
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 November 2020
Sec. Crop and Product Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.554158
This article is part of the Research Topic Regulation of Fruit Ripening and Senescence View all 13 articles
 Ryohei Nakano1†
Ryohei Nakano1† Takashi Kawai2†
Takashi Kawai2† Yosuke Fukamatsu2‡
Yosuke Fukamatsu2‡ Kagari Akita2
Kagari Akita2 Sakine Watanabe2
Sakine Watanabe2 Takahiro Asano2
Takahiro Asano2 Daisuke Takata3
Daisuke Takata3 Mamoru Sato3
Mamoru Sato3 Fumio Fukuda2*
Fumio Fukuda2* Koichiro Ushijima2*
Koichiro Ushijima2*The postharvest properties of two ultra-late maturing peach cultivars, “Tobihaku” (TH) and “Daijumitsuto” (DJ), were investigated. Fruit were harvested at commercial maturity and held at 25°C. TH exhibited the characteristics of normal melting flesh (MF) peach, including rapid fruit softening associated with appropriate level of endogenous ethylene production In contrast, DJ did not soften at all during 3 weeks experimental period even though considerable ethylene production was observed. Fruit of TH and DJ were treated with 5,000 ppm of propylene, an ethylene analog, continuously for 7 days. TH softened rapidly whereas DJ maintained high flesh firmness in spite of an increase in endogenous ethylene production, suggesting that DJ but not TH lacked the ability to be softened in response to endogenous and exogenous ethylene/propylene. DNA-seq analysis showed that tandem endo-polygalacturonase (endoPG) genes located at melting flesh (M) locus, Pp-endoPGM (PGM), and Pp-endoPGF (PGF), were deleted in DJ. The endoPG genes at M locus are known to control flesh texture of peach fruit, and it was suggested that the non-softening property of DJ is due to the lack of endoPG genes. On the other hand, TH possessed an unidentified M haplotype that is involved in determination of MF phenotype. Structural identification of the unknown M haplotype, designated as M0, through comparison with previously reported M haplotypes revealed distinct differences between PGM on M0 haplotype (PGM-M0) and PGM on other haplotypes (PGM-M1). Peach M haplotypes were classified into four main haplotypes: M0 with PGM-M0; M1 with both PGM-M1 and PGF; M2 with PGM-M1; and M3 lacking both PGM and PGF. Re-evaluation of M locus in association with MF/non-melting flesh (NMF) phenotypes in more than 400 accessions by using whole genome shotgun sequencing data on database and/or by PCR genotyping demonstrated that M0 haplotype was the common haplotype in MF accessions, and M0 and M1 haplotypes were dominant over M2 and M3 haplotypes and co-dominantly determined the MF trait. It was also assumed on the basis of structural comparison of M haplotypes among Prunus species that the ancestral haplotype of M0 diverged from those of the other haplotypes before the speciation of Prunus persica.
Fruit firmness is an important quality that influences consumer preference, damage during distribution, and shelf life. Studies associated with the decrease in fruit firmness after harvest have been conducted with an eye toward reducing distribution loss and prolonging shelf life and thus, supplying high-quality fruit to consumers (Nimmakayala et al., 2016; Moggia et al., 2017; Tucker et al., 2017; Fernandez et al., 2018; Liu et al., 2018; Carrasco-Valenzuela et al., 2019). Fruit can be classified as climacteric or non-climacteric depending on their respiration and ethylene production patterns during ripening (Biale and Young, 1981). In climacteric fruit, ethylene is acknowledged to play an important role in controlling ripening- and senescence-related phenomena including fruit softening, due to the fact that massive ethylene production commences at the onset of ripening; exogenously applied ethylene and/or ethylene analog, propylene, induces ripening and senescence; ethylene inhibitors retard the progress of fruit ripening and senescence; and mutants and transgenic lines defective in ethylene production ability exhibit suppressed fruit ripening, especially softening (Gapper et al., 2013; Minas et al., 2015; Tucker et al., 2017).
Peach [Prunus persica (L.) Batsch] is generally known to belong to the climacteric type and to exhibit dramatic increases in respiration and ethylene production during ripening (Tonutti et al., 1991). In melting flesh (MF) peaches, the increased ethylene stimulates fruit softening principally through cell wall modification (Brummell et al., 2004; Hayama et al., 2006, 2008; Liu et al., 2018). MF peaches are highly perishable, softening rapidly after harvest. The increasing interest in improving peach shelf life has sparked investigations and resulted in findings of peach strains with long shelf lives. Those studies have demonstrated phenotypic variability associated with fruit softening and identified the possible causal genes for peach shelf life, as described below.
Intensively studied peaches that have long shelf lives are the stony-hard (SH) and slow-ripening (SR) peaches (Brecht and Kader, 1984; Haji et al., 2005; Bassi and Monet, 2008). The SH is determined by Hdhd gene and fruit with SH flesh bear the hdhd genotype (Haji et al., 2005). SH peaches are characterized by the absence of ethylene production and high firmness during postharvest storage, which are caused by the reduced expression of ethylene biosynthesis related gene PpACS1 encoding 1-aminocyclopropane-1-carboxylic acid synthase (Tatsuki et al., 2006). YUCCA flavin mono-oxygenase gene PpYUC11, which is involved in the auxin biosynthesis pathway, has been proposed as a candidate for causal gene for this phenotype (Pan et al., 2015; Tatsuki et al., 2018). SR peaches are known to show delayed maturation on the tree, thereby resulting in late harvest. In SR peaches harvested earlier than the optimum harvest date, flesh firmness decreased slowly (Brecht and Kader, 1984). Its genetic base was characterized and a deletion mutation in a gene encoding the NAC transcription factor was reported to be responsible for the SR phenotype (Eduardo et al., 2015; Nuñez-Lillo et al., 2015; Meneses et al., 2016).
Another peach strain that shows high flesh firmness during postharvest ripening is non-melting flesh (NMF) peaches (Fishman et al., 1993; Yoshioka et al., 2011). Whereas MF peaches soften dramatically and bear melting texture during the final stage of ripening called “melting phase,” NMF fruit appear to lack this “melting phase” of softening and remain relatively firm during ripening not only on the tree and but also after harvest (Fishman et al., 1993; Yoshioka et al., 2011). The MF/NMF phenotypes segregate as a single locus (M) that is linked tightly to the stone adhesion locus (Bailey and French, 1949; Monet, 1989). MF is dominant over NMF and the recessive allele determines the NMF character (Bailey and French, 1949; Monet, 1989). In NMF peaches, solubilization of cell wall pectin and enzymatic activity and protein accumulation of endo-polygalacturonase (endoPG), a pectin hydrolase, are markedly reduced compared with MF peaches (Pressey and Avants, 1978; Fishman et al., 1993; Lester et al., 1996; Yoshioka et al., 2011). Studies aimed at demonstrating endoPG(s) as a candidate gene for M locus have shown suppressed or undetectable expression of endoPG(s) in NMF peaches and polymorphisms in endoPG genes coinciding with MF/NMF phenotypes (Lester et al., 1994, 1996; Callahan et al., 2004; Peace et al., 2005, 2007; Morgutti et al., 2006, 2017; Gu et al., 2016). M locus is located at 3.5 cM interval on the bottom of linkage group 4 of the peach map, the position within which a genomic region with clusters of endoPG genes exists (Cao et al., 2016; Gu et al., 2016). Two tandem endoPG genes in that region, Pp-endoPGM (PGM) and Pp-endoPGF (PGF), corresponding to sequences Prupe.4G262200 in v2.0 of peach genome (ppa006857m in v1.0) and Prupe.4G261900 (ppa006839m in v1.0), respectively, were found to be responsible for determining the MF/NMF phenotypes (Gu et al., 2016). Gu et al. (2016) proposed a scenario where M locus has three allelic copy number variants of endoPG genes designated by H1 (possessing PGF and PGM), H2 (only PGM), and H3 (null). Accessions harboring either H1 and/or H2 haplotype (H1H1, H1H2, H1H3, H2H2, and H2H3) exhibit MF phenotype whereas those harboring homozygous recessive H3 (H3H3) show NMF phenotype (Gu et al., 2016). It was also speculated that H2 is the ancestral haplotype whereas H1 and H3 haplotypes are two variants due to the duplication and deletion of PGM, respectively, (Gu et al., 2016). However, research on different NMF peach germplasms suggested that mutations in endoPG gene(s) could be of more than one type arising from more than one source (Lester et al., 1996; Callahan et al., 2004; Peace et al., 2005; Morgutti et al., 2006, 2017) and some NMF accessions seemed to be incompatible with the model proposed by Gu et al. (2016). Much more comprehensive evaluation of M locus in association with flesh textural traits is required. Pursuing M locus evolution with much broader genetic resources covering Prunus species is also necessary for precise judgment.
Peach is known to have high diversity with regard to not only flesh texture but also fruit maturation date (Elsadr et al., 2019). In Japan, MF peaches reaching maturation stage in early July through September are mainly produced. Recently, because of the increasing demand for fresh peach in late autumn, ultra-late maturing cultivars whose optimum harvest dates are October and November are gathering the attention of growers. However, the postharvest properties of some of these rare cultivars have not yet been characterized.
In this study, first, two postharvest properties, namely, ethylene production and fruit softening, of two extremely late harvest cultivars, “Tobihaku” (TH) and “Daijumitsuto” (DJ), were investigated. It was revealed that TH showed normal MF peach ripening properties, whereas DJ possessed unique properties in that the fruit did not soften at all in spite of significant endogenous ethylene production and exogenous propylene treatment. Second, DNA-seq analysis of these cultivars demonstrated that DJ was a homozygote of an allele lacking PGM and PGF at M locus, whereas TH possessed a previously structurally unidentified haplotype that contained one endoPG. Third, accessing database sequences at M locus in more than 400 peach accessions and Prunus species indicated that the newly identified haplotype was an important allele that distributed widely within MF accessions, determined MF phenotype, and seemed to have been diverse from the other haplotypes before the speciation of P. persica. The scenario is discussed in which not three but four allelic variants at M locus are associated with the flesh texture and the newly identified haplotype is one of the two dominant determinants of MF texture.
Ultra-late maturing peach (Prunus persica) cultivars “Tobihaku” (TH) and “Daijumitsuto” (DJ), whose genetic backgrounds are unknown, were examined. TH fruit were harvested on November 7, 2018, the commercial harvest date, from two trees grown in a commercial orchard in Okayama Prefecture located in southwestern Japan. DJ fruit were harvested on October 12, 2018, the commercial harvest date, from two trees grown in the Research Farm of Okayama University. For each cultivar, 26 fruit without any disease and injuries were selected and used for investigation described below. As regards DJ, 16 fruit from a different production area, namely, Fukushima Prefecture, which is located in northeastern Japan, were harvested on October 22, 2018, the commercial harvest date, and 12 fruit without any disease and injuries after transfer to Okayama were used to investigate the effects of growing conditions and harvest maturity. Climate conditions in the peach production areas, which were obtained from the website of the Japan Meteorological Agency1, are listed in Supplementary Table S1. Regardless of cultivar or growing area, fruit were grown under suitable climate conditions for peach production by skilled growers using conventional growing techniques in Japan, including fruit thinning and bagging by the end of June. The fruit were harvested on the basis of skilled growers’ visual evaluation using de-greening of fruit ground color as harvest index. It is equivalent to color chip No. 3 (color meter score of L: 76.8, a∗: −8.16, b∗: 29.7) of de-greening harvest index reported in Yamazaki and Suzuki (1980). After harvest, 16 out of the 26 fruit of TH and DJ harvested from Okayama Prefecture were ripened at 25°C for 3 weeks and ethylene production rate, flesh firmness, soluble solids content (SSC), and juice pH were measured on days 0, 7, 14, and 21 at 25°C. DJ fruit harvested from Fukushima Prefecture were packed carefully in a corrugated cardboard box and transported by vehicle at ambient temperature to Okayama University 2 days after harvest, where 12 fruit without any disease and injuries were ripened at 25°C.
Ten out of 26 fruit of TH and DJ harvested from Okayama Prefecture were used for experiment with continuous propylene treatment. Six fruits per each cultivar, excluding those used for measurement of values at day 0, were treated with 5,000 ppm of propylene, an ethylene analog, continuously for 7 days at 25°C, as described previously (McMurchie et al., 1972; Hiwasa et al., 2004). Ethylene production rates and flesh firmness were measured on treatment days 0, 3, and 7.
Ethylene production rate, flesh firmness, SSC, and juice pH were measured as described previously (Kawai et al., 2018; Nakano et al., 2018). For the measurement of ethylene production rate, individual fruit were incubated in a 1.3 L plastic container at room temperature for 30 min. Headspace gas withdrawn from the container was injected into a gas chromatograph (GC8 CMPF; Shimadzu, Kyoto, Japan) equipped with a flame ionization detector (set at 200°C) and an activated alumina column (ϕ 4 mm × 1 m) set at 80°C. For flesh firmness, the cheek parts of each fruit were cut and peeled, and flesh penetration force was measured using a rheometer (FUDOH RTC Rheometer; RHEOTECH, Tokyo, Japan) with a 3-mm-diameter cylindrical plunger and expressed in Newton per plunger area (N/mm2). The relationships between the flesh penetration force measured by this system and fruit maturity indexes are listed in Supplementary Table S2. SSC and juice pH in the cheek parts of each fruit were measured with a refractometer (PR-1; Atago, Tokyo, Japan) and a pH meter (B-712; HORIBA, Kyoto, Japan), respectively.
Three to four fruit were used as biological replicates at each measurement point. In order to evaluate postharvest changing patterns in flesh firmness and ethylene production in different cultivars or production areas under the condition with and without propylene, Tukey’s multiple comparison test was conducted after one-way ANOVA. The different letters shown in each figure indicate significant differences among measurement days by Tukey’s test (p < 0.05).
Genomic DNA was isolated from the leaves of DJ, TH, and “Benihakuto” (BH) by Nucleon PhytoPure (Cytiva). DNA-seq analysis was performed by Novogen and 9G data of PE150 reads, corresponding to approx. 30 times coverage, were obtained. We further searched SRA (Sequence Read Archive) database to obtain whole genome shotgun sequencing (WGS) data for doubled haploid “Lovell” (dhLL), “Dr. Davis” (DD), and “Big Top” (BT). Illumina WGS reads were mapped to peach reference genome (ver. 2.0) (Verde et al., 2017) by CLC Genomics Workbench or minimap2 (Li, 2018). SNP, indel, and structural variants were called by CLC Genomics Workbench.
