
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Plant Sci. , 02 November 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.578739
This article is part of the Research Topic Understanding C4 Evolution and Function View all 12 articles
C4 photosynthesis evolved over 65 times, with around 24 origins in the eudicot order Caryophyllales. In the Caryophyllales family Nyctaginaceae, the C4 pathway is known in three genera of the tribe Nyctagineae: Allionia, Okenia and Boerhavia. Phylogenetically, Allionia and Boerhavia/Okenia are separated by three genera whose photosynthetic pathway is uncertain. To clarify the distribution of photosynthetic pathways in the Nyctaginaceae, we surveyed carbon isotope ratios of 159 species of the Nyctaginaceae, along with bundle sheath (BS) cell ultrastructure, leaf gas exchange, and C4 pathway biochemistry in five species from the two C4 clades and closely related C3 genera. All species in Allionia, Okenia and Boerhavia are C4, while no C4 species occur in any other genera of the family, including three that branch between Allionia and Boerhavia. This demonstrates that C4 photosynthesis evolved twice in Nyctaginaceae. Boerhavia species use the NADP-malic enzyme (NADP-ME) subtype of C4 photosynthesis, while Allionia species use the NAD-malic enzyme (NAD-ME) subtype. The BS cells of Allionia have many more mitochondria than the BS of Boerhavia. Bundle sheath mitochondria are closely associated with chloroplasts in Allionia which facilitates CO2 refixation following decarboxylation by mitochondrial NAD-ME. The close relationship between Allionia and Boerhavia could provide insights into why NADP-ME versus NAD-ME subtypes evolve, particularly when coupled to analysis of their respective genomes. As such, the group is an excellent system to dissect the organizational hierarchy of convergent versus divergent traits produced by C4 evolution, enabling us to understand when convergence is favored versus when divergent modifications can result in a common phenotype.
C4 photosynthesis is a complex trait that arises following modifications to hundreds if not thousands of individual genes within a genome (Gowik et al., 2011). Despite this, it is one of the most convergent of evolutionary phenomena in the biosphere, with over 65 independent origins (Conway-Morris, 2003; Sage, 2016; Heyduk et al., 2019). Evolutionary convergence, however, does not necessarily reflect convergence throughout the hierarchy of traits that give rise to a complex phenotype, because multiple mechanisms can support a common function (Losos, 2011). This is well illustrated in the case of C4 photosynthesis and crassulacean acid metabolism (CAM), each of which have been repeatedly assembled using disparate enzymes and structural modifications (Sage et al., 2012; Christin and Osborne, 2013; Edwards, 2019). While examples of evolutionary convergence are many, the mechanisms of convergence remain a major question in the life sciences, particularly in the cases where complex traits such as C4 photosynthesis repeatedly evolve (Blount et al., 2018). Because the complexity of the C4 system is well-understood, as well as the phylogenetic distribution of the many C4 clades, C4 photosynthesis represents an excellent system to understand the mechanics of convergent evolution, and its implication for the rise of C4-dominated biomes over the past 30 million years (Christin and Osborne, 2013; Heyduk et al., 2019).
C4 photosynthesis first captures CO2 at low concentration in an outer mesophyll (M) compartment via the activity of phosphoenolpyruvate (PEP) carboxylase (PEPCase), and then concentrates it into an internal compartment, typically a layer of cells around the leaf vasculature termed the bundle sheath (BS; Edwards and Walker, 1983; Hatch, 1987).1 The C4 pathway begins with the conversion of CO2 to bicarbonate (HCO3–) by carbonic anhydrase (CA) in M cells, followed by PEP carboxylation (Figure 1). These two steps occur in all C4 plants, and thus are universally convergent traits in C4 photosynthesis; however, different paralogs have been recruited to carry out the PEPCase and CA functions, demonstrating evolutionary flexibility at a lower level of organization (Christin et al., 2010b, 2013a; Ludwig, 2016a; Heyduk et al., 2019). The product of PEP carboxylation, oxaloacetate (OAA), is too labile to safely move between M and BS cells, so it must be converted to a stable metabolite (Edwards and Walker, 1983). Metabolite transport between M and BS cells is by diffusion, which necessitates that metabolites form steep concentration gradients to support rapid flux, and thus must be stable at high concentration (Bräutigam and Weber, 2011). The solution to the challenge presented by OAA instability is to convert it to the stable metabolites malate or aspartate. This highlights another fundamental feature of evolutionary convergence, in that it occurs where there are strict physiochemical constraints such as OAA lability. A common step (OAA conversion) is accomplished via divergent metabolic solutions (formation of malate versus aspartate). The selected transport metabolite, as it turns out, reflects the enzyme that catalyzes the decarboxylation step.
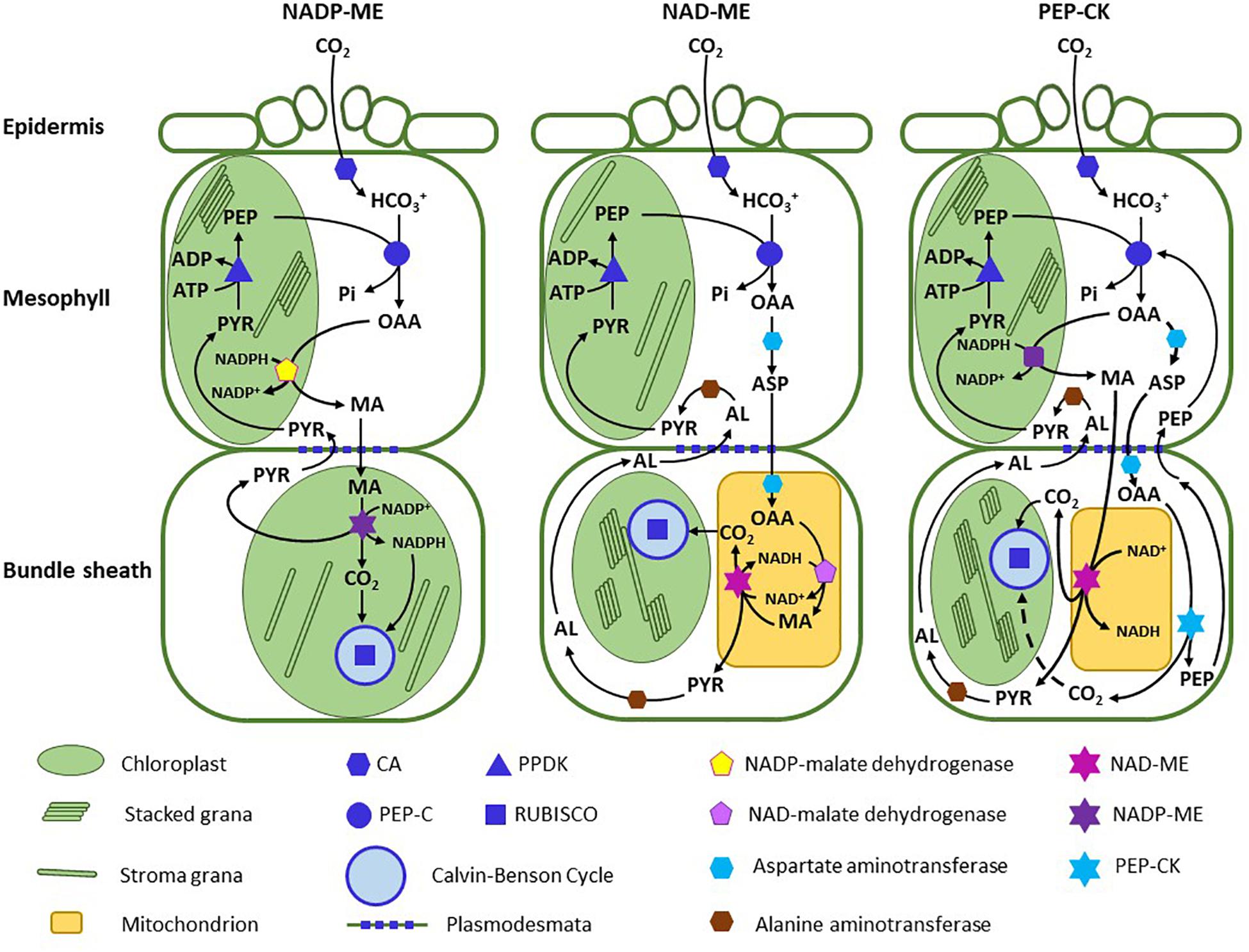
Figure 1. A schematic outlining the C4 pathway in NADP-ME type species, NAD-ME species, and PEP carboxykinase species. The diagram represents the classical depiction of the pathway (as per Hatch, 1987 and Furbank, 2011) assuming malate or aspartate predominate in their respective pathways. For clarity, nitrogen exchange reactions between aspartate and pyruvate are not shown in the NAD-ME and PCK subtypes. Thylakoid stacking is indicated by multi-layered rectangles in the chloroplasts. AL, alanine; ASP, aspartate; CA, carbonic anhydrase; MA, malate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PEP-C, PEP carboxylase; PEP-CK, PEP carboxykinase; PPDK, pyruvate, phosphate dikinase; PVA, pyruvate.
There are three major decarboxylating enzymes co-opted for the decarboxylation step in C4 photosynthesis: NADP-malic enzyme (NADP-ME), NAD-malic enzyme (NAD-ME) and PEP carboxykinase (PCK; Hatch, 1987). Most C4 lineages use NADP-ME as the primary decarboxylating enzyme, about 1/3 use NAD-ME and five lineages predominantly use PCK (Sage R. F. et al., 2011). The consequence of the decarboxylase selection affects many aspects of C4 photosynthesis, to include the pathway biochemistry, BS and M ultrastructure, M to BS transport processes, leaf energetics, and photosynthetic efficiency (Hatch, 1987; Drincovich et al., 2011; Ghannoum et al., 2011). To recognize the suite of traits associated with each decarboxylation mode, three subtypes of C4 photosynthesis have been delineated – the NADP-ME subtype, the NAD-ME subtype, and the PCK subtype (Figure 1). Although each subtype conducts C4 photosynthesis, they represent three evolutionary mechanisms to concentrate CO2 that are derived from a distinct set of biochemical, structural and transport traits (Rao and Dixon, 2016). While each subtype represents a divergent means of concentrating CO2 around Rubisco, traits associated within each subtype reflect strong convergence in response to constraints imposed by the decarboxylating enzyme. The NADP-ME enzyme co-opted by the C4 cycle is located in the BS chloroplasts, where it uses malate and NADP+ to produce CO2, NADPH and pyruvate, with the NADPH directly supporting reduction of PGA produced by Rubisco. NAD-ME is mitochondrial, and uses malate and NAD+ to produce CO2, NADH and pyruvate. Because it is active in the BS mitochondria, substantial mitochondrial volume is needed to meet the metabolic requirements of the C4 pathway, and consistently, species of the NAD-ME subtype often have more and/or larger mitochondria in BS cells than NADP-ME species (Dengler and Nelson, 1999; Edwards and Voznesenskaya, 2011). The PCK enzyme is located in the cytosol, and uses ATP and OAA to produce PEP, CO2, and ADP + Pi. Large amounts of ATP are needed for the PCK reaction, and hence there is a large investment in BS mitochondrial volume to meet the ATP requirement (Yoshimura et al., 2004; Voznesenskaya et al., 2006). The decarboxylation type also constrains the selection of the transport metabolite. In NADP-ME species, NADPH produced by malate oxidation in the BS chloroplast is rapidly consumed in the metabolism of PGA generated by Rubisco, so there is no feedback onto malate flux. In NAD-ME species, the use of malate as a transport molecule would be problematic. NADH cannot readily exit the mitochondria, and there is an insufficient energy sink in the mitochondria to use the large amount of NADH that would be generated if NAD-ME oxidized malate imported from the M cells. The utilization of aspartate avoids these issues because there is no net import of reducing power into the BS mitochondria (Kanai and Edwards, 1999). Once in the BS mitochondria, aspartate forms OAA, which is reduced to malate via malate dehydrogenase using the NADH generated by NAD-ME (Figure 1).
PEP carboxykinase directly uses OAA, and hence to avoid flooding the BS cytosol with reducing power, aspartate is also imported from M cells. However, the PCK reaction requires large amounts of ATP to support rapid photosynthesis, and this can be generated in the mitochondria by oxidizing NADH created by NAD-ME using malate directly imported into the BS mitochondria from the M cells (Leegood and Walker, 1999). PEP carboxykinase species are thought to use NAD-ME at about a fourth to a third of the rate of the PCK reaction, in order to supply sufficient NADH for ATP production (Leegood and Walker, 1999). Consistently, PCK species have many mitochondria in the BS (Dengler and Nelson, 1999).
To meet the energy requirements imposed by the decarboxylating enzymes and associated transport systems, a distinct arrangement of chloroplast membranes occurs in each of the three subtypes (Hatch, 1987; Edwards and Voznesenskaya, 2011; Rao and Dixon, 2016). NADP-ME species have low grana stacking in the BS chloroplasts, yet high stacking in the M chloroplasts. More grana stacking in M cells increases the PSII content in the thylakoids, and hence the potential to generate NADPH by linear electron transport. Because NADP-ME species import reducing power into the BS with malate, the demand for NADPH to reduce PGA in the BS chloroplasts of NADP-ME plants is halved.; hence less PSII and granal stacking are required (Kanai and Edwards, 1999). Chloroplasts in the M cells of NADP-ME species are well stacked to support malate reduction following PEP carboxylation. NAD-ME species, by contrast, have low grana stacking in M chloroplasts because their primary energetic function is to produce ATP for PEP regeneration. The thylakoids of the BS exhibit pronounced stacking in NAD-ME species, to supply sufficient NADPH to potentially reduce all the PGA generated by the C3 cycle. However, there can be variation on these general chloroplast phenotypes, depending upon the degree to which multiple decarboxylases are employed, whether PGA is exported to the M tissue for reduction, or if aspartate is imported into the chloroplast in lieu of malate, as suggested to occur in NADP-ME Flaveria species (Leegood and Walker, 1999; Furbank, 2011).
The theoretical pattern that emerges in C4 evolution is thus one of universal convergence in terms of overall outcome (CO2 concentration into a BS-like compartment), with some steps being universal (CO2 conversion to bicarbonate, PEP carboxylation), and others being divergent (decarboxylation, transport, chloroplast energetics, and possibly how PEP is regenerated). Convergence occurs within the subtype pathways where specific biophysical constraints are present, for example, in transport metabolites associated with the distinct decarboxylating enzymes. An organizational hierarchy between convergence and divergence can thus be envisioned for complex traits that scales from the level of the genes up through to the composite phenotype. The challenge for research on convergent evolution is to describe this hierarchy and to understand why and under what constraints convergent patterns emerge (Losos, 2011). To do this, one needs effective research systems, such as fast cycling microbes, or in terrestrial plants, multiple lineages where convergence has occurred, such as the dozens of clades that have independently evolved C4 photosynthesis (Blount et al., 2018; Heyduk et al., 2019). However, most closely related C4 clades are of the same subtype (Sage R. F. et al., 2011), which restricts the ability to examine sub-type impacts on convergent versus divergent solutions to creating a C4 pathway. One potentially strong study system occurs in Portulaca, where both NADP-ME and NAD-ME species are present (Voznesenskaya et al., 2010; Ocampo et al., 2013). Another possibility occurs in the Nyctagineae tribe of the Nyctaginaceae. Here, three related genera of the tribe Nyctagineae - Allionia, Boerhavia and Okenia - contain C4 species while their closest relatives in the genera Anulocaulis, Cyphomeris, Commicarpus, and Nytctaginia are not known to have any C4 species (Sage R. F. et al., 2011). In the phylogeny of Douglas and Manos (2007), Okenia and Boerhavia form a common C4 clade, while Allionia forms a distinct C4 clade that is separated from the Okenia/Borehavia lineage by the Anulocaulis/Nyctaginia complex. However, there has been no systematic survey of the occurrence of C3 and C4 photosynthesis in the Nyctaginaceae, so it is unclear whether C4 may exist in Anulocaulis, Commicarpus, Cyphomeris, and Nyctaginia, or even whether Allionia, Okenia, and Boerhavia are completely C4, as opposed to also containing C3 and C3–C4 intermediate species. Boerhavia and Allionia are listed as being NADP-ME, although this is based on enzyme assays for Boerhavia only (Muhaidat et al., 2007). Intriguingly, our preliminary TEM images show many mitochondria in the BS ultrastructure of Allionia, suggesting it is NAD-ME. If so, then the Allionia and Okenia/Boerhavia clade could join Portulaca in forming a robust study system for addressing evolutionary convergence as influenced by C4 subtype.
