- 1Key Laboratory of Saline-alkali Vegetation Ecology Restoration, Ministry of Education, College of Life Science, Northeast Forestry University, Harbin, China
- 2College of Life Science, Northeast Forestry University, Harbin, China
- 3Institute for Sustainable Agroecosystem Services, Graduate School of Agriculture and Life Science, The University of Tokyo, Tokyo, Japan
In the aerial plant organs, cuticular wax forms a hydrophobic layer that can protect cells from dehydration, repel pathogen attacks, and prevent organ fusion during development. The MIXTA gene encodes an MYB-like transcription factor, which is associated with epicuticular wax biosynthesis to increase the wax load on the surface of leaves. In this study, the AmMIXTA-homologous gene EgMIXTA1 was functionally characterized in the Eustoma grandiflorum. EgMIXTA1 was ubiquitously, but highly, expressed in leaves and buds. We identified the Eustoma MIXTA homolog and developed the plants for overexpression. EgMIXTA1-overexpressing plants had more wax crystal deposition on the leaf surface compared to wild-type and considerably more overall cuticular wax. In the leaves of the overexpression line, the cuticular transpiration occurred more slowly than in those of non-transgenic plants. Analysis of gene expression indicated that several genes, such as EgCER3, EgCER6, EgCER10, EgKCS1, EgKCR1, and EgCYP77A6, which are known to be involved in wax biosynthesis, were induced by EgMIXTA1-overexpression lines. Expression of another gene, WAX INDUCER1/SHINE1, encoding a transcription factor that stimulates the production of cutin, was also significantly higher in the overexpressors than in wild-type. However, the expression of a lipid-related gene, EgABCG12, did not change relative to the wild-type. These results suggest that EgMIXTA1 is involved in the biosynthesis of cuticular waxes.
Introduction
Eustoma grandiflorum (Raf.) Shinn. is a perennial, herbaceous, ornamental plant, which originated from the southern part of North America (Wang et al., 2011). E. grandiflorum, one of the most common potted plants, has very high economic and ornamental value due to its large, attractive, long-stalked flowers, long-lasting in vase flowers (Ecker et al., 1994; Zaccai and Edri, 2002; Uddin et al., 2004; Lena et al., 2009). E. grandiflorum, used to produce cut flowers, is vulnerable to plant diseases, insect pest attacks, and require large amounts of water. Hence, considerable efforts were dedicated to screening for drought tolerance, disease, and insect pest resistance.
Cuticular waxes are composed of long chain fatty acids (VLCFAs) and their derivatives, including aldehydes, alkanes, esters, and primary and secondary alcohols (Kunst and Samuels, 2009; Bernard and Joubes, 2013). The cuticular waxes form the outermost barrier to non-stomatic loss of water and play a significant role in responding to environmental stress. Composition, secretion, and synthesis of wax and cutin are modulated during cell expansion (Suh et al., 2005) and vary in different tissues (Yonghua et al., 2007, 2009). Many genes involved in cuticular wax biosynthesis and transportation have been characterized by forward and reverse genetic approaches for understanding the metabolism of plant wax (Hooker et al., 2002; Aharoni et al., 2004; Kim et al., 2013). Some transcription factors (TFs) were also shown to regulate the biosynthesis of cuticle. For example, Arabidopsis thaliana WAX INDUCER1/SHINE1 (WIN1/SHN1), which belongs to the TF family of AP2/EREBP type, activates the genes of cuticular wax and cutin biosynthesis (Kannangara et al., 2007). DECREASE WAX BIOSYNTHESIS (DEWAX), another AP2 TF, represses the expression of cutic wax biosynthesis genes to negatively regulate wax production in Arabidopsis (Go et al., 2014).
The MIXTA gene, first identified in Antirrhinum majus, encodes a TF an R2R3 MYB, which is a key regulator for the differentiation of epidermal cells into conical cells (Noda et al., 1994). MIXTA and MIXTA-like TFs have been demonstrated to be critical regulators for epidermal cell differentiation across multiple species of plants (Brockington et al., 2013). The MIXTA/MIXTA-like TFs were documented to function as positive regulators for the formation of conical epidermal cells (Noda et al., 1994; Stilio et al., 2010), production of cotton fibers (Walford et al., 2011), trichomes (Gilding and Marks, 2010; Plett et al., 2010), and cuticle development (Oshima et al., 2013; Lashbrooke et al., 2015). The Arabidopsis MIXTA-like orthologs AtMYB16 and AtMYB106 have been shown to control the formation of cuticles, MYB106, and MYB16; loss- and gain-of-function research has shown that they control the expression of the genes of cutin biosynthesis (Oshima et al., 2013). AtMYB96 regulates cutin biosynthesis by directly binding to the promoters of genes involved in cuticular wax biosynthesis (Seo et al., 2011). Solanum lycopersicum MIXTA-like (SlMIXTA-like) positively regulates both cuticle and conical epidermal cell formation, and links cutin polymer formation, cuticle assembly, and epidermal cell patterning in the fruit (Lashbrooke et al., 2015). The Artemisia annua MIXTA-like protein AaMIXTA1 is a positive regulator of trichome initiation and cuticle development; it forms a regulatory complex leading to increased transcription of AaHD1 and cuticle development genes (Yan et al., 2018). These studies indicate the involvement of MIXTA-like MYBs in cuticle development.
In this study, we functionally characterize the E. grandiflorum MIXTA-like 1 (EgMIXTA1) gene by investigating the transgenic lines that overexpress EgMIXTA1. Our result indicated that EgMIXTA1 participates in the regulation of cuticular wax biosynthesis genes in E. grandiflorum. Moreover, we suggest an essential role for an EgMIXTA-like gene and investigate the regulatory relationships between EgMIXTA1 and other genes involved in the biosynthesis of cuticular wax.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of E. grandiflorum cv. No. 2003-2-2, were used to generate the MIXTA1 overexpression lines (OX-1, OX-2, and OX-3) The seeds were rinsed with sterilized water for 10 min, rinsed twice with 70% alcohol for 1 min each, surface sterilized with 3% (v/v) sodium hypochlorite for 10 min, and rinsed with sterilized water five times for 5 min each before sowing on 1.5% agar plates containing Murashige and Skoog (MS) medium. After sowing, the plates were kept at 4°C for 2 days and then incubated under aseptic conditions at 24°C with a photoperiod of 16 h white light (2,000 l×) In order to examine tissue-specific expression patterns, seeds were plated on MS medium with 1.5% sucrose and stratified in darkness for 2 days at 4°C before being transferred to the growth chamber (16 h of light and 8 h of darkness, 22°C). Roots, stems, leaves, petals, and buds were collected and stored at −80°C for further processing.
