
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 September 2020
Sec. Technical Advances in Plant Science
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.501358
Agrobacterium tumefaciens-mediated gene transfer is the most commonly used method for plant genetic engineering. However, during the period of A. tumefaciens culture, the effects of Agrobacterium culture before inoculation on genetic transformation are poorly understood. In the present study, we investigated the factors that affect the genetic transformation efficiency during Agrobacterium culture using Tamarix hispida as transgenic plant material. Agrobacterium treatment with spermidine (Spe), azacitidine (5-AzaC), dithiothreitol (DTT), or acetosyringone (AS) alone all significantly improved the efficiency of T-DNA transfer. Treatment with 5-AzaC reduced DNA methylation in Agrobacterium to induce the expression of virulence (vir) family genes, including virA, virB1, virC1, virD2, virD4 virE2, and virG. Spe treatment significantly induced the expression of all the studied genes, including virA, virB1, virC1, virD1, virD2, virD4, virE2, and virG. DTT treatment decreased reactive oxygen species accumulation. AS treatment activated the expression of the genes virA, virB1, virC1, virD1, virD2, virD4 and virG. All these effects resulted in increased T-DNA transfer. Additionally, combined Spe, 5-AzaC, DTT, and AS treatment improve Agrobacterium infection to a greater extent compared with their use alone, increasing T-DNA transfer by more than 8-fold relative to no treatment. Therefore, to improve genetic transformation, pretreatment of Agrobacterium during the culture period is important for improving genetic transformation efficiency.
Genetic transformation is a method that transfers DNA of interest into the cell of an organism resulting in genetic alteration, and has been used in many areas of biotechnology, such as studies of gene function, genetic improvement, and molecular breeding. To deliver foreign genes into host plants, three methods are mainly used: Agrobacterium tumefaciens- or Agrobacterium rhizogenes-mediated plant transformation (Hinchee et al., 1988), protoplast transformation (Fischer and Hain, 1995), and particle bombardment (McCabe et al., 1988). Among these methods, Agrobacterium-mediated plant genetic transformation has low cost, is the best choice for plant transformation, and is the predominant method used to generate genetically modified plants. Most transgenic plants (85%) were generated by using Agrobacterium-mediated transformation (Yu et al., 2010). This genetic transformation method has the advantages of simple operation, high reproducibility, low copy number of transgene in the host genome, easier manipulation, and transfer of larger fragments of DNA (Kohli et al., 2003; Ishizaki et al., 2008). However, the efficiency of Agrobacterium-mediated plant genetic transformation could be improved. Transgene delivery mediated by Agrobacterium is affected by many factors, such as explant type, Agrobacterium strain, Agrobacterium concentrations, Agrobacterium suspension medium, the physiological status of the host plant species, and co-cultivation conditions. To improve the efficiency of transgene delivery, multiple factors involved in transformation need to be considered. For instance, phenolic compounds can activate the vir genes to regulate T-DNA transport, and several studies have shown that the addition of a phenolic compound such as acetosyringone (AS), into the co-cultivation medium could highly improve transgenic efficiency (Rashid et al., 1996; Wenck et al., 1999; Rohini and Rao, 2000). During the Agrobacterium infection period, the infected plants will induce a pathogen defense response to generate reactive oxygen species (ROS), and supplementation with of ROS scavenging factors in the co-cultivation medium, such as dithiothreitol (DTT), sodium thiosulfate (STS), and L-cysteine (L-Cys), can reduce ROS accumulation and increase the efficiency of transgene delivery. For instance, Sivanandhan et al. (2015) added thiol compounds, including DTT at 75 mg/l, STS at 125 mg/l, and L-Cys at 100 mg/l, to the co-cultivation medium, resulting in increased rates of genetic transformation. In addition, to improve T-DNA transfer, vacuum infiltration can be used, which could infiltrate plants in a short period of time and is more robust (Chen et al., 2014).
Su et al. (2018) added Spe to the culture medium during Vitis vinifera somatic embryo growth, which increased transgenic resistant somatic embryos induction rate, suggesting that Spe plays a positive role in T-DNA transfer. The above studies significantly improved the efficiency of T-DNA transfer and indicated that the importance of activating the T-DNA transfer capability of Agrobacterium.
However, most previous studies on improvement of T-DNA transfer assessed the period of the transformation and co-cultivation. The treatments were mainly conducted during co-cultivation time or were added to the co-cultivation medium. In the present study, we identified and studied factors that might affect the infection capability of Agrobacterium. Our results also indicated that treatment of Agrobacterium in culture before inoculation is relatively important for T-DNA transfer efficiency and should be considered in Agrobacterium-mediated transformation studies.
The seeds of T. hispida were collected from the Turpan Botanical Garden of the Chinese Academy of Sciences. The seeds of T. hispida were surface-sterilized in 33% (v/v) sodium hypochlorite and grown on Murashige-Skoog (MS) solid medium containing 2% sucrose for germination. Two-month-old seedlings were used for transient transformation investigation.
A single colony of Agrobacterium tumefaciens (EHA105) harboring pCAMBIA1301 was picked and cultured in LB liquid culture supplied with 100 μg/ml rifampicin and 50 μg/ml Kanamycin at 28°C until the OD600 reached 0.8 (about 24 h). The culture was diluted 100-fold to a volume of 50 ml and used for the following treatments. To determine the effects of Spe, the diluted cultures were supplied with 0, 1, 3, 5, and 7 mM Spe, and then cultured at 28°C until the OD600 reached 0.6–0.7 (about 7–8 h). For 5-AzaC treatment, the diluted cultures were added with 0, 10, 20, and 30 μM5-AzaC, and cultured at 28°C until the OD600 reached 0.6–0.7 (about 7–8 h). To test the effect of AS, the diluted cultures containing 0, 100, 120, 150, and 200 μM AS were cultured at 28°C until the OD600 reached 0.6–0.7 (about 7–8 h). For DTT supplementation, the diluted cultures were supplied with 0, 50, 100, and 150 μM DTT, and cultured at 28°C until the OD600 reached 0.6–0.7 (about 7–8 h). After cultivation, the cultures were centrifuged at 2,800 × g to pellet the Agrobacterium cells, and then used for transient transformation.
