- 1Life Science Collegue, Taizhou University, Taizhou, China
- 2National Key Laboratory for Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
SKIP, a component of the spliceosome, is involved in numerous signaling pathways. However, there is no direct genetic evidence supporting the function of SKIP in defense responses. In this paper, two SKIPs, namely, SlSKIP1a and SlSKIP1b, were analyzed in tomato. qRT-PCR analysis showed that the SlSKIP1b expression was triggered via Pseudomonas syringae pv. tomato (Pst) DC3000 and Botrytis cinerea (B. cinerea), together with the defense-associated signals. In addition, the functions of SlSKIP1a and SlSKIP1b in disease resistance were analyzed in tomato through the virus-induced gene silencing (VIGS) technique. VIGS-mediated SlSKIP1b silencing led to increased accumulation of reactive oxygen species (ROS), along with the decreased expression of defense-related genes (DRGs) after pathogen infection, suggesting that it reduced B. cinerea and Pst DC3000 resistance. There was no significant difference in B. cinerea and Pst DC3000 resistance in TRV-SlSKIP1a-infiltrated plants compared with the TRV-GUS-silencing counterparts. As suggested by the above findings, SlSKIP1b plays a vital role in disease resistance against pathogens possibly by regulating the accumulation of ROS as well as the expression of DRGs.
Introduction
The splicing process is completed by spliceosome, which can be classified as two types, including major spliceosome and minor spliceosome (Moore and Proudfoot, 2009; Turunen et al., 2013). SKIP, one of the splicing factors and important components of spliceosome, has possessed several conserved domains (including the SNW/SKI-interacting protein, SKIP) and the specific motifs (Folk et al., 2004; Bres et al., 2009; Chen et al., 2011; Wang et al., 2012). The C terminal plays a vital role in protein stability, while the SNW domain is necessary for the biochemical activity. In Arabidopsis, the SNW domain can integrate into spliceosome in the meantime of interacting with the Paf1 complex (Li et al., 2016). The interaction of SMP1/2 with SKIP facilitates the recruitment of second-step splicing factors into the Arabidopsis spliceosome (Liu et al., 2016).
SKIP is involved in splicing in an either direct or indirect way. In mammals and yeast, SKIP is a component of the 35S-U5snRNP complex, which participates in the common RNA splicing directly (Albers et al., 2003). Growing evidence shows that SKIP is involved in transcription regulation and RNA splicing through interacting with different proteins and thus takes parts in regulating several signaling pathways. For example, SKIP protein participates in at least five signaling pathways in human, including the steroid hormone (Zhang et al., 2001, 2003), TGF-β (Barry et al., 2003), MyoD (Kim et al., 2001), Notch (Zhou et al., 2000; Laduron et al., 2004), and E2F/pRb (Prathapam et al., 2002) pathways.
In addition to RNA splicing, SKIP exerts its functions in numerous steps, such as transcription elongation and transport of mature mRNA. Many species harbor the homologous SKIP protein in nucleus that is between 60 and 80 kDa. However, SKIP in different species has different functions. For instance, in yeast, the weak mutation of the SKIP homolog Prp45 has defects in the splicing of ACTIN and other genes, which leads to the fatal potent mutation of Prp45 and the growth with temperature sensitivity (Figueroa and Hayman, 2004; Bessonov et al., 2008; Gahura et al., 2009). In yeast, SKIP, one of the transcription factors (TFs), helps to modulate gene expression patterns (Lim et al., 2010). In drosophila, Bx42 involved in transcription regulates ecdyson, the Notch signaling pathway, nervous system development, and organ formation (Wieland et al., 1992; Negeri et al., 2002; Ivanov et al., 2004). In nematode, CeSKIP exerts an essential role in individual survival and embryonic development (Kostrouchova et al., 2002; Piano et al., 2002; Kamath et al., 2003; Simmer et al., 2003; Rual et al., 2004; Sonnichsen et al., 2005). Recently, the SKIP functions have been gradually explored, but mainly in Arabidopsis. SKIP shows physical interaction with SR45 (an SR protein specific to plant) for regulating the biological clock. Mutations of the skip-1 gene will lead to a phenotype of a prolonged clock period through changing alternative splicing (AS) in PSEUDO-RESPONSE REGULATOR 7 (PRR7) together with PRR9, both of which are related to the oscillator morning loop. As indicated by this result, SKIP participates in regulating the genes associated with the biological clock of Arabidopsis at the post-transcription level (Wang et al., 2012). Apart from the defects of the biological clock, the skip-1 plant also exhibits the pleiotropic phenotype, such as the early blossoming. Additionally, it is still unclear about how SKIP suppresses floral transition at a molecular or biochemical level, although there are several hypotheses. Cao et al. (2015) reported that, in Arabidopsis, SKIP activated the transcription of FLC to modulate its blossoming through interacting with the Paf1 complex. In Arabidopsis, SKIP regulates the blossoming time by AS of SEF pre-mRNA (Cui et al., 2017), while AtSKIP plays a role of an adjuster between the light signal transduction pathway and cytokinin and thus regulates the cytokinin-associated leaf growth (Zhang X. et al., 2014).
Nowadays, it has been reported that SKIP plays a certain role in the response to stress in a variety of plant, such as Arabidopsis, rice, and maize. The expression level of SKIP can be triggered via salt, abscisic acid (ABA), and mannitol. In the germination process of Arabidopsis, SKIP overexpression leads to abiotic stress tolerance, while SKIP downregulation reduces the abiotic stress tolerance. AtSKIP participates in the ABA signal transduction pathway, which renders resistance to salt or osmotic stress by controlling gene AS of Arabidopsis (Lim et al., 2010; Feng et al., 2015). OsSKIPa, which results in fatal defect in the SKIP homolog knockout mutant of yeast, shows positive modulation on cell growth, viability, and resistance to stress in rice through regulating diverse genes associated with stress at the transcription level (Hou et al., 2009). Besides, interaction of OsSKIP and OsCYP18-2 also exerts a vital part in regulating genes associated with stress at the post-transcription or transcription level and enhancing drought resistance (Lee et al., 2015). In addition, the ZmSKIP overexpression plants with increased ABA content show significantly increased resistance to drought compared with the controls, suggesting that ZmSKIP is involved in the regulation of drought resistance by modulating drought-associated gene expression (Niu, 2012; Wei et al., 2015). GhSKIP35 has a certain function in the resistance to verticillium wilt in Gossypium hirsutum (Liu, 2015). GmGBP1, the human ski interacting protein homolog of soybean, can modulate blossoming together with stress resistance of Arabidopsis by regulating the scavenging activity for reactive oxygen species (ROS) (Zhang et al., 2013).
This research aimed to analyze the functions of SKIP genes in tomato to resist against Botrytis cinerea as well as Pseudomonas syringae pv. tomato (Pst) DC3000, the necrotrophic fungal and the (hemi)-biotrophic bacterial pathogens, separately, using virus-induced gene silencing (VIGS). It was shown in this study that the VIGS-mediated silencing of SlSKIP1b resulted in the accumulation of more ROS, but decreased the levels of defense-related genes (DRGs) in the case of pathogen infection, thereby attenuating B. cinerea and Pst DC3000 resistance. These findings demonstrated that the SKIP genes play vital parts in regulating the anti-pathogen response of tomato.
Materials and Methods
Plant Cultures as Well as Treatments
Two tomato varieties (Solanum lycopersicum), Suhong2003 as well as MicroTom, were employed in this research. Suhong2003 was used in all experiments except for the whole plant disease assays against B. cinerea that used the cultivar MicroTom. Tomato seedlings were grown in a material mixture within a greenhouse. Besides, seedlings of 2 and weeks old were utilized to carry out VIGS assays and to analyze gene expression after pathogen inoculation and treatments with defense-related signal molecules, respectively. Typically, the treatments with defense-related signal molecules were done by spraying MeJA, ACC, and SA (all at 100 μM and from Sigma-Aldrich), and water was used as control. At the designated time points after treatment, the leaves were collected.
Pathogen Infection Together With Disease Assays
In this study, B. cinerea infection in tomato plants was completed by two approaches (Li et al., 2014; Zhang et al., 2020). In brief, after collecting spores, their densities were tuned to 1 × 105 spores/mL. For the detached leaf disease assays, leaf samples with full expansion were collected from the 6-week-old VIGS agroinfiltrated plants, and put on the cheesecloth pre-immerged within the distill sterilized water in trays. Each side of the leaves was inoculated by a drop of 2.5 μL spore suspension, followed by disease development in high humidity. At 4 days later, the lesion size in those infected leaf samples was recorded. In the whole plant disease assays, spore suspension was sprayed onto tomato plants until it evenly covered the leaf surface. Afterward, those infected plants were then put into a high-humidity environment. At 4 days after inoculation, photographs were taken for the phenotype. Then, after collecting leaves at the designated time points, the fungus quantity and gene levels were analyzed. qRT-PCR was adopted to define fungal growth by B. cinerea BcActinA gene expression.
Plants were inoculated with Pst DC3000 according to the following steps (Li et al., 2014): after harvesting and resuspending bacteria into MgCl2 (10 mM) to OD600 = 0.0002, all leaves were immersed into the bacterial suspension using the 0.04% Silwet L-77, followed by 1.5 min of negative pressure treatment at −40 kPa. The phenotype was photographed at 4 days following infection. Besides, leaves were harvested to analyze specific gene levels as well as bacterial growth. To measure the bacterial growth, 70% ethanol was used to sterilize leaf discs for 10 s, then sterile water was utilized to wash them for thrice, followed by grinding within the 10 mM MgCl2 solution (200 μL) until a homogenate was obtained. Later, the homogenate was diluted with 10 mM MgCl2 at a ratio of 1:10 to different concentrations, cultured in the King’s B solid medium for 3 days, and the colonies were recorded.
