- 1Department of Horticulture, Zhejiang University, Hangzhou, China
- 2College of Forestry, Henan University of Science and Technology, Luoyang, China
- 3Wenzhou Vocational College of Science & Technology, Wenzhou, China
Plants intensely modulate respiration when pathogens attack, but the function of mitochondrial respiration-related genes in plant–bacteria interaction is largely unclear. Here, the functions of α-ketoglutarate dehydrogenase (α-kGDH) E2 subunit and alternative oxidase (AOX) were investigated in the interaction between tomato and the virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst). Pst inoculation suppressed the transcript abundance of α-kGDH E2, but enhanced AOX expression and salicylic acid (SA) accumulation. Gene silencing and transient overexpression approaches revealed that plant susceptibility to Pst was significantly reduced by silencing α-kGDH E2 in tomato, but increased by overexpressing α-kGDH E2 in Nicotiana benthamiana, whereas silencing or overexpressing of AOX1a did not affect plant defense. Moreover, silencing octanoyltransferase (LIP2), engaged in the lipoylation of α-kGDH E2, significantly reduced disease susceptibility and hydrogen peroxide accumulation. Use of transgenic NahG tomato plants that cannot accumulate SA as well as the exogenous SA application experiment evidenced that α-kGDH E2 acts downstream of SA defense pathway. These results demonstrate tomato α-kGDH E2 plays a negative role in plant basal defense against Pst in an AOX-independent pathway but was associated with lipoylation and SA defense pathways. The findings help to elucidate the mechanisms of mitochondria-involved plant basal immunity.
Introduction
Plants growing in diverse environments are constantly challenged by a wide range of microbial pathogens and herbivorous insects that often result in growth inhibition and crop yield losses, leading to a significant risk to global agriculture. A plant mitochondrion is a membrane-bound organelle that occurred in the cytoplasm that primarily functions in the respiration to generate large quantities of energy in the form of ATP and carbon skeletons required for numerous plant biosynthetic processes, cellular maintenance, and active transport (Daloso et al., 2015). Many studies have suggested that leaf respiration is one of the most important metabolic processes in immune response (Colombatti et al., 2014). However, our understanding of the roles and mechanisms of mitochondria proteins during the immune response is largely unclear.
During evolution, plants have acquired a sophisticated immune system to mitigate the adverse effects of pathogen attack, i.e., effector-triggered immunity (ETI) and pattern-triggered immunity (PTI). ETI usually occurs in incompatible plant–pathogen interactions, in which plant Resistance (R) proteins detect the presence of avirulent pathogen effectors that are delivered inside plant cells (Jones and Dangl, 2006). In contrast, PTI often occurs in the compatible plant–pathogen interaction, which relies on the recognition of pathogen/damage-associated molecular patterns by cell surface pattern recognition receptors (Couto and Zipfel, 2016). For the susceptible host plants, PTI effectively repels most virulent pathogens, contributing to basal immunity (Couto and Zipfel, 2016).
Plant mitochondrial respiration is known to consist of glycolysis, the tricarboxylic acid (TCA) cycle, and mitochondrial electron transport chain (miETC). TCA cycle is a hub for cellular metabolism; it channels electrons from reduced substrates to the membrane-bound miETC for efficient energy conversion. A central enzyme of the TCA cycle is α-ketoglutarate dehydrogenase (α-kGDH), which catalyzes the oxidative decarboxylation reaction converting α-ketoglutarate, coenzyme A, and NAD+ to succinyl-CoA, CO2, and NADH, supplying the reducing equivalents for miETC in the form of NADH (Condori-Apfata et al., 2019). Prosthetic lipoyl groups are required for the function of α-kGDH, and the E2 subunit of α-kGDH protein was reported to be lipoylated by octanoyltransferase (LIP2) in Arabidopsis (Ewald et al., 2014). The plant miETC supporting oxidative phosphorylation branches at ubiquinone (UQ). The main pathway of respiration that flows from UQ through the usual cytochrome (Cyt) pathway operates via the four-electron reduction of O2 to H2O is the Cyt pathway, which leads to proton translocation at respiratory complexes I to IV, resulting in ATP production. In addition to this main pathway, plants also have a non-phosphorylating electron transport pathway that involves a protein called alternative oxidase (AOX) that catalyzes the direct oxidation of UQ and reduction of O2 to H2O, and bypass two-proton translocation at the sites of complexes III and IV, thus limiting ATP production (Vanlerberghe et al., 2020). Alternative respiration is widespread in plants, fungi, and some prokaryotes, but not in higher animals.
During plant–pathogen interactions, salicylic acid (SA) acts as a major defense hormone in plants, and it is required for the activation of the immune system and the development of systemic acquired resistance. Tissue SA levels increase in response to biotrophic and hemibiotrophic pathogens, and SA has been demonstrated to play a crucial role in plant defenses against a broad spectrum of pathogens, including viruses, oomycetes, fungi, and bacteria (López-Gresa et al., 2017; Qi et al., 2018). Intensive research efforts have been focused on the identification of SA-binding proteins (SABPs) and/or receptors. The non-expresser of pathogenesis-related gene 1 (NPR1) is a master regulator of SA signaling (Chen et al., 2019), which binds SA with a high affinity (Wu et al., 2012; Manohar et al., 2015). Two other SABPs, NPR3 and NPR4, were recently shown to act as negative regulators that fine tune SA signaling responses (Ding et al., 2018). The function of SA in plant defense has also been linked to mitochondrial metabolism or signaling (Poór, 2020). In the mitochondria, the E2 unit of α-kGDH protein was identified as SABPs in Arabidopsis and tomato (Tian et al., 2012; Liao et al., 2015). An in-depth study in tomato demonstrated that binding by α-kGDH E2 of SA acts upstream and affects the activity of the miETC, which helps to limit tobacco mosaic virus (TMV)-induced systemic viral inoculation (Liao et al., 2015). Similarly, the AOX pathway can also be induced by treatments with exogenous SA (Lee et al., 2011). Notably, the alternative pathway has also been demonstrated to be associated with disease resistance in plants. Evidence supporting this notion comes from some interesting findings—that overexpression of AOX in transgenic tomato and petunia significantly lowered the levels of tomato spotted wilt virus (TSWV) (Ma et al., 2011). Works on Cyt pathway inhibitors and salicylhydroxamic acid (SHAM) have further led to the proposition that the AOX pathway plays a key role in the resistance of tobacco plants to virus inoculation (Murphy et al., 1999; Ma et al., 2011).
Nevertheless, studies of the role of α-KGDH and AOX in biotic stress resistance are mainly limited to viruses, and it is still unclear in other kinds of pathogens. For example, tobacco resistance to TMV, potato virus X (PVX), and cucumber mosaic virus (CMV) can be stimulated by activation of the AOX pathway (e.g., antimycin A and potassium cyanide [KCN]) or repressed by inhibition of the AOX pathway by SHAM (Naylor et al., 1998; Mayers et al., 2005). In contrast, SHAM does not inhibit the induction of the pathogenesis-related protein 1 (PR1) or induction of resistance to another pathogen species Eminia carotovora by SA. Similarly, in Arabidopsis, SA-mediated resistance to turnip crinkle virus and SA-, or antimycin A-induced resistance to turnip vein clearing virus, as well as the induction of the AOX (the potential target for the chemicals), still remain active in npr1 mutants (Kachroo et al., 2000; Wong et al., 2002). But the same mutation leads to loss of resistance to isolates of Peronospora parasitica and Pseudomonas syringae (Glazebrook et al., 1996; Rairdan and Delaney, 2002). These studies indicate that the signaling pathways for virus defense appear to branch below SA from that for fungi and bacteria, which involve AOX but are independent of NPR1 (Murphy et al., 1999). However, AOX expression also responds strongly to bacterial inoculation (Cvetkovska and Vanlerberghe, 2012). Transgenic AOX knockdown Nicotiana tabacum plants inoculated with avirulent P. syringae pv. maculicola display a delayed reactive oxygen species (ROS) burst that manifests itself in a delayed hypersensitive response (HR) in an incompatible ETI response (Cvetkovska and Vanlerberghe, 2013). However, in another study of incompatible plant–pathogen interaction, transgenic tobacco (Nicotiana attenuata) plants silenced in the expression of AOX (irAOX) accumulate higher levels of ROS and HR after P. syringae pv. tomato DC3000 (Pst) inoculation (Zhang et al., 2012). These studies suggest that AOX is implicated during ETI response. But, to date, there has been little study to establish the role of AOX, as well as α-KGDH E2 in the plant basal immunity.
Plant basal defense, which provides broad-spectrum defenses in compatible plant–pathogen interactions, is particularly important in crop cultivars. Tomato is one of the most economically important vegetable crops throughout the world, which often suffers from diseases and economic losses. In this study, we used virulent pathogens, Pst, as bacterial agents and explored the roles of mitochondrial α-KGDH E2 and AOX in the plant basal defense. Our data indicate that α-kGDH E2 in the TCA cycle, but not the miETC AOX, is involved in tomato basal defense against Pst. The defense functions of tomato α-kGDH E2 are associated with lipoylation and SA signaling. This information helps to elucidate the mechanism(s) of mitochondria-involved plant defense.
