- 1Laboratory of Modern Biotechnology, School of Forestry and Landscape Architecture, Anhui Agricultural University, Hefei, China
- 2National Engineering Laboratory of Crop Stress Resistance Breeding, Anhui Agricultural University, Hefei, China
CCCH zinc finger proteins are a class of important zinc-finger transcription factors and have functions in various plant growth and stress responses, but their functions in moso bamboo (Phyllostachys edulis) are unclear. In this current study, we main investigated the structures, phylogenetic relationships, promoter elements and microsynteny of PeC3Hs. In this research, 119 CCCH zinc finger proteins (PeC3H1-119) identified genes in moso bamboo were divided into 13 subfamilies (A-M) based on phylogenetic analysis. Meanwhile, moso bamboo were treated with abscisic acid (ABA), methyl jasmonate (Me-JA) and gibberellic acid (GA) and 12 CCCH genes expression levels were assayed using qRT-PCR. In the three hormone treatments, 12 genes were up-regulated or down-regulated, respectively. In addition, PeC3H74 was localized on the cytomembrane, and it had self-activation activities. Phenotypic and physiological analysis showed that PeC3H74 (PeC3H74-OE) conferred drought tolerance of transgenic Arabidopsis, including H2O2 content, survival rate, electrolyte leakage as well as malondialdehyde content. Additionally, compared with wild-type plants, transgenic Arabidopsis thaliana seedling roots growth developed better under 10 μM ABA; Moreover, the stomatal of over-expressing PeC3H74 in Arabidopsis changed significantly under ABA treatment. The above results suggest that PeC3H74 was quickly screened by bioinformatics, and it may enhanced drought tolerance in plants through the ABA-dependent signaling pathway.
Introduction
Moso bamboo is one of the most important forest types in China. Moso bamboo plays an important role not only in the ecological environment but also in China’s rural economy (Zhang X. P. et al., 2019). Transcription factors (TFs) not only regulate plant growth and development but also regulate the biosynthesis of plant secondary metabolites (Zhang et al., 2009). Zinc finger transcription factors, as one of the largest transcription factor (TF) families in plants, play a role in many biological processes, such as morphogenesis (Stege et al., 2002), signal transduction and environmental stress responses (Takatsuji, 1998). Nowadays, many zinc finger proteins have been found in plants, such as CCCH-type (Yang et al., 2020), ERF (Nakano et al., 2006), WRKY (Cheng et al., 2020), and DOF (Lijavetzky et al., 2003).
CCCH zinc finger protein contains a typical C3H-type motif (three cysteines and one histidine), and form a class of proteins that mainly exist in diverse eukaryotic organisms (Blackshear, 2002). So far, CCCH zinc finger family genes have been identified in many organisms, such as humans (55) (Liang et al., 2008), mouse (58) (Liang et al., 2008), Arabidopsis thaliana (68) (Wang D. et al., 2008), rice (Oryza sativa) (67) (Wang D. et al., 2008), poplar (Populus trichocarpa) (91) (Chai et al., 2012), Brassica rapa (103) (Pi et al., 2018), and tomato (Solanum lycopersicum) (80) (Xu, 2014). In rice and A. thaliana, based on the specially ordered cysteines and histidine in the CCCH domain, the sequence of the motif was determined to be C-X4–15-C-X4–6-C-X3-H (X for any amino acid) and CCCH protein contains at least one, and at most six, CCCH zinc finger domains (Wang D. et al., 2008). In maize, the sequence for these motifs was defined as C-X4–17-C-X4–6-C-X3-H, meanwhile, ZmC3H17 contains 7 CCCH zinc finger domains (Peng et al., 2012). In many plants, C-X7/8-C-X5-C-X3-H is the most common domain (Pi et al., 2018).
CCCH zinc finger proteins are involved in developmental processes in plants, including seed germination (Kim et al., 2008), flowering and senescence (Yan et al., 2017), embryo development (Li and Thomas, 1998), and secondary wall synthesis (Zhang et al., 2018). For example, KHZ1 (AtC3H36) and KHZ2 (AtC3H52) participate in flowering and regulating leaf senescence in A. thaliana (Yan et al., 2017). In rice, OsTZF1 regulates leaf-delayed senescence by regulating stress-response genes (Jan et al., 2013). In poplar, PdC3H17 and PdC3H18 positively regulate secondary wall formation (Chai et al., 2014). CCCH genes not only contribute to plant developmental processes, they also regulate biotic and abiotic stress responses. In A. thaliana, AtC3H17, a non-tandem zinc finger gene, positively regulates salt tolerance through an abscisic acid (ABA)-dependent pathway (Seok et al., 2018). In addition, AtTZF4, 5 and 6 play important roles in seed germination by regulating ABA, light and gibberellic acid (GA) (Bogamuwa and Jang, 2013). IbC3H18, which is a nuclear transcriptional activator, regulates the expression of abiotic stress response genes associated with ABA signaling (Zhang H. et al., 2019).
CCCH proteins have been studied in A. thaliana, rice, poplar and maize (Zea mays L.) (Chen et al., 2017). A previous study showed that CCCH plays a role in growth and responses to various stresses (Yan et al., 2017). However, they have not been studied in moso bamboo. In the current study, we investigated the structures, phylogenetic relationships, promoter elements, scaffold locations and microsynteny of 119 identified PeC3Hs. Meanwhile, we investigated 12 genes responses to ABA, Me-JA and GA treatments. In addition, the subcellular localizations and transcriptional activities of PeC3H74 were analyzed. Then, this gene was over expressed in Arabidopsis. PeC3H74 promoted root growth under ABA condition at seedling stages in transgenic Arabidopsis. PeC3H74 increased the drought-stress tolerance and decreased the ABA sensitivity of transgenic plants. Thus, rapid screening of bioinformatics to PeC3H74, and it might participated in the positive regulation of drought-stress responses through an ABA-dependent pathway in plants.
Materials and Methods
CCCH Gene Identification
The GIGADB Dateset1 provided the basic data, including protein numbers, cDNA sequence numbers and open reading frame lengths, as well as the genomic sequences and their scaffold positions. The published rice CCCH proteins were originally used as probes to search for CCCH members in the moso bamboo genome with BLASTP and TBLASTN having an E-value cutoff set as 1e-005, and the sequences were detected again with SMART2 (Letunic et al., 2004) and Pfam3 (Finn et al., 2008) to ensure the existence of the CCCH domain. ExPASY provided information about molecular weights and isoelectric points (Gasteiger et al., 2003).
Phylogenetic Analysis and Gene Structure
The ClustalX (version 1.83) program (Chai et al., 2012) was used for the multiple sequence alignment of CCCH sequences of moso bamboo. The neighbor-joining method of MEGA 6.0 software4 was used to construct the phylogenetic trees. Bootstrapping of each branch was performed using 1,000 replications. MEGA 6.0 and ClustalX programs were used to construct the integrated phylogenetic tree of moso bamboo, maize and rice. The Gene Structure Display Server program (Han et al., 2014) was used to study the intron/exon structure of PeC3H genes.
The MEME5 program was used to determine the conserved motifs of the 119 amino acid sequences of the PeC3H genes with the following parameters: the maximum number of motifs was 15 and the optimum width was 18–200 residues (Ma et al., 2014). In addition, sequence logos of the CCCH zinc finger motifs were produced using online WebLogo software (Crooks et al., 2004).
Gene Duplication and Ka/Ks Analysis
The genomic location of PeC3H genes were identified according to the information provided by moso Bamboo SMRT database, and the collinearity of CCCH gene family of bamboo were analyzed by using the default parameters of MCScanX (Wang et al., 2012) and shown by Circos (Krzywinski et al., 2009). To study relationships between orthologous PeC3H genes and other selected species, the homolinear analysis maps were constructed using Dual Synteny Plotter software6. DnaSP 5 software was used to calculate the PeC3H’s Ka and Ks values. The duplication time (T) was calculated as T = Ks/2λ × 10–6 million years ago (MYA) (λ = 6.5 × 10–9 for grasses) (Gaut et al., 1996).
Cis-element Analysis
To study the cis-elements of PeC3H genes’ promoter sequences, the cis-elements upstream of the transcription initiation site (2,000 bp of genomic DNA sequence) were analyzed using the PlantCARE program (Liu et al., 2009).
Plant Materials and Stress Treatments
Bamboo seedlings gathered from Guilin City, Guangxi Province, China and were cultured in an artificial climate incubator for 7 weeks (26 ± 2°C, 16 h light) in preparation for subsequent experiments. Sow the moso bamboo seeds in a round pot (diameter 20.5 cm, height 19 cm). Afterward, the seedlings were sprayed independently with H2O, 100-μM ABA, 100-μM Me-JA, and 100-μM GA solutions (10 ml) at 0 h. Untreated seedlings served as the controls. Leaf samples were taken at different times (1, 3, 6, 12, and 24 h). Subsequently, total RNAs of these samples were extracted using the TRIzol method. RNA was reversed transcribed into cDNA using a PrimeScript® RT Reagent Kit (TaKaRa, Dalin, China).
Quantitative Real-Time PCR (qRT-PCR)
To study the expression levels of PeC3Hs under different hormone-treatment conditions, 12 genes were selected for qRT-PCR analysis. The specific primers for the 12 selected PeC3H genes were designed using Primer 5.0 software, and the intrinsic membrane protein 41 (TIP41) was used as the reference gene (Fan et al., 2013). The program settings for the qRT-PCR were as follows: 95°C for 30 s; 40 cycles of 95°C for 5 s, 55–60°C for 10 s, and 72°C for 10 s. The relative expression levels of relevant genes were calculated using the following formula: 2–ΔΔCT [ΔCT = CT, Target − CT,TIP41. ΔΔCT = ΔCT − ΔCT, H2O]. (Schmittgen and Livak, 2008). GraphPad5 was used to analyze the data.
Subcellular Localization and Transactivation Activity
The full-length coding sequence of PeC3H74 was obtained using specific primers (F: ATGAC TCCCTTGACTGGTTTCTTGACTGTT, R: GATAAGCAGA TTGGTAGCCTAGCA ACATAG) and cloned into pCAMBIA1305, containing CAMV35S and green fluorescent protein (GFP), using XbaI and Sam I restriction sites. PeC3H74-GFP was introduced into Agrobacterium tumefaciens EHA105, then injected into Nicotiana tabacum and observed using a confocal laser scanning microscope (Carl Zeiss LSM710, Germany) (Zhou et al., 2013).