DNA-seq analysis of BH was further performed by Macrogen to obtain 46G data of PE150 reads, which correspond to approx. 170 times coverage of P. persica genome. Illumina reads were assembled by ABySS 2.0 (Jackman et al., 2017). Contigs encompassing M locus were detected by Blastn analysis using PG1, PG2, PGM/F, NADH, and F-box genes, which were located in the M locus region, as query. M0 and M1 haplotype sequences (see section “Results”) were compared by nucmer (Kurtz et al., 2004) and their relationship was drawn by Circos2. We generated new reference sequence set “PpREF20 + M0,” in which the M0 haplotype sequence was added to P. persica reference genome. Illumina short reads of DJ, TH, BH, dhLL, DD, and BT were mapped to PpREF20 + M0 by minimap2. Coverage was analyzed from BAM file by samtools and drawn by Circos.
Genomic DNA was extracted from leaves of peach accessions. Genotyping by PCR was conducted with six sets of primers described in Supplementary Table S3. PCR was performed with BIOTAQ DNA Polymerase (Bioline, United Kingdom) using the following program: 30–35 cycles at 95°C for 20 s, annealing for 15 s, and extension at 72°C an initial denaturation at 95°C for 3 min, and a final extension at 72°C for 7 min. Annealing temperature and extension time are described under “PCR conditions” in Supplementary Table S3. PGM/F products were separated on a 15% acrylamide gel and others were separated on an agarose gel. PCR products were stained with UltraPower DNA Safedye (Gellex International Co., Ltd., Japan).
Postharvest changes in ethylene production rate, flesh firmness, SSC, and juice pH were investigated in two ultra-late maturing peach cultivars, TH and DJ. In TH, as much as 22 nl⋅g–1⋅h–1 of ethylene was produced at harvest and the amount increased gradually during storage at 25°C (Figure 1A). Flesh firmness was 0.7 N/mm2 at harvest but decreased dramatically to less than 0.2 N/mm2 by day 7 at 25°C (Figure 1D). Thereafter, the decrease became slight until the last day of experiment (day 21). SSC and juice pH did not change remarkably during storage (Supplementary Figure S1). In DJ harvested from the Research Farm of Okayama University on October 12, ethylene production rate was almost negligible at harvest. Thereafter, ethylene production rate increased and peaked on day 7, reaching more than 10 nl⋅g–1⋅h–1 (Figure 1B). Flesh firmness was around 0.85 N/mm2 at harvest (Figure 1E). In spite of considerable ethylene production, flesh firmness showed no significant decrease during storage at 25°C and was almost unchanged until the last day of experiment (day 21). SSC slightly increased during storage, reaching a peak of 18 °Brix on day 14, and juice pH was maintained at around pH 4.0 during storage (Supplementary Figure S1).
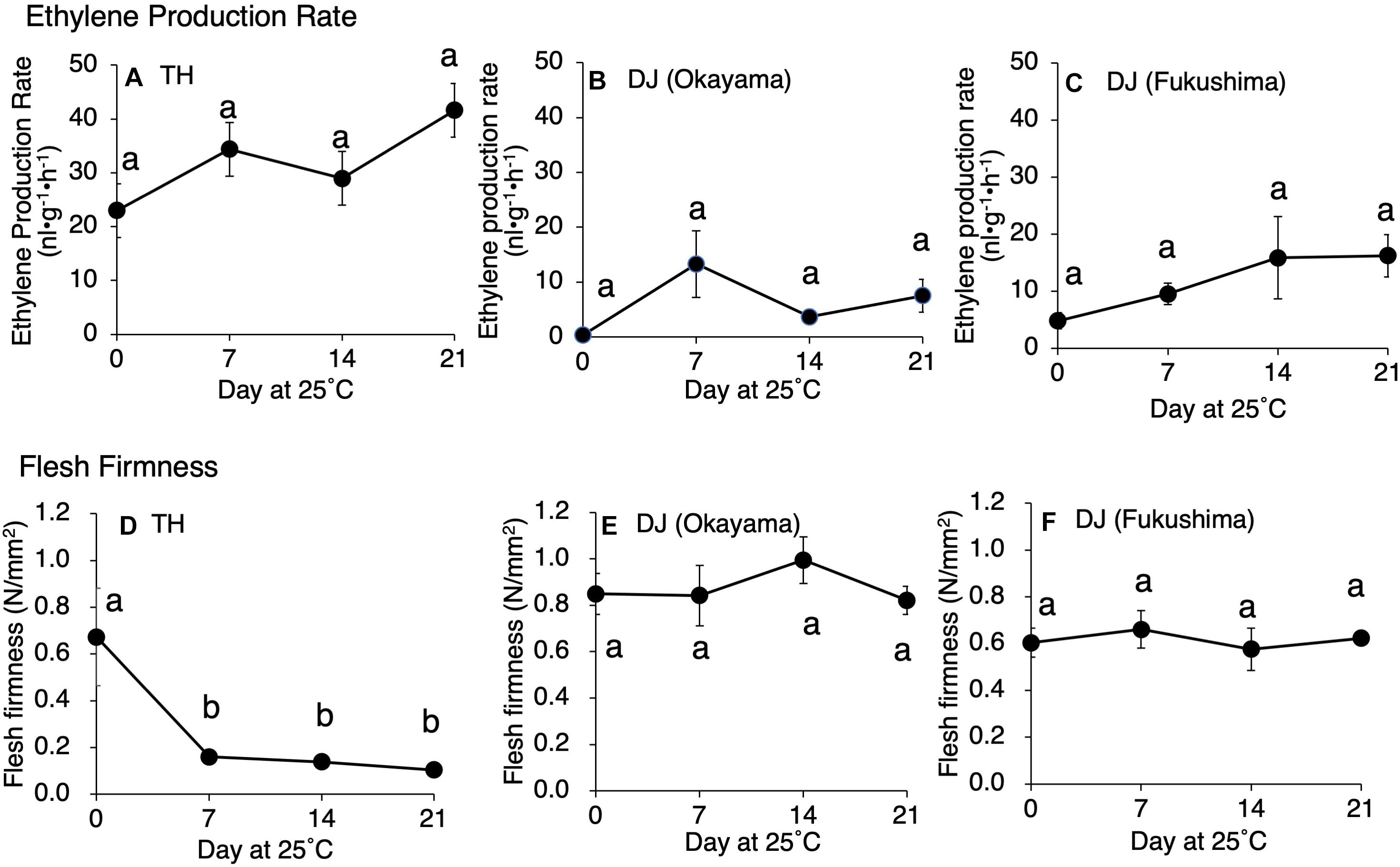
Figure 1. Postharvest changes in (A–C) ethylene production rate and (D–F) flesh firmness in “Tobihaku” (TH) and “Daijumitsuto” (DJ) fruit grown in Okayama Prefecture and DJ grown in Fukushima Prefecture Japan. In (A,D), TH were harvested on November 7 from a commercial orchard in Okayama Prefecture, Japan and held at 25°C for 21 days. In (B,E), DJ from Okayama were harvested on October 12 from the Research Farm of Okayama University, Japan and held at 25°C for 21 days. In (C,F), DJ from Fukushima were harvested on October 22 from a commercial orchard in Fukushima Prefecture, Japan, followed by 2 days transport at ambient temperature to Okayama University, where fruit were held at 25°C for 21 days. Fruit were harvested at commercial maturity. Each point in (A,B,D,E) and in (C,F) represents the mean value of three and four fruits, respectively. Vertical bars indicate ± SE (n = 3–4). Statistical analysis was conducted by Tukey’s multiple comparison test after one-way ANOVA. Different letters indicate significant differences among measurement days by Tukey’s test (p < 0.05).
To confirm that the unique characteristics of DJ were not due to climatic effects and/or misestimated harvest maturity, DJ fruit harvested from a different production area were investigated. In the case of DJ harvested from a commercial orchard in Fukushima Prefecture on October 22, fruit delivered to Okayama University two days after harvest had an ethylene production rate of as high as 5.2 nl⋅g–1⋅h–1 and flesh firmness of 0.6 N/mm2 (Figures 1C,F). Substantially high level of ethylene production was maintained during storage at 25°C, scoring 16 nl⋅g–1⋅h–1 on day 21. On the other hand, flesh firmness did not change significantly during storage; flesh firmness on day 21 was almost the same as that at harvest. SSC and juice pH did not change remarkably during storage (Supplementary Figure S1). Figure 2 shows DJ fruit on day 14. The external appearance showed no significant deterioration; however, the longitudinal sections showed flesh browning and slight flesh breakdown around the stones.
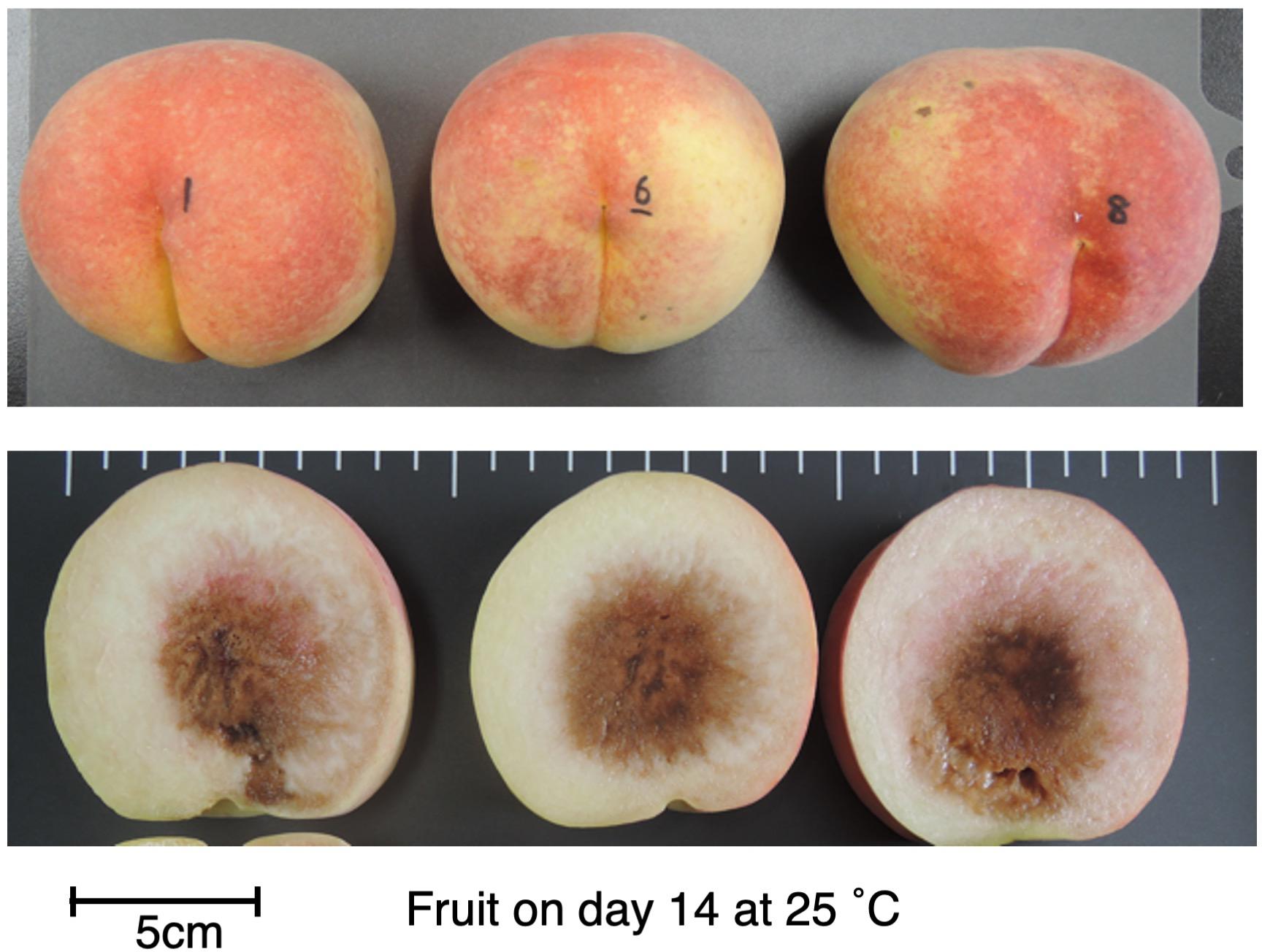
Figure 2. DJ on day 14 at 25°C. Fruit were harvested from a commercial orchard in Fukushima Prefecture and held at 25°C for 21 days, as shown in Figure 1. Appearance (top) and longitudinal section (bottom) of each fruit are shown. Bar indicates 5 cm.
Although ethylene production rate during storage was higher in TH than DJ, generally speaking, the amount of ethylene produced in DJ is sufficient to induce physiological effects on fruit ripening including flesh softening (Mathooko et al., 2001; Hiwasa et al., 2004; Nishiyama et al., 2007; Hayama et al., 2008; Liu et al., 2018). In order to confirm that the non-softening characteristic of DJ is not due to the low ethylene production rate of this cultivar, TH and DJ were treated with 5,000 ppm of propylene, an ethylene analog, continuously for 7 days. This concentration of propylene is equivalent to 50 ppm of ethylene and is sufficient to induce ethylene response in climacteric fruits (Burg and Burg, 1967; McMurchie et al., 1972). Ethylene production rates of TH and DJ were increased rapidly by the propylene treatment, reaching 45 and 22 nl⋅g–1⋅h–1 on day 3 of treatment, respectively (Supplementary Figure S2). Flesh firmness of TH decreased rapidly whereas that of DJ decreased slightly, and high firmness was maintained in DJ even after 7 days of continuous propylene treatment (Figure 3). These results suggest that regardless of the differences in the ethylene production rate, DJ lacked the ability to be softened in response to ethylene.
