
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 November 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.591457
This article is part of the Research Topic Reproductive Barriers and Gene Introgression in Rice Species View all 16 articles
 Jegadeesan Ramalingam1,2*
Jegadeesan Ramalingam1,2* Chandavarapu Raveendra1,3
Chandavarapu Raveendra1,3 Palanisamy Savitha2
Palanisamy Savitha2 Venugopal Vidya2
Venugopal Vidya2 Thammannagowda Lingapatna Chaithra2
Thammannagowda Lingapatna Chaithra2 Senthilvel Velprabakaran1,3
Senthilvel Velprabakaran1,3 Ramasamy Saraswathi4
Ramasamy Saraswathi4 Ayyasamy Ramanathan5
Ayyasamy Ramanathan5 Madhavan Pillai Arumugam Pillai6
Madhavan Pillai Arumugam Pillai6 Samudrakani Arumugachamy7
Samudrakani Arumugachamy7 Chockalingam Vanniarajan3
Chockalingam Vanniarajan3Bacterial blight, blast, and sheath blight are the commonest diseases causing substantial yield loss in rice around the world. Stacking of broad-spectrum resistance genes/QTLs into popular cultivars is becoming a major objective of any disease resistance breeding program. The varieties ASD 16 and ADT 43 are the two popular, high yielding, and widely grown rice cultivars of South India, which are susceptible to bacterial blight (BB), blast, and sheath blight diseases. The present study was carried out to improve the cultivars (ASD 16 and ADT 43) through introgression of bacterial blight (xa5, xa13, and Xa21), blast (Pi54), and sheath blight (qSBR7-1, qSBR11-1, and qSBR11-2) resistance genes/QTLs by MABB (marker-assisted backcross breeding). IRBB60 (xa5, xa13, and Xa21) and Tetep (Pi54; qSBR7-1, qSBR11-1, and qSBR11-2) were used as donors to introgress BB, blast, and sheath blight resistance into the recurrent parents (ASD 16 and ADT 43). Homozygous (BC3F3 generation), three-gene bacterial blight pyramided (xa5 + xa13 + Xa21) lines were developed, and these lines were crossed with Tetep to combine blast (Pi54) and sheath blight (qSBR7-1, qSBR11-1, and qSBR11-2) resistance. In BC3F3 generation, the improved pyramided lines carrying a total of seven genes/QTLs (xa5 + xa13 + Xa21 + Pi54 + qSBR7-1 + qSBR11-1 + qSBR11-2) were selected through molecular and phenotypic assay, and these were evaluated for resistance against bacterial blight, blast, and sheath blight pathogens under greenhouse conditions. We have selected nine lines in ASD 16 background and 15 lines in ADT 43 background, exhibiting a high degree of resistance to BB, blast, and sheath blight diseases and also possessing phenotypes of recurrent parents. The improved pyramided lines are expected to be used as improved varieties or used as a potential donor in breeding programs. The present study successfully introgressed Pi54, and qSBR QTLs (qSBR7-1, qSBR11-1, and qSBR11-2) from Tetep and major effective BB-resistant genes (xa5, xa13, and Xa21) from IRBB60 into the commercial varieties for durable resistance to multiple diseases.
Rice (Oryza sativa L.) is considered a major staple food crop for billions of population across the globe, and it provides 23% of calories shared by different food crops (Sharma et al., 2012). Exponential growth of the world population demands an increase in rice production by 26% to fulfill calorie requirements (Khush, 2013). According to the Food and Agricultural Organization, the global rice production would have to increase by 42% over the present-day production to meet the growing population by 2050 (Ray et al., 2013). However, the yield potential is frequently threatened by various biotic stresses, mostly fungi, and bacteria. To address these problems and to increase production, developing cultivars with durable resistance is a prerequisite. The host-plant resistance can be ideally improved through pyramiding of major R-genes/QTLs for multiple diseases and biotic stress factors.
Bacterial blight (BB) caused by Xanthomonas oryzae pv. oryzae (Xoo) is a major destructive disease of rice, causing a yield loss of up to 80% depending on the severity (Kumar et al., 2012). Improving the host-plant resistance is the most efficient and eco-friendly approach, as chemical control of BB is not effective (Lee et al., 2003). The infection chain starts by entering into the plant through the hydathodes, and it reaches to xylem vessels, where the infection became systemic. Till date, 46 resistance genes have been identified from the different sources of rice (Chen et al., 2020). Of these, the Xa4, xa5, Xa7, xa13, Xa21, Xa33, and Xa38 genes are most frequently utilized in hybridization programs for developing BB-resistant cultivars (Hsu et al., 2020). Natural allelic variations in Xoo challenges the resistance levels conferred by a single gene; hence, pyramiding of two or more effective resistance genes is highly essential for broad-spectrum and durable resistance to Xoo at field conditions. Xa21, a major dominant resistant gene, originated from African wild species, Oryza longistaminata, was observed to confer the resistance to many Xoo isolates (Nguyen et al., 2018). The encoding proteins of Xa21 gene carries both leucine-rich repeats (LRR) and serine-threonine kinases, and these complexes perceives the presence of pathogen ligand on the cell surface and activates the subsequent intracellular defense response R-proteins (Song et al., 1995). The Xa21 gene was physically mapped on the long arm of chromosome 11, and a highly efficient PCR-based co-dominant molecular marker (pTA 248) was developed for marker-assisted selection of Xa21 (Ronald et al., 1992). A unique, fully recessive gene, xa13, was first identified in cultivar BJ1 and physically mapped on the long arm of chromosome 8 (Zhang et al., 1996). Mutations in the promoter region of dominant allele (Xa13) resulted in a recessive gene, xa13, which does not encode for a modulator for pathogen (Chu et al., 2006). Another broad-spectrum recessive resistant gene, xa5, was identified and mapped on the subtelomeric region of chromosome number 5 (Blair et al., 2003). Unlike other R-genes, the xa5 gene encodes for a gamma transcription factor-like protein (TFIIAγ). Pyramiding of xa5 gene with other dominant genes gives durable resistance to Xoo than the plants with single BB-resistant gene (Huang et al., 1997).
Rice blast, caused by Magnaporthe oryzae (Teleomorph: Pyricularia oryzae), is one of the devastating diseases of rice growing areas across the world. Yield loss is estimated to be more than 50% when it occurs in epidemic proportions (Babujee and Gnanamanickam, 2000). Similar to BB, developing of host-plant resistance is the most effective strategy for management of blast disease (Sharma et al., 2012). So far, about 100 resistance genes have been identified, and 37 of them were cloned (Zhang et al., 2019). Although several blast resistance genes have been identified, only a few of them were used in breeding programs for blast disease management in India (Singh et al., 2011). Among them, the Pi54 gene located on chromosome 11 provides stable and durable resistance to diverse strains of M. oryzae collected across India (Thakur et al., 2015). The predicted proteins of the Pi54 gene contains NBS-LRR proteins along with unique zinc finger domain (Sharma et al., 2005). During the host–pathogen interaction, the Pi54 gene induces the synthesis of callose (β-1,3-glucan), which acts as a physical barrier by blocking the penetration of fungal hyphae (Gupta et al., 2012). A functional marker has been developed for the Pi54 gene and used in maker-assisted selection for developing blast-resistant cultivars (Ramkumar et al., 2011).
Rhizoctonia solani Kühn, the causative agent of rice sheath blight disease (ShB), poses a significant impact on both yield and quality (Singh et al., 2019). Introduction of high yielding varieties and application of high doses of nitrogenous fertilizers resulted in a steep rise in incidence of sheath blight disease (Savary et al., 1997). R. solani Kühn is a soil-borne facultative parasite, survives as sclerotia or mycelium, or rarely as basidiospores. No varieties resistant to sheath blight were reported till date (Channamallikarjuna et al., 2010). Breeding for resistance to ShB is quite unsuccessful owing to inability to identify effective resistance sources from the available germplasm, wide-ranging host compatibility, high genetic variability, and capability of the pathogen to survive from season to season in the form of dormant sclerotia, makes additional complications in controlling the disease (Molla et al., 2020). Though qualitative resistance to ShB was not found, quantitative resistance was reported in some landraces viz., Tetep, Teqing, Jasmine 85, etc., (Channamallikarjuna et al., 2010; Wang et al., 2012; Yadav et al., 2015). Up to now, 50 QTLs conferring moderate resistance to rice sheath blight have been identified from the different sources of rice (Zhang et al., 2019). Among these, qSBR7-1, qSBR11-1, and qSBR11-2 were identified in the background of Tetep (Channamallikarjuna et al., 2010) and pyramided in Pusa 6B (Singh et al., 2015). We have used these three QTLs (qSBR7-1, qSBR11-1, and qSBR11-2) for pyramiding in our recurrent parents to improve sheath blight resistance.