Materials and Methods
Plant Growth and Chemical Treatment
The tomato (Solanum lycopersicum) lines used in most of the studies were mainly in the Zheza 809 wild-type background. The NahG transgenic line that cannot accumulate SA was in Money maker (MM) background and was obtained from the laboratory of J.D.G. Jones (Sainsbury Laboratory, Norwich, United Kingdom). Tomato plants were grown in trays and then transplanted to pots containing a growth substrate (peat:vermiculite, 3:1, vol/vol) in a plant growth room, under 12-h light/12-h dark photoperiod, 25°C/19°C day/night temperature, 500 μmol m–2 s–1 photosynthetic photon flux density, and 88% relative humidity. Tomato plants at the five-leaf stage were used for experiments. Nicotiana benthamiana plants were grown under similar conditions and used for transient overexpression assay after 6-week germination. For the chemical agent treatment, unless otherwise noted, 2 mM SA or water as control was sprayed onto plant leaves on both the adaxial and abaxial surfaces at 12 h prior to Pst inoculation.
Virus-Induced Gene Silencing (VIGS) and Transient Overexpression
To generate virus-induced gene-silenced tomato plants, cDNA fragments of α-kGDH E2 (380 bp) and AOX1a (594 bp) and LIP2 (495 bp) were selected according to VIGS tool1. The polymerase chain reaction (PCR) fragments were inserted into pTRV2, and the primers used are shown in Supplementary Table S1. VIGS was performed by Agrobacterium-mediated transformation as previously described (Zhang et al., 2018). After viral inoculation, plants were maintained at 21°C before use in experiments.
α-kGDH E2 and AOX1a gene fragments were amplified using primers shown in Supplementary Table S1 and cloned into pFGC5941 vector with an hemagglutinin (HA) tag and pAC402 vector with a green fluorescent protein (GFP) tag, respectively. After sequence confirmation, the plasmids were shuttled into Agrobacterium tumefaciens GV3101 strain. The transient overexpression in N. benthamiana was performed as previously described (Liao et al., 2015). Samples were collected 48 h later to assess expression by Western blot using an anti-HA and GFP antibody. The A. tumefaciens-infiltrated leaves were further used for pathogen inoculation experiments.
Pathogen Inoculation and Disease Symptom Assays
Wild-type Pst was used for tomato plants to study the basal immunity, as there are no R proteins in Zheza 809 tomato. To avoid activation of the ETI response in N. benthamiana, Pst hrcC, which is deficient in the type III secretion system, was used for N. benthamiana inoculation. Pst or Pst hrcC were grown at 28°C in King’s B medium (10 g/L of peptone, 1.5 g/L of K2HPO4, 15 g/L of glycerol, and 5 g/L of MgSO4) with 50 mg/mL rifampicin. Bacterial cells were collected by centrifugation and dissolved in 10 mM MgCl2 to OD600 = 0.5. The bacterial inoculation of Pst was carried out by spraying the 10-time diluted bacterial suspension with 0.02% Silwet-L77 on the whole leaves of tomato plants. Leaves of N. benthamiana plants were infiltrated with Pst hrcC at the final concentration of OD600 = 0.01 using a syringe without a needle.
After pathogen inoculation, disease severity was assessed by the maximal quantum yield of PSII (Fv/Fm), trypan blue staining, or bacterial growth according to previously described methods (Zhang et al., 2015). The trypan blue staining was also quantified by ImageJ software based on the rate of dying area. As A. tumefaciens-carrying plasmids were infiltrated to N. benthamiana leaves for transient overexpression, which have common antibiotic resistance similar to bacterial Pst pathogens, the Pst bacterial growth was not measured to avoid potential confusion.
SA Quantification
Endogenous SA content was measured in tomato leaves by HPLC-MS/MS (Agilent 6460; Agilent Technologies) with D4-SA (OlChemlm) as internal standards using the same method described previously (Zhang et al., 2018).
RNA Isolation and RT-qPCR Assay
Total RNA was extracted from leaves using RNA simple Total RNA Kit (Tiangen, China), followed by DNase digestion, and reverse transcribed using a ReverTra Ace quantitative (qPCR) RT kit (Toyobo, Japan). Real time-quantitative PCR (RT-qPCR) assays were performed using SYBR Green PCR Master Mix Kit (Takara, Japan) on a Light Cycler 480 II detection system (Roche, Germany). Actin2 was used as the appropriate reference gene. Gene-specific primers for RT-qPCR are listed in Supplementary Table S1.
Respiration Analysis
Leaf respiration was assessed following the method of Millenaar et al. (2002) using a Clark-type oxygen electrode (Hansatech, King’s Lynn, United Kingdom). Total respiration was measured without any treatments. CN-resistant respiration and SHAM-resistant respiration were measured in the presence of 1 mM KCN and 3 mM SHAM, respectively.
Diaminobenzidine Staining
Diaminobenzidine (DAB) staining was performed as previously described with minor changes (Xia et al., 2009). Leaf samples were vacuum infiltrated with 1 mg mL–1 DAB in 50 mM Tris-HCl (pH 3.8) and incubated overnight in dark at room temperature. Then, leaf samples were rinsed in 80% (vol/vol) ethanol for 10 min at 70°C, finally mounted in destaining solution (lactic acid/phenol/water, 1:1:1, vol/vol/vol). Pictures were taken under a microscope (Zeiss, Germany).
Statistical Analysis
At least three independent biological replicates were performed for each determination. Unless otherwise stated, each biological replicate contained an independent sample that was pooled of two leaves, each from a different plant. The experiments were independently performed three times. The obtained data were subjected to analysis of variance using SAS software, version 8 (SAS Institute), and tested for significance using Tukey test at the 5% level.
Results
Effects of Pst Inoculation on Gene Transcripts, Mitochondrial Respiration, and SA Content
To examine the changes in the expression of genes involved in mitochondrial respiration, the transcript levels of 24 relevant genes were assayed at 8 and 12 h post inoculation with Pst (hpi). Among these, the SA biosynthesis- and signaling-related genes, including phenylalanine ammonia-lyase (PAL2/4/6), enhanced disease susceptibility 1 (EDS1), peptidyl arginine deiminase 4 (PAD4), NPR1, and PR family members, are up-regulated in response to Pst inoculation, which were also reported in a previous independent study (Yang et al., 2015). The transcripts of NAD-dependent isocitrate dehydrogenase (ICDH) in the TCA cycle and several miETC-related genes, including genes encoding succinate dehydrogenase of complex II (SDH1and SDH2), AOX family members (AOX1a and AOX1b), type II NAD(P)H dehydrogenases (NDA1and NDB2), and uncoupling proteins (UCP1 and UCP2), were also significantly increased, especially at 12 hpi. By contrast, other seven genes clustered in another group were down-regulated in response to Pst inoculation, including TCA cycle-related genes α-kGDH E2, lipoyl synthase (LIP1), octanoyltransferase (LIP2) and mitochondrial β-ketoacyl-acyl carrier protein synthase (mtKAS), miETC-related genes cytochrome c oxidase subunit (COX1), and AOX1c, as well as SA biosynthesis-related gene isochorismate synthase (ICS).
Changes in the rate of mitochondrial respiration and the endogenous SA content were further detected upon Pst inoculation. Pst inoculation caused a 12.58% decrease in total respiration rate at 8 hpi, followed by a significant decline at 12 hpi. A similar change occurred in the SHAM-resistant respiration rate, which decreased by 21.18% and 20.88% at 8 and 12 hpi, receptively. By contrast, Pst inoculation caused a 76.88% increase in the CN-resistant respiration rate at 8 hpi and a 41.39% increase at 12 hpi (Figure 1B). Meanwhile, consistent with previous studies on tomato (Yang et al., 2015; López-Gresa et al., 2017), the SA content was significantly induced and dramatically increased up to 10-fold at 12 hpi (Figure 1C).