To evaluate the potential of the Nyctaginaceae to become a model for studying evolutionary convergence, we present here a detailed study of the distribution of the C3 and C4 pathways in the Nyctaginaceae. We present a survey of carbon isotope ratios from 560 herbarium specimens, in addition to an anatomical/ultrastructural study using five representative species of Allionia, Anulocaulis, Boerhavia, Commicarpus, and Nyctaginia. We also examine climate data and geographic distributions of species in Allionia, Anulocaulis, Boerhavia, Commicarpus, Cyphomeris, and Nyctaginia to evaluate ecological factors contributing to C4 origins in the Nyctaginaceae. The biochemical sub-types of C4 species in Allionia and Boerhavia were determined, and we present a transcriptome-based phylogeny that updates the phylogenies of Douglas and Manos (2007) and Douglas and Spellenberg (2010). We also use transcriptome data to evaluate whether there has been convergence in the gene sequences of the major C4 pathway enzymes. For comparative purposes, we also present TEM images of leaf tissues from two Portulaca (Portulaceae) species previously shown to be NADP-ME (Portulaca pilosa) or NAD-ME (Portulaca oleracea). Through these efforts, we present an initial hierarchical assessment of how divergence occurs during the convergent evolution of C4 photosynthesis.
Seeds of Allionia incarnata, Anulocaulis gypsogenus, Boerhavia burbigeana, Boerhavia coccinea, Commicarpus scandens, and Nyctaginia capitata were collected from naturally occurring field populations in Southwestern North America and Australia between 2000 and 2016 (see Supplementary Table 1 for collection and voucher information). Portulaca pilosa seeds were a gift from Gerry Edwards and Elena Vosnesenskaya (Washington State University), while P. oleracea grew as a weed in our greenhouse. Seeds were sown directly into 10 or 20 L pots containing a sandy-loam mixture and grown in the University of Toronto greenhouse complex housed on the roof of the Earth’s Sciences Centre. Plants were watered as needed to avoid soil drying (daily in high summer, two to three times weekly in cooler weather), and fertilized bimonthly with a Miracle-Grow commercial fertilizer mix (All-Purpose Brand, 24-8-16) amended with 4 mM calcium nitrate and 1 mM magnesium sulfate. Greenhouse conditions were 27 to 33°C daytime temperature, 20–24°C night temperature, and a peak photon flux density (PFD) of 1500 μmol photons m–2 s–1 on clear days. We used 400 W sodium vapor lamps to supplement natural daylight to maintain a minimum PFD on cloudy days of 250 μmol m–2 s–1 over a >13 h photoperiod. Unless otherwise indicated, three to five plants were sampled for gas exchange, biochemical assay, leaf structural properties and transcriptomics between May and October of 2010 to 2017. Care was taken to sample tissues under the same environmental conditions in the greenhouse (full sun exposure, a leaf temperature near 27°C, at a time between 9 am and 2 pm) to minimize year to year and month to month variation. For the characteristics examined here, subtle variation that may exist between sampling dates is not known to affect results pertinent to our hypotheses. For example, C3 versus C4 expression patterns are largely constitutive, and sun to shade variation in phenotype would not be present at the bright light intensities present in our greenhouses during summer.
To determine the distribution of the C3 and C4 pathways in species of the Nyctaginaceae, 560 herbarium specimens representing 23 genera and 159 species were sampled from the herbaria at Kew gardens (Richmond, London, United Kingdom), Missouri Botanical Gardens (St Louis, MO United States), and New York Botanical Gardens (The Bronx, New York, NY, United States; Supplementary Tables 2, 3). Species were sampled from all genera in the Nyctagineae with the exception of the monospecific genus Cuscatlainia. Approximately 50% (Mirabilis) to 100% (Okenia, Allionia) of the known species from the sampled Nyctagineae genera are present in the survey. We also sampled species from 12 genera occurring in each of the other six tribes of the Nyctaginaceae (Table 1 and Supplementary Table 2). To measure the carbon isotope ratio (δ13C) of each herbarium specimen, 2–4 mg of leaf or stem material were sampled from herbarium sheets and assayed for δ13C by the Washington State University Stable Isotope Core.2 δ13C was determined for at least two distinct plant specimens if available in the herbaria. Carbon isotope ratios between −10 and −16‰ correspond to C4 values, while carbon isotope ratios between −23 and −32‰ correspond to C3 values. Species that are evolutionary intermediates between C3 and C4 photosynthesis exhibit δ13C ratios that are similar to C3 values except where a strong C4 metabolic cycle has been engaged; such C4-like plants typically exhibit δ13C values between −16 and −22‰ (Monson et al., 1988; Von Caemmerer, 1992).
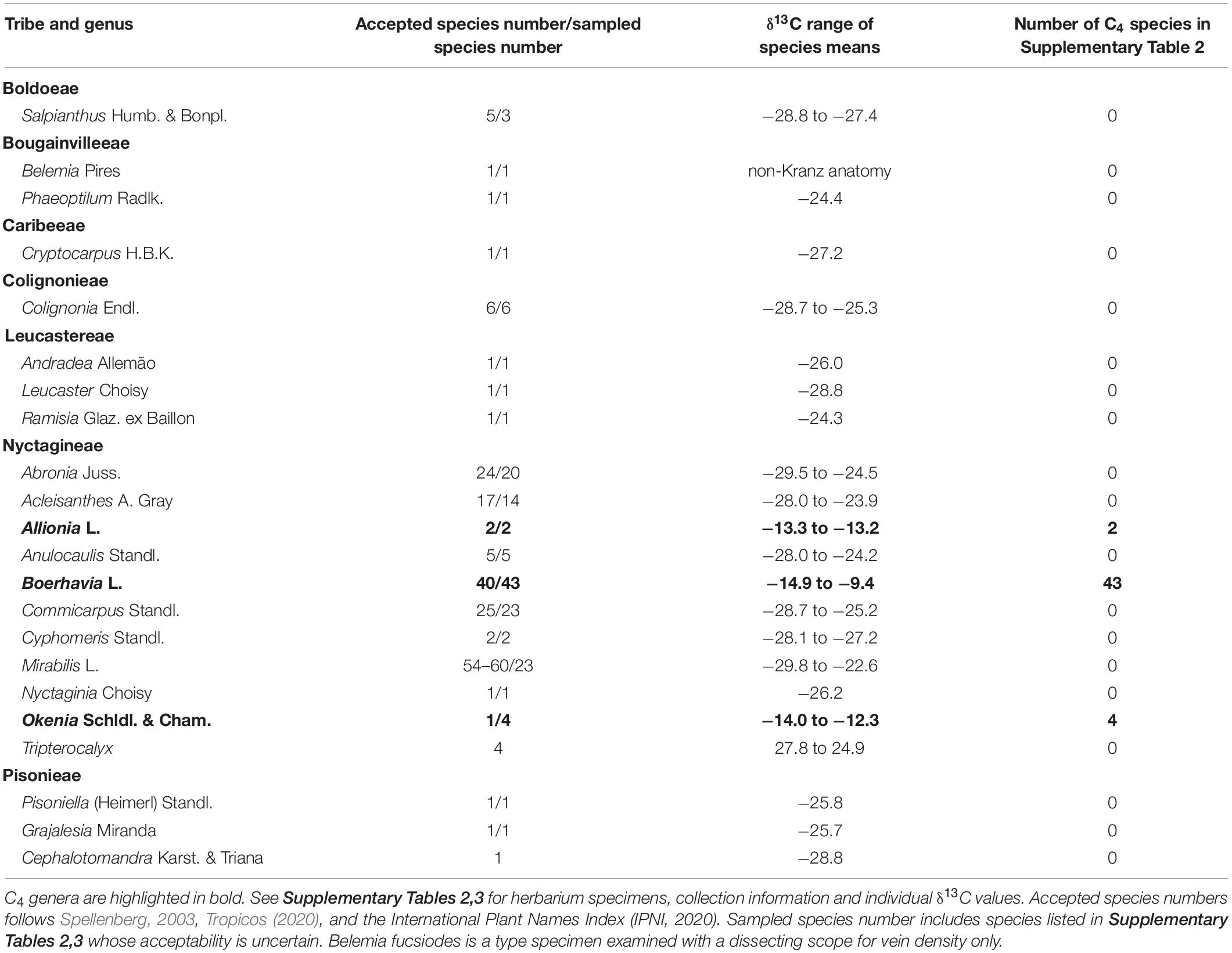
Table 1. δ13C data for sampled species from the Nyctaginaceae showing number of accepted species for each genus and the number of species sampled, the range of δ13C for species means and number of C4 species we identified.
The response of net carbon assimilation rate (A) to intercellular CO2 concentration (Ci) was measured for Allionia incarnata, B. coccinea, and N. capitata using a Li-COR 6400 photosynthetic gas analyzer at 33°C, a vapor pressure differences of 2.0–2.5 kPa and a photosynthetic PFD of 1800 μmol m–2 s–1. The most recent, fully mature leaves were first equilibrated in the leaf cuvette at an ambient CO2 concentration of 400 μmol mol–1 and 1800 μmol photons m–2 s–1. After equilibration and measurement, the ambient CO2 was increased to 1200 μmol mol–1 and then gas exchange parameters were measured after equilibration. The CO2 concentration was then reduced to near the CO2 compensation point in approximately 13 steps, with steady-state gas exchange values determined at each step. The initial slope of the A versus Ci response was estimated as the linear slope of the measurements below a Ci of 100 μmol mol–1. Intrinsic water use efficiency (WUE) was determined as the ratio of A to stomatal conductance (gs) at an ambient CO2 of 400 ppm, and the CO2-saturated rate of A (A1200) was measured at 1200 μmol mol–1 CO2.
The activities of PEPC, NADP-malic enzyme, and NAD-malic enzyme were assayed at 30°C using a coupled-enzyme assay that measured oxidation/reduction rate of NADP(H) or NAD(H) at a wavelength of 340 nm using a Hewitt-Packard 8230 spectrophotometer following procedures in Ashton et al. (1990) as modified by Sage T. L. et al. (2011). Two to three cm2 of recent, fully-mature leaves of A. incarnata, B. coccinea, and N. capitata were sampled under full illumination in the greenhouse and then rapidly ground using a glass tissue homogenizer in an extraction buffer (100 mM HEPES – pH 7.6, 5 mM MgCl2, 10 mM KHCO3, 2 mM EDTA, 10 mM 6-aminocaproic acid, 2 mM benzamide, 1 mM phenylmethylsulfonyl fluoride, 1% (w/v) PVPP, 2% (w/v) PVP, 0.5% Triton X-100, 2% (w/v) BSA, 5 mM DTT, 1% (w/v) casein). After removing two aliquots for chlorophyll assay (in 80% acetone at 645 and 663 nm; Arnon, 1949), the extract was centrifuged 30 s and divided into three aliquots and put on ice. The aliquot used for NAD-ME assay was immediately treated with sufficient MnCl2 to give a 2 mM solution. PEPC was assayed by coupling the production of OAA to NADH oxidation via malate dehydrogenase in an assay buffer containing 50 mM Bicine (pH 8.0), 1 mM EDTA, 5 mM MgCl2, 2 mM NaHCO3, 2 mM DTT, 1 mM glucose 6-phosphate, 2 units ml–1 malate dehydrogenase, and 0.2 mM NADH. The reaction was initiated by the addition of PEP to give 5mM. NADP-ME was assayed by following NADP+ reduction at 340 nm. The assay buffer contained 25 mM Tricine (pH 8.2), 1 mM EDTA, 20 mM MgCl2, 2 mM DTT, 0.5 mM NADP. The reaction was initiated by the addition of malic acid to give 5 mM. NAD-ME was assayed by measuring NAD+ reduction at 340 nm. The reaction mixture contained 25 mM Hepes (pH 7.2), 0.2 mM EDTA, 8 mM ammonium sulfate, 1 unit ml–1 malate dehydrogenase, 5 mM malic acid, 0.025 mM NADH, and 2 mM NAD+. The reaction was initiated by adding MnCl2 to give 5 mM.
For all anatomical and ultrastructural imaging, the middle portion of a leaf blade equidistant between the mid-rib and leaf margin were sampled from recent, fully expanded leaves. Leaf pieces approximately 2 mm2 were collected between 9:00–11:00 am and were prepared for light and transmission electron microscopy as described previously (Khoshravesh et al., 2017). Cell and organelle features of BS cells were quantified using Image J software (Schneider et al., 2012). One leaf per plant was sampled from three to five plants. Each mean value per plant was compiled from measurements of 5–10 imaged cells from the adaxial region of the leaf. All measurements were conducted on planar images of cross (=tranverse) sections. Cells measured were randomly selected from the pool of cells where the plane of section passed through the central region of a BS cell, rather than the periphery. Parameters measured include BS cell area; number of chloroplasts per BS cell; number of mitochondria per BS cell; area of individual chloroplasts and mitochondria; and% BS cell area covered by all chloroplasts and mitochondria in a BS cell.
To prepare a phylotranscriptome of the Nyctaginaceae, we used publicly availably RNA-sequence data from the NCBI Short Read Archive (Supplementary Table 4).3 FASTQ read data was trimmed using Trimmomatic (Bolger et al., 2014) with minimum leading and trailing quality cutoffs at 30, a 5-base sliding window cutoff of 30, and 70bp minimum trimmed length. Transcriptomes were de novo assembled using Trinity (Grabherr et al., 2011) with default settings including in silico read normalization. Open reading frames were predicted and translated using the getorf program from the EMBOSS package (Rice et al., 2000), and the longest open reading frame for each putative locus identified by Trinity was selected using a Python script. OrthoFinder (Emms and Kelly, 2015) was used to predict groups of orthologous genes using 10 reference proteomes with MapMan annotations (Thimm et al., 2004) and 10 with all other annotation types available from the Phytozome v12 (Supplementary Table 5).4 Single- or low-copy orthogroups were selected with a Python script based on the following criteria: a minimum of 40 of 53 species present; no more than 10% of the species having multiple sequences; and a minimum alignment length of 100 amino acids. For species with multiple sequences, a Python script was used to concatenate all orthogroup sequences from a given species if none overlapped by more than 10% of the shortest sequence length, or remove them all if they did. Sequences were assumed to represent fragmented assemblies in the former case and paralogs in the latter. This selection process resulted in 1084 genes for phylogenetic analysis. Initial protein alignments were produced with mafft (Katoh and Standley, 2013) and DNA codon alignments were generated from these using pal2nal (Suyama et al., 2006). The codon alignments were trimmed using trimAl (Capella-Gutiérrez et al., 2009) with a gap threshold of 0.5, then concatenated into a partitioned super-matrix using a Python script written by co-author M. Stata. Phylogenetic inference was conducted using RaxML (Stamatakis, 2014). This was conducted using separate evolutionary models for all partitions, rapid bootstrap analysis with a search for the best tree in one run (command line option -f a), and bootstrap convergence testing (autoMRE). Convergence testing showed that 50 bootstrap replicates were sufficient, but in order to be certain of support values we conducted 200. The species tree was visualized using FigTree5. All Python scripts are available at github.com/MattStata/Nyctaginaceae_Scripts.