In cognizance of the above reports, the present study was formulated with the following objectives: (i) introgression of BB (xa5, xa13, and Xa21), blast (Pi54), and sheath blight (qSBR7-1, qSBR11-1, and qSBR11-2) resistance genes/QTLs in the backgrounds of ASD 16 and ADT 43; (ii) analysis of recurrent parent genome recovery (RPG) with a set of polymorphic SSR (simple sequence repeats) markers; (iii) evaluation of improved pyramided lines for physical resistance against BB, blast, and sheath blight diseases; and (iv) evaluation of agro-morphological and quality traits of improved pyramided lines in comparison with parents.
The two recurrent parents, ASD 16 (ADT 31 × CO 39) and ADT 43 (IR 50 × Improved White Ponni) are popular, high yielding, and widely grown rice cultivars of South India. ASD 16 has short bold grains, while ADT 43 has medium slender fine grains. Though both cultivars are high yielding, they are highly susceptible to BB, blast, and sheath blight diseases. A bacterial blight-resistant genotype, IRBB60 harboring xa5 + xa13 + Xa21 was used as donor for BB resistance genes. Tetep, a Vietnamese indica land race possessing blast resistance (Pi54) (Ramkumar et al., 2011) and moderately resistant to sheath blight, harboring qSBR QTLs (qSBR7-1, qSBR11-1, and qSBR11-2) (Channamallikarjuna et al., 2010) was also used as one of the donors for targeted transfer of blast and sheath blight resistance into the background of recurrent parents (ASD 16 and ADT 43).
Two independent crosses (ASD 16 × IRBB60 and ADT 43 × IRBB60) were made between recipient parents and IRBB60 for targeted gene transfer of xa5, xa13, and Xa21. The hybridity of the F1 plants were confirmed by functional/linked PCR-based co-dominant molecular markers of xa5, xa13, and Xa21 (Table 1). The heterozygous plants for xa5, xa13, and Xa21 were backcrossed with respective recurrent parents (ASD 16 and ADT 43) to generate BC1F1. The BC1F1 hybrids were screened for targeted genes (xa5, xa13, and Xa21), and the confirmed plants were assessed with a set of polymorphic SSR markers to recover the maximum percentage of RPG (Sundaram et al., 2008). The solitary plant with targeted genes (xa5, xa13, and Xa21) and maximum recovery of RPG was selected and backcrossed to generate BC2F1, and this procedure was repeated till BC3F1. At every backcross, foreground and background selections were carried out to forward the plant with targeted genes and maximum recovery of RPG. Pedigree-based breeding strategy with marker-assisted selection was followed after BC3F1 to generate BC3F2 and BC3F3 populations. In BC3F2 population, the homozygous plants for targeted genes were identified through foreground selection and self-pollinated to generate BC3F3 population. The homozygous plants at BC3F3 were evaluated for resistance against BB as well as for key agronomic traits, and the best lines were pooled to generate three-gene bacterial blight (xa5, xa13, and Xa21) pyramided lines of ASD 16, and ADT 43. The three-gene bacterial blight pyramided lines were crossed with Tetep to introgress blast and sheath blight resistance. The plants showing heterozygous allele for all the targeted genes/QTLs were backcrossed with respective recurrent parent (three-gene bacterial blight pyramided lines of ASD 16 and ADT 43) to generate BC1F1. The BC1F1 hybrids were screened with molecular markers for all the targeted traits in the study (Table 1) followed by background selection with polymorphic SSR markers. The “positive” plants with maximum recovery of RPG were again backcrossed with recurrent parents to produce BC2F1 and BC3F1. In the BC3F2 population, foreground selection was carried out to identify the plants carrying all the targeted genes (xa5 + xa13 + Xa21 + Pi54) and targeted QTLs (qSBR7-1 + qSBR11-1 + qSBR11-2) in homozygous condition, and the identified plants were self-pollinated to generate BC3F3 population. The plants carrying all the targeted traits in homozygous condition at BC3F3 generation were evaluated for resistance against bacterial blight, blast, and sheath blight diseases under greenhouse conditions and also assessed for key agronomic as well as grain quality traits. The detailed plan of program for marker-assisted gene pyramiding of bacterial blight, blast, and sheath blight resistance genes/QTLs is depicted in Figure 1.
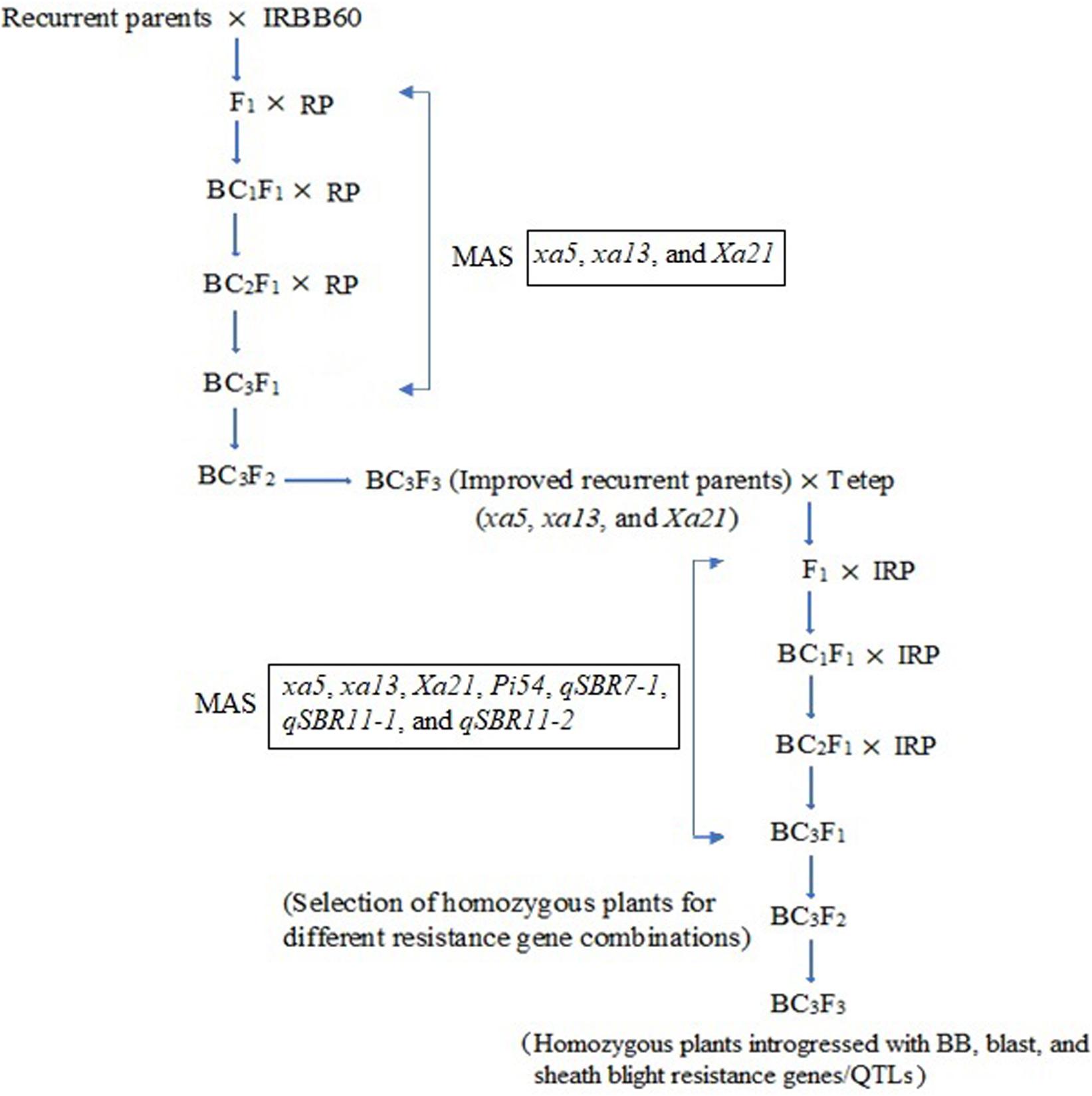
Figure 1. Schematic illustration of marker-assisted gene pyramiding of bacterial blight, blast, and sheath blight resistance genes/QTLs in the backgrounds of ASD 16 and ADT 43. RP, recurrent parents (ASD 16 and ADT 43), IRP, improved recurrent parents (ASD 16 and ADT 43 introgressed with xa5, xa13, and Xa21 genes); MAS, marker-assisted selection.
