
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 17 June 2020
Sec. Plant Membrane Traffic and Transport
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00756
This article is part of the Research Topic Highlights of ENPER2019 - European Network for Plant Endomembrane Research Meeting View all 11 articles
Autophagy is a highly conserved system for degrading and recycling cytoplasmic components. The identification of autophagy-related (ATG) genes, required for autophagosome formation, has led to numerous studies using atg mutants. These studies have revealed the physiological significance of autophagy in various functions of diverse organisms. In land plants, autophagy is required for higher-order functions such as stress responses and development. Although defective autophagy does not result in any marked defect in the reproductive processes of Arabidopsis thaliana under laboratory conditions, several studies have shown that autophagy plays a pivotal role in male reproduction in several land plants. In this review, we aim to summarize information on the role of autophagy in male reproductive processes in land plants.
Autophagy is a highly conserved system for degrading and recycling cytoplasmic components, including organelles, in the vacuole or lysosome. Among the various modes of autophagy reported thus far, macroautophagy, hereafter referred to as autophagy, has been the most intensively studied. This type of autophagy begins with the formation of a membrane sac called the isolation membrane (also known as the phagophore), which extends, engulfing cytoplasmic components, to form a double membrane-bound autophagosome. The outer membrane of this autophagosome fuses with the vacuolar membrane, releasing the inner membrane-bound autophagic body into the vacuolar lumen, to be degraded by vacuolar hydrolases (Figure 1A; Takeshige et al., 1992; Baba et al., 1994). In the 1990s, a gene set required for autophagosome formation, hereafter referred to as core autophagy-related (ATG) genes, was identified by forward genetics in Saccharomyces cerevisiae (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1995; Klionsky et al., 2003). The core ATG genes encode a group of Atg proteins that form several functional units: the Atg1 complex, the phosphatidylinositol 3-kinase (PI3K) complex, Atg9, the Atg2-Atg18 complex, and two ubiquitin-like conjugation system complexes (Nakatogawa et al., 2009; Mizushima et al., 2011). One of the core Atg proteins, Atg8, is conjugated to phosphatidylethanolamine by the ubiquitin-like conjugation systems (Figure 1B; Mizushima et al., 1998, 1999; Kirisako et al., 1999, 2000; Shintani et al., 1999; Ichimura et al., 2000; Hanada et al., 2007). Since lipidated Atg8 localizes to the isolation membrane from the beginning until after completion of autophagosome formation, it is commonly used as an autophagosome marker in various organisms (Kirisako et al., 1999; Kabeya et al., 2000; Yoshimoto et al., 2004). Reverse genetic approaches have unraveled the physiological roles of autophagy in a wide range of biological functions, including metabolic adaptation, intracellular quality control, and development (Mizushima and Komatsu, 2011; Mizushima, 2018).
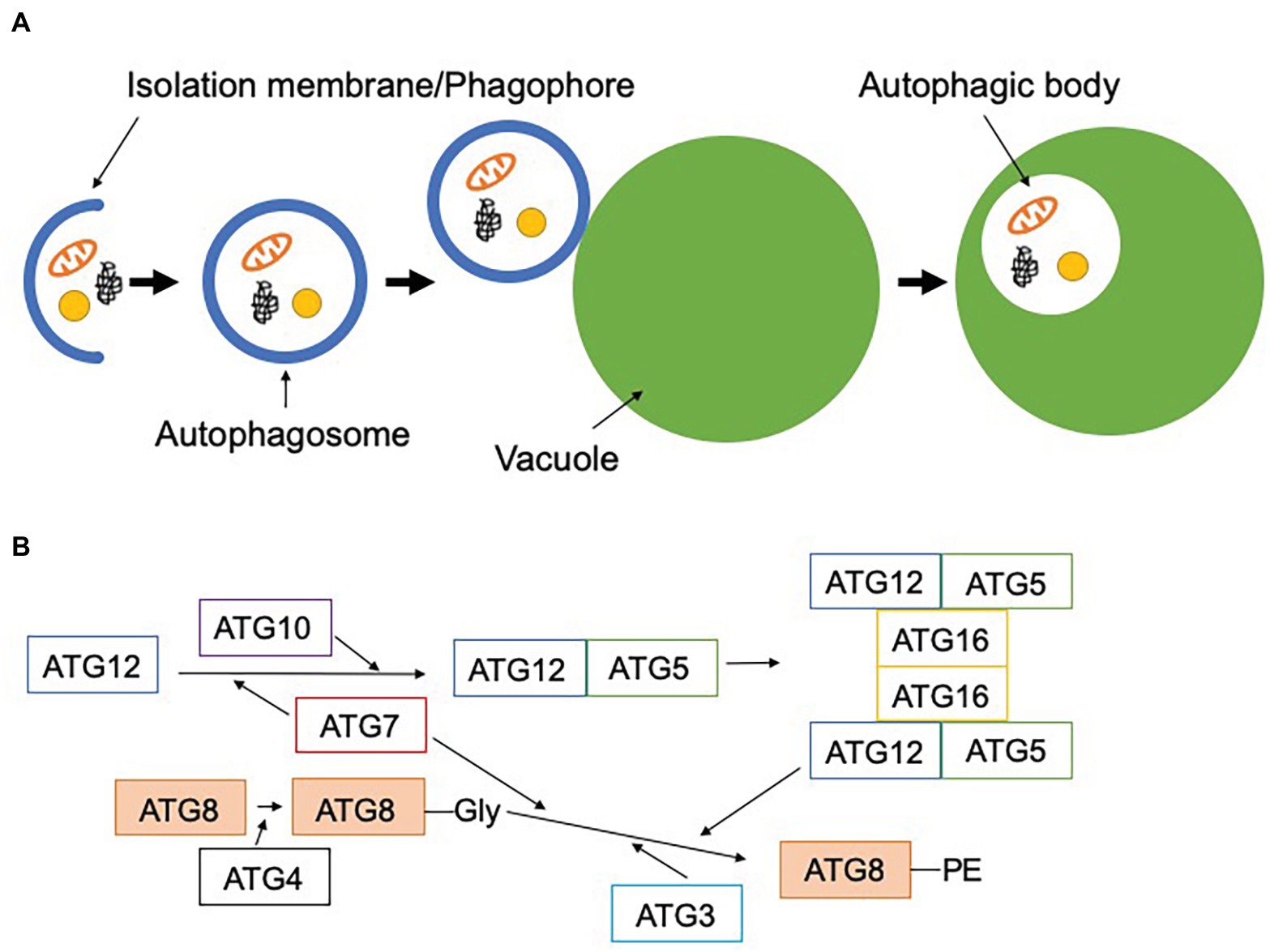
