- 1Department of Agriculture, Food and Environment (Di3A), University of Catania, Catania, Italy
- 2Department of Agriculture, University Mediterranea of Reggio Calabria, Reggio Calabria, Italy
- 3Council for Agricultural Research and Agricultural Economy Analysis, Research Centre for Olive, Citrus and Tree Fruit-Rende CS (CREA-OFA), Rende, Italy
Decoding the mechanisms of plant defense against plant pathogens in a scenario where antagonistic activity and the plant growth-promoting effects of useful organisms intervene simultaneously is a new frontier of plant pathology. Here, we demonstrated that (i) two selected strains of Trichoderma asperellum and Trichoderma atroviride promoted tomato (Solanum lycopersicum) growth and reduced the severity of disease caused by the oomycete Phytophthora nicotianae and (ii) the genetic patterns of the components of the experimental model system tomato–Trichoderma spp.–P. nicotianae were differentially expressed. The beneficial effects in both the promotion of the growth of host plant and the biological control of the pathogen by two selected strains of different Trichoderma species were tested both in planta and in vitro. In both respects, T. atroviride demonstrated to be more effective than T. asperellum. Additionally, the simultaneous transcriptional reprogramming of several plant defense-related genes, pathogen effectors, and mycoparasitism-related genes in tomato, P. nicotianae, and Trichoderma spp., respectively, was evaluated during the three-component interaction. Results support the hypothesis that Trichoderma spp. elicit the expression of plant defense-related genes. As expected, a mycoparasitism-related gene was significantly up-regulated in Trichoderma-colonizing tomato plants infected by P. nicotianae. Finally, a marked up-regulation of the genes encoding two necrosis-inducing effectors was observed in P. nicotianae infecting tomato plants colonized by Trichoderma. In conclusion, this study is a contribution toward understanding the genetic pathways related with the ability of Trichoderma spp. to counteract the challenge of P. nicotianae infections on tomato. Additionally, the experiments revealed the beneficial effects in the tomato growth promotion of a new T. atroviride strain and its good antagonistic effectiveness in the biological control of root and crown rot incited by P. nicotianae, confirming that Trichoderma spp. can be a powerful tool in integrated pest management strategies of Phytophthora diseases of horticultural crops.
Introduction
In the last years, the biological control agents of plant pathogens, also known as antagonists, have inspired several research projects and the development of new strategies in the management of plant diseases (Ghazanfar et al., 2018).
The use of antagonistic microorganisms in plant protection is a low-risk practice for human health; moreover, the combination of these organisms with reduced levels of fungicides promotes a degree of disease suppression similar to that achieved with treatment using fungicide at normal doses (Monte, 2001; Benítez et al., 2004; Gilardi et al., 2020).
Among plant pathogens, phytopathogenic soil-borne fungi and oomycetes stand out since they are a threat to plant productivity on a global scale for a broad range of crops (Erwin and Ribeiro, 1996; Benítez et al., 2004; Mammella et al., 2011; Cacciola and Gullino, 2019).
The majority of applications of antagonistic microorganisms in the control of soil-borne plant diseases caused by fungal and oomycete pathogens have been conducted by selected strains of Trichoderma species (Benítez et al., 2004; Woo et al., 2014; Guzmán-Guzmán et al., 2019). The high effectiveness of Trichoderma spp. as biological control agents is due to both their antagonistic activity (Benítez et al., 2004) and the efficiency of these organisms in promoting plant growth and defense mechanisms (Harman et al., 2004; Verma et al., 2007; Shoresh et al., 2010; Singh and Islam, 2010; Tucci et al., 2011). The antagonistic activity of Trichoderma spp. can be considered the final result of different mechanisms, direct and indirect, acting synergistically to achieve disease control (Howell, 2003; Benítez et al., 2004). The indirect mechanisms include the competition for nutrients and space and the ability to produce metabolites that either inhibit spore germination, kill the cells (antibiosis), or modify the pH of rhizosphere. The direct interaction between antagonist and pathogen, usually indicated as mycoparasitism, includes both physical contact and the synthesis of hydrolytic enzymes, toxic compounds, and/or antibiotics that act synergistically to kill the pathogen (Benítez et al., 2004). Among hydrolytic enzymes, chitinases are the most relevant in mycoparasitism (Carsolio et al., 1994; Osorio-Hernández et al., 2016). High levels of these enzymes produced by Trichoderma spp. have been positively correlated with the inhibition in the growth of both fungi and oomycetes (Osorio-Hernández et al., 2016).
The colonization of the rhizosphere by Trichoderma spp. also produces direct positive effects on plants, promoting their growth and activating their defense mechanisms. It is well known that the interaction with microorganisms triggers two main defense mechanisms in plants that protect them against the infection (Shoresh et al., 2005). The first is known as systemic acquired resistance (SAR); this mechanism, which is considered to be triggered by local infection, can provide long-term resistance throughout the plant to subsequent infection by different pathogens. It is correlated with the synthesis of pathogenesis-related (PR) proteins, which is mediated by the up-regulation of genes encoding enzymes involved in the biosynthesis of salicylic acid (SA) (Zhang et al., 2010). The second mechanism, known as induced systemic resistance (ISR) and initially described in plants colonized by non-pathogenic rhizobacteria (Segarra et al., 2007), is correlated with the synthesis of jasmonic acid (JA) and ethylene (ET), which are mediated by the transcription factors MYC2 and ERF (Shoresh et al., 2005). This kind of resistance induces a primed state which enhances defense gene expression in the plant upon subsequent pathogen attack (Segarra et al., 2007). Since Trichoderma spp. are able to activate both kinds of resistances, the current hypothesis is that plants initially perceive their root colonization as a potential pathogen attack and then react with the activation of ISR mechanisms (Shoresh et al., 2005; Tucci et al., 2011).
Plant defensive mechanisms also comprise the synthesis of protective molecules acting directly against the pathogen; these include plant defensins, a family of small cationic peptides widely distributed among all plant families (Cui et al., 2018). These antimicrobial peptides proved to be very effective in the inhibition on the growth of pathogens (Lay and Anderson, 2005; Seo et al., 2014). Antifungal defensins have been identified in radish (Terras et al., 1992), pea (Mendenhall and Hodge, 1998; Almeida et al., 2000; Lobo et al., 2007; Gonçalves et al., 2012), and tomato (Cui et al., 2018). The sequencing of the whole genome of tomato (Sato et al., 2012) makes this plant a good model system for the study of plant-microorganisms interaction (Cui et al., 2018). With more than 75,000 ha, tomato is the most widely cultivated vegetable crop in Italy (ISTAT, 2020). Globally, among horticultural products, tomato ranks third for volumes of production–after potato and sweet potato–and first in terms of processing volumes (Brasesco et al., 2019). Tomato is susceptible to numerous diseases (Jones et al., 2016), among which root and crown rot incited by Phytophthora species represent one of the most important causes of yield losses (Erwin and Ribeiro, 1996; Jones et al., 2016). Several Phytophthora spp., including Phytophthora capsici, Phytophthora cryptogea, Phytophthora drechsleri, Phytophthora infestans, and Phytophthora nicotianae, have been reported to infect tomato worldwide (Pane et al., 2000; Jones et al., 2016). In Italy, P. nicotianae is the main species associated with the disease (Garibaldi and Gullino, 2010). P. nicotianae is worldwide recognized as one of the most devastating oomycete plant pathogens with a very broad host range of more than 255 plant species, including model plants such as Nicotiana tabacum and Arabidopsis thaliana (Kamoun et al., 2015; Panabières et al., 2016). The progress in the knowledge of the genomics of P. nicotianae makes it a suitable model to understand the molecular basis of pathogenesis of oomycete plant pathogens (Meng et al., 2014).
Plant pathogens secrete arsenals of proteins (effectors) that enable parasitic infection and reproduction (Birch et al., 2006; Stassen and Van den Ackerveken, 2011). Plants recognize the initial pathogen-associated molecular pattern (PAMPs) signals and activate pattern-triggered immunity (PTI) to counteract the further colonization by the pathogen. Successful pathogens have developed wide effector repertoires that not only function directly as toxins to induce plant cell death but can also suppress PTI and trigger susceptibility of the plant (Jones and Dangl, 2006). Phytophthora species also secrete a large array of effectors during infection of the plant hosts (Stam et al., 2013). Effectors of several species of Phytophthora have been identified (Chepsergon et al., 2020; McGowan et al., 2020). Among these, the Crinkler (CRN) proteins are a family of effectors that cause necrosis in the cells of the host and also induce further intracellular effectors that target the host nucleus during infection (Stam et al., 2013). The necrosis-inducing Phytophthora protein 1 (NPP1) is another important Phytophthora effector which has been associated with the induction of necrosis in parsley, A. thaliana, and potato (Fellbrich et al., 2002; Gijzen and Nürnberger, 2006). After artificial infiltration, this protein has also been observed to induce the transcription of PR-genes in A. thaliana leaves (Fellbrich et al., 2002). An additional important group of effectors includes cell wall glycoproteins named cellulose-binding elicitor lectin (CBEL); they have been found localized in the inner and outer layers of the Phytophthora mycelium cell walls and are present in close contact with the host cell during infection (Séjalon et al., 1995). Previous studies carried out on tobacco demonstrated that artificial infiltration with CBEL results in local necrosis of the infiltrated area and the induction of an array of defense responses (Séjalon-Delmas et al., 1997).
As a consequence of the restrictions in the use of synthetic fungicides due to their toxicity to humans and animals as well as to their environmental impact, there has been growing interest in alternative approaches to chemical control of P. nicotianae, including the application of biocontrol agents (Singh and Islam, 2010; Gilardi et al., 2014a, b). Although the genomics of the infection process of host plants by Phytophthora spp. as well as the plant root colonization process and the mycoparasitism of fungal pathogens by Trichoderma spp. has been extensively investigated (Kubicek et al., 2011; Atanasova et al., 2013; Meng et al., 2014; Crutcher et al., 2015; Guzmán-Guzmán et al., 2017; Köhl et al., 2019; Morán-Diez et al., 2019; Ramírez-Valdespino et al., 2019; Chepsergon et al., 2020; Pachauri et al., 2020; Wang et al., 2020), there is limited information on the complex interaction of plant-beneficial antagonistic microorganism-pathogen, as analyzed in a comprehensive experimental workflow considering together physiological effects, metabolic pathways, and genes involved in this tripartite interaction (Figure 1).
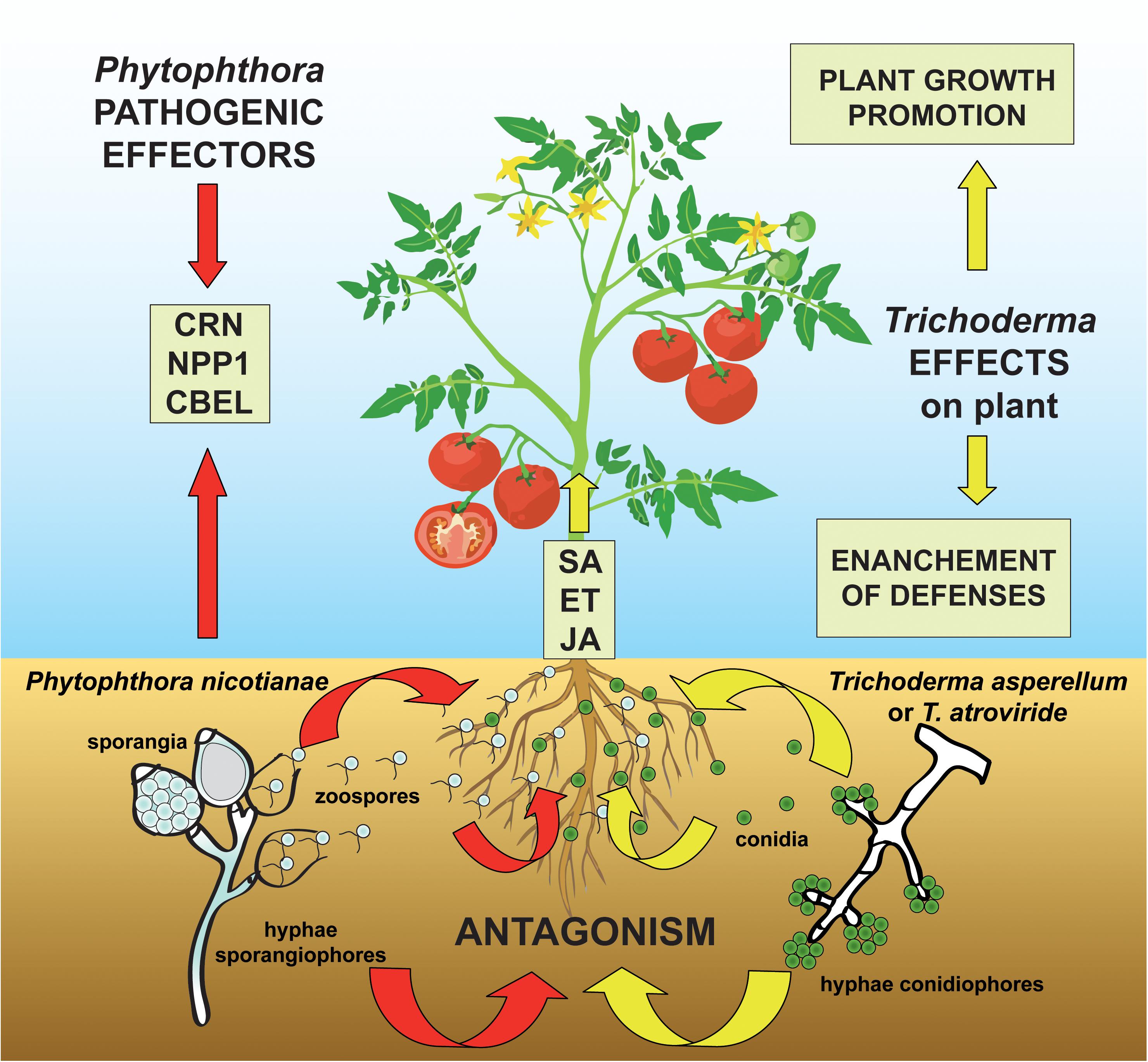
Figure 1. Proposed model for the three-way system plant-pathogen-antagonist showing how Trichoderma species can modulate the molecular signaling in the challenge between the oomycete pathogen Phytophthora nicotianae and the host plant tomato. The colonization of the tomato rhizosphere by Trichoderma spp. triggers both growth promotional effects and plant defense mechanisms by the elicitation of salicylic acid (SA)-, ethylene (ET)-, and jasmonic acid (JA)-dependent processes. At the same time, the P. nicotianae-parasitic infection process is mediated by the secretion of pathogenic effectors (including Crinkler–CRN–proteins, the necrosis-inducing Phytophthora protein 1–NPP1, and cellulose-binding elicitor lectin–CBEL–glycoproteins) which act to suppress the ET- and JA-plant response and whose synthesis is modified by the antagonistic interaction with Trichoderma spp.
