- 1Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Forestry University, Beijing, China
- 2National Engineering Laboratory for Tree Breeding, Beijing Forestry University, Beijing, China
- 3College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
Salt stress is an adverse environmental factor for plant growth and development. Under salt stress, plants can activate the selective autophagy pathway to alleviate stress. However, the regulatory mechanism of selective autophagy in response to salt stress remains largely unclear. Here, we report that the selective autophagy receptor PagNBR1 (neighbor of BRCA1) is induced by salt stress in Populus. Overexpression of PagNBR1 in poplar enhanced salt stress tolerance. Compared with wild type (WT) plants, the transgenic lines exhibited higher antioxidant enzyme activity, less reactive oxygen species (ROS), and higher net photosynthesis rates under salt stress. Furthermore, co-localization and yeast two-hybrid analysis revealed that PagNBR1 was localized in the autophagosome and could interact with ATG8 (autophagy-related gene). PagNBR1 transgenic poplars formed more autophagosomes and exhibited higher expression of ATG8, resulting in less accumulation of insoluble protein and insoluble ubiquitinated protein compared to WT under salt stress. The accumulation of insoluble protein and insoluble ubiquitinated protein was similar under the treatment of ConA in WT and transgenic lines. In summary, our results imply that PagNBR1 is an important selective autophagy receptor in poplar and confers salt tolerance by accelerating antioxidant system activity and autophagy activity. Moreover, the NBR1 gene is an important potential molecular target for improving stress resistance in trees.
Introduction
Soil salinity is an adverse environmental factor, which has a serious impact on plant growth, development, and productivity (Zorb et al., 2005). About 6% of the world’s total land area and nearly 20% of irrigated land are exposed to excessive salt concentrations, and the problem is becoming progressively worse (Munns and Tester, 2008). Therefore, understanding the mechanism of plant tolerance and breeding salt-tolerant plants is of great biological significance for the amelioration and utilization of salinated soil. Salt stress is triggered by excessive amounts of sodium (Na+) and chloride ions in soil (Cl−; Ismail et al., 2014), which puts plants under multi-component stress, including ion stress, osmotic stress, and oxidative stress (Munns and Tester, 2008; Cabot et al., 2014). Osmotic, ionic, and oxidative stress can induce serious damage, which includes the disruption of cellular structures and macromolecules (e.g., proteins, DNA, and lipids; Wang et al., 2009; Golldack et al., 2014; Genisel et al., 2015). An important strategy for plants to cope with salt stress is to maintain the water potential and K+/Na+ homeostasis by synthesizing several classes of osmolytes (e.g., proline, polyamine, and sugar alcohol) and regulating the activity of ion channel proteins, such as SOS1 (Deinlein et al., 2014; Yang and Guo, 2018). Another important strategy is scavenging reactive oxygen species through the antioxidant system, such as superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), and ascorbate-glutathione (AsA-GSH) cycle (Raja et al., 2017; Yang and Guo, 2018).
In recent years, autophagy, which means “self-eating,” has become an important strategy for eukaryotes in resisting stress conditions (Avin-Wittenberg et al., 2012; Avin-Wittenberg, 2019). Autophagy can be classified into different types, macro-autophagy, micro-autophagy (Cvt), and chaperone-mediated autophagy (CMA). The autophagy-related genes (ATG) discovered in yeast have been identified and studied in other eukaryotes (Cebollero and Reggiori, 2009; Wen and Klionsky, 2016), implying that autophagy is an evolutionarily conserved process in eukaryotes.
Compared with the selective CMA and Cvt pathways, the most important role of the macro-autophagy pathway is initially considered to degrade bulk cytoplasm through the non-selective pathway. Recent research has revealed that selective autophagy mechanisms are mediated by specific cargo receptor proteins (Ichimura et al., 2008; Johansen and Lamark, 2011). Several receptors have been identified in mammalian organisms, such as nuclear dot protein 52 kDa (NDP52; von Muhlinen et al., 2010), optineurin (OPTN; Liu et al., 2014), sequestosome-1 (P62/SQSTM1; Ichimura et al., 2008), and neighbor of BRCA1 gene 1 (NBR1; Kirkin et al., 2009). p62/SQSTM1 is an important selective receptor in human and mammalian cells. Previous research has largely focused on human p62/SQSTM1 (Lamark et al., 2009; Lee et al., 2017). The p62/SQSTM1 protein mainly binds and facilitates the clearance of ubiquitinated proteins that accumulate in various chronic, toxic, and degenerative diseases (Liu et al., 2016; Carroll et al., 2018). The p62/SQSTM1 protein also participates in ROS cleaning processes in human cells. NBR1 was also found in numerous metazoan species for the degradation of ubiquitinated proteins that share similar domain architecture to p62/SQSTM1 (Kirkin et al., 2009; Kenific and Debnath, 2016). In addition, both proteins are involved in the degradation of ubiquitinated proteins via PB1-dependent polymerization. However, the PB1 domain in p62/SQSTM1 and NBR1 differs. The PB1 domain in the p62/SQSTM1 protein harbors two OPCA (OPR/PC/AID) motifs and can oligomerize with itself; meanwhile, the NBR1 protein harbors only one OPCA (OPR/PC/AID) motif and binds with p62/SQSTM1 in a PB1-PB1 fashion (Kirkin et al., 2009; Mizushima, 2015).
Autophagy studies in plants benefit from the functional conservation of ATG proteins and autophagy has been shown to participate in multiple biological processes in plants, which include biotic stress, abiotic stress, growth, and development (Inoue et al., 2006; Zhou et al., 2015; Luo et al., 2017). In Arabidopsis, atg mutants exhibit early senescence and chlorosis phenotype, as well as nitrogen hypersensitivity or carbon nutrient deficiency (Masclaux-Daubresse, 2016). Autophagy also regulates salicylic acid signaling to inhibit programmed cell death and senescence under pathogen infection (Yoshimoto et al., 2009). Thus, autophagy is a functionally important process for plants in response to stress. Although studies on autophagy in plants have provided great progress in recent years (Li et al., 2015; Luo et al., 2017), our knowledge of selective autophagy remains limited. and the adapter of selective autophagy still needs further research.
Plant NBR1 (neighbor of BRCA1), which is a selective receptor, is homologous with mammalian p62/SQSTM1 and NBR1 proteins (Svenning et al., 2011; Zientara-Rytter et al., 2011). They share conserved domains such as the N-terminal PB1 (Phlox and Bem1), ZZ-type zinc finger domain, LIR (LC3-interacting region) motif, and C-terminal UBA (ubiquitin-associated) domain (Svenning et al., 2011). In contrast to p62/SQSTM1, the plant NBR1 protein contains four specific tryptophan (FW) residues, which are highly conserved in the homologs of NBR1 proteins (Svenning et al., 2011). Research indicates that the homopolymerized characterization of NBR1 protein in plants was similar to p62/SQSTM1, thereby suggesting that plant NBR1 possesses hybrid properties of mammalian NBR1 and p62/SQSTM1 proteins (Svenning et al., 2011; Zientara-Rytter and Sirko, 2014). Meanwhile, p62/SQSTM1 protein is confined to metazoans, and only NBR1 protein is detected throughout the plant kingdom. In Arabidopsis, catalases were highly aggregated proteins in the nbr1 mutant suggesting the ital roles of NBR1 in the cleaning of ROS (Zhou et al., 2014). Thus, this study speculated that the plant NBR1 could be involved in the degradation of ubiquitinated insoluble proteins and the cleaning of ROS under stress conditions.
Populus is one of the main forestation tree species that exhibits fast growth, adaptability, and worldwide distribution. This species has important economic and ecologic value for the quick recovery of vegetation and soil remediation (Yannikos et al., 2014; Radojčić Redovniković et al., 2017). Soil salinity has seriously affected the growth of trees and timber yields. Populus is a model plant for tree research (Tuskan et al., 2006), and understanding the mechanisms of salt stress resistance in Populus is of practical significance for the amelioration and utilization of salinated soil.
Selective autophagy mechanism in response to abiotic stress remains unreported in Populus. However, the homologs of selective autophagy receptors were found in Arabidopsis (AtNBR1), tobacco (Joka2/NBR1), and tomato (NBR1). Previous research has focused on heat stress (Zhou et al., 2013), chilling stress (Chi et al., 2020), and sulfur deficiency (Zientara-Rytter et al., 2011). Only a few studies have emphasized the response of NBR1-mediated selective autophagy to salt stress in plants, and the functional mechanism of plant NBR1 is still largely unclear. Our previous studies showed that expression levels of the NBR1 gene in Populus is induced by salt stress and oxidative stress. In the current study, we hypothesized that the poplar NBR1 protein may function in response to salt stress through the degradation of ubiquitinated proteins and activation of antioxidant enzymes. To explore this we isolated an NBR1 gene from Poplar 84 K clone (Populus alba × P. tremula var. glandulosa; PagNBR1), a hybrid poplar introduced from Korea that has been extensively cultivated in northern China (Qiu et al., 2019). Transgenic poplars were generated to evaluate the potential tolerance mechanism mediated by the NBR1 gene in response to salt stress.
Experimental Procedures
Plant Material and Treatments
Poplar 84 K hybrids (Populus alba × P. tremula var. glandulosa) have been commonly used in molecular biological studies (He et al., 2018). The 84 K saplings were approximately 5 cm high culturing on the rooting medium. They were transplanted in seed pots (10 cm × 10 cm), in the phytotron artificial climate chamber with a 16/8-h day/night cycle (22–25°C, 60% humidity, 54 μmol m−2 s−1) in Beijing Forestry University, Beijing, China. The plants were watered with a nutrient solution every 3 days for 60 days before treatment.
The expression level of PagNBR1 was analyzed in different tissues of poplar 84 K including roots (R), xylem (X), phloem (P), mature leaves (ML), old leaves (OL), and young leaves. For salt stress, 84 K saplings were watered with 400 mM NaCl solution. For oxidative stress, 100 μM MV (Sigma, 856177) or 0.5 mM H2O2 solution was sprayed once on the leaves of 84 K. For plant hormone treatment, the 84 K saplings were sprayed once on leaves with a solution containing 100 μM abscisic acid (ABA, SigmaA1049), 5 mM ethephon (ET, Sigma 45473), 2 mM salicylic acid (SA, SigmaS5922), 100 μM Methyl Jasmonate (MeJA, Sigma, 392707) or 500 μM Gibberellin (GA3, Sigma 48870). For the control groups, 84 K saplings were watered or sprayed with water. Mature leaves (from 7th to 11th) were collected after 0 h, 1 h, 2 h, 4 h, 6 h, 12 h, 24 h of treatment and immediately put into liquid nitrogen and stored at −80°C for use.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from the Control and treatment materials using the cetyltrimethylammonium bromide (CTAB) method (He et al., 2018). Total RNA quality and integrity were detected using the NanoDrop 2000 spectrophotometer (Thermal Fisher Scientific, West Palm Beach, FL) and agarose gel electrophoresis. 2 μg RNA was used for reverse transcription using the Prime Script™ RT reagent Kit with gDNA Eraser (Perfect Real Time, Takara, RR047Q) following the manufacturer’s guidance. The qRT-PCR was performed as previously methods (He et al., 2018) using the TB Green® Premix Ex Taq™ GC (Perfect Real Time, Takara, RR071Q). The relative expression level of genes was calculated using 2-∆∆Ct as described previously (Wang et al., 2014, 2015b). Primers were designed using Primer 5.0 (Sigma-Aldrich Corp., St. Louis, MO) and listed in Supplementary Table S1. Three biological replicates and four technical replicates were carried out in the experiment.
Isolation and Bio-Informatics Analysis of PagNBR1
The complete coding sequence (CDS) of PagNBR1 was amplified using high-fidelity thermostable DNA polymerize Prime Star (Takara, R045A). The primer used in this study is listed in Supplementary Table S1. The multiple sequence alignment for the proteins of AtNBR1, PtrNBR1, and PagNBR1 were performed with DNAMAN V6. The conserved domains were analyzed using the conserved domain database.1 The physicochemical property of PagNBR1 protein was predicted using ProtParam database.2
Subcellular Location of PagNBR1
To confirm the subcellular localization of the PagNBR1 protein, the PagNBR1 and AtATG8a genes were inserted into a plant binary expression vector 35S-GFP which was reconstructed from pCAMBIA1300-GUS under the control of CaMV35S promoter. The constructed vectors 35S::PagNBR1::GFP and 35S::mCherry::AtATG8a were transiently transformed into tobacco protoplast using the methods previously described (Yoo et al., 2007; Chupeau et al., 2013). The 35S::mCherry::AtATG8a was used as an autophagosome marker and constructed as previous methods (Ren et al., 2014). The 35S-GFP was used as vector control. The signal of GFP and mCherry were detected using a confocal microscope (Leica TCS SP8; Leica, Wetzlar, Germany) after being transformed in tobacco protoplast for12-16 h. The excitation wavelength was 488 nm (GFP), 580 nm (mCherry), and the emission wavelengths 507 nm (GFP) and 610 nm (mCherry) were used to detected fluorescence signal.
