- Division of Applied Life Science (BK21 Plus Program), Plant Molecular Biology and Biotechnology Research Center, Gyeongsang National University, Jinju, South Korea
Lateral root development is known to be regulated by Aux/IAA-ARF modules in Arabidopsis thaliana. As components, several Aux/IAAs have participated in these Aux/IAA-ARF modules. In this study, to identify the biological function of IAA15 in plant developments, transgenic plant overexpressing the gain-of-function mutant of IAA15 (IAA15P75S OX) under the control of dexamethasone (DEX) inducible promoter, in which IAA15 protein was mutated by changing Pro-75 residue to Ser at the degron motif in conserved domain II, was constructed. As a result, we found that IAA15P75S OX plants show a decreased number of lateral roots. Coincidently, IAA15 promoter-GUS reporter analysis revealed that IAA15 transcripts were highly detected in all stages of developing lateral root tissues. It was also verified that the IAA15P75S protein is strongly stabilized against proteasome-mediated protein degradation by inhibiting its poly-ubiquitination, resulting in the transcriptional repression of auxin-responsive genes. In particular, transcript levels of LBD16 and LBD29, which are positive regulators of lateral root formation, dramatically repressed in IAA15P75S OX plants. Furthermore, it was elucidated that IAA15 interacts with ARF7 and ARF19 and binds to the promoters of LBD16 and LBD29, strongly suggesting that IAA15 represses lateral root formation through the transcriptional suppression of LBD16 and LBD29 by inhibiting ARF7 and ARF19 activity. Taken together, this study suggests that IAA15 also plays a key negative role in lateral root formation as a component of Aux/IAA-ARF modules.
Introduction
Lateral root development is an auxin-regulated developmental process that maximizes the ability of the root system to absorb water and nutrients from the soil (Péret et al., 2009). The phytohormone auxin has been known to promote lateral root development (Torrey, 1950; Blakely et al., 1988). Exogenous auxin promotes cell division in the root pericycle, thereby inducing lateral root formation, whereas inhibitors of auxin transport inhibit lateral root initiation (Casimiro et al., 2001; Alarcón et al., 2019). In Arabidopsis, lateral root development begins with the auxin-dependent local activation of pericycle cells at the two protoxylem poles (Beeckman et al., 2001; Casimiro et al., 2001; Casimiro et al., 2003). These pericycle-derived cells undergo a series of cell division and differentiation processes, leading to the formation of lateral root (Laskowski et al., 1995).
Auxin regulates almost every aspect of plant growth and development, including embryo patterning, root and stem growth, organ senescence, vascular differentiation, seed germination, and flower development (Gälweiler et al., 1998; Friml et al., 2002; Ellis et al., 2005; Weijers et al., 2006; Wang et al., 2013; Hussain et al., 2020). The auxin-signaling pathway is highly conserved in all higher plants (Reed, 2001; Guilfoyle and Hagen, 2007; Mockaitis and Estelle, 2008). Three components primarily control auxin signaling: the auxin receptors TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX PROTEINs (AFBs), transcriptional repressors AUXIN/INDOLE-3-ACETIC ACIDs (Aux/IAAs), and transcription factors AUXIN RESPONSE FACTORs (ARFs). Under low auxin levels, Aux/IAA proteins recruit the co-repressor TOPLESS to inhibit the transcription of auxin-responsive genes by repressing the activities of their interaction partners ARFs. ARFs directly bind to the promoters of their target genes and activate or inhibit their transcription (Weijers et al., 2005; Szemenyei et al., 2008). Under high auxin levels, auxin is perceived by the auxin receptors TIR1/AFBs, and the SCFTIR1/AFBs E3 ubiquitin-ligase complex targets Aux/IAA repressors for degradation via poly-ubiquitination (Dharmasiri et al., 2005; Mockaitis and Estelle, 2008). Once Aux/IAAs are degraded by the 26S proteasome, the free ARFs initiate the transcription of auxin-responsive genes (Ulmasov et al., 1997; Weijers et al., 2005).
The Aux/IAAs family in Arabidopsis were first identified based on their strong induction by auxin at the transcriptional level. Most Aux/IAA proteins contain four conserved domains (I, II, III, and IV) (Liscum and Reed, 2002). Domain I contributes to the transcriptional repression of Aux/IAA targets by recruiting the co-repressor, TOPLESS. Domain II contains a highly conserved degron motif (VGWPPV), the docking site of the auxin-activated SCFTIR1/AFBs ubiquitin E3 ligase complex, leading to the poly-ubiquitination of the target proteins. Domain III and IV are involved in homo- or hetero-interactions with other Aux/IAAs or ARFs. When the conserved amino acid in domain II of an Aux/IAA protein harbors a point mutation, the protein becomes stable because it cannot interact with SCFTIR1/AFBs, even in the presence of auxin. Stabilized Aux/IAA mutants are thought to constitutively interact with and inactivate ARFs during various biological processes, thereby inhibiting auxin signaling. Many biological functions of Aux/IAAs have been identified by investigating various gain-of-function mutants of Aux/IAA, such as iaa1/axr5, iaa3/shy2, iaa6/shy1, iaa7/axr2, iaa8, iaa12/bdl, iaa14/slr, iaa16, iaa17/axr3, iaa18, iaa19/msg2, and iaa28 (Kim et al., 1996; Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001; Hamann et al., 2002; Tatematsu et al., 2004; Yang et al., 2004; Fukaki et al., 2006; Ploense et al., 2009; De Rybel et al., 2010; Rinaldi et al., 2012; Wang et al., 2013). These mutations result in auxin-related phenotypes, including altered developments of hypocotyl, root, leaf, embryo, and tropisms.
Many genetic studies using Arabidopsis mutants have reported that lateral root development is primarily regulated by several Aux/IAAs, ARFs, and LATERAL ORGAN BOUNDARIES-DOMAINs (LBDs). For example, gain-of-function mutants of Aux/IAA, such as iaa3/shy2, iaa14/slr, iaa19/msg2, and iaa28, show severely reduced lateral root phenotypes due to inhibited ARF activity (Tian and Reed, 1999; Rogg et al., 2001; Tatematsu et al., 2004; Fukaki et al., 2006). Analysis of the arf7, arf19, and arf7 arf19 mutants showed that ARF7 and ARF19 play positive roles in lateral root formation (Okushima et al., 2005; Wilmoth et al., 2005). Furthermore, LBD16 and LBD29 play a positive role in lateral root formation downstream of ARF7 and ARF19 (Okushima et al., 2007; Goh et al., 2012). These studies provided the molecular signaling pathway of how Aux/IAA-ARF modules-mediated auxin signaling is involved in lateral root formation.
It was reported that IAA15 modulates auxin homeostasis that controls gravitropic responses and stem cell differentiation (Yan et al., 2013). However, the biological functions of IAA15 in other developmental processes were not extensively studied. In the current study, we generated transgenic Arabidopsis plants overexpressing a gain-of-function mutant form of IAA15 (IAA15P75S OX) with a mutation in a conserved amino acid in domain II. The IAA15P75S OX plants showed auxin-deficient phenotypes, including shortened primary roots and decreased lateral roots compared to the wild type. When we investigated the expression of IAA15 gene by using IAA15pro::GUS reporter construct, IAA15 transcripts were mainly detected in lateral root primordia and mature lateral root tips. Stabilized IAA15P75S mutant protein inhibits the transcription of auxin-responses genes including two LBD genes. Moreover, we showed that IAA15 interacted with ARF7 and ARF19 and bound to the promoters of LBD16 and LBD29.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana lines used in this study were in the Columbia background. For surface sterilization, seeds were soaked for 1 min in 70% EtOH, followed by 10 min in 1/10-diluted commercial bleach (0.4% NaOCl) and three washes with sterile distilled water. Surface-sterilized seeds were sown on agar plates containing half-strength Murashige-Skoog (MS) salts and vitamins (Murashige and Skoog, 1962), 2.0% sucrose, and 0.8% agar. The plates were incubated for 3 d at 4°C in the dark, and then at 22°C in a growth chamber under a 16 h light/8 h dark photoperiod with a light intensity of ~120 μmol m-2 s-1. Ten- to twelve-day-old seedlings were transferred to soil and grown under the same conditions. To explore the plant response to exogenous auxin, IAA15P75S OX plants were vertically germinated and grown on MS medium containing 0, 10, 25, or 50 nM NAA with or without 50 μM DEX and photographed at 14 d after germination. For stress treatments, IAA15P75S OX plants were vertically germinated and grown on MS medium containing 50 μM DEX plus NaCl (50 or 100 mM), mannitol (200 or 300 mM), or ABA (0.1 or 0.2 μM). The response to stress was estimated by measuring primary root length at 14 d after germination.
