
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 July 2020
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00957
Excessive cadmium (Cd) damages plants by causing cell death. The present study discusses the function of natural resistance-associated macrophage protein (NRAMP) on cell death caused by Cd in Malus hupehensis. MhNRAMP1 was isolated from M. hupehensis roots, and its protein was located in the cell membrane as a transmembrane protein characterized by hydrophobicity. MhNRAMP1 expression in the roots was induced by Cd stress and calcium (Ca) deficiency. MhNRAMP1 overexpression increased Cd concentration in yeasts and enhanced their sensitivity to Cd. Phenotypic comparisons of plants under Cd stress revealed that the growth of transgenic tobacco and apple calli overexpressing MhNRAMP1 was worse than that of the wild type (WT). The Cd2+ influx of transgenic tobacco roots and apple calli was higher, and the recovery time of the Cd2+ influx to a stable state in transgenic apple calli was longer than that of the WT. Cd accumulation and the percentage of apoptotic cells in transgenic lines were higher. Correspondingly, the caspase-1-like and vacuolar processing enzyme (VPE) activities and MdVPEγ expression were higher in transgenic apple calli, but the expression levels of genes that inhibit cell death were lower than those in the WT under Cd stress. Moreover, the Cd translocation from the roots to leaves was increased after MhNRAMP1 overexpression, but the Cd translocation from the leaves to seeds was not affected. These results suggest that MhNRMAP1 exacerbated Cd-induced cell death, which was accomplished by mediating Cd2+ uptake and accumulation, as well as stimulating VPE.
Cadmium (Cd) is a toxic heavy metal and widespread pollutant found in natural environments. When exposed to an environment containing Cd, the Cd concentration in plants and animals easily exceeds the normal range due to the enrichment of Cd, which seriously threatens human health though the food chain (He et al., 2011). Cd in soil is easily taken up by plant roots and causes severe phytotoxicity to plants (Zare et al., 2018). Excessive Cd restricts nutrient uptake (Yamaguchi et al., 2016; Wang et al., 2017; Cengiz et al., 2019; Wu et al., 2019), decreases photosynthetic efficiency (Pereira de Araújo et al., 2017), delays seed germination (Zhang et al., 2014), and impairs plant growth and development (Bruno et al., 2017; Sofo et al., 2017), resulting in serious production reductions (Abbas et al., 2018; Guo et al., 2018; Chen et al., 2019). The current study shows that some orchard soils and even fruits have been contaminated by Cd, which threatens the growth of fruit trees and the edible safety of fruits (Cheng et al., 2015).
The entry of Cd into plant cells often accompanies the uptake of some essential micronutrients, such as iron (Fe), zinc (Zn), and manganese (Mn), because plant cells do not have a specific channel that binds to Cd only (Himeno et al., 2002; Verbruggen et al., 2009; Cai et al., 2019; Zou et al., 2020). The natural resistance-associated macrophage protein (NRAMP) family is one type of transporter responsible for metal uptake and translocation, such as Cd, Zn, Fe, and Mn (Cai et al., 2019; Wang et al., 2019c; Zou et al., 2020). OsNRAMP6 in rice functions as a Fe and Mn transporter (Peris-Peris et al., 2017). TpNRAMP3 in polish wheat transports Cd and Mn, but not Fe or Zn (Peng et al., 2018). AhNRAMP1 is responsible for Mn and Zn uptake in peanut (Wang et al., 2019c). Due to the response to metal uptake and transport, NRAMPs regulate many environmental stress responses. NRAMP2 is required for Arabidopsis root growth under Mn deficiency (Gao et al., 2018). Knockout of OsNRAMP5 produces low Cd-accumulating indica rice without compromising yield (Tang et al., 2017). OsNRAMP1 regulates rice aluminum (Al) tolerance by controlling Al uptake (Li et al., 2014). OsNRAMP6 contributes to disease resistance in rice by transporting Fe and Mn (Peris-Peris et al., 2017). AtNRAMP6 possesses the ability to transport Cd and maintain intracellular Fe homeostasis in Arabidopsis under Fe-deficient conditions (Cailliatte et al., 2009; Li et al., 2019). NRAMPs have been well studied in rice because of their good control of Cd accumulation in grain; however, NRAMP in fruit trees is rarely reported. Previously, two NRAMPs were cloned from Malus. MbNRAMP1 in M. baccata is responsible for Cd accumulation (Xiao et al., 2008). MxNRAMP1 in M. xiaojingensis promotes Fe absorption and increases plant resistance to Fe deficiency (Pan et al., 2015). Although MbNRAMP1 increased yeast’s sensitivity to Cd, the mechanism has not been elucidated. The impact on fruit safety has not been reported.
Cell death is a stress response to Cd in plants. The excessive accumulation of reactive oxygen species (ROS) caused by Cd exacerbates DNA damage and increases cell membrane permeability and the loss of transmembrane potential (Li et al., 2003; Michelle et al., 2014; Li et al., 2017; Huybrechts et al., 2019; Wang et al., 2019b). Vacuolar rupture is a unique and important factor causing cell death in higher plants, which is mediated by vacuolar processing enzyme (VPE) (Yamada et al., 2020). VPE causes vacuolar rupture and initiates the proteolytic cascade leading to cell death and also possesses peptide ligation activity for producing cyclic peptides, which support developmental and environmental responses in plants (Jiang et al., 2019; Yamada et al., 2020). VPE is therefore recognized as a moderator in response to environmental tolerances. Expression of IbVPE1 from sweet potato in Arabidopsis affects leaf development, flowering time and chlorophyll catabolism (Jiang et al., 2019). Aluminum-induced cell death requires upregulation of NtVPE1 in tobacco (Kariya et al., 2018). The suppression of OsVPE3 enhances salt tolerance by attenuating vacuole rupture during programmed cell death in rice (Lu et al., 2016). Our previous study showed that the expression of MhVPEγ exacerbated cell death in Arabidopsis under high temperature stress (Su et al., 2015). Correspondingly, cell death suppressors are also needed in plant responses to cell death, such as BAX inhibitor-1 (BI-1), Bcl-2-associated athanogene (BAG), and a defender against apoptosis death (DAD). BI-1 is a BCL-2- and BCL-XL-interacting anti-apoptotic protein capable of inhibiting BAX-mediated cell death induced by ROS in plants (Ishikawa et al., 2013; Ramiro et al., 2016). Plant BAG proteins function in cytoprotection under stress conditions (Kabbage and Dickman, 2008). DAD regulates light-induced cell death antagonistically through jasmonate and salicylate levels (Beaugelin et al., 2019). The expression of MhBAG and MhBI-1 was associated with root cell death in Malus hupehensis (Fan et al., 2013). The molecular machinery underlying cell death in plants is complex, and cell death caused by Cd has rarely been reported.
Malus. hupehensis Rehd. var. pingyiensis Jiang is a unique plant germplasm resource found in China that exhibits apomictic characteristics (Yang et al., 2008). It has waterlogging tolerance and is often used as an apple root stocks. In this study, the full-length of MhNRAMP1 was obtained from M. hupehensis roots. The effects of MhNRAMP1 on Cd-induced cell death, and Cd2+ uptake and accumulation were investigated using MhNRAMP1-overexpressing transgenic tobacco and apple calli. Phenotypic comparisons were made between wild type (WT) and transgenic plants under Cd stress. Moreover, the Cd translocation from roots to leaves and from leaves to seeds, and the expression of genes related to cell death were analyzed to investigate the possible regulatory mechanisms of MhNRAMP1 involved in Cd-induced cell death and Cd translocation to seeds.
M. hupehensis var. pingyiensis Jiang, tobacco (Nicotiana tabacum NC89), and apple calli (“Orin” cultivar) were used in this study. The germinated M. hupehensis seeds after stratification were cultivated in plastic pots filled with soil and grown in incubators under a 14/10 h light/dark photoperiod at 25/22°C. Seedlings with eight leaves were used in the experiment.
WT tobacco seedlings were grown in soil for 40 d, and the leaves were selected for genetic transformation. Both the WT and transgenic tobacco seedlings were grown in Murashige and Skoog’s (MS) agar medium and cultured in incubators under a 16 h light and 8 h dark photoperiod at 25°C.
The WT apple calli were used for genetic transformation. Both the WT and transgenic apple calli were grown on MS agar plates containing 0.5 mg L-1 IAA and 1.5 mg L-1 6-BA at 24°C and were subcultured in 16 d intervals.
The complete mRNA sequence of MhNRAMP1 was obtained by searching for MdNRAMP1 (NCBI accession No.: XP_008354189.2) in the NCBI database and was amplified using MhNRAMP1-F and MhNRAMP1-R primers provided in Table S1, and then registered as MhNRAMP1 in GenBank (accession No.: MT035802.1). Complementary DNA (cDNA) was produced by reverse-transcription using the total RNA from M. hupehensis roots (Xu et al., 2012; Zhang et al., 2019a). PCR amplification was performed as follows: 5 min at 94°C, 35 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 10 min. The products were checked by sequencing (BGI, Shenzhen, China).
