- 1Faculty of Biotechnology and Food Sciences, Institute of Fermentation Technology and Microbiology, Lodz University of Technology, Lodz, Poland
- 2Research Centre for Agriculture and Environment, Council for Agricultural Research and Economics, Rome, Italy
- 3Faculty of Biotechnology and Food Sciences, Institute of Molecular and Industrial Biotechnology, Lodz University of Technology, Lodz, Poland
- 4 Institute for Biological Systems, Council of National Research of Italy (CNR), Rome, Italy
In extreme environments, the relationships between species are often exclusive and based on complex mechanisms. This review aims to give an overview of the microbial ecology of saline soils, but in particular of what is known about the interaction between plants and their soil microbiome, and the mechanisms linked to higher resistance of some plants to harsh saline soil conditions. Agricultural soils affected by salinity is a matter of concern in many countries. Soil salinization is caused by readily soluble salts containing anions like chloride, sulphate and nitrate, as well as sodium and potassium cations. Salinity harms plants because it affects their photosynthesis, respiration, distribution of assimilates and causes wilting, drying, and death of entire organs. Despite these life-unfavorable conditions, saline soils are unique ecological niches inhabited by extremophilic microorganisms that have specific adaptation strategies. Important traits related to the resistance to salinity are also associated with the rhizosphere-microbiota and the endophytic compartments of plants. For some years now, there have been studies dedicated to the isolation and characterization of species of plants’ endophytes living in extreme environments. The metabolic and biotechnological potential of some of these microorganisms is promising. However, the selection of microorganisms capable of living in association with host plants and promoting their survival under stressful conditions is only just beginning. Understanding the mechanisms of these processes and the specificity of such interactions will allow us to focus our efforts on species that can potentially be used as beneficial bioinoculants for crops.
Introduction
Soil salinization is a process of localised accumulation of soluble salts. This phenomenon today is unanimously considered a severe threat to agricultural lands as it directly undermines the value and quality of soil (Ammari et al., 2013; Daliakopoulos et al., 2016). The soil is a complex system in continuous evolution and dynamic equilibrium with the other environmental components, sensitive to the effects of climate change and human activities (Smith et al., 2012). Still, it constitutes a substantially non-renewable resource in the sense that the rate of its degradation is potentially rapid (Zewdu et al., 2017), while the soil formation and regeneration processes are extremely slow.
Globally it has been estimated that 33% of the irrigated agricultural land and over 20% of the total cultivated land of the world is saline. If the current trend of salinization is maintained, then by 2050 we will see an approximately 30% increase in the cultivated land salinity. This means that agricultural productivity will be reduced due to the decline in cultivable land, with a consequent increase in the number of people suffering from hunger. Plants are the first in the chain of food production to be hit by salinity stress, which hampers their basic physiological and biochemical processes, such as water absorption and photosynthesis, thus resulting in overall reduced growth (Vaishnav et al., 2016). However, plants develop various morphological, physiological, biochemical, and molecular strategies in response to salinity in their environment (Meng et al., 2018).
In recent decades, significant overviews about mechanisms of salinity tolerance in plants have been shared by several authors (Munns and Tester, 2008; Deinlein et al., 2014; Gupta and Huang, 2014; Meng et al., 2018). In general, two types of plant adaptation to high salt concentration in the soil are distinguished: avoidance and tolerance strategy (Dajic, 2006). The first consisting of creating physical, physiological or metabolic barriers that counteract the penetration of stress factors in the plant, which causes morphological and physiological changes at the whole plant level. The tolerance strategy is based instead on plant ability to survive in stress conditions through cellular, molecular, and biochemical modifications aimed at minimising stress effects (Bahmani et al., 2015). Interestingly, those plants have a strong influence on shaping the rhizosphere and endorhiza microbiome (Abedinzadeh et al., 2019).
In particular, some plant growth-promoting rhizobacteria (PGPR) may exert a direct stimulation on plants’ growth and development by providing them with fixed nitrogen, phytohormones (IAA, GB), iron (Egamberdieva et al., 2019), and soluble phosphate (Shrivastava and Kumar, 2015) that help to overcome the effects of salinity stress. Another mechanism that minimizes the impact of salinity consists in the production of substances (proline, trehalose, glycine, betaine) that work as osmoprotectant for plants’ cells (Kushwaha et al., 2019). Moreover, under various environmental stresses, plants typically produce ethylene, from the precursor 1-aminocyclopropane-1-carboxylic acid (ACC). It is a hormone that limits plants’ growth in conditions of high salinity, high temperatures, drought, or at the presence of toxic metals or organic pollutants. PGPRs produce a particular enzyme that degrades ethylene (ACC-deaminase), allowing the plants’ roots to develop despite the environmental stress (Glick, 2012).
When salt meddles between soil, plant and microorganisms in and around the rhizosphere, a unique extreme environment is created, that provides a scene for mutualistic relationships. In this system, plant-endophytes and plant-microbiome interactions play a crucial role in the activation or stimulation of different adaptation mechanisms for survival in saline soil (Acosta-Motos et al., 2017). In this review, we provide information about natural and anthropic causes of soil salinity and discuss the plant’s strategies for stress management. An overview of the known interactions occurring between endophytic communities and host plants is provided along with a discussion on the potential of plant growth-promoting rhizobacteria (PGPR) for increasing plant’s salt stress tolerance.
Natural and Anthropic Causes of Soil Salinity Affecting Agricultural Land Use
All soils naturally contain a mixture of salts soluble in water, and some of these are essential for plant development. Their origin is mainly from the meteoric alteration of the igneous rocks of the lithosphere. Following the hydrological and climatic events, in past ages, there has been the deposition of large quantities of salts in sedimentary rocks, in surface and underground waters, in seas and oceans (Daliakopoulos et al., 2016; Zaman et al., 2018). From these deposits, through various mechanisms, the salts reach the soil. In particular conditions, the formation of a water evaporation front in the soil moves water by evaporation rather than by percolation, so the salts remain in the soil and accumulate (Datta and de Jong, 2002).
Soil salinity can be distinguished in primary and secondary. The first depends on factors related mostly to the lithology of the substrate (in particular hydrological characteristics), morphological characteristics of the area, intrinsic chemistry of soils and climatic factors (Schofield and Kirkby, 2003). If soil parent rock contains carbonate minerals or feldspar, physical or chemical weathering operated by water bring salts in solution, increasing their concentration in groundwater and consequently on the wetted topsoil layer. Soil porosity, texture and mineral composition influence soil hydrological properties which also depend on the salts’ accumulation on the soil surface. The amount of saline precipitates is in turn modulated by soil transpiration and the extent and characteristics of the capillary fringe. This kind of accumulation process is reported in different European areas (Schofield and Kirkby, 2003; Kováčová and Velísková, 2012; Gkiougkis et al., 2015; Daliakopoulos et al., 2016).
Another group of naturally saline soils originate from salty marine seawater in the coastal regions. This phenomenon can occur under short- or long-term periods. In short term periods, soil salinization can be caused by stochastic rapid events like floods or tsunamis that cause salinization on the surface of the geographical area beaten by the waves (Central Water Commission, 2017). In contrast, in a geological time scale, prolonged high tides can cause the formation of marine deposits. Soils of the coastal areas can be therefore easily affected by salinization, due to the intrusion of the marine seawater wedge inside the fresh groundwater (Daliakopoulos et al., 2016). Despite the saltwater in a natural environment usually stays below the fresh, because of its higher density, in many cases the water extraction by wells for different human uses could interfere by breaking this balance, inducing the recall of the saline wedge with rising saltwater and the consequent formation of an area of brackish water near the ground surface. This anthropic cause is mainly due to inadequate agricultural water management practices and could induce “secondary soil salinization” along time by increasing the salt content in the irrigation waters (Napoli and Vanino, 2011; Greggio et al., 2012).
Agriculture plays an essential role as one of the leading causes of secondary salinization given by the use of poor-quality water, often worsened by the presence of soil components that limit the leaching of salts, such as the presence of impermeable horizons and an unfavorable physiographic position.
Secondary salinity could also depend by the overuse of fertilizers, insecticides and fungicides, unsustainable use of the land, excessive drainage of the water tables (Datta and de Jong, 2002).
Salinization caused by overuse of fertilizers is due to different phenomena. The excessive use of nitrogen fertilizers both chemicals or organic can cause a rise in nitrate content in the soil that can be detected as a rise in Electrical Conductivity (EC) or interpreted through signs of crop disease often associated with high soil salinity (Gkiougkis et al., 2015). The use of fertilizers with a high content of potassium and sodium can cause soil degradation by the accumulation of these salts in particular conditions (Chang et al., 2018). The application of incorrect irrigation practices added to an excess of fertilization, together with particular climatic conditions sometimes may favor the accumulation of salts (Daliakopoulos et al., 2016). Poorly permeable silty-clay soils in climatic conditions with limited rainfall and high temperatures favor high evapotranspiration and therefore the quick accumulation of salts in the first layers of the soil (Chang et al., 2018)
It should be considered, however, that even in areas irrigated using “good” quality water, moderate levels of salinity were detected as a consequence of the irrigation methods applied and local arid climatic conditions. Vice versa, the phenomenon of salinization may not occur on lands irrigated for several years with waters rich in salts. These examples indicate that each area is characterized by different and peculiar balances, which influence its possible evolution. In certain soils, drained and with particular thermo-pluviometric trends, the accumulation of salts in the soils could be only temporary. A balance could quickly be established between accumulation and leaching that would allow over time, and with due care, to maintain agricultural activity (Machado and Serralheiro, 2017).
Secondary soil salinization caused by incorrect irrigation strategies is also associated with the use of wastewaters. When the use of poor quality water exceeds the natural buffering effect of the soil, a whole series of substances, such as insecticides and fungicides remain on the ground which, as a result, cause an increase in salinity (Kováčová and Velísková, 2012; Rodríguez-Liébana et al., 2014). Another common cause of secondary salinization phenomenon is the consequence of the replacement of spontaneous vegetation (polyannual and primarily arboreal species) with crops (annual and exclusively herbaceous), characterized by superficial, less deep root systems than the pristine vegetation. This artificial change of vegetation causes a drastic modification of the delicate hydrological balance. In essence, the reduced root systems of the new plant species, requiring less water than the original tall trees leads, over time, to a progressive rise of the water table and lower solubilization of the salts in the subsoil, causing them to rise together with the water until it affects the layer occupied by the roots of the crops (Bui, 2013). The water absorption from the roots and the evapotranspiration process inevitably causes a gradual accumulation of the salts on the surface, making the soil progressively inhospitable to agricultural plants and unsuitable for agriculture (Hanson et al., 1993; Machado and Serralheiro, 2017). The saline sources in secondary salinization could also come from the use of soil improvers that are themselves saline (i.e. gypsum or elemental sulphur), from manure and chemical fertilizers (Wallender and Tanji, 2012).
Classification and Extension of Saline Soils
As reported by Bui in 2013, the definition of saline soil is confusing. Are considered saline the soils in which salt concentration can interfere with the capability of plants to absorb water, affecting their growth or more specifically a soil with an electric conductivity (EC) on a saturated soil paste extract >4 dS m-1 (Bui, 2013; Shrivastava and Kumar, 2015; Zaman et al., 2018). Salts concentration and the osmotic pressure of a saline soil also depend on soil texture and relative water characteristics more than only on the salt content (Darwish et al., 2005; Daliakopoulos et al., 2016; Zaman et al., 2018).
In literature, there are two most accepted classifications for saline soil. One is the US Salinity Laboratory Staff Classification in which it is most commonly used the term “salt-affected soil” to indicate saline, saline-sodic and sodic soils. In this classification, a saline soil has an EC ≥4 dS m-1, exchangeable sodium percentage (ESP) <15 and pH <8.5 while saline-sodic differs only for ESP ≥15 and pH ≥8.5. Sodic soils have EC <4 dS m-1, ESP ≥15 and pH >8.5. These soils are also characterized by the loss of permeability to water caused by the disruption of soil aggregate operated mostly by the Na+ ions (Daliakopoulos et al., 2016; Machado and Serralheiro, 2017; Zaman et al., 2018) and the ensuing collapse of soil structure in the Natric Horizon.
According to the classification approach of World Reference Base Classification (IUSS Working Group WRB, 2014), salt-affected soils are divided into two classes: solonchacks (saline soils with ECe >15 dSm-1 in the top 125 cm) and solonetz (are sodium-rich soils with an ESP > 15), both divided into subclasses (Zaman et al., 2018).
The Reference Group of Solonchacks is quite widespread in all the arid and semi-arid areas of North Africa, Near East, Central Asia, India, Iran and Iraq, Australia and the Americas. The Reference Group of Solonetz, mainly located in the steppe climatic regimes and flat landscapes with poor drainage, were mapped in Ukraine, Russian Federation, Eastern Europe, China, India, USA, Canada, Southern and Eastern Africa and Australia. Last studies and maps at world level reported an estimated area of about 260 Mha and 135 Mha covered by Solonchacks and Solonetz, respectively (Cherlet et al., 2018).
Apart from mapping the soils classified as “saline”, the exact extension and localisation of salinity based on its causes were hardly addressed geographically. Primary (natural) salinization was estimated to be slightly under 1 billion ha worldwide, secondary (artificially induced by human activities) occurs on around 77 Mha, mostly in intensively cultivated and irrigated areas of India, Pakistan, China, Iraq, and Iran. Because of the human activities linked to wrong water management, large areas of the Mediterranean Basin, Australia, Central Asia, the Middle East and Northern Africa are interested in the risk of salinization (Cherlet et al., 2018).
In Europe, secondary salinization affects approximately 4 Mha of European soils (Daliakopoulos et al., 2016) and focusing on the Mediterranean region. Soil salinization affects 25% of irrigated agricultural land. Along the Mediterranean coasts, soil salinity is the primary cause of desertification due to human activities. Artificially induced salinization caused by irrigated agriculture is affecting significant parts of southern Italy (Napoli and Vanino, 2011), Spain (e.g. the Ebro Valley), Hungary (e.g. Great Alfold), Greece, Cyprus, Portugal, France (West coast), the Dalmatian coast, Slovakia and Romania. Besides, North Europe countries (e.g. Denmark, Poland, Latvia, and Estonia) are facing similar issues (Daliakopoulos et al., 2016).
Limitation to Agricultural Uses
Salinization represents a critical threat for agriculture because it can cause the alteration of the delicate balance of ecological processes occurring in soil.
The phenomenon of salinization represents only one of the multiple aspects that the accumulation of salts in soil causes (Shrivastava and Kumar, 2015; Nouri et al., 2017). The loss of soil occurs in terms of both surface subtraction (Peng et al., 2019), and the alteration of its chemical characteristics Nouri et al., 2017). The salinized soil readily undergoes to a further series of degradation processes, such as erosion (Ishida et al., 2009), a decrease of organic matter content (de Souza and Fay, 2014), local or widespread contamination (Hanson et al., 1993), sealing, compaction, and decline of biodiversity (Nouri et al., 2017). These processes contribute to compromising the quality of the soil and its ability to interact with the ecosystem to maintain biological productivity, environmental quality and promote the health of all living organisms (Salvati and Ferrara, 2015; Zaman et al., 2018).
Saline and sodic soils reduce the value and productivity of large areas around the world (Qadir et al., 2014; Acuña-Rodríguez et al., 2019). It is estimated that every day in the world 2000 hectares of arable land are lost due to salinization, and this problem can cause loss of yield for many crops of 10-25% and in exceptional conditions can lead to desertification (Zaman et al., 2018).
Geographical areas affected by salinity problems have an impact on both the environmental and socioeconomic sphere. These areas face the loss of productivity (Datta and de Jong, 2002) with a different degree of magnitude depending on the crops (Qadir et al., 2014) seeing their farmers sometimes reduced to poverty or forced to migrate searching for a new source of income (Ammari et al., 2013; Salvati and Ferrara, 2015; Zaman et al., 2018). Soil can become saline at first, causing a decrease in agricultural yield and then, in the long run, it can progressively turn into completely sterile (desertification) (Gorji et al., 2017; Cuevas et al., 2019).
The pollution from chemicals causes the water that returns to the water cycle to be of poor quality with negative consequences on agriculture and health, giving rise to a vicious cycle Zewdu et al., 2017). Soil salinization has a significant impact on the environment, causing landscape and consequently, ecosystem fragmentation (Cramer and Hobbs, 2002). A poor vegetative growth leads to the reduction of the protective role of the plants cover, enhancing soil degradation and erosion. The presence of chemical residues on the soil surface, together with salts, cause the elevation of toxic clouds of dust (Zewdu et al., 2017).
The enrichment of the soil with organic matter, humic substances, the application of biofertilizer containing microorganism and the application of fertilizers through irrigation water (fertigation) could help to alleviate the negative effect of salt accumulation on salt-sensitive crops (Darwish et al., 2005).
Given the importance of agricultural production, it is crucial to understand the impacts of salinization on different crops and to find effective strategies to reduce economic losses in salt-affected areas.
Effects of Salinity Stress on Plant Growth
Soil salinity affects (directly or indirectly) both growth and reproduction of plants as a consequence of complex interactions between physicochemical properties of soil (salt content, poor aeration, an increase of crusting, hard setting, reduced infiltration, reduction of water uptake, and difficult root penetration) and plants’ morphological and physiological features (Rogers et al., 2005; Akbarimoghaddam et al., 2011).
