
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 02 September 2020
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.512844
This article is part of the Research TopicPrimary Metabolism in FruitsView all 31 articles
α-Farnesene, a sesquiterpene volatile compound plays an important role in plant defense and is known to be associated with insect attraction and with superficial scald of apple and pear fruits during cold storage. But the mechanism whereby transcription factors regulate apple α-farnesene biosynthesis has not been clarified. Here, we report that two transcription factors, MdMYC2 and MdERF3 regulated α-farnesene biosynthesis in apple fruit. Dual-luciferase assays and Y1H assays indicated that MdMYC2 and MdERF3 effectively trans-activated the MdAFS promoter. EMSAs showed that MdERF3 directly binds the DRE motif in the MdAFS promoter. Subsequently, overexpression of MdMYC2 and MdERF3 in apple calli markedly activated the transcript levels of MdHMGR2 and MdAFS. Furthermore, transient overexpression of MdMYC2 and MdERF3 in apple fruit significantly increased MdAFS expression and hence, α-farnesene production. These results indicate that MdMYC2 and MdERF3 are positive regulators of α-farnesene biosynthesis and have important value in genetic engineering of α-farnesene production.
Plants produce a large number of metabolites that are essential for ecological interactions and terpenes are natural products of the largest and structurally most diverse class (Reddy et al., 2017). Terpenes play an important role in the communication between plants and the environment, between plants and animals, and between plants and plants; they can attract pollinators and seed spreaders, and they can act as defense agents against herbivores and pathogens. In addition, they protect plants from abiotic stress phenomena, such as high light intensity, high temperature, and oxidative stress, among others. Terpenes are also useful nutrients in human diet and are used as chemotherapeutic agents for their antitumor activities (Dudareva et al., 2006).
α-Farnesene is an abundant compound in apple peel, which has an important function in plant defense (Nieuwenhuizen et al., 2014). The oxidative product of α-farnesene, are widely considered as a principal cause of scald, the physiological disorder of apple and pear, which intensify when fruits are transferred to ambient temperature for transportation to the market (Huelin and Coggiola, 1970; Ingle and D’Souza, 1989; Rowan et al., 1995). Overexpression of α-farnesene synthase in pathogen-sensitive soybeans enhances plant antimicrobial activity and α-farnesene has also been considered as a potential biofuel precursor (Lin et al., 2017).
There are three kinds enzymes involved in α-farnesene metabolism: 1) 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), which initiates synthesize sesquiterpenes (Rupasinghe et al., 2001; Zhang et al., 2020); 2) farnesyl diphosphate synthase (FPS), which catalyzes the conversion of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) to farnesyl diphosphate (FPP), the substrate of α-farnesene synthesis; 3) α-farnesene synthase (AFS), which catalyzes the final rate-limiting step in α-farnesene biosynthesis (Pechous and Whitaker, 2004; Gapper et al., 2006); But in plant the regulatory mechanisms of α-farnesene biosynthesis remains largely unknown.
The synthesis of terpenes in plants is regulated by many transcription factors, including MYC, AP2/ERF, bZIP, WRKY. MYC2 is important for JA response to secondary metabolite accumulation. AtMYC2 binds the promoters of AtTPS11 and AtTPS21 for regulating the synthesis of sesquiterpenes in Arabidopsis thaliana (Hong et al., 2012). In Artemisia annua, AaMYC2 binds the G-boxlike motifs within the promoters of genes CYP71AV1 and DBR2 (Shen et al., 2016). In Salvia miltiorrhiza, SmMYC2a and SmMYC2b play important roles in regulating the biosynthesis of phenolic acids (Zhou et al., 2016). Ethylene response factors have also been well characterized for their roles in regulating the production of terpenes. AaERF1 and AaERF2 bound to the promoter of AaADS for inducing artemisinin synthesis (Yu et al., 2012). TcERF15, respectively, act as activator of tasy gene of taxol biosynthesis in Taxus chinensis (Zhang et al., 2015). In Newhall sweet orange fruit, the transcription factor CitERF71 directly binds the CitTPS16 promoter, therefore probably has a function in the transcriptional regulation of E-geraniol production (Li et al., 2017).
Although transcription factors related to the metabolic pathway of terpenes have been reported in recent years, there are few reports on transcription factors related to the synthesis of terpenes in apples. At present, regardless of the metabolic engineering research of α-farnesene or the biological research of apple superficial scald, transcription factors regulating the mechanism of α-farnesene biosynthesis have not been reported. By screening transcription factors involved in secondary metabolic regulation, we studied whether these transcription factors are participated in regulating the expression of α-farnesene synthase, thereby affecting the biosynthesis of α-farnesene.
Our results revealed that transcription factors MdMYC2 and MdERF3 effectively activated the promoter region of MdAFS, which is the terminal enzyme gene in the α-farnesene biosynthesis pathway; additionally, they activated the expression of the MdAFS gene, and ultimately promoted the accumulation the α-farnesene.
Leaves obtained from five-year-old apple (Malus domestica Borkh. cv. White winter pearmain) trees were used in this study. The apple trees were cultivated in a culture room at Shandong Agriculture University. The “Orin” apple calli were used for genetic transformation and were grown at 24°C under dark conditions.
“White winter pearmains” apple leaves were treated with 100 μM MeJA and 50 mg L-1 Ethephon (Sigma-Aldrich), with 0.1% (v/v) ethanol as the mock. Samples were taken after 0, 2, 4, 6, and 12 h to analyze gene expression.
Fruits harvested at 140 days after full bloom were divided into four groups. The first group was not treated. The second group was treated with methyl jasmonate for 5 min. The third group was used for Ethephon treatments for 30 s. The fourth group was treated with 1-MCP for 12 h. All fruits were stored at room temperature (24°C) and 0°C for 4 weeks, with sampling every week during the storage period. The samples were frozen immediately in liquid nitrogen and then stored at -80°C.
Total RNA was isolated and first-strand cDNAs were synthesized, respectively, using a total RNA isolation system and First-strand cDNA Synthesis Kit (Tiangen, Beijing, China). All qRT-PCR assays were used in a CFX96 Real-time system (BIO-RAD) according to manufacturer instructions. Three independent biological replicates were carried out for each sample. Primer sequences used for real-time quantitative PCR were described in Supplementary Table S1.
The ERF3 and MYC2 transgenic apple calli were obtained from Professor Hao’s laboratory (An et al., 2016; An et al., 2018). The constructed recombinant plasmids were introduced into Agrobacterium tumefaciens LBA4404. 15-day-old “Orin” apple calli were infected with A. tumefaciens for 20 min which were carrying recombinant plasmids, and the apple calli were cultured on agar solidified MS medium for 2 days at 24 °C in darkness. Then, the apple calli were transferred to selective medium containing 35 mg L−1 hygromycin and 300 mg L−1 carbenicillin.
Full-length MdMYC2 and MdERF3 sequences were amplified with the primers described in Supplementary Table S1 and were inserted into pBI 121 vectors. The promoter of MdAFS (1,500 bp) was constructed in the pGreenII 0800-LUC vector. All constructs were individually transformed into Agrobacterium GV3101 and stored as glycerol stocks at -80°C. Agrobacterium cultures were prepared with infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6) to an OD600 of 1.0. The mixtures of transcription factor and promoter were infiltrated into tobacco leaves by needleless syringes. A living imaging apparatus was used for luminescence detection. For each transcription factor-promoter interaction, at least three independent experiments were performed with four replicates in each experiment.