Figure 1. Scheme of macroautophagy. (A) Macroautophagy starts with the formation of the isolation membrane (phagophore) in the cytosol. This engulfs cytoplasmic components and forms the double membrane-bound autophagosome. The outer membrane of the autophagosome fuses with the vacuolar membrane to release a single membrane-bound autophagic body into the vacuole. (B) Two ubiquitin-like conjugation systems are involved in the lipidation of ATG8. First, ATG12 is conjugated to ATG5 by ATG7 (E1-like) and ATG10 (E2-like), and ATG12-ATG5 forms a complex with ATG16. ATG8 is cleaved by ATG4, resulting in the exposure of glycine at its carboxyl terminus. This processed ATG8 is conjugated to phosphatidylethanolamine by ATG7 (E1-like), ATG3 (E2-like), and the dimeric ATG12-ATG5-ATG16 complex (E3-like). Lipidated ATG8 can be localized to the autophagosomal membrane.
In land plants (embryophytes), core ATG genes are highly conserved, and their functions are shown to be similar to homologs in yeast and mammals (Avin-Wittenberg et al., 2012; Yoshimoto, 2012; Norizuki et al., 2019). Studies of Arabidopsis thaliana atg mutants have demonstrated that autophagy is involved in responses to abiotic and biotic stressors such as nutrient starvation and pathogen attacks (Marshall and Vierstra, 2018). Furthermore, recent studies have shown that autophagy plays a critical role in male reproduction in various species including Oryza sativa, Nicotiana tabacum, Marchantia polymorpha, and Physcomitrella patens (Kurusu et al., 2014; Minamino et al., 2017; Sanchez-Vera et al., 2017; Zhao et al., 2020). In this review, we will briefly outline male reproduction in angiosperms and bryophytes, then summarize the physiological roles of autophagy in these processes.
Various organisms reproduce through a sexual process in which haploid male and female gametes fuse with each other to generate diploid zygotes. Male gametes in land plants are roughly classified into two types based on the presence or absence of flagella. In angiosperms and the majority of gymnosperms, male gametes lack a flagellum and are therefore immotile, requiring transportation to egg cells via pollen tubes to accomplish fertilization. Conversely, bryophytes, lycophytes, monilophytes, and some gymnosperms such as ginkgoes and cycads utilize motile male gametes called spermatozoids, which are equipped with two or more flagella, for sexual reproduction (Southworth and Cresti, 1997; Renzaglia and Garbary, 2001). In both cases, drastic reorganization of cellular components occurs during male gamete development (Hackenberg and Twell, 2019). In the angiosperm, A. thaliana, four haploid microspores are produced by meiosis of a diploid pollen mother cell. Each microspore divides asymmetrically to form vegetative and generative cells, and each generative cell undergoes symmetrical division to form two sperm cells (Figure 2A; Southworth and Russell, 2001; Berger and Twell, 2011). Once pollen grains are attached to the surface of stigmas, they germinate to produce pollen tubes, which are precisely guided to female gametes to deliver sperm cells (Figure 2A; Higashiyama and Takeuchi, 2015; Zheng et al., 2018). Pollen grains are covered by an outer cell wall called the exine, which provides chemical and physical protection against stressors. The tapetum surrounding pollen grains plays a pivotal role in the synthesis of the exine by supplying nutrients and metabolites to pollen grains (Ariizumi and Toriyama, 2011).
