- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
- 2College of Life Sciences, Northwest A&F University, Yangling, China
Rac/Rop proteins play important roles in the regulation of cell growth and plant defense responses. However, the function of Rac/Rop proteins in wheat remains largely unknown. In this study, a small G protein gene, designated as TaRac6, was characterized from wheat (Triticum aestivum) in response to Puccinia striiformis f. sp. tritici (Pst) and was found to be highly homologous to the Rac proteins identified in other plant species. Transient expression analyses of the TaRac6-GFP fusion protein in Nicotiana benthamiana leaves showed that TaRac6 was localized in the whole cell. Furthermore, transient expression of TaRac6 inhibited Bax-triggered plant cell death (PCD) in N. benthamiana. Transcript accumulation of TaRac6 was increased at 24 h post-inoculation (hpi) in the compatible interaction between wheat and Pst, while it was not induced in an incompatible interaction. More importantly, silencing of TaRac6 by virus induced gene silencing (VIGS) enhanced the resistance of wheat (Suwon 11) to Pst (CYR31) by producing fewer uredinia. Histological observations revealed that the hypha growth of Pst was markedly inhibited along with more H2O2 generated in the TaRac6-silenced leaves in response to Pst. Moreover, transcript levels of TaCAT were significantly down-regulated, while those of TaSOD and TaNOX were significantly up-regulated. These results suggest that TaRac6 functions as a potential susceptibility factor, which negatively regulate the reactive oxygen species (ROS) burst in the wheat–Pst interaction.
Introduction
Small GTP-binding proteins are proteins that have a molecular weight of 20–40 kd. They constitute a superfamily with five families—Ras, Ran, Rab, Rho, and Arf—which includes more than 100 members (Takai et al., 2001). The Rho family in animals is further divided into three subfamilies: Rho, Rac, and CDC42. However, because the Rho family in plants more closely resembles the Rac subfamily in animals, they are also called Rac-like or Rop-like (Rac/Rop) proteins (Winge et al., 2000).
The Rac/Rop protein family contains five highly conserved G-boxes and a C-terminal motif (Wennerberg et al., 2005). One G-box is the binding region of downstream effectors. The function of the other four G-boxes is to bind GTP/GDP and to hydrolyze GTP to GDP. The C-terminal motifs are related to the function of GTPase and the subcellular localization (Williams, 2003). Based on its C-terminal motifs, Rac/Rop proteins may be divided into two types (Winge et al., 2000): Type I have a conserved CaaL motif (a: aliphatic amino acid), while Type II lack the CaaL motif (Lavy et al., 2002) but have a cysteine domain to the membrane (Kawano et al., 2014). All type-I Rac/Rop proteins are prenylated (Lavy et al., 2002). Prenylation is required for membrane attachment and function of type I Rops, while type II Rops with the cysteine domain are attached to the plasma membrane by S-acylation. The prenylation of Rops determines their stable distribution between the plasma membrane and cytoplasm but has little effect on the dynamics of membrane interaction. In addition, the prenylation of type I Rops has only a small effect on ROP function. The mechanism of type II ROP S-acylation and membrane attachment is unique to plants and likely responsible for the viability of plants in the absence of CaaL prenylation activity. Type I ROPs affect the cell structure, primarily on the adaxial side, while type II ROPs induce a novel cell division phenotype (Sorek et al., 2011).
There are two states of Rac/Rop: the GTP-bound state Rac/Rop and GDP-bound state Rac/Rop, with the former being active and the latter being inactive (Vetter and Wittinghofer, 2001). GTPase-activating proteins (GAPs) reconvert the active Rac/Rop to an inactive state by promoting GTPase activity. The guanine nucleotide dissociation inhibitor (GDI) inhibits GDP-bound to GTP-bound, and guanine nucleotide exchange factors (GEFs) release GDP from Rac/Rop and bind Rac/Rop to GTP. The active Rac/Rop are able to interact with downstream effectors to function.
The Rac/Rop family is an important signal transduction regulator in plants, participating in various key life processes, including plant cell polarity, cell growth, morphological development, cytoplasmic division, signal transduction of hormones, and resistance to adversity (Schiene et al., 2000; Vernoud et al., 2003; Berken, 2006; Kawano et al., 2010a, b; Kawano and Shimamoto, 2013). However, the functioning of Rac/Rop family members in the interaction between plants and pathogens are still largely unknown. In rice (Oryza sativa), seven Rac/Rop family genes have been isolated (Miki et al., 2005). Among them, OsRac1 plays a positive role in blast resistance but overexpressed transgenic plants of OsRac4, OsRac5, and OsRac6 showed greater susceptibility to rice blast, whereas OsRac3 and OsRac7 may not participate in plant disease responses (Jung et al., 2006; Chen et al., 2010). OsRac1 contributes to disease resistance by regulating reactive oxygen species (ROS) and the biosynthesis of chitin and lignin (Wong et al., 2004; Kawasaki et al., 2006; Akamatsu et al., 2013). Additionally, several proteins, such as OsMAPK6, CERK1, GEF1, and SPL11, were found to be associated with Rac/Rop proteins participating in the interaction between plants and their pathogens (Lieberherr et al., 2005; Akamatsu et al., 2013; Liu et al., 2015). In barley (Hordeum vulgare), six Rac/Rop family genes were isolated, of which HvRacB was confirmed to be able to promote the susceptibility of barley to Blumeria graminis f. sp. hordei (Bgh) (Schultheiss et al., 2002, 2003). Furthermore, HvRacB was shown to affect barley’s resistance to Bgh by modulating the reorganization of actin (Opalski et al., 2005). Thus, different members of the same Rac/Rop family can play distinct roles in shaping how plants respond to pathogenic attacks and infection. Hence, it is of great significance to explore the mechanisms underpinning the Rac/Rop family genes’ involvement in plant responses to pathogens.
Wheat stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is among the most devastating diseases afflicting wheat (Chen et al., 2014), having become one the most important biotic problems threatening wheat production worldwide (Schwessinger, 2017). A better understanding of host–pathogen interactions will lay a theoretical foundation to formulate new strategies for the sustainable control of stripe rust. Analysis of cDNA library data revealed a Rac/Rop homologous gene in wheat that was up regulated in a compatible interaction (Ma et al., 2009). Yet, the function of this Rac/Rop gene in wheat’s response to Pst is still unknown. In this study, we report on this Rac/Rop family gene, designated as TaRac6, which was located to the whole cell and inhibited cell death induced by Bax. The function of TaRac6 was further analyzed using VIGS (virus induced gene silencing), which demonstrated that TaRac6 could regulate the resistance of wheat to Pst negatively by affecting the ROS burst. These results lay a foundation to explore the functioning of plant Rac/Rop proteins under pathogen infection.
Materials and Methods
Preparation of Plant Materials, Wheat Stripe Rust and Bacterial
Wheat and tobacco (Nicotiana benthamiana) plants were planted at 16°C and 23°C, respectively, under 60% relative humidity. The Pst isolates CYR31 and CYR23 were cultured on wheat cultivars ‘Suwon11’ and ‘Mingxian169,’ respectively (Kang et al., 2002). The Escherichia coli strain JM109 was cultured in Luria-Bertani (LB) culture medium overnight at 37°C in the dark. The Agrobacterium tumefaciens strain GV3101 was cultured in LB at 30°C in the dark for 1–2 days.
Sequence Analysis of TaRac6
The protein features were predicted in NCBI1. Protein molecular weight was predicted by ExPASy2. SignalP 4.13 was used to predict protein signal peptide. TMHMM Server v. 2.04 was used to predict transmembrane helices in proteins. PSORT5 was used to predict subcellular localization. cNLS Mapper (nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi#opennewwindow) was used to predict nuclear location signal. The software DNAMAN6.0 was used to align multiple sequences. The Phylogenetic tree was produced with the MEGA5 using the neighbor-joining approach. The fragment used for VIGS was aligned with the whole genome information of wheat6 and Pst7 to ensure sequence specificity.
Plasmid Construction
Primers used for plasmid construction are listed in Supplementary Table S1. The ORF sequence of TaRac6 was cloned into the pCAMBIA-1302 and pBinGFP2 vectors to verify its subcellular localization. pCAMBIA-1302 vector was used to express GFP at the C-terminus of TaRac6 (TaRac6-GFPC) and pBinGFP2 at the N-terminus (GFPN-TaRac6). To silence TaRac6, a specific 183-bp fragment containing a 13-bp untranslated region and a 170-bp fragment of a translated region was constructed into the BSMV-γ vector. To overexpress TaRac6 in N. benthamiana, the ORF sequence was inserted into the PVX vector pGR106.
RNA Extraction and Quantitative RT-PCR
The fresh urediniospores of CYR23 (incompatible interaction) and CYR31 (compatible interaction) were inoculated on the first leaves of 7-day-old wheat seedlings (Suwon11) with an inoculation needle. After inoculation, wheat seedlings were cultured in dark for 24 h with 100% humidity, and then transferred to a greenhouse at 15°C with a 16 h photoperiod. The leaves inoculated with CYR23 and CYR31 were sampled at 0, 12, 24, 48, 72, and 120 h post-inoculation (hpi), respectively. Total RNA from each sample was extracted using the MiniBEST Universal RNA Extraction Kit (TaKaRa, Kusatsu, Japan). The quality of obtained RNA was checked by electrophoresis. As described by Feng et al. (2011), first-strand cDNA was synthesized using Oligo dT Primer.
