
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 18 August 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01267
This article is part of the Research Topic Understanding C4 Evolution and Function View all 12 articles
Global climate change is a challenge for efforts to ensure food security for future generations. It will affect crop yields through changes in temperature and precipitation, as well as the nutritional quality of crops. Increased atmospheric CO2 leads to a penalty in the content of proteins and micronutrients in most staple crops, with the possible exception of C4 crops. It is essential to understand the control of nutrient homeostasis to mitigate this penalty. However, despite the importance of mineral nutrition for plant performance, comparably less is known about the regulation of nutrient uptake and homeostasis in C4 plants than in C3 plants and mineral nutrition has not been a strong focus of the C4 research. Here we review what is known about C4 specific features of nitrogen and sulfur assimilation as well as of homeostasis of other essential elements. We identify the major knowledge gaps and urgent questions for future research. We argue that adaptations in mineral nutrition were an integral part of the evolution of C4 photosynthesis and should be considered in the attempts to engineer C4 photosynthetic mechanisms into C3 crops.
As global population continues to increase, crop yields must increase proportionally to meet the future demand for food. However, the quantity of food is not the only threat to food security, but also the nutritional quality of the food produced (Myers et al., 2017). Indeed, micronutrient deficiencies are estimated to affect over 2 billion people worldwide (Amoroso, 2016). Thus, micronutrient deficiencies impinge on agricultural production, food security, and human health. Global climate change is another factor negatively influencing crop nutritional quality. Many crops grown under the predicted elevated atmospheric CO2 concentration show an increase in yield, but a decrease in micronutrients (zinc, iron) and proteins (as nitrogen) (Loladze, 2014; Zhu et al., 2018; Ujiie et al., 2019). This decrease is partly due to an increased synthesis of carbohydrates at the expense of proteins, often referred to as the carbon dilution effect. However, it is also caused by the immobilization of nitrogen in vegetative tissues and soil (Luo et al., 2004) and by direct reduction in nitrate assimilation by elevated CO2 (Bloom et al., 2010). Interestingly, at least for rice, the decreased protein and nitrogen content observed is not completely due to a general carbon dilution, but due to differential responses of the superior grains (derived from early flowers) and the inferior grains (derived from late flowers) to elevated CO2 (Zhang et al., 2013). Nitrogen content decreases in superior grains, but it does not change in inferior grains. However, inferior grains are frequently lost during harvest, which further decreases the total grain protein yield (Zhang et al., 2013). Decreased protein content in crops means sulfur will also be less available for human nutrition as plant proteins are the primary source of the essential sulfur-containing amino acid methionine (Parcell, 2002). Indeed, independent FACE (free‐air CO2 enrichment) experiments in wheat showed a 7% decrease in total grain sulfur and an 8% decrease in methionine and cysteine content (Hogy et al., 2009; Fernando et al., 2012). This nutrient penalty has been observed for multiple crops, with one notable exception—C4 crops (Myers et al., 2014). Presumably, because C4 crops profit much less from elevated CO2 as carbon uptake in C4 plants is saturated at ambient CO2 levels (Von Caemmerer and Furbank, 2003), no carbon dilution effect occurs, and the elevated CO2 does not affect protein and micronutrient levels. Thus, C4 crops have great potential to deliver sufficient nutrients for human food and health. However, more effort is needed to understand the control of nutrient fluxes and homeostasis in C4 plants to ensure that this will also be true in the coming decades.
Compared to C3 crops, such as rice, wheat, or oil-seed rape, less is known about specific alterations in mineral nutrition of C4 plants, despite substantial differences in the organization of nitrate and sulfate assimilation (Jobe et al., 2019). Therefore, in this review, we summarize what is known about C4 specific features of nitrogen and sulfur metabolism as well as of homeostasis of other essential elements. To identify the major knowledge gaps and urgent questions for future research, we relate the current knowledge of plant mineral nutrition in C3 vs. C4 plants with future needs for human nutrition and health and with the predicted changes in atmospheric CO2 levels. Finally, we discuss future directions and approaches to prevent additional declines in the nutritional quality of crops, mainly engineering C4 photosynthetic mechanisms into C3 crops.
Rubisco, the enzyme responsible for assimilating CO2 into reduced carbon compounds, is an inefficient catalyst under the current atmospheric conditions (Parry et al., 2013; Pottier et al., 2018; Ashida et al., 2019). This inefficiency arises because the carboxylase function of Rubisco can be competitively inhibited by atmospheric oxygen. Thus, many photosynthetic organisms have evolved CO2 concentrating mechanisms to boost the efficiency of Rubisco by increasing the concentration of CO2 at the site of carboxylation. Plants using the C4 photosynthetic pathway accomplish this by dividing the photosynthetic process into two specialized cell types (Figure 1). Within mesophyll cells (MC), the initial CO2 fixation step occurs via carboxylation of phosphoenolpyruvate using the enzyme phosphoenolpyruvate carboxylase (PEPC) (Hatch and Slack, 1966; Slack and Hatch, 1967). This is an essential step because PEPC is not inhibited by atmospheric oxygen. The product of this reaction is a four-carbon organic acid that then moves into the bundle sheath (BS) cells, where it is decarboxylated, releasing CO2 for Rubisco. Because of the low oxygen environment in the BS cells, Rubisco can operate near its maximal efficiency. This pathway has evolved independently in angiosperms at least 66 times, representing three families of monocots and 16 families of dicots (Von Caemmerer and Furbank, 2003; Sage et al., 2012).
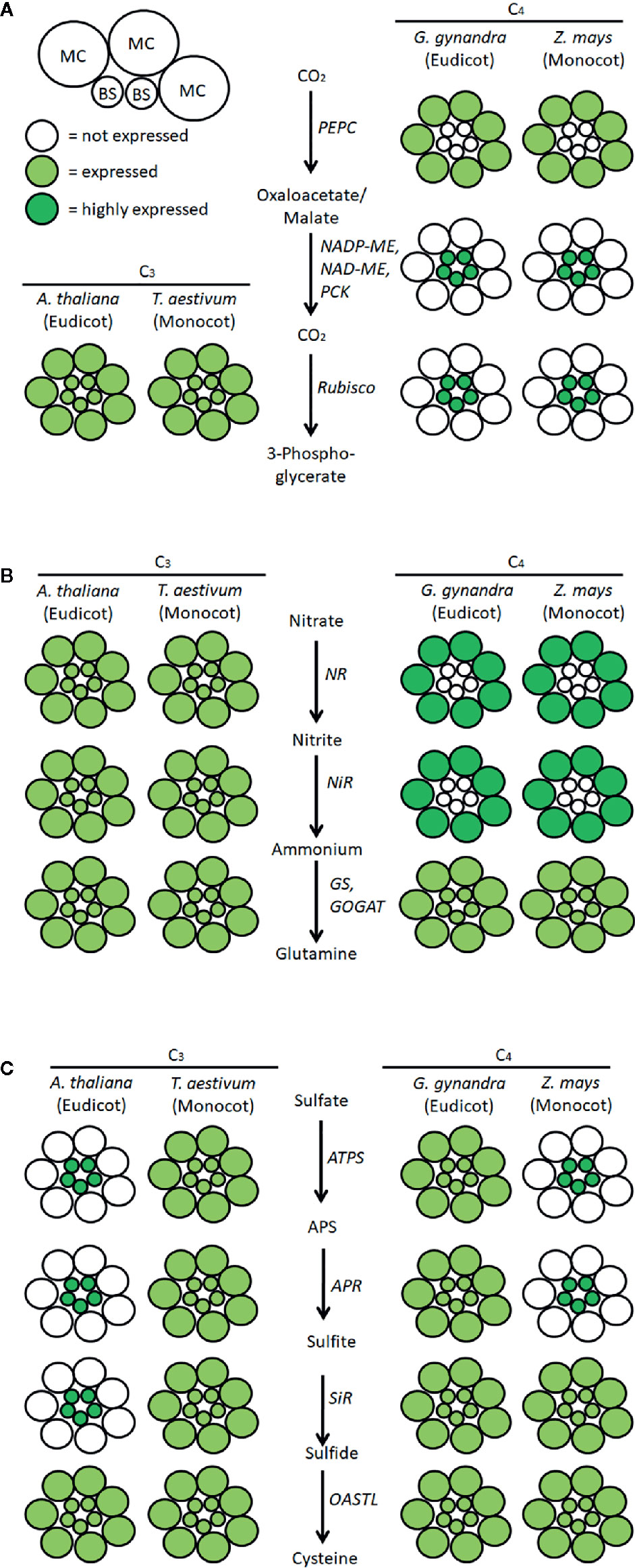
Figure 1 Scheme of localization of the pathways of carbon, nitrate, and sulfate assimilation in exemplary C3 and C4 plants. Schematic localization of key enzymes of (A) CO2, (B) nitrate, and (C) sulfate assimilation in mesophyll cells (MC) and bundle sheath cells (BS) of representative species for C3 and C4 monocots and dicots was compiled from the literature described in the manuscript. MC, mesophyll cells; BS, bundle sheath cells; PEPC, phosphoenolpyruvate carboxylase; ME, malic enzyme; PCK, phosphoenolpyruvate carboxykinase; NR, nitrate reductase; NiR, nitrite reductase, GS, glutamine synthetase; GOGAT, glutamate synthase; ATPS, ATP sulfurylase; APS, adenosine 5´-phosphosulfate; APR, APS reductase; SiR, sulfite reductase; OASTL, O-acetylserine (thiol)lyase.
