
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 June 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00703
 Aditya Dutta1†‡
Aditya Dutta1†‡ Pratibha Choudhary1†‡
Pratibha Choudhary1†‡ Pallavi Gupta-Bouder1†
Pallavi Gupta-Bouder1† Snigdha Chatterjee1†
Snigdha Chatterjee1† Po-Pu Liu2†
Po-Pu Liu2† Daniel F. Klessig2
Daniel F. Klessig2 Ramesh Raina1*
Ramesh Raina1*Salicylic acid (SA) and reactive oxygen species (ROS) are known to be key modulators of plant defense. However, mechanisms of molecular signal perception and appropriate physiological responses to SA and ROS during biotic or abiotic stress are poorly understood. Here we report characterization of SMALL DEFENSE-ASSOCIATED PROTEIN 1 (SDA1), which modulates defense against bacterial pathogens and tolerance to oxidative stress. sda1 mutants are compromised in defense gene expression, SA accumulation, and defense against bacterial pathogens. External application of SA rescues compromised defense in sda1 mutants. sda1 mutants are also compromised in tolerance to ROS-generating chemicals. Overexpression of SDA1 leads to enhanced resistance against bacterial pathogens and tolerance to oxidative stress. These results suggest that SDA1 regulates plant immunity via the SA-mediated defense pathway and tolerance to oxidative stress. SDA1 encodes a novel small plant-specific protein containing a highly conserved seven amino acid (S/G)WA(D/E)QWD domain at the N-terminus that is critical for SDA1 function in pathogen defense and tolerance to oxidative stress. Taken together, our studies suggest that SDA1 plays a critical role in modulating both biotic and abiotic stresses in Arabidopsis (Arabidopsis thaliana) and appears to be a plant-specific stress responsive protein.
Plants frequently encounter pathogenic microbes, yet most of these interactions do not lead to disease. Generally, plants have two levels of defense against pathogens. The first level of defense is activated upon recognition of pathogen-associated molecular pattern (PAMP) that are highly conserved molecules such as flagellin and lipopolysaccharide, commonly present on the surface of several types of bacterial pathogens. Plants upon sensing these PAMPs with receptor kinases on the surface of the plasma membrane initiate PAMP-triggered immunity (PTI), which in turn leads to activation of several downstream defense-associated genes via MAP kinase signaling leading to basal resistance. However, basal defenses are not sufficient to prevent pathogen growth and disease progression. Over time, pathogens have developed mechanisms to suppress PTI by interfering with recognition at the plasma membrane or by secreting effector proteins (also known as virulence factors) directly into the plant cell. In response, plants have evolved resistance (R) proteins, which directly or indirectly detect the presence of effector proteins. This detection results in the induction of a more rapid and robust defense response known as R gene-mediated response or effector–triggered immunity (ETI) (Chisholm et al., 2006; Bent and Mackey, 2007; Zipfel, 2009; Dodds and Rathjen, 2010).
A frequent outcome of ETI is activation of the hypersensitive response (HR). HR is a form of programmed cell death (PCD) induced at the site of pathogen infection. HR is usually accompanied by the synthesis of antimicrobial compounds, strengthening of the cell walls, activation of several defense genes, production of reactive oxygen species (ROS), changes in ion fluxes and protein phosphorylation, expression of pathogenesis-related (PR) genes, and the eventual death of the infected cell and the pathogen (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Gust et al., 2012; Boyd et al., 2013; Wirthmueller et al., 2013; Mauch-Mani et al., 2017). In most cases, HR is followed by the activation of a long-lasting, broad-spectrum resistance termed as systemic acquired resistance (SAR), which is characterized by elevated levels of salicylic acid (SA) and expression of PR genes in the systemic tissues. In plants, SA is produced from chorismate primarily by two known biosynthetic pathways. In the first case, chorismate is converted to SA via phenylalanine, whereas in the second case, chorismate is converted to SA via isochorismate. In Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), and tomato (Solanum lycopersicum) the bulk of the pathogen-induced SA is synthesized via chorismate. In plants, SA is likely to be produced in chloroplasts and the bulk of it is stored in vacuoles as inactive SA O-β-glucoside (SAG), which can be converted back to active SA (Wildermuth, 2006; Chen et al., 2009; Vlot et al., 2009). Additionally, SA is also present as a conjugate with amino acids (Staswick et al., 2002, 2005; Okrent et al., 2009).
SA and its derivatives play a critical role in modulating local and systemic resistance against biotrophic and hemi-biotrophic microbial pathogens. Plants that are defective in biosynthesis and accumulation of SA or signaling mediated by SA are compromised in resistance against pathogens (Durrant and Dong, 2004; Vlot et al., 2009). For example, transgenic Arabidopsis and tobacco plants expressing a bacterial salicylate hydroxylase gene (NahG) are unable to accumulate SA and develop SAR, and therefore display enhanced susceptibility to virulent pathogens (Gaffney et al., 1993; Delaney et al., 1994). SA has also been shown to be a key modulator of PTI (Bigeard et al., 2015), given that disruption of SA-mediated signaling affects bacterial flagellin-mediated responses. Arabidopsis mutants such as eds1, pad4, sid2, sid1/eds5, gdg1/pbs3/win3, and eps1 are compromised in their ability to accumulate SA in response to pathogen infection and display enhanced susceptibility to pathogens (Glazebrook et al., 1996, 1997; Parker et al., 1996; Rogers and Ausubel, 1997; Nawrath and Metraux, 1999; Dewdney et al., 2000; Feys et al., 2001; Wildermuth et al., 2001; Nawrath et al., 2002; Jagadeeswaran et al., 2007; Lee et al., 2007; Nobuta et al., 2007; Zheng et al., 2009). Additionally, Arabidopsis npr1/nim1/sai1 mutants are defective in transmitting SA signals, and are also unable to develop SAR and display enhanced susceptibility to virulent pathogens (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997).
Here, we describe identification and characterization of the SMALL DEFENSE-ASSOCIATED PROTEIN 1 (SDA1) gene of Arabidopsis thaliana (hereafter Arabidopsis). We show that sda1 mutants are compromised in activation of Pathogenesis-Related (PR) genes, accumulation of SA, resistance against bacterial pathogen Pseudomonas syringae (P. syringae), and in tolerance against ROS-inducing agents. The SDA1 gene is predicted to encode a small 86 amino acid long plant-specific protein, which contains a highly conserved seven amino acid (S/G)WA(D/E)QWD domain at the N-terminus. Site-directed mutagenesis revealed the requirement of this domain in modulating bacterial defense and tolerance against oxidative stress in Arabidopsis. Our results show that SDA1 defines a novel class of small plant-specific proteins involved in modulating pathogen defense responses and oxidative stress tolerance.
Growth of bacterial pathogens, plant infections, and in planta pathogen growth assays were performed as described previously (Dutta et al., 2017). Plants were grown and treated with salicylic acid (SA) as described previously (Jagadeeswaran et al., 2007). For pathogen growth assay, bacterial pathogens were grown at 28°C on LB agar plates or in liquid medium supplemented with 100 μg/mL rifampicin and 50 μg/mL kanamycin. Bacterial cultures were prepared by resuspending the overnight grown cells in 10 mM MgCl2 to a titer of 5 × 105 cfu/mL (OD600; 1 OD600 = 109 cfu/mL). The bacterial suspension was pressure-infiltrated on the abaxial side of the leaves using a 1-mL syringe. Alternatively, whole rosettes were dipped into the bacterial suspension containing 0.02% Silwet L-77 (surfactant) and swirled for 30 s to evenly coat the leaves. For each genotype, eight plants were analyzed individually. Three leaf discs (0.5 cm in diameter) from each plant were collected at indicated times and placed in 1 mL of 10 mM MgCl2 in a microfuge tube. Tubes were shaken at 200 rpm for 1 h. Serial dilutions were plated on LB agar plates supplemented with appropriate antibiotics. Plates were incubated at 28°C for 2 days to determine the number of colony-forming units.
T-DNA insertion lines sda1-1 (SAIL_390_F08) and sda1-2 (GABI_283D01) were obtained from the Syngenta T-DNA collection (ABRC) (Alonso et al., 2003) and GABI-Kat collection (NASC), respectively. Genomic DNA flanking the T-DNA insertion site in sda1-1 was amplified using SDA1 gene-specific forward primer and T-DNA reverse primer as listed in Supplementary Table S1. Genomic DNA flanking the T-DNA insertion in sda1-2 was amplified using SDA1 gene specific forward primer and T-DNA reverse primer as listed in Supplementary Table S1. Amplified PCR products were sequenced to determine the site of insertion of the T-DNA. Homozygous null mutants were identified by northern blot analysis. To test the ability of SA to restore resistance to Pst DC3000 and Psm ES4326 in sda1 mutants, sda1 mutants and the corresponding Col-0 were sprayed with water or 1 mM SA 24 h prior to treatment with 5 × 105 cfu/mL Pst DC3000 and Psm ES4326 suspension prepared in 10mM MgSO4 solution, and growth of the pathogen was determined 3 days post-inoculation (dpi), and disease development was monitored for 5 dpi.
Full length SDA1 cDNA was amplified using Platinum Taq High Fidelity DNA Polymerase (Invitrogen, United States), with forward and reverse primers as listed in Supplementary Table S1, using U11998 SSP pUNI clone (ABRC) as template, and was cloned into pCR8/GW/TOPO TA cloning vector (Invitrogen, United States). Sequence of the amplified product was compared with the mRNA sequences available in the public databases to confirm that correct full-length cDNA was amplified. SDA1 cDNA was cloned in the gateway system pMDC32 vector (ABRC) (Curtis and Grossniklaus, 2003) under control of a strong 35S promoter to engineer SDA1 overexpression mutants. β-estradiol dose-dependent inducible overexpression of SDA1 was engineered by cloning of SDA1 cDNA in the gateway vector pMDC7 (Curtis and Grossniklaus, 2003). All of these constructs were then introduced into wildtype Col-0 plants via Agrobacterium-mediated floral dip transformation method (Clough and Bent, 1998). Transgenic plants were selected on solid Peters media or half-strength MS media with 1% sucrose and agar supplemented with 40 μg/mL hygromycin.
β-estradiol (Sigma, United States) treatment at indicated concentrations in 0.1% ethanol for inducible overexpression of SDA1, were syringe infiltrated into the rosette leaves of 4-week-old plants and tissue samples were harvested for RNA isolation at indicated times.
The SDA1 promoter was isolated as a 2 kb 5’-region upstream of the translation start site of SDA1 by PCR amplification of the genomic DNA using forward and reverse primers as listed in Supplementary Table S1. The amplified product was cloned into pCR8/GW/TOPO TA cloning vector (Invitrogen, United States), and sequenced to confirm the sequence of the amplified product. The promoter was cloned upstream of the GUS gene in the gateway system pMDC163 vector (ABRC) to create a transcriptional fusion SDA1 promoter::GUS. This construct was introduced into Col-0 plants via Agrobacterium-mediated transformation (Clough and Bent, 1998). Transgenic plants were selected on solid Peters media supplemented with 40 μg/mL hygromycin. Histochemical analyses for GUS activity were carried out in 6–8 independent transgenic lines for each tissue. Tissue samples were incubated overnight at 37°C in GUS assay buffer (10 mM phosphate buffer, pH 7; 0.5% Triton X-100; 2 mM potassium ferricyanide; 1 mg/mL X-Gluc) and were cleared in 70% ethanol for 2–3 days.