The primers used for the qRT-PCR can be found in Supplementary Table S1. Elongation factor 1α (EF-1α) of wheat was selected as the inner reference gene (Ling et al., 2007). The procedure of qRT-PCR followed that of Feng et al. (2011). The results were analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2012), with three independent biological replicates.
Transient Expression Assays for Subcellular Localization
GV3101 carrying pCAMBIA-1302-TaRac6-GFPC, pCAMBIA-1302-GFP, pBinGFP2-GFPN-TaRac6 or 35S-mCherry plasmids were cultured in LB (50 μg/mL kanamycin and 50 μg/mL rifampicin) for 1–2 days. The cells were collected and suspended as described by Zhao et al. (2018). GFP, GFPN-TaRac6, TaRac6-GFPC of A. tumefaciens were co-injected into leaves of tobacco plants 4–6 weeks old with mCherry, respectively. Two days later plant tissue samples were harvested. The GFP images were taken under a LSM510 Confocal Microscope (Zeiss, Germany) with 488 nm laser lines. The mCherry images were taken under a LSM510 Confocal Microscope (Zeiss, Germany) with 584 nm laser lines.
The expression of TaRac6-GFPC and GFPN-TaRac6 were further confirmed by western blot. The total proteins of injected tobacco leaves were extracted using the Native lysis buffer (Solarbio, Beijing, China). Specifically, 10 μL PMSF (100 mM) and 10 μL protease inhibitor cocktail (EDTA-Free, 100 × in DMSO) were added per ml of lysate. The extraction of total protein and the western blot procedure used are described in Zhao et al. (2018).
Inhibition Assay of PCD Induced by Bax
The pGR106-TaRac6, pGR106-eGFP (negative control), and pGR106-Avr1b (positive control, Dou et al., 2008) were respectively transformed into GV3101 (Hellens et al., 2000). Details on the treatment of the positive transformant can be found in Zhao et al. (2018). The A. tumefaciens cell suspensions of pGR106-TaRac6, pGR106-eGFP, and pGR106-Avr1b were injected separately into N. benthamiana leaves using sterile syringes. Then, 24 h later, the agrobacterium suspension containing the Bax gene was injected again at the same location. The tobacco leaves were sampled at 3 days post inoculation (dpi). The total RNA and cDNA of all samples at 3 dpi with Bax were obtained using the procedural methods described above. qRT-PCR was used to detect the transcription levels of N. benthamiana defense-related genes (PR1α, PR2, and PR5). The N. benthamiana housekeeping gene NbActin was selected as the inner reference gene. The results were analyzed by the 2–ΔΔCT method (Livak and Schmittgen, 2012), using three independent biological replicates. Symptoms were observed 5–7 days later. Leaves were decolorized by ethanol/glacial acetic acid (v/v, 1:1).
BSMV-Mediated TaRac6 Gene Silencing
The VIGS (virus induced gene silencing) system was implemented as described by Holzberg et al. (2002). ‘Suwon11’ was the cultivar used for the experiment. To silence TaRac6, BSMV: α + β + γ-TaRac6 was used to inoculate wheat seedlings. BSMV: α + β + γ was used as the control. About 30 seedlings were inoculated with each treatment. Ten days after virus inoculation, fresh CYR31 urediniospores were inoculated onto the fourth leaf. The wheat seedlings were cultured as described by Zhao et al. (2018). Their fourth leaves were sampled at 24 hpi and 48 hpi, to detect the gene-silencing efficiency and to observe the hyphae lengths and H2O2 accumulation at the histological level. The RNA of the fourth leaves inoculated with BSMV: α + β + γ and BSMV: α + β + γ-TaRac6 were isolated, and the qRT-PCR was used to assess the silencing efficiency and expression of the TaCAT, TaSOD and TaNOX genes. Cytological analyses of Pst growth and the host response in the control and TaRac6-silenced wheat plants were carried out as described by Zhao et al. (2018). Thirty-five infection sites from three leaves per treatment were used to calculate the hyphal length and H2O2 accumulation. Only the infected sites with substomatal vesicles under the stomata were considered to be successfully infected. Wheat germ agglutinin conjugated to Alexa Fluor 488 (Invitrogen, Carlsbad, CA, United States) was used to stain the Pst infection structures as described in Ayliffe et al. (2011). The length of hyphae and accumulation of H2O2 were each observed under an Olympus BX-51 microscope. The wheat phenotypes were observed 14 days after the Pst inoculation (dpi). For each treatment, six inoculated leaves were used to observe the phenotype. The phenotype was quantified by calculating the uredinium number within 1 cm2 area for one leaf. To avoid bias among the leaf samples, test points were randomly selected from the six treated plants. To estimate changes in the fungal biomass, DNA quantification of the single-copy target genes PsEF1 (from Pst) and TaEF1 (from wheat) was further measured using qRT-PCR as previously described (Panwar et al., 2013; Liu et al., 2016). Three independent biological replicates were performed.
Results
Sequence Analysis of TaRac6
An up-regulated transcript in the cDNA library of the compatible interaction between wheat and Pst was isolated (Ma et al., 2009), and designated as TaRac6 based on the Blast results in NCBI (see foot note 1). BlastN analyses in the Triticum aestivum genome sequence showed that there are three copies of this gene in the wheat genome, located on 6A, 6B, and 6D. The cDNA sequence of the three copies obtained in Suwon11 are highly similar (Supplementary Figure S1) and encodes the same proteins (Supplementary Figure S2). The three copies of TaRac6 encode the same 197 amino acids, which showed high homology with the Rac/Rop proteins from other plants (Supplementary Figure S3). TaRac6 has no signal peptide or transmembrane domain predicted by the SignalP 4.1 and TMHMM Server. It was predicted to be located in the plasma membrane, cytoplasm, and Chloroplast and has a nuclear localization signal predicted by the PSORT and cNLS Mapper. The protein features analysis indicated that TaRac6 contained a Rop-like domain (Supplementary Figure S4). Phylogenetic analysis indicated that TaRac6 and other plant Rho-related GTPases clustered together (Figure 1A). As a protein in the Rac/Rop GTPase family, TaRac6 contains five G boxes and a CxxL motif, which is the typical motif of the Rac/Rop protein belonging to Type I (Figure 1B).
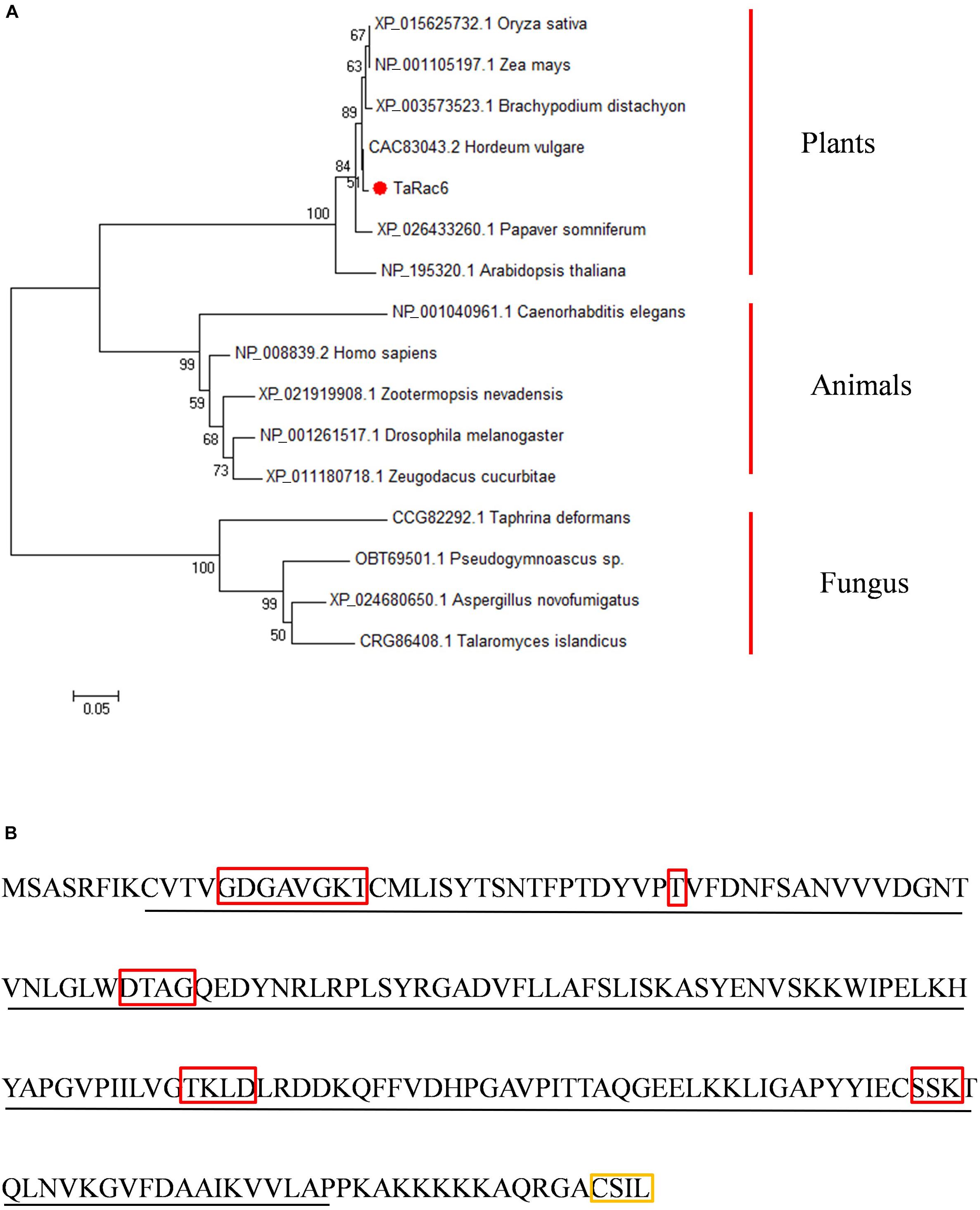
Figure 1. Amino acid sequences analysis and characterization. (A) The Phylogenetic tree of TaRac6 and RAC/ROP proteins from various eukaryotes was carried out using the MEGA5 by neighbor-joining approach. Branches are labeled with GenBank accession numbers and the corresponding name of each eukaryotic species. XP_015625732.1 (OsRac6), NP_001105197.1 (ZmRop9), XP_003573523.1 (BdRac6), CAC83043.2 (HvRopB), XP_026433260.1 (PsRac6), NP_195320.1 (AtRac6), NP_001040961.1 (CeRac2), NP_008839.2 (HsRac1), XP_021919908.1 (ZnRac1), NP_001261517.1 (DmRac2), XP_011180718.1 (ZcRac1), and CCG82292.1 (TdRho2). (B) Conserved domain of the TaRac6 protein. The red boxes indicate G1–G5 boxes. The yellow box indicates the CxxL motif. The sequence underlined with a black line indicates the Rho domain.
