
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 August 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01229
 Xingdan Liu1†
Xingdan Liu1† Fengfeng Fan2†
Fengfeng Fan2† Manman Liu2
Manman Liu2 Weixiong Long2
Weixiong Long2 Yajie Yu2
Yajie Yu2 Huanran Yuan2
Huanran Yuan2 Guojing Pan2
Guojing Pan2 Nengwu Li2
Nengwu Li2 Shaoqing Li2*
Shaoqing Li2* Jianfeng Liu1*
Jianfeng Liu1*Mineral elements play an extremely important role in human health, and are worthy of study in rice grain. Wild rice is an important gene pool for rice improvement including grain yield, disease, and pest resistance as well as mineral elements. In this study, we identified 33 quantitative trait loci (QTL) for Fe, Zn, Se, Cd, Hg, and As contents in wild rice Oryza longistaminata. Of which, 29 QTLs were the first report, and 12 QTLs were overlapped to form five clusters as qSe1/qCd1 on chromosome 1, qCd4.2/qHg4 on chromosome 4, qFe5.2/qZn5.2 on chromosome 5, qFe9/qHg9.2/qAs9.2 on chromosome 9, and qCd10/qHg10 on chromosome 10. Importantly, qSe1/qCd1, can significantly improve the Se content while reduce the Cd content, and qFe5.2/qZn5.2 can significantly improve both the Fe and Zn contents, they were delimited to an interval about 53.8 Kb and 26.2 Kb, respectively. These QTLs detected from Oryza longistaminata not only establish the basis for subsequent gene cloning to decipher the genetic mechanism of mineral element accumulation, but also provide new genetic resource for rice quality improvement.
As one of the most important staple crops, rice provides more than 40% of the daily calories for the world’s population and is also an important source for protein, vitamins and minerals (Khush, 2001; Parengam et al., 2010). Mineral elements play an extremely important role on human health. However, micronutrient malnutrition, a major human health problem in the world, particularly among women and children in developing countries, is mainly resulted from a dietary deficiency in iron (Fe) and zinc (Zn) (Zeng et al., 2005; Liu et al., 2006). Micronutrient deficiencies can cause a range of problems, such as Fe deficiency leading to anaemia, Zn deficiency leading to stunted growth and intellectual disability, and selenium (Se) deficiency increasing the risk of cancer (Clark et al., 1996; Umeta et al., 2000; Finley et al., 2001). Therefore, increasing the content of mineral elements in rice has important nutritional value for people, especially for those who live rice as the staple food. However, as we know that not all mineral elements are beneficial to human health. Some heavy metals such as cadmium (Cd), mercury (Hg), and arsenic (As), pose a serious threat to human health (Takahashi et al., 2011; Zhang et al., 2014; Huang et al., 2015). Accumulation of the heavy metals in rice grains will seriously affect their edible value, thus, breeding rice varieties with high trace elements and less heavy metal are supposed to improve the health of people.
Molecular marker-assisted selection and genetic engineering are important and effective methods to improve rice quality (Masuda et al., 2013; Fan et al., 2017). Thus, exploring and identifying new genes or quantitative trait loci (QTL) for mineral element accumulation in Oryza species becomes the premise to breed mineral element-rich rice. For these reasons, more and more researches tend to explore genes or genetic loci for mineral element accumulation in rice. To date, there are 14 QTLs for Fe and Zn contents in unpolished rice grains having been identified by using RILs, and one of the main QTL was further analyzed to obtain ten candidate genes (Anuradha et al., 2012). Twenty QTLs for Fe, Zn, Se, Cd, and lead (Pb) content in brown rice have been identified from 378 accessions, and some of them are co-located, such as Cd and Pb on chromosome 5, Zn and Pb on chromosome 7, and Se and Pb on chromosome 11 (Huang et al., 2015). Moreover, some genes such as OsNRAMP1 for Cd (Takahashi et al., 2011), OsLCT1 for Cd (Uraguchi et al., 2011), OsABCC1 for As (Song et al., 2014), and OsHMA3 for Cd and Zn (Cai et al., 2019; Sui et al., 2019) have been cloned, they promote or repress mineral element accumulation through different pathways of mineral element uptake or transport, implying the complexity of genetic mechanism of mineral element accumulation in rice.
Wild rice, as an important gene pool for rice yield, quality, and resistance improvement, is also an essential genetic carrier for mineral element in nature (Londo et al., 2006; Garcia-Oliveira et al., 2009; Huang et al., 2013; Fan et al., 2019). Previous studies have identified some QTLs for mineral elements content in wild rice Oryza rufipogon, these QTLs have great potential in the subsequent gene cloning and breeding applications (Garcia-Oliveira et al., 2009). Oryza longistaminata, an ancient Africa wild species of AA genome, less genes have been explored for rice breeding apart from Xa21 in the last 1990s. Here, we analyzed the QTLs for Fe, Zn, Se, Cd, Hg, and As contents using 127 backcross-inbred lines (BILs) derived from O. longistaminata. Totally, 40 QTLs for different minerals were identified, of which, 13 QTLs for trace elements Fe, Zn, and Se, and 16 QTLs for heavy metal elements Pb, Cd and As from O. longistaminata were the firstly reported, which will greatly help to breed high quality rice beneficial to human health-care.
9311, an elite cultivated rice, was used as the recurrent parent for BIL population construction. O. longistaminata, a wild rice with a variety of excellent traits, was used as the donor parent. The population comprising 127 BILs was derived from a distant cross between O. longistaminata and 9311. Then, 9311 was used as the recurrent parent to backcross the hybrid 2 times, obtaining BC2F1. Finally, a set of 127 BC2F16 lines was constructed. The recurrent parent 9311 and 127 BILs were planted in the experimental field of Wuhan university experimental base in the summer of 2014 (Huashan, N30.54°, E114.52°) and 2015 (Ezhou, N30.40°, E114.88°), respectively. Each BIL and the recurrent parent consisted of 50 plants planted in five rows of 10 plants each adopting a uniform spacing of 20 cm between rows and 15 cm between plants. Three replications were performed by randomized complete block design.
After harvesting, rice grains were husked to obtain brown rice using five randomly-selected plants in each replicate. The unpolished grains were dried in hot air oven at 80°C for 24 h. Then ground to fine powder with a grinder, and filtered through a stainless-steel 0.3 mm mesh screen. To ensure consistency in elements analyses, seeds from every lot were analyzed as three replicates. Then 0.2 g was digested in 5 ml of HNO3–H2O2 (4:1 v/v) mixture. The Fe and Zn content in the digest solutions were determined with flame atomic absorption spectrometry (FLAA) (ContrAA 700, Analytik Jena AG, Germany). The Cd content in the digest solutions were determined with graphite furnace atomic absorption spectrometry (GFAA) (ContrAA 700, Analytik Jena AG, Germany). The Hg content in the digest solutions were determined with Atomic fluorescence spectrum (AFS) (PF6-2, Beijing General Analysis Instrument Co. LTD, China). Then 1.0 g was digested in 15 ml of HNO3–HClO4–HCl (10:1:4 v/v) mixture. The Se and As content in the digest solutions were determined with AFS.
