- 1Graduate School of Science, The University of Tokyo, Tokyo, Japan
- 2Exploratory Research Center on Life and Living Systems, National Institutes of Natural Sciences, Okazaki, Japan
Members of the genus Monophyllaea are unique in that they produce no new organ during the vegetative phase in the shoot; instead, one of the cotyledons grows indeterminately. The mechanism of this unique trait is unclear, in part because of the lack of suitable assessment techniques. We therefore established a whole-mount in situ hybridization technique, a powerful means of examining spatial patterns in gene expression, for Monophyllaea glabra. By using this, we examined the expression pattern of a SHOOT MERISTEMLESS (STM) ortholog, which is indispensable for the formation and maintenance of the shoot apical meristem (SAM) in typical angiosperms. Expression was confined to the groove meristem (GM), which corresponds to the SAM. We also assessed the expression pattern of ANGUSTIFOLIA3 (AN3), a key promoter for cell division in the leaf meristem. It was expressed not only in the basal meristem (BM) tissue with active cell division in the basal part of the growing cotyledon but also in the GM. The findings suggest that the unusual gene expression pattern of the GM underpins the fuzzy morphogenesis of Monophyllaea.
Introduction
Plants can produce new organs from the meristem throughout their lives, whereas animals complete most morphogenesis during the embryonic stages (Steeves and Sussex, 1989; Graham et al., 2000; Wolpert and Tickle, 2011). The aerial part of typical seed plants, the shoot, is composed of repeating phytomere units (Gray, 1879), each of which consists of a leaf, stem, and axillary bud. All of these components are produced from the indeterminate meristem, the shoot apical meristem (SAM), at the tip of the shoot. The indeterminate nature of the SAM is dependent on the maintenance of stem cells. Because of the indeterminate nature of the SAM, the shoot system is indeterminate. SHOOT MERISTEMLESS (STM) is indispensable for the formation and maintenance of the indeterminate SAM in model plants; moreover, loss-of-function mutants of this gene lack a SAM (Endrizzi et al., 1996; Long et al., 1996). STM encodes a class I KNOX transcription factor. In Arabidopsis thaliana four class I KNOX genes, STM, KNAT1, KNAT2, and KNAT6 support the maintenance of a meristematic state (Hay and Tsiantis, 2010). STM and KNAT1 are expressed only in the SAM (Lincoln et al., 1994; Long and Barton, 1998). WUSCHEL (WUS) is important for the maintenance of a stem cell niche in the SAM, and loss-of-function mutants lack a SAM (Laux et al., 1996; Mayer et al., 1998; Lenhard et al., 2002).
Leaf primordia are initiated from the flanking region of the SAM and have a determinate meristem, known as the leaf meristem (LM) (Ichihashi and Tsukaya, 2015) that generates leaf lamina cells; therefore, leaves are determinate organs. Genes supporting the LM function have been identified in model plants. ANGUSTIFOLIA3 (AN3)/GRF-INTERACTING FACTOR1 (GIF1) regulates cell division in the leaf meristem by functioning as a transcriptional co-activator of transcription factors, such as GROWTH-REGULATING FACTOR5 (GRF5), in A. thaliana (Horiguchi et al., 2005; Lee et al., 2009; Kim and Tsukaya, 2015). The leaf of the loss-of-function mutant an3 has around 30% of the cells of the wild type; the cell number of cotyledons is also decreased (Horiguchi et al., 2005; Lee et al., 2009). In A. thaliana and Oryza sativa, AN3 is expressed in the basal part of the leaf primordia but not in the SAM (Horiguchi et al., 2011; Shimano et al., 2018). In Zea mays, the AN3 ortholog is expressed from the bottom to the center of the SAM but not at the tip (Zhang et al., 2018).
AN3 protein can move between cells (Kawade et al., 2013; Kawade et al., 2017). Therefore, although the area of AN3 expression is smaller than that of actively cell dividing area, it matches the meristematic area in leaf primordia, which suggests that it is a determinant thereof. This intercellular movement of AN3 protein is necessary for the proper regulation of leaf meristem activity; in one study, immobilized AN3 protein fused to three GFP molecules did not fully complement the reduced number of leaf cells in an an3 mutant (Kawade et al., 2013).
One-leaf plants have a developmental system unlike that of typical seed plants such as the model plant, A. thaliana. They lack a typical shoot system and instead have one indeterminately growing cotyledon and do not produce other new organs, such as stems or foliage leaves, until the reproductive phase (Jong, 1970; Jong and Burtt, 1975; Kinoshita and Tsukaya, 2019). Because they are eudicots, they develop two cotyledons of identical size immediately after germination (isocotyledonous stage), both of which grow. However, after some time, one of the cotyledons (the microcotyledon) stops growing or wither away, whereas the other (the macrocotyledon) continues growing as the sole photosynthetic organ (anisocotyledonous stage), leading to the appearance of harboring a single leaf (Crocker, 1860; Ridley, 1906; Tsukaya, 1997; Nishii et al., 2017). Studies of one-leaf plants have focused on the genera Monophyllaea and Streptocarpus in Gesneriaceae, which, based on their phylogenetic position, evolved independently. In fact, these genera belong to two different tribes: Epithemateae and Trichosporeae (Jong and Burtt, 1975; Burtt, 1978; Smith, 1996; Smith et al., 1997; Möller et al., 2009; Weber et al., 2013).
Because one-leaf plants have an indeterminate shoot-like character in their macrocotyledon and yet the macrocotyledon is a planar, photosynthetic organ, similar to the leaf of typical plants, the plant body system can be interpreted as “fuzzy morphology” (Rutishauser and Isler, 2001). The term “phyllomorph” has been proposed for this fuzzy morphological unit, which consists of a stem-/petiole-like structure, a petiolode, and an indeterminately growing lamina. One-leaf plants are composed of this single unit (Jong, 1970; Jong and Burtt, 1975; Rutishauser and Sattler, 1985). The growth of the phyllomorph is supported by three meristems: the groove meristem (GM), the basal meristem (BM), and the petiolode meristem (PM). The GM is located at the junction between the macrocotyledon and the petiolode (Jong, 1970; Jong and Burtt, 1975) and is thought to correspond to the SAM because of its position, its tunica-corpus structure (reminiscent of the SAM; Jong and Burtt, 1975; Imaichi et al., 2000; Ayano et al., 2005), and its ability to produce inflorescence (Imaichi et al., 2000; Ayano et al., 2005) although one-leaf plants do not produce new organs in the vegetative phase. The BM is positioned in the basal part of the lamina of the macrocotyledon, which is laterally adjacent to the GM and contributes to lamina growth by active cell division. The BM remains active indeterminately, whereas the LM, which is cell proliferative area in leaf primordia (Ichihashi and Tsukaya, 2015), disappears at a certain developmental stage (Kazama et al., 2010). Because of the indeterminate meristem, the cotyledon in most one-leaf plant species grows for several years; in some Monophyllaea species, the inflorescence-bearing mature cotyledon retains the BM (Hilliard and Burtt, 1971; Imaichi et al., 2001). By contrast, the activity of the leaf meristem of A. thaliana is maintained for only ~1 week (Kazama et al., 2010). The PM is positioned immediately below the GM (Imaichi et al., 2000; Imaichi et al., 2001) or below the two cotyledons (Ayano et al., 2005) and contributes to petiolode growth.
Because the GM does not produce new organs during the vegetative phase, it has been hypothesized that although the GM is corresponding tissue to the SAM, some aspects of SAM functions are lost/suppressed at the GM (Cronk and Möller, 1997; Tsukaya, 1997; Tsukaya, 2000). By contrast, the BM has been hypothesized to have a similar regulatory system as the SAM based on its indeterminate cell division activity (Cronk and Möller, 1997).
