- 1Department of Botany and Biodiversity Research Centre, University of British Columbia, Vancouver, BC, Canada
- 2Thünen Institute of Forest Genetics, Grosshansdorf, Germany
A well-established hypothesis for the evolution of dioecy involves two genes linked at a sex-determining region (SDR). Recently there has been increased interest in possible single gene sex determination. Work in Populus has finally provided direct experimental evidence for single gene sex determination in plants using CRISPR-Cas9 to knock out a single gene and convert individuals from female to male. In poplar, the feminizing factor popARR17 acts as a “master regulator”, analogous to the mammalian masculinizing factor SRY. The production of fully functional males from females by a simple single gene knockout is experimental evidence that an antagonistic male-determining factor does not exist in Populus. Mammals have a “default sex” (female), as do poplar trees (Populus), although the default sex in poplars is male. The occurrence of single gene sex determination with a default sex may be much commoner in plants than hitherto expected, especially when dioecy evolved via monoecy. The master regulator does not even need to be at the SDR (although it may be). In most poplars the feminizing factor popARR17 is not at the SDR, but instead a negative regulator of it. So far there is little information on how high-level regulators are connected to floral phenotype. A model is presented of how sex-determining genes could lead to different floral morphologies via MADS-box floral developmental genes.
Introduction: the Two-Gene and Single-Gene Models
The “two-gene model” for the evolution of dioecy put forward in 1978 has been enormously influential, the original paper being cited over 1,000 times (Charlesworth and Charlesworth, 1978). This model posits that two mutations (most probably in two separate genes, a male-sterility factor and a female-sterility factor) are likely to be involved in the evolution of dioecy, and further that these two genes are usually brought together in linkage. This model was developed in the context of the evolution of dioecy through the gynodioecy pathway, and in this context it is well supported. Silene, for instance, has iconic status as a model system for the two-gene system (e.g. Westergaard, 1958; Kazama et al., 2016).
However, dioecy also evolves through the monoecy pathway, and here the model may be less useful. For instance, in a monoecious plant it is likely that there has been a mutation to allow spatiotemporal factors (such as hormone gradients) to interact with development to produce male or female flowers, depending on the position within the plant or on developmental timing. Floral developmental pathways in monoecy could therefore evolve to be under the overall control of a single high-level regulator with two states (on vs off) determining the two phenotypes. If this high-level regulator is segregating in the population (present vs absent), then dioecy, involving only a single regulatory gene, could readily evolve from monoecy. This is the alternative “single-gene model” (Renner, 2016) which this perspective argues is likely to be most commonly involved in the evolution of dioecy through the monoecy pathway.
Two final points should be mentioned here: first, discussion of “single-gene sex determination” or “two-gene sex determination” refers not to the total numbers of genes involved in sexual differentiation: the pathway that leads to the suppression of stamens or carpels may, of course, have multiple genes involved. Rather it refers to the numbers of genes needed for the segregation of sex at the sex-determining region (SDR) of the genome. Secondly, the SDR may be a genomic block subject to recombination suppression, in which case the chromosome may be characterized as a sex chromosome, and the other chromosomes as autosomes. However, the sex chromosome/autosome distinction is complicated if the region of recombination suppression is small (as in many plants) or even non-existent. In the pufferfish, Takifugu rubripes, sex determination is effected by one single nucleotide polymorphism (Kamiya et al., 2012). Here we refer to regions of the genome as sex-determining (SDR) or “autosomal” (outside the SDR).
