- Department of Biological Production, Faculty of Bioresource Sciences, Akita Prefectural University, Akita, Japan
Iron (Fe) is an essential nutrient for all living organisms but can lead to cytotoxicity when present in excess. Fe toxicity often occurs in rice grown in submerged paddy fields with low pH, leading dramatical increases in ferrous ion concentration, disrupting cell homeostasis and impairing growth and yield. However, the underlying molecular mechanisms of Fe toxicity response and tolerance in plants are not well characterized yet. Microarray and genome-wide association analyses have shown that rice employs four defense systems to regulate Fe homeostasis under Fe excess. In defense 1, Fe excess tolerance is implemented by Fe exclusion as a result of suppression of genes involved in Fe uptake and translocation such as OsIRT1, OsYSL2, OsTOM1, OsYSL15, OsNRAMP1, OsNAS1, OsNAS2, OsNAAT1, OsDMAS1, and OsIRO2. The Fe-binding ubiquitin ligase, HRZ, is a key regulator that represses Fe uptake genes in response to Fe excess in rice. In defense 2, rice retains Fe in the root system rather than transporting it to shoots. In defense 3, rice compartmentalizes Fe in the shoot. In defense 2 and 3, the vacuolar Fe transporter OsVIT2, Fe storage protein ferritin, and the nicotinamine synthase OsNAS3 mediate the isolation or detoxification of excess Fe. In defense 4, rice detoxifies the ROS produced within the plant body in response to excess Fe. Some OsWRKY transcription factors, S-nitrosoglutathione-reductase variants, p450-family proteins, and OsNAC4, 5, and 6 are implicated in defense 4. These knowledge will facilitate the breeding of tolerant crops with increased productivity in low-pH, Fe-excess soils.
Introduction
Iron Acquisition by Rice
Iron (Fe) is a transition metal essential for the survival of virtually all living organisms. In plants, Fe participates in several important metabolic processes such as photosynthesis, chloroplast development, chlorophyll biosynthesis, electron transport, and redox reactions (Marschner, 1995). Graminaceous plants, including rice, acquire Fe from soil by chelation (strategy II) method (Römheld and Marschner, 1986). Rice synthesizes the Fe chelator 2′-deoxymugineic acid (DMA), a phytosiderophore of the mugineic acid (MA) family (Takagi, 1976), and secretes it into to the rhizosphere via the MA transporter (TOM1) (Nozoye et al., 2011). The secreted DMAs chelate and solubilize insoluble Fe (III) in the rhizosphere to form Fe (III)-DMA complexes. These Fe complexes are taken up into root cells by yellow stripe 1 (YS1) and yellow stripe-like (YSL) transporters, OsYSL15 (Curie et al., 2001; Inoue et al., 2009). Rice is typically grown in submerged paddy field conditions, where highly soluble ferrous ion (Fe2+) is abundant. Under such conditions, unlike other gramineous plants, rice directly takes up ferrous ions via iron-regulated transporter 1 and 2 (OsIRT1 and OsIRT2) (Ishimaru et al., 2006) together with phytosiderophores-based Fe uptake.
Iron Toxicity and its Damage to Rice Plants
The Fe2+ ions are abundant in paddy fields and absorption of them by rice roots causes severe Fe toxicity. The rhizosphere of acid sulfate soils with pH less than 5 contains the massive amount of Fe2+ about 10–2,000 mg kg−1 (da Silveira et al., 2007). The Fe2+ ions are highly soluble in water (Ksp = [Fe2+] [OH-]2 = 8 × 10-16) compared to ferric ions (Fe3+) (Ksp = [Fe3+] [OH-]3 = 1 × 10-36) at 25°C (Stumm and Lee, 1961). Thus, Fe3+ ions merely dissolve in water (~1 × 10-9 mol L-1 at pH 5) while Fe2+ ions can dissolve proficiently (~800 mol L-1 at pH 5). Therefore, it states that all Fe2+ ions which exist in soil can be soluble in low pH (pH 5 or below). Among soil types, mainly acid sulfate soil, acid clay soil, and peat soil cause Fe toxicity (Becker and Asch, 2005). Acid soils occupy approximately 3,950 million hectares or 30% of the total landmass, representing more than 50% of potentially arable land (von Uexküll and Mutert, 1995). Thus, Fe toxicity is one of the major determinants of crop yield and quality, particularly in China, India, and Southeast Asia (NRCS, 2005; Mahender et al., 2019), west and central Africa (Gridley et al., 2006), and Brazil (Crestani et al., 2009).
Iron overload in plants causes tissue damage and disrupts cellular homeostasis. The sequential effects of Fe toxicity in rice plants are shown in Figure 1. Ferrous toxicity inhibits cell division and elongation of the primary roots and subsequently the growth of lateral roots (Li et al., 2015). High Fe absorption by the roots and its transport into the leaves by xylem via the transpiration stream lead to cellular Fe overload in plant tissues (Briat and Lobréaux, 1997).
