- 1Department of Biotechnology, Guru Nanak Dev University, Amritsar, India
- 2Department of Bioinformatics, Hans Raj Mahila Maha Vidyalaya, Jalandhar, India
- 3William Harvey Heart Centre, Queen Mary University of London, London, United Kingdom
Cyclophilins constitute a family of ubiquitous proteins that bind cyclosporin A (CsA), an immunosuppressant drug. Several of these proteins possess peptidyl-prolyl cis-trans isomerase (PPIase) activity that catalyzes the cis-trans isomerization of the peptide bond preceding a proline residue, essential for correct folding of the proteins. Compared to prokaryotes and other eukaryotes studied until now, the cyclophilin gene families in plants exhibit considerable expansion. With few exceptions, the role of the majority of these proteins in plants is still a matter of conjecture. However, recent studies suggest that cyclophilins are highly versatile proteins with multiple functionalities, and regulate a plethora of growth and development processes in plants, ranging from hormone signaling to the stress response. The present review discusses the implications of cyclophilins in different facets of cellular processes, particularly in the context of plants, and provides a glimpse into the molecular mechanisms by which these proteins fine-tune the diverse physiological pathways.
Introduction
A peptide bond in a folded protein can attain either cis or trans conformation, with the latter being favored due to geometrical and thermodynamic parameters (Ramachandran and Sasisekharan, 1968). However, the peptide bond preceding a proline (Pro) residue tends to adopt the cis configuration since its cyclic five-membered ring imposes rigid constraints on rotation about the N-Cα bond (Schulz and Schirmer, 2013). Hence, about 10–15% of peptidyl-prolyl bonds tend to adopt the cis conformation (Brandts et al., 1975). The presence of cis-proline peptide bonds has many structural implications as these tend to introduce bends in a protein and decrease stability. Therefore, cis to trans isomerization of peptide bonds, a rate-limiting process, is essential for the proper folding of proteins. Peptidyl-prolyl cis-trans isomerases (PPIases) are the only enzymes known that can catalyze cis-trans transition (Fischer et al., 1989). Unlike chaperones which require energy, the PPIases are typical enzymes that follow the Michaelis-Menten kinetics (Schmid et al., 1993; Fanghänel and Fischer, 2004).
The PPIases belong to three major classes of proteins viz., cyclophilins, FK506-binding proteins or FKBPs, and parvulins. While cyclophilins bind cyclosporin A (CsA), FKBPs and parvulins show interaction with FK506 (tacrolimus)/rapamycin (sirolimus) and juglone (5-hydroxy-1, 4-naphthoquinone), respectively. CsA and FK506 and its structural analog, rapamycin, are immunosuppressive drugs that are used for preventing graft rejection after allogeneic transplants (Göthel and Marahiel, 1999). These drugs block T-cell activation by interfering with the signal transduction pathways (Schreiber, 1991). The target of CsA was first detected in the bovine thymus as an 18 kDa protein, while the receptor for FK506 was identified as a protein of 12 kDa which was later also shown to bind to rapamycin (Handschumacher et al., 1984; Harding et al., 1989; Siekierka et al., 1989). The parvulins (Latin: parvulus, very small) were first identified in E. coli as a protein of 92 amino acid residues (Rahfeld et al., 1994). The PPIase activity of these proteins is sensitive only to juglone and is not affected by either CsA or FK506. Though cyclophilins and FKBPs are collectively referred to as immunophilins (Schreiber, 1991), members of these families show characteristics and conserved sequence features that differ between the two classes (He et al., 2004). Two new classes of PPIases viz., FCBPs (FK506 and CsA-binding proteins) that contain both cyclophilin and FKBP domains (Adams et al., 2005), and Protein Phosphatase 2A Phosphatase Activator (PTPA; Jordens et al., 2006) have also been discovered. While the FCBPs have not been reported in plants (Geisler and Bailly, 2007; Barik, 2018), the PTPA orthologs, though encoded by the plant genomes, have not been characterized yet for their PPIase activity (Chen et al., 2014).
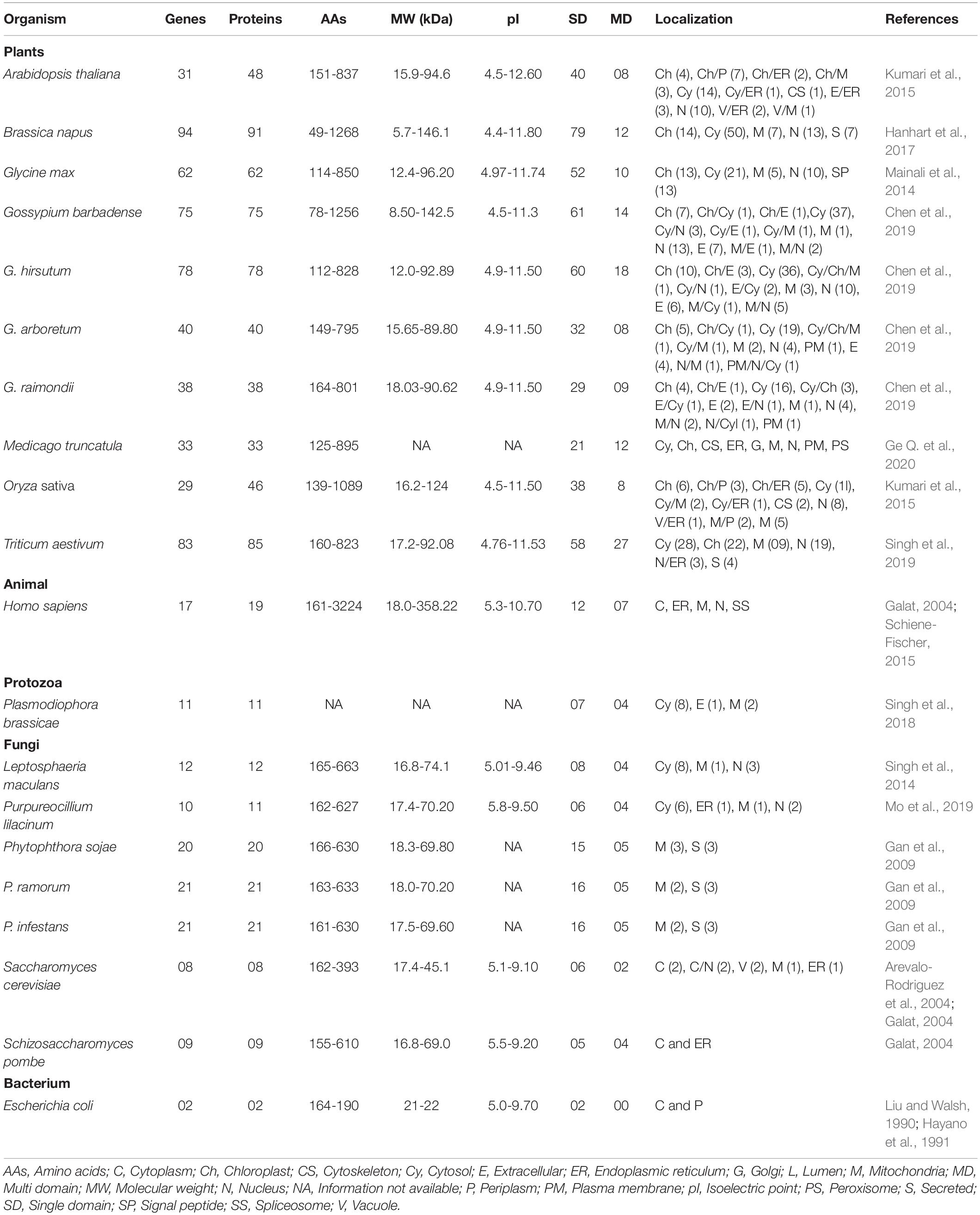
Cyclophilins are ubiquitous proteins and are present in a wide range of organisms including viruses, bacteria, fungi, mammals and plants (Galat, 2003; Thai et al., 2008). Besides PPIase activity, a few members of this family also demonstrate chaperone activity, implying their multifaceted properties (Rinfret et al., 1994; Mayr et al., 2000; Marín-Menéndez et al., 2012). Recent advances in genome and transcriptome sequencing have revealed that relative to other organisms, the cyclophilin gene families show dramatic expansion in plants. The smallest and largest cyclophilin families with 29 and 94 genes have been reported in Oryza sativa and Brassica napus, respectively (Table 1). These proteins exhibit intra- and inter-specific differences in size (5.7 – 358.22 kDa) and pI values (4.4 – 12.6) (Table 1), suggesting divergence in their roles (Galat, 2004; Pemberton and Kay, 2005; Singh et al., 2019). Although inter- and intra-specific diversity of cyclophilins in plants indicates that these proteins may be performing distinct cellular functions (Table 2), with few of the roles being species-specific, the physiological significance of the majority of these proteins in plants is still a matter of conjecture. In the present article, we have attempted to summarize the different structural and functional aspects of cyclophilins in plants and their likely implications in different facets of growth and development.
Structural Analyses of Cyclophilin Genes and Proteins in Plants
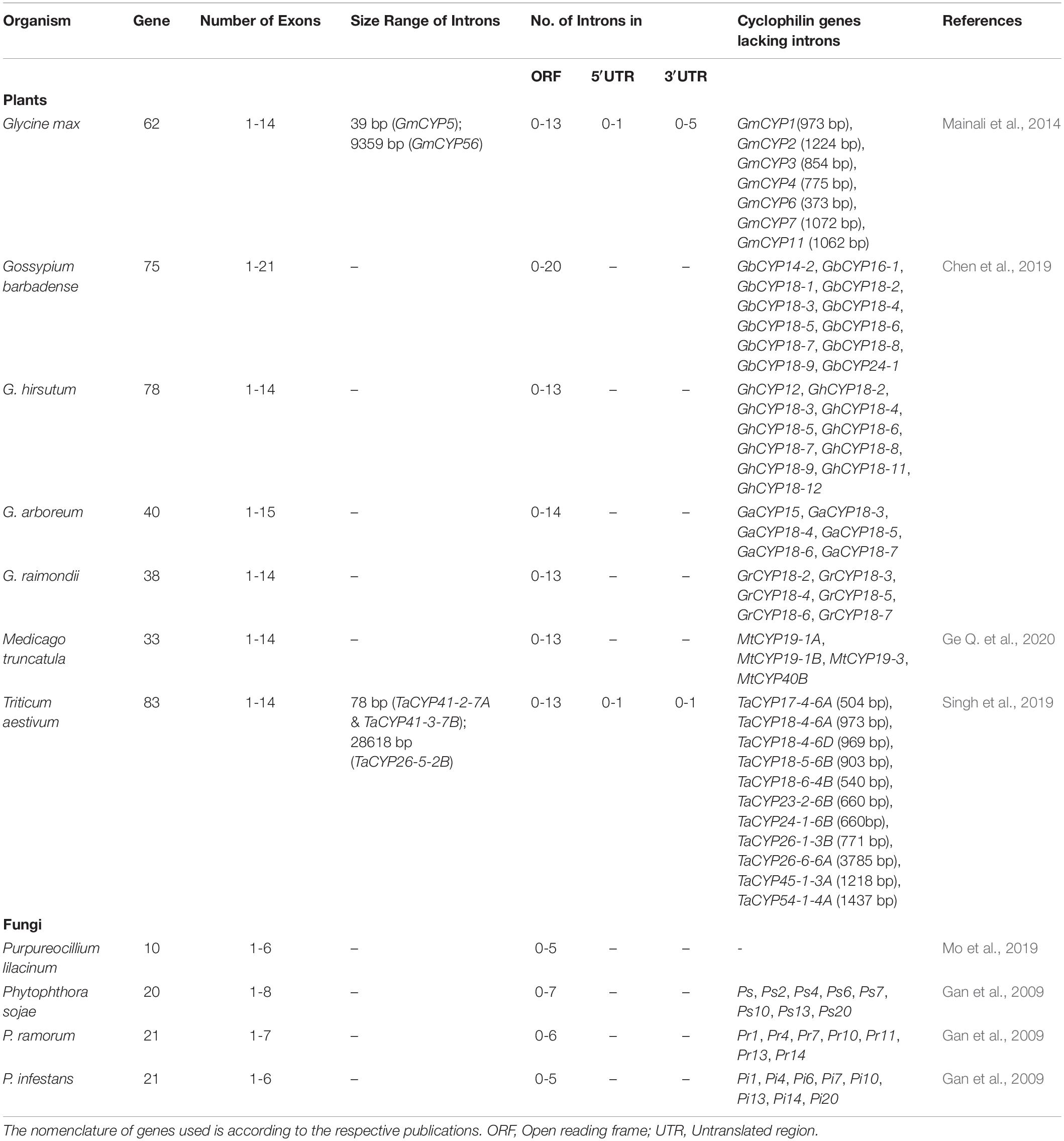
Genome-wide analyses revealed that the distribution of cyclophilin genes on different chromosomes in plants is uneven (Table 3). The cyclophilin genes in allopolyploids such as B. napus and wheat occur in pairs, with each member originating from one progenitor chromosomal set. These pairs are highly identical and share localization patterns (Hanhart et al., 2017; Singh et al., 2019). Structural analysis of cyclophilin genes in plants has been carried out for soybean, cotton, wheat and Medicago truncatula (Mainali et al., 2014; Chen et al., 2019; Singh et al., 2019; Ge L. et al., 2020). These studies revealed considerable variability in the distribution and size of introns in the open reading frames (ORFs) and untranslated regions (UTRs) as compared to other organisms (Table 4). The cyclophilin genes with the highest number of introns include cotton (20 in GbCYP142; Chen et al., 2019), wheat (13 each in TaCYP64-1-7A, TaCYP64-2-7B, and TaCYP64-3-7D; Singh et al., 2019) and soybean (13 each in GmCYP56 and GmCYP59; Mainali et al., 2014). The largest intron (28618 bp) was observed in TaCYP26-5-2B, while the smallest (39 bp) was noticed in GmCYP5 (Table 4). Information about variations in the structure of cyclophilin genes in rice, Arabidopsis and Brassica, which is lacking, may provide further insights into the evolution of these families in plants. Loss or gain of introns, an important aspect of structural variation, is vital for gene evolution (Roy and Gilbert, 2006). The intron size may be correlated with the genome size and longer introns have been proposed to confer a selective advantage by improving the recombination, and also by counterbalancing the mutational bias towards deletions (Carvalho and Clark, 1999; McLysaght et al., 2000). Thus, the variability in introns in plant cyclophilins may have important implications in their functionalization which needs to be explored further. Since 5′ and 3′ UTRs are structurally important and regulate the expression of eukaryotic genes (Wilkie et al., 2003), differences in these regions may likely enable differential regulation of plant cyclophilins, leading to divergence in their physiological roles.
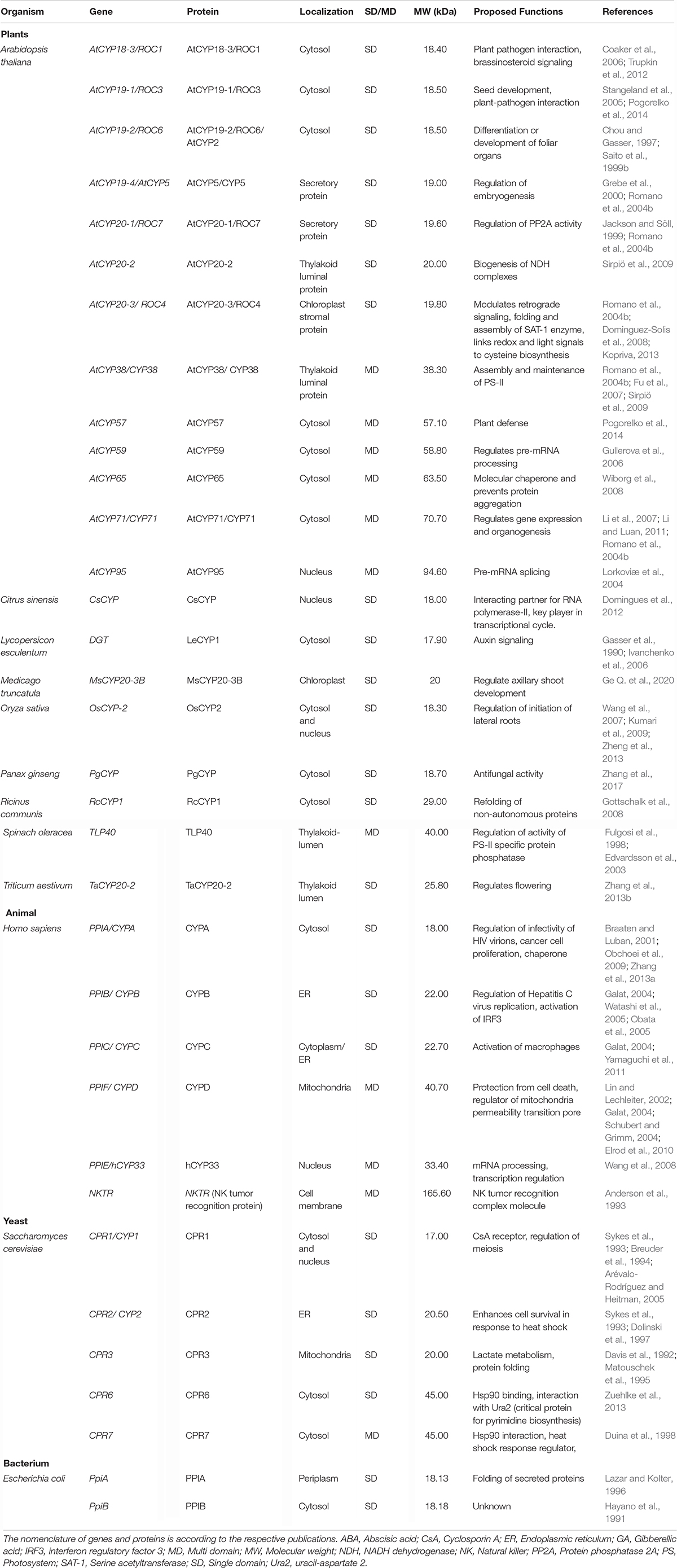
The cyclophilins in plants and other organisms, though predominantly cytosolic, are also predicted to be localized in the chloroplast, nucleus, mitochondria, extracellular/secretory and plasma membrane (Table 1). The presence of cyclophilins in different organelles of plants signifies their specific and distinct roles in the cell (Tables 1, 2). Based on domain organization, the cyclophilins are classified as single- (SD) or multi-domain (MD) forms (Table 1). The SD cyclophilins possess the characteristic cyclophilin-like domain (CLD), while the MD cyclophilins also contain additional specific functional domains (Table 5). Analysis of CLD in the typical human cyclophilin, hCYPA (hCYP18-A/CYPA), demonstrated that the residues Arg55, Phe60, Met61, Glu63, Ala101, Phe113, Trp121, Leu122 and His126 are essential for PPIase activity (Zydowsky et al., 1992b; Ke et al., 1994; Zhao et al., 1997; Howard et al., 2003; Davis et al., 2010). Arg55, in particular, plays a critical role in PPIase functions, whereas Trp121, though not involved in cis-trans isomerization, is essential for CsA binding (Liu et al., 1991; Zydowsky et al., 1992b; Howard et al., 2003). Interestingly, in the plant MD cyclophilins, the TPR and WD40 repeats are observed more commonly compared to other domains (Table 5). The domains such as TPR, WD40, F-box, coiled-coil, etc., have been reported to facilitate protein-protein interactions in the cell (Lamb et al., 1995; Craig and Tyers, 1999; Van Nocker and Ludwig, 2003; Liu et al., 2006). Hence, the cyclophilins consisting of these motifs may be acting as platforms for assembling protein complexes or mediate transient interactions among other proteins, further indicating their functional versatility (Bandziulis et al., 1989; Van Nocker and Ludwig, 2003; Stirnimann et al., 2010; Earley and Poethig, 2011). Compared with yeast and human cyclophilins, the presence of various additional domains such as PsbQ-like, F-box, Helical bundle, ATPase and PAN_4 domain in the plant MD cyclophilins (Figure 1 and Table 5) signifies divergence of their roles that are yet to be explored completely (Dornan et al., 2003; Romano et al., 2004b; Mainali et al., 2014; Kumari et al., 2015; Hanhart et al., 2017; Chen et al., 2019; Singh et al., 2019).
