
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 September 2020
Sec. Crop and Product Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.543747
This article is part of the Research Topic Regulation of Fruit Ripening and Senescence View all 13 articles
Fruit ripening is a highly coordinated developmental process driven by a complex hormonal network. Ethylene is the main regulator of climacteric fruit ripening. However, a putative role of other key phytohormones in this process cannot be excluded. We previously observed an increasing level of auxin during the post-harvest ripening of the durian fruit, which occurred concomitantly with the rise in the climacteric ethylene biosynthesis. Herein, we connect the key auxin signaling component, auxin response factors (ARFs), with the regulatory network that controls fruit ripening in durian through the identification and functional characterization of a candidate ripening-associated ARF. Our transcriptome-wide analysis identified 15 ARF members in durian (DzARFs), out of which 12 were expressed in the fruit pulp. Most of these DzARFs showed a differential expression, but DzARF2A had a marked ripening-associated expression pattern during post-harvest ripening in Monthong, a commercial durian cultivar from Thailand. Phylogenetic analysis of DzARF2A based on its tomato orthologue predicted a role in ripening through the regulation of ethylene biosynthesis. Transient expression of DzARF2A in Nicotiana benthamiana leaves significantly upregulated the expression levels of ethylene biosynthetic genes, pointing to a ripening-associated role of DzARF2A through the transcriptional regulation of ethylene biosynthesis. Dual-luciferase reporter assay determined that DzARF2A trans-activates durian ethylene biosynthetic genes. We previously reported significantly higher auxin level during post-harvest ripening in a fast-ripening cultivar (Chanee) compared to a slow-ripening one (Monthong). DzARF2A expression was significantly higher during post-harvest ripening in the fast-ripening cultivars (Chanee and Phuangmanee) compared to that of the slow-ripening ones (Monthong and Kanyao). Thus, higher auxin level could upregulate the expression of DzARF2A during ripening of a fast-ripening cultivar. The auxin-induced expression of DzARF2A confirmed its responsiveness to exogenous auxin treatment in a dose-dependent manner, suggesting an auxin-mediated role of DzARF2A in fruit ripening. We suggest that high DzARF2A expression would activate ARF2A-mediated transcription of ethylene biosynthetic genes, leading to increased climacteric ethylene biosynthesis (auxin-ethylene crosstalk) and faster ripening. Hence, we demonstrated DzARF2A as a new component of the regulatory network possibly mediating durian fruit ripening through transcriptional regulation of ethylene biosynthetic genes.
Fruit ripening is a genetically programmed and coordinated developmental process involving dramatic physiological transformations, some of which include changes in the aroma, texture, color, and nutritional value of the flesh (Alexander and Grierson, 2002; Adams-Phillips et al., 2004). This process is controlled by transcriptional regulatory networks with transcription factors (TFs) acting as pivotal regulators. Depending on the mechanism, the ripening of fleshy fruits can be divided into climacteric and non-climacteric ripening (Lelievre et al., 1997). Climacteric ripening is associated with an autocatalytic increase in ethylene biosynthesis and the key role played by ethylene in this process (as the main trigger of fruit ripening) has been well documented (Barry et al., 2000). Consistently, various genes involved in ethylene biosynthesis and signaling were shown to be vital for ripening (Ayub et al., 1996; Xie et al., 2006; Wang et al., 2009), but the involvement of other plant hormones in this process (together with ethylene) seems also likely. The possibility that fruit ripening is regulated by a complex hormonal network has already been formulated and suggested by the existing literature. Auxin (IAA; indole-3-acetic acid) is a crucial phytohormone which regulates numerous aspects of plant growth and development (Woodward and Bartel, 2005), including ripening (Jones et al., 2002). Along with significant increases in the climacteric ethylene, elevated auxin levels of have also been detected during the ripening of tomato (Gillaspy et al., 1993), peach (Miller et al., 1987), and durian (Khaksar et al., 2019), suggesting a possible ripening-associated role of auxin. Auxin response factor (ARF) TFs represent the core of auxin signaling. These proteins mediate the auxin-dependent gene expression via binding to the auxin response elements (AuxREs; TGTCTC), located in the promoter regions of the auxin responsive genes, in a dose-dependent manner (Hagen and Guilfoyle, 2002). ARFs are generally characterized by a highly conserved DNA binding domain (DBD) at their N terminus. They also contain a middle region, responsible for the transactivation or repression of target genes and a carboxyl-terminal dimerization domain (CTD) that is involved in the protein-protein interactions (Guilfoyle and Hagen, 2007; Guilfoyle and Hagen, 2012; Nanao et al., 2014; Guilfoyle, 2015). The domain architecture of ARFs is known to play a key role in their specificity. Since the identification and characterization of the first Arabidopsis ARF (AtARF1) (Ulmasov et al., 1997), numerous studies have identified members of the ARF gene family in plant species, such as tomato (Kumar et al., 2011), rice (Wang et al., 2007), Medicago (Shen et al., 2015), banana (Hu et al., 2015), apple (Luo et al., 2014), maize (Xing et al., 2011), chickpea (Singh et al., 2017), physic nut (Tang et al., 2018), grape (Wan et al., 2014), and papaya (Liu et al., 2015). Functional characterization of some ARFs based on the phenotypes of the loss-of-function and gain-of-function mutants have also been carried out. For instance, the Arabidopsis arf1 and arf2 mutants showed abnormal abscission of the floral organs (Ellis et al., 2005), while mutation in the AtARF7 gene diminished hypocotyl response to blue light and auxin (Harper et al., 2000). The AtARF8 loss-of-function mutation, on the other hand, impaired hypocotyl elongation and auxin homeostasis (Goetz et al., 2006), while the tomato SlARF3 was found to participate in the formation of trichomes and epidermal cells (Zhang et al., 2015). In addition to their roles in plant growth and development, ARFs are also involved in the responses to abiotic stressors. OsARF16, for example, has been found to participate in the adaptive response to phosphate starvation in rice (Shen et al., 2013), while another member (OsARF12) has been found to play role in iron homeostasis (Qi et al., 2013). Although the links between ARFs and plant growth and development have been extensively studied, our actual knowledge regarding the possible roles of ARFs in fruit ripening remains poorly understood and limited only to a few studies in tomato and papaya. In tomato, the SlARF2A and SlARF2B (two ARF2 paralogs) displayed a ripening-associated expression pattern. Down-regulation of either SlARF2A or SlARF2B led to ripening defects, while the arf2a/arf2b double mutation resulted in a severe inhibition of the ripening process (Hao et al., 2015). Moreover, a significant decrease in the climacteric ethylene biosynthesis and the expression levels of ethylene biosynthetic genes was observed in the SlARF2AB-RNAi fruits (Hao et al., 2015). Breitel et al. (2016) identified SlARF2A, an auxin signaling component that controls ripening. They found SlARF2A expression to be ripening-regulated and responsive to exogenous ethylene and auxin treatments. In papaya, Liu et al. (2015) profiled the expression pattern of 11 ARFs (CpARFs) during the different fruit developmental stages and found that the expression of most CpARFs underwent significant changes with ripening. Specifically, the expression of CpARF1 and CpARF2 increased, while that of CpARF7 and CpARF11 decreased.
Durian (Durio zibethinus Murr.) is a tropical fruit crop of rising economy value endemic to Southeast Asia, with an ever-growing popularity on the international market. Durian is distinctive for its strong and unique odor and formidable spiny husk. With over 200 existing cultivars, Thailand is the top durian exporter across the Southeast Asian region. However, only a few of these cultivars are commercially cultivated and in high demand both locally and on the international market. These include Monthong (D. zibethinus Murr. cv. “Monthong”), Chanee (D. zibethinus Murr. cv. “Chanee”), Phuangmanee (D. zibethinus Murr. cv. “Phuangmanee”), and Kanyao (D. zibethinus Murr. cv. “Kanyao”). Among these, Monthong—due to its mild odor and creamy texture—is of great interest (Pinsorn et al., 2018). Notably, these cultivars harbor different post-harvest ripening behaviors; Monthong and Kanyao are slow-ripening cultivars that need around 5 days to fully ripen after being harvested at mature stage. Chanee and Phuangmanee, on the other hand, are fast-ripening cultivars that ripen 3 days after harvesting at mature stage. Durian is a climacteric fruit that has a strikingly limited shelf life after harvesting. Hence, the fast and slow ripening processes are agronomic traits of great economic importance for agricultural industry. Offering durian fruit with a pleasant flavor and nutritional value, while also harboring longer shelf life remains a challenge for the industry. Over recent decades, investigations have focused on extending shelf life. However, our actual knowledge on the molecular mechanisms underlying the ripening process is still limited, especially with regard to fast- and slow-ripening cultivars. In our previous study, we observed an increasing level of auxin production during the post-harvest ripening of the durian fruit in both Monthong and Chanee cultivars, suggesting a ripening-associated role of auxin during fruit ripening (Khaksar et al., 2019). Here, considering the major role of ARFs in the auxin signaling pathway, we performed a transcriptome-wide analysis on the durian genome and identified 15 members of the ARF gene family. Comprehensive gene expression profiling of the durian ARFs (DzARFs) during the post-harvest ripening revealed significant differential expression of some ARFs and suggested putative ripening-associated ones. We then functionally characterized a candidate ripening-associated TF (DzAFR2A). Our findings positioned DzARF2A as a key component of the regulatory network behind the post-harvest ripening of the durian. To our knowledge, this is the first report on the identification and characterization of the ARF gene family in this fruit. Findings from this study can be further exploited towards the development of molecular markers for breeding new durian varieties with longer shelf lives.
Durian (Durio zibethinus Murr.) cv. Monthong, Chanee, and Phuangmanee were collected from commercial durian plantations located in the eastern part of Thailand. Fruit samples having similar size and weight (~3–4 kg each) were harvested at mature stage which occurs at 90 days (for Chanee and Phuangmanee) and 105 days (for Monthong) after anthesis. Samples were kept at room temperature (30°C) until peeling. For these cultivars, three types of samples during post-harvest ripening (unripe, midripe, and ripe) were used. For unripe samples, fruits harvested at the mature stage of all three cultivars were kept at room temperature for one day and then peeled. For midripe samples, fruits harvested at the mature stage were kept at room temperature for two days (for Chanee and Phuangmanee) and three days (for Monthong) and then peeled. For ripe samples, fruits harvested at the mature stage were kept at room temperature for three days (for Chanee and Phuangmanee) and five days (for Monthong) and then peeled. In this study, another cultivar called Kanyao was also used. However, for Kanyao cultivar, we only had access to the ripe fruit which was bought at the ripe stage and peeled. After peeling, two of the central pulps were collected from each fruit sample, following the method described in Pinsorn et al. (2018). To ensure samples of the different cultivars were compared at the same ripening stage, the first pulp was collected along with a seed, and was used to measure fruit firmness with a texture analyzer following our previous study (Khaksar et al., 2019). Midripe and ripe pulps had a mean [± standard deviation (SD)] firmness of 3.4 ± 0.81 and 1.55 ± 0.45 N, respectively in all cultivars. After this test, the second pulp was collected without a seed, immediately frozen in liquid nitrogen, and stored at -80°C for RNA extraction.