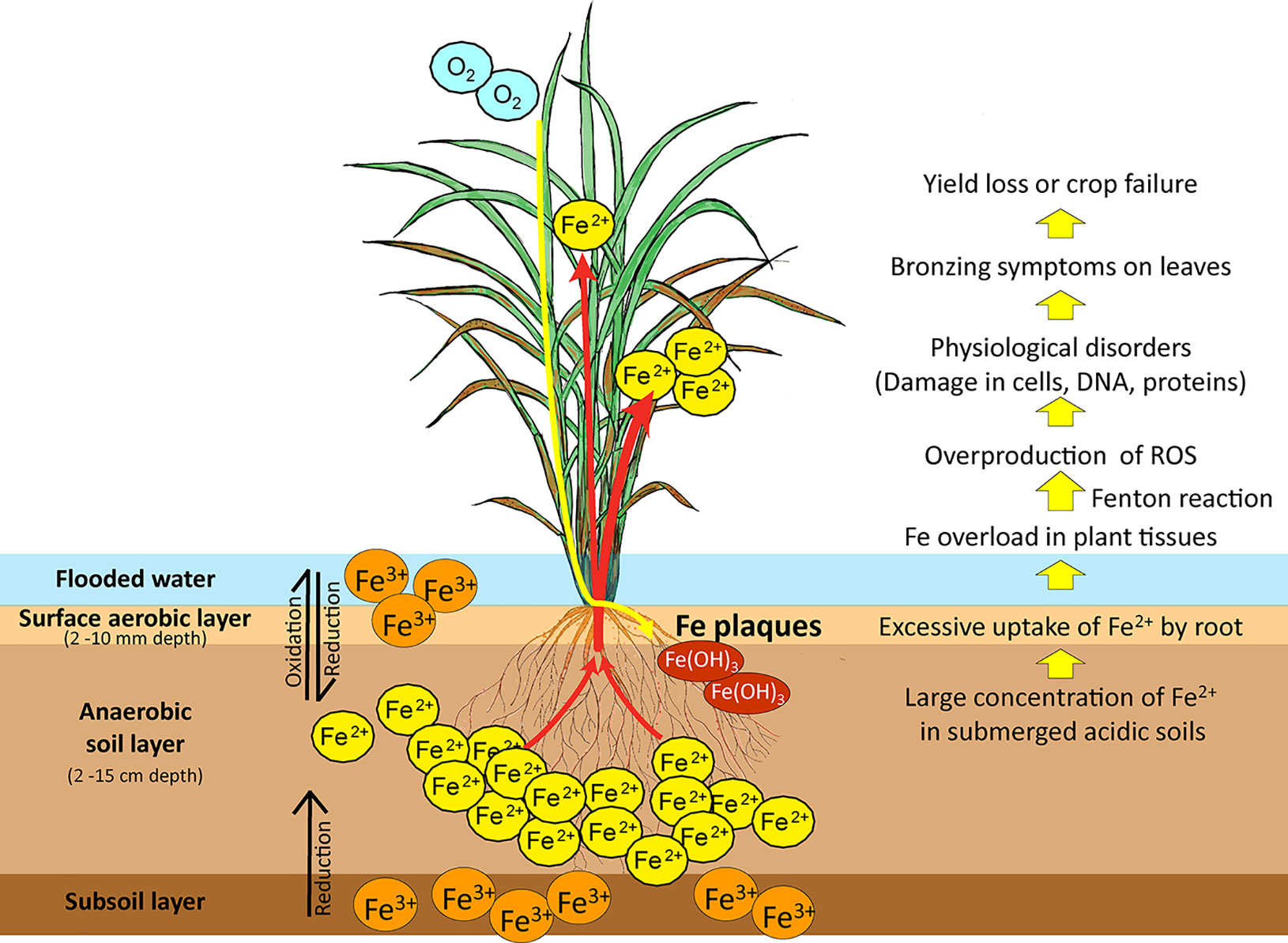
Figure 1 Iron reduction in submerged low-pH soils and the effects of Fe toxicity on rice plants. In submerged soils with anaerobic and low pH conditions, ferric ion (Fe3+) is reduced to the more soluble ferrous ion (Fe2+). Excess ferrous ion is absorbed by the roots and transported by the xylem to the leaves, causing Fe overload in plant tissues. Excessive Fe accumulation in plant tissues causes overproduction of ROS by the Fenton reaction. These damage cellular structures, membranes, DNA, and proteins and impair physiological processes. Fe toxicity causes bronzing symptoms in leaves, followed by loss of rice yield or complete crop failure. Fe3+, sparingly soluble ferric ion; Fe2+, soluble ferrous ion; ROS, reactive oxygen species. Red arrows: Fe uptake and transport by rice; Yellow arrow: Rhizospheric oxidation of Fe2+ to Fe3+ by oxygen transport from shoot to root, leading to formation of Fe plaques on the root surface.
Free Fe ions caused by Fe overload has a high affinity to bind with oxygen, leading to high oxygen tension and excess Fe accumulation in plant tissues. In plants, the Fe solubility varies in different cellular compartments and their pH is one of the influential factors. For examples, in typical xylem sap at pH 5.5 (Ariga et al., 2014), the solubility of Fe3+ is ~3 ×10-10 mol L-1 and that of Fe2+ is ~80 mol L-1; in the apoplast at pH 6 (Geilfus, 2017), the solubility of Fe3+ is ~1 × 10-12 mol L-1 and that of Fe2+ is ~8 mol L-1; in the cytosol at pH 8 (Martinière et al., 2013), the solubility of Fe3+ is 1 × 10-18 mol L-1 and that of Fe2+ is ~8 × 10-4 mol L-1. The Fe2+ ions can be oxidized to Fe3+ ions, and then those Fe3+ ions can readily be precipitated in most of the plant tissues where it becomes immobilized, less reactive, and thus less cellular toxicity. On the other hand, Fe2+ ions exist as a highly soluble and reactive form in the plant tissues with Fe overload and even numerous Fe2+ ions might exist as a free form in the cytosol if without any chelation or isolation, causing toxic reactions such as Fenton reaction. The Fenton reaction causes overproduction of reactive oxygen species (ROS), particularly the cytotoxic hydroxyl radical (•OH) (Becana et al., 1998; Thongbai and Goodman, 2000). Plant Fe toxicity disturbs photosystem II and elevates the cytochrome b6/f content of thylakoids, lowering the photosynthetic rate and accelerating oxygen production (Suh et al., 2002). Fe overload in chloroplasts results in oxidative damage caused by ROS (Balk and Schaedler, 2014). This leads to irreversible damage to cellular structures, membranes, DNA, and proteins (Briat et al., 1995; Stein et al., 2009a). Fe excess in cells impairs biological processes and leads to bronzing symptoms on leaves as a result of cell death. Leaf bronzing induced by Fe toxicity is closely associated with yield loss (Wu et al., 2014).
In this review, we summarize recent progress in the physiological and molecular mechanisms of Fe toxicity in rice, as well as its strategies to maintain Fe homeostasis.
Physiological and Molecular Defense Mechanisms Against Fe Toxicity
Rice responds to Fe toxicity by means of four defense mechanisms (Defense 1, 2, 3 and 4). A model of the mechanisms of tolerance to excess Fe is shown in Figure 2.