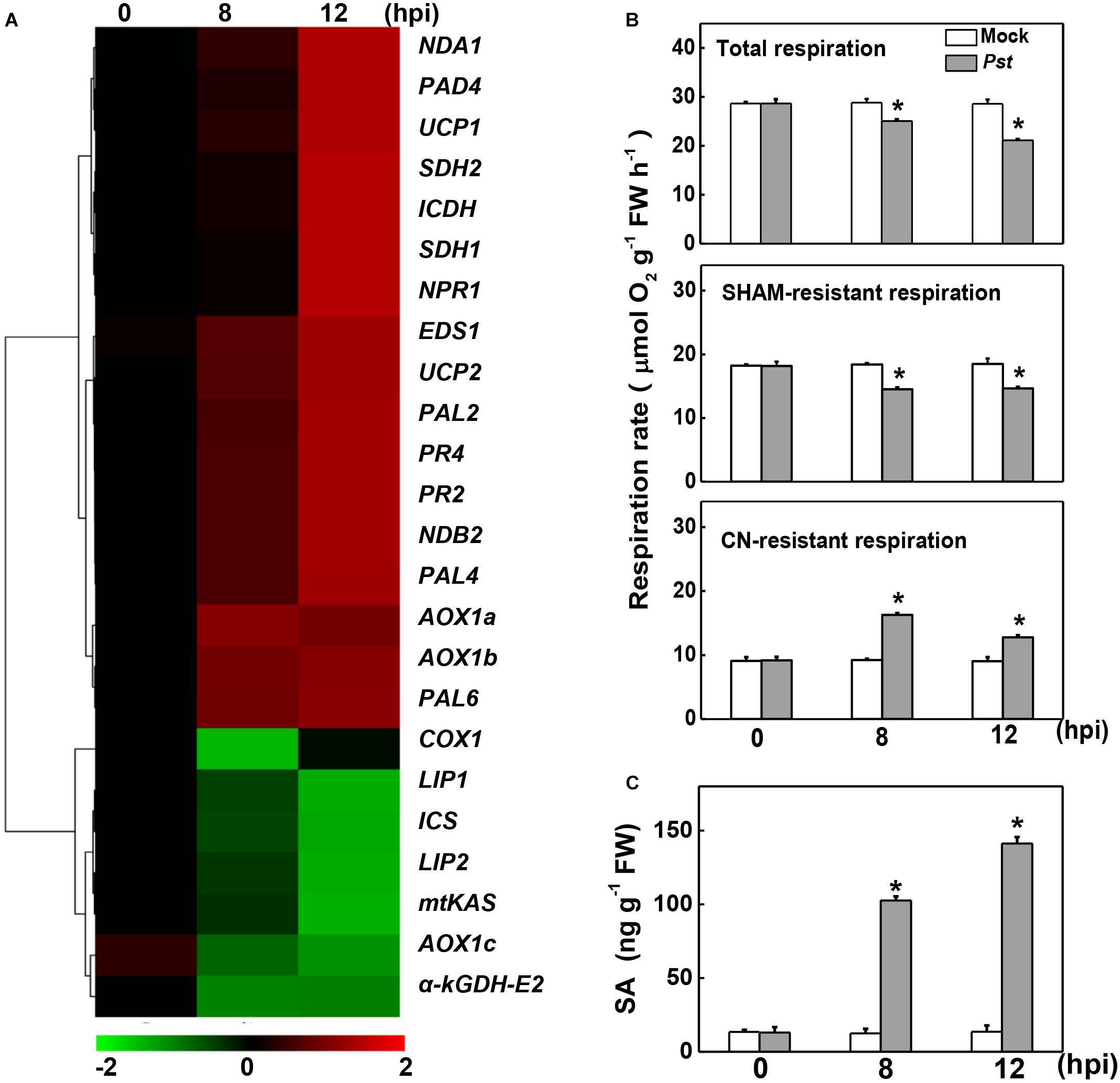
Figure 1. Effects of Pseudomonas syringae pv. tomato DC3000 (Pst) inoculation on gene expression, mitochondrial respiration, and salicylic acid (SA) content in tomato. (A) Changes in the gene transcripts 8- and 12-h post inoculation (hpi) with Pst based on cluster analysis. The intensity of the red or green color represents the degree of up- or down-regulation of the tested genes, respectively. The transcript levels of genes were determined by real-time quantitative PCR, with the Actin2 gene serving as an internal control. (B) Effects of Pst inoculation on the rate of total respiration, salicylhydroxamic acid (SHAM)-resistant respiration and cyanide (CN)-resistant respiration at indicated time. (C) Effects of Pst inoculation on endogenous SA content in tomato leaves. The asterisk indicates a significant difference between treatments (P < 0.05, Tukey test). The results in panels (B,C) are presented as mean values ± SD; n = 3. The above experiments were repeated three times with similar results.
α-kGDH E2, but Not AOX Functions in Plant Basal Immune Response to Pst
To study the roles of tomato α-kGDH E2 and AOX in the defense against Pst, we generated α-kGDH E2- and AOX1a-silenced plants via VIGS approach. RT-PCR was used to examine the efficiency of VIGS. Notably, under Pst-inoculated condition, the transcript levels of α-kGDH E2 and AOX1a significantly decreased by 63.76% and 65.01%, in α-kGDH E2-silenced (TRV: α-kGDH E2) and AOX1a-silenced plants (TRV: AOX1a), respectively, as compared with the empty-vector control (TRV:00) (Supplementary Figures S1A,B). At 3 days post inoculation (dpi) with Pst, TRV: α-kGDH E2 plants exhibited less reduction in the Fv/Fm and cell death, as well as increased PR1, PR2, and PR4 transcripts at 12 hpi (Figures 2A–C and Supplementary Figure S1C). In contrast, AOX1a-silencing did not result in a significant difference compared with TRV:00 control. Also, α-kGDH E2/AOX1a gene cosilencing had similar Fv/Fm value, cell death quantification, and PRs transcripts level when compared with a-kGDH E2 single gene-silenced plants. These parameters were corroborated with the proliferation of Pst in tomato leaves; bacterial growth in α-kGDH E2-silenced but not that of AOX1a-silenced leaves was significantly lower than that in the leaves of TRV:00 at 3 dpi (Figure 2D). Based on these findings, we assumed that the α-kGDH E2 negatively regulated plant basal defense against bacterial pathogens.
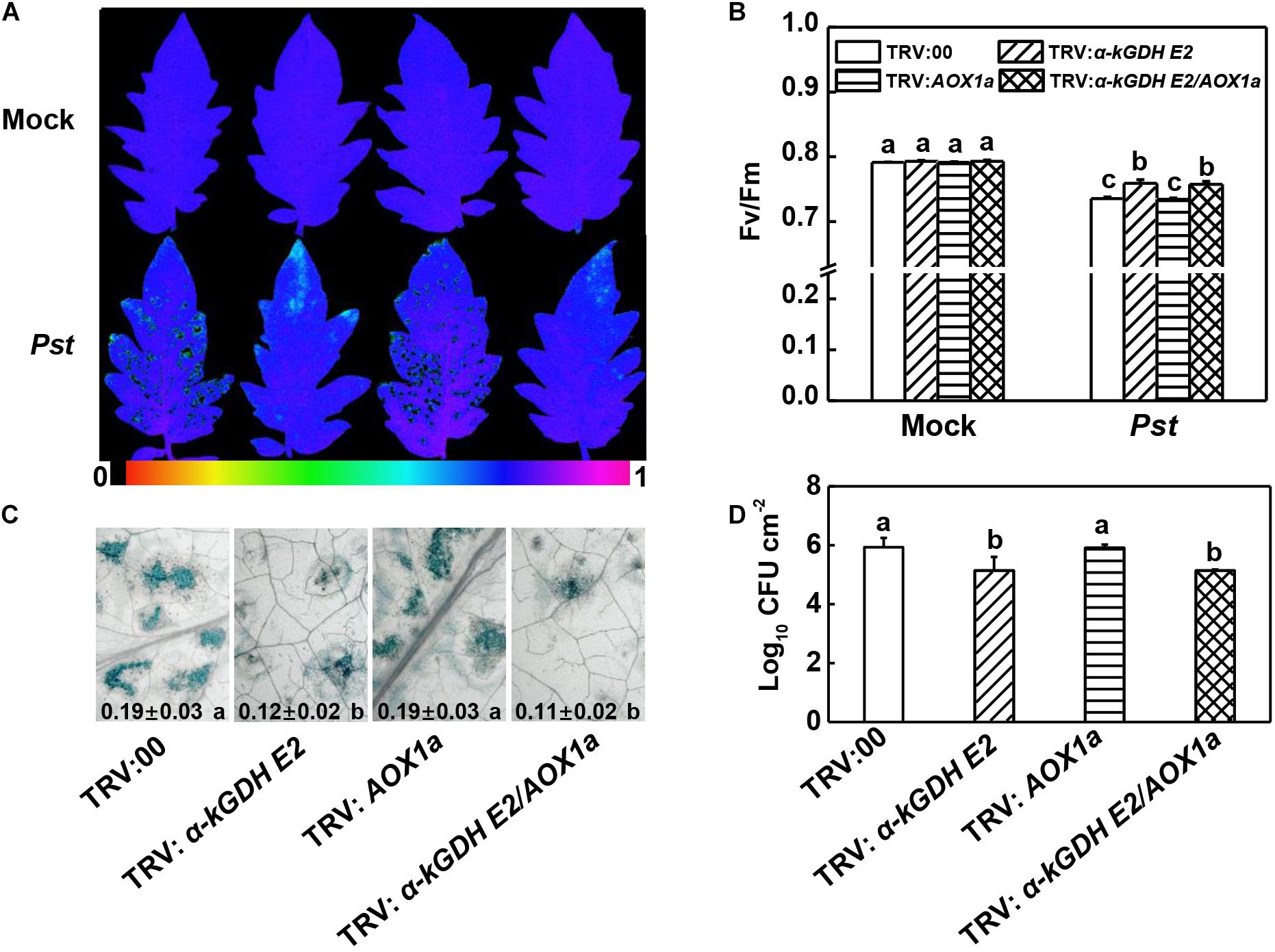
Figure 2. Effects of α-kGDH E2 and AOX1a silencing on plant defense against Pseudomonas syringae pv. tomato DC3000 (Pst) inoculation in tomato. (A) Representative images of leaf maximum quantum yield of PSII (Fv/Fm) at 3 days post inoculation (dpi) with Pst. The color gradient scale at the bottom indicates the magnitude of the fluorescence signal represented by each color. (B) Fv/Fm values at 3 dpi. (C) Representative images for trypan blue staining of Pst-inoculated leaves at 3 dpi. Quantificative data are shown in each figure. (D) Bacterial growth at 3 dpi. The results in panels (B–D) are presented as mean values ± SD; n = 5. Different letters indicate significant a differences between treatments (P < 0.05, Tukey test). The above experiments were repeated three times with similar results.