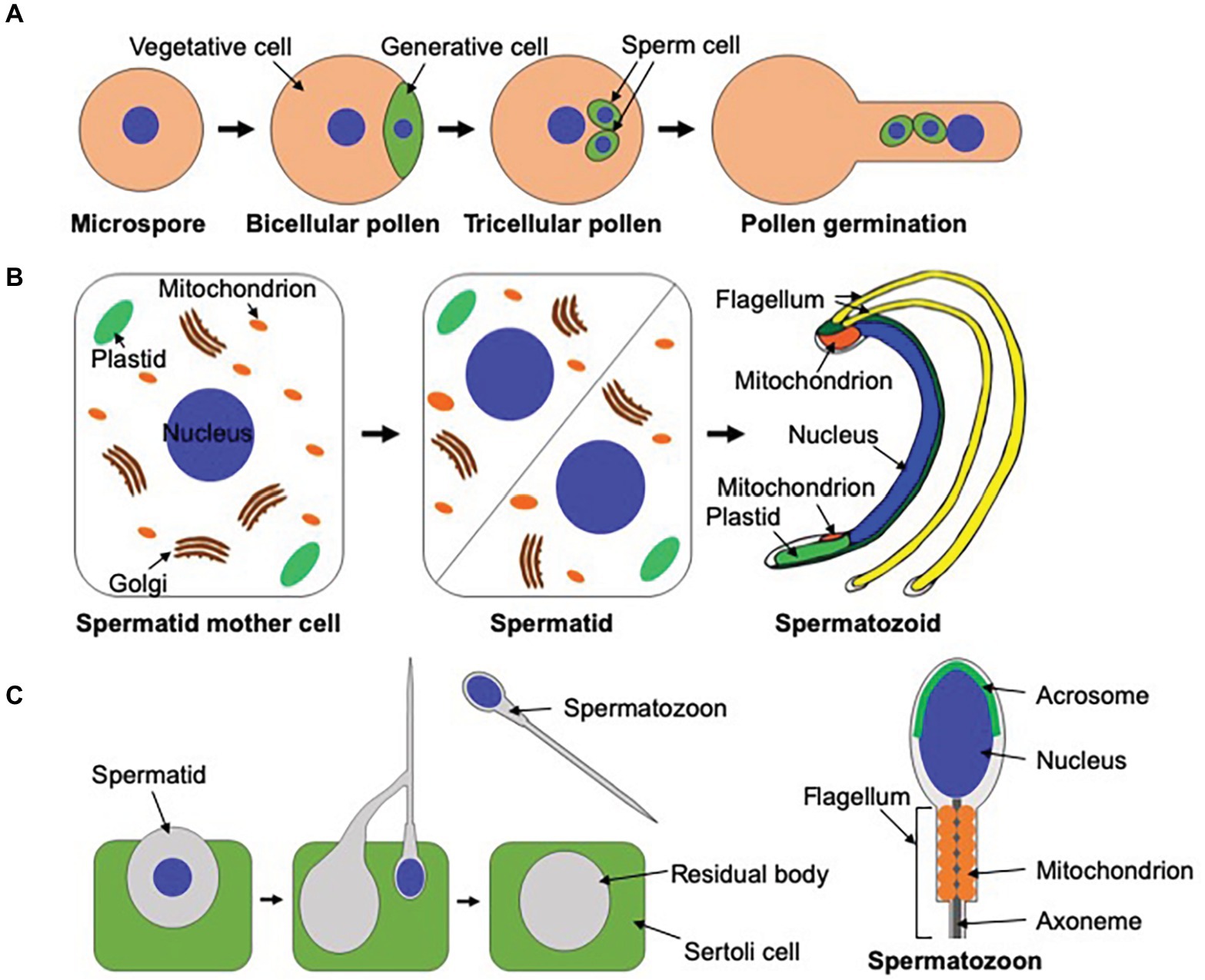
Figure 2. Male gametogenesis in Arabidopsis thaliana, Marchantia polymorpha, and mammals. (A) In A. thaliana, microspores generated from meiosis of pollen mother cells undergo asymmetrical cell division to form vegetative and generative cells. Each generative cell divides symmetrically to yield two immotile sperm cells. Once pollen grains are attached to the surface of stigmas, they germinate to produce pollen tubes, which transport male gametes to female gametes. This figure is illustrated based on figures in Berger and Twell (2011) and Hackenberg and Twell (2019). (B) In M. polymorpha, spermatids are formed by the diagonal cell division of spermatid mother cells. Spermatids undergo a dynamic morphogenetic transformation called spermiogenesis to form spermatozoids. This figure was illustrated based on a figure in Shimamura (2016). (C) Dynamic cellular reorganization also takes place during mammalian spermiogenesis. Just before the release of spermatozoa, unnecessary cytoplasmic components are excluded from their cell bodies as the residual body, which is phagocytosed and degraded by the neiboring Sertoli cell.
In contrast to angiosperms, in which the sporophytic generation is dominant in the life cycle, the gametophytic generation is dominant in bryophytes, and spermatozoids are generated without meiosis. In the liverwort, M. polymorpha, spermatids are produced by the diagonal division of spermatid mother cells. Spermatids then differentiate into motile spermatozoids through a dynamic morphological conversion called spermiogenesis. This process includes de novo synthesis of the locomotory apparatus, chromatin condensation, nuclear elongation, a decrease in the number of mitochondria, and exclusion of a major part of the cytoplasm (Figure 2B). Spermatozoids move toward female gametes in water to accomplish fertilization (Shimamura, 2016). Although the molecular mechanisms of male reproduction in angiosperms are well-documented, molecular mechanisms of spermatozoid formation in basal land plants remain mostly ambiguous (Hackenberg and Twell, 2019).
