- 1Institute of Crop Science, National Agriculture and Food Research Organization (NARO), Tsukuba, Japan
- 2Graduate School of Horticulture, Chiba University, Matsudo, Chiba, Japan
Seed dispersal among wild species belonging to the tribe Triticeae is typically achieved by the formation of a brittle rachis. The trait relies on the development of a disarticulation layer, most frequently above the rachis node (resulting in wedge type dispersal units), but in some species below the rachis node (resulting in barrel type dispersal units). The genes responsible for the former type are the complementary pair Btr1 and Btr2, while the genetic basis of the latter type has yet to be determined. Aegilops tauschii forms barrel type dispersal units and previous study showed this species lacked an intact copy of Btr1. Here it has been demonstrated that Ae. tauschii carries two of Btr2; and that Btr2 transcript is present in a region below the rachis node where the abscission zone forms. The implication is that in this species, the Btr2 product is involved in the formation of barrel type dispersal units.
Introduction
Many wild grass species ensure the dispersal of their seed by their inflorescence becoming brittle at physiological maturity; the trait requires the formation of one or multiple abscission zones (Cousens et al., 2008). The anatomy and positioning of the abscission zone varies from species to species, although not in a way which necessarily correlates with phylogeny (Yu et al., 2020b). Among species belonging to the Triticeae tribe, the zone forms either in the rachis or the rachilla (Sakuma et al., 2011). Rachis disarticulation most frequently occurs above the node, but in a few cases (as in, for example, Aegilops tauschii) it can occur below the node (Zohary et al., 2012).
Disarticulation above the node results in the formation of wedge type dispersal units, each consisting of a single spikelet attached to a short proximal section of the rachis. The presence on chromosome arm 3HS of the dominant, complementary and tightly linked genes Btr1 and Btr2 is necessary for the expression of the trait in wild forms of Hordeum vulgare (barley; Pourkheirandish et al., 2015). In Triticum and Aegilops species, Btr1 and Btr2 map to the short arm of the group 3 homeolog (Avni et al., 2017; Zeng et al., 2020b). The products of this pair of genes are presumed to determine the unique mode of disarticulation exhibited by the Aegilops longissima spike, in which only one or two of the central nodes are brittle (Zeng et al., 2020a). An alteration in the Btr1 coding region sequence is thought to be responsible for the non-brittle nature of the spikes formed by both einkorn (Triticum monococcum; Pourkheirandish et al., 2018; Zhao et al., 2019) and emmer (Triticum dicoccum; Avni et al., 2017) wheat. The transcription of Btr2 above the rachis node has been suggested as a critical determinant of the formation of a disarticulation zone in both barley (Pourkheirandish et al., 2015) and Ae. longissima (Zeng et al., 2020a).
The barrel type dispersal unit consists of a single spikelet attached to a short distal rachis segment; among Aegilops species, this form is only found in species harboring the D genome (van Slageren, 1994). According to Chen (2001), F1 hybrids formed by crossing a weedy form of Triticum aestivum forming wedge type dispersal units with Triticum spelta (barrel type units) disarticulate both above and below the rachis node, suggesting that different gene(s) underlie the formation of the two forms of dispersal unit. The genetic basis of the barrel type unit was found to be a locus mapping to chromosome arm 3DL (Li and Gill, 2006; Zhang et al., 2015). The locus harbored a homolog of rice gene qSH1, TaqSH1-D (Katkout et al., 2015), which encodes a BELL1-type homeobox protein implicated in the shattering phenotype (Konishi et al., 2006). An insertion present in the TaqSH1-D 3'-UTR has been proposed to be the genetic basis of rachis non-brittleness, although this hypothesis awaits verification. As yet, the genetic basis of the barrel type dispersal unit has yet to be identified.
The geographical range of the diploid species Ae. tauschii, the donor of the D genome harbored by bread wheat (T. aestivum), stretches across western and central Asia into south eastern Europe (van Slageren, 1994). Two phylogenetic lineages (L1 and L2) of the species have been recognized (Wang et al., 2013): the former includes accessions of ssp. tauschii, while the latter includes both ssp. tauschii and ssp. strangulata accessions. As it has been suggested that the L2 type is the one more closely related to the bread wheat D genome (Wang et al., 2013), and L2 representative has been selected for whole genome sequencing (Jia et al., 2013; Luo et al., 2017).
In both rice and sorghum, the products of multiple genes are known to determine seed shattering (Yu and Kellogg, 2018; Di Vittori et al., 2019). A comparison of the Btr1 and Btr2 sequences harbored by a range of grass species has shown that while Ae. tauschii lacks an intact copy of Btr1, two copies of Btr2 are present, both of which map along chromosome arm 3DS (Zeng et al., 2020b). Since Btr1 and Btr2 act as a complementary gene pair in most Triticeae species, Ae. tauschii represents an anomaly, raising the question as to whether the products of the two Btr2 genes underly the formation of the species’ barrel-type disarticulation units. The present experiments sought to establish the involvement, if any, of the Btr2 product in the formation of barrel-type units by Ae. tauschii.
Materials and Methods
Plant Materials
Accessions of each of Ae. tauschii ssp. tauschii (AE 956) and Ae. longissima ssp. longissima (AE 417) were obtained from the IPK Genebank.1 Plants were grown in a glasshouse at the National Institute of Crop Science (Tsukuba, Japan).