The pGBKT7 vector (Clontech, Palo Alto, CA, United States) was used to study the transcriptional activity of the PeC3H74 protein in yeast. The full-length PeC3H74 was PCR amplified using gene-specific primers (F: GGAATTCATGACTCCCTTGACTGGTTT; R: ACGCGTCGACGATAAGCAGATTGGTAGCCT) and cloned into pGBKT7 vector containing a GAL4 DNA-binding domain. The empty plasmids (pGBKT7) and pGBKT7-53 + pGADT7-T antigen as negative and positive experimental controls. pGBKT7-PeC3H74, pGBKT7 and pGBKT7-53 + pGADT7-T were transformed in yeast strain by the lithium acetate method, respectively. The transformants were dropped on various SD selective media, namely SD/–Trp and SD/–Trp/–His/–Ade/X-α-GAL and incubated at 30°C for 3–5 days.
Phenotypic Analysis of Transgenic Plants
To determine drought tolerance, 3-week-old plants (OE-6, OE-7, OE-9, WT) were withholding water for a week. The electrolyte leakage was measured according to previous studies (McKersie et al., 1996). MDA and H2O2 content was measured using Biochemical Assay Kit (BC0025 and BC3595, Liandong U Valley A85, Tongzhou District, Beijing, China). According to the method provided by (Kumar et al., 2014), DAB staining of the leaves after drought treatment. PeC3H74-OE transgenic Arabidopsis seedlings were grown on 1/2 MS medium for 3 days, and then transferred to 1/2 MS medium containing 10 μM ABA for vertical cultivation for 7 days, and the change in root length was observed.
ABA Sensitivity Test
The mature Arabidopsis leaves were soaked in (0 and 1 μM) ABA for 6 h, and then transferred to a solution containing 25% propanetriol and 2 g/mL chloral hydrate for 4 days. After removing chlorophyll, observe and measure under a fluorescence microscope. The stomata ratios of width to length were >0.5 (open), 0.5–0.2 (partially closed) and <0.2 (closed), respectively.
Statistical Analysis
Statistical significance was determined using a paired Student’ s t-test9. The mean ± standard error from the mean (SE) of at least three replicates are presented, and significant differences relative to controls are indicated at ∗p < 0.05 and ∗∗p < 0.01.
Results
Identification of the PeC3H Genes
Through a BLASTP preliminary screening, 123 putative CCCH-encoding genes of moso bamboo were obtained. The present of the CCCH domain was determined by Pfam and SMART, and 119 potential CCCH sequences were ultimately identified as CCCH genes (PeC3H1-119) (Liu et al., 2011). Detailed information of PeC3H genes are given in Table 1. The identified PeC3H genes encoded proteins ranging from 73 to 2,039 amino acids, with a mean length of 533 amino acids. The greatest and lowest molecular weights were 224.86 and 8.12 kDa, respectively.
We compared the sequences of 150 C-X8-C-X5-C-X3-H and 62 C-X7-C-X5-C-X3-H motifs separately, and then generated the sequence logos (Figure 1). To study the characteristics of the CCCH motifs of the PeC3H genes, the sequences were compared with CCCH motifs of rice and A. thaliana. In addition, the compound sequence logos of the two motifs were generated in the same way, and the motif sequences were highly conserved. Four of the amino acids in the three patterns were completely conserved, similar to the sequence diagrams provided by Pfam and SMART. There were also some differences in the sequence logos among the three plants. For example, lysine occurred more often at C1 + 6 than arginine in both moso bamboo and rice B motif logos, while the reverse case was observed in the A. thaliana B motif logo.
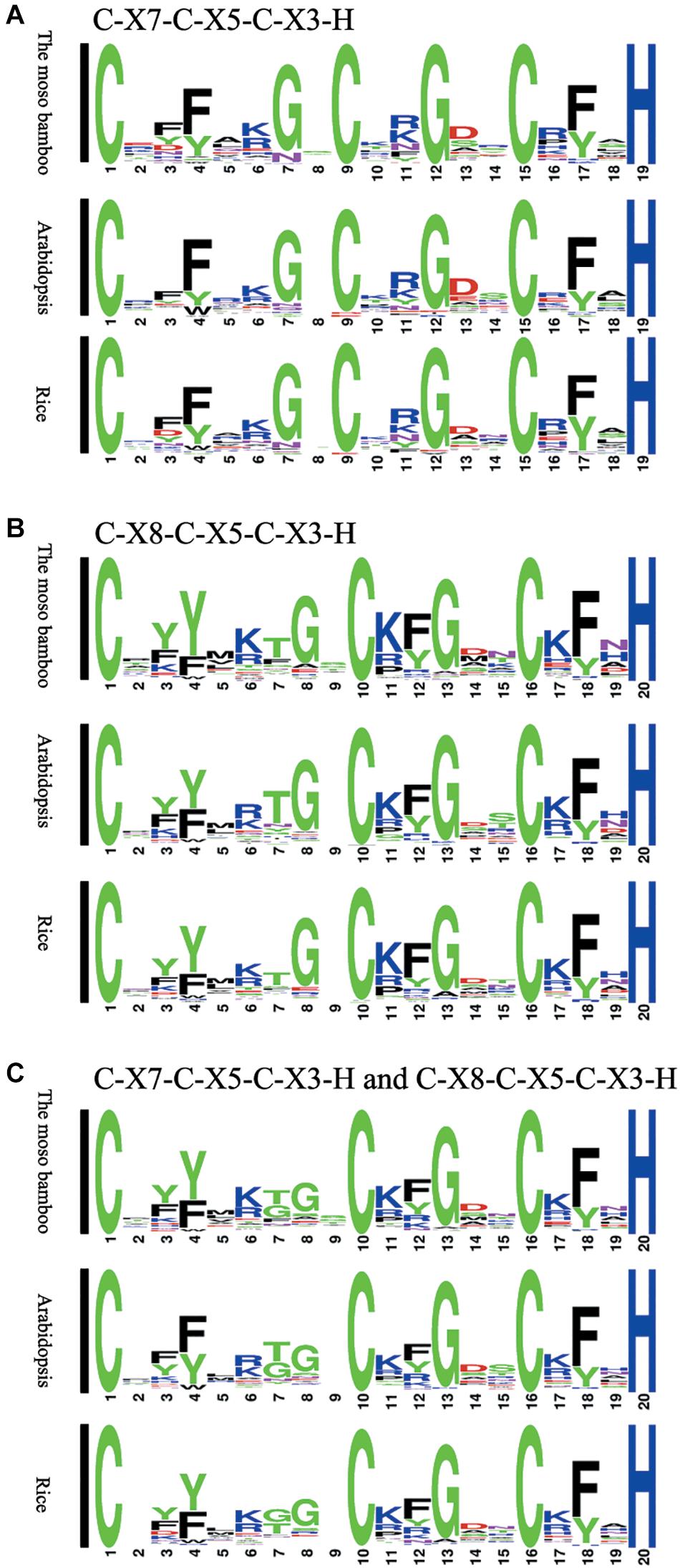
Figure 1. Sequence logos for common CCCH zinc finger motifs. (A) C-X7-C-X5-C-X3-H motifs of moso bamboo, rice and Arabidopsis thaliana. (B) C-X8-C-X5-C-X3-H motifs of moso bamboo, rice and A. thaliana. (C) C-X7-C-X5-C-X3-H and C-X8-C-X5-C-X3-H motifs of moso bamboo, rice and A. thaliana. The conservativeness of amino acids in different positions is indicated by the size of the characters.
Phylogenetic Relationships and Gene Architecture
To evaluate the phylogenetic relationships among the 119 predicted moso bamboo CCCH zinc finger proteins, we used the 119 putative protein sequences to construct an unrooted phylogenetic tree with 1,000 bootstrap replicates (Wang D. et al., 2008) (Supplementary Figure 1). We divided them into 13 subfamilies (CCCH A to M) based on bootstrap values above 100. However, the Pe3H24 gene was not included in any of the 13 subfamilies because its bootstrap values with other genes were less than 100, which is a phenomenon that also occurs among the CCCH genes of other plants (Chai et al., 2012). Among the 13 subfamilies, subfamily L contained the greatest number of CCCH (29), followed by subfamilies C (25), F (15), A (12), and I (9). Both G and K subfamilies had six members, H and J had four members, and the B, D, E, and M had only two members each.
The MEME server identified 15 conserved motifs in the CCCH protein of moso bamboo (Supplementary Figure 2), and details of conserved amino acids are provided in Supplementary Table 1. Motif 1, 4, and 15 are typical CCCH motifs, and each PeC3H gene contains at least one CCCH motif (motif 1, 4, and 15). PeC3H11, −15, −28, −33, −51, and −81 in I subfamily all contain SWIB motif (motif 13), and the SWI/SNF protein is required for proper protein complex formation in yeast. However, there are also some genetic differences within the same subfamily. In subfamily A, only PeC3H110 contained motif 6, meanwhile, motif 5 only appears in PeC3H4 and -8.
By analyzing the Gene Structure Display Server diagram (Supplementary Figure 3), we found that there were differences in the numbers of introns in the different genes, ranging from 0 to 13, with an average of 4. In total, 15 PeC3H genes had no introns, while 37 genes contained only one or two introns. Some subfamilies had similar numbers of introns in each member, but some subfamilies had significant differences. For example, PeC3H9, −28, and −111 contain 1, 7 and 11 introns, respectively, and all they are all members of the I subfamily.