SDA1 cDNA-pCR8/GW/TOPO TA construct was used as a template to engineer four SDA1-mutant constructs (D8A, Q9R, V27A, and K52A) by using a site-directed mutagenesis kit (Quickchange Kit, Stratagene). Point mutations were confirmed by DNA-sequencing using oligonucleotide primers listed in Supplementary Table S1. These four mutants were further sub-cloned into pMDC32 (under cauliflower mosaic virus 35S promoter) and pMDC7 (under a β-estradiol-inducible G1090::XVE promoter) destination vectors by using LR Clonase enzyme (Life Technologies). These constructs were introduced into homozygous sda1-1 mutant plants via Agrobacterium-mediated transformation using the floral dip method. Transgenic plants were screened for hygromycin resistance. The transformants in following generations were established as homozygous lines for further analysis. Supplementary Table S1 lists all of the primers used to confirm the positive transformants.
Tissue samples were collected from soil-grown plants or seedlings grown on Peters/MS media plates (United Industries Corporation, United States) at the indicated time points, followed by flash-freezing in liquid nitrogen. Total RNA was isolated using TRIzol reagent according to manufacturer’s protocol (Invitrogen, Carlsbad, CA, United States). Northern blot analysis was performed as described previously (Dutta et al., 2017). RT-qPCR was used to check transcript levels of SDA1 and PR1 in selected plant lines. First-strand cDNA was synthesized using the SuperScript II RT Kit (Invitrogen). RT-qPCR reactions were performed in Bio-Rad CFX connect real time system using SYBR Green mix (Bio-Rad), as instructed by the manufacturer. All PCR reactions were performed in triplicates, normalized using internal control UBC, and averaged. For qPCR analysis. 2–ΔΔCT method (Livak and Schmittgen, 2001) was used, and fold enrichments for WT genes were set to one. The data represent mean ± SEM of three independent biological replicates. All primers used in RT-qPCR analysis are listed in Supplementary Table S1.
For SA analyses, rosette leaves of 4-week-old plants of indicated genotypes were inoculated with 5 × 107 cfu/mL Pst DC3000 and Pst DC3000 (avrRpm1). The tissue samples were collected at indicated time points and flash frozen. Free SA and SAG were extracted and quantified by HPLC as described previously (Jagadeeswaran et al., 2007).
Abiotic stress treatment was essentially performed as described previously (Dutta et al., 2015). For salinity stress treatment, 7-day old seedlings were grown on MS media plates and transferred to Whatman paper soaked in 200 mM sodium chloride for 12 h. For heat treatment, 7-day old seedlings were transferred to 37°C constant temperature room for 24 h. For cold treatment, 7-day old seedlings were transferred to 4°C constant temperature incubator for 24 h. For drought treatment, 4-week-old plants were deprived of water for 10 days. Tissue samples were harvested following the above mentioned abiotic stress treatments and used for RNA blot analysis.
Seed germination and root growth assays were essentially performed as described previously (Dutta et al., 2015). WT, sda1-1, sda1-2, pER8::SDA1 transgenic lines, and amino acid mutant seeds were surface sterilized using 20% bleach for 5 min, followed by washing three times with sterile water in a sterile hood environment. Sterilized seeds were plated in MS medium in square petri plates. ROS-inducing agents (Paraquat, H2O2, NaCl, and 3-AT) were added to the MS medium. pER8::SDA1 and amino acid mutant seeds were induced in the presence of 55 μm β-estradiol in MS media. These plates were assayed for percent-seed germination and compared with control MS medium (no treatment). Root growth was assayed with a digital camera (Nikon D70) and root length was calculated with ImageJ software. These experiments were repeated twice with similar results.
SDA1 gene (At1g19020) was identified in a large microarray screen as a gene that is strongly induced in response to bacterial pathogens (Appel et al., 2014). In order to further understand the role of SDA1 in pathogen defense, we analyzed accumulation of SDA1 transcript in wild-type Col-0 (WT) plants in response to bacterial pathogens. Rosette leaves of 4-week-old WT plants were inoculated with virulent bacterial pathogen P. syringae pv. tomato DC3000 (Pst DC3000) and avirulent bacterial pathogen Pst DC3000 expressing avrRpm1 [Pst DC3000 (avrRpm1)], and accumulation of SDA1 transcript was monitored by northern blot analysis. Control plants were infiltrated with 10mM MgSO4. In response to 10 mM MgSO4 (and distilled water), the SDA1 gene was induced transiently within 1 h (Figure 1A). This transient expression of SDA1 could be the result of mechanical stress resulting from syringe infiltration. To test this possibility, we infected plants by dip inoculation method and analyzed the kinetics of SDA1 expression. There was no detectable induction of SDA1 at 1 and 4 h post-dipping, confirming that induction observed at 1 and 4 h was the result of mechanical stress induced by syringe infiltration (Figure 1B). Induction in response to Pst DC3000 started at 8 hours post-inoculation (hpi) and peaked by 12 hpi (Figure 1A). However, in response to Pst DC3000 (avrRpm1), SDA1 transcript started to accumulate as early as four hpi and peaked by eight hpi. This pattern of SDA1 expression in response to bacterial pathogens is similar to that of the PR1 gene, a widely used defense marker gene. However, SDA1 expression precedes PR1 gene expression. Expression of SDA1 was also induced in response to another virulent bacterial pathogen P. syringae maculicola ES4326 (Psm) and avirulent bacterial pathogens expressing avrRpt2 or avrRps4, suggesting that SDA1 functions downstream of several R genes (Figure 1C).
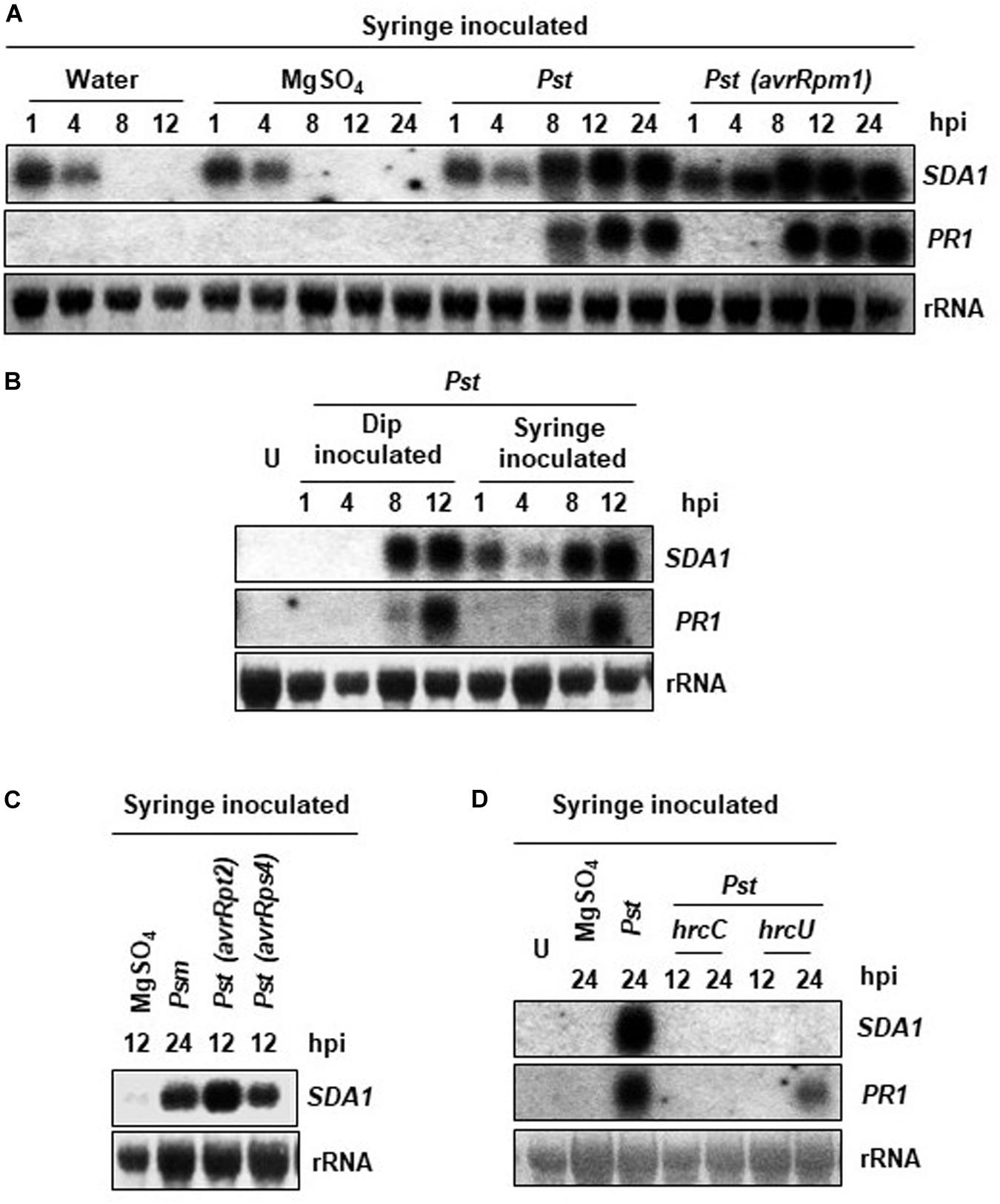
Figure 1. Expression analysis of the SDA1 gene in response to bacterial pathogens. (A–D) Leaves of 4-week-old WT Col-0 plants were either syringe or dip inoculated with the indicated pathogens at a titer of 5 × 107 cfu/mL in 10 mM MgSO4. Mock treatment was performed with 10 mM MgSO4. Tissue samples were harvested at the indicated hpi (hours post-inoculation). RNA was isolated and transcript levels were determined by RNA gel-blot analysis. Blots were stained with methylene blue to show the relative amounts of RNA in each lane (28S rRNA). Blots were probed with the SDA1 gene probe, stripped and re-probed with the PR1 gene probe. All experiments were repeated at least twice and similar results were obtained. Pst, Pst DC3000; Psm, Psm ES4326; avrRpm1, Pst DC3000 (avrRpm1); avrRps4, Pst DC3000 (avrRps4); hrcC, hrp mutant of Pst with a mutation in hrcC gene; hrcU, hrp mutant of Pst with mutation in hrcU gene; U, uninoculated.
Bacterial pathogens employ the type III secretion system (TTSS) to deliver the effector proteins into the host cells to cause disease. TTSS is encoded by the hrp genes; hrcC and hrcU mutants of Pst DC3000 are defective in delivering virulence factors into plant cells and therefore are non-pathogenic (Collmer et al., 2000). To determine if functional TTSS is required for expression of SDA1 during pathogen infection, we analyzed expression of SDA1 in response to hrcC and hrcU mutants. No SDA1 transcript was detected at 12 or 24 hpi by northern blot analysis suggesting that the transfer of effector proteins is required for expression of SDA1 (Figure 1D).