Characterization of SlSKIP Genes
Using the BlastP program, the tomato genome database was searched at http://solgenomics.net using those featured Arabidopsis AtSKIP as queries. Afterward, those obtained sequences of SlSKIPs nucleotides and amino acids (AAs) were downloaded.
RNA Extraction Along With qRT-PCR
The Trizol reagent (Invitrogen, Shanghai, China) was used to extract total RNA according to specific protocol. The PrimeScript RT reagent kit (TaKaRa, Dalian, China) was used for reverse transcription following specific instructions to synthesize cDNAs, which served as the templates for PCR and qRT-PCR. In this study, the CFX96 real-time PCR assay system (Bio-Rad, Hercules, CA, United States) was used for qRT-PCR. Dissociation curves were used to verify that the amplified production was single in PCR. Target gene transcript expression was shown as relative transcript expression to an Actin gene in tomato. The 2–△ △ CT approach was applied in calculating the relative gene expression level according to previous description. Table 1 lists those gene-specific primers adopted for qRT-PCR.
Construction of the VIGS Vector and Agroinfiltration
Fragments of 300–400 bp in sizes for SlSKIPs were amplified by PCR with respective pairs of gene-specific primers (Table 1). The amplified PCR products were digested with corresponding restriction enzymes (XbaI/XhoI) and cloned into TRV2, yielding recombinant plasmids TRV-SlSKIP1a and TRV-SlSKIP1b. After confirmation by sequencing, the correct recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation and positive clones were selected by colony PCR for VIGS assays. Agrobacteria carrying TRV-SlSKIP1a or TRV- SlSKIP1b were grown in YEP liquid medium with 50 μg/mL kanamycin, 50 μg/mL rifampicin, and 25 μg/mL gentamicin in a shaker until OD600 reached to 0.8∼1.0. Agrobacterial cells were collected by centrifugation and resuspended in infiltration buffer containing 10 mM MgCl2, 10 mM MES (pH5.7) and 200 μM acetosyringone, and the bacterial concentrations in suspensions were adjusted to OD600 = 1.5. The agrobacteria carrying TRV-SlSKIP1a or TRV-SlSKIP1b were mixed with the same volume of agrobacteria carrying TRV1, and the mixtures were incubated for 3 h at room temperature. The mixed agrobacterial suspension was separately infiltrated into the abaxial surface of the 2-week-old seedlings using a 1-mL needleless syringe (Liu et al., 2002). A group of tomato seedlings were infiltrated with agrobacteria harboring a construct of TRV-PDS (Phytoene desaturase) and used as positive controls for silencing evaluation of the VIGS procedure (data showed in Supplementary Material). The agroinfiltrated plants were allowed to grow for 4 weeks in a growth room under the same conditions as mentioned above and then used for different experiments.
Detection and Measurement of H2O2
The DAB staining method was utilized to detect H2O2 accumulation within the leaf tissues. After B. cinerea (0 and 24 h) and Pst DC3000 (0 and 48 h) inoculation, leaves were harvested, respectively. After 3 h of immersion into the 1-mg/ml DAB solution (pH 3.8), the leaf samples were boiled in 95% ethanol until the chlorophyll was completely removed. Finally, a digital camera was utilized to visualize H2O2 accumulation in those stained leaf samples. The H2O2 measurement was done by an H2O2 Kit (Jiancheng, Nanjing, China). The content of H2O2 was calculated by the formula: (OD value of the measured sample − OD value of the blank)/(OD value of a standard solution − OD value of the blank × 163 mmol/L)/concentration of the protein of tissue.
Experiment Design and Data Analysis
All experiments were repeated independently three times. More than 10 plants were used in each of independent experiments such as disease assay with B. cinerea or Pst DC3000. Data obtained from three independent experiments were subjected to statistical analysis according to the Student’s t-test. The probability values of p < 0.05 were considered as significant difference between treatments and their corresponding controls.
Results
Characterization of SlSKIP Genes in Tomato
Using the characterized Arabidopsis AtSKIP genes as queries, a tomato genomic database was searched by Blastp analysis, and two loci were identified in tomato genome, which were named as SlSKIP1a (XM_004251580.4) and SlSKIP1b (XM_004250540.4) (the information about SlSKIPs can be seen in Supplementary Data). Moreover, ESTs together with the potential full-length cDNAs of SlSKIPs were discovered against tomato genomic database as well as NCBI GenBank database, separately, which indicated the constitutive expression of SlSKIPs in tomato. The ORFs of SlSKIPs were cloned and sequenced, which found the totally same SlSKIPs ORF sequences with those predicted ORF sequences.
Expression Models of SlSKIPs With Pathogen Inoculation and Treatments With Defense-Related Signal Molecules
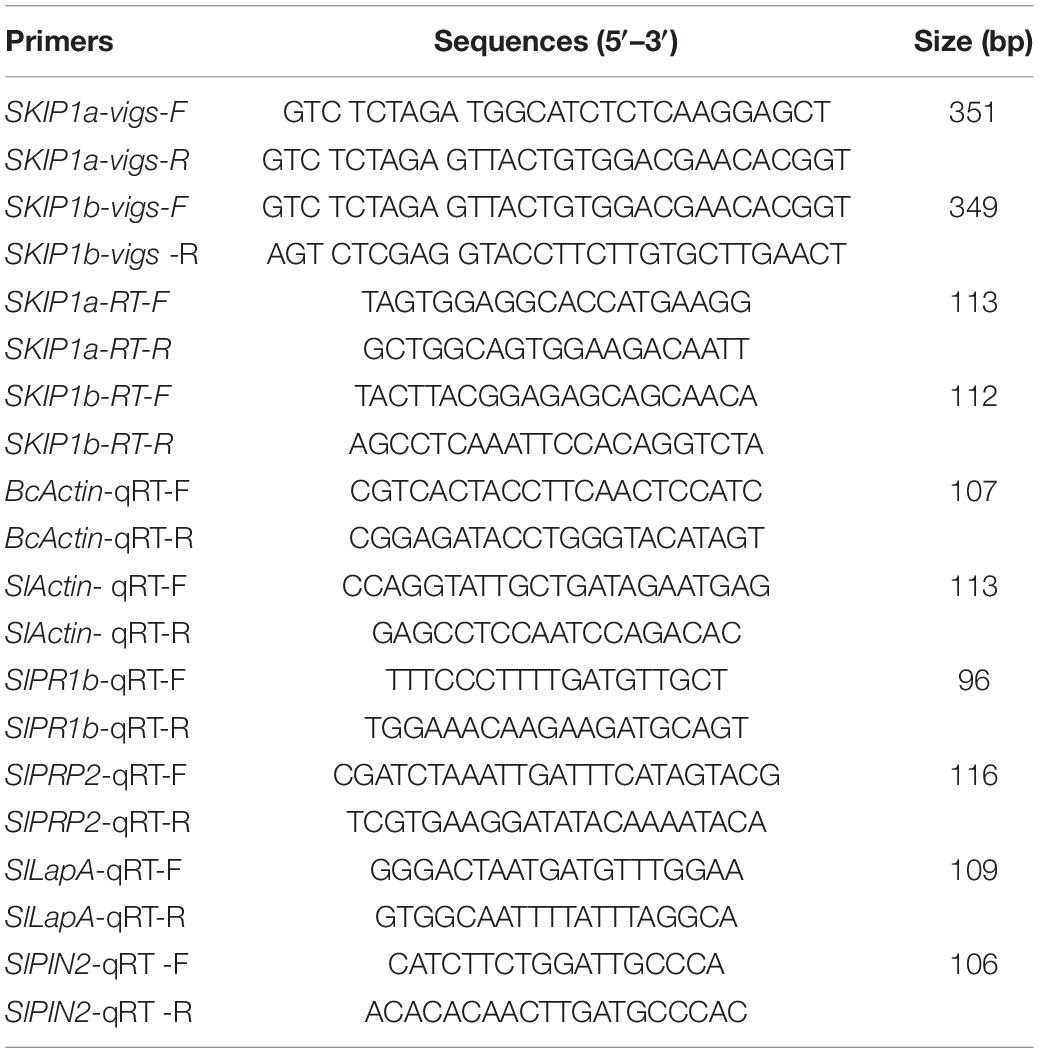
The function analysis of SlSKIPs was completed to reveal their probable biological roles in resisting against disease. First, this study investigated the SlSKIP1a and SlSKIP1b expression models responding to the inoculation of Pst DC3000 and B. cinerea, together with the treatment of defense-related signal molecules, including 1-amino cyclopropane-1-carboxylic acid (ACC, the ET precursor), methyl jasmonate (MeJA), and salicylic acid (SA) in tomato plants. After 72 h of Pst DC3000 infection, the SlSKIP1b level was notably upregulated by about 6.3 times relative to that in control plants with mimic inoculation, whereas the SlSKIP1a level showed no difference after Pst DC3000 infection (Figure 1A). In the case of inoculation with B. cinerea, the condition was very similar to that in inoculation with Pst DC3000. After 48 h of B. cinerea infection, the SlSKIP1b level prominently increased by about 7.1-folds relative to that in control plants with mimic inoculation, but the SlSKIP1a level showed no dramatic difference (Figure 1B). Moreover, none of our selected signal molecules associated with defense affected the SlSKIP1a level, while all of those signal molecules triggered SlSKIP1b expression (Figures 2A,B). In addition, the SlSKIP1b level significantly increased at 12 h after ACC and JA treatments, and its expression was the maximal at 24 h after SA treatment (Figure 2B). As suggested by these data, SlSKIP expression might be triggered via B. cinerea and Pst DC3000 as well as the defense-related signal molecules.