To characterize the meristem of one-leaf plants, data on the spatiotemporal expression of the aforementioned key genes are needed. Although genes implicated in the formation and maintenance of meristems in phyllomorphs have been investigated in Streptocarpus, no such analyses by in situ hybridization have been performed in the genus Monophyllaea because of methodological limitations (Ishikawa et al., 2017). Here we established a whole-mount in situ hybridization (WMISH) technique for Monophyllaea glabra for the first time and investigated the expression of STM and AN3 orthologs to evaluate the GM and the BM of one-leaf plants of the genus Monophyllaea.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of Monophyllaea glabra were originally collected at Srakaew Cave, Thailand (Ishikawa et al., 2017). The strain was maintained by cultivation in growth chambers. Seeds were sown on one third MS medium with 0.8% (w/v) agar, and plants were grown at 22–23°C under white light provided by fluorescent lamps or metal halide lamps under continuous light or short-day (SD; 8 h light and 16 h dark) conditions (Kinoshita and Tsukaya, 2019). The light intensity was ~45 µmol m−2 s−1.
Calcofluor Staining and Confocal Microscopy
M. glabra individuals of 17 DAS fixed and dehydrated for WMISH were rehydrated and immersed in 50% (v/v) calcofluor white stain (Sigma-Aldrich) and 5% (w/v) KOH for ~16 h. The petiolode of M. glabra was excised, and the samples were mounted with ClearSee solution (Kurihara et al., 2015) and observed with a confocal microscope Fluoview FV10i (Olympus).
Isolation and Characterization of CYCB1 and AN3 Orthologs From M. glabra
Total RNA was extracted from inflorescences of M. glabra with an RNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s protocol. First-strand cDNA was synthesized from total RNA with the SuperScript III First-Strand Synthesis Kit (Invitrogen, USA) according to the manufacturer’s protocol.
Primers for isolating CYCB1 and AN3 homologs were designed based on de novo assembled sequences obtained from mRNA-seq of M. glabra (Kinoshita et al., unpublished). The primers for cloning Mg-AN3 were Mg-AN3_clon-F1 (5′-TTATTACATTACAATCTCGCAGCAC-3′) and Mg-AN3_clon-R2 (5′-AAAAGTGTGACGAAACAAGATCACT-3′). Mg-CYCB1_clon-F1 (5′-CTTCTCAATGGCTTCAAAACAAGT-3′) and Mg-CYCB1_clon-R1 (5′-CAATTAACTGCTAAGTGAAGAAGAAGA-3′) were used to clone Mg-CYCB1. The amplicons were ligated to EcoRV-digested pZErO-2 plasmids (Thermo Fisher Scientific) and were introduced to Escherichia coli to be amplified. We sequenced the plasmids or conducted direct colony sequencing to confirm the sequences of the amplicons. The nucleotide sequences were deposited in DDBJ under accession number LC536022 for MgAN3-1, LC536023 for MgAN3-2, LC536024 for Mg-CYCB1-1, and LC536025 for Mg-CYCB1-2.
Since cyclin degradation at a particular cell cycle phase is important for progressing into next cell cycle, the destruction box motif, the key region for the regulation of the cyclin degradation (Glotzer et al., 1991), we referred to Hemerly et al. (1992) to determine the destruction box and used Pfam (https://pfam.xfam.org/; Cyclin_N, PF00134; Cyclin_C, PF02984) for the other domains to characterize Mg-CYCB1. For Mg-AN3, Kim and Tsukaya (2015) was referred for the SNH domain.
Molecular Phylogenetic Analyses
The amino acid sequences other than from M. glabra were obtained from the following databases: Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) for Amborella trichopoda (version 1.0), Oryza sativa (version 7.0), Zea mays (version 5.0), and Solanum lycopersicum (ITAG 2.4), Snapdragon Genome Database (http://bioinfo.sibs.ac.cn/Am) for Antirrhinum majus (version 3.0) and TAIR10 (https://www.arabidopsis.org/) for Arabidopsis thaliana. GeneBank (https://www.ncbi.nlm.nih.gov/genbank/) (Accession number: AJ250315.1) for CYCB1;1 of Petunia hybrida. The amino acid sequences from M. glabra were inferred from the longest open reading frames in the cDNA sequences. Amino acid sequences were aligned by MAFFT v. 7.407 in auto mode (Katoh and Standley, 2013), and poorly aligned sequences were trimmed with trimAl v. 1.4. rev. 15 (Capella-Gutiérrez et al., 2009) in automated1 mode. RAxML v. 8.2.12 (Stamatakis, 2014) was used to analyze the phylogenetic relationship with the maximum likelihood method. Bootstrap analysis (Felsenstein, 1985) with 100 replicates was performed with the same software, and phylogenetic trees were generated with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Whole-Mount In Situ Hybridization
After confirming the plasmid sequences, we conducted PCR using M13 forward (5′-GTAAAACGACGGCCAGT-3′) and M13 reverse (5′-CAGGAAACAGCTATGAC-3′) primers. The amplicons were used as the template for generating DIG-labelled antisense and sense probes for Mg-CYCB1-1 and Mg-AN3-2 by SP6 or T7 polymerase (Roche) using DIG RNA Labeling Mix (Roche). For WMISH, we slightly modified the protocol of Rozier et al. (2014) in order to facilitate the permeabilization of cells. First, we used 4% (w/v) paraformaldehyde (PFA) with 15% (v/v) dimethyl sulfoxide (DMSO) in phosphate-buffered saline with 0.1% (v/v) Tween-20 (PBST) as the fixative unless stated otherwise. Second, the cell wall enzyme treatment was performed for 30 or 60 min with six times diluted stock solution of 1.2% (w/v) Macerozyme R10 (Yakult), 0.5% (w/v) Cellulase Y-C (Kyowa Chemical), and 0.25% (w/v) Pectolyase Y23 (Kyowa Chemical) in PBST. Hybridization was performed at 50°C or 55°C for 3 days. DIG was detected with a DIG Detection Kit (Roche) with a 1:2,000 dilution of the anti-digoxigenin antibody (Roche).
Results
Determining the Precise Position of the GM and the BM
We first defined the GM and the BM anatomically. To determine precisely the position of the GM and the BM from the top of the phyllomorph, we stained the cell walls of anisocotyledonous-stage individuals (17 DAS; Figures 1B, C) with calcofluor white and take an image of a section including cells of 2nd and 3rd layers in the macrocotyledon with a confocal microscope (Figure 1A). Two positionally distinct meristems, the GM and the BM, were evident. One meristem resided in the most proximal part of the macrocotyledon around 100 µm from the mediolateral axis and adjacent to five rows of differentiated cells in the distal part of the meristem. This meristem is the GM—inflorescence was produced at this position, as evidenced by the presence of a bulge (Figures 1D, E and 3I). The other meristem was laterally adjacent to the GM, and its smaller cells were distributed more widely than the GM, both laterally and distally. In the basal part of macrocotyledon, changes in contour were observed at certain points (Figure 1A). Hereafter we regard the basal part of the tissue inside these points as the GM and the basal part of the tissue outside it as the BM.

Figure 1 The meristem position in anisocotyledonous-stage M. glabra. (A) Paradermal view confocal micrograph of the tissue structure in an anisocotyledonous individual stained with calcofluor white. The upper right leaf is the macrocotyledon, and the lower left leaf is the microcotyledon. The positions of the GM and the BM are colored pink and pale blue, respectively with an image processing software. The red arrowheads show the position where changes in contour in the macrocotyledon were observed. (B) An anisocotyledonous-stage (17 DAS) individual in the vegetative phase grown under continuous light. (C) The basal part of the macrocotyledon of the individual in (B). (D) An anisocotyledonous-stage (32 DAS) individual in the reproductive phase grown under short-day conditions. (E) The basal part of the macrocotyledon of the individual in (D). Black arrowhead, bulging inflorescence produced from the basal part of the macrocotyledon. BM, basal meristem; GM, groove meristem; inf, inflorescence; mc, microcotyledon; Mc, macrocotyledon. Bar = 50 µm in (A), 1 mm in (B, D), 100 µm in (C, E).
Isolation and Characterization of Orthologs of CYCB1
CYCB1, a marker of the G2/M phase of the cell cycle in A. thaliana and other model plants, is expressed scattered in tissue with actively dividing cells, such as leaf primordia or floral leaf primordia (Donnelly et al., 1999; Porceddu et al., 1999). We used this gene as the positive control for WMISH of M. glabra. We isolated cDNA of the CYCB1 orthologs Mg-CYCB1-1 and Mg-CYCB1-2 (Figure 2A), the nucleotide and amino acid sequences of which showed 98.3 and 98.8% similarity to each other, respectively. Both harbored a destruction box, a Cyclin_N domain, and a Cyclin_C domain (Figure 2B).
