- Key Laboratory of Plant Resources Conservation and Utilization, College of Biological Resources and Environmental Sciences, Jishou University, Jishou, China
The subfamily Nolinoideae of Asparagaceae is an extremely morphologically heterogeneous group, which is comprised of seven lineages, formerly known as Eriospermaceae, Polygonateae, Ophiopogoneae, Convallarieae, Ruscaceae s.s., Dracaenaceae, and Nolinaceae from different families or even orders. Their drastically divergent morphologies and low level of molecular resolution have hindered our understanding on their evolutionary history. To resolve reliable and clear phylogenetic relationships of the Nolinoideae, a phylogenetic study was conducted based on transcriptomic sequencing of 15 species representing all the seven lineages. A dataset containing up to 2,850,331 sites across 2,126 genes was analyzed using both concatenated and coalescent methods. Except for Eriospermum as outgroup, the transcriptomic data strongly resolved the remaining six lineages into two groups, one is a paraphyletic grade including the woody lineages of dracaenoids, ruscoids, and nolinoids and a monophyletic herbaceous clade. Within the herbaceous group, the Ophiopogoneae + Theropogon is sister to a clade that is composed of Convallarieae and the monophyletic Polygonateae. Our work provides a first robust deep relationship of the highly heterogeneous Nolinoideae and paves the way for further investigations of its complex evolution.
Introduction
Phylogenetic studies have greatly changed our understanding on plant classification and evolution. To accord with molecular phylogenetic results, for example, Asparagales has been recircumscribed as the largest order in the monocotyledons with seven redefined or newly erected families (Chase et al., 1995; Fay et al., 2000; Seberg et al., 2012; Chen et al., 2013). Among them, Asparagaceae s.l. is greatly expanded as a highly complicated group with little shared characters, including seven subfamilies, i.e., Agavoideae, Aphyllanthoideae, Asparagoideae, Brodiaeoideae, Lomandroideae, Nolinoideae, and Scilloideae (Chase et al., 2009). Nolinoideae, formerly known as Ruscaceae s.l. or Convallariaceae s.l., is also a complex group comprised of seven heterogeneous lineages, traditionally known as Eriospermaceae, Polygonateae, Ophiopogoneae, Convallarieae, Ruscaceae s.s., Dracaenaceae, and Nolinaceae, and many of them have been traditionally placed in different families or even orders (Chase et al., 2009; Kim et al., 2010; APGIV, 2016). To avoid taxonomic confusions, Ruscaceae s.s., Dracaenaceae, and Nolinaceae are referred to as the ruscoids, the dracaenoids, and the nolinoids following Rudall et al. (2000) in this study, respectively.
The seven lineages in the Nolinoideae are extremely diverse in morphology, four of which are herbaceous and three are woody-like (Rudall et al., 2000; Yamashita and Tamura, 2000b; Kim et al., 2010). The first includes only Eriospermum Jacq. ex Willd. with about 80 species occurring mostly in southern Africa, which are perennial herbs with dormant structures of tubers, free perianth parts, and the epidermal hairs of the seed, traditionally assigned to its own monotypic family (Dahlgren et al., 1985). The APGIII (2009) system treated Eriospermum as a sister to the remainder in the Nolinoideae. The other three herbaceous groups (i.e., Polygonateae, Ophiopogoneae, and Convallarieae) had been well-known as Convallariaceae s.s. together separated from traditional Liliaceae (Rudall et al., 2000). They are usually rhizomatous and perennial herbs that occurred in the Northern Hemisphere and especially abundant in eastern and southeastern Asia (Dahlgren et al., 1985; Rudall et al., 2000).
Polygonateae is usually recognized into two lineages, one is the axillary-flowered group, including Polygonatum Mill., Disporopsis Hance and Heteropolygonatum M.N. Tamura & Ogisu, and the other is the terminal-flowered of Maianthemum F.H. Wigg. (Meng et al., 2008, 2014). Many published multi-locus analyses have demonstrated the monophyly of the tribe and its inclusion within Nolinoideae (Rudall et al., 2000; Meng et al., 2014), but there are still many controversies, especially the position of Maianthemum lineage (Kim et al., 2010; Floden and Schilling, 2018). The Ophiopogoneae includes Ophiopogon Meisn., Liriope Lour., and Peliosanthes Andrews, usually characterized with a sympodial rhizome, fruits dehisced at an early stage, seeds with sarcotesta, and basic chromosome number of x = 18 (Dahlgren et al., 1985). Molecular phylogenetic analyses strongly supported the monophyly of Ophiopogoneae within Nolinoideae (Kim et al., 2010; Wang et al., 2014; Wang and Yang, 2018). The tribe Convallarieae was expanded to include Aspidistreae (Conran and Tamura, 1998; Yamashita and Tamura, 2000b; Kim et al., 2010). They can be distinguished from the other herbaceous taxa by a short stem with monopodial rhizome and usually berries but rarely drupes (Tricalistra Ridl.) and usually basic chromosome numbers of x = 19 but rarely x = 18 (some Aspidistra Ker Gawl. spp.) (Yamashita and Tamura, 2004).
The woody taxa includes the ruscoids, the dracaenoids, and the nolinoids, usually recognized as different families. The dracaenoids include the genera of Dracaena L. and Sansevieria Thunb. mainly from tropical and subtropical Asia and Africa, which share the synapomorphic characters of berries, have no oils in guard cells, and have mucilage-filled cells with crystal raphides in their vegetative parts (Lu and Morden, 2014; Takawira-Nyenya et al., 2018). The ruscoids are comprised of three small genera from the Mediterranean and West Asia (Ruscus L., Danae Medik., and Semele Kunth) characterized by scale-like leaves, woody stems, berries, and a basic chromosome number of x = 20 (Rudall and Campbell, 1999). The nolinoids were previously represented as Nolinaceae including the four genera Beaucarnea Lem., Calibanus Rose, Dasylirion Zucc., and Nolina Michx. from the southern states of the United States through Mexico into Guatemala (Rojas-Piña et al., 2014). They were excluded from Agavaceae and placed close to Dracaenaceae, featured with arborescent, anomalously woody plants; terminal rosette leaves; and indehiscent fruits (Dahlgren et al., 1985).
The Nolinoideae is proved to be an extremely morphologically heterogeneous group with very few distinguishable synapomorphic characters from the other asparagoid members except for the absence of phytomelan in the seed coat (Rudall et al., 2000; Kim et al., 2010). The taxonomic classification in this group with drastically divergent morphologies has been problematic. Although molecular evidence indicated a well-supported group of the subfamily with the inclusion of the seven lineages as stated above, most internal branches among these seven groups have weak supports, except for Eriospermum that is strongly supported as a sister group to the remaining taxa (Rudall et al., 2000; Kim et al., 2010; Seberg et al., 2012). Low support values have also been observed in the other analyses and are the main argument for grouping all of them into one large family in the APG systems (Pires et al., 2006; Kim et al., 2010).
In a word, Nolinoideae is a well-supported group but without obvious synapomorphies and remains poorly resolved for its deep evolutionary relationships. However, it seems that traditional multi-locus approaches are unable to resolve a reliable and highly confident phylogenetic backbone of the subfamily. Rapid development of the next-generation sequencing technology has made large dataset accessible, allowing high-throughput selection of low or single-copy nuclear genes as phylogenetic markers (Wen et al., 2015a). De novo sequencing of transcriptome among many species has been tested recently as effective phylogenetic approaches (Wickett et al., 2011; Wen et al., 2015a; Ran et al., 2018). Coalescent analyses for species-tree estimation are becoming a dominant approach for reconstructing species histories over multi-locus data for recently diverged species. Recent studies used transcriptome datasets to successfully reconstruct phylogenies of various scales from various genera to angiosperm-wide or even land plants (Wen et al., 2013; Wickett et al., 2014; Zeng et al., 2017; Ran et al., 2018).
This study aims to reconstruct deep phylogenetic relationships of Nolinoideae and provide phylogenetic placement of some uncertain genera using both concatenated and coalescent analyses. Phylogenetic analyses were conducted based on transcriptomic data from representatives of the seven lineages. To avoid stochastic and systematic errors, we sequenced the transcriptomes of all sampled species and used OrthoFinder for orthology prediction, followed with multiple filter procedures.