The DNA extraction for PCR amplification was carried out by the CTAB method (Varghese et al., 1997). The PCR protocol for marker-assisted selection of targeted genes/QTLs was followed according to the earlier reports (Chu et al., 2006; Iyer-Pascuzzi and McCouch, 2007; Sundaram et al., 2008; Channamallikarjuna et al., 2010; Ramkumar et al., 2011; Table 1). Ten microliters of PCR reaction mixture contains 4 μl of DreamTaq green 2× PCR master mix (Thermo scientific, United States), 4 μl of water, 50 ng of template DNA, and 30 ng each of forward and reverse primers. To find the specific allelic pattern of xa5 allele, 5–10 μl of PCR product is digested with BsrI (5 units of enzyme) at 65°C for 4 h with 2 μl of 10×PCR buffer (Iyer-Pascuzzi and McCouch, 2007). The amplified PCR products were separated by 2.5% agarose gel stained with ethidium bromide and visualized on UV light in gel documentation system (Bio-Rad Laboratories., United States). Background selection was carried out with a set of 463 SSR markers1; 69 and 68 markers were found to be polymorphic for ADT 43 and ASD 16 cross combinations, respectively (Supplementary Table S1), with a wide coverage of all the 12 chromosomes (five to six polymorphic markers per chromosome). Additional polymorphic markers, i.e., 10 polymorphic markers were employed on chromosome number 11, which carries Xa21, Pi54, qSBR11-1, and qSBR11-2 to minimize the linkage drag.
The selected IPLs (improved pyramided lines) and parents (IRBB60 as resistant check and ASD 16 and ADT 43 as susceptible checks) were tested for resistance against the DX-027 of Xoo isolate under greenhouse conditions. In addition to the selected IPLs, single- and two-gene BB pyramided lines were also evaluated to check the effectiveness of three-gene BB pyramided lines. Three replications were maintained with 30 plants per replication. The top leaves were clipped off, and bacterial suspension was inoculated with a density of 109 cells/ml by clip inoculation method at maximum tillering stage (Kauffman et al., 1973). Eight leaves per plant were inoculated, and mean lesion length was taken on six leaves to measure the accurate disease reaction. Symptoms were measured 21 days post-inoculation, and observations were recorded based on visual score and lesion length (LL). The plants with an average lesion length of <5 cm were considered as resistant, and those with >5 cm were considered as susceptible (International Rice Research Institute [IRRI], 2002).
The IPLs and parents (Tetep as resistant check; ASD 16 and ADT 43 as susceptible checks) were evaluated for leaf blast resistance against IS (KUL)-6, a virulent local isolate of M. oryzae. Five-week-old seedlings were individually transplanted into mud pots (19 cm × 22 cm × 22 cm) with three replications and inoculated with M. oryzae IS (KUL)-6 isolate at a spore density of 5 × 105 spores/ml. Disease reaction was recorded 9 days post-inoculation and evaluated based on blast lesion type (BLT) according to the 0–9 scale of SES (Standard Evaluation System) (International Rice Research Institute [IRRI], 2002). The plants with a score of 0–3 were rated as resistant and those with more than a score of 4 were rated as susceptible.
A pure culture of R. solani collected from the Tamil Nadu Rice Research Institute (TRRI), Aduthurai, Tamil Nadu, India, was used for testing the resistance of IPLs and parents against sheath blight. The pathogen was multiplied on sterilized shoot bits of water sedge, Typha angustata. Infected Typha shoot bits with mycelium and sclerotia were used as source of inoculum and placed carefully between the tillers of rice hills with the help of forceps at 80 days after sowing. The inoculated portion was covered with wet cotton and aluminum foil to avoid the moisture loss in the inoculated portion. The observations were recorded 25 days post-inoculation on randomly selected plants consisting of three infected tillers in each of three replications. Disease reaction was measured based on RLH% (relative lesion height), and scoring (0–9) was given as per SES (International Rice Research Institute [IRRI], 2002) [0 (immune), 1–20% (resistant), 21–30% (moderately resistant), 31–45% (moderately susceptible), 46–65% (susceptible), and >65% (highly susceptible)].
The 30-day old seedlings of recurrent parents and selected IPLs were transplanted to an experimental plot at the Agricultural College and Research Institute, Tamil Nadu Agricultural University, Madurai, India, with a spacing of 15 cm × 20 cm. The experimental plot was arranged in a randomized block design (RBD) with four blocks, and three replications were maintained in each block. Standard agronomic practices were followed as prescribed by TNAU, Coimbatore, India2, to raise the healthy crop. Observations were recorded on five plants in each line for key agronomic traits viz., days to 50% flowering (DFF), plant height (PH) (cm), number of productive tillers per plant (NPT), panicle length (PL) (cm), number of grains per panicle (NGP), flag leaf length (FL) (cm), flag leaf width (FW) (cm), 1,000-grain weight (1,000-GW) (g), grain-L/B ratio (cm), and single plant yield (SPY) (g). In addition, the quality traits viz., hulling percentage (HP%), milling percentage (MP%), head rice recovery (HRR%), kernel length (KL) (mm), kernel breadth (KB) (mm), kernel length breadth ratio (KLBR), milled rice length (MRL) (mm), milled rice breadth (MRB) (mm), kernel length after cooking (KLAC) (mm), kernel breadth after cooking (KBAC) (mm), and linear elongation ratio (LER) were analyzed in the homozygous improved pyramided lines.
Statistical analysis was done with SPSS software to complement the ANOVA (analysis of variance) to determine the significant variation among the improved pyramided lines. The coefficient of genetic distance among the selected pyramided lines and parents was calculated based on 10 morphological characters and used for generating the dendrogram using the “R” software (R Core Team, 2013).
The hybrids (F1) from the crosses of ASD 16 × IRBB60 and ADT 43 × IRBB60 were analyzed with functional/linked molecular markers of xa5, xa13, and Xa21, i.e., xa5-1, xa-13 prom and pTA 248, respectively. A total of 12/138 plants in ASD 16 × IRBB60 cross combination and 15/157 plants in ADT 43 × IRBB60 cross combination were found to be “positive” for all the targeted genes (xa5, xa13, and Xa21). These positive plants were backcrossed with respective recurrent parents (ASD 16 and ADT 43) to generate the BC1F1 population. We have found that 9/134 plants in ASD 16 background and 13/150 plants in ADT 43 background were heterozygous for xa5, xa13, and Xa21, and these plants were screened with polymorphic SSR markers to assess the recovery of RPG. Background assay revealed that three plants in ASD 16 combination viz., IL-1-2-2, IL-1-5-6, and IL-1-8-18 with an RPG% recovery of 75.60, 76.82, and 76.56, respectively, and two plants in ADT 43 combination viz., IL-2-1-33 and IL-2-2-53 with a recovery of 77.08% and 76.04% were identified (Supplementary Table S2). These plants were backcrossed alone with respective recurrent parents to produce BC2F1. In BC2F1, 10/154 plants in ASD 16 background and 12/166 plants in ADT 43 background were found to be heterozygous for the targeted genes. In these BC2F1 populations, we have identified two plants in ASD 16 combination and three plants in ADT 43 combination with background genome recovery ranging from 87.5 to 89.02% (Supplementary Table S2), and these plants were backcrossed to produce BC3F1. In BC3F1, we have found that 11/92 in ASD 16 background and 17/110 in ADT 43 background were shown to be triple heterozygous, and background assay indicates that three plants with RPG recovery ranged from 93.85 to 94.96% in ASD 16 combination, and two plants with a recovery of 94.26% and 94.04% were observed in ADT 43 combination (Supplementary Table S2). Self-pollination was carried out in the selected plants to generate BC3F2 followed by BC3F3. Homozygous plants for xa5, xa13, and Xa21 with key agro-morphological traits were identified through genotypic and phenotypic assays, and the best lines (six lines in ASD 16 combination and nine lines in the ADT 43 combination) were constituted to generate triple-gene (xa5, xa13, and Xa21) pyramided lines of ASD 16 and ADT 43 for bacterial blight resistance.