Y1H was used to detect verification of interaction between transcription factor and AFS as described by An et al. (2016). The MdMYC2 and MdERF3 gene was cloned into the pGADT7 vector and the promoter fragment of MdAFS were inserted into the pAbAi vector.
EMSAs were performed as described previously (Li et al., 2017). The LightShift™ Chemiluminescent EMSA Kit (Thermo, USA) was used in EMSA experiment. Oligonucleotide probes were synthesized and labeled with biotin. Biotin-labeled probes were incubated with MdERF3-GST protein in a binding buffer for 25 min, and the free and bound DNAs were separated in an acrylamide gel.
Fruit injection assays were carried out as described previously (Li et al., 2012). The overexpression viral vectors MdMYC2-IL60-2 and MdMERF3-IL60-2 were generated and the mixed vectors were injected into the fruit peels. Two days after infiltration, the peel near the infiltration point was collected for volatile analysis.
Volatile analysis was carried out as in our previous study (Deng et al., 2015). The fresh apple peels were ground in liquid nitrogen and 0.3 g was extracted with 5 ml of extraction buffer in a sealed container. 10 μl of 3-Nonanone (0.4 g L-1) was an internal standard. The volatile compounds were collected by solid phase microextraction (SPME) and analyzed using GCMS-QP2010 with a FID detector (Shimadzu, Tokyo, Japan).
Statistical analysis of the data was performed with SPSS. Data points represent the mean values ± standard deviation of three biological replicates. Differences were considered statistically significant when * P < 0.05 and ** P < 0.01.
In order to study the mechanism of regulation of α-farnesene synthesis, we first analyzed the promoter of MdAFS, which is the terminal enzyme gene in the α-farnesene synthesis pathway. In the MdAFS promoter, many potential cis-acting elements associated with hormone-related responses were identified, such as MeJA, Ethylene, and ABA (Supplementary Table S2). JA has been widely used in regulating plant growth and secondary metabolism, and significant progress has been achieved in regulating the accumulation of terpenoid secondary metabolites by its use. Ethylene is an important hormone and ethylene treatment reportedly involves in volatile synthesis and ethephon facilitates the release of the ethylene. To examine whether the MdAFS and α-farnesene were induced by phytohormones, “White winter pearmains” leaves and fruits were treated with MeJA and Ethephon. The results indicated that the expression of MdAFS was significantly upregulated after these treatments. When apple leaves were treated with MeJA, the maximum expression level of MdAFS occurred at 6 h post treatment (Figure 1A). The response of MdAFS to Ethephon treatment was peaked within 12 h post treatment (Figure 1A) and the α-farnesene content in the apple leaves increased with the increase of treatment time, reaching the highest in 12 h (Supplementary Figure 1A). Concomitantly, α-farnesene content was markedly higher than in controls following MeJA and Ethephon treatments, but markedly lower treated with 1-MCP relative (Figures 1B, C).
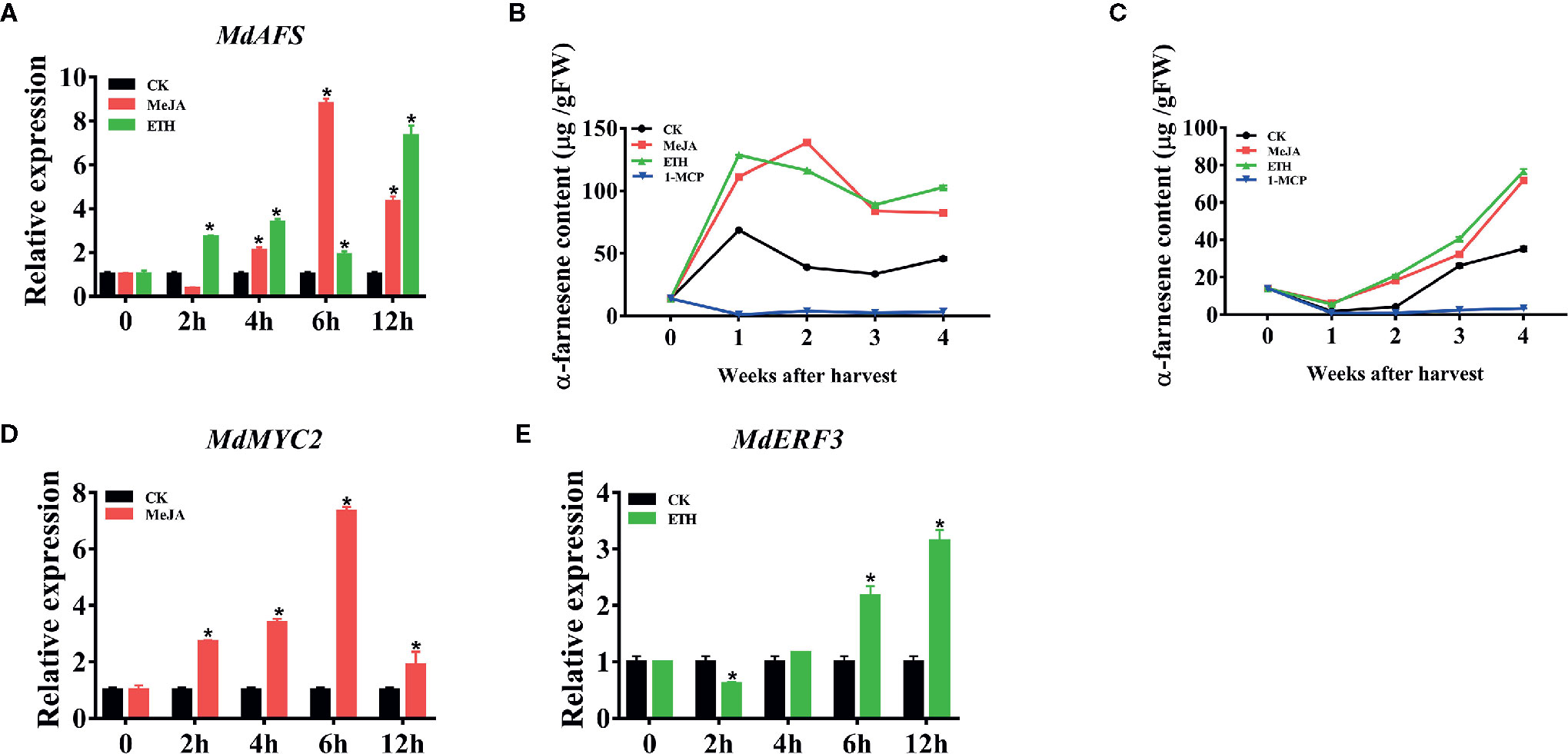
Figure 1 (A) Expression of the MdAFS gene in apple leaves under MeJA and ETH treatments. (B, C) Effects of MeJA, ETH and 1-MCP on production of α-farnesene. White Winter Pearmain apples were treated with JA, ETH, and 1-MCP, and stored at room temperature and 0℃ for 4 weeks to detect the content of α-farnesene. (D, E) Effect of MeJA and ETH treatments on MdMYC2 and MdERF3 expression in apple leaves. Data points represent the mean values ± standard deviation of three biological replicates * indicates P < 0.05 compared to CK.
