- Bavarian State Research Centre for Agriculture, Institute for Crop Science and Plant Breeding, Freising, Germany
Wheat (Triticum aestivum L.) is a self-pollinating crop whose hybrids offer the potential to provide a major boost in yield. Male sterility induced by the cytoplasm of Triticum timopheevii is a powerful method for hybrid seed production. Hybrids produced by this method are often partially sterile, and full fertility restoration is crucial for wheat production using hybrid cultivars. To identify the genetic loci controlling fertility restoration in wheat, we produced two cytoplasmic male-sterile (CMS) backcross (BC1) mapping populations. The restorer lines Gerek 79 and 71R1203 were used to pollinate the male-sterile winter wheat line CMS-Sperber. Seed set and numbers of sterile spikelets per spike were evaluated in 340 and 206 individuals of the populations derived from Gerek 79 and 71R1203, respectively. Genetic maps were constructed using 930 and 994 single nucleotide polymorphism (SNP) markers, spanning 2,160 and 2,328 cM over 21 linkage groups in the two populations, respectively. Twelve quantitative trait loci (QTL) controlled fertility restoration in both BC1 populations, including a novel restorer-of-fertility (Rf) locus flanked by the SNP markers IWB72413 and IWB1550 on chromosome 6AS. The locus was mapped as a qualitative trait in the BC1 Gerek 79 population and was designated Rf9. One hundred-nineteen putative candidate genes were predicted within the QTL region on chromosome 6AS. Among them were genes encoding mitochondrial transcription termination factor and pentatricopeptide repeat-containing proteins that are known to be associated with fertility restoration. This finding is a promising step to better understand the functions of genes for improving fertility restoration in hybrid wheat.
Introduction
Since the discovery of male sterility and restoration systems in the 1960s, hybrid wheat triggered attention due to its potential for improved grain and straw productivity and yield stability particularly under harsh and marginal environments (Longin et al., 2012). The major gains of hybrid vs. line varieties are improved trait values due to heterosis (Castillo et al., 2014). Hybrid wheat has been reported to provide uniform plant establishment and tolerance against frost, lodging, and diseases such as leaf rust, stripe rust, Septoria tritici blotch, and powdery mildew (Gupta et al., 2019).
To harness yield gains associated with hybrid vigor, the cytoplasmic male sterility (CMS) system provides a cost-effective tool for efficient hybrid seed production (Chen and Liu, 2014). CMS in plants is based on the rearrangements of mitochondrial DNA that lead to chimaeric genes and a condition under which a plant is unable to produce fertile pollen (Eckardt, 2006; Whitford et al., 2013). CMS evades the need for manual removal of anthers, thus facilitating a technology to produce unlimited numbers of hybrid plants. It has been successfully used in crops such as rye, rice, maize, and sunflower (Castillo et al., 2015). The use of T. timopheevii cytoplasm in bread and durum wheat creates male sterility, whereas female fertility is not impaired. Wilson and Ross (1962) were the first to describe a workable cytoplasmic male sterile of T. timopheevii with largely neutral effects on the agronomic and quality characteristics. This CMS system has gained widespread use due to the deleterious effects of other cytoplasms of the genera Triticum and Aegilops on various traits, and because no advantage existed over the T. timopheevii system (Virmani and Edwards, 1983). It uses three different breeding lines: a CMS line, maintainer line, and restorer line. The CMS line is used as the female parent with at least one CMS-causing gene in the T. timopheevii-derived cytoplasm and lacking functional nuclear-encoded Restorer-of-fertility (Rf) genes (Schnable and Wise, 1998). The maintainer line serves as the male parent in crosses for the propagation and maintenance of the CMS line, with the same nuclear genome as the CMS line but a normal fertile T. aestivum cytoplasm. The restorer line retains (a) functional Rf genes and acts as the male parent to cross with the CMS line to produce the F1 hybrid seeds. In F1 plants, the Rf genes restore male fertility, and the combination of the nuclear genomes from the CMS line and the restorer line produces hybrid vigor. For commercial hybrid seed production, a male-sterile line has to be crossed with a line carrying dominant restorer alleles and suitable pollinator qualities (Whitford et al., 2013). While fertility restoration is a crucial trait in hybrid breeding, hybrids produced using this method are often partially sterile due to the complex interaction between the mitochondrial and nuclear genes controlling male specificity and restoration of fertility (Chen and Liu, 2014). Therefore, incomplete fertility restoration poses a major bottleneck for hybrid wheat breeding, as it compromises the heterotic gain for grain yield and the uniformity or quality of end-use products.
Fertility restoration is a genetically complex process and is mainly controlled by the mitochondrial genome in interaction with Rf genes (Eckardt, 2006). Besides, it is known that fertility restoration is influenced by environmental factors including photoperiod, water stress, light intensity, and temperature (Johnson et al., 1967). The nuclear encoded gene families that act in the mitochondria produce proteins that share the common structural organization of similar repeated helical motifs and include pentatricopeptide repeat (PPR) proteins and mitochondrial transcription termination factors (mTERFs) (Pan et al., 2019). The effect of cytoplasmic male sterility can be suppressed by preventing the accumulation of mitochondrial encoded CMS-conferring gene products through the function of a class of Rf genes, generally belonging to a large family of genes that encode organelle-targeted PPR proteins (Hu et al., 2012). mTERF genes are widely distributed in metazoans, plants, and green alga. They regulate transcription, translation, and DNA replication of mitochondrial genes in metazoans while regulating gene expression in chloroplasts and mitochondria in plants (Pan et al., 2019). In wheat, the presence of eight major loci (Rf1-Rf8) for timopheevii-based cytoplasmic male sterility is known and assigned to the chromosomes 1A, 7D, 1B, 6B, 6D, 5D, 7B, and 2D, respectively (Mukai and Tsunewaki, 1979; Sinha et al., 2013; Gupta et al., 2019). Rf1 and Rf3 are the most effective genes for achieving restoration in wheat (Geyer et al., 2016, 2018; Würschum et al., 2017). Previous studies have indicated that combinations of two or three major Rf genes and restorer genes with small effect or low penetrance (modifier loci) can modify the degree of fertility restoration (Ma et al., 1995; Ahmed et al., 2001; Zhou et al., 2005; Stojałowski et al., 2013). Consequently, attempts are made to pyramid multiple dominant or partially dominant alleles of the most favorable genes or quantitative trait loci (QTL), including those involved in epistatic interactions to achieve complete fertility restoration in hybrid wheat (Gupta et al., 2019). Understanding the genetic mechanisms underlying restoration of fertility and developing elite restorer lines are crucial to overcome the intricate barriers in hybrid breeding programs. For this reason, our objective was to identify new genetic loci controlling fertility restoration that can be employed in hybrid wheat breeding. Here, we developed two CMS-based backcross (BC) mapping populations, which we used for QTL mapping and identification of candidate genes. Our study identified a new Rf locus (Rf9) and novel QTL for seed set and number of sterile spikelets on chromosomes 1DS, 2AL, 4AL, 5BL, and 6AS. Putative candidate genes located in the target regions are discussed.
Materials and Methods
Plant Materials and Population Development
Two BC1 mapping populations were developed using Gerek 79 and 71R1203 as fertility-restoring parental lines. The winter wheat cultivar Gerek 79 (PI 559560, pedigree: Mentana/Mayo-48//4-11/3/Yayla-305) originated in 1979 from the Transitional Zone Agricultural Research Institute, Anadolu ARI, Eskisehir, Turkey1. The restoration capacity of Gerek 79 was found in initial screening experiments (unpublished) when pollinating CMS-Sperber with cultivars from various regions and testing the hybrids for self-fertility in the greenhouse at the Bavarian State Research Center for Agriculture (LfL). The winter wheat restorer line 71R1203 (PI 473552, pedigree: NB542437/CI 13438//2∗Burt/3/NB542437/2∗CI13438) was developed in 1982 by the USDA-ARS and Washington State University. The specific fertility restoration loci possessed by each of the two sources have not been previously determined, but 71R1203 was known to potentially carry restorer loci Rf1 and Rf2 that are present in NB542437 (Allan and Rubenthaler, 1984). The variety Sperber (registered 1983) and CMS-Sperber are maintained at the LfL. Seeds of Gerek 79 were kindly provided by Prof. Friedrich Zeller (Technical University of Munich, Freising, Germany) and are available at the Germplasm Resources Information Network (GRIN), U.S. National Plant Germplasm System. Seeds of the line 71R1203 were obtained from the National Small Grains Collection, US. Gerek 79 and 71R1203 were used as restorer lines in crosses with the male-sterile winter wheat line CMS-Sperber. The hybrids were then backcrossed with the maintainer line Sperber to develop the mapping populations CMS-Sperber/Gerek 79//Sperber (BC1 Gerek 79) and CMS-Sperber/71R1203//Sperber (BC1 71R1203).