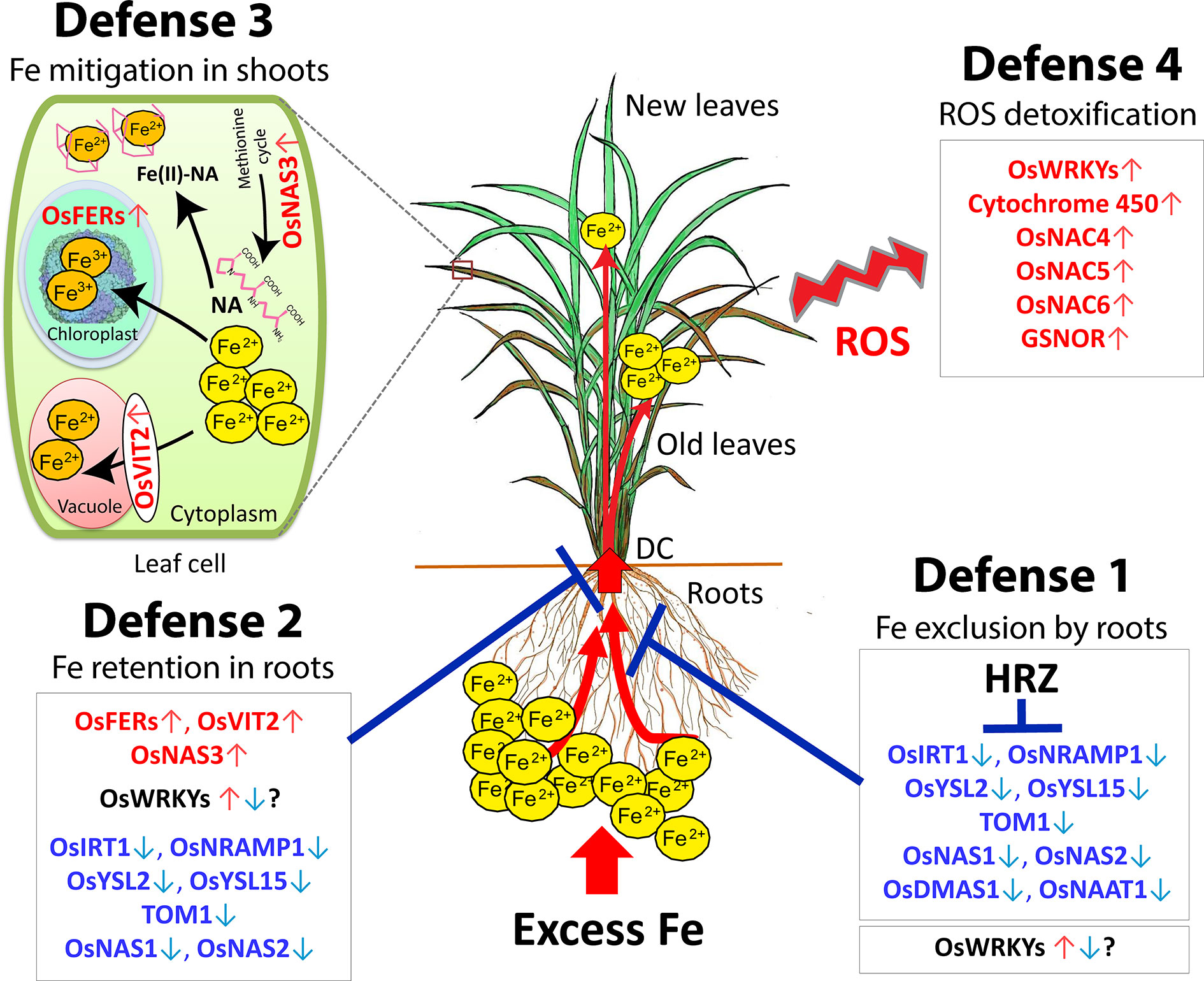
Figure 2 Hypothetical model of the four defense mechanisms of rice against excess Fe. Defense 1: Fe excess tolerance by Fe exclusion in the roots. Defense 2: Fe-excess-tolerance by Fe retention in root and avoidance of Fe translocation to shoot. Defense 3: Fe excess tolerance by Fe compartmentalization in the shoot. Defense 4: Fe excess tolerance by ROS detoxification in the plant. DC, Discrimination center; NA, nicotinamine. Red letters, highly induced genes; Blue letters, highly suppressed genes. This figure is modified from the Supplemental figure of Aung et al., 2018b. The ferritin image was provided by Dr. David S. Goodsell (Scripps Research Institute, La Jolla, CA) and the RCSB PDB.
Defense 1 (Fe Exclusion from Roots)
Under Fe excess condition, rice plants employ a defense mechanism of Fe exclusion, a root-based tolerant mechanism through inhibition of root Fe uptake. Rhizospheric oxidation of Fe2+ to Fe3+ by oxygen transport from shoots to roots leads to the formation of Fe plaques on the root surface due to Fe3+ precipitation, which acts as a barrier to the uptake of excess Fe2+ into root tissues (Becker and Asch, 2005). Tolerant genotype with Fe excluder may display the high formation of Fe plaque on the root surface (Mahender et al., 2019). Together with a larger diameter of the pith cavity of the shoots, the genotype with Fe excluder has high root oxidation power by an increase in aerenchyma volume and the number of lateral roots that favors the internal oxygen movement (Wu et al., 2014). Silicon supply mitigates the effect of Fe toxicity on rice by reducing Fe uptake (Ma and Takahashi, 1990). Also, its supply can strengthen the casparian bands in the root epidermis, which serves as a diffusion barrier and inhibits Fe flux into roots under excess Fe (Hinrichs et al., 2017; Becker et al., 2020). Therefore, rice genotypes with enhanced ability to exclude Fe such as high root oxidation power, large aerenchyma, several lateral roots, and strong casparian bands may be superior in Fe excess tolerance.
Microarray analyses have indicated that the expression of Fe uptake- and transport-related genes, such as OsIRT1, OsIRT2, OsYSL2, OsYSL15, and OsNRAMP1, is highly suppressed in roots in the presence of mild to high Fe excess levels (Aung et al., 2018b). high Fe excess level (Quinet et al., 2012), or very high Fe level (Finatto et al., 2015), suggesting that rice prevents Fe uptake and transport under Fe excess stress. Moreover, the expression of the genes involved in the biosynthesis of MAs, such as OsNAS1, OsNAS2, OsNAAT1, and OsDMAS1, is highly suppressed in roots, suggesting that plants restrain DMA release into the rhizosphere under excess Fe (Aung et al., 2018b). Therefore, rice utilizes defense 1 Fe exclusion to prevent Fe uptake by roots under Fe excess condition (Figure 2).
Role of HRZ in Fe Excess Tolerance (in Defense 1)
The HRZ, a Hemerythrin motif-containing Really Interesting New Gene- and Zinc-finger protein, is one of the key Fe homeostasis regulators. Kobayashi et al. (2013) identified two Fe-binding ubiquitin ligases, OsHRZ1 and OsHRZ2, which negatively regulate Fe deficiency response in rice. HRZ-knockdown rice lines are hypersensitive to Fe toxicity, demonstrating severe growth defects and leaf bronzing since even under mild Fe excess conditions (Aung et al., 2018a). In addition, disruption of HRZ causes 5- to 10-fold hyperaccumulation of Fe in leaves, and the expression of Fe uptake- and transport-related genes including OsIRT1, OsYSL2, OsYSL15, TOM1, OsNAS1, OsNAS2 and OsIRO2 is severely elevated in roots of knockdown rice subjected to Fe excess stress (Aung et al., 2018a). Therefore, HRZ is an important protein to protect plant cells from Fe toxicity and maintain Fe homeostasis in plants under Fe excess by repressing the genes involved in Fe uptake and translocation (Aung et al., 2018a, Figure 2).