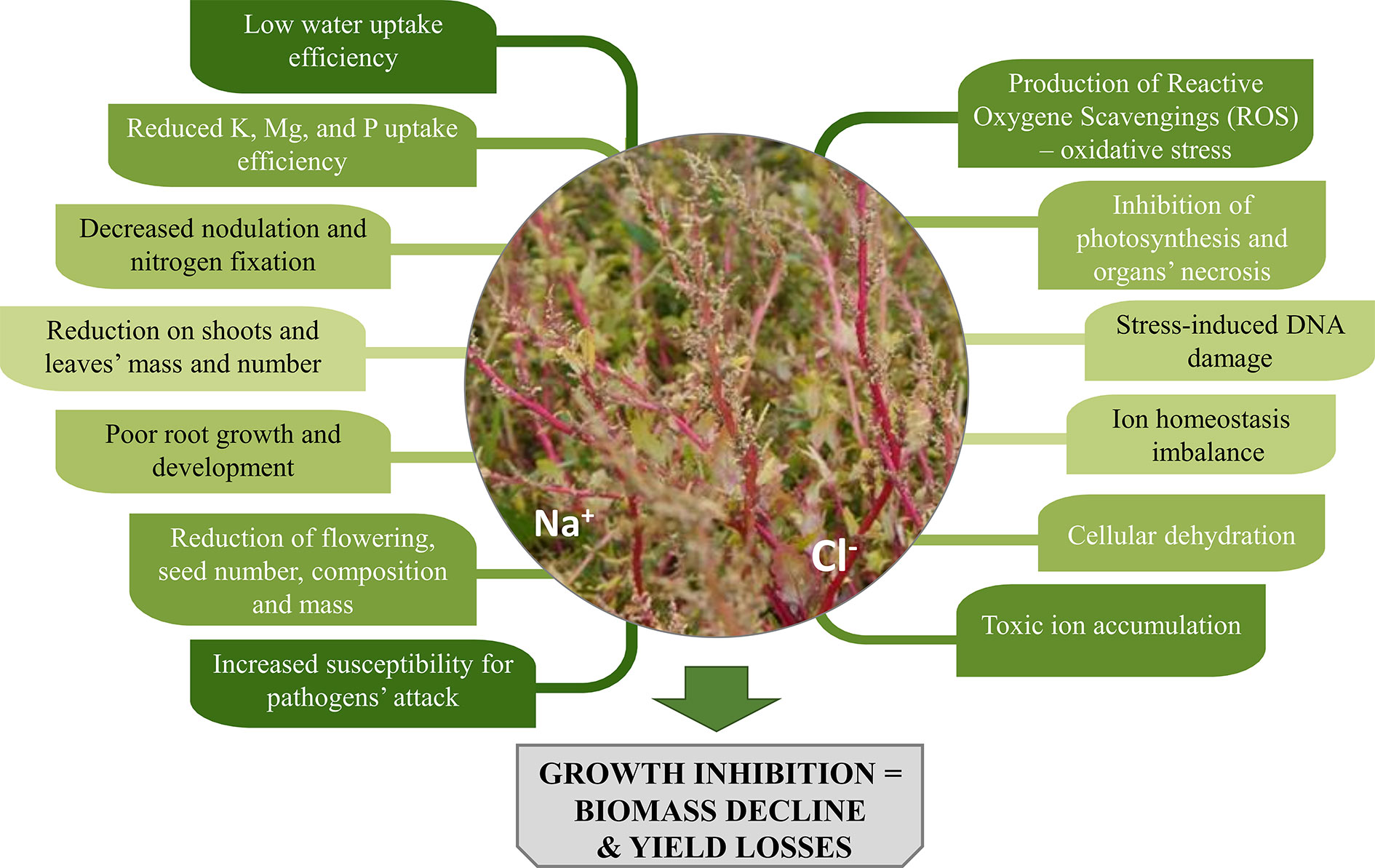
Salinity causes low water potential in the soil, which negatively affects plants’ water and nutrients uptake. Plants collect salts simultaneously with the water they use and often accumulate Na+ and Cl- ions, that result toxic to plants’ cells due to ion imbalance mechanisms. What is more, enzymatic activity in cells may be disturbed. These factors trigger different responses in plants, manifested by a variety of symptoms both at cell and organ’s level (Figure 1).
A reduction in respiration characterises stressed plants, which also show altered assimilates distribution, inhibited photosynthesis process and lower production of new leaves. Simultaneously, increased morphological changes of organs (leaf thickening and succulence, a decrease of internode lengths), wilting, drying and even necrosis of organs and entire plants are observed (Parida and Das, 2005; Kumar and Verma, 2018). Not to mention that cultivation of crops in saline areas can adversely affect their palatability. Some plants under salinity stress might accumulate higher amounts of compounds such as oxalate and tannins, which are bitter, whereas for others, higher sugar content, and an improved flavour was observed (Masters et al., 2005; Rahneshan et al., 2018).
Different models may describe plants’ salt tolerance. One of the most popular is the Maas-Hoffman model and its modifications (van Straten et al., 2019). This mathematical tool reflects the relationship between crop yield in response to soil salinity, and for most crops, this follows a sigmoidal function. The breakpoint between the first part of the plot, being horizontal, and the second, which is sloping downward, is known as a threshold (ECt) or salt tolerance. Based on threshold and slope values, a classification in sensitive, moderately sensitive, moderately tolerant and tolerant crops can be made. There is a large variation in salt tolerance between plants’ species, from the extremely sensitive (like chickpea) to more tolerant (like cotton or beet). Among agricultural species, greens are particularly sensitive throughout the whole ontogeny of the plant, whereas the majority of cereals are highly resistant to salt impact (Munns and Rawson, 1999; Machado and Serralheiro, 2017).
In each crop, salt tolerance or sensitivity depend on the ability to uptake water and nutrients from saline soils avoiding at the same time an excessive accumulation of salt ions in the tissues. The absolute sensitivity and tolerance of plants to salinity, vary in a wide range, depending upon species, climate, soil type and its features, and agricultural practices including water management, e.g. irrigation or waterlogging methods, their frequency and intensity (Shrivastava and Kumar, 2015 and references therein).
The plants are not susceptible to salt for the whole life since their sensitivity changes during the various growth stages. In general, it was found that plants are more sensitive to salt stress when they are in their early growth stages (seedling or establishment) compared to later development phases (Machado and Serralheiro, 2017).
As mentioned, it is believed that the most sensitive to salinity stage of the plant’s growth is the germination and seedling (Läuchli and Grattan, 2007). Plants, which are better adapted to soil salinity must either have a high tolerance to salt during germination or can delay germination. Moreover, tolerance to salinity differs widely among crops without always correlating to salt-tolerances based on yield-response functions. Cotton, for instance, which is considered a salt-tolerant crop, based on lint yields, showed to be susceptible to poor stands when growing in fields irrigated with saline-sodic water (Grattan and Oster, 2003).
With maturation, plants acquire a higher tolerance to salinity. However, a prolonged exposure to salinity causes a reduction in biomass, due to lower stems numbers and smaller leaf area. A decreased leaf area commonly expresses salt-affected reduction in shoot growth, which is crucial for water uptake by the plant (Munns and Tester, 2008). In sugar beet leaves, cells’ elongation was found to be more salt-sensitive in comparison to leaf initiation related to cells’ division (Rozema et al., 2015). Hu and Schmidhalter (2007) reported that cells’ division of grass leaves was reduced by salinity. Khan et al. (2017) found in chickpea (Cicer arietinum L.) a reproductive failure due to reduced supply of assimilates to reproductive tissues, decreased leaf area and reduced photosynthesis, water restriction and hormonal imbalances.
Roots are the organs directly exposed to the saline environment, and they control the uptake and internal translocation of water, nutrients and salts. The anoxic situation often present in saline soils can have a more significant impact on radical architecture than salinity itself. In the case of root systems, oxygen deficiency contributes significantly to poor uptake of nutrient ions and decreased ability to toxic ions (such as Na+) removal (Barrett-Lennard, 2003). It is believed that roots are generally affected by excess salinity but commonly still less than aboveground organs (Rahneshan et al., 2018).
Bacterial and Fungal Communities Associated With Roots and Rhizosphere in Saline Soils
According to Matilla et al. (2007), two selective forces of different nature are essential for microorganisms to colonise the rhizosphere: stress adaptation and the availability of particular nutrients. In saline soils, microbial communities associated with the rhizosphere, phyllosphere, and endosphere of halophytes comprehend members of Archaea and Bacteria domains and kingdom Fungi (Mukhtar et al., 2019). These communities are directly or indirectly involved in the osmoregulation of halophytes that allows them to survive under salinity stress conditions. Endophytic bacteria and fungi are those organisms whose life-cycle take place partly or entirely inside a plant. These organisms can live in the intercellular spaces of different tissues and plant’s organs (Kandel et al., 2017) without causing visible external sign of infection or a negative effect of the host (Weyens et al., 2009). Endophytes’ population in a specific environment or within a single plant may differ with tissue type, plant growth stage and dimension of the ecological niches. Several studies comparing rhizosphere and endophytic microbial communities showed how species assemblages are significantly different but also that endophytic bacteria and their communities have some peculiar, commune, traits (Kushwaha et al., 2020).
Endophytic bacterial species, for example, have larger genomes as compared to rhizosphere bacteria (Pini et al., 2011). Moreover, the diversity, richness and evenness of rhizosphere bacterial communities seem higher compared to endophytic ones (Huang, 2018). Bacterial phyla are different when considering rhizospheric soil and the plant’s tissues, with a predominance of Proteobacteria and Chloroflexi in the former and Acidobacteria, Bacteroidetes, and Planctomycetes in the latter (Kandel et al., 2017; Huang, 2018). However, Compant et al. (2010) pointed out that the majority of plant-associated bacteria derives from the soil environment. Some of them can penetrate plants’ roots thanks to specific mechanisms that are responsible for rhizosphere and endophytic competence. Chemotaxis, quorum sensing, flagella, antibiotic secretion, siderophore production are only some of the tools that rhizosphere bacteria need to be able to inhabit some specific plant-associated niches (Matilla et al., 2007; Compant et al., 2010). Liu et al. (2017) suggested that plant roots act as “gatekeepers” because they select soil bacteria from the rhizosphere and rhizoplane. This result in an endophytic root microbiome dominated by Proteobacteria, Actinobacteria and to a lower degree, by Firmicutes and Bacteroidetes, but with Acidobacteria and Gemmatimonadetes resulting almost depleted (Liu et al., 2017). The traits that characterise the bacteria that can successfully colonise and establish in endophytic niches are motility, reactive oxygen species scavenging, plant cell-wall degradation abilities. The salinity is in itself a critical environmental filter that selects species with very particular characteristics, which are probably upstream of the further selection that the rhizosphere first, and the tissues of the plant then, exert in soil microbial communities. In some studies, it has also emerged a decisive role of salinity in defining the type of endophyte associations that a plant establishes. Qin et al. (2016) provided further evidence that plants gain a higher advantage from association with a diverse microbial community (microbiome) compared with the interaction with single members of a community.
In addition to bacterial communities, also mycorrhizal symbioses showed a fundamental role in the improvement of plant nutrition, especially at the presence of environmental stresses (Qin et al., 2017). Plant-associated mycobiota belong to arbuscular mycorrhizal fungi (AMF), ectomycorrhizal fungi (EMF), non-mycorrhizal basidiomycetous fungi (NMF), and a consistent number of ascomycetous species (Zuccaro et al., 2014). Different groups, mostly based on host colonisation pattern and type of transmission, have been delimited in plant-fungal endophytes. Some fungal endophytes exhibit a vertical transmission through the host seeds or a horizontal transfer with soil- or air-borne spores (Rodriguez et al., 2009). Moreover, habitat-adapted symbiosis has been observed in some groups of fungal endophytes, and they impart a host-specific tolerance to stress in limiting environments (Qin et al., 2017). Certain groups of soil fungi have only recently been associated with the rhizosphere and endophytic situations, particularly in extreme environments or where one or more forms of stress exist (Khidir et al., 2010). Like some bacterial species, that live both as saprophytes in the soil and as endophytes, many fungal species can be isolated from both the free soil, far from the roots of plants and in close association with them (Maciá-Vicente et al., 2012). However, it must be said that several studies, conducted at different functional scales, have shown that in saline soils, the gradient of salinity as well as the presence of a high spatial heterogeneity favor the presence of species with very different functional traits and an extremely uneven distribution of communities (Maciá-Vicente et al., 2012). A similar phenomenon has been observed in the distribution of communities of bacteria in Mediterranean saline soil. Canfora et al. (2014) showed apparent differences in bacterial community distribution, diversity and composition, according to an increasing degree of soil salinity, as a consequence of a multi-scale spatial variability. A patchy distribution of vegetation structure and soil chemical properties coincided with a heterogeneous distribution of many bacterial groups. Coversely, some bacterial phyla resulted spread in the whole study area, along with the occurrence of a significant number of “salinity unrelated” phyla (Nitrospira, Spirochaetes). Canfora et al. (2014) hypothesized that a patchy saline environment could be “compared to a set of islands that allow the formation of different communities, separated from each other by the discontinuity of the limiting and stress factors”. Therefore, a patchy saline environment would contain not a homogeneous microbial community developed to tolerate an extreme environment, but a whole set of different communities. By comparing this evidence to the rhizosphere, it is also possible to imagine that the roots and tissues of halophilic or halotolerant plants in a saline soil represent islands of biodiversity and constitute a complicated system in which processes, at the microscale level, are particularly relevant. The living conditions within the tissues of a plant in an extreme environment such as a saline soil can reasonably be less limiting than those an organism can experience when living free in the soil. Therefore, it can be hypothesized that in saline soils, it is possible to find species as endophytes that in other systems are more easily found free in the soil. Kearl et al. (2019) isolated bacteria from halophytes (Salicornia and Allenrolfea) and observed that there were different populations in samples collected at different times of the year, with a majority of the genera, however, present independent of when the samples were collected.
Thiem et al. (2018) analysed the community structure of plant-associated endophytes of Alnus glutinosa, that is a dual mycorrhizal tree that forms ectomycorrhizal (EM) and arbuscular (AM) root structures, and can typically associate also with nitrogen-fixing actinomycetes. The authors sampled the plant’s root microbiome present at two forest test sites (saline and non-saline). They found that the dominant type of root microsymbionts of alder were ectomycorrhizal fungi, whose distribution depended on the site (salinity). In contrast, representatives of fungal saprotrophs or endophytes displayed the opposite tendency.
Same applies to fungi, as the Pleosporalean taxa (i.e. Pleosporales order, within the class Dothideomycetes, Ascomycota) and other generalist endophytes and epiphytes that seem particularly present in high salinity environments (Quin et al., 2017). Pleosporalean fungi like those belonging to the genera Pleospora, Alternaria, and Phoma, are very frequent colonisers in halophytes (Quin et al., 2017 and references therein), and in plants from arid environments (Khidir et al., 2010), and have common traits of endophytes from other adverse environments. More recent studies based on a comprehensive molecular analysis involving both fungal and bacterial communities have also highlighted a close relationship between the two. Furtado et al. (2019) examined the microbiome of the non-mycorrhizal halophyte Salicornia europaea and showed a significant influence of the Salicornia bacterial community on the fungal one, but not the other way around. They also found that the sampling season was not influencing the biodiversity. Seasonality did not appear to be an essential factor in shaping endophytic microbial communities in saline soils also from other studies (Thiem et al., 2018).
Recent studies illustrated some main mechanisms that emerged as capable of supporting directly or indirectly plant growth under saline stress (Kushwaha et al., 2020). Plants’ strategy to survive under salinity conditions comprises the synthesis and accumulation of osmolytes, as free amino acids (i.e. proline) and sugars to sustain an adequate osmotic cellular pressure needed for the functioning of cellular metabolism (Kushwaha et al., 2020). Endophytes in plants living in saline soils proved to help them accumulating osmolytes and antioxidant compounds (Vaishnav et al., 2019).
Strategies for Increasing Salt Stress Tolerance
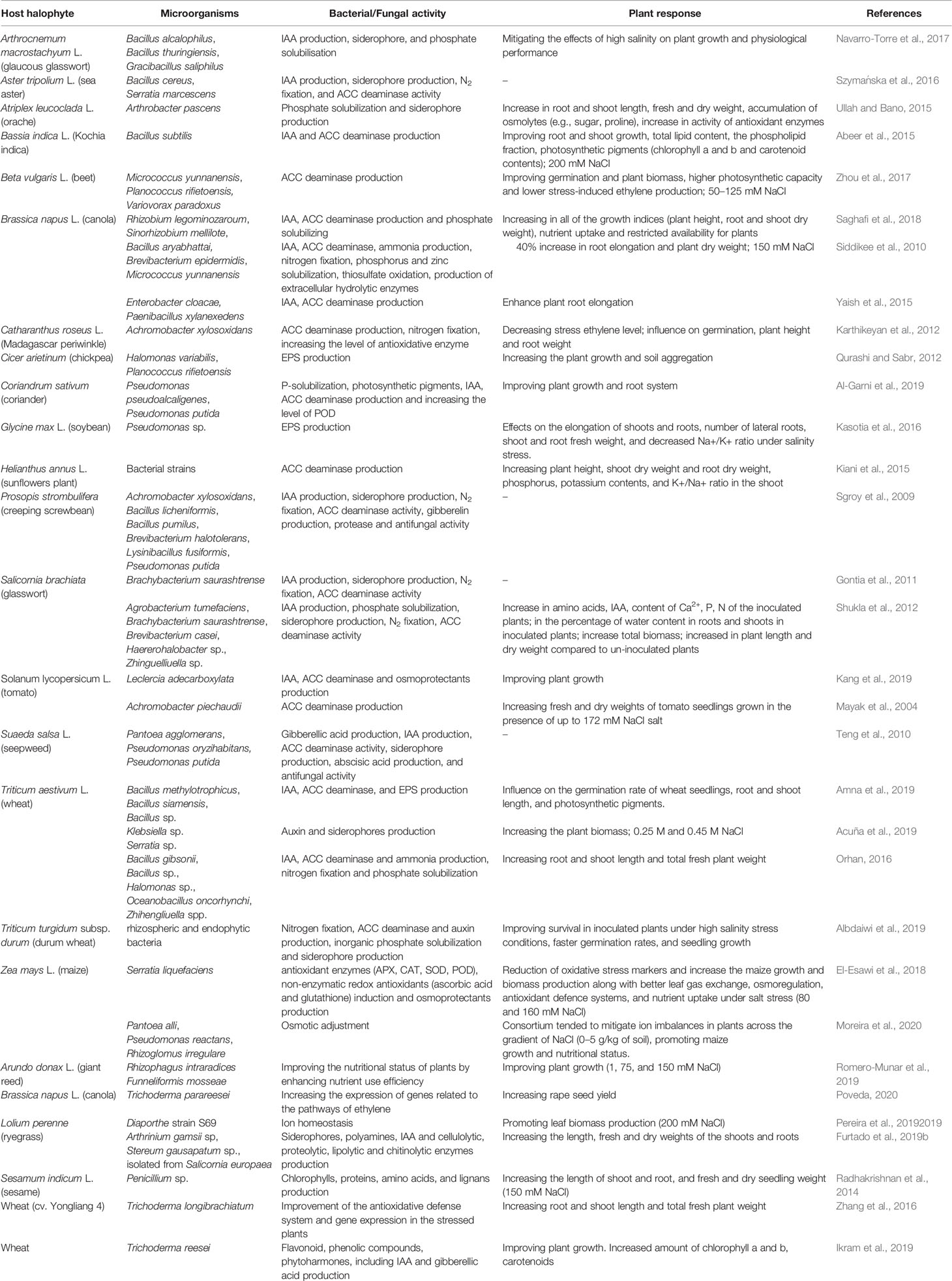
Halophytes are considered model plants, enabling the study of adaptive mechanisms like the induction of enzymes with antioxidant functions, the accumulation of toxic ions in their vacuoles, the storage of compatible soluble substances, occurring in the cell in response to cellular stress. Consequently, the salt-resistance genes involved in the above processes can be expressed in conventional crops, increasing their resistance to environmental salinity. So far, however, this strategy has proved to be inefficient and was implemented mainly under laboratory conditions. An equally costly and environmentally unfriendly approach in the production of salinity resistant plants is the pre-treatment of biological materials with specific, selective, chemicals, e.g. ascorbic acid, nitric oxide, H2O2, Ca2+, K+, paraquat and glutamate, silicon, phosphorus and humic acid, glycine betaine, jasmonates and salicylic acid, 5-aminolevulinic acid (El-Esawi et al., 2018) or with physical effectors like UV-B irradiation (Dhanya Thomas et al., 2020). However, these methods are not recommended for sustainable agriculture. Instead, the use of soil bacterial and fungal community colonizing plant’s roots and stimulating plant’s growth under stress conditions are promising for the increase of agricultural productivity in saline areas. Kloepper and Schroth (1978) introduced the term “rhizobacteria” to describe this microbial community collectively. Three years later, the same authors expanded the term to “plant growth-promoting rhizobacteria” (PGPR). PGPRs can be distinguished between “extracellular plant growth-promoting rhizobacteria (ePGPR)” and “intracellular plant growth-promoting rhizobacteria (iPGPR)” (Martinez-Viveros et al., 2010). The ePGPRs are bacteria that can have their niche in the rhizosphere, on the rhizoplane, or in the interstitial microenvironments of the root cortex. The iPGPRs live inside specialised nodular structures of root cells (Bhattacharyya and Jha, 2012). Some ePGPRs and iPGPRs show tolerance to a high concentration of salts and thus can also grow in saline soils and be associated to halophytes, or more in general, in niches characterized by low water potentials due to salt stress or to a dry climate (Khan et al., 2019) (Supplementary Material Table 1). Salt-tolerant PGPRs (ST-PGPR) include mainly bacteria of the genus Bacillus, Pseudomonas, Enterobacter, Agrobacterium, Streptomyces, Klebsiella and Ochrobactrum (Sharma et al., 2016). Whipps (2001) roughly distinguished between three strategies of interaction of the rhizobacteria with the plants: neutral, negative, or positive. Neutral interaction means that the rhizobacteria, although living in the plant’s rhizosphere have no visible effect on the growth and metabolism of the host (Beattie, 2006). Positive interactions comprise those ST-PGPRs (salt-tolerant plant growth promoting rhizobacteria) that have a promoting effect on plant physiology and tolerance to salinity (Figure 2). The ST-PGPRs, in turn, from a utilitarian point of view, can be subdivided into biofertilizer microorganisms, phytostimulators, and biopesticides. Negative interactions comprehend all those situations where the bacteria are phytopathogenic or produce substances toxic to plants (i.e. hydrogen cyanide or ethylene). However, sometimes, the secretion of the toxic substance, such as hydrogen cyanide by PGPRs, is also beneficial as it is one of the many ways to protect the plant from phytopathogens. ST-PGPR can improve plant’s growth and tolerance to salinity stress through the accumulation of osmolytes (proline, trehalose, and glycine betaines) (Supplementary Material Table 2), the production of phytohormones (auxins, gibberellins, cytokinins), ion homeostasis, the improvement of nutrients uptake (N2 fixation, solubilising of P, K, Zn, and Si), the activation of plant’s antioxidative enzymes, and the synthesis of ACC deaminase, indole-3-acetic acid (IAA), exopolysaccharides, and siderophores (Egamberdieva et al., 2019).