Next, we constructed vectors carrying α-kGDH E2 with an HA tag or AOX1a with a GFP tag and infiltrated them into N. benthamiana leaves for transient overexpression. Western blotting showed that α-kGDH E2 and AOX1a were abundantly expressed (Figures 3A,B). These leaves were then inoculated with Pst hrcC, which is deficient in the type III secretion system and can avoid triggering ETI response. Results in Figure 3 showed that overexpression of α-kGDH E2 in N. benthamiana rendered more susceptibility to Pst hrcC inoculation, as reflected by significantly decreased Fv/Fm (Figures 3C,D). However, overexpression of AOX1a did not affect the defense against Pst inoculation. Taken together, these results indicate that tomato α-kGDH E2 plays a negative role in plant basal immune response, but AOX1a seems to have no apparent function in the basal defense against Pst.
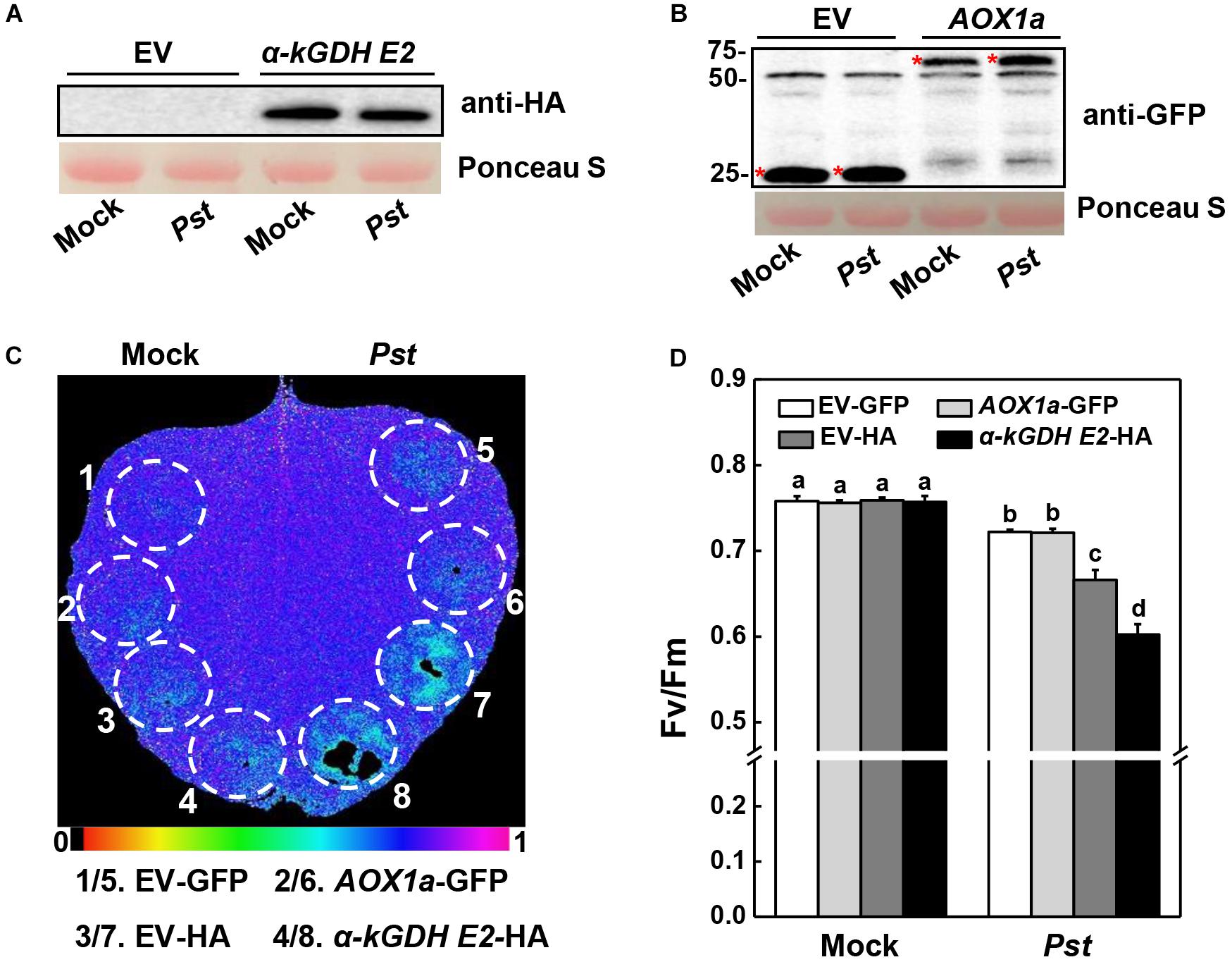
Figure 3. Effects of tomato gene α-kGDH E2 and AOX1a-transient overexpression on plant defense against Pseudomonas syringae pv. tomato DC3000 hrcC (Pst hrcC) inoculation in N. benthamiana inoculation. (A,B) Tomato α-kGDH E2 and AOX1a were transiently overexpressed in N. benthamiana, 48 h later, leaf samples were collected for proteins detection by Western blotting with an anti-HA antibody (A) and anti-GFP antibody (B). Protein loading was verified by Ponceau S staining. (C) Representative image of gene overexpressed leaf maximum quantum yield of PSII (Fv/Fm) at 3 days post inoculation (dpi) with Pst hrcC. The color gradient scale at the bottom indicates the magnitude of the fluorescence signal represented by each color. (D) Fv/Fm values at 3 dpi. The results in panel (D) are presented as mean values ± SD; n = 5. Different letters indicate significant differences between treatments (P < 0.05, Tukey test). The above experiments were repeated three times with similar results.
Lipoylation of LIP2 to α-kGDH E2 Is Involved in Plant Basal Immunity Against Pst
The lipoylation of α-kGDH E2 is essential for its catalytic activity, and α-kGDH E2 was previously reported to be lipoylated by LIP2 (Ewald et al., 2014). Therefore, changes in the lipoylation of α-kGDH E2 in response to Pst inoculation were examined, which showed that the lipoylation level significantly decreased in response to Pst at 12 hpi (Figure 4A). We then generated LIP2- and LIP2/α-kGDH E2- cosilenced plants (Supplementary Figures S2A,B). Silencing of LIP2 in tomato plants did result in a noticeable reduction in the lipoylation (Figure 4B).