Grain elements content were computed as parts per million (ppm). The statistical analyses and correlation analysis were performed with SPSS Statistics 20 (IBM, United States) using the mean values of each trait. Correlation among traits was computed at P < 0.05 and P < 0.01, respectively.
The 127 BILs were subjected to whole-genome sequencing, and a high-quality bin map of ultrahigh-density SNPs was constructed based on the sequencing data (unpublished data). The DNA extraction, SNP screening, genotyping, bin map and genetic linkage map construction all adopt the previous reported methods (Jin et al., 2018; Fan et al., 2019). A total of 2,432 bins were obtained for the subsequent QTL analysis. The QTL mapping analysis was assembled from the genotype and phenotypic data using QTL IciMapping Version 3.3 software, and the interval mapping method of BIL module was selected (Meng et al., 2015). The logarithm of the odds (LOD) values (95% confidence interval) for the six traits were obtained through 1,000 permutations, and the average LOD threshold was 2.39. So the LOD score beyond 2.5 was defined as one QTL. The QTL were named as qXY (the X indicate the elements, the Y indicate the chromosome number).
A putative gene in the QTL region was predicted by referring to the Rice Information GateWay (http://rice.hzau.edu.cn/rice/). The protein sequences of candidate genes were separately submitted to SMART (http://smart.embl-heidelberg.de/) to predict the functional domains using the domain annotation tool.
The mean, standard deviation (SD), coefficient of variation (CV), and range of the mineral elements for a total of 127 BILs were evaluated as presented in Table 1. Significant differences in mineral element contents were found. The means for trace elements such as Fe, Zn, and Se content were 24.10 mg/kg, 22.48 mg/kg and 0.043 mg/kg, respectively. The means for heavy metal elements such as Cd, Hg, and As content were 0.063 mg/kg, 0.015 mg/kg, and 0.266 mg/kg, respectively. At the same time, we also observed that the variation coefficient and range of each mineral element in the BILs were large, indicating a rich genetic diversity of the BILs (Table 1). Phenotypic values of the BILs were found being continuous and exhibited normal or skewed distribution patterns in all elements (Figure 1). It can be seen that the content of Fe, Zn, and As in the soil of Ezhou experimental base in 2015 was significantly higher than that of Huashan experimental base in 2014, while the content of Hg presented an opposite trend (Table S1). As a result, the content of Fe, Zn, and As in BIL group in 2015 was higher than that in 2014, while the content of Hg was lower than that in 2014. Although the contents of these mineral elements varied in different years, the overall trend remained consistent, which mean that this BIL population can be well used for the further QTL analysis.
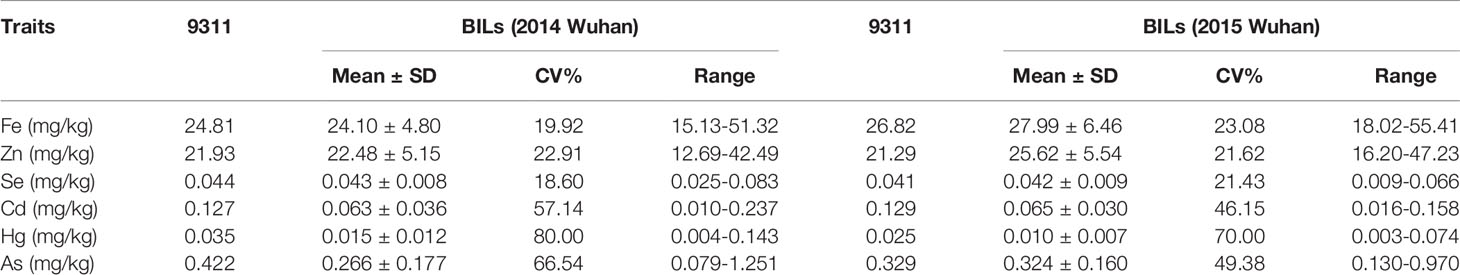
Table 1 Phenotypic variation of the mineral elements in brown rice of 9311 and BIL lines in different environments.
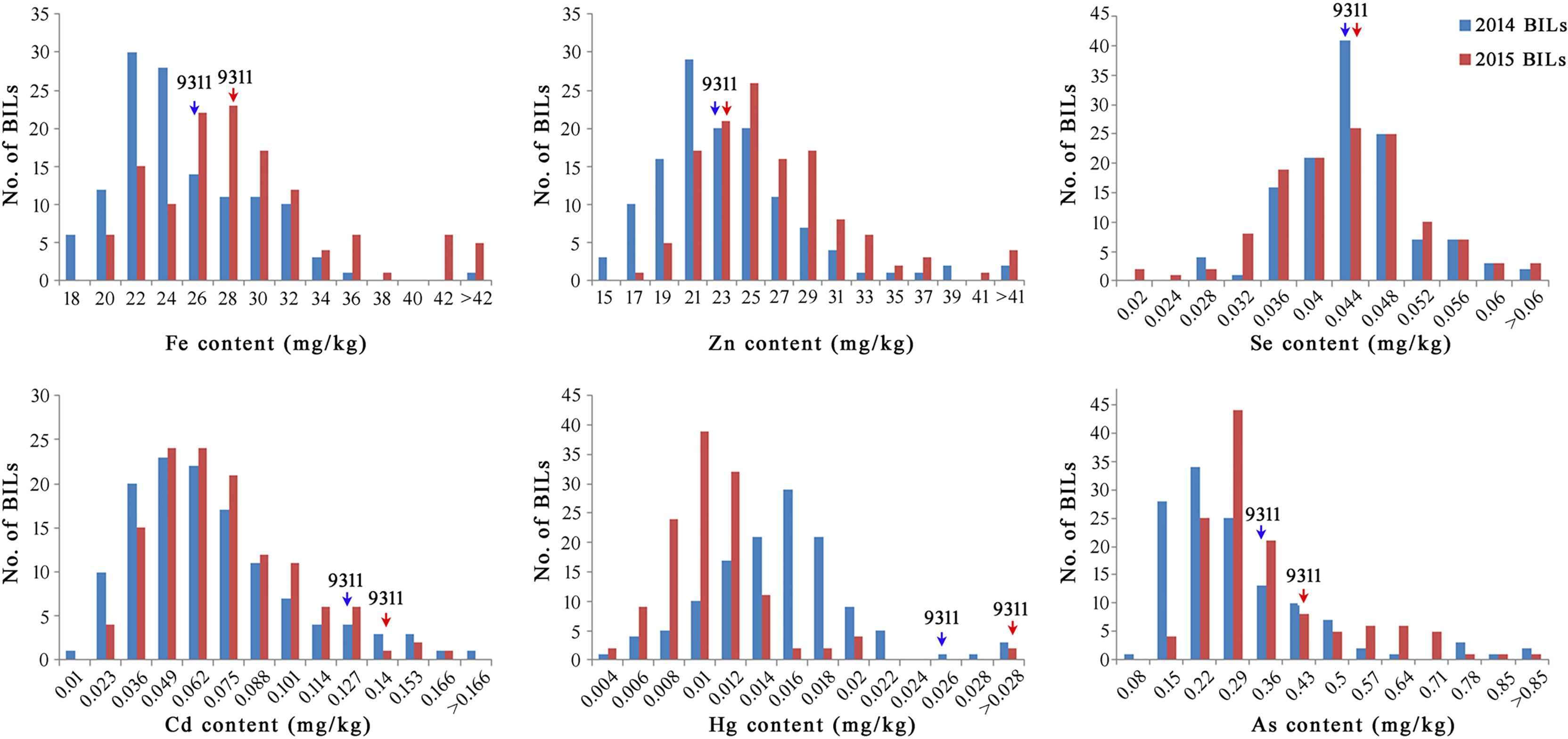
Figure 1 Distribution of six mineral element contents of BIL population in 2 years. Arrow head represents the content of 9311.