All homologs of PEPC, NADPME, NADME, and PPDK were identified in the Nyctaginaceae transcriptomes in the NCBI Short Read Archive (Supplementary Table 4) using BLASP with Arabidopsis sequences as queries. Matches were aligned using mafft (Katoh and Standley, 2013) and preliminary trees were inferred with FastTree (Price et al., 2010) in order to identify the number of copies present in Nyctaginaceae. For each copy, sequences from the Nyctagineae clade were extracted from the alignment and a consensus sequence was generated using Geneious (www.geneious.com) to create full-length consensus references for each copy onto which RNA-seq data for all Nyctagineae species in the NCBI SRA were mapped using HiSat2 (Kim et al., 2019) with the scoring argument – score-min L,0,-1.4. Read mappings were scrutinized manually using the software Geneious to be certain there were no additional paralogs beyond those we had detected, which would be evident as paralogous reads mapped onto the closest reference. Consensus sequences based on mapped reads were generated in Geneious. Gene trees were inferred using MrBayes (Huelsenbeck and Ronquist, 2001) with two runs, 40 chains, 2 million generations, and a heating factor of 0.05. About 10,000 trees from the end of each run where the average SD of split frequencies remained flat at or below 0.01, were used to generate the final tree and infer posterior probabilities using the consense program included with ExaBayes (Aberer et al., 2014). Trees were visualized using FigTree.5 Gene expression values were calculated as reads per kb of transcript per million reads (RPKM) using the SAM files produced by HiSat and a Python script. Only species from the 1KP project6 submitted by our lab were used for gene expression as these represent both C4 lineages and a closely-related C3 outgroup, and were grown and sampled identically. For these, the newest fully expanded leaves were sampled during a sunny day from plants grown in a glass house at the University of Toronto between 9 am and 1 pm. Because complete mappings to all transcripts were not conducted, numbers of total sequenced reads were used in RPKM calculations rather than total mapped reads. Scripts written by M Stata are available at github.com/MattStata/Nyctaginaceae_Scripts.
Biogeographic distributions for species of the tribe Nyctagineae were obtained from the Global Biodiversity Information Facility website (GBIF).7 Duplicate data points and those lacking herbarium records or corresponding to marine coordinates were removed (maptools, Bivand and Lewin-Koh, 2013). The remaining 15,870 observations represented 75 species within Allionia (two species), Boerhavia (40 species), Anulocaulis (five species), Commicarpus (25 species), Cyphomeris (two species), and Nyctaginia (one species). Bioclimate and monthly minimum and maximum temperature parameters (2.5 min resolution) were downloaded from the WorldClim 2.0 dataset8 (Fick and Hijmans, 2017). Monthly potential evapotranspiration and Global Aridity Indexes (AI) were downloaded from CGIAR-CSI GeoPortal.9 Values per observation for 19 bioclimatic variables in the Worldclim dataset, plus minimum and maximum temperatures and AI were then extracted using the extract function in the R raster package (Hijmans and van Etten, 2012). Median values per variable per species were calculated and normalized to Z-scores for use in subsequent analyses. Climate variables that significantly predicted the occurrence of photosynthetic type at p < 0.05 were selected using stepwise regression. Mixed-effect models (R package lme4, Bates et al., 2015) were built using the selected bioclimatic variables and photosynthetic subtypes (NAD-ME or NADP-ME) as the main effect and genus and species as random effects. These models were compared by ANOVA and Akaike’s Information Criteria (AIC). The best model was selected based on the lowest AIC and p values <0.05. A principal component analysis was performed by R package FactoMineR (Lê et al., 2008) to evaluate species distribution across the multivariate predictors. A subset of the data for which we had phylogenetic data was also used to run a phylogenetically corrected ANOVA using phytools in R (Revell, 2012).
For the δ13C survey, we sampled herbarium specimens from all genera of the Tribe Nyctagineae, except for one monospecific genus from El Salvador (Cuscatlainia vulcanicola; Table 1). All sampled species from the genera Allionia, Boerhavia, and Okenia exhibited C4 δ13C values (−9 to −15‰) (Table 1 and Supplementary Tables 2,3). All other species exhibited C3 δ13C values (−22 to −30‰), including all assayed species in the Nyctagineae genera Abronia, Acleisanthes, Anulocaulis, Commicarpus, Cyphomeris, Mirabilis, and Tripterocalyx. The δ13C survey provides little evidence for C3–C4 intermediate species in the Nyctagineae. Values from all species were clearly C4 or within the more negative range of δ13C values typical of C3 plants, with two possible exceptions – the arid zone species Acleisanthes angustifolia (δ13C = −23.9) and Mirabilis polyphylla (δ13C = −22.6). While high for a typical C3 δ13C value, these exceptions are within the range of values observed in arid-zone C3 species with high WUE (Farquhar et al., 1989).
Both Allionia incarnata and B. coccinea exhibited typical C4 photosynthetic parameters. The CO2 compensation point of photosynthesis (Γ) was below 5 μmol mol–1 in both species, the ratio of intercellular to ambient CO2 concentration (Ci/Ca) ratio was 0.31–0.35, and their carboxylation efficiency of photosynthesis, measured as the initial slope of the photosynthetic response to intercellular CO2 concentration (A/Ci response), was 5–7 times greater than the carboxylation efficiency in the C3 N. capitata (Figure 2 and Table 2). Allionia exhibited a steeper initial slope of the A/Ci response than Boerhavia. When relativized by dividing by the maximum net CO2 assimilation rate at high CO2 (A1200) to correct for variation in photosynthetic capacity, the normalized carboxylation efficiency in Allionia was 50% greater than in Boerhavia (Table 2).
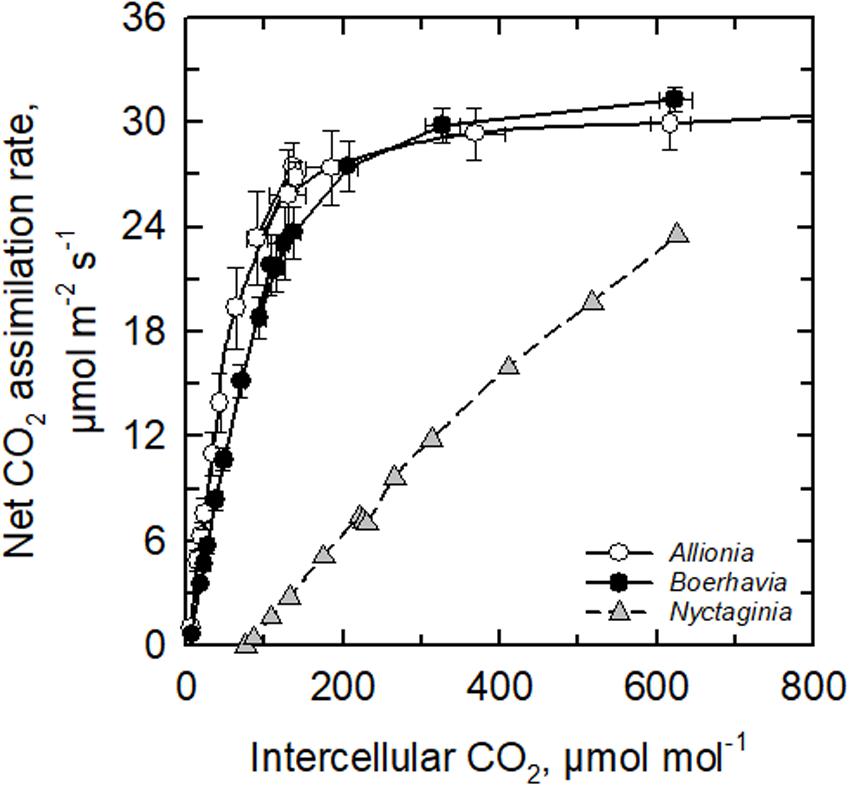
Figure 2. The response of the net CO2 assimilation rate to intercellular CO2 concentration in Allionia incarnata (C4), Boerhavia coccinea (C4) and Nyctaginia capitata (C3) at 30°C and a light intensity of 1800 μmol photons m–2 s–1. Means ± SE, N = 4 (for the C4 species) or 1 (for the C3 species).
The in vitro activity of PEPC was high in the C4 species relative to the C3 N. capitata, and similar on a leaf area basis in the two C4 species; however, on a chlorophyll basis, the PEPC activity is 63% higher in Allionia than Boerhavia (Table 2). NADP-ME activity was high in Boerhavia and not detected in Allionia, while NAD-ME activity was negligible in Boerhavia and high in Allionia (Table 2). Chlorophyll content was 43% higher in Boerhavia than Allionia, and chlorophyll a/b ratio was 21% higher in Boerhavia. Observed enzyme activities are considered robust because they are equivalent to or greater than the observed A values in each species. From these results, we conclude that Boerhavia belongs to the NADP-ME subtype, while Allionia is of the NAD-ME subtype.
Allionia incarnata and B. coccinea exhibited typical C4-Atriplicoid Kranz anatomy, which is characterized by large BS cells in planar cross sections with centripetal organelle arrangements (Figures 3, 4; Edwards and Voznesenskaya, 2011). Notably, enlarged BS cells in both Allionia and Boerhavia do not wrap around the entire vascular bundle, mimicking patterns observed in Atriplex but not many other C4 eudicots classified as having Atriplicoid Kranz anatomy (Muhaidat et al., 2007). A. gypsogenus, C. scandens, N. capitata, and Mirabilis jalapa have characteristic C3 leaf anatomy (Figures 3, 4 and Supplementary Figure 1). Planar areas of the BS cells are comparable between the C3 and C4 species, indicating similar dimensions of the BS tissue in a radial direction with respect to the vasculature (Table 3). Both C4 species exhibit greater coverage of the BS cell by chloroplasts than the C3 species, because there are more chloroplasts per BS cell and greater mean area per chloroplast in the C4 species (Figure 4; Table 3). In A. incarnata, the planar area of BS cells covered by mitochondria is significantly greater than in C3 species and the C4 B. coccinea, due to the presence of larger mitochondria in planar section, and more mitochondria per cell area (Table 3; Figures 4A,C). The percent mitochondrial coverage of the BS cells is similar in B. coccinea and the C3 species (Table 3; Figures 4B,D–F).
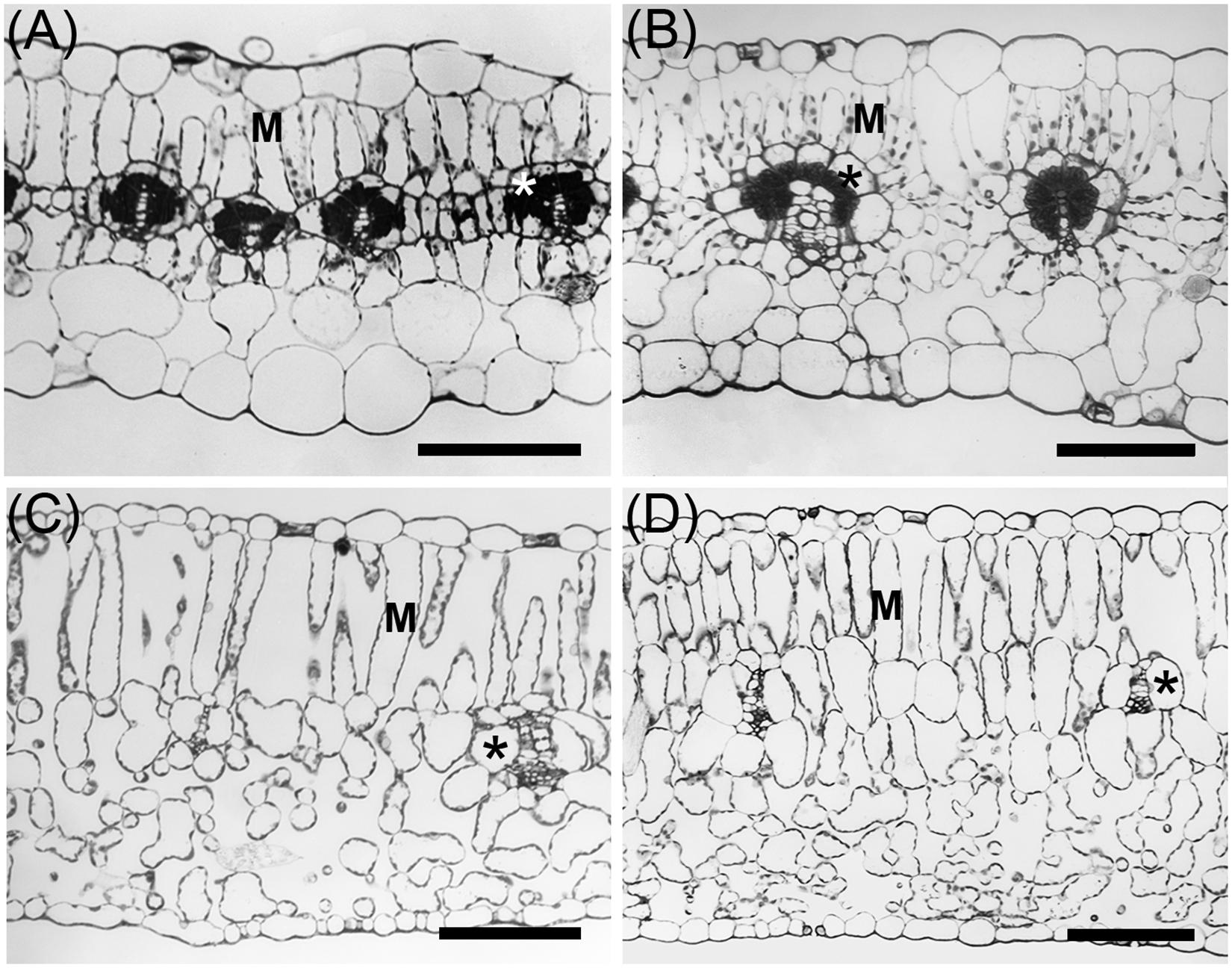
Figure 3. Light microscope images of leaves from C3 and C4 species of Nyctaginaceae, tribe Nyctagineae. (A) Allionia incarnata, (B) Boerhavia coccinea, (C) Commicarpus scandens, (D) Nyctaginia capitata. M, mesophyll cells; *Asterisks mark bundle sheath cells. Scale bars = 100 μm.
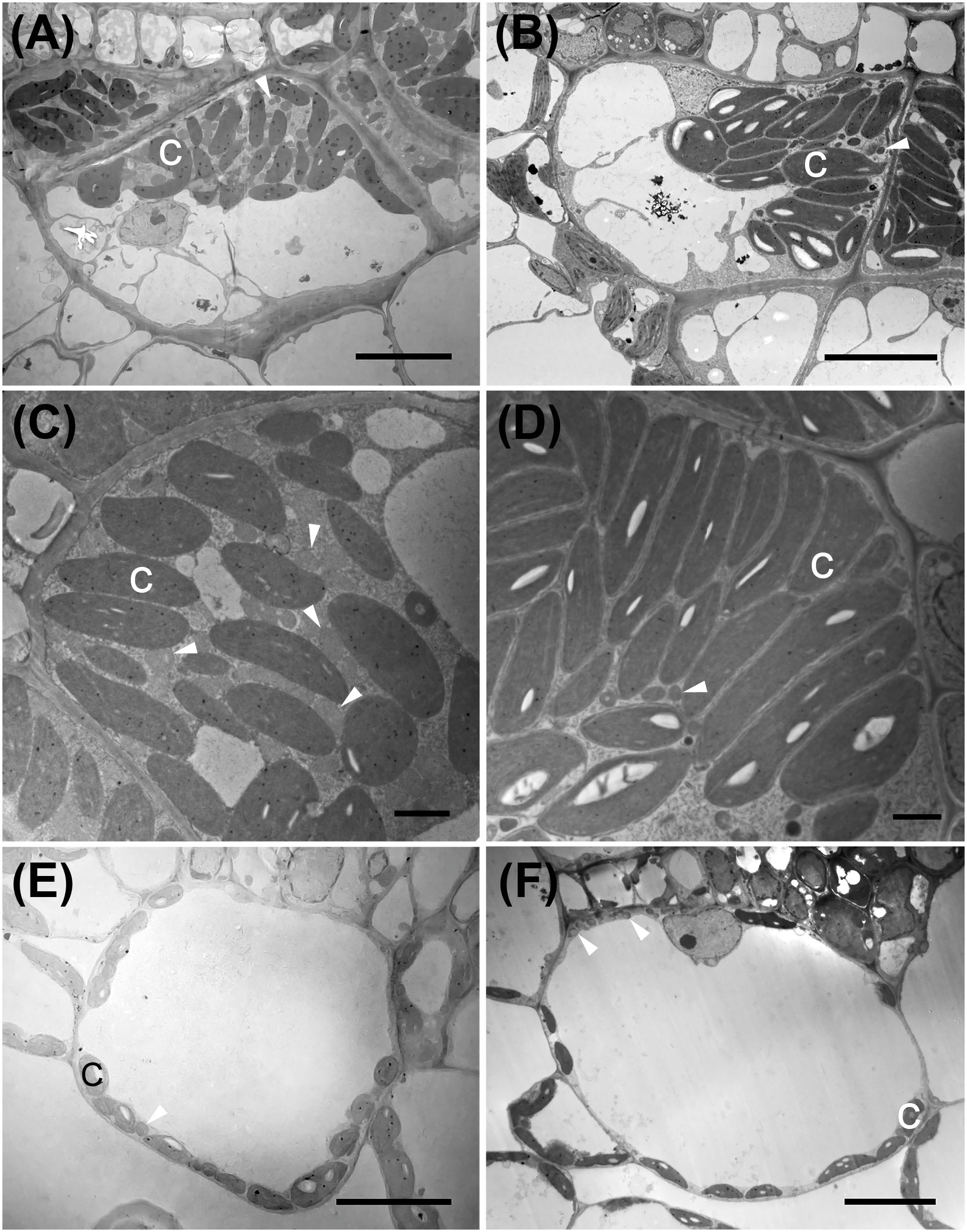
Figure 4. Transmission electron micrographs illustrating bundle sheath cells of C3 and C4 Nyctaginaceae species from the tribe Nyctagineae. (A–D) C4 species; (A,C) Allionia incarnata, (B,D); Boerhavia coccinea; (C,D) high magnification images of the bundle sheath cells. Note high numbers of mitochondria in Allionia incarnata (C). (E,F) The C3 species Commicarpus scandens (E), and Nyctaginia capitata (F). C, chloroplast; arrowheads mark mitochondria. Scale bars = 10 μm.