While these independent C4 lineages share many characteristics, there are also significant differences in the C4-acid decarboxylase enzymes. These differences allow us to classify C4 plants into three biochemical subgroups. Plants that use NAD malic enzyme (NAD-ME) decarboxylate C4 acids in the BS mitochondria, while plants using NADP malic enzyme (NADP-ME) decarboxylate C4 acids in the BS chloroplasts. The third C4 subtype uses phosphoenolpyruvate carboxykinase (PCK) to decarboxylate C4 acids primarily in the cytosol of the BS cells. While all of these result in enhanced Rubisco efficiency, these biochemical subtleties reflect differences in the genetic prerequisites for C4 evolution as well as differences in the selective pressures that favored one subtype over another (Pinto et al., 2016; Sonawane et al., 2017). For example, within C4 grasses, NADP-ME plants increase in abundance geographically with increasing rainfall, while the number of NAD-ME grasses decreases in these conditions (Taub, 2000; Cabido et al., 2008). Thus, since the discovery of the C4 photosynthetic pathway, many studies have focused on identifying differences between C3 and C4 plants and between different C4 subtypes to unravel the genetics and evolution of C4 photosynthesis. Interestingly, nitrogen appears to be an essential component in many of these studies.
Early research in Poaceae noted that C4 grasses contained less total nitrogen in their leaves and produced more dry matter per unit of nitrogen fertilizer applied than C3 grasses. These observations quickly led to the hypothesis that C4 species utilize nitrogen more efficiently than C3 species (Brown, 1978). The obvious explanation was the lower investment of nitrogen in Rubisco in C4 plants (Sage et al., 1987). While this hypothesis is broadly accepted, recent studies suggest minor refinements are justified. For example, Ghannoum et al. (2005) evaluated combinations of various NAD-ME and NADP-ME grass species under high and low nitrogen treatments. They found that while the net CO2 assimilation rates were similar between these two C4 subtypes, NAD-ME plants contained more leaf nitrogen than NADP-ME plants with comparable CO2 assimilation rates. By measuring the total nitrogen in the MC and BS cells, Ghannoum et al. (2005) also showed that in the NAD-ME species, BS cells contained approximately 60% of the nitrogen and chlorophyll. In comparison, only 35% of the total nitrogen and chlorophyll were found in the BS of NADP-ME plants. Analysis of N partitioning suggested that NAD-ME plants invest more nitrogen into the production of Rubisco and other soluble proteins than NADP-ME plants. This seemed to be compensated by significantly greater kcat values of Rubisco in NADP-ME than in NAD-ME species (Ghannoum et al., 2005). Furthermore, a systematic evaluation of several lineages of C4 grasses encompassing all three biochemical C4 subtypes found that the nitrogen use efficiency of C4 grasses is highly correlated with the biochemical subtype with NADP-ME and PCK grasses having higher nitrogen use efficiency than NAD-ME counterparts (Pinto et al., 2016). Thus, while C4 plants have a higher photosynthetic nitrogen use efficiency (PNUE) than C3 plants, different biochemical C4 subtypes vary in their PNUE.
Nitrogen assimilation in C4 plants differs from C3 plants not only in PNUE but also in the intercellular compartmentalization of nitrogen assimilation enzymes (Figure 1; Kopriva, 2011; Jobe et al., 2019). Already at the onset of C4 photosynthesis research, it was shown that the activity of nitrate reductase, the key enzyme of nitrate assimilation, is localized mainly in the MC of maize, Sorghum sudanense, and Gomphrena globosa (Mellor and Tregunna, 1971). Further studies including all three C4 metabolic subtypes revealed that nitrate reductase was coordinately localized with nitrite reductase, glutamine synthetase, and glutamate synthase, in MC of maize, Sorghum bicolor, Digitaria sanguinalis, and Panicum miliaceum, while in Panicum maximum, nitrite reductase was present both in MC and BS (Rathnam and Edwards, 1976). Other studies confirmed the predominant localization of nitrate reductase and nitrite reductase activities in MC, but glutamine synthetase and glutamate synthase were mostly found in both MC and BS (Harel et al., 1977; Moore and Black, 1979). Immunogold labeling confirmed the exclusive localization of maize nitrate reductase in the cytosol of MC (Vaughn and Campbell, 1988) and glutamine synthetase and glutamate synthase in both cell types (Becker et al., 2000). However, whether the spatial distribution of nitrate assimilation in C4 plants contributes to their PNUE is unknown.
These observations prompted researchers to evaluate the potential role of nitrogen use efficiency and nitrate assimilation as drivers for the evolution of C4 photosynthesis. Classical schematic models of C4 evolution suggest that ancestral C3 plants progressed through a series of discrete stages on the path to C4 photosynthesis (Edwards et al., 2001; Heckmann et al., 2013; Schluter and Weber, 2016). The first stage is an increase in the BS : MC ratio driven by CO2 limitation or other environmental factors and a reallocation of glycine decarboxylase (GDC) expression from the MC to the BS. Next is the establishment of C2 photosynthesis (Mallmann et al., 2014). Next, there is an upregulation of the photorespiratory genes in both the BS and MC, a decrease in Rubisco expression in the MC, and an upregulation of PEPC in the MC (Schluter and Weber, 2016). In the final evolutionary stages, the expression of Rubisco and the photorespiratory genes become confined to the BS. Recent advances in constraint-based modeling have enabled researchers to examine the selective pressures that lead to C4 photosynthesis in silico (Blatke and Brautigam, 2019). These analyses suggested that while light and light distribution were the main drivers governing choice of decarboxylation enzymes, they also predicted that nitrogen limitation might have contributed to C4 evolution under high levels of photorespiration (Blatke and Brautigam, 2019).
What advantages do these evolutionary adaptations give to C4 plants over C3 plants as atmospheric CO2 increases? Nitrate assimilation was shown to be inhibited by elevated CO2 in a number of C3 species but not C4 plants (Hocking and Meyer, 1991; Bloom et al., 2012). Elevated CO2 increased PNUE of wheat but not maize, particularly at lower nitrate input, due to enhancing growth, however, at the expense of N accumulation in leaves (Hocking and Meyer, 1991). Nitrate reductase activity was inhibited by the elevated CO2 in wheat and not in maize (Hocking and Meyer, 1991). Nitrate assimilation can be quantified in vivo by an assimilatory quotient, the ratio of net CO2 consumed over net O2 evolved (Bloom et al., 2012). Plants assimilating nitrate increase net O2 evolution while CO2 consumption is constant, therefore, the assimilatory quotient is low in plants reducing nitrate (Bloom et al., 1989). The quotient is usually determined in comparison with ammonium nutrition after the addition of nitrate as a ΔAQ. In a number of C3 plants ΔAQ was high at low CO2 concentrations, but rapidly diminished with increasing CO2 in accordance with inhibition of nitrate reductase by elevated CO2 (Bloom et al., 2012). In contrast, in three C4 species analyzed, the ΔAQ was lower at low CO2 levels but remained constant with increasing CO2. Interestingly, in C3–C4 intermediate plants the response of ΔAQ to CO2 was intermediate between C3 and C4. Accordingly, FACE experiments have consistently shown that increasing CO2 negatively impacts nitrogen levels in C3 plants. This is true for leaves, where often, but not always, Rubisco content diminishes (Bowes, 1991) and for seeds and grains. A recent meta-analysis showed that the average differential effect of increased CO2 on C3 plants is—4% (Ebi and Loladze, 2019). In a comparison between several C3 crops, Myers et al. (2014) found no significant changes in nitrogen content in maize grown under elevated CO2. While it remains unclear if nitrogen limitation contributed to C4 evolution, the rising CO2 levels do not pose a threat for a reduction in nitrogen in C4 plants as they are already saturated at current CO2 levels (Von Caemmerer and Furbank, 2003). Although the lower abundance of Rubisco and the identity of the decarboxylation enzyme were shown to impact nitrogen use efficiency, less is known regarding the significance of confining nitrate reduction to the MC. However, it highlights the extensive metabolic rewiring that accompanies C4 evolution and suggests that multiple mechanisms contribute to enhanced nitrogen use efficiency in C4 plants. Taken together, both recent and historical studies show that C4 plants require less total nitrogen, have higher nitrogen use efficiency, and maintain nitrogen levels under elevated CO2 conditions.
Sulfur is an essential macronutrient for all living organisms, with organic S-compounds representing an important class of metabolites in plant physiology. Sulfate assimilation by plants and microorganisms constitute the entry point of this element into organic molecules in the global sulfur cycle and also in human nutrition. Sulfate is the primary source of S available in nature, and specific H+/sulfate co-transporters from the SULTR family mediate sulfate uptake and mobilization within the plant (reviewed in Takahashi et al., 2011a; Gigolashvili and Kopriva, 2014). Once inside the plant cell, sulfate is initially activated by ATP sulfurylase (ATPS), producing adenosine 5′-phosphosulfate (APS). In primary S-metabolism, APS undergoes two subsequent reduction reactions catalyzed by APS reductase (APR) to generate sulfite and sulfite reductase (SiR) to produce sulfide. Finally, in a two-step process, serine acetyltransferase catalyzes the transfer of an acetyl moiety from acetyl Coenzyme A to serine resulting in O-acetyl-L-serine (OAS). OAS is then used as a substrate for O-acetylserine(thiol)lyase (OASTL), which replaces the acetyl group of OAS with sulfide to produce cysteine, the first organic form of sulfur (reviewed in Takahashi et al., 2011b). Cys is the source of reduced S for other metabolites, such as methionine or the tripeptide glutathione (GSH), an essential part of plant redox homeostasis and stress defense (Noctor et al., 2012).
