
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 October 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.570862
Early detection of plant diseases is a crucial factor to prevent or limit the spread of a rising infection that could cause significant economic loss. Detection test on plant diseases in the laboratory can be laborious, time consuming, expensive, and normally requires specific technical expertise. Moreover, in the developing countries, it is often difficult to find laboratories equipped for this kind of analysis. Therefore, in the past years, a high effort has been made for the development of fast, specific, sensitive, and cost-effective tests that can be successfully used in plant pathology directly in the field by low-specialized personnel using minimal equipment. Nucleic acid-based methods have proven to be a good choice for the development of detection tools in several fields, such as human/animal health, food safety, and water analysis, and their application in plant pathogen detection is becoming more and more common. In the present review, the more recent nucleic acid-based protocols for point-of-care (POC) plant pathogen detection and identification are described and analyzed. All these methods have a high potential for early detection of destructive diseases in agriculture and forestry, they should help make molecular detection for plant pathogens accessible to anyone, anywhere, and at any time. We do not suggest that on-site methods should replace lab testing completely, which remains crucial for more complex researches, such as identification and classification of new pathogens or the study of plant defense mechanisms. Instead, POC analysis can provide a useful, fast, and efficient preliminary on-site screening that is crucial in the struggle against plant pathogens.
Plant pathogens represent one of the major threats for agriculture worldwide (Narayanasamy, 2011; Aslam et al., 2017; Pallas et al., 2018). In the last centuries, in parallel with the continuing growth of the human population, the percentage of the land surface covered by crops has increased constantly, with ca. 50% of the current habitable world land dedicated to agriculture1. In 2016, it was estimated that approximately 540 million hectares were planted worldwide to only three major crop species: maize, rice, and wheat (McDonald and Stukenbrock, 2016). Plant pathogens can cause substantial reduction of crop productivity. Considering five of the major crops (maize, rice, wheat, potatoes, and soybean), it was shown that the losses, associated with pathogens and pests at a global level, ranged between 17 and 30% annually (Ramakrishnan et al., 2019; Savary et al., 2019). In agriculture and forestry, most of the damage is due to the accidental introduction of invasive alien pathogen species into new areas, as a consequence of global trade and transport (Ghelardini et al., 2017). Moreover, the extension of the distribution range of the pathogens due to human-mediated activities facilitates hybridization and horizontal gene transfer, leading to the emergence of new pathogens (Gluck-Thaler and Slot, 2015). Also, the introduction of arthropod vectors into new areas can bring the establishment of novel association with introduced or native pathogen species (Wingfield et al., 2016). Well known examples of introduction of invasive pathogens exist both in agriculture and forestry. The wheat blast pathogen Pyricularia graminis-tritici appeared recently in Asia for the first time, maybe coming from South America, and devastated more than 15,000 ha of crops in Bangladesh (Callaway, 2016). In Italy, since 2013, Xylella fastidiosa is causing a severe loss of olive trees in the southern part of the country. Genetic analysis of the Italian strains showed similarity to isolates from Central America (Marcelletti and Scortichini, 2016; Giampetruzzi et al., 2017). Cronartium ribicola, the causal agent of white pine blister rust migrated from Europe to North America along with plant material, while the subspecies americana of the Dutch elm disease fungus Ophiostoma novo-ulmi arrived in Europe from North America with rock elm logs (Ghelardini et al., 2017).
Another factor that influences plant–pathogen interaction is climate change. Increased temperature, climate extremes, as well as differences in quantity and pattern of annual precipitation, can support the spread of plant diseases in agriculture (Schmidhuber and Tubiello, 2007) and forests (La Porta et al., 2008). Finally, the great majority of the cultivated soil is planted to monocultures or even to only one genotype (fruit trees, grape, and potato), thus creating a homogeneous genetic environment that can easily select host-specialized crop pathogens.
In order to control, and when possible to prevent, plant diseases and the spread of plant pathogens into new areas, it is mandatory to develop fast, efficient, and inexpensive methods for early detection of pathogens. Traditional methods for fungi and bacteria identification rely on symptom observation and culture-based methods (Alahi and Mukhopadhyay, 2017). This implies the necessity of growing the pathogen on specific media in controlled conditions, a process that usually takes days or even weeks. Moreover, the morphological identification of the pathogen by microscopy requires specifically trained personnel.
Other widely used phyto-diagnostic methods are based on immunological techniques such as enzyme-linked immunosorbent assay (ELISA) (Cho and Irudayaraj, 2013), immunofluorescent staining (Khater et al., 2017), immunoblot (Novakova et al., 2006), and lateral flow immunoassays (LFIA) (Burnham-Marusich et al., 2018; Cassedy et al., 2020). Nonetheless, the production of monoclonal antibodies may be expensive, and in some cases, problems of low specificity have been reported (Gorris et al., 1994; Martinelli et al., 2015). Since the nineties, more laboratories started to adopt DNA-based methods for pathogen detection and identification, using polymerase chain reaction (PCR) and its variants, such as quantitative PCR (qPCR), nested PCR, multiplex PCR, and digital PCR (ddPCR) (Martinelli et al., 2015). Such methods are usually very specific, relatively fast, and cheap but still present some disadvantages, as in most cases, specific equipment is required as well as trained personnel. Since the first few years of this century, new technologies are emerging based on isothermal amplification of DNA, such as the loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), helicase-dependent amplification (HDA), and others (Lau and Botella, 2017), capable of overcoming some of the drawbacks of PCR-based methods (Zhong and Zhao, 2018). All these approaches, even if based on different principles, share the main characteristic of amplifying target DNA at constant temperature, therefore eliminating the need of a thermal cycler.
Due to the rapid evolution of methods for pathogen detection, in many fields such as human health and food safety, it is dawning the possibility to develop new tools that can be taken directly to the desired analysis site, also called point-of-care (POC) (Chen et al., 2019; Vidic et al., 2019). Ideally, such tools should consist of robust, portable, user-friendly, and inexpensive equipment capable of performing all the steps required for a complete analysis in a short time and with a minimal number of steps, in order to be used also by low-trained personnel. All these requirements have been summarized for the first time by the World Health Organization (WHO) in the ASSURED (Affordable, Sensitive, Specific, User-friendly, Robust and rapid, Equipment-free, Deliverable) guidelines for POC testing (Peeling et al., 2006). The potential advantages of POC analysis are numerous (Table 1), going from the speed of the analysis to the lower risk of sample contamination and pathogen dispersion. As a matter of fact, the possibility to detect a pathogen (Radhakrishnan et al., 2019) directly in the field would be of great interest in agriculture and forestry. It would allow extensive and fast screening of plants in order to prevent disease spreading. Moreover, imported plants and products could be analyzed directly on-site before they can even enter a country. Two important applications of POC analysis are the study of emergent non-native introduced pathogens (Tomlinson et al., 2010; Buhlmann et al., 2013; Blaser et al., 2018; Aglietti et al., 2019; Rani et al., 2019) and chronic disease monitoring (De Boer and Lopez, 2012; Zheng et al., 2013).

Table 1. Main advantages of point-of-care (POC) analysis when compared to standard laboratory testing.
The DNA-based analysis methods are proving to be good candidates for the development of POC analysis protocols, especially since the advent of the new isothermal technologies. Nevertheless, providing a portable tool capable of performing a complete test in the field is not an easy task as there are at least three main steps that should be performed: (a) sample preparation/DNA extraction; (b) DNA amplification; (c) signal detection (Figure 1). In the following sections, we analyze the progress made in the recent years in order to exploit DNA-based technologies for the development of portable tools suitable for plant pathogen detection, considering all the main steps of the analysis and with a particular focus on their application in agriculture and forestry.
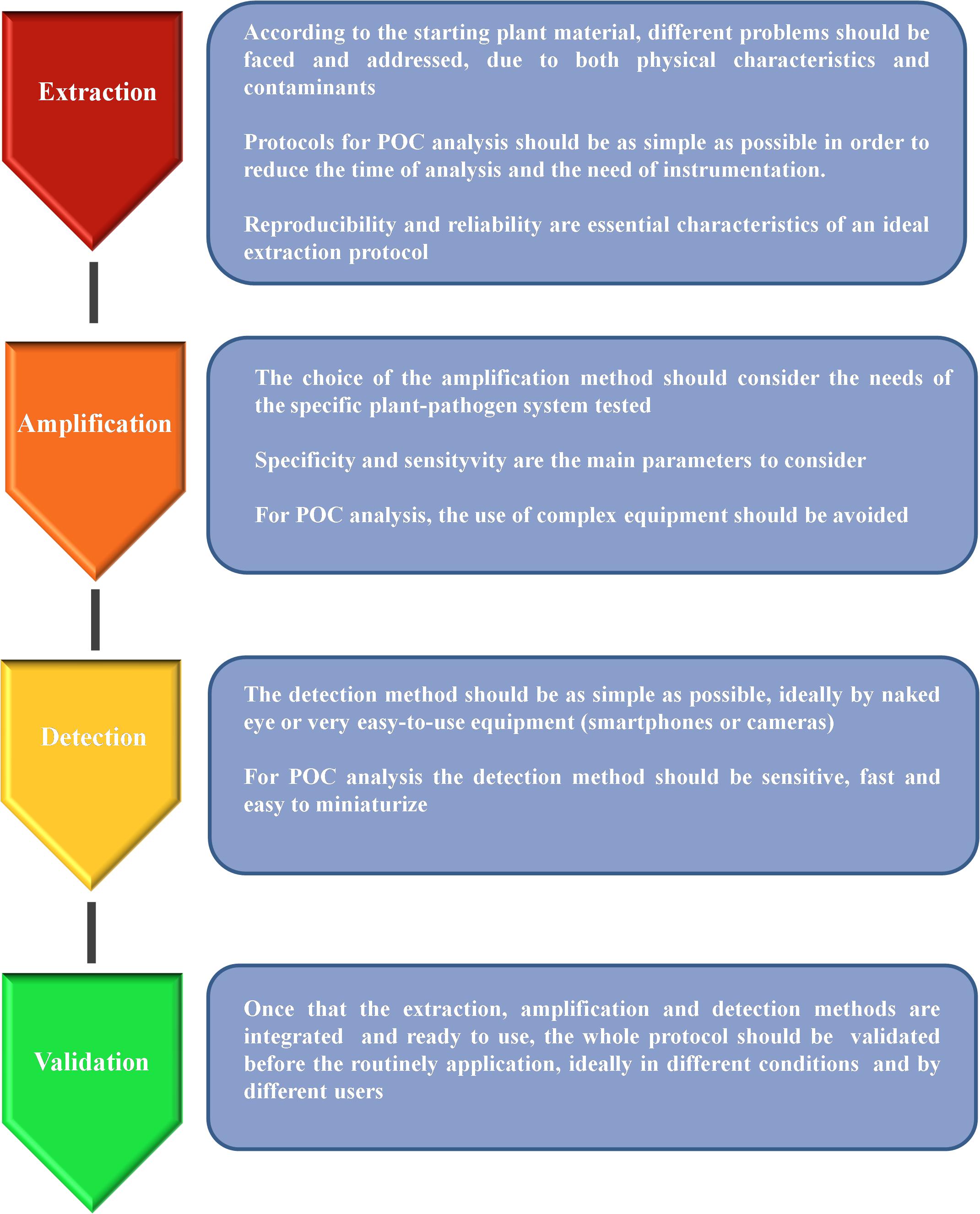
Figure 1. Schematic representation of the main steps needed for the development of a reliable protocol suitable for point-of-care (POC) analysis.
Despite the significant progress made to adapt the DNA amplification technologies to the needs of POC analysis of plant pathogens (Lau and Botella, 2017; Donoso and Valenzuela, 2018; Dairawan and Shetty, 2020), one of the main obstacles is plant sample preparation and nucleic acid extraction. As a matter of fact, a high-quality DNA or RNA is always necessary in order to obtain reliable and reproducible results, irrespective of the method used for nucleic acid amplification. Even if several methods have been developed for the fast extraction of nucleic acids from bacteria, animal cells, and food (Cheng and Jiang, 2006; Kim et al., 2009; Cawthorn et al., 2011; Abdalhai, 2016), such technologies are often difficult to apply to plant tissues. If we consider a leaf, the most common plant tissue used for nucleic acid extraction, cells are surrounded by several protective structures, such as cell wall and waxy cuticle layers. All these structures must be degraded in order to extract the genetic material inside, therefore adding a level of complexity to the protocol. Moreover, depending on the different plant species and tissues, a lot of plant contaminant products can be present, such as proteins, polysaccharides, polyphenolics, and a great number of secondary metabolites (Varma et al., 2007). All these contaminants should be removed from the genetic material in order to obtain a high-quality template suitable for amplification. Several strategies can be applied, such as the removal of cellular proteins by organic solvents (typically phenol and chloroform), but all these steps are quite difficult to include in a completely portable device.
