
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 September 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.556039
 Yu Maebara1†‡
Yu Maebara1†‡ Masanori Tamaoki2‡
Masanori Tamaoki2‡ Yuka Iguchi1†
Yuka Iguchi1† Naoyuki Nakahama3,4
Naoyuki Nakahama3,4 Takaaki Hanai5
Takaaki Hanai5 Atsushi Nishino1†
Atsushi Nishino1† Daisuke Hayasaka1*
Daisuke Hayasaka1*Among invasive species, aquatic plants pose serious threats to local biodiversity and ecosystem functions. Spartina alterniflora Loisel. (Poaceae), native to the eastern United States, was introduced unintentionally into Japan (Aichi and Kumamoto Prefectures) at around 2010. This invasive species could easily and rapidly spread to estuarine areas of Japan via vigorous trade and transport, making the prediction of its future invasion necessary. Here, the distribution and structure of the genetic variation of S. alterniflora in Japan were examined using chloroplast DNA (cpDNA) and microsatellite genotyping analyses for clarifying its invasion route and process. According to the cpDNA analysis, S. alterniflora populations in Japan had a single haplotype (haplotype C4) that is the most dominant genotype around the Florida Peninsula, the region of its origin, and is also widely found in the introduced populations in the East Asia. Microsatellite analysis also showed a loss of genetic diversity in Japanese S. alterniflora populations (allelic richness (AR) = 1.20–1.39) compared with that in its native region (AR = 4.58–4.59), suggesting a founder effect on S. alterniflora that might have occurred after invasion of the species into Japan. The principal coordinate analysis and The STRUCTURE analysis indicated that no gene mixing among Japanese local populations (Aichi, northern and southern Kumamoto) was observed, indicating that Spartina invasion occurred independently into these regions. Among the three regions, trading between the ports of northern Kumamoto and the U.S. was obviously lower than trading with China. We concluded that invasive S. alterniflora might have independently invaded Japan at different times through an East Asia route, particularly via China (i.e., secondary introduction). Therefore, it is important to strengthen the quarantine control on the importation of commodities, especially of transport vehicles at potential donor spots (i.e., border control/border biosecurity system), and to share information networks on invasive species between each region/port for minimizing further risks of biological species such as Spartina
Invasive species are extremely harmful to native ecosystems and thus are regarded as one of the major threats of biodiversity loss (Pyšek and Richardson, 2010; Vilà et al., 2011; Pyšek et al., 2012). International trade serves as one of the driving factors for the widespread invasion of the invasive species (Elton, 1958; Lockwood et al., 2007; Davis, 2009; Richardson, 2011). Many empirical and theoretical studies on biological invasions have been conducted on various taxonomic groups for resolving this worldwide concern (Lee, 2002). Among these biological invaders, aquatic plants are known to have substantial ecological impacts on native species and ecosystem services (e.g., Hayasaka et al., 2018), as well as subsequent huge economic losses. Therefore, ecological knowledge that may lead to urgent control and/or eradication of invasive aquatic plants are imperative to conserve a biological diversity (Koncki and Aronson, 2015).
Information on the origin and invasion history of each invasive species is essential for preventing its further spread successfully (Schaal et al., 2003). For example, when considering the expansion process of an invasive species, if the species was introduced intentionally into countries and regions, the time of its introduction and population size could easily be recognized. In contrast, it is very difficult to obtain such information on biological invaders when due to unintentional introductions. The invasion history of invasive species, especially plants, are estimated directly, for example, using published literature, aerial photographs, and herbarium collections in order to determine the date and place of its first record. On the other hand, molecular genetic data including population genetic structure and diversity can provide a great deal of information, such as the origin of the targeted species and the route of its propagation, as well as the process of the range expansion, which indirectly contributes to the elucidation of its invasion history (Lowe et al., 2004; Prentis et al., 2009; Hoos et al., 2010; Lombaert et al., 2010). Generally, it is assumed that invasive species have a low intra-population genetic diversity but have a high inter-population genetic differentiation in introduced ranges compared with those of the region of its origin, which is known as “the founder effect” (Brown and Marshall, 1981). However, the degree of genetic diversity and differentiation of introduced populations obviously varies for each invasion event (e.g., Amsellem et al., 2000; McCauley et al., 2003; Provan et al., 2005). For example, Bossdorf et al. (2005) indicated that multiple introductions of invasive populations appear to be the rule rather than the exception, while other researchers have reported that the frequency of introductions may greatly contribute to the decrease of genetic diversity in these populations if a highly competitive species has invaded a region rich in genetic diversity, and to the relief from inbreeding depression over the short run (years to decades) (e.g., Frankham et al., 2002; Saltonstall, 2002; Dlugosch and Parker, 2008). In addition, the genetic characterization of a population is largely associated with the ability of distribution expansion (Lee, 2002). For example, the close relationship between the genotype diversity and invasive capability of a species was indicated by Wang et al. (2012). Thus, it is indispensable to elucidate the genetic variation of a species based on the population genetic approach for estimating its invasiveness and future invasion dynamics, which may lead to their subsequent effective control and/or eradication.
In Japan, Spartina alterniflora Loisel (smooth cordgrass), a plant native to the Atlantic coast of North America and the Gulf of Mexico, was first detected in 2008 in Aichi Prefecture and in 2009 in Kumamoto Prefecture, followed by identification in multiple rivers and tidal flats in both prefectures (i.e., unintentional introduction) (Tamaoki and Takizaki, 2015). However, the reason why S. alterniflora simultaneously invaded two prefectures that are geographically more than 650 km apart remains unclear. The Spartina spp. (cordgrass) (Bortolus et al., 2019) greatly alter brackish and estuarine salt marsh environments via changes in the sediment properties of the tidal flats with growth, resulting in its subsequent further population expansion (e.g., Howes and Teal, 1994; Neira et al., 2005). In addition, serious ecological impacts of Spartina species on native aquatic ecosystems through competitive exclusion (Goss-Custard and Moser, 1988; Wan et al., 2009; Zhou et al., 2009; Morgan and Systma, 2010) and changes in community and trophic structures (Simenstad and Thom, 1995; Levin et al., 2006; Bortolus et al., 2015) were found due to their expansion. Accordingly, Spartina anglica C.E. Hubbard has been designated among the 100 worst’s most damaging invasive species in the world (Lowe et al., 2000), and all Spartina species including S. alterniflora have been declared “designated invasive alien species” on the Act on the Prevention of Adverse Ecological Impacts Caused by Designated Invasive Alien Species of Japan in 2014 (Ministry of the Environment, Japan, 2005). To achieve control and/or eradication of invasive S. alterniflora and prevent its future invasion successfully, knowledge about the current status of S. alterniflora in Japan through a population genetic approach is thought indispensable.
There are some studies that compared the genetic variation of S. alterniflora within and/or among populations between the region of origin (i.e. the Atlantic coast of the United States) (Blum et al., 2007) and the introduced (China) or invaded (i.e., the Pacific coast of the U.S. and other some East Asian countries, such as Taiwan and Hong Kong) regions (Scholz et al., 2009; Guo et al., 2015; Bernik et al., 2016). For example, the most likely invasion pathways of S. alterniflora in Willapa Bay, Washington, on the Pacific coast of the U.S. was the transport and translocation of oysters for cultivation via interstate railroad after the 1890s (Civille et al., 2005). This fact suggests that S. alterniflora populations in the Willapa Bay might be derived from multiple populations on the Atlantic coast around New York (i.e., mid-Atlantic source) (Blum et al., 2007). Also, Blum et al. (2007) estimated that S. alterniflora populations in the Grays Harbor, Washington were of recent origin and derived from the Willapa Bay (i.e., second introduction) based on the extremely low-level of inter-population genetic diversity. On the other hand, populations of this species in the San Francisco Bay, California, and China, which were introduced intentionally, had a relatively high genetic diversity (Blum et al., 2007; Bernik et al., 2016).
Here, the genetic structure of invasive S. alterniflora in Japan and its origin were assessed by analyzing the degree of genetic diversity and genetic mixing in Japanese populations, using chloroplast and nuclear molecular markers. Then, the genetic variance of S. alterniflora was compared between populations in the region of origin (the eastern U.S.) and those in several introduced regions (the Pacific coast of the U.S. and some East Asian countries). This study will elucidate whether actual invasion route of S. alterniflora into Japan was derived from the region of origin (i.e., primary introduction) or from a secondary introduction via introduced regions. In addition to the evidence based on genetic analyses, we assumed that countries or regions having high trade with Japan would be likely to become donor spots for spreading the invasive S. alterniflora irrespective of intentional/unintentional pathways. Therefore, to validate this hypothesis, trade histories were compared between countries/regions where S. alterniflora has grown naturally (the United States, the East Asian countries) and Japan (Aichi and Kumamoto Prefectures). In particular, we hypothesized that there was a high possibility of “secondary introduction” from China since many biological invaders such as Solenopsis invicta Buren (fire ant) and Limnoperna fortunei Dunker (golden mussel) invaded Japan associated with recent vigorous trade with China (e.g., Magara et al., 2001; Murakami, 2018).