Synteny Analysis of Moso Bamboo CCCH Genes
The strongly conserved microsynteny of the CCCH gene was observed by comparing CCCH genes with other genes in moso bamboo (Figure 2), the synteny pairs details were shown in Supplementary Table 2. Pe_Scaffold1 did not contain any genes and is, therefore, not shown in Figure 2. In total, 63 collinear gene pairs were identified. We found that of the 119 CCCH genes, a pair of genes (PeC3H88/-89) in scaffold 6 were at a distance of less than 100 kb, which may be caused by a tandem duplication (Indicated by the red line) (Figure 2) (Lin et al., 2014). Both PeC3H88 and PeC3H89 belonged to the C subfamily, and their bootstrap values reach 1,000, indicating a highly conserved. The analysis of the collinear gene pairs of CCCH genes showed that 62 gene pairs remained in the conserved positions in the segmental duplicated blocks, indicating that gene duplication played an important role in the CCCH gene expansion of moso bamboo.
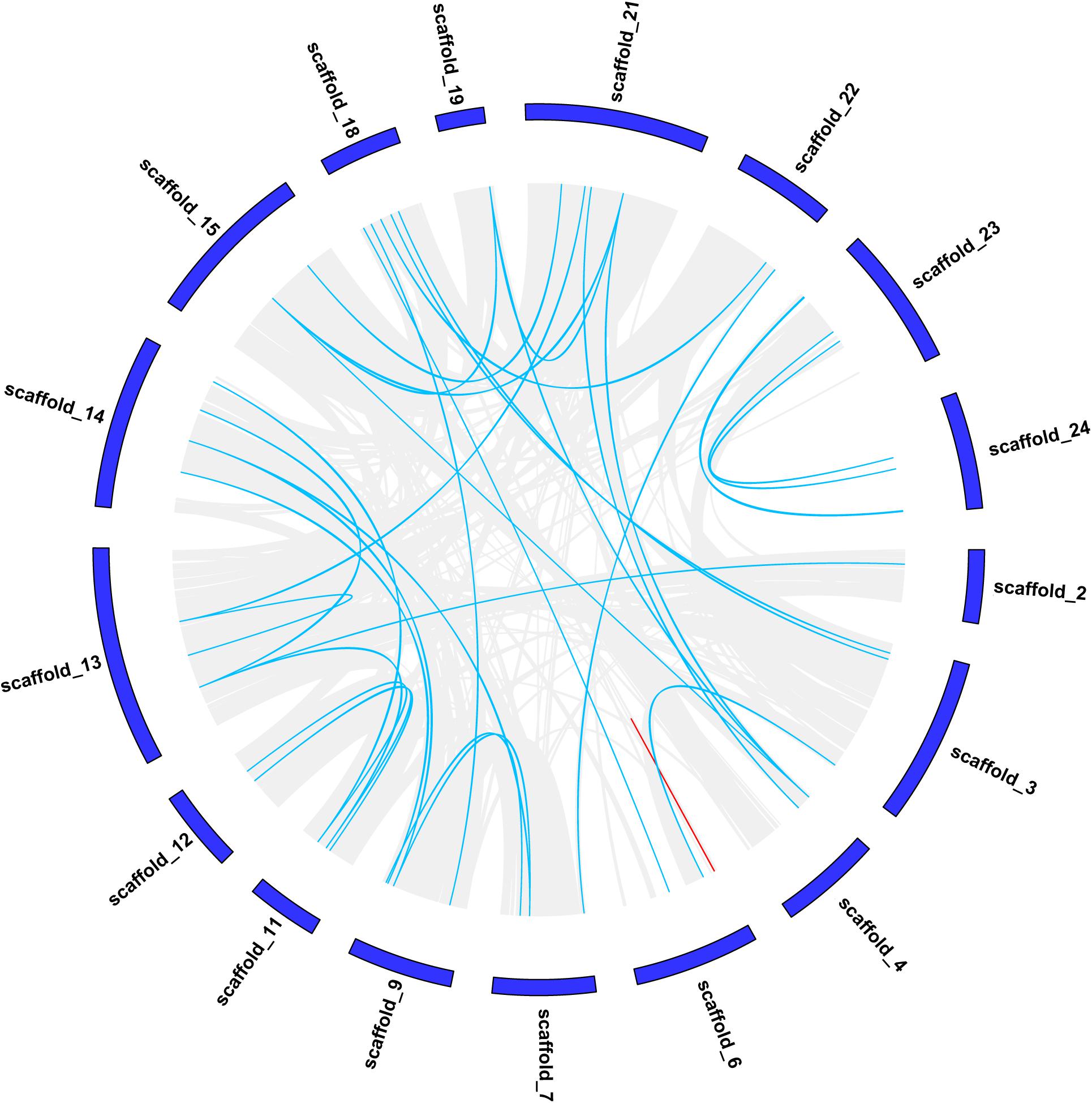
Figure 2. Chromosomal localizations and duplications of CCCH genes on moso bamboo chromosomes. The duplicated CCCH genes are indicated with blue lines and tandemly duplicated genes are marked with red.
The gene duplication mechanisms of CCCH gene family in moso bamboo was studied by constructing a comparative syntenic maps with four representative species [one dicots (Arabidopsis) (Supplementary Figure 4A) and three monocots (Brachypodium distachyon, rice and maize) (Supplementary Figure 4B)]. The syntenic relationship between CCCH genes of bamboo and four species, the most is maize (118), followed by rice (107), B. distachyon (93) and Arabidopsis (2), indicating that in comparison with monocotyledonous plants, CCCH genes of moso bamboo show a high evolution divergence with dicotyledonous plants. PeC3H4 and PeC3H8 have syntenic pairs in all four plants, and these two genes may play a key role in the evolution of CCCH genes family. Through to Ka/Ks ratios calculations of the CCCH genes synteny pairs (Supplementary Tables 2–6), Ka/Ks < 1 of most of the synteny pairs, show that the moso bamboo CCCH genes family during evolution may experience a strong purifying selection pressure, and the PeC3H genes experienced a large-scale duplication event, probably 5.13–271.79 million years ago (MYA) by the formula T = Ks/2λ (λ = 6.5 × 10–9) (Gaut et al., 1996).
Cis-element Analysis
The promoter region of a gene usually contains multiple cis-elements that play key roles in responses to different stresses (Dung Tien et al., 2012). Cis-elements directly influence gene regulation involved in stress-responsive gene expression (Bilas et al., 2016). Various interactions between cis-acting elements and transcription factors function as molecular switches for transcription to determine transcription initiation events (Bilas et al., 2016). Therefore, identifying cis-acting elements in promoter region is very important to understand the role of transcription factors in stress response. The cis-acting elements in promoter regions of 119 PeC3H genes were detected to prepare study their regulatory mechanism. We focused on three types of cis-elements (ABA, Me-JA, and GA) (Figure 3 and Supplementary Table 7), and there were a large number of cis-elements related to these three hormones among the PeC3Hs. ABA-responsive elements (ABREs), the cis-acting elements of ABA, existed in many PeC3H genes. Therefore, we speculated that most PeC3H genes were regulated by ABA stress responses. In total, 85.7% (102/119) of PeC3H gene promoter regions contained an ABA-responsive element (Figure 3 and Supplementary Table 7). Meanwhile, we found 440 ABRE elements in PeC3Hs, and the largest number of three elements. The CGTCA/TGACG-elements are cis-acting elements of the Me-JA response and have regulatory effects on plant leaf senescence (He et al., 2002). The CGTCA/TGACG-elements were found in promoter regions of 101 (84.8%) PeC3Hs (Supplementary Table 7), and 300 CGTCA/TGACG-elements in PeC3Hs were discovered. The GARE/P-box/TATC-box-element associated with GA was present in 62 of 119 (52.1%) promoter regions of PeC3Hs, and 104 GARE/P-box/TATC-box-element in PeC3Hs were discovered. In addition, none of the three cis-acting elements was found in PeC3H1. The analysis of cis-acting elements in the PeC3Hs will aid in further studies of the tolerance of moso bamboo.
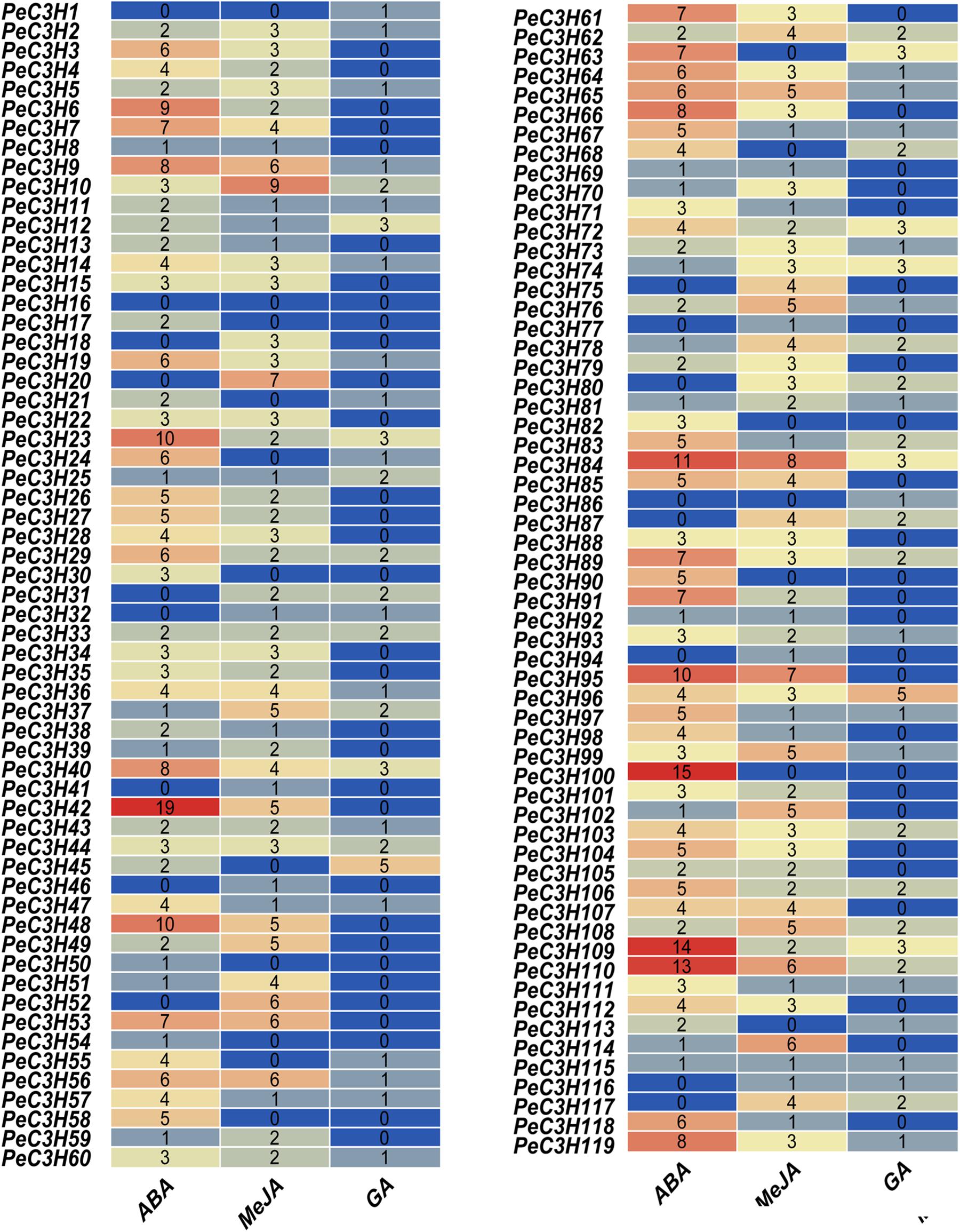
Figure 3. The position of moso bamboo CCCH genes abscisic acid (ABA), Me-JA, and GA in promoter. ABA was shown in red. Me-JA was shown in yellow. GA was shown in blue.