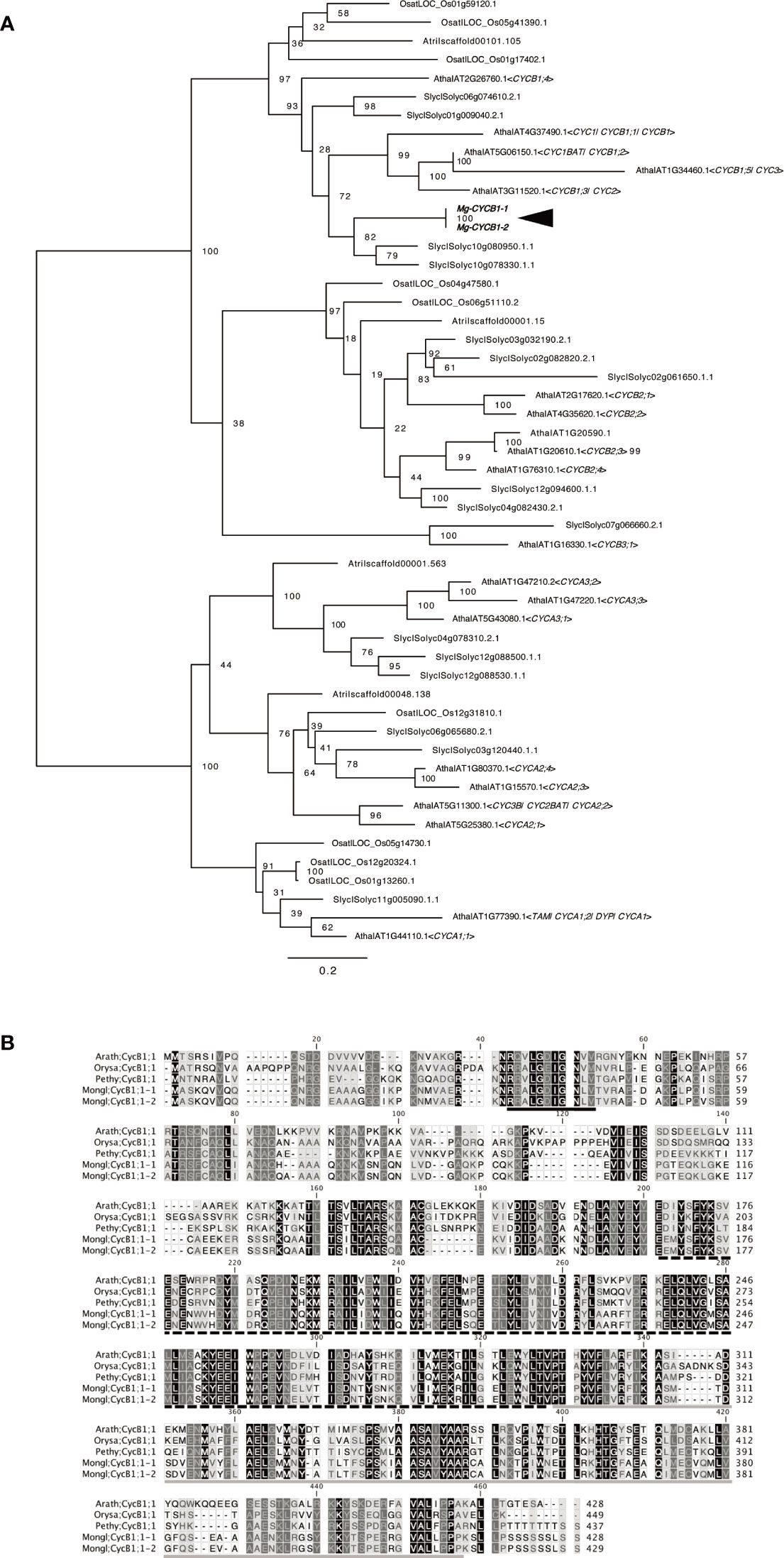
Figure 2 Molecular characterisation of Mg-CYCB1 (Mongl;CycB1;1-1, Mongl;CycB1;1-2). (A) Maximum likelihood tree of cyclin amino acid sequences. Bootstrap values are shown at the branches. Amaj, Antirrhinum majus; Atha, Arabidopsis thaliana; Atri, Amborella trichopoda; Osat, Oryza sativa; Slyc, Solanum lycopersicum; Zmay, Zea mays. (B) Amino acid sequence alignment of CYCB1 orthologs; dashes indicate gaps. The destruction box, cyclin C-terminal domain, and cyclin N-terminal domain are indicated by black, black hatched, and gray underlining, respectively. The darker background color indicates an amino acid conserved among the five sequences. Arath, Arabidopsis thaliana; Orysa, Orya sativa; Pethy, Petunia hybrida; Mongl, Monophyllaea glabra.
Optimization of WMISH Conditions Based on the Mg-CYCB1 Expression Pattern
At 15–20 days after sowing, the two cotyledons of M. glabra were of different sizes. Using individuals at this stage, we optimized the WMISH conditions based on Rozier et al. (2014). Because Tsukaya (1997) showed that BrdU is incorporated into the basal part of the macrocotyledon but not the microcotyledon, we regarded patchy Mg-CYCB1 signals in the basal part of the macrocotyledon but not in the microcotyledon as a success. It has been known that fixation and permeabilization are key factors for the success in in situ hybridization because both factors affect the accessibility of RNA probes and antibodies to target molecules inside cells (Engler et al., 1998; Fuentes and Fernández, 2014). In particular, in terms of the success in the WMISH of plants, permeabilization of cell wall is essential because unlike the classical sectioning in situ hybridization, cells are not cut open, so it is more difficult for probes and antibodies to reach the inside cells (Engler et al., 1998; Rozier et al., 2014). Considering above, we tested 4% PFA with 15% DMSO in PBST (Figures 3A, D), 4% PFA with 1.25% glutaraldehyde (GA) in PBST (Figures 3B, E), and 4% PFA with 1.25% GA and 15% DMSO (Figures 3C, F) as fixatives with cell wall enzymatic solution treatment (CWES) for 1 h (Figures 3A–C) and 3 h (Figures 3D–F). DMSO was added to the fixative intending to increase permeabilization (Brodelius and Nilsson, 1983). Fixation with 4% PFA with 15% DMSO in PBST and CWES for 1 h at room temperature was optimal for detecting Mg-CYCB1 in the basal part of the macrocotyledon (Figure 3A) with low background signal. In addition, CWES for 30 and 60 min yielded comparable results, so 30 min CWES was used in subsequent experiments. Whereas anisocotyledonous samples treated with the antisense or sense probe exhibited a pale purple background, patchy dark purple signals were observed only when the antisense probe was used (Figures 3G, H). In addition, the Mg-CYCB1 signal was denser in the BM than in the GM (Figure 3G). Mg-CYCB1 was expressed in both cotyledons at the isocotyledonous stage (Figures 3J–O), consistent with the report of Tsukaya (1997) that BrdU is incorporated into both cotyledons immediately after germination. Moreover, the signal was detected in the inner tissue of the tip of the petiolode (Figures 3J, K) from which the first root newly emerges as reported in Monphyllaea singularis by Imaichi et al. (2001) and in M. glabra by Ayano et al. (2005). Therefore, the signals should be from the proliferative cells which start to form the root primordia. We also performed in situ hybridization using individuals in the reproductive phase that had started to produce inflorescence meristems (Figure 3I). The inflorescence meristem exhibited more signals than the vegetative-phase GM, confirming the suitability of the WMISH condition.