It is reported that MYC2 and ERF play a significant roles in ethylene and JA signaling pathways. Combined with the previous promoter analysis and PlantCare software analysis results, we found that promoters contained MYCCONSENSUSAT (MYC), G-box, DRE and ERE motifs, which were reported to be the binding sites of MYC2 and ERF transcription factors, implying these transcription factors might involve in transcriptional regulation of MdAFS. Therefore, we selected transcription factors MdMYC2 and MdERF3 to test their correlation to MdAFS. As shown in Figures 1D, E, MdMYC2 and MdERF3 showed the same accumulation pattern as MdAFS in response to both MeJA and Ethephon treatments. The expression levels of MdMYC2 promoted at 2 h after MeJA treatment and peaked at 6 h; Further, the expression of MdERF3 peaked at 12 h. Gene expression analysis of MdAFS, MdMYC2, and MdERF3 during room temperature storage of apple fruits under MeJA, ETH, and 1-MCP treatments showed the same results (Supplementary Figures 1B–D).
We conducted a firefly luciferase (Luc) complementation imaging assay to test if MdMYC2 and MdERF3 could regulate the expression of MdAFS. As predicted, these two transcription factors showed trans-activation effects on the MdAFS promoter (Figure 2). Studies have shown that transcription factors MdMYC2 and MdERF3 can bind G-box and DRE elements to regulate the expression of downstream genes (Li et al., 2016; Li et al., 2017). Y1H assays were performed to test whether MdMYC2 and MdERF3 could bind promoters of MdAFS. Thus, G-box and four repeated DRE motifs were integrated into yeast cells. We found that, indeed, the two transcription factors were capable of binding both the G-box and the DRE motifs. In addition, to conform the binding results, we performed an EMSA with the MdERF3 together with 25 bp promoter fragments of MdAFS containing the DRE motif. The DRE motif of the MdAFS promoter was recognized by MdERF3 (Figure 3). These results indicated that MdMYC2 and MdERF3 effectively activated the α-farnesene biosynthetic gene MdAFS, and MdERF3 directly bound the DRE motif in the MdAFS promoter.
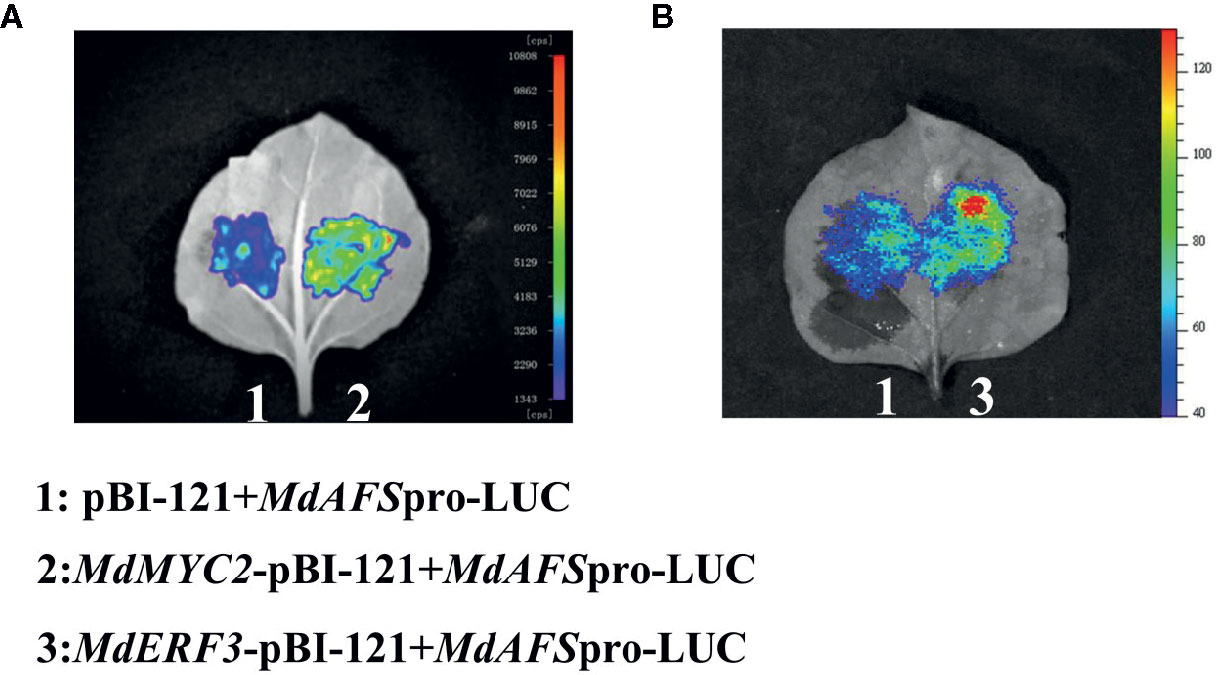
Figure 2 The firefly luciferase (Luc) complementation imaging assays. Agrobacterium GV3101 strain harboring different constructs was infiltrated into tobacco leaves. Luminescence signals in the infiltrated region were measured 48 h after infiltration. (A) 1: pBI-121+MdAFSpro-LUC, 2: MdMYC2-pBI-121+MdAFSpro-LUC. (B) 1: pBI-121+MdAFSpro-LUC, 2: MdERF3-pBI-121+MdAFSpro-LUC.
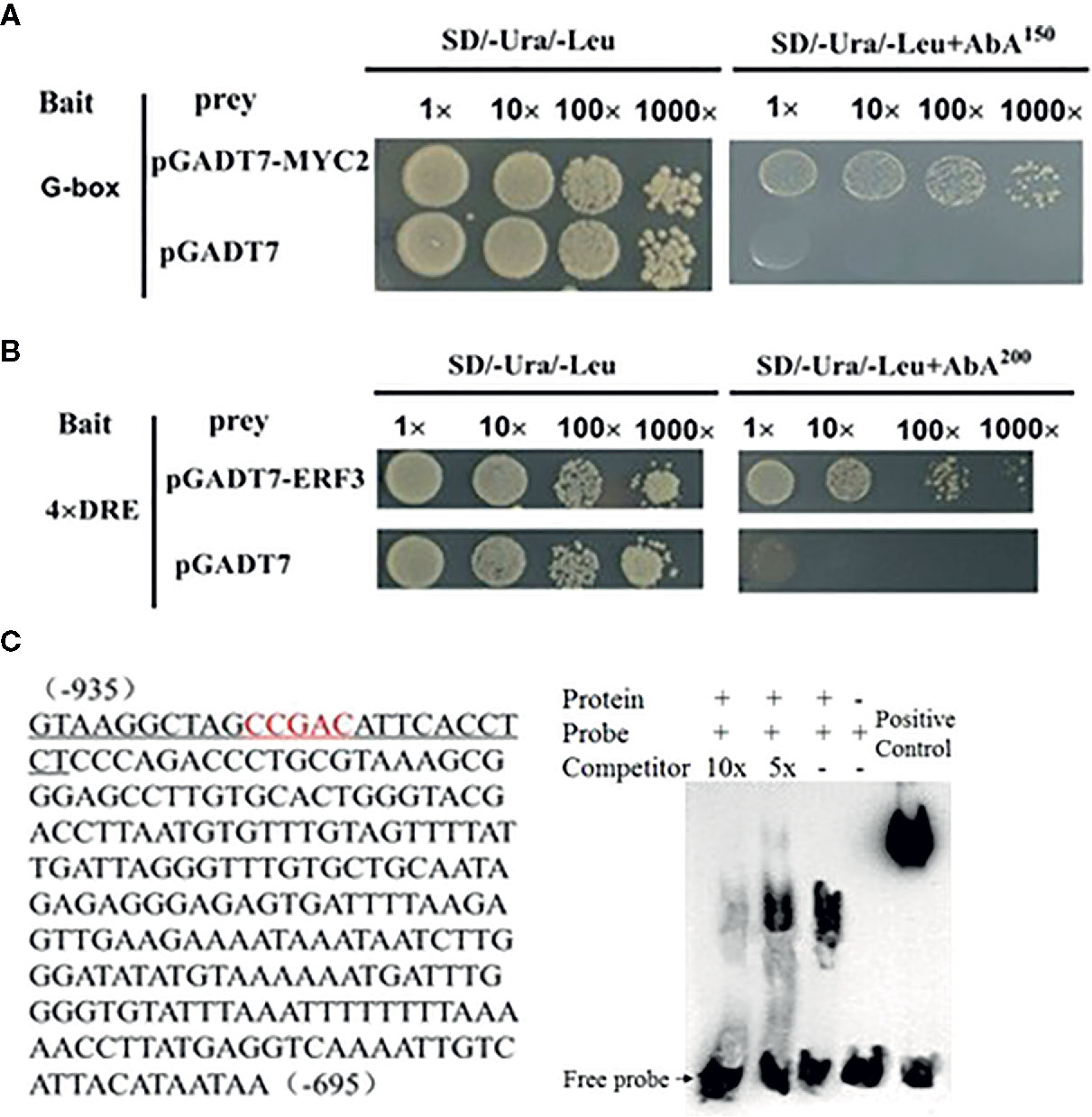
Figure 3 MdMYC2 and MdERF3 bind to the MdAFS promoter. (A, B) Yeast one-hybrid analysis of MdMYC2 and MdERF3 binding to MdAFS promoter. (C) EMSA indicating that MdERF3 directly bound to the MdAFS promoter at CCGAC in vitro. Biotin-labeled probes were incubated with MdERF3 protein and then separated in an acrylamide gel. The competitors were unlabeled probes.