In order to understand the potential relationships between the six mineral elements, correlation analysis was performed for the mineral elements in the 127 BILs (Table 2). Results showed that a significantly positive correlation was detected between Fe and Zn in 2 years, and an extremely significant correlation was observed between Zn and Se in both 2014 and 2015. The other elements showed a weak correlation with each other.
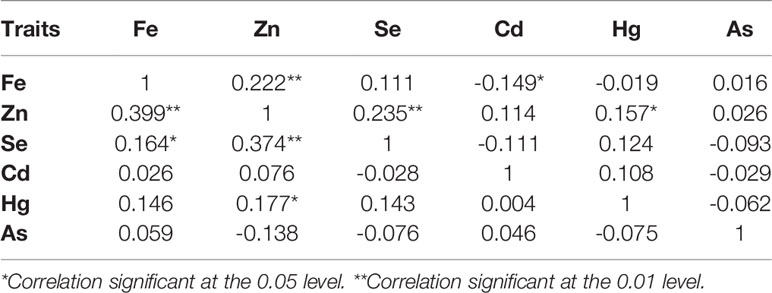
Table 2 Correlation coefficients among elements of BILs in 2014 (upper right) and 2015 (lower left).
Interval mapping revealed that a total of 40 QTL for six mineral element contents were identified on all chromosomes except chromosome 7 (Figure 2). Of which, 15 QTL for trace element contents and 18 QTL for heavy metal element contents were derived from wild rice O. longistaminata (Table S2).
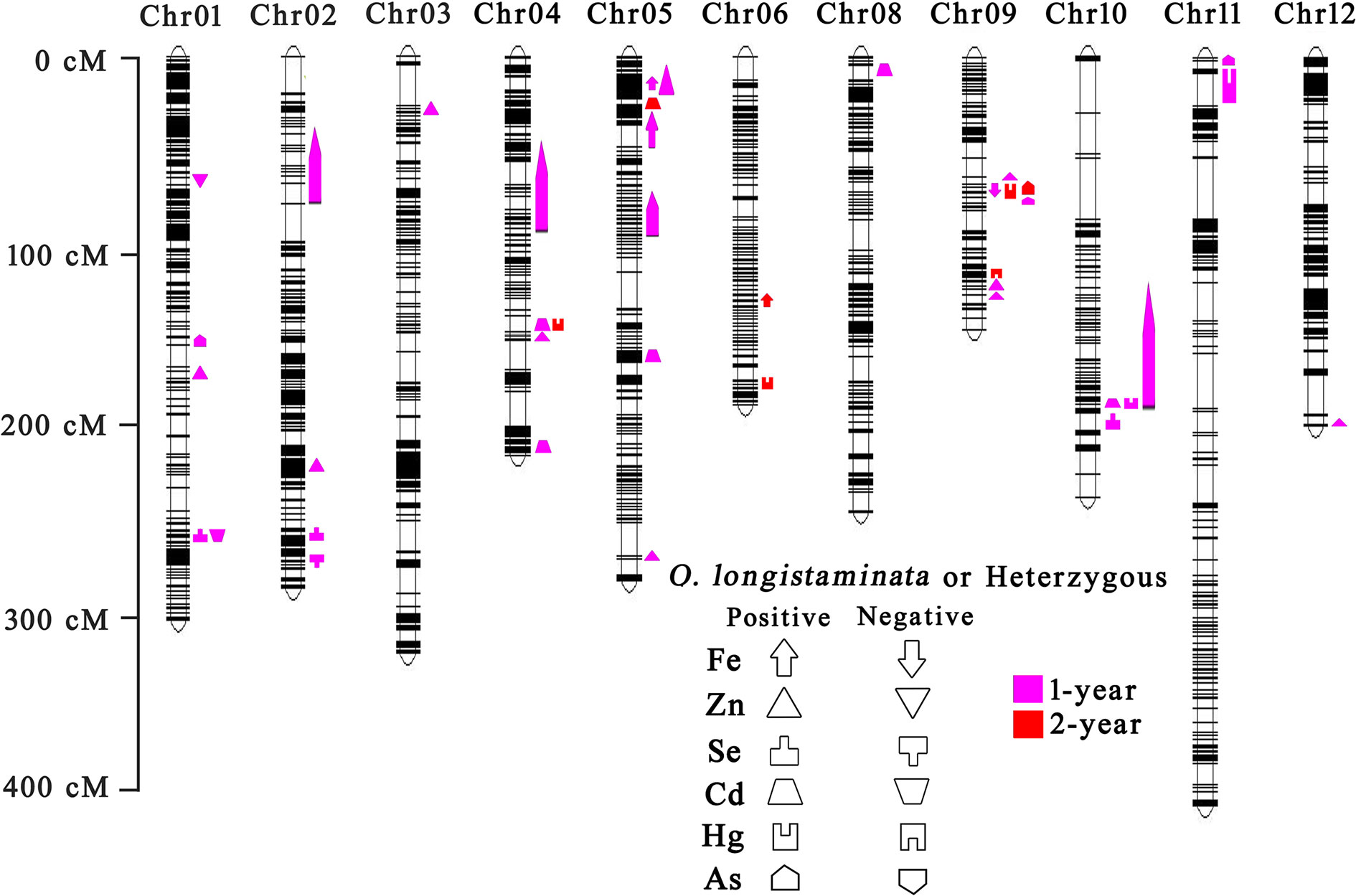
Figure 2 QTL analysis of six mineral element contents of BIL population in different years. Left is the scale for the genetic length of each chromosome. The upward direction indicates the O. longistaminata parental allele or heterozygous genotype for each locus showing positive effects. The downward direction indicates the O. longistaminata allele or heterozygous genotype for each locus showing negative effects. The pink indicates QTLs detected in 1 year. The red indicates QTLs detected in 2 years.