Studies of A. thaliana atg mutants have not detected a marked effect of atg mutations on sexual reproduction under normal experimental conditions, whereas these mutations affect vegetative growth in this species (Doelling et al., 2002; Hanaoka et al., 2002; Marshall and Vierstra, 2018). Mutation in ATG6 is the only exception; the atg6 mutant exhibits a defect in pollen germination (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008). However, this defect might not be a result of defective autophagy. In yeast, Atg6 is also known as Vps30 and forms a complex with Vps34 and Vps15 to produce phosphatidylinositol 3-phosphate (PI3P) from phosphatidylinositol (Kihara et al., 2001). Because the A. thaliana vps15 mutant also exhibits a defect in pollen germination and PI3P is also required for various cellular reactions beyond autophagy, defective pollen germination in the atg6 mutant could result from a deficiency independent of autophagy (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Xu et al., 2011; Wang et al., 2012). Thus, ATG-dependent autophagy should be dispensable for male reproduction in A. thaliana. In addition, Zea mays atg mutants are fertile under normal experimental conditions (Li et al., 2015). However, autophagy is indispensable for male reproduction in O. sativa (Kurusu et al., 2014). In this species, autophagy is highly activated in the tapetum during microspore development (Kurusu et al., 2014; Hanamata et al., 2019). The tapetum undergoes programmed cell death to supply metabolites and nutrients to developing microspores, which is essential for pollen maturation and pollen tube elongation (Ku et al., 2003; Kawanabe et al., 2006; Li et al., 2006; Zhang et al., 2008). The atg7 mutant exhibits limited anther dehiscence, and its pollen maturation and germination are severely compromised, resulting in markedly reduced male fertility (Kurusu et al., 2014; Sera et al., 2019). This could be explained by the fact that the atg7 mutant exhibits defective programmed cell death of the tapetum, which could result in an insufficient supply of metabolites and nutrients to developing microspores (Kurusu et al., 2014). Given that autophagy executes programmed cell death during tracheary element differentiation in A. thaliana and embryogenesis in Picea abies (Kwon et al., 2010; Minina et al., 2013), autophagy could directly induce programmed cell death in the tapetum of O. sativa. Alternatively, autophagy might indirectly affect programmed cell death by regulating the metabolism of phytohormones. The phytohormone gibberellin plays an essential role in the development of tapeta and pollen in O. sativa (Chhun et al., 2007; Aya et al., 2009). Gibberellin accumulation is reduced in the anther of the atg7 mutant, and treatment with active gibberellin (GA4) fully and partially repairs the defect in pollen maturation and germination, respectively. This suggests that autophagy regulates the development of male reproductive tissues via the metabolism of gibberellin to some extent (Kurusu et al., 2017). The different effects of defective autophagy on male fertility between O. sativa and A. thaliana might result from differences in the structure of the tapetum, lipidic components of pollen grains, or both (Hanamata et al., 2014). Further study will be needed to clarify why autophagy is particularly required during male reproductive processes in O. sativa.
Cellular and molecular reorganization during pollen germination and pollen tube elongation also involve autophagy. In addition to the essential role of autophagy in O. sativa pollen germination described above (Kurusu et al., 2014), a similar process in N. tabacum also requires autophagy (Zhao et al., 2020). In this species, autophagy is highly activated during the initial stage of pollen germination, and autophagosomes accumulate around the germination aperture. ATG2, ATG5, and ATG7 RNAi N. tabacum lines exhibit reduced rates of pollen germination, and in these lines, unlike in wild-type plants, a convex layer of the cytoplasm containing mitochondria remains at the germination aperture. Furthermore, a mitochondrial marker and the autophagosome marker ATG8 partially colocalize, and cardiolipin, a mitochondria-specific phospholipid, accumulates in the ATG RNAi lines. This information suggests that mitochondria are a target of autophagy in N. tabacum pollen grains (Zhao et al., 2020). In contrast, atg mutants of A. thaliana exhibit no detectable abnormality in pollen germination (Zhao et al., 2020). Vacuolar degradation systems other than ATG-dependent autophagy might contribute to reorganization of intracellular components during pollen germination in A. thaliana; this should be verified in future studies.
The spermatozoids of most bryophytes consist of two flagella and a cell body, which comprises an elongated spiral nucleus, one plastid, two mitochondria, and trace amounts of cytosol (Figure 2B; Renzaglia and Garbary, 2001; Shimamura, 2016). Although reorganization of intracellular structures during bryophyte spermiogenesis has been intensively observed by transmission electron microscopy (TEM; Renzaglia and Garbary, 2001), the dynamics of intracellular reorganization remain unclear. The moss, P. patens, and the liverwort, M. polymorpha, are model plants associated with genetic studies (Rensing et al., 2008; Strotbek et al., 2013; Ishizaki et al., 2016; Bowman et al., 2017). Taking advantage of various organelle markers established in M. polymorpha (Kanazawa et al., 2016; Minamino et al., 2018), Minamino et al. (2017) observed the dynamics of organelles during spermiogenesis by confocal microscopy. They found that the size of the vacuole increases during spermiogenesis, and proteins in various organelles, including the plasma membrane, Golgi apparatus, and multivesicular endosomes, are transported to the luminal space of the vacuole during spermiogenesis. These findings indicate that the vacuole plays a major role in the removal and degradation of cellular components, including organelles, during M. polymorpha spermiogenesis (Minamino et al., 2017). Multivesicular endosomes and autophagosomes, which are involved in endocytic degradation of membrane proteins and degradation of cytoplasmic components, respectively, are frequently observed in spermatids undergoing spermiogenesis. The number of autophagosomes increases during spermiogenesis, and autophagic body-like structures are observed inside the vacuole, suggesting that autophagy is activated during spermiogenesis. These findings suggest that both autophagy and endocytic degradation play important roles during M. polymorpha spermiogenesis.