Plasmid Construction, Site-Directed Mutagenesis, and Plant Transformation
To generate the DEXpro::3XFLAG-IAA15WT and DEXpro::3XFLAG-IAA15P75S constructs, the coding sequence of IAA15 was amplified by PCR from a cDNA library prepared from Arabidopsis seedlings using gene-specific primers (Supplementary Table 1) and cloned into the T-blunt vector (Solgent, Korea). The amino acid substitution (Pro-75 to Ser-75) in full-length IAA15 was produced with the primers listed in Supplementary Table 1 using a QuikChange Site-directed Mutagenesis kit (Stratagene, USA). IAA15WT and IAA15 P75S were subcloned into the BamHI and SpeI sites of the pBlueScript II KS (+) vector containing 3XFLAG. 3XFLAG-IAA15WT and 3XFLAG-IAA15P75S were cloned into the pTA7002 binary vectors under the control of DEX-inducible promoter. For IAA15pro::GUS, a 2.4 kb genomic region upstream of the ATG start codon was cloned into the HindIII and SalI sites of pBlueScript II KS (+) containing GUS. IAA15pro::GUS was cloned into the HindIII and SacI sites of the pPZP211 binary vector. All constructs were confirmed by sequencing and transformed into Arabidopsis (Col-0) using Agrobacterium strain GV3101 (pMP90) by the floral dip method (Clough and Bent, 1998). Transformants were selected on MS medium containing the proper antibiotics. T3 homozygous progeny of transgenic plants expressing high levels of IAA15 were used for all experiments.
Extraction and Immunoblot Analysis of Arabidopsis Proteins
To extract total plant proteins, tissues were ground in liquid nitrogen and extracted in protein extraction buffer (50 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 1 mM Na3VO4, 25 mM NaF, 50 mM-glycerophosphate, 2 mM DTT, 2 mM PMSF, 5% glycerol, 1% Triton X-100, and protease inhibitor). After two rounds of centrifugation at 12,000 × g for 10 min, the supernatants were transferred to clean tubes and stored at -80°C until use. Protein concentrations were determined using a Bio-Rad Protein Assay kit (Bio-Rad) with BSA as a standard. For immunoblot analysis, 50 µg total protein samples were separated by 10% SDS-PAGE and transferred to PVDF membranes. Proteins were detected using mouse anti-Flag (1:5,000; Sigma, USA) as a primary antibody and horseradish peroxidase-conjugated anti-mouse as a secondary antibody (1:5,000) and visualized using an ECL kit (Bio-Rad, USA).
Histochemical GUS Assays
To investigate the expression pattern of IAA15, GUS activity in IAA15pro::GUS plants was detected histochemically as described by Jefferson et al. (1987) with slight modifications. Briefly, the tissue was incubated in 2 mM 5-bromo-4-chloro-3-indolyl- β-D-glucuronic acid in 50 mM phosphate buffer, pH 7.0, containing 0.5 mM K3Fe(CN)6 and 0.5 mM K4Fe(CN)6 for 6 h at 37°C. The tissue was rinsed with 50 mM phosphate buffer, fixed, and cleared overnight in ethanol (100%): acetic acid (9:1, v/v) at room temperature. The sample was photographed under a Nikon SMZ1000 stereoscopic microscope equipped with an OLYMPUS C-5050 ZOOM digital camera.
In Vivo Turnover and Ubiquitination Assays
For the in vivo turnover assay of IAA15 proteins, 10-day-old seedlings overexpressing IAA15WT or IAA15P75S were pretreated with 50 μM DEX and 10 μM MG132 for 24 h, and then treated with 10 μM NAA in the presence of 1 mM cycloheximide (CHX) for 4 h. 50 µg total protein samples were separated by SDS-PAGE, and immunoblot analysis was performed using mouse anti-Flag (Sigma, USA). For the ubiquitination assay, 10-day-old DEX-treated IAA15WT OX and IAA15P75S OX seedlings were pretreated with 10 μM MG132 for 2 h, and then treated with or without 10 μM NAA for 24 h. Total plant protein (1 mg) was immunoprecipitated using anti-Flag antibody coupled to agarose beads (Sigma, St. Louis, MO, USA) for 4 h at 4°C. The beads were recovered by centrifugation, washed five times with extraction buffer, and eluted with 30 μL SDS-PAGE sample buffer. The eluted samples were separated by SDS-PAGE, and immunoblot analysis was performed using a monoclonal anti-ubiquitin antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) as described (Miura et al., 2011).
Quantitative RT-PCR
Total RNA was extracted from the samples using an RNA purification kit (Macherey-Nagel, Germany), and 5 μg total RNA was reverse transcribed in a 100 μl reaction volume using SuperScript II RNase-Reverse Transcriptase (Invitrogen, USA). Quantitative RT-PCR was performed in a 10 μl reaction volume containing 1 μl RT products, 10 pmol of gene-specific primers, and 5 μl SsoFast EvaGreen Supermix (Bio-Rad, USA) using the CFX384 Real-Time System (Bio-Rad, USA). The reaction conditions included an initial 5 min pre-incubation at 94°C, 45 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s, followed by melting curve analysis via 90 cycles at 55°C, increasing by 0.5°C/cycle, and a final cooling step for 10 min at 72°C. The primers used for PCR are shown in Supplementary Table 2.
Yeast Two-Hybrid Analysis
The full-length open reading frames of IAA15 and ARF7/ARF29 were amplified using gene-specific primers (Supplementary Table 1) and cloned into pGAD424 (AD) and pAS2-1 (BD), respectively. The AD and BD plasmids were co-transformed into yeast strain pJ69-4A for interaction analysis, and colonies were selected after 2–3 days of growth at 30°C on SD-LEU/TRP medium. The transformants were tested for positive bait-prey interactions by monitoring the activation of HIS3 reporter genes, as described in the Clontech yeast protocol handbook.
Chromatin Immunoprecipitation (ChIP) Assay
Two-week-old IAA15P75S OX seedlings were treated with or without 50 μM DEX for 24 h, and the samples were collected. ChIP was carried out as described (Bastow et al., 2004; Tian et al., 2005) using mouse polyclonal anti-Flag antibody (1:3,000; Sigma, USA). PCR amplification was performed quantitatively using the CFX384 Real-Time System (Bio-Rad, USA). The immunoprecipitation was replicated three times, and each sample was quantified at least in triplicate. The ChIP primers are listed in Supplementary Table 2.
Results
Transgenic Plants Expressing an IAA15 Gain-of-Function Mutant Show Defects in Lateral Root Development
Almost all Aux/IAA gain-of-function mutants having point mutation at the degron motif (VGWPP) of these proteins showed auxin deficient phenotypes (Reed, 2001). However, to date, a gain-of-function mutant of IAA15 has not been isolated. To identify the biological function of IAA15, in this study, we constructed transgenic plants overexpressing wild-type IAA15 (IAA15WT) and a gain-of-function mutant of IAA15 (IAA15P75S) under the control of the 35S promoter. IAA15P75S contains a Pro-75 to Ser-75 substitution in domain II of IAA15. Three independent lines of homozygous transgenic plants overexpressing IAA15WT (IAA15WT OX) were selected (Supplementary Figure 1). However, we failed to generate homozygous transgenic plants overexpressing IAA15P75S (IAA15P75 OX), perhaps because they were all embryonically lethal. Therefore, we generated transgenic plants expressing IAA15WT and IAA15P75S under the control of a DEX-inducible promoter. Three independent lines of homozygous transgenic plants with high levels of IAA15 transcripts were selected after DEX treatment (Figure 1).