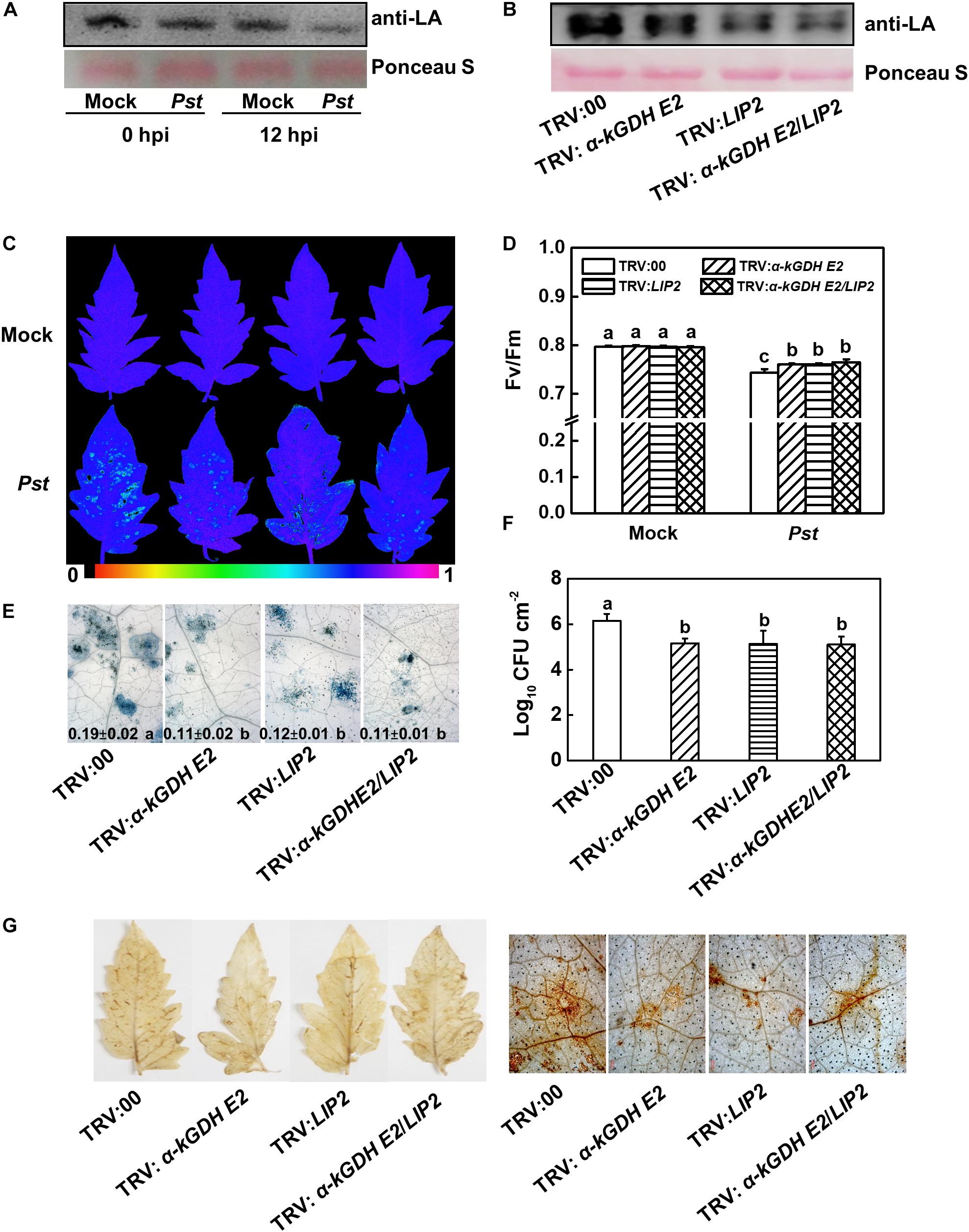
Figure 4. Effects of lipoylation of LIP2 to α-kGDH E2 on the plant defense against Pseudomonas syringae pv. tomato DC3000 (Pst) in tomato. (A,B) Effects of Pst inoculation and silencing LIP2 on lipoylation of α-kGDH E2. (A) The lipoylation of α-kGDH E2 was detected by immunoblot analysis with an anti-lipoic acid (LA) antibody in tomato plants at 0 and 12 h post inoculation (hpi) with Pst. (B) Effects of silencing α-kGDH E2 and LIP2 on the lipoylation of α-kGDH E2 in tomato plants. Protein loading was verified by Ponceau S staining. (C) Representative images of leaf maximum quantum yield of PSII (Fv/Fm) at 3 days post inoculation (dpi) with Pst. The color gradient scale at the bottom indicates the magnitude of the fluorescence signal represented by each color. (D) Fv/Fm values at 3 dpi. (E) Representative images for trypan blue staining of Pst-inoculated leaves at 3 dpi. Quantificative data are shown in each figure. (F) Bacterial growth at 3 dpi. (G) Hydrogen peroxide (H2O2) contents at 24 hpi. The results in panels (D–F) are presented as mean values ± SD; n = 5. Different letters indicate significant differences between treatments (P < 0.05, Tukey test). The above experiments were repeated three times with similar results.
We compared plant basal defense between α-kGDH E2- and LIP2-silenced plants. Based on the data of Fv/Fm, cell death, bacterial growth, and PRs gene expression (PR1, PR2, and PR4), silencing LIP2 reduced plant susceptibility and increased the PRs transcripts to a similar level of α-kGDH E2-silenced plants. Notably, cosilencing of these two genes did not result in a further change in plant defense (Figures 4C–F and Supplementary Figure S2C), suggesting that α-kGDH E2 and LIP2 work in the same pathway in the basal defense response.
We then analyzed the accumulation of hydrogen peroxide (H2O2) via DAB staining in the empty-vector control and target gene-silenced leaves at 24 hpi. Silencing of α-kGDH E2, LIP2, or gene cosilencing dramatically suppressed the accumulation of H2O2 to a similar level (Figure 4G). Overall, these results suggest that lipoylation of LIP2 to α-kGDH E2 plays a negative role in plant basal immunity.
α-kGDH E2 Acts in Downstream of SA Defense Pathway Against Pst Inoculation
Tomato α-kGDH E2 has been reported to be an SABP (Liao et al., 2015). We thus determined the relationship between α-kGDH E2 and SA in the basal defense against Pst. In the TRV:00 control plants, the Pst inoculation-induced PSII damage and bacterial growth were significantly alleviated by pretreatment with exogenous SA. Silencing of α-kGDH E2 again diminished Pst inoculation-induced PSII damage or cell death, but they were not further reduced by exogenous SA pretreatment (Figures 5A–D). Similarly, exogenous SA pretreatment did not lead to further induction of PR1 gene transcripts in a-kGDH E2-silenced plants (Supplementary Figure S3).
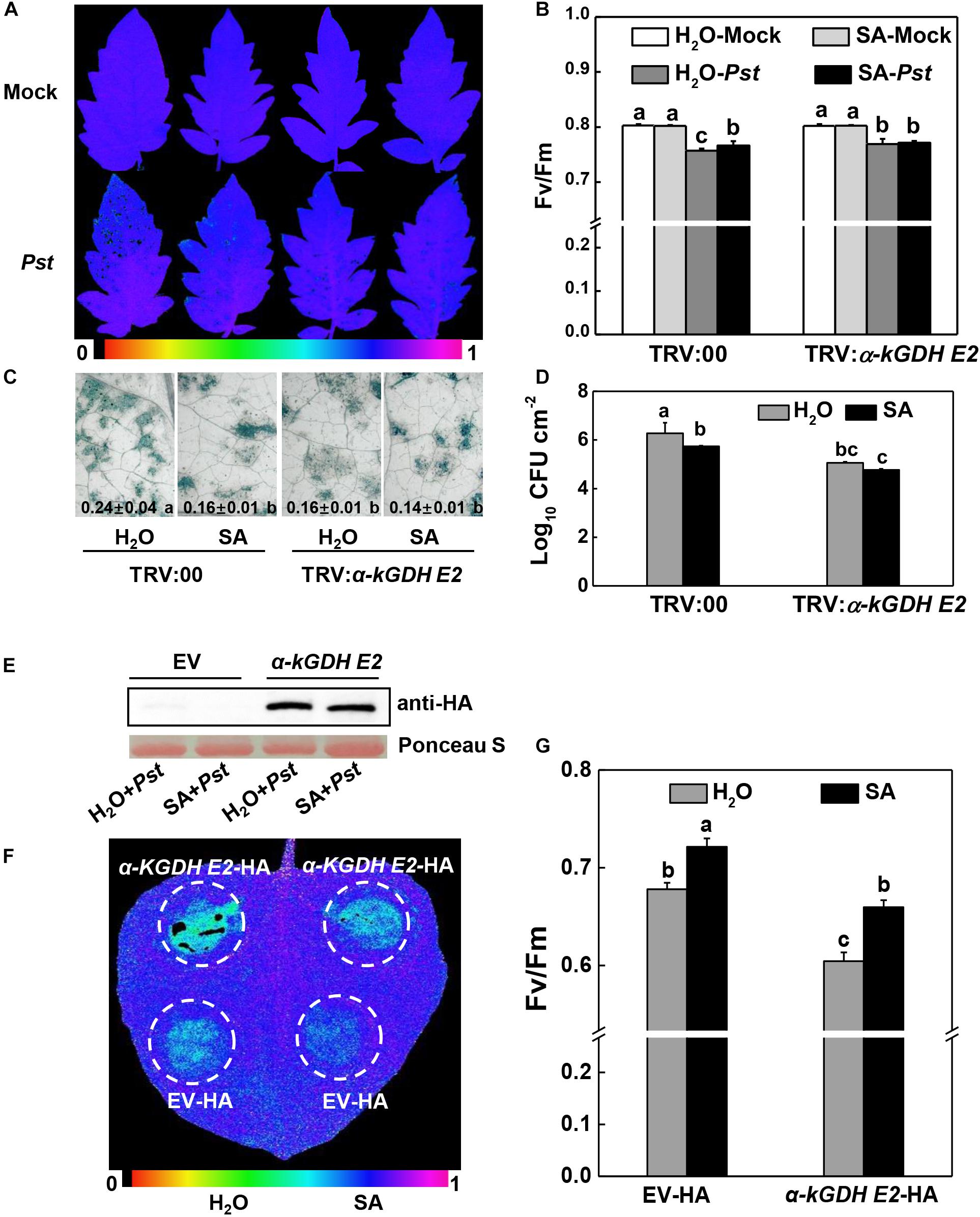
Figure 5. Effects of SA application on plant defense against Pseudomonas syringae pv. tomato DC3000 (Pst) inoculation in α-kGDH E2-silenced tomato or transiently overexpressed N. benthamiana plants. (A–D) Effects of SA application on the resistance to Pst in α-kGDH E2-silenced tomato plants. 2 mM SA or water as control was sprayed onto tomato leaves at 12 h prior to Pst inoculation. (A) Representative images of leaf maximum quantum yield of PSII (Fv/Fm) at 3 days post inoculation (dpi) with Pst. The color gradient scale at the bottom indicates the magnitude of the fluorescence signal represented by each color. (B) Fv/Fm values at 3 dpi. (C) Representative images for trypan blue staining of Pst-inoculated leaves at 3 dpi. Quantificative data are shown in each figure. (D) Bacterial growth at 3 dpi. (E–G) Effects of SA application on the resistance to Pst hrcC in α-kGDH E2 transient overexpression N. benthamiana plants. (E) Tomato α-kGDH E2 was transiently overexpressed in N. benthamiana; 48 h later, leaf samples were collected for proteins detection by Western blotting with an anti-HA antibody. Protein loading was verified by Ponceau S staining. 2 mM SA or water as control was sprayed onto leaves at 12 h prior to Pst hrcC inoculation. (F) Representative image of leaf Fv/Fm at 3 dpi. The color gradient scale at the bottom indicates the magnitude of the fluorescence signal represented by each color. (G) Fv/Fm values at 3 dpi. The data are shown in panels (B–D,G) as means ± SD (n = 5), and different letters indicate significant differences between treatments (P < 0.05, Tukey test). The above experiments were repeated three times with similar results.