Like nitrate assimilation, sulfate assimilation is differentially localized in MC and BS of C4 plants. In a number of C4 species spanning all three C4 subtypes, most of the total leaf ATPS activity is confined to BS chloroplasts (Gerwick et al., 1980; Passera and Ghisi, 1982). Similar to nitrate assimilation, not all enzymes of the pathway are coordinately expressed. While APR was also found almost exclusively in BS of maize (Schmutz and Brunold, 1984; Burgener et al., 1998), the activities of SiR and OASTL were detected at comparable levels in MC and BS (Passera and Ghisi, 1982; Schmutz and Brunold, 1985). Reduced sulfur needed in MC is transported from maize BS in the form of cysteine (Burgener et al., 1998). Interestingly, GSH synthesis and homeostasis are also differently organized in MC and BS. In maize, GSH synthetase activity is higher in MC than in BS, in line with the export of Cys from BS (Burgener et al., 1998). This results in a higher accumulation of GSH in MC, possibly connected to higher H2O2 levels in MC than in BS (Doulis et al., 1997). Given the importance of GSH for maintaining cellular redox potential, it is surprising that glutathione reductase, the key element of the glutathione redox cycle, was also found exclusively in MC of maize (Doulis et al., 1997; Pastori et al., 2000). However, not all C4 plants follow the same pattern. In the C4 species of the dicot genus Flaveria, APR and ATPS are expressed in both MC and BS (Koprivova et al., 2001). Since the C4 species analyzed previously were all monocots, BS-exclusive localization of sulfate assimilation could be a trait of C4 monocots but not C4 eudicots (Figure 1; Koprivova et al., 2001; Kopriva and Koprivova, 2005). Indeed, numerous RNA-seq analyses of MC and BS transcripts showed BS localization of transcripts for ATPS and APR in different C4 monocots (maize, sorghum, Setaria viridis) but a similar transcript abundance in MC and BS of the eudicot C4 species Gynandropsis gynandra (Aubry et al., 2014a; John et al., 2014; Doring et al., 2016; Denton et al., 2017). Thus, the localization of sulfate assimilation in BS cannot be a general C4 trait. This conclusion was unexpectedly confirmed by experiments with the C3 model plant, Arabidopsis thaliana. In an analysis aimed at discerning the function of the BS cell layer in C3 plants using a translatome approach, Aubry et al. (2014b) found an enrichment of transcripts for sulfate assimilation genes in the BS. Transcripts of ATPS, APR, SiR, as well as other components of Cys synthesis, in addition to sulfate transporters and genes for synthesizing the sulfur-rich secondary compounds glucosinolates were all overrepresented in RNA from BS compared to the whole leaf (Aubry et al., 2014b). Three obvious questions arise from this study. First, what is the ancestral localization of the sulfate assimilation pathway? Secondly, what is the metabolic significance of the various relocations? Finally, in the C4 lineages with relocated sulfate assimilation enzymes, was the relocation of sulfate assimilation a prerequisite for C4 evolution or a consequence of C4 evolution? These remain key open questions in plant sulfur research.
An analysis of sulfate assimilation in the eudicot genus Flaveria revealed another intriguing result. A gradient in the accumulation of leaf Cys and GSH was observed with higher concentrations in the leaves of C4 species than in C3 and C3-C4 intermediate species (Koprivova et al., 2001; Gerlich et al., 2018). This gradient is sustained through a similar gradient in sulfate uptake, reduction rate, transcript levels, and activity of APR (Koprivova et al., 2001; Weckopp and Kopriva, 2014; Gerlich et al., 2018). Interestingly, expression analyses suggested that sulfate reduction and GSH synthesis are preferentially localized in the roots of C4 Flaveria species. Interspecies grafts of C3 F. robusta and C4 F. bidentis were created to test this hypothesis. The results of this experiment showed that the high GSH accumulation in C4 leaves is indeed controlled by the roots (Gerlich et al., 2018). While it is plausible that the importance of roots for Cys and GSH synthesis in C4 Flaveria is connected to serine synthesis, which is preferentially synthesized in the roots of C4 plants through the phosphorylated pathway (Gerlich et al., 2018), this hypothesis should be tested in more C4 species.
Sulfur is much less abundant in the plant body than nitrogen making it unlikely to be the driving force behind the metabolic adaptations leading to the evolution of C4 photosynthesis. However, it is possible that the gradient of higher sulfate assimilation flux with increasing C4 photosynthesis in Flaveria is a result of the adaptation to dry and warm habitats typical for C4 plants. Thus, the higher GSH contents in C4 Flaveria might be a mechanism to cope with increased oxidative stress caused by such environmental conditions. This is consistent with the critical role of GSH in chilling tolerance in maize (Kocsy et al., 2001). However, the importance of the BS-localization of sulfate assimilation in C4 monocots and possibly in the roots of C4 dicots is still elusive.
In addition to carbon, nitrogen, and sulfur, phosphorus is a macronutrient crucial for plant growth and development. As an essential player in cellular energy conversion, an enzymatic substrate, as well as a regulatory factor of enzyme activity, phosphate plays many crucial roles in cellular biochemistry. Moreover, phosphate is responsible for the acidic nature of nucleic acids and is a vital constituent of phospholipid membranes. Plants employ several morphological and physiological adaptations to mitigate phosphorus deficiency, including interconnections with the rhizosphere and soil microbes and diverse molecular mechanisms (Lopez-Arredondo et al., 2014). Phosphate is taken up by various phosphate transporters as an inorganic anion. However, unlike nitrate and sulfate, phosphate is not reduced and remains in its oxidized state as either a free anion or is incorporated into organic compounds via phosphate esters. Disruptions in phosphate homeostasis have intensive footprints on plants. Thus, shoot phosphate concentrations are tightly regulated by systemic control of phosphate uptake and allocation (Bari et al., 2006; Ham et al., 2018; Kopriva and Chu, 2018). Control of phosphate homeostasis is coordinated with the regulation of other nutrients, particularly nitrate and sulfate (Rouached et al., 2011; Hu et al., 2019; Medici et al., 2019).
Phosphate has a vital role in photosynthesis. The metabolic energy of the cell and the energy generated during the light reactions of photosynthesis are stored in phosphate esters and energy-rich pyrophosphate bonds. Inorganic phosphate in the chloroplast regulates the partitioning of photosynthates between starch synthesis and export to the cytosol (Heldt et al., 1977). Moreover, phosphate is indispensable for the function of the triose-phosphate/phosphate translocator (TPT), an antiporter in the inner membrane of the chloroplast (Lee et al., 2017). The TPT exchanges phosphate from the cytosol with triose-phosphates synthesized in the Calvin cycle (Fliege et al., 1978). In C4 plants, the TPT is even more highly abundant in envelopes of MC chloroplasts as the flux through this transporter is higher in C4 plants than in C3 plants (Brautigam et al., 2008). In addition, C4 plants possess another abundant phosphate driven transporter, the phosphoenolpyruvate phosphate translocator (PPT), which is essential for the transport of PEP from the chloroplast in MC (Brautigam et al., 2008; Majeran et al., 2008). Also, the activities of the critical enzymes involved in C4 carbon assimilation, such as PEPC, PCK, and pyruvate phosphate dikinase, are modulated by reversible phosphorylation (Ashton and Hatch, 1983; Jiao and Chollet, 1991; Chao et al., 2014).
Although phosphate demand to facilitate transport processes in C4 plants is high, C4 specific features of phosphate homeostasis or possible differences in (photosynthetic) phosphate use efficiency (PUE) have not been described. Phosphate deficiency was shown to decrease Rubisco activity in sunflower, but Rubisco activity was not affected by phosphate deficiency in maize (Jacob and Lawlor, 1992). Similarly, C4 grasses produced higher forage yields on phosphate-limited soil than C3 grasses (Morris et al., 1982). Accordingly, in a comparative survey of photosynthetic and growth responses to phosphate deficiency in 12 species with diverse photosynthetic characteristics, C3 species showed more substantial growth retardation in comparison to C4 species (Halsted and Lynch, 1996). However, no photosynthesis type-dependent changes in photosynthetic PUE could be determined. Although the CO2 exchange rate was decreased less by phosphate deficiency in C4 plants than in C3 ones, due to higher foliar phosphate concentration, the photosynthetic PUE remained unchanged (Halsted and Lynch, 1996). Interestingly, the monocot species were less sensitive to low phosphate stress than dicots irrespective of photosynthesis type, due to a lower phosphate content in the leaf and better maintenance of growth (Halsted and Lynch, 1996). In an independent study focusing on monocots, the response of CO2 assimilation rates to leaf phosphate concentration was saturated in C4 species but not in their C3 relatives (Ghannoum et al., 2008). It seems, therefore, that although C4 plants require higher amounts of phosphate than C3 plants, their CO2 assimilation is less sensitive to phosphate limitation.