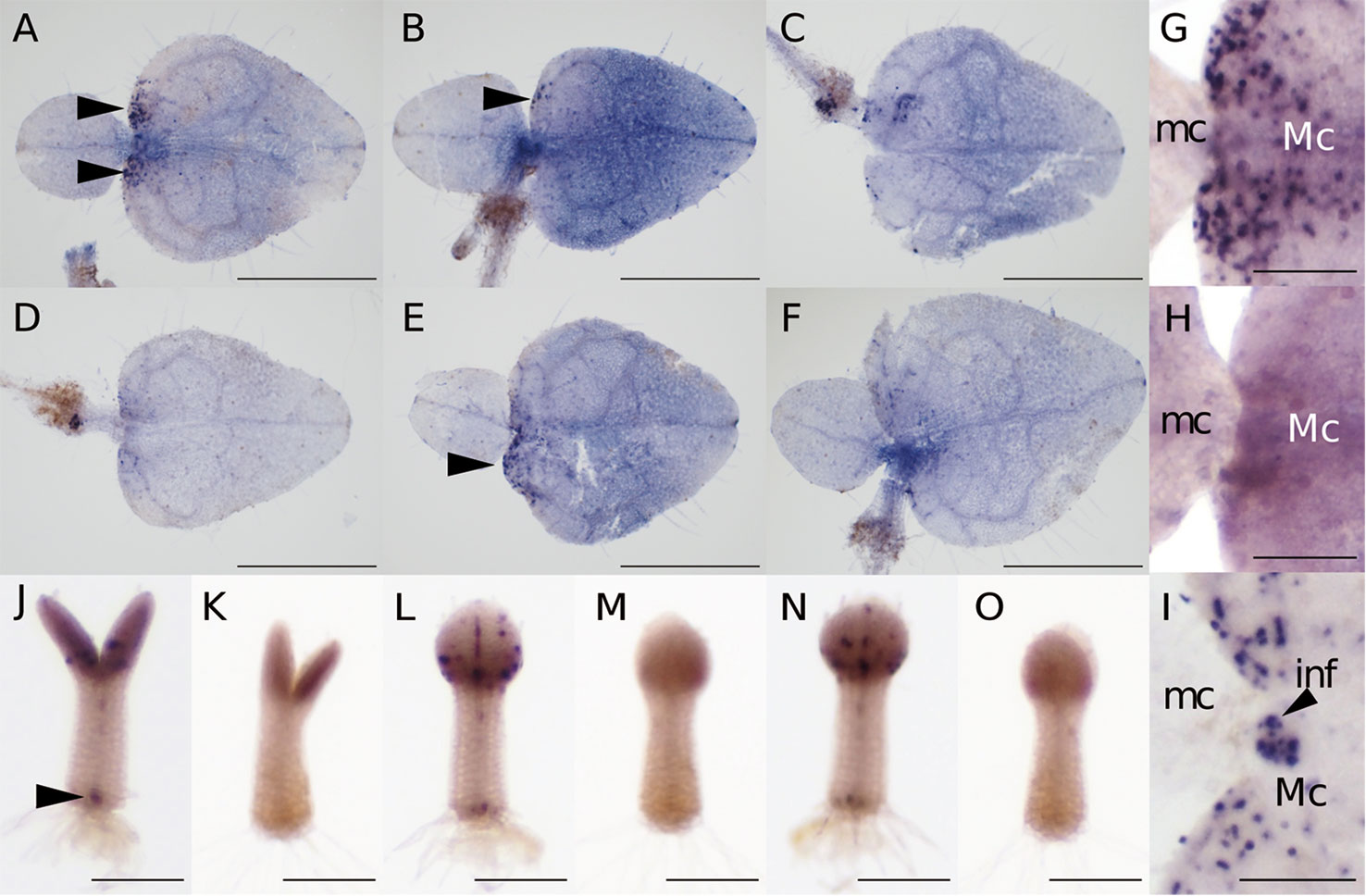
Figure 3 Mg-CYCB1 expression pattern and meristem positions. (A–F) WMISH of Mg-CYCB1 using an antisense probe with fixation in 4% PFA/15% DMSO (A, D), 4% PFA/1.25% glutaraldehyde (B, E), or 4% PFA/1.25% glutaraldehyde/15% DMSO in PBST, respectively. Cell wall enzyme treatment was performed for 1 h (A–C) or 3 h (D–F). Black arrowheads indicate signals in the lamina of macrocotyledons. (G–O) Mg-CYCB1 expression pattern at different developmental stages. (G, H) Proximal part of the macro- and microcotyledon in an anisocotyledonous individual (17 DAS). (G) Antisense probe. (H) Sense probe. (I) Proximal part of the macro- and microcotyledon at the reproductive stage (38 DAS). (J–O) Whole plants at 7 DAS with identical cotyledon size. (J, L, N) Antisense probe. The black arrowhead indicates signals in the distal part of the petiolode. (K, M, O) Sense probe. (J, K) Frontal view. (L–O) Side view. inf, inflorescence; mc, microcotyledon; Mc, macrocotyledon. Bar = 1 mm in (A–F), 200 µm in (G–O).
Expression of Mg-STM in M. glabra
Because the GM has a tunica-corpus structure similar to the SAM and the inflorescence meristem develops from it, the GM has been hypothesized to be a suppressed SAM that lacks the gene expression necessary for functioning as the SAM (Cronk and Möller, 1997; Tsukaya, 1997; Tsukaya, 2000). To investigate this, Ishikawa et al. (2017) isolated the GM and surrounding tissue by laser microdissection and found that Mg-STM expression was higher in the GM than in other tissue. However, they arbitrarily defined the border between the GM and BM meristem, so the spatial pattern of expression of Mg-STM was unclear. Therefore, we examined by WMISH the expression pattern of Mg-STM at the isocotyledonous and anisocotyledonous stages (7 and 18 DAS, respectively).
At the isocotyledonous stage, Mg-STM was expressed between the two cotyledons on the central axis, showing no apparent bias to left or right (Figures 4A–D). At the anisocotyledonous stage, Mg-STM was expressed only in the most basal part on the midrib of the macrocotyledon, the GM (Figures 4E–H).
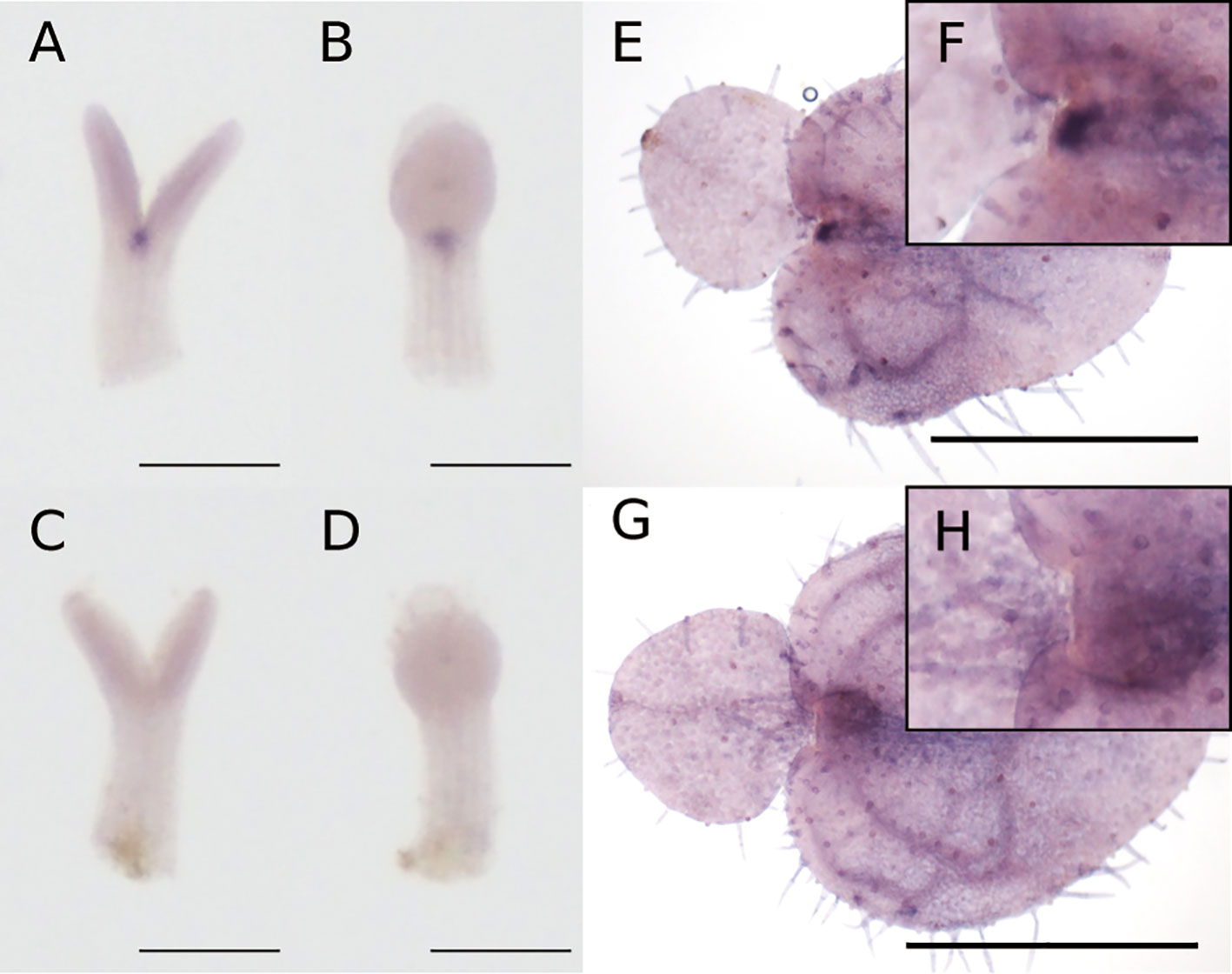
Figure 4 WMISH of Mg-STM-B at the isocotyledonous stage (7 DAS) (A–D) and anisocotyledonous stage (18 DAS) (E–H). (A, B, E, F) Antisense probe. (C, D, G, H) Sense probe. (A, C) Frontal view. (B, D) Side view. (F) Magnified image of the GM in (E). (H) Magnified image of the GM in (G). Bar = 200 µm in (A–D), 1 mm in (E, G).