Materials and Methods
Taxon Sampling and Transcriptome Sequencing
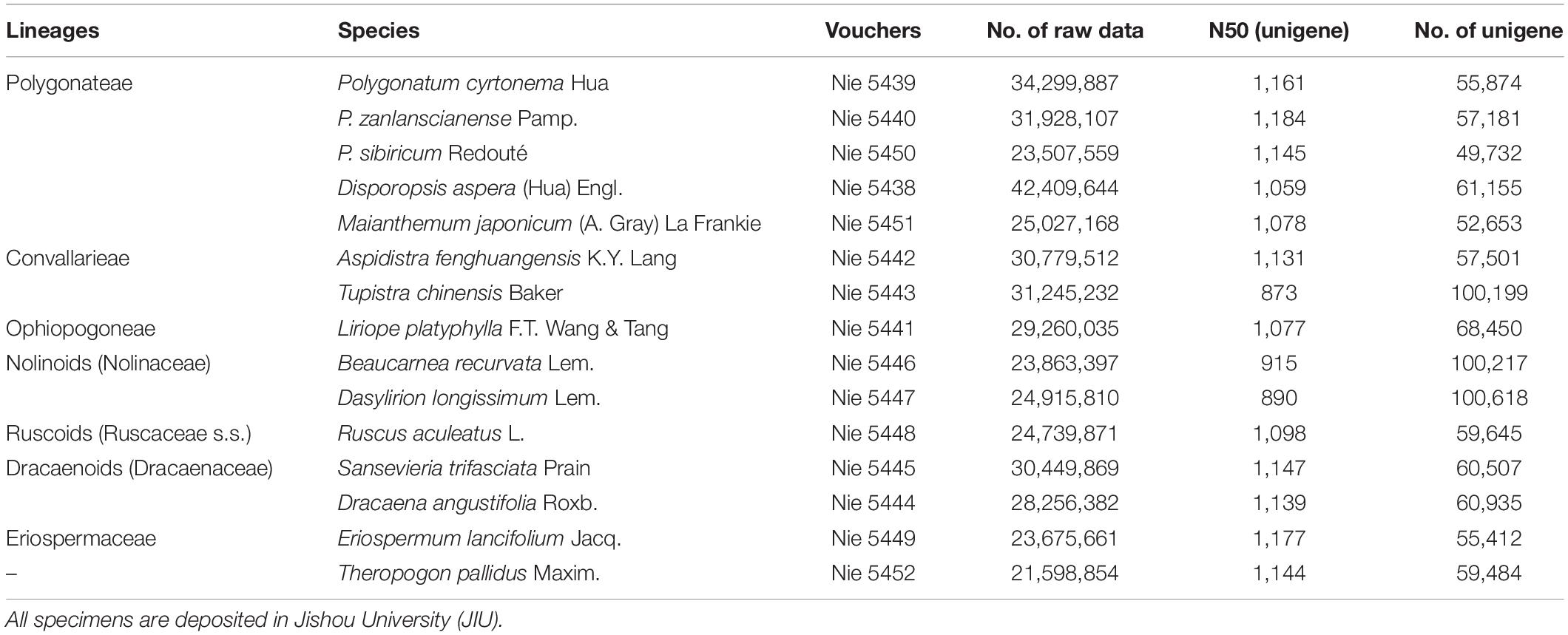
We provide transcriptomic sequencing from all seven lineages within Nolinoideae, including five species from the two lineages of Polygonateae, two from Convallarieae, one of Ophiopogoneae, two from the nolinoids, one of the ruscoids, two of the dracaenoids, and one from Eriospermum (Table 1). In addition, we sequenced Theropogon pallidus collected from southern Xizang of China, which is phylogenetically uncertain by traditional molecular data (Kim et al., 2010). Eriospermum was selected as outgroup as its sister relationship to the rest of the taxa of the subfamily is robustly supported by all previous studies (Fay et al., 2000; Jang and Pfosser, 2002; Kim et al., 2010; Wang et al., 2016). All samples for RNA sequencing were collected from the wild or botanical gardens and cultivated in a greenhouse of Jishou University (Table 1).
Fresh juvenile leaf tissues of each sample were flash-frozen in liquid nitrogen. Total RNA was extracted using the RNA plant Plus Reagent (Tiangen, Beijing, China) and digested by DNase I (Promega, Madison, WI, United States). RNA concentration was measured using NanoDrop 2000 (Thermo), and integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States), following the manufacturer’s recommendations. The transcriptome library was constructed using NEBNext®UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) following the manufacturer’s protocols, and Illumina sequencing was performed by BioMarker (Beijing, China).
Data Assembly and Identification of Orthologs
Raw reads were firstly checked with FastQC1 and trimmed using fastp with a quality filtering cutoff of 20 (Chen et al., 2018). The clean reads were de novo assembled with Trinity version 2.8.4 (Grabherr et al., 2011) using default parameters. Redundancy reduction was done for the raw assemblies, using CD-HIT with a threshold of 20 (Li and Godzik, 2006). For each transcriptome, transcripts were translated into peptides using default settings in TransDecoder version 0.362. The quality and completeness of the final transcriptomes (unigene sets) for all samples were also benchmarked with BUSCO version 3.0.2 according to conserved ortholog content (Waterhouse et al., 2017). The analysis was carried out using the Liliopsida odb10 plant-specific reference database following default parameters of the software (Kriventseva et al., 2018) to obtain single-copy gene dataset with more strict criteria.
The conserved orthogroups (OG) were identified from the sets of translated proteins using OrthoFinder version 2.2.6 (Emms and Kelly, 2015) and further filtered using the following criteria: (i) sequences are missed in at least one species; (ii) the average copy number of an OG is greater than five; and (iii) the median copy number of an OG is greater than two. We used a rigorous comprehensive methodology for quantifying multiple sequence alignment uncertainties with GUIDANCE2 (Sela et al., 2015). The remaining OG sequences that are longer than 100 aligned amino acids (AA) and present in all species were then used to reconstruction preliminary maximum likelihood (ML) trees using IQ-TREE version 1.6.12 (Nguyen et al., 2014). PhyloTreePruner was used to identify the maximally inclusive subtree with each taxon represented by one sequence (Kocot et al., 2013). The predicted AA sequences were aligned with MAFFT version 7 (Katoh and Standley, 2013). The nucleotide coding sequences (CDS) were then aligned by using PAL2NAL according to the corresponding AA alignment (Suyama et al., 2006).
Phylogenetic and Coalescent Analyses
To reconstruct the phylogeny of Nolinoideae, we employed maximum likelihood (ML), Bayesian inference (BI), and coalescent-based methods. CDS and AA sequences of all OGs were concatenated using FASconCAT-G version 1.02 (Kück and Longo, 2014). ML analyses were performed using the parallel version of IQ-TREE. ModelFinder was used in IQ-TREE to select the best model with free-rate heterogeneity based on BIC and AICc scores (Kalyaanamoorthy et al., 2017). Branch support was evaluated with 1,000 ultrafast bootstraps and SH-like approximate likelihood ratio tests using 1,000 replicates (Guindon et al., 2010; Hoang et al., 2017). In the BI analysis, parameters were set to aamodelpr = mixed and rates = gamma for AA sequences and nst = 6 and rates = gamma for nuclear sequences. Four chains were run for 1,000,000 generations and sampled every 1,000 generations with the first ca. 15% of the samples discarded as burn-in.
We used ASTRAL-III version 5.6.3 (Zhang et al., 2018), a quartet-based method under the multispecies coalescent, to estimate the species tree from nuclear gene trees. Each nuclear gene tree was generated with IQ-TREE, including 100 bootstrap replicates using the same parameter setting as above. Supports on the ASTRAL phylogeny were assessed using multi-locus bootstrap with 100 times of the gene tree bootstrap phylogenies and the more recently developed local posterior probability method, which estimates relative quartet support on each branch. In order to reflect uncertainty in gene tree estimates, another ASTRAL analysis was performed using the gene tree nodes collapsed with <50% bootstrap support (BS) with SumTrees.py from DendroPy 4.4.0 (Sukumaran and Holder, 2010).
Because of the debate of Maianthemum being close to Polygonatum group and the phylogenetic uncertainty of Theropogon within the subfamily, an approximately unbiased (AU) test (Shimodaira, 2002) was conducted to test alternative placements of each of them. The AU test compares log-likelihood scores among alternative trees using the CONSEL version 0.20 (Shimodaira and Hasegawa, 2001).
Analyses of Conflicts
The presence of gene tree conflicts and concordance in the pseudocoalescence analyses was checked using PhyParts (Smith et al., 2015). This method allowed us to assess how many genes support or conflict with individual bipartitions within a species tree: if there is a dominant tree topology in the gene trees or, if there is conflict, whether this stems from an alternative tree topology, or from low-frequency alternative gene topologies, or lack of support for conflicting bipartitions. Gene trees used as input for ASTRAL and the resulting species tree generated by the program were rooted to be used as input in PhyParts, which was done using the program pxrr in the package phyx (Brown et al., 2017). Species trees were rooted having Eriospermum as outgroup. We ran PhyParts with the -b option set to 50 so that branches with less than 50% BS in the gene trees would not be considered. The results from PhyParts were used as input in the phypartspiecharts.py script3, to generate a species tree with pie charts in each node showing the proportion of concordant gene trees and conflicting topologies.