The three-gene bacterial blight (xa5, xa13, and Xa21) pyramided lines of ASD 16 and ADT 43 were used as recipient parents for the targeted introgression of blast (Pi54) and sheath blight QTLs (qSBR7-1, qSBR11-1, and qSBR11-2) from the Tetep. The bacterial blight pyramided lines of ASD 16 and ADT 43 harboring xa5, xa13, and Xa21 were crossed with Tetep, and the F1 plants were analyzed for all the targeted traits (Table 1). The hybrids with xa5 + xa13 + Xa21 + Pi54 + qSBR7-1 + qSBR11-1 + qSBR11-2 in heterozygous condition were selected and backcrossed with ASD 16 and ADT 43 introgressions with the three-gene bacterial blight genes (xa5, xa13, and Xa21) to recover the original RPG. Up to BC3F1, all the foreground and background selections were made similar to pyramiding of bacterial blight-resistant genes, as explained above. Foreground selection of BC3F1 hybrids revealed five plants in ASD 16 background and seven plants in ADT 43 background possessing all the targeted genes and QTLs (xa5 + xa13 + Xa21 + Pi54 + qSBR7-1 + qSBR11-1 + qSBR11-2) in heterozygous condition. Background selection of these positive plants revealed two plants with RPG recovery ranging from 93.88 to 94.92% in both combinations (ASD 16, and ADT 43) (Supplementary Table S2). Selfing was done in “positive” BC3F1 hybrids with high RPG recovery to generate BC3F2. In BC3F2 generation, 11 plants in the ASD 16 background and 14 plants in the ADT 43 background were homozygous for targeted genes (xa5 + xa13 + Xa21 + Pi54) and targeted QTLs (qSBR7-1 + qSBR11-1 + qSBR11-2), and these were selfed to produce BC3F3. These homozygous-improved gene pyramided lines at BC3F3 generation were evaluated for physical expression of resistance against bacterial blight, blast, and sheath blight pathogens under greenhouse conditions. Out of 1,500 plants screened in both the crosses at field conditions, we found nine lines in ASD 16 background and 15 lines in ADT 43 background performing on par with the recurrent parents and, moreover, exhibiting a high level of resistance to all the three major diseases (Tables 2,3 and Figures 2–5).
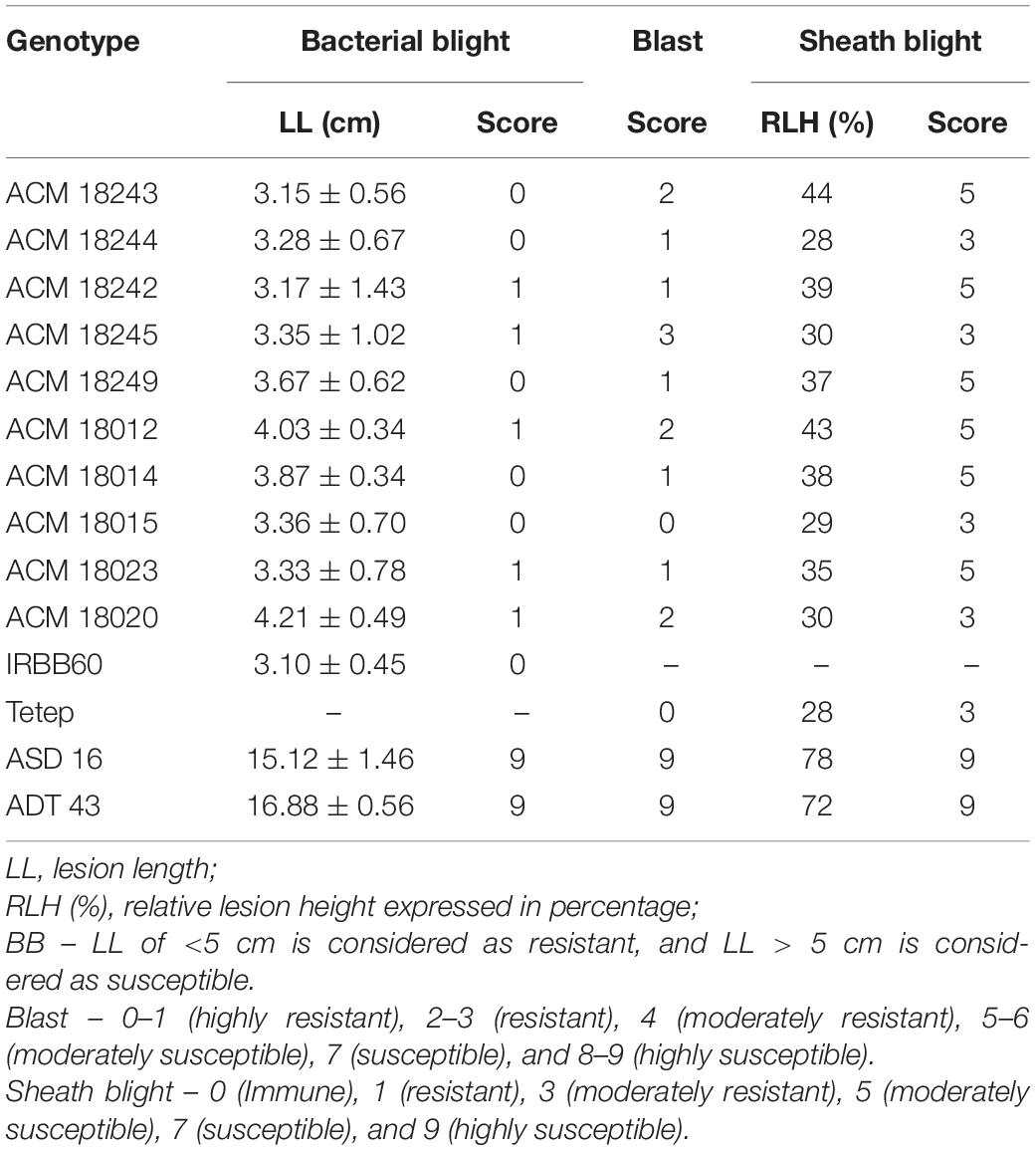
Table 2. Phenotypic reaction and disease score of improved pyramided lines against bacterial blight (BB), blast, and sheath blight diseases.

Figure 2. Phenotypic screening of improved pyramided lines against DX-027 isolate of bacterial blight (BB). P1 – ASD 16, P2 – ADT 43, P3 – IRBB60; serially numbered 1, 2, 3, 4, 5, and 6 lines indicate improved pyramided lines of recurrent parents harboring xa5, xa13, and Xa21 genes.