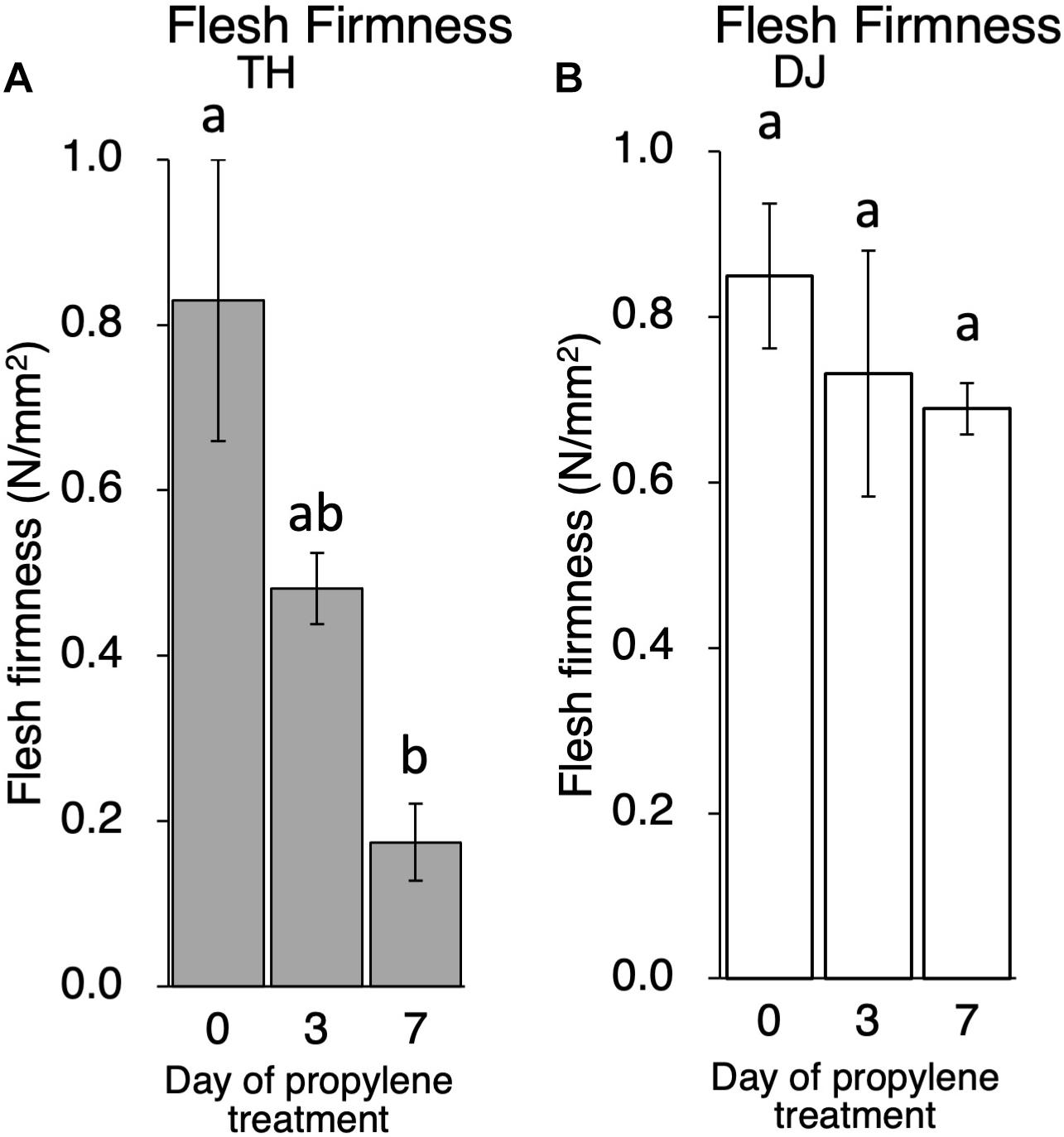
Figure 3. Effect of propylene treatment on postharvest fruit softening in (A) TH and (B) DJ. Harvested fruit were treated with 5,000 ppm of propylene continuously for 7 days. Flesh firmness was measured on days 0, 3, and 7. For DJ, fruit harvested on October 12 from the Research Farm of Okayama University were used. Each point on day 0, 3 and 7 represents the mean value of four and three fruits, respectively. Vertical bars indicate ± SE (n = 3–4). Statistical analysis was conducted by Tukey’s multiple comparison test after one-way ANOVA. Different letters indicate significant differences among measurement days by Tukey’s test (p < 0.05).
M locus is involved in the regulation of peach flesh texture (Bailey and French, 1949; Monet, 1989; Bassi and Monet, 2008). Four polygalacturonase genes, PG1, PG2, PGM, and PGF; three NADHs; and one F-box gene were annotated in the M locus region of peach reference genome (Figure 4A). Gu et al. (2016) walked the chromosome of three accessions by using PCR based on sequence information of reference genome, and proposed three haplotypes, H1, H2, and H3. The sequence of H1 haplotype was identical to reference genome, in which tandem duplication of endoPG genes, PGM and PGF, was observed. H2 haplotype possessed only PGM and lacked PGF. The 70 kbp region containing PG2, PGM, PGF, and NADHs was deleted in H3 haplotype. Gu et al. (2016) hypothesized that H1 and H2 haplotypes were dominant over H3 haplotype, PGM conferred the melting texture, and PGF, which was present only in H1 haplotype, was involved in freestone phenotype. The lack of both PGM and PGF in H3 homozygote could result in the non-melting texture. Further investigations are required to confirm the function of PGM in H2 and H1 haplotypes and the roles of PGM and PGF in fruit trait.
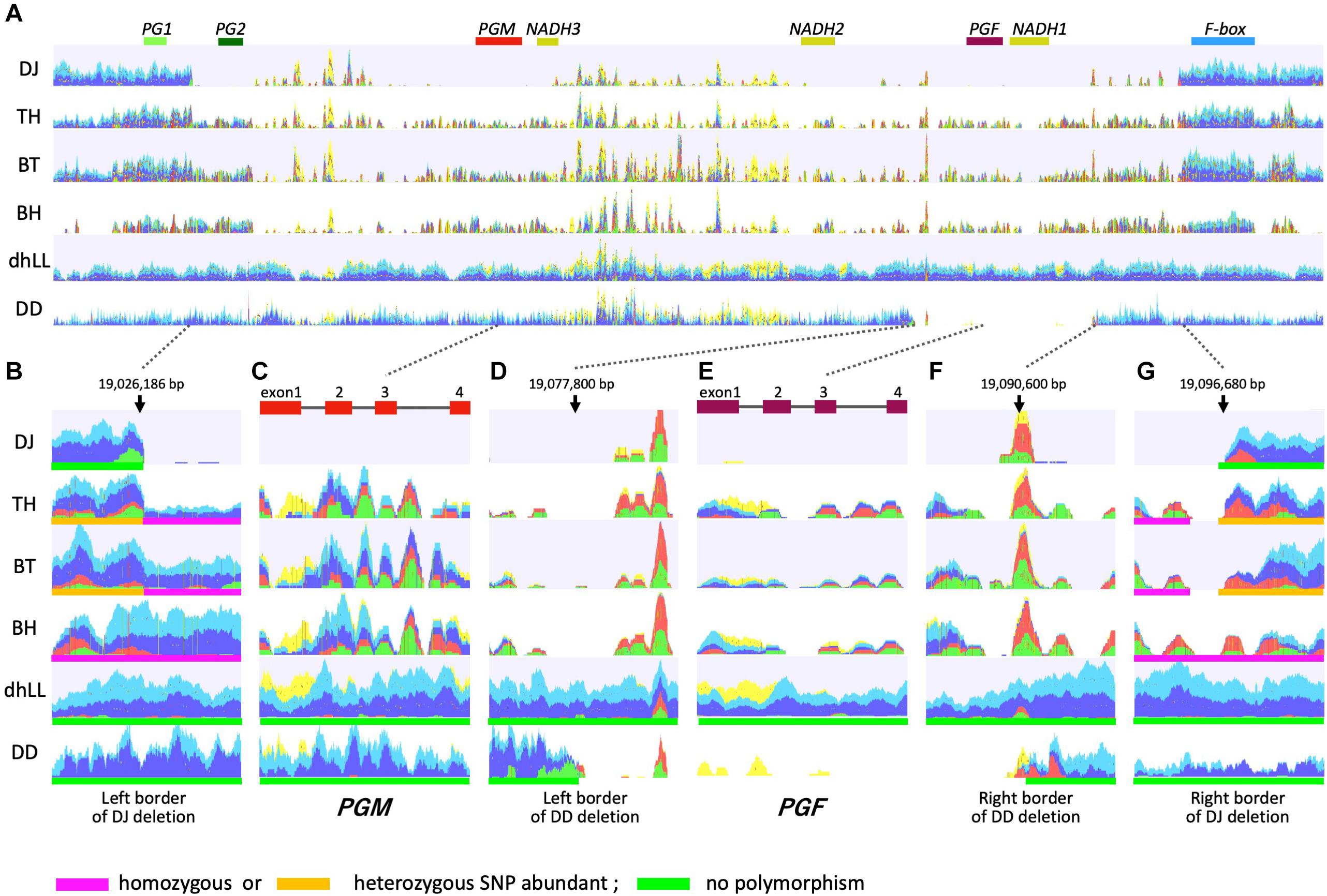
Figure 4. Mapping DNA-seq reads to M1 haplotype. Illumina short reads of DJ, TH, “Benihakuto” (BH), “Big Top” (BT), “Dr. Davis” (DD), and doubled haploid “Lovell” (dhLL) were mapped to reference genome of P. persica (ver 2.0) (Verde et al., 2017) by CLC Genomics Workbench. Properly paired reads that are in the correct orientation and distance are shown in blue and light blue. Green and red reads are broken pairs. Non-specific matches are shown in yellow. (A) Overview of mapping graph in the M locus region. PG1 to F-box above graph are genes named according to Gu et al. (2016). (B,G) show the region around the junction of M3 deletion. (D,F) are those of M2 deletion. (C,E) are PGM and PGF, respectively.
WGS of DJ and TH by Illumina was performed to investigate the M locus region (19.0–19.1 Mbp of chromosome 4) (Figure 4), and the mapping patterns were compared with those of three accessions with different flesh textures: dhLL of MF accession, DD of NMF accession, and BT of MF accession, which is categorized in slow softening peaches (Bassi and Monet, 2008; Ghiani et al., 2011; Morgutti et al., 2017). dhLL, an H1 homozygote, was used for peach genome sequencing and the reads were mapped uniformly to M locus. The reads of DD were also mapped to the entire M locus region except the ∼12 kbp region including PGF and NADH1. No or less reads were mapped to the ∼12 kbp region, indicating that the region was deleted in DD (Figures 4D,F). The position and length indicated that the DD deletion unexpectedly corresponded to the region deleted in H2 haplotype in Gu et al. (2016). The mapping patterns of DJ and TH were not the same as those of dhLL and DD, suggesting that DJ and TH possessed neither H1 nor H2 haplotype.
TH and BT showed similar mapping patterns and thus were expected to possess the same haplotypes (Figure 4). The DJ mapping results indicated that the genotype of DJ was not identical to those of TH and BT, but the three accessions were expected to share one haplotype. Mapping the pair reads of DJ to peach reference genome revealed many broken pairs around 19,026,186 and 19,096,680 bp (Figures 4A,B,G). Only a few reads were mapped between them, except for the many non-specific reads from other chromosomal regions that were mapped between NADH3 and NADH2, the region named M1 insertion in this study (see below). Structural mutation analysis by CLC Genomics Workbench indicated that the region from 19,026,186 to 19,096,680 bp was deleted in DJ, suggesting that DJ is homozygote for the deleted haplotype identical to that reported as H3 by Gu et al. (2016). Many broken pairs were found at the same position as DJ when mapped with TH and BT pair reads, indicating that one of the two haplotypes was H3 (Figures 4A,B,G). The other haplotype was presumed to be an unidentified novel haplotype that cannot be predicted directly by mapping to reference genome. In TH and BT, SNPs were heterozygous in the outer region of H3 deletion, whereas they were homologous in the inner region of H3 deletion (Figures 4B,G and Supplementary Table S4). All SNPs in this region were homozygous in BH, a Japanese MF accession. In contrast, no SNP was detected in either dhLL or DD. In the region of PGM and PGF, the coverage of TH, BT, and BH was low and many SNPs and broken pair reads were observed (Figures 4C,E). Throughout M locus and its surrounding region, SNPs were hardly observed in dhLL, DD, and DJ, whereas many mutations were detected in TH, BT, and BH (Figure 4 and Supplementary Table S4), suggesting the existence of an unidentified haplotype in TH, BT, and BH.
Sequence polymorphism and mapping pattern indicated that the sequence and structure of the unidentified haplotype of TH, BT and BH was highly diverged from those of the reference sequence (M1). We decided to obtain the unidentified haplotype sequence by de novo assembly. However, polymorphism shown in the M locus of TH had a negative effect on de novo assembly. As we found that BH was a homozygote of the unknown haplotype, de novo assembly with PE150 reads of BH was conducted and two contigs covering M locus were obtained. PG1, PG2, PGM, and NADH were located in one of the two contigs. The F-box gene was found in another contig. The haplotype composed of the two contigs was defined as M0 according to the locus name. Haplotypes H1, H2, and H3 in Gu et al. (2016) were renamed M1, M2, and M3, respectively, based on haplotype features, to avoid confusion and define the haplotypes precisely.
When the genome structure of M0 was compared with M1 sequence, the region corresponding to PGF was not found in M0 and only one NADH was located on M0 (Figure 5). The sequences were conserved among the two haplotypes but M1 specific sequences were found in the region from PG2 to PGM and the downstream region of NADH3. Many non-specific reads were mapped to the region spanning from NADH3 to NADH2 of M1 haplotype, the region corresponding to M1 insertion described above (Figures 4, 5). The M1 insertion was assumed to be translocated from another chromosomal region and inserted into NADH to disrupt it, because both NADH3 and NADH2 were partial and their structures appeared to be generated from one NADH gene divided at the third intron by the M1 insertion (Supplementary Figures S3, S4 and NADH3/2 of Supplementary Figure S5).