Field Trials and Phenotyping
The BC1 Gerek 79 population was vernalized in a climate chamber at 6°C for 8 weeks and planted in spring 2019 in an LfL field at Freising (48°24′12.64″N, 11°44′55.54″E), Germany. The BC1 71R1203 population was sown in autumn 2018 in the field at KWS LOCHOW GMBH in Bergen (52°48′30.13″N, 9°57′49.46″E), Germany. We used non-replicated trials for assessing the fertility restoration due to having mortal mapping populations. Four emerging spikes from the main tillers of each BC1 line were covered before anthesis using glassine bags. After ripening, the spikes were harvested and the seed set (as the restored fertility trait) and number of sterile spikelets per spike (as the non-restored fertility trait) were counted. The seed set of a plant was calculated as the number of kernels divided by the number of spikelets, averaged over all four bagged spikes per individual. Plants were considered fertile if they had at least one seed per spike and male sterile when no seed was produced. Observed ratios of fertile to sterile plants in each mapping population were tested against the expected segregation pattern using the chi-squared goodness-of-fit test. Statistical analyses including descriptive statistics, correlation, and frequency distribution of the traits were conducted in the SigmaPlot (Systat Software, San Jose, CA, United States).
Genotyping and Linkage Analysis
Genomic DNA of parental lines and BC1 progenies was extracted from young leaf tissues following the procedure of Plaschke et al. (1995). Based on the fertility restoration data, the DNA of 273 and 184 individuals from BC1 Gerek 79 and BC1 71R1203, respectively, were selected for genotyping using a bead chip comprising 16,762 single nucleotide polymorphism (SNP) markers selected from the 90K iSelect® array (Wang et al., 2014). SNP genotyping was done by KWS SAAT S & Co., KGaA, Einbeck, Germany. The raw SNP data were analyzed as described by Geyer et al. (2018). Briefly, all monomorphic SNPs and those with more than 10% missing values and a minor allele frequency of less than 10% were discarded from further analysis using the synbreed package V0.12-6 (Wimmer et al., 2012) in R (R Core Team, 2017). Linkage analysis was done using JoinMap® (Kyazma BV, Wageningen, Netherlands). The Kosambi mapping function (Kosambi, 1944) was used to convert the recombination frequencies into centimorgans (cM).
To determine whether Gerek 79 and 71R1203 carried Rf3, they were genotyped with SNP marker IWB72107, earlier shown to have a high potential for predicting Rf3 (Geyer et al., 2016). The SNP-containing sequence for IWB72107 was retrieved from The Triticeae Toolbox (T32) and converted to a Kompetitive Allele Specific Polymerase (KASP) chain reaction marker assay (Supplementary Table S1). Plants were genotyped according to the manufacturer’s instructions (LGC Genomics, Hoddeson, United Kingdom). Each KASP reaction was prepared in a volume of 10 μL with 5 μL DNA and 5 μL of the genotyping master mix. Amplification was carried out using the CFX96 Touch Real-Time PCR SNP Detection System (Bio-Rad, Hercules, CA, United States), starting with 15 min at 94°C, followed by 40 cycles of PCR with 94°C for 20 s and 65°C for 1 min and 10 cycles of touch down PCR where the annealing temperature was gradually reduced by 0.8°C per cycle. Endpoint analysis and allelic discrimination related to SNP calls were accomplished using the CFX96 TouchTM software (Bio-Rad, Hercules, CA, United States). The DNA of the restorer line Primepi was used as a reference control for Rf3 (Geyer et al., 2016).
QTL Mapping
To detect the QTL controlling seed set and number of sterile spikelets per spike in BC1 populations, composite interval mapping with a 5-cM window and a maximum of 10 marker cofactors per model was carried out using the Windows QTL Cartographer version 2.5 (Wang et al., 2012). Tests were performed at 1-cM intervals, and cofactors were selected by the forward-backward stepwise regression Model 6 (Shahinnia et al., 2009; Wang et al., 2012). Genome-wide, trait-specific threshold values (α = 0.05) of the likelihood ratio test statistic for declaring the presence of a significant QTL was determined by 2,000 permutations (Churchill and Doerge, 1994). The additive effect of an allelic substitution at each QTL and the phenotypic variation explained by a QTL (R2) conditioned by the composite interval mapping cofactors involved in the model was calculated at the most likely QTL position. The LOD peak of each significant QTL was reflected as the QTL location on the linkage map. To identify markers associated with trait variation located in the confidence interval of a target QTL, single marker analysis was performed using Wald statistics (Kenward and Roger, 1997; Shahinnia et al., 2016). QTL designation followed the recommended rules for wheat3. QTL nomenclature (Qphenotype.lab-chromosome.Qnumber) included “lfl,” “Rf,” and “StS” representing ‘Bayerische Landesanstalt für Landwirtschaft’ (LfL), seed set, and number of sterile spikelets per spike, respectively. The Qnumber after the chromosome designation refers to overlapping QTL identified on the same chromosome in the two BC1 populations.
Physical Mapping and Identification of Candidate Genes
The sequences of the QRf.lfl-6AS.1 flanking markers IWB72428 (3.8 cM) and IWB841 (6.5 cM) within the detected region, that were up to 2 LOD drops from the maximum likelihood value of the selected QTL, were aligned to the reference sequence of Chinese Spring (IWGSC RefSeq v.1.0, Appels et al., 2018) by BLASTN through the URGI portal4 to identify the physical position of the QTL. The protein sequences of the genes in the QTL interval were obtained from Ensembl Plants5 and used for BLASTP homology search (Adamski et al., 2020). Descriptions for the wheat predicted genes based on the IWGSC RefSeq v.1.1 were obtained from BioMart6.
In silico expression values for tissue series of the wheat spike, root, leaf, grain, and stem organs at different developmental stages (Zadoks et al., 1974) were obtained through the WheatExp7 (Pearce et al., 2015) and a bread wheat tissue series RNA-Seq data set8 in POTAGE (Suchecki et al., 2017). Fragments per kb per million reads (FPKM) were used to show the gene expression quantity, thus avoiding the influence of sequencing length and differences on expression values.
Results
Evaluation of Fertility Restoration
The values for seed set and number of sterile spikelets per spike in BC1 Gerek 79 and BC1 71R1203 are presented in Table 1. Whereas a 1:1 segregation ratio for fertile to sterile lines was observed in both BC1 Gerek 79 (174:166) and BC1 71R1203 (100:106), the average seed set was higher in BC1Gerek 79 (0.5) than in BC1 71R1203 (0.3). Number of sterile spikelets per spike showed a negative correlation with seed set in both BC1 Gerek 79 (r = −0.65) and BC1 71R1203 (r = −0.87). Frequency distribution of the traits ranged between 0–2.5 and 0–2.1 for seed set (Figures 1A,B), whereas a range between 5.5–29.5 and 3.7–23.0 was observed for number of sterile spikelets per spike (Figures 1C,D) in BC1 Gerek 79 and BC1 71R1203, respectively.