Cloning of the Full-Length MIXTA1 Gene
Total RNA was extracted from leaves of the aseptic seedlings using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), then treated with DNase I (TaKaRa, Otsu, Shiga, Japan) and re-extracted with TRIzol reagent. Reverse transcription PCR (RT-PCR) was performed using the SuperScript III Reverse Transcriptase (TaKaRa, Otsu, Shiga, Japan). PCR products were purified using the High Pure PCR Product Purification Kit (Invitrogen). A degenerate primer set was designed from protein domains of MIXTA conserved across proteins from related species, namely, A. majus (AY821655, AJ006292, X79108, AY661654), Malus sieversii (DQ074464), Gossypium hirsutum (AF336283), and Petunia hybrida (Z13996). Partial fragments of EgMIXTA1 were amplified from the cDNA library by PCR. Then 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE were performed with gene- and vector-specific primers [MIXTA1F/R, Primer5′ 1st (GSP1) Primer5′ 2nd (GSP2), Primer3′ 1st (GSP1), Primer3′ 2nd (GSP2), Adaptor, Anchor, Primer LF (5′ GSP), Primer LR (3′ GSP), MIXTA1-F/R] (Supplementary Table 1).
Generation of Transgenic Plants
The coding sequence of MIXTA1 from E. grandiflorum cv. No. 2003-2-2 cDNA was amplified using a standard RT-PCR protocol. The full-length coding region of MIXTA1 was cloned into the binary vector pH7WG2D to generate an MIXTA1 overexpression vector in which MIXTA1 expression was driven by the CaMV35S promoter. The constructs were introduced into Agrobacterium tumefaciens strain LBA4404. Seedlings of E. grandiflorum (wild-type) were transformed using the leaf disc transformation method and screened on 0.8% agar plates containing diluted (50% v/v) MS medium and 20 mg/L kanamycin sulfate. Transgenic lines were selected based on kanamycin resistance.
Real-Time Fluorescence Quantitative PCR Analysis
Total RNA was extracted from E. grandiflorum plants using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). Two micrograms of total RNA pooled from three replicate extractions was used for reverse transcription with the PrimeScript First Strand cDNA Synthesis kit (TaKaRa, Tokyo, Japan). The cDNA was then diluted 20-fold, and expression was quantified using the Power SYBR Green PCR Master Mix with the 7500 Real-Time PCR System according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA, United States). Amplification was done using the gene-specific primers listed in Supplementary Table 1. The quantitative RT-PCR (qRT-PCR) conditions used were 30 s at 95°C, followed by 40 cycles of 5 s at 95°C, 34 s at 60°C, and 15 s at 95°C. The relative quantification method (2–ΔΔCT) was used to evaluate quantitative variation between replicates. Data were normalized against the EgActin gene. The reactions were performed in biological triplicate using RNA samples extracted from three independent plant samples.
Scanning Electron Microscopy
To observe the cuticular waxes in the transgenic aseptic seedlings, images were captured with a QUANTA 200 scanning electron microscopy (FEI Co., Hillsboro, OR, United States). When the seedlings were at least 5 weeks old, the third true leaves from each transgenic line were analyzed for cuticular wax. Before the observations, the leaves were cut into small pieces (1 cm × 1 cm) and fixed to sample holders, then dehydrated to make the surface of the leaves clean and water free. The samples were sputter-coated with gold particles using an SCD 005 Sputter Coater (Leica Co., Solms, Germany) and examined by scanning electron microscopy at an accelerating voltage of 10 kV.
Immunostaining Analysis
Immunostaining was performed as previously described (Xuhong et al., 2007). Leaves were ground to a fine powder and fixed in ice-cold fixation buffer (4% formaldehyde, 10 mM Tris–HCl, pH 7.5, 10 mM EDTA, and 100 mM NaCl). Then, the sample was suspended in sorting buffer (100 mM Tris–HCl, pH 7.5, 50 mM KCl, 2 mM MgCl, 0.05% Tween 20, and 5% sucrose). Nuclei were sorted using a Falcon® 40 μm Cell Strainer. Nuclei were incubated with antiEgMIXTA1 antibody, washed (10 mM sodium phosphate, pH 7.0, and 143 mM NaCl), incubated with Rhodamine Red-x-conjugated donkey anti-rabbit IgG (1/500 dilution; Alexa Fluor), and washed thoroughly. Images were captured and analyzed using Zeiss Axioskop 2 Plus, and processed using Adobe Photoshop software.
Water Loss Assay
The rate of water loss was estimated using leaves from well-watered 5-week-old plants. The plants were acclimated in the dark for 6 h, then soaked in water for 1 h. Three of the third true leaves were then dried and weighed gravimetrically using a microbalance at the time points indicated.
Results
Identification and Sequence Analysis of the EgMIXTA1 Gene
The experiments were performed to find the TFs that regulate the synthesis of cuticular waxes and the differentiation of epidermal cells in E. grandiflorum. We looked for the MIXTA-like gene encoding sequences in A. majus (AY821655, AJ006292, X79108, AY661654), M. sieversii (DQ074464), G. hirsutum (AF336283), and P. hybrida (Z13996) in GeneBank. Based on the conserved domains and RACE technology, we identified the E. grandiflorum homolog of the MIXTA-like gene and named it EgMIXTA1. EgMIXTA1 has an ORF of 1,128 nucleotides, encoding a polypeptide of 375 amino acids.
Furthermore, the molecular phylogenetic analysis of EgMIXTA1 and Arabidopsis R2R3domain proteins showed that EgMIXTA1 is also closely related to AtMYB16 and AtMYB106 (Figure 1A). In previous phylogenetic analyses, it was found that AtMYB16 and AtMYB106 belong to the R2R3 subgroup 9 clade (SBG9); SBG9 members have a well-established relationship with trichome and papillate cell formation (Brockington et al., 2013). Compared with other MIXTA-like genes, EgMIXTA1 and S. lycopersicum MIXTA-like are more closely related; EgMIXTA1 shares 59% amino acid identity with S. lycopersicum MIXTA-like (Weng et al., 2011; Ying et al., 2019; Figure 1B). We were interested in whether EgMIXTA1 has same role in the differentiation of epidermal cells or some other function. Consequently, we examined the function of EgMIXTA1 in E. grandiflorum in greater detail.
