
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 November 2020
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.577676
This article is part of the Research Topic Cytokinins as Central Regulators of Plant Growth and Development View all 14 articles
 Maria V. Yamburenko1†
Maria V. Yamburenko1† Jennifer M. Worthen1†
Jennifer M. Worthen1† Asyia Zeenat1,2
Asyia Zeenat1,2 Beenish J. Azhar1,2
Beenish J. Azhar1,2 Swadhin Swain1
Swadhin Swain1 Adam R. Couitt1
Adam R. Couitt1 Samina N. Shakeel1,2
Samina N. Shakeel1,2 Joseph J. Kieber3
Joseph J. Kieber3 G. Eric Schaller1*
G. Eric Schaller1*The phytohormone cytokinin plays a critical role in regulating growth and development throughout the life cycle of the plant. The primary transcriptional response to cytokinin is mediated by the action of the type-B response regulators (RRs), with much of our understanding for their functional roles being derived from studies in the dicot Arabidopsis. To examine the roles played by type-B RRs in a monocot, we employed gain-of-function and loss-of-function mutations to characterize RR22 function in rice. Ectopic overexpression of RR22 in rice results in an enhanced cytokinin response based on molecular and physiological assays. Phenotypes associated with enhanced activity of RR22 include effects on leaf and root growth, inflorescence architecture, and trichome formation. Analysis of four Tos17 insertion alleles of RR22 revealed effects on inflorescence architecture, trichomes, and development of the stigma brush involved in pollen capture. Both loss‐ and gain-of-function RR22 alleles affected the number of leaf silica-cell files, which provide mechanical stability and improve resistance to pathogens. Taken together, these results indicate that a delicate balance of cytokinin transcriptional activity is necessary for optimal growth and development in rice.
The phytohormone cytokinin plays a critical role in regulating growth and development throughout the life cycle of the plant, including the regulation of cell proliferation, shoot and root architecture, seed yield, senescence, and stress responses (Sakakibara, 2006; Hwang et al., 2012; Kieber and Schaller, 2014; Jameson and Song, 2016). Much of our understanding of the mechanisms that underlie metabolism and perception of cytokinin come from studies of the model dicot Arabidopsis (Hwang et al., 2012; Kieber and Schaller, 2014). More recently, such studies have been extended to monocots due to their agronomic importance, with rice being a preferred monocot species for such studies in part because of its small genome and ease of transformation (Devos and Gale, 2000). Of particular interest has been the discovery that cytokinin plays a key role in regulating the development and architecture of the rice inflorescence, alterations in cytokinin levels accounting for differences in the yield for several rice varieties (Ashikari et al., 2005; Kurakawa et al., 2007; Gu et al., 2015; Yeh et al., 2015).
The cytokinin signal is transduced through a multi-step phosphorelay that incorporates cytokinin receptors with histidine-kinase (HK) activity, histidine-containing phosphotransfer proteins (AHPs), and type-B response regulators (RRs; Ito and Kurata, 2006; Pareek et al., 2006; Du et al., 2007; Schaller et al., 2007; Werner and Schmülling, 2009; Tsai et al., 2012; Kieber and Schaller, 2014). Activation of the receptors initiates the transfer of phosphate from one signaling element to the next, thereby relaying the cytokinin signal from membrane to nucleus. Within the nucleus, the type-B RRs function as transcription factors to regulate the expression of cytokinin primary-response genes. Among the primary-response genes are a second family of RRs, the type-A RRs, which function as negative feedback regulators to desensitize the plant to cytokinin (To et al., 2004; Kieber and Schaller, 2014). The KMD family of F-box proteins targets the type-B RRs for degradation (Kim et al., 2013a,b). In rice, as in most plants, these signaling elements are encoded by multi-gene families, there being 13-type-B RRs and 13 type-A RRs in the rice genome (Pareek et al., 2006; Du et al., 2007; Schaller et al., 2007; Tsai et al., 2012).
According to this model for the cytokinin signal transduction pathway, the type-B RRs play a key role in regulating the initial transcriptional response to cytokinin. The type-B RRs contain two conserved signaling motifs: a receiver domain that is phosphorylated on a conserved aspartate residue to regulate their activity, and a long C-terminal extension with a Myb-like DNA-binding domain (Imamura et al., 1999; Hosoda et al., 2002). Multiple lines of evidence support the role of type-B RRs as transcription factors (Sakai et al., 2000, 2001; Hwang and Sheen, 2001; Imamura et al., 2001, 2003; Lohrmann et al., 2001; Hosoda et al., 2002; Mason et al., 2004, 2005; Rashotte et al., 2006; Liang et al., 2012; Tsai et al., 2012), and our recent analyses using protein-binding microarrays reveal similar DNA-binding motifs for the Myb-like domains of the type-B RRs from rice and Arabidopsis (Raines et al., 2016; Zubo et al., 2017). The type-B RRs fall into five subfamilies based on phylogenetic analysis of rice and Arabidopsis (Tsai et al., 2012), with subfamily-1 being the only subfamily to contain type-B RR members from both rice and Arabidopsis. Genetic analysis in Arabidopsis indicates that subfamily-1 plays the most prominent role in cytokinin signaling (Mason et al., 2005; Yokoyama et al., 2007; Argyros et al., 2008; Ishida et al., 2008).
We have employed several approaches to functionally characterize the role of the rice type-B RRs. Initially, we determined that the rice RR22 of subfamily-1 can complement an Arabidopsis type-B RR mutant (arr1/12), consistent with rice subfamily-1 also mediating cytokinin responses (Tsai et al., 2012). More recently, we employed a CRISPR/Cas9 gene editing approach to target the four most abundant type-B RRs of rice subfamily-1: RR21, RR22, RR23, and RR24. Results from this analysis revealed functional overlap as well as subfunctionalization within the type-B RR gene family, the rr21/22/23 triple mutant exhibiting decreased cytokinin sensitivity and a variety of defects in growth and development, including effects on shoot and root growth, panicle architecture, flower development, and trichome formation (Worthen et al., 2019). Here, we employ a gain-of-function approach to characterize the effects of ectopic overexpression of RR22 in rice. To complement this gain-of-function approach, we also examined loss-of-function Tos17 insertion alleles of RR22. Results from these analyses indicate that a delicate balance of cytokinin transcriptional activity is necessary for optimal growth and development in rice.
For overexpression of RR22, a GFP-tagged version was driven by the ZmUbi promoter (Mann et al., 2012) in the pCAMBIA1300 vector (GenBank Accession Number AF234296). For this purpose, sGFP along with the NOS terminator were amplified by PCR from the plasmid pGWB5 (Nakagawa et al., 2007), and this sGFP-tNOS fragment inserted into the Pst I and Hind III sites of pCambia1300 by an In-Fusion reaction according to the manufacturer (Takara Bio United States, Inc.), creating the vector pCambia1300-MCS-GFP. The ZmUbi1 promoter was amplified from the pANIC6A vector (Mann et al., 2012), and a genomic version of RR22 amplified from rice genomic DNA, and both fragments cloned into the Xma I and Pst I sites of the pCambia1300-MCS-GFP vector by an In-Fusion reaction. Primers used for cloning are listed in Supplementary Table 1.The resulting pZmUbi1:RR22-GFP:tNOS vector was confirmed by restriction digest and sequencing. Transformation of Oryza sativa ssp. japonica cv. Kitaake rice with pZmUbi1:RR22-GFP: tNOS vector in the EHA105 strain of Agrobacterium was performed by the Plant Transformation Facility at Iowa State University.1 Three RR22-OX transgenic lines, characterized by a single T-DNA insertion and high expression of the transgene, were selected for further characterization. Wild-type (WT) siblings isolated from segregating populations for each of the transgenic lines were retained as controls.
For isolation of RR22 lines containing Tos17 insertions, seed was obtained from the Tos17 Rice Insertion Mutant (TRIM) collection at the National Institute of Agrobiological Sciences (NIAS) in Japan2 (Miyao et al., 2003; Hirochika et al., 2004). Four Tos17 insertion alleles for RR22 designated rr22-1 (NF0017), rr22-2 (ND3038), rr22-3 (NF6804), and rr22-4 (NG4931) were obtained for Oryza sativa ssp. japonica cv. Nipponbare. PCR-based screening of DNA isolated from mature leaves by the CTAB extraction method (Clarke, 2009) was used to identify homozygous rr22 mutant lines and their WT siblings, using the primers listed in Supplementary Table 1. The locations for the Tos17 insertions were confirmed by sequencing (Supplementary Table 2). The rr21/22/23 CRISPR-Cas9 derived line has been previously described (Worthen et al., 2019).
Plants were grown in a greenhouse at 30°C during the day and 25°C at night, using a 10-h light/14-h dark cycle as described (Worthen et al., 2019). Panicle parameters were quantified as described, using the largest panicle from each plant characterized (Worthen et al., 2019). To visualize stigma brush hairs and trichomes, spikelets and hulls were dissected from plants right before anthesis and images captured using a Leica MZ16 microscope with a Spot Idea camera. To quantify stigma brush hair length for each line, we imaged 10 stigma brushes, with two or three stigmas isolated from each of four plants, and measured the five longest branches on each stigma brush using ImageJ. To quantify length of hull trichomes for each line, we imaged 10 hull tips, with two or three hulls imaged from each of four plants after anthesis, and measured the length of the five longest trichomes on each image.
To characterize leaf epidermal cells, flag leaves from the central tillers of mature plants were chosen, and nail polish impressions made of the abaxial side from the central region of the leaf. Approximately 1.5 cm of the leaf surface from one side of the central vein was brushed to cover with a single layer of a transparent nail polish, left on the living plant for 16 h, and then removed with a scalpel. Impressions were made of two leaves from each of five plants for each line characterized. Impressions were imaged with a Nikon 90i microscope in DIC mode and a 20x objective. Measurements of cell length and width were made with ImageJ software for 50 epidermal long cells (LC1; two different cell files from each of five specimens, with five consecutive cells analyzed per cell file; Luo et al., 2012). Cell file counts for the silica/cork cell files were performed using a Zeiss Axioscope, and all files present in the impressions were counted.