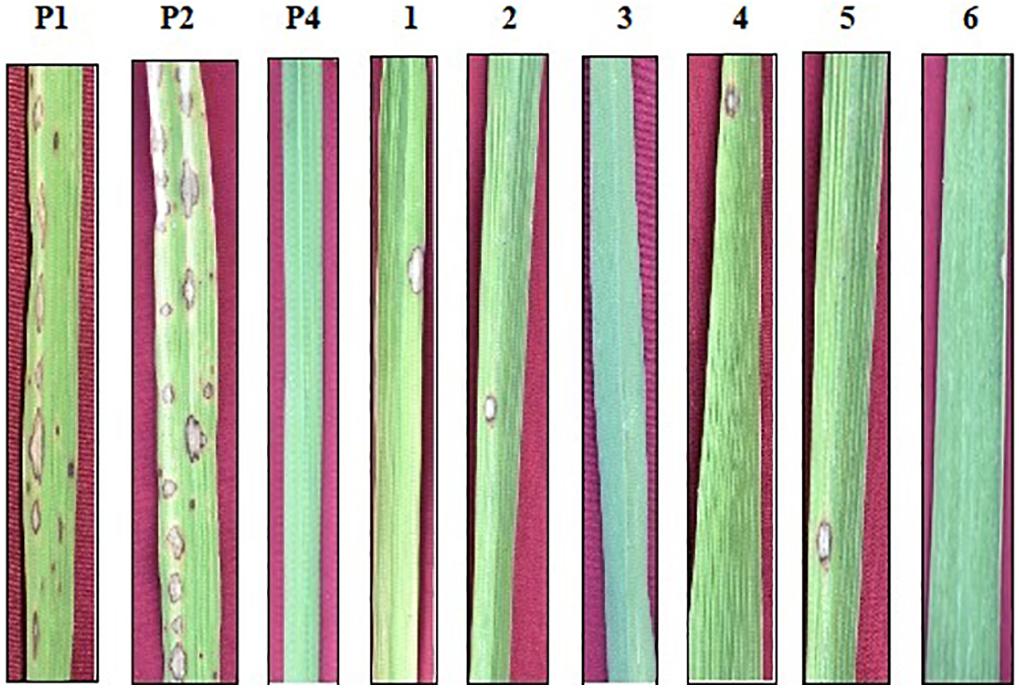
Figure 3. Phenotypic screening of improved pyramided lines against IS (KUL)-6 isolate of Magnaporthe oryzae for leaf blast. P1 – ASD 16, P2 – ADT 43, P4 – Tetep; serially numbered 1, 2, 3, 4, 5, and 6 lines indicate improved pyramided lines of recurrent parents harboring the Pi54 gene.

Figure 4. Phenotypic screening of improved pyramided lines against sheath blight resistance. P1 – ASD 16, P2 – ADT 43, P4 – Tetep; serially numbered 1, 2, 3, and 4 lines indicate improved pyramided lines harboring qSBR7-1, qSBR11-1, and qSBR11-2 QTLs.
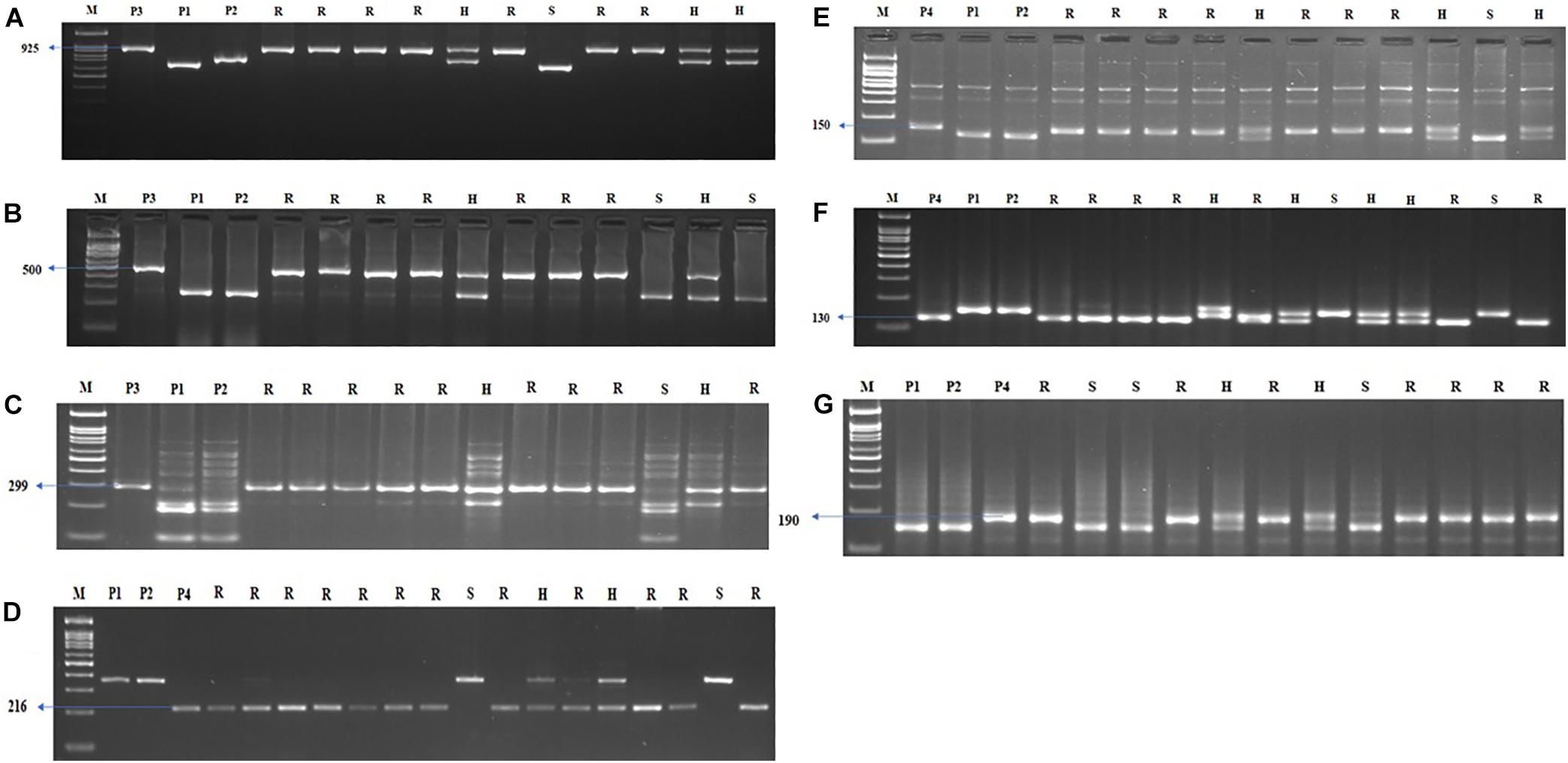
Figure 5. Agarose gel electrophoresis images illustrating the presence of (A) Xa21, (B) xa13, (C) xa5, (D) Pi54, (E) qSBR11-2, (F) qSBR11-I, and (G) qSBR7-1 alleles. P1 – ASD 16, P2 – ADT 43, P3 – IRBB60, P4 – Tetep, M – 100-bp ladder, R, resistant; H, heterozygote; S, susceptible.
The resistant parent, IRBB60, possessing xa5, xa13, and Xa21 exhibited a mean lesion length of 3.10 ± 0.45 with a disease reaction score of 0–1 (highly resistant) against the isolate of DX-027. Similarly, the identified homozygous IPLs were also expressed, and the similar mean lesion length ranged from 3.15 ± 0.56 to 4.21 ± 0.49 with the same disease reaction score as the resistant parent. The two-gene pyramided lines (xa5 + xa13, xa5 + Xa21, and xa13 + Xa21) were shown a mean lesion length of 4.58 ± 1.15 with a score of 1–2 (resistant). Evaluation of single-gene pyramided lines showed that Xa21 pyramided lines expressed a mean lesion length of 4.93 ± 0.22, xa13 lines with 6.45 ± 0.79, and xa5 lines with 6.85 ± 0.64 lesion lengths. This indicates that the lines that harbor the Xa21 component exhibited a high level of resistance than the other components in the disease reaction (data not shown), while the recurrent parents (ASD 16 and ADT 43) have shown an average lesion length of more than 15 cm with a disease reaction score of 9 (highly susceptible) for the same isolate of BB (Table 2 and Figure 2).