Interestingly, HRZ expression is highly induced in response to Fe deficiency (Kobayashi et al., 2013). However, several Fe-uptake genes are strongly induced in that condition. These results suggest that Fe uptake mechanism by plant should be strictly regulated, and thus HRZ is ready to regulate like a break to Fe uptake genes since under Fe deficiency condition. The hemerythrin domains of HRZs, likewise its homolog BRUTUS (BTS) in Arabidopsis (Selote et al., 2015; Hindt et al., 2017), are predominantly bound by Fe and zinc (Zn) (Kobayashi et al., 2013), and direct binding of the Fe to the key regulators including HRZ or BTS, may be responsible for intracellular Fe-sensing and signaling events (Kobayashi, 2019). Thus, it is conceivable that HRZ may play a major role as an Fe-binding sensor in the earliest response system to excess Fe.
In addition to defense 1 in roots, HRZs might have additional roles in other tissues or organs under Fe excess condition. Further studies are desirable to clarify HRZ-mediated regulations in other defenses.
Defense 2 (Fe Retention in Roots and Suppression of Fe Translocation to Shoots)
Rice can retain Fe in the root tissues and decrease Fe translocation from roots to shoots (Tadano, 1975) to avoid excessive Fe accumulation in leaves and maintain homeostasis (Becker and Asch, 2005; da Silveira et al., 2007). In the Fe-toxic field experiment, the tolerant genotype (EPAGRI 108) retains a higher Fe concentration in root symplast (7 mg g−1 dry weight) compared to the sensitive genotype (BR IRGA 409 with 1.93 mg g−1 dry weight) (Stein et al., 2014). Moreover, the tolerant genotype precipitates approximately 94% of the Fe in its root apoplast during Fe excess. Thus, tolerant genotypes likely to cope with an elevated intracellular and apoplastic Fe concentrations in roots. Hence, protecting aboveground parts from excessive Fe overload is crucial in determining plants’ tolerance level.
In graminaceous plants, various metals absorbed by roots accumulate in the discrimination center (DC) in the basal part of the shoot, and the DC regulates mineral distribution to other plant parts (Mori, 1998). Protecting shoots, particularly the newest leaf, from Fe toxicity is important for the survival of rice plants, and the DC preferentially transports Fe to old leaves rather than new leaves (Aung et al., 2018b). Therefore, the DC may play an important role in Fe partitioning to aerial parts of plants and also act as a barrier to root-to-shoot transport of excessive Fe. Furthermore, Fe transport-related genes such as OsIRT1, OsYSL2, OsTOM1, OsNRAMP1, and OsYSL15 are highly suppressed in the DC, as in roots under Fe excess, suggesting that these genes likely distribute less Fe to shoots, instead retaining Fe in roots and the DC (Figure 2).
Defense 3 (Fe Compartmentalization in Shoots)
When defense 1 and defense 2 systems are insufficient, plants may allow Fe transport from the roots through DC for the subsequent avoidance of Fe toxicity by Fe compartmentalization, disposal, or storage inside shoots. In fact, some tolerant genotypes accumulate more Fe in aerial shoots, thus maintaining healthy shoot growth (Onaga et al., 2013). Indonesian rice genotypes, such as Siam Saba, Mahsuri, Margasari, and Pokkali, reportedly exhibit shoot-based tolerance, as evidenced by a high shoot Fe concentration, reduced leaf bronzing, and a high grain yield (Nugraha et al., 2016).
Notably, the Fe concentration in roots increased nine-fold in the presence of 20-fold Fe excess treatment and then reached to constant at higher Fe treatments (Aung et al., 2018b). By contrast, the concentration in shoots increased in proportion to the level of Fe excess. Therefore, rice has a capacity to retain a limited amount of Fe in root tissues. At Fe levels greater than that can be coped with by defense 2, rice translocates excess Fe from roots to shoots. Interestingly, the higher the Fe excess levels, the more Fe was preferentially translocated to old leaves, which showed bronzing earlier than new leaves. The citrate efflux transporter, OsFRDL1 is required for root-to-shoot Fe translocation and its expression in nodes is required for Fe distribution to rice grains (Yokosho et al., 2016). The expression of OsFRDL1 and its homolog OsFRDL2 is upregulated in roots and the DC at high Fe levels by microarray analyses (Aung et al., 2018b). Thus, OsFRDL1 and 2 may be important in dividing Fe in DC under Fe excess, presumably for distributing Fe to old leaves than new leaves. To prevent Fe excess-mediated damage to new leaves, rice plants may have a mechanism to partition excess Fe to old leaves to prevent damage to the newest leaves.
Role of Ferritin and OsVIT2 in Fe Excess Tolerance (in Defense 2 and 3)
The Fe2+ ions, once in the cells, can be associated into proteins or stored in cellular compartments such as ferritins and vacuoles. Ferritin is a ubiquitous Fe storage protein that stores up to 4,000 Fe atoms in a complex (Theil, 2003). Ferritin genes are transcriptionally induced in response to excess Fe in both monocot and dicot plants (Stein et al., 2009b; Briat et al., 2010). By microarray analyses, the rice ferritin genes, OsFER1 and OsFER2, are strongly upregulated in roots and shoots by excess ferrous treatments (Quinet et al., 2012; Finatto et al., 2015; Aung et al., 2018b). Therefore, excess Fe is isolated into ferritin in a safe and bioavailable form by OsFER1 and OsFER2 in response to Fe overload as a part of defense mechanism (Briat and Lobréaux, 1997; Silveira et al., 2009).
Vacuoles are important cellular organelle for Fe isolation to prevent cellular toxicity. The vacuolar Fe transporters, OsVIT1 and OsVIT2, are responsible for the sequestration of Fe into vacuoles via the tonoplast (Zhang et al., 2012). OsVIT1 is less responsive to Fe abundance (Zhang et al., 2012) and its expression is suppressed in all tissues mainly in roots (Aung et al., 2018b). On the other hand, OsVIT2 is highly responsive to Fe abundance (Zhang et al., 2012) and its expression is strongly upregulated in all tissues subjected to excess Fe (Quinet et al., 2012; Finatto et al., 2015; Aung et al., 2018b). Therefore, OsVIT2 might be important for the sequestration of excess Fe into vacuoles in a non-toxic form.