To further examine the regulation of MdAFS by MdMYC2 and MdERF3, overexpression and antisense constructs were transformed into “Orin” apple calli. The expression levels of MdAFS gene in the wildtype and transgenic calli were analyzed by qRT-PCR. As shown in Figure 4, we found that when MdMYC2 expression was upregulated by ~15-fold, MdAFS expression was upregulated by ~10-fold in the calli overexpression MdMYC2. The expression levels of MdAFS were significantly lower in MdMYC2-antisense calli than in control calli. Similarly, MdERF3-overexpressing calli significantly upregulated expression of the MdAFS gene, which was increased over 13-fold. The α-farnesene content of calli were measured, and the results showed that the content of over-expressed MdMYC2 and MdERF3 calli were significantly higher than that of wildtype, and the α-farnesene content in antisense calli was the lowest (Supplementary Figure 2). These results showed that MdMYC2 and MdERF3 could positively regulate the expression of MdAFS.
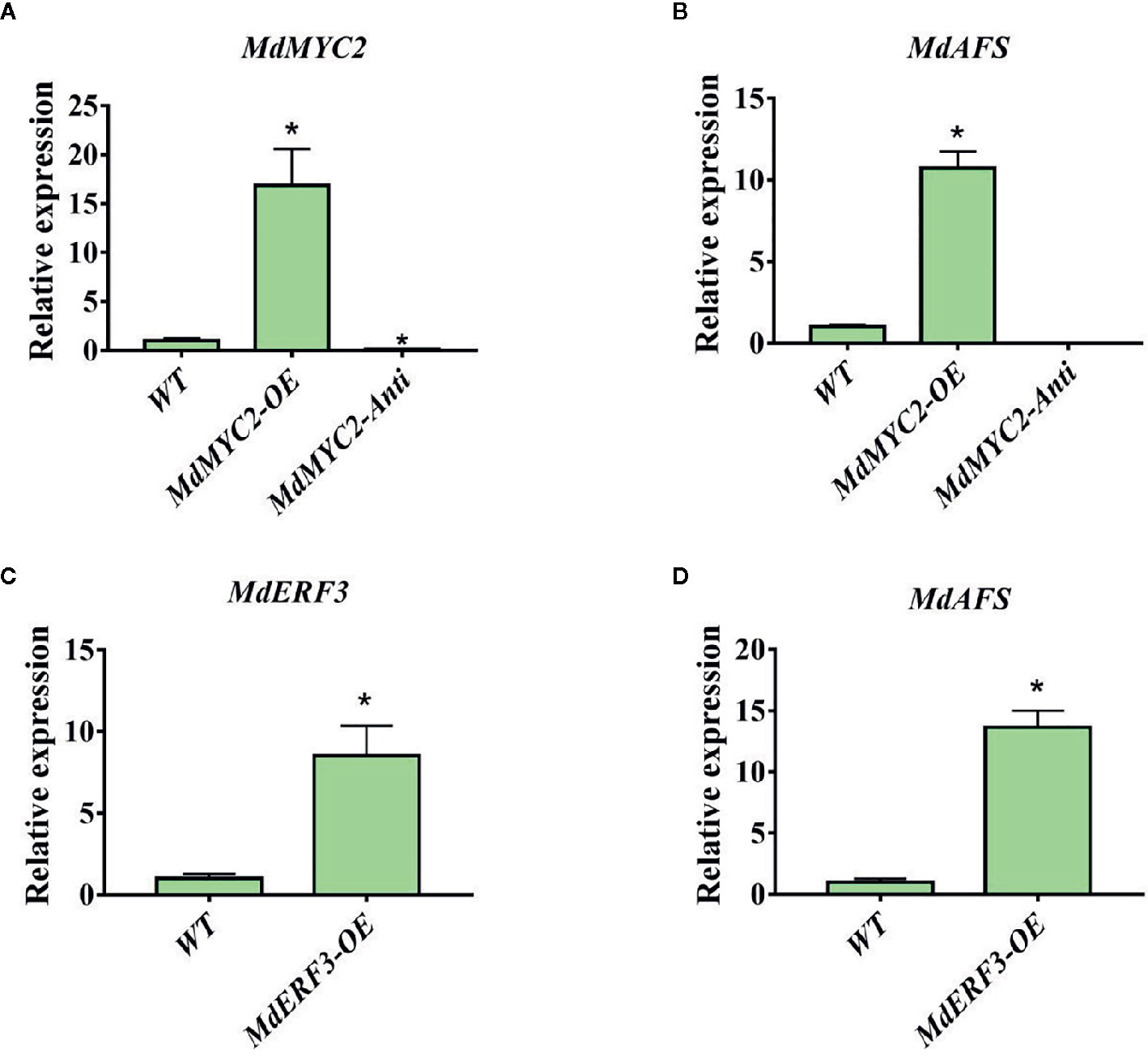
Figure 4 Expression analysis of MdAFS, MdMYC2, and MdERF3 in transgenic apple calli. (A) Expression level of MdMYC2 in apple calli overexpressing and silencing MdMYC2. (B) Expression level of MdAFS in apple calli overexpressing and silencing MdMYC2. (C) Expression level of MdERF3 in apple calli overexpressing MdERF3. (D) Expression level of MdAFS in apple calli overexpressing MdERF3. Standard errors were calculated from three sets of biological replicates. * indicates P < 0.05 compared to WT.
