
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 July 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01014
This article is part of the Research TopicUnderstanding C4 Evolution and FunctionView all 12 articles
C4 photosynthesis results from a set of anatomical features and biochemical components that act together to concentrate CO2 within the leaf and boost productivity. This complex trait evolved independently many times, resulting in various realizations of the phenotype, but in all C4 plants the primary fixation of atmospheric carbon is catalyzed by phosphoenolpyruvate carboxylase. Comparisons of C4 and non-C4 PEPC from a few closely related species suggested that the enzyme was modified to meet the demands of the C4 cycle. However, very few C4 groups have been investigated, hampering general conclusions. To test the hypothesis that distant C4 lineages underwent convergent biochemical changes, we compare the kinetic variation between C4 and non-C4 PEPC from a previously assessed young lineage (Flaveria, Asteraceae) with those from an older lineage found within the distantly related grass family (Panicum). Despite the evolutionary distance, the kinetic changes between the non-C4 and C4 PEPC are qualitatively similar, with a decrease in sensitivity for inhibitors, an increased specificity (kcat/Km) for bicarbonate, and a decreased specificity (kcat/Km) for PEP. The differences are more pronounced in the older lineage Panicum, which might indicate that optimization of PEPC for the C4 context increases with evolutionary time.
C4 photosynthesis is a CO2-concentrating mechanism that boosts productivity in tropical conditions (Atkinson et al., 2016). The higher efficiency of C4 plants results from the increased concentration of CO2 around ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco), the entry enzyme of the Calvin–Benson cycle (Sage et al., 2012). Rubisco has a tendency to confuse CO2 and O2 (Tcherkez et al., 2006). The reaction of O2 produces compounds that need to be recycled in the energetically costly photorespiration pathway (Nisbet et al., 2007). In C3 plants, Rubisco is in direct contact with atmospheric gases, and photorespiration can become consequential in conditions that decrease the relative concentration of CO2, including high temperature, aridity and salinity (Ehleringer and Bjorkman, 1977; Skillman, 2007). C4 plants tackle this problem by segregating primary carbon fixation from the enzyme Rubisco into two cell types (Hatch, 1987; Sage, 2004; Sage et al., 2012). In C4 plants, atmospheric CO2 in the form of bicarbonate is initially fixed by the enzyme phosphoenolpyruvate carboxylase (PEPC) (Hatch, 1987). PEPC produces the four-carbon acid oxaloacetate, which is rapidly converted into the more stable four-carbon acids malate and/or aspartate (Bräutigam et al., 2014). These acids are shuttled to a cell isolated from the atmosphere in which Rubisco is localized, and CO2 is released. This biochemical pumping of CO2 leads to an increase of the relative concentration of CO2 by a factor of 10 when compared to a non-C4 cell, and a dramatic increase of photosynthetic efficiency at high temperature (Ehleringer and Bjorkman, 1977; von Caemmerer and Furbank, 2003; Sage, 2004; Sage et al., 2012).
The C4 photosynthetic mechanism is a classic example of convergent evolution, having evolved more than 60 times independently in various groups of flowering plants (Sage et al., 2011). As all known C4 enzymes exist in C3 plants, the evolution of C4 photosynthesis involved the co-option of genes and proteins essential for the cycle followed by adaption of their expression levels and, at least in some cases, their kinetic properties (Blasing et al., 2002; Tausta et al., 2002; Ghannoum et al., 2005; Aubry et al., 2011; Christin et al., 2013; Heckmann et al., 2013; Kulahoglu et al., 2014; Huang et al., 2017; Moreno-Villena et al., 2018; Alvarez et al., 2019; Niklaus and Kelly, 2019). In particular, the transcript level, enzyme abundance and activity of PEPC are massively increased in all C4 lineages screened so far (Engelmann et al., 2003; Marshall et al., 2007; Bräutigam et al., 2014; Christin et al., 2015; Moreno-Villena et al., 2018). In contrast, the kinetic behavior of the PEPC enzyme has received less attention and has been investigated mainly in a few systems of eudicot plants that contain closely related C4 and non-C4 species, such as the Flaveria genus [Asteraceae, (McKown et al., 2005)]. In Flaveria, the C4 PEPC has a ten-fold lower specificity for phosphoenolpyruvate (PEP), an increased sensitivity to activators such as glucose-6-phosphate, and a decreased sensitivity to feedback inhibition from malate and aspartate (Svensson et al., 1997; Engelmann et al., 2003; Svensson et al., 2003; Paulus et al., 2013a; DiMario and Cousins, 2018). Comparison of PEPCs from C3 to C4 intermediate species in Flaveria further suggested that C4 properties of the enzyme were gradually acquired during the diversification of the genus (Engelmann et al., 2003). Investigations of PEPC in Amaranthaceae, a distantly related family of eudicots that contains multiple C4 origins, have shown that PEP specificity evolved convergently in the two groups of C4 eudicots (Gowik et al., 2006). In contrast, kinetics of PEPC from grasses (Poaceae), the group that contains the largest number of C4 species, and the most productive and ecologically successful ones (Cerling et al., 1997; Osborne and Beerling, 2006; Sage et al., 2011), remain poorly known. Indeed, previous investigations of PEPC from grass species have used whole leaf preparations, which report on the behavior of mixtures of isoforms and not on well defined, pure enzymes (Huber and Edwards, 1975; Holaday and Black, 1981). PEPC isoforms are encoded by a multi-gene family, with at least six highly divergent gene lineages in most grasses (Christin et al., 2007). The kinetic behaviors have been compared among distant grass paralogs (Dong et al., 1998), but comparisons of closely related C4 and non-C4 orthologs in the family are missing.
According to molecular dating, the origins of C4 photosynthesis are spread throughout the last 35 million years (Christin et al., 2008; Christin et al., 2011). The genus Flaveria represents one of the most recent C4 origins, its different photosynthetic types having diverged in the last 3 million years, with the emergence of fully C4 plants 1–2 million years ago (Christin et al., 2011). While old C4 groups exist in eudicots, the previously investigated Alternanthera (Gowik et al., 2006) is only slightly older than Flaveria, having evolved the C4 trait 5–10 million years ago (Christin et al., 2011). With more than 22 C4 origins spanning a recent past up to 35 million years ago, the grass family contains the oldest and largest C4 lineages (Christin et al., 2008; Christin et al., 2011). In terms of C4 PEPC evolution, grasses and eudicots co-opted different genes (Christin et al., 2015). Genes encoding C4-specific PEPC evolved under positive selection in several C4 groups, but the identity and quantity of fixed amino acid changes varies among families (Besnard et al., 2009; Rosnow et al., 2015). In particular, more of these amino acid changes are observed among grasses than in Flaveria (Christin et al., 2007), which might result from the longer divergence between the photosynthetic types. Alternatively, the genes co-opted for C4 photosynthesis in grasses might have been less fit for the C4 function, requiring therefore more adaptive changes (Christin et al., 2010). Testing these hypotheses requires generating kinetic data for orthologous non-C4 and C4 PEPC genes from grasses. The PEPCs from Flaveria are well-studied (Svensson et al., 1997; Svensson et al., 2003; Paulus et al., 2013a; DiMario and Cousins, 2018) and make an excellent starting point for a detailed comparison with other non-characterized PEPCs.
In this work, we characterize the enzymes encoded by orthologous non-C4 and C4 genes from two grass species belonging to the same tribe (the C4 Panicum queenslandicum and the C3 Panicum pygmaeum from the tribe Paniceae) and compare them to non-C4 and C4 PEPC from Flaveria to test the hypotheses that (i) despite very different starting points, qualitatively similar changes happened in C4 PEPC from Flaveria and grasses, and (ii) the kinetic changes differ more between C4 and non-C4 PEPC in grasses than in Flaveria due to an expanded period of adaptive evolution. We describe the changes in specificity for both substrates (bicarbonate and PEP) as well as the nature of inhibition by aspartate and malate. Overall, out work sheds new light on the impacts of evolutionary time and distance on the convergent evolution of enzyme kinetics.