Many different solutions have been proposed (Table 2), the simpler of which is the use of plant crude extract (ground plant material with an extraction buffer) directly for amplification. Munawar et al. (2019) used strawberry crude crown tissue macerated with a standard ELISA grinding buffer coupled with RPA for the detection of Phytophthora cactorum infecting strawberry. Successful detection of the pathogen was achieved, after testing several concentrations of the grinding buffer, in order to optimize amplification also in plants showing early or no symptoms. Several works showed comparison in amplification efficiency between purified templates and crude extracts. For the detection of Xanthomonas fragariae, the causal agent of angular leaf spot of strawberry, a test was carried out using two types of templates: pure bacterial DNA and crude extract (Getaz et al., 2017). LAMP DNA amplification was successful with crude tissue extract as well as with the pure bacterial DNA control, after optimization of the protocol. The optimized protocol with phenol inhibitors was also successfully tested in naturally infected strawberry leaves, even those with weak symptoms, therefore suggesting that this method could be a suitable POC analysis. In another work, different plant sample preparation methods were compared, using sap obtained after grinding apricot (Prunus armeniaca) plant tissues as a starting material for the detection of Candidatus phytoplasma prunorum (Minguzzi et al., 2016). Similarly, several grinding buffers were compared for Fragaria ananassa crude sample preparation for the rapid detection of Phytophthora spp. (Miles et al., 2015). All the above reported results seem to indicate that the use of plant crude extract for the direct detection of pathogens with nucleic acid amplification technologies can be effective and therefore used for in-field application. The advantage of crude extracts is essentially due to the very simple and fast extraction protocols, while the main drawback is the possible lack of sensitivity. Therefore, a preliminary testing of the method is always necessary, together with an appropriate optimization according to the plant species, tissue used as starting material, and specific primers designed for the amplification step.
Sensitivity and specificity of pathogen detection protocols can be improved by purifying target DNA. There are a lot of commercial kits available for this purpose, but most of them require to be used in a laboratory with dedicated equipment. Nonetheless, several commercial kits have been tested, and some of them showed characteristics that could be suitable for POC analysis, especially those based on the use of magnetic beads for nucleic acid purification (DeShields et al., 2018). Several works can be found in literature exploiting this extraction principle on various plant species and pathogens, such as detection of the fungus Leptosphaeria maculans, causing Phoma stem canker (black leg) of Brassica napus (Lei et al., 2019). Other examples include detection of soil-borne pathogens of potatoes (DeShields et al., 2018); Alternaria panax Whetz infecting ginseng (Wei et al., 2018); Puccinia striiformis f.sp. tritici (wheat yellow rust) infecting wheat (Radhakrishnan et al., 2019); Phytophthora kernoviae infecting Rhododendron ponticum (Schwenkbier et al., 2015b) and several other species of Phytophthora (Julich et al., 2011; Schwenkbier et al., 2014). It must be noted that in some of the above reported cases, nucleic acid extraction was performed from cultivated pathogens and not directly from infected plants, therefore suggesting that optimization of the protocol could be necessary before its routine application in the field. A magnetic bead-based RNA extraction method was used to develop a microfluidic chip for the detection of viruses infecting Phalaenopsis orchid (Chang et al., 2013; Lin et al., 2015). The main advantage of the magnetic bead-based extraction method is reliability, as most of the more commonly used protocols exploit robust commercial kits already available on the market. Moreover, it is possible to extract genetic material without the need of centrifugation steps, but simply using a magnet that can be easily transported and used on site even when electricity supply is not available. One possible drawback is that most of the commercial kits are not 100% user friendly, and some training is still necessary to use them properly and avoid contamination. In order to overcome this problem, some authors tried to simplify the extraction protocol, still using commercial kits but introducing some modifications. The first report describing a complete protocol for plant pathogen detection that could be performed directly in the field dates back to 2005, and the DNA was extracted modifying a commercial kit in order to avoid the use of liquid nitrogen and laboratory equipment, such as centrifuge and vortex (Tomlinson et al., 2005). Similarly, other authors proposed a modification to standard protocols to simplify the extraction of genetic material suitable for amplification with various techniques (Li et al., 2011; Koo et al., 2013).
A very simple method proposed for DNA or RNA extraction is the so-called “toothpick method,” in which a sterile toothpick is used to pierce the sample tissue several times. The tip of the toothpick is then dipped directly in the amplification reaction mix, releasing target genetic material (Fukuta et al., 2005; Suzuki et al., 2016; Wilisiani et al., 2019). Despite the extreme simplicity, this method needs to be tested and optimized for each specific need; an example is provided in the next lines. A modern version of the “toothpick method” was recently proposed using disposable polymeric microneedle patches (MP) to pierce plant leaves. These patches are made of polyvinyl alcohol (PVA), a water-absorbing polymer that showed a good combination of mechanical strength, chemical resistance, and biocompatibility (Paul et al., 2019, 2020). The use of MP to detect Phytophthora infestans from tomato leaves showed a slightly lower sensitivity in comparison to the standard CTAB extraction method, especially during the very early stages of infection. On the other hand, the extraction time was reduced to 1 min, instead of 3–4 h of the CTAB method, suggesting that MP could be easily used for in-field applications.
In the last few years, the development of new nucleic acid extraction methods for POC analysis has increased, exploring different approaches. Zou et al. (2017) tested a variety of materials for their ability to capture nucleic acids and found that Whatman No. 1 cellulose-based filter paper can efficiently entrap and retain DNA without the need of any chemical treatment. Filter paper-based extraction methods were positively tested on different plant species, such as wheat, rice, tomato, soybean, tobacco, mandarin, and lemon, showing good sensitivity and reproducibility. Similarly, Flinders Technology Associates (FTA® Card, WhatmanTM, catalog number: WB120205, Merck KGaA, Darmstadt, Germany) cards were used for extraction of DNA or RNA from different starting materials, including plants, to provide a versatile POC pathogen analysis method (Lu et al., 2016). Paper-based microfluidics device has emerged as a multiplexable POC platform, which might be useful in resource-limited settings (Yetisen et al., 2013). This device was applied in health care, veterinary medicine, food safety, and environmental and crop monitoring.
A rapid technique for the extraction of viral RNA from rose plants was also proposed, based on the direct virus absorption onto PCR tubes at 4°C (Babu et al., 2017). This extraction method provided viral RNA suitable for molecular analysis in as few as 5 min. This method is very fast and simple, but more studies are still necessary to understand if it could be applicable routinely to different pathogens and plant species.
Another option for the rapid extraction of nucleic acids from plant tissue is the use of lateral-flow devices (LFDs). A method was developed allowing the extraction of amplifiable DNA from plant tissue using LFD in as few as 5 min without the use of any equipment (Danks and Barker, 2000). A simplified version of this method was described and applied to the detection of Phytophthora ramorum and Phytophthora kernoviae (Tomlinson et al., 2010).
Once that the DNA or RNA is extracted, the next step is the amplification of the genetic material using specific primers designed to recognize target sequences of the pathogen. Several approaches can be used and are listed in Table 3. The most popular methods for laboratory nucleic acid amplification are still PCR-based ones, including many variants such as quantitative real-time PCR (qPCR), nested PCR, and digital PCR. Nevertheless, when rapid and simple POC amplification is required, PCR may present some drawbacks, as it usually requires a thermal cycler, a device that is not easy to miniaturize and integrate in a portable tool. Still, some attempts in this direction have been made for POC application in agriculture. Julich et al. (2011) developed a lab-on-a-chip device for rapid nucleic acid-based detection of Phytophthora species. Temperature management in the PCR and hybridization zones relies on two independent Peltier elements. The system is completed by a compact portable pump and power supply for use on site. A similar approach was proposed by Schwenkbier et al. (2014) who described a lab-on-a-chip device for on-site detection of Phytophthora species, exploiting linear-after-the-exponential PCR (LATE-PCR) as amplification method.
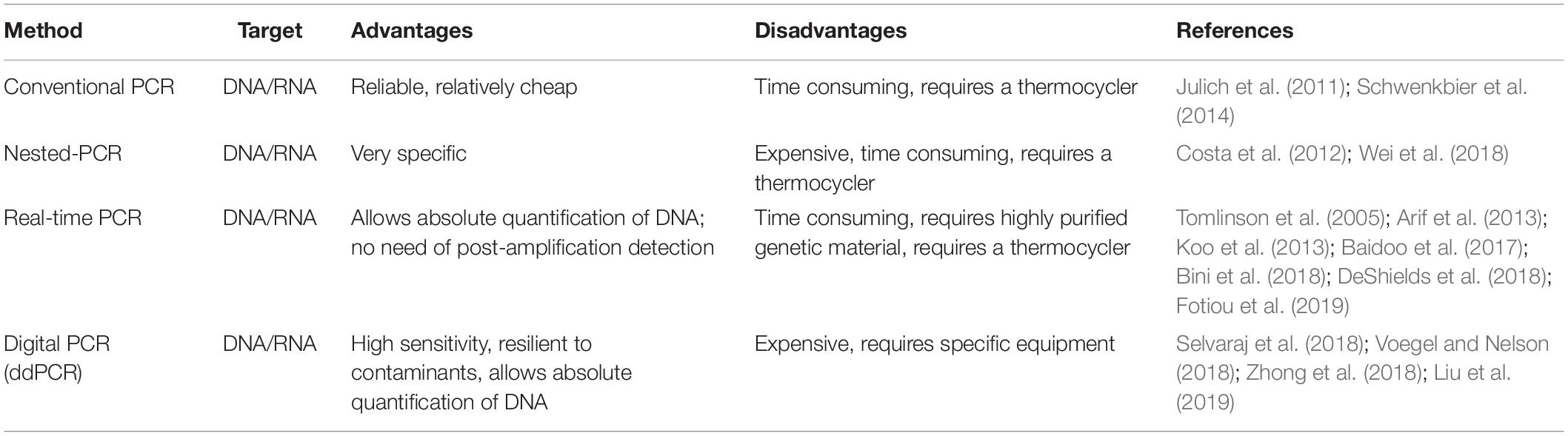
Table 3. List of the main polymerase chain reaction (PCR)-based amplification methods applied for POC plant pathogen detection.
Also, qPCR was often proposed as suitable for POC analysis. One of the first reports dates back to 2002 (Schaad et al., 2002) when a protocol for 1-h on-site diagnosis of Xylella fastidiosa was described using a portable Smart Cycler. Other examples of qPCR-based methods for POC pathogen detection include a system for the detection of Phymatotrichopsis omnivora, in alfalfa and other dicot plants (Arif et al., 2013) and a protocol for the detection of Phytophthora ramorum, an invasive plant pathogen listed under quarantine in many countries and capable of infecting over 180 forest trees species (Tomlinson et al., 2005). Papers describing fast protocols for plant pathogen detection via qPCR that could be potentially used for in-field applications have been published for several species such as Plum pox virus (Fotiou et al., 2019), Spongospora subterranea (DeShields et al., 2018), Austropuccinia psidii (Bini et al., 2018), and Pratylenchus penetrans (Baidoo et al., 2017). In some cases, real-time-based protocols were proposed for multiplex detection of plant pathogens (Papayiannis, 2014; Khan et al., 2015; Yang et al., 2015; Osman et al., 2017; Nikitin et al., 2018). A recent study proposed an In Situ Processing and Efficient Environmental Detection (iSPEED) kit to detect pests and pathogens of forest trees using POC qPCR (Capron et al., 2020). Targeted diseases included the poplar canker pathogen Sphaerulina musiva, the white pine blister rust Cronartium ribicola, the comandra blister rust of pines C. comandrae, the Port Orford cedar pathogen Phytophthora lateralis, and the agent responsible for sudden oak and sudden larch death P. ramorum. The iSPEED kit could be carried in a small backpack and showed results comparable to those obtained with standard laboratory equipment. The advantages of the iSPEED kit are the existence of a great amount of published and validated real-time PCR assays, multiple enzymes and chemistry for qPCR, as well as a growing range of portable instruments, cost effectiveness, and flexibility of design. Another variant of conventional PCR that can be used to increase specificity and sensitivity is nested-PCR. The use of two pairs of specific primers usually make this approach more time consuming and expensive than conventional PCR. Nevertheless, some researchers have developed a single-tube nested-PCR approach that potentially eliminates the drawbacks of this technique, such as contamination of the sample (Costa et al., 2012; Wei et al., 2018).