Results
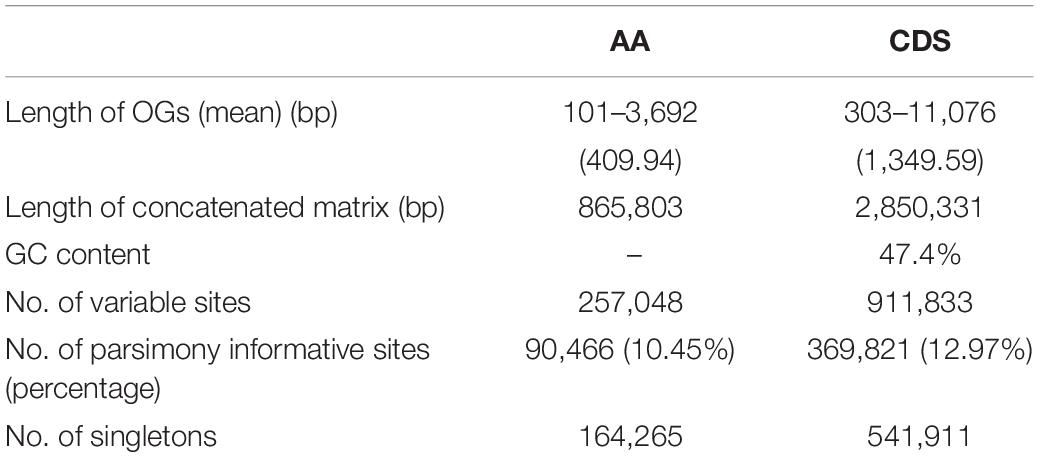
A summary of the assembly statistics of the 15 transcriptomic data is shown in Table 1. After ORF prediction and redundancy reduction, 35,232–58,558 unigenes were retained. We obtained 2,126 putative one-to-one orthogroups (each taxon has only one sequence) after further stringent filtering. The mean proportion of missing data was 2.06% (0–20.77% in each OG) (Figure 1). The aligned length of the 2,126 concatenated AA sequences was 865,803 bp with 257,048 variable sites and 90,466 parsimony informative sites, and the aligned CDS sequences contain 2,850,331 bp with 911,833 variable sites and 369,821 parsimony informative sites (Table 2).
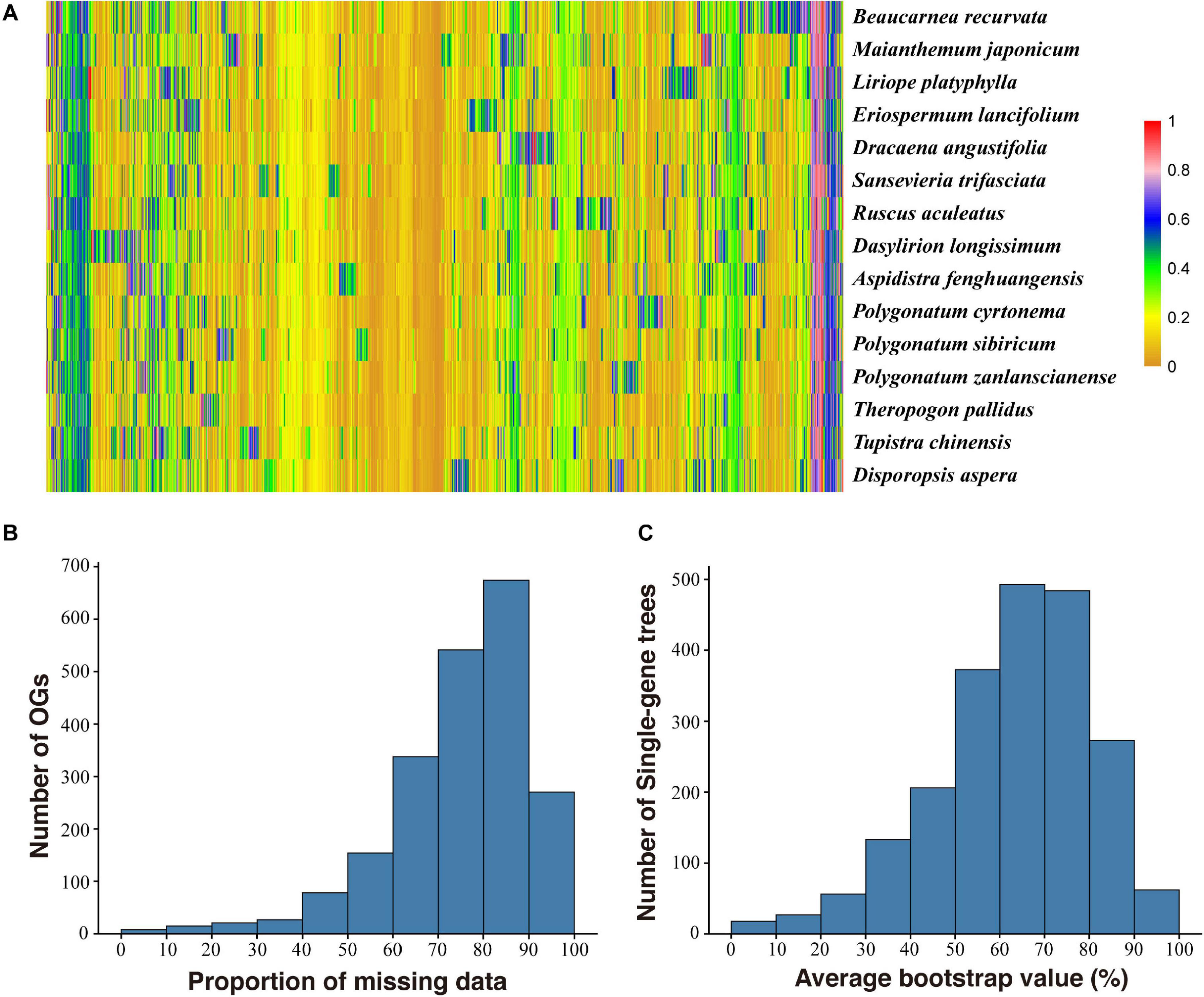
Figure 1. Frequency of missing data of each orthogroup (OG) in each taxon (A), distribution of missing data in the OGs (B), and average bootstrap value of single gene tree (C). Each row corresponds to a taxon and each column corresponds to an OG in (A).
Both the concatenation based on CDS and AA matrices yielded similar topologies (Figure 2), with all nodes supported by 100% BS values and posterior probability (PP) of 1.00, respectively (Supplementary Figures 1–3). The sister relationship between Theropogon and Ophiopogoneae has been robustly supported with BS = 100% and PP = 1.00, but with weak SH-like supports in both AA and CDS datasets (Supplementary Figures 1, 2). The coalescent analyses from ASTRAL produced similar topologies to those of concatenation results with difference from the placement of Theropogon (Supplementary Figures 4, 5). A total of 760 OGs were produced using the BUSCO approach. The ML tree of the concatenated sequences and the species tree (Supplementary Figures 6, 7) are similar to those of the large dataset with 2,126 OGs.