Molecular Characterization and Expression Pattern of Mg-AN3
Next, we evaluated AN3 expression in M. glabra to assess the leaf-meristem-like nature of the BM. We isolated the cDNA of two AN3 orthologs, Mg-AN3-1 and Mg-AN3-2 (Figure 5A). The two had 97.4 and 99.5% sequence similarity at the nucleotide and amino acid levels, respectively. The only difference was residue 72, which was methionine or leucine. The putative AN3 protein of M. glabra possessed an SNH domain (Figure 5B), which is conserved among known AN3 orthologs and is necessary for interaction with GRF transcription factors (Kim and Kende, 2004; Horiguchi et al., 2005; Kim and Tsukaya, 2015).
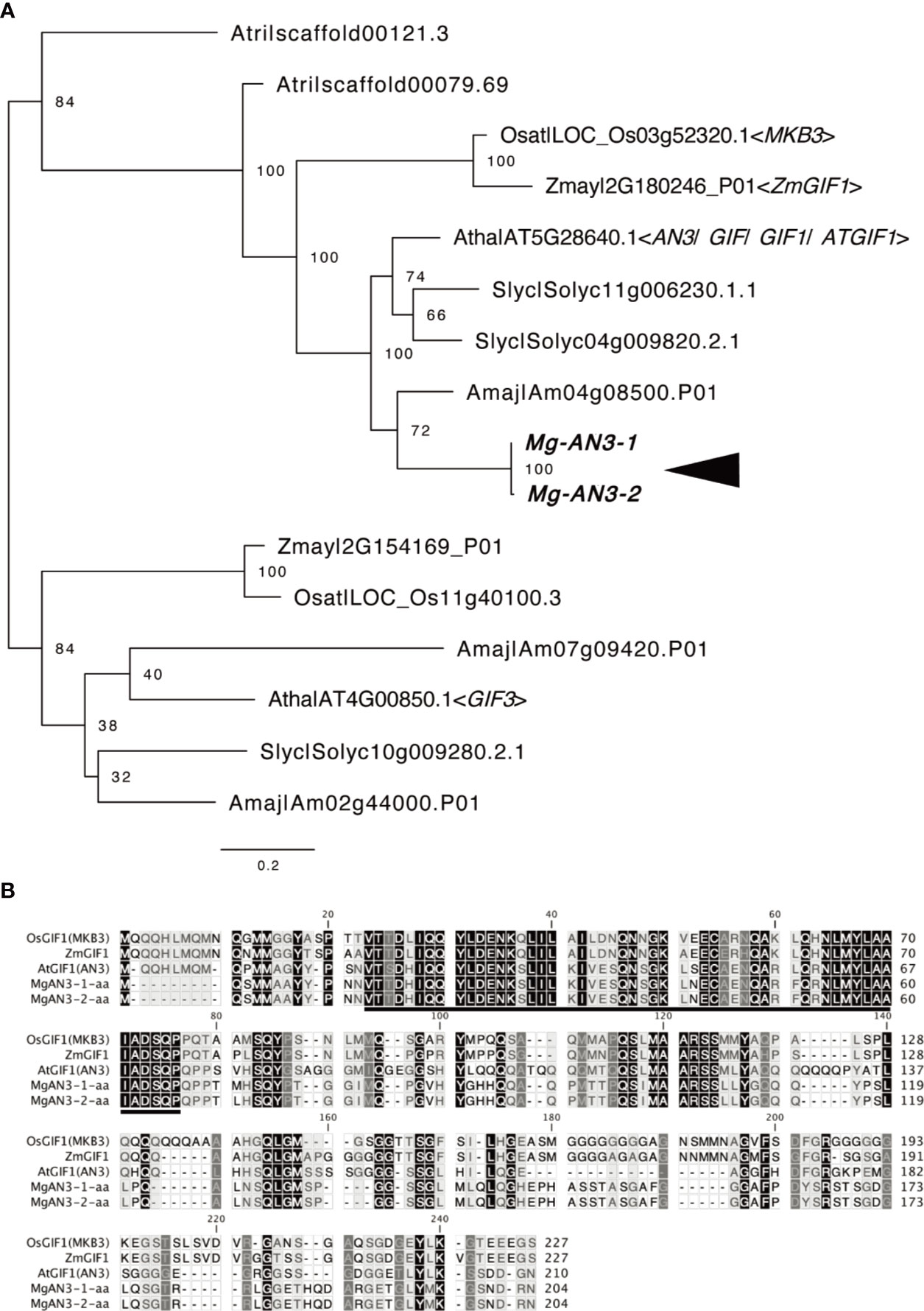
Figure 5 Molecular characterisation of Mg-AN3. (A) Partial ML phylogenetic tree of the GIF family. Bootstrap values are shown at the branches. Amaj, Antirrhinum majus; Atha, Arabidopsis thaliana; Atri, Amborella trichopoda; Osat, Oryza sativa; Slyc, Solanum lycopersicum; Zmay, Zea mays. (B) Amino acid sequence alignment of the AN3 homologs Mg-AN3-1, Mg-AN3-2, AtAN3 (Arabidopsis thaliana AT5G28640.1), ZmAN3 (Zea mays: Zhang et al., 2018), and OsAN3 (Oryza sativa: Shimano et al., 2018).
WMISH revealed that Mg-AN3 was expressed in the basal part of both cotyledons at the isocotyledonous stage (Figures 6A–D) and in the basal part of the macrocotyledon at the anisocotyledonous stage (Figures 6E–H) in both the GM and the BM.