Transient transformation mediated by Agrobacterium was performed according to the method described by Zang et al. (2017). Briefly, the pellet of A. tumefaciens EHA105 cells were centrifuged at 2,800 × g for 10 min, and then adjusted to an OD600 of 0.8 using a solution comprising 1/2 MS +10 mM CaCl2+.5% (w/v) sucrose + 100 μM acetosyringone + Tween20 (0.02%, v/v), pH 5.8 (transformation solution). The plants were soaked in the transformation solution with shaking at 90 rpm for 2 h at 25°C. The plants were then washed quickly with distilled water for 10 s to remove the excessive A. tumefaciens cells. After washing, the plants were planted vertically on 1/2 MS solid medium [1/2 MS+1% (w/v) sucrose+100 μM acetosyringone, pH 5.8]. After culture for 72 h, the plants were washed with distilled water and wiped with sterile filter paper to remove excessive water. These plants were harvested for GUS staining and GUS activity assays. Each sample contains at least 10 seedlings, and three biological repeats were performed.
GUS activity was measured following the method described by Jefferson et al. (1987). Briefly, the samples were ground into a fine powder under liquid nitrogen, and then homogenized in an extraction buffer (50 mM NaH2PO4-Na2HPO4, pH 7.4, 10 mM β-mercaptoethanol, 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine). Then, 1 mM 4-methylumbelliferyl-β-d-glucuronide (MUG) was added into the extraction buffer, the enzyme reaction was performed at 37°C, and was stopped by adding 500 μl of 0.2 M Na2CO3. After stopping the enzyme reaction, the fluorescence of 4-methylumbelliferone was checked using a DyNA Quant fluorometer (Hoefer Pharmacia Biotech Inc., San Francisco, CA), and a protein standard curve was generated using the Bradford assay to calculate the GUS activity.
The ROS content was determined using a Hydrogen Peroxide Content measuring kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer’s protocol. The DNA methylation content was determined using Methylated DNA Quantification Kit (Fluorometric) according to the manufacturer’s protocol (Epigentek Group Inc., China). Three independent biological replications were performed.
Total RNA from T. hispida or Agrobacterium cells was isolated using the Trizol reagent (Promega) and treated with DNaseI (RNase free) to remove DNA contamination. The integrality of RNA was analyzed using agarose gel electrophoresis. For the synthesis of T. hispida cDNA, 1 μg of total RNA from each sample was reverse-transcribed using a random primer (9 mer) as synthesis primers with a PrimeScript™ RT reagent Kit (Takara, China) in volume of 10 μl according to its instruction. The products were subsequently diluted to 100 μl and used as the template for qRT-PCR. For T. hispada, two internal references were used, including β-tubulin (GenBank number: FJ618519) and α-tubulin (GenBank number: FJ618518). For Agrobacterium cells, 16S rRNA was used as internal reference. All the primers used are shown as Table S1. qRT-PCR was performed on a qTOWER2.0 (Analytic Jena, Germany). The qRT-PCR reaction mixture contained 10 μl of SYBR Green Real-time PCR Master Mix (Takara), forward and reverse primers (0.5 μM each), and cDNA template (2 μl cDNA that is equivalent to the transcript from total RNA 20 ng) in a volume of 20 μl. qRT-PCR was conducted according to the following cycling parameters: 94°C for 30 s; followed by 40 cycles at 94°C for 10 s, 59°C for 30 s, and 72°C for 40 s. Three independent biological replicates were performed, and the expression levels were calculated from the Ct (cycle threshold) using the 2-ΔΔCt method (Livak and Schmittgen, 2001)
Statistical analyses were carried out using the SPSS 16.0 software package (SPSSInc, Chicago, IL, USA). data were compared using one-way analysis of variance (ANOVA) and differences were considered statistically significant at P < 0.05.
Agrobacterium cells treated with 5-AzaC before transformation significantly elevated β-glucuronidase (GUS) expression and activity in transient transformed T. hispida plants (Figures 1A, B). GUS expression and activity was not significantly activated by 10 μM 5-AzaC treatment but was by 20 and 30 μM AzaC treatment. The expression and activity of GUS reached peak levels after treatment with 20 μM 5-AzaC (Figures 1A, B). 5-AzaC is a DNA demethylation agent; therefore, treatment with 5-AzaC may decrease DNA methylation in Agrobacterium cells. The level of DNA methylation was investigated in Agrobacterium cells after 5-AzaC treatment. The results showed that 20 and 30 μM 5-AzaC treatment significantly decreased DNA methylation; however, 20 μM 5-AzaC reduced DNA methylation to a greater extent than 30 μM AzaC (Figure 1C), which is consistent with the trend of T-DNA transfer efficiency (Figure 1A).