Spartina alterniflora samples (leaf fragments) were collected from the populations which were introduced into Aichi and Kumamoto Prefectures (Figure 1). In Aichi Prefecture, 27 samples (i.e., one individual per colony) were collected from multiple colonies along and around the Umeda River (N34° 42′, E 137° 18′) facing the Mikawa Bay. In Kumamoto Prefecture, 20 and 19 S. alterniflora samples were randomly collected from multiple colonies in the Tsuboi River (N 32° 46′, E 130° 37′) facing the Ariake Sea (northern Kumamoto) and the Oono River (N32° 37′, E 130° 39′) facing the Yatsushiro Sea (southern Kumamoto), respectively. In contrast, only three samples from three colonies were collected in the estuary of the Shirakawa River (N 32° 46′, E 130° 36′) facing the Ariake Sea (northern Kumamoto) because almost all the populations had been eradicated by 2012 to 2015 by drawing out and backhoe dredger (Figure 1). The sample collection was carried out following the method in Blum et al. (2007), who indicated that samples should be collected from colonies that are at least about 2.5 m apart from each other (Supplementary Table 1). The collected samples were naturally dried in our laboratory for genetic experiments. Total DNA was extracted from a 0.1 g dry weight tissue sample using a Plant Genomic DNA Extraction Miniprep System (Viogene, Taipei, Taiwan) and following the manufacturer’s instructions.
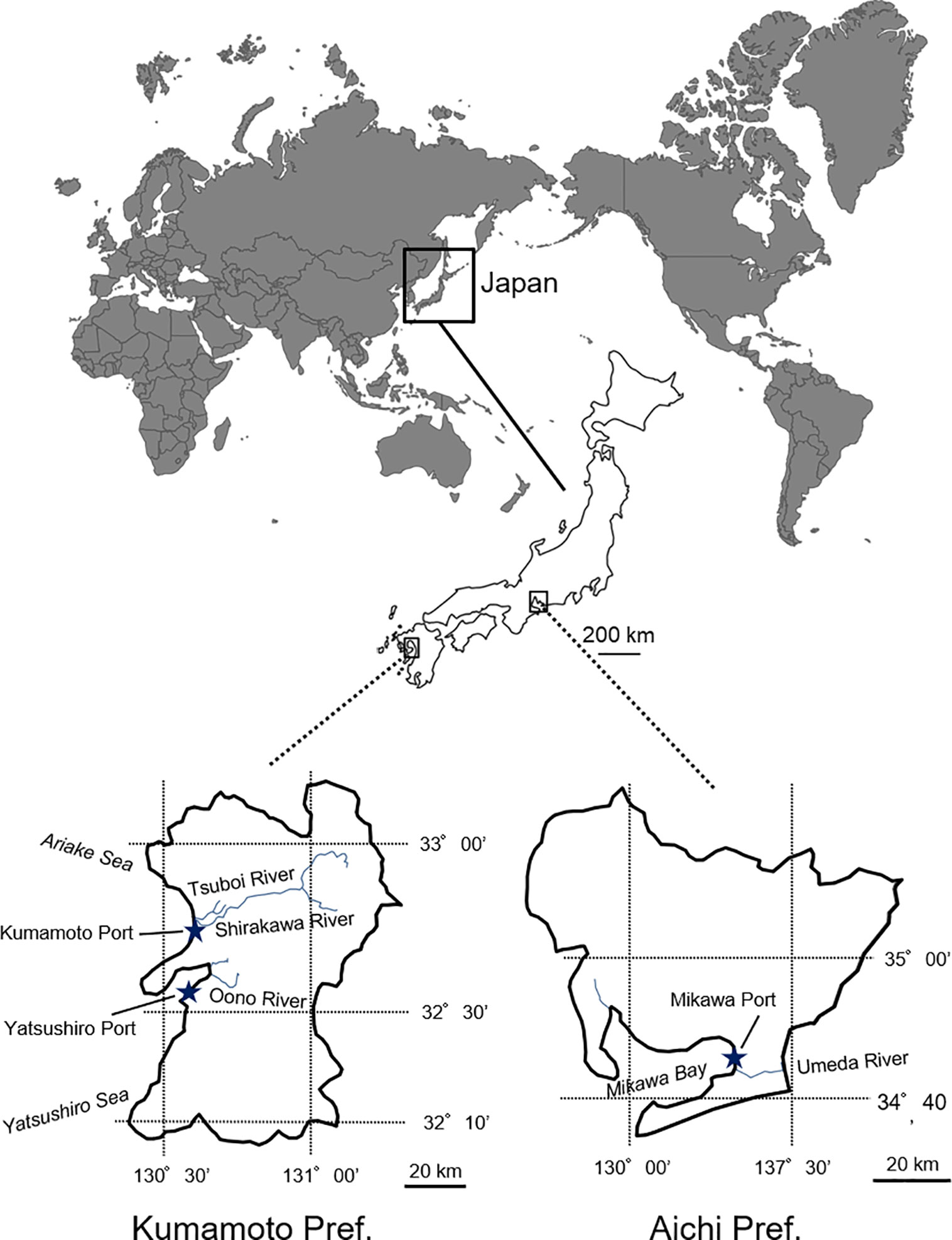
Figure 1 Invasion areas (Aichi and Kumamoto Prefectures) of invasive Spartina alterniflora in Japan.
To compare the chloroplast DNA (cpDNA) haplotypes of S. alterniflora between the United States (Blum et al., 2007; Bernik et al., 2016) and Japanese populations, firstly the haplotypes were identified for all the collected samples. For polymerase chain reaction (PCR) amplification and sequencing of the trnT–trnF region of cpDNA, two primer pairs were used: Tab A (5′-CAT TAC AAA TGC GAT GCT CT-3′) and Tab B (5′-TCT ACC GAT TTC GCC ATA TC-3′) targeting the trnT–trnL region; and Tab C (5′-CGA AAT CGG TAG ACG CTA CG-3′) and Tab F (5′-ATT TGA ACT GGT GAC ACG AG-3′) targeting the trnL–trnF region were used (Taberlet et al., 1991). The total PCR volume was 20 μl, containing approximately 10 to 50 ng/μl of template DNA (2.0 μl), 10× NH4 reaction Buffer (2.0 μl), 10 mM dNTP mix (1.6 μl), 50 mM MgCl2 (1.6 μl), 0.2 μl of each 100 pM primer pair, and 5 U/µl of Biotaq™ DNA polymerase (0.1 μl) (Nippon Genetics, Tokyo, Japan) were used. The temperature conditions of Blum et al. (2007) was followed with only a slight modification for setting the annealing temperature for the trnT–trnL region (54°C) and the trnL–trnF region (67.5°C). A TaKaRa PCR Thermal Cycler (TaKaRa BIO, Shiga, Japan) was used for the PCR assay. PCR products were purified using NucleoSpin Extract II (Macherey–Nagel, Düren, Germany) and then were used as a template for the cycle sequencing reaction. The cycle sequencing reaction assay was conducted by Macrogen Japan Corporation (Kyoto, Japan) and analyzed using a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). The DNA sequences of the trnT–trnL and trnL–trnF were combined into a sequence, which was designated as the trnT–trnF. The base sequence of the trnT–trnL obtained in this study was compared with the existing 42 haplotypes in S. alterniflora (accession numbers AY927278–AY927299 and DQ486839–DQ486858) (Blum et al., 2007) in order to determine its haplotype. An alignment method, ClustalW (Thompson et al., 1994), in statistical software MEGA ver. 6.0 was used for competitive multiple sequence alignment (MSA) (Tamura et al., 2013). All names of the haplotypes obtained in this study were assigned according to the method of Blum et al. (2007).
Microsatellite analysis was conducted using 11 microsatellite markers (SPR1, SPR2, SPR3, SPR4, SPR5, SPR6, SPR7, SPR8, SPR9, SPR10, SPR11), developed by Blum et al. (2004). The PCR amplification were carried out in a total volume of 20 μl, consisting of approximately 10 to 50 ng/μl template DNA (4.0 μl), 10× Buffer (2.0 μl), 2 mM dNTP mixture (2.0 μl), 0.2 μl of each 100 pM primer pair, and 2.5 U/μl of Blend Taq (0.5 μl) (TOYOBO, Osaka, Japan). The forward primer was fluorescently labeled with 5′-FAM, TAMRA, and 5′-JOE. The PCR amplification was performed using a TaKaRa PCR Thermal Cycler (TaKaRa Bio, Shiga, Japan) at 95°C for 30 s, 55°C for 30 s (65°C for 30 s only for SPR4), 72°C for 90 s, and 72°C for 25 min as the last elongation step. Fragment analysis was conducted by Macrogen (Seoul, South Korea). Based on the microsatellite analysis, individuals which had an exact genotype match were considered as “clones” and then were excluded from the analyses. The genotypes of S. alterniflora populations in Japan were identified using 11 different microsatellite markers (Supplementary Table 2). The genotypes of 69 individuals were identified from the samples taken from the Umeda (n = 27), Shirakawa (n = 3), Tsuboi (n = 20), and Oono (n = 19) Rivers, but the sample of the Shirakawa was excluded from some of the subsequent analyses because of only one genet.
