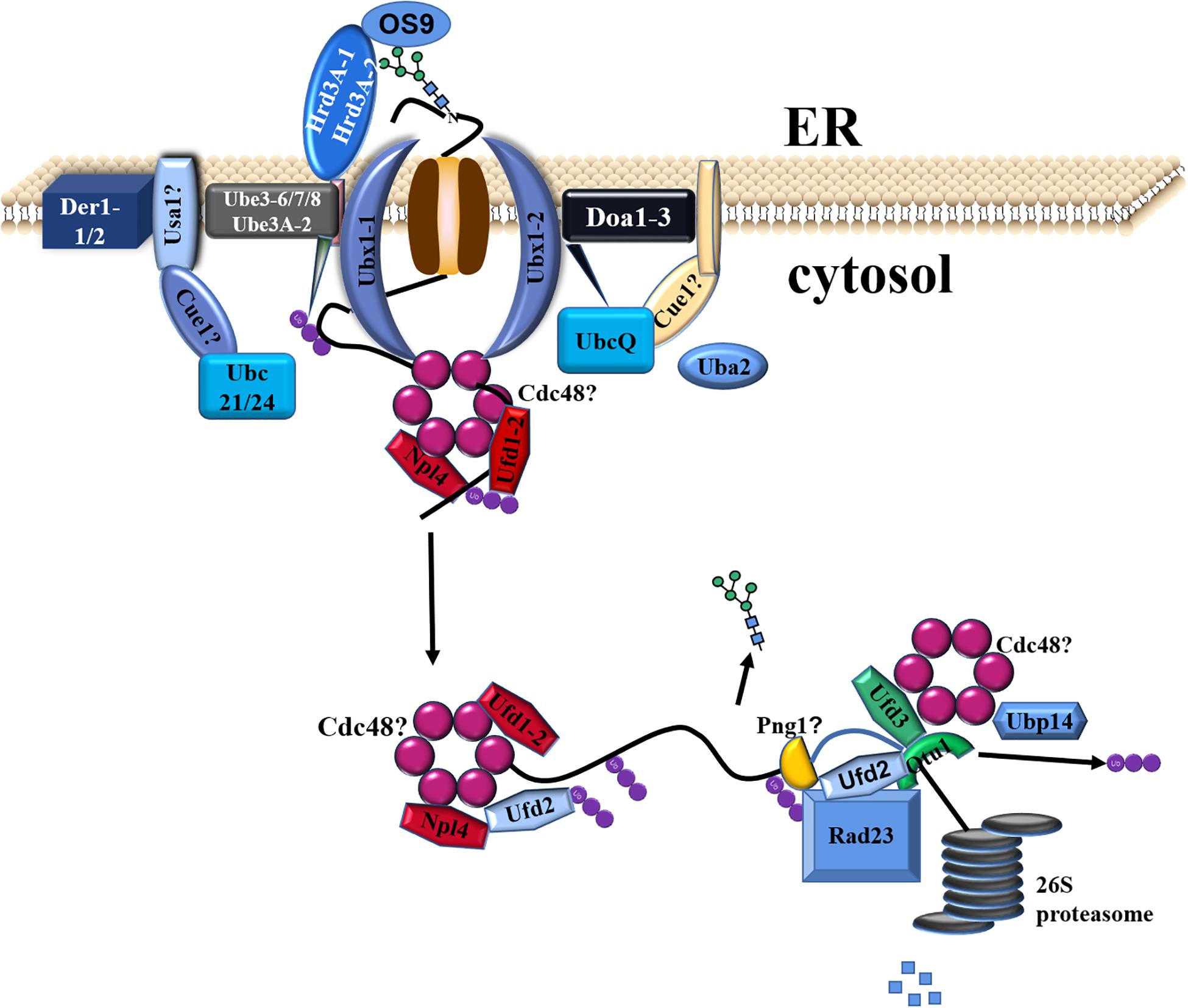
Incomplete/misfolded proteins from the CNX/CRT cycle were finally eliminated in the ERAD pathway through step-wise procedures (Smith et al., 2011). Incomplete/misfolded proteins were increasingly accumulated to cause ER stress under biotic and/or abiotic stresses. Subsequently, UPR was activated to alleviate ER stress via different mechanisms, e.g., inhibition of protein synthesis, suppression of entry of other proteins into the ER, enhancement of protein folding, and degradation (Grootjans et al., 2016; Vincenz-Donnelly and Hipp, 2017). In this study, genes involved in ER stress and UPR were not found to be differentially expressed in C. reinhardtii under the lead stress. However, genes related to ERAD (e.g., Uba, Ubc, Ube, Ubx, Ufd, Ubp, and Otu) were significantly upregulated, and a putative working model was proposed, as shown in Figure 6. The first step is the recruitment of ERAD substrates. The C. reinhardtii genome has three OS9/Hrd3 homologous, CrOS9, CrHrd3A-1, and CrHrd3A-2 (Figure 6). Hrd3 could recognize and bind hydrophobic amino acid residues of misfolded proteins and make the initial selection of a potential ERAD substrate (Hirsch et al., 2009). OS9 was certificated to specifically recognize and bind N-glycans with alpha-1,6-Man residue (Hosokawa et al., 2010). After these recognitions, only terminally incomplete/misfolded proteins were recruited for degradation. However, these three genes were not differentially expressed in this study. The second step is the retrotranslocation of ERAD substrates from the ER lumen to the cytosolic surface of the ER membrane for their ubiquitination (Figure 6). This step is almost unknown in plants and algae. The third step is the ubiquitination of chosen ERAD substrates. The typical ubiquitination reaction was performed by a three-step process, including the activation by a ubiquitin-activating enzyme (E1/Uba), conjugation by the ubiquitin-conjugating enzyme (E2/Ubc), and ligation catalyzed by E3 ligase (Ube) (Pickart, 2004). In this study, one E1 (Uba2), three E2 (Ubc21, Ubc24, and UbcQ), and four E3 (Ube3-6, Ube3-7, Ube3-8, and Ube3A-2) genes were upregulated, indicating