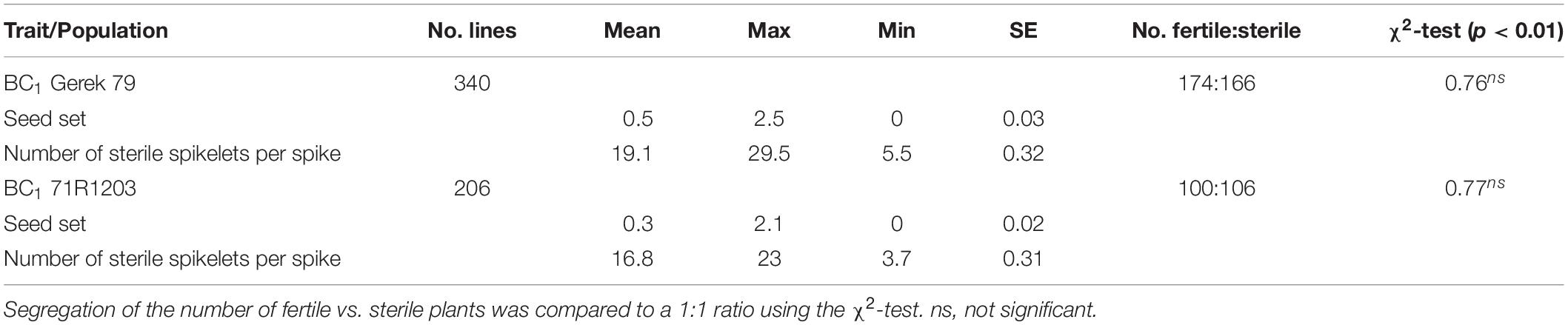
Table 1. Descriptive statistics for seed set and number of sterile spikelets per spike in BC1 Gerek 79 and BC1 71R1203.
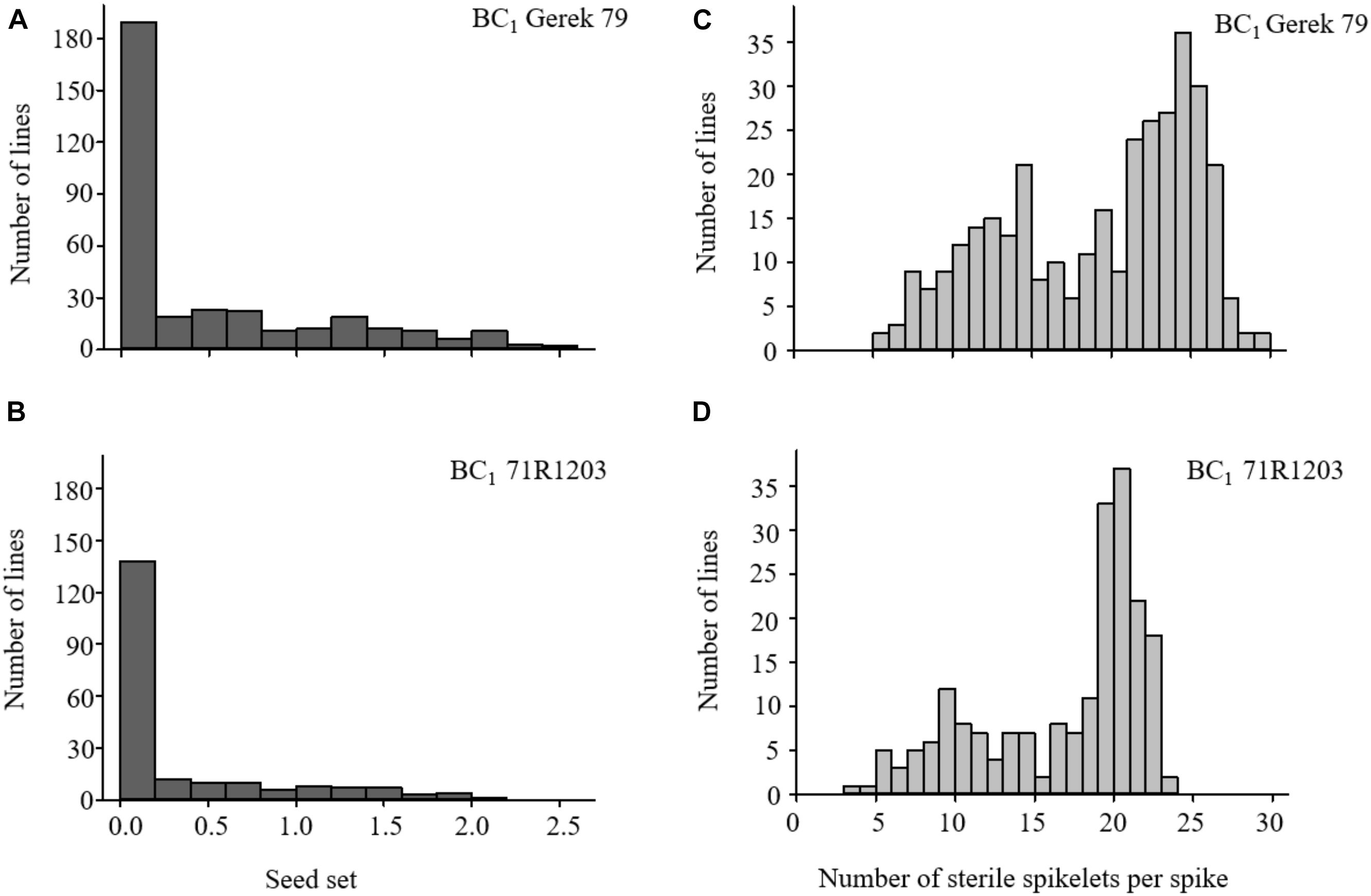
Figure 1. Frequency distribution of phenotypes for (A,B) seed set and (C,D) number of sterile spikelets per spike in BC1 Gerek 79 (n = 340 lines) and BC1 71R1203 (n = 206 lines), respectively. Data are shown based on average overall four spikes (n = 4) per BC1 line.
Construction of Genetic Maps
Following filtration of 16,762 SNPs used for genotyping of BC1 lines, the resulting genetic base maps consisted of 929 and 994 unique SNP loci, spanning 2,160 and 2,328 cM over 21 linkage groups in BC1 Gerek 79 and BC1 71R1203, respectively. The average distance (2.4 cM) between two unique loci was similar in both linkage maps (Supplementary Tables S2, S3).
Using the categorical fertility phenotypes (completely sterile or fertile), a new restorer locus was mapped as a qualitative (monogenically inherited) trait between SNP markers IWB72413 (4.3 cM) and IWB1550 (4.7 cM) in the subtelomeric region of chromosome 6AS in BC1 Gerek 79 (Figure 2A). The newly dissected locus was designated Rf9 following the Catalog of Gene Symbols for Restorers for Cytoplasmic Male Sterility in wheat9. No restorer locus underlying the binary phenotype could be genetically mapped in BC1 71R1203.