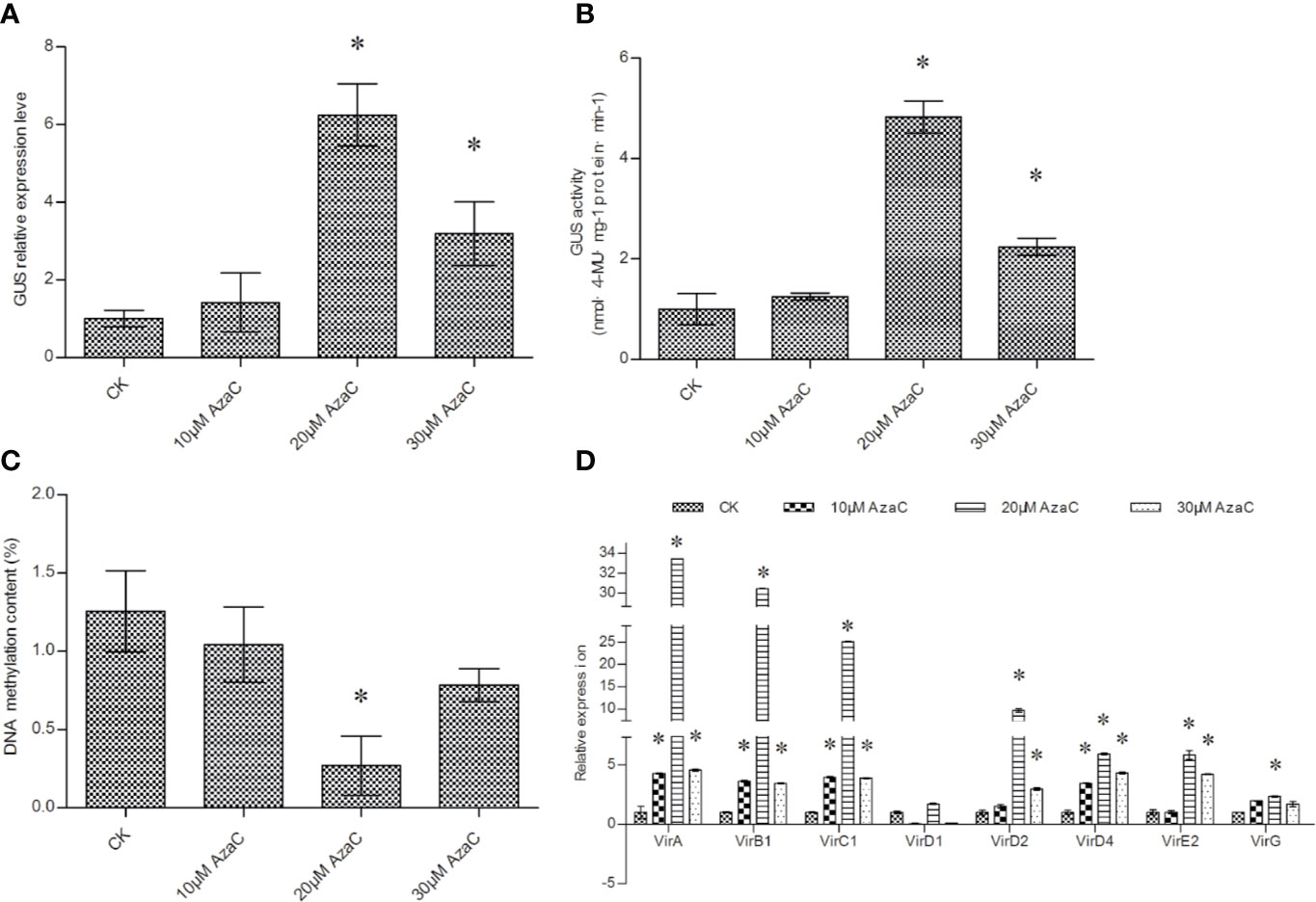
Figure 1 The efficiency of T-DNA transfer induced by 5-AzaC treatment. (A) Analysis of GUS expression to determine the effects of 5-AzaC treatment of Agrobacterium tumefaciens cells. (B) GUS activity analysis of T. hispida plants transiently transformed using Agrobacterium cells treated with 5-AzaC. (C) The effects of 5-AzaC on total DNA methylation in Agrobacterium cells. DNA methylation was analyzed on Agrobacterium cells treated with different concentrations of 5-AzaC. (D) The relative expression of vir family genes in Agrobacterium cells in response to 5-AzaC supplements. The relative expression was calculated as the transcript level at different treatments divided by the transcript level without treatment (0 μM). LB medium was added with 0, 10, 20, or 30 mM 5-AzaC to culture Agrobacterium cells before genetic transformation, and qRT-PCR was performed to determine the expression of the vir genes. Data are means ± SD from three independent experiments. (*) indicates a significant difference with the wild-type (WT) at P < 0.05.
DNA methylation usually correlates with gene expression; therefore, we further studied the expression of different vir genes in response to 5-AzaC treatment. The results showed that 10, 20, and 30 μM 5-AzaC all significantly induced the expression vir genes. However, 20 μM 5-AzaC treatment activated the expression of vir genes more potently than 10 and 30 μM 5-AzaC The virA, virB1, virE2, virC1, virD2, virD4, and virG genes were all significantly activated by 20 μM 5-AzaC treatment (Figure 1D).
To study the effect Spe treatment on T-DNA transfer, different concentrations of Spe were supplied in Luria-Bertani (LB) medium for Agrobacterium cultivation. The cultures were harvested for transient transformation. After transient transformation, T-DNA transfer efficiency was analyzed by determining GUS expression in the transiently transformed plants. Compared with the control (transient transformation with untreated Agrobacterium cells), supplementation with 5 and 7 mM Spe significantly increased GUS expression and activity (Figures 2A, B). GUS expression and activity increased to a greater extent under 5 mM Spe than under 7 mM Spe (Figures 2A, B). These results suggested that supplementation with Spe at the appropriate level can significantly improve genetic transformation.