In defense 2 and 3, plants allow internal Fe distribution or Fe sequestration in roots and shoots. To detoxify excess Fe in other compartments or tissues, plants transform excess Fe into non-toxic form by accumulation in ferritin and/or disposal of it in a vacuole via OsVIT2, or by exclusion from the symplast and efflux into the apoplast (Figure 2).
Role of OsNAS3 in Fe Excess Tolerance (in Defense 2 and 3)
Nicotianamine (NA) is a major chelator of metal cations in higher plants. It is a chelate compound involved in Fe transport within the plant body. Among the three NA synthase genes in rice, OsNAS1 and OsNAS2 are involved in Fe deficiency response (Inoue et al., 2003). On the other hand, the expression of OsNAS3 is strongly induced in response to Fe excess in various tissues including root, DC, stem, and old and newest leaves (Aung et al., 2018b). Also, the expression of the methionine cycle-related genes is not completely suppressed in Fe excess roots and other tissues. Thus, it is thought that methionine cycle works under excess Fe as well in producing NA along with OsNAS3 (Figure 2).
Promoter-GUS (β-glucuronidase) analyses have revealed high-level OsNAS3 expression in the vascular bundle, epidermis, and exodermis of the root, stem, and old leaf under Fe excess (Aung et al., 2019). Moreover, disruption of OsNAS3 by T-DNA knockout increases sensitivity to excess Fe, reduces growth, causes severe leaf bronzing, and promote Fe accumulation in leaves. These findings provide the evidence that OsNAS3 is crucial to mitigate excess Fe. NA synthesized by OsNAS3 contributes to chelate excess Fe in roots and shoots, then it can be stored as a safe form since Fe chelated with NA does not cause the Fenton reaction. It may also protect the photosynthesis and water loss in leaves during Fe excess stress. NA involves in Fe-buffering to maintain Fe homeostasis under Fe excess (Pich et al., 2001) and plays a role in scavenging Fe and protecting the cells from oxidative damage (von Wirén et al., 1999). Therefore, it suggests NA synthesized by OsNAS3 chelates excess Fe and contributes efficient Fe translocation and sequestration, exhibiting its important role in mitigation of excess Fe in defense mechanisms 2 and 3, and in Fe excess tolerance in rice.
Defense 4 (ROS Detoxification)
The fourth defense mechanism is Fe inclusion with tolerance to cellular Fe overload and damage in leaves, which is mediated by enzymatic detoxification in the symplast (Becker and Asch, 2005; Stein et al., 2009a), scavenging of ROS by antioxidants (e.g., ascorbic acid and reduced glutathione) (Fang et al., 2001; Gallie, 2013), and protection from ROS damage by the action of antioxidant enzymes (e.g., superoxide dismutase, peroxidase, and catalase) (Bode et al., 1995; Fang and Kao, 2000). Cultivation with various Fe excess intensities and microarray analyses revealed that the molecular defense mechanism of the defenses 1, 2, and 3 works since mild to moderate Fe excess conditions that do not seriously affect rice growth yet. However, the genes contribute to defense 4 work in molecular level in much severe Fe excess condition, which causes severe leaf bronzing and remarkably reduces the plant growth (Aung et al., 2018b).
Under severe Fe excess, plants induce the expression of genes associated with oxidative stress, oxygen and electron transfer, and those encoding cytochrome P450 family proteins (Quinet et al., 2012; Aung et al., 2018b), or transcription factors related to cell death and stress responses (OsNAC4, OsNAC5, and OsNAC6) (Finatto et al., 2015; Aung et al., 2018b). to alleviate the damage caused by excess Fe-induced ROS and other abiotic stresses caused by Fe excess.
Li et al. (2019) reported that S-nitrosoglutathione-reductase (GSNOR) promotes root tolerance to Fe toxicity by inhibiting Fe-dependent nitrosative and oxidative cytotoxicity in plants via the nitric oxide pathway. In Arabidopsis, knockout mutants display a root growth defect in the presence of high Fe (250 µM), demonstrating that GSNOR is important for root tolerance to Fe toxicity. Then, WRKY transcription factors are important superfamilies and key regulators of several processes associated with numerous abiotic and biotic stress tolerance in plants. Most WRKYs genes are critical in regulation of various contrasting stresses simultaneously, and also involved in ROS defense mechanism to various stresses including Fe excess stress (Figure 2).
Possible Working Model of Four Defenses
Given the roles of each Fe excess defense mechanism, the hypothesis is outlined in the model shown in Figure 2. Whenever the plant receives the Fe excess stress, defense 1 (Fe exclusion) works to inhibit root Fe absorption by the suppression of Fe uptake genes. This defense might protect well the plant in conditions of weak Fe toxicity. When Fe excess is more sever, defense 1 may no longer prevent the roots from excessive Fe uptake because some metal transporters also absorb Fe at the same time in the process of absorbing other essential metals such as Zn, copper (Cu) or manganese (Mn). In such a condition, defense 2 (Fe retention in roots and suppression of Fe translocation to shoots) works by accumulating Fe in a non-toxic form in safety compartments of roots that leads an increase in root Fe concentration, and also prevents the root-to-shoot Fe translocation by the suppression of the related genes. It is likely that defense 1 and 2 mechanisms constitutively drive since prior to the onset of visual damage in leaves and throughout the stress from low to severe Fe excess levels.
When defense 2 is insufficient to sequester excess Fe in root tissues, the excess Fe is translocated from root to shoot. In this case, defense 3 (Fe compartmentalization in shoots) begins to employ the functions of chelation, isolation and/or sequestration of excess Fe in a safe form in the shoot that leads an increase in shoot Fe concentration. Lastly, when free divalent Fe cannot be completely sequestered in safety form under severe Fe conditions, it becomes largely accumulated in the cytosol and causes Fenton reaction that generates ROS stress. In this condition, the corresponding genes in defense 4 (ROS detoxification) are induced to mitigate ROS stress in plants. Overall, these four defense mechanisms might work cooperatively and consecutively for Fe detoxification in rice depending on the degree of Fe excess progression.
Future Prospects For Crops Tolerant To Excess Fe
Gene Response and Regulatory Mechanism to the Fe Toxicity in Plants
There has been progress in our understanding of Fe excess defense and tolerance mechanisms in plants. Fe deficiency response and its regulation network in plants is well characterized and widely understood (Kobayashi et al., 2019). However, the Fe excess-signaling factor, transcription factors and regulatory genes conserved in the molecular mechanism of the Fe toxicity response are not well identified yet. Some genes involved in defense system may start to work after receiving a direct Fe excess signal mediated by ROS. Also, HRZ-mediated regulation to Fe-homeostasis-related genes is crucial in Fe excess as described above. But the other potent regulators and the genes participated in the Fe-excess responsive pathway are still largely unknown.