Multiple sequence alignments were performed using DNAMAN software (Lynnon Biosoft, USA). The full-length amino acid sequences of NRAMPs (Figure S1) were used for phylogenetic analysis. All of the acquired sequences were first aligned by ClustalX (version 1.83) software with the default parameters (Thompson et al., 1997). A neighbor-joining phylogenetic tree was constructed using MEGA6.0 software after 1,000 bootstraps (Tamura et al., 2007). ExPASy software was used to compute the isoelectric points (pI) and molecular weights (MW) (https://web.expasy.org/compute_pi/), as well as predict hydrophobicity (https://web.expasy.org/protscale/)and possible transmembrane helices (http://www.ch.embnet.org/software/TMPRED_form.html).
The open reading frame sequence of MhNRAMP1MhNRAMP1 was obtained using the NR1-F and NR1-R primers (Table S1) and was then cloned into the pROKII-GFP vector. Fluid from Agrobacterium containing the MhNRAMP1-GFP fusion vector or pROKII-GFP control vector was infiltrated into the WT tobacco leaves (Sheludko et al., 2007). The infiltrated tobacco was cultured in the dark for 2 d, and then the infected leaves were cut and observed under a Fluo-View FV1000confocal laser scanning microscope (Olympus, Japan).
The cultured M. hupehensis seedlings were irrigated with 1/2 Hoagland’s nutrient solution containing 50 μM CdSO4 for 14 d. The control M. hupehensis seedlings were irrigated with 1/2 Hoagland’s nutrient solution alone. Roots, stems, and leaves were collected after 0.5 h, 6 h, 12 h, 24 h, 48 h, 7 d, and 14 d. For Mn, Zn, Fe, and Ca stress responses, M. hupehensis seedlings were irrigated with 1/2 Hoagland’s nutrient solution containing 8 mM MgCl2, 8 mM ZnSO4, 8 mM FeCl3, or 8 mM CaNO3 for 24 h. For Mn-, Zn-, Fe-, and Ca-deficiency responses, M. hupehensis seedlings were irrigated with 1/2 Hoagland’s nutrient solution containing 0 mM Mn, 0 mM Zn, 0 mM Fe, or 0 mM Ca for 14?A3B2 show [#,32] ?> d. Lateral roots were subsequently sampled. Afterward, MhNRAMP1 expression was measured.
The MhNRAMP1 ORF was amplified using NRAMP1-YF and NRAMP1-YR and then subcloned into the pYES2 empty vector. The MhNRAMP1-pYES2 recombinant vector and pYES2 empty vector were transformed into the BY4741 WT yeast strain. The transformed yeasts were selected on SD solid medium containing ampicillin. The transformed yeasts after sequence confirmation were cultured in liquid SD medium at 30°C until OD600 = 0.8. Each cell suspension was diluted to five sequential dilutions: 1, 0.1, 0.01, 0.001, and 0.0001. Five mL of each diluted cell suspension was spotted on plates with SD medium containing CdSO4 (0 or 10 μM) with 2% galactose. All plates were incubated at 30°C for 3 d to observe growth.
To further confirm metal sensitivity, 5 μL of yeasts (OD600 = 0.8) were cultured in 50 mL of SD liquid medium with 2% galactose and 5 μm CdSO4. OD600 values were measured every 6 h using a microplate spectrophotometer (Fisher Scientific, USA) with three replicates.
The Cd was extracted from the yeasts grown in SD liquid medium containing 5 μM CdSO4 for 84 h and was then determined by inductively coupled plasma-mass spectrometry (ICP-MS) (Fisher Scientific, USA).
The above PCR products were subcloned into the pROKII vector, and then the CaMV35S-MhNRAMP1 confusion vector was subsequently transformed into the LBA4404 A. tumefaciens strain. Then, the transgenic tobacco generation was obtained using the leaf disk transformation (Horsh et al., 1985). Three tobacco lines were selected for further analysis.
The sterilized tobacco seeds were sown on MS agar medium for 10 d, then transferred to plates containing 50 μm CdSO4, cultured for 14 d, and then photographed. Some of the seedlings were used for the Cd2+ flux analysis. The other seedlings were cultured until they were 80 d old and were then transferred to pots. The seedlings in pots were irrigated with 1/2 Hoagland’s nutrient solution (control) or 1/2 Hoagland’s solution containing 50 μM CdSO4 for 14 d. Then, the lateral roots, leaves, and seeds were collected to measure the Cd contents.
The CaMV35S-MhNRAMP1 overexpression vector was used, and the MhNRAMP1-overexpression apple calli, NR1 and NR2, were generated through Agrobacterium-mediated genetic transformation (An et al., 2018).
The 8-day-old WT and transgenic apple calli (NR1, NR2) were transferred to MS agar medium containing CdSO4 and were photographed after 10 d. The fresh weight, Cd2+ flux, Cd contents, fluorescence, and gene expression levels were measured.
The cDNA was obtained as described above in 2.2. The relative expressions of MhNRAMP1, MdVPEγ (NCBI accession No.: XM_008379705.2), MdBI-1 (NCBI accession No.: XM_008358248.3), MdBAG (NCBI accession No.: XM_008378307.3), and MdDAD (NCBI accession No.: NM_001293917.1) were analyzed by quantitative real-time PCR (qRT-PCR) using SYBR Premix Ex Taq II (TaKaRa, Beijing, China). The expression levels of MhNRAMP1 were normalized to the Mh18S gene, and the expression levels of MdBI-1, MdBAG MdDAD, and MdVPEγ were normalized to the MdACTIN gene. Primers are provided in Table S1.
The net Cd2+ flux of tobacco and apple calli was measured using non-invasive micro-test technology (NMT100 series) (YoungerUSA LLC, Amherst, MA, USA) [Xuyue (Beijing) Science & Technology Co., Ltd., Beijing, China]. The pre-pulled and silanized microsensor was made following the methods of Zhang et al. (2019a). The Cd2+ influx of tobacco roots was measured in fresh measuring solution for 25 min according to the methods described by Zhang et al. (2019a). The Cd2+ influx of apple calli was recorded for 25 min using the methods of Zhang et al. (2019b). imFluxes 2.0 software (YoungerUSA, LLC, Amherst, MA, USA) was used for data recording. Each treatment was repeated three times and all tests were repeated six times.
Dried tobacco samples and apple calli were digested with nitric acid in a MARS 6 microwave (CEM Microwave Technology, Ltd., Matthews, NC, USA). The Cd concentrations were determined using a NexION 300×, ICP-MS instrument (PerkinElmer, USA).
The translocation factor (TF) is defined as the transferability of Cd from one organ to another. TFs were calculated as the ratio of Cd concentration in the root, leaf or seed using the following equations:
where C[leaf], C[root], and C[seed] are the Cd contents of the leaves, roots, and seeds, respectively.
The distribution of Cd2+ in apple calli cells was detected by a fluorescence probe Leadmium™ Green AM (Invitrogen, Carlsbad, CA, USA) according to Zhang et al. (2019b).
Apoptotic activities were analyzed by determining the amount of annexin V positive cells for apoptosis or the amount of pI-positive cells for necrosis using a flow cytometer (PA, Partec, Germany) (Zhao et al., 2011). The apoptosis rate was expressed as the percentage of apoptotic cells (%).
Caspase 1-like activities were measured using a Caspase 1 Activity Assay kit and Micro- Bradford assay kit (Beyotime, Shanghai, China) following the manufacturer’s instructions. Total protein extracts (20 mg) were incubated consecutively for 2 h at 37°C with the synthetic tetrapeptide Ac-YVAD-pnitroaniline (pNA), during which, the addition of the substrate resulted in a signal caused by the caspase 1-dependent cleavage of the chromophore pNA from the labelled substrate. Caspase 1-like activities were measured at 405 nm. Enzymatic activities were expressed as the percentage of activity present in control extracts (%). Each measurement was conducted with three independent experiments.
VPE activities were measured following the methods with minor modifications described by Zhang et al. (2019a).
The data were analyzed by an analysis of variance (ANOVA) using SPSS v16.0 software (IBM, Chicago, IL, USA). All experiments were conducted in triplicate and the data are expressed as the mean ± standard deviation (SD). The confidence level for statistical significance was p ≤ 0.05. Figures were plotted using Origin v9.0 software (OriginLab, Northampton, MA, USA).
The ORF of MhNRAMP1 was 1,638 bp encoding a 545-amino-acid peptide. The deduced amino acid sequence was a 59.12 kDa polypeptide with a pI of 8.46. The MhNRAMP1 protein was predicted to be hydrophobic and had 11 strong transmembrane helices, implying that it was likely a transmembrane protein (Figures S1A, B).