Defense against pathogens is usually associated with activation of PR genes. To determine if SDA1 is involved in activation of defense genes in response to pathogen infections, we determined expression of PR1 in sda1 mutants in response to bacterial pathogens. We identified two T-DNA insertion lines, sda1-1 (SAIL_390_F08, Syngenta Arabidopsis Insertion Library collection) (Sessions et al., 2002) and sda1-2 (GABI_283D01, GABI-Kat T-DNA insertional collection) (Rosso et al., 2003; Figure 2A). Sites of T-DNA insertion in these lines were confirmed by determining the sequence of the genomic DNA flanking the T-DNA. The T-DNA was inserted between nucleotides 85 and 86 of the open reading frame in sda1-1 and between nucleotides 149 and 150 of the open reading frame in sda1-2. To determine if these T-DNA insertion lines represent null alleles of SDA1, expression of SDA1 in response to Pst DC3000 was analyzed in these lines by northern blot analysis. No transcript was detected, confirming that these lines represent null alleles of SDA1 (Figures 2A,B). No visible phenotypic differences were observed in the sda1 mutants compared to the WT plants. Expression of PR genes in sda1 mutant plants was analyzed 8 and 12 hpi by northern blot analysis. As expected, the PR1 gene was strongly induced in the WT plants in response to Pst DC3000. However, expression in both sda1 mutants was significantly compromised and delayed. Similarly, expression of two other defense genes, PR2 and PR5 was also compromised in sda1 mutants (Figure 2B). Taken together, these results suggest that full pathogen induction of PR1, PR2, PR5, and likely other defense-related genes requires SDA1.
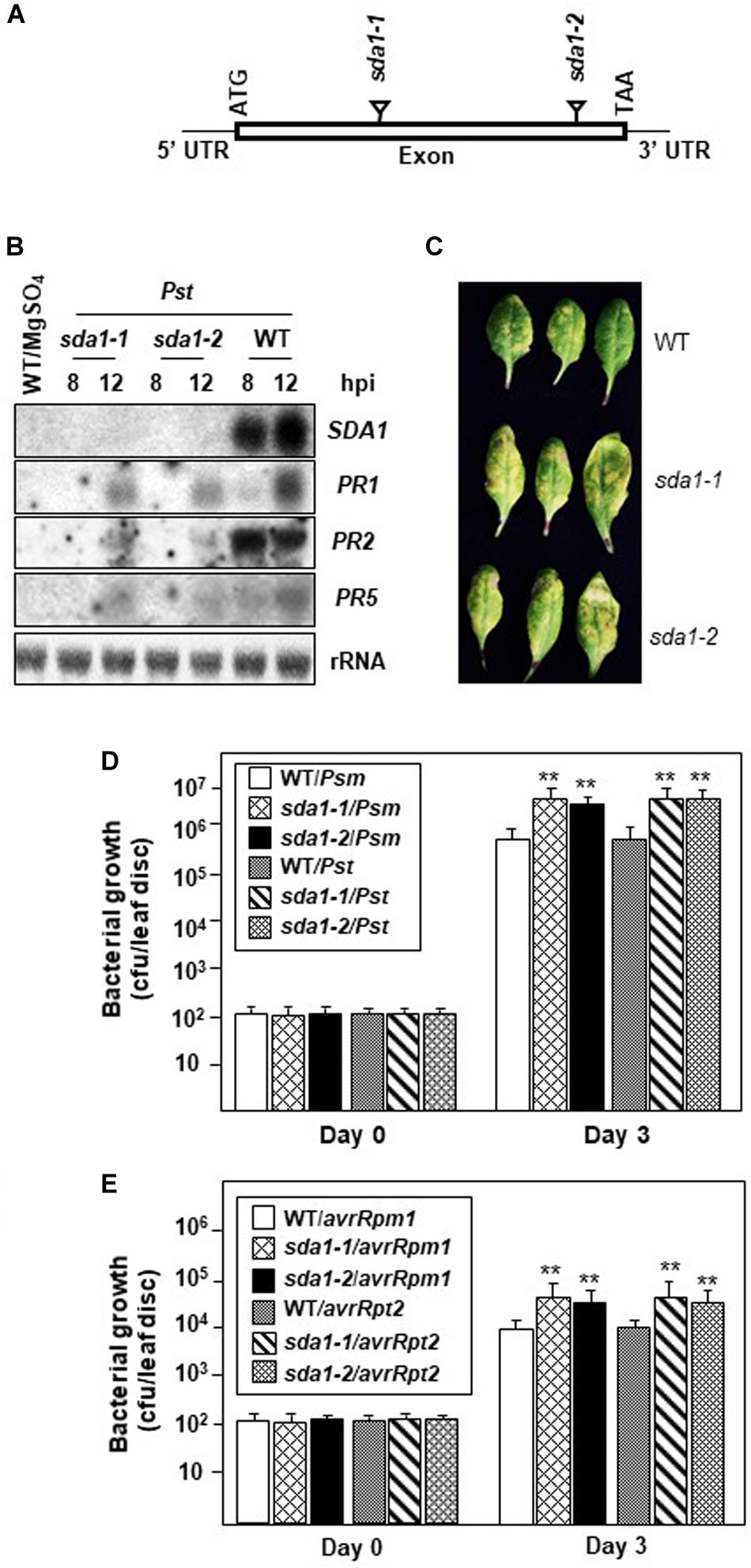
Figure 2. PR gene expression and resistance against bacterial pathogen is compromised in sda1 mutants. (A) Diagram depicting site of insertion of T-DNA in sda1-1 and sda1-2 mutants. The Rectangle indicates exon and black lines indicate 5’ and 3’ UTRs. (B) Total RNA was isolated from the leaves of 4-week-old plants of indicated genotypes inoculated with virulent bacterial pathogen Pst DC3000 (Pst) at a titer of 5 × 107 cfu/mL in 10 mM MgSO4. Tissue samples were harvested at indicated hpi. Transcript levels were determined by RNA gel-blot analysis. Blots were stained with methylene blue to show the relative amounts of RNA in each lane (28S rRNA). Blots were probed with SDA1 gene probe, stripped and re-probed with PR gene probes as indicated. (C) Rosette leaves of 4-week-old plants of indicated genotypes were inoculated with Pst DC3000 at a titer of 5 × 105 cfu/mL and leaves were photographed at 3 dpi (days post-inoculation). (D,E) Rosette leaves of 4-week-old plants of the indicated genotypes were inoculated with indicated pathogen at a titer of 5 × 105 cfu/mL. Eight plants for each genotype were analyzed individually. Data are reported as mean bacterial count (cfu per leaf disc) ± SD. Asterisks indicate statistically significant differences Statistical analysis was performed by one-way ANOVA followed by Dunnett’s test with reference to WT control. Asterisks indicate statistically significant differences (**p < 0.01). This experiment was repeated two more times with similar results. Psm, Psm ES4326; Pst, Pst DC3000; avrRpm1, Pst DC3000 expressing avrRpm1; avrRpt2, Pst DC3000 expressing avrRpt2.
To determine the role of SDA1 in defense against bacterial pathogens, we assessed the growth of virulent and avirulent bacterial pathogens in the sda1 knockout lines. Virulent Pst DC3000 was inoculated into the rosette leaves of sda1 and WT plants, and development of disease symptoms and growth of the pathogen was monitored for 3 days post-inoculation (dpi). sda1 mutants developed more disease symptoms and supported ∼5–7-fold more pathogen growth compared to the wild-type. Similar results were obtained with another virulent pathogen Pseudomonas syringae pv. maculicola ES4326 (Psm) (Figures 2C,D). These results suggest that SDA1 is required for basal resistance against a variety of virulent bacterial pathogens.
To test whether sda1 mutants are compromised in R gene-mediated resistance (ETI), rosette leaves of 4-week-old sda1 mutants were inoculated with Pst DC3000 expressing avrRpm1 [Pst DC3000 (avrRpm1)] or avrRpt2 [Pst DC3000 (avrRpt2)], and pathogen growth was determined 3 dpi. Both avirulent pathogens grew ∼3–4-fold more in the sda1 mutants compared to the WT (Figure 2E). These results suggest that SDA1 also contributes to R gene-mediated resistance.
To further determine the role of SDA1 in pathogen defense, we constructed transgenic plants expressing SDA1 under the control of a β-estradiol-inducible XVE system (pER8::SDA1) in the pMDC7 vector (Zuo et al., 2000; Curtis and Grossniklaus, 2003). Upon treatment with β-estradiol, these lines accumulated SDA1 transcript in an β-estradiol concentration-dependent manner (Supplementary Figure S1A). The resulting elevated levels of SDA1 transcript correlated with increasing PR gene expression (Supplementary Figure S1A). Together, these results suggest that SDA1 positively modulates expression of PR genes and likely other defense genes. To assess whether overexpression of SDA1 would confer enhanced resistance against virulent bacterial pathogens, growth of Pst DC3000 in these lines was tested. Pathogen growth was reduced ~5–7-fold in the SDA1 inducible overexpression lines compared to that in the WT plants (Supplementary Figure S1B).
To determine the epistatic relationship of SDA1 with other components of the defense-signaling network, we analyzed expression of SDA1 in response to Pst DC3000 in several Arabidopsis mutants compromised in defense against pathogens. Rosette leaves of 4-week-old WT and mutant plants were inoculated with Pst DC3000 and expression of SDA1 was determined 24 hpi. Expression of SDA1 was compromised in pad4 (Glazebrook et al., 1996), eds1 (Parker et al., 1996), pbs3/gdg1/win3 (Jagadeeswaran et al., 2007; Lee et al., 2007; Nobuta et al., 2007), and ndr1 (Century et al., 1995) mutants (Figure 3A). In contrast, mutation in sid2 (Wildermuth et al., 2001) and npr1 (Cao et al., 1994) had negligible effect on SDA1 expression. Taken together, these results suggest that SDA1 is downstream of PAD4, EDS1, GDG1 and NDR1 but is either upstream of SID2 and NPR1 or is in another signaling pathway. However, epistatic analysis across mutants of each indicated gene (or groups of genes) will be required to establish these relationships. Furthermore, expression of SDA1 was blocked in NahG plants, suggesting that SA is required for expression of SDA1. To test if SA is sufficient to induce the expression of SDA1, SA was applied externally to the WT plants. Expression of SDA1 was induced within 6 h, and peaked by 12 h after SA application, suggesting that SA alone is sufficient to induce expression of SDA1 (Figure 3B). Expression of SDA1 was also assessed in these mutants in response to Pst DC3000 (avrRpm1) but no change in SDA1 expression was detected (Figure 3C). These results suggest that R gene-mediated induction of SDA1 is independent of the SA-mediated defense pathway or that in response to avirulent pathogens signals other than SA may be involved in SDA1 activation. Alternatively, the threshold level of SA-mediated signaling required to activate SDA1 expression may be sufficiently low such that the robust R gene-mediated signal, despite being compromised by the various mutants, is still above this threshold.
