
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 July 2020
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00997
This article is part of the Research Topic Plant Glycobiology - A Sweet World of Glycans, Glycoproteins, Glycolipids, and Carbohydrate-Binding Proteins View all 33 articles
 Bussarin Wachananawat1‡
Bussarin Wachananawat1‡ Takeshi Kuroha2†‡
Takeshi Kuroha2†‡ Yuto Takenaka1,3
Yuto Takenaka1,3 Hiroyuki Kajiura1†
Hiroyuki Kajiura1† Satoshi Naramoto4
Satoshi Naramoto4 Ryusuke Yokoyama2
Ryusuke Yokoyama2 Kimitsune Ishizaki5
Kimitsune Ishizaki5 Kazuhiko Nishitani6
Kazuhiko Nishitani6 Takeshi Ishimizu1,3*
Takeshi Ishimizu1,3*Rhamnogalacturonan I (RG-I) comprises approximately one quarter of the pectin molecules in land plants, and the backbone of RG-I consists of a repeating sequence of [2)-α-L-Rha(1-4)-α-D-GalUA(1-] disaccharide. Four Arabidopsis thaliana genes encoding RG-I rhamnosyltransferases (AtRRT1 to AtRRT4), which synthesize the disaccharide repeats, have been identified in the glycosyltransferase family (GT106). However, the functional role of RG-I in plant cell walls and the evolutional history of RRTs remains to be clarified. Here, we characterized the sole ortholog of AtRRT1–AtRRT4 in liverwort, Marchantia polymorpha, namely, MpRRT1. MpRRT1 had RRT activity and genetically complemented the AtRRT1-deficient mutant phenotype in A. thaliana. However, the MpRRT1-deficient M. polymorpha mutants showed no prominent morphological changes and only an approximate 20% reduction in rhamnose content in the cell wall fraction compared to that in wild-type plants, suggesting the existence of other RRT gene(s) in the M. polymorpha genome. As expected, we detected RRT activities in other GT106 family proteins such as those encoded by MpRRT3 in M. polymorpha and FRB1/AtRRT8 in A. thaliana, the deficient mutant of which affects cell adhesion. Our results show that RRT genes are more redundant and diverse in GT106 than previously thought.
Rhamnogalacturonan I (RG-I), along with homogalacturonan (HG) and RG-II, constitutes the major part of the pectin in cell walls of land plants, which include charophytes, bryophytes, and vascular plants (Kulkarni et al., 2012; O’Raurke et al., 2015). RG-I consists of a ramified backbone composed of the disaccharide repeating unit [2)-α-L-Rha(1-4)-α-D-GalUA(1-] (Lau et al., 1985), which is branched at the O4 or O3 position of Rha residues with arabinan, galactan, or arabinogalactan (Lau et al., 1987). Some of these side chains are further modified by fucose and glucose residues at the site close to the backbone (Nakamura et al., 2001) or terminal ferulic acid (Ishii, 1997; Ralet et al., 2005).
The contents and/or branching pattern of RG-I depend on the plant species, tissues of the same plants, and developmental stages of the same tissues. The side chains of RG-I are developmentally or spatially regulated in pea cotyledons (McCartney et al., 2000), Arabidopsis thaliana inflorescence stems (Phyo et al., 2017), G-layers in poplar tension wood (Gorshkova et al., 2015; Guedes et al., 2017), seed mucilage (Dean et al., 2007; Macquet et al., 2007), and fruit softening (Orfila et al., 2002; Paniagua et al., 2016; Wang et al., 2019). These studies showed that RG-I is related to the maturation and mechanical properties of some tissues; however, the associated molecular mechanisms remain unclear. The functional roles of pectin in cell wall mechanics are considered more important than previously thought (Cosgrove, 2018; Haas et al., 2020); however, functional analyses of RG-I are insufficient to determine its roles because appropriate RG-I-deficient mutants are not easily created.
To address the functional roles of RG-I polysaccharides, it is vital to identify and analyze corresponding biosynthetic genes. An analysis of A. thaliana mutants deficient in ARABINAN DEFICIENT 1 (ARAD1) and/or ARAD2, which are responsible for arabinan elongation, previously revealed that arabinan has a role in the mechanical properties of inflorescence stems (Harholt et al., 2006; Verherbruggen et al., 2013). Further, loss-of-function mutants in GALACTAN SYNTHASE 1 (GALS1) to GALS3, which are involved in galactan elongation, do not show any obvious phenotypic change, and analyses of their double or triple deficient mutants have not been reported to date (Liwanag et al., 2012; Ebert et al., 2018). Four backbone-synthetic RG-I rhamnosyltransferases (AtRRT1 to AtRRT4) in the glycosyltransferase family (GT 106) were recently identified in A. thaliana (Takenaka et al., 2018). The AtRRT1-deficient mutant was observed to have reduced content of RG-I in seed mucilage; however, the RG-I-deficient mutant (deficient in all four AtRRT genes) has not been analyzed to date. Thus, it is not easy to prepare RG-I-deficient mutants and analyze the functions of RG-I polysaccharides because the genes encoding its biosynthetic enzymes are redundant.
The genes encoding cell wall polysaccharide biosynthetic-enzymes of vascular plants are also found in bryophytes and charophytes (Mikkelsen et al., 2014; Bowman et al., 2017), suggesting that typical cell walls of land plants are required for plant terrestrialization. Among the various plant species, Marchantia polymorpha, a liverwort in bryophyte, is an attractive plant species for the functional analysis of cell walls because it exhibits low genetic redundancy compared to other land plants (Bowman et al., 2017). Bryophyte plants also have RG-I in their cell walls (Kulkarni et al., 2012; Roberts et al., 2012; McCarthy et al., 2014). Vascular plants including A. thaliana have two or more RG-I-synthetic RRT genes in their genomes, whereas the M. polymorpha genome has only one gene homologous to AtRRT genes (Bowman et al., 2017; Takenaka et al., 2018). An analysis of the M. polymorpha mutant deficient in this gene is an attractive approach to elucidate the functional and evolutionary roles of RG-I in the cell wall, even though its life cycles, reproduction system, and the presence of xylem are to some extent distinct from those of vascular plants. The biochemical and functional analyses of RRT in M. polymorpha in this study were performed to provide profound knowledge of RRT genes in land plants, including their evolutionary history and gene redundancy in plant genomes.
The A. thaliana T-DNA insertion lines for Atrrt1 (SALK_022924 and SALK_042968) were obtained from the Arabidopsis Biological Resource Center. The seeds of wild-type (Col-0) and Atrrt1 strains were grown on MS medium at 22°C under a 16-h light/8-h dark cycle. After 10 days, the seedlings were transferred to compounded soil under the same conditions. Homozygous insertion mutant lines were identified by PCR analysis (Takenaka et al., 2018). The Nicotiana benthamiana seeds were sown in the soil and incubated with a 16-h light/8-h dark cycle at 22°C for 2 weeks. The seedlings were transferred to each pot under the same conditions for 6 weeks.
A male accession of liverwort (M. polymorpha), Takaragaike-1 (Tak-1), was used in this study and asexually maintained via the transplantation and growth of gemmae (Ishizaki et al., 2008). Liverworts were cultured on a Petri dish using half-strength Gamborg’s B5 (1/2 B5) medium (Gamborg et al., 1968) containing 1% agar (Nacalai Tesque), under continuous 50 to 60 μmol/m2·s white fluorescent light at 22°C. To induce the reproductive phase, thalli were transferred to plastic cases with 1/2 B5 medium containing 1% agar and grown under continuous 50 to 60 μmol/m2·s white light supplemented with 10 to 20 μmol m−2 s−1 far-red light irradiation at 22°C.
The cDNA of the MpRRT1 (Mapoly0033s0138.1) gene was amplified by PCR with specific primers, MpRRT1_F and MpRRT1_R (Supplementary Table 1), using M. polymorpha cDNA as a template, and the resulting PCR product was cloned into the pWAT202 vector (Kumakura et al., 2013) using the in-fusion HD cloning kit (Clontech). The resulting construct was transformed into the Atrrt1-1 (SALK_022924) mutant via Agrobacterium tumefaciens (GV3101 strain)–mediated transformation (Holsters et al., 1978). Transgenic plants were selected on MS agar plates containing 10 μg/ml bialaphos. The seed mucilage phenotype was observed with a stereo microscope (Olympus MVX10). The mature dry seeds were soaked in water for 2 h and stained with 0.01% ruthenium red solution for 1 h and the color was washed out with water (McFarlane et al., 2014). The volume of seed mucilage was calculated by assuming that the seeds or seeds containing mucilage were spheroid. The length of each axis was calculated using ImageJ software.
The full-length MpRRT1 and MpRRT3 (Mapoly0014s0149.1) open reading frames were amplified with sets of gene-specific primers, [MpRRT1-FLAG_F and MpRRT1-FLAG_R] and [MpRRT3-FLAG_F and MpRRT3-FLAG_R] (Supplementary Table 1), respectively, using M. polymorpha cDNA as a template. The full length FRB1/AtRRT8 open reading frame was amplified with gene-specific primers, AtRRT8-FLAG_F and AtRRT8-FLAG_R (Supplementary Table 1), using A. thaliana cDNA as a template. The amplified DNA was cloned into the pBI121 vector using the in-fusion HD cloning kit (Clontech). The recombinant RRT plasmids or the empty pBI121 vector were transformed into A. tumefaciens GV3101 strain using a freeze-thaw method (Holsters et al., 1978). The transformed A. tumefaciens were selected on an LB agar plate containing 50 µg/ml kanamycin, 50 µg/ml gentamycin, and 50 µg/ml rifampicin. The transformed A. tumefaciens cells (OD600 0.5) were suspended in 10 mM MES-KOH buffer, pH 5.8, containing 10 mM MgSO4 and inoculated into N. benthamiana leaves via the vacuum infiltration method (Leuzinger et al., 2013). After infiltration, the plants were grown under 16-h light/8-h dark conditions at 22°C for 3 days. The tobacco leaves were ground with liquid nitrogen and homogenized with 25 mM Tris-HCl buffer, pH 7.0, containing 10 mM MgCl2, 2 mM dithiothreitol, 250 mM sucrose, 2 µM leupeptin, and 0.1 mM PMSF. The homogenate filtered through miracloth was centrifuged at 3,000 × g for 10 min at 4°C. The supernatant was centrifuged at 100,000 × g for 1 h at 4°C to pellet the microsomal fraction. The microsomal proteins (approximately, 1.2 mg) were solubilized in 100 µl of the buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1.0% Triton X-100. The solubilized proteins were suspended with 10 µl of an anti-FLAG M2 affinity gel (Sigma-Aldrich) on a rotating wheel at 4°C for 1 h. The gel beads were then washed three times with 50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl and 0.01% Triton X-100. The amount of protein used for enzyme assays was approximately 1.5 µg. The recombinant proteins were detected by western blotting with a monoclonal anti-FLAG M2 antibody conjugated with alkaline phosphatase (1:2,000; Sigma-Aldrich) using Immobilon Forte Western HRP Substrate (Merck Millipore). Affinity-bound proteins from the microsomal fraction of tobacco leaves infiltrated with the empty pBI121 vector were used as a control.