Expression Analysis of PeC3H Genes by qRT-PCR
There are some CCCH genes were positively or negatively regulated by ABA, GA and Me-JA in rice (Huang et al., 2012). Twelve genes (PeC3H2, −7,−11, −20, −21, −26, −56, −34, −74, −99, −100, and −110) were selected to study the expression level using qRT-PCR. The specific primers used in qRT-PCR analysis of these genes are shown in Supplementary Table 8. At the same time, the expression levels of these genes under water treatment were analyzed (Figure 4). Under water treatment, although the gene expression level changes, it is basically low expression.
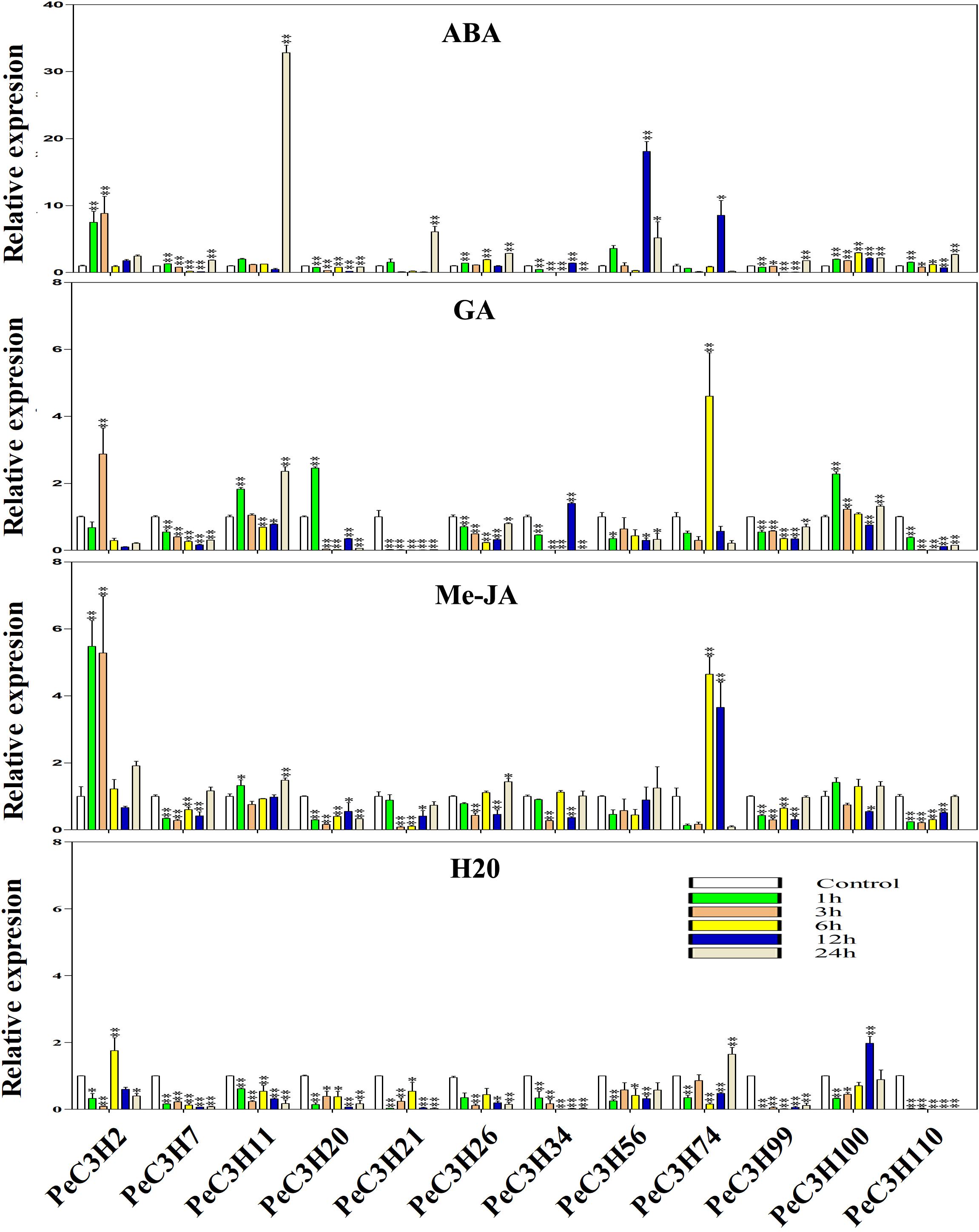
Figure 4. Expression levels of PeC3Hs under ABA, Me-JA, GA and H2O treatment by qRT-PCR. The Y-axis and X-axis indicate the relative expression levels and the time courses of plant hormone treatments, respectively. The experiment was performed using three biological and technical replicates each, and asterisks indicate significant difference compared to the transcription level of control groups, as determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01). Bars indicate standard error of the mean (SE).
The ABA treatment of moso bamboo, 11 genes reached their maximum expression levels at different times, except PeC3H20 was inhibited compared with the control (Figure 4). In particular, gene PeC3H11 had an expression level that was 32-fold greater than that of the control at 24 h (Figure 4).
After the GA treatment, the expression profiles of five genes (PeC3H7, −21, −26, −56, and −99) were suppressed, while those of other PeC3H genes were up-regulated, but the expression levels were lower than those of the control at some time. For example, PeC3H74 reached its peak at 6 h and was greater than the control; however, its expression levels at other times were lower than the control (Figure 4). After the GA treatment, the expression levels of PeC3H genes were not significantly different from those of the control. For example, the expression levels of PeC3H2 and PeC3H56 were initially up-regulated, reaching their maximum levels, which were no more than fivefold those of the control. Furthermore, the expression levels of five genes (PeC3H7, −11, −20, −100, and −110) were greatest at 1 h.
Next, the expression levels of CCCH genes after Me-JA treatment was analyzed, and 4 genes (PeC3H20, −21, −99, and −110) were suppressed, while those of other PeC3H genes were up-regulated (Figure 4). At 24 h, the expression levels of four genes (PeC3H7, −11, −26, and −56) were the greatest. In addition, PeC3H2 and PeC3H100 reached the highest expression levels at 1 h. PeC3H34 and PeC3H74 reached their highest expression levels at 6 h.
Under ABA treatment, only the expression of PeC3H20 was suppressed, but under the other two hormone treatments, at least 4 genes were suppressed. Only PeC3H74 gene under the treatment of three hormones, the highest expression level exceeds fivefold, suggesting that this gene plays a role in resisting stress during plant growth and development. Thus, most PeC3H genes were up-regulated under stress treatments, indicating that they play key roles in abiotic and biotic stress responses.
Subcellular Localization of PeC3H74 and Transcriptional Activity
To study the subcellular localization of PeC3Hs, a PeC3H-GFP vector was constructed and transiently expressed in N. tabacum leaves. 35S:GFP served as a control (Figure 5A). PeC3H74 gene expression had higher induction under ABA, GA, and Me-JA treatment, so it was further analyzed. Based on the GFP signal, PeC3H74 was localized to the cytomembrane.