A critical role of autophagy in spermiogenesis has been identified in P. patens (Sanchez-Vera et al., 2017). Autolysosome-like structures are frequently observed in spermatids undergoing spermiogenesis; these may be formed by fusion between autophagosomes and the vacuole. An elevated expression level of GFP-PpATG8e has also been detected in P. patens spermiogenesis, suggesting upregulated autophagy during this process. Furthermore, spermatozoids of the autophagy-defective atg5 mutant are sterile and possess a wide spectrum of morphological abnormalities such as a larger amount of cytoplasm and an abnormally shaped nucleus. TEM observation has also revealed that the atg5 mutation impairs decreasing the number of mitochondria and plastids, and flagellar formation during spermiogenesis (Sanchez-Vera et al., 2017). Thus, autophagy plays an important role in male gametogenesis in bryophytes, whose molecular regulatory mechanisms would be interesting targets to study.
Mammalian sexual reproduction also utilizes motile male gametes with a flagellum (spermatozoa; Figure 2C), whose composition of intracellular structures is different from that of bryophytes. A mammalian spermatozoon possesses a nucleus at its head and a flagellum at the tail, and a mitochondrial helical sheath surrounds the axoneme at the midpiece (Figure 2C; Toure et al., 2020). Although the exclusion of the cytoplasm takes place during spermiogenesis in both mammals and bryophytes, their molecular mechanisms must be not the same. Although the residual body released from mammalian spermatids, which contain unnecessary cytoplasmic components, is removed by the phagocytic activity of the neighboring Sertoli cell during mammalian spermiogenesis (O’Donnell et al., 2011), phagocytosis by neighboring cells cannot take place in bryophytes due to the surrounding rigid cell wall (Figure 2C). Nevertheless, autophagy also plays indispensable roles during mammalian spermiogenesis. The germ cell-specific ATG7 knockout in mice results in male sterility, exhibiting multiple defects in spermiogenesis, such as defective biogenesis of the acrosome (Wang et al., 2014). The acrosome, which is not present in the male gametes of plants, is a lysosome-related organelle required for fertilization (Figure 2C; Moreno and Alvarado, 2006; Ikawa et al., 2010; Khawar et al., 2019). LC3, which is homologous to yeast Atg8, is localized on the proacrosomal vesicles in an ATG7-dependent manner. These proacrosomal vesicles accumulate near the nucleus without fusing with each other in the atg7 mutant, suggesting that autophagy is required for the biogenesis of the acrosome (Wang et al., 2014). Another marked defect in the mouse atg7 mutant is the abnormal reorganization of microtubules during spermiogenesis. Irregular cytoskeletal structures are observed in autophagy-defective mouse embryonic fibroblasts (MEFs). PDLIM1, a regulator of cytoskeletons, accumulates in atg7 MEFs and spermatids, and knockdown of PDLIM1 partially suppresses cytoskeletal defects in atg7 MEFs. These results suggest that autophagy regulates cytoskeletal organization by degrading PDLIM1 (Shang et al., 2016). The P. patens atg5 mutant also exhibits defective microtubule organization during flagella formation, which may reflect a similar mechanism of cytoskeletal regulation by autophagy during spermiogenesis. Further investigation to identify targets of autophagy during spermiogenesis would be needed to understand the precise functions of autophagy during plant spermiogenesis.
As described above, autophagy is involved in distinct male reproductive processes in land plants. However, the regulatory networks and precise targets of autophagy remain almost unknown. The first step to address this would be to determine whether autophagic degradation during male reproduction in each plant species is devoted to bulk degradation of the cytoplasm or selective degradation of certain targets. Recent studies have revealed that a wide range of targets, including organelles and proteins, are selectively degraded by autophagy in various organisms, including A. thaliana (Marshall and Vierstra, 2018; Johansen and Lamark, 2020). Selective autophagy appears to operate during spermiogenesis in plants because organelles unnecessary for spermatozoids seem to be removed through autophagic degradation (Minamino et al., 2017; Sanchez-Vera et al., 2017). Bryophyte spermatozoids only retain two mitochondria and a plastid in the cell body, potentially resulting from selective removal of unneeded organelles by autophagy. Furthermore, in germinating pollen of N. tabacum, mitochondrial markers are colocalized with an autophagosome marker, implying selective autophagic degradation of mitochondria (mitophagy) (Zhao et al., 2020). However, the existence of mitophagy is not firmly demonstrated in plants thus far (Broda et al., 2018) and detailed electron microscopic or super-resolution microscopic observation of phagophores and autophagosomes is needed to be conclusive. Genetic or pharmacological inhibition of autophagic body degradation in the vacuole would also be effective in investigating the targets of autophagic degradation during male reproduction. Another promising approach is to identify proteins that interact with ATG8, since ATG8 is involved in cargo recognition in selective autophagy as well as in the formation and transport of autophagosomes in various organisms (Nakamura and Yoshimori, 2017; Marshall and Vierstra, 2018; Mizushima, 2019; Stephani and Dagdas, 2019; Johansen and Lamark, 2020). Since many land plants possess multiple ATG8 genes, each of which could play a specialized function (Kellner et al., 2017), it would be also informative to examine whether any of ATG8 genes are highly and/or specifically expressed during male reproductive development.