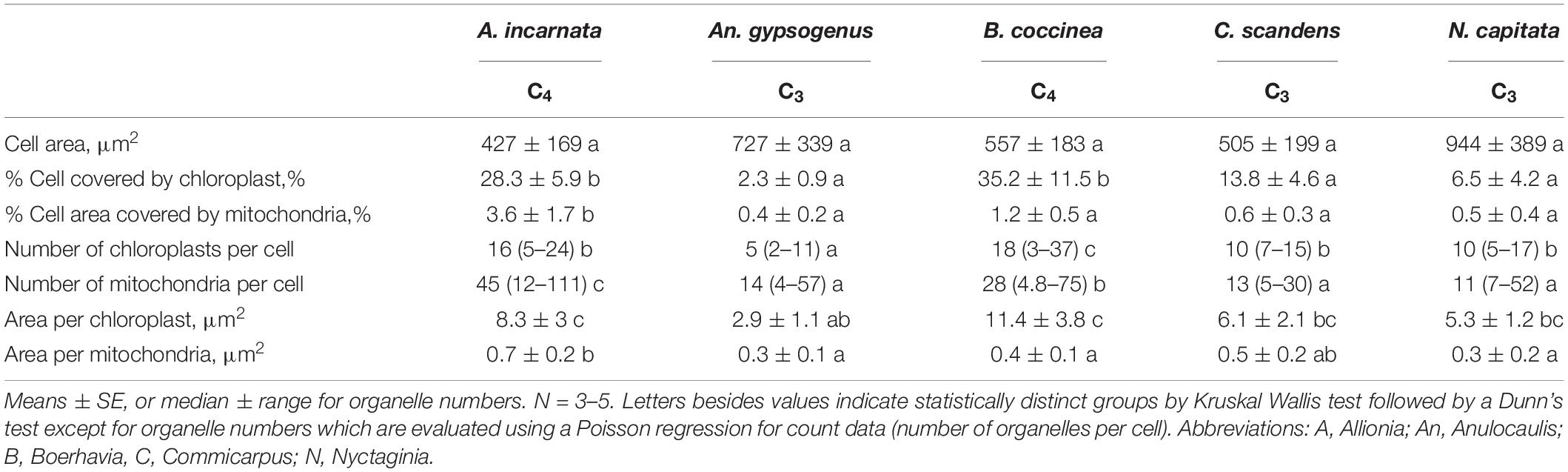
Table 3. Ultrastructural parameters for the bundle sheath cells in cross section of two C4 species and three C3 species from the Nyctagineae tribe of the Nyctaginaceae.
The BS cells in species with C3 δ13C values have smaller and fewer chloroplasts than the C4 species of the Nyctagineae, and these are positioned mostly along the outer BS wall exposed to intercellular airspace (Figures 4E,F and Supplementary Figure 1). None of the three C3 species examined exhibit traits associated with C3–C4 intermediacy or even the incipient intermediate state termed “proto-Kranz”; there was not a greater mitochondrial number, nor a repositioning of mitochondria and chloroplasts toward the inner side of the BS cells. Chloroplasts in the BS of Boerhavia and the M tissue of Allionia are largely agranal, whereas distinct levels of grana stacking are apparent in the thylakoids of the BS chloroplasts in Allionia, and the M chloroplasts of Boerhavia (Figure 5)
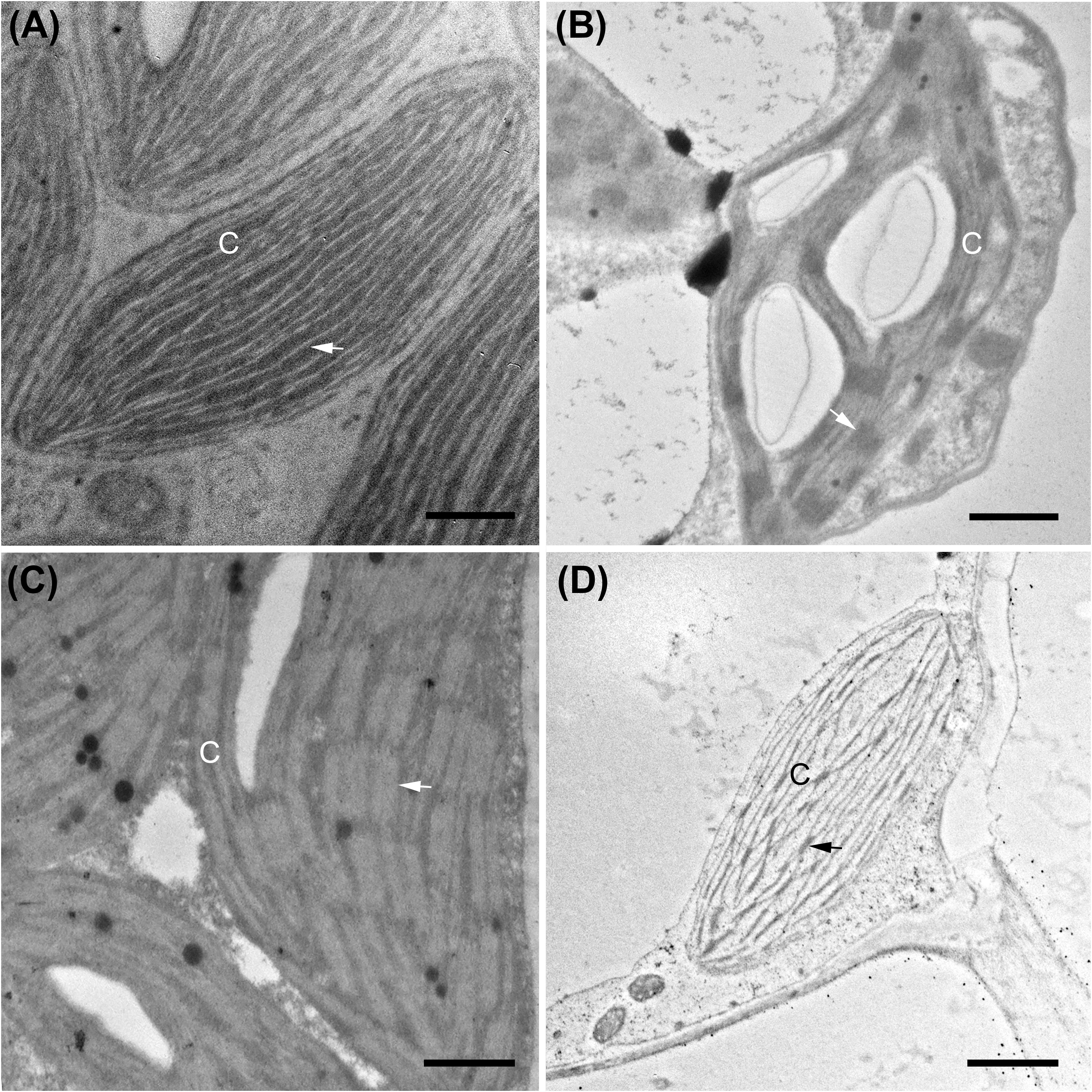
Figure 5. Transmission electron micrographs illustrating chloroplast fine structure. (A) Boerhavia burbidgeana bundle sheath cell; (B) B. burbidgeana mesophyll cell; (C) Allionia incarnata bundle sheath cell; (D) A. incarnata mesophyll cell. In panels (A,C), thylakoids are lighter stacks and striations against a dark stroma. In panels (B,D), thylakoids are dark stacks and striations against a lighter-staining stroma. Scale bars, 0.5 μm. C, chloroplast; arrows mark stacked thylakoids. Scale bars = 0.5 μm.
In P. oleracea (NAD-ME), chloroplasts with pronounced grana stacks cluster in the inner BS (Figure 6A). Many mitochondria are interspersed between the chloroplasts, and exhibit distinct structural connections with nearby chloroplasts (Figure 6B). In P. pilosa (NADP-ME), chloroplasts also occur in the inner BS where they form clusters with no discernable thylakoid stacks (Figures 6C,D). Mitochondria are largely absent between chloroplasts, although some mitochondria occur along the inner wall of the BS in a pattern that is commonly observed in C3–C4 intermediate species (Figure 6C).
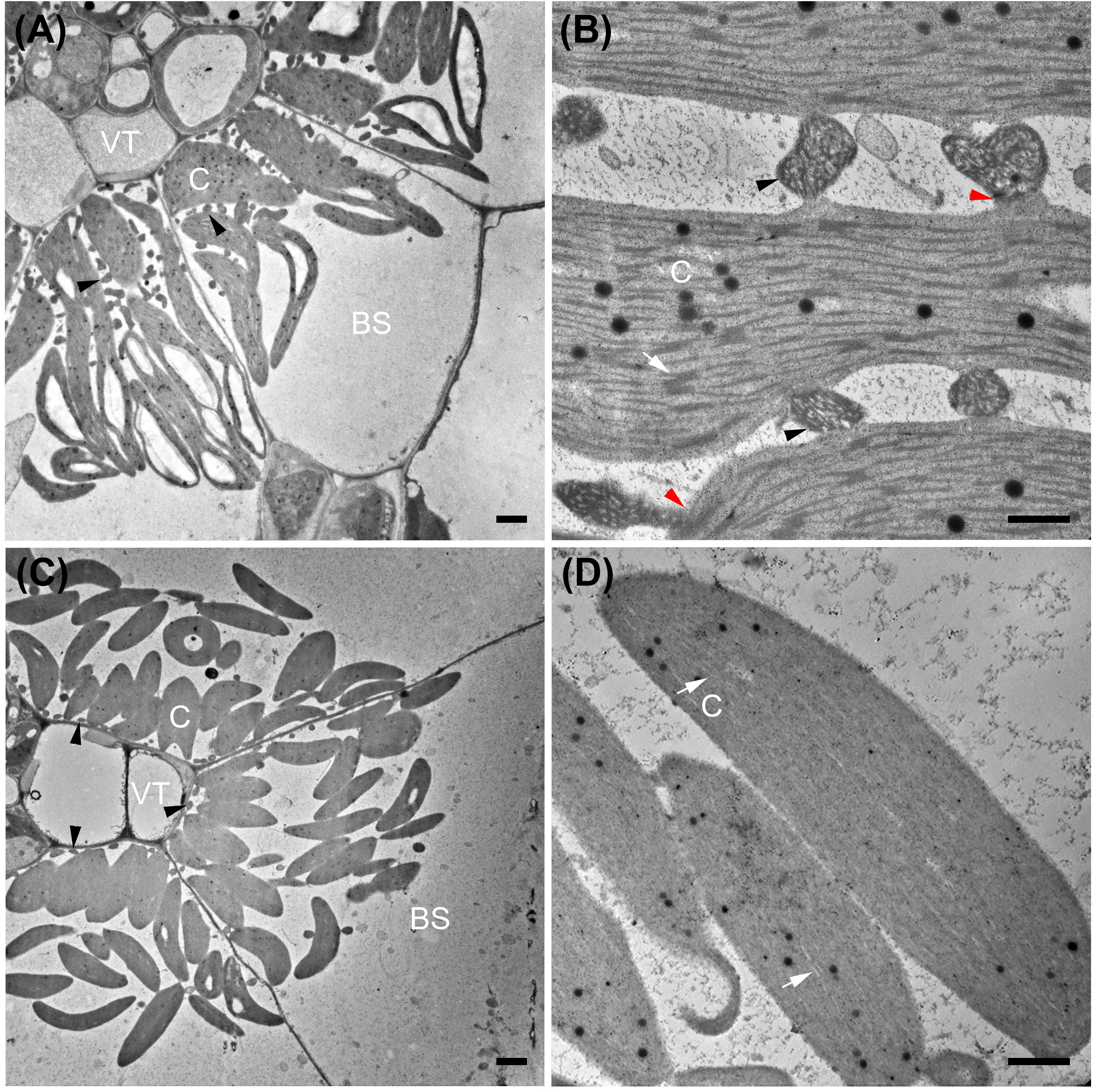
Figure 6. Transmission electron micrographs of bundle sheath cells (A,C) and bundle sheath chloroplasts (B,D) from NAD-ME subtype Portulaca oleracea (A,B) and NADP-ME subtype Portulaca pilosa (C,D). Red arrowheads mark mitochondria to chloroplasts connection structures in P. oleracea (B). BS, bundle sheath; C, chloroplast; VT, vascular tissue; black arrowheads mark mitochondria; white arrows mark thylakoids. Bars = 2 μm (A,C) or 0.5 μm (B,C).
We update the molecular phylogeny of the Nyctagineae using transcriptome sequence data available at the NCBI short read archive (Figure 7). The tree largely replicates the phylogenies of Douglas and Manos (2007) and Douglas and Spellenberg (2010) showing the C4 and C3 clades of the Nyctagineae correspond to a clade of xerophytic herbs and shrubs that Douglas and Spellenberg term the North American Xeric (NAX) clade. C4 Allionia branches with Cyphomeris at the base of a clade that includes the isotopically C3 Anulocaulis and Nyctaginia species, and the C4 Okenia and Boerhavia species. Anulocaulis and Nyctaginia form a clade that branches between Allionia/Cyphomeris and Boerhavia/Okenia. C. scandens branches at the base of the clade containing the C4 species and their immediate non-C4 sister clades, with Mirabilis species branching just below C. scandens. Unlike Douglas and Spellenberg (2010) who predicted Bougainvilleeae + Pisonieae to be sister to the Nyctagineae, we predict only Bougainvilleeae branches in a sister position, with maximal support. Also, the branching order of Phytolaccaceae, Sarcobataceae, Gisekiaceae, and Nyctaginaceae is unclear in the literature and in the APG Caryophllalyes tree, where they form a polytomy.10 Our tree resolves them with 100% support, improving the phylogenetic context for the family.