To compare the degree of genetic diversity of S. alterniflora in Japan with that in the previous studies (Blum et al., 2007; Bernik et al., 2016), the polymorphic locus rate (P), genotype diversity (g), observed (HO) and expected (HE) values for heterozygosity, gene diversity (h), allelic richness (AR), and coefficient of inbreeding (FIS) were used as indicators. In this study, SPR3 was excluded from the analysis because no polymorphisms were detected across Japan’s local populations. The polymorphic locus rate (P) was calculated for each local population. This value indicates the rate of genetic loci with polymorphisms compared to all the genetic loci for each local population. Presence/absence of the multilocus genotype matches in among individual polymorphic gene loci was analyzed using Software GENALEX ver. 6.5 (Peakall and Smouse, 2012). The genotype diversity (g) for each S. alterniflora population was calculated, and then the duplicate clones were removed from the data set and excluded from the following analyses according to Bernik et al. (2016). The value of g indicates the rate of the individuals with duplicate clones removed in each local population. The observed (HO) and expected (HE) values for heterozygosity were calculated using GenAlEx ver. 6.5 (Peakall and Smouse, 2012). The gene diversity (h), allelic richness (AR), and coefficient of inbreeding (FIS), and its confidence intervals were calculated using FSTAT ver. 2.9.3 (Goudet, 2001). Tests for deviation from Hardy–Weinberg equilibrium (HWE) were also performed using FSTAT ver. 2.9.3 (Goudet, 2001).
To verify whether a genetic bottleneck had been formed in each local population in Japan, BOTTLENECK analysis (Piry et al., 1999) was conducted using Wilcoxon’s heterozygosity excess test (Luikart et al., 1998a) and the mode shift test (Luikart et al., 1998b). This is because Piry et al. (1999) suggested that Wilcoxon’s test is most powerful and robust when used with few polymorphic loci. Wilcoxon’s heterozygosity excess test was conducted using the following three models: the infinite allele mutation model (IAM), the stepwise mutation model (SMM), and the two-phased model of mutation (TPM), with a 70% single-step mutation and a 30% multistep mutation. Regarding the genetic differences among the individuals, the pairwise co-dominant genotypic distances in each Japanese population were calculated using GenAlEx ver. 6.5 and then evaluated by principal coordinate analysis (PCoA) (Peakall and Smouse, 2012).
To evaluate the genetic structure in the individuals, analysis with STRUCTURE allocated all individuals to K clusters by Bayesian’s clustering and was conducted to maximize the linkage disequilibrium and Hardy-Weinberg’s disequilibrium. Software STRUCTURE ver. 2.3.4 (Pritchard et al., 2000) was used for this analysis. Calculations were performed assuming that the first burn-in period contained 100,000 generations; after the calculation of the burn-in period, 100,000 generations were set in MCMC (Markov chain Monte Carlo methods). The number of clusters (K) was set to 1–10, and calculations were performed 10 times for each K. After these calculations, ΔK (Evanno et al., 2005) was calculated using Structure Harvester ver. 0.6.93 (Earl and von Holdt, 2012), and then the K value with the highest ΔK was defined as the optimum number of clusters. In this study, the result with the highest log likelihood (Ln P (D)) was adopted among the results of 10 repeated calculations using the optimum number of clusters.
The sequences of trnT–trnF region from chloroplast DNA were identified from all S. alterniflora individuals sampled in both prefectures and regions: the Umeda River (Aichi), the Shirakawa River and Tsuboi River (northern Kumamoto), and Oono River (southern Kumamoto). The sequences of trnT–trnF region from Japanese populations revealed that all S. alterniflora populations in Japan had a single haplotype (accession number: LC565815): the haplotype C4 (accession number: KJ499448, Guo et al., 2015; MG201950, Qiao et al., 2019) (Figure 2, Table 1). Haplotype C4 has been identified as widespread in the Atlantic coast of the U.S., especially around the Florida Peninsula. Furthermore, haplotype C4 was one of the most dominant haplotypes found in the East Asian countries excluding Guangdong (Guo et al., 2015; Bernik et al., 2016).
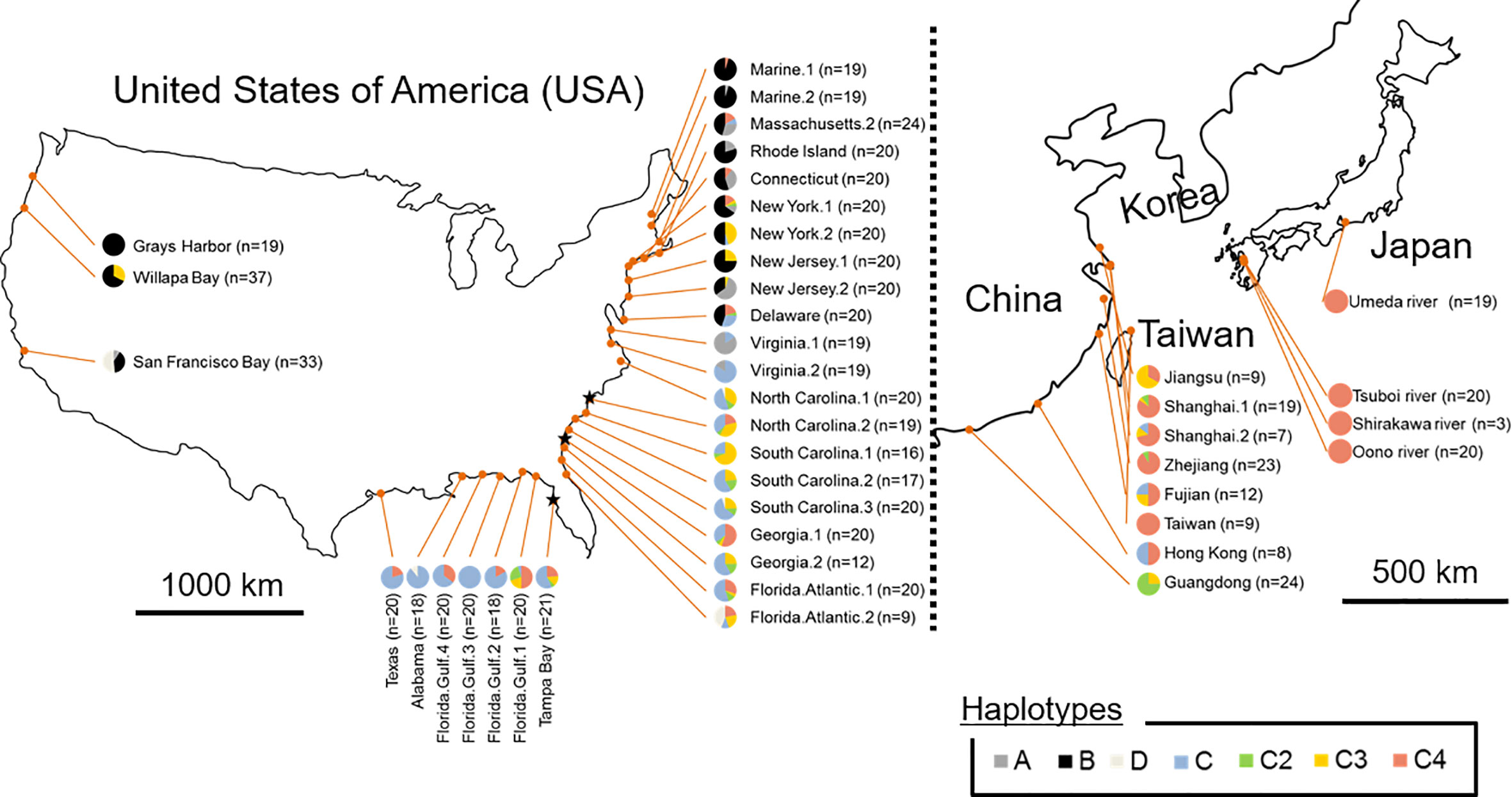
Figure 2 Frequency and distribution of chloroplast DNA (cpDNA) haplotypes in the region of origin (the eastern Unites States) and in the regions where Spartina alterniflora had been introduced intentionally and/or unintentionally (the Pacific coast of the U.S. and the East Asian countries). Groups A, B, and D consisting of a single haplotype are shown in dark grey, black and light grey, respectively. Haplotype C2, C3, and C4 of Group C consisting of multiple haplotypes are shown in green, yellow, and pink, respectively, and other C members are shown in blue. ★ indicates the region estimated as the place that S. alterniflora was initially introduced into China, according to Bernik et al. (2016).