Recent Results on the Genetic and Molecular Basis of Sex Determination
Very rapid progress is being made in understanding the genetic and molecular basis of dioecy in flowering plants with important papers in a diverse array of systems including papaya (Carica: Liu et al., 2004) and more recently, kiwifruit (Actinidia: e.g. Akagi et al., 2019), persimmon (Diospyros: Akagi et al., 2020), date palm (Phoenix: Torres et al., 2018), grape (Vitis: Massonnet et al., 2020), asparagus (Asparagus: Harkess et al., 2017), and poplar (Populus: Geraldes et al., 2015; Müller et al., 2020). The first five of these are fruit crops in which dioecy has obvious economic importance in fruit production. Asparagus and poplar are vegetable and tree crops, respectively, and are also important in the role they have in shedding light on dioecy. So far, molecular work in most economically important species has provided support for the two-gene model, for instance in kiwifruit (Akagi et al., 2019), Vitis (Massonnet et al., 2020), Phoenix (Torres et al., 2018), and asparagus (Harkess et al., 2020). However, in two other systems for which dioecy is well characterized at the molecular level, persimmon and poplar, the results seem to point at a one-gene model at the SDR.
In Diospyros lotus (Akagi et al., 2014), there is an autosomal feminizing gene MeGI, giving rise to females. Males are produced when a Y-specific suppressor, OGI, inhibits MeGI expression. Notably, transcriptional regulation of MeGI is also involved in the control of sex expression in monoecious persimmon (D. kaki: Akagi et al., 2016).
In poplar (Müller et al., 2020) there is an autosomal (i.e. not associated with the sex-determining region, or SDR) feminizing gene popARR17 (the poplar homolog of Arabidopsis Response Regulator 17). There is also, critically, a Y-specific suppressor of ARR17 at the SDR (ΨARR17-IR) (Figure 1). In poplar, ΨARR17-IR is an inverted repeat of the target gene that forms hairpin RNA, generating sRNA and silences popARR17 by RNA-directed DNA methylation (RdDM) (Figure 1). In one species of poplar (Populus alba: Müller et al., 2020) and apparently in several willow species (Salix: Almeida et al., 2020; Zhou et al., 2020) the same basic sex-determining system appears to be used (discussed in Müller et al., 2020), except here the “master regulator” ARR17 is hemizygous at the SDR on the W chromosome: it is inferred that females develop when it is present (ZW), males when it is absent (ZZ), implying that males are the “default sex” (Figure 1).
