- 1College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, China
- 2State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
Vacuolar processing enzymes (VPEs) play an important role in stress resistance and development of plants. Despite their diverse roles, little information is available in apple (Malus × domestic). This study firstly presents the genome-wide identification of VPE family genes in apple, resulting in 20 family members those are unevenly distributed across six out of the 17 chromosomes. Phylogenetic analysis assigned these genes into four groups. Analysis of exon–intron junctions and motifs of each candidate gene revealed high levels of conservation within and between phylogenetic groups. Cis-element including w box, ABRE, LTR, and TC-rich repeats were found in promoters of MdVPEs. NCBI-GEO database shown that the expression of MdVPEs exhibited diverse patterns in different tissues as well as the infection of Pythium ultimum and Apple Stem Grooving Virus. Furthermore, qRT-PCR showed that MdVPE genes were responsive to salt, cadmium, low-temperature, and drought. Overexpression of MDP0000172014, which was strongly induced by salt and drought stress, significantly decreased Arabidopsis tolerance to salt stress. The genome-wide identification and characterization of MdVPEs in apple provided basic information for the potential utilization of MdVPEs in stress resistance.
Introduction
Programmed cell death (PCD), which plays an important role in developmental processes and stress resistance of eukaryotes, is a genetically regulated physiological process (Hatsugai et al., 2015). In animals, the performed of PCD requires the caspase enzymes with cysteine-dependent aspartyl protease activity (Fagundes et al., 2015), and caspase enzymes could hydrolyze the substrates with specific aspartic acid residues (Lamkanfi et al., 2002; De Pinto et al., 2012). Unfortunately, its protein sequences are not conserved in the genomes of plants (Lam and Zhang, 2012). However, caspase-like enzymatic activity has been shown that is essential in many forms PCD of plants (Del Pozo and Lam, 1998; Woltering et al., 2002; Baskett, 2012). And some caspase-like proteases including subtilases (Vartapetian et al., 2011), metacaspases (Tsiatsiani et al., 2011) and legumains (Rojo et al., 2004) had been identified one after another.
Legumain, which is referred to as vacuolar processing enzymes (VPEs), has the characteristics of caspase-1-like activity (Hatsugai et al., 2015). VPEs is conversed in plants, and their conversed functional domain is peptidase_C13 (Pfam ID: PF01650), which composed of CASc superfamily motifs (Vorster et al., 2019). Based on the expression and functional characteristics, VPEs could be divided into three sub-families: vegetative VPEs, seed VPEs, and uncharacterized VPEs. Vegetative VPEs including γVPE and δVPE, are essential for stress resistance of plants, while seed VPEs including αVPE and βVPE, play an important role in developmental process (Christoff et al., 2014), and the functions of uncharacterized VPEs have not been identified.
As a key regulator of plant cell death, VPEs were involved in many plant developmental processes. For example, NtTPE8, a VPE-like protease from tobacco, only expressed in the integumentary tapetum of tobacco seeds, and its downregulation induced seeds abortion (Wang et al., 2019). Heterologous expression of VvβVPE, a βVPE from Vitis vinifera, was essential for ovule maturation and the increased germination of seeds in Arabidopsis (Gong et al., 2018). Expression of IbVPE1 from sweet potato in Arabidopsis affected leaf development, flowering time and chlorophyll catabolism (Jiang et al., 2019). Previous study also reported VPEs were involved in the regulation of seed size in barley (Radchuk et al., 2018; Yamada et al., 2019), the breakdown of apical bud dominance in potato tubers (Teper-Bamnolker et al., 2012), the development and senescence of root nodules (Van Wyk et al., 2014; Cilliers et al., 2017), and the floral bud abortion of radish (Zhang et al., 2013).
VPEs were also involved in the resistance of plants biotic and abiotic stress. VPEs has been identified to be essential for TMV-induced hypersensitive cell death of Nicotiana benthamiana by VIGS (virus-induced gene silencing) (Hatsugai et al., 2004) and mycotoxin-induced cell death in Arabidopsis (Kuroyanagi et al., 2005). γVPE promoted the heat shock-induced cell death in Arabidopsis leaves (Li et al., 2012). VPEs were also involved in aluminum-induced cell death (Kariya et al., 2013, 2018) and H2O2-induced cell death (Deng et al., 2011; Kim et al., 2014). Besides, our lab also found that MhVPEγ, a γVPE isolated from apple rootstock-Malus hupenensis Rhed., was involved in cadmium-induced cell death (Ran et al., 2014; Zhang et al., 2019) and high temperature-induced cell death (Su et al., 2015).
Consider that so many important roles of VPEs, they have been analyzed at the whole genome scale in various model plants and crops (Vorster et al., 2019). However, the study related to them in apple, one of the fruits with the largest production and consumption in the world, is little. Fresh roots of apple have abundant vacuole, an organelle plays an important role in the stress resistance of plants, and VPE is essential for function performed of the vacuole (Shimada et al., 2018). Furthermore, genome-wide identification and characterization of a gene family can provide basic information for the potential utilization of the gene function. Since apple genome sequencing (Velasco et al., 2010), more and more gene family of apple had been identified (Meng et al., 2016; Mao et al., 2017), whereas the whole information of VPE family genes in apple is limited.
In this study, 20 VPE genes were identified in the apple (Malus × domestic) genome, and their expression profiles were determined in different tissues, and during the infection of Pythium ultimum and Apple Stem Grooving Virus (ASGV). qRT-PCR was also used to detect the expression levels in response to salt, cadmium, drought and low-temperature treatment. Overexpression of MDP0000172014, which was strongly induced by salt and drought stress, significantly decreased Arabidopsis tolerance to salt stress and promoted salt stress-induced roots cell death. Our widely identified and expression analysis of MdVPEs in apple filled the blank of woody plants VPE information and contributed to the further exploration of VPE functions.
Materials and Methods
Plant Materials and Stress Treatment
“Royal gala” tissue culture seedlings were used in this study. Consistent growth tissue culture seedlings were selected and rooted with Murashige and Skoog (MS) agar medium which contain 30 g/L sucrose and 0.3 mg/L IBA. After grown for 30 days, the rooted tissue culture seedlings were transferred to 1/2 Hoagland’s nutrient solution for stress treatment. For salt stress, 1/2 Hoagland’s nutrient solution with 200 mM NaCl was used for plant treatment; for cadmium stress, 1/2 Hoagland’s nutrient solution with 200 μM CdSO4 was used for plants treatment; for low-temperature stress, illumination incubator with 4°C and 1/2 Hoagland nutrient solution was used for plants treatment, and for drought stress, 1/2 Hoagland’s with 5% PEG 6000 (W/V) was used for plants treatment. The plants treated with 1/2 Hoagland’s nutrient at room temperature (RT) was as the control of each group treatment. Each group treatment was treated for 24 h. After 24 h of such pre-cultivation, stress treatments were initiated. All collected tissue samples had treated for 24 h were frozen quickly in liquid nitrogen and stored at −80°C for RNA extraction.
Identification of Vacuolar Processing Enzyme Family in Apple
The Hidden Markov Model (HMM, PF01650) of VPEs was downloaded from the Pfam database1 and used to search for the apple genome database by HMM 3.0. To avoid the loss of members due to the incomplete domain of VPE, local BLAST-P searches were performed in Phytozome2 plant genome database using the protein sequences of Arabidopsis (AT1G62710.1, BETA-VPE; AT2G25940.1, ALPHA-VPE; AT3G20210.1, DELTA-VPE, and AT4G32940.1, GAMMA-VPE), which downloaded from TAIR.3 And then combined the two-part results and deleted the repeated sequences. The results were submitted to SMART,4 NCBI–CDD5 (Marchler-Bauer et al., 2017) and Pfam to identify the protein structure domain. The sequences which do not contain the peptidase_C13 domain and no complete Open Reading Frame (ORF) were deleted.
Sequence Annotation and Structural Analysis
All the high confidence sequences were submitted to the ExPaSy Proteomics Server online tools6 for predicting molecular weight (MW), isoelectric point (PI) and amino acid (AA) number. The chromosomal location and exons quantity information were downloaded from the Phytozome. The exon-intron structures of MdVPE genes were visualized via Gene Structure Display Server 2.07 (GSDS) (Hu et al., 2015) and the conserved motifs of MdVPEs were analyzed by the MEME program version 5.0.2,8 with the parameter as 6 × 50 and the number of motifs as 10. ProtComp version 9.09 was used to predict subcellular localization of MdVPEs.
Phylogenetic Analysis, Chromosome Localization and Gene Duplication of MdVPEs
Multiple sequence alignment of AA sequences in Arabidopsis and apple were used the clustal W version 2.0 (Thompson et al., 1997) and the MEGA 7.0 (Kumar et al., 2016) was used to generate phylogenetic trees by neighbor-joining (NJ) method, with Bootstrap parameters were tested for 1,000 times. Chromosome location was shown by Map Draw version 2.1. Gene duplication of MdVPEs including tandem duplicated genes and segmental duplicated genes were identified by TBtools (Chen et al., 2018) with the E-value is 1e−10.
Analysis of Cis-Acting Element in MdVPEs’ Promoters
The upstream sequences (2 kb) of the MdVPE-coding sequences were retrieved from the Phytozome plant genome database and then submitted to Plant CARE10 to identify four regulatory elements, abscisic acid (ABA)-responsive elements (ABRE); low-temperature responsive elements (LRE); TC-rich repeats, which were involved in defense and stress response; and w box, which is the binding site of WRKY transcription factor-play an important role in defensing to drought stress.
Expression Profile Analysis of MdVPEs in Different Tissue and During P. ultimum and ASGV via NCBI-GEO Database
Expression profile database (GSE42873, for different tissues; GSE62103, for P. ultimum; GSE53825, for ASGV) were downloaded from NCBI-GEO database.11 MdVPE sequences were used BLAST for probe database (GPL16374, for different tissues; GPL18137, for P. ultimum and ASGV) and the matched probes were selected to represent MdVPEs. Heatmaps showed the expression profiles of MdVPEs were constructed by TBtools (Chen et al., 2018) and RPKM (reads per kilobase of exon model per million mapped reads) values of each gene obtained in the NCBI-GEO were used to compare their expression levels between different tissues and during the infection of P. ultimum and ASGV.
Total RNA Extraction and qRT-PCR
Total RNA was extracted from “royal gala” seedlings roots via polysaccharide rich polyphenols total RNA extraction kits (TianGen, China). First-strand cDNA was obtained by Prime ScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan). qRT-PCR was performed three independent biological replicates on LightCycler® 96 (Roche) using the TB Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa, Japan) according to the specification. All primer sequences for qRT-PCR were given in Supplementary Table 1, and Md-actin was used as the internal control gene. The data were analyzed by 2–ΔΔCt method (Livak and Schmittgen, 2001).