that the ubiquitin of substrates was activated under the high lead stress in C. reinhardtii. Cue1 recruited E2 to the ER membrane (Hirsch et al., 2009), and Usa1 could regulate the stability and/or oligomerization of E3 and recruited Der1 to E3 (Carroll and Hampton, 2010). However, blast search failed to find homolog of these two genes in C. reinhardtii genome, suggesting that some alternative genes exist but not yet known are functionally similar to these two genes. The last step is the extraction, processing, delivery, and degradation of ERAD substrates (Figure 6; Liu and Li, 2014). (CDC48)6-Npl4-ATPase-Ufd1 complex participated in the extraction of incomplete/misfolded proteins from ER lumen. Ubx was used to recruit (CDC48)6-Npl4-ATPase-Ufd1 complex to Hrd1/Doa10 E3 complexes on the ER membrane. After the extraction, ERAD proteins were further processed by Ufd2/3-Otu1 complex. Ufd2 is a U-box-containing E4 multiubiquitination enzyme. Otu1 is a deubiquitylating enzyme. Finally, the processed ERAD proteins were delivered to the cytosolic proteasome through Cdc48-Rad23-Dsk2 complex for the final degradation. In this study, Ufd1-2 gene in extraction, Ubx1-1 and Ubx1-2 genes in recruitment, Ufd2 and Otu1 genes in processing, and Ubp gene in degradation were all upregulated under the high lead stress. The upregulated expression of genes in the ERAD pathway suggested that the ERAD pathway was activated under the Pb stress. It is very likely that Pb stress decreased the efficiency of protein folding and increased the accumulation of incomplete/misfolded proteins in the ER, which required a highly efficient and activated ERAD pathway to remove the proteins to maintain ER homeostasis (Liu and Li, 2014). In conclusion, these changes in molecular level explained the significantly reduced content of protein under the high lead stress.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01443/full#supplementary-material. Sequence reads are available by the NCBI sequence read archives [SRR10269729 (control), SRR10269730 (control), SRR10269727 (low lead stress), SRR10269728 (low lead stress), SRR10269731 (high lead stress), and SRR10269732 (high lead stress)].