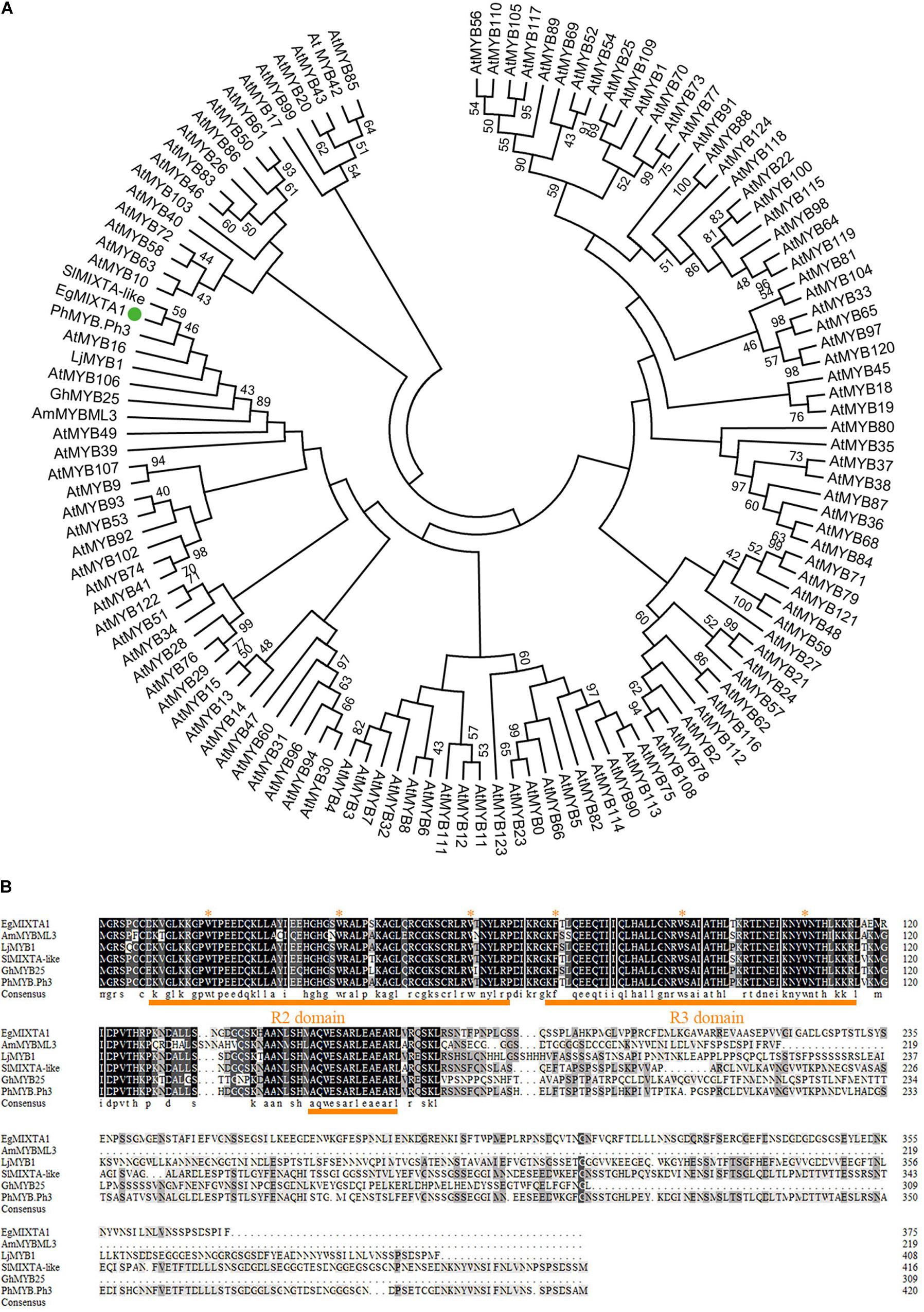
Figure 1. Sequence analysis of EgMIXTA1 homologs. (A) Phylogenetic analysis of EgMIXTA1, Arabidopsis R2R3 domain TFs, and other species of MIXTA-like proteins. ClustalW and MEGA7 were used to align the proteins and compute the neighbor-joining tree with significance percentages (bootstrap values = 1,000). (B) Amino acid alignment showing the conserved R2R3 MYB domain and subgroup 9 motifs underlined in orange. The highly conserved amino acid residues are marked with red stars.
Expression Patterns of EgMIXTA1 in E. grandiflorum
In order to investigate the spatial expression pattern of EgMIXTA1, the levels of EgMIXTA1 mRNA in different tissues were analyzed. In our study, qRT-PCR revealed that EgMIXTA1 was ubiquitously expressed in roots, leaves, petals, stems, and flower buds (Figure 2A).
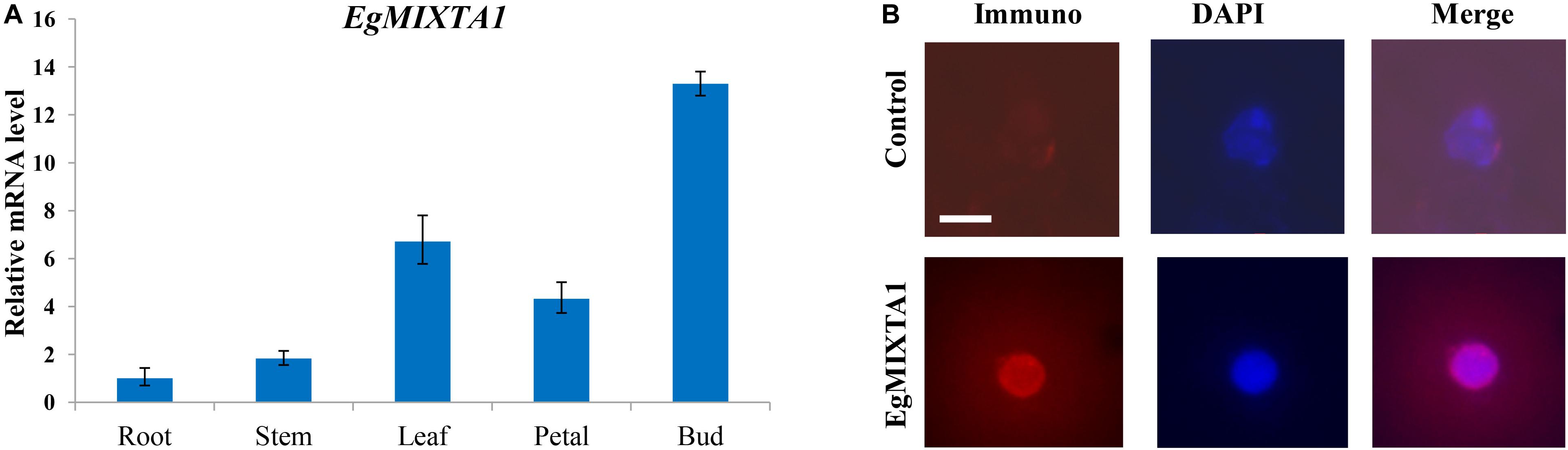
Figure 2. Gene expression pattern of EgMIXTA1 and nuclear localization of EgMIXTA1 (A). The expression levels of EgMIXTA1 in roots, stems, leaves, buds, and petals of E. grandiflorum were measured by quantitative RT-PCR (qRT-PCR). Actin was used as an internal reference. Data are presented as mean ± SD (n = 3). (B) Nuclei isolated from the leaves of 5-week seedlings of E Eustoma grandiflorum were probed with anti-EgMIXTA1 or a solution lacking anti-EgMIXTA1 (control), and visualized by staining with DAPI (blue) and Rhodamine Red-x-conjugated donkey-anti-rabbit IgG. Scale bar, 5 μm.