Unless otherwise stated, reagents and components were from Sigma, protein purification equipment was from GE Healthcare and both enzymes for cloning and E. coli strains were from NEB.
Genes that encode the Flaveria trinervia PEPC gene and the Flaveria pringlei PEPC gene in the pTrc 99A plasmid were provided by Peter Westhoff (Dusseldorf). The PEPC genes were sub cloned into the pET-1B His6 TEV LIC vector plasmid, provided by Scott Gradia (Berkley; Addgene plasmid #29653). Genes were sub cloned using the ligation independent cloning method with Q5 DNA polymerase and T4 DNA polymerase (NEB). Cloned plasmids were isolated using a Miniprep DNA kit (Qiagen). Plasmids were Sanger sequenced to confirm the sequence identity (GATC Biotech).
Leaf samples were collected from P. queenslandicum at midday in full daylight and flash frozen in liquid nitrogen. Leaf samples were homogenized with a pestle and mortar in liquid nitrogen. RNA was extracted from ground leaves using the RNeasy Kit (Qiagen). Libraries of cDNA were generated with SuperScript II Reverse Transcriptase (Thermo Fischer Scientific). The PEPC from P. queenslandicum was amplified using the primers PquFor1B and PquRev1B (Supplementary Table 1), and Q5 polymerase. The amplified gene was Sanger sequenced (GATC Biotech) with the PCR primers and with the primers Pqu_1323_Seq_For and Pqu_1752_Seq_Rev (Primers synthesized by Sigma, summarized in Supplementary Table 1). The gene was then cloned into the pET-1B His6 TEV LIC vector plasmid as above.
Because non-C4 PEPC from C4 grasses generally represent distant paralogs resulting from ancient duplications that predate the origin of the family (Christin et al., 2007), the most closely related non-C4 PEPC are in most cases those from related C3 species. We consequently selected a gene from a C3 species from the same tribe as P. queenslandicum. The sequence for PEPC from P. pygmeaum has been previously obtained via leaf transcriptome sequencing (Dunning et al., 2017). The sequence was codon optimized for expression in E. coli and synthesized by GenArt Gene Synthesis in the pTRCC Plasmid. The synthesized gene was sub-cloned into the pET-1B His6 TEV LIC plasmid and verified by Sanger sequencing.
For protein expression the BL21λ (DE3) strain of E. coli (NEB) was used. Chemically competent E. coli cells were transformed with each of the plasmids. Eight liters of cultures were grown in LB media at 37°C to OD600 0.8. Cultures were cooled to 4°C for 1 h prior to recombinant protein induction with 0.5 mM IPTG (Fischer). Cultures were then incubated at 18°C for 18 h. Cells were harvested by centrifugation at 5,400×g for 25 min and stored at −80°C.
Cells were suspended in IMAC buffer (25 mM Tris, 0.5 M NaCl, 0.3 M glycerol, 20 mM imidazole (Acros Scientific)), 10 ml per 2 L of culture with 50 µl of 50 mgml−1 DNase I and 100 µl of 100 mgml−1 Pefabloc. Cells were passed twice through a cell disruptor (Constant Systems) before centrifugation at 276,000×g for 40 min. The supernatant was passed through a 0.45 µm pore filter (Elkay Labs.). PEPC was separated from soluble protein with a prepacked 1 ml nickel affinity column using an ÄKTA Pure 25 L Chromatography System. The loaded column was washed with 50 column volumes of IMAC buffer, then 50 column volumes of IMAC buffer containing 150 mM imidazole. Pure PEPC was eluted with 10 column volumes of IMAC buffer containing 400 mM imidazole.
Protein eluted from IMAC purification was loaded onto a Sephadex G50 desalting column (Amersham Biosciences) and rebuffered in storage buffer (20 mM Tris, 5% v/v glycerol, 150 mM KCl, 1 mM DTT (AnaSpec. Inc.)). Protein was aliquoted and frozen at −80°C until use.
PEPC enzyme concentration was quantified by absorption at 280 nm. Enzyme extinction coefficients were calculated using the ExPASy protein parameter tool and corrected by determining the absorbance of the protein denatured in 6 M guanidine hydrochloride (Gill and von Hippel, 1989). The extinction coefficients for the F. trinervia, F. pringlei, P. queenslandicum and P. pygmaeum PEPC were 120,480 M−1cm−1, 117,030 M−1cm−1, 105,810 M−1cm−1 and 111,510 M−1cm−1, respectively. Gel based protein quantification was not used.
Protein samples were analyzed for purity using SDS PAGE analysis. Samples of cell lysate or pure protein (25 µg or 5 µg protein respectively; BCA assay from Pierce) were denatured in 2 × SDS PAGE loading dye (200 mM Tris HCl pH 6.8, 2% SDS, 20% Glycerol, 0.01% Bromophenol blue (BDH Laboratory Supplies) and 7% β-mercaptoethanol). Protein was loaded onto an 8% acrylamide SDS gel with 2 µl of Blue Prestained Protein Standard Broad Range (11–190 kDa) (NEB). Gels were run for 50 min at 200 V with 1 × Tris/Glycine/SDS running buffer (Geneflow). Gels were stained with InstantBlue (Expedeon) and imaged with a ChemiDoc MP (BioRad).
PEPC activity was measured spectroscopically at 340 nm by coupling to NADH-malate dehydrogenase. Assays with a high fixed concentration of bicarbonate were observed using a FLUOstar plate reader (BMG Labtech) through a 340 nm ± 5 nm filter in absorbance mode. These assays were conducted in a reaction volume of 150 µl at 25°C. A typical reaction mixture contained 50 mM Tricine.KOH pH 8.0, 10 mM MgCl2 (Fluka), 5 mM KHCO3, 0.2 mM NADH (Fischer) and 0.1 U µl−1 malate dehydrogenase. Assays were initiated with the addition of PEPC enzyme. Rates were calculated with a NADH calibration curve; this method takes account of the short pathlength in microtiter plates.
Assays at a range of bicarbonate concentrations were observed with a Cary spectrophotometer (Agilent Technologies) in the same reaction buffer, in a total reaction volume of 600 µl. Initial rates were calculated using the Cary analysis software. To remove background bicarbonate, the water and tricine buffer were sparged with nitrogen for 18 h prior to use in assays. These assays were constructed under a nitrogen flow and performed in a sealed cuvette. The reaction was initiated with the addition of 50 nM PEPC, delivered with a gastight syringe (Hamilton). Bicarbonate concentrations were controlled with the addition of freshly prepared potassium bicarbonate.
The background bicarbonate was determined using an endpoint assay with no potassium bicarbonate (30 min). This procedure determines the total concentration of dissolved and hydrated CO2, (i.e. CO2 (aq), H2CO3, HCO3− and CO32−), at this pH over 97% is in the form of bicarbonate. Reported bicarbonate concentrations are the sum of the background and the added bicarbonate.
Kinetic parameters were evaluated by non-linear regression analysis in Igor Pro (Version 7.0.8.1; Wavemetrics Inc., Lake Oswego, Oregon). In all cases, the enzyme was assumed to be fully active. Primary plots were analyzed using Equation (1).
Analysis of secondary plots (i.e. of or vs [PEP]) with Equation (2) allowed determination of the steady-state kinetic parameters, and and kcat.
Where k and kapp are the true and apparent values of kcat or and K is or .
In the case of inhibition data, secondary plots were analyzed using Equation (3), where in the case of competitive inhibition kapp is and Ki is the competitive inhibition constant Kic or in the case of non-competitive inhibition kapp is and Ki is the non-competitive inhibition constant Kiu.