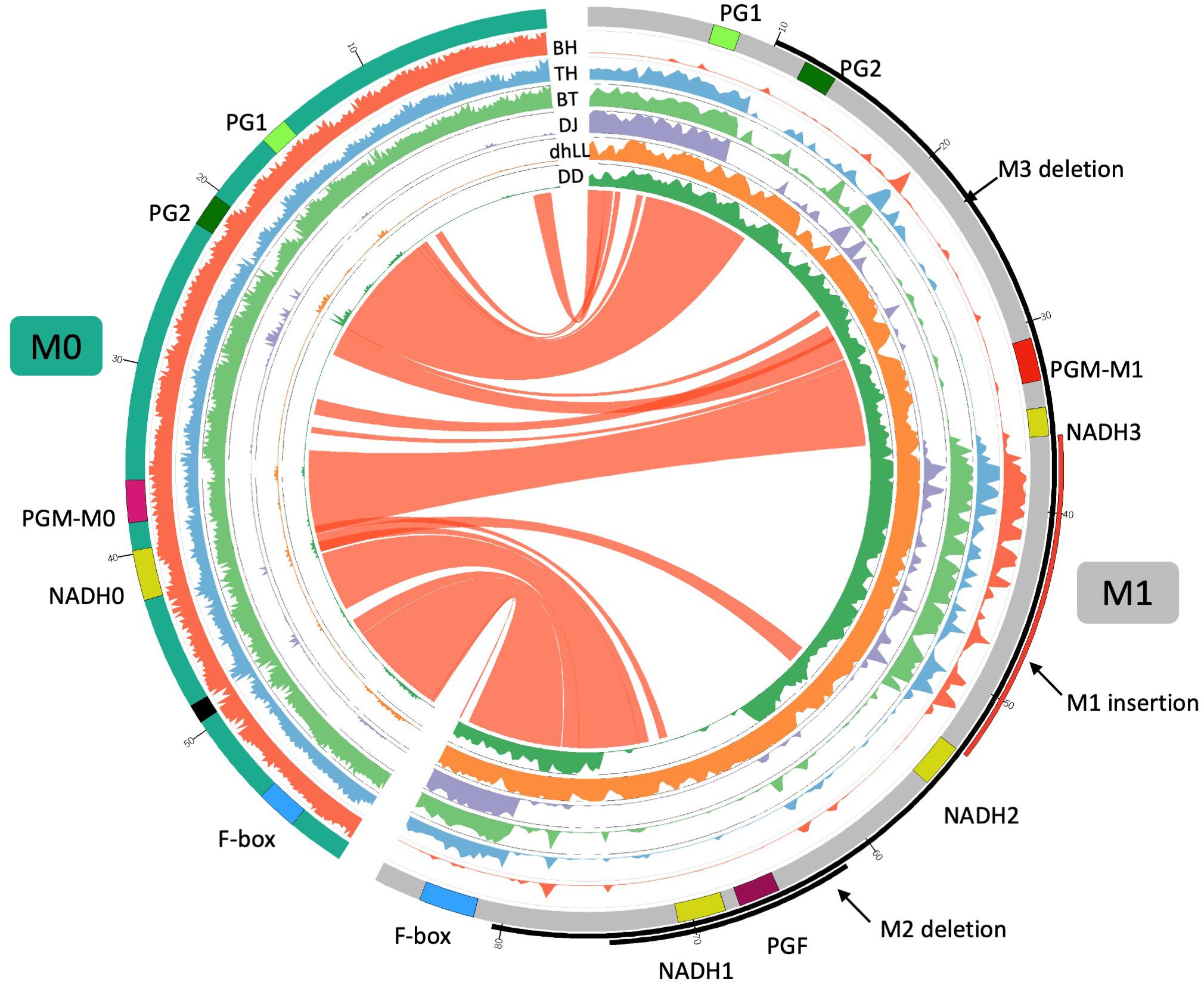
Figure 5. Comparison of M0 and M1 haplotypes. Inner six tracks are coverage graphs of DNA-seq reads of BH TH, BT, DJ, dhLL, and DD. Ribbons link similar regions between two haplotypes. M0 haplotype consists of two contigs connected by N (black in the chromosome track). The nucleotide at 19,016,188 bp of Pp04 chromosome is positioned as + 1 in M1 haplotype. PGM of M0 haplotype is similar to PGM of M1 but not identical. We named those PGMs PGM-M0 and PGM-M1, respectively. NADH0 is NADH of M0 haplotype and seems to be an intact gene as described in Supplementary Figures S3–S5.
The CDS sequence of PGM of M0 (PGM-M0) showed 99% similarity to those of PGM of M1 (PGM-M1) and PGF of M1. One amino acid substitution between PGM-M0 and PGM-M1 was found at residue 49, where Ser was substituted to Phe in PGM-M1 (Supplementary Figure S6). The Ser at residue 49 of PGM-M0 was conserved in other Prunus species. Amino acid sequence comparison between PGM-M0 and PGF showed substitution of Ser for Thr at residue 269 in PGF, although it was not considered to have a significant effect on the function of the PG protein. The S49F substitution between f and fl alleles was reported by Peace et al. (2005) and Morgutti et al. (2017). However, because they did not perform resequencing analysis, it was not possible to understand the entire structure of the f haplotype and its origin. It was probable that f haplotype was the same as M0 in this study. We further demonstrated that this M0 haplotype is not a specific haplotype but a widely found haplotype in various peach accessions, and plays an important role in MF phenotype determination.
DNA-seq reads of BH, TH, DJ, dhLL, DD, and BT were mapped to the reference “PpREF20 + M0,” in which the M0 haplotype sequence was added to peach genome sequence (Figure 5). Coverage graphs show that the reads of M0, M1, M2, and M3 haplotypes could be classified clearly. The reads of BH (M0M0) were mapped only to M0 haplotype and those of dhLL (M1M1) were also mapped only to M1 haplotype. DJ (M3M3) and DD (M2M2) reads were mapped to M1 haplotype except for the deleted region, as shown in Figure 4. The reads of TH (M0M3) and BT (M0M3) were mapped to both M0 and M1 haplotypes except for M3 deletion.
It was demonstrated that WGS data were useful for the precise genotyping of M locus. Many WGS data of peach accessions were registered in the SRA database and mapped to “PpREF20 + M0” to identify M genotype. We finally predicted M genotypes of 412 accessions from the WGS mapping pattern and/or by PCR genotyping. M0 and M1 haplotypes were the most popular in the peach accessions analyzed (Figure 6 and Supplementary Table S5). On the other hand, the frequency of M2 haplotype was very low. In addition to the four main haplotypes, M0, M1, M2, and M3, we found variant-type haplotypes and chimeric haplotypes. The former haplotypes exhibited deletion in the region different from those found in M2 and M3, whereas the latter haplotypes appeared to be generated by the recombination between M0 and M1 (Figure 6). In total, 11 haplotypes were structurally identified at M locus (Figure 7). They were first classified into M0 to M3 on the basis of the existence of PGM-M0, PGM-M1, and PGF; the haplotype containing only PGM-M0 is M0; the haplotype containing PGM-M1 and PGF was M1; the haplotype containing only PGM-M1 was M2; and the haplotype containing neither PGM nor PGF was M3. Furthermore, variants and recombinant types of haplotypes were identified from structural variations and such characters as b, c… or r1, r2… were added to their names, respectively.
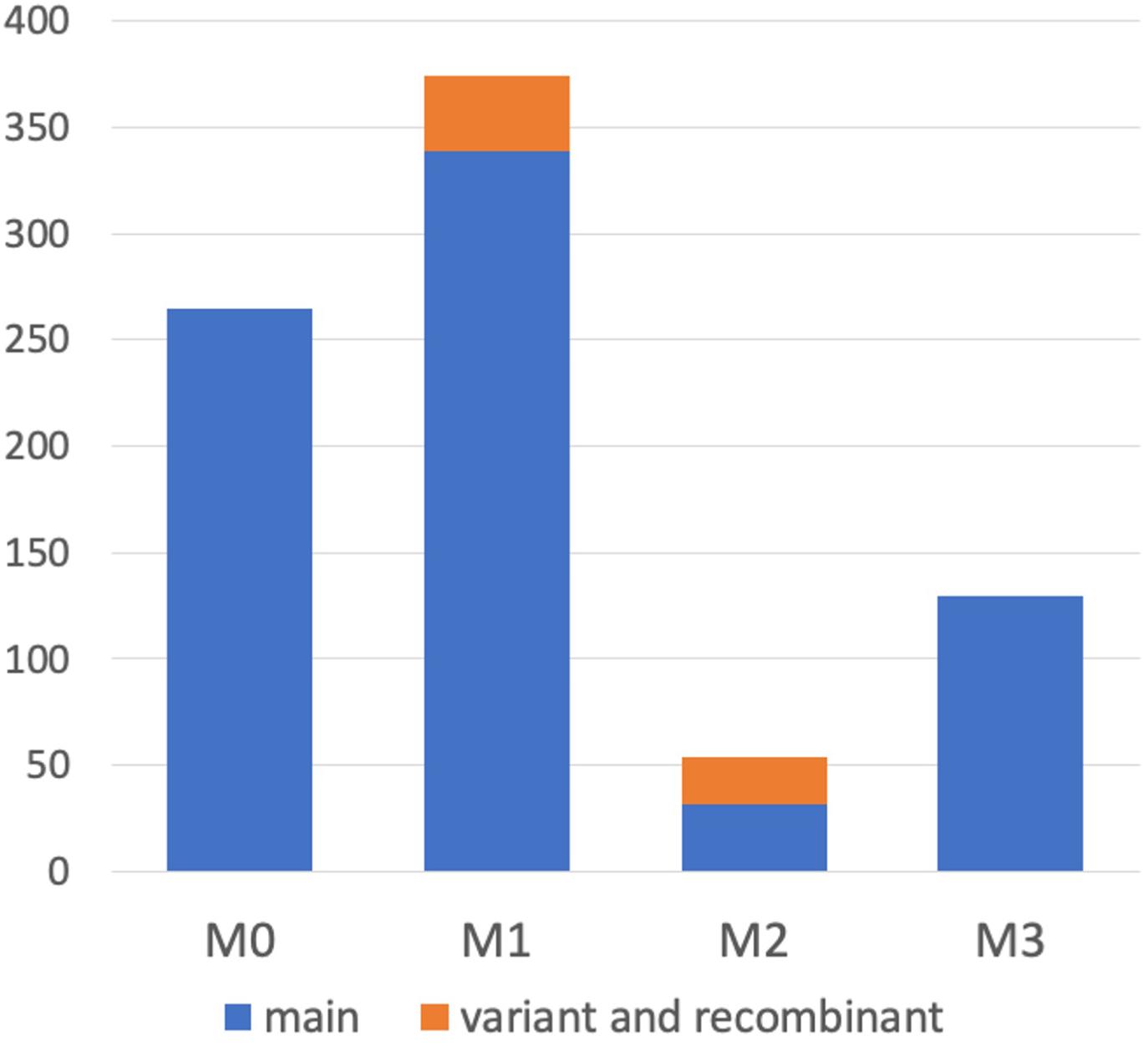
Figure 6. Haplotype frequency at M locus in peach accessions. We classified M haplotypes into 11 haplotypes (Figure 7) in 412 peach accessions, on the basis of the coverage graph patterns and/or PCR genotyping (Figure 8 and Supplementary Figure S9).
Although no significant structural change in gene composition was detected, the sequence variation was found in the accessions with M0 haplotype, when analyzed on the basis of the mapping patterns of DNA-seq reads of TH. The insertion, whose length was unknown, was found in the 1.8 kbp upstream region of PGM-M0, and SNPs were also detected in PGM-M0, one of which was located in CDS and led to a synonymous substitution (Supplementary Figure S7). It was expected that M0 was also diversified. However, we regarded both types of M0 as M0 in this study because no changes in gene composition (structural feature as haplotype) and no amino acid substitutions were found.
We designed primers for PCR genotyping on the basis of the sequence variations at the third intron of PGM and PGF and the M haplotype structural differences (Figure 8, Supplementary Figure S8, and Supplementary Table S3). The structural variations and the PGM and PGF compositions of M haplotypes identified in this study indicated that the four main haplotypes could be classified using three primer sets, PGM/F, M2D, and M3D. PGM/F primer set detected the indel at the third intron of PGM and PGF, and three different fragment sizes were amplified (Figure 8). The upper fragment was derived from PGM-M1, the lower one was from PGF, and the middle one was from PGM-M0. When the middle fragment was amplified, the accession possessed M0 haplotype. From M1 haplotype, both the upper and the lower fragments should be amplified. When the lower fragment was not amplified and only the upper one was amplified, this meant that only PGM-M1 was amplified, showing that the accession had M2 haplotype. The genotyping results obtained with the PGM/F primer set could be confirmed by amplification with the M2D primer set that detects the deletion on M2. Because no fragment was amplified from M3 with the PGM/F primer set, the M3D primer set, which amplifies the junction of M3 deletion, should be useful to identify M3 haplotype.