The World Health Organization (WHO) defines malnutrition as deficiencies, excesses, or imbalances in a person´s energy intake and/or nutrient intake (https://www.who.int/news-room/fact-sheets/detail/malnutrition) and recognizes three broad groups of malnutrition conditions - undernutrition, micronutrient-related malnutrition, and overnutrition and noncommunicable diseases. Over the past 60–70 years, plant biologists and plant breeders have focused their attention on alleviating undernutrition by dramatically increasing crop yields by improving plant genetics and intensifying agricultural production systems. However, by focusing on yield, changes in the nutritional value of our food have been largely neglected, especially regarding micronutrient content. Thus, micronutrient levels in plants have decreased for two main reasons. First, intensive agricultural practices have depleted micronutrients from the soil, and second, rising atmospheric carbon dioxide negatively affects the nutrient profiles of C3 crop plants (Loladze, 2014).
Micronutrient-related malnutrition, sometimes called hidden hunger, is caused by poorly diversified diets that meet the caloric but not the nutritional needs of an individual and is primarily associated with micronutrient deficiency (Myers et al., 2017). In addition to the well documented adverse effects of increasing atmospheric CO2 on macronutrients in C3 crops (see above), there is evidence that the effects are equally adverse, or in some cases, much worse for micronutrient levels. For example, a study on the impact of elevated CO2 on nine diverse rice cultivars showed that growth at elevated CO2 decreased the manganese (Mn) content in the body of rice plants by 53% (Ujiie et al., 2019). In this same study, the Mn content in the brown rice decreased by 7%, while the polished rice showed a 20.5% decrease in Mn when grown under elevated CO2 (Ujiie et al., 2019). The vast differences observed in Mn content in different tissues is a significant finding as rice is becoming an important forage crop in some regions of the world (Cheng et al., 2018). While Mn deficiency in forage animals is considered rare, such a significant decrease in micronutrients in the body of the plant suggests that forage animal nutrition will also suffer as a result of rising CO2. Thus, to accurately assess all the potential impacts of CO2-induced nutrient depletion on human health, it is crucial to measure nutrients in multiple plant tissues.
Interestingly, a more extensive meta-analysis of 130 plant species/cultivars was unable to detect a significant decrease in Mn content among C3 crops (Loladze, 2014). However, this study did identify significant decreases in many other micronutrients, namely iron (Fe) and zinc (Zn). Iron is of particular interest as at least 2 billion people currently suffer from Fe deficiency, making anemia a leading cause of maternal mortality (Micronutrient_Initiative, 2009). Zinc deficiency is also widespread, with approximately 30% of the world population at risk. Zinc deficiency can cause compromised immune responses, stunting during childhood, and increased risk of child mortality (Micronutrient_Initiative, 2009; Livingstone, 2015). While crosstalk between Fe, Zn, P, and S signaling in plants is recognized, not much is known in C3 or C4 plants regarding the mechanistic integration of these signaling networks (Mendoza-Cozatl et al., 2019; Xie et al., 2019). However, it was recently proposed that Fe, Zn, P, and S signaling are integrated in a PHR1 dependent manner in the C3 plant Arabidopsis (Briat et al., 2015). Interestingly, the rice homolog of PHR1, OsPHR2, was also shown to play a role in the integration of P and N signaling networks in rice (Hu et al., 2019). Thus, in C3 plants, it seems PHR proteins may be essential network hubs integrating signaling from multiple nutrients. When viewed from this perspective, the changes in micronutrient levels observed in C3 plants under elevated CO2 could be pleiotropic effects caused by disruption of N and/or P signaling. It remains unknown if these signaling networks are conserved between C3 and C4 plants.
Additionally, the genetic diversity in the C3 crops has a large impact on the effects of elevated CO2. The variation within species may even exceed the variation between species. For example, a study with 17 rice cultivars grown under controlled conditions in normal or 664 ppm CO2 showed 10–265% increase in total biomass and even greater—10–350% variation in response of grain yield (Ziska et al., 1996). This is true also for qualitative traits; protein content in grains of 18 field-grown rice cultivars cultivated at ca., 585 ppm CO2 decreased by 5–20%, whereas grain Zn and Fe concentrations decreased on average, but actually increased in four and two genotypes, respectively, and were not affected in another variety, Nipponbare (Zhu et al., 2018). Similar variation was observed in other species and, interestingly, modern varieties of oat, wheat, or soybean seem to be less responsive to elevated CO2 than varieties from the 1920s (Ziska and Blumenthal, 2007). There might, therefore, be a potential for the selection of new crop varieties for response to elevated CO2 levels (Shimono et al., 2018).
The question thus arises, can C4 crops help to alleviate “hidden hunger”? There are currently only five economically important C4 food crops—maize, sorghum, sugar cane, onion, and pearl millet. While the list of C4 crops is small, they account for a large proportion of global crop production. For example, the average annual production of maize from 2008–2010 was 750 million metric tons representing 27% of cereal area, 34% of cereal production and 8% of the value of all primary crop production (Shiferaw et al., 2011). The nutritional quality of these C4 crops is at best average, e.g., due to low lysine content in maize proteins or poor digestibility of sorghum and millet proteins (Millward, 1999; Galili and Amir, 2013). However, there are also several regionally important C4 crops, often called orphan crops, that have more desirable nutritional traits for combating hidden hunger. Notable orphan crops include grain amaranth, teff (Eragrostis tef), foxtail millet (Setaria italica), finger millet (Eleusine coracana), and proso millet (Panicum miliaceum).
These data suggest that while C4 crops do not show a CO2-induced nutritional penalty, the current staple C4 crops may not be best suited to address dietary deficits and hidden hunger. However, significant genetic advances have been made to improve the nutritional quality of maize and sorghum. For example, a recent genome-wide association study on 923 maize lines identified 46 QTLs significantly associated with seed Zn and Fe concentrations (Hindu et al., 2018). Introgressing favorable alleles of these QTLs into commercial varieties could improve both Zn and Fe levels in maize kernels. Additionally, researchers have developed quality protein maize (QPM), having almost twice the amount of lysine and tryptophan as traditional varieties, and maize lines with enhanced levels of provitamin-A or methionine (Wurtzel et al., 2012; Galili and Amir, 2013; Planta et al., 2017). Thus, biofortification is a viable approach to enhance the nutritional value of C4 crops and address hidden hunger.
In addition to food crops, there are eight C4 crops grown for turf, forage, or bioenergy. These include Miscanthus x giganteus, Panicum virgatum (switchgrass), Chloris gayana (Rhodes grass), Cynodon dactylon (Bermuda grass), Melinis minutifolia (molasses grass), Panicum maximum, Cenchrus purpureus (Napier grass), and Zoysia japonica. Collectively, these crops are all known for their high productivity and demonstrate the potential of C4 plants. Similar to food crops, a nutritional comparison of C3 and C4 forage grasses grown under high and low CO2 levels found that the C3 grasses had higher levels of protein, nonstructural carbohydrates, and water, but lower levels of fiber when grown under elevated CO2 compared to the C4 species (Barbehenn et al., 2004).
Under current environmental conditions, the staple C4 crops show superior productivity compared to C3 crops, and some of the C4 orphan crops seem to have the same or even better nutritional quality (Table 1). While the productivity gap can be expected to narrow down, due to elevated atmospheric CO2 that fertilizes C3 crops but not C4 crops, the relative nutritional value of the current C4 crops may improve because of the lack of the carbon nutrient penalty. Also the rise in temperatures may favor C4 crops in the future, or at least extend their cultivation areas. However, hidden hunger cannot be combatted without investment into further crop improvement, specifically targeting nutritional quality of staple C4 crops and improving the productivity of selected local crops with high nutritional value, such as pearl millet.
To improve the nutritional value of C4 crops for human food, it is necessary to understand more about the control of their nutrient homeostasis. While some progress has been made, e.g., in the biofortification of maize (Wurtzel et al., 2012; Galili and Amir, 2013; Planta et al., 2017), many questions on mineral nutrition of C4 plants are still open (Figure 2). Probably the biggest set of fundamental questions concerns the drivers and the consequences of the spatial separation of nitrate and sulfate assimilation in C4 monocots. Does the MC localization of nitrate reductase contribute to the improved nitrogen use efficiency of C4 plants? Have C3–C4 intermediate plants improved nitrogen use efficiency compared to C3 plants? Are there any differences in sulfur use efficiency between C3 and C4 plants? Is the gradient in the accumulation of sulfur compounds found in Flaveria conserved in other genera with C3 and C4 photosynthesis? Are the pathways of nitrate and sulfate assimilation differently regulated in C3 and C4 plants? Why is sulfate assimilation differently localized in C4 monocots and C4 dicots?
The other set of questions concerns other nutrients. Is there a gradient similar to that of sulfur compounds in Flaveria in accumulation of other nutrients between closely related C3 and C4 plants? Does the high flux through TPT and PPT in C4 plants affect their phosphate needs and homeostasis? Is phosphate homeostasis affected by elevated CO2? Is there a different need for Fe or Cu in C4 and C3 plants given the different arrangements of photosynthetic apparatus?
All these fundamental unknowns lead to one overarching question: Were adaptations in nutrient pathways necessary for the evolution of C4 photosynthesis? This question has major practical implications for the efforts to improve C4 crops, but particularly for engineering C4 photosynthesis to C3 crops.