To gain a better understanding of the events involved in the plant-microorganisms interaction, this study focuses on (i) the evaluation of the ability of two selected strains of Trichoderma asperellum and Trichoderma atroviride to promote the growth of Solanum lycopersicum and control root and crown rot incited by P. nicotianae and (ii) the identification of the main differentially expressed genes and metabolic pathways activated as a consequence of tripartite interaction in the experimental system tomato–Trichoderma spp.–P. nicotianae.
Materials and Methods
Fungal Isolates
The pathogen P. nicotianae, strain Ph_nic, was sourced from roots of a symptomatic plant of a local cultivar of S. lycopersicum in a nursery in Sicily. For the isolation, infected roots were firstly separated from the stem of the plant and washed with distilled water; then, washed roots were blotted dry and plated on selective PARPNH V8-agar (Jung et al., 2019) and examined under a stereomicroscope for the presence of Phytophthora-like coenocytic hyphae after 48 h of incubation at 28°C in the dark. Then, pieces cut from the advancing margins of the colony were sub-cultured on V8-agar Petri dishes and incubated at 28°C in the dark for a week. Purified cultures were finally obtained by single hyphal culture on V8-agar.
The antagonist T. asperellum strain IMI393899 (Puglisi et al., 2012; Cacciola et al., 2015), previously identified as Trichoderma harzianum, belonged to the collection of the Molecular Plant Pathology Laboratory (MPPL); Department of Agriculture, Food, and Environment (Di3A); University of Catania, while the T. atroviride, isolate TS, was obtained from the parasitized basidiocarp of a specimen of Ganoderma lucidum collected in Apulia (southern Italy). For the isolation of T. atroviride, infected tissues from the parasitized basidiocarp of G. lucidum were excised in 5-mm fragments, disinfected with 1% NaClO for 2 min, rinsed in sterile distilled water, and plated on Potato Dextrose Agar (PDA) amended with streptomycin sulfate at the concentration of 0.25 g/l. After 24 h of incubation at 25°C in the dark, growing colonies were sub-cultured on PDA plates. Purified cultures were finally obtained by single spore culture on PDA medium.
Molecular Identification of Fungal and Oomycete Isolates
The identification of the isolates of P. nicotianae and T. atroviride was carried out by the amplification and analysis of the internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA). In this study, the DNA was extracted by using PowerPlant® Pro DNA Isolation Kit following the manufacturer’s instructions. The amplifications of the ITS regions of the rDNA of Phytophthora and Trichoderma isolates were performed by using the Taq DNA polymerase, recombinant (InvitrogenTM) with the universal primer pairs ITS-6 (5′-GAAGGTGAAGTCGTAA CAAGG-3′) (Cooke et al., 2000) and ITS-4 (5′-TCCTCCGCTT ATTGATATGC-3′) (White et al., 1990) and ITS-5 (5′-GGAAGT AAAAGTCGTAACAAGG-3′) (White et al., 1990) and ITS-4, respectively. The PCR amplifications were carried out in a 25-μl reaction mix containing PCR buffer (1×), dNTP mix (0.2 mM), MgCl2 (1.5 mM), forward and reverse primers (0.5 μM each), Taq DNA polymerase (1 U), and 100 ng of DNA. The thermo-cycler conditions were as follows: 94°C for 3 min; followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and then 72°C for 10 min. Obtained amplicons were detected in 1% agarose gel and sequenced in both directions by an external service (Macrogen). Obtained sequences were analyzed by using FinchTV v.1.4.01. For species identification, blast searches in GenBank2 were performed.
Two representative isolates, namely, Ph_nic (P. nicotianae) and TS (T. atroviride), were randomly selected for further experimentations. T. asperellum strain IMI393899 had been previously identified (Puglisi et al., 2012; Cacciola et al., 2015) with the same procedure used for T. atroviride and stored in the collection of MPPL, Di3A.
Treatment of Tomato Plants With Trichoderma Strains
In order to investigate the plant growth-promoting effect and induction of resistance to the pathogen infection by the symbiotic interaction with Trichoderma spp., tomato plants were grown in association with the selected Trichoderma strains at the root system.
The Trichoderma-tomato interaction was established in accordance with the method described by Tucci et al. (2011). Seeds of S. lycopersicum cv. Cuor di bue (Vilmorin Italia S.R.L., Funo, Bologna, Italy) were sterilized in 2% NaClO for 20 min, rinsed in sterile distilled water, and incubated in a conidial suspension (106 conidia/ml) of either IMI393899 or TS; control seeds were suspended in water. Both treated and control seeds were air dried for 24 h and then sown in sterilized universal soil (©Cifo Srl, Giorgio di Piano, Bologna, Italy) in 40-well polystyrene trays and maintained in a growth chamber at 23°C, 80% relative humidity, and a photoperiod of 16 h of light and 8 h of dark. After 22 days, seedlings were transplanted in 200-cm3 plastic pots in sterilized universal soil. The positive-root colonization by Trichoderma was verified by re-isolation on PDA from roots of additional tomato control plants; the identity of the purified cultures of the Trichoderma strains was confirmed by PCR amplification and sequencing of their ITS region. These tomato plants will be called “Trichoderma-treated plants” in the text.
Growth Promotion of Tomato Plants
Twenty-two-day-old tomato seedlings were grown with either strain IMI393899 of T. asperellum or strain TS of T. atroviride, which colonized the root systems and were grown for 12 weeks in a growth chamber at 23°C, 80% relative humidity, and a photoperiod of 16 h of light and 8 h of dark. Untreated seedlings (controls) were grown in the same conditions. A normal weekly irrigation was also provided.
The experimental set-up consisted of three treatments with 10 repetitions each: (i) untreated tomato plants (controls), (ii) tomato plants grown with T. asperellum strain IMI393899, and (iii) tomato plants grown with T. atroviride strain TS.
The promotion of the plant growth of “Trichoderma-treated plants” was then evaluated as follows: (i) weekly stem growth rate (cm/week), (ii) seedling total length, (iii) fresh root weight, and (iv) length at the end of the test (i.e., 12 weeks after the transplanting). All data were analyzed by using one-way ANOVA followed by Tukey’s honestly significant difference (HSD) test as a post hoc test (R software). Differences at P ≤ 0.05 were considered significant.
At the end of the test, T. asperellum strain IMI393899 and T. atroviride strain TS were re-isolated from roots of tomato plants from the respective treatments and then sequenced.
Biological Control of Phytophthora nicotianae
In vitro Antagonistic Ability
The selected strains of T. asperellum and T. atroviride were screened for their ability to inhibit the mycelial growth (%) of P. nicotianae strain Ph_nic by in vitro dual culture assays. The formula applied was as follows:
where,
X = growth of pathogen alone without antagonist (control)
and
Y = growth of pathogen along with the antagonist.
The dual culture test was carried out in 90-mm Petri dishes with 20 ml of PDA by placing 5-mm diameter agar plugs of the pathogen and antagonists taken from the margin of 1-week-old colonies grown on V8-agar and PDA, respectively. The dual culture was set by placing the pathogen and the antagonist 4 cm apart from each other. Furthermore, since the daily radial growth rate of the P. nicotianae strain Ph_nic was significantly lower than that of Trichoderma isolates, P. nicotianae was plated 72 h before the Trichoderma sp. Single cultures of the pathogen were used as control. Plates were incubated at 28°C in the dark, and radial mycelial growth was measured when P. nicotianae mycelium of control cultures covered the whole Petri dishes (namely, 13 days after incubation). Overall, the experimental set-up consisted of the following three treatments (including controls) made of 10 replicates each: (i) P. nicotianae isolate Ph_nic, (ii) P. nicotianae isolate Ph_nic + T. asperellum strain IMI393899, and (iii) P. nicotianae isolate Ph_nic + T. atroviride strain TS.
Data from the inhibition of the growth (%) of P. nicotianae 13 days from the beginning of the trial were analyzed by using one-way ANOVA followed by Tukey’s HSD test as a post hoc test (R software). Differences at P ≤ 0.05 were considered significant.
In planta Antagonistic Ability
The in planta antagonistic ability of T. asperellum strain IMI393899 and T. atroviride strain TS toward P. nicotianae, strain Ph_nic, was demonstrated on tomato plants in a soil infestation test. Inoculum consisted of 12-day-old culture of the isolate Ph_nic grown at 25°C in a 750-ml jar containing 140 ml autoclaved V8-juice broth (200 ml/l juice and 800 ml/l distilled water amended with 3 g/l CaCO3) (Jung et al., 1996) and 170 ml of millet seeds. This experimental trial was carried out in two different steps which included a double treatment with the Trichoderma strains.
First Step
For the test, 4-month-old potted plants of S. lycopersicum cv. Cuor di bue, grown in sterilized universal soil with either T. asperellum strain IMI393899 or T. atroviride strain TS, in association with the root system (“Trichoderma-treated tomato seedlings”), were transplanted into 1,000-cm3 pots filled with a mixture of sterilized soil and the inoculum of Ph_nic prepared as described above (20 cm2 of inoculum per 1,000 cm3 of potting mixture). Untreated plants (controls) were not grown in association with Trichoderma strains; they were transplanted into pots filled with a mixture of sterilized soil and non-infested millet seed/V8-juice medium at the same rate.
Second Step
After transplanting, additional 100 ml of a conidial suspension (106 conidia/ml) from T. asperellum IMI393899 and T. atroviride TS were provided to each plant from respective Trichoderma pre-treatment. All plants were then irrigated and maintained in a growth chamber at 23°C, 80% relative humidity, and a photoperiodic lighting of 16 h of light and 8 h of dark; a normal irrigation was also provided twice per week.
Overall, the experimental assay consisted of the following six treatments: (i) untreated tomato plants transplanted in a non-infested potting mixture (NI-PM); (ii) untreated tomato plants inoculated with infested potting mixture (I-PM); (iii) tomato plants treated with T. asperellum strain IMI393899 and transplanted in NI-PM; (iv) tomato plants treated with T. asperellum strain IMI393899 and transplanted in I-PM; (v) tomato plants treated with T. atroviride strain TS and transplanted in NI-PM; and (vi) tomato plants treated with T. atroviride strain TS and transplanted in I-PM. Each treatment included 10 replicates. The test was considered completed when plants of treatment (ii) showed severe symptoms of decay (i.e., 15 days after inoculation).
Plant damage was assessed on the basis of three different parameters: (i) wilting severity, visually evaluated in accordance with the empirical scale reported by Engelbrecht et al. (2007); (ii) fresh root weight; and (iii) fresh root length; the last two parameters were determined by separating the root system from the rest of the plant. The empirical scale used to rate the severity of wilting included the following values: 1 = normal (not wilted)–no signs of wilting or drought stress; 2 = slightly wilted–slight leaf angle changes but no folding, rolling, or changes in leaf surface structure; 3 = wilted–strong leaf angle change or protrusion of veins on the leaf surface but no cell death; 4 = severely wilted–very strong change of leaf angle or protrusion of veins on the leaf surface with initial necrosis; 5 = nearly dead–most leaves necrotic, some young leaves still green near the midrib, and leaf angles mostly near 0; 6 = dead–all above-ground parts dead.
At the end of the test, T. asperellum strain IMI393899, T. atroviride strain TS, and P. nicotianae isolate Ph_nic were re-isolated from plants of respective treatments and their identity was confirmed by PCR amplification and sequencing of their ITS region.
Data were analyzed by using one-way ANOVA followed by Tukey’s HSD test as a post hoc test (R software). Differences at P ≤ 0.05 were considered significant.
Gene Expression in the Three-Way System Tomato–Trichoderma spp.–Phytophthora nicotianae Assay
Fungal Isolates
Trichoderma asperellum strain IMI393899 and T. atroviride strain TS were cultured on PDA for 7 days at 25°C, while P. nicotianae isolate Ph_nic was cultured in Petri dishes on V8-agar for 1 week at 28°C in the dark.
Tomato Plants
Tomato seeds (S. lycopersicum cv. Cuor di bue–Vilmorin Italia S.R.L., Funo, Bologna, Italy) were sterilized in 2% NaClO for 20 min, rinsed in sterile distilled water, and sown in an alveolar tray containing sterile vermiculite soaked in a nutrient solution (NS) prepared in accordance with Guérin et al. (2014) and Lebreton et al. (2018) with the following modifications: fertilizer 20-20-20 (Asso di Fiori-Cifo, S. Giorgio di Piano, Bologna, Italy) (0.1634 g/l), MgSO4 × 7H2O (0.15 g/l), FeNa-EDTA (40 mg/l). Trays were kept for 3 days in the dark at 23°C and 80% relative humidity; then, seedlings were transferred to a photoperiodic lighting (16 h of light:8 h of dark) and kept at the same temperature conditions and relative humidity for 30 days. Moreover, 30 ml of NS were provided once a week to renew the content of mineral salts; tomato plantlets were also watered twice a week. Seedlings were then transferred into plastic tubes containing 30 ml of NS.