Histological and Anatomical Analysis
Spikelets were sectioned longitudinally by hand and stained by immersion for 3 h in 0.01% w/v acridine orange in distilled water, then rinsed in 10x PBS buffer (pH7; Acridine orange stains lignin green and anionic polysaccharides red, see Li and Reeve (2005) and Houtman et al. (2016). The sections were illuminated with a 488 nm diode laser and the output was captured using a confocal laser scanning microscope (LSM 700, Carl Zeiss, Tokyo, Japan) equipped with ZEN 2009 Light Edition CLSM software, scanning at 505–530 nm (green) and >600 nm (red). The surface of the rachis separation layer in mature dispersal units was characterized using a TM3000 scanning electron microscope (SEM; Hitachi, Tokyo, Japan), set to deliver an accelerating voltage of 1 kV.
DNA Sequence Analysis
Genomic DNA was extracted from fresh leaves following Komatsuda et al. (1998). The subsequent PCR amplification and DNA sequencing procedures followed those given by Pourkheirandish et al. (2015). CLC Sequences Viewer v8 software was used to align sequences.2
Quantitative RT-PCR
Three replicates, each consisting of three 0.5–1.5 cm long spikes at the white anther stage, were sampled (Kirby and Appleyard, 1981), and RNA was extracted from these using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States). After treatment with RNase-free DNase (Takara Bio, Kusatsu, Japan) and quantification using a NanoDrop 1,000 device (Thermo Scientific, Tokyo, Japan), a 1 μg aliquot was reverse-transcribed using SuperScript III (Invitrogen), and the resulting ss cDNA used as template for quantitative PCRs (qPCRs) based on the Thunderbird SYBR qPCR Mix kit (TOYOBO, Tokyo, Japan) and a CFX96 real-time PCR system (Bio-Rad, Tokyo, Japan). Each sample was represented by a minimum of three technical replicates.
RNA in situ Hybridization
Both an anti-sense and sense version of a segment of the Btr2 sequences were generated by PCR from Ae. longissima accession AE 417 (Zeng et al., 2020a). The PCR amplicons were purified using a QIAquick PCR purification kit (Qiagen, Germantown, MD, United States). One of the probes for Btr2-Lo-1 (identical to Btr2-D-1) covered 150 bp of the Btr2 coding region and 200 bp of its 3'-UTR, while the other one for Btr2-Lo-2 (identical to Btr2-D-2) covered 150 bp of the Btr2 coding region and 150 bp of its 3'-UTR. Adaptor sequences including promoter of T7 RNA polymerases (CGCGCGTAATACGACTCACTATAGGG) were added to the 5' end of the primers. The amplicons were validated by DNA sequencing before use. Hybridization probes were prepared using T7 RNA polymerase. The RNA in situ hybridization procedure, based on digoxigenin-labeled RNA probes, was performed on samples harvested at the white anther stage, following methods given by Komatsuda et al. (2007).
Results
Disarticulation in the Mature Spikes of Aegilops tauschii and Aegilops longissima
The mature Ae. tauschii spike was brittle (Figure 1A), disarticulating below each rachis node, thereby generating barrel type dispersal units (Figure 1B). In contrast, disarticulation in the Ae. longissima spike occurred above two nodes in the central portion of the rachis, resulting in the production of wedge type units (Figures 1C,D). The surface of the separation layer in the Ae. tauschii spike was smooth, as a result of all but the non-vascular cells having been capped by an intact cell wall (Figures 2A,C). In Ae. longissima, by way of contrast, although the surface of separation layer was also smooth, few cells were capped by a cell wall (Figures 2B,D). A typical LSM image of a stained longitudinal section of Ae. tauschii is shown in Figure 3A. No autofluorescence was observed in either the green or the red channel (data not shown), but following staining with acridine orange, there was evidence of lignification in cells lying below the rachis node (Figure 3A). In particular, lignin accumulated in about four layers of small cells within the abscission zone (Supplementary Figure S1A), while the accumulation of anionic polysaccharides in this zone was relatively limited (Supplementary Figure S1C). The lignin was preferentially deposited in the secondary wall (Figure 3B). No such cells were observed in the Ae. longissima disarticulation layer (Figures 3C,D), even in a higher magnification (Supplementary Figure S2), implying the absence of a distinct abscission zone.