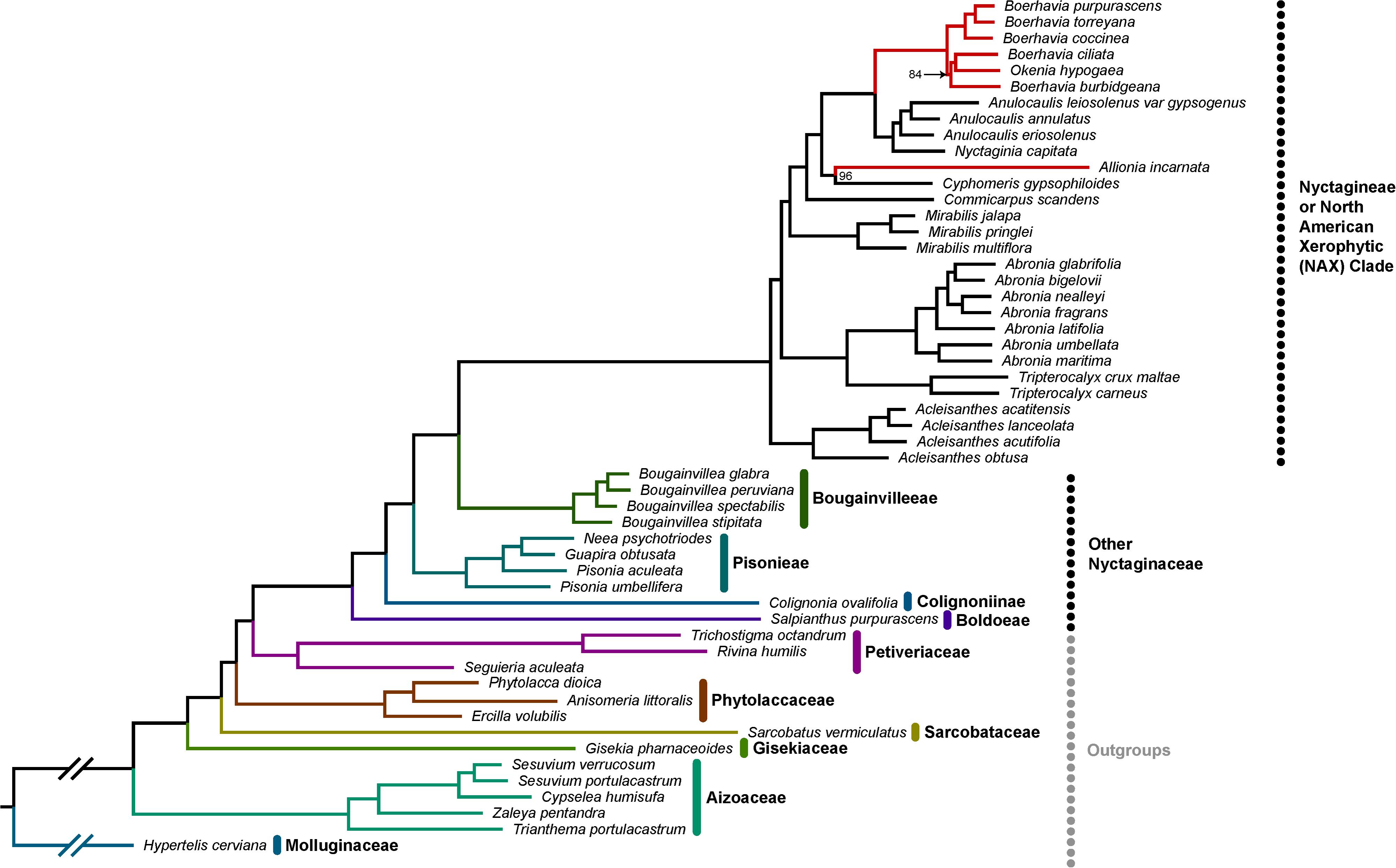
Figure 7. Phylotranscriptomic reconstruction of Nyctaginaceae. A maximum likelihood phylogeny of the Nyctaginaceae family and six outgroup families with previously uncertain topology. This tree is based on a concatenated super-matrix of 847 codon-aligned single-copy orthologs totaling 834,018 sites. All bootstrap support values are 100% except for the two nodes with values indicated, based on 200 bootstrap replicates. Within the Nyctagineae (identified as the North American Xerophytic clade by Douglas and Spellenberg, 2010), C4 lineages are denoted with red branches. The tree is rooted on Hypertelis cerviana and gapped branches here were shortened for visual convenience.
We examined the trees of four important C4 cycle genes – PEPCase, NADP-ME, NAD-ME, and PPDK (Figure 8 and Supplementary Figures 2–5). For each gene, we assumed that the functional copy in the C4 pathway was the one with the highest expression in the transcriptome analysis. In this analysis, we numbered gene copies based on branching order from the base of the gene tree; the numbers assigned are not meant to imply direct orthology with gene copies from any other lineage. For Allionia incarnata and two Boerhavia species, the PEPC1 gene is the copy used in the C4 pathway because it shows an expression strength that is orders of magnitude greater than PEPC2 and PEPC3 (Table 4). By similar logic, NAD-ME3 is the main decarboxylase gene in the C4 pathway of Allionia, and NADP-ME2 is the main decarboxylase gene in Boerhavia. Allionia also exhibited significant expression of NADP-ME1, approaching that of NAD-ME3 (Table 4). The expression of the PEP carboxykinase gene was minimally detectable in both C4 clades (not shown). In the gene trees for PEPC1, PPDK and NADP-ME1, the respective orthologs from numerous C3 species of the Nyctagineae branch between the two C4 lineages, with strong support (Figure 8 and Supplementary Figures 2–5). This indicates the C4 genes arose independently from an ancestral C3 copy, rather than by lateral transfer between clades. If one or more of the genes had moved laterally between the C4 clades, the orthologs from both C4 lineages would form a common clade.
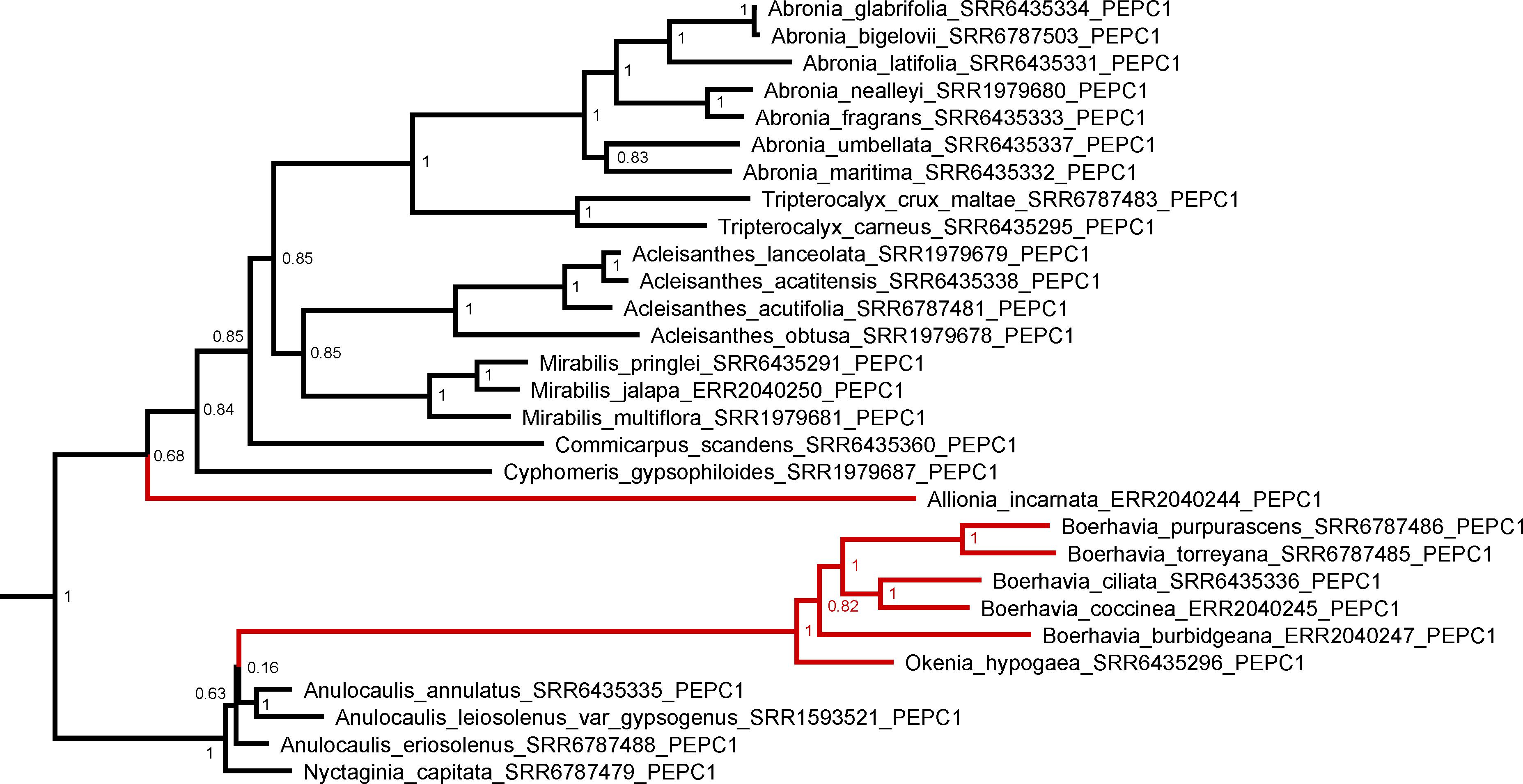
Figure 8. A Baysian gene phylogeny for the PEPCase gene copy co-opted for C4 function in the Nyctaginaceae. Node values are posterior probabilities. Red branches denote C4 species. A tree with all three PEPC paralogs is presented in Supplementary Figure 2.
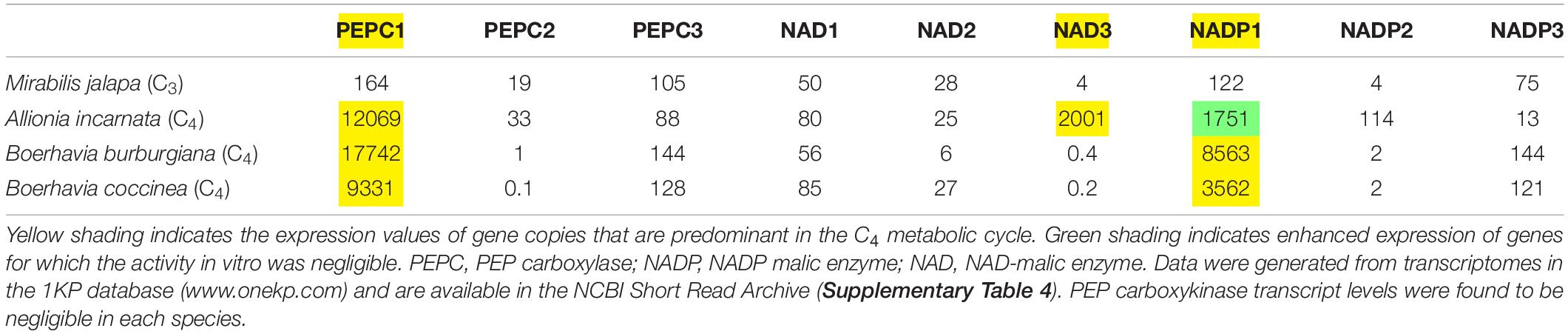
Table 4. RNA Transcript Expression of C4 cycle enzymes from four selected species of the Nyctaginaceae.
We examined PEPC1 sequences for evidence of convergence in the C4 isoforms (Table 5). Christin et al. (2008) showed that 21 amino acid sites were under positive selection in the PEPC1 of various C4 grasses, with the best-known example of sequence convergence being a serine for alanine substitution near the maize 780 position in the PEPC1 sequence (position 774 in the Flaveria sequence; Gowik and Westhoff, 2011). We did not find evidence for convergence with other C4 lineages at position 780 in Allionia nor Boerhavia, as they both exhibited an alanine, which is typical of C3 isoforms (Table 5 and Supplementary Figure 6). There was consistent convergence at sites 572, 761, and 807, where all C4 species exhibited the same amino acids as those under positive selection in C4 grasses. Convergence was observed in both C4 lineages at site 813, but in only one of the six Boerhavia/Okenia species in the database (Table 5). Boerhavia also converged on the same amino acids as the C4 grass species at site 733 and 863, and partially at position 502 (B. purpurescens and B. torreyana only). In total, of the 21 C3 to C4 amino acid switches repeatedly observed in the grass PEPCase sequences (Christin et al., 2007), about a third were replicated in the Boerhavia lineage and a fifth in the Allionia lineage (Table 5). At one site, position 577, the C3 and C4 Nyctaginaceae species share a serine with the C4 grass Zea mays. In the grasses, most C3 species examined by Christin et al. (2007, 2012b) exhibit an alanine at this site. It is possible that C4 evolution in Nyctaginaceae did not require this substitution as the ancestral C3 state was already a serine.
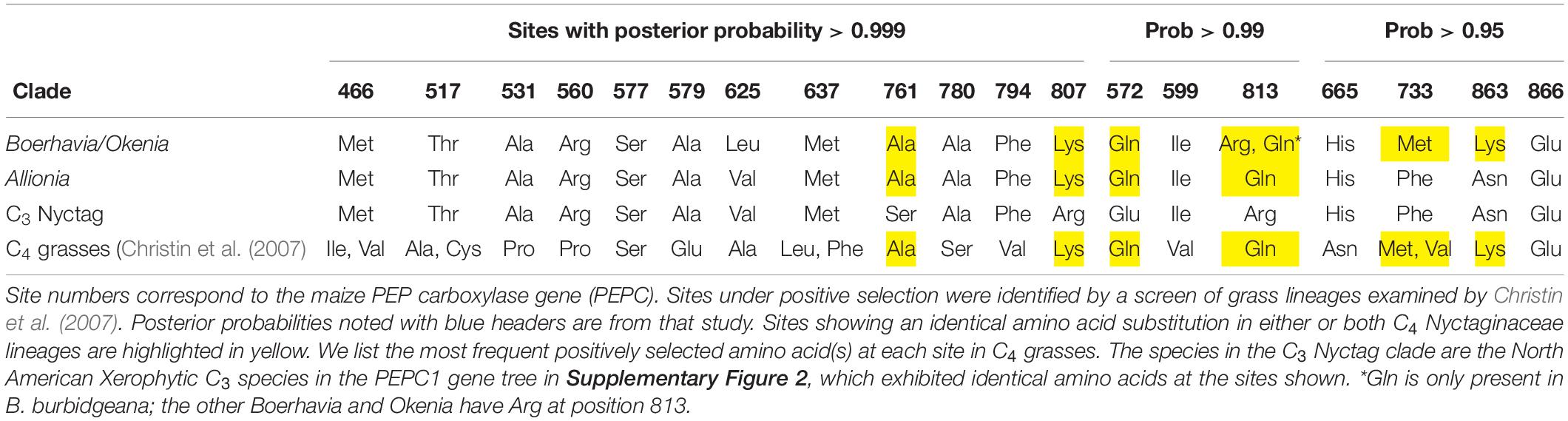
Table 5. Comparison of amino acids in the C4 PEP carboxylases of the Allionia and Okenia/Boerhavia C4 clades with PEP carboxylase sites under positive selection in C4 grasses.
Nyctagineae species from the genera Allionia, Boerhavia, Anulocaulis, Commicarpus, Cyphomeris, and Nyctaginia are distributed in tropical and subtropical regions of similar climate (Supplementary Figures 7–9). Six bioclimatic variables significantly predicted (p < 0.05) the C3 and C4 distribution using a stepwise regression model (Supplementary Figure 8 and Supplementary Table 6). To address whether the variation in distribution of C3 and C4 species could be species or genera dependent, GLMM models were performed with and without considering taxa as random effects. The model was significantly improved (p < 2e–16) when genus was added as the random effect (Supplementary Table 6); however, after this addition, none of the main effects significantly predicted the pattern of photosynthetic pathway distribution. A phylogenetically-corrected ANOVA also failed to detect a significant difference between the C3 and C4 species in response to six variables (Supplementary Table 7). These analyses show that the distribution of Nyctagineae species in climate and geographic space follows taxonomic and phylogenetic affinities rather than photosynthetic pathway.