TaRac6 Is Localized in Plasma Membrane, Cytoplasm, and Nucleus
The control GFP, the TaRac6-GFPC, or the GFPN-TaRac6 were transiently expressed in tobacco leaves with 35S-mCherry, respectively. The fluorescence of TaRac6-GFPC was observed in the plasma membrane, cytoplasm, and nuclear region of N. benthamiana. Similarly, the signal of TaRac6-GFPN was also detected in the whole cell of N. benthamiana (Figure 2A). Western blot assays indicated that the TaRac6-GFPC and TaRac6-GFPN fusion proteins were successfully expressed in N. benthamiana (Figure 2B).
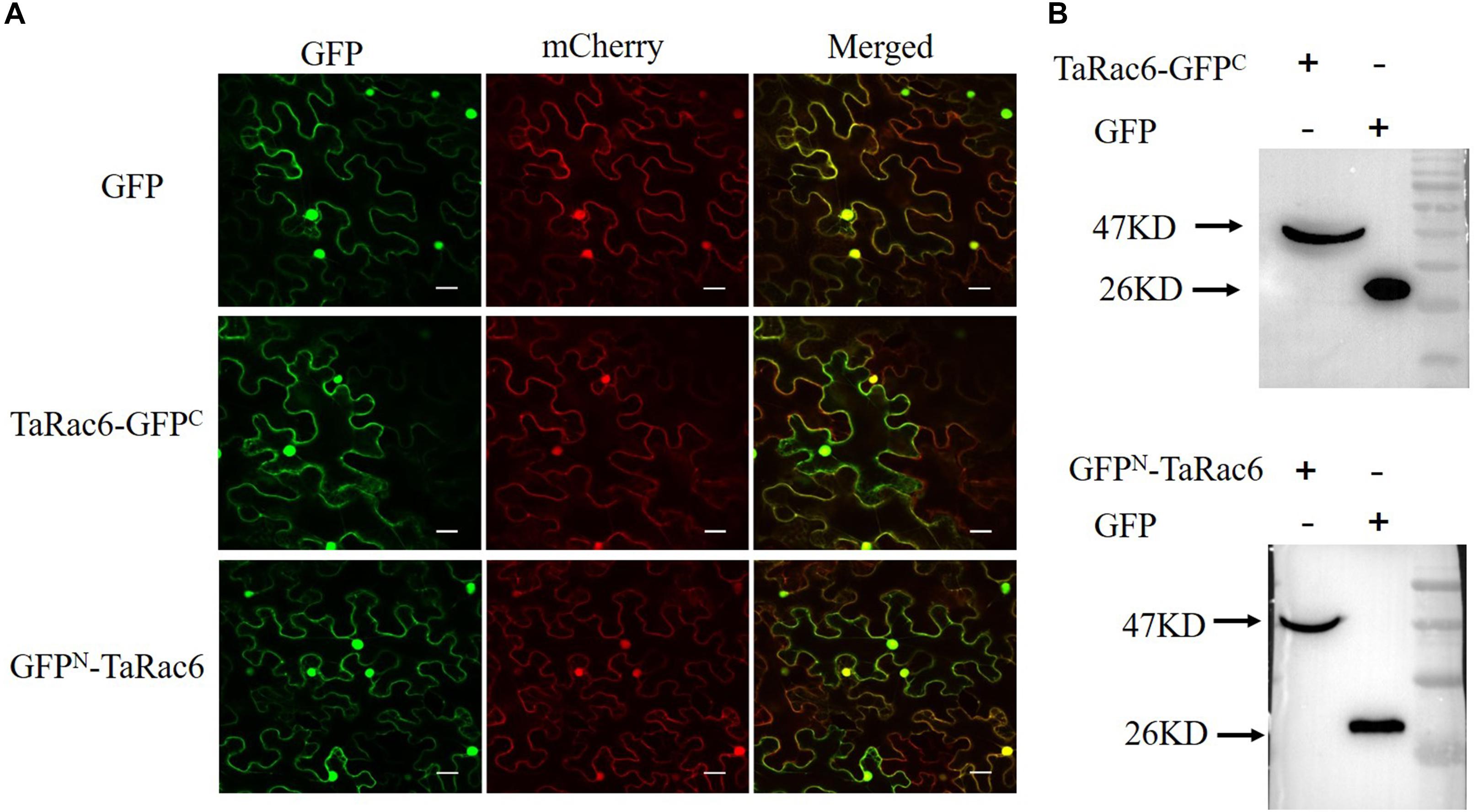
Figure 2. Subcellular localization of TaRac6 in tobacco leaves. (A) The green fluorescent protein (GFP), TaRac6-GFPC, and the GFPN-TaRac6 fusion protein were transiently expressed in Nicotiana benthamiana with mCherry, respectively. Signals of both were detected throughout the cell. (B) Western blot was used to analyze the GFP, TaRac6-GFPC, and GFPN-TaRac6 fusion protein. Bars = 20 μm.
Transient Expression of TaRac6 Inhibits Cell Death in Tobacco
Cell death is associated with plant resistance to invasion and spread by pathogens (Van Doorn et al., 2011). Bax is a death-promoting member of the Bcl-2 family of proteins which trigger cell death when expressed in plants (Lacomme and Santa Cruz, 1999). Bax-triggered cell death has similar physiological characteristics to plant hypersensitive responses (Lacomme and Santa Cruz, 1999). To determine whether TaRac6 could induce cell death or inhibit Bax-induced cell death to affect plant defense response, TaRac6 was transiently overexpressed in tobacco leaves with the Bax system. When Bax was expressed in tobacco leaves, the cells showed obvious necrosis. By contrast, cell death induced by Bax was starkly inhibited when TaRac6 and Bax were co-expressed. However, no cell death was observed in the leaves injected with Avr1b, which served as the positive control. The transcription levels of PR1α, PR2, and PR5 were reduced in tobacco leaves when Bax was co-expressed with pGR106-TaRac6 or pGR106-Avr1b, compared to pGR106-eGFP (Figure 3). These results indicated that TaRac6 could play an important role in inhibiting cell death.
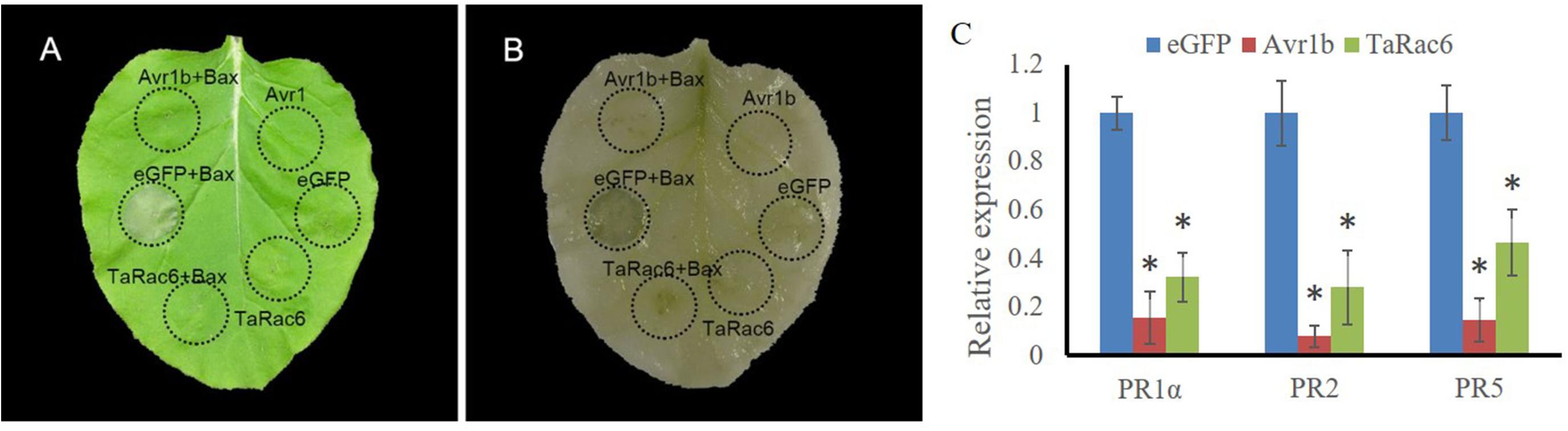
Figure 3. Transient expression of TaRac6 inhibited Bax-induced plant cell death in tobacco. (A) Tobacco leaves were injected with Agrobacterium tumefaciens cells containing pGR106-eGFP (negative control), pGR106-TaRac6, or pGR106-Avr1b (positive control); 24 h later, A. tumefaciens cells containing Bax were injected into the tobacco leaves. Photos were taken 5 days later. (B) Leaves were decolorized by ethanol/glacial acetic acid (1:1, v/v). (C) Transcription level changes of N. benthamiana defense-related genes PR1α, PR2, and PR5 in tobacco leaves when Bax was co-expressed with pGR106-eGFP, pGR106-TaRac6, or pGR106-Avr1b at 3dpi. NbActin was selected as the inner reference gene. Three independent biological replications were performed to calculate each of the mean values. Vertical bars represent the standard deviation. *P < 0.05.