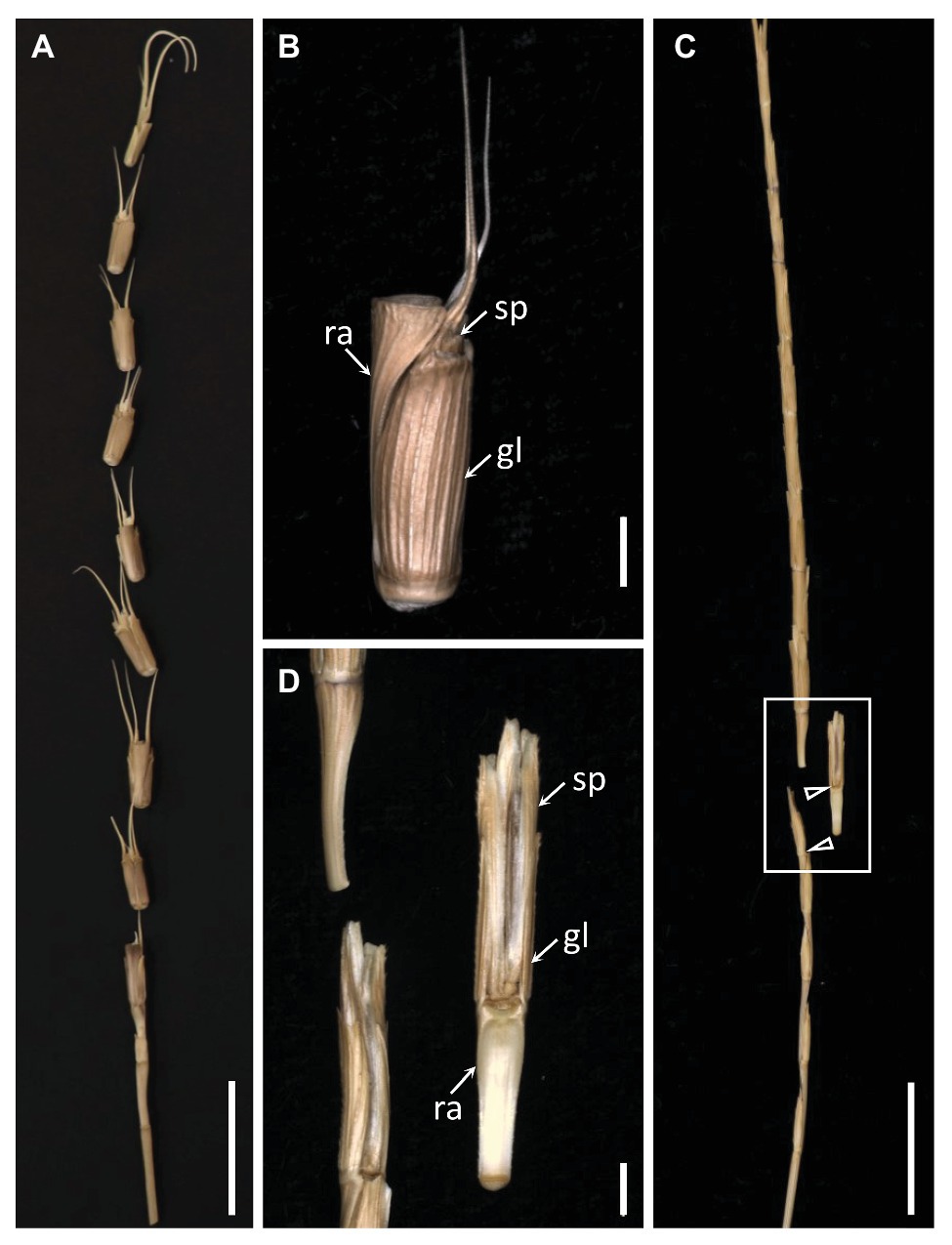
Figure 1. The dispersal units of Aegilops tauschii and Aegilops longissima. (A) The mature Ae. tauschii spike, which produces (B) barrel type dispersal units comprising one spikelet attached to a distal rachis segment. (C) The mature Ae. longissima spike produces (D) wedge type dispersal units comprising one spikelet attached to a proximal rachis segment. Arrowheads indicate brittle positions. sp, spikelet; gl, glume; ra, rachis. Scale bars: 2 cm in (A) and (C), 2 mm in (B) and (D).
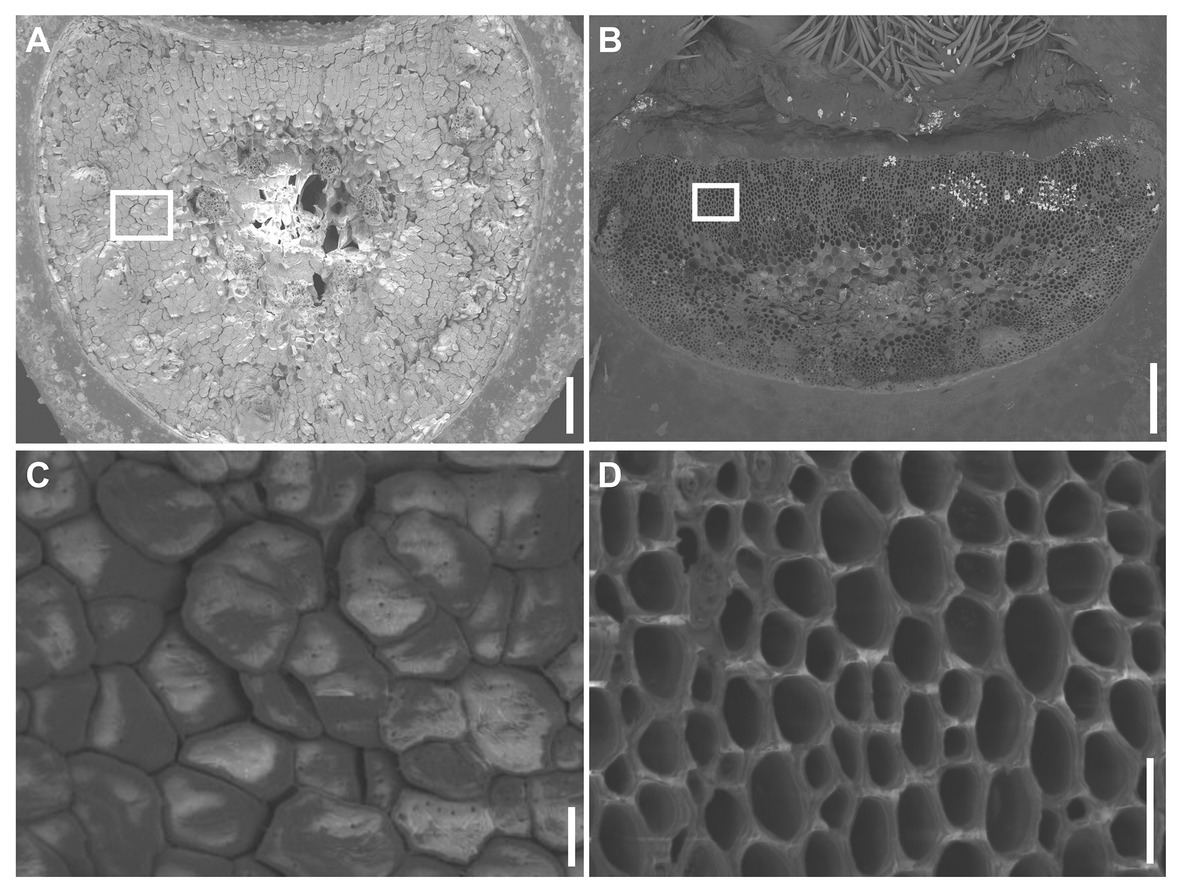
Figure 2. SEM visualization of the disarticulation surface. (A) The disarticulation surface below the rachis node in Ae. tauschii has a smooth surface, with all but the vascular cells being capped by intact cell wall (C). (B) The disarticulation surface above a rachis node in Ae. longissima is smooth, but all the cells are uncapped by cell walls (D). Scale bars: 200 μm in (A) and (B), 20 μm in (C) and (D).