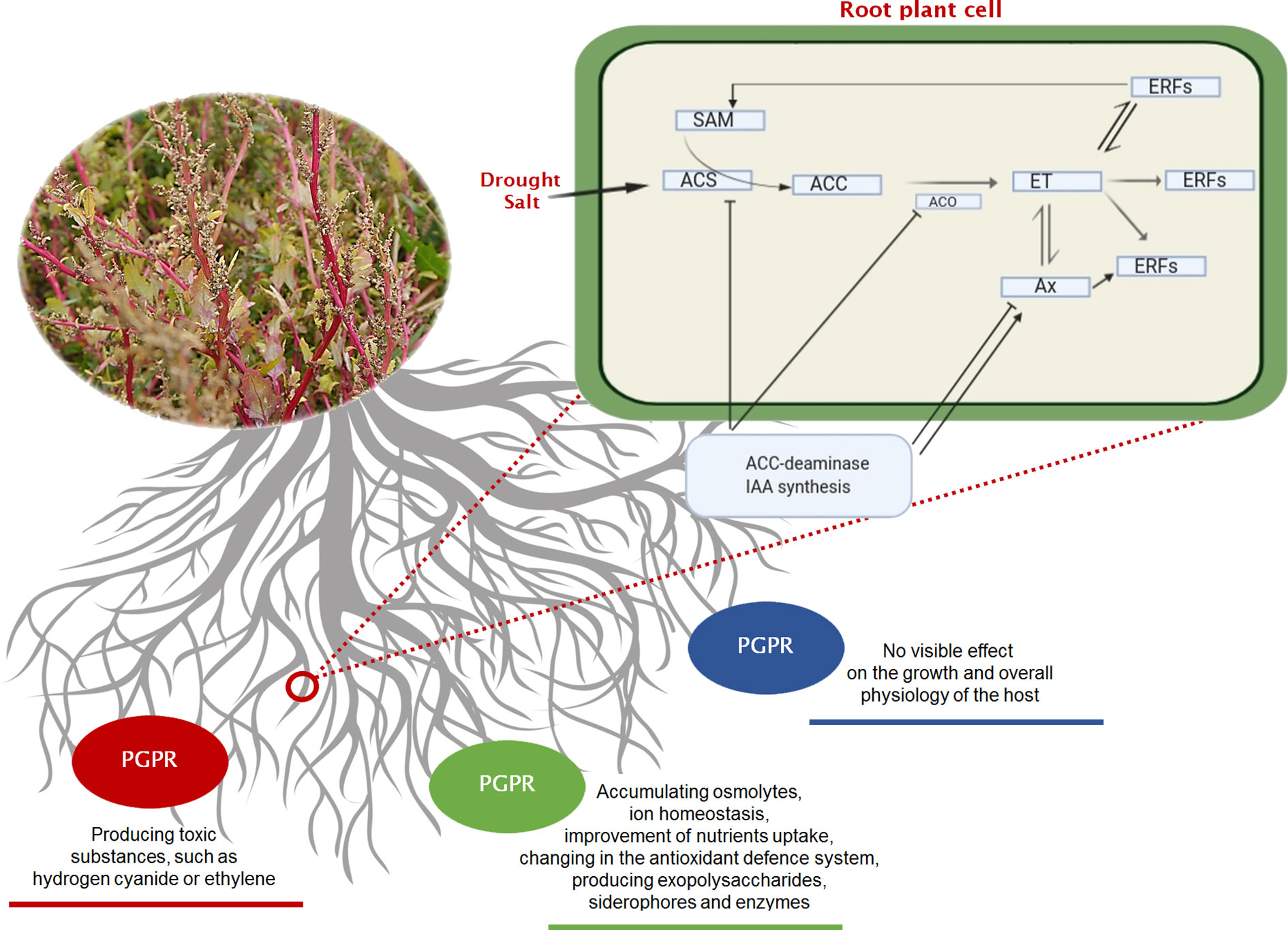
Figure 2 Three basic strategies of interaction (neutral—blue line, negative—red line, positive—green line) existing between the rhizobacteria and growing plants. In the negative interaction, it is highlighted the pathway of ethylene (ET) that occurs under soil stress, and the effect of mitigation that can be played by PGPRs on a plant root’s cell. Regulation of ethylene signalling and plant stress response. Ethylene pathway in plants. ACC (1-aminocyclopropane-1-carboxylic acid) the amino acid methionine is converted to SAM (S-adenosylmethionine) by the action of ACC synthase enzyme (ACS). ACC is then converted to ethylene by the enzyme ACC oxidase (ACO), triggering different ethylene response factors (ERFs). Plant growth-promoting bacteria can alter all steps of ethylene signalling. Some bacteria species can increase the ethylene levels by producing ACC oxidase (microbial ethylene-forming enzyme), by inducing ACC synthase in plant or by affecting other plant hormones indirectly. They can also modulate ethylene response by producing plant hormones that interact with ethylene signalling. Other microorganisms can also decrease ethylene production by cleaving its precursor ACC. ACC, 1-aminocyclopropane-1-carboxylate; ACS, ACC synthase; ACO, ACC oxidase; Ax, Auxine; ET, Ethylene; ERFs, Ethylene Response Factors; SAM, S-adenosyl methionine.
Plant growth-promoting fungi (PGPFs) are soil-borne, non-pathogenic, saprophytic microorganisms. This group contains several fungal taxa, and among the most quoted genera, especially in saline environments, there are Penicillium spp., Fusarium spp., Alternaria spp., Aspergillus spp., Sclerotium spp., and Phoma spp., which colonise plants roots and form symbiotic interactions with them (Gangwar et al., 2017; Naziya et al., 2020). Pereira et al. (2019), for example, surveyed the culturable endophytic mycobiota of Festuca rubra, a perennial grass diffused in coastal environments with low nutrient availability, wind, and salinity. Taxa belonging to Fusarium, Diaporthe, Helotiales, Drechslera, Slopeiomyces, and Penicillium were constant inhabitants of the plant’s roots (occurred in more than 20% of the plants the authors analysed). Seventy-one point eight percent of the strains they could culture were halotolerant. When the authors used the isolates to inoculate the grass Lolium perenne, a Diaporthe strain increased leaf biomass production under both normal and saline watering regimes (200 mM NaCl). PGPFs enhance the plant growth directly and indirectly by the production of IAA, siderophore, cellulase, chitinase, gibberellins, and increasing phosphorus solubilisation and availability (Mishra et al., 2017; Naziya et al., 2020).
High salinity increases susceptibility to various phytopathogens and promotes some fungal soil-borne diseases in plants. Crops protection against pathogens is extremely difficult, because the use of chemicals agents e.g. fungicides, bactericides and nematicides has a negative impact on the environment and organisms while increases the costs of cultivation (Egamberdieva et al., 2017; Sharma et al., 2017). An alternative to the use of chemicals is the use of PGPRs and PGPFs, which could synthesise the aforementioned lytic enzymes, siderophores, IAA, and antibiotics. These compounds could reduce and inhibit phytopathogens that cause plant infections (Labuschagne et al., 2010; Murali et al., 2012; Gangwar et al., 2017). Trichoderma isolates widely used in bio-fungicidal formulations can be ineffective at high salinity conditions, since most of these fungal species have a low osmotolerance (Mohamed and Haggag, 2006; Bheemaraya et al., 2013). However, some studies targeted halotolerant biological control fungi. This is the case of the experiments by Gal-Hemed et al. (2011) who isolated T. atroviride and T. asperelloides from the Mediterranean sponge Psammocinia sp. and used them to reduce Rhizoctonia solani disease in beans, and more recently the study by Sánchez-Montesinos et al. (2019). They isolated Trichoderma strains from the seagrass Posidonia oceanica and evaluated its capacity to control the disease caused by Pythium ultimum in melon seedlings under various levels of salt stress.
Osmoprotectants
In response to salinity stress, ST-PGPRs accumulate many metabolites called compatible (organic) solutes, among which amino acids and derivatives (e.g., glutamate, proline, peptides, and N-acetylated amino acids), quaternary amines (e.g., glycine betaine and carnitine), sugars (e.g., sucrose and trehalose), and tetrahydro pyrimidines (ectoines). These metabolites enhance the stability of protein conformation, the balance of cell redox condition, cytosolic pH, complex II electron transport, membrane integrity, and the activity of enzymes such as ribulose bisphosphate carboxylase/oxygenase (RUBISCO) (Saghafi et al., 2019). Moreover, the storage of osmolytes represents a successful stress response mechanism that helps the bacteria to limit water loss, increasing their cytoplasmic concentration of K+. In some halophilic bacteria, the internal concentration of osmoprotectants due to salt stress may reach up to 1 M (Saum and Müller, 2007). Rodriguez-Salazar et al. (2009) observed that Azospirillum spp. accumulates proline, trehalose, and glycine betaine as a mechanism for protection against osmotic stress. Kushwaha et al. (2019) found in Halomonas sp. that accumulation of betaine suppresses the de novo synthesis of ectoine at low NaCl concentrations, however, at higher NaCl concentrations the amount of ectoine is significantly larger than betaine. It means that the salinity stress transcriptionally up-regulates ectoine accumulation. The expression of the genes proH, proJ, and proA involved in proline biosynthesis was induced in some of the halophilic bacteria at higher salt concentration, leading to the highest accumulation of proline (Saum and Müller, 2007). Also, several reports determined the link between proline accumulation and pyrroline-5-carboxylate synthase (P5CS) gene expression level after PGPR inoculation and hypothesized that bacterial treatment upregulates the P5CS gene expression in plant roots, causing intracellular storage of free proline (Kim et al., 2007; Kumari et al., 2015a; Kumari et al., 2015b). Many ST-PGPRs showed a high expression of genes implicated in trehalose biosynthetic pathways (e.g. trehalose 6-phosphate gene) (Qin et al., 2018). Trehalose is an osmoprotectant, and its role in salt-stress tolerance has been well documented (Garg et al., 2019; Orozco-Mosqueda et al., 2019; Shim et al., 2019). Figueiredo et al. (2008) reported that Rhizobium tropici and Paenibacillus polymyxa were modified to overexpress trehalose 6-phosphate gene and were co-inoculated in Phaseolus vulgaris plants resulting beneficial for plants grown under saline stress, with higher nodulation and N content. A differential gene expression analysis of the tissues of the nodules compared to normal roots revealed upregulation of stress tolerance genes, which suggested that extracellular trehalose works as an osmoprotectant, including tolerance to salinity (Figueiredo et al., 2008).
Ion Homeostasis
One strategy used by bacteria to limit salt uptake, also by plants, is by trapping cations in their exopolysaccharide matrix. This mechanism results in an altered root structure with the formation of extensive rhizo-sheaths (agglutinated soil that adheres to roots when they are removed from the pot or field). Moreover, at the rhizosphere level, it has been found differential regulation of the expression of genes involved in ion affinity transporters. PGPR often impact the mineral nutrient exchange of both macro and micronutrients as a strategy to react to nutrient imbalance due to a higher uptake of Na+ and Cl− ions (Saghafi et al., 2019). Both fungi and bacteria can help plants to keep cellular ion homeostasis and sustainable K+/Na+ ratios in shoots. This has been documented as a mechanism that reduces Na+ and Cl− accumulation in leaves, by increasing Na+ exclusion via roots and boosting the activity of high-affinity K+ transporters (Ilangumaran and Smith, 2017). Zhang et al. (2008a) reported that the inoculation of Arabidopsis thaliana with B. subtilis moderated the adverse effects of salinity by regulating HKT1 potassium transporter. This bacterium also stimulated the overexpression of the AtHKT1 gene, expressing for a high-compatibility transporter for potassium ion, in Arabidopsis under conditions of salt stress. Puccinellia tenuiflora, a salt-excluding halophytic grass, when inoculated with B. subtilis showed lower levels of Na+ accumulation. The plant at the same time upregulated plasma membrane Na+/H+ transporters SOS1, and HKT-type protein and tonoplast Na+/H+ antiporters genes. However, one of the HKT genes was downregulated in roots under high salt concentrations (Zhang et al., 2008a; Niu et al., 2016). This showed how the bacterium synergistically regulated Na+ homeostasis by controlling Na+ transport systems at the whole-plant level under both lower and higher salt conditions, differentiating the mechanisms at play in case of high or mild salinity conditions. According to Rojas-Tapias et al. (2012), the inoculation of auxin-producing strains of Azotobacter in maize plants exposed to saline stress resulted in better K+ uptake and Na+ exclusion from plant’s tissues.
Moreover, the authors showed that after PGPR inoculation chlorophyll, proline, and polyphenol contents in maize leaves increased along with a better general plant stress response (Rojas-Tapias et al., 2012). Ilangumaran and Smith (2017) reported that in many studies on the interaction between plants and PGPR species, the genes involved in ion homeostasis showed differential expression under saline stress. For example, in an experiment where Arabidopsis thaliana was treated with Burkholderia phytofirmans the expression of both bacterial and plant’s genes involved in ion homeostasis (KT1, HKT1, NHX2, and SOS1) was rapidly altered as a result of an imposed saline stress (Pinedo et al., 2015).
Also, fungi can play a decisive role in a plant’s ability to improve nutrients’ uptake and regulate the osmotic balance in soils affected by salinity. Arbuscular mycorrhizal fungi are plants symbionts that increase root phosphorus uptake and confer the mycorrhizal plants’ tolerance to salinity. Romero-Munar et al. (2019) demonstrated that in Arundo donax a commercial inoculum containing the arbuscular mycorrhizal Rhizophagus intraradices and Funneliformis mosseae improved the nutritional status by enhancing nutrient use efficiency. The authors suggested that increased use efficiency of phosphorus could have improved ion (Na+ and K+) uptake and allocation. Arbuscular mycorrhizal fungi have also been reported to enhance the ability of wheat plants to modulate the reactive oxygen scavenging system when coping with salinity stress (Talaat and Shawky, 2011) (Table 1).
Symbiotic fungi can modulate gene expression of the host plant to modify its phenotype in order to improve the tolerance to abiotic stress factors caused by soil salinity (Khan et al., 2013). Molina-Montenegro et al. (2020) showed that inoculation of plants’ roots with Antarctic fungal endophytes improves growth and survival by changing the expression of a gene responsible for Na+/H+ antiporters proteins integrated with vacuolar membranes. In particular, the NHX proteins are involved in the maintenance of cell turgor through ionic balance control and are associated with the capacity of accumulating Na+ inside vacuoles.