The non-competitive inhibition constant (Kiu) was determined by the secondary plot of against inhibitor concentration. The competitive inhibition constant (Kic) was determined by the secondary plot of against inhibitor concentration.
All data points shown on plots of initial rate against substrate concentration are individual measurements. Standard errors are provided for every parameter estimate. In secondary plots of apparent kinetic parameters against substrate or inhibitor concentration the standard error of those parameter estimates are shown. These standard errors are provided directly by the nonlinear regression analysis routine implemented within Igor Pro.
Four PEPC isoforms were characterized. In grasses, the C4 and non-C4 forms of ppc-1P3 genes were isolated from the C4 P. queenslandicum and synthesized based on the sequence of the C3 species P. pygmaeum, respectively. The cloned genes were 962 and 969 codons long, respectively. They have an 86.2% identity in amino acids and a 93.2% similarity, including on the two positions that have been linked in C4 Flaveria to and decreased inhibition (positions 774 and 884, respectively; Blasing et al., 2000; DiMario and Cousins, 2018). In Flaveria, the C4 and non-C4 ppc-1E2 genes corresponding to the C4 F. trinervia and the C3 species F. pringlei were analyzed [ppcA as described in Svensson et al. (1997)]. The two genes are both 967 codons long, with a 94.7% identity and a 97.5% similarity. The orthologous relationships between these pairs of genes were confirmed by phylogenetic analyses (Supplementary Figure 1).
All four genes were prepared in vectors for over-expression in E. coli with an N-terminal His tag. In all cases, expressed protein was purified to >95% purity as assessed by SDS PAGE with a single immobilized metal column (Supplementary Figure 2).
Assays at saturating bicarbonate and variable concentrations of PEP (Supplementary Figure 3) showed that both His tagged Flaveria PEPCs behaved similarly to untagged proteins previously described (Svensson et al., 1997; Blasing et al., 2000; Jacobs et al., 2008). Specifically, at pH 8.0, 10 mM MgCl2, 10 mM KHCO3, coupled to malate dehydrogenase, the His6-PEPC from F. trinervia catalyses the formation of oxaloacetate with a of 0.61 ± 0.05 mM and a kcat of 47.99 ± 1.22 s−1. Literature values are ranging from 0.278 to 0.652 mM and Vmax of 29 U mg−1, allowing for the different protein concentration, our kcat would be equivalent to a Vmax of 25.56 U mg−1 (Svensson et al., 1997; Blasing et al., 2000). Under the same conditions, the His6-PEPC from F. pringlei catalyses the formation of oxaloacetate with a of 0.05 ± 0.01 mM and a kcat of 52.65 ± 1.37 s−1; literature values are ranging from 0.029 to 0.061 mM and Vmax of 27 U mg−1, and allowing for the different protein concentration, our kcat would be equivalent to a Vmax of 28.02 U mg−1 (Svensson et al., 1997; Blasing et al., 2000). This confirms previous reports (Paulus et al., 2013a) that the presence of an N-terminal poly-histidine tag does not adversely affect the activity of these proteins.
The specificity for bicarbonate of all four enzymes was determined using a gas-tight assay system. Background bicarbonate was reduced to ca. 50 µM by sparging with nitrogen gas. Assays were performed at five PEP concentrations, while varying the concentration of bicarbonate (Figure 1). The analysis of secondary plots (Supplementary Figures 4 and 5) provided estimates of kcat and the specificity constant, kcat/Km, for both substrates (Table 1).
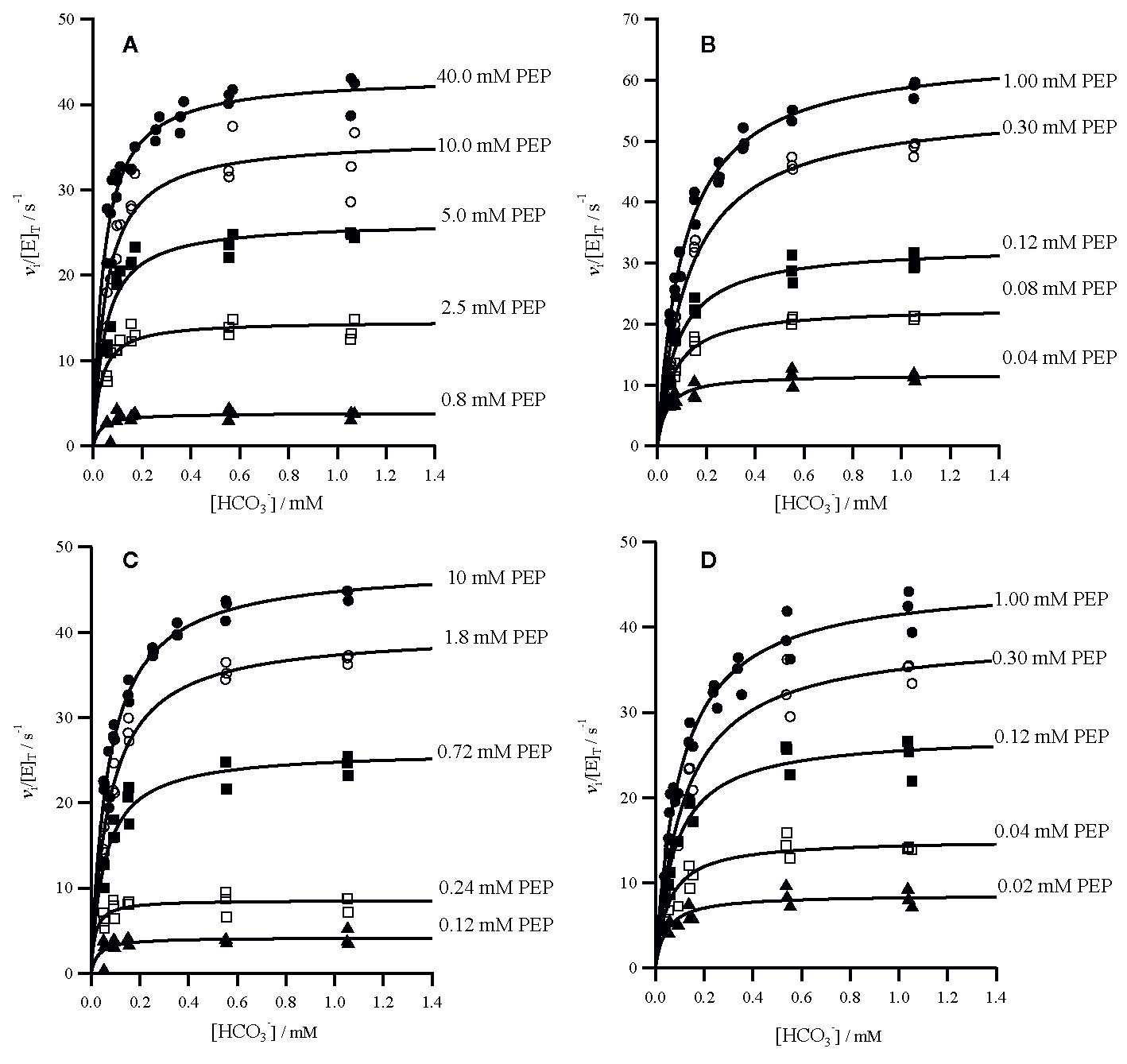
Figure 1 Initial rates of oxaloacetate formation catalyzed by PEPC. Assay conditions were 50 mM Tricine.KOH pH 8.0, 10 mM MgCl2, 0.2 mM NADH, 0.01 Uμl−1 malate dehydrogenase and 50 nM PEPC at 25°C, the concentration of PEP also varied as shown. Assays were repeated (n = 3) at each concentration. Individual data points are shown for the following PEPC (A) Panicum queenslandicum (B) Panicum pygmaeum (C) Flaveria trinervia and (D) Flaveria pringlei PEPC. Kinetic parameters are summarized in Table 1.