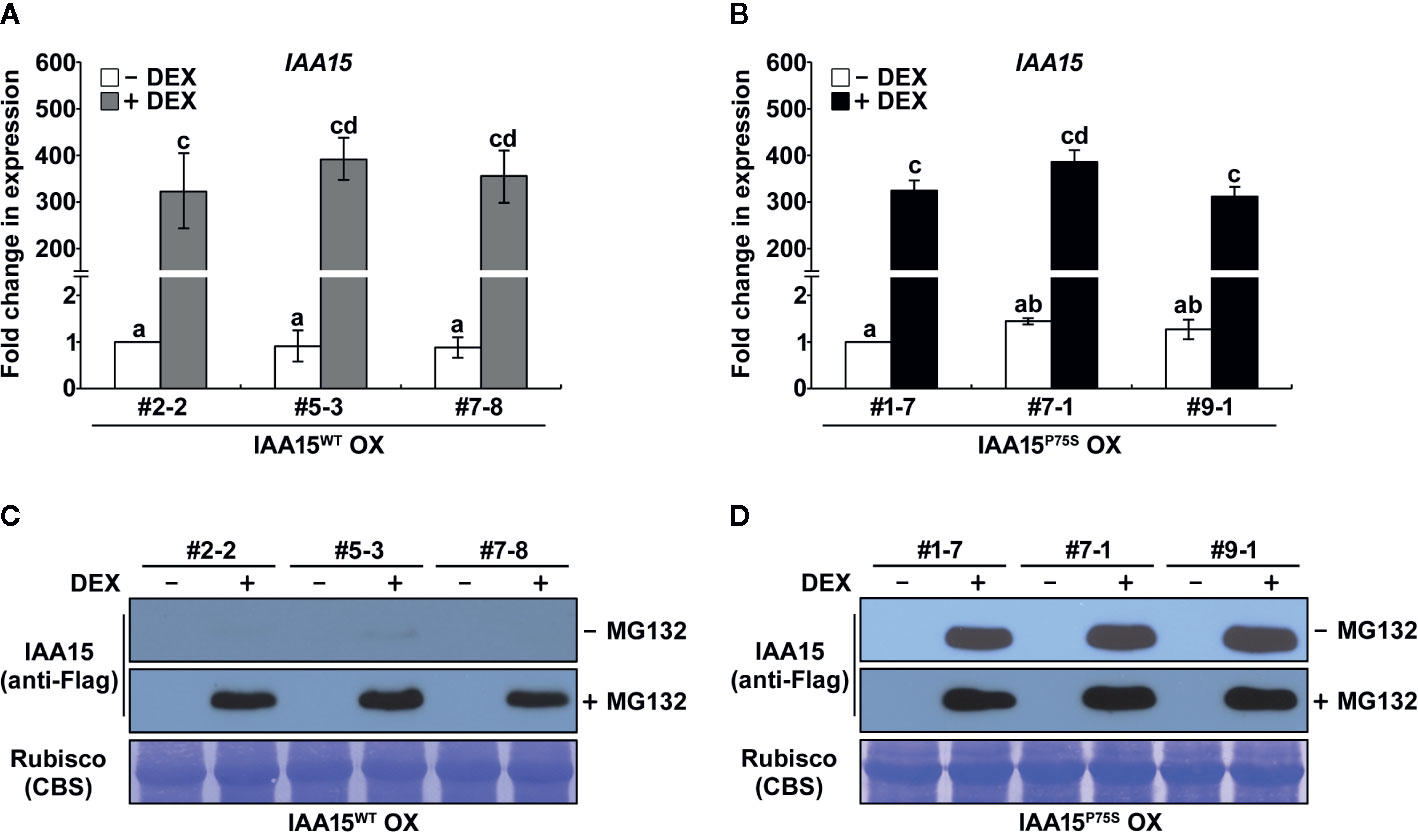
Figure 1 IAA15P75S protein is stable. (A, B) Analysis of IAA15 transcript levels via qRT-PCR using total RNA isolated from 2-week-old seedlings of three independent IAA15WT OX (A) and IAA15P75S OX (B) lines with or without 50 μM DEX induction of IAA15WT and IAA15P75S expression. The bars indicate the mean ± S.D. (n = 3). Different letters represent significant differences. (C, D) Immunoblot and Coomassie blue staining (CBS) of a polyacrylamide gel containing total proteins isolated from the IAA15WT OX (C) and IAA15P75S OX (D) seedlings with or without 50 μM DEX in the absence and presence of MG132. The gel blot was probed with anti-Flag antibody (anti-Flag). The Rubisco band detected by CBS shows the amount of protein loaded in each well.
We investigated the auxin-related phenotypes of these transgenic plants. The treatment of IAA15WT OX plants with or without DEX showed similar phenotypes to WT in terms of plant growth and development (Figures 2A, C, D and Supplementary Figure 2). DEX-untreated IAA15P75S OX plants also showed no different phenotypes compared to WT, while DEX-treated IAA15P75S OX plants showed defects in root development (Figures 2B–D and Supplementary Figure 3). In particular, the primary root length and lateral root number in DEX-treated IAA15P75S OX plants were reduced by approximately 40% and 90%, respectively, compared to the WT (Figures 2C, D), suggesting that the IAA15P75S protein is negatively involved in primary and lateral root development. Because each of three independent lines of IAA15WT OX and IAA15P75S OX showed similar expression patterns and root growth phenotype, we mixed seeds from these lines together and used these for further studies, and hereafter refer as IAA15WT OX and IAA15P75S OX, respectively.
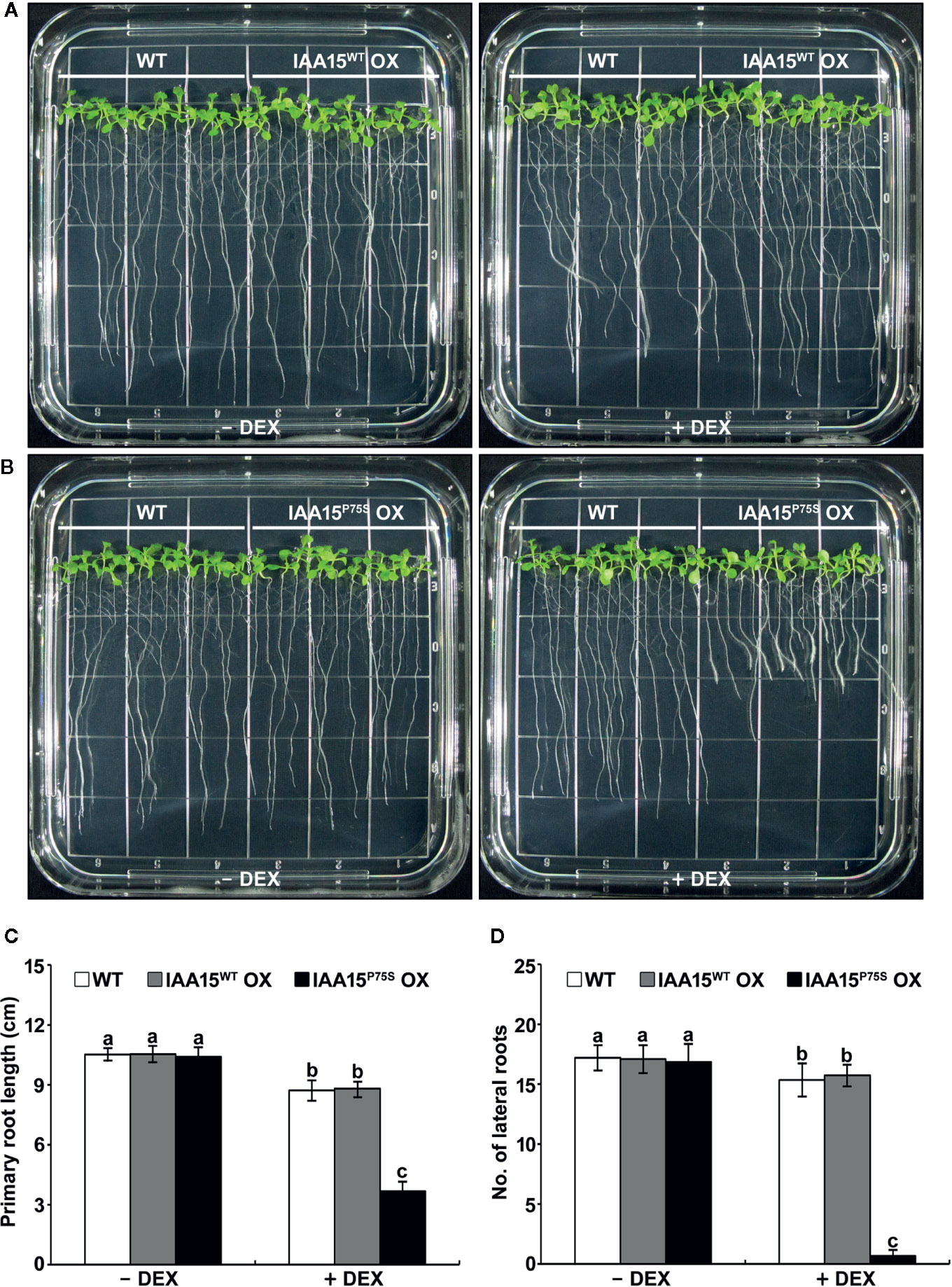
Figure 2 Primary root growth and lateral root formation are inhibited in IAA15P75S OX plants. (A, B) Root growth phenotypes of wild-type (WT), IAA15WT OX (A), and IAA15P75S OX (B) with and without DEX. Shown are two-week-old seedlings grown on MS medium with or without 50 μM DEX. (C, D) Primary root length (C) and lateral root number (D) of 2-week-old seedlings of IAA15WT OX and IAA15P75S OX grown on MS medium with or without 50 μM DEX. The bars indicate the mean ± S.D. (n = 20 to 25). Different letters indicate significant differences (P < 0.05) among lines that were explored through Tukey’s multiple comparisons tests.