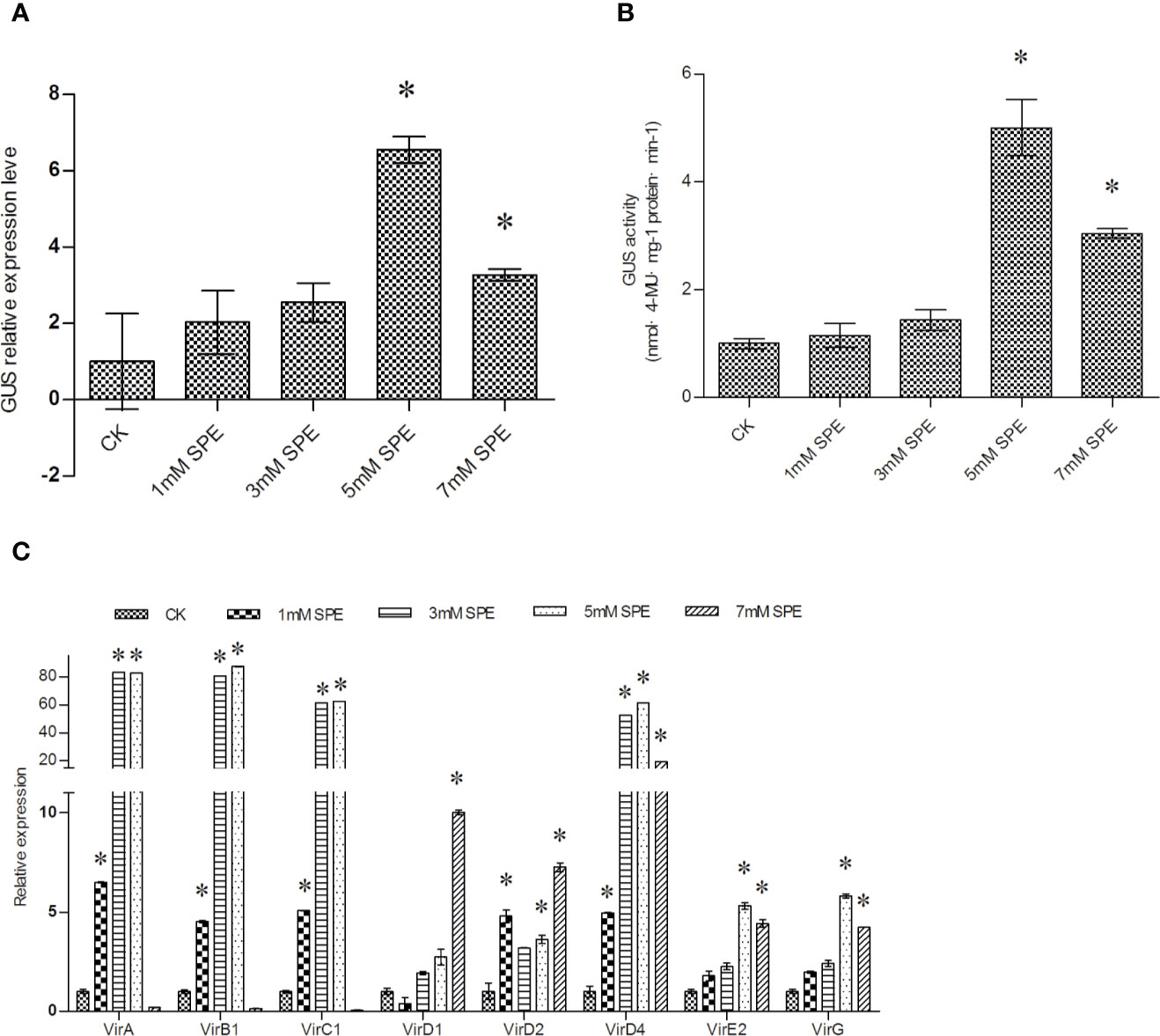
Figure 2 T-DNA transfer efficiency was improved when Agrobacterium cells were treated with spermidine (Spe). (A) The effect of Spe treatment on the efficiency of T-DNA transfer was determined via GUS expression analysis; (B) GUS activity analysis on the transformed T. hispida plants using Agrobacterium cells treated with Spe. (C) The relative expression of vir genes in Agrobacterium cells when treated with different concentrations of Spe. The relative expression was calculated as the transcript level at different treatments divided by the transcript level without treatment (0 mM Spe). For A. tumefaciens treatment, 0, 1, 3, 5, or 7 mM Spe was added into LB medium before genetic transformation, and qRT-PCR was performed to determine the expression of the vir genes. Data are means ± SD from three independent experiments. (*) indicates a significant difference with the wild-type (WT) at P < 0.05.
To further study the mechanism of the improved genetic transformation mediated by Spe treatment, the expression levels of different vir genes were studied. The results showed that except for virD1, all the studied vir genes were significantly induced by treatment with 5 mM Spe (Figure 2C). These results indicated that Spe treatment could induce the expression of certain vir family genes, leading to improved T-DNA transfer efficiency.
Different concentrations of DTT were used in the culture of Agrobacterium before transient transformation. GUS expression and activity measurements showed that supplementation with DTT could significantly improve T-DNA transfer. DTT at concentrations from 50 to 150 μM could significantly increase T-DNA transfer. The transformation efficiency reached a peak at 100 μM DTT (Figures 3A, B).
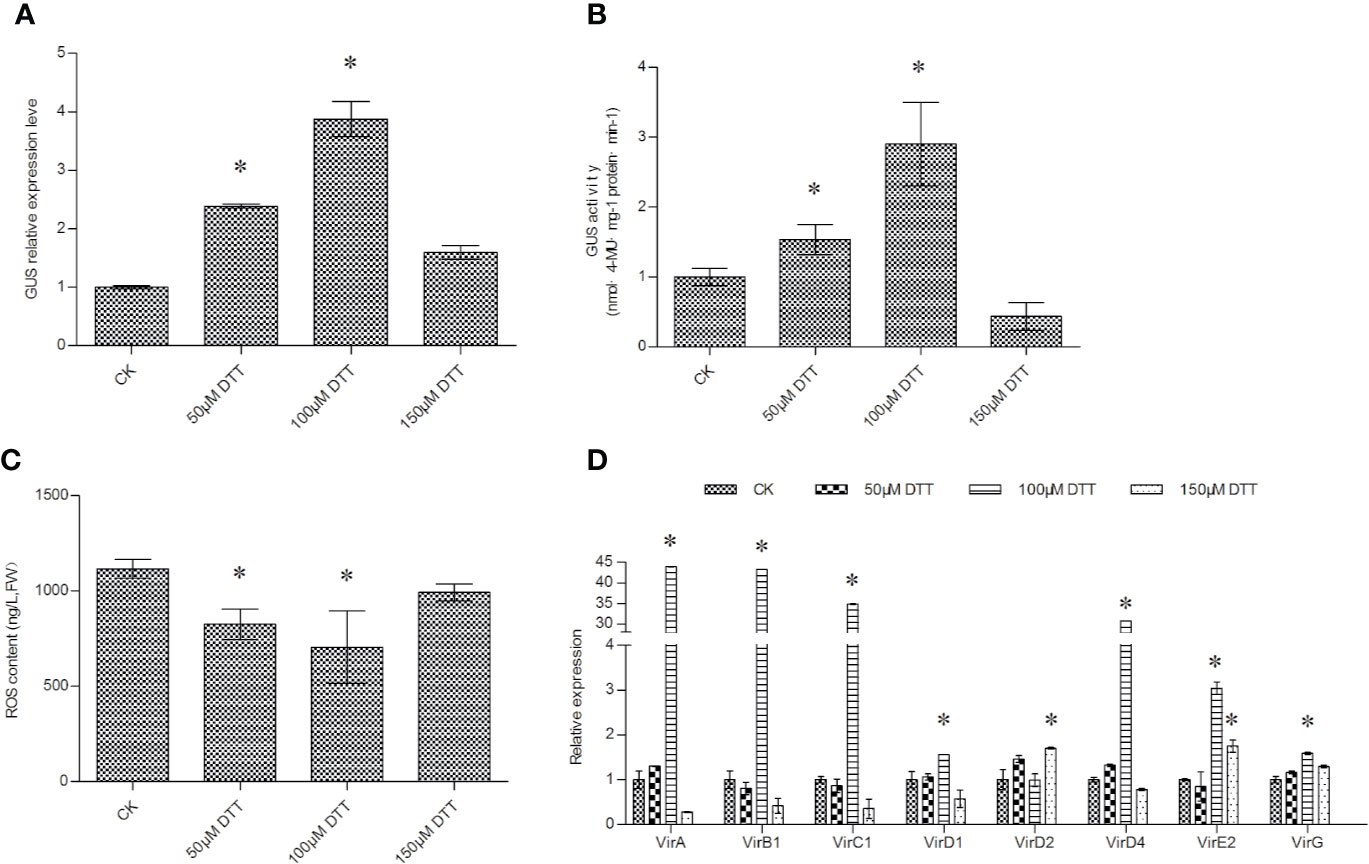
Figure 3 DTT treatment of Agrobacterium cells improves the efficiency of T-DNA transfer. (A) The effect of DTT treatment on the efficiency of T-DNA transfer was determined via GUS expression analysis; (B) GUS activity analysis on the transiently transformed T. hispida plants. (C) ROS accumulation in Agrobacterium cells in response to treatment with different concentrations of DTT. (D) The relative expression of vir genes in Agrobacterium cells when treated with different concentrations of DTT. The relative