The oligosaccharides derived from RG-I were labeled with 2-aminopyridine at their reducing ends (Uehara et al., 2017) and used as acceptor substrates of RG-I rhamnosyltransferase. The structures and abbreviations for these oligosaccharides are listed in Supplementary Table 2. UDP-rhamnose was enzymatically synthesized as described previously (Ohashi et al., 2016). The RG-I rhamnosyltransferase assay was carried out in a reaction mixture (total volume, 10 µL) containing the enzyme, 50 mM HEPES-KOH buffer, pH 7.0, 25 mM KCl, 0.2 M sucrose, 0.05% bovine serum albumin, 0.25% Triton X-100, 0.5 mM UDP-Rha, and 50 μM GR8-PA at 30°C for 1 h. The reaction was terminated by heating at 100°C for 3 min. The enzyme product was separated using a TSKgel-DEAE-5PW column (7.5 mm × 75 mm) as described previously (Uehara et al., 2017). It was then detected based on its fluorescence (Ex 320 nm, Em 400 nm) and quantified from the peak area on the chromatogram.
The sequence information of the M. polymorpha genome portal site MarpolBase (http://marchantia.info; JGI 3.1) was used for plasmid construction. Constructs for the CRISPR/Cas9-target mutagenesis of MpRRT1 were generated by annealing oligonucleotide pairs with MpRRT1_CR1_F and MpRRT1_CR1_R (Supplementary Table 1) and the ligation of these annealed products into the BsaI site of a pMpGE_En03 vector (Addgene) (Sugano et al., 2018) to produce plasmids En03_MpRRT1_CR1 including gRNA expression cassettes. The sequence between attL1 and attL2 within En03_MpRRT1_CR1 was inserted into the binary vector pMpGE011 (Addgene) (Sugano et al., 2018) via an LR reaction using Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific). The resulting construct, GE011_MpRRT1_CR1, was introduced into Tak-1 using A. tumefaciens strain GV2260 as described previously (Kubota et al., 2013).
To construct reporter plasmids, a DNA fragment of the 5′-putative MpRRT1 promoter sequence was amplified by PCR using Prime STAR MAX DNA polymerase (Takara Bio) with MpRRT1-4217_F and MpRRT1_0_R primers (Supplementary Table 1) from the Tak-1 genome. The fragment was cloned into a pENTR/D-TOPO entry vector (Thermo Fisher Scientific). We found that the tandem duplication of a 1.0-kbp sequence in the MpRRT1 promoter did not exist in the PCR product sequence based on the results of Sanger sequencing and the fragment size of the cloned vector digested with the restriction enzyme NcoI. Therefore, we used this 3,217-bp fragment of the MpRRT1 promoter for further analyses. The resulting construct, pENTR_proMpRRT1, was inserted into the destination vectors pMpGWB316 and pMpGWB304 (Addgene) (Ishizaki et al., 2015) via the LR reaction using LR Clonase II Enzyme Mix to produce proMpRRT1:tdTOMATO and proMpRRT1:GUS plasmids, respectively.
For subcellular localization analysis of the MpRRT1-tagRFP fusion protein, the cDNA sequence of MpRRT1 without a stop codon (TGA) was amplified by PCR using Prime STAR MAX DNA polymerase (Takara Bio) with MpRRT1_1_F and MpRRT1+3107_nostop_R primers (Supplementary Table 1), followed by cloning into the pENTR/D-TOPO entry vector. The MpRRT1 cDNA sequence of the resulting construct, pENTR_cMpRRT1ns, was inserted into the destination vector pMpGWB328 (Addgene) (Ishizaki et al., 2015) via an LR reaction to produce the pro35S:MpRRT1-tagRFP plasmid. The plasmids pro35S:MpUSE1A-Citrine, pro35S:MpSYP3-Citrine, and pro35S:MpSYP4-Citrine were obtained from Ueda, T. and Kanazawa, T. (National Institute for Basic Biology) (Kanazawa et al., 2016).
The transformation of liverwort using regenerating thalli was performed according to previously described methods (Kubota et al., 2013). Transformants were selected on plates containing 0.5 µM chlorsulfuron or 10 µg/ml hygromycin. For analyses of the subcellular localization of the MpRRT1-tagRFP protein, pro35S:MpUSE1A-Citrine, pro35S:MpSYP3-Citrine, and pro35S:MpSYP4-Citrine constructs were first transformed into Tak-1 thalli, followed by selection with hygromycin. Transformants were confirmed by observing Citrine fluorescence with a laser scanning confocal microscope (FV1000-D BX61, Olympus). Each established line was used for the second transformation with pro35S:MpRRT1-tagRFP and selection with chlorsulfuron.
For plasmid construction, the Tak-1 DNA was extracted from thalli using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s protocol. For genotyping after genome editing, DNA was extracted based on the following method. Approximately 5-mm2 thallus samples were grinded with a Tissuelyser II (Qiagen) with 150 µl of DNA extraction buffer [1.5 M Tris-HCl (pH 8.8), 10% (w/v) SDS, 10 M LiCl, 1.5 M EDTA (pH8.0)], and 150 µl phenol:chloroform:isoamyl alcohol (25:24:1) for 5 min at 20 Hz. Resulting lysates were centrifuged at 20,000 × g for 10 min at 4°C. The supernatant was mixed with the same volume of isopropanol. After another centrifuge step (20,000 × g; 10 min; 4°C), the supernatant was removed and the pellet was washed with 160 µl of 70% ethanol. After further centrifuging (20,000 × g; 10 min; 4°C), the supernatant was removed and the pellet was dried at room temperature for at least 5 min. The dried pellet was dissolved in 50 µl TE buffer and used as the DNA sample for genotyping. For PCR reactions, Tks Gflex DNA Polymerase (Takara Bio) was used according to the manufacturer’s protocol. Primers for genotyping after genome editing are described in Supplementary Table 1.
The morphology of 9-day-old thalli and GUS-stained thalli was imaged using a stereomicroscope (Leica M205 FA). For reporter analyses with proMpRRT:tdTOMATO, tdTOMATO was excited at 546 nm under a fluorescence microscope (Leica DM5500 B). Confocal laser scanning microscopy observations were conducted using a Zeiss LSM880. Citrine and tagRFP were excited with 488-nm and 543-nm laser wavelengths, respectively. Samples were observed with LD-LCI Plan-Apochromat 40x/1.2 NA multi-immersion objectives.
Histochemical assays for GUS activity were performed as described previously (Ishizaki et al., 2012).
For cell wall monosaccharide analysis, the cell wall samples were fractionated as described previously with minor modifications (Nishitani and Masuda, 1979). The 0- to 5-mm region from the apical notch of 3-week-old thalli were subjected to extraction with 80% ethanol at 80°C and a methanol/chloroform mixture at 25°C. Samples were next washed with 100% ethanol three times, followed by three washes with acetone. After removing acetone, the samples were hydrolyzed with 2 N trifluoroacetic acid for 1 h at 121°C. After evaporation and subsequent dissolution in water, monosaccharides were analyzed using a high-performance anion exchange chromatograph (DionexICS 5000) equipped with a CarboPak PA1 column. Detailed conditions for this analysis were described previously (Takenaka et al., 2018).
The A. thaliana genome possesses 34 genes predicted to encode glycosyltransferases of GT106 proteins. Four of the 34 genes (AtRRT1 to AtRRT4) have been identified as encoding RRT enzymes responsible for RG-I synthesis (Takenaka et al., 2018). In contrast, M. polymorpha of the bryophytes possesses 13 genes predicted to encode GT106 proteins, and only a single M. polymorpha gene (Mapoly0033s0138.1) was found to be homologous to the four AtRRTs. This gene was named MpRRT1 and further characterized in this study.
To examine RG-I rhamnosyltransferase activity of MpRRT1 using the fluorescent-labeled RG-I oligosaccharide (Figure 1A), we expressed recombinant MpRRT1 as a FLAG-tagged fusion protein in N. benthamiana leaves and purified it with an anti-FLAG M2 affinity gel. The recombinant protein was detected by immunoblotting as a 61-kDa protein, corresponding to its calculated molecular mass (61 kDa; Figure 1B). A 1-h reaction of the recombinant protein with the RG-I oligosaccharide, GR8-PA (Supplementary Table 2), and UDP-Rha quantitatively produced GR9-PA, a rhamnosyl residue-adduct of GR8-PA (Figure 1C), whereas the protein fraction prepared by the same procedure from N. benthamiana leaves transformed with the empty vector had no enzyme activity. These results show that MpRRT1 exhibits RG-I rhamnosyltransferase activity, acts on RG-I oligosaccharides with a degree of polymerization (DP) more than 5 as acceptor substrates, and prefers those with DP more than 8 (Figure 1D). The enzyme activity did not depend on divalent cations (Figure 1E). Its optimum pH and temperature were approximately 7.0 and 20°C, respectively, under the conditions used in this study. These characteristics are similar to those of AtRRTs (Takenaka et al., 2018).