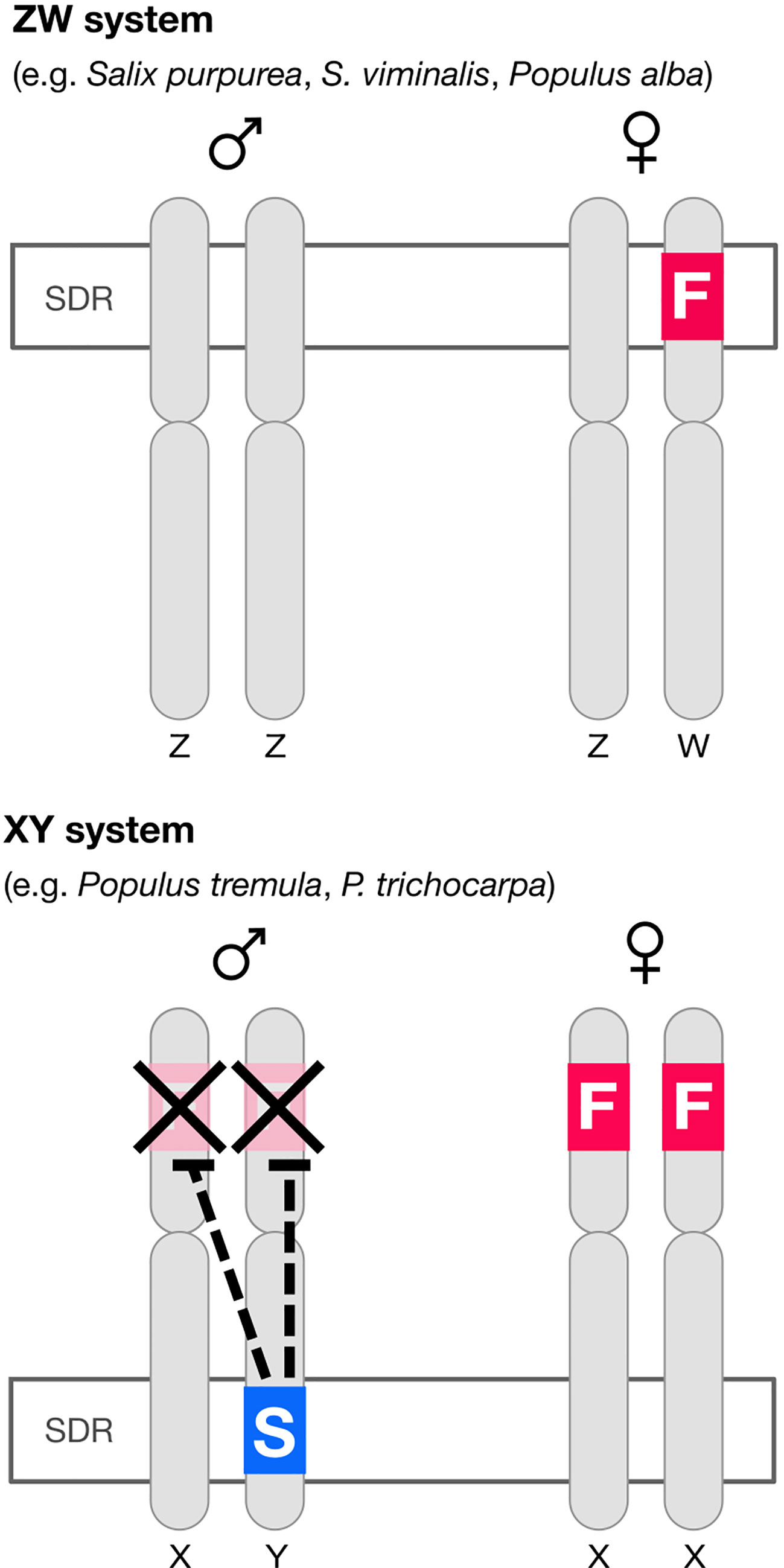
Figure 1 Diagram of the architecture of sex determination in willow and poplar. In at least some willow and one poplar species (P. alba) the default developmental pathway is male but a feminizing factor F (the homolog of the cytokinin response regulator ARR17) segregates, giving a ZW sex determination system. In most poplar species the default developmental pattern is again male, but both sexes have a copy of the feminizing factor. A “suppressor of F” factor (S) segregates, giving an XY system of sex determination. The “suppressor of F” consists of an ARR17 pseudogene inverted repeat (ΨARR17-IR). This structure leads to the production of sRNA and thence to RNA-directed DNA methylation (RdDM) silencing ARR17 (Müller et al., 2020). The feminizing factor ARR17 is therefore analogous to the masculinizing factor SRY in mammals (except sex-reversed).
The Concept of a Default Sex and a “Master Regulator”
The sex-determination system of mammals (specifically eutherians and marsupials) is a well-worked out single-gene system with a master regulator (SRY) and a default sex (female). The mammalian system now appears to provide a surprisingly useful analogous system for sex determination in some plants.
Even before the discovery of SRY, the notion of female as the mammalian “default sex” was established experimentally. An inferred gene, testes determining factor (TDF) was shown to act dominantly in mammalian development. If testes are removed from young XY embryos, they develop as females. Whereas if ovaries are removed from young XX embryos, they still develop as females (Jost, 1970), implying that a specific signal was required for male development, but not for female development. For this reason females were considered to be the default sex (Graves, 2017). Later, the gene ‘Sex-determining Region on the Y’ or SRY was discovered (Sinclair et al., 1990) which fitted the requirements of the TDF master regulator that could divert default female development into male development. If SRY is mutated to be non-functional, female development ensues despite the presence of a Y chromosome (Berta et al., 1990). Conversely if SRY is introduced transgenically into XX females, males develop (Koopman et al., 1991). However, it should be noted that mammalian sex determination is complex (e.g. Zhao et al., 2017) and monotremes are very different (Veyrunes et al., 2008).