There are technically and experimentally challenging in generation and testing of transgenic apple fruit (Kotoda et al., 2006). The assay was chosen to examine the role of MdMYC2 and MdERF3 in the biosynthesis of α-farnesene in apple fruits. The α-farnesene content of the peels infiltrated with MdMYC2 was 8.97 μg g–1, respectively, representing a marked increase relative to the peels (7.4 μg g–1) infiltrated with empty vector (Figure 5). At the same time, apple peels infiltrated with MdERF3 exhibited a significant increase by ~1.7-fold in the level of α-farnesene content (Figure 6).
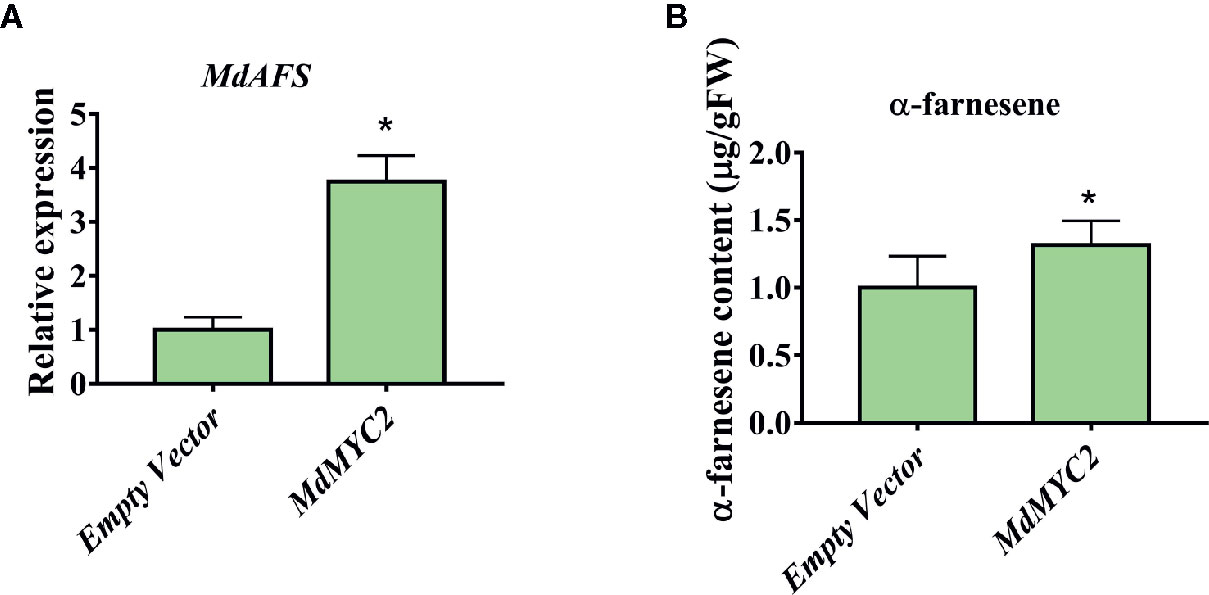
Figure 5 Transient expression of MdMYC2 in apple peel. (A) Relative MdAFS gene expression in apple peel injected with Empty Vector and MdMYC2. (B) α-farnesene content in peel infiltrated with MdMYC2. More than three apples were injected for determination, and the data were the average value obtained. * indicates P < 0.05 compared to Empty Vector.
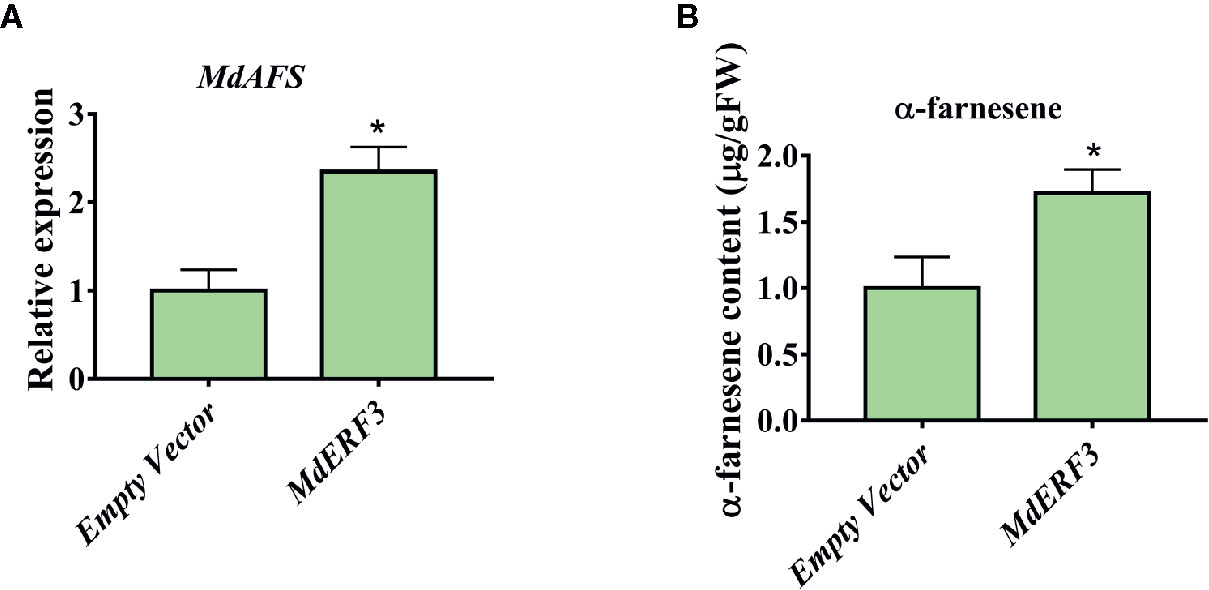
Figure 6 Transient expression of MdERF3 in apple peel. (A) Relative MdAFS gene expression in apple peel injected with Empty Vector and MdERF3. (B) α-farnesene content in peel infiltrated with MdERF3. More than three apples were injected for determination, and the data were the average value obtained. * indicates P < 0.05 compared to Empty Vector.
Transcription factors not only regulate single enzyme genes in a pathway, they also regulate the co-expression of multiple genes, thereby regulating the synthesis of specific secondary metabolites. It has been reported that the expression level of the MdHMGR2 gene is positively correlated with α-farnesene and ethylene production in apples during low temperature storage (Rupasinghe et al., 2001). Nevertheless, studies on the FPPS gene in apples are scarce. Only two genes, MdFPPS1 and MdFPPS2, have been cloned from apples (Yuan et al., 2013), and the similarity between them is as high as 99%. In addition, the regulation of MdFPPS on the synthesis of α-farnesene has not been clarified. However, our results demonstrated that overexpression of MdMYC2 and MdERF3 maybe affect the expression of the α-farnesene synthesis pathway genes MdHMGR2 and MdFPPS (Figure 7).