The failure of SA to improve defense in α-kGDH E2-silenced plants might be attributable to the already lower α-kGDH E2 gene expression in these plants. We therefore tested the role of exogenous SA in the α-kGDH E2 overexpressed N. benthamiana plants. Strikingly, Pst-caused damage of the Fv/Fm was evidently attenuated by the exogenous SA pretreatment in α-kGDH E2-overexpressed leaves (Figures 5E–G).
We asked whether α-kGDH E2 functions in the defense response of NahG plants that are impaired in SA accumulation. The results showed that NahG plants were more susceptible to Pst inoculation compared with control WT plants, whereas silencing α-kGDH E2 gene led to higher defense against Pst in both WT and NahG backgrounds (Figure 6).
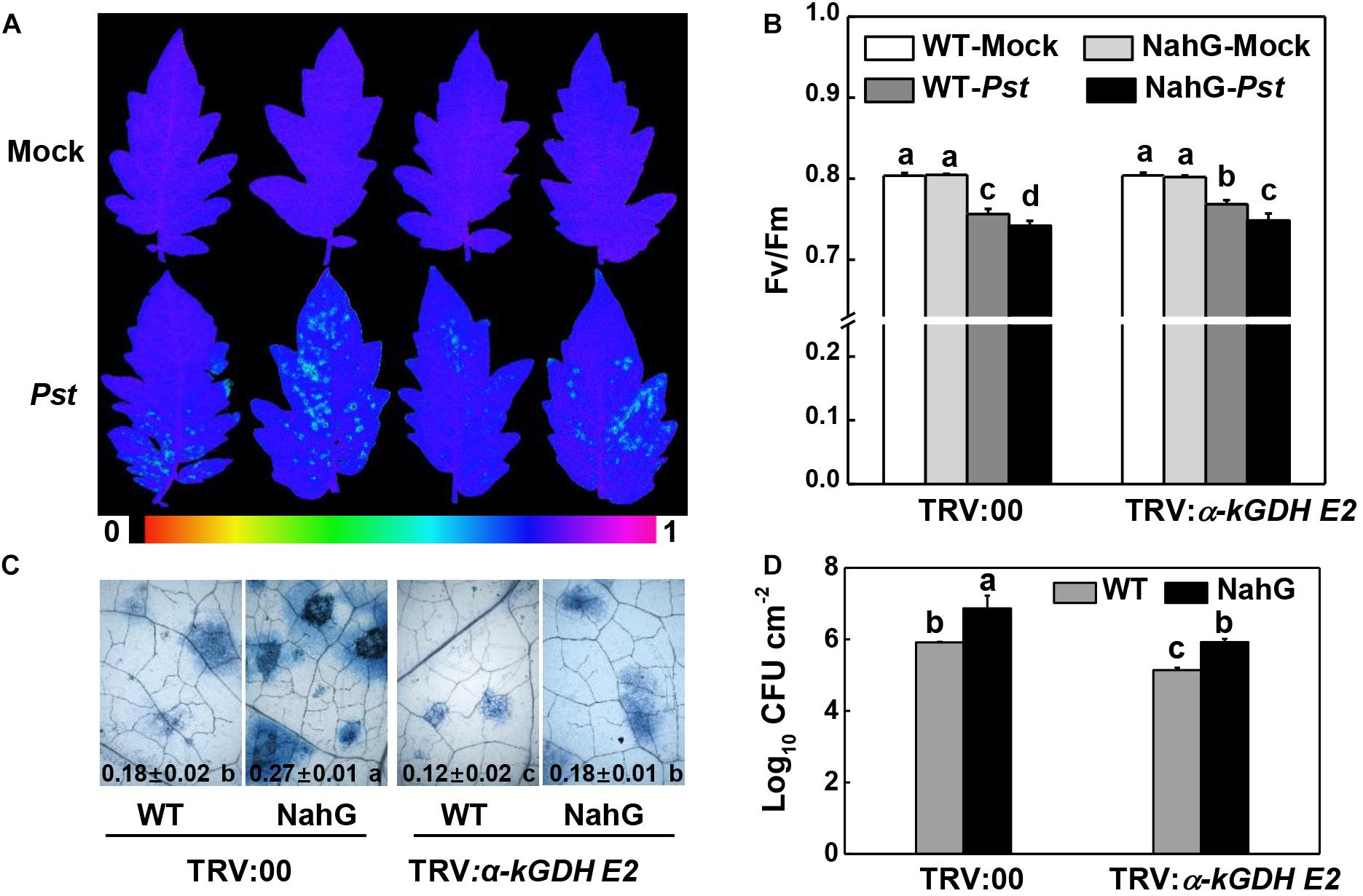
Figure 6. Effects of silencing α-kGDH E2 on plant defense against Pseudomonas syringae pv. tomato DC3000 (Pst) inoculation in SA accumulation- defective-transgenic NahG and wild-type tomato plants. (A) Representative images of leaf maximum quantum yield of PSII (Fv/Fm) at 3 days post inoculation (dpi) with Pst. The color gradient scale at the bottom indicates the magnitude of the fluorescence signal represented by each color. (B) Fv/Fm values at 3 dpi. (C) Representative images for trypan blue staining of Pst-inoculated leaves at 3 dpi. Quantitative data are shown in each figure. (D) Bacterial growth at 3 dpi. The results in panels (B–D) are presented as mean values ± SD; n = 5. Different letters indicate significant differences between treatments (P < 0.05, Tukey test). The above experiments were repeated twice with similar results.
Discussion
Mitochondrial respiration is an important metabolic process in plant innate immunity (Colombatti et al., 2014). Particularly, α-KGDH E2 of the TCA cycle and AOX pathway of miETC participate in plant defense, mostly against viral pathogens in plants. In this study, we showed that tomato α-kGDH E2 but not AOX negatively regulated plant basal defense to bacterial pathogen Pst in an SA-dependent process.
In this study, the SHAM-resistant Cyt pathway respiration and expression of α-kGDH E2 were repressed, whereas the CN-resistant AOX pathway was significantly induced by the Pst inoculation (Figure 1). These results are in accordance with previous reports in which application of elicitor harpin (virulence factors produced by bacterial pathogens such as P. syringae) inhibited mitochondrial respiration and causing a strong induction of AOX in Arabidopsis cells (Krause and Durner, 2004). Several studies have described AOX1a as the most stress-responsive AOX gene, whereas AOX1c expression is relatively stable or even decreased in response to stresses or elicitor treatment (Clifton et al., 2006; Czobor et al., 2019).
VIGS and transient overexpression approaches were used to further understand the biological functions of α-kGDH E2 and AOX in the defense against virulent Pst. We found that the susceptibility to Pst was significantly reduced by silencing α-kGDH E2 in tomato plants, but increased by overexpressing α-kGDH E2 in N. benthamiana plants. In contrast, silencing or overexpressing of AOX1a gene did not have significant effects as compared with that of the control counterpart (Figures 2, 3). To the best of our knowledge, this is the first evidence that α-kGDH E2 acts as a negative regulator in plant defense against bacterial pathogen Pst. Previously, α-kGDH E2 was reported to function in the plant defense against TMV through modulating the miETC AOX pathway and the associated mitochondrial oxidative phosphorylation in tomato (Liao et al., 2015). However, in contrast to the plant defense to viral pathogens, the tomato defense against Pst is independent on AOX pathway as evidenced by both gene silencing and overexpression experiments (Figures 2, 3). Thus, the function of AOX in plant defense seems to be pathogen species-specific. For instance, N. attenuata AOX contributes to resistance to piercing-sucking insects but not to Manduca sexta larvae (Zhang et al., 2012). In the incompatible plant–bacterial pathogen interaction, AOX knockdown N. tabacum displayed a delayed ROS and HR in response to P. syringae pv. Maculicola (Cvetkovska and Vanlerberghe, 2013), but showed higher levels of ROS and HR after Pst inoculation (Zhang et al., 2012). The present study indicates that α-kGDH E2, but not AOX, is a key component of basal immunity, which warrants further investigation in broader compatible plant–bacteria pathogen systems.