What is the equivalent in persimmon, poplar and willow? In Populus alba and in ZW willow species, ARR17 is present only on the W chromosome, apparently as a dominant feminizer, giving rise to ZW females, and in its absence, to default ZZ males. The parallels to SRY are obvious (except sex-reversed) and imply that ARR17 is the master regulator and male the default sex. This is complicated in other poplars however, where the ARR17 “master regulator” is autosomal and is toggled on/off by a Y-specific suppressor at the SDR. But it is not helpful to consider the suppressor the master regulator: the fundamental regulation by ARR17 remains the same, as does the default sex (male). It is only the regulation of ARR17 that differs (toggled on/off by a repressor in XY poplar and regulated by the presence/absence in ZW willow).
The first experimental evidence in dioecious plants of both the existence of a master regulator (ARR17) and a default sex (male) was obtained in poplar by CRISPR-Cas9 knock-out of ARR17 (Müller et al., 2020) which converted XX females to males, reminiscent of SRY knock-outs in mammals (but sex-reversed). The fact that in poplar a single gene is necessary and sufficient for female development, and without it males develop, is powerful evidence for a single master regulator and a default sex. The CRISPR result shows that an antagonistic male determining factor is not necessary in poplar. Instead the system is driven by a single gene, the feminizing factor popARR17.
The situation in Diospyros seems very similar and can be interpreted as an autosomal feminizing master regulator (MeGI) capable of redirecting default male development to female and toggled on–off by the presence of a suppressor at the SDR. We thus have two potential models for the molecular control of dioecy: (1) a segregating hemizygous single gene master regulator at the SDR (Salix, Populus alba) or (2) an autosomal single gene master regulator, toggled on–off by a segregating suppressor at the SDR (Diospyros, Populus).
Monoecious plant species also need to control unisexual flower development, only in different parts of the same plant instead of different individuals. Interestingly, transcriptional regulation of a feminizing master regulator appears to control this differential sexual development in diverse monoecious species as well. In monoecious maize and cucumber, for example, the genes Silkless and ACS11, respectively, trigger female development, and mutations lead to purely male plants (androecy) (Jones, 1934; Boualem et al., 2015). It is simple to imagine an adoption of this mechanism in the evolution of dioecy by limiting expression of the feminizing master regulator to female individuals instead of creating an expression gradient within one plant. As the unisexual separation of floral developmental pathways has already occurred in the monoecious ancestor, the evolution of dioecy from monoecy may be a simple matter of separating pre-existing epistatic regulatory architecture to different individuals, which may be accomplished by a single segregating gene. For this reason, we might expect the evolution of single gene-sex determination systems to be associated with monoecious lineages. Willow and poplar appear to be examples of this, as the family Salicaceae (to which they belong) is largely composed of monoecious or “polygamous” (i.e. mixing hermaphrodite and unisexual flowers) species (Cronk, 2005; Cronk et al., 2015).
Enter the Mads-Box Genes
Despite the rapid advances in understanding the high-level molecular mechanisms for dioecy, there has been little progress on how those high-level regulators are connected to phenotypes through gene regulatory cascades. There is likely to be a fundamental difference in the development between dioecious plants in which organs of the opposite sex are deleted and those where they are vestigial. So, in dioecious members of the Rosaceae it is common to find vestigial stamens in female flowers, whereas in the poplar there is no sign whatever of stamens in female flowers (apparently at any stage of development): they are cleanly deleted. Floral MADS-box genes are required to specify the identity of floral organ types, according to the well-known ABC model (Litt and Kramer, 2010). If the required combinations of MADS-box genes are not expressed, the relevant organs will not form. Therefore MADS-box genes are candidates for clean deletion of stamens or carpels. However, in cases of dioecy where floral organs are vestigial, it is unlikely that MADS-box genes are involved as they must be expressed correctly if the relevant organs (even if vestigial) are formed. Instead it is likely that genes downstream of the MADS-box gene pathway are blocking development of those organs once formed.