Yeast Two-Hybrid Analysis
To evaluate the interactions of PagNBR1 and ATG8 proteins in 84 K (PagATG8), a yeast two-hybrid system was prepared. The CDS of PagNBR1 and PagATG8 (ATG8a, ATG8e, ATG8g, ATG8h) genes were inserted into pGBKT7 and pGADT7 vectors to generate BK-PagNBR1 and AD-PagATG8s vectors, respectively. Then the vectors were co-transformed into yeast competent AH109 cells. The AD/BK, AD/BK-NBR1, BK/AD-ATG8s were performed as a negative control. The positive transformants were selected on the SD/-Leu-Trp medium and then serial dilution assay including 1, 1/10, 1/100, and 1/1000 was performed on SD/-Trp-Leu-His-Ade medium at 30°C for 4–6 d.
Generation of Transgenic Plants
The CDS of PagNBR1 was inserted into the 35S-GFP vector before the GFP with termination codon deleted under the control of the CaMV35S promoter. The fusion recombinant vector was transformed into Agrobacterium tumefactions strain EHA105 before infecting poplar leaves. The Agrobacterium-mediated transformation was performed using previously described methods (He et al., 2018) and the detailed information was supported in the Supplement Methods.
Total DNA was isolated using the CTAB method (He et al., 2018) and materials were collected from the saplings selected using hygromycin B. PCR was performed using 20 bp 35S promoter as the forward primer and 21 bp GFP as reserve primer to validate the transgenic plants at the DNA level. qRT-PCR was also used to analyze the expression level of the PagNBR1 gene in wild type (WT) and transgenic lines. The reference gene UBQ was selected based on a previous result (Wang et al., 2015b). The primers used are listed in Supplementary Table S1.
Assessment of Salt Stress Tolerance in Transgenic Lines
Twenty-day asexual-propagation seedlings were transplanted to seedling pots (10 cm length × 10 cm width × 10 cm height) with the same seedling substrate (turfy soil: vermiculite =1:1) and then the culture in a phytotron artificial climate chamber with a16/8-h day/night cycle (25°C, 60% humidity, 54 μmol m−2 s−1). To assess the tolerance of salt stress at the whole-plant level, 60-day WT and transgenic lines were subjected to 400 mM NaCl solution until damaged spots were visible on the leaves of the plants. For the control group, WT and transgenic lines were cultured in the same conditions but replaced the NaCl solution with water. After NaCl treatment, the plants were harvested for enzyme activity, relative water content, MDA content, Na+ content, K+ content Fresh weight (FW), and Dry weight (DW) shoots and roots. The treatment was performed with four biological replicates and three times technological replicates, respectively. The detailed treatment methods were listed in the Supplement Methods.
To further verify whether autophagy is involved in the process of the degradation of insoluble protein and the insoluble ubiquitinated proteins among WT and transgenic lines under salt stress, the leaves of WT and transgenic lines were prepared in small rounds and treated with water (Control group), 200 mM NaCl and 200 mM NaCl with 0.5 μM Concanamycin A (ConA, Sigma-Aldrich, United States) and harvested after 12 h of treatment.
Malonaldehyde Concentration, Electrolyte Leakage, and Relative Water Content
Malonaldehyde (MDA) concentration and Electrolyte leakage (EL) were analyzed in WT and transgenic plants after treatment. MDA was measured using thiobarbituric acid (TBA)-reactive substances (Li et al., 2012). Electrolyte leakage was measured according to a previous protocol (Shi et al., 2013). In brief, nine leaf discs with 0.5 cm diameter were immersed in 10 ml deionized water for 8 h for detecting initial conductivity (Ci). Then, the samples were incubated at 95°C for 30 min to destroy the leaf tissues and release the whole electrolyte. After cooling the solution to room temperature, the max conductivity (Cmax) was detected. The EL was estimated as the formula EL(%) = (Ci/Cmax) × 100%.
Relative water content (RWC) was also analyzed in WT and transgenic plants after treatment. In brief, the fresh weight of leaves was recorded as FW. Then, the samples were incubated in distilled water for 24 h to measure the saturated weight (WW). In the end, they were drying at 80°C and dry weights were measured. The relative water content was estimated following the formula RWC (%) = (FW-DW)/(WW-DW) × 100%. 27 replicates containing nine biological and three technical replicates were conducted per experiment.
Measurement of Photosynthetic Gas Exchange and Chlorophyll Fluorescence Parameters
The Li-6400 Photosynthesis System (Li-Cor, Lincoln, NE, United States) was used in this study. Fully expanded mature leaves (from 7th to 11th) were selected to detect the net photosynthetic rate (Pn), Transpiration rate (Tr), stomatal conductance (Cond), and intercellular CO2 concentration (Ci) with 400 μmol/l CO2 concentration and 800 μmol m−2 s−1 photon flux density. For Quantum yield of photochemical energy conversion in PSII (Y (II)) and non-photochemical Quenching (qN), the same leaves were detected with a Dual-PAM-100 (Walz Heinz GmbH, Effeltrich, Germany).
Determination of Na+ and K+ Content
Root, stem, and leaves were collected from WT and transgenic poplars after treatment. They were heated at 120°C for 15 min and kept at 80°C until constant weight. Then the materials were ground into powder for the determination of Na+ and K+. 0.2 g materials were digested with concentrated sulfuric acid and titrated with H2O2. Na+ and K+ contents were determined with an atomic absorption spectrophotometer as described previously (Yang et al., 2015).
Measurement of ROS Accumulation, Antioxidant System Activity
Poplar 84 K leaves (0.2 g) were ground in liquid nitrogen and an ice-cold phosphate buffer solution (PBS, pH 7.8) was used to extract antioxidant enzyme contain SOD, CAT, POD. The homogenate was centrifuged, and the supernatant was used to measure antioxidant enzyme activity.
SOD activity was measured using the inhibition of photochemical nitro blue tetrazolium reduction methods (He et al., 2018). POD activity was measured by detecting the oxidation of guaiacol (He et al., 2018). CAT activity was measured using the ultraviolet absorption method by determining the decrease of H2O2 (Patra et al., 1978).
For the detection of O2− and H2O2 accumulation, histochemical staining methods were performed by immersing the leaves in nitro blue tetrazolium (NBT, Sigma, 11585029001) and diaminobenzidine (DAB, Sigma, D7679) respectively. The content of H2O2 in leaves was detected by monitoring ferrous ion oxidation using xylenol orange as an indicator (Zhang et al., 2017).
The content of AsA, DHA, GSH, and GSSG was analyzed according to the method described by Wang et al. (2012).
Isolation of Soluble, Insoluble Proteins, and Insoluble Ubiquitinated Proteins
The poplar 84 K leaves collected from the control and NaCl treatment groups were prepared. The samples were homogenized in protein extraction buffer (100 mM Tris/HCl, pH 8.0, 10 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.2% ß-mercaptoethanol). Soluble and insoluble proteins were isolated by low-speed centrifugation as previously described (Zhou et al., 2013). The contents of soluble and insoluble proteins were examined using Brilliant Blue G-250 dye (Bradford, 1976). Ubiquitinated protein in insoluble protein was detected using Plant ubiquitin (Ub) ELISA Kit (Shanghai renjie biotechnology co., ltd, Cat. No RJ217773) according to the manufacturer’s instructions. In brief, purified plant Ub antibody was used to coat microtiter plate wells and to make a solid-phase antibody, then Ub solution was added to the wells. It was combined with an antibody labeled with HRP and became an antibody-antigen-enzyme-antibody complex. After washing completely, a TMB substrate solution was added to the microtiter plate wells. The TMB substrate then turned a blue color under the catalyzation of the HRP enzyme. The reaction was terminated by a sulfuric acid solution and the color change was measured spectrophotometrically at a wavelength of 450 nm. The concentration of Ub in the samples was then determined by comparing the O.D. of the samples to the standard curve.
Autophagy Analysis
To analyze whether autophagy was induced in WT and transgenic lines under salt stress, leaves were excised and immediately immersed with 100 μM MDC (Sigma-Aldrich, 30432) for 30 min and then washed with phosphate-buffered saline. The fluorescence signal was detected at 400-508 nm according to Wang et al. (2015a).
TEM analysis was performed following previous methods. After 4 days of NaCl treatment, the mature leaves excised from poplar 84 K were used for observation. Small strips of leaves (1 mm × 2 mm, 10 pieces from each plant) were immediately mixed with 2.5% glutaraldehyde and fixed under dark conditions at 4°C for 12 h. PBS was used to wash the pieces before being fixed in 1% osmium tetroxide. Subsequently, different concentration gradients of ethanol (30–100%) were used. Fixed materials were packaged with Epon812. The ultrathin section was prepared using an ultra-microtome (Leica ULTRACUT, Wetzlar, Germany). Serial sections were examined using a JEOL-1230 transmission electron microscope (Hitachi, Tokyo, Japan) for autophagosome observation.
Statistics
All data were subjected to Statistical Product and Service Solutions v. 17.0 (SPSS, Chicago, IL) for statistical analysis. LSD multiple range tests were used to detect the significant differences between individual means. Differences at the 5% level were considered significant and denoted by lowercase letters or asterisk among different groups.
Results
Molecular Characterization of PagNBR1
NBR1, a selective autophagy receptor protein, is conserved in both animal and plant kingdoms. The target gene identified from Poplar 84 K contains 2301 bases which encode 767 amino acids. Protein primary structure analysis showed that the molecular weight of the protein is 83.5 KDa, and its theoretical pI is 4.94. Multiple sequence alignment analysis revealed that the protein sequence shares 42.1% identity with AtNBR1 protein and 96.6% identify with NBR1 protein from P. trichocarpa (PtrNBR1). The protein contains an FW domain (or named NBR1 domain), which is also highly conserved in homologs of NBR1 proteins. The N-terminal PB1 domain, ZZ-type zinc finger, and C-terminal UBA-domain (Figure 1A and Supplementary Figure S1) were also found in the protein, and all the three domains are highly conserved in the mammalian proteins of p62/SQSTM1 and NBR1 and plant NBR1 protein (Svenning et al., 2011). These results indicated that the protein is highly homologous with AtNBR1 protein suggesting that they may have a similar biological function. Thus, we named it PagNBR1 protein.