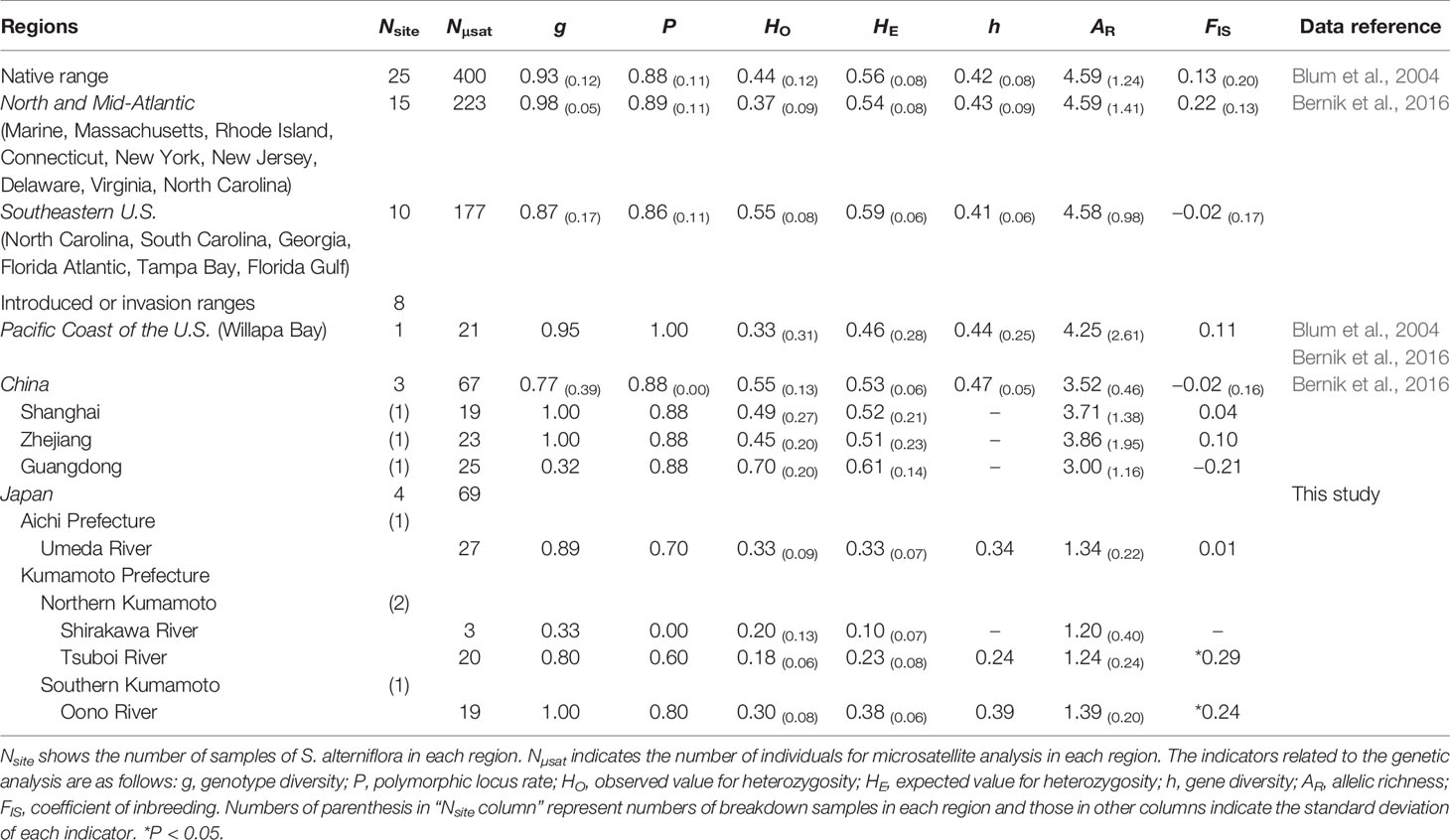
Table 1 Information on the genetic diversity of invasive Spartina alterniflora based on the microsatellite loci in Japan.
Results of the genetic analysis of Japanese S. alterniflora samples collected using the different markers demonstrated that the number of alleles of S. alterniflora individual stands in each river was less than or equal to 2, except for one sample from the Tsuboi River (Supplementary Table 2). Among the 11 microsatellite markers, no genetic polymorphisms were detected from the locus SPR3. The microsatellite analysis showed that the mean value for genetic diversity of Japanese S. alterniflora samples were as follows; the Umeda River (h = 0.34, AR = 1.34 ± 0.22), Tsuboi River (h = 0.24, AR = 1.24 ± 0.24), and Oono River (h = 0.39, AR = 1.39 ± 0.20).
The genotype diversity (g) was extremely high at 0.93 ± 0.12 in samples from the Atlantic coast of the U.S. (native range), and similar tendencies were also found in other regions where S. alterniflora invaded (Table 1). The g values of Japanese S. alterniflora populations, excluding the Shirakawa, lay within 0.80 to 1.00, almost equivalent to those in China (g = 0.77 ± 0.39) and the introduced in Willapa Bay (the Pacific coast of the U.S.) (g = 0.95). On the other hand, low g values were found in samples from the Shirakawa River (g = 0.33) and Guangdong province in China (g = 0.32), where almost all analyzed samples had the same genotype. The inbreeding coefficient (FIS) of each population in Japan indicated that estimated FIS values of samples from the Tsuboi (FIS = 0.29) and Oono (FIS = 0.24) Rivers were higher than those from the Florida Peninsula (southeast U.S.) (FIS = −0.02 ± 0.17) and China (FIS = −0.02 ± 0.16), suggesting the significantly excessive homozygosity (P<0.05). However, the FIS values of samples from the Umeda River (FIS = 0.01) did not deviate from the Hardy-Weinberg equilibrium (Table 1). The significant excessive homozygosity on Japanese S. alterniflora populations was observed in the infinite allele mutation model (IAM), the stepwise mutation model (SMM), and the two-phased model of mutation (TPM) (P<0.05). In addition, the mode shift test suggested that a bottleneck (i.e., shifted mode) would have been formed in these populations in recent years (Table 2).
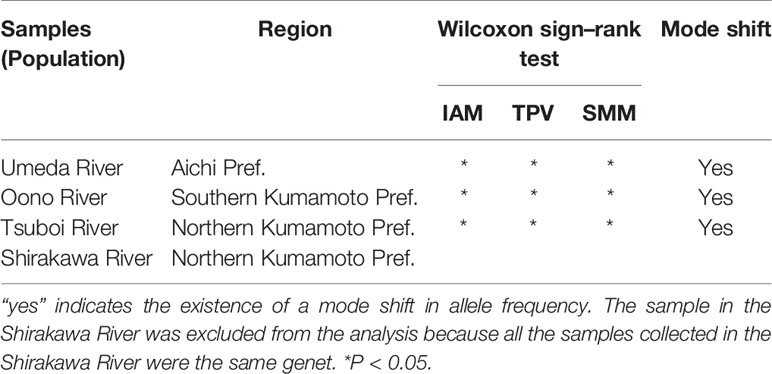
Table 2 Bottleneck analysis of Spartina alterniflora populations in Japan using three models: IAM, SMM, and TPM.
Principal coordinate analysis (PCoA) based on co-dominant genotypic distances revealed that genetic distances of S. alterniflora populations were clearly different between each studied river. The proportions for Axis 1 and Axis 2 were 41.2% and 23.3%, respectively. Although S. alterniflora populations in the Shirakawa and Tsuboi Rivers were placed in the same position, those in the Oono and Umeda Rivers were clearly separated along Axis 1 (Figure 3), suggesting that there were at least three S. alterniflora local populations in Japan.