The specificity for bicarbonate (kcat/Km) of the C4 P. queenslandicum PEPC is 1.09 × 106 M−1s−1, almost twice as large as that of the non-C4 P. pygmaeum enzyme (Table 1). The specificity of this non-C4 enzyme is comparable to that of the C4 PEPC of Flaveria at 0.69 × 106 M−1s−1 (Table 1), which again is slightly higher than that of the Flaveria non-C4 PEPC (Table 1). In both cases the specificity constant for PEP is smaller in the C4 form of the enzyme (Table 1). In terms of bicarbonate Km values these are within the range previously reported for C4 and non-C4 plant PEPC isoforms in work with reasonably careful control of background bicarbonate (O'Leary, 1982; Bauwe, 1986; Janc et al., 1992; Dong et al., 1998; DiMario and Cousins, 2018).
For both non-C4 and C4 enzymes, we investigated inhibition by the two feedback inhibitors, malate (Supplementary Figure 6) and aspartate (Supplementary Figure 7) across a range of PEP concentrations. These two structurally related inhibitors show different kinetic characteristics; unlike aspartate, malate remains an inhibitor at saturating concentration of PEP (Table 2).
The C4 cycle of Flaveria produces both malate and aspartate (Moore and Edwards, 1986; Meister et al., 1996), while Panicum species are expected to produce mainly aspartate around PEPC (Rao and Dixon, 2016). The two molecules have however been shown to inhibit PEPC in a variety of C4 species (Huber and Edwards, 1975). In our analyses, all the PEPC enzymes are inhibited by malate at both limiting and saturating concentrations of PEP, and malate is a mixed inhibitor (Figures 2 and 3). This mixed inhibition can be characterized by two inhibition constants; at limiting PEP and at saturating PEP. In all cases, which means that malate can be viewed as a predominantly competitive inhibitor. The two C4 forms of the enzyme are both less sensitive to malate than the two non-C4 forms (Table 2). Unlike malate, aspartate is solely a competitive inhibitor for all of these enzymes (Figure 4). Increasing concentrations of aspartate do not affect kcat (Supplementary Figure 8). Once again, the two C4 forms of the enzyme are much less sensitive to aspartate than the two non-C4 forms (Table 2). Overall, our analyses indicate that the C4 forms are much less sensitive to both inhibitors, independently of the taxonomic group and C4 subtype, confirming previous reports (Huber and Edwards, 1975).
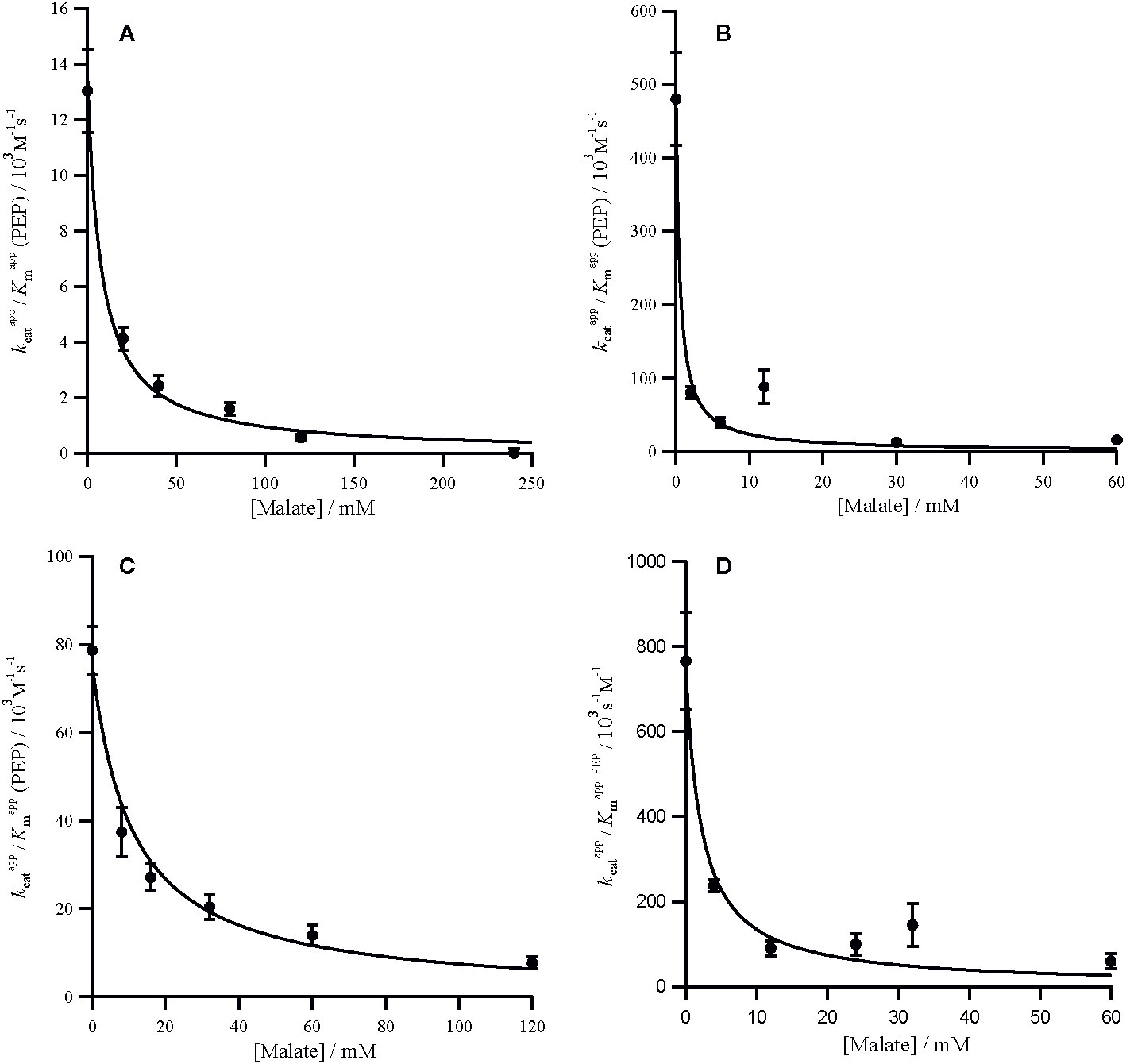
Figure 2 Competitive inhibition of PEPC by malate. Markers represent from assays in the presence of malate (Supplementary Figure 6) and error bars represent the standard errors. against malate concentration with inhibition curves characterized by Equation (3) and a Kic for the following PEPC (A) Panicum queenslandicum (= 7.51 ± 1.17 mM), (B) Panicum pygmaeum (= 0.52 ± 0.22 mM), (C) Flaveria trinervia (= 10.96 ± 1.55 mM), and (D) Flaveria pringlei (= 2.14 ± 0.62 mM). Inhibition parameters are summarized in Table 2.