TaRac6 Is Highly Expressed in the Compatible Wheat–Pst Interaction
To determine whether TaRac6 participate in wheat-Pst interactions, qRT-PCR was used to detect the expression of TaRac6 in the compatible and incompatible interaction of wheat–Pst. The expression of TaRac6 was up-regulated in the compatible interaction of wheat–Pst (CYR31). The transcript level of TaRac6 at 24 hpi was approximately 7.2-fold that of the control (0 hpi). However, the transcript level of TaRac6 was almost unchanged in the incompatible interaction (Figure 4). The result indicated that TaRac6 played an important role in the compatible interaction between wheat and Pst.
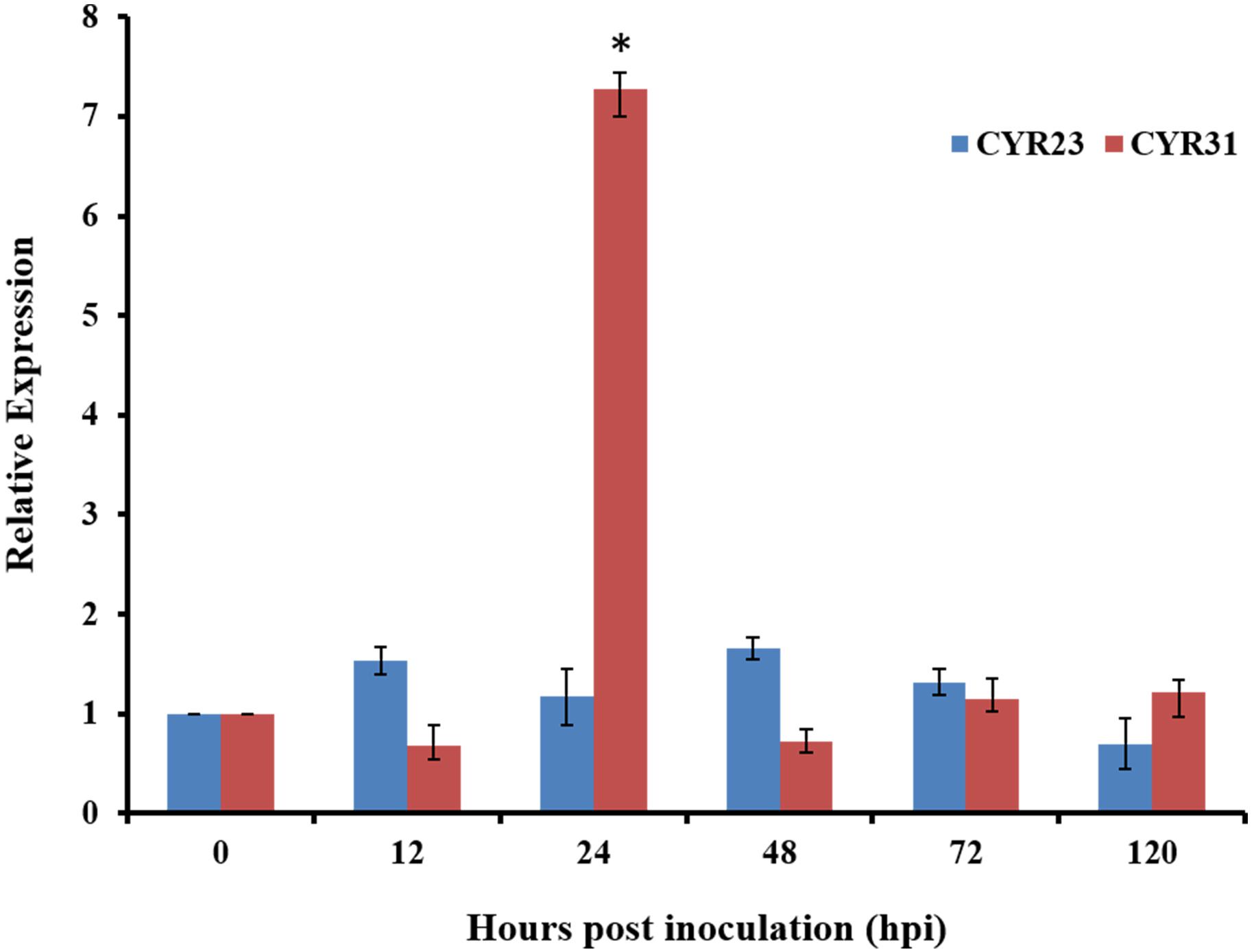
Figure 4. Transcript profiles of TaRac6 in wheat inoculated with Pst. Expression pattern analyses of TaRac6 during the different developing stages of the incompatible interaction (CYR23) and compatible interaction (CYR31) were calculated using the 2–ΔΔCT method. Three independent biological replications were performed to calculate each of the mean values. The wheat gene TaEF-1α was used to normalize the qRT-PCR data. Vertical bars represent the standard deviation. *P < 0.05.
Silencing BSMV-TaRac6 Increased the Resistance of Wheat
The BSMV-VIGS system was used to silence the expression of TaRac6 and thereby to characterize its function in the wheat–Pst interaction. Compared with the control leaves inoculated with FES-buffer, leaves inoculated with the vector of BSMV-γ and BSMV-TaRac6 displayed chlorotic striping at 10 days post-virus inoculation (dpvi) (Figures 5Aa–c). A bleaching phenotype was observed in PDS-silenced plants (Figures 5A,d), which suggested that the BSMV-VIGS system was effective. Race CYR31 was inoculated on the fourth leaf of wheat (Suwon 11), and 14 days later the leaves pre-inoculated with the FES-buffer and control leaves continued to display the typical compatible phenotype (Figures 5A,e,f); however, the susceptibility level of TaRac6-silenced leaves was significantly decreased (Figures 5A,g). The uredinia number in TaRac6-silenced leaves was reduced by approximately 30% relative to the control (BSMV-γ) (Figure 5B). Fungal DNA content was used as a proxy for Pst biomass in the leaves. The Pst DNA content was significantly reduced in TaRac6-silenced leaves indicating that fungal growth was inhibited (Figure 5C). To determine whether the phenotypic changes were caused by TaRac6’s silencing, qRT-PCR was used to detect the silencing efficiency compared with leaves inoculated with BSMV-γ. The transcript level of TaRac6 was reduced by 67% and 71% at 24 hpi and 48 hpi, respectively (Figure 5D). This result indicated that the TaRac6 gene had been successfully silenced. To determine whether the phenotypic changes between control and TaRac6-silenced plants are associated with fungal growth and development, Pst infection structures were stained and observed by microscopy. Histological analysis revealed a shorter hyphal length in TaRac6-silenced plants than that in control leaves (Figure 6). The accumulation of reactive oxygen species is considered to be the earliest inducing event in the plant-pathogens interaction, which controls and inhibits the growth of pathogens (Camejo et al., 2016). Therefore, to clarify whether the silencing of TaRac6 led to the changes in host resistance level to Pst, DAB staining was used to detect the accumulation of H2O2 in leaves, the H2O2 accumulation area was significantly increased in TaRac6-silenced plants when compared with the control (BSMV-γ) at both 24 hpi and 48 hpi (Figures 7A,B).