Another enigma is how autophagy is regulated during male reproduction in plants. As described above, autophagic activity is highly activated in certain male reproductive processes. Autophagic activity can be regulated at several distinct levels, for example, at the transcriptional and post-transcriptional levels, as reported in S. cerevisiae and mammals (Fullgrabe et al., 2016; Corona Velazquez and Jackson, 2018). In A. thaliana, the expression of core ATG genes is spatiotemporally regulated, and the transcription factor TGA9 has been shown to positively regulate ATG8 expression and autophagic activity (Slavikova et al., 2005; Rose et al., 2006; Wang et al., 2019). Post-transcriptional regulation has also been reported in A. thaliana, which is exemplified by that the TOR and SnRK1 complexes catalyze phosphorylation of and SINAT proteins mediate ubiquitylation of the ATG1 complex responding to the nutrient status (Chen et al., 2017; Huang et al., 2019; Van Leene et al., 2019; Qi et al., 2020). It would be useful to explore whether these regulations have a role in male reproduction in plants. Transcription factors responsible for the differentiation of male gametes have been identified in various organisms, including M. polymorpha (Hackenberg and Twell, 2019; Hisanaga et al., 2019). It would be worthwhile to study whether these transcription factors also regulate autophagic activities in order to understand the genetic regulation of autophagy during male reproduction.
TN and TU drafted the manuscript. NM edited the manuscript.
This study was financially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [grant nos. 19H05675, 19H05760, and 18H02470 (to TU)], and a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) (to TN, grant no. 19J13751).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ariizumi, T., and Toriyama, K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62, 437–460. doi: 10.1146/annurev-arplant-042809-112312
Avin-Wittenberg, T., Honig, A., and Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Aya, K., Ueguchi-Tanaka, M., Kondo, M., Hamada, K., Yano, K., Nishimura, M., et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21, 1453–1472. doi: 10.1105/tpc.108.062935
Baba, M., Takeshige, K., Baba, N., and Ohsumi, Y. (1994). Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913. doi: 10.1083/jcb.124.6.903
Berger, F., and Twell, D. (2011). Germline specification and function in plants. Annu. Rev. Plant Biol. 62, 461–484. doi: 10.1146/annurev-arplant-042110-103824
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287.e15–304.e15. doi: 10.1016/j.cell.2017.09.030
Broda, M., Millar, A. H., and Van Aken, O. (2018). Mitophagy: a mechanism for plant growth and survival. Trends Plant Sci. 23, 434–450. doi: 10.1016/j.tplants.2018.02.010
Chen, L., Su, Z. Z., Huang, L., Xia, F. N., Qi, H., Xie, L. J., et al. (2017). The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 8:1201. doi: 10.3389/fpls.2017.01201
Chhun, T., Aya, K., Asano, K., Yamamoto, E., Morinaka, Y., Watanabe, M., et al. (2007). Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19, 3876–3888. doi: 10.1105/tpc.107.054759
Corona Velazquez, A. F., and Jackson, W. T. (2018). So many roads: the multifaceted regulation of autophagy induction. Mol. Cell. Biol. 38, e00303–e00318. doi: 10.1128/MCB.00303-18
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Fujiki, Y., Yoshimoto, K., and Ohsumi, Y. (2007). An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143, 1132–1139. doi: 10.1104/pp.106.093864
Fullgrabe, J., Ghislat, G., Cho, D. H., and Rubinsztein, D. C. (2016). Transcriptional regulation of mammalian autophagy at a glance. J. Cell Sci. 129, 3059–3066. doi: 10.1242/jcs.188920
Hackenberg, D., and Twell, D. (2019). The evolution and patterning of male gametophyte development. Curr. Top. Dev. Biol. 131, 257–298. doi: 10.1016/bs.ctdb.2018.10.008
Hanada, T., Noda, N. N., Satomi, Y., Ichimura, Y., Fujioka, Y., Takao, T., et al. (2007). The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302. doi: 10.1074/jbc.C700195200
Hanamata, S., Kurusu, T., and Kuchitsu, K. (2014). Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 5:457. doi: 10.3389/fpls.2014.00457
Hanamata, S., Sawada, J., Toh, B., Ono, S., Ogawa, K., Fukunaga, T., et al. (2019). Monitoring autophagy in rice tapetal cells during pollen maturation. Plant Biotechnol. 36, 99–105. doi: 10.5511/plantbiotechnology.19.0417a
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Harding, T. M., Morano, K. A., Scott, S. V., and Klionsky, D. J. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602. doi: 10.1083/jcb.131.3.591
Harrison-Lowe, N. J., and Olsen, L. J. (2008). Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4, 339–348. doi: 10.4161/auto.5629
Higashiyama, T., and Takeuchi, H. (2015). The mechanism and key molecules involved in pollen tube guidance. Annu. Rev. Plant Biol. 66, 393–413. doi: 10.1146/annurev-arplant-043014-115635
Hisanaga, T., Yamaoka, S., Kawashima, T., Higo, A., Nakajima, K., Araki, T., et al. (2019). Building new insights in plant gametogenesis from an evolutionary perspective. Nat. Plants 5, 663–669. doi: 10.1038/s41477-019-0466-0