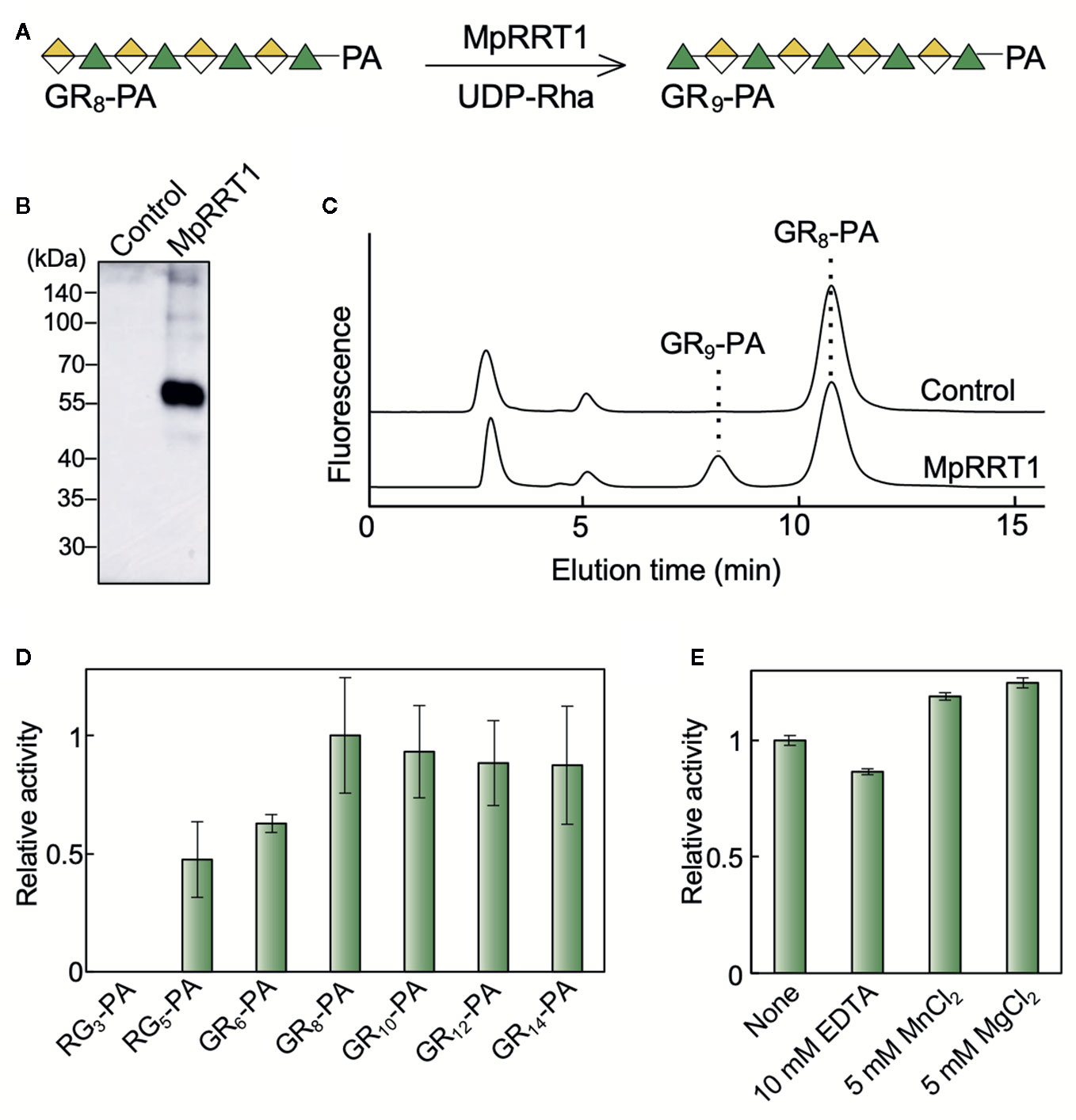
Figure 1 MpRRT1 has RG-I rhamnosyltransferase activity. (A) Reaction scheme for MpRRT1. Green triangles and yellow-divided diamonds represent Rha and GalUA residues, respectively. The fluorescent pyridylamino group (PA) is covalently linked to the reducing-end of the oligosaccharides. (B) SDS-PAGE of the recombinant MpRRT1 protein with a C-terminal FLAG tag expressed in tobacco leaves. The protein was detected by immunoblotting using an anti-FLAG antibody. The control proteins were prepared from tobacco leaves infiltrated with the empty pBI121 vector with the same procedure as that used to prepare MpRRT1. (C) Rhamnosyltransferase activity of MpRRT1. The recombinant protein was reacted with 50 µM GR8-PA and 0.5 mM UDP-Rha at 30°C for 1 h. The upper and lower chromatograms represent after the enzyme reaction with the fraction from control proteins and MpRRT1, respectively. The enzyme product GR9-PA was detected after the enzyme reaction with MpRRT1. (D) Acceptor substrate specificity of MpRRT1. The structures of acceptor oligosaccharides are shown in Supplementary Table 2. (E) Divalent cation-dependence of MpRRT1. Values are presented as the mean of three biologically independent samples with SD.
To functionally characterize MpRRT1 in planta, we next investigated whether the overexpression of MpRRT1 cDNA complements the A. thaliana loss-of-function mutant, in which T-DNA was inserted into the AtRRT1 gene (Figure 2). Whereas the Atrrt1 mutants exhibited an approximate 28% reduction in seed mucilage volume (Supplementary Figure S1 and Figures 2B, C; Takenaka et al., 2018), the pro35S:MpRRT1 transgenic lines in the Atrrt1-1 mutant background showed restored reductions in mucilage production characteristic of the Atrrt1-1 mutant (Figures 2B, C). These results suggest that MpRRT1 functions in the production of RG-I in planta, as is the case with AtRRT1 (Takenaka et al., 2018).
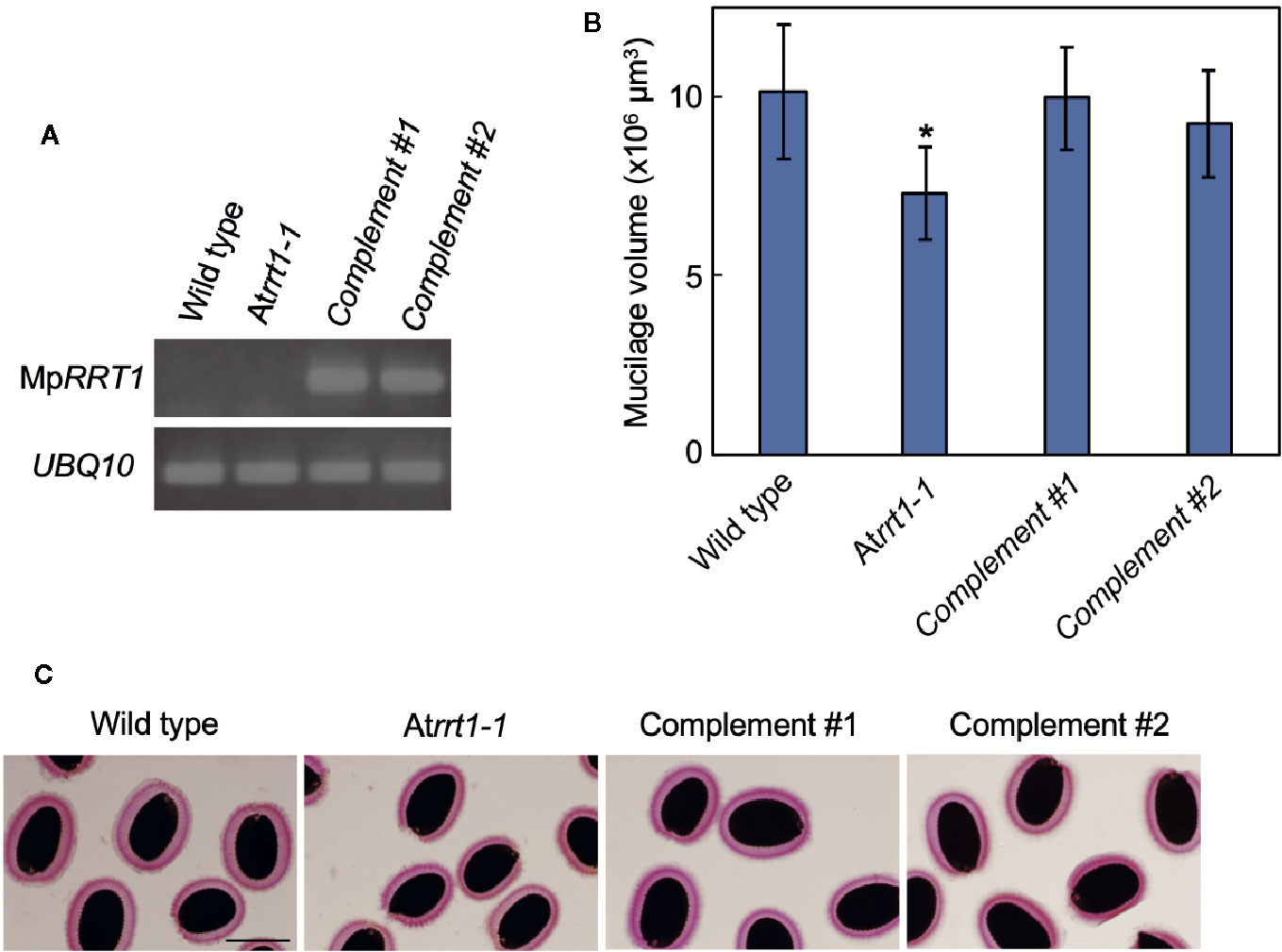
Figure 2 MpRRT1 complements the reduction in seed mucilage production in the Atrrt1-1 mutant. (A) RT-PCR analysis of the MpRRT1 transcript in the wild-type, Atrrt1-1 (SALK_022924), and two complemented Atrrt1-1 (pro35S:MpRRT1/Atrrt1-1) strains. UBQ10 was used as a loading control. (B) Mucilage volumes of seed coats in the wild-type, Atrrt1-1, and two complemented Atrrt1-1 strains. The mucilage volumes are presented as the mean values of 20 seeds with SD. The asterisk indicates that the value was significantly different from those of the wild-type and complemented Atrrt1-1 strains by Student’s t-test: *P < 0.05. (C) Arabidopsis seed mucilage of the wild-type, Atrrt1-1, and two complemented Atrrt1-1 strains, stained with ruthenium red. Scale bar, 250 μm.
Pectin biosynthesis occurs in the Golgi apparatus and AtRRT1 has been shown to localize to this organelle (Takenaka et al., 2018). We next compared the subcellular localization of MpRRT1 with those of marker proteins MpUSE1A, MpSYP4, and MpSYP3, of which the localizations are the endoplasmic reticulum, trans-Golgi network, and cis-Golgi, respectively (Kanazawa et al., 2016). Each marker protein fused with Citrine (green) was co-transformed with pro35S:MpRRT1-tagRFP into thalli of liverworts. The red fluorescence of MpRRT1-tagRFP was distributed in a dot-like pattern (Figures 3B, E, H). This localization did not correspond to that of Citrine-MpUSE1A (Figures 3A, C). The dot-like patterns were also observed for Golgi-localized proteins, specifically Citrine-MpSYP4 (Figure 3D) and Citrine-MpSYP3 (Figure 3G). Among them, Citrine-MpSYP3 co-localized with the MpRRT1-tagRFP (Figure 3I), indicating that MpRRT1 localizes at the cis-Golgi in liverwort.