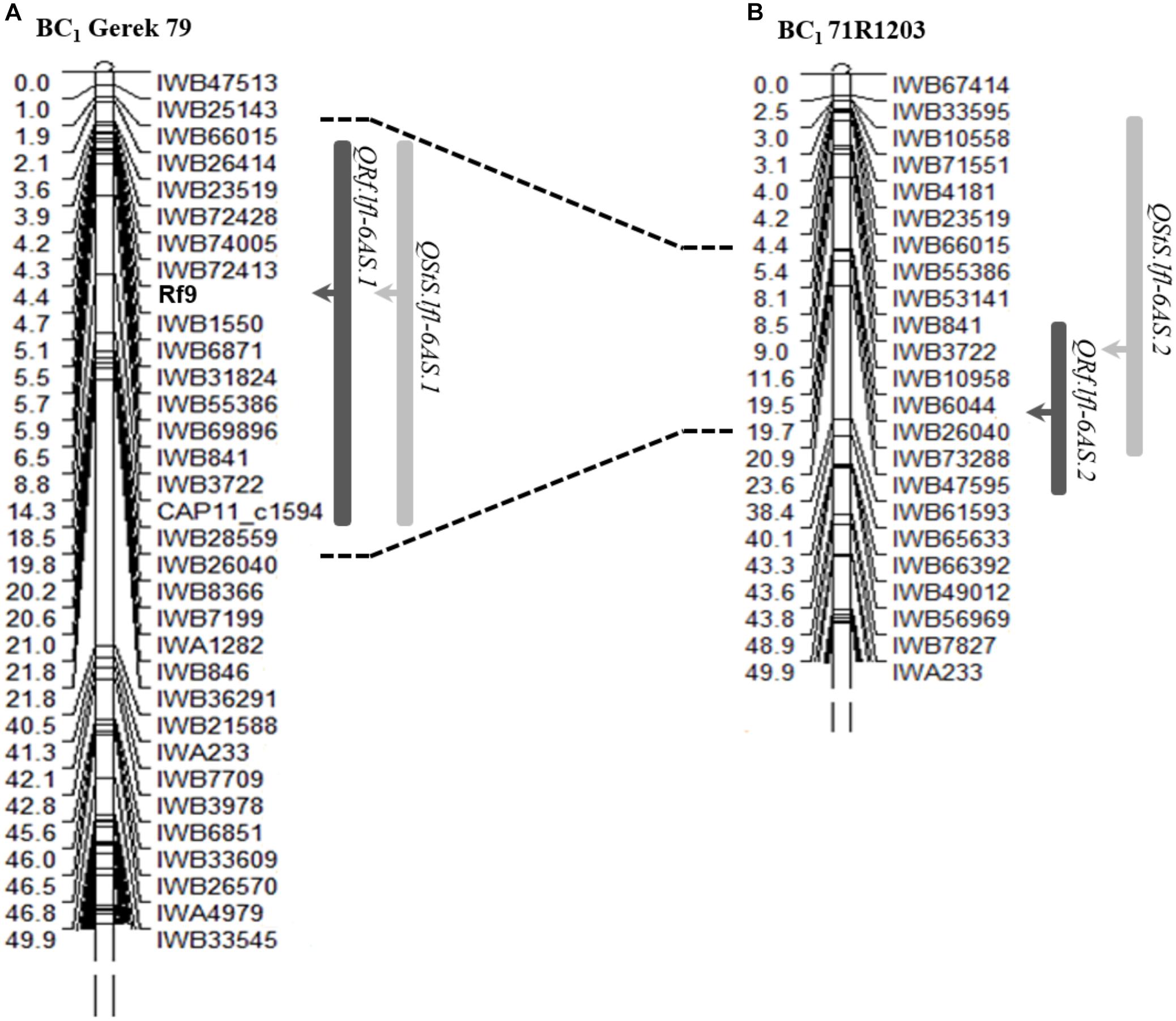
Figure 2. Genetic position of the gene Rf9 and co-location of QTL detected for seed set (QRf, dark bars) and number of sterile spikelets per spike (QStS, light bars) on chromosome 6AS in (A) BC1 Gerek 79 and (B) BC1 71R1203. The peak of each QTL is shown with arrows. Correspondence interval of IWB66015-IWB26040 common markers in two maps is shown with dot lines. A partial linkage map of the short arm of chromosome 6A is presented.
Identification of QTL in BC1 Gerek 79 and BC1 71R1203
Composite interval mapping in BC1 Gerek 79 (Table 2) detected two QTL for seed set on chromosomes 6AS (QRf.lfl-6AS.1) and 4AL (QRf.lfl-4AL) that explained 18 and 14% of the phenotypic variation, respectively. At both loci, the parental line Sperber contributed with negative additive effects indicating that the Gerek 79 alleles increased seed set values. In this population, three QTL for number of sterile spikelets per spike were identified on chromosomes 6AS (QStS.lfl-6AS.1), 6BS (QStS.lfl-6BS), and 2AL (QStS.lfl-2AL). Of these, QStS.lfl-6AS.1, located close to IWB72428, showed the highest LOD score (46.3) and explained 53% of the total phenotypic variation for number of sterile spikelets per spike. The QTL allele that increased the number of sterile spikelets was inherited from the parental line Sperber (Table 2).
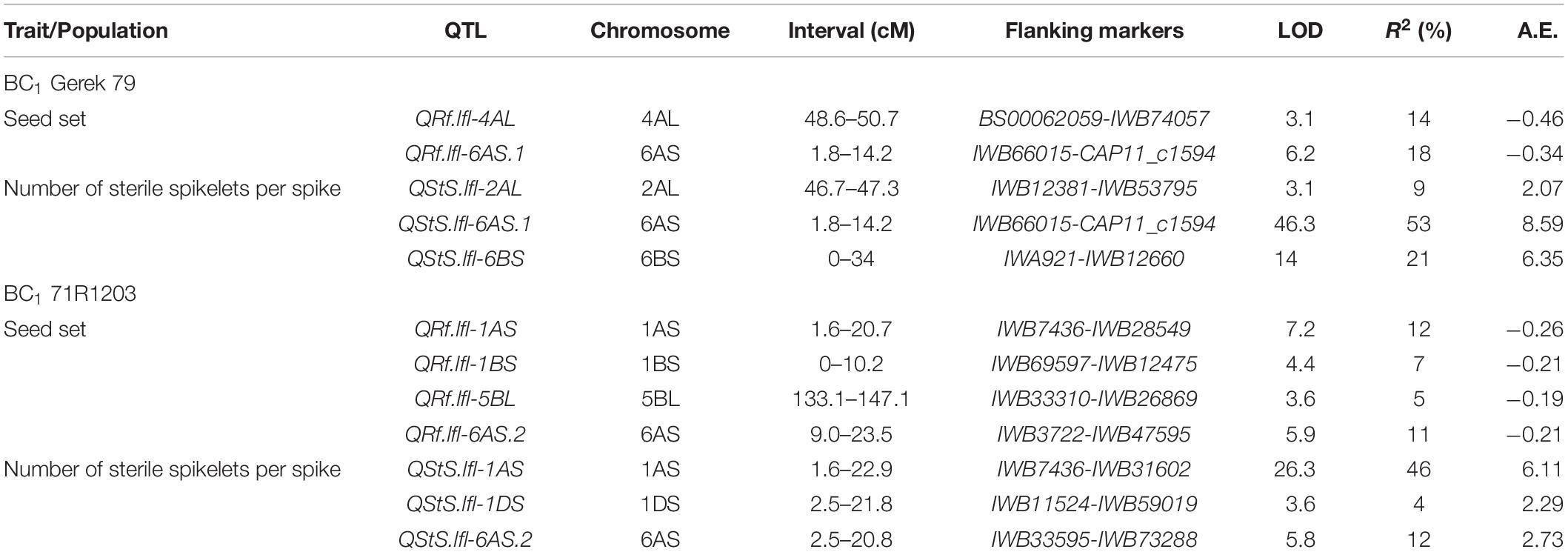
Table 2. Chromosomal locations, map intervals, flanking markers, LOD scores, additive effects (A.E.) of Sperber, and percentage of explained variance by QTL detected for seed set (QRf) and number of sterile spikelets per spike (QStS) in BC1 Gerek 79 and BC1 71R1203.
Four QTL for seed set in BC1 71R1203 population (Table 2) were identified on chromosomes 1AS (QRf.lfl-1AS), 1BS (QRf.lfl-1BS), 5BL (QRf.lfl-5BL), and 6AS (QRf.lfl-6AS.2), explaining 12, 7, 5, and 11% of the total phenotypic variation, respectively. A higher seed set was conferred by the 71R1203 allele at all loci. The most significant QTL for number of sterile spikelets per spike with a LOD score of 26.3 was detected on chromosome 1AS (QStS.lfl-1AS) near to IWB7436 with a positive additive effect derived from Sperber. This QTL, together with two QTL on chromosomes 1DS (QStS.lfl-1DS) and 6AS (QStS.lfl-6AS.2), explained 46, 4, and 12%, respectively, of the total phenotypic variation for the trait (Table 2).
The QTL hotspot for seed set and number of sterile spikelets per spike on chromosome 6AS found in both populations was mapped to the same genomic region (Figures 2A,B). In BC1 Gerek 79, the QTL were identified in the interval between IWB66015 (1.8 cM) and CAP11_c1594 (14.2 cM) for controlling seed set (QRf.lfl-6AS.1) and number of sterile spikelets per spike (QStS.lfl-6AS.1) with the opposite allelic effect of Sperber (−0.34 and 8.59, respectively) (Table 2). Remarkably, the peak of both QTL harbored Rf9, located 4.4 cM proximal to the subtelomeric region in BC1 Gerek 79 (Figure 2A). The magnitudes and directions of allelic effects at Rf9 and SNP loci IWB72413 (4.3 cM) and IWB1550 (4.7 cM) showed a highly significant effect for seed set (Figure 3A) and number of sterile spikelets per spike (Figure 3B), with the favorable allele derived from Gerek 79. In BC1 71R1203, the QTL for seed set and number of sterile spikelets per spike (Figure 2B) revealed negative (−0.21 for QRf.lfl-6AS.2) and positive (2.73 for QStS.lfl-6AS.2) allelic effects of Sperber for seed set and number of sterile spikelets per spike, respectively.