Trichoderma spp. Colonization Assay
Thirty-day-old tomato seedlings growing in the aforementioned plastic tubes were treated with 300 μl of a suspension of germinated conidia (100 conidia/ml) of T. asperellum strain IMI393899 and T. atroviride strain TS. The suspension of germinated conidia of Trichoderma spp. was prepared as reported in Yedidia et al. (1999) with the following modifications: two flasks containing 100 ml each of a synthetic medium consisting of the aforementioned NS amended with 15 g/l of sucrose were autoclaved and then inoculated with 1 ml of conidial suspension (106 conidia/ml) of each Trichoderma obtained from 7-day-old cultures grown on PDA medium; flasks were then shaken at 150 rpm for 24 h at 25°C to allow spore germination; after 24 h, tubes containing tomato seedlings were inoculated with 300 μl of the suspension of germinated conidia. Controls were inoculated with the NS amended with 15 g/l of sucrose.
Phytophthora nicotianae Infection Assay
Forty-eight hours after the treatment with Trichoderma spp., tomato seedlings were inoculated with zoospores of P. nicotianae (concentration: 100 zoospores/ml). P. nicotianae inoculum was prepared as follows: mycelial plugs from a 7-day-old culture of P. nicotianae grown on V8-agar were flooded with 20 ml of sterile distilled water and incubated at 25°C for 48 h under a constant fluorescent light. Zoospores were released in sterile distilled water by mature sporangia by placing mycelial plugs at 6°C for 1 h followed by another hour at 25°C. Zoospore concentration was measured by using a hemocytometer. Controls were inoculated with sterile distilled water.
Experimental Assay
Overall, the experimental assay consisted of the following six treatments: (i) untreated tomato seedlings; (ii) untreated tomato seedlings inoculated with P. nicotianae isolate Ph_nic; (iii) “Trichoderma-treated tomato seedlings” with T. asperellum strain IMI393899; (iv) “Trichoderma-treated tomato seedlings” with T. atroviride strain TS; (v) “Trichoderma-treated tomato seedlings” with T. asperellum strain IMI393899 and inoculated with P. nicotianae Ph_nic; and (vi) “Trichoderma-treated tomato seedlings” with T. atroviride strain TS and inoculated with P. nicotianae isolate Ph_nic. Each treatment was made up of six replicates. The test was considered completed (7 days after the inoculation of P. nicotianae) when seedlings of treatment (ii) showed severe symptoms of disease.
At the end of the test, seedlings from each treatment were collected and immediately frozen in liquid nitrogen and stored at −80°C. At the end of the test, T. asperellum strain IMI393899, T. atroviride strain TS, and P. nicotianae isolate Ph_nic were re-isolated and then sequenced, from additional seedlings from respective treatments.
RNA Isolation From Colonized Tomato Seedlings and cDNA Synthesis
Total RNA was extracted by using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) from frozen stem and roots from tomato seedlings (100 mg) ground to a fine powder with liquid nitrogen, following the protocol of the manufacturer and treated with TURBO DNA-freeTM Kit. RNA concentration was then adjusted to 200 ng/μl, and its quality was verified by performing a denaturing RNA electrophoresis gel in TAE agarose (Masek et al., 2005). Reverse transcription was performed by using High-Capacity cDNA Reverse Transcription Kit (Applied BiosystemsTM, Foster City, CA, United States) following the manufacturer’s instructions.
Selection of Genes and Development of Specific Primers
Several genes from tomato, Trichoderma spp., and P. nicotianae involved in the tripartite interaction plant-antagonist-pathogen were selected (Supplementary Table S1). Both housekeeping and target genes from tomato and P. nicotianae were selected from previous studies (Tucci et al., 2011; Cui et al., 2018; Dalio et al., 2018). An NCBI nucleotide database3 search was carried out to select specific sequences from both endochitinase and housekeeping genes in Trichoderma spp. In order to obtain the highest primer specificity, sequences of genes LOC101262163, PR1b1, TomLoxA, SlyDF2, PpCRN4, PpCBEL4, PpNPP1.1, PpNPP1.3, PpNPP1.4, EF-1α, chi42, Gp_dh_N, and CHI18-5 (Supplementary Table S1) were directly derived from the respective genomic region as reported in the GenBank “whole genome shotgun sequencing project” of the respective organism (GenBank accession numbers: AEKE00000000.3–Tomato; MBGH00000000.1–T. asperellum; ABDG00000000.2–T. atroviride; AVGE00000000.1–P. nicotianae), and respective primer pairs were designed by using the Primer BLAST NCBI tool4; specificity of all selected primers was tested both by in silico (by using the Primer BLAST NCBI tool) and PCR amplification and sequencing of the target region.
Quantitative Real-Time PCR Analysis of Gene Expression
Amplifications were performed by using the iCycler iQTM Real-Time PCR Detection System (Bio-Rad). Reactions were performed in a total volume of 20 μl by mixing 10 ng of cDNA with 1 μl of 10 μM of each primer and 10 μl of PowerUpTM SYBRTM Green Master Mix (2×) (Applied Biosystems). Quantitative real-time PCR (qRT-PCR) experiments were carried out in triplicate. The thermo-cycling conditions were 2 min at 50°C (UDG activation) and 2 min at 95°C (Dual-LockTM DNA polymerase) followed by 40 cycles of two steps: 95°C for 15 s (denaturation) and 59°C or 60°C (annealing/extension) for 1 min. The quantification of gene expression was carried out by using the 2–ΔΔCt method (Livak and Schmittgen, 2001). For each organism involved in the experiments, calibrator samples were represented by six replicates of the following: (i) untreated tomato seedling control samples and 7-day-old cultures of (ii) P. nicotianae isolate Ph_nic, (iii) T. asperellum strain IMI393899, and (iv) T. atroviride strain TS grown on NS-agarized medium (16 g/l of agar) amended with 15 g/l of sucrose.
The PCR efficiency was checked by standard curve Ct values vs. log (cDNA dilution). Curves were constructed by serial 10-fold dilution of cDNA for each primer pair; linear equations, determination coefficients (R2), and reaction efficiencies are given in Supplementary Table S2.
Data on gene expression were analyzed by using one-way ANOVA followed by Dunnett’s multiple comparisons test by using R software. Differences at P ≤ 0.05 were considered significant.
Results
Growth Promotion Assay
Results from growth promotion test showed that the growth of plants treated with T. atroviride strain TS was significantly stimulated compared with T. asperellum IMI393899-treated plants and untreated control plants (Figure 2). Overall, the weekly shoot growth of plants treated with T. atroviride strain TS was on average ca. 0.5 cm more than both the untreated and T. asperellum-treated plants, while the weekly shoot growth of T. asperellum-treated plants did not differ statistically from the untreated control (Figure 2).
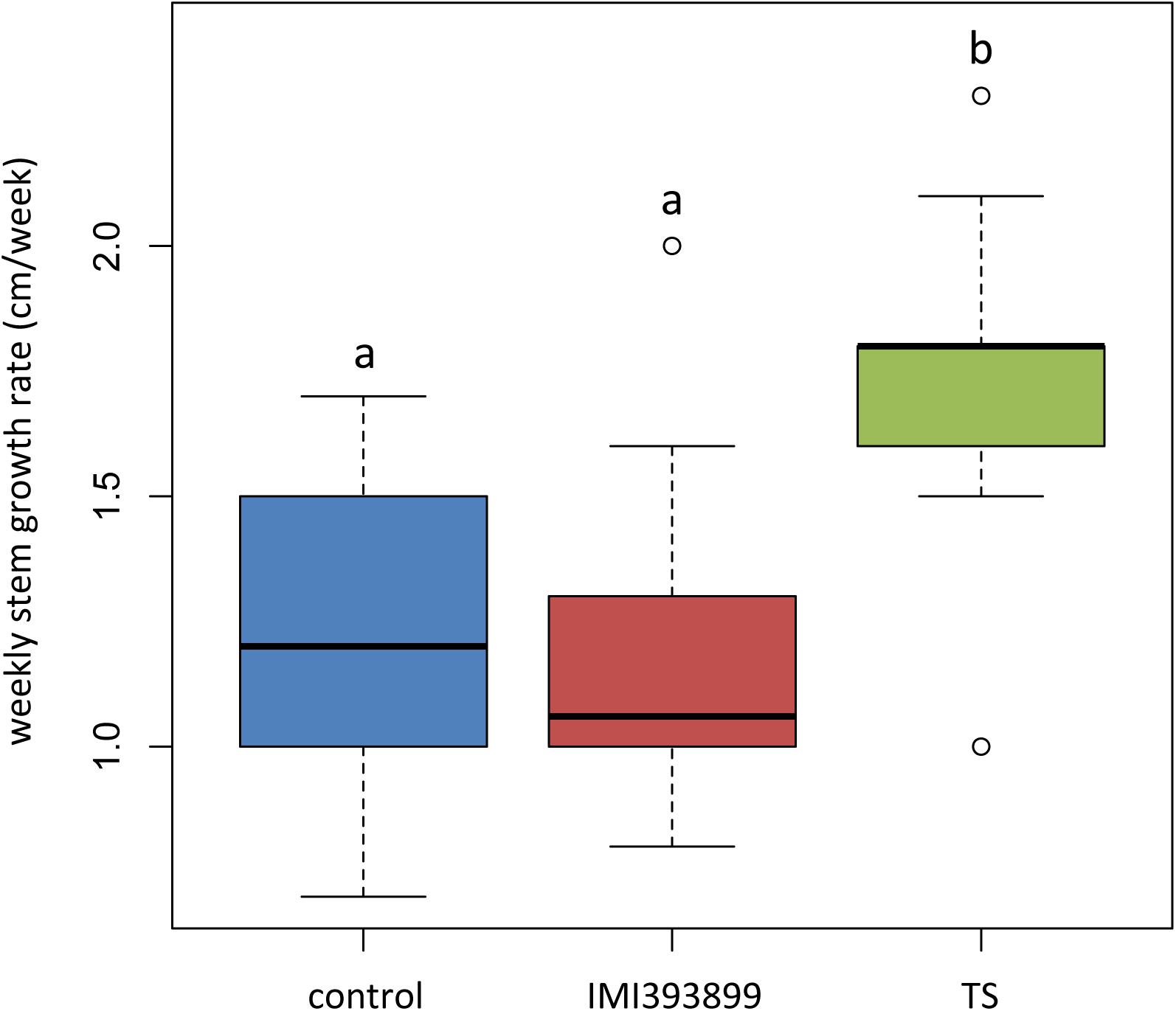
Figure 2. Effects of Trichoderma asperellum IMI393899 and Trichoderma atroviride TS treatments on the growth of Solanum lycopersicum cv. Cuor di bue seedlings. Weekly stem growth rate. Open and gray circles represent outliers and mean value data, respectively. Values sharing same letters are not statistically different according to Tukey’s honestly significant difference (HSD) test (P ≤ 0.05).
The same attitude in growth promotion was also confirmed by values of root length and fresh weight (Figures 3A,B). Overall, the treatment with T. asperellum IMI393899 reduced the growth of the root system of the plants, while plants treated with T. atroviride strain TS did not differ from the untreated control. The same trend was also observed for the total length of seedlings (Figure 4).
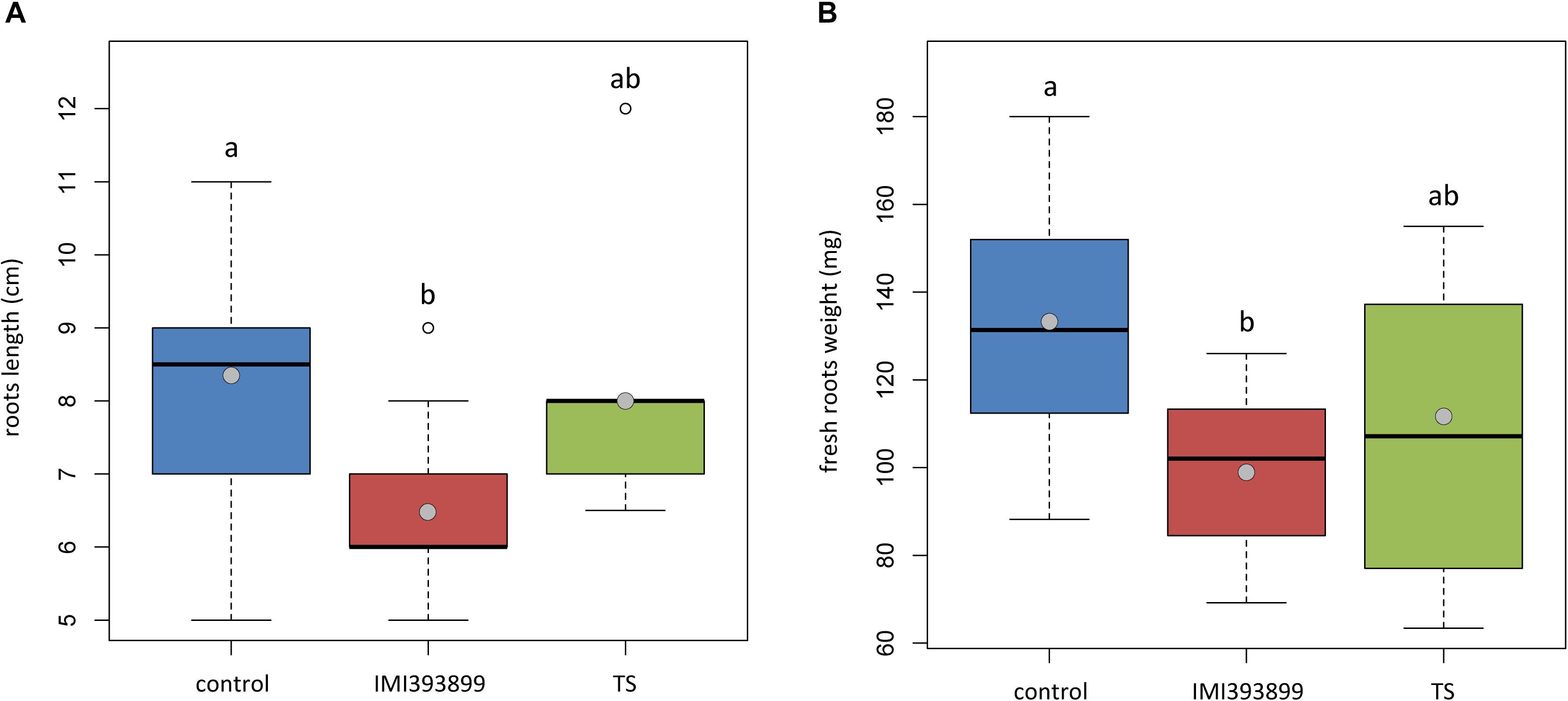
Figure 3. Effects of Trichoderma asperellum IMI393899 and Trichoderma atroviride TS treatments on the growth of Solanum lycopersicum cv. Cuor di bue seedlings. (A) Stem length and (B) fresh root weight at the end of the test. Open and gray circles represent outliers and mean value data, respectively. Values sharing same letters are not statistically different according to Tukey’s honestly significant difference (HSD) test (P ≤ 0.05).