Arabidopsis Transformation and Stress Treatments
Col-0 Arabidopsis were used for genetic transformation. Full-length MDP0000172014 was cloned from cDNA by using primers MDP0000172014-F/R (Supplementary Table 1), and its coding region was inserted into the pBI121 vector under the control of the 35S promoter through the In-Fusion Cloning (C112, Vazyme, China). The recombinant vector named pBI121-35S-MDP0000172014 was transformed into GV3101, an Agrobacterium tumefaciens strain. Inflorescence infection was used for the transformation of Col-0 Arabidopsis (Clough and Bent, 1998). The transgenic Arabidopsis seeds were sterilized and selected for 50 mg/L kanamycin resistance on MS agar medium and finally three independent third homozygous (T3) were generated.
The T3 and Col-0 Arabidopsis seeds were sterilized and growth on MS agar medium for 7 days, and then transferred onto MS agar medium containing 100 mM NaCl and cultured in an incubator for another 10 days. The seedlings were then photographed. The root length was recorded based on at least 10 seedlings. Seven-days-old-seedlings growth on standard MS agar medium were transferred onto a matrix containing 30% turf, 30% vermiculite, and 10% perlite and cultured in illumination incubator with culture conditions of 24°C, light cycle for 16 h light/dark 8 h, watered once every 4 days. Forty-days-old seedlings growth on a matrix which were watered with 1/2 Hoagland’s nutrient solution (as in the control) or 1/2 Hoagland’s solution containing 200 mM NaCl. The roots treated for 48 h and then sampled to measured MDA, H2O2 content and other physiological indexes.
Measurement of H2O2 and MDA
H2O2 content was determined by sulfuric acid precipitation (Patterson et al., 1984). The MDA content was performed according to the thiobarbituric method and the absorbance of the mixture was measured at 532, 600, and 450 nm (Landi, 2017).
Measurement of Cell Death Number and Vacuolar Processing Enzyme Activity
The measurement of cell death followed the Evans blue staining (Zhang et al., 2019). A total of 0.1 g roots were washed and placed in 0.25% (W/V) Evans blue solution. After cultured for 24 h, the roots were then rinsed with deionized water until no more blue stain eluted and put into a centrifuge tube containing 5 mL 1% SDS solution for 24 h to extract. A total of 1% SDS solution with unstained roots was used as negative control and the roots completely killed in boiling water 1 h were used as the positive control. The extracting solution was measured according to the absorbance of at 600 nm.
Vacuolar processing enzyme activity was measured following Wang et al. (2009) with slight modifications. VPE-specific substrate Ac-ESEN-MCA (Ac–Glu–Ser–Glu–Asn–MCA (Peptide Institute, Japan)) was used to measure the VPE activity. 0.1 g roots were used to make crude protein extract, and the extract was incubated with 100 μM Ac-ESEN-MCA in an acidic buffer (100 mM dithiothreitol, 100 mM sodium acetate, and pH 5.5) for 2 h at 20°C. The fluorescence was monitored under an excitation wavelength of 380 nm and an emission wavelength of 460 nm (Shimada et al., 2003; Wang et al., 2009).
Statistical Analysis
SPSS software version 19.0 (IBM, Chicago, IL, United States) was used to analyze variance (ANOVA). All experiments were carried out in triplicate (n = 3) and expressed as mean value ± standard deviation. Duncan’s new complex range method was used for significance analysis with the test level was 5% (P < 0.05), and origin 2017 was used to draw the chart.
Results
Identification and Characterization of Vacuolar Processing Enzyme Gene Family in Apple
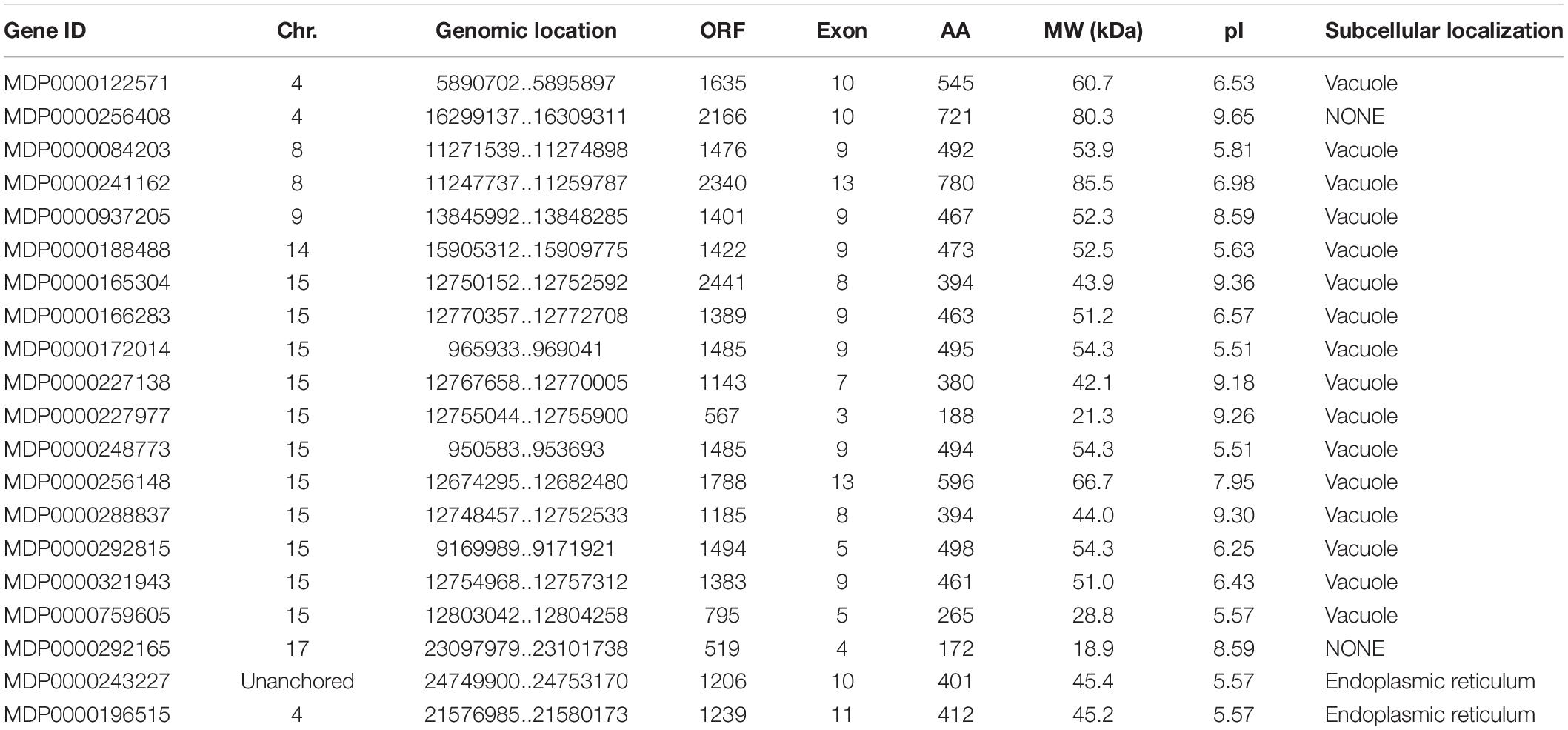
To identify VPE genes in the apple genome, the HMM of VPEs was downloaded from the Pfam database and used to search proteins database of apple by hmm search 3.0. Moreover, Arabidopsis VPE protein sequences were downloaded from the TAIR database and used to BLAST-P in the apple proteins database. Based on the SMART, NCBI-CDD, and Pfam, all of the MdVPE proteins without peptidase_C13 domain or complete ORF were removed. Finally, a total of 20 MdVPE genes in apple were obtained and annotated (Table 1). As shown in the Table 1, the size of these proteins ranges from 118 (MDP0000258293) to 780 (MDP0000241162) AAs residues, with the MW varies from 18.9 kDa (MDP0000292165) to 85.5 kDa (MDP0000241162) and the PI distribution varies from 5.51 (MDP0000172014 and MDP0000248773) to 9.65 (MDP0000256408). Subcellular locations were also predicted of MdVPE proteins by ProtComp version 9.0. Most of the MdVPEs were predicted to be vacuole proteins, whereas MDP0000243227 and MDP0000196515 were predicted to be endoplasmic reticulum (ER) membrane proteins. Unfortunately, MDP0000256408 and MDP0000292165 were predicted to be free of sub-cellular localization proteins.
Sequence Alignment and Phylogenetic Analysis of MdVPE Genes
To predict the potential function of MdVPEs, we constructed an unrooted phylogenetic tree by MEGA 7.0 following the NJ method. with the imported Arabidopsis VPE proteins as the reference proteins, MdVPE proteins were divided into four groups, and each group contains diverse MdVPEs family members (Figure 1). Group 4 had the most members (seven), followed by group 1 (six), while group 3 had four. Group 2 has the smallest, only three members.