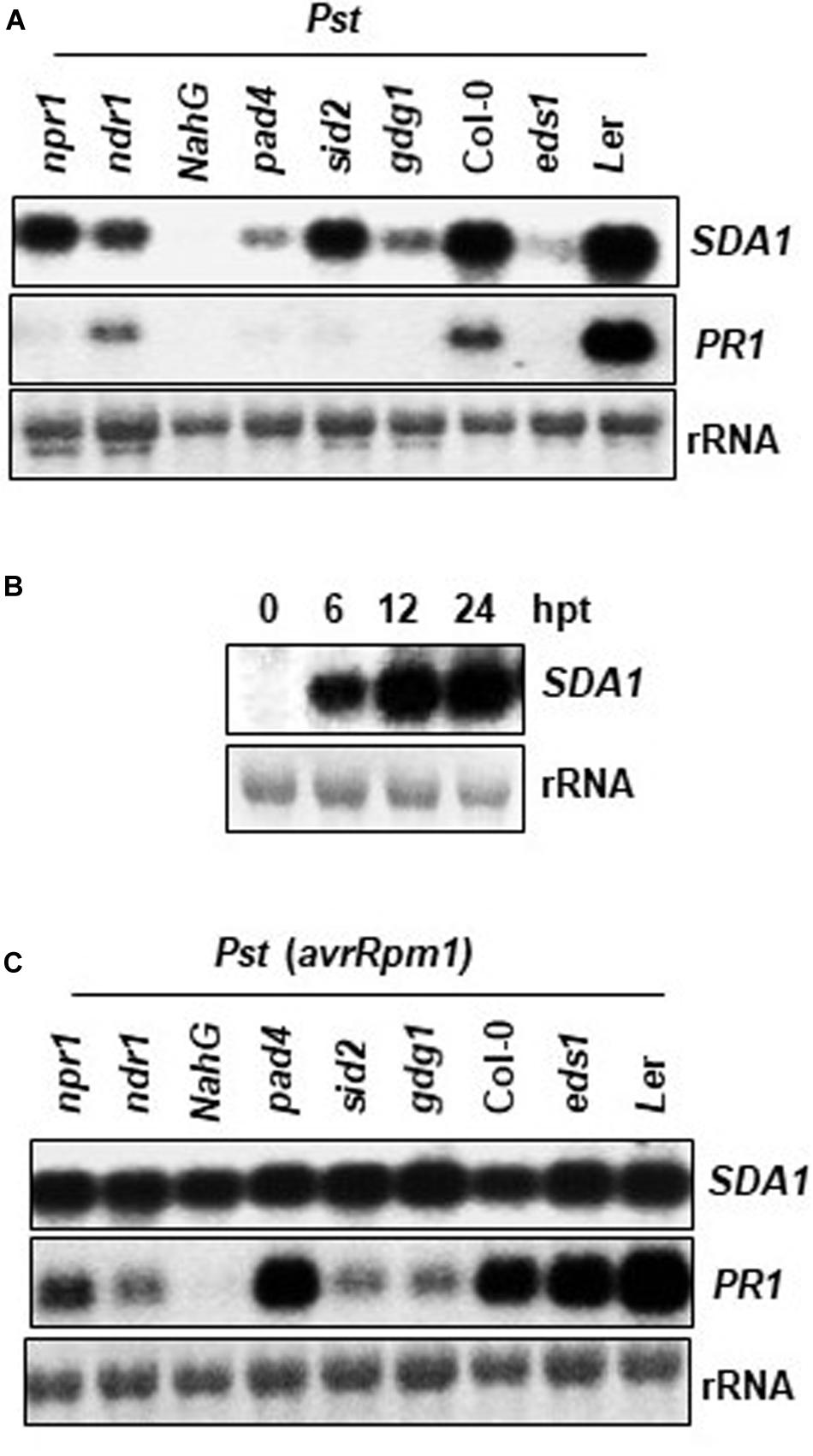
Figure 3. Expression analysis of SDA1 in defense mutants and in response to SA. (A,C) Mutant and wild type plants (4-week-old) were treated with Pst DC3000 (A) and Pst DC3000 (avrRpm1) (C) at a concentration of 5 × 107 cfu/mL and tissue samples were harvested at 24 hpi (hours post-inoculation). Northern gel blot analysis was employed to determine transcript levels. Blots were stained with methylene blue to show the relative amounts of RNA in each lane (rRNA). Blots were probed with SDA1 gene probe, stripped and re-probed with PR1 gene probe. All mutants are in the Col-0 background, except eds1, which is in the Ler background. (B) Four-week-old wild-type (Col-0) plants were sprayed with 1 mM SA. Rosette leaves were collected at the indicated hours post-treatment (hpt) for RNA isolation. Transcript levels were determined by RNA gel-blot analysis. Blots were stained with methylene blue to show the relative amounts of RNA in each lane (28S rRNA). Blots were probed with the SDA1 gene probe. All experiments were repeated at least twice and similar results were obtained. Pst, Pst DC3000; Pst (avrRpm1), Pst DC3000 (avrRpm1).
Resistance to biotrophic bacterial pathogens is typically dependent on SA. Mutants compromised in SA biosynthesis (e.g., sid2), metabolism (e.g., pbs3/gdg1/win3), or signaling (e.g., npr1) are compromised in resistance against a variety of bacterial, fungal and viral pathogens. To determine if sda1 mutants are compromised in SA signaling, PR1 expression was assessed in sda1 mutants in response to external application of SA. PR1 expression was induced to similar levels in the sda1 mutant and the WT plants (Supplementary Figure S2A). Additionally, an application of SA also rescued resistance against Pst DC3000 (Supplementary Figure S2B) in sda1 mutants. Together these results demonstrate that SA-mediated signaling is not compromised in the sda1 mutants.
The above results suggest that SA perception and signaling is not impaired in sda1 mutants. To determine if SDA1 has any role to play in the accumulation of SA in response to pathogens, we determined levels of free SA and SA glucoside (SAG) in sda1 mutants in response to Pst DC3000 (Figures 4A,B) and Pst DC3000 (avrRpm1) (Figures 4C,D). Accumulation of free SA and SAG in response to Pst DC3000 was reduced ∼2–3-fold in sda1 mutants compared to the WT plants (Figure 4). The reduced levels of SA and SAG accumulation in response to infection by Pst DC3000 might explain the enhanced susceptibility of sda1 mutants to virulent P. syringae.
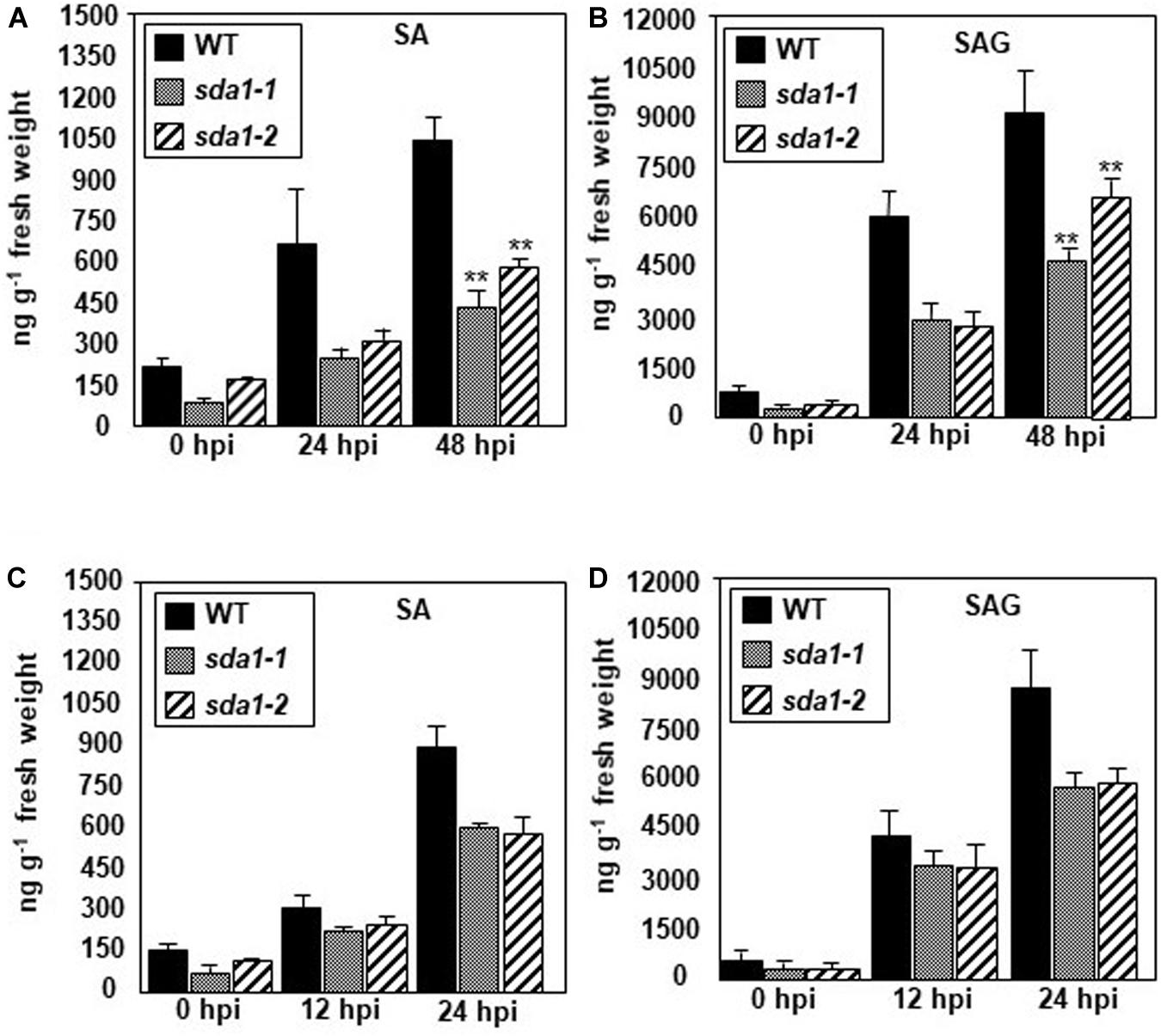
Figure 4. sda1 mutant is compromised in accumulating pathogen-inducible SA and SAG. Rosette leaves of 4-week-old plants of wild-type (WT) and sda1 mutants were inoculated with Pst DC3000 (A,B) and Pst DC3000 (avrRpm1) (C,D) at a titer of 5 × 107 cfu/mL in 10 mM MgSO4. Tissue samples were collected at the indicated hpi and free SA (SA) and glucose-conjugated SA (SAG) were extracted. The values are means ± SD of three replicates, each consisting of leaves from 10 plants per genotype. Statistical analysis was performed by one-way ANOVA followed by Dunnett’s test with reference to WT control. Asterisks indicate statistically significant differences (**p < 0.05).