Multiple alignments showed that the MhNRAMP1 protein was highly homologous to MdNRAMP1 and MdNRAMP6 from apple (99.08% identity), PbNRAMP6 from Pyrus bretschneideri (96.70%), PpNRAMP6 from Prunus persica (92.66%), and AtNRAMP1 from Arabidopsis thaliana (77.66%) (Figure S1C). The phylogenetic tree showed that MhNRAMP1 was homologous to a number of NRAMPs, but was most closely related to MdNRAMP1 and MdNRAMP6 (Figure S1D). The subcellular location indicated that MhNRAMP1 was localized in the cell membrane (Figure S1E).
The expression levels of MhNRAMP1 were examined in different plant tissues and treatments. Under normal conditions, MhNRAMP1 was highly expressed in the roots (Figure 1A), but the expression varied greatly in different tissues as the Cd-treatment period progressed. MhNRAMP1 in the roots was induced by Cd stress and was highly expressed after 24 h (Figure 1B). Despite starting at a low level, MhNRAMP1 expression in the stems was slightly induced at 24 h under Cd stress (Figure 1C). However, MhNRAMP1 in the leaves was inhibited under Cd stress (Figure 1D). These results indicated that MhNRAMP1 was preferentially expressed in the roots, which were more sensitive to Cd than the stems or leaves.
The responses of MhNRAMP1 in the roots to the over-supplementation or deficiency of nutrient metals, including Fe, Mn, Zn, and Ca, were also investigated. MhNRAMP1 expression was significantly upregulated by Fe and was slightly upregulated by the supply of Mn, Zn, and Ca (Figure 1E). In contrast, MhNRAMP1 expression was downregulated by Fe, Mn, and Zn deficiency, but was induced by Ca deficiency (Figure 1F).
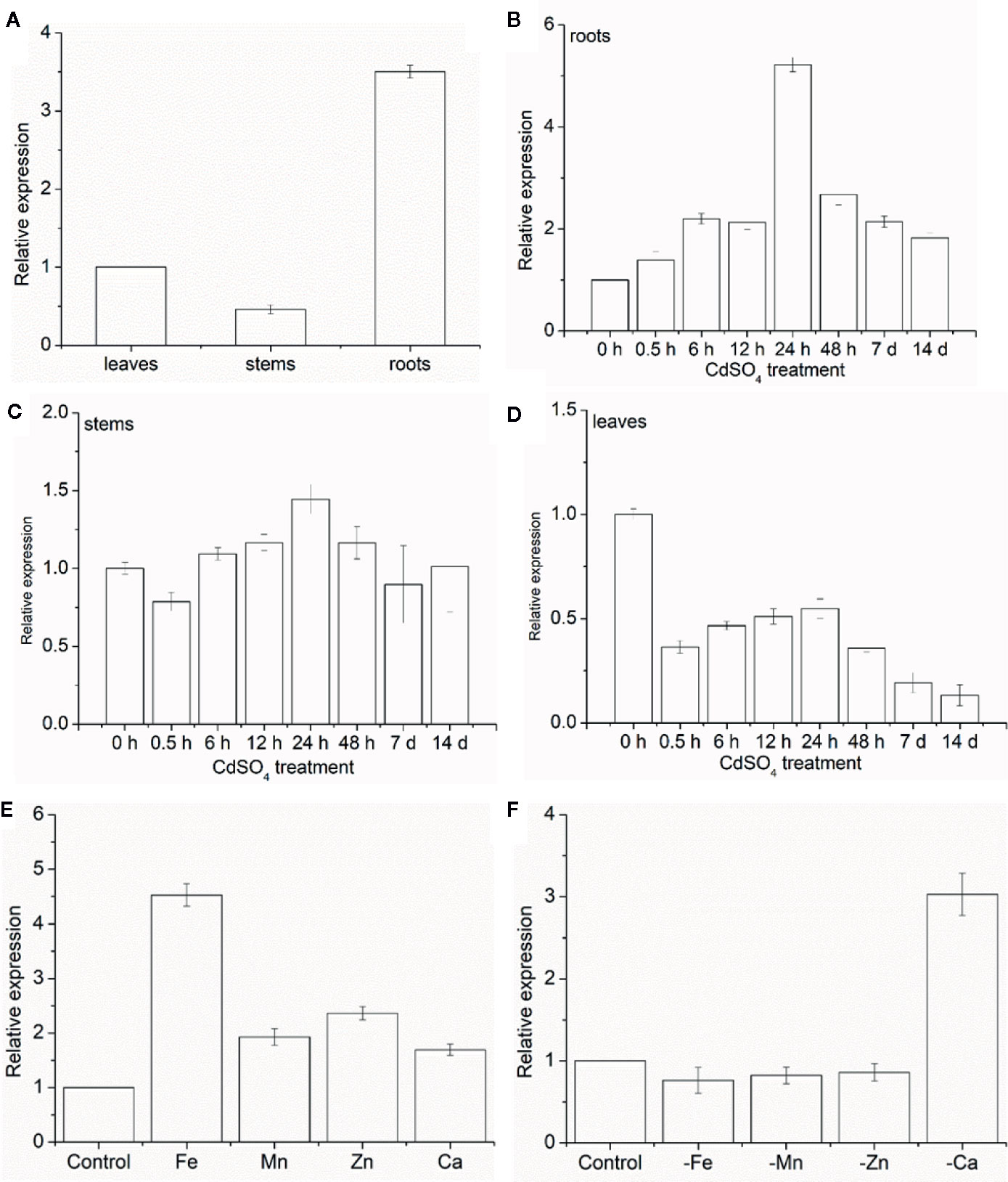
Figure 1 Expression pattern analysis of MhNRAMP1. (A–D) The relative expression of MhNRAMP1 in M. hupehensis roots, stems, and leaves under normal (A) and 50 μm CdSO4 conditions (B–D). (E, F) The relative expression of MhNRAMP1 in M. hupehensis roots under over-supplementation (E) and deficiency (F) of nutrient metals, including Fe, Mn, Zn, and Ca. Data are presented as the mean ± standard derivation (SD) of three replicates; error bars represent the SD.
To understand the response of MhNRAMP1 to Cd, the transgenic yeasts, tobacco and apple calli were used to measure the Cd uptake and accumulation. Figure 2A illustrates that the transformed yeasts with MhNRAMP1 had higher Cd concentrations than the yeasts with the empty vector. The Cd contents in the roots, leaves, and seeds of transgenic tobacco lines (TR1, TR2, TR3) that overexpressed MhNRAMP1 were higher than those of the WT tobacco (Figure 2B). The transgenic apple calli (NR1 and NR2) that overexpressed MhNRAMP1 also had higher Cd contents than the WT apple calli (Figure 2C). The Cd2+ distribution, as indicated by a fluorescent probe, showed that the cells of transgenic apple calli had stronger green fluorescent spots than the WT (Figure 2D).