Breeding material with high nutritional value is available for maize (Newell et al., 2014; Wang et al., 2019) and thus breeding for improved nutritional value is feasible. A few approaches for maintaining the nutritional levels of crops under elevated CO2 have been proposed. For example, the negative effect of elevated CO2 on nitrate assimilation and nitrogen content might be attenuated by increasing the proportion of ammonium as the nitrogen source (Bloom et al., 2010). However, crop species differ in their tolerance to ammonium, therefore, as discussed above, the most straightforward approach is to incorporate FACE studies into modern breeding programs. This approach would be useful for both C3 and C4 crops and would allow us to accomplish two goals. First, we could screen specifically for traits that improve the nutritional levels of crops under elevated CO2 and select for these traits in future cultivars. Secondly, we could ensure that traits selected to meet other breeding goals (i.e., pathogen resistance traits or drought resistance traits) are not negatively affected by elevated CO2 levels and do not further decrease the nutritional standards of our crops. While this approach would be technically challenging for breeding programs due to the expense and large space requirements associated with field-scale FACE studies, it has a high likelihood of success in the short term. As noted by Ujiie et al. (2019), carbohydrates, nitrogen, and sulfur resources are all transported through the phloem during nutrient reallocation and grain filling. Thus, improving nutrient translocation or the strength of the sink organ could counteract the nutritional decrease in crops grown under elevated CO2. These goals are well within the scope of modern breeding programs.
The conversion of C3 crops to full C4 photosynthesis is a long-standing goal of plant biologists, and significant advances have been made with the help of both systems biology and synthetic biology (Schuler et al., 2016; Ermakova et al., 2020). To achieve this, at least five major milestones have been identified that are necessary to convert C3 crops to C4 photosynthesis: 1) induction of higher-order veins, 2) increase BS:M ratio, 3) adaptation of BS morphology, 4) engineering of dimorphic chloroplasts in BS and M cells, and 5) compartmentalization of the photosynthetic enzymes between BS and M cells (reviewed in Schuler et al., 2016). However, significant hurdles remain, especially in identifying a suitable C3 chassis for engineering, establishing Kranz anatomy, and the establishment of a carbon concentrating mechanism (Hennacy and Jonikas, 2020). Despite these challenges, consortiums like the C4 Rice Project, a global collaboration between leading researchers in photosynthesis, aim to engineer C4 photosynthesis into rice. Increasing rice yield and decreasing water and nitrogen fertilization requirements would significantly increase the sustainability of rice, a staple crop for 50% of the world population (see c4rice.com). Furthermore, additional C3 and C4 plant species are being developed for comparative studies to better understand the evolution of C4 traits. Potential model species of interest include the C3 panicoid grass Dichanthelium oligosanthes, which diverged from the C4 species Setaria viridis approximately 15 million years ago, representing a more recent divergence than most other C3 and C4 panicoid grasses (Studer et al., 2016).
An alternative to engineering C4 photosynthesis into C3 plants is using synthetic biology for improving photosynthesis (Kubis and Bar-Even, 2019). Possible mechanisms include engineering carbon concentrating mechanisms (Long et al., 2018), exploiting CAM mechanisms (DePaoli et al., 2014), or manipulating photorespiration (Maurino, 2019). Another possibility is to increase the performance of C4 crops directly. Indeed, it was possible to increase CO2 assimilation in maize by overexpressing Rubisco together with a chaperon, RUBISCO ASSEMBLY FACTOR 1 (RAF1), which resulted in fresh weight gain of the transgenic plants (Salesse-Smith et al., 2018). Alternatively, CO2 assimilation was increased by overexpression of Rieske FeS protein of the Cytochrome b6f complex in Setaria viridis (Ermakova et al., 2019). These efforts, however, concentrate fully on carbon fixation and do not consider the nutritional aspects, neither with respect to the crop nutritional value nor the mineral nutrient homeostasis and use efficiency of the new crops. Nevertheless, while engineering C4 crops is a very active area of research, it is unlikely to contribute significantly to food security or improved crop nutrition in the short term.
Recently, Lundgren (2020) presented a compelling case for engineering C2 photosynthesis into C3 crop plants to improve photosynthetic performance in the face of climate change. The main argument made in favor of this approach is that C2 photosynthesis is a stable intermediate physiological state between C3 and C4 metabolism that increases net carbon assimilation under high temperatures (Monson, 1989; Bellasio and Farquhar, 2019). But, importantly, C2 photosynthesis does not require the complex anatomical changes associated with C4 photosynthesis (Lundgren, 2020). This strategy could be useful in improving crop yields (or in mitigating yield declines) in the medium term. However, it is unclear how C2 engineering will impact the nutritional status of crops, particularly under elevated CO2, and the photosynthetic nutrient use efficiency. To the best of our knowledge, there are no FACE experiments evaluating the effects of elevated CO2 on the yield or nutritional status of C2 plants. Despite these limitations, this approach seems feasible for two reasons. First, C2 engineering appears to be a necessary step toward C4 engineering, suggesting that these efforts will not be wasted in the long term. Secondly, even if initial C2 engineering has a negative impact on plant nutrition, when combined with traditional breeding approaches and additional engineering efforts, there is a high likelihood that these can be reverted. Thus, C2 engineering of C3 crops is likely to increase yield while maintaining or improving nutritional quality.
Of the approximately 150 commonly cultivated crops worldwide, humans obtain almost 50% of their calories from just three crops - rice, wheat, and maize (Ross-Ibarra et al., 2007). This is in stark contrast to preagricultural humans who had significantly more diverse diets and achieved some level of domestication in approximately 2,500 plant species (Khoury et al., 2014; Smykal et al., 2017). Recent advances in genome editing technology have made de novo domestication of wild plants a viable option to design ideal crops for the future (Fernie and Yan, 2019). For example, a recent study targeting a small number of critical genes in the orphan Solanaceae crop “groundcherry” (Physalis pruinosa) was able to rapidly improve plant architecture and productivity (Lemmon et al., 2018). Because groundcherry is a semi-domesticated orphan crop in the same family as tomato, researchers quickly identified homologues of two domestication genes—SELF PRUNING 5 and CLAVATA1. Using genome editing techniques to induce mutations in these genes resulted in an increased fruit size of over 20% and improved plant architecture (more compact growth), making groundcherry easier to grow and harvest. Furthermore, advances in multiplexing platforms that allow simultaneous genome editing of six or more genes in a single transformation open the door for similar improvements to be made quickly in wild species (Zhang et al., 2016). Considering the small number of C4 plant species that have been domesticated and the growing list of known domestication genes to target, there is good reason to believe the weeds of today could be the nutritious and sustainable foods of tomorrow.
C4 crops play an essential role in human nutrition, and this role will probably be even stronger in the future. They are characterized by high productivity and adaptability to warm and dry climates and by their better water and nitrogen use efficiency than C3 crops. While their yields will not directly benefit from elevated CO2, their nutritional value is not predicted to be negatively affected. However, to unlock the full potential of C4 crops for the future, more fundamental knowledge on the connection between mineral nutrition and C4 photosynthesis needs to be generated. As outlined above, in particular nitrogen metabolism underwent significant alterations in the course of evolution of C4 photosynthesis and might have been one of the evolutionary drivers. The increasing number and availability of new genomic and genetic resources and tools will enable us to extend the investigations of plant nutrition to a wider variety of C4 and C3–C4 intermediate species, and at the same time, to include investigations of nutrient homeostasis in the general framework of C4 photosynthesis research.
All authors contributed to the article and approved the submitted version.