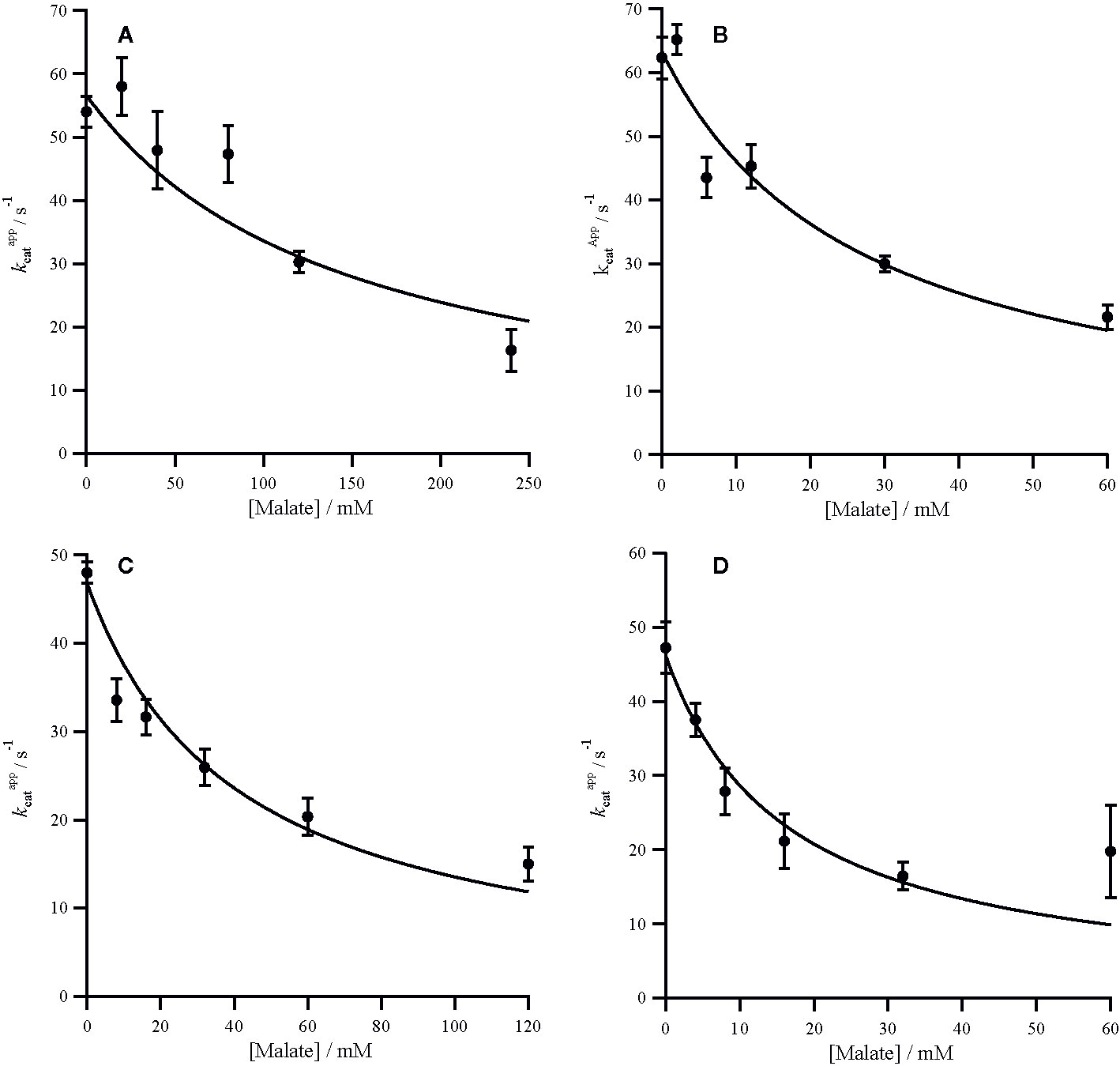
Figure 3 Non-competitive inhibition of PEPC by malate. Markers represent the from assays in the presence of malate (Supplementary Figure 6) and error bars represent the standard errors. against malate concentration with inhibition curves characterized by Equation (3) and a Kiu for the following PEPC (A) Panicum queenslandicum (= 146.08 ± 20.40 mM), (B) Panicum pygmaeum (= 31.23 ± 0.65 mM), (C) Flaveria trinervia (= 40.72 ± 4.59 mM) and (D) Flaveria pringlei ( = 4.56 ± 1.72 mM). Inhibition parameters are summarized in Table 2.
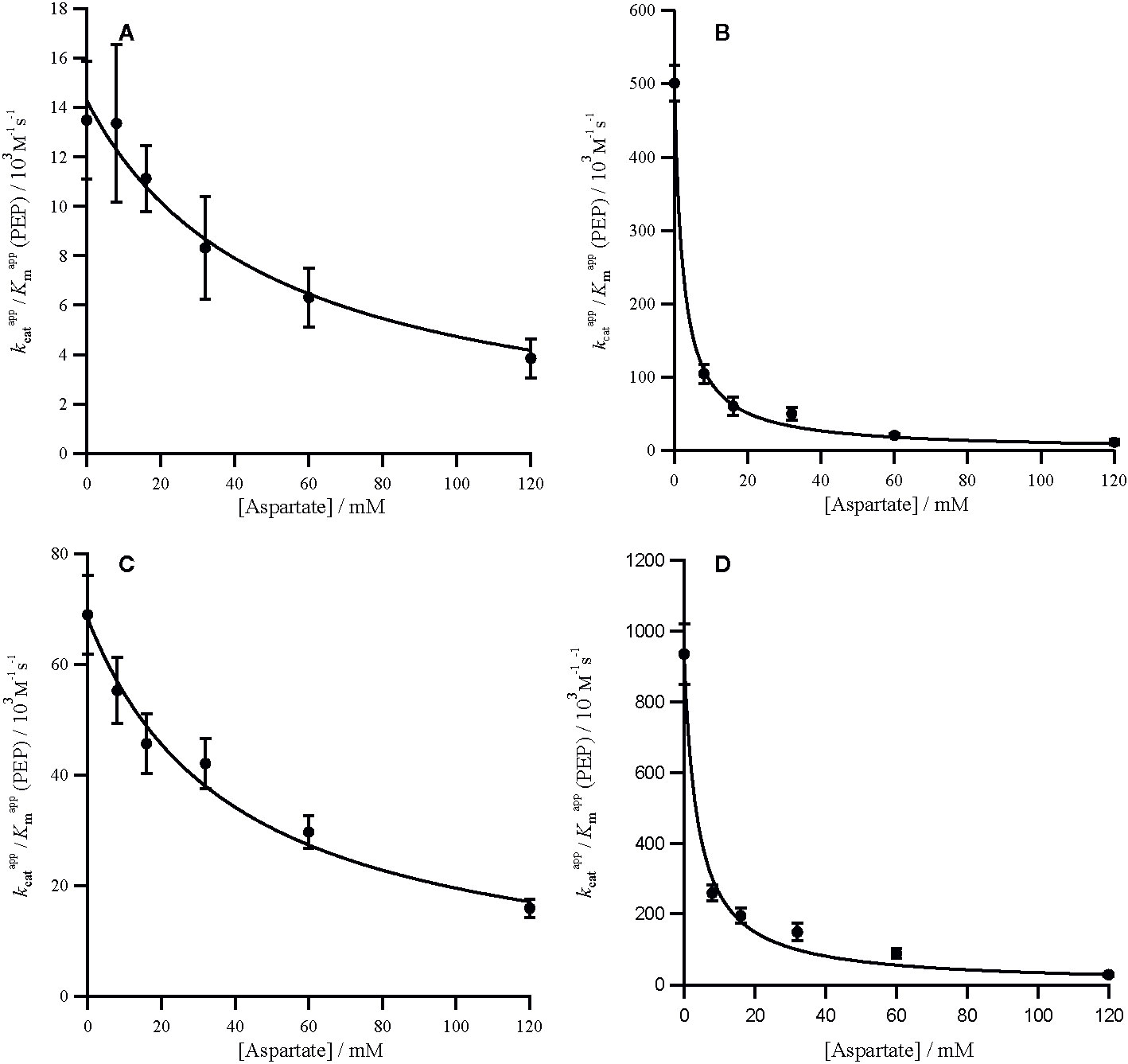
Figure 4 Competitive inhibition of PRPC by aspartate. Markers represent the from assays in the presence of aspartate (Supplementary Figure 7) and error bars represent the standard errors. against aspartate concentration with inhibition curves characterized by Equation (3) and a Kic for the following PEPC (A) Panicum queenslandicum (= 49.44 ± 7.86 mM), (B) Panicum pygmaeum (= 2.27 ± 0.02 mM), (C) Flaveria trinervia (= 40.02 ± 6.49 mM) and (D) Flaveria pringlei (= 4.31 ± 0.60 mM). Inhibition parameters are summarized in Table 2.