Carbon isotope ratios demonstrate that all examined species in Allionia, Boerhavia and Okenia are C4, while all examined species in other Nyctaginaceae genera are not, including species in Anulocaulis, Commicarpus, Cyphomeris, and Nyctaginia that branch sister to the C4 clades. The number of C4 species in the Nyctaginaceae is 43–46 based on recent assessments that conclude there are 40 Boerhavia, one to four Okenia, and two Allionia species (Spellenberg, 2003; Tropicos, 2020). The typically C3 δ13C values in the 31 examined species of Anulocaulis, Commicarpus, Cyphomeris, and Nyctaginia indicate that none of the examined species have a type of C3–C4 intermediacy termed C4-like, where a strong C4 metabolic cycle has been engaged. As a C4 cycle becomes engaged, the values of δ13C increase from C3 toward C4 values, becoming distinct from typical C3 values when δ13C rises above −22‰ (Von Caemmerer, 1992; Alonso-Cantabrana and von Caemmerer, 2016). This occurs because PEPCase does not discriminate against the 13C isotope to the degree that Rubisco does (Farquhar et al., 1989). Carbon isotope screens cannot differentiate between C3 plants and a type of C3–C4 intermediacy utilizing C2 photosynthesis, a metabolic pathway that concentrates CO2 into the BS using photorespiratory metabolites to shuttle CO2 from M to BS cells. In C2 photosynthesis, the photorespiratory enzyme glycine decarboxylase is expressed only in BS mitochondria, which forces photorespiratory glycine to diffuse from the M cells where it is formed to the BS cell for metabolism (Sage et al., 2012). The released CO2 from glycine decarboxylation accumulates in the BS cells, where it can be refixed by Rubisco in adjacent chloroplasts with high efficiency. C2 photosynthesis improves carbon gain at low CO2 concentrations, but because all CO2 is fixed by Rubisco, the δ13C values of C2 plants reflect those of C3 plants (Von Caemmerer, 1992). Anatomically, C2 plants have increased numbers of mitochondria and chloroplasts in BS cells, typically in a centripetal position against the BS wall facing the vascular tissue (Khoshravesh et al., 2016). This characteristic identifies candidate C2 species, with low CO2 compensation points of photosynthesis (Γ) confirming the presence of the C2 physiology. Our examination of the BS structure in Anulocaulis, Commicarpus and Nyctaginia, and leaf gas exchange in Nyctaginia, showed no evidence of C2 photosynthesis as the structural characteristics and Γ were typically C3. Hence, we conclude that the sister clades to the Allionia and Boerhavia/Okenia clades are most likely comprised of only C3 plants. Gas exchange data and anatomical studies demonstrate strong C4 features in selected species of Allionia and Boerhavia, which along with the consistently C4 values of δ13C in these clades, lead us to conclude they are completely C4 rather than C4-like. C4-like species have a fully functional C4 cycle but retain limited C3 photosynthesis in M tissues, and exhibit features suggesting they are newly evolved C4 plants that have not yet optimized the C4 pathway (Moore et al., 1989).
In the phylogeny, Anulocaulis and Nyctaginia species occur in a clade that branches sister to the Boerhavia/Okenia clade, while Cyphomeris branches sister to Allionia, supporting a hypothesis of two independent C4 origins, one in ancestral Allionia and a second in ancestors to the Okenia/Boerhavia clade. An alternative possibility is a single C4 origin followed by reversions to C3 in ancestors of Cyphomeris and the Anulocaulis/Nyctaginia clade. This option is less parsimonious, requiring three changes – one acquisition of C4 and two reversions – rather than two. It is also not favored because reversions are considered unlikely and have never been confirmed (Christin et al., 2010a; Oakley et al., 2014; Bräutigam and Gowik, 2016). In addition, the two C4 clades exhibit different biochemical and structural subtypes. The high degree of structural, transport, and biochemical specialization associated with each subtype indicates switching subtypes would be difficult, and consistently, no instances of subtype switching have been documented. Gene sequences of C4 pathway enzymes can also be used to assess reversal possibilities. If a reversion had occurred, the derived ortholog of a C4-pathway enzyme in any C3 progeny might retain sequences from their ancestral C4 function (Christin et al., 2010a; Khoshravesh et al., 2020). In Anulocaulis, Cyphomeris, and Nyctaginia, the C4-type orthologs of PEPCase, PPDK, and both decarboxylases exhibited no signatures of prior C4 function. For example, the C3 species in the Nyctagineae share only one positively selected amino acid substitution with the C4 Nyctaginaceae and Zea mays in the PEPC1 sequence, a serine at position 577. C3 species outside the clade where an ancestral C4 transition could have occurred also share this serine, indicating it is not a relic from a C4-state. Based on these points, we conclude that Allionia and Boerhavia represent independent yet closely related C4 clades that have diverged in terms of biochemical subtype. With other examples of subtype divergence in closely related species of Portulaca and possibly other clades (Salsola and Sesuvioideae; Voznesenskaya et al., 1999; Bohley et al., 2015), comparative methods of evolutionary biology can be used to address questions of convergence and divergence during C4 evolution.
In contrast to a prior conclusion that both Boerhavia and Allionia are NADP-ME (Muhaidat et al., 2007), the biochemical assays demonstrate species in these two clades are different biochemical subtypes of C4 photosynthesis. Consistently, Allionia and Boerhavia showed pronounced differences in BS and M ultrastructure that support their designation as NAD-ME and NADP-ME subtypes, respectively. In both B. coccinea (high NADP-ME activity, low NAD-ME activity) and A. incarnata (high NAD-ME activity, nil NADP-ME activity), chloroplasts and mitochondria occupy the inner half of the BS cells, as is typical in eudicot C4 species (Edwards and Voznesenskaya, 2011). In A. incarnata, there is an abundance of mitochondria scattered among the elongated chloroplasts, while in B. coccinea, BS mitochondria are much less frequent and not commonly interspersed within the chloroplast cluster. In NAD-ME species, the juxtaposition of chloroplasts and mitochondria facilitates rapid movement of CO2 released from mitochondria into adjacent chloroplasts, while the tight packing of chloroplasts and mitochondria reduces the chance of CO2 escape. This tight packing is a major means by which CO2 is trapped in species lacking a suberized layer around the BS wall, as is the case in eudicots (von Caemmerer and Furbank, 2003). In Portulaca species, the same patterns hold but with one important variation that is not reported in the literature, notably, mitochondria in the BS of NAD-ME P. oleracea form distinct attachment structures to adjacent chloroplasts that are not obvious in Allionia and other NAD-ME type C4 species (for example, Anticharis, Cleome, Salsola, and Suaeda; Voznesenskaya et al., 1999, 2018; Khoshravesh et al., 2012). Such structures may facilitate rapid CO2 diffusion between mitochondria and chloroplasts, enhancing refixation efficiency.
The structural characteristics observed here are consistent with patterns in other C4 clades. NADP-ME species among C4 grasses, sedges and eudicots (as shown in Flaveria, Euphorbia, Gomphrena, Heliotropium, Salsola, and Tribulus) share with Boerhavia and Portulaca pilosa the pattern of enlarged chloroplasts with weakly developed grana stacks, and few BS mitochondria (Carolin et al., 1978; Kim and Fisher, 1990; Ueno, 1996, 2013; Voznesenskaya et al., 1999; Yoshimura et al., 2004; Muhaidat et al., 2011; Sage T. L. et al., 2011; Lauterbach et al., 2019). NAD-ME eudicots in Amaranthus, Atriplex, Anticharis, Cleome, Gisekia, Salsola, Suaeda, and Tecticornia share with Allionia and P. oleracea the pattern of enlarged chloroplasts with well-developed grana and large numbers of interspersed mitochondria (Carolin et al., 1978; Voznesenskaya et al., 1999, 2007, 2008; Khoshravesh et al., 2012; Bissinger et al., 2014; Oakley et al., 2014). In grasses, Dengler and Nelson (1999) describe NAD-ME species as having 5-to 20-fold more mitochondria than NADP-ME species, and NAD-ME mitochondria are often larger with greater internal membrane surface area in the BS. The differences between C4 subtypes hold even when different tissue layers are co-opted as the site of CO2 concentration, whether it is the mestome sheath as in grasses, an inner chlorenchyma layer as occurs in Salsola and other succulent chenopods, or single-cell type of C4 photosynthesis as shown in the NAD-ME chenopods Bienertia cycloptera and Suaeda aralocaspica (Ueno, 1996, 2013; Voznesenskaya et al., 1999; Pyankov et al., 2000; Edwards and Voznesenskaya, 2011; Khoshravesh et al., 2020). In the middle of M cells of Bienertia cycloptera, for example, there are many mitochondria within a ball of Rubisco-containing chloroplasts where CO2 is concentrated (Voznesenskaya et al., 2002). The central chloroplasts in B. cycloptera have well-stacked grana, while peripheral chloroplast that are functionally equivalent to M chloroplasts of typical C4 species do not.
Lateral transfer of genes encoding C4 pathway elements has been observed in closely related grass clades within Alloteropsis and Neurachne (Christin et al., 2012a,b; Dunning et al., 2017; Khoshravesh et al., 2020). Lateral gene transfer is relevant to discussions of evolutionary convergence because the acquisition of previously evolved C4 genes by a non-C4 relative could facilitate C4 evolution in the receptive clade, possibly creating a false impression of convergence. When we examined the phylogenies of C4 pathway genes in the Nyctagineae, we found no evidence of lateral transfer. The gene trees consistently showed the two C4 lineages do not share a common gene copy of a C4-adapted isoforms of PEPC, PPDK, or either malic enzyme, because the respective orthologs from related C3 species branch between the C4 lineages with high support (Figure 8 and Supplementary Figures 2–5). This evidence is consistent with a hypothesis that the two C4 clades in the Nyctaginaceae arose de novo from a completely C3 state, rather than via assisted origins involving gene introgression from a pre-existing C4 clade (Christin et al., 2012a,b; Dunning et al., 2017).
With sequencing data becoming widely available, it may be possible to identify C4-subtypes using transcriptomics (Lauterbach et al., 2019). Such an approach, however, could produce errors if not coupled with enzyme activity assays. Here, the transcriptomes consistently showed high expression of the copies we designate as NADP-ME1 in Boerhavia and NAD-ME3 in Allionia. Allionia also exhibited significant expression of NADP-ME1 transcripts which approach expression of its NAD-ME3 gene. This data by itself would suggest co-function of NADP-ME and NAD-ME in Allionia; however, no NADP-ME activity was detected in Allionia, leading us to conclude its NADP-ME1 transcripts are not translated into functional enzyme. The possibility of error in this conclusion is unlikely because the BS ultrastructure in Allionia is consistent with patterns generally seen in NAD-ME subtypes. The high transcript level of both NAD-ME and NADP-ME may be a relic from ancestral Allionia species that may have utilized both decarboxylases during an early phase of C4 photosynthesis. A chance selection event may have subsequently enhanced NAD-ME activity, after which leaf structure and energetics were optimized for the NAD-ME subtype.
One of the notable features of C4 photosynthesis is that it is concentrated in relatively few orders of higher plants. The Poideae (grasses, sedges) and Caryophyllales, for example, account for over 90% of all C4 species and about 50 of the estimated 65 independent origins of C4 photosynthesis (Sage, 2016). The ability of these clades to repeatedly evolve the C4 pathway is hypothesized to reflect the presence of enabling traits that facilitate C4 evolution (Sage, 2004; Christin et al., 2013b). Wider BS cells in C3 species, for example, could be a structural enabling trait (Muhaidat et al., 2011; Christin et al., 2013b; Griffiths et al., 2013), while greater numbers of organelles in BS tissues may reflect an activation of photosynthetic physiology which enables establishment of photorespiratory glycine shuttles (Sage et al., 2013; Schulze et al., 2013). Photosynthetic activation of the BS can facilitate the rise of photorespiratory glycine shuttles because mitochondria and some chloroplasts can reposition to the inner BS region, forcing glycine formed in centrifugal chloroplasts to migrate to the inner BS for processing by glycine decarboxylase (Sage et al., 2013). To evaluate whether enabling traits are present in close C3 relatives of the C4 Nyctaginaceae clades, we examined their BS structure and organelle characteristics. In the C3 species of Anulocaulus, Cyphomeris, and Nyctaginia, BS cells had similar cross-sectional areas as the C4 BS cells, and exhibited numerous chloroplasts along the outer wall, indicating photosynthetic activation. We thus conclude the close C3 relatives of Allionia and Boerhavia exhibit numerous enabling traits for C4 evolution, indicating they were present in ancestral C3 taxa from which the C4 clades arose.
The results indicate C4 photosynthesis evolved in Nyctaginaceae species from hot and dry climates of the New World, most likely in the arid-to-semi-arid regions of Southwestern North America where the center of diversity occurs for the North American Xerophytic clade of the Nyctaginaceae (Douglas and Manos, 2007). Christin et al. (2011) estimate that the C4 pathway appeared in the Boerhavia/Okenia clade about 4.7 million years ago, and 6.1 million years ago in Allionia, based on molecular phylogenies. This was during a period of aridification in Southwestern North American and reduced atmospheric CO2 (Sage et al., 2018). The environmental habitat of Allionia versus Boerhavia did not differ much from each other or their C3 ancestors, indicating the C3 ancestors were adapted to the same kind of hot, dry environments where the C4 species are common. This supports a hypothesis that C4 arose in these sorts of environments, consistent with a model that high levels of photorespiration brought on by heat, drought and/or salinity promoted C4 evolution in C3 plants pre-adapted to such harsh environments (Sage et al., 2018).
Surveys of the floristic distribution of NADP-ME versus NAD-ME grasses have identified a trend where NADP-ME species predominate in grass floras of wetter climates while NAD-ME species predominate in drier climates (Vogel et al., 1978; Hattersley, 1992; Ghannoum et al., 2011). The reasons for this trend have not been clarified, although it has been hypothesized that the pattern reflects phylogenetic ancestry where NAD-ME species are more likely to arise in clades from drier areas while NADP-ME species evolve in wetter climate zones (Taub, 2000). Explanations for habitat differences between C4 subtypes can be evaluated using closely related NADP-ME and NAD-ME clades. In A. incarnata and B. coccinea, we observed similar photosynthetic capacities and intrinsic WUE (A/gs), indicating no obvious differences in photosynthetic parameters that might explain subtype segregation along a moisture gradient. Allionia did exhibit a steeper initial slope of the A/Ci response than Boerhavia, indicating a stronger C4 metabolic cycle in the NAD-ME plant. Consistently, the NAD-ME clade in Allionia had greater PEPCase activity per unit chlorophyll than Boerhavia (NADP-ME clade). If a stronger metabolic cycle is an inherent feature of NAD-ME relative to NADP-ME species, there could be a photosynthetic advantage under low intercellular CO2 concentrations that commonly occur where drought and low humidity reduce stomatal aperture. A comparative analysis using closely related species of differing subtype in the Nyctagineae, Portulaca and other lineages could evaluate this possibility.
As a highly convergent, complex trait, C4 photosynthesis represents an excellent system to dissect evolutionary convergence, particularly the degree of convergence versus divergence in the mix of traits that give rise to a composite phenotype (Heyduk et al., 2019). Numerous studies have previously examined convergent properties of C4 components, such as convergence in genes for C4 enzymes (Christin et al., 2007, 2009, 2010b; Emms et al., 2016); regulatory components (Gowik and Westhoff, 2011; John et al., 2014; Reyna-Llorens and Hibberd, 2017), structural features (Yoshimura et al., 2004; Kadereit et al., 2014; Stata et al., 2014; Danila et al., 2018); and biochemical sub-types (Gutierrez et al., 1974; Hatch et al., 1975; Sage R. F. et al., 2011; Ludwig, 2016b). However, the assembly of multiple datasets into a hierarchical framework has not been attempted in a C4 context. In Table 6, we present a preliminary hierarchy of convergent and divergent traits observed in comparative studies of the many C4 clades, thereby enabling deeper assessments of when and why convergence is favored, versus when divergent solutions to CO2 concentration can occur. Convergence within C4 photosynthesis is apparent in two key steps: the carboxylation of PEP by PEPCase, and the conversion of CO2 to bicarbonate by CA. The ubiquitous use of PEPCase is probably due to three factors. First, it is readily available for co-option, because it is widely used in C3 plants for processes such as pH control, metabolite generation for the Krebs cycle and nitrogen assimilation, and shuttling reducing power between cellular compartments (Aubry et al., 2011; Mallmann et al., 2014). Second, there are no obvious alternatives in vascular plants that may be co-opted to provide the carboxylation function of a C4 cycle. While numerous carboxylases are active in prokaryotes, few occur in higher plants (Erb, 2011). One obvious candidate to recruit into a C4 cycle is pyruvate carboxylase, which produces OAA from pyruvate and bicarbonate in bacteria, animals and algae, but it is not known to occur in plants (Tsuji et al., 2012). Third, it is worth considering the chain of events giving rise to C4 photosynthesis. The evolutionary rise of C2 photosynthesis in C3–C4 intermediate species establishes a Kranz-like leaf anatomy and M to BS transport systems that facilitate the subsequent upregulation of PEPCase and a C4 metabolic cycle (Sage et al., 2012). A leading hypothesis to explain this upregulation is PEPCase provides carbon skeletons to support re-assimilation of photorespiratory ammonia (Mallmann et al., 2014). If so, then the ubiquitous use of PEPCase may arise out of its pre-exiting role supporting N metabolism in C3 leaves, which make it the ready candidate for optimizing the performance of C2 photosynthesis, which in turn positions it to be further upregulated as the benefits of the nascent C4 cycle improve fitness (Heckmann et al., 2013). The convergence upon enhanced CA activity then becomes necessary to enable PEPCase to have enough bicarbonate to maintain rapid activity.