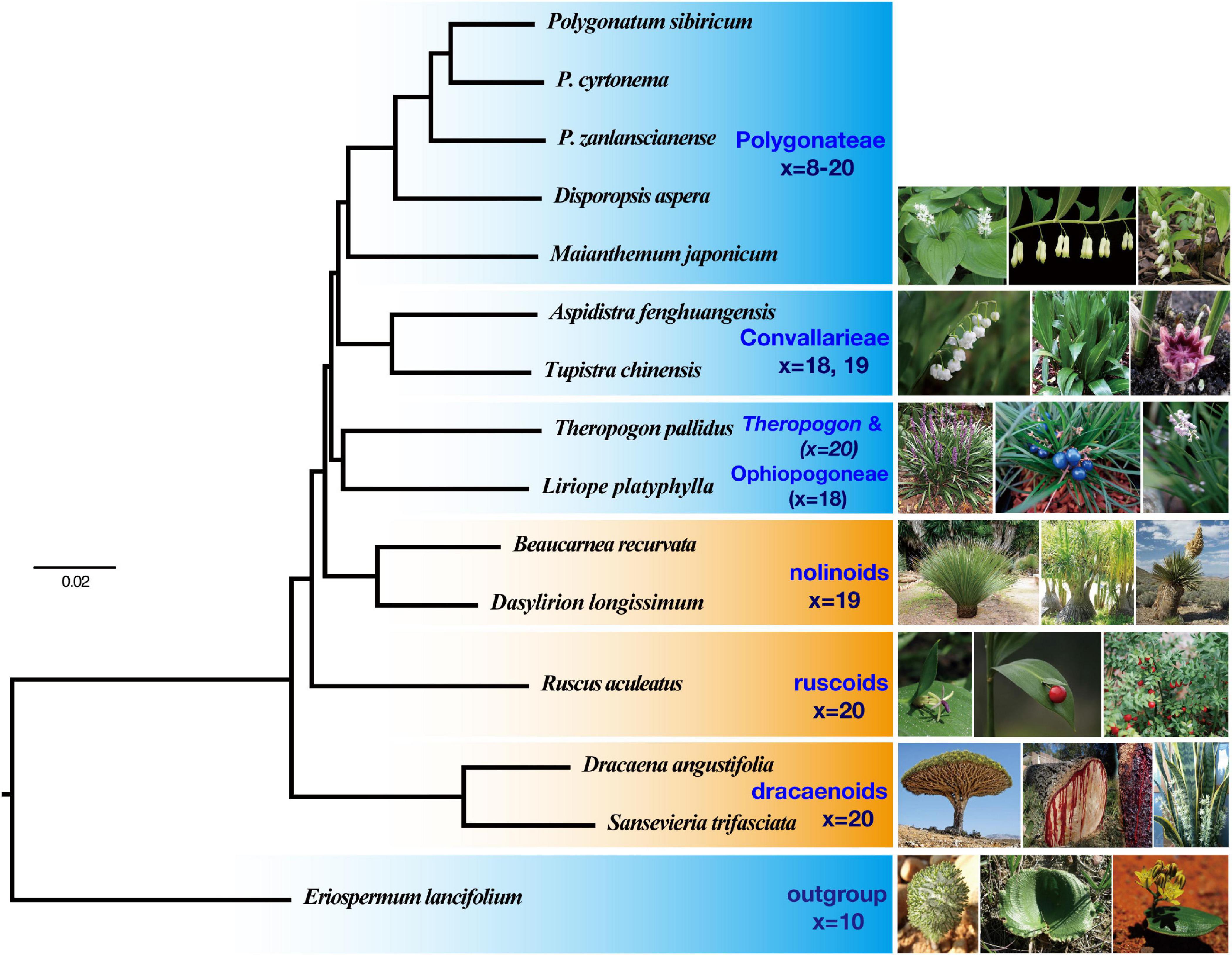
Figure 2. A partitioned maximum likelihood tree of Nolinoideae reconstructed from the concatenated alignment of 2,126 coding sequences. All branches are supported by 100% bootstrap values and posterior probability of 1.00.
In our phylogenetic results, the dracaenoid clade (represented by Sansevieria and Dracaena) is supported to be sister to a clade including the ruscoids further sister to a clade including the nolinoids and the herbaceous Convallariaceae clade (Figure 2). In the second ASTRAL species tree, the ruscoids are sister to the dracaenoids (Figure 3). Within the traditional Convallariaceae group, Ophiopogoneae + Theropogon were recovered as the first lineage sister to Convallarieae + Polygonateae. Based on the AU test, Maianthemum sister to Polygonatum group and Theropogon sister to the Ophiopogoneae were the best topologies, and other topologies about the phylogenetic placements of Maianthemum and Theropogon were rejected (p-values < 0.005). The best topologies suggested by the AU test were also supported by more single-gene trees (Figure 4). After stripping out the single-gene trees with low average support (BP < 50%) to reduce topological uncertainty, 24% and 14% of 785 gene trees supported a sister relationship between Maianthemum and Polygonatum clade and Theropogon as sister to the Ophiopogoneae, respectively (Figure 4).
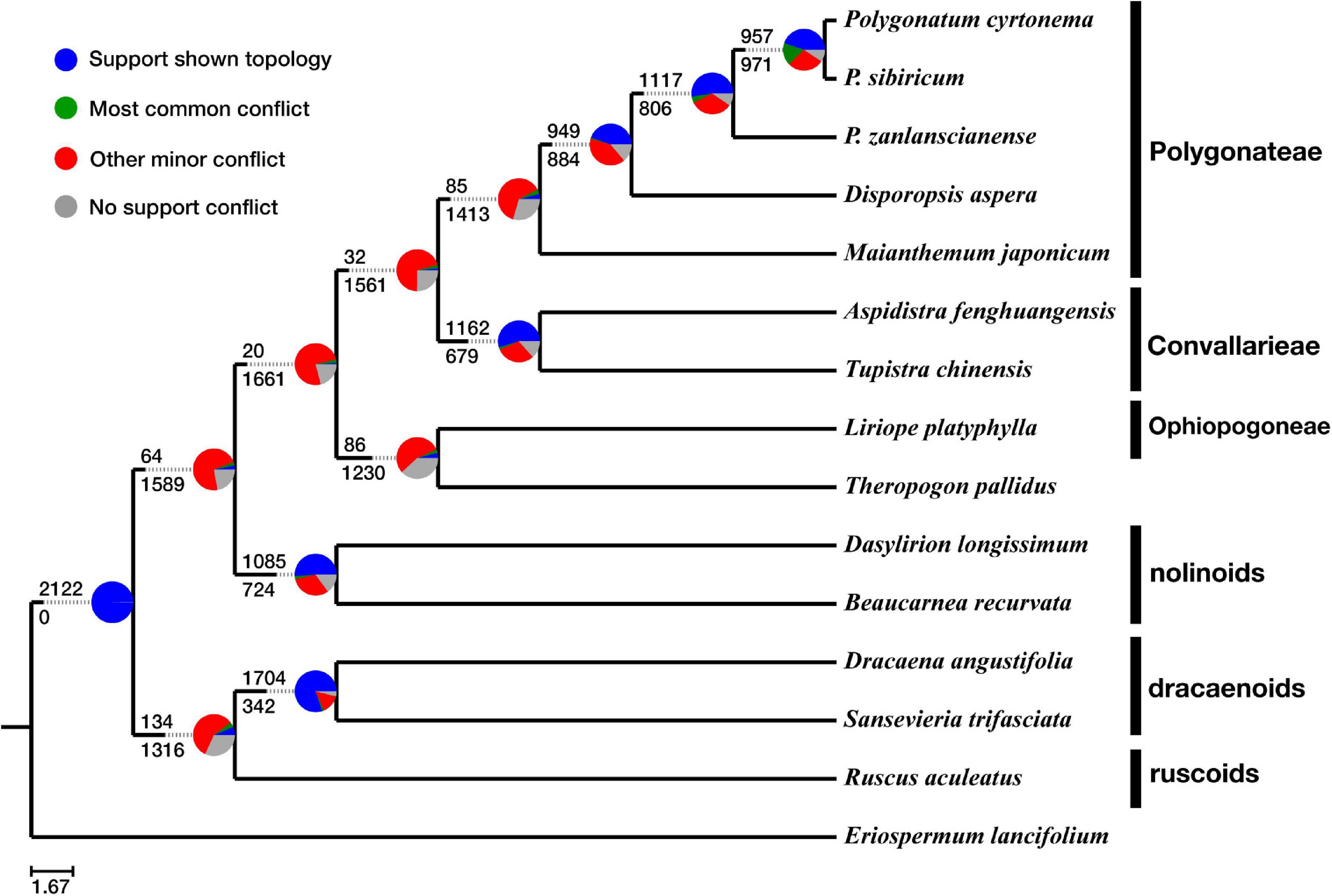
Figure 3. ASTRAL-III tree shows gene–tree conflict with pie chart at each node as the following: proportion of gene trees in concordance (blue), in conflict with the dominant alternative topology (green), in conflict with all other topologies (red), and unsupported with less than 50% bootstrap scores (gray).