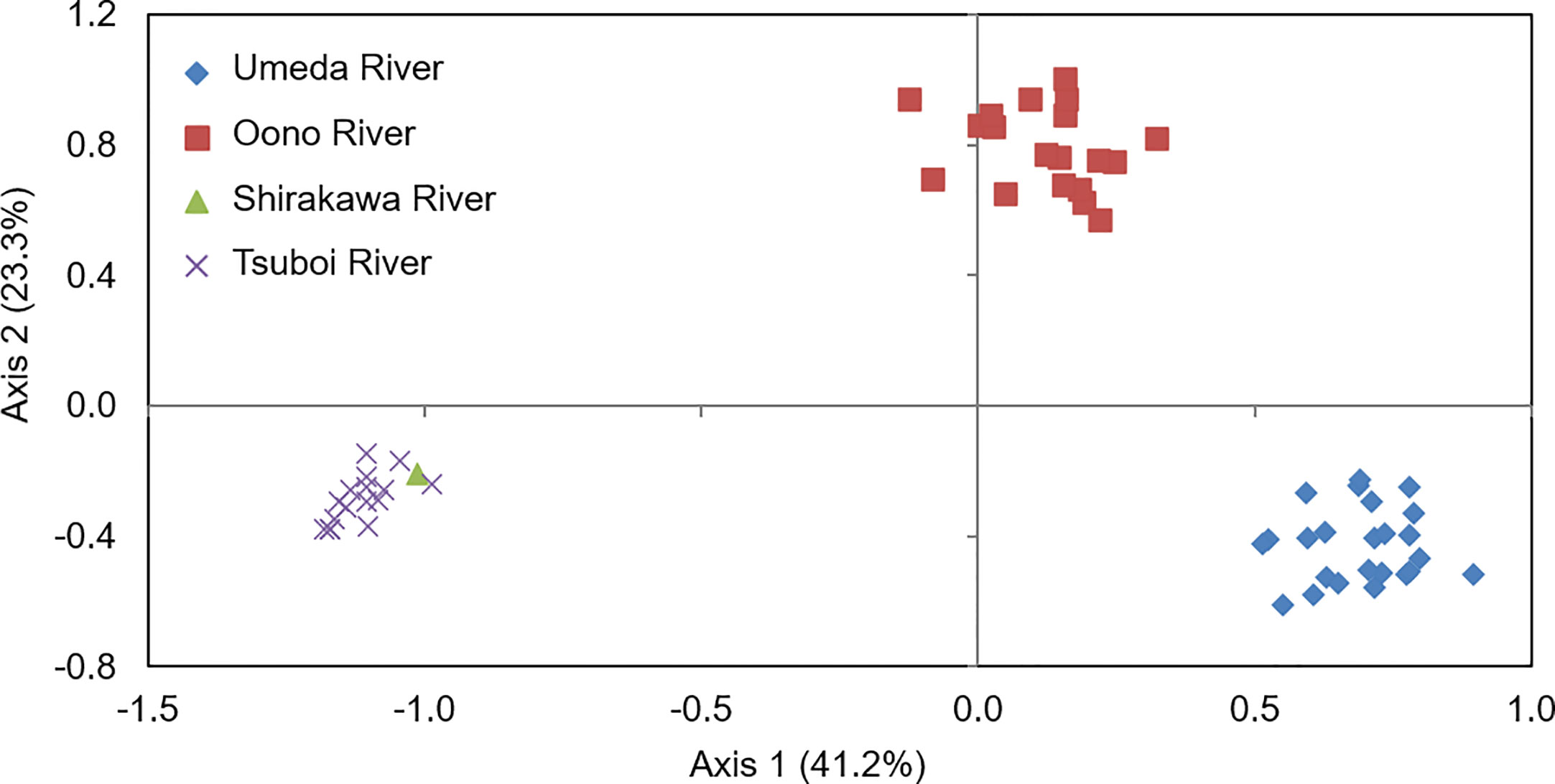
Figure 3 Results of a principal coordinate analysis (PCoA) of Spartina alterniflora local populations in Japan based on co-dominant genotypic distances. Axis 1 and Axis 2 account for 41.2% and 23.3% of the variance, respectively. Symbols are as follows: rhomboid, populations in Umeda River (Aichi); square, in Oono River (southern Kumamoto); triangle, in Shirakawa River (northern Kumamoto); cross, in Tsuboi River (northern Kumamoto).
The STRUCTURE analysis indicated that the studied populations were divided into distinct genetic clusters. The ΔK value was clearly the highest at K = 3 (Figure 4A). As a result, S. alterniflora populations of Japan were classified into three groups: 1) Umeda River (Aichi), 2) Shirakawa and Tsuboi Rivers (northern Kumamoto), and 3) Oono River (southern Kumamoto) (Figure 4B). In addition, each group was practically unmixed with any other group.
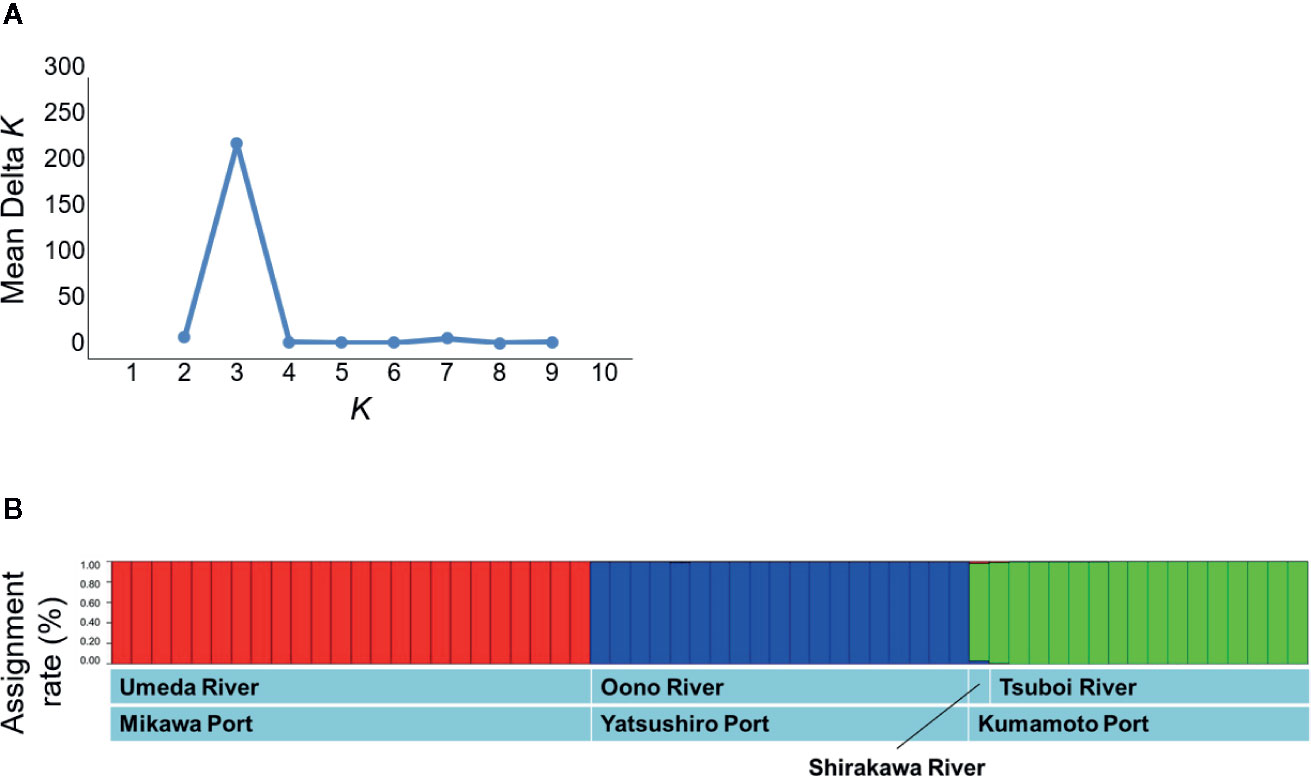
Figure 4 Population structures based on the microsatellite mutation among the genes sampled from Spartina alterniflora populations in Japan using Bayesian estimation. (A) The estimation of the optimum number of clusters based on ΔK. (B) The assignment of each individual into the clusters using STRUCTURE analysis. The vertical axis in panel B shows attributed rates of each sample collected in S. alterniflora local populations.
Genetic analysis of cpDNA revealed that all S. alterniflora populations in Japan had a single haplotype (haplotype C4) (Figure 2, Table 1). Within the region of its origin, haplotype C4 was widely observed in the Atlantic coast of the U.S. Also, this haplotype was the most dominant in the East Asian countries where S. alterniflora has been introduced intentionally (China, see An et al., 2007) or unintentionally (Taiwan and Hong Kong e.g., Scholz et al., 2009; Guo et al., 2015). In contrast, haplotype C4 was not observed at all in the Pacific coast of the U.S. (Blum et al., 2007; Guo et al., 2015; Bernik et al., 2016). Therefore, this finding suggests that S. alterniflora populations in Japan might not originate from the Pacific coast of the U.S. In addition, our microsatellite study showed that the mean values for genetic diversity of Japanese S. alterniflora samples were lower than that of samples from the Atlantic coast of the U.S. (h = 0.42 ± 0.08, AR = 4.59 ± 1.24) and the Florida Peninsula (southeast U.S.) (h = 0.41 ± 0.06, AR = 4.58 ± 0.98), the region of its origin (Blum et al., 2007; Bernik et al., 2016), and China (h = 0.47 ± 0.05, AR = 3.52 ± 0.46) (Bernik et al., 2016) and Willapa Bay (h = 0.44 ± 0.25, AR = 4.25 ± 2.61) located in the Pacific coast of the U.S. (Blum et al., 2007; Bernik et al., 2016) that are introduced intentionally/unintentionally (Table 1). It should be noted that no information has been reported on S. alterniflora populations with such low genetic diversity so far (Blum et al., 2007; Bernik et al., 2016). Indeed, values of AR in Japanese S. alterniflora were very low in comparison to those in the U.S. and China populations, with significant excessive homozygosity detected in three mutation models. In addition, the formation of a bottleneck (i.e., shifted mode) was expected by the mode shift test in S. alterniflora population in Japan. Therefore, these results reveal that the founder effect might have occurred in Japanese S. alterniflora population. Such low genetic diversities associated with a founder effect were also found in other Spartina species such as S. versicolor Fabre introduced in Europe (Baumel et al., 2016) and S. densiflora Brongn. invading the Pacific coast of the U.S. (Castillo et al., 2018). Since the cause of a lower genetic diversity among invasive Spartina species is of great interest, we discuss below the reason why S. alterniflora populations had lower genetic diversity when invading Japan.