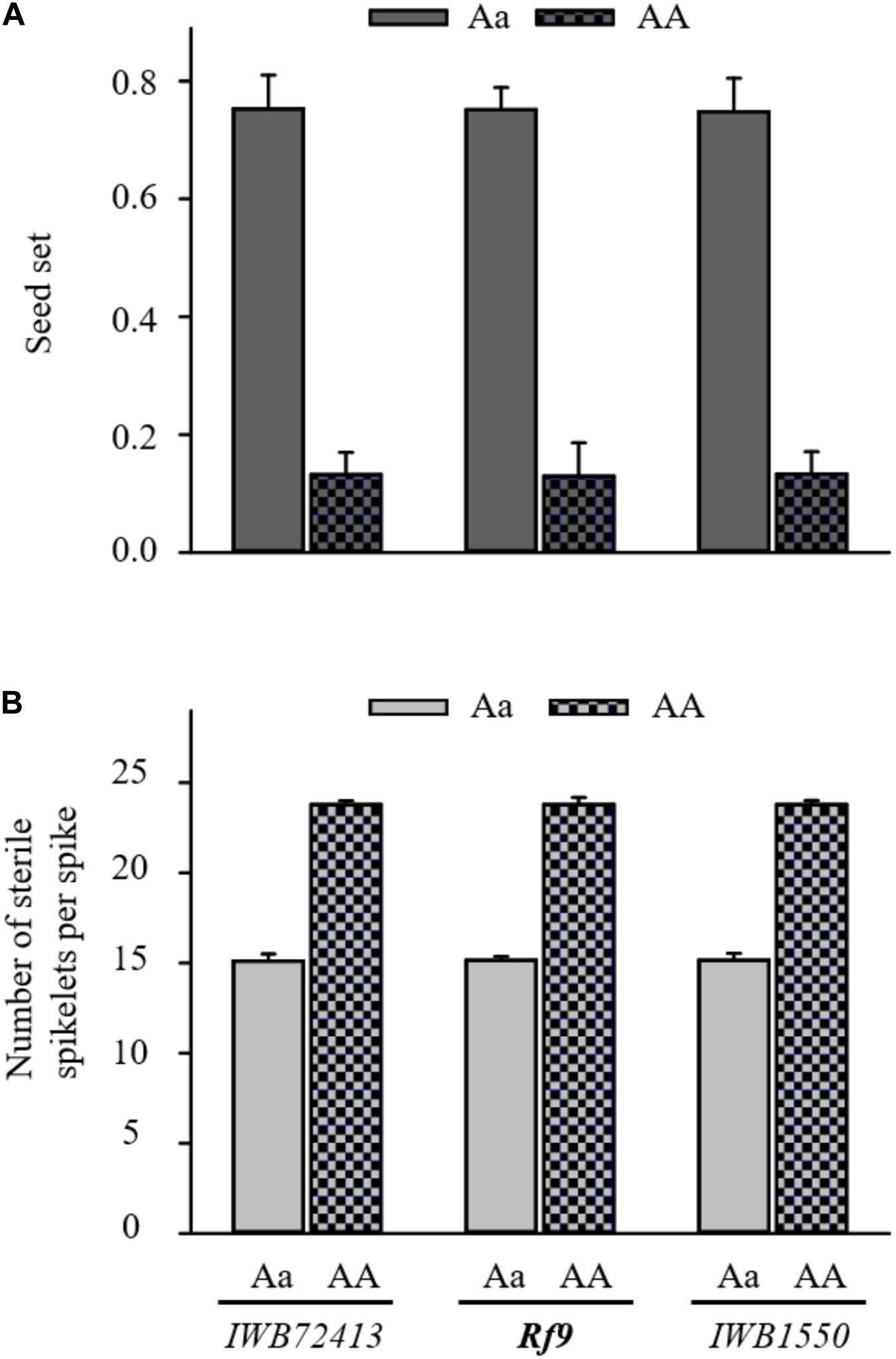
Figure 3. Comparison of allele effects for Rf9 locus and two flanking SNP loci (IWB72413 and IWB1550) at the peak of QRf.lfl-6AS.1 and QStS.lfl-6AS.1 on (A) seed set and (B) number of sterile spikelets per spike in BC1 Gerek 79. Dotted bars refer to the alleles of recurrent parent Sperber (AA) vs. heterozygous BC1F1 lines (Aa) in grey. T-tests based on the mean value of each trait showed highly significant differences (p < 0.001) between the allele effects of Sperber compared to Gerek 79 for all loci.
Candidate Genes Associated With QRf.lfl-6AS.1
To further investigate the chromosomal region associated with QRf.lfl-6AS.1, harboring Rf9, SNP markers IWB72428 (6A: 6.59 Mbp) and IWB841 (6A: 12.39 Mbp) were used to search for putative candidate genes (Supplementary Table S4). The search resulted in 119 gene sequences physically located in nearly 5.8 Mbp on chromosome 6A (6.59–12.38 Mbp). Among those, 23 and 9 genes belonged to the mTERF and PPR family, respectively. The genes TraesCS6A02G019500, TraesCS6A02G019600, TraesCS6A02G019800, TraesCS6A02G019900, and TraesCS6A02G020000 encoded for mTERF family physically (9.3 Mbp) located in the peak of QRf.lfl-6AS.1 close to IWB1550, the flanking marker of Rf9. The in silico expression analysis of these genes showed a wide range of expression in different organs and at three developmental stages (Figure 4). The gene TraesCS6A02G019800 was highly expressed (4.4 FPKM) at Zadoks 65 (full flowering: 50% of anthers matured) in spikes of wheat. The expression of TraesCS6A02G020000 in spikes was higher (3.1 FPKM) at Zadoks 39 (flag leaf ligule visible). The highest expression in grain was observed for TraesCS6A02G019800 (2.5 FPKM) at Zadoks 71 (kernel water ripe, no starch) and for TraesCS6A02G020000 (4.1 FPKM) at Zadoks 85 (kernel soft dough) (Figure 4). Their expression patterns indicated that they could have biological roles in spike and grain, and more likely, spikelet development in wheat.