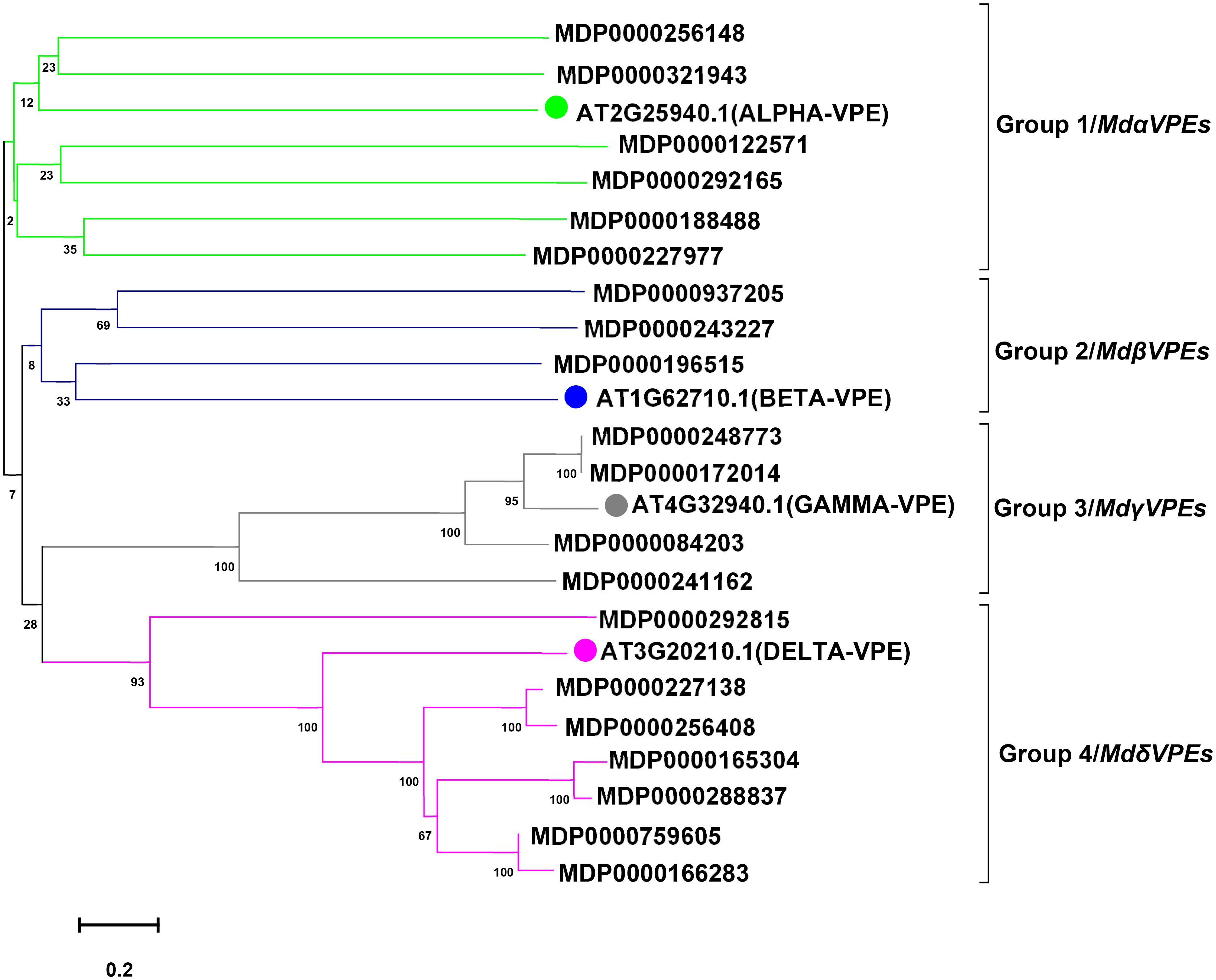
Figure 1. Phylogenetic analysis of apple and Arabidopsis VPE proteins. A total of 20 VPE proteins from apple and four from Arabidopsis were aligned using Clustal W. The phylogenetic tree was constructed using the MEGA 7.0 program by the neighbor-joining (NJ) method with bootstrap values 1,000 using protein sequences. According to Arabidopsis VPE protein sequcences, MdVPE proteins were devided into four groups. Different colors represent different groups.
Structural Characterization Analysis of MdVPEs
To identify the structure difference between MdVPEs, the gene and protein structure of MdVPEs were visualization by GSDS 2.0 and the MEME program. Most of the MdVPEs have the exons above five (including five) (90%), with only two members have less than five exons (MDP0000227977, three; MDP0000292165, four), and all the members contain introns (Figure 2B). The similar exon-intron structure of MdVPEs suggested that they has a closely evolutionary relationship and similar functions.
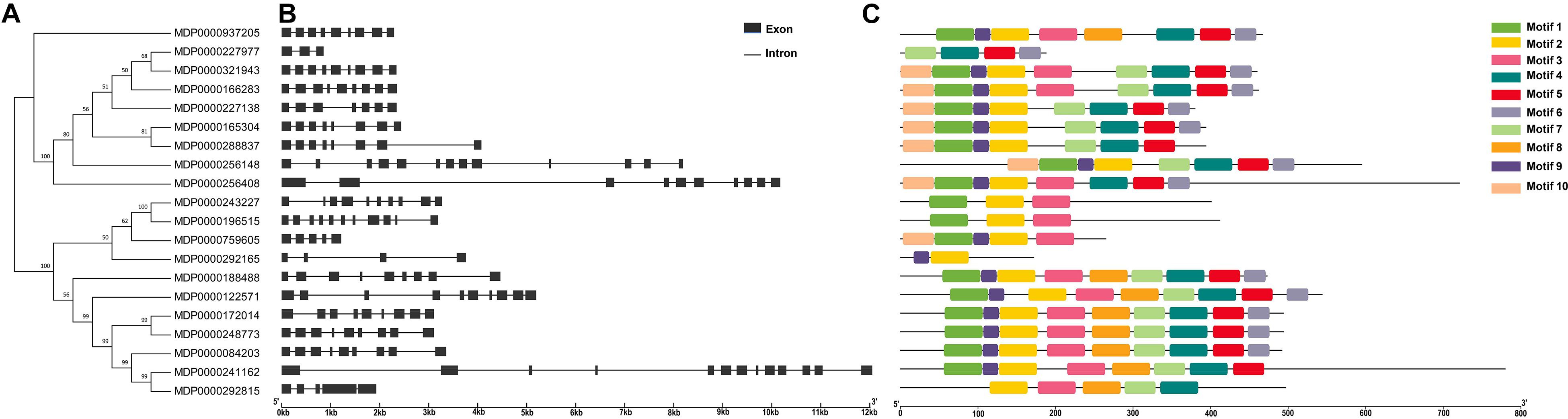
Figure 2. Phylogenetic relationship, gene structure, conserved motif and proteins structure analysis of MdVPE genes. (A) Phylogenetic tree of 20 MdVPE proteins. The unrooted neighbor-joining phylogenetic tree was constructed with MEGA 7.0 using full-length amino acid sequences of 20 MdVPE proteins, and the bootstrap test replicate was set as 1,000 times. (B) The exon-intron structure of MdVPE genes. Black boxes represent exons and black lines represent introns. (C) Distributions of conserved motifs in MdVPE genes. Ten putative motifs are indicated in different colored boxes. For details of motifs refer to Table 2.
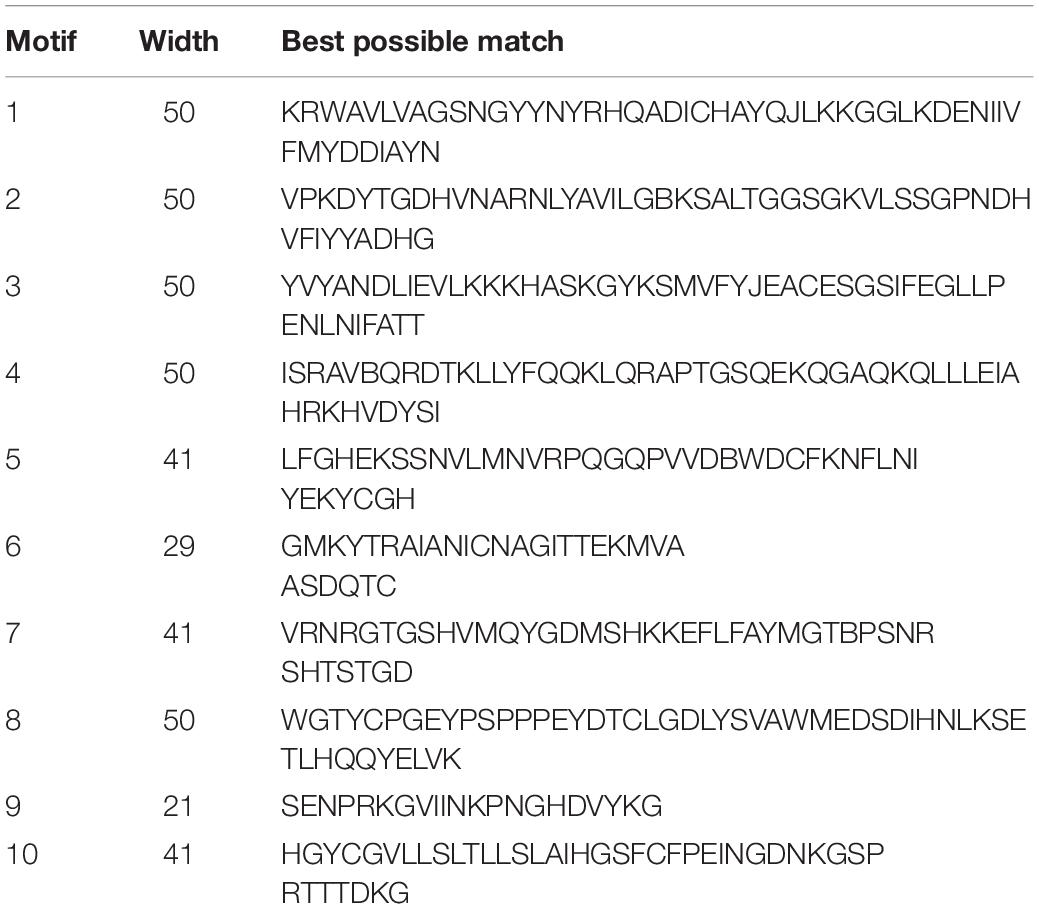
Ten conserved motifs of MdVPEs were identified by the MEME program, whose size ranged between 29 to 50 AA residues. Based on the NCBI-CDD, except the motif four, five and six is unknown, the others belong to the CASc superfamily, which is an important part of the Peptidase C_13 domain. Among the MdVPEs proteins, motifs five and seven near the N terminal, while motifs three and six near the C terminal and the motifs of all MdVPEs proteins are highly conserved (Figure 2C). In addition, all motifs details were listed in Table 2.
Chromosomal Locations and Genes Duplicated of the MdVPEs
To determine the distribution of MdVPE genes in 17 apple chromosomes, we analyzed their chromosomal location by MapDraw version 2.1. The MdVPE genes were spread among six chromosomes of the apple, including chromosome 4, 8, 9, 14, 15, and 17. Among them, chromosome 15 contains the most of MdVPE genes, followed by chromosome 4 (three) and chromosome 8 (two), whereas chromosome 9, 14, and 17 had only one MdVPEs gene (Figure 3A). The genetic distance ofMdVPEs genes was shown in Figure 3B and the MdVPE genes cluster were formed in chromosome 15 (black boxes). The uneven distribution of MdVPE genes on chromosomes indicated that genetic variations exist in the evolutionary process of apple.