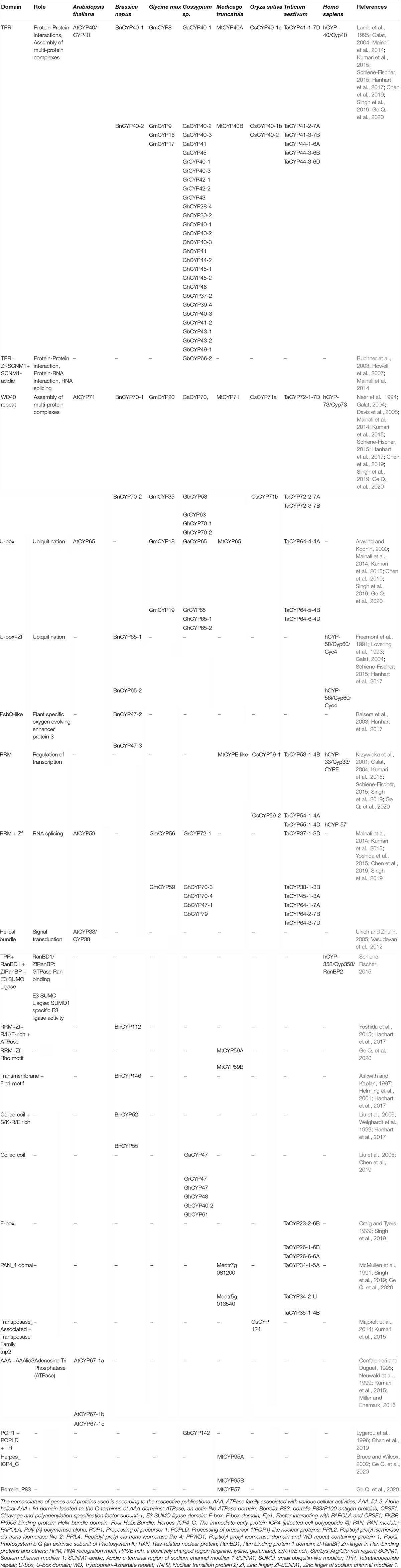
Table 5. Comparative analysis of functional domains (other than cyclophilin-like domain) in the different multi-domain cyclophilins.
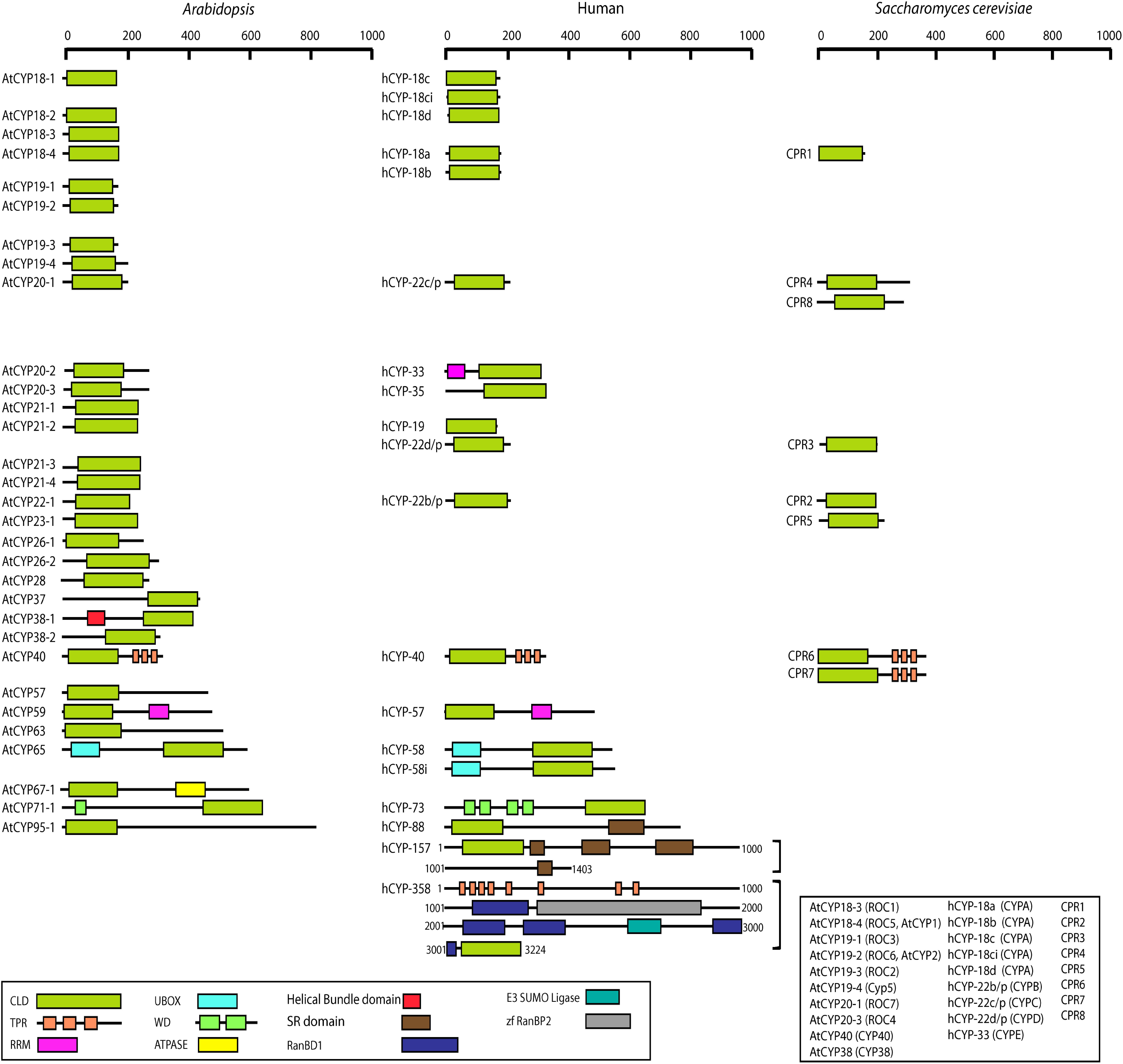
Figure 1. Comparative analysis of domain architecture of Arabidopsis cyclophilins with their orthologs in human and Saccharomyces cerevisiae. The amino acid residues that define the protein domains are designated according to Galat (2004), Wang and Heitman (2005), Kumari et al. (2015), and Schiene-Fischer (2015). For Arabidopsis cyclophilins that may have alternatively spliced forms, the domain architecture is shown for only a single variant. CLD, cyclophilin-like domain; RRM, RNA recognition motif; TPR, tetratricopeptide repeat; U-box, U box domain; WD40, WD40 repeat; RanBDl, Ran binding protein 1 domain; zf RanBP, Zn-finger, Ran-binding; SR, Serine arginine rich domain. The nomenclature and alternative protein names are given in the box. Scale bar represents the length of amino acid sequence.
So far, only five different plant cyclophilins viz., TaCYPA-1 (Sekhon et al., 2013), CsCYP (Campos et al., 2013), Catharanthus roseus Cat r 1 (Ghosh et al., 2014), BnCYP19-1 (Hanhart et al., 2019) and AtCYP38 or CYP38 (Vasudevan et al., 2012) have been characterized for their crystal structures. While the former four are single-domain proteins and show PPIase activity, the AtCYP38 is a MD cyclophilin that lacks cis-trans isomerization capability (Vasudevan et al., 2012). The crystal structures of TaCYPA-1, CsCYP, BnCYP19-1 and CLD of AtCYP38 are similar to “archetypal” human cyclophilin hCYPA, and consist of eight-stranded antiparallel β-barrel capped at either end by two α-helices (Vasudevan et al., 2012; Campos et al., 2013; Sekhon et al., 2013; Hanhart et al., 2019). However, Cat r 1 (PDB: 2MC9) shows variability in its structure since the β-barrel in this protein consists of seven antiparallel β-strands instead of eight (Ghosh et al., 2014). The CsA-binding site in hCYPA and other such cyclophilins is composed of seven aromatic and other hydrophobic residues that constitute the hydrophobic core within the barrel (Kallen et al., 1991). The topology of this β-barrel structure is unique in the sense that it remains occupied with a set of closely packed aromatic groups making no room for binding of either CsA or the Pro containing peptides (Ke, 1992). Therefore, the CsA and other substrates bind to an active site that is formed by amino acid residues located on the outer face of the β sheet. The active sites consist of 13 residues which are identical in CsCYP, TaCYPA-1, BnCYP19-1 and hCYPA (Ke et al., 1994; Campos et al., 2013; Sekhon et al., 2013; Hanhart et al., 2019). However, the electrostatic surface map studies indicated that despite conservation of all the 13 active site residues, the active site pocket in Cat r 1 appears to be slightly broader and is more acidic in nature, which might be imparting precision for binding of peptides with a specific amino acid composition (Ghosh et al., 2014). While the conservation of CLD structure in cyclophilins underlines its fundamental role in the cell, the remarkable diversity in their domain architecture could have subtle or profound effects on the structure of these proteins which may, in turn, affect their biochemical activities differently, enabling them to perform a wide variety of roles in different cellular processes. Elucidation of crystal structures of different cyclophilins and identification of their interacting proteins is, thus, imperative to gain further insights into their specific functions.
Regulation of PPIase Activity of Cyclophilins
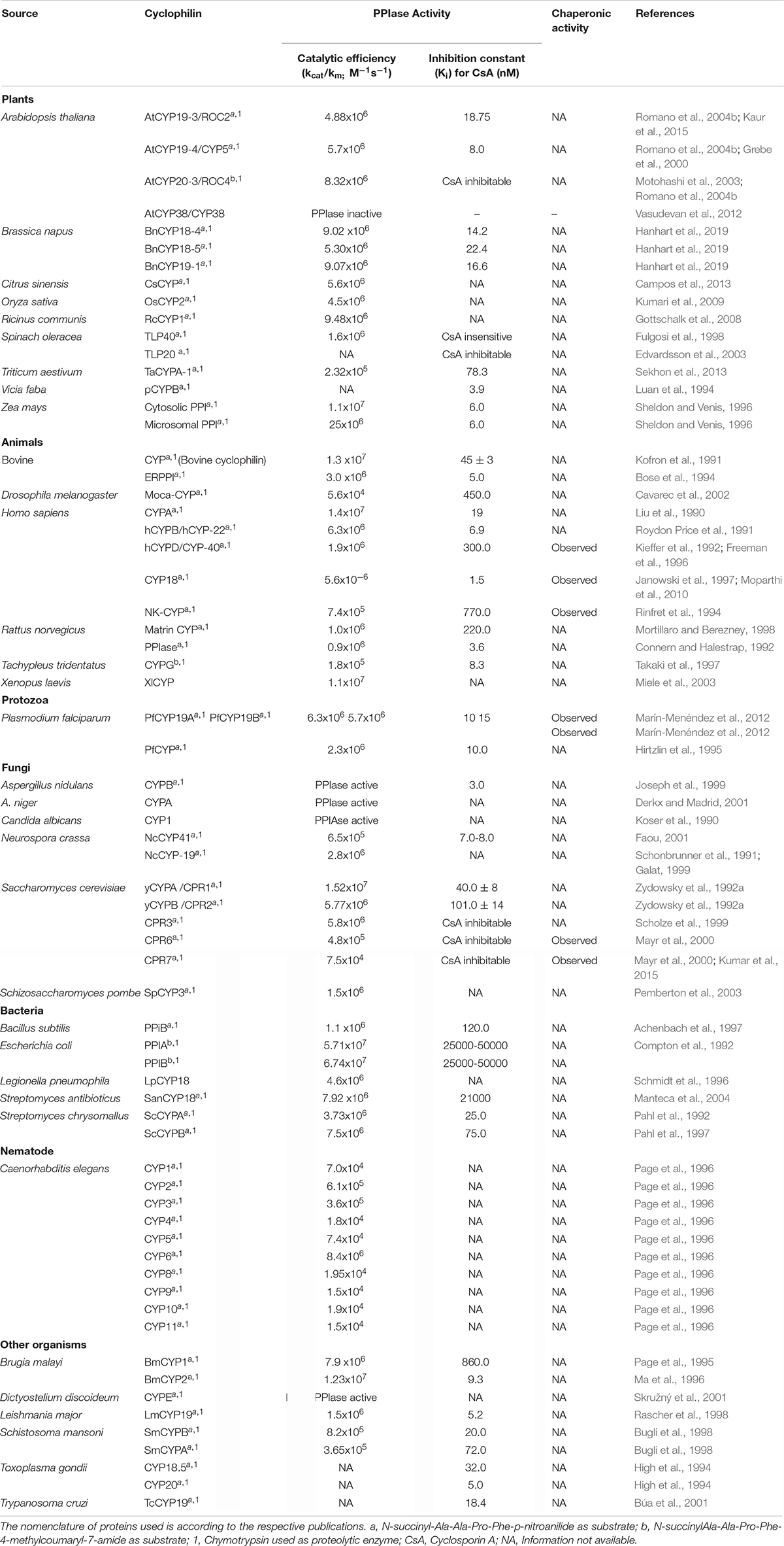
The PPIase activity of immunophilins is assayed by several in vitro methods viz., isomer-specific cleavage of the peptide with chymotrypsin, protease-free assay, NMR-based methods, protein folding/unfolding and fluorescence-based assays (Fischer et al., 1984; Janowski et al., 1997; Davis et al., 2010). The recent development of an in vivo method provides a useful tool to study the regulation of PPIase activity by temporal, spatial and environmental factors in the living cells (Jiang et al., 2018). Cyclophilins have been characterized biochemically from several organisms (Table 6), some of which were reviewed earlier (Fanghänel and Fischer, 2004). As observed for cyclophilins in other organisms, the plant cyclophilins also exhibit variability in their kinetic parameters and sensitivity to CsA (Table 6). Whereas the catalytic constants (kcat/km) of the different plant cyclophilins reported until now vary between 105 to 107 M –1s–1 for the suc-AAPF-pNA oligopeptide substrate, the inhibition constants for CsA range between 6.0 (ZmCYP18) to 78.3 nM (TaCYPA-1). The implications of diversity in biochemical attributes of cyclophilins in modulating the physiological response in plants are not understood and need to be investigated by overexpressing mutant cyclophilins that exhibit graded cis-trans isomerization capabilities.
Contingent upon the presence of an extra loop of four or more amino acid residues present at residue 50 corresponding to hCYPA, the cyclophilins are classified as divergent or non-divergent (Dornan et al., 1999). The divergent loop cyclophilins such as TaCYPA-1 (Sekhon et al., 2013), CsCYP (Campos et al., 2013) and Cat r 1 (Ghosh et al., 2014) are similar to hCYPA in their active site composition and CsA binding characteristics except for the presence of a characteristic additional loop (consensus sequence XXGKXLH corresponding to amino acid residues 48–54 in TaCYPA-1), two conserved Cys residues (Cys40 and Cys168) and a conserved glutamate (Glu83) residue (Kaur et al., 2015; Vasudevan et al., 2015). On the contrary, the non-divergent cyclophilins such as hCYPA, SmCYPA and AtCYP20-3 or ROC4 (Rotamase Cyclophilin 4) lack the additional loop and are characterized by two conserved Cys residues at positions 122 and 126 (Gourlay et al., 2007; Laxa et al., 2007). AtCYP38, however, is a unique kind of non-divergent cyclophilin since it lacks both the characteristic divergent loop as well as the Cys amino acids observed in other plant non-divergent cyclophilins (Vasudevan et al., 2012).
The PPIase activity of cyclophilins, in general, is regulated in a redox-dependent or independent manner. Contrary to the E. coli cyclophilin PPIB, that is regulated by redox-independent mechanisms (Hayano et al., 1991; Kaur et al., 2015), the PPIase activity of AtCYP19-3 (ROC2), AtCYP20-3, SmCYPA, CsCYP and TaCYPA-1 is subject to redox regulation (Motohashi et al., 2003; Gourlay et al., 2007; Laxa et al., 2007; Campos et al., 2013; Kaur et al., 2015, 2017). Furthermore, the redox-regulatory mechanisms observed in different cyclophilins are also distinct. For instance, the regulation of non-divergent cyclophilins hCYPA and AtCYP20-3 involves glutathionylation and thioredoxin-mediated thiol-disulfide exchange, respectively. Whereas glutathionylation of Cys residues in hCYPA renders the protein inactive under oxidative conditions, deglutathionylation through reduction of thiol groups by intracellular pH changes or in response to reducing environment restores its activity (Ghezzi et al., 2006; Townsend, 2007; Dalle-Donne et al., 2009). On the contrary, the activity of AtCYP20-3 is modulated by thioredoxin (Trx)-mediated thiol-disulphide exchange (Motohashi et al., 2003; Laxa et al., 2007). Under oxidizing conditions, the formation of two disulphide pairs in AtCYP20-3 (Cys53-Cys70 and Cys128-Cys175) abrogates the PPIase activity, while Trx-mediated reduction results in restoration of the catalytic function.