Author Contributions
XL and HD designed the experiments and supervised the project. XL, CZ, XX, MA, WW, and YY performed the experiments. XL, HD, and MA wrote the original draft of the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages.
Funding
This work was supported by the National Natural Science Foundation of China (41806168 and 41976125), Start-Up funding of Shantou University (no. NTF18004), and Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0606).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All authors acknowledge the help given by Prof. Edmond Sanganyado (a native English speaker) for professional language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.598552/full#supplementary-material
Footnotes
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://pfam.xfam.org/
- ^ http://smart.embl.de/
- ^ https://www.ncbi.nlm.nih.gov/cdd/
- ^ http://plants.ensembl.org/index.html
- ^ http://meme.nbcr.net/meme3/mme.html
- ^ http://www.ebi.ac.uk/interpro
- ^ http://topcons.cbr.su.se/
- ^ http://www.cbs.dtu.dk/services/TMHMM-2.0/
- ^ http://dgpred.cbr.su.se/index.php?p=fullscan
- ^ http://www.cbs.dtu.dk/services/SignalP-3.0/
- ^ https://open.predictprotein.org/about
References
Aebi, M., Bernasconi, R., Clerc, S., and Molinari, M. (2010). N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82. doi: 10.1016/j.tibs.2009.10.001
Ahsan, N., Lee, D. G., Alam, I., Kim, P. J., Lee, J. J., Ahn, Y. O., et al. (2008). Comparative proteomic study of arsenic-induced differentially expressed proteins in rice roots reveals glutathione plays a central role during as stress. Proteomics 8, 3561–3576. doi: 10.1002/pmic.200701189
Ahsan, N., Lee, D. G., Lee, S. H., Kang, K. Y., Lee, J. J., Kim, P. J., et al. (2007a). Excess copper induced physiological and proteomic changes in germinating rice seeds. Chemosphere 67, 1182–1193. doi: 10.1016/j.chemosphere.2006.10.075
Ahsan, N., Lee, S. H., Lee, D. G., Lee, H., Lee, S. W., Bahk, J. D., et al. (2007b). Physiological and protein profiles alternation of germinating rice seedlings exposed to acute cadmium toxicity. C. R. Biol. 330, 735–746. doi: 10.1016/j.crvi.2007.08.001
Anelli, T., Ceppi, S., Bergamelli, L., Cortini, M., Masciarelli, S., Valetti, C., et al. (2007). Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 26, 4177–4188. doi: 10.1038/sj.emboj.7601844
Blanco-Herrera, F., Moreno, A. A., Tapia, R., Reyes, F., Araya, M., D’Alessio, C., et al. (2015). The UDP-glucose: glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. BMC Plant Biol. 15:127. doi: 10.1186/s12870-015-0525-2
Branlard, G., Faye, A., Rhazi, L., Tahir, A., Lesage, V., and Aussenac, T. (2020). Genetic and environmental factors associated to glutenin polymer characteristics of wheat. Foods 9, 23–27. doi: 10.3390/foods9050683
Caramelo, J. J., and Parodi, A. J. (2008). Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283, 10221–10225. doi: 10.1074/jbc.R700048200
Carfagna, S., Lanza, N., Salbitani, G., Basile, A., Sorbo, S., and Vona, V. (2013). Physiological and morphological responses of lead or cadmium exposed chlorella sorokiniana 211-8K (Chlorophyceae). Springerplus 2:147. doi: 10.1186/2193-1801-2-147
Carroll, S. M., and Hampton, R. Y. (2010). Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J. Biol. Chem. 285, 5146–5156. doi: 10.1074/jbc.M109.067876
Dao, L. H. T., and Beardall, J. (2016). Effects of lead on growth, photosynthetic characteristics and production of reactive oxygen species of two freshwater green algae. Chemosphere 147, 420–429. doi: 10.1016/j.chemosphere.2015.12.117
Dempski, R. E., and Imperiali, B. (2002). Oligosaccharyl transferase: gatekeeper to the secretory pathway. Curr. Opin. Chem. Biol. 6, 844–850.