The Improvement of Phosphorus, Potassium, and Zinc Solubilisation
Phosphorus (P) is one of the essential macronutrients for plants, although its availability is limited due to its low solubility. P is required as an essential nutritional element for photosynthesis, energy transfer, biosynthesis of macromolecules and respiration (Fernandez et al., 2007). The average content of phosphorus ions in the soil is 0.05% (w/w). Still, often only 0.1% of the total P is available to plants because of its precipitation in soil (Etesami and Beattie, 2018). Salinity leads to depletion and sedimentation of absorbable phosphorus. Phosphate-solubilising halotolerant PGPRs (Bacillus, Pseudomonas, Achromobacter, Alcaligenes, Brevibacterium, Serratia, Xanthomonas, and Rhizobium) provide an opportunity to enhance P availability to plants without deteriorating soil salinity levels. These microorganisms can hydrolyse inaccessible phosphorus forms into absorbable forms via various mechanisms like chelation, ion exchange, and acidification by secreting low molecular weight organic acids, such as gluconic acid, citric acid, succinic acid, propionic acid, and lactic acid (Choudhary, 2012; Etesami and Beattie, 2018; Saghafi et al., 2018). In salt-affected soils, the inoculation of wheat with Bacillus aquimaris increased plant P content under salinity stress (Upadhyay and Singh, 2015). Khan et al. (2019) identified highly salt stress-tolerant strains of Arthrobacter woluwensis, Microbacterium oxydans, Arthrobacter aurescens, Bacillus megaterium, and Bacillus aryabhattai, which showed to increase phosphate uptake in several plants: Artemisia princeps, Chenopodium ficifolium, Echinochloa crus-galli, and Oenothera biennis (Khan et al., 2019). Salcedo et al. (2014) identified seven best phosphate-solubilising actinobacteria strains out of the 57 strains that they isolated from soil. El-Tarabily and Youssef (2010) screened the mangrove A. marina rhizosphere and identified 129 bacterial strains capable of solubilising rock phosphate. In particular, Oceanobacillus picturae showed to be able to mobilize 97% of the available mineral P. Many other genera of bacteria isolated from halophytes (i.e. Arthrobacter, Bacillus, Azospirillum, Vibrio, Phyllobacterium) were found capable to implement P absorption in halophytes under salinity stress (Banerjee et al., 2010; Yasmin and Bano, 2011). Bashan et al. (2000) showed that in the leaves of halophytes inoculated with halotolerant PGPRs, as species of the genera Azospirillum, Vibrio, Bacillus, and Phyllobacterium, the P content increased. Vaishnav et al. (2016) used a hydroponic system to demonstrate how insoluble phosphate-solubilising bacteria that solubilise sedimentary phosphorous actively increased the availability of assimilable P to plants in salinity stress conditions. Yadav et al. (2011) reported the contribution of Aspergillus niger, Penicillium citrinum, and Trichoderma harzianum in phosphate solubilisation and their beneficial effects on chickpea growth. Amongst studied fungal genera, the highest P-solubilizing ability was attributed to Aspergillus and Trichoderma species. Ceci et al. (2018) showed that many saprotrophic fungi could mobilize P from insoluble forms according to a variety of mechanisms, with strains of Rhizopus stolonifer var. stolonifer, Aspergillus niger and Alternaria alternata among the best performing strains in terms of amounts of insoluble phosphate solubilisation.
Apart from phosphorus, another essential nutrient ingredient for plants is potassium (K). This element plays a vital role in plant metabolism and improves the quality of the crop production due to its role in grain filling, and in promoting disease resistance, leading to a higher resistance of plants to stress. The concentration of potassium in the soil solution is usually 1%–2% (Sindhu et al., 2010). The potential of soil application of potassium solubilising microorganisms (KSBs) is widely studied, especially in saline soils where this element is even less available to plants. These bacteria solubilise potassium-containing minerals (mica and orthoclase) by producing tartaric, succinic, citric, oxalic, and alpha-ketogluconic acids (Saghafi et al., 2019). Singh et al. (2010) reported that Bacillus mucilaginosa, Azotobacter chroococcum, and Rhizobium sp. were able to increase potassium absorption by wheat and corn.
Several authors showed that also fungi, especially ectomycorrhizal species, can weather silicate minerals to extract nutrients like P actively, K, Ca, Mg, and Fe, in particular under conditions of nutrient limitation. Mycorrhizal (ecto- and endo-) contribution to K+ acquisition by plants has also been demonstrated (Benito and Gonzalez-Guerrero, 2014). Fungi are already used at industrial level to mobilize or precipitate also other metals, like Cu, Mn, Zn, even though they are poorly applied for plant nutrition. There are studies, however, for the application of fungi and bacteria as solubilizers of specific nutrients, and some bacteria are already applied in fertilization treatments to this aim.
Zinc deficiency, for example, is a significant problem for the plant, especially in saline arid and semi-arid soils. Plants absorb Zn mainly in the form of Zn2+, zinc hydrate, and organic zeolite and use it in biochemical reactions, for the stability of biological membranes, the activity of oxidative and carbonic anhydrase enzymes and the synthesis of the enzyme auxin (Broadley et al., 2007; Alaghemand et al., 2018). The most important method to provide plants with Zn is the application of rhizobacteria together with Zn-containing fertilizers. These bacteria can increase the solubility of poorly soluble Zn compounds by employing different mechanisms, such as chelation by siderophore (Tariq et al., 2007), reduction of soil pH by the production of organic acids (2-ketogluconic acid, gluconic acid) and proton secretion (Subramanian et al., 2009). Abaid-Ullah et al. (2015) reported that Serratia sp. could increase wheat yield through solubilisation of ZnO under different climates.
Biological Nitrogen Fixation by PGPRs
Halophytic crop species used in agriculture can be limited by the lack of available nitrogen often affecting saline soils. PGPRs can fix nitrogen through symbiotic and non-symbiotic mechanisms (Saghafi et al., 2019). The first method involves the formation of nodes in the host roots by bacteria which results in nitrogen content of approximately 65% of the total nitrogen assimilation by plants (Rajwar et al., 2013). The other group of nitrogen-fixing bacteria, including Azospirillum, Azotobacter, Burkholderia, Herbaspirillum, Bacillus, and Paenibacillus is not plant-specific (Goswami et al., 2015). Salt-tolerant N2-fixing PGPRs are an essential source of available N in saline soils, and the amount of nitrogen fixed by these bacteria has been estimated as 20-30 kg h-1 year-1 (Oberson et al., 2013). The potential benefits of nitrogen-fixing strains to halophytes and salt-sensitive crops underline the need of implementing the studies on N2-fixing halotolerant PGPRs to be used as boosters in saline soil-based agriculture (Goswami et al., 2015; Ilangumaran and Smith, 2017).
Siderophore Production
Some strains of bacteria produce siderophores, especially in the rhizosphere, this increases plant growth and prevent phytopathogens from proliferation by inhibiting them from accumulating iron (Scavino and Pedraza, 2013). Siderophores are Fe(III)-chelating compounds, usually small and with high-affinity so that plants, in need for iron nutrient, quickly access the iron-siderophore complexes. Iron is an integral part and cofactor of enzymes involved in plants’ respiration, photosynthesis, N2 fixation and many other biochemical processes (Abbas et al., 2015; Etesami and Beattie, 2018). Bacterial siderophores have a higher affinity for iron than fungal pathogens, which require iron for their metabolism and plants’ infecting mechanisms (Miethke and Marahiel, 2007). Many halotolerant PGPRs and PGPFs, particularly those isolated from halophytes (Table 1) produce iron siderophores. Among biocontrol agents, the strains belonging to Pseudomonas sp. secreting non-fluorescent and fluorescent siderophores such as pyochelins and pseudobactins are most effective competitors of Fe3+. The potential uses of siderophore producing bacterial strains have been reported in the suppression of fungal pathogens of rice and wheat (Labuschagne et al., 2010). Moreover, pyoverdine synthetised by P. aeruginosa under low-iron stress condition could inhibit the growth of Aspergillus flavus, A. oryzae, F. oxysporum, and Sclerotium rolfsii (Manwar et al., 2004).
Also, fungi, however, produce siderophores that can act as protectants against plants’ pathogens. Fungal strains belonging to Aureobasidium and Emericellopsis genera synthesise siderophores, which are part of the biocontrol strategies occurring in Salicornia plants (Furtado et al., 2019).
IAA Production
Indole-3-acetic acid (IAA) is the most common plant hormone of the auxin class, and it regulates various aspects of plant growth and development. IAA acts as an effector molecule between bacteria and IAA producing plants, and in bacterial-bacterial interactions (Spaepen and Vanderleyden, 2011). It is involved in many processes such as seed germination, root system development, or increasing plant tolerance to stress conditions (Aeron et al., 2011). IAA-producing microorganisms increase the root growth and root length of plants, which contributes to a greater root surface area enabling the plant to acquire more nutrients from the soil (Boiero et al., 2007). Tryptophan is the IAA precursor in most biosynthetic pathways. However, several reports indicate the possibility of IAA synthesis in tryptophan-independent reactions (Sitbon et al., 2000; Saghafi et al., 2018).
The positive impact of IAA produced by PGPRs on the growth of various plants in conditions of salinity stress has been determined. For example, IAA-producing, halotolerant and halophilic bacteria significantly affected root and shoot elongation and freshly available mass of Triticum aestivum plants under salt stress conditions (Orhan, 2016). Also, Brassica napus L. seedlings inoculated with IAA producing Rhizobium bacteria showed an improved growth rate under salt stress, regarding especially plant height, as well as root and shoot dry weight (Saghafi et al., 2018). It has been confirmed that the salt-tolerant B. subtilis promotes the growth and fitness of Indian bassia plants (Bassia indica) under salt stress by providing an additional supply of IAA, and induces salt stress resistance by reducing ethylene levels. Inoculation of unstressed and salt-stressed Indian bassia with B. subtilis has significantly improved root and shoot growth, total lipid content, the phospholipid fraction, the content of photosynthetic pigments and also increased oleic, linoleic, and linolenic acids in plant leaves, as compared to uninoculated plants (Abeer et al., 2015). IAA-producing bacteria are also involved in suppression of plant disease-causing pathogenic fungi. Pseudomonas extremorientalis and P. aureantiaca were successfully used for control cucumber root infection caused by F. solani (Egamberdieva et al., 2014). Other examples are given in Table 1. Sodium chloride (NaCl) induces a decline in the IAA level in rice seedlings (Sakhabutdinova et al., 2003). Auxin activates the transcription of various genes known as primary auxin response genes in Arabidopsis, rice and soybeans (Hagen and Guilfoyle, 2002). Auxin negatively regulates the expression of the rice gene adenosine phosphate isopentenyltransferase (OsIPT) that encodes a key enzyme in CTK biosynthesis in nodes, thus inhibiting the growth of tiller buds in rice (Liu et al., 2011). Therefore, the identification of new genes that respond to high saline conditions offers investigators the opportunity to develop new approaches to select varieties with different mechanisms of tolerance to salinity stress (Zhu, 2002).
ACC Deaminase Production
The production of ACC deaminase enzyme is an essential mechanism for the direct promotion of plant growth by PGPRs. Bacterial IAA affects the level of ethylene in plants by increasing the activity of ACC deaminase, catalysing the hydrolysis of 1-amino-cyclopropane-1-carboxylic acid (ACC), an ethylene precursor, to ammonia and α-ketobutyric acid (Glick, 2005; Etesami et al., 2015). In fact, under stress conditions, including salinity, ethylene level increases in the plant and 1-aminocyclopropane-1-carboxylate (ACC) is more consistently enzymatically converted into ethylene (Figures 2, 3). Ethylene is a plant growth regulator and stress hormone which plays a key role in causing physiological changes in plants at the molecular level (Pierik et al., 2009). Ethylene interferes with plant growth under salinity stress by inhibiting roots elongation (Glick, 2005), causing defoliation, premature senescence (Shaharoona et al., 2006; Bari and Jones, 2009). To increase resistance against the harmful effects of ethylene, plants are commonly treated with ACC deaminase-producing bacteria (Glick, 2005; Etesami et al., 2015).
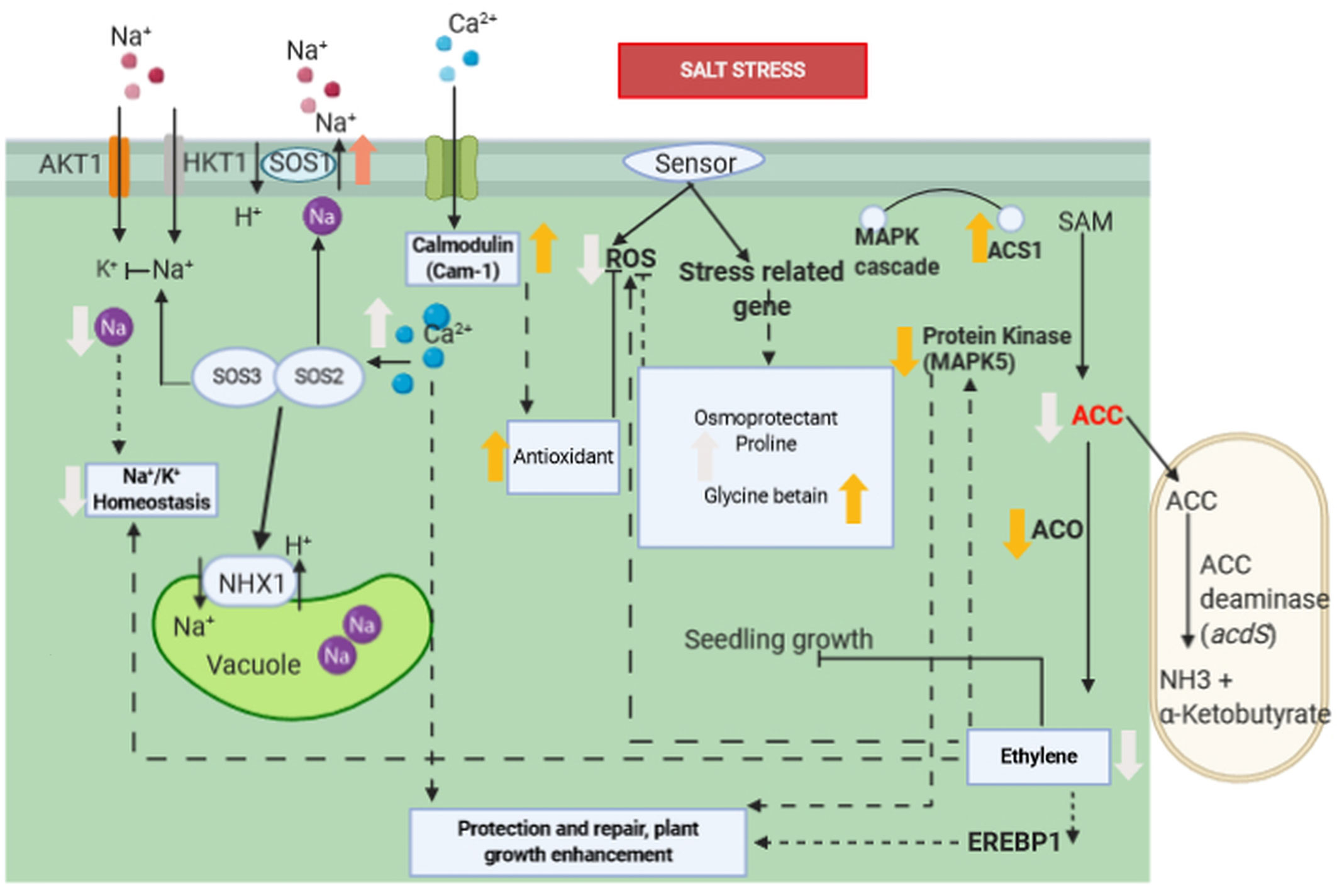
Figure 3 Molecular interaction of ACCD-producing endophytic bacteria associated with plant roots under saline stress. Salt stress induces the ethylene biosynthesis pathway by upregulation of ACS1. However, ACC is consumed as a result of the activation of acdS gene encoding ACCD of PGPR, whereas ACO1 and EREBP1 are down-regulated and ethylene production is reduced as a consequence. The reduction of ethylene induces a lower expression of MAPK5 and reduces the accumulation of ROS. Increase in proline, betaine, and glycine improves salt tolerance in plant’s roots. Ca2+content is increased, and Na+/K+ ratio is decreased, which are correlated with up-regulation of Cam1, SOS1, and NHX1 genes. Ca2+ signal activates the SOS3/SOS2 protein kinase complex, which negatively regulates the activity of Na+ ion channel. Association of Ca2+ and calmodulin activates antioxidant enzymes which subsequently inhibits ROS. Bold orange arrow indicates gene regulation, bold white arrow indicates plant physiological regulation, black arrow indicates positive regulation, dashed arrow indicates indirect positive regulation, black line with bar-end indicates inhibition and dashed line with bar-end indicates indirect inhibition.
So far, many researchers focused on the performance of PGPR with ACC deaminase activity to mitigate the adverse effects of elevated ethylene levels caused by salinity stress (Table 1). Siddikee et al. (2010) reported that 25 out of 140 halotolerant bacterial strains isolated from coastal soils of the South Korean Yellow Sea showed ACC deaminase activity. In particular, three of them Brevibacterium epidermidis, Micrococcus yunnanensis, and Bacillus aryabhattai generated more than 40% increase in root elongation and plants’ dry weight when compared to uninoculated salt-stressed canola seedlings (Siddikee et al., 2010). In the study by Kiani et al. (2015), sunflower plants were inoculated with ACC deaminase-producing bacteria, which resulted in better growth in terms of plants’ height and dry weight of shoots and roots. It has also been confirmed that Achromobacter piechaudii, an ACC deaminase-containing PGPR, can significantly increase the fresh and dry masses of tomato seedlings (Mayak et al., 2004). The study by Amna et al. (2019) investigated the role of halotolerant ACC deaminase-producing Bacillus spp. strains to help wheat seeds germination and seedling growth at different NaCl levels. There are pieces of evidence that ACC deaminase producing PGPRs enhance uptake of essential nutrients like N, P, and K, which consequently increase K+/Na+ ratios in the stressed plants (Nadeem et al., 2009).
Some fungi use similar mechanisms to alleviate plants stress. Some Trichoderma strains produce the enzyme 1-aminocyclopropane-1-carboxylate deaminase that regulates plants’ endogenous 1-aminocyclopropane-1-carboxylic acid (ACC), which is the direct precursor of the plant hormone ethylene. The ACC level regulates the plant’s tolerance to abiotic stress (Zhang et al., 2019). Poveda (2020) showed an impact of the fungi on plant’s hormones as well. The author determined the role that the enzyme chorismate mutase plays in Trichoderma parareesei ability to promote tolerance to salinity and drought in plants. This enzyme is at the base of a mechanism that increases the expression of genes related to the hormonal pathways of abscisic acid (ABA) under drought stress, and ethylene (ET) under salt stress.