Regulation of another non-divergent cyclophilin SmCYPA from Schistosoma mansoni is attributed to oxidation-induced disulfide bond formation between Cys122 and Cys126 that results in loss of activity (Gourlay et al., 2007). On the contrary, the regulation of a divergent cyclophilin from Citrus sinensis, CsCYP, involves both disulphide bond formation between Cys40 and Cys168 as well as loop displacement (Campos et al., 2013). Our earlier studies revealed that the wheat divergent cyclophilin, TaCYPA-1, has an additional Cys126 residue corresponding to the residue 126 in non-divergent SmCYP (Gourlay et al., 2007; Kaur et al., 2015). Site-directed mutagenesis studies provided evidence that PPIase activity of TaCYPA-1 is regulated through a dual mechanism involving loop displacement (Kaur et al., 2017), as observed in the divergent cyclophilin CsCYP (Campos et al., 2013), and also by the interaction between Cys122 and Cys126, as reported for the non-divergent SmCYPA (Supplementary Figure 1; Gourlay et al., 2007), with the latter mechanism playing a predominant role (Kaur et al., 2017). These observations make TaCYPA-1 unique since despite being a divergent cyclophilin its activity is also subject to regulation by mechanisms that are more common to the non-divergent cyclophilins. In silico studies in our lab revealed that several other wheat cyclophilins may also follow similar regulation (Singh et al., 2019), the significance of which is not understood yet. It is evident that despite the conservation of active sites in cyclophilins, distinct regulatory mechanisms have evolved for the regulation of these proteins, possibly to impart versatility to these proteins to regulate diverse cellular processes. However, the physiological implication of different regulatory mechanisms of cyclophilins in plants is a matter of conjecture and merits further investigations.
Cyclophilins as Protein Folding Catalysts
Evidence for in vivo role of cyclophilins in protein folding was first provided by analysis of Drosophila melanogaster ninaA (Neither inactivation nor after potential protein A) protein, which is a tissue-specific integral membrane protein required for the proper synthesis of the visual pigment rhodopsin 1 (Rh1; Stamnes et al., 1991). In D. melanogaster, Rh1 is synthesized in the ER and is transported to rhabdomeres via the secretory pathway where it performs phototransduction. Mutation in ninaA blocks this transportation and results in accumulation of rhodopsin in the ER, leading to its degradation and consequently impaired visual function (Colley et al., 1991). The CPR3 in yeast also catalyzes protein folding in vivo, as isolated mitochondria from Δcpr3 (yeast strain mutated in CPR3 gene) showed a reduced rate of protein folding (Matouschek et al., 1995). The chaperonic function of an Arabidopsis cyclophilin AtCYP40 (CYP40) was shown to be independent of PPIase activity since the enzymatically inactive mutants of AtCYP40 were able to facilitate the assembly of RNA induced silencing complex (RISC; Iki et al., 2012). Evidence for the chaperonic role of RcCYP1, a highly active PPIase abundant in companion cell sieve element complex of Ricinus communis, was provided by microinjection studies (Gottschalk et al., 2008). These authors observed that RcCYP1 is involved in auto-cell to cell trafficking via interaction with plasmodesmata special proteins and performs unique functions by assisting their refolding. Studies carried out in our laboratory demonstrated that PPIase activity in the wheat grains is associated with the deposition of grain storage proteins or prolamines (Dutta et al., 2011). Since prolamines are rich in prolyl residues (10–15%; Shewry et al., 2002), the PPIases might be involved in the folding of these proteins. Plants have diverse cyclophilins, but information on biochemical properties and chaperonic activities of these proteins is rather scarce. Therefore, molecular analysis and biochemical characterization of different cyclophilins in plants are imperative for gaining insights into their physiological roles which might further lead to the development of crops with improved agronomic traits.
Roles of Cyclophilins in Chloroplast
The CsA-sensitive PPIase activity in chloroplasts was first demonstrated in pea by Breiman et al. (1992). Since the characterization of TLP40, a 40 kDa thylakoid lumen cyclophilin from spinach chloroplasts (Fulgosi et al., 1998), proteomics and bioinformatics approaches resulted in the identification of 11 FKBPs and 5 cyclophilins in the chloroplast lumen of Arabidopsis (Edvardsson et al., 2007; Trivedi et al., 2012). TLP40 is a multi-domain cyclophilin that shows PPIase activity and acts as a negative regulator of the thylakoid membrane protein phosphatase (Fulgosi et al., 1998; Vener et al., 1999). This protein plays an essential role in the growth and development of plants since mutations in its Arabidopsis ortholog, AtCYP38, resulted in impaired development of chloroplasts, retarded plant growth, hypersensitivity to light, and enhanced degradation of D1 and D2 components of PSII under high light conditions (Fu et al., 2007; Sirpiö et al., 2008; Vasudevan et al., 2012; Vojta et al., 2019). Together with other immunophilins such as FKBP13 and FKBP20-2, that are required for accumulation of the cytochrome b6f complex and PSII supercomplexes, respectively (Gupta et al., 2002; Lima et al., 2006), AtCYP38, despite lacking PPIase activity, appears to be indispensable for proper biogenesis and maintenance of photosynthetic complexes. On the contrary, impaired functioning of AtCYP20-2, a highly active PPIase and orthologous to the spinach cyclophilin TLP20, had no apparent phenotypic effect, suggesting redundancy in the function of these proteins (Fulgosi et al., 1998; Sirpiö et al., 2009). It has been proposed that while TLP40 performs specialized regulatory function(s), TLP20 might act as a general protein folding catalyst (Edvardsson et al., 2003). The chloroplast stromal protein AtCYP20-3, 65.64 % identical to AtCYP20-2, facilitates the folding of serine acetyltransferase (SAT) that catalyzes the ultimate step in Cys biosynthesis which is important for glutathione formation. The PPIase and folding activities of AtCYP20-3, sensitive to photooxidation and stress-induced ROS, were restored following reduction by photoreduced Trx (Laxa et al., 2007). Mutation in AtCYP20-3 resulted in hypersensitivity to oxidative stress in Arabidopsis (Dominguez-Solis et al., 2008), implying that it enables the Cys-based thiol biosynthesis pathway to adjust to light and stress conditions. Isothermal titration microcalorimetry and gel overlay assays further indicated that AtCYP20-3 interacts with thiol based peroxidases, 2-Cysteine peroxiredoxins (2-CysPrx), which can exist as either dimer or decamer. The dimer form is favored under oxidizing conditions whereas the decamer is formed under reducing conditions. High affinity of AtCYP20-3 for the dimer leads to a decrease in the free dimer concentration. Thus it appears that AtCYP20-3 regulates the critical transition concentration (concentration responsible for dimer-decameric form transition) value of 2-CysPrx, suggesting redox-dependent conformational dynamics of this protein (Liebthal et al., 2016).
Roles of Cyclophilins in Growth and Development of Plants
Various studies have substantiated the role of cyclophilins in the regulation of different aspects of plant growth and development. Whereas, a CsA-inhibitable PPIase in Arabidopsis, AtCYP19-4 (CYP5), was proposed to determine cell-polarity and regulate embryogenesis, the cytosolic SD cyclophilin AtCYP19-1 (ROC3) was implicated in seed development (Grebe et al., 2000; Stangeland et al., 2005). Cyclophilins also appear to affect organogenesis in Arabidopsis since the loss of function of a nuclear-localized MD protein, AtCYP71, resulted in compromised lateral organ formation and apical meristem activity (Li et al., 2007). Chromatin remodeling and transcriptional regulation were proposed as the likely mechanisms of action for AtCYP71 because this protein exhibited interaction with FAS1 (a subunit of Chromatin Assembly factor-1) and LHP1 (a heterochromatin protein) (Li et al., 2007; Li and Luan, 2011).
Another cytosolic cyclophilin, AtCYP40, was identified as a regulator of vegetative growth in Arabidopsis. Mutation (sqn) in this gene (SQUINT) resulted in a decrease in the number of juvenile leaves (Berardini et al., 2001). The mutated plants exhibited attenuated ARGONAUTE1 (AGO1) function that decreased the miRNA activity, resulting in enhanced expression of miR156-sensitive squamosa promoter binding protein-like family (SPL) of transcription factors (Smith et al., 2009). Even though reproductive maturation was not affected in the sqn mutants, later studies revealed that AtCYP40, along with REBELOTE (RBL; protein of unknown function) and ULTRAPET ALA (ULT1; a putative transcription factor), is important for floral developmental homeostasis (Prunet et al., 2008). AtCYP40 is a multidomain cyclophilin and contains TPR domain at its C-terminus which mediates its interaction with cytoplasmic HSP90, a feature also conserved for its orthologs in animals and S. cerevisiae (Berardini et al., 2001; Wandinger et al., 2008; Earley and Poethig, 2011; Blackburn et al., 2015). AtCYP40 facilitates the formation of miRISC assembly by mediating the interaction of HSP90-AGO1 complex with a small RNA duplex that leads to the formation of mature RISC. Though the interaction of AtCYP40 with HSP90-AGO 1 complex, imperative for RISC assembly, is sensitive to CsA, the role of PPIase activity in this process is still elusive (Iki et al., 2012).
Recent studies have demonstrated that regulation of growth and development in plants by cyclophilins may also be isoform-dependent (Jung et al., 2020). The Golgi-localized cyclophilin in rice, OsCYP21, exists in four different isoforms viz., OsCYP21-1, OsCYP21-2, OsCYP21-3 and OsCYP21-4. Despite the conservation of active site residues, these isoforms differ in their activity. While OsCYP21-1 and OsCYP21-2 are enzymatically active, the latter two lack PPIase activity. The isoforms OsCYP21-1 and OsCYP21-2 were implicated in the regulation of growth and development through modulation of ABA pathway genes. The significance of PPIase activity in this role needs to be corroborated by generating plants with mutated OsCYP21-1 and OsCYP21-2 that are deficient in PPIase function. Thus, it is evident that the regulation of various facets of growth and development by different cyclophilins entails distinct mechanisms that further signifies their functional versatility.
Implications of Cyclophilins in Hormone Signaling
Recent studies have provided evidence for the involvement of cyclophilins in several hormone-mediated responses in plants. Brassinosteroids and gibberellic acid (GA) are key regulators of plant stem elongation, and defects in the biosynthetic or signaling pathways of these hormones result in dwarf phenotype (Wang and Li, 2008). Genes contributing to dwarfness are of agronomic importance due to their potential for developing crops that are resistant to lodging under water-logging and strong wind conditions. DELLA proteins (named after conserved N-terminal D-E-L-L-A amino acid sequence) are inhibitors of stem growth and have been implicated in dwarf phenotype in Arabidopsis, B. napus and peach (Lawit et al., 2010; Zhao et al., 2017; Cheng et al., 2019). GA degrades DELLA proteins via the ubiquitin-proteasome pathway to promote stem growth (Sun, 2008, 2010). Mutations in the DELLA domain that abrogate interaction with F-box containing proteins SLY1, GID1 and GID2 prevent their GA-dependent degradation (Dill et al., 2004; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Lou et al., 2016). Functional impairment of DELLA proteins was reported to result in the dominant GA-insensitive dwarf phenotype (gaid) in wheat and B. rapa (Brrga1-d) (Ho et al., 1981; Muangprom et al., 2005). The gaid phenotype in wheat was also associated with higher levels of a 20 kDa cyclophilin, TaCYP20-2, overexpression of which in the wild-type wheat lead to gaid-like phenotype (Li et al., 2010), implying that this protein plays an essential role in maintaining GA homoeostasis by regulating the DELLA proteins. However, elucidation of the precise mechanism of action requires further intense experimentations.
The inhibition of hypocotyl growth and the expansion of cotyledons by light after the emergence of shoot from the soil in Arabidopsis is regulated by the photoreceptors phytochromes (PHYA to PHYE) and cryptochromes (CRY1 and CRY2) (Cashmore et al., 1999; Quail, 2005). Screening of the transgenic Arabidopsis 35S-cDNA lines for defective de-etiolation under a combination of blue and far-red light resulted in the isolation of a mutant (roc1-1D) that depicted enhanced expression of a cytoplasmic cyclophilin, AtCYP18-3 (ROC1, Rotamase Cyclophilin 1). The roc1-1D plants exhibited long hypocotyls and poorly unfolded cotyledons under blue and far-red light, and lower anthocyanin under far-red or blue light (Trupkin et al., 2012). Further analysis revealed that the mutant plants were hypersensitive to brassinosteroids in light but not in the dark. Inhibition of brassinosteroid synthesis and mutations in the genes responsible for brassinosteroid signaling abolished the mutant phenotype, implying that AtCYP18-3 links cryptochrome and phytochrome to brassinosteroid sensitivity (Trupkin et al., 2012).
Subsequent studies also provided evidence that functionality of AtCYP18-3 is highly sensitive to single amino acid substitution, since plants which over-expressed its variant containing phenylalanine instead of serine at position 58 exhibited reduced height, increase in shoot branching and higher sensitivity to photoperiod and temperature (Ma et al., 2013). The wild type AtCYP18-3 though does not appear to control stem elongation, likely conformation changes due to amino acid substitution might have resulted in the identification of new targets, thereby, affecting the stem growth. Therefore, structural analysis and identification of interacting proteins are imperative to understand the molecular mechanisms by which the mutated AtCYP18-3 controls growth and development in plants. Further, whether the mutated AtCYP18-3 can facilitate cross-talk between brassinosteroid signaling and photoreceptors is also a subject of future studies.
Besides brassinosteroid and GA signaling, cyclophilins have also been demonstrated to mediate auxin response. At low levels of auxin, the expression of auxin-responsive genes is kept in check by the unstable transcriptional repressors Aux/IAA proteins that bind to and inhibit the activity of auxin response factors (ARFs), a family of transcriptional activators (Figure 2; Theologis et al., 1985; Ainley et al., 1988; Conner et al., 1990; Yamamoto et al., 1992; Guilfoyle et al., 1993; Abel et al., 1995). The Aux/IAA genes are also induced by IAA and control the auxin response through a negative feedback loop (Reed, 2001). The Aux/IAA proteins consist of four highly conserved domains I-IV and bind to the ARFs either directly or through recruitment of transcriptional corepressor such as TOPLESS (TPL), the interactions being mediated by domain I that contains Leu-rich motif (Tiwari et al., 2004; Szemenyei et al., 2008). At high levels, the auxin binds to its receptor TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEINS (TIR1/AFBs), an F-box containing protein, and the auxin-responsive genes are activated through auxin-dependent proteasomal degradation of Aux/IAA proteins that require ubiquitination (Wang and Estelle, 2014). The ubiquitination of proteins is catalyzed by a cascade of three enzymes viz., the Ub-activating enzyme (E1), the Ub-conjugating enzyme (E2) and the Ub-protein ligase (E3). The SCF (Skp1-Cul1-F box) E3, one of the four different types of E3s described in plants, is a complex of four different polypeptides viz., SKP1 (a member of an ASK family in plants), CDC53 or Cullin (Cul1), an F Box protein and RBX. The Cul1 acts as a central scaffold protein, while the SKP1 interacts with the F-box protein that further binds to the substrate proteins (Smalle and Vierstra, 2004). Transfer of Ub from Ub-E2 to the substrate protein is catalyzed by the fourth subunit (RBX1, ROC, or Hrt1) of the SCF complex (Petroski and Deshaies, 2005). The TIR1 interacts with SKP1 to form the SCFTIR1 complex (Ruegger et al., 1998; Gray et al., 1999). Auxin acts as a molecular glue and after binding to TIR1, it enhances the interaction of the latter with the highly conserved ‘degron’ motif GWPPV in domain II of Aux/IAAs, leading to ubiquitination and proteolytic degradation of the latter (Figure 2; Gray et al., 2001; Reed, 2001; Tan et al., 2007). The Aux/IAA proteins bind to SCFTIR1-Auxin complex only when the ‘degron’ motif GWPPV is in the cis W-P isomer (Tan et al., 2007; Acevedo et al., 2019). Recent studies have provided insights into the implications of cyclophilin-associated PPIase activity in mediating the interaction of Aux/IAA with the SCFTIR1-Auxin complex. The LATERAL ROOTLESS 2 (LRT2) in rice encodes a cyclophilin PPIase OsCYP2, and disruption of this gene leads to an auxin-resistant phenotype and defective development of lateral roots (Kang et al., 2013; Zheng et al., 2013). The OsCYP2 was demonstrated to physically interact with the rice OsAux/IAA and TIR proteins, and catalyze the cis-trans isomerization of the OsIAA11 degron motif (Jing et al., 2015). These findings, thus, imply that the equilibrium of cis to trans populations of Aux/IAA proteins acts as a molecular timer to regulate auxin signal transduction (Acevedo et al., 2019). Since transcription of genes responsive to jasmonic acid, GA and strigolactone is also dependent on proteasome-mediated degradation of their specific repressors, the involvement of PPIases in controlling regulatory circuits of other hormones cannot be ruled out and should be the subject of future studies.
