
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 17 December 2020
Sec. Aquatic Photosynthetic Organisms
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.589026
This article is part of the Research Topic Metabolic Regulation of Diatoms and other Chromalveolates View all 12 articles
Diatoms can accumulate high levels of triacylglycerols (TAGs) under nitrogen depletion and have attracted increasing attention as a potential system for biofuel production. In Phaeodactylum tricornutum, a model diatom, about 40% of lipid is synthesized from the breakdown of cellular components under nitrogen starvation. Our previous studies indicated that carbon skeletons from enhanced branched-chain amino acid (BCAA) degradation under nitrogen deficiency contribute to TAG biosynthesis in P. tricornutum. In this review, we outlined the catabolic pathways of all 20 amino acids based on the genome, transcriptome, proteome, and metabolome data. The contribution of these amino acid catabolic pathways to TAG accumulation was also analyzed.
Diatoms are a group of unicellular eukaryotic algae and an important component of marine phytoplankton. Although thousands of genes were of green algal derivation (Moustafa et al., 2009), diatoms are believed to emerge as the result of a secondary endosymbiotic event between two eukaryotes, a red alga and an oomycete (Medlin et al., 2000). Therefore, diatoms possess some unique features in comparison with other photosynthetic eukaryotes, including the presence of hundreds of genes from bacteria, the Entner–Doudoroff pathway, and urea cycle unfound in plants and green algae (Bowler et al., 2008; Allen et al., 2011; Fabris et al., 2012; Singh et al., 2015). Phaeodactylum tricornutum is one of the model diatoms with short generation time, and routine and simple genetic manipulation is available (Zhang and Hu, 2014; Karas et al., 2015; Falciatore et al., 2020). This diatom has the capacity to accumulate eicosapentaenoic acid, fucoxanthin, and neutral lipids (mostly triacylglycerols, TAGs) and thus is perceived as a microalgal cell factory and a potential system for biofuel production (Butler et al., 2020).
Nitrogen, accounting for over 7% of cellular mass in marine microorganisms, is one of the major constituents of both proteins and nucleic acids (Geider and La Roche, 2002). Many nutrients and nitrogen in particular are restricted in the open ocean (Moore et al., 2013), though seasonal inputs of nitrate (NO3–) can cause diatom-dominated phytoplankton blooms in coastal ecosystems (Kudela and Dugdale, 2000). Diatoms are capable of assimilating dissolved nitrogen sources of different forms, including inorganic ones such as nitrate (NO3–), nitrite (NO2–), and ammonium (NH4+) and organic ones such as urea and amino acids (Jauffrais et al., 2016). Amino acids can be taken up by cells and intracellularly metabolized as diatom genomes containing plasma membrane amino acid transporters (Armbrust et al., 2004; Sipler and Bronk, 2015), and they can also be oxidized by extracellular L-amino acid oxidase to produce α-keto acid, NH4+, and hydrogen peroxide (Palenik and Morel, 1990; Rees and Allison, 2006; Contreras and Gillard, 2020). Under nitrogen stress, cellular protein content decreases and amino acid degradation occurs (Guerra et al., 2013). Acetyl-CoA, a product of the metabolism of some amino acids, enters the tricarboxylic acid (TCA) cycle and is shunted toward fatty acid biosynthesis (Hockin et al., 2012; Ge et al., 2014; Levitan et al., 2015). It is indicated that carbon skeletons from enhanced branched-chain amino acid (BCAA) degradation under nitrogen deficiency feed into the TCA cycle and contribute to TAG biosynthesis in P. tricornutum (Ge et al., 2014; Pan et al., 2017) and Chlamydomonas reinhardtii (Liang et al., 2019). However, few studies have examined the interaction between metabolic pathways of the other amino acids and TAG biosynthesis. Although amino acid biosynthesis pathways have been reviewed (Bromke, 2013), catabolic pathways of amino acids in P. tricornutum have not been summarized. In plants, amino acid catabolism and regulation have received considerable attention (Hildebrandt et al., 2015), and amino acid catabolism is important not only during normal senescence but also in stress tolerance. The capacity of diatoms to use dissolved amino acids has been considered to help diatoms survive in blooms or in light-impenetrable sediments (Admiraal and Peletier, 1979). In response to nitrogen deprivation, amino acid degradation could promote the redistribution of carbon and nitrogen flow in diatom cells (Alipanah et al., 2015).
In this review, we outlined the catabolic pathways of all 20 amino acids and provided the subcellular localization information of related enzymes according to the prediction (Supplementary Material) from genome annotation in P. tricornutum. Based on published transcriptomes (Levitan et al., 2015; Matthijs et al., 2016, 2017; Remmers et al., 2018; Smith et al., 2019), proteomes (Remmers et al., 2018), and metabolomes (Ge et al., 2014), we arranged the expression levels of related enzymes and the content of amino acids to interpret the contribution of amino acid degradation to TAG accumulation. In addition, the homologous genes and their transcription levels (Bender et al., 2014) involved in the catabolic pathways of amino acids in Thalassiosira pseudonana were also provided.
Branched-chain amino acid (leucine, valine, and isoleucine) content decreased in P. tricornutum cells during TAG accumulation (Supplementary Figure 1). The catabolism of BCAAs has been mostly unraveled in our previous studies (Figures 1A,B; Ge et al., 2014; Pan et al., 2017). The initial steps in the degradation pathways of BCAAs are catalyzed by branched-chain amino acid transaminase (BCAT), which also catalyzes the final step in BCAA synthesis (Figures 1A,B). Many copies of BCATs have been identified in both plants and humans. In Arabidopsis, there are seven isoforms of BCAT localized in different compartments, and the mitochondrial isoform BCAT2 has been shown to be especially relevant to degradation (Angelovici et al., 2013). Six BCATs were annotated in P. tricornutum genome, and all BCATs could be up-regulated during nitrogen limitation except BCAT1 (Figure 2A and Supplementary Table 1). The function of BCATs depends on their localizations (Campbell et al., 2001), and thus, it is reasonable that the chloroplast-localized P. tricornutum BCAT1, which may be mainly responsible for BCAA synthesis, is down-regulated during nitrogen limitation. Transcriptional and/or protein levels of the other genes involved in BCAA catabolism were also found to be up-regulated during nitrogen limitation in our previous studies (Ge et al., 2014; Pan et al., 2017). The roles of methylcrotonyl-CoA carboxylase (MCC), propionyl-CoA carboxylase (PCC), 3-hydroxyisobutyryl-CoA hydrolase (HIBCH), and branched-chain α-keto acid dehydrogenase (BCKDH) in TAG accumulation have been demonstrated by genetic manipulation (Ge et al., 2014; Pan et al., 2017; Liang et al., 2019). Knockdown of MCC or knockout of BCKDH led to decreased TAG accumulation (Ge et al., 2014; Liang et al., 2019), and overexpression of HIBCH or knockdown of PCC increased TAG accumulation (Pan et al., 2017). Carbon skeletons from BCAA degradation enter the TCA cycle through acetyl-CoA in the mitochondria. The single-copy HIBCH in P. tricornutum has been proved to be localized in the mitochondria (Pan et al., 2017). Moreover, subcellular localization prediction shows that single-copy β-subunit of BCKDH (BCKDH2), α-subunit of MCC (MCC1), α-subunit of PCC (PCC1), 3-hydroxyisobutyrate dehydrogenase (HIBADH), and methylmalonyl-CoA mutase (MCM) together with two aldehyde dehydrogenases (ALDHs) are located in the mitochondria (Supplementary Table 1). Different from P. tricornutum, the whole BCAA degradation in plants takes place in both the mitochondria and peroxisome (Zolman et al., 2001).