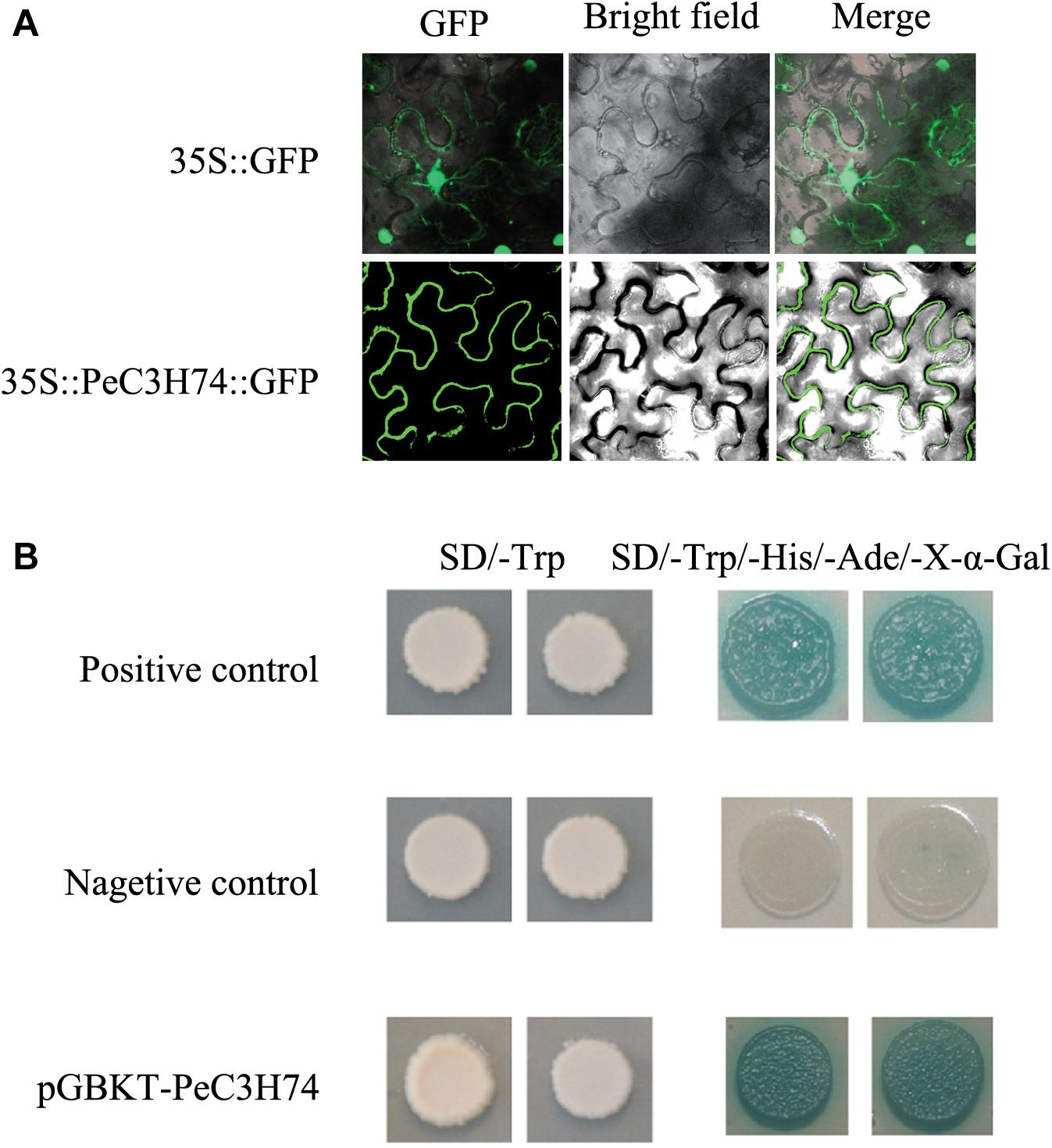
Figure 5. Subcellular localization of PeC3H74 and transactivational analyses of PeC3H proteins in yeast Y2HGold strain. (A) The PeC3H74-GFP fusion proteins and GFP as a control were transiently expressed in N. tabacum leaves and observed under a fluorescence microscope. (B) The positive constructs, negative constructs, and fusion constructs were transformed into yeast Y2HGold strain and successively incubated in SD/–Trp media and SD–His/–Ade/–Trp plate supplemented with X-α-GAL.
The Y2H yeast strain was used to study the transcriptional activities of PeC3Hs. The pGBKT7-PeC3H74, positive control plasmids pGBKT7-53 and pGADT7-T, and pGBKT7 (the negative control plasmid) transformed into the Y2H yeast strain, independently. The transformants were cultured on SD/–Trp medium, and they all produced white colonies (Figure 5B). On the SD–Ade/–His/–Trp/X-α-GAL medium, strains containing PeC3H74 and positive control turned blue, while the negative control did not grow (Figure 5B). These results suggest that PeC3H74 can function as a transcriptional activator.
Overexpression of PeC3H74 Enhanced Drought Tolerance in Arabidopsis
Because the transgenic technology of moso bamboo is still immature, we transferred the PeC3H74 gene into A. thaliana and studied whether PeC3H74-OE was related to drought stress through transgenic Arabidopsis strains (OE-6, OE-7, and OE-9). Leaves of transgenic Arabidopsis strains grown for 2 weeks drive GUS activity (Supplementary Figure 5). After 6 days of cultivation on 1/2 MS medium, Arabidopsis seedlings were placed on 1/2 MS medium containing different concentrations of ABA (0 and 10 μM). The root lengths of wild type and PeC3H74-OE on 0 μM ABA were not significantly different, while the root length of PeC3H74-OE on 10 μM ABA was significantly different from that of wild type (Figures 6A,B).
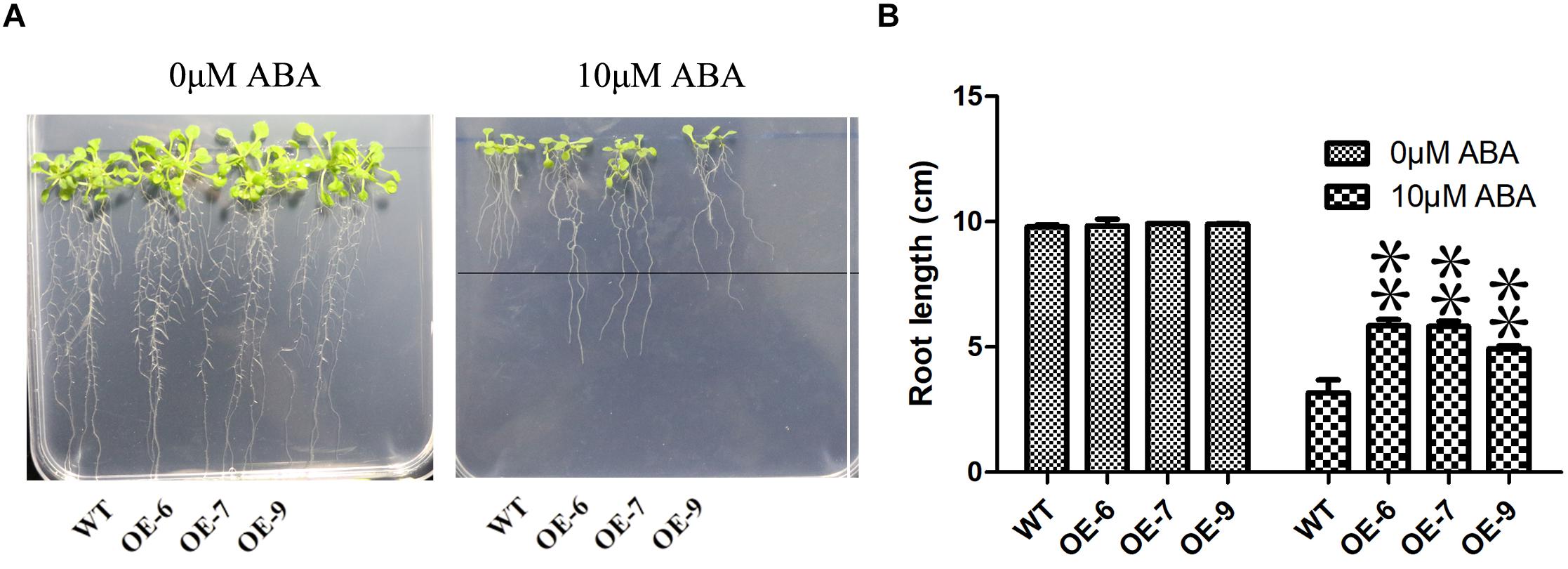
Figure 6. The PeC3H74 gene improves resistance to ABA in Arabidopsis thaliana. (A) Root growth of WT and transgenic plants exposed to ABA. The seedlings were grown vertically in 1/2 MS for 4 days and then transferred to the 1/2 MS containing different concentrations of ABA. After 7 days, the representative images were taken and the root length in each line was measured. (B) Measurements of root lengths in ABA condition. The experiment was performed using three biological and technical replicates each, and asterisks indicate significant difference compared to the transcription level of control groups, as determined by Student’ s t-test (∗p < 0.05, ∗∗p < 0.01). Bars indicate standard error of the mean (SE).
Subsequently, we examined the drought tolerance of PeC3H74-OE strain. 3 weeks of WT and PeC3H74-OE plants were not irrigated for 7 days, WT withered more than PeC3H74-OE plants (Figure 7A). After re-watering for 3 days, all PeC3H74-OE plants were survived, the survival rate of the WT was only 16.7 percent. In addition, before drought treatment, the electrolyte leakage (EL) and malondialdehyde (MDA) of WT and PeC3H74-OE plants are not much different (Figures 7B,C). After 7 days of drought, the content of EL and MDA of WT Increased, significantly different from PeC3H74-OE plants. The results showed that after drought treatment, PeC3H74-OE plants suffered less membrane damage than WT. Drought stress can lead to accumulation of reactive oxygen species (ROS). Before drought treatment, DAB staining showed that H202 accumulated less in WT and PeC3H74-OE plants (Figure 7D). After 10 days of drought treatment, the accumulation of H2O2 in PeC3H74-OE plants was significantly less than that of WT. The detection of H2O2 content before and after treatment showed that after treatment, the difference in H2O2 between WT and PeC3H74-OE plants was significant, which was consistent with DAB staining results (Figure 7E). These results indicate that PeC3H74 enhances the drought tolerance of Arabidopsis.