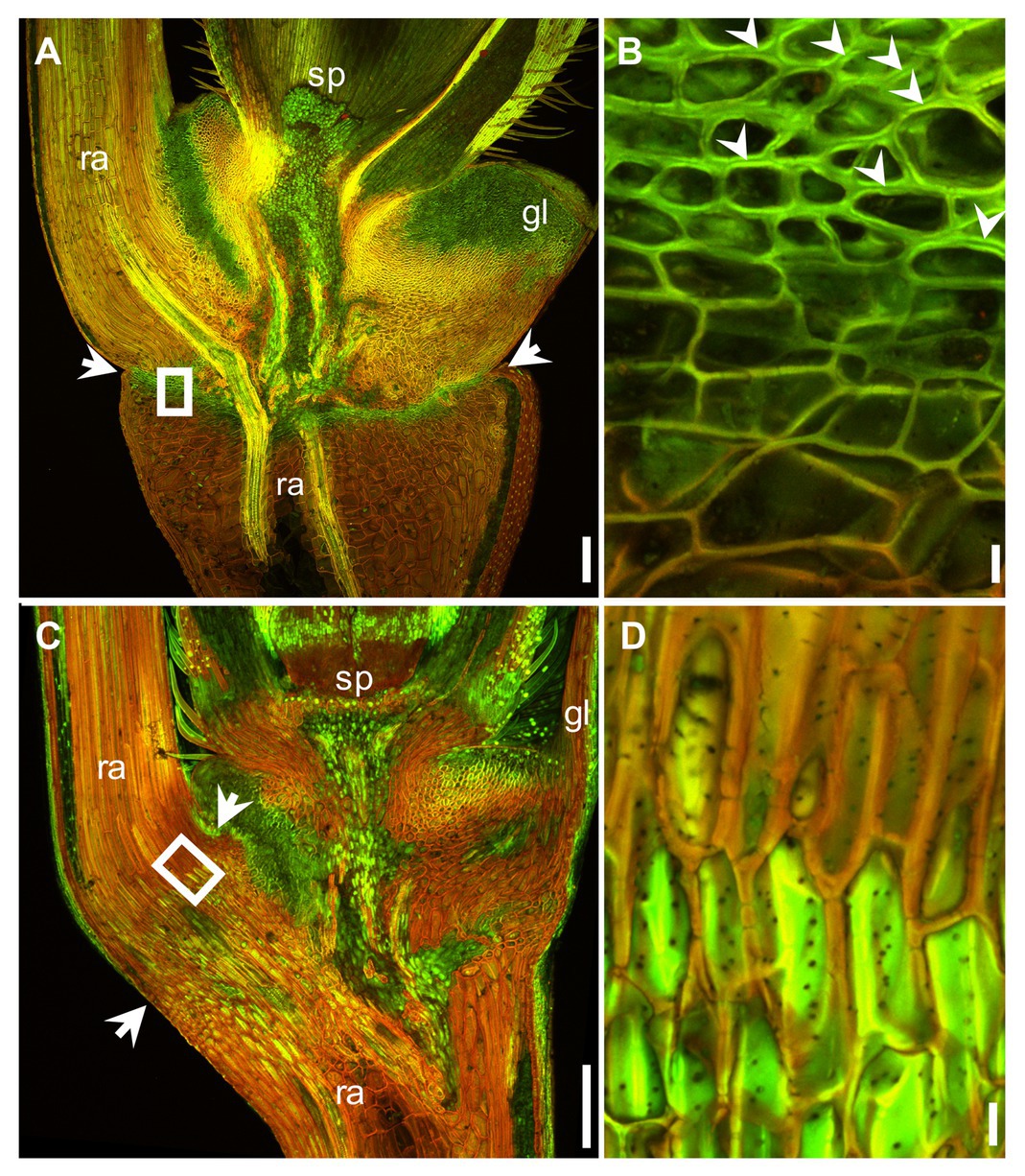
Figure 3. Acridine orange staining of the region around the abscission zone imaged at anthesis using LSM. The output of the green (505–530 nm) and red (>600 nm) channels have been merged. Lignin appears green and anionic polysaccharides red. Longitudinal section across the basal part of a floret and a section of the rachis in Ae. tauschii (A) and Ae. longissima (C). The abscission zone is arrowed. (B) and (D) A magnified view of the region shown in (A) and (C) by a white box. The preferential deposition of lignin in the secondary wall of the abscission zone is indicated by the arrowheads shown in (B). sp.: spikelet, gl: glume, ra: rachis. Scale bars: 200 μm in (A) and (C), 10 μm in (B) and (D).