expression was calculated as the transcript level at different treatments divided by the transcript level without treatment (0 μM DTT). For treatment, 0, 50, 100, or 150 μM of DTT was added into LB medium to culture A. tumefaciens before genetic transformation, and qRT-PCR was performed to determine the expression of the GUS gene. Data are means ± SD from three independent experiments. (*) indicates a significant difference with the wild-type (WT) at P < 0.05.
DTT plays a role in ROS scavenging; therefore, the ROS concentration in Agrobacterium cells was determined. The results showed that 50, 100, and 150 μM DTT all significant reduced ROS accumulation in Agrobacterium cells (Figure 3C). The decreased ROS accumulation would reduce the damage to Agrobacterium cells, which might contribute to increased T-DNA transfer. In addition, supplement with DTT during culture period can induce the expression of all studied vir genes, especially 100 μM DTT was added (Figure 3D), which is consistent with the results that 100 μM DTT could highly activate T-DNA transformation.
AS was added into LB medium to culture Agrobacterium cells before transient transformation. Supplementation with AS from 100 to 200 μM could significantly improve T-DNA transfer, with 120 μM AS treatment producing the largest effect (Figures 4A, B). The expression of vir genes induced by AS were further studied. The results showed that compared with other AS concentrations, 120 μM AS treatment could significantly induce the expression of all vir genes, except virE2. In particular, 120 μM AS treatment induced vir gene expression to a greater extent than 100 and 150 μM AS. However, 200 μM AS treatment only induced high expression of virD1 and virD2 compared with 150 μM AS treatment (Figure 4C). These results indicated that AS treatment during Agrobacterium cell culture could induce the expression of vir genes to improve T-DNA transfer.
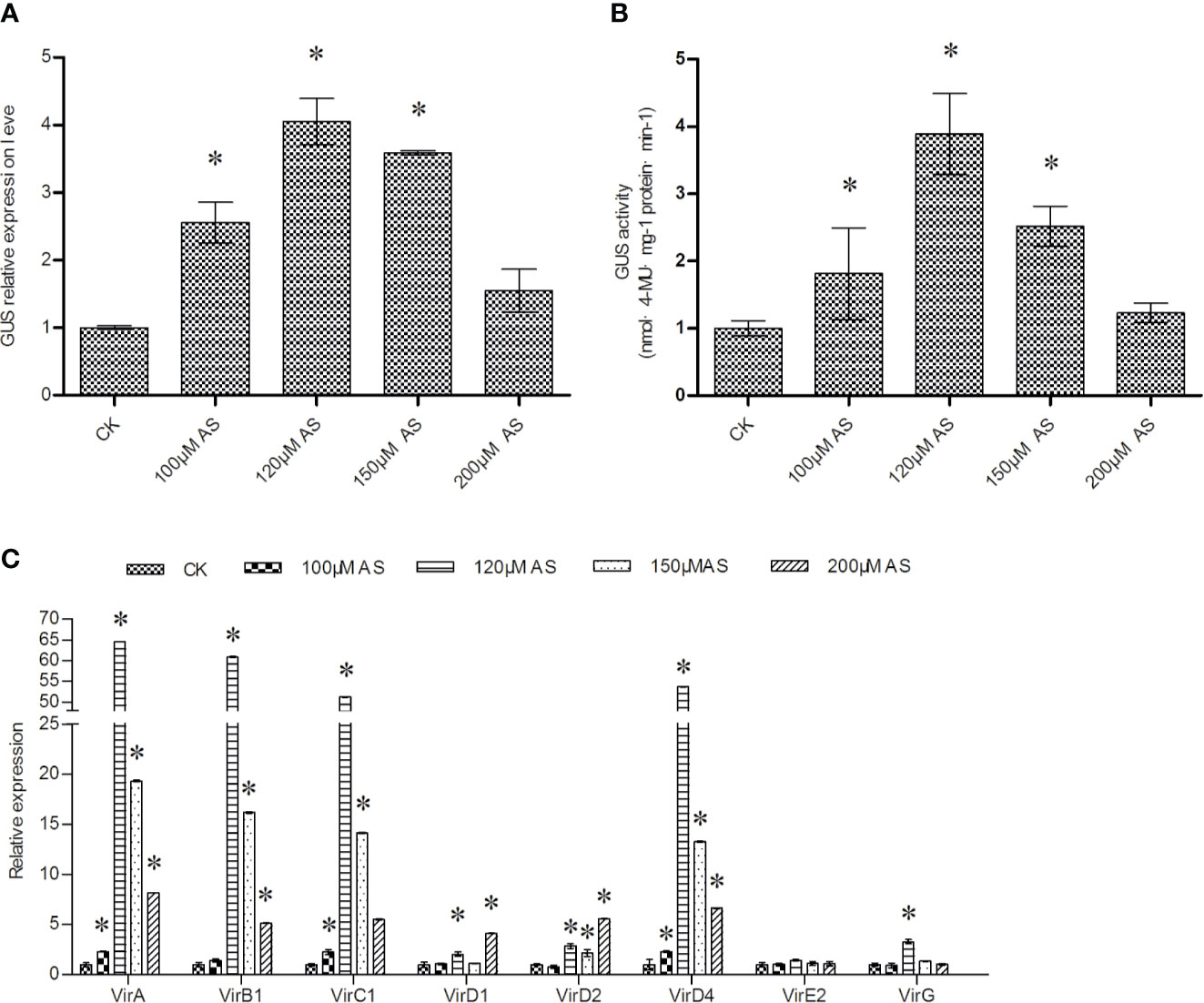
Figure 4 The effects of AS treatment on T-DNA transfer efficiency. (A) Analysis of GUS expression to determine the effects of AS treatment of Agrobacterium cells; (B) GUS activity analysis in T. hispida plants transiently transformed with Agrobacterium treated with AS. (C) The relative expression of vir family genes in Agrobacterium cells treated with