For growth of seedlings, seeds were sterilized and germinated as described (Worthen et al., 2019), then transferred to plastic mesh floating on Yoshida medium (Yoshida et al., 1976) supplemented with 50 μg/ml carbenicillin to control bacteria contamination in a dark plastic container. Seedlings were grown for 8 days at 30/28°C with a 12-h light/12-h dark cycle (light = 400 μE). To assess cytokinin sensitivity, the Yoshida medium was supplemented with NaOH as a vehicle control, 10 nM BA, or 100 nM BA.
For RNA sample preparation from shoots of RR22-OX lines and their corresponding WT siblings, germinated seeds were grown in sterile Kimura media as described (Worthen et al., 2019) in Solo Sundae cups (TS5R) with lids (DLR100) at 30/28°C with a 12-h light/12-h dark cycle (light = 100 μE) for 7 days. Seedlings were then removed and treated with 1 μM BA or a NaOH vehicle control for 1 h in liquid Kimura media by placing roots into the solution. Sample preparation from the Tos17 rr22 carpels and seedlings was as described (Worthen et al., 2019). Samples were flash frozen in liquid nitrogen, ground with a tissue homogenizer (Mixer Mill 400, Retsch), and total RNA extracted using the E.Z.N.A Plant RNA Kit (Omega Bio-Tek) as described (Worthen et al., 2019). DNase treatment, first-strand cDNA synthesis, and RT-qPCR with three biological replicates, along with technical replicates, were performed as described (Worthen et al., 2019). Primers used for qRT-PCR are listed in Supplementary Table 1.
Leaf samples were flash frozen in liquid nitrogen, ground with a tissue homogenizer (Mixer Mill 400, Retsch), and total protein isolated by resuspending 50 mg of leaf powder in 500 μl of SDS-PAGE loading buffer composed of 0.12 M Tris-HCl (pH 6.8), 0.1 M EDTA, 4% (w/v) SDS, 10% (v/v) ß-mercaptoethanol, 5% (v/v) glycerol, and 0.005% (w/v) bromophenol blue. The suspension was incubated at 90°C for 10 min, centrifuged at 14,000 g for 2 min to pellet insoluble plant debris, proteins in the supernatant fractionated by SDS-PAGE, then electroblotted to Immobilon-P PVDF membrane (MilliporeSigma) and immundetection performed as described (Gao et al., 2008). Ponceau S staining of the nylon membrane was used to visualize proteins as a loading control (Salinovich and Montelaro, 1986). An anti-GFP antibody (B-2) from Santa Cruz Biotechnology was used as the primary antibody for detection of the RR22-GFP fusion proteins.
ANOVA-based analyses were performed using an online calculator.3 T-tests were performed in Excel.
To functionally characterize the role of a type-B RR in rice, we focused on RR22 based on its relatively high expression level among the rice type-B RRs, prior indication that RR22 plays a role in mediating cytokinin signaling, and the availability of insertion alleles through public resources (Kim et al., 2012; Tsai et al., 2012; Yamburenko et al., 2017; Worthen et al., 2019). Lines for the ectopic overexpression of RR22 (RR22-OX lines) were generated by expressing a genomic copy of the RR22 coding region from the Zea mays UBI1 promoter. Sequence encoding a GFP tag was incorporated at the 3'-end of the RR22 gene to allow for in planta visualization and assessment of relative protein levels of the RR22-OX transgenes (Figure 1A). Out of 15 independent transgenic lines obtained for this construct in Oryza sativa ssp. japonica cv. Kitaake, we selected three lines (lines L4, L5, and L9) for further characterization, based on their having a single T-DNA insertion and also exhibiting a high level of protein expression (Figure 1C). We note that the RR22-GFP transgenes had a tendency to decrease in expression and silence in subsequent generations, and so following their protein levels was important for maintaining ectopic overexpression in these lines. From the segregating populations, we isolated WT siblings for each of the RR22-OX lines to serve as controls. Based on fluorescence microscopy, RR22-GFP was expressed in root and in shoot tissues, and the protein localized in nuclei, consistent with what was observed following transient expression in rice protoplasts and with its function as a transcription factor (Figure 1D; Supplementary Figure 1A; Tsai et al., 2012).
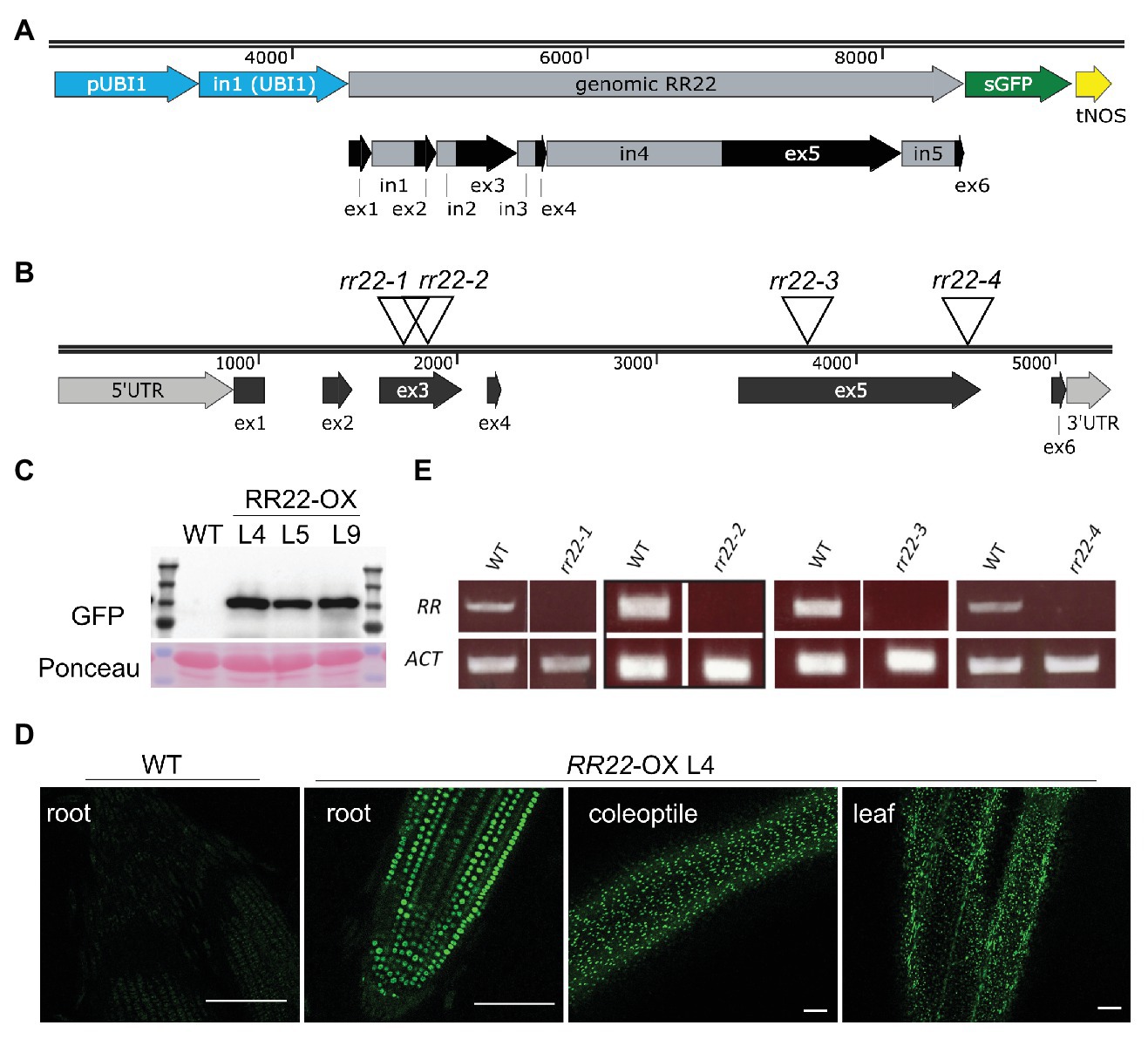
Figure 1. RR22 gain-of-function and loss-of-function lines. (A) Structural design of the RR22-OX cassette. A genomic version of RR22 was expressed from the Zea mays UBI1 promoter incorporating the first intron of the UBI1 gene. A sGFP fluorescent tag was encoded at the 3'-end of RR22. The intron/exon structure of the RR22 gene is shown below the cassette diagram. (B) Insertional positions for the four Tos17 alleles of rr22. (C) Relative protein expression levels of RR22-GFP in three independent RR22-OX lines as assessed by immunoblot using an anti-GFP antibody. A non-transgenic wild-type (WT) extract serves as a negative control. Ponceau S staining was used as a protein loading control. (D) Distribution of RR22-GFP fluorescent signal in root, dark-grown coleoptile, and leaf of RR22-OX L4. The WT root is a negative control. Scale bar = 100 μm. (E) Transcript levels of rr22 and ACT1 in the Tos17 rr22 mutant lines and corresponding WT siblings assessed by semiquantitative qPCR.
To assist in the functional characterization of RR22, we obtained Tos17 insertional mutant alleles from publicly available sources (Miyao et al., 2003; Hirochika et al., 2004; Figure 1B). The Tos17 lines are in Oryza sativa ssp. japonica cv. Nipponbare, prior loss-of-function analyses having been performed through the CRISPR-Cas9 approach in Oryza sativa ssp. japonica cv. Kitaake (Worthen et al., 2019). Four independent Tos17 alleles for RR22 were brought to homozygosity from the segregating populations, and their WT siblings isolated as controls. The locations for the Tos17 insertions were confirmed by sequencing (Figure 1B; Supplementary Table 2). All four alleles disrupt the coding region of RR22: the Tos17 insertions for rr22-1 and rr22-2 are in exon 3, and for rr22-3 and rr22-4 are in exon 5 (Figure 1B). None of the four Tos17 rr22 mutants produced full-length transcripts, consistent with their being loss-of-function alleles (Figure 1E).