Aegilops tauschii Harbors Two Copies of Btr2
The Ae. tauschii accession AE 956 harbored two copies of Btr2, namely Btr2-D-1 (GenBank accession MT920643) and Btr2-D-2 (GenBank accession MT920644). Btr2-D-1 of AE 956 corresponds to the Ae. tauschii accession AL8/78 sequence jcf7190000128337: 33-629, and Btr2-D-2 of AE 956 corresponds to the Ae. tauschii accession AL8/78 sequence 3D: 59425339-59424743. The level of homology between Btr2-D-1 of AE 956 and its AL8/78 equivalent was 99.5%, and the homology between Btr2-D-2 of AE 956 and its AL8/78 equivalent was 100% (Supplementary Figure S3). Both AE 956 genes included a 597 nt coding region (Supplementary Figure S3) encoding a 198 residue protein (Figure 4). The two genes differed from one another at 29 nucleotide positions (Supplementary Figure S3), resulting in 19 polymorphisms at the polypeptide level (Figure 4). The Triticeae Btr2 sequences most closely related to Btr2-D-1 and -D-2 are, respectively, the Ae. longissima accession AE 417 homologs Btr2-Lo-1 (94% homology) and Btr2-Lo-2 (97% homology; Zeng et al., 2020a). A recent duplication in the genome of the common ancestor of Ae. longissima and Ae. tauschii ssp. strangulata is thought to be responsible for the presence of two copies of Btr2 (Zeng et al., 2020b). Both Btr2 copies of Ae. tauschii ssp. strangulata are almost identical to those in Ae. tauschii ssp. tauschii except for two amino acid substitutions (Figure 4). The D genome of bread wheat (hexaploid; Alaux et al., 2018) has retained both Btr2 copies; their sequences in the model cultivar Chinese Spring are identical to those in Ae. tauschii ssp. strangulata (Figure 4). Inspection of the genome sequence of 10 other bread wheat cultivars available at https://webblast.ipk-gatersleben.de/wheat_ten_genomes/viroblast.php shows the same result. The origin of Btr2 and Btr2-like dates back to an ancient duplication event (Pourkheirandish et al., 2015), which occurred in the common ancestor of the Triticeae tribe (Zeng et al., 2020b).

Figure 4. Alignment of the deduced polypeptide sequences encoded by Btr2s of the Ae. tauschii ssp. tauschii, ssp. strangulata and a Triticum aestivum. Keys below indicate conserved sequence (*), conservative mutations (:), semi-conservative mutations (.), and non-conservative mutations ().
The Profile of Btr2 Transcription in Aegilops tauschii
The primer sequences employed for qPCR were based on those targeting the Btr2 locus present (Supplementary Figure S3), and barley Actin (GenBank accession AK362208) was used as the reference sequence. According to a qPCR assay, transcript of both Btr2-D-1 and -D-2 was present in spikes sampled at the white anther stage (Figure 5). When RNA in situ hybridization was employed using an antisense version of the Btr2-D-1 sequence as probe, signal was observed below the rachis node and spikelets, and also just above the rachis node (Figure 6A); equivalent signals were not detected when the sense probe was employed (Figure 6B). The same experiment using the Btr2-D-2 sequence as probe produced the same overall result (Figures 6C,D).
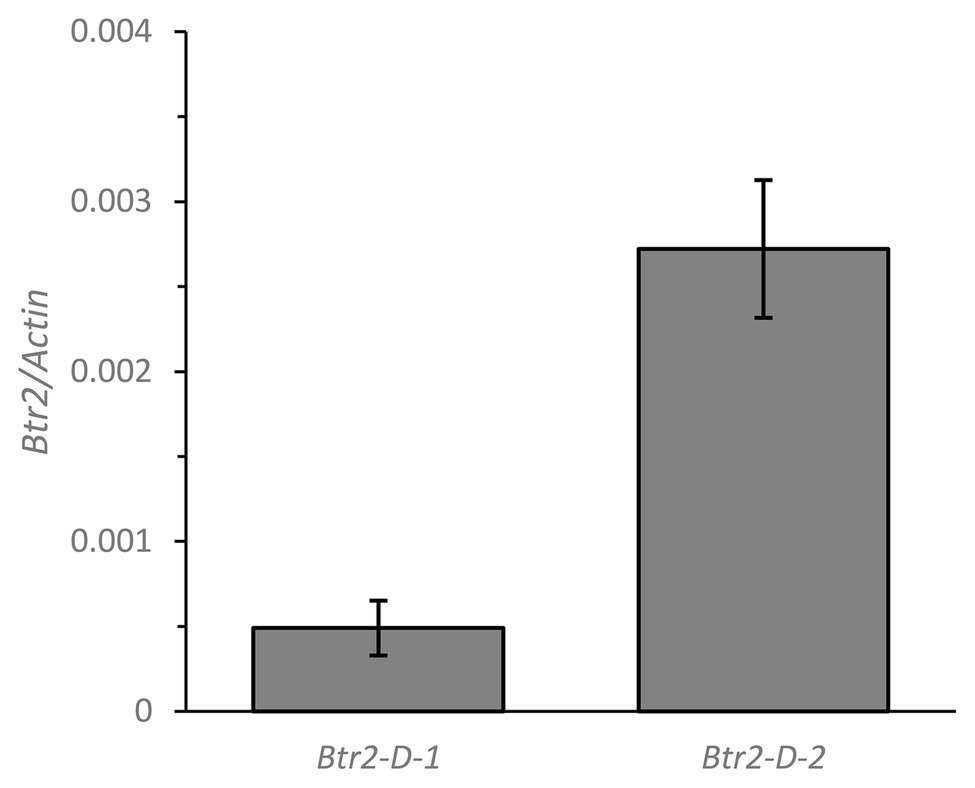
Figure 5. The abundance of Btr2 transcript, estimated from a quantitative PCR (qPCR) assay, in the spike of Ae. tauschii sampled at the white anther stage. Data shown in the form mean ± SE (n = 3).