Du, H., Liang, H., Jiang, Y., Qu, X., Yan, H., and Liu, X. (2018). Proteome responses of Gracilaria lemaneiformis exposed to lead stress. Mar. Pollut. Bull. 135, 311–317. doi: 10.1016/j.marpolbul.2018.07.030
Filonova, A., Haemsch, P., Gebauer, C., Weisheit, W., and Wagner, V. (2013). Protein disulfide isomerase 2 of Chlamydomonas reinhardtii is involved in circadian rhythm regulation. Mol. Plant 6, 1503–1517. doi: 10.1093/mp/sst048
Frank, J., Kaulfürst-Soboll, H., Rips, S., Koiwa, H., and Von Schaewen, A. (2008). Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiol. 148, 1354–1367. doi: 10.1104/pp.108.127027
Gillet, S., Decottignies, P., Chardonnet, S., and Le Maréchal, P. (2006). Cadmium response and redoxin targets in Chlamydomonas reinhardtii: a proteomic approach. Photosynth. Res. 89, 201–211. doi: 10.1007/s11120-006-9108-2
Grootjans, J., Kaser, A., Kaufman, R. J., and Blumberg, R. S. (2016). The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 16, 469–484. doi: 10.1038/nri.2016.62
Harris, E. H. (2013). The Chlamydomonas Sourcebook Volume1: Introduction to Chlamydomonas and Its Laboratory Use. Oxford: Elsevier Academic Press.
Harrison, E. P., Willingham, N. M., Lloyd, J. C., and Raines, C. A. (1997). Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 204, 27–36. doi: 10.1007/s004250050226
Hasan, M. K., Cheng, Y., Kanwar, M. K., Chu, X.-Y., Ahammed, G. J., and Qi, Z.-Y. (2017). Responses of plant proteins to heavy metal stress—a review. Front. Plant Sci. 8:1492. doi: 10.3389/fpls.2017.01492
Hassler, C. S., Slaveykova, V. I., and Wilkinson, K. J. (2004). Some fundamental (and often overlooked) considerations underlying the free ion activity and biotic ligand models. Environ. Toxicol. Chem. 23, 283–291. doi: 10.1897/03-149
Häweker, H., Rips, S., Koiwa, H., Salomon, S., Saijo, Y., Chinchilla, D., et al. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636. doi: 10.1074/jbc.M109.063073
Hirsch, C., Gauss, R., Horn, S. C., Neuber, O., and Sommer, T. (2009). The ubiquitylation machinery of the endoplasmic reticulum. Nature 458, 453–460. doi: 10.1038/nature07962
Hosokawa, N., Kamiya, Y., and Kato, K. (2010). The role of MRH domain-containing lectins in ERAD. Glycobiology 20, 651–660. doi: 10.1093/glycob/cwq013
Hutner, S. H. (1990). The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Elizabeth H. Harris. Q. Rev. Biol. 65, 86–86. doi: 10.1086/416634
Kajiura, H., Seki, T., and Fujiyama, K. (2010). Arabidopsis thaliana ALG3 mutant synthesizes immature oligosaccharides in the ER and accumulates unique N-glycans. Glycobiology 20, 736–751. doi: 10.1093/glycob/cwq028
Kamoda, S., Nakano, M., Ishikawa, R., Suzuki, S., and Kakehi, K. (2005). Rapid and sensitive screening of N-glycans as 9-fluorenylmethyl derivatives by high-performance liquid chromatography: a method which can recover free oligosaccharides after analysis. J. Proteome Res. 4, 146–152. doi: 10.1021/pr049825o
Kaulfürst-Soboll, H., Rips, S., Koiwa, H., Kajiura, H., Fujiyama, K., and Von Schaewen, A. (2011). Reduced immunogenicity of Arabidopsis hgl1 mutant N-glycans caused by altered accessibility of xylose and core fucose epitopes. J. Biol. Chem. 286, 22955–22964. doi: 10.1074/jbc.M110.196097
Kellokumpu, S., Hassinen, A., and Glumoff, T. (2016). Glycosyltransferase complexes in eukaryotes: long-known, prevalent but still unrecognized. Cell. Mol. Life Sci. 73, 305–325. doi: 10.1007/s00018-015-2066-0