Viana et al. (2017) reported that the cis-regulatory elements involved in responses to abiotic stresses, such as light and salicylic acid pathway, participate in the molecular signaling involved in the response to Fe excess. However, promoter analyses of the functional cis-acting elements for the core Fe excess-responsive in rice still needs to be identified. Some bHLH transcription factors have important roles for Fe homeostasis in plants. OsIRO3 (OsbHLH063) acts as a negative regulator of the Fe deficiency response in rice (Zheng et al., 2010) and also involves in shoots-to-roots signal transmission which is important to prevent Fe toxicity in plants (Wang et al., 2020). Moreover, some WRKY transcription factors may also involve in Fe homeostasis in plants. Ricachenevsky et al. (2010) firstly reported the Fe-excess induced transcription factor, OsWRKY80 in rice, which is also regulated by the dark-induced senescence- and drought- stress. In that study, OsWRKY80 transcript level was increased up to threefold in roots and shoots after 9 days of Fe excess. Furthermore, microarray analyses have yielded 19 upregulated WRKYs in Fe-excess rice (Finatto et al., 2015). Then, OsWRKY55-like, OsWRKY46, OsWRKY64, and OsWRKY113 are upregulated in an Fe-sensitive genotype, and OsWRKY55-like may be involved in the early stress response (Viana et al., 2017). Considering these results, some WRKY genes may also involve in defense mechanisms 1, 2 and 3 in addition to defense 4 (Figure 2). Therefore, transcription factors, such as some bHLHs and WRKYs, may involve in Fe excess response mechanism of the plants. Further comprehensive studies are required for the discovery of their specific roles in Fe excess. Therefore, exploration of Fe-excess responsive cis-acting elements, transcription factors, other potent regulators and candidate genes becomes a prerequisite for the deeper understanding of the mechanism of the excess Fe response and the development of rice varieties tolerant to Fe toxicity.
Developing Fe Excess Tolerant Rice via Molecular Breeding
Based on the tolerant rice lines selected by screening, the introduction of target genes into tolerant genotypes will increase further tolerance to Fe toxicity stress. As described above, the previous- and new-finding candidate genes encoding ferritin, VIT, NAS3, HRZ, GSNOR, and WRKYs may contribute to Fe excess tolerance in plants. In addition to these genes, there is a number of genes associated to the response to excess Fe; for example, overexpression of Arabidopsis Fe transporter, AtNramp1, leads to resistance to Fe toxicity (Curie et al., 2000). Use of these promising target genes and/or other candidate Fe homeostasis-related genes in molecular-breeding approaches, such as marker-assisted breeding, transformation, or genome editing methods, will facilitate the development of genotypes that are well adapted to Fe toxicity and contribute to increasing rice productivity in hotspot sites of soils with excess Fe. In addition, Fe excess-responsive genes have potential for addressing the global issue of Fe biofortification in grains (Masuda et al., 2020).
Conclusion
Understanding of the molecular mechanisms of Fe excess tolerance in rice is essential for varietal development. There is a progress in discovery of candidate genes involved in the physiological and molecular response of rice to Fe overload, and their roles in Fe homeostasis, Fe detoxification, and Fe excess tolerance are becoming clear. These genes could be promising targets of genetic resources for the development of rice genotypes with increased Fe toxicity tolerance by breeding and to increase rice productivity in regions prone to Fe toxicity to meet the food demand for increasing population.
Author Contributions
MSA and HM designed and wrote the manuscript. MSA led the writing of the manuscript. HM edited and improved the manuscript.
Funding
This publication was supported by a Grant-in-Aid for Young Scientists (B) (Grant No. 18K14367) from Japan Society for the Promotion of Sciences, JSPS KAKENHI (to MSA).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ariga, T., Hazama, K., Yanagisawa, S., Yoneyama, T. (2014). Chemical forms of iron in xylem sap from graminaceous and non-graminaceous plants. Soil Sci. Plant Nutr. 60, 460–469. doi: 10.1080/00380768.2014.922406
Aung, M. S., Kobayashi, T., Masuda, H., Nishizawa, N. K. (2018a). Rice HRZ ubiquitin ligases are crucial for response to excess iron. Physiol. Plant 163, 282–296. doi: 10.1111/ppl.12698
Aung, M. S., Masuda, H., Kobayashi, T., Nishizawa, N. K. (2018b). Physiological and transcriptomic analysis of responses to different levels of iron excess stress in various rice tissues. Soil Sci. Plant Nutr. 64, 370–385. doi: 10.1080/00380768.2018.1443754
Aung, M. S., Masuda, H., Nozoye, T., Kobayashi, T., Jeon, J.-S., An, G., et al. (2019). Nicotianamine synthesis by OsNAS3 is important for mitigating iron excess stress in rice. Front. Plant Sci. 10, 660. doi: 10.3389/fpls.2019.00660
Balk, J., Schaedler, T. A. (2014). Iron cofactor assembly in plants. Annu. Rev. Plant Biol. 65, 125–153. doi: 10.1146/annurev-arplant-050213-035759
Becana, M., Moran, J. F., Iturbe-Ormaetxe, I. (1998). Iron dependent oxygen free radical generation in plants subjected to environmental stress: toxicity and antioxidant protection. Plant Soil 201, 137–147. doi: 10.1023/A:1004375732137
Becker, M., Asch, F. (2005). Iron toxicity in rice - conditions and management concepts. J. Plant Nutr. Soil Sci. 168, 558–573. doi: 10.1002/(ISSN)1522-2624
Becker, M., Ngo, N. S., Schenk, M. K. A. (2020). Silicon reduces the iron uptake in rice and induces iron homeostasis related genes. Sci. Rep. 10, 5079. doi: 10.1038/s41598-020-61718-4
Bode, K., Döring, O., Lüthje, S., Neue, H. U., Böttger, M. (1995). The role of active oxygen in iron tolerance of rice (Oryza sativa L.). Protoplasma 184, 249–255. doi: 10.1007/BF01276928