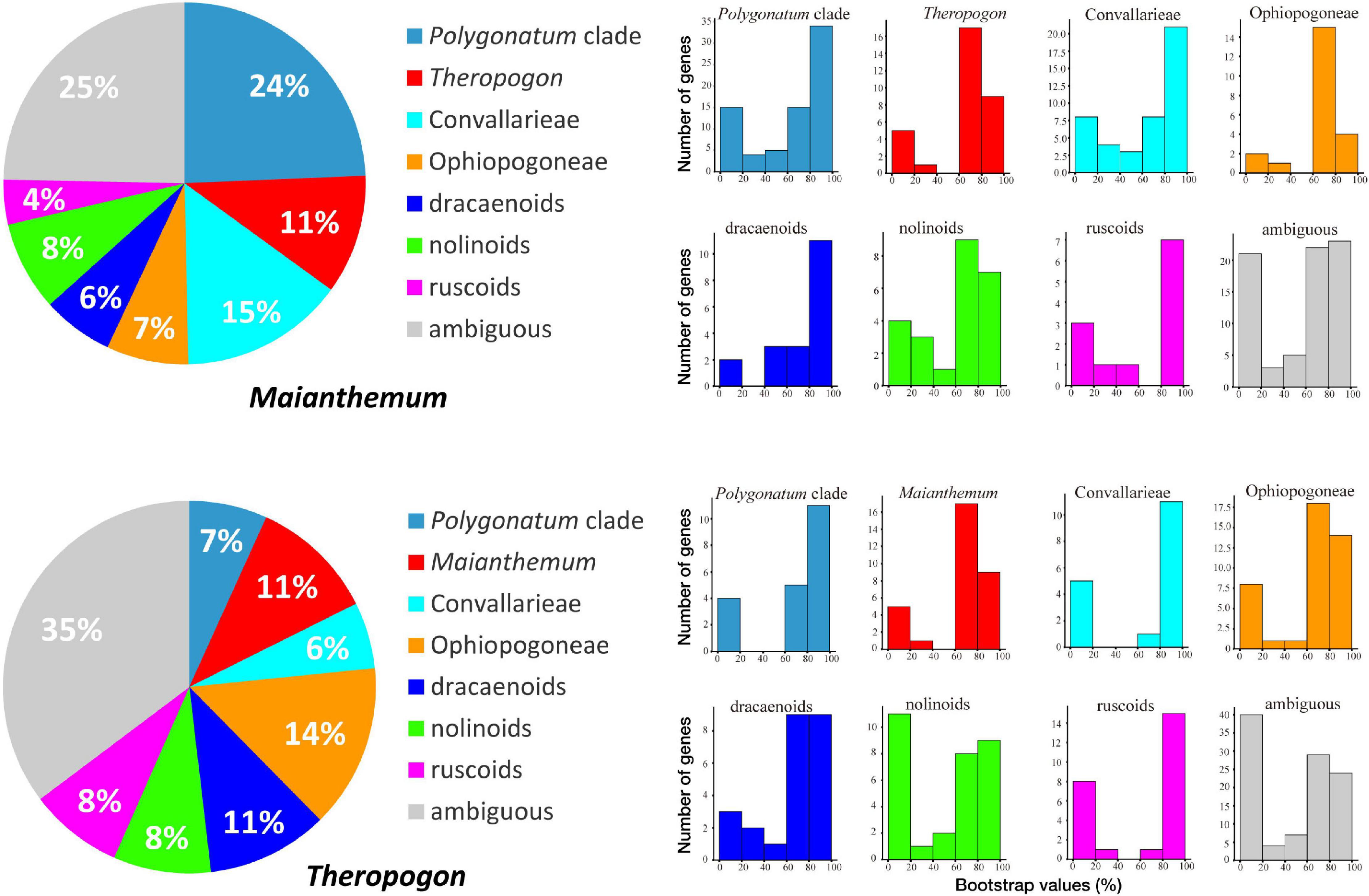
Figure 4. Numbers of individual gene trees supporting different relationships. Pie graphs show the proportions of gene trees with different topologies. Histograms indicate distributions of maximum likelihood (ML) bootstrap supports for the corresponding topologies.
Conflicts of gene trees against species tree are shown in Figure 3. A high level of concordance (>50%) was observed for the monophyly of Convallarieae, the nolinoids, and the dracaenoids, respectively. In the ASTRAL tree, the only well-supported clade in the gene trees was the node next to the root (the Nolinoideae excluding Eriospermum lineage), supported by 2,122 (∼99.8%) of the 2,126 loci tree topologies, and only 4 (0.2%) of the gene trees have no supported conflicts (Figure 3). We found only 134 of the 2,126 gene trees concordant with the species tree surrounding the sister relationships between the ruscoids and dracaenoids. The monophyly of the major lineages was supported by different numbers of gene trees (Figure 4). More than 80% of gene trees supported the monophyly of the dracaenoids and less gene trees (ca. 50%) supported the monophyly of the nolinoids and tribe Convallarieae, respectively (Figure 3). The sister relationship between Maianthemum and Polygonatum lineages was only supported by 85 gene trees (4.0%). The same pattern was observed in the node linking the Theropogon and the tribe Ophiopogoneae (Figure 3).
Discussion
Deep Relationships Within the Subfamily
Previous phylogenetic works have greatly improved our understanding of the relationships of Nolinoideae (Rudall et al., 2000; Jang and Pfosser, 2002; Kim et al., 2010), but robust resolution of their early divergences has proven to be a formidable task. Here, with our transcriptomic dataset, we reconstructed most relationships with strong supports (Figure 2). The higher supports were found for deeper-level relationships that previously had weak supports (e.g., Polygonateae and Convallarieae), as well as for new hypotheses (e.g., Theropogon as sister to Ophiopogoneae).
The tree topology of Nolinoideae reconstructed in this study was largely congruent with previous results (Rudall et al., 2000; Jang and Pfosser, 2002; Kim et al., 2010). Regardless of which datasets were analyzed and what methods were utilized, all phylogenetic analyses consistently support the dividing of whole Nolinoideae into seven major lineages: (1) Eriospermum, (2) dracaenoids, (3) ruscoids, (4) nolinoids, (5) Ophiopogoneae, (6) Convallarieae, and (7) Polygonateae (Figures 2, 3). In addition, the uncertain Theropogon is suggested to be close to Ophiopogoneae. These seven groups were also recognized by the previous works based on both molecular and/or morphological evidences (Rudall et al., 2000; Yamashita and Tamura, 2000b; Pires et al., 2006; Kim et al., 2010). In particular, most nodes in this transcriptome-based phylogeny are robustly supported by BS = 100% and PP of 1.00 (Figure 2), in contrast to previous studies that most internal relationships among these seven clades have extremely low supports (Rudall et al., 2000; Jang and Pfosser, 2002; Kim et al., 2010; Seberg et al., 2012). Furthermore, except for the Eriospermum lineage as outgroup, transcriptomic results could largely classify the remaining six lineages into two groups, one is a paraphyletic grade predominantly characterized by woody habits and a monophyletic group, previously recognized as the Convallariaceae, uniformly characterized by herbaceous life forms (Figure 2).
The three woody lineages of dracaenoids, ruscoids, and nolinoids are not sister related but form a paraphyletic grade with the dracaenoids sister to the ruscoids and then the nolinoids (Figure 2). These woody taxa share the tenuinucellate parietal cells and the same basic number of x = 19–20 (Rudall et al., 2000).
The dracaenoids have a complex taxonomic history and their taxonomic placement has been changed several times, including classification in Liliaceae (Brown, 1915), Agavaceae (Cronquist, 1981; Bogler and Simpson, 1996), Dracaenaceae (Dahlgren et al., 1985), Ruscaceae (Chase et al., 1995), Convallariaceae (Yamashita and Tamura, 2000b), and, finally, Asparagaceae subfamily Nolinoideae (APGIII, 2009). Within Nolinoideae, the phylogenetic relationship of the dracaenoids is still unresolved. The dracaenoids are differed from the nolinoids in having berries, no oils in guard cells, and mucilage-filled cells with crystal raphides. Some previous studies have suggested the dracaenoids have a close relationships with the nolinoids, but with very low supports (Rudall et al., 1997; Kim et al., 2010). Our results suggested that the dracaenoids have a closer relationship with the ruscoids than the nolinoids (Figure 2). Chromosome numbers are probably more similar between the dracaenoids (x = 20) and the ruscoids (x = 20) than between the dracaenoids and the nolinoids (x = 19). Interestingly, the ruscoids were suggested to be directly sister to the dracaenoids in the second ASTRAL species tree (Figure 3).
The ruscoids have once been considered the most closely related to the traditional Asparagaceae by many botanists (Dahlgren et al., 1985; Takhtajan, 1997). However, analysis of molecular sequence data indicated a close relationship with Convallariaceae taxa (Chase et al., 1995). Karyotypes are probably more similar between ruscoids (x = 20) and some Convallariaceae s.l. (mainly x = 18, 19) than between Ruscaceae and Asparagus (x = 10) (Tamura, 1995; Rudall et al., 2000). Ruscoids lack phytomelan in the seed coat, a relatively consistent apomorphy shared with Convallariaceae s.l., and are serologically closer to Convallariaceae s.l. than to Asparagus (Chupov and Cutjavina, 1980; Rudall and Campbell, 1999). Our study suggested a relatively basal position within the subfamily as the second woody lineages between the dracaenoids and the nolinoids (Figure 2).