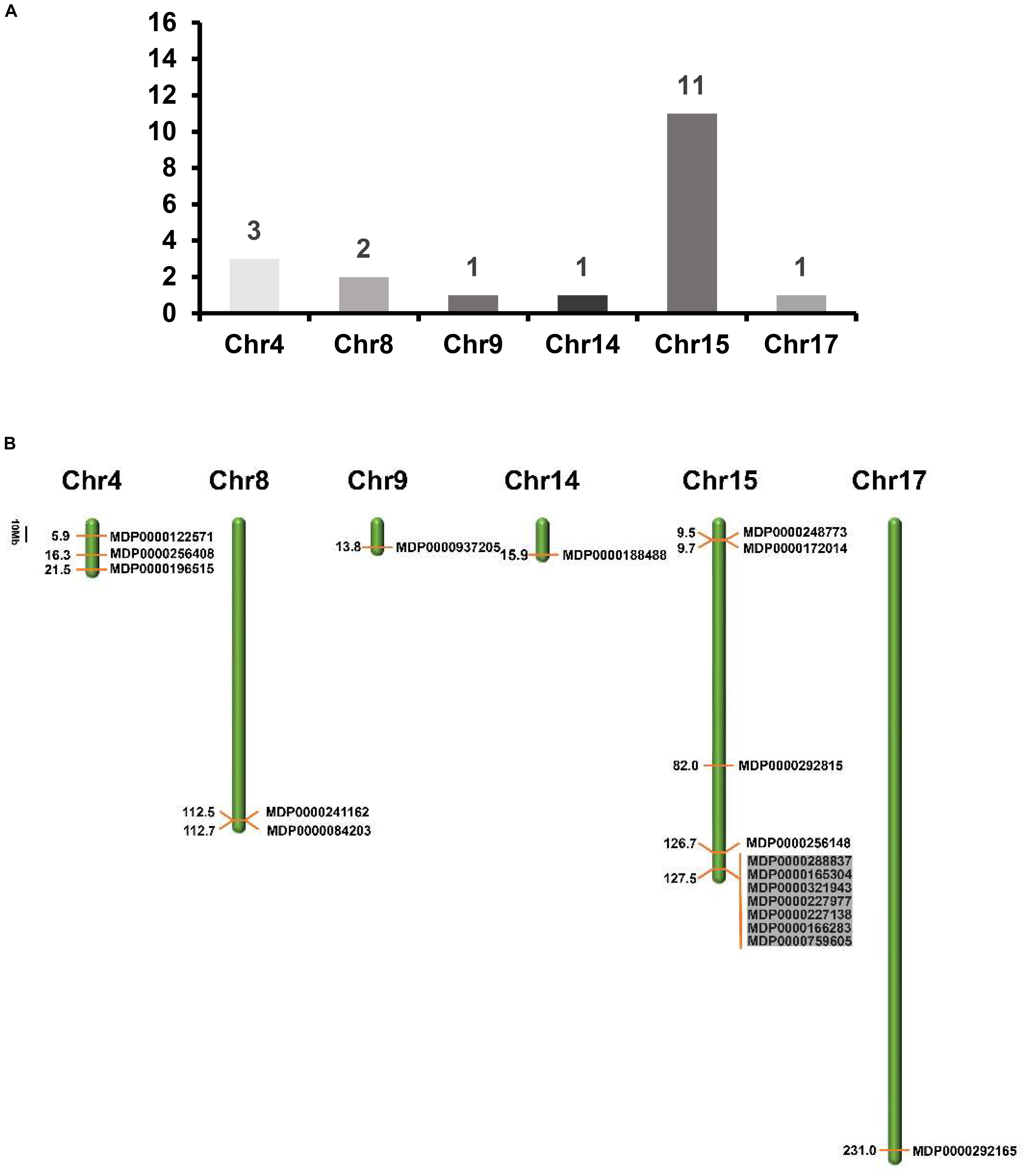
Figure 3. The distribution of MdVPE genes in apple chromosomes. (A) The number of MdVPE genes in each chromosome. (B) MdVPEs were mapped onto apple chromosomes. The black boxes represented clusters of genes.
During the evolutionary process of apple, genes duplication contributed to the generation of a gene family. Among them, segmental duplication played the most important role, followed by tandem duplication (Cannon et al., 2004). Thus, we analyzed the genes duplicated of the MdVPEs. Unfortunately, we have not found segmental duplication or segmental duplication in the MdVPE genes family. This suggested the expansion of the MdVPE genes family is slow.
Stress-Related Cis-Elements in MdVPEs Promoters
To predict the potential functions of MdVPEs under stresses, cis-element including w box, ABRE, LTR, and TC-rich repeats were analyzed by PlantCARE. All MdVPEs possessed at least one stress-response-related cis-element except MDP0000165304, MDP0000937205, and MDP0000292815, suggesting that the expressions of MdVPEs were associated with these abiotic stresses. In total, 14 MdVPEs (70.0%) had two or more ABREs, which suggested potential ABA response of MdVPEs, 10 MdVPEs (50.0%) had one or more w box, and this indicated MdVPEs may be involved in salt and drought stress. One or two LTRs existed in 11 MdVPEs, and one TC-rich repeats were found in two MdVPEs (Figure 4). The cis-element analysis indicated that MdVPEs may be involved in different abiotic stresses.
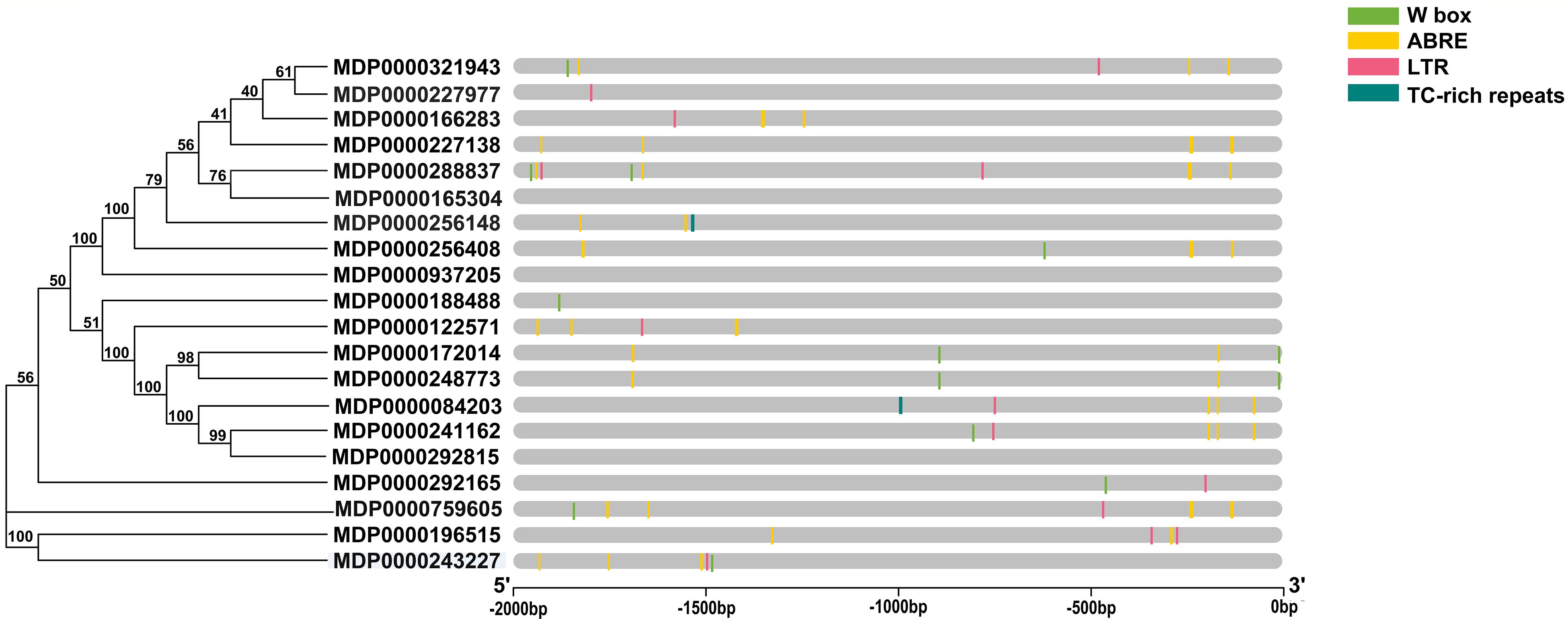
Figure 4. Predicted stress-related cis-elements in MdVPE promoters. Promoter sequences (–2,000 bp) of MdVPEs are analyzed by Plant CARE. Different color boxes represent different cis-element. According to the bottom scale, the upstream length to the translation start site can be inferred.
Expression Profile of MdVPEs in Different Tissues
Consider that any given gene functions is closely related to its expression patterns. Therefore, to provide more information for the study of MdVPEs potential function, we analyzed the expression patterns of 20 MdVPEs in different tissues including flowers, fruits, leaves, roots, stems, seeds, and seedlings based on NCBI-GEO (GSE42873) database. In the heatmap (Figure 5), red was used to represent a relatively high expression level, while the green was used to represent a relatively weak gene expression level. Some MdVPEs had similar expression patterns in various tissues. MDP0000241162, MDP0000288837, MDP0000292815, MDP0000188488, MDP0000165304, MDP0000256408, and MDP0000256418 has relatively higher expression levels in flowers, fruits, and leaves, but relatively lower in roots, stems, seeds, and seedlings. Although MDP0000122571, MDP0000227977, MDP0000166283, MDP0000227138, and MDP0000258293 also were relatively higher expression levels in flowers, fruits, and leaves, they had more higher expression levels in stems (MDP0000227977, MDP0000166283, MDP0000227138, and MDP0000258293) or seeds and seedlings (MDP0000122571). Additionally, MDP0000122571 and MDP0000321943 had not much different expression levels in various tissues. Furthermore, some MdVPE genes had higher expression levels in specific tissues. For example, MDP0000227977, MDP0000166283, MDP0000227138, and MDP0000258293 have the highest expression levels in stems, this suggested MDP0000227977, MDP0000166283, MDP0000227138, and MDP0000258293 may contributed to the function exertion of stem. Additionally, MDP0000122571 has the highest expression levels in seeds and seedlings, this implied MDP0000122571 may play a central role in the development of seeds and seedlings in apple. Interestingly, MDP0000084203, MDP0000172014, and MDP0000248773 (all belong to MdγVPEs) have the highest expression levels in roots, and this indicated they may be involved in the response to soil stresses or the development of roots. In summary, the different expression patterns of MdVPEs in various tissues suggested different MdVPEs played a different role in different tissues of apple.