To further validate the putative ripening-associated DzARFs in Monthong cultivar, three different ripening treatments were applied as follows: natural, ethephon-induced, and 1-methylcyclopropene (1-MCP)-delayed ripening. Durian samples were collected at mature stage and treated with either ethephon (48% 2-chloroethylphosphonic acid; Alpha Agro Tech Co., Ltd., Thailand) or 1-MCP (0.19% 1-MCP tablet; BioLene Co., Ltd., China) for ethephon-induced and 1-MCP-delayed ripening, respectively following the method described in our previous study (Khaksar et al., 2019). After treatments, control and treated samples were kept at room temperature (30°C) until the ethephon-induced samples ripened. As soon as ethephon-treated samples ripened, all samples from the other ripening conditions were peeled. Two central pulps were collected from each sample and processed as mentioned previously. In this study, for each type of sample (unripe, midripe, and ripe, as well as natural ripening, ethephon-induced ripening, and 1-MCP-delayed ripening) of each cultivar, three independent biological replicates were used. Each biological replicate is one durian fruit harvested from a separate tree.
For transient expression of DzAFR2A in Nicotiana benthamiana leaves, N. benthamiana seeds were sown in pots containing peat moss and were grown under controlled conditions (temperature 25°C and 16/8 h light/dark photoperiod; artificial light of 4,500 Lux). Two-week-old seedlings were transplanted individually into pots and were grown under similar conditions.
For exogenous auxin treatment, young leaves of Monthong cultivar were soaked in different concentrations (10, 20, and 40 μM) of indole-3-acetic acid (IAA) (Duchefa Biochemie, The Netherlands) for 1 and 2 h. As control, leaves were soaked in distilled water without IAA. For reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis, leaves were immediately frozen in liquid nitrogen and stored at -80°C until total RNA extraction.
In order to obtain the assembled transcriptome data, Illumina sequencing reads from RNA-Seq study of durian (Musang King cultivar) were retrieved from a public repository database (SRA, Sequence Read Archive) with the following accession numbers: SRX3188225 (root tissue), SRX3188222 (stem tissue), SRX3188226 (leaf tissue), and SRX3188223 (aril/pulp tissue). To obtain the de novo assembled transcriptome, we followed the method described in our previous study (Khaksar et al., 2019). We then followed Hidden Markov Model (HMM) search to identify ARF gene family members. First, we obtained the ARF conserved domain (PF06507) from the Pfam protein database (http://pfam.xfam.org) and used as a query to search against the assembled transcriptome database using the HMMER software (http://hmmer.org/) following e-value cut-off ≤1e-5. This approach enabled us to identify 15 non-redundant ARF gene family members. Further, the amino acid sequences of durian ARFs (DzARFs) were checked in SMART database for the presence of the ARF domain. The expression levels of DzARFs in different tissues, including root, stem, leaf, and aril (pulp) were analyzed using the de novo assembled transcriptome. Abundance estimation tool in Trinity package was used to align the input reads to the de novo assembled transcriptome to obtain raw counts of each contig. The raw read counts were then merged in a single read count matrix which was then normalized to generate a Trimmed Mean of M-values (TMM) normalized matrix. For generating heatmap, the normalized total read counts were used as queries in MetaboAnalyst 4.0, an open source R-based program (Chong et al., 2018). For the heatmap construction, the values were sum normalized, log2 transformed and autoscaled.
To study evolutionary relationships among DzARFs and ARFs from Arabidopsis and tomato, a neighbor-joining (NJ) phylogenetic tree was constructed with 1,000 bootstrap replicates using MEGA X software (with a JTT model and pairwise gap deletion) by aligning their full-length protein sequences (Kumar et al., 2018).
MEME program (http://meme-suite.org) was used for identification of the conserved motifs (Bailey et al., 2009) with the following parameter settings: motif length = 6–100; motif sites = 2–120; maximum number of motifs = 10; and the distribution of one single motif was “any number of repetitions”. Exon/intron organization of the durian ARF genes was determined using Gene Structure Display Server 2.0 (with default parameters) (http://gsds.cbi.pku.edu.cn).
Total RNA isolation from durian fruit pulp samples was carried out using PureLink Plant RNA Reagent (Thermo Fisher Scientific™) following the manufacturer’s instructions. Genomic DNA was removed with DNase I (Thermo Fisher Scientific™) treatment. The quality and quantity of RNA samples were examined using agarose gel electrophoresis and an Eppendorf BioPhotometer D30 with A260/280 and A260/230 ratios between 1.8 to 2.0 and 2.0 to 2.2, respectively following the standard guidelines described in Bustin et al. (2009). For reverse transcription, one microgram of total RNA was used to generate cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific™), following the manufacturer’s recommended protocol and the standard guidelines of reverse transcription described in Bustin et al. (2009). Primer 3 online (http://primer3.ut.ee/) was used to design the primers used in this study which are presented in Supplementary Table S1. To measure the transcript levels of DzARFs at three stages (unripe, midripe, and ripe) during post-harvest ripening of Monthong and compare the expression level of candidate ripening-associated DzARF among different cultivars, RT-qPCR was performed. The PCR reaction was performed in a total volume of 10 μl containing 1 μl of diluted cDNA (1 ng of cDNA), 5 μl of 2x QPCR Green Master Mix LRox (biotechrabbit, Berlin, Germany), and 200 nM of each gene-specific primer. A CFX95 Real-time System (Bio-Rad Laboratories Inc., California, USA) was used under the following conditions: initial activation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s, then annealing at 58–60°C for 30 s, and finally extension at 72°C for 20 s. Three independent biological replicates were used for each RT-qPCR experiment. The elongation factor 1 alpha (EF-1α; XM_022889169.1) and actin (ACT; XM_022897008.1) genes of durian were selected as reference genes for the normalization of RT-qPCR data according to our in-house transcriptome data of durian fruit from different cultivars which confirmed the invariant expression levels of EF-1α and ACT under different experimental conditions. In addition, to further confirm this, we used Genorm software (Vandesompele et al., 2002) to check the stability of our reference genes (see Supplementary Table S1 for primers).
Statistical analysis of RT-qPCR data was carried out using the method described in Steibel et al. (2009) which consists of the analysis of cycles to threshold values (Ct) for the target and reference genes according to a linear mixed model [Model I of the report of Steibel et al. (2009)]. Treatments, genes, and their interactions were the fixed factors whereas the obtained Ct values comprised the dependent variables of the model. Samples were the random effects of the model. The normalization of RT-qPCR expression data was carried out using normalization factors; the geometric mean of the two reference genes calculated with Genorm software (Vandesompele et al., 2002). For graphical illustration, the expression data are presented as fold change (2-ΔΔCt) (Livak and Schmittgen, 2001). P < 0.05 was considered statistically significant. R programming language (https://www.R-project.org/) was used for the analysis.
The full-length cDNA of DzARF2A was amplified from the cDNA of Monthong cultivar (at midripe stage) using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) (primers are listed in Supplementary Table S2). After that, the PCR amplicon was inserted into the entry vector pDONR207 using Gateway® BP Clonase® II (Thermo Fisher Scientific, Waltham, MA, USA). After sequencing, DzARF2A in Monthong cultivar was confirmed to be identical to the DzARF2A in Musang King cultivar (XM_022883347.1). From the resultant pDONR207-DzARF2A, the putative DzARF2A was inserted in to the pGWB2 expression vector using Gateway® LR Clonase® II (Thermo Fisher Scientific, Waltham, MA, USA) to produce pGWB2-DzARF2A. The resulting pGWB2-DzARF2A as well as pGWB2-GFP (control) were individually introduced into the Agrobacterium tumefaciens strain GV3101 by electroporation. The positive A. tumefaciens colony harbouring each construct was further grown in 25 ml of LB broth containing 25 mg L-1 gentamycin and 25 mg L-1 rifampicin with shaking at 250 rpm overnight at 30°C. Cells were harvested by centrifugation at 4,000×g for 10 min and resuspended in MM buffer (10 mM MES and 10 mM MgCl2, pH 5.6) to reach an optical density of 0.6 (OD600). For the agroinfiltration experiment, the agrobacterium solution containing either pGWB2-DzARF2A or pGWB2-GFP (control) was mixed with the agrobacterium solution harbouring the gene encoding the silencing inhibitor protein p19 at a ratio of 1:1. Then, acetosyringone was added at a concentration of 100 mg L-1 and the mixture was incubated at room temperature for 3 h under dark conditions. Then, the solution was infiltrated into the abaxial surface of three individual leaves per plant using a needleless 1 ml syringe. Three independent biological replicates (three 4-week-old plants) were used for each construct at each timepoint. At three and five days after infiltration, samples of the infiltrated leaves were collected, frozen in liquid nitrogen and ground into a fine powder with a mortar and pestle. This powder was then used for RNA isolation and cDNA synthesis. The cDNA was further used as the template for RT-qPCR analysis. The elongation factor 1 alpha (EF-1α; XM_016613715.1) and actin (ACT; XM_016618072.1) genes of Nicotiana were used as reference genes for the normalization of RT-qPCR data. In order to design primers for N. benthamiana ethylene biosynthetic genes, the Nicotiana tabacum genome [i.e., 1-aminocyclopropane-1-carboxylic acid (ACC) synthase; ACS (NM_001326261.2) and ACC oxidase; ACO (NM_001325823.1)] were used to blast against the genome sequence of N. benthamiana (SGN Nicotiana benthamiana ftp site; https://solgenomics.net/organism/Nicotiana_benthamiana/genome). Primers are listed in Supplementary Table S2. The 2000-bp upstream promoter regions of these genes were scanned for the auxin response elements (AuxREs; TGTCTC) using the online tool PLACE (http://www.dna.affrc.go.jp/) (Liu et al., 2017).
