
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 16 July 2020
Sec. Aquatic Photosynthetic Organisms
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01033
This article is part of the Research Topic Metabolic Regulation of Diatoms and other Chromalveolates View all 12 articles
Diatoms belong to a major, diverse and species-rich eukaryotic clade, the Heterokonta, within the polyphyletic chromalveolates. They evolved as a result of secondary endosymbiosis with one or more Plantae ancestors, but their precise evolutionary history is enigmatic. Nevertheless, this has conferred them with unique structural and biochemical properties that have allowed them to flourish in a wide range of different environments and cope with highly variable conditions. We review the effect of pH, light and dark, and CO2 concentration on the regulation of carbon uptake and assimilation. We discuss the regulation of the Calvin-Benson-Bassham cycle, glycolysis, lipid synthesis, and carbohydrate synthesis at the level of gene transcripts (transcriptomics), proteins (proteomics) and enzyme activity. In contrast to Viridiplantae where redox regulation of metabolic enzymes is important, it appears to be less common in diatoms, based on the current evidence, but regulation at the transcriptional level seems to be widespread. The role of post-translational modifications such as phosphorylation, glutathionylation, etc., and of protein-protein interactions, has been overlooked and should be investigated further. Diatoms and other chromalveolates are understudied compared to the Viridiplantae, especially given their ecological importance, but we believe that the ever-growing number of sequenced genomes combined with proteomics, metabolomics, enzyme measurements, and the application of novel techniques will provide a better understanding of how this important group of algae maintain their productivity under changing conditions.
The chromalveolates are a polyphyletic eukaryote supergroup that includes many photosynthetic lineages including the cryptomonads, dinoflagellates, haptophytes, and heterokonts (also called stramenopiles) (Keeling, 2009). The phylogeny of diatoms and their allied groups is complicated (Dorrell et al., 2017; Falciatore et al., 2020) and while the chromalveolates are not now regarded as a natural group we have retained the name here as a convenient and widely-used term. It has been estimated that over 50% of all formally described protists are chromalveolates (Cavalier-Smith, 2004; Cavalier-Smith and Chao, 2006). Within the diverse clade Heterokonta, diatoms (Bacillariophyceae) are photosynthetic microalgae that comprise between 30,000 and 100,000 species (Mann and Vanormelingen, 2013). They evolved about 250 Myrs ago (Medlin, 2016), are found today in all aquatic environments, and contribute about 20% to global primary production (Falkowski et al., 1998). Like other heterokonts, diatoms originated via serial endosymbioses (Stiller et al., 2014) and their chloroplasts derive from a red and a green algal endosymbiosis and also contain genes from prokaryotes, their eukaryotic host, and genes acquired by horizontal transfer (Moustafa et al., 2009; Deschamps and Moreira, 2012; Dorrell et al., 2017). Consequently, diatom genomes are enriched in genes from different origins and this combination has gifted them with unique metabolic features. In addition to the metabolism needed to produce a silica cell wall (Hildebrand et al., 2018) a functioning urea cycle is present (Allen et al., 2011; Nonoyama et al., 2019). Diatoms have an Entner-Doudoroff glycolytic pathway (Fabris et al., 2012) but lack the oxidative pentose phosphate (OPP) pathway in their chloroplast (Wilhelm et al., 2006; Kroth et al., 2008; Gruber et al., 2009). Their principal storage compound is a polysaccharide, chrysolaminarin (β-1,3 linked glucan) that is located in the vacuole rather than the chloroplast (Huang et al., 2018). Diatoms also have a large diversity of the metalloenzyme carbonic anhydrase (CA) that interconverts CO2 and HCO3−. They possess seven of the eight known CA sub-classes, some of which can make use of metal cations other than the canonical zinc (Jensen et al., 2019a; Alissa et al., 2020; Morel et al., 2020). In diatoms, both the large and the small subunits of ribulose bisphosphate carboxylase-oxygenase (RuBisCO) are encoded by the chloroplast genome, in contrast to Viridiplantae where the small subunit is a nuclear encoded protein (Oudot-Le Secq et al., 2007). Moreover, most diatom plastid genomes, unlike those in Viridiplantae and the diatom Seminavis robusta, lack introns (Brembu et al., 2014). Also in diatoms, RuBisCO activation is mediated by the protein CbbX (Mueller-Cajar et al., 2011) that does not possess the cysteine residues found in RuBisCO activase (RCA) found in Viridiplantae, and thus cannot be redox regulated (Jensen et al., 2017). In addition, diatoms also have a pigment composition that substantially differs from plants (Carreto and Catoggio, 1976; Falkowski and Owens, 1980; Gilstad et al., 1993; Kuczynska et al., 2015). The most important accessory pigments in diatoms are fucoxanthin and chlorophyll c rather than chlorophyll b in Viridiplantae (Green, 2011). Also, like all photosynthetic eukaryotes and cyanobacteria, they contain xanthophylls that are derived from β carotene but in contrast, lack the α-carotene pathway. Diatoms are able to acclimate to a broad range of light irradiance and nutrient concentrations by adjusting their physiology and biochemical activity (Schoefs et al., 2017; Heydarizadeh et al., 2019). This requires a variety of mechanisms for balancing energy harvesting and light-energy consuming metabolic processes including carbon fixation (Wilhelm et al., 2006). In contrast to Viridiplantae, diatoms have a very low cyclic electron flow. To equilibrate the ratio of ATP to NADPH required for optimal photosynthesis, the chloroplast and the mitochondrion, that are physically in contact, exchange these compounds (Bailleul et al., 2015) (Figure 1).
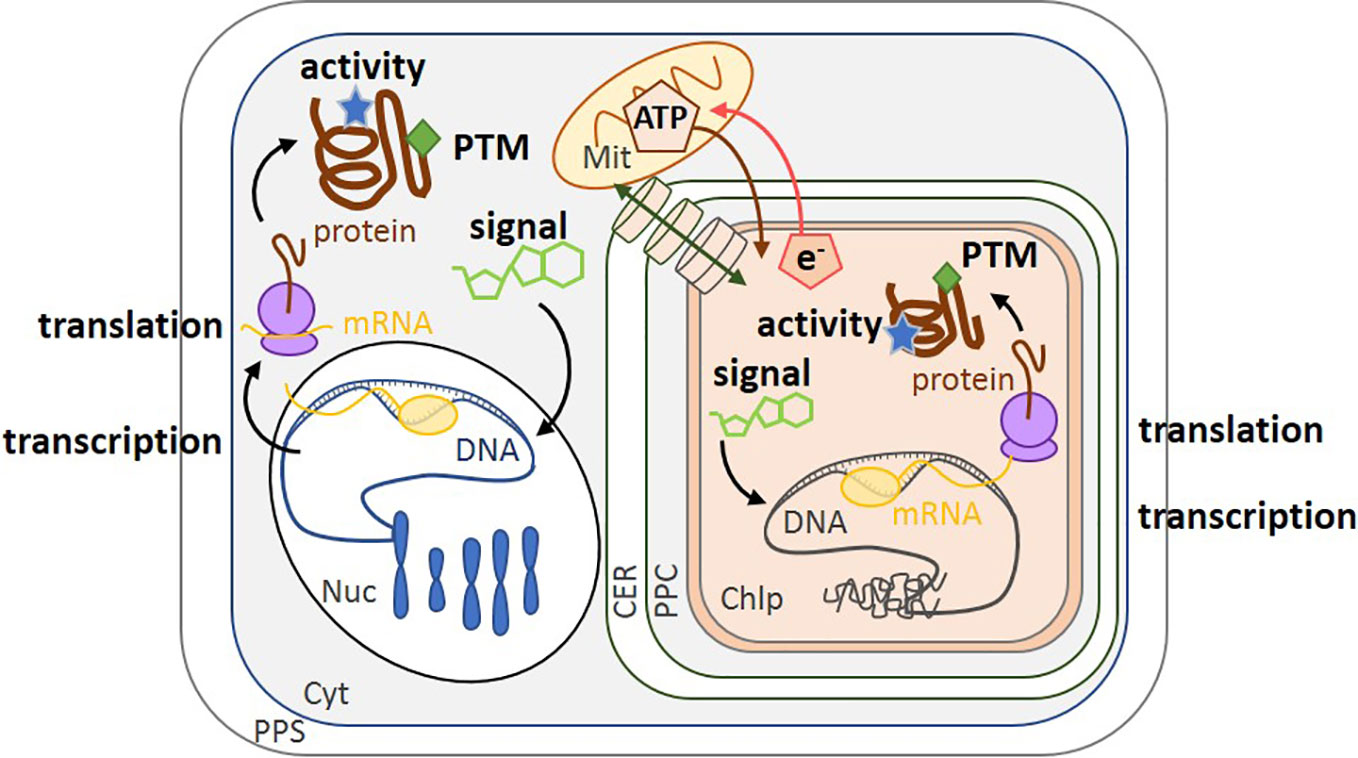
Figure 1 Schematic of the different levels of regulation in diatoms. Transcription of the genome by mRNA polymerase (yellow circle) converts nuclear DNA (blue) and chloroplast DNA (gray) into mRNA (yellow). Translation by the ribosome (purple) converts mRNA into protein (brown). Regulation can also be modulated by enzyme activity (blue stars), post-translational modification (green diamonds) and small molecules such as co-factors or metabolites (green carbon skeletons). The transport of molecules between the cytoplasm and the chloroplast is represented by a green double-headed arrow across cylinders. ATP synthesized in the mitochondrion can be transported into the chloroplast (brown arrow), while the reducing power (e.g., NADPH, represented by e-) of the chloroplast is transported to the mitochondrion (red arrow) (Bailleul et al., 2015). PPS, Periplasmic Space; CER, Chloroplast Endoplasmic Reticulum; PPC, Periplastidial Compartment; Mit, Mitochondrion; Chlp, Chloroplast; Cyt, Cytoplasm; PTM, Post-Translational Modification; Nuc, Nucleus.
Regulation can act on, and be studied at, a cascade of different levels from DNA (genomics), mRNA (transcriptomics), proteins (proteomics and post-translational modifications, PTMs), metabolites (metabolomics), and enzyme activity, because each approach provides different types of information (Figure 1). Genomes are powerful resources to determine if a specific gene is present while transcriptomics detect under what conditions it is expressed. Fully annotated diatom genome sequences are only available for Thalassiosira pseudonana, a marine centric diatom and Phaeodactylum tricornutum, a marine or coastal pennate diatom (Armbrust et al., 2004; Bowler et al., 2008). Other sequences are also available for Fragilariopsis cylindrus (Mock et al., 2017) and Pseudo-nitzschia multiseries (https://genome.jgi.doe.gov/portal/pages/tree-of-life.jsf) and there are further published genomes that are not yet publicly available (Tanaka et al., 2015; Traller et al., 2016; Basu et al., 2017; Villain et al., 2017; Ogura et al., 2018). Lauritano et al. (2019) reviewed the current development of omics approaches on microalgae. Of the 26 eukaryotic microalgal genomes they listed, 15 were from chromalveolates and of these, 8 were from diatoms. However, regulation should also be analyzed at the level of enzyme activity and/or metabolites (metabolomics), as these represent the final outcome of gene expression and activity (Prosser et al., 2014). The lifetime of an active enzyme, or of a metabolite, is related to its rate of synthesis and turnover. While the synthesis rates are on the order of 4–6 amino acids per second for enzymes (Stein and Frydman, 2019) and several seconds for metabolites (Nikolaev and Lohse, 2006), their turnover rates can vary from a few seconds to extended periods respectively. Enzyme activity is also modulated by PTMs, such as acetylation, phosphorylation, methylation, glycosylation and formation/dissociation of disulfide bonds. These types of modulation are very fast (rate on the order of few per second), reversible, and are the most flexible regulatory responses at the protein level (Prabakaran et al., 2012). In diatom RuBisCO, a number of post-translational modifications of the large subunit are present, including 4-hydroxyproline, β-hydroxyleucine, hydroxylated and nitrosylated cysteine, mono- and dihydroxylated lysine, and trimethylated lysine (Valegard et al., 2018). Nevertheless, in order to understand the full scope of regulation by post-translational modifications (Grabsztunowicz et al., 2017) in chromalveolates, more studies are needed on PTM and proteomics. Regulation of gene expression is itself dependent on earlier response regulators (for example, PTMs of histones and transcription factors) and on metabolite productions (for example, cAMP). As a consequence, upon environmental changes, regulation of gene expression occurs over a longer timescale of several minutes to hours (Chauton et al., 2013).