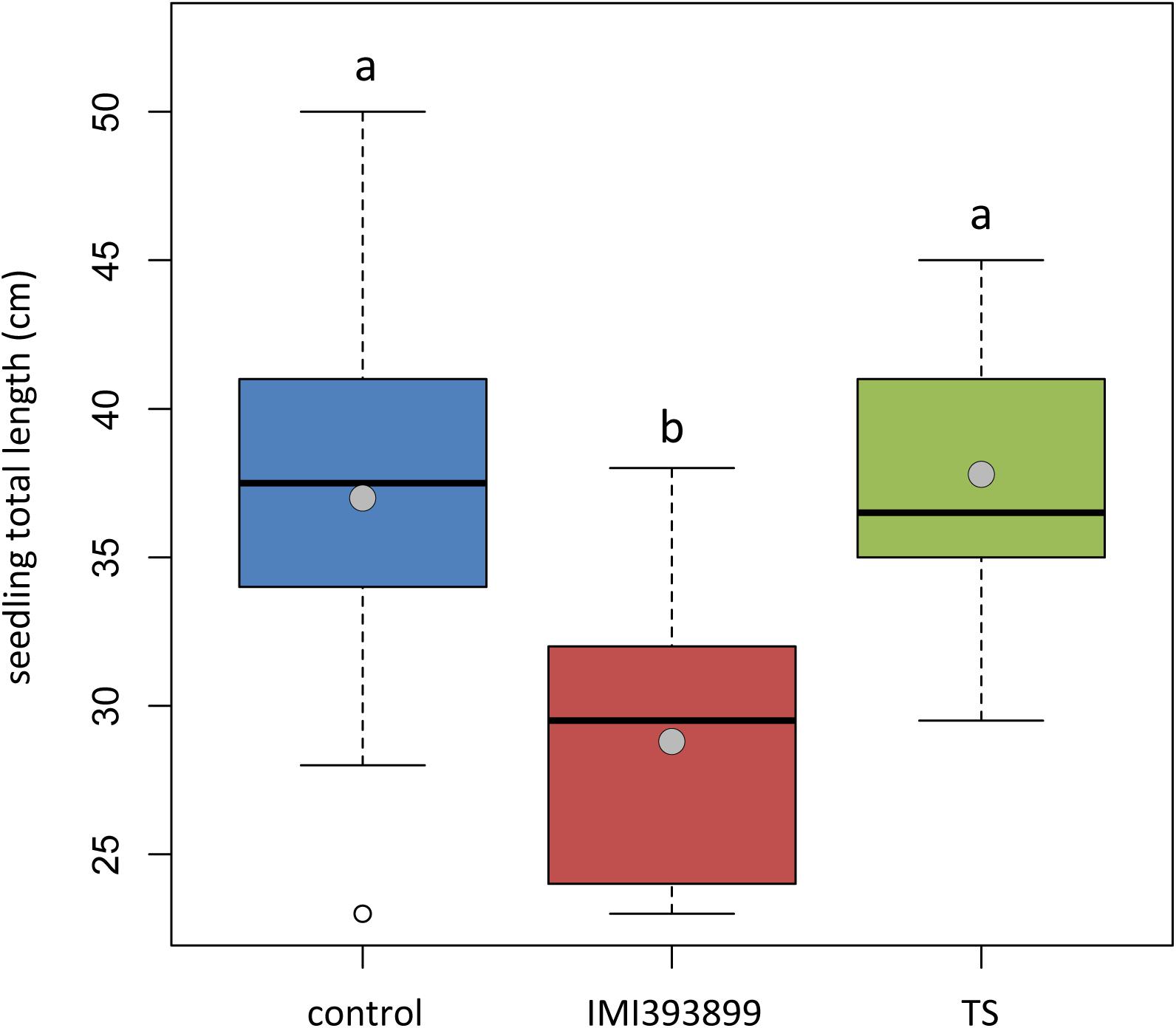
Figure 4. Effects of Trichoderma asperellum IMI393899 and Trichoderma atroviride TS treatments on the growth of Solanum lycopersicum cv. Cuor di bue seedlings. Seedling total length at the end of the test. Open and gray circles represent outliers and mean value data, respectively. Values sharing same letters are not statistically different according to Tukey’s honestly significant difference (HSD) test (P ≤ 0.05).
Biological Control of Phytophthora nicotianae
In vitro Antagonistic Ability
Results from dual culture trial showed that both Trichoderma-tested isolates had an antagonistic effect on the growth of P. nicotianae (Table 1). In particular, the T. atroviride strain TS was more effective and inhibited the P. nicotianae growth by 63.50%, while T. asperellum strain IMI393899 inhibited the growth of P. nicotianae by 58.77% (differences between means were significant).
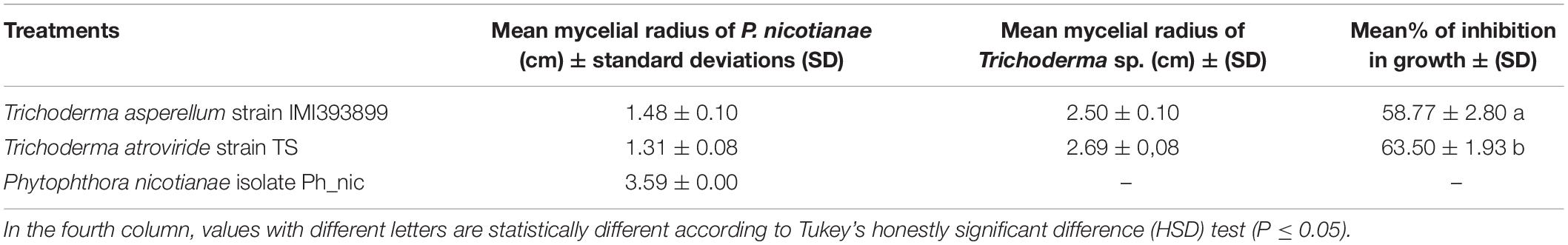
Table 1. In vitro antagonistic ability of Trichoderma asperellum IMI393899 and Trichoderma atroviride test strains against Phytophthora nicotianae isolate Ph_nic.
In planta Antagonistic Ability
At the end of the trial, both untreated plants and plants treated with Trichoderma spp. and transplanted into non-infested potting mixture (NI-PM), namely, those from treatments (i), (iii), and (v), were substantially asymptomatic showing a mean rating of wilting of 1.60, 1.60, and 1.30, respectively (Figure 5) (differences among means were not significant). They also showed a healthy root system, with the only exception of plants treated with T. asperellum IMI393899 that showed a statistically significant reduction of fresh weight and length compared with untreated controls (Figures 6A,B).
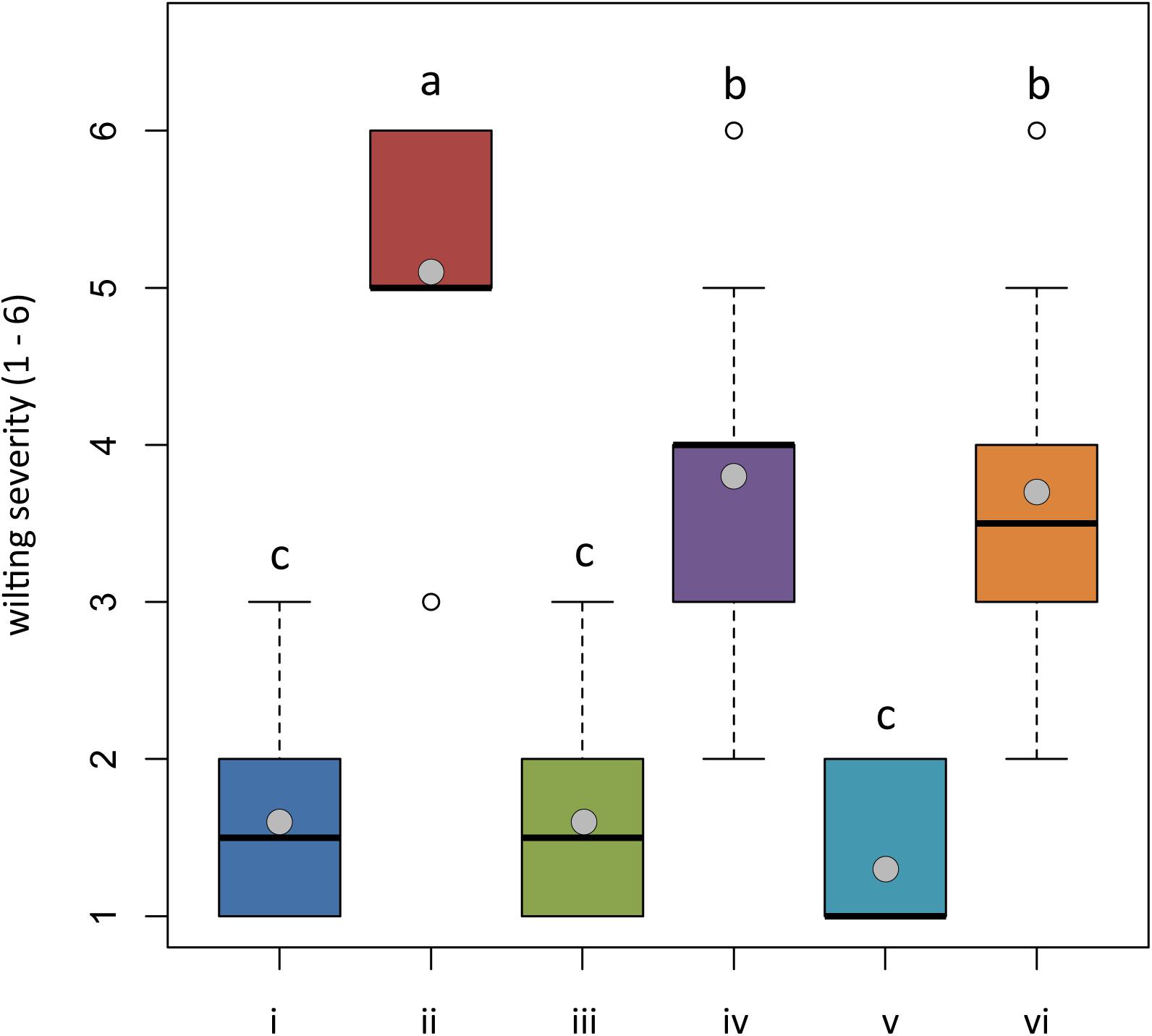
Figure 5. In planta antagonistic effectiveness of Trichoderma test strains against Phytophthora nicotianae. Wilting severity of 4-month-old Solanum lycopersicum cv. Cuor di bue developed from the following: (i) untreated tomato plants transplanted in a non-infested potting mixture (NI-PM); (ii) untreated tomato plants inoculated with infested potting mixture (I-PM); (iii) tomato plants treated with Trichoderma asperellum strain IMI393899 and transplanted in NI-PM; (iv) tomato plants treated with T. asperellum strain IMI393899 and transplanted in I-PM; (v) tomato plants treated with Trichoderma atroviride strain TS and transplanted in NI-PM; and (vi) tomato plants treated with T. atroviride strain TS and transplanted in I-PM. Open and gray circles represent outliers and mean value data, respectively. Values sharing same letters are not statistically different according to Tukey’s honestly significant difference (HSD) test (P ≤ 0.05).
