
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 July 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00894
This article is part of the Research Topic Nematodes in Phytobiomes View all 21 articles
Rice (Oryza sativa L.) is one of the major staple food crops of the world. The productivity of rice is considerably affected by the root-knot nematode, Meloidogyne graminicola. Modern nematode management strategies targeting the physiological processes have established the potency of use of neuromotor genes for their management. Here, we explored the utility of two FMRFamide like peptide coding genes, Mg-flp-1 and Mg-flp-12 of M. graminicola for its management through host-induced gene silencing (HIGS) using Agrobacterium-mediated transformation of rice. The presence and integration of hairpin RNA (hpRNA) constructs in transgenic lines were confirmed by PCR, qRT-PCR, and Southern and Northern hybridization. Transgenic plants were evaluated against M. graminicola, where phenotypic effect of HIGS was pronounced with reduction in galling by 20–48% in the transgenic plants. This also led to significant decrease in total number of endoparasites by 31–50% for Mg-flp-1 and 34–51% for Mg-flp-12 transgenics. Likewise, number of egg masses per plant and eggs per egg mass also declined significantly in the transgenics, ultimately affecting the multiplication factor, when compared to the wild type plants. This study establishes the effectiveness of the two M. graminicola flp genes for its management and also for gene pyramiding.
Rice (Oryza sativa L.) is the staple cereal “global grain” constituting everyday meal of more than three billion people around the world. Although rice is popularly grown across different parts of the world, the tropical parts of Asia and South-East Asian countries produce approximately 90% of the global rice output (Mantelin et al., 2017). In the present era of modern farming, plant-parasitic nematodes (PPNs) pose a major threat to the agricultural food production and rice is also attacked by a wide array of nematodes (Prasad et al., 1992; Jones et al., 2013). Among the major PPN species attacking rice, the root-knot nematode (RKN) Meloidogyne graminicola Golden and Birchfield, 1965 alone has been reported to inflict up to 50% yield loss under different conditions (Lorenzana et al., 1998; Bridge et al., 2005). This economically important RKN species is widespread in almost all the rice growing areas in the world, and has been well documented to cause extensive yield and quality losses (Dutta et al., 2012; Kyndt et al., 2012). The infective pre-parasitic second-stage juveniles (J2s) of M. graminicola enter the roots and develop permanent feeding cells (giant cells) resulting in typical hook shaped galls (De Waele and Elsen, 2007).
Various strategies are used for management of PPNs, and the traditional practices are mostly relied on the use of cultural/physical methods and chemotherapeutics. However, due to the adverse effects of most of the nematicidal chemicals on environment, non-target organisms and human health, they were either discontinued or restricted for use in agricultural fields (Bridge et al., 2005; De Waele et al., 2013). Hence, there has been a continuous demand for development of environmentally benign target-specific nematode management approach, and genetic engineering based techniques have gained promising popularity in this regard (Roderick et al., 2018; Dutta et al., 2019). Genetic improvement of rice against PPNs through breeding programs faces a major challenge due to scant availability of suitable resistant sources (Soriano et al., 1999; Cabasan et al., 2012). Considerable natural resistance against M. graminicola has been reported in Oryza glaberrima and O. longistaminata, but limited resistant source has been reported in O. sativa (Soriano et al., 1999; Cabasan et al., 2012; Kumari et al., 2016; Kumari et al., 2017; Hatzade et al., 2020). The recent advancements in PPN genomics and transcriptomics have enabled us to identify the propitious molecular targets in nematodes that can be exploited for their management and also for drug designing (Danchin et al., 2013; Taylor et al., 2013; Shivakumara et al., 2019). The availability of draft genome can also offer a supportive platform to identify and validate the candidate genes responsible for rice–M. graminicola interaction (Somvanshi et al., 2018).
Nematode neuropeptides play a key role in controlling and modulating the physiological processes like host recognition, navigation, infection, secretion, reproduction etc., and have been proved to be potential drug targets (Maule et al., 2002; Kimber et al., 2007; Warnock et al., 2017). The FMRFamide-like peptides (FLPs) constitute a large and diverse group of neuropeptides in nematodes, governing the basic behavioral functions by coupling to the G-protein coupled receptors (GPCRs) (McVeigh et al., 2006; Atkinson et al., 2013). Similar results have also been reported due to silencing of flp genes that resulted in locomotory defects, reduced penetration, aberrant behavior and reduced reproduction in Meloidogyne incognita and Globodera pallida (Dalzell et al., 2010; Dong et al., 2013; Papolu et al., 2013; Banakar et al., 2015). However, scant knowledge is available to date with respect to performance of the flp gene repertoire against M. graminicola in rice through HIGS. Of late, HIGS has emerged as an effective and successful strategy to silence the genes in plant-parasitic nematodes for functional validation (Fairbairn et al., 2007; Danchin et al., 2013). In the present study, we have selected two M. graminicola FLP coding genes, Mg-flp-1 and Mg-flp-12, for evaluation based on their reported effect on juvenile penetration and infectivity in rice. Previously, Rao et al. (2013) found that the translated sequence of Mg-flp-12 putatively contains a N-terminal secretion signal peptide indicating its involvement in extra cellular signal transduction functioning like G protein coupled receptor (GO: 0004930); and similar finding was noted for Mg-flp-1, which contains the highly conserved LFRGR motif. These observations putatively indicated the potential of the genes as molecular target(s) against M. graminicola. Subsequently, Kumari et al. (2017) characterized nine flp genes from M. graminicola including flp-1 and flp-12, and in vitro silencing of the said genes resulted in significantly reduced penetration of J2s and their infection potential in rice. Additionally, Masler (2008) showed that disruption of flp-1 and flp-12 results in various neuromuscular dysfunctions in other nematodes, and flp-12 has also been proved to be a potential target against pine wood nematode, Bursaphelenchus xylophilus (Huang et al., 2010). Hence, to explore the potential of flp-1 and flp-12 as molecular targets, transgenic rice plants were developed expressing the dsRNA constructs of the two genes, and their effectiveness were evaluated against M. graminicola using soil-less Pluronic media and soil system. Thus, the present study strengthens our knowledge on the effect of M. graminicola flp-1 and flp-12 in nematode reproduction and plant parasitism potential, when applied through HIGS in rice.
Pure culture of an Indian isolate of M. graminicola Golden and Birchfield, 1965 was maintained on rice (O. sativa cv. PB 1121) in a glasshouse at ICAR–Indian Agricultural Research Institute, New Delhi, India. Galls were handpicked from washed infected roots, and infective second-stage juveniles (J2s) were hatched using modified Baermann’s assembly (Whitehead and Hemming, 1965). The freshly hatched J2s were used for all the experiments.
Previously, flp-1 and flp-12 were reported from M. graminicola by Rao et al. (2013) and subsequently Kumari et al. (2017) performed their molecular characterization and showed in vitro silencing effect of the genes on M. graminicola against rice. The partial sequence of Mg-flp-1 (214 bp) and Mg-flp-12 (299 bp) were then subjected to BlastN search against GenBank nonredundant (nr) databank and M. graminicola genome database (Altschul et al., 1990). Further, off-target sites in the targeted dsRNA were investigated at http://dsCheck.RNAi.jp/ for their presence, if any (Naito et al., 2005).