different concentrations of AS. The relative expression was calculated as the transcript level at different treatments divided by the transcript level without treatment (0 μM AS). To culture A. tumefaciens, 0, 100, 120, 150, or 200 μM AS were added separately into LB medium before genetic transformation, and qRT-PCR was performed to determine the expression of the vir genes. Data are means ± SD from three independent experiments. (*) indicates a significant difference with the wild-type (WT) at P < 0.05.
The effects of a combination of Spe, 5-AzaC, DTT and AS on T-DNA transfer were investigated. The mixture of 100 μM DTT, 20 μM 5-AzaC, 5 mM SPE, and 120 μM AS were added into LB medium to culture Agrobacterium cells before transformation. Both GUS expression and GUS activity measurements showed that the combined treatment could increase the efficiency of T-DNA transfer to a greater extent than any of the reagents used alone, and could improve GUS expression by more than eight-fold (Figures 5A, B). This result indicated that DTT, Azac, SPE, and AS could work coordinately to activate T-DNA transfer.
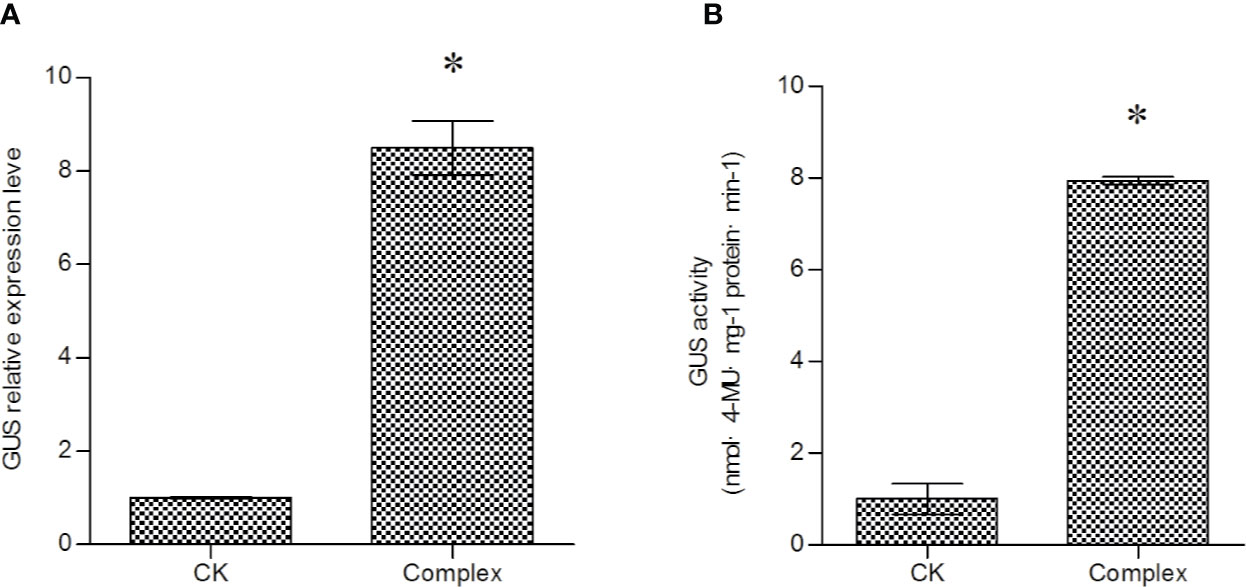
Figure 5 The combined effect of 5-AzaC, DTT, AS, and Spe on T-DNA transfer. (A) Investigation of T-DNA transfer efficiency induced by combined treatment with 5-AzaC, DTT, AS, and Spe; the expression of the GUS gene was analyzed to determine T-DNA transfer efficiency. (B) Analysis of GUS activity in transiently transformed T. hispida plants. For A. tumefaciens treatment, 20 mM 5-AzaC, 120 μM AS, 5 mM Spe, and 100 μM DTT were added together into LB medium to culture A. tumefaciens before genetic transformation, and qRT-PCR was performed to determine the expression of the GUS gene. Data are means ± SD from three independent experiments. (*) indicates a significant difference with the wild-type (WT) at P < 0.05.
Our results showed that treatment of Agrobacterium cells before genetic transformation could significantly increase the T-DNA transfer efficiency, and 5-AzaC, Spe, DTT, and AS all exert positive roles to improve T-DNA transfer. Combined use of 5-AzaC, Spe, DTT, and AS improved T-DNA transfer by more than eight-fold, suggesting that treatment of Agrobacterium cells before genetic transformation is important and should be used in genetic transformation.