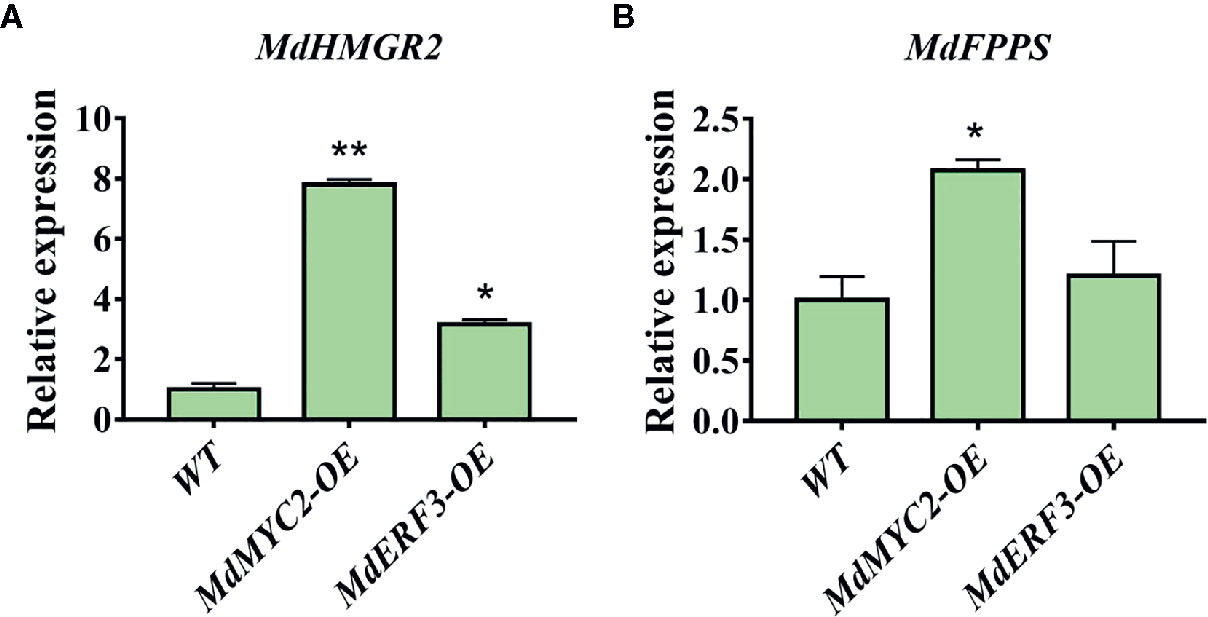
Figure 7 Expression analyses of MdHMGR2 and MdFPPS in apple calli. (A) Expression analysis of MdHMGR2 in overexpressing MdMYC2 and MdERF3 apple calli. (B) Expression analysis of MdFPPS in overexpressing MdMYC2 and MdERF3 apple calli. Each value is expressed as mean ± SE (n = 3). * indicates P < 0.05 compared to WT and ** indicates P < 0.01 compared to WT.
Terpenes are the largest and most diverse class of chemicals among the volatile compounds produced by plants (Tholl, 2015). In addition to their phyto-ecological benefits, terpenes are also of great economic value to humans, as they can be widely used in flavors, agriculture and in the chemical industry (Bouvier et al., 2006). α-farnesene was first found in apple and was found to play a role in plant defense. Farnesene is the precursor of the biofuel farnesane, which has broad market value, as it has attracted extensive attention of the society in recent years (Peralta-Yahya et al., 2011). However, sesquiterpenes including α-farnesene are naturally produced in limited quantities (Wallaart et al., 1999). Therefore, metabolic engineering of organisms is an alternative and attractive way to produce these rare and valuable compounds.
Currently, heterologous production of α-farnesene can be carried out in the host of Escherichia coli and Yarrowia lipolytica. In E. coli, the heterologous expression of α-farnesene synthase from fruits made production of α-farnesene in bacterial (Zhu et al., 2014). Recently, the feasibility of producing α-farnesene in metabolically engineered Y. lipolytica was demonstrated for the first time (Yang et al., 2016).
In recent years, there have been many studies on transcriptional regulation, as the use of transcription factors is one of the effective methods to increase the yield of terpenoid secondary metabolites. Moreover, transcription factors can activate or inhibit the expression of several key enzymes in plant secondary metabolic biosynthetic pathways by interacting with cis-acting elements in target gene promoters, thus, they effectively start or close secondary metabolic biosynthetic pathways, and regulate the biosynthetic process of specific secondary metabolites, thereby effectively affecting their accumulation.
MYC transcription factors are the most widely separated and thoroughly studied bHLH transcription factors. Their conserved domains regulate the expression of target genes by combining with E-Box (CANNTG) or G-Box (CACGTG) elements of target promoters (Pires and Dolan, 2010). At the same time, MYC family members participate in plant growth and development, resistance to environmental stress, JA and other signal transduction processes, and also in the regulation of secondary metabolic pathways (Gao et al., 2015). MYC transcription factors have been isolated from species such as Catharanthus roseus, Taxus, Artemisinin, Arabidopsis thaliana, tomato, and apple. Among MYC transcription factors found in plants, MYC2 is the most in-depth studied that plays an important role in the JA-mediated signal regulation pathway in plants. MYC2 can enhance plant resistance to insects by positively regulating JA-induced insect-resistant genes. In Arabidopsis thaliana, MYC2 regulates JA-mediated resistance to insect pests, and tolerance to oxidative stress by enhancing ascorbic acid redox cycle and flavonoid biosynthesis (Dombrecht et al., 2007). In addition, MYC2 is a positive regulator of JAs-mediated secondary metabolite synthesis. In tobacco, NtMYC2a and NtMYC2b positively regulate the JA response gene PMT to promote nicotine formation (Zhang et al., 2012). In Salvia sclarea, WRKY and MYC2 transcription factors which are controlled by MJ elicitation, coactivate MEP-biosynthetic genes and accumulation of abietane diterpenes (Alfieri et al., 2018).
APETALA2/Ethylene (AP2/ERF) transcription factors play an important role in regulating plant growth, development and maturation, responses to biotic and abiotic stress, and secondary metabolism (Agarwal et al., 2006). The ERF subfamily responds primarily to abiotic stress either dependently or independently of plant hormones such as ethylene and various biotic stress phenomena, such as pathogens and insect attack (Lai et al., 2014). ERF transcription factors reportedly can bind specifically GCC-box or DRE/CRT elements. The terpenes can be regulated by transcription factors, including AP2/ERF. ZmERF58 is able to directly bind the promoter of ZmTPS10 to synthesize E-β-farnesene and E-α-bergamot in maize (Li et al., 2015). NtERF32 and related ERF genes are important non-NIC2 locus related to transcriptional regulators of nicotine and total alkaloid formation (Sears et al., 2014). Further, ORA59, which is the AP2/ERF-domain transcription factor, and two GCC box binding sites that enables the PDF1.2 gene to respond to the JA and ET signaling pathways (Zarei et al., 2011).
However, although there are several transcription factors being involved in terpene biosynthesis, the transcriptional regulation of the α-farnesene remains unclear. Similarly, it has been reported that jasmonic acid and ethylene, two important plant hormones, coordinate to regulate plant growth, development and tolerance to pathogens. Indeed, EIN3/EIL1, two important transcription factors in ethylene, reportedly mediate the signal interaction between jasmonic acid and ethylene; hence, EIN3/EIL1 is also a positive regulator of the jasmonic acid signaling pathway that regulates plant root development and resistance responses (Zhu et al., 2011). Thus, the question may be asked, is there a common transcription factor regulating α-farnesene synthesis in the JA and ET signaling pathways?