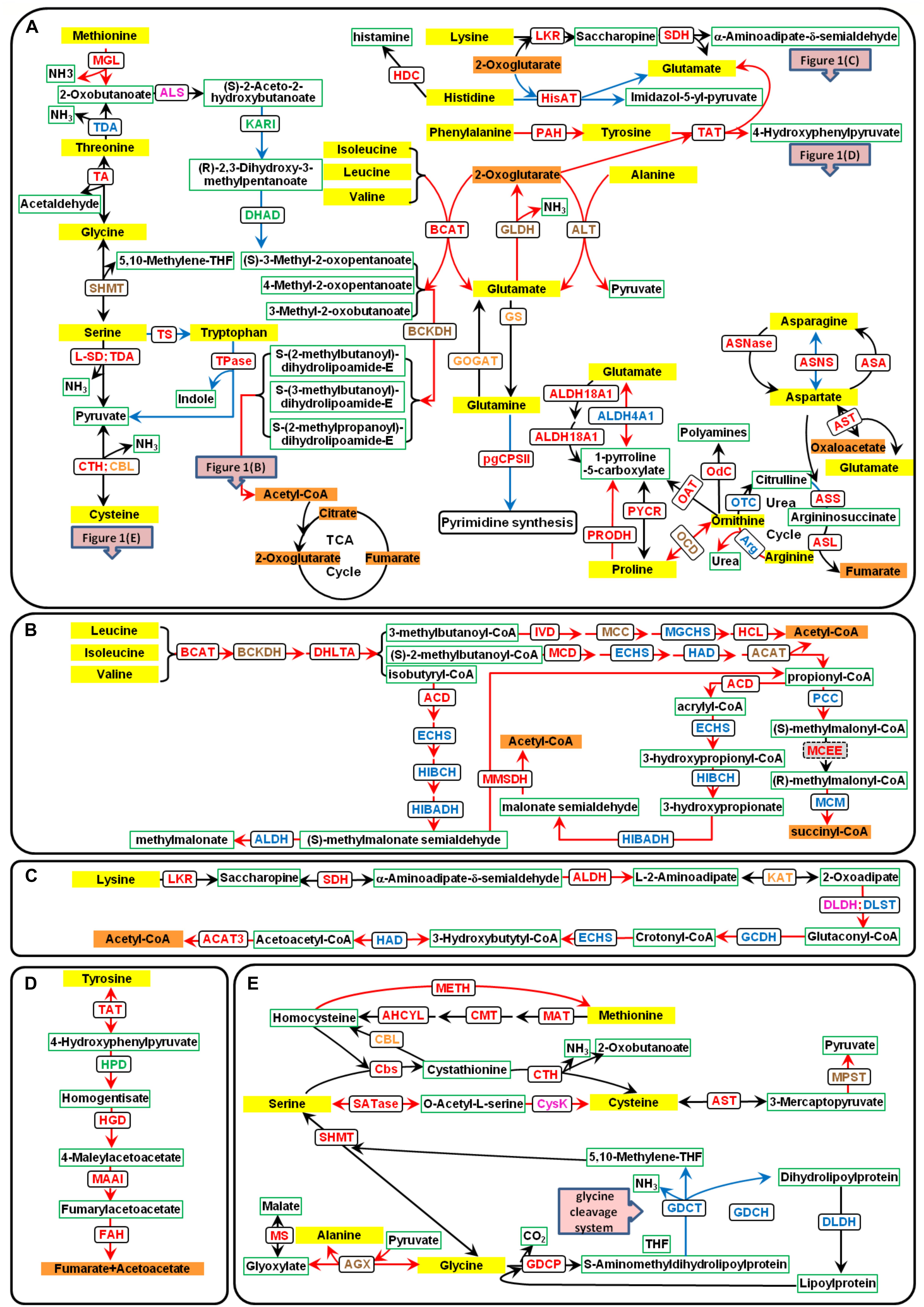
Figure 1. Amino acid catabolic pathways in P. tricornutum. The yellow text box represents amino acids, the orange text boxes indicate metabolites that could enter the TCA cycle, the white text boxes with green borders indicate other metabolites, and the white text boxes with black borders indicate the enzymes. The different font colors of enzymes represent different predicted subcellular locations: plastid localization in green font; mitochondria location in blue font; multiple isoenzymes with the plastid and mitochondria localization in orange font; those with mitochondria and other location in brown font; those with plastid and other localization in purple font; and those with plastid, mitochondria, and other localization in red font. Up-regulated genes are indicated with red lines, and down-regulated genes with blue lines. (A) Catabolic pathways of all 20 amino acids. (B) Catabolic pathways of BCAAs. (C) Catabolic pathways of lysine. (D) Catabolic pathways of tyrosine. (E) Catabolic pathways of serine, methionine, cysteine, and glycine. ACAT, acetyl-CoA C-acyltransferase (EC2.3.1.16); ACD, acyl-CoA dehydrogenase (EC1.3.8.1); AGX, alanine–glyoxylate aminotransferase (EC:2.6.1.44); AHCYL, adenosylhomocysteinase (EC3.3.1.1); ALDH, aldehyde dehydrogenase (EC1.2.1.31); ALDH18A1, delta-1-pyrroline-5-carboxylate synthetase (EC2.7.2.11 and EC1.2.1.41); ALDH4A1, 1-pyrroline-5-carboxylate dehydrogenase (EC1.2.1.88); ALS, acetolactate synthase (EC2.2.1.6); ALT, alanine transaminase (EC2.6.1.2); Arg, arginase (EC3.5.3.1); ASA, aspartate–ammonia ligase (EC6.3.1.1); ASL, argininosuccinate lyase (EC4.3.2.1); ASNase, asparaginase (EC3.5.1.1); ASNS, asparagine synthase (EC6.3.5.4); ASS, argininosuccinate synthase (EC6.3.4.5); AST, aspartate aminotransferase (EC2.6.1.1); BCAT, branched-chain amino acid transaminase (EC2.6.1.42); BCKDH, branched-chain α-keto acid dehydrogenase (EC1.2.4.4); CBL, cysteine-S-conjugate beta-lyase (EC4.4.1.13); Cbs, cystathionine beta-synthase (EC4.2.1.22); CMT, DNA (cytosine-5)-methyltransferase (EC2.1.1.37); CPSII, carbamoyl-phosphate synthase II (EC6.3.5.5); CTH, cystathionine gamma-lyase (EC4.4.1.1); CysK, cysteine synthase (EC2.5.1.47); DHAD, dihydroxy-acid dehydratase (EC4.2.1.9); DHLTA, dihydrolipoyllysine-residue (2-methylpropanoyl) transferase (EC2.3.1.168); DLDH, dihydrolipoyl dehydrogenase (EC1.8.1.4); DLST, dihydrolipoamide