Relationships between mRNA level and protein expression can be observed though this might be influenced by biological (e.g., properties of mRNA and proteins, cell cycle status) and by technical problems (accurate quantification of these two biological molecules) (Maier et al., 2009; Ponnala et al., 2014). Therefore, there are discrepancies in the literature as regard to the extent of correlation between them. Net mRNA levels can be a major contributor to protein abundance, and for instance, positive relationship has been observed in yeast (Fournier et al., 2010), in the green alga Chlamydomonas reinhardtii (Castruita et al., 2011) and specifically in the diatom T. pseudonana (Clement et al., 2017b). Nevertheless, there are additional mechanisms, that control protein abundance including translational control and differential protein and mRNA degradation rates (Ponnala et al., 2014). However, since data on protein expression and activity are scarce, we have supplemented this type of information with data on gene regulation as a first step to assess how diatoms respond to environmental change, even though there is not always a direct and positive relationship between mRNA level, protein expression and finally, metabolic activity (Figure 1).
Changing light levels affect many processes, including cell division, and diatoms can acclimate efficiently to light variation by altering the expression of different cell cycle genes such as cyclins and cyclin-dependent kinases genes (Huysman et al., 2013). Here, we focus on the best-studied effect of light, the regulation of photosynthesis, although there is much less information for diatoms than for the Viridiplantae (Jensen et al., 2017). In the Viridiplantae, that includes the Embryophyta, carbon fixation by the Calvin-Benson-Bassham (CBB) cycle is well known to be fine-tuned by dark-light transitions, involving regulation by pH (Werdan et al., 1975), Mg2+ (Portis and Heldt, 1976), metabolite concentration (Anderson, 1973; Pupillo and Giulianipiccari, 1975; Gardemann et al., 1983; Baalmann et al., 1994), and primarily by the redox state of key enzymes (Buchanan et al., 1980; Schurmann and Jacquot, 2000). Non-covalently bound “small molecules” or metabolites also affect the rates of redox-interconversion of each redox-regulated enzyme in Viridiplantae and this fine-tuning regulation is well-described in a review from Knuesting and Scheibe (2018).
In chloroplasts from Embryophyta, dark-to-light transitions are accompanied by a shift of the chloroplast internal pH from 7 in the dark to 8 in the light (Werdan and Heldt, 1973; Hauser et al., 1995). These changes directly regulate photosynthesis since many key chloroplastic enzymes have optimal activity at pH 8 and are much less active at pH 7 [reviewed in Gontero et al. (2007)]. In diatoms, pH responses have mainly been studied for external/environmental, rather than internal, pH. External pH can affect growth rate, silicon metabolism and biomineralization of Conticribra weissflogii (formerly known as Thalassiosira weissflogii) as well as its intracellular/cytoplasmic pH homeostasis (Herve et al., 2012). For other photosynthetic organisms, it is not the extracellular pH, but the intracellular pH in the chloroplast that is the critical factor for regulation of carbon acquisition, transport capacity and other metabolic processes. To our knowledge, internal pH has only been measured for a small number of diatoms. For P. tricornutum and Cyclotella sp. the pH was around 7 in the dark and 7.5 in the light (Colman and Rotatore, 1995), and for Navicula pelliculosa, it was 7.4 in the dark and 7.6 in the light (Colman and Rotatore, 1988). We found no published values for the pH within diatom chloroplasts stroma. One of the few studies of the effect of pH on enzyme activity in chromalveolates is for the chloroplastic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that catalyzes the reversible reduction and dephosphorylation of 1,3-bisphosphoglycerate to produce glyceraldehyde-3-phosphate and inorganic phosphate. Avilan et al. (Avilan et al., 2012) compared the optimal pH of GAPDH in the freshwater diatom, Asterionella formosa, the freshwater eustigmatophyte, Pseudocharaciopsis ovalis, and the model green alga, C. reinhardtii. In A. formosa, GAPDH was still active at the pH occurring in the dark, assuming that the internal pH (pH 7) reflects the one in the chloroplast. This suggests that GAPDH is regulated by factors other than pH in this diatom, unlike in the green algal enzyme that was down-regulated at the pH that occurs in the dark. The response of GAPDH from the eustigmatophyte P. ovalis was similar to that of the green alga C. reinhardtii. We do not know the internal chloroplast pH for P. ovalis but if the dark-to-light pH transition in this species is similar to that of C. reinhardtii, GAPDH could be partly regulated by pH under dark-light transitions. The different regulation of GAPDH by pH in the two heterokonts, A. formosa and P. ovalis, might be the result of the diverse evolutionary history of chromalveolates. Another example of regulation by pH is the lumenal enzyme violaxanthin de-epoxidase [VDE, (Lavaud et al., 2012)] that is involved in dissipating excess light energy (Lohr and Wilhelm, 1999).
Beyond photosynthesis, the carbon metabolism of the marine diatom Skeletonema costatum is regulated by the pH of the growth medium (Taraldsvik and Myklestad, 2000). The content of the carbohydrate storage compound, chrysolaminarin (β-1,3 linked glucan) decreased from 7.1 mg.L−1 at pH 6.5 to 0.2 mg.L−1 at pH 9.4 and concomitantly, the total organic carbon as glucan also decreased from 60 to 10%. The total amino acid content also decreased from 7.41 to 2.51 fmol.cell−1 when the pH of the growth medium increased (Taraldsvik and Myklestad, 2000). It is unclear if these are direct effects on carbon and nitrogen metabolism of external or internal pH or indirect effects linked to the greater external concentration of CO2 at pH 6.5 than at pH 9.4. Nevertheless, to relate these physiological responses to enzyme activity regulation, the authors report results from a Norwegian PhD thesis (Kirkvold, 1994) that showed that the specific activity of glutamine synthetase, a key enzyme in the metabolic pathway of glutamine and glutamate synthesis, also decreased with increasing pH when measured in vitro.
Studies on the effect of pH on activity should be expanded to more enzymes and their optimal pH compared to the internal pH in dark and light in order to determine if enzyme activity is regulated by internal pH. The difficulty of working with enzymes from diatoms and from chromalveolates in general, is probably responsible for the lack of data for this important group. For instance, in order to extract proteins from diatoms, litres of culture are required and it is not always easy to measure activity. Expressing recombinant diatom enzymes in heterologous systems is also challenging with many enzymes found in the insoluble fractions (B. Gontero, personal communication). Measurement of internal pH is also an experimental tour de force. Colman and Rotatore used the 5,5-dimethyl-2,4-oxazolidinadione distribution method that distributes between the medium and the cell as a function of their respective pH (Colman and Rotatore, 1988; Colman and Rotatore, 1995). However, this method does not distinguish between the pH in the cytoplasm, chloroplast stroma or thylakoid lumen.
The redox control of enzyme activity in the Viridiplantae is primarily mediated by small proteins, thioredoxins, that are oxidized in the dark and reduced in the light (Buchanan et al., 1980; Buchanan, 2017). This regulation avoids futile cycles between the CBB and the OPP pathway, since both occur within the chloroplast, with enzymes from the CBB being active in the light and those from OPP being active in the dark. In contrast, in diatom plastids the OPP is incomplete, and presumably lacking (Kroth et al., 2008), and accordingly the regulation of their metabolism is different (Jensen et al., 2017). Moreover, diatoms have a high stromal reductant pressure and in contrast to Viridiplantae, metabolic activity in long dark periods leads to an enhanced reduction state of the plastoquinone pool. In the dark, since the plastoquinone pool is reduced, it may regulate redox-sensitive enzymes as is the case for algal nitrate reductase (Giordano et al., 2005). This avoids reducing equivalents to accumulate maintaining cellular redox poise (Wilhelm et al., 2006). In Viridiplantae in contrast, oxidizing conditions prevail in the dark, therefore suggesting that redox control may be different. Because of this unusual redox control, the redox regulation of diatom enzymes has been questioned (Wilhelm et al., 2006). However, diatoms possess many different thioredoxins, each encoded by a specific gene and located in different compartments, including the chloroplast. Most thioredoxins contain the regulatory cysteine residue in the conserved motif, WCGPC (Weber et al., 2009), thus they are likely to have specific regulation targets and some targets have been identified such as two CAs in P. tricornutum (Kikutani et al., 2012). The relatively few targets of thioredoxins currently identified in diatoms, contrasts with the 1188 targets found by combining qualitative and quantitative proteomic analyses in the C. reinhardtii thioredoxome (Perez-Perez et al., 2017).
Using a redox proteomics approach on P. tricornutum, Rosenwasser et al. identified the “redoxome”, or in other words the redox-sensitive proteins, and demonstrated its involvement in photosynthesis, photorespiration, lipid biosynthesis, and nitrogen metabolism (Rosenwasser et al., 2014). In that case, however, the redox-sensitivity is a response to oxidative stress rather than light-dark transition even though reactive oxygen species are photo-induced, and increase in parallel to glutathione (GSH). GSH is a low-molecular-weight tripeptide that consists of cysteine (Cys), glutamic acid (Glu), and glycine (Gly) and is present in microorganisms, plants, and mammals (Zaffagnini et al., 2012). It can regulate protein activity by forming a mixed disulfide bridge between the thiol group of its Cys and an accessible free thiol on a protein, a process known as protein S-glutathionylation (Zaffagnini et al., 2012; Marri et al., 2014). This post-translational modification can protect specific Cys residues from irreversible oxidation but can also modulate protein activities (Zaffagnini et al., 2012; Marri et al., 2014; Thieulin-Pardo et al., 2015). In T. pseudonana, a diurnal redox-related pattern has been observed in which GSH accumulates in the light, and decreases upon darkness, (Dupont et al., 2004) (Figure 2). However, direct regulation of the enzyme activities by glutathionylation in diatoms, or in other chromalveolates, in contrast to Viridiplantae, has not yet been studied, to the best of our knowledge.
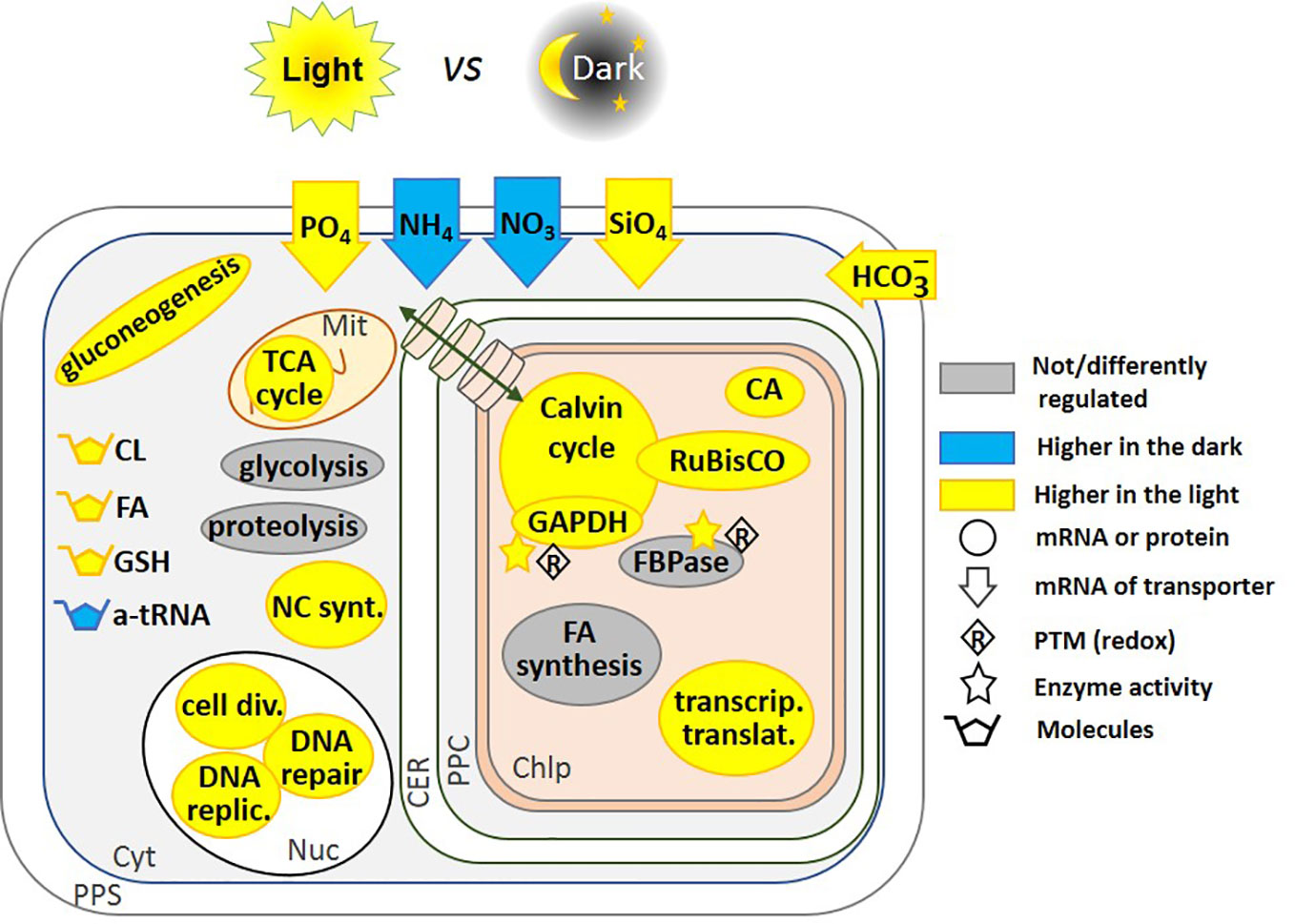
Figure 2 Regulation of pathways by light and dark. This schematic includes regulatory pathways from P. tricornutum (Chauton et al., 2013; Bai et al., 2016), and T. pseudonana (Ashworth et al., 2013). Where up-regulation of the mRNA transcription preceded the start of the photoperiod, we have represented it as being higher in the light, this is the case for RuBisCO expression in T. pseudonana (Ashworth et al., 2013). Glycolysis and proteolysis pathways are up-regulated in the light in T. pseudonana, but down-regulated in P. tricornutum. In contrast, fatty acid synthesis is down-regulated in T. pseudonana but up-regulated in P. tricornutum. Fatty acid content increased after 3 h of light in P. tricornutum (Chauton et al., 2013), we have thus represented the molecule as being higher in the light. FBPase gene expression is not regulated, but its activity is redox-regulated (Michels et al., 2005; Mekhalfi et al., 2012). Redox regulation of enzyme activity has been shown on the isolated proteins in vitro, and by analogy with the Viridiplantae lineage, this could be related to light-to-dark transitions. The transport of molecules between the cytoplasm and the chloroplast is represented by a green double-headed arrow across cylinders. CL, Chrysolaminarin; FA, Fatty Acid; GSH, Glutathione; a-tRNA, aminoacyl-transfer RNA; CA, Carbonic Anhydrase; NC synt., Nucleotide synthesis; cell div., Cell division; DNA replic., DNA replication; PPS, Periplasmic Space; CER, Chloroplast Endoplasmic Reticulum; PPC, Periplastidial Compartment; Mit, Mitochondrion; Chlp, Chloroplast; Cyt, Cytoplasm; Nuc, Nucleus.