As the most heavily studied enzyme in C4 photosynthesis, the sequence of the PEPCase gene serves as a model of how convergence versus divergence can operate at the gene sequence level. Convergence is apparent in that PEPC1 was separately co-opted for C4 function in the Allionia and Okenia/Boerhavia lineages, while divergence is apparent in the sequence of PEPC1 in these clades, as indicated by the distinct branches on the gene trees and variation at specific sites in the amino acid sequences. Is this sequence divergence random, or could it reflect divergent solutions for optimizing PEPcase function in C4 leaves? In C4 plants, the affinity of PEPCase for its substrate bicarbonate should be increased to compensate for subsaturating bicarbonate concentrations in M tissues (Gowik and Westhoff, 2011; Di Mario and Cousins, 2019). Also, C4 orthologs of PEPCase require reduced sensitivity to malate and aspartate, because these allosteric inhibitors of PEPCase must accumulate to high concentration in M cells to drive rapid diffusion into the BS (Jacobs et al., 2008; Gowik and Westhoff, 2011). Numerous studies have documented similar changes in the amino acid sequence of PEPCase in a pattern that is associated with changes in sensitivity to malate, aspartate and PEP, supporting hypotheses of convergent optimization of PEPCase for the C4 leaf (Engelmann et al., 2003; Gowik et al., 2006; Christin et al., 2007). Convergence is indicated by a widely noted substitution of a serine for alanine at a homologous site in the PEPC sequence, position 780 in maize, which is proposed to alter PEP and possibly bicarbonate sensitivity (Christin et al., 2007; Gowik and Westhoff, 2011; Di Mario and Cousins, 2019). Christin et al. (2007) also observed common sequence substitutions in numerous distinct clades of C4 grasses, some of which occur in gene regions controlling sensitivity to malate. However, transcriptome surveys of numerous chenopods indicate the sequence convergence observed in grasses is less common in eudicots, suggesting divergent solutions to meeting the kinetic and regulatory requirements of a C4 PEPCase (Rosnow et al., 2014, 2015). Our results with Boerhavia and Allionia support a hypothesis of flexibility in how PEPCase is modified for the C4 leaf. Both clades lack the serine at the position near 780, instead sharing an alanine with their C3 sisters. They also exhibit differences in half of the sequence positions where Christin et al. (2007) noted convergence in grasses, but do exhibit some of the same substitutions as C4 grasses and certain C4 eudicots (at positions 572, 577, 761, and 807; Table 5). These patterns raise a number of possibilities. On the one hand, these convergent substitutions may sufficiently alter kinetics to allow the C4 PEPCase to efficiently function. Alternatively, other sequence shifts may be able to accomplish the necessary change in sensitivities. A third possibility is the C4 Nyctaginaceae species may simply have a non-optimized PEPCase for the C4 function, in which case other mechanisms may compensate for inefficiencies. One compensation mechanism may be environmental, for example, high temperature in the natural habitat may override a need for convergence in PEPCase sequences by stimulating catalytic capacity. Where convergence is beneficial, but not accomplished, the consequences and compensation mechanisms become as interesting as the convergence itself.
We next consider the evolutionary transition at the opposite end of the C4 pathway, where there is clear divergence in decarboxylase function. The divergence arises because there are enzymatic alternatives to meeting the decarboxyation imperative, with NADP-ME, NAD-ME, and PCK having significant roles in numerous metabolic pathways in C3 plants (Aubry et al., 2011). Elevated activities of NADP-ME, NAD-ME, and PCK have been detected in vascular tissues and BS cells of C3 species, where they metabolize organic acids used in long-distance transport from the roots, and may have a role in pH homeostasis, coordinating carbon and nitrogen metabolism, and providing metabolites for numerous biosynthetic pathways (Hibberd and Quick, 2002; Aubry et al., 2011). The mechanism by which one decarboxylase is selected over another is not known, but since all three are active in BS tissues, it is possible that each may function in a nascent C4 pathway, perhaps to support recovery of photorespired nitrogen in the BS (Mallmann et al., 2014). It may be that chance determines which of the three decarboxylases is upregulated as the C4 pathway strengthens, with convergence on the distinctive subtype characteristics occurring afterward, as the C4 pathway is optimized for the selected decarboxylase. Alternatively, pre-existing traits in the C3 or C2 ancestors may determine which decarboxylase is selected and also influence the characteristics of each C4 subtype.
To close, we note that comparative studies of evolutionary convergence and divergence in C4 plants show that convergence is greatest where constraints are high and biochemical options are limited, while divergence occurs where the constraints are low and multiple alternative solutions can be selected during the evolutionary process. What remains unclear is the influence that chance, ancestry, and environment have over divergent possibilities. By demonstrating multiple sets of closely related clades differing in C4 subtype, we have identified replicated examples with which to address these issues using comparative methods of evolutionary biology (Harvey and Pagel, 1991). C4 photosynthesis can thus become a powerful tool to unravel the intricacies of convergence in complex trait evolution.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RK led the imaging efforts and their quantification and ecological data analysis. MS conducted the phylotranscriptomic and gene sequence analyses. SA performed gas exchange and enzyme assays. TS and RS directed the labs where the work was conducted and supplied funding. RS wrote the manuscript with input from each co-author. All authors contributed to the article and approved the submitted version.
This paper is dedicated to the memory of Dr. Udo Gowik (1971–2020), a fun-loving friend, helpful to all, a pioneer in C4 plant biology and a fearsome political debater.
This research was supported by Canadian Natural Science and Engineering Research Council grants RGPIN-04878-2015 to TS and RGPIN-2017-06476 to RS. The contribution of SA was supported by a travel grant from the Institute of Global Innovation Research, Tokyo University of Agriculture and Technology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Nina Esmail and Perlina Lim for assistance in plant growth and the fixation and embedding of leaves for microscopic imaging. We also thank Gerry Edwards and Elena Vosnesenskaya for the gift of Portulaca pilosa seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.578739/full#supplementary-material
Aberer, A. J., Kobert, K., and Stamatakis, A. (2014). ExaBayes: massively Bayesian tree inference for the whole-genome era. Mol. Biol. Evol. 31, 2553–2556. doi: 10.1093/molbev/msu236
Alonso-Cantabrana, H., and von Caemmerer, S. (2016). Carbon isotope discrimination as a diagnostic tool for C4 photosynthesis in C3–C4 intermediate species. J. Exp. Bot. 67, 3109–3121. doi: 10.1093/jxb/erv555
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Ashton, A. R., Burnell, J. N., Furbank, R. T., Jenkins, C. L. D., and Hatch, M. D. (1990). “Enzymes of C4 photosynthesis,” in Methods in Plant Biochemistry, Vol. 7A, ed. P. J. Lea, (London: Academic Press), 39–72. doi: 10.1016/B978-0-12-461013-2.50010-1
Aubry, S., Brown, N. J., and Hibberd, J. M. (2011). The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot. 62, 3049–3059. doi: 10.1093/jxb/err012
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Bissinger, K., Khoshravesh, R., Kotrade, J. P., Oakley, J., Sage, T. L., Sage, R. F., et al. (2014). Gisekia (Gisekiaceae): phylogenetic relationships, biogeography, and ecophysiology of a poorly known C4 lineage in the Caryophyllales. Am. J. Bot. 101, 499–509. doi: 10.3732/ajb.1300279
Bivand, R., and Lewin-Koh, N. (2013). maptools: Tools for Reading and Handling Spatial Objects. R Package Version 0.8–27. Available online at: https://cran.r-project.org/web/packages/maptools/index.html (accessed August 24, 2020).
Blount, Z. D., Lenski, R. E., and Losos, J. B. (2018). Contingency and determinism in evolution: replaying life’s tape. Science 362:eaam5979. doi: 10.1126/science.aam5979
Bohley, K., Joos, O., Hartmann, H., Sage, R. F., Liede-Schumann, S., and Kadereit, G. (2015). phylogeny of sesuvioideae (Aizoaceae)-biogeography, leaf anatomy and the evolution of C4photosynthesis. Perspect. Plant Ecol. 17, 116–130.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bräutigam, A., and Gowik, U. (2016). Photorespiration connects C3 and C4 photosynthesis. J. Exp. Bot. 67, 2953–2962.
Bräutigam, A., and Weber, A. P. M. (2011). “Transport processes: connecting the reactions of C4 photosynthesis,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage, (Dordrecht: Springer Netherlands), 277–300.
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carolin, R., Jacobs, S., and Vesk, M. (1978). Kranz cells and mesophyll in the Chenopodiales. Aust. J. Bot. 26:683. doi: 10.1071/BT9780683
Christin, P.-A., Besnard, G., Samaritani, E., Duvall, M. R., Hodkinson, T. R., Savolainen, V., et al. (2008). Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18, 37–43. doi: 10.1016/j.cub.2007.11.058
Christin, P.-A., Boxall, S. F., Gregory, R., Edwards, E. J., Hartwell, J., and Osborne, C. P. (2013a). Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. Evol. 5, 2174–2187. doi: 10.1093/gbe/evt168
Christin, P.-A., Edwards, E. J., Besnard, G., Boxall, S. F., Gregory, R., Kellogg, E. A., et al. (2012a). Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Curr. Biol. 22, 445–449. doi: 10.1016/j.cub.2012.01.054
Christin, P.-A., Freckleton, R. P., and Osborne, C. P. (2010a). Can phylogenetics identify C4 origins and reversals? Trends Ecol. Evol. 25, 403–409. doi: 10.1016/j.tree.2010.04.007
Christin, P.-A., and Osborne, C. P. (2013). The recurrent assembly of C4 photosynthesis, an evolutionary tale. Photosynth. Res. 117, 163–175. doi: 10.1007/s11120-013-9852-z
Christin, P.-A., Osborne, C. P., Chatelet, D. S., Columbus, J. T., Besnard, G., Hodkinson, T. R., et al. (2013b). Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc. Natl. Acad. Sci. U.S.A. 110, 1381–1386. doi: 10.1073/pnas.1216777110
Christin, P. A., Osborne, C. P., Sage, R. F., Arakaki, M., and Edwards, E. J. (2011). C4 eudicots are not younger than C4 monocots. J. Exp. Bot. 62, 3171–3181. doi: 10.1093/jxb/err041
Christin, P.-A., Salamin, N., Savolainen, V., Duvall, M. R., and Besnard, G. (2007). C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr. Biol. 17, 1241–1247. doi: 10.1016/j.cub.2007.06.036
Christin, P.-A., Samaritani, E., Petitpierre, B., Salamin, N., and Besnard, G. (2009). Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol. Evol. 1, 221–230. doi: 10.1093/gbe/evp020
Christin, P.-A., Wallace, M. J., Clayton, H., Edwards, E. J., Furbank, R. T., Hattersley, P. W., et al. (2012b). Multiple photosynthetic transitions, polyploidy, and lateral gene transfer in the grass subtribe Neurachninae. J. Exp. Bot. 63, 6297–6308. doi: 10.1093/jxb/ers282
Christin, P. A., Weinreich, D. M., and Besnard, G. (2010b). Causes and evolutionary significance of genetic convergence. Trends Genet. 26, 400–405. doi: 10.1016/j.tig.2010.06.005
Conway-Morris, S. (2003). Life’s Solutions: Inevitable Humans in a Lonely Universe. Cambridge: Cambridge University Press.
Danila, F. R., Quick, W. P., White, R. G., Kelly, S., von Caemmerer, S., and Furbank, R. T. (2018). Multiple mechanisms for enhanced plasmodesmata density in disparate subtypes of C4 grasses. J. Exp. Bot. 69, 1135–1145. doi: 10.1093/jxb/erx456
Dengler, N. G., and Nelson, T. (1999). “Leaf structure and development in C4 plants,” in C4 Plant Biology, eds R. F. Sage and R. K. Monson, (San Diego: Academic Press), 133–172.
Di Mario, R. J., and Cousins, A. B. (2019). A single serine to alanine substitution decreases bicarbonate affinity of phosphoenol pyruvate carboxylase in C4 Flaveria trinervia. J. Exp. Bot. 70, 995–1004. doi: 10.1093/jxb/ery403
Douglas, N. A., and Manos, P. S. (2007). Molecular phylogeny of Nyctaginaceae: taxonomy, biogeography, and characters associated with a radiation of xerophytic genera in North America. Am. J. Bot. 94, 856–872. doi: 10.3732/ajb.94.5.856
Douglas, N. A., and Spellenberg, R. (2010). A new tribal classification of Nyctaginaceae. Taxon 59, 905–910. doi: 10.1002/TAX.593018
Drincovich, M. F., Lara, M. V., Andreo, C. S., and Maurino, V. G. (2011). “C4 decarboxylases: different solutions for the same biochemical problem, the provision of CO2 to Rubisco in the bundle sheath cells,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage, (Dordrecht: Springer Netherlands), 277–300.
Dunning, L. T., Lundgren, M. R., Moreno-Villena, J. J., Namaganda, M., Edwards, E. J., Nosil, P., et al. (2017). Introgression and repeated co-option facilitated the recurrent emergence of C4 photosynthesis among close relatives: complex transitions among photosynthetic types. Evolution 71, 1541–1555. doi: 10.1111/evo.13250
Edwards, E. J. (2019). Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytol. 223, 1742–1755. doi: 10.1111/nph.15851
Edwards, G. E., and Voznesenskaya, E. V. (2011). “C4 photosynthesis: kranz forms and single-cell C4 in terrestrial plants,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage, (Dordrecht: Springer Netherlands), 29–61.
Edwards, G. E., and Walker, D. A. (1983). C3, C4: Mechanism, and Cellular and Environmental Regulation, of Photosynthesis. Oxford: Blackwell Scientific Publications.
Emms, D. M., Covshoff, S., Hibberd, J. M., and Kelly, S. (2016). Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol. Biol. Evol. 33, 1796–1806. doi: 10.1093/molbev/msw057
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Engelmann, S., Bläsing, O. E., Gowik, U., Svensson, P., and Westhoff, P. (2003). Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria? A gradual increase from C3 to C4 characteristics. Planta 217, 717–725. doi: 10.1007/s00425-003-1045-0
Erb, T. J. (2011). Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 77, 8466. doi: 10.1128/AEM.05702-11
Farquhar, G. D., Ehleringer, J. R., and Hubick, K. T. (1989). Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 40, 503–537. doi: 10.1146/annurev.pp.40.060189.002443
Fick, S. E., and Hijmans, R. J. (2017). Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315. doi: 10.1002/joc.5086
Furbank, R. T. (2011). Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108. doi: 10.1093/jxb/err080
Ghannoum, O., Evans, J. R., and Caemmerer, S. (2011). “Nitrogen and water use efficiency of C4 plants”,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage, (Dordrecht: The Netherlands: Springer), 129–146.
Gowik, U., Bräutigam, A., Weber, K. L., Weber, A. P. M., and Westhoff, P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23, 2087–2105. doi: 10.1105/tpc.111.086264
Gowik, U., Engelmann, S., Bläsing, O. E., Raghavendra, A. S., and Westoff, P. (2006). Evolution of C4 phosphoenolpyruvate carboxylase in the genus Alternanthera: gene families and the enzymatic characterisitics of the C4 isozyme and its orthologues in C3 and C4 Alternanthera. Planta 223, 359–368. doi: 10.1007/s00425-005-0085-z
Gowik, U., and Westhoff, P. (2011). “C4-phosphoenolpyruvate carboxylase,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage, (Dordrecht: Springer Netherlands), 257–275.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Griffiths, H., Weller, G., Toy, L. F. M., and Dennis, R. J. (2013). You’re so vein: bundle sheath physiology, phylogeny and evolution in C3 and C4 plants: origins and function of bundle sheath in C3 plants. Plant Cell Environ. 36, 249–261. doi: 10.1111/j.1365-3040.2012.02585
Gutierrez, M., Gracen, V. E., and Edwards, G. E. (1974). Biochemical and cytological relationships in C4 plants. Planta 119, 279–300. doi: 10.1007/BF00388331
Harvey, P. H., and Pagel, M. D. (1991). The Comparative Method in Evolutionary Biology. Oxford: Oxford Press.