succinyltransferase; ECHS, enoyl-CoA hydratase (EC4.2.1.17); FAH, fumarylacetoacetase (EC3.7.1.2); GCDH, glutaryl-CoA dehydrogenase (EC1.3.8.6); GDCH, glycine cleavage system H protein; GDCP, glycine decarboxylase p-protein (EC1.4.4.2); GDCT, glycine decarboxylase t-protein (EC2.1.2.10); GLDH, glutamate dehydrogenase (EC1.4.1.2 and EC1.4.1.4); GOGAT, glutamine 2-oxoglutarate aminotransferase (EC1.4.1.13, EC1.4.1.14, and EC1.4.7.1); GS, glutamine synthetase (EC6.3.1.2); HAD, 3-hydroxyacyl-CoA dehydrogenase (EC1.1.1.35); HCL, hydroxymethylglutaryl-CoA lyase (EC4.1.3.4); HDC, histidine decarboxylase (EC:4.1.1.22); HGD, homogentisate 1,2-dioxygenase (EC1.13.11.5); HIBADH, 3-hydroxyisobutyrate dehydrogenase (EC1.1.1.31); HIBCH, 3-hydroxyisobutyryl-CoA hydrolase (EC3.1.2.4); HisAT, histidine transaminase (EC2.6.1.38); HPD, 4-hydroxyphenylpyruvate dioxygenase (EC1.13.11.27); IVD, isovaleryl-CoA dehydrogenase (EC1.3.8.4); KARI, ketol-acid reductoisomerase (EC1.1.1.86); KAT, kynurenine aminotransferase (EC2.6.1.39); LKR, lysine-2-oxoglutarate reductase (EC1.5.1.8); L-SD, L-serine ammonia-lyase (EC4.3.1.17); MAAI, maleylacetoacetate isomerase (EC5.2.1.2); MAT, S-adenosylmethionine synthetase (EC2.5.1.6); MCC, methylcrotonyl-CoA carboxylase (EC6.4.1.4); MCD, 2-methylacyl-CoA dehydrogenase (EC1.3.99.12); MCEE, methylmalonyl-CoA epimerase (EC5.1.99.1); MCM, methylmalonyl-CoA mutase (EC5.4.99.2); METH, methionine synthase (EC2.1.1.13); MGCHS, methylglutaconyl-CoA hydratase (EC4.2.1.18); MGL, methionine gamma-lyase (EC4.4.1.11); MMSDH, methylmalonate semialdehyde dehydrogenase (EC1.2.1.27); MPST, 3-mercaptopyruvate sulfurtransferase (EC2.8.1.2); MS, malate synthase (EC2.3.3.9); OAT, ornithine aminotransferase (EC2.6.1.13); OCD, ornithine cyclodeaminase (EC4.3.1.12); Odc, ornithine decarboxylase (EC4.1.1.17); OTC, ornithine carbamoyltransferase (EC2.1.3.3); PAH, phenylalanine hydroxylase (EC1.14.16.1); PCC, propionyl-CoA carboxylase (EC6.4.1.3); PRODH, proline dehydrogenase (EC1.5.5.2); PYCR, pyrroline-5-carboxylate reductase (EC1.5.1.2); SATase, serine O-acetyltransferase (EC2.3.1.30); SDH, saccharopine dehydrogenase (EC1.5.1.9); SHMT, serine hydroxymethyltransferase (EC2.1.2.1); TA, threonine aldolase (EC4.1.2.5); TAT, tyrosine aminotransferase (EC2.6.1.5); TDA, threonine deaminase (EC4.3.1.19); Tpase, tryptophanase (EC4.1.99.1); TS, tryptophan synthase (EC4.2.1.20)..