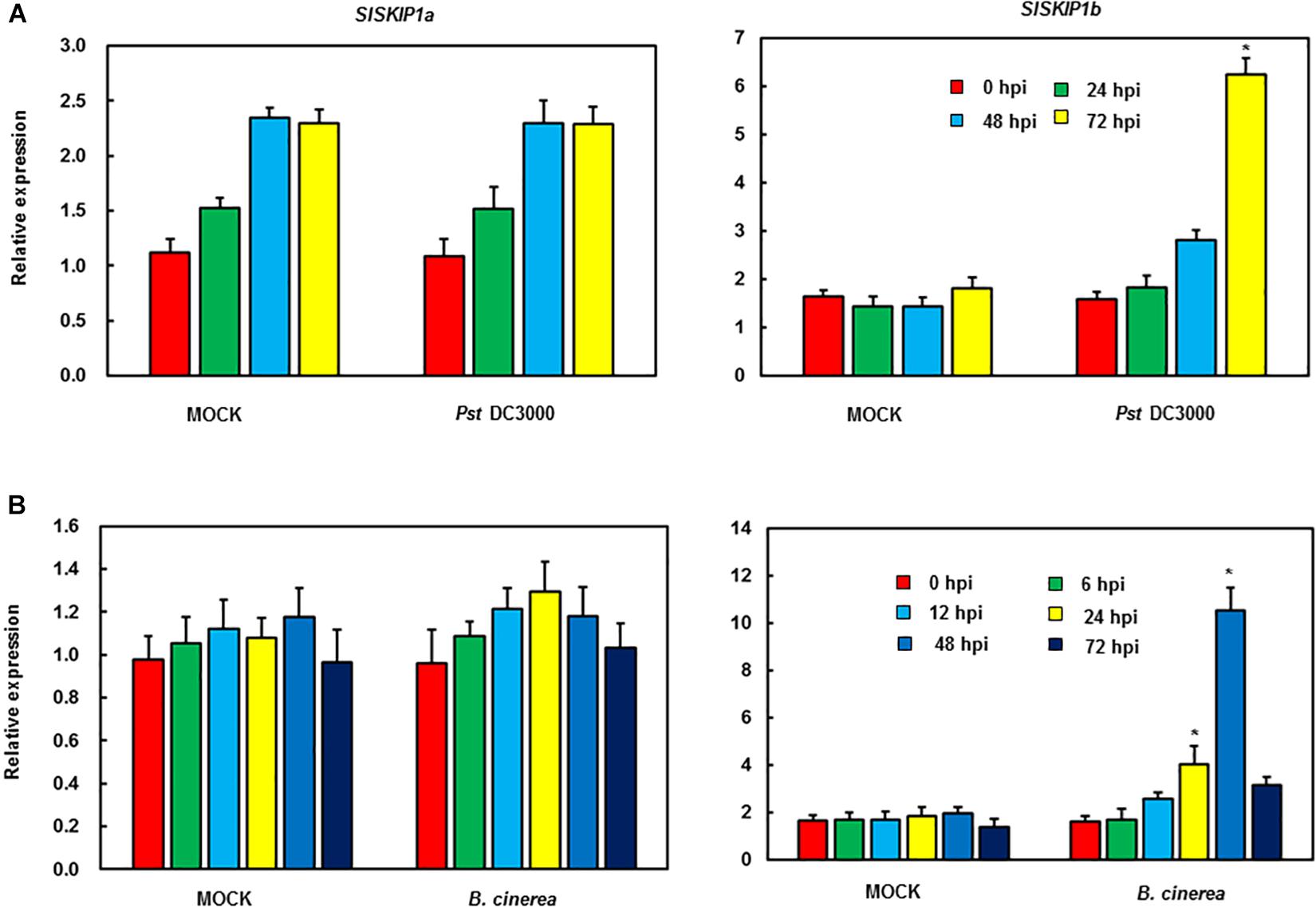
Figure 1. SlSKIP expression responding to B. cinerea together with Pst DC3000 infection. (A) SlSKIP gene expression patterns responding to Pst DC3000 infection. (B) SlSKIP gene expression patterns responding to B. cinerea infection. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
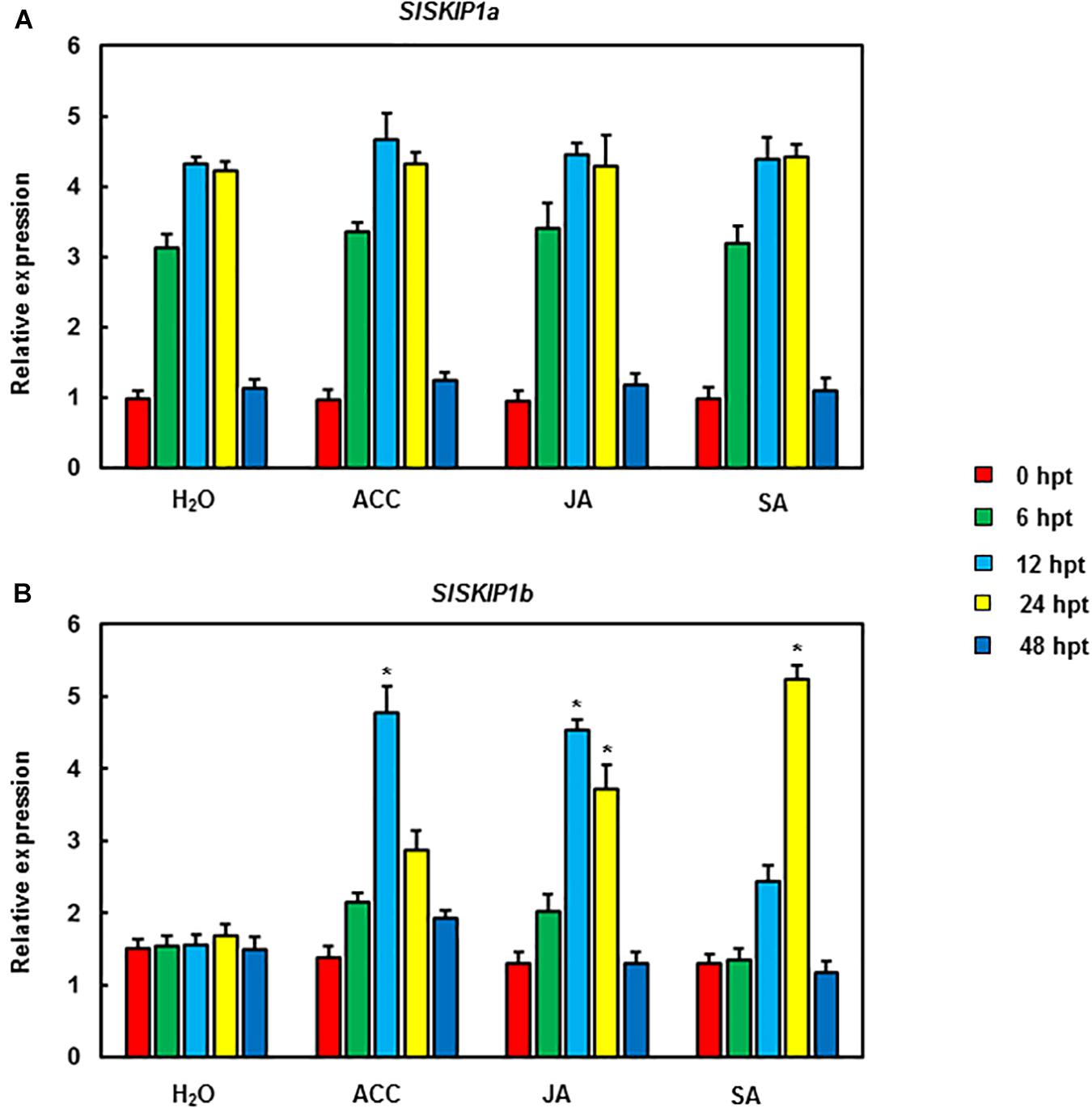
Figure 2. SlSKIP expression levels responding to treatments with defense-related signal molecules. SlSKIP gene expression models responding to defense-related signal molecules treatments were analyzed. To be specific, 100 μM of SA, MeJA, and ACC was used to treat tomato plants by foliar spraying; alternatively, a similar amount of solution with no abovementioned signal molecules was used as control. Later, the leaves were harvested at the indicated time points to analyze gene expression through qRT-PCR. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level. (A) The expression level of SlSKIP1a with the treatments of defense-related signal molecules. (B) The expression level of SlSKIP1b with the treatments of defense-related signal molecules.
SlSKIP Silencing of Tomato
For analyzing SlSKIP effects on the resistance against disease, the VIGS approach was utilized to manage the endogenous SlSKIP expression. Therefore, the SlSKIP gene silencing efficiency was checked at first. Then, the normal VIGS protocol was adopted for the 2-week-old tomato seedlings. At 4 weeks later, the silencing efficiency was measured, with plants transfected using the TRV-PDS construct being the positive controls. The silencing efficiency of SlSKIP genes was evaluated to be 65%, which was used for further functional studies (Figures 3A,B).