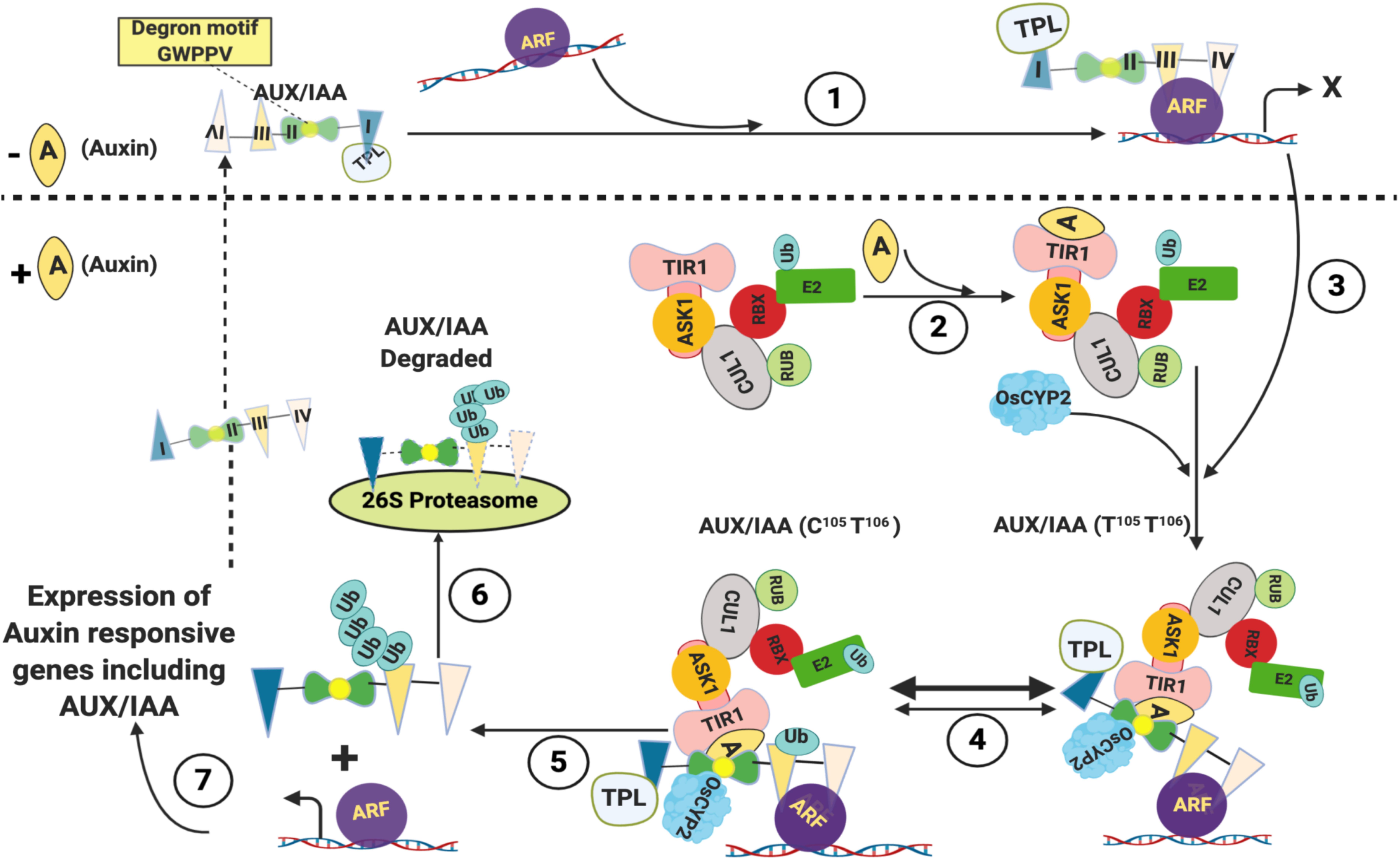
Figure 2. Role of cyclophilins in regulation of Auxin-responsive genes. At low levels of Auxin, Aux/IAA proteins bind to auxin response factors (ARFs) directly or through recruitment of transcriptional corepressor such as TOPLESS (TPL) and inhibit their activity (1). When present at high levels, the auxin binds to its receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1) and enhances its interaction with the highly conserved ‘degron’ motif GWPPV in domain II of Aux/IAAs (2 and 3). The Aux/IAA proteins bind to SCFTIR1-Auxin complex only when W104-P105 isomer in the ‘degron’ motif GWPPV (residues 103, 104, 105, 106 and 107, respectively, in rice) is in cis conformation. The trans conformer of W-P (T105-T106) in the ‘degron’ motif is catalyzed to cis form (c105-T106) by OsCYP2 (4). The Aux/IAA-SCFTIR1 complex leads to ubiquitination of Aux/IAA proteins (5), which are then degraded by 26S Proteasome (6), leading to expression of Auxin responsive genes including Aux/IAA (7) (Adapted from Tan et al., 2007; Mockaitis and Estelle, 2008; Jing et al., 2015; Created with BioRender.com).
Given the diversity of PPIases in plants, it is likely that parallel regulatory mechanisms may be operating for several other processes in plants that, nonetheless, are yet to be identified. The presence of different functional domains, several of which facilitate protein-protein interactions, may enable the cyclophilins to identify a multitude of proteins as targets, thereby controlling complex regulatory circuits that enable the plants to respond to various developmental and environmental cues. It is apparent that, as proposed earlier for several biological processes such as cell division, gene expression, immune response and neural functions in animals (Lu et al., 2002, 2007), the PPIase catalyzed cis-trans conversion may act as a molecular switch in plants as well.
Roles of Cyclophilins in Transcriptional and Post-transcriptional Gene Regulation
Transcript turnover and translational control are important post-transcriptional mechanisms of regulation of gene expression. Several cyclophilins have been reported to contain RNA Recognition Motif (RRM), a 90 amino acid long conserved RNA binding motif that is a characteristic feature of RNA-interacting proteins known to actively participate in pre-mRNA processing events (Kenan et al., 1991; Birney et al., 1993). This group of proteins is popularly known as cyclophilin-RNA interacting proteins (CRIPs). The first gene belonging to this group, KIN241, was identified in Paramecium and demonstrated to play an essential role in cell morphogenesis, cortical organization and nuclear reorganization (Krzywicka et al., 2001). The Arabidopsis cyclophilin AtCYP59, which besides PPIase domain also contains an RRM motif, a Zn-knuckle and a charged C-terminal domain consisting of RS/RD (arginine/serine and arginine/aspartate) repeats, was proposed to regulate transcription through its interaction with the immature mRNA (Gullerova et al., 2006; Bannikova et al., 2012). However, contrary to human RRM-containing cyclophilin hCYP33 (CYPE), that showed enhanced PPIase activity after binding to RNA (Wang et al., 2008), the catalytic activity of AtCYP59 was repressed by RNA, indicating a possible negative feedback loop. The physiological significance of this observation in plants is, however, still to be established. Though the presence of RRM along with other domains is also observed in other cyclophilins viz., BnCYP52, BnCYP55 and BnCYP112 in B. napus, and TaCYP37-1-3D, TaCYP38-1-3B, TaCYP45-1-3A, TaCYP53-1-4B, TaCYP54-1-4A, TaCYP55-1-4D, TaCYP64-1-7A, TaCYP64-2-7B and TaCYP64-3-7D in wheat (Hanhart et al., 2017; Singh et al., 2019), the precise role of these proteins in RNA processing or transcriptional regulation is only speculative. A multi-domain cyclophilin, BnCYP146, the largest cyclophilin in B. napus, exhibits the presence of a putative Fip1 domain that has not been identified earlier in any of the cyclophilins. As Fip1 is a transmembrane motif involved in polyadenylation of mRNAs via interaction with the poly(A) polymerase (Hanhart et al., 2017), BnCYP146 might have a role in the stabilization of target RNA molecules and, hence, in the regulation of translation. This, however, requires further validation.
Implications of Cyclophilins in Abiotic Stress Response
The expression of cyclophilins in plants and other organisms is regulated by several different stress conditions (Table 7), supporting their role in the adaptation process (Marivet et al., 1994; Godoy et al., 2000; Sharma et al., 2003; Sekhar et al., 2010; Kumari et al., 2013, 2015). Our studies on sorghum were the first in plants to demonstrate that stress-induced PPIase activity is associated with drought tolerance (Sharma and Singh, 2003a,b; Sharma et al., 2003). Since then, conclusive evidence for the role of cyclophilins in the adaptation of plants to abiotic stress has been provided by several transgenic studies (Table 7). Heterologous expression of pigeonpea (CcCYP) and Golgi-localized rice (OsCYP21-4) cyclophilins imparted tolerance against salt and oxidative stress in Arabidopsis and rice (Oryza sativa), respectively (Sekhar et al., 2010; Lee et al., 2015a). Ectopic expression of a cold-induced cyclophilin PPIase, OsCYP19-4, in transgenic rice resulted in a significant increase in the number of tillers, spikes, grain weight, and was associated with cold resistance (Yoon et al., 2016). Due to high similarity (70 %) to AtCYP19-4 (Ahn et al., 2010), that interacts with guanine nucleotide exchange factor (GNOM protein) which is involved in polar localization of the auxin efflux carrier PIN1, the enhanced performance of OsCYP19-4 overexpressing plants was ascribed to alteration in auxin homeostasis (Yoon et al., 2016). Determination of the auxin levels is required to support the proposed mechanism.
The ability to confer tolerance against a broad range of abiotic stress conditions was also observed for the rice cyclophilin OsCYP2 (Table 7), which is localized to cytosol and nucleus, and shares 62.79 % and 32.08 % identity with OsCYP19-4 and OsCYP21-4, respectively (Kumari et al., 2013, 2015). The OsCYP2-induced tolerance to stress in transgenic tobacco plants was attributed to the regulation of ion homeostasis due to an enhanced K+/Na+ ratio (Kumari et al., 2015). The drought tolerance in the OsCYP18-2 over-expressing transgenic Arabidopsis, on the contrary, was ascribed to reduced transpiration rate due to a decrease in stomatal aperture (Lee et al., 2015b). Though OsCYP18-2 was also shown to interact with the Ski interacting protein (OsSKIP) in rice (Lee et al., 2015b), the role of this interaction in stress tolerance is not understood. The abrogation of this interaction by engineering OsCYP18-2 and OsSKIP will provide further insights into its functional significance.
The plastidic cyclophilins have also been demonstrated to impart protection against stress. Ectopic expression of the thylakoid localized cyclophilins, OsCYP20-2 and AtCYP38, resulted in enhanced tolerance to various abiotic stresses in the transgenic Arabidopsis and tobacco plants (Kim et al., 2012; Wang et al., 2015; Ge Q. et al., 2020). While the OsCYP20-2-induced-tolerance was ascribed to higher chloroplast PPIase activity and maintenance of NADH dehydrogenase-like complex that protects the stroma against over-reduction under stress conditions, the AtCYP38-stimulated protection against high light intensity was due to inhibition of PsbO2 activity which is an important component of photosystem II (Wang et al., 2015). Recent studies have demonstrated the presence of two different variants of OsCYP20-2 in rice, and the two isoforms contribute to chilling stress tolerance through different mechanisms (Ge Q. et al., 2020). While the chloroplast-localized OsCYP2 contributes to scavenging of ROS by enhancing the activity of a superoxide dismutase, OsFSD2, the nuclear-localized isoform, generated following truncation of the chloroplast signal peptide, interacts with a DELLA protein, SLENDER RICE1, and stimulates its degradation to promote growth. These studies, hence, highlight the crucial role of OsCYP20-2 in integrating plant growth and abiotic stress response. As observed in transgenic tobacco plants that constitutively expressed GjCYP-1, a cyclophilin gene from red alga Griffithsia japonica, the PPIase-induced stress tolerance might also be associated with adverse effects on growth and yield (Cho and Kim, 2008), thereby, necessitating the use of stress-inducible promoters.
Though molecular processes underlying the cyclophilin-induced stress tolerance are not fully understood for the majority of the cyclophilins, prevention of protein aggregation, as reported for GjCYP-1, may be one of the protective mechanisms (Cho et al., 2005). The heat stress tolerance in E. coli that overexpressed a redox-regulated wheat cytosolic cyclophilin, TaCYPA-1, was however attributed to its PPIase activity (Kaur et al., 2016, 2017). Since the redox status of plants undergoes reversible changes under stress conditions (Jubany-Mari et al., 2010), application of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status (Brossa et al., 2013) is needed to explore the role of TaCYPA-1 in the maintenance of redox homeostasis in the cell under stress conditions. Further, our studies also demonstrated that TaCYPA-1 and AtCYP19-3, that are 74 % identical, interact with calmodulin (CaM) in a Ca2+-dependent fashion (Popescu et al., 2007; Kaur et al., 2015). As Ca2+ is a transducer of stress signals (Snedden and Fromm, 2001; Virdi et al., 2015), cyclophilins may likely constitute an important component of Ca2+-CaM signaling pathway. Whether interaction with CaM is a property shared by all cyclophilins is still a matter of speculation and requires further investigations for elucidating the role of these proteins in CaM-mediated responses
The expression of cyclophilin genes is also regulated by CO2 and nitrogen. Transcript levels of a tobacco cyclophilin gene were reported to increase under low nitrogen conditions (Yang et al., 2013), but the physiological implication of this observation is yet to be ascertained. Due to the competitive nature of ribulose-1, 5-bisphosphate carboxylase oxygenase (Rubisco) catalyzed carboxylation and oxygenation reactions, the photosynthetic activity is low in plants and algae. Hence, under low CO2, the CO2-concentrating mechanism (CCM) is induced in several algae such as Chlamydomonas reinhardtii (Moroney and Ynalvez, 2007). CCM leads to a high ratio of CO2 to O2 at the site of Rubisco and stimulates the carboxylation reaction under depleted CO2 conditions (Badger et al., 1980; Moroney and Mason, 1991). The establishment of CCM under low CO2 conditions in C. reinhardtii was reported to coincide with a transient increase in expression of a cyclophilin gene, indicating its likely role in this mechanism (Somanchi and Moroney, 1999). It was conjectured that this cyclophilin may be required for protecting the proteins against photodamage since CO2 is an electron receptor and a decrease in CO2 concentration at the same light imposes photooxidative stress. Similar roles cannot be ruled out for other cyclophilins, particularly the chloroplast-localized ones, and warrants further experimentation.
The cyclophilins from extremophiles such as Piriformospora indica and Thellungiella halophila also offer an attractive alternative to improve stress tolerance in crop plants (Table 7) (Chen et al., 2007; Trivedi et al., 2013b,c). PiCYPA cloned from the xerophytic fungus P. indica, despite lacking the canonical RRM, demonstrated interaction with RNA. It is likely that protection against stress in the PiCYPA-overexpressing transgenic E. coli and tobacco plants might be due to its role in the stabilization of RNA transcripts (Trivedi et al., 2013b). Induction of a 17.5 kDa cyclophilin PmCYP in dinoflagellate algae Prorocentrum minimum in response to different pollutants viz., copper and polychlorinated biphenyl (Ponmani et al., 2015) further suggests that the role of cyclophilins as stress proteins is conserved. The role of cyclophilins as universal stress proteins is also substantiated by studies on Brucella, an intracellular bacterial pathogen in humans and cows which causes the disease brucellosis (Young, 1995). Comparative proteomic analysis in B. abortus resulted in the identification of two cyclophilins (CYPA and CYPB) which were differentially expressed and implicated in bacterial intracellular adaptation (Roset et al., 2013). Studies employing ΔcypAB mutants revealed that these genes were essential for virulence and tolerance to various abiotic stresses such as oxidative, acidic pH and detergents (Roset et al., 2013).
It is apparent that despite being distinct, protection against stress-induced damage is a property common to several cyclophilins (Table 7), suggesting an overlap of their roles. However, the precise mechanisms by which these proteins protect the cellular machinery against stress-induced damage are still elusive for the majority of these proteins. Although except for AtCYP38, all the cyclophilins implicated in stress tolerance are SD proteins, similar roles for the MD cyclophilins cannot be ruled out and should be the subject of future studies. Further investigations are therefore necessary to unravel the physiological implications of cyclophilins in plants that will enable their applications in crop improvement through biotechnological interventions or conventional breeding.
Future Prospects
The characterization of cyclophilins in plants is revealing new insights into their physiological relevance. The presence of large gene families suggests that these cyclophilins might have overlapping yet distinct functions which are still speculative. As signified by analyses of available genomic data, the cyclophilin genes in plants display substantial variability in their structure, particularly in the context of the distribution of introns. Since introns play a role in regulating gene expression, rigorous studies are required to understand the implications of these differences in the regulation of cyclophilin genes. These studies are likely to provide insight into their physiological role. Despite the presence of conserved CLD, the presence of different domains such as TPR, WD, RRM, etc., in the MD cyclophilins indicate the acquisition of novel functions. However, the role of these domains in imparting specific functionalities to cyclophilins is still conjectural for the majority of these proteins. Therefore, it is imperative to carry out the targeted deletion of different motifs in MD cyclophilins of plants and analyze the effect thereof on various facets of growth and development. Despite high sequence similarity, variability in the structure of cyclophilins has been reported to result in dramatic changes in their biochemical properties. Given the diversity in plant cyclophilins, it is imperative to elucidate the structure of these proteins by using different biophysical approaches such as X-ray diffraction and nuclear magnetic resonance to identify their mechanism of action. Both PPIase active and inactive (AtCYP38) cyclophilins have been reported to play specific roles in plants, thus, rendering the role of PPIase activity in plants a matter of speculation. Hence, the expression of site-directed mutants that show graded PPIase activity might illustrate the precise function of this biochemical attribute in the plants. Since PPIase activity of several cyclophilins is regulated by different redox mechanisms, and several of these proteins are induced by stress that affects the redox status of the cell, investigations should also be undertaken to comprehend their role in the maintenance of redox-homeostasis. Though the cyclophilin-induced stress tolerance in plants has been attributed to their chaperonic functions, the detailed cellular mechanisms, with few exceptions, are yet to be deciphered. The chaperonic activities (holdase and foldase) of cyclophilins can be independent of PPIase function, due to which concerted efforts are required to characterize the different biochemical activities of plant cyclophilins and their implications in stress tolerance. The multifaceted nature of cyclophilins warrants multipronged approaches to delineate their mechanisms of action in plants.
Conclusion
Compared with prokaryotes and animals, the cyclophilin gene families in plants have undergone dramatic expansion, implying functional diversification and their importance for different growth and developmental processes. Being sessile, the divergence of cyclophilins may enable the plants to respond and adapt to adverse environmental conditions since several of these genes are responsive to different abiotic and biotic stressors. It is evident that though the roles of majority of the cyclophilins in plants are obscure, these proteins by virtue of their PPIase and chaperonic activities are likely to regulate diverse aspects of growth and development. Furthermore, presence of additional functional domains such as WD, F-box, RRM, and Zn-knuckle might enable these proteins to facilitate assembly of multiprotein complexes and modulation of cellular processes through transcriptional, post-transcriptional, translational and post-translational regulation of gene expression, thereby, enabling them to play multifaceted roles in the cell. Studies carried out so far also reveal that the enzymatic activity of cyclophilins is regulated through diverse mechanisms that might be redox-dependent or independent, the physiological significance of which is still a matter of speculation. The implications of cyclophilins such as LeCYP, TaCYP20-2 and AtCYP18-3 in auxin, GA and brassinosteroid signaling further underline their functional versatility and indispensability for the plants. The studies carried out until now have though provided novel insights into the functional and regulatory aspects of plant cyclophilins, the physiological significance of the majority of these proteins is still a matter of conjecture. Therefore, concerted efforts are imperative to understand the importance of different cyclophilins in plants so that these genes can be used for the improvement of different traits in the crop plants.