After 1979, seeds and individuals of S. alterniflora were intentionally introduced into China from multiple areas of the Atlantic coast of the U.S. (e.g., North Carolina, Georgia, and Florida) for eco-engineering purposes (i.e., reclamation of tideland) (Xu and Zhou, 1985; Wan et al., 2009). The coverage of S. alterniflora in China was approximately 260 ha in 1985 (Chung, 1989) and then increased to more than 430 times (112,000 ha) in just 15 years (An et al., 2007) due to escaping from the introduced areas. These facts suggest that S. alterniflora expanding in East Asian countries originates from populations (found) in the southeast U.S., especially around the Florida Peninsula. Populations of S. alterniflora in the Grays Harbor, Washington (haplotype B) and Taiwan (haplotype C4), which had only a single haplotype as well as Japan (Figure 2), were unintentionally and secondarily introduced from the Willapa Bay, Washington (the Pacific coast of the U.S.) (Civille et al., 2005) and the vicinity of Fujian (China) (Lin et al., 2015), respectively.
In this study, we assumed that countries or regions having high trade with Japan would be likely to become donor spots for spreading the invasive S. alterniflora irrespective of intentional/unintentional pathways as mentioned before. Undoubtedly, this can be generalized regardless of taxonomic groups. Thus, to validate this hypothesis, trade histories were compared between countries/regions where S. alterniflora has grown (the United States, China, Taiwan, Hong Kong) (Blum et al., 2007; Guo et al., 2015; Bernik et al., 2016) and the ports nearest to each studied river in Japan (i.e., Kumamoto Port, Yatsushiro Port, and Mikawa Port) using historical trade data from the 2003 to 2013 in the Global Trade Atlas (https://www.gtis.com/gta/). The findings revealed that when compared the amount of trade between the Yatsushiro Port (southern Kumamoto), which includes the Oono River and the U.S. ($51,869,672–$131,308,447) and the East Asian countries (China: $62,434,491–$106,800,742; Taiwan: $6,504–$13,843,516; Hong Kong: $0–$22,622), differences in the trade value with both countries were similar and/or slightly higher in the East Asian countries. Similar trend on the amount of trade with U.S. ($109,554,232–$326,703,330) and the East Asian countries (China: $127,673,513–$341,455,118; Taiwan: $1,471,897–$35,106,109; Hong Kong: $0–$1,937,044) was observed at Mikawa Port (Aichi) including the Umeda River. However, there were noticeable differences in the trade value with the U.S. ($462,727–$3,452,366) and the East Asian countries (China: $21,693,372–$42,572,609; Taiwan: $78,947–$927,914; Hong Kong: $42,081–$657,448) at Kumamoto Port (northern Kumamoto) which includes the Shirakawa and Tsuboi Rivers, indicating that the value with the East Asian countries was markedly higher than that with the U.S. These data suggest that the route through which invasive S. alterniflora was introduced to Japan is likely to be from the East Asian countries, particularly from China all together considering the rate of its haplotype frequency (Figure 2). The East Asian countries are one of the largest supply sources on young shellfish and seedlings for cultivation in tidal flats of Japan (Okoshi, 2007), and thus the contamination of multiple species of organisms is often observed with the imports. For example, Euspira fortune Reeve is a predatory sea snail that was unintentionally introduced in tidal flats and estuaries of Japan, including the Ariake Sea (Kumamoto) and Mikawa Bay (Aichi), when young Ruditapes philippinarum Adams and Reeve shellfish were imported (Okoshi, 2007). Similarly, S. alterniflora in the western U.S. was also introduced unintentionally from the eastern U.S. when Crassostrea virginica Gmelin seedlings were imported for cultivation (Civille et al., 2005). From these facts, we cannot deny the possibility that S. alterniflora was introduced unintentionally into Japan through the importation of cultured shellfishes. Therefore, further research on the genetic characteristics of the invasive S. alterniflora should be carried out worldwide for estimating its global spread and future invasion risks.
Comparison of microsatellite data among S. alterniflora local populations in Japan for estimating the route through which S. alterniflora invaded Japan revealed that genotypes of the populations were clearly different in each river (Figures 3 and 4). These results suggest that there is no exchange of S. alterniflora genome among the four rivers in Japan. Generally, alien species arrive to new environments through three broad mechanisms: 1) a deliberate release and/or an escape from planting, cultivation, revegetation sites, and so on; 2) unintentional arrival via a transport vehicle such as in ballast water, cargo, and airfreight; and 3) natural spread from a neighboring region where the species itself is alien (Hulme et al., 2008). Among these invasive mechanisms, the possibility of S. alterniflora invasion in Japan via intentional introductions is almost impossible, since Japan has no such imports for the reclamation of tidal flats. Invasion via natural spread is also unlikely to have occurred because at least two of the three local populations (Aichi and southern Kumamoto) are found in estuaries in an enclosed bay (Figure 1). Therefore, the most likely invasion route may have been the arrival through a transport vehicle (i.e., stowaway) (Hulme et al., 2008).
Results of the microsatellite analysis made it clear that some S. alterniflora individuals (St. 13, 15, 16, and 18) in the Tsuboi River (northern Kumamoto) had a heterozygous at only one locus, while two individuals growing sympatrically (St. 14 and 17) had a homozygous at all of the loci (Supplementary Table 2). Spartina alterniflora rapidly spread their populations via both sexual (seed) (Hayasaka et al., 2020) and asexual (clonal) reproduction and then form a high-density single colony (Somers and Grant, 1981; Davis et al., 2004). It is suggested that although these individuals have actually grown via seed propagation (i.e., sexual reproduction), they may be considered as clones with exactly the same genotype due to the extreme homozygosity. The number of alleles per marker on each S. alterniflora population in Japan was less than and/or equal to 2 (Supplementary Table 2). Again, values of AR in S. alterniflora populations in Japan were lower than those in the U.S. and China populations, and the formation of a bottleneck was expected in Japanese populations. Therefore, these facts indicate that the founder effect might have occurred in S. alterniflora populations in Japan. In addition, no S. alterniflora population was found in Japan before 2008 (Tamaoki and Takizaki, 2015). In other words, only a few individuals of S. alterniflora might have successfully invaded Japan. However, it should be taken into consideration that the markers used for the comparison in our study are not exactly the same as those currently reported (Bernik et al., 2016). Nevertheless, it suggests that only one S. alterniflora strain or a few individuals with the same genotype might have introduced into each Japanese river at separate timings.
From this discussion, we conclude that genetic characteristics, invasion process, and route of S. alterniflora populations in Japan were as follows: 1) all S. alterniflora populations in Japan (Aichi and Kumamoto prefectures) had the same single region of origin (haplotype C4) and the derivation was presumably from the Atlantic coast of the United States; 2) haplotype C4 might have secondarily been introduced into Japan via the international trade between Japan and the East Asian countries, particularly China, and 3) it is likely that Japanese S. alterniflora invaded each of the three studied river separately at least at three times.
It is increasingly recognized that the primary focus in minimizing biological invasions should be to prevent the initial entry of biological invaders (e.g., Williams and West, 2000; Saccaggi et al., 2016). Therefore, it is important to strengthen the quarantine control on the importation of commodities, especially of transport vehicles at potential donor spots (i.e., border control/border biosecurity system), to decrease further risks of various biological invaders (Chornesky and Randall, 2003; Xu et al., 2006) including that of Spartina species (Castillo et al., 2018; Gallego-Tévar et al., 2019). Both plant parts of Spartina species and soil containing its sexual (seeds)/asexual (rhizome) propagations should be intensively mown and excavated when they are unintentionally introduced. Simultaneously, it is also important that S. alterniflora is detected and eliminated at an early invasion stage in order to minimize its invasion. In this study, we predicted the low frequency of S. alterniflora invasion. The reason was that the number of alleles per marker on each S. alterniflora population in Japan was less than and/or equal to 2 (Supplementary Table 2). Despite this, it took approximately 6 years from its first detection to the start at the eradication project. As mentioned above, trade with China is extremely large. Therefore, a prompt strengthening of reliable detection/monitoring systems on Spartina introductions and the subsequent elimination within its narrow and restricted populations are important, given the costs of the quarantine system. For this purpose, it is essential to continue monitoring areas where S. alterniflora has already invaded. Furthermore, given that ports that trade with China are all over Japan, the strengthening of shared information networks on introduced species between each region/port would lead to minimize their biological invasion risks.
The datasets generated for this study can be found in the DNA Data Bank of JAPAN (DDJB), accession number: LC565815.
YM, MT, and DH designed and coordinated the research. YM, TH, AN, and DH collected samples. YM, MT, and YI analyzed the data. YM and DH drafted the paper with the input of NN. All authors contributed to the article and approved the submitted version.