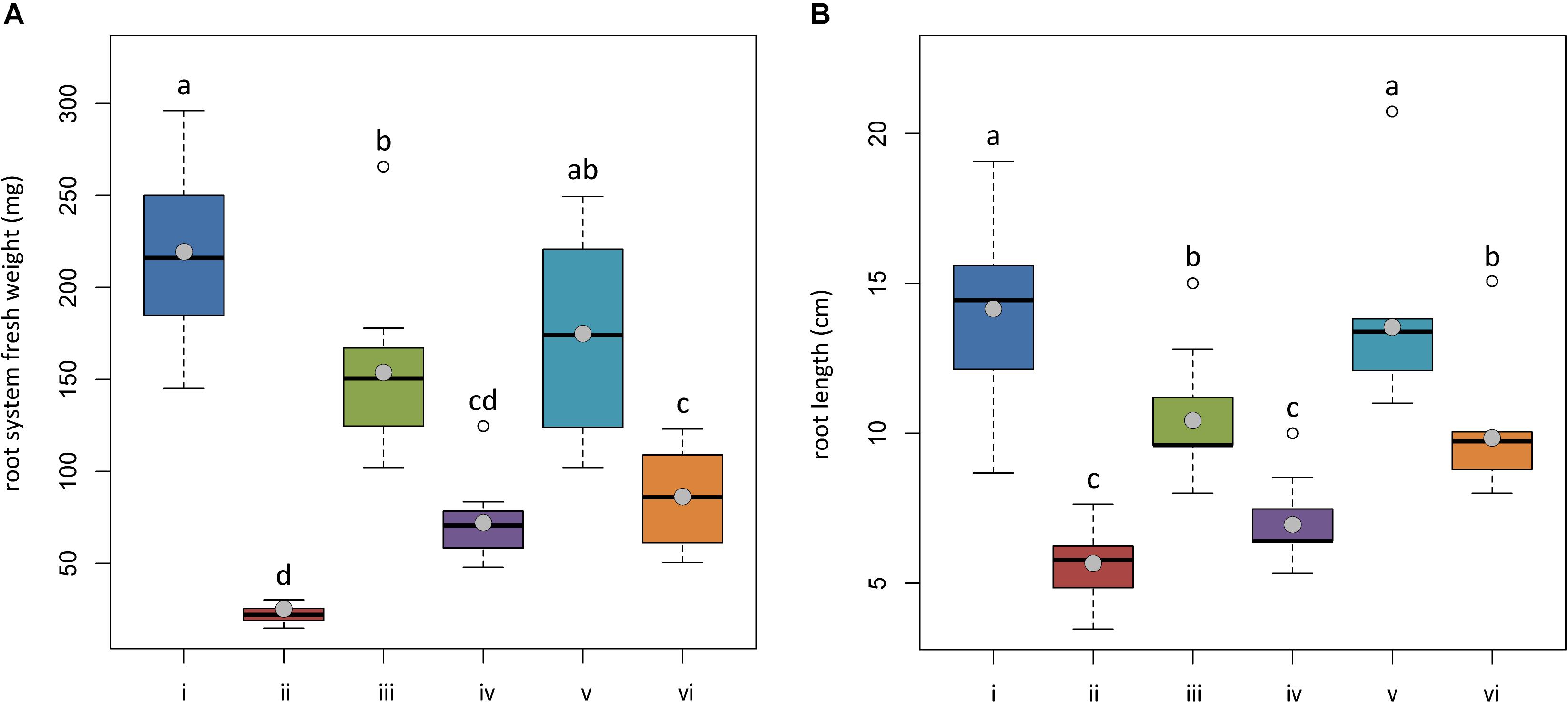
Figure 6. In planta antagonistic effectiveness of Trichoderma test strains against Phytophthora nicotianae. (A) Fresh root weight and (B) length of 4-month-old Solanum lycopersicum cv. Cuor di bue developed from the following: (i) untreated tomato plants transplanted in a non-infested potting mixture (NI-PM); (ii) untreated tomato plants inoculated with infested potting mixture (I-PM); (iii) tomato plants treated with Trichoderma asperellum strain IMI393899 and transplanted in NI-PM; (iv) tomato plants treated with T. asperellum strain IMI393899 and transplanted in I-PM; (v) tomato plants treated with Trichoderma atroviride strain TS and transplanted in NI-PM; (vi) tomato plants treated with T. atroviride strain TS and transplanted in I-PM. Open and gray circles represent outliers and mean value data, respectively. Values sharing same letters are not statistically different according to Tukey’s honestly significant difference (HSD) test (P ≤ 0.05).
Plants inoculated with infested potting mixture (I-PM) showed severe symptoms of wilting (Figure 5) and a substantial reduction of root system (Figures 6A,B). However, the treatment with T. atroviride TS and T. asperellum IMI393899, with 40 and 60% of plant mortality, respectively, significantly reduced the mortality over untreated control plants (90% of mortality). Similarly, plants treated with T. atroviride TS and T. asperellum IMI393899 showed significant higher values of fresh root weight and length than untreated controls. However, overall, T. atroviride TS was more effective than T. asperellum IMI393899 in preventing root rot (Figures 6A,B).
Gene Expression Levels in the Tripartite Interaction Tomato–Trichoderma spp.–Phytophthora nicotianae
Differences in the Expression of Tomato Defense-Related Genes
The defense mechanisms activated by tomato plants upon the simultaneous colonization of the root system by a root pathogen (P. nicotianae) and biocontrol agents (Trichoderma spp.) were evaluated on the basis of the expression of genes involved in the main plant defense pathways, namely, SA (i.e., PR proteins–PR1b1 and PR-P2-encoding genes), JA (i.e., lipoxygenases enzymes–TomLoxC and TomLoxA-encoding genes), and a tomato plant defensin protein (i.e., SlyDF2-encoding gene) usually strongly involved in the tomato-Phytophthora sp. infection process (Cui et al., 2018; Figure 7).
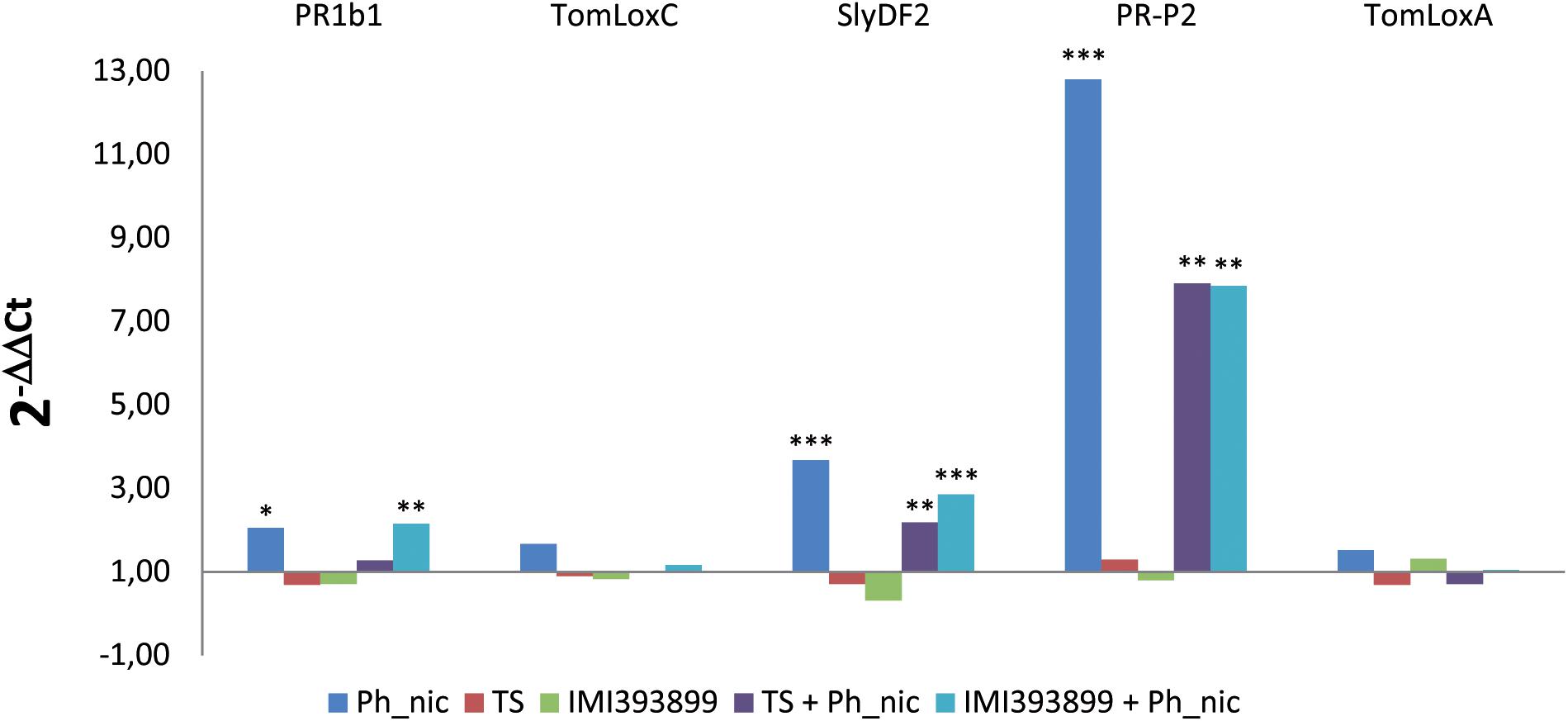
Figure 7. Differences in the expression levels of PR1b, TomLoxC, SlyDF2, PR-P2, and TomLoxA-encoding genes (GenBank accession numbers: Y08804.1, U37839.1, NM_001346524.1, X58548.1 and U09026.1, respectively) from 7-day-old Trichoderma-treated or non-treated Solanum lycopersicum cv. Cuor di bue seedlings inoculated or non-inoculated with Phytophthora nicotianae. Columns with asterisks are statistically different according to Dunnett’s test (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001), compared to their calibrator.
Statistically significant reprogramming in the gene expression was observed for PR1b1, PR-P2, and SlyDF2 in treatments that included the inoculation with the pathogen. PR1b1 was up-regulated in treatments (ii) (i.e., untreated tomato seedlings inoculated with P. nicotianae isolate Ph_nic) and (vi) (i.e., tomato seedlings treated with T. atroviride strain TS and inoculated with P. nicotianae isolate Ph_nic). PR-P2 was strongly up-regulated in treatment (ii), while showed a lower up-regulation in treatments (v) (i.e., tomato seedlings treated with T. asperellum strain IMI393899 and inoculated with P. nicotianae Ph_nic) and (vi). Similarly, the tomato defensin SlyDF2 was up-regulated only in treatments (ii), (v), and (vi). Both lipoxygenase-encoding genes (i.e., TomLoxC and TomLoxA) were expressed at similar levels in all treatments.
Differences in the Expression of Phytophthora nicotianae Pathogenic Effectors
The effector expression of P. nicotianae was evaluated as differences in the relative expression levels of effector genes from different families: CRinkling and Necrosis effector PpCRN4; CBEL PpCBEL4; and three different members of the NEP1-like necrosis-inducing proteins PpNPP1.1, PpNPP1.3, and PpNPP1.4 (Figure 8).
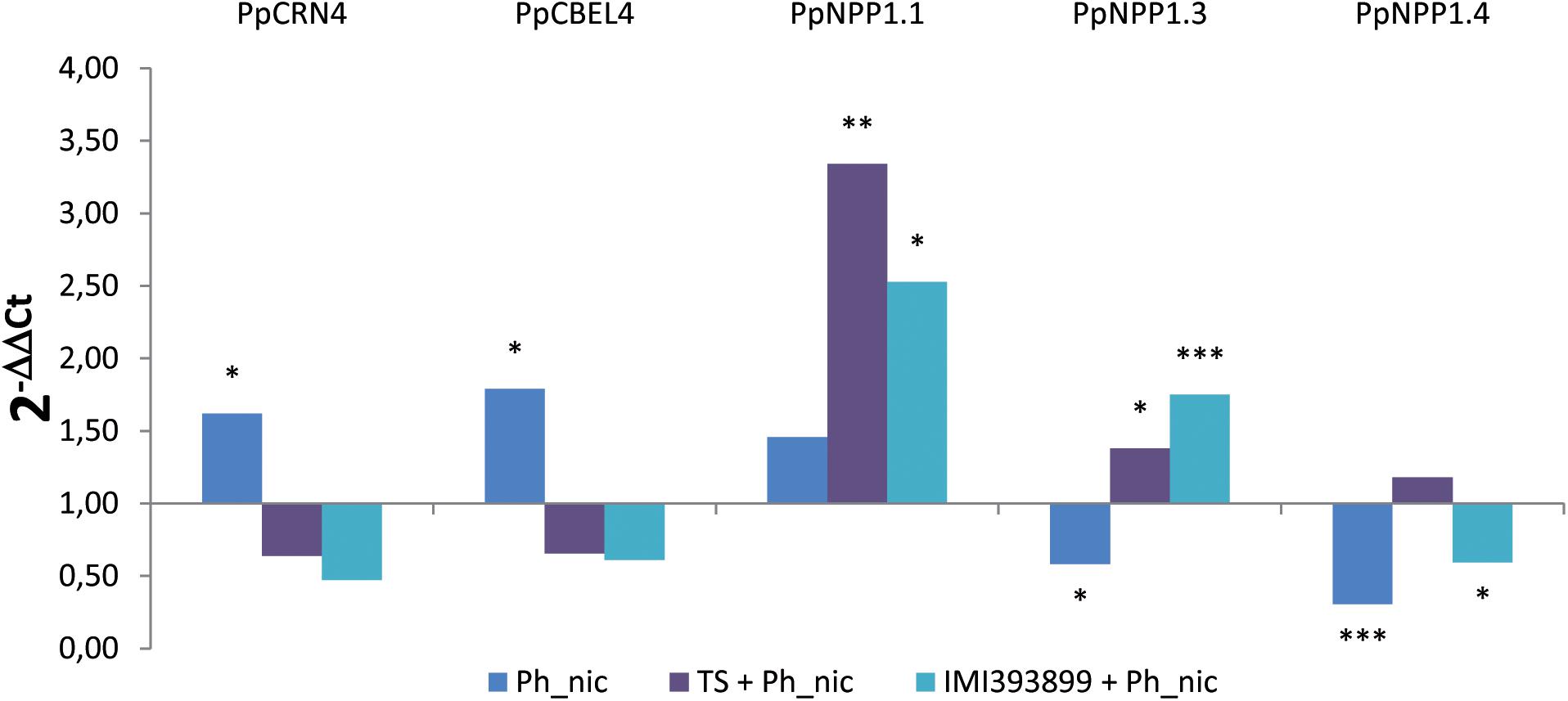
Figure 8. Differences in the expression levels of PpCRN4, PpCBEL4, PpNPP1.1, PpNPP1.3, and PpNPP1.4-encoding genes (GenBank accession numbers: ETM55095.1, ETM43740.1, ETM52620.1, ETM39327.1 and ETM36738.1, respectively) in Phytophthora nicotianae isolate Ph_nic from 7-day-old Trichoderma-treated or non-treated Solanum lycopersicum cv. Cuor di bue P. nicotianae-inoculated seedlings. Columns with asterisks are statistically different according to Dunnett’s test (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001), compared to their calibrator.