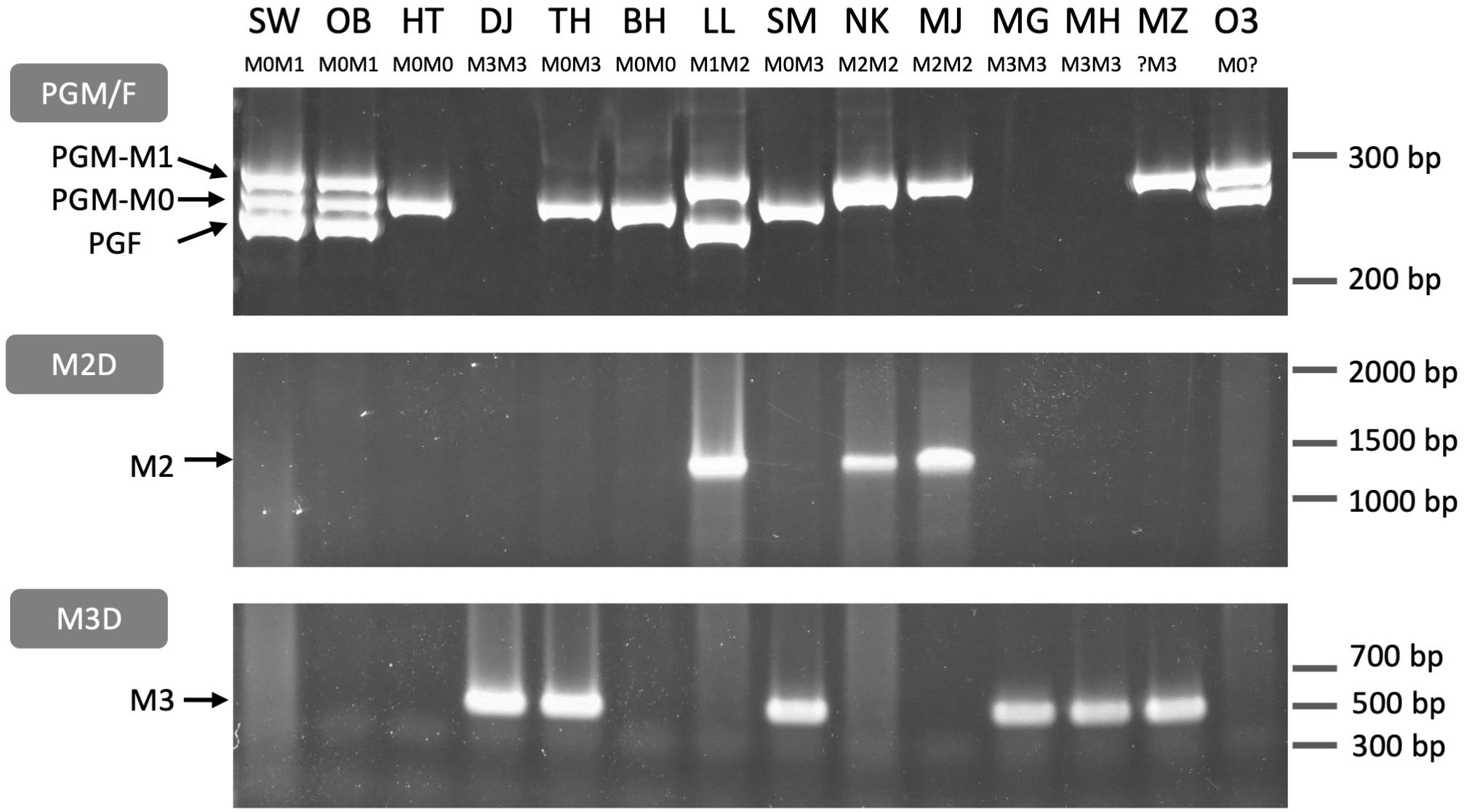
Figure 8. PCR genotyping of M locus. Three primer sets PGM/F, M2D, and M3D (Supplementary Figure S8 and Supplementary Table S3) were used to distinguish M haplotypes. PGM/F and M2D/M3D fragments were separated on an acrylamide gel and an agarose gel, respectively. PGM/F primer set amplified three fragments derived from PGM-M1, PGM-M0, and PGF, respectively. M2D and M3D detected M2 and M3 haplotypes, respectively. All genotypes under each cultivar name except MZ and O3 were identified from these fragment patterns (see Supplementary Figure S8). SW, SunagoWase; OB, Okubo; HT, Hakuto; DJ, Daijumitsuto; TH, Tobihaku; BH, BeniHakuto; LL, Lovell; SM, ShimizuHakuto; NK, Nishiki; MJ, Myojo; MG, Meigetsu; MH, Meiho; MZ, Mochizuki; O3, Okayama-3.
The three primer sets were used to genotype 14 accessions. The M genotypes of six accessions, “Okubo” (OB), DJ, TH, BH, LL, and “Myojo” (MJ), were predicted from the mapping patterns of WGS data, which were re-confirmed by PCR genotyping. In the other eight accessions, we found inconsistent amplification patterns in “Mochizuki” (MZ) and “Okayama-3” (O3). These two accessions were expected to have M2 haplotype judging from the result that the PGM/F primer set amplified PGM-M1 but not PGF. However, no amplification was observed in M2D. These results suggested that M2 haplotype of MZ and O3 was a variant type of M2. Indeed, an additional primer set showed that they have M2b (Supplementary Figure S9).
“Hakuto” (HT), a progeny of “Chinese Cling” (CC), was mainly used as germplasm for MF peach breeding in Japan (Supplementary Figure S10A; Yamamoto et al., 2003a,b). PCR genotyping showed that HT was a homozygote of M0, indicating the possibility that M0 haplotype had been spread among Japanese peaches. All the major Japanese cultivars tested in this study were found to have M0 haplotype (Supplementary Figure S10B). All cultivars except OB and “ShimizuHakuto” (SM) were homozygotes of M0. SM was a typical MF cultivar in Japan (Yamamoto et al., 2003b), and its genotype was M0M3, the same as that of TH (Figure 8).
DJ was an M3 homozygote and did not have PGM and PGF. Thirty seven M3 homozygotes were identified in this study. Flesh textural phenotypes of 16 out of 37 accessions were reported, and 12 accessions were reported as NMF except those whose flesh texture was only reported as Supplementary Data in Cao et al. (2016) (Table 1). This may support our conclusion that M3 haplotype was likely involved in the determination of non-softening postharvest property in DJ. M2 haplotype, in which only PGM-M1 was located, also appeared to confer the NMF phenotype, unlike M0 and M1 haplotypes. In this study, we found 22 homozygotes for M2 including variant types, 11 of which had fruit textural report(s), and all 11 were reported as NMF (see reference in Table 1). Only NJF16 accession had M2M3 combination as well as phenotype report. NJF16 was reported to be NMF peach (Clark and Finn, 2010). MZ was M2bM3 that had only PGM-M1 as confirmed by PCR genotyping, and is a well-known Japanese NMF cultivar (Yoshioka et al., 2011). All together, these findings suggest that PGM-M1 may not be functional. On the other hand, PGM-M0 and PGF seemed to confer the MF phenotype dominantly because flesh texture of M0M2 (“Tsukuba 86”), M0M2b (O3), M0M3 (TH, SM, BT, and CC), M1M2 (LL), and M1M3 [“Georgia Bell” (GB)] was MF (Table 1). “Early Gold” (EG) was identified as M1M1 on the basis of the mapping pattern of reads from the SRA database (Supplementary Table S6), despite that the flesh texture was considered NMF (Yoshida, 1981). We suspected that the reads of EG registered in SRA were confused with those of the other accessions. This is because “Nishiki” (NK) (M2M2), an NMF accession, was the parent of EG (Yoshida, 1981) and one of EG haplotypes was supposed to be M2.
The structure of M locus was identified from reference genome sequences of other Prunus species, including P. mira, P. kansuensis, almond (P. dulcis), apricot (P. armeniaca), Japanese apricot (P. mume), sweet cherry (P. avium), and Yoshino cherry (P. x yedoensis; called “Sakura” in Japan), and compared with the four main haplotypes of peach structurally identified in this study (Figure 9 and Supplementary Figure S11). PGM and PGF were found in the “Lauranne” genome (PdLN) of almond, which belongs to subgenus Amygdalus together with peach, indicating that the PdLN M haplotype was similar to peach M1 haplotype. As shown in M1 of peach, NADH located downstream of PGM in PdLN was broken by an insertion sequence, although this insertion sequence was not similar to the M1 insertion. F-box and NADH were pseudogenes, probably due to multiple genome rearrangements occurring in almond “Texas” genome (PdTX). One endoPG was found in addition to PG1 and PG2, and it was expected to be PGF, because PGFs in PdLN and PdTX shared amino acid substitutions specific to them and genomic sequence similarity was found not only in the gene region, but also in the intergenic region flanking them. A frameshift mutation was found in PdLN-PGF, indicating that PdLN-PGF did not function. The absence of this mutation in PdTX suggests that the mutation in PdLN-PGF occurred after the haplotype diverged. No tandem duplications of PGM, PGF, and M1 insertions were observed in P. mira or P. kansuensis. NADH appeared to be a fragmented structure in P. kansuensis, but because disrupted exons were located at different contigs in the draft genome, their actual relationship was unclear. PGM, PGF, and NADH were not present in P. mira genome.
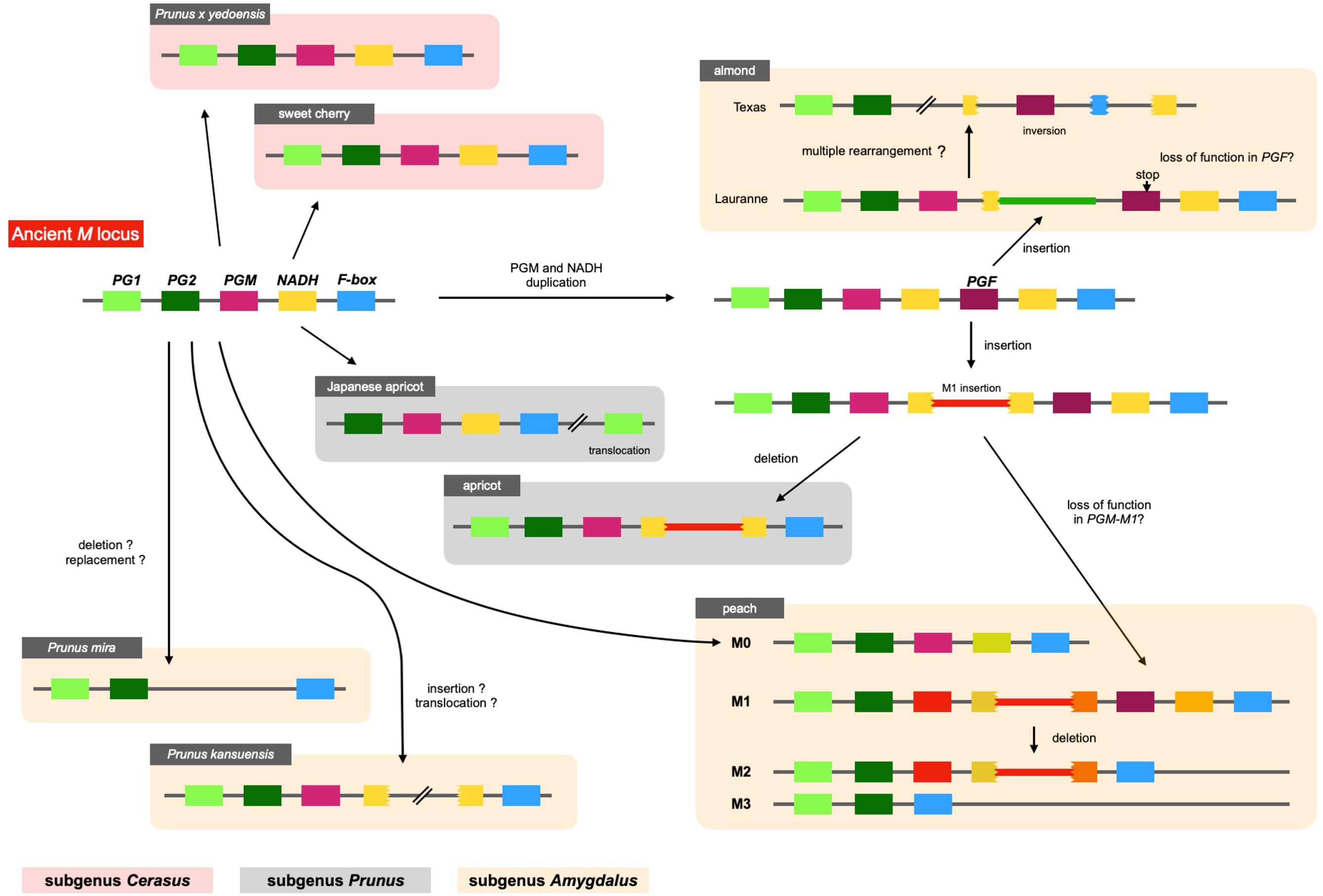
Figure 9. Diversification of M haplotypes in genus Prunus. We investigated M loci of Prunus species by Blastn analysis using their reference genomes. Five genes, PG1, PG2, PGM, NADH, and F-box genes, were shared among Prunus and speculated to be located at an ancient M haplotype. The structure of the ancient haplotype was conserved in peach M0, P. kansuensis, Japanese apricot, sweet cherry, and P. x yedoensis. No PGM and NADH were observed in P. mira. M1 type structure, which possessed PGF or M1 insertion, was found in peach M1 and M haplotypes of almond and apricot. This haplotype was speculated to diverge from ancient haplotype by duplication of PGM (and NADH), followed by M1 or almond insertion, which disrupted NADH gene located downstream of PGM.
In subgenus Amygdalus, the M1 insertion was found only in peach. However, a sequence similar to the M1 insertion was found in apricot of subgenus Prunus. The insertion also disrupted NADH, which was located downstream of PGM. These findings implied that the two insertions originated from the same event (haplotype), and that this insertion event occurred before the divergence of subgenera Amygdalus and Prunus. A structure similar to M0 was found in Japanese apricot (P. mume) of subgenus Prunus. PG2, PGM, intact NADH, and F-box were clustered in P. mume haplotype, but PG1 was translocated to the downstream region to M locus. Sweet cherry and Yoshino cherry belong to subgenus Cerasus, which is far from subgenus Amygdalus (Chin et al., 2014). Their M haplotype structure and gene composition were similar to those of peach M0 haplotype.
In this study, first, the postharvest properties of ultra-late maturing peach cultivars TH and DJ were investigated. Generally, peach is considered to be a climacteric fruit in which ripening-related phenomena, such as accelerated endogenous ethylene biosynthesis and fruit softening, were reported to be controlled by ethylene (Liguori et al., 2004; Hayama et al., 2006; Liu et al., 2018). TH exhibited the characteristics of normal MF peach fruit, including rapid fruit softening leading to MF texture associated with appropriate level of endogenous ethylene production. In contrast, DJ did not soften at all even though significant ethylene production was observed. From their sugar contents and pH-values at day 0, it was confirmed that DJ fruit used in this study were not harvested at too early maturity stage to be ripen normally (Supplementary Figure S1). It seemed that DJ possessed the ability to produce ripening-related ethylene but lacked the ability to be softened in response to the ethylene. The lack of softening ability in response to ethylene in DJ was supported by the continuous propylene treatment. Propylene, instead of ethylene, treatment has been used to monitor endogenous ethylene production in parallel to other ripening-related changes (McMurchie et al., 1972). As much as 5,000 ppm of propylene was used for treatment in this study. According to previous reports, this concentration of propylene is equivalent to 50 ppm of ethylene and is sufficient to induce ethylene response in climacteric fruits (Burg and Burg, 1967; McMurchie et al., 1972). Indeed, in peach fruit treated with 500–5,000 ppm of propylene, induction of autocatalytic ethylene production and dramatic fruit softening were reported (Yoshioka et al., 2010; Liu et al., 2018). In this study, DJ exhibited only a slight decrease in flesh firmness and maintained almost similar firmness to that at harvest even after 7 days of continuous propylene treatment. As TH showed dramatic softening by the propylene treatment, it is suggested that the propylene treatment in this study is capable of inducing ethylene response in peach and that DJ has a non-softening characteristic in response to both endogenous and exogenous ethylene/propylene.