The non-C4 genes encoding the PEPC enzymes of the C3 plants P. pygmaeum and F. pringlei diverged about 150 million years ago and since then have accumulated numerous mutations and undergone multiple gene duplications (Christin et al., 2007; Christin et al., 2015). They share an 83.5% identity and a 91.2% similarity, and greater than 93% similarity with their respective C4 proteins. While the exact function of each non-C4 isoform is unknown, they are transcribed at similarly moderate levels (Moreno-Villena et al., 2018). Our investigation shows that the two non-C4 enzymes characterized here exhibit functionally similar kinetic characteristics, including high sensitivity to competitive inhibition by malate and aspartate and a similar sensitivity to bicarbonate. However, the two non-C4 isoforms differ in their which is three-fold lower in the F. pringlei enzyme (Table 1). While systematic screens of non-C4 PEPC are missing, those of Alternanthera and a distant root paralog from Z. mays have a similar to the F. pringlei enzyme (Dong et al., 1998; Gowik et al., 2006). These data suggest that despite hundreds of million years of divergence, non-C4 PEPC are generally associated with high sensitivity to inhibitors and low (<0.2 mM) Km for both substrates. These properties are likely required for a tight regulation and fast response of isoforms involved in anaplerotic functions, where the concentrations of substrates and products are low.
In both Flaveria and Panicum, the C4 PEPC shows a markedly reduced sensitivity to both malate and aspartate as compared with the non-C4 ortholog (Table 2). This reduction in sensitivity, reported before in Flaveria (Blasing et al., 2002; Paulus et al., 2013b; DiMario and Cousins, 2018) and a variety of grasses from different C4 subtypes (Huber and Edwards, 1975), is observed at all concentrations of PEP (Figures 2–4). Our observations are thus consistent with the conclusion that the same selective pressures act in C4 eudicots and at least some grasses to decrease the sensitivity to the inhibitors malate and aspartate. In C4 plants the concentration of malate and aspartate are high, so this reduced sensitivity prevents PEPC being inhibited by downstream metabolites (Arrivault et al., 2017). The respective amounts of malate and aspartate vary among C4 species (Moore and Edwards, 1986; Meister et al., 1996; Rao and Dixon, 2016), and concerted reduction of inhibition by both species is consistent with them sharing a binding site (Paulus et al., 2013a).
The adaptation of PEPC to the demands of the C4 pathway involved qualitatively similar changes in substrate specificity between Flaveria and the grasses considered here (Table 1). In both cases the specificity for PEP decreases and the specificity for bicarbonate increases. The C4 form of Zea mays, an independent C4 origin within grasses, has an affinity for PEP that is similar to P. queenslandicum (Dong et al., 1998). In addition, changes of Km for PEP in the same direction have been reported in Alternanthera (Gowik et al., 2006), suggesting that decreases in PEP Km happened convergently across C4 origins. The functional value of these changes remains speculative and might be a side-effect of adaptation of other properties of the enzyme or a direct target of selection for tighter regulation when PEP concentrations are higher (Svensson et al., 2003). The differences in Km for bicarbonate are less marked than those of PEP (Table 1). The Km for bicarbonate parameter is much higher in the non-C4 root isoform from Z. mays (Dong et al., 1998), indicating it varies tremendously among non-C4 PEPC. Data from more species are needed to determine whether the qualitative convergence observed here between Flaveria and Panicum is universal, or depends on the co-opted gene or the details of the C4 phenotype (e.g. biochemical and anatomical subtypes). Indeed, the cellular concentration of bicarbonate depends on the action of the enzyme carbonic anhydrase, in addition to the cell pH, and it is thus possible that variation in these factors changes the adaptive value of bicarbonate affinity.
While differences in substrate specificity and sensitivity to inhibitors are qualitatively convergent between Flaveria and the two grasses considered here, they are more marked in the latter (Table 1). These quantitative differences might be linked to the contrast between the length of time spent as C4 in each lineage, from more than 16 million years for P. queenslandicum to less than three for Flaveria (Christin et al., 2008; Christin et al., 2011). The C4 PEPCs share a 76.5% identity and an 88.1% similarity. Indeed, the kinetic properties observed in the PEPC of extant taxa result from adaptive changes accumulated since the initial origin of C4 photosynthesis. According to current models, an initial C4 pathway can evolve via enzyme upregulation and limited modifications of the proteins (Sage et al., 2012; Heckmann et al., 2013; Dunning et al., 2019; Heyduk et al., 2019), as observed in C3–C4 intermediates (Svensson et al., 2003; Dunning et al., 2017). Once a C4 pathway is in place, selection will act to improve its efficiency (Heckmann et al., 2013), and variation among members of the same C4 lineage indicates that such process can take protracted periods of selection on novel mutations (Heyduk et al., 2019).
Because these are likely necessary for a function of PEPC in C4 cells with high concentrations of metabolites, we suggest that relaxed sensitivity to inhibitors happens early during the evolution of C4 PEPC. This hypothesis is supported by the fact that changes in sensitivity to inhibitors are observed in intermediates from Flaveria (Engelmann et al., 2003). It has moreover been shown that one single amino acid replacement is sufficient to generate a large decrease in sensitivity (Paulus et al., 2013a). The C4-specific residue at this site is observed in multiple C4 lineages of both grasses and eudicots (Paulus et al., 2013a; Paulus et al., 2013b), suggesting that a rapid decrease of inhibition is involved in many origins of C4 PEPC.
Other properties of C4-specific PEPC might represent secondary adaptations to the C4 context, which might happen either to strengthen the early trends or in response to other changes of the plant biochemical phenotype. Over time, sustained diversifying selection on C4 PEPC would have led to stronger differences between P. queenslandicum and the C3 grasses. This view is supported by the similar kinetic parameters between the C4 PEPC of P. queenslandicum and Zea mays, two grass lineages of similar age, as well as similar kinetic parameters observed between the C4 PEPC in Alternanthera and Flaveria, two comparatively young lineages (Christin et al., 2011). It is however possible that secondary PEPC adaptations vary among and maybe even within old C4 lineages, as different biochemical and anatomical C4 subtypes evolved. Data from more lineages are needed to test the hypothesis that such diversifying PEPC secondary adaptation happened.
The molecular basis of the C4 specific properties reported here are not well understood. Analysis of the evolution of the amino acid sequence of C4 PEPC has shown that at least 22 sites underwent positive selection in grasses and sedges (Christin et al., 2007). Of these sites, three are also observed in C4 Flaveria (Christin et al., 2007; Besnard et al., 2009). Some of these mutations have been shown to be responsible for key C4 specific kinetic properties. Of these, a mutation for alanine to serine at position 774 (Flavaria numbering) has been identified as an important determinant of the low specificity for PEP of the C4 form of the enzyme (Blasing et al., 2000); interestingly, the effect of this position on bicarbonate specificity depends on the rest of the sequence and concentrations of allosteric regulators (DiMario and Cousins, 2018). Additionally, a mutation at position 884 (Flaveria numbering), in the allosteric inhibitor binding site, has been shown to have a notable effect on the IC50 for malate. An arginine residue in this position, as seen in the non-C4 form of the enzyme is well placed to directly interact with the inhibitor, increasing the susceptibility to inhibition (Paulus et al., 2013a). These amino acid changes are observed in the C4 PEPC of Panicum but not its non-C4 ortholog, and presumably contribute to the kinetic differences between the two. The specific role of other grass mutations has yet to be identified, a task that will be complicated by the large amount of variation among grass PEPC and possible epistasy among sites. These factors make it difficult to associate specific kinetic changes with specific amino acid replacements. Here, we compared the characteristics of PEPC from old, diverse lineages; these efforts now need to be expanded to other C4 lineages, with the well characterized isoforms of Flaveria continuing to serve as a model to assess the effect of specific sites.