Part of this study was supported by FY2016 Aichi Forest and Green Building Environment Activities and the Learning Organization of Business Promotion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Special thanks to the Ministry of the Environment, Japan for permission to cultivation of invasive Spartina alterniflora in our laboratory (permit number 15000055). We would like to thank Dr. Tadao Kitagawa, Dr. Takuo Sawahata, Dr. Kaori Kochi, Takahiro Kusaka (Kindai University), Kano Koide (Japan Wildlife Research Center), and Reiko Ito (Kyushu Kaihatsu Engineering Co., Ltd.) for their helpful suggestions and supports regarding this manuscript. The authors also wish to thank Moe Nakagawa, Ryu Ikeda, Kota Kohara and Yoshinori Taruma (Kindai University) for helping with S. alterniflora sampling. We thank Dr. Francisco Sánchez-Bayo (The University of Sydney), Dr. Jean Beran Tanangonan, and Robert John Sheridan (Kindai University) for English editing of the original manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.556039/full#supplementary-material
Amsellem, L., Noyer, J. L., Le Bourgeois, T., Hossaert-Mckey, M. (2000). Comparison of genetic diversity of the invasive weed Rubus alceifolius Poir. (Rosaceae) in its native range and in areas of introduction, using amplified fragment length polymorphism (AFLP) markers. Mol. Ecol. 9 (4), 443–455. doi: 10.1046/j.1365-294x.2000.00876.x
An, S. Q., Gu, B. H., Zhou, C. F., Wang, Z. S., Deng, Z. F., Zhi, Y. B., et al. (2007). Spartina invasion in China: implications for invasive species management and future research. Weed Res. 47 (3), 183–191. doi: 10.1111/j.1365-3180.2007.00559.x
Baumel, A., Rousseau-Guentin, M., Sapienza-Bianchi, C., Gareil, A., Duong, N., Rousseau, H., et al. (2016). Spartina versicolor Fabre: Another case of Spartina trans-Atlantic introduction? Biol. Invasions 18, 2123–2135. doi: 10.1007/s10530-016-1128-z
Bernik, B. M., Li, H., Blum, M. J. (2016). Genetic variation of Spartina alterniflora intentionally introduced to China. Biol. Invasions 18 (5), 1485–1498. doi: 10.1007/s10530-016-1096-3
Blum, M. J., Sloop, C. M., Ayres, D. R., Strong, D. R. (2004). Characterization of microsatellite loci in Spartina species (Poaceae). Mol. Ecol. Notes 4 (1), 39–42. doi: 10.1046/j.1471-8286.2003.00556.x
Blum, M. J., Bando, K. J., Katz, M., Strong, D. R. (2007). Geographic structure, genetic diversity and source tracking of Spartina alterniflora. J. Biogeogr. 34 (12), 2055–2069. doi: 10.1111/j.1365-2699.2007.01764.x
Bortolus, A., Carlton, J. T., Schwindt, E. (2015). Reimagining South American coasts: unveiling the hidden invasion history of an iconic ecological engineer. Divers. Distrib. 21 (11), 1267–1283. doi: 10.1111/ddi.12377
Bortolus, A., Adam, P., Adams, J. B., Ainouche, M. L., Ayres, D., Bertness, M. D., et al. (2019). Supporting Spartina: interdisciplinary perspective shows Spartina as a distinct solid genus. Ecology 100 (11), e02863. doi: 10.1002/ecy.2863
Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E., Siemann, E., Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144 (1), 1–11. doi: 10.1007/s00442-005-0070-z
Brown, A. H. D., Marshall, D. R. (1981). “Evolutionary changes accompanying colonization in plants,” in Evolution today. Eds. Scudder, G. G. E., Reveal, J. L. (Pittsburgh, PA: Carnegie–Mellon University), 351–363.
Castillo, J. M., Gallego-Tévar, B., Figueroa, E., Grewell, B. J., Vallet, D., Rousseau, H., et al. (2018). Low genetic diversity contrasts with high phenotypic variability in heptaploid Spartina densiflora populations invading the Pacific coast of North America. Ecol. Evol. 8 (10), 4992–5007. doi: 10.1002/ece3.4063
Chornesky, E. A., Randall, J. M. (2003). The threat of invasive alien species to biological diversity: setting a future course. Ann. Mo. Bot. Gard. 90 (1), 67–76. doi: 10.2307/3298527
Chung, C. H. (1989). “Ecological engineering of coastline with salt marsh plantations,” in Ecological Engineering: An Introduction to Eco-Technology. Eds. Mitsch, W. J., Jorgensen, S. A. (New York, NY: Wiley & Sons), 255–289.
Civille, J. C., Sayce, K., Smith, S. D., Strong, D. R. (2005). Reconstructing a century of Spartina alterniflora invasion with historical records and contemporary remote sensing. Ecoscience 12 (3), 330–338. doi: 10.2980/i1195-6860-12-3-330.1
Davis, H. G., Taylor, C. M., Lambrinos, J. G., Strong, D. R. (2004). Pollen limitation causes an allee effect in a wind-pollinated invasive grass (Spartina alterniflora). Proc. Natl. Acad. Sci. U. S. A. 101 (38), 13804–13807. doi: 10.1073/pnas.0405230101
Dlugosch, K. M., Parker, I. M. (2008). Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17 (1), 431–449. doi: 10.1111/j.1365-294x.2007.03538.x
Earl, D. A., von Holdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4 (2), 359–361. doi: 10.1007/s12686-011-9548-7
Elton, C. S. (1958). The Ecology of Invasions by Animals and Plants (Chicago, IL: University of Chicago Press).
Evanno, G., Regnaut, S., Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 (8), 2611–2620. doi: 10.1111/j.1365-294x.2005.02553.x
Frankham, R., Briscoe, D. A., Ballou, J. D. (2002). Introduction to conservation genetics (Cambridge, UK: Cambridge university press).
Gallego-Tévar, B., Infante-Izquierdo, M. D., Figueroa, E., Nieva, F. J. J., Muñoz-Rodriguez, A. F., Grewell, B. J., et al. (2019). Some like it hot: maternal-switching with climate change modifies formation of invasive Spartina hybrids. Front. Plant Sci. 10, 484. doi: 10.3389/fpls.2019.00484
Goss-Custard, J. D., Moser, M. E. (1988). Rates of change in the numbers of dunlin, Calidris alpina, wintering in British estuaries in relation to the spread of Spartina anglica. J. Appl. Ecol. 25 (1), 95–109. doi: 10.2307/2403612
Goudet, J. (2001). FSTAT (version 2.9.3), a program to estimate and test gene diversity and fixation indices (Lausanne, Switzerland: Lausanne University). Available at: http://www2.unil.ch/popgen/softwares/fstat.htm (Accessed March 18, 2018).
Guo, W., Qiao, S., Wang, Y., Shi, S., Tan, F., Huang, Y. (2015). Genetic diversity, population structure, and genetic relatedness of native and non–native populations of Spartina alterniflora (Poaceae, Chloridoideae). Hydrobiologia 745 (1), 313–327. doi: 10.1007/s10750-014-2117-9
Hayasaka, D., Fujiwara, S., Uchida, T. (2018). Impacts of invasive Iris pseudacorus L. (yellow flag) establishing in an abandoned urban pond on native semi-wetland vegetation. J. Integr. Agric. 17 (8), 1881–1887. doi: 10.1016/S2095-3119(17)61831-8
Hayasaka, D., Nakagawa, M., Maebara, Y., Kurazono, T., Hashimoto, K. (2020). Seed germination characteristics of invasive Spartina alterniflora Loisel in Japan: implications for its effective management. Sci. Rep. 10, 2116. doi: 10.1038/s41598-020-58879-7
Hoos, P. M., Miller, A. W., Ruiz, G. M., Vrijenhoek, R. C., Geller, J. B. (2010). Genetic and historical evidence disagree on likely sources of the Atlantic amethyst gem clam Gemma gemma (Totten 1834) in California. Divers. Distrib. 16 (4), 582–592. doi: 10.1111/j.1472-4642.2010.00672.x
Howes, B. L., Teal, J. M. (1994). Oxygen loss from Spartina alterniflora and its relationship to salt marsh oxygen balance. Oecologia 97 (4), 431–438. doi: 10.1007/BF00325879
Hulme, P. E., Bacher, S., Kenis, M., Klotz, S., Kühn, I., Minchin, D., et al. (2008). Grasping at the routes of biological invasions: a framework for integrating pathways into policy. J. Appl. Ecol. 45 (2), 403–414. doi: 10.1111/j.1365-2664.2007.01442.x
Koncki, N. G., Aronson, M. F. J. (2015). Invasion risk in a warmer world: modeling range expansion and habitat preferences of three nonnative aquatic invasive plants. Invasive Plant Sci. Manage. 8 (4), 436–450. doi: 10.1614/IPSM-D-15-00020.1
Lee, C. E. (2002). Evolutionary genetics of invasive species. Trends Ecol. Evol. 17 (8), 386–391. doi: 10.1016/S0169-5347(02)02554-5
Levin, L. A., Neira, C., Grosholz, E. D. (2006). Invasive cordgrass modifies wetland trophic function. Ecology 87 (2), 419–432. doi: 10.1890/04-1752
Lin, H.-J., Hsu, C.-B., Liao, S.-H., Chen, C.-P., Hsieh, H. L. (2015). Effects of Spartina alterniflora invasion on the abundance and community of meiofauna in a subtropical wetland. Wetlands 35 (3), 547–556. doi: 10.1007/s13157-015-0643-5
Lockwood, J. L., Hoopes, M. F., Marchetti, M. P. (2007). Invasion Ecology. 2nd edn (Oxford, UK: Blackwell Publishing).
Lombaert, E., Guillemaud, T., Cornuet, J.-M., Malausa, T., Facon, B., Estoup, A. (2010). Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PloS One 5 (3), e9743. doi: 10.1371/journal.pone.0009743
Lowe, S., Browne, M., Boudjelas, S. (2000). 100 of the world"s worst invasive alien species (Auckland, NZ: IUCN-ISSG).