Finally, droplet digital PCR (ddPCR) is a new technology allowing absolute quantification of target DNA, which is partitioned into approximately 20,000 droplets. The amplification reaction is carried out independently within each droplet, and a reader can estimate the amount of amplified DNA in each droplet by fluorescence measurement (Hindson et al., 2011). ddPCR offers many advantages when compared to conventional PCR and even qPCR, as multiple target genes can be detected in a single reaction using different fluorescence signals. Moreover, no standard curve is required for quantification, and the sensitivity of ddPCR was shown to be higher than quantitative PCR when measuring low copy-number genes (Hayden et al., 2013). Another advantage is the high resilience of ddPCR to contaminants, that can present a major problem when plant samples are analyzed (Racki et al., 2014; Blaya et al., 2016). Although ddPCR has not been used for POC analysis in agriculture yet, some approaches using this method have been described (Selvaraj et al., 2018; Zhong et al., 2018; Liu et al., 2019), and in one case, the ddPCR protocol was proposed to be applied for use in national clean plant programs to prevent the import of infected nursery stock (Voegel and Nelson, 2018).
In recent years, new technologies for isothermal amplification of nucleic acids have been developed, and their use is rapidly spreading (Table 4). The first and most commonly used is loop-mediated isothermal amplification (LAMP) (Notomi et al., 2000). The main advantages of LAMP when compared to PCR-based approaches are the possibility to perform the amplification reaction at constant temperature, the short reaction time, high amplification efficiency, and sensitivity, combined to a relatively low cost, as LAMP requires very simple equipment to be performed (Zhao et al., 2015; Lai et al., 2018). Several portable devices for POC detection of plant pathogen have been described exploiting LAMP technology. A rapid and simple method to identify three different species of begomovirus infecting Cucurbitaceae and Solanaceae plants was recently published, using a commercial portable device that is battery powered, user friendly, handheld, and waterproof (Wilisiani et al., 2019). Another example of portable device suitable for agriculture and forestry application is a micropipette tip-based nucleic acid test (MTNT) capable of detecting both DNA and RNA from different plant crude extracts (Lu et al., 2016). An increasing number of fast LAMP protocols for POC detection of plant pathogens are being developed for several species including forest trees (Tomlinson et al., 2010; Yaseen et al., 2015; Aglietti et al., 2019) as well as herbaceous plants (Chang et al., 2013; Lin et al., 2015; Ammour et al., 2017). All these protocols combined fast nucleic acid extraction methods and signal detection to LAMP in order to perform all the required steps directly in the field. It must be noted that, although LAMP is actually a very promising technique for POC analysis (Notomi et al., 2015), it still presents some limitations, such as the design of the specific primers, that sometimes can be difficult. Several authors have developed fast LAMP protocols that, even if not yet incorporated in a specific portable device, were evaluated to be simpler, faster, cheaper, and with higher sensitivity when compared with traditional methods (Kikuchi et al., 2009; Tomlinson et al., 2010; Villari et al., 2013). By the optimization of RT-LAMP assay, a protocol was developed for the detection of B. xylophilus, a pine nematode. Such protocol can distinguish between living and dead nematodes, by detecting the presence of mRNA encoding an expansin gene as a viability marker (Leal et al., 2014). Other examples of optimized LAMP-based protocols include a tool for the early detection of Heterobasidion irregulare (Sillo et al., 2018); a fast, easy, and in-field deployable method for detection of Pepino mosaic virus and Potato spindle tuber viroid (Mehle et al., 2017); a LAMP protocol starting from crude plant materials for the detection of Xanthomonas fragariae (Getaz et al., 2017); a protocol for the detection of three different phytoplasmas infecting fruit trees (De Jonghe et al., 2017); and an assay for the rapid detection of Pectobacterium atrosepticum (Li et al., 2011). Similar to PCR-based approaches, multiplexing of the samples is possible with LAMP (Denschlag et al., 2014; Yasuhara-Bell et al., 2018; Feng et al., 2019).
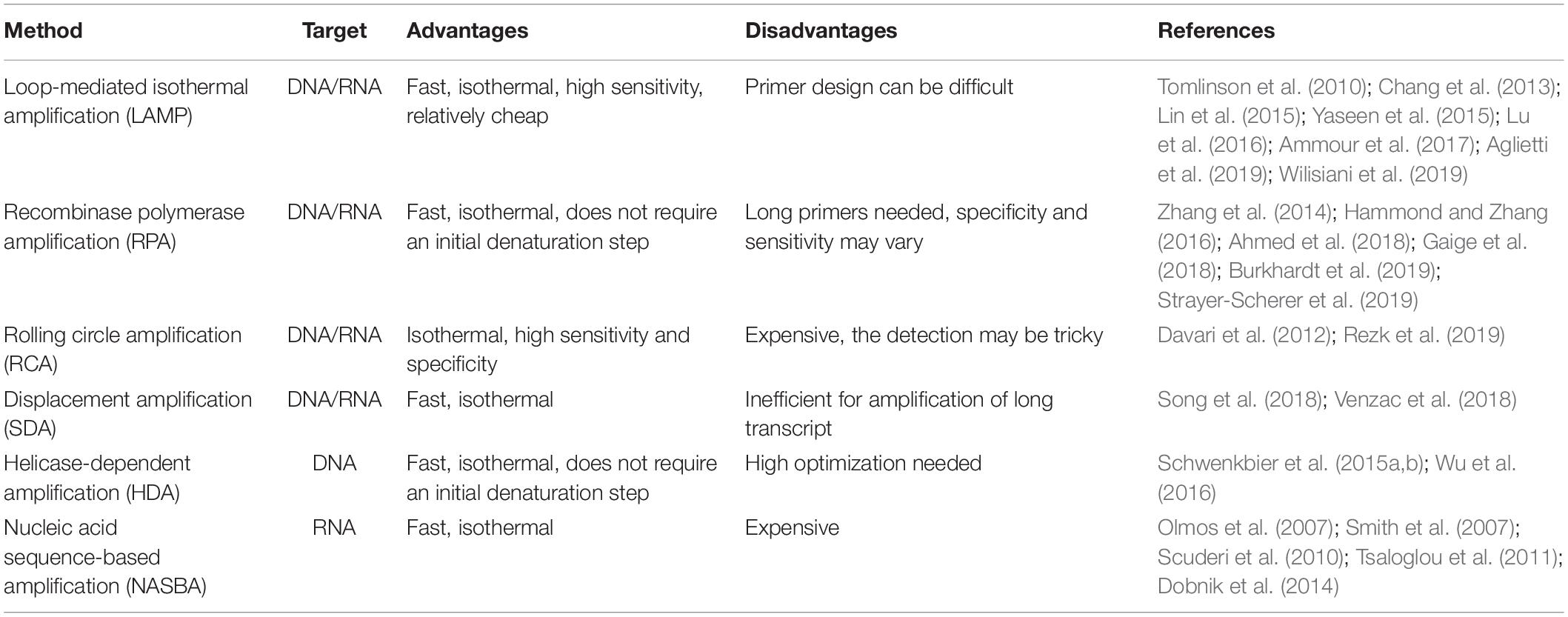
Table 4. List of the main isothermal amplification methods applied for POC plant pathogen detection.
Another recently developed isothermal amplification technique is the so-called recombinase polymerase amplification (RPA) (Piepenburg et al., 2006). RPA is becoming a common choice when POC analysis is required for application in agriculture (Ahmed et al., 2018; Burkhardt et al., 2019; Strayer-Scherer et al., 2019) and forestry (Cha et al., 2020), as it presents several advantages when compared to PCR and even to LAMP. In fact, RPA does not require an initial heating step for DNA denaturation, as it exploits enzymatic activity to separate the double strand. Moreover, the reaction temperature is quite low (37 to 42°C), and the reaction time is usually very short (Kappagantu et al., 2017; Munawar et al., 2019). When RPA was compared to RT-LAMP as a detection method using a small (150 mm × 200 mm × 35 mm) and light (400 g) battery-mounted portable optical isothermal device (Cha et al., 2020), the detection limit of RPA was shown to be 10 times lower than RT-LAMP. All these characteristics make RPA a very easy-to-use approach, especially in developing countries when fast analysis in low resource environments is needed (Wambua et al., 2017; Ghosh et al., 2018; Silva et al., 2018). The possibility to multiplex the reaction can further increase the speed and reduce the costs of RPA (Lau et al., 2016; Lei et al., 2019). Several RPA-based portable devices are already available for POC detection of human pathogens (Ereku et al., 2018; Tsaloglou et al., 2018), and assays for plant pathogen detection are rapidly developing (Zhang et al., 2014; Hammond and Zhang, 2016; Gaige et al., 2018). An integrated cheap prototype platform termed POCKET (Point-Of-Care Kit for the Entire Test) was recently developed demonstrating that combining 3D printing, microfluidics, RPA, and a smartphone, it is possible to create an ultraportable, inexpensive, and versatile device for analyzing multiple types of DNA from clinics to environment to food to agriculture in a sample-to-answer manner (Xu et al., 2020). The POCKET device is less than 100 g and smaller than 25 cm in length.
However, RPA still presents some limitations: it allows amplification of only small DNA fragments (<500 bp) with long primers (30–35 nt), resulting sometimes in non-specific amplification (Lau and Botella, 2017) and a highly variable sensitivity (Miles et al., 2015; Babu et al., 2017; Rojas et al., 2017).
Several other techniques for the isothermal amplification of nucleic acid exist such as rolling circle amplification (RCA), strand displacement amplification (SDA), helicase-dependent amplification (HDA), and nucleic acid sequence-based amplification (NASBA). All such techniques have characteristics that can be suitable for POC analysis in agriculture, and even if at present only a few examples of their use in field conditions exist, they are evolving rapidly. RCA was recently proposed for rapid diagnostic use in human health (Cao et al., 2019; Soares et al., 2019; Zhang et al., 2019), and some portable devices have been developed (Yao et al., 2018). Several examples of the use of RCA for plant pathogen detection exist (Davari et al., 2012; Rezk et al., 2019), but no portable devices have been described yet for application in agriculture. For SDA, some portable devices have been recently developed for human health care (Song et al., 2018; Venzac et al., 2018), so it is more than conceivable that in the next few years, this technique will be applied also for POC analysis in plant pathogen detection.
Another promising isothermal technique for amplification of genetic material is HDA, which was used in agriculture for the detection of tomato spotted wilt virus (Wu et al., 2016) and Phytophthora species (Schwenkbier et al., 2015a,b). Finally, NASBA has already found application in plant pathogen detection (Olmos et al., 2007; Scuderi et al., 2010; Dobnik et al., 2014), but it has not yet become a standard method for this kind of analysis, especially when POC pathogen detection is required, even though several portable devices have been developed and described (Smith et al., 2007; Tsaloglou et al., 2011). Some of the possible reasons could be that NASBA is not yet a widely known technique and the relatively high cost of the reactions (Honsvall and Robertson, 2017).
In order to detect the amplification products efficiently, various methods can be employed (Table 5). In particular, for POC tests, the detection systems should require as few equipment as possible to facilitate the analysis in the field. Most of the devices proposed for POC analysis use optical or visual tools (Zhang et al., 2019). Different principles can be exploited for the optical detection of amplicons, such as fluorescence, chemiluminescence, and colorimetric visualization among others. The signal can be read by the naked eye or by the use of standard optical instruments including CCD cameras, smartphones, and light detectors.
The fluorescence-based detection methods have been widely employed for many years for the visualization of qPCR amplification products, using SYBR green or TaqMan probes to generate the signal (Holland et al., 1991). Simplicity and high sensitivity are the main advantages of fluorescence-based detection methods, together with the possibility of coupling fluorescence detection tools to microfluidic devices (Neethirajan et al., 2011). Several examples of portable devices exploiting fluorescence detectors for POC plant pathogen analysis can be found. Such devices combine fluorescence detectors with several amplification methods, such as qPCR (Koo et al., 2013; DeShields et al., 2018; Fotiou et al., 2019), LAMP (Larrea-Sarmiento et al., 2018; Tahzima et al., 2019; Xia et al., 2019), RCA (Zhang et al., 2019), and RPA (Lei et al., 2019). A possible alternative to the classical fluorescence-based detection methods is the so-called surface-enhanced Raman scattering (SERS), a technique that exploits the signal enhancement given by metal nanoparticles surfaces upon laser excitation (Kneipp et al., 1997). SERS has the potential to guarantee higher performance than fluorescence-based approaches, especially for multiplex analysis, due to its narrow and distinct spectral peaks and the possibility to use different labeled Raman reporters (Laing et al., 2016). Indeed SERS has been proposed as a technique of choice for the realization of a POC tool for multiplex plant pathogen detection (Lau et al., 2016). SERS was used in combination with RPA for the early detection of several plant pathogens infecting Arabidopsis thaliana before any visible symptom or at early stages of infection (Lau et al., 2016).