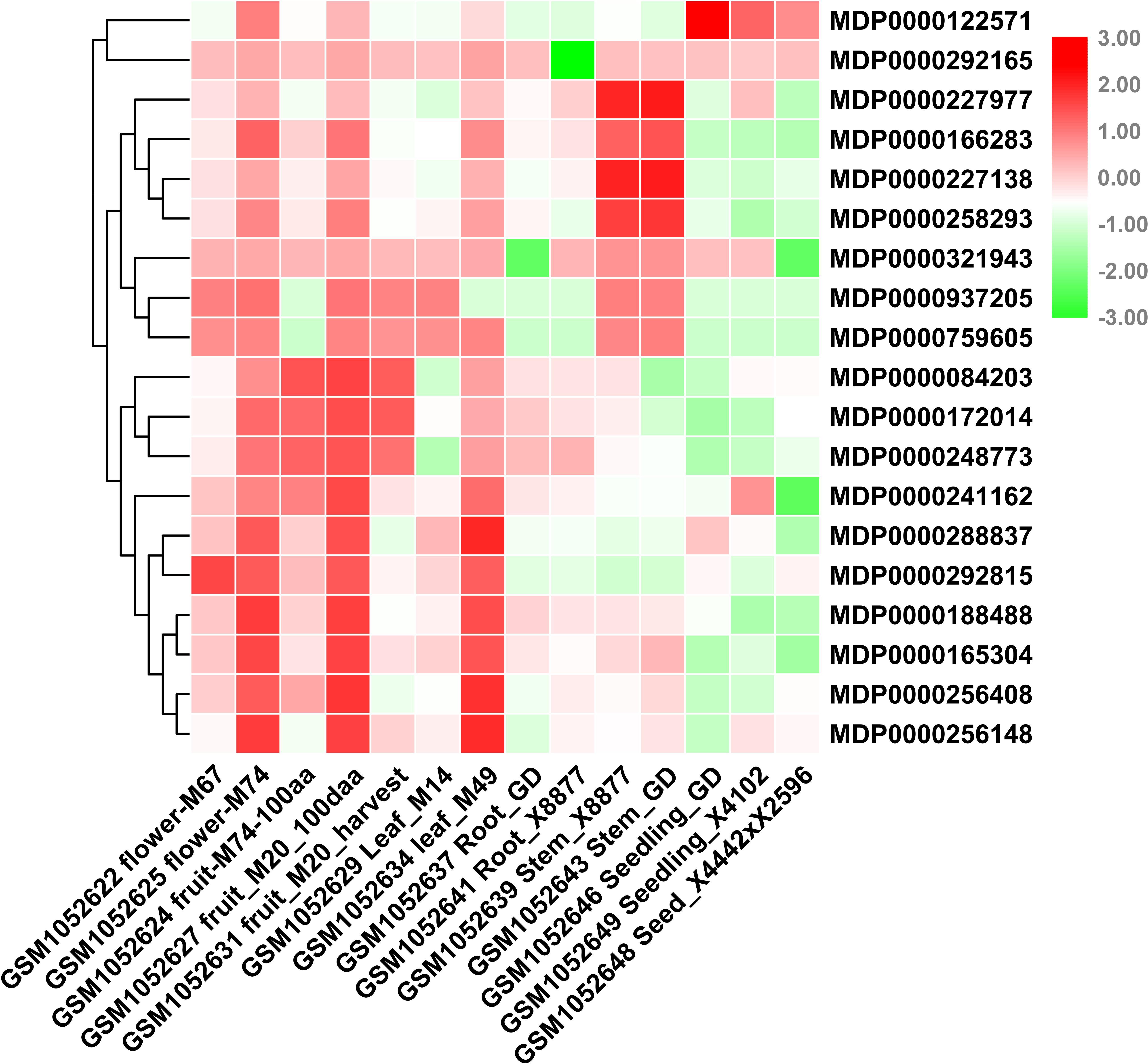
Figure 5. Expression profiles of MdVPE genes in different tissues of apple. NCBI-GEO data (GSE42873) were downloaded for the expression profile analysis. GSM1052622 flower-M67 and GSM1052625 flower-M74 represent the whole flower of M67 and the whole flower of M74, respectively. GSM1052624 fruit-M74-100aa, GSM1052627 fruit-M20-100aa, and GSM1052631 fruit-M20-harvest represent the 100 days after anthesis-fruit of M74, the 100 days after anthesis-fruit of M20 and fruit flesh at harvest of M20, respectively. GSM1052629 leaf-M14 and GSM1052634 leaf-M49 represents the whole leaf of M14, the whole leaf of M49, respectively. GSM1052637 Root-GD and GSM1052641 Root-X8877 represents the in vitro root of Golden Delicious, the in vitro root of X8877, respectively. GSM1052639 stem-X8877 and GSM1052643 stem-GD represent the fully developed-stem of X8877, the fully developed-stem of Golden Delicious, respectively. GSM1052646 seeding-GD and GSM1052649 seeding-X41002 represents the 10 days old-seedling of Golden Delicious, the 10 days old-seedling of X41002, respectively. GSM1052648 seed-X4442 × X2596 represents the dormant seed from cross X4442 and X2596. M67, M74, M20, M14, M49, X8877, Golden Delicious, X41002, X4442, and X2596 represent apple cultivars. RPKM values of MdVPE genes were transformed by log2 and the heatmap was constructed by TBtools.
Expression Profile of MdVPEs With the Infection of P. ultimum and Apple Stem Grooving Virus
VPEs had been reported play an important role in disease resistance of plants (Vorster et al., 2019). P. ultimum seriously threatened to the quality and yield of apples. To understand the potential involvement of MdVPE genes in response to P. ultimum, we analyzed the changes of transcription levels of VPE family genes in response to P. ultimum in apple via NCBI-GEO data (GSE62103). The expression levels of MDP0000172104, MDP0000288837, MDP0000188488, MDP0000196515, MDP0000243227, and MDP0000241162 were down-regulated, while the expression levels of MDP0000321943, MDP0000292165, MDP0000759605, MDP0000166283, and MDP0000256408 were up-regulated during the infection of P. ultimum (Figure 6A).
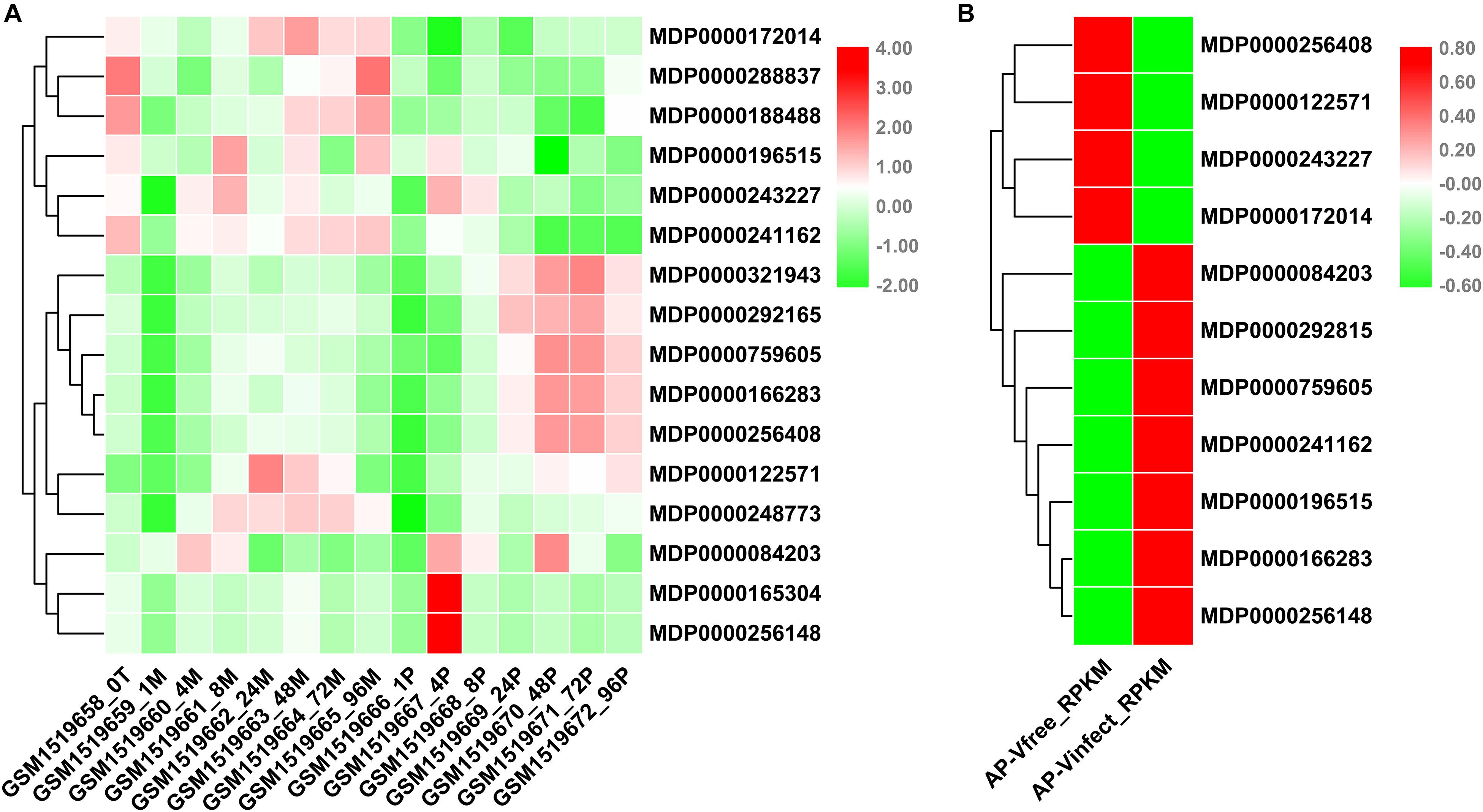
Figure 6. Expression profile of MdVPEs under P. ultimum and Apple Stem Grooving Virus (ASGV) stress. (A) Expression profile of MdVPEs under Pythium ultimum stress. NCBI-GEO data (GSE62103) were downloaded for the expression profile analysis. GSM1519658 represents the apple root organization at 0 h, GSM1519662, GSM1519663, GSM1519664, and GSM1519665 represents the simulated inoculation mold 24, 48, 72, and 96 h, and the apple root organization, GSM1519669, GSM1519670, GSM1519671, and GSM1519672 represents apple root tissue when inoculated with humus 24, 48, 72, and 96 h, respectively. (B) Expression profile of MdVPEs under ASGV stress. NCBI-GEO data (GSE53825) were downloaded for the expression profile analysis. AP-Vinfect represents ASGV-infected asymptomatic apple plantlets and AP-Vfree represents virus-free apple plantlets. RPKM values of MdVPE genes were transformed by log2 and the heatmap was constructed by TBtools.