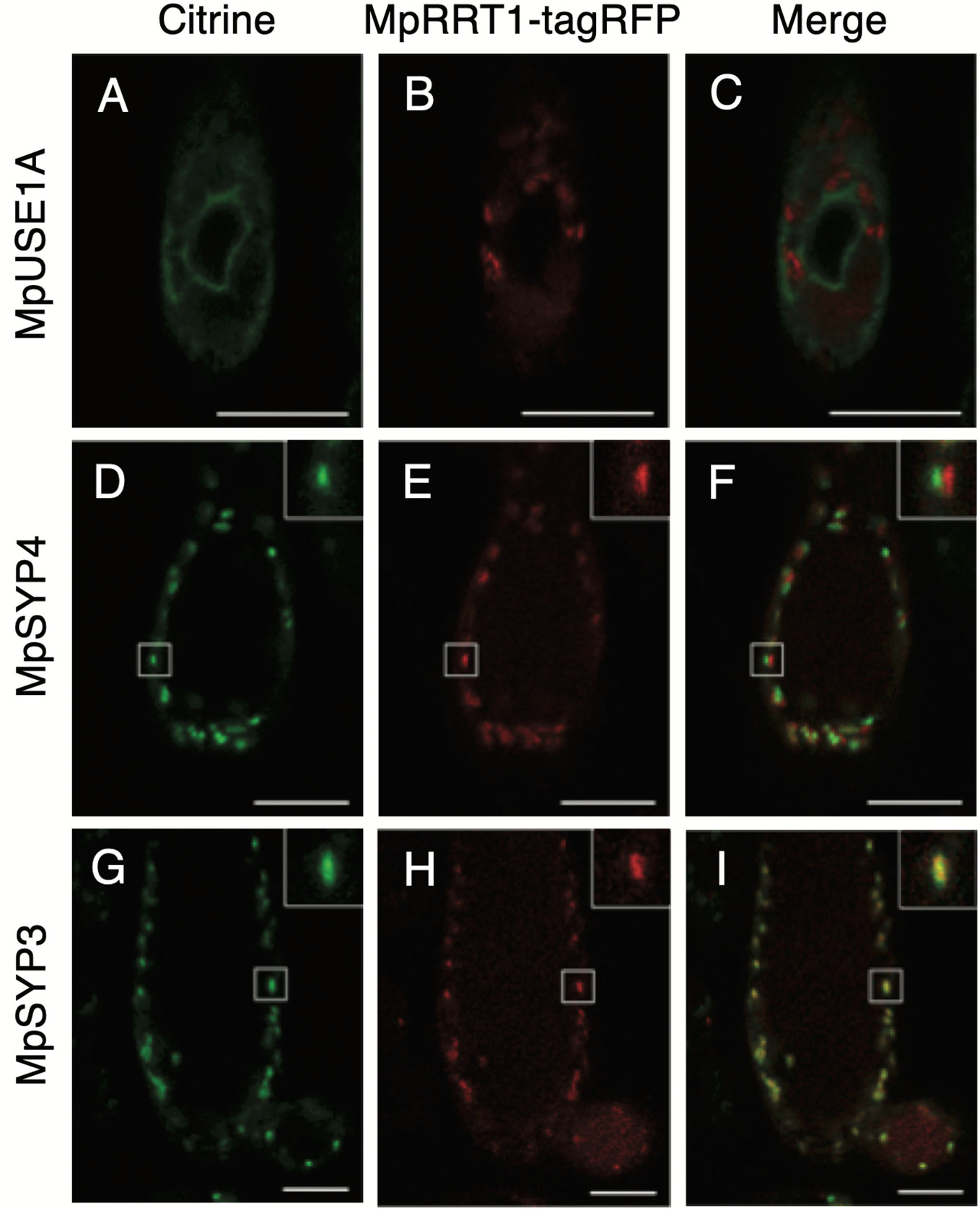
Figure 3 Subcellular localization of MpRRT1-tagRFP protein in liverwort. Thallus cells in 2-week-old gemmalings of transgenic pro35S:MpRRT1-tagRFP liverworts co-overexpressing Citrine-MpUSE1A (A–C), Citrine-MpSYP4 (D–F), or Citrine-MpSYP3 (G–I) were observed with a laser scanning confocal microscope. The localization of the Citrine-proteins (green) (A, D, G), tagRFP florescence (B, E, H), and their merged images (C, F, I) are shown. In D–I, small white squares are shown at an increased magnification at the upper right. Scale bars, 25 μm.
A previous RNA-seq analysis of M. polymorpha tissues (Bowman et al., 2017) showed that MpRRT1 was expressed in all tissues investigated (Figure 4A). To explore the expression pattern of MpRRT1 at a higher spatial resolution, we generated promotor:tdTOMATO and promotor:GUS lines for MpRRT1. The predicted promotor region of MpRRT1 (proMpRRT1, 4,217 bp) was extracted from MarpolBase (http://marchantia.info). Although this region contained duplicated 1,000-bp nucleotide sequences (Supplementary Figure S2), we confirmed that this duplicated region was missing in the M. polymorpha wild-type genome by Sanger sequencing (Supplementary Figure S3). The structure of the MpRRT1 gene is shown in Figure 4B. The upstream promoter region (proMpRRT, 3,217 bp) was used for reporter analysis. According to MarpolBase, a predicted gene (Mapoly0033s0139.1) was located −0.7- to −3.9-kbp upstream of the MpRRT gene (Figure 4B). We excluded the transcriptional start site of Mapoly0033s0139.1 (−3.9-kbp upstream) from the proMpRRT1 region to avoid the unexpected transcription of this gene in transformants. Next, proMpRRT1 was fused with the tdTOMATO gene (proMpRRT1:tdTOMATO) or GUS gene (proMpRRT1:GUS). Each resulting construct was then transformed into thalli of wild-type liverwort. The fluorescence of tdTOMATO was concentrated in the notch of the gemma (Figures 4E, H). Similarly, the strong signal at the apical notch was also observed by GUS-staining 11-day-old thalli in the proMpRRT1:GUS transformants (Figures 4I, J). In addition, GUS staining was observed in the region ~1 mm from the apical notch in 11-day-old thalli (Figure 4I). This region corresponds to the area in which thallus growth is decreased or ceases (Solly et al., 2017). The region of GUS staining was spread out in the 19-day-old thalli except for the region ~1 mm from the apical notch (Figure 4K). The region showing weak GUS staining was previously shown to correspond to the area exhibiting higher aerial growth rates than others (Solly et al., 2017). In the antheridiophore, strong GUS staining was observed in the lobed disc but not in the elongating stalk (Figure 4L). These expression patterns imply that MpRRT1 is predominantly expressed in the meristematic and maturation stage of development in liverwort tissues.
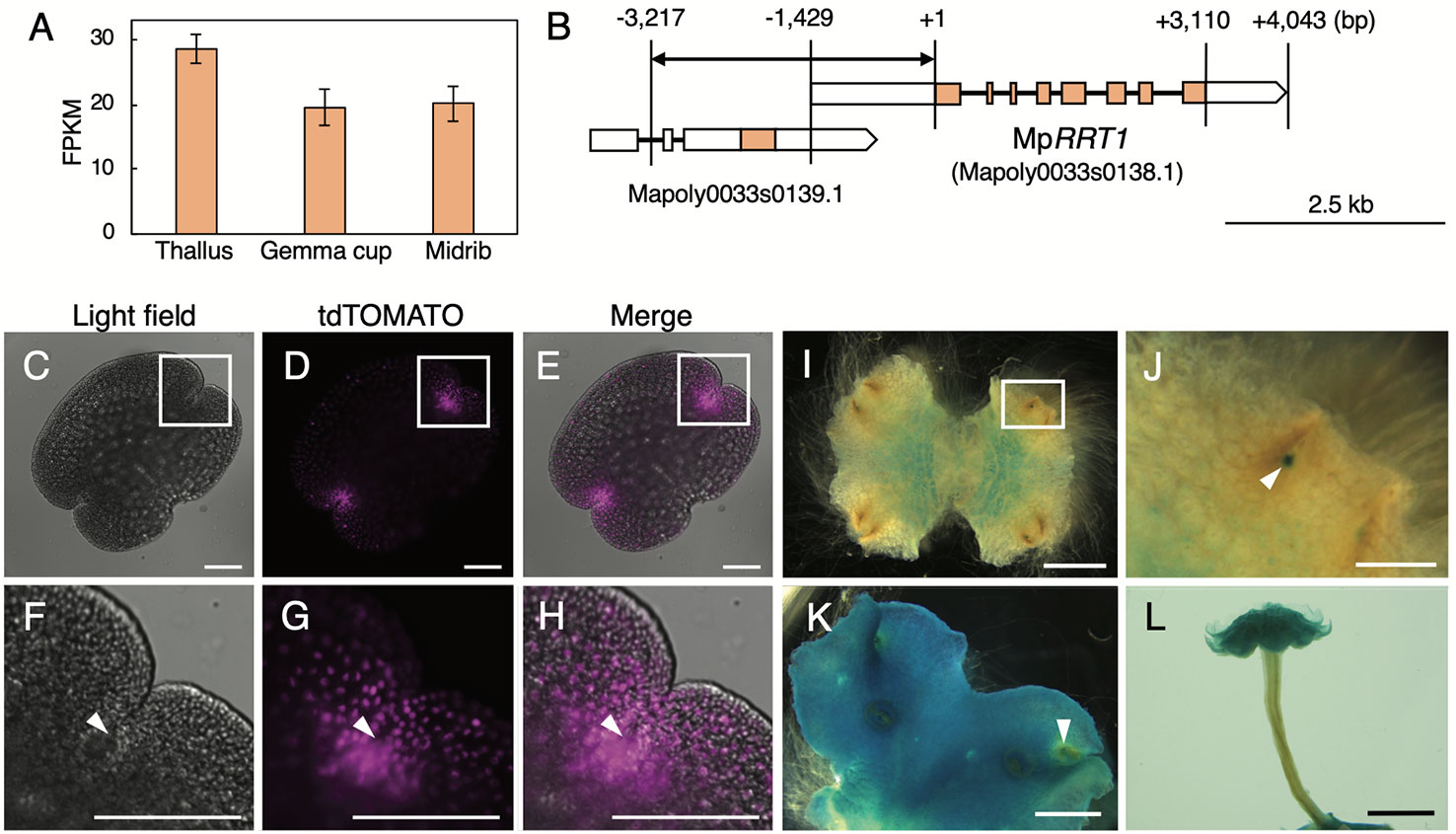
Figure 4 Expression profiles of MpRRT1 in liverwort. (A) Expression profile of MpRRT1 based on RNA-Seq data. Expression levels of MpRRT1 in the thallus, gemma cup, and midrib were estimated based on the relative abundances of transcripts with the unit fragments per kilobase million (FPKM). The values are presented as the mean of three biologically independent samples with SD. (B) Schematic representation of the structure of the Tak-1 MpRRT1 (Mapoly0033s0138.1) and the adjacent gene locus based on Marpolbase and sequencing results in this study. Orange and white boxes represent predicted exons and UTRs, respectively. Black lines represent introns. A bidirectional arrow indicates the promoter region used in reporter analyses. (C–H) Expression pattern of proMpRRT1:tdTOMATO in gemma. Images of light field (C, F), fluorescence images of tdTOMATO (D, G), and their merged images (E, H) are shown. Magnified images of the white squares in the upper panels are presented in the images in lower panels. White arrowheads indicate the apical notch. (I–L) GUS staining of proMpRRT1:GUS in 11-day-old thalli (I, J), 19-day-old thalli (K), and antheridiophores (L). (J) is a magnified image of the area of the white squares of (I). White arrowhead indicates GUS staining at the apical notch. Scale bars, 100 μm (C–H), 500 μm (J), 2 mm (I, K), and 5 mm (L).