Statistically significant differences were observed in the levels of all the effectors; both the PpCRN4 and PpCBEL4 genes were up-regulated only in untreated and inoculated seedlings [i.e., treatment (ii)], while were normally expressed on seedlings inoculated with the pathogen after being treated with TS of T. atroviride and IMI393899 of T. asperellum [i.e., treatments (v) and (vi), respectively]. Referring to the necrosis-inducing Phytophthora protein-encoding genes, PpNPP1.1 showed a strong up-regulation in treatments (v), “Trichoderma-treated seedlings” with IMI393899 and inoculated with Ph_nic, and (vi), tomato seedlings treated with T. atroviride strain TS and inoculated with P. nicotianae isolate Ph_nic. On the contrary, PpNPP1.3 was down-regulated in treatment (ii) and up-regulated in treatments (v) and (vi). Finally, PpNPP1.4 was down-regulated in untreated seedlings and in treatment (v), “Trichoderma-treated seedlings” with IMI393899 and inoculated with Ph_nic.
Differences in the Expression of Trichoderma Antagonistic-Related Gene
The mycoparasitism of both Trichoderma test strains was assessed based on the differential expression of gene encoding for chitinases (i.e., CHI18-5-encoding gene for T. atroviride; chi42-encoding gene for T. asperellum). As expected, both Trichoderma strains up-regulated the respective selected endochitinase-encoding gene exclusively in the treatment with Phytophthora-inoculated seedlings (Figure 9).
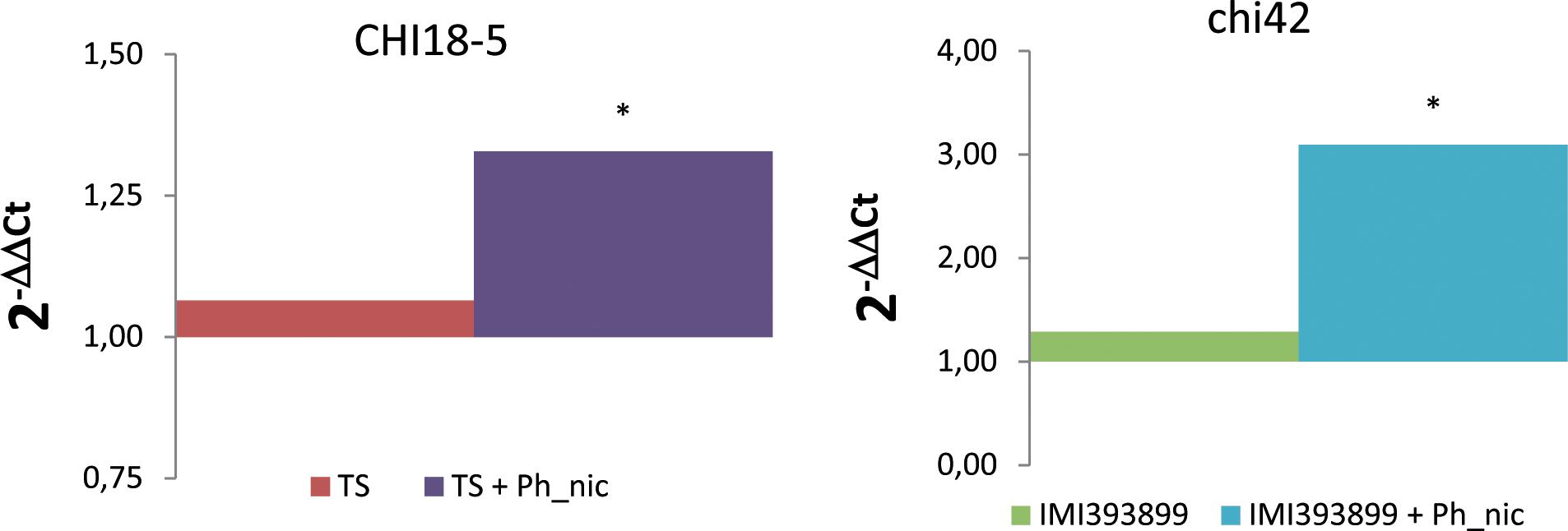
Figure 9. Differences in the expression levels of CHI18-5 (GenBank accession number: XM_014088210) and chi42 (GenBank accession number: HM191684.1)-encoding genes, respectively, in Trichoderma atroviride strain TS (on the left) and Trichoderma asperellum strain IMI393899 (on the right) from 7-day-old Phytophthora nicotianae-inoculated or non-inoculated Solanum lycopersicum cv. Cuor di bue Trichoderma-treated seedlings. Columns with asterisks are statistically different according to Dunnett’s test (∗P < 0.05), compared to their calibrator.
Discussion
This study provides a comprehensive assessment of physiological and molecular mechanisms involved in the complex three-way plant-antagonist-pathogen interaction. Firstly, the effectiveness in the promotion of growth of the model plant S. lycopersicum cv. Cuor di bue by two selected strains of T. asperellum and T. atroviride was compared. Previous studies revealed that different cultivated lines of tomato have a differential response in the promotion of growth by commercial strains of Trichoderma spp. (Tucci et al., 2011); the strains T. atroviride P1 and T. harzianum T22 induced statistically significant improvements in the development of the stem only in one and two (respectively) out of four different tomato lines (i.e., Corbarino, TA209, M82, and SM36), compared with untreated controls. A similar trend was also observed in the root weight with also a significant decrement for one variety (i.e., M82) treated with T. harzianum T22 (Tucci et al., 2011). In general, our results indicated that between the two Trichoderma species, only T. atroviride was significantly able to promote the weekly growth rate of the tomato cv. Cuor di bue. This result confirms the variability of the effects induced by Trichoderma spp. in the promotion of the growth of different tomato varieties. In order to acquire a complete evaluation of the antagonistic activity of the selected T. asperellum IMI393899 and T. atroviride TS strains against the P. nicotianae Ph_nic isolate virulent toward tomato seedlings, in this study, a dual culture test and an in planta antagonistic trial were carried out. Various Trichoderma spp., including T. harzianum, Trichoderma viride, Trichoderma virens, T. asperellum, Trichoderma gamsii, Trichoderma longibrachiatum, and T. atroviride, showed a good antagonistic activity both in vitro and in vivo against several soil-borne fungal pathogens (Smith, 1990; Al-mughrabi, 2008; Mastouri et al., 2010; Singh and Islam, 2010; Haggag and El-Gamal, 2012; Widmer, 2014; Yao et al., 2015; Ghazanfar et al., 2018). Results obtained here from in vitro tests show that both Trichoderma spp. tested strongly inhibited the growth of P. nicotianae isolate Ph_nic, with a more marked inhibition by T. atroviride TS over T. asperellum IMI393899. Similarly, the in planta antagonistic trial showed that T. atroviride TS was more effective than T. asperellum IMI393899 in the reduction of disease severity, even if none of them provided a complete control of the disease. These results are in agreement with those obtained in other similar pathogen-plant systems. Positive in vitro and in planta effectiveness by Trichoderma spp. was reported against virulent Phytophthora strains from pepper (Ezziyyani et al., 2017) and potato (Al-mughrabi, 2008). In accordance with previous studies, our results confirm that selected strains from Trichoderma spp. can represent a valid support in the integrated pest management strategies of Phytophthora diseases.
Decrypting the genetic pathways of plant defense mechanisms to counteract the plethora of effectors deployed by pathogens to develop the infection, concomitant with both the antagonistic activity and the growth-promoting effects of plant beneficial organisms, nowadays represents one of the main topics of modern plant pathology. Many studies have already investigated the modulation of the expression of protective molecules, pathogenic metabolites, and mucolytic enzymes in plants, pathogens, and antagonists, respectively, during a simplified two-way interaction (i.e., plant-antagonist, plant-pathogen, and antagonist-pathogen) (Carsolio et al., 1994; Tucci et al., 2011; Dalio et al., 2018). However, this compartmentalized approach does not clarify how the real mutual and simultaneous three-way interaction between the main actors involved in the biological control can reprogram their respective metabolic responses. In the present study, it has been investigated how the mutual gene-induced metabolic response of S. lycopersicum cv. Cuor di bue, P. nicotianae, and Trichoderma spp. is modified under the influence of the infection by the pathogen as well as of both the mycoparasitic and plant-beneficial activity of the antagonistic beneficial microorganism. To this aim, the modulation of the genetic pathways related with SA-dependent SAR (PR1b1 and PR-P2), ISR (TomLoxC and TomLoxA), and antifungal defensins (SlyDF2)-encoding genes were evaluated in tomato plants under P. nicotianae infection and the simultaneous Trichoderma spp. root colonization. The expression levels were compared with both Trichoderma-untreated–P. nicotianae-inoculated and Trichoderma-treated–P. nicotianae-non-inoculated tomato plants.
Among (SA)-dependent SAR and ISR-related genes, a statistically significant increment of transcripts was observed only in PR1b1 and PR-P2 transcript levels, while both analyzed TomLoxC and TomLoxA were normally expressed in all treatments. Previous studies have already demonstrated a significant variability between different Trichoderma-treated tomato varieties, including a widespread normal expression, in the levels of PR and ISR-related genes (Tucci et al., 2011). Considering also that root colonization by Trichoderma spp. activates only transiently the expression of defense-related genes (Yedidia et al., 1999, 2003; Shoresh et al., 2005; Segarra et al., 2009), results obtained here contribute to support the hypothesis that the colonization of roots by Trichoderma spp. could markedly take place only during the first phases of the interaction and then run out after a short time. Interestingly, the PR protein-encoding gene PR-P2 was activated more strongly in Phytophthora-inoculated and Trichoderma-untreated plants over treated and inoculated ones. The PR-P2 is a PR4-encoding gene induced both by SA and wounding (Tornero et al., 1997; Bertini et al., 2003; Tucci et al., 2011). The expression of PR protein-encoding genes in Trichoderma-treated and pathogen-inoculated plants seems to be a mechanism characterized by a high spectrum of responses (Shoresh et al., 2005). A decreasing trend in the expression of PR-encoding genes was reported from different lines of tomato after the inoculation with Botrytis cinerea (Tucci et al., 2011); at the same time, proteomic studies on levels of PR proteins in Trichoderma-treated plants reported increments in pepper under P. capsici infection (Ezziyyani et al., 2017), in cucumber and maize (inoculated with Pythium ultimum and Colletotrichum graminicola, respectively) (Harman et al., 2004), and decrements in bean under leaf infection by B. cinerea and Rhizoctonia solani (Marra et al., 2006). By comparing the results obtained here with previous studies, it could then be speculated that the promotion of plant defenses by Trichoderma spp. is a mechanism affected by a variability of factors that could depend both on plant species and pathogens.
Additional significant increments of transcripts were observed in the levels of the tomato defensin SlyDF2-encoding gene. In this study, the evaluated tomato defensin gene was up-regulated only in Phytophthora-inoculated plants, including both Trichoderma-treated and untreated ones, while it was slightly down-regulated in Trichoderma-treated plants which did not receive Phytophthora inoculum. Plant defensins play a crucial role in the resistance of plants to pathogens (Penninckx et al., 1996). Antifungal activities were reported for several plant defensins, including RsAFP1 and RsAFP2 from radish (Terras et al., 1992), MsDef1 and MtDef4 from Medicago spp. (Gao et al., 2000), NaD1 from tobacco (Lay et al., 2003a, b), and Psd1 from pea (Almeida et al., 2000). However, in the last few years, the research has been mainly focused on the study of the potentiality of transgenic up-regulating defensin plants in the protection from fungal infections (Breen et al., 2015), resulting in a significant lack of knowledge about the modulation in the induction of defensins in planta under the pathogen/antagonist interaction. Results obtained here from the two-way interactions Trichoderma-tomato and Phytophthora-tomato agree with previous studies which described the induction of plant defensins by Trichoderma spp. in A. thaliana (Poveda et al., 2019) and the modulation of plant defensins by Phytophthora infection in tomato plants (Cui et al., 2018), respectively. However, the expression levels of defensins in tomato plants under the simultaneous Trichoderma root colonization and Phytophthora infection were not previously reported. In this study, the generalized reduction in the expression levels of the SlyDF2-encoding gene in Trichoderma-treated and Phytophthora-inoculated tomato plants over the untreated and Phytophthora-inoculated ones make it possible to speculate that the presence of Trichoderma control agents could reduce the intensity of this particular plant response.