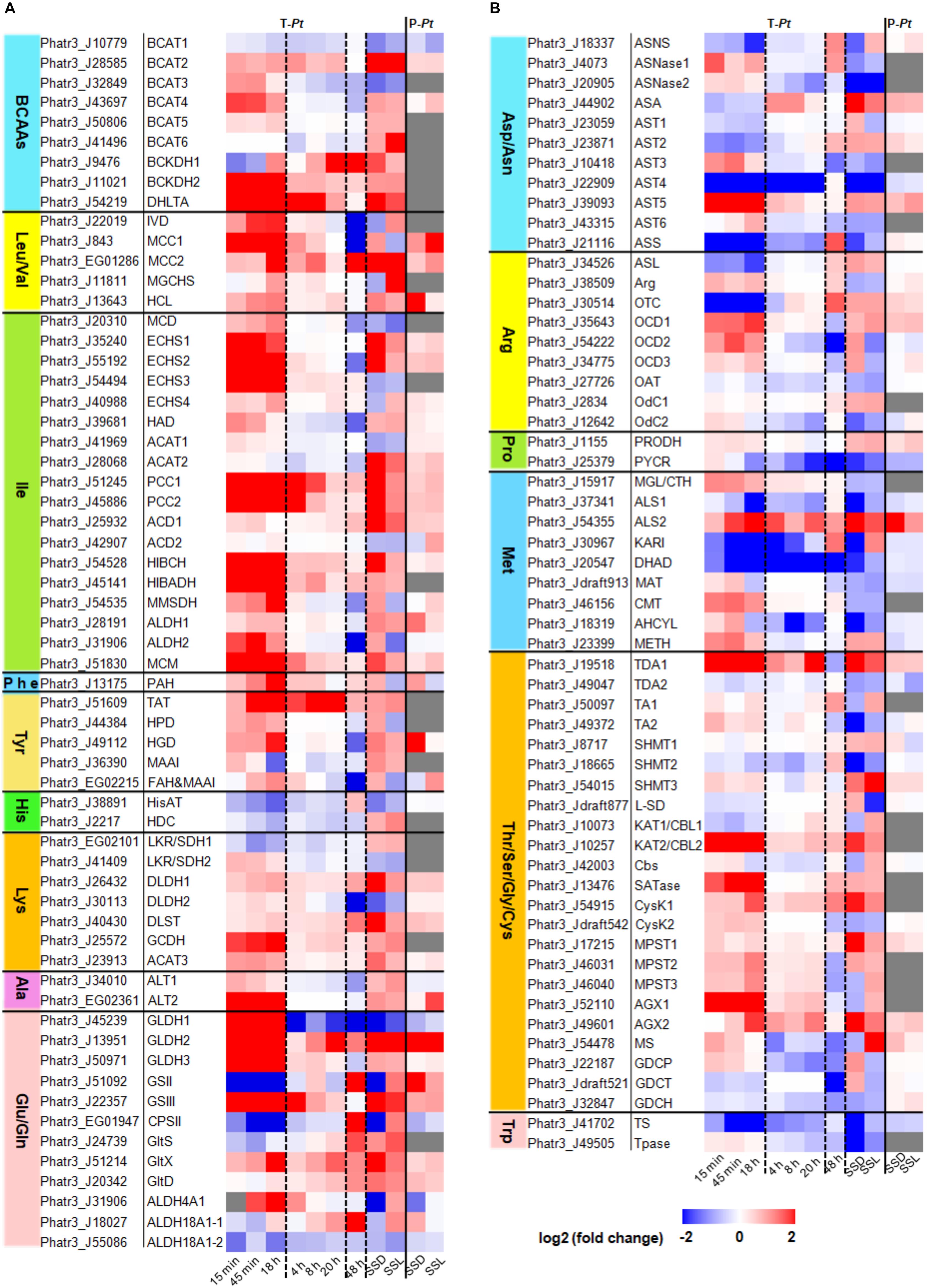
Figure 2. Expression levels of amino acid catabolism-related genes in P. tricornutum. Transcriptome data of 15 min, 45 min, and 18 h were cited from Smith et al. (2019). Fold changes of 15 min, 45 min, and 18 h were re-calculated by N-4, N-5, and N-6 contrasting with pre_3, respectively. Transcriptome data of 4, 8, and 20 h were cited from Matthijs et al. (2016, 2017). Transcriptome data of 48 h were cited from Levitan et al. (2015). The transcriptome data and the proteome data of SSL (nitrogen stress to steady state in the light period) and SSD (nitrogen stress to steady state in the dark period) were cited from Remmers et al. (2018). T-Pt, transcriptome of P. tricornutum; P-Pt, proteome of P. tricornutum. The homologous genes in T. pseudonana and the fold changes and RPKM data were shown in Supplementary Tables 1, 2, respectively. All the experimental conditions were shown in the Supplementary Material. The abbreviations of related enzymes are the same as those in Figure 1.
Histidine (His) is converted to glutamate (Glu) by four enzymatic steps in animals (Litwack, 2018). However, this pathway has not yet been investigated in plants. In P. tricornutum, there is a homologous histidine transaminase (HisAT), which could convert His to Glu and imidazol-5-yl-pyruvate using 2-oxoglutarate as a carbon skeleton (Figure 1A). The degradation pathway of imidazol-5-yl-pyruvate is not clear yet in P. tricornutum. Free His content was very low in P. tricornutum cells and was almost undetectable during nitrogen deficiency in our previous studies (Supplementary Figure 1; Ge et al., 2014; Pan et al., 2017). The expression of HisAT was down-regulated during nitrogen limitation (Figure 2A), which suggests that His might mainly be metabolized to histamine by the action of histidine decarboxylase (HDC) in the mitochondria (Figure 1A). It is likely that His degradation has no contribution to TAG accumulation.
Lysine (Lys) is catabolized via the α-amino adipic acid pathway identical to that in plants (Hildebrandt et al., 2015). It is converted to saccharopine and subsequently to α-aminoadipate-δ-semialdehyde by a bifunctional enzyme with two functionally independent domains, namely, lysine-ketoglutarate reductase (LKR) and saccharopine dehydrogenase (SDH), using 2-oxoglutarate as substrate to produce Glu (Figure 1C). The response of the bifunctional LKR/SDH enzyme encoding genes (Phatr3_EG02101 and Phatr3_J41409) to nitrogen limitation was different between nitrogen-starved batch cultures and continuous cultures (Figure 2A). In Arabidopsis, the expression of LKR/SDH was regulated by abscisic acid (ABA), jasmonate, sugar starvation, and/or nitrogen starvation (Stepansky and Galili, 2003). Some enzymes in the catabolic pathways of Lys are shared by other amino acids, including enoyl-CoA hydratase (ECHS) and 3-hydroxyacyl-CoA dehydrogenase (HAD) involved in the catabolic pathways of isoleucine (Ile), and kynurenine aminotransferase (KAT, also annotated as cysteine-S-conjugate beta-lyase, CBL) in cysteine (Cys) metabolism (Figures 1A,E). Almost all the genes involved in Lys degradation were up-regulated with the decrease of Lys under nitrogen limitation (Figure 2A and Supplementary Figure 1). The early steps of Lys degradation (from Lys to α-aminoadipate-d-semialdehyde) may take place in the cytoplasm and the later steps (from α-aminoadipate-d-semialdehyde to acetyl-CoA) in the mitochondria according to subcellular localization prediction (Supplementary Table 1). The reaction of 2-oxoadipate oxidatively decarboxylated to glutaryl-CoA is equivalent to the oxidative decarboxylation of 2-oxoglutarate in the TCA cycle. The subsequent degradation reaction was similar to that of Ile in the mitochondria. Since one of the end degradation products of Lys is acetyl-CoA (Figure 1C) and the genes involved in Lys degradation are up-regulated, it is likely that Lys catabolism participates in the regulation of carbon/nitrogen partitioning and TAG accumulation.