Figure 3. SlSKIP gene silencing efficiency along with specificity of plants inoculated with VIGS. (A) The SlSKIP1a and SlSKIP1b silencing efficiency of plants inoculated with TRV-SlSKIP1a and those inoculated with TRV-SlSKIP1b, respectively. (B) SlSKIP1a silencing specificity of plants inoculated with TRV-SlSKIP1a. (C) SlSKIP1b silencing specificity of plants inoculated with TRV-SlSKIP1b. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
Silencing of SlSKIP1b Led to Reduced B. cinerea Tolerance
For studying those potential SlSKIP genes’ functions to resist B. cinerea, this study applied two distinct approaches, namely, detached leaf and whole plant disease assays for preliminary and further confirmation, respectively. The seedlings of TRV-SlSKIPs- and TRV-GUS-infiltrated plants were compared for their disease phenotypes and fungal quantity, for the sake of confirming disease phenotype. As obtained from detached leaf disease assays, the leaf lesion size in TRV-SlSKIP1b-infiltrated plants prominently elevated by about 54% at 3 days after infection (dpi) (Figure 4A), compared with that in TRV-GUS-infiltrated counterparts (Figure 4B). Meanwhile, the leaf lesion size in TRV-SlSKIP1a-infiltrated plants (3 dpi) was not significantly different from that of the TRV-GUS-infiltrated counterparts (Figures 4A,B).
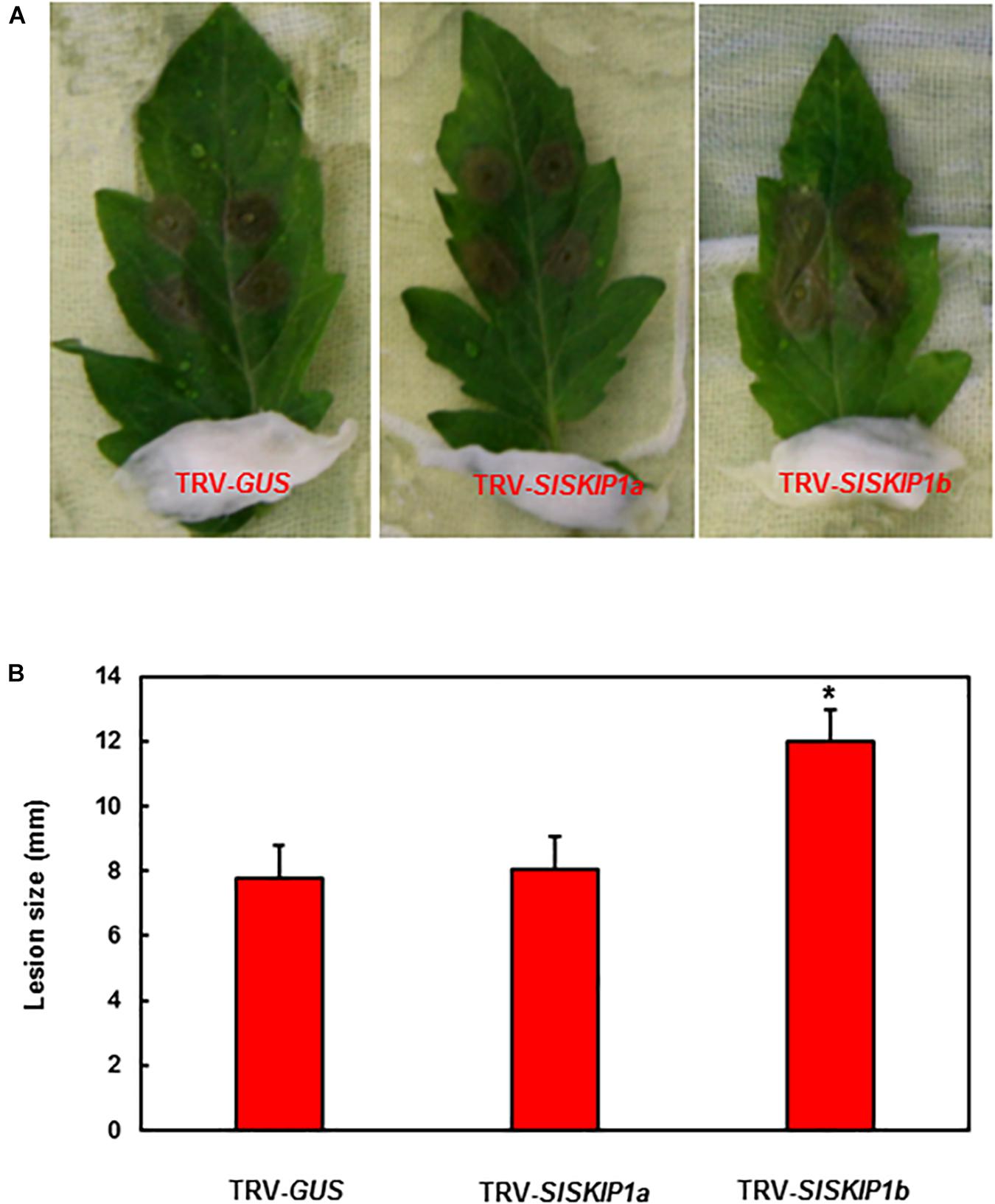
Figure 4. SlSKIP1b silencing led to the weakened B. cinerea tolerance verified through detached leaf disease assays. Agrobacteria that carried TRV-SlSKIPs and TRV-GUS constructs were transfected into the tomato plants of 10 days old, and then leaves were harvested following agroinfiltration for four weeks. (A) Disease symptoms of typical leaf samples collected based on plants inoculated with TRV-SlSKIP and those inoculated with TRV-GUS. (B) Leaf lesion size in plants inoculated with TRV-SlSKIP and those inoculated with TRV-GUS. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
For further confirming the above finding, whole-plant disease assays were conducted to estimate disease phenotype and test B. cinerea fungal growth in planta of plants inoculated with TRV-SlSKIP. According to Figure 5A, plants infiltrated with TRV-GUS showed mild disease symptom compared with those inoculated with TRV-SlSKIP1b, while those inoculated with TRV-SlSKIP1a displayed no difference from those inoculated with TRV-GUS at 5 dpi. At 24 and 48 hpi, the B. cinerea growth in planta, which was expressed as B. cinerea BcActinA gene transcript level, notably elevated by threefold in leaves of plants inoculated with TRV-SlSKIP1b compared with those inoculated with TRV-GUS (Figure 5B). While B. cinerea growth for plants inoculated with TRV-SlSKIP1a did not show any significant difference compared with plants inoculated with TRV-GUS (Figure 5B). Collectively, the above findings suggested that SlSKIP1b silencing reduced B. cinerea tolerance in tomato plants, with excessive B. cinerea growth of TRV-SlSKIP1b-infiltrated plants.