The genetic background of DJ and TH and its relationships with other cultivars having long storability and/or shelf lives are unknown. One of the peach strains reported to have a long shelf life is SH peach (Haji et al., 2005). SH peaches, however, are characterized by the absence of ethylene production (Tatsuki et al., 2006). It is reported that ethylene sensing is normal in SH peaches and the fruit soften rapidly with exogenous ethylene or propylene treatment (Haji et al., 2003; Yoshioka et al., 2010), in contrast to DJ. A similar ripening characteristic observed in DJ has been reported in early harvest fruit of SR peaches (Brecht and Kader, 1984). In SR peaches harvested at an earlier date than the optimum harvest date, autocatalytic ethylene production was induced with or without propylene treatment whereas flesh firmness decreased quite slowly. However, DJ is phenotypically different from SR peaches in that it bears large fruit with red coloration, as shown in Figure 2, whereas SR fruit do not show normal ripening in terms of fruit size and coloration (Giné-Bordonaba et al., 2020). In agreement with these phenotypical differences between DJ and SH and/or SR peaches, the genomic sequences of DJ and TH did not exhibit any significant differences and/or mutations in the candidate causal genes for these specific strains, YUCCA flavin mono-oxygenase (Pan et al., 2015; Tatsuki et al., 2018) and NAC transcription factor (Eduardo et al., 2015; Nuñez-Lillo et al., 2015; Meneses et al., 2016) genes (data not shown).
On the other hand, significant differences between DJ and TH were found with regard to genomic sequences at M locus, which has been reported to control MF and NMF textures (Bassi and Monet, 2008; Gu et al., 2016). M locus is composed of two tandem endoPG genes, PGM and PGF (Gu et al., 2016). Resequencing analysis revealed that DJ is a homozygote of the haplotype that lacks both PGM and PGF, designated as M3 in this study, whereas TH is a heterozygote of M3 and a structurally uncharacterized haplotype that possesses PGM (PGM-M0) but not PGF, designated as M0 in this study (Figures 4, 5). Many reports have demonstrated that high level of enzymatic activity, protein accumulation, and gene expression of endoPG are observed only in MF, and supported the involvement of endoPG in the determination of the flesh texture (reviewed in Bassi and Monet, 2008). Thus, it is considered that DJ is a member of NMF peaches. NMF peaches are known to be firm at maturity and to soften slowly during ripening without melting. Different from DJ, it was reported that softening progressed steadily during postharvest ripening in NMF peaches (Fishman et al., 1993; Yoshioka et al., 2011) and thus, the possible involvement of a number of factors in the non-softening property of DJ other than the lack of endoPG genes cannot be excluded. Nevertheless, it was indicated that the non-softening postharvest property of DJ is attributed to the lack of endoPG genes at M locus. The rapid softening property of TH is due to the presence of newly characterized M0 haplotype (PGM-M0 gene) in TH genome. This was further confirmed by the re-evaluation of M haplotype in relation to flesh textural phenotypes in 412 accessions, as described below.
In this study, we revealed that M0 was not only a unique M haplotype as shown in TH, but also a widely spread haplotype in MF accessions, particularly popular peach cultivars grown in Japan, and was responsible for the MF phenotype in these accessions (Figure 6, Supplementary Figure S10B, and Supplementary Table S5). Based on the results obtained from the re-evaluation of M locus in 412 accessions in relation to flesh textural traits, we proposed the scenario in which four M alleles/haplotypes, M0 to M3, were involved in the determination of flesh texture, with M0 and M1 dominantly controlling MF texture over M2 and M3.
Various alleles/haplotypes have been proposed for M locus. The correspondence of M alleles/haplotypes in previous studies to those in this study is summarized in Supplementary Tables S7, S8. Peace et al. (2005) classified endoPGs at M locus into four alleles, F, f1, f, and null, on the basis of the results obtained from germplasm derived from peach cultivars GB and DD with f allele being hypothesized to be segregated via outcross from an unknown origin. Morgutti et al. (2017) assumed the same four haplotypes F, f1, f, and fnull by referring to Peace’s classification and further found two variations, PGSH and PGBT, in f haplotype. The haplotype structures of H1, H2, and H3 reported by Gu et al. (2016) corresponded to those of F, f1, and fnull alleles, respectively, although the haplotype corresponding to f was not reported by Gu et al. (2016). In this study, we structurally identified four main haplotypes M0 to M3. Judging from S49F substitution and indel at the third intron detected between PGM-M0 and PGM-M1, M0 appeared to be identical to f haplotype. However, the derivation of f(M0) haplotype assumed in this study was different from that in Morgutti et al. (2017), in which f (M0) was expected to be derived from F(M1) via fl(M2). We detected sequence diversifications and large structural differences between M0 and M1/M2 and assumed that M0 was not derived from M1/M2 directly. This assumption was supported by a structural comparison of M loci of other Prunus species (Figure 9). A similar specific structure to peach M1, such as M1 insertion and/or tandem duplication of endoPG, was found in M haplotypes of almond and apricot. On the other hand, there was no insertion to disrupt NADH in peach M0 or M haplotypes of the Japanese apricot, sweet cherry, and P. x yedoensis. These findings suggested that the ancestral haplotypes of M0 and M1 diverged relatively early, before subgenus divergence, and evolved independently of each other. On the other hand, we could not find any SNP-level variation among M1, M2, and M3 (Supplementary Table S4), suggesting that the divergence of M1 into M2 and M3 occurred relatively recently after P. persica speciation. It was also suggested that M2 did not lead to M1 but rather M2 was derived from M1. This might be supported by the fact that the frequencies of M2 and M3 were much lower than that of M1 at least in the accessions investigated in this study (Figure 6). In this study, we showed 11 haplotypes in total (Figure 7), but more haplotypes are expected to exist. We only examined reference genomes in Prunus species other than P. persica. Considering the divergence of M haplotype before speciation, it would not be surprising to find other species harboring haplotypes similar to both M0 and M1 haplotypes.
Gu et al. (2016) did not consider the presence of PGM-M0 (f allele) and defined H2 as the sole haplotype harboring PGM but not PGF because they attempted to distinguish each haplotype on the basis of copy number of endoPG genes quantified by qPCR. Therefore, not only M2 but also M0 was genotyped as H2 in Gu et al. (2016). This misgenotyping of M0 as H2 produced results that included incongruity between genotype and phenotype as M0 (PGM-M0) and M2 (PGM-M1) were likely to have different effects on flesh texture. For example, the genotyping in Gu et al. (2016) identified that SM and “Hakuho” (HH), both of which are popular MF cultivars in Japan, were H2H3 and H2H2, respectively. Supposing PGM-M1 on H2 is not functional, as assumed in this study, SM and HH should be NMF. Conversely, supposing H2 (M2) is a dominant haplotype that determines MF texture, as assumed by Gu et al. (2016), the H2H2 genotype shown in DD, “OroA” (OA), and MZ cannot explain their NMF phenotypes (Peace et al., 2005; Morgutti et al., 2006, 2017; Yoshioka et al., 2011).
Re-evaluation of M locus in association with MF/NMF phenotypes in this study revealed that M0 and M1 were likely to function dominantly over M2 and M3. To our knowledge, M0M3 was linked to MF, as shown in TH, SM, and BT. LL (M1M2) was also reported to exhibit MF phenotype (Font i Forcada et al., 2013), whereas NJF16 (M2M3) and MZ (M2bM3) had NMF phenotype (Clark and Finn, 2010; Yoshioka et al., 2011; Table 1). Although PGM-M1 was present in M2, its expression level seemed to be suppressed as reported in OA, whose genotype was determined as M2M2 in this study (Morgutti et al., 2006). Thus, the low expression of PGM-M1 was consistent with the feature of M2, namely, recessive against M1 and M0, and comparable to M3. The hypothesis cannot be excluded that M2 haplotypes in NMF accessions are specific haplotypes possessing additional mutation(s) that result in the disruption of M2 function. The sequences of PGM-M1 and its surrounding region on M2 and M1 haplotypes were identical with each other. As it was predicted that M2 was generated from M1 by deletion of the region including PGF, it seemed reasonable to consider that PGM-M1 had lost its function before the emergence of M2 haplotype and, thus M2 haplotype in general was not functional. This prediction was supported by Morgutti et al. (2017), who reported that all accessions harboring flfl (M2M2) or flfnull (M2M3) exhibited NMF phenotype, as well as previous reports showing the existence of accessions exhibiting NMF phenotype but not completely lacking endoPG genes (Lester et al., 1994, 1996; Callahan et al., 2004; Peace et al., 2005; Morgutti et al., 2006).
It was hypothesized that two tandem endoPG genes at M locus, PGM, and PGF, were responsible for peach flesh texture regulation (Gu et al., 2016). In this study, we suggested that PGM-M0 and PGF in particular would affect flesh textural quality whereas PGM-M1 would have no effect. Considering the lack of sequence diversification and the relatively recent divergence between M1 and M2, it would not be possible that PGM-M1 on M1 retains its function. Although 11 haplotypes were characterized in this study, it might not be necessary to identify correctly all the haplotypes in order to estimate flesh textural quality in a breeding program. Only an analysis to confirm the presence of PGM-M0 and PGF should be sufficient. This means we only need to test whether the PGM/F primer set (Figure 8 and Supplementary Figures S9, S10B) amplifies PGM-M0 or PGF fragments to estimate MF/NMF phenotypes in individual accessions and progenies.
M locus is strongly linked to freestone/clingstone (F) locus. It seems that PGM-M0 is not responsible for the determination of F trait because most M0M0 and M0M3 accessions have clingstones. Gu et al. (2016) hypothesized that not PGM but only PGF on H1 (M1) haplotype is associated with the freestone phenotype. We re-evaluated the relationship between M genotypes and reported freestone/clingstone phenotypes. We found some accessions harboring M1 haplotype but being reported to have not freestone but clingstone phenotype (data not shown). Further studies are required to unravel the role of PGF in the regulation of stone adhesion.
The classification of M haplotypes based on genomic structure and the re-evaluation of M locus in association with flesh melting traits in this study are expected to provide valuable information for studies on controlling fruit softening and textural quality. For example, BT is a well-known cultivar having slow softening behavior, but reaches MF texture at full maturity (Bassi and Monet, 2008; Ghiani et al., 2011). The M genotype of BT was found to be M0M3 in this study, which was the same as that of SM, a famous Japanese MF cultivar that softens rapidly (Yamamoto et al., 2003b). Therefore, we suggest that the slow softening behavior of BT is controlled by loci other than M locus. Even with the re-evaluated genotypes in this study, there are few incongruities between M genotypes and MF/NMF phenotypes (Supplementary Table S6). These incongruities may be due to different conditions and definitions for phenotyping flesh texture among experiments and/or specific mutation(s) that occurred in particular accessions, as well as the involvement of other loci. Further studies addressing the reason for these incongruities are expected to uncover mechanism(s) that determine flesh texture and fruit softening and to improve our understanding of factors related to long shelf life.
We found that two ultra-late maturing cultivars, DJ and TH, showed different postharvest properties. DJ did not soften at all during ripening in spite of significant ethylene production, whereas TH showed rapid fruit softening leading to MF texture. Resequencing analyses of DJ and TH demonstrated that DJ was a homozygote of M haplotype designated as M3 and lacked two tandem endoPG genes, PGM and PGF, at M locus. On the other hand, TH was a heterozygote of M3 and a structurally unidentified haplotype designated as M0 that consisted of only PGM-M0 and was responsible for determining MF texture. Further classification of M haplotypes in 412 peach accessions revealed four main haplotypes: M0; M1 consisting of PGM-M1 and PGF; M2 consisting of PGM-M1 and M3; and M0 was widely spread among MF accessions. We proposed the scenario that combinations of M0 to M3 determined flesh texture, and M0 and M1 dominantly controlled MF texture over M2 and M3. These suggested the possibility that PGM-M0 and PGF could confer MF phenotype and PGM-M1 of M1 and M2 haplotypes may have lost its function. This scenario was supported by the evolution history of each M haplotype assumed from the structural features of M locus in Prunus species, in which the ancestral haplotypes of M0 and M1 diverged before subgenus divergence and evolved independently of each other, whereas M2 and M3 were assumed to be derived from M1 in recent age by deletion mutations.
The datasets analyzed for this study can be found in the DDBJ Sequenced Read Archive database (https://www.ddbj.nig.ac.jp/dra/index-e.html), accession numbers DRR248809-DRR248811 and DRR249197-DRR249201. The contigs A and B of M0 haplotype will appear in the DDBJ/EMBL/GenBank databases under the accession numbers LC592228 and LC592229, respectively.
RN, TK, KU, and FF designed the study and drafted the manuscript. YF, DT, and MS performed sampling and phenotyping. RN, TK, YF, KA, and SW investigated postharvest ethylene production and flesh firmness. KU, RN, TK, and TA analyzed the genomic sequences of peach accessions and determined the genotypes of M locus in various peach accessions. All authors have contributed to manuscript revision and have read and approved the submitted version.
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid for Scientific Research No. 18H02200 to RN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.554158/full#supplementary-material
Supplementary Figure 1 | Soluble solids contents and juice pH in postharvest TH and DJ. Postharvest changes in (A–C) soluble solids content and (D–F) juice pH in TH and DJ fruit grown in Okayama Prefecture and DJ grown in Fukushima Prefecture Japan. In (A,D), TH were harvested on November 7 from a commercial orchard in Okayama Prefecture, Japan and held at 25°C for 21 days. In (B,E), DJ from Okayama were harvested on October 12 from the Research Farm of Okayama University, Japan and held at 25°C for 21 days. In (C,F) DJ from Fukushima were harvested on October 22 from a commercial orchard in Fukushima Prefecture, Japan, followed by 2-day transport at ambient temperature to Okayama University, where fruit were held at 25°C for 21 days. Fruit were harvested at commercial maturity. Each point in (A,B,D,E) and in (C,F) represents the mean value of three and four fruits, respectively. Vertical bars indicate ± SE (n = 3–4). Statistical analysis was conducted by Tukey’s test after one-way ANOVA. Different letters indicate significant differences among measurement days by Tukey’s multiple comparison test (p < 0.05).