The resistant check, Tetep, possessing the Pi54 gene, exhibited a high level of resistance to leaf blast with no lesions observed on the leaf with a disease score of 0–1 (highly resistant) against the isolate of IS (KUL)-6. Similarly, the selected IPLs harboring the Pi54 gene displayed small pin-point size brown specks to slightly elongated necrotic patches with a sporulating center with a disease score of 1–3 (highly resistant to resistant). While, the recurrent parents (ASD 16 and ADT 43) displayed spindle-shaped lesions with brown margin with a disease reaction score of 9 (highly susceptible) (Table 2 and Figure 3). With regard to the sheath blight, Tetep possessing qSBR QTLs (qSBR7-1, qSBR11-1, and qSBR11-2) exhibited an RLH up to 28% with a disease score of 3 (moderately resistant). The susceptible checks, both ASD 16 and ADT 43, expressed an RLH of 78% and 71%, respectively, with disease score of 9 (highly susceptible). While the selected IPLs possessing qSBR7-1, qSBR11-1, and qSBR11-2 have expressed an RLH ranging from 28 to 45% with a disease score of 3 to 5 (moderately resistant to moderately susceptible) (Table 2 and Figure 4).
All the selected improved pyramided lines (nine lines in ASD 16 background and 15 lines in ADT 43 background) have similar agro-morphological and quality traits as recurrent parents. Some promising lines have shown superior agro-morphological and quality traits and, moreover harboring BB, blast, and sheath blight resistance. Significant differences for plant height were observed among the few improved pyramided lines (BC3F3 generation), which were shown as taller than recurrent parents viz., ACM 18012, ACM 18245, ACM 18242, ACM 18243, ACM 18244, ACM 18013, and ACM 18014 (Table 3). The DFF for recurrent parents (ASD 16 and ADT 43) and donor parents (IRBB60 and Tetep) were 85 and 97 days, respectively. The selected improved pyramided lines at BC3F3 generation harboring xa5 + xa13 + Xa21 + Pi54 + qSBR7-1 + qSBR11-1 + qSBR11-2 also showed a similar range for DFF, i.e., 78–95 days. Two lines in the background of ASD 16 viz., ACM 18242 and ACM 18246 were significantly flowered ∼13–18 days earlier than its respective recurrent parent, ASD 16 (Table 3). The mean values of recurrent parents for the number of grains per panicle ranged from 191 ± 14.3 to 201 ± 11.2. Several improved pyramided lines viz., ACM 18012, ACM 18245, ACM 18242, ACM 18243, ACM 18017, ACM 18016, ACM 18247, and ACM 18015 have shown significantly higher number of grains per panicle than both recurrent parents (Table 3). The mean 1,000-grain weight (g) of recurrent parents ranged from 15.6 ± 0.4 to 23.2 ± 0.5 for ADT 43 and ASD 16, respectively. The selected improved pyramided lines, ACM 18245, ACM 18244, ACM 18013, ACM 18016, ACM 18017, and ACM 18050 have shown significantly higher 1,000-grain weight than both the recurrent parents ASD 16 and ADT 43 (Table 3). The HRR% of recurrent parents ranged from 59.87 to 62.14 for ADT 43 and ASD 16, respectively, while the selected homozygous lines HRR% are also on par with the recurrent parents and some of the lines have recovered more HRR% than both the recurrent parents (Supplementary Table S3). Some of the promising lines, ACM 18012, ACM 18014, and ACM 18015 in the background of ADT 43 and ACM 18244, ACM 18245, and ACM 18249 in the background of ASD 16 have shown superior quality traits than both the recurrent parents (Supplementary Table S3).
The coefficient of genetic distance on 10 morphological traits of 24 pyramided lines and four parents revealed that all the pyramided lines were very similar with their respective recurrent parent. It was observed that two solitary clusters were formed, one with Tetep alone and another with improved pyramided lines, IRBB60 and recurrent parents (ASD 16, and ADT 43). Cluster II was subdivided into two clusters, i.e., one subcluster with ASD 16 and its respective pyramided lines, another subcluster with IRBB60, ADT 43 parent, and its respective pyramided lines. Obviously, all the improved pyramided lines were being clubbed into their respective recurrent parent cluster (Figure 6).
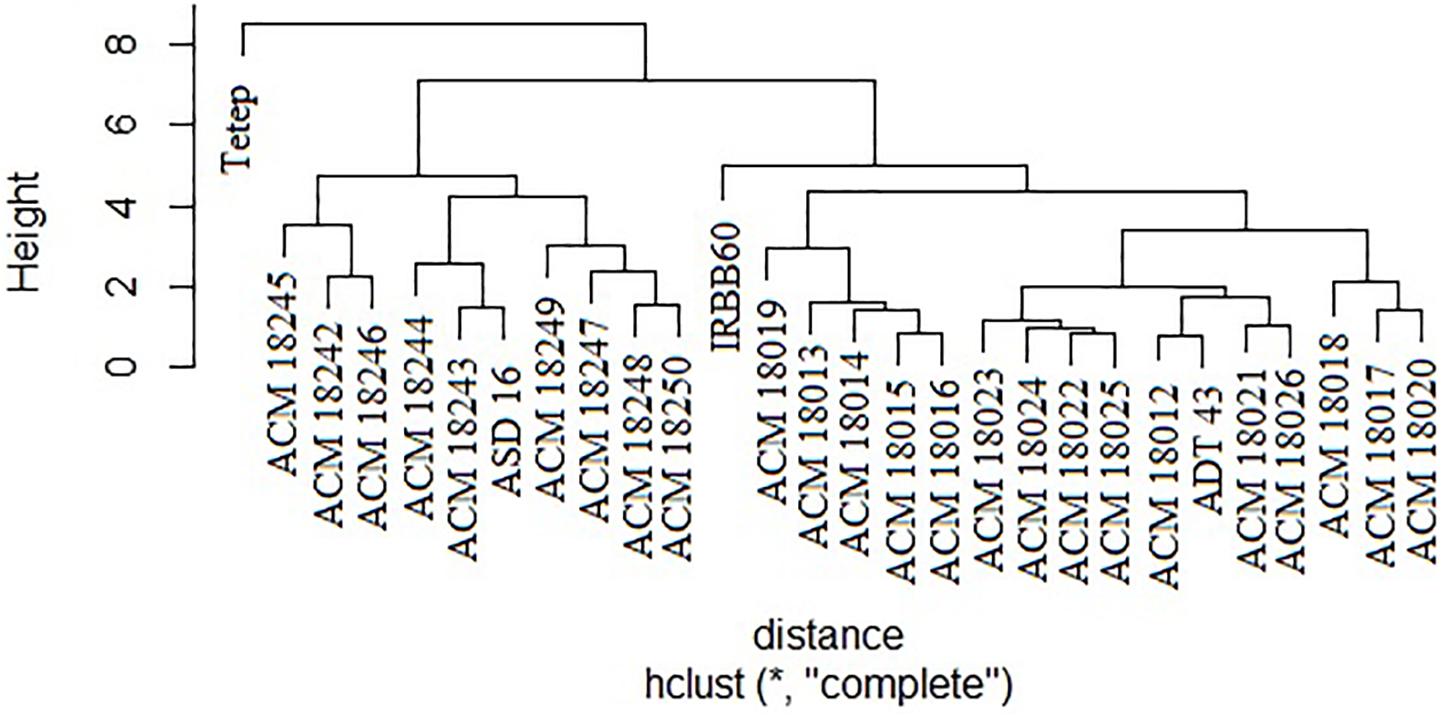
Figure 6. Clustering of 24 selected improved pyramided lines based on 10 morphological characters. ∗Euclidean distance.
More than half of the population of the globe is consuming rice to meet their dietary requirements. The demand for rice production is increasing, and many studies have reported that global rice production need to be doubled to meet the demands of the growing population (Ray et al., 2013). Besides the growing population, many biotic and abiotic stresses are affecting both yield and quality of rice crop. To address these constraints and to increase rice production, development of high-yielding cultivars enriched with resistant genes will enhance the yield as well as host-plant resistance. Conventional backcross method is the primary approach to develop resistant cultivars for single gene resistance, but phenotypic selection is a difficult and time-consuming process when multiple genes are involved in disease resistance (Crossa et al., 2017). The use of marker-assisted selection with stringent phenotypic selection enhances the efficiency and precision of breeding program for developing multiple disease-resistant varieties.