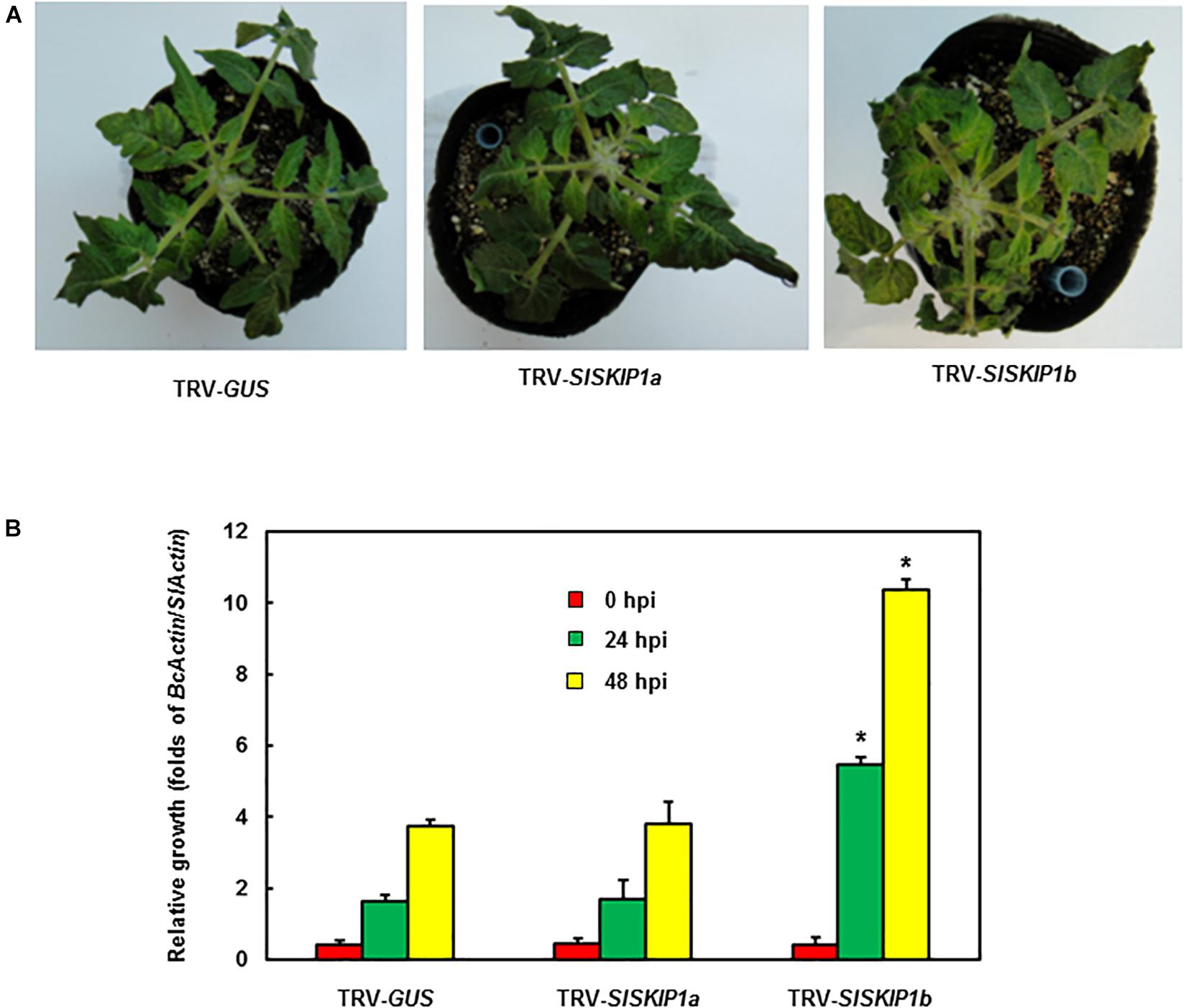
Figure 5. Silencing of SlSKIP1b resulted in the weakened B. cinerea tolerance evidenced by whole-plant disease assays. (A) Disease phenotypes in typical plant leaves inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS. After four days of inoculation, the photographs were taken. (B) B. cinerea growth in planta of plant leaf samples inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
To gain insights into the probable mechanism by which SlSKIP1b silencing led to weakened B. cinerea tolerance, this study analyzed the ROS accumulation together with the expression levels of DRGs. Before B. cinerea infection, no obvious H2O2 accumulation was observed in plants inoculated with TRV-SlSKIP1b or those inoculated with TRV-GUS, but H2O2 accumulation significantly increased at 24 h following B. cinerea inoculation (Figure 6A). The H2O2 concentration was further measured. The results showed that the H2O2 concentration in plants inoculated with TRV-SlSKIP1b was much higher than that of the ones inoculated with TRV-GUS after B. cinerea infection, but there was no significant difference before B. cinerea infection (Figure 6B). Similarly, SlPRP2 and SlPR1b (the DRGs responding to the SA signaling) and SlLapA and SlPIN2 (DRGs responding to the JA/ET signaling) were almost the same in plants inoculated with TRV-SlSKIP1b as those inoculated with TRV-GUS prior to B. cinerea inoculation (Figure 6C). B. cinerea infection was the primary cause inducing the expression of the above four DRGs, relative to those of uninfected controls. However, at 24 hpi, SlPR1b and SlPRP2 expression slightly decreased, whereas SlPIN2 and SlLapA expression notably reduced in plants inoculated with TRV-SlSKIP1b, relative to those TRV-GUS-infected counterparts (Figure 6C). Collectively, the above results suggested that SlSKIP1b silencing resulted in reduced accumulation of ROS, as well as decreased levels of DRGs responding to the JA/ET signaling after B. cinerea inoculation.
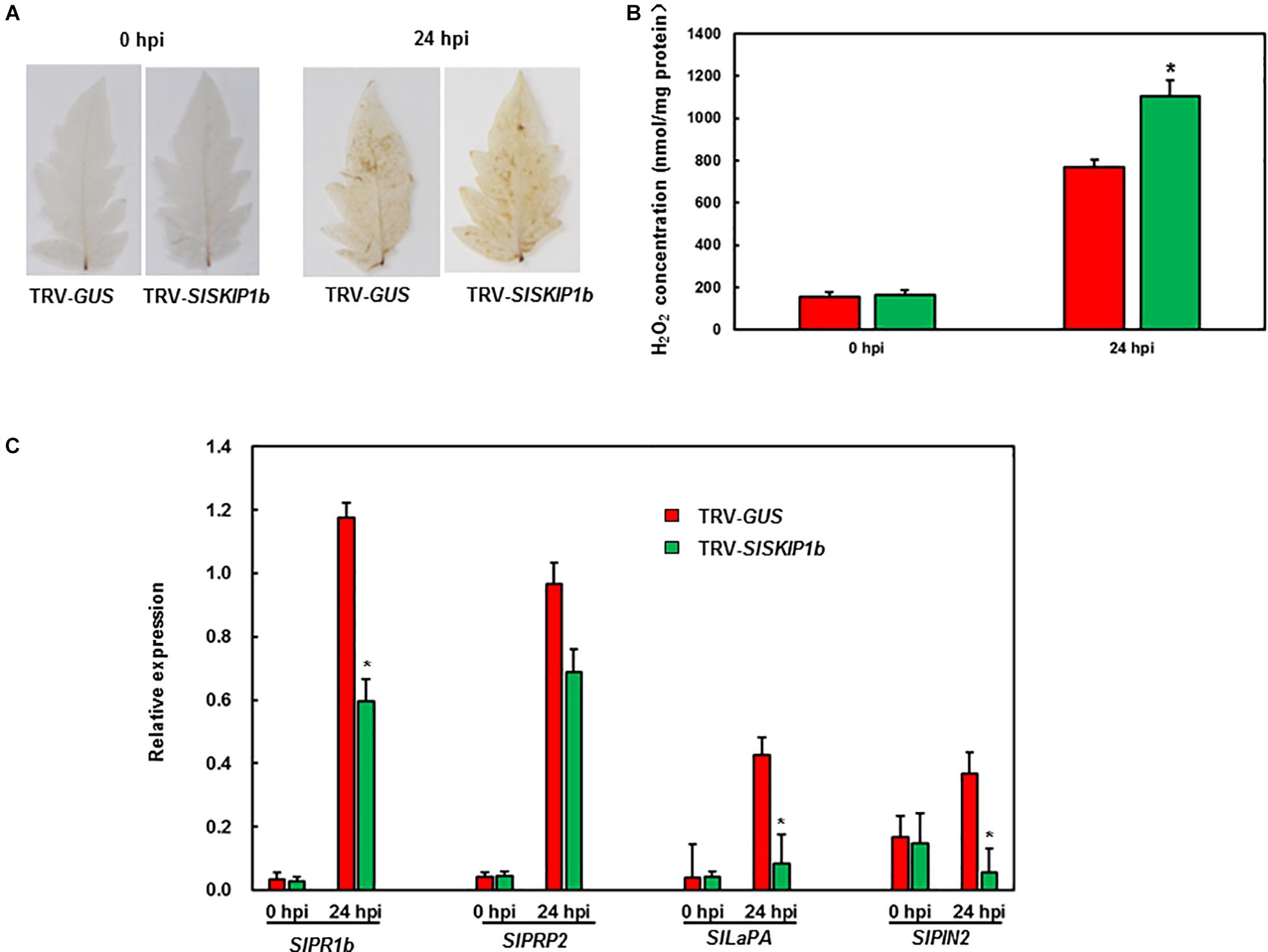
Figure 6. Silencing of SlSKIP1b elevated H2O2 accumulation but downregulated the JA/ET signaling-responsive DRG expression following B. cinerea inoculation. At 4 weeks following VIGS inoculation, 2 × 105 spores/mL pore suspension was used to spray on the leaf surface through whole-plant disease assays, then leaves were harvested at 24 h following infection. (A) H2O2 accumulation in plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS revealed through DAB staining following B. cinerea inoculation. (B) The H2O2 concentration in plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS before and after B. cinerea inoculation. The H2O2 concentration was measured using an H2O2 kit. (C) Specific DRG expression levels in plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS following B. cinerea inoculation. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
SlSKIP1b Silencing Led to Weakened Pst DC3000 Tolerance
The potential SlSKIP1b functions in the resistance against Pst DC3000 was further studied. The disease phenotype and in planta bacterial quantity of plants inoculated with TRV-SlSKIPs were compared with those of plants inoculated with TRV-GUS. Differences in disease symptom and bacterial growth at 3 dpi were not significant between plants inoculated with TRV-SlSKIP1a and those inoculated with TRV-GUS (Figures 7A,B), indicating that SlSKIP1a did not possibly participate in the Pst DC3000 tolerance. Plants inoculated with TRV-SlSKIP1b showed serious disease symptom relative to those inoculated with TRV-GUS (Figure 7A), and the bacterial population at 4 dpi was about 20 times higher than that in control (Figure 7B). The above findings demonstrated the effect of SlSKIP1b silencing on reducing Pst DC3000 tolerance of tomato.