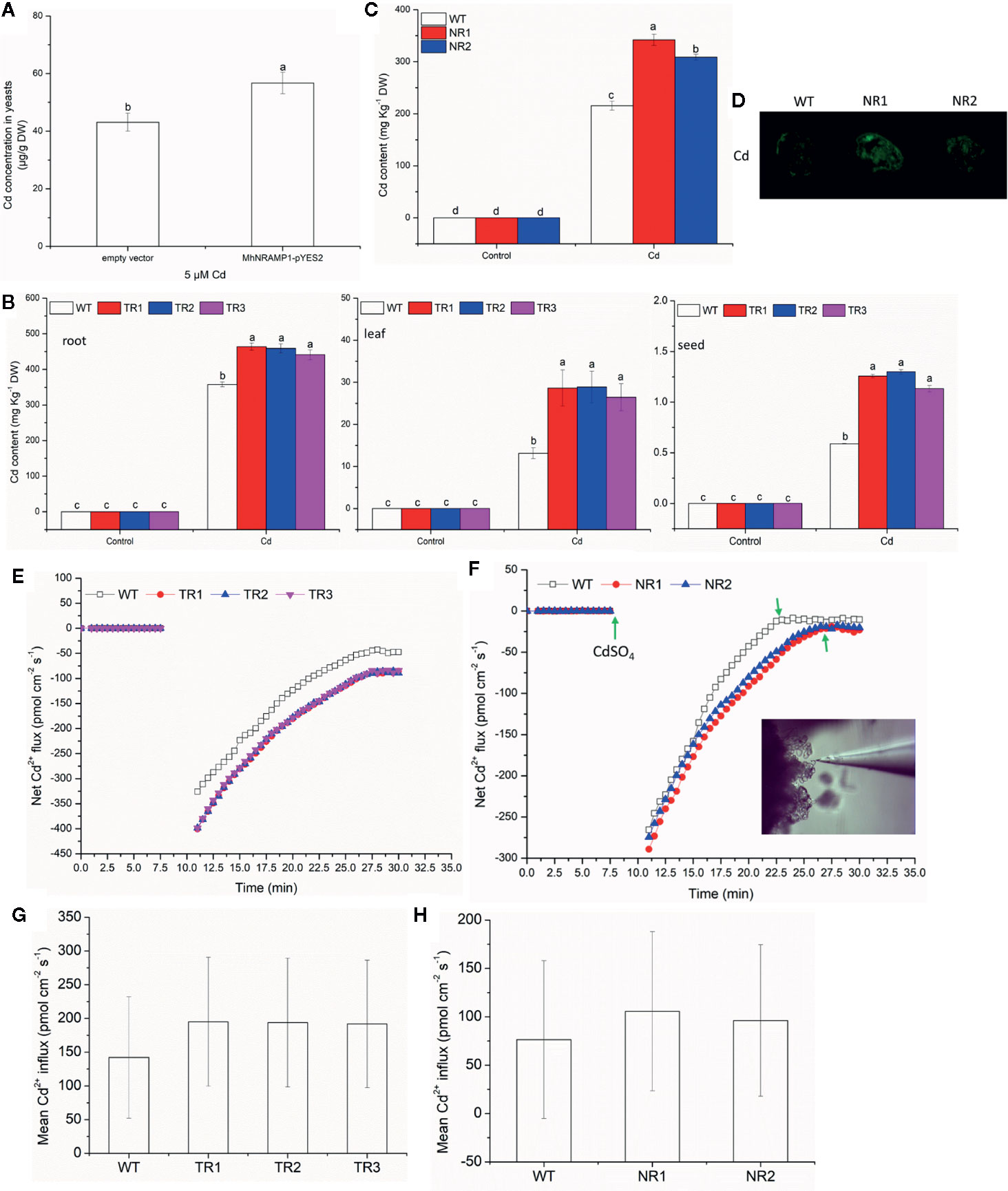
Figure 2 Effects of the overexpression of MhNRAMP1 on Cd accumulation and Cd2+ uptake. (A) Cd concentrations in yeasts under CdSO4 treatment for 48 h. (B) the Cd contents in the root, leaf and seeds of WT and transgenic tobacco in the normal control and 50 μm CdSO4 treatment. (C) the Cd contents in the WT and transgenic apple calli under normal conditions and 50 μm CdSO4 treatment. (D) Cd distribution of the WT and transgenic apple calli in the 50 μm CdSO4 treatment. (E) the transient Cd2+ influx into the roots of WT and transgenic tobacco in the 50 μm CdSO4 treatment. (F) the transient Cd2+ influx into the WT and transgenic apple calli under normal conditions and the 50 μm CdSO4 treatment. (G, H) the mean Cd2+ influx in tobacco (G) and apple calli (H) in the 50 μm CdSO4 treatment. Data are presented as the mean ± SD of 3 independent measurements of three individual plants. Different letters above the columns indicate significant differences (p < 0.05).
The dynamic curves illustrating the net Cd2+ influx showed that the net Cd2+ influx into transgenic tobacco roots was higher than that of the WT tobacco (Figure 2E). The net Cd2+ influx into transgenic apple calli was similar to that of the WT before Cd addition (Figure 2F). After the addition of CdSO4, the net Cd2+ influx of transgenic apple calli immediately increased and was higher than that in the WT, and the net Cd2+ influx of transgenic apple calli reached a stable state after 27 min, which was later than the 23 min for the WT (Figure 2F). The mean Cd2+ influx of both transgenic tobacco and transgenic apple calli was also higher than that of their corresponding WTs (Figures 2G, H). Additionally, the Cd transport factor from roots to leaves (TFleaf/root) of the transgenic tobacco was higher compared to the WT; however, the TF from leaves to seeds (TFseed/leaf) of the transgenic tobacco was not different from the WT (Table 1). These results suggested that the overexpression of MhNRAMP1 promoted Cd uptake and accumulation but did not affect Cd transport from the leaves to the seeds.
The growth of yeasts, tobacco, and apple calli was measured to explore the role of MhNRAMP1 in Cd sensitivity. No growth differences were observed between the transformed yeasts with MhNRAMP1 and yeasts with the empty vector under 0 μm Cd stress (Figure 3A), but their growth was inhibited by 10 μm Cd, and the growth of transformed yeasts with MhNRMAP6 was worse compared to the yeasts with the empty vector (Figure 3A). The growth of yeasts in liquid SD medium containing 5 μm Cd showed similar results (Figure 3B). Similarly, the transgenic tobacco seedlings under Cd stress displayed smaller sizes (Figure 3C) and lower fresh weight (Figure 3D). Notably, the NR1 and NR2 transgenic apple calli displayed slightly larger tissues than those of the WT under normal conditions; however, the transgenic apple calli were smaller than the WT under Cd stress (Figure 3E). The fresh weights of NR1 and NR2 under normal conditions were 1.26 and 1.17 times those of the WT, respectively. Under Cd stress, the fresh weights were 0.71 and 0.82 times those of the WT, respectively (Figure 3F). These results suggested that overexpression of MhNRAMP1 enhanced plant sensitivity to Cd, was not conducive to growth under Cd stress.
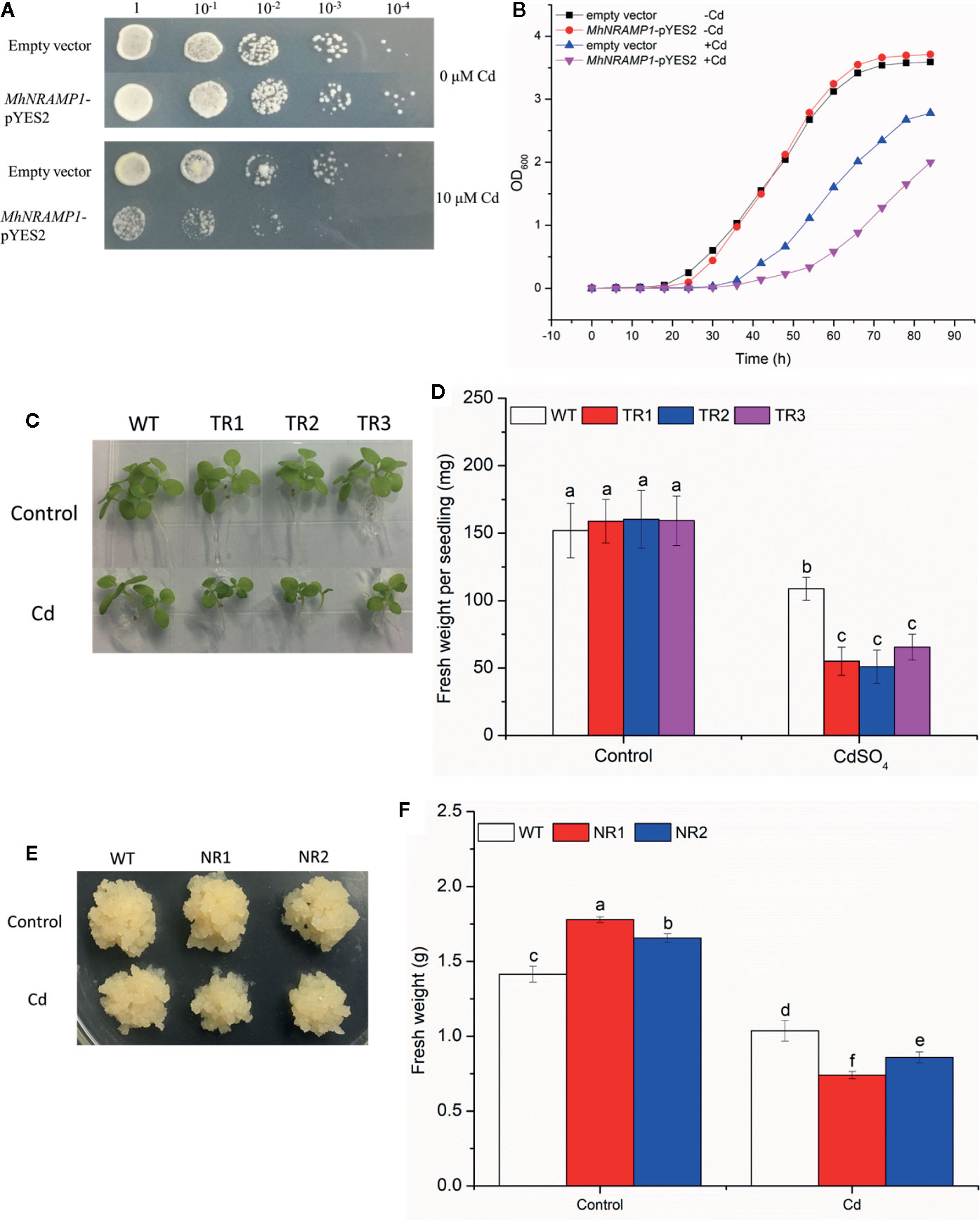
Figure 3 Effects of overexpression of MhNRAMP1 on the growth of yeasts, tobacco, and apple calli. (A) the growth of yeasts on plates in SD medium with CdSO4. (B) the growth curves of yeasts in liquid SD medium with CdSO4. (C) phenotype comparisons between the WT and MhNRAMP1-overexpressing transgenic tobacco lines (TR1, TR2, and TR3) under normal conditions and CdSO4 stress. (D) the fresh weight of tobacco under normal conditions and 50 μm CdSO4 stress. (E) phenotype comparisons between the WT and MhNRAMP1-overexpressing transgenic apple calli (NR1 and NR2) and their fresh weight (F) under normal conditions and 50 μm CdSO4 stress. Data are presented as the mean ± SD of three independent measurements from three individual plants. Different letters above the columns indicate significant differences (p < 0.05).