To examine the binding activity of DzARF2A to the promoters of ethylene biosynthetic genes, dual-luciferase assay was performed following the method described in Feng et al. (2016). Briefly, genomic DNA was extracted from durian leaves using the cetyltrimethyl ammonium bromide (CTAB)-based method according to Abdel-Latif and Osman (2017). The 2,000-bp promoter regions of ethylene biosynthetic genes of durian; DzACS (XM_022901720.1) and DzACO (XM_022903265.1) were amplified using the primers listed in Supplementary Table S2 and cloned into the pGreenII 0800-LUC double-reporter vector (Hellens et al., 2005). For generating the effector construct, the coding sequence of DzARF2A was cloned into the pGWB2 expression vector. A. tumefaciens (GV3101) harbouring each of the reporter or effector constructs were then co-infiltrated into N. benthamiana leaves. After three days, firefly (LUC) and Renilla (REN) luciferase activities were measured by a dual-luciferase reporter assay system (Promega, E1910) according to the manufacturer’s instructions on a Tecan Infinite 200 PRO microplate reader (TECAN) and the relative LUC/REN ratios were calculated. Four independent biological replicates were used.
Based on the bioinformatics analysis of the publicly available transcriptomic data from different tissues (stem, root, leaf, and pulp) of the durian cv. Musang King, we identified 15 genes encoding putative ARFs (Table 1). The deduced DzARFs ranged from 576 (ARF17) to 1142 (ARF7) amino acids in length and varied from 63.65 (ARF17) to 128.00 (ARF7) kDa in molecular weight. Their isoelectric points were between 5.10 (ARF5A) and 8.50 (ARF10), suggesting that they could function in various microenvironments.
To examine the evolutionary relationships between DzARFs and the previously-reported ARFs from the model plants Arabidopsis and tomato, we constructed a NJ phylogenetic tree based on the full-length amino acid sequence similarity and topology of 23 Arabidopsis ARFs (AtARFs) and 18 tomato ARFs (SlARFs), by aligning their full-length protein sequences with 1,000 bootstrap replicates using MEGA X. Our phylogenetic analysis classified DzARFs into three major clades (A, B, and C), which were further classified into 12 subclades (A1, A2, B1, B2, B3, C1, C2, C3, C4, C5, C6, and C7) (Figure 1A). Subclades C2, C6, and C7 lacked any DzARFs and harbored AtARFs and/or SlARFs. Group C6 was the biggest subclade and was comprised of eight members, whereas groups C2 and C7 were the smallest ones and harbored only two members in each group.
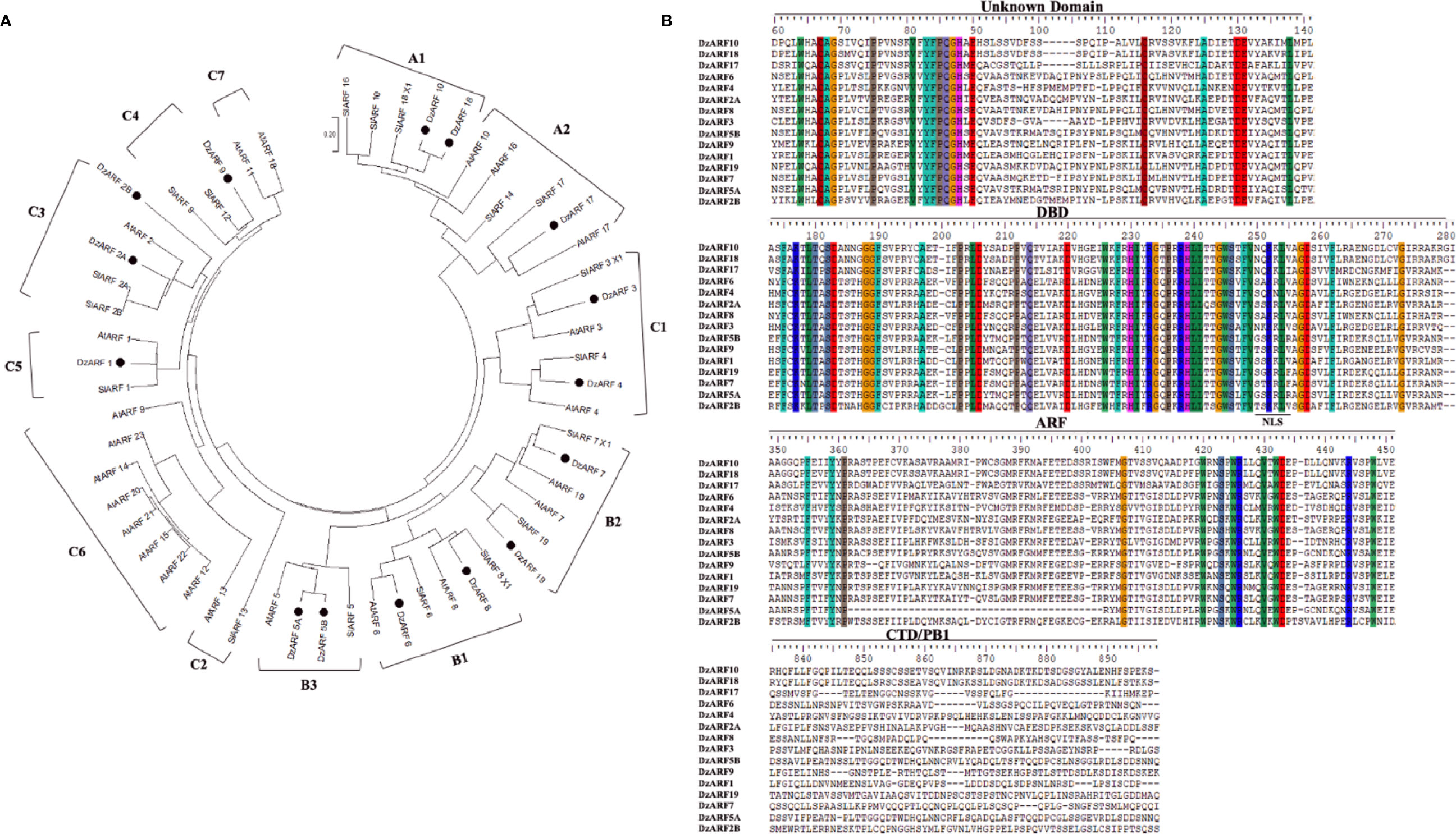
Figure 1 Phylogenetic tree and multiple sequence alignment of the amino acid sequences of the DzARFs. (A) The deduced full-length protein sequences of durian ARFs (DzARFs) were aligned with Arabidopsis thaliana (AtARFs) and Solanum lycopersicum (SlARFs) protein sequences to construct the phylogenetic tree using MEGA X software and the neighbor-joining method (with 1,000 bootstrap replicates, a JTT model and pairwise gap deletion using a bootstrap test of phylogeny with the minimum evolution test and default parameters). (B) Multiple sequence alignment analysis was carried out using ClutalW. Identical amino acids are highlighted by color. Only important domains are presented (DBD, DNA binding domain; ARF, auxin response factor domain; CTD/PB1, carboxyl-terminal dimerization/Phox and Bem 1 domain). A nuclear localization signal (NLS) was identified at the end of DBD in all DzARFs.
Multiple peptide sequence alignment of DzARFs revealed a highly conserved DBD of around 80 amino acids at the N-terminal region (Figure 1B). Moreover, these DzARFs contained a middle region that functions as a transcriptional activation or repression domain and a CTD responsible for the protein-protein interactions. Notably, we also identified an unknown domain located at the early N-terminal region of the DzARF protein sequences (Figure 1B). Moreover, our in silico analyses revealed the presence of a conserved putative mono-partite nuclear localization signal (NLS) at the end of the DBD of all DzARFs which confirmed their localization to the nucleus (Figure 1B). Our observation is in line with previous studies which have well documented the occurrence of NLS in members of the ARF TF family (Shen et al., 2010; Yu et al., 2014; Niu et al., 2018; Diao et al., 2020).
By using MEME, we analyzed the protein sequence features of DzARFs and identified at least 10 conserved motifs (Supplementary Figure S1). Among these, two novel motifs (1 and 2) were associated with the N-terminal unknown domain. Motifs 3, 4, and 5 were found to be associated with the DBD. The ARF domain corresponded to motifs 6, 7, and 8 and the CTD corresponded to motifs 9 and 10. Notably, the DzARFs in the phylogenetic tree harbored similar motif organization and clustered together (Supplementary Figure S1). We also analyzed the exon-intron organization of DzARFs and found no introns within the DzARFs (Supplementary Figure S2). This observation was similar to members of the Dof gene family in durian (DzDofs), which harbored no introns (Khaksar et al., 2019).
We analyzed the publicly available transcriptomic data on the different tissues (stem, root, leaf, and pulp) from durian cv. Musang King and observed that 12 out of the 15 DzARFs (DzARF1, DzARF2A, DzARF2B, DzARF3, DzARF5A, DzARF5B, DzARF6, DzARF7, DzARF8, DzARF10, DzARF17, and DzARF19) were expressed in the fruit pulp (Figure 2). This suggests a possible role of these DzARFs in the post-harvest ripening of the durian fruit. The three remaining DzARFs, including DzARF4, DzARF9, and DzARF18 were expressed in other tissues, but not in the fruit pulp (Figure 2), suggesting that their roles in fruit ripening seem unlikely.