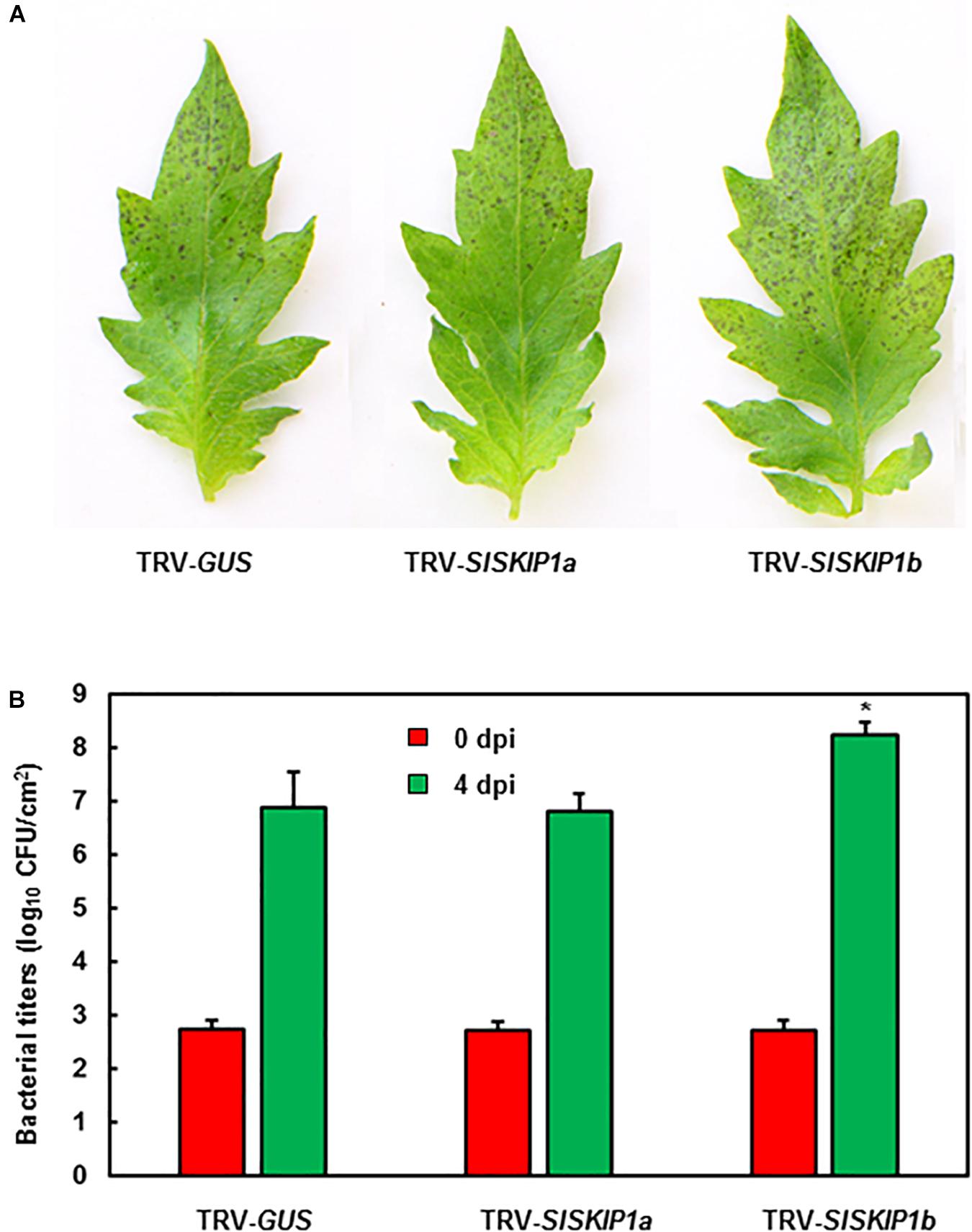
Figure 7. SlSKIP1b silencing weakened Pst DC3000 tolerance. Agrobacteria that carried TRV-SlSKIP and TRV-GUS constructs were transfected into the tomato plants 10 days old, at 4 weeks later, the disease assays were performed. (A) Disease symptoms in typical plant leaf samples inoculated with TRV-SlSKIPs and those inoculated with TRV-GUS at 4 days following Pst DC3000 infection. (B) Bacterial quantity in plant leaf samples inoculated with TRV-SlSKIPs and those inoculated with TRV-GUS. Leaves were harvested at 0 and 4 days following inoculation to measure the bacterial quantity. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
For exploring the potential mechanism of action by which SlSKIP1b silencing affected Pst DC3000 tolerance, the ROS accumulation together with DRGs expression was examined subsequently. Prior to Pst DC3000 infection, there was no distinct H2O2 accumulation observed in leaves from plants inoculated with TRV-SlSKIP1b and TRV-GUS (Figure 8A). Compared with controls, plants inoculated with TRV-SlSKIP1b showed obvious H2O2 accumulation at 3 dpi (Figure 8A). We also measured the H2O2 concentration. As shown in Figure 8B, the H2O2 concentration in TRV-SlSKIP1b seedlings was much higher than that of TRV-GUS after pathogen infection. There was no significant difference in SlPRP2, SlPR1b, SlPIN2, or SlLapA expression in plants inoculated with TRV-SlSKIP1b relative to those inoculated with TRV-GUS prior to Pst DC3000 inoculation (Figure 8C). Besides, relative to plants inoculated with TRV-GUS at 2 dpi, those inoculated with TRV-SlSKIP1b showed decreased SlPR1b together with SlPRP2 expression (Figure 8C). However, SlLapA or SlPIN2 expression of plants inoculated with TRV-SlSKIP1b showed no prominent change relative to TRV-GUS-infiltrated counterparts at 2 days following Pst DC3000 infection (Figure 8C). The above results suggested the effect of SlSKIP1b silencing on reducing the SA signaling-responsive DRGs levels by Pst DC3000 inoculation.
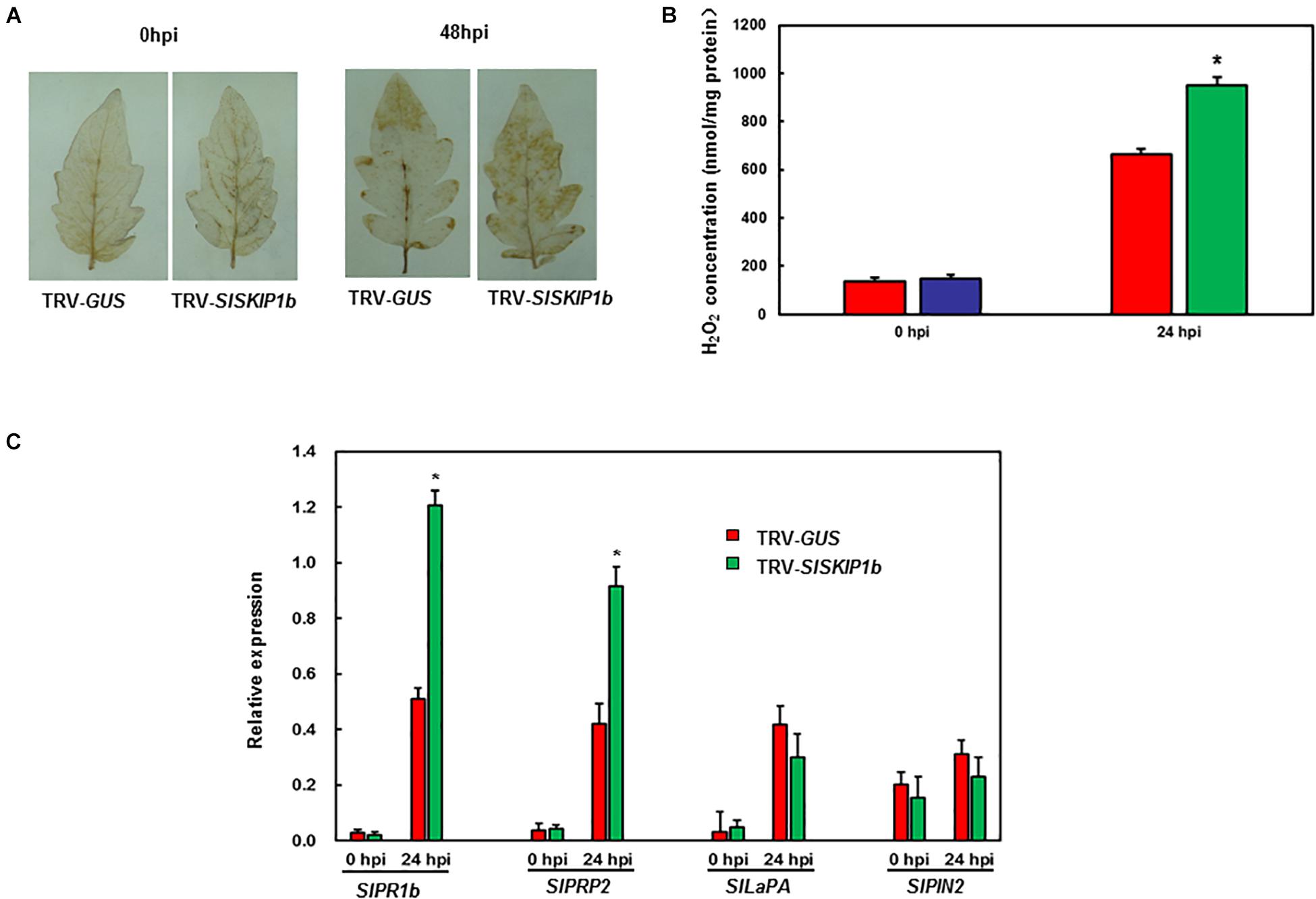
Figure 8. Silencing of SlSKIP1b affected H2O2 accumulation and SA signaling-responsive DRG expression following Pst DC3000 inoculation. The Pst DC3000 suspension (OD600 = 0.0002) was used to infect plants at negative pressure in the whole-plant disease assays 4 weeks following VIGS inoculation. Leaves were harvested to analyze the H2O2 accumulation together with DRG expression following infection. (A) H2O2 accumulation in plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS following Pst D3000 inoculation measured through DAB staining. (B) The H2O2 concentration in plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS before and after Pst DC3000 inoculation. The H2O2 concentration was measured using an H2O2 kit. (C) Specific DRG expression in plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS following Pst DC3000 inoculation. Data presented are the means ± SD from three independent experiments with biological distinct samples and * above the columns indicate significant differences at p < 0.05 level.