Research in SK’s lab is funded by the Deutsche Forschungsgemeinschaft (DFG) under Germany´s Excellence Strategy – EXC 2048/1 – project 390686111. IZ is supported by a DAAD fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Amoroso, L. (2016). The Second International Conference on Nutrition: Implications for Hidden Hunger. World Rev. Nutr. Diet 115, 142–152. doi: 10.1159/000442100
Ashida, H., Mizohata, E., Yokota, A. (2019). Learning RuBisCO’s birth and subsequent environmental adaptation. Biochem. Soc. Trans. 47, 179–185. doi: 10.1042/BST20180449
Ashton, A. R., Hatch, M. D. (1983). Regulation of C4 photosynthesis: regulation of pyruvate, Pi dikinase by ADP-dependent phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 115, 53–60. doi: 10.1016/0006-291x(83)90967-1
Aubry, S., Kelly, S., Kumpers, B. M., Smith-Unna, R. D., Hibberd, J. M. (2014a). Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10, e1004365. doi: 10.1371/journal.pgen.1004365
Aubry, S., Smith-Unna, R. D., Boursnell, C. M., Kopriva, S., Hibberd, J. M. (2014b). Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 78, 659–673. doi: 10.1111/tpj.12502
Barbehenn, R. V., Chen, Z., Karowe, D. N., Spickard, A. (2004). C3 grasses have higher nutritional quality than C4 grasses under ambient and elevated atmospheric CO2. Global Change Biol. 10, 1565–1575. doi: 10.1111/j.1365-2486.2004.00833.x
Bari, R., Datt Pant, B., Stitt, M., Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999. doi: 10.1104/pp.106.079707
Becker, T. W., Carrayol, E., Hirel, B. (2000). Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211, 800–806. doi: 10.1007/s004250000355
Bellasio, C., Farquhar, G. D. (2019). A leaf-level biochemical model simulating the introduction of C2 and C4 photosynthesis in C3 rice: gains, losses and metabolite fluxes. New Phytol. 223, 150–166. doi: 10.1111/nph.15787
Blatke, M. A., Brautigam, A. (2019). Evolution of C4 photosynthesis predicted by constraint-based modelling. Elife 8, e49305. doi: 10.7554/eLife.49305
Bloom, A. J., Caldwell, R. M., Finazzo, J., Warner, R. L., Weissbart, J. (1989). Oxygen and carbon dioxide fluxes from barley shoots depend on nitrate assimilation. Plant Physiol. 91, 352–356. doi: 10.1104/pp.91.1.352
Bloom, A. J., Burger, M., Rubio Asensio, J. S., Cousins, A. B. (2010). Carbon dioxide enrichment inhibits nitrate assimilation in wheat and Arabidopsis. Science 328, 899–903. doi: 10.1126/science.1186440
Bloom, A. J., Asensio, J. S., Randall, L., Rachmilevitch, S., Cousins, A. B., Carlisle, E. A. (2012). CO2 enrichment inhibits shoot nitrate assimilation in C3 but not C4 plants and slows growth under nitrate in C3 plants. Ecology 93, 355–367. doi: 10.1890/11-0485.1
Bowes, G. (1991). Growth at elevated CO2: photosynthetic responses mediated through Rubisco. Plant Cell Environ. 14, 795–806. doi: 10.1111/j.1365-3040.1991.tb01443.x
Brautigam, A., Hoffmann-Benning, S., Weber, A. P. (2008). Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 148, 568–579. doi: 10.1104/pp.108.121012
Briat, J. F., Rouached, H., Tissot, N., Gaymard, F., Dubos, C. (2015). Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front. Plant Sci. 6:290. doi: 10.3389/fpls.2015.00290
Brown, R. H. (1978). A difference in the nitrogen use efficiency of C3 and C4 plants and its implications in adaptation and evolution. Crop Sci. 18, 93–98. doi: 10.2135/cropsci1978.0011183X001800010025x
Burgener, M., Suter, M., Jones, S., Brunold, C. (1998). Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiol. 116, 1315–1322. doi: 10.1104/pp.116.4.1315
Cabido, M., Pons, E., Cantero, J. J., Lewis, J. P., Anton, A. (2008). Photosynthetic pathway variation among C4 grasses along a precipitation gradient in Argentina. JBiogeog 35, 131–140. doi: 10.1111/j.1365-2699.2007.01760.x
Caselato-Sousa, V. M., Amaya-Farfán, J. (2012). State of knowledge on amaranth grain: a comprehensive review. J. Food Sci. 77, R93–R104. doi: 10.1111/j.1750-3841.2012.02645.x
Chao, Q., Liu, X. Y., Mei, Y. C., Gao, Z. F., Chen, Y. B., Qian, C. R., et al. (2014). Light-regulated phosphorylation of maize phosphoenolpyruvate carboxykinase plays a vital role in its activity. Plant Mol. Biol. 85, 95–105. doi: 10.1007/s11103-014-0171-3
Cheng, W., Kimani, S. M., Kanno, T., Tang, S., Oo, A. Z., Tawaraya, K., et al. (2018). Forage rice varieties Fukuhibiki and Tachisuzuka emit larger CH4 than edible rice Haenuki. Soil Sci. Plant Nutr. 64, 77–83. doi: 10.1080/00380768.2017.1378569
Denton, A. K., Mass, J., Kulahoglu, C., Lercher, M. J., Brautigam, A., Weber, A. P. (2017). Freeze-quenched maize mesophyll and bundle sheath separation uncovers bias in previous tissue-specific RNA-Seq data. J. Exp. Bot. 68, 147–160. doi: 10.1093/jxb/erw463
DePaoli, H. C., Borland, A. M., Tuskan, G. A., Cushman, J. C., Yang, X. (2014). Synthetic biology as it relates to CAM photosynthesis: challenges and opportunities. J. Exp. Bot. 65, 3381–3393. doi: 10.1093/jxb/eru038
Doring, F., Streubel, M., Brautigam, A., Gowik, U. (2016). Most photorespiratory genes are preferentially expressed in the bundle sheath cells of the C4 grass Sorghum bicolor. J. Exp. Bot. 67, 3053–3064. doi: 10.1093/jxb/erw041
Doulis, A. G., Debian, N., Kingston-Smith, A. H., Foyer, C. H. (1997). Differential Localization of Antioxidants in Maize Leaves. Plant Physiol. 114, 1031–1037. doi: 10.1104/pp.114.3.1031
Ebi, K. L., Loladze, I. (2019). Elevated atmospheric CO2 concentrations and climate change will affect our food’s quality and quantity. Lancet Planet Health 3, e283–e284. doi: 10.1016/S2542-5196(19)30108-1
Edwards, G. E., Furbank, R. T., Hatch, M. D., Osmond, C. B. (2001). What does it take to be C4? Lessons from the evolution of C4 photosynthesis. Plant Physiol. 125, 46–49. doi: 10.1104/pp.125.1.46
Ermakova, M., Lopez-Calcagno, P. E., Raines, C. A., Furbank, R. T., Von Caemmerer, S. (2019). Overexpression of the Rieske FeS protein of the Cytochrome b 6 f complex increases C4 photosynthesis in Setaria viridis. Commun. Biol. 2, 314. doi: 10.1038/s42003-019-0561-9
Ermakova, M., Danila, F. R., Furbank, R. T., Von Caemmerer, S. (2020). On the road to C4 rice: advances and perspectives. Plant J. 101, 940–950. doi: 10.1111/tpj.14562
Fernando, N., Panozzo, J., Tausz, M., Norton, R., Fitzgerald, G., Seneweera, S. (2012). Rising atmospheric CO2 concentration affects mineral nutrient and protein concentration of wheat grain. Food Chem. 133, 1307–1311. doi: 10.1016/j.foodchem.2012.01.105
Fernie, A. R., Yan, J. (2019). De Novo Domestication: An Alternative Route toward New Crops for the Future. Mol. Plant 12, 615–631. doi: 10.1016/j.molp.2019.03.016
Fliege, R., Flugge, U. I., Werdan, K., Heldt, H. W. (1978). Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim. Biophys. Acta 502, 232–247. doi: 10.1016/0005-2728(78)90045-2
Galili, G., Amir, R. (2013). Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol. J. 11, 211–222. doi: 10.1111/pbi.12025
Gerlich, S. C., Walker, B. J., Krueger, S., Kopriva, S. (2018). Sulfate Metabolism in C4 Flaveria Species Is Controlled by the Root and Connected to Serine Biosynthesis. Plant Physiol. 178, 565–582. doi: 10.1104/pp.18.00520
Gerwick, B. C., Ku, S. B., Black, C. C. (1980). Initiation of sulfate activation: a variation in c4 photosynthesis plants. Science 209, 513–515. doi: 10.1126/science.209.4455.513
Ghannoum, O., Evans, J. R., Chow, W. S., Andrews, T. J., Conroy, J. P., Von Caemmerer, S. (2005). Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiol. 137, 638–650. doi: 10.1104/pp.104.054759
Ghannoum, O., Paul, M. J., Ward, J. L., Beale, M. H., Corol, D. I., Conroy, J. P. (2008). The sensitivity of photosynthesis to phosphorus deficiency differs between C3 and C4 tropical grasses. Func. Plant Biol. 35, 213–221. doi: 10.1071/FP07256
Gigolashvili, T., Kopriva, S. (2014). Transporters in plant sulfur metabolism. Front. Plant Sci. 5:442. doi: 10.3389/fpls.2014.00442
Halsted, M., Lynch, J. (1996). Phosphorus responses of C3 and C4 species. J. Exp. Bot. 47, 497–505. doi: 10.1093/jxb/47.4.497
Ham, B. K., Chen, J., Yan, Y., Lucas, W. J. (2018). Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49, 1–9. doi: 10.1016/j.copbio.2017.07.005
Harel, E., Lea, P. J., Miflin, B. J. (1977). The localisation of enzymes of nitrogen assimilation in maize leaves and their activities during greening. Planta 134, 195–200. doi: 10.1007/BF00384971
Hatch, M. D., Slack, C. R. (1966). Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101, 103–111. doi: 10.1042/bj1010103
Heckmann, D., Schulze, S., Denton, A., Gowik, U., Westhoff, P., Weber, A. P., et al. (2013). Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. doi: 10.1016/j.cell.2013.04.058