IAA15 Is Expressed During Lateral Root Initiation and Development
The investigation of the expression patterns is helpful in estimating the unknown physiological function of a specific gene. To explore the expression pattern of IAA15, we generated transgenic plants expressing the β-glucuronidase (GUS) reporter gene under the control of the IAA15 promoter. GUS activities were detected in the shoot apical meristem, petioles, veins, tips of cotyledons, primary root tips, lateral root primordia, and lateral root tips (Figure 3A). GUS activities were also highly detected at all stages in morphologically recognizable lateral root primordia and highly in primordium tips after maturation (Figure 3B). To examine the presence of specific cis-regulatory elements in the promoter, we performed in silico analysis on the IAA15 promoter. We found that IAA15 promoter includes many root-specific cis- regulatory elements, such as Sorlip1, AS1-Box, and FaRB7_root-specific (Supplementary Figure 4) (Puzio et al., 2000; Jiao et al., 2005; Vaughan et al., 2006; Kakrana et al., 2017). These cis-elements were also found on the IAA14 gene promoter, which plays role in lateral root development. Taken together, these results suggest that IAA15 plays a functional role in lateral root development.
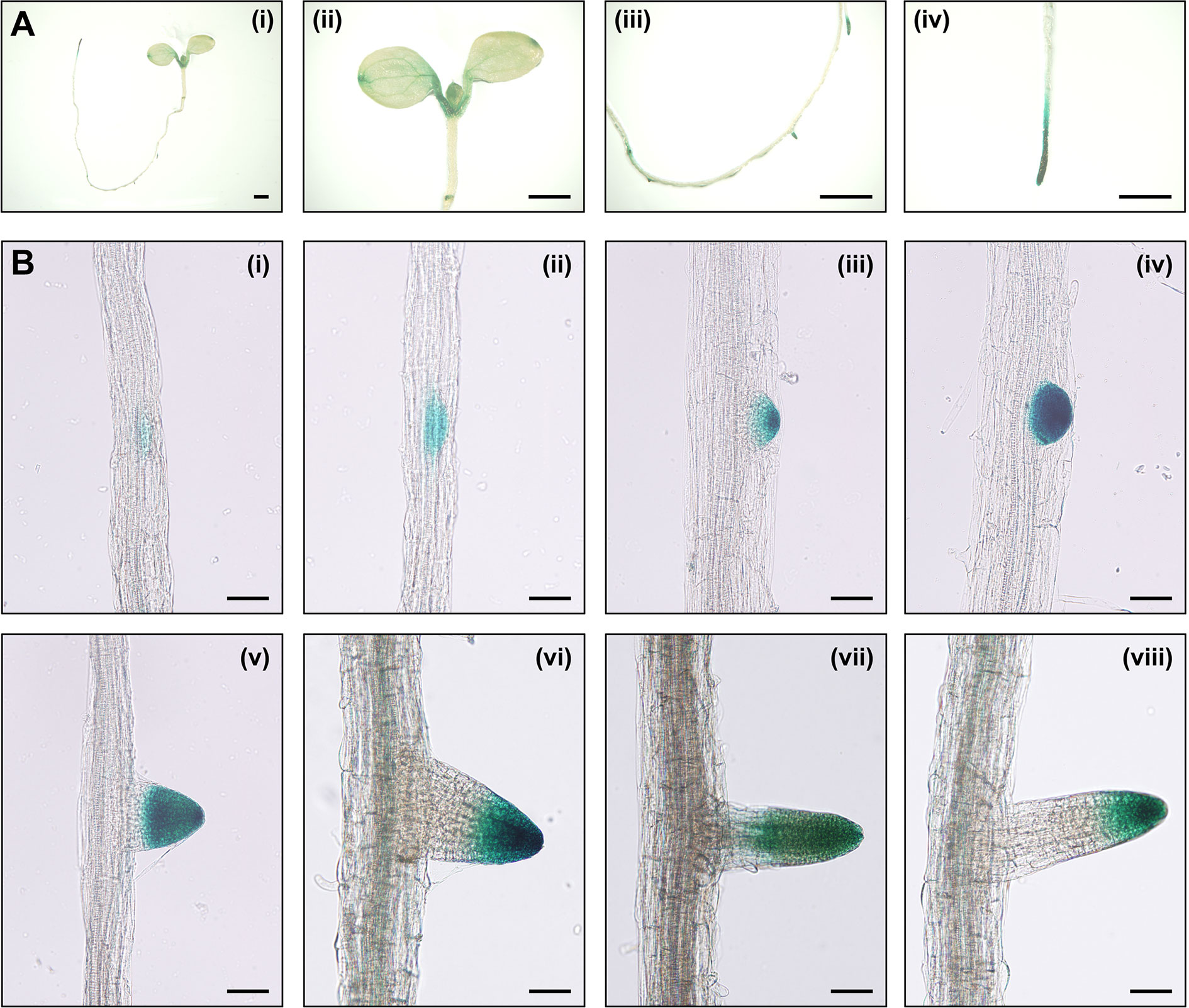
Figure 3 The expression patterns of IAA15 in Arabidopsis tissues. (A) Tissue-specific expression of IAA15 in IAA15pro::GUS transgenic plants, including a 7-day-old seedling (i), cotyledons, and shoot apical region (ii), lateral root and primordia in the root elongation zone (iii), and primary root tip (iv). Bars = 1 mm. (B) Expression pattern of IAA15 during different stages of lateral root formation. Seven-day-old seedlings were incubated for 24 h in 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc) for GUS staining. Bars = 50 μm.
Mutation of the Degron Motif of IAA15 Increases Protein Stability by Inhibiting Poly-Ubiquitination
Aux/IAAs are short-lived proteins that are rapidly degraded by the ubiquitin-proteasome mediated pathway (Santner et al., 2009). Gain-of-function mutations of Aux/IAA genes confer auxin-related phenotypes by stabilizing Aux/IAA proteins (Reed, 2001). As expected, IAA15 proteins were not detected in IAA15WT OX plants because this protein is unstable in normal condition (Figure 1C and Supplementary Figure S1). By contrast, IAA15 highly accumulated in response to treatment with the proteasome inhibitor MG132. Interestingly, high levels of IAA15 were also detected in IAA15P75S OX plants in response to DEX treatment, even in the absence of MG132 (Figure 1D). This result indicates that IAA15P75S become stable in normal growth condition.
To confirm whether IAA15P75S is indeed more stable than IAA15WT, we measured the degradation rate of these proteins in IAA15WT OX and IAA15P75S OX plants. We accumulated IAA15WT and IAA15P75S proteins in these transgenic plants by treatment with MG132 and DEX for 24 h, and then measured the amount of proteins by Western blots in the presence or absence of NAA plus CHX for 4 h. CHX was used to prevent de novo synthesis of proteins in plants. As a result, IAA15WT was rapidly degraded within 2 h by the treatment with or without NAA and almost undetectable after 4 h (Figure 4A). In contrast, IAA15P75S was stable even in the presence of NAA (Figure 4B), indicating that IAA15P75S is more stable than IAA15WT.