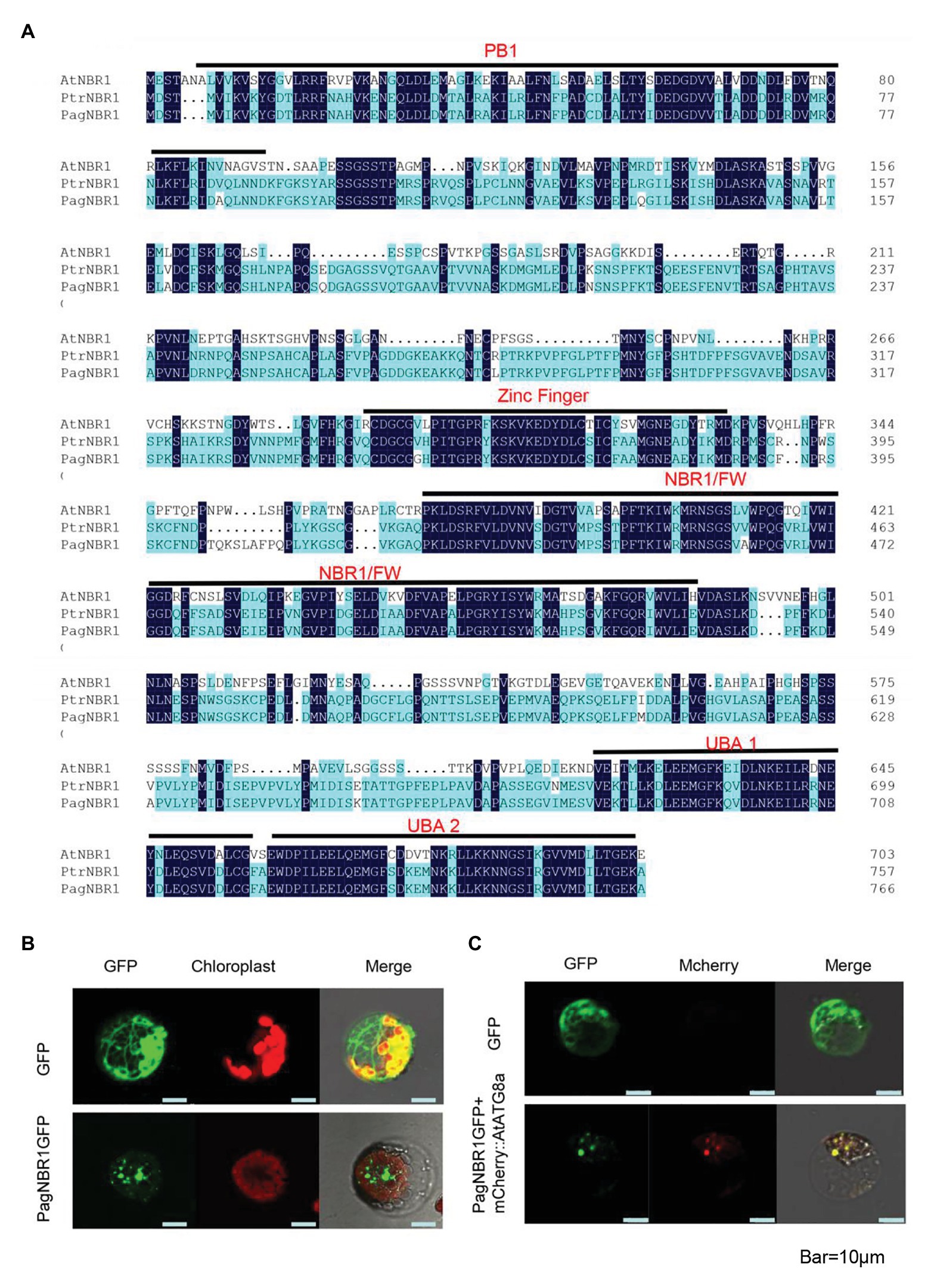
Figure 1. Sequence alignments of NBR1 in Arabidopsis thaliana and poplars and subcellular location of PagNBR1. (A) Sequence alignments of AtNBR1, PtrNBR1, and PagNBR1. Identical amino acids are shown with a dark blue background. The domains contained in sequences are shown with a black line and label. (B) Confocal laser scanning microscopy images of poplar mesophyll protoplast that transiently expresses PagNBR1:GFP and GFP (C) Confocal laser scanning microscopy images of poplar mesophyll protoplast that transiently expresses PagNBR1::GFP and mCherry::AtATG8a in tobacco protoplast. Bar = 10 um.
PaNBR1 Localizes in Autophagosome
Previous studies have demonstrated that NBR1 functions as an autophagy receptor, which facilitates specific substrate degradation by autophagy (Deosaran et al., 2013; Zhou et al., 2014). To confirm the detailed subcellular location of PagNBR1, we transiently transformed the recombinant vector 35S: NBR1: GFP into tobacco protoplast (Figure 1B). AtATG8a protein is used as an autophagosome-localized marker (Yoshimoto et al., 2004) and the mCherry::AtATG8a, which is restricted in our research, was co-transformed with PagNBR1::GFP to confirm its targeting position. Figures 1B,C show that the green fluorescence of PagNBR1 was localized in punctuated cytosolic bodies, and the red fluorescence of AtATG8a was localized in punctuated cytosolic bodies with overlapping of green fluorescence. This suggests that the PagNBR1 protein is located in the autophagosomes.
Expression Patterns of PagNBR1 in Different Tissues and in Response to Abiotic Stress
To illuminate the potential roles of the PagNBR1 gene, the expression patterns in different tissues, including the root (R), xylem (X), phloem (P), young leaf (YL), mature leaf (ML), and old leaf (OL), were analyzed by qRT-PCR under normal conditions. The accumulation of PagNBR1 was relatively high in the xylem, mature leaves, and old leaves (Figure 2A). In addition, the expression levels of PagNBR1 increased with leafage adding among young leaves, mature leaves, and old leaves implying that it may play important roles during plant senescence.
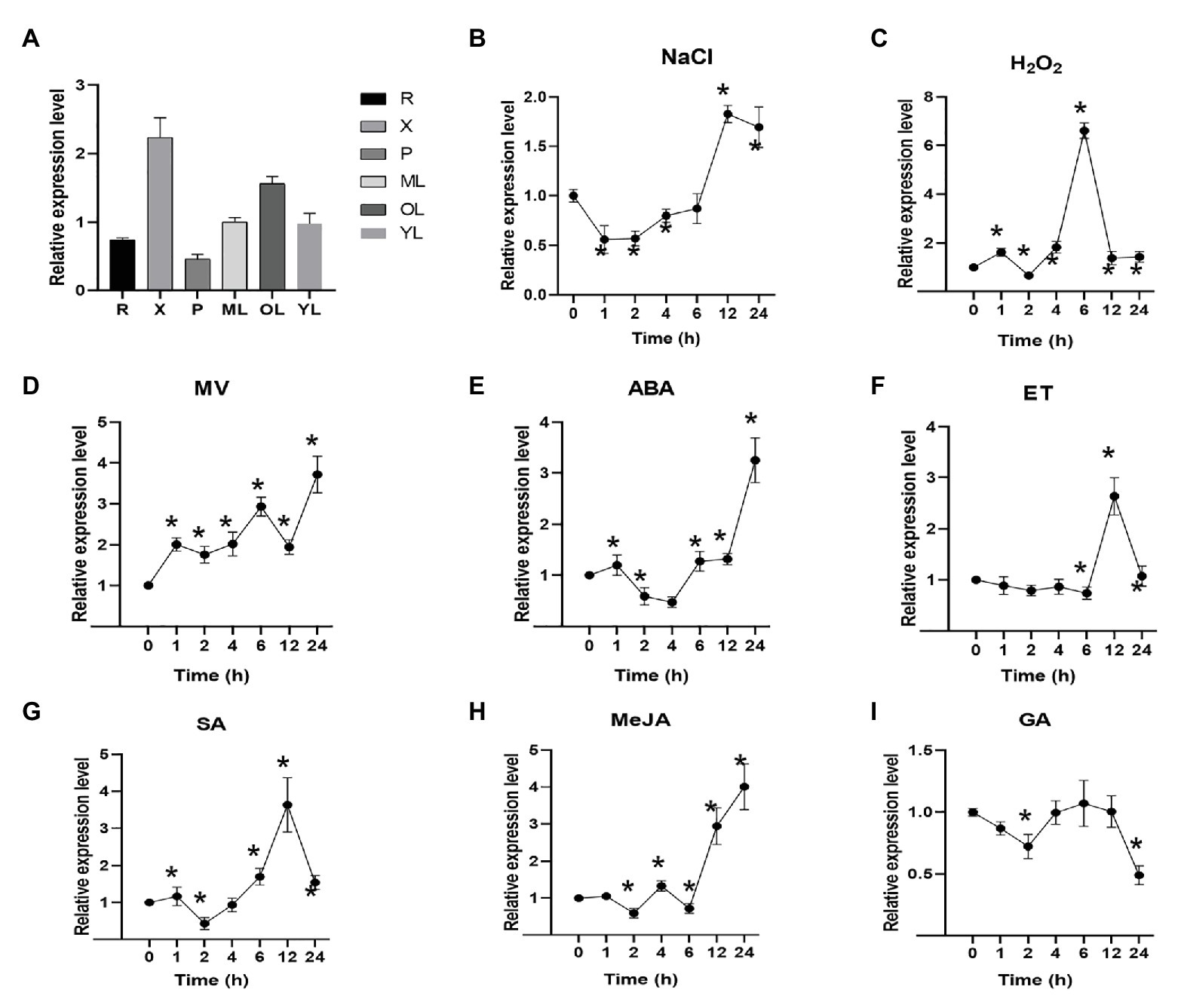
Figure 2. PagNBR1 expression patterns in different tissues and under different treatments. (A) Relative PagNBR1 expression levels in different tissues; Relative PagNBR1 expression levels in response to different treatment, NaCl (B), H2O2 (C), MV (D), ABA (E), ET (F), SA (G), Me-JA (H), and GA (I). R, root; YL, young leaves; X, xylem; P, phloem; ML, Mature leaf; OL, old leaf. Total RNA was isolated from leaf samples collected at indicated times. Each treatment was performed with three biological replicates (n = 3) and values are means ± SE. Asterisks indicate that values were significantly different from the control at p < 0.05.
Plant hormones are involved in many processes of plant development and stress tolerance (Verma et al., 2016). To illuminate whether PagNBR1 were induced by different plant hormones, the plant hormones such as ABA, ET, Me-JA, SA, and GA3, were sprayed on the leaves of WT plants. PagNBR1 gene showed similar expression patterns after ABA and Me-JA treatment (Figures 2E,H). It was downregulated by 0.5-fold after 2 (ABA) or 4 h (Me-JA) of treatment and upregulated by three-fold to four-fold after 24 h of treatment. In the case of ET treatment, the expression of the PagNBR1 gene was largely induced two-fold to three-fold after 6–12 h of treatment. The expression level of PagNBR1 reached a peak after 12 h treatment of ET (Figure 2F). SA treatment downregulated the transcription level of PagNBR1 during the first 2 h and was induced by more than three-fold of the control after 12 h of treatment (Figure 2G). GA treatment downregulated the expression levels of PagNBR1 after 2 h and 24 h of treatment, but it was similar after 4 to 12 h of treatment (Figure 2I).
To further analyze the expression levels of PagNBR1 during different stress conditions, WT plants were treated with NaCl, MV, and H2O2. Materials were collected after 0, 1, 2, 4, 6, 12, and 24 h. Our results showed that PagNBR1 mRNA level decreased to the minimum value after 1 h of salt treatment and then increased after 2 h of treatment and peaked to about two-fold of the control after 12 h of treatment (Figure 2B). H2O2 treatment dramatically increased the transcription level of the PagNBR1 gene by more than six-fold within 6 h of treatment and subsequently decreased and recovered to the basal level from 12 to 24 h (Figure 2C). MV treatment exhibited different expression patterns at an early stage (i.e., 0–2 h) with NaCl and H2O2. The PagNBR1 mRNA level increased with MV treatment duration and it was induced by more than three-fold of the control after 24 h of treatment (Figure 2D). In summary, our results showed that salt stress, oxidative stress, and plant hormones (which included ABA, ET, Me-JA, and SA) could induce PagNBR1 gene expression suggesting PagNBR1 maybe play important roles in resisting different external environmental stress.
PagNBR1 Overexpression Improves Salt Stress Tolerance in Poplar
Abiotic stress such as salt stress, oxidative stress, and plant hormone treatment promoted the expression of the PagNBR1 gene (Figure 2). Thus, we hypothesized that PagNBR1 may be involved in plant tolerance to salt stress. Transgenic poplar 84 K was generated and identified at the DNA and transcription levels (Supplementary Figure S2). Before formal experiments for collecting data, we performed a pre-experiment to calibrate the concentration of NaCl solution using two-month-old WT and transgenic lines. Results showed that the leaves of WT or transgenic lines did not exhibit obvious phenotype and wilted after 10 days of treatment with 200 mM NaCl (Supplementary Figure S4). This might be because watering the NaCl solution more than once could cause the enrichment of NaCl. The leaves of WT wilted after 4 days of treatment with 400 mM NaClD. During the treatment process, we watered the seedlings only once. We used 400 mM of NaCl solution and used it to simulate salt stress conditions. The seedlings treated with 400 mM NaCl were used in the treatment group, whereas the control group was replaced with water. The leaves of WT were more wilted than those of transgenic poplars, which suggested that WT suffered more serious damage than transgenic poplars after treatment (Figure 3A).