Accumulation of reactive oxygen species (ROS) is one of the earliest plant defense responses against pathogen infection (Torres et al., 2006). ROS function as antimicrobial compounds and also act as signaling molecules for defense activation. To determine if ROS might play a role in activation of SDA1, we determined the expression of SDA1 in response to H2O2, 3-AT (3-amino-1,2,4-triazole), and paraquat (methyl viologen, N,N’-Dimethyl-4,4’-bipyridinium dichloride) over a 48 h period by northern blot analysis. 3-AT is an irreversible inhibitor of catalase, which scavenges H2O2 from the photorespiration system in leaves, and thus an application of 3-AT results in accumulation of intracellular H2O2 (Wang et al., 1999). Paraquat is widely used as source of intracellular superoxide radicals (Babbs et al., 1989). Treatment with H2O2 resulted in a rapid but very transient expression of SDA1. This is consistent with the previous report that SDA1 is induced in response to H2O2 (Davletova et al., 2005). In contrast, 3-AT induced a slower but prolonged induction of SDA1 expression; while SDA1 induction by paraquat was more modest and not as prolonged as with 3-AT (Supplementary Figure S3A). Together these results suggest that activation of SDA1 in response to pathogen might be modulated via intracellular ROS.
Several lesion mimic mutants constitutively express defense-related genes, accumulate high levels of ROS, and have enhanced resistance against several virulent pathogens. We determined expression of SDA1 in some of these lesion mimic mutants by northern blot analysis. All tested lesion mimic mutants hrl1, acd2, dll1, and lsd1 (Dietrich et al., 1994; Greenberg et al., 1994; Devadas et al., 2002; Pilloff et al., 2002; Dutta et al., 2015) accumulate high levels of SDA1 transcript indicating that high levels of ROS in these mutants might be responsible for activation of SDA1 (Supplementary Figure S3B).
Whole genome expression analysis has indicated induction of SDA1 in response to multiple abiotic stresses (Davletova et al., 2005; Dinneny et al., 2008; Luhua et al., 2008). To determine if SDA1 might play any role in modulating abiotic stress in a temporal context, we determined expression of SDA1 in response to salinity, drought, cold stress and heat stress. Salt, cold and heat stress treatments were performed on 7-day-old seedlings, and drought treatment was performed on 4-week-old-plants as described in the Experimental Procedures. All tested stress treatments induced expression of SDA1 (Supplementary Figures S3C,D). Taken together, these results suggest that SDA1 might be a stress response protein involved in modulating both biotic and abiotic stresses.
To analyze temporal and spatial expression of SDA1, we constructed transgenic plants containing SDA1 promoter::GUS fusion construct. A 2000 bp region containing the 5‘ UTR region of SDA1 was fused with the β-glucuronidase (GUS) gene and was used to transform wild-type plants (Figure 5A). GUS analysis was carried out during various stages of plant development. SDA1 promoter activity was detected in 10-day old seedlings, flowers, trichomes, stem, root, and at the tip of the siliques (Figures 5Ba–h). Faint GUS activity was also detected in rosette and cauline leaves of 4-week-old plants. These results suggest that SDA1 is developmentally regulated. To determine the spatial response of the SDA1 promoter, rosette leaves of these plants were inoculated with Pst DC3000. GUS staining was detected 12 hpi and was limited to the region of infiltration (Figures 5Bi,j). In addition, consistent with results that SDA1 is induced by mechanical injury (see Figure 1B); strong GUS activity was detected at the site on wounding (Figure 5Bk). This is consistent with the gene expression analysis results in Figures 1A,B.
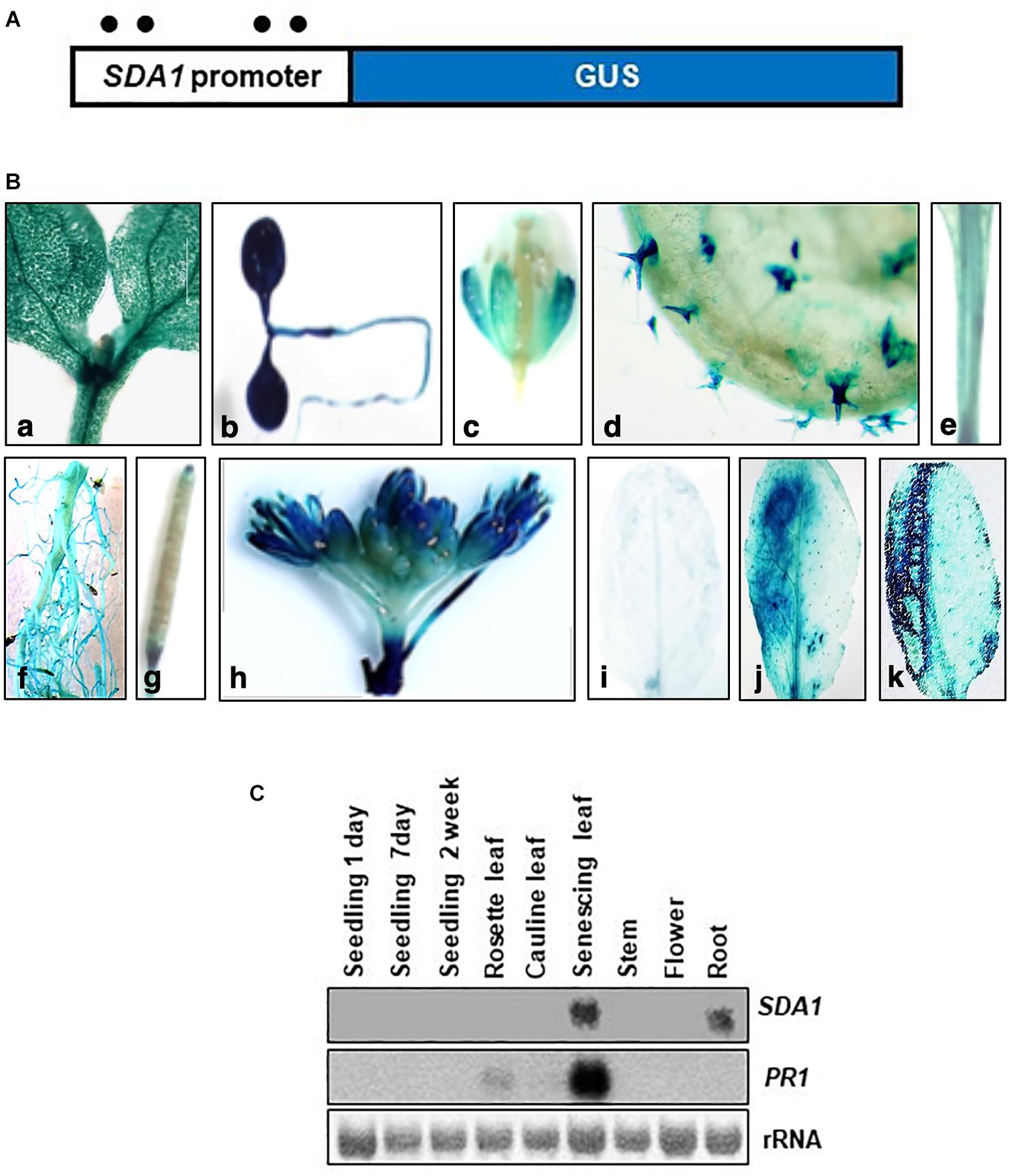
Figure 5. Histochemical analysis of GUS expression in transgenic plants expressing the SDA1 promoter::GUS construct. (A) Schematic representation of the SDA1 promoter::GUS fusion construct. A 2 kb promoter region of the SDA1 gene was used to drive expression of the GUS gene. Filled circles show position of W-boxes. (B) GUS expression in transgenic plants expressing the SDA1 promoter::GUS: (a,b) 10-day old seedling, (c) flower, (d) trichome, (e) stem, (f) root, (g) silique, (h) inflorescence, (i) untreated rosette leaf, (j) rosette leaf inoculated (left half) with Pst DC3000 at titer of 5 × 107 cfu/mL, (k) rosette leaves wounded (left half with serrated forceps). This analysis was carried out in five to seven independent transgenic lines and similar results were observed. (C) Northern gel blot analysis was employed to determine transcript levels of SDA1 in indicated tissue samples. Blot was stained with methylene blue to show the relative amounts of RNA in each lane (rRNA) and was hybridized with indicated gene probes.
Expression of SDA1 in these tissues and in senescing leaves was also analyzed by northern blot analysis. SDA1 transcript was detected only in roots and in senescing leaves suggesting that transcript levels in seedlings, leaves, stems, and flowers are not high enough to be detected by northern blot analysis. Furthermore, since senescence is a type of programmed cell death, expression of SDA1 in senescing leaves suggests that SDA1 might have a role in programmed cell death as well. Consistent with previous reports, PR1 expression was also detected in senescing leaves (Figure 5C). Analysis of the 2 kb upstream region of SDA1 for stress and pathogen responsive elements identified four W-boxes (TTGAC) (Figure 5A). W-boxes are the binding sites for WRKY transcription factors known to modulate defense-related genes (Pandey and Somssich, 2009).
SDA1 (At1g19020) is predicted to encode a small protein of 86 amino acids (calculated mass 9.15 kDa, pI 10.17). The Arabidopsis genome contains a homolog of SDA1, At3g48180 (SDA2), which is predicted to encode a 77 amino acid-long protein that shares 52% identity and 63% similarity with SDA1. Homologs of SDA1 are present in several plant species but no other organism (Figure 6A). All these proteins have a highly conserved novel seven amino acid (S/G)WA(D/E)QWD domain at the N-terminus (Figure 6A). To the best of our knowledge, this domain has not been previously reported. Given that the founding member of this family, SDA1, is involved in pathogen defense, we call this family “small defense-associated (SDA)” protein family and the seven-amino acid domain, the “SWAD” domain.

Figure 6. Conserved D8 and Q9 amino acid of SDA1 are functionally important for proper growth. (A) Alignment of Small Defense-Associated (SDA) protein family members in plants. An Alignment was done using MultAlin. Conserved (S/G)WA(D/E)QWD domain is underlined. All engineered amino acid mutants are indicated by asterisks. Arabidopsis lyrate (related to Arabidopsis), Populus trichocarpa (black cottonwood), Ricinus communis (castor bean), Glycine max (soybean), Nicotiana benthamiana (related to tobacco), Vitis vinifera (common grape vine), Picea sitchensis (sitka spruce), Physcomitrella patens (spreading earthmoss), Medicago truncatula (barrelclover), Pisum sativum (pea), Oryza sativa (asian rice), Sorghum bicolor (sorghum). (B) Growth of T1 and T2 generation of SDA1 D8A, Q9R, V27A, K52A mutant and WT (wild type) Col-0 plants. Plants were photographed at 25 days after sowing in day neutral light (12 h light:12 h dark). (C) Flowering of SDA1 D8A, Q9R, V27A, K52A mutant and WT Col-0 plants. Plants are photographed at 70 days after sowing (DAS) in day neutral light (12 h light: 12 h dark).