For trace elements, a total of 19 QTLs were detected in 2 years (Figure 2). Among these, 15 QTLs with positive effects were detected from wild rice O. longistaminata (Table S2). In details, three QTLs for Fe content were mapped on chromosomes 5, 5 and 6, and explained 6.8%, 10.0%, and 32.4% of the phenotypic variation, respectively. Ten QTLs for Zn content explained the phenotypic variation ranging from 4.7% to 16.6%. Two QTLs for Se content were mapped on chromosomes 1 and 2, and explained about 10.3% and 13.9% of the variance, respectively (Table S2). As for heavy metal elements, 21 QTLs were identified in 2 years, and 18 QTLs with positive effects were detected from O. longistaminata (Figure 2 and Table S2). Of which, five QTLs for Cd content were mapped on chromosomes 4, 4, 5, 5, and 10, and explained 8.4%, 25.1%, 21.0%, 33.4%, and 23.9% of the phenotypic variations, respectively. Five QTLs, qHg4, qHg6, qHg9.2, qHg10, and qHg11 were detected for Hg content, and explained the phenotypic variations ranging from 15.0% to 27.7%. Eight QTLs were detected for As content and explained 25.6%~54.6% of the phenotypic variation (Table S2).
Notably, the qFe6 for Fe content, qZn4 for Zn content, qCd5.2 for Cd content, qAs9.2 for As content and qHg4, qHg6, qHg9.2 for Hg content were repeatedly identified from O. longistaminata in 2 years, meaning their potentially great value in rice breeding practice.
Co-localization of QTL for different element content in grains is common in rice (Garcia-Oliveira et al., 2009; Zhang et al., 2014; Huang et al., 2015). In our study, five cases of QTL colocalization, i.e. Se and Cd on chromosome 1, Cd and Hg on chromosome 4, Fe, and Zn on chromosome 5, Fe, Hg, and As on chromosome 9 and Cd, Hg on chromosome 10, were observed (Figure 2). Among them, the qSe1/qCd1 site explained 10.3% of the phenotypic variance for Se content and 6.9% for Cd content in the BIL population. The qFe5.2/qZn5.2 site explained 10.0% of the phenotypic variance for Fe content and 13.2% for Zn content. The qCd4.2/qHg4, qFe9/qHg9.2/qAs9.2 and qCd10/qHg10 site explained the phenotypic variation ranging from 12.8% to 29.2%.
In order to further validate the functions of the newly detected QTLs, we analyzed the genetic effects of the qSe1/qCd1 and qFe5.2/qZn5.2. Results showed that the Bin1-161 and Bin1-162 well defines the core interval of qSe1/qCd1 contributing to the Se and Cd contents in the BIL lines (Figure 3), and the Se and Cd content of the lines containing qSe1/qCd1 from O. longistaminata were about 21% higher and 35% lower than those lines with 9311 fragments, respectively (Figure 3B). The other site, qFe5.2/qZn5.2, was delimited by the tightly linked marker Bin5-196 (Figure 4A). Surprisingly, the Fe and Zn content in the lines with qFe5.2/qZn5.2 were about 28% and 23% higher than those no qFe5.2/qZn5.2 lines, respectively (Figure 4B). These results indicate that qSe1/qCd1 and qFe5.2/qZn5.2 from O. longistaminata could significantly improve the nutritional value of mineral elements in rice.
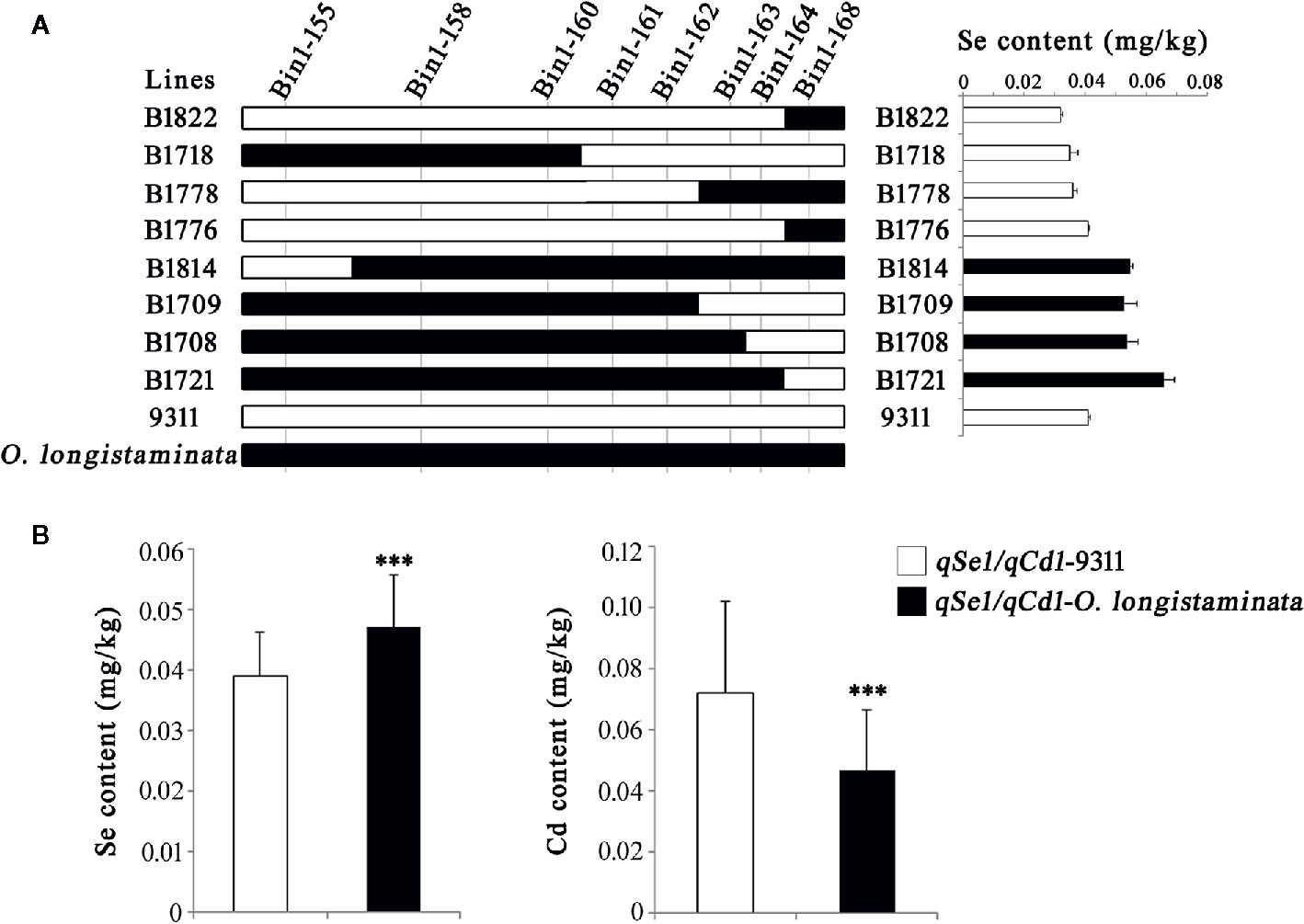
Figure 3 Validation of the function of qSe1/qCd1. (A) Progeny testing. (B) Analysis of the Se and Cd content of the BIL population. ***p < 0.001.