The primary role established for type-B RRs is in mediating the transcriptional response to cytokinin (Imamura et al., 1999; Hosoda et al., 2002; Raines et al., 2016; Zubo et al., 2017). Therefore, increased expression of RR22 is predicted to result in cytokinin hypersensitivity, which can include increased induction of cytokinin-inducible genes. To this end, we examined the expression of RR9/10 and CKX5, which we previously identified as potential cytokinin primary response genes based on their rapid induction by cytokinin in multiple tissues as well as the presence of extended type-B RR binding motifs in their promoters (Raines et al., 2016; Worthen et al., 2019). We compared gene expression in shoots for the RR22-OX lines to their WT siblings following a 1-h treatment with 1 μM BA or a vehicle control. These treatment conditions were chosen because they result in only a modest increase in cytokinin-dependent gene expression in the WT siblings, such that the response is not saturated under these conditions (Figure 2A). All three RR22-OX lines exhibited enhanced induction of expression of RR9/10 and CKX5 in response to cytokinin as compared to their WT siblings, consistent with a heightened transcriptional response to cytokinin.
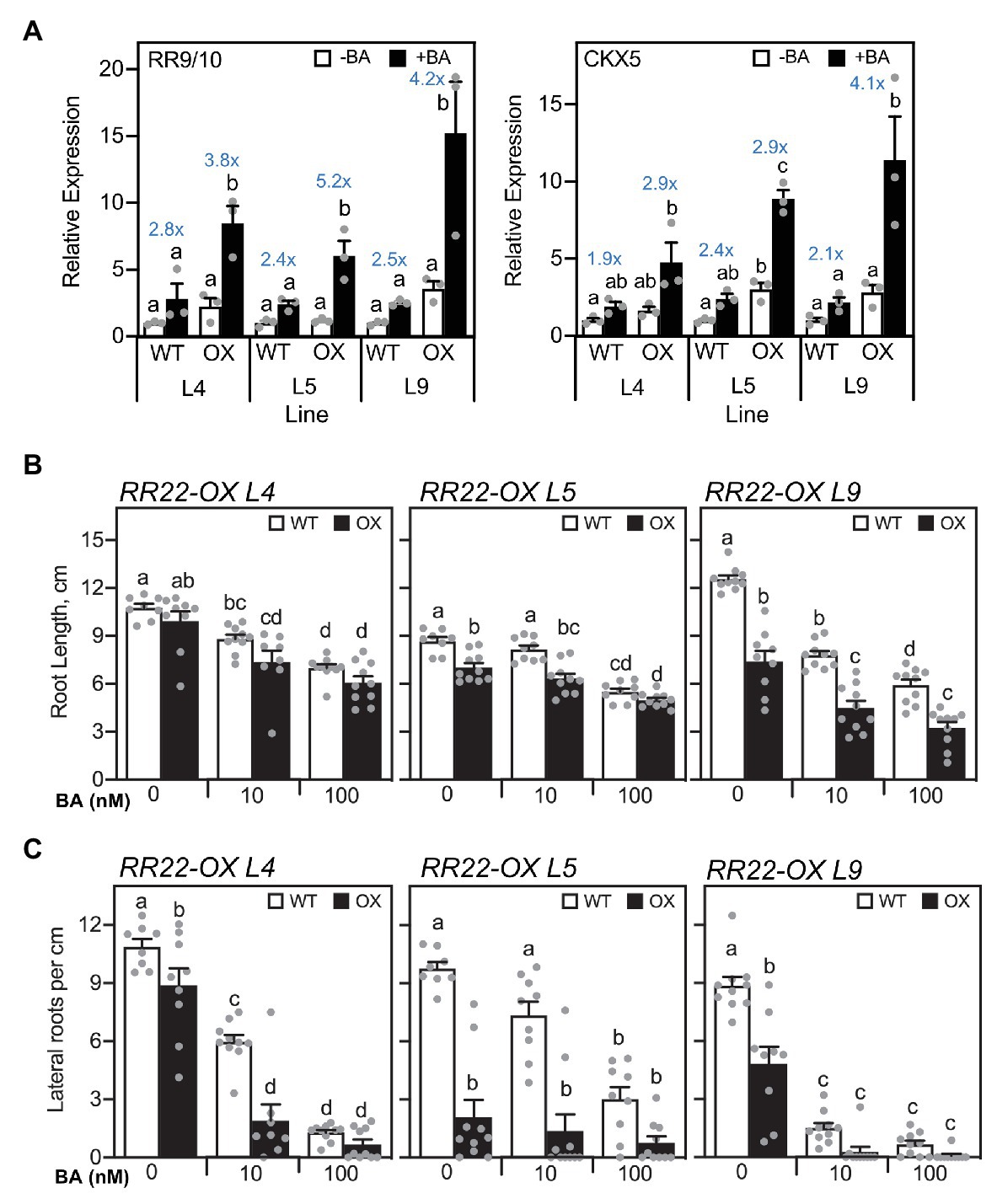
Figure 2. Cytokinin hypersensitive response of RR22-OX lines. (A) Expression of cytokinin-dependent genes based on qRT-PCR. Seven-day-old seedlings were treated with 1 μM BA or a NaOH vehicle control for 1 h, and gene expression examined in the shoots of the RR22-OX lines and their WT siblings (n = 3). Expression of the ACT1 gene was used for normalization. The fold change in gene expression is indicated in blue lettering for each BA-treated to untreated sample. (B,C) Root growth response to cytokinin. Seminal root length (B) and lateral root density (C) were determined for 7-day-old seedlings grown on Yoshida hydroponics medium supplemented with a vehicle control, 10, or 100 nM BA (n ≥ 8). For statistical analyses, ANOVA with a post hoc Holm multiple comparison calculation was performed for each line, involving RR22-OX and the WT sibling of the line. Different letters indicate significant differences at p < 0.05.
The enhanced transcriptional response of the RR22-OX lines to cytokinin suggests that these lines should also exhibit an enhanced response to cytokinin in growth assays. For this purpose, we examined the root growth response to cytokinin, both for seminal root length as well as for lateral root production, both of which are inhibited to various extents by cytokinin (Rani Debi et al., 2005). As shown in Figure 2B, cytokinin significantly inhibits growth of the WT seminal root, when comparing the untreated to 100 nM BA-treated roots. The roots of all three RR22-OX lines are shorter than their WT siblings in the absence and presence of BA, lines L5 and L9 significantly so, a result most consistent with an enhanced response to endogenous cytokinin. A more pronounced difference between the RR22-OX lines and their WT siblings is observed when examining lateral root production (Figure 2C). All three RR22-OX lines exhibit significantly reduced production of lateral roots in the absence of added BA, by 18, 78, and 45% for RR22-OX lines L4, L5, and L9, respectively, consistent with an enhanced response to endogenous cytokinin. In addition, lines L4 and L9 also exhibit an enhanced responsiveness to exogenous cytokinin, their production of lateral roots being reduced by 68 and 81%, respectively, when compared to their WT siblings when grown in the presence of 10 nM BA. The enhanced transcriptional response to cytokinin of the RR22-OX lines, based on gene expression and growth assays, is consistent with the abundance of rice type-B RRs as being rate-limiting for cytokinin signal transduction.
When grown on soil, mature RR22-OX lines were dwarfed in comparison to their WT siblings (Figure 3). On average, the plant height for the three RR22-OX lines decreased 43% compared to their WT siblings. As noted earlier, the RR22-GFP transgene had a tendency toward silencing, and we observed a correlation between the level of RR22-GFP protein and the stature of the RR22-OX plants, a higher level of RR22-GFP protein being observed in the plants that exhibited the most substantial dwarfing (Supplementary Figure 1B). The RR22-OX lines also exhibited an elevated number of tillers (Figures 3A,B), and we observed a delay of approximately 2 weeks in flowering time compared to their WT siblings.
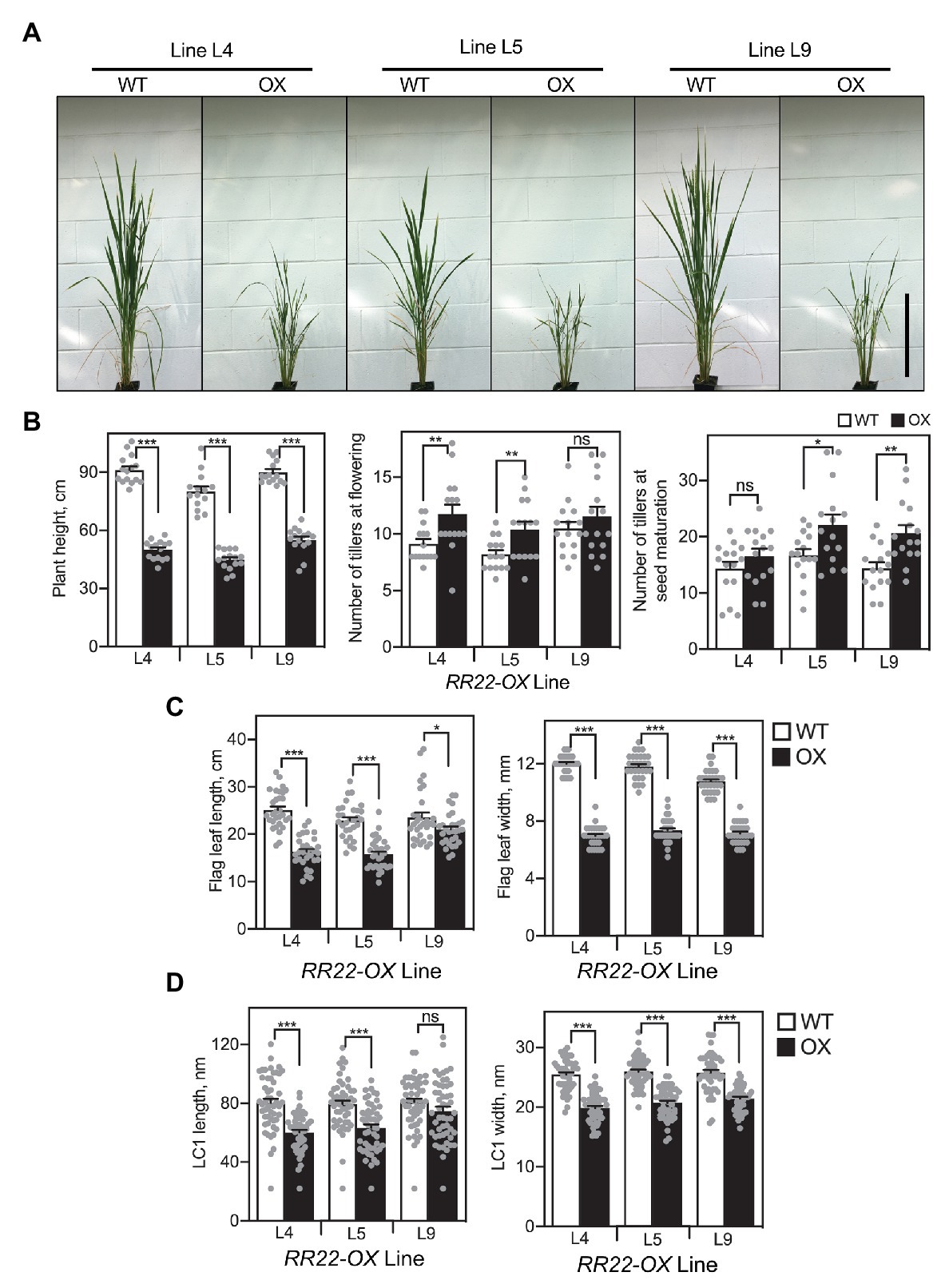
Figure 3. Shoot growth phenotypes of RR22-OX lines. (A) Shoots of RR22-OX lines and their WT siblings, taken at the initiation of flowering in the WT plants. Scale bar = 30 cm. (B) Plant height at flowering, number of tillers at flowering and at seed maturation. (C) Flag leaf length and width. (D) Epidermal long cell 1 (LC1) length and width of the flag leaves. Error bars show SE. The T-test was used for statistical comparison of each RR22-OX line to its WT sibling (*p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant).