Huang, X., Zheng, C., Liu, F., Yang, C., Zheng, P., Lu, X., et al. (2019). Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31, 2973–2995. doi: 10.1105/tpc.19.00066
Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. doi: 10.1038/35044114
Ikawa, M., Inoue, N., Benham, A. M., and Okabe, M. (2010). Fertilization: a sperm’s journey to and interaction with the oocyte. J. Clin. Invest. 120, 984–994. doi: 10.1172/JCI41585
Ishizaki, K., Nishihama, R., Yamato, K. T., and Kohchi, T. (2016). Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 57, 262–270. doi: 10.1093/pcp/pcv097
Johansen, T., and Lamark, T. (2020). Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103. doi: 10.1016/j.jmb.2019.07.016
Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728. doi: 10.1093/emboj/19.21.5720
Kanazawa, T., Era, A., Minamino, N., Shikano, Y., Fujimoto, M., Uemura, T., et al. (2016). SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 57, 307–324. doi: 10.1093/pcp/pcv076
Kawanabe, T., Ariizumi, T., Kawai-Yamada, M., Uchimiya, H., and Toriyama, K. (2006). Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 47, 784–787. doi: 10.1093/pcp/pcj039
Kellner, R., De la Concepcion, J. C., Maqbool, A., Kamoun, S., and Dagdas, Y. F. (2017). ATG8 expansion: a driver of selective autophagy diversification? Trends Plant Sci. 22, 204–214. doi: 10.1016/j.tplants.2016.11.015
Khawar, M. B., Gao, H., and Li, W. (2019). Mechanism of acrosome biogenesis in mammals. Front. Cell Dev. Biol. 7:195. doi: 10.3389/fcell.2019.00195
Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530. doi: 10.1083/jcb.152.3.519
Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., et al. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. doi: 10.1083/jcb.147.2.435
Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., et al. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276. doi: 10.1083/jcb.151.2.263
Klionsky, D. J., Cregg, J. M., Dunn, W. A. Jr., Emr, S. D., Sakai, Y., Sandoval, I. V., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. doi: 10.1016/S1534-5807(03)00296-X
Ku, S., Yoon, H., Suh, H. S., and Chung, Y. Y. (2003). Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217, 559–565. doi: 10.1007/s00425-003-1030-7
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Kurusu, T., Koyano, T., Kitahata, N., Kojima, M., Hanamata, S., Sakakibara, H., et al. (2017). Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 12:e1365211. doi: 10.1080/15592324.2017.1365211
Kwon, S. I., Cho, H. J., Jung, J. H., Yoshimoto, K., Shirasu, K., and Park, O. K. (2010). The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 64, 151–164. doi: 10.1111/j.1365-313X.2010.04315.x
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X. X., Liang, W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Marshall, R. S., and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Minamino, N., Kanazawa, T., Era, A., Ebine, K., Nakano, A., and Ueda, T. (2018). RAB GTPases in the basal land plant Marchantia polymorpha. Plant Cell Physiol. 59, 845–856. doi: 10.1093/pcp/pcy027
Minamino, N., Kanazawa, T., Nishihama, R., Yamato, K. T., Ishizaki, K., Kohchi, T., et al. (2017). Dynamic reorganization of the endomembrane system during spermatogenesis in Marchantia polymorpha. J. Plant Res. 130, 433–441. doi: 10.1007/s10265-017-0909-5
Minina, E. A., Filonova, L. H., Fukada, K., Savenkov, E. I., Gogvadze, V., Clapham, D., et al. (2013). Autophagy and metacaspase determine the mode of cell death in plants. J. Cell Biol. 203, 917–927. doi: 10.1083/jcb.201307082
Mizushima, N. (2018). A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527. doi: 10.1038/s41556-018-0092-5
Mizushima, N. (2019). The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 63, 1–10. doi: 10.1016/j.ceb.2019.12.001
Mizushima, N., and Komatsu, M. (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. doi: 10.1016/j.cell.2011.10.026
Mizushima, N., Noda, T., and Ohsumi, Y. (1999). Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18, 3888–3896. doi: 10.1093/emboj/18.14.3888
Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M. D., et al. (1998). A protein conjugation system essential for autophagy. Nature 395, 395–398. doi: 10.1038/26506
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Moreno, R. D., and Alvarado, C. P. (2006). The mammalian acrosome as a secretory lysosome: new and old evidence. Mol. Reprod. Dev. 73, 1430–1434. doi: 10.1002/mrd.20581
Nakamura, S., and Yoshimori, T. (2017). New insights into autophagosome-lysosome fusion. J. Cell Sci. 130, 1209–1216. doi: 10.1242/jcs.196352
Nakatogawa, H., Suzuki, K., Kamada, Y., and Ohsumi, Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467. doi: 10.1038/nrm2708
Norizuki, T., Kanazawa, T., Minamino, N., Tsukaya, H., and Ueda, T. (2019). Marchantia polymorpha, a new model plant for autophagy studies. Front. Plant Sci. 10:935. doi: 10.3389/fpls.2019.00935
O’Donnell, L., Nicholls, P. K., O’Bryan, M. K., McLachlan, R. I., and Stanton, P. G. (2011). Spermiation: the process of sperm release. Spermatogenesis 1, 14–35. doi: 10.4161/spmg.1.1.14525