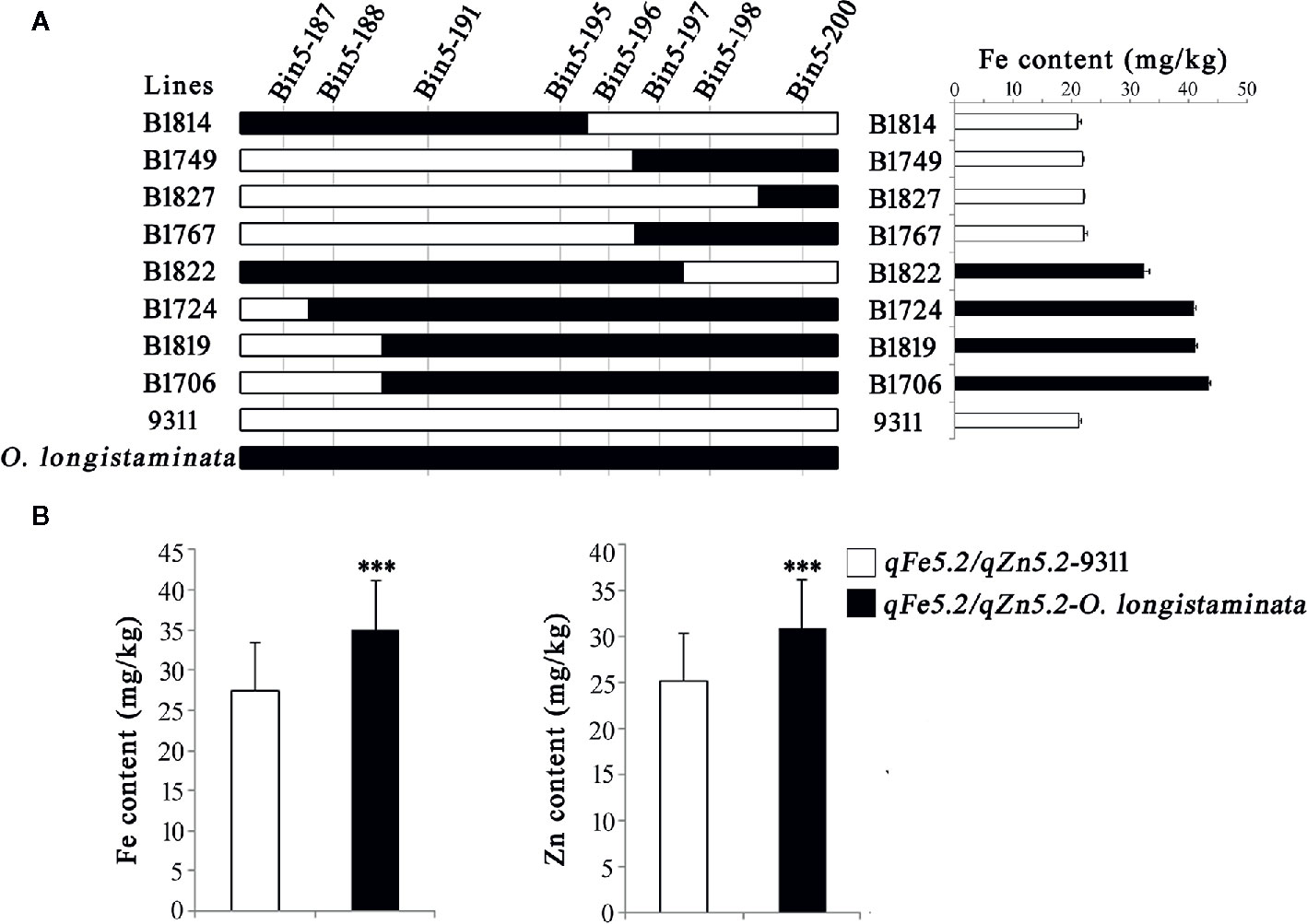
Figure 4 Validation of the function of qFe5.2/qZn5.2. (A) Progeny testing. (B) Analysis of the Fe and Zn content of the BIL population. ***p < 0.001.
Although rice is not considered as a main source of mineral elements, this does not exclude the importance to breed novel rice varieties with high content of the trace elements including Fe, Zn, and Se (Zhang et al., 2014; Huang et al., 2015). Recently, more and more attention has been paid to the research of Zn- and Se-rich rice, because of a variety of health-care functions of both Zn and Se on human health (Liu et al., 2015; Xu et al., 2016; Yuan et al., 2017; D’Amato et al., 2018). On the other hand, with the aggravation of the industrialization process, the enrichment of heavy metals in rice has become increasingly prominent, especially the enrichment of Cd, which has seriously threatened the health of people living with rice (Takahashi et al., 2011; Ueno et al., 2011). Therefore, breeding of rice varieties with high trace elements and low heavy metals, especially those rich in Zn/Se and low in Cd, is becoming the aim of breeders.