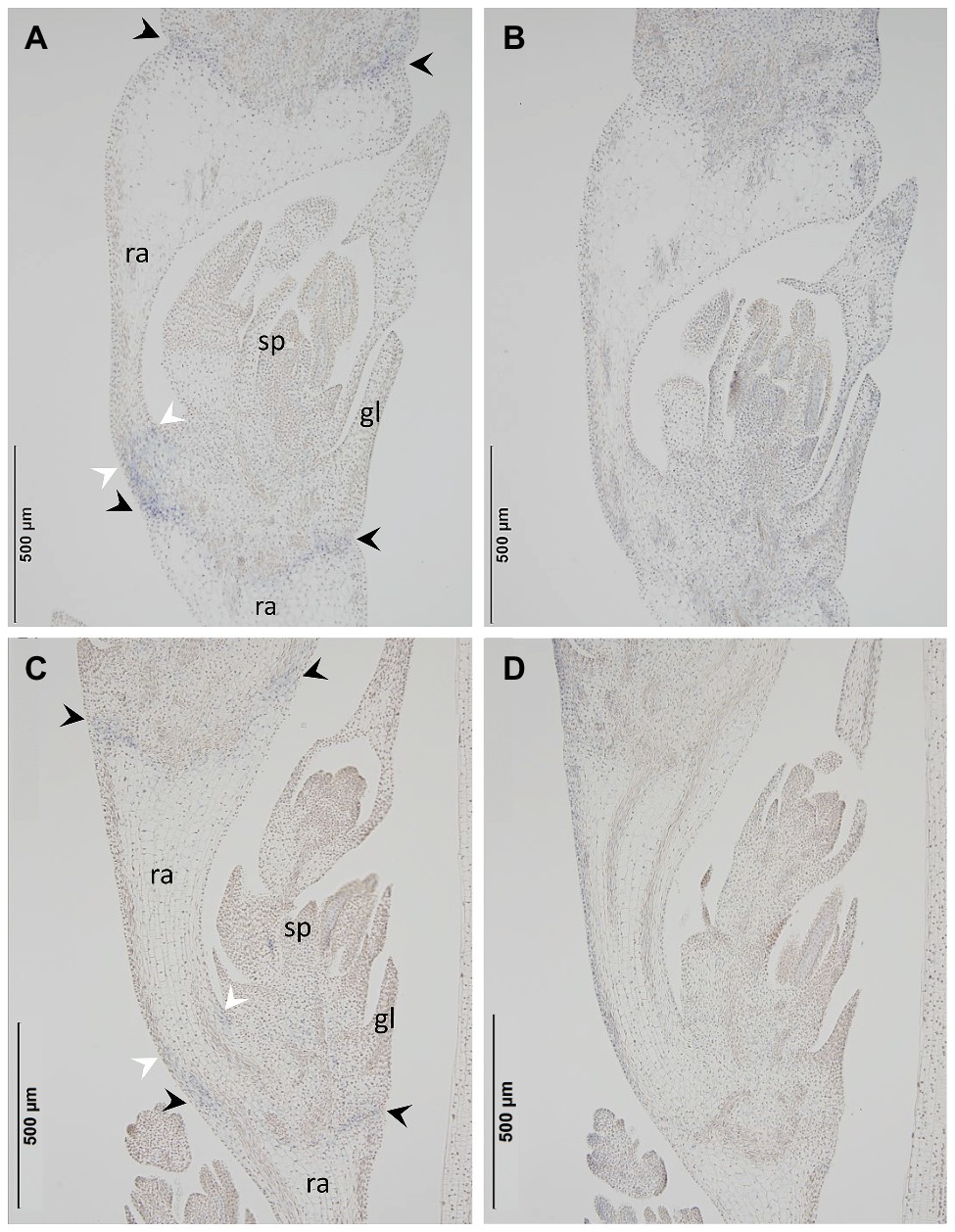
Figure 6. The site in the Ae. tauschii spike where Btr2 transcript accumulates, as assayed using RNA in situ hybridization. The probes used were (A) Btr2-D-1 antisense, (B) Btr2-D-1 sense, (C) Btr2-D-2 antisense, (D) Btr2-D-2 sense. Signal was detected between the spikelet and the rachis node (black arrows) and above the rachis node (white arrows) in (A) and (C), but not in (B) or (D). sp, spikelet; gl, glume; ra, rachis.
Discussion
The Abscission Zone in Aegilops tauschii Forms Below the Rachis Node
Aegilops tauschii produces barrel type dispersal units by disarticulating below the rachis node. The point of breakage is ensured by the formation of an abscission zone in which an array of lignified cells is formed. A similar structure has been described in the spikes of both Elymus sibiricus (Zhao et al., 2017) and Brachypodium distachyon (Yu et al., 2020a), whereas the abscission zone cells in the panicles of both wild rice (Oryza nivara; Li et al., 2006) and wild sorghum (Sorghum virgatum; Lin et al., 2012) are not lignified. In wild Hordeum spp., by contrast, which form wedge type units, disarticulation is achieved by the formation of a layer of thin-walled cells in the separation zone (Pourkheirandish et al., 2015). As suggested by (Zhao et al., 2019), the mechanistic basis of rachis brittleness in the genus Triticum lies in repressing the synthesis of cell walls. The disarticulation surface in these species is characterized by cells not capped by a cell wall. In Ae. tauschii, the separation surface was similarly smooth, but, with the exception of vascular elements, the cells at the surface were capped by an undamaged cell wall. This phenotype resembles that of the separation layer seen in the rachilla of Elymus sibiricus (Zhao et al., 2017) and at the base of the pedicel in Oryza spp. (Konishi et al., 2006).