Nuclear localization is necessary for TFs to execute their functions, so we used nuclear immunostaining to determine whether EgMIXTA1 accumulates in the nuclei of E. grandiflorum leaf tissues. Nuclei isolated from E. grandiflorum leaves were stained with DAPI and anti-EgMIXTA1 or a control solution lacking antibodies, followed by incubation in Rhodamine Red-x-conjugated secondary antibody. The Rhodamine Red-x and DAPI signals overlapped in the nuclei stained with anti-EgMIXTA1, indicating that EgMIXTA1 is localized in the nucleus (Figure 2B).
Overexpression of EgMIXTA1 Increases Cuticular Wax Accumulation
To further dissect the biological function of EgMIXTA1, transgenic E. grandiflorum plants overexpressing EgMIXTA1 under the control of the 35S promoter were generated (Figure 3A). Three independent transgenic plants were obtained, and significantly increased levels of EgMIXTA1 gene expression were detected in independent transgenic lines by qRT-PCR (Figure 3B). Scanning electron microscopy analysis showed obvious deposition of epicuticular wax crystals on the leaves of EgMIXTA1 overexpressing lines than wild-type (Figure 3C).
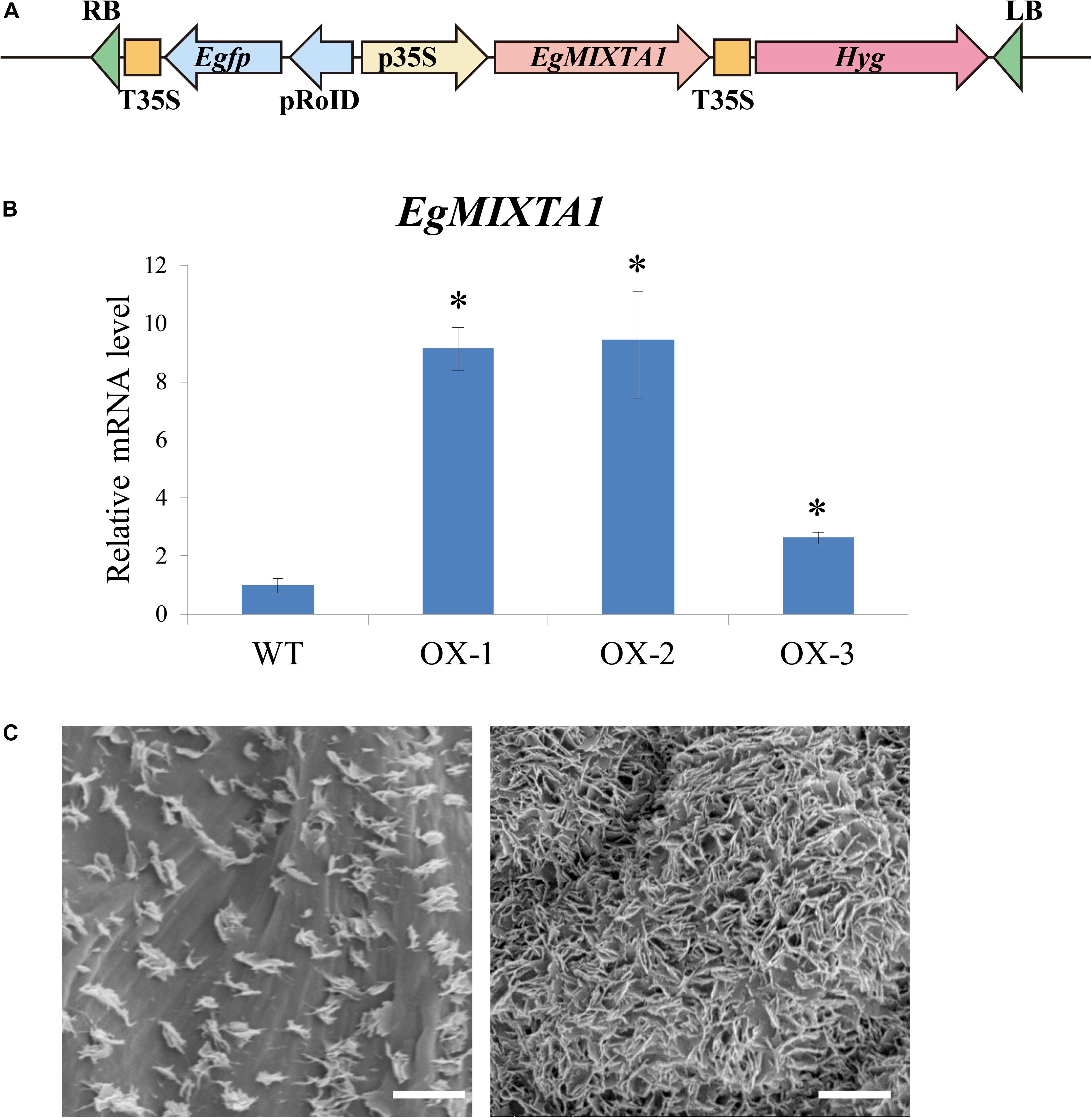
Figure 3. Cuticular wax accumulation in E. grandiflorum leaves with altered EgMIXTA1 expression. (A) Schematic diagram of EgMIXTA1 in the pH7WG2D vector under the control of the CaMV 35S promoter. p35S, CaMV 35S promoter; T-35S, CaMV 35S terminator; pRoID, rolD promoter; Egfp, enhanced green-fluorescent protein; Hyg, hygromycin-resistant gene; RB, T-DNA right border sequence; LB, T-DNA left border sequence. (B) Expression levels of EgMIXTA1 in EgMIXTA1-overexpressing plants were measured by qRT-PCR. Total RNA was isolated from leaves of WT and MIXTA1 overexpression lines (OX-1, OX-2, and OX-3), and subjected to qRT-PCR analysis. Actin was used as an internal reference. Student’s t-test: ∗P < 0.01. (C) Scanning electron microscopy images of epicuticular wax crystals on the leaves of 3-week-old wild-type (left) and MIXTA1 overexpression lines (right). Scale bar, 5 μm.