In order to achieve this goal, predecessors have done a lot of preliminary work. Some QTLs related to mineral element accumulation identified from cultivated rice and common wild rice have been successfully applied in production (Garcia-Oliveira et al., 2009; Nawaz et al., 2015). In this study, we focused on wild rice O. longistaminata, an Africa-derived important genetic resource that less favorable genes were explored (Zhang et al., 2015; Fan et al., 2017; Jin et al., 2018; Fan et al., 2019). Here, we firstly identified 33 QTLs for content of six mineral elements from O. longistaminata. Apart from the the qFe5.1, qFe6, qAs4, and qAs10 were overlapped with the Zinc-iron transporter gene OsZIP7a, OsZIP12 (Yang et al., 2009; Liu et al., 2019), and the arsenate reductase gene OsHAC1.2 and OsACR2.1 (Duan et al., 2007; Shi et al., 2016), respectively (Table S2), the other 29 QTLs found in O. longistaminata are different from the others. These results indicate that O. longistaminata contains abundant genetic resources for mineral element content improvement.
In addition, we also found that these QTLs tend to act in cluster. Among them, qSe1/qCd1, a QTL cluster improve the Se and Cd contents in the BILs, was delimited to an interval about 53.8 Kb on chromosome 1 (Figure 3). This locus contains three hypothetical protein and seven function genes including MH01g0727800 (Table 3). The MH01g0727800 encodes a CHD3-type chromatin-remodeling factor PICKLE, which is belonging to the CHD3 family (Hu et al., 2013). CHD3 protein can interact with H3K4me2 and H3K27me3 by the chromodomains and plant homeodomain finger, respectively (Hu et al., 2012). Many studied have demonstrated that histone methylation modification plays numerous important roles in the growth and development of rice (Cui et al., 2013; Li et al., 2013; Yan et al., 2015). Therefore, we speculate that this gene may play an important role in regulating the Se and Cd contents. Another QTL cluster, qFe5.2/qZn5.2, which can significantly improve the content of both Fe and Zn in rice grains, was located in an interval about 26.2 Kb on chromosome 5 (Figure 4). This locus containing only four predicted function genes (Table 3). Of which, MH05g0033700 encoding a phosphatidylinositol-specific phospholipase C, which effectively influences the metal ion signal transduction (Petersen and Wakui, 1990; Meldrum et al., 1991). MH05g0033800, containing a serine/threonine protein kinases domain and plays an important role in metal ion transport (Johnson and Wu, 2016). The discovery will help us to further understand the molecular mechanisms of mineral element accumulation in rice seeds.
Pyramiding multiple favorable genes in a single material is considered to be an important method of rice breeding (Brunner et al., 2010; Fan et al., 2017; Zeng et al., 2017). In order to further study the application value of these new QTLs, we conducted a comprehensive analysis of the BILs. Firstly, we divided the BIL population into four different subpopulations: BILs-A containing both of qSe1/qCd1 and qFe5.2/qZn5.2, BILs-B containing only qSe1/qCd1, BILs-C containing only qFe5.2/qZn5.2 and BILs-D containing no any of these favorable QTL clusters. We further analyzed the Fe, Zn, Se, and Cd content of different groups. Results showed that the Fe, Zn and Se content of BILs-A were higher than other groups, while the Cd content was lower than other groups (Figure 5). These results indicated that the BILs pyramided with qSe1/qCd1 and qFe5.2/qZn5.2 exhibit an ideal state of high trace elements and low heavy metals, which showed great application potential in rice nutritional quality improvement.
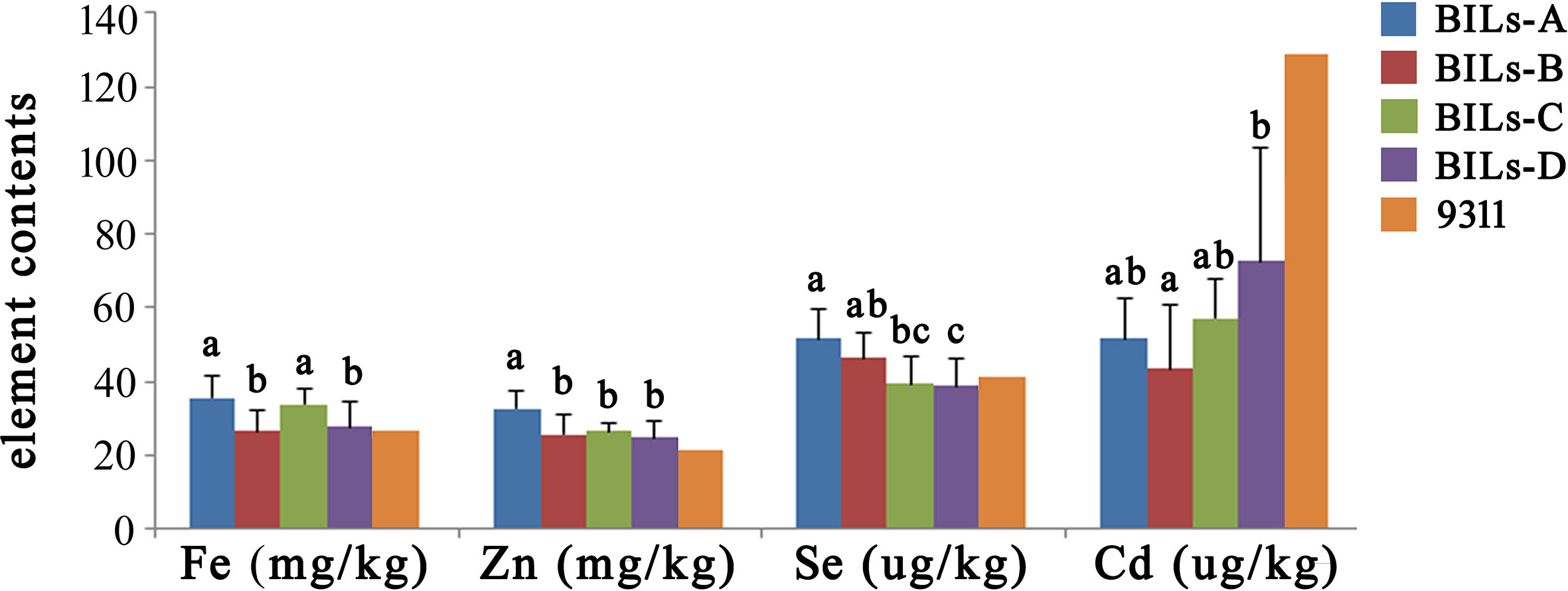
Figure 5 Genotype analysis and element content performance of different subpopulations. BILs-A containing both of qSe1/qCd1 and qFe5.2/qZn5.2, BILs-B containing only qSe1/qCd1, BILs-C containing only qFe5.2/qZn5.2, BILs-D containing no any of these favorable QTL clusters. 9311 was used as a control. The letters at the top of bars represent significant differences (P < 0.05) as determined by Student’s t test.