In order to determine if the SWAD domain is critical for SDA1 function, we generated four point-mutant constructs by site-directed mutagenesis. Two of these mutations were introduced in the SWAD domain to change aspartic acid at position 8 to alanine (SDA1D8A), and glutamine at position 9 to arginine (SDA1Q9R). The third mutation was introduced in one of the conserved lysine residues at position 52 to change it to alanine (SDA1K52A), and the fourth one was introduced in a non-conserved valine at position 27 to change it to alanine (SDA1V27A) (Figure 6A). A constitutive 35S promoter was used to drive the expression of these engineered constructs and they were introduced into WT plants. For each construct, we analyzed 18 independent transgenic lines. Nine 35S::SDA1D8A transgenic lines were mildly compromised in their overall growth compared to wild-type plants. Ten 35S::SDA1Q9R transgenic lines were severely stunted and were significantly delayed in flowering (Figures 6B,C). Subsequently, these stunted lines had a poor seed set and some of them failed to set any seeds at all. All 35S::SDA1K52A and 35S::SDA1V27A transgenic lines grew normally and showed no visible phenotypic differences compared to WT. These developmental phenotypes of 35S::SDA1D8A and 35S::SDA1Q9R transgenic lines are likely due to dominant negative effects of these mutations.
To determine if the SWAD domain has any role to play in modulating SDA1-mediated defense in Arabidopsis, we analyzed transcript levels of SDA1 and PR1 by reverse transcriptase-quantitative PCR (RT-qPCR) in two constitutively overexpressing independent transgenic lines of each of the amino acid mutants (Supplementary Figure S4A). While SDA1 levels were relatively similar in all the transgenic lines, PR1 levels were significantly reduced only in 35S::SDA1D8A and 35S::SDA1Q9R transgenic lines. These results suggest that aspartic acid at position 8 and glutamine at position 9, and most likely the entire SWAD domain is required for SDA1-mediated expression of PR1 and most likely other defense genes.
As described above (Figure 6), 35S::SDA1D8A and 35S::SDA1Q9R transgenic lines have severe developmental defects, and therefore are not suitable for bacterial pathogen growth assays (especially 35S::SDA1Q9R). Therefore, in order to determine if the SWAD domain has any role in defense against pathogens, we constructed transgenic lines expressing SDA1 point mutants from an β-estradiol-inducible promoter (pER8::SDA1D8A/Q9R/V27A/K52A) in the sda1-1 mutant background. In the absence of external β-estradiol, no transgenic line showed any visible phenotype. In response to β-estradiol application, all transgenic lines induced expression of the transgene (Supplementary Figure S4B). However, expression of PR1 was significantly induced only in transgenic lines expressing pER8::SDA1K52A and pER8::SDA1V27A but not in the transgenic lines expressing pER8::SDA1D8A and pER8::SDA1Q9R (Supplementary Figure S4B). These results are consistent with the expression of PR1 in transgenic lines expressing SDA1 point mutants from the constitutive 35S promoter (Supplementary Figure S4A). To test for response of these transgenic lines to virulent bacterial pathogens, 4-week-old rosette leaves of WT, sda1-1, mutant lines expressing β-estradiol-inducible transgenes, and pER8::SDA1 were induced by β-estradiol. After 24 h, all plants were inoculated with Pst DC3000 and pathogen growth was determined 3 dpi. Pathogen growth in pER8::SDA1K52A and pER8::SDA1V27A transgenic lines was similar to that in the pER8::SDA1 line, however, growth in the pER8::SDA1D8A and pER8::SDA1Q9R transgenic lines was similar to the sda1 mutant (Figure 7). These results suggest that the SWAD domain is critical for SDA1-mediated resistance against pathogens. Together, these results suggest that conserved amino acids aspartic acid at position 8, glutamine at position 9, and possibly the entire SWAD domain are functionally important for SDA1 function in defense against bacterial pathogen Pst DC3000.
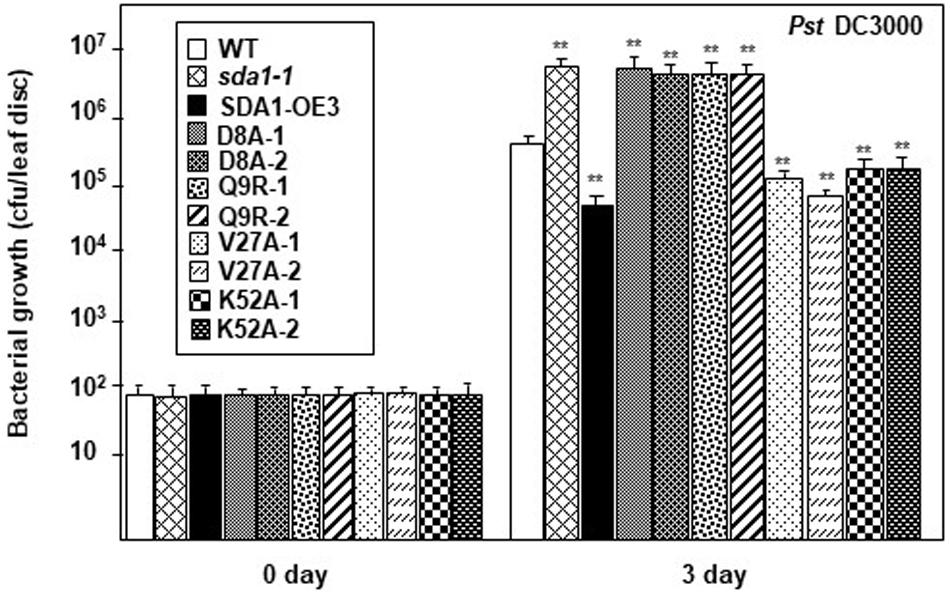
Figure 7. SDA1 D8A and Q9R amino-acid mutants are susceptible against Pst DC3000. Rosette leaves of 4-week-old plants were treated with 55 μm of β-estradiol and 24 h later the leaves were inoculated with Pst DC3000 at a titer of 5 × 105 cfu/mL and pathogen growth was determined at indicated days after inoculation. Eight plants for each genotype were analyzed individually. Data are reported as mean bacterial count (cfu per leaf disc) ± SD. Statistical analysis was performed by one-way ANOVA followed by Dunnett’s test with reference to WT control. Asterisks indicate statistically significant differences (**p < 0.01). Statistical analysis is with reference to WT. This experiment was repeated two more times with similar results. SDA1-OE3, SDA1 expressed from β-estradiol-inducible XVE system (pER8::SDA1, line # 3); D8A, Q9R, V27A, K52A are various point mutants of SDA1 expressed from pER8 promoter.
The enhanced SDA1 expression in response to oxidative stress suggested that SDA1 may be involved in modulating tolerance to oxidative stress. Oxidative stress has been shown to affect both seed germination (Bailly et al., 2008) and seedling root growth (Tsukagoshi, 2016) in Arabidopsis. We analyzed seed germination of sda1 mutants in the presence of ROS-producing agents including paraquat (Apel and Hirt, 2004), H2O2 (Apel and Hirt, 2004), NaCl (Borsani et al., 2001), and 3-AT (Ozgur et al., 2015). Under normal growth conditions, seed germination and seedling root growth of sda1 mutants were not significantly different from the WT (Figure 8 and Supplementary Figure S5). However, when subjected to exogenous oxidative stress, sda1 seeds germinate poorly in the presence of paraquat but not H2O2, NaCl and 3-AT (Figure 8A and Supplementary Figure S5A). Furthermore, germination rate of two independent lines of pER8::SDA1 was higher than the WT and sda1 mutants in the presence of all oxidative stress generating agents (Figure 8A). Seed germination of pER8::SDA1K52A and pER8::SDA1V27A transgenic lines was similar to the pER8::SDA1 lines, however, germination of pER8::SDA1D8A and pER8::SDA1Q9R transgenic lines was comparable to the sda1 mutant.
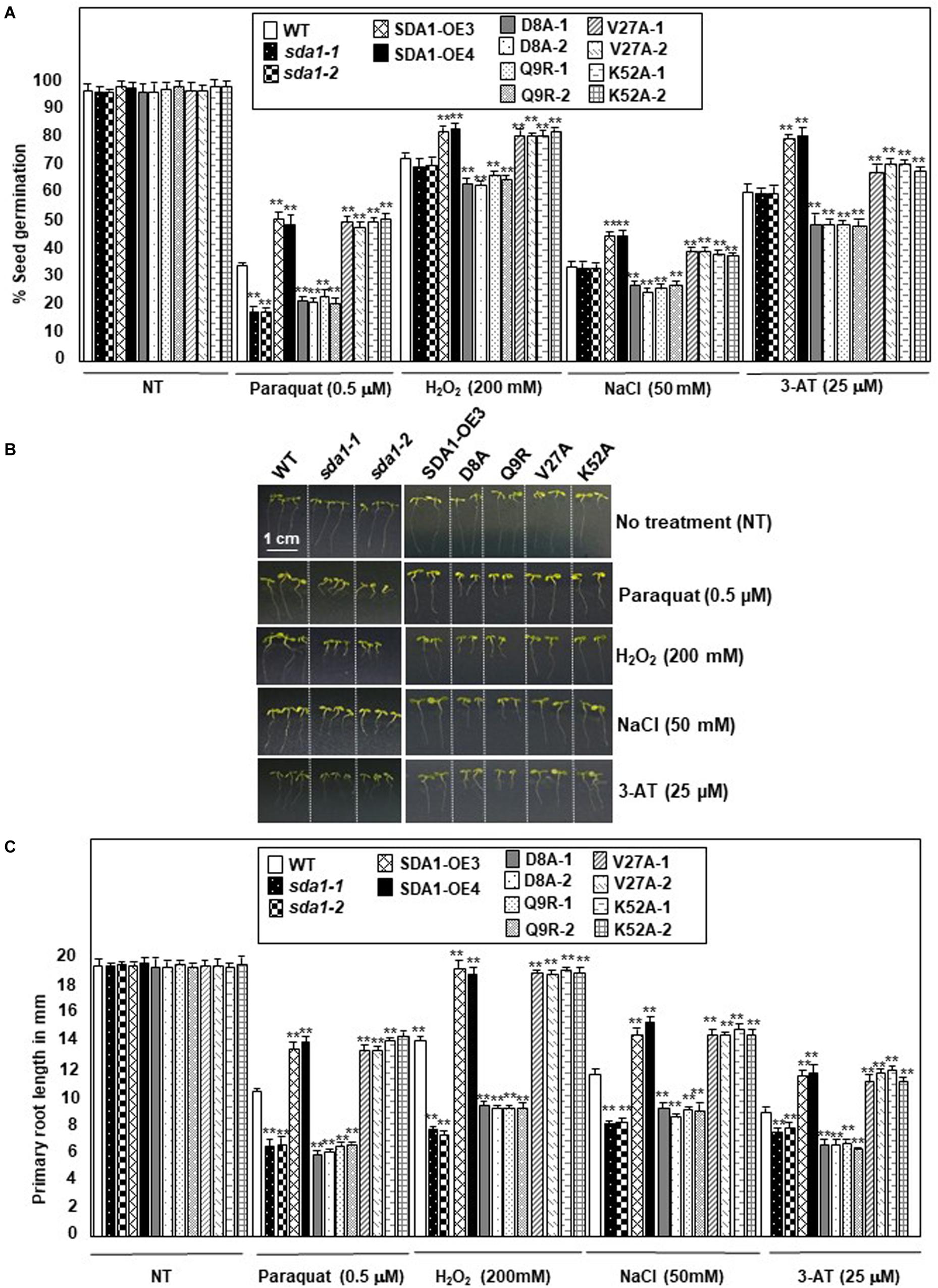
Figure 8. SDA1 is required for tolerance to oxidative stress. (A) Percent seed germination of WT, sda1 mutants, pER8::SDA1 plants, and various amino acids mutants of SDA1 expressed from pER8 promoter in the presence of oxidative stress inducing chemicals. (B) Representative 5-days-old seedlings of genotypes in (A) grown in the presence of indicated oxidative stress inducing chemicals. Scale bar is 1 cm for all samples. (C) Mean root length of seedlings in (B). (n = 25–35; Student’s t-test; **p < 0.01). Statistical analysis was performed by one-way ANOVA followed by Dunnett’s test with reference to respective WT control of each treatment. Asterisks indicate statistically significant differences (**p < 0.01). All experiments were repeated twice and similar results were obtained. SDA1-OE3 and SDA1-OE4, SDA1 expressed from β-estradiol-inducible XVE system (pER8::SDA1, line # 3 and # 4); D8A, Q9R, V27A, K52A are various point mutants of SDA1 expressed from pER8 promoter.