These results indicated that high amounts of cuticular waxes accumulated in the leaves of transgenic E. grandiflorum plants overexpressing MIXTA1.
Expression of Wax Synthesis-Related Genes Is Altered in Leaves of Transgenic E. grandiflorum Plants Overexpressing EgMIXTA1
Because epicuticular wax deposition was significantly increased in EgMIXTA1-overexpressing plants (Figure 3C), we asked whether the expression of wax biosynthetic genes was altered in these plants. The biosynthesis of cuticular wax is a complex process, and a number of enzymes related to cuticular wax biosynthesis and transport are involved in wax biosynthesis. C16 and C18 fatty acids are synthesized in the plastids then exported to the cytoplasm, where they are further elongated to VLCFAs through sequential addition of two-carbon units in a reaction catalyzed by fatty acid elongase complexes at the endoplasmic reticulum (Lee and Suh, 2013; Fich et al., 2016; Xue et al., 2017). We investigated the expression patterns of some important genes functioning in wax biosynthetic pathways. The E. grandiflorum genes were obtained by performing homologous sequence alignments with Arabidopsis genes. We selected six genes that might be involved in cuticular wax biosynthesis: EgCER3, EgCER6, EgCER10, EgKCS1, EgKCR1, and EgCYP77A6 (Figure 4). We also checked the expression of a gene (EgABCG12) putatively involved in lipid transport. A previous study confirmed that the homolog of this gene in Arabidopsis, ABCG12, which encodes an ATP-binding cassette (ABC) transporter, is a key component of the export pathway for cuticular lipids (Bird et al., 2007; McFarlane et al., 2010). In addition, we investigated the expression pattern of WIN1/SHN1, a TF that induces cutin production in Arabidopsis (Kannangara et al., 2007). Real-time RT-PCR results showed that EgCER3, EgCER6, EgCER10, EgKCS1, EgKCR1, EgCYP77A6, and EgWIN1 were upregulated in EgMIXTA1-overexpressing plants compared with non-transgenic plants. However, no significant changes in the transcript levels of EgABCG12 were observed. This result revealed that overexpression of EgMIXTA1 activated the expression of E. grandiflorum wax biosynthetic genes, which consequently caused an increase in cuticular wax biosynthesis and accumulation in transgenic E. grandiflorum plants.
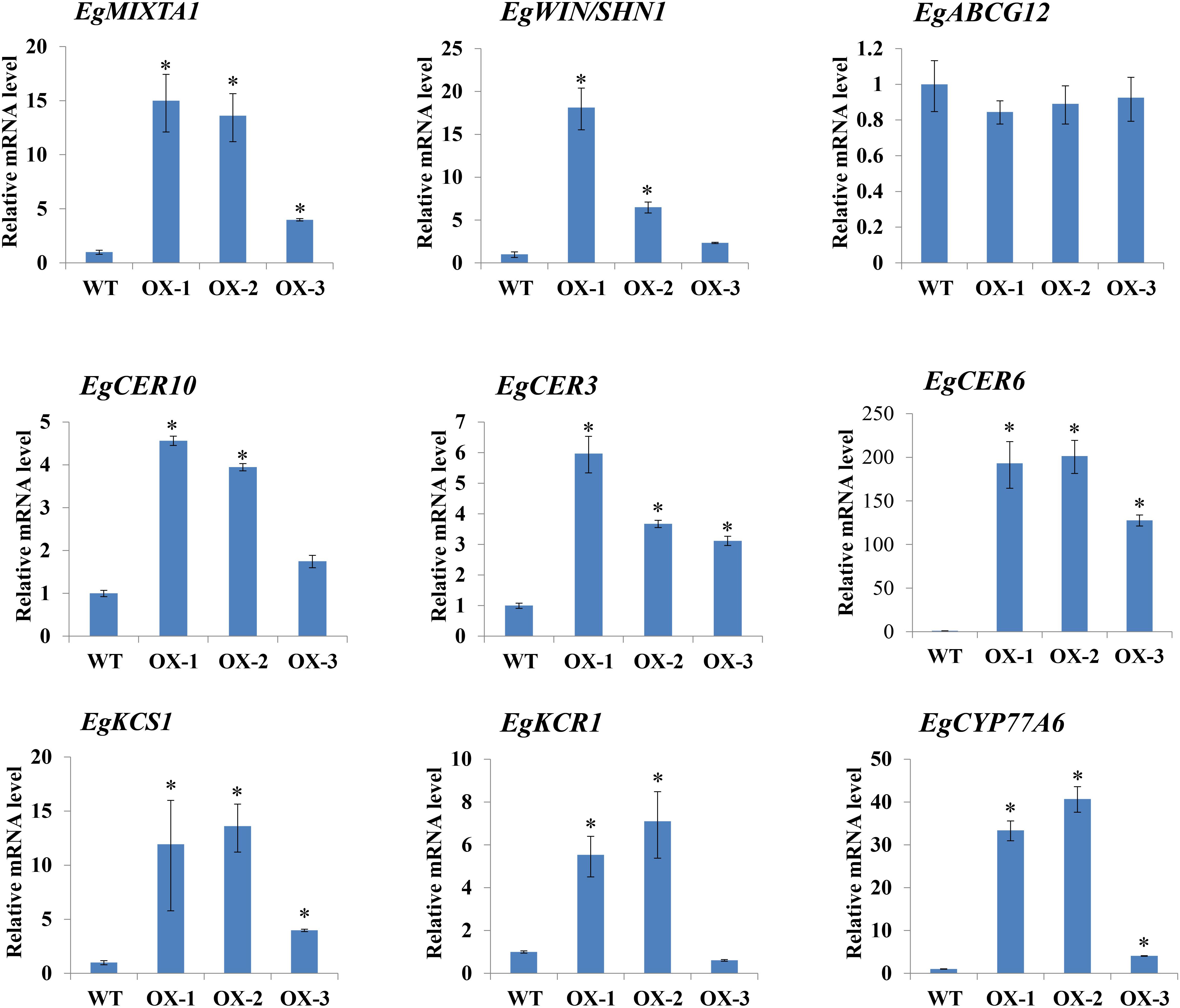
Figure 4. Expression of cuticular wax biosynthetic genes in EgMIXTA1-overexpressing plants. qRT-PCR analysis of the expression of genes involved in cutin and wax biosynthesis in the leaves of wild-type (WT) and EgMIXTA1-overexpressing plants (OX-1, OX-2, and OX-3). Expression level in the wild-type is set as 1. Actin was used as an internal reference. Student’s t-test: ∗P < 0.01.