In the TCA cycle, the lipoylation of α-kGDH E2 is essential for its catalytic activity, and it was reported to be lipoylated by LIP2 in Arabidopsis (Ewald et al., 2014). The deficiency in the lipoylation of α-kGDH E2 and pyruvate dehydrogenase leads to an early onset of fatal lactic acidosis in humans (Tort et al., 2014). Whether the lipoylation of LIP2 to α-kGDH E2 functions in plant basal immunity remains unclear. In this study, the transcripts of LIP2 greatly decreased in response to Pst inoculation (Figure 1A). Also, Pst inoculation caused a significant decrease in the lipoylation of α-kGDH E2 (Figure 4B). Most importantly, silencing LIP2 contributed to an enhanced resistance to Pst, as well as higher level of SA-dependent marker gene PR1, PR2, and PR4 to the same level with α-kGDH E2-silenced plants (Figure 4 and Supplementary Figure S2), indicating that the lipoylation of LIP2 to α-kGDH E2 plays a negative role in plant defense against Pst. Excessive accumulation of ROS causes severe oxidative damage to plants, and control of oxidative damage is essential for plants to survive under and recover from stresses (Samsatly et al., 2018). Previous reports have suggested that α-kGDH is not only a target of ROS but also could significantly contribute to the control of ROS accumulation in the mitochondria (Tretter and Adam-Vizi, 2005). In this study, silencing α-kGDH E2 or LIP2 both significantly reduced the accumulation of H2O2 (Figure 4G). Therefore, these results suggest that silencing α-kGDH E2 or LIP2 may alleviate the H2O2 accumulation to prevent the plant from oxidative damage during Pst inoculation. However, we cannot exclude a possibility that α-kGDH E2 in plants is associated with physiological responses such as nutritional shift and carbon partitioning (Araújo et al., 2012), which affect pathogen growth as well. In addition, 2-oxoglutarate (2-OG), a key organic acid of the TCA cycle, is an obligatory substrate for α-kGDH (Bailey and Nathan, 2018). Previous evidence has indicated that 2-OG plays an important role in the metabolism of glucosinolate, flavonoid, and alkaloid (Araújo et al., 2014), which are widely distributed secondary metabolites with different biological functions in plants, including the defense against pathogen inoculation (Hunziker et al., 2020; Nabavi et al., 2020). Thus, it is tempting to speculate that the down-regulation of α-kGDH E2 and LIP2 may also induce the accumulation of 2-OG, which may play a role in basal immune responses.
Salicylic acid-dependent signaling controls the activation of complex plant defense responses to combat microbial pathogens (Zhang and Li, 2019). In agreement with previous studies (Yang et al., 2015; López-Gresa et al., 2017), the SA content dramatically increased in response to Pst inoculation, which was accompanied by a significant increase in the transcripts of SA biosynthesis- and signaling-related genes (Figure 1). Tomato α-kGDH E2 was proven to be SABP, and α-kGDH E2 acted as a negative regulator of SA-dependent defense to TMV (Liao et al., 2015). In this study, SA treatment and silencing α-kGDH E2 both increased resistance to Pst. SA did not further enhance defense against Pst in α-kGDH E2-silenced tomato plants, but did reduce the susceptibility in N. benthamiana plants transiently overexpressing α-kGDH E2 (Figure 5). Additionally, silencing α-kGDH E2 gene led to higher defense against Pst in both WT or NahG backgrounds (Figure 6). Thus, tomato α-kGDH E2 was proposed to function as a negative regulator of SA-dependent defense against Pst. By the way, the AtICS is the most important gene for SA biosynthesis in Arabidopsis upon pathogen inoculation (Garcion et al., 2008). Unlike that in Arabidopsis, in this study, the PAL expression but not that of ICS was induced by Pst inoculation (Figure 1). The expression profile of ICS was in agreement with previous genome-wide studies in tomato–Pst interactions (Yang et al., 2015). Similarly, in a study on pepper plants, Obuda pepper virus inoculation markedly induced the expression of three PAL genes, whereas that of ICS gene was not modified (Dziurka et al., 2016). Also, multiple pathogen infections with virulent or avirulent strains of the Pst pathogens on soybean were associated with suppression of ICS gene expression (Shine et al., 2016). Thus, pathogen-induced SA biosynthesis may use different pathways in different species, which need further studies.
In summary, the study reveals a novel function of a mitochondrial TCA cycle enzyme α-kGDH E2, but not miETC AOX, in the orchestration of plant basal immunity in a compatible plant–pathogen interaction. The study not only provides new insights into the function of mitochondrial respiration against bacterial pathogens, but also identifies new targets and markers for the development of improved cultivars that are better equipped to combat pathogens.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
KS designed the experiments. QM and YL performed most of experiments and analyzed the data. HF, PW, and WZ assisted in experiments and discussed the results. QM and KS wrote the manuscript. KS and GA edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province for Distinguished Young Scholar (LR19C150001), the National Natural Science Foundation of China (31772355 and 31822046), and the Fundamental Research Funds for the Central Universities.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.579772/full#supplementary-material
Supplementary Figure 1 | Real-time RT-PCR analysis of α-kGDH E2, AOX1a and PR genes expression in virus-induced genes silencing (VIGS) tomato plants as infected by Pseudomonas syringae pv. tomato DC3000 (Pst). (A,B) The efficiency of VIGS was examined in α-kGDH E2- (A) and AOX1a- (B) silenced tomato plants. Samples were collected at 12 hours post inoculation (hpi) with Pst. (C) The transcript expression of PR1, PR2 and PR4 at 0 and 12 hpi. The Actin2 gene was used as the internal control. The control sample was normalized to 1. Data are presented as mean values ± SD; n = 3. Different letters indicate a significant differences between treatments (P < 0.05, Tukey’s test).
Supplementary Figure 2 | Real-time RT-PCR analysis of α-kGDH E2, AOX1a and PR genes expression in virus-induced genes silencing (VIGS) tomato plants as infected by Pseudomonas syringae pv. tomato DC3000 (Pst). (A,B) The efficiency of VIGS was examined in α-kGDH E2- (A) and LIP2- (B) silenced tomato plants. Samples were collected at 12 hours post inoculation (hpi) with Pst. (C) The transcript expression of PR1, PR2 and PR4 at 12 hpi. The Actin2 gene was used as the internal control. The control sample was normalized to 1. Data are presented as mean values ± SD; n = 3. Different letters indicate a significant differences between treatments (P < 0.05, Tukey’s test).
Supplementary Figure 3 | Real-time RT-PCR analysis of PR1 expression in empty-vector control and α-kGDH E2-silenced tomato plants with exogenous SA application as infected by Pseudomonas syringae pv. tomato DC3000 (Pst). The Actin2 gene was used as the internal control. The control sample was normalized to 1. Data are presented as mean values ± SD; n = 3. Different letters indicate a significant differences between treatments (P < 0.05, Tukey’s test).
Footnotes
References
Araújo, W. L., Martins, A. O., Fernie, A. R., and Tohge, T. (2014). 2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 5:552. doi: 10.3389/fpls.2014.00552
Araújo, W. L., Tohge, T., Osorio, S., Lohse, M., and Balbo, I. (2012). Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24, 2328–2351. doi: 10.1105/tpc.112.099002
Bailey, P., and Nathan, J. (2018). Metabolic regulation of hypoxia-inducible transcription factors: the role of small molecule metabolites and iron. Biomedicines 6:60. doi: 10.3390/biomedicines6020060
Chen, J., Mohan, R., Zhang, Y., Li, M., and Chen, H. (2019). NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 181, 289–304. doi: 10.1104/pp.19.00124
Clifton, R., Millar, A. H., and Whelan, J. (2006). Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta Bioenerget. 1757, 730–741. doi: 10.1016/j.bbabio.2006.03.009
Colombatti, F., Gonzalez, D. H., and Welchen, E. (2014). Plant mitochondria under pathogen attack: sigh of relief or a last breath? Mitochondrion 19, 238–244. doi: 10.1016/j.mito.2014.03.006
Condori-Apfata, J. A., Batista-Silva, W., Medeiros, D. B., Vargas, J. R., Valente, L. M. L., Heyneke, E., et al. (2019). The Arabidopsis E1 subunit of the 2-oxoglutarate dehydrogenase complex modulates plant growth and seed production. Plant Mol. Biol. 101, 183–202. doi: 10.1007/s11103-019-00900-3
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Cvetkovska, M., and Vanlerberghe, G. C. (2012). Coordination of a mitochondrial superoxide burst during the hypersensitive response to bacterial pathogen in Nicotiana tabacum. Plant Cell. Environ. 35, 1121–1136. doi: 10.1111/j.1365-3040.2011.02477.x
Cvetkovska, M., and Vanlerberghe, G. C. (2013). Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ. 36, 721–732. doi: 10.1111/pce.12009
Czobor, Á., Hajdinák, P., Németh, B., Piros, B., Németh, Á., and Szarka, A. (2019). Comparison of the response of alternative oxidase and uncoupling proteins to bacterial elicitor induced oxidative burst. PLoS One 14:e210592. doi: 10.1371/journal.pone.0210592
Daloso, D. M., Muller, K., Obata, T., Florian, A., and Tohge, T. (2015). Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 1392–1400. doi: 10.1073/pnas.1424840112
Ding, Y., Sun, T., Ao, K., Peng, Y., and Zhang, Y. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Dziurka, M., Janeczko, A., Juhász, C., Gullner, G., and Oklestková, J. (2016). Local and systemic hormonal responses in pepper leaves during compatible and incompatible pepper-tobamovirus interactions. Plant Physiol. Biochem. 109, 355–364. doi: 10.1016/j.plaphy.2016.10.013
Ewald, R., Hoffmann, C., Florian, A., Neuhaus, E., and Fernie, A. R. (2014). Lipoate-protein ligase and octanoyltransferase are essential for protein lipoylation in mitochondria of Arabidopsis. Plant Physiol. 165, 978–990. doi: 10.1104/pp.114.238311
Garcion, C., Lohmann, A., Lamodière, E., Catinot, J., and Buchala, A. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi: 10.1104/pp.108.119420
Glazebrook, J., Rogers, E. E., and Ausubel, F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982.