All three lines of the RR22-OX plants have shorter and narrower leaves compared to their WT siblings (Figure 3C), accounting at least in part for the reduced stature of the RR22-OX plants. To determine whether the decrease in leaf size of the RR22-OX lines was due to changes in cell proliferation and/or cell size, we examined epidermal long cells (LC1) on the abaxial surface of the flag leaf. Both length and width of the cells are reduced in the RR22-OX lines compared to their WT siblings (Figure 3D). The decrease in cell length (~18%) closely parallels the reduction of flag leaf length (~26%), suggesting that this decrease is primarily due to the decrease in cell length rather than any reduction in cell proliferation. Although a decrease in cell width also occurs in the RR22-OX lines (~20%), the decrease is not as proportionately great as that observed in flag leaf width (~38%), suggesting that other factors such as cell proliferation may also play a role in this leaf dimension.
In addition to changes in cell size, we also observed effects of the RR22-OX lines on the development of specific cell types. The abaxial side of the rice leaf epidermis has a distinct pattern consisting of several different cell files (Luo et al., 2012). We observed effects on the number of files that contain the silica and phellem (cork) cells in the RR22-OX lines. In the silica/cork cell files, dumbbell-shaped silica cells alternate with cork cells and provide mechanical stability for rice leaves and improve rice resistance to pathogens (Kaufman et al., 1985). In WT plants, the silica/cork cells are typically arranged in a single file (Figure 4). However, we observed much greater variability in the number of adjacent silica/cork cell files in the RR22-OX lines: generally, the rows of adjacent files increased (Figure 4), although we also observed instances in which the file was no longer present (Figure 4B). In addition, we observed effects on the number of stomata files (Figure 4A; Supplementary Figure 2). In the WT flag leaves, stomata were typically arranged in two adjacent files. However, in the flag leaves of the RR22-OX lines, stomata were typically arranged in a single file (Figure 4A; Supplementary Figure 2), although in some cases, we also observed the sporadic appearance of stomata in a neighboring file.
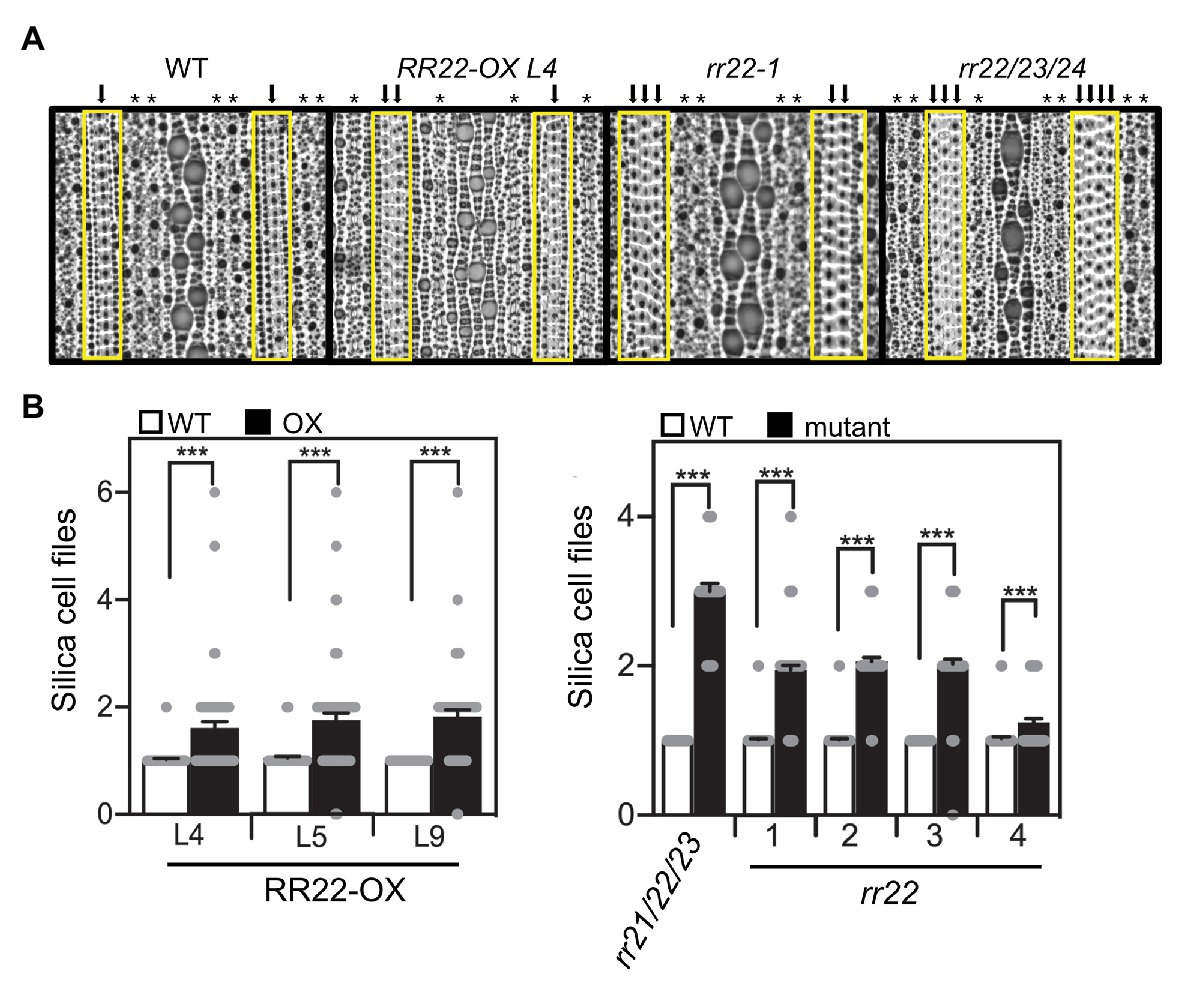
Figure 4. Leaf cell file phenotypes of RR22-OX, rr22, and rr21/22/23 mutant lines. (A) Representative images of the abaxial epidermis of the flag leaf. The region containing the silica/cork cell files is highlighted with a yellow box, and each silica/cork cell file indicated with a black arrow. Each stomata file is indicated with an asterisk (*). (B) Quantification for the number of rows of adjacent silica/cork cell files. Impressions from five plants (two per plant) were examined and the number of rows of adjacent silica/cork cell files determined (n > 47). Error bars show SE. The T-test was used for statistical comparison of each mutant line to its WT sibling (***p < 0.001).
Overexpression of RR22 also altered development of the rice inflorescence. Rice produces a panicle-type inflorescence, which is determinate and branched, with a defined architecture derived from the organization of the reproductive meristem (Itoh et al., 2005; Prusinkiewicz et al., 2007). The rice panicle consists of a central axis with several primary and secondary branches, each of which produces spikelets that encapsulate the rice flower. Panicles of the RR22-OX lines were smaller and had reduced branching compared to those of their WT siblings (Figure 5). In this respect, all three of the RR22-OX lines exhibited significant reductions in panicle length, primary branch number, and especially secondary branch number. The effects of the RR22-OX lines on panicle architecture resulted in a significant decrease in the number of spikelets per panicle (Figure 5). Seed set was also reduced in the RR22-OX lines and this, combined with the decreased number of spikelets, severely reduced yield based on the number of grains per panicle (Figure 5).