The nolinoids were originally placed in the broadly defined and traditional family of Liliaceae and then in the tribe Dracaeneae or Nolinaceae (Rudall et al., 2000). All taxa from Nolinaceae, Dracaeneae, and Yuccoideae were placed in Agavaceae based on their fibrous leaves and anomalous woody growth of a secondary thickening meristem (Hutchinson, 1934). However, this placement was not supported by the other morphological evidence (flowers, fruits, and seeds) and the chromosome data (Sharma and Chaudhuri, 1964; Rudall et al., 2000). Several other studies proposed a close relationship to Convallariaceae and Dracaenaceae (Bogler et al., 1995; Bogler and Simpson, 1996), particularly Ophiopogoneae (Rudall et al., 2000; Yamashita and Tamura, 2000b), but there are no obvious morphological evidence to support this affinity. More recently, the nolinoids were suggested to be close to Convallariaceae–Dracaenaceae–Ruscaceae s.s. in the maximum parsimony analysis while close to the Aspidistreae–Convallarieae group in the Bayesian analysis (Kim et al., 2010). In the present study, the nolinoids are suggested to be closer to the herbaceous Convallariaceae group than other woody lineages (Figure 2). It is difficult to find any obvious synapomorphies of them, but chromosome data provide possible insights as the nolinoids usually with x = 19 and the Convallariaceae group dominantly featured with x = 18–19 (Figure 2).
The herbaceous clade, consists of three tribes sharing sympodially or monopodially branching rhizomes, is confirmed with strong BS and PP supports as revealed by some previous analyses (Rudall et al., 2000; Yamashita and Tamura, 2000b). Based on the phylogenomic tree, our data suggest Ophiopogoneae + Theropogon diverged first, sister to a clade that is composed of Convallarieae (including Aspidistra and Tupistra) and the monophyletic Polygonateae (Figure 2).
The Ophiopogoneae was treated as a well monotypic member of Convallarieae s.s., but its phylogenetic relationship to the other herbaceous tribes has long been in uncertainty within the traditional Convallariaceae (Rudall et al., 2000; Yamashita and Tamura, 2000b; Kim et al., 2010; Wang and Yang, 2018). In our analyses, Ophiopogoneae is represented by Liriope platyphylla, together with Theropogon, which is suggested to be sister to Convallarieae + Polygonateae (Figure 2). Based on comparative plastid genomic data, Floden and Schilling (2018) provided similar results and suggested that Ophiopogoneae was close to a clade including the Polygonateae and Aspidistreae (treated as Convallarieae).
Convallarieae share some synapomorphic characters such as the chromosome basic number (x = 19), usually berries, and non-septal nectaries (Dahlgren et al., 1985). Rudall et al. (2000) demonstrated close relationships between Convallarieae and the ruscoids, but the latter is distinguished with their basic number (x = 20) and septal nectaries from Convallarieae. The present study suggested Convallarieae is close to Polygonateae (Figure 2). Moreover, based on comparative plastid genomics, Floden and Schilling (2018) regarded Aspidistra from Convallarieae as a separate lineage close to Polygonateae.
Phylogenetic Placement of Maianthemum and Theropogon
The monophyly of Polygonateae with two lineages has been recovered by our transcriptome data, similar to most other molecular studies (Rudall et al., 2000; Yamashita and Tamura, 2000b; Meng et al., 2008, 2014). However, a few other phylogenetic studies have recovered the non-monophyly of the tribe, such as in the plastid matK + rbcL and rDNA 18S ML tree (Kim et al., 2010) and a combined six-gene matrix (Seberg et al., 2012). A more recent study based on the whole plastid genomes suggested that the terminal-flowered Maianthemum is sister to Ophiopogoneae other than the other members of the Polygonateae (Floden and Schilling, 2018). Here, we tested the phylogenetic status of Maianthemum using topological statistics from single-gene trees. Except for the uncertainty, the results indicate that the largest number of single-gene trees (24%) supports the close relationship with Polygonatum lineage (Figure 4). Only 11% single-gene trees support the close relationship between Maianthemum and Ophiopogoneae (Figure 4).
The large dataset generated in our study provide a unique insight into the sources of this topological instability, especially evaluating phylogenetic placement of Theropogon. It was treated as a member of Convallarieae by morphological characters (Vaikos et al., 1989). However, all previous analyses of molecular data have failed to support the placement of Theropogon in Convallarieae clade or herbaceous lineage, but always showing close relationship to the ruscoids or dracaenoids, all of which also shared similar basic chromosome numbers (x = 20) (Rudall et al., 2000; Yamashita and Tamura, 2000a, b). Our transcriptome data based on concatenated ML, BI, and one ASTRAL analysis indicate that it is sister to Ophiopogoneae (Figures 2, 3), but collapsed in the other coalescent ASTRAL analyses (Supplementary Figures 4, 5). Except for the uncertainty, the highest number of single-gene trees (14%) supports a close relationship with Ophiopogoneae (Figure 4). Our results suggested a possible reticulate evolution had occurred in the early origin of Theropogon as evidenced by the complicated distributions of single-gene trees (Figure 4). The discordance could result from horizontal gene transfer, incomplete lineage sorting, and/or ancient hybridization between different ancestral lineages.
Concordance and Conflicts Among Gene Trees
Even though the backbone nodes showed BS = 100% and PP = 1.00 in the super-matrix approach (Figure 2), the PhyParts analysis on the ASTRAL species tree showed a high degree of gene–tree conflicts (Figure 3). High gene–tree conflicts (>75%) were prevalent across many relationships including the position of the nolinoids, the relationship of Theropogon pallidus and Ophiopogoneae, the relationship of the Convallarieae and the Polygonateae, and the relationship of the genus Maianthemum with respect to the Polygonatum and Disporopsis (Figure 3).
Only 1.5% gene trees support and dominance of other minor conflicting bipartitions (73.4%) were found for the sister relationship between Polygonateae and Convallarieae. The nolinoids as sister to the herbaceous group was supported only by 64 (3.0%) gene trees, and the ruscoids as sister to the dracaenoids by 134 (6.3%) gene trees (Figure 3). Low gene tree supports and dominance of other conflicting bipartitions were also detected in the other clades. Similar patterns were observed for grouping the Ophiopogoneae–Theropogon and Convallarieae–Polygonateae in a single clade (Figure 3). Although a very low number of gene trees supported the deeper relationships within the subfamily, there was no dominant alternative topology found among the conflicting topologies (indicated in green in the pie charts of Figure 3), but they exhibited the dominance for other minor conflicting topologies as indicated in red in the pie charts of Figure 3.
Similar to other studies on plants and animals with multi-locus datasets, it seems to be unnecessary that the analyses of multiple gene copies have to be in concordance or high support of the topologies resulted from coalescent-based methods of species tree estimations (Jeffroy et al., 2006; Sun et al., 2015; Tang et al., 2015; Siniscalchi et al., 2019). On the other hand, individual gene trees with various topologies are commonly found in many taxa on different levels, suggesting that incomplete lineage sorting, hybridization, gene duplication, and horizontal gene transfer are pervasive phenomena and could be significant causes of these topological conflicts or discordances (Yu et al., 2013; Sun et al., 2015; Tang et al., 2015; Mallo and Posada, 2016; Arcila et al., 2017; Bogarín et al., 2018).
Incomplete lineage sorting might cause strong conflicts in taxa with closely related species with fast diversifications due to the alleles within a population without enough time to coalesce (Degnan and Rosenberg, 2009; Meyer et al., 2016; Arcila et al., 2017). Except for incomplete lineage sorting and early speciation, discordance could also be resulted from estimation mistakes in the sequence alignments of individual genes, such as missing sequences, phylogenetic noise, or long-branch attractions (Mallo and Posada, 2016; Mirarab et al., 2016). Including gene alignments without missing sequences did obtain some difference in topologies, but the major differences are mostly from the position of some taxa, such as Theropogon and Maianthemum as shown in Figure 4. Additional investigation with plastid genomic data and comprehensive analyses of both nuclear and plastid data are required to clarify the complex evolution history within the subfamily.
Conclusion
The utility of transcriptome phylogenetics is demonstrated by the reconstruction of the relationships within Nolinoideae. Rapid diversification or complex evolution is an obstacle to well-resolved relationships, which can be remedied with increased sequence data (Xi et al., 2012; Wen et al., 2015b). Relationships within the subfamily have long been obscured by a rapid diversification and now the phylogeny is becoming more resolved. The three woody lineages of dracaenoids, ruscoids, and nolinoids are not sister relationship but are forming a paraphyletic grade with dracaenoids as the first diverged lineage followed with ruscoids and then nolinoids (Figure 2). The monophyly of herbaceous clade is confirmed with strong BS, which consists of three tribes sharing sympodially or monopodially branching rhizomes (Rudall et al., 2000; Yamashita and Tamura, 2000b). Our study further revealed a low level of gene trees supported the backbone relationships within the Nolinoideae and most of the conflicts were located at deeper nodes along the phylogeny (Figure 3), indicating that incomplete lineage sorting, hybridization, and gene duplication are all possible causes of these topological incongruences. Further works are necessary to uncover their complex evolution using more advanced genomic data such as genomic skimming or target enrichment technologies (Mamanova et al., 2010; Wen et al., 2015b). This work paves the way for investigations on evolutionary history of the highly morphological heterogeneous Nolinoideae on the genomic level.