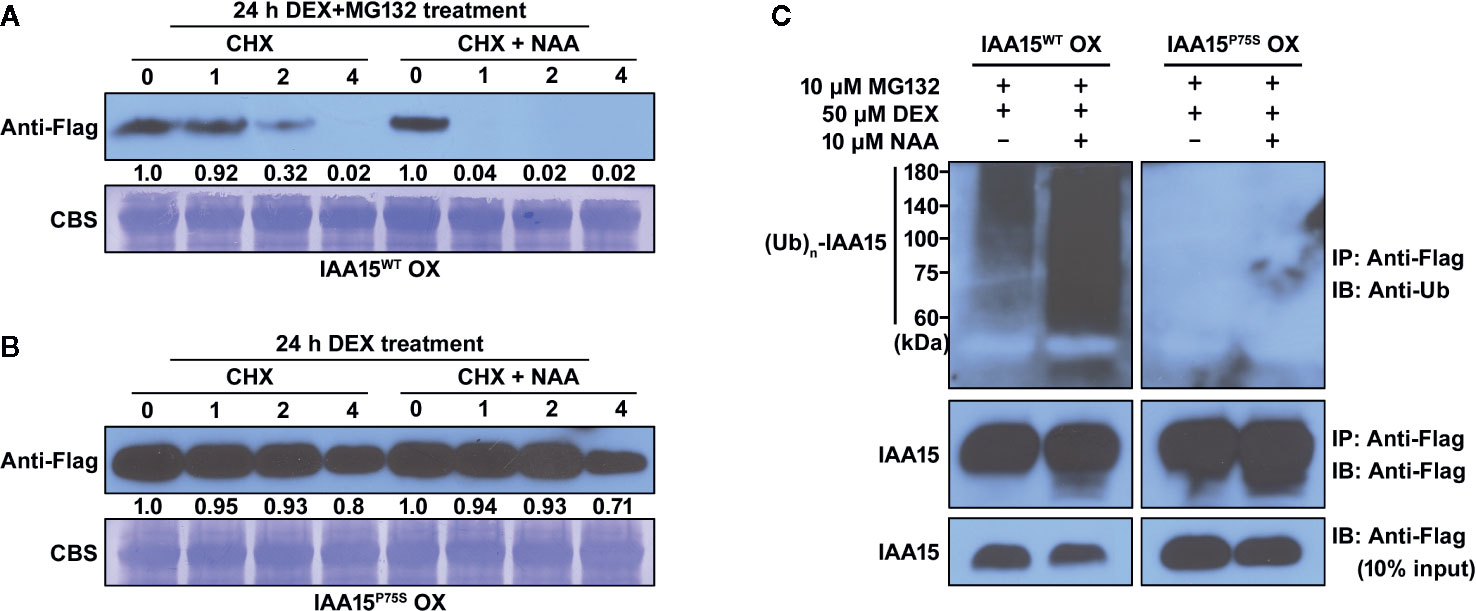
Figure 4 IAA15P75S protein has greater stability than IAA15WT. (A, B) Stability of IAA15WT and IAA15P75S in vivo. To induce accumulation of IAA15WT and IAA15P75S, 14-day-old seedlings of IAA15WT OX (A) and IAA15P75S OX (B) were treated with 50 μM DEX and 10 μM MG132 for 24 h, followed by 1 mM cycloheximide (CHX) with or without 10 μM NAA. Images show polyacrylamide gels of total proteins isolated from the seedlings after 0, 1, 2, and 4 h of CHX or CHX + NAA treatment. IAA15WT and IAA15P75S were detected with anti-Flag antibody. Rubisco band detected by CBS shows the amount of protein loaded in each well. (C) Auxin-dependent poly-ubiquitination of IAA15WT and IAA15P75S. IAA15 poly-ubiquitination was detected by immunoblotting (IB) with anti-ubiquitin antibody after immunoprecipitation (IP) with anti-Flag antibody from DEX-treated IAA15WT OX and IAA15P75S OX plants treated with MG132 in the absence or presence of NAA (upper panel). IAA15 proteins were detected by immunoblotting with anti-Flag antibody (lower panel, 10% input).
Next, to examine whether the Pro-75 to Ser point mutation of the degron domain would affect the poly-ubiquitination of IAA15 by the SCFTIR1/AFBs complex, we measured the ubiquitination efficiencies of IAA15WT and IAA15P75S proteins. To block the auxin-induced degradation of ubiquitinated IAA15, we treated DEX-pretreated IAA15WT OX and IAA15P75S OX plants with MG132 in the absence or presence of NAA. The accumulated IAA15 proteins were immunoprecipitated from total extracts of IAA15WT OX and IAA15P75S OX plants with anti-Flag antibody and immunoblotted with anti-ubiquitin antibody to measure the levels of ubiquitinated IAA15 protein, as described (Miura et al., 2011). Inputs of IAA15 proteins in IAA15WT OX and IAA15P75S OX plants were similarly detected by treatments with DEX and MG132 (Figure 4C, lower panel). As expected, poly-ubiquitinated IAA15WT was highly detected in IAA15WT OX plant by treatment with NAA, while those of IAA15P75S were less detected in IAA15P75S OX plant in both the presence and the absence of NAA (Figure 4C, upper panel). Taken together, these results suggest that the point mutation in the degron motif of IAA15 increases its protein stability by the inhibition of poly-ubiquitination and the subsequent proteasome-mediated degradation.
IAA15 Negatively Regulates Auxin-Responsive Genes Including LBD16 and LBD29
To examine the effect of stabilization of IAA15 protein on auxin response, we analyzed root growth in IAA15P75S OX plants in response to exogenous auxin (Supplementary Figure 4). As expected, DEX-untreated IAA15P75S OX seedlings showed inhibited primary root growth and increased lateral root number in response to 10 and 25 nM NAA. In DEX-treated IAA15P75S OX seedlings, however, the primary root growth was only slightly reduced, and there was no significant difference in lateral root number in the presence of 10 and 25 nM NAA. This insensitivity of IAA15P75S OX seedlings to auxin was no longer detected in a high auxin level (over 50 nM) (Supplementary Figure 4). These results indicate that the stabilized IAA15 protein represses auxin response.
Because the stabilization of IAA15 resulted in impairment of auxin-related phenotypes, we hypothesized that the accumulation of IAA15 inhibits the expression of auxin-responsive genes. To test this hypothesis, DEX-untreated and -treated IAA15P75S OX plants have analyzed the expression of auxin-responsive genes such as IAA5, IAA14, GH3.3, and SAUR10 in the absence and presence of NAA. The expression of these genes was enhanced by treatment with NAA alone, while the induction of these genes by NAA was significantly suppressed by the accumulation of IAA15P75S (Figure 5A). In addition, we also analyzed the expression of LBD genes, which are critical regulators of lateral root formation. As expected, LBD16, LBD29, and LBD33 were highly induced by NAA treatment (Figure 5B). By the accumulation of IAA15P75S, however, the inductions of LBD16 and LBD29 genes were significantly reduced, and that of LBD33 was slightly reduced (Figure 5B). These results indicate that the gain-of-function mutation of IAA15 suppresses the transcription of auxin-responsive genes including LBD16 and LBD29.