We dissected the genetic basis for mineral element contents of Fe, Zn, and Se, and heavy mental contents of Cd, Pb, and As in African wild rice Oryza longistaminata and identified 29 novel QTL for these traits. Among the new QTLs, qSe1/qCd1 and qFe5.2/qZn5.2 largely contributed to Fe, Zn, Se, and Cd contents in BILs, and provide a basis for subsequent breeding applications and gene cloning. This work provided new ideas to explain the genetic mechanism of mineral element accumulation
All datasets generated for this study are included in the article/Supplementary Material.
JL, SL, and FF designed the research. XL, FF, ML, WL, HY, GP, and NL performed field experiment and QTL analysis. XL, FF, and SL analyzed data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was partly granted from the National Key Research and Development Program (2016YFD0100903), National Natural Science Foundation (31870322) of China and Huanghe Patent Program of Wuhan City.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01229/full#supplementary-material
Anuradha, K., Agarwal, S., Rao, Y. V., Rao, K. V., Viraktamath, B. C., Sarla, N. (2012). Mapping QTLs and candidate genes for iron and zinc concentrations in unpolished rice of MadhukarxSwarna RILs. Gene 508, 233–240. doi: 10.1016/j.gene.2012.07.054
Brunner, S., Hurni, S., Streckeisen, P., Mayr, G., Albrecht, M., Yahiaoui, N., et al. (2010). Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J. 64, 433–445. doi: 10.1111/j.1365-313X.2010.04342.x
Cai, H., Huang, S., Che, J., Yamaji, N., Ma, J. F. (2019). The tonoplast-localized transporter OsHMA3 plays an important role in maintaining Zn homeostasis in rice. J. Exp. Bot. 70, 2717–2725. doi: 10.1093/jxb/erz091
Clark, L. C., Combs, G. F., Jr., Turnbull, B. W., Slate, E. H., Chalker, D. K., Chow, J., et al. (1996). Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional prevention of cancer study group. JAMA 276, 1957–1963. doi: 10.1001/jama.1996.03540240035027
Cui, X., Jin, P., Cui, X., Gu, L., Lu, Z., Xue, Y., et al. (2013). Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. U. S. A. 110, 1953–1958. doi: 10.1073/pnas.1217020110
D’Amato, R., Fontanella, M. C., Falcinelli, B., Beone, G. M., Bravi, E., Marconi, O., et al. (2018). Selenium biofortification in rice (Oryza sativa L.) sprouting: effects on Se yield and nutritional traits with focus on phenolic acid profile. J. Agric. Food Chem. 66, 4082–4090. doi: 10.1021/acs.jafc.8b00127
Duan, G. L., Zhou, Y., Tong, Y. P., Mukhopadhyay, R., Rosen, B. P., Zhu, Y. G. (2007). A CDC25 homologue from rice functions as an arsenate reductase. New Phytol. 174, 311–321. doi: 10.1111/j.1469-8137.2007.02009.x
Fan, F., Li, N., Chen, Y., Liu, X., Sun, H., Wang, J., et al. (2017). Development of elite BPH-resistant wide-spectrum restorer lines for three and two line hybrid rice. Front. Plant Sci. 8, 986. doi: 10.3389/fpls.2017.00986
Fan, F., Long, W., Liu, M., Yuan, H., Pan, G., Li, N., et al. (2019). Quantitative trait locus mapping of the combining ability for yield-related traits in wild rice Oryza longistaminata. J. Agric. Food Chem. 67, 8766–8772. doi: 10.1021/acs.jafc.9b02224
Finley, J. W., Ip, C., Lisk, D. J., Davis, C. D., Hintze, K. J., Whanger, P. D. (2001). Cancer-protective properties of high-selenium broccoli. J. Agric. Food Chem. 49, 2679–2683. doi: 10.1021/jf0014821
Garcia-Oliveira, A. L., Tan, L., Fu, Y., Sun, C. (2009). Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J. Integr. Plant Biol. 51, 84–92. doi: 10.1111/j.1744-7909.2008.00730.x
Hu, Y., Liu, D., Zhong, X., Zhang, C., Zhang, Q., Zhou, D. X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U. S. A. 109, 5773–5778. doi: 10.1073/pnas.1203148109
Hu, Y., Zhu, N., Wang, X., Yi, Q., Zhu, D., Lai, Y., et al. (2013). Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol. Biochem. 70, 33–42. doi: 10.1016/j.plaphy.2013.05.001
Huang, D., Qiu, Y., Zhang, Y., Huang, F., Meng, J., Wei, S., et al. (2013). Fine mapping and characterization of BPH27, a brown planthopper resistance gene from wild rice (Oryza rufipogon Griff.). Theor. Appl. Genet. 126, 219–229. doi: 10.1007/s00122-012-1975-7
Huang, Y., Sun, C., Min, J., Chen, Y., Tong, C., Bao, J. (2015). Association mapping of quantitative trait loci for mineral element contents in whole grain rice (Oryza sativa L.). J. Agric. Food Chem. 63, 10885–10892. doi: 10.1021/acs.jafc.5b04932
Jin, J., Long, W., Wang, L., Liu, X., Pan, G., Xiang, W., et al. (2018). QTL mapping of seed vigor of backcross inbred lines derived from Oryza longistaminata under artificial aging. Front. Plant Sci. 9, 1909. doi: 10.3389/fpls.2018.01909
Johnson, A. J., Wu, M. J. (2016). The new role for an old kinase: protein kinase CK2 regulates metal ion transport. Pharmaceuticals (Basel) 9, 80. doi: 10.3390/ph9040080
Khush, G. (2001). Green revolution: The way forward. Nat. Rev. Genet. 2, 815–822. doi: 10.1038/35093585
Li, T., Chen, X., Zhong, X., Zhao, Y., Liu, X., Zhou, S., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Liu, Z. H., Wang, H. Y., Wang, X. E., Zhang, G. P., Chen, P. D., Liu, D. J. (2006). Genotypic and spike positional difference in grain phytase activity, phytate, inorganic phosphorus, iron, and zinc contents in wheat (Triticum aestivum L.). J. Cereal Sci. 44, 212–219. doi: 10.1016/j.jcs.2006.06.001
Liu, K., Zhao, Y., Chen, F., Fang, Y. (2015). Purification and identification of Se-containing antioxidative peptides from enzymatic hydrolysates of Se-enriched brown rice protein. Food Chem. 187, 424–430. doi: 10.1016/j.foodchem.2015.04.086