Although the effect of glutathionylation on enzymes has not been studied in diatoms, the effect of other reducing agents such as dithiothreitol has been investigated, though understudied as compared to other photosynthetic organisms such as Cyanobacteria, Chlorophyta, Rhodophyta, and Embryophyta. The chloroplastic phosphoglycerate kinase belonging to the CBB cycle, catalyzes the ATP-Mg2+-dependent phosphorylation of 3-phosphoglycerate (3-PGA) to 1,3-bisphosphoglycerate, in a reversible reaction and was redox-regulated in P. tricornutum (Belen Bosco et al., 2012). However, in our hands, PGK was not redox-regulated, as was also the case in T. pseudonana, in Navicula pelliculosa grown with sea water and fresh water medium and in a freshwater diatom, A. formosa (Jensen et al., 2019b). In contrast to the Viridiplantae, two enzymes that are unique to the CBB were not redox regulated (Michels et al., 2005; Maberly et al., 2010; Jensen et al., 2017). These include, sedoheptulose 1,7-bisphosphatase that irreversibly catalyzes the dephosphorylation of sedoheptulose-1,7-bisphosphate producing sedoheptulose-7-phosphate, and phosphoribulokinase (PRK) that irreversibly catalyzes the ATP-Mg2+-dependent phosphorylation of ribulose-5-phosphate into ribulose-1,5-phosphate. The general lack or weak redox regulation of PRK in the chromalveolates (diatoms and other groups) seems to be related to its sequence, where the connectivity between two regulatory cysteine residues is crucial [at position 16 and 55 in C. reinhardtii (Maberly et al., 2010)]. In many photosynthetic organisms PRK can also be sequestered, and inactivated, in a PRK-GAPDH-CP12 complex, that has not yet been found in diatoms. The absence of the ternary complex with GAPDH in diatoms has been attributed to the absence of two cysteine residues on PRK (at position 243 and 249 numbered from the enzyme from C. reinhardtii) that are present in Cyanobacteria, Chlorophyta, Rhodophyta, and Embryophyta where the complex has been identified (Thieulin-Pardo et al., 2015). In contrast however, a ferredoxin-NADP reductase (FNR)-GAPDH-CP12 complex has been found in A. formosa (Mekhalfi et al., 2014). For the chloroplastic GAPDH, the regulation is more complex as discussed above in the pH regulation section, but in many diatoms, this enzyme seems to be redox regulated (Maberly et al., 2010; Mekhalfi et al., 2012; Mekhalfi et al., 2014; Jensen et al., 2019b).
Regulation at the transcriptional level by light-dark transitions, occurs in Viridiplantae, and also in diatoms (Sun et al., 2003; Fey et al., 2005). In T. pseudonana, after 12 h of light, 1,859 genes were upregulated compared to cells exposed to 12 hours of dark, and inversely, after 12h of dark, 1,326 genes were up-regulated compared to cells exposed to 12 h of light (Ashworth et al., 2013). Among the most highly expressed genes after 12 h of light were the ones encoding enzymes for cell division, DNA replication and repair, carbon metabolism and oxidative phosphorylation while after 12 h of dark, the most highly expressed genes were those encoding ribosomal biogenesis, aminoacyl-tRNA and key photosynthetic enzymes (Figure 2). Some genes, such as that encoding RuBisCO, anticipates the diurnal cycle and is up-regulated before the onset of light. The dark-light expression pattern of genes was affected by growth phase (exponential vs. stationary). In the stationary phase, the expression of only a few genes fluctuated under dark-light transitions (Ashworth et al., 2013). One of these genes encodes a putative pyruvate carboxylase suggesting a switch toward other types of metabolism such as gluconeogenesis and lipid biosynthesis. This might be explained by the hypothesis raised by Norici et al. in S. marinoi (Norici et al., 2011) who postulated that the diatom re-routes its metabolism toward lipid biosynthesis, because of the relatively high volume-based energy content of lipids in an organism in which size decreases with vegetative cell divisions, thus requiring carbon allocation into more energy-compact compounds.
In the light, more than 4,500 transcripts were differentially expressed in P. tricornutum, including genes such as the one encoding for pyruvate transporter that had never been previously described in this organism (Chauton et al., 2013). This work shows that transcriptional regulation of carbohydrate and lipid metabolism occurs in diatoms (Figure 2). Indeed, the content of soluble glucans and lipids decreased in the dark and fatty acid biosynthesis genes were up-regulated within 30 min of a switch from dark to light. Fatty acid biosynthesis and the tricarboxylic acid (TCA) cycle are also tightly co-ordinated (Chauton et al., 2013). During the day, carbon skeletons are produced within the chloroplast while in the night these carbon-rich compounds are broken down in the mitochondria and the cytosol.
Interestingly, four carbon fixation enzymes were co-regulated in P. tricornutum: PGK, GAPDH, triose phosphate isomerase/GAPDH and PRK. Their mRNAs were all highest at the beginning of the light period (dawn) and lowest at the beginning of the dark period (dusk). Bai et al. (2016) showed however that the expression of PRK increased after 4 days of dark treatment using a proteomic approach. Similarly, the expression of phosphoribose isomerase, the enzyme that produces PRK substrate, as well as the expression of transaldolase and glyoxylase, increased after dark treatment.
Redox regulation of PRK is observed and well-studied in Viridiplantae, but in contrast in diatoms and other chromalveolates, as mentioned above, the activity of PRK was affected neither by oxidizing nor by reducing conditions (Maberly et al., 2010). Therefore, for enzymes that are not redox-regulated, regulation at the transcriptional level could allow a dark-light regulation of the CBB. The expression pattern of the gene of fructose-1,6-bisphosphatase (FBPase) that catalyzes the dephosphorylation of fructose-1,6-bisphosphate into fructose-6-phosphate and inorganic phosphate, was not induced to the same degree by light as other CBB enzymes. Since this enzyme can be redox regulated (Michels et al., 2005; Mekhalfi et al., 2012), its regulation could occur both at the level of activity and transcription.
Carbon acquisition is also stopped at night since in P. tricornutum, mRNA encoding for bicarbonate transporters of the Solute Carrier family 4 (SLC4) and for alpha-carbonic anhydrases, especially α-CA-VII, were much less abundant in the dark than in the light (Chauton et al., 2013). Recently, in the same organism, the pattern of mRNA levels at a photon irradiance of 30,300, 1,000 µmol photon m−2 s−1 differed at the lag, exponential and stationary phases of growth (Heydarizadeh et al., 2019).
Dark-treated P. tricornutum cells preferentially utilize carbon and nitrogen obtained from protein breakdown to increase lipid cell quotas at low cost (Bai et al., 2016). Long-term dark stress inhibited several key proteins involved in nitrogen assimilation and in the synthesis of the photosynthetic machinery. Simultaneously, key enzymes of glycolysis and the synthesis of fatty acids were induced apparently to assimilate the excess of C and N from protein breakdown. Uptake of other resources for growth are also light-regulated: transporters for uptake of phosphate and silica are higher in the light and nitrate and ammonium transporters are higher in the dark (Ashworth et al., 2013) (Figure 2). Data from the literature showing differential expression of proteins, transcripts and metabolites in light and dark are summarized in Figure 2.
Gene expression gives important clues on how light (quality, irradiance, duration) affects metabolism. However, it is also necessary to measure enzyme activity and metabolite concentration as these are the ultimate response to environmental change. For instance, an excess of light modifies lipid biosynthesis in the coastal marine diatom, Skeletonema marinoi (Norici et al., 2011). In S. costatum, carbohydrate increased with irradiance (Hitchcock, 1980) while lipids increased in Chaetoceros calcitrans (Harrison et al., 1990) and therefore the carbon allocation seems to be different and species-specific. Under different light-regimes, different species behave differently and the amount of essential fatty acids with growth irradiance is also species-specific. These examples illustrate that although enzyme activities have been measured, the mechanism underlying the change in activity is unknown. For example, PEPCK that converts oxaloacetate into PEP and CO2 in gluconeogenesis, increased 2.5-fold in cells of S. marinoi grown under low light (25 µmol.photon.m−2.s−1) vs. high light (250 µmol.photon.m−2.s−1). The authors suggested that this enzyme might be involved in the conversion of lipid to carbohydrates especially under low light (Norici et al., 2011). In contrast, since energy demand for lipid synthesis is much higher than for carbohydrate synthesis (Raven et al., 2005; Subramanian et al., 2013) under excess light, lipids represent a better sink for excess energy. As a consequence, lipid accumulation in high irradiance was observed in S. marinoi, although this is not always the case. In this species, other enzymes are probably not regulated by light since their activity remains unchanged. For instance, the activity of PEPC, that catalyzes the addition of bicarbonate (HCO3−) to PEP to produce oxaloacetate, was similar for cells grown at low or high light. This enzyme is involved in C4 metabolism and in anaplerotic reactions. Similarly, the activity of glutamine synthetase that is involved in photorespiration did not change in cells grown at low or high light.
Like other algae, diatoms exhibit a range of responses to varying CO2 concentration, including effects on photophysiology, rate of photosynthesis and growth, chemical and pigment composition, and community species composition, but there are large species- and context-specific variations in the magnitude and sign of response (Boelen et al., 2011; Torstensson et al., 2012; Gao and Campbell, 2014; Dutkiewicz et al., 2015; Bach and Taucher, 2019; Jensen et al., 2019b). At the ocean surface, the air-equilibrium concentration of CO2 (Dickson, 2010) varies between 5 and 25 µM depending on temperature (Raven, 1994; Tortell, 2000; Kim et al., 2006; Matsuda et al., 2011; Maberly and Gontero, 2017). This CO2 concentration is insufficient to saturate the carboxylating enzyme, RuBisCO (Young et al., 2016) and may not saturate rates of diatom growth or photosynthesis (Riebesell et al., 1993; Dutkiewicz et al., 2015). This is mitigated by CO2-concentrating mechanisms (CCMs) (Hopkinson et al., 2011) that are facultative and increase the concentration of CO2 around RuBisCO, and are present in many phytoplankton. CCMs can involve biophysical or biochemical processes (Reinfelder et al., 2000; Reinfelder, 2011; Hopkinson et al., 2016) although the latter is controversial in diatoms (Clement et al., 2017a). The CCM regulation in diatoms is highly dependent on light as well as CO2 concentration (Harada et al., 2006). However, the major determinant of the extent of CCM expression in P. tricornutum is CO2 concentration, as it is in green algae (Matsuda and Kroth, 2014) and many components of CCM are suppressed under elevated CO2 concentrations and induced at atmospheric levels or lower. The CO2 concentration affects expression, and consequently the activity, of not only CCMs components but also that of enzymes from metabolic pathways such as the CBB cycle and glycolysis, though this is still understudied. Below, we compile information on the regulation by CO2 at the transcriptional and/or the enzyme activity levels of enzymes involved in the CCM and other metabolic pathways.