The colorimetric methods represent an interesting alternative to the fluorescence-based detection. Colorimetric sensing can be exploited for a number of applications as it presents many advantages, such as low costs, low equipment requirements, possibility of multiplexing, and easy integration with microfluidic devices. Moreover, it is possible to detect the signal by the naked eye or using very common tools as smartphones and cameras (Ong and Poljak, 2020). The change in color, brightness, or intensity can be obtained using different methods that generally include a reaction triggered by the presence of the target amplification product. In agriculture, the application of colorimetric testing has been proposed for POC analysis in several species and using different methods. In some cases, amplification products are labeled using biotinylated primers and specifically detected by simple devices as lateral flow dipsticks (Tomlinson et al., 2010; Zhang et al., 2013; Rigano et al., 2014; Kolm et al., 2015; Wei et al., 2018). Another colorimetric method proposed for application in POC plant pathogen detection exploits the formation of a large amount of pyrophosphate ions as a byproduct of LAMP reactions (Chang et al., 2013; Lin et al., 2015). Several other methods have been exploited to perform POC colorimetric detection of nucleic acids for plant or human pathogen detection, as for example, the use of hydroxyl naphthol blue (HNB) (Harper et al., 2010), the activation of peroxidase-like deoxyribozyme (PDz) (Reed et al., 2019), pH-sensitive dyes (Hamidi and Perreault, 2019), and silver enhancement (Dharanivasan et al., 2019). Despite many advantages, the colorimetric detection methods still present some drawbacks, mainly the lack of sensitivity and specificity.
Another principle that can be applied for nucleic acid detection is bioluminescence. In particular, a recently described bioluminescence assay in real time (BART) can detect continuously the exponential increase in inorganic pyrophosphate (PPi) produced during an isothermal amplification reaction such as LAMP (Gandelman et al., 2010). The big advantage of BART is that it allows to simplify greatly data interpretation and hardware requirements. This characteristic make BART a good candidate for POC detection of amplified nucleic acids. A combination of LAMP and BART reactions was used for GMO detection, showing a high sensitivity level and good tolerance to inhibitors, proving to be suitable for field application (Kiddle et al., 2012).
A less developed, but still interesting, method for nucleic acid detection is based on electrochemistry (Hnaiein et al., 2008; Deiss et al., 2011). This approach showed high sensitivity and the possibility of miniaturizing all the components of the device very easily, making the electrochemical detection method a good candidate for the development of portable tools for POC analysis of plant pathogens. Another advantage of the electrochemical approach is the independence from background light or sample color, even though electrochemically active samples may influence the readout. Examples of portable devices for the electrochemical detection of amplified DNA from plant pathogens are already available (Julich et al., 2011; Schwenkbier et al., 2014, 2015a). The method exploits the enzymatic-triggered silver deposition on the electrode to read the signal both visually and electrochemically.
It is worth to mention magnetic sensing as a promising technique that could be potentially applied to POC analysis of amplified genetic material. This technique requires the labeling of the target sequence with magnetic nanoparticles and the detection of their stray field by the use of highly sensitive magnetic sensors (Lee et al., 2014; Orlov et al., 2016; Khater et al., 2017). The main advantages of magnetic sensing techniques are the high sensitivity, fast analysis time, and the possibility to be integrated onto handheld, portable on-chip systems and microfluidic devices (Devkota et al., 2015; Giouroudi and Kokkinis, 2017).
When the etiological agent is not known, the pathogen identification must rely on the use of an untargeted screening method by extracting, preparing, and sequencing all of the genomic material in a particular sample at once (Yeh et al., 2019). Even though POC sequencing is not routinely applied for plant pathogens yet, in the last few years, some portable sequencers have been developed that allow to perform amplicon sequencing directly in the field. The application value of portable sequencers was illustrated for the first time during emergent outbreaks of the Ebola and Zika virus in West Africa (Quick et al., 2016, 2017). Genome sequencing of the virus was carried out in situ by the first portable MinION sequencer [Oxford Nanopore Technologies (ONT) Ltd., Didcot, United Kingdom]. MinION is a portable, real-time device for DNA and RNA sequencing. Each consumable flow cell can generate as much as 30 Gb of DNA sequence data or 7–12 million reads when analyzing RNA. Ultra-long reads (hundreds of kb) are possible. Data are streamed in real time so that analysis can be performed during the experiment, and workflows are fully versatile. The MinION weighs under 100 g and plugs into a PC or laptop using a high-speed USB 3.0 cable, or used alongside the MinIT device for real-time analyses. The MinION sequencer is commercially available on the market2 with a price that is lower than 1,000 USD (Solares et al., 2018). Using DNA barcodes, useful for species discovery and species identification, it was estimated that up to 1,000 barcodes can be generated in one flow cell and that the cost per barcode can be <2 USD (Srivathsan et al., 2018). Speed is an important characteristic of MinION, as it is possible to perform the entire workflow from sample preparation, through DNA extraction, sequencing, bioinformatics, and interpretation in 2.5 h (Loit et al., 2019). Another advantage of MinION is that it can be easily coupled with different techniques to perform both targeted and untargeted analysis. For targeted analysis, both PCR-based and isothermal amplification can be used (Yamagishi et al., 2017; Radhakrishnan et al., 2019). Whole-genome amplification can be obtained by multiple displacement amplification (MDA), a technique that uses the high-fidelity phi29 polymerase combined with random hexamer primers to amplify DNA in isothermal reaction (Spits et al., 2006). Due to its portability, affordability, and speed in data production, the MinION sequencer found many applications, and it was used on a mountain, in a jungle, in the arctic, and on the International Space Station (Castro-Wallace et al., 2017). Recently, it was successfully applied for sequencing and assembly of a human genome with ultralong reads (Jain et al., 2018). It was shown to be suitable for COVID-19 fast diagnosis, and this platform is going to be further extended for diagnosing other viruses and pathogens (Wang et al., 2020).
In plants, an open- source workflow for long-read sequences was established in Eucalyptus. It was able to reliably and repeatedly obtain >6.5 Gb of long−read sequencing data with a mean read length of 13 kb and an N50 of 26 kb (Schalamun et al., 2019). Other examples of MinION application in agriculture and forestry include detection of pathogenic fungi (Cui et al., 2019; Hu et al., 2019; Rajarammohan et al., 2019; Srivastava et al., 2019; Purushotham et al., 2020), bacteria (Badial et al., 2018; Krehenwinkel et al., 2019; Zervas et al., 2019; Fujiyoshi et al., 2020), viruses (van der Merwe et al., 2017; Filloux et al., 2018; Boykin et al., 2019; Della Bartola et al., 2020), and nematodes (Eccles et al., 2018; Knot et al., 2020).
Despite the great success, the MinION technique still presents some drawbacks. In fact, the portability of MinION comes at the expense of sequencing accuracy, as sequencing errors often range between 5 and 15% (Rang et al., 2018). Nevertheless, recent improvements in chemistry, as well as the use of consensus calling using bioinformatics tools, allowed to increase sequencing accuracy up to 97%, comparable with standard lab equipment (Vaser et al., 2017; van Dijk et al., 2018). A recently developed base caller, DeepNano-blitz (Boza et al., 2020), enables real-time data analysis without requirement of a powerful IT facility, increasing the possibility to deploy MinION sequencing in the field. However, the optimization, establishment, and standardization of methods for the quantitative evaluation of microbial composition in the environment are inevitable. PCR-based 16S rRNA analysis of bacterial community structure was shown to be subject to biases from the PCR-related conditions. These include the template concentration, DNA polymerase choice, number of cycles used, amplification reaction time, and the reaction temperature. When MinION sequencing was used after an accurate optimization of PCR condition, it was able to obtain bacterial community structures that were comparable in quality with MiSeq (Fujiyoshi et al., 2020).
Point-of-care sequencing is a still evolving technology that has shown a very fast development in portability, sequencing accuracy, and ease of operation. All these characteristics could make POC sequencing the gold standard of pathogen diagnosis in the next few years.
The development of affordable and reliable methods for POC detection and identification of plant pathogens is not an easy task. There is an increasing demand for portable devices requiring minimal instrumentation that could perform a complete analysis directly in the field, even when used by personnel with only minimal training. This could bring great benefits to agriculture worldwide, as it would be possible to efficiently monitor wide cultivated areas for the presence of dangerous pathogens and detect them even before any symptom is actually visible. In the same way, it would be relatively easy to analyze imported plants and crops in order to prevent the spreading of pests and pathogens to new areas, where they could cause enormous damage to the local agriculture. There is also the need of pathogen monitoring in forestry and environmental applications, which often requires sampling in remote areas far away from laboratory facilities.
The nucleic acid-based methods are proving to be good candidates for POC pathogen detection, especially since new isothermal amplification protocols have been developed. These methods are not widely used for field screening or for the analysis of imported plant material, probably because technical difficulties still exist, especially when trying to develop a protocol that can be routinely used for the detection of different pathogens in different plant species. Also, the relatively high cost of some of the methods, especially when applied to large-scale analysis, can be a problem. Nevertheless, PCR-based and isothermal nucleic acid amplification protocols coupled with fast extraction methods and simple detection devices are being adopted for some pathogens and crops. The development of new portable and sensitive nucleic acid detection methods is still a growing field of research that, in the next years, will provide exciting possibilities to both researchers and farmers. In many cases, POC analysis can provide a fast disease detection, have minimal risk, low cost, and provide a high-quality diagnostic experience for the operator. However, traditional laboratory testing is generally more advanced and allows a more accurate analysis, validation, and recording of the results. There are clearly situations where each methodology excels. Therefore, the two methodologies are going to merge and integrate by the development of new technologies. The main challenges that need to be addressed for POC testing are the optimization of reliable methods for all the main pathogens threatening agriculture worldwide, the adaptation of the existing methods to the specific conditions that a given plant or pathogen requires, a robust comparison to traditional technologies, and encouraging the final users to replace the traditional detection methods with novel ones offering significant benefits.
Both authors conceived and prepared the manuscript, performed literature search, wrote and edited the manuscript. Both authors contributed equally to the article and approved the submitted version.
The study was carried out by the Grant P1611034I and P1611005I, provided by the Research and Innovation Centre, Fondazione Edmund Mach.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdalhai, M. (2016). Comparative study of rapid DNA extraction methods of pathogenic bacteria. Am. J. Biosci. Bioeng. 4, 1–8. doi: 10.11648/j.bio.20160401.11
Aglietti, C., Luchi, N., Pepori, A. L., Bartolini, P., Pecori, F., Raio, A., et al. (2019). Real-time loop-mediated isothermal amplification: an early-warning tool for quarantine plant pathogen detection. AMB Express 9:50. doi: 10.1186/s13568-019-0774-9
Ahmed, F. A., Larrea-Sarmiento, A., Alvarez, A. M., and Arif, M. (2018). Genome-informed diagnostics for specific and rapid detection of Pectobacterium species using recombinase polymerase amplification coupled with a lateral flow device. Sci. Rep. 8:15972. doi: 10.1038/s41598-018-34275-0
Alahi, M. E. E., and Mukhopadhyay, S. C. (2017). Detection methodologies for pathogen and toxins: a review. Sensors 17:1885. doi: 10.3390/s17081885
Ammour, M. S., Bilodeau, G. J., Tremblay, D. M., Van der Heyden, H., Yaseen, T., Varvaro, L., et al. (2017). Development of real-time isothermal amplification assays for on-site detection of Phytophthora infestans in potato leaves. Plant Dis. 101, 1269–1277. doi: 10.1094/pdis-12-16-1780-re
Arif, M., Fletcher, J., Marek, S. M., Melcher, U., and Ochoa-Corona, F. M. (2013). Development of a rapid, sensitive, and field-deployable razor ex biodetection system and quantitative PCR assay for detection of phymatotrichopsis omnivora using multiple gene targets. Appl. Environ. Microbiol. 79, 2312–2320. doi: 10.1128/aem.03239-12
Aslam, S., Tahir, A., Aslam, M. F., Alam, M. W., Shedayi, A. A., and Sadia, S. (2017). Recent advances in molecular techniques for the identification of phytopathogenic fungi - a mini review. J. Plant Interact. 12, 493–504. doi: 10.1080/17429145.2017.1397205
Babu, B., Washburn, B. K., Ertek, T. S., Miller, S. H., Riddle, C. B., Knox, G. W., et al. (2017). A field based detection method for Rose rosette virus using isothermal probe-based Reverse transcription-recombinase polymerase amplification assay. J. Virol. Methods 247, 81–90. doi: 10.1016/j.jviromet.2017.05.019
Badial, A. B., Sherman, D., Stone, A., Gopakumar, A., Wilson, V., Schneider, W., et al. (2018). Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis. 102, 1648–1652. doi: 10.1094/pdis-04-17-0488-re
Baidoo, R., Yan, G. P., Nagachandrabose, S., and Skantar, A. M. (2017). Developing a real-time PCR assay for direct identification and quantification of Pratylenchus penetrans in soil. Plant Dis. 101, 1432–1441. doi: 10.1094/pdis-01-17-0117-re
Bini, A. P., Quecine, M. C., da Silva, T. M., Silva, L. D., and Labate, C. A. (2018). Development of a quantitative real-time PCR assay using SYBR Green for early detection and quantification of Austropuccinia psidii in Eucalyptus grandis. Eur. J. Plant Pathol. 150, 735–746. doi: 10.1007/s10658-017-1321-7
Blaser, S., Diem, H., von Felten, A., Gueuning, M., Andreou, M., Boonham, N., et al. (2018). From laboratory to point of entry: development and implementation of a loop-mediated isothermal amplification (LAMP)-based genetic identification system to prevent introduction of quarantine insect species. Pest. Manag. Sci. 74, 1504–1512. doi: 10.1002/ps.4866
Blaya, J., Lloret, E., Santisima-Trinidad, A. B., Ros, M., and Pascual, J. A. (2016). Molecular methods (digital PCR and real-time PCR) for the quantification of low copy DNA of Phytophthora nicotianae in environmental samples. Pest. Manag. Sci. 72, 747–753. doi: 10.1002/ps.4048
Boykin, L. M., Sseruwagi, P., Alicai, T., Ateka, E., Mohammed, I. U., Stanton, J. A. L., et al. (2019). Tree lab: portable genomics for early detection of plant viruses and pests in sub-saharan Africa. Genes 10:632. doi: 10.3390/genes10090632
Boza, V., Peresini, P., Brejova, B., and Vinař, T. (2020). DeepNano-blitz: a fast base caller for MinION nanopore sequencers. Bioinformatics 36, 4191–4192.