The transgenic apple calli and tobacco roots showed no significant difference in the percentage of apoptotic cells compared to their corresponding WTs under control conditions; however, the percentage of apoptotic cells under Cd stress in the transgenic apple calli and the transgenic tobacco roots was higher (Table 2). The gene expression and enzymes activities involved with cell death were measured in apple calli. Because MhVPEγ was found to have similar characteristics as caspase-1 in plants (Su et al., 2015), both the caspase-1-like activities and VPE activities of apple calli were detected and found to be higher in NR1 and NR2 transgenic apple calli under Cd stress, but they were not affected by the overexpression of MhNRAMP1 under normal conditions (Figure 4A). The gene expression showed that the transgenic apple calli under Cd stress had higher MdVPEγ expression and lower expression levels of genes inhibiting cell death, including MdBI-1, MdBAG, and MdDAD, than the WT (Figures 4A, B). Furthermore, these gene expressions were not different between transgenic apple calli and the WT under normal conditions (Figures 4A, B).
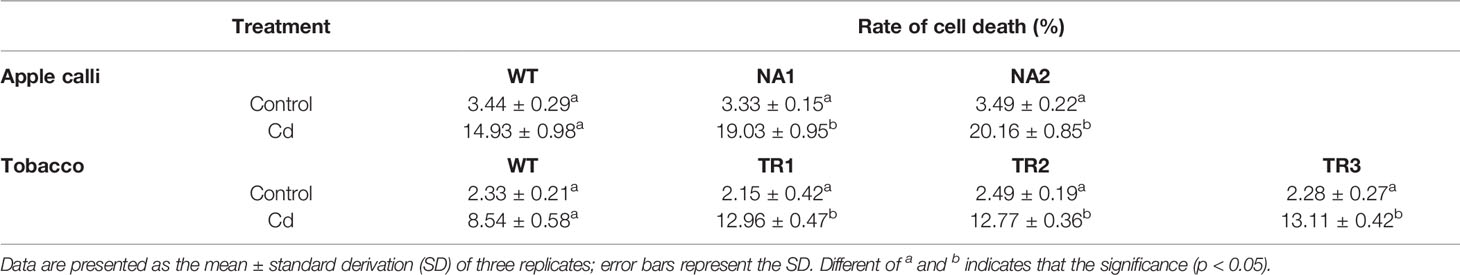
Table 2 The percentages of apoptotic cells in apple calli and tobacco roots under normal conditions (control) and CdSO4 treatment.
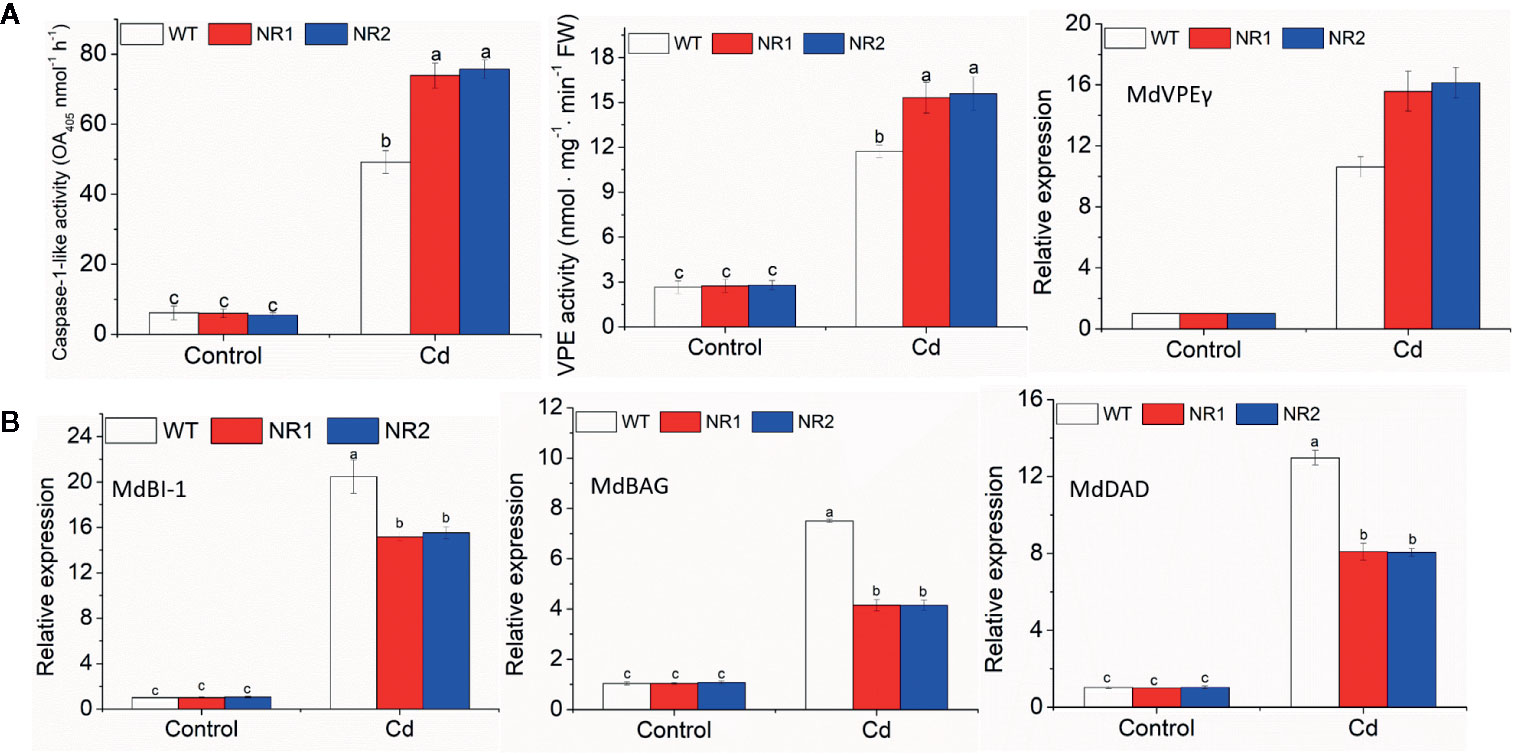
Figure 4 The caspase-1-like activities, VPE activities, and expression of MdVPEγ (A), and the expression of MdBI-1, MdBAG, and MdDAD (B) in the WT and transgenic apple calli under Cd stress and normal conditions. Data are presented as the mean ± SD of three independent measurements from three individual plants. Different letters above the columns indicate significant differences (p < 0.05).
NRAMPs regulate plant stress responses and have been cloned from apple rootstocks, including M. baccata (Xiao et al., 2008) and M. xiaojingensis (Pan et al., 2015). In the present study, MhNRAMP1 was isolated from M. hupehensis roots and was found to be highly homologous to MdNRAMP1 and MdNRAMP6 from apple (Figure S1). As a metal transporter, NRAMP genes were found to be induced by metals, including Fe (Peris-Peris et al., 2017), Mn (Wu et al., 2016), and Cd (Peng et al., 2018). MhNRAMP1 was significantly induced by the oversupply of Fe and only slightly induced by the oversupply of Mn, Zn, or Ca (Figure 1), suggesting that MhNRAMP1 might be more sensitive to Fe stress than to Mn, Zn, or Ca. When exposed to Ca deficiency, MhNRAMP1 expression was upregulated, which contrasted its downregulation due to Fe, Mn, or Zn deficiency (Figure 1). Similarly, Picard et al. (2000) demonstrated that NRAMPs do not transport Ca2+, as the amino acid of NRAMPs has low effective selectivity on Ca (Bozzi et al., 2016). Thus, it was inferred that the significant increase of MhNRAMP1 expression levels may be caused by its participation in Ca deficiency stress responses.