In C4 metabolism, PEPC is the first carboxylating enzyme and traps bicarbonate into a C4 carbon compound. This compound is subsequently cleaved by a decarboxylase enzyme to provide a 3-carbon compound and CO2, near the active site of RuBisCO (Sage, 2004). As mentioned above, the presence of C4 or biochemical CCM in diatoms does not seem to be universal. For instance, there is evidence for it in C. weissflogii (Reinfelder et al., 2000; Reinfelder, 2011; Hopkinson et al., 2016) but it is absent in P. tricornutum (Haimovich-Dayan et al., 2013; Clement et al., 2017a; Ewe et al., 2018). In the eustigmatophyte Nannochloropsis oceanica, a novel type of C4-based CCM was proposed to occur when cells were shifted from high CO2 (50,000 ppm) to low CO2 (100 ppm) (Wei et al., 2019). In this C4-based CCM, PEPC and PEPCK have been proposed to be involved in the primary inorganic carbon fixation steps in mitochondria, and not in chloroplasts. Subsequent decarboxylation of malate by a malic enzyme in the chloroplast enriches CO2 in the vicinity of RuBisCO (Figure 3). Transcripts levels of some C4 enzymes were not altered by CO2 concentration and do not suggest a classic C4 metabolism, but activities of PEPC and malic enzyme increased under low CO2 (Wei et al., 2019). More work is required to confirm this interesting possibility.
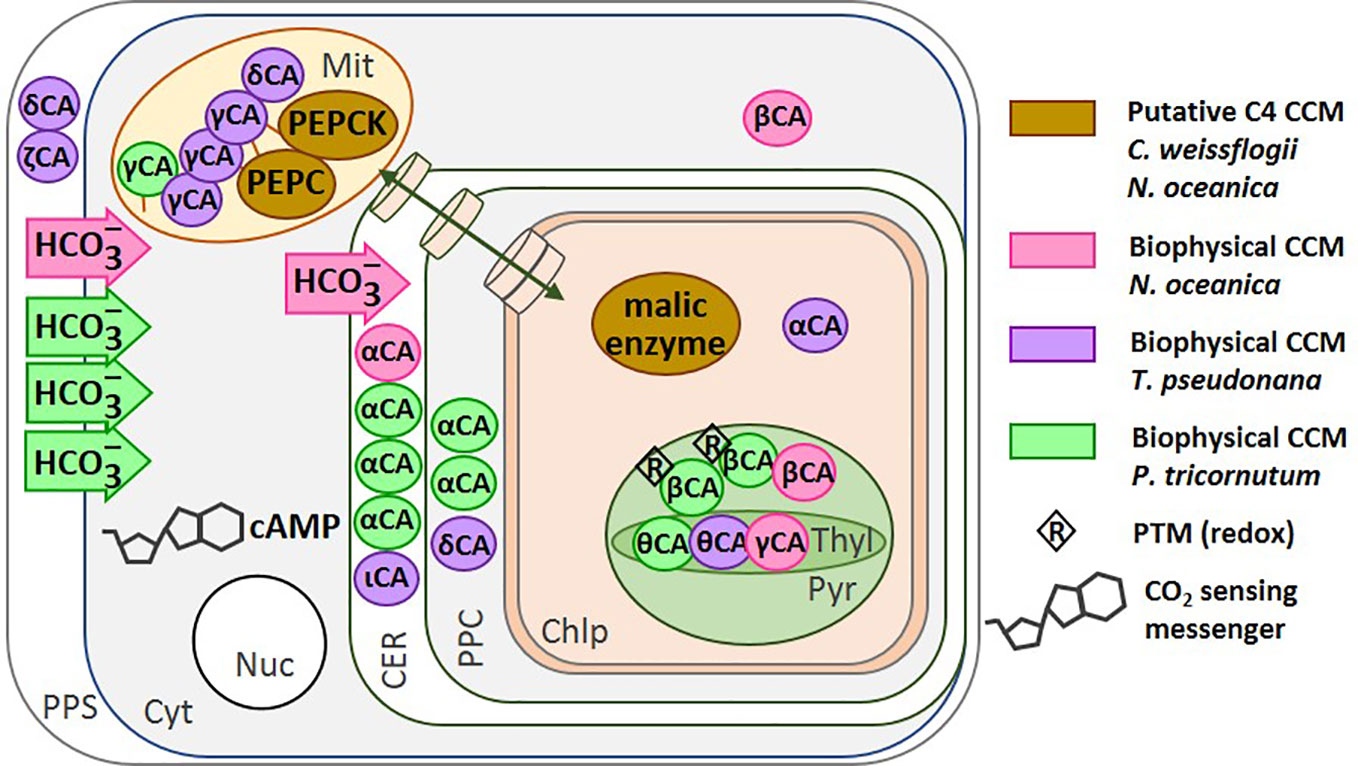
Figure 3 Location of the CCM components in different diatom species. The carbonic anhydrases (circles) and bicarbonate transporters (arrows) are shown for: N. oceanica (Wei et al., 2019) (magenta), T. pseudonana (Samukawa et al., 2014; Jensen et al., 2020) (purple), and P. tricornutum (Jensen et al., 2020) (green). The putative C4-CCM components proposed for N. oceanica (Wei et al., 2019) and C. weissflogii (Reinfelder et al., 2000; Ohno et al., 2012; Hopkinson et al., 2016; Tanaka et al., 2016). The transport of molecules between the cytoplasm and the chloroplast is represented by a green double-headed arrow across cylinders. Redox regulation of CA activity has been shown on the isolated P. tricornutum β-CA in vitro. PEPC, Phosphoenolpyruvate Carboxylase; PEPCK, Phosphoenolpyruvate Carboxykinase; CA, Carbonic Anhydrase; PPS, Periplasmic Space; CER, Chloroplast Endoplasmic Reticulum; PPC, Periplastidial Compartment; Mit, Mitochondrion; Chlp, Chloroplast; Cyt, Cytoplasm; Nuc, Nucleus. Figure adapted from Jensen et al. (2020).
In the genomes of P. tricornutum and T. pseudonana, nine and thirteen CA gene sequences have been found respectively (Tachibana et al., 2011; Samukawa et al., 2014). In P. tricornutum, the two chloroplastic pyrenoidal β-CAs PtCA1 and 2, responded to CO2 (Satoh et al., 2001; Harada and Matsuda, 2005; Harada et al., 2005) and later this was confirmed by Tachibana et al. by semi-quantitative RT-PCR (Tachibana et al., 2011). The activation of both PtCA1 and PtCA2 under CO2 limitation involves three cis-regulatory elements, TGACGT, ACGTCA, and TGACGC, at a region minus 86 to minus 42 upstream of the transcription start site. These elements, CCRE1 to 3, are critical for the transcriptional response to ambient CO2 via the level of the second messenger cAMP (Ohno et al., 2012; Tanaka et al., 2016). The sensing of CO2 mediated by cAMP has been reported in cyanobacteria, fungi and mammals (Matsuda et al., 2011) and also in T. pseudonana (Hennon et al., 2015; Young and Morel, 2015). The transcriptional activation of PtCA2 in response to the decrease in CO2 concentration was strongly light-dependent, such that either CO2 or the absence of light can down-regulate the promoter. In contrast, CO2 concentration and light have additive effects on the regulation of PtCA1 (Tanaka et al., 2016). It is worth remarking that both PtCA1 and PtCA2 were post translationally regulated by redox modifications via thioredoxins (Kikutani et al., 2012) (Figure 3).
Tachibana et al. also showed that three putative CA genes in T. pseudonana, CA-1, 3, and 7 (α-CA, ζ-CA, and δ-CA, respectively) were induced by decreasing CO2, and function in CO2-limited environments (Tachibana et al., 2011). Similarly, in C. weissflogii, both CO2 and HCO3− uptake increased in response to a CO2 decrease and this was accompanied by an increase in both internal and external CA activity (Burkhardt et al., 2001). A recent proteomic study on T. pseudonana acclimated to low CO2 (50 ppm) revealed a new uncharacterized protein, later called LCIP63, for “low-CO2-inducible protein of 63 kDa” that was up-regulated under low CO2 (50 ppm) or at atmospheric CO2 (400 ppm) (Clement et al., 2017a) but down-regulated when nitrogen, phosphorus or silicon were limiting conditions (Lin et al., 2017; Chen et al., 2018). LCIP63 was up-regulated in T. pseudonana growing at 300 ppm vs. 1,000 ppm CO2 (Valenzuela et al., 2018). Recently, this protein was identified as a new CA [designed as iota CA, (Jensen et al., 2019a)] that uses Mn2+ as a co-factor instead of the more common divalent cation Zn2+ (Tsuji et al., 2017; Dimario et al., 2018). The gene of iota CA is also present in bacterial genomes (Jensen et al., 2019a) and recently, the gene encoding this enzyme from the gram negative bacterium Burkholderia territorii was cloned and the purified recombinant protein showed a CA activity using Zn2+ instead of Mn2+ (Del Prete et al., 2020) indicating that the use of Mn2+ as a co-factor could be a feature of diatoms.
In N. oceanica, transcriptomic, proteomic and metabolomic data are available for cells grown at high CO2 (50,000 ppm) and low CO2 (100 ppm); three of the CA transcripts were up-regulated (β-CA-2, β-CA-4 and α-CA-5) under low CO2 (Wei et al., 2019). In addition, the transcripts of bicarbonate transporters of the SLC4 family and several of the ABC transporter family were also more abundant at low CO2, indicating an active biophysical CCM in this organism. Similarly, in P. tricornutum, beside the regulation of numerous CAs by CO2, three out of seven SLC4 genes were induced by low CO2 and were highly inhibited by the anion exchange inhibitor 4,4'-diisothiocyanatostilbene 2,2'-disulphonate (Nakajima et al., 2013). In T. pseudonana, genes homologous to those in P. tricornutum have been found (Matsuda et al., 2017); however, their functionality at the protein level has not yet been studied. Two chloroplast transporters of the bestrophin family of anion channels that are permeable to HCO3− were also more abundant at low CO2 and may play a role in the biophysical CCM of this diatom (Kustka et al., 2014). These data are summarized in Figure 3.
Depending on future CO2 emission scenarios, atmospheric levels of CO2 are likely to reach 800 ppm by 2,100 (IPCC, 2014; Gattuso et al., 2015). This relates to a temperature-dependent dissolved CO2 concentration at the ocean surface of 25 to 50 μM. At this concentration, cAMP plays a crucial role in down-regulating CCM genes in T. pseudonana, in particular those encoding the chloroplastic δCA3, some transporters and some involved in photorespiration (e.g., glycolate oxidase). These photorespiration genes and CCM genes interestingly, belong to a single CO2-responsive cluster that shares the same upstream cis-regulatory sequences found in P. tricornutum that is also responsible for down-regulation of the β-CA gene upon increased CO2 (Ohno et al., 2012). Similarly, genes involved in photosynthesis, the TCA cycle, oxidative phosphorylation and protein degradation were down-regulated, while in contrast other genes involved in signalling mechanisms were up-regulated at 800 ppm compared to 400 ppm CO2 (Hennon et al., 2015). Not all these genes contain the upstream regulatory region, though they were highly affected. This regulation is likely an indirect effect linked to the impact of high CO2 on the genes of structural maintenance of chromosomes (SMC), transcription factors, and histones.
Sensing a change, either an increase or a decrease, in external CO2 concentrations through cAMP seems to be a general rule of gene regulation rather than an exception even in diatoms (Young and Morel, 2015).