The Ectopic Expression of Btr2 May Explain the Formation of the Abscission Zone Below the Rachis Node
The co-expression of Btr1 and Btr2 is required for disarticulation to occur above the rachis node (Pourkheirandish et al., 2015; Zeng et al., 2020a). The expression of Btr2 above the rachis node determines where the disarticulation layer forms. Btr2 is not expressed below the rachis node in species which form wedge type disarticulation units (Pourkheirandish et al., 2015; Zeng et al., 2020a). The genome sequence of Ae. tauschii ssp. strangulata (Luo et al., 2017) lacks a copy of Btr1, and the assumption is that this is similarly the case in Ae. tauschii ssp. tauschii. The reason why Ae. tauschii fails to form a disarticulation layer above the rachis node is presumed to be the absence of an intact copy of Btr1, while its formation of a disarticulation layer below the rachis node reflects the operation of a cell-cell separation mechanism, as demonstrated here. The RNA in situ hybridization experiment confirmed that there was an abundance of Btr2 transcript in the abscission zone below the rachis node, which implies that the ectopic expression of Btr2 is required for the formation of this structure.
The genome of Ae. longissima (which does not form barrel type disarticulation units) harbors two copies of Btr2, neither of which is transcribed below the rachis node (Zeng et al., 2020a). The implication is that the unusual site of Btr2 transcription shown by Ae. tauschii is a recently acquired trait. An intriguing question is how this product can operate, given that its function is to ensure that disarticulation occurs below the rachis node. A further possibility is the formation of the abscission zone requires not just the product of Btr2 but also that of a gene(s) such as the ortholog of the rice locus qSH1 to achieve cell-cell separation in the abscission zone (Konishi et al., 2006; Katkout et al., 2015). Abscission zone is formed above glumes in Brachypodium, above the rudimentary glumes and below the empty glumes (sterile lemmas) in rice (Yoshida and Nagato, 2011; Zhou et al., 2012), and below glumes in Setaria (Yu et al., 2020a). Though they share distinct abscission zone, qSH1 is transcribed in each site corresponded to the abscission zone (Yu et al., 2020a). Thus, qSH1 appears to be a strong candidate to collaborate with Btr2 to form the abscission zone in Ae. tauschii.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
XZ and TK planned and designed the research and drafted the manuscript. XZ performed the morphological analysis, the DNA sequencing and the qPCRs, while AT and XZ performed the RNA in situ hybridization experiments. HS, SK, and TK monitored the acquisition of data. Each of the authors has read and agreed to the submitted version of the manuscript.
Funding
This research was financially supported in part by a grant-in-aid from the Japan Society for the Promotion of Science (JSPS) to TK (18H02184). XZ is grateful for the award of a Monbukagakusho scholarship given by from Government of Japan.
Conflict of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships which could be construed as a potential conflict of interest.
Acknowledgments
We thank F. Blattner (IPK, Gatersleben, Germany) for the gift of Ae. tauschii and Ae. longissima germplasm; T. Koba, T. Oikawa, K. Mishina, and M. Ohta for advice on the conduct of the experiments; and T. Kawakatsu for the use of SEM facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.582622/full#supplementary-material
Footnotes
References
Alaux, M., Rogers, J., Letellier, T., Flores, R., Alfama, F., Pommier, C., et al. (2018). Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 19:111. doi: 10.1186/s13059-018-1491-4
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Chen, Q. F. (2001). Inheritance of disarticulation derived from some hexaploid brittle rachis wheat. Genet. Resour. Crop. Evol. 48, 21–26. doi: 10.1023/a:1011291914213
Cousens, R., Dytham, C., and Law, R. (2008). Dispersal in plants: A population perspective. New York, NY: Oxford University Press.
Di Vittori, V., Gioia, T., Rodriguez, M., Bellucci, E., Bitocchi, E., Nanni, L., et al. (2019). Convergent evolution of the seed shattering trait. Gen. Dent. 10:68. doi: 10.3390/genes10010068
Houtman, C. J., Kitin, P., Houtman, J. C. D., Hammel, K. E., and Hunt, C. G. (2016). Acridine orange indicates early oxidation of wood cell walls by fungi. PLoS One 11:e0159715. doi: 10.1371/journal.pone.0159715
Jia, J., Zhao, S., Kong, X., Li, Y., Zhao, G., He, W., et al. (2013). Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496, 91–95. doi: 10.1038/nature12028
Katkout, M., Sakuma, S., Kawaura, K., and Ogihara, Y. (2015). TaqSH1-D, wheat ortholog of rice seed shattering gene qSH1, maps to the interval of a rachis fragility QTL on chromosome 3DL of common wheat (Triticum aestivum). Genet. Resour. Crop. Evol. 62, 979–984. doi: 10.1007/s10722-015-0301-z
Kirby, E. J. M., and Appleyard, M. (1981). Cereal development guide. Stoneleigh, Warwickshire, UK: Cereal Unit, National Agricultural Centre.