Exopolysaccharides Production
The biofilm formation and exopolysaccharide (EPS) production by soil bacteria constitute important strategies to assist metabolism during stress imposed by salinity. EPS produced by PGPRs have a significant impact on plant growth and stress tolerance, such as drought or high salt concentration. They are hydrating compounds that are available for use before the decay of roots or germinating seeds. Bacteria produce polymeric biofilms on a variety of surfaces such as roots and soil, cementing particles, and forming aggregates. This can improve crop performance and soil physicochemical properties (Qurashi and Sabr, 2012; Amna et al., 2019). Salt-tolerant Halomonas variable and Planococcus rifietoensis strains can improve plant growth and aggregation of soil. These strains showed the formation of a biofilm and accumulated exopolysaccharides as a result of increasing salt stress (Qurashi and Sabr, 2012). Bacterial EPS can help to alleviate salinity stress by reducing the Na+ content available for plant uptake (Upadhyay et al., 2011). The kind of EPS produced by Pseudomonas spp. (Kasotia et al., 2016), and Bacillus spp. (Amna et al., 2019) helped in the binding of free Na+ from the soil, thus making Na+ unavailable to the soybean and wheat plants, respectively.
Increased Antioxidant Activity
High soil salinity is also responsible for increased production of reactive oxygen species (ROS) by plants, such as superoxide radical (O2−), hydrogen peroxide (H2O2), hydroxyl radical (OH·), and alkaline radicals which have a negative impact on proteins, DNA, lipids, and other biomolecules and cause oxidative effects including plant cell damage and premature senescence or necrosis (Møller et al., 2007; Miller et al., 2010; Habib et al., 2016; Zhang et al., 2018). ROS are produced at a low level in organelles (chloroplasts, mitochondria, peroxisomes) under optimal plant growth conditions. However, under stress conditions, their concentration increases significantly (Miller et al., 2010). A critical system responsible for the production of ROS is the plasma membrane-bound NADPH oxidase (RBOH), which controls cellular redox homeostasis under salinity stress (Hossain and Dietz, 2016; Hossain et al., 2017). Many other plant cell components also play a role in regulating intracellular ROS levels. Among them, the most important are antioxidant enzymes such as peroxidases (POD), catalase (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione reductase (GR), dehydroascorbate reductase (DHAR), monodehydroascorbate reductase (MDAR), glutathione peroxidases (GPX), or glutathione s-transferase (GST) (Yan et al., 2013; Hossain and Dietz, 2016; Sukweenadhi et al., 2018). Non-enzymatic components include glutathione, ascorbic acid, tocopherol, phenolic or polyphenolic compounds (Yan et al., 2013; El-Sayed et al., 2014; Fatma et al., 2014; El-Esawi et al., 2018a). To some species of endophytic fungi or ectomycorrhizal fungi are attributed antioxidant properties that could improve plant’s resistance to plant’s endogenous reactive oxygen species. Trichoderma is a genus of soilborne filamentous fungi that comprehend species capable of triggering plants’ defensive mechanisms and inducing tolerance to abiotic stress. Plants’ roots colonised by T. harzianum increased the production of antioxidant enzymes (Zehra et al., 2017). Also, Mastouri (2010) showed that plants colonised by T. harzianum showed a lower accumulation of lipid peroxides and a higher production of antioxidant compounds such as glutathione, resulting in a mechanism based on the control of the accumulation of reactive oxygen species that occur in stressed plants. T. harzianum can accelerate seeds germination while reducing the adverse effects caused to seeds and seedlings by thermal, osmotic, saline, and water stress. Examples of PGPRs and PGPFs, increasing the level of antioxidant enzymes and non-enzymatic redox antioxidants are listed in Table 1.
Biosynthesis of Hydrolytic Enzymes
One of the main indirect mechanisms of plant’s pathogens biocontrol by PGPRs is the production of cell wall degrading enzymes, like chitinases, glucanases, proteases and cellulases that cause lysis of the fungal cell walls (Siddikee et al., 2010; Berrada et al., 2012; Goswami et al., 2014). Teng et al. (2010) reported that halotolerant Pseudomonas sp. strain isolated from Suaeda salsa is a source of proteinases active against phytopathogenic fungi like Fusarium oxysporum (Teng et al., 2010). Two bacterial strains (B. halotolerans and B. pumilus) isolated from the halophyte Prosopis strombulifera were able to produce proteinases inhibiting the growth of Alternaria (Sgroy et al., 2009). B. cereus and B. thuringiensis isolated from salty Tunisian soils were able to produce N-acetyl-β-D-glucosaminidases, chitobiosidases, endochitinases, and they were active against F. roseum (Sadfi et al., 2001). The extracellular chitinases of Serratia marcescens and Enterobacter agglomerans have been indicated as biocontrol agents against Sclerotium rolfsii. The ability to suppress Fusarium oxysporum and Rhizoctonia solani was found in Peanibacillus spp., Bacillus spp., and Streptomyces spp., which synthesise β-1,3-glucanase that are lytic enzymes able to destroy the cell walls of some fungi (Compant et al., 2005; Labuschagne et al., 2010).
Other Extracellular Molecules
Bacteria secrete many extracellular molecules such as lipo-chitooligosaccharides, bacteriocins, polyamines and volatile organic compounds (Supplementary Material Table 2). It has been demonstrated that these molecules often control metabolic pathways and have a role in regulatory functions that increase the plant’s defence and stimulate its growth, stress tolerance, and disease resistance.
Lipo-chitooligosaccharides (LCOs), in particular, are produced by rhizobia and have been found to initiate nodule formation in response to root exudates and flavonoids (Ilangumaran and Smith, 2017). The LCOs molecules have a conserved core and a variable N-Acetyl chain length, with different substitutions (sulfation or glycosylation) and degree of saturation, which account for host specificity (Oldroyd, 2013). Miransari and Smith (2009) reported how Bradyrhizobium japonicum, when inoculated in soybean under different salinity levels (from 36 to 61 mM NaCl), enhanced nodulation and growth of plants with the effects becoming more consistent with time.
Rhizobacteria secrete bacteriocins, that are small proteinaceous or peptidic toxins. These act as bactericidal or bacteriostatic agents against competing bacteria and indirectly promote microbial diversity under salinity stress. Some bacteriocins showed a role in plant’s resistance to stress. Bacillus thuringiensis is a soybean endosymbiont that, in vitro, produces “thuricin 17”, a bacteriocin that is capable of manipulating plant proteome profile, enhancing its tolerance to salinity (Subramanian et al., 2016).
Polyamines (Pas) such as spermidine, spermine, and putrescine, consist of low molecular weight aliphatic amines that can have antioxidant activity. These compounds are present practically in all living organisms and impact reactive oxygen species by scavenging free radicals and inducing the expression of genes related to cellular antioxidant mechanisms. Among all, spermidine which is secreted by Bacillus megaterium showed to increase the cellular accumulation of polyamines in plants. The mechanism in Arabidopsis involved osmotic stress tolerance via the activation of polyamines- mediated cellular signalling, which resulted in greater biomass, higher antioxidant enzyme activity and high photosynthetic capacity in the inoculated plant, compared to the untreated control (Zhou et al., 2016; Ilangumaran and Smith, 2017). Some authors used the definition “systemic induced resistance (SIR)” to cover several bio-protecting mechanisms induced by the PGPRs in plants and that act on multiple functions, once activated at the presence of a pathogenic infection (Numan et al., 2018).
Volatile organic compounds (VOC) are low molecular weight compounds, such as aldehydes, alcohols, ketones and hydrocarbons, which can enter the atmosphere as vapours due to significantly high vapour pressure. They are released from by PGPRs and stimulate plant growth, resulting in increased shoot biomass, and modulated stress responses (Ilangumaran and Smith, 2017). The role of VOCs in the biocontrol of plant’s pathogens and antibiosis is not fully understood, but some of the mechanisms at play gained attention in the last decades and will require further research (Bailly and Weisskopf, 2012). Paraburkholderia phytofirmans produced VOCs such as 2-undecanone, 7-hexanol, 3-methylbutanol that stimulate plant growth and induce salinity stress tolerance as demonstrated both in vitro and in soil. Growth parameters of Arabidopsis plants treated with these VOCs and measured as rosette area, fresh weight, and primary root length were higher than in the control plants (Ledger et al., 2016; Ilangumaran and Smith, 2017). VOCs emitted by Bacillus subtilis can stimulate many different hormonal signals in Arabidopsis thaliana, which includes cytokinins, salicylic acid, gibberellin, auxin and brassinosteroids (Zhang et al., 2007; Zhang et al., 2008b). Almost 600 genes related to metabolism, auxin homeostasis, cell wall modification and stress response were identified, and these studies showed that VOCs could play an essential role in plant growth and development. PGPRs VOCS can stimulate many chemical and physical changes, some of which could be addressed to improve plants’ tolerance towards abiotic stress (Mantelin and Touraine, 2004; Zhang et al., 2008a).
Several antimicrobial metabolites are the basis of biocontrol mechanisms activated by PGPMs against other microorganisms, and also phytopathogenic species. Strains of Arthrobacter spp., Pseudomonas spp., Bacillus spp., Streptomyces spp., can synthesise one or several types of bioactive compounds including amphisin, bacillomycin, 2,4-diacetylphloroglucinol (DAPG), fengycin, hydrogen cyanide, iturin, macrolactin, phenazine-1-carboxylic acid (PCA), pyoluteorin, pyrrolnitrin, surfactin, tensin, tropolone, and viscosinamide (Compant et al., 2005; Hinarejos et al., 2016). However, among bacterial biocontrol agents, the most cited are Bacillus spp. (B. amyloliquefaciens, B. cereus, B. licheniformis, B. pumilus, B. subtilis) and Pseudomonas spp. (P. chlororaphis, P. fluorescens, P. putida). Their antagonistic properties against bacterial (Ralstonia solanacearum, Xanthomonas axonopodis) and fungal (F. oxysporum, F. culmorum, P. ultimum, Rhizoctonia solani) phytopathogens of barley, chickpea, maize, peanut, rice, and wheat were widely proven (Raaijmakers et al., 2002; Yuttavanichakul et al., 2012). Moreover, due to multifaceted mechanisms of action in preventing pathogens’ infections, some strains of Bacillus sp. have been commercialized and used for improving crop production (Radhakrishnan et al., 2017; Nakkeeran et al., 2020). The formation of a biofilm around plant’s roots by some Bacillus species and the secretion of antagonistic metabolites inhibit pathogenic communities and reduce the occurrence and frequency of the diseases in plants (Radhakrishnan et al., 2017).
One of the antibiosis mechanisms adopted by Pseudomonas spp. is the production of hydrogen cyanide. which inhibits the therminal cytochrome c oxidase in the respiratory chain and binds to metalloenzyme (Ramette et al., 2003). However, hydrogen cyanide antagonistic potential against phytopathogens, mainly fungi, is still a matter of discussion (Rijavec and Lapanje, 2016). Ramette et al. (2003) showed a broad spectrum of antifungal activity, whereas Rudrappa et al. (2008) and Blom et al. (2011) reported that hydrogen cyanide is unlikely a biocontrol agent. The authors indicated that pigments and other antibiotic substances are more effective against fungi. Interestingly, Rijavec and Lapanje (2016) proved that HCN regulates phosphate availability of PGPRs and host plants.
The Interplay Between Halophytes and Their Microbiome: A Glimpse Into the Future
For many years, a wide range of PGPRs and PGPFs have been studied, and some bacterial and fungal isolates, including species of the genera Pseudomonas, Bacillus, Enterobacter, Klebsiella, Azobacter, Variovorax, Azosprillum, Serratia and Trichoderma, Aspergillus, Penicillium, Phoma have been used in commercial products (Glick, 2012; Jahagirdar et al., 2019). Nevertheless, the application of PGPRs and PGPFs in the agricultural industry is only a small part of agricultural practice worldwide (Bashan et al., 2014). Disadvantages regarding the utilization of bacteria are connected with properties of the inoculated PGPRs and chiefly depend on their survival in soil, their interaction within indigenous soil microflora, and other complex environmental factors (Martinez-Viveros et al., 2010). Moreover, the modes of action of PGPRs are incredibly varied, and not all rhizobacteria have the same effects with identical mechanisms (Choudhary, 2012; Arora et al., 2020). Little is known about PGPFs compared to bacteria regarding their effectiveness in the plant growth-promoting processes. However, many researchers reported beneficial effects of PGPFs application to plants growth by activation of induced systemic resistance (ISR) (Murali et al., 2012; Naziya et al., 2020). Another frontier is the exploitation of interactions between several microorganisms, as it is often from the interaction of several species that the production of bioactive compounds is obtained. Complex interactions between different mycorrhizal species were documented, for instance. Poveda et al. (2019) showed, for example, that plant roots’ colonization by Trichoderma harzianum biocontrol strain increases the colonization of the same host by arbuscular mycorrhizal fungal species. The authors, analysing the expression profile of defence-related marker genes, suggested that the phytohormone salicylic acid could play a key role in the modulation of the roots’ colonization process when both fungi are jointly applied.
According to Gangwar et al. (2017) and Egamberdieva et al. (2019), an ideal plant growth-promoting microorganisms (PGPMs) should possess a high rhizosphere competence, enhance plant growth capabilities, have a broad spectrum of action, be safe to the environment, be compatible with other rhizobacteria, and be tolerant to heat, UV radiation, and oxidizing agents. So far, organisms with interesting properties have been isolated, and some possess more than one of the qualities required for a perfect PGPR, however, imagining such an ideal organism capable of accomplishing all the necessary actions, is a kind of extreme. Nevertheless, the direction is the right one because by continuing to search and experimenting, it is possible to find different organisms that together can work with complementary mechanisms. The research on PGPMs as biofertilizers is the most natural and realistic aspiration to face a global agricultural productivity requirement, capable of feeding the world’s population, which is going to escalate to 9 billion people by 2050.
Author Contributions
All authors contributed to the article and approved the submitted version.
Funding
This research was conducted in the frame of the Programme “Canaletto, Bilateral Exchange of Scientists Italy-Poland” supported by Polish National Agency for Academic Exchange (project no. PPN/BIL/2018/2/00038/U/00001) and Italian Ministry of Foreign Affairs and International Cooperation (project no. M03375).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.553087/full#supplementary-material
References
Abaid-Ullah, M., Hassan, M. N., Jamil, M., Brader, G., Shah, M. K., Sessitsch, A., et al. (2015). Plant growth promoting rhizobacteria: an alternate way to improve yield and quality of wheat (Triticum aestivum). Int. J. Agric. Biol. 17, 51–60.
Abbas, G., Saqib, M., Akhtar, J. (2015). Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 178, 306–311. doi: 10.1002/jpln.201400358
Abedinzadeh, M., Etesami, H., Alikhani, H. A. (2019). Characterization of rhizosphere and endophytic bacteria from roots of maize (Zea mays L.) plant irrigated with wastewater with biotechnological potential in agriculture. Biotechnol. Rep. 21, e00305. doi: 10.1016/j.btre.2019.e00305
Abeer, H., Abdallah, E. F., Alqarawi, A. A., Alhuqail, A., Alshalawi, S. R. M., Wirth, S., et al. (2015). Impact of plant growth promoting Bacillus subtilis on growth and physiological parameters of Bassia indica (Indian Bassia) grown under salt stress. Pak. J. Bot. 47 (5), 1735–1741.
Acosta-Motos, J. R., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sanchez-Blanco, M. J., Hernandez, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy 7, 18–56. doi: 10.3390/agronomy7010018
Acuña, J. J., Campos, M., de la Luz Mora, M., Jaisi, D. P., Jorquera, M. A. (2019). ACCD producing rhizobacteria from an Andean Altiplano native plant (Parastrephia quadrangularis) and their potential to alleviate salt stress in wheat seedlings. Appl. Soil Ecol. 136, 184–190. doi: 10.1016/j.apsoil.2019.01.005
Acuña-Rodríguez, I. S., Hansen, H., Gallardo-Cerda, J., Atala, C., Molina-Montenegro, M. A. (2019). Antarctic Extremophile: Biotechnological Alternative to Crop Productivity in Saline Soils. Front. Bioeng. Biotechnol. 7, 22. doi: 10.3389/fbioe.2019.00022
Aeron, A., Kumar, S., Pandey, P., Maheshwari, D. K. (2011). “Emerging role of plant growth promoting rhizobacteria in agrobiology”, in Bacteria in agrobiology: crop ecosystems. Ed. Maheshwari, D. K. (Berlin Heidelberg: Springer), 1–36.
Akbarimoghaddam, H., Galavi, M., Ghanbari, A., Panjehkeh, N. (2011). Salinity effects on seed germination and seedling growth of bread wheat cultivars. Trakia J. Sci. 9 (1), 43–50.
Alaghemand, A., Khaghani, S., Bihamta, M. R., Gomarian, M., Ghorbanpour, M. (2018). Green synthesis of zinc oxide nanoparticles using Nigella Sativa L. extract: the effect on the height and number of branches. J. Nanostruct. 8 (1), 82–88. doi: 10.22052/JNS.2018.01.010
Albdaiwi, R. N., Khyami-Horani, H., Ayad, J. Y., Alananbeh, K. M., Al-Sayaydeh, R. (2019). Isolation and characterization of halotolerant plant growth promoting rhizobacteria from durum wheat (Triticum turgidum subsp. durum) cultivated in saline areas of the Dead Sea Region. Front. Microbiol. 10, 1639. doi: 10.3389/fmicb.2019.01639FSha
Al-Garni, S. M. S., Khan, M. M. A., Bahieldin, A. (2019). Plant growth-promoting bacteria and silicon fertilizer enhance plant growth and salinity tolerance in Coriandrum sativum. J. Plant Interact. 14 (1), 386–396. doi: 10.1080/17429145.2019.1641635
Ammari, T. G., Tahhan, R., Abubaker, S., Al-zu’bi, Y., Tahboub, A., Ta’any, R., et al. (2013). Soil salinity changes in the Jordan valley potentially threaten sustainable irrigated agriculture. Pedosphere 23, 376–384. doi: 10.1016/S1002-0160(13)60029-6
Amna, Ud D., Sarfraz, S., Xia, Y., Kamran, M. A., Javed, M. T., Sultan, T., et al. (2019). Mechanistic elucidation of germination potential and growth of wheat inoculated with exopolysaccharide and ACC-deaminase producing Bacillus strains under induced salinity stress. Ecotoxicol. Environ. Saf. 183, 109466. doi: 10.1016/j.ecoenv.2019.109466
Arora, N. K., Fatima, T., Mishra, J., Mishra, I., Verma, S., Verma, R., et al. (2020). Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Advanced Res. doi: 10.1016/j.jare.2020.07.003
Bahmani, K., Noori, S. A. S., Darbandi, A., II, Akbari, A. (2015). Molecular mechanisms of plant salinity tolerance: A review. Aust. J. Crop Sci. 9, 321–336.