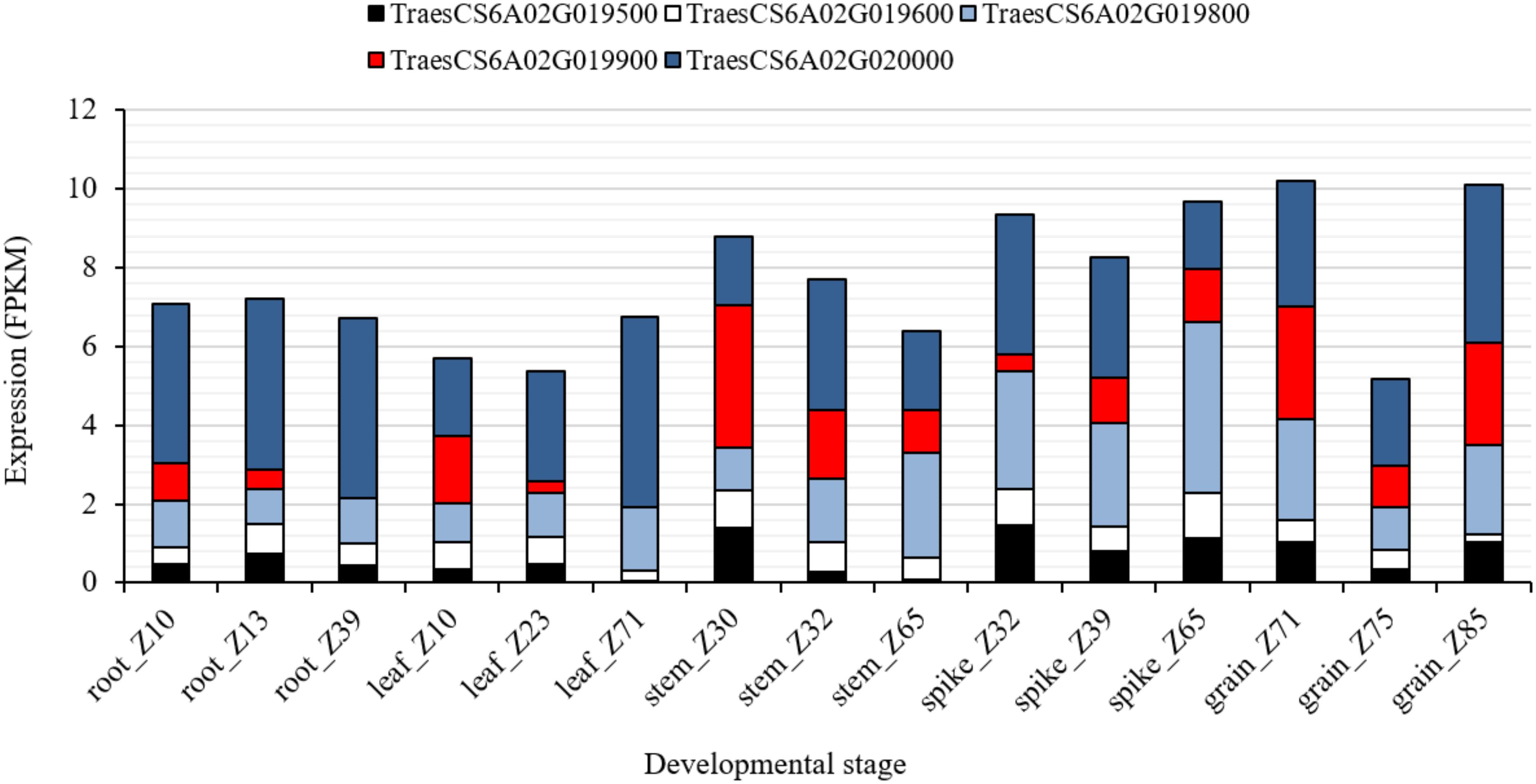
Figure 4. In silico expression analysis of the candidate genes TraesCS6A02G019500, TraesCS6A02G019600, TraesCS6A02G019800, TraesCS6A02G019900, and TraesCS6A02G020000 encoded for mTERF in different wheat plant organs at different developmental stages according to Zadoks et al. (1974).
Discussion
Yield gains associated with heterosis in wheat have been achieved at a slower pace than in other major crops such as maize and rice. The lack of an efficient system for producing hybrid seed is the major bottleneck impairing the competitiveness of hybrid breeding over line-breeding. At the breeding level, it hinders the efficient development of large numbers of test crosses required for the developing of heterotic pools. The CMS hybridization system based on sterility induced by the cytoplasm of T. timopheevii has been proven as a potentially efficient strategy for hybrid seed production. However, further genes and QTL controlling fertility restoration have to be identified and tagged with breeder-friendly molecular markers for utility in a more efficient breeding process. In wheat, this was severely constrained by its large complex genome of about 17 Gb, until the recent release of the reference sequence of the bread wheat variety Chinese Spring (IWGSC RefSeq v1.0, Appels et al., 2018). QTL for general fertility restoration in common wheat have been identified on chromosomes 1BS, 2AL, 2BS, 4BS, and 6AS (Ahmed et al., 2001), 1BS, 5A, and 7D (Zhou et al., 2005), 2DS (Dou et al., 2009), 1AS, 1BS, and 6BS (Geyer et al., 2018), and 2DS, 4BS, and 7AL (Yuan et al., 2020). In the present study, we performed QTL analyses in two populations segregating for restoration capacity contributed by 71R1203 and Gerek 79, for which a novel restorer locus on chromosome 6AS (Rf9) was identified, together with QTL on chromosomes 1AS, 1BS, 1DS, 2AL, 4AL, 5BL, and 6BS.
Gerek 79 was the most widely grown winter wheat cultivar in Turkey in the 1990s. It is a tall wheat derived from a Turkish landrace “Yala,” which has resistance to common bunt caused by Tilletia caries and T. foetida (Bonjean et al., 2001). 71R1203, developed from a cross of winter wheat “NB542437,” was one of the best restorer lines identified in field trials conducted in Eastern Washington during 1974–1978 (Allan and Rubenthaler, 1984). In BC1 populations derived from Gerek 79 and 71R1203, we found 12 QTL for seed set and number of sterile spikelets per spike. Four of these QTL (QRf.lfl-6AS.1, QStS.lfl-6AS.1, QRf.lfl-6AS.2, QStS.lfl-6AS.2) overlapped at the distal end of chromosome 6AS. Since the development of an immortal population which is segregating for restorer loci is hardly feasible in the cytoplasm of T. timopheevii (Geyer et al., 2018), QTL mapping in this study was performed using non-replicated trials, which limits the estimation of phenotypic variance components and testing of the QTL stability across diverse environments. This approach was also demonstrated efficiently and was successful for mapping QTL and major genes Rf1 (Geyer et al., 2018) and Rf8 (Sinha et al., 2013). Ahmed et al. (2001) detected a minor QTL for seed fertility on chromosome 6AS, but located much closer to the centromere using recombinant inbred lines derived from a cross of T. aestivum cv. ‘Chinese Spring’ and T. spelta var. duhamelianum.
It seems the new restorer locus Rf9 that coincided with the QTL peaks of QRf.lfl-6AS.1 and QStS.lfl-6AS.1 in the subtelomeric region of chromosome 6AS (Figure 2A) is new as no Rf gene has been previously reported in the region. Putatively annotated high confidence genes located between the flanking markers of the gene Rf9 showed possible associations with the candidate genes coding for mTERF proteins with higher expression patterns in the spikes and grains (Figure 4). Members of the mTERF family seem to be involved in fertility restoration in cereals. Pan et al. (2019) identified the candidate gene Zea mays small kernel 3 (Zmsmk3), which contained two mTERF motifs and was required for the intron splicing of mitochondrial nad4 and nad1 genes and kernel development. Genome-wide association studies in a multiparental mapping population in hybrid barley detected two mTERF proteins linked to the restorer locus Rfm3 on the short arm of chromosome 6H (Bernhard et al., 2019). A group of candidate genes belonging to the PPR family was also identified in the physical region associated with QRf.lfl-6AS.1. PPR genes have been cloned and well-characterized as essential components for fertility restoration in rice (Hu et al., 2012) and sorghum (Klein et al., 2005). Rizzolatti et al. (2017) identified Rfm1 in barley as a major restorer gene that was described for the CMS system msm1 derived from Hordeum vulgare ssp. spontaneum. The annotation of the nucleotide sequence for the Rfm1 restorer allele showed that the locus carries tandemly repeated genes encoding for PPR proteins of the PLS-DYW subfamily. This group of PPR genes is known to be involved in RNA editing in plant organelles but has not been identified as restorer genes yet. Since the predicted restorer proteins are based on the reference sequence of Chinese Spring, which may not reflect gene content in tested restorer lines, functional characterization of the Rf9 locus identified in BC1 Gerek 79 deserves additional investigations. Molecular cloning of Rf genes could improve our understanding of cytoplasmic nuclear interactions and provide molecular tools to facilitate the development of novel restorer lines. Furthermore, targeted screening of recombinant protein fragments (Edfors et al., 2019) could be used for dissecting the potential role of each candidate and their reciprocal interactions in mitochondrial transcript processing in future studies.
In BC1 71R1203, a genome-wide QTL scan revealed two overlapping major QTL, QRf.lfl-1AS and QStS.lfl-1AS, within the interval between IWB7436 (1.6 cM) and IWB31602 (22.9 cM) on chromosome 1AS (Figure 2B). The identified QTL probably corresponds to Rf1 in line R3, which was reported by Livers (1964), Yen et al. (1969), and Geyer et al. (2018).
Composite interval mapping also detected minor QTL QRf.lfl-4AL, QStS.lfl-2AL, and QStS.lfl-6BS in BC1 Gerek 79 and QRf.lfl-1BS, QRf.lfl-5BL, and QStS.lfl-1DS in BC1 71R1203 (Table 2). The two QTL on chromosomes 1BS and 6BS may be identical to previously identified minor QTL (Ahmed et al., 2001; Zhou et al., 2005; Geyer et al., 2018). A KASP assay developed based on the SNP IWB72107 was used to determine whether QTL QRf.lfl-1BS could be associated with Rf3 located on chromosome 1BS (Geyer et al., 2018). No genetic polymorphism at this SNP locus was found between the parental lines when compared to the marker genotype of Primepi as a reference for the Rf3 allele (data not shown). This indicates that Rf3 was not present in BC1 Gerek 79 or BC1 71R1203. A comparison of flanking SNP markers and genetic distances also showed that none of these minor QTL was linked to a region associated with a major Rf gene previously detected (Ma et al., 1995; Ahmed et al., 2001; Zhou et al., 2005; Stojałowski et al., 2013).