RNAi Gateway vector ph7GWIWG2(II) was obtained from VIB-UGent Center for Plant Systems Biology, Ghent University, Ghent, Belgium. Partial sequence of Mg-flp-1 (214 bp), Mg-flp-12 (299 bp) and an unrelated gene gfp (375 bp; green fluorescent protein used as non-native negative control) were PCR amplified from the corresponding recombinant pGEM-T clones (Accessions: KC250005, KC250006 and HF675000), sub-cloned into pDONR 221 entry vector, followed by subsequent cloning into ph7GWIWG2(II) destination vector using the LR recombination in sense and antisense orientation, using GATEWAY recombination cloning kit (Invitrogen, Carlsbad, CA, USA). The recombinant vectors were transformed into E. coli (DH5α), mobilized into A. tumefaciens (LBA4404), and confirmed the orientation of target genes by PCR using gene specific primers, CaMV 35S promoter and attB2, CaMV 35S terminator and attB2, hptII marker specific primers. Primer details are given in Table S1.
For Agrobacterium primary culture preparation, the positive clones were inoculated into 5 ml of liquid Yeast Extract Mannitol (YEM) medium (Sambrook et al., 1989) supplemented with 100 mg L–1 spectinomycin (Sigma Aldrich, St. Louis, Missouri, USA) and 30 mg L–1 rifampicin (Sigma Aldrich, St. Louis, Missouri, USA), and incubated at 28°C for 48 h at 200 rpm in an incubator shaker (New Brunswick Innova 44; Eppendorf, Hamburg, Germany). Primary culture was re-inoculated into 100 ml of antibiotic supplemented YEM medium, incubated overnight at aforesaid conditions, until the A600 (Agrobacterium suspension cells) reaches at 0.8. The culture was centrifuged for 20 min at 5,000×g to pellet the cells which were re-suspended in Murashige–Skoog (MS) medium (MS salt 4.41 g L–1, sucrose 1.5%; pH 5.4) containing 150 μM of acetosyringone and used for co-cultivation.
O. sativa cv. Taipei 309 was used for Agrobacterium- mediated transformation. The seeds were procured from Division of Genetics, ICAR-Indian Agricultural and Research Institute, New Delhi, India. Healthy seeds were de-husked and surface sterilized with 70% ethanol for 90 s, followed by washing with 50% (v/v) sodium hypochlorite solution for 20 min. Seeds were then washed with autoclaved double distilled water thrice until traces of sodium hypochlorite vanished completely. About 50 surface sterilized rice seeds were inoculated aseptically on callus induction medium (MCI) and incubated at 26 ± 2°C in dark for development of embryogenic calli. The calli were then co-cultivated by immersing them in Agrobacterium suspension for 20 min, excess moisture removed using a sterilized filter paper (Whatman Grade 4, Whatman International, UK), transferred on to the co-cultivation medium (MCCM, pH 5.8) containing 150 μM of acetosyringone and incubated in dark for 48 h.
After co-cultivation, the infected calli were washed with sterilized double distilled water for 25 min, containing 300 mg L–1 cefotaxime (HIMEDIA, Mumbai, India) and 200 mg L–1 ticarcillin (Sigma Aldrich, St. Louis, Missouri, USA). Calli were then transferred onto the selection medium-I (MSM I) and incubated for 15 days in dark. After primary selection, the healthy calli were transferred to fresh selection medium twice at 14 days interval for further proliferation. The newly proliferated macro-calli were grown on regeneration medium-I (MSRM-I, pH 5.8) for 7 days at 26 ± 2°C in dark and subsequently sub-cultured on fresh MSRM-I before exposing to light (light:dark photoperiod 16:8 h). Thereafter, the healthy regenerated shoots were transferred to rooting medium (RM) for root development, and later rooted plants were grown in ½ strength Yoshida’s nutrient solution (Yoshida et al., 1976) for 7 days. Plants with well-established roots were hardened in Soilrite-mix and moved to the green house for further development and seed setting. T0 seeds were harvested and used for further studies by raising T1 generation plants.
Genomic DNA (gDNA) was isolated from young leaf tissues of primary transformants and wild type (WT) plants using Nucleospin Plant II DNA extraction kit (Macherey-Nagel, Düren, Germany). Presence of transgenes was primarily confirmed by PCR using target genes specific primers. T1 generation plants were raised in autoclaved soil and DNA was isolated from the fresh leaves as stated above and PCR confirmation of the transgenes was done using different sets of primers (Table S1).
For determination of T-DNA integration and transgene copy number, gDNA was isolated from young leaf tissues of T1 of both sets of transgenics expressing Mg-flp-1; Mg-flp-12 and WT plants; and Southern hybridization was performed. About 12 μg of gDNA was digested with SacI (20 U μl–1) (New England Biolabs, Massachusetts, USA) at 37°C for 16 h. Digested DNA was then resolved on 0.8% high resolution Meta Phor agarose gel and transferred to nitrocellulose membrane (Bio-Rad, Hercules, California, USA). Probe synthesis (Mg-flp-1: 214 bp and Mg-flp-12: 299 bp), digoxigenin (DIG) labeling, hybridization, detection and blot development was carried out as described previously (Papolu et al., 2013).
Following Southern blot analysis, Northern hybridization was performed to check the presence of siRNA in the transgenics. For this, total RNA (small and large RNA in single fraction) was extracted from young leaves of T1 plants using NucleoSpin® miRNA Kit (Macherey-Nagel, Düren, Germany). Separation of RNA, membrane transfer, probe preparation (Mg-flp-1: 214 bp and Mg-flp-12: 299 bp), DIG labeling, hybridization and detection was carried out as described earlier (Papolu et al., 2013).
Transgenic plants, after being confirmed by PCR and Southern blotting, from different lines of the two genes were subjected to quantitative real-time PCR (qRT-PCR) to analyze the transcript abundance of Mg-flp-1 and Mg-flp-12. Total RNA was extracted from young leaves of T1 plants using Nucleospin plant II RNA kit (Macherey-Nagel, Düren, Germany), and assessed for quality and quantity using Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Approximately 500 ng of purified RNA was reverse transcribed (Superscript VILO, Invitrogen, Carlsbad, CA, USA), and qRT-PCR was carried out using gene specific primers in realplex2 thermal cycler (Eppendorf, Hamburg, Germany). Gene expression was normalized using O. sativa 18S rRNA (Accession: AF069218). Three biological and three technical replicates were kept for each sample, and data were analyzed by 2–ΔΔCt method (Livak and Schmittgen, 2001).
For analysis of expression abundance of Mg-flp-1 and Mg-flp-12 in the nematodes feeding on T1 transgenic lines, mature females were extracted from the transgenic and WT plants. RNA extraction, cDNA preparation and qRT-PCR analysis were done as described earlier (Shivakumara et al., 2019). All analyses were based on three biological and three technical replicates and nematode 18S rRNA (Accession: HE667742) was used for normalization.
The T1 plants expressing dsRNA constructs of Mg-flp-1, Mg-flp-12, and the gfp and WT controls were initially evaluated against M. graminicola using Pluronic gel medium, PF-127 (Wang et al., 2009; Kumari et al., 2016). Four day-old rice seedlings (T1, WT and reference control plants) were used for evaluation. Approximately, 30 M. graminicola J2s were inoculated at the root tip of each seedling, and assays were performed as described earlier (Kumari et al., 2016). There were five replicates for each event and the parameters like total number of galls, endoparasites, number of egg masses and eggs per egg mass per plant were used for disease scoring. Nematode multiplication factor was derived as described earlier (Kumari et al., 2017).
The transgenic lines, after being evaluated on PF-127, were further screened in soil system under greenhouse conditions. Five replicates were kept for each event keeping the WT and gfp-control plants as reference. Twenty day-old seedlings raised singly in pots filled with autoclaved soil were inoculated at the rate of 2 J2s per g of soil and plants were grown in greenhouse as described earlier (Hatzade et al., 2020). Plants were uprooted carefully at 45 days post inoculation (dpi), roots washed free of soil and scored for total number of galls, endoparasites, number of egg masses and eggs per egg mass. Nematode multiplication factor was derived as described earlier (Kumari et al., 2017). Photographs were taken using a ZEISS SteREO Discovery V20 microscope.