In this study, we began with the regulation of these two hormones on α-farnesene. We found that whether stored at room temperature or at low temperature, JA and ET increased and 1-MCP decreased α-farnesene biosynthesis in apple fruit (Figures 1B, C). ERF and MYC2 are not only the key regulators in JA signaling pathway, but also the nodal factors connecting JA, ET, and other signal hormones. In addition, most steroid and alkaloid biosynthesis regulation is related to ERF and MYC2. Consistently with previous studies, we selected MdMYC2 and MdERF3 transcription factors for research, finding that these two transcription factors showed a similar expression pattern to MdAFS (Figure 1). This suggested that transcription factors MdMYC2 and MdERF3 might share a putative common regulatory mechanism of α-farnesene biosynthesis in plants. Further investigation of the transcriptional regulation of MdAFS gene by MdMYC2 and MdERF3, together with results of the dual-luciferase assay, the Y1H assay, and EMSA, led to the conclusion that MdMYC2 and MdERF3 is participated in transcriptional regulation of the MdAFS gene (Figures 2 and 3). In addition, to study the role of MdMYC2 and MdERF3, transient overexpression experiments were carried out. Apple peel infiltrated with MdMYC2 and MdERF3 showed a marked increase α-farnesene (Figures 5 and 6). Additionally, we found that transcription factors MdMYC2 and MdERF3 could regulate the expression of key enzyme genes MdAFS, MdHMGR2, and MdFPPS in the synthesis pathway of α-farnesene in stable transgenic apple calli (Figures 4 and 7). This indicated that MdMYC2 and MdERF3 effectively improved the synthesis of α-farnesene by activating the co-expression of multiple genes in the α-farnesene biosynthetic pathway. Consistently, it has been reported that MdMYC2 enhanced the transcription of MdERF3 by binding its promoter (Xu et al., 2017). We analyzed the transcript level of MdERF3 in apple calli after overexpressing MdMYC2, and the results showed that the expression of MdERF3 was increased in the overexpressing MdMYC2 apple calli (Supplementary Figure 3). Although we failed to verify that MdMYC2 binds directly the promoter of gene MdAFS, our studies showed that transcription factor MdMYC2 promoted the synthesis of α-farnesene by regulating the expression of MdAFS. Based on these results and earlier studies, we propose the hypothetical working model shown in Figure 8 to explain the synthesis of α-farnesene and the involvement of MdMYC2 and MdERF3 in its regulation.
It has been suggested that superficial scald development in apple and pear is related to α-farnesene and its oxidation products, namely, conjugated trienols (CTols), which accumulate progressively in the fruit peel during storage (Bordonaba et al., 2013). Previous investigations showed that l-MCP treatment induced scald resistance while inhibiting ethylene-dependent ripening. This indicated that the ethylene metabolic pathway may also be involved in the process of superficial scald (Karagiannis et al., 2018). Additionally, analysis of “Granny Smith” apples suggested that ethylene-related transcription factors might regulate AFS transcription during low temperature and then regulate the synthesis of α-farnesene and the occurrence of fruit superficial scald (Busatto et al., 2018). This study provided new insights into the regulation of α-farnesene and the mechanism of superficial scald.
In summary, this is the first report on the mechanism of regulation of the biosynthesis of α-farnesene at the transcriptional level. Our results indicate that MdMYC2 and MdERF3 exert their regulatory effects as positive regulators of α-farnesene biosynthesis-related genes. Further, our study identified key candidate genes and new strategies for using metabolic engineering methods to achieve high yields of α-farnesene.
The datasets generated for this study are available on request to the corresponding authors.
Designed the experiments: QW, YH, and YZ. Performed the experiments: QW and HL. Analyzed the data: QW, HL, MZ, and SL. Wrote the paper: QW, HL.
This study was financially supported by the National Natural Science Foundation of China (No. 31370359).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Jian-Ping An (Shandong Agricultural University) for providing transgenic apple calli and vectors.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.512844/full#supplementary-material
Supplementary Figure 1 | (A) α-farnesene content in apple leaves under MeJA and ETH treatments. (B) Expression of the MdAFS gene in apples stored at room temperature for 4 weeks under MeJA, ETH and 1-MCP treatments. (C) Expression of the MdMYC2 gene in apples stored at room temperature for 4 weeks under MeJA, ETH and 1-MCP treatments. (D) Expression of the MdERF3 gene in apples stored at room temperature for 4 weeks under MeJA, ETH, and 1-MCP treatments.
Supplementary Figure 2 | α-farnesene content in apple calli overexpressing MdMYC2, MdERF3 and silencing MdMYC2.
Supplementary Figure 3 | Analysis of MdERF3 transcript level in apple calli after overexpression MdMYC2.
Agarwal, P. K., Agarwal, P., Reddy, M. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. doi: 10.1007/s00299-006-0204-8
Alfieri, M., Vaccaro, M. C., Cappetta, E., Ambrosone, A., Tommasi, N. D., Leone, A., et al. (2018). Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 8, 11009. doi: 10.1038/s41598-018-29389-4
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X., et al. (2016). The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Bioch. 108, 24–31. doi: 10.1016/j.plaphy.2016.06.032
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., et al. (2018). EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation. Plant Physiol. 178, 808–832. doi: 10.1104/pp.18.00068
Bordonaba, J. G., Matthieu-Hurtiger, V., Westercamp, P., Coureau, C., Dupille, E., Larrigaudière, C. (2013). Dynamic changes in conjugated trienols during storage may be employed to predict superficial scald in ‘Granny Smith’ apples. LWT-Food Sci. Technol. 54, 535–541. doi: 10.1016/j.lwt.2013.06.025
Bouvier, F., Rahier, A., Camara, B. (2006). Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44, 357–429. doi: 10.1016/j.plipres.2005.09.003
Busatto, N., Farneti, B., Commisso, M. (2018). Apple fruit superficial scald resistance mediated by ethylene inhibition is associated with diverse metabolic processes. Plant J. 93, 270–285. doi: 10.1111/tpj.13774
Deng, S., Cheng, N. N., Ding, R. R. (2015). Cloning, Sequence Alignment and Functional Analysis of AFS Gene Promoter in Apple and Pear Fruits. Acta Hortic. Sin. 42, 2353–2361. doi: 10.16420/j.issn.0513-353x.2015-0554
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Dudareva, N., Negre, F., Nagegowda, D. A., Orlova, I. (2006). Plant volatiles: recent advances and future perspectives. Crit. Rev. Plant Sci. 25, 417–440. doi: 10.1080/07352680600899973
Gao, K., Wang, L., Wu, S. R., Sui, C. (2015). Advances in studies on transcriptional factor regulation of biosynthesis of active components in medicinal plants. Chin. Tradit. Herb. Drugs 46, 3100–3108. doi: 10.7501/j.issn.0253-2670.2015.20.024