Like that in animals, phenylalanine (Phe) is hydroxylated to tyrosine (Tyr) by phenylalanine hydroxylase (PAH) prior to degradation in P. tricornutum (Figure 1A). However, the catabolic pathway remains largely unknown since no PAH homolog has been found in plants (Hildebrandt et al., 2015). The complete degradation pathway of Tyr has been demonstrated in plants based on the degradation pathway in mammals (Dixon and Edwards, 2006). The amino group is transferred to Glu by tyrosine aminotransferase (TAT), and the product was finally degraded into fumarate and acetoacetate by four enzymatic steps. The homologous enzymes for each step have been found in P. tricornutum (Figure 1D). Phatr3_EG02215 annotated as fumarylacetoacetase has two annotations in UniProt, namely, fumarylacetoacetase and glutathione S-transferase (GST). Peptides of the latter annotation of protein Phatr3_EG02215 are homologous with human glutathione S-transferase zeta 1 (GSTZ1), which was also described as maleylacetoacetate isomerase (MAAI). The amino acid sequence of GST in P. tricornutum shares 48 and 53% identity with mammalian and T. pseudonana MAAI, respectively, and all the three sequences contain the conserved motif and active site of MAAI (Polekhina et al., 2001; Supplementary Figure 2). This means that Phatr3_EG02215, annotated as one gene (fumarylacetoacetase encoding gene) in Ensembl Protists, may actually be a compound of two genes (fumarylacetoacetase encoding gene and MAAI). In addition, Phatr3_J36390 was annotated as MAAI in the studies of Levering et al. (2016, 2017), though it is more similar to glutathione S-transferase alpha (GSTA) than GSTZ1 by BlastP analysis. The concentration of Phe and Tyr was decreased during nitrogen limitation in P. tricornutum (Supplementary Figure 1). The expression levels of genes involved in Phe and Tyr degradation were up-regulated accordingly (Figure 2A). Phe and Tyr catabolic pathways may take place in the cytoplasm according to subcellular localization prediction (Supplementary Table 1). Although fumarate, the final product of Phe and Tyr degradation, is an intermediate of the TCA cycle, the contribution of the catabolic pathways of the two amino acids to TAG accumulation still needs to be investigated.
Alanine (Ala) can be directly converted to pyruvate by alanine aminotransferases (ALT). Glutamate dehydrogenase (GLDH) catalyzes oxidative deamination of Glu to produce 2-oxoglutarate (Figure 1A). The glutamine synthetase (GS)/glutamine 2-oxoglutarate aminotransferase (GOGAT) pathway is important for ammonium assimilation. GS catalyzes Glu and ammonium to produce glutamine (Gln), and Glu also provides α-amino group for all other amino acid biosynthesis directly or indirectly. Glu and Gln are the most important amino acids as donors for the biosynthesis of major N-containing compounds, including amino acids, nucleotides, chlorophylls, polyamines, and alkaloids (Ireland and Lea, 1999). GS is found, as multiple isoenzyme forms, located both in the cytosol (GS1) and chloroplast/plastid (GS2) and plays distinct roles in most of the higher plants (Ireland and Lea, 1999; Lancien et al., 2000). In P. tricornutum, one GS is located in the plastid (GSII, Phatr3_J51092) and the other one in the mitochondria (GSIII, Phatr3_J22357) (Smith et al., 2019). GOGAT transfers the amide-nitrogen of Gln to 2-oxoglutarate, thus providing two molecules of Glu (Forde and Lea, 2007). There are three forms of GOGAT in P. tricornutum (Alipanah et al., 2015): one that uses reduced ferredoxin as the electron donor (Fd-GOGAT/GltS, EC 1.4.7.1, Phatr3_J24739), one that uses NADH as the electron donor (NADH-GOGAT/GltX, EC 1.4.1.14, Phatr3_J51214), and the third that uses NADPH as the electron donor (NADPH-GOGAT/GltD, EC 1.4.1.13, Phatr3_J20342). The first two GOGAT present in plants are located in the chloroplast or plastid (Oliveira et al., 1997), and the third is found in bacteria (Reitzer, 1987). The subcellular localization of GOGAT in P. tricornutum is not the same to that in plants, which may be contributed to their different origins (Smith, 2018). GltS is located in the plastid, and GltX and GltD are predicated to be in the mitochondria. Thus, the plastidial GSII and GltS are responsible for the assimilation of ammonium produced by nitrate reduction, while mitochondrial GSIII, GltX, and GltD may catalyze the assimilation of Gln from ammonium derived from cytosolic catabolic reactions, e.g., deamination and hydrolysis of organic N (Hockin et al., 2012; Alipanah et al., 2015). pgCPSII uses Gln to perform the first committed step of pyrimidine synthesis (Allen et al., 2011). Besides, Glu can also be converted to 1-pyrroline-5-carboxylate, the intermediate product of proline (Pro) and ornithine (Orn) degradation, by one-step catalytic reaction of 1-pyrroline-5-carboxylate dehydrogenase (ALDH4A1) or by two-step catalytic reaction of delta-1-pyrroline-5-carboxylate synthetase (ALDH18A1) (Figure 1A).
The contents of Ala, Glu, and Gln dropped sharply, and Gln was even undetectable during nitrogen limitation in P. tricornutum (Supplementary Figure 1). The two ALT enzymes and the three GLDH enzymes were up-regulated under nitrogen-starved batch cultures or/and continuous cultures (Figure 2A). The mitochondrial GS/GOGAT was up-regulated during nitrogen limitation to assimilate ammonium derived from cytosolic catabolic reactions. The plastidial GS/GOGAT was up-regulated only under nitrogen-starved continuous cultures, suggesting that nitrate reduction was activated in plastid during nitrogen-starved continuous cultures. On the contrary, little ammonium was produced via nitrate reduction in plastid under nitrogen-starved batch cultures. Glu may not be used for pyrimidine synthesis during nitrogen limitation as pgCPSII was down-regulated. The mutual conversion of Glu to 1-pyrroline-5-carboxylate was mainly catalyzed by ALDH4A1 in the mitochondria due to the up-regulation of ALDH4A1 and almost unchanged expression level of ALDH18A1 according to the transcriptome data. In sum, based on the up-regulated expression of ALT gene and the products of metabolism of Ala together with the important roles of Glu and Gln in nitrogen metabolism, it seems that Ala, Glu, and Gln catabolism may contribute to TAG accumulation during nitrogen limitation.