Transient transformation can accurately reflect the efficiency of T-DNA transfer. In addition, because transfer of T-DNA into cells is the first step to obtain stable transformation, determining of the efficiency of transient transformation could reflect the efficiency of stable genetic transformation. Wild type of A. tumefaciens is a gram-negative bacterium that can transfer genes into host plants, which leads to tumors. The Ti plasmid in A. tumefaciens contains different vir family genes and a transfer DNA (T-DNA) region. VirA and virG families serve as transcriptional activators for vir gene transfer and transformation of higher plants (Gao and Lynn, 2005). The virB proteins and the virD4 protein can form a membrane complex to mediate the transfer of T-DNA to plant cells (Vergunst et al., 2005). VirG serve as a positive regulatory protein that can activate vir gene expression together with plant molecules (Chen et al., 1991). Michielse et al. (2005) showed that during gene transformation, virC and virD excise the T-DNA region from the Ti plasmid, and then the excised T-DNA forms a T-DNA complex with virD and virE2 to enter host plant cells. The T-DNA finally enters the plant nucleus, and the virD and virE2 proteins are stripped from T-DNA complex when the T-DNA enters the cell nucleus (Nonaka et al., 2017). In the present study, the results showed that during the period of Agrobacterium cell culture, treatment with 5-AzaC, Spe, and AS all highly induce vir gene expression (Figures 1–4), suggesting that these factors might improve T-DNA transfer by directly activating the vir genes.
Azacytidine is a DNA demethylating agent that can reduce or inhibit DNA methylation. Palmgren et al. (1993) showed that Agrobacterium cells treated with 5-AzaC showed significantly increased transient expression efficiency, and AzaC also inhibits the methylation-dependent inactivation of the transformed gene. These results suggested that 5-AzaC treatment could improve the genetic transformation efficiency. The results of the present study showed that 5-AzaC treatment could reduce the methylation of Agrobacterium DNA (Figure 1), which leads to the increased expression of vir genes, which ultimately improved the transformation efficiency. Therefore, reduction of methylation of Agrobacterium DNA is important for enhancing T-DNA transfer.
Kumar and Rajam (2005) showed that treatment of Agrobacterium cells with polyamine prior to transformation, including putrescine and Spe, could induce T-DNA transfer. These results showed that Spe plays a role in inducing genetic transformation. In the present study, Spe treatment of Agrobacterium cells highly induced T-DNA transfer in T. hispida (Figure 2A); at the same time, all the studied vir genes were significantly induced by Spe (Figure 2C). These results suggested that Spe could improve T-DNA transfer by inducing the expression of virA, virB1, virC1, virD1, virD2, virD4, virE2, and virG gene.
DTT is an antioxidant that efficiently prevents oxidation of nucleic acids by phenolic compounds and has been used to stabilize enzymes and other proteins containing sulfhydryl groups (Thanh et al., 2009; Chen et al., 2011). Supplement with DTT during inoculation period could improve genetic transformation (Ahn et al., 2013; Li et al., 2017). Our results showed that DTT treatment could significantly reduce ROS accumulation in Agrobacterium cells, and the ROS level was negatively correlated with the transformation efficiency (Figure 3C). These results suggested that ROS accumulation might damage Agrobacterium cells to decrease their infection capability. Therefore, reducing ROS accumulation by DTT treatment would improve T-DNA transfer efficiency in T. hispida. Therefore, DTT is also an important genetic transformation inducer.
Our results showed that treatment with 5-AzaC or Spe increased T-DNA transfer compared with treatments DTT and AS (Figures 1–4). However, AS is widely used to induce efficient transformation in A. tumefaciens-mediated genetic transformation. These results indicated that 5-AzaC and Spe might be superior to AS in the induction of T-DNA transfer during Agrobacterium cell culture. 5- AzaC at 20 μM or 5 mM Spe are the most suitable concentrations to improve T-DNA transfer (Figures 1A, 2A), and 120 μM AS is also the most suitable concentration for improving T-DNA transfer (Figure 4A). However, 20 μM AzaC and 5 mM Spe both improved T-DNA transfer to a greater extent than 120 μM AS (Figures 1A, 4A). In addition, treatment with 5-AzaC seems to be more efficient in inducing T-DNA transfer, because 20 μM AzaC treatment could induce T-DNA transformation more efficiently than 120 μM AS in T. hispida (Figures 1D, 4C). Therefore, 5-AzaC should be considered for use in A. tumefaciens-mediated transformation rather than AS.
The datasets generated for this study are available on request to the corresponding author.