Koiwa, H. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15, 2273–2284. doi: 10.1105/tpc.013862
Kopittke, P. M., Asher, C. J., and Menzies, N. W. (2008). Prediction of Pb speciation in concentrated and dilute nutrient solutions. Environ. Pollut. 153, 548–554. doi: 10.1016/j.envpol.2007.09.012
Lannoo, N., and Van Damme, E. J. M. (2015). Review/N-glycans: the making of a varied toolbox. Plant Sci. 239, 67–83. doi: 10.1016/j.plantsci.2015.06.023
Li, G. K., Gao, J., Peng, H., Shen, Y. O., Ding, H. P., Zhang, Z. M., et al. (2016). Proteomic changes in maize as a response to heavy metal (lead) stress revealed by iTRAQ quantitative proteomics. Genet. Mol. Res. 15:gmr7254. doi: 10.4238/gmr.15017254
Liu, C., Niu, G., Zhang, H., Sun, Y., Sun, S., Yu, F., et al. (2018). Trimming of N-glycans by the golgi-localized a -1, 2-mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol. Plant 11, 678–690. doi: 10.1016/j.molp.2018.01.006
Liu, J. X., and Howell, S. H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942. doi: 10.1105/tpc.110.078154
Liu, Y. C., Fujimori, D. G., and Weissman, J. S. (2016). Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proc. Natl. Acad. Sci. U. S. A. 113, E4015–E4024. doi: 10.1073/pnas.1608795113
Liu, Y., and Li, J. (2014). Endoplasmic reticulum-mediated protein quality control in Arabidopsis. Front. Plant Sci. 5:162. doi: 10.3389/fpls.2014.00162
Lizak, C., Gerber, S., Numao, S., Aebi, M., and Locher, K. P. (2011). X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–356. doi: 10.1038/nature10151
Lucas, P. L., Dumontier, R., Loutelier-Bourhis, C., Mareck, A., Afonso, C., Lerouge, P., et al. (2018). User-friendly extraction and multistage tandem mass spectrometry based analysis of lipid-linked oligosaccharides in microalgae. Plant Methods 14:107. doi: 10.1186/s13007-018-0374-8
Lucas, P. L., Mathieu-Rivet, E., Song, P. C. T., Oltmanns, A., Loutelier-Bourhis, C., Plasson, C., et al. (2019). Multiple xylosyltransferases heterogeneously xylosylate protein N-linked glycans in Chlamydomonas reinhardtii. Plant J. 102, 230–245. doi: 10.1111/tpj.14620
Mamedov, T., and Yusibov, V. (2011). Green algae Chlamydomonas reinhardtii possess endogenous sialylated N-glycans. FEBS Open Bio 1, 15–22. doi: 10.1016/j.fob.2011.10.003
Mathieu-rivet, E., Scholz, M., Arias, C., Dardelle, F., Schulze, S., Mauff, L., et al. (2013). Exploring the N -glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol. Proteomics 12, 3160–3183. doi: 10.1074/mcp.M113.028191
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi: 10.1126/science.1143609
Nagashima, Y., von Schaewen, A., and Koiwa, H. (2018). Function of N-glycosylation in plants. Plant Sci. 274, 70–79. doi: 10.1016/j.plantsci.2018.05.007
Nakano, M., Higo, D., Arai, E., Nakagawa, T., Kakehi, K., Taniguchi, N., et al. (2009). Capillary electrophoresis-electrospray ionization mass spectrometry for rapid and sensitive N-glycan analysis of glycoproteins as 9-fluorenylmethyl derivatives. Glycobiology 19, 135–143. doi: 10.1093/glycob/cwn115
Nekrasov, V., Li, J., Batoux, M., Roux, M., Chu, Z. H., Lacombe, S., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438. doi: 10.1038/emboj.2009.262
Pedersen, C. T., Loke, I., Lorentzen, A., Wolf, S., Kamble, M., Kristensen, S. K., et al. (2017). N-glycan maturation mutants in Lotus japonicus for basic and applied glycoprotein research. Plant J. 91, 394–407. doi: 10.1111/tpj.13570