Briat, J. F., Lobréaux, S. (1997). Iron transport and storage in plants. Trends Plant Sci. 2, 187–193. doi: 10.1016/S1360-1385(97)85225-9
Briat, J. F., Fobis-Loisy, I., Grignon, N., Lobréaux, S., Pascal, N., Savino, G., et al. (1995). Cellular and molecular aspects of iron metabolism in plants. Biol. Cell. 84, 69–81. doi: 10.1016/0248-4900(96)81320-7
Briat, J. F., Ravet, K., Arnaud, N., Duc, C., Boucherez, J., Touraine, B., et al. (2010). New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. 105, 811–822. doi: 10.1093/aob/mcp128
Crestani, M., da Silva, J. A. G., da Souza, V. Q., Hartwig, I., Luche, H. S., de Sousa, R. O., et al. (2009). Irrigated rice genotype performance under excess iron stress in hydroponic culture. Crop Breed. Appl. Biotechnol. 9, 87–95. doi: 10.12702/1984-7033.v09n01a12
Curie, C., Alonso, J. M., Le Jean, M., Ecker, J. R., Briat, J. F. (2000). Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem. J. 347, 749–755. doi: 10.1042/bj3470749
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 409, 346–349. doi: 10.1038/35053080
da Silveira, V. C., de Oliveira, A. P., Sperotto, R. A., Espindola, L. S., Amaral, L., Dias, J. F., et al. (2007). Influence of iron on mineral status of two rice (Oryza sativa L.) cultivars. Braz. J. Plant Physiol. 19, 127–139. doi: 10.1590/S1677-04202007000200005
Fang, W. C., Kao, C. H. (2000). Enhanced peroxidase activity in rice leaves in response to excess iron, copper and zinc. Plant Sci. 158, 71–76. doi: 10.1016/S0168-9452(00)00307-1
Fang, W. C., Wang, J. W., Lin, C. C., Kao, C. H. (2001). Iron induction of lipid peroxidation and effects on antioxidative enzyme activities in rice leaves. Plant Growth Regul. 35, 75–80. doi: 10.1023/A:1013879019368
Finatto, T., Oliveira, A. C., Chaparro, C., Maia, L. C., Farias, D. R., Woyann, L. G., et al. (2015). Abiotic stress and genome dynamics: specific genes and transposable elements response to iron in rice. Rice 8, 13. doi: 10.1186/s12284-015-0045-6
Gallie, D. R. (2013). The role of l-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 64, 433–443. doi: 10.1093/jxb/ers330
Geilfus, C. M. (2017). The pH of the Apoplast: Dynamic Factor with Functional Impact Under Stress. Mol. Plant. 10, 1371–1386. doi: 10.1016/j.molp.2017.09.018
Gridley, H. E., Efisue, A., Tolou, B., Bakayako, T. (2006). Breeding for Tolerance to Iron Toxicity at WARDAl (Cotonou, Benin: Africa Rice Center (WARDA)), 96–111.
Hindt, M. N., Akmakjian, G. Z., Pivarski, K. L., Punshon, T., Baxter, I., Salt, D. E., et al. (2017). BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9, 876–890. doi: 10.1039/c7mt00152e
Hinrichs, M., Fleck, A. T., Biedermann, E., Ngo, N. S., Schreiber, L., Schenk, M. K. (2017). An ABC Transporter Is Involved in the Silicon-Induced Formation of Casparian Bands in the Exodermis of Rice. Front. Plant Sci. 8, 671. doi: 10.3389/fpls.2017.00671
Inoue, H., Higuchi, K., Takahashi, M., Nakanishi, H., Mori, S., Nishizawa, N. K. (2003). Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 36, 366–381. doi: 10.1046/j.1365-313X.2003.01878.x
Inoue, H., Kobayashi, T., Nozoye, T., Takahashi, M., Kakei, Y., Suzuki, K., et al. (2009). Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284, 3470–3479. doi: 10.1074/jbc.M806042200
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+- phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Kobayashi, T., Nagasaka, S., Senoura, T., Itai, R. N., Nakanishi, H., Nishizawa, N. K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4, 2792. doi: 10.1038/ncomms3792
Kobayashi, T. (2019). Understanding the Complexity of Iron Sensing and Signaling Cascades in Plants. Plant and Cell Physiol. 60, 1440–1446. doi: 10.1093/pcp/pcz038
Kobayashi, T., Nozoye, T., Nishizawa, N. K. (2019). Iron transport and its regulation in plant. Free Radic. Biol. Med. 133, 11–20. doi: 10.1016/j.freeradbiomed.2018.10.439
Li, G., Song, H., Li, B., Kronzucker, H. J., Shi, W. (2015). Auxin resistant1 and PIN-FORMED2 protect lateral root formation in Arabidopsis under iron stress. Plant Physiol. 169, 2608–2623. doi: 10.1104/pp.15.00904
Li, B., Sun, L., Huang, J., Gösch, C., Shi, W., Chory, J., et al. (2019). GSNOR provides plant tolerance to iron toxicity via preventing iron-dependent nitrosative and oxidative cytotoxicity. Nat. Commun. 10, 3896. doi: 10.1038/s41467-019-11892-5
Ma, J. F., Takahashi, E. (1990). Effect of silicon on the growth and phosphorus uptake of rice. Plant Soil 126, 115–119. doi: 10.1007/BF00041376
Mahender, A., Swamy, B. P. M., Anandan, A., Ali, J. (2019). Tolerance of iron-deficient and -toxic soil conditions in rice. Plants 8, 31. doi: 10.3390/plants8020031
Martinière, A., Desbrosses, G., Sentenac, H., Paris, N. (2013). Development and properties of genetically encoded pH sensors in plants. Front. Plant Sci. 4, 523. doi: 10.3389/fpls.2013.00523
Masuda, H., Aung, M. S., Kobayashi, T., Nishizawa, N. K. (2020). “Iron Biofortification: The gateway to overcoming hidden hunger,” in The Future of Rice Demand: Quality Beyond Productivity. Eds. de Oliveira, A. C., Pegoraro, C., Viana, V. E. (Switzerland: Springer Nature), 149–177. doi: 10.1007/978-3-030-37510-2
Mori, S. (1998). “Iron transport in graminaceous plants,” in Iron Transport and Storage in Microorganisms, Plants and Animals. Vol. 35, Metal Ions in Biological Systems. Eds. Sigel, A., Sigel, H. (New York: Marcel Dekker), 215 – 237.
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Uozume, N., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
NRCS (2005). Global soil regions map, National Resources Conservation Services (NRCS) (Washington, DC: US Department of Agriculture (USDA), Soil Conservation Service). https://www.nrcs.usda.gov/wps/portal/nrcs/detail/soils/use/?cid=nrcs142p2_054013.