The bioassay data were subjected to one way analysis of variance (ANOVA) using completely randomized design (CRD), and statistical significance was determined at P = 0.05 and P = 0.01. Values of mean of total replications from each treatment were taken for statistical analyses for individual assays.
The amplified sequences of Mg-flp-1 (214 bp) and Mg-flp-12 (299 bp) neither showed similarity with any sequence at nr database nor showed homology with any M. graminicola sequence. The target dsRNA sequences were queried in dsCheck database to identify the potential off-target sites, and no exact match could be found for the processed siRNAs (19 nucleotides) in the existing database (Figure S1). Therefore, silencing of the aforesaid genes may not induce any off-target effects on other organisms.
RNAi Gateway vector ph7GWIWG2(II) was used to clone Mg-flp-1, Mg-flp-12 and gfp genes flanked by attB1 and attB2 sites in sense and antisense orientation (Figure S2). PCR analysis confirmed the orientation of the target genes (Figure S3).
The embryogenic calli of rice cv. Taipei 309 were used for Agrobacterium-mediated genetic transformation with hairpin constructs of Mg-flp-1, Mg-flp-12 and gfp, separately. Agrobacterium-infected calli were proliferated, histo-differentiated and transferred to MSM-I (Table S2). Following incubation, some calli remained healthy-creamish, whereas others turned brownish. The healthy calli were transferred to fresh MSM-II for further proliferation and later-transferred to MSRM-I to induce shoot development (Figure S4). The regenerated plants were hardened in soilrite-mix. A regeneration frequency of 47% was observed in the transformed calli (Table S3).
Ten primary transgenic events (T0) of each of Mg-flp-1 and Mg-flp-12 were genotyped by PCR using gene specific, CaMV 35S promoter and attB2, CaMV 35S terminator and attB2, and hptII primers. No amplification was observed in the WT plants while transgenic plants yielded expected amplicons (Figures S5A, B). Independent transformed events were confirmed by presence of transgenes conferring hygromycin resistance with an overall transformation efficiency of 6.4% (Table S3).
The T1 progeny plants were generated by selfing the selected T0 plants and re-validated by PCR using the aforesaid pair of primers, which showed amplification of expected fragments in all the tested lines (Figures S6A, B).
In order to confirm the integration and copy number of transgenes in various transgenic lines, PCR positive T1 plants were analyzed by Southern blot hybridization. It was found that the lines A2-3, A3-5, A5-3, A6-1, A9-1, and A10-3 for Mg-flp-1, and lines B2-1, B5-7, B6-4, B7-1, B9-2, and B11-3 for Mg-flp-12 showed positive integration. WT plants were used as control (Figures 1A, B).

Figure 1 Southern and Northern blot analyses for transgenic lines harboring dsRNA constructs of Mg-flp-1 and Mg-flp-12. (A) Southern blot analysis of Mg-flp-1 expressing lines; A1-5, A2-3, A3-5, A4-1, A5-3, A6-1, A8-2, A9-1, A10-3, A11-7, and (B) Mg-flp-12 expressing lines; B2-1, B4-4, B5-7, B6-4, B7-1, B8-6, B9-2, B10-1, B11-3. (C) Northern blot analysis for Mg-flp-1-specific sRNA in selected transgenic lines; A2-3, A3-5, A5-3, A6-1, A9-1, A10-3, and (D) Mg-flp-12-specific sRNA in selected transgenic lines; B2-1, B5-7, B6-4, B7-1, B8-6, B9-2, B11-3. (PC, positive control; NC, negative control; B, blank; WT, wild type plant).
As a key component of HIGS, expression of siRNAs of Mg-flp-1 and Mg-flp-12 was established by Northern blot analysis in the transgenic lines. The siRNAs were detected in the representative samples of Mg-flp-1 (A3-5, A5-3, A10-3) and Mg-flp-12 (B2-1, B5-7, B6-4, B11-3) transgenic lines (Figures 1C, D), confirming the possibility for HIGS.
Transgenic plants expressing Mg-flp-1 and Mg-flp-12 dsRNA constructs were further confirmed at mRNA level by qRT-PCR. The results indicated variable level of expression of the transgenes in all the selected lines. The line A5-3 of Mg-flp-1 and B5-7 of Mg-flp-12 had the highest expression, whereas line A10-3 of Mg-flp-1 and B8-6 of Mg-flp-12 showed least expression (Figure S7). However, no expression was detected in the WT plants.
In order to investigate the effect of HIGS in suppressing target genes in nematode, qRT-PCR was performed with adult females of M. graminicola extracted from the transgenic plants of both genes. Mg-flp-1 extracted females exhibited down regulation in the range of 0.63 ± 0.2–1.71 ± 0.2 fold while that of Mg-flp-12 showed 0.04 ± 0.1–1.80 ± 0.2 fold reduction (P ≤0.05) (Figure 2).
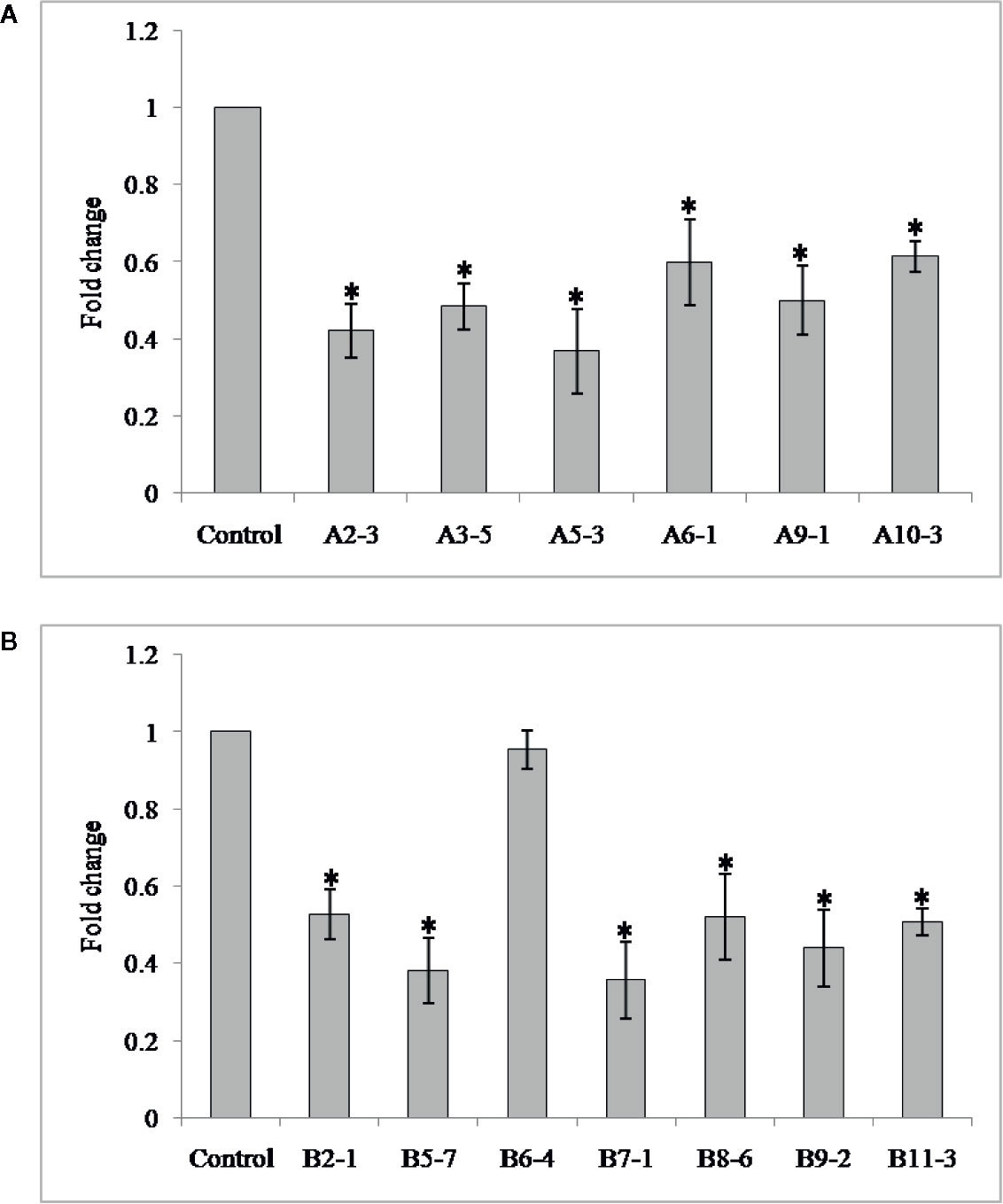
Figure 2 Transcript levels of Mg-flp-1 and Mg-flp-12 in M. graminicola females extracted from dsRNA expressing transgenic lines. (A) Mg-flp-1 lines; A2-3, A3-5, A5-3, A6-1, A9-1, A10-3, (B) Mg-flp-12 lines; B2-1, B5-7, B6-4, B7-1, B8-6, B9-2, B11-3. Expression was quantified as fold change values calculated by 2–ΔΔCT method, and 18S rRNA gene was used as reference. Each bar represents the mean ± SE of n = 3, and asterisks indicate significant difference at P < 0.05.