Aspartate (Asp) and asparagine (Asn) can be converted into each other by asparagine synthase (ASNS), using Gln and Glu as substrates, respectively. Besides, Asp can be converted to Asn by aspartate–ammonia ligase (ASA) with the absorption of ammonia, and Asn can be converted to Asp by asparaginase (ASNase) with the release of ammonia. Asp can be converted to 2-oxoglutarate by aspartate aminotransferase (AST), producing oxaloacetate and Glu (Figure 1A). There are six AST encoding genes in P. tricornutum that are predicted to be localized in various chambers of cells. Since AST was up-regulated, Asp may be converted to oxaloacetate during nitrogen limitation. The aspartate–argininosuccinate shunt established an association between the ornithine-urea cycle (OUC) and the TCA cycle (Morris, 2002; Allen et al., 2011). In addition, arginine (Arg), Orn, and Pro are directly connected to the OUC, which has been well elaborated in P. tricornutum (Allen et al., 2011). Although the contents of Asp, Asn, Arg, Pro, and Orn were decreased during nitrogen limitation (Supplementary Figure 1), neither the expression level of genes responsible for the conversion between Asp and Asn nor that of those genes related to OUC was significantly up-regulated (Figure 2B). Orn can be metabolized by ornithine carbamoyltransferase (OTC), ornithine decarboxylase (OdC), ornithine aminotransferase (OAT), or ornithine cyclodeaminase (OCD), and only OCD is markedly up-regulated during nitrogen limitation. Arg can be converted to Orn by arginase, which was up-regulated during nitrogen limitation. Therefore, it is very likely that the degradation of Arg produces Pro subsequently converted to 1-pyrroline-5-carboxylate by proline dehydrogenase (PRODH). Then, 1-pyrroline-5-carboxylate is converted to Glu to provide nitrogen in cells for growth. Based on the final degradation products of these four amino acids, their catabolism may contribute little to TAG accumulation.
The pathway for methionine (Met) degradation that converts Met to 2-oxobutanoate by methionine gamma-lyase (MGL) has been identified in plants but is absent in animals (Rébeillé et al., 2006). 2-Oxobutanoate is a precursor for Ile synthesis, and similar to Ile, it can also be degraded to acetyl-CoA via oxidation (Figure 1A). An alternative pathway that converts Met to homocysteine, which is subsequently converted to Cys in animals and plants, is catalyzed by three enzymes (Figure 1E). Homocysteine can be directly converted to Met, which is catalyzed by methionine synthase (METH). The expression of genes for Met degradation through 2-oxobutanoate was down-regulated, except for one acetolactate synthase (ALS) encoding gene (Figure 2B). ALS catalyzes the degradation of 2-oxobutanoate, an intermediate of threonine (Thr) catabolism. Genes involved in other Met degradation pathways were slightly up-regulated during nitrogen limitation with the decrease of Met (Supplementary Figure 1). It is not clear which pathway was dominant for the degradation of Met during nitrogen limitation in P. tricornutum.
The catabolic pathways of the five amino acids are very complicated, and some involved enzymes are also present in other amino acid degradation pathways. As mentioned above, Thr can be converted to 2-oxobutanoate by threonine deaminase (TDA). It can also be interconverted with glycine (Gly) by threonine aldolase (TA). Gly can also be interconverted with serine (Ser) by serine hydroxymethyltransferase (SHMT). The easy interconversion of these three amino acids indicates that these reactions can be relevant for the synthesis of the product amino acids and the degradation of the substrate amino acids as well (Figure 1A; Hildebrandt et al., 2015).
Cysteine and tryptophan (Trp) can be produced from Ser by two and one enzymatic reactions, respectively (Figure 1A). The conversion of the former can be completed through the intermediate product cystathionine or O-acetyl-L-serine (Figure 1E). Ser, Trp, and Cys can be directly degraded into pyruvate with the release of ammonia or indole. In addition, Cys can also be degraded into pyruvate through a two-step reaction (Hildebrandt et al., 2015; Figure 1E). The Gly cleavage system (GCS), which is a mitochondrial multi-enzyme system (also named glycine decarboxylase or glycine dehydrogenase system), comprises four proteins, three enzymes (P-protein, T-protein, and L-protein), and a small lipoylated protein known as H-protein. GCS, an essential and ubiquitous step of both photorespiration and primary metabolism in plants, is responsible for the interconversion of Gly and Ser (Bauwe and Kolukisaoglu, 2003). Although the H-protein has no catalytic activity itself, it acts as a substrate for the P-, T-, and L-proteins and increases the GCS activity (Hasse et al., 2009). Gly can also be transaminated by alanine–glyoxylate aminotransferase (AGX), and the resulting glyoxylate can be acetylated in the peroxisomes to produce malate in plants (Mazelis, 1980). Homologs of genes mentioned above are present in the P. tricornutum genome (Figure 1E), although the locations of the encoded proteins are not exactly the same as those in plants (Supplementary Table 1). Thr and Trp contents decreased and Ser content showed no marked difference, while Gly content increased during nitrogen limitation in P. tricornutum (Supplementary Figure 1). Cys was not detected in our previous studies (Ge et al., 2014; Pan et al., 2017). In general, Thr and Trp may be degraded mainly through the catalysis of TDA and tryptophanase (Tpase), respectively. The conversion of Ser to Cys and the degradation of Cys may be active during nitrogen limitation according to gene expression levels (Figure 2B). However, Ser might not be converted to Trp but was interconverted with Gly frequently. Since GCS-related genes were not up-regulated during nitrogen limitation, Gly was not degraded mainly by GCS. Up-regulated AGX and malate synthase (MS) indicated that Gly was transaminated and acetylated under nitrogen starvation, which may help to explain why the content of Gly did not decrease and even increased during nitrogen limitation. In brief, the contribution of the degradation of these five amino acids to TAG accumulation remains unclear.
Amino acids, as protein constituents and essential metabolites, play critical roles in living organisms. Some amino acids (e.g., serine, proline, and leucine) have been shown to act as signaling molecules in plants (Szabados and Savouré, 2010; Häusler et al., 2014; Ros et al., 2014). Therefore, pool sizes of amino acids are of critical importance and are adjusted by amino acid catabolism. The degradation pathways of amino acids in P. tricornutum were not identical with those in plants and mammals. In particular, the metabolic pathway of BCAAs in the diatom is different from that in animals, and the subcellular locations of related enzymes are not exactly the same with those in plants. In addition, the metabolic pathways of His and Phe and the OUC in P. tricornutum are similar to those in animals, but no related enzymes are found in plants. The mutual transformation pathway of essential amino acids in P. tricornutum does not exist in animals. Considering the end degradation products of amino acids and the expression levels of related enzymes in the metabolic pathways during nitrogen limitation, BCAAs, Lys, Ala, Glu, and Gln may contribute to TAG accumulation. Furthermore, to fully understand the catabolic pathways and their regulatory mechanisms, genetic manipulation and a combination of post-genomic approaches (transcriptome, proteome, and metabolome) are necessary for the analyses of mutant and wild-type diatoms.
YP, CY, CL, and TH arranged the data under the guidance of HH. YP and HH prepared and wrote the manuscript. FH reviewed and revised this manuscript. All authors contributed to the article and approved the submitted version.