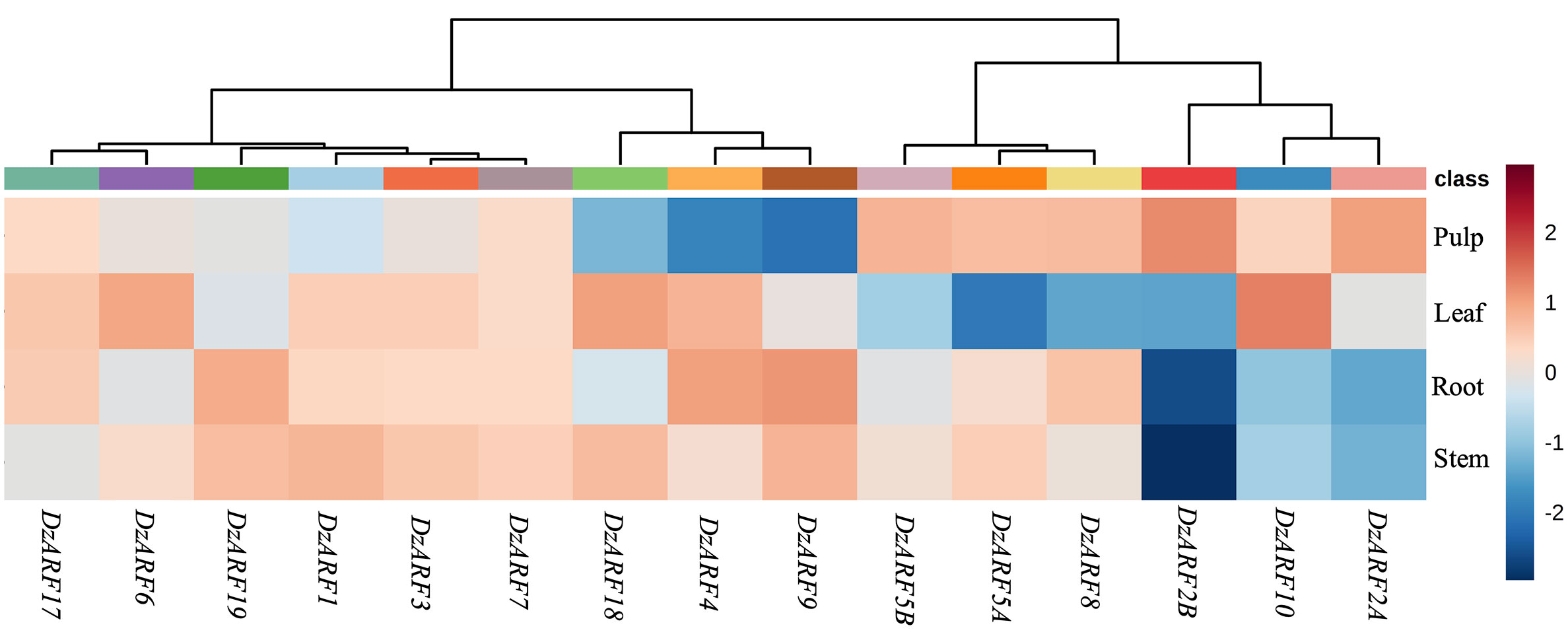
Figure 2 Tissue-specific expression profile of DzARFs in Musang King cultivar at ripe stage. The publicly available Illumina RNA-seq data were used to analyze the expression levels of DzARFs in root, stem, leaf, and fruit pulp tissues. The higher expression for each gene was presented in red; otherwise, blue was used. Heatmap was generated using MetaboAnalyst 4.0, an open source R-based program. Data were sum normalized, log transformed, and auto scaled.
We used RT-qPCR to verify the expression patterns of DzARFs in three stages of the post-harvest ripening (unripe, midripe, and ripe) and identified 12 pulp-expressed DzARFs that were similar to those found in the Musang King cultivar. Notably, eight out of these 12 DzARFs (DzARF2A, DzARF2B, DzARF3, DzARF5A, DzARF5B, DzARF6, DzARF7, and DzARF19) harbored a ripening-associated expression pattern and were considered as putative ripening-associated TFs. The expression levels of the remaining four DzARFs (DzARF1, DzARF8, DzARF10, and DzARF17) did not vary significantly over the course of post-harvest ripening (Figure 3) and therefore these four DzARFs were not considered as ripening-associated TFs. Among the eight ripening-associated DzARF candidates, the DzARF2A and DzARF19 transcripts were up-, while the DzARF2B, DzARF3, DzARF5A, DzARF5B, DzARF6, and DzARF7 transcripts were down-regulated during post-harvest ripening. The expression levels of these eight putative ripening-associated DzARFs were further investigated under ethylene treatment.
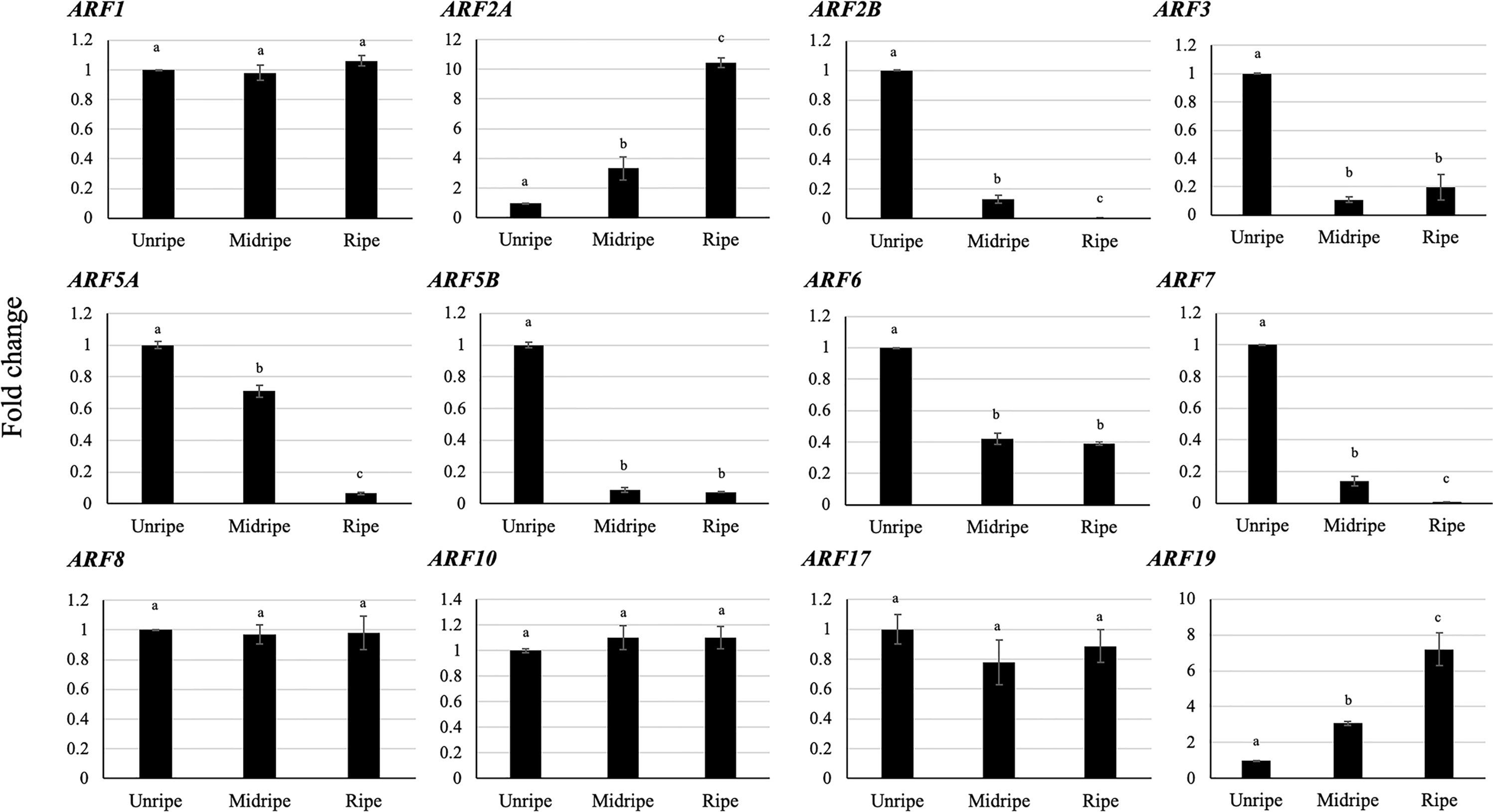
Figure 3 Fold changes in expression levels of DzARFs at three different stages (unripe, midripe, and ripe) during post-harvest ripening of durian fruit (Monthong cultivar). The relative gene expression levels were calculated by using the 2-ΔΔCt method and levels were normalized by the geometric mean of reference genes and the unripe stage as control. Three independent biological replicates were used. Bars with different letters show significant differences (P < 0.05).
Since ethylene plays a key role in climacteric fruit ripening, we analyzed the expression levels of the eight putative ripening-associated DzARFs under three different ripening conditions: ethephon-induced, natural, and 1-methylcyclopropane (1-MCP)-delayed ripening. Among these, the expression levels of DzARF2A and DzARF19 were significantly induced under ethephon and suppressed under 1-MCP treatment, compared to those under natural ripening (control). However, the expression levels of DzARF3 and DzARF6 were significantly induced by 1-MCP and suppressed by ethephon relative to the control (Figure 4). Taken together, these results provide compelling evidence on the ripening-associated roles of these four DzARFs during post-harvest ripening in the durian fruit. Notably, the expression levels of DzARF2B, DzARF5A, and DzARF5B did not vary significantly under ethephon and 1-MCP treatments (compared to the control), so they were not considered as ripening-associated TFs. DzARF7, on the other hand, was not expressed under any examined condition, and thus it was also not considered as a ripening-associated TF. Among the validated ripening-associated DzARFs, DzARF2A displayed a dramatic increase of transcript accumulation under ethephon treatment and it was expressed increasingly during the post-harvest ripening of the durian fruit, suggesting a potential role as a transcriptional activator of ripening. This marked ripening-associated expression pattern of DzARF2A prompted its functional characterization to gain a better understanding of its physiological significance during fruit ripening in durian.