In this study, the modulation of the transcription levels of the P. nicotianae genes encoding the effectors PpCRN4, PpCBEL4, PpNPP1.1, PpNPP1.3, and PpNPP1.4 was evaluated in Phytophthora-inoculated and Trichoderma-treated tomato plants and compared with untreated controls. Even though the role of Phytophthora effectors in the plant-pathogen interaction was previously reported (Séjalon et al., 1995; Séjalon-Delmas et al., 1997; Fellbrich et al., 2002; Gijzen and Nürnberger, 2006; Stam et al., 2013), this is the first study where the modulation of P. nicotianae effectors in a three-way tomato–Trichoderma spp–Phytophthora system has been evaluated.
Interestingly, in this study, among NPP1-encoding genes, a marked up-regulation was observed in the levels of PnNPP1.1 and PnNPP1.3 of P. nicotianae from Trichoderma-treated tomato plants over the untreated controls. The P. nicotianae necrosis-inducing Phytophthora protein 1 (NPP1) has been associated with the induction of necrosis in parsley and A. thaliana (Fellbrich et al., 2002), and a significant up-regulation of NPP1-encoding genes was reported for P. nicotianae during the infection of Citrus sunki and Poncirus trifoliata (Dalio et al., 2018). By comparing the expression levels from Trichoderma-untreated and -treated tomato plants, it could be speculated, therefore, that the antagonistic activity of Trichoderma spp. toward P. nicotianae could hamper the infection process of the plant, resulting in an up-regulation of the transcripts in this particular genetic pathway, which, in principle, should weaken the plant defenses, making the invasion of the host possible.
In this study, the PpCRN4 and PpCBEL4 genes showed both a generalized down-regulation in Trichoderma-treated tomato plants over the untreated ones. PpCRN4 is a gene encoding a clinker (CRN) protein, while PpCBEL4 encodes a cell wall glycoprotein named CBEL (Dalio et al., 2018). Both groups of effectors (i.e., CRN and CBEL) induce necrosis in plant tissue (Séjalon-Delmas et al., 1997; Stam et al., 2013). Dalio et al. (2018) observed that 3 days after inoculation, the expression levels of the P. nicotianae PpCRN4-encoding gene manifested differences that depended on the host plant, as the gene down-regulated and normally expressed by the pathogen in C. sunki and P. trifoliata, respectively. In the same plant-pathogen systems, PpCBEL4-encoding gene was up-regulated both in C. sunki and P. trifoliata. As already observed by other authors (Woo et al., 2006), the different response in the expression level of the pathogenic effectors of P. nicotianae could be explained by the great variety of molecular weapons of Trichoderma spp. These mechanisms can be activated in different combinations or at different steps of the infection process; they can be also triggered depending on the pathogen they are confronting or the plant they are colonizing. In particular, the up-regulation of PnNPP1.1 and PnNPP1.3 pathogenic effectors could be a response of P. nicotianae to the induction of the plant resistance mechanisms by Trichoderma spp. An analogous pattern has been already demonstrated by Marra et al. (2006) who observed, using a proteomic approach, an over production of the superoxide dismutase by B. cinerea in the three-way system B. cinerea-bean-Trichoderma. Conversely, the down-regulation of PpCRN4 and PpCBEL4 in P. nicotianae in the presence of Trichoderma spp. could be due to the antagonistic interaction between the microorganisms as well as the result of the enhancement of specific plant defenses by Trichoderma spp., which, in turn, could trigger in tomato plants a response similar to that constitutively activated by C. sunki plants infected by P. nicotianae (Dalio et al., 2018), thus determining the down-regulation of the PpCRN4 gene. However, these pathogen genetic responses deserve to be further investigated.
Finally, in accordance with other authors (Osorio-Hernández et al., 2016), in this study, the level of endochitinase from both T. asperellum and T. atroviride was higher in Trichoderma-treated and Phytophthora-inoculated plants over the non-inoculated ones.
Conclusion
This study provides the basis for understanding the complex and often unpredictable genetic interactions in a tripartite system, plant/beneficial organism/pathogen, instead of two, plant/pathogen, as in most systems studied so far. The experimental approach, including the individual components of the system, the host plant tomato, the oomycete P. nicotianae, i.e., the challenging pathogen, and the beneficial fungus Trichoderma, in all possible two-way combinations and the comparison with the three-way combinations made it possible to confirm or verify genetic mechanisms involved in the host-pathogen, host-growth-promoting beneficial organism, and pathogen-antagonistic beneficial organism interactions. Moreover, a better insight on how reciprocal interactions are modulated in more complex systems has been obtained. In particular, in this tripartite system, it was observed the simultaneous transcriptional reprogramming of plant defense-related genes, pathogen effectors, and mycoparasitism-related genes. Results support the hypothesis that Trichoderma spp. elicit the expression of plant defense-related genes and, as expected, a mycoparasitism-related gene was significantly up-regulated in Trichoderma-colonizing tomato plants infected by P. nicotianae.
Moreover, for the first time, it was observed that the Trichoderma treatment of tomato plants induced a marked up-regulation of the P. nicotianae pathogenic effectors PnNPP1.1 and PnNPP1.3 and, at the same time, a slight down-regulation of PpCRN4 and PpCBEL4.
The findings that both the two- and three-way interactions vary with different Trichoderma species and the selection of a T. atroviride strain showing both a direct antagonistic activity against P. nicotianae and a growth-promoting effect on tomato plants are other interesting achievements of this study that have practical implications in the development and design of sustainable disease management strategies based on the application of biocontrol agents.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
SOC, FLS, and AP conceptualized the study, analyzed the results, and reviewed and edited the draft. FLS, CS, and MR did the investigation and formal analysis and performed the experiments. SOC and AP were responsible for funding acquisition and supervised the study. FLS wrote the original draft. All authors reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This study was funded by grant awarded to SOC by MIUR–FFABR 2017 (n°. 5A725192051) and to AP and SOC by the University of Catania–Program n°. 5A722192134: “Emergent Pests and Pathogens and Relative Sustainable Management Strategies (DIFESA).” FLS was supported by a Ph.D. fellowship funded by “PON Ricerca e Innovazione” 2014–2020 (CCI 2014IT16M2OP005).
Acknowledgments
We wish to thank Prof. Sophien Kamoun for his encouragement and helpful suggestions and Dr. Rossella Labarile for her technical advice and useful discussions. We are grateful to Mrs. Ann Davies for the English revision.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.583539/full#supplementary-material
Footnotes
- ^ https://digitalw orldbiology.com/FinchTV
- ^ http://www.ncbi.nlm.nih.gov/BLAST/
- ^ https://www.ncbi.nlm.nih.gov/nucleotide/
- ^ https://www.ncbi.nlm.nih.gov/tools/primer-blast/
References
Almeida, M. S., Cabral, K. M. S., Zingali, R. B., and Kurtenbach, E. (2000). Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Archiv. Biochem. Biophys. 378, 278–286. doi: 10.1006/abbi.2000.1824
Al-mughrabi, K. (2008). Biological control of Phytophthora infestans of potatoes using Trichoderma atroviride. Pest Technol. 2, 104–108.
Atanasova, L., Crom, S. Le, Gruber, S., Coulpier, F., Seidl-Seiboth, V., Kubicek, C. P., et al. (2013). Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genom. 14:121. doi: 10.1186/1471-2164-14-121
Benítez, T., Rincón, A. M., Limón, M. C., and Codón, A. C. (2004). Biocontrol mechanisms of Trichoderma strains. Intern. Microbiol. 7, 249–260. doi: 10.2436/im.v7i4.9480
Bertini, L., Leonardi, L., Caporale, C., Tucci, M., Cascone, N., Di Berardino, I., et al. (2003). Pathogen-responsive wheat PR4 genes are induced by activators of systemic acquired resistance and wounding. Plant Sci. 164, 1067–1078. doi: 10.1016/S0168-9452(03)00112-2
Birch, P. R. J., Rehmany, A. P., Pritchard, L., Kamoun, S., and Beynon, J. L. (2006). Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 14, 8–11. doi: 10.1016/j.tim.2005.11.007
Brasesco, F., Asgedom, D., and Casari, G. (2019). Strategic Analysis and Intervention Plan for Fresh and Industrial Tomato in the Agro-Commodities Procurement Zone of the Pilot Integrated Agro-Industrial Park in Central-Eastern Oromia, Ethiopia. Addis Ababa: FAO.
Breen, S., Solomon, P. S., Bedon, F., and Vincent, D. (2015). Surveying the potential of secreted antimicrobial peptides to enhance plant disease resistance. Front. Plant Sci. 6:900. doi: 10.3389/fpls.2015.00900
Cacciola, S. O., and Gullino, M. L. (2019). Emerging and re-emerging fungus and oomycete soil-borne plant diseases in Italy. Phytopathol. Med. 58, 451–472. doi: 10.14601/PHYTO-10756
Cacciola, S. O., Puglisi, I., Faedda, R., Sanzaro, V., Pane, A., Lo Piero, A. R., et al. (2015). Cadmium induces cadmium-tolerant gene expression in the filamentous fungus Trichoderma harzianum. Mol. Biol. Rep. 42, 1559–1570. doi: 10.1007/s11033-015-3924-4
Carsolio, C., Gutiérrez, A., Jiménez, B., Van Montagu, M., and Herrera-Estrella, A. (1994). Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc. Natl. Acad. Sci. U.S.A. 91, 10903–10907. doi: 10.1073/pnas.91.23.10903
Chepsergon, J., Motaung, T. E., Bellieny-Rabelo, D., and Moleleki, L. N. (2020). Organize, don’t agonize: strategic success of Phytophthora species. Microorganisms 8:917. doi: 10.3390/microorganisms8060917
Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G., and Brasier, C. M. (2000). A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30, 17–32. doi: 10.1006/fgbi.2000.1202
Crutcher, F. K., Moran-Diez, M. E., Ding, S., Liu, J., Horwitz, B. A., Mukherjee, P. K., et al. (2015). A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 119, 476–486. doi: 10.1016/j.funbio.2015.01.004
Cui, J., Jiang, N., Meng, J., Hou, X., Yang, G., and Luan, Y. (2018). Identification and characterization of defensin genes conferring Phytophthora infestans resistance in tomato. Physiol. Mol. Plant Pathol. 103, 28–35. doi: 10.1016/J.PMPP.2018.04.003
Dalio, R. J. D., Máximo, H. J., Oliveira, T. S., Azevedo, T., de, M., Felizatti, H. L., et al. (2018). molecular basis of Citrus sunki susceptibility and Poncirus trifoliata resistance upon Phytophthora parasitica attack. Mol. Plant Microb. Interact. 31, 386–398. doi: 10.1094/MPMI-05-17-0112-FI
Engelbrecht, B. M. J., Tyree, M. T., and Kursar, T. A. (2007). Visual assessment of wilting as a measure of leaf water potential and seedling drought survival. J. Trop. Ecol. 23, 497–500. doi: 10.1017/S026646740700421X
Erwin, D. C., and Ribeiro, O. K. (1996). Phytophthora Diseases Worldwide. St. Paul, MI: American Phytopathological Society.
Ezziyyani, M., Hamdache, A., Egea-Gilabert, C., Requena, M. E., Candela, M. E., and Mater, J. (2017). Production of Pathogenesis-Related proteins during the induction of resistance to Phytophthora capsici in pepper plants treated with Burkholderia cepacia and Trichoderma harzianum in combination compatible. J. Mater. Environ. Sci. 8, 4785–4795.
Fellbrich, G., Romanski, A., Varet, A., Blume, B., Brunner, F., Engelhardt, S., et al. (2002). NPP1, a Phytophthora -associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390. doi: 10.1046/j.1365-313X.2002.01454.x
Gao, A. G., Hakimi, S. M., Mittanck, C. A., Wu, Y., Woerner, B. M., Stark, D. M., et al. (2000). Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18, 1307–1310. doi: 10.1038/82436
Garibaldi, A., and Gullino, M. L. (2010). Emerging soilborne diseases of horticultural crops and new trends in their management. Acta Hortic. 883, 37–48. doi: 10.17660/actahortic.2010.883.2
Ghazanfar, M. U., Raza, M., Raza, W., and Qamar, M. I. (2018). Trichoderma as potential biocontrol agent, its exploitation in agriculture: a review. Plant Protect. 2, 109–135.
Gijzen, M., and Nürnberger, T. (2006). Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67, 1800–1807. doi: 10.1016/j.phytochem.2005.12.008
Gilardi, G., Demarchi, S., Gullino, M. L., and Garibaldi, A. (2014a). Control of Phytophthora nicotianae of tomato by using non-conventional strategies. Acta Hortic. 8, 325–330. doi: 10.17660/ActaHortic.2014.1044.42
Gilardi, G., Demarchi, S., Gullino, M. L., and Garibaldi, A. (2014b). Managing Phytophthora crown and root rot on tomato by pre-plant treatments with biocontrol agents, resistance inducers, organic and mineral fertilizers under nursery conditions. Phytopathol. Med. 53, 205–215. doi: 10.14601/Phytopathol_Mediterr-12361
Gilardi, G., Pugliese, M., Gullino, M. L., and Garibaldi, A. (2020). Effect of biocontrol agents and potassium phosphite against Phytophthora crown rot, caused by Phytophthora capsici, on zucchini in a closed soilless system. Sci. Hortic. 265:207. doi: 10.1016/j.scienta.2020.109207
Gonçalves, S., Teixeira, A., Abade, J., De Medeiros, L. N., Kurtenbach, E., and Santos, N. C. (2012). Evaluation of the membrane lipid selectivity of the pea defensin Psd1. Biochim. Biophys. Acta Biomembr. 1818, 1420–1426. doi: 10.1016/j.bbamem.2012.02.012
Guérin, V., Lebreton, A., Phytologie, D., and De Laval, U. (2014). A zoospore inoculation method with Phytophthora sojae to assess the prophylactic role of silicon on soybean cultivars. Plant Dis. 98, 1632–1638. doi: 10.1094/PDIS-01-14-0102-RE
Guzmán-Guzmán, P., Alemán-Duarte, M. I., Delaye, L., Herrera-Estrella, A., and Olmedo-Monfil, V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18:16. doi: 10.1186/s12863-017-0481-y
Guzmán-Guzmán, P., Porras-Troncoso, M. D., Olmedo-Monfil, V., and Herrera-Estrella, A. (2019). Trichoderma species: versatile plant symbionts. Phytopathology 109, 6–16. doi: 10.1094/PHYTO-07-18-0218-RVW
Haggag, H., and El-Gamal, G. (2012). In vitro study on Fusarium solani and Rhizoctonia solani isolates causing the damping off and root rot diseases in tomatoes. Nat. Sci. 10, 16–25.