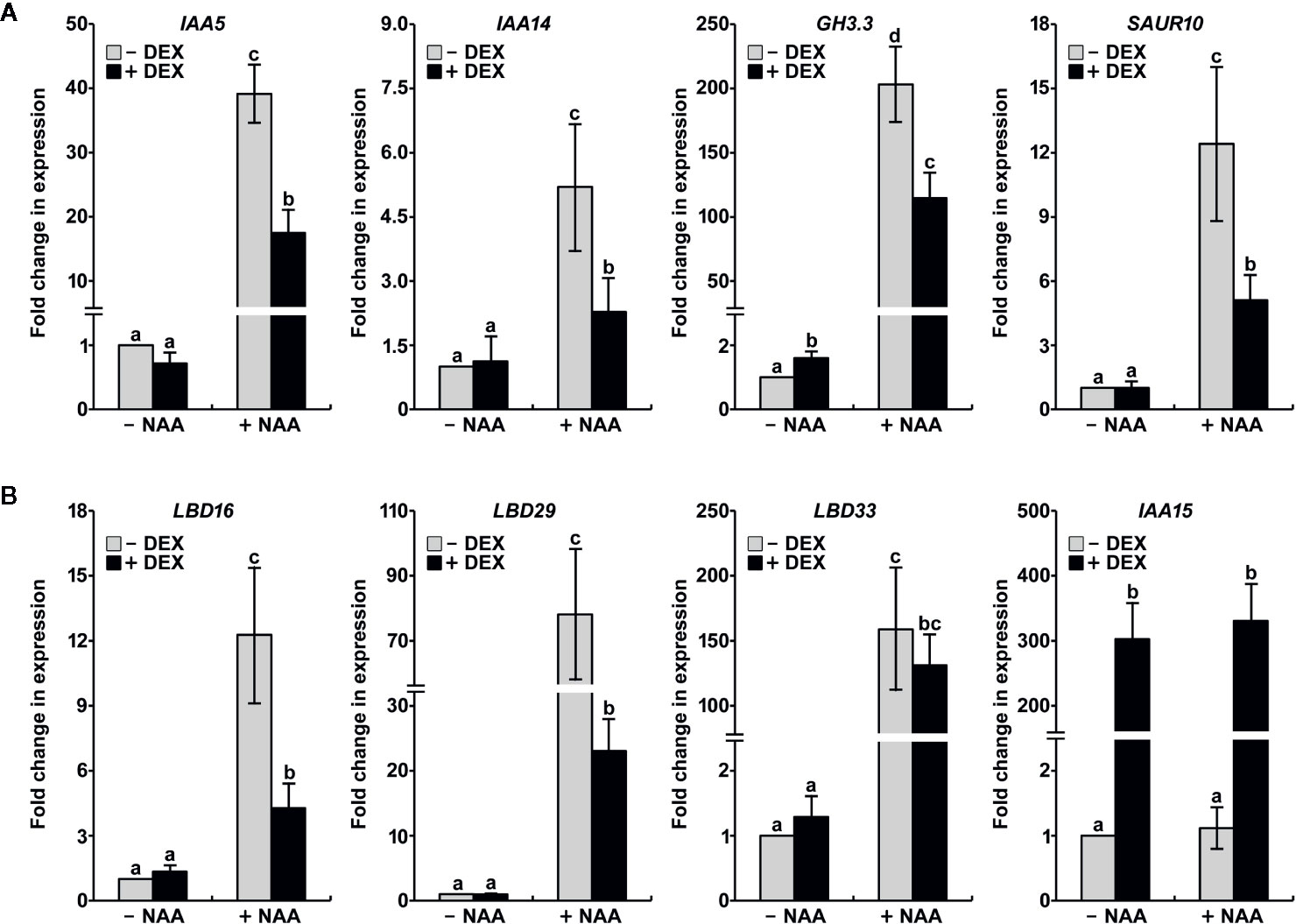
Figure 5 IAA15P75S inhibits auxin-responsive gene expression. Total RNA was extracted from IAA15P75S OX plants treated with or without 10 μM NAA and/or 50 μM DEX. (A) Transcript levels of auxin-responsive genes are reduced in IAA15P75S OX plants. IAA5, IAA14, GH3.3, and SAUR10 transcript levels were measured by qRT-PCR using specific primers. The bars indicate the mean ± S.D. (n = 3). Different letters indicate significant differences (P < 0.05) among lines that were explored through Tukey’s multiple comparisons tests. (B) The mutation of IAA15 inhibits the expression of lateral root-related LBDs. IAA15, LBD16, LBD29, and LBD33 transcript levels were measured by qRT-PCR using specific primers. The bars indicate the mean ± S.D. (n = 3). Different letters indicate significant differences (P < 0.05) among lines that were explored through Tukey’s multiple comparisons tests.
IAA15 Binds to the Promoters of LBD16 and LBD29
Although IAA15 negatively regulates the expression of LBD16 and LBD29, it is likely to repress auxin-responsive transcription by interacting with the partner ARF proteins because Aux/IAA repressors do not have a DNA-binding domain (Guilfoyle, 2015; Powers and Strader, 2020). To know the molecular mechanism to explain how IAA15 inhibits the expression of LBD16 and LBD29 genes, we first investigated whether IAA15 interacts with ARF7 or ARF19, which are direct and positive regulating transcription factors in the expression of LBD16 and LBD29 genes (Okushima et al., 2007). IAA15 was previously shown to interact with the C-terminal fragments (Domain III and IV) of ARF7 and ARF19 in a yeast two-hybrid assay (Vernoux et al., 2011). Therefore, we examined the interactions of IAA15 with the full-length ARF7 and ARF19 in yeast cells. As expected, IAA15 physically interacts with full-length ARF7 and ARF19 (Figure 6A).
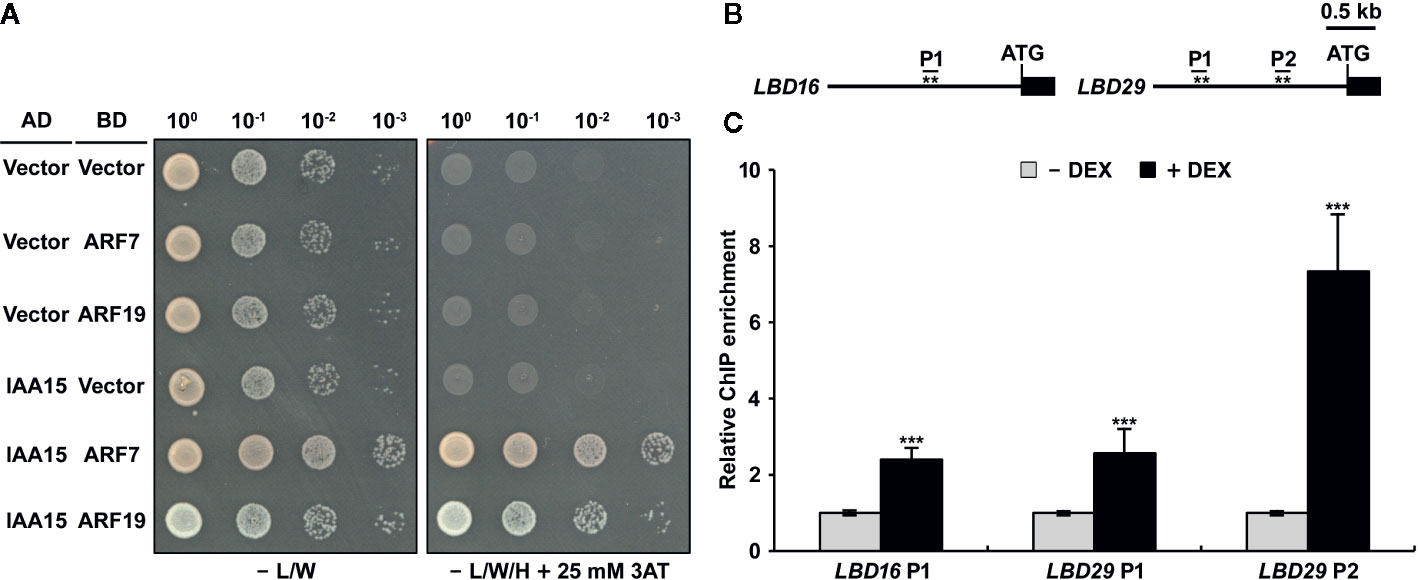
Figure 6 IAA15 interacts with ARF7 and ARF19 and binds to the promoters of LBD16 and LBD29. (A) Yeast two-hybrid analysis showing the interaction of IAA15 with ARF7 and ARF19. Various combinations of plasmids were co-transformed into yeast strain pJ69-4A. Shown are growth tests of the transformants on selective SD medium lacking Leu and Trp (-L/W) (control) and on SD medium lacking Leu, Trp, and His (-L/W/H) supplemented with 25 mM 3-amino-1,2,4-triazole (3-AT) to demonstrate activation of the HIS3 reporter gene. (B) Diagram of the LBD16 and LBD29 promoters, which contain target sequences of ARF7 and ARF19. The asterisks (**) show the positions of primers used for ChIP-qPCR. (C) ChIP assay of the binding of IAA15 to the ARF7- and ARF19-response elements in the LDB16 and LBD29 promoters in vivo. ChIP assays were performed with chromatin prepared from IAA15P75S OX treated with or without 50 μM DEX by using anti-Flag antibody. ChIP-DNA was applied to RT-qPCR using primers specifically targeting to promoter regions of LBD16 and LBD29. The bars indicate the mean ± S.D. (n = 3). Significant differences were analyzed by Student’s t-test (***P < 0.01).