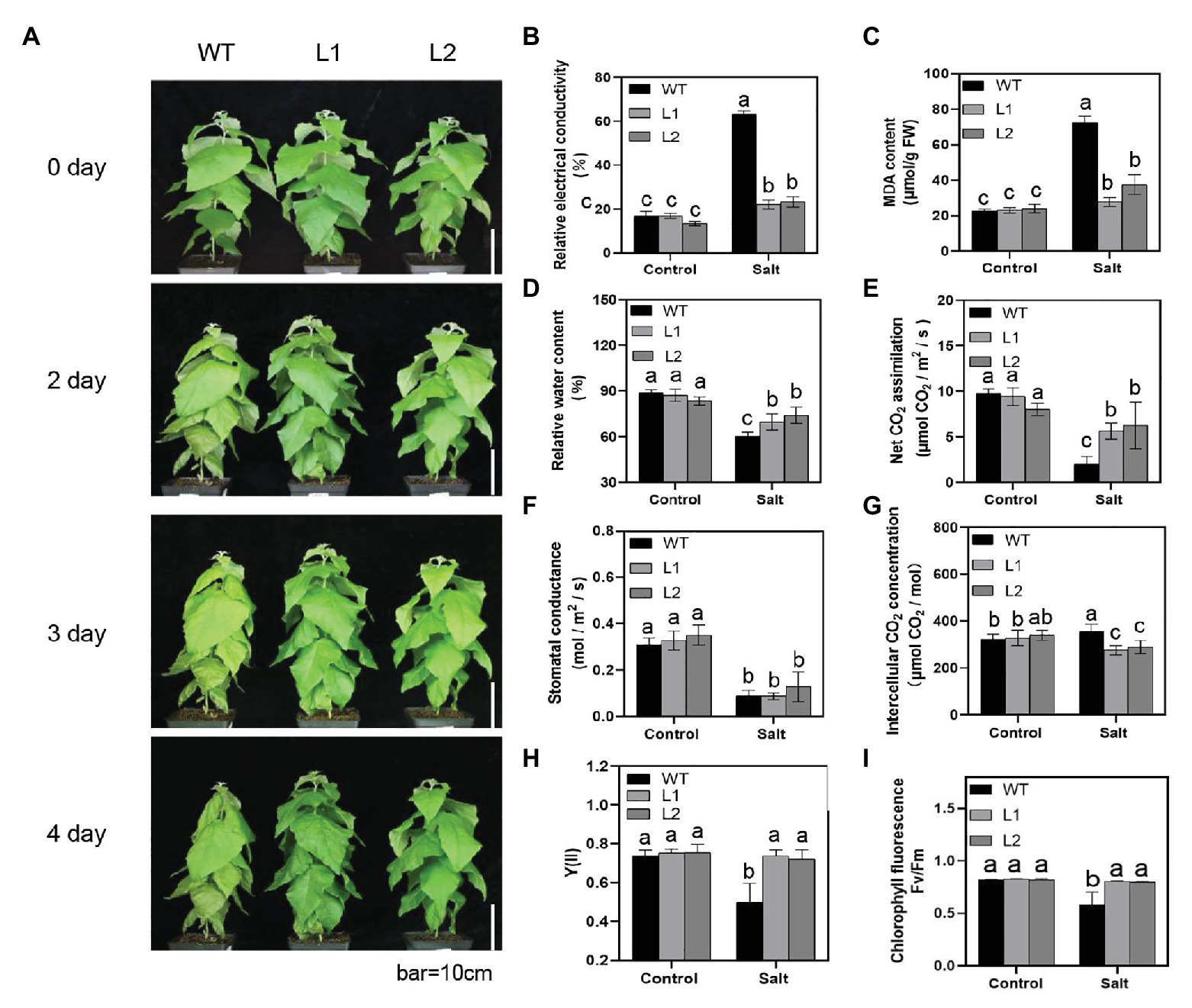
Figure 3. PagNBR1 overexpression enhances salt tolerance in transgenic poplars. (A) Effects of salt stress on the growth of wild-type (WT) plants and overexpressed transgenic plants (L1 and L2). About 60-day-old plants were treated with 0 or 400 mM NaCl for 4 days and photographed before and after treatment. Bar = 10 cm; (B) Relative electrical conductivity. (C) MDA content. (D) Relative water content (RWC). (E) Net CO2 assimilation. (F) Stomatal conductance. (G) Intercellular CO2 concentration. (H) Quantum yield of photochemical energy conversion in PSII (Y [II]). (I) Fv/Fm. These data were measured after 4 days of treatment. Each treatment was performed with three biological replicates (n = 3) and values are means ± SE. Data were analyzed through LSD multiple range tests in the ANOVA program of SPSS (IBM SPSS17.0). Different letters indicate significant differences between treatments at a level of p < 0.05.
The potential and permeability of the cell membrane are sensitive to stress conditions. An increase in membrane lipid peroxidation and membrane permeability results in increased EL and MDA. Thus, EL and MDA are important indicators that reflect the integrity of the cell membrane under stress conditions. To evaluate the degree of membrane damage, we detected the EL and MDA levels in WT and transgenic plants under control and salt stress conditions (Figures 3B,C). Compared with the control group, salt stress significantly increased the MDA level in WT and transgenic poplars. After 4 days of NaCl treatment, the MDA content in transgenic plants increased from 23 to 23.7 μmol/g FW to 27.7–37.3 μmol/g FW. However, WT plants showed a more than three-fold increase from 22.5 μmol/g FW to 72.4 nmol/g FW (Figure 3C). Moreover, the EL increased between WT and transgenic plants after salt stress. The WT plants exhibited a three-fold increase from 16.8 to 63.3%, whereas transgenic poplar leaves presented a one-fold increase from 13.4–16.9% to 22.1–23% under salt stress (Figure 3B). The relative water content (RWC) also showed lower values in WT than transgenic plants after treatment (Figure 3D). The results of FW/DW (Supplementary Figure S6) supported the RWC results. In summary, these results indicated that overexpression of PagNBR1 enhanced tolerance of salt stress in transgenic lines.
Photosynthetic Gas Exchange and Chlorophyll Fluorescence Parameters
To analyze the effect of PagNBR1 on the characteristics of photosynthetic physiology during salt stress conditions, we determined the photosynthetic parameters using the Lico-6,400 Portable Photosynthesis System. Under normal conditions, the net photosynthetic rates were not observably different between WT and transgenic poplars (Figure 3E). However, a significant difference was found between WT and transgenic poplars after salt stress (Figure 3E). After salt stress, the photosynthetic capacity of WT was about 10% of that in the control group. However, the transgenic poplars declined to nearly 50% of that in the control group (Figure 3E). Similarly, the inter-cellular CO2 concentration showed a dramatic decline in transgenic plants under stress conditions but not in WT (Figure 3G). The variation trend of stomatal conductance showed no notable difference between WT and transgenic plants under stress conditions (Figure 3F).
Chlorophyll fluorescence is an indicator of the photosynthetic efficiency of plants (An et al., 2014). Y(II) is an indicator of the conversion efficiency of PSII, and the FV/Fm presents the maximum optical quantum yield of PSII which reflects the potential maximum light energy conversion efficiency in plants (Alpuerto et al., 2016). Therefore, Y(II) and Fv/Fm were measured using PAM100. Salt stress was found to cause a decrease in Y(II) from 0.75 to 0.49 in WT plants. However, no significant decrease was observed in transgenic poplars (Figure 3H). Salt stress decreased Fv/Fm levels in WT. Compared with transgenic poplars, WT had a lower value of Fv/Fm under salt stress (Figure 3I). In general, these results showed that transgenic poplars had better tolerance to salt stress than WT.
PagNBR1 Regulates the Distribution of Na+ and K+ in Different Tissues
To explain the underlying mechanism of salt tolerance caused by PagNBR1 through the regulation of Na+ and K+ homeostasis, the Na+ and K+ distribution in different tissues (such as roots, stems, and leaves) were measured. Under NaCl stress, the Na+ concentrations dramatically increased in WT and transgenic poplars (Figures 4A–C). Compared with transgenic lines, the Na+ concentration in leaves of WT increased observably higher about ten-fold after 4 days of treatment (Figure 4A). The Na+ concentration in roots of WT also increased and was observably higher than transgenic lines, but there was no significant difference in stems among WT and transgenic lines after 4 days of treatment (Figures 4B,C). Under salt stress, the dynamic balance of K+/Na+ contributed to maintaining ion balance and promoting plant development. Figures 4D–F show that all plants maintained a high K+/Na+ ratio in leaves, stems, and roots under normal conditions. The K+/Na+ ratio between WT and transgenic lines was not remarkably different in leaves in the control group (Figure 4D). Under salt stress, the K+/Na+ ratio in transgenic poplars was significantly higher than that in WT (Figure 4D). In stems, overexpressed PagNBR1 did not significantly change the ratio of K+/Na + under normal and stress conditions (Figure 4E). In roots, the K+/Na + ratio was higher in transgenic poplars than in WT under stress conditions (Figure 4F). To further illuminate the role of PagNBR1 under salt stress, we analyzed SOS1 expression (Supplementary Figure S3). The SOS1 expression levels in the treatment group were upregulated, but the transgenic poplars exhibited 1.8-fold higher expression than the WT at the transcription level (Supplementary Figure S3). Thus, overexpression of PagNBR1 enhanced tolerance of salt tolerance by regulating redistribution of Na+ and K+ under salt stress.
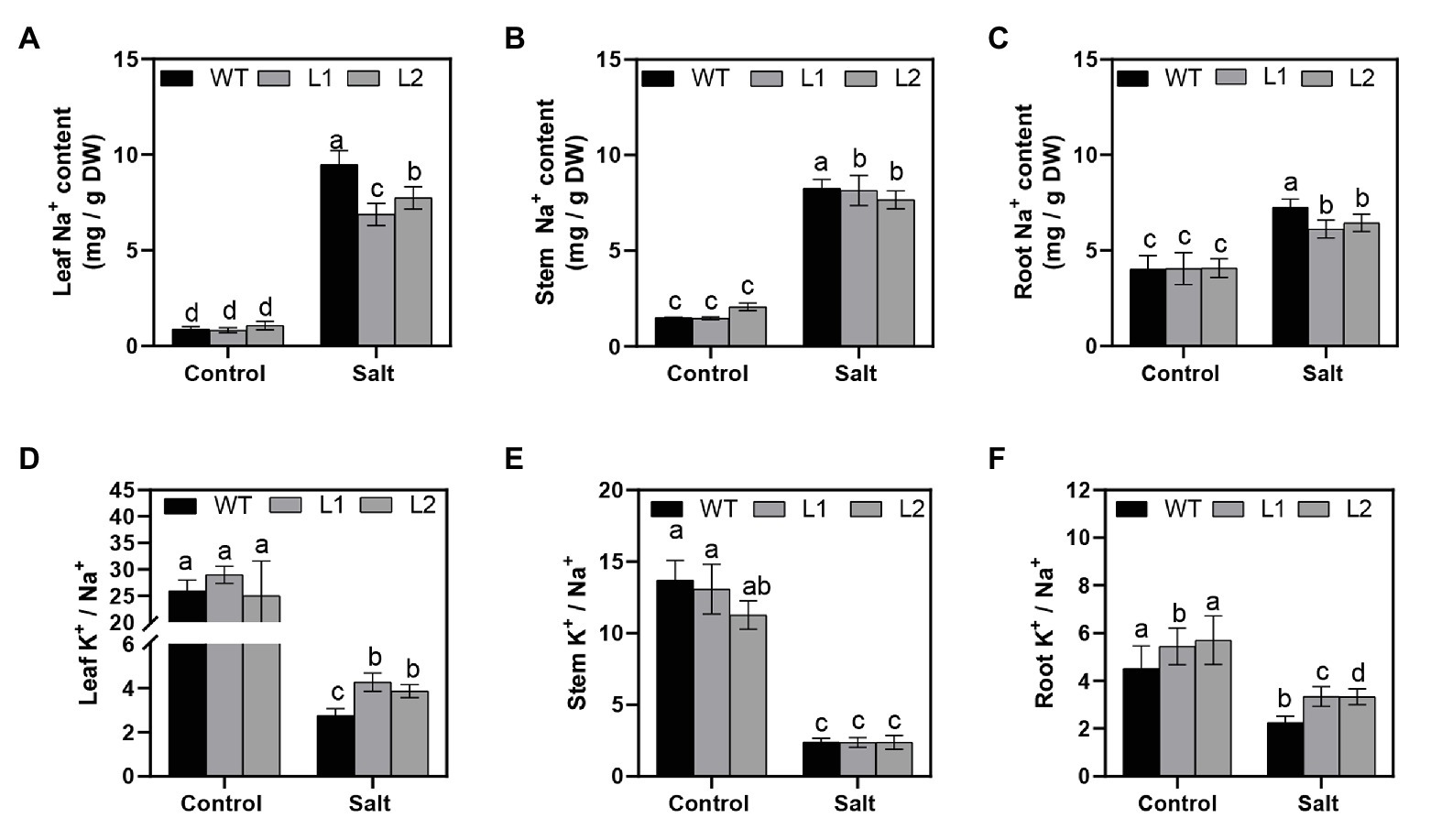
Figure 4. Na+ and K+ contents in different tissues of wild-type (WT) and transgenic plants (L1 and L2). After NaCl treatment, plant materials were collected and divided into leaves, stems, and roots for the estimation of Na+ and K+ contents. (A) Na+ contents of leaves, (B) Na+ contents of stems, (C) Na+ contents of roots, (D) K+/Na+ of leaves, (E) K+/Na+ of stems, and (F) K+/Na+ of roots. These data were measured after 4 days of treatment. Values are means (±SE) of three independent experiments (n = 3). Data were analyzed through LSD multiple range tests in the ANOVA program of SPSS (IBM SPSS17.0). Different letters indicate significant differences between treatments at a level of p < 0.05.