Peharec Štefanić, P., Šikić, S., Cvjetko, P., and Balen, B. (2012). Cadmium and zinc induced similar changes in protein and glycoprotein patterns in tobacco (Nicotiana tabacum L.) seedlings and plants. Arh. Hig. Rada Toksikol. 63, 321–335. doi: 10.2478/10004-1254-63-2012-2173
Pickart, C. M. (2004). Back to the future with ubiquitin. Cell 116, 181–190. doi: 10.1016/S0092-8674(03)01074-2
Qiao, X., Shi, G., Chen, L., Tian, X., and Xu, X. (2013). Lead-induced oxidative damage in steriled seedlings of Nymphoides peltatum. Environ. Sci. Pollut. Res. 20, 5047–5055. doi: 10.1007/s11356-013-1475-6
Rahman, M. A., Alam, I., Kim, Y. G., Ahn, N. Y., Heo, S. H., Lee, D. G., et al. (2015). Screening for salt-responsive proteins in two contrasting alfalfa cultivars using a comparative proteome approach. Plant Physiol. Biochem. 89, 112–122. doi: 10.1016/j.plaphy.2015.02.015
Sánchez-Marín, P., Fortin, C., and Campbell, P. G. C. (2014). Lead (Pb) and copper (Cu) share a common uptake transporter in the unicellular alga Chlamydomonas reinhardtii. BioMetals 27, 173–181. doi: 10.1007/s10534-013-9699-y
Sarry, J. E., Kuhn, L., Ducruix, C., Lafaye, A., Junot, C., Hugouvieux, V., et al. (2006). The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6, 2180–2198. doi: 10.1002/pmic.200500543
Scheidegger, C., Sigg, L., and Behra, R. (2011). Characterization of lead induced metal-phytochelatin complexes in Chlamydomonas reinhardtii. Environ. Toxicol. Chem. 30, 2546–2552. doi: 10.1002/etc.654
Selles, B., Jacquot, J. P., and Rouhier, N. (2011). Comparative genomic study of protein disulfide isomerases from photosynthetic organisms. Genomics 97, 37–50. doi: 10.1016/j.ygeno.2010.10.001
Shanab, S., Essa, A., and Shalaby, E. (2012). Bioremoval capacity of three heavy metals by some microalgae species (Egyptian Isolates). Plant Signal. Behav. 7, 392–399. doi: 10.4161/psb.19173
Sharma, P., and Dubey, R. S. (2005). Lead toxicity in plants. Braz. J. Plant Physiol. 17, 35–52. doi: 10.1590/s1677-04202005000100004
Shin, Y. J., Vavra, U., Veit, C., and Strasser, R. (2018). The glycan-dependent ERAD machinery degrades topologically diverse misfolded proteins. Plant J. 94, 246–259. doi: 10.1111/tpj.13851
Smith, M. H., Ploegh, H. L., and Weissman, J. S. (2011). Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090. doi: 10.1126/science.1209235
Stewart, T. J., Szlachetko, J., Sigg, L., Behra, R., and Nachtegaal, M. (2015). Tracking the temporal dynamics of intracellular lead speciation in a Green Alga. Environ. Sci. Technol. 49, 11176–11181. doi: 10.1021/acs.est.5b02603
Strasser, R. (2020). Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plants Biol. 69, 147–172. doi: 10.1146/annurev-arplant-042817-040331.Protein
Szivák, I., Behra, R., and Sigg, L. (2009). Metal-induced reactive oxygen species production in Chlamydomonas reinhardtii (chlorophyceae). J. Phycol. 45, 427–435. doi: 10.1111/j.1529-8817.2009.00663.x
Takano, S., Matsuda, S., Funabiki, A., Furukawa, J. I., Yamauchi, T., Tokuji, Y., et al. (2015). The rice RCN11 gene encodes β1, 2-xylosyltransferase and is required for plant responses to abiotic stresses and phytohormones. Plant Sci. 236, 75–88. doi: 10.1016/j.plantsci.2015.03.022
Van Hoewyk, D. (2013). A tale of two toxicities: malformed selenoproteins and oxidative stress both contribute to selenium stress in plants. Ann. Bot. 112, 965–972. doi: 10.1093/aob/mct163