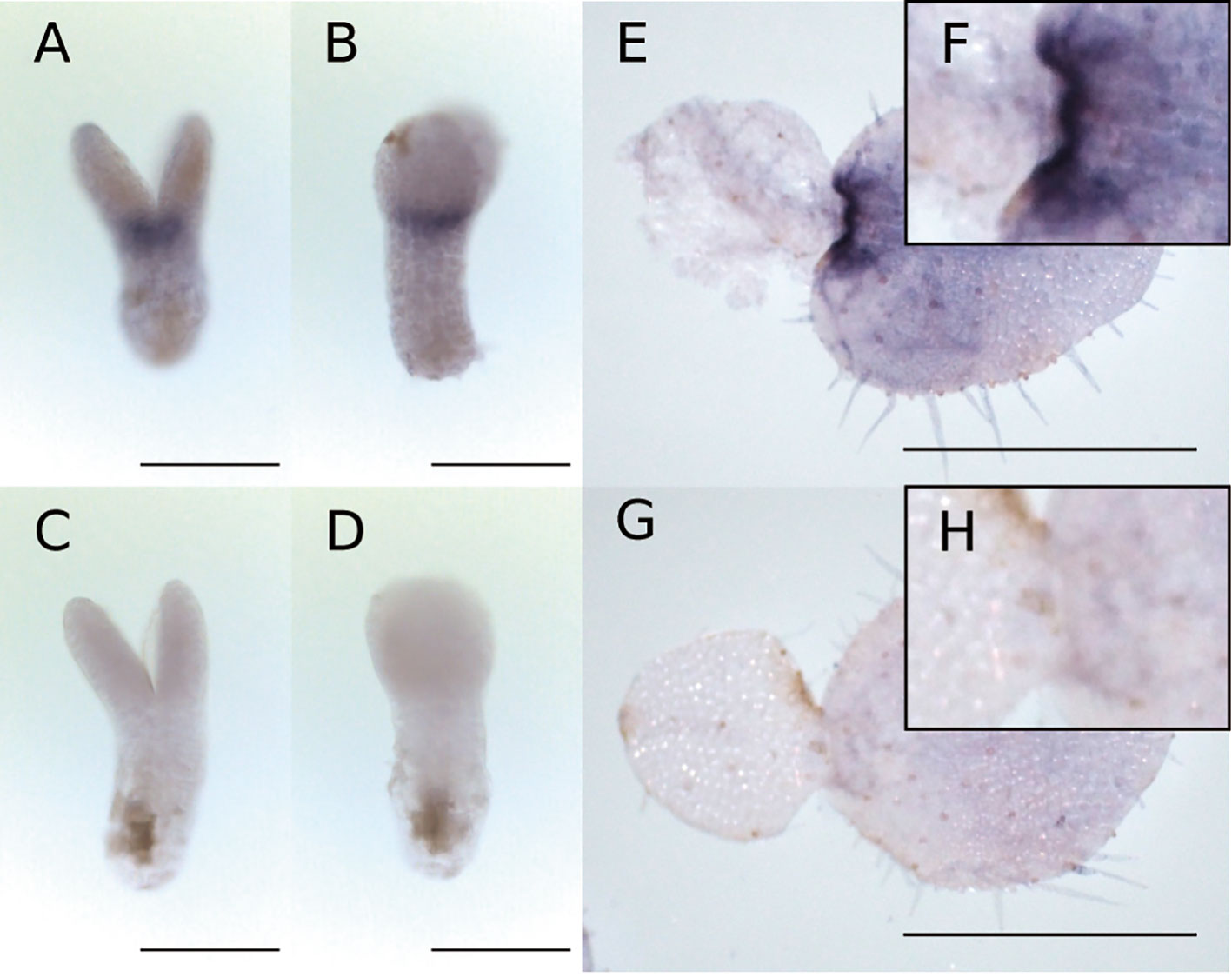
Figure 6 WMISH of Mg-AN3 at the at the isocotyledonous stage (8 DAS) (A–D) and anisocotyledonous stage (17 DAS) (E–H). (A, B, E, F) Antisense probe. (C, D, G, H) Sense probe. (A, C) Frontal view. (B, D) Side view. (F) Magnified image of the GM in (E). (H) Magnified image of the GM in (G). Bar = 200 µm in (A–D), 1 mm in (E, G).
Discussion
Position of the GM and the BM and Leaf Contour
The leaf contour changed in the proximal part of the macrocotyledon, the position of which was co-terminus with the border between two different kinds of tissue consisting of small cells: one in the proximal centerline of the macrocotyledon and the other laterally adjacent but toward the distal part. Based on this, we defined the former tissue as the GM and the latter tissue as the BM. The change in leaf contour between these meristems might result partly from their different cell division rates. As shown by WMISH of Mg-CYCB1 at the anisocotyledonous stage, the BM had more signal than the GM, which suggests greater cell division activity. This may explain the heart shape of the leaf (i.e., the protrusion of the lamina toward the proximal part, leaving behind the GM).
Establishment of a WMISH Technique for M. glabra
We established a WMISH technique for M. glabra, as confirmed by patchy Mg-CYCB1 expression in mitotic regions in cotyledons, the petiolode, and the inflorescence meristem at the isocotyledonous and anisocotyledonous stages. The technique is rapid because it does not require laborious embedding or sectioning. Moreover, spatial patterns of expression can be easily evaluated because the three-dimensional structure is retained. We could identify gene expression in the GM and/or the BM in a paradermal view, which is difficult with traditional in situ hybridization.
WMISH is used less frequently in plant research compared to animal research (Tautz and Pfeifle, 1989; Hemmati-Brivanlou et al., 1990; Herrmann, 1991) and has rarely been used in studies of the photosynthetic organ of plants other than A. thaliana (Althoff et al., 2014). Our WMISH technique will facilitate further studies of Monophyllaea and other non-model organisms.
Mg-STM Expression Pattern
STM is essential for the formation and maintenance of the SAM. The loss-of-function mutant stm lacks SAM; therefore, no new organ is formed after the cotyledons unfold. This phenotype is similar to that of one-leaf plants (Cronk and Möller, 1997; Tsukaya, 1997; Tsukaya, 2000). STM expression and other class I KNOX protein accumulation have been investigated in some phyllomorphs. Harrison et al. (2005) reported that in the one-leaf plant Streptocarpus dunni, KNOX I protein was detected in the GM during the reproductive phase but not the vegetative phase, whose result seems to be consistent with the hypothesis that SAM formation/maintenance system is lost/suppressed in the vegetative GM (Cronk and Möller, 1997; Tsukaya, 1997; Tsukaya, 2000). SdSTM1, an ortholog of STM, is not expressed in aboveground parts of S. dunni during the vegetative phase. The rosulate species Streptocarpus has repeating phyllomorphs (Jong, 1970; Jong and Burtt, 1975) because the GM can produce new phyllomorphs even in the vegetative phase (Nishii and Nagata, 2007). In a rosulate species S. rexii, SrSTM1 expression varies according to the stage of the GM and correlates with the production of new phyllomorphs (Mantegazza et al., 2007). Therefore, STM expression in the GM is correlated with additional organ formation in Streptocarpus.
In this study, Mg-STM expression was detected in the proximal part of the future midrib in M. glabra, consistent with Ishikawa et al. (2017), which suggests that no organ is formed in the GM irrespective of STM expression. Moreover, Mg-STM was expressed in the area between the two cotyledons at the isocotyledonous stage. This suggests that Mg-STM might be necessary for the activity of the GM before macrocotyledon differentiation and is involved in GM formation. Therefore, the molecular mechanism underlying GM formation and maintenance may differ between Streptocarpus and Monophyllaea. Alternatively, because the class I KNOX gene, KNAT1, is expressed in the GM at the no-organ-producing stage in S. rexii (Nishii et al., 2010), and KNAT1 is functionally similar to STM (Kim et al., 2003), it may replace STM in the flat-stage GM of Streptocarpus.
Regarding the indeterminacy of the BM, Cronk and Möller (1997) hypothesized that misexpression of SAM-organising genes may cause the indeterminate growth of the cotyledon because these genes maintain the meristematic state. In S. rexii, STM and KNAT1 orthologs are expressed in the BM. Ishikawa et al. (2017) reported that Mg-STM might be expressed in the BM of M. glabra, causing indeterminate growth. However, our findings suggest that this is unlikely because Mg-STM was not expressed in the BM according to our positional definition. Therefore, the mechanisms of phyllomorph growth likely differ between Streptocarpus and Monophyllaea.
Prolonged Cell Division Activity in Cotyledons Coincides With Mg-AN3 Expression
AN3, a transcription co-activator expressed in the basal part of leaf primordia, promotes the division of leaf meristem cells in A. thaliana, O. sativa, and Z. mays (Kim and Kende, 2004; Horiguchi et al., 2005; Shimano et al., 2018; Zhang et al., 2018). In isocotyledonous-stage M. glabra, cell division occurs in both cotyledons, as evidenced by the Mg-CYCB1 expression pattern. At this stage, AN3 was expressed in the basal part of each cotyledon. At the anisocotyledonous stage, Mg-CYCB1 was expressed in the basal part of macrocotyledon but not in the microcotyledon, which indicates that cell division is confined to this area. Mg-AN3 expression is also confined to the basal part of the area of cell division in the BM, as in leaf primordia of A. thaliana (Kawade et al., 2017). This suggests that the cell division activity of the BM is, at least in part, supported by Mg-AN3. The greater area of Mg-CYCB1 than Mg-AN3 expression could be caused by intercellular diffusion of Mg-AN3 proteins, as in A. thaliana (Kawade et al., 2017). Therefore, the BM is equivalent to the leaf meristem in terms of the AN3 and STM expression patterns.