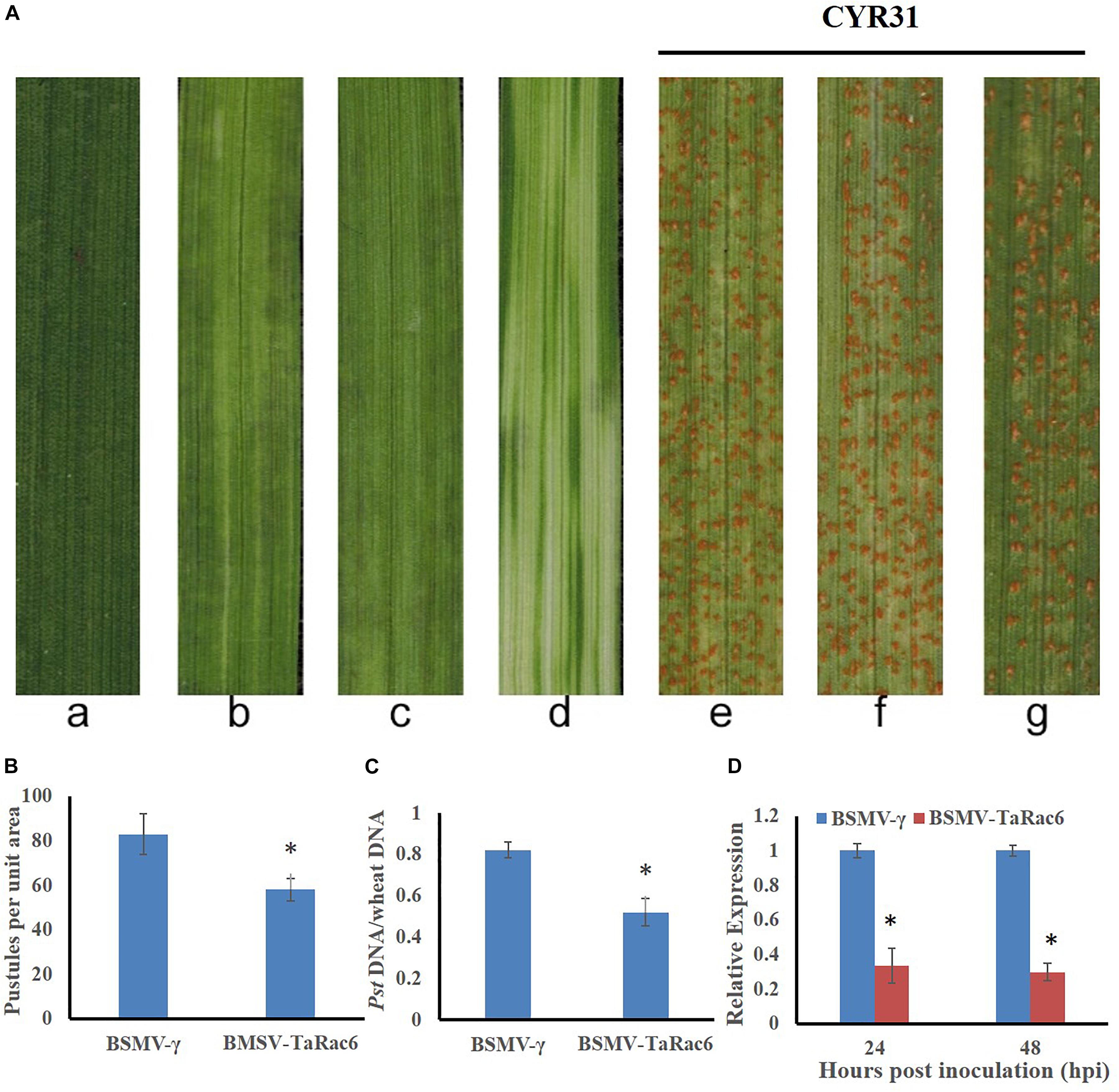
Figure 5. Functions of TaRac6 silencing by virus induced gene silencing (VIGS) in the compatible interaction between wheat (Suwon 11) and stripe rust pathogen (CYR31). (A) Phenotypes of the fourth leaves inoculated with the FES-buffer (a), BSMV-γ (b), BSMV-TaRac6(c), or BSMV-PDS (d) at 12 days post-virus treatment. Phenotypes of the fourth leaves inoculated with CYR31 at 14-day post inoculation that had been pre-inoculated with the FES-buffer (e), BSMV-γ (f), or BSMV-TaRac6 (g). (B) Quantification of the uredinium density at 14 dpi with Pst. Means and standard deviations were calculated from three independent replicates. Six treated leaves were used to calculate the uredinium number per replicate. ∗P < 0.05. (C) Fungal biomass was measured with total genomic DNA extracted from control and TaRac6-silenced wheat plants using qRT-PCR. Means and standard deviations were calculated from three independent replicates. Samples were taken at 14-day post inoculation with Pst. ∗P < 0.05. (D) Silencing efficiency in the TaRac6-knockdown plants inoculated with CYR31 were calculated using the 2–ΔΔCT method. Three independent biological replications were performed to calculate the standard deviations and mean values. The wheat gene TaEF-1α was used to normalize the qRT-PCR data. Vertical bars represent the standard deviation. ∗P < 0.05.
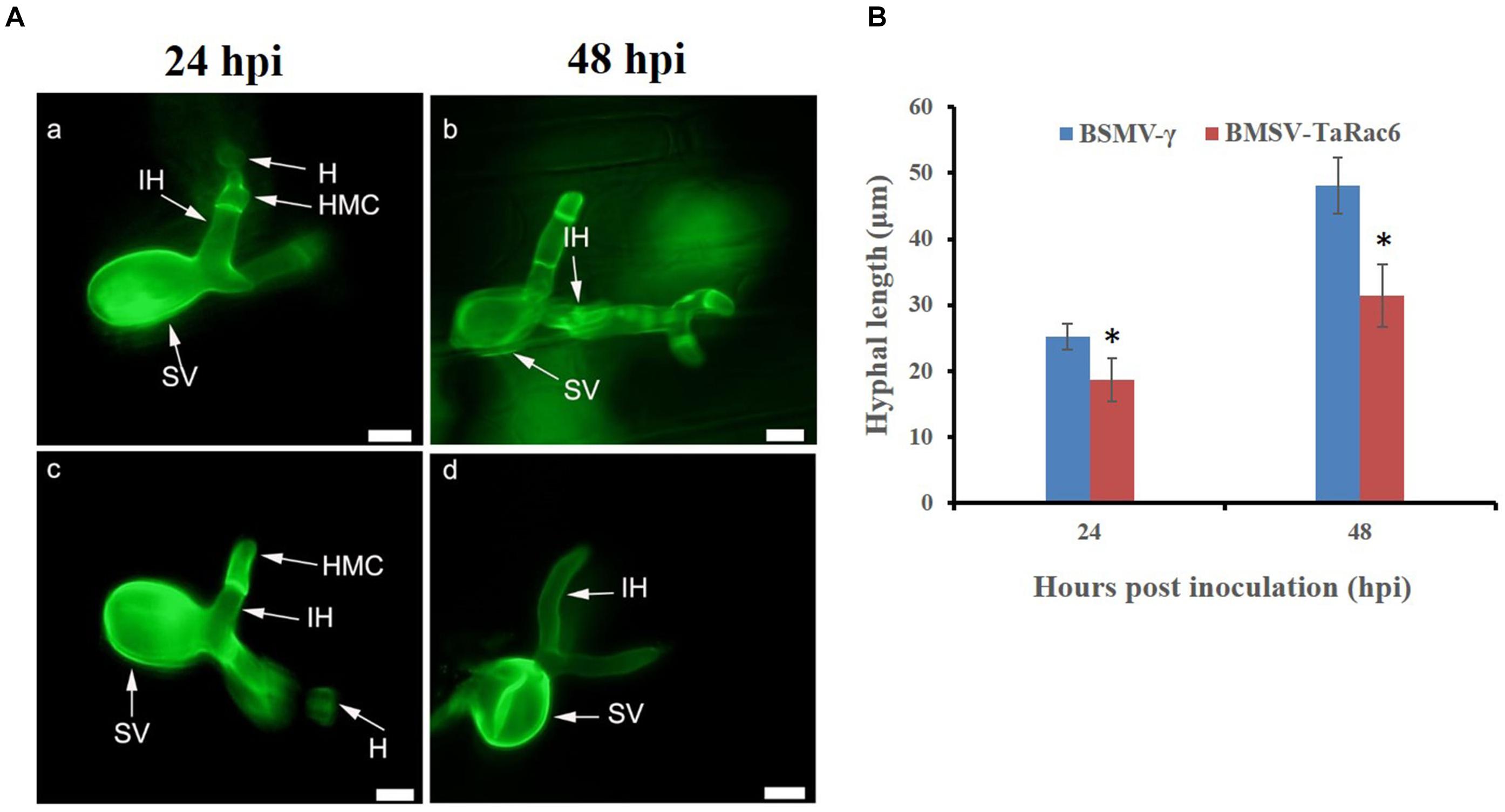
Figure 6. Histological observation of fungal growth in the TaRac6-knockdown plants inoculated with CYR31. (A) Microscopic observation of wheat pre-inoculated with BSMV-γ (a,b) and BSMV-TaRac6 (c,d). Wheat leaves inoculated with race CYR31 at 24 hpi and 48 hpi, respectively. The histological changes were stained with wheat germ agglutinin (WGA) and observed under a fluorescent microscope. H, haustoria; HMC, haustoria mother cell; IH, infection hypha; SV, substomatal vesicle. (B) Histological statistical analysis of hyphae lengths in the TaRac6-knockdown plants inoculated with CYR31 compared to BSMV-γ at 24 hpi and 48 hpi. The average distance was calculated from the substomatal vesicles to hyphal tips from 35 infection sites of three leaves. Means and standard deviations were calculated from three independent replicates. *P < 0.05. Bars = 10 μm.