Bailly, A., Weisskopf, L. (2012). The modulating effect of bacterial volatiles on plant growth: current knowledge and future challenges. Plant Signal. Behav. 7, 79–85. doi: 10.4161/psb.7.1.18418
Banerjee, S., Palit, R., Sengupta, C., Standing, D. (2010). Stress induced phosphate solubilization by Arthrobacter sp. and Bacillus sp. isolated from tomato rhizosphere. Aust. J. Crop Sci. 4, 378–383.
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Barrett-Lennard, E. G. (2003). The interaction between waterlogging and salinity in higher plants: causes, consequences and implications. Plant Soil 253, 35–54. doi: 10.1023/A:1024574622669
Bashan, Y., Moreno, M., Troyo, E. (2000). Growth promotion of the seawater irrigated oilseed halophyte Salicornia bigelovii inoculated with mangrove rhizosphere bacteria and halotolerant Azospirillum spp. Biol. Fertil. Soils 32, 265–272. doi: 10.1007/s003740000246
Bashan, Y., de-Bashan, L. E., Prabhu, S. R., Hernandez, J.-P. (2014). Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil. 378, 1–33. doi: 10.1007/s11104-013-1956-x
Beattie, G. A. (2006). “Plant-associated bacteria: survey, molecular phylogeny, genomics and recent advances”, in Plant-associated bacteria. Ed. Gnanamanickam, S. S. (The Netherlands: Springer), 1–56.
Benito, B., Gonzalez-Guerrero, M. (2014). Unravelling potassium nutrition in ectomycorrhizal associations. New Phytol. 201, 707–709. doi: 10.1111/nph.12659
Berrada, I., Benkhemmar, O., Swings, J., Bendaou, N., Amar, M. (2012). Selection of halophilic bacteria for biological control of grey tomato mould caused by Botrytis cinerea. Phytopathol. Mediterr 51, 625–630. doi: 10.14601/Phytopathol_Mediterr-1062
Bhattacharyya, P. N., Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Bheemaraya, P. M. B., Ramesh, Y. S. T., Amaresh, Y. S., Naik, M. K. (2013). Salinity stress tolerance in native Trichoderma isolates. Environ. Ecol 31, 727–729.
Blom, D., Fabbri, C., Eberl, L., Weisskopf, L. (2011). Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly due to hydrogen cyanide. Appl. Environ. Microbiol. 77, 1000–1008. doi: 10.1128/AEM.01968-10
Boiero, L., Perrig, D., Masciarelli, O., Penna, C., Cassan, F., Luna, V. (2007). Phytohormone production by three strains of Bradyrhizobium japonicum and possible physiological and technological implications. Appl. Microbiol. Biotechnol. 74, 874–880. doi: 10.1007/s00253-006-0731-9
Broadley, M. R., White, P. J., Hammond, J. P., Zelko, I., Lux, A. (2007). Zinc in plants. New Phytol. 173 (4), 677–702. doi: 10.1111/j.1469-8137.2007.01996.x
Bui, E. N. (2013). Soil salinity: A neglected factor in plant ecology and biogeography. J. Arid. Environ. 92, 14–25. doi: 10.1016/j.jaridenv.2012.12.014
Canfora, L., Bacci, G., Pinzari, F., Lo Papa, G., Dazzi, C., Benedetti, A. (2014). Salinity and Bacterial Diversity: To What Extent Does the Concentration of Salt Affect the Bacterial Community in a Saline Soil? PloS One 9 (11), e106662. doi: 10.1371/journal.pone.0114658
Ceci, A., Pinzari, F., Russo, F., Maggi, O., Persiani, A. M. (2018). Saprotrophic soil fungi to improve phosphorus solubilisation and release: In vitro abilities of several species. Ambio 47, 30–40. doi: 10.1007/s13280-017-0972-0
Central Water Commission (2017). Problems of Salination of Land in Coastal Areas of India and Suitable Protection Measures. (New Delhi, India: Hydrological Studies Organization, Central Water Commission), Vol. 346. Available at: http://cwc.gov.in/sites/default/files/salinity-report-hydrology-cwc.pdf.
Chang, T. T., Zhang, Y. J., Xu, H. L., Shao, X. H., Xu, Q. C., Li, F. L., et al. (2018). Osmotic adjustment and up-regulation expression of stress-responsive genes in tomato induced by soil salinity resulted from nitrate fertilization. Int. J. Agric. Biol. Eng. 11, 126–136. doi: 10.25165/j.ijabe.20181103.2952
Cherlet, M., Hutchinson, C., Reynolds, J., Hill, J., Sommer, S., von Maltitz, G. (2018). World Atlas of Desertification (Luxembourg: Publication Office of the European Union).
Choudhary, D. K. (2012). Microbial rescue to plant under habitat imposed abiotic and biotic stresses. Appl. Microbiol. Biotechnol. 96 (5), 1137–1155. doi: 10.1007/s00253-012-4429-x
Compant, S., Duffy, B., Nowak, J., Clément, C., Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Compant, S., Clément, C., Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42 (5), 669–678. doi: 10.1016/j.soilbio.2009.11.024
Cramer, V. A., Hobbs, R. J. (2002). Ecological consequences of altered hydrological regimes in fragmented ecosystems in southern Australia: Impacts and possible management responses. Austral Ecol. 27, 546–564. doi: 10.1046/j.1442-9993.2002.01215.x
Cuevas, J., Daliakopoulos, I. N., Moral, F., Hueso, J. J., Tsanis, I. K. (2019). A Review of Soil-Improving Cropping Systems for Soil Salinization. Agronomy 9, 295. doi: 10.3390/agronomy9060295
Dajic, Z. (2006). “Salt stress”, in Physiology and Molecular Biology of Stress Tolerance in Plants. Eds. Rao, K. V. M., Raghavendra, A. S., Reddy, K. J. (The Netherlands: Springer), 41–99.
Daliakopoulos, I. N., Tsanis, I. K., Koutroulis, A., Kourgialas, N. N., Varouchakis, A. E., Karatzas, G. P., et al. (2016). The threat of soil salinity: A European scale review. Sci. Total Environ. 573, 727–739. doi: 10.1016/j.scitotenv.2016.08.177
Darwish, T., Atallah, T., El Moujabber, M., Khatib, N. (2005). Salinity evolution and crop response to secondary soil salinity in two agro-climatic zones in Lebanon. Agric. Water Manage. 78, 152–164. doi: 10.1016/j.agwat.2005.04.020
Datta, K. K., de Jong, C. (2002). Adverse effect of waterlogging and soil salinity on crop and land productivity in northwest region of Haryana, India. Agric. Water Manage. 57, 223–238. doi: 10.1016/S0378-3774(02)00058-6
de Souza, S. M., Fay, F. E. (2014). “Effect of Salinity on Soil Microorganisms”, in Soil Health and Land Use Management. Ed. Hernandez-Soriano, M. C. (Rijeka, Croatia: IntechOpen), 177–198. doi: 10.5772/28613
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J., II (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Dhanya Thomas, T. T., Dinakar, C., Puthur, J. T. (2020). Effect of UV-B priming on the abiotic stress tolerance of stress-sensitive rice seedlings: Priming imprints and cross-tolerance. Plant Physiol. Biochem. 147, 21–30. doi: 10.1016/j.plaphy.2019.12.002
de Souza, S. M., Faye, F. E. (2014). “Effect of Salinity on Soil Microorganisms” in Soil Health and Land Use Management Ed. Hernandez-Soriano, M. C. (Rijeka, Croatia: IntechOpen) 177–198. doi: 10.5772/28613
Egamberdieva, D., Hashem, A., Abd-Allah, E. F. (2014). “Emerging Technologies and Management of Crop Stress Tolerance”, in Biological Control of Fungal Disease by Rhizobacteria under Saline Soil Conditions. Eds. Ahmad, P., Rasool, S. (Oxford, UK: Elsevier Inc.), 161–173. doi: 10.1016/B978-0-12-800875-1.00007-7
Egamberdieva, D., Davranov, K., Wirth, S., Hashem, A., Abd-Allah, E. F. (2017). Impact of soil salinity on the plant-growth – promoting and biological control abilities of root associated bacteria. Saudi J. Biol. Sci. 24 (7), 1601–1608. doi: 10.1016/j.sjbs.2017.07.004
Egamberdieva, D., Wirth, S., Belligrath-Kimura, S. D., Mishra, J., Arora, N. K. (2019). Salt tolerant plant growth promoting Rhizobacteria for enhancing crop productivity of saline soils. Front. Microbiol. 18, 2791 (10), 2791. doi: 10.3389/fmicb.2019.02791
El-Esawi, M. A., Alaraidh, I. A., Alsahli, A. A., Ali, H. M., Alayafi, A. A., Witczak, J., et al. (2018). Genetic variation and alleviation of salinity stress in barley (Hordeum vulgare L.). Molecules 23, 10. doi: 10.3390/molecules23102488
El-Esawi, M. A., Alaraidh, I. A., Alsahli, A. A., Alzahrani, S. M., Ali, H. M., Alayafi, A. A., et al. (2018a). Serratia liquefaciens KM4 improves salt stress tolerance in maize by regulating redox potential, ion homeostasis, leaf gas exchange and stress-related gene expression. Int. J. Mol. Sci. 19 (11), 3310. doi: 10.3390/ijms19113310
El-Sayed, O. M., El-Gammal, O. H. M., Salama, A. S. M. (2014). Effect of ascorbic acid, proline and jasmonic acid foliar spraying on fruit set and yield of Manzanillo olive trees under salt stress. Sci. Hortic. 176, 32–37. doi: 10.1016/j.scienta.2014.05.031
El-Tarabily, K. A., Youssef, T. (2010). Enhancement of morphological, anatomical and physiological characteristics of seedlings of the mangrove Avicennia marina inoculated with a native phosphate solubilizing isolate of Oceanobacillus picturae under greenhouse conditions. Plant Soil 332, 147–162. doi: 10.1007/s11104-010-0280-y
Etesami, H., Beattie, G. A. (2018). Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 9, 148. doi: 10.3389/fmicb.2018.00148
Etesami, H., Alikhani, H. A., Hosseini, H. M. (2015). “Indole-3-acetic acid and 1-aminocyclopropane-1-carboxylate deaminase: bacterial traits required in rhizosphere, rhizoplane and/or endophytic competence by beneficial bacteria”, in Bacterial metabolites in sustainable agroecosystem. Ed. Maheshwari, D. K. (New York: Springer International), 183–258.
Fatma, M., Asgherm, M., Masood, A., Khan, N. A. (2014). Excess sulfur supplementation improves photosynthesis and growth in mustard under salt stress through increased production of glutathione. Environ. Exp. Bot. 107, 55–63. doi: 10.1016/j.envexpbot.2014.05.008
Fernandez, L. A., Zalba, P., Gomez, M. A., Sagardoy, M. A. (2007). Phosphate solubilization activity of bacterial strains in soil and their effect on soybean growth under greenhouse conditions. Biol. Fertil. Soil 43, 805–809. doi: 10.1007/s00374-007-0172-3
Figueiredo, M. V. B., Burity, H. A., Martinez, C. R., Chanway, C. P. (2008). Alleviation of drought stress in the common bean (Phaseolus vulgaris L.) by coinoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl. Soil Ecol. 40, 182–188. doi: 10.1016/j.apsoil.2008.04.005
Furtado, B. U., Gołebiewski, M., Skorupa, M., Hulisz, P., Hrynkiewicz, K. (2019). Bacterial and fungal endophytic microbiomes of Salicornia europaea. App. Environ. Microbiol. 85 (13), e00305–e00319. doi: 10.1128/AEM.00305-19
Furtado, B. U., Szymańska, S., Hrynkiewicz, K. A. (2019b). A window into fungal endophytism in Salicornia europaea: deciphering fungal characteristics as plant growth promoting agents. Plant Soil. 445, 577. doi: 10.1007/s11104-019-04315-3
Gal-Hemed, I., Atanasova, L., Komon-Zelazowska, M., Druzhinina, I. S., Viterbo, A., Yarden, O. (2011). Marine isolates of Trichoderma spp. as potential halotolerant agents of biological control for arid-Zone agriculture. Appl. Environ. Microbiol. 77, 5100–5109. doi: 10.1128/AEM.00541-11
Gangwar, M., Saini, P., Nikhanj, P., Kaur, S. (2017). “Plant Growth-Promoting Microbes (PGPM) as Potential Microbial Bio-Agents for Eco-Friendly Agriculture,” in Advances in Soil Microbiology: Recent Trends and Future Prospects. Eds. Adhya, T., Mishra, B., Annapurna, K., Verma, D., Kumar, U. (Singapore: Springer Nature Singapore Pte Ltd), 37–55.
Garg, N., Bharti, A., Sharma, A., Bhalla, S. (2019). “Plant-Mycorrhizal and Plant-Rhizobial Interfaces: Underlying Mechanisms and Their Roles in Sustainable Agroecosystems,” in Plant Microbe Interface. Eds. Varma, A., Tripathi, S., Prasad, R. (Cham: Springer), 27–67.
Gkiougkis, I., Kallioras, A., Pliakas, F., Pechtelidis, A., Diamantis, V., Diamantis, I., et al. (2015). Assessment of soil salinization at the eastern Nestos River Delta, N.E. Greece. Catena 128, 238–251. doi: 10.1016/j.catena.2014.06.024
Glick, B. R. (2005). Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7. doi: 10.1016/j.femsle.2005.07.030
Glick, B. R. (2012). Plant Growth-Promoting Bacteria: Mechanisms and Applications (Hindawi Publishing Corporation Scientifica), 2012, 15. doi: 10.6064/2012/963401
Gontia, I., Kavita, K., Schmid, M., Hartmann, A., Jha, B. (2011). Brachybacterium saurashtrense sp. nov., a halotolerant root-associated bacterium with plant growth-promoting potential. Int. J. Syst. Evol. Microbiol. 61, 2799–2804. doi: 10.1099/ijs.0.023176-0
Gorji, T., Sertel, E., Tanik, A. (2017). Recent Satellite Technologies for Soil Salinity Assessment With Special Focus on Mediterranean Countries. Fresenius Environ. Bull. 26, 196–203.
Goswami, D., Dhandhukia, P., Patel, P., Thakker, J. N. (2014). Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 169, 66–75. doi: 10.1016/j.micres.2013.07.004
Goswami, D., Parmar, S., Vaghela, H., Dhandhukia, P., Thakker, J. (2015). Describing Paenibacillus mucilaginosus strain N3 as an efficient plant growth promoting rhizobacteria (PGPR). Cogent. Food Agric. 1, 1000714. doi: 10.1080/23311932.2014.1000714
Grattan, S. R., Oster, J. D. (2003). “Use and reuse of saline-sodic waters for irrigation of crops”, in Crop Production in Saline Environments: Global and Integrative Perspectives. Eds. Goyal, S. S., Sharma, S. K., Rains, D. W. (New York: Haworth Press), 131–162.
Greggio, N., Mollema, P., Antonellini, M., Gabbianelli, G. (2012). ““Irrigation Management in Coastal Zones to Prevent Soil and Groundwater Salinization”,” in Resources Management of Sustainable Agriculture. Ed. Abrol, V. (Rijeka, Croatia: IntechOpen). doi: 10.5772/50534
Gupta, B., Huang, B. (2014). Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics. 2014, 18 doi: 10.1155/2014/701596
Gururaja Rao, G., Kanani, A. D., Purohit, D., Waghela, D. (2019). “Coastal Saline Soils of Gujarat (India): Problems, Reclamation Measures and Management Strategies”, in Research Developments in Saline Agriculture. Eds. Dagar, J. C., Yadav, R. K., Sharma, P. C. (Singapore: Springer), 629–651. doi: 10.1007/978-981-13-5832-6_21
Habib, S. H., Kausar, H., Saud, H. M. (2016). Plant growth-promoting Rhizobacteria enhance salinity stress tolerance in Okra through ROS-scavenging enzymes. Biomed. Res. Int. 2016, 6284547. doi: 10.1155/2016/6284547
Hagen, G., Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Hanson, B. R., Grattan, S. R., Fulton, R. (1993). Agricultural Salinity and Drainage (Water Management Series, USA: University of California, Division of Agriculture and Natural Resources).