Hatch, M., Kagawa, T., and Craig, S. (1975). Subdivision of C4-pathway species based on differing C4 acid decarboxylating systems and ultrastructural features. Funct. Plant Biol. 2:111. doi: 10.1071/PP9750111
Hatch, M. D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta Rev. Bioenerget. 895, 81–106. doi: 10.1016/S0304-4173(87)80009-5
Hattersley, P. W. (1992). “C4 photosynthetic pathway variation in grasses (Poaceae): its significance for arid and semi-arid lands,” in Desertified Grasslands: their Biology and Management, ed. G. P. Chapman, (London: Academic Press), 181–212.
Heckmann, D., Schulze, S., Denton, A., Gowik, U., Westhoff, P., Weber, A. P. M., et al. (2013). Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. doi: 10.1016/j.cell.2013.04.058
Heyduk, K., Moreno-Villena, J. J., Gilman, I. S., Christin, P.-A., and Edwards, E. J. (2019). The genetics of convergent evolution: insights from plant photosynthesis. Nat. Rev. Genet. 20, 485–493. doi: 10.1038/s41576-019-0107-5
Hibberd, J. M., and Quick, W. P. (2002). Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454.
Hijmans, R. J., and van Etten, J. (2012). raster: Geographic Analysis and Modeling with Raster Data. R package Version 2.0-12. Available online at: http://CRAN.R-project.org/package=raster (accessed July 17, 2020).
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jacobs, B., Engelmann, S., Westhoff, P., and Gowik, U. (2008). Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria: determinants for high tolerance towards the inhibitor L-malate. Plant Cell Environ. 31, 793–803. doi: 10.1111/j.1365-3040.2008.01796
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S., and Hibberd, J. M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75. doi: 10.1104/pp.114.238667
Kadereit, G., Ackerly, D., and Pirie, M. D. (2012). A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proc. R. Soc. B. 279, 3304–3311.
Kadereit, G., Lauterbach, M., Pirie, M. D., Arafeh, R., and Freitag, H. (2014). When do different C4 leaf anatomies indicate independent C4 origins? Parallel evolution of C4 leaf types in Camphorosmeae (Chenopodiaceae). J. Exp. Bot. 65, 3499–3511. doi: 10.1093/jxb/eru169
Kanai, R., and Edwards, G. (1999). “The biochemistry of C4 photosynthesis,” in C4 Plant Biology, eds R. F. Sage and R. K. Monson, (San Diego, CA: Academic Press), 49–87.
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khoshravesh, R., Akhani, H., Sage, T. L., Nordenstam, B., and Sage, R. F. (2012). Phylogeny and photosynthetic pathway distribution in Anticharis Endl. (Scrophulariaceae). J. Exp. Bot. 63, 5645–5658. doi: 10.1093/jxb/ers218
Khoshravesh, R., Lundsgaard-Nielsen, V., Sultmanis, S., and Sage, T. L. (2017). “Light microscopy, transmission electron microscopy, and immunohistochemistry protocols for studying photorespiration,” in Photorespiration: Methods and Protocols, eds A. R. Fernie, H. Bauwe, and A. P. M. Weber, (New York, NY: Springer), 243–270.
Khoshravesh, R., Stata, M., Busch, F. A., Saladié, M., Castelli, J. M., Dakin, N., et al. (2020). The evolutionary origin of C4 photosynthesis in the grass subtribe Neurachninae. Plant Physiol. 182, 566–583. doi: 10.1104/pp.19.00925
Khoshravesh, R., Stinson, C. R., Stata, M., Busch, F. A., Sage, R. F., Ludwig, M., et al. (2016). C3–C4 intermediacy in grasses: organelle enrichment and distribution, glycine decarboxylase expression, and the rise of C2 photosynthesis. J. Exp. Bot. 67, 3065–3078. doi: 10.1093/jxb/erw150
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim, I., and Fisher, D. G. (1990). Structural aspects of the leaves of seven species of Portulaca growing in Hawaii. Can. J. Bot. 68, 1803–1811. doi: 10.1139/b90-233
Lauterbach, M., Zimmer, R., Alexa, A. C., Adachi, S., Sage, R., Sage, T., et al. (2019). Variation in leaf anatomical traits relates to the evolution of C4 photosynthesis in Tribuloideae (Zygophyllaceae). Perspect. Plant Ecol. 39:125463. doi: 10.1016/j.ppees.2019.125463
Lê, S., Josse, J., and Husson, F. (2008). FactoMineR: a package for multivariate analysis. J. Stat. Softw. 25, 1–18. doi: 10.18637/jss.v025.i01
Leegood, R. C., and Walker, R. O. (1999). “Regulation of the C4 pathway,” in C4 Plant Biology, eds R. F. Sage and R. K. Monson, (San Diego, CA: Academic Press), 89–131.
Losos, J. B. (2011). Convergence, adaptation, and constraint. Evolution 65, 1827–1840. doi: 10.1111/j.1558-5646.2011.01289
Ludwig, M. (2016a). Evolution of carbonic anhydrases in C4 plants. Curr. Opin. Plant Sci. 31, 16–22. doi: 10.1016/j.pbi.2016.03.003
Ludwig, M. (2016b). The roles of organic acids in C4 photosynthesis. Front. Plant Sci. 7:647. doi: 10.3389/fpls.2016.00647
Mallmann, J., Heckmann, D., Bräutigam, A., Lercher, M. J., Weber, A. P., Westhoff, P., et al. (2014). The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 3:e02478. doi: 10.7554/eLife.02478
Monson, R. K., Teeri, J. A., Ku, M. S. B., Gurevitch, J., Mets, L. J., and Dudley, S. (1988). Carbon-isotope discrimination by leaves of Flaveria species exhibiting different amounts of C3–and C4-cycle co-function. Planta 174, 145–151. doi: 10.1007/BF00394765
Moore, B. D., Ku, M. S. B., and Edwards, G. E. (1989). Expression of C4-like photosynthesis in several species of Flaveria. Plant Cell Environ. 12, 541–549. doi: 10.1111/j.1365-3040.1989.tb02127
Muhaidat, R., Sage, R. F., and Dengler, N. G. (2007). Diversity of Kranz anatomy and biochemistry in C4 eudicots. Am. J. Bot. 94, 362–381. doi: 10.3732/ajb.94.3.362
Muhaidat, R., Sage, T. L., Frohlich, M. W., Dengler, N. G., and Sage, R. F. (2011). Characterization of C3–C4 intermediate species in the genus Heliotropium L. (Boraginaceae): anatomy, ultrastructure and enzyme activity. Plant Cell Environ. 34, 1723–1736. doi: 10.1111/j.1365-3040.2011.02367
Oakley, J. C., Sultmanis, S., Stinson, C. R., Sage, T. L., and Sage, R. F. (2014). Comparative studies of C3 and C4 Atriplex hybrids in the genomics era: physiological assessments. J. Exp. Bot. 65, 3637–3647. doi: 10.1093/jxb/eru106
Ocampo, G., Koteyeva, N. K., Voznesenskaya, E. V., Edwards, G. E., Sage, T. L., Sage, R. F., et al. (2013). Evolution of leaf anatomy and photosynthetic pathways in Portulacaceae. Am. J. Bot. 100, 2388–2402. doi: 10.3732/ajb.1300094
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Pyankov, V. I., Voznesenskaya, E. V., Kuz’min, A. N., Ku, M. S. B., Ganko, E., Franceschi, V. R., et al. (2000). Occurrence of C3 and C4 photosynthesis in cotyledons and leaves of Salsola species (Chenopodiaceae). Photosynth. Res. 63, 69–84. doi: 10.1023/A:1006377708156
Rao, X., and Dixon, R. A. (2016). The differences between NAD-ME and NADP-ME subtypes of C4 photosynthesis: more than decarboxylating enzymes. Front. Plant Sci. 7:1525. doi: 10.3389/fpls.2016.01525
Revell, L. J. (2012). phytools: an R package for phylogenetic comparative biology (and other things).” Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169
Reyna-Llorens, I., and Hibberd, J. M. (2017). Recruitment of pre-existing networks during the evolution of C4 photosynthesis. Phil. Trans. R. Soc. B 372:20160386. doi: 10.1098/rstb.2016.0386
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277.
Rosnow, J. J., Edwards, G. E., and Roalson, E. H. (2014). Positive selection of Kranz and non-Kranz C4 phosphoenolpyruvate carboxylase amino acids in Suaedoideae (Chenopodiaceae). J. Exp. Bot. 65, 3595–3607. doi: 10.1093/jxb/eru053
Rosnow, J. J., Evans, M. A., Kapralov, M. V., Cousins, A. B., Edwards, G. E., and Roalson, E. H. (2015). Kranz and single-cell forms of C4 plants in the subfamily Suaedoideae show kinetic C4 convergence for PEPC and Rubisco with divergent amino acid substitutions. J. Exp. Bot. 66, 7347–7358. doi: 10.1093/jxb/erv431
Sage, R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. doi: 10.1111/j.1469-8137.2004.00974
Sage, R. F. (2016). A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and Hall of Fame. J. Exp. Bot. 67, 4039–4056. doi: 10.1093/jxb/erw156
Sage, R. F., Christin, P.-A., and Edwards, E. J. (2011). The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169. doi: 10.1093/jxb/err048
Sage, R. F., Khoshravesh, R., and Sage, T. L. (2014). From proto-Kranz to C4 Kranz: building the bridge to C4 photosynthesis. J. Exp. Bot. 65, 3341–3356. doi: 10.1093/jxb/eru180
Sage, R. F., Monson, R. K., Ehleringer, J. R., Adachi, S., and Pearcy, R. W. (2018). Some like it hot: the physiological ecology of C4 plant evolution. Oecologia 187, 941–966. doi: 10.1007/s00442-018-4191-6
Sage, R. F., Sage, T. L., and Kocacinar, F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47. doi: 10.1146/annurev-arplant-042811-105511
Sage, T. L., Busch, F. A., Johnson, D. C., Friesen, P. C., Stinson, C. R., Stata, M., et al. (2013). Initial events during the evolution of C4 photosynthesis in C3 species of Flaveria. Plant Physiol. 163, 1266–1276. doi: 10.1104/pp.113.221119
Sage, T. L., Sage, R. F., Vogan, P. J., Rahman, B., Johnson, D. C., Oakley, J. C., et al. (2011). The occurrence of C2 photosynthesis in Euphorbia subgenus Chamaesyce (Euphorbiaceae). J. Exp. Bot. 62, 3183–3195. doi: 10.1093/jxb/err059
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schulze, S., Mallmann, J., Burscheidt, J., Koczor, M., Streubel, M., Bauwe, H., et al. (2013). Evolution of C4 photosynthesis in the genus Flaveria: establishment of a photorespiratory CO2 pump. Plant Cell 25, 2522–2535. doi: 10.1105/tpc.113.114520
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Stata, M., Sage, T. L., Rennie, T. D., Khoshravesh, R., Sultmanis, S., Khaikin, Y., et al. (2014). Mesophyll cells of C4 plants have fewer chloroplasts than those of closely related C3 plants: C3 versus C4 mesophyll chloroplasts. Plant Cell Environ. 37, 2587–2600. doi: 10.1111/pce.12331
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
Taub, D. R. (2000). Climate and the U.S. distribution of C4 grass subfamilies and decarboxylation variants of C4 photosynthesis. Am. J. Bot. 87, 1211–1215. doi: 10.2307/2656659
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313x.2004.02016.x
Tropicos (2020). Tropicos.org. Missouri Botanical Garden http://www.tropicos.org (accessed October 20, 2020).
Tsuji, Y., Suzuki, I., and Shiraiwa, Y. (2012). Enzymological evidence for the function of a plastid-located pyruvate carboxylase in the haptophyte alga Emiliania huxleyi: a novel pathway for the production of C4 compounds. Plant Cell Physiol. 53, 1043–1052. doi: 10.1093/pcp/pcs045
Ueno, O. (1996). Structural characterization of photosynthetic cells in an amphibious sedge, Eleocharis vivipara, in relation to C3 and C4 metabolism. Planta 199, 382–393. doi: 10.1007/BF00195730
Ueno, O. (2013). Ultrastructure and carbon isotope ratios of leaves in C4 species of Rhynchospora (Cyperaceae) that differ in the location of Kranz cells. Int. J. Plant Sci. 174, 702–709. doi: 10.1086/669912
Vogel, J. C., Fuls, A., and Ellis, R. P. (1978). The geographical distribution of Kranz grasses in South Africa. S. Afr. J. Sci. 74, 209–215.
Von Caemmerer, S. (1992). Carbon isotope discrimination in C3−C4 intermediates. Plant Cell Environ. 15, 1063–1072. doi: 10.1111/j.1365-3040.1992.tb01656
von Caemmerer, S., and Furbank, R. T. (2003). The C4 pathway: an efficient CO2 pump. Photosynth. Res. 77, 191–207. doi: 10.1023/A:1025830019591
Voznesenskaya, E. V., Akhani, H., Koteyeva, N. K., Chuong, S. D. X., Roalson, E. H., Kiirats, O., et al. (2008). Structural, biochemical, and physiological characterization of photosynthesis in two C4 subspecies of Tecticornia indica and the C3 species Tecticornia pergranulata (Chenopodiaceae). J. Exp. Bot. 59, 1715–1734. doi: 10.1093/jxb/ern028
Voznesenskaya, E. V., Chuong, S. D. X., Koteyeva, N. K., Franceschi, V. R., Freitag, H., and Edwards, G. E. (2007). Structural, biochemical, and physiological characterization of C4 photosynthesis in species having two vastly different types of Kranz anatomy in genus Suaeda (Chenopodiaceae). Plant Biol. 9, 745–757. doi: 10.1055/s-2007-965579
Voznesenskaya, E. V., Franceschi, V. R., Chuong, S. D. X., and Edwards, G. E. (2006). Functional characterization of phosphoenolpyruvate carboxykinase-type C4 leaf anatomy: immuno-, cytochemical and ultrastructural analyses. Ann. Bot. 98, 77–91. doi: 10.1093/aob/mcl096
Voznesenskaya, E. V., Franceschi, V. R., Kiirats, O., Artyusheva, E. G., Freitag, H., and Edwards, G. E. (2002). Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J. 31, 649–662. doi: 10.1046/j.1365-313X.2002.01385
Voznesenskaya, E. V., Franceschi, V. R., Pyankov, V. I., and Edwards, G. E. (1999). Anatomy, chloroplast structure and compartmentation of enzymes relative to photosynthetic mechanisms in leaves and cotyledons of species in the tribe Salsoleae (Chenopodiaceae). J. Exp. Bot. 50, 1779–1795. doi: 10.1093/jxb/50.341.1779
Voznesenskaya, E. V., Koteyeva, N. K., Cousins, A., and Edwards, G. E. (2018). Diversity in structure and forms of carbon assimilation in photosynthetic organs in Cleome (Cleomeaceae). Funct. Plant Biol. 45:983. doi: 10.1071/FP17323
Voznesenskaya, E. V., Koteyeva, N. K., Edwards, G. E., and Ocampo, G. (2010). Revealing diversity in structural and biochemical forms of C4 photosynthesis and a C3–C4 intermediate in genus Portulaca L. (Portulacaceae). J. Exp. Bot. 61, 3647–3662. doi: 10.1093/jxb/erq178
Keywords: Allionia, Boerhavia, C4 photosynthesis, convergent evolution, Nyctaginaceae phylogeny, PEP carboxylase
Citation: Khoshravesh R, Stata M, Adachi S, Sage TL and Sage RF (2020) Evolutionary Convergence of C4 Photosynthesis: A Case Study in the Nyctaginaceae. Front. Plant Sci. 11:578739. doi: 10.3389/fpls.2020.578739
Received: 01 July 2020; Accepted: 06 October 2020;
Published: 02 November 2020.
Edited by:
Tingshuang Yi, Kunming Institute of Botany, Chinese Academy of Sciences, ChinaReviewed by:
Isabel Larridon, Royal Botanic Gardens, Kew, United KingdomCopyright © 2020 Khoshravesh, Stata, Adachi, Sage and Sage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rowan F. Sage, ci5zYWdlQHV0b3JvbnRvLmNh
†ORCID: Roxana Khoshravesh, orcid.org/0000-0002-1766-8993; Matt Stata, orcid.org/0000-0002-5744-4898; Shunsuke Adachi, orcid.org/0000-0003-0471-3369; Tammy L. Sage, orcid.org/0000-0002-7061-832X; Rowan F. Sage, orcid.org/0000-0001-6183-9246
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.