Performance of the transgenic plants was initially assessed against M. graminicola on Pluronic F-127 medium. Number of galls, endoparasites and egg masses developed per plant, and eggs per egg masses were counted at 18 dpi. The results showed reduction in average galling by 13‒44% for Mg-flp-1 (Figure 3A) and 30‒48% for Mg-flp-12 in different transgenic lines (Figure 4A). This observation was corroborated with reduced number of endoparasites which was 28‒46% for Mg-flp-1 and 36‒57% for Mg-flp-12 transgenic lines. Likewise, a significant reduction was seen in number of egg masses (25–55%) and eggs per egg mass (19–35%) in Mg-flp-1 transgenics. Similarly, Mg-flp-12 silencing decreased number of egg masses by 34–60% and eggs per egg mass by 29–47%. Finally, derived multiplication factor was reduced in Mg-flp-1 and Mg-flp-12 silenced plants by 40‒68% and 53‒78%, respectively when compared to the WT and negative control plants (Figure 5A).
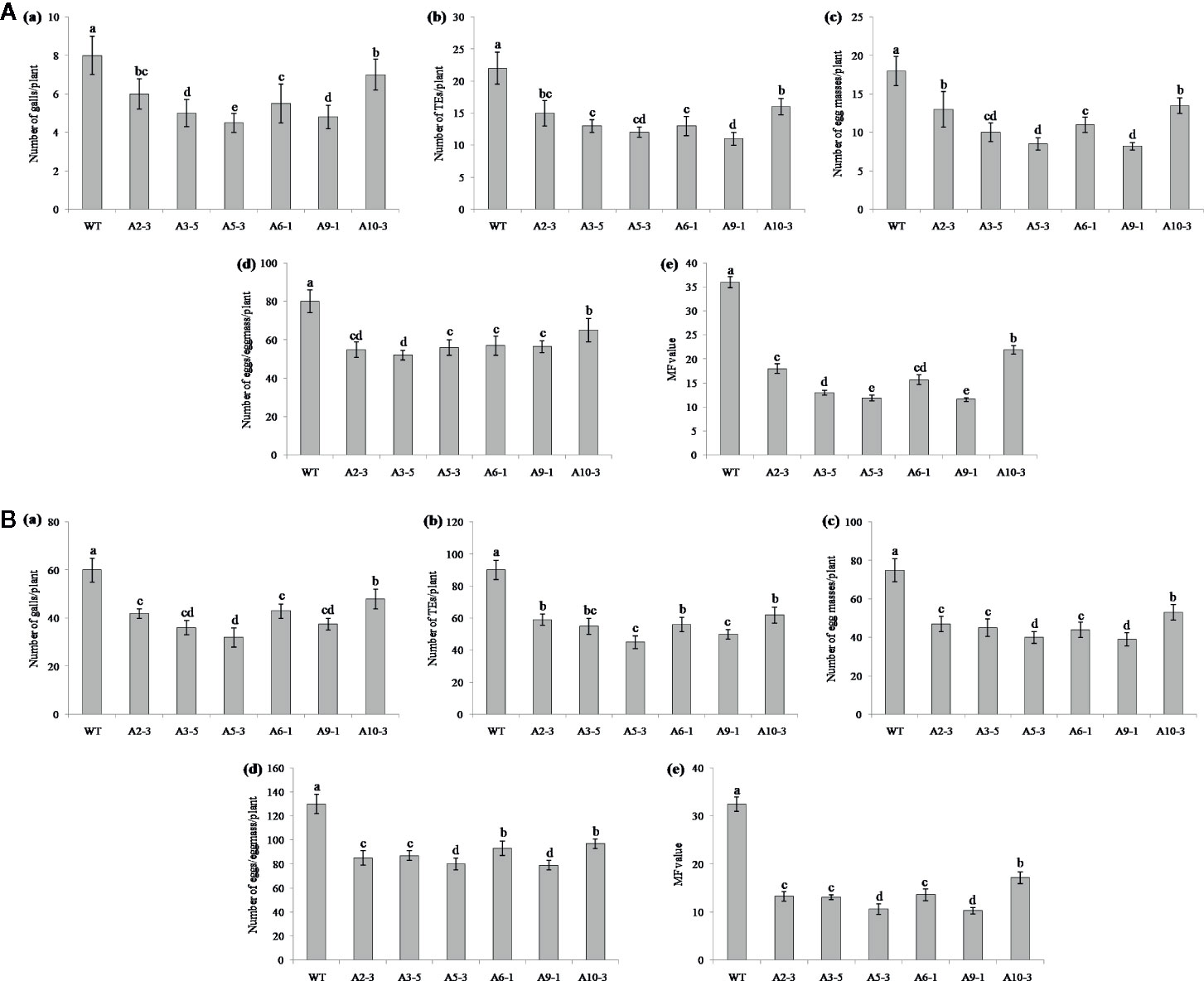
Figure 3 Effect of host induced silencing of Mg-flp-1 on development and reproduction of M. graminicola. Performance of transgenic rice plants was assessed on (A) Pluronic F-127, and (B) soil medium. Relative number of galls (a), total number of endoparasites (b), egg masses (c), eggs per egg mass (d), and the respective multiplication factor (e) were determined in different transgenic lines (A2-3, A3-5, A5-3, A6-1, A9-1, A10-3) and wild type plants at 18 and 45 dpi. Each bar represents the mean ± SE of n = 5, and bars with different letters (within each parameter) denote significant difference at P < 0.05.