Heldt, H. W., Chon, C. J., Maronde, D., Herold, A., Stankovic, Z. S., Walker, D. A., et al. (1977). Role of orthophosphate and other factors in the regulation of starch formation in leaves and isolated chloroplasts. Plant Physiol. 59, 1146–1155. doi: 10.1104/pp.59.6.1146
Hennacy, J. H., Jonikas, M. C. (2020). Prospects for Engineering Biophysical CO2 Concentrating Mechanisms into Land Plants to Enhance Yields. Annu. Rev. Plant Biol. 71, 461–485. doi: 10.1146/annurev-arplant-081519-040100
Hindu, V., Palacios-Rojas, N., Babu, R., Suwarno, W. B., Rashid, Z., Usha, R., et al. (2018). Identification and validation of genomic regions influencing kernel zinc and iron in maize. Theor. Appl. Genet. 131, 1443–1457. doi: 10.1007/s00122-018-3089-3
Hocking, P. J., Meyer, C. P. (1991). Enrichment and Nitrogen Stress on Growth, and Partitioning of Dry Matter and Nitrogen in Wheat and Maize. Austr. J. Plant Physiol. 18, 339–356. doi: 10.1071/PP9910339
Hogy, P., Wieser, H., Kohler, P., Schwadorf, K., Breuer, J., Franzaring, J., et al. (2009). Effects of elevated CO2 on grain yield and quality of wheat: results from a 3-year free-air CO2 enrichment experiment. Plant Biol. (Stuttg) 11 (Suppl 1), 60–69. doi: 10.1111/j.1438-8677.2009.00230.x
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Jacob, J., Lawlor, D. W. (1992). Dependence of photosynthesis of sunflower and maize leaves on phosphate supply, ribulose-1,5-bisphosphate carboxylase/oxygenase activity, and ribulose-1,5-bisphosphate pool size. Plant Physiol. 98, 801–807. doi: 10.1104/pp.98.3.801
Jiao, J. A., Chollet, R. (1991). Posttranslational regulation of phosphoenolpyruvate carboxylase in c(4) and crassulacean Acid metabolism plants. Plant Physiol. 95, 981–985. doi: 10.1104/pp.95.4.981
Jobe, T. O., Zenzen, I., Rahimzadeh Karvansara, P., Kopriva, S. (2019). Integration of sulfate assimilation with carbon and nitrogen metabolism in transition from C3 to C4 photosynthesis. J. Exp. Bot. 70, 4211–4221. doi: 10.1093/jxb/erz250
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S., Hibberd, J. M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75. doi: 10.1104/pp.114.238667
Khoury, C. K., Bjorkman, A. D., Dempewolf, H., Ramirez-Villegas, J., Guarino, L., Jarvis, A., et al. (2014). Increasing homogeneity in global food supplies and the implications for food security. Proc. Natl. Acad. Sci. U. S. A. 111, 4001–4006. doi: 10.1073/pnas.1313490111
Kocsy, G., Galiba, G., Brunold, C. (2001). Role of glutathione in adaptation and signalling during chilling and cold acclimation in plants. Physiol. Plant 113, 158–164. doi: 10.1034/j.1399-3054.2001.1130202.x
Kopriva, S., Chu, C. (2018). Are we ready to improve phosphorus homeostasis in rice? J. Exp. Bot. 69, 3515–3522. doi: 10.1093/jxb/ery163
Kopriva, S., Koprivova, A. (2005). Sulfate assimilation and glutathione synthesis in C-4 plants. Photosynt. Res. 86, 363–372. doi: 10.1007/s11120-005-3482-z
Kopriva, S. (2011). “Nitrogen and Sulfur Metabolism in C-4 Plants,” In: C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Advances in Photosynthesis and Respiration. Raghavendra A., Sage R. (eds), (Dordrecht: Springer), vol 32, pp. 109–128. doi: 10.1007/978-90-481-9407-0_7
Koprivova, A., Melzer, M., Von Ballmoos, P., Mandel, T., Brunold, C., Kopriva, S. (2001). Assimilatory sulfate reduction in C-3, C-3-C-4, and C-4 species of Flaveria. Plant Physiol. 127, 543–550. doi: 10.1104/pp.010144
Kubis, A., Bar-Even, A. (2019). Synthetic biology approaches for improving photosynthesis. J. Exp. Bot. 70, 1425–1433. doi: 10.1093/jxb/erz029
Lee, Y., Nishizawa, T., Takemoto, M., Kumazaki, K., Yamashita, K., Hirata, K., et al. (2017). Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity. Nat. Plants 3, 825–832. doi: 10.1038/s41477-017-0022-8
Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., et al. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Livingstone, C. (2015). Zinc: physiology, deficiency, and parenteral nutrition. Nutr. Clin. Pract. 30, 371–382. doi: 10.1177/0884533615570376
Loladze, I. (2014). Hidden shift of the ionome of plants exposed to elevated CO(2)depletes minerals at the base of human nutrition. Elife 3, e02245. doi: 10.7554/eLife.02245
Long, B. M., Hee, W. Y., Sharwood, R. E., Rae, B. D., Kaines, S., Lim, Y. L., et al. (2018). Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9, 3570. doi: 10.1038/s41467-018-06044-0
Lopez-Arredondo, D. L., Leyva-Gonzalez, M. A., Gonzalez-Morales, S. I., Lopez-Bucio, J., Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
Lundgren, M. R. (2020). C2 photosynthesis: a promising route towards crop improvement? New Phytol. in press. doi: 10.1111/nph.16494
Luo, Y., Su, B., Currie, W. S., Dukes, J. S., Finzi, A., Hartwig, U., et al. (2004). Progressive Nitrogen Limitation of Ecosystem Responses to Rising Atmospheric Carbon Dioxide. BioScience 54, 731–739. doi: 10.1641/0006-3568(2004)054[0731:PNLOER]2.0.CO;2
Majeran, W., Zybailov, B., Ytterberg, A. J., Dunsmore, J., Sun, Q., Van Wijk, K. J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell Proteomics 7, 1609–1638. doi: 10.1074/mcp.M800016-MCP200
Mallmann, J., Heckmann, D., Brautigam, A., Lercher, M. J., Weber, A. P., Westhoff, P., et al. (2014). The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 3, e02478. doi: 10.7554/eLife.02478
Maurino, V. G. (2019). Using energy-efficient synthetic biochemical pathways to bypass photorespiration. Biochem. Soc. Trans. 47, 1805–1813. doi: 10.1042/BST20190322
Medici, A., Szponarski, W., Dangeville, P., Safi, A., Dissanayake, I. M., Saenchai, C., et al. (2019). Identification of Molecular Integrators Shows that Nitrogen Actively Controls the Phosphate Starvation Response in Plants. Plant Cell 31, 1171–1184. doi: 10.1105/tpc.18.00656
Mellor, G. E., Tregunna, E. B. (1971). The localization of nitrate-assimilating enzymes in leaves of plants with the C4-pathway of photosynthesis. Can. J. Bot. 49, 137–142. doi: 10.1139/b71-024
Mendoza-Cozatl, D. G., Gokul, A., Carelse, M. F., Jobe, T. O., Long, T. A., Keyster, M. (2019). Keep talking: crosstalk between iron and sulfur networks fine-tunes growth and development to promote survival under iron limitation. J. Exp. Bot. 70, 4197–4210. doi: 10.1093/jxb/erz290
Micronutrient_Initiative (2009). Investing in the future: A united call to action on vitamin and mineral deficiencies (Ottawa: Micronutrient Initiative).
Millward, D. J. (1999). The nutritional value of plant-based diets in relation to human amino acid and protein requirements. Proc. Nutr. Soc. 58, 249–260. doi: 10.1017/s0029665199000348
Monson, R. K. (1989). The relative contributions of reduced photorespiration, and improved water-and nitrogen-use efficiencies, to the advantages of C3-C4 intermediate photosynthesis in Flaveria. Oecologia 80, 215–221. doi: 10.1007/BF00380154
Moore, R., Black, C. C. (1979). Nitrogen Assimilation Pathways in Leaf Mesophyll and Bundle Sheath Cells of C(4) Photosynthesis Plants Formulated from Comparative Studies with Digitaria sanguinalis (L.) Scop. Plant Physiol. 64, 309–313. doi: 10.1104/pp.64.2.309
Morris, R. J., Fox, R. H., Jung, G. A. (1982). Growth, P uptake and quality of warm and cool season grasses on a low available P soil. Agron. J. 74, 125–129. doi: 10.2134/agronj1982.00021962007400010032x
Myers, S. S., Zanobetti, A., Kloog, I., Huybers, P., Leakey, A. D., Bloom, A. J., et al. (2014). Increasing CO2 threatens human nutrition. Nature 510, 139–142. doi: 10.1038/nature13179
Myers, S. S., Smith, M. R., Guth, S., Golden, C. D., Vaitla, B., Mueller, N. D., et al. (2017). Climate Change and Global Food Systems: Potential Impacts on Food Security and Undernutrition. Annu. Rev. Public Health 38, 259–277. doi: 10.1146/annurev-publhealth-031816-044356
Newell, M. A., Vogel, K. E., Adams, M., Aydin, N., Bodnar, A. L., Ali, M., et al. (2014). Genetic and biochemical differences in populations bred for extremes in maize grain methionine concentration. BMC Plant Biol. 14:49. doi: 10.1186/1471-2229-14-49
Niro, S., D’Agostino, A., Fratianni, A., Cinquanta, L., Panfili, G. (2019). Gluten-Free Alternative Grains: Nutritional Evaluation and Bioactive Compounds. Foods 8, E208. doi: 10.3390/foods8060208
Noctor, G., Mhamdi, A., Chaouch, S., Han, Y., Neukermans, J., Marquez-Garcia, B., et al. (2012). Glutathione in plants: an integrated overview. Plant Cell Environ. 35, 454–484. doi: 10.1111/j.1365-3040.2011.02400.x
Parcell, S. (2002). Sulfur in human nutrition and applications in medicine. Altern. Med. Rev. 7, 22–44.