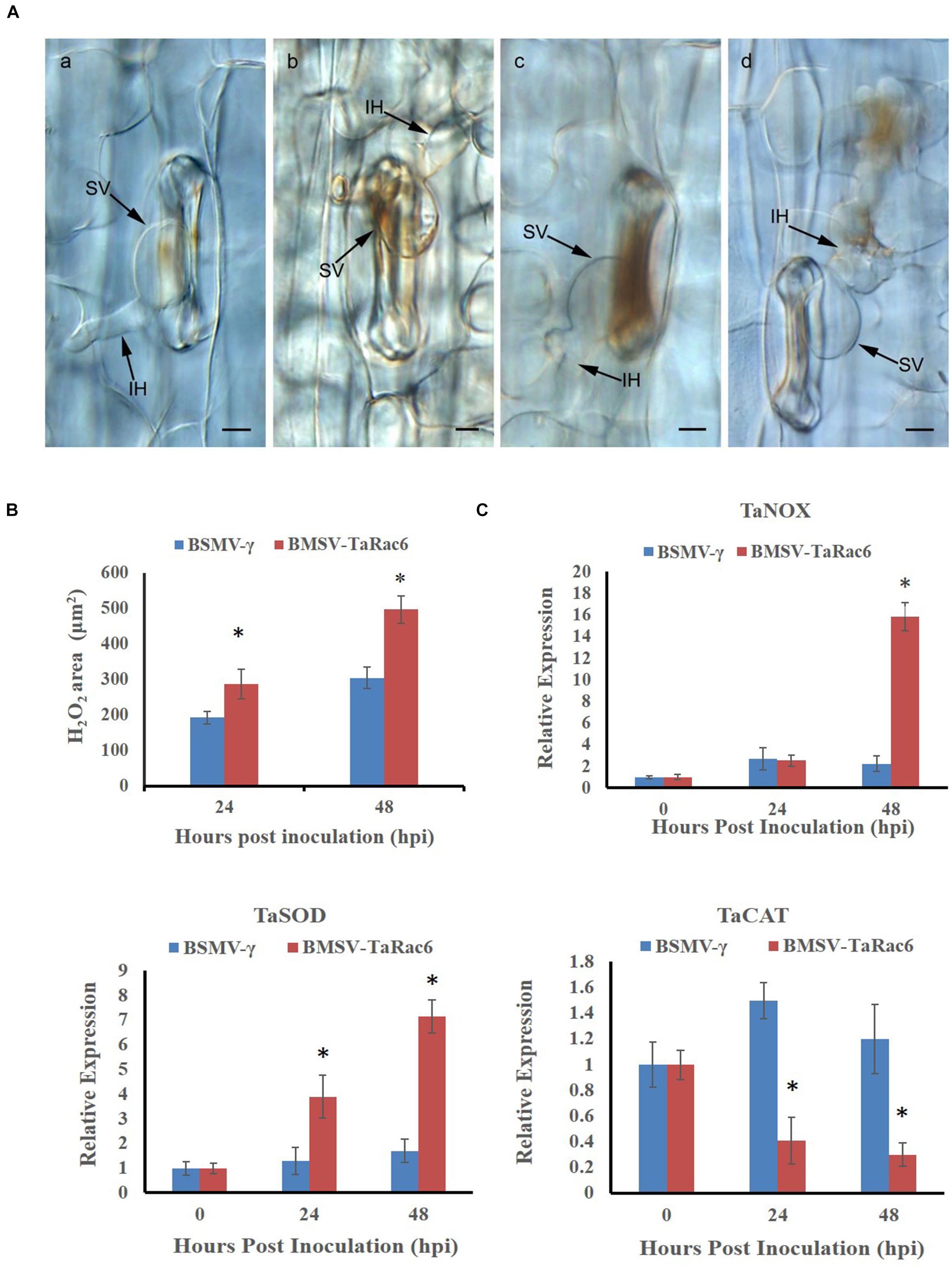
Figure 7. Histological observation of H2O2 accumulation by the 3, 3′-diaminobenzidine (DAB) staining. (A) (a,b) shows the histological of H2O2 accumulation in the control leaves at 24 hpi and 48 hpi, respectively; (c,d) shows the histological H2O2 accumulation in TaRac6-silenced leaves at 24 hpi and 48 hpi, respectively. IH, infection hypha; SV, substomatal vesicle. Bars = 10 μm. (B) Histological statistical analysis of H2O2 areas in the TaRac6-knockdown plants inoculated with CYR31 compared to BSMV-γ at 24 hpi or 48 hpi, respectively. The production of H2O2 was stained by the DAB and the average staining area was calculated at 35 infection sites. Statistical differences were assessed using Student’s t tests. *P < 0.05. (C) The qRT-PCR analysis of TaCAL, TaSOD and TaNOX in the TaRac6-knockdown plants inoculated with CYR31 using the 2–ΔΔCT method. TaCAT, catalase (AKP21073.1); TaSOD, superoxide dismutase (JX398977.1). TaNOX, NADPH oxidase RBOHa (BK010636.1). BSMV-γ pre-inoculation plants served as the control. Three independent biological replications were performed to calculate the standard deviations and mean values. The wheat gene TaEF-1α was used to normalize the qRT-PCR data. Vertical bars represent the standard deviation. *P < 0.05.
To investigate the expression of genes known to control ROS accumulation, we selected TaNOX, TaSOD, and TaCAT. NOX enzymes are known to generate H2O2 (Lambeth, 2004), SOD catalyzes the conversion of superoxide anion to O2 and H2O2 (Fukai and Ushio-Fukai, 2011), and CAT is the major H2O2 scavenging enzyme (Yang and Poovaiah, 2002). To explore the reason of the H2O2 accumulation in TaRac6-silenced plants, qRT-PCR was used to analyze the transcript level of these ROS-related genes in comparison with the control plants. In the TaRac6-silenced plants, the expression of TaSOD and TaNOX increased compared with that in control leaves (BSMV-γ), while the transcript level of TaCAT was down-regulated in the TaRac6-knockdown wheat (Figure 7C). Altogether, the above results indicated that TaRac6 might increase wheat susceptibility to Pst by inhibiting the production of H2O2.
Discussion
The Rac/Rop signaling pathway has a significant role in regulating many organism activities (Takai et al., 2001). The Rac/Rop proteins of rice, Medicago sativa, and barley (Hordeum vulgare) are known to be critical for the establishment of those plants’ defense systems (Schiene et al., 2000; Schultheiss et al., 2003; Kawasaki et al., 2006; Chen et al., 2010). As in animals, such proteins can regulate the production of H2O2 by activating the NADPH oxidase at the plasma membrane (Park et al., 2000; Jones et al., 2007). Yet the function and mechanisms of similar Rac/Rop members in the response of plants to their pathogens remains largely understudied.
In this study, TaRac6 was isolated and characterized from wheat plants, and the involvement of TaRac6 in wheat’s response to Pst was experimentally investigated. Sequence analysis showed that the TaRac6 contains a CxxL motif at its C-terminal. According to the C-terminal motif, Rac/Rop GTPases comprise two types. Type I proteins have a CxxL motif, while Type II possess a cysteine motif to anchor the membrane (Winge et al., 2000). On the basis of this protein domain, TaRac6 constituted a Type I protein, with further analysis showing that it occurred in the whole cell. In earlier work, Type II proteins of A. thaliana were found mainly localized at the plasma membrane (Lavy et al., 2002). Generally, however, unlike type II proteins, type I Rac/Rop proteins are more often detected in the whole cell, including its plasma membrane, cytoplasm, and nucleus (Chen et al., 2010).
Our phylogenetic analysis indicated that TaRac6 encodes nearly the same amino acids as HvRacB and OsRac6. HvRacB was identified as a negative regulator of barley defense to Bgh (Schultheiss et al., 2003), and the expression of constitutively activated HvRacB made barley more susceptible to Bgh (Schultheiss et al., 2005; Pathuri et al., 2008). The RNAi lines of HvRacB markedly induced barley’s resistance to Bgh by restricting the formation of haustoria (Hoefle et al., 2011). Finally, overexpression of OsRac6 enhanced susceptibility of rice to blast (Jung et al., 2006). Thus, in light of those findings, our results strongly suggest that TaRac6 is a potential susceptibility factor in wheat.
Bax-triggered plant cell death has similar physiological characteristics to plant HR. The cell death promoting function of Bax in plants correlated with accumulation of the defense-related protein PR1, suggesting that Bax activated an endogenous cell-death program in plants (Lacomme and Santa Cruz, 1999). This system has been used to successfully determine gene functioning as related to HR (Abramovitch et al., 2003; Wang et al., 2011). In our study, TaRac6 was able to inhibit the cell death induced by Bax and the transcription levels of PR1α, PR2, and PR5 were reduced in tobacco leaves when Bax was co-expressed with pGR106-TaRac6 or pGR106-Avr1b, compared to pGR106-eGFP, which indicated that TaRac6 could inhibit the Bax-triggered cell-death.