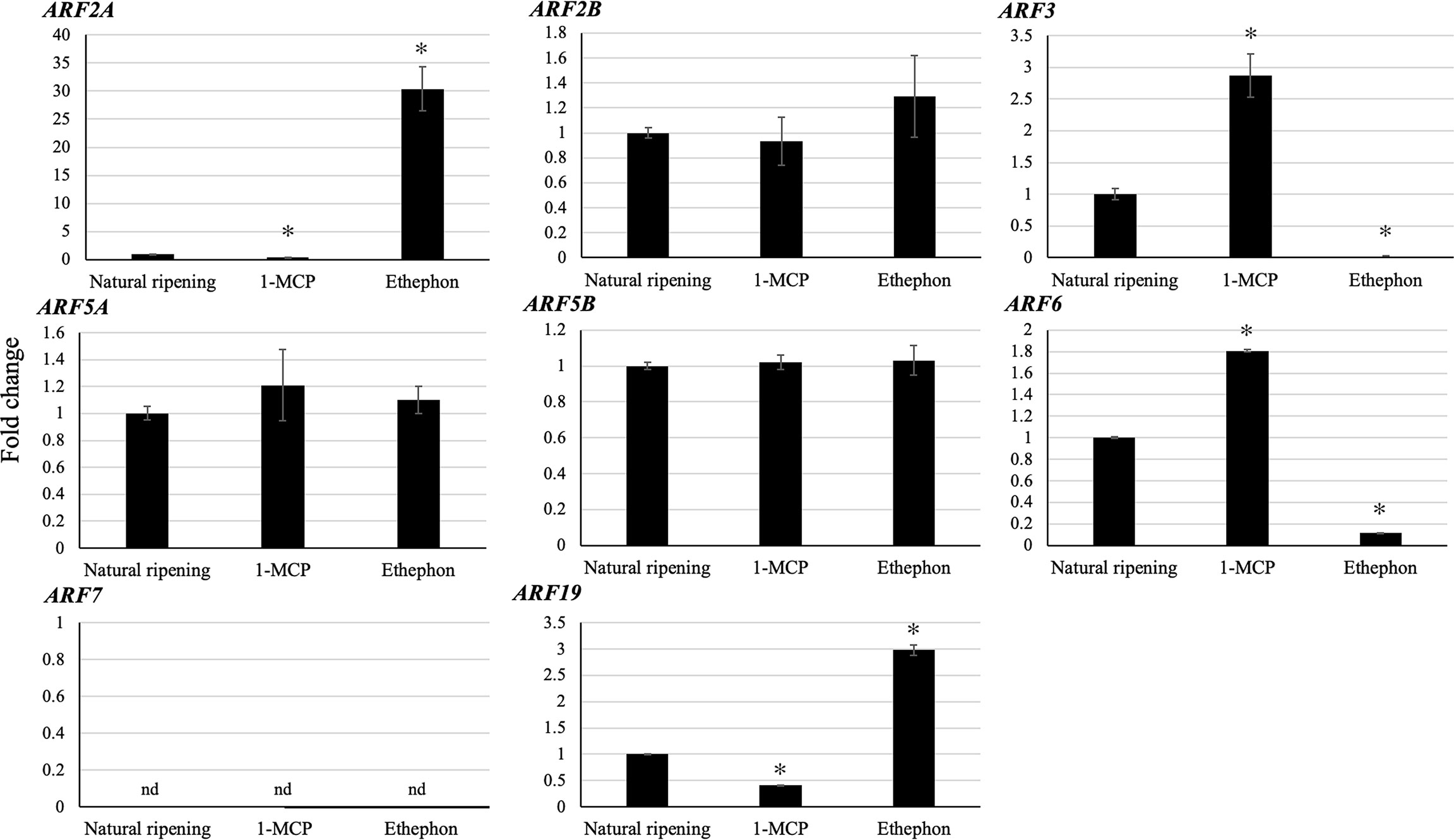
Figure 4 Fold changes in expression levels of eight candidate ripening-associated DzARFs under three different ripening conditions. The relative expression levels of candidate ripening-associated DzARFs were quantified under three different ripening conditions; natural (control), ethylene-induced, and 1-MCP-delayed ripening by using the 2-ΔΔCt method and levels were normalized by the geometric mean of reference genes and the natural ripening condition as control. Three independent biological replicates were used. An asterisk (*) above the bar indicates a significant difference (P < 0.05) (nd = not detected).
According to our phylogenetic analysis, DzARF2A was paired with ARF2 from tomato (SlARF2A and 2B) in subclade C3 (Figure 1A). Therefore, SlARF2A seemed to be DzARF2A’s closest tomato orthologue. Since down-regulation of SlARF2A resulted in ripening defects, decreased levels of climacteric ethylene production, and a decreased expression of the ethylene biosynthetic genes in tomato (Hao et al., 2015), it raised the possibility of a similar role by DzARF2A in the transcriptional regulation of ethylene biosynthetic genes during fruit ripening in durian.
To gain insights into the in planta function of DzARF2A during the post-harvest ripening of durian, we transiently expressed it in N. benthamiana leaves. We found a significantly higher expression levels of ethylene biosynthetic genes (NbACS and NbACO) in the leaves infiltrated with DzARF2A compared to the control at day 3 and 5 post-infiltration (Figure 5A). In addition, our in silico analyses of the 2-kb promoter regions located upstream of the translation start site of NbACS and NbACO confirmed the existence of cis-regulatory binding sites (TGTCTC) for ARFs (Supplementary Figure S3). Notably, the 2-kb upstream region of the transcription start site should cover the putative promoter region of a gene (Jo and Choi, 2019; Liu et al., 2020). We compared the expression levels of DzARF2A, NbACS, and NbACO in the DzARF2A-infiltrated leaves between day 3 and 5 and found a strong positive correlation between the expression levels of DzARF2A and ethylene biosynthetic genes (Figure 5A). Taken together, our results strengthen the possible ripening-associated role of DzARF2A during durian fruit ripening through the transcriptional regulation of ethylene biosynthetic genes. To further confirm this role, the transcriptional activity of DzARF2A was investigated using dual-luciferase reporter assay.
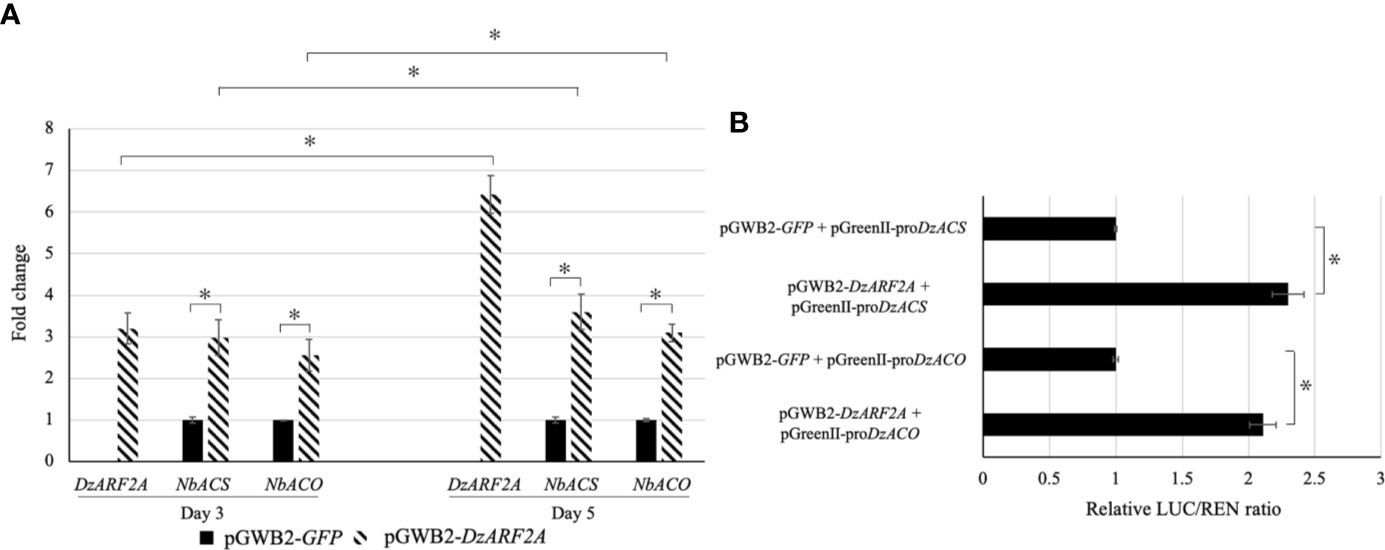
Figure 5 Fold changes in expression levels of DzARF2A and ethylene biosynthetic genes in Nicotiana benthamiana leaves transiently expressing DzARF2A and in vivo transcriptional activity of DzARF2A in N. benthamiana leaves. (A) The relative expression levels of DzARF2A and ethylene biosynthetic genes from N. benthamiana; ACC synthase (NbACS) and ACC oxidase (NbACO) were measured in N. benthamiana leaves infiltrated with pGWB2-DzARF2A (treatment) and pGWB2-GFP (control) at three and five days after infiltration by using the 2-ΔΔCt method. The levels were then normalized by the geometric mean of reference genes and the pGWB2-GFP as control. Three independent biological replicates were used. An asterisk (*) above the bars indicates a significant difference (P < 0.05). In the DzARF2A-infiltrated leaves, comparisons were also made between day 3 and day 5. An asterisk (*) above the bars indicates a significant difference between day 3 and day 5 (P < 0.05). (B) After three days of infiltration, firefly luciferase (LUC) and Renilla luciferase (REN) activities were measured and presented as relative LUC/REN ratio. The LUC/REN ratio in the control leaves co-infiltrated with the pGWB2-GFP and pGreenII-proDzACS or pGreenII-proDzACO was set as 1 and was used as a calibrator. Error bars represent means ± standard deviations (SD) among four independent biological replicates. An asterisk (*) above the bars indicates a significant difference compared to the control (Student’s t-test, P < 0.05).
We performed dual-luciferase assay to evaluate the transcriptional activity of DzARF2A in vivo. Our effector plasmid contained the DzARF2A under the control of a CaMV35S promoter (pGWB2-DzARF2A) whereas our double-reporter vector harbored an internal control REN driven by the CaMV35S promoter and LUC driven by the 2000-bp promoter region of ACS (pGreenII-proDzACS) or ACO (pGreenII-proDzACO) (Supplementary Figure S4). The relative LUC/REN ratios in the leaves co-expressing both effector (pGWB2-DzARF2A) and reporter (pGreenII-proDzACS or pGreenII-proDzACO) constructs were compared to the control leaves co-infiltrated with pGWB2-GFP and reporter (pGreenII-proDzACS or pGreenII-proDzACO) plasmids. As shown in Figure 5B, compared to the control, the co-expression of DzARF2A with each of the two promoters significantly increased the relative LUC/REN ratio, suggesting that DzARF2A trans-activates ethylene biosynthetic genes.
It was our great interest to investigate any possible correlation between the expression level of candidate ripening-associated DzARF2A during ripening and the ripening behaviors of different durian cultivars. Therefore, we analyzed the expression level of DzARF2A in four cultivars; two fast-post-harvest ripening (Chanee and Phuangmanee) and two slow-post-harvest ripening (Monthong and Kanyao) cultivars. We observed a significantly higher expression level of DzARF2A in the fast-ripening cultivars compared to the slow-ripening ones (Figure 6 and Supplementary Figure S5). This higher DzARF2A expression could therefore enhance the climacteric ethylene biosynthesis through the transcriptional regulation of ethylene biosynthetic genes and thus contribute to the faster ripening of Chanee and Phuangmanee, compared to the Monthong and Kanyao cultivars.