PagNBR1 Enhances Antioxidant System Activity Under Salt Stress
Reactive oxygen species (ROS) can be induced under stress conditions and play a major role in plants under abiotic stress (Choudhury et al., 2013). In the above analysis, we realized that salt stress resulted in more serious cellular membrane damage in WT than in transgenic lines (Figures 3B,C), which suggested that WT plants accumulated more ROS than transgenic poplars under salt stress. Therefore, 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) was used to detect H2O2 and O2− in situ, respectively (Figures 5A,B). Figure 4A shows that WT greatly enriched more H2O2 and O2− than transgenic plants during salt stress. We also measured the H2O2 content by monitoring ferrous ion oxidation using xylenol orange. The results were consistent with those in situ in WT and transgenic poplars (Figure 5B). SOD, POD, and CAT play main roles in scavenging ROS and contribute to maintaining ROS homeostasis in plants (Xing et al., 2018; He et al., 2019). After 4 days of salt stress, the enzyme activities of SOD, POD, and CAT in transgenic lines were dramatically higher than those in WT. However, the enzyme activities between WT and transgenic lines were similar under normal conditions (Figures 5C–E).
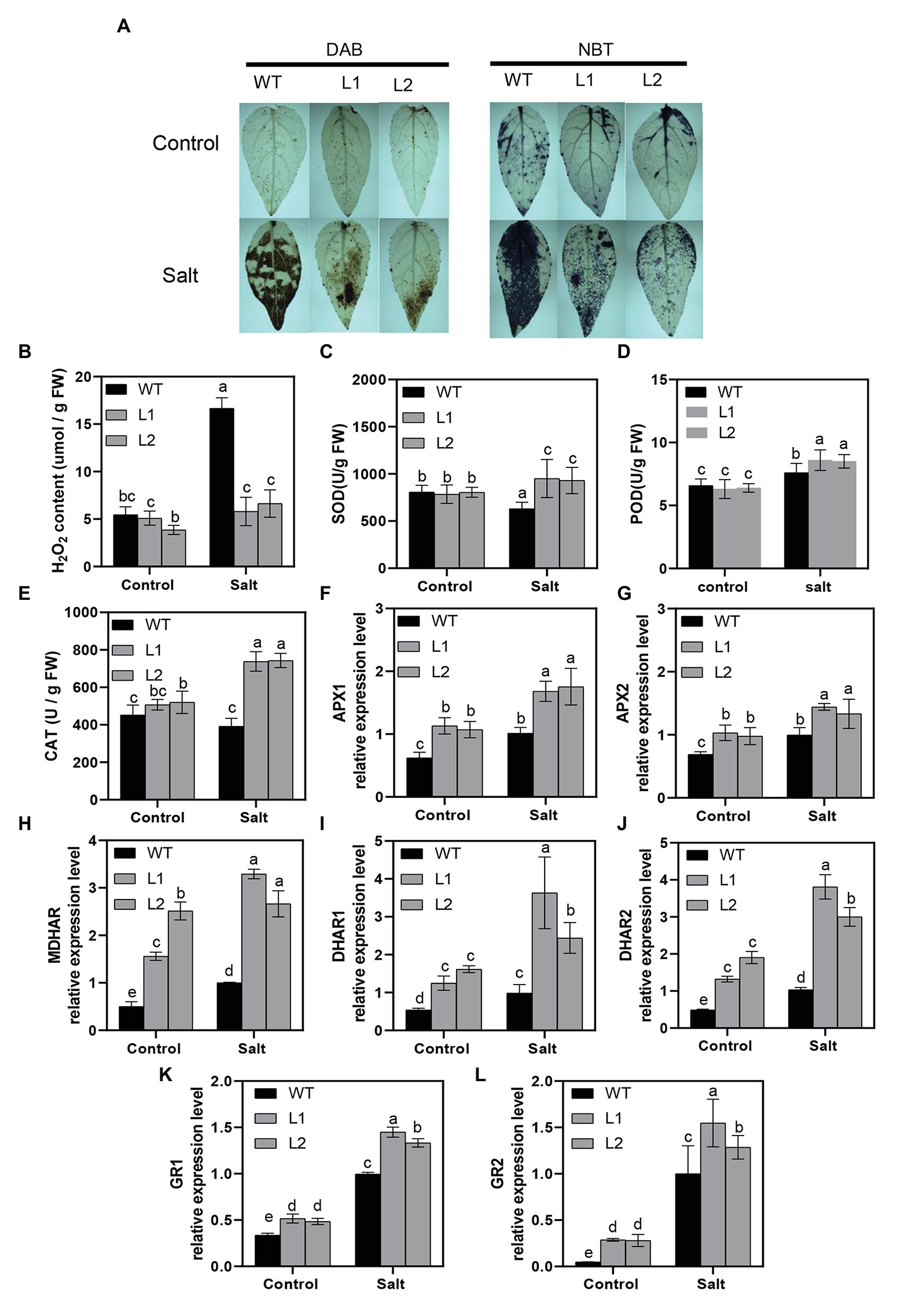
Figure 5. Activity of H2O2, O2−, and antioxidant system in WT and transgenic plants. (A) Histochemical staining with DAB and NBT for the detection of H2O2 and O2−, respectively, in WT transgenic plants before and after treatment. (B) Accumulation of H2O2 in WT and transgenic plants, (C) SOD activity, (D) POD activity, (E) CAT activity, (F,G) relative expression of APXs, (H) relative expression of MDHAR, (I,J) relative expression of DHARs, and (K,L) relative expression of GRs in WT and transgenic plants (L1, L2) under control and treatment conditions. For H2O2 and O2−, leaves were excised from lines after 4 days of treatment and immediately immersed in DAB (H2O2) and NBT(O2−). Data were measured after 4 days of treatment. Values are means (±SE) of three independent experiments (n = 3). Data were analyzed through LSD multiple range tests in the ANOVA program of SPSS (IBM SPSS17.0). Different letters indicate significant differences between treatments at a level of p < 0.05.
Ascorbic acid (AsA) and glutathione (GSH) cycling are important factors in scavenging H2O2 and O2− for plants to main ROS homeostasis under stress conditions (Wang et al., 2012; Sun et al., 2018a,b). To further reveal the function of AsA-GSH cycling in scavenging ROS, we detected the crucial genes APX, MDHAR, DHAR, and GR at the transcription levels in WT and transgenic lines. As shown in Figures 5F–L, the transcription level of these genes increased under salt stress conditions in WT and transgenic poplars; however, transgenic poplars demonstrated significantly higher transcription levels compared with WT. Similar results were obtained in the control group (Figures 5F–L). We also analyzed the content of AsA + DHA and GSH + GSSG and results showed that transgenic poplars kept higher levels at metabolic levels than that of WT poplars (Supplementary Figure S6). In general, PagNBR1 could enhance tolerance of salt stress via activating the antioxidant system.
PagNBR1 Can Interact With PagATG8s
Previous studies have revealed that the AtNBR1 protein can interact with the AtATG8 protein to facilitate the degradation of insoluble ubiquitinated protein aggregates through selective autophagy (Zhou et al., 2013). In the present study, to identify a similar interaction between the proteins of PagNBR1 and PagATG8, we performed a two-hybrid experiment in yeast. PagATG8 (ATG8a ATG8e ATG8h ATG8g) was fused with pGADT7, and PagNBR1 was fused with pGBKT7. The recombination vector pGBK-NBR1 and pGAD-ATG8 were co-expressed in yeast AH109 cells. A negative control was also prepared (pGBK-NBR1/pGADT7; pGBKT7/pGAD-ATG8s). Figure 6 shows that the yeast containing pGBK-NBR1 and pGAD-ATG8 grew normally on SD/−leu-Trp and SD/-Ade-His-Leu-Trp medium. In the negative control, normally grown yeast was observed on SD/-Leu-Trp, and no yeast was observed on SD/-Ade-His-Leu-Trp medium. Notably, our results showed that PagNBR1 could interact with PagATG8, which suggested that PagNBR1 is a receptor and may have functional characteristics similar to AtNBR1.
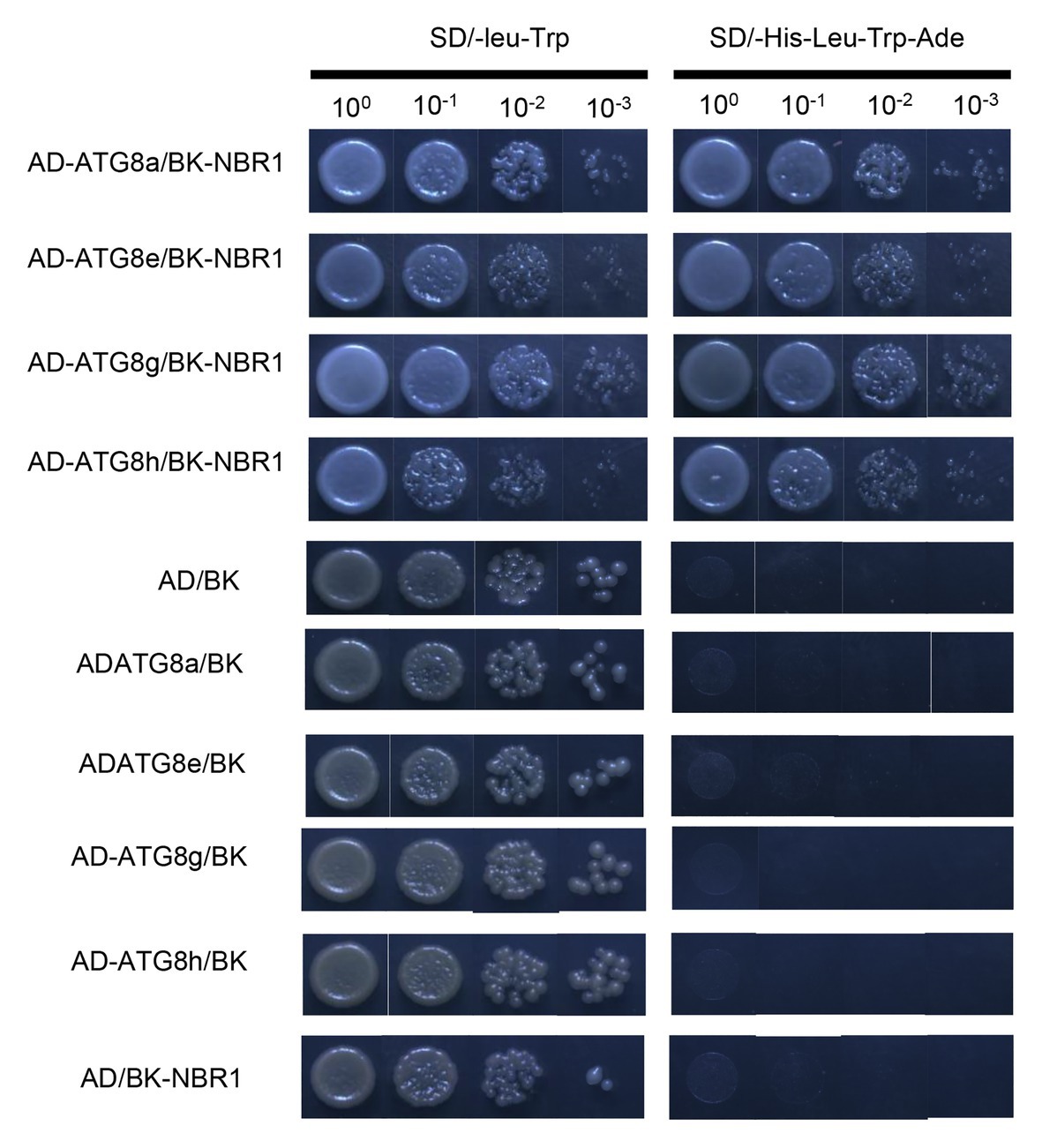
Figure 6. Interaction analysis of PagNBR1 and ATG8s using a yeast two-hybrid system. pGBK-NBR1 co-transformation with pGAD-ATG8a, pGAD-ATG8e, pGAD-ATG8h, pGAD-ATG8g, and pGAD-T7. Negative control of the interaction of PagNBR1 and ATG8s. Co-transformation of pGAD-T7/pGBK-T7, pGBK-T7/pGAD-ATG8a/8e/8g/8h, and pGADT7/PGBK-NBR1. All plasmids were transformed into yeast AH109 competent cells and cultured on SD/-leu-Trp and SD/-Ade-His-Leu-Trp medium. Data are from three independent experiments (three yeast lines measured in each independent experiment). One representative image of them is shown.