Both BC1 populations exhibited moderate restoration potential leading to incomplete fertility in most of the individuals. Nevertheless, genetic components of restorer parental lines could improve fertility restoration when combined with other restorer loci. Robertson and Curtis (1967) identified modifier genes located on chromosomes 1B, 2A, 3D, 6A, and 6B in Chinese Spring wheat that prevented full restoration, even in the presence of Rf1 and Rf2. Geyer et al. (2018) reported a significant effect of modifier loci located on chromosomes 1BS and 6BL on the penetrance of Rf1, however, such interactions between major genes and modifier loci were proposed in several studies as essential for full fertility restoration (Ahmed et al., 2001; Zhou et al., 2005; Stojałowski et al., 2013; Würschum et al., 2017).
Besides the need to change the autogamous reproduction of wheat to an outcrossing system, the production of commercial hybrid seed will require a stable and efficient hybridization system comprising an easily pollinated female parent and a male parent with the ability to completely restore fertility under most field conditions. Selection for male characteristics such as plant height, anther size, pollen viability and longevity, general and specific combining ability with the female parent(s), and genetic diversity is important for exploring the best pollinators. However, the emphasis should be on the integration of genetic factors that control fertility restoration comprising QTL and Rf genes for developing males through repeated backcrossing. Selection of males segregating for fertility restoration loci in T. timopheevii cytoplasm could be performed by test crossing and evaluation of the plants, particularly under conditions that might inhibit the expression of fertility restoration such as heat and drought (Bonjean et al., 2001). To reach an acceptable level of hybrid fertility, a combination of two to three genes should be sufficient. Therefore, restorer lines with effective major QTL and genes such as Rf1, Rf3, and Rf9 could be tested for maximum fertility restoration. Minor QTL for fertility restoration could be used for improving the female lines. Understanding the mode of action between the candidate genes and their activity in suppressing CMS-inducing open reading frames may help in the development of functional and predictive markers for tracking fertility restoration loci toward increasing the efficiency of the breeding processes. The results of our study could be used for developing new genetic resources of restorer lines and marker-assisted breeding in hybrid wheat. Also, it provides new insights into the genetic mechanisms controlling fertility restoration in wheat with the cytoplasm of T. timopheevii and lays the groundwork for gene characterization, cloning, and manipulation in future fundamental studies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
FS constructed the genetic maps, performed QTL mapping, and wrote the manuscript. FS and AB collected the phenotypic data and carried out the genetic analysis. MG and AB conducted the preliminary experiments. MG and VM supported the genetic analysis and designing of the project. LH conceived the study, supervised the project, and gained funding. All authors discussed the results, read, edited and approved the final manuscript for publication.
Funding
The project was supported by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) under the innovation support programme.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Sabine Schmidt, Petra Greim, Mahira Duran, and the working group Wheat and Oat Breeding Research of the Bavarian State Research Center for Agriculture for excellent technical assistance. We are thankful to Drs. Bianca Büttner, Günther Schweizer, and Theresa Albrecht for the helpful discussions. We gratefully acknowledge valuable support and cooperation from Drs. Julia Rudloff and Erhard Ebmeyer, KWS LOCHOW, Germany in this project, especially for phenotyping the BC1 71R1203 population. We appreciate Prof. Robert McIntosh for providing critical information on the gene symbols and reviewing the manuscript. This manuscript has been released as a pre-print at bioRxiv 2020.06.20.162644 (Shahinnia et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.577475/full#supplementary-material
Supplementary Table 1 | Primer sequences designed for KASP_IWB72107 marker associated with the gene Rf3.
Supplementary Table 2 | Position of the markers (cM) in 21 linkage groups of wheat in BC1 Gerek 79 mapping population.
Supplementary Table 3 | Position of the markers (cM) in 21 linkage groups of wheat in BC1 71R1203 mapping population.
Supplementary Table 4 | List of the potential candidate genes associated with QTL QRf.lfl-6AS.1, harboring Rf9, including flanking SNP markers, physical position, strand, gene ID, and functional description.
Footnotes
- ^ http://wheatpedigree.net/sort/show/21747
- ^ https://triticeaetoolbox.org/wheat/
- ^ https://wheat.pw.usda.gov/ggpages/wgc/98/Intro.htm
- ^ https://urgi.versailles.inra.fr/blast_iwgsc/blast.php
- ^ http://plants.ensembl.org/Triticum_aestivum/Info/Index
- ^ https://plants.ensembl.org/biomart/martview/714673a041bd810256b7d4eb91aafc2d
- ^ https://wheat.pw.usda.gov/WheatExp/
- ^ http://urgi.versailles.inra.fr/files/RNASeqWheat/
- ^ https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassListAction.do?geneClassificationId=68
References
Adamski, N. M., Borrill, P., Brinton, J., Harrington, S. A., Marchal, C., Bentley, A. R., et al. (2020). A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat. Elife. 9:e55646. doi: 10.7554/eLife.55646
Ahmed, T. A., Tsujimoto, H., and Sasakuma, T. (2001). QTL analysis of fertility-restoration against cytoplasmic male sterility in wheat. Genes Genet. Syst. 76, 33–38. doi: 10.1266/ggs.76.33
Allan, R. E., and Rubenthaler, G. L. (1984). Registration of male fertility restoration wheat germplasm. Crop Sci. 24:1220. doi: 10.2135/cropsci1984.0011183X002400060069x
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Bernhard, T., Koch, M., Snowdon, R. J., Friedt, W., and Wittkop, B. (2019). Undesired fertility restoration in msm1 barley associates with two mTERF genes. Theor. Appl. Genet. 132, 1335–1350. doi: 10.1007/s00122-019-03281-9
Bonjean, A. P., Angus, W. J., and van Ginkel, M. (2001). The world wheat book: A history of wheat breeding. Lavoisier: Tec & Doc intercept Ltd.