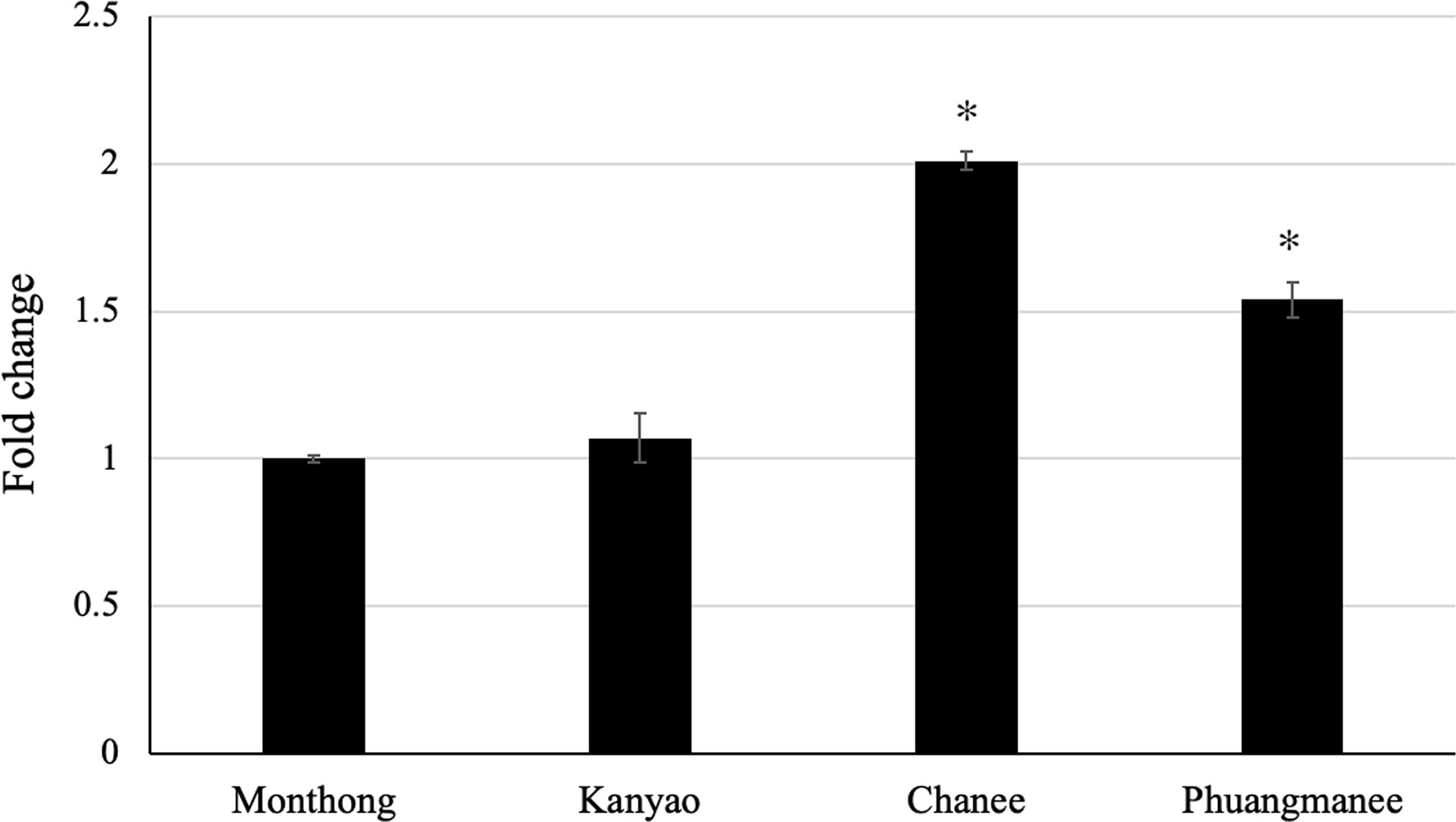
Figure 6 Fold changes in expression level of DzARF2A at ripe stage of different durian cultivars. The relative expression level of DzARF2A was analyzed in fruit pulps of two fast-ripening (Chanee and Phuangmanee) and two slow-ripening (Monthong and kanyao) cultivars at ripe stage by using the 2-ΔΔCt method and levels were normalized by the geometric mean of reference genes and the Monthong cultivar as control. Three independent biological replicates were used. An asterisk (*) above the bar indicates a significant difference (P < 0.05).
In our previous study, we observed an increasing level of auxin during fruit ripening in both Monthong and Chanee cultivars with a significantly higher auxin level in Chanee compared to Monthong (Khaksar et al., 2019). This observation highlighted the possible ripening-associated role of auxin during durian fruit ripening and prompted us to further investigate whether there is a positive correlation between auxin levels and DzARF2A expression. Accordingly, we aimed to investigate the auxin inducibility of DzARF2A expression. Indeed, the accumulation of DzARF2A transcript was induced by exogenous auxin in the leaves of Monthong cultivar. We observed a significantly higher transcript abundance of DzARF2A in the leaves with increasing concentration of auxin and expression was significantly higher than that of the control (mock treatment) (Figure 7). Among the three auxin treatments (10, 20, and 40 µMu), exogenous auxin treatment at 40 µM elicited the highest expression of DzARF2A (Figure 7). Notably, auxin-induced expression of DzARF2A also depended on induction time and the highest expression level occurred at 40 µM after 2 h auxin exposure (Figure 7). This finding confirmed the auxin responsiveness of DzARF2A in a dose- and time-dependent manner. Moreover, it strengthened the possibility that DzARF2A could mediate the post-harvest ripening of durian fruit in harmony with auxin.
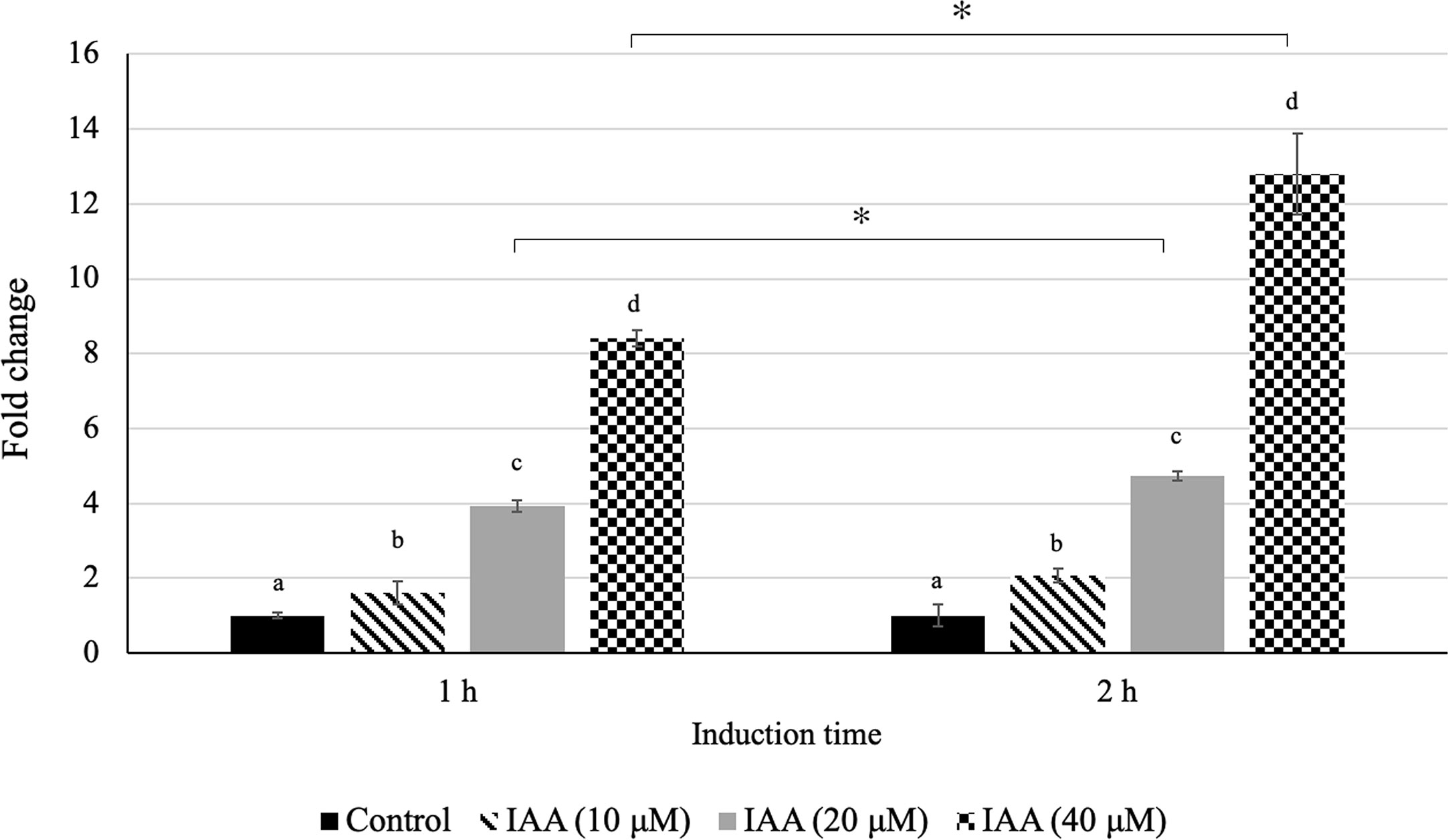
Figure 7 Auxin responsiveness of DzARF2A. Fold changes in expression level of DzARF2A in young leaves of Monthong cultivar treated with 0 (control), 10, 20, and 40 μM IAA for 1 and 2 h were calculated by using the 2-ΔΔCt method and levels were normalized by the geometric mean of reference genes and the control samples (0 μM IAA). Three independent biological replicates were used for each condition at each timepoint. Different letters indicate significant differences (P < 0.05). Comparisons were also made for each condition between the two timepoints; an asterisk (*) above the bars indicates a significant difference for that condition between 1 and 2 h (P < 0.05).
We found an increasing expression of DzARF2A in the leaves treated with different concentrations of exogenous auxin for 2 h. To further examine any correlation between the expression levels of DzARF2A and ethylene biosynthetic genes of durian, we profiled the expression levels of DzACS and DzACO in those leaves. The expression levels of DzACS and DzACO were positively correlated with increase in the expression level of DzARF2A (Supplementary Figure S6), consistent with our observation in N. benthamiana leaves (Figure 5A).