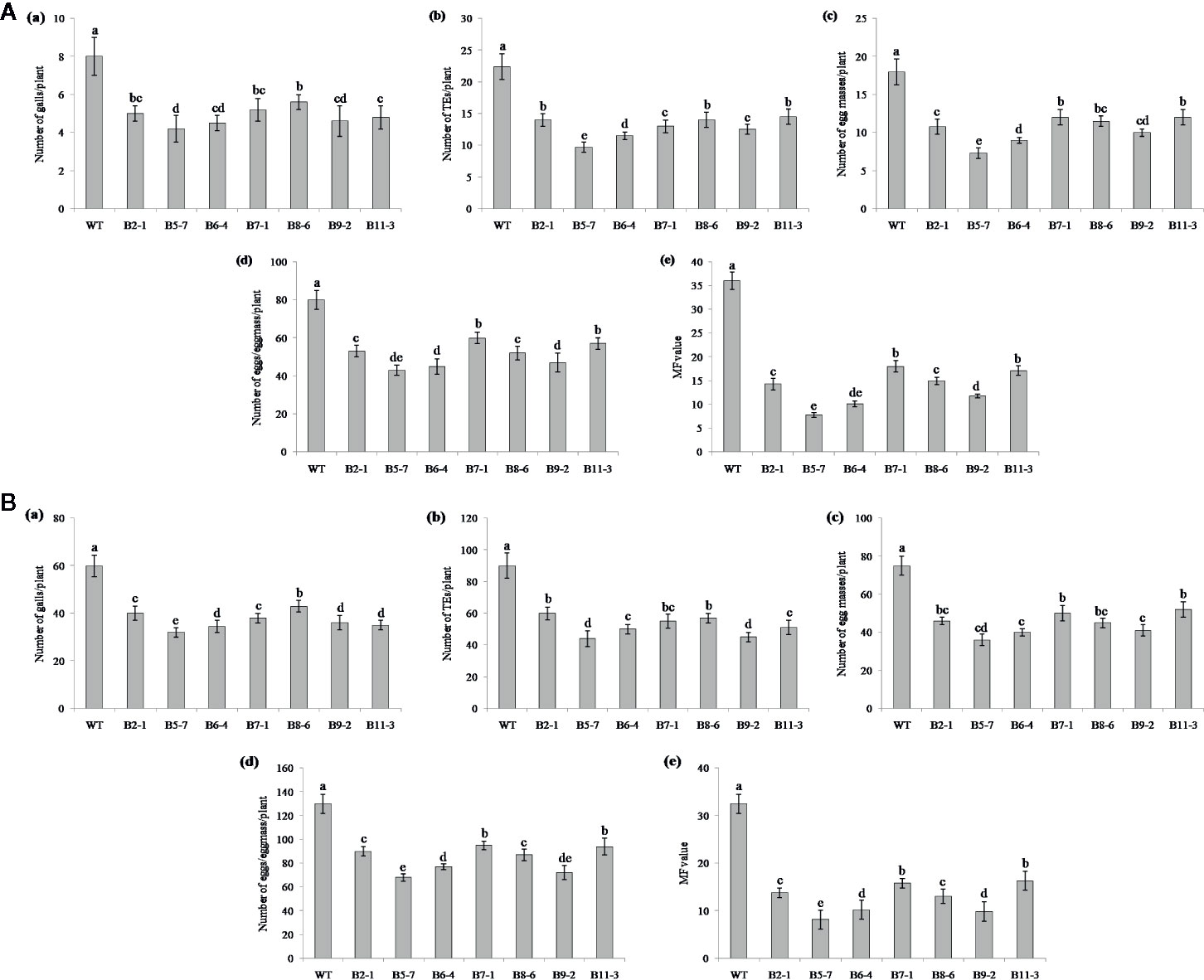
Figure 4 Effect of host induced silencing of Mg-flp-12 on development and reproduction of M. graminicola. Performance of the transgenic rice plants was assessed on (A) Pluronic F-127, and (B) soil medium. Relative number of galls (a), total number of endoparasites (b), egg masses (c), eggs per egg mass (d), and the respective multiplication factor (e) were determined in different transgenic lines (B2-1, B5-7, B6-4, B7-1, B8-6, B9-2, B11-3) and WT plants at 18 and 45 dpi. Each bar represents the mean ± SE of n = 5, and bars with different letters (within each parameter) denote significant difference at P < 0.05.
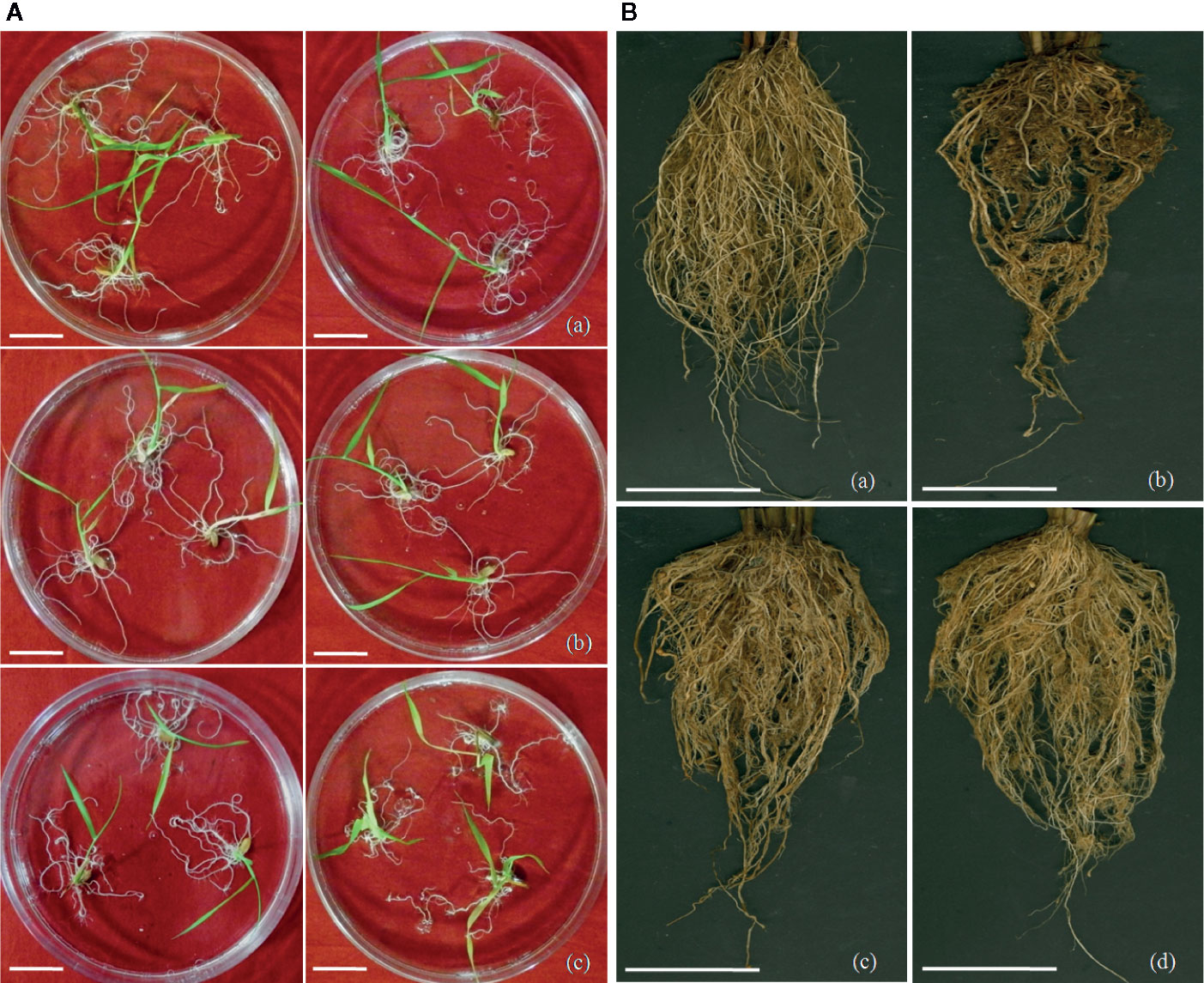
Figure 5 Screening of transgenic rice lines for nematode infection. (A) Screening of M. graminicola infected plants in PF-127 medium at 18 dpi; representative nematode infected plants of (a) Mg-flp-1 transgenic lines, (b) Mg-flp-12 transgenic lines, (c) wild type plants. Typical hook-like galls on roots with greater number of them in WT compared to transgenic lines; scale bar = 0.5 cm, (B) Comparison of M. graminicola infection in transgenic plant roots and WT plants at 45 dpi; representative roots of (a) healthy WT plants, (b) nematode infected WT plants, (c) nematode infected Mg-flp-1 transgenics, and (d) nematode infected Mg-flp-12 transgenics. Intensity of galling was comparatively higher in WT plants than the transgenics; scale bar = 5 cm.