To investigate the significance of RG-I in M. polymorpha plants, we generated the MpRRT1-deficient mutants by genome editing using the CRISPR/Cas9 system. These mutants were expected to be RG-I-deficient because MpRRT1 was considered the sole RG-I rhamnosyltransferase-encoding gene found in the M. polymorpha genome. We transformed a construct encoding a guide RNA that targeted the fourth exon of MpRRT1 (Supplementary Figure S4A) and detected >8 events of genome editing around the fourth exon of MpRRT1 in the genome at the T1 generation. Among them, we chose two mutants, Mprrt1-1 with a 155-bp deletion and a 16-bp insertion resulting in deletion of amino acids (ΔD158 to R188) (Supplementary Figure S4B) and Mprrt1-2 with a 5-bp deletion resulting in a frameshift that introduced a premature stop codon (Supplementary Figure S4C) for further analyses (Supplementary Figure S5). The percentages of rhamnose residues among cell wall monosaccharides of Mprrt1-1 and Mprrt1-2 were only 20% and 21% lower than wild-type levels, respectively (Figure 5A). There was also no significant difference in galacturonic acid contents between wild-type and the mutant strains (Figure 5A). We next investigated the morphological differences between the wild-type and the Mprrt1-1 mutant based on 9-day-old gemmalings, 21-day-old gemmalings, and antheridiophores. However, contrary to expectations, prominent morphological differences were not observed between wild-type and mutant strains (Figures 5B–G). These results suggested that MpRRT1 is not the sole RRT gene in M. polymorpha and that this species has other RRT genes in its genome.
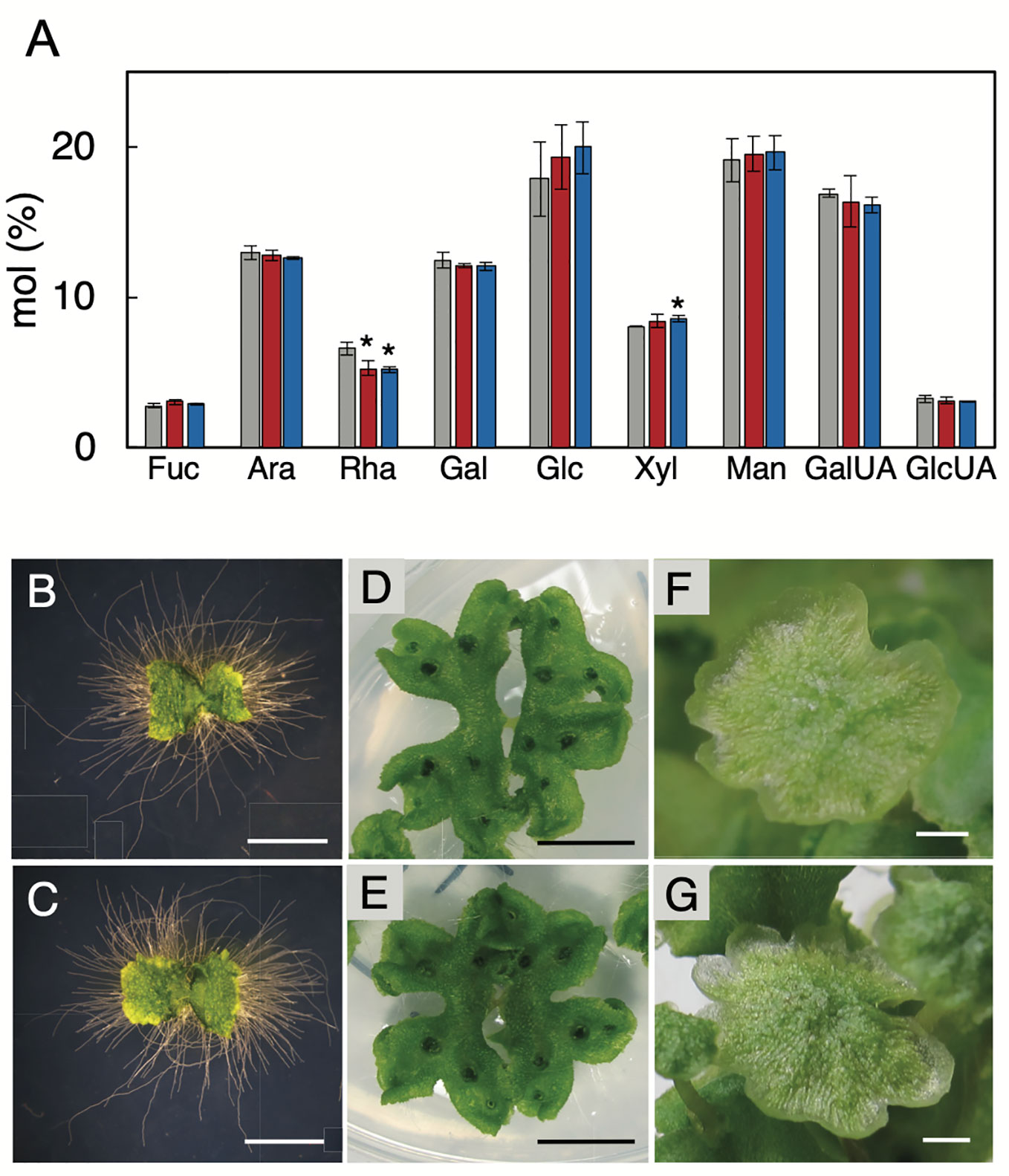
Figure 5 Chemotype and phenotype of the MpRRT1 genome-edited lines. (A) Monosaccharide compositions of thalli in the MpRRT1 genome-edited lines. Trifluoroacetic acid-hydrolysate non-cellulosic fractions from 21-day-old gemmalings of Tak-1 (gray), Mprrt1-1 (red), and Mprrt1-2 (blue) strains were subjected to ion-chromatography. The monosaccharide contents are shown as the percentages of the total amounts of monosaccharides. Values represent means of three biologically independent samples with SD. Asterisks indicate that the value was significantly different from that of Tak-1 by Student’s t-test: *P < 0.05. Ara, arabinose; Rha, rhamnose; Fuc, fucose; Xyl, xylose; Man, mannose; Gal, galactose; Glc, glucose; GalUA, galacturonic acid; GlcUA, glucuronic acid. Nine-day-old gemmalings of Tak-1 (B) and the Mprrt1-1 mutant (C). Gemmalings were grown on medium covered with cellophane to clearly observe rhizoids. Twenty-one-day-old gemmalings of Tak-1 (D) and the Mprrt1-1 mutant (E). Antheridiophores of Tak-1 (F) and the Mprrt1-1 mutant (G). Bars, 2 mm (B, C, F, G) and 1 cm (D, E).
Next, we examined the RG-I rhamnosyltransferase activity of GT106 proteins other than AtRRT1, AtRRT2, AtRRT3, AtRRT4 (Takenaka et al., 2018), and MpRRT1 (this study). First, we examined MpRRT3 (Mapoly0014s0149.1), which is most closely related to the clade containing the RRTs identified in the phylogenetic tree of GT106 (Figure 6). We expressed a recombinant MpRRT3 fused with a FLAG-tag as an 85-kDa protein, for which the molecular mass was calculated to be 81 kDa, in N. benthamiana leaves (Figure 7A). MpRRT3 exhibited the same RG-I rhamnosyltransferase activity as MpRRT1 (Figure 7C), as GR8-PA was used as an acceptor substrate.