Hunziker, P., Ghareeb, H., Wagenknecht, L., Crocoll, C., and Halkier, B. A. (2020). De novo indol-3-ylmethyl glucosinolate biosynthesis, and not long-distance transport, contributes to defence of Arabidopsis against powdery mildew. Plant Cell Environ. 43, 1571–1583. doi: 10.1111/pce.13766
Kachroo, P., Yoshioka, K., Shah, J., Dooner, H. K., and Klessig, D. F. (2000). Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 12, 677–690. doi: 10.1105/tpc.12.5.677
Krause, M., and Durner, J. (2004). Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol. Plant Microbe Interact. 17, 131–139. doi: 10.1094/MPMI.2004.17.2.131
Lee, W. S., Fu, S. F., Verchot-Lubicz, J., and Carr, J. P. (2011). Genetic modification of alternative respiration in Nicotiana benthamiana affects basal and salicylic acid-induced resistance to potato virus X. BMC Plant Biol. 11:41. doi: 10.1186/1471-2229-11-41
Liao, Y., Tian, M., Zhang, H., Li, X., and Wang, Y. (2015). Salicylic acid binding of mitochondrial alpha-ketoglutarate dehydrogenase E2 affects mitochondrial oxidative phosphorylation and electron transport chain components and plays a role in basal defense against tobacco mosaic virus in tomato. New Phytol. 205, 1296–1307. doi: 10.1111/nph.13137
López-Gresa, M. P., Lisón, P., Campos, L., Rodrigo, I., and Rambla, J. L. (2017). A non-targeted metabolomics approach unravels the VOCs associated with the tomato immune response against Pseudomonas syringae. Front. Plant Sci. 8:1188. doi: 10.3389/fpls.2017.01188
Ma, H., Song, C., Borth, W., Sether, D., and Melzer, M. (2011). Modified expression of alternative oxidase in transgenic tomato and petunia affects the level of tomato spotted wilt virus resistance. BMC Biotechnol. 11:96. doi: 10.1186/1472-6750-11-96
Manohar, M., Tian, M., Moreau, M., Park, S., and Choi, H. W. (2015). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5:777. doi: 10.3389/fpls.2014.00777
Mayers, C. N., Lee, K. C., Moore, C. A., Wong, S. M., and Carr, J. P. (2005). Salicylic acid-induced resistance to cucumber mosaic virus in squash and Arabidopsis thaliana: contrasting mechanisms of induction and antiviral action. Mol. Plant Microbe Interact. 18, 428–434. doi: 10.1094/MPMI-18-0428
Millenaar, F. F., Gonzalez-Meler, M. A., Siedow, J. N., Wagner, A. M., and Lambers, H. (2002). Role of sugars and organic acids in regulating the concentration and activity of the alternative oxidase in Poa annua roots. J. Exp. Bot. 53, 1081–1088. doi: 10.1093/jexbot/53.371.1081
Murphy, A. M., Chivasa, S., Singh, D. P., and Carr, J. P. (1999). Salicylic acid-induced resistance to viruses and other pathogens: a parting of the ways? Trends Plant Sci. 4, 155–160. doi: 10.1016/S1360-1385(99)01390-4
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., and Russo, D. (2020). Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnol. Adv. 38:107316. doi: 10.1016/j.biotechadv.2018.11.005
Naylor, M., Murphy, A. M., Berry, J. O., and Carr, J. P. (1998). Salicylic acid can induce resistance to plant virus movement. Mol. Plant Microbe Interact. 11, 860–868. doi: 10.1094/mpmi.1998.11.9.860
Poór, P. (2020). Effects of salicylic acid on the metabolism of mitochondrial reactive oxygen species in plants. Biomolecules 10:341. doi: 10.3390/biom10020341
Qi, G., Chen, J., Chang, M., Chen, H., and Hall, K. (2018). Pandemonium breaks out: disruption of salicylic acid-mediated defense by plant pathogens. Mol. Plant 11, 1427–1439. doi: 10.1016/j.molp.2018.10.002
Rairdan, G. J., and Delaney, T. P. (2002). Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161, 803–811.
Samsatly, J., Copley, T. R., and Jabaji, S. H. (2018). Antioxidant genes of plants and fungal pathogens are distinctly regulated during disease development in different Rhizoctonia solani pathosystems. PLoS One 13:192682. doi: 10.1371/journal.pone.0192682
Shine, M. B., Yang, J. W., El-Habbak, M., Nagyabhyru, P., and Fu, D. Q. (2016). Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 212, 627–636. doi: 10.1111/nph.14078
Tian, M., von Dahl, C. C., Liu, P., Friso, G., and van Wijk, K. J. (2012). The combined use of photoaffinity labeling and surface plasmon resonance-based technology identifies multiple salicylic acid-binding proteins. Plant J. 72, 1027–1038. doi: 10.1111/tpj.12016
Tort, F., Ferrer-Cortès, X., Thió, M., Navarro-Sastre, A., and Matalonga, L. (2014). Mutations in the lipoyltransferase LIPT1 gene cause a fatal disease associated with a specific lipoylation defect of the 2-ketoacid dehydrogenase complexes. Hum. Mol. Genet. 23, 1907–1915. doi: 10.1093/hmg/ddt585
Tretter, L., and Adam-Vizi, V. (2005). Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos. Trans. R. Soc. B 360, 2335–2345. doi: 10.1098/rstb.2005.1764
Vanlerberghe, G. C., Dahal, K., Alber, N. A., and Chadee, A. (2020). Photosynthesis, respiration and growth: a carbon and energy balancing act for alternative oxidase. Mitochondrion 52, 197–211. doi: 10.1016/j.mito.2020.04.001
Wong, C. E., Carson, R. A. J., and Carr, J. P. (2002). Chemically induced virus resistance in Arabidopsis thaliana is independent of pathogenesis-related protein expression and the NPR1 gene. Mol. Plant Microbe Interact. 15, 75–81. doi: 10.1094/MPMI.2002.15.1.75
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., and Wang, Y. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Xia, X., Wang, Y., Zhou, Y., Tao, Y., and Mao, W. (2009). Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 150, 801–814. doi: 10.1104/pp.109.138230
Yang, Y., Wang, M., Yin, Y., Onac, E., and Zhou, G. (2015). RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genomics 16:120. doi: 10.1186/s12864-015-1228-7
Zhang, H., Hu, Z., Lei, C., Zheng, C., and Wang, J. (2018). A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 30, 652–667. doi: 10.1105/tpc.17.00537
Zhang, L., Oh, Y., Li, H., Baldwin, I. T., and Galis, I. (2012). Alternative oxidase in resistance to biotic stresses: Nicotiana attenuata AOX contributes to resistance to a pathogen and a piercing-sucking insect but not Manduca sexta Larvae. Plant Physiol. 160, 1453–1467. doi: 10.1104/pp.112.200865
Zhang, S., Li, X., Sun, Z., Shao, S., and Hu, L. (2015). Antagonism between phytohormone signalling underlies the variation in disease susceptibility of tomato plants under elevated CO2. J. Exp. Bot. 66, 1951–1963. doi: 10.1093/jxb/eru538
Keywords: mitochondrion, α-ketoglutarate dehydrogenase E2, alternative oxidase, lipoylation, salicylic acid, Solanum lycopersicum, Pst DC3000, basal immunity
Citation: Ma Q, Liu Y, Fang H, Wang P, Ahammed GJ, Zai W and Shi K (2020) An Essential Role of Mitochondrial α-Ketoglutarate Dehydrogenase E2 in the Basal Immune Response Against Bacterial Pathogens in Tomato. Front. Plant Sci. 11:579772. doi: 10.3389/fpls.2020.579772
Received: 03 July 2020; Accepted: 18 September 2020;
Published: 30 October 2020.
Edited by:
Víctor Flors, University of Jaume I, SpainReviewed by:
Purificación Lisón, Universitat Politècnica de València, SpainSang-Wook Han, Chung-Ang University, South Korea
Copyright © 2020 Ma, Liu, Fang, Wang, Ahammed, Zai and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Shi, a2Fpc2hpQHpqdS5lZHUuY24=
†These authors have contributed equally to this work
 Qiaomei Ma
Qiaomei Ma Yaru Liu1†
Yaru Liu1† Hanmo Fang
Hanmo Fang Ping Wang
Ping Wang Golam Jalal Ahammed
Golam Jalal Ahammed Kai Shi
Kai Shi