PISTILLATA and APETALA3 (PI/AP3) are a pair of MADS-box genes required for the specification of stamens (class-B MADS-box genes), and where they are expressed carpels do not form (Figure 2). Constitutive overexpression of PI/AP3 is therefore expected to produce stamen-only flowers as seen in male poplars (Cronk et al., 2020). The action of the feminizing factor ARR17 therefore could be to locally block the expression of PI/AP3, inhibiting stamens and allowing carpels to form (as in female flowers). A hypothetical model of this is presented in Figure 2. However, the pathway between ARR17 and PI/AP3 (if it exists) is completely unknown. This should be a target for future research. ARR17 is a response regulator implicated in regulation of the cytokinin pathway, but its mode of action in floral development at present remains elusive. In considering the potential involvement of PI it is of significance that the sex determinant MeGI in persimmon (which acts as a repressor of the androecium) functions via its ability to regulate PI during early androecium development (Yang et al., 2019). In addition, Nicotiana tabacum transgenic for MeGI showed significant downregulation of PI (Akagi et al., 2020).
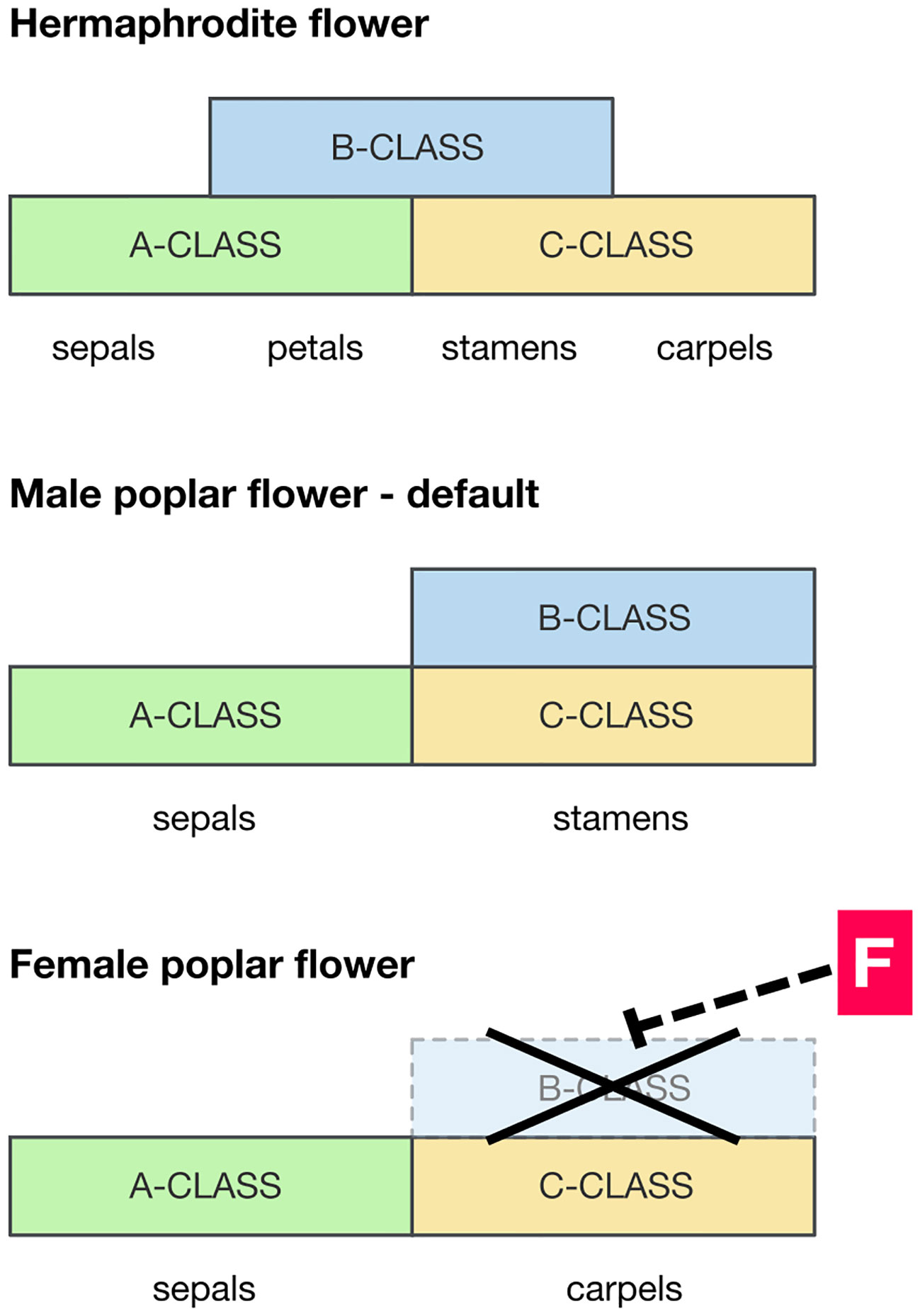
Figure 2 Hypothetical scenario for the developmental pathway leading to male and female floral development in poplar and willow. Diagrams of the expression domains of MADS-box floral developmental genes relative to floral organ development. Top panel: pattern typical of a hermaphrodite eudicot flower (e.g. Arabidopsis), for reference. Bottom panels: suggested MADS-box gene expression domains in the formation of male and female poplar flowers. Where the B-class MADS-box genes PISTILLATA and APETALA3 (PI/AP3) are expressed, along with C-class genes, stamens develop. A wide expression domain of PI/AP3 is therefore expected in males, and this is likely to be the default in both sexes. However, in females the presence of a feminizing factor F downregulates PI/AP3 diverting development to a female developmental pathway.
Conclusions