The previous investigation reported that ARF7 and ARF19 directly bind to auxin-responsive elements (AuxREs) of LBD16 and LBD29 promoters. (Figure 6B) (Okushima et al., 2007). To explore whether IAA15 binds to these AuxREs of LBD promoters, therefore, we performed a chromatin immunoprecipitation (ChIP) assay in DEX-untreated and -treated IAA15P75S OX plants using anti-Flag antibody. The enrichment of these AuxREs was highly increased by the induction of IAA15P75S (Figure 6C). These results strongly suggest that IAA15 binds to the promoters of LBD16 and LBD29, presumably indirectly through interactions with an ARF7 and ARF19.
Discussion
The Stabilized IAA15 Inhibits the Lateral Root Growth
Almost all gain-of-function mutations of Aux/IAA occurring within conserved domain II result in defective plant growth due to the stabilization of Aux/IAA. In the current study, we found that IAA15P75S OX plants exhibited auxin-related abnormal developmental phenotypes such as short primary roots and reduced lateral root formation (Figure 2 and Supplementary Figures 2 and 3). These developmental phenotypes of IAA15P75S OX plants were caused by the inhibition of auxin signaling through an accumulation of IAA15P75S (Figures 1 and 5 and Supplementary Figure 4). The accumulation of IAA15P75S was due to the inhibited poly-ubiquitination by the ubiquitin E3 ligase complex (Figure 4). Interestingly, these auxin-related root defective phenotypes of IAA15P75S OX plants are also found in gain-of-function mutants of other Aux/IAAs such as iaa3/shy2, iaa14/slr1-1, iaa19/msg2, and iaa28 (Reed, 2001; Liscum and Reed, 2002; Mockaitis and Estelle, 2008). The similar root phenotypes of those mutants suggest that several Aux/IAA proteins may be redundantly involved in plant root development. The next possible speculation is that similar root defective phenotypes by several gain-of-function mutants can be caused by over-accumulation of corresponding Aux/IAA proteins. Over-accumulated Aux/IAA proteins in those mutants may cause non-specific interactions with ARF partner proteins, which result in unusual auxin-related root development phenotypes. To determine which Aux/IAA genes are bona fide involved in root development, the more detailed experimental investigations by using new transgenic plants expressing AUX/IAAs mutant under the control of their promoters and loss-of-function mutant of Aux/IAAs should be performed.
IAA15 Is a Component of Aux/IAA-ARF Modules Inhibiting Lateral Root Formation
The molecular model explaining the mechanism of lateral root formation is well established based on many studies (Tian and Reed, 1999; Rogg et al., 2001; Tatematsu et al., 2004; Okushima et al., 2005; Weijers et al., 2005; Wilmoth et al., 2005; Fukaki et al., 2006; Li et al., 2006). According to this model, auxin de-represses the activity of the transcriptional activators ARF7 and ARF19 through the degradation of Aux/IAAs such as IAA3, IAA14, IAA19, and IAA28, which directly activates the transcription of downstream genes including LBD16 and LBD29 to induce lateral root formation (Okushima et al., 2007). The lateral root formation is redundantly regulated by multiple Aux/IAA-ARF modules such as IAA3/SHY2-ARFs, IAA14/SLR-ARF7 and -ARF19, IAA19/MSG2-ARF7, and IAA28-ARFs. In the current study, we found that in addition to other IAAs, IAA15 also negatively regulates lateral root formation through the transcriptional repression of LBD16 and LBD29 (Figures 1 and 4). In addition, we showed that IAA15 directly interacts with ARF7 and ARF19 and binds to the promoters of LBD16 and LBD29. These findings suggest that the roles of the IAA15-ARF7 and -ARF19 modules in lateral root development are similar to those of other known IAA-ARF modules.
The identification of several IAA-ARF modules with redundant roles in lateral root formation raises the question of why multiple Aux/IAAs redundantly regulate this process. We propose two possibilities. First, several Aux/IAA-ARF modules could be specifically involved in various stages of lateral root formation. Lateral root formation is a complex process involving three major steps: lateral root initiation, lateral root primordia development, and lateral root emergence (Malamy and Benfey, 1997; Jung and McCouch, 2013). For example, IAA28-ARF modules are involved in the specification of lateral root founder cells in the basal root meristem during the early stage of lateral root formation (De Rybel et al., 2010). IAA14/SLR-ARF7 or -ARF19 modules are involved in the asymmetric division of lateral root founder cells for lateral root initiation (Fukaki et al., 2002; Okushima et al., 2005; Okushima et al., 2007). The IAA12/BDL-ARF5 module also participates in lateral root initiation, and IAA3/SHY2-ARF modules regulate lateral root primordia development and emergence after lateral root initiation (De Smet et al., 2010). IAA15-ARF7 or -ARF19 modules may be involved in specific stages of lateral root formation. However, IAA15-ARF modules may be involved in all stages of this process because IAA15 is expressed in all stages in lateral root initiation and development. Second, specific Aux/IAA-ARF modules could be responsive to particular environmental stresses. Abiotic stresses such as salt stress, drought stress, and nutrient depletion affect root development, including lateral root formation (Xiong et al., 2006; Zolla et al., 2010; Sun et al., 2017; Huang et al., 2018). In response to specific environmental stresses, specific Aux/IAA may be stabilized to inhibit lateral root formation, which contributes plants to survive. This hypothesis may explain how Aux/IAAs regulate growth and defense trade-off in plants. In the future, detailed studies elucidating the biological functions of IAAs under stress conditions should be performed.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
SK and WC designed, planned, and organized the experiments. SK, SB, JA, SH, NN, and HD generated all Arabidopsis transgenic lines and all plant materials used in this study. SK, SB, JA, and JH performed the research. SK, J-YK, JH, and WC wrote the article.
Funding
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ01325401) funded by the Rural Development Administration, by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2018R1A6A3A11042628 and 2020R1A6A1A03044344), by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. 2019R1A2C1009932).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01239/full#supplementary-material
References
Alarcón, M. V., Salguero, J., Lloret, P. G. (2019). Auxin Modulated Initiation of Lateral Roots Is Linked to Pericycle Cell Length in Maize. Front. Plant Sci. 2019 10:11:11. doi: 10.3389/fpls.2019.00011
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Beeckman, T., Burssens, S., Inze, D. (2001). The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52, 403–411. doi: 10.1093/jexbot/52.suppl_1.403
Blakely, L. M., Blakely, R. M., Colowit, P. M., Elliott, D. S. (1988). Experimental Studies on Lateral Root Formation in Radish Seedling Roots: II. Analysis of the Dose-Response to Exogenous Auxin. Plant Physiol. 87 (2), 414–419. doi: 10.1104/pp.87.2.414
Casimiro, I., Marchant, A., Bhalerao, R. P., Beeckman, T., Dhooge, S., Swarup, R., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. doi: 10.1105/tpc.13.4.843
Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., et al. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8, 165–171. doi: 10.1016/S1360-1385(03)00051-7
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
De Rybel, B., Vassileva, V., Parizot, B., Demeulenaere, M., Grunewald, W., Audenaert, D., et al. (2010). A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706. doi: 10.1016/j.cub.2010.09.007
De Smet, I., Lau, S., Voss, U., Vanneste, S., Benjamins, R., Rademacher, E. H., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 107, 2705–2710. doi: 10.1073/pnas.0915001107
Dharmasiri, N., Dharmasiri, S., Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Ellis, C. M., Nagpal, P., Young, J. C., Hagen, G., Guilfoyle, T. J., Reed, J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 Regulate Senescence and Floral Organ Abscission in Arabidopsis Thaliana. Development 132, 4563–4574. doi: 10.1242/dev.02012
Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., et al. (2002). AtPIN4 Mediates Sink-Driven Auxin Gradients and Root Patterning in Arabidopsis. Cell 108, 661–673. doi: 10.1016/s0092-8674(02)00656-6
Fukaki, H., Tameda, S., Masuda, H., Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. doi: 10.1046/j.0960-7412.2001.01201.x
Fukaki, H., Taniguchi, N., Tasaka, M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48, 380–389. doi: 10.1111/j.1365-313X.2006.02882.x
Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., et al. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. doi: 10.1126/science.282.5397.2226
Goh, T., Joi, S., Mimura, T., Fukaki, H. (2012). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139, 883–893. doi: 10.1242/dev.071928
Guilfoyle, T. J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33–43. doi: 10.1105/tpc.114.132753
Guilfoyle, T. J., Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Hamann, T., Benkova, E., Baurle, I., Kientz, M., Jurgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. doi: 10.1101/gad.229402
Huang, K. L., Ma, G. J., Zhang, M. L., Xiong, H., Wu, H., Zhao, C. Z., et al. (2018). The ARF7 and ARF19 Transcription Factors Positively Regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis Roots. Plant Physiol. 178, 413–427. doi: 10.1104/pp.17.01713
Hussain, S., Kim, S. H., Bahk, S., Ali, A., Nguyen, X. C., Yun, D. J., et al. (2020). The Auxin Signaling Repressor IAA8 Promotes Seed Germination Through Down-Regulation of ABI3 Transcription in Arabidopsis. Front. Plant Sci. 11:111:111. doi: 10.3389/fpls.2020.00111
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jiao, Y., Ma, L., Strickland, E., Deng, X. W. (2005). Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17, 3239–3256. doi: 10.1105/tpc.105.035840
Jung, J. K., McCouch, S. (2013). Getting to the roots of it: Genetic and hormonal control of root architecture. Front. Plant Sci. 4:186:186. doi: 10.3389/fpls.2013.00186
Kakrana, A., Kumar, A., Satheesh, V., Abdin, M. Z., Subramaniam, K., Bhattacharya, R. C., et al. (2017). Identification, Validation and Utilization of Novel Nematode-Responsive Root-Specific Promoters in Arabidopsis for Inducing Host-Delivered RNAi Mediated Root-Knot Nematode Resistance. Front Plant Sci. 8, 2049. doi: 10.3389/fpls.2017.02049
Kim, B. C., Soh, M. C., Kang, B. J., Furuya, M., Nam, H. G. (1996). Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 9, 441–456. doi: 10.1046/j.1365-313x.1996.09040441.x
Laskowski, M. J., Williams, M. E., Nusbaum, H. C., Sussex, I. M. (1995). Formation of lateral root meristems is a two-stage process. Development 121, 3303–3310.