Qi, H., Li, J., Xia, F. N., Chen, J. Y., Lei, X., Han, M. Q., et al. (2020). Arabidopsis SINAT proteins control autophagy by mediating ubiquitylation and degradation of ATG13. Plant Cell 32, 263–284. doi: 10.1105/tpc.19.00413
Qin, G., Ma, Z., Zhang, L., Xing, S., Hou, X., Deng, J., et al. (2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17, 249–263. doi: 10.1038/cr.2007.7
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Renzaglia, K. S., and Garbary, D. J. (2001). Motile gametes of land plants: diversity, development, and evolution. Crit. Rev. Plant Sci. 20, 107–213. doi: 10.1080/20013591099209
Rose, T. L., Bonneau, L., Der, C., Marty-Mazars, D., and Marty, F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 98, 53–67. doi: 10.1042/BC20040516
Sanchez-Vera, V., Kenchappa, C. S., Landberg, K., Bressendorff, S., Schwarzbach, S., Martin, T., et al. (2017). Autophagy is required for gamete differentiation in the moss Physcomitrella patens. Autophagy 13, 1939–1951. doi: 10.1080/15548627.2017.1366406
Sera, Y., Hanamata, S., Sakamoto, S., Ono, S., Kaneko, K., Mitsui, Y., et al. (2019). Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci. Rep. 9:18544. doi: 10.1038/s41598-019-54361-1
Shang, Y., Wang, H., Jia, P., Zhao, H., Liu, C., Liu, W., et al. (2016). Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 12, 1575–1592. doi: 10.1080/15548627.2016.1192750
Shimamura, M. (2016). Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 57, 230–256. doi: 10.1093/pcp/pcv192
Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T., and Ohsumi, Y. (1999). Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234–5241. doi: 10.1093/emboj/18.19.5234
Slavikova, S., Shy, G., Yao, Y., Glozman, R., Levanony, H., Pietrokovski, S., et al. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56, 2839–2849. doi: 10.1093/jxb/eri276
Southworth, D., and Cresti, M. (1997). Comparison of flagellated and nonflagellated sperm in plants. Am. J. Bot. 84:1301. doi: 10.2307/2446056
Southworth, D., and Russell, S. (2001). “Male gametogenesis” in Current trends in the embryology of angiosperms. eds. S. S. Bhojwani and W.-Y. Soh (Dordrecht: Springer Netherlands), 1–16.
Stephani, M., and Dagdas, Y. (2019). Plant selective autophagy—still an uncharted territory with a lot of hidden gems. J. Mol. Biol. 432, 63–79. doi: 10.1016/j.jmb.2019.06.028
Strotbek, C., Krinninger, S., and Frank, W. (2013). The moss Physcomitrella patens: methods and tools from cultivation to targeted analysis of gene function. Int. J. Dev. Biol. 57, 553–564. doi: 10.1387/ijdb.130189wf
Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311. doi: 10.1083/jcb.119.2.301
Thumm, M., Egner, R., Koch, B., Schlumpberger, M., Straub, M., Veenhuis, M., et al. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280. doi: 10.1016/0014-5793(94)00672-5
Toure, A., Martinez, G., Kherraf, Z. E., Cazin, C., Beurois, J., Arnoult, C., et al. (2020). The genetic architecture of morphological abnormalities of the sperm tail. Hum. Genet. doi: 10.1007/s00439-020-02113-x [Epub ahead of print].
Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. doi: 10.1016/0014-5793(93)80398-E
Van Leene, J., Han, C., Gadeyne, A., Eeckhout, D., Matthijs, C., Cannoot, B., et al. (2019). Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 5, 316–327. doi: 10.1038/s41477-019-0378-z
Wang, P., Nolan, T. M., Yin, Y., and Bassham, D. C. (2019). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 16, 123–139. doi: 10.1080/15548627.2019.1598753
Wang, H., Wan, H., Li, X., Liu, W., Chen, Q., Wang, Y., et al. (2014). Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 24, 852–869. doi: 10.1038/cr.2014.70
Wang, W. Y., Zhang, L., Xing, S., Ma, Z., Liu, J., Gu, H., et al. (2012). Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J. Genet. Genomics 39, 81–92. doi: 10.1016/j.jgg.2012.01.002
Xu, N., Gao, X. Q., Zhao, X. Y., Zhu, D. Z., Zhou, L. Z., and Zhang, X. S. (2011). Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol. Biol. 77, 251–260. doi: 10.1007/s11103-011-9806-9
Yoshimoto, K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 53, 1355–1365. doi: 10.1093/pcp/pcs099
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Zhang, D. S., Liang, W. Q., Yuan, Z., Li, N., Shi, J., Wang, J., et al. (2008). Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 1, 599–610. doi: 10.1093/mp/ssn028
Zhao, P., Zhou, X. M., Zhao, L. L., Cheung, A. Y., and Sun, M. X. (2020). Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy 30, 1–13. doi: 10.1080/15548627.2020.1719722
Keywords: autophagy, male reproductive processes, tapetum, pollen germination, spermiogenesis
Citation: Norizuki T, Minamino N and Ueda T (2020) Role of Autophagy in Male Reproductive Processes in Land Plants. Front. Plant Sci. 11:756. doi: 10.3389/fpls.2020.00756
Received: 03 April 2020; Accepted: 12 May 2020;
Published: 17 June 2020.
Edited by:
Erika Isono, University of Konstanz, GermanyReviewed by:
Kohki Yoshimoto, Meiji University, JapanCopyright © 2020 Norizuki, Minamino and Ueda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Ueda, dHVlZGFAbmliYi5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.