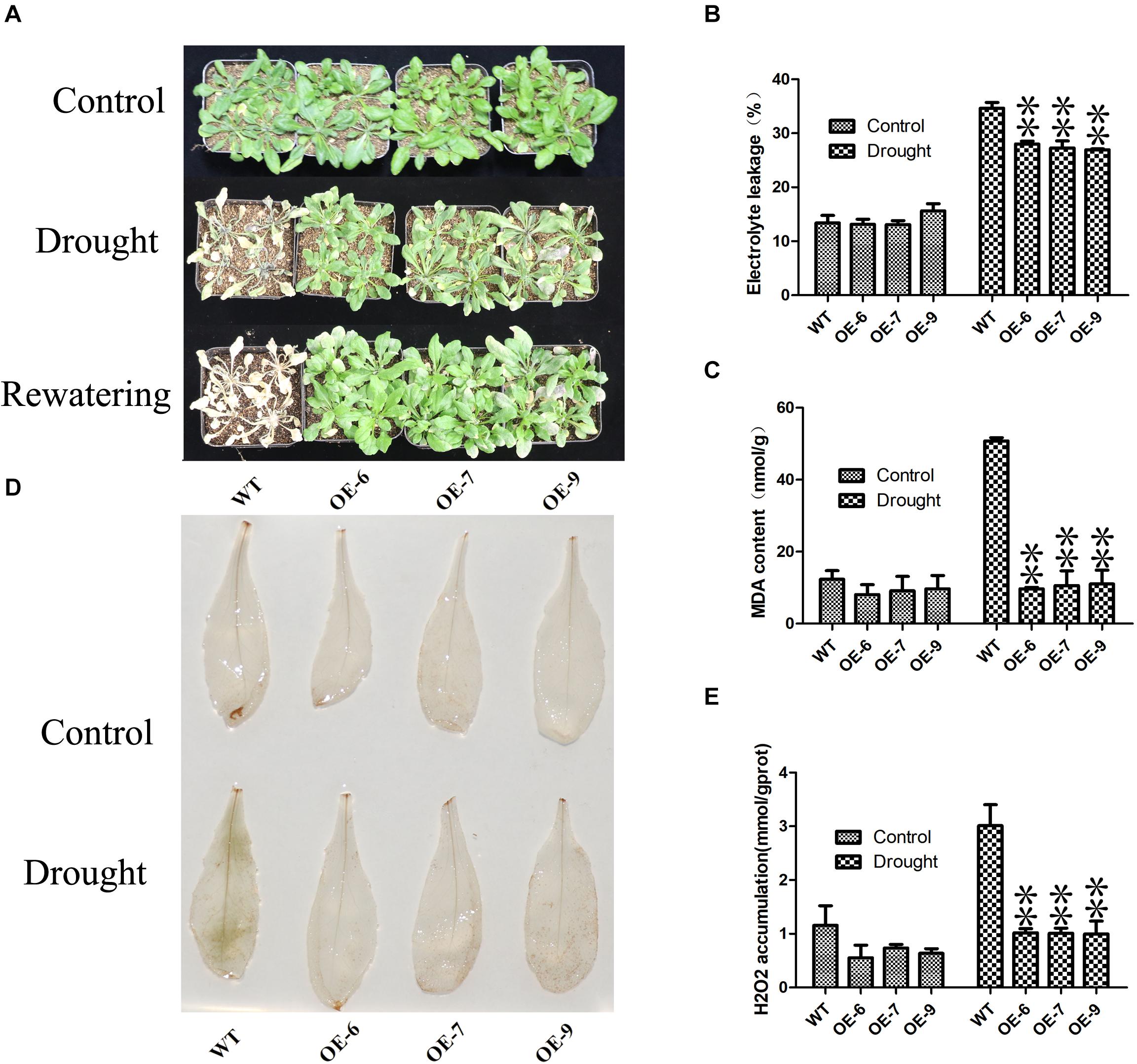
Figure 7. The PeC3H74 gene improves drought resistance in A. thaliana. 3 weeks WT and transgenic plants were withheld water for 7 days to induce dehydration. After dehydration for 7 days, the representative images were taken (A), Electrolyte leakage (B), MDA content (C), and H2O2 accumulation (D,E) were determined. The experiment was performed using three biological and technical replicates each, and asterisks indicate significant difference compared to the transcription level of control groups, as determined by Student’ s t-test (∗p < 0.05, ∗∗p < 0.01). Bars indicate standard error of the mean (SE).
ABA Sensitivity Analysis
The opening and closing of stomatal are affected by ABA, and the loss of water on the leaf surface is closely related to the regulation of stomatal (Osakabe et al., 2014). In order to determine the regulating effect of PeC3H74-OE on plant stomatal size. Observe the changes in stomatal size of WT and overexpressing strains without treatment or 1 μM ABA treatment, respectively. Observation by fluorescence microscopy revealed that the stomatal size of WT and overexpressing strains were not significantly different under untreated conditions. After 6 h of 1 μM ABA treatment, the rate of stomatal closure and partial closure of PeC3H74-OE plants was significantly higher than that of WT (Figures 8A–C). The results showed that PeC3H74 may induce stomatal closure through ABA to achieve the purpose of drought resistance.
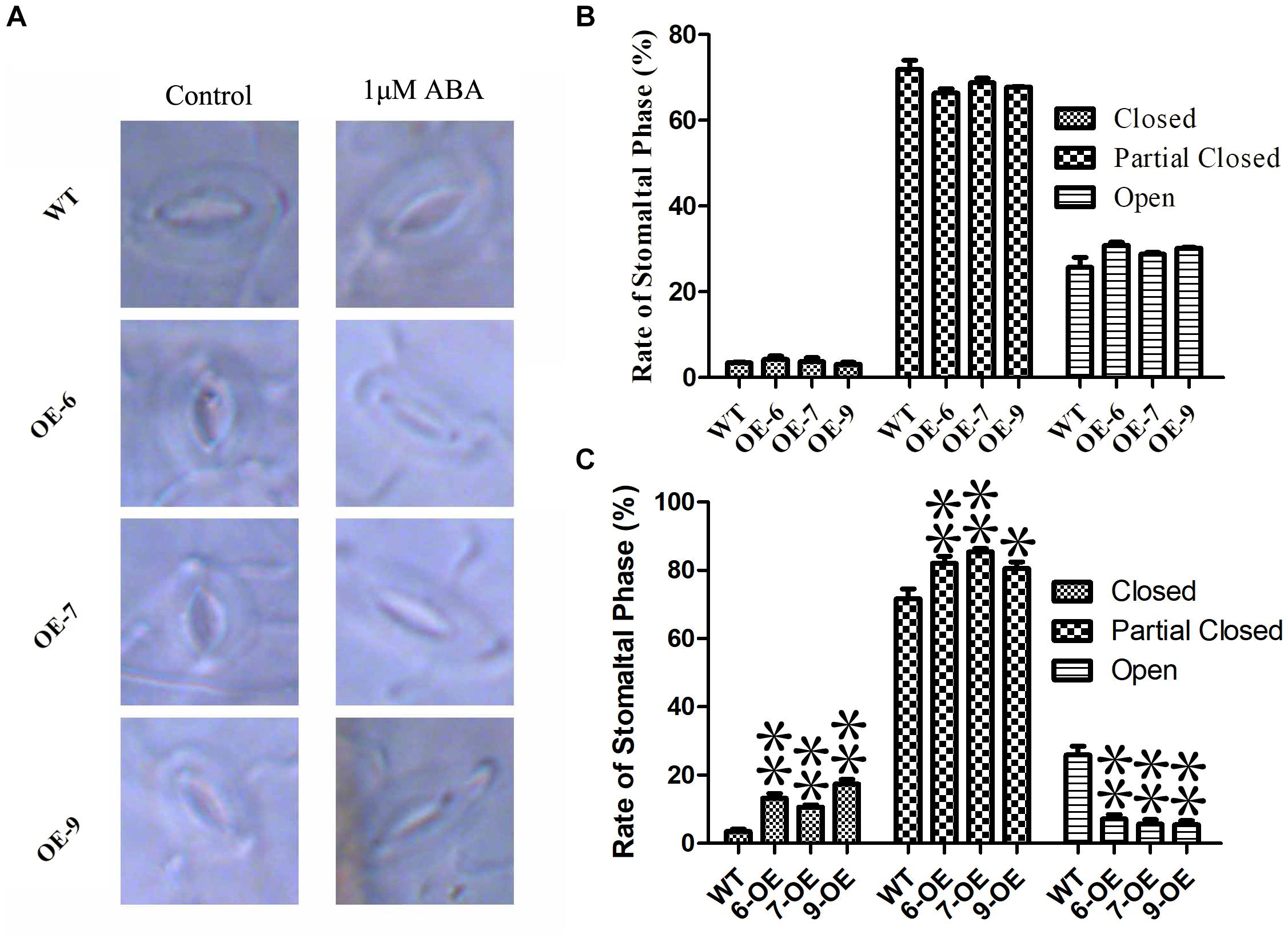
Figure 8. Abscisic acid sensitive and stomatal regulation of PeC3H74 overexpression in Arabidopsis. (A) Typical phenotype of stomatal opening phase in transgenic lines and WT with or without ABA treatment. Percentages of the three types of stomata in transgenic lines and WT plants are calculated in stomatal-induced liquid with (B) or without (C) ABA treatment. The experiment was performed using three biological and technical replicates each, and asterisks indicate significant difference compared to the transcription level of control groups, as determined by Student’ s t-test (∗p < 0.05, ∗∗p < 0.01). Bars indicate standard error of the mean (SE).
Discussion
CCCH zinc finger proteins have been systematically analyzed in the model plants A. thaliana and rice (Wang D. et al., 2008). However, moso bamboo had not been studied. Here, 119 CCCH zinc finger protein were identified, which was a greater number than in A. thaliana and rice, and divided them into 13 subfamilies (A–M). Bioinformatics analyses found that the CCCH of moso bamboo had some characteristics that were similar to those of other species and some novel characteristics.
The sequence of the CCCH motif was highly conserved in moso bamboo. However, the PeC3H genes had different numbers of CCCH motifs, and the adjacent sequences and cysteine and histidine sequences of the motif members were also different. Previously, the CCCH family was defined as a group of zinc finger proteins having a C-X6–14-C-X4–5-C-X3-H motif, and CCCH proteins contain 1–6 copies of the CCCH zinc finger motif (Berg and Shi, 1996). However, there were different CCCH gene sequences in some plants. In A. thaliana and rice, the CCCH motifs were C-X4–15-C-X4–6-C-X3-H (Wang D. et al., 2008). In maize, ZmC3H17, which is a novel motif, was defined as C-X17-C-X6-C-X3-H, and we found a highly conserved CCCH zinc finger motif with 7 copies (Peng et al., 2012). In moso bamboo, PeC3H13 and PeC3H52 were defined as C-X17-C-X6-C-X3-H. In this study, moso bamboo, as in the previous result, has no more than six conserved CCCH motifs. In moso bamboo, 73.5% of all identified CCCH motifs are C-X7–8-C-X5-C-X3-H, and there were less of this particular motif than in A. thaliana (82.2%), rice (78.6%), maize (79.4%), and poplar (82.0%) CCCH genes families. In total, 287 CCCH zinc finger motifs were identified, which was more than in A. thaliana (152) and rice (150) (Wang D. et al., 2008), and the results were similar to the number of CCCH proteins.