Buhlmann, A., Pothier, J. F., Rezzonico, F., Smits, T. H., Andreou, M., Boonham, N., et al. (2013). Erwinia amylovora loop-mediated isothermal amplification (LAMP) assay for rapid pathogen detection and on-site diagnosis of fire blight. J. Microbiol. Methods 92, 332–339. doi: 10.1016/j.mimet.2012.12.017
Burkhardt, A., Henry, P. M., Koike, S. T., Gordon, T. R., and Martin, F. (2019). Detection of Fusarium oxysporum f. sp. fragariae from infected strawberry plants. Plant Dis. 103, 1006–1013. doi: 10.1094/pdis-08-18-1315-re
Burnham-Marusich, A. R., Hubbard, B., Kvam, A. J., Gates-Hollingsworth, M., Green, H. R., Soukup, E., et al. (2018). Conservation of mannan synthesis in fungi of the zygomycota and ascomycota reveals a broad diagnostic target. mSphere 3:e00094-18. doi: 10.1128/mSphere.00094-18
Callaway, E. (2016). Devastating wheat fungus appears in Asia for first time. Nature 532, 421–422. doi: 10.1038/532421a
Cao, H. M., Zhou, X., and Zeng, Y. (2019). Microfluidic exponential rolling circle amplification for sensitive microRNA detection directly from biological samples. Sens. Actuators B Chem. 279, 447–457. doi: 10.1016/j.snb.2018.09.121
Capron, A., Stewart, D., Hrywkiw, K., Allen, K., Feau, N., Bilodeau, G., et al. (2020). In Situ processing and efficient environmental detection (iSPEED) of tree pests and pathogens using point-of-use real-time PCR. PLoS One 15:e0226863. doi: 10.1371/journal.pone.0226863
Cassedy, A., Mullins, E., and O’Kennedy, R. (2020). Sowing seeds for the future: the need for on-site plant diagnostics. Biotechnol. Adv. 39:107358. doi: 10.1016/j.biotechadv.2019.02.014
Castro-Wallace, S. L., Chiu, C. Y., John, K. K., Stahl, S. E., Rubins, K. H., McIntyre, A. B. R., et al. (2017). Nanopore DNA sequencing and genome assembly on the international space station. Sci. Rep. 7:18022. doi: 10.1038/s41598-017-18364-0
Cawthorn, D. M., Steinman, H. A., and Witthuhn, R. C. (2011). Comparative study of different methods for the extraction of DNA from fish species commercially available in South Africa. Food Control 22, 231–244. doi: 10.1016/j.foodcont.2010.07.003
Cha, D., Kim, D., Choi, W., Park, S., and Han, H. (2020). Point-of-care diagnostic (POCD) method for detecting Bursaphelenchus xylophilus in pinewood using recombinase polymerase amplification (RPA) with the portable optical isothermal device (POID). PLoS One 15:e0227476. doi: 10.1371/journal.pone.0227476
Chang, W. H., Yang, S. Y., Lin, C. L., Wang, C. H., Li, P. C., Chen, T. Y., et al. (2013). Detection of viruses directly from the fresh leaves of a Phalaenopsis orchid using a microfluidic system. Nanomed. Nanotechnol. Biol. Med. 9, 1274–1282. doi: 10.1016/j.nano.2013.05.016
Chen, H., Liu, K. K., Li, Z., and Wang, P. (2019). Point of care testing for infectious diseases. Clin. Chim. Acta 493, 138–147. doi: 10.1016/j.cca.2019.03.008
Cheng, H. R., and Jiang, N. (2006). Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 28, 55–59. doi: 10.1007/s10529-005-4688-z
Cho, I. H., and Irudayaraj, J. (2013). In-situ immuno-gold nanoparticle network ELISA biosensors for pathogen detection. Int. J. Food Microbiol. 164, 70–75. doi: 10.1016/j.ijfoodmicro.2013.02.025
Costa, J., Mafra, I., Kuchta, T., and Oliveira, M. (2012). Single-tube nested real-time PCR as a new highly sensitive approach to trace hazelnut. J. Agric. Food Chem. 60, 8103–8110. doi: 10.1021/jf302898z
Cui, C. M., Herlihy, J. H., Bombarely, A., McDowell, J. M., and Haak, D. C. (2019). Draft assembly of Phytophthora capsici from long-read sequencing uncovers complexity. Mol. Plant Microbe Interact. 32, 1559–1563. doi: 10.1094/mpmi-04-19-0103-ta
Dairawan, M., and Shetty, P. J. (2020). The evolution of DNA extraction methods. Am. J. Biomed. Sci. Res. 8, 39–45.
Danks, C., and Barker, I. (2000). On-site detection of plant pathogens using lateral-flow devices. EPPO Bull. 30, 421–426. doi: 10.1111/j.1365-2338.2000.tb00922.x
Davari, M., van Diepeningen, A. D., Babai-Ahari, A., Arzanlou, M., Najafzadeh, M. J., van der Lee, T. A. J., et al. (2012). Rapid identification of Fusarium graminearum species complex using rolling circle amplification (RCA). J. Microbiol. Methods 89, 63–70. doi: 10.1016/j.mimet.2012.01.017
De Boer, S. H., and Lopez, M. M. (2012). New grower-friendly methods for plant pathogen monitoring. Annu. Rev. Phytopathol. 50, 197–218. doi: 10.1146/annurev-phyto-081211-172942
De Jonghe, K., De Roo, I., and Maes, M. (2017). Fast and sensitive on-site isothermal assay (LAMP) for diagnosis and detection of three fruit tree phytoplasmas. Eur. J. Plant Pathol. 147, 749–759. doi: 10.1007/s10658-016-1039-y
Deiss, F., Laurent, S., Descamps, E., Livache, T., and Sojic, N. (2011). Opto-electrochemical nanosensor array for remote DNA detection. Analyst 136, 327–331. doi: 10.1039/c0an00501k
Della Bartola, M., Byrne, S., and Mullins, E. (2020). Characterization of potato virus Y isolates and assessment of nanopore sequencing to detect and genotype potato viruses. Viruses Basel 12:478. doi: 10.3390/v12040478
Denschlag, C., Rieder, J., Vogel, R. F., and Niessen, L. (2014). Real-time loop-mediated isothermal amplification (LAMP) assay for group specific detection of important trichothecene producing Fusarium species in wheat. Int. J. Food Microbiol. 177, 117–127. doi: 10.1016/j.ijfoodmicro.2014.02.010
DeShields, J. B., Bomberger, R. A., Woodhall, J. W., Wheeler, D. L., Moroz, N., Johnson, D. A., et al. (2018). On-site molecular detection of soil-borne phytopathogens using a portable real-time PCR system. Jove J. Vis. Exp. 132:e56891. doi: 10.3791/56891
Devkota, J., Kokkinis, G., Berris, T., Jamalieh, M., Cardoso, S., Cardoso, F., et al. (2015). A novel approach for detection and quantification of magnetic nanomarkers using a spin valve GMR-integrated microfluidic sensor. RSC Adv. 5, 51169–51175. doi: 10.1039/c5ra09365a
Dharanivasan, G., Jesse, D. M. I., Rajamuthuramalingam, T., Rajendran, G., Shanthi, S., and Kathiravan, K. (2019). Scanometric detection of tomato leaf curl new delhi viral DNA using mono- and bifunctional AuNP-conjugated oligonucleotide probes. ACS Omega 4, 10094–10107. doi: 10.1021/acsomega.9b00340
Dobnik, D., Morisset, D., Lenarcic, R., and Ravnikar, M. (2014). Simultaneous detection of RNA and DNA targets based on multiplex isothermal amplification. J. Agric. Food Chem. 62, 2989–2996. doi: 10.1021/jf5002149
Donoso, A., and Valenzuela, S. (2018). In-field molecular diagnosis of plant pathogens: recent trends and future perspectives. Plant Pathol. 67, 1451–1461. doi: 10.1111/ppa.12859
Eccles, D., Chandler, J., Camberis, M., Henrissat, B., Koren, S., Le Gros, G., et al. (2018). De novo assembly of the complex genome of Nippostrongylus brasiliensis using MinION long reads. BMC Biol. 16:6. doi: 10.1186/s12915-017-0473-4
Ereku, L. T., Mackay, R. E., Craw, P., Naveenathayalan, A., Stead, T., Branavan, M., et al. (2018). RPA using a multiplexed cartridge for low cost point of care diagnostics in the field. Anal. Biochem. 547, 84–88. doi: 10.1016/j.ab.2018.02.010
Feng, W. Z., Hieno, A., Kusunoki, M., Suga, H., and Kageyama, K. (2019). LAMP detection of four plant-pathogenic oomycetes and its application in lettuce fields. Plant Dis. 103, 298–307. doi: 10.1094/pdis-05-18-0858-re
Filloux, D., Fernandez, E., Loire, E., Claude, L., Galzi, S., Candresse, T., et al. (2018). Nanopore-based detection and characterization of yam viruses. Sci. Rep. 8:17879. doi: 10.1038/s41598-018-36042-7
Fotiou, I. S., Pappi, P. G., Efthimiou, K. E., Katis, N. I., and Maliogka, V. I. (2019). Development of one-tube real-time RT-qPCR for the universal detection and quantification of Plum pox virus (PPV). J. Virol. Methods 263, 10–13. doi: 10.1016/j.jviromet.2018.10.006
Fujiyoshi, S., Muto-Fujita, A., and Maruyama, F. (2020). Evaluation of PCR conditions for characterizing bacterial communities with full-length 16S rRNA genes using a portable nanopore sequencer. Sci. Rep. 10:12580. doi: 10.1038/s41598-020-69450-9
Fukuta, S., Anai, N., Kato, M., Yoshimura, Y., Fukaya, M., Yabe, K., et al. (2005). Simplification of template DNA preparation for the loop-mediated isothermal amplification (LAMP) method to detect tomato yellow leaf curl virus. Annu. Rep. Kansai Plant Protect. Soc. 47, 37–41. doi: 10.4165/kapps.47.37
Gaige, A. R., Dung, J. K. S., and Weiland, J. E. (2018). A rapid, sensitive and field-deployable isothermal assay for the detection of Verticillium alfalfae. Can. J. Plant Pathol. 40, 408–416. doi: 10.1080/07060661.2018.1474262
Gandelman, O. A., Church, V. L., Moore, C. A., Kiddle, G., Carne, C. A., Parmar, S., et al. (2010). Novel bioluminescent quantitative detection of nucleic acid amplification in real-time. PLoS One 5:e14155. doi: 10.1371/journal.pone.0014155
Getaz, M., Buhlmann, A., Schneeberger, P. H. H., Van Malderghem, C., Duffy, B., Maes, M., et al. (2017). A diagnostic tool for improved detection of Xanthomonas fragariae using a rapid and highly specific LAMP assay designed with comparative genomics. Plant Pathol. 66, 1094–1102. doi: 10.1111/ppa.12665
Ghelardini, L., Luchi, N., Pecori, F., Pepori, A. L., Danti, R., Della Rocca, G., et al. (2017). Ecology of invasive forest pathogens. Biol. Invasions 19, 3183–3200. doi: 10.1007/s10530-017-1487-0
Ghosh, D. K., Kokane, S. B., Kokane, A. D., Warghane, A. J., Motghare, M. R., Bhose, S., et al. (2018). Development of a recombinase polymerase based isothermal amplification combined with lateral flow assay (HLB-RPA-LFA) for rapid detection of “Candidatus Liberibacter asiaticus”. PLoS One 13:e0208530. doi: 10.1371/journal.pone.0208530
Giampetruzzi, A., Saponari, M., Loconsole, G., Boscia, D., Savino, V. N., Almeida, R. P. P., et al. (2017). Genome-wide analysis provides evidence on the genetic relatedness of the emergent Xylella fastidiosa genotype in Italy to isolates from Central America. Phytopathology 107, 816–827. doi: 10.1094/phyto-12-16-0420-r
Giouroudi, I., and Kokkinis, G. (2017). Recent advances in magnetic microfluidic biosensors. Nanomaterials 7:171. doi: 10.3390/nano7070171
Gluck-Thaler, E., and Slot, J. C. (2015). Dimensions of horizontal gene transfer in eukaryotic microbial pathogens. PLoS Pathog. 11:e1005156. doi: 10.1371/journal.ppat.1005156
Gorris, M. T., Alarcon, B., Lopez, M. M., and Cambra, M. (1994). Characterization of monoclonal-antibodies specific for erwinia-carotovora subsp atroseptica and comparison of serological methods for its sensitive detection on potato-tubers. Appl. Environ. Microbiol. 60, 2076–2085.