The independent T1 lines harboring RNAi constructs were further assessed for performance in soil by inoculating 20-day old plants with freshly hatched J2s. The plants were harvested after completion of two successive life cycles of nematode. The results demonstrated that transgenic plants of different lines showed significant reduction (P ≤0.05) in gall number compared to WT plants and control plants. Line A10-3 of Mg-flp-1 (Figure 3B) and B8-6 of Mg-flp-12 (Figure 4B) showed highest number of galls but also showed significant reduction in total endoparasites (31–37%), egg masses (30–40%) and eggs per egg mass (25–33%). The derived multiplication factor was found to be reduced by 47–60% (Figure 5B).
In the present study, we have evaluated HIGS of two FMRFamide-like peptide (FLP) genes of M. graminicola, Mg-flp-1 and Mg-flp-12 in rice. Nematode neuropeptides, especially the FLPs are associated with numerous physiological functions including host recognition, feeding, sensory perception, navigation, reproduction and parasitism (Kimber et al., 2007; Morris et al., 2017; Warnock et al., 2017). Hence, these neuropeptides could be potential targets for designing safe and specific nematode management schedule (Peymen et al., 2014; Warnock et al., 2017). Several FLPs have been identified from the PPNs, and FMRFamide-like immune reactivity has also been observed in the nervous systems of some PPNs like G. pallida and G. rostochiensis (Kimber et al., 2001). To date, 19 flp genes have been identified in the most notorious nematode species M. incognita, and HIGS of flp-14 and flp-18 in tobacco provided significant reduction in reproductive potential and parasitism of the nematode (Papolu et al., 2013). Rice production is largely affected by M. graminicola (Jones et al., 2013), and earlier observations demonstrated that in vitro silencing of nine flp genes (flp-1, flp-3, flp-6, flp-7, flp-11, flp-12, flp-14, flp-16 and flp-18) in M. graminicola J2s significantly retarded their penetration and reproduction potential while infecting rice (Kumari et al., 2017). Further, results involving G. pallida, G. rostochiensis, Heterodera glycies, H. schachtii, M. incognita, M. javanica, Pratylenchus spp., etc. suggest that HIGS of parasitism genes provide a better platform for nematode management and also strengthens the findings of in vitro RNAi (Urwin et al., 2002; Chen and Roberts, 2003; Vanholme et al., 2004; Dubreuil et al., 2007; Adam et al., 2008; Bakhetia et al., 2008; Tan et al., 2013; Chaudhary et al., 2019; Shivakumara et al., 2019). In this regard, the present study convincingly supports the earlier findings of Kumari et al. (2017), and also validates the efficiency of M. graminicola flp-1 and flp-12, as potential targets for nematode management through HIGS in Asian rice O. sativa.
Molecular analyses by PCR and Southern hybridization revealed the insertion, integration and inheritance of the T-DNA harboring dsRNA constructs of Mg-flp-1 and Mg-flp-12 independently in most of the transformed lines. However, the expression of hpRNA coding transgenes quantified by qRT-PCR was found to be variable among different lines. This suggests that the transgenes were randomly integrated at diverse transcriptionally active sites in the rice genome. Additionally, the nematode gene sequences showed no similarity or homology with any short stretches of similar sequences in rice genome, which reduces the risk of any non-target effect.
The results of nematode bioassays revealed significant reduction in development and reproduction in most of the transgenics lines, containing dsRNA construct of Mg-flp-1 and Mg-flp-12, when compared to the WT plants. Simultaneously, galling and multiplication factor also declined significantly in the transgenic plants. No apparent morphological variation was observed in transgenic lines compared to the WT plants, indicating the focused RNAi effect. In addition, the detection of target gene specific siRNAs in transgenic lines provided authentic evidence for the HIGS of Mg-flp-1 and Mg-flp-12 genes, which resulted in reduced virulence and reproduction of the nematode species. A previous study has showed that in vitro silencing of both these genes reduced penetration and infection of the M. graminicola juveniles (Kumari et al., 2017). This finding can be corroborated with some other studies (Kimber et al., 2007; Papolu et al., 2013), where silencing of flp genes also showed similar results. dsRNA mediated silencing of flp-1, flp-6, flp-12, flp-14 and flp-18 genes in G. pallida, led to aberrant behavioral phenotypes (Kimber et al., 2007). Host induced silencing of flp-14 and flp-18 in tobacco affected development of M. incognita (Papolu et al., 2013). In B. xylophilus, flp-4 and flp-18 might coordinate with NPR-4 receptor, and flp-3, flp-18, flp-7 and flp-11 activate NPR-10 and FRPR-3 receptors, thus forming an integral part of complex neuronal network controlling nematode movement through motor and sensory neurons (Kimber et al., 2007). The reduced development, reproduction and parasitism as a result of silencing Mg-flp-1 and Mg-flp-12 might also involve a complex neuromuscular system in M. graminicola that affects the rice-nematode interaction. Studies with Ascaris suum revealed that silencing of flp-14 and 15 other flps inhibit nematode ovijection by shortening oviduct length, relaxing the circular muscles and cessation of contractile activity simultaneously (Moffett et al., 2003). Similar physiological anomalies might take place upon silencing of Mg-flp-1 and Mg-flp-12, which possibly result in reduction in nematode’s egg laying capacity. Similar findings were also recorded in the model nematode Caenorhabditis elegans, where loss of function mutants of flp-1 showed altered locomotion and egg laying (Chang et al., 2015).
In order to assess the HIGS of Mg-flp-1 and Mg-flp-12, the adult females were extracted from the T1 transgenic lines and WT plants and qRT-PCR was performed to assess the transcript abundance. A significant reduction in gene expression (up to 1.8 folds) was observed in the nematode females extracted from the transformed lines. These findings unequivocally support that HIGS of both flp genes in rice could effectively control the RKN species M. graminicola independently, and are adequately substantiated by the inclusion of negative controls during in planta RNAi studies.
Although there are several reports of efficacy of flp genes disrupting the neuromotor functions in nematodes, this is an established report to demonstrate the effectiveness of Mg-flp-1 and Mg-flp-12 genes in the management of M. graminicola using HIGS in rice. The significant reduction of nematode multiplication factor in the transgenic plants has proved the potential of RNAi silencing of both genes. Based on the environmental effects, agronomic conditions and biology of the nematodes, absolute resistance is very hard to achieve against these pests (Fuller et al., 2008; Rosso et al., 2009). In this direction, Mg-flp-1 and Mg-flp-12 genes would be highly effective to withstand the initial population toll in the field, and will also help bringing down the population build up that will help in reducing the initial nematode pressure in soil for successive crop(s). Thus the early crop growth won’t get affected. Further, sustainable management can also be achieved by pyramiding few promising flp genes together to efficiently interfere different physiological processes required for successful nematode parasitism.
All datasets generated for this study are included in the article/Supplementary Material.