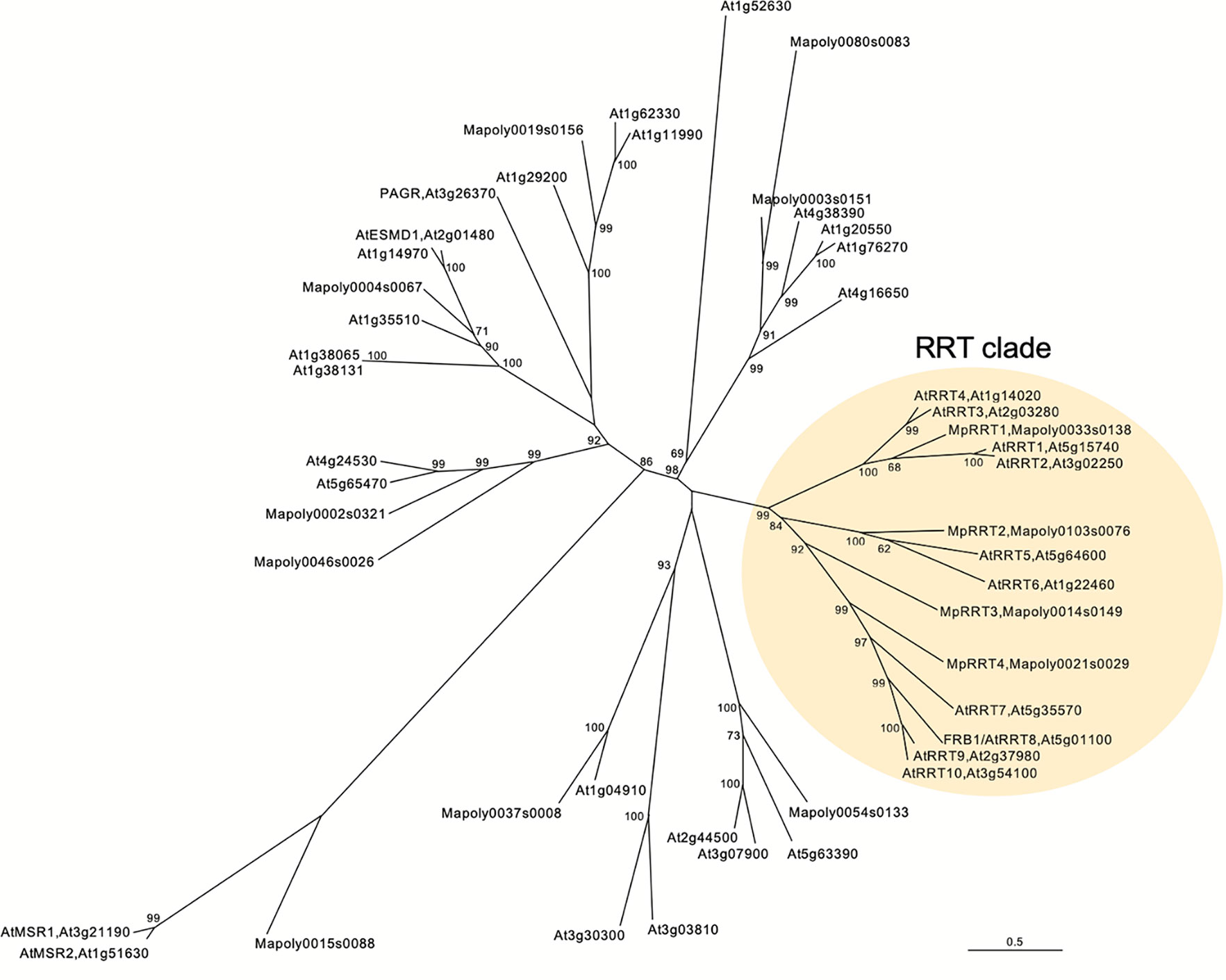
Figure 6 A neighbor joining phylogenetic tree of Marchantia polymorpha and Arabidopsis thaliana GT106 proteins. Amino acid sequences of 34 Arabidopsis and 13 Marchantia GT106 proteins were aligned using ClustalW, and bootstrap analyses were performed with 1,000 replicates. The RG-I rhamnosyltransferase (RRT) clade is highlighted in pale yellow. Scale bar indicates the number of amino acid substitutions per site.
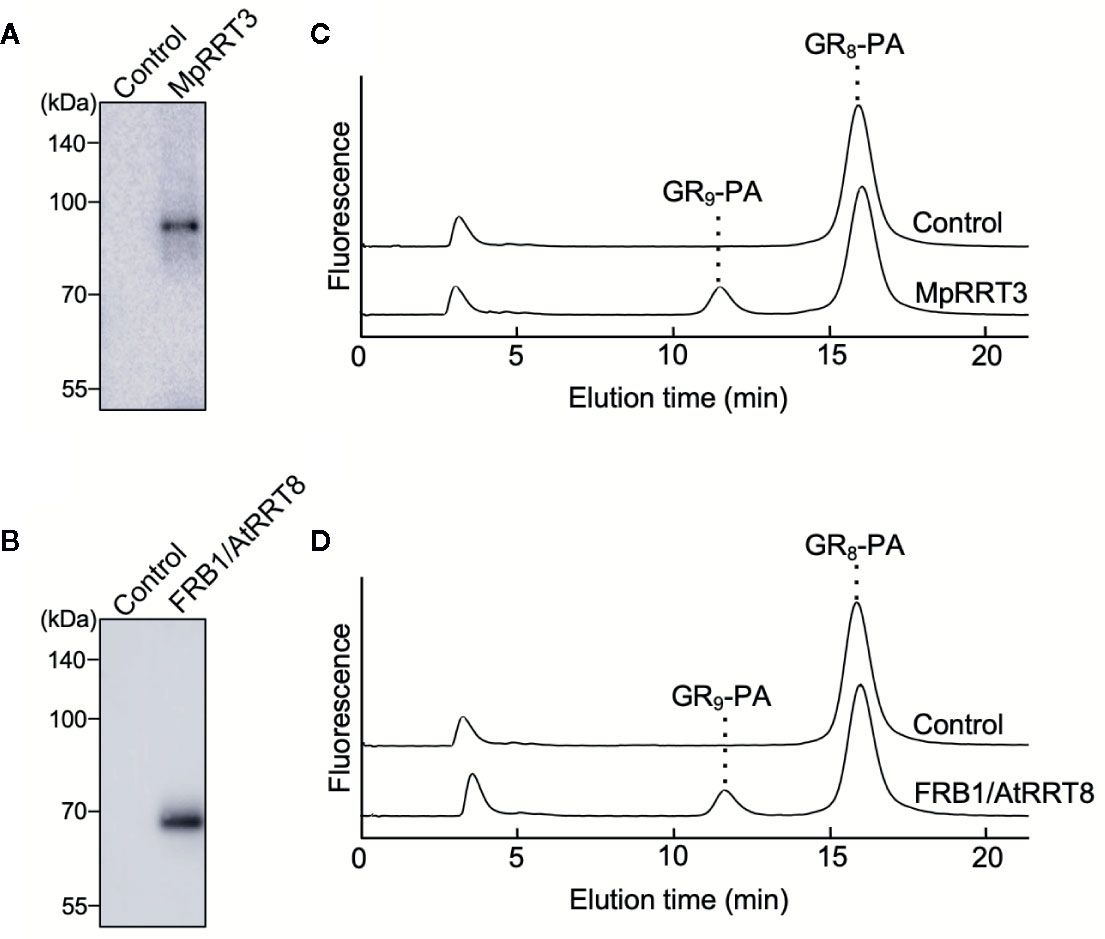
Figure 7 MpRRT3 and FRB1/AtRRT8 have RG-I rhamnosyltransferase activity. (A, B) SDS-PAGE of the recombinant (A) MpRRT3 and (B) FRB1/AtRRT8 proteins with a C-terminal FLAG tag expressed in tobacco leaves. The proteins were detected by immunoblotting using an anti-FLAG antibody. (C, D) Rhamnosyltransferase activity of (C) MpRRT3 and (D) FRB1/AtRRT8. The recombinant proteins were reacted with 50 µM GR8-PA and 0.5 mM UDP-Rha at 30°C for 1 h. The upper and lower chromatograms represent after enzyme reactions with the fraction from control proteins and the recombinant proteins.
The clade containing MpRRT3 in GT106 includes several A. thaliana putative glycosyltransferases (Figure 6). FRB1/AtRRT8, one A. thaliana putative enzyme, was shown to be involved in cell adhesion, but enzyme activity had not yet been characterized (Neumetzler et al., 2012). Accordingly, we produced a recombinant FRB1/AtRRT8 protein as a 68-kDa protein (calculated value, 72 kDa) in N. benthamiana leaves (Figure 7B) and detected its RG-I rhamnosyltransferase activity using GR8-PA as an acceptor substrate (Figure 7D).
These results extended the clade for pectin-synthetic RG-I rhamnosyltransferases in GT family 106. All proteins in the RRT clade defined as shown in Figure 6 appear to have RG-I rhamnosyltransferase activity. Streptophyte plants have several RRT genes in their genomes (Table 1). A. thaliana (angiosperm, eudicot), Oryza sativa (angiosperm, monocot), Picea abies (gymnosperm), Selaginella moellendorffii (lycophyte), M. polymorpha (bryophyte), and Klebsormidium flaccidum (charophyte) were found to have 10, 11, 15, 7, 4, and 2 RRT genes in their genomes, respectively (Table 1).
The first rhamnosyltransferase responsible for the biosynthesis of the RG-I backbone, named RRT, was identified in eudicot plants (Uehara et al., 2017; Takenaka et al., 2018). In this study, we demonstrated the existence of the same enzyme activity in the liverwort M. polymorpha using a recombinant MpRRT1 protein expressed in N. benthamiana (Figure 1). We also observed that the enzyme functions to produce RG-I in vivo (Figure 2). These results biochemically verified the evolutionary view that RRT exists in the genome of Streptophyta plants including the liverwort M. polymorpha (Takenaka et al., 2018).
Pectin biosynthesis occurs in the lumen of the Golgi apparatus (Driouich et al., 2012). Some RG-I biosynthetic glycosyltransferases have been shown to localize to the Golgi endomembrane system (Harholt et al., 2012; Liwanag et al., 2012; Takenaka et al., 2018). This study showed that MpRRT1 localizes to the cis-Golgi (Figure 4). The epitopes of LM5, which recognize the RG-I side chain galactan have been shown to be present mostly in the trans-Golgi and trans-Golgi network of flax root cells (Vicré et al., 1998). These results support the idea that the biosynthesis of the RG-I backbone and side chains occurs in early and late Golgi compartments, respectively.
The developmental (spatial and temporal) regulation of MpRRT1 expression was also observed (Figures 3 and 4). Reporter analysis suggested that MpRRT1 might function at the meristematic and maturation stage rather than the elongation phase in the development of thalli and antheridiophores (Figure 4). The Mprrt1-deficient lines did not show prominent phenotypic changes (Figure 5), although this might require further detailed analysis. These results can be explained by the redundancy of RRT genes in the M. polymorpha genome (Figure 6). Each MpRRT was expressed in all organs investigated (Supplementary Figure S6), showing that these glycosyltransferases cooperatively synthesize RG-I in each plant organ. This fits with the observation that the MpRRT1-deficient mutants did not show complete elimination of RG-I in their cell walls. However, the biological significance of RG-I polysaccharides in M. polymorpha could not be solved in this study. This plant species still has a relative technological advantage over A. thaliana because of its lower redundancy with respect to RRTs (Table 1). The analysis of M. polymorpha mutants deficient in multiple RRT genes is the next subject for RG-I-related biology.