Genes on corresponding scaffolds (syntenic genes) and corresponding sequences (collinear genes) were preserved to a certain extent in eukaryotic genomes during evolution. Synteny mainly refers to the similarity of arrangements in different genes. Microsynteny has been studied in both monocotyledonous and dicotyledonous plants (Deleu et al., 2007). There were 63 genes in moso bamboo that had no microsyntenic relationships with other genes, indicating that either they were ancient genes without detectable linkage to other CCCH genes or that they were formed through complete transposition and loss of their primogenitors (Wang Y. et al., 2015). Gene duplication is an important mechanism of biological evolution and plays an important role in allowing organism to cope with adverse environments (Bowers et al., 2003). Segmental duplication, tandem duplication and retro-positioning are the main mechanisms of gene duplication (Kong et al., 2007). Here, we found that most CCCH genes were distributed in duplicated blocks, indicating that segmental duplication was involved in CCCH gene amplification in moso bamboo.
Studying cis-elements in the upstream region of genes can be helpful (Lin et al., 2011a) to further understand and predict their transcriptional regulation (Ibraheem et al., 2010). ABA was produced in the vegetative tissue in the absence of water; therefore, under drought conditions, it promotes the expression of related genes (Yamaguchi-Shinozaki and Shinozaki, 2005). In rice, OsC3H47 alters the drought resistance of rice by regulating ABA sensibilities (Wang W. et al., 2015). AtTZF1 regulates ABA-mediated growth and stress responses by affecting gene expression (Lin et al., 2011b). ABREs play important roles in ABA-dependent gene expression (Yoshida et al., 2014). A promoter region analysis showed that several PeC3Hs possess ABRE cis-elements (Figure 3 and Supplementary Table 7) (Yamauchi et al., 2007). For the 12 genes, except for the lower expression level of PeC3H20 than the control, the maximum expression levels of other genes is higher than the control, so ABA sensitivity may be a common phenomenon of PeC3H genes.
GAs are tetracyclic diterpenes, which play roles in the growth and developmental stages of many plants, especially during the germination of seeds (Yamauchi et al., 2007). Ga1-3 and Ga2-1 are GA-deficient mutants with obvious seed germination defects (Yamauchi et al., 2007). In A. thaliana, the loss-of-function of SOMNUS (AtTZF4) results in elevated GA levels (Kim et al., 2008). AtTZF1 regulates the GA-dependent growth of plants by affecting gene expression, which is a negative regulatory GA response (Lin et al., 2011a). Interestingly, the expression levels of five genes (PeC3H7, −21, −26, −56, and −99) were inhibited after GA treatments, and they may also have negative regulatory effects on GA levels (Figure 4). There may be both positive and negative regulation of PeC3H genes under GA treatment.
Me-JA is involved in plant immunity, and leaf senescence is regulated by JA (He et al., 2002). Transcriptome sequencing showed that the JA pathway is significantly active in age-dependent, dark-induced and starvation-induced leaf senescence (He et al., 2002). In rice, OsDOS can delay leaf senescence through the JA pathway (Kong et al., 2007). The expression of GhTZF1 was significantly up-regulated after Me-JA treatments (Zhou et al., 2014). In the promoter analysis, we found that almost all CCCH genes contained a CGTCA or TGACG motif, which are Me-JA-responsive elements. qRT-PCR revealed that some CCCH genes in moso bamboo (PeC3H2, −7, −11, −26, −34, −74, and −100) had significantly increased expression levels at different times during the Me-JA treatment, indicating that they were positively regulated. PeC3H genes were positively regulated under Me-JA treatment, which may be a common phenomenon.
However, the expression levels of some genes treated with ABA, Me-JA, and GA were not as predicted (Figure 4). For example, the promoter regions of PeC3H100 did not contain Me-JA-related cis-elements, but their expression levels increased during the Me-JA treatment. Thus, gene expression is a complex process that requires further study.
Some CCCH proteins localize in the nucleus, such as OsDOS, PEI1, AtSZF1, and SOMNUS, while some localize in the cytomembrane, such as HUA1, AtC3H49/AtTZF3, and AtC3H20/AtTZF2 (Lee et al., 2012). PeC3H74 localized to the cytomembrane. In rice, OsLIC, a Novel CCCH-Type Zinc Finger Protein, displays transcriptional activation activity in yeast (Wang L. et al., 2008). Meanwhile, AtC3H17, in A. thaliana, showed transcriptional activation activity in yeast (Seok et al., 2018). In our research, the transactivation activity experiments with PeC3H74 in a yeast system revealed that it was a transcriptional gene in yeast.
In plants, the CCCH gene plays an important (Ibraheem et al., 2010) role in all stages of growth and development, for example, seed germination (Kim et al., 2008), embryonic development (Li and Thomas, 1998) and secondary wall synthesis (Zhang et al., 2018), etc. However, there were few reports on the research of moso bamboo on abiotic stress. After we treated the transgenic A. thaliana for 3 weeks under drought conditions, PeC3H74-OE plants achieved a higher survival rate, as well as lower EL, MDA and H2O2 contents (Figures 7A–E). In addition, the roots of PeCEH74-OE Arabidopsis seedlings grew better on 10 μM ABA 1/2 MS medium (Figures 6A,B). ABA was a key factor in stomatal regulation. OsC3H47, an ABA-induced CCCH tandem zinc finger protein, regulates drought stresses by promoting ABA sensitivity in rice (Wang W. et al., 2015). Our research shows that under ABA treatment, transgenic A. thaliana contains more closure and partial closure stomatal than WT (Figures 8A–C). The above results show that the PeC3H74 gene was quickly screened by bioinformatics. In addition, it may play a drought resistance function in plants through ABA-dependent pathways.
Conclusion
In summary, the characteristics of CCCH gene families had been reported in some plants, such as A. thaliana, rice, tomato (Solanum lycopersicum) and poplar. However, no CCCH gene family studies had been reported in moso bamboo. Here, we identified 119 CCCH genes, and their phylogenetics, WebLogos, conserved motifs, divergence times, genetic structures and cis-acting components were analyzed. In addition, the subcellular localization and transcriptional activity of PeC3H74 in moso bamboo were studied. The PeC3H74 gene was quickly screened through bioinformatics. In addition, analysis of the phenotype and physiological and biochemical indicators of transgenic plants showed that PeC3H74 gene may rely on the ABA pathway to play a positive role in regulating plant drought stress.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FC carried out most of the experiments and bioinformatics analysis, and completed the main part of the manuscript. H-LL and KW guided some experiments. MW advised on the manuscript. Y-MG provided assistance with software usage. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 31670672) and the 2020 Graduate Innovation Fund of Anhui Agricultural University (Grant No. 2020ysj-16).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.579255/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of CCCH in moso bamboo.
Supplementary Figure 2 | Schematic representation of the15 conserved motifs in PeC3Hs. Conserved motifs of the PeC3Hs were identified using the online MEME program based on 119 full-length amino acid sequences with the following parameters: maximum number of motifs, 10; maximum width, 100. The lengths and positions of different motifs in the protein sequences are identified by the lengths and positions of the different color blocks.
Supplementary Figure 3 | Gene structures of CCCH in moso bamboo. Gene structures were performed using the Gene Structure Display Server online tool. Exons, introns, and untranslated regions (UTRs) are indicated by yellow rectangles, gray lines, and blue rectangles, respectively.
Supplementary Figure 4 | Synteny analysis of CCCH genes between moso bamboo and (A) dicotyledonous plant Arabidopsis thaliana, (B) maize, rice and Brachypodium distachyon. Gray lines in the background indicate the collinear blocks within moso bamboo and other plant genomes, while the red lines highlight the syntenic CCCH gene pairs. Orange or green bars represent the chromosomes. The chromosome number is labeled at the top or bottom of each chromosome.
Supplementary Figure 5 | Analysis of β-glucuronidase (GUS) activity driven of PeC3H74 in overexpression Arabidopsis leaves.
Supplementary Table 1 | Detailed information on conserved amino acid sequences and motif lengths.
Supplementary Table 2 | Ka/Ks value for duplicate CCCH genes in moso bamboo.
Supplementary Table 3 | Ka/Ks value for duplicate CCCH genes between moso bamboo and Arabidopsis.
Supplementary Table 4 | Ka/Ks value for duplicate CCCH genes between moso bamboo and Brachypodium distachyon.
Supplementary Table 5 | Ka/Ks value for duplicate CCCH genes between moso bamboo and rice.
Supplementary Table 6 | Ka/Ks value for duplicate CCCH genes between moso bamboo and maize.
Supplementary Table 7 | Kinds and numbers of known stress-related elements in the upstream regions of CCCH genes in moso bamboo.
Supplementary Table 8 | The 12 gene primer sequences.
Footnotes
- ^ http://gigadb.org/
- ^ http://smart.embl-heidelberg.de/
- ^ https://pfam.sanger.ac.uk/
- ^ http://www.megasoftware.net/
- ^ http://meme.sdsc.edu/meme/itro.html
- ^ https://github.com/CJ-Chen/TBtools
References
Berg, J. M., and Shi, Y. (1996). The galvanization of biology: a growing appreciation for the roles of zinc. Science 271, 1081–1085. doi: 10.1126/science.271.5252.1081
Bilas, R., Szafran, K., Hnatuszko-Konka, K., and Kononowicz, A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. 127, 269–287. doi: 10.1007/s11240-016-1057-7
Blackshear, P. J. (2002). Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30, 945–952. doi: 10.1042/bst0300945
Bogamuwa, S., and Jang, J.-C. (2013). The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid- mediated regulation of seed germination. Plant Cell Environ. 36, 1507–1519. doi: 10.1111/pce.12084
Bowers, J. E., Chapman, B. A., Rong, J. K., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Chai, G., Hu, R., Zhang, D., Qi, G., Zuo, R., Cao, Y., et al. (2012). Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics 13:253. doi: 10.1186/1471-2164-13-253
Chai, G., Qi, G., Cao, Y., Wang, Z., Yu, L., Tang, X., et al. (2014). Poplar PdC3H17 and PdC3H18 are direct targets of PdMYB3 and PdMYB21, and positively regulate secondary wall formation in Arabidopsis and poplar. New Phytol. 203, 520–534. doi: 10.1111/nph.12825
Chen, D., Chen, Z., Wu, M., Wang, Y., Wang, Y., Yan, H., et al. (2017). Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Moso Bamboo (Phyllostachys edulis). J. Plant Growth Regul. 36, 323–337. doi: 10.1007/s00344-016-9642-x
Cheng, W., Jiang, Y., Peng, J. T., Guo, J. W., Lin, M. L., Jin, C. T., et al. (2020). The transcriptional reprograming and functional identification of WRKY family members in pepper’s response to Phytophthora capsici infection. BMC Plant Biol. 20:256. doi: 10.1186/s12870-020-02464-7
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Deleu, W., González, V., Monfort, A., Bendahmane, A., Puigdomènech, P., Arús, P., et al. (2007). Structure of two melon regions reveals high microsynteny with sequenced plant species. Mol. Genet. Genomics 278, 611–522.