Current molecular results on sex-regulation in Populus and Diospyros provide evidence for two related pathways, neither of which require two linked genes at the SDR. The first involves an autosomal master regulator (ARR17, MeGI), toggled on–off by a suppressor at the SDR (ΨARR17-IR, OGI). The second involves a hemizygous master regulator at the SDR (ARR17 in P. alba and Salix). Many more examples will need to be worked through in other plants before we can determine with certainty whether this is the norm, or whether the model of two linked loci at the SDR (as seems to be the case in Asparagus or kiwifruit) is more usual. More work on other systems is therefore a priority. It may be, however, that a single gene at the SDR is all that is needed to regulate sex, and this will be much more common than currently thought (especially in the monoecy–dioecy pathway where floral dimorphism has already evolved).
Furthermore, the pathway connecting these high-level regulators to floral phenotype is largely unknown, and work in this area is urgently required if we are to fully understand dioecy. In this regard, attention is drawn to two very different developmental outcomes: deleted organs vs. vestigial organs. The first is likely to involve changes in floral MADS-box gene expression (especially PI/AP3), whereas the second is likely to involve genes involved in organ growth and maturation downstream of the MADS-box gene pathway.
Data Availability Statement
The original contributions presented in the study are published (see citations); further inquiries can be directed to the corresponding author.
Author Contributions
QC and NM developed the ideas and wrote the manuscript.
Funding
Work in the laboratory of QC is funded by the Natural Sciences and Engineering Research Council of Canada (NSERC). NM acknowledges the support of the Deutsche Forschungsgemeinschaft (DFG: MU 4357/1-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the editors of the research topic “Dioecy in Fruit Crops: The Gender Rise and Decline and Its Agronomic Impact” for organizing this collection, and the two reviewers for their insightful comments.