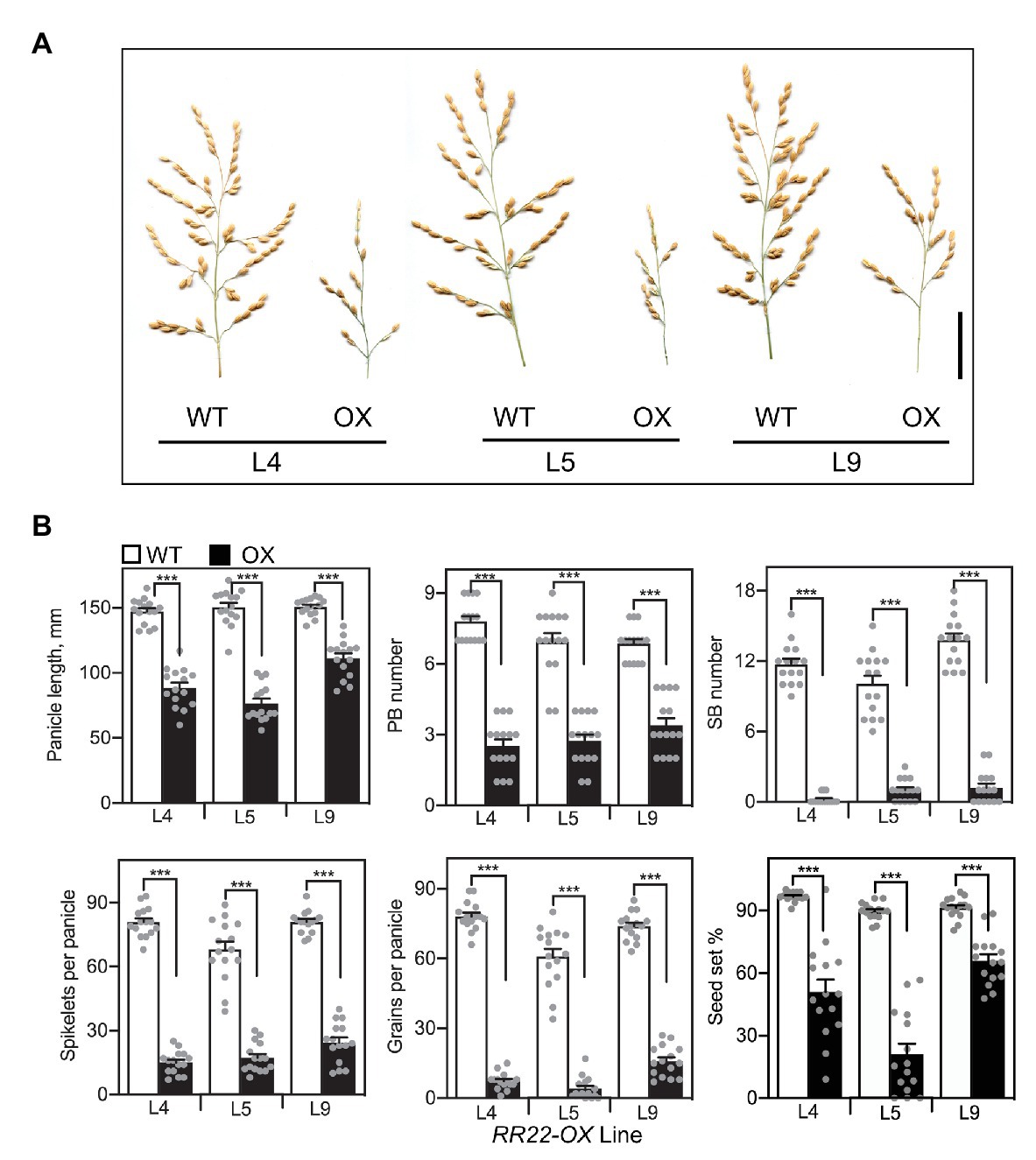
Figure 5. Panicle phenotypes of RR22-OX lines. (A) Panicles of RR22-OX lines and their WT siblings. Scale bar = 5 cm. (B) Yield parameters of the RR22-OX lines, including panicle length, primary branch (PB) and secondary branch (SB) number, spikelets and grains per panicle, and percent seed set (n = 15). Error bars show SE. The T-test was used for statistical comparison of each RR22-OX line to its WT sibling (***p < 0.001).
Cytokinin and the rice type-B RRs have also been implicated in trichome initiation and elongation (Maes et al., 2008; Maes and Goossens, 2010; Worthen et al., 2019). The RR22-OX lines produced shorter trichomes on the grain hulls than their WT siblings (Supplementary Figure 3). On the other hand, the RR22-OX lines had no apparent effect on the production and growth of the stigma brush hairs (Supplementary Figure 3), which are trichome-related structures involved in pollen capture whose development has been shown to be affected by cytokinin signaling (Worthen et al., 2019).
Four Tos17 insertion mutations of RR22 (rr22-1, rr22-2, rr22-3, and rr22-4) were identified (Figure 1A) and characterized. These did not exhibit any prominent growth phenotypes for the shoots and roots, nor a consistent change in sensitivity for the induction of cytokinin primary response genes (Supplementary Figure 4). This is likely due to functional redundancy of RR22 with other members of the type-B RR family of rice, as we previously found that the triple mutant rr21/22/23 is of reduced stature and exhibits a reduced capacity for the induction of cytokinin primary response genes (Worthen et al., 2019). However, as described below, we were able to uncover effects of the single rr22 mutants on the development of silica/cork files, panicle architecture, and trichomes.
As shown in Figure 4, WT leaves generally produce a single file containing alternating silica and cork cells. In contrast, the single mutants rr22-1, rr22-2, and rr22-3 produced two adjacent files of silica/cork cells; rr22-4 was more similar to the WT phenotype, but still produced several instances with two adjacent files of silica/cork cells. Analysis of the triple mutant rr21/22/23 revealed an even greater increase in the rows of adjacent silica/cork files, there being three rows on average (Figure 4). Based on the number of these cell files in rr21/22/23, along with additional internal silica/cork cells appearing within these expanded rows, the increase in cell files appears due to increased latitudinal cell proliferation.
The panicles of the rr22 mutants were smaller than those of their WT siblings, primarily due to a reduced branching complexity (Figure 6). Analysis of various parameters associated with panicle architecture revealed that three out of four rr22 alleles exhibited significant decreases in panicle length. The rr22 mutants produced similar numbers of primary branches to their WT siblings but, strikingly, all four rr22 mutant alleles exhibited a significant reduction in secondary branch number (Figure 6). These effects on panicle architecture result in a significant decrease in the total number of spikelets per panicle compared to WT. Due to fewer spikelets per panicle, coupled to a decrease in seed set, the rr22 mutants produce fewer grains per panicle than their WT siblings (Figure 6).
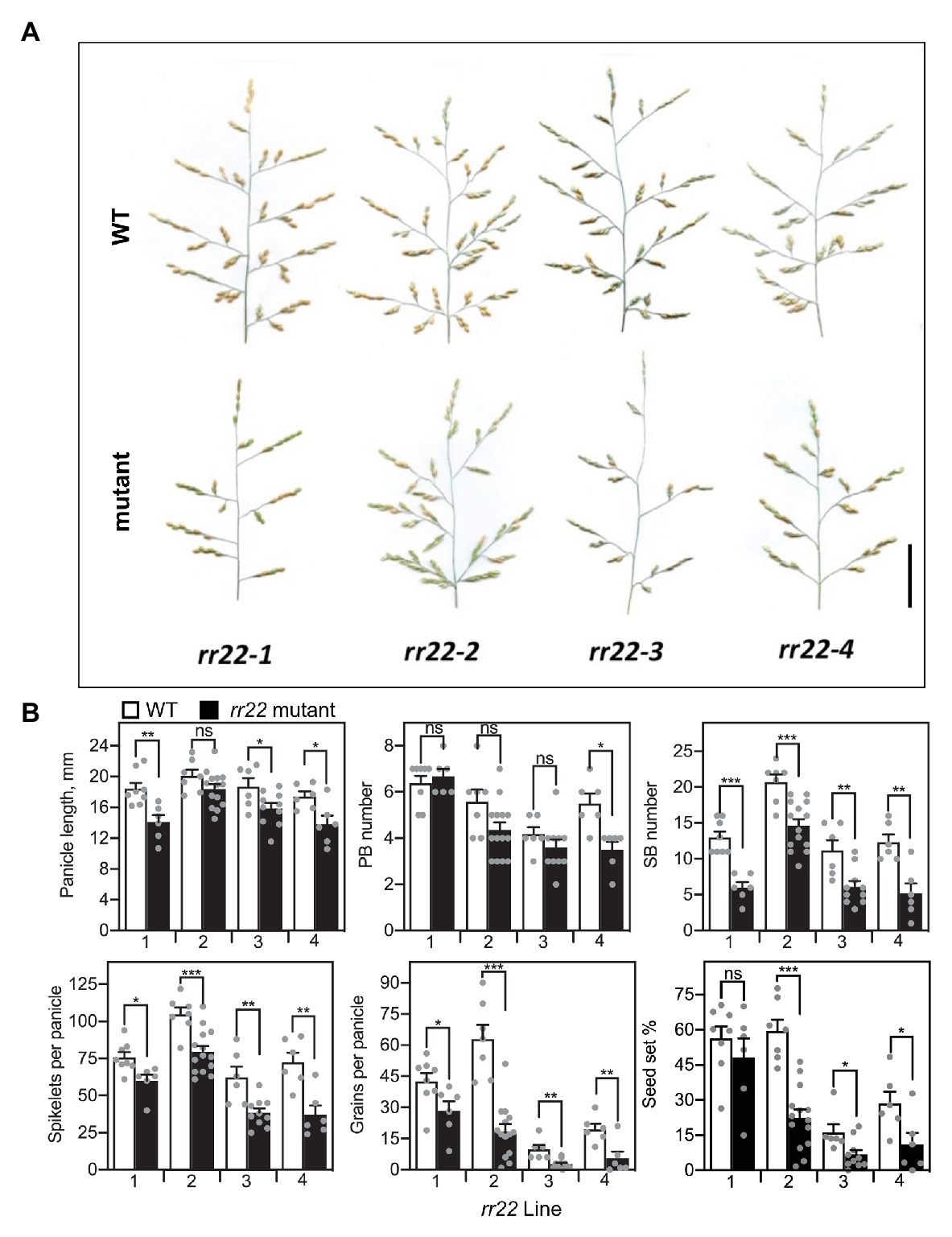
Figure 6. Panicle phenotypes of Tos17 rr22 lines. (A) Panicles of rr22-1, rr22-2, rr22-3, and rr22-4 lines and their WT siblings. Scale bar = 5 cm. (B) Yield parameters of the rr22 lines, including panicle length, PB and SB number, spikelets and grains per panicle, and percent seed set (n ≥ 6). The T-test was used for statistical comparison of each rr22 line to its WT sibling (*p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant).