Cd is a preferred substrate of NRAMPs (Bozzi et al., 2016), which lead to long-term, low-dose Cd that could induce NRAMP responses. Previous studies have shown that NRAMP genes were induced by Cd (Ullah et al., 2018; Meena et al., 2018). In the present study, MhNRAMP1 in the roots was induced by Cd, and its expression after 24 h was more sensitive to Cd (Figure 1). The effects of Cd on MhNRAMP1 expression in the stems was not significant, but Cd inhibited the expression of MhNRAMP1 in the leaves (Figure 1), implying that MhNRAMP1 may play a more important role in the roots.
Many NRAMPs have Cd transport activity besides Fe and Mn. MbNRAMP of M. baccata was induced by Cd and was able to increase Cd accumulation and Cd sensitivity in yeast cells (Xiao et al., 2008). Previous studies reported that NRAMP6 overexpression enhanced Cd accumulation in plants (Wang et al., 2019a; Chen et al., 2017). In the present study, MhNRAMP1 overexpression increased Cd accumulation in yeasts (Figure 2) and also increased Cd accumulation in transgenic tobacco and apple calli. Correspondingly, the growth of transgenic yeasts was worse, and the transgenic tobacco and apple calli displayed smaller sizes and lower fresh weights under Cd stress (Figures 2 and 3). Therefore, the overexpression of MhNRAMP1 increased Cd sensitivity in yeasts, tobacco, and apple calli by stimulating Cd accumulation. Importantly, the net Cd2+ influx in both tobacco roots and apple calli was increased by the overexpression of MhNRAMP1 (Figure 2), which directly demonstrated that MhNRAMP1 controlled Cd2+ uptake. Moreover, MhNRAMP1 overexpression delayed the stabilization of the Cd influx to a relatively low level, which may explain the greater Cd2+ uptake observed in transgenic seedlings (Figure 2). These findings suggested that the sensitivities of transgenic yeasts, tobacco, and apple calli were likely due to the increase in Cd uptake regulated by MhNRAMP1.
Cell death is a typical physiological characteristic of Cd-damaged plants (Huybrechts et al., 2019). The percentage of apoptotic cells in tobacco roots and apple calli increased after Cd stress, and transgenic tobacco and apple calli overexpressing MhNRAMP1 had higher percentages of cell death (Table 2). These higher percentages of cell death were certainly caused by higher Cd accumulation in transgenic lines. However, the contribution mediated by VPE should not be ignored. Both the VPE activities and capase-1-like activities exhibited higher levels in transgenic apple calli under Cd stress (Figure 4). The expression levels of MdBI-1, MdDAD, and MdBAG, which inhibit cell death, were down-regulated in transgenic apple calli (Figure 4). The regulation of enzyme activities and gene expression was probably caused by higher Cd accumulation in transgenic apple calli under Cd stress because the levels of enzyme activities and gene expressions in transgenic apple calli were similar to those in the WT under normal conditions (Figure 4). However, the effect of the stimulation of overexpression of MhNRAMP1 on enzymes activities and gene expression that regulate cell death cannot be excluded. MhNRAMP1 promoted Cd-induced cell death mediated by VPE by accelerating Cd uptake.
The overexpression of MhNRAMP1 in tobacco is due to the Cd transport from roots to leaves. Due to the increase of environmental Cd, the Cd contents of tobacco roots, seeds, and leaves increased in the present study (Figure 2). However, the Cd TF from leaves to seeds and from roots to leaves was different from that from leaves to seeds. The Cd TF from roots to leaves in transgenic tobacco was higher than the WT; however, the Cd TF from leaves to seeds in transgenic tobacco was similar to that of the WT (Figure 2). These results reflected the fact that MhNRAMP1 played a role in Cd transport from the roots to leaves but did not influence the Cd transport from leaves to seeds, which agreed with the previous inference that MhNRAMP1 tends to play a more important role in the roots. Therefore, we infer that MhNRAMP1 might be similar to OsNRAMP1 in assisting with Cd uptake and xylem loading for root to shoot mobilization, thus minimizing the Cd content in grain (Tiwari et al., 2014). Based on these effects, MhNRAMP1 may be used in Cd enrichment without threatening the food safety of apple fruits.
In the present study, MhNRAMP1 was isolated from M. hupehensis roots and was mainly expressed in roots. MhNRAMP1 overexpression increased Cd accumulation in yeasts and enhanced their sensitivity to Cd. Transgenic tobacco and apple calli overexpressing MhNRAMP1 displayed greater Cd2+ uptake and Cd accumulation, and exhibited worse growth under Cd stress. Transgenic tobacco and apple calli also had higher cell death. The overexpression of MhNRAMP1 in apple calli up-regulated VPE activities and MdVPEγ expression and inhibited the expression of genes involved in anti-cell death. These findings are the first to suggest that NRAMPs from Malus regulate Cd2+ uptake in plant roots, thereby controlling Cd accumulation in plants. This preliminary exploration into the molecular mechanisms underlying Cd-induced cell death in M. hupehensis is an important finding for future studies. However, the manner in which MhNRAMP1 regulates VPE activities or its expression requires further investigation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/MT035802.1/, MT035802.1, https://www.ncbi.nlm.nih.gov/protein/XP_008354189.2, XP_008354189.2, https://www.ncbi.nlm.nih.gov/nuccore/XM_008379705.2, XM_008379705.2, https://www.ncbi.nlm.nih.gov/nuccore/XM_008358248.3, XM_008358248.3, https://www.ncbi.nlm.nih.gov/nuccore/XM_008378307.3, XM_008378307.3, https://www.ncbi.nlm.nih.gov/nuccore/NM_001293917.1, NM_001293917.1.
WZ designed the experiments and drafted the manuscript. SY, MX, and JS conducted the gene cloning and transformations, and performed the ion flux and ion content experiments. SY and MH analyzed the data. WZ and HY were the project leaders.
This work was financially supported by the National Natural Science Foundation of China (grant No. 31801838), the National Key R&D Program of China (grant No. 2019YFD1000103), and the Shandong Provincial Natural Science Foundation of China (grant No. ZR2017BC024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00957/full#supplementary-material
Figure S1 | The sequence analysis and subcellular location of MhNRAMP1. A–B, analysis of hydrophobicity (A) and transmembrane (B) of the MhNRAMP1 protein. C, multiple alignment of MhNRAMP1-deduced amino acid sequences and several NRAMP proteins from other plants, including apples (MdNRAMP6, XP_008354189.1), Pyrus bretschneideri (PbNRAMP6, XP_009354189.1), Prunus persica (PpNRAMP6, XP_007217247.1), Prunus avium (PaNRAMP6, XP_021821838.1), Citrus sinensis (CsNRAMP6, XP_006493353.1), Fragaria vesca subsp. vesca (FvsNRAMP6, XP_004303243.1), and Arabidopsis thaliana (AtNRAMP6, NP_173048.3). D, Phylogenetic tree construction of MhNRAMP1 in Malus hupehensis with NRAMPs from 18 other plant species, including apples (MdNRAMP2, XP_008351981.1; MdNRAMP3, XP_008361431.1; MdNRAMP5, XP_008361001.1; MdNRAMP6, XP_008354189.1), Arabidopsis thaliana (AtNRAMP1, NP_178198.1; AtNRAMP2, NP_175157.1; AtNRAMP3, NP_179896.1; AtNRAMP4, NP_201534.1; AtNRAMP5, OAO96785.1; AtNRAMP6, NP_173048.3), Cajanus cajan (CcNRAMP6, XP_020232276.1), Citrus sinensis (CsNRAMP6, XP_006493353.1), Fragaria vesca subsp. vesca (FvsNRAMP6, XP_004303243.1), Glycine max (GmNRAMP6, XP_003543701.1), Glycine soja (GsNRAMP6, KHN31883.1), Juglans regia (JrNRAMP6, XP_018846317.1), Medicago truncatula (MtNRAMP6, XP_013465598.1), Prunus avium (PaNRAMP6, XP_021821838.1), Pyrus bretschneideri (PbNRAMP6, XP_009354189.1), Prunus mume (PmNRAMP6, XP_008228792.1), Prunus persica (PpNRAMP6, XP_007217247.1), Populus trichocarpa (PtNRAMP6, XP_006368514.1), Rosa chinensis (RcNRAMP6, XP_024185381.1), and Theobroma cacao (TcNRAMP6, XP_017977974.1). E, subcellular location of MhNRAMP1.
Table S1 | Primer sequences used for cloning, subcellular localization, vector construction, and expression analysis.