Data Availability Statement
Raw data in the fastq format are deposited at the NCBI Sequence Read Archive, under BioProjects PRJNA608213, and other data can be found at https://data.mendeley.com/datasets/ncy4td2hb9.
Author Contributions
RM designed the study, collected samples, conducted lab work, conducted data analyses, and wrote the manuscript. L-YL and D-GZ participated in field collections and provided samples. YM and Z-LN helped to design the study, wrote the Discussion section, and reviewed and commented on the manuscript. J-YZ provided samples, collaborated on lab work, conducted data analyses, and reviewed and commented on the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31760055), the Natural Science Foundation of Hunan Province (2019JJ40233), the Traditional Chinese Medicine Public Health Special Project [(2011)76, 201207002], the Comprehensive Scientific Investigation of Biodiversity from the Wuling Mountains (2014FY110100), and the Hunan Provincial Innovation Foundation for Postgraduate (CX2018B724).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.584981/full#supplementary-material
Footnotes
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- ^ https://github.com/TransDecoder/TransDecoder/releases
- ^ https://github.com/mossmatters/phyloscripts/tree/master/phypartspiecharts
References
APGIII (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
APGIV (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 18, 1–20. doi: 10.1111/boj.12385
Arcila, D., Ortí, G., Vari, R., Armbruster, J. W., Stiassny, M. L. J., Ko, K. D., et al. (2017). Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat. Ecol. Evol. 1, 0020. doi: 10.1038/s41559-016-0020
Bogarín, D., Pérez-Escobar, O. A., Groenenberg, D., Holland, S. D., Karremans, A. P., Lemmon, E. M., et al. (2018). Anchored hybrid enrichment generated nuclear, plastid and mitochondrial markers resolve the Lepanthes horrida (Orchidaceae: Pleurothallidinae) species complex. Mol. Phylogenet. Evol. 129, 27–47. doi: 10.1016/j.ympev.2018.07.014
Bogler, D. J., Neff, J. L., and Simpson, B. B. (1995). Multiple origins of the yucca-yucca moth association. Proc. Natl. Acad. Sci. U.S.A. 92, 6864–6867. doi: 10.1073/pnas.92.15.6864
Bogler, D. J., and Simpson, B. B. (1996). Phylogeny of Agavaceae based on ITS rDNA sequence variation. Am. J. Bot. 83, 1225–1235. doi: 10.1002/j.1537-2197.1996.tb13903.x
Brown, J. W., Walker, J. F., and Smith, S. A. (2017). Phyx: phylogenetic tools for unix. Bioinformatics 33, 1886–1888. doi: 10.1093/bioinformatics/btx063
Brown, N. E. (1915). Sansevieria, a monograph of all the known species. Bull. Misc. Inform. 5, 185–261. doi: 10.2307/4107465
Chase, M. W., Duvall, M. R., Hills, H. G., Conran, J. G., Cox, A. V., Eguiarte, L. E., et al. (1995). “Molecular phylogenetic of Lilianae,” in Monocotyledons: Systematics and Evolution, eds P. J. Rudall, P. J. Cribb, D. F. Cutler, and C. J. Humphries (Kew: Royal Botanic Gardens), 109–137.
Chase, M. W., Reveal, J. L., and Fay, M. F. (2009). A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot. J. Linn. Soc. 161, 132–136. doi: 10.1111/j.1095-8339.2009.00999.x
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, S. C., Kim, D. K., Chase, M. W., and Kim, J. H. (2013). Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLoS One 8:e59472. doi: 10.1371/journal.pone.0059472
Chupov, V., and Cutjavina, N. (1980). Phylogeny of Some Groups of Liliales based on the Data of Serological Analysis. Systematics and Evolution of Higher Plants. Leningrad: Nauka, 101–110.
Conran, J. G., and Tamura, M. N. (1998). “Convallariaceae,” in The Families and Genera of Vascular Plants, Vol. 3, ed. K. Kubitzki (Berlin: Springer-Verlag).
Cronquist, A. (1981). An Integrated System of Classification of Flowering Plants. New York, NY: Columbia University Press.
Dahlgren, R. M. T., Clifford, H. T., and Yeo, P. F. (1985). The Families of the Monocotyledons. Berlin: Springer-Verlag.
Degnan, J. H., and Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340. doi: 10.1016/j.tree.2009.01.009
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157. doi: 10.1186/s13059-015-0721-2
Fay, M. F., Rudall, P. J., Sullivan, S., Stobart, K. L., Bruijn, A. Y., Qamaruz, Z. F., et al. (2000). “Phylogenetic studies of Asparagales based on four plastid DNA loci,” in Monocots: Systematics and Evolution, eds K. L. Wilson and D. A. Morrison (Melbourne, VIC: CSIRO), 360–371.
Floden, A., and Schilling, E. E. (2018). Using phylogenomics to reconstruct phylogenetic relationships within tribe Polygonateae (Asparagaceae), with a special focus on Polygonatum. Mol. Phylogenet. Evol. 129, 202–213. doi: 10.1016/j.ympev.2018.08.017
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2017). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hutchinson, J. (1934). The Families of Flowering Plants. Vol. 2. Monocotyledons. Oxford: Clarendon Press.
Jang, C. G., and Pfosser, M. (2002). Phylogenetics of Ruscaceae sensu lato based on plastid rbcL and trnL-F DNA sequences. Stapfia 80, 333–348.
Jeffroy, O., Brinkmann, H., Delsuc, F., and Philippe, H. (2006). Phylogenomics: the beginning of incongruence? Trends Genet. 22, 225–231. doi: 10.1016/j.tig.2006.02.003
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, J.-H., Kim, D.-K., Forest, F., Fay, M. F., and Chase, M. W. (2010). Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Ann. Bot. 106, 775–790. doi: 10.1093/aob/mcq167
Kocot, K. M., Citarella, M. R., Moroz, L. L., and Halanych, K. M. (2013). PhyloTreePruner: a phylogenetic tree-based approach for selection of orthologous sequences for phylogenomics. Evol. Bioinform. 9, 429–435. doi: 10.4137/EBO.S12813
Kriventseva, E. V., Kuznetsov, D., Tegenfeldt, F., Manni, M., Dias, R., Simão, F. A., et al. (2018). OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47, D807–D811. doi: 10.1093/nar/gky1053
Kück, P., and Longo, G. C. (2014). FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 11:81. doi: 10.1186/s12983-014-0081-x
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lu, P.-L., and Morden, C. W. (2014). Phylogenetic relationships among Dracaenoid genera (Asparagaceae: Nolinoideae) inferred from chloroplast DNA loci. Syst. Bot. 39, 90–104. doi: 10.1600/036364414X678035
Mallo, D., and Posada, D. (2016). Multilocus inference of species trees and DNA barcoding. Philos. Trans. R. Soc. B Biol. Sci. 371, 20150335. doi: 10.1098/rstb.2015.0335
Mamanova, L., Coffey, A. J., Scott, C. E., Kozarewa, I., Turner, E. H., Kumar, A., et al. (2010). Target-enrichment strategies for next-generation sequencing. Nat. Methods 7, 111–118. doi: 10.1038/nmeth.1419
Meng, Y., Nie, Z.-L., Deng, T., Wen, J., and Yang, Y.-P. (2014). Phylogenetics and evolution of phyllotaxy in the Solomon’s seal genus Polygonatum (Asparagaceae: Polygonateae). Bot. J. Linn. Soc. 176, 435–451. doi: 10.1111/boj.12218
Meng, Y., Wen, J., Nie, Z.-L., Sun, H., and Yang, Y.-P. (2008). Phylogeny and biogeographic diversification of Maianthemum (Ruscaceae: Polygonatae). Mol. Phylogenet. Evol. 49, 424–434. doi: 10.1016/j.ympev.2008.07.017
Meyer, B. S., Matschiner, M., and Salzburger, W. (2016). Disentangling incomplete lineage sorting and introgression to refine species-tree estimates for lake tanganyika cichlid fishes. Syst. Biol. 66, 531–550. doi: 10.1093/sysbio/syw069
Mirarab, S., Bayzid, M. S., and Warnow, T. (2016). Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Syst. Biol. 65, 366–380. doi: 10.1093/sysbio/syu063
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Pires, J. C., Maureira, I. J., Givnish, T. J., Sytsma, K. J., Seberg, O., Petersen, G., et al. (2006). Phylogeny, genome size, and chromosome evolution of Asparagales. Aliso 22, 287–304. doi: 10.5642/aliso.20062201.24
Ran, J.-H., Shen, T.-T., Wu, H., Gong, X., and Wang, X.-Q. (2018). Phylogeny and evolutionary history of Pinaceae updated by transcriptomic analysis. Mol. Phylogenet. Evol. 129, 106–116. doi: 10.1016/j.ympev.2018.08.011
Rojas-Piña, V., Olson, M. E., Alvarado-Cárdenas, L. O., and Eguiarte, L. E. (2014). Molecular phylogenetics and morphology of Beaucarnea (Ruscaceae) as distinct from Nolina, and the submersion of Calibanus into Beaucarnea. Taxon 63, 1193–1211. doi: 10.12705/636.31
Rudall, P. J., and Campbell, G. (1999). Flower and pollen structure of Ruscaceae in relation to Aspidistreae and other Convallariaceae. Flora 194, 201–214. doi: 10.1016/S0367-2530(17)30908-8
Rudall, P. J., Conran, J. G., and Chase, M. W. (2000). Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Bot. J. Linn. Soc. 134, 73–92. doi: 10.1006/bojl.2000.0365
Rudall, P. J., Furness, C. A., Chase, M. W., and Fay, M. F. (1997). Microsporogenesis and pollen sulcus type in Asparagales (Lilianae). Can. J. Bot. 75, 408–430. doi: 10.1139/b97-044
Seberg, O., Petersen, G., Davis, J. I., Pires, J. C., Stevenson, D. W., Chase, M. W., et al. (2012). Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. Am. J. Bot. 99, 875–889. doi: 10.3732/ajb.1100468
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Sharma, A. K., and Chaudhuri, U. (1964). Cytological studies as an aid in assessing the status of Sansevieria, Ophiopogon and Curculigo. Nucleus 7, 43–58.
Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508. doi: 10.1080/10635150290069913
Shimodaira, H., and Hasegawa, M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247. doi: 10.1093/bioinformatics/17.12.1246
Siniscalchi, C. M., Loeuille, B., Funk, V. A., Mandel, J. R., and Pirani, J. R. (2019). Phylogenomics yields new insight into relationships within Vernonieae (Asteraceae). Front. Plant Sci. 10:1224. doi: 10.3389/fpls.2019.01224
Smith, S. A., Moore, M. J., Brown, J. W., and Yang, Y. (2015). Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol. 15:150. doi: 10.1186/s12862-015-0423-0
Sukumaran, J., and Holder, M. T. (2010). DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571. doi: 10.1093/bioinformatics/btq228
Sun, M., Soltis, D. E., Soltis, P. S., Zhu, X., Burleigh, J. G., and Chen, Z. (2015). Deep phylogenetic incongruence in the angiosperm clade Rosidae. Mol. Phylogenet. Evol. 83, 156–166. doi: 10.1016/j.ympev.2014.11.003
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Suppl. 2), W609–W612. doi: 10.1093/nar/gkl315
Takawira-Nyenya, R., Mucina, L., Cardinal-Mcteague, W. M., and Thiele, K. R. (2018). Sansevieria (Asparagaceae, Nolinoideae) is a herbaceous clade within Dracaena : inference from non-coding plastid and nuclear DNA sequence data. Phytotaxa 376, 22.
Takhtajan, A. (1997). Diversity and Classification of Flowering Plants. New York, NY: Columbia University Press.
Tamura, M. N. (1995). A karyological review of the orders Asparagales and Liliales (Monocotyledonae). Feddes Repertorium 106, 83–111. doi: 10.1002/fedr.4921060118
Tang, L., Zou, X.-H., Zhang, L.-B., and Ge, S. (2015). Multilocus species tree analyses resolve the ancient radiation of the subtribe Zizaniinae (Poaceae). Mol. Phylogenet. Evol. 84, 232–239. doi: 10.1016/j.ympev.2015.01.011
Vaikos, N. P., Markandeya, S. K., and Pai, R. M. (1989). Floral anatomy of the Liliaceae: Tribe Convallarieae. Proc. Plant Sci. 99, 91–95. doi: 10.1007/BF03053520
Wang, G.-Y., Meng, Y., Huang, J.-L., and Yang, Y.-P. (2014). Molecular phylogeny of ophiopogon (Asparagaceae) inferred from nuclear and plastid DNA sequences. Syst. Bot. 39, 776–784. doi: 10.1600/036364414X682201
Wang, G. Y., and Yang, Y. P. (2018). Hypothesizing the origin, migration routes, and distribution patterns of Ophiopogon (Asparagaceae) in East and Southeast Asia. J. Syst. Evol. 56, 194–201. doi: 10.1111/jse.12304
Wang, J.-J., Yang, Y.-P., Sun, H., Wen, J., Deng, T., Nie, Z.-L., et al. (2016). The biogeographic south-north divide of Polygonatum (Asparagaceae tribe Polygonateae) within eastern Asia and its recent dispersals in the Northern Hemisphere. PLoS One 11:e0166134. doi: 10.1371/journal.pone.0166134
Waterhouse, R. M., Seppey, M., Simão, F. A., Manni, M., Ioannidis, P., Klioutchnikov, G., et al. (2017). BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. doi: 10.1093/molbev/msx319
Wen, J., Egan, A. N., Dikow, R. B., and Zimmer, E. A. (2015a). “Utility of transcriptome sequencing for phylogenetic inference and character evolution,” in Next-Generation Sequencing in Plant Systematics, eds E. Hörandl and M. S. Appelhans (Bratislava: International Association for Plant Taxonomy), 51–91.
Wen, J., Liu, J., Ge, S., Xiang, Q.-Y., and Zimmer, E. A. (2015b). Phylogenomic approaches to deciphering the tree of life. J. Syst. Evol. 53, 369–370. doi: 10.1111/jse.12175
Wen, J., Xiong, Z., Nie, Z.-L., Mao, L., Zhu, Y., Kan, X.-Z., et al. (2013). Transcriptome sequences resolve deep relationships of the grape family. PLoS One 8:e74394. doi: 10.1371/journal.pone.0074394
Wickett, N. J., Honaas, L. A., Wafula, E. K., Das, M., Huang, K., Wu, B. A., et al. (2011). Transcriptomes of the parasitic plant family orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr. Biol. 21, 2098–2104. doi: 10.1016/j.cub.2011.11.011
Wickett, N. J., Mirarab, S., Nguyen, N., Warnow, T., Carpenter, E., Matasci, N., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868. doi: 10.1073/pnas.1323926111
Xi, Z. X., Ruhfel, B. R., Schaefer, H., Amorim, A. M., Sugumaran, M., Wurdack, K. J., et al. (2012). Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. U.S.A. 109, 17519–17524. doi: 10.1073/pnas.1205818109
Yamashita, J., and Tamura, M. N. (2000a). Karyotype analysis of two species of the Chinese epiphytic genus Heteropolygonatum (Convallariaceae-Polygonateae). Acta Phytotaxonom. Geobot. 51, 147–153.
Yamashita, J., and Tamura, M. N. (2000b). “Molecular phylogeny of the Convallariaceae (Asparagales),” in Monocots: Systematics and Evolution, eds K. L. Wilson and D. A. Morrison (Melbourne, VIC: CSIRO Publishing), 387–400.
Yamashita, J., and Tamura, M. N. (2004). Phylogenetic analyses and chromosome evolution in Convallarieae (Ruscaceae sensu lato), with some taxonomic treatments. J. Plant Res. 117, 363–370. doi: 10.1007/s10265-004-0169-z
Yu, W.-B., Huang, P.-H., Li, D.-Z., and Wang, H. (2013). Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS One 8:e74828. doi: 10.1371/journal.pone.0074828
Zeng, L., Zhang, N., Zhang, Q., Endress, P. K., Huang, J., and Ma, H. (2017). Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 214, 1338–1354. doi: 10.1111/nph.14503
Keywords: Asparagaceae, Nolinoideae, transcriptome, deep relationship, phylogeny
Citation: Meng R, Luo L-Y, Zhang J-Y, Zhang D-G, Nie Z-L and Meng Y (2021) The Deep Evolutionary Relationships of the Morphologically Heterogeneous Nolinoideae (Asparagaceae) Revealed by Transcriptome Data. Front. Plant Sci. 11:584981. doi: 10.3389/fpls.2020.584981
Received: 19 July 2020; Accepted: 14 December 2020;
Published: 14 January 2021.
Edited by:
Tingshuang Yi, Kunming Institute of Botany, Chinese Academy of Sciences, ChinaReviewed by:
Domingos Cardoso, Federal University of Bahia, BrazilYan Yu, Sichuan University, China
Xiu-Qun Liu, Huazhong Agricultural University, China
Copyright © 2021 Meng, Luo, Zhang, Zhang, Nie and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Meng, bWVuZ3lpbmdAanN1LmVkdS5jbg==
 Ran Meng
Ran Meng Ze-Long Nie
Ze-Long Nie