Author Contributions
HS: methodology, visualization, data curation, and writing-original draft preparation. KK: data curation, validation, visualization, and writing-original draft preparation. MS and GK: writing-original draft preparation and validation. PS: conceptualization, supervision, methodology, and reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
We gratefully acknowledge the financial support from the Department of Biotechnology, Government of India, New Delhi for carrying out this research work. Thanks are also due to the Department of Science and Technology, Government of India, for supporting the Department of Bioinformatics, Hans Raj Mahila Maha Vidyalaya, Jalandhar with the “Fund for Improvement of Science and Technology Infrastructure” grant for the development of computational resources (Grant No.: - D. O. No. SR/FST/PG College/2009). We are also thankful to the University Grant Commission, New Delhi, Government of India for providing Rajiv Gandhi National Fellowship to KK (Number and date of award letter: F1-17.1/2017-18/RGNF-2017-18-SC-PUN-44307/)Sa-III/website), 28-07-2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.585212/full#supplementary-material
References
Abassi, S., Wang, H., Park, B. S., Park, J. W., and Ki, J. S. (2017). A novel cyclophilin b gene in the red tide dinoflagellate Cochlodiniumpolykrikoides: Molecular characterizations and transcriptional responses to environmental stresses. Biomed. Res. Int. 2017, 4101580. doi: 10.1155/2017/4101580
Abel, S., Nguyen, M. D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. doi: 10.1006/jmbi.1995.0454
Acevedo, L. A., Kwon, J., and Nicholson, L. K. (2019). Quantification of reaction cycle parameters for an essential molecular switch in an auxin-responsive transcription circuit in rice. Proc. Natl. Acad. Sci. U.S.A. 116, 2589–2594. doi: 10.1073/pnas.1817038116
Achenbach, T. V., Göthel, S. F., and Marahiel, M. A. (1997). Histidine 109 in peptidyl-prolyl cis-trans isomerase of Bacillus subtilis plays an important role in catalysis and in cyclosporin A binding. FEMS Microbiol. Lett. 154, 139–144. doi: 10.1016/S0378-1097(97)00314-5
Adams, B., Musiyenko, A., Kumar, R., and Barik, S. (2005). A novel class of dual-family immunophilins. J. Biol. Chem. 280, 24308–24314. doi: 10.1074/jbc.M500990200
Ahn, J. C., Kim, D. W., You, Y. N., Seok, M. S., Park, J. M., Hwang, H., et al. (2010). Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress. BMC Plant Biol. 10:253. doi: 10.1186/1471-2229-10-253
Ainley, W. M., Walker, J. C., Nagao, R. T., and Key, J. L. (1988). Sequence and characterization of two auxin-regulated genes from soybean. J. Biol. Chem. 263, 10658–10666.
Anderson, S. K., Gallinger, S., Roder, J., Frey, J., Young, H. A., and Ortaldo, J. R. (1993). A cyclophilin-related protein involved in the function of natural killer cells. Proc. Natl. Acad. Sci. U.S.A. 90, 542–546. doi: 10.1073/pnas.90.2.542
Aravind, L., and Koonin, E. V. (2000). The U box is a modified RING finger – a common domain in ubiquitination. Curr. Biol. 10, 132–134. doi: 10.1016/S0960-9822(00)00398-5
Arévalo-Rodríguez, M., and Heitman, J. (2005). Cyclophilin A is localized to the nucleus and controls meiosis in Saccharomyces cerevisiae. Eukaryot. Cell 4, 17–29. doi: 10.1128/EC.4.1.17-29.2005
Arevalo-Rodriguez, M., Wu, X., Hanes, S. D., and Heitman, J. (2004). Prolyl isomerases in yeast. Front. Biosci. 9:2420–2446. doi: 10.2741/1405
Askwith, C., and Kaplan, J. (1997). An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272, 401–405. doi: 10.1074/jbc.272.1.401
Badger, M. R., Kaplan, A., and Berry, J. A. (1980). Internal inorganic carbon pool of Chlamydomonas reinhardtii. Plant Physiol. 66, 407–413. doi: 10.1104/pp.66.3.407
Balsera, M., Arellano, J. B., Gutiérrez, J. R., Heredia, P., Revuelta, J. L., and De Las Rivas, J. (2003). Structural analysis of the PsbQ protein of photosystem II by fourier transform infrared and circular dichroic spectroscopy and by bioinformatic methods. Biochemistry 42, 1000–1007. doi: 10.1021/bi026575l
Bandziulis, R. J., Swanson, M. S., and Dreyfuss, G. (1989). RNA-binding proteins as developmental regulators. Genes Dev. 3, 431–437. doi: 10.1101/gad.3.4.431
Bannikova, O., Zywicki, M., Marquez, Y., Skrahina, T., Kalyna, M., and Barta, A. (2012). Identification of RNA targets for the nuclear multidomain cyclophilin AtCyp59 and their effect on PPIase activity. Nucleic Acids Res. 41, 1783–1796. doi: 10.1093/nar/gks1252
Barik, S. (2018). Dual-family peptidyl-prolyl isomerases (Immunophilins) of select monocellular organisms. Biomolecules 8:148. doi: 10.3390/biom8040148
Berardini, T. Z., Bollman, K., Sun, H., and Scott Poethig, R. (2001). Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291, 2405–2407. doi: 10.1126/science.1057144
Birney, E., Kumar, S., and Krainer, A. R. (1993). Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21, 5803–5816. doi: 10.1093/nar/21.25.5803
Blackburn, E. A., Wear, M. A., Landre, V., Narayan, V., Ning, J., Erman, B., et al. (2015). Cyclophilin 40 isomerase activity is regulated by a temperature-dependent allosteric interaction with Hsp90. Biosci. Rep. 35, 1–12. doi: 10.1042/BSR20150124
Bose, S., Mucke, M., and Freedman, R. B. (1994). The characterization of a cyclophilin-type peptidyl-prolyl cis-trans isomerase from the endoplasmic-reticulum lumen. Biochem. J. 300, 871–875. doi: 10.1042/bj3000871
Braaten, D., and Luban, J. (2001). Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20, 1300–1309. doi: 10.1093/emboj/20.6.1300
Brandts, J. F., Halvorson, H. R., and Brennan, M. (1975). Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 14, 4953–4963. doi: 10.1021/bi00693a026
Breiman, A., Fawcett, T. W., Ghirardi, M. L., and Mattoo, A. K. (1992). Plant organelles contain distinct peptidyl-prolyl cis- trans-isomerases. J. Biol. Chem. 25, 21293–21296.
Breuder, T., Hemenway, C. S., Movva, N. R., Cardenas, M. E., and Heitman, J. (1994). Calcineurin is essential in cyclosporin A and FK506 sensitive yeast strains. Proc. Natl. Acad. Sci. U.S.A. 91, 5372–5376. doi: 10.1073/pnas.91.12.5372
Brossa, R., Pintó-Marijuan, M., Jiang, K., Alegre, L., and Feldman, L. J. (2013). Assessing the regulation of leaf redox status under water stress conditions in Arabidopsis thaliana: Col-0 ecotype (wild-type and vtc-2), expressing mitochondrial and cytosolic roGFP1. Plant Signal. Behav. 8:e24781. doi: 10.4161/psb.24781
Bruce, J. W., and Wilcox, K. W. (2002). Identification of a motif in the c terminus of herpes simplex virus regulatory protein ICP4 that contributes to activation of transcription. J. Virol. 76, 195–207. doi: 10.1128/jvi.76.1.195-207.2002
Búa, J., Åslund, L., Pereyra, N., García, G. A., Bontempi, E. J., and Ruiz, A. M. (2001). Characterisation of a cyclophilin isoform in Trypanosoma cruzi. FEMS Microbiol. Lett. 200, 43–47. doi: 10.1016/S0378-1097(01)00193-8
Buchner, D. A., Trudeau, M., and Meisler, M. H. (2003). SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301, 967–969. doi: 10.1126/science.1086187
Bugli, F., Khattab, A., Vigneti, E., Butler, R., Cioli, D., and Klinkert, M. Q. (1998). Expression cloning and biochemical characterizations of recombinant cyclophilin proteins from Schistosoma mansoni. Protein Expr. Purif. 12, 340–346. doi: 10.1006/prep.1997.0852
Campos, B. M., Sforça, M. L., Ambrosio, A. L. B., Domingues, M. N., De Souza, T., de, A. C. B., et al. (2013). A Redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins. Plant Physiol. 162, 1311–1323. doi: 10.1104/pp.113.218339
Carvalho, A. B., and Clark, A. G. (1999). Genetic recombination: Intron size and natural selection. Nature 401:344. doi: 10.1038/43827
Cashmore, A. R., Jarillo, J. A., Wu, Y. J., and Liu, D. (1999). Cryptochromes: blue light receptors for plants and animals. Science 284, 760–765. doi: 10.1126/science.284.5415.760
Cavarec, L., Kamphausen, T., Dubourg, B., Callebaut, I., Lemeunier, F., Métivier, D., et al. (2002). Identification and characterization of Moca-cyp: a Drosophila melanogaster nuclear cyclophilin. J. Biol. Chem. 277, 41171–41182. doi: 10.1074/jbc.M203757200
Chen, A. P., Wang, G. L., Qu, Z. L., Lu, C. X., Liu, N., Wang, F., et al. (2007). Ectopic expression of ThCYP1, a stress-responsive cyclophilin gene from Thellungiella halophila, confers salt tolerance in fission yeast and tobacco cells. Plant Cell Rep. 26, 237–245. doi: 10.1007/s00299-006-0238-y
Chen, J., Hu, R., Zhu, Y., Shen, G., and Zhang, H. (2014). Arabidopsis PHOSPHOTYROSYL PHOSPHATASE ACTIVATOR is essential for PROTEIN PHOSPHATASE 2A holoenzyme assembly and plays important roles in hormone signaling, salt stress response, and plant development. Plant Physiol. 3, 1519–1534. doi: 10.1104/pp.114.250563
Chen, Q., Chen, Q. J., Sun, G. Q., Zheng, K., Yao, Z. P., Han, Y. H., et al. (2019). Genome-wide identification of cyclophilin gene family in cotton and expression analysis of the fibre development in Gossypium barbadense. Int. J. Mol. Sci. 20:349. doi: 10.3390/ijms20020349
Cheng, J., Zhang, M., Tan, B., Jiang, Y., Zheng, X., Ye, X., et al. (2019). A single nucleotide mutation in GID1c disrupts its interaction with DELLA1 and causes a GA-insensitive dwarf phenotype in peach. Plant Biotechnol. J. 17, 1723–1735. doi: 10.1111/pbi.13094
Cho, E. K., and Kim, M. (2008). A red algal cyclophilin has an effect on development and growth in Nicotiana tabacum. Plant Physiol. Biochem. 46, 868–874. doi: 10.1016/j.plaphy.2008.05.013
Cho, E. K., Lee, Y. K., and Hong, C. B. (2005). A cyclophilin from Griffithsia japonica has thermoprotective activity and is affected by CsA. Mol. Cells 20, 142–150.
Chou, I. T., and Gasser, C. S. (1997). Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 6, 873–892. doi: 10.1023/A:1005930024796
Coaker, G., Zhu, G., Ding, Z., Van Doren, S. R., and Staskawicz, B. (2006). Eukaryotic cyclophilin as a molecular switch for effector activation. Mol. Microbiol. 61, 1485–1496. doi: 10.1111/j.1365-2958.2006.05335.x
Colley, N. J., Baker, E. K., Stamnes, M. A., and Zuker, C. S. (1991). The cyclophilin homolog ninaA is required in the secretory pathway. Cell 67, 255–263. doi: 10.1016/0092-8674(91)90177-Z
Compton, L. A., Davis, J. M., Macdonald, J. R., and Bächinger, H. P. (1992). Structural and functional characterization of Escherichia coli peptidyl-prolyl cis-trans isomerases. Eur. J. Biochem. 206, 927–934. doi: 10.1111/j.1432-1033.1992.tb17002.x
Confalonieri, F., and Duguet, M. (1995). A 200−amino acid ATPase module in search of a basic function. BioEssays 17, 639–650. doi: 10.1002/bies.950170710
Conner, T. W., Goekjian, V. H., LaFayette, P. R., and Key, J. L. (1990). Structure and expression of two auxin-inducible genes from Arabidopsis. Plant Mol. Biol. 15, 623–632. doi: 10.1007/BF00017836
Connern, C. P., and Halestrap, A. P. (1992). Purification and N-terminal sequencing of peptidyl-prolyl cis-trans-isomerase from rat liver mitochondrial matrix reveals the existence of a distinct mitochondrial cyclophilin. Biochem. J. 284, 381–385. doi: 10.1042/bj2840381
Craig, K. L., and Tyers, M. (1999). The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72, 299–328. doi: 10.1016/S0079-6107(99)00010-3
Dalle-Donne, I., Rossi, R., Colombo, G., Giustarini, D., and Milzani, A. (2009). Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96. doi: 10.1016/j.tibs.2008.11.002
Davis, E. S., Becker, A., Heitman, J., Hall, M. N., and Brennan, M. B. (1992). A yeast cyclophilin gene essential for lactate metabolism at high temperature. Proc. Natl. Acad. Sci. U.S.A. 89, 11169–11173. doi: 10.1073/pnas.89.23.11169
Davis, T. L., Walker, J. R., Campagna-Slater, V., Finerty, P. J., Finerty, P. J., Paramanathan, R., et al. (2010). Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 8:e1000439. doi: 10.1371/journal.pbio.1000439
Davis, T. L., Walker, J. R., Ouyang, H., MacKenzie, F., Butler−Cole, C., Newman, E. M., et al. (2008). The crystal structure of human WD40 repeat−containing peptidylprolyl isomerase (PPWD1). FEBS J. 9, 2283–2295. doi: 10.1111/j.1742-4658.2008.06381.x
Derkx, P. M. F., and Madrid, S. M. (2001). The Aspergillus niger cypA gene encodes a cyclophilin that mediates sensitivity to the immunosuppressant cyclosporin A. Mol. Genet. Genomics 266, 527–536. doi: 10.1007/s004380100586
Dill, A., Thomas, S. G., Hu, J., Steber, C. M., and Sun, T. P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405. doi: 10.1105/tpc.020958
Dolinski, K., Muir, S., Cardenas, M., and Heitman, J. (1997). All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 13093–13098. doi: 10.1073/pnas.94.24.13093
Domingues, M. N., Campos, B. M., Oliveira, M. L. P., Mello, U. Q., and Benedetti, C. E. (2012). TAL effectors target the C-terminal domain of RNA polymerase II (CTD) by inhibiting the prolyl-isomerase activity of a CTD-associated cyclophilin. PLoS One 7:41553. doi: 10.1371/journal.pone.0041553
Dominguez-Solis, J. R., He, Z., Lima, A., Ting, J., Buchanan, B. B., and Luan, S. (2008). A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 105, 16386–16391. doi: 10.1073/pnas.0808204105
Dornan, J., Page, A. P., Taylor, P., Wu, S. Y., Winter, A. D., Husi, H., et al. (1999). Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans. J. Biol. Chem. 274, 34877–34883. doi: 10.1074/jbc.274.49.34877
Dornan, J., Taylor, P., and Walkinshaw, M. (2003). Structures of immunophilins and their ligand complexes. Curr. Top. Med. Chem. 3, 1392–1409. doi: 10.2174/1568026033451899
Duina, A. A., Kalton, H. M., and Gaber, R. F. (1998). Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273, 18974–18978. doi: 10.1074/jbc.273.30.18974
Dutta, T., Kaur, H., Singh, S., Mishra, A., Tripathi, J. K., Singh, N., et al. (2011). Developmental changes in storage proteins and peptidyl-prolyl cis-trans isomerase activity in grains of different wheat cultivars. Food Chem. 128, 450–457. doi: 10.1016/j.foodchem.2011.03.052
Earley, K. W., and Poethig, R. S. (2011). Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 286, 38184–38189. doi: 10.1074/jbc.M111.290130
Edvardsson, A., Eshaghi, S., Vener, A. V., and Andersson, B. (2003). The major peptidyl-prolyl isomerase activity in thylakoid lumen of plant chloroplasts belongs to a novelcyclophilin TLP20. FEBS Lett. 542, 137–141. doi: 10.1016/S0014-5793(03)00366-1
Edvardsson, A., Shapiguzov, A., Petersson, U. A., Schröder, W. P., and Vener, A. V. (2007). Immunophilin AtFKBP13 sustains all peptidyl-prolyl isomerase activity in the thylakoid lumen from Arabidopsis thaliana deficient in AtCYP20-2. Biochemistry 46, 9432–9442. doi: 10.1021/bi700426q
Elrod, J. W., Wong, R., Mishra, S., Vagnozzi, R. J., Sakthievel, B., Goonasekera, S. A., et al. (2010). Cyclophilin D controls mitochondrial pore – dependent Ca2+ exchange, metabolic flexibility, and propensity for heart failure in mice. J. Clin. Invest. 120, 3680–3687. doi: 10.1172/JCI43171
Fanghänel, J., and Fischer, G. (2004). Insights into the catalytic mechanism of peptidyl prolyl cis-trans isomerases. Front. Biosci. 9:78. doi: 10.2741/1494
Faou, P. (2001). NcCyP41, a Two Domain Neurospora crassa Cyclophilin: Characterization of its peptidyl-prolyl cis-trans isomerase Activity; Isolation and Functional Analysis of Two Novel NcCyP41-Binding Proteins. Freiburg: Verlag nichtermittelbar.