References
Akagi, T., Henry, I. M., Tao, R., Comai, L. (2014). A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346 (6209), 646–650. doi: 10.1126/science.1257225
Akagi, T., Kawai, T., Tao, R. (2016). A male determinant gene in diploid dioecious Diospyros, OGI, is required for male flower production in monoecious individuals of Oriental persimmon (D. kaki). Scientia Horticulturae 213, 243–251.
Akagi, T., Pilkington, S. M., Varkonyi-Gasic, E., Henry, I. M., Sugano, S. S., Sonoda, M., et al. (2019). Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 5 (8), 801–809. doi: 10.1038/s41477-019-0489-6
Akagi, T., Shirasawa, K., Nagasaki, H., Hirakawa, H., Tao, R., Comai, L., et al. (2020). The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PloS Genet. 16 (2), e1008566. doi: 10.1371/journal.pgen.1008566
Almeida, P., Proux-Wera, E., Churcher, A., Soler, L., Dainat, J., Pucholt, P., et al. (2020). Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion. BMC Biol. 18 (1), 1–18. doi: 10.1186/s12915-020-00808-1
Berta, P., Hawkins, J. B., Sinclair, A. H., Taylor, A., Griffiths, B. L., Goodfellow, P. N., et al. (1990). Genetic evidence equating SRY and the testis-determining factor. Nature 348, 448–450. doi: 10.1038/348448A0
Boualem, A., Troadec, C., Camps, C., Lemhemdi, A., Morin, H., Sari, M. A., et al. (2015). A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350 (6261), 688–691. doi: 10.1126/science.aac8370
Charlesworth, B., Charlesworth, D. (1978). A model for the evolution of dioecy and gynodioecy. Am. Nat. 112 (988), 975–997. doi: 10.1086/283342
Cronk, Q. C. (2005). Plant eco-devo: the potential of poplar as a model organism. New Phytol. 166 (1), 39–48. doi: 10.1111/j.1469-8137.2005.01369.x
Cronk, Q. C., Needham, I., Rudall, P. J. (2015). Evolution of catkins: inflorescence morphology of selected Salicaceae in an evolutionary and developmental context. Front. Plant Sci. 6, 1030. doi: 10.3389/fpls.2015.01030
Cronk, Q. C., Soolanayakanahally, R., Braäutigam, K. (2020). Gene expression trajectories during male and female reproductive development in balsam poplar (Populus balsamifera L.). Sci. Rep. 10 (1), 1–14. doi: 10.1038/s41598-020-64938-w
Geraldes, A., Hefer, C. A., Capron, A., Kolosova, N., Martinez-Nuñez, F., Soolanayakanahally, R. Y., et al. (2015). Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol. Ecol. 24 (13), 3243–3256. doi: 10.1111/mec.13126
Graves, J. A. M. (2017). How Australian mammals contributed to our understanding of sex determination and sex chromosomes. Aust. J. Zool. 64 (4), 267–276. doi: 10.1071/ZO16054
Harkess, A., Zhou, J., Xu, C., Bowers, J. E., Van der Hulst, R., Ayyampalayam, S., et al. (2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 8 (1), 1–10.
Harkess, A., Huang, K., van der Hulst, R., Tissen, B., Caplan, J. L., Koppula, A., et al. (2020). Sex determination by two Y-linked genes in garden Asparagus. Plant Cell. 32, 1790–1796. doi: 10.1105/tpc.19.00859
Jones, D. F. (1934). Unisexual maize plants and their bearing on sex differentiation in other plants and in animals. Genetics 19 (6), 552.