This work was partially supported by the Featured Institute Service Projects from the Institute of Hydrobiology, the Chinese Academy of Sciences (grant number: Y85Z061601) and the National Natural Science Foundation of China (no. 31800302).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.589026/full#supplementary-material
Admiraal, W., and Peletier, H. (1979). Influence of organic compounds and light limitation on the growth rate of estuarine benthic diatoms. Br. Phycol. J. 14, 197–206. doi: 10.1080/00071617900650211
Alipanah, L., Rohloff, J., Winge, P., Bones, A. M., and Brembu, T. (2015). Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 66, 6281–6296. doi: 10.1093/jxb/erv340
Allen, A. E., Dupont, C. L., Oborník, M., Horák, A., Nunes-Nesi, A., McCrow, J. P., et al. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207. doi: 10.1038/nature10074
Angelovici, R., Lipka, A. E., Deason, N., Gonzalez-Jorge, S., Lin, H., Cepela, J., et al. (2013). Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25, 4827–4843. doi: 10.1105/tpc.113.119370
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Bauwe, H., and Kolukisaoglu, Ü. (2003). Genetic manipulation of glycine decarboxylation. J. Exp. Bot. 54, 1523–1535. doi: 10.1093/jxb/erg171
Bender, S. J., Durkin, C. A., Berthiaume, C. T., Morales, R. L., and Armbrust, E. (2014). Transcriptional responses of three model diatoms to nitrate limitation of growth. Front. Mar. Sci. 1:3. doi: 10.3389/fmars.2014.00003
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Bromke, M. A. (2013). Amino acid biosynthesis pathways in diatoms. Metabolites 3, 294–311. doi: 10.3390/metabo3020294
Butler, T., Kapoore, R. V., and Vaidyanathan, S. (2020). Phaeodactylum tricornutum: a diatom cell factory. Trends Biotechnol. 38, 606–622. doi: 10.1016/j.tibtech.2019.12.023
Campbell, M. A., Patel, J. K., Meyers, J. L., Myrick, L. C., and Gustin, J. L. (2001). Genes encoding for branched-chain amino acid aminotransferase are differentially expressed in plants. Plant Physiol. Biochem. 39, 855–860. doi: 10.1016/S0981-9428(01)01306-7
Contreras, J. A., and Gillard, J. T. (2020). Asparagine-based production of hydrogen peroxide triggers cell death in the diatom Phaeodactylum tricornutum. Bot. Lett. 1–12. doi: 10.1080/23818107.2020.1754289
Dixon, D. P., and Edwards, R. (2006). Enzymes of tyrosine catabolism in Arabidopsis thaliana. Plant Sci. 171, 360–366. doi: 10.1016/j.plantsci.2006.04.008
Fabris, M., Matthijs, M., Rombauts, S., Vyverman, W., Goossens, A., and Baart, G. J. (2012). The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner–Doudoroff glycolytic pathway. Plant J. 70, 1004–1014. doi: 10.1111/j.1365-313X.2012.04941.x
Falciatore, A., Jaubert, M., Bouly, J. P., Bailleul, B., and Mock, T. (2020). Diatom molecular research comes of age: model species for studying phytoplankton biology and diversity. Plant Cell 32, 547–572. doi: 10.1105/tpc.19.00158
Forde, B. G., and Lea, P. J. (2007). Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 58, 2339–2358. doi: 10.1093/jxb/erm121
Ge, F., Huang, W., Chen, Z., Zhang, C., Xiong, Q., Bowler, C., et al. (2014). Methylcrotonyl-CoA carboxylase regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum. Plant Cell 26, 1681–1697. doi: 10.1105/tpc.114.124982
Geider, R., and La Roche, J. (2002). Redfield revisited: variability of C:N:P in marine microalgae and its biochemical basis. Eur. J. Phycol. 37, 1–17. doi: 10.1017/S0967026201003456
Guerra, L. T., Levitan, O., Frada, M. J., Sun, J. S., Falkowski, P. G., and Dismukes, G. C. (2013). Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenerg. 59, 306–315. doi: 10.1016/j.biombioe.2013.10.007
Hasse, D., Mikkat, S., Hagemann, M., and Bauwe, H. (2009). Alternative splicing produces an H-protein with better substrate properties for the P-protein of glycine decarboxylase. FEBS J. 276, 6985–6991. doi: 10.1111/j.1742-4658.2009.07406.x
Häusler, R. E., Ludewig, F., and Krueger, S. (2014). Amino acids – A life between metabolism and signaling. Plant Sci. 229, 225–237. doi: 10.1016/j.plantsci.2014.09.011
Hildebrandt, T. M., Nunes Nesi, A., Araújo, W. L., and Braun, H. P. (2015). Amino acid catabolism in plants. Mol. Plant 8, 1563–1579. doi: 10.1016/j.molp.2015.09.005
Hockin, N. L., Mock, T., Mulholland, F., Kopriva, S., and Malin, G. (2012). The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 158, 299–312. doi: 10.1104/pp.111.184333
Ireland, R. J., and Lea, P. J. (1999). “The enzymes of glutamine, glutamate, asparagine, and aspartate metabolism,” in Plant Amino Acids. Biochemistry and Biotechnology, ed. B. K. Singh (New York, NY: Marcel Dekker), 49–109.
Jauffrais, T., Jesus, B., Méléder, V., Turpin, V., Arnaldo, D., Raimbault, P., et al. (2016). Physiological and photophysiological responses of the benthic diatom Entomoneis paludosa (Bacillariophyceae) to dissolved inorganic and organic nitrogen in culture. Mar. Biol. 163:115. doi: 10.1007/s00227-016-2888-9
Karas, B. J., Diner, R. E., Lefebvre, S. C., McQuaid, J., Phillips, A. P., Noddings, C. M., et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6:6925. doi: 10.1038/ncomms7925
Kudela, R. M., and Dugdale, R. C. (2000). Nutrient regulation of phytoplankton productivity in Monterey Bay, California. Deep Sea Res. Part II Top. Stud. Oceanogr. 47, 1023–1053. doi: 10.1016/S0967-0645(99)00135-6
Lancien, M., Gadal, P., and Hodges, M. (2000). Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol. 123, 817–824. doi: 10.1104/pp.123.3.817
Levering, J., Broddrick, J., Dupont, C. L., Peers, G., Beeri, K., Mayers, J., et al. (2016). Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PLoS One 11:e0155038. doi: 10.1371/journal.pone.0155038
Levering, J., Dupont, C. L., Allen, A. E., Palsson, B. O., and Zengler, K. (2017). Integrated regulatory and metabolic networks of the marine diatom Phaeodactylum tricornutum predict the response to rising CO2 levels. mSystems 2:e00142-16. doi: 10.1128/mSystems.00142-16
Levitan, O., Dinamarca, J., Zelzion, E., Lun, D. S., Guerra, L. T., Kim, M. K., et al. (2015). Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc. Natl. Acad. Sci. U.S.A. 112, 412–417. doi: 10.1073/pnas.1419818112
Liang, Y., Kong, F., Torres-Romero, I., Burlacot, A., Cuine, S., Legeret, B., et al. (2019). Branched-chain amino acid catabolism impacts triacylglycerol homeostasis in Chlamydomonas reinhardtii. Plant Physiol. 179, 1502–1514. doi: 10.1104/pp.18.01584
Litwack, G. (ed.) (2018). “Metabolism of amino acids,” in Human Biochemistry, (Boston, MA: Academic Press), 359–394.