Expression of Mg-AN3 in the GM
In A. thaliana and O. sativa, AN3 is not expressed in the SAM (Horiguchi et al., 2011; Shimano et al., 2018). In maize, AN3 is expressed from the bottom to the center of the SAM but not in the tip (Zhang et al., 2018). In this study, an AN3 ortholog in M. glabra was expressed not only in the BM but also in the GM, together with an STM ortholog. Such complete overlapping of STM with AN3 has not been reported in plants to date. It suggests that the GM has a leaf-meristem-like as well as a SAM-like nature, which may explain the fuzzy plant-body system of one-leaf plants. Moreover, Ishikawa et al. (2017) reported that Mg-AS1 and Mg-STM expression is not mutually exclusive, which suggests that Mg-STM and Mg-AS1 are co-expressed in the BM or GM of M. glabra. In the present study, we found that Mg-STM is not expressed in the BM in WMISH, but Ishikawa et al. (2017) reported that Mg-STM was expressed in tissue defined as the BM in their study. This suggests that tissue defined as the BM included a portion of the GM in their study. Nevertheless, Mg-AS1 expression was detected in the GM suggesting that both Mg-AS1 and Mg-STM are expressed in the GM. The genes that maintain SAM function are believed to repress genes that promote differentiation. For example, in model plants with simple leaves, the SAM-maintaining STM suppresses AS1 (Byrne et al., 2000; Byrne et al., 2002) to maintain an undifferentiated SAM. In addition, the SAM stem cell niche gene WUS represses genes that promote differentiation, such as KANADI1 (Yadav et al., 2013). Thus, the suppression of genes that promote differentiation by genes that maintain the SAM might be impaired in vegetative-stage Monophyllaea. This suppression might explain the expression of Mg-AN3 in the GM. In summary, Mg-AN3 expression in the GM suggests that it has a leaf-like, as well as a SAM-like, nature.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession numbers can be found in the main text.
Author Contributions
AK and HT designed the experiments. AK performed the experiments. AK and HK analyzed the data. AK and HT wrote the manuscript to which HK contributed.
Funding
This research was supported by a Grant-in-Aid for JSPS Fellows (AK, #19J14140), a Grand-in-Aid for Scientific Research on Innovation Areas (HT, #25113002 and 19H05672) from MEXT and the Graduate Program for Leaders in Life Innovation (GPLLI)/World-leading Innovative Graduate Study Program for Life Science and Technology (WINGS-LST) of the University of Tokyo (AK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Naoko Ishikawa for providing M. glabra seed stock and assistance with plant cultivation.
References
Althoff, F., Kopischke, S., Zobell, O., Ide, K., Ishizaki, K., Kohchi, T., et al. (2014). Comparison of the MpEF1α and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Res. 23, 235–244. doi: 10.1007/s11248-013-9746-z
Ayano, M., Imaichi, R., Kato, M. (2005). Developmental morphology of the Asian one-leaf plant, Monophyllaea glabra (Gesneriaceae) with emphasis on inflorescence morphology. J. Plant Res. 118, 99–109. doi: 10.1007/s10265-005-0195-5
Burtt, B. L. (1978). Studies in the Gesneriaceae of the Old World. XLV. A preliminary revision of Monophyllaea. Notes R. Bot. Gard. Edinburgh 37, 1–59.
Brodelius, P., Nilsson, K. (1983). Permeabilization of immobilized plant cells, resulting in release of intracellularly stored products with preserved cell viability. Eur. J. Appl. Microbiol. Biotechnol. 17, 275–280. doi:10.1007/BF00508020
Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. doi: 10.1038/35050091
Byrne, M. E., Simorowski, J., Martienssen, R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965.
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Crocker, C. W. (1860). Notes on the Germination of certain species of Cyrtandreae. J. Proc. Linn. Soc. London. Bot. 5, 65–67. doi: 10.1111/j.1095-8312.1860.tb01039.x
Cronk, Q., Möller, M. (1997). Strange morphogenesis - organ determination in Monophyllaea. Trends Plant Sci. 2, 327–328. doi: 10.1016/S1360-1385(97)84614-6
Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E., Dengler, N. G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. doi: 10.1006/dbio.1999.9443
Endrizzi, K., Moussian, B., Haecker, A., Levin, J. Z., Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 967–979. doi: 10.1046/j.1365-313X.1996.10060967.x
Engler, J. de A., Montagu, M. V., Engler, G. (1998). “Whole-Mount In Situ Hybridization in Plants”. in Arabidopsis Protocols. Methods in Molecular Biology. (Totowa, NJ: Humana Press), 373–384. doi: 10.1385/0-89603-391-0:373
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evol. (N. Y). 39, 783. doi: 10.2307/2408678
Fuentes, R., Fernández, J. (2014). “Fixation/permeabilization procedure for mRNAin situ hybridization of zebrafish whole-mount oocytes, embryos, andlarvae,” in In Situ Hybridization Protocol, ed. Nielsen, B.S. (New York: Humana Press), 1–13. doi: 10.1007/978-1-4939-1459-3_1
Glotzer, M., Murray, A. W., Kirschner, M. W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature doi: 10.1038/349132a0
Graham, L. E., Cook, M. E., Busse, J. S. (2000). The origin of plants: Body plan changes contributing to a major evolutionary radiation. Proc. Natl. Acad. Sci. U. S. A. 97, 4535–4540. doi: 10.1073/pnas.97.9.4535
Gray, A. (1879). Botanical text-book: Structural botany, or organography on the basis of morphology. To which is added the principles of taxonomy and phytography, and a glossary of botanical terms (New York and Chicago: Ivision, Blakeman & Company). doi: 10.5962/bhl.title.30210
Harrison, J., Möller, M., Langdale, J., Cronk, Q., Hudson, A. (2005). The role of KNOX genes in the evolution ofmorphological novelty in Streptocarpus. Plant Cell 17, 430–443. doi: 10.1105/tpc.104.028936
Hay, A., Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049
Hemmati-Brivanlou, A., Frank, D., Bolce, M. E., Brown, B. D., Sive, H. L., Harland, R. M. (1990). Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development 110, 325–330.
Hemerly, A., Bergounioux, C., Van Montagu, M., Inzé, D., Ferreira, P. (1992). Genes regulating the plant cell cycle: Isolationof a mitotic-like cyclin from Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 89, 3295–3299. doi:10.1073/pnas.89.8.3295
Herrmann, B. G. (1991). Expression pattern of the Brachyury gene in whole-mount T(Wis)/T(Wis) mutant embryos. Development 113, 913–917.
Hilliard, O. M., Burtt, B. L. (1971). Streptocarpus, an African plant study (Pietermaritzburg: University of Natal Press).
Horiguchi, G., Kim, G. T., Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Horiguchi, G., Nakayama, H., Ishikawa, N., Kubo, M., Demura, T., Fukuda, H., et al. (2011). ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol. 52, 112–124. doi: 10.1093/pcp/pcq178
Ichihashi, Y., Tsukaya, H. (2015). Behavior of leaf meristems and their modification. Front. Plant Sci. 6:1060. doi: 10.3389/fpls.2015.01060
Imaichi, R., Nagumo, S., Kato, M. (2000). Ontogenetic anatomy of Streptocarpus grandis (Gesneriaceae) with implications for evolution of monophylly. Ann. Bot. 86, 37–46. doi: 10.1006/anbo.2000.1155
Imaichi, R., Inokuchi, S., Kato, M. (2001). Developmental morphology of one-leaf plant Monophyllaea singularis (Gesneriaceae). Plant Syst. Evol. 229, 171–185. doi: 10.1007/s006060170010
Ishikawa, N., Takahashi, H., Nakazono, M., Tsukaya, H. (2017). Molecular bases for phyllomorph development in a one-leaf plant, Monophyllaea glabra. Am. J. Bot. 104, 233–240. doi: 10.3732/ajb.1600303
Jong, K., Burtt, B. L. (1975). The evolution of morphological novelty exemplified in the growth patterns of some Gesneriaceae. New Phytol. 75, 297–311. doi: 10.1111/j.1469-8137.1975.tb01400.x
Jong, K. (1970). Developmental aspects of vegetative morphology in Streptocarpus. [Ph. D dissertation] (The University of Edinburgh). Available at: http://hdl.handle.net/1842/12318.