We also tested the effect of these oxidative stress conditions on root growth for sda1, pER8::SDA1, and pER8::SDA1 point-mutant transgenic lines. Roots of sda1 mutants grew significantly shorter compared to the WT in the presence of paraquat, H2O2, and NaCl (Figures 8B,C and Supplementary Figure S5B). A more modest but statistically significant difference in root length was observed in the sda1 mutant in the presence of 3-AT compared to the WT. Furthermore, pER8::SDA1 lines grew longer roots in presence of oxidative stress than WT (Figures 8B,C). Similar to the seed-germination phenotype, root-length of pER8::SDA1D8A and pER8::SDA1Q9R transgenic lines was comparable to the sda1 mutant. However, root-length of pER8::SDA1K52A and pER8::SDA1V27A transgenic lines was similar to pER8::SDA1 lines. Consistent with our results that overexpression of SDA1 confers resistance to oxidative stress, a previous paper reported that overexpression of SDA1 leads to sensitivity to salt stress (Luhua et al., 2008). The reason for this discrepancy to salt stress is not known at this time. Nevertheless, taken together, these results suggest a role for SDA1 and its SWAD domain in tolerance to oxidative stress.
Here, we report the characterization of the SDA1 gene of Arabidopsis (Arabidopsis thaliana). Several lines of evidence suggest that SDA1 is required for resistance against the bacterial pathogen P. syringae. First, SDA1 is induced rapidly and to high levels in response to several virulent and avirulent strains of P. syringae. Like a typical defense gene, SDA1 induction in response to avirulent strain is faster and to higher levels compared to the virulent strains. Second, pathogen-mediated induction of SDA1 requires functional TTSS, suggesting that virulence factors must be delivered into the plant cells for induction of SDA1. Third, pathogen-mediated induction of SDA1 is compromised in several defense mutants. Finally, induction of defense genes and resistance against virulent and avirulent strains of Pst DC3000 is compromised in sda1 knockout mutants.
sda1 mutation compromises resistance against virulent pathogens more significantly than resistance against avirulent pathogens. This may be the result of a significantly higher loss in SA/SAG accumulation in sda1 mutant in response to virulent pathogens as compared to avirulent pathogens. Furthermore, while expression of SDA1 is significantly compromised in defense signaling mutants in response to virulent pathogen, it is not affected in response to avirulent pathogen. This could be because the threshold level of SA-mediated signaling required to activate SDA1 expression may be sufficiently lower than that required for robust R gene-mediated signaling.
SDA1 is predicted to code for a small 86-amino acid protein. It does not have homology to any protein of known function in the public databases. The Arabidopsis Information Resource (TAIR, Araport 11) indicates predicted function as a CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase, an enzyme that participates in glycerophospholipid metabolism. Glycerophospholipids have been described as key regulators of stress response and salicylic acid signaling (Janda et al., 2013; Janda and Ruelland, 2015; Gujas and Rodriguez-Villalon, 2016; Verma and Agrawal, 2017). Homologs of SDA1 are present in several plant species including bean, grape, poplar, tobacco, medicago (alfalfa), soybean and rice. However, no homologs in other plant species or any other organism have been reported suggesting that SDA1 is a plant-specific protein. All homologs of SDA1 have a novel small seven amino acid (S/G)WA(D/E)QWD conserved domain at the N-terminus of the protein; this may suggest that this domain is critical for protein function. We identified critical roles for the conserved domain in SDA1 function specifically for bacterial defense and ROS tolerance, which further highlights the importance of this novel domain.
In addition to pathogens, expression of SDA1 is also induced in response to a variety of biotic and abiotic stresses, including aphid (Myzus persicae), insect larvae (Spodoptera), and oxidative stress induced by glucose-glucose oxidase (Appel et al., 2014). SDA1 expression is also induced in response to abiotic stresses such as drought, salt, heat, wounding, and ROS-generating chemicals. All these abiotic stresses are known to generate ROS. sda1 mutants are hypersensitive while overexpressing pER8::SDA1 lines are more tolerant to oxidative stress-inducing agents such as paraquat, H2O2, NaCl, 3-AT, which suggests a role in oxidative stress tolerance. Overall, these results suggest that SDA1 functions as a broad-range, stress–response plant protein.
PR gene expression is delayed and compromised in sda1 mutants in response to virulent bacterial pathogen Pst DC3000. External application of SA rescues both PR1 expression and resistance to pathogens in sda1 mutants. These results suggest that the sda1 mutant is not compromised in sensing or signaling mediated by SA and that SDA1 probably functions upstream of SA. Additionally, accumulation of SA and SAG in response to Pst DC3000 is compromised in sda1 mutants, further suggesting that SDA1 acts upstream of SA. However, external application of SA induces expression of SDA1, suggesting a SA-mediated positive amplification loop that might modulate expression of SDA1 in response to pathogen infections. Similar SA-mediated positive amplification loops have been proposed previously for other defense genes, such as PAD4 and GDG1 (Jirage et al., 1999; Jagadeeswaran et al., 2007).
To determine the epistatic relationship of SDA1 with other defense genes in the SA-mediated defense-signaling pathway, we analyzed expression of Pst DC3000-mediated induction of SDA1 in several defense mutants. SDA1 expression was compromised in ndr1, eds1, pad4, gdg1/pbs3/win3 mutants, and NahG expressing plants. However, SDA1 expression was not affected in the sid2 mutant. This is similar to a previous report that virulent bacterial pathogen-mediated expression of some genes is compromised in the gdg1 mutant but not in the sid2 mutant (Wang et al., 2008). Interestingly, both gdg1 and sid2 mutants are compromised in Pst DC3000-mediated SA accumulation. SID2 codes for isochorismate synthase, an important SA biosynthetic enzyme (Wildermuth et al., 2001). PBS3/GDG1/WIN3 has been shown to catalyze the conjugation of specific amino acids to SA acyl substrates and has been suggested to allow for coordination of flux through diverse chorismate-derived pathways (Okrent et al., 2009). Our results suggest that SDA1 expression might be modulated by one of these chorismate-derived signals. SA modulates expression of SDA1 by positive feedback regulation, possibly via GDG1/PBS3/WIN3 because SA has been shown to modulate GDG1/PBS3/WIN3 via positive feedback regulation (Jagadeeswaran et al., 2007). Figure 9 shows a proposed model for the role of SDA1 in pathogen defense signaling.
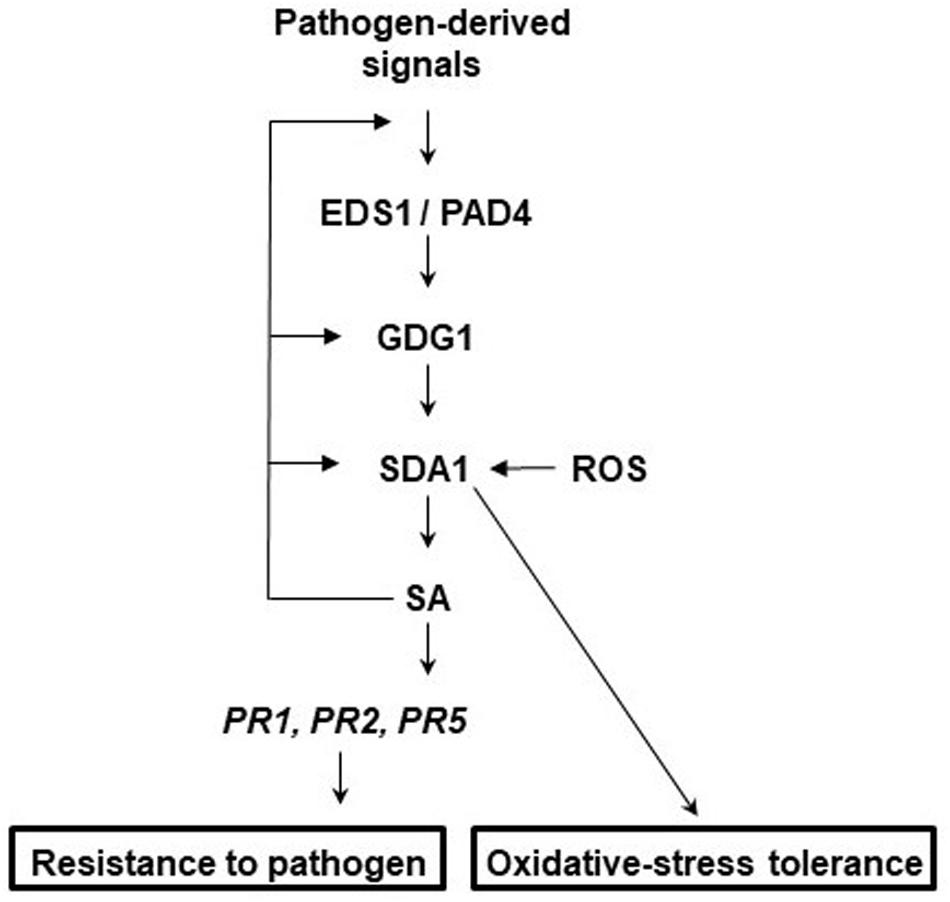
Figure 9. Proposed model for the role of SDA1 in defense signaling and oxidative-stress tolerance. In response to virulent pathogens, expression of SDA1 is regulated via EDS1, PAD4, and GDG1 (also known as PBS3 and WIN3) but not SID2. SDA1 is at least in part required for pathogen-induced SA biosynthesis, which in turn regulates expression of PR genes and defense via NPR1. SA regulates expression of SDA1 by positive feedback controls, possibly via GDG1. Additionally, control via GDG1 may be dependent on positive feedback to GDG1 at low SA levels and negative feedback at high SA levels. In addition, SDA1 modulates tolerance to oxidative stress.