All datasets presented in this study are included in the article/Supplementary Material.
All authors contributed to the article and approved the submitted version. NM carried out all experimental work.
P-AC is supported by a Royal Society University Research Fellowships (URF\R\180022), JR is supported by the Biotechnology and Biological Sciences Research Council (BBSRC, UK; award number BB/M000265/1). NM was supported by the Grantham Centre for Sustainable Futures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Peter Westhoff (Heinrich Heine University, Dusseldorf) for supplying expression plasmids for the Flaveria trinervia PEPC and Flaveria pringlei PEPC, Luke Dunning for his help in constructing a maximum likelihood tree, and Scott Gradia (Berkley) for providing the expression vector pET-1B His6 TEV LIC vector plasmid (Addgene plasmid #29653).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01014/full#supplementary-material
Alvarez, C. E., Bovdilova, A., Höppner, A., Wolff, C.-C., Saigo, M., Trajtenberg, F., et al. (2019). Molecular adaptations of NADP-malic enzyme for its function in C4 photosynthesis in grasses. Nat. Plants 5 (7), 755–765. doi: 10.1038/s41477-019-0451-7
Arrivault, S., Obata, T., Szecowka, M., Mengin, V., Guenther, M., Hoehne, M., et al. (2017). Metabolite pools and carbon flow during C4 photosynthesis in maize: 13CO2 labeling kinetics and cell type fractionation. J. Exp. Bot. 68 (2), 283–298. doi: 10.1093/jxb/erw414
Atkinson, R. R., Mockford, E. J., Bennett, C., Christin, P. A., Spriggs, E. L., Freckleton, R. P., et al. (2016). C4 photosynthesis boosts growth by altering physiology, allocation and size. Nat. Plants 2 (5), 16038. doi: 10.1038/nplants.2016.38
Aubry, S., Brown, N. J., Hibberd, J. M. (2011). The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot. 62 (9), 3049–3059. doi: 10.1093/jxb/err012
Bauwe, H. (1986). An efficient method for the determination of Km values for HCO3- of phosphoenolpyruvate carboxylase. Planta 169 (3), 356–360. doi: 10.1007/BF00392131
Besnard, G., Muasya, A. M., Russier, F., Roalson, E. H., Salamin, N., Christin, P. A. (2009). Phylogenomics of C4 Photosynthesis in Sedges (Cyperaceae): Multiple Appearances and Genetic Convergence. Mol. Biol. Evol. 26 (8), 1909–1919. doi: 10.1093/molbev/msp103
Blasing, O., Westhoff, P., Svensson, P. (2000). Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275 (36), 27917–27923. doi: 10.1074/jbc.M909832199
Blasing, O. E., Ernst, K., Streubel, M., Westhoff, P., Svensson, P. (2002). The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia - implications for the evolution of C4 photosynthesis. Planta 215 (3), 448–456. doi: 10.1007/s00425-002-0757-x
Bräutigam, A., Schliesky, S., Külahoglu, C., Osborne, C. P., Weber, A. P. M. (2014). Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 65 (13), 3579–3593. doi: 10.1093/jxb/eru100
Cerling, T. E., Harris, J. M., MacFadden, B. J., Leakey, M. G., Quade, J., Eisenmann, V., et al. (1997). Global vegetation change through the Miocene/Pliocene boundary. Nature 389 (6647), 153–158. doi: 10.1038/38229
Christin, P. A., Salamin, N., Savolainen, V., Duvall, M. R., Besnard, G. (2007). C4 Photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr. Biol. 17 (14), 1241–1247. doi: 10.1016/j.cub.2007.06.036
Christin, P. A., Besnard, G., Samaritani, E., Duvall, M. R., Hodkinson, T. R., Savolainen, V., et al. (2008). Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18 (1), 37–43. doi: 10.1016/j.cub.2007.11.058
Christin, P. A., Weinreich, D. M., Besnard, G. (2010). Causes and evolutionary significance of genetic convergence. Trends Genet. 26 (9), 400–405. doi: 10.1016/j.tig.2010.06.005
Christin, P. A., Osborne, C. P., Sage, R. F., Arakaki, M., Edwards, E. J. (2011). C4 eudicots are not younger than C4 monocots. J. Exp. Bot. 62 (9), 3171–3181. doi: 10.1093/jxb/err041
Christin, P. A., Boxall, S. F., Gregory, R., Edwards, E. J., Hartwell, J., Osborne, C. P. (2013). Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. Evol. 5 (11), 2174–2187. doi: 10.1093/gbe/evt168
Christin, P. A., Arakaki, M., Osborne, C. P., Edwards, E. J. (2015). Genetic enablers underlying the clustered evolutionary origins of C4 photosynthesis in angiosperms. Mol. Biol. Evol. 32 (4), 846–858. doi: 10.1093/molbev/msu410
DiMario, R. J., Cousins, A. B. (2018). A single serine to alanine substitution decreases bicarbonate affinity of phosphoenolpyruvate carboxylase in C4 Flaveria trinervia. J. Exp. Bot. 70 (3), 995–1004. doi: 10.1093/jxb/ery403
Dong, L. Y., Masuda, T., Kawamura, T., Hata, S., Izui, K. (1998). Cloning, Expression, and Characterization of a Root-Form Phosphoenolpyruvate Carboxylase from Zea mays: Comparison with the C4-Form Enzyme. Plant Cell Physiol. 39 (8), 865–873. doi: 10.1093/oxfordjournals.pcp.a029446
Dunning, L. T., Lundgren, M. R., Moreno-Villena, J. J., Namaganda, M., Edwards, E. J., Nosil, P., et al. (2017). Introgression and repeated co-option facilitated the recurrent emergence of C4 photosynthesis among close relatives. Evolution 71 (6), 1541–1555. doi: 10.1111/evo.13250
Dunning, L. T., Moreno-Villena, J. J., Lundgren, M. R., Dionora, J., Salazar, P., Adams, C., et al. (2019). Key changes in gene expression identified for different stages of C4 evolution in Alloteropsis semialata. J. Exp. Bot. 70 (12), 3255–3268. doi: 10.1093/jxb/erz149
Ehleringer, J., Bjorkman, O. (1977). Quantum Yields for CO2 Uptake in C3 and C4 Plants: Dependence on Temperature, CO2, and O2 Concentration. Plant Physiol. 59 (1), 86–90. doi: 10.1104/pp.59.1.86
Engelmann, S., Blasing, O., Gowik, U., Svensson, P., Westhoff, P. (2003). Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria–a gradual increase from C3 to C4 characteristics. Planta 217 (5), 717–725. doi: 10.1007/s00425-003-1045-0
Ghannoum, O., Evans, J. R., Chow, W. S., Andrews, T. J., Conroy, J. P., von Caemmerer, S. (2005). Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiol. 137 (2), 638–650. doi: 10.1104/pp.104.054759
Gill, S. C., von Hippel, P. H. (1989). Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182 (2), 319–326. doi: 10.1016/0003-2697(89)90602-7
Gowik, U., Engelmann, S., Bläsing, O., Raghavendra, A., Westhoff, P. (2006). Evolution of C4 phosphoenolpyruvate carboxylase in the genus Alternanthera: gene families and the enzymatic characteristics of the C4 isozyme and its orthologues in C3 and C3/C4 Alternantheras. Planta 223 (2), 359–368. doi: 10.1007/s00425-005-0085-z
Hatch, M. D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895 (2), 81–106. doi: 10.1016/s0304-4173(87)80009-5