Fischer, G., Bang, H., Berger, E., and Schellenberger, A. (1984). Conformational specificity of chymotrypsin toward proline-containing substrates. Biochim. Biophys. Acta Protein Struct. Mol. 791, 87–97. doi: 10.1016/0167-4838(84)90285-1
Fischer, G., Wittmann-Liebold, B., Lang, K., Kiefhaber, T., and Schmid, F. X. (1989). Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337, 476–478. doi: 10.1038/337476a0
Freeman, B. C., Toft, D. O., and Morimoto, R. I. (1996). Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science 274, 1718–1720. doi: 10.1126/science.274.5293.1718
Freemont, P. S., Hanson, I. M., and Trowsdale, J. (1991). A novel gysteine-rich sequence motif. Cell 64, 483–484. doi: 10.1016/0092-8674(91)90229-R
Fu, A., He, Z., Hye, S. C., Lima, A., Buchanan, B. B., and Luan, S. (2007). A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 15947–15952. doi: 10.1073/pnas.0707851104
Fulgosi, H., Vener, A. V., Altschmied, L., Herrmann, R. G., and Andersson, B. (1998). A novel multi-functional chloroplast protein: Identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17, 1577–1587. doi: 10.1093/emboj/17.6.1577
Galat, A. (1999). Variations of sequences and amino acid compositions of proteins that sustain their biological functions: an analysis of the cyclophilin family of proteins. Arch. Biochem. Biophys. 371, 149–162. doi: 10.1006/abbi.1999.1434
Galat, A. (2003). Peptidyl-prolyl cis-trans isomerases (immunophilins): biological diversity – targets - functions. Curr. Top. Med. Chem. 3, 1315–1347. doi: 10.2174/1568026033451862
Galat, A. (2004). Function-dependent clustering of orthologues and paralogues of cyclophilins. Proteins Struct. Funct. Genet. 56, 808–820. doi: 10.1002/prot.20156
Gan, P. H. P., Shan, W., Blackman, L. M., and Hardham, A. R. (2009). Characterization of cyclophilin-encoding genes in Phytophthora. Mol. Genet. Genomics 281, 565–578. doi: 10.1007/s00438-009-0431-0
Gasser, C. S., Gunning, D. A., Budelier, K. A., and Brown, S. M. (1990). Structure and expression of cytosolic cyclophilin peptidyl-prolyl cis-trans isomerase of higher plants and production of active tomato cyclophilin in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 9519–9523. doi: 10.1073/pnas.87.24.9519
Ge, L., Zhang, K., Cao, X., Weng, Y., Liu, B., Mao, P., et al. (2020). Sequence characteristics of Medicago truncatula cyclophilin family members and function analysis of MsCYP20-3B involved in axillary shoot development. Mol. Biol. Rep. 47, 907–919. doi: 10.1007/s11033-019-05183-x
Ge, Q., Zhang, Y., Xu, Y., Bai, M., Luo, W., Wang, B., et al. (2020). Cyclophilin OsCYP20−2 with a novel variant integrates defense and cell elongation for chilling response in rice. New Phytol. 225:2453. doi: 10.1111/nph.16324
Geisler, M., and Bailly, A. (2007). Tete-a-tete: the function of FKBPs in plant development. Trends Plant Sci. 10, 465–473. doi: 10.1016/j.tplants.2007.08.015
Ghezzi, P., Casagrande, S., Massignan, T., Basso, M., Bellacchio, E., Mollica, L., et al. (2006). Redox regulation of cyclophilin A by glutathionylation. Proteomics 6, 817–825. doi: 10.1002/pmic.200500177
Ghosh, D., Mueller, G. A., Schramm, G., Edwards, L. L., Petersen, A., London, R. E., et al. (2014). Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin Cat r 1. J. Biol. Chem. 289, 21374–21385. doi: 10.1074/jbc.M114.559971
Godoy, A. V., Lazzaro, A. S., Casalongué, C. A., and San Segundo, B. (2000). Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Sci. 152, 123–134. doi: 10.1016/S0168-9452(99)00211-3
Göthel, S. F., and Marahiel, M. A. (1999). Peptidyl-prolyl cis-trams isomerases, a superfamily of ubiquitous folding catalysts. Cell. Mol. Life Sci. 55, 423–436. doi: 10.1007/s000180050299
Gottschalk, M., Dolgener, E., Xoconostle-Cázares, B., Lucas, W. J., Komor, E., and Schobert, C. (2008). Ricinus communis cyclophilin: Functional characterisation of a sieve tube protein involved in protein folding. Planta 228, 687–700. doi: 10.1007/s00425-008-0771-8
Gourlay, L. J., Angelucci, F., Baiocco, P., Boumis, G., Brunori, M., Bellelli, A., et al. (2007). The three-dimensional structure of two redox states of cyclophilin A from Schistosoma mansoni: evidence for redox regulation of peptidyl-prolyl cis-trans isomerase activity. J. Biol. Chem. 282, 24851–24857. doi: 10.1074/jbc.M702714200
Gray, W. M., Del Pozo, J. C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., et al. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. doi: 10.1101/gad.13.13.1678
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414, 271–276. doi: 10.1038/35104500
Grebe, M., Gadea, J., Steinmann, T., Kientz, M., Rahfeld, J. U., Salchert, K., et al. (2000). A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12, 343–356. doi: 10.1105/tpc.12.3.343
Guilfoyle, T. J., Hagen, G., Li, Y., Ulmasov, T., Zhanbin, L. S., and Gee, M. (1993). Auxin-regulated transcription. Aust. J. Plant Physiol. 20, 489–502. doi: 10.1071/pp9930489
Gullerova, M., Barta, A., and Lorković, Z. J. (2006). AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA 12, 631–643. doi: 10.1261/rna.2226106
Gupta, R., Mould, R. M., He, Z., and Luan, S. (2002). A chloroplast FKBP interacts with and affects the accumulation of rieske subunit of cytochrome bf complex. Proc. Natl. Acad. Sci. U.S.A. 99, 15806–15811. doi: 10.1073/pnas.222550399
Handschumacher, R. E., Harding, M. W., Rice, J., Drugge, R. J., and Speicher, D. W. (1984). Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226, 544–547. doi: 10.1126/science.6238408
Hanhart, P., Falke, S., Garbe, M., Rose, V., Thieß, M., Betzel, C., et al. (2019). Enzyme activity and structural features of three single-domain phloem cyclophilins from Brassica napus. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-45856-y
Hanhart, P., Thieß, M., Amari, K., Bajdzienko, K., Giavalisco, P., Heinlein, M., et al. (2017). Bioinformatic and expression analysis of the Brassica napus L. cyclophilins. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-01596-5
Harding, M. W., Galat, A., Uehling, D. E., and Schreiber, S. L. (1989). A receptor for the immuno-suppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341, 758–760. doi: 10.1038/341758a0
Hayano, T., Takahashi, N., Kato, S., Maki, N., and Suzuki, M. (1991). Two distinct forms of peptidyl-prolyl cis-trans isomerase are expressed separately in periplasmic and cytoplasmic compartments of Escherichia coli cells. Biochemistry 30, 3041–3048. doi: 10.1021/bi00226a009
He, Z., Li, L., and Luan, S. (2004). Immunophilins and parvulins. Superfamily of peptidyl-prolyl isomerases in Arabidopsis. Plant Physiol. 134, 1248–1267. doi: 10.1104/pp.103.031005
Helmling, S., Zhelkovsky, A., and Moore, C. L. (2001). Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 21, 2026–2037. doi: 10.1128/mcb.21.6.2026-2037.2001
High, K. P., Joiner, K. A., and Handschumacher, R. E. (1994). Isolation, cDNA sequences, and biochemical characterization of the major cyclosporin-binding proteins of Toxoplasma gondii. J. Biol. Chem. 269, 9105–9112.
Hirtzlin, J., Färber, P. M., Franklin, R. M., and Bell, A. (1995). Molecular and biochemical characterization of a Plasmodium falciparum cyclophilin containing a cleavable signal sequence. Eur. J. Biochem. 232, 765–772. doi: 10.1111/j.1432-1033.1995.tb20871.x
Ho, T.-H. D., Nolan, R. C., and Shute, D. E. (1981). Characterization of a gibberellin-insensitive dwarf wheat, D6899. Plant Physiol. 67, 1026–1031. doi: 10.1104/pp.67.5.1026
Howard, B. R., Vajdos, F. F., Li, S., Sundquist, W. I., and Hill, C. P. (2003). Structural insights into the catalytic mechanism of cyclophilin A. Nat. Struct. Biol. 10, 475–481. doi: 10.1038/nsb927
Howell, V. M., Jones, J. M., Bergren, S. K., Li, L., Billi, A. C., Avenarius, M. R., et al. (2007). Evidence for a direct role of the disease modifier SCNM1 in splicing. Hum. Mol. Genet. 16, 2506–2516. doi: 10.1093/hmg/ddm206
Iki, T., Yoshikawa, M., Meshi, T., and Ishikawa, M. (2012). Cyclophilin 40 facilitates Hsp90-mediated RISC assembly in plants. EMBO J. 31, 267–278. doi: 10.1038/emboj.2011.395
Ivanchenko, M. G., Coffeen, W. C., Lomax, T. L., and Dubrovsky, J. G. (2006). Mutations in the Diageotropica (Dgt) gene uncouple patterned cell division during lateral root initiation from proliferative cell division in the pericycle. Plant J. 46, 436–447. doi: 10.1111/j.1365-313X.2006.02702.x
Jackson, K., and Söll, D. (1999). Mutations in a new Arabidopsis cyclophilin disrupt its interaction with protein phosphatase 2A. Mol. Gen. Genet. 262, 830–838. doi: 10.1007/s004380051147
Janowski, B., Wöllner, S., Schutkowski, M., and Fischer, G. (1997). A protease-free assay for peptidyl-prolyl cis-trans isomerases using standard peptide substrates. Anal. Biochem. 252, 299–307. doi: 10.1006/abio.1997.2330
Jia, Z., Niu, J., Huan, L., Wu, X., Wang, G., and Hou, Z. (2013). Cyclophilin participates in responding to stress situations in Porphyrahaitanensis (Bangiales, Rhodophyta). J. Phycol. 49, 194–201. doi: 10.1111/j.1529-8817.2012.01234.x
Jiang, Q., Li, X. R., Wang, C. K., Cheng, J., Tan, C., Cui, T. T., et al. (2018). A fluorescent peptidyl substrate for visualizing peptidyl-prolyl: Cis-trans isomerase activity in live cells. Chem. Commun. 54, 1857–1860. doi: 10.1039/c7cc09135d
Jing, H., Yang, X., Zhang, J., Liu, X., Zheng, H., Dong, G., et al. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6, 1–10. doi: 10.1038/ncomms8395
Jordens, J., Janssens, V., Longin, S., Stevens, I., Martens, E., Bultynck, G., et al. (2006). The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis-trans isomerase. J. Biol. Chem. 281, 6349–6357. doi: 10.1074/jbc.M507760200
Joseph, J. D., Heitman, J., and Means, A. R. (1999). Molecular cloning and characterization of Aspergillus nidulans cyclophilin B. Fungal Genet. Biol. 27, 55–66. doi: 10.1006/fgbi.1999.1131
Jubany-Mari, T., Alegre-Batlle, L., Jiang, K., and Feldman, L. J. (2010). Use of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status in Arabidopsis thaliana water-stressed plants. FEBS Lett. 584, 889–897. doi: 10.1016/j.febslet.2010.01.014
Jung, H., Jo, S. H., Park, H. J., Lee, A., Kim, H. S., Lee, H. J., et al. (2020). Golgi-localized cyclophilin 21 proteins negatively regulate ABA signalling via the peptidyl-prolyl isomerase activity during early seedling development. Plant Mol. Biol. 102, 19–38. doi: 10.1007/s11103-019-00928-5
Kallen, J., Spitzfaden, C., Zurini, M. G. M., Wider, G., Widmer, H., Wüthrich, K., et al. (1991). Structure of human cyclophilin and its binding site for cyclosporin A determined by X-ray crystallography and NMR spectroscopy. Nature 353, 276–279. doi: 10.1038/353276a0
Kang, B., Zhang, Z., Wang, L., Zheng, L., Mao, W., Li, M., et al. (2013). OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 74, 86–97. doi: 10.1111/tpj.12106
Kaur, G., Singh, H., Kaur, K., Roy, S., Pareek, A., and Singh, P. (2017). Role of cysteine residues in regulation of peptidyl-prolyl cis-trans isomerase activity of wheat cyclophilin TaCYPA-1. Protein Pept. Lett. 24, 551–560. doi: 10.2174/0929866524666170417165823
Kaur, G., Singh, S., Dutta, T., Kaur, H., Singh, B., Pareek, A., et al. (2016). The peptidyl-prolyl cis-trans isomerase activity of the wheat cyclophilin. TaCYPA-1, is essential for inducing thermotolerance in Escherichia coli. Biochim. Open 2, 9–15. doi: 10.1016/j.biopen.2015.11.003
Kaur, G., Singh, S., Singh, H., Chawla, M., Dutta, T., Kaur, H., et al. (2015). Characterization of peptidyl-prolyl cis-trans isomerase- and calmodulin-binding activity of a cytosolic Arabidopsis thaliana Cyclophilin AtCyp19-3. PLoS One 10:e0136692. doi: 10.1371/journal.pone.0136692
Ke, H. (1992). Similarities and differences between human cyclophilin A and other β-barrel structures. Structural refinement at 1.63 Å resolution. J. Mol. Biol. 228, 539–550. doi: 10.1016/0022-2836(92)90841-7
Ke, H., Mayrose, D., Belshaw, P. J., Alberg, D. G., Schreiber, S. L., Chang, Z. Y., et al. (1994). Crystal structures of cyclophilin A complexed with cyclosporin A and N-methyl-4-[(E)-2-butenyl]-4,4-dimethylthreonine cyclosporin A. Structure 2, 33–44. doi: 10.1016/S0969-2126(00)00006-X
Kenan, D. J., Query, C. C., and Keene, J. D. (1991). RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 16, 214–220. doi: 10.1016/0968-0004(91)90088-D
Kieffer, L. J., Thalhammer, T., and Handschumacher, R. E. (1992). Isolation and characterization of a 40-kDa cyclophilin-related protein. J. Biol. Chem. 267, 5503–5507.
Kim, I. S., Kim, H. Y., Shin, S. Y., Kim, Y. S., Lee, D. H., Park, K. M., et al. (2010). A cyclophilin A CPR1 overexpression enhances stress acquisition in Saccharomyces cerevisiae. Mol. Cells 29, 567–574. doi: 10.1007/s10059-010-0071-6
Kim, S. K., You, Y. N., Park, J. C., Joung, Y., Kim, B. G., Ahn, J. C., et al. (2012). The rice thylakoid lumenal cyclophilin OsCYP20-2 confers enhanced environmental stress tolerance in tobacco and Arabidopsis. Plant Cell Rep. 31, 417–426. doi: 10.1007/s00299-011-1176-x
Kofron, J. L., Kuzmic, P., Kishore, V., Colon-Bonilla, E., and Rich, D. H. (1991). Erratum: determination of kinetic constants for peptidyl-prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry 30, 6127–6134. doi: 10.1021/bi00239a007
Kong, H. Y., Lee, S. C., and Hwang, B. K. (2001). Expression of pepper cyclophilin gene is differentially regulated during the pathogen infection and abiotic stress conditions. Physiol. Mol. Plant Pathol. 59, 189–199. doi: 10.1006/pmpp.2001.0356
Kopriva, S. (2013). 12-oxo-phytodienoic acid interaction with cyclophilin CYP20-3 is a benchmark for understanding retrograde signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 110, 9197–9198. doi: 10.1073/pnas.1307482110
Koser, P. L., Livi, G. P., Levy, M. A., Rosenberg, M., and Bergsma, D. J. (1990). A Candida albicans homolog of a human cyclophilin gene encodes a peptidyl-prolyl cis-trans isomerase. Gene 96, 189–195. doi: 10.1016/0378-1119(90)90252-M
Krzywicka, A., Beisson, J., Keller, A. M., Cohen, J., Jerka-Dziadosz, M., and Klotz, C. (2001). KIN241: A gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol. Microbiol. 42, 257–267. doi: 10.1046/j.1365-2958.2001.02634.x
Küllertz, G., Liebau, A., Rücknagel, P., Schierhorn, A., Diettrich, B., Fischer, G., et al. (1999). Stress-induced expression of cyclophilins in proembryonic masses of Digitalis lanata does not protect against freezing/thawing stress. Planta 208, 599–605. doi: 10.1007/s004250050598
Kumar, N., Gaur, D., Gupta, A., Puri, A., and Sharma, D. (2015). Hsp90-associated immunophilin homolog Cpr7 is required for the mitotic stability of [URE3] prion in Saccharomyces cerevisiae. PLoS Genet. 11:e1005567. doi: 10.1371/journal.pgen.1005567
Kumari, S., Joshi, R., Singh, K., Roy, S., Tripathi, A. K., Singh, P., et al. (2015). Expression of a cyclophilin OsCyp2-P isolated from a salt-tolerant landrace of rice in tobacco alleviates stress via ion homeostasis and limiting ROS accumulation. Funct. Integr. Genomics 15, 395–412. doi: 10.1007/s10142-014-0429-5
Kumari, S., Roy, S., Singh, P., Singla-Pareek, S. L., and Pareek, A. (2013). Cyclophilins: proteins in search of function. Plant Signal. Behav. 8:e22734. doi: 10.4161/psb.22734
Kumari, S., Singh, P., Singla-Pareek, S. L., and Pareek, A. (2009). Heterologous expression of a salinity and developmentally regulated rice cyclophilin gene (OsCyp2) in E. coli and S. cerevisiae confers tolerance towards multiple abiotic stresses. Mol. Biotechnol. 42, 195–204. doi: 10.1007/s12033-009-9153-0
Lamb, J. R., Tugendreich, S., and Hieter, P. (1995). Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20, 257–259. doi: 10.1016/S0968-0004(00)89037-4
Lawit, S. J., Wych, H. M., Xu, D., Kundu, S., and Tomes, D. T. (2010). Maize della proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 51, 1854–1868. doi: 10.1093/pcp/pcq153
Laxa, M., König, J., Dietz, K. J., and Kandlbinder, A. (2007). Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis-trans isomerase and redox-related functions. Biochem. J. 401, 287–297. doi: 10.1042/BJ20061092
Lazar, S. W., and Kolter, R. (1996). SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178, 1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996
Lee, H. N., Kim, S. H., Han, Y. J., Im, S., Jeong, W. J., Park, E. J., et al. (2017). PsCYP1 of marine red alga Pyropiaseriata (Bangiales, Rhodophyta) confers salt and heat tolerance in Chlamydomonas. J. Appl. Phycol. 29, 617–625. doi: 10.1007/s10811-016-0934-0
Lee, S. S., Park, H. J., Jung, W. Y., Lee, A., Yoon, D. H., You, Y. N., et al. (2015a). OsCYP21-4, a novel golgi-resident cyclophilin, increases oxidative stress tolerance in rice. Front. Plant Sci. 6:797. doi: 10.3389/fpls.2015.00797