YW conceived and directed the project. HZ performed the experiments. YC and YJ performed the overall data analysis. HZ and YW wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (No. 31770704). Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team) and the Overseas Expertise Introduction Project for Discipline Innovation (B16010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.501358/full#supplementary-material
Ahn, Y., Yoon, M., Jeon, J. (2013). Development of an efficient Agrobacterium-mediated transformation system and production of herbicide-resistant transgenic plants in garlic (Allium sativum L.). Mol. Cells 36, 158–162. doi: 10.1007/s10059-013-0142-6
Chen, C. Y., Wang, L., Winans, S. C. (1991). Characterization of the supervirulent virG gene of the Agrobacterium tumefaciens plasmid pTiBo542. Mol. Gen. Genet. 230, 302–309. doi: 10.1007/BF00290681
Chen, C. H., Chang, Y. J., Ku, M. S., Chung, K. T., Yang, J. T. (2011). Enhancement of temozolomide-induced apoptosis by valproic acid in human glioma cell lines through redox regulation. J. Mol. Med. (Berl). 89, 303–315. doi: 10.1007/s00109-010-0707-1
Chen, Q., Lai, H., Hurtado, J., Stahnke, J., Leuzinger, K., Dent, M. (2014). Agroinfiltration as an effective and scalable strategy of gene delivery for production of pharmaceutical proteins. Adv. Tech. Biol. Med. 1, 103. doi: 10.4172/atbm.1000103
Fischer, R., Hain, R. (1995). Tobacco Protoplast Transformation and Use for Functional Analysis of Newly Isolated Genes and Gene Constructs. Methods Cell Biol. 50, 401–410. doi: 10.1016/S0091-679X(08)61046-8
Gao, R., Lynn, D. G. (2005). Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J. Bacteriol. 187, 2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005
Hinchee, M. A. W., Connor-Ward, D. V., Newell, C. A., McDonnell, R. E., Sato, S. J., Gasser, C. S., et al. (1988). Production of transgenic soybean plants using agrobacterium-mediated DNA transfer. Nat. Biotechnol. 6, 915–922. doi: 10.1038/nbt0888-915
Ishizaki, K., Chiyoda, S., Yamato, K. T., Kohchi, K. (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091. doi: 10.1093/pcp/pcn085
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 13, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Kohli, A., Twyman, R. M., Abranches, R., Wegel, E., Stoger, E., Christou, P. (2003). Transgene integration, organization and interaction in plants. Plant Mol. Biol. 52, 247–258. doi: 10.1023/A:1023941407376
Kumar, S. V., Rajam, M. V. (2005). Polyamines enhance Agrobacterium tumefaciens vir gene induction and T-DNA transfer. Plant Sci. 168, 475–480. doi: 10.1016/j.plantsci.2004.09.018
Li, S. X., Cong, Y. H., Liu, Y. P., Wang, T. T., Shuai, Q., Chen, N. N., et al. (2017). Optimization of Agrobacterium-Mediated Transformation in Soybean. Front. Plant Sci. 24, 8–246. doi: 10.3389/fpls.2017.00246
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)). Method 4, 402–408. doi: 10.1006/meth.2001.1262
McCabe, D. E., Swain, W. F., Martinell, B. J., Christou, P. (1988). Stable Transformation of Soybean (Glycine Max) by Particle Acceleration. Nat. Biotechnol. 6, 923–926. doi: 10.1038/nbt0888-923
Michielse, C. B., Arentshorst, M., Ram, A. F. J., van den Hondel, C.A.M.J.J. (2005). Agrobacterium -mediated transformation leads to improved gene replacement efficiency in Aspergillus awamori. Fungal Genet. Biol. 42, 9–19. doi: 10.1016/j.fgb.2004.06.009
Nonaka, S., Someya, T., Zhou, S., Takayama, M., Nakamura, K., Ezura, H. (2017). An Agrobacterium tumefaciens Strain with Gamma-Aminobutyric Acid Transaminase Activity Shows an Enhanced Genetic Transformation Ability in Plants. Sci. Rep. 7, 426–449. doi: 10.1038/srep42649
Palmgren, G., Mattson, O., Okkels, F. T. (1993). Treatment of Agrobacterium or leaf disks with 5-azacytidine increases transgene expression in tobacco. Plant Mol. Biol. 21, 429–435. doi: 10.1007/BF00028801
Rashid, H., Yokoi, S., Toriyama, K., Hinata, K. (1996). Transgenic plant production mediated by Agrobacterium in Indica rice. Plant Cell Rep. 15, 727–730. doi: 10.1007/BF00232216
Rohini, V., Rao, K. S. (2000). Transformation of peanut (Arachis hypogaea L.): a non-tissue culture based approach for generating transgenic plants. Plant Sci. 150, 41–49. doi: 10.1016/S0168-9452(99)00160-0
Sivanandhan, G., Kapil, D. G., Theboral, J., Selvaraj, N., Ganapathi, A., Manickavasagam, M. (2015). Sonication, Vacuum infiltration and thiol compounds enhance the Agrobacterium-Mediated transformation frequency of Withania somnifera (L.) Dunal. PLoS One. doi: 10.1371/journal.pone.0124693. eCollection 2015.
Su, H., Jiao, Y. T., Wang, F. F., Liu, Y. E., Niu, W. L., Liu, G. T., et al. (2018). Overexpression of VpPR10.1 by an efficient transformation method enhances downy mildew resistance in V. vinifera. Plant Cell Rep. 37, 819–832. doi: 10.1007/s00299-018-2271-z
Thanh, T., Omar, H., Abdullah, M. P., Chi, V. T., Noroozi, M., Ky, H., et al. (2009). Rapid and effective method of RNA isolation from green microalga Ankistrodesmus convolutus. Mol. Biotechnol. 43, 148–153. doi: 10.1007/s12033-009-9182-8
Vergunst, A. C., van Lier, M. C. M., Dulk-Ras, A. D., Grosse Stüve, T. A., Ouwehand, A., Hooykaas, P. J. J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. 102, 832–837. doi: 10.1073/pnas.0406241102
Wenck, A. R., Quinn, M., Whetten, R. W., Pullman, G., Sederoff, R. (1999). High-efficiency Agrobacterium-mediated transformation of Norway spruce (Picea abies) and loblolly pine (Pinus taeda). Plant Mol. Biol. 39, 407–416. doi: 10.1023/A:1006126609534
Yu, Y., Liang, H. Z., Wang, S. F., Lian, Y., Wei, Y. L., Wang, T. F. (2010). Research progress and commercialization on transgenic soybean in China. Soybean Sci. 29, 143–150. doi: 10.11861/j.issn.1000-9841.2010.01.0143
Keywords: Agrobacterium-mediated transformation method, spermidine (Spe), azacitidine (5-AzaC), dithiothreitol (DTT), acetosyringone (AS), T-DNA transfer, Agrobacterium infection
Citation: Zhao H, Jia Y, Cao Y and Wang Y (2020) Improving T-DNA Transfer to Tamarix hispida by Adding Chemical Compounds During Agrobacterium tumefaciens Culture. Front. Plant Sci. 11:501358. doi: 10.3389/fpls.2020.501358
Received: 02 October 2019; Accepted: 10 September 2020;
Published: 29 September 2020.
Edited by:
Roger Deal, Emory University, United StatesReviewed by:
Sadiye Hayta, John Innes Centre, United KingdomCopyright © 2020 Zhao, Jia, Cao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yucheng Wang, d2FuZ3l1Y2hlbmdAbXMueGpiLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.