Marker-assisted backcross breeding (MABB) is a fastidious method for introgression of two or more targeted genes in the elite cultivars for improving the deficient trait. The prime aim of MABB is to transfer the targeted genes into the background of elite cultivars and to recover the RP genome as quickly as possible with a limited number of backcrosses. MABB also focuses on gradual reduction of donor parent genome as much as possible to avoid the undesirable effects on agronomical, yield, and quality traits. The recurrent parents (ASD 16 and ADT 43) in this study are the popular and high-yielding cultivars of South India. Bacterial blight, blast, and sheath blight diseases are the major diseases of South India causing huge losses to the crop.
Many studies have focused on developing resistance to one or two diseases, as developing resistance to more than two diseases is a complex and time-consuming process. There are a few reports on developing multiple disease-resistant cultivars. Singh et al. (2012) introgressed blast and sheath blight resistance genes/QTLs in the background of Improved Pusa Basmati 1, harboring xa13, and Xa21 genes. Das and Rao (2015) introgressed blast (Pi2 and Pi9), submergence (Sub1), gall midge (Gm1, Gm4), and salinity (Saltol1) genes/QTLs in the background of Improved Lalat harboring Xa4, xa5, xa13, and Xa21. Arunakumari et al. (2016) has introgressed bacterial blight and blast resistance genes into the popular cultivar MTU 1010. As a part of sustainable management, we have planned and executed the introgression of BB (xa5, xa13, and Xa21), blast (Pi54), and sheath blight (qSBR7-1, qSBR11-1, and qSBR11-2) resistance genes/QTLs in the backgrounds of ASD 16 and ADT 43 for achieving multiple disease resistance and to offer IPL to farmers.
In this study, background screening with polymorphic SSR markers was used to recover the maximum percentage of RPG in early generations. At BC3F1, we have achieved RPG recovery in both ASD 16 and ADT 43 backgrounds. The background recovery rate in this study was at par with the theoretical recovery rate. We have observed inheritance of some unfavorable characters (plant height and grain qualities) along with favorable resistant traits while introgressing genes/QTLs from Tetep. Nevertheless, we have identified superior segregates with minimal residual effect from the Tetep genome by assessing greater population size (1,500 plants in both crosses). Sundaram et al. (2008), reported that introgression of Xa21, xa13, and xa5 genes from SS1113 exercises a “pull” through inheritance of undesirable loci from a donor segment. Singh et al. (2015) introgressed qSBR7-1, qSBR11-1, and qSBR11-2 into Pusa 6B, but they have not observed the inheritance of undesirable loci from Tetep. The observed undesirable effects of Tetep on the agronomic characters might be due to the linkage drag or environmental influence on agronomic characters.
The agronomic performance of 24 selected improved pyramided lines of both ASD 16 and ADT 43 backgrounds at BC3F3 generation revealed that most of the agro-morphological traits were on par with their recurrent parents (ASD 16 and ADT 43) and also showed durable resistance to BB, blast, and sheath blight diseases. The higher yield and superior quality traits of improved pyramided lines were probably due to the inheritance of yield contributing traits from the recurrent parents. The yield, agro-morphological, and quality traits were normally controlled by polygenes, and these are distributed throughout the genome. Employment of a greater number of background markers accelerated the recovery of RPG in the early generations. Cluster analysis of selected pyramided lines and parents based on 10 morphological characters revealed that all the selected pyramided lines were clustered in their recurrent parent’s cluster. This is due to the similar morphological characters of pyramided lines with their recurrent parent’s. Similar results were also obtained by Pradhan et al. (2015) and Hsu et al. (2020), while introgressing BB resistance genes, which supports the present study results.
The homozygous improved pyramided lines (BC3F3 generation) harboring xa5 + xa13 + Xa21 + Pi54 + qSBR7-1 + qSBR11-1 + qSBR11-2 were assessed for physical resistance under greenhouse conditions. The results of bioassays suggest that pyramiding three BB-resistant genes exhibited higher resistance levels than the lines with one or two genes. In addition, the present study results also reveal that the gene combination with Xa21 component expressed a shorter lesion length than the remaining gene combinations. These results were consistent with the results of previous studies (Peng et al., 2015; Pradhan et al., 2015; Ramalingam et al., 2017; Yugander et al., 2018; Hsu et al., 2020). The improved pyramided lines harboring the Pi54 gene and ShB-resistant QTLs have expressed a similar disease reaction as donor parent, Tetep. This is due to the transfer of resistant alleles from the donor parent, which was confirmed by functional/linked molecular markers as well as phenotypic screening methods.
The field evaluation of improved pyramided lines at BC3F3 generation demonstrated that the candidate lines of both recurrent parents had equivalent expression of yield, agro-morphological, and quality traits and, more importantly, with pyramided genes for BB, blast, and sheath blight. The higher levels of resistance to multiple diseases, without any yield penalty, is an integrated approach of genotypic and phenotypic selection methods. Developing broad-spectrum resistance to multiple diseases is a challenging task due to rich diversity of agro-climatic conditions in India along with the existence of genetically distinct virulent strains of different plant pathogens. Pyramiding of multiple or effective resistant genes/QTLs for different biotic stresses can contribute broad-spectrum and durable resistance to multiple diseases to the rice regions in India. The present study results prove that MABB is an effective tool for pyramiding major genes/QTLs to obtain improved plant lines in a quick time frame.
Introgression of BB resistance genes from IRBB60, blast and sheath blight resistance genes/QTLs from Tetep into the commercial cultivars is a significant achievement for obtaining durable resistance to multiple diseases. Introgression of identified effective resistance genes from the wild relatives or landraces into the commercial cultivars gradually improves the host-plant resistance to different biotic and abiotic stresses for attaining food and nutritional security. In conclusion, we have introgressed xa5, xa13, Xa21, Pi54, and qSBR QTLs (qSBR7-1, qSBR11-1, and qSBR11-2) in the backgrounds of ASD 16 and ADT 43 to improve host-plant resistance to BB, blast, and sheath blight diseases. The improved pyramided lines can be further tested in multilocational trails and could be released as improved variety or used as a potential donor in hybridization programs for developing multiple disease-resistant cultivars in rice.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JR designed the experiment. ChR, PS, RS, VV, and TC were involved in the experiments. JR, ChR, RS, and PS developed the improved pyramided lines. SV and MA were involved in a part of the experiment. JR, ChR, VV, and AR were involved in the screening of improved pyramided lines. JR, SA, and CV were involved in the revision of the final version of the manuscript. JR and all the authors prepared and approved the final version of the manuscript.
This study was financially supported by DBT under a ATGC research grant (127-SP32889), New Delhi, India. The first author acknowledges the DBT for providing assistance for research programme at Tamil Nadu Agricultural University, Coimbatore, part of which is reported in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.591457/full#supplementary-material
Arunakumari, K., Durgarani, C. V., Satturu, V., Sarikonda, K. R., Chittoor, P. D. R., and Vutukuri, B. (2016). Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into Indian rice variety MTU1010. Rice Sci. 23, 306–316. doi: 10.1016/j.rsci.2016.04.005
Babujee, L., and Gnanamanickam, S. S. (2000). Molecular tools for characterization of rice blast pathogen (Magnaporthe grisea) population and molecular marker-assisted breeding for disease resistance. Curr. Sci. 78, 248–257.
Blair, M. W., Garris, A. J., Iyer, A. S., Chapman, B., Kresovich, S., and McCouch, S. R. (2003). High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 107, 62–73. doi: 10.1007/s00122-003-1231-2
Channamallikarjuna, V., Sonah, H., Prasad, M., Rao, G. J. N., Chand, S., and Upreti, H. C. (2010). Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol. Breed. 25, 155–166. doi: 10.1007/s11032-009-9316-5
Chen, S., Wang, C., Yang, J., Chen, B., Wang, W., and Su, J. (2020). Identification of the novel bacterial blight resistance gene Xa46(t) by mapping and expression analysis of the rice mutant H120. Sci. Rep. 10:12642. doi: 10.1038/s41598-020-69639-y
Chu, Z., Fu, B., Yang, H., Xu, C., Li, Z., and Sanchez, A. (2006). Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 112, 455–461. doi: 10.1007/s00122-005-0145-6
Crossa, J., Pérez-Rodríguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., and de los Campos, G. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011
Das, G., and Rao, G. J. N. (2015). Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6:698. doi: 10.3389/fpls.2015.00698
Gupta, S. K., Rai, A. K., Kanwar, S. S., Chand, D., Singh, N. K., and Sharma, T. R. (2012). The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J. Exp. Bot. 63, 757–772. doi: 10.1093/jxb/err297
Hsu, Y. C., Chiu, C. H., Yap, R., Tseng, Y. C., and Wu, Y. P. (2020). Pyramiding bacterial blight resistance genes in tainung82 for broad-spectrum resistance using marker-assisted selection. Int. J. Mol. Sci. 21:1281. doi: 10.3390/ijms21041281
Huang, N., Angeles, E. R., Domingo, J., Magpantay, G., Singh, S., and Zhang, G. (1997). Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor. Appl. Genet. 95, 313–320. doi: 10.1007/s001220050565
International Rice Research Institute [IRRI] (2002). Standard Evaluation System (SES). Los Baños: IRRI.