Hamidi, S. V., and Perreault, J. (2019). Simple rolling circle amplification colorimetric assay based on pH for target DNA detection. Talanta 201, 419–425. doi: 10.1016/j.talanta.2019.04.003
Hammond, R. W., and Zhang, S. L. (2016). Development of a rapid diagnostic assay for the detection of tomato chlorotic dwarf viroid based on isothermal reverse-transcription-recombinase polymerase amplification. J. Virol. Methods 236, 62–67. doi: 10.1016/j.jviromet.2016.06.013
Harper, S. J., Ward, L. I., and Clover, G. R. G. (2010). Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 100, 1282–1288. doi: 10.1094/phyto-06-10-0168
Hayden, R. T., Gu, Z., Ingersoll, J., Abdul-Ali, D., Shi, L., Pounds, S., et al. (2013). Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 51, 540–546. doi: 10.1128/jcm.02620-12
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J., et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610. doi: 10.1021/ac202028g
Hnaiein, M., Hassen, W. M., Abdelghani, A., Fournier-Wirth, C., Coste, J., Bessueille, F., et al. (2008). A conductometric immunosensor based on functionalized magnetite nanoparticles for E. coli detection. Electrochem. Commun. 10, 1152–1154. doi: 10.1016/j.elecom.2008.04.009
Holland, P. M., Abramson, R. D., Watson, R., and Gelfand, D. H. (1991). Detection of specific polymerase chain reaction product by utilizing the 5’—-3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 88, 7276–7280. doi: 10.1073/pnas.88.16.7276
Honsvall, B. K., and Robertson, L. J. (2017). From research lab to standard environmental analysis tool: will NASBA make the leap? Water Res. 109, 389–397. doi: 10.1016/j.watres.2016.11.052
Hu, Y. H., Green, G. S., Milgate, A. W., Stone, E. A., Rathjen, J. P., and Schwessinger, B. (2019). Pathogen detection and microbiome analysis of infected wheat using a portable DNA sequencer. Phytobiomes J. 3, 92–101. doi: 10.1094/pbiomes-01-19-0004-r
Jain, M., Koren, S., Miga, K. H., Quick, J., Rand, A. C., Sasani, T. A., et al. (2018). Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–349. doi: 10.1038/nbt.4060
Julich, S., Riedel, M., Kielpinski, M., Urban, M., Kretschmer, R., Wagner, S., et al. (2011). Development of a lab-on-a-chip device for diagnosis of plant pathogens. Biosens. Bioelectron. 26, 4070–4075. doi: 10.1016/j.bios.2011.03.035
Kappagantu, M., Villamor, D. E. V., Bullock, J. M., and Eastwell, K. C. (2017). A rapid isothermal assay for the detection of Hop stunt viroid in hop plants (Humulus lupulus), and its application in disease surveys. J. Virol. Methods 245, 81–85. doi: 10.1016/j.jviromet.2017.04.002
Khan, S., Mackay, J., Liefting, L., and Ward, L. (2015). Development of a duplex one-step RT-qPCR assay for the simultaneous detection of Apple scar skin viroid and plant RNA internal control. J. Virol. Methods 221, 100–105. doi: 10.1016/j.jviromet.2015.04.032
Khater, M., de la Escosura-Muniz, A., and Merkoci, A. (2017). Biosensors for plant pathogen detection. Biosens. Bioelectron. 93, 72–86. doi: 10.1016/j.bios.2016.09.091
Kiddle, G., Hardinge, P., Buttigieg, N., Gandelman, O., Pereira, C., McElgunn, C. J., et al. (2012). GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 12:15. doi: 10.1186/1472-6750-12-15
Kikuchi, T., Aikawa, T., Oeda, Y., Karim, N., and Kanzaki, N. (2009). A rapid and precise diagnostic method for detecting the Pinewood nematode Bursaphelenchus xylophilus by loop-mediated isothermal amplification. Phytopathology 99, 1365–1369. doi: 10.1094/phyto-99-12-1365
Kim, J., Johnson, M., Hill, P., and Gale, B. K. (2009). Microfluidic sample preparation: cell lysis and nucleic acid purification. Integr. Biol. 1, 574–586. doi: 10.1039/b905844c
Kneipp, K., Wang, Y., Kneipp, H., Perelman, L. T., Itzkan, I., Dasari, R., et al. (1997). Single molecule detection using surface-enhanced Raman scattering (SERS). Phys. Rev. Lett. 78, 1667–1670. doi: 10.1103/PhysRevLett.78.1667
Knot, I. E., Zouganelis, G. D., Weedall, G. D., Wich, S. A., and Rae, R. (2020). DNA barcoding of nematodes using the MinION. Front. Ecol. Evol. 8:100. doi: 10.3389/fevo.2020.00100
Kolm, C., Mach, R. L., Krska, R., and Brunner, K. (2015). A rapid DNA lateral flow test for the detection of transgenic maize by isothermal amplification of the 35S promoter. Anal. Methods 7, 129–134. doi: 10.1039/c4ay01997k
Koo, C., Malapi-Wight, M., Kim, H. S., Cifci, O. S., Vaughn-Diaz, V. L., Ma, B., et al. (2013). Development of a real-time microchip PCR system for portable plant disease diagnosis. PLoS One 8:e82704. doi: 10.1371/journal.pone.0082704
Krehenwinkel, H., Pomerantz, A., and Prost, S. (2019). Genetic biomonitoring and biodiversity assessment using portable sequencing technologies: current uses and future directions. Genes 10:858. doi: 10.3390/genes10110858
La Porta, N., Capretti, P. I. M. T., Kasanen, R., Hietala, A., and Kaviya, V. (2008). Forest pathogens with higher damage potential due to climate change in Europe. Can. J. Plant Pathol. 30, 177–195. doi: 10.1080/07060661.2008.10540534
Lai, D. W., Zhang, Y. L., Huang, Q. X., Yin, G. H., Pennerman, K. K., Liu, Z. X., et al. (2018). Reverse transcription loop-mediated isothermal amplification to rapidly detect Rice ragged stunt virus. Saudi J. Biol. Sci. 25, 1577–1584. doi: 10.1016/j.sjbs.2016.02.024
Laing, S., Gracie, K., and Faulds, K. (2016). Multiplex in vitro detection using SERS. Chem. Soc. Rev. 45, 1901–1918. doi: 10.1039/c5cs00644a
Larrea-Sarmiento, A., Dhakal, U., Boluk, G., Fatdal, L., Alvarez, A., Strayer-Scherer, A., et al. (2018). Development of a genome-informed loop-mediated isothermal amplification assay for rapid and specific detection of Xanthomonas euvesicatoria. Sci. Rep. 8:14298. doi: 10.1038/s41598-018-32295-4
Lau, H. Y., and Botella, J. R. (2017). Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci. 8:2016. doi: 10.3389/fpls.2017.02016
Lau, H. Y., Wang, Y. L., Wee, E. J. H., Botella, J. R., and Trau, M. (2016). Field demonstration of a multiplexed point-of-care diagnostic platform for plant pathogens. Anal. Chem. 88, 8074–8081. doi: 10.1021/acs.analchem.6b01551
Leal, I., Allen, E., Foord, B., Anema, J., Reisle, C., Uzunovic, A., et al. (2014). Detection of living Bursaphelenchus xylophilus in wood, using reverse transcriptase loop-mediated isothermal amplification (RT-LAMP). Forest Pathol. 45, 134–148. doi: 10.1111/efp.12149
Lee, H., Xu, L. F., Koh, D., Nyayapathi, N., and Oh, K. W. (2014). Various on-chip sensors with microfluidics for biological applications. Sensors 14, 17008–17036. doi: 10.3390/s140917008
Lei, R., Kong, J., Qiu, Y. H., Chen, N. Z., Zhu, S. F., Wang, X. Y., et al. (2019). Rapid detection of the pathogenic fungi causing blackleg of Brassica napus using a portable real-time fluorescence detector. Food Chem. 288, 57–67. doi: 10.1016/j.foodchem.2019.02.089
Li, X., Nie, J. B., Ward, L. J., Nickerson, J., and De Boer, S. H. (2011). Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection and identification of Pectobacterium atrosepticum. Can. J. Plant Pathol. 33, 447–457. doi: 10.1080/07060661.2011.603751
Liau, Y., Cree, S. L., Maggo, S., Miller, A. L., Pearson, J. F., Gladding, P. A., et al. (2019). A multiplex pharmacogenetics assay using the MinION nanopore sequencing device. Pharmacogenet. Genomics 29, 207–215. doi: 10.1097/FPC.0000000000000385
Lin, C. L., Chang, W. H., Wang, C. H., Lee, C. H., Chen, T. Y., Jan, F. J., et al. (2015). A microfluidic system integrated with buried optical fibers for detection of Phalaenopsis orchid pathogens. Biosens. Bioelectron. 63, 572–579. doi: 10.1016/j.bios.2014.08.013
Liu, Y. J., Wang, Y. L., Wang, Q., Zhang, Y. H., Shen, W. X., Li, R. H., et al. (2019). Development of a sensitive and reliable reverse transcription droplet digital PCR assay for the detection of citrus yellow vein clearing virus. Arch. Virol. 164, 691–697. doi: 10.1007/s00705-018-04123-7
Liu, Z., Lei, S., Zou, L., Li, G., and Ye, B. (2020). Grafting homogenous electrochemical biosensing strategy based on reverse proximity ligation and Exo III assisted target circulation for multiplexed communicable disease DNA assay. Biosens. Bioelectron. 167:112487. doi: 10.1016/j.bios.2020.112487
Loit, K., Adamson, K., Bahram, M., Puusepp, R., Anslan, S., Kiiker, R., et al. (2019). Relative performance of MinION (oxford nanopore technologies) versus sequel (pacific biosciences) third-generation sequencing instruments in identification of agricultural and forest fungal pathogens. Appl. Environ. Microbiol. 85:e01368-19. doi: 10.1128/aem.01368-19
Lu, W. J., Wang, J. D., Wu, Q., Sun, J. S., Chen, Y. P., Zhang, L., et al. (2016). High-throughput sample-to-answer detection of DNA/RNA in crude samples within functionalized micro-pipette tips. Biosens. Bioelectron. 75, 28–33. doi: 10.1016/j.bios.2015.08.016
Marcelletti, S., and Scortichini, M. (2016). Xylella fastidiosa CoDiRO strain associated with the olive quick decline syndrome in southern Italy belongs to a clonal complex of the subspecies pauca that evolved in Central America. Microbiol. SGM 162, 2087–2098. doi: 10.1099/mic.0.000388
Martinelli, F., Scalenghe, R., Davino, S., Panno, S., Scuderi, G., Ruisi, P., et al. (2015). Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 35, 1–25. doi: 10.1007/s13593-014-0246-1
McDonald, B. A., and Stukenbrock, E. H. (2016). Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20160026. doi: 10.1098/rstb.2016.0026
Mehle, N., Kogovsek, P., Racki, N., Jakomin, T., Gutierrez-Aguirre, I., Kramberger, P., et al. (2017). Filling the gaps in diagnostics of Pepino mosaic virus and Potato spindle tuber viroid in water and tomato seeds and leaves. Plant Pathol. 66, 1191–1201. doi: 10.1111/ppa.12710
Miles, T. D., Martin, F. N., and Coffey, M. D. (2015). Development of rapid isothermal amplification assays for detection of Phytophthora spp. in plant tissue. Phytopathology 105, 265–278. doi: 10.1094/phyto-05-14-0134-r
Minguzzi, S., Terlizzi, F., Lanzoni, C., Pollini, C. P., and Ratti, C. (2016). A rapid protocol of crude RNA/DNA extraction for RT-qPCR detection and quantification of ’Candidatus Phytoplasma prunorum’. PLoS One 11:e0146515. doi: 10.1371/journal.pone.0146515
Munawar, M., Toljamo, A., Martin, F., and Kokko, H. (2019). Recombinase Polymerase Amplification Assay for fast, sensitive and on-site detection of Phytophthora cactorum without DNA extraction. Eur. J. Hortic. Sci. 84, 14–19. doi: 10.17660/eJHS.2019/84.1.2
Narayanasamy, P. (2011). Microbial Plant Pathogens-Detection and Disease Diagnosis Bacterial and Phytoplasmal Pathogens. Berlin: Springer.