EgMIXTA1-Overexpressing Plants Lose Water More Slowly Than the Wild-Type
Cuticular wax is the outermost layer that protects plants from water loss, and there is a lot of evidence showing that cuticular wax accumulation is closely associated with the water loss rate (Aharoni et al., 2004; Wang et al., 2018). We examined whether cuticular wax accumulation is linked to water loss by measuring cuticular transpiration. Five-week-old plants were dark acclimated for 6 h to ensure stomatal closure and soaked in water for 1 h in the dark to equilibrate the water content. Compared with wild-type leaves, transpiration occurred more slowly in the leaves of EgMIXTA1-overexpressing plants (Figure 5). The results showed that the elevated accumulation of cuticular waxes contributes to a reduction in water loss.
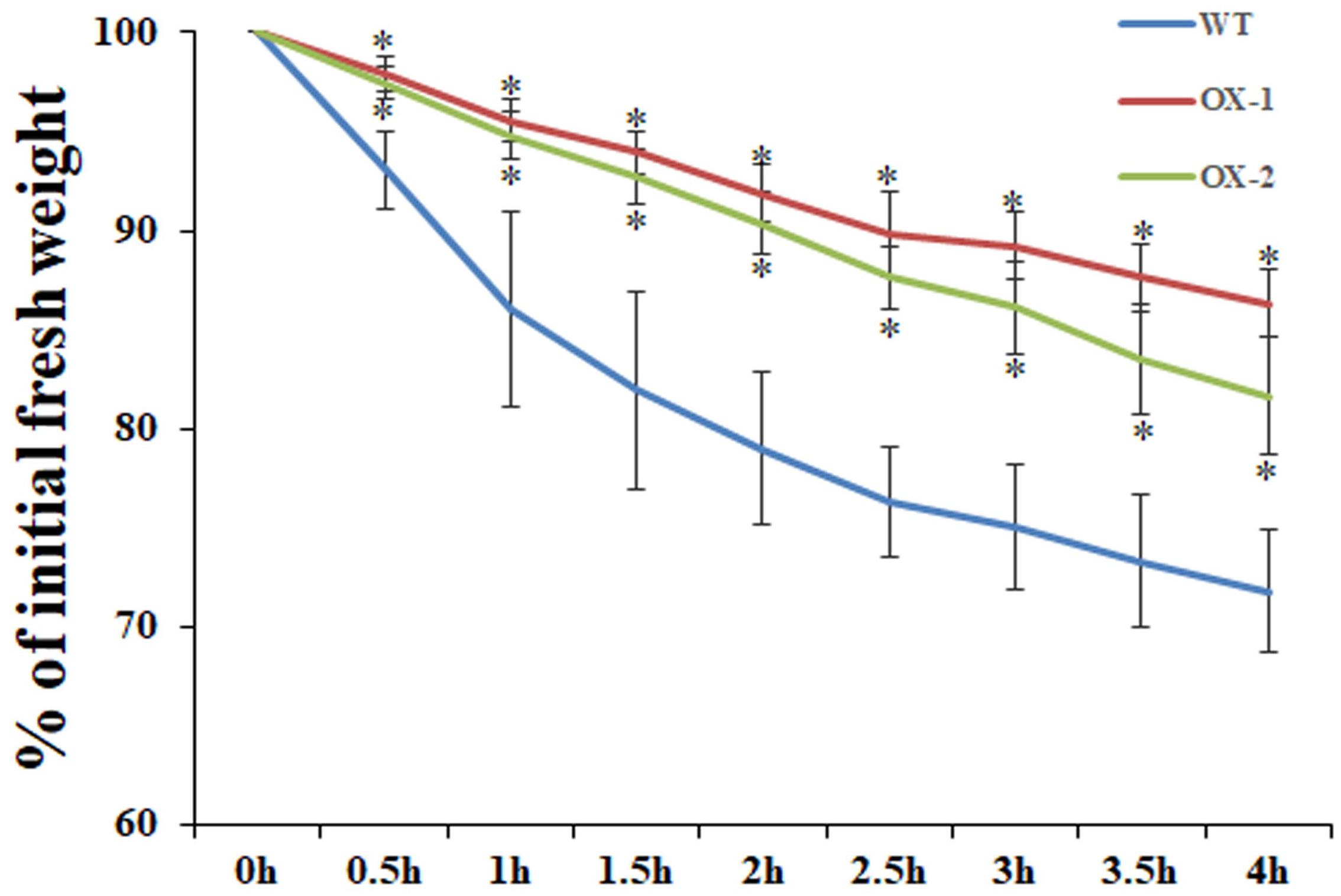
Figure 5. Water loss rate in the leaves of wild-type (WT), 35S:MIXTA1 (OX-1), and 35S:MIXTA1 (OX-2) plants. Leaves at the same developmental stage were excised and weighed at various time points after detachment. The water loss rate is shown as the percentage of initial fresh weight at each time point. The bars indicate the SE of three individual plants per genotype, and the asterisks denote significant differences from the wild-type (Student’s t-test, ∗P < 0.05).