Many related metabolic pathways are affected by CO2 in addition to CCM expression. Gamma CA and the NADH-ubiquinone oxidoreductase complex (C1) are associated in many organisms (Cardol, 2011) and in N. oceanica, at very low [CO2] (100 ppm), the genes coding for these two mitochondrial enzymes were up-regulated (Figure 4). Since they can facilitate transport of CO2 produced by the TCA cycle and photorespiration toward the chloroplast, in the form of bicarbonate, they contribute to what is called a basal CCM (Wei et al., 2019). At the enzyme activity level, T. pseudonana cells grown at high CO2 concentration (20,000 ppm) vs. atmospheric (400 ppm) displayed higher NADPH-dependent GAPDH and FBPase activity (Figure 4) indicating that the CBB cycle could be affected even though PRK activity was unaltered (Clement et al., 2017b). The activity of pyruvate kinase, a glycolytic enzyme, was also strongly stimulated when cells were grown at 20,000 ppm (Figure 4) while two other glycolytic enzymes, NADH-dependent GAPDH and glucose-6-phosphate dehydrogenase (G6PDH) were unaffected (Clement et al., 2017b). In this diatom, a model summarizing the effect of inorganic carbon limitation, based both on activities and protein expression profiles has been elaborated (Clement et al., 2017b). This model shows the remodelling of metabolism with a diversion of energy and resources toward carbon metabolism at high CO2 and toward carbon capture at low CO2. An increase in δ-CA gene expression, to capture as much CO2 as possible, was also observed by Kutska et al. (2014) in T. pseudonana. Metabolism enzymes, as well as enzymes responsible for pigment synthesis and indirectly in light capture (Figure 4), were less abundant, while enzymes involved in reactive oxygen species (ROS) defence increased in order to avoid accumulation of ROS that might occur when energy is in excess. The increased activity of pyruvate kinase (Figure 4), observed in T. pseudonana seems to be shared by other diatoms, as it was observed in many diatoms from freshwater to seawater species grown under high CO2 (20,000 ppm) vs. atmospheric CO2 (400 ppm) (Jensen et al., 2019b). In addition, a modification of gene expression has been described that allows synthesis of either PEP or pyruvate under carbon shortage, indicating that pyruvate is an important hub in these organisms (Heydarizadeh et al., 2017; Heydarizadeh et al., 2019) (Figure 4).
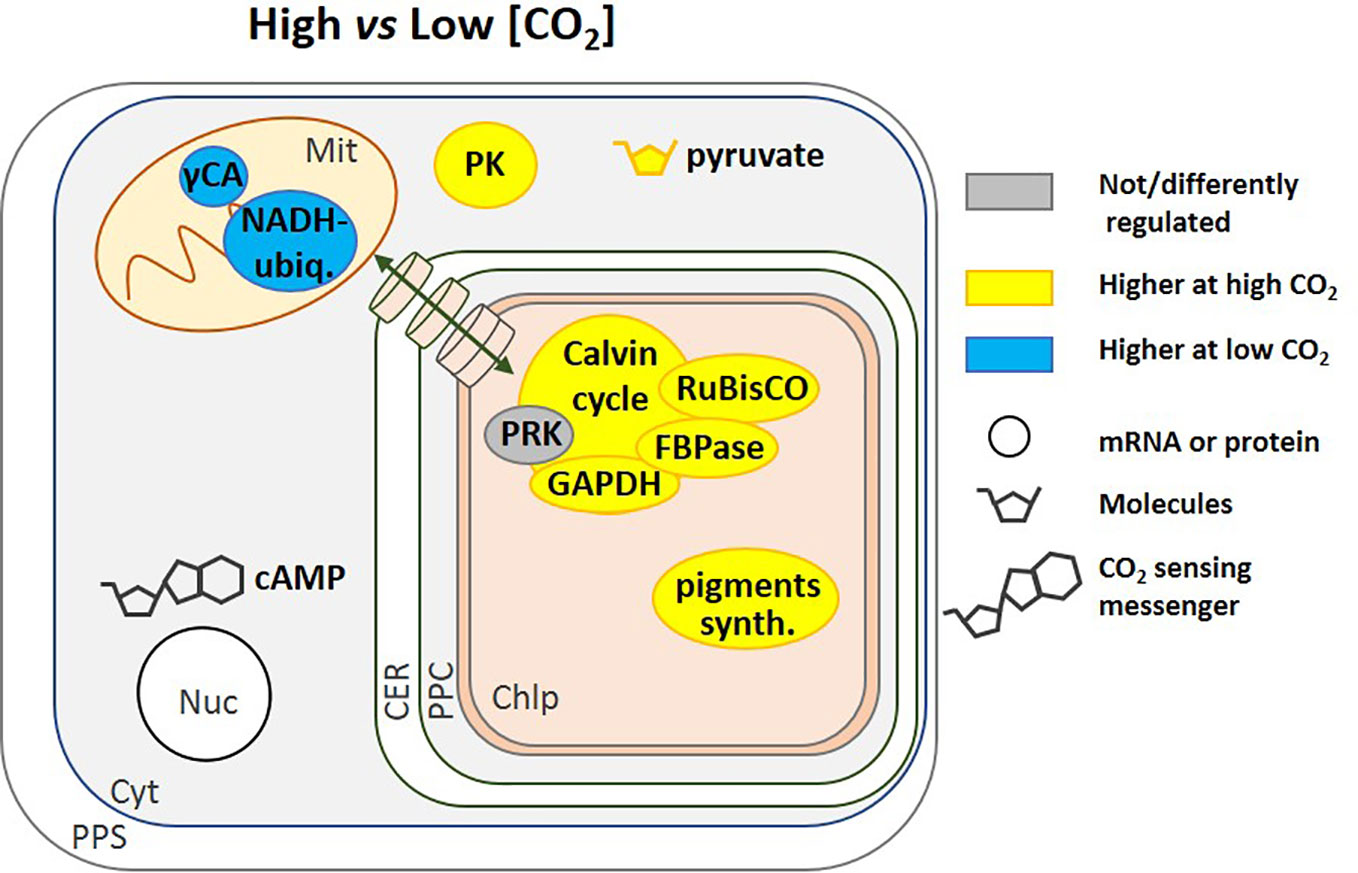
Figure 4 Regulation of pathways by [CO2]. This schematic includes regulatory pathways from different species, including N. oceanica (Wei et al., 2019) and T. pseudonana (Clement et al., 2017b; Jensen et al., 2019b). Cyclic AMP (cAMP) is a general [CO2] signalling molecule that regulates gene expression (Ohno et al., 2012; Hennon et al., 2015; Young and Morel, 2015). High CO2 corresponds to 20,000 to 50,000 ppm but in Hennon et al. (2015) it is 800 ppm and low CO2 is 50 to 400 ppm. The transport of molecules between the cytoplasm and the chloroplast is represented by a green double-headed arrow across cylinders. NADH ubiq., NADH-ubiquinone oxidoreductase complex; PK, Pyruvate Kinase; pigments synth., pigments synthesis; CA, Carbonic Anhydrase; PPS, Periplasmic Space; CER, Chloroplast Endoplasmic Reticulum; PPC, Periplastidial Compartment; Mit, Mitochondrion; Chlp, Chloroplast; Cyt, Cytoplasm; Nuc, Nucleus.
Physiological and genomic data are available for the response of some chromalveolates, especially diatoms, to light and CO2. They reveal the multitude and complexity of mechanisms that these organisms have evolved to cope with environmental variation. However, chromalveolates are still understudied compared to the Viridiplantae and more research is needed to unravel fully how this important group of algae maintain their productivity under changing conditions. There is a particular lack of information in diatoms on internal pH, especially in the chloroplast, on the identity of redox-regulated enzymes, on regulation by post-translational modification and on protein-protein interactions. There are existing and new methods that could be employed to tackle these knowledge gaps. For example, a range of pH-sensitive fluorescent probes are available to measure internal pH (Loiselle and Casey, 2010), although their low ability to penetrate the cell and organelle can be challenging. However, internal pH has also been measured successfully using the inorganic phosphate (31P) nuclear magnetic resonance frequency in other organisms such as fungi (Hesse et al., 2000) and the anammox bacterium, Kuenenia stuttgartiensis (Van Der Star et al., 2010), and this could be applied to chromalveolates.
There are a growing number of studies taking advantage of proteomics to study diatom responses to stress, e.g. Muhseen et al. (2015) or Chen et al. (2018). Proteomics approaches have been used successfully in Viridiplantae to identify candidates for thioredoxin interactions (Montrichard et al., 2009; Perez-Perez et al., 2017). This could be extended to diatoms where there is a real challenge to assign specific targets to each of the many thioredoxins found in diatoms (Weber et al., 2009). Biotin-based proximity labelling approaches, coupled to quantitative proteomics, such as APEX BioID, are emerging tools for the study of protein-protein interactions (Santin et al., 2018; Beganton et al., 2019) that could be developed for chromalveolates. There is evidence for unusual PTMs involved in the regulation of RuBisCO from an arctic diatom (Valegard et al., 2018) and proteomics could also be a powerful approach to analyse these modifications. For instance, a phosphoproteomic analysis in P. tricornutum confirmed that phosphorylation occurs in many metabolic pathways (Chen et al., 2014).
Over-expressing or silencing a gene is starting to be applied to diatoms in order to determine their metabolic role (Hildebrand et al., 2017; Huang and Daboussi, 2017; Falciatore et al., 2020). Recently, tailored TALEN endonucleases and the CRISPR/Cas9 system have been utilized in diatoms (Daboussi et al., 2014; Hopes et al., 2016; Nymark et al., 2016), allowing knockout strains with targeted genetic modifications to be produced. An overview of the genetic toolbox currently available for performing stable genetic modifications in diatoms is reviewed in Kroth et al. and Falciatore et al. (Kroth et al., 2018; Falciatore et al., 2020).
It is clear that the techniques mentioned above in combination with genome sequencing, “omics” and targeted approaches, will allow the biology of diatoms and chromalveolates to be understood better. However, since many responses seem to be species-specific, a wider range of species need to be studied, especially those from non-marine systems, to produce a more complete picture of the functioning in this supergroup with a mosaic of multi-lineage genomes.
All authors contributed to the article and approved the submitted version.
CNRS and Aix Marseille are our current funders and the ANR-15-CE05-0021-03 allows us to work on diatom project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
BG and HL were supported by the Centre National de la Recherche Scientifique (CNRS) and Aix-Marseille Université. BG is also supported by by the Agence Nationale de la Recherche (ANR-15-CE05-0021-03, SignauxBioNRJ). SM was supported by the UK Centre for Ecology & Hydrology. WH was supported by a fellowship for a visiting scholar from the Chinese Academy of Sciences.