Lee, S. S., Park, H. J., Yoon, D. H., Kim, B. G., Ahn, J. C., Luan, S., et al. (2015b). Rice cyclophilin OsCYP18-2 is translocated to the nucleus by an interaction with SKIP and enhances drought tolerance in rice and Arabidopsis. Plant Cell Environ. 38, 2071–2087. doi: 10.1111/pce.12531
Li, B., Xu, W., Xu, Y., Zhang, Y., Wang, T., Bai, Y., et al. (2010). Integrative study on proteomics, molecular physiology, and genetics reveals an accumulation of cyclophilin-like protein, Tacyp20-2, leading to an increase of rht protein and dwarf in a novel ga-insensitive mutant (gaid) in wheat. J. Proteome Res. 9, 4242–4253. doi: 10.1021/pr100560v
Li, H., He, Z., Lu, G., Sung, C. L., Alonso, J., Ecker, J. R., et al. (2007). A WD40 domain cyclophilin interacts with histone H3 and functions in gene repression and organogenesis in Arabidopsis. Plant Cell 19, 2403–2416. doi: 10.1105/tpc.107.053579
Li, H., and Luan, S. (2011). The cyclophilin AtCYP71 Interacts with CAF-1 and LHP1 and functions in multiple chromatin remodeling processes. Mol. Plant 4, 748–758. doi: 10.1093/mp/ssr036
Liebthal, M., Strüve, M., Li, X., Hertle, Y., Maynard, D., Hellweg, T., et al. (2016). Redox-dependent conformational dynamics of decameric 2-cysteine peroxiredoxin and its interaction with cyclophilin 20-3. Plant Cell Physiol. 57, 1415–1425. doi: 10.1093/pcp/pcw031
Lima, A., Lima, S., Wong, J. H., Phillips, R. S., Buchanan, B. B., and Luan, S. (2006). A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 103, 12631–12636. doi: 10.1073/pnas.0605452103
Lin, D. T., and Lechleiter, J. D. (2002). Mitochondrial targeted cyclophilin D protects cells from cell death by peptidylprolyl isomerization. J. Biol. Chem. 277, 31134–31141. doi: 10.1074/jbc.M112035200
Liu, J., Chen, C. M., and Walsh, C. T. (1991). Human and Escherichia coli cyclophilins: sensitivity to inhibition by the immunosuppressant cyclosporin a correlates with a specific tryptophan residue. Biochemistry 30, 2306–2310. doi: 10.1021/bi00223a003
Liu, J., Mark, W. A., Chih, M. C., Stuart, L. S., and Christopher, T. W. (1990). Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 6, 2304–2308. doi: 10.1073/pnas.87.6.2304
Liu, J., and Walsh, C. T. (1990). Peptidyl-prolyl cis-trans isomerase from Escherichia coli: a periplasmic homolog of cyclophilin that is not inhibited by cyclosporin A. Proc. Natl. Acad. Sci. U.S.A. 87, 4028–4032. doi: 10.1073/pnas.87.11.4028
Liu, J., Zheng, Q., Deng, Y., Cheng, C. S., Kallenbach, N. R., and Lu, M. (2006). A seven-helix coiled coil. Proc. Natl. Acad. Sci. U.S.A. 103, 15457–15462. doi: 10.1073/pnas.0604871103
Liu, J., He, M., Liu, C., Liao, X., Li, X., Wang, L., et al. (2020). Saline-alkaline resistance analysis of rice overexpressing the CsCYP1A gene of alkaline Chlorella. J. Agric. Sci. 158, 80–87. doi: 10.1017/S0021859620000283
Lorkoviæ, Z. J., Lopato, S., Pexa, M., Lehner, R., and Barta, A. (2004). Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleo protein-specific proteins suggest their involvement in pre-mRNA splicing. J. Biol. Chem. 279, 33890–33898. doi: 10.1074/jbc.M400270200
Lou, X. Y., Li, X., Li, A. X., Pu, M. Y., Shoaib, M., Liu, D. C., et al. (2016). Molecular characterization of three GIBBERELLIN-INSENSITIVE DWARF2 Homologous genes in common wheat. PLoS One 11:e0157642. doi: 10.1371/journal.pone.0157642
Lovering, R., Hanson, I. M., Borden, K. L. B., Martin, S., O’Reilly, N. J., Evan, G. I., et al. (1993). Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. U.S.A. 90, 2112–2116. doi: 10.1073/pnas.90.6.2112
Lu, K. P., Finn, G., Lee, T. H., and Nicholson, L. K. (2007). Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 3:619. doi: 10.1038/nchembio.2007.35
Lu, K. P., Liou, Y. C., and Zhou, X. Z. (2002). Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 12, 164–172. doi: 10.1016/S0962-8924(02)02253-5
Luan, S., Lane, W. S., and Schreiber, S. L. (1994). pCyP B: A chloroplast-localized, heat shock-responsive cyclophilin from fava bean. Plant Cell 6, 885–892. doi: 10.1105/tpc.6.6.885
Lygerou, Z., Pluk, H., van Venrooij, W. J., and Séraphin, B. (1996). hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 15, 5936–5948. doi: 10.1002/j.1460-2075.1996.tb00980.x
Ma, D., Hong, X., Raghavan, N., Scott, A. L., McCarthy, J. S., Nutman, T. B., et al. (1996). A cyclosporin A-sensitive small molecular weight cyclophilin of filarial parasites. Mol. Biochem. Parasitol. 79, 235–241. doi: 10.1016/0166-6851(96)02654-0
Ma, X., Song, L., Yang, Y., and Liu, D. (2013). A gain-of-function mutation in the ROC1 gene alters plant architecture in Arabidopsis. New Phytol. 197, 751–762. doi: 10.1111/nph.12056
Mainali, H. R., Chapman, P., and Dhaubhadel, S. (2014). Genome-wide analysis of cyclophilin gene family in soybean (Glycine max). BMC Plant Biol. 14:282. doi: 10.1186/s12870-014-0282-7
Majorek, K. A., Dunin-Horkawicz, S., Steczkiewicz, K., Muszewska, A., Nowotny, M., Ginalski, K., et al. (2014). The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 42, 4160–4179. doi: 10.1093/nar/gkt1414
Manteca, A., Kamphausen, T., Fanghanel, J., Fischer, G., and Sanchez, J. (2004). Cloning and characterization of a Streptomyces antibioticus ATCC11891 cyclophilin related to gram negative bacteria cyclophilins. FEBS Lett. 572, 19–26. doi: 10.1016/j.febslet.2004.06.091
Marín-Menéndez, A., Monaghan, P., and Bell, A. (2012). A family of cyclophilin-like molecular chaperones in Plasmodium falciparum. Mol. Biochem. Parasitol. 184, 44–47. doi: 10.1016/j.molbiopara.2012.04.006
Marivet, J., Margis-Pinheiro, M., Frendo, P., and Burkard, G. (1994). Bean cyclophilin gene expression during plant development and stress conditions. Plant Mol. Biol. 26, 1181–1189. doi: 10.1007/BF00040698
Matouschek, A., Rospert, S., Schmid, K., Glick, B. S., and Schatz, G. (1995). Cyclophilin catalyzes protein folding in yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 92, 6319–6323. doi: 10.1073/pnas.92.14.6319
Mayr, C., Richter, K., Lilie, H., and Buchner, J. (2000). Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275, 34140–34146. doi: 10.1074/jbc.M005251200
McLysaght, A., Enright, A. J., Skrabanek, L., and Wolfe, K. H. (2000). Estimation of synteny conservation and genome compaction between pufferfish (Fugu) and human. Yeast 17, 22–36. doi: 10.1002/(sici)1097-0061(200004)17:1<22::aid-yea5>3.0.co;2-s
McMullen, B. A., Fujikawa, K., and Davie, E. W. (1991). Location of the disulfide bonds in human plasma prekallikrein: the presence of four novel apple domains in the amino-terminal portion of the molecule. Biochemistry 30, 2050–2056. doi: 10.1021/bi00222a007
Meza-Zepeda, L. A., Baudo, M. M., Palva, E. T., and Heino, P. (1998). Isolation and characterization of a cDNA corresponding to a stress-activated cyclophilin gene in Solanum commersonii. J. Exp. Bot. 49, 1451–1452. doi: 10.1093/jxb/49.325.1451
Miele, R., Borro, M., Mangoni, M. L., Simmaco, M., and Barra, D. (2003). A peptidyl-prolyl cis-trans isomerase from Xenopus laevis skin: cloning, biochemical characterization and putative role in the secretion. Peptides 24, 1713–1721. doi: 10.1016/j.peptides.2003.07.024
Miller, J. M., and Enemark, E. J. (2016). Fundamental characteristics of AAA+ protein family structure and function. Archaea 2016:Article ID 9294307, 12 pages. doi: 10.1155/2016/9294307
Mo, C., Xie, C., Wang, G., Liu, J., Hao, Q., Xiao, X., et al. (2019). Genome-wide identification and characterization of the cyclophilin gene family in the nematophagous fungus Purpureocilliumlilacinum. Int. J. Mol. Sci. 20:241. doi: 10.3390/ijms20122978
Mockaitis, K., and Estelle, M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80. doi: 10.1146/annurev.cellbio.23.090506.123214
Moparthi, S. B., Fristedt, R., Mishra, R., Almstedt, K., Karlsson, M., Hammarstr̈om, P., et al. (2010). Chaperone activity of Cyp18 through hydrophobic condensation that enables rescue of transient misfolded molten globule intermediates. Biochemistry 49, 1137–1145. doi: 10.1021/bi901997q
Moroney, J. V., and Mason, C. B. (1991). The role of the chloroplast in inorganic carbon acquisition by Chlamydomonas reinhardtii. Can. J. Bot. 69, 1017–1024. doi: 10.1139/b91-131
Moroney, J. V., and Ynalvez, R. A. (2007). Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell. 6, 1251–1259. doi: 10.1128/EC.00064-07
Mortillaro, M. J., and Berezney, R. (1998). Matrin CYP, an SR-rich cyclophilin that associates with the nuclear matrix and splicing factors. J. Biol. Chem. 273, 8183–8192. doi: 10.1074/jbc.273.14.8183
Motohashi, K., Koyama, F., Nakanishi, Y., Ueoka-Nakanishi, H., and Hisabori, T. (2003). Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J. Biol. Chem. 278, 31848–31852. doi: 10.1074/jbc.M304258200
Muangprom, A., Thomas, S. G., Sun, T. P., and Osborn, T. C. (2005). A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol. 137, 931–938. doi: 10.1104/pp.104.057646
Nakajima, M., Shimada, A., Takashi, Y., Kim, Y. C., Park, S. H., Ueguchi-Tanaka, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889. doi: 10.1111/j.1365-313X.2006.02748.x
Neer, E. J., Schmidt, C. J., Nambudripad, R., and Smith, T. F. (1994). Erratum: the ancient regulatory-protein family of WD-repeat proteins. Nature 371:812. doi: 10.1038/371812b0
Neuwald, A. F., Aravind, L., Spouge, J. L., and Koonin, E. V. (1999). AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43. doi: 10.1101/gr.9.1.27
Obata, Y., Yamamoto, K., Miyazaki, M., Shimotohno, K., Kohno, S., and Matsuyama, T. (2005). Role of cyclophilin B in activation of interferon regulatory factor-3. J. Biol. Chem. 280, 18355–18360. doi: 10.1074/jbc.M501684200
Obchoei, S., Wongkhan, S., Wongkham, C., Li, M., Yao, Q., and Chen, C. (2009). Cyclophilin A: potential functions and therapeutic target for human cancer. Med. Sci. Monit. 15, 221–232.
Page, A. P., Landry, D., Wilson, G. G., and Carlow, C. K. S. (1995). Molecular characterization of a Cyclosporin A-Insensitive cyclophilin from the parasitic nematode Brugiamalayi. Biochemistry 34, 11545–11550. doi: 10.1021/bi00036a030
Page, A. P., MacNiven, K., and Hengartner, M. O. (1996). Cloning and biochemical characterization of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans. Biochem. J. 317, 179–185. doi: 10.1042/bj3170179
Pahl, A., Gewies, A., and Keller, U. (1997). ScCypB is a novel second cytosolic cyclophilin from Streptomyces chrysomalluswhich is phylogenetically distant from ScCypA. Microbiology 143, 117–126. doi: 10.1099/00221287-143-1-117
Pahl, A., Ühlein, M., Bang, H., Schlumbohm, W., and Keller, U. (1992). Streptomycetespossess peptidyl−prolyl cis−trans isomerases that strongly resemble cyclophilins from eukaryotic organisms. Mol. Microbiol. 6, 3551–3558. doi: 10.1111/j.1365-2958.1992.tb01790.x
Pemberton, T. J., Rulten, S. L., and Kay, J. E. (2003). Identification and characterisation of Schizosaccharomyces pombe cyclophilin 3, a cyclosporin A insensitive orthologue of human USA-CyP. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 786, 81–91. doi: 10.1016/S1570-0232(02)00738-9
Pemberton, T. J., and Kay, J. E. (2005). The cyclophilin repertoire of the fission yeast Schizosaccharomyces pombe. Yeast 22, 927–945. doi: 10.1002/yea.1288
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 1, 9–20. doi: 10.1038/nrm1547
Pogorelko, G. V., Mokryakova, M., Fursova, O. V., Abdeeva, I., Piruzian, E. S., and Bruskin, S. A. (2014). Characterization of three Arabidopsis thaliana immunophilin genes involved in the plant defense response against Pseudomonas syringae. Gene 538, 12–22. doi: 10.1016/j.gene.2014.01.029
Ponmani, T., Guo, R., and Ki, J. S. (2015). A novel cyclophilin gene from the dinoflagellate Prorocentrum minimum and its possible role in the environmental stress response. Chemosphere 139, 260–267. doi: 10.1016/j.chemosphere.2015.06.036
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Seay, M., Gerstein, M., et al. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 104, 4730–4735. doi: 10.1073/pnas.0611615104
Prunet, N., Morel, P., Thierry, A. M., Eshed, Y., Bowman, J. L., Negrutiu, I., et al. (2008). Rebelote, Squint, and Ultrapetala1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20, 901–919. doi: 10.1105/tpc.107.053306
Quail, P. H. (2005). “Phytochrome overview,” in Light Sensing in Plants, eds M. Wada, K. Shimazaki, and M. Iino (Cham: Springer), 21–35. doi: 10.1007/4-431-27092-2_2
Rahfeld, J. U., Rücknagel, K. P., Schelbert, B., Ludwig, B., Hacker, J., Mann, K., et al. (1994). Confirmation of the existence of a third family among peptidyl-prolyl cis-trans isomerases Amino acid sequence and recombinant production of parvulin. FEBS Lett. 352, 180–184. doi: 10.1016/0014-5793(94)00932-5
Ramachandran, G. N., and Sasisekharan, V. (1968). “Conformation of polypeptides and proteins,” in Advances in Protein Chemistry, eds C. B. Anfinsen, M. L. Anson, J. T. Edsall, F. M. Richards, and D. S. Eisenberg (New York, NY: Academic Press), 283–437. doi: 10.1016/S0065-3233(08)60402-7
Rascher, C., Pahl, A., Pecht, A., Brune, K., Solbach, W., and Bang, H. (1998). Leishmania major parasites express cyclophilin isoforms with an unusual interaction with calcineurin. Biochem. J. 334, 659–667. doi: 10.1042/bj3340659
Reed, J. W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. doi: 10.1016/S1360-1385(01)02042-8
Rinfret, A., Collins, C., Ménard, R., and Anderson, S. K. (1994). The N-terminal cyclophilin-homologous domain of a 150-kilodalton tumor recognition molecule exhibits both peptidyl-prolyl cis-trans isomerase and chaperone activities. Biochemistry 33, 1668–1673. doi: 10.1021/bi00173a008
Romano, P. G. N., Edvardsson, A., Ruban, A. V., Andersson, B., Vener, A. V., Gray, J. E., et al. (2004a). Arabidopsis AtCYP20-2 is a light-regulated cyclophilin-type peptidyl-prolyl cis-trans isomerase associated with the photosynthetic membranes. Plant Physiol. 134, 1244–1247. doi: 10.1104/pp.104.041186
Romano, P. G. N., Horton, P., and Gray, J. E. (2004b). The Arabidopsis cyclophilin gene family. Plant Physiol. 134, 1268–1282. doi: 10.1104/pp.103.022160
Roset, M. S., Fernández, L. G., DelVecchio, V. G., and Briones, G. (2013). Intracellularly induced cyclophilins play an important role in stress adaptation and virulence of Brucella abortus. Infect. Immun. 81, 521–530. doi: 10.1128/IAI.01125-12
Roy, S. W., and Gilbert, W. (2006). The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–213. doi: 10.1038/nrg1807
Roydon Price, E., Zydowsky, L. D., Jin, M., Hunter Baker, C., Mckeon, F. D., and Walsh, C. T. (1991). Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc. Natl. Acad. Sci. U.S.A. 88, 1903–1907. doi: 10.1073/pnas.88.5.1903
Ruan, S. L., Ma, H. S., Wang, S. H., Fu, Y. P., Xin, Y., Liu, W. Z., et al. (2011). Proteomic identification of OsCYP2, a rice cyclophilin that confers salt tolerance in rice (Oryza sativa L.) seedlings when overexpressed. BMC Plant Biol. 11:34. doi: 10.1186/1471-2229-11-34
Ruegger, M., Dewey, E., Gray, W. M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207. doi: 10.1101/gad.12.2.198
Saito, T., Niwa, Y., Ashida, H., Tanaka, K., Kawamukai, M., Matsuda, H., et al. (1999a). Expression of a gene for cyclophilin which contains an amino-terminal endoplasmic reticulum-targeting signal. Plant Cell Physiol. 40, 77–87. doi: 10.1093/oxfordjournals.pcp.a029477
Saito, T., Tadakuma, K., Takahashi, N., Ashida, H., Tanaka, K., Kawamukai, M., et al. (1999b). Two cytosolic cyclophilin genes of Arabidopsis thaliana differently regulated in temporal and organ-specific expression. Biosci. Biotechnol. Biochem. 63, 632–637. doi: 10.1271/bbb.63.632
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Schiene-Fischer, C. (2015). Multidomain peptidyl-prolyl cis-trans isomerases. Biochim. Biophys. Acta Gen. Subj. 10, 2005–2016. doi: 10.1016/j.bbagen.2014.11.012
Schmid, F. X., Mayr, L. M., Mucke, M., and Schonbrunner, E. R. (1993). Prolyl isomerases: role in protein folding. Adv. Protein Chem. 44, 25–66. doi: 10.1016/S0065-3233(08)60563-X
Schmidt, B., Tradler, T., Rahfeld, J. U., Ludwig, B., Jain, B., Mann, K., et al. (1996). A cyclophilin-like peptidyl-propyl cis-trans isomerase from Legionella pneumophila – characterization, molecular cloning and overexpression. Mol. Microbiol. 21, 1147–1160. doi: 10.1046/j.1365-2958.1996.00061.x
Scholze, C., Peterson, A., Diettrich, B., and Luckner, M. (1999). Cyclophilin isoforms from Digitalis lanata. Sequences and expression during embryogenesis and stress. J. Plant Physiol. 155, 212–219. doi: 10.1016/S0176-1617(99)80009-1
Schonbrunner, E. R., Mayer, S., Tropschug, M., Fischer, G., Takahashi, N., and Schmid, F. X. (1991). Catalysis of protein folding by cyclophilins from different species. J. Biol. Chem. 266, 3630–3635.