ASGV, a kind of latent viruses, is one of the most difficult viruses to remove, in spite of the method of tissue culture were used. Through the analysis of NCBI-GEO database (GSE58293), we found that the expression levels of MDP0000256408, MDP0000122571, MDP0000243227 and MDP0000172014 were down-regulated, and the expression levels of MDP0000084203, MDP0000292815, MDP0000759605, MDP0000241162, MDP0000196515, MDP0000166283, and MDP0000256148 were up-regulated (Figure 6B). Above all, the MDP0000243227 and MDP0000172014 had similar expression patterns under biotic stress including P. ultimum and ASGV, while MDP0000759605 and MDP0000166283 were up-regulated. This results indicated MdVPE family genes have a diverse roles in responses to biotic stress and MDP0000243227, MDP0000172014, MDP0000759605, and MDP0000166283 may be broad-spectrum disease-resistant genes.
Expression Analysis of the Selected Genes in Response to Abiotic by qRT-PCR
Increasing studies reported that VPEs played an important role in plants response to abiotic stress (Li et al., 2012; Misas-Villamil et al., 2013; Kariya et al., 2018; Locato and De Gara, 2018). Salt, cadmium, low-temperature, and drought stress seriously effected the apple trees growth and development. To reveal the potential function of MdVPEs in response to abiotic stress, the expression levels of 18 selected MdVPE genes were measured under abiotic stress by qRT-PCR. Considered that the root is the most direct and sensitive organs to feel the changes in the soil environment than the other organs of plants. Vegetative vacuole played a more important role than seed vacuole in the response to stress (Christoff and Rogerio, 2014), and among vegetable organs, fresh roots are rich in vacuoles. Furthermore, most VPEs were predicted to be vacuolar proteins (Table 1). Based on the above views, we selected fresh roots as the samples to investigate the expression patterns of MdVPE genes in response to abiotic stress. The expression of all MdVPEs were up-regulated under salt stress. Except the down-regulation of MDP0000243227 and the insensitivity of MDP0000188488, MDP0000937205, and MDP0000227977, the other MdVPEs were up-regulated under drought stress. Except MDP0000256408, MDP000084203, and MDP0000241162, most of MdVPEs were sensitive to cadmium stress. Furthermore, except MDP0000256408, MDP0000241162, MDP0000188488, MDP0000759605, and MDP0000196515, the other MdVPEs were sensitive to low-temperature stress. Interestingly, MDP0000084203, MDP0000241162, and MDP0000172014, three MdVPE genes belong to MdγVPEs (Figure 1), have similar expression patterns, that were more strongly response to drought and salt stress than cadmium and low-temperature stress (Figure 7).
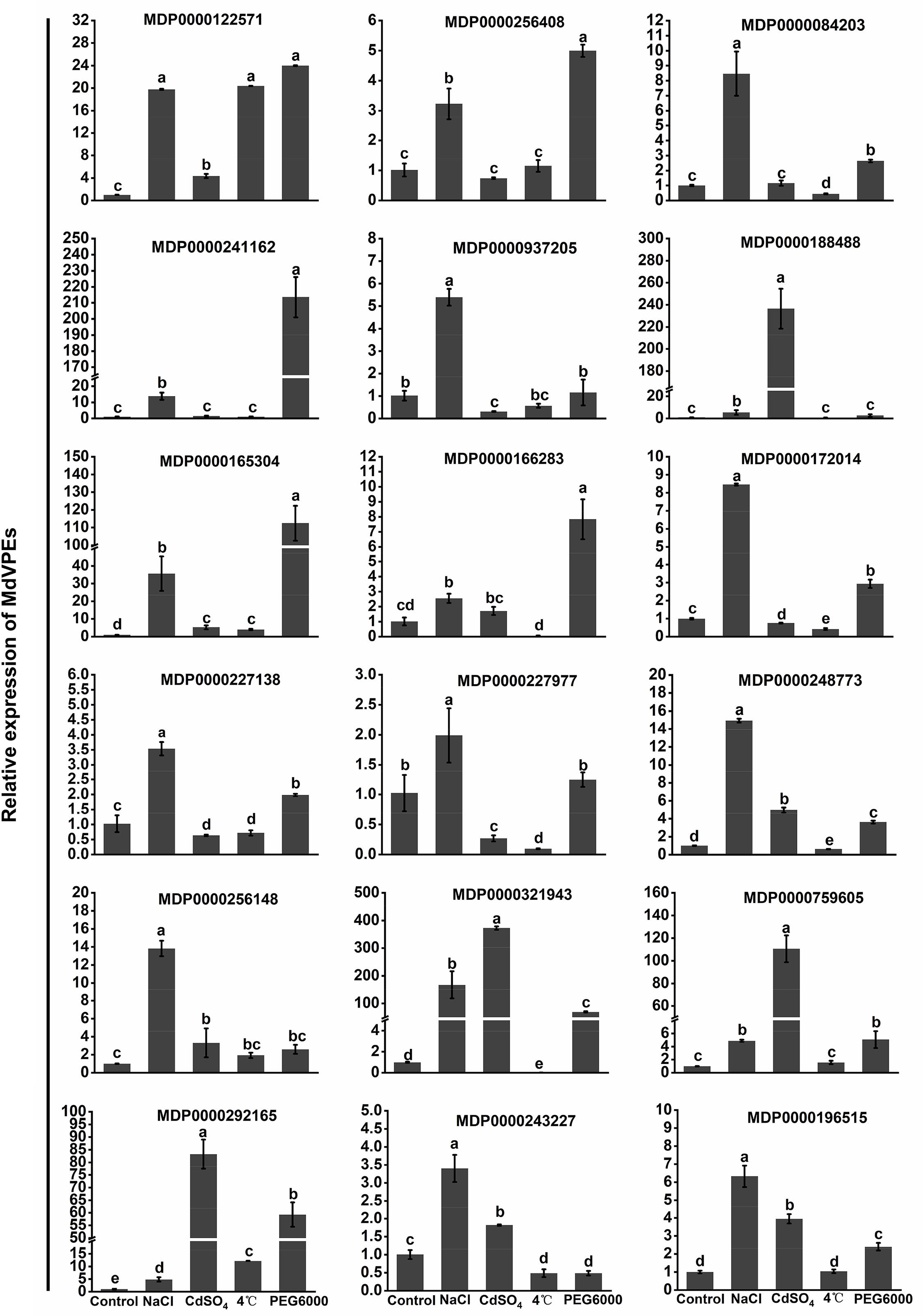
Figure 7. Expression profiles of MdVPE genes under salt, cadmium, cold, and drought stresses. Quantitative RT-PCR was used to investigate the expression levels of each MdVPE genes in apple roots. The data are presented as means ± standard deviations (SD) of three independent measurements from three individual plants. Means with different small letters in the column are significantly different at P < 0.05.
Overexpression of MDP0000172014 Decreased Arabidopsis thaliana Tolerance to Salt Stress
Considered that (1) the salt is the most seriously abiotic stress threated growth and development of apple trees; (2) previous studies had reported that γVPE played important role in biotic and abiotic stress resistance (Hatsugai et al., 2004; Christoff et al., 2014; Ran et al., 2014; Su et al., 2015) and the MDP0000172014 belongs to γVPE via the analysis of phylogenetic trees (Figure 1); (3) the MDP0000172014 were strongly induced by salt and drought stress, two stress all caused osmotic stress for plants (Figure 7); (4) the cis-acting element including w box and ABRE were found in the promoter of MDP0000172014 (Figure 4). To further reveal the MDP0000172014 functions in response to salt stress, we construsted the pBI121-35S-MDP0000172014 vector and transformed it into Arabidopsis. The results of Kanamycin screening in T1 and expression analysis of T3 showed that the pBI121-35S-MDP0000172014 vector had been successfully transferred to Col-1 Arabidopsis thaliana (Supplementary Figure 2). Moreover, T1 seeds were further screened to T3 on kanamycin resistance medium. Transgenetic lines showed shorter roots length (Figures 8A,B) as well as significantly higher MDA content (Figure 8C), H2O2 content (Figure 8D) and cell death number (Figure 8E) than control lines. These results suggested that overexpression of MDP0000172014 decreased Arabidopsis tolerance to salt stress and promoted salt stress-induced roots cell death.