Alissa, S. A., Alghulikah, H. A., Alothman, Z. A., Osman, S. M., Del Prete, S., Capasso, C., et al. (2020). Inhibition survey with phenolic compounds against the delta- and eta-class carbonic anhydrases from the marine diatom thalassiosira weissflogii and protozoan Plasmodium falciparum. J. Enzym. Inhib. Med. Ch. 35, 377–382. doi: 10.1080/14756366.2019.1706089
Allen, A. E., Dupont, C. L., Obornik, M., Horak, A., Nunes-Nesi, A., Mccrow, J. P., et al. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207. doi: 10.1038/nature10074
Anderson, L. E. (1973). Regulation of pea leaf ribulose-5-phosphate kinase-activity. Biochim. Biophys. Acta 321, 484–488. doi: 10.1016/0005-2744(73)90190-3
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Ashworth, J., Coesel, S., Lee, A., Armbrust, E. V., Orellana, M. V., Baliga, N. S. (2013). Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana. Proc. Natl. Acad. Sci. U.S.A. 110, 7518–7523. doi: 10.1073/pnas.1300962110
Avilan, L., Maberly, S. C., Mekhalfi, M., Plateau, J., Puppo, C., Gontero, B. (2012). Regulation of glyceraldehyde-3-phosphate dehydrogenase in the eustigmatophyte Pseudocharaciopsis ovalis is intermediate between a chlorophyte and a diatom. Eur. J. Phycol. 47, 207–215. doi: 10.1080/09670262.2012.687459
Baalmann, E., Backhausen, J. E., Kitzmann, C., Scheibe, R. (1994). Regulation of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase-activity in spinach-chloroplasts. Bot. Acta 107, 313–320. doi: 10.1111/j.1438-8677.1994.tb00801.x
Bach, L. T., Taucher, J. (2019). CO2 effects on diatoms: a synthesis of more than a decade of ocean acidification experiments with natural communities. Ocean Sci. 15, 1159–1175. doi: 10.5194/os-15-1159-2019
Bai, X., Song, H., Lavoie, M., Zhu, K., Su, Y., Ye, H., et al. (2016). Proteomic analyses bring new insights into the effect of a dark stress on lipid biosynthesis in Phaeodactylum tricornutum. Sci. Rep. 6, 25494. doi: 10.1038/srep25494
Bailleul, B., Berne, N., Murik, O., Petroutsos, D., Prihoda, J., Tanaka, A., et al. (2015). Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. doi: 10.1038/nature14599
Basu, S., Patil, S., Mapleson, D., Russo, M. T., Vitale, L., Fevola, C., et al. (2017). Finding a partner in the ocean: molecular and evolutionary bases of the response to sexual cues in a planktonic diatom. New Phytol. 215, 140–156. doi: 10.1111/nph.14557
Beganton, B., Coyaud, E., Mange, A., Solassol, J. (2019). New approaches for protein-protein interaction study. Med. Sci. 35, 223–231. doi: 10.1051/medsci/2019035
Belen Bosco, M., Cristina Aleanzi, M., Alvaro Iglesias, A. (2012). Plastidic phosphoglycerate kinase from Phaeodactylum tricornutum: on the critical role of cysteine residues for the enzyme function. Protist 163, 188–203. doi: 10.1016/j.protis.2011.07.001
Boelen, P., De Poll, W. H. V., Van Der Strate, H. J., Neven, I. A., Beardall, J., Buma, A. G. J. (2011). Neither elevated nor reduced CO2 affects the photophysiological performance of the marine Antarctic diatom Chaetoceros brevis. J. Exp. Mar. Biol. Ecol. 406, 38–45. doi: 10.1016/j.jembe.2011.06.012
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Brembu, T., Winge, P., Tooming-Klunderud, A., Nederbragt, A. J., Jakobsen, K. S., Bones, A. M. (2014). The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genom. 16, 17–27. doi: 10.1016/j.margen.2013.12.002
Buchanan, B. B. (1980). Role of light in the regulation of chloroplast nzymes. Ann. Rev. Plant Physiol. 31, 341–374. doi: 10.1146/annurev.pp.31.060180.002013
Buchanan, B. B. (2017). The path to thioredoxin and redox regulation beyond chloroplasts. Plant Cell Physiol. 58, 1826–1832. doi: 10.1093/pcp/pcx119
Burkhardt, S., Amoroso, G., Riebesell, U., Sultemeyer, D. (2001). CO2 and HCO3- uptake in marine diatoms acclimated to different CO2 concentrations. Limnol. Oceanogr. 46, 1378–1391. doi: 10.4319/lo.2001.46.6.1378
Cardol, P. (2011). Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: a highly conserved subunit composition highlighted by mining of protein databases. Biochim. Biophys. Acta 1807, 1390–1397. doi: 10.1016/j.bbabio.2011.06.015
Carreto, J. I., Catoggio, J. A. (1976). Variations in pigment contents of the diatom Phaeodactylum tricornutum during growth. Mar. Biol. 36, 105–112. doi: 10.1007/BF00388433
Castruita, M., Casero, D., Karpowicz, S. J., Kropat, J., Vieler, A., Hsieh, S. I., et al. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292. doi: 10.1105/tpc.111.084400
Cavalier-Smith, T., Chao, E. E. Y. (2006). Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 62, 388–420. doi: 10.1007/s00239-004-0353-8
Cavalier-Smith, T. (2004). “Chromalveolate diversity and cell megaevolution: interplay of membranes, genomes and cytoskeleton,” in Organelles, genomes and eukaryote phylogeny: an evolutionary synthesis in the age of genomics. Eds. Hirt, R. P., Horner, D. S. (London, UK: CRC Press LLC), 75–108.
Chauton, M. S., Winge, P., Brembu, T., Vadstein, O., Bones, A. M. (2013). Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 161, 1034–1048. doi: 10.1104/pp.112.206177
Chen, Z., Yang, M. K., Li, C. Y., Wang, Y., Zhang, J., Wang, D. B., et al. (2014). Phosphoproteomic analysis provides novel insights into stress responses in Phaeodactylum tricornutum, a model diatom. J. Proteome. Res. 13, 2511–2523. doi: 10.1021/pr401290u
Chen, X.-H., Li, Y.-Y., Zhang, H., Liu, J.-L., Xie, Z.-X., Lin, L., et al. (2018). Quantitative proteomics reveals common and specific responses of a marine diatom Thalassiosira pseudonana to different macronutrient deficiencies. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02761
Clement, R., Jensen, E., Prioretti, L., Maberly, S. C., Gontero, B. (2017a). Diversity of CO2-concentrating mechanisms and responses to CO2 concentration in marine and freshwater diatoms. J. Exp. Bot. 68, 3925–3935. doi: 10.1093/jxb/erx035
Clement, R., Lignon, S., Mansuelle, P., Jensen, E., Pophillat, M., Lebrun, R., et al. (2017b). Responses of the marine diatom Thalassiosira pseudonana to changes in CO2 concentration: a proteomic approach. Sci. Rep. 7. doi: 10.1038/srep42333
Colman, B., Rotatore, C. (1988). Uptake and accumulation of inorganic carbon by a fresh-water diatom. J. Exp. Bot. 39, 1025–1032. doi: 10.1093/jxb/39.8.1025
Colman, B., Rotatore, C. (1995). Photosynthetic inorganic carbon uptake and accumulation in two marine diatoms. Plant Cell Environ. 18, 919–924. doi: 10.1111/j.1365-3040.1995.tb00601.x
Daboussi, F., Leduc, S., Marechal, A., Dubois, G., Guyot, V., Perez-Michaut, C., et al. (2014). Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5, 3831. doi: 10.1038/ncomms4831
Del Prete, S., Nocentini, A., Supuran, C. T., Capasso, C. (2020). Bacterial iota-carbonic anhydrase: a new active class of carbonic anhydrase identified in the genome of the Gram-negative bacterium Burkholderia territorii. J. Enzyme Inhib. Med. Chem. 35, 1060–1068. doi: 10.1080/14756366.2020.1755852
Deschamps, P., Moreira, D. (2012). Reevaluating the green contribution to diatom genomes. Genome Biol. Evol. 4, 795–800. doi: 10.1093/gbe/evs053
Dickson, A. G. (2010). “The carbon dioxide system in seawater: equilibrium chemistry and measurements,” in Guide to best practices for ocean acidification research and data reporting. Eds. Riebesell, U., Fabry, V. J., Hansson, L., Gattuso, J. P. (Luxembourg: Publications Office of the European Union), 17–40.
Dimario, R. J., Machingura, M. C., Waldrop, G. L., Moroney, J. V. (2018). The many types of carbonic anhydrases in photosynthetic organisms. Plant Sci. 268, 11–17. doi: 10.1016/j.plantsci.2017.12.002
Dorrell, R. G., Gile, G., Mccallum, G., Meheust, R., Bapteste, E. P., Klinger, C. M., et al. (2017). Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. Elife 6, e23717. doi: 10.7554/eLife.23717
Dupont, C. L., Goepfert, T. J., Lo, P., Wei, L. P., Ahnerz, B. A. (2004). Diurnal cycling of glutathione in marine phytoplankton: field and culture studies. Limnol. Oceanogr. 49, 991–996. doi: 10.4319/lo.2004.49.4.0991
Dutkiewicz, S., Morris, J. J., Follows, M. J., Scott, J., Levitan, O., Dyhrman, S. T., et al. (2015). Impact of ocean acidification on the structure of future phytoplankton communities. Nat. Clim. Change 5, 1002–1006. doi: 10.1038/nclimate2722
Ewe, D., Tachibana, M., Kikutani, S., Gruber, A., Bartulos, C. R., Konert, G., et al. (2018). The intracellular distribution of inorganic carbon fixing enzymes does not support the presence of a C4 pathway in the diatom Phaeodactylum tricornutum. Photosynth. Res. 137, 263–280. doi: 10.1007/s11120-018-0500-5
Fabris, M., Matthijs, M., Rombauts, S., Vyverman, W., Goossens, A., Baart, G. J. E. (2012). The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 70, 1004–1014. doi: 10.1111/j.1365-313X.2012.04941.x
Falciatore, A., Jaubert, M., Bouly, J.-P., Bailleul, B., Mock, T. (2020). Diatom molecular research comes of age: model species for studying phytoplankton biology and diversity. Plant Cell 32, 547–572. doi: 10.1105/tpc.19.00158
Falkowski, P. G., Owens, T. G. (1980). Light-shade adaptation - 2 strategies in marine-phytoplankton. Plant Physiol. 66, 592–595. doi: 10.1104/pp.66.4.592
Falkowski, P. G., Barber, R. T., Smetacek, V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. doi: 10.1126/science.281.5374.200
Fey, V., Wagner, R., Brautigam, K., Pfannschmidt, T. (2005). Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 56, 1491–1498. doi: 10.1093/jxb/eri180
Fournier, M. L., Paulson, A., Pavelka, N., Mosley, A. L., Gaudenz, K., Bradford, W. D., et al. (2010). Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell. Proteomics 9, 271–284. doi: 10.1074/mcp.M900415-MCP200
Gao, K., Campbell, D. A. (2014). Photophysiological responses of marine diatoms to elevated CO2 and decreased pH: a review. Funct. Plant Biol. 41, 449–459. doi: 10.1071/FP13247
Gardemann, A., Stitt, M., Heldt, H. W. (1983). Control of CO2 fixation. Regulation of spinach ribulose-5-phosphate kinase by stromal metabolite levels. BBA-Bioenergetics 722, 51–60. doi: 10.1016/0005-2728(83)90156-1
Gattuso, J. P., Magnan, A., Billé, R., Cheung, W. W. L., Howes, E. L., Joos, F., et al. (2015). Contrasting futures for ocean and society from different anthropogenic CO2 emissions scenarios. Science 349, aac4722. doi: 10.1126/science.aac4722780
Gilstad, M., Johnsen, G., Sakshaug, E. (1993). Photosynthetic parameters, pigment composition and respiration rates of the marine diatom Skeletonema-costatum grown in continuous light and a 12/12 h light-dark cycle. J. Plankton Res. 15, 939–951. doi: 10.1093/plankt/15.8.939
Giordano, M., Chen, Y. B., Koblizek, M., Falkowski, P. G. (2005). Regulation of nitrate reductase in Chlamydomonas reinhardtii by the redox state of the plastoquinone pool. Eur. J. Phycol. 40, 345–352. doi: 10.1080/09670260500334263
Gontero, B., Avilan, L., Lebreton, S. (2007). “Control of carbon fixation in chloroplasts,” in Annual plant reviews. Eds. Plaxton, W. C., Mcmanus, M. T. (Oxford: Blackwell Publishing), 187–218. doi: 10.1002/9780470988640.ch7
Grabsztunowicz, M., Koskela, M. M., Mulo, P. (2017). Post-translational modifications in regulation of chloroplast function: recent advances. Front. Plant Sci. 8, 1–12. doi: 10.3389/fpls.2017.00240
Green, B. R. (2011). After the primary endosymbiosis: an update on the chromalveolate hypothesis and the origins of algae with Chl c. Photosynth. Res. 107, 103–115. doi: 10.1007/s11120-010-9584-2
Gruber, A., Weber, T., Bártulos, C. R., Vugrinec, S., Kroth, P. G. (2009). Intracellular distribution of the reductive and oxidative pentose phosphate pathways in two diatoms. J. Basic Microb. 49, 58–72. doi: 10.1002/jobm.200800339
Haimovich-Dayan, M., Garfinkel, N., Ewe, D., Marcus, Y., Gruber, A., Wagner, H., et al. (2013). The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol. 197, 177–185. doi: 10.1111/j.1469-8137.2012.04375.x
Harada, H., Matsuda, Y. (2005). Identification and characterization of a new carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Can. J. Bot. 83, 909–916. doi: 10.1139/b05-078
Harada, H., Nakatsuma, D., Ishida, M., Matsuda, Y. (2005). Regulation of the expression of intracellular beta-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 139, 1041–1050. doi: 10.1104/pp.105.065185
Harada, H., Nakajima, K., Sakaue, K., Matsuda, Y. (2006). CO2 sensing at ocean surface mediated by cAMP in a marine diatom. Plant Physiol. 142, 1318–1328. doi: 10.1104/pp.106.086561
Harrison, P. J., Thompson, P. A., Calderwood, G. S. (1990). Effects of nutrient and light limitation on the biochemical composition of phytoplankton. J. Appl. Phycol. 2, 45–56. doi: 10.1007/BF02179768
Hauser, M., Eichelmann, H., Oja, V., Heber, U., Laisk, A. (1995). Stimulation by light of rapid pH regulation in the chloroplast stroma in vivo as indicated by CO2 solubilization in leaves. Plant Physiol. 108, 1059. doi: 10.1104/pp.108.3.1059
Hennon, G. M. M., Ashworth, J., Groussman, R. D., Berthiaume, C., Morales, R. L., Baliga, N. S., et al. (2015). Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression. Nat. Clim. Change 5, 761–765. doi: 10.1038/nclimate2683
Herve, V., Derr, J., Douady, S., Quinet, M., Moisan, L., Lopez, P. J. (2012). Multiparametric analyses reveal the pH-dependence of silicon biomineralization in diatoms. PloS One 7, e46722. doi: 10.1371/journal.pone.0046722
Hesse, S. J. A., Ruijter, G. J. G., Dijkema, C., Visser, J. (2000). Measurement of intracellular (compartmental) pH by 31P NMR in. Aspergillus Niger J. Biotechnol. 77 , 5–15. doi: 10.1016/S0168-1656(99)00203-5
Heydarizadeh, P., Boureba, W., Zahedi, M., Huang, B., Moreau, B., Lukomska, E., et al. (2017). Response of CO2-starved diatom Phaeodactylum tricornutum to light intensity transition. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160396. doi: 10.1098/rstb.2016.0396
Heydarizadeh, P., Veidl, B., Huang, B., Lukomska, E., Wielgosz-Collin, G., Couzinet-Mossion, A., et al. (2019). Carbon orientation in the diatom Phaeodactylum tricornutum: the effects of carbon limitation and photon flux density. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00471
Hildebrand, M., Manandhar-Shrestha, K., Abbriano, R. (2017). Effects of chrysolaminarin synthase knockdown in the diatom Thalassiosira pseudonana: Implications of reduced carbohydrate storage relative to green algae. Algal Res. 23, 66–77. doi: 10.1016/j.algal.2017.01.010
Hildebrand, M., Lerch, S. J. L., Shrestha, R. P. (2018). Understanding diatom cell wall silicification—moving forward. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00125
Hitchcock, G. L. (1980). Diel variation in chlorophyll a, carbohydrate and protein content of the marine diatom Skeletonema costatum. Mar. Biol. 57, 271–278. doi: 10.1007/BF00387570
Hopes, A., Nekrasov, V., Kamoun, S., Mock, T. (2016). Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 12, 49. doi: 10.1186/s13007-016-0148-0
Hopkinson, B. M., Dupont, C. L., Allen, A. E., Morel, F. M. M. (2011). Efficiency of the CO2-concentrating mechanism of diatoms. Proc. Natl. Acad. Sci. U.S.A. 108, 3830–3837. doi: 10.1073/pnas.1018062108
Hopkinson, B. M., Dupont, C. L., Matsuda, Y. (2016). The physiology and genetics of CO2 concentrating mechanisms in model diatoms. Curr. Opin. Plant Biol. 31, 51–57. doi: 10.1016/j.pbi.2016.03.013
Huang, W., Daboussi, F. (2017). Genetic and metabolic engineering in diatoms. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160411. doi: 10.1098/rstb.2016.0411
Huang, W., Haferkamp, I., Lepetit, B., Molchanova, M., Hou, S., Jeblick, W., et al. (2018). Reduced vacuolar β-1,3-glucan synthesis affects carbohydrate metabolism as well as plastid homeostasis and structure in Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. U.S.A. 115, 4791–4796. doi: 10.1073/pnas.1719274115
Huysman, M. J. J., Fortunato, A. E., Matthijs, M., Costa, B. S., Vanderhaeghen, R., Van Den Daele, H., et al. (2013). AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25, 215–228. doi: 10.1105/tpc.112.106377
IPCC (2014). “Climate Change. Synthesis Report,” in Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, vol. 151. Eds. Core Writing Team, Pachauri, R. K., Meyer, L. A. (Geneva, Switzerland: IPCC).