UR conceived, designed, and supervised the experiments. AH and CK performed the experiments. DS help in plant transformation. AH analyzed the data, and wrote the draft of the manuscript. UR, VP, and VC reviewed and wrote the final draft of the manuscript. All authors contributed to the article and approved the submitted version.,
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors acknowledge funding from the IARI outreach project (IARI : ORP:NEM:09:04) and Department of Biotechnology (DBT) COE project BT/PR-18924/COE/34/48/2017. Authors also acknowledge the financial support received from NAHEP-CAAST, ICAR-IARI (Grant No. NAHEP/CAAST/2018-19/07).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00894/full#supplementary-material
FLPs, FMRFamide-like peptides; HIGS, Host-induced gene silencing; dsRNA, Double-stranded RNA; hpRNA, Hairpin RNA; PPNs, Plant-parasitic nematodes; RKN, Root-knot nematode; MF, Multiplication factor.
Adam, M. A. M., Phillips, M. S., Jones, J. T., Block, V. C. (2008). Characterization of the cellulose-binding proteins Mj-cbp-1 of the root knot nematode. Meloidogyne Javan. Physiol. Mol. Plant P. 72, 21–28. doi: 10.1016/j.pmpp.2008.05.002
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Atkinson, L. E., Stevenson, M., McCoy, C. J., Marks, N. J., Fleming, C., Zamanian, M., et al. (2013). Flp-32 ligand/receptor silencing phenocopy faster plant pathogenic nematodes. PloS Pathog. 9 (2), e1003169. doi: 10.1371/journal.ppat.1003169
Bakhetia, M., Urwin, P. E., Atkinson, H. J. (2008). Characterisation by RNAi of pioneer genes expressed in the dorsal pharyngeal gland cell of Heterodera glycines and the effects of combinatorial RNAi. Int. J. Parasitol. 38 (13), 1589–1997. doi: 10.1016/j.ijpara.2008.05.003
Banakar, P., Sharma, A., Lilley, C. J., Gantasala, N. P., Kumar, M., Rao, U. (2015). Combinatorial in vitro RNAi of two neuropeptide genes and a pharyngeal gland gene on. Meloidogyne Incogn. Nematol. 17 (2), 155–167. doi: 10.1163/15685411-00002859
Bridge, J., Plowright, R. A., Peng, D. (2005). “Nematode parasites of rice,” in Plant-Parasitic Nematodes in Subtropical and Tropical Agriculture. Eds. Luc, M., Sikora, R. A., Bridge, J. (Wallingford, UK: CABI International), 87–130.
Cabasan, M. T. N., Kumar, A., De Waele, D. (2012). Comparison of migration, penetration, development and reproduction of Meloidogyne graminicola on susceptible and resistant rice genotypes. Nematology 14 (4), 405–415. doi: 10.1163/156854111X602613
Chang, Y. J., Burton, T., Ha, L., Huang, Z., Olajubelo, A., Li, C. (2015). Modulation of locomotion and reproduction by FLP neuropeptides in the nematode. Caenorhabditis Elegans PloS One 10 (9), e0135164. doi: 10.1371/journal.pone.0135164
Chaudhary, S., Dutta, T. K., Tyagi, N., Shivakumara, T. N., Papolu, P. K., Chobhe, K. A., et al. (2019). Host-induced silencing of Mi-msp-1 confers resistance to root-knot nematode Meloidogyne incognita in eggplant. Transgenic Res. 28, 327–340. doi: 10.1007/s11248-019-00126-5
Chen, P., Roberts, P. A. (2003). Genetic analysis of (a) virulence in Meloidogyne hapla to resistance in bean (Phaseolus vulgaris). Nematology 5 (5), 687–697. doi: 10.1163/156854103322746869
Dalzell, J. J., McMaster, S., Fleming, C. C., Maule, A. G. (2010). Short interfering RNA-mediated gene silencing in Globodera pallida and Meloidogyne incognita infective stage juveniles. Int. J. Parasitol. 40 (1), 91–100. doi: 10.1016/j.ijpara.2009.07.003
Danchin, E. G. J., Arguel, M. J., Campan-Fournier, A., Perfus-Barbeoch, L., Magliano, M., Rosso, M.-N., et al. (2013). Identification of novel targetgenes for safer and more specific control of root-knot nematodes from a pan-genome mining. PloS Pathog. 9, e1003745. doi: 10.1371/journal.ppat.1003745
De Waele, D., Elsen, A. (2007). Challenges in tropical plant nematology. Annu. Rev. Phytopathol. 45, 457–485. doi: 10.1146/annurev.phyto.45.062806.094438
De Waele, D., Das, K., Zhao, D., Tiwari, R., Shrivastava, D., Vera-Cruz, C., et al. (2013). Host response of rice genotypes to the rice root-knot nematode (Meloidogyne graminicola) under aerobic soil conditions. Arch. Phytopathol. PFL. 46 (6), 670–681. doi: 10.1080/03235408.2012.749702
Dong, L., Li, X., Huang, L., Gao, Y., Zhong, L., Zheng, Y., et al. (2013). Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J. Exp. Bot. 65 (1), 131–141. doi: 10.1093/jxb/ert356
Dubreuil, G., Magliano, M., Deleury, E., Abad, P., Rosso, M. N. (2007). Transcriptome analysis of root knot nematode functions induced in early stage of parasitism. New Phytol. 176, 426–436. doi: 10.1111/j.1469-8137.2007.02181.x
Dutta, T. K., Ganguly, A. K., Gaur, H. S. (2012). Global status of rice root-knot nematode, Meloidogyne graminicola. Afr. J. Microbiol. Res. 6 (31), 6016–6021. doi: 10.20546/ijcmas.2017.608.376
Dutta, T. K., Khan, M. R., Phani, V. (2019). Plant-parasitic nematode management via biofumigation using brassica and nonbrassica plants: current status and future prospects. Curr. Plant Biol. 17, 17–32. doi: 10.1016/j.cpb.2019.02.001
Fairbairn, D. J., Cavallaro, A. S., Bernard, M., Mahalinga-Iyer, J., Graham, M. W., Botella, J. R. (2007). Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226, 1525–1533. doi: 10.1007/s00425-007-0588-x
Fuller, V. L., Lilley, C. J., Urwin, P. E. (2008). Nematode resistance. New Phytol. 180 (1), 27–44. doi: 10.1111/j.1469-8137.2008.02508.x
Golden, A. M., Birchfield, W. (1965). Meloidogyne graminicola (Heteroderidae), a new species of root-knot nematode from grass. P. Helm. Soc Wash. 32 (2), 228–231.
Hatzade, B., Singh, D., Phani, V., Kumbhar, S., Rao, U. (2020). Profiling of defense responsive pathway regulatory genes in Asian rice (Oryza sativa) against infection of Meloidogyne graminicola (Nematoda:Meloidogynidae). 3 Biotech. 10 (2), 60. doi: 10.1007/s13205-020-2055-3
Huang, Q. X., Cheng, X. Y., Mao, Z. C., Wang, Y. S., Zhao, L. L., Yan, X., et al. (2010). MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PloS One 5 (10), e13271. doi: 10.1371/journal.pone.0013271
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Kimber, M. J., Fleming, C. C., Bjourson, A. J., Halton, D. W., Maule, A. G. (2001). FMRFamide-related peptides in potato cyst nematodes. Mol. Biochem. Parasitol. 116 (2), 199–208. doi: 10.1016/s0166-6851(01)00323-1
Kimber, M. J., McKinney, S., McMaster, S., Day, T. A., Fleming, C. C., Maule, A. G. (2007). flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J. 21 (4), 1233–1243. doi: 10.1096/fj.06-7343com
Kumari, C., Dutta, T. K., Banakar, P., Rao, U. (2016). Comparing the defence-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Sci. Rep. 6:22846. doi: 10.1038/srep22846
Kumari, C., Dutta, T. K., Chaudhary, S., Banakar, P., Papolu, P. K., Rao, U. (2017). Molecular characterization of FMRFamide-like peptides in Meloidogyne graminicola and analysis of their knockdown effect on nematode infectivity. Gene 619, 50–60. doi: 10.1016/j.gene.2017.03.042
Kyndt, T., Nahar, K., Haegeman, A., De Vleesschauwer, D., Hofte, M., Gheysen, G. (2012). Comparing systemic defence-related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biol. 14 (s1), 73–82. doi: 10.1111/j.1438-8677.2011
Livak, K., Schmittgen, T. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2-DDCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorenzana, O. J., Matamis, P. P., Mallinin, C. B., Jose, O. L., De-leon, D. S. (1998). “Cultural management practices to control rice root knot nematode,” in 1998 Regional Research and Development Symposia 15 Jul-11 Sep 1998.