PagNBR1 Promotes the Autophagic Clearance of Ubiquitinated Protein Aggregates
NBR1 is considered a kind of selective autophagy receptor to clear ubiquitinated protein aggregates and ub-tagged organelle via interacting with ATG8 in plants and mammals (Svenning et al., 2011; Jung et al., 2019). Our results indicated that the proteins of PagNBR1 and PagATG8 could interact with each other, which is consistent with the interaction between the proteins of AtNBR1 and AtATG8 in Arabidopsis. These results suggested that the function is conserved between AtNBR1 and PagNBR1 (Figure 6). Thus, we hypothesized that the overexpression of PagNBR1 may promote the degradation of the insoluble ubiquitinated protein. Therefore, the content of insoluble protein and ubiquitinated protein in insoluble protein was measured in WT and transgenic plant leaves under normal and salt stress conditions (Figures 7A–C). In Figure 7, the insoluble protein was observably enriched in WT compared with that in transgenic poplars under salt stress. However, no significant difference was found between WT and transgenic poplars under normal conditions (Figure 7A). The content of soluble protein also supported the above-mentioned results (Figure 7C). Meanwhile, the content of insoluble ubiquitinated protein was higher in WT than in transgenic plants. Furthermore, the content of insoluble protein and insoluble ubiquitinated protein was observably higher in the NaCl+ConA group than the NaCl group, the inhibition of autophagy resulted in the accumulation of insoluble protein and insoluble ubiquitinated proteins (Supplementary Figure S7). These results revealed that the overexpression of PagNBR1 enhanced salt tolerance by promoting the degradation of insoluble ubiquitinated protein in transgenic poplars under salt stress.
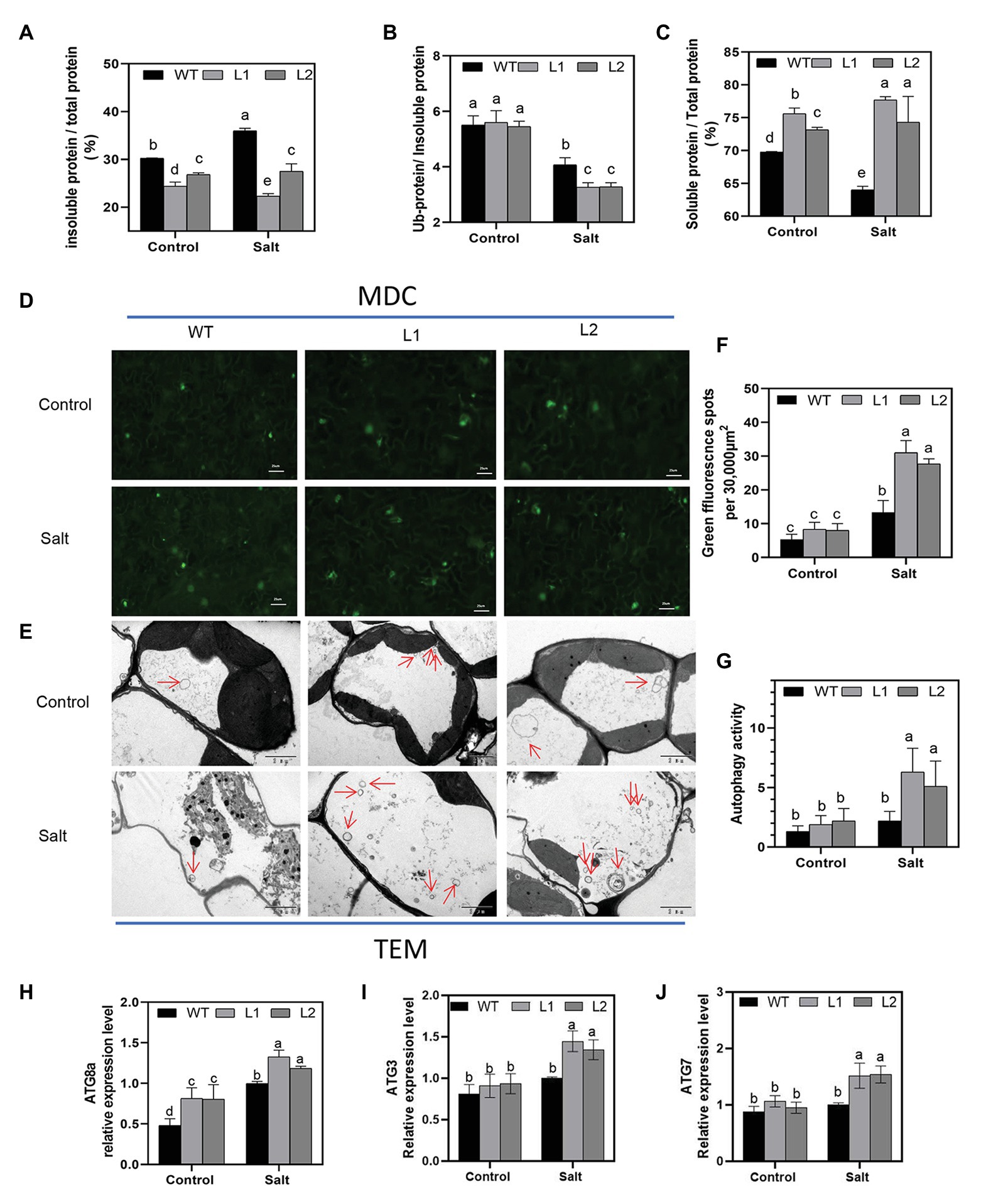
Figure 7. Accumulation of insoluble protein, insoluble ubiquitinated protein, the autophagosome, and ATG expression levels in WT and transgenic plants under control and treatment conditions. (A) Proportion of insoluble protein in total protein, (B) proportion of ubiquitinated protein in insoluble protein and soluble protein in total protein, (C) proportion of soluble protein in total protein, (D) MDC-stained autophagosomes are shown in green fluorescence signal (bar = 25um), (E) representative TEM images of autophagosome in WT and transgenic plant leaves, (F) number of green fluorescence signal per 30,000um2 area in (D), (G) quantified autophagosomes in (E) and (H–J) relative expression of ATG8a, AT3, and ATG7. More than 10 cells were used to quantify autophagic structures in mesophyll cells in the TEM results. These data were measured after 4 days of treatment. Values are means (±SE) of three independent experiments (n = 3). Data were analyzed through LSD multiple range tests in the ANOVA program of SPSS (IBM SPSS17.0). Different letters indicate significant differences between treatments at a level of p < 0.05.
Transcriptional levels of ATG genes have generally been used as an acceptable indicator to analyze the induction of autophagy under stress conditions in plants (Zhou et al., 2014; Chen et al., 2016; Sun et al., 2018a,b). To clarify whether autophagy is induced under salt stress, the transcriptional level of ATG3, ATG7, and ATG8 (a-g) genes were used to evaluate whether autophagy was induced under stress conditions (Figures 7H–J; Supplementary Figure S8) and the results showed that ATG genes (ATG8a-g, ATG3, and ATG7) increased to different degrees in WT and transgenic plants after salt stress compared with the control group. ATG gene expression levels in transgenic plants were observably higher than those in WT under salt stress. MDC has always been used as a marker of autophagosome in previous studies (Wang et al., 2015a). Our study showed that salt stress induced autophagy and that transgenic lines obtained higher autophagy activity under salt stress (Figures 7D,F). TEM is a valid and acceptable method used in plants for autophagosome detection (Sun et al., 2018a,b; Chi et al., 2020). To further investigate the induction of autophagy under salt stress conditions, TEM observation was performed, and the leaves of WT and transgenic plants under the control and treatment groups were used. Figures 7E,G show that the transgenic line accumulated more autophagosomes in the vacuole compared with WT lines after 4 days of treatment. Overall, these results demonstrated that overexpression of the PagNBR1 gene enhanced tolerance of salt stress but induced the accumulation of autophagosomes in transgenic lines.
Discussion
NBR1 Is Evolutionarily Conserved in Eukaryotes
Selective autophagy is considered a specific process regulated by autophagy cargo receptors through interaction with ATG8 proteins (Jin et al., 2013). In mammals, the proteins of p62/SQSTM1 and NBR1-mediated selective autophagy targets ubiquitinated proteins and aggregate-prone proteins to degrade (Johansen and Lamark, 2011). The specific protein was also found because of the similar domain architectures and function with the metazoarl NBR1 and p62/SQSTM1 proteins (Svenning et al., 2011). The PagNBR1 gene was cloned from Poplar 84 K. Analysis of the amino acid sequence revealed that the PagNBR1 protein was highly homologous with AtNBR1 protein and harbored the characteristic FW domain in NBR1 homologous proteins (Figure 1A and Supplementary Figure S1). The PB1-ZZ-UBA domain organizations harbored in mammal NBR1,p62/SQSTM1 and AtNBR1 were also found in the PagNBR1 protein. Meanwhile, the sub-cellular localization pattern of the PagNBR1 protein in tobacco protoplast (Figures 1B,C) and yeast two-hybrid analysis (Figure 6) revealed that the PagNBR1 protein was localized in autophagosomes and could interact with ATG8 proteins. These findings were consistent with AtNBR1 and suggest that PagNBR1 could be a candidate receptor of selective autophagy.
PagNBR1 Is a Crucial Candidate Gene for Breeding Salt-Tolerant Poplars
Autophagy contributes to the enhancement of tolerance of drought, salt, heat, and oxidative stress through degrading damaged proteins and organelles (Liu et al., 2009; Zhai et al., 2016). Previous studies have focused on the functional analysis of ATG genes, which are conserved genes for autophagosome formation in plants (Batoko et al., 2017). Our understanding of selective autophagy in response to abiotic stress remains limited. The AtNBR1 protein is the first selective autophagy receptor isolated from plants (Avin-Wittenberg, 2019). The nbr1 mutant exhibits hypersensitivity to heat stress (Zhou et al., 2013), but its potential function in breeding stress-tolerant plants is unclear. Thus, we speculated that PagNBR1 may be applied as a target gene for breeding stress-tolerant poplars. As the PagNBR1 is a candidate for the selective autophagy receptor, the increased expression levels of PagNBR1 with leafage adding among young leaves, mature leaves and old leaves imply that it could play important roles in degrading and recycling damaged proteins and organelle during plant senescence. In addition, the expression levels of the PagNBR1 gene were induced by stress-response plant hormones such as ABA, ET, SA, and Me-JA (Figures 2E–I), suggesting that the PagNBR1 gene might be involved in stress tolerance. To further elucidate the potential breeding value in resisting salt stress and oxidative stress, we analyzed the expression pattern of the PagNBR1 gene after treatment with NaCl, MV, or H2O2, and the transgenic poplar to estimate tolerance to salt stress. Our results showed that salt stress or oxidative stress increased the transcriptional level of the PagNBR1 gene (Figures 2B–D). Under salt stress, PagNBR1 overexpression in poplars exhibited enhanced capacity of salt stress than WT, and reduced damage to the cell membrane (Figures 3A–C), and maintained higher RWC (Figure 3D) and photosystem activity (Figures 3E–I). In addition, the FW/DW value of transgenic lines was higher than that of WT plants under salt stress but it was similar under normal conditions (Supplementary Figure S6). Therefore, PagNBR1 overexpression conferred the tolerance of salt stress in transgenic poplars. In brief, PagNBR1 is a crucial candidate gene for breeding salt-tolerant plants.
PagNBR1 Enhances Salt Stress Tolerance by Activating the Antioxidant System
Stress conditions always trigger the excessive accumulation of ROS, which leads to cell dysfunction and cell damage and death in plants (Raja et al., 2017; Yang and Guo, 2018). Thus, it is important for plants to maintain ROS homeostasis during stress conditions. In an apple, the overexpression of the ATG18a gene increases the activity of the antioxidant system, enhancing its tolerance to drought and alkaline stress (Sun et al., 2018a; Li et al., 2020). In Arabidopsis, heat stress triggered the accumulation of highly aggregate-prone proteins in nbr1 mutant lines such as catalases, suggesting the important role of the NBR1 protein in regulating the antioxidant system (Zhou et al., 2014). Our results showed that PagNBR1 overexpression in poplars reduced ROS levels by increasing enzyme activity including SOD, POD, CAT, transcriptional levels of genes involving ASA-GSH cycling (Figures 5C–L) and the content of AsA + DHA and GSH + GSSG (Supplementary Figure S6) contributed to relieving the damage of plant membranes, as evidenced by the results of MDA, electrolyte leakage, and higher photosynthetic capacity. These findings suggest that the PagNBR1 protein enhanced salt stress tolerance by activating the oxygen scavenging system.