Jensen, E., Clement, R., Maberly, S. C., Gontero, B. (2017). Regulation of the Calvin-Benson-Bassham cycle in the enigmatic diatoms: biochemical and evolutionary variations on an original theme. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160401. doi: 10.1098/rstb.2016.0401
Jensen, E. L., Clement, R., Kosta, A., Maberly, S. C., Gontero, B. (2019a). A new widespread subclass of carbonic anhydrase in marine phytoplankton. ISME J. 13, 2094–2106. doi: 10.1038/s41396-019-0426-8
Jensen, E. L., Yanguez, K., Carriere, F., Gontero, B. (2019b). Storage compound accumulation in diatoms as response to elevated CO2 concentration. Biology 9, 5. doi: 10.3390/biology9010005
Jensen, E. L., Maberly, S. C., Gontero, B. (2020). Insights on the functions and ecophysiological relevance of the diverse carbonic anhydrases in microalgae. Int. J. Mol. Sci. 21, 2922. doi: 10.3390/ijms21082922
Keeling, P. J. (2009). Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 56, 1–8. doi: 10.1111/j.1550-7408.2008.00371.x
Kikutani, S., Tanaka, R., Yamazaki, Y., Hara, S., Hisabori, T., Kroth, P. G., et al. (2012). Redox regulation of carbonic anhydrases via thioredoxin in chloroplast of the marine diatom Phaeodactylum tricornutum. J. Biol. Chem. 287, 20689–20700. doi: 10.1074/jbc.M111.322743
Kim, J.-M., Lee, K., Shin, K., Kang, J.-H., Lee, H.-W., Kim, M., et al. (2006). The effect of seawater CO2 concentration on growth of a natural phytoplankton assemblage in a controlled mesocosm experiment. Limnol. Oceanogr. 51, 1629–1636. doi: 10.4319/lo.2006.51.4.1629
Knuesting, J., Scheibe, R. (2018). Small molecules govern thiol redox switches. Trends Plant Sci. 23, 769–782. doi: 10.1016/j.tplants.2018.06.007
Kojadinovic-Sirinelli, M., Villain, A., Puppo, C., Sing, S. F., Prioretti, L., Hubert, P., et al. (2018). Exploring the microbiome of the “star” freshwater diatom Asterionella formosa in a laboratory context. Environ. Microbiol. 20, 3601–3615. doi: 10.1111/1462-2920.14337
Kroth, P. G., Chiovitti, A., Gruber, A., Martin-Jezequel, V., Mock, T., Parker, M. S., et al. (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 3, e1426. doi: 10.1371/journal.pone.0001426
Kroth, P. G., Bones, A. M., Daboussi, F., Ferrante, M. I., Jaubert, M., Kolot, M., et al. (2018). Genome editing in diatoms: achievements and goals. Plant Cell. Rep. 37, 1401–1408. doi: 10.1007/s00299-018-2334-1
Kuczynska, P., Jemiola-Rzeminska, M., Strzalka, K. (2015). Photosynthetic pigments in diatoms. Mar. Drugs 13, 5847–5881. doi: 10.3390/md13095847
Kustka, A. B., Milligan, A. J., Zheng, H., New, A. M., Gates, C., Bidle, K. D., et al. (2014). Low CO2 results in a rearrangement of carbon metabolism to support C4 photosynthetic carbon assimilation in Thalassiosira pseudonana. New Phytol. 204, 507–520. doi: 10.1111/nph.12926
Lauritano, C., Ferrante, M. I., Rogato, A. (2019). Marine natural products from microalgae: an -omics overview. Mar. Drugs 17, 269. doi: 10.3390/md17050269
Lavaud, J., Materna, A. C., Sturm, S., Vugrinec, S., Kroth, P. G. (2012). Silencing of the violaxanthin de-epoxidase gene in the diatom Phaeodactylum tricornutum reduces diatoxanthin synthesis and non-photochemical quenching. PloS One 7, e36806. doi: 10.1371/journal.pone.0036806
Lin, Q., Liang, J. R., Huang, Q. Q., Luo, C. S., Anderson, D. M., Bowler, C., et al. (2017). Differential cellular responses associated with oxidative stress and cell fate decision under nitrate and phosphate limitations in Thalassiosira pseudonana: Comparative proteomics. PloS One 12, e0184849. doi: 10.1371/journal.pone.0184849
Lohr, M., Wilhelm, C. (1999). Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc. Natl. Acad. Sci. U.S.A. 96, 8784–8789. doi: 10.1073/pnas.96.15.8784
Loiselle, F. B., Casey, J. R. (2010). “Measurement of intracellular pH,” in Membrane transporters in drug discovery and development: methods and protocols. Ed. Yan, Q. (Totowa, NJ: Humana Press), 311–331.
Maberly, S. C., Gontero, B. (2017). Ecological imperatives for aquatic CO2-concentrating mechanisms. J. Exp. Bot. 68, 3797–3814. doi: 10.1093/jxb/erx201
Maberly, S. C., Courcelle, C., Groben, R., Gontero, B. (2010). Phylogenetically-based variation in the regulation of the Calvin cycle enzymes, phosphoribulokinase and glyceraldehyde-3-phosphate dehydrogenase, in algae. J. Exp. Bot. 61, 735–745. doi: 10.1093/jxb/erp337
Maier, T., Güell, M., Serrano, L. (2009). Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973. doi: 10.1016/j.febslet.2009.10.036
Mann, D. G., Vanormelingen, P. (2013). An inordinate fondness? The number, distributions, and origins of diatom species. J. Eukaryot. Microbiol. 60, 414–420. doi: 10.1111/jeu.12047
Marri, L., Thieulin-Pardo, G., Lebrun, R., Puppo, R., Zaffagnini, M., Trost, P., et al. (2014). CP12-mediated protection of Calvin-Benson cycle enzymes from oxidative stress. Biochimie 97, 228–237. doi: 10.1016/j.biochi.2013.10.018
Matsuda, Y., Kroth, P. G. (2014). ““Carbon fixation in diatoms,”,” in The structural basis of biological energy generation. Advances in photosynthesis and respiration (including bioenergy and related processes). Ed. Hohmann-Marriott, M. F. (Dordrecht: Springer Netherlands), 335–362.