To define the potential role of TaRac6 in the wheat–Pst interaction, qRT-PCR was used to detect the transcript level of TaRac6. This showed that TaRac6 was highly induced in the compatible interaction, especially at 24 hpi, which is a critical time-point in the compatible interaction, marked by the formation of haustoria (Kang et al., 2002). The VIGS results also showed increased resistance when TaRac6 was silenced. According to the results of histological observation and expression analysis of ROS-related enzymes, we speculated that silencing TaRac6 drove an increase in H2O2 production. More ROS limited the normal expansion of hyphal at the infected sites, resulting in decreased sporulation. Thus, TaRac6 could affect the susceptibility of wheat to Pst by inhibiting the cell death triggered by the ROS burst. In rice and other plants, Rac proteins can regulate the production of H2O2 (Park et al., 2000; Jones et al., 2007).
As an important signaling molecule in plant cells, ROS is not only involved in programmed cell death, but also more importantly related to the formation of plant defense (Neill et al., 2002). In plants, Rac proteins have been shown to affect hydrogen peroxide production by regulating the activity of NADPH oxidase, which is necessary for the production of ROS (Bokoch and Diebold, 2002). Under hypoxic conditions, Rops are rapidly activated in Arabidopsis, resulting in ROP-dependent H2O2 production (Baxter-Burrell et al., 2002). In soybean cells, the Rac protein participated in the regulation of ROS production (Park et al., 2000). Overexpression of cotton GhRac13 promoted the production of H2O2 and then affected the formation of secondary walls of cotton cells (Potikha et al., 1999). It was speculated that Rac/Rops in dicotyledon may contribute to the ROS generation, however, the function of Rac/Rops to ROS generation in monocotyledon varies. In this study, TaRAC6 was demonstrated to play a negative role in wheat to Pst by inhibiting the H2O2 accumulation. Similarly, the mutant of OsRac1 promotes ROS accumulation and cell death to increase the rice resistance (Kawasaki et al., 1999; Ono et al., 2001). The production of H2O2 mediated by CA-OsRac1 could be inhibited by DPI (NADPH oxidase inhibitor), indicating that NADPH oxidase downstream of OsRac1 regulated the production of H2O2 (Kawasaki et al., 1999). OsRac1 could interact with the NLR protein Pit to generate ROS and HR. The results showed that OsRac1 is required for Pit-mediated resistance to rice blast fungus (Kawano et al., 2010b). However, Rac genes in Zea mays could induce the production of ROS (Hassanain et al., 2000). What is more, constitutively activated mutant HvRacB has no significant effect on ROS production; it partially inhibited F-actin polarization distribution to Bgh invasion sites to prevent the invasion (Opalski et al., 2005). Another study further inferred that HvRacB could activate a ROP-binding protein kinase HvRBK1, which functioned in basal resistance to powdery mildew by affecting microtubule organization (Huesmann et al., 2012). In Arabidopsis, overexpression of AtRac1 blocked the depolymerization of actin filaments during stomatal closure (Lemichez et al., 2001). In Barley, HvRacB was proven not only to regulate the reorganization of actin filaments to enhance susceptibility to Bgh, but also to affect stomata closure in ABA response (Schultheiss et al., 2005). Moreover, stomatal closure could be induced by ABA signaling in guard cells, which requires ROS formation to interact with Ca2+-channels (Kwak et al., 2006; Li et al., 2006). These results indicate that the plant Rac protein not only participates in regulating the production of ROS, but also affects the opening and closing of stoma. In this study, silencing of TaRac6 could reduce ROS accumulation of wheat to improve the infection of Pst. As we know, Pst infects wheat leaves through stoma (Wang et al., 2007). Whether TaRac6 plays a role in stomatal opening and closing and whether it affects the infection of Pst by affecting stomatal opening and closing needs be further explored.
Conclusion
In conclusion, TaRac6 was characterized in wheat’s response to Pst. As a type I Rac/Rop GTPase, TaRac6 was located in the whole cell, where it could inhibit the cell death induced by Bax. More importantly, TaRac6 plays a role in governing the level of wheat susceptibility to Pst by affecting the ROS burst. This finding is of great significance in advancing the full functional exploration of Rac/Rop in plant responses to pathogens, and it lays a foundation for breeding disease-resistance in wheat by modifying its susceptibility genes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ZK and XW designed the experiments. QZ performed most of the experiments, analyzed the data, and wrote the manuscript. XZ, RZ, ZW, WS, XW, and ZK assisted in the experiments and discussed the results. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the “111 Project” from the Ministry of Education of China (B07049) and the National Natural Science Foundation of China (No. 31620103913) to ZK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Hao Feng for providing suggestions on how to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00716/full#supplementary-material
FIGURE S1 | Multiple alignment of the cDNA sequence of the three copies of TaRac6 isolated from wheat cultivar “Suwon11.”
FIGURE S2 | Multiple alignment of the encoding sequence of the three copies of TaRac6.
FIGURE S3 | Multiple alignment of TaRac6 against homologous amino acid sequences from other plant species. The red boxes indicate the G1–G5 boxes. The yellow box indicates the CxxL motif. Ta, Triticum aestivum; Os, Oryza sativa; Zm, Zea mays; At, Arabidopsis thaliana; Zm, Zea mays; Bd, Brachypodium distachyon. Different colors indicate homology levels of amino acids. The pink shading indicates at least 75% amino acid homology. Light blue shading indicates at least 50% amino acid homology.
FIGURE S4 | The protein feature of TaRac6.
FIGURE S5 | The original image file for western blot of TaRac6.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov
- ^ https://web.expasy.org/compute_pi/
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://www.cbs.dtu.dk/services/TMHMM-2.0/#opennewwindow
- ^ https://wolfpsort.hgc.jp/
- ^ http://plants.ensembl.org/Triticum_aestivum/Info/Index
- ^ http://fungi.ensembl.org/Puccinia_graminis/Info/Index
References
Abramovitch, R. B., Kim, Y. J., Chen, S., Dickman, M. B., and Martin, G. B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22, 60–69. doi: 10.1093/emboj/cdg006
Akamatsu, A., Wong, H. L., Fujiwara, M., Okuda, J., Nishide, K., Uno, K., et al. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of Chitin-induced rice immunity. Cell Host Microbe. 13, 465–476. doi: 10.1016/j.chom.2013.03.007
Ayliffe, M., Devilla, R., Mago, R., White, R., Talbot, M., Pryor, A., et al. (2011). Nonhost resistance of rice to rust pathogens. Mol. Plant Microbe Interact. 24:1143. doi: 10.1094/MPMI-04-11-0100
Baxter-Burrell, A., Yang, Z., Springer, P., and Bailey-Serres, J. (2002). RopGAP4-dependent rop GTpase rheostat control of arabidopsis oxygen deprivation tolerance. Science 296, 2026–2028. doi: 10.1126/science.1071505
Berken, A. (2006). ROPs in the spotlight of plant signal transduction. Cell Mol. Life Sci. 63, 2446–2459. doi: 10.1007/s00018-006-6197-1
Bokoch, G., and Diebold, B. (2002). Current molecular models for NADPH oxidase regulation by Rac GTPase. Blood 100, 2692–2695. doi: 10.1182/blood-2002-04-1149
Camejo, D., Guzmán-Cedeño, Á, and Moreno, A. (2016). Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 103, 10–23. doi: 10.1016/j.plaphy.2016.02.035
Chen, L., Shiotani, K., Togashi, T., Miki, D., Aoyama, M., Wong, H. L., et al. (2010). Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 51, 585–595. doi: 10.1093/pcp/pcq024
Chen, W., Wellings, C., Chen, X., Kang, Z., and Liu, T. (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 15, 433–446. doi: 10.1111/mpp.12116
Dou, D., Kale, S., Wang, X., Jiang, R., Bruce, N., Arredondo, F., et al. (2008). RXLR-Mediated entry of Phytophthora sojae effector Avr1b into Soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947. doi: 10.1105/tpc.107.056093
Feng, H., Wang, X., Sun, Y., Wang, X., Chen, X., Guo, J., et al. (2011). Cloning and characterization of a calcium binding EF-hand protein gene TaCab1 from wheat and its expression in response to Puccinia striiformis f. sp. tritici and abiotic stresses. Mol. Biol. Rep. 38, 3857–3866. doi: 10.1007/s11033-010-0501-8
Fukai, T., and Ushio-Fukai, M. (2011). Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox. Signal. 15, 1583–1606. doi: 10.1089/ars.2011.3999
Hassanain, H., Sharma, Y., Moldovan, L., Khramtsov, V., Berliner, L., Duvick, J., et al. (2000). Plant Rac proteins induce superoxide production in mammalian cells. Biochem. Biophys. Res. Commun. 272, 783–788. doi: 10.1006/bbrc.2000.2791
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/a:1006496308160
Hoefle, C., Huesmann, C., Schultheiss, H., Börnke, F., Hensel, G., Kumlehn, J., et al. (2011). A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell 23, 2422–2439. doi: 10.1105/tpc.110.082131
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313x.2002.01291.x
Huesmann, C., Reiner, T., Hoefle, C., Preuss, J., Jurca, M., Domoki, M., et al. (2012). Barley ROP binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus. Plant Physiol. 159, 311–320. doi: 10.1104/pp.111.191940
Jones, M. A., Raymond, M. J., Yang, Z., and Smirnoff, N. (2007). NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58, 1261–1270. doi: 10.1093/jxb/erl279
Jung, Y. H., Agrawal, G. K., Rakwal, R., Kim, J. A., Lee, M. O., Choi, P. G., et al. (2006). Functional characterization of OsRacB GTPase-a potentially negative regulator of basal disease resistance in rice. Plant Physiol. Biochem. 44, 68–77. doi: 10.1016/j.plaphy.2005.12.001
Kang, Z., Huang, L., and Buchenauer, H. (2002). Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis. J. Plant Dis. Prot. 109, 25–37.