Neethirajan, S., Kobayashi, I., Nakajima, M., Wu, D., Nandagopal, S., and Lin, F. (2011). Microfluidics for food, agriculture and biosystems industries. Lab Chip 11, 1574–1586. doi: 10.1039/c0lc00230e
Nikitin, M., Deych, K., Grevtseva, I., Girsova, N., Kuznetsova, M., Pridannikov, M., et al. (2018). Preserved microarrays for simultaneous detection and identification of six fungal potato pathogens with the use of real-time PCR in matrix format. Biosens. Basel 8:129. doi: 10.3390/bios8040129
Notomi, T., Mori, Y., Tomita, N., and Kanda, H. (2015). Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J. Microbiol. 53, 1–5. doi: 10.1007/s12275-015-4656-9
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. doi: 10.1093/nar/28.12.e63
Novakova, S., Klaudiny, J., Kollerova, E., and Subr, Z. W. (2006). Expression of a part of the Potato virus A non-structural protein P3 in Escherichia coli for the purpose of antibody preparation and P3 immunodetection in plant material. J. Virol. Methods 137, 229–235. doi: 10.1016/j.jviromet.2006.06.020
Olmos, A., Bertolini, E., and Cambra, M. (2007). Isothermal amplification coupled with rapid flow-through hybridisation for sensitive diagnosis of Plum pox virus. J. Virol. Methods 139, 111–115. doi: 10.1016/j.jviromet.2006.09.012
Ong, D. S. Y., and Poljak, M. (2020). Smartphones as mobile microbiological laboratories. Clin. Microbiol. Infect 26, 421–424. doi: 10.1016/j.cmi.2019.09.026
Orlov, A. V., Bragina, V. A., Nikitin, M. P., and Nikitin, P. I. (2016). Rapid dry-reagent immunomagnetic biosensing platform based on volumetric detection of nanoparticles on 3D structures. Biosens. Bioelectron. 79, 423–429. doi: 10.1016/j.bios.2015.12.049
Osman, F., Dang, T., Bodaghi, S., and Vidalakis, G. (2017). One-step multiplex RT-qPCR detects three citrus viroids from different genera in a wide range of hosts. J. Virol. Methods 245, 40–52. doi: 10.1016/j.jviromet.2017.03.007
Pallas, V., Sanchez-Navarro, J. A., and James, D. (2018). Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 9:2087. doi: 10.3389/fmicb.2018.02087
Papayiannis, L. C. (2014). Diagnostic real-time RT-PCR for the simultaneous detection of Citrus exocortis viroid and Hop stunt viroid. J. Virol. Methods 196, 93–99. doi: 10.1016/j.jviromet.2013.11.001
Paul, R., Ostermann, E., Gu, Z., Ristaino, J., and Wei, Q. (2020). DNA extraction from plant leaves using a microneedle patch. Curr. Protoc. Plant Biol. 5:e20104. doi: 10.1002/cppb.20104
Paul, R., Saville, A. C., Hansel, J. C., Ye, Y. Q., Ball, C., Williams, A., et al. (2019). Extraction of plant DNA by microneedle patch for rapid detection of plant diseases. ACS Nano 13, 6540–6549. doi: 10.1021/acsnano.9b00193
Peeling, R. W., Holmes, K. K., Mabey, D., and Ronald, A. (2006). Rapid tests for sexually transmitted infections (STIs): the way forward. Sex. Trans. Infect. 82, V1–V6. doi: 10.1136/sti.2006.024265
Piepenburg, O., Williams, C. H., Stemple, D. L., and Armes, N. A. (2006). DNA detection using recombination proteins. PLoS Biol. 4:1115–1121. doi: 10.1371/journal.pbio.0040204
Purushotham, N., Jones, A., Poudel, B., Nasim, J., Adorada, D., Sparks, A., et al. (2020). Draft genome resource for Macrophomina phaseolina associated with charcoal rot in sorghum. Mol. Plant Microbe Interact. 33, 724–726. doi: 10.1094/mpmi-12-19-0356-a
Quick, J., Grubaugh, N. D., Pullan, S. T., Claro, I. M., Smith, A. D., Gangavarapu, K., et al. (2017). Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 12, 1261–1276. doi: 10.1038/nprot.2017.066
Quick, J., Loman, N. J., Duraffour, S., Simpson, J. T., Severi, E., Cowley, L., et al. (2016). Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232. doi: 10.1038/nature16996
Racki, N., Dreo, T., Gutierrez-Aguirre, I., Blejec, A., and Ravnikar, M. (2014). Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 10:42. doi: 10.1186/s13007-014-0042-6
Radhakrishnan, G. V., Cook, N., Bueno Sancho, V., Lewis, C., Persoons, A., Mitiku, A., et al. (2019). MARPLE, a point-of-care, strain-level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biol. 17:65. doi: 10.1186/s12915-019-0684-y
Rajarammohan, S., Paritosh, K., Pental, D., and Kaur, J. (2019). Comparative genomics of Alternaria species provides insights into the pathogenic lifestyle of Alternaria brassicae - a pathogen of the Brassicaceae family. BMC Genomics 20:1036. doi: 10.1186/s12864-019-6414-6
Ramakrishnan, B., Venkateswarlu, K., Sethunathan, N., and Megharaj, M. (2019). Local applications but global implications: can pesticides drive microorganisms to develop antimicrobial resistance? Sci. Total Environ. 654, 177–189. doi: 10.1016/j.scitotenv.2018.11.041
Rang, F. J., Kloosterman, W. P., and de Ridder, J. (2018). From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19:90. doi: 10.1186/s13059-018-1462-9
Rani, A., Donovan, N., and Mantri, N. (2019). Review: the future of plant pathogen diagnostics in a nursery production system. Biosens. Bioelectron. 145:111631. doi: 10.1016/j.bios.2019.111631
Reed, A. J., Connelly, R. P., Williams, A., Tran, M., Shim, B. S., Choe, H., et al. (2019). Label-free pathogen detection by a deoxyribozyme cascade with visual signal readout. Sens. Actuat. B Chem. 282, 945–951. doi: 10.1016/j.snb.2018.11.147
Rezk, A. A., Sattar, M. N., Alhudaib, K. A., and Soliman, A. M. (2019). Identification of watermelon chlorotic stunt virus from watermelon and zucchini in Saudi Arabia. Can. J. Plant Pathol. 41, 285–290. doi: 10.1080/07060661.2019.1567590
Rigano, L. A., Malamud, F., Orce, I. G., Filippone, M. P., Marano, M. R., do Amaral, A. M., et al. (2014). Rapid and sensitive detection of Candidatus Liberibacter asiaticus by loop mediated isothermal amplification combined with a lateral flow dipstick. BMC Microbiol. 14:86. doi: 10.1186/1471-2180-14-86
Rojas, J. A., Miles, T. D., Coffey, M. D., Martin, F. N., and Chilvers, M. I. (2017). Development and application of qPCR and RPA genus- and species-specific detection of Phytophthora sojae and P. sansomeana root rot pathogens of soybean. Plant Dis. 101, 1171–1181. doi: 10.1094/pdis-09-16-1225-re
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schaad, N. W., Opgenorth, D., and Gaush, P. (2002). Real-time polymerase chain reaction for one-hour on-site diagnosis of Pierce’s disease of grape in early season asymptomatic vines. Phytopathology 92, 721–728. doi: 10.1094/phyto.2002.92.7.721
Schalamun, M., Nagar, R., Kainer, D., Beavan, E., Eccles, D., Rathjen, J. P., et al. (2019). Harnessing the MinION: an example of how to establish long-read sequencing in a laboratory using challenging plant tissue from Eucalyptus pauciflora. Mol. Ecol. Resour. 19, 77–89. doi: 10.1111/1755-0998.12938
Schmidhuber, J., and Tubiello, F. N. (2007). Global food security under climate change. Proc. Natl. Acad. Sci. U.S.A. 104, 19703–19708. doi: 10.1073/pnas.0701976104
Schwenkbier, L., Konig, S., Wagner, S., Pollok, S., Weber, J., Hentschel, M., et al. (2014). On-site detection of Phytophthora spp.-single-stranded target DNA as the limiting factor to improve on-chip hybridization. Microchimica Acta 181, 1669–1679. doi: 10.1007/s00604-013-1107-3
Schwenkbier, L., Pollok, S., Konig, S., Urban, M., Werres, S., Cialla-May, D., et al. (2015a). Towards on-site testing of Phytophthora species. Anal. Methods 7, 211–217. doi: 10.1039/c4ay02287d
Schwenkbier, L., Pollok, S., Rudloff, A., Sailer, S., Cialla-May, D., Weber, K., et al. (2015b). Non-instrumented DNA isolation, amplification and microarray-based hybridization for a rapid on-site detection of devastating Phytophthora kernoviae. Analyst 140, 6610–6618. doi: 10.1039/c5an00855g
Scuderi, G., Golmohammadi, M., Cubero, J., Lopez, M. M., Cirvilleri, G., and Llop, P. (2010). Development of a simplified NASBA protocol for detecting viable cells of the citrus pathogen Xanthomonas citri subsp citri under different treatments. Plant Pathol. 59, 764–772. doi: 10.1111/j.1365-3059.2010.02305.x
Selvaraj, V., Maheshwari, Y., Hajeri, S., Chen, J. C., McCollum, T. G., and Yokomi, R. (2018). Development of a duplex droplet digital PCR assay for absolute quantitative detection of “Candidatus Liberibacter asiaticus”. PLoS One 13:e0197184. doi: 10.1371/journal.pone.0197184
Shaffer, L. (2019). “Inner workings: portable dna sequencer helps farmers stymie devastating viruses,” in Proceedings of the National Academy of Sciences, 116, 3351–3353. doi: 10.1073/pnas.1901806116
Sillo, F., Giordano, L., and Gonthier, P. (2018). Fast and specific detection of the invasive forest pathogen Heterobasidion irregulare through a Loop-mediated isothermal AMPlification (LAMP) assay. Forest Pathol. 48:e12396. doi: 10.1111/efp.12396
Silva, G., Oyekanmi, J., Nkere, C. K., Bomer, M., Kumar, P. L., and Seal, S. E. (2018). Rapid detection of potyviruses from crude plant extracts. Anal. Biochem. 546, 17–22. doi: 10.1016/j.ab.2018.01.019
Smith, M. C., Steimle, G., Ivanov, S., Holly, M., and Fries, D. P. (2007). An integrated portable hand-held analyser for real-time isothermal nucleic acid amplification. Anal. Chim. Acta 598, 286–294. doi: 10.1016/j.aca.2007.07.036
Soares, R. R. G., Neumann, F., Caneira, C. R. F., Madaboosi, N., Ciftci, S., Hernandez-Neuta, I., et al. (2019). Silica bead-based microfluidic device with integrated photodiodes for the rapid capture and detection of rolling circle amplification products in the femtomolar range. Biosens. Bioelectron. 128, 68–75. doi: 10.1016/j.bios.2018.12.004
Solares, E. A., Chakraborty, M., Miller, D. E., Kalsow, S., Hall, K., Perera, A. G., et al. (2018). Rapid low-cost assembly of the Drosophila melanogaster reference genome using low-coverage, long-read sequencing. G3 (Bethesda) 8, 3143–3154. doi: 10.1534/g3.118.200162
Song, J. Z., Liu, C. C., Mauk, M. G., Peng, J., Schoenfeld, T., and Bau, H. H. (2018). A multifunctional reactor with dry-stored reagents for enzymatic amplification of nucleic acids. Anal. Chem. 90, 1209–1216. doi: 10.1021/acs.analchem.7b03834
Spits, C., Le Caignec, C., De Rycke, M., Van Haute, L., Van Steirteghem, A., Liebaers, I., et al. (2006). Whole-genome multiple displacement amplification from single cells. Nat. Protoc. 1, 1965–1970. doi: 10.1038/nprot.2006.326
Srivastava, S., Abad, Z. G., Knight, L. M., Rivera, Y., Zeller, K. A., Mavrodieva, V. A., et al. (2019). High-throughput sequencing of Phytophthora ramorum Ex-type using long-read MinION nanopore sequencing. Phytopathology 109, 186–186.