The rr22 mutants exhibited defects in the development of several types of trichomes (Figure 7). We previously found that the triple type-B RR mutant rr21/22/23 was defective in the development of macro trichomes on leaves and grain hulls, as well as in the production of the stigma brush hairs (Worthen et al., 2019). We therefore examined the length of hull trichomes and stigma brush hairs in the Tos17 rr22 lines, finding these significantly reduced in length when compared to those of their WT siblings (Figures 7A, B). Additionally, stigma-enriched genes exhibited reduced expression in carpels of the rr22 lines when compared to their WT siblings (Figure 7C; Supplementary Figure 5). Expression of GL3A, which encodes a rice homolog of the transcription factor GLABRA3 of Arabidopsis, and of EXPA6 was reduced in all four rr22 lines (Figure 7C). Expression of two other stigma-enriched genes (GH3.1 and WDA1) was less consistently reduced (Supplementary Figure 5). All four of these genes were previously found to be significantly downregulated in the rr21/22/23 mutant, which exhibits a more severe stigma brush hair defect than the rr22 single mutants; the more variable effects on downregulation of these genes in the Tos17 rr22 lines are thus likely due to the intermediate mutant phenotype for the brush hairs and functional overlap with the other type-B RRs in the regulation of their development.
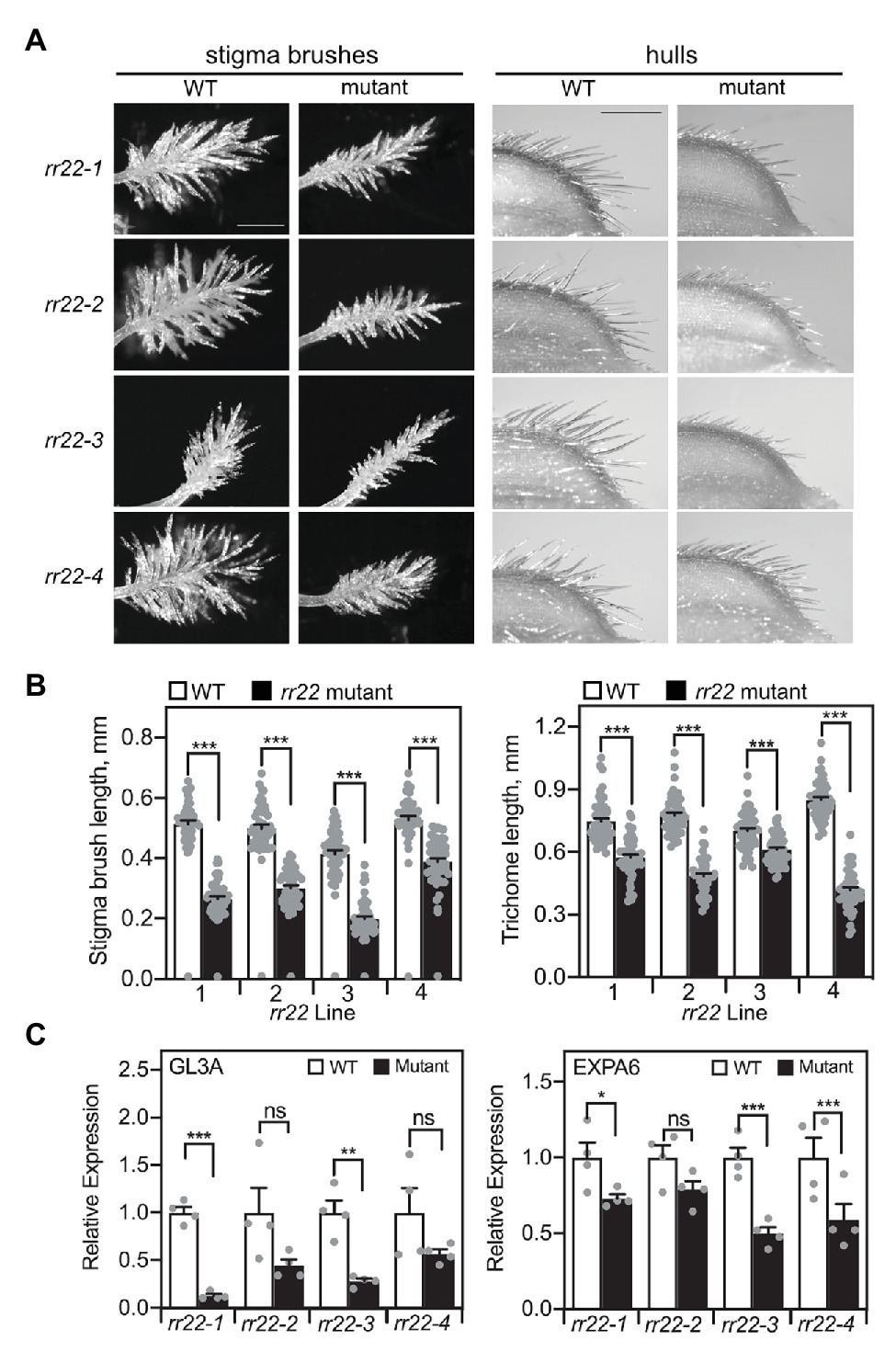
Figure 7. Trichome-related phenotypes of Tos17 rr22 lines. (A) Stigma brushes (scale bar = 0.5 mm) and trichomes on grain hulls (scale bar = 1 mm). (B) Stigma brush hair length and hull trichome length, with the five longest hairs or trichomes measured per stigma or hull, respectively (n = 50). (C) Expression analysis by qRT-PCR of the stigma-enriched genes GL3A and EXPA6 in carpels of rr22 mutants and their WT siblings (n = 4). Expression of the UBQ5 gene was used for normalization. Error bars are for SE. The T-test was used for statistical comparison of each rr22 line to its WT sibling (*p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant).
We recently employed a CRISPR/Cas9 gene editing approach to target the four most abundant type-B RRs of rice (RR21, RR22, RR23, and RR24), results from that analysis confirming a role in cytokinin signaling and revealing various roles in the growth and development (Worthen et al., 2019). As a complement to this CRISPR/Cas9-based loss-of-function approach, we have now employed a gain-of-function approach to characterize the effects of ectopic overexpression of RR22 in rice, as well as characterized four Tos17 loss-of-function alleles of RR22. Below, we discuss these results in relationship to cytokinin sensitivity, inflorescence architecture, grain yield, the production of trichome-related structures, and the growth and development of leaves and roots.
Genetic analysis of the type-B RRs indicates that they can be rate-limiting for cytokinin signal transduction in rice and Arabidopsis. There is functional overlap among members of the type-B RR family such that the overall abundance of the type-B RRs correlates reasonably well with the effect of loss-of-function mutations on cytokinin-dependent gene expression as well as on some physiological responses in both rice (Tsai et al., 2012; Yamburenko et al., 2017; Worthen et al., 2019) and Arabidopsis (Mason et al., 2005; Argyros et al., 2008; Hill et al., 2013). Furthermore, increased expression of RR22 in rice, as we show here, and of the Arabidopsis type-B RRs ARR1 and ARR10 can confer cytokinin hypersensitivity on plants (Sakai et al., 2001; Zubo et al., 2017). However, although cytokinin sensitivity can be modulated by altering the expression levels of type-B RRs, the degree to which these alterations affect growth and development can vary depending on the process, tissue, and organ involved, as discussed in more detail below.
Prior studies emphasize the critical role that cytokinin levels play in regulating meristematic activity and inflorescence architecture in rice (Ashikari et al., 2005; Kurakawa et al., 2007; Gu et al., 2015; Yeh et al., 2015). Rice and Arabidopsis both produce inflorescences; however, the architecture of these inflorescences is quite different and arises from differences in the development of their inflorescence meristems (Itoh et al., 2005; Prusinkiewicz et al., 2007). Rice produces a panicle-type inflorescence, which is determinate and branched, whereas Arabidopsis produces a simple raceme-type inflorescence, which is indeterminate and unbranched. The rice panicle consists of a central stem with several primary and secondary branches, each of which produces the floral structures, called spikelets (Itoh et al., 2005). The branches are produced by the primary and secondary branch meristems and the spikelets by spikelet meristems. Disruption of the LOG gene, involved in cytokinin biosynthesis, produces a smaller panicle and reduced grain yield due to a failure to maintain meristematic cells in both inflorescence and spikelet meristems (Kurakawa et al., 2007; Gu et al., 2015). Of particular interest has been the discovery that a reduction in the expression of the cytokinin oxidase gene CKX2 results in a larger panicle due to an elevation in cytokinin levels, suggesting that grain yield can be manipulated by modulating cytokinin activity (Ashikari et al., 2005; Yeh et al., 2015).
Our analyses of loss‐ and gain-of-function type-B RR mutants of rice emphasize the sensitivity of panicle architecture to alterations in cytokinin signal transduction. RR21, RR22, and RR23 are all expressed during the initial stages of panicle development when primary and secondary branch meristems are produced (Yamburenko et al., 2017), and we find that the loss of a single type-B RR compromises the panicle architecture. This is evident in all four Tos17 rr22 loss-of-function mutants, the most pronounced effect being a reduction in secondary branch production, a similar architectural defect also occurring in CRISPR/Cas9 generated loss-of-function mutants of RR21 and RR23 (Worthen et al., 2019). Interestingly, we did not observe such a reduction in panicle size for the CRISPR/Cas9 generated mutant of RR22, this mutant in fact exhibiting an increase in secondary branch number (Worthen et al., 2019). This phenotypic difference between the Tos17 and CRISPR/Cas9 alleles may arise due differences in the rice varieties examined (Nipponbare for the Tos17 lines; Kitaake for the CRISPR-Cas9 lines) and/or functional compensation of the single rr22 mutant by another type-B RR family member. The latter is a possibility because the triple rr21/22/23 CRISPR/Cas9-generated mutant exhibited substantially smaller panicles with a reduction in both primary and secondary branch production, indicating functional overlap among the type-B RRs in the regulation of panicle architecture (Worthen et al., 2019).