Hinarejos, E., Castellano, M., Rodrigo, I., Belles, J. M., Conejero, V., Lopez-Gresa, M. P., et al. (2016). Bacillus subtilis IAB/BS03 as a potential biological control agent. Eur. J. Plant Pathol. 146, 597–608. doi: 10.1007/s10658-016-0945-3
Hossain, M. S., Dietz, K. J. (2016). Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front. Plant Sci. 7, 548. doi: 10.3389/fpls.2016.00548
Hossain, M. S., El Sayed, A., II, Moore, M., Dietz, K.-J. (2017). Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. J. Exp. Bot. 68 (5), 1283–1298. doi: 10.1093/jxb/erx019Advance
Hu, Y., Schmidhalter, U. (2007). Effect of salinity on the composition, number and size of epidermal cells along the mature blade of wheat leaves. J. Integr. Plant Biol. 49 (7), 1016–1023. doi: 10.1016/S2095-3119(17)61731-3
Huang, Y. (2018). Comparison of rhizosphere and endophytic microbial communities of Chinese leek through high-throughput 16S rRNA gene Illumina sequencing. J. Integr. Agric. 17, 359–367. doi: 10.1016/S2095-3119(17)61731-3
IIangumaran, G., Smith, D. L. (2017). Plant growth promoting Rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 8, 1768. doi: 10.3389/fpls.2017.01768
Ikram, M., Ali, N., Jan, G., Iqbal, A., Hamayun, M., Jan, F. G., et al. (2019). Trichoderma reesei improved the nutrition status of wheat crop under salt stress. J. Plant Interact. 14 (1), 590. doi: 10.1080/17429145.2019.1684582
Ishida, T. A., Nara, K., Ma, S. (2009). Ectomycorrhizal fungal community in alkaline-saline soil in northeastern China. Mycorrhiza 19, 329–335. doi: 10.1007/s00572-008-0219-9
IUSS Working Group WRB (2014). World reference base for soil resources 2014 (International soil classification system for naming soils and creating legends for soil maps). (Rome: FAO). doi: 10.1017/S0014479706394902
Jahagirdar, S., Kambrekar, D. N., Navi, S. S., Kunta, M. (2019). “Plant Growth-Promoting Fungi: Diversity and Classification”, in Bioactive Molecules in Plant Defense. Signaling in Growth and Stress. Eds. Jogaiah, S., Abdelrahman, M. (Switzerland AG: Springer International Publishing), 25–35. doi: 10.1007/978-3-030-27165-7
Kandel, S. L., Joubert, P. M., Doty, S. L. (2017). Bacterial endophyte colonization and distribution within plants. Microorganisms 5 (4), E77. doi: 10.3390/microorganisms5040077
Kang, S. M., Shahzad, R., Bilal, S., Khan, A. L., Park, Y. G., Lee, K. E., et al. (2019). Indole-3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 19 (1), 80. doi: 10.1186/s12866-019-1450-6
Karthikeyan, B., Joe, M. M., Islam, M. R., Sa, T. (2012). ACC deaminase containing diazotrophic endophytic bacteria ameliorate salt stress in Catharanthus roseus through reduced ethylene levels and induction of antioxidative defence systems. Symbiosis 56 (2), 77–86. doi: 10.1007/s13199-012-0162-6
Kasotia, A., Varma, A., Tuteja, N., Choudhary, D. K. (2016). Amelioration of soybean plant from saline-induced condition by exopolysaccharide producing Pseudomonas-mediated expression of high affinity K+-transporter (HKT1) gene. Curr. Sci. 111 (12), 25. doi: 10.18520/cs/v111/i12/1961-1967
Kearl, J., McNary, C., Lowman, J. S., Mei, C., Aanderud, Z. T., Smith, S. T., et al. (2019). Salt-tolerant halophyte rhizosphere bacteria stimulate growth of alfalfa in salty soil Front. Microbiol 10, 1849. doi: 10.3389/fmicb.2019.01849
Khan, A. L., Waqas, M., Khan, A. R., Hussain, J., Kang, S.-M., Gilani, S. A., et al. (2013). Fungal endophyte Penicillium janthinellum LK5 improves growth of ABA-deficient tomato under salinity. World J. Microbiol. Biotechnol. 29, 2133–2144. doi: 10.1007/s11274-013-1378-1
Khan, H. A., Siddique, K., Colmer, T. D. (2017). Vegetative and reproductive growth of salt-stressed chickpea are carbon-limited: sucrose infusion at the reproductive stage improves salt tolerance. J. Experiment Bot. 68 (8), 2001–2011. doi: 10.1093/jxb/erw177
Khan, M. A., Asaf, S., Khan, A. L., Adhikari, A., Rahmatullah, J., Sajid, A., et al. (2019). Halotolerant Rhizobacterial strains mitigate the adverse effects of NaCl stress in soybean seedings. Biomed. Res. Int. 2019, 9530963. doi: 10.1155/2019/9530963
Khidir, H. H., Eudy, D. M., Porras-Alfaro, A., Herrera, J., Natvig, D. O., Sinsabaugh, R. L. (2010). A general suite of fungal endophytes dominates the roots of two dominant grasses in a semiarid grassland. J. Arid. Environ. 74, 35–42. doi: 10.1016/j.jaridenv.2009.07.014
Kiani, M. Z., Ali, A., Sultan, T., Ahmad, R., Hydar, I. S. (2015). Plant Growth Promoting Rhizobacteria having 1-aminocyclopropane-1-carboxylic acid deaminase to induce salt tolerance in sunflower (Helianthus annus L.) Nat. Res 6, 391–397. doi: 10.4236/nr.2015.66037
Kim, D. W., Shibato, J., Agrawal, G. K., Fujihara, S., Iwahashi, H., Kim, D. H. (2007). Gene transcription in the leaves of rice undergoing salt induced morphological changes (Oryza sativa L.). Mol. Cell 24, 45–59.
Kloepper, J. W., Schroth, M. N. (1978). Plant growth promoting rhizobacteria on radishes. Proc. 4th Int. Conf. Plant Path. Bact. Angers. 879–882.
Kováčová, V., Velísková, Y. (2012). The risk of the soil salinization of the eastern part of Žitný Ostrov. J. Hydrol. Hydromechanics 60, 57–63. doi: 10.2478/v10098-012-0005-4
Kumar, A., Verma, J. P. (2018). Does plant-microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 207, 41–52. doi: 10.1016/j.micres.2017.11.004
Kumari, A., Das, P., Parida, A. K., Agarwal, P. K. (2015a). Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front. Plant Sci. 6, 537. doi: 10.3389/fpls.2015.00537
Kumari, S., Vaishnav, A., Jain, S., Varma, A., Choudhary, D. K. (2015b). Bacterial-mediated induction of systemic tolerance to salinity with expression of stress alleviating enzymes in soybean (Glycine max L. Merrill). J. Plant Growth Regul. 34, 558–573. doi: 10.1007/s00344-015-9490-0
Kushwaha, B., Jadhav, I., Verma, H. N., Geethadevi, A., Parashar, D., Jadhav, K. (2019). Betaine accumulation suppresses the de-novo synthesis of ectoine at a low osmotic concentration in Halomonas sp. SBS 10, a bacterium with broad salinity tolerance. Mol. Biol. Rep. 46, 4779 – 4786. doi: 10.1007/s11033019-04924-2
Kushwaha, P., Kashyap, P. L., Bhardwaj, A. K., Kuppusamy, P., Srivastava, A. K., Tiwari, R. K. (2020). Bacterial endophyte mediated plant tolerance to salinity: growth responses and mechanisms of action. World J. Microbiol. Biotech. 36 (2), 26. doi: 10.1007/s11274-020-2804-9
Labuschagne, N., Pretorius, T., Idris, A. H. (2010). “Plant Growth Promoting Rhizobacteria as Biocontrol Agents Against Soil-Borne Plant Diseases”, in Plant Growth and Health Promoting Bacteria. Ed. Maheshwari, D. K. (Berlin Heidelberg: Springer-Verlag), 211–229. doi: 10.1007/978-3-642-13612-2_9
Läuchli, A., Grattan, S. (2007). “Plant growth and development under salinity stress”, in Advances in molecular breeding toward drought and salt tolerant crops. Eds. Jenks, M. A., Hasegawa, P. M., Jain, S. M. (Dordrecht: Springer), 1–32.
Ledger, T., Rojas, S., Timmermann, T., Pinedo, I., Poupin, M. J., Garrido, T., et al. (2016). Volatile-mediated effects predominate in Paraburkholderia phytofirmans growth promotion and salt stress tolerance of Arabidopsis thaliana. Front. Microbiol. 7, 1838. doi: 10.3389/fmicb.2016.01838
Liu, Y., Xu, J., Ding, Y., Wang, Q., Li, G., Wang, S. (2011). Auxin inhibits the outgrowth of tiller buds in rice (Oryza sativa L.) by downregulating OsIPT expression and cytokinin biosynthesis in nodes. Aust. J. Crop Sci. 5, 169–174.
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8, :2552. doi: 10.3389/fmicb.2017.02552
Machado, R. M. A., Serralheiro, R. P. (2017). Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 3 (2), 30. doi: 10.3390/horticulturae3020030
Maciá-Vicente, J. G., Ferraro, V., Burruano, S., Lopez-Llorca, L. V. (2012). Fungal assemblages associated with roots of halophytic and non-halophytic plant species vary differentially along a salinity gradient. Microb. Ecol. 64, 668–679. doi: 10.1007/s00248-012-0066-2
Mantelin, S., Touraine, B. (2004). Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J. Exp. Bot. 55 (394), 27–34. doi: 10.1093/jxb/erh010
Manwar, A. V., Khandelwal, S. R., Chaudhari, B. L., Meyer, J. M., Chincholkar, S. B. (2004). Siderophore production by a marine Pseudomonas aeruginosa and its antagonistic action against phytopathogenic fungi. Appl. Biochem. Biotechnol. 118, 243–252. doi: 10.1385/ABAB:118:1-3:243
Martinez-Viveros, O., Jorquera, M. A., Crowley, D. E., Gajardo, G., Mora, M. L. (2010). Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 10, 293–319. doi: 10.4067/S0718-95162010000100006
Masters, D. G., Norman, H. C., Barrett-Lennard, E. G. (2005). Agricultural systems for saline soil: The potential role of livestock. Asian-Aust. J. Anim. Sci. 18, 296–300. doi: 10.5713/ajas.2005.296
Mastouri, F. (2010). Use of Trichoderma spp. to improve plant performance under abiotic stresses. [dissertation]. (Ithaca, NY: Cornell University).
Matilla, M. A., Espinosa-Urgel, M., Rodríguez-Herva, J. J., Ramos, J. L., Ramos González, M., II. (2007). Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8, R179. doi: 10.1186/gb-2007-8-9-r179
Mayak, S., Tirosh, T., Glick, B. R. (2004). Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 42, 565–572. doi: 10.1016/j.plaphy.2004.05.009
Meng, X., Zhou, J., Sui, N. (2018). Mechanisms of salt tolerance in halophytes: Current understanding and recent advances. Open Life Sci. 13, 149–154. doi: 10.1515/biol-2018-0020
Miethke, M., Marahiel, M. A. (2007). Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 71 (3), 413–451. doi: 10.1128/MMBR.00012-07
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell. Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Miransari, M., Smith, D. L. (2009). Alleviating salt stress on soybean (Glycinemax (L.) Merr.) - Bradyrhizobium japonicum symbiosis, using signal molecule genistein. Eur. J. Soil Biol. 45, 146–152. doi: 10.1016/j.ejsobi.2008.11.002
Mishra, J., Singh, R., Arora, N. K. (2017). “Plant Growth-Promoting Microbes: Diverse Roles in Agriculture and Environmental Sustainability”, in Probiotics and Plant Health. Eds. Kumar, V., Kumar, M., Sharma, S., Prasad, R. (Singapore: Springer Nature Singapore Pte Ltd.), 71–111. doi: 10.1007/978-981-10-3473-2
Mohamed, H., Haggag, W. (2006). Biocontrol potential of salinity tolerant mutants of Trichoderma harzianum against Fusarium oxysporum. Braz. J. Microbiol. 37, 181–191. doi: 10.1590/S1517-83822006000200016
Molina-Montenegro, M. A., Acuna-Rodriguez, I. S., Diaz, C. T., Gundel, P. E., Dreyer, I. (2020). Antarctic root endophytes improve physiological performance and yield in crops under salt stress by enhanced energy production and Na+ sequestration. Sci. Rep. 10, 5819. doi: 10.1038/s41598-020-62544-4
Møller, I. M., Jensen, P. E., Hansson, A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481. doi: 10.1146/annurev.arplant.58.032806.103946
Moreira, H., Pereira, S., II, Vega, A., Castro, P. M. L., Marques, A. P. G. C. (2020). Synergistic effects of arbuscular mycorrhizal fungi and plant growth-promoting bacteria benefit maize growth under increasing soil salinity. J. Environ. Manage. 257, 109982. doi: 10.1016/j.jenvman.2019.109982
Mukhtar, S., Malik, K. A., Mehnaz, S. (2019). Microbiome of halophytes: Diversity and importance for plant health and productivity. Korean J. Microbiol. Biotechnol. 47 (1), 1–10. doi: 10.4014/mbl.1804.04021
Munns, R., Rawson, H. M. (1999). Effect of salinity on salt accumulation and reproductive development in the apical meristem of wheat and barley. Aust. J. Plant Physiol. 26, 459–464. doi: 10.1071/PP99049
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Murali, M., Amrutthesh, K. N., Sudisha, J., Niranjana, S. R., Shetty, H. S. (2012). Screening for plant growth promoting fungi and their ability for growth promotion and induction of resistance in pearl millet against downy mildew disease. J. Phyt. 4 (5), 30–36.
Nadeem, S. M., Zahir, Z. A., Naveed, M., Arshad, M. (2009). Rhizobacteria containing ACC-deaminase confer salt tolerance in maize grown on salt-affected fields. Can. J. Microbiol. 55, 1302–1309. doi: 10.1139/W09-092
Nakkeeran, S., Surya, T., Vinodkumar, S. (2020). Antifungal Potential of Plant Growth Promoting Bacillus Species Against Blossom Blight of Rose. J. Plant Growth Reg. 39, 99–111. doi: 10.1007/s00344-019-09966-1
Napoli, R., Vanino, S. (2011). Valutazione del rischio di salinizzazione dei suoli e di intrusione marina nelle aree costiere delle regioni meridionali in relazione agli usi irrigui (Roma: INEA).
Navarro-Torre, S., Barcia-Piedras, J. M., Mateos-Naranjo, E., Redondo-Gómez, S., Camacho, M., Caviedes, M. A., et al. (2017). Assessing the role of endophytic bacteria in the halophyte Arthrocnemum macrostachyum salt tolerance. Plant Biol. 19, 249–256. doi: 10.1111/plb.12521
Naziya, B., Murali, M., Amruthesh, K. N. (2020). Plant Growth-Promoting Fungi (PGPF) Instigate Plant Growth and Induce Disease Resistance in Capsicum annuum L. upon Infection with Colletotrichum capsici (Syd.) Butler & Bisby. Biomolecules 10, 41. doi: 10.3390/biom10010041
Niu, S. Q., Li, H. R., Pare, P. W., Aziz, M., Wang, S. M., Shi, H. Z., et al. (2016). Induced growth promotion and higher salt tolerance in the halophyte grass Puccinellia tenuiflora by beneficial rhizobacteria. Plant Soil 407, 217–230. doi: 10.1007/s11104-015-2767-z
Nouri, H., Chavoshi Borujeni, S., Nirola, R., Hassanli, A., Beecham, S., Alaghmand, S., et al. (2017). Application of green remediation on soil salinity treatment: A review on halophytoremediation. Process Saf. Environ. Prot. 107, 94–107. doi: 10.1016/j.psep.2017.01.021
Numan, M., Bashir, S., Khan, Y., Mumtaz, R., Shinwari, Z. K., Khan, A. L., et al. (2018). Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol. Res. 209, 21–32. doi: 10.1016/j.micres.2018.02.003
Oberson, A., Frossard, E., Bühlmann, C., Mayer, J., Mader, P., Luscher, A. (2013). Nitrogen fixation and transfer in grass clover leys under organic and conventional cropping systems. Plant Soil 371, 237– 255. doi: 10.1007/s11104-013-1666-4
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Orhan, F. (2016). Alleviation of salt stress by halotolerant and halophilic plant growth-promoting bacteria in wheat (Triticum aestivum). Braz. J. Microbiol. 47 (3), 621–627. doi: 10.1016/j.bjm.2016.04.001
Orozco-Mosqueda, M. D. C., Duan, J., Di Bernardo, M., Zetter, E., Campos-García, J., Glick, B. R., et al. (2019). The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 10, 1392. doi: 10.3389/fmicb.2019.01392
Parida, A. K., Das, A. B. (2005). Salt tolerance and salinity effects on plant: a review. Ecotoxicol. Environ. Safe. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Peng, J., Biswas, A., Jiang, Q., Zhao, R., Hu, J., Hu, B., et al. (2019). Estimating soil salinity from remote sensing and terrain data in southern Xinjiang Province, China. Geoderma 337, 1309–1319. doi: 10.1016/j.geoderma.2018.08.006
Pereira, E., Vázquez de Aldana, B. R., San Emeterio, L., Zabalgogeazcoa, I. (2019). A Survey of Culturable Fungal Endophytes From Festuca rubra subsp. pruinosa, a Grass From Marine Cliffs, Reveals a Core Microbiome. Front. Microbiol. 9, 3321. doi: 10.3389/fmicb.2018.03321
Pierik, R., Djakovic-Petrovic, T., Keuskamp, D. H., de Wit, M., Voesenek, L. A. C. J. (2009). Auxin and Ethylene Regulate Elongation Responses to Neighbor Proximity Signals Independent of Gibberellin and DELLA Proteins in Arabidopsis. Plant Physiol. 149 (4), 1701–1712. doi: 10.1104/pp.108.133496
Pinedo, I., Ledger, T., Greve, M., Poupin, M. J. (2015). Burkholderia phytofirmans PsJN induces long-term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front. Plant Sci. 6, 466. doi: 10.3389/fpls.2015.00466
Pini, F., Galardini, M., Brzzicalupo, M., Mengoni, A. (2011). Plant-bacteria association and symbiosis: Are there common genomic traits in Alpha proteobacteria? Genes (Basel) 2 (4), 1017–1032. doi: 10.3390/genes2041017
Poveda, J., Hermosa, R., Monte, E., Nicolás, C. (2019). Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-48269-z
Poveda, J. (2020). Trichoderma parareesei Favors the Tolerance of Rapeseed (Brassica napus L.) to Salinity and Drought Due to a Chorismate Mutase. Agronomy 10 (1), 118. doi: 10.3390/agronomy10010118
Qadir, M., Quillérou, E., Nangia, V., Murtaza, G., Singh, M., Thomas, R. J., et al. (2014). Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 38, 282–295. doi: 10.1111/1477-8947.12054
Qin, Y., Druzhinina, I. S., Pan, X., Yuan, Z. (2016). Microbially Mediated Plant Salt Tolerance and Microbiome-based Solutions for Saline Agriculture. Biotech. Adv. 34 (7), 1245–1259. doi: 10.1016/j.biotechadv.2016.08.005
Qin, Y., Pan, X., Kubicek, C., Druzhinina, I., Chenthamara, K., Labbe, J., et al. (2017). Diverse Plant-Associated Pleosporalean Fungi from Saline Areas: Ecological Tolerance and Nitrogen-Status Dependent Effects on Plant Growth. Front. Microbiol. 8, 158. doi: 10.3389/fmicb.2017.00158
Qin, S., Feng, W. W., Zhang, Y. J., Wang, T. T., Xiong, Y. W., Xing, K. (2018). Diversity of bacterial microbiota of coastal halophyte Limonium sinense and amelioration of salinity stress damage by symbiotic plant growth-promoting actinobacterium Glutamicibacter halophytocola KLBMP 5180. Appl. Environ. Microbiol. 84, e1533–e1518. doi: 10.1128/AEM.01533-18
Qurashi, A. W., Sabr, A. N. (2012). Bacterial exopolysaccharide and biofilm formation stimulate Chickpea growth and soil aggregation under salt stress. Braz. J. Microbiol. 43, 1183–1191. doi: 10.1590/S1517-838220120003000046
Raaijmakers, J. M., Vlami, M., de Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agent. Anton Van Leeuwenhoek 81, 537–547. doi: 10.1023/A:1020501420831
Radhakrishnan, R., Kang, S.-M., Baek, I.-Y., Lee, I.-J. (2014). Characterization of plant growth-promoting traits of Penicillium species against the effects of high soil salinity and root disease. J. Plant Interact. 9 (1), 754. doi: 10.1080/17429145.2014.930524
Radhakrishnan, R., Hashem, A., Abd-Allah, E. F. (2017). Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 8, 667. doi: 10.3389/fphys.2017.00667
Rahneshan, Z., Nasibi, F., Ahmadi Moghadam, A. (2018). Effects of salinity stress on some growth, physiological, biochemical parameters and nutrients in two pistachio (Pistacia vera L.) rootstocks. J. Plant Int. 13, 73–82. doi: 10.1080/17429145.2018.1424355
Rajwar, A., Sahgal, M., Johri, B. N. (2013). Legume rhizobia symbiosis and interactions in agro ecosystems. Plant microbe symbiosis: fundamentals and advances (New Delhi: Springer).