Mantelin, S., Bellafiore, S., Kyndt, T. (2017). Meloidogyne graminicola: a major threat to rice agriculture. Mol. Plant Pathol. 18 (1), 3–15. doi: 10.1111/mpp.12394
Masler, E. P. (2008). Digestion of invertebrate neuropeptides by preparations from the free-living nematode. Panagrellus Redivivus. J. Helminthol. 82 (3), 279–285. doi: 10.1017/S0022149X08982596
Maule, A. G., Mousley, A., Marks, N. J., Day, T. A., Thompson, D. P., Geary, T. G., et al. (2002). Neuropeptide signaling systems-potential drug targets for parasite and pest control. Curr. Top. Med. Chem. 2 (7), 733–758. doi: 10.2174/1568026023393697
McVeigh, P., Geary, T. G., Marks, N. J., Maule, A. G. (2006). The FLP-side of nematodes. Trends. Parasitol. 22 (8), 385–396. doi: 10.1016/j.pt.2006.06.010
Moffett, C., Beckett, A., Mousley, A., Geary, T., Marks, N., Halton, D., et al. (2003). The ovijector of Ascaris suum: multiple response types revealed by Caenorhabditis elegans FMRFamide-related peptides. Int. J. Parasitol. 33 (8), 859–876. doi: 10.1016/s0020-7519(03)00109-7
Morris, R., Wilson, L., Sturrock, M., Warnock, N. D., Carrizo, D., Cox, D., et al. (2017). A neuropeptide modulates sensory perception in the entomopathogenic nematode Steinernema carpocapsae. PloS Pathog. 13 (3), e1006185. doi: 10.1371/journal.ppat.1006185
Naito, Y., Yamada, T., Matsumiya, T., Ui-Tei, K., Saigo, K., Morishita, S. (2005). dsCheck: highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res. 33, W589–W591. doi: 10.1093/nar/gki419
Papolu, P. K., Gantasala, N. P., Kamaraju, D., Banakar, P., Sreevathsa, R., Rao, U. (2013). Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode. Meloidogyne Incogn. PloS One 8 (11), e80603. doi: 10.1371/journal.pone.0080603
Peymen, K., Watteyne, J., Frooninckx, L., Schoofs, L., Beets, I. (2014). The FMRFamide-like peptide family in nematodes. Front. Endocrinol. 5:90:90. doi: 10.3389/fendo.2014.00090
Prasad, J. S., Panwar, M. S., Rao, Y. S. (1992). “Nematode pests of rice,” in Nematode pests of crops. Eds. Bhatti, D. S., Walia, R. K. (NewDelhi: CBS Publishers and Distributors), 43–61.
Rao, U., Thakur, P. K., Prasad, N., Banakar, P., Kumar, M. (2013). Identification of neuropeptides, flp-1 and flp-12 targeting neuromuscular system of rice root knot nematode (RRKN). Meloidogyne Graminicola. Bioinform. 9 (4), 182–186. doi: 10.6026/97320630009182
Roderick, H., Urwin, P. E., Atkinson, H. J. (2018). Rational design of biosafe crop resistance to a range of nematodes using RNA interference. Plant Biotechnol. J. 16, 520–529. doi: 10.1111/pbi.12792
Rosso, M. N., Jones, J. T., Abad, P. (2009). RNAi and functional genomics in plant parasitic nematodes. Annu. Rev. Phytopathol. 47, 207–232. doi: 10.1146/annurev.phyto.112408.132605
Sambrook, J., Fristch, E. F., Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. 2nd ed. (Cold Spring Harbor, NY: CSH Laboratory Press).
Shivakumara, T. N., Somvanshi, V. S., Phani, V., Chaudhary, S., Hada, A., Budhwar, R., et al. (2019). Meloidogyne incognita (Nematoda: Meloidogynidae) sterol-binding protein Mi-SBP-1 as a target for its management. Int. J. Parasitol. 49 (13-14), 1061–1073. doi: 10.1016/j.ijpara.2019.09.002
Somvanshi, V. S., Ghosh, O., Budhwar, R., Dubay, B., Shukla, R. N., Rao, U. (2018). A comprehensive annotation for the root-knot nematode Meloidogyne incognita proteome data. Data Brief. 19, 1073–1079. doi: 10.1016/j.dib.2018.05.131
Soriano, I. R., Schmit, V., Brar, D. S., Prot, J. C., Reversat, G. (1999). Resistance to rice root-knot nematode Meloidogyne graminicola identified in Oryza longistaminata and. O. Glaberrima. Nematol. 1 (4), 395–398. doi: 10.1163/156854199508397
Tan, J. A., Jones, M. G., Fosu-Nyarko, J. (2013). Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 133, 166–178. doi: 10.1016/j.exppara.2012.11.011
Taylor, C. M., Wang, Q., Rosa, B. A., Huang, S. C.-C., Powell, K., Schedl, T., et al. (2013). Discovery of anthelmintic drug targets and drugs using chokepoints in nematode metabolic pathways. PloS Pathog. 9, e1003505. doi: 10.1371/journal.ppat.1003505
Urwin, P. E., Lilley, C. J., Atkinson, H. J. (2002). Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 15, 747–752. doi: 10.1094/MPMI.2002.15.8.747
Vanholme, B., De Meutter, J., Tytgat, T., Van Montagu, M., Coomans, A., Gheysen, G. (2004). Secretions of plant-parasitic nematodes: a molecular update. Gene 332, 13–27. doi: 10.1016/j.gene.2004.02.024
Wang, C., Lower, S., Williamson, V. M. (2009). Application of Pluronic gel to the study of root-knot nematode behaviour. Nematology 11 (3), 453–464. doi: 10.1163/156854109X447024
Warnock, N. D., Wilson, L., Patten, C., Fleming, C. C., Maule, A. G., Dalzell, J. J. (2017). Nematode neuropeptides as transgenic nematicides. PloS Pathog. 13 (2), e1006237. doi: 10.1371/journal.ppat.1006237
Whitehead, A. G., Hemming, J. R. (1965). A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann. Appl. Biol. 55, 25–38. doi: 10.1111/j.1744-7348.1965.tb07864.x
Keywords: root-knot nematodes, FMRFamide-like peptide, RNAi, Oryza sativa, transgenics
Citation: Hada A, Kumari C, Phani V, Singh D, Chinnusamy V and Rao U (2020) Host-Induced Silencing of FMRFamide-Like Peptide Genes, flp-1 and flp-12, in Rice Impairs Reproductive Fitness of the Root-Knot Nematode Meloidogyne graminicola. Front. Plant Sci. 11:894. doi: 10.3389/fpls.2020.00894
Received: 27 February 2020; Accepted: 02 June 2020;
Published: 17 July 2020.
Edited by:
Sylvia Schleker, University of Bonn, GermanyReviewed by:
Lee Robertson, National Institute of Agricultural and Food Research and Technology, SpainCopyright © 2020 Hada, Kumari, Phani, Singh, Chinnusamy and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uma Rao, dW1hcmFvQGlhcmkucmVzLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.