The results from CKX2 mutants (Ashikari et al., 2005; Yeh et al., 2015), in which an increase in cytokinin levels resulted in a larger more branched panicles might suggest that increased activity of signal transduction components could phenocopy this effect. However, this was not the case based on our analysis of the RR22-OX lines. Instead, the RR22-OX lines exhibited substantially smaller panicles due to reductions in both primary branch and secondary branch production, the production of secondary branches being almost eliminated. This may suggest a heightened cytokinin response due to overexpression is inhibitory, and/or that an expanded zone of cytokinin activity due to ectopic expression results in a lack of proper spatial temporal coordination of signaling within the meristematic tissues of the panicle.
Grain yield per panicle is reduced in the rr22 loss-of-function mutants due to a reduction in spikelet production coupled to a reduction in seed set. The reduced seed set is likely due in part to a defect in stigma brush development, because the stigma brushes of grasses play an important role in pollen capture, hydration, and guidance of the pollen tube toward the ovary (Heslop-Harrison et al., 1984; Heslop-Harrison and Reger, 1988). Each brush hair in rice is multicellular and branched, and our prior analysis indicates that type-B RR mutants severely affect cell proliferation of the brush hairs but not their cell expansion (Worthen et al., 2019). Our results from the Tos17 rr22 mutants confirm a predominant role for RR22 in mediating this effect on stigma brush development. The stigma brush is an epidermal trichome-related structure and we also observed that the Tos17 rr22 mutants exhibited reduced growth of trichomes on their seed hulls, consistent with a general role for RR22 in modulating trichome growth and development. Interestingly, the RR22-OX lines affected growth of the hull trichomes, but had no apparent effect on growth and development of the stigma brush structure.
Alterations in type-B RR activity also affect the growth and development of rice leaves. A recent study reported that the overexpression of the rice type-B RR ORR2, which is equivalent to RR22 from our study (Schaller et al., 2007), reduced plant height by approximately 20% but reported no additional effects on growth and development (Shi et al., 2020). We observed a substantially greater decrease in plant height of over 40% in our overexpression lines, potentially due to higher levels of RR22 expression in our study. The decrease in plant height of the over-expression lines, based on our study, can be largely attributed to a decrease in leaf growth, this primarily arising due to a decrease in cell size rather than cell proliferation. This effect of type-B RR overexpression contrasts with that found in the triple mutant rr21/22/23 of rice, which like the RR22-OX lines also has shorter flag leaves, but due to a decrease in longitudinal cell proliferation rather than a decrease in cell size (Worthen et al., 2019). Interestingly, ectopic overexpression of RR22 in rice did not recapitulate what was found from the ectopic overexpression of the type-B RR ARR10 of Arabidopsis, which induced leaf cell proliferation, although this was also sometimes accompanied by a decrease in cell size (Zubo et al., 2017).
Alterations in type-B RR activity affected the development of rice leaves along with affecting their growth. The architecture of grass leaves differs from that of dicots such as Arabidopsis, with various cell files serving distinct functions related to the development of diagnostic cell types (Kaufman et al., 1985; Luo et al., 2012). Both loss‐ and gain-of-function type-B RR mutants affected the number of silica/cork cell files, the rr mutants resulting in an increase in the number of adjacent files, likely due to an increase in lateral cell proliferation, whereas ectopic overexpression of RR22 resulted in a greater variability in the number of adjacent cell files. Ectopic overexpression of RR22 also resulted in a decrease in the number of adjacent stomatal cell files. This effect on stomata development may involve the genes SHR and SCR, because these partner transcription factors positively regulate the formation of stomatal cell files in rice (Schuler et al., 2018; Buckley et al., 2019; Wu et al., 2019) and, based on studies in Arabidopsis, cytokinin can inhibit SCR expression (Zhang et al., 2013). These results suggest that, in addition to affecting cell size and proliferation, that alterations in the cytokinin transcriptional response can also affect the developmental architecture of the rice leaf.
Our results are also consistent with a role for cytokinin in regulating root architecture in rice. Overexpression of RR22 inhibited seminal root growth and had an even more pronounced effect on lateral root formation. Such an effect is consistent with cytokinin and the type-B RRs serving as negative regulators of primary root growth and lateral root development, based on studies in dicots such as Arabidopsis and tobacco as well as the monocot rice (Mason et al., 2005; Dello Ioio et al., 2007; Werner et al., 2010; Hill et al., 2013; Zubo et al., 2017). We did not note any obvious developmental abnormalities in roots of the rr22 mutants, although RR22 is one of the more abundantly expressed members of the type-B RR family in the root, shoot, and inflorescence of rice (Tsai et al., 2012; Yamburenko et al., 2017). The lack of rr22 root phenotypes is likely due to functional overlap of RR22 with the other type-B RRs based on our prior analyses of the CRISPR/Cas9-generated type-B RR mutants (Worthen et al., 2019), that study indicating that a triple rr21/22/23 mutant exhibited defects in root development as well as root growth sensitivity to exogenous cytokinin. The rr22 single mutant phenotypes we do observe in inflorescence architecture, stigma brush development, and silica/cork cell production may arise due to the sensitivity of those tissues to small changes in cytokinin activity and/or a relatively greater role for RR22 compared to other type-B RRs in their developmental regulation.
In conclusion, altered expression of the type-B RRs, mediated either by loss-of-function or ectopic overexpression, results in compromised growth and development in rice. Ectopic overexpression of RR22 in rice appears to hamper growth and development more severely than is the case when a similar study was performed in Arabidopsis (Zubo et al., 2017). The ectopic overexpression of ARR10 in Arabidopsis closely parallels expectations for increased cytokinin signaling, resulting in increased cell proliferation in the shoot and the inhibition of root growth, the opposite of what is found with loss-of-function mutants of the type-B RRs, and with no major defects in inflorescence development. In rice, on the other hand, ectopic overexpression compromises growth of the leaves and the inflorescence in a manner reminiscent to what is observed in type-B RR loss-of-function mutations (Worthen et al., 2019). Two possibilities, not mutually exclusive, may explain these phenotypic observations. First, that an optimal level of cytokinin activity is required to mediate these responses, with either a decrease or increase in activity compromising growth and development. Second, due to the substantial differences in inflorescence and leaf development between rice and Arabidopsis, rice is more sensitive to expanded zones of cytokinin activity arising from the ectopic expression of the type-B RR. Regardless, these studies demonstrate that a delicate balance of cytokinin transcriptional activity is necessary for optimal growth and development in rice.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
GS, JK, MY, and JW designed the research. GS and SNS provided support and oversaw the research. MY, JW, AZ, BA, and AC performed the experiments. MY, JW, AZ, BA, and SS analyzed the data. GS, MY, and JW wrote the manuscript with contributions from all authors. All authors contributed to the article and approved the submitted version.
This work was supported by the National Science Foundation grant nos. IOS-1238051 and MCB-1856248 (GS and JK), by the Agriculture and Food Research Initiative competitive grant nos. 2019-67013-29191 (GS) and 2016-67011-24723 (JW) from the USDA National Institute of Food and Agriculture, and the International Research Support Initiative Program of Higher Education Commission of Pakistan (AZ).