Ramette, A., Frapolli, M., Défago, G., Moënne-Loccoz, Y. (2003). Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant Microbe Interact. 16 (6), 525–535. doi: 10.1094/MPMI.2003.16.6.525
Rijavec, T., Lapanje, A. (2016). Hydrogen Cyanide in the Rhizosphere: Not Suppressing Plant Pathogens, but Rather Regulating Availability of Phosphate. Front. Microbiol. 7, 1785. doi: 10.3389/fmicb.2016.01785
Rodriguez, R. J., White, J. F., Jr., Arnold, A. E., Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Rodríguez-Liébana, J. A., ElGouzi, S., Mingorance, M. D., Castillo, A., Peña, A. (2014). Irrigation of a Mediterranean soil under field conditions with urban wastewater: Effect on pesticide behaviour. Agric. Ecosyst. Environ. 185, 176–185. doi: 10.1016/j.agee.2013.12.026
Rodríguez-Salazar, J., Suárez, R., Caballero-Mellado, J., Iturriaga, G. (2009). Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett. 296, 52–59. doi: 10.1111/j.1574-6968.2009.01614.x
Rogers, M. E., Craig, A. D., Munns, R. E., Colmer, T. D., Nichols, P. G. H., Malcolm, C. V., et al. (2005). The potential for developing fodder plants for the salt-affected areas of southern and eastern Australia: an overview. Austral. J. Exp. Agric. 45, 301–329. doi: 10.1071/EA04020
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Romero-Munar, A., Baraza, E., Gulias, J., Cabot, C. (2019). Arbuscular Mycorrhizal Fungi Confer Salt Tolerance in Giant Reed (Arundo donax L.) Plants Grown Under Low Phosphorus by Reducing Leaf Na+ Concentration and Improving Phosphorus Use Efficiency. Front. Plant Sci. 10, 843. doi: 10.3389/fpls.2019.00843
Rozema, J., Cornelisse, D., Zhang, Y., Li, H., Bruning, B., Katschnig, D., et al. (2015). Comparing salt tolerance of beet cultivars and their halophytic ancestor: consequences of domestication and breeding programmes. AoB Plants 7, plu083. doi: 10.1093/aobpla/plu083
Rudrappa, T., Splaine, R. E., Biedrzycki, M. L., Bais, H. P. (2008). Cyanogenic pseudomonads influence multitrophic interactions in the rhizosphere. PLoSONE 3, e2073. doi: 10.1371/journal.pone.0002073
Sadfi, N., Chérif, M., Fliss, I., Boudabbous, A., Antoun, H. (2001). Evaluation of bacterial isolates from salty soils and Bacillus thuringiensis strains for the biocontrol of Fusarium dry rot of potato tubers. J. Plant Pathol. 83, 101–117. doi: 10.4454/jpp.v83i2.1118
Saghafi, D., Ghorbanpour, M., Asgari Lajayer, B. (2018). Efficiency of Rhizobium strains as plant growth promoting rhizobacteria on morpho-physiological properties of Brassica napus L. under salinity stress. J. Soil Sci. Plant Nutr. 18 (1), 253–268. doi: 10.4067/S0718-95162018005000903
Saghafi, D., Delangiz, N., Lajayer, B. A., Ghorbanpour, M. (2019). An overview on improvement of crop productivity in saline soils by halotolerant and halophilic PGPRs. 3 Biotech. 9, 261. doi: 10.1007/s13205-019-1799-0
Sakhabutdinova, A. R., Fatkhutdinova, D. R., Bezrukova, M. V., Shakirova, F. M. (2003). Salicylic acid prevents the damaging action of stress factors on wheat plants. Bulg. J. Plant Physiol. 2003, 314–319.
Salcedo, L. D. P., Prieto, C., Correa, M. F. (2014). Screening phosphate solubilizing actinobacteria isolated from the rhizosphere of wild planta from the Eastern Cordillera of the Colombian Andes. Afr. J. Microbiol. Res. 8, 734–742. doi: 10.5897/AJMR2013.5940
Salvati, L., Ferrara, C. (2015). The local-scale impact of soil salinization on the socioeconomic context: An exploratory analysis in Italy. Catena 127, 312–322. doi: 10.1016/j.catena.2015.01.008
Sánchez-Montesinos, B., Diánez, F., Moreno-Gavira, A., Gea, F. J., Santos, M. (2019). Plant growth promotion and biocontrol of Pythium ultimum by saline tolerant Trichoderma isolates under salinity stress. Int. J. Environ. Res. Public Health 16, 2053. doi: 10.3390/ijerph16112053
Saum, S. H., Müller, V. (2007). Salinity-dependent switching of osmolyte strategies in a moderately halophilic bacterium: glutamate induces proline biosynthesis in Halobacillus halophilus. J. Bacteriol. 189, 6968–6975. doi: 10.1128/jb.00775-07
Scavino, A. F., Pedraza, R. O. (2013). “The role of siderophores in plant growth promoting bacteria”, in Bacteria in agrobiology: crop productivity. Eds. Maheshwari, D. K., Saraf, M., Aeron, A. (Berlin Heidelberg: Springer-Verlag), 265–285. doi: 10.1007/978-3-642-37241-4
Schofield, R. V., Kirkby, M. J. (2003). Application of salinization indicators and initial development of potential global soil salinization scenario under climatic change. Global Biogeochem. Cycles 17, 1078. doi: 10.1029/2002gb001935
Sgroy, V., Cassán, F., Masciarelli, O., Del Papa, M. F., Lagares, A., Luna, V. (2009). Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 85, 371–381. doi: 10.1007/s00253-009-2116-3
Shaharoona, B., Arshad, M., Zahir, Z. A. (2006). Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett. Appl. Microbiol. 42 (2), 155–159. doi: 10.1111/j.1472-765X.2005.01827.x
Sharma, S., Kulkarni, J., Jha, B. (2016). Halotolerant Rhizobacteria promote growth and enhance salinity tolerance in peanut. Front. Microbiol. 13, 2791 (7), 2791. doi: 10.3389/fmicb.2019.02791
Sharma, M., Tarafdar, A., Ghosh, R., Gopalakrishanan, S. (2017). “Biological Control as a Tool for Eco-friendly Management of Plant Pathogens”, in Advances in Soil Microbiology: Recent Trends and Future Prospects. Eds. Adhaya, T. K., Mishra, B. B., Annapurna, K., Verma, D. K., Kumar, U. (Singapore: Springer Nature Singapore), 153–189. doi: 10.1007/978-981-10-7380-9
Shim, J. S., Seo, J.-S., Seo, J. S., Kim, Y., Koo, Y., Do Choi, Y., et al. (2019). Heterologous expression of bacterial trehalose biosynthetic genes enhances trehalose accumulation in potato plants without adverse growth effects. Plant Biotechnol. Rep. 13, 409–418. doi: 10.1007/s11816-019-00554-z
Shrivastava, P., Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Shukla, P. S., Agarwal, P. K., Jha, B. (2012). Improved salinity tolerance of Arachis Hypogaea (L.) by the interaction of halotolerant plant growth-promoting rhizobacteria. J. Plant Growth Regul. 31, 195–206. doi: 10.1007/s00344-011-9231-y
Siddikee, M. A., Chauhan, P. S., Anandham, R., Han, G. H., Sa, T. (2010). Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 20 (11), 1577–1584. doi: 10.4014/jmb.1007.07011
Singh, G., Biswas, D. R., Marwaha, T. S. (2010). Mobilization of potassium from waste mica by plant growth promoting rhizobacteria and its assimilation by maize (Zea mays) and wheat (Triticum aestivum L.): a hydroponics study under phytotron growth chamber. J. Plant Nutr. 33 (8), 1236–1251. doi: 10.1080/01904161003765760
Sitbon, F., Åstot, C., Edlund, A., Crozier, A., Sanderg, G. (2000). The relative importance of tryptophan-dependent and tryptophan-independent biosynthesis of indole-3-acetic acid in tobacco during vegetative growth. Planta 211, 715–721. doi: 10.1007/s004250000338
Smith, P., Davies, C. A., Ogle, S., Zanchi, G., Bellarby, J., Bird, R. N., et al. (2012). Towards an integrated global framework to assess the impacts of land use and management change on soil carbon: Current capability and future vision. Glob. Change Biol. 18, 2089–2101. doi: 10.1111/j.1365-2486.2012.02689.x
Spaepen, S., Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 1, 3(4). doi: 10.1101/cshperspect.a001438
Subramanian, K. S., Tenshia, V., Jayalakshmi, K., Ramachandran, V. (2009). Role of arbuscular mycorrhizal fungus (Glomus intraradices) - (fungus aided) in zinc nutrition of maize. Agric. Biotechnol. Sust. Dev. 1, 29–38.
Subramanian, S., Ricci, E., Souleimanov, A., Smith, D. L. (2016). A proteomic approach to lipo-chitooligosaccharide and thuricin 17 effects on soybean germination unstressed and salt stress. PloS One 11, e0160660. doi: 10.1371/journal.pone.0160660
Sukweenadhi, J., Balusamy, S. R., Kim, Y. J., Lee, C. H., Kim, Y. J., Koh, S. C., et al. (2018). A growth-promoting bacteria, Paenibacillus yonginensis DCY84T enhanced salt stress tolerance by activating defense-related systems in Panax ginseng. Front. Plant Sci. 9, 813. doi: 10.3389/fpls.2018.00813
Szymańska, S., Płociniczak, T., Piotrowska-Seget, Z., Złoch, M., Ruppel, S., Hrynkiewicz, K. (2016). Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol. Res. 182, 68–79. doi: 10.1016/j.micres.2015.09.007
Talaat, N. B., Shawky, B. T. (2011). Influence of arbuscular mycorrhizae on yield, nutrients, organic solutes, and antioxidant enzymes of two wheat cultivars under salt stress. J. Plant Nutr. Soil Sci. 174, 283–291. doi: 10.1002/jpln.201000051
Tariq, M., Hameed, S., Malik, K. A., Hafeez, F. Y. (2007). Plant root associated bacteria for zinc mobilization in rice. Pak. J. Bot. 39 (1), 245–253.
Teng, S., Liu, Y., Zhao, L. (2010). Isolation, identification and characterization of ACC deaminase-containing endophytic bacteria from halophyte Suaeda salsa. Wei Sheng Wu Xue Bao 50, 1503–1509.
Thiem, D., Gołebiewski, M., Hulisz, P., Piernik, A., Hrynkiewicz, K. (2018). How does salinity shape bacterial and fungal microbiomes of Alnus glutinosa roots? Front. Microbiol. 9, 651. doi: 10.3389/fmicb.2018.00651
Ullah, S., Bano, A. (2015). Isolation of plant-growth-promoting rhizobacteria from rhizospheric soil of halophytes and their impact on maize (Zea mays L.) under induced soil salinity. Can. J. Microbiol. 61, 307–313. doi: 10.1139/cjm-2014-0668
Upadhyay, S. K., Singh, D. P. (2015). Effect of salt-tolerant plant growth promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 17, 288–293. doi: 10.1111/plb.12173
Upadhyay, S. K., Singh, J. S., Singh, D. P. (2011). Exopolysaccharide plant growth promoting Rhizobacteria under salinity condition. Pedosphere 21 (2), 214–222. doi: 10.1016/S1002-0160(11)60120-3
Vaishnav, A., Varma, A., Tuteja, N., Choudhary, D. K. (2016). “PGPR-Mediated Amelioration of Crops Under Salt Stress”, in Plant-Microbe Interaction: An Approach to Sustainable Agriculture. Eds. Choudhary, D., Varma, A., Tuteja, N. (Singapore: Springer), 205–226.
Vaishnav, A., Shukla, A. K., Sharma, A., Kumar, R., Choudhary, D. K. (2019). Endophytic Bacteria in Plant Salt Stress Tolerance: Current and Future Prospects. J. Plant Growth Regul. 38 (2), 650–668. doi: 10.1007/s00344-018-9880-1
van Straten, G., de Vos, A. C., Rozema, J., Bruning, B., van Bodegom, M. (2019). An improved methodology to evaluate crop salt tolerance from field trials. Agric. Water Manage. 213, 375–387. doi: 10.1016/j.agwat.2018.09.008
Wallender, W. W., Tanji, K. K. (2012). Agricultural Salinity Assessment and Management. 2nd ed. (Reston: ASCE).
Weyens, N., van der Lelie, D., Taghavi, S. (2009). Exploiting plant-microbe partnerships to improve biomass production and remediation. Trend. Biotechnol. 27, 591–598. doi: 10.1016/j.tibtech.2009.07.006
Whipps, J. M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. doi: 10.1093/jexbot/52.suppl_1.487
Yadav, J., Verma, J. P., Tiwari, K. N. (2011). Plant Growth Promoting Activities of Fungi and their Effect on Chickpea Plant Growth. Asian J. Biol. Sci. 4 (3), 291–299. doi: 10.3923/ajbs.2011.291.299
Yaish, M. W., Antony, I., Glick, R. (2015). Isolation and characterization of endophytic plant growth promoting bacteria from date palm tree (Phoenix dactylifera L.) and their potential role in salinity tolerance. Antonie Van Leeuwenhoek 107, 1519–1532. doi: 10.1007/s10482-015-0445-z
Yan, K., Shao, H., Shao, C. H., Chen, P., Zhao, S. H., Brestic, M., et al. (2013). Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiol. Plant 35, 2867–2878. doi: 10.1007/s11738-013-1325-7
Yasmin, H., Bano, A. (2011). Isolation and characterization of phosphate solubsilizing bacteria from rhizosphere soil of weeds of khewra salt range and attock. Pak. J. Bot. 43, 1663–1668.
Yuttavanichakul, W., Lawongsa, P., Wongkaew, S., Teaumroong, N., Boonkerd, N., Nomura, N., et al. (2012). Improvement of peanut rhizobial inoculant by incorporation of plant growth promoting rhizobacteria (PGPR) as biocontrol against the seed borne fungus, Aspergillus niger. Biol. Control 63 (2), 87–97. doi: 10.1016/j.biocontrol.2012.06.008
Zaman, M., Shahid, S. A., Heng, L. (2018). Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques (Switzerland AG: Springer International Publishing). doi: 10.1007/978-3-319-96190-3
Zehra, A., Meena, M., Dubey, M. K., Aamir, M., Upadhyay, R. S. (2017). Synergistic effects of plant defence elicitors and Trichoderma harzianum on enhanced induction of antioxidant defence system in tomato against Fusarium wilt disease. Bot. Stud. 58 (1), 44. doi: 10.1186/s40529-017-0198-2
Zewdu, S., Suryabhagavan, K. V., Balakrishnan, M. (2017). Geo-spatial approach for soil salinity mapping in Sego Irrigation Farm, South Ethiopia. J. Saudi Soc Agric. Sci. 16, 16–24. doi: 10.1016/j.jssas.2014.12.003
Zhang, H., Kim, M.-S., Krishnamachari, V., Payton, P., Sun, Y., Grimson, M., et al. (2007). Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226 (4), 839–851. doi: 10.1007/s00425-007-0530-2
Zhang, H., Xie, X., Kim, M. S., Kornyeyev, D. A., Holaday, S., Paré, P. W. (2008a). Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J. 56 (2), 264–273. doi: 10.1111/j.1365-313X.2008.03593.x
Zhang, H., Kim, M.-S., Sun, Y., Dowd, S. E., Shi, H., Paré, P. W. (2008b). Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant Microbe Interact. 21 (6), 737–744. doi: 10.1094/MPMI-21-6-0737
Zhang, S., Gan, Y., Xu, B. (2016). Application of Plant-Growth-Promoting Fungi Trichoderma longibrachiatum T6 Enhances Tolerance of Wheat to Salt Stress through Improvement of Antioxidative Defense System and Gene Expression. Front. Plant Sci. 7, 1405. doi: 10.3389/fpls.2016.01405
Zhang, X., Chen, L., Wang, J., Wang, M., Yang, S., Zhao, C. (2018). Photosynthetic acclimation to long-term high temperature and soil drought stress in two spruce species (Picea crassifolia and P.wilsonii) used for afforestation. J. For. Res. 29 (2), 363–372. doi: 10.1007/s11676-017-0468-6
Zhang, S., Gan, Y., Xu, B. (2019). Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 19, 22. doi: 10.1186/s12870-018-1618-5
Zhou, C., Ma, Z., Zhu, L., Xiao, X., Xie, Y., Zhu, J., et al. (2016). Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. Int. J. Mol. Sci. 17, 976. doi: 10.3390/ijms17060976
Zhou, N., Zhao, A., Tian, C.-Y. (2017). Effect of halotolerant rhizobacteria isolated from halophytes on the growth of sugar beet (Beta vulgaris L.) under salt stress. FEMS Microbiol. Lett. 364, fnx091. doi: 10.1093/femsle/fnx091
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: extreme environment, halophilic microorganisms, endophytes, PGPR—plant growth-promoting rhizobacteria, agriculture, microbial metabolism, salty soils
Citation: Otlewska A, Migliore M, Dybka-Stępień K, Manfredini A, Struszczyk-Świta K, Napoli R, Białkowska A, Canfora L and Pinzari F (2020) When Salt Meddles Between Plant, Soil, and Microorganisms. Front. Plant Sci. 11:553087. doi: 10.3389/fpls.2020.553087
Received: 17 April 2020; Accepted: 31 August 2020;
Published: 16 September 2020.
Edited by:
Nikolay Vassilev, University of Granada, SpainReviewed by:
Jorge Poveda, Biological Mission of Galicia (MBG), SpainDilfuza Egamberdieva, Leibniz Center for Agricultural Landscape Research (ZALF), Germany
Copyright © 2020 Otlewska, Migliore, Dybka-Stępień, Manfredini, Struszczyk-Świta, Napoli, Białkowska, Canfora and Pinzari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Flavia Pinzari, ZmxhdmlhLnBpbnphcmlAY25yLml0
 Anna Otlewska
Anna Otlewska Melania Migliore
Melania Migliore Katarzyna Dybka-Stępień
Katarzyna Dybka-Stępień Andrea Manfredini
Andrea Manfredini Katarzyna Struszczyk-Świta
Katarzyna Struszczyk-Świta Rosario Napoli
Rosario Napoli Aneta Białkowska
Aneta Białkowska Loredana Canfora
Loredana Canfora Flavia Pinzari
Flavia Pinzari