Abbas, T., Rizwan, M., Ali, S., Adrees, M., Mahmood, A., Zia-Ur-Rehman, M., et al. (2018). Biochar application increased the growth and yield and reduced cadmium in drought stressed wheat grown in an aged contaminated soil. Ecotoxicol. Environ. Saf. 148, 825–833. doi: 10.1016/j.ecoenv.2017.11.063
An, J., Li, R., Qu, F., You, C., Wang, X., Hao, Y. (2018). An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 221, 74–80. doi: 10.1016/j.jplph.2017.12.009
Beaugelin, I., Chevalier, A., D’Alessandro, S., Ksas, B., Novák, O., Strnad, M., et al. (2019). OXI1 and DAD regulate light-induced cell death antagonistically through jasmonate and salicylate levels. Plant Physiol. 180 (3), 1691–1708. doi: 10.1104/pp.19.00353
Bozzi, A. T., Bane, L. B., Weihofen, W. A., McCabe, A. L., Singharoy, A., Chipot, C. J., et al. (2016). Conserved methionine dictates substrate preference in Nramp-family divalent metal transporters. Proc. Natl. Acad. Sci. U. S. A. 113 (37), 10310–10315. doi: 10.1073/pnas.1607734113
Bruno, L., Pacenza, M., Forgione, I., Lamerton, L. R., Greco, M., Chiappetta, A., et al. (2017). Arabidopsis thaliana cadmium impact on the growth of primary root by altering SCR expression and auxin-cytokinin cross-talk. Front. Plant Sci. 8, 1323. doi: 10.3389/fpls.2017.01323
Cai, Y., Xu, W., Wang, M., Chen, W., Li, X., Li, Y., et al. (2019). Mechanisms and uncertainties of Zn supply on regulating rice Cd uptake. Environ. Pollut. 253, 959–965. doi: 10.1016/j.envpol.2019.07.077
Cailliatte, R., Lapeyre, B., Briat, J., Mari, S., Curie, C. (2009). The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem. J. 422, 217–228. doi: 10.1042/BJ20090655
Cengiz, K., Mustafa, O., Ferhat, U., Mohammed, N. A., Muhammad, A., Parvaiz, A. H. (2019). Melatonin-mediated nitric oxide improves tolerance to cadmium toxicity by reducing oxidative stress in wheat plants. Chemosphere 225, 627–638. doi: 10.1016/j.chemosphere.2019.03.026
Chen, S., Han, X., Fang, J., Lu, Z., Qiu, W., Liu, M., et al. (2017). Sedum alfredii SaNramp6 metal transporter contributes to cadmium accumulation in transgenic Arabidopsis thaliana. Sci. Rep. 7, 13318. doi: 10.1038/s41598-017-13463-4
Chen, J., Huang, X., Salt, D., Zhao, F. (2019). Mutation in OsCADT1 enhances cadmium tolerance and enriches selenium in rice grain. New Phytol. 226, 838–850. doi: 10.1111/nph.16404
Cheng, J., Ding, C., Li, X., Zhang, T., Wang, X. (2015). Heavy metals in navel orange orchards of Xinfeng Country and their transfer from soils to navel oranges. Ecotoxicol. Environ. Saf. 122, 153–158. doi: 10.1016/j.ecoenv.2015.07.022
Fan, S., Yang, H. Q., Ran, K., Shen, W., Zhang, W. W. (2013). Differential expression of anti-apoptotic genes in extensive and absorbing roots of Malus hupehensis and their response to 2,4-D. Acta Hortic. Sin. 40, 1225–1232. doi: 10.16420/j.issn.0513-353x.2013.07.002
Gao, H., Xie, W., Yang, C., Xu, J., Li, J., Wang, H., et al. (2018). Nramp2, a trans-Golgi network-localized manganese transporter, is required for Arabidopsis root growth under manganese deficiency. New Phytol. 217, 179–193. doi: 10.1111/nph.14783
Guo, F., Ding, C., Zhou, Z., Huang, G., Wang, X. (2018). Effects of combined amendments on crop yield and cadmium uptake in two cadmium contaminated soils under rice-wheat rotation. Ecotoxicol. Environ. Saf. 148, 303–310. doi: 10.1016/j.ecoenv.2017.10.043
He, X., Chen, W., Huang, Q. (2011). Surface display of monkey metallothionein α tandem repeats and EGFP fusion protein on Pseudomonas putida X4 for biosorption and detection of cadmium. Appl. Microbiol. Biotechnol. 95, 1605–1613. doi: 10.1007/s00253-011-3768-3
Himeno, S., Yanagiya, T., Enomoto, S., Kondo, Y., Imura, N. (2002). Cellular cadmium uptake mediated by the transport system for manganese. Tohoku J. Exp. Med. 196 (1), 43–50. doi: 10.1620/tjem.196.43
Horsh, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. C., Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Huybrechts, M., Cuypers, A., Deckers, J., Iven, V., Vandionant, S., Jozefczak, M., et al. (2019). Cadmium and plant development: an agony from seed to seed. Int. J. Mol. Sci. 20, 3971. doi: 10.3390/ijms20163971
Ishikawa, T., Uchimiya, H. I., Kawai-Yamada, M. (2013). Chapter thirteen - the role of plant bax inhibitor-1 in suppressing H2O2-induced cell death. Method. Enzymol. 527, 239–256. doi: 10.1016/B978-0-12-405882-8.00013-1
Jiang, J., Hu, J., Tan, R., Han, Y., Li, Z. (2019). Expression of IbVPE1 from sweet potato in Arabidopsis affects leaf development, flowering time and chlorophyll catabolism. BMC Plant Biol. 19, 184. doi: 10.1186/s12870-019-1789-8
Kabbage, M., Dickman, M. B. (2008). The BAG proteins: a ubiquitous family of chaperone regulators Cell. Mol. Life Sci. 65, 1390–1402. doi: 10.1007/s00018-008-7535-2
Kariya, K., Tsuchiya, Y., Sasaki, T., Yamamoto, Y. (2018). Aluminium-induced cell death requires upregulation of NtVPE1 gene coding vacuolar processing enzyme in tobacco (Nicotiana tabacum L.). J. Inorg. Biochem. 181, 152–161. doi: 10.1016/j.jinorgbio.2017.09.008
Li, M., Xia, T., Jiang, C., Li, L., Zhou, Z. (2003). Cadmium directly induced the opening of membrane permeability pore of mitochondria which possibly involved in cadmium-triggered apoptosis. Toxicology 194, 19–33. doi: 10.1016/S0300-483X(03)00327-5
Li, J., Liu, J., Dong, D., Jia, X., McCouch, S., Kochian, L. (2014). Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. U. S. A. 111, 6503–6508. doi: 10.1073/pnas.1318975111
Li, R., Lub, X., Zhu, Y., Zhao, L., Li, L., Peng, Q., et al. (2017). ATM signals to AMPK to promote autophagy and positively regulate DNA damage in response to cadmium-induced ROS in mouse spermatocytes. Environ. Pollut. 231, 1560–1568. doi: 10.1016/j.envpol.2017.09.044
Li, J., Wang, Y., Zheng, L., Li, Y., Zhou, X., Li, J., et al. (2019). The Intracellular transporter AtNRAMP6 is involved in Fe homeostasis in Arabidopsis. Front. Plant Sci. 10:, 1124. doi: 10.3389/fpls.2019.01124
Lu, W., Deng, M., Guo, F., Wang, M., Zeng, Z., Han, N., et al. (2016). Suppression of OsVPE3 enhances salt tolerance by attenuating vacuole rupture during programmed cell death and affects stomata development in rice. Rice 9, 65. doi: 10.1186/s12284-016-0138-x
Meena, M., Aamir, M., Kumar, V., Swapnil, P., Upadhyay, R. S. (2018). Evaluation of morpho-physiological growth parameters of tomato in response to Cd induced toxicity and characterization of metal sensitive NRAMP3 transporter protein. Environ. Exp. Bot. 148, 144–167. doi: 10.1016/j.envexpbot.2018.01.007
Michelle, S., Li, M., Filice, F. P., Ding, Z. (2014). A time course study of cadmium effect on membrane permeability of single human bladder cancer cells using scanning electrochemical microscopy. J. Inorg. Biochem. 136, 177–183. doi: 10.1016/j.jinorgbio.2014.02.009
Pan, H., Wang, Y., Zha, Q., Yuan, M., Yin, L., Wu, T., et al. (2015). Iron deficiency stress can induce MxNRAMP1 protein endocytosis in M. xiaojinensis. Gene 567 (2), 225–234. doi: 10.1016/j.gene.2015.05.002
Peng, F., Wang, C., Zhu, J., Zeng, J., Kang, H., Fan, X., et al. (2018). Expression of TpNRAMP5, a metal transport from polish wheat (Triticum polonicum L.), enhances the accumulation of Cd, Co and Mn in transgenic Arabidopsis plants. Planta 247, 1395–1406. doi: 10.1007/s00425-018-2872-3