Parry, M. A., Andralojc, P. J., Scales, J. C., Salvucci, M. E., Carmo-Silva, A. E., Alonso, H., et al. (2013). Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 64, 717–730. doi: 10.1093/jxb/ers336
Passera, C., Ghisi, R. (1982). ATP sulphurylase and O-acetylserine sulphydrylase in isolated mesophyll protoplasts and bundle sheath strands of S-deprived maize leaves. J. Exp. Bot. 33, 432–438. doi: 10.1093/jxb/33.3.432
Pastori, G. M., Mullineaux, P. M., Foyer, C. H. (2000). Post-transcriptional regulation prevents accumulation of glutathione reductase protein and activity in the bundle sheath cells of maize. Plant Physiol. 122, 667–675. doi: 10.1104/pp.122.3.667
Pinto, H., Powell, J. R., Sharwood, R. E., Tissue, D. T., Ghannoum, O. (2016). Variations in nitrogen use efficiency reflect the biochemical subtype while variations in water use efficiency reflect the evolutionary lineage of C4 grasses at inter-glacial CO2. Plant Cell Environ. 39, 514–526. doi: 10.1111/pce.12636
Planta, J., Xiang, X., Leustek, T., Messing, J. (2017). Engineering sulfur storage in maize seed proteins without apparent yield loss. Proc. Natl. Acad. Sci. U. S. A. 114, 11386–11391. doi: 10.1073/pnas.1714805114
Pottier, M., Gilis, D., Boutry, M. (2018). The Hidden Face of Rubisco. Trends Plant Sci. 23, 382–392. doi: 10.1016/j.tplants.2018.02.006
Rathnam, C. K., Edwards, G. E. (1976). Distribution of Nitrate-assimilating Enzymes between Mesophyll Protoplasts and Bundle Sheath Cells in Leaves of Three Groups of C(4) Plants. Plant Physiol. 57, 881–885. doi: 10.1104/pp.57.6.881
Ross-Ibarra, J., Morrell, P. L., Gaut, B. S. (2007). Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. U. S. A. 104 (Suppl 1), 8641–8648. doi: 10.1073/pnas.0700643104
Rouached, H., Secco, D., Arpat, B., Poirier, Y. (2011). The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 11:19. doi: 10.1186/1471-2229-11-19
Sage, R. F., Pearcy, R. W., Seemann, J. R. (1987). The Nitrogen Use Efficiency of C(3) and C(4) Plants : III. Leaf Nitrogen Effects on the Activity of Carboxylating Enzymes in Chenopodium album (L.) and Amaranthus retroflexus (L.). Plant Physiol. 85, 355–359. doi: 10.1104/pp.85.2.355
Sage, R. F., Sage, T. L., Kocacinar, F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47. doi: 10.1146/annurev-arplant-042811-105511
Saleh, A. S., Zhang, Q., Chen, J., Shen, Q. (2013). Millet grains: nutritional quality, processing, and potential health benefits. Compr. Rev. Food Sci. Food Saf. 12, 281–295. doi: 10.1111/1541-4337.12012
Salesse-Smith, C. E., Sharwood, R. E., Busch, F. A., Kromdijk, J., Bardal, V., Stern, D. B. (2018). Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 4, 802–810. doi: 10.1038/s41477-018-0252-4
Schluter, U., Weber, A. P. (2016). The Road to C4 Photosynthesis: Evolution of a Complex Trait via Intermediary States. Plant Cell Physiol. 57, 881–889. doi: 10.1093/pcp/pcw009
Schmutz, D., Brunold, C. (1984). Intercellular Localization of Assimilatory Sulfate Reduction in Leaves of Zea mays and Triticum aestivum. Plant Physiol. 74, 866–870. doi: 10.1104/pp.74.4.866
Schmutz, D., Brunold, C. (1985). Localization of nitrite and sulfite reductase in bundle sheath and mesophyll cells of maize leaves. Physiol. Plant 64, 523–528. doi: 10.1111/j.1399-3054.1985.tb08533.x
Schuler, M. L., Mantegazza, O., Weber, A. P. (2016). Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 87, 51–65. doi: 10.1111/tpj.13155
Shiferaw, B., Prasanna, B. M., Hellin, J., Bänziger, M. (2011). Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 3, 307–327. doi: 10.1007/s12571-011-0140-5
Shimono, H., Farquhar, G., Brookhouse, M., Busch, F. A., Grady, A. O., Tausz, M., et al. (2018). Prescreening in large populations as a tool for identifying elevated CO2-responsive genotypes in plants. Funct. Plant Biol. 46, 1–14. doi: 10.1071/FP18087
Slack, C. R., Hatch, M. D. (1967). Comparative studies on the activity of carboxylases and other enzymes in relation to the new pathway of photosynthetic carbon dioxide fixation in tropical grasses. Biochem. J. 103, 660–665. doi: 10.1042/bj1030660
Smykal, P., Hradilova, I., Trneny, O., Brus, J., Rathore, A., Bariotakis, M., et al. (2017). Genomic diversity and macroecology of the crop wild relatives of domesticated pea. Sci. Rep. 7, 17384. doi: 10.1038/s41598-017-17623-4
Sonawane, B. V., Sharwood, R. E., Von Caemmerer, S., Whitney, S. M., Ghannoum, O. (2017). Short-term thermal photosynthetic responses of C4 grasses are independent of the biochemical subtype. J. Exp. Bot. 68, 5583–5597. doi: 10.1093/jxb/erx350
Studer, A. J., Schnable, J. C., Weissmann, S., Kolbe, A. R., Mckain, M. R., Shao, Y., et al. (2016). The draft genome of the C3 panicoid grass species Dichanthelium oligosanthes. Genome Biol. 17, 223. doi: 10.1186/s13059-016-1080-3
Takahashi, H., Buchner, P., Yoshimoto, N., Hawkesford, M. J., Shiu, S. H. (2011a). Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2:119. doi: 10.3389/fpls.2011.00119
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., Hell, R. (2011b). Sulfur Assimilation in Photosynthetic Organisms: Molecular Functions and Regulations of Transporters and Assimilatory Enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Taub, D. R. (2000). Climate and the U.S. distribution of C4 grass subfamilies and decarboxylation variants of C4 photosynthesis. Am. J. Bot. 87, 1211–1215. doi: 10.2307/2656659
Ujiie, K., Ishimaru, K., Hirotsu, N., Nagasaka, S., Miyakoshi, Y., Ota, M., et al. (2019). How elevated CO2 affects our nutrition in rice, and how we can deal with it. PLoS One 14, e0212840. doi: 10.1371/journal.pone.0212840
Vaughn, K. C., Campbell, W. H. (1988). Immunogold localization of nitrate reductase in maize leaves. Plant Physiol. 88, 1354–1357. doi: 10.1104/pp.88.4.1354
Von Caemmerer, S., Furbank, R. T. (2003). The C(4) pathway: an efficient CO(2) pump. Photosynth. Res. 77, 191–207. doi: 10.1023/A:1025830019591
Wang, W., Niu, S., Dai, Y., Zhai, X., Wang, M., Ding, Y., et al. (2019). Molecular Mechanisms Underlying Increase in Lysine Content of Waxy Maize through the Introgression of the opaque2 Allele. Int. J. Mol. Sci. 20, E684. doi: 10.3390/ijms20030684
Weckopp, S. C., Kopriva, S. (2014). Are changes in sulfate assimilation pathway needed for evolution of C4 photosynthesis? Front. Plant Sci. 5:773. doi: 10.3389/fpls.2014.00773
Wurtzel, E. T., Cuttriss, A., Vallabhaneni, R. (2012). Maize provitamin a carotenoids, current resources, and future metabolic engineering challenges. Front. Plant Sci. 3:29. doi: 10.3389/fpls.2012.00029
Xie, X., Hu, W., Fan, X., Chen, H., Tang, M. (2019). Interactions Between Phosphorus, Zinc, and Iron Homeostasis in Nonmycorrhizal and Mycorrhizal Plants. Front. Plant Sci. 10:1172. doi: 10.3389/fpls.2019.01172
Zhang, G., Sakai, H., Tokida, T., Usui, Y., Zhu, C., Nakamura, H., et al. (2013). The effects of free-air CO(2) enrichment (FACE) on carbon and nitrogen accumulation in grains of rice (Oryza sativa L.). J. Exp. Bot. 64, 3179–3188. doi: 10.1093/jxb/ert154
Zhang, Z., Mao, Y., Ha, S., Liu, W., Botella, J. R., Zhu, J. K. (2016). A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 35, 1519–1533. doi: 10.1007/s00299-015-1900-z
Zhu, C., Kobayashi, K., Loladze, I., Zhu, J., Jiang, Q., Xu, X., et al. (2018). Carbon dioxide (CO2) levels this century will alter the protein, micronutrients, and vitamin content of rice grains with potential health consequences for the poorest rice-dependent countries. Sci. Adv. 4, eaaq1012. doi: 10.1126/sciadv.aaq1012
Ziska, L. H., Blumenthal, D. M. (2007). Empirical selection of cultivated oat in response to rising atmospheric carbon dioxide. Crop Sci. 47, 1547–1552. doi: 10.2135/cropsci2006.09.0616
Keywords: sulfur, nitrogen, phosphorus, C4 photosynthesis, maize, Flaveria, hidden hunger
Citation: Jobe TO, Rahimzadeh Karvansara P, Zenzen I and Kopriva S (2020) Ensuring Nutritious Food Under Elevated CO2 Conditions: A Case for Improved C4 Crops. Front. Plant Sci. 11:1267. doi: 10.3389/fpls.2020.01267
Received: 19 May 2020; Accepted: 03 August 2020;
Published: 18 August 2020.
Edited by:
Martha Ludwig, University of Western Australia, AustraliaReviewed by:
Daniel Marino, University of the Basque Country, SpainCopyright © 2020 Jobe, Rahimzadeh Karvansara, Zenzen and Kopriva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stanislav Kopriva, c2tvcHJpdmFAdW5pLWtvZWxuLmRl
†Present address: Parisa Rahimzadeh Karvansara, Centre Algatech, Institute of Microbiology of the Czech Academy of Sciences, Třeboň, Czechia
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.