Heckmann, D., Schulze, S., Denton, A., Gowik, U., Westhoff, P., Weber, A. P., et al. (2013). Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153 (7), 1579–1588. doi: 10.1016/j.cell.2013.04.058
Heyduk, K., Moreno-Villena, J. J., Gilman, I. S., Christin, P. A., Edwards, E. J. (2019). The genetics of convergent evolution: insights from plant photosynthesis. Nat. Rev. Genet. 20 (8), 485–493. doi: 10.1038/s41576-019-0107-5
Holaday, A. S., Black, C. C. (1981). Comparative characterization of phosphoenolpyruvate carboxylase in C3, C4, and C3-C4 intermediate Panicum species. Plant Physiol. 67 (2), 330–334. doi: 10.1104/pp.67.2.330
Huang, P., Studer, A. J., Schnable, J. C., Kellogg, E. A., Brutnell, T. P. (2017). Cross species selection scans identify components of C4 photosynthesis in the grasses. J. Exp. Bot. 68 (2), 127–135. doi: 10.1093/jxb/erw256
Huber, S. C., Edwards, G. E. (1975). Inhibition of phosphoenolpyruvate carboxylase from C4 plants by malate and aspartate. Can. J. Bot. 53 (17), 1925–1933. doi: 10.1139/b75-216
Jacobs, B., Engelmann, S., Westhoff, P., Gowik, U. (2008). Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria: determinants for high tolerance towards the inhibitor L-malate. Plant Cell Environ. 31 (6), 793–803. doi: 10.1111/j.1365-3040.2008.01796.x
Janc, J., O'Leary, M., Cleland, W. (1992). A kinetic investigation of phosphoenolpyruvate carboxylase from Zea mays. Biochemistry 31 (28), 6421–6426. doi: 10.1021/bi00143a009
Kulahoglu, C., Denton, A. K., Sommer, M., Mass, J., Schliesky, S., Wrobel, T. J., et al. (2014). Comparative Transcriptome Atlases Reveal Altered Gene Expression Modules between Two Cleomaceae C3 and C4 Plant Species. Plant Cell 26 (8), 3243–3260. doi: 10.1105/tpc.114.123752
Marshall, D. M., Muhaidat, R., Brown, N. J., Liu, Z., Stanley, S., Griffiths, H., et al. (2007). Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesis. Plant J. 51 (5), 886–896. doi: 10.1111/j.1365-313x.2007.03188.x
McKown, A. D., Moncalvo, J. M., Dengler, N. G. (2005). Phylogeny of Flaveria (Asteraceae) and inference of C4 photosynthesis evolution. Am. J. Bot. 92 (11), 1911–1928. doi: 10.3732/ajb.92.11.1911
Meister, M., Agostino, A., Hatch, M. D. (1996). The roles of malate and aspartate in C4 photosynthetic metabolism of Flaveria bidentis (L.). Planta 199 (2), 262–269. doi: 10.1007/BF00196567
Moore, B. D., Edwards, G. E. (1986). Photosynthetic Induction in a C4 Dicot, Flaveria trinervia: II. Metabolism of Products of CO2 Fixation after Different Illumination Times. Plant Physiol. 81 (2), 669–673. doi: 10.1104/pp.81.2.669
Moreno-Villena, J. J., Dunning, L. T., Osborne, C. P., Christin, P. A. (2018). Highly Expressed Genes Are Preferentially Co-Opted for C4 Photosynthesis. Mol. Biol. Evol. 35 (1), 94–106. doi: 10.1093/molbev/msx269
Niklaus, M., Kelly, S. (2019). The molecular evolution of C4 photosynthesis: opportunities for understanding and improving the world's most productive plants. J. Exp. Bot. 70 (3), 795–804. doi: 10.1093/jxb/ery416
Nisbet, E. G., Grassineau, N. V., Howe, C. J., Abell, P. I., Regelous, M., Nisbet, R. E. R. (2007). The age of Rubisco: the evolution of oxygenic photosynthesis. Geobiology 5 (4), 311–335. doi: 10.1111/j.1472-4669.2007.00127.x
O'Leary, M. H. (1982). Phosphoenolpyruvate Carboxylase: An Enzymologist's View. Annu. Rev. Plant Physiol. 33 (1), 297–315. doi: 10.1146/annurev.pp.33.060182.001501
Osborne, C. P., Beerling, D. J. (2006). Nature's green revolution: the remarkable evolutionary rise of C4 plants. Philos. Trans. R. Soc. B.: Biol. Sci. 361 (1465), 173–194. doi: 10.1098/rstb.2005.1737
Paulus, J. K., Schlieper, D., Groth, G. (2013). Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4, 1518. doi: 10.1038/ncomms2504
Paulus, J. K., Niehus, C., Groth, G. (2013). Evolution of C4 Phosphoenolpyruvate Carboxylase: Enhanced Feedback Inhibitor Tolerance Is Determined by a Single Residue. Mol. Plant 6 (6), 1996–1999. doi: 10.1093/mp/sst078
Rao, X., Dixon, R. A. (2016). The Differences between NAD-ME and NADP-ME Subtypes of C4 Photosynthesis: More than Decarboxylating Enzymes. Front. Plant Sci. 7, 1525. doi: 10.3389/fpls.2016.01525
Rosnow, J. J., Evans, M. A., Kapralov, M. V., Cousins, A. B., Edwards, G. E., Roalson, E. H. (2015). Kranz and single-cell forms of C4 plants in the subfamily Suaedoideae show kinetic C4 convergence for PEPC and Rubisco with divergent amino acid substitutions. J. Exp. Bot. 66 (22), 7347–7358. doi: 10.1093/jxb/erv431
Sage, R. F., Christin, P. A., Edwards, E. J. (2011). The C4 plant lineages of planet Earth. J. Exp. Bot. 62 (9), 3155–3169. doi: 10.1093/jxb/err048
Sage, R. F., Sage, T. L., Kocacinar, F. (2012). Photorespiration and the Evolution of C4 Photosynthesis. Annu. Rev. Plant Biol. 63 (1), 19–47. doi: 10.1146/annurev-arplant-042811-105511
Sage, R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161 (2), 341–370. doi: 10.1111/j.1469-8137.2004.00974.x
Skillman, J. B. (2007). Quantum yield variation across the three pathways of photosynthesis: not yet out of the dark. J. Exp. Bot. 59 (7), 1647–1661. doi: 10.1093/jxb/ern029
Svensson, P., Blasing, O., Westhoff, P. (1997). Evolution of the enzymatic characteristics of C4 phosphoenolpyruvate carboxylase–a comparison of the orthologous PPCA phosphoenolpyruvate carboxylases of Flaveria trinervia (C4) and Flaveria pringlei (C3). Eur. J. Biochem. 246 (2), 452–460. doi: 10.1111/j.1432-1033.1997.t01-1-00452.x
Svensson, P., Blasing, O., Westhoff, P. (2003). Evolution of C4 phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys. 414 (2), 180–188. doi: 10.1016/S0003-9861(03)00165-6
Tausta, S. L., Coyle, H. M., Rothermel, B., Stiefel, V., Nelson, T. (2002). Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol. Biol. 50 (4-5), 635–652. doi: 10.1023/A:1019998905615
Tcherkez, G., Farquhar, G., Andrews, T. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. 103 (19), 7246–7251. doi: 10.1073/pnas.0600605103
Keywords: C4 photosynthesis, carbon fixation, enzyme evolution, feedback inhibition, kinetics, phosphoenolpyruvate carboxylase
Citation: Moody NR, Christin P-A and Reid JD (2020) Kinetic Modifications of C4 PEPC Are Qualitatively Convergent, but Larger in Panicum Than in Flaveria. Front. Plant Sci. 11:1014. doi: 10.3389/fpls.2020.01014
Received: 22 March 2020; Accepted: 22 June 2020;
Published: 03 July 2020.
Edited by:
Sarah Covshoff, Independent Researcher, Las Vegas, NV, United StatesReviewed by:
Robert Edward Sharwood, Australian National University, AustraliaCopyright © 2020 Moody, Christin and Reid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James D. Reid, ai5yZWlkQHNoZWZmaWVsZC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.