Komatsuda, T., Nakamura, I., Takaiwa, F., and Oka, S. (1998). Development of STS markers closely linked to the vrs1 locus in barley, Hordeum vulgare. Genome 41, 680–685. doi: 10.1139/g98-069
Komatsuda, T., Pourkheirandish, M., He, C., Azhaguvel, P., Kanamori, H., Perovic, D., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. U. S. A. 104, 1424–1429. doi: 10.1073/pnas.0608580104
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Li, W., and Gill, B. S. (2006). Multiple genetic pathways for seed shattering in the grasses. Funct. Integr. Genomics 6, 300–309. doi: 10.1007/s10142-005-0015-y
Li, K., and Reeve, D. W. (2005). Fluorescent labeling of lignin in the wood pulp fiber wall. J. Wood Chem. Technol. 24, 169–181. doi: 10.1081/wct-200026572
Li, C., Zhou, A., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi: 10.1126/science.1123604
Lin, Z., Li, X., Shannon, L. M., Yeh, C. T., Wang, M. L., Bai, G., et al. (2012). Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44, 720–724. doi: 10.1038/ng.2281
Luo, M. C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Pourkheirandish, M., Dai, F., Sakuma, S., Kanamori, H., Distelfeld, A., Willcox, G., et al. (2018). On the origin of the non-brittle rachis trait of domesticated einkorn wheat. Front. Plant Sci. 8:2031. doi: 10.3389/fpls.2017.02031
Pourkheirandish, M., Hensel, G., Kilian, B., Senthil, N., Chen, G., Sameri, M., et al. (2015). Evolution of the grain dispersal system in barley. Cell 162, 527–539. doi: 10.1016/j.cell.2015.07.002
Sakuma, S., Salomon, B., and Komatsuda, T. (2011). The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol. 52, 738–749. doi: 10.1093/pcp/pcr025
van Slageren, M. W. (1994). Wild wheats: A monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Wageningen: Wageningen Agricultural University.
Wang, J., Luo, M. C., Chen, Z., You, F. M., Wei, Y., Zheng, Y., et al. (2013). Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 198, 925–937. doi: 10.1111/nph.12164
Yoshida, H., and Nagato, Y. (2011). Flower development in rice. J. Exp. Bot. 62, 4719–4730. doi: 10.1093/jxb/err272
Yu, Y., Hu, H., Doust, A. N., and Kellogg, E. A. (2020a). Divergent gene expression networks underlie morphological diversity of abscission zones in grasses. New Phytol. 225, 1799–1815. doi: 10.1111/nph.16087
Yu, Y., and Kellogg, E. A. (2018). “Inflorescence abscission zones in grasses: diversity and genetic regulation” in Annual plant reviews online. Vol. 2. ed. J. Roberts (John Wiley & Sons, Ltd), 1–35.
Yu, Y., Leyva, P., Tavares, R. L., and Kellogg, E. A. (2020b). The anatomy of abscission zones is diverse among grass species. Am. J. Bot. 107, 549–561. doi: 10.1002/ajb2.1454
Zeng, X. X., Chen, G., Wang, L., Tagiri, A., Kikuchi, S., Sassa, H., et al. (2020a). The unique disarticulation layer formed in the rachis of Aegilops longissima probably results from the spatial co-expression of Btr1 and Btr2. Ann. Bot. doi: 10.1093/aob/mcaa147 [Epub ahead of print]
Zeng, X. X., Mishina, K., Jia, J. Q., Distelfeld, A., Maughan, P. J., Kikuchi, S., et al. (2020b). The brittle rachis trait in species belonging to the Triticeae and its controlling genes Btr1 and Btr2. Front. Plant Sci. 11:1000. doi: 10.3389/fpls.2020.01000
Zhang, Z., Zhu, H., Gill, B. S., and Li, W. (2015). Fine mapping of shattering locus Br2 reveals a putative chromosomal inversion polymorphism between the two lineages of Aegilops tauschii. Theor. Appl. Genet. 128, 745–755. doi: 10.1007/s00122-015-2469-1
Zhao, Y., Xie, P., Guan, P., Wang, Y., Li, Y., Yu, K., et al. (2019). Btr1-A induces grain shattering and affects spike morphology and yield-related traits in wheat. Plant Cell Physiol. 60, 1342–1353. doi: 10.1093/pcp/pcz050
Zhao, X., Xie, W., Zhang, J., Zhang, Z., and Wang, Y. (2017). Histological characteristics, cell wall hydrolytic enzymes activity and candidate genes expression associated with seed shattering of Elymus sibiricus accessions. Front. Plant Sci. 8:606. doi: 10.3389/fpls.2017.00606
Zhou, Y., Lu, D. F., Li, C. Y., Luo, J. H., Zhu, B. F., Zhu, J. J., et al. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 24, 1034–1048. doi: 10.1105/tpc.111.094383
Keywords: triticeae, barrel type dispersal units, lignin, cell wall, abscission zone, ectopic expression
Citation: Zeng X, Tagiri A, Kikuchi S, Sassa H and Komatsuda T (2020) The Ectopic Expression of Btr2 in Aegilops tauschii Switches the Disarticulation Layer From Above to Below the Rachis Node. Front. Plant Sci. 11:582622. doi: 10.3389/fpls.2020.582622
Edited by:
Dianella G. Howarth, St. John’s University, United StatesReviewed by:
Yunqing Yu, Donald Danforth Plant Science Center, United StatesMohammad Pourkheirandish, The University of Melbourne, Australia
Copyright © 2020 Zeng, Tagiri, Kikuchi, Sassa and Komatsuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takao Komatsuda, dGFrYW9AYWZmcmMuZ28uanA=
 Xiaoxue Zeng
Xiaoxue Zeng Akemi Tagiri1
Akemi Tagiri1 Takao Komatsuda
Takao Komatsuda