Gapper, N. E., Bai, J., Whitaker, B. D. (2006). Inhibition of ethylene-induced ɑ-farnesene synthase gene PcAFS1 expression in ‘d’Anjou’ pears with 1-MCP reduces synthesis and oxidation of ɑ-farnesene and delays development of superficial scald. Postharvest Biol. Tec. 41, 225–233. doi: 10.1016/j.postharvbio.2006.04.014
Hong, G. J., Xue, X. Y., Mao, Y. B., Wang, L. J., Chen, X. Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648. doi: 10.2307/23264480
Huelin, F. E., Coggiola, I. M. (1970). Superficial scald, a functional disorder of stored apples VI.-Evaporation of ɑ-farnesene from the fruit. J. Sci. Food Agr. 21, 82–86. doi: 10.1002/jsfa.2740210207
Ingle, M., D’Souza, M. C. (1989). Physiology and control of superficial scald of apples: a review. Hortscience 24, 28–31. doi: 10.1016/0304-4238(89)90078-2
Karagiannis, E., Michailidis, M., Tanou, G. (2018). Ethylene-dependent and-independent superficial scald resistance mechanisms in ‘Granny Smith’ apple fruit. Sci. Rep. 8, 11436. doi: 10.1038/s41598-018-29706-x
Kotoda, N., Iwanami, H., Takahashi, S., Abe, K. (2006). Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J. Am. Soc Hortic. Sci. 131, 74–81. doi: 10.21273/JASHS.131.1.74
Lai, Y., Dang, F., Lin, J., Yu, L., Lin, J., Lei, Y., et al. (2014). Overexpression of a pepper CaERF5 gene in tobacco plants enhances resistance to Ralstonia solanacearum infection. Funct. Plant Biol. 41, 758–767. doi: 10.1071/FP13305
Li, Y. Y., Mao, K., Zhao, C., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 Ubiquitin E3 Ligases Interact with MdMYB1 to Regulate Light-Induced Anthocyanin Biosynthesis and Red Fruit Coloration in Apple. Plant Physiol. 160, 1011–1022. doi: 10.2307/41694818
Li, S., Wang, H., Li, F., Chen, Z., Li, X., Zhu, L., et al. (2015). The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles. Plant J. 84, 296–308. doi: 10.1111/tpj.12994
Li, T., Jiang, Z., Zhang, L., Tan, D., Wang, A. (2016). Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 88, 735–748. doi: 10.1111/tpj.13289
Li, X., Xu, Y., Shen, S., Yin, X., Klee, H., Zhang, B., et al. (2017). Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 68, 4929–4938. doi: 10.1093/jxb/erx316
Lin, J., Wang, D., Chen, X., Kollner, T. G., Mazarei, M., Guo, H., et al. (2017). An (E, E)-ɑ-farnesene synthase gene of soybean has a role in defence against nematodes and is involved in synthesizing insect-induced volatiles. Plant Biotechnol. J. 15, 510–519. doi: 10.1111/pbi.12649
Nieuwenhuizen, N. J., Green, S., Atkinson, R. G. (2014). Floral sesquiterpenes and their synthesis in dioecious kiwifruit. Plant Signal. Behav. 5, 61–63. doi: 10.4161/psb.5.1.10161
Pechous, S. W., Whitaker, B. D. (2004). Cloning and functional expression of an (E, E)-ɑ-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219, 84–94. doi: 10.1007/s00425-003-1191-4
Peralta-Yahya, P. P., Ouellet, M., Chan, R., Mukhopadhyay, A., Keasling, J. D., Lee, T. S., et al. (2011). Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2, 483. doi: 10.1038/ncomms1494
Pires, N., Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Reddy, V. A., Wang, Q., Dhar, N., Kumar, N., Sarojam, R. (2017). Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (Ms GPPS. LSU). Plant Biotechnol. J. 15, 1105–1119. doi: 10.1111/pbi.12701
Rowan, D. D., Allen, J. M., Fielder, S., Spicer, J. A., Brimble, M. A. (1995). Identification of conjugated triene oxidation products of ɑ-farnesene in apple skin. J. Agr. Food Chem. 43, 2040–2045. doi: 10.1021/jf00056a016
Rupasinghe, H. P. V., Almquist, K. C., Paliyath, G., Murr, D. P. (2001). Cloning of hmg1 and hmg2 cDNAs encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase and their expression and activity in relation to ɑ-farnesene synthesis in apple. Plant Physiol. Bioch. 39, 933–947. doi: 10.1016/S0981-9428(01)01316-X
Sears, M. T., Zhang, H., Rushton, P. J., Wu, M., Timko, M. P. (2014). NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol. Biol. 84, 49–66. doi: 10.1007/s11103-013-0116-2
Shen, Q., Lu, X., Yan, T., Fu, X., Tang, K. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210, 1269–1281. doi: 10.1111/nph.13874
Tholl, D. (2015). Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biot. 148, 63. doi: 10.1007/10_2014_295
Wallaart, T. E., Pras, N., Beekman, A. C., Quax, W. J. (1999). Seasonal variation of artemisinin and its biosynthetic precursors in plants of Artemisia annua of different geographical origin: proof for the existence of chemotypes. Planta Med. 66, 57–62. doi: 10.1055/s-1999-14094
Xu, Y. H., Liao, Y. C., Lv, F. F., Zhang, Z., Sun, P. W., Gao, Z. H. (2017). Transcription factor AsMYC2 controls the jasmonate-responsive expression of ASS1 regulating sesquiterpene biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 58, 1924–1933. doi: 10.1093/pcp/pcx161
Yang, X., Nambou, K., Wei, L., Hua, Q. (2016). Heterologous production of α-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresour. Technol. 216, 1040–1048. doi: 10.1016/j.biortech.2016.06.028
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Yuan, K., Wang, C., Xin, L., Zhang, A., Ai, C. (2013). Genomic organization and expression analysis of a farnesyl diphosphate synthase gene (FPPS2) in apples (Malus domestica Borkh.). Gene 524, 90–94. doi: 10.1016/j.gene.2013.04.062
Zarei, A., Körbes, A. P., Younessi, P., Montiel, G., Champion, A., Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Zhang, H. B., Bokowiec, M. T., Rushton, P. J. (2012). Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant 5, 73–84. doi: 10.1093/mp/ssr056
Zhang, M., Li, S., Nie, L., Chen, Q., Xu, X., Yu, L., et al. (2015). Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 89, 463–473. doi: 10.1007/s11103-015-0382-2
Zhang, M., Liu, H., Wang, Q., Liu, S. H., Zhang, Y. H. (2020). The 3-hydroxy-3-methylglutaryl-coenzyme A reductase 5 gene from Malus domestica enhances oxidative stress tolerance in Arabidopsis thaliana. Plant Physiol. Bioch. 146, 269–277. doi: 10.1016/j.plaphy.2019.11.031
Zhou, Y., Sun, W., Chen, J., Tan, H., Xiao, Y., Li, Q. (2016). SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 6, 22852. doi: 10.1038/srep22852
Zhu, Z., An, F., Feng, Y., Li, P., Xue, L. A. M., Jiang, Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. P. Natl. Acad. Sci. U.S.A. 108, 12539–12544. doi: 10.1073/pnas.1103959108
Keywords: apple, α-farnesene, α-farnesene synthase, transcriptional regulation, MdMYC2, MdERF3, genetic engineering, terpenes biosynthesis
Citation: Wang Q, Liu H, Zhang M, Liu S, Hao Y and Zhang Y (2020) MdMYC2 and MdERF3 Positively Co-Regulate α-Farnesene Biosynthesis in Apple. Front. Plant Sci. 11:512844. doi: 10.3389/fpls.2020.512844
Received: 18 November 2019; Accepted: 18 August 2020;
Published: 02 September 2020.
Edited by:
Richard Charles Leegood, The University of Sheffield, United KingdomReviewed by:
Raimund Nagel, University of Leipzig, GermanyCopyright © 2020 Wang, Liu, Zhang, Liu, Hao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujin Hao, aGFveXVqaW5Ac2RhdS5lZHUuY24=; Yuanhu Zhang, eXloemhhbmdAc2RhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.