Streptophyte plants have several RRT genes in their genome (Table 1), meaning that approximately one-third of GT family106 exhibits RRT activity. The presence of a large number of RRT genes in the genomes of Streptophyta indicates that the evolution and diversification of RG-I biosynthesis was a critical event during the terrestrialization of Streptophyta.
The A. thaliana genome has 10 RRT genes in total (Figure 6). This number is comparable with that of GAUT (GT8), as 15 genes can be found in its genome (Atmodjo et al., 2013). GAUT is the enzyme responsible for biosynthesis of the HG backbone. This suggests that the biological significance of RG-I is similar to that of HG. The functional analysis of each GAUT has not been straightforward because of redundancy in this gene (Caffall et al., 2009). Therefore, it can be easily seen that the functional analysis of each RRT gene is difficult because all 10 RRT genes are expressed in all tissues and their expression levels and patterns are diverse based on the Arabidopsis eFP browser (Winter et al., 2007). However, frb1/Atrrt8 single-knockout mutations change the biochemical properties of the cell wall and middle lamella and affect cell-cell adhesion (Neumetzler et al., 2012). These pleiotropic effects of this mutation made it to be difficult to specify the glycosyltransferase activity of FRB1/AtRRT8. Taking the results of this study into account, RG-I synthesized by FRB1/AtRRT8 is involved in cell adhesion in the cell wall and/or middle lamella of A. thaliana. FRB1/AtRRT8 affects the contents of monosaccharide residues constituted of RG-I (Neumetzler et al., 2012). Further, it affects the abundance of arabinose and galactose residues rather than the reduction of rhamnose residues (Neumetzler et al., 2012). In this single RRT-deficient A. thaliana, the RG-I structure in the limited region associated with adhesion appears to be affected, and a reduction in rhamnose residues in the frb1/Atrrt8-mutant could not be detected because other RRTs compensate for this to some extent. The changes in arabinose and galactose abundances in this mutant appear to be due to multifaceted effects caused by FRB1/AtRRT8 deficiency. Thus, pectin RG-I synthetic-RRTs are more redundant and diverse in GT106 (Figure 6 and Table 1) than previously thought (Takenaka et al., 2018). A functional study of these RRTs will further contribute to the understanding of the biological roles of RG-I in land plants.
Approximately 30% of the enzymes in the GT106 family were found to be RRTs (Figure 6 and Table 1). However, the biochemical characteristics of other enzymes belonging to the GT106 family appear to be quite different from those of RRTs. This is because previous mutant analyses of the genes belonging to GT106 have shown that they might be involved in the biosynthesis of other polysaccharides, including mannan (Stonebloom et al., 2016; Verger et al., 2016; Smith et al., 2018; Voiniciuc et al., 2019). These GT106 proteins would be undoubtedly glycosyltransferases, as their in vivo functions were lost due to substitutions of amino acid that appear to be catalytic residues (Smith et al., 2018; Voiniciuc et al., 2019). The next step for the study of GT106 is to detect glycosyltransferase activity and analyze biochemical characteristics for each enzyme.
In this study, we concluded that MpRRT1, MpRRT3, and FRB1/AtRRT8 have rhamnosyltransferase activity. This conclusion was highly probable because it was drawn from multiple lines of biochemical evidence (Figures 1, 2, 5, and 7). However, we cannot completely exclude the possibility that the recombinant RRTs indirectly mediate rhamnosyltransferase activity because they were not completely purified from plant microsomal fractions. We did not show an image of the SDS-PAGE gel stained with Coomassie brilliant blue for the protein fractions used for glycosyltransferase assays but did show the western blotting pattern (Figures 1 and 7) because a protein band corresponding to the recombinant protein could not be detected by Coomassie brilliant blue staining. The question of whether RRT alone or a complex containing RRT has rhamnosyltransferase activity is an issue that needs to be clarified in the future.
We extended our knowledge of the RRTs involved in RG-I main chain biosynthesis. However, it has not been determined which galacturonosyltransferase is involved in biosynthesis of the RG-I main chain. Candidates for this include GATL5, GAUT11, and MUCI70, for which deletion mutants show severe phenotypes of reduced seed mucilage (Kong et al., 2013; Voiniciuc et al., 2018). Biochemical analysis of the enzymes encoded by these genes is eagerly awaited.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
BW, TK, KN, and TI designed the research. BW, TK, YT, HK, SN, RY, and KI performed the research. BW, TK, YT, and KI analyzed the data. BW, TK, KN, and TI wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a Grant-in-Aid for Scientific Research (No. 18H05495 and 19H03252 to TI, and 19K16173 to YT) from the Ministry of Education, Culture, Sports, Science, and Technology and a JICA Innovative Asia Scholarship (No. D1707016 to BW) from Japan International Cooperation Agency. It was also supported by the Mizutani Foundation for Glycoscience, the Novartis Foundation (Japan) for the Promotion of Science, and the Program for the Third-Phase R-GIRO Research from the Ritsumeikan Global Innovation Research Organization, Ritsumeikan University to TI.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Konan Ishida for assistance with the construction of plasmid vectors and transformation. We thank Takayuki Kohchi and Ryuichi Nishihama (University of Kyoto) for providing vectors, specifically pMpGE_En03 and pMpGE011, and for suggestions on the transformation and genome editing of liverwort. We also thank Takashi Ueda and Takehiko Kanazawa (National Institute for Basic Biology) for providing the constructs pro35S:MpUSE1A-Citrine, pro35S:MpSYP3-Citrine, and pro35S:MpSYP4-Citrine. We are grateful to Masahiro Kasahara and Atsushi Takeda for providing the Marchantia cDNA and pWAT202 vector, respectively.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00997/full#supplementary-material
Ara, arabinose; DP, degree of polymerization; FPKM, fragments per kilobase million; Fuc, fucose; Gal, galactose; GalUA, galacturonic acid; Glc, glucose; GlcUA, glucuronic acid; HG, homogalacturonan; Man, mannose; Rha, rhamnose; RG-I, rhamnogalacturonan I; RRT, RG-I rhamnosyltransferase; Xyl, xylose.
Atmodjo, M. A., Hao, Z., Mohnen, D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64, 747–779. doi: 10.1146/annurev-arplant-042811-105534
Bowman, J. L., Kohchi, T., Yamato, K., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marcantia polymorpha genome. Cell 171, 287–304. doi: 10.1016/j.cell.2017.09.030
Caffall, K. H., Pattathil, S., Phillips, S. E., Hahn, M. G., Mohnen, D. (2009). Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol. Plant 2, 1000–1014. doi: 10.1093/mp/ssp062
Cosgrove, D. J. (2018). Diffuse growth of plant cell walls. Plant Physiol. 176, 16–27. doi: 10.1104/pp.17.01541
Dean, G. H., Zheng, H., Tewari, J., Huang, J., Young, D. S., Hwang, Y. T., et al. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19, 4007–4021. doi: 10.1105/tpc.107.050609
Driouich, A., Follet-Gueye, M.-L., Bernard, S., Kousar, S., Chevalier, L., Vicré-Gibouin, M., et al. (2012). Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 3, 1–15. doi: 10.3389/fpls.2012.00079
Ebert, B., Birdseye, D., Liwanag, A. J. M., Laursen, T., Rennie, E. A., Guo, X., et al. (2018). The three members of the Arabidopsis glycosyltransferase family 92 are functional β-1,4-galactan synthases. Plant Cell Physiol. 59, 2624–2636. doi: 10.1093/pcp/pcy180
Gamborg, O. L., Miller, R. A., Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Gorshkova, T., Mokshina, N., Chernova, T., Ibragimova, N., Salnikov, V., Mikshina, P., et al. (2015). Aspen tension wood fibers contain β-(1→4)-galactans and acidic arabinogalactans retained by cellulose microfibrils in gelatinous walls. Plant Physiol. 169, 2048–2063. doi: 10.1104/pp.15.00690
Guedes, E. T. P., Laurans, F., Quemener, B., Assor, C., Lainé-Prade, V., Boizot, N., et al. (2017). Non-cellulosic polysaccharide distribution during G-layer formation in poplar tension wood fiber: abundance of rhamnogalacturonan I and arabinogalactan proteins but no evidence of xyloglucan. Planta 246, 857–878. doi: 10.1007/s00425-017-2737-1
Haas, K. T., Wightman, R., Meyerowitz, E. M., Peaucelle, A. (2020). Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science 367, 1003–1007. doi: 10.1126/science.aaz5103
Harholt, J., Jensen, J. K., Sørensen, S. O., Orfila, C., Pauly, M., Scheller, H. V. (2006). ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 140, 49–58. doi: 10.1104/pp.105.072744
Harholt, J., Jensen, J. K., Verhertbruggen, Y., Søgaard, C., Bernard, S., Nafisi, M., et al. (2012). ARAD proteins associated with pectic arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236, 115–128. doi: 10.1007/s00425-012-1592-3
Holsters, M., de Waele, D., Depicker, A., Messens, E., van Montagu, M., Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. doi: 10.1007/bf00267408
Ishii, T. (1997). Structure of functions of feruloylated polysaccharides. Plant Sci. 127, 111–127. doi: 10.1016/S0168-9452(97)00130-1
Ishizaki, K., Chiyoda, S., Yamato, K. T., Kohchi, T. (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091. doi: 10.1093/pcp/pcn085
Ishizaki, K., Nonomura, M., Kato, H., Yamato, K. T., Kohchi, T. (2012). Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J. Plant Res. 125, 643–651. doi: 10.1007/s10265-012-0477-7