Schreiber, S. L. (1991). Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251, 283–287. doi: 10.1126/science.1702904
Schubert, A., and Grimm, S. (2004). Cyclophilin D, a component of the permeability transition-pore, is an apoptosis repressor. Cancer Res. 64, 85–93. doi: 10.1158/0008-5472.CAN-03-0476
Schulz, G. E., and Schirmer, R. H. (2013). Principles of Protein Structure. Berlin: Springer Science & Business Media.
Sekhar, K., Priyanka, B., Reddy, V. D., and Rao, K. V. (2010). Isolation and characterization of a pigeonpea cyclophilin (CcCYP) gene, and its over-expression in Arabidopsis confers multiple abiotic stress tolerancepce. Plant Cell Environ. 33, 1324–1338. doi: 10.1111/j.1365-3040.2010.02151.x
Sekhon, S. S., Kaur, H., Dutta, T., Singh, K., Kumari, S., Kang, S., et al. (2013). Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCYPA-1. Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 555–563. doi: 10.1107/S0907444912051529
Shapiguzov, A., Edvardsson, A., and Vener, A. V. (2006). Profound redox sensitivity of peptidyl-prolyl isomerase activity in Arabidopsis thylakoid lumen. FEBS Lett. 580, 3671–3676. doi: 10.1016/j.febslet.2006.05.054
Sharma, A. D., and Singh, P. (2003a). Comparative studies on drought-induced changes in peptidyl prolyl cis-trans isomerase activity in drought-tolerant and susceptible cultivars of Sorghum bicolor. Curr. Sci. 84, 911–918.
Sharma, A. D., and Singh, P. (2003b). Effect of water stress on expression of a 20 kD cyclophilin-like protein in drought susceptible and tolerant cultivars of sorghum. J. Plant Biochem. Biotechnol. 12, 77–80. doi: 10.1007/bf03263165
Sharma, A. D., Wajapeyee, N., Yadav, V., and Singh, P. (2003). Stress-induced changes in peptidyl-prolyl cis-trans isomerase activity of Sorghum bicolor seedlings. Biol. Plant 47, 367–371. doi: 10.1023/B:BIOP.0000023879.74558.48
Sheldon, P. S., and Venis, M. A. (1996). Purification and characterization of cytosolic and microsomal cyclophilins from maize (Zea mays). Biochem. J. 315, 965–970. doi: 10.1042/bj3150965
Shewry, P. R., Halford, N. G., Belton, P. S., and Tatham, A. S. (2002). The structure and properties of gluten: an elastic protein from wheat grain. Philos. Trans. R. Soc. B Biol. Sci. 357, 133–142. doi: 10.1098/rstb.2001.1024
Siekierka, J. J., Staruch, M. J., Hung, S. H. Y., and Sigal, N. H. (1989). FK-506, a potent novel immunosuppressive agent, binds to a cytosolic protein which is distinct from the cyclosporin A-binding protein, cyclophilin. J. Immunol. 143, 1580–1583.
Singh, H., Kaur, K., Singh, S., Kaur, P., and Singh, P. (2019). Genome-wide analysis of cyclophilin gene family in wheat and identification of heat stress responsive members. Plant Gene 19:100197. doi: 10.1016/j.plgene.2019.100197
Singh, K., Tzelepis, G., Zouhar, M., Ryšánek, P., and Dixelius, C. (2018). The immunophilin repertoire of Plasmodiophorabrassicae and functional analysis of PbCYP3 cyclophilin. Mol. Genet. Genomics 293, 381–390. doi: 10.1007/s00438-017-1395-0
Singh, K., Zouhar, M., Mazakova, J., and Rysanek, P. (2014). Genome wide identification of the immunophilin gene family in Leptosphaeria maculans: a causal agent of blackleg disease in oilseed rape (Brassica napus). OMICS 18, 645–657. doi: 10.1089/omi.2014.0081
Sirpiö, S., Holmström, M., Battchikova, N., and Aro, E. M. (2009). AtCYP20-2 is an auxiliary protein of the chloroplast NAD(P)H dehydrogenase complex. FEBS Lett. 583, 2355–2358. doi: 10.1016/j.febslet.2009.06.031
Sirpiö, S., Khrouchtchova, A., Allahverdiyeva, Y., Hansson, M., Fristedt, R., Vener, A. V., et al. (2008). AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55, 639–651. doi: 10.1111/j.1365-313X.2008.03532.x
Skružný, M., Ambrozková, M., Fuková, I., Martånková, K., Blahùšková, A., Hamplová, L., et al. (2001). Cyclophilins of a novel subfamily interact with SNW/SKIP coregulator in Dictyosteliumdiscoideum and Schizosaccharomyces pombe. Biochim. Biophys. Acta Gene Struct. Expr. 1521, 146–151. doi: 10.1016/S0167-4781(01)00301-3
Smalle, J., and Vierstra, R. D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. doi: 10.1146/annurev.arplant.55.031903.141801
Smith, M. R., Willmann, M. R., Wu, G., Berardini, T. Z., Möller, B., Weijers, D., et al. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5424–5429. doi: 10.1073/pnas.0812729106
Snedden, W. A., and Fromm, H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151, 35–66. doi: 10.1046/j.1469-8137.2001.00154.x
Somanchi, A., and Moroney, J. V. (1999). As Chlamydomonas reinhardtii acclimates to low-CO2 conditions there is an increase in cyclophilin expression. Plant Mol. Biol. 40, 1055–1062. doi: 10.1023/A:1006262123918
Stamnes, M. A., Shieh, B. H., Chuman, L., Harris, G. L., and Zuker, C. S. (1991). The cyclophilin homolog ninaA is a tissue-specific integral membrane protein required for the proper synthesis of a subset of Drosophila rhodopsins. Cell 65, 219–227. doi: 10.1016/0092-8674(91)90156-S
Stangeland, B., Nestestog, R., Grini, P. E., Skrbo, N., Berg, A., Salehian, Z., et al. (2005). Molecular analysis of Arabidopsis endosperm and embryo promoter trap lines: reporter-gene expression can result from T-DNA insertions in antisense orientation, in introns and in intergenic regions, in addition to sense insertion at the 5’ end of genes. J. Exp. Bot. 56, 2495–2505. doi: 10.1093/jxb/eri242
Stirnimann, C. U., Petsalaki, E., Russell, R. B., and Müller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574. doi: 10.1016/j.tibs.2010.04.003
Sun, T. P. (2008). Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6:e0103. doi: 10.1199/tab.0103
Sun, T. P. (2010). Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 154, 567–570. doi: 10.1104/pp.110.161554
Sykes, K., Gething, M. J., and Sambrook, J. (1993). Proline isomerases function during heat shock. Proc. Natl. Acad. Sci. U.S.A. 90, 5853–5857. doi: 10.1073/pnas.90.12.5853
Szemenyei, H., Hannon, M., and Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. doi: 10.1126/science.1151461
Takaki, Y., Muta, T., and Iwanaga, S. (1997). A peptidyl-prolyl cis-transisomerase (cyclophilin g) in regulated secretory granules. J. Biol. Chem. 272, 28615–28621. doi: 10.1074/jbc.272.45.28615
Tan, X., Calderon-Villalobos, L. I. A., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Thai, V., Renesto, P., Fowler, C. A., Brown, D. J., Davis, T., Gu, W., et al. (2008). Structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the Mimivirus. J. Mol. Biol. 378, 71–86. doi: 10.1016/j.jmb.2007.08.051
Theologis, A., Huynh, T. V., and Davis, R. W. (1985). Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J. Mol. Biol. 183, 53–68. doi: 10.1016/0022-2836(85)90280-3
Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. doi: 10.1105/tpc.017384
Townsend, D. M. (2007). S-glutathionylation: Indicator of cell stress and regulator of the unfolded protein response. Mol. Interv. 7:313. doi: 10.1124/mi.7.6.7
Trivedi, D. K., Ansari, M. W., Dutta, T., Singh, P., and Tuteja, N. (2013c). Molecular characterization of cyclophilin A-like protein from Piriformosporaindica for its potential role to abiotic stress tolerance in E. coli. BMC Res. Notes 6:555. doi: 10.1186/1756-0500-6-555
Trivedi, D. K., Ansari, M. W., and Tuteja, N. (2013a). Multiple abiotic stress responsive rice cyclophilin: (OsCYP-25) mediates a wide range of cellular responses. Commun. Integr. Biol. 6:e25260. doi: 10.4161/cib.25260
Trivedi, D. K., Bhatt, H., Pal, R. K., Tuteja, R., Garg, B., Johri, A. K., et al. (2013b). Structure of RNA-interacting Cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants. Sci. Rep. 3:3001. doi: 10.1038/srep03001
Trivedi, D. K., Yadav, S., Vaid, N., and Tuteja, N. (2012). Genome wide analysis of cyclophilin gene family from rice and Arabidopsis and its comparison with yeast. Plant Signal. Behav. 7, 1653–1666. doi: 10.4161/psb.22306
Trupkin, S. A., Mora-García, S., and Casal, J. J. (2012). The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 71, 712–723. doi: 10.1111/j.1365-313X.2012.05013.x
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Ulrich, L. E., and Zhulin, I. B. (2005). Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics 21, iii45–iii48. doi: 10.1093/bioinformatics/bti1204
Van Nocker, S., and Ludwig, P. (2003). The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics. 4:50. doi: 10.1186/1471-2164-4-50
Vasudevan, D., Fu, A., Luan, S., and Swaminathan, K. (2012). Crystal structure of Arabidopsis cyclophilin38 reveals a previously uncharacterized immunophilin fold and a possible autoinhibitory mechanism. Plant Cell 24, 2666–2674. doi: 10.1105/tpc.111.093781
Vasudevan, D., Gopalan, G., Kumar, A., Garcia, V. J., Luan, S., and Swaminathan, K. (2015). Plant immunophilins: a review of their structure-function relationship. Biochim. Biophys. Acta Gen. Subj. 1850, 2145–2158. doi: 10.1016/j.bbagen.2014.12.017
Vener, A. V., Rokka, A., Fulgosi, H., Andersson, B., and Hermann, R. G. (1999). A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38, 14955–14965. doi: 10.1021/bi990971v
Virdi, A. S., Singh, S., and Singh, P. (2015). Abiotic stress responses in plants: roles of calmodulin-regulated proteins. Front. Plant Sci. 6:809. doi: 10.3389/fpls.2015.00809
Vojta, L., TomašićPaić, A., Horvat, L., Rac, A., Lepeduš, H., and Fulgosi, H. (2019). Complex lumenal immunophilin AtCYP38 influences thylakoid remodelling in Arabidopsis thaliana. J. Plant Physiol. 243:153048. doi: 10.1016/j.jplph.2019.153048
Wandinger, S. K., Richter, K., and Buchner, J. (2008). The Hsp90 chaperone machinery. J. Biol. Chem. 283, 18473–18477. doi: 10.1074/jbc.R800007200
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, P., and Heitman, J. (2005). The cyclophilins. Genome Biol. 7, 1–6. doi: 10.1186/gb-2005-6-7-226
Wang, Q., Wang, Y., Chai, W., Song, N., Wang, J., Cao, L., et al. (2017). Systematic analysis of the maize cyclophilin gene family reveals ZmCYP15 involved in abiotic stress response. Plant Cell. Tissue Organ Cult. 128, 543–561. doi: 10.1007/s11240-016-1132-0
Wang, R., and Estelle, M. (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51–58. doi: 10.1016/j.pbi.2014.06.006
Wang, Y., Han, R., Zhang, W., Yuan, Y., Zhang, X., Long, Y., et al. (2008). Human CyP33 binds specifically to mRNA and binding stimulates PPIase activity of hCyP33. FEBS Lett. 582, 835–839. doi: 10.1016/j.febslet.2008.01.055
Wang, Y., and Li, J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
Wang, Y., Zeng, L., and Xing, D. (2015). ROS-mediated enhanced transcription of CYP38 promotes the plant tolerance to high light stress by suppressing GTPase activation of PsbO2. Front. Plant Sci. 6:777. doi: 10.3389/fpls.2015.00777
Watashi, K., Ishii, N., Hijikata, M., Inoue, D., Murata, T., Miyanari, Y., et al. (2005). Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19, 111–122. doi: 10.1016/j.molcel.2005.05.014
Weighardt, F., Cobianchi, F., Cartegni, L., Chiodi, I., Villa, A., Riva, S., et al. (1999). A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 112, 1465–1476.
Wiborg, J., O’Shea, C., and Skriver, K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413, 447–457. doi: 10.1042/BJ20071568
Wilkie, G. S., Dickson, K. S., and Gray, N. K. (2003). Regulation of mRNA translation by 5’- and 3’-UTR-binding factors. Trends Biochem. Sci. 28, 182–188. doi: 10.1016/S0968-0004(03)00051-3
Yamaguchi, R., Hosaka, M., Torii, S., Hou, N., Saito, N., Yoshimoto, Y., et al. (2011). Cyclophilin C-associated protein regulation of phagocytic functions via NFAT activation in macrophages. Brain Res. 1397, 55–65. doi: 10.1016/j.brainres.2011.03.036
Yamamoto, K. T., Mori, H., and Imaseki, H. (1992). cDNA cloning of indole-3-acetic acid-regulated genes: Aux22 and SAUR from mung bean (Vigna radiata) hypocotyl tissue. Plant Cell Physiol. 33, 93–97. doi: 10.1093/oxfordjournals.pcp.a078225
Yan, H., Bo, Z., Wei, H., Yuzhe, N., and Yuhua, L. (2018). Expression characterisation of cyclophilin BrROC1 during light treatment and abiotic stresses response in Brassica rapa subsp. rapa ‘Tsuda’. Funct. Plant Biol. 12, 1223–1232. doi: 10.1071/fp18029
Yang, H., Xu, L., Cui, H., Zhong, B., Liu, G., and Shi, H. (2013). Low nitrogen-induced expression of cyclophilin in Nicotiana tabacum. J. Plant Res. 126, 121–129. doi: 10.1007/s10265-012-0499-1
Yoon, D. H., Lee, S. S., Park, H. J., Lyu, J., Chong, W. S., Liu, J. R., et al. (2016). Overexpression of OsCYP19-4 increases tolerance to cold stress and enhances grain yield in rice (Oryza sativa). J. Exp. Bot. 67, 69–82. doi: 10.1093/jxb/erv421
Yoshida, H., Park, S. Y., Oda, T., Akiyoshi, T., Sato, M., Shirouzu, M., et al. (2015). A novel 3’ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 29, 1649–1660. doi: 10.1101/gad.267104.115
Young, E. J. (1995). An overview of human brucellosis. Clin. Infect. Dis. 21, 283–290. doi: 10.1093/clinids/21.2.283
Zhang, H., Wang, J., Li, S., Wang, S., Liu, M., Wang, W., et al. (2017). Molecular cloning, expression, purification and functional characterization of an antifungal cyclophilin protein from Panax ginseng. Biomed. Rep. 7, 527–531. doi: 10.3892/br.2017.998
Zhang, X. C., Wang, W. D., Wang, J. S., and Pan, J. C. (2013a). PPIase independent chaperone-like function of recombinant human Cyclophilin A during arginine kinase refolding. FEBS Lett. 587, 666–672. doi: 10.1016/j.febslet.2013.01.028
Zhang, Y., Li, B., Xu, Y., Li, H., Li, S., Zhang, D., et al. (2013b). The cyclophilin CYP20-2 modulates the conformation of BRASSINAZOLE-RESISTANT1, which binds the promoter of FLOWERING LOCUS D to regulate flowering in Arabidopsis. Plant Cell 25, 2504–2521. doi: 10.1105/tpc.113.110296
Zhao, B., Li, H., Li, J., Wang, B., Dai, C., Wang, J., et al. (2017). Brassica napus DS-3, encoding a DELLA protein, negatively regulates stem elongation through gibberellin signaling pathway. Theor. Appl. Genet. 130, 727–741. doi: 10.1007/s00122-016-2846-4
Zhao, Y., Yongquan, C., Mike, S., Gunter, F., and Hengming, K. (1997). Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity. Structure 1, 139–146. doi: 10.1016/S0969-2126(97)00172-X
Zheng, H., Li, S., Ren, B., Zhang, J., Ichii, M., Taketa, S., et al. (2013). LATERAL ROOTLESS2, a cyclophilin protein, regulates lateral root initiation and auxin signaling pathway in rice. Mol. Plant. 6, 1719–1721. doi: 10.1093/mp/sst052
Zhu, C., Wang, Y., Li, Y., Bhatti, K. H., Tian, Y., and Wu, J. (2011). Overexpression of a cotton cyclophilin gene (GhCyp1) in transgenic tobacco plants confers dual tolerance to salt stress and Pseudomonas syringaepv. tabaci infection. Plant Physiol. Biochem. 49, 1264–1271. doi: 10.1016/j.plaphy.2011.09.001
Zuehlke, A. D., Wren, N., Tenge, V., and Johnson, J. L. (2013). Interaction of heat shock protein 90 and the co-chaperone Cpr6 with Ura2, a bifunctional enzyme required for pyrimidine biosynthesis. J. Biol. Chem. 288, 27406–27414. doi: 10.1074/jbc.M113.504142
Zydowsky, L. D., Etzkorn, F. A., Chang, H. Y., Ferguson, S. B., Stolz, L. A., Ho, S. I., et al. (1992b). Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1, 1092–1099. doi: 10.1002/pro.5560010903
Keywords: cyclophilin, FKBP, hormones, immunophilins, peptidyl-prolyl cis-trans isomerase, stress
Citation: Singh H, Kaur K, Singh M, Kaur G and Singh P (2020) Plant Cyclophilins: Multifaceted Proteins With Versatile Roles. Front. Plant Sci. 11:585212. doi: 10.3389/fpls.2020.585212
Received: 20 July 2020; Accepted: 22 September 2020;
Published: 22 October 2020.
Edited by:
Markus Geisler, Université de Fribourg, SwitzerlandReviewed by:
Wayne Snedden, Queen’s University, CanadaDerek Gingerich, University of Wisconsin–Eau Claire, United States
Copyright © 2020 Singh, Kaur, Singh, Kaur and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prabhjeet Singh, singhprabhjeet62@gmail.com
†These authors have contributed equally to this work
 Harpreet Singh
Harpreet Singh Kirandeep Kaur
Kirandeep Kaur Mangaljeet Singh
Mangaljeet Singh Gundeep Kaur1,3†
Gundeep Kaur1,3† Prabhjeet Singh
Prabhjeet Singh