Pereira de Araújo, R., Furtado de Almeida, A. A., Silva Pereira, L., Mangabeira, P. A. O., Olimpio Souza, J., Pirovani, C. P., et al. (2017). Photosynthetic, antioxidative, molecular and ultrastructural responses of young cacao plants to Cd toxicity in the soil. Ecotoxicol. Environ. Saf. 144, 148–157. doi: 10.1016/j.ecoenv.2017.06.006
Peris-Peris, C., Serra Cardona, A., Sánchez Sanuy, F., Campo, S., Ariño, J., San Segundo, B. (2017). Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol. Plant-Microbe Interact. 30, 385–398. doi: 10.1094/MPMI-01-17-0005-R
Picard, V., Govoni, G., Jabado, N., Gross, P. (2000). NRAMP2 (DCT1/DMT1) expressed at the plasma membrane transports iron and other divalent cations into a calcein-accessible cytoplasmic pool. J. Biol. Chem. 275, 35738–35745. doi: 10.1074/jbc.M005387200
Ramiro, D. A., Melotto-Passarin, D. M., Barbosa, M. A., Santos, F., Gomez, S. G. P., Júnior, N. S. M., et al. (2016). Expression of Arabidopsis Bax Inhibitor-1 in transgenic sugarcane confers drought tolerance. Plant Biotech. J. 14 (9), 1826–1837. doi: 10.1111/pbi.12540
Sheludko, Y., Sindarovska, Y., Gerasymenko, I., Bannikova, M., Kuchuk, N. (2007). Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol. Bieng. 96, 608–614. doi: 10.1002/bit.21075
Sofo, A., Bochicchio, R., Amato, M., Rendina, N., Vitti, A., Nuzzaci, M., et al. (2017). Plant architecture, auxin homeostasis and phenol content in Arabidopsis thaliana grown in cadmium- and zinc-enriched media. J. Plants Physiol. 216, 174–180. doi: 10.1016/j.jplph.2017.06.008
Su, Q., Ran, K., Men, X., Zhang, W., Fan, S., Yan, L., et al. (2015). Response of vacuolar processing enzyme in Malus hupehensis and MhVPEγ-overexpressing Arabidopsis to high temperature stress. Acta Physiol. Plant 37, 82. doi: 10.1007/s11738-015-1830-y
Tamura, K., Stecher, G., Paterson, D., Filipski, A., Kumar, S. (2007). Mega6: molecular revolutionary genetics analysis software version 6.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7, 14438. doi: 10.1038/s41598-017-14832-9
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). Theclustal_x windows interface: flexible strategies for multiple sequencealignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tiwari, M., Sharma, D., Dwivedi, S., Singh, M., Tripathi, R. D., Trivedi, P. K. (2014). Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance. Plant Cell Environ. 37 (1), 140–152. doi: 10.1111/pce.12138
Ullah, I., Wang, Y., Eide, D. J., Dunwell, J. (2018). Evolution, and functional analysis of Natural Resistance-Associated Macrophage Proteins (NRAMPs) from Theobroma cacao and their role in cadmium accumulation. Sci. Rep. 8 (1), 14412. doi: 10.1038/s41598-018-32819-y
Verbruggen, N., Hermans, C., Schat, H. (2009). Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 12, 364–372. . doi: 10.1016/j.pbi.2009.05.001
Wang, Y., Wang, X., Wang, C., Peng, F., Wang, R., Xiao, X., et al. (2017). Transcriptomic profiles reveal the interactions of Cd/Zn in dwarf polish wheat (Triticum polonicum L.) roots. Front. Physiol. 8, 168. doi: 10.3389/fphys.2017.00168
Wang, C., Chen, X., Yao, Q., Long, D., Fan, X., Kang, H., et al. (2019a). Overexpression of TtNRAMP6 enhances the accumulation of Cd in Arabidopsis. Gene 696, 225–232. doi: 10.1016/j.gene.2019.02.008
Wang, H., Cao, Q., Zhao, Q., Arfan, M., Liu, W. (2019b). Mechanisms used by DNA MMR system to cope with Cadmium-induced DNA damage in plants. Chemosphere 246, 125614. doi: 10.1016/j.chemosphere.2019.125614
Wang, N., Qiu, W., Dai, J., Guo, X., Lu, Q., Wang, T., et al. (2019c). AhNRAMP1 Enhances Manganese and Zinc Uptake in Plants. Front. Plant Sci. 10, 415. doi: 10.3389/fpls.2019.00415
Wu, D., Yamaji, N., Yamane, M., Kashino-Fujii, M., Sato, K., Ma, J. (2016). The HvNRAMP5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol. 172, 1899–1910. doi: 10.1104/pp.16.01189
Wu, C., Dun, Y., Zhang, Z., Li, M., Wu, G. (2019). Foliar application of selenium and zinc to alleviate wheat (Triticum aestivum L.) cadmium toxicity and uptake from cadmium-contaminated soil. Ecotoxicol. Environ. Saf. 190, 110091. doi: 10.1016/j.ecoenv.2019.110091
Xiao, H., Yin, L., Xu, X., Li, T., Han, Z. (2008). The iron-regulated transporter, MbNRAMP1, isolated from Malus baccata is involved in Fe, Mn and Cd trafficking. Ann. Bot. 102, 881–889. doi: 10.1093/aob/mcn178
Xu, Y. T., Feng, S. Q., Jiao, Q. Q., Liu, C. C., Zhang, W. W., Chen, W. Y., et al. (2012). Comparison of MdMYB1 sequence and expression of anthocyanin biosynthetic and regulatory genes between Malus dometica Borkh. cultivar ‘Ralls’ and its blushed sport. Euphytica 185, 157–170. . doi: 10.1007/s10681-011-0494-y
Yamada, K., Basak, A. K., Goto-Yamada, S., Tarnawska-Glatt, K., Hara-Nishimura, I. (2020). Vacuolar processing enzymes in the plant life cycle. New Phytol. 226 (1), 21–31. doi: 10.1111/nph.16306
Yamaguchi, C., Takimoto, Y., Ohkama-Ohtsu, N., Hokura, A., Shinano, T., Nakamura, T., et al. (2016). Effects of cadmium treatment on the uptake and translocation of sulfate in Arabidopsis thaliana. Plant Cell Physiol. 57 (11), 2353–2366. doi: 10.1093/pcp/pcw156
Yang, H. Q., Duan, K. X., Zhang, W. (2008). Biology and physiology of Malus hupehensis for the apopgamic plant resource. Acta Hortic. 769, 441–447. doi: 10.17660/ActaHortic.2008.769.63
Zare, A. A., Khoshgoftarmanesh, A. H., Malakouti, M. J., Bahrami, H. A., Chaney, R. L. (2018). Root uptake and shoot accumulation of cadmium by lettuce at various Cd:Zn ratios in nutrient solution. Ecotoxicol. Environ. Saf. 148, 441–446. doi: 10.1016/j.ecoenv.2017.10.045
Zhang, W., Yang, H., You, S., Xu, Y., Ran, K., Fan, S. (2014). Cloning, characterization and functional analysis of the role MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd., plays in plant tolerance to osmotic and Cd2+ stresses. Plant Soil 381, 143–160. doi: 10.1007/s11104-014-2120-y
Zhang, W. W., Song, J. F., Yue, S. Q., Duan, K. X., Yang, H. Q. (2019a). MhMAPK4 from Malus hupehensis Rehd. decreases cell death in tobacco roots by controlling Cd2+ uptake. Ecotoxicol. Environ. Saf. 168:, 230–240. doi: 10.1016/j.ecoenv.2018.09.126
Zhang, W. W., Wang, Z. P., Song, J. F., Yue, S. Q., Yang, H. Q. (2019b). Cd2+ uptake inhibited by MhNCED3 from Malus hupehensis alleviates Cd induced cell death. Environ. Exp. Bot. 166:, 103802. doi: 10.1016/j.envexpbot.2019.103802
Zhao, Y., Shen, X., Chao, X., Ho, C. C., Cheng, X., Zhang, Y., et al. (2011). Ergosta-4,6,8(14), 22 tetraen-3-one induces G2/M cell cycle arrest apoptosis in human hepatocellular carcinoma HepG2 cells. Biochem. Biophys. Acta 1810, 384–390. doi: 10.1016/j.bbagen.2010.12.005
Keywords: cadmium, uptake, Malus hupehensis, natural resistance-associated macrophage protein, cell death, vacuolar processing enzyme
Citation: Zhang W, Yue S, Song J, Xun M, Han M and Yang H (2020) MhNRAMP1 From Malus hupehensis Exacerbates Cell Death by Accelerating Cd Uptake in Tobacco and Apple Calli. Front. Plant Sci. 11:957. doi: 10.3389/fpls.2020.00957
Received: 24 April 2020; Accepted: 10 June 2020;
Published: 07 July 2020.
Edited by:
Elizabeth R. Waters, San Diego State University, United StatesReviewed by:
Matthew John Milner, National Institute of Agricultural Botany (NIAB), United KingdomCopyright © 2020 Zhang, Yue, Song, Xun, Han and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Zhang, emhhbmd3d0BzZGF1LmVkdS5jbg==; Hongqiang Yang, aHF5YW5nQHNkYXUuZWR1LmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.