Kawano, Y., Akamatsu, A., Hayashi, K., Housen, Y., Okuda, J., Yao, A., et al. (2010a). Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7, 362–375. doi: 10.1016/j.chom.2010.04.010
Kawano, Y., Chen, L., and Shimamoto, K. (2010b). The function of Rac small GTPase and associated proteins in rice innate immunity. Rice 3, 112–121. doi: 10.1007/s12284-010-9049-4
Kawano, Y., Kaneko-Kawano, T., and Shimamoto, K. (2014). Rho family GTPase-dependent immunity in plants and animals. Front. Plant Sci. 5:522. doi: 10.3389/fpls.2014.00522
Kawano, Y., and Shimamoto, K. (2013). Early signaling network in rice PRR and R-mediated immunity. Curr. Opin. Plant Biol. 16, 496–504. doi: 10.1016/j.pbi.2013.07.004
Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., et al. (1999). The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. U.S.A. 96, 10922–10926. doi: 10.1073/pnas.96.19.10922
Kawasaki, T., Koita, H., Nakatsubo, T., Hasegawa, K., Wakabayashi, K., Takahashi, H., et al. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 230–235. doi: 10.1073/pnas.0509875103
Kwak, J., Nguyen, V., and Schroeder, J. (2006). The role of reactive oxygen species in hormonal responses. Plant Physiol. 141, 323–329. doi: 10.1104/pp.106.079004
Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Lambeth, J. D. (2004). NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4, 181–189. doi: 10.1038/nri1312
Lavy, M., Bracha-Drori, K., Sternberg, H., and Yalovsky, S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14, 2431–2450. doi: 10.1105/tpc.005561
Lemichez, E., Wu, Y., Sanchez, J. P., Mettouchi, A., Mathur, J., and Chua, N. H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15, 1808–1816. doi: 10.1101/gad.900401
Li, S., Assmann, S., and Albert, R. (2006). Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4:e312. doi: 10.1371/journal.pbio.0040312
Lieberherr, D., Thao, N. P., Nakashima, A., Umemura, K., Kawasaki, T., and Shimamoto, K. (2005). A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-Protein in rice. Plant Physiol. 138, 1644–1652. doi: 10.1104/pp.104.057414
Ling, P., Wang, M., Chen, X., and Campbell, K. G. (2007). Construction and characterization of a full-length cDNA library for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici). BMC Genomics 8:145. doi: 10.1186/1471-2164-8-145
Liu, J., Guan, T., Zheng, P., Chen, L., Yang, Y., Huai, B., et al. (2016). An extracellular Zn-only superoxide dismutase from Puccinia striiformis confers enhanced resistance to host-derived oxidative stress. Environ. Microbiol. 18:4118. doi: 10.1111/1462-2920.13451
Liu, J., Park, C., He, F., Nagano, M., Wang, M., Bellizzi, M., et al. (2015). The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog. 11:e1004629. doi: 10.1371/journal.ppat.1004629
Livak, K. J., and Schmittgen, T. D. (2012). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J., Huang, X., Wang, X., Chen, X., Qu, Z., Huang, L., et al. (2009). Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library. BMC Genomics 10:586. doi: 10.1186/1471-2164-10-586
Miki, D., Itoh, R., and Shimamoto, K. (2005). RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 138, 1903–1913. doi: 10.1104/pp.105.063933
Neill, S., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signaling. Curr. Opin. Plant Biol. 5, 388–395. doi: 10.1016/S1369-5266(02)00282-0
Ono, E., Wong, H., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. U.S.A. 98, 759–764. doi: 10.1073/pnas.021273498
Opalski, K. S., Schultheiss, H., Kogel, K. H., and Hückelhoven, R. (2005). The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J. 41, 291–303. doi: 10.1111/j.1365-313X.2004.02292.x
Panwar, V., Mccallum, B., and Bakkeren, G. (2013). Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73:521. doi: 10.1111/tpj.12047
Park, J., Choi, H. J., Lee, S., Lee, T., Yang, Z., and Lee, Y. (2000). Rac-related GTP-binding protein in elicitor-induced reactive oxygen generation by suspension-cultured soybean cells. Plant Physiol. 124, 725–732. doi: 10.1104/pp.124.2.725
Pathuri, I. P., Zellerhoff, N., Schaffrath, U., Hensel, G., Kumlehn, J., Kogel, K., et al. (2008). Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Rep. 27, 1877–1887. doi: 10.1007/s00299-008-0607-9
Potikha, T., Collins, C., Johnson, D., Delmer, D., and Levine, A. (1999). The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119, 849–858. doi: 10.1104/pp.119.3.849
Schiene, K., Pühler, A., and Niehaus, K. (2000). Transgenic tobacco plants that express an antisense construct derived from a Medicago sativa cDNA encoding a Rac-related small GTP-binding protein fail to develop necrotic lesions upon elicitor infiltration. Mol. Gen. Genet. 263, 761–770. doi: 10.1007/s004380000248
Schultheiss, H., Dechert, C., Kogel, K. H., and Hückelhoven, R. (2002). A small GTP binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 128, 1447–1454. doi: 10.1104/pp.010805
Schultheiss, H., Dechert, C., Kogel, K. H., and Hückelhoven, R. (2003). Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J. 36, 589–601. doi: 10.1046/j.1365-313x.2003.01905.x
Schultheiss, H., Hensel, G., Imani, J., Broeders, S., Sonnewald, U., Kogel, K. H., et al. (2005). Ectopic expression of constitutively activated RACB in barley enhances susceptibility to powdery mildew and abiotic stress. Plant Physiol. 139, 353–362. doi: 10.1104/pp.105.066613
Schwessinger, B. (2017). Fundamental wheat stripe rust research in the 21st century. New Phytol. 213, 1625–1631. doi: 10.1111/nph.14159
Sorek, N., Gutman, O., Bar, E., Abu-Abied, M., Feng, X., Runing, M., et al. (2011). Differential effects of prenylation and S-acylation on type I and II ROPS membrane interaction and function. Plant Physiol. 155, 706–720. doi: 10.1104/pp.110.166850
Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153–208. doi: 10.1152/physrev.2001.81.1.153
Van Doorn, W. G., Beers, E. P., Dangl, J. L., Tong, V. E. F., Gallois, P., Nishimura, I. H., et al. (2011). Morphological classification of plant cell deaths. Cell Death. Differ 18, 1241–1246. doi: 10.1038/cdd.2011.36
Vernoud, V., Horton, A. C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily in Arabidopsis. Plant Physiol. 131, 1191–1208. doi: 10.1104/pp.013052
Vetter, I. R., and Wittinghofer, A. (2001). The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304. doi: 10.1126/science.1062023
Wang, C., Huang, L., Buchenauer, H., Han, Q., Zhang, H., and Kang, Z. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2- and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 71, 230–239. doi: 10.1016/j.pmpp.2008.02.006
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional Programming and Functional Interactions within the Phytophthora sojae RXLR Effector Repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wennerberg, K., Rossman, K. L., and Der, C. J. (2005). The Ras superfamily at a glance. J. Cell Sci. 118, 843–846. doi: 10.1242/jcs.01660
Williams, C. L. (2003). The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell. Signal. 15, 1071–1080. doi: 10.1016/s0898-6568(03)00098-6
Winge, P., Brembu, T., Kristensen, R., and Bones, A. M. (2000). Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 156, 1959–1971.
Wong, H. L., Sakamoto, T., Kawasaki, T., Umemura, K., and Shimamoto, K. (2004). Down-regulation of Metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 135, 1447–1456. doi: 10.1104/pp.103.036384
Yang, T., and Poovaiah, B. W. (2002). Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc. Natl. Acad. Sci. U.S.A. 99, 4097–4102. doi: 10.1073/pnas.052564899
Keywords: TaRac6, wheat, Puccinia striiformis f. sp. tritici, reactive oxygen species, susceptibility factor
Citation: Zhang Q, Zhang X, Zhuang R, Wei Z, Shu W, Wang X and Kang Z (2020) TaRac6 Is a Potential Susceptibility Factor by Regulating the ROS Burst Negatively in the Wheat–Puccinia striiformis f. sp. tritici Interaction. Front. Plant Sci. 11:716. doi: 10.3389/fpls.2020.00716
Received: 24 October 2019; Accepted: 06 May 2020;
Published: 30 June 2020.
Edited by:
Silvia Proietti, University of Tuscia, ItalyReviewed by:
Oswaldo Valdes-Lopez, National Autonomous University of Mexico, MexicoMatthew Rouse, United States Department of Agriculture-Agricultural Research Service, United States
Steven Whitham, Iowa State University, United States
Copyright © 2020 Zhang, Zhang, Zhuang, Wei, Shu, Wang and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojie Wang, d2FuZ3hpYW9qaWVAbndzdWFmLmVkdS5jbg==; Zhensheng Kang, a2FuZ3pzQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Qiong Zhang
Qiong Zhang Xinmei Zhang
Xinmei Zhang Rui Zhuang
Rui Zhuang Zetong Wei
Zetong Wei Weixue Shu
Weixue Shu Xiaojie Wang
Xiaojie Wang Zhensheng Kang
Zhensheng Kang