Matthijs, M., Fabris, M., Broos, S., Vyverman, W., and Goossens, A. (2016). Profiling of the early nitrogen stress response in the diatom Phaeodactylum tricornutum reveals a novel family of RING-domain transcription factors. Plant Physiol. 170, 489–498. doi: 10.1104/pp.15.01300
Matthijs, M., Fabris, M., Obata, T., Foubert, I., Franco-Zorrilla, J. M., Solano, R., et al. (2017). The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J. 36, 1559–1576. doi: 10.15252/embj.201696392
Mazelis, M. (1980). “15 - Amino acid catabolism,” in Amino Acids and Derivatives, ed. B. J. Miflin (Cambridge, MA: Academic Press), 541–567. doi: 10.1016/b978-0-12-675405-6.50021-8
Medlin, L., Kooistra, W., and Schmid, A. M. (2000). “A review of the evolution of the diatoms-a total approach using molecules, morphology and geology,” in The Origin and Early Evolution of the Diatoms: Fossil, Molecular and Biogeographical Approaches, eds A. Witkowski and J. Sieminska (Krakow: Polish Academy of Sciences), 13–35.
Moore, C. M., Mills, M. M., Arrigo, K. R., Berman-Frank, I., Bopp, L., Boyd, P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Morris, S. M. (2002). Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 22, 87–105. doi: 10.1146/annurev.nutr.22.110801.140547
Moustafa, A., Beszteri, B., Maier, U. G., Bowler, C., Valentin, K., and Bhattacharya, D. (2009). Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726. doi: 10.1126/science.1172983
Oliveira, I. C., Lam, H. M., Coschigano, K., Melo-Oliveira, R., and Coruzzi, G. (1997). Molecular-genetic dissection of ammonium assimilation in Arabidopsis thaliana. Plant Physiol. Biochem. 35, 185–198.
Palenik, B., and Morel, F. M. (1990). Comparison of cell-surface L-amino acid oxidases from several marine phytoplankton. Mar. Ecol. Prog. Ser. 59, 195–201. doi: 10.3354/meps059195
Pan, Y., Yang, J., Gong, Y., Li, X., and Hu, H. (2017). 3-Hydroxyisobutyryl-CoA hydrolase involved in isoleucine catabolism regulates triacylglycerol accumulation in Phaeodactylum tricornutum. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160409. doi: 10.1098/rstb.2016.0409
Polekhina, G., Board, P. G., Blackburn, A. C., and Parker, M. W. (2001). Crystal structure of maleylacetoacetate isomerase/glutathione transferase zeta reveals the molecular basis for its remarkable catalytic promiscuity. Biochemistry 40, 1567–1576. doi: 10.1021/bi002249z
Rébeillé, F., Jabrin, S., Bligny, R., Loizeau, K., Gambonnet, B., Van Wilder, V., et al. (2006). Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc. Natl. Acad. Sci. U.S.A. 103, 15687–15692. doi: 10.1073/pnas.0606195103
Rees, T. A. V., and Allison, V. J. (2006). Evidence for an extracellular L–amino acid oxidase in nitrogen-deprived Phaeodactylum tricornutum (Bacillariophyceae) and inhibition of enzyme activity by dissolved inorganic carbon. Phycologia 45, 337–342. doi: 10.2216/04-92.1
Reitzer, L. J. (1987). “Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, L-alanine and D-alanine,” in Escherichia Coli and Salmonella typhimurium. Cellular and Molecular Biology, Vol. 2, eds F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (Washington, DC: American Society for Microbiology), 302–320.
Remmers, I. M., D’Adamo, S., Martens, D. E., de Vos, R. C., Mumm, R., America, A. H., et al. (2018). Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res. 35, 33–49. doi: 10.1016/j.algal.2018.08.012
Ros, R., Muñoz-Bertomeu, J., and Krueger, S. (2014). Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci. 19, 564–569. doi: 10.1016/j.tplants.2014.06.003
Singh, D., Carlson, R., Fell, D., and Poolman, M. (2015). Modelling metabolism of the diatom Phaeodactylum tricornutum. Biochem. Soc. Trans. 43, 1182–1186. doi: 10.1042/BST20150152
Sipler, R. E., and Bronk, D. A. (2015). “Dynamics of dissolved organic nitrogen,” in Biogeochemistry of Marine Dissolved Organic Matter, ed. D. A. H. A. Carlson (Boston, MA: Academic Press), 127–232. doi: 10.1016/B978-0-12-405940-5.00004-2
Smith, S. (2018). Figshare: Nitrogen Gene Phylogenies. Available online at: https://doi.org/10.6084/m9.figshare.6233198 (accessed October 15, 2020).
Smith, S. R., Dupont, C. L., McCarthy, J. K., Broddrick, J. T., Oborník, M., Horák, A., et al. (2019). Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom. Nat. Commun. 10:4552. doi: 10.1038/s41467-019-12407-y
Stepansky, A., and Galili, G. (2003). Synthesis of the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase enzyme of lysine catabolism is concertedly regulated by metabolic and stress-associated signals. Plant Physiol. 133, 1407–1415. doi: 10.1104/pp.103.026294
Szabados, L., and Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Zhang, C., and Hu, H. (2014). High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genomics 16, 63–66. doi: 10.1016/j.margen.2013.10.003
Keywords: diatom, amino acid, catabolism, nitrogen, triacylglycerols
Citation: Pan Y, Hu F, Yu C, Li C, Huang T and Hu H (2020) Amino Acid Catabolism During Nitrogen Limitation in Phaeodactylum tricornutum. Front. Plant Sci. 11:589026. doi: 10.3389/fpls.2020.589026
Received: 07 August 2020; Accepted: 23 November 2020;
Published: 17 December 2020.
Edited by:
Miroslav Obornik, Institute of Parasitology, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Jeroen T. F. Gillard, California State University, Bakersfield, United StatesCopyright © 2020 Pan, Hu, Yu, Li, Huang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanhua Hu, aGFuaHVhaHVAaWhiLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.