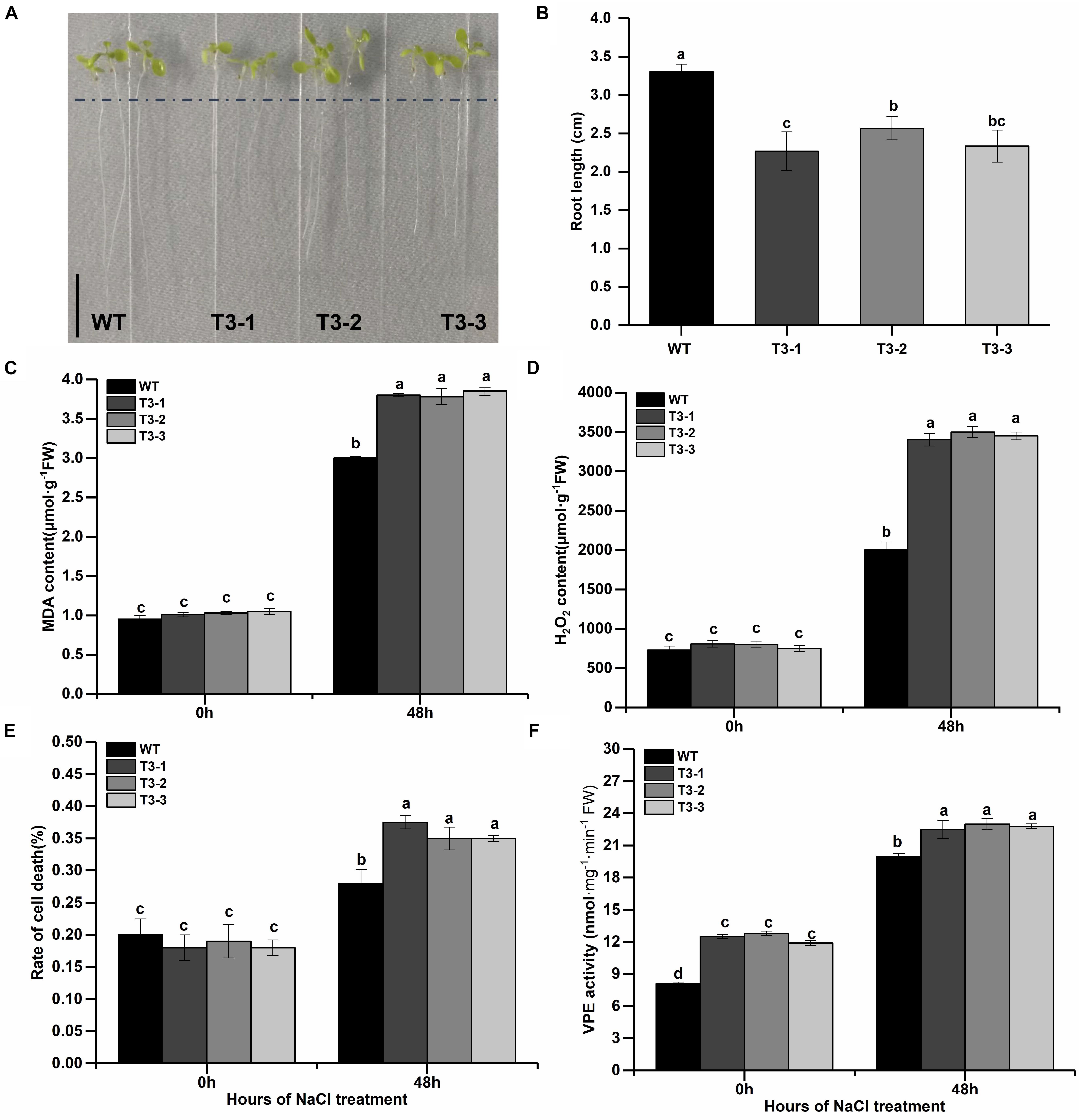
Figure 8. Overexpression of MDP0000172014 decreased the tolerance of Arabidopsis to salt stress. (A) Phenotype comparisons (B) and root length between the wild type (WT) and overexpressed MDP0000172014 transgenic Arabidopsis seedlings (T3-1, T3-2, and T3-3) after 100 mM NaCl treatment, and the length of the black line represents 1 cm. (C) The MDA, (D) H2O2, (E) rate of cell death, and (F) VPE activity of WT and transgenic Arabidopsis after 200 mM NaCl treatment. The data are presented as means ± standard deviations (SD) of three independent measurements from three individual plants. Means with different small letters in the column are significantly different at P < 0.05.
Discussion
Increasing studies showed that PCD played a central role in plants developmental process and stress responses (Hatsugai et al., 2015). In animals, the performed of PCD requires caspase cascade including caspase-1 as the initiator and caspase-3 as the executor (Fagundes et al., 2015). Because of VPEs with caspase-1-like activity, it had been known as the key activator of PCD in plants (Hatsugai et al., 2015). Additionally, VPEs composed of a N-terminal propeptides (NTPP), a C-terminal propeptides (CTPP) and a peptidase_C13 domain (Kuroyanagi et al., 2002), and VPEs also shared high identity in AA sequence (Kuroyanagi et al., 2002). By following criteria, we identified 20 VPEs genes in the apple genome (Table 1). Apple has more members than Arabidopsis (four), barley (eight), and tomato (fourteen) (Vorster et al., 2019). Based on ProtComp version 9.0 program, all subcellular localizations of MdVPE proteins were predicted, interestingly, MDP0000243227 and MDP0000196515 were located in ER which different from the others, which were located in vacuole. This may suggest that MDP0000243227 and MDP0000196515 had different sites of action from others.
According to homology and functional characteristics, VPEs has been divided into three sub-families, including vegetative VPEs (γVPE and δVPE), seed VPEs (αVPE and βVPE) and uncharacterized VPEs (Christoff et al., 2014). The seed VPEs mainly play an important role in plant development and growth, while vegetative VPEs mainly performed functions in stress responses. The NJ phylogenetic trees were constructed for MdVPEs with Arabidopsis, rice, soybean, and tobacco VPEs, which demonstrated their evolutionary relationships and potential similarities of function (Figure 1, Supplementary Figure 1). Following it, we divided the MdVPEs into four groups, including MdαVPEs, MdβVPEs, MdγVPEs, and MdδVPEs, and these phylogenetic trees would be the basic information for future study of MdVPEs. Considered that genes function were related to their protein structure, the intron-exon structure, motif, and protein structure of MdVPEs were analyzed, and these are highly conserved in MdVPEs. For example, the homology of MDP0000321943, MDP0000166283, and MDP0000937205 reached over 90%. Moreover, almost all MdVPEs have the same motifs in N terminal and C terminal, which belong to CASc superfamily via the analysis of NCBI-CDD and SMART (Figures 2A–C). However, different and similar motif numbers present in the MdVPEs may indicate diverse and similarities functions between various MdVPEs.
Increasing studies reported that VPEs involved in plants growth and development, such as participation in tomato sucrose accumulation (Wang et al., 2016), the radish bud abortive (Zhang et al., 2013), and the lack of advantage at the top of the potato tuber (Teper-Bamnolker et al., 2012, 2017). GEO-database (GSE42873) analysis showed the different expression patterns of MdVPEs in different tissues, and some of them had similar expression patterns. This may suggest that MdVPEs played different roles in different tissues of apple trees and some of them had similar functions. Additionally, some MdVPEs had relatively higher expression level in specific tissues (Figure 5), this may indicate them contributed to exercise of function in specific tissues. The important functions of VPE in response to biotic and abiotic stress have been widely reported in pervious study. For example, VPE increased tobacco tolerance to TMV via promoting hypersensitive death (HR) (Hatsugai et al., 2004), and promoted the cell death in plants under heavy mental stress (Kariya et al., 2013, 2018; Ran et al., 2014; Cai et al., 2018). MPK6 promoted PCD of Arabidopsis with heat stock treatment though VPEs (Li et al., 2012), Moreover, GmNAC30 and GmNAC81 integrate the ER stress-induced cell death responses through γVPE (Mendes et al., 2013). Here, GEO database were used to analyze the expression patterns of MdVPEs during the infection of P. ultimum and ASGV, two diseases hardly removed from apple trees, we found that except MDP0000084203, MDP0000165304, and MDP0000256148, the other MdVPEs were involved in P. ultimum and ASGV stress response. Among them, MdγVPEs, including MDP0000172014 and MDP0000243227 were down-regulated, while MDP0000759605 and MDP0000166283 were up-regulated under the infection of P. ultimum and ASGV (Figure 6), and this suggested them may be broad-spectrum disease-resistant genes. Analysis of cis-element showed in the promoter of MdVPEs had w box, ABRE, LTR, and TC-rich repeats (Figure 4), which indicated MdVPEs may response to drought, ABA and low-temperature. Additionally, expression analysis of 18 selected MdVPEs showed except MDP0000243227, MDP0000188488, MDP0000937205, and MDP0000227977, the other MdVPEs were up-regulated under salt and drought stress. Most MdVPEs were sensitive to cadmium and low-temperature stress (Figure 7), which indicated MdVPEs may perform important roles in abiotic stress response including salt, cadmium, low-temperature, and drought stress. Mendes et al. (2013) had reported GmNAC30 and GmNAC81 mediated osmotic stress-induced cell death through γVPE. Interestingly, MDP0000084203, MDP0000241162, and MDP0000172014, three MdVPE genes belong to MdγVPEs (Figure 1), have similar expression patterns, that were more strongly response to drought and salt stress (all construsted osmotic stress) for plants than cadmium and low-temperature stress (Figure 7). This indicated MdγVPEs may play a more central role than other kinds of MdVPEs in deafsing to drought and salt stress in apple.
Salt stress seriously threaten the quality and yield of apple. MdγVPEs were found may play a more important function under salt stress, and the ABRE was also found in the promoter of MDP0000172014, a MdγVPE gene. Therefore, we selected MDP0000172014 as the representative, which was used to reveal the function of MDP0000172014 in response to salt stress. The transgenic Arabidopsis of MDP0000172014 were significantly sensitive to salt than wildtype Arabidopsis (Figure 8), suggesting that MdγVPEs enhanced the sensibility of apple trees to salt stress.
In conclusion, our comprehensive analysis including the identified of MdVPEs’ characteristics and structure, and the expression analysis under stress, filling the blank of woody plants VPE information and laid a foundation for the further exploration of VPE functions in apple stress resistance. However, the resistance molecular mechanism of them also needed to be further explored.
Data Availability Statement
The datasets presented in this study can be found in NCBI (https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE42873, GSE62103, and GSE53825.
Author Contributions
JS and WZ designed the experiment. FY and MX performed the MDA and H2O2 content experiments. LX and XT performed the data analysis. JS drafted the manuscript. WZ revised the manuscript. HY was the project leader. All authors read and approved its final version.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31772251), National Key R&D Program of China (No. 2019YFD1000103), Shandong Province Natural Science Foundation (No. ZR2018ZC08N3), and Shandong Province Major Science and Technology Innovation Projects, China (No. 2018CXGC0207).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00626/full#supplementary-material
Footnotes
- ^ http://pfam.xfam.org/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ https://www.arabidopsis.org/
- ^ http://smart.embl-heidelberg.de/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://web.expasy.org/protparam/
- ^ http://gsds.cbi.pku.edu.cn/index.php
- ^ http://alternate.meme-suite.org/tools/meme
- ^ http://linux.softberry.com/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html
- ^ https://www.ncbi.nlm.nih.gov/geo/
References
Baskett, A. J. (2012). A Type II Metacaspase Interacts With RPS1-K-2 in Soybean and Analysis of the Soybean Metacaspase Gene Family. Dissertations Gradworks, Londonderry.
Cai, Y. M., Yu, J., Ge, Y., Mironov, A., and Gallois, P. (2018). Two proteases with caspase-3-like activity, cathepsin B and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis. New Phytol. 218, 1143–1155. doi: 10.1111/nph.14676
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chen, C. J., Xia, R., Chen, H., and He, Y. H. (2018). TBtools, a toolkit for biologists integrating various HTS-data2 handling tools with a user-friendly interface. BioRxiv [Preprint], doi: 10.1101/069187
Christoff, A. P., and Rogerio, M. (2014). The diversity of rice phytocystatins. Mol. Gen. Genomics 289, 1321–1330.
Christoff, A. P., Turchetto-Zolet, A. C., and Margis, R. (2014). Uncovering legumain genes in rice. Plant Sci. 215-216, 100–109.