Castillo, A., Atienza, S. G., and Martin, A. C. (2014). Fertility of CMS wheat is restored by two Rf loci located on a recombined acrocentric chromosome. J. Exp. Bot. 65, 6667–6677. doi: 10.1093/jxb/eru388
Castillo, A., Rodríguez-Suárez, C., Martín, A. C., and Pistón, F. (2015). Contribution of chromosomes 1HchS and 6HchS to fertility restoration in the wheat msH1 CMS system under different environmental conditions. PLoS One 10:e0121479. doi: 10.1371/journal.pone.0121479
Chen, L., and Liu, Y.-G. (2014). Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Churchill, G. A., and Doerge, R. W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. doi: 10.1534/genetics.107.080101
Dou, B., Hou, B., Xu, H., Lou, X., Chi, X., Yang, J., et al. (2009). Efficient mapping of a female sterile gene in wheat (Triticum aestivum L.). Genet. Res. 91, 337–343. doi: 10.1017/S0016672309990218
Eckardt, N. A. (2006). Cytoplasmic male sterility and fertility restoration. Plant Cell 18, 515–517. doi: 10.1105/tpc.106.041830
Edfors, F., Forsström, B., Vunk, H., Kotol, D., Fredolini, C., Maddalo, G., et al. (2019). Screening a resource of recombinant protein fragments for targeted proteomics. J. Proteome Res. 18, 2706–2718. doi: 10.1021/acs.jproteome.8b00924
Geyer, M., Albrecht, T., Hartl, L., and Mohler, V. (2018). Exploring the genetics of fertility restoration controlled by Rf1 in common wheat (Triticum aestivum L.) using high-density linkage maps. Mol. Genet. Genomics 293, 451–462. doi: 10.1007/s00438-017-1396-z
Geyer, M., Bund, A., Albrecht, T., Hartl, L., and Mohler, V. (2016). Distribution of the fertility-restoring gene Rf3 in common and spelt wheat determined by an informative SNP marker. Mol. Breed 36:167. doi: 10.1007/s11032-016-0592-6
Gupta, P. K., Balyan, H. S., Gahlaut, V., Saripalli, G., Pal, B., Basnet, B. R., et al. (2019). Hybrid wheat: past, present and future. Theor. Appl. Genet. 132, 2463–2483. doi: 10.1007/s00122-019-03397-y
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122. doi: 10.1105/tpc.111.093211
Johnson, V. A., Schmidt, J. W., and Mattern, P. J. (1967). Hybrid wheat in the United States. Plant Food Hum. Nutr. 14, 193–211. doi: 10.1007/BF02419924
Kenward, M. G., and Roger, J. H. (1997). Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53, 983–997. doi: 10.2307/2533558
Klein, R. R., Klein, P. E., Mullet, J. E., Minx, P., Rooney, W. L., and Schertz, K. F. (2005). Fertility restorer locus Rf1 [corrected] of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet 111, 994–1012. doi: 10.1007/s00122-005-2011
Kosambi, D. D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Livers, R. W. (1964). Fertility restoration and its inheritance in cytoplasmic male-sterile wheat. Science 144:420. doi: 10.1126/science.144.3617.420
Longin, C. F. H., Mühleisen, J., Maurer, H. P., Zhang, H., Gowda, M., and Reif, J. C. (2012). Hybrid breeding in autogamous cereals. Theor. Appl. Genet. 125, 1087–1096. doi: 10.1007/s00122-012-1967-7
Ma, Z. Q., Sorrells, M. E., and Zhao, Y. H. (1995). Inheritance and chromosomal locations of male fertility restoring gene transferred from Aegilops umbellulata Zhuk. to Triticum aestivum L. Mol. Gen. Genet. 247, 351–357. doi: 10.1007/BF00293203
Mukai, Y., and Tsunewaki, K. (1979). Basic studies on hybrid wheat breeding. Theor. Appl. Genet. 54, 153–160. doi: 10.1007/bf00263045
Pan, Z., Ren, X., Zhao, H., Liu, L., Tan, Z., and Qiu, F. (2019). A mitochondrial transcription termination factor, ZmSmk3, is required for nad1 intron4 and nad4 intron1 splicing and kernel development in maize. G3 9, 2677–2686. doi: 10.1534/g3.119.400265
Pearce, S., Vazquez-Gross, H., Herin, S. Y., Hane, D., Wang, Y., Gu, Y. Q., et al. (2015). WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 15:1–8. doi: 10.1186/s12870-015-0692-1
Plaschke, J., Ganal, M. W., and Röder, M. S. (1995). Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor. Appl. Genet. 91, 1001–1007. doi: 10.1007/BF00223912
R Core Team (2017). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Rizzolatti, C., Bury, P., Tatara, E., Pin, P. A., Rodde, N., Bergès, H., et al. (2017). Map-based cloning of the fertility restoration locus Rfm1 in cultivated barley (Hordeum vulgare). Euphytica 213:276. doi: 10.1007/s10681-017-2056-4
Robertson, L. D., and Curtis, B. C. (1967). Monosomic analysis of fertility restoration in common wheat (Triticum aestivum L.). Crop Sci. 7, 493–495. doi: 10.2135/cropsci1967.0011183X000700050026x
Schnable, P. S., and Wise, R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. doi: 10.1016/S1360-1385(98)01235-7
Shahinnia, F., Geyer, M., Block, A., Mohler, V., and Hartl, L. (2020). Identification of a new gene Rf9 and unravelling the genetic complexity for controlling fertility restoration in hybrid wheat. bioRxiv doi: 10.1101/2020.06.20.162644
Shahinnia, F., Roy, J., Le, Laborde, B., Sznajder, B., Kalambettu, P., et al. (2016). Genetic association of stomatal traits and yield in wheat grown in low rainfall environments. BMC Plant Biol. 16:150. doi: 10.1186/s12870-016-0838-9
Shahinnia, F., Sayed-Tabatabaei, B. E., Sato, K., Pourkheirandish, M., and Komatsuda, T. (2009). Mapping of QTL for intermedium spike on barley chromosome 4H using EST-based markers. Breed. Sci. 59, 383–390. doi: 10.1270/jsbbs.59.383
Sinha, P., Tomar, S. M. S., Vinod, Singh, V. K., and Balyan, H. S. (2013). Genetic analysis and molecular mapping of a new fertility restorer gene Rf8 for Triticum timopheevi cytoplasm in wheat (Triticum aestivum L.) using SSR markers. Genetica 141, 431–441. doi: 10.1007/s10709-013-9742-5
Stojałowski, S., Bobrowska, A., Hanek, M., and Myśków, B. (2013). The importance of chromosomes from the sixth homeologic group in the restoration of male fertility in winter triticale with Triticum timopheevii cytoplasm. J. Appl. Genet. 54, 179–184. doi: 10.1007/s13353-013-0144-2
Suchecki, R., Watson-Haigh, N. S., and Baumann, U. (2017). POTAGE: A visualisation tool for speeding up gene discovery in wheat. Sci. Rep. 7, 1–8. doi: 10.1038/s41598-017-14591-7
Virmani, S. S., and Edwards, I. B. (1983). Current status and future prospects for breeding hybrid rice and wheat. Adv. Agron. 36, 145–214. doi: 10.1016/S0065-2113(08)60354-5
Wang, S., Basten, C. J., and Zeng, Z. B. (2012). Windows QTL Cartographer 2.5. Raleigh, NC: North Carolina State University.
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Whitford, R., Fleury, D., Reif, J. C., Garcia, M., Okada, T., Korzun, V., et al. (2013). Hybrid breeding in wheat: Technologies to improve hybrid wheat seed production. J. Exp. Bot. 64, 5411–5428. doi: 10.1093/jxb/ert333
Wilson, J. A., and Ross, W. M. (1962). Male sterility interaction of the Triticum aestivum nucleus and Triticum timopheevii cytoplasm. Wheat Inf. Serv. 14, 29–30.
Wimmer, V., Albrecht, T., Auinger, H. J., and Schön, C. C. (2012). synbreed : a framework for the analysis of genomic prediction data using R. Bioinformatics 28, 2086–2087. doi: 10.1093/bioinformatics/bts335
Würschum, T., Leiser, W. L., Weissmann, S., and Maurer, H. P. (2017). Genetic architecture of male fertility restoration of Triticum timopheevii cytoplasm and fine-mapping of the major restorer locus Rf3 on chromosome 1B. Theor. Appl. Genet. 130, 1253–1266. doi: 10.1007/s00122-017-2885-5
Yen, F. S., Evans, L. E., and Larter, E. H. (1969). Monosomic analysis of fertility restoration in three restorer lines of wheat. Can. J. Genet. Cytol. 11, 531–546. doi: 10.1139/g69-062
Yuan, S.-H., Bai, J.-F., Guo, H.-Y., Duan, W.-J., Liu, Z.-H., Zhang, F.-T., et al. (2020). QTL mapping of male sterility-related traits in a photoperiod and temperature-sensitive genic male sterile wheat line BS366. Plant Breed. 139, 498–507. doi: 10.1111/pbr.12811
Zadoks, J. C., Chang, T. T., and Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Keywords: cytoplasmic male sterility, seed set, sterile spikelet, Triticum aestivum, Triticum timopheevii
Citation: Shahinnia F, Geyer M, Block A, Mohler V and Hartl L (2020) Identification of Rf9, a Gene Contributing to the Genetic Complexity of Fertility Restoration in Hybrid Wheat. Front. Plant Sci. 11:577475. doi: 10.3389/fpls.2020.577475
Received: 29 June 2020; Accepted: 14 October 2020;
Published: 10 December 2020.
Edited by:
Marcelino Perez De La Vega, Universidad de León, SpainReviewed by:
Ryan Whitford, University of Adelaide, AustraliaJoanna Melonek, University of Western Australia, Australia
Copyright © 2020 Shahinnia, Geyer, Block, Mohler and Hartl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenz Hartl, bG9yZW56LmhhcnRsQGxmbC5iYXllcm4uZGU=
 Fahimeh Shahinnia
Fahimeh Shahinnia Manuel Geyer
Manuel Geyer Annette Block
Annette Block Volker Mohler
Volker Mohler Lorenz Hartl
Lorenz Hartl