Jost, A. (1970). Hormonal factors in the sex differentiation of the mammalian foetus. Philos. Trans. R. Soc. London Ser. B: Biol. Sci. 259, 119–131. doi: 10.1098/rstb.1970.0052
Kamiya, T., Kai, W., Tasumi, S., Oka, A., Matsunaga, T., Mizuno, N., et al. (2012). A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PloS Genet. 8 (7), e1002798. doi: 10.1371/journal.pgen.1002798
Kazama, Y., Ishii, K., Aonuma, W., Ikeda, T., Kawamoto, H., Koizumi, A., et al. (2016). A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 6, 1–10. doi: 10.1038/srep18917
Koopman, P., Gubbay, J., Vivian, N., Goodfellow, P., Lovell-Badge, R. (1991). Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121. doi: 10.1038/351117a0
Litt, A., Kramer, E. M. (2010). The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 21, 129–137. doi: 10.1016/j.semcdb.2009.11.019
Liu, Z., Moore, P. H., Ma, H., Ackerman, C. M., Ragiba, M., Yu, Q., et al. (2004). A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427 (6972), 348–352. doi: 10.1038/nature02228
Massonnet, M., Cochetel, N., Minio, A., Vondras, A. M., Lin, J., Muyle, A., et al. (2020). The genetic basis of sex determination in grapes. Nat. Commun. 11 (1), 1–12. doi: 10.1038/s41467-020-16700-z
Müller, N., Kersten, B., Leite Montalvão, A. P., Mähler, N., Bernhardsson, C., Bräutigam, K., et al. (2020). A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants. 6, 630–637. doi: 10.1038/s41477-020-0672-9
Renner, S. S. (2016). Pathways for making unisexual flowers and unisexual plants: Moving beyond the “two mutations linked on one chromosome” model. Am. J. Bot. 103 (4), 587–589. doi: 10.3732/ajb.1600029
Sinclair, A. H., Berta, P., Palmer, M. S., Hawkins, J. R., Griffiths, B. L., Smith, M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244. doi: 10.1038/346240a0
Torres, M. F., Mathew, L. S., Ahmed, I., Al-Azwani, I. K., Krueger, R., Rivera-Nuñez, D., et al. (2018). Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 9 (1), 1–9. doi: 10.1038/s41467-018-06375-y
Veyrunes, F., Waters, P. D., Miethke, P., Rens, W., McMillan, D., Alsop, A. E., et al. (2008). Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18, 965–973. doi: 10.1101/gr.7101908
Westergaard, M. (1958). The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 9, 217–281. doi: 10.1016/S0065-2660(08)60163-7
Yang, H. W., Akagi, T., Kawakatsu, T., Tao, R. (2019). Gene networks orchestrated by MeGI: a single-factor mechanism underlying sex determination in persimmon. Plant J. 98 (1), 97–111. doi: 10.1111/tpj.14202
Zhao, F., Franco, H. L., Rodriguez, K. F., Brown, P. R., Tsai, M. J., Tsai, S. Y., et al. (2017). Elimination of the male reproductive tract in the female embryo is promoted by COUP-TFII in mice. Science 357 (6352), 717–720. doi: 10.1126/science.aai9136
Keywords: dioecy, monoecy, Populus, ARR17, Diospyros, OGI, SRY, MADS-box
Citation: Cronk Q and Müller NA (2020) Default Sex and Single Gene Sex Determination in Dioecious Plants. Front. Plant Sci. 11:1162. doi: 10.3389/fpls.2020.01162
Received: 28 May 2020; Accepted: 16 July 2020;
Published: 29 July 2020.
Edited by:
Sarah M. Pilkington, The New Zealand Institute for Plant and Food Research Ltd, New ZealandReviewed by:
Michael Nicolas, National Center for Biotechnology (CNB), SpainMaria Manuela Ribeiro Costa, University of Minho, Portugal
Copyright © 2020 Cronk and Müller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quentin Cronk, cXVlbnRpbi5jcm9ua0B1YmMuY2E=
 Quentin Cronk
Quentin Cronk Niels A. Müller2
Niels A. Müller2