Discussion
In this research, two SlSKIP genes were identified in tomato, while only one SKIP gene was discovered in Arabidopsis. Although it is already known that SKIP is involved in transcription regulation and RNA splicing, thus leading to the regulation of several signaling pathways. There is no direct genetic proof for SKIP functions in the disease resistance of plants.
In our experiment, the SlSKIP target gene silencing efficiency was predicted as around 65% (Figure 3A), close to that obtained in our prior works (Li et al., 2014, 2015; Liu et al., 2014; Zhang Y. et al., 2014; Zhang et al., 2016). It is previously reported that pathogen infection may induce SKIP expression (Liu, 2015). As found in the present work, both B. cinerea and Pst DC3000 triggered SlSKIP1b expression (Figure 1). Also, SlSKIP expression was triggered in response to signal molecules associated with defense (Figure 2), which was consistent with previous study reporting that OsSKIPa expression was triggered upon a variety of phytohormone treatments as well as abiotic stress conditions (Hou et al., 2009). The SlSKIP genes showed different responses to B. cinerea or Pst DC3000 inoculation, together with the signal molecules associated with defense, which suggested the potential functions in the B. cinerea as well as Pst DC3000 tolerance.
The VIGS-based method was adopted to analyze the SlSKIP functions in terms of disease resistance. As a result, the silencing of SlSKIP1b led to weakened B. cinerea (Figures 4, 5) along with Pst DC3000 (Figure 7) tolerance. Typically, plants silenced by SlSKIP1b displayed the serious disease symptom, together with excessive pathogen growth, which confirmed their reduced B. cinerea tolerance (Figures 4, 5). At the same time, the SlSKIP1b-silenced plants displayed more severe disease symptoms, along with more bacterial growth, which confirmed the reduced Pst DC3000 tolerance (Figure 7). These results were consistent with a previous report that SKIP did have a certain function in the resistance to biotic stress. Moreover, GhSKIP35 has certain functions in the resistance to verticillium wilt in G. hirsutum (Liu, 2015).
In this study, the alterations of ROS accumulation and certain specific DRG expression were analyzed to explore the cause of the decreased B. cinerea and Pst DC3000 tolerance of SlSKIP1b-silenced plants. In this experiment, plants silenced by SlSKIP1b showed more H2O2 accumulation following B. cinerea and Pst DC3000 inoculation (Figures 6, 8). It is known that late-stage ROS accumulation facilitates disease development resulting from the necrotrophic pathogens (like B. cinerea) and (hemi) biotrophic pathogens (like Pst DC3000) (Govrin and Levine, 2000; Govrin et al., 2006; Temme and Tudzynski, 2009; Ishiga et al., 2012; Mengiste, 2012). Therefore, the increased ROS content resulting from SlSKIP1b silencing was possibly related to the weakened tolerance to B. cinerea as well as Pst DC3000 of plants silenced by SlSKIP1b.
Besides, plants silenced by SlSKIP1b had decreased levels of SlRP1b (SA-related gene), SlLapA (JA-related gene), and SlPIN2 (JA-related gene) following B. cinerea inoculation and increased levels of SlRP1b and SlRPP2 (SA-related gene) following Pst DC3000 inoculation (Figures 6C, 8C). It is known that SA-regulated defense responses are good for the infection of necrotrophic pathogen B. cinerea, while JA-regulated defense responses are involved in restricting the disease. The inverse model is proposed for (hemi) biotrophic pathogen Pst DC3000 (Glazebrook, 2005; Pieterse et al., 2009). We speculated that SKIP1b may not mediate in the antagonistic effects between SA- and JA-signaling pathways. Instead, SKIP1b may involve in these two pathways. So the expression levels of SA-dependent genes and lower JA-dependent genes changed in TRV-SlSKIP1b inoculated plants.
The reduced resistance of the SlSKIP1b-silenced plants might be induced by the increased ROS accumulation together with changed DRGs levels. However, further physiological and biochemical experiments are required to find out the mechanisms responsible for the altered disease resistance observed in the SlSKIP1b-silenced plants.
Our results showed that the silencing of SKIP1b led to increased susceptibility to both Botrytis and Pst DC3000. It consists with previous reports that the silencing of genes will reduce or increase resistance to both Botrytis and Pseudomonas (Li et al., 2015; Wang et al., 2018,2020).
It is suggested that SKIP plays a role in abiotic stress tolerance. SKIP is involved in the ABA signaling and renders the osmotic resistance in the case of salt stress through regulating AS genes of Arabidopsis (Lim et al., 2010; Feng et al., 2015). OsSKIPa positively modulates the stress tolerance of rice by regulating different genes associated with stress in rice at the transcription level (Hou et al., 2009). The interaction of OsSKIP with OsCYP18-2 is essential for regulating genes associated with stress at both transcription and post-transcription levels and for enhancing drought resistance (Lee et al., 2015). The ZmSKIP overexpression plants with increased ABA contents exhibit significantly enhanced resistance to drought compared with controls, which suggested that ZmSKIP was involved in the regulation of drought resistance by regulating specific gene levels (Wei et al., 2015). Nonetheless, no experiment was carried out to examine the abiotic stress tolerance in this study, and no difference was found between plants inoculated with TRV-SlSKIP1b and those inoculated with TRV-GUS during the vegetative growth process. This might be consistent with previous report that SKIP participated in regulating certain reproductive stage gene expression at the post-transcription level in Arabidopsis thaliana (Wang et al., 2012; Cao et al., 2015; Cui et al., 2017). Unfortunately, this study did not conduct an experiment on plants that had entered the reproductive phase. In our future studies, experiments should be performed to examine the resistance to abiotic stress and plants of the reproductive stag.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
HZ, LY, MJ, and FS carried out most of the experiments. MJ and HZ designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Taizhou Municipal Science and Technology Project (20ny18), Science Foundation for Distinguished Young Scholars of Taizhou University (2019JQ001), and Natural Science Foundation of Zhejiang Province (LY19C150004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.593267/full#supplementary-material
References
Albers, M., Diment, A., Muraru, M., Russell, C. S., and Beggs, J. D. (2003). Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9, 138–150. doi: 10.1261/rna.2119903
Barry, J. B., Leong, G. M., Church, W. B., Issa, L. L., Eisman, J. A., and Gardiner, E. M. (2003). Interactions of SKIP/NCoA-62, TFIIB, and retinoid X receptor with vitamin D receptor helix H10 residues. J. Biol. Chem. 278, 8224–8228. doi: 10.1074/jbc.c200712200
Bessonov, S., Anokhina, M., Will, C. L., Urlaub, H., and Luhrmann, R. (2008). Isolation of an active step I spliceosome and composition of its RNP core. Nature 452, 846–850. doi: 10.1038/nature06842
Bres, V., Yoshida, T., Pickle, L., and Jones, K. A. (2009). SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol. Cell 36, 75–87. doi: 10.1016/j.molcel.2009.08.015
Cao, Y., Wen, L., Wang, Z., and Ma, L. (2015). SKIP interacts with the Paf1 complex to regulate flowering via the activation of FLC transcription in Arabidopsis. Mol. Plant 8, 1816–1819. doi: 10.1016/j.molp.2015.09.004
Chen, Y., Zhang, L., and Jones, K. A. (2011). SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 25, 701–716. doi: 10.1101/gad.2002611
Cui, Z., Tong, A., Huo, Y., Yan, Z., Yang, W., Yang, X., et al. (2017). SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 15:80. doi: 10.1186/s12915-017-0422-2
Feng, J., Li, J., Gao, Z., Lu, Y., Yu, J., Zheng, Q., et al. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8, 1038–1052. doi: 10.1016/j.molp.2015.01.011
Figueroa, J. D., and Hayman, M. J. (2004). The human Ski-interacting protein functionally substitutes for the yeast PRP45 gene. Biochem. Biophys. Res. Commun. 319, 1105–1109. doi: 10.1016/j.bbrc.2004.05.096
Folk, P., Puta, F., and Skruzny, M. (2004). Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell Mol. Life Sci. 61, 629–640. doi: 10.1007/s00018-003-3215-4
Gahura, O., Abrhamova, K., Skruzny, M., Valentova, A., Munzarova, V., Folk, P., et al. (2009). Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J. Cell Biochem. 106, 139–151. doi: 10.1002/jcb.21989
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi: 10.1016/s0960-9822(00)00560-1
Govrin, E. M., Rachmilevitch, S., Tiwari, B. S., Solomon, M., and Levine, A. (2006). An elicitor from Botrytis cinerea induces the hypersensitive response in Arabidopsis thaliana and other plants and promotes the gray mold disease. Phytopathology 96, 299–307. doi: 10.1094/PHYTO-96-0299
Hou, X., Xie, K., Yao, J., Qi, Z., and Xiong, L. (2009). A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. U.S.A. 106, 6410–6415. doi: 10.1073/pnas.0901940106
Ishiga, Y., Ishiga, T., Wangdi, T., Mysore, K. S., and Uppalapati, S. R. (2012). NTRC and chloroplast-generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis. Mol. Plant Microbe Interact. 25, 294–306. doi: 10.1094/MPMI-05-11-0130
Ivanov, A. I., Rovescalli, A. C., Pozzi, P., Yoo, S., Mozer, B., Li, H. P., et al. (2004). Genes required for Drosophila nervous system development identified by RNA interference. Proc. Natl. Acad. Sci. U.S.A. 101, 16216–16221. doi: 10.1073/pnas.0407188101
Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237.