Supplementary Figure 2 | Ethylene production in propylene treated TH and DJ. Effect of propylene treatment on postharvest ethylene production in (A) TH and (B) DJ fruit. Harvested fruit were treated with 5,000 ppm of propylene continuously for 7 days. Ethylene production was measured on days 0, 3, and 7. For (B) DJ, fruit harvested on October 12 from the Research Farm of Okayama University were used. Each point on days 0, 3, and 7 represents the mean value of four and three fruits, respectively. Vertical bars indicate ± SE (n = 3–4). Statistical analysis was conducted by Tukey’s multiple comparison test after one-way ANOVA. Different letters indicate significant differences among measurement days by Tukey’s test (p < 0.05).
Supplementary Figure 3 | Amino acid sequence comparison of PGM-M0, PGM-M1, and PGF. Amino acid sequences of PGM-M0, PGM-M1, and PGF of peach and PGM/F of almond “Texas” (PdTX), “Lauranne” (PdLN), P. kansuensis (Pkan), apricot (Parm), Japanese apricot (Pmum), sweet cherry (Pavi), and P. x yedoensis (Pyed) were aligned by CLC Genomics Workbench. Red and blue arrowheads denote amino acid substitution in PGM-M1 and PGF, respectively. The C-terminal region of PdLN_PGF was truncated because of the frameshift at third exon. Only one PGM/F was found in reference genome of almond “Texas,” but it could be orthologous to PGF of reference genome of “Lauranne.” PGFs of “Texas” and “Lauranne” shared some amino acid substitutions that were not conserved in PGM of “Lauranne.”
Supplementary Figure 4 | Schematic representation of NADH genes. We compared the gene structures of NADH0–3, NADH0/2 of M2r1, and NADH3/1 of M2b. Boxes are exons. Gray exons are gene-specific sequences. Other region sequences are homologous to other genes. Sequence comparison with Arabidopsis NADH indicated that the original NADH could be composed of five exons as shown in NADH3/1 of M2b and NADH of Japanese apricot (Supplementary Figures S5, S6).
Supplementary Figure 5 | Amino acid sequence comparison of NADH. Putative amino acid sequences of peach NADHs and Arabidopsis NADH (AtNADH; AT3G03080) were aligned by CLC Genomics Workbench.
Supplementary Figure 6 | Comparison of NADH gene structures among Prunus species. Gene structures of 11 NADHs from eight Prunus species were compared. Considering exon composition, NADH3 and NADH2 of peach were regarded as one gene (NADH3/2), although they were annotated as different genes in reference genome. Pkan NADH gene was divided into different contigs whose linkages were unknown. Disrupted structures were found in five NADHs: Pkan_NADH, NADH3/2, Parm_NADH, PdLN_NADH3/2, and PdTX_NADH. The others were likely intact structures. The M1 insertion was found at third intron of NADH3/2 and Parm_NADH.
Supplementary Figure 7 | Nucleotide sequence comparison of PGM-M0 and -M0b. Sequence comparison showed six mutations in PGM of TH (designated as PGM-M0b in this figure). Furthermore, a large insertion was predicted at the upstream region of PGM-M0b. Of the six mutations, only one was located in CDS region and it was a synonymous substitution. AAC and AAT at 346–348 encode Asn. Therefore, we did not discriminate PGM-M0 and PGM-M0b in this study. In the study of Morgutti et al. (2017), PGM-M0b was detected by CAPS analysis using BstXI. This restriction enzyme site was caused by the nucleotide substitution at 348 bp of M0b haplotype and the fragment from PGM-M0 was expected to be insensitive to BstXI. PGM-M0b could confer the MF phenotype because flesh texture was melting in both TH (M0bM3) and BT (M0bM3). Morgutti et al. (2017) proposed four alleles, PG_M, PGm, PGSH, and PGBT, at M locus from OA (M2M2), “Bolero” (M1M1), “Yumyeong” (M0M0), “Ghiaccio” (M0M0), and BT (M0bM03). Based on Figure 7, our classification suggested that PG_M and PGm were derived from M1 or M2 haplotype and these were designated as PGM-M1 and PGF, respectively, in this study (Supplementary Table S7). PGSH and PGBT could correspond to PGM-M0 and PGM-M0b, respectively. These might be strictly different alleles because of the nucleotide substitution, but flesh texture was melting in both SM (M0M3) and TH (M0bM3), suggesting that their effects on flesh texture were the same and they were not different functionally.
Supplementary Figure 8 | Primers for PCR genotyping. Three primer sets were designed for the discrimination of four main haplotypes, M0, M1, M2, and M3. (A) Primer position at M1 haplotype. (B) Position of PGM/F primer set.
Supplementary Figure 9 | MZ and O3 possessed M2b haplotype. MZ and O3 possessed PGM-M1 and no PGF. This pattern indicated the M2 haplotype, but no amplification to detect the M2 deletion was observed by PCR (Figure 8). In addition to M2 haplotype, we found the same combination in M2b and M2r1 haplotypes, which were rare haplotypes compared with M2. To determine the genotypes of MZ and O3, we designed three primer sets to discriminate M2, M2b, and M2r1 haplotypes on the basis of the sequences of NADH genes (Supplementary Figure S4). M2D2 amplified one fragment from M0, M2b or M2r1 haplotype. NDP1 was expected to amplify two fragments: upper for M0, and lower for M1, M2, M2b or M2r1. NDP2 was also expected to amplify two fragments: upper for M1 or M2b, and lower for M0 or M2r1. The amplification of M2D2 fragment indicated MZ had M0, M2b or M2r1 but not M1 and M2. The lower NDP1 fragment in MZ excluded the possibility of M0. Furthermore, the upper NPD2 fragment was amplified in PCR, indicating that MZ possessed M2b haplotype. O3 also had M2b, but two fragments were amplified in NDP1 and NDP2 because O3 had M0 haplotype (Figure 8).
Supplementary Figure 10 | Peach cultivars bred in Japan shared M0 haplotype. HT, Hakuto; HH, Hakuho; AK, Akatsuki; BH, Benihakuto; YZ, Yuzora; TS, Tosui; KN, KawanakajimaHakuto; OB, Okubo; SM, ShimizuHakuto; O3, Okayama-3; HK, HikawaHakuho; LL, Lovell; CC, Chinese Cling (Shanhai Suimitsuto); TW, Tachibanawase; UNK, unknown cultivar. (A) Genealogy of peach cultivars in Japan. Green cultivars were the five leading cultivars in Japan in 2016 (e-stat Japan, https://www.e-stat.go.jp/). Their cultivation areas accounted for more than 60% of the total peach cultivation area in Japan. Haplotypes in parentheses were presumed from haplotypes of offspring and another parent. (B) PCR haplotyping. CC was imported from China to Japan in the late nineteenth century. HT was reported to be found and selected as a chance seedling of CC in 1899 (Yamamoto et al., 2003a). HT was frequently used as seed parent in breeding programs and many Japanese peach cultivars were the progeny of HT, as described Supplementary Figure S10A. We selected 11 Japanese MF cultivars and carried out PCR genotyping to determine the genotypes of their M loci. All cultivars shared M0 haplotype and all cultivars except OB and SM were M0 homozygous. OB was M0M1 and SM was M0M3. SM was found as a chance seedling at a mixed orchard of HT and O3. SSR analysis supported the hypothesis that SM was a progeny of HT (Yamamoto et al., 2003a). Because of male sterility of HT, O3 had been regarded as the pollen parent of SM. M0 haplotype of SM was inherited from HT and seed parent should possess M3 haplotype, because HT was M0 homozygous. These indicated that O3 was not the parent of SM.
Supplementary Figure 11 | Structural comparison of M loci among Prunus species. Circos plots show sequence similarities among M0, M1, and M haplotypes of other Prunus species. Ribbons link homologous regions between haplotypes. ∗Indicates genes that did not have intact CDS sequence. PG1 of P. mume was translocated to the > 2 Mbp downstream region. To determine the M locus region, reference genomes of almond (“Lauranne” and “Texas”), P. kansuensis, P. mira, Japanese apricot, apricot, sweet cherry, and P. x yedoensis were searched by Blastn analysis using PG1, PG2, PGM/F, NADH, and F-box genes as query. M locus was found at the right arm of chromosome 4 or LG3 in all Prunus species except P. kansuensis, in which no pseudomolecule was released. Nucleotide sequences of M locus were compared by nucmer and the relationships were drawn by Circos.
Supplementary Table 1 | Climate conditions in peach production areas in Okayama and Fukushima Prefectures, Japan in 2018.
Supplementary Table 2 | Relationships between flesh penetration force measured by the system used in this study and other fruit maturity indexes.
Supplementary Table 3 | Primers used in this study.
Supplementary Table 4 | Comparison of SNP number in regions from PG1 to PG2. Intergenic region 1 was from end of PG1 to 19,026,185 bp (outside M3 deletion region). Intergenic region 2 was from 19,026,186 bp (inside M3 deletion region) to start of PG2. Values in parentheses are those of heterozygous SNPs.
Supplementary Table 5 | M genotypes of 412 peach accessions.
Supplementary Table 6 | Differences in flesh texture predicted by genotype and reported phenotype. ∗The reported phenotype matched the predicted one in this study. The 11 accessions showed differences between predicted and reported phenotypes. All except “Early Gold” (EG) were reported by Yoon et al. (2006) and Cao et al. (2016). EG was a canning peach and its parent was “Nishiki” (NK). EG should possess at least one M2 haplotype because NK was an M2 homozygote, but resequencing analysis showed that EG was M1M1.
Supplementary Table 7 | Correlation of PGM/F genes in this study with those in previous reports. PGBT in Morgutti et al. (2017) was BstXI-sensitive (Supplementary Figure S6).
Supplementary Table 8 | Correlation of haplotypes in this study with those in previous reports. ∗M0 was misidentified as H2 haplotype in Gu et al. (2016). f haplotype structure in Morgutti et al. (2017) was postulated from M1 haplotype. PG gene composition was correct but the genome structure was not the same as M0 haplotype in this study. PGM-M0 and PGM-M1 were allelic.
M locus, Melting flesh locus; MF, melting flesh; NMF, non-melting flesh; SH, stony-hard; SR, slow-ripening; SSC, soluble solids content; endo-PG, endo-polygalacturonase; NADH, nicotinamide adenine dinucleotide dehydrogenase; SRA, sequence read archive; WGS, whole genome shotgun sequencing; SNP, single nucleotide polymorphism; indel, insertion/deletion.
Bailey, J. S., and French, H. P. (1949). The inheritance of certain fruit and foliage characters in peach. Mass. Agr. Expt. Sta. Bul. 452, 2–31.
Bassi, D., and Monet, R. (2008). “Botany and taxonomy,” in The Peach: Botany, Production and Uses, eds D. R. Layne and D. Bassi (Wallingford: CAB International), 1–36. doi: 10.1079/9781845933869.0001
Biale, J. B., and Young, R. E. (1981). “Respiration and ripening in fruits- retrospect and prospect,” in Recent Advances in the Biochemistry of Fruits and Vegetables, eds J. Friend and M. J. C. Rhodes (London: Academic Press), 1–39. doi: 10.4324/9781315130590-1
Brecht, J. K., and Kader, A. A. (1984). Ethylene production by fruit of some slow-ripening nectarine genotypes. J. Am. Soc. Hort. Sci. 109, 763–767.