Lowe, A., Harris, S., Ashton, P. (2004). Ecological Genetics: Design, Analysis, and Application (Malden, MA: Blackwell Publishing).
Luikart, G., Sherwin, W. B., Steele, B. M., Allendorf, F. W. (1998a). Usefulness of molecular markers for detecting population bottlenecks via monitoring genetic change. Mol. Ecol. 7 (8), 963–974. doi: 10.1046/j.1365-294x.1998.00414.x
Luikart, G., Allendorf, F. W., Cornuet, J.-M., Sherwin, W. B. (1998b). Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 89 (3), 238–247. doi: 10.1093/jhered/89.3.238
Magara, Y., Matsui, Y., Goto, Y., Yuasa, A. (2001). Invasion of the non-indigenous nuisance mussel, Limnoperna fortunei, into water supply facilities in Japan. J. Water Supply 50 (3), 113–124. doi: 10.2166/aqua.2001.0011
McCauley, D. E., Smith, R. A., Lisenby, J. D., Hsieh, C. (2003). The hierarchical spatial distribution of chloroplast DNA polymorphisms across the introduced range of Silene vulgaris. Mol. Ecol. 12 (12), 3227–3235. doi: 10.1046/j.1365-294X.2003.01992.x
Ministry of the Environment, Japan (2005). List of regulated living organisms under the Invasive Alien Species Act. Available at: https://www.env.go.jp/nature/intro/2outline/files/siteisyu_list_e.pdf (Accessed April 16, 2020).
Morgan, V. H., Systma, M. (2010). Alaska Spartina Prevention, Detection and Response Plan (Juneau, AK: National Marine Fisheries Service Alaska Region).
Murakami, T. (2018). Three case studies for control of invasive alien ant species, fire ant (Solenopsis invicta, Formicidae) in Japan. Ketsudan Kagaku 4, 33–42.
Neira, C., Levin, L. A., Grosholz, E. D. (2005). Benthic macrofaunal communities of three sites in San Francisco Bay invaded by hybrid Spartina, with comparison to uninvaded habitats. Mar. Ecol. Prog. Ser. 292, 111–126. doi: 10.3354/meps292111
Okoshi, K. (2007). Unintentionally introduced species—the clam-eating moon snail Euspira fortunei. Nippon Suisan Gakkaishi 73 (6), 1129–1132. doi: 10.2331/suisan.73.1129 (in Japanese
Peakall, R., Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28 (19), 2537–2539. doi: 10.1093/bioinformatics/bts460
Piry, S., Luikart, G., Cornuet, J.-M. (1999). BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 90 (4), 502–503. doi: 10.1093/jhered/90.4.502
Prentis, P. J., Sigg, D. P., Raghu, S., Dhileepan, K., Pavasovic, A., Lowe, A. J. (2009). Understanding invasion history: genetic structure and diversity of two globally invasive plants and implications for their management. Divers. Distrib. 15 (5), 822–830. doi: 10.1111/j.1472-4642.2009.00592.x
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 (2), 945–959.
Provan, J., Murphy, S., Maggs, C. A. (2005). Tracking the invasive history of the green alga Codium fragile ssp. tomentosoides. Mol. Ecol. 14 (1), 189–194. doi: 10.1111/j.1365-294X.2004.02384.x
Pyšek, P., Richardson, D. M. (2010). Invasive species, environmental change and management, and health. Annu. Rev. Environ. Resour. 35, 25–55. doi: 10.1146/annurev-environ-033009-095548
Pyšek, P., Jarošik, V., Hulme, P. E., Pergl, J., Hejda, M., Schaffner, U., et al. (2012). A global assessment of invasive plant impacts on resident species, communities and ecosystems: the interaction of impact measures, invading species’ traits and environment. Glob. Change Biol. 18 (5), 1725–1737. doi: 10.1111/j.1365-2486.2011.02636.x
Qiao, H., Liu, W., Zhang, Y., Zhang, Y.-Y., Li, Q. Q. (2019). Genetic admixture accelerates invasion via provisioning rapid adaptive evolution. Mol. Ecol. 28 (17), 4012–4027. doi: 10.1111/mec.15192
Richardson, D. M. (2011). Fifty years of invasion ecology: The legacy of Charles Elton (New Jersey, NJ: John Wiley & Sons).
Saccaggi, D. L., Karsten, M., Robertson, M. P., Kumschick, S., Somers, M. J., Wilson, J. R. U., et al. (2016). Methods and approaches for the management of arthropod boader incursions. Biol. Invasions 18 (4), 1057–1075. doi: 10.1007/s10530-016-1085-6
Saltonstall, K. (2002). Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc. Natl. Acad. Sci. U.S.A. 99 (4), 2445–2449. doi: 10.1073/pnas.032477999
Schaal, B. A., Gaskin, J. F., Caicedo, A. L. (2003). Phylogeography, haplotype trees, and invasive plant species. J. Hered. 94 (3), 197–204. doi: 10.1093/jhered/esg060
Scholz, H., Chen, C.-W., Jung, M.-J. (2009). Supplements to the Grassess (Poaceae) in Taiwan (II). Taiwania 54 (2), 168–174. doi: 10.6165/tai.2009.54(2).168
Simenstad, C. A., Thom, R. M. (1995). Spartina alterniflora (smooth cordgrass) as an invasive halophyte in Pacific Northwest Estuaries. Hortus Northwest 6, 9–12, 38-40.
Somers, G. F., Grant, D. (1981). Influence of seed source upon phenology of flowering of Spartina alterniflora Loisel. and the likelihood of cross pollination. Am. J. Bot. 68 (1), 6–9. doi: 10.1002/j.1537-2197.1981.tb06349.x
Taberlet, P., Gielly, L., Pautou, G., Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi: 10.1007/BF00037152
Tamaoki, M., Takizaki, Y. (2015). Current status and environmental effects of Spartina spp. in Japan. J. Jap. Soc Water Environ. 38 (2), 61–66. in Japanese with English Abstract.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 (22), 4673–4680. doi: 10.1093/nar/22.22.4673
Vilà, M., Espinar, J. L., Hejda, M., Hulme, P. E., Jarošik, V., Maron, J. L., et al. (2011). Ecological impacts of invasive alien plants: a meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 14 (7), 702–708. doi: 10.1111/j.1461-0248.2011.01628.x
Wan, S., Qin, P., Liu, J., Zhou, H. (2009). The positive and negative effects of exotic Spartina alterniflora in China. Ecol. Eng. 35 (4), 444.452. doi: 10.1016/j.ecoleng.2008.05.020
Wang, X. Y., Shen, D. W., Jiao, J., Xu, N. N., Yu, S., Zhou, X. F., et al. (2012). Genotypic diversity enhances invasive ability of Spartina alterniflora. Mol. Ecol. 21 (10), 2542–2551. doi: 10.1111/j.1365-294x.2012.05531.x
Williams, J. A., West, C. J. (2000). Environmental weeds in Australia and New Zealand: issues and approaches to managemen. Austral Ecol. 25 (5), 425–444. doi: 10.1046/j.1442-9993.2000.01081.x
Xu, G. W., Zhou, R. Z. (1985). Preliminary studies of introduced Spartina alterniflora Loisel in China (I). J. Nanjing Univ. 40 (2), 212–225. in Chinese with English Abstract.
Xu, H., Qiang, S., Han, Z., Guo, J., Huang, Z., Sun, H., et al. (2006). Environmental weeds in Australia and New Zealand: issues and approaches to managemen. Biodivers. Conserv. 15 (9), 2893–2904. doi: 10.1007/s10531-005-2575-5
Keywords: biological invasion, chloroplast DNA, founder effect, genetic structure, microsatellite, secondary introduction, smooth cordgrass, trade history
Citation: Maebara Y, Tamaoki M, Iguchi Y, Nakahama N, Hanai T, Nishino A and Hayasaka D (2020) Genetic Diversity of Invasive Spartina alterniflora Loisel. (Poaceae) Introduced Unintentionally Into Japan and Its Invasion Pathway. Front. Plant Sci. 11:556039. doi: 10.3389/fpls.2020.556039
Received: 27 April 2020; Accepted: 18 August 2020;
Published: 07 September 2020.
Edited by:
Nina Rønsted, National Tropical Botanical Garden, United StatesReviewed by:
Petr Koutecký, University of South Bohemia, CzechiaCopyright © 2020 Maebara, Tamaoki, Iguchi, Nakahama, Hanai, Nishino and Hayasaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisuke Hayasaka, aGF5YXNha2FAbmFyYS5raW5kYWkuYWMuanA=; YXdheW90b3VAaG90bWFpbC5jb20=
†Present addresses: Yu Maebara, Nagoya Branch Office, Nippon Koei Co. Ltd., Aichi, Japan
Yuka Iguchi, Research Division 2, Japan Wildlife Research Center (JWRC), Tokyo, Japan
Atsushi Nishino, Daiichi Fukken Co. Ltd., Fukuoka, Japan
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.