Liu, X. S., Feng, S. J., Zhang, B. Q., Wang, M. Q., Cao, H. W., Rono, J. K., et al. (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 19, 283. doi: 10.1186/s12870-019-1899-3
Londo, J. P., Chiang, Y. C., Hung, K. H., Chiang, T. Y., Schaal, B. A. (2006). Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. U. S. A. 103, 9578–9583. doi: 10.1073/pnas.0603152103
Masuda, H., Aung, M. S., Nishizawa, N. K. (2013). Iron biofortification of rice using different transgenic approaches. Rice (N. Y.) 6, 40. doi: 10.1186/1939-8433-6-40
Meldrum, E., Parker, P. J., Carozzi, A. (1991). The PtdIns-PLC superfamily and signal transduction. Biochim. Biophys. Acta 1092, 49–71. doi: 10.1016/0167-4889(91)90177-Y
Meng, L., Li, H. H., Zhang, L. Y., Wang, J. K. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Nawaz, Z., Kakar, K. U., Li, X. B., Li, S., Zhang, B., Shou, H. X., et al. (2015). Genome-wide association mapping of quantitative trait loci (QTLs) for contents of eight elements in brown rice (Oryza sativa L.). J. Agric. Food Chem. 63, 8008–8016. doi: 10.1021/acs.jafc.5b01191
Parengam, M., Judprasong, K., Srianujata, S., Jittinandana, S., Laoharo¬janaphand, S., Arporn, B. (2010). Study of nutrients and toxic minerals in rice and legumes by instrumental neutron activation analysis and graphite furnace atomic absorption spectrophotomerty. J. Food Comp. Anal. 23, 340–345. doi: 10.1016/j.jfca.2009.12.012
Petersen, O. H., Wakui, M. (1990). Oscillating intracellular Ca2+ signals evoked by activation of receptors linked to inositol lipid hydrolysis: mechanism of generation. J. Membr. Biol. 118, 93–105. doi: 10.1007/BF01868467
Shi, S., Wang, T., Chen, Z., Tang, Z., Wu, Z., Salt, D. E., et al. (2016). OsHAC1;1 and OsHAC1;2 function as arsenate reductases and regulate arsenic accumulation. Plant Physiol. 172, 1708–1719. doi: 10.1104/pp.16.01332
Song, W. Y., Yamaki, T., Yamaji, N., Ko, D., Jung, K. H., Fujii-Kashino, M., et al. (2014). A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. U. S. A. 111, 15699–15704. doi: 10.1073/pnas.1414968111
Sui, F., Zhao, D., Zhu, H., Gong, Y., Tang, Z., Huang, X. Y., et al. (2019). Map-based cloning of a new total loss-of-function allele of OsHMA3 causes high cadmium accumulation in rice grain. J. Exp. Bot. 70, 2857–2871. doi: 10.1093/jxb/erz093
Takahashi, R., Ishimaru, Y., Senoura, T., Shimo, H., Ishikawa, S., Arao, T., et al. (2011). The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 62, 4843–4850. doi: 10.1093/jxb/err136
Ueno, D., Koyama, E., Yamaji, N., Ma, J. F. (2011). Physiological, genetic, and molecular characterization of a high-Cd-accumulating rice cultivar, Jarjan. J. Exp. Bot. 62, 2265–2272. doi: 10.1093/jxb/erq383
Umeta, M., West, C. E., Haidar, J., Deurenberg, P., Hautvast, J. G. (2000). Zinc supplementation and stunted infants in Ethiopia: a randomised controlled trial. Lancet 355, 2021–2026. doi: 10.1016/S0140-6736(00)02348-5
Uraguchi, S., Kamiya, T., Sakamoto, T., Kasai, K., Sato, Y., Nagamura, Y., et al. (2011). Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. U.S.A. 108, 20959–20964. doi: 10.1073/pnas.1116531109
Xu, Z., Fang, Y., Chen, Y., Yang, W., Ma, N., Pei, F., et al. (2016). Protective effects of Se-containing protein hydrolysates from Se-enriched rice against Pb(2+)-induced cytotoxicity in PC12 and RAW264.7 cells. Food Chem. 202, 396–403. doi: 10.1016/j.foodchem.2016.02.021
Yan, D., Zhang, X., Zhang, L., Ye, S., Zeng, L., Liu, J., et al. (2015). Curved chimeric palea 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 82, 12–24. doi: 10.1111/tpj.12784
Yang, X., Huang, J., Jiang, Y., Zhang, H. S. (2009). Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.). Mol. Biol. Rep. 36, 281–287. doi: 10.1007/s11033-007-9177-0
Yuan, H., Wang, W., Chen, D., Zhu, X., Meng, L. (2017). Effects of a treatment with Se-rich rice flour high in resistant starch on enteric dysbiosis and chronic inflammation in diabetic ICR mice. J. Sci. Food Agric. 97, 2068–2074. doi: 10.1002/jsfa.8011
Zeng, Y. W., Shen, S. Q., Wang, L. X., Liu, J. F., Pu, X. Y., Du, J., et al. (2005). Correlation of plant morphological and grain quality traits with mineral element contents in Yunnan rice. Rice Sci. 12, 101–106.
Zeng, D., Tian, Z., Rao, Y., Dong, G., Yang, Y., Huang, L., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3, 17031. doi: 10.1038/nplants.2017.31
Zhang, M., Pinson, S. R., Tarpley, L., Huang, X. Y., Lahner, B., Yakubova, E., et al. (2014). Mapping and validation of quantitative trait loci associated with concentrations of 16 elements in unmilled rice grain. Theor. Appl. Genet. 127, 137–165. doi: 10.1007/s00122-013-2207-5
Keywords: wild rice, Oryza longistaminata, mineral element, quantitative trait loci, backcross-inbred lines
Citation: Liu X, Fan F, Liu M, Long W, Yu Y, Yuan H, Pan G, Li N, Li S and Liu J (2020) Quantitative Trait Loci Mapping of Mineral Element Contents in Brown Rice Using Backcross Inbred Lines Derived From Oryza longistaminata. Front. Plant Sci. 11:1229. doi: 10.3389/fpls.2020.01229
Received: 20 March 2020; Accepted: 27 July 2020;
Published: 12 August 2020.
Edited by:
Junhua Peng, Huazhi Rice Bio-Tech Co., Ltd., ChinaReviewed by:
Chengdao Li, Murdoch University, AustraliaCopyright © 2020 Liu, Fan, Liu, Long, Yu, Yuan, Pan, Li, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoqing Li, c2hhb3FpbmdsaUB3aHUuZWR1LmNu; Jianfeng Liu, bGl1amlhbmZlbmdAaHVuYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.