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment softwareversion 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi:10.1093/molbev/mst010
Kawade, K., Horiguchi, G., Usami, T., Hirai, M. Y., Tsukaya, H. (2013). ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr. Biol. 23, 788–792. doi: 10.1016/j.cub.2013.03.044
Kawade, K., Tanimoto, H., Horiguchi, G., Tsukaya, H. (2017). Spatially different tissue-scale diffusivity shapes ANGUSTIFOLIA3 gradient in growing leaves. Biophys. J. 113, 1109–1120. doi: 10.1016/j.bpj.2017.06.072
Kazama, T., Ichihashi, Y., Murata, S., Tsukaya, H. (2010). The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 51, 1046–1054. doi: 10.1093/pcp/pcq051
Kim, J. H., Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Kim, J. H., Tsukaya, H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66, 6093–6107. doi: 10.1093/jxb/erv349
Kim, J. Y., Yuan, Z., Jackson, D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130, 4351–4362. doi: 10.1242/dev.00618
Kinoshita, A., Tsukaya, H. (2019). One-leaf plants in the Gesneriaceae: Natural mutants of the typical shoot system. Dev. Growth Differ. 61, 25–33. doi: 10.1111/dgd.12582
Kurihara, D., Mizuta, Y., Sato, Y., Higashiyama, T. (2015). ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179. doi: 10.1242/dev.127613
Laux, T., Mayer, K. F. X., Berger, J., Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Lee, B. H., Ko, J. H., Lee, S., Lee, Y., Pak, J. H., Kim, J. H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668. doi: 10.1104/pp.109.141838
Lenhard, M., Jürgens, G., Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206.
Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. doi: 10.1105/tpc.6.12.1859
Long, J. A., Barton, M. K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035.
Long, J. A., Moan, E., II, Medford, J., II, Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. doi: 10.1038/379066a0
Mantegazza, R., Möller, M., Jill Harrison, C., Fior, S., De Luca, C., Spada, A. (2007). Anisocotyly and meristem initiation in an unorthodox plant, Streptocarpus rexii (Gesneriaceae). Planta 225, 653–663. doi: 10.1007/s00425-006-0389-7
Mayer, K. F. X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/S0092-8674(00)81703-1
Möller, M., Pfosser, M., Jang, C.-G., Mayer, V., Clark, A., Hollingsworth, M. L., et al. (2009). A preliminary phylogeny of the “Didymocarpoid Gesneriaceae” based on three molecular data sets: Incongruence with available tribal classifications. Am. J. Bot. 96, 989–1010. doi: 10.3732/ajb.0800291
Nishii, K., Huang, B. H., Wang, C. N., Möller, M. (2017). From shoot to leaf: step-wise shifts in meristem and KNOX1 activity correlate with the evolution of a unifoliate body plan in Gesneriaceae. Dev. Genes Evol. 227, 41–60. doi: 10.1007/s00427-016-0568-x
Nishii, K., Nagata, T. (2007). Developmental analyses of the phyllomorphformation in the rosulate species Streptocarpus rexii (Gesneriaceae). Plant Syst. Evol. 265, 135–145. doi: 10.1007/s00606-007-0515-4
Nishii, K., Möller, M., Kidner, C., Spada, A., Mantegazza, M., Wang, C. N. (2010). A complex case of simple leaves: Indeterminateleaves co-express ARP and KNOX1 genes. Dev. Genes Evol. 220, 25–40. doi: 10.1007/s00427-010-0326-4
Porceddu, A., Reale, L., Lanfaloni, L., Moretti, C., Sorbolini, S., Tedeschini, E., et al. (1999). Cloning and expression analysis of a Petunia hybrida flower specific mitotic-like cyclin. FEBS Lett. 462,211–215. doi: 10.1016/S0014-5793(99)01484-2
Ridley, H. N. (1906). Note on the foliar organs of Monophyllaea. Ann. Bot. 20, 213–214. doi: 10.1093/oxfordjournals.aob.a089094
Rozier, F., Mirabet, V., Vernoux, T., Das, P. (2014). Analysis of 3D gene expression patterns in plants using whole-mount RNA in situ hybridization. Nat. Protoc. 9, 2464–2475. doi: 10.1038/nprot.2014.162
Rutishauser, R., Isler, B. (2001). Developmental genetics and morphologicalevolution of flowering plants, especially bladderworts (Utricularia): Fuzzy Arberian morphologycomplements classical morphology. Ann. Bot. 88, 1173–1202. doi: 10.1006/anbo.2001.1498
Rutishauser, R., Sattler, R. (1985). Complementarity and heuristic value of contrasting models in structural botany. Bot. Jahrbücher fur Syst. 107, 415–455.
Shimano, S., Hibara, K., II, Furuya, T., Arimura, S., II, Tsukaya, H., Itoh, J., II (2018). Conserved functional control, but distinct regulation, of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-INTERACTING FACTOR 1 orthologs. Development 145, dev159624. doi: 10.1242/dev.159624
Smith, J. F., Wolfram, J. C., Brown, K. D., Carroll, C. L., Denton, D. S. (1997). Tribal relationships in the Gesneriaceae: Evidence from DNA sequences of the chloroplast gene ndhF. Ann. Missouri Bot. Gard. 84, 50–66. doi: 10.2307/2399953
Smith, J. F. (1996). Tribal relationships within Gesneriaceae: A cladistic analysis of morphological data. Syst. Bot. 21, 497–513. doi: 10.2307/2419611
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steeves, T. A., Sussex, I. M. (1989). Patterns in Plant Development. 2nd ed. (New York: Cambridge University Press). doi: 10.1017/cbo9780511626227
Tautz, D., Pfeifle, C. (1989). A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85. doi: 10.1007/BF00291041
Tsukaya, H. (1997). Determination of the unequal fate of cotyledons of a one-leaf plant, Monophyllaea. Development 124, 1275–1280.
Tsukaya, H. (2000). The role of meristematic activities in the formation of leaf blades. J. Plant Res. 113, 119–126. doi: 10.1007/PL00013921
Weber, A., Clark, J., Möller, M. (2013). A new formal classification of Gesneriaceae. Selbyana 31, 68–94.
Yadav, R. K., Perales, M., Gruel, J., Ohno, C., Heisler, M., Girke, T., et al. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9, 654. doi: 10.1038/msb.2013.8
Keywords: ANGUSTIFOLIA3, class I KNOX, Gesneriaceae, indeterminate growth, leaf meristem, Monophyllaea, shoot apical meristem, whole-mount in situ hybridization
Citation: Kinoshita A, Koga H and Tsukaya H (2020) Expression Profiles of ANGUSTIFOLIA3 and SHOOT MERISTEMLESS, Key Genes for Meristematic Activity in a One-Leaf Plant Monophyllaea glabra, Revealed by Whole-Mount In Situ Hybridization. Front. Plant Sci. 11:1160. doi: 10.3389/fpls.2020.01160
Received: 12 April 2020; Accepted: 16 July 2020;
Published: 12 August 2020.
Edited by:
Hongzhi Kong, Chinese Academy of Sciences, ChinaReviewed by:
Chun-Neng Wang, National Taiwan University, TaiwanChuanen Zhou, Shandong University, China
Copyright © 2020 Kinoshita, Koga and Tsukaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirokazu Tsukaya, dHN1a2F5YUBicy5zLnUtdG9reW8uYWMuanA=
 Ayaka Kinoshita
Ayaka Kinoshita Hiroyuki Koga
Hiroyuki Koga Hirokazu Tsukaya
Hirokazu Tsukaya