Dung Tien, L., Nishiyama, R., Watanabe, Y., Vankova, R., Tanaka, M., Seki, M., et al. (2012). Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS One 7:e0042411. doi: 10.1371/journal.pone.0042411
Fan, C., Ma, J., Guo, Q., Li, X., Wang, H., and Lu, M. (2013). Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8:e0056573. doi: 10.1371/journal.pone.0056573
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H.-R., et al. (2008). The Pfam protein families database. Nucleic Acids Res. 36, D281–D288. doi: 10.1093/nar/gkm960
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gaut, B. S., Morton, B. R., McCaig, B. C., and Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. U.S.A. 93, 10274–10279. doi: 10.1073/pnas.93.19.10274
Han, G., Wang, M., Yuan, F., Sui, N., Song, J., and Wang, B. (2014). The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 86, 237–253. doi: 10.1007/s11103-014-0226-5
He, Y. H., Fukushige, H., Hildebrand, D. F., and Gan, S. S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
Huang, P., Ju, H.-W., Min, J.-H., Zhang, X., Chung, J.-S., Cheong, H.-S., et al. (2012). Molecular and physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signaling pathway. Plant Cell Physiol. 53, 193–203. doi: 10.1093/pcp/pcr162
Ibraheem, O., Botha, C. E. J., and Bradley, G. (2010). In silico analysis of cis-acting regulatory elements in 5’ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 34, 268–283. doi: 10.1016/j.compbiolchem.2010.09.003
Jan, A., Maruyama, K., Todaka, D., Kidokoro, S., Abo, M., Yoshimura, E., et al. (2013). OsTZF1, a CCCH-Tandem Zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161, 1202–1216. doi: 10.1104/pp.112.205385
Kim, D. H., Yamaguchi, S., Lim, S., Oh, E., Park, J., Hanada, A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20, 1260–1277. doi: 10.1105/tpc.108.058859
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and dePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, D., Yusuf, M. A., Singh, P., Sardar, M., and Sarin, N. B. (2014). Histochemical detection of superoxide and H2O2 accumulation in Brassica juncea seedlings. Bio Protoc. 4:1.
Lee, S.-J., Jung, H. J., Kang, H., and Kim, S. Y. (2012). Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are Involved in ABA and JA Responses. Plant Cell Physiol. 53, 673–686. doi: 10.1093/pcp/pcs023
Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., et al. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32, D142–D144. doi: 10.1093/nar/gkh088
Li, Z., and Thomas, T. L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10, 383–398. doi: 10.1105/tpc.10.3.383
Liang, J., Song, W., Tromp, G., Kolattukudy, P. E., and Fu, M. (2008). Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One 3:2880. doi: 10.1371/journal.pone.0002880
Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3:17. doi: 10.1186/1471-2148-3-17
Lin, P.-C., Pomeranz, M. C., Jikumaru, Y., Kang, S. G., Hah, C., Fujioka, S., et al. (2011). The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 65, 253–268. doi: 10.1111/j.1365-313X.2010.04419.x
Lin, Y., Cheng, Y., Jin, J., Jin, X., Jiang, H., Yan, H., et al. (2014). Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS One 9:e0102825. doi: 10.1371/journal.pone.0102825
Liu, Q., Wang, H., Zhang, Z., Wu, J., Feng, Y., and Zhu, Z. (2009). Divergence in function and expression of the NOD26-like intrinsic proteins in plants. BMC Genomics 10:313. doi: 10.1186/1471-2164-10-313
Liu, S. Q., Liu, X. H., and Jiang, L. W. (2011). Genome-wide identification, phylogeny and expression analysis of the lipoxygenase gene family in cucumber. Genet. Mol. Res. 10, 2613–2636. doi: 10.4238/2011.October.25.9
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., and Zhang, B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4:6645. doi: 10.1038/srep06645
McKersie, B. D., Bowley, S. R., Harjanto, E., and Leprince, O. (1996). Water-deficit tolerance and field performance of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiol. 111, 1177–1181. doi: 10.1104/pp.111.4.1177
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Lam-Son Phan, T. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Peng, X., Zhao, Y., Cao, J., Zhang, W., Jiang, H., Li, X., et al. (2012). CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One 7:e0040120. doi: 10.1371/journal.pone.0040120
Pi, B., He, X., Ruan, Y., Jang, J.-C., and Huang, Y. (2018). Genome-wide analysis and stress-responsive expression of CCCH zinc finger family genes in Brassica rapa. BMC Plant Biol. 18:373. doi: 10.1186/s12870-018-1608-7
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seok, H.-Y., Linh Vu, N., Park, H.-Y., Tarte, V. N., Ha, J., Lee, S.-Y., et al. (2018). Arabidopsis non-TZF gene AtC3H17 functions as a positive regulator in salt stress response. Biochem. Biophys. Res. Commun. 498, 954–959. doi: 10.1016/j.bbrc.2018.03.088
Stege, J. T., Guan, X., Ho, T., Beachy, R. N., and Barbas, C. F. (2002). Controlling gene expression in plants using synthetic zinc finger transcription factors. Plant J. 32, 1077–1086. doi: 10.1046/j.1365-313X.2002.01492.x
Takatsuji, H. (1998). Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54, 582–596. doi: 10.1007/s000180050186
Wang, D., Guo, Y., Wu, C., Yang, G., Li, Y., and Zheng, C. (2008). Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9:44. doi: 10.1186/1471-2164-9-44
Wang, L., Xu, Y., Zhang, C., Ma, Q., Joo, S.-H., Kim, S.-K., et al. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation. Mediates rice architecture via brassinosteroids signaling. PLoS One 3:3521. doi: 10.1371/journal.pone.0003521
Wang, W., Liu, B., Xu, M., Jamil, M., and Wang, G. (2015). ABA-induced CCCH tandem zinc finger protein OsC3H47 decreases ABA sensitivity and promotes drought tolerance in Oryza sativa. Biochem. Biophys. Res. Commun. 464, 33–37. doi: 10.1016/j.bbrc.2015.05.087
Wang, Y., Feng, L., Zhu, Y., Li, Y., Yan, H., and Xiang, Y. (2015). Comparative genomic analysis of the WRKY III gene family in populus, grape, Arabidopsis and rice. Biol. Direct. 10:48. doi: 10.1186/s13062-015-0076-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Xu, R. (2014). Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Solanum lycopersicum. Mol. Genet. Genomics 289, 965–979. doi: 10.1007/s00438-014-0861-1
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yamauchi, Y., Takeda-Kamiya, N., Hanada, A., Ogawa, M., Kuwahara, A., Seo, M., et al. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48, 555–561. doi: 10.1093/pcp/pcm023
Yan, Z., Jia, J., Yan, X., Shi, H., and Han, Y. (2017). Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol. Biol. 95, 549–565. doi: 10.1007/s11103-017-0667-8
Yang, J. I., Shin, Y., Jung, I., and Chun, J. S. (2020). A role of zinc finger ccch type containing 12a (zc3h12a) in osteoarthritis pathogenesis. Osteoarthr. Cartil. 28, S100–S100.
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Zhang, D., Xu, Z., Cao, S., Chen, K., Li, S., Liu, X., et al. (2018). An uncanonical CCCH-tandem zinc-finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mo. Plant 11, 163–174. doi: 10.1016/j.molp.2017.11.004
Zhang, H., Gao, X., Zhi, Y., Li, X., Zhang, Q., Niu, J., et al. (2019). A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 223, 1918–1936. doi: 10.1111/nph.15925
Zhang, X. P., Gao, G. B., Wu, Z. Z., Wen, X., Zhong, H., Zhong, Z. K., et al. (2019). Agroforestry alters the rhizosphere soil bacterial and fungal communities of moso bamboo plantations in subtropical China. Appl. Soil Ecol. 143, 192–200. doi: 10.1016/j.apsoil.2019.07.019
Zhang, L. Y., Bai, M. Y., Wu, J. X., Zhu, J. Y., Wang, H., Zhang, Z. G., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. doi: 10.1105/tpc.109.070441
Zhou, T., Yang, X., Wang, L., Xu, J., and Zhang, X. (2014). GhTZF1 regulates drought stress responses and delays leaf senescence by inhibiting reactive oxygen species accumulation in transgenic Arabidopsis. Plant Mol. Biol. 85, 163–177. doi: 10.1007/s11103-014-0175-z
Keywords: CCCH zinc finger proteins, moso bamboo (Phyllostachys edulis), PeC3H74, drought tolerance, stomatal
Citation: Chen F, Liu H-L, Wang K, Gao Y-M, Wu M and Xiang Y (2020) Identification of CCCH Zinc Finger Proteins Family in Moso Bamboo (Phyllostachys edulis), and PeC3H74 Confers Drought Tolerance to Transgenic Plants. Front. Plant Sci. 11:579255. doi: 10.3389/fpls.2020.579255
Received: 24 August 2020; Accepted: 12 October 2020;
Published: 09 November 2020.
Edited by:
Ayako N. Sakamoto, National Institutes for Quantum and Radiological Science and Technology, JapanReviewed by:
Jian Gao, International Centre for Bamboo and Rattan, ChinaKazuo Nakashima, Japan International Research Center for Agricultural Sciences (JIRCAS), Japan
Copyright © 2020 Chen, Liu, Wang, Gao, Wu and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Xiang, eGlhbmd5YW5haGF1QHNpbmEuY29t
 Feng Chen1
Feng Chen1 Yan Xiang
Yan Xiang