Iyer-Pascuzzi, A. S., and McCouch, S. R. (2007). Functional markers for xa5-mediated resistance in rice (Oryza sativa L.). Mol. Breed. 19, 291–296. doi: 10.1007/s11032-006-9055-9
Kauffman, H. E., Reddy, A. P. K., Hsieh, S. P. Y., and Merca, S. D. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Khush, G. S. (2013). Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breed. 132, 433–436. doi: 10.1111/pbr.1991
Kumar, P. N., Sujatha, K., Laha, G. S., Rao, K. S., Mishra, B., and Viraktamath, B. C. (2012). Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathology 102, 222–228. doi: 10.1094/Phyto-03-11-0075
Lee, K. S., Rasabandith, S., Angeles, E. R., and Khush, G. S. (2003). Inheritance of resistance to bacterial blight in 21 cultivars of rice. Phytopathology 93:147. doi: 10.1094/phyto.2003.93.2.147
Molla, K. A., Karmakar, S., Molla, J., Bajaj, P., Varshney, R. K., and Datta, S. K. (2020). Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol. J. 18, 895–915. doi: 10.1111/pbi.13312
Nguyen, H. T., Vu, Q. H., Van Mai, T., Nguyen, T. T., Vu, L. D., and Nguyen, T. T. (2018). Marker-assisted selection of Xa21 conferring resistance to bacterial leaf blight in Indica rice cultivar LT2. Rice Sci. 25, 52–56. doi: 10.1016/j.rsci.2017.08.004
Peng, H., Chen, Z., Fang, Z., Zhou, J., Xia, Z., and Gao, L. (2015). Rice Xa21 primed genes and pathways that are critical for combating bacterial blight infection. Sci. Rep. 5, 1–12. doi: 10.1038/srep12165
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., and Pandit, E. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 8:19. doi: 10.1186/s12284-015-0051-8
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ramalingam, J., Savitha, P., Alagarasan, G., Saraswathi, R., and Chandrababu, R. (2017). Functional marker-assisted improvement of stable cytoplasmic male sterile lines of rice for bacterial blight resistance. Front. Plant Sci. 8:1131. doi: 10.3389/fpls.2017.01131
Ramkumar, G., Srinivasarao, K., Mohan, K. M., Sudarshan, I., Sivaranjani, A. K. P., and Gopalakrishna, K. (2011). Development and validation of functional marker targeting an InDel in the major rice blast disease resistance gene Pi54 (Pikh). Mol. Breed. 27, 129–135. doi: 10.1007/s11032-010-9538-6
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e066428. doi: 10.1371/journal.pone.0066428
Ronald, P. C., Albano, B., Tabien, R., Abenesl, L., Wu, K., McCouch, S. R., et al. (1992). Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Genet. Genom. 236, 113–120. doi: 10.1007/bf00279649
Savary, S., Willocquet, L., and Teng, P. S. (1997). Modelling sheath blight epidemics on rice tillers. Agric. Syst. 55, 359–384. doi: 10.1016/s0308-521x(97)00014-0
Sharma, T. R., Madhav, M. S., Singh, B. K., Shanker, P., Jana, T. K., and Dalal, V. (2005). High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 274, 569–578. doi: 10.1007/s00438-005-0035-2
Sharma, T. R., Rai, A. K., Gupta, S. K., Vijayan, J., Devanna, B. N., and Ray, S. (2012). Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52. doi: 10.1007/s40003-011-0003-5
Singh, A. K., Gopalakrishnan, S., Singh, V. P., Prabhu, K. V., Mohapatra, T., and Singh, N. K. (2011). Marker assisted selection: a paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 71, 120–128.
Singh, A. K., Singh, V. K., Singh, A., Ellur, R. K., Pandian, R. T. P., and Gopala Krishnan, S. (2015). Introgression of multiple disease resistance into a maintainer of Basmati rice CMS line by marker-assisted backcross breeding. Euphytica 203, 97–107. doi: 10.1007/s10681-014-1267-1
Singh, A. K., Singh, V. K., Singh, S. P., Pandian, R. T. P., Ellur, R. K., and Singh, D. (2012). Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012:pls029. doi: 10.1093/aobpla/pls029
Singh, P., Mazumdar, P., Harikrishna, J. A., and Babu, S. (2019). Sheath blight of rice: a review and identification of priorities for future research. Planta 250, 1387–1407. doi: 10.1007/s00425-019-03246-8
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., and Holsten, T. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Sundaram, R. M., Vishnupriya, M. R., Biradar, S. K., Laha, G. S., Reddy, G. A., and Rani, N. S. (2008). Marker-assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160, 411–422. doi: 10.1007/s10681-007-9564-6
Thakur, S., Singh, P. K., Das, A., Rathour, R., Variar, M., and Prashanthi, S. K. (2015). Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 6:345. doi: 10.3389/fpls.2015.00345
Varghese, Y. A., Knaak, C., Sethuraj, M. R., and Ecke, W. (1997). Evaluation of random amplified polymorphic DNA (RAPD) markers in Hevea brasiliensis. Plant Breed. 116, 47–52. doi: 10.1111/j.1439-0523
Wang, Y., Pinson, S. R. M., Fjellstrom, R. G., and Tabien, R. E. (2012). Phenotypic gain from introgression of two QTL, qSB9-2 and qSB12-1, for rice sheath blight resistance. Mol. Breed. 30, 293–303. doi: 10.1007/s11032-011-9619-1
Yadav, S., Anuradha, G., Kumar, R. R., Vemireddy, L. R., Sudhakar, R., and Donempudi, K. (2015). Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). Springerplus 4:175. doi: 10.1186/s40064-015-0954-2
Yugander, A., Sundaram, R. M., Singh, K., Ladhalakshmi, D., Subba Rao, L. V., and Madhav, M. S. (2018). Incorporation of the novel bacterial blight resistance gene Xa38 into the genetic background of elite rice variety Improved Samba Mahsuri. PLoS One 13:e0198260. doi: 10.1371/journal.pone.0198260
Zhang, G., Angeles, E. R., Abenes, M. L. P., Khush, G. S., and Huang, N. (1996). RAPD and RFLP mapping of the bacterial blight resistance gene xa-13 in rice. Theor. Appl. Genet. 93, 65–70. doi: 10.1007/BF00225728
Keywords: rice, multiple disease resistance, marker-assisted backcross breeding, gene pyramiding, phenotyping
Citation: Ramalingam J, Raveendra Ch, Savitha P, Vidya V, Chaithra TL, Velprabakaran S, Saraswathi R, Ramanathan A, Arumugam Pillai MP, Arumugachamy S and Vanniarajan C (2020) Gene Pyramiding for Achieving Enhanced Resistance to Bacterial Blight, Blast, and Sheath Blight Diseases in Rice. Front. Plant Sci. 11:591457. doi: 10.3389/fpls.2020.591457
Received: 04 August 2020; Accepted: 06 October 2020;
Published: 19 November 2020.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Qinzhong Yang, Yunnan Academy of Agricultural Sciences, ChinaCopyright © 2020 Ramalingam, Raveendra, Savitha, Vidya, Chaithra, Velprabakaran, Saraswathi, Ramanathan, Arumugam Pillai, Arumugachamy and Vanniarajan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jegadeesan Ramalingam, cmFtYWxpbmdhbS5qQHRuYXUuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.