Ishizaki, K., Nishihama, R., Ueda, M., Inoue, K., Ishida, S., Nishimura, Y., et al. (2015). Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PloS One 10, e0138876. doi: 10.1371/journal.pone.0138876
Kanazawa, T., Era, A., Minamino, N., Shikano, Y., Fujimoto, M., Uemura, T., et al. (2016). SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 57, 307–324. doi: 10.1093/pcp/pcv076
Kong, Y., Zhou, G., Abdeen, A. A., Schafhauser, J., Richardson, B., Atmodjo, M. A., et al. (2013). GALACTURONOSYLTRANSFERASE-LIKE5 is involved in the production of Arabidopsis seed coat mucilage. Plant Physiol. 163, 1203–1217. doi: 10.1104/pp.113.227041
Kubota, A., Ishizaki, K., Hosaka, M., Kohchi, T. (2013). Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci. Biotechnol. Biochem. 77, 167–172. doi: 10.1271/bbb.120700
Kulkarni, A. R., Peña, M. J., Avci, U., Mazumder, K., Urbanowicz, B. R., Pattathil, S., et al. (2012). The ability of land plants to synthesize glucuronoxylans predates the evolution of tracheophytes. Glycobiology 22, 439–451. doi: 10.1093/glycob/cwr117
Kumakura, N., Otsuki, H., Tsuzuki, M., Takeda, A., Watanabe, Y. (2013). Arabidopsis AtRRP44A is the functional homolog of Rrp44/Dis3, an exosome component, is essential for viability and is required for RNA processing and degradation. PloS One 11, e79219. doi: 10.1371/journal.pone.0079219
Lau, J. M., McNeil, M., Darvill, A. G., Albersheim, P. (1985). Structure of the backbone of rhamnogalacturonan I, a pectin polysaccharide in the primary cell walls of plants. Carbohydr. Res. 137, 111–125. doi: 10.1016/0008-6215(85)85153-3
Lau, J. M., McNeil, M., Darvill, A. G., Albersheim, P. (1987). Treatment of rhamnogalacturonan I with lithium in ethylendiamine. Carbohydr. Res. 168, 245–274. doi: 10.1016/0008-6215(87)80029-0
Leuzinger, K., Dent, M., Hurtado, J., Stahnke, J., Lai, H., Zhou, X., et al. (2013). Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins. J. Vis. Exp. 77, e50521. doi: 10.3791/50521
Liwanag, A. J. M., Ebert, B., Verhertbruggen, Y., Rennie, E. A., Rautengarten, C., Oikawa, A., et al. (2012). Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell 24, 5024–5036. doi: 10.1105/tpc.112.106625
Macquet, A., Ralet, M.-C., Loudet, O., Kronenberger, J., Mouille, G., Marion-Poll, A., et al. (2007). A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19, 3990–4006. doi: 10.1105/tpc.107.050179
McCarthy, T. W., Der, J. P., Honaas, L. A., de Pamphilis, C. W., Anderson, C. T. (2014). Phylogenetic analysis of pectin-related gene families in Physcomitrella patens and nine other plant species yields evolutionary insights into cell walls. BMC Plant Biol. 14, 79. doi: 10.1186/1471-2229-14-79
McCartney, L., Ormerod, A. P., Gidley, M. J., Knox, J. P. (2000). Temporal and spatial regulation of pectic (1→4)-β-D-galactan in cell walls of developing pea cotyledons: implications for mechanical properties. Plant J. 22, 105–113. doi: 10.1046/j.1365-313x.2000.00719.x
McFarlane, H. E., Gendre, D., Western, T. L. (2014). Seed coat ruthenium red staining assay. Bio-Protocal 4, e1096. doi: 10.21769/BioProtoc.1096
Mikkelsen, M. D., Harholt, J., Ulskov, P., Johansen, I. E., Frangel, J. U., Doblin, M. S., et al. (2014). Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae. Ann. Bot. 114, 1217–1236. doi: 10.1093/aob/mcu171
Nakamura, A., Furuta, H., Maeda, H., Nagamatsu, Y., Yoshimoto, A. (2001). Analysis of structural components and molecular construction of soybean soluble polysaccharides by stepwise enzymatic degradation. Biosci. Biotechnol. Biochem. 65, 2249–2258. doi: 10.1271/bbb.65.2249
Neumetzler, L., Humphrey, T., Lumba, S., Snyder, S., Yeats, T. H., Usadel, B., et al. (2012). The FRIABLE1 gene product affects cell adhesion in Arabidopsis. PloS One 7, e42914. doi: 10.1371/journal.pone.0042914
Nishitani, K., Masuda, Y. (1979). Growth and cell wall changes in azuki bean epicotyls I. Changes in wall polysaccharides during intact growth. Plant Cell Physiol. 20, 63–74. doi: 10.1093/oxfordjournals.pcp.a075806
O’Raurke, C., Gregson, T., Murray, L., Sadler, I. H., Fry, S. C. (2015). Sugar composition of the pectic polysaccharides of charophytes, the closest algal relatives of land-plants: presence of 3-O-methyl-D-galactose residues. Ann. Bot. 116, 225–236. doi: 10.1093/aob/mcv089
Ohashi, T., Hasegawa, Y., Misaki, R., Fujiyama, K. (2016). Substrate preference of citrus naringenin rhamnosyltransferases and their application to flavonoid glycoside production in fission yeast. Appl. Microbiol. Biotechnol. 199, 687–696. doi: 10.1007/s00253-015-6982-6
Orfila, C., Huisman, M. M., Willats, W. G., van Alebeek, G. J., Schols, H. A., Seymour, G. B., et al. (2002). Altered cell wall disassembly during ripening of Cnr tomato fruit: implications for cell adhesion and fruit softening. Planta 215, 440–447. doi: 10.1007/s00425-002-0753-1
Paniagua, C., Blanco-Portales, R., Barceló-Muñoz, M., García-Gago, J. A., Waldron, M. A., Barceló-Muñoz, J., et al. (2016). Antisense down-regulation of the strawberry β-galactosidase gene FaβGal4 increases cell wall galactose levels and reduces fruit softening. J. Exp. Bot. 67, 619–631. doi: 10.1093/jxb/erv462
Phyo, P., Wang, T., Kiemle, S. N., O’Neill, H., Pingali, S. V., Hong, M., et al. (2017). Gradients in wall mechanics and polysaccharides along growing inflorescence stems. Plant Physiol 175, 1593–1607. doi: 10.1104/pp.17.01270
Ralet, M. C., André-Leroux, G., Quéméner, B., Thibault, J. F. (2005). Sugar beet (Beta vulgaris) pectins are covalently cross-linked through diferulic bridges in the cell wall. Phytochemistry 66, 2800–2814. doi: 10.1016/j.phytochem.2005.09.039
Roberts, A. W., Roberts, E. M., Haigler, C. H. (2012). Moss cell walls: structure and biosynthesis. Front. Plant Sci. 3, 166. doi: 10.3389/fpls.2012.00166
Smith, D. K., Jones, D. M., Lau, J. B. R., Cruz, E. R., Brown, E., Harper, J. F., et al. (2018). A putative protein O-fucosyltransferase facilitates pollen tube penetration through the stigma–style interface. Plant Physiol. 176, 2804–2818. doi: 10.1104/pp.17.01577
Solly, J. E., Cunniffe, N. J., Harrison, C. J. (2017). Regional growth rate differences specified by apical notch activities regulate liverwort thallus shape. Curr. Biol. 27, 16–26. doi: 10.1016/j.cub.2016.10.056
Stonebloom, S., Ebert, B., Xiong, G., Pattathil, S., Birdseye, D., Lao, J., et al. (2016). A DUF-246 family glycosyltransferase-like gene affects male fertility and the biosynthesis of pectic arabinogalactans. BMC Plant Biol. 16, 90. doi: 10.1186/s12870-016-0780-x
Sugano, S. S., Nishihama, R., Shirakawa, M., Takagi, J., Matsuda, Y., Ishida, S., et al. (2018). Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PloS One 13, e0205117. doi: 10.1371/journal.pone.0205117
Takenaka, Y., Kato, K., Ogawa-Ohnishi, M., Tsuruhama, K., Kajiura, H., Yagyu, K., et al. (2018). Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat. Plants 4, 669–676. doi: 10.1038/s41477-018-0217-7
Uehara, Y., Tamura, S., Maki, Y., Yagyu, K., Mizoguchi, T., Tamiaki, H., et al. (2017). Biochemical characterization of rhamnosyltransferase involved in biosynthesis of pectic rhamnogalacturonan I in plant cell wall. Biochem. Biophys. Res. Commun. 486, 130–136. doi: 10.1016/j.bbrc.2017.03.012
Verger, S., Chabout, S., Gineau, E., Mouille, G. (2016). Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development 143, 2536–2540. doi: 10.1242/dev.132308
Verherbruggen, Y., Marcus, S. E., Chen, J., Knox, J. P. (2013). Cell wall pectin arabinans influence the mechanical properties of Arabidopsis thaliana inflorescence stems and their response to mechanical stress. Plant Cell Physiol. 54, 1278–1288. doi: 10.1093/pcp/pct074
Vicré, M., Jauneau, A., Knox, J. P., Driouich, A. (1998). Immunolocalization of β(1-4) and β (1-6)-D-galactan epitope in the cell wall and Golgi stacks of developing flax root tissues. Protoplasma 203, 26–34. doi: 10.1007/BF01280584
Voiniciuc, C., Engle, K. A., Günl, M., Dieluweit, S., Schmidt, M. H.-W., Yang, J.-Y., et al. (2018). Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol. 178, 1045–1064. doi: 10.1104/pp.18.00584
Voiniciuc, C., Dama, M., Gawenda, N., Stritt, F., Pauly, M. (2019). Mechanistic insights from plant heteromannan synthesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 116, 522–527. doi: 10.1073/pnas.1814003116
Wang, D., Samsulrizal, N. H., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2019). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 170, 544–557. doi: 10.1104/pp.18.01187
Keywords: glycosyltransferase, GT106, Marchantia polymorpha, pectin, rhamnogalacturonan I, rhamnosyltransferase
Citation: Wachananawat B, Kuroha T, Takenaka Y, Kajiura H, Naramoto S, Yokoyama R, Ishizaki K, Nishitani K and Ishimizu T (2020) Diversity of Pectin Rhamnogalacturonan I Rhamnosyltransferases in Glycosyltransferase Family 106. Front. Plant Sci. 11:997. doi: 10.3389/fpls.2020.00997
Received: 07 April 2020; Accepted: 17 June 2020;
Published: 02 July 2020.
Edited by:
Georg J. Seifert, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Olga A. Zabotina, Iowa State University, United StatesCopyright © 2020 Wachananawat, Kuroha, Takenaka, Kajiura, Naramoto, Yokoyama, Ishizaki, Nishitani and Ishimizu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeshi Ishimizu, aXNoaW1penVAZmMucml0c3VtZWkuYWMuanA=
†Present addresses: Takeshi Kuroha, Institute of Agrobiological Sciences, National Agriculture and Food Research Organization, Tsukuba, Japan
Hiroyuki Kajiura, International Center for Biotechnology, Osaka University, Suita, Japan
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.