Fruit ripening is described as a well-orchestrated coordination of several steps, marked by major physiological and biochemical changes, and regulated by a complex hormonal network. Ethylene is regarded as the major regulator of climacteric fruit ripening. However, without minimizing the role of ethylene, the putative role of other phytohormones in regulating the climacteric ripening process remains elusive. Auxin, the most ubiquitous hormone in plants, is a crucial phytohormone involved in the regulation of various growth and developmental processes. In recent years, an increasing body of evidence has pointed towards a possible ripening-associated role of auxin in climacteric fruit ripening, with a number of emerging studies in peach, tomato, and durian. Auxin signaling is known to regulate the expression of early/primary auxin response genes through ARFs. Previous studies have already identified members of the ARF family in crops with economic values (Wang et al., 2007; Kumar et al., 2011; Xing et al., 2011; Luo et al., 2014; Wan et al., 2014; Shen et al., 2015; Hu et al., 2015; Liu et al., 2015; Singh et al., 2017; Tang et al., 2018). According to the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn), 4,578 members of the ARF gene family exist in the genomes of different plant species in varying numbers; papaya (10), tomato (22), Arabidopsis (37), banana (50), and durian (77). Even though genetic and molecular studies have begun documenting the important roles of ARF gene family members in several aspects of plant growth and development, such as AtARF1, AtARF2, AtARF7, and AtARF8 in Arabidopsis and SlARF3 of tomato, the actual knowledge on the role of ARFs in fruit ripening remains strikingly limited. Durian is a typical climacteric fruit with restricted shelf life after the initiation of ripening. This limits the handling and transportation of the fruit, which can cause significant economic losses to both farmers and consumers. Therefore, a deeper understanding of the molecular mechanisms behind ripening and the different ripening behaviors of durian cultivars (e.g., fast and slow post-harvest ripening) is critical to improve their post-harvest life. Our transcriptome-wide analysis of durian (Musang King cultivar) identified 15 non-redundant members of the ARF gene family. Out of these, our gene expression analysis identified 12 fruit pulp-expressed DzARFs during the post-harvest ripening of the Monthong cultivar, from which eight ARFs harbored ripening-associated expression patterns (Figure 3). The expression levels of these putative ripening-associated DzARFs were further examined under three different ripening conditions. The expression levels of DzARF2A and DzARF19 were induced under ethephon and suppressed under 1-MCP (Figure 4). The expression of these ethylene-induced DzARFs showed an increasing trend during the post-harvest ripening of durian and are speculated to act as transcriptional activators of ripening. Similar to our observation, Hao et al. (2015) identified a member of the ARF gene family in tomato (SlARF2A) which was ethylene-induced and acted as a transcriptional activator of ripening. In contrast, the expression levels of DzARF3 and DzARF6 were significantly suppressed under ethylene but induced under 1-MCP and showed a decreasing trend during ripening. According to our knowledge, this is the first report on the inducing effect of 1-MCP on the expression of an ARF gene. Since the inhibition of ethylene biosynthesis (Kieber et al., 1993; Tatsuki and Endo, 2006) and signaling (Hall et al., 2000) by 1-MCP is well documented, a plausible explanation for the upregulation of DzARF3 and DzARF6 by 1-MCP through the inhibition of ethylene biosynthesis and signaling is possible since the expression of these two genes was negatively regulated by ethylene. Thus, these two ethylene-repressed DzARFs possibly act as transcriptional repressors of ripening. A similar observation was made in our previous study, where we identified both ethylene-induced and ethylene-repressed Dof TFs in durian (Khaksar et al., 2019).
From the validated ripening-associated and ethylene-induced DzARFs, the dramatic increase in the expression level of DzARF2A under ethylene and its significantly higher expression level during fruit ripening, compared to that of DzARF19 (Figure 2), motivated us to characterize it further. Transient expression of DzARF2A in N. benthamiana leaves significantly upregulated the expression levels of the ethylene biosynthetic genes (NbACS and NbACO) (Figure 5A), confirming the transcriptional regulation of these genes by DzARF2A. This observation is in line with the results of previous studies on ARF2. Specifically, the expression of ACS genes was downregulated in the Arabidopsis arf2 mutant (Okushima et al., 2005) and Hao et al. (2015) showed that tomato fruits under-expressing SlARF2A/B produced less climacteric ethylene and exhibited a dramatic down-regulation of the ethylene biosynthetic genes. Our in silico analyses of the ACS and ACO promoter regions from durian and N. benthamiana revealed the existence of cis-regulatory binding sites (TGTCTC) that are needed to interact with ARFs (Supplementary Figures S7 and S3). Using dual-luciferase reporter assay, we then confirmed the transcriptional activation ability of DzARF2A (Figure 5B). Taken together, these observations strengthen the possible role of DzARF2A in the regulation of fruit ripening through the transcriptional regulation of ethylene biosynthetic genes in durian.
In our previous study, we profiled the expression levels of ethylene biosynthetic genes (DzACS and DzACO) during durian fruit ripening of Monthong and Chanee cultivars. Both genes were found to express increasingly over the course of ripening in both cultivars (Khaksar et al., 2019). Notably, the increasing expression levels of DzACS and DzACO were consistent with the expression level of DzARF2A during ripening. In addition, we observed significantly higher expression levels of DzACS and DzACO during post-harvest ripening in Chanee (a fast-ripening cultivar), compared to Monthong (a slow-ripening cultivar) (Khaksar et al., 2019). This observation prompted us to further examine any possible correlation between the expression levels of DzARF2A and ethylene biosynthetic genes during durian fruit ripening. We observed a strong positive correlation between the expression levels of DzARF2A and ethylene biosynthetic genes (Figure 5A and Supplementary Figure S6). Moreover, the expression level of DzARF2A was positively correlated with the faster post-harvest ripening of durian fruit (Figure 6). Thus, higher DzARF2A expression would upregulate climacteric ethylene biosynthesis and lead to a faster ripening of the fast-ripening cultivars (Chanee and Phuangmanee) compared to those of the slow-ripening ones (Monthong and Kanyao). To the best of our knowledge, this is the first report on the role of a ripening-associated TF that positively regulates the post-harvest ripening of a climacteric fruit in a cultivar-dependent manner; in other words, the higher the DzARF2A expression (in fast-ripening cultivars), the faster the ripening. It has been postulated that the regulatory network mediating fast- and slow-ripening cultivars would involve not only DzARF2A and ethylene as master regulators, but also other TFs and hormones. We observed an exogenous auxin-induced expression pattern of DzARF2A in a concentration-dependent manner (Figure 7), which was similar to that of SlARF2A in tomato (Breitel et al., 2016) and CpARF2 of papaya (Liu et al., 2015), suggesting that ARF2 has an evolutionarily conserved function in regulating fruit ripening. Thus, the auxin responsiveness of ARF2 in these climacteric fruits strengthened the active involvement of auxin during climacteric fruit ripening.
In our previous study, we observed an increasing level of auxin during ripening in both Monthong and Chanee cultivars and a significantly higher auxin level during post-harvest ripening in Chanee over Monthong. This phenomenon coincided with the significantly higher expression levels of auxin biosynthetic genes in Chanee compared to those of Monthong (Khaksar et al., 2019). In addition, we identified the cultivar-dependent DzDof2.2, a member of the Dof TF family, which was expressed at a significantly higher level during ripening in Chanee compared to Monthong. Functional characterization of DzDof2.2 suggested its role in controlling fruit ripening through the transcriptional regulation of the auxin biosynthetic genes (Khaksar et al., 2019). Based on these previous findings and the results of our current study, DzARF2A might act jointly with DzDof2.2 in a regulatory network to control post-harvest ripening in durian. In the fast-ripening cultivars, higher DzDof2.2 expression would enhance auxin biosynthesis. Higher auxin level would consequently upregulate the expression of DzARF2A and activate the ARF2A-mediated transcription of ethylene biosynthetic genes, leading to a higher climacteric ethylene biosynthesis (auxin-ethylene crosstalk) and faster ripening.
The present study documents for the first time a regulatory network that controls fast and slow post-harvest ripening in different cultivars of a climacteric fruit, in which DzARF2A interconnects signals from ethylene and auxin to fine-tune and to coordinate the initiation of ripening. The relationship between ethylene and auxin in plant growth, development, and fruit senescence has been previously documented (Iqbal et al., 2017). In addition, Bleecker and Kende (2000) pointed out that auxin can stimulate climacteric ethylene biosynthesis through its inductive action on the expression of ethylene biosynthetic genes. Besides the already known role of auxin to induce climacteric ethylene synthesis and thereby to influence ripening, the assumption that auxin would play a direct role during climacteric ripening, even though previously formulated, due to the lack of clear experimental evidence is still somewhat uncertain. A study by El-Sharkawy et al. (2016) suggested a possibly direct role of auxin in plum fruit ripening through the upregulation of several genes encoding cell-wall metabolism-related proteins. However, the intriguing question about which ripening-associated genes could be auxin responsive remains open and needs further investigation.
Often, different TFs control the expression of a particular gene and the fine-tuning of these different TFs occurs through the formation of enhanceosome or repressosome complexes that affect protein–protein and protein–DNA interactions (Martinez and Rao, 2012). Various ripening-associated TFs have been investigated. However, only a few studies have indicated their involvement in fruit ripening, such as the studies in tomato (Shima et al., 2013; Fujisawa et al., 2014) and in banana (Feng et al., 2016). To the best of our knowledge, there has only been one study that reported a possible ARF interactor in the regulation of ripening. Breitel et al. (2016) identified the ASR1 (ABA STRESS RIPENING-INDUCED 1) protein as a putative ARF2A interactor in tomato fruit. They suggested the possibility that these two proteins could be coordinating the interaction between ethylene and ABA during tomato ripening. The possible interaction between DzARF2A and other TFs, such as DzDof2.2 in controlling fruit ripening in durian could be the subject of a further investigation.
In summary, out of the 15 identified DzARFs, 12 were expressed in the fruit pulp. Gene expression analysis revealed differential expression of the fruit pulp-expressed DzARFs during post-harvest ripening in the Monthong cultivar. A total of eight DzARFs harbored a ripening-associated expression pattern, suggesting a potential role in fruit ripening. Among those, the appealing ripening-associated expression pattern of DzARF2A prompted its further functional characterization and confirmed its role in ripening through binding to the promoters of ethylene biosynthetic genes and transcriptionally regulating their expression levels. We observed a positive correlation between the expression level of DzARF2A and ripening speed in the fast-ripening cultivars (Chanee and Phuangmanee). Taken together, our results propose DzARF2A as a component of the regulatory network underlying fruit ripening in durian. Thus, the present study on durian suggests another layer in the complex regulatory networks behind fruit ripening and it strengthens the concept that the ripening process relies on the interplay between different transcription factors and hormones. Further functional analysis of DzARF2A in fruits would provide more insights into its ripening-associated role during durian fruit ripening.
The datasets presented in this study can be found in online repositories. Illumina sequencing reads from RNA-Seq study of durian (Musang King cultivar) were retrieved from a public repository database (SRA, Sequence Read Archive) with the following accession numbers: SRX3188225 (root tissue), SRX3188222 (stem tissue), SRX3188226 (leaf tissue), and SRX3188223 (aril/pulp tissue).