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species — opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Howell, C. R. (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87, 4–10. doi: 10.1094/PDIS.2003.87.1.4
ISTAT (2020). Istat.it Agriculture. Available online at: https://www.istat.it/en/agriculture?data-and-indicators (accessed January 22, 2020).
Jones, J. B., Zitter, T. A., Momol, T. M., and Miller, S. A. (2016). Compendium of Tomato Diseases and Pests, 2nd Edn, Saint Paul, MI: American Phytopathological Society.
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jung, T., Blaschke, H., and Neumann, P. (1996). Isolation, identification and pathogenicity of Phytophthora species from declining oak stands. Eur. J. For. Pathol. 26, 253–272. doi: 10.1111/j.1439-0329.1996.tb00846.x
Jung, T., La Spada, F., Pane, A., Aloi, F., Evoli, M., Horta Jung, M., et al. (2019). Diversity and distribution of Phytophthora species in protected natural areas in Sicily. Forests 10:259. doi: 10.3390/f10030259
Kamoun, S., Furzer, O., Jones, J. D. G., Judelson, H. S., Ali, G. S., Dalio, R. J. D., et al. (2015). The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., Druzhinina, I. S., Thon, M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. doi: 10.1186/gb-2011-12-4-r40
Lay, F., and Anderson, M. (2005). Defensins - components of the innate immune system in plants. Curr. Protein Pept. Sci. 6:29. doi: 10.2174/1389203053027575
Lay, F. T., Brugliera, F., and Anderson, M. A. (2003a). Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol. 131:26. doi: 10.1104/pp.102.016626
Lay, F. T., Schirra, H. J., Scanlon, M. J., Anderson, M. A., and Craik, D. J. (2003b). The three-dimensional solution structure of NaD1, a new floral defensin from Nicotiana alata and its application to a homology model of the crop defense protein alfAFP. J. Mol. Biol. 325, 175–188. doi: 10.1016/S0022-2836(02)01103-8
Lebreton, A., Labbé, C., De Ronne, M., Xue, A. G., Marchand, G., and Bélanger, R. R. (2018). Development of a simple hydroponic assay to study vertical and horizontal resistance of soybean and pathotypes of Phytophthora sojae. Plant Dis. 102, 114–123. doi: 10.1094/PDIS-04-17-0586-RE
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lobo, D. S., Pereira, I. B., Fragel-Madeira, L., Medeiros, L. N., Cabral, L. M., Faria, J., et al. (2007). Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 46, 987–996. doi: 10.1021/bi061441j
Mammella, M. A., Cacciola, S. O., Martin, F., and Schena, L. (2011). Genetic characterization of Phytophthora nicotianae by the analysis of polymorphic regions of the mitochondrial DNA. Fungal Biol. 115, 432–442. doi: 10.1016/j.funbio.2011.02.018
Marra, R., Ambrosino, P., Carbone, V., Vinale, F., Woo, S. L., Ruocco, M., et al. (2006). Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr. Genet. 50, 307–321. doi: 10.1007/s00294-006-0091-0
Masek, T., Vopalensky, V., Suchomelova, P., and Pospisek, M. (2005). Denaturing RNA electrophoresis in TAE agarose gels. Analyt. Biochem. 336, 46–50. doi: 10.1016/J.AB.2004.09.010
Mastouri, F., Björkman, T., and Harman, G. E. (2010). Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology 100, 1213–1221. doi: 10.1094/PHYTO-03-10-0091
McGowan, J., O’hanlon, R., Owens, R. A., and Fitzpatrick, D. A. (2020). Comparative genomic and proteomic analyses of three widespread Phytophthora species: Phytophthora chlamydospora, Phytophthora gonapodyides and Phytophthora pseudosyringae. Microorganisms 8:653. doi: 10.3390/microorganisms8050653
Mendenhall, M. D., and Hodge, A. E. (1998). Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1191–1243.
Meng, Y., Zhang, Q., Ding, W., and Shan, W. (2014). Phytophthora parasitica: a model oomycete plant pathogen. Mycology 5, 43–51. doi: 10.1080/21501203.2014.917734
Monte, E. (2001). Understanding Trichoderma: between biotechnology and microbial ecology. Intern. Microbiol. 4, 1–4. doi: 10.1007/s101230100001
Morán-Diez, M. E., Carrero-Carrón, I., Belén Rubio, M., Jiménez-Díaz, R. M., Monte, E., and Hermosa, R. (2019). Transcriptomic analysis of Trichoderma atroviride overgrowing plant-wilting verticillium dahliae reveals the role of a new m14 metallocarboxypeptidase CPA1 in biocontrol. Front. Microbiol. 10:1120. doi: 10.3389/fmicb.2019.01120
Osorio-Hernández, E., Hernández-Morales, J., Conde-Martínez, V., Michel-Aceves, A. C., Lopez-Santillan, J. A., and Torres-Castillo, J. A. (2016). In vitro activities of Trichoderma species against Phytophthora parasitica and Fusarium oxysporum. Afr. J. Microbiol. Res. 10, 521–527. doi: 10.5897/ajmr2016.7958
Pachauri, S., Sherkhane, P. D., Kumar, V., and Mukherjee, P. K. (2020). Whole genome sequencing reveals major deletions in the genome of M7, a gamma ray-induced mutant of Trichoderma virens that is repressed in conidiation, secondary metabolism, and Mycoparasitism. Front. Microbiol. 11:1030. doi: 10.3389/fmicb.2020.01030
Panabières, F., Ali, G. S., Allagui, M. B., Dalio, R. J. D., Gudmestad, N. C., Kuhn, M. L., et al. (2016). Phytophthora nicotianae diseases worldwide: new knowledge of a long-recognised pathogen. Phytopathol. Med. 55, 20–40. doi: 10.14601/Phytopathol_Mediterr-16423
Pane, A., Agosteo, G. E., and Cacciola, S. O. (2000). Phytophthora species causing crown and root rot of tomato in southern Italy. EPPO Bull. 30, 251–255. doi: 10.1111/j.1365-2338.2000.tb00890.x
Penninckx, I. A. M. A., Eggermont, K., Terras, F. R. G., Thomma, B. P. H. J., De Samblanx, G. W., Buchala, A., et al. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. doi: 10.2307/3870470
Poveda, J., Hermosa, R., Monte, E., and Nicolás, C. (2019). Trichoderma harzianum favours the access of Arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity. Sci. Rep. 9:11650. doi: 10.1038/s41598-019-48269-z
Puglisi, I., Faedda, R., Sanzaro, V., Lo Piero, A. R., Petrone, G., and Cacciola, S. O. (2012). Identification of differentially expressed genes in response to mercury I and II stress in Trichoderma harzianum. Gene 506, 325–330. doi: 10.1016/j.gene.2012.06.091
Ramírez-Valdespino, C. A., Casas-Flores, S., and Olmedo-Monfil, V. (2019). Trichoderma as a model to study effector-like molecules. Front. Microbiol. 10:1030. doi: 10.3389/pmic.2019.01030
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Segarra, G., Casanova, E., Bellido, D., Odena, M. A., Oliveira, E., and Trillas, I. (2007). Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics 7, 3943–3952. doi: 10.1002/pmic.200700173
Segarra, G., Van Der Ent, S., Trillas, I., and Pieterse, C. M. J. (2009). MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96. doi: 10.1111/j.1438-8677.2008.00162.x
Séjalon, N., Dargent, R., Villalba, F., Bottin, A., Rickauer, M., and Esquerré-Tugayé, M. T. (1995). Characterization of a cell-surface antigen isolated from the plant pathogen Phytophthora parasitica var. nicotianae. Can. J. Bot. 73, 1104–1108. doi: 10.1139/b95-365
Séjalon-Delmas, N., Villalba Mateos, F., Bottin, A., Rickauer, M., Dargent, R., and Esquerré-Tugayé, M. T. (1997). Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology 87, 899–909. doi: 10.1094/PHYTO.1997.87.9.899
Seo, H. H., Park, S., Park, S., Oh, B. J., Back, K., Han, O., et al. (2014). Overexpression of a defensin enhances resistance to a fruit-specific anthracnose fungus in pepper. PLoS One 9:e97936. doi: 10.1371/journal.pone.0097936
Shoresh, M., Harman, G. E., and Mastouri, F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. doi: 10.1146/annurev-phyto-073009-114450
Shoresh, M., Yedidia, I., and Chet, I. (2005). Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95, 76–84. doi: 10.1094/PHYTO-95-76
Singh, A., and Islam, M. N. (2010). In vitro evaluation of Trichoderma spp. against Phytophthora nicotianae. Intern. J. Exper. Agric. 1, 1923–7766.
Smith, V. L. (1990). Potential for biological control of Phytophthora root and crown rots of apple by Trichoderma and Gliocladium spp. Phytopathology 80, 880–885. doi: 10.1094/phyto-80-880
Stam, R., Jupe, J., Howden, A. J. M., Morris, J. A., Boevink, P. C., Hedley, P. E., et al. (2013). Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS One 8:e59517. doi: 10.1371/journal.pone.0059517
Stassen, J. H. M., and Van den Ackerveken, G. (2011). How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14, 407–414. doi: 10.1016/j.pbi.2011.05.002
Terras, F. R. G., Schoofs, H. M. E., De Bolle, M. F. C., Van Leuven, F., Rees, S. B., Vanderleyden, J., et al. (1992). Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 267, 15301–15309.
Tornero, P., Gadea, J., Conejero, V., and Vera, P. (1997). Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol. Plant Microb. Interact. 10, 624–634. doi: 10.1094/MPMI.1997.10.5.624
Tucci, M., Ruocco, M., De Masi, L., De Palma, M., and Lorito, M. (2011). The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12, 341–354. doi: 10.1111/j.1364-3703.2010.00674.x
Verma, M., Brar, S. K., Tyagi, R. D., Surampalli, R. Y., and Valéro, J. R. (2007). Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem. Eng. J. 37, 1–20. doi: 10.1016/J.BEJ.2007.05.012
Wang, K., Der Borrego, E. J., Kenerley, C. M., and Kolomiets, M. V. (2020). Oxylipins other than jasmonic acid are xylem-resident signals regulating systemic resistance induced by Trichoderma virens in Maize. Plant Cell 32, 166–185. doi: 10.1105/tpc.19.00487
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press Inc), 315–322.
Widmer, T. L. (2014). Screening Trichoderma species for biological control activity against Phytophthora ramorum in soil. Biol. Control 79, 43–48. doi: 10.1016/j.biocontrol.2014.08.003
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126. doi: 10.2174/1874437001408010071
Woo, S. L., Scala, F., Ruocco, M., and Lorito, M. (2006). The molecular biology of the interactions between Trichoderma spp., Phytopathogenic Fungi, and Plants. Phytopathology 96, 181–185. doi: 10.1094/PHYTO-96-0181
Yao, Y., Li, Y., Chen, Z., Zheng, B., Zhang, L., Niu, B., et al. (2015). Biological control of potato late blight using isolates of Trichoderma. Am. J. Potato Res. 93, 33–42. doi: 10.1007/s12230-015-9475-3
Yedidia, I., Benhamou, N., and Chet, I. (1999). Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65, 1061–1070.
Yedidia, I., Shoresh, M., Kerem, Z., Benhamou, N., Kapulnik, Y., and Chet, I. (2003). Concomitant induction of systemic resistance to Pseudomonas syringae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of Phytoalexins. Appl. Environ. Microbiol. 69, 7343–7353. doi: 10.1128/AEM.69.12.7343-7353.2003
Keywords: gene expression, antagonism, Trichoderma asperellum, Trichoderma atroviride, biological control, root rot, crown rot
Citation: La Spada F, Stracquadanio C, Riolo M, Pane A and Cacciola SO (2020) Trichoderma Counteracts the Challenge of Phytophthora nicotianae Infections on Tomato by Modulating Plant Defense Mechanisms and the Expression of Crinkler, Necrosis-Inducing Phytophthora Protein 1, and Cellulose-Binding Elicitor Lectin Pathogenic Effectors. Front. Plant Sci. 11:583539. doi: 10.3389/fpls.2020.583539
Received: 15 July 2020; Accepted: 06 October 2020;
Published: 04 November 2020.
Edited by:
Katharina Pawlowski, Stockholm University, SwedenReviewed by:
Carlos Lucena, University of Córdoba, SpainJianfei Wang, Anhui University of Science and Technology, China
Copyright © 2020 La Spada, Stracquadanio, Riolo, Pane and Cacciola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Pane, YXBhbmVAdW5pY3QuaXQ=; Santa Olga Cacciola, b2xnYS5jYWNjaW9sYUB1bmljdC5pdA==
 Federico La Spada
Federico La Spada Claudia Stracquadanio
Claudia Stracquadanio Mario Riolo
Mario Riolo Antonella Pane
Antonella Pane Santa Olga Cacciola
Santa Olga Cacciola