Nugraha, Y., Utami, D. W., Rosdianti, I. D. A., Ardie, S. W., Ghulammahdi, M., Suwarno, S., et al. (2016). Markers-traits association for iron toxicity tolerance in selected Indonesian rice varieties. Biodivers. 17, 753–763. doi: 10.13057/biodiv/d170251
Onaga, G., Edema, R., Asea, G. (2013). Tolerance of rice germplasm to iron toxicity stress and the relationship between tolerance, Fe2+, P and K content in the leaves and roots. Arch. Agron. Soil Sci. 59, 213–229. doi: 10.1080/03650340.2011.622751
Pich, A., Manteuffel, R., Hillmer, S., Scholz, G., Schmidt, W. (2001). Fe homeostasis in plant cells: does nicotianamine play multiple roles in the regulation of cytoplasmic Fe concentration? Planta 213, 967–976. doi: 10.1007/s004250100573
Quinet, M., Vromman, D., Clippe, A., Bertin, P., Lequeux, H., Dufey, I., et al. (2012). Combined transcriptomic and physiological approaches reveal strong differences between short- and long-term response of rice (Oryza sativa) to iron toxicity. Plant Cell Env. 35, 1837–1859. doi: 10.1111/j.1365-3040.2012.02521.x
Römheld, V., Marschner, H. (1986). Evidence for a specific uptake system for iron phytosiderophore in roots of grasses. Plant Physiol. 80, 175–180. doi: 10.1104/pp.80.1.175
Ricachenevsky, F. K., Sperotto, R. A., Menguer, P. K., Fett, J. P. (2010). Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol. Biol. Rep. 37, 3735–3745. doi: 10.1007/s11033-010-0027-0
Selote, D., Samira, R., Matthiadis, A., Gillikin, J. W., Long, T. A. (2015). Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 167, 273–286. doi: 10.1104/pp.114.250837
Silveira, V. C., Fadanelli, C., Sperotto, R. A., Stein, R. J., Basso, L. A., Vaz Junior, I. S., et al. (2009). Role of ferritin in the rice tolerance to iron overload. Sci. Agric. 66, 549–555. doi: 10.1590/S0103-90162009000400019
Stein, R. J., Duarte, G. L., Spohr, M. G., Lopes, S., II, Fett, J. P. (2009a). Distinct physiological responses of two rice cultivars subjected to iron toxicity under field conditions. Ann. Appl. Biol. 154, 269–277. doi: 10.1111/j.17447348.2008.00293.x
Stein, R. J., Ricachenevsky, F. K., Fett, J. P. (2009b). Differential regulation of the two rice ferritin genes (OsFER1 and OsFER2). Plant Sci. 177, 563–569. doi: 10.1016/j.plantsci.2009.08.001
Stein, R. J., Lopes, S., II, Fett, J. P. (2014). Iron toxicity in field-cultivated rice: contrasting tolerance mechanisms in distinct cultivars. Theor. Exp. Plant Physiol. 26, 135–146. doi: 10.1007/s40626-014-0013-3
Stumm, W., Lee, G. F. (1961). Oxygenation of ferrous iron. Ind. Eng. Chem. 53, 143–146. doi: 10.1021/ie50614a030
Suh, H. J., Kim, C. S., Lee, J. Y., Jung, J. (2002). Photodynamic effect of iron excess on photosystem II function in pea plants. Photochem. Photobiol. 75, 513–518. doi: 10.1562/0031-8655(2002)075<0513:PEOIEO>2.0.CO;2
Tadano, T. (1975). Devices of rice roots to tolerant high iron concentrations in growth media. Japan Agric. Res. Q. 9, 34 –39.
Takagi, S. (1976). Naturally occurring iron-chelating compounds in oat- and rice root washing. I: activity measurement and preliminary characterization. Soil Sci. Plant Nutr. 22, 423–433. doi: 10.1080/00380768.1976.10433004
Theil, E. C. (2003). Ferritin: at the crossroads of iron and oxygen metabolism. J. Nutr. 133, 1549–1553. doi: 10.1093/jn/133.5.1549S
Thongbai, P., Goodman, B. A. (2000). Free radical generation and post anoxic injury in an iron toxic soil. J. Plant Physiol. 23, 1887–1990. doi: 10.1080/01904160009382151
Viana, V. E., Marini, N., Finatto, T., Ezquer, I., Busanello, C., Dos Santos, R. S., et al. (2017). Iron excess in rice: From phenotypic changes to functional genomics of WRKY transcription factors. Genet. Mol. Res. 16 (3):gmr16039694. doi: 10.4238/gmr16039694
von Uexküll, H. R., Mutert, E. (1995). “Global extent, development and economic impact of acid soils,” in Plant–Soil Interactions at Low pH: Principles and Management. Eds. Date, R. A., Grundon, N. J., Raymet, G. E., Probert, M. E. (Dordrecht, The Netherlands: Kluwer Academic Publishers), 5–19.
von Wirén, N., Klair, S., Bansal, S., Briat, J. F., Khodr, H., Shioiri, T., et al. (1999). Nicotianamine chelates both Fe-III and Fe-II. Implications for metal transport in plants. Plant Physiol. 119, 1107–1114. doi: 10.1104/pp.119.3.1107
Wang, F., Itai, R. N., Nozoye, T., Kobayashi, T., Nishizawa, N. K., Nakanishi, N. (2020). The bHLH protein OsIRO3 is critical for plant survival and iron (Fe) homeostasis in rice (Oryza sativa L.) under Fe-deficient conditions. Soil Sci. Plant Nutr. doi: 10.1080/00380768.2020.1783966
Wu, L., Shhadi, M. Y., Gregorio, G., Matthus, E., Becker, M., Frei, M. (2014). Genetic and physiological analysis of tolerance to acute iron toxicity in rice. Rice 7, 8. doi: 10.1186/s12284-014-0008-3
Yokosho, K., Yamaji, N., Ma, J. F. (2016). OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. J. Exp. Bot. 67, 5485–5494. doi: 10.1093/jxb/erw314
Zhang, Y., Xu, Y. H., Yi, H. Y., Gong, J. M. (2012). Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 72, 400–410. doi: 10.1111/j.1365-313X.2012.05088.x
Keywords: iron excess, rice, OsNAS3, HRZ, OsVIT2, ROS, iron homeostasis, tolerant mechanism
Citation: Aung MS and Masuda H (2020) How Does Rice Defend Against Excess Iron?: Physiological and Molecular Mechanisms. Front. Plant Sci. 11:1102. doi: 10.3389/fpls.2020.01102
Received: 27 March 2020; Accepted: 03 July 2020;
Published: 07 August 2020.
Edited by:
Louis Grillet, Academia Sinica, TaiwanReviewed by:
Karl Ravet, Colorado State University, United StatesPing Lan, Institute of Soil Science (CAS), China
Hannetz Roschzttardtz, Pontificia Universidad Católica de chile, Chile
Copyright © 2020 Aung and Masuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: May Sann Aung, bWF5YXVuZ0Bha2l0YS1wdS5hYy5qcA==
 May Sann Aung
May Sann Aung Hiroshi Masuda
Hiroshi Masuda