Srivathsan, A., Baloglu, B., Wang, W., Tan, W. X., Bertrand, D., Ng, A. H. Q., et al. (2018). A MinION-based pipeline for fast and cost-effective DNA barcoding. Mol. Ecol. Resour. 18, 1035–1049. doi: 10.1111/1755-0998.12890
Strayer-Scherer, A., Jones, J. B., and Paret, M. L. (2019). Recombinase polymerase amplification assay for field detection of tomato bacterial spot pathogens. Phytopathology 109, 690–700. doi: 10.1094/phyto-03-18-0101-r
Suzuki, R., Fukuta, S., Matsumoto, Y., Hasegawa, T., Kojima, H., Hotta, M., et al. (2016). Development of reverse transcription loop-mediated isothermal amplification assay as a simple detection method of Chrysanthemum stem necrosis virus in chrysanthemum and tomato. J. Virol. Methods 236, 29–34. doi: 10.1016/j.jviromet.2016.07.005
Tahzima, R., Foucart, Y., Peusens, G., Belien, T., Massart, S., and De Jonghe, K. (2019). New sensitive and fast detection of Little cherry virus 1 using loop-mediated isothermal amplification (RT-LAMP). J. Virol. Methods 265, 91–98. doi: 10.1016/j.jviromet.2018.12.019
Tomlinson, J. A., Boonham, N., Hughes, K. J. D., Griffen, R. L., and Barker, I. (2005). On-site DNA extraction and real-time PCR for detection of Phytophthora ramorum in the field. Appl. Environ. Microbiol. 71, 6702–6710. doi: 10.1128/aem.71.11.6702-6710.2005
Tomlinson, J. A., Dickinson, M. J., and Boonham, N. (2010). Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology 100, 143–149. doi: 10.1094/phyto-100-2-0143
Tsaloglou, M. N., Bahi, M. M., Waugh, E. M., Morgan, H., and Mowlem, M. (2011). On-chip real-time nucleic acid sequence-based amplification for RNA detection and amplification. Anal. Methods 3, 2127–2133. doi: 10.1039/c1ay05164d
Tsaloglou, M. N., Nemiroski, A., Camci-Unal, G., Christodouleas, D. C., Murray, L. P., Connelly, J. T., et al. (2018). Handheld isothermal amplification and electrochemical detection of DNA in resource-limited settings. Anal. Biochem. 543, 116–121. doi: 10.1016/j.ab.2017.11.025
van der Merwe, M., Jukes, M. D., Rabalski, L., Knox, C., Opoku-Debrah, J. K., Moore, S. D., et al. (2017). Genome analysis and genetic stability of the Cryptophlebia leucotreta granulovirus (CrleGV-SA) after 15 Years of commercial use as a biopesticide. Int. J. Mol. Sci. 18:2327. doi: 10.3390/ijms18112327
van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi: 10.1016/j.tig.2018.05.008
Varma, A., Padh, H., and Shrivastava, N. (2007). Plant genomic DNA isolation: an art or a science. Biotechnol. J. 2, 386–392. doi: 10.1002/biot.200600195
Vaser, R., Sovic, I., Nagarajan, N., and Sikic, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Venzac, B., Diakite, M. L., Herthnek, D., Cisse, I., Bockelmann, U., Descroix, S., et al. (2018). On-chip conductometric detection of short DNA sequences via electro-hydrodynamic aggregation. Analyst 143, 190–199. doi: 10.1039/c7an00798a
Vidic, J., Vizzini, P., Manzano, M., Kavanaugh, D., Ramarao, N., Zivkovic, M., et al. (2019). Point-of-need DNA testing for detection of foodborne pathogenic bacteria. Sensors 19:1100. doi: 10.3390/s19051100
Villari, C., Tomlinson, J., Battisti, A., Boonham, N., Capretti, P., and Faccoli, M. (2013). Use of loop-mediated isothermal amplification for detection of Ophiostoma clavatum, the primary blue stain fungus associated with Ips acuminatus. Appl. Environ. Microbiol. 79, 2527–2533. doi: 10.1128/aem.03612-12
Voegel, T. M., and Nelson, L. M. (2018). Quantification of agrobacterium vitis from grapevine nursery stock and vineyard soil using droplet digital PCR. Plant Dis. 102, 2136–2141. doi: 10.1094/pdis-02-18-0342-re
Wambua, L., Schneider, B., Olcvvaro, A., Wanga, J. O., Imali, O., Wambua, P. N., et al. (2017). Development of field-applicable tests for rapid and sensitive detection of Candidatus Phytoplasma oryzae. Mol. Cell. Probes 35, 44–56. doi: 10.1016/j.mcp.2017.06.004
Wang, M., Fu, A. S., Hu, B., Tong, Y. Q., Liu, R., Liu, Z., et al. (2020). Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small 16:2002169. doi: 10.1002/smll.202002169
Wei, S. Q., Sun, Y. J., Xi, G. S., Zhang, H. J., Xiao, M. Y., and Yin, R. (2018). Development of a single-tube nested PCR-lateral flow biosensor assay for rapid and accurate detection of Alternaria panax Whetz. PLoS One 13:e0206462. doi: 10.1371/journal.pone.0206462
Wilisiani, F., Tomiyama, A., Katoh, H., Hartono, S., Neriya, Y., Nishigawa, H., et al. (2019). Development of a LAMP assay with a portable device for real-time detection of begomoviruses under field conditions. J. Virol. Methods 265, 71–76. doi: 10.1016/j.jviromet.2018.10.005
Wingfield, M. J., Garnas, J. R., Hajek, A., Hurley, B. P., de Beer, Z. W., and Taerum, S. J. (2016). Novel and co-evolved associations between insects and microorganisms as drivers of forest pestilence. Biol. Invasions 18, 1045–1056. doi: 10.1007/s10530-016-1084-7
Wu, X. H., Chen, C. F., Xiao, X. Z., and Deng, M. J. (2016). Development of reverse transcription thermostable helicase-dependent DNA amplification for the detection of tomato spotted wilt virus. J. AOAC Int. 99, 1596–1599. doi: 10.5740/jaoacint.16-0132
Xia, H., Zhao, N., Zhao, L., Wang, Y. J., Zhao, W., and Yuan, Z. M. (2019). Rapid detection of Banna virus by reverse transcription-loop -mediated isothermal amplification (RT-LAMP). Int. J. Infect. Dis. 78, 93–98. doi: 10.1016/j.ijid.2018.10.025
Xu, H., Xia, A., Wang, D., Zhang, Y., Deng, S., Lu, W., et al. (2020). An ultraportable and versatile point-of-care DNA testing platform. Sci. Adv. 6:eaaz7445. doi: 10.1126/sciadv.aaz7445
Yamagishi, J., Runtuwene, L. R., Hayashida, K., Mongan, A. E., Thi, L. A. N., Thuy, L. N., et al. (2017). Serotyping dengue virus with isothermal amplification and a portable sequencer. Sci. Rep. 7:3510. doi: 10.1038/s41598-017-03734-5
Yang, L. L., Sun, L. X., Ruan, X. L., Qiu, D. Y., Chen, D. H., Cai, X. Q., et al. (2015). Development of a single-tube duplex real-time fluorescence method for the rapid quantitative detection of Fusarium oxysporum f. sp cubense race 1 (FOC1) and race 4 (FOC4) using TaqMan probes. Crop Protect. 68, 27–35. doi: 10.1016/j.cropro.2014.11.004
Yao, Q., Wang, Y. Q., Wang, J., Chen, S. M., Liu, H. Y., Jiang, Z. R., et al. (2018). An ultrasensitive diagnostic biochip based on biomimetic periodic nanostructure-assisted rolling circle amplification. ACS Nano 12, 6777–6783. doi: 10.1021/acsnano.8b01950
Yaseen, T., Drago, S., Valentini, F., Elbeaino, T., Stampone, G., Digiaro, M., et al. (2015). On-site detection of Xylella fastidiosa in host plants and in “spy insects” using the real-time loop-mediated isothermal amplification method. Phytopathol. Mediterranea 54, 488–496.
Yasuhara-Bell, J., Pedley, K. F., Farman, M., Valent, B., and Stack, J. P. (2018). Specific detection of the wheat blast pathogen (Magnaporthe oryzae Triticum) by loop-mediated isothermal amplification. Plant Dis. 102, 2550–2559. doi: 10.1094/pdis-03-18-0512-re
Yeh, K. B., Wood, H., Scullion, M., Russell, J. A., Parker, K., Gnade, B. T., et al. (2019). Molecular detection of biological agents in the field: then and now. mSphere 4:e00695-19. doi: 10.1128/mSphere.00695-19
Yetisen, A. K. A., Muhammad, S., and Lowe, C. R. (2013). Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 13, 2210–2251.
Zervas, A., Zeng, Y. H., Madsen, A. M., and Hansen, L. H. (2019). Genomics of aerobic photoheterotrophs in wheat phyllosphere reveals divergent evolutionary patterns of photosynthetic genes in Methylobacterium spp. Genome Biol. Evol. 11, 2895–2908. doi: 10.1093/gbe/evz204
Zhang, S. L., Ravelonandro, M., Russell, P., McOwen, N., Briard, P., Bohannon, S., et al. (2014). Rapid diagnostic detection of plum pox virus in Prunus plants by isothermal AmplifyRP (R) using reverse transcription-recombinase polymerase amplification. J. Virol. Methods 207, 114–120. doi: 10.1016/j.jviromet.2014.06.026
Zhang, X., Zhang, H., Pu, J. J., Qi, Y. X., Yu, Q. F., Xie, Y. X., et al. (2013). Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Fusarium oxysporum f. sp cubense tropical race 4 in soil. PLoS One 8:e82841. doi: 10.1371/journal.pone.0082841
Zhang, Z. Y., Wang, Y. F., Li, Y. L., Yu, D. S., Chen, H. Y., Cai, Y., et al. (2019). Portable fluorescence-based microRNA detection system based on isothermal signal amplification technology. Biotechnol. Appl. Biochem. 66, 82–90. doi: 10.1002/bab.1699
Zhao, L. M., Li, G., Gao, Y., Zhu, Y. R., Liu, J., and Zhu, X. P. (2015). Reverse transcription loop-mediated isothermal amplification assay for detecting tomato chlorosis virus. J. Virol. Methods 213, 93–97. doi: 10.1016/j.jviromet.2014.11.013
Zheng, Y., Luo, Y., Zhou, Y.-L., Zeng, X., Duan, X., Cao, X., et al. (2013). Real-time PCR quantification of latent infection of wheat powdery mildew in the field. Eur. J. Plant Pathol. 136, 565–575. doi: 10.1007/s10658-013-0188-5
Zhong, J. L., and Zhao, X. H. (2018). Isothermal amplification technologies for the detection of foodborne pathogens. Food Anal. Methods 11, 1543–1560. doi: 10.1007/s12161-018-1177-2
Zhong, X., Liu, X. L., Lou, B. H., Zhou, C. Y., and Wang, X. F. (2018). Development of a sensitive and reliable droplet digital PCR assay for the detection of ’Candidatus Liberibacter asiaticus’. J. Integr. Agric. 17, 483–487. doi: 10.1016/s2095-3119(17)61815-x
Keywords: biosensor portable devices, in field detection, qPCR, LAMP, RPA, RCA, SDA, MinION nanopore sequencing
Citation: Baldi P and La Porta N (2020) Molecular Approaches for Low-Cost Point-of-Care Pathogen Detection in Agriculture and Forestry. Front. Plant Sci. 11:570862. doi: 10.3389/fpls.2020.570862
Received: 09 June 2020; Accepted: 29 September 2020;
Published: 28 October 2020.
Edited by:
Sébastien Duplessis, INRA Centre Nancy-Lorraine, FranceReviewed by:
Nicolas Feau, University of British Columbia, CanadaCopyright © 2020 Baldi and La Porta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paolo Baldi, cGFvbG8uYmFsZGlAZm1hY2guaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.