Brummell, D. A., Dal Cin, V., Crisosto, C. H., and Labavitch, J. M. (2004). Cell wall metabolism during maturation, ripening and senescence of peach fruit. J. Exp. Bot. 55, 2029–2039. doi: 10.1093/jxb/erh227
Burg, S. P., and Burg, E. A. (1967). Molecular requirements for the biological activity of ethylene. Plant Physiol. 42, 144–152. doi: 10.1104/pp.42.1.144
Callahan, A. M., Scorza, R., Bassett, C., Nickerson, M., and Abeles, F. B. (2004). Deletions in an endopolygalacturonase gene cluster correlate with non-melting flesh texture in peach. Funct. Plant Biol. 31, 159–168. doi: 10.1071/FP03131
Cao, K., Zhou, Z., Wang, Q., Guo, J., Zhao, P., Zhu, G., et al. (2016). Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 7:13246. doi: 10.1038/ncomms13246
Carrasco-Valenzuela, T., Muñoz-Espinoza, C., Riveros, A., Pedreschi, R., Arús, P., Campos-Vargas, R., et al. (2019). Expression QTL (eQTLs) analyses reveal candidate genes associated with fruit flesh softening rate in peach [Prunus persica (L.) Batsch]. Front. Plant Sci. 10:1581. doi: 10.3389/fpls.2019.01581
Chin, S. W., Shaw, J., Haberle, R., Wen, J., and Potter, D. (2014). Diversification of almonds, peaches, plums and cherries–molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogenet. Evol. 76, 34–48. doi: 10.1016/j.ympev.2014.02.024
Clark, J. R., and Finn, C. E. (2010). Register of new fruit and nut cultivars List 45. HortScience 45, 716–756. doi: 10.21273/HORTSCI.45.5.716
Davis, L. (1937). The gumming of Phillips cling peaches. Hilgardia 11, 1–34. doi: 10.3733/hilg.v11n01p001
Eduardo, I., Picañol, R., Rojas, E., Batlle, I., Howad, W., Aranzana, M. J., et al. (2015). Mapping of a major gene for the slow ripening character in peach: co-location with the maturity date gene and development of a candidate gene-based diagnostic marker for its selection. Euphytica 205, 627–636. doi: 10.1007/s10681-015-1445-9
Elsadr, H., Sherif, S., Banks, T., Somers, D., and Jayasankar, S. (2019). Refining the genomic region containing a major locus controlling fruit maturity in peach. Sci. Rep. 9:7522. doi: 10.1038/s41598-019-44042-4
Fernandez, I., Marti, A., Saski, C. A., Manganaris, G. A., Gasic, K., and Crisosto, C. H. (2018). Genomic sequencing of Japanese plum (Prunus salicina Lindl.) mutants provides a new model for Rosaceae fruit ripening studies. Front. Plant Sci. 9:21. doi: 10.3389/fpls.2018.00021
Fishman, M. L., Levaj, B., Gillespie, D., and Scorza, R. (1993). Changes in the physicochemical properties of peach fruit pectin during on-tree ripening and storage. J. Amer. Soc. Hort. Sci. 118, 343–349. doi: 10.21273/JASHS.118.3.343
Font i Forcada, C., Oraguzie, N., Igartua, E., Ángeles Moreno, M., and Gogorcena, Y. (2013). Population structure and marker–trait associations for pomological traits in peach and nectarine cultivars. Tree Genet. Genomes 9, 331–349. doi: 10.1007/s11295-012-0553-0
Gapper, N. E., McQuinn, R. P., and Giovannoni, J. J. (2013). Molecular and genetic regulation of fruit ripening. Plant Mol. Biol. 82, 575–591. doi: 10.1007/s11103-013-0050-3
Ghiani, A., Negrini, N., Morgutti, S., Baldin, F., Nocito, F. F., Spinardi, A., et al. (2011). Melting of “Big Top” nectarine fruit: some physiological, biochemical, and molecular aspects. J. Am. Soc. Hort. Sci. 136, 61–68. doi: 10.21273/JASHS.136.1.61
Giné-Bordonaba, J., Eduardo, I., Arús, P., and Cantín, C. M. (2020). Biochemical and genetic implications of the slow ripening phenotype in peach fruit. Sci. Hortic. 259:108824. doi: 10.1016/j.scienta.2019.108824
Gu, C., Wang, L., Wang, W., Zhou, H., Ma, B. Q., Zheng, H. Y., et al. (2016). Copy number variation of a gene cluster encoding endopolygalacturonase mediates flesh texture and stone adhesion in peach. J. Exp. Bot. 67, 1993–2005. doi: 10.1093/jxb/erw021
Haji, T., Yaegaki, H., and Yamaguchi, M. (2003). Softening of stony hard peach by ethylene and the induction of endogenous ethylene by 1-aminocyclopropene-1-carboxylic acid (ACC). J. Japan Soc. Hort. Sci. 72, 212–217. doi: 10.2503/jjshs.72.212
Haji, T., Yaegaki, H., and Yamaguchi, M. (2005). Inheritance and expression of fruit texture melting, non-melting and stony hard in peach. Sci. Hortic. 105, 241–248. doi: 10.1016/j.scienta.2005.01.017
Hayama, H., Shimada, T., Fujii, H., Ito, A., and Kashimura, Y. (2006). Ethylene-regulation of fruit softening and softening-related genes in peach. J. Exp. Bot. 57, 4071–4077. doi: 10.1093/jxb/erl178
Hayama, H., Tatsuki, M., and Nakamura, Y. (2008). Combined treatment of aminoethoxyvinylglycine (AVG) and 1-methylcyclopropene (1-MCP) reduces melting-flesh peach fruit softening. Postharvest Biol. Technol. 50, 228–230. doi: 10.1016/j.postharvbio.2008.05.003
Hiwasa, K., Nakano, R., Hashimoto, A., Matsuzaki, M., Murayama, H., Inaba, A., et al. (2004). European, Chinese and Japanese pear fruits exhibit differential softening characteristics during ripening. J. Exp. Bot. 55, 2281–2290. doi: 10.1093/jxb/erh250
Infante, R., Contador, L., Rubio, P., Aros, D., and Penþa-Neira, A. ì (2011). Postharvest sensory and phenolic characterization of ‘Elegant Lady’ and ‘Carson’ peaches. Chilean J. Agric. Res. 71, 445–451. doi: 10.4067/S0718-58392011000300016
Jackman, S. D., Vandervalk, B. P., Mohamadi, H., Chu, J., Yeo, S., Hammond, S. A., et al. (2017). ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27, 768–777. doi: 10.1101/gr.214346.116
Kawai, T., Matsumori, F., Akimoto, H., Sakurai, N., Hirano, K., Nakano, R., et al. (2018). Nondestructive detection of split-pit peach fruit on trees with an acoustic vibration method. Hort. J. 87, 499–507. doi: 10.2503/hortj.UTD-012
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. doi: 10.1186/gb-2004-5-2-r12
Lester, D. R., Sherman, W. B., and Atwell, B. J. (1996). Endopolygalacturonase and the melting flesh (M) locus in peach. J. Amer. Soc. Hort. Sci. 121, 231–235. doi: 10.21273/JASHS.121.2.231
Lester, D. R., Speirs, G., Orr, G., and Brady, C. J. (1994). Peach (Prunus persica) endo-PG cDNA isolation and mRNA analysis in melting and non-melting peach cultivars. Plant Physiol. 105, 225–231. doi: 10.1104/pp.105.1.225
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Liguori, G., Weksler, A., Zutahi, Y., Lurie, S., and Kosto, I. (2004). Effect of 1-methylcyclopropene on ripening of melting flesh peaches and nectarines. Postharvest Biol. Technol. 31, 263–268. doi: 10.1016/j.postharvbio.2003.09.007
Liu, H., Qian, M., Song, C., Li, J., Zhao, C., Li, G., et al. (2018). Down-regulation of PpBGAL10 and PpBGAL16 delays fruit softening in peach by reducing polygalacturonase and pectin methylesterase activity. Front. Plant Sci. 9:1015. doi: 10.3389/fpls.2018.01015
Mathooko, F. M., Tsunashima, Y., Owino, W. Z. O., Kubo, Y., and Inaba, A. (2001). Regulation of genes encoding ethylene biosynthetic enzymes in peach (Prunus persica L.) fruit by carbon dioxide and 1-methylcyclopropene. Postharvest Biol. Technol. 21, 265–281. doi: 10.1016/S0925-5214(00)00158-7
McMurchie, E. J., McGlasson, W. B., and Eaks, I. L. (1972). Treatment of fruit with propylene gives information about bio-genesis of ethylene. Nature 237, 235–236. doi: 10.1038/237235a0
Meneses, C., Ulloa-Zepeda, L., Cifuentes-Esquivel, A., Infante, R., Cantin, C. M., Batlle, I., et al. (2016). A codominant diagnostic marker for the slow ripening trait in peach. Mol. Breed. 36:77. doi: 10.1007/s11032-016-0506-7
Minas, I. S., Font i Forcada, C., Dangl, G. S., Gradziel, T., Dandekar, A. M., and Crisosto, C. H. (2015). Discovery of non-climacteric and suppressed-climacteric bud sport mutations originating from a climacteric Japanese plum cultivar (Prunus salicina lindl.). Front. Plant Sci. 6:316. doi: 10.3389/fpls.2015.00316
Moggia, C., Graell, J., Lara, I., González, G., and Lobos, G. A. (2017). Firmness at harvest impacts postharvest fruit softening and internal browning development in mechanically damaged and non-damaged highbush blueberries (Vaccinium corymbosum L.). Front. Plant Sci. 8:535. doi: 10.3389/fpls.2017.00535
Monet, R. (1989). Peach genetics: past, present and future. Acta Hortic. 254, 49–57. doi: 10.17660/ActaHortic.1989.254.8
Morgutti, S., Negrini, N., Ghiani, A., Baldin, F., Bassi, D., and Cocucci, M. (2017). Endopolygalacturonase gene polymorphisms: asset of the locus in different peach accessions. Am. J. Plant Sci. 8, 941–957. doi: 10.4236/ajps.2017.84063
Morgutti, S., Negrini, N., Nocito, F. F., Ghiani, A., Bassi, D., and Cocucci, M. (2006). Changes in endopolygalacturonase levels and characterization of a putative endo-PG gene during fruit softening in peach genotypes with nonmelting and melting flesh fruit phenotypes. New Phytol. 171, 315–328. doi: 10.1111/j.1469-8137.2006.01763.x
Nakano, R., Akimoto, H., Fukuda, F., Kawai, T., Ushijima, K., Fukamatsu, Y., et al. (2018). Nondestructive detection of split pit in peaches using an acoustic vibration method. Hort. J. 87, 281–287. doi: 10.2503/hortj.OKD-094
Nimmakayala, P., Tomason, Y. R., Abburi, V. L., Alvarado, A., Saminathan, T., Vajja, V. G., et al. (2016). Genome-wide differentiation of various melon horticultural groups for use in GWAS for fruit firmness and construction of a high resolution genetic map. Front. Plant Sci. 7:1437. doi: 10.3389/fpls.2016.01437
Nishiyama, K., Guis, M., Rose, J. K., Kubo, Y., Bennett, K. A., Wangjin, L., et al. (2007). Ethylene regulation of fruit softening and cell wall disassembly in Charentais melon. J. Exp. Bot. 58, 1281–1290. doi: 10.1093/jxb/erl283
Nuñez-Lillo, G., Cifuentes-Esquivel, A., Troggio, M., Micheletti, D., Infante, R., Campos-Vargas, R., et al. (2015). Identification of candidate genes associated with mealiness and maturity date in peach [Prunus persica (L.) Batsch] using QTL analysis and deep sequencing. Tree Genet. Genomes 11:86. doi: 10.1007/s11295-015-0911-9
Pan, L., Zeng, W., Niu, L., Lu, Z., Liu, H., Cui, G., et al. (2015). PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 66, 7031–7044. doi: 10.1093/jxb/erv400
Peace, C. P., Callahan, A., Ogundiwin, E. A., Potter, D., Gradziel, T. M., Bliss, F. A., et al. (2007). Endopolygalacturonase genotypic variation in Prunus. Acta Hortic. 738, 639–646. doi: 10.17660/actahortic.2007.738.83
Peace, C. P., Crisosto, C. H., and Gradziel, T. M. (2005). Endopolygalacturonase: a candidate gene for freestone and melting flesh in peach. Mol. Breed. 16, 21–31. doi: 10.1007/s11032-005-0828-3
Pressey, R., and Avants, J. K. (1978). Differences in polygalacturonase composition of clingstone and freestone peaches. J. Food Sci. 43, 1415–1423. doi: 10.1111/j.1365-2621.1978.tb02507.x
Tatsuki, M., Haji, T., and Yamaguchi, M. (2006). The involvement of 1−aminocyclopropane−1−carboxylic acid synthase isogene, Pp−ACS1, in peach fruit softening. J. Exp. Bot. 57, 1281–1289. doi: 10.1093/jxb/erj097
Tatsuki, M., Soeno, K., Shimada, Y., Sawamura, Y., Suesada, Y., Yaegaki, H., et al. (2018). Insertion of a transposon−like sequence in the 5’−flanking region of the YUCCA gene causes the stony hard phenotype. Plant J. 96, 815–827. doi: 10.1111/tpj.14070
Tonutti, P., Casson, P., and Ramina, A. (1991). Ethylene biosynthesis during peach fruit development. J. Amer. Soc. Hort. Sci. 116, 274–279. doi: 10.21273/JASHS.116.2.274
Tucker, G., Yin, X., Zhang, A., Wang, M., Zhu, Q., Liu, X., et al. (2017). Ethylene and fruit softening. Food Qual. Safe 1, 253–267. doi: 10.1093/fqsafe/fyx024
Verde, I., Jenkins, J., Dondini, L., Micali, S., Pagliarani, G., Vendramin, E., et al. (2017). The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 18:225. doi: 10.1186/s12864-017-3606-9
Wang, Z.-H., and Lu, Z.-X. (1992). Advances of peach breeding in China. HortScience 27, 729–732. doi: 10.21273/HORTSCI.27.7.729
Yamamoto, T., Mochida, K., and Hayashi, T. (2003a). Shanhai suimitsuto, one of the origins of Japanese peach cultivars. J. Japan. Soc. Hort. Sci. 72, 116–121. doi: 10.2503/jjshs.72.116
Yamamoto, T., Mochida, K., Imai, T., Haji, T., Yaegaki, H., Yamaguchi, M., et al. (2003b). Parentage analysis in Japanese peaches using SSR markers. Breed. Sci. 53, 35–40. doi: 10.1270/jsbbs.53.35
Yamazaki, T., and Suzuki, K. (1980). Color charts: useful guide to evaluate the fruit maturation. I. Colorimetric specifications of color charts for Japanese pear, apple, peach, grape, kaki and citrus fruits. (In Japanese with English summary). Bull. Fruit Tree Res. Sta. A 19–44.
Yoon, J., Liu, D., Song, W., Liu, W., Zhang, A., and Li, S. (2006). Genetic diversity and ecogeographical phylogenetic relationships among peach and nectarine cultivars based on simple sequence repeat (SSR) markers. J. Am. Soc. Hort. Sci. 131, 513–521. doi: 10.21273/JASHS.131.4.513
Yoshioka, H., Hayama, H., Tatsuki, M., and Nakamura, Y. (2010). Cell wall modification during development of mealy texture in the stony-hard peach “Odoroki” treated with propylene. Postharvest Biol. Technol. 55, 1–7. doi: 10.1016/j.postharvbio.2009.08.005
Yoshioka, H., Hayama, H., Tatsuki, M., and Nakamura, Y. (2011). Cell wall modifications during softening in melting type peach “Akatsuki” and non-melting type peach “Mochizuki”. Postharvest Biol. Technol. 60, 100–110. doi: 10.1016/j.postharvbio.2010.12.013
Zhang, X., Su, M., Du, J., Zhou, H., Li, X., Li, X., et al. (2019). Comparison of phytochemical differences of the pulp of different peach [Prunus persica (L.) Batsch] cultivars with alpha-glucosidase inhibitory activity variations in China using UPLC-Q-TOF/MS. Molecules 24:1968. doi: 10.3390/molecules24101968
Keywords: fruit, softening, ethylene, Prunus persica, melting flesh locus, endoPG, postharvest
Citation: Nakano R, Kawai T, Fukamatsu Y, Akita K, Watanabe S, Asano T, Takata D, Sato M, Fukuda F and Ushijima K (2020) Postharvest Properties of Ultra-Late Maturing Peach Cultivars and Their Attributions to Melting Flesh (M) Locus: Re-evaluation of M Locus in Association With Flesh Texture. Front. Plant Sci. 11:554158. doi: 10.3389/fpls.2020.554158
Received: 23 April 2020; Accepted: 28 October 2020;
Published: 26 November 2020.
Edited by:
Cai-Zhong Jiang, USDA-ARS, United StatesReviewed by:
Carlos H. Crisosto, University of California, Davis, United StatesCopyright © 2020 Nakano, Kawai, Fukamatsu, Akita, Watanabe, Asano, Takata, Sato, Fukuda and Ushijima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fumio Fukuda, ZmZ1a3VkYUBva2F5YW1hLXUuYWMuanA=; Koichiro Ushijima, dXNoaWppbWFAY2Mub2theWFtYS11LmFjLmpw
†These authors have contributed equally to this work
‡Present address: Yosuke Fukamatsu, Research Institute for Biological Sciences, Okayama Prefectural Technology Center for Agriculture, Forestry, and Fisheries, Okayama, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.