PagNBR1 Enhances Salt Stress Tolerance Through the Maintenance of K+/Na+ Homeostasis
K+ is an important nutrient factor, and maintenance of a high K+/Na+ ratio is favorable for plant growth and development (Barragan et al., 2012; Almeida et al., 2017). However, under salt stress, Na+ accumulates in the cytoplasm, resulting in a decrease in K+/Na+. This phenomenon perturbs enzymatic functions and leads to plant cell damage (Almeida et al., 2017; Yang and Guo, 2018). Therefore, the maintenance of relatively high levels of the K+/Na+ ratio is important for enabling plants to resist salt stress. In our studies, the selective autophagy receptor PagNBR1 was verified to confer salt tolerance in transgenic lines. The transgenic poplars accumulated lower content of Na+ and maintained higher K+/Na+ than WT in leaves and roots under salt stress (Figures 4A–F). These results indicated the involvement of the PagNBR1 gene in the redistribution of Na+ and K+. SOS1, which is a Na+/H+ antiporter, is responsible for extruding excessive Na+ from the cytoplasm to the extracellular environment (Yang et al., 2015). Research has shown that low levels of ROS increased the expression levels of SOS1 and contributed to the maintenance of K+/Na+ homeostasis under salt stress (Wu et al., 2007; Chung et al., 2008). In our results, the lower levels of ROS in the transgenic line could act as a signal molecular, increasing the transcription level of SOS1 (Supplementary Figure S3) and contributing to maintaining a higher K+/Na+ ratio under salt stress (Figure 8).
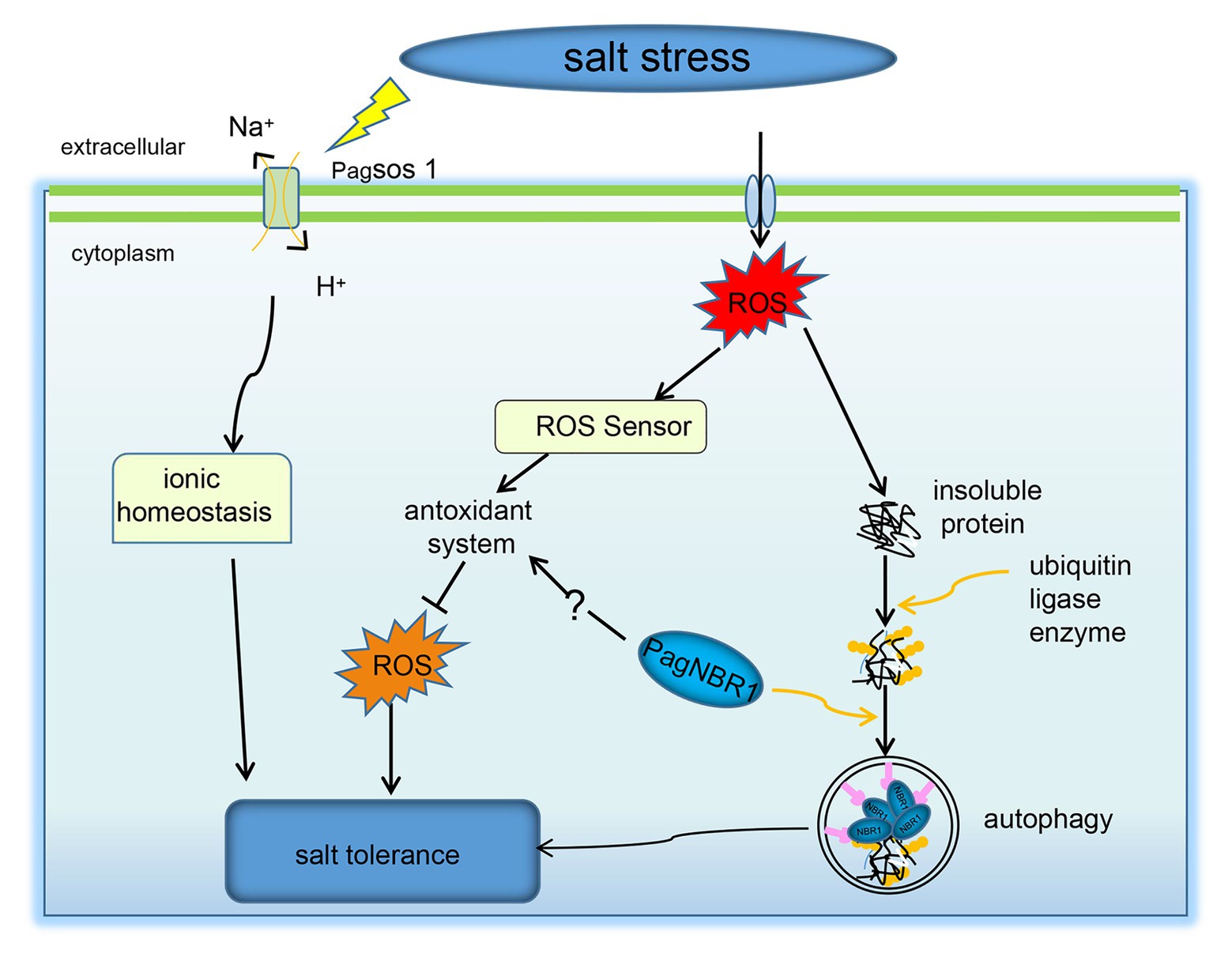
Figure 8. A possible model for NBR1-mediated stress resistance pathways. Under salt stress, the upregulated PagNBR1 involved in the selective autophagy pathway facilitates the degradation of insoluble ubiquitinated protein, which contributes to the quality control of proteins. The upregulated PagNBR1 may also mediate the activity of the antioxidant system, which maintains the homeostasis of ROS and K+/Na+ through the non-ubiquitinated pathway.
PagNBR1 Enhances Salt Stress Tolerance by Activating Autophagy
Excess ROS leads to proteins and organelles damage in plants, which is toxic to the survival of plants. Selective autophagy is considered to contribute to the degradation of insoluble ubiquitinated protein aggregates and organelles for recycling during imposed stress conditions (Johansen and Lamark, 2011; Avin-Wittenberg, 2019). In Arabidopsis, the sensitive phenotype characteristics of nbr1 mutant in response to heat stress are related to the excessive accumulation of insoluble ubiquitinated protein aggregates (Zhou et al., 2013). Thus, we hypothesized that the increased tolerance of salt stress in transgenic poplars is associated with the degradation of insoluble ubiquitinated protein aggregates by inducing an autophagy system. In line with this hypothesis, the transgenic poplars accumulated less insoluble proteins and insoluble ubiquitinated proteins in contrast to WT lines under salt stress (Figures 7B,C). According to the results reported by Chen et al. (2016), the transcriptional levels of ATG genes and autophagic flux were used to investigate the involvement of autophagy in the submergence response and their results showed the upregulation of ATG genes associated with the increased autophagic flux, suggesting that the transcriptional levels of ATGs are an important indicator in investigating the induction of autophagy under stress conditions. The expression level of ATGs has been used as an important indicator to analyze the induction of autophagy under stress conditions in plants (Chen et al., 2016; Sun et al., 2018a,b). The results obtained from MDC-staining showed that autophagosomes were induced under salt stress and it accumulated more in transgenic lines than in WT, suggesting that autophagy was enhanced in the transgenic line compared with WT during salt stress (Figures 7D,F). In woody plants, such as apple, autophagy was well studied in terms of enhancing tolerance of stress conditions and the expression of ATG genes (eg., ATG3, ATG5, ATG7, ATG8 et al.) have been widely used as an indicator for the induction of autophagy. It has been used together with the TEM to evaluate the autophagy activity in WT and transgenic lines (Sun et al., 2018a,b; Huo et al., 2020; Li et al., 2020). Thus, we detected the transcription level of ATG3, ATG7, and ATG8 (a-g) genes using the qRT-PCR method, and results (Figures 7H–G, Supplementary Figure S8) revealed that overexpression of the PagNBR1 gene promoted the upregulation of ATG genes, suggesting that autophagy was induced under salt stress. Furthermore, our results revealed that transgenic poplars formed more vesicles in the vacuole in transgenic lines than that of WT lines (Figures 7E,G) which is similar to the previous results reported by Sun et al. (2018a,b), Huo et al. (2020), and Li et al. (2020) suggesting that overexpression of PagNBR1 increased autophagy activity in transgenic lines under salt stress. Furthermore, inhibiting autophagy with ConA in WT and transgenic lines resulted in the similar accumulation of insoluble proteins and insoluble ubiquitinated proteins (Supplementary Figure S8) suggesting that the activated autophagy promoted the degradation of insoluble ubiquitinated proteins. In summary, the overexpression of PagNBR1 confers the tolerance of salt stress in transgenic poplars through enhancing autophagy activity (Figure 8).
Conclusion
Our results showed that PagNBR1 is a selective autophagy receptor that is homologous with AtNBR1. External environment factors such as salt stress, oxidative stress, and plant hormones (ABA, ET, SA, GA, me-JA) induced an increase in PagNBR1. Overexpression of PagNBR1 in poplars enhanced the tolerance of salt stress via promoting degradation of insoluble ubiquinitated protein, which was associated with enhanced autophagy activity. In addition, the overexpression of PagNBR1 enhanced the scavenging of ROS, which may be associated with maintaining higher K+/Na+ by increasing the expression levels of SOS1.
Data Availability Statement
All datasets generated for this study are included in the article.
Author Contributions
WS and XX conceived and designed the experiments. WS, YL, YB, SW, DW, and XY performed the experiments and analyzed the experimental data. WS wrote the article. WS, CL, WY, and XX revised the article. All authors contributed to the article and approved the submitted version.
Funding
This study was supported financially by the National Science and Technology Major Project for Water Pollution Control and Treatment (No. 2017ZX07101002) and National Natural Science Foundation of China (32071734 and 31770649).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.568411/full#supplementary-material
Abbreviations
NBR1, neighbor of BRCA1 gene 1; ATG8, Autophagy-related gene 8; SOD, superoxide dismutase; CAT, Catalase; POD, Peroxidase and AsA, ascorbate; GSH, glutathione cycle; UBA, ubiquitin-associated; ROS, Reactive oxygen species; MDA, Malonaldehyde; EL, Electrolyte leakage.