Li, X., Mo, X., Shou, H., Wu, P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47, 1112–1123. doi: 10.1093/pcp/pcj082
Liscum, E., Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1093/pcp/pcj082
Malamy, J. E., Benfey, P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44.
Miura, K., Nakazawa, M., Ono, M., Hasegawa, M. (2011). ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J. 67, 269–279. doi: 10.1111/j.1365-313X.2011.04589.x
Mockaitis, K., Estelle, M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev Biol. 24, 55–80. doi: 10.1146/annurev.cellbio.23.090506.123214
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nagpal, P., Walker, L. M., Young, J. C., Sonawala, A., Timpte, C., Estelle, M., et al. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–574. doi: 10.1104/pp.123.2.563
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Péret, B., De Rybel, B., Casimiro, I., Benková, E., Swarup, R., Laplaze, L., et al. (2009). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408. doi: 10.1016/j.tplants.2009.05.002
Ploense, S. E., Wu, M. F., Nagpal, P., Reed, J. W. (2009). A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136, 1509–1517. doi: 10.1242/dev.025932
Powers, S. K., Strader, L. C. (2020). Regulation of auxin transcriptional responses. Dev. Dyn. 249, 483–495. doi: 10.1002/dvdy.139
Puzio, P., Lausen, J., Heinen, P., Grundler, F. (2000). Promoter analysis of pyk20, a gene from Arabidopsis thaliana. Plant Sci. 157, 245–255. doi: 10.1016/S0168-9452(00)00287-9
Reed, J. W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. doi: 10.1016/s1360-1385(01)02042-8
Rinaldi, M. A., Liu, J., Enders, T. A., Bartel, B., Strader, L. C. (2012). A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol. Biol. 79, 359–373. doi: 10.1007/s11103-012-9917-y
Rogg, L. E., Lasswell, J., Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. doi: 10.1105/tpc.13.3.465
Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. doi: 10.1126/science.279.5355.1371
Santner, A., Calderon-Villalobos, L., Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Sun, C. H., Yu, J. Q., Hu, D. G. (2017). Nitrate: A Crucial Signal during Lateral Roots Development. Front. Plant Sci. 8:485:485. doi: 10.3389/fpls.2017.00485
Szemenyei, H., Hannon, M., Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. doi: 10.1126/science.1151461
Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M. K., Harper, R. M., et al. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393. doi: 10.1105/tpc.018630
Tian, Q., Reed, J. W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721.
Tian, L., Fong, M. P., Wang, J. J., Wei, N. E., Jiang, H., Doerge, R. W., et al. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169, 337–345. doi: 10.1534/genetics.104.033142
Torrey, J. G. (1950). The induction of lateral roots by indoleacetic acid and root decapitation. Am. J. Bot. 37, 257–263. doi: 10.1002/j.1537-2197.1950.tb12192.x
Ulmasov, T., Murfett, J., Hagen, G., Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Vaughan, S. P., James, D. J., Lindsey, K., Massiah, A. J. (2006). Characterization of FaRB7, a near root-specific gene from strawberry (Fragaria x ananassa Duch.) and promoter activity analysis in homologous and heterologous hosts. J. Exp. Bot. 57, 3907–3910. doi: 10.1093/jxb/erl185
Vernoux, T., Brunoud, G., Farcot, E., Morin, V., Van den Daele, H., Legrand, J., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst/Biol., 7, 508. doi: 10.1038/msb.2011.39
Wang, J., Yan, D. W., Yuan, T. T., Gao, X., Lu, Y. T. (2013). A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol. Biol. 82, 71–83. doi: 10.1007/s11103-013-0039-y
Weijers, D., Benkova, E., Jäger, K. E., Schlereth, A., Hamann, T., Kientz, M., et al. (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24, 1874–1885. doi: 10.1038/sj.emboj.7600659
Weijers, D., Schlereth, A., Ehrismann, J. S., Schwank, G., Kientz, M., Jürgens, G. (2006). Auxin Triggers Transient Local Signaling for Cell Specification in Arabidopsis Embryogenesis. Dev. Cell 10, 265–270. doi: 10.1016/j.devcel.2005.12.001
Wilmoth, J. C., Wang, S., Tiwari, S. B., Joshi, A. D., Hagen, G., Guilfoyle, T. J., et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130. doi: 10.1111/j.1365-313X.2005.02432.x
Xiong, L., Wang, R. G., Mao, G., Koczan, J. M. (2006). Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol. 142, 1065–1074. doi: 10.1104/pp.106.084632
Yan, D. W., Wang, J., Yuan, T. T., Hong, L. W., Gao, X., Lu, Y. T. (2013). Perturbation of Auxin Homeostasis by Overexpression of Wild-Type IAA15 Results in Impaired Stem Cell Differentiation and Gravitropism in Roots. PloS One 8, e58103. doi: 10.1371/journal.pone.0058103
Yang, X., Lee, S., So, J. H., Dharmasiri, S., Dharmasiri, N., Ge, L., et al. (2004). The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 40, 772–782. doi: 10.1111/j.1365-313X.2004.02254.x
Keywords: auxin, Aux/IAA, gain-of-function, lateral root, LBD genes, repressor
Citation: Kim SH, Bahk S, An J, Hussain S, Nguyen NT, Do HL, Kim J-Y, Hong JC and Chung WS (2020) A Gain-of-Function Mutant of IAA15 Inhibits Lateral Root Development by Transcriptional Repression of LBD Genes in Arabidopsis. Front. Plant Sci. 11:1239. doi: 10.3389/fpls.2020.01239
Received: 23 April 2020; Accepted: 28 July 2020;
Published: 12 August 2020.
Edited by:
Stephan Pollmann, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Dipak Kumar Sahoo, Iowa State University, United StatesShucai Wang, Linyi University, China
Copyright © 2020 Kim, Bahk, An, Hussain, Nguyen, Do, Kim, Hong and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo Sik Chung, Y2h1bmd3c0BnbnUuYWMua3I=
 Sun Ho Kim
Sun Ho Kim Shah Hussain
Shah Hussain Jae-Yean Kim
Jae-Yean Kim Woo Sik Chung
Woo Sik Chung