Vanier, G., Lucas, P. L., Loutelier-Bourhis, C., Vanier, J., Plasson, C., Walet-Balieu, M. L., et al. (2017). Heterologous expression of the N-acetylglucosaminyltransferase I dictates a reinvestigation of the N-glycosylation pathway in Chlamydomonas reinhardtii. Sci. Rep. 7:10156. doi: 10.1038/s41598-017-10698-z
Varki, A., Cummings, R. D., Esko, J. D., Freeze, H. H., Stanley, P., Marth, J. D., et al. (2009). Symbol nomenclature for glycan representation. Proteomics 9, 5398–5399. doi: 10.1002/pmic.200900708
Vincenz-Donnelly, L., and Hipp, M. S. (2017). The endoplasmic reticulum: a hub of protein quality control in health and disease. Free Radic. Biol. Med. 108, 383–393. doi: 10.1016/j.freeradbiomed.2017.03.031
von Schaewen, A., Frank, J., and Koiwa, H. (2008). Role of complex N -glycans in plant stress tolerance. Plant Signal. Behav. 3, 871–873. doi: 10.4161/psb.3.10.6227
Walliwalagedara, C., Keulen, H., Willard, B., and Wei, R. (2012). Differential proteome analysis of Chlamydomonas reinhardtii response to arsenic exposure. Am. J. Plant Sci. 03, 764–772. doi: 10.4236/ajps.2012.36092
Wang, Y., Xu, L., Chen, Y., Shen, H., Gong, Y., Limera, C., et al. (2013). Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLoS One 8:e66539. doi: 10.1371/journal.pone.0066539
Xu, H., Xu, W., Xi, H., Ma, W., He, Z., and Ma, M. (2013). The ER luminal binding protein (BiP) alleviates Cd2+-induced programmed cell death through endoplasmic reticulum stress-cell death signaling pathway in tobacco cells. J. Plant Physiol. 170, 1434–1441. doi: 10.1016/j.jplph.2013.05.017
Yuen, C. Y. L., Wong, K., and Christopher, D. A. (2016). Phylogenetic characterization and promoter expression analysis of a novel hybrid protein disulfide isomerase/cargo receptor subfamily unique to plants and chromalveolates. Mol. Genet. Genomics 291, 455–469. doi: 10.1007/s00438-015-1106-7
Zhang, M., Henquet, M., Chen, Z., Zhang, H., Zhang, Y., Ren, X., et al. (2009). LEW3, encoding a putative-1, 2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J. 60, 983–999. doi: 10.1111/j.1365-313X.2009.04013.x
Zheng, C., Aslam, M., Liu, X., Du, H., Xie, X., Jia, H., et al. (2020). Impact of Pb on Chlamydomonas reinhardtii at physiological and transcriptional levels. Front. Microbiol. 11:1443. doi: 10.3389/fmicb.2020.01443
Keywords: protein level, lead, N-glycan, ER-mediated protein quality control, ER-associated degradation pathway
Citation: Du H, Zheng C, Aslam M, Xie X, Wang W, Yang Y and Liu X (2021) Endoplasmic Reticulum-Mediated Protein Quality Control and Endoplasmic Reticulum-Associated Degradation Pathway Explain the Reduction of N-glycoprotein Level Under the Lead Stress. Front. Plant Sci. 11:598552. doi: 10.3389/fpls.2020.598552
Received: 25 August 2020; Accepted: 04 December 2020;
Published: 13 January 2021.
Edited by:
Philippe Juneau, Université du Québec à Montréal, CanadaReviewed by:
Tse-Min Lee, National Sun Yat-sen University, TaiwanZhaoqing Chu, Shanghai Chenshan Plant Science Research Center (CAS), China
Copyright © 2021 Du, Zheng, Aslam, Xie, Wang, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojuan Liu, bGl1eGlhb2p1YW5Ac3R1LmVkdS5jbg==
†These authors have contributed equally to this work
 Hong Du
Hong Du Canqi Zheng1,2†
Canqi Zheng1,2† Yingquan Yang
Yingquan Yang Xiaojuan Liu
Xiaojuan Liu