SS conceived the research. GK performed the experiments. GK and SS designed the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research is supported by the Thailand Research Fund (RSA6080021) and Chulalongkorn University (GRU 6203023003-1) (to SS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research is supported by Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University (to GK). We sincerely thank Tsuyoshi Nakagawa (Shimane University, Japan) for providing the Gateway® expression vector pGWB2, Sophien Kamoun (Sainsbury Laboratory, UK) for providing A. tumefaciens GV3101 carrying pJL3:p19, Roger P Hellens (HortResearch, Mt Albert Research Centre, New Zealand) for providing pGreenII 0800-LUC double-reporter vector, Lalida Sangpong for assisting in plant material collection, and Pinnapat Pinsorn and Kittiya Tantisuwanichkul for assisting in genomic DNA extraction.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.543747/full#supplementary-material
Abdel-Latif, A., Osman, G. (2017). Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 13, 1.
Adams-Phillips, L., Barry, C., Giovannoni, J. (2004). Signal transduction systems regulating fruit ripening. Trends Plant Sci. 9, 331–338.
Alexander, L., Grierson, D. (2002). Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055.
Ayub, R., Guis, M., Ben Amor, M., Gillot, L., Roustan, J. P., Latché, A., et al. (1996). Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nat. Bitechnol. 14, 862–866.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). “MEME SUITE: tools for motif discovery and searching”. Nucleic Acids Res. 37, 202–208.
Barry, C. S., Llop-Tous, M., II, Grierson, D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986.
Bleecker, A. B., Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Ann. Rev. Cell Dev. Biol. 16, 1–18.
Breitel, D. A., Chappell-Maor, L., Meir, S., Panizel, I., Puig, C. P., Hao, Y., et al. (2016). AUXIN RESPONSE FACTOR 2 intersects hormonal signals in the regulation of tomato fruit ripening. PloS Genet. 12, e1005903.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622.
Chong, J., Soufan, O., Li, C., Caraus, I., Li, S., Bourque, G., et al. (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, 486–494.
Diao, D., Hu, X., Guan, D., Wang, W., Yang, H., Liu, Y. (2020). Genome-wide identification of the ARF (auxin response factor) gene family in peach and their expression analysis. Mol. Biol. Rep. 47, 4331–4344.
Ellis, C. M., Nagpal, P., Young, J. C., Hagen, G., Guilfoyle, T. J., Reed, J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574.
El-Sharkawy, I., Sherif, S., Qubbaj, T., Sullivan, A. J., Jayasankar, S. (2016). Stimulated auxin levels enhance plum fruit ripening, but limit shelf-life characteristics. Postharvest Biol. Tec. 112, 215–223.
Feng, B. H., Han, Y. C., Xiao, Y. Y., Kuang, J. F., Fan, Z. Q., Chen, J. Y., et al. (2016). The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 67, 2263–2275.
Fujisawa, M., Shima, Y., Nakagawa, H., Kitagawa, M., Kimbara, J., Nakano, T., et al. (2014). Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell. 26, 89–101.
Gillaspy, G., Ben-David, H., Gruissem, W. (1993). Fruits: a develop- mental perspective. Plant Cell. 5, 1439–1451.
Goetz, M., Vivian-Smith, A., Johnson, S. D., Koltunow, A. M. (2006). AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell. 18, 1873–1886.
Guilfoyle, T. J., Hagen, G. (2012). Getting a grasp on domain III/IV responsible for auxin response factor–IAA protein interactions. Plant Sci. 190, 82–88.
Guilfoyle, T. J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33–43.
Hagen, G., Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385.
Hall, A. E., Findell, J. L., Schaller, G. E., Sisler, E. C., Bleecker, A. B. (2000). Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 123, 1449–1457.
Hao, Y., Hu, G., Breitel, D., Liu, M., Mila, I., Frasse, P., et al. (2015). Auxin Response Factor SlARF2 Is an Essential Component of the Regulatory Mechanism Controlling Fruit Ripening in Tomato. PloS Genet. 11, e1005649.
Harper, R. M., Stowe-Evans, E. L., Luesse, D. R., Muto, H., Tatematsu, K., Watahiki, M. K., et al. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 12, 757–770.
Hellens, R., Allan, A., Friel, E., Bolitho, K., Grafton, K., Templeton, M., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13.
Hu, W., Zuo, J., Hou, X., Yan, Y., Wei, Y., Liu, J., et al. (2015). The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 6, 742.
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., Khan, M., II (2017). Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 8, 475.
Jo, S. S., Choi, S. S. (2019). Enrichment of rare alleles within epigenetic chromatin marks in the first intron. Genomics Inform. 17, e9.
Jones, B., Frasse, P., Olmos, E., Zegzouti, H., Li, Z. G., Latché, A., et al. (2002). Down-regulation of DR12, an auxin- response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 32, 603–613.
Khaksar, G., Sangchay, W., Pinsorn, P., Sangpong, L., Sirikantaramas, S. (2019). Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent Dof transcription factors. Sci. Rep. 9, 12109.
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., Ecker, J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441.
Kumar, R., Tyagi, A. K., Sharma, A. K. (2011). Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Gen. Genet. 285, 245–260.
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Lelievre, J.-M., Latche, A., Jones, B., Bouzayen, M., Pech, J.-C. (1997). Ethylene and fruit ripening. Physiol. Plant 101, 727–739.
Liu, K., Yuan, C., Li, H., Lin, W., Yang, Y., Shen, C., et al. (2015). Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genomics 16, 901.
Liu, K., Yuan, C., Feng, S., Zhong, S., Li, H., Zhong, J., et al. (2017). Genome-wide analysis and characterization of Aux/IAA family genes related to fruit ripening in papaya (Carica papaya L.). BMC Genomics 18, 351.
Liu, P., Wang, S., Wang, X., Yang, X., Li, Q., Wang, C., et al. (2020). Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.). Plant Physiol. Biochem 151, 197–213.
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 4, 402–408.
Luo, X. C., Sun, M. H., Rui, X. U., Shu, H. R., Wang, J. W., Zhang, S. Z. (2014). Genome wide identification and expression analysis of the ARF gene family in apple. J. Genet. 93, 785–797.
Martinez, G. J., Rao, A. (2012). Immunology. Cooperative transcription factor complexes in control. Science 338, 891–892.
Miller, A. N., Walsh, C. S., Cohen, J. D. (1987). Measurement of indole- 3-acetic acid in peach fruits (Prunus persica L. Batsch cv. Redhaven) during development. Plant Physiol. 84, 491–494.
Nanao, M. H., Vinos-Poyo, T., Brunoud, G., Thévenon, E., Mazzoleni, M., Mast, D., et al. (2014). Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 5, 3617.
Niu, J., Bi, Q., Deng, S., Chen, H., Yu, H., Wang, L., et al. (2018). Identification of AUXIN RESPONSE FACTOR gene family from Prunus sibirica and its expression analysis during mesocarp and kernel development. BMC Plant Biol. 18, 21.
Okushima, Y., Mitina, I., Quach, H., Theologis, A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46.
Pinsorn, P., Oikawa, A., Watanabe, M., Sasaki, R., Ngamchuachit, P., Hoefgen, R., et al. (2018). Metabolic variation in the pulps of two durian cultivars: Unraveling the metabolites that contribute to the flavor. Food Chem. 268, 118–125.
Qi, Y., Wang, S., Shen, C., Zhang, S., Chen, Y., Xu, Y., et al. (2013). OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 193, 109–120.
Shen, C., Wang, S., Bai, Y., Wu, Y., Zhang, S., Chen, M., et al. (2010). Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J. Exp. Bot. 61, 3971–3981.
Shen, C., Wang, S., Zhang, S., Xu, Y., Qian, Q., Qi, Y., et al. (2013). OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 36, 607–620.
Shen, C., Yue, R., Sun, T., Zhang, L., Xu, L., Tie, S., et al. (2015). Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front. Plant Sci. 6, 73.
Shima, Y., Kitagawa, M., Fujisawa, M., Nakano, T., Kato, H., Kimbara, J., et al. (2013). Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 82, 427–438.
Singh, V. K., Rajkumar, M. S., Garg, R., Jain, M. (2017). Genome-wide identification and co-expression network analysis provide insights into the roles of auxin response factor gene family in chickpea. Sci. Rep. 7, 10895.
Steibel, J. P., Poletto, R., Coussens, P. M., Rosa, G. J. M. (2009). A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 94, 146–152.
Tang, Y., Bao, X., Liu, K., Wang, J., Zhang, J., Feng, Y., et al. (2018). Genome-wide identification and expression profiling of the auxin response factor (ARF) gene family in physic nut. PloS One 13, e0201024.
Tatsuki, M., Endo, A. (2006). Analyses of expression patterns of ethylene receptor genes in apple (Malus domestica Borkh.) fruits treated with or without 1-methylcyclopropene (1-MCP). J. Japan Soc Hortic. Sci. 75, 481–487.
Ulmasov, T., Hagen, G., Guilfoyle, T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT- PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034.
Wan, S., Li, W., Zhu, Y., Liu, Z., Huang, W., Zhan, J. (2014). Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 33, 1365–1375.
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24.
Wang, A., Yamakake, J., Kudo, H., Wakasa, Y., Hatsuyama, Y., Igarashi, M., et al. (2009). Null mutation of the MdACS3 gene, coding for a ripening-specific 1-aminocyclopropane-1-carboxylate synthase, leads to long shelf life in apple fruit. Plant Physiol. 151, 391–399.
Woodward, A. W., Bartel, B. (2005). Auxin, regulation, action, and interaction. Ann. Bot. 95, 707–735.
Xie, Y. H., Zhu, B. Z., Yang, X. L., Zhang, H. X., Fu, D. Q., Zhu, H. L., et al. (2006). Delay of post-harvest ripening and senescence of tomato fruit through virus-induced LeACS2 gene silencing. Postharvest Biol. Technol. 42, 8–15.
Xing, H., Pudake, R. N., Guo, G., Xing, G., Hu, Z., Zhang, Y., et al. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12, 178.
Yu, H., Soler, M., Mila, I., San Clemente, H., Savelli, B., Dunand, C., et al. (2014). Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PloS One 9, e108906.
Keywords: auxin, auxin response factor, durian, ethylene, fruit ripening, transcriptional regulation
Citation: Khaksar G and Sirikantaramas S (2020) Auxin Response Factor 2A Is Part of the Regulatory Network Mediating Fruit Ripening Through Auxin-Ethylene Crosstalk in Durian. Front. Plant Sci. 11:543747. doi: 10.3389/fpls.2020.543747
Received: 18 March 2020; Accepted: 24 August 2020;
Published: 09 September 2020.
Edited by:
Carlos R. Figueroa, University of Talca, ChileReviewed by:
Jian-ye Chen, South China Agricultural University, ChinaCopyright © 2020 Khaksar and Sirikantaramas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Supaart Sirikantaramas, c3VwYWFydC5zQGNodWxhLmFjLnRo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.