Discussion
Plant cuticular wax covers the outer surface of aerial plant tissues. Many different studies have demonstrated that the plant cuticle plays an important role in limiting water loss and protecting plants from ultraviolet radiation and pathogen attacks (Kunst, 2003; Fich et al., 2016). Many Arabidopsis genes involved in wax synthesis, transport, and export, and in regulating the biosynthetic pathway have been identified. For example, KCR1 encodes a β-ketoacyl-CoA reductase enzyme and is a candidate fatty acid elongase (Beaudoin et al., 2009), and ECERIFERUM3 (CER3) is involved in the alkane-forming pathway (Chen et al., 2003). There are several lines of evidence from previous reports indicating that cuticular wax biosynthesis is regulated at the transcriptional level. For example, significant upregulation of the WSD1, KCS2/DAISY, CER1, CER2, FAR3, and ECR genes was observed in the leaves of transgenic Arabidopsis plants overexpressing MYB94 (Lee and Suh, 2015). In the activation-tagged myb96-1D mutant, a group of genes encoding cuticular wax biosynthetic enzymes, including KCS1, KCS2, KCS6, KCR1, CER1, and CER3, were greatly upregulated. In addition, the dominant active form of MYB106 (35S:MYB106-VP16) increased the expression of WIN1/SHN1 and wax biosynthetic genes. In this study, we cloned the EgMIXTA1 gene, which encodes a MIXTA-like R2R3-MYB TF, and generated transgenic plants overexpressing this gene. Overexpression of EgMIXTA1 increased the expression of the wax biosynthetic genes (Figure 4).
MIXTA and MIXTA-like TFs have been shown to be key regulators of epidermal cell differentiation and cuticle biosynthesis. A. majus, a mixta mutant, in which conical cells become flat, shows alterations in petal color intensity (Noda et al., 1994). In Arabidopsis, the MIXTA-like genes AtMYB16 and AtMYB106 were shown to regulate epidermal cell morphology and cuticle development (Oshima et al., 2013). In this experiment, through scanning electron microscopy pictures, we observed that no epidermal hair was observed on the wild-type leaves of E. grandiflorum, and the same results were also shown after overexpression. At the same time, no changes were found in the morphology of the epidermal cells. In addition to the increase in cuticular wax, it is possible that EgMIXTA1 in the E. grandiflorum leaves mainly regulates the biosynthesis and transport of wax crystal. Unfortunately, we failed to observe changes in the morphological differentiation status and cuticular wax of E. grandiflorum petal epidermal cells after overexpression. In our study, EgMIXTA1 was demonstrated to function quite similarly to the MIXTA-like MYB TFs in determining cuticular wax formation in E. grandiflorum leaves (Figure 3C). It has been documented that biosynthesis and accumulation of cuticular waxes are closely linked with drought resistance responses. Under water deficit conditions, cuticular wax accumulation significantly increases. In roses, drought stress caused a 9–15% increase in wax load on leaves exposed to drought during development (Jenks et al., 2001). In tobacco, the total wax load increased 1.5- to 2.5-fold under periodic dehydration stress. In addition, a high-wax-producing Dianthus spiculifolius mutant with increased cuticular wax accumulation exhibited stronger drought resistance (Zhou et al., 2018). Our results demonstrate that EgMIXTA1 regulates cuticular wax deposition and decrease water loss (Figure 5).
Eustoma grandiflorum, which belongs to the Gentianaceae family, is becoming increasingly popular for use in cut flower production because it has large flowers, long stems, and an extended vase life (Davies et al., 1993). We found that elevated accumulation of cuticular waxes contributes to a reduction in water loss in E. grandiflorum (Figure 5). We did not conduct an experiment to elucidate the phenotype change and drought stress tolerance in the naturally growing conditions or the fresh-cut vase life. Our subsequent research will also verify this. The subsequent research will help in production and application. Based on this observation, increasing the accumulation of cuticular waxes may be important in reducing water loss and possibly delaying senescence and extending the vase life of cut E. grandiflorum flowers.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
LW and JW carried out the experiments and data analysis and drafted the manuscript. YL and YZ provided guidance on experimental design and drafting the manuscript. WX, XL, JL, LX, and SK provided help in carrying out the experiments and modifying the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by a grant from the National Natural Science Foundation of China (Grant Nos. 31772339, and 31872139), Natural Science Foundation of Heilongjiang Province of China (Grant No. C2016008), the Fundamental Research Fund for Central Universities (Grant No. 2572019AA24), and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.524947/full#supplementary-material
Supplementary Table 1 | Primer sequences used in this paper.
References
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., Van, A. G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the Arabidopsis β-ketoacyl-coenzyme a reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Bernard, A., and Joubes, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52, 110–129. doi: 10.1016/j.plipres.2012.10.002
Bird, D., Beisson, F., Brigham, A., Shin, J., Greer, S., Jetter, R., et al. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52, 485–498. doi: 10.1111/j.1365-313X.2007.03252.x
Brockington, S. F., Alvarezfernandez, R., Landis, J. B., Alcorn, K., Walker, R. H., Thomas, M. M., et al. (2013). Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Mol. Biol. Evol. 30, 526–540. doi: 10.1093/molbev/mss260
Chen, X., Goodwin, S. M., Boroff, V. L., Liu, X., and Jenks, M. A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15, 1170–1185. doi: 10.1105/tpc.010926
Davies, K. M., Bradley, J. M., Schwinn, K. E., Markham, K. R., and Podivinsky, E. (1993). Flavonoid biosynthesis in flower petals of five lines of lisianthus (Eustoma grandiflorum Grise.). Plant Sci. 95, 67–77. doi: 10.1016/0168-9452(93)90080-j
Ecker, R., Barzilay, A., and Osherenko, E. (1994). The genetic relations between length of time to germination and seed dormancy in lisianthus (Eustoma grandiflorum). Euphytica 80, 125–128. doi: 10.1007/bf00039307
Fich, E. A., Segerson, N. A., and Rose, J. K. (2016). The plant polyester cutin: biosynthesis, structure, and biological roles. Annu. Rev. Plant Biol. 67, 207–233. doi: 10.1146/annurev-arplant-043015-111929
Gilding, E. K., and Marks, M. D. (2010). Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 64, 304–317. doi: 10.1111/j.1365-313x.2010.04329.x
Go, Y. S., Kim, H., Kim, H. J., and Suh, M. C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26, 1666–1680. doi: 10.1105/tpc.114.123307
Hooker, T. S., Millar, A. A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129, 1568–1580. doi: 10.1104/pp.003707
Jenks, M. A., Andersen, L., Teusink, R. S., and Williams, M. H. (2001). Leaf cuticular waxes of potted rose cultivars as affected by plant development, drought and paclobutrazol treatments. Physiol. Plant 112, 62–70. doi: 10.1034/j.1399-3054.2001.1120109.x
Kannangara, R., Branigan, C., Liu, Y., Penfield, T., Rao, V., Mouille, G., et al. (2007). The transcription factor WIN1/SHN1 regulates Cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19, 1278–1294. doi: 10.1105/tpc.106.047076
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162, 567–580. doi: 10.1104/pp.112.210450
Kunst, L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 51–80. doi: 10.1016/s0163-7827(02)00045-0
Kunst, L., and Samuels, L. (2009). Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727. doi: 10.1016/j.pbi.2009.09.009
Lashbrooke, J., Adato, A., Lotan, O., Alkan, N., Tsimbalist, T., Rechav, K., et al. (2015). The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol. 169, 2553–2571. doi: 10.1104/pp.15.01145
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6, 246–249. doi: 10.1093/mp/sss159
Lee, S. B., and Suh, M. C. (2015). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56, 48–60. doi: 10.1093/pcp/pcu142
Lena, S., Dalia Rav, D., Irit, D., Uri, Y., Shimon, P., Rahel, L., et al. (2009). Cultural methods and environmental conditions affecting gray mold and its management in lisianthus. Phytopathology 99, 557–570.