Kim, Y. J., Noguchi, S., Hayashi, Y. K., Tsukahara, T., Shimizu, T., and Arahata, K. (2001). The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum. Mol. Genet. 10, 1129–1139. doi: 10.1093/hmg/10.11.1129
Kostrouchova, M., Housa, D., Kostrouch, Z., Saudek, V., and Rall, J. E. (2002). SKIP is an indispensable factor for Caenorhabditis elegans development. Proc. Natl. Acad. Sci. U.S.A. 99, 9254–9259. doi: 10.1073/pnas.112213799
Laduron, S., Deplus, R., Zhou, S., Kholmanskikh, O., Godelaine, D., De Smet, C., et al. (2004). MAGE-A1 interacts with adaptor SKIP and the deacetylase HDAC1 to repress transcription. Nucleic Acids Res. 32, 4340–4350. doi: 10.1093/nar/gkh735
Lee, S. S., Park, H. J., Yoon, D. H., Kim, B. G., Ahn, J. C., Luan, S., et al. (2015). Rice cyclophilin OsCYP18-2 is translocated to the nucleus by an interaction with SKIP and enhances drought tolerance in rice and Arabidopsis. Plant Cell Environ. 38, 2071–2087. doi: 10.1111/pce.12531
Li, X., Huang, L., Hong, Y., Zhang, Y., Liu, S., Li, D., et al. (2015). Co-silencing of tomato S-adenosylhomocysteine hydrolase genes confers increased immunity against Pseudomonas syringae pv. tomato DC3000 and enhanced tolerance to drought stress. Front. Plant Sci. 6:717. doi: 10.3389/fpls.2015.00717
Li, X., Huang, L., Zhang, Y., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 114:286. doi: 10.1186/s12870-014-0286-3
Li, Y., Xia, C., Feng, J., Yang, D., Wu, F., Cao, Y., et al. (2016). The SNW domain of SKIP is required for its integration into the spliceosome and its interaction with the Paf1 complex in Arabidopsis. Mol. Plant 9, 1040–1050. doi: 10.1016/j.molp.2016.04.011
Lim, G. H., Zhang, X., Chung, M. S., Lee, D. J., Woo, Y. M., Cheong, H. S., et al. (2010). A putative novel transcription factor, AtSKIP, is involved in abscisic acid signaling and confers salt and osmotic tolerance in Arabidopsis. New Phytol. 185, 103–113. doi: 10.1111/j.1469-8137.2009.03032.x
Liu, B., Ouyang, Z., Zhang, Y., Li, X., Hong, Y., Huang, L., et al. (2014). Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS One 9:e102067. doi: 10.1371/journal.pone.0102067
Liu, K. (2015). Cloning and functional analysis of GhSKIP35 gene responsible for resistance to Verticillium wilt in Gossypium hirsutum L. Chinese Acad. Agric. Sci. 2015:62. (in chinese).
Liu, L., Wu, F., and Zhang, W. (2016). A conserved interaction between SKIP and SMP1/2 aids in recruiting the second-step splicing factors to the spliceosome in Arabidopsis. Mol. Plant 9, 1660–1663. doi: 10.1016/j.molp.2016.09.007
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Moore, M. J., and Proudfoot, N. J. (2009). Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700. doi: 10.1016/j.cell.2009.02.001
Negeri, D., Eggert, H., Gienapp, R., and Saumweber, H. (2002). Inducible RNA interference uncovers the Drosophila protein Bx42 as an essential nuclear cofactor involved in Notch signal transduction. Mech. Dev. 117, 151–162. doi: 10.1016/S0925-4773(02)00193-4
Niu, J. B. (2012). Cloning and the Expression Analysis of ZmSKIP in Corn. Hefei: Anhui Agricultural University.
Piano, F., Schetter, A. J., Morton, D. G., Gunsalus, K. C., Reinke, V., and Kemphues, K. J. (2002). Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12, 1959–1964. doi: 10.1016/S0960-9822(02)01301-5
Pieterse, C. M., Leno-Reyes, A., Van, D. E., and Van, W. S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 5, 308–316. doi: 10.1038/nchembio.164
Prathapam, T., Kühne, C., and Banks, L. (2002). Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity. Nucleic Acids Res. 30, 5261–5268. doi: 10.1093/nar/gkf658
Rual, J. F., Ceron, J., Koreth, J., Hao, T., Nicot, A. S., Hirozane-Kishikawa, T., et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168. doi: 10.1101/gr.2505604
Simmer, F., Moorman, C., van der Linden, A. M., Kuijk, E., van den Berghe, P. V., Kamath, R. S., et al. (2003). Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:E12. doi: 10.1371/journal.pbio.0000012
Sonnichsen, B., Koski, L. B., Walsh, A., Marschall, P., Neumann, B., Brehm, M., et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469. doi: 10.1038/nature03353
Temme, N., and Tudzynski, P. (2009). Does Botrytis cinerea ignore H2O2-induced oxidative stress during infection? Characterization of Botrytis activator protein 1. Mol. Plant-Microbe Interact. 22, 987–998. doi: 10.1094/MPMI-22-8-0987
Turunen, J. J., Niemela, E. H., Verma, B., and Frilander, M. J. (2013). The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 4, 61–76. doi: 10.1002/wrna.1141
Wang, L., Liu, W. D., and Wang, Y. J. (2020). Heterologous expression of chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to Botrytis cinerea. Plant Sci. 293:110421. doi: 10.1016/j.plantsci.2020.110421
Wang, M., Zhu, Y. X., Han, R., Yin, W. C., Guo, C. L., Li, C. L., et al. (2018). Expression of Vitis amurensis VaERF20 in Arabidopsis thaliana improves resistance to Botrytis cinerea and Pseudomonas syringae pv. Tomato DC3000. Int. J. Mol. Sci. 9:696. doi: 10.3390/ijms19030696
Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., Yue, Y., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295. doi: 10.1105/tpc.112.100081
Wei, W., Niu, J. B., Li, Y. H., and Cheng, B. J. (2015). Molecular cloning and transformation of ZmSKIP in maize. J. Anhui Agric. Univ. 5, 666–673.
Wieland, C., Mann, S., von Besser, H., and Saumweber, H. (1992). The Drosophila nuclear protein Bx42, which is found in many puffs on polytene chromosomes, is highly charged. Chromosoma 101, 517–525. doi: 10.1007/BF00352475
Zhang, C., Baudino, T. A., Dowd, D. R., Tokumaru, H., Wang, W., and MacDonald, P. N. (2001). Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 276, 40614–40620. doi: 10.1111/j.1365-3091.2006.00792.x
Zhang, C., Dowd, D. R., Staal, A., Gu, C., Lian, J. B., van Wijnen, A. J., et al. (2003). Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278, 35325–35336. doi: 10.1074/jbc.m305191200
Zhang, H., Hong, Y., Huang, L., Liu, S., Tian, L., Dai, Y., et al. (2016). Virus-induced gene silencing-based functional analyses revealed the involvement of several putative trehalose-6-phosphate synthase/phosphatase genes in disease resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 in tomato. Front. Plant Sci. 7:1176. doi: 10.3389/fpls.2016.01176
Zhang, H., Yan, M., Deng, R., Song, F., and Jiang, M. (2020). The silencing of DEK reduced disease resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 based on virus-induced gene silencing analysis in tomato. Gene 727:144245. doi: 10.1016/j.gene.2019.144245
Zhang, X., Min, J. H., Huang, P., Chung, J. S., Lee, K. H., and Kim, C. S. (2014). AtSKIP functions as a mediator between cytokinin and light signaling pathway in Arabidopsis thaliana. Plant Cell Rep. 33, 401–409. doi: 10.1007/s00299-013-1540-0
Zhang, Y., Liu, B., Li, X., Ouyang, Z., Huang, L., Hong, Y., et al. (2014). The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol. Plant Microbe Interact. 27, 688–699. doi: 10.1094/mpmi-01-14-0020-r
Zhang, Y., Zhao, L., Li, H., Gao, Y., Li, Y., Wu, X., et al. (2013). GmGBP1, a homolog of human ski interacting protein in soybean, regulates flowering and stress tolerance in Arabidopsis. BMC Plant Biol. 13:21. doi: 10.1186/1471-2229-13-21
Keywords: SKIP, B. cinerea, Pst DC3000, resistance, ROS
Citation: Zhang H, Yin L, Song F and Jiang M (2020) SKIP Silencing Decreased Disease Resistance Against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 in Tomato. Front. Plant Sci. 11:593267. doi: 10.3389/fpls.2020.593267
Received: 10 August 2020; Accepted: 19 November 2020;
Published: 14 December 2020.
Edited by:
Harold Meijer, Wageningen University and Research, NetherlandsReviewed by:
Ana María Laxalt, National University of Mar del Plata, ArgentinaZhao Zhang, China Agricultural University, China
Copyright © 2020 Zhang, Yin, Song and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Jiang, amlhbmdtaW5nMTk3M0AxMzkuY29t
 Huijuan Zhang
Huijuan Zhang Longfei Yin1
Longfei Yin1 Fengming Song
Fengming Song Ming Jiang
Ming Jiang