The reviewer JH is currently organizing a Research Topic with one of the authors GS. The review process met the standards of a fair and objective review.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.577676/full#supplementary-material
Argyros, R. D., Mathews, D. E., Chiang, Y. -H., Palmer, C. M., Thibault, D. M., Etheridge, N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116. doi: 10.1105/tpc.108.059584
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Buckley, C. R., Caine, R. S., and Gray, J. E. (2019). Pores for thought: can genetic manipulation of stomatal density protect future rice yields? Front. Plant Sci. 10:1783. doi: 10.3389/fpls.2019.01783
Clarke, J. D. (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc 2009:pdb.prot5177. doi: 10.1101/pdb.prot5177
Dello Ioio, R., Linhares, F. S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., et al. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678–682. doi: 10.1016/j.cub.2007.02.047
Devos, K. M., and Gale, M. D. (2000). Genome relationships: the grass model in current research. Plant Cell 12, 637–646. doi: 10.1105/tpc.12.5.637
Du, L., Jiao, F., Chu, J., Jin, G., Chen, M., and Wu, P. (2007). The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics 89, 697–707. doi: 10.1016/j.ygeno.2007.02.001
Gao, Z., Wen, C. -K., Binder, B. M., Chen, Y. -F., Chang, J., Chiang, Y. -H., et al. (2008). Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J. Biol. Chem. 283, 23081–23810. doi: 10.1074/jbc.M800641200
Gu, B., Zhou, T., Luo, J., Liu, H., Wang, Y., Shangguan, Y., et al. (2015). An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol. Plant 8, 1635–1650. doi: 10.1016/j.molp.2015.08.001
Heslop-Harrison, Y., and Reger, B. J. (1988). Tissue organisation, pollen receptivity and pollen tube guidance in normal and mutant stigmas of the grass Pennisetum typhoides (Burm.) Stapf et Hubb. Sex. Plant Reprod. 1, 182–193. doi: 10.1007/BF00193749
Heslop-Harrison, Y., Reger, B. J., and Heslop-Harrison, J. (1984). The pollen-stigma interaction in the grasses. 5. Tissue organization and cytochemistry of the stigma (‘silk’) of Zea mays L. Acta Bot. Neerl. 33, 81–99. doi: 10.1111/j.1438-8677.1984.tb01774.x
Hill, K., Mathews, D. E., Kim, H. J., Street, I. H., Wildes, S. L., Chiang, Y. H., et al. (2013). Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol. 162, 212–224. doi: 10.1104/pp.112.208736
Hirochika, H., Guiderdoni, E., An, G., Hsing, Y. I., Eun, M. Y., Han, C. D., et al. (2004). Rice mutant resources for gene discovery. Plant Mol. Biol. 54, 325–334. doi: 10.1023/B:PLAN.0000036368.74758.66
Hosoda, K., Imamura, A., Katoh, E., Hatta, T., Tachiki, M., Yamada, H., et al. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14, 2015–2029. doi: 10.1105/tpc.002733
Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. doi: 10.1038/35096500
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., et al. (1999). Compilation and characterization of Arabidopsis thaliana response regulators implicated in his-asp phosphorelay signal transduction. Plant Cell Physiol. 40, 733–742. doi: 10.1093/oxfordjournals.pcp.a029600
Imamura, A., Kiba, T., Tajima, Y., Yamashino, T., and Mizuno, T. (2003). In vivo and in vitro characterization of the ARR11 response regulator implicated in the his-to-asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44, 122–131. doi: 10.1093/pcp/pcg014
Imamura, A., Yoshino, Y., and Mizuno, T. (2001). Cellular localization of the signaling components of Arabidopsis his-to-asp phosphorelay. Biosci. Biotechnol. Biochem. 65, 2113–2117. doi: 10.1271/bbb.65.2113
Ishida, K., Yamashino, T., Yokoyama, A., and Mizuno, T. (2008). Three type-B response regulators, ARR1, ARR10, and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49, 47–57. doi: 10.1093/pcp/pcm165
Ito, Y., and Kurata, N. (2006). Identification and characterization of cytokinin-signalling gene families in rice. Gene 382, 57–65. doi: 10.1016/j.gene.2006.06.020
Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., et al. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46, 23–47. doi: 10.1093/pcp/pci501
Jameson, P. E., and Song, J. (2016). Cytokinin: a key driver of seed yield. J. Exp. Bot. 67, 593–606. doi: 10.1093/jxb/erv461
Kaufman, P. B., Dayanandan, P., Franklin, C. I., and Takeoka, Y. (1985). Structure and function of silica bodies in the epidermal system of grass shoots. Ann. Bot. 55, 487–507. doi: 10.1093/oxfordjournals.aob.a086926
Kieber, J. J., and Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12:e0168. doi: 10.1199/tab.0168
Kim, H. J., Chiang, Y. H., Kieber, J. J., and Schaller, G. E. (2013a). SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. U. S. A. 110, 10028–10033. doi: 10.1073/pnas.1300403110
Kim, H. J., Kieber, J. J., and Schaller, G. E. (2012). Overlapping and lineage-specific roles for the type-B response regulators of monocots and dicots. Plant Signal. Behav. 7, 1110–1113. doi: 10.4161/psb.21293
Kim, H. J., Kieber, J. J., and Schaller, G. E. (2013b). The rice F-box protein KISS ME DEADLY2 functions as a negative regulator of cytokinin signalling. Plant Signal. Behav. 8:e26434. doi: 10.4161/psb.26434
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Liang, Y., Wang, X., Hong, S., Li, Y., and Zuo, J. (2012). Deletion of the initial 45 residues of ARR18 induces cytokinin response in Arabidopsis. J. Genet. Genomics 39, 37–46. doi: 10.1016/j.jgg.2011.12.004
Lohrmann, J., Sweere, U., Zabaleta, E., Baurle, I., Keitel, C., Kozma-Bognar, L., et al. (2001). The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Gen. Genomics. 265, 2–13. doi: 10.1007/s004380000400
Luo, L., Zhou, W. -Q., Liu, P., Li, C. -X., and Hou, S. -W. (2012). The development of stomata and other epidermal cells on the rice leaves. Biol. Plant. 56, 521–527. doi: 10.1007/s10535-012-0045-y
Maes, L., and Goossens, A. (2010). Hormone-mediated promotion of trichome initiation in plants is conserved but utilizes species‐ and trichome-specific regulatory mechanisms. Plant Signal. Behav. 5, 205–207. doi: 10.4161/psb.5.2.11214
Maes, L., Inze, D., and Goossens, A. (2008). Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148, 1453–1464. doi: 10.1104/pp.108.125385
Mann, D., Lafayette, P., Abercrombie, L., King, Z., Mazarei, M., Halter, M., et al. (2012). Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10, 226–236. doi: 10.1111/j.1467-7652.2011.00658.x
Mason, M. G., Li, J., Mathews, D. E., Kieber, J. J., and Schaller, G. E. (2004). Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 135, 927–937. doi: 10.1104/pp.103.038109
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., et al. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17, 3007–3018. doi: 10.1105/tpc.105.035451
Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., et al. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15, 1771–1780. doi: 10.1105/tpc.012559
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Pareek, A., Singh, A., Kumar, M., Kushwaha, H. R., Lynn, A. M., and Singla-Pareek, S. L. (2006). Whole genome analysis of Oryza sativa L. reveals similar architecture of two-component-signaling-machinery with Arabidopsis. Plant Physiol. 142, 380–397. doi: 10.1104/pp.106.086371
Prusinkiewicz, P., Erasmus, Y., Lane, B., Harder, L. D., and Coen, E. (2007). Evolution and development of inflorescence architectures. Science 316, 1452–1456. doi: 10.1126/science.1140429
Raines, T., Blakley, I. C., Tsai, Y. C., Worthen, J. M., Franco-Zorrilla, J. M., Solano, R., et al. (2016). Characterization of the cytokinin-responsive transcriptome in rice. BMC Plant Biol. 16:260. doi: 10.1186/s12870-016-0932-z
Rani Debi, B., Taketa, S., and Ichii, M. (2005). Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J. Plant Physiol. 162, 507–515. doi: 10.1016/j.jplph.2004.08.007
Rashotte, A. M., Mason, M. G., Hutchison, C. E., Ferreira, F. J., Schaller, G. E., and Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. U. S. A. 103, 11081–11085. doi: 10.1073/pnas.0602038103
Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24, 703–711. doi: 10.1046/j.1365-313x.2000.00909.x
Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., et al. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521. doi: 10.1126/science.1065201
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Salinovich, O., and Montelaro, R. C. (1986). Reversible staining and peptide mapping of proteins transferred to nitrocellulose after separation by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Anal. Biochem. 156, 341–347. doi: 10.1016/0003-2697(86)90263-0
Schaller, G. E., Doi, K., Hwang, I., Kieber, J. J., Khurana, J. P., Kurata, N., et al. (2007). Letter to the editor: nomenclature for two-component signaling elements of Oryza sativa. Plant Physiol. 143, 555–557. doi: 10.1104/pp.106.093666
Schuler, M. L., Sedelnikova, O. V., Walker, B. J., Westhoff, P., and Langdale, J. A. (2018). SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency. Plant Physiol. 176, 757–772. doi: 10.1104/pp.17.01005
Shi, F., Wang, M., and An, Y. (2020). Overexpression of a B-type cytokinin response regulator (OsORR2) reduces plant height in rice. Plant Signal. Behav. 15:1780405. doi: 10.1080/15592324.2020.1780405
To, J. P., Haberer, G., Ferreira, F. J., Deruere, J., Mason, M. G., Schaller, G. E., et al. (2004). Type-a Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671. doi: 10.1105/tpc.018978
Tsai, Y. C., Weir, N. R., Hill, K., Zhang, W., Kim, H. J., Shiu, S. H., et al. (2012). Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158, 1666–1684. doi: 10.1104/pp.111.192765
Werner, T., Nehnevajova, E., Kollmer, I., Novak, O., Strnad, M., Kramer, U., et al. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920. doi: 10.1105/tpc.109.072694
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538. doi: 10.1016/j.pbi.2009.07.002
Worthen, J. M., Yamburenko, M. V., Lim, J., Nimchuk, Z. L., Kieber, J. J., and Schaller, G. E. (2019). Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 146:dev174870. doi: 10.1242/dev.174870
Wu, Z., Chen, L., Yu, Q., Zhou, W., Gou, X., Li, J., et al. (2019). Multiple transcriptional factors control stomata development in rice. New Phytol. 223, 220–232. doi: 10.1111/nph.15766
Yamburenko, M. V., Kieber, J. J., and Schaller, G. E. (2017). Dynamic patterns of expression for genes regulating cytokinin metabolism and signaling during rice inflorescence development. PLoS One 12:e0176060. doi: 10.1371/journal.pone.0176060
Yeh, S. Y., Chen, H. W., Ng, C. Y., Lin, C. Y., Tseng, T. H., Li, W. H., et al. (2015). Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice 8:36. doi: 10.1186/s12284-015-0070-5
Yokoyama, A., Yamashino, T., Amano, Y., Tajima, Y., Imamura, A., Sakakibara, H., et al. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48, 84–96. doi: 10.1093/pcp/pcl040
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1976). Laboratory manual for physiological studies of Rice. Los Banos, Laguna, Philippines: The International Rice Research Institute.
Zhang, W. J., Swarup, R., Bennett, M., Schaller, G. E., and Kieber, J. J. (2013). Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 23, 1979–1989. doi: 10.1016/J.Cub.2013.08.008
Zubo, Y. O., Blakley, I. C., Yamburenko, M. V., Worthen, J. M., Street, I. H., Franco-Zorrilla, J. M., et al. (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, E5995–E6004. doi: 10.1073/pnas.1620749114
Keywords: type-B response regulator, cytokinin, rice, panicle, grain yield, trichome, root architecture, silica cell
Citation: Yamburenko MV, Worthen JM, Zeenat A, Azhar BJ, Swain S, Couitt AR, Shakeel SN, Kieber JJ and Schaller GE (2020) Functional Analysis of the Rice Type-B Response Regulator RR22. Front. Plant Sci. 11:577676. doi: 10.3389/fpls.2020.577676
Received: 29 June 2020; Accepted: 14 October 2020;
Published: 10 November 2020.
Edited by:
Patrick H. Masson, University of Wisconsin-Madison, United StatesReviewed by:
Jan Hejatko, Central European Institute of Technology (CEITEC), CzechiaCopyright © 2020 Yamburenko, Worthen, Zeenat, Azhar, Swain, Couitt, Shakeel, Kieber and Schaller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: G. Eric Schaller, Z2VvcmdlLmUuc2NoYWxsZXJAZGFydG1vdXRoLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.