In conclusion, the studies reported here suggest that SDA1 represents a novel class of plant genes required for defense against bacterial pathogens that is part of the SA-mediated defense pathway. Furthermore, SDA1 might be involved in modulating a variety of biotic and abiotic stress responses in addition to response against pathogens. We propose that SDA1 might act to coordinate crosstalk across development, biotic and abiotic stress, ROS and salicylic acid signaling.
All datasets generated for this study are included in the article/Supplementary Material.
AD and RR conceived the original screening and research plans. RR and DK supervised the experiments. AD and PC performed most of the experiments. SC provided technical assistance. AD, PC, PG-B, P-PL, DK, and RR designed the experiments and analyzed the data. AD, PC, and RR wrote the manuscript with contributions of all the authors. RR supervised and completed the writing and agreed to serve as the author responsible for contact and ensures communication.
This work was supported by the NSF Grant (IBN-0313492) and funds from Syracuse University to RR, and NSF Grants IOS-0820405 and IOS-0525360 to DK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Arabidopsis Biological Resource Center (ABRC) for pad4, CS60000, and SAIL_390_F08 T-DNA insertion line, Nottingham Arabidopsis Stock Centre for GABI_283D01 T-DNA insertion line, Dr. Mary Wildermuth (UC Berkeley) for sid2-2 seeds, and Syngenta Biotechnology Inc. for NahG transgenic seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00703/full#supplementary-material
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Appel, H. M., Maqbool, S. B., Raina, S., Jagadeeswaran, G., Acharya, B. R., Hanley, J. C., et al. (2014). Transcriptional and metabolic signatures of Arabidopsis responses to chewing damage by an insect herbivore and bacterial infection and the consequences of their interaction. Front. Plant Sci. 5:441. doi: 10.3389/fpls.2014.00441
Babbs, C. F., Pham, J. A., and Coolbaugh, R. C. (1989). Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol. 90, 1267–1270. doi: 10.1104/pp.90.4.1267
Bailly, C., El-Maarouf-Bouteau, H., and Corbineau, F. (2008). From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C. R. Biol. 331, 806–814. doi: 10.1016/j.crvi.2008.07.022
Bent, A. F., and Mackey, D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. doi: 10.1146/annurev.phyto.45.062806.094427
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Borsani, O., Valpuesta, V., and Botella, M. A. (2001). Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 126, 1024–1030. doi: 10.1104/pp.126.3.1024
Boyd, L. A., Ridout, C., O’Sullivan, D. M., Leach, J. E., and Leung, H. (2013). Plant-pathogen interactions: disease resistance in modern agriculture. Trends Genet. 29, 233–240. doi: 10.1016/j.tig.2012.10.011
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Century, K. S., Holub, E. B., and Staskawicz, B. J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. U.S.A. 92, 6597–6601. doi: 10.1073/pnas.92.14.6597
Chen, Z., Zheng, Z., Huang, J., Lai, Z., and Fan, B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496.
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Collmer, A., Badel, J. L., Charkowski, A. O., Deng, W. L., Fouts, D. E., Ramos, A. R., et al. (2000). Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 8770–8777.
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Davletova, S., Rizhsky, L., Liang, H., Shengqiang, Z., Oliver, D. J., Coutu, J., et al. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281. doi: 10.1105/tpc.104.026971
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., et al. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250.
Devadas, S. K., Enyedi, A., and Raina, R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. doi: 10.1046/j.1365-313x.2002.01300.x
Dewdney, J., Reuber, T. L., Wildermuth, M. C., Devoto, A., Cui, J., Stutius, L. M., et al. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. doi: 10.1046/j.1365-313x.2000.00870.x
Dietrich, R. A., Delaney, T. P., Uknes, S. J., Ward, E. R., Ryals, J. A., and Dangl, J. L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. doi: 10.1016/0092-8674(94)90218-6
Dinneny, J. R., Long, T. A., Wang, J. Y., Jung, J. W., Mace, D., Pointer, S., et al. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. doi: 10.1126/science.1153795
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209.
Dutta, A., Chan, S. H., Pauli, N. T., and Raina, R. (2015). Hypersensitive response-like lesions 1 codes for AtPPT1 and regulates accumulation of ROS and defense against bacterial pathogen Pseudomonas syringae in Arabidopsis thaliana. Antioxid. Redox. Signal. 22, 785–796. doi: 10.1089/ars.2014.5963
Dutta, A., Choudhary, P., Caruana, J., and Raina, R. (2017). JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. Plant J. 91, 1015–1028. doi: 10.1111/tpj.13623
Feys, B. J., Moisan, L. J., Newman, M. A., and Parker, J. E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. doi: 10.1093/emboj/20.19.5400
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Glazebrook, J., Rogers, E. E., and Ausubel, F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982.
Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E. E., Crute, I. R., et al. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392.
Greenberg, J. T., Guo, A., Klessig, D. F., and Ausubel, F. M. (1994). Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. doi: 10.1016/0092-8674(94)90217-8
Gujas, B., and Rodriguez-Villalon, A. (2016). Plant phosphoglycerolipids: the gatekeepers of vascular cell differentiation. Front. Plant Sci. 7:103. doi: 10.3389/fpls.2014.00103
Gust, A. A., Willmann, R., Desaki, Y., Grabherr, H. M., and Nurnberger, T. (2012). Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502. doi: 10.1016/j.tplants.2012.04.003
Jagadeeswaran, G., Raina, S., Acharya, B. R., Maqbool, S. B., Mosher, S. L., Appel, H. M., et al. (2007). Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 51, 234–246. doi: 10.1111/j.1365-313x.2007.03130.x
Janda, M., Planchais, S., Djafi, N., Martinec, J., Burketova, L., Valentova, O., et al. (2013). Phosphoglycerolipids are master players in plant hormone signal transduction. Plant Cell Rep. 32, 839–851. doi: 10.1007/s00299-013-1399-0
Janda, M., and Ruelland, E. (2015). “Magical mystery tour: salicylic acid signalling,” in Plant Signalling Mechanisms In Response To The Environment, eds E. Ruelland and J. Farrant (Amsterdam: Elsevier), 1–158.
Jirage, D., Tootle, T. L., Reuber, T. L., Frost, L. N., Feys, B. J., Parker, J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 13583–13588. doi: 10.1073/pnas.96.23.13583
Lee, M. W., Lu, H., Jung, H. W., and Greenberg, J. T. (2007). A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol. Plant Microb. Interact. 20, 1192–1200. doi: 10.1094/mpmi-20-10-1192
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luhua, S., Ciftci-Yilmaz, S., Harper, J., Cushman, J., and Mittler, R. (2008). Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 148, 280–292. doi: 10.1104/pp.108.124875
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense priming: an adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512. doi: 10.1146/annurev-arplant-042916-041132
Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J. P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. doi: 10.1105/tpc.010376
Nawrath, C., and Metraux, J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. doi: 10.1105/tpc.11.8.1393
Nobuta, K., Okrent, R. A., Stoutemyer, M., Rodibaugh, N., Kempema, L., Wildermuth, M. C., et al. (2007). The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156. doi: 10.1104/pp.107.097691
Okrent, R. A., Brooks, M. D., and Wildermuth, M. C. (2009). Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284, 9742–9754. doi: 10.1074/jbc.m806662200
Ozgur, R., Uzilday, B., Sekmen, A. H., and Turkan, I. (2015). The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in arabidopsis. Ann. Bot. 116, 541–553. doi: 10.1093/aob/mcv072
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Parker, J. E., Holub, E. B., Frost, L. N., Falk, A., Gunn, N. D., and Daniels, M. J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. doi: 10.1105/tpc.8.11.2033
Pilloff, R. K., Devadas, S. K., Enyedi, A., and Raina, R. (2002). The Arabidopsis gain-of-function mutant dll1 spontaneously develops lesions mimicking cell death associated with disease. Plant J. 30, 61–70. doi: 10.1046/j.1365-313x.2002.01265.x
Rogers, E. E., and Ausubel, F. M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. doi: 10.1105/tpc.9.3.305
Rosso, M. G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. doi: 10.1023/b:plan.0000009297.37235.4a
Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994.
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78. doi: 10.1094/mpmi.1997.10.1.69
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. doi: 10.1105/tpc.104.026690
Staswick, P. E., Tiryaki, I., and Rowe, M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. doi: 10.1105/tpc.000885
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Tsukagoshi, H. (2016). Control of root growth and development by reactive oxygen species. Curr. Opin. Plant Biol 29, 57–63. doi: 10.1016/j.pbi.2015.10.012
Verma, K., and Agrawal, S. B. (2017). “Salicylic acid-mediated defence signalling in respect to its perception, alteration and transduction,” in Salicylic Acid: A Multifaceted Hormone, eds R. Nazar, N. Iqbal, and N. Khan (Singapore: Springer), 97–122. doi: 10.1007/978-981-10-6068-7_6
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, J., Zhang, H., and Allen, R. D. (1999). Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol. 40, 725–732. doi: 10.1093/oxfordjournals.pcp.a029599
Wang, L., Mitra, R. M., Hasselmann, K. D., Sato, M., Lenarz-Wyatt, L., Cohen, J. D., et al. (2008). The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol. Plant Microb. Interact 21, 1408–1420. doi: 10.1094/mpmi-21-11-1408
Wildermuth, M. C. (2006). Variations on a theme: synthesis and modification of plant benzoic acids. Curr. Opin. Plant Biol. 9, 288–296. doi: 10.1016/j.pbi.2006.03.006
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wirthmueller, L., Maqbool, A., and Banfield, M. J. (2013). On the front line: structural insights into plant-pathogen interactions. Nat. Rev. Microbiol. 11, 761–776. doi: 10.1038/nrmicro3118
Zheng, Z., Qualley, A., Fan, B., Dudareva, N., and Chen, Z. (2009). An important role of a BAHD acyl transferase-like protein in plant innate immunity. Plant J. 57, 1040–1053. doi: 10.1111/j.1365-313x.2008.03747.x
Zipfel, C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. doi: 10.1016/j.pbi.2009.06.003
Keywords: SMALL DEFENSE-ASSOCIATED PROTEIN 1, SDA1, salicylic acid, bacterial pathogen, defense signaling, Arabidopsis thaliana, ROS, abiotic stress
Citation: Dutta A, Choudhary P, Gupta-Bouder P, Chatterjee S, Liu P-P, Klessig DF and Raina R (2020) Arabidopsis SMALL DEFENSE-ASSOCIATED PROTEIN 1 Modulates Pathogen Defense and Tolerance to Oxidative Stress. Front. Plant Sci. 11:703. doi: 10.3389/fpls.2020.00703
Received: 10 February 2020; Accepted: 05 May 2020;
Published: 03 June 2020.
Edited by:
Péter Poór, University of Szeged, HungaryCopyright © 2020 Dutta, Choudhary, Gupta-Bouder, Chatterjee, Liu, Klessig and Raina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramesh Raina, cmFyYWluYUBzeXIuZWR1
†Present address: Aditya Dutta, Department of Animal and Food Sciences, University of Delaware, Newark, DE, United States; Pratibha Choudhary, Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN, United States; Pallavi Gupta-Bouder, SUNY Polytechnic Institute, Utica, NY, United States; Snigdha Chatterjee, Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States; Po-Pu Liu, RiceTec Inc., Alvin, TX, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.