Matsuda, Y., Nakajima, K., Tachibana, M. (2011). Recent progresses on the genetic basis of the regulation of CO2 acquisition systems in response to CO2 concentration. Photosynth. Res. 109, 191–203. doi: 10.1007/s11120-011-9623-7
Matsuda, Y., Hopkinson, B. M., Nakajima, K., Dupont, C. L., Tsuji, Y. (2017). Mechanisms of carbon dioxide acquisition and CO2 sensing in marine diatoms: a gateway to carbon metabolism. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160403. doi: 10.1098/rstb.2016.0403
Medlin, L. K. (2016). Evolution of the diatoms: major steps in their evolution and a review of the supporting molecular and morphological evidence. Phycologia 55, 79–103. doi: 10.2216/15-105.1
Mekhalfi, M., Avilan, L., Lebrun, R., Botebol, H., Gontero, B. (2012). Consequences of the presence of 24-epibrassinolide, on cultures of a diatom, Asterionella formosa. Biochimie 94, 1213–1220. doi: 10.1016/j.biochi.2012.02.011
Mekhalfi, M., Puppo, C., Avilan, L., Lebrun, R., Mansuelle, P., Maberly, S. C., et al. (2014). Glyceraldehyde-3-phosphate dehydrogenase is regulated by ferredoxin-NADP reductase in the diatom Asterionella formosa. New Phytol. 203, 414–423. doi: 10.1111/nph.12820
Michels, A. K., Wedel, N., Kroth, P. G. (2005). Diatom plastids possess a phosphoribulokinase with an altered regulation and no oxidative pentose phosphate pathway. Plant Physiol. 137, 911–920. doi: 10.1104/pp.104.055285
Mock, T., Otillar, R. P., Strauss, J., Mcmullan, M., Paajanen, P., Schmutz, J., et al. (2017). Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540. doi: 10.1038/nature20803
Montrichard, F., Alkhalfioui, F., Yano, H., Vensel, W. H., Hurkman, W. J., Buchanan, B. B. (2009). Thioredoxin targets in plants: the first 30 years. J. Proteomics 72, 452–474. doi: 10.1016/j.jprot.2008.12.002
Morel, F. M. M., Lam, P. J., Saito, M. A. (2020). Trace metal substitution in marine phytoplankton. Annu. Rev. Earth Planet. Sci. 48, 491–517. doi: 10.1146/annurev-earth-053018-060108
Moustafa, A., Beszteri, B., Maier, U. G., Bowler, C., Valentin, K., Bhattacharya, D. (2009). Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726. doi: 10.1126/science.1172983
Mueller-Cajar, O., Stotz, M., Wendler, P., Hartl, F. U., Bracher, A., Hayer-Hartl, M. (2011). Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199. doi: 10.1038/nature10568
Muhseen, Z. T., Xiong, Q., Chen, Z., Ge, F. (2015). Proteomics studies on stress responses in diatoms. Proteomics 15, 3943–3953. doi: 10.1002/pmic.201500165
Nakajima, K., Tanaka, A., Matsuda, Y. (2013). SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc. Natl. Acad. Sci. U.S.A. 110, 1767–1772. doi: 10.1073/pnas.1216234110
Nikolaev, V. O., Lohse, M. J. (2006). Monitoring of cAMP synthesis and degradation in living cells. Physiology 21, 86–92. doi: 10.1152/physiol.00057.2005
Nonoyama, T., Kazamia, E., Nawaly, H., Gao, X., Tsuji, Y., Matsuda, Y., et al. (2019). Metabolic innovations underpinning the origin and diversification of the diatom chloroplast. Biomolecules 9, 322. doi: 10.3390/biom9080322
Norici, A., Bazzoni, A. M., Pugnetti, A., Raven, J. A., Giordano, M. (2011). Impact of irradiance on the C allocation in the coastal marine diatom Skeletonema marinoi Sarno and Zingone. Plant Cell Environ. 34, 1666–1677. doi: 10.1111/j.1365-3040.2011.02362.x
Nymark, M., Sharma, A. K., Sparstad, T., Bones, A. M., Winge, P. (2016). A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6, 24951. doi: 10.1038/srep24951
Ogura, A., Akizuki, Y., Imoda, H., Mineta, K., Gojobori, T., Nagai, S. (2018). Comparative genome and transcriptome analysis of diatom, Skeletonema costatum, reveals evolution of genes for harmful algal bloom. BMC Genomics 19, 765. doi: 10.1186/s12864-018-5144-5
Ohno, N., Inoue, T., Yamashiki, R., Nakajima, K., Kitahara, Y., Ishibashi, M., et al. (2012). CO2-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-Like basic zipper domain in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 158, 499–513. doi: 10.1104/pp.111.190249
Oudot-Le Secq, M. P., Grimwood, J., Shapiro, H., Armbrust, E. V., Bowler, C., Green, B. R. (2007). Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277, 427–439. doi: 10.1007/s00438-006-0199-4
Perez-Perez, M. E., Mauries, A., Maes, A., Tourasse, N. J., Hamon, M., Lemaire, S. D., et al. (2017). The deep thioredoxome in Chlamydomonas reinhardtii: new insights into redox regulation. Mol. Plant 10, 1107–1125. doi: 10.1016/j.molp.2017.07.009
Ponnala, L., Wang, Y., Sun, Q., Van Wijk, K. J. (2014). Correlation of mRNA and protein abundance in the developing maize leaf. Plant J. 78, 424–440. doi: 10.1111/tpj.12482
Portis, A. R., Heldt, H. W. (1976). Light-dependent changes of Mg2+ concentration in stroma in relation to Mg2+ dependency of CO2 fixation in intact chloroplasts. BBA-Bioenergetics 449, 434–446. doi: 10.1016/0005-2728(76)90154-7
Prabakaran, S., Lippens, G., Steen, H., Gunawardena, J. (2012). Post-translational modification: nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wires Syst. Biol. Med. 4, 565–583. doi: 10.1002/wsbm.1185
Prosser, G. A., Larrouy-Maumus, G., De Carvalho, L. P. (2014). Metabolomic strategies for the identification of new enzyme functions and metabolic pathways. EMBO Rep. 15, 657–669. doi: 10.15252/embr.201338283
Pupillo, P., Giulianipiccari, G. (1975). The reversible depolymerization of spinach chloroplast glyceraldehyde-phosphate dehydrogenase-interaction with nucleotides and dithiothreitol. Eur. J. Biochem. 51, 475–482. doi: 10.1111/j.1432-1033.1975.tb03947.x
Raven, J., Brown, K., Mackay, M., Beardall, J., Giordano, M., Granum, E., et al. (2005). “Iron, nitrogen, phosphorus and zinc cycling and consequences for primary productivity in the oceans,” in Micro-organisms and earth systems-advances in geomicrobiology. Eds. Gadd, G., Semple, K., Lappin-Scott, H. (Cambridge, UK: Cambridge University Press), 247–272.
Raven, J. A. (1994). Carbon fixation and carbon availability in marine phytoplankton. Photosynth. Res. 39, 259–273. doi: 10.1007/BF00014587
Reinfelder, J. R., Kraepiel, A. M. L., Morel, F. M. M. (2000). Unicellular C4 photosynthesis in a marine diatom. Nature 407, 996–999. doi: 10.1038/35039612
Reinfelder, J. R. (2011). “Carbon concentrating mechanisms in eukaryotic marine phytoplankton,” in Annu. Rev. Mar. Sci. Eds. Carlson, C. A., Giovannoni, S. J. (Palo Alto, USA: Annual Reviews Publisher), 291–315.
Riebesell, U., Wolfgladrow, D. A., Smetacek, V. (1993). Carbon dioxide limitation of marine phytoplankton growth rates. Nature 361, 249–251. doi: 10.1038/361249a0
Rosenwasser, S., Graff Van Creveld, S., Schatz, D., Malitsky, S., Tzfadia, O., Aharoni, A., et al. (2014). Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc. Natl. Acad. Sci. U.S.A. 111, 2740–2745. doi: 10.1073/pnas.1319773111
Sage, R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. doi: 10.1111/j.1469-8137.2004.00974.x
Samukawa, M., Shen, C., Hopkinson, B. M., Matsuda, Y. (2014). Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth. Res. 121, 235–249. doi: 10.1007/s11120-014-9967-x
Santin, Y. G., Doan, T., Lebrun, R., Espinosa, L., Journet, L., Cascales, E. (2018). In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat. Microbiol. 3, 1304–1313. doi: 10.1038/s41564-018-0234-3
Satoh, D., Hiraoka, Y., Colman, B., Matsuda, Y. (2001). Physiological and molecular biological characterization of intracellular carbonic anhydrase from the marine diatom Phaeodactylum tricornutum. Plant Physiol. 126, 1459–1470. doi: 10.1104/pp.126.4.1459
Schoefs, B., Hu, H., Kroth, P. G. (2017). The peculiar carbon metabolism in diatoms. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160405. doi: 10.1098/rstb.2016.0405
Schurmann, P., Jacquot, J. P. (2000). Plant thioredoxin systems revisited. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371–400. doi: 10.1146/annurev.arplant.51.1.371
Stein, K. C., Frydman, J. (2019). The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J. Biol. Chem. 294, 2076–2084. doi: 10.1074/jbc.REV118.002814
Stiller, J. W., Schreiber, J., Yue, J., Guo, H., Ding, Q., Huang, J. (2014). The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 5, 5764. doi: 10.1038/ncomms6764
Subramanian, S., Barry, A. N., Pieris, S., Sayre, R. T. (2013). Comparative energetics and kinetics of autotrophic lipid and starch metabolism in chlorophytic microalgae: implications for biomass and biofuel production. Biotechnol. Biofuels 6, 150. doi: 10.1186/1754-6834-6-150
Sun, N., Ma, L. G., Pan, D. Y., Zhao, H. Y., Deng, X. W. (2003). Evaluation of light regulatory potential of Calvin cycle steps based on large-scale gene expression profiling data. Plant Mol. Biol. 53, 467–478. doi: 10.1023/B:PLAN.0000019071.12878.9e
Tachibana, M., Allen, A. E., Kikutani, S., Endo, Y., Bowler, C., Matsuda, Y. (2011). Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth. Res. 109, 205–221. doi: 10.1007/s11120-011-9634-4
Tanaka, T., Maeda, Y., Veluchamy, A., Tanaka, M., Abida, H., Marechal, E., et al. (2015). Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27, 162–176. doi: 10.1105/tpc.114.135194
Tanaka, A., Ohno, N., Nakajima, K., Matsuda, Y. (2016). Light and CO2/cAMP signal cross talk on the promoter elements of chloroplastic beta-carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 170, 1105–1116. doi: 10.1104/pp.15.01738
Taraldsvik, M., Myklestad, S. M. (2000). The effect of pH on growth rate, biochemical composition and extracellular carbohydrate production of the marine diatom Skeletonema costatum. Eur. J. Phycol. 35, 189–194. doi: 10.1080/09670260010001735781
Thieulin-Pardo, G., Remy, T., Lignon, S., Lebrun, R., Gontero, B. (2015). Phosphoribulokinase from Chlamydomonas reinhardtii: a Benson-Calvin cycle enzyme enslaved to its cysteine residues. Mol. Biosyst. 11, 1134–1145. doi: 10.1039/C5MB00035A
Torstensson, A., Chierici, M., Wulff, A. (2012). The influence of increased temperature and carbon dioxide levels on the benthic/sea ice diatom Navicula directa. Polar Biol. 35, 205–214. doi: 10.1007/s00300-011-1056-4
Tortell, P. D. (2000). Evolutionary and ecological perspectives on carbon acquisition in phytoplankton. Limnol. Oceanogr. 45, 744–750. doi: 10.4319/lo.2000.45.3.0744
Traller, J. C., Cokus, S. J., Lopez, D. A., Gaidarenko, O., Smith, S. R., Mccrow, J. P., et al. (2016). Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol. Biofuels 9, 258. doi: 10.1186/s13068-016-0670-3
Tsuji, Y., Nakajima, K., Matsuda, Y. (2017). Molecular aspects of the biophysical CO2-concentrating mechanism and its regulation in marine diatoms. J. Exp. Bot. 68, 3763–3772. doi: 10.1093/jxb/erx173
Valegard, K., Andralojc, P. J., Haslam, R. P., Pearce, F. G., Eriksen, G. K., Madgwick, P. J., et al. (2018). Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications. J. Biol. Chem. 293, 13033–13043. doi: 10.1074/jbc.RA118.003518
Valenzuela, J. J., Garcia De Lomana, A. L., Lee, A., Armbrust, E. V., Orellana, M. V., Baliga, N. S. (2018). Ocean acidification conditions increase resilience of marine diatoms. Nat. Commun. 9, 2328. doi: 10.1038/s41467-018-04742-3
Van Der Star, W. R., Dijkema, C., De Waard, P., Picioreanu, C., Strous, M., Van Loosdrecht, M. C. (2010). An intracellular pH gradient in the anammox bacterium Kuenenia stuttgartiensis as evaluated by 31P NMR. Appl. Microbiol. Biot. 86, 311–317. doi: 10.1007/s00253-009-2309-9
Villain, A., Kojadinovic, M., Puppo, C., Prioretti, L., Hubert, P., Zhang, Y., et al. (2017). Complete mitochondrial genome sequence of the freshwater diatom Asterionella formosa. Mitochondrial DNA B 2, 97–98. doi: 10.1080/23802359.2017.1285210
Weber, T., Gruber, A., Kroth, P. G. (2009). The presence and localization of thioredoxins in diatoms, unicellular algae of secondary endosymbiotic origin. Mol. Plant 2, 468–477. doi: 10.1093/mp/ssp010
Wei, L., El Hajjami, M., Shen, C., You, W., Lu, Y., Li, J., et al. (2019). Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol. Biofuels 12, 168. doi: 10.1186/s13068-019-1506-8
Werdan, K., Heldt, H. W. (1973). Measurement of pH value in stroma and thylakoid space of intact chloroplasts. Hoppe Seylers Z. Physiol. Chem. 354, 223–224.
Werdan, K., Heldt, H. W., Milovancev, M. (1975). The role of pH in regulation of carbon fixation in chloroplast stroma. Studies on CO2 fixation in light and dark. BBA-Bioenergetics 396, 276–292. doi: 10.1016/0005-2728(75)90041-9
Wilhelm, C., Buechel, C., Fisahn, J., Goss, R., Jakob, T., Laroche, J., et al. (2006). The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae. Protist 157, 91–124. doi: 10.1016/j.protis.2006.02.003
Young, J. N., Morel, F. M. M. (2015). The CO2 switch in diatoms. Nat. Clim. Change 5, 722–723. doi: 10.1038/nclimate2691
Young, J. N., Heureux, A. M. C., Sharwood, R. E., Rickaby, R. E. M., Morel, F. M. M., Whitney, S. M. (2016). Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J. Exp. Bot. 67, 3445–3456. doi: 10.1093/jxb/erw163
Keywords: Calvin cycle, CO2 concentrating mechanism, Phaeodactylum tricornutum, redox regulation, Thalassiosira pseudonana
Citation: Launay H, Huang W, Maberly SC and Gontero B (2020) Regulation of Carbon Metabolism by Environmental Conditions: A Perspective From Diatoms and Other Chromalveolates. Front. Plant Sci. 11:1033. doi: 10.3389/fpls.2020.01033
Received: 29 April 2020; Accepted: 23 June 2020;
Published: 16 July 2020.
Edited by:
Benoit Schoefs, Le Mans Université, FranceReviewed by:
Tore Brembu, Norwegian University of Science and Technology, NorwayCopyright © 2020 Launay, Huang, Maberly and Gontero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brigitte Gontero, Ym1ldW5pZXJAaW1tLmNucnMuZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.