McFarlane, H. E., Shin, J. J., Bird, D. A., and Samuels, A. L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22, 3066–3075. doi: 10.1105/tpc.110.077974
Noda, K., Glover, B. J., Linstead, P., and Martin, C. (1994). Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369:661. doi: 10.1038/369661a0
Oshima, Y., Shikata, M., Koyama, T., Ohtsubo, N., Mitsuda, N., and Ohme-Takagi, M. (2013). MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25, 1609–1624. doi: 10.1105/tpc.113.110783
Plett, J. M., Wilkins, O., Campbell, M. M., Ralph, S. G., and Regan, S. (2010). Endogenous overexpression of Populus MYB186 increases trichome density, improves insect pest resistance, and impacts plant growth. Plant J. 64, 419–432. doi: 10.1111/j.1365-313x.2010.04343.x
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Stilio, V. S., Di Cathie, M., Schulfer, A. F., and Connelly, C. F. (2010). An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol. 183, 718–728. doi: 10.1111/j.1469-8137.2009.02945.x
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., et al. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139, 1649–1665. doi: 10.1104/pp.105.070805
Uddin, A. F. M. J., Hashimoto, F., Miwa, T., Ohbo, K., and Sakata, Y. (2004). Seasonal variation in pigmentation and anthocyanidin phenetics in commercial Eustoma flowers. Sci. Horticult. 99, 103–115. doi: 10.1016/j.scienta.2003.07.002
Walford, S. A., Wu, Y., Llewellyn, D. J., and Dennis, E. S. (2011). GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797. doi: 10.1111/j.1365-313x.2010.04464.x
Wang, Q., Zhang, Y., Kawabata, S., and Li, Y. (2011). Double fertilization and embryogenesis of Eustoma grandiflorum. J. Japanese Soc. Horticult. Sci. 80, 351–357. doi: 10.2503/jjshs1.80.351
Wang, Z., Tian, X., Zhao, Q., Liu, Z., Li, X., Ren, Y., et al. (2018). The E3 Ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in Rice. Plant Cell 30, 228–244. doi: 10.1105/tpc.17.00823
Weng, L., Tian, Z., Feng, X., Li, X., Xu, S., Hu, X., et al. (2011). Petal development in Lotus japonicus. J. Integr. Plant Biol. 53, 770–782. doi: 10.1111/j.1744-7909.2011.01072.x
Xue, D., Zhang, X., Lu, X., Chen, G., and Chen, Z. H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8:621. doi: 10.3389/fpls.2017.00621
Xuhong, Y., John, K., Xiaoying, Z., Dror, S., Maskit, M., Hongyun, Y., et al. (2007). Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19, 3146–3156. doi: 10.1105/tpc.107.053017
Yan, T., Li, L., Xie, L., Chen, M., Shen, Q., Pan, Q., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 218, 567–578. doi: 10.1111/nph.15005
Ying, S., Su, M., Wu, Y., Zhou, L., and Zhang, Y. (2019). Trichome regulator SlMX1 directly manipulates primary metabolism in tomato fruit. Plant Biotechnol. J. 18, 354–363. doi: 10.1111/pbi.13202
Yonghua, L., Fred, B., Koo, A. J. K., Isabel, M., Mike, P., and John, O. (2007). Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. U.S.A. 104, 18339–18344. doi: 10.1073/pnas.0706984104
Yonghua, L. B., Mike, P., Vincent, S., Franck, P., John, O., and Fred, B. (2009). Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. U.S.A. 106, 22008–22013. doi: 10.1073/pnas.0909090106
Zaccai, M., and Edri, N. (2002). Floral transition in lisianthus (Eustoma grandiflorum). Sci. Horticult. 95, 333–340. doi: 10.1016/s0304-4238(02)00057-2
Keywords: cuticular wax, lipid biosynthesis, Eustoma grandiflorum, MIXTA genes, transgenic plants, water loss
Citation: Wang L, Xue W, Li X, Li J, Wu J, Xie L, Kawabata S, Li Y and Zhang Y (2020) EgMIXTA1, a MYB-Type Transcription Factor, Promotes Cuticular Wax Formation in Eustoma grandiflorum Leaves. Front. Plant Sci. 11:524947. doi: 10.3389/fpls.2020.524947
Received: 07 January 2020; Accepted: 22 September 2020;
Published: 22 October 2020.
Edited by:
Shiwen Wang, Northwest A&F University, ChinaReviewed by:
Takashi Nakatsuka, Shizuoka University, JapanDaoqian Chen, Fujian Agriculture and Forestry University, China
Copyright © 2020 Wang, Xue, Li, Li, Wu, Xie, Kawabata, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhua Li, bHloc2hlbkAxMjYuY29t; Yang Zhang, c3VtbWVyemhhbmdAMTI2LmNvbQ==
 Lishan Wang
Lishan Wang Wanjie Xue1,2
Wanjie Xue1,2 Saneyuki Kawabata
Saneyuki Kawabata Yuhua Li
Yuhua Li