Footnotes
References
Almeida, D. M., Oliveira, M. M., and Saibo, N. J. M. (2017). Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 40, 326–345. doi: 10.1590/1678-4685-gmb-2016-0106
Alpuerto, J. B., Hussain, R. M., and Fukao, T. (2016). The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 39, 672–684. doi: 10.1111/pce.12661
An, Y., Han, X., Tang, S., Xia, X., and Yin, W. (2014). Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. 119, 313–327. doi: 10.1007/s11240-014-0536-y
Avin-Wittenberg, T. (2019). Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 42, 1045–1053. doi: 10.1111/pce.13404
Avin-Wittenberg, T., Honig, A., and Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Barragan, V., Leidi, E. O., Andres, Z., Rubio, L., De Luca, A., Fernández, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142. doi: 10.1105/tpc.111.095273
Batoko, H., Dagdas, Y., Baluska, F., and Sirko, A. (2017). Understanding and exploiting autophagy signaling in plants. Essays Biochem. 61, 675–685. doi: 10.1042/EBC20170034
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cabot, C., Sibole, J. V., Barcelo, J., and Poschenrieder, C. (2014). Lessons from crop plants struggling with salinity. Plant Sci. 226, 2–13. doi: 10.1016/j.plantsci.2014.04.013
Carroll, B., Otten, E. G., Manni, D., Stefanatos, R., Menzies, F. M., Smith, G. R., et al. (2018). Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat. Commun. 9:256. doi: 10.1038/s41467-017-02746-z
Cebollero, E., and Reggiori, F. (2009). Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413–1421. doi: 10.1016/j.bbamcr.2009.01.008
Chen, L., Liao, B., Qi, H., Xie, L., Huang, L., Tan, W., et al. (2016). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Chi, C., Li, X., Fang, P., Xia, X., Shi, K., Zhou, Y., et al. (2020). Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J. Exp. Bot. 71, 1092–1106. doi: 10.1093/jxb/erz466
Choudhury, S., Panda, P., Sahoo, L., and Panda, S. K. (2013). Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 8:e23681. doi: 10.4161/psb.23681
Chung, J. S., Zhu, J. K., Bressan, R. A., Hasegawa, P. M., and Shi, H. (2008). Reactive oxygen species mediate Na+−induced SOS1 mRNA stability in Arabidopsis. Plant J. 53, 554–565. doi: 10.1111/j.1365-313X.2007.03364.x
Chupeau, M., Granier, F., Pichon, O., Renou, J., Gaudin, V., and Chupeau, Y. (2013). Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25, 2444–2463. doi: 10.1105/tpc.113.109538
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Deosaran, E., Larsen, K. B., Hua, R., Sargent, G., Wang, Y., Kim, S., et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952. doi: 10.1242/jcs.114819
Genisel, M., Erdal, S., and Kizilkaya, M. (2015). The mitigating effect of cysteine on growth inhibition in salt-stressed barley seeds is related to its own reducing capacity rather than its effects on antioxidant system. Plant Growth Regul. 75, 187–197. doi: 10.1007/s10725-014-9943-7
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
He, F., Li, H.-G., Wang, J.-J., Su, Y., Wang, H.-L., Feng, C.-H., et al. (2019). PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 17, 2169–2183. doi: 10.1111/pbi.13130
He, F., Wang, H. L., Li, H. G., Su, Y., Li, S., Yang, Y., et al. (2018). PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 16, 1514–1528. doi: 10.1111/pbi.12893
Huo, L., Sun, X., Guo, Z., Jia, X., Che, R., Sun, Y., et al. (2020). MdATG18a overexpression improves basal thermotolerance in transgenic apple by decreasing damage to chloroplasts. Hortic. Res. 7:21. doi: 10.1038/s41438-020-0243-2
Ichimura, Y., Kumanomidou, T., Sou, Y. S., Mizushima, T., Ezaki, J., Ueno, T., et al. (2008). Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857. doi: 10.1074/jbc.M802182200
Inoue, Y., Suzuki, T., Hattori, M., Yoshimoto, K., Ohsumi, Y., and Moriyasu, Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47, 1641–1652. doi: 10.1093/pcp/pcl031
Ismail, A., Takeda, S., and Nick, P. (2014). Life and death under salt stress: same players, different timing? J. Exp. Bot. 65, 2963–2979. doi: 10.1093/jxb/eru159
Jin, M., Liu, X., and Klionsky, D. J. (2013). SnapShot: selective autophagy. Cell 152, 368–368.e2. doi: 10.1016/j.cell.2013.01.004
Johansen, T., and Lamark, T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. doi: 10.4161/auto.7.3.14487
Jung, H., Lee, H. N., Marshall, R. S., Lomax, A. W., Yoon, M. J., Kim, J., et al. (2019). NBR1 mediates selective autophagy of defective proteins in Arabidopsis. J. Exp. Bot. 71, 73–89. doi: 10.1093/jxb/erz404
Kenific, C. M., and Debnath, J. (2016). NBR1-dependent selective autophagy is required for efficient cell-matrix adhesion site disassembly. Autophagy 12, 1958–1959. doi: 10.1080/15548627.2016.1212789
Kirkin, V., Mcewan, D. G., Novak, I., and Dikic, I. (2009). A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269. doi: 10.1016/j.molcel.2009.04.026
Lamark, T., Kirkin, V., Dikic, I., and Johansen, T. (2009). NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8, 1986–1990. doi: 10.4161/cc.8.13.8892
Lee, H., Kim, M. N., and Ryu, K. Y. (2017). Effect of p62/SQSTM1 polyubiquitination on its autophagic adaptor function and cellular survival under oxidative stress induced by arsenite. Biochem. Biophys. Res. Commun. 486, 839–844. doi: 10.1016/j.bbrc.2017.03.146
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, Y., Liu, C., Sun, X., Liu, B., Zhang, X., Liang, W., et al. (2020). Overexpression of MdATG18a enhances alkaline tolerance and GABA shunt in apple through increased autophagy under alkaline conditions. Tree Physiol. 40:tpaa075. doi: 10.1093/treephys/tpaa075
Li, D., Song, S., Xia, X., and Yin, W. (2012). Two CBL genes from Populus euphratica confer multiple stress tolerance in transgenic triploid white poplar. Plant Cell Tissue Organ Cult. 109, 477–489. doi: 10.1007/s11240-011-0112-7
Liu, Z., Chen, P., Gao, H., Gu, Y., Yang, J., Peng, H., et al. (2014). Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 26, 106–120. doi: 10.1016/j.ccr.2014.05.015
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Liu, W. J., Ye, L., Huang, W. F., Guo, L. J., Xu, Z. G., Wu, H. L., et al. (2016). P62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 21:29. doi: 10.1186/s11658-016-0031-z
Luo, L., Zhang, P., Zhu, R., Fu, J., Su, J., Zheng, J., et al. (2017). Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 8:1459. doi: 10.3389/fpls.2017.01459
Masclaux-Daubresse, C. (2016). Autophagy controls carbon, nitrogen, and redox homeostasis in plants. Autophagy 12, 896–897. doi: 10.4161/auto.36261
Mizushima, N. (2015). Nbr1, a receptor for ESCRT-dependent endosomal microautophagy in fission yeast. Mol. Cell 59, 887–889. doi: 10.1016/j.molcel.2015.09.004
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Patra, K. H., Kar, M., and Mishra, D. (1978). Catalase activity in leaves and cotyledons during plant development and senescence1. Biochem. Physiol. Pflanz. 172, 385–390. doi: 10.1016/S0015-3796(17)30412-2
Qiu, D., Bai, S., Ma, J., Zhang, L., Shao, F., Zhang, K., et al. (2019). The genome of Populus alba x Populus tremula var. Glandulosa clone 84K. DNA Res. 26, 423–431. doi: 10.1093/dnares/dsz020
Radojčić Redovniković, I., De Marco, A., Proietti, C., Hanousek, K., Sedak, M., Kang, T., et al. (2017). Poplar response to cadmium and lead soil contamination. Ecotox. Environ. Safe. 144, 482–489. doi: 10.1016/j.ecoenv.2017.06.011
Raja, V., Majeed, U., Kang, H., Andrabi, K. I., and John, R. (2017). Abiotic stress: interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 137, 142–157. doi: 10.1016/j.envexpbot.2017.02.010
Ren, C., Liu, J., and Gong, Q. (2014). Functions of autophagy in plant carbon and nitrogen metabolism. Front. Plant Sci. 5:301. doi: 10.3389/fpls.2014.00301
Shi, H., Ye, T., Chen, F., Cheng, Z., Wang, Y., Yang, P., et al. (2013). Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 64, 1367–1379. doi: 10.1093/jxb/ers400
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018b). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Sun, X., Wang, P., Jia, X., Huo, L., Che, R., and Ma, F. (2018a). Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557. doi: 10.1111/pbi.12794
Svenning, S., Lamark, T., Krause, K., and Johansen, T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. doi: 10.4161/auto.7.9.16389
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 1–10. doi: 10.1186/s12870-016-0771-y
von Muhlinen, N., Thurston, T., Ryzhakov, G., Bloor, S., and Randow, F. (2010). NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy 6, 288–289. doi: 10.4161/auto.6.2.11118
Wang, Y., Cai, S., Yin, L., Shi, K., Xia, X., Zhou, Y., et al. (2015a). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 11, 2033–2047. doi: 10.1080/15548627.2015.1098798
Wang, H.-L., Chen, J., Tian, Q., Wang, S., Xia, X., and Yin, W. (2014). Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol. Plant. 152, 529–545. doi: 10.1111/ppl.12206
Wang, W.-B., Kim, Y.-H., Lee, H.-S., Kim, K.-Y., Deng, X.-P., Kwak, S.-S., et al. (2009). Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 47, 570–577. doi: 10.1016/j.plaphy.2009.02.009
Wang, H.-L., Li, L., Tang, S., Yuan, C., Tian, Q., Su, Y., et al. (2015b). Evaluation of appropriate reference genes for reverse transcription-quantitative PCR studies in different tissues of a desert poplar via comparision of different algorithms. Int. J. Mol. Sci. 16, 20468–20491. doi: 10.3390/ijms160920468
Wang, P., Yin, L., Liang, D., Li, C., Ma, F., and Yue, Z. (2012). Delayed senescence of apple leaves by exogenous melatonin treatment: toward regulating the ascorbate-glutathione cycle. J. Pineal Res. 53, 11–20. doi: 10.1111/j.1600-079X.2011.00966.x
Wen, X., and Klionsky, D. J. (2016). An overview of macroautophagy in yeast. J. Mol. Biol. 428, 1681–1699. doi: 10.1016/j.jmb.2016.02.021
Wu, Y. X., Ding, N., Zhao, X., Zhao, M. G., Chang, Z. Q., Liu, J., et al. (2007). Molecular characterization of PeSOS1: the putative Na+/H+ antiporter of Populus euphratica. Plant Mol. Biol. 65:1. doi: 10.1007/s11103-007-9170-y
Xing, C., Liu, Y., Zhao, L., Zhang, S., and Huang, X. (2018). A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 42, 832–845. doi: 10.1111/pce.13387
Yang, Y., and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y., Tang, R.-J., Jiang, C.-M., Li, B., Kang, T., Liu, H., et al. (2015). Overexpression of the PtSOS2 gene improves tolerance to salt stress in transgenic poplar plants. Plant Biotechnol. J. 13, 962–973. doi: 10.1111/pbi.12335
Yannikos, N., Leinweber, P., Helgason, B. L., Baum, C., Walley, F. L., Van Rees, K., et al. (2014). Impact of Populus trees on the composition of organic matter and the soil microbial community in Orthic gray Luvisols in Saskatchewan (Canada). Soil Biol. Biochem. 70, 5–11. doi: 10.1016/j.soilbio.2013.11.025
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, S., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. doi: 10.1105/tpc.109.068635
Zhai, Y., Guo, M., Wang, H., Lu, J., Liu, J., Zhang, C., et al. (2016). Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum l. Front. Plant Sci. 7:131. doi: 10.3389/fpls.2016.00131
Zhang, M., Zhang, G., Kang, H., Zhou, S., and Wang, W. (2017). TaPUB1, a putative E3 ligase gene from wheat, enhances salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Physiol. 58, 1673–1688. doi: 10.1093/pcp/pcx101
Zhou, J., Wang, J., Cheng, Y., Chi, Y.-J., Fan, B., Yu, J.-Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9:e1003196. doi: 10.1371/journal.pgen.1003196
Zhou, J., Zhang, Y., Qi, J., Chi, Y., Fan, B., Yu, J.-Q., et al. (2014). E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10:e1004116. doi: 10.1371/journal.pgen.1004116
Zhou, X., Zhao, P., Wang, W., Zou, J., Cheng, T., Peng, X., et al. (2015). A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 22, 245–257. doi: 10.1093/dnares/dsv012
Zientara-Rytter, K., Lukomska, J., Moniuszko, G., Gwozdecki, R., Surowiecki, P., Lewandowska, M., et al. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7, 1145–1158. doi: 10.4161/auto.7.10.16617
Zientara-Rytter, K., and Sirko, A. (2014). Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front. Plant Sci. 5:13. doi: 10.3389/fpls.2014.00013
Keywords: salt stress, selective autophagy, poplars, NBR1, antioxidant system activity, ubiquitinated proteins
Citation: Su W, Bao Y, Lu Y, He F, Wang S, Wang D, Yu X, Yin W, Xia X and Liu C (2021) Poplar Autophagy Receptor NBR1 Enhances Salt Stress Tolerance by Regulating Selective Autophagy and Antioxidant System. Front. Plant Sci. 11:568411. doi: 10.3389/fpls.2020.568411
Edited by:
Andy Pereira, University of Arkansas, United StatesReviewed by:
Lingaraj Sahoo, Indian Institute of Technology Guwahati, IndiaZhuo Renying, Chinese Academy of Forestry, China
Qinglin Tang, Southwest University, China
Copyright © 2021 Su, Bao, Lu, He, Wang, Wang, Yu, Yin, Xia and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Liu, bGl1Y2hhbzEzMDZAYmpmdS5lZHUuY24=
 Wanlong Su1,2,3
Wanlong Su1,2,3 Yingying Lu
Yingying Lu Dongli Wang
Dongli Wang Weilun Yin
Weilun Yin Xinli Xia
Xinli Xia Chao Liu
Chao Liu