Cilliers, M., Wyk, S. G. V., Kunert, K. J., and Vorster, B. J. (2017). Identification and changes of the drought-induced cysteine protease transcriptome in soybean (Glycine max) root nodules. Environ. Exp. Bot. 148:S0098847217303118.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
De Pinto, M. C., Locato, V., and De Gara, L. (2012). Redox regulation in plant programmed cell death. Plant Cell Environ. 35, 234–244. doi: 10.1111/j.1365-3040.2011.02387.x
Del Pozo, O., and Lam, E. (1998). Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol. 8:R896. doi: 10.1016/s0960-9822(07)00555-6
Deng, M. J., Bian, H. W., Xie, Y. K., Kim, Y. H., Wang, W. Z., Lin, E., et al. (2011). Bcl-2 suppresses hydrogen peroxide-induced programmed cell death via OsVPE2 and OsVPE3, but not via OsVPE1 and OsVPE4, in rice. FEBS J. 278, 4797–4810. doi: 10.1111/j.1742-4658.2011.08380.x
Fagundes, D., Bohn, B., Cabreira, C., Leipelt, F., Dias, N., Bodanese-Zanettini, M. H., et al. (2015). Caspases in plants: metacaspase gene family in plant stress responses. Funct. Integr. Genomics 15, 639–649. doi: 10.1007/s10142-015-0459-7
Gong, P. J., Li, Y., Tang, Y. J., Wei, R., Zhu, H. Z., Wang, Y. J., et al. (2018). Vacuolar processing enzyme (VvbetaVPE) from Vitis vinifera, processes seed proteins during ovule development, and accelerates seed germination in VvbetaVPE heterologously over-expressed Arabidopsis. Plant Sci. 274, 420–431. doi: 10.1016/j.plantsci.2018.06.023
Hatsugai, N., Kuroyanagi, M., Yamada, K., Meshi, T., Tsuda, S., Kondo, M., et al. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305, 855–858. doi: 10.1126/science.1099859
Hatsugai, N., Yamada, K., Goto-Yamada, S., and Hara-Nishimura, I. (2015). Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 6:234. doi: 10.3389/fpls.2015.00234
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jiang, J. J., Hu, J. Z., Tan, R. J., Han, Y. H., and Li, Z. Y. (2019). Expression of IbVPE1 from sweet potato in Arabidopsis affects leaf development, flowering time and chlorophyll catabolism. BMC Plant Biol. 19:184. doi: 10.1186/s12870-019-1789-8
Kariya, K., Demiral, T., Sasaki, T., Tsuchiya, Y., Turkan, I., Sano, T., et al. (2013). A novel mechanism of aluminum-induced cell death involving vacuolar processing enzyme and vacuolar collapse in tobacco cell line BY-2. J. Inorg. Biochem. 128, 196–201. doi: 10.1016/j.jinorgbio.2013.07.001
Kariya, K., Tsuchiya, Y., Sasaki, T., and Yamamoto, Y. (2018). Aluminium-induced cell death requires upregulation of NtVPE1 gene coding vacuolar processing enzyme in tobacco (Nicotiana tabacum L.). J. Inorg. Biochem. 181, 152–161. doi: 10.1016/j.jinorgbio.2017.09.008
Kim, Y., Wang, M. Q., Bai, Y., Zeng, Z. H., Guo, F., Han, N., et al. (2014). Bcl-2 suppresses activation of VPEs by inhibiting cytosolic Ca(2)(+) level with elevated K(+) efflux in NaCl-induced PCD in rice. Plant Physiol. Biochem. 80, 168–175. doi: 10.1016/j.plaphy.2014.04.002
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuroyanagi, M., Nishimura, M., and Hara-Nishimura, I. (2002). Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 43, 143–151. doi: 10.1093/pcp/pcf035
Kuroyanagi, M., Yamada, K., Hatsugai, N., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2005). Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 280, 32914–32920. doi: 10.1074/jbc.M504476200
Lam, E., and Zhang, Y. (2012). Regulating the reapers: activating metacaspases for programmed cell death. Trends Plant Sci. 17, 487–494. doi: 10.1016/j.tplants.2012.05.003
Lamkanfi, M., Declercq, W., Kalai, M., Saelens, X., and Vandenabeele, P. (2002). Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 9, 358–361. doi: 10.1038/sj.cdd.4400989
Landi, M. (2017). Commentary to: Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds by Hodges et al., Planta (1999) 207:604-611. Planta 245:1067. doi: 10.1007/s00425-017-2699-3
Li, Z., Yue, H. Y., and Xing, D. (2012). MAP Kinase 6-mediated activation of vacuolar processing enzyme modulates heat shock-induced programmed cell death in Arabidopsis. New Phytol. 195, 85–96. doi: 10.1111/j.1469-8137.2012.04131.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Locato, V., and De Gara, L. (2018). Programmed cell death in plants: an overview. Methods Mol. Biol. 1743, 1–8. doi: 10.1007/978-1-4939-7668-3_1
Mao, K., Dong, Q. L., Li, C., Liu, C. H., and Ma, F. W. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant. Sci. 8:480. doi: 10.3389/fpls.2015.00480
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Mendes, G. C., Reis, P. A., Calil, I. P., Carvalho, H. H., Aragao, F. J., and Fontes, E. P. (2013). GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. U.S.A. 110, 19627–19632. doi: 10.1073/pnas.1311729110
Meng, D., Li, Y. Y., Bai, Y., Li, M. J., and Cheng, L. L. (2016). Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 103, 71–83. doi: 10.1016/j.plaphy.2016.02.006
Misas-Villamil, J. C., Toenges, G., Kolodziejek, I., Sadaghiani, A. M., Kaschani, F., Colby, T., et al. (2013). Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 73, 689–700. doi: 10.1111/tpj.12062
Patterson, B. D., Macrae, E. A., and Ferguson, I. B. (1984). Estimation of hydrogen peroxide in plant extracts using titanium(IV). Anal. Biochem. 139, 487–492. doi: 10.1016/0003-2697(84)90039-3
Radchuk, V., Tran, V., Radchuk, R., Diaz-Mendoza, M., Weier, D., Fuchs, J., et al. (2018). Vacuolar processing enzyme 4 contributes to maternal control of grain size in barley by executing programmed cell death in the pericarp. New Phytol. 218, 1127–1142. doi: 10.1111/nph.14729
Ran, K., Yang, H. Q., Sun, X. L., Li, Q., Jiang, Q. Q., Zhang, W. W., et al. (2014). Isolation, characterization, and structure analysis of a vacuolar processing enzyme gene (MhVPEgamma) from Malus hupehensis (Pamp) Rehd. Appl. Biochem. Biotechnol. 173, 579–595. doi: 10.1007/s12010-014-0867-5
Rojo, E., Martin, R., Carter, C., Zouhar, J., Pan, S., Plotnikova, J., et al. (2004). VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 14, 1897–1906. doi: 10.1016/j.cub.2004.09.056
Shimada, T., Takagi, J., Ichino, T., Shirakawa, M., and Hara-Nishimura, I. (2018). Plant Vacuoles. Annu. Rev. Plant Biol. 69, 123–145.
Shimada, T., Yamada, K., Kataoka, M., Nakaune, S., Koumoto, Y., Kuroyanagi, M., et al. (2003). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278, 32292–32299. doi: 10.1074/jbc.M305740200
Su, Q., Ran, K., Men, X. J., Zhang, W. W., Fan, S. L., Yan, L. J., et al. (2015). Response of vacuolar processing enzyme in Malus hupehensis and MhVPEγ -overexpressing Arabidopsis to high temperature stress. Acta Physiol. Plant. 37:82.
Teper-Bamnolker, P., Buskila, Y., Belausov, E., Wolf, D., Doron-Faigenboim, A., Ben-Dor, S., et al. (2017). Vacuolar processing enzyme activates programmed cell death in the apical meristem inducing loss of apical dominance. Cell 40, 2381–2392. doi: 10.1111/pce.13044
Teper-Bamnolker, P., Buskila, Y., Lopesco, Y., Ben-Dor, S., Saad, I., Holdengreber, V., et al. (2012). Release of apical dominance in potato tuber is accompanied by programmed cell death in the apical bud meristem. Plant Physiol. 158, 2053–2067. doi: 10.1104/pp.112.194076
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tsiatsiani, L., Van Breusegem, F., Gallois, P., Zavialov, A., Lam, E., and Bozhkov, P. V. (2011). Metacaspases. Cell Death. Differ. 18, 1279–1288.
Van Wyk, S. G., Du Plessis, M., Cullis, C. A., Kunert, K. J., and Vorster, B. J. (2014). Cysteine protease and cystatin expression and activity during soybean nodule development and senescence. BMC Plant Biol. 14:294. doi: 10.1186/s12870-014-0294-3
Vartapetian, A. B., Tuzhikov, A. I., Chichkova, N. V., Taliansky, M., and Wolpert, T. J. (2011). A plant alternative to animal caspases: subtilisin-like proteases. Cell Death. Differ. 18, 1289–1297. doi: 10.1038/cdd.2011.49
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839. doi: 10.1186/s12864-015-2096-x
Vorster, B. J., Cullis, C. A., and Kunert, K. J. (2019). Plant vacuolar processing enzymes. Front. Plant Sci. 10:479. doi: 10.3389/fpls.2015.00479
Wang, N., Duhita, N., Ariizumi, T., and Ezura, H. (2016). Involvement of vacuolar processing enzyme SlVPE5 in post-transcriptional process of invertase in sucrose accumulation in tomato. Plant Physiol. Biochem. 108, 71–78. doi: 10.1016/j.plaphy.2016.06.037
Wang, W., Xiong, H. Y., Lin, R. X., Zhao, N. T., Zhao, P., and Sun, M. X. (2019). A VPE-like protease NtTPE8 exclusively expresses in the integumentary tapetum and is involved in seed development. J. Integr. Plant Biol. 61, 598–610. doi: 10.1111/jipb.12766
Wang, Y. H., Zhu, S. S., Liu, S. J., Jiang, L., Chen, L. M., Ren, Y. L., et al. (2009). The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 58, 606–617. doi: 10.1111/j.1365-313X.2009.03801.x
Woltering, E. J., Van Der Bent, A., and Hoeberichts, F. A. (2002). Do plant caspases exist? Plant Physiol. 130, 1764–1769. doi: 10.1104/pp.006338
Yamada, K., Basak, A. K., Goto-Yamada, S., Tarnawska-Glatt, K., and Hara-Nishimura, I. (2019). Vacuolar processing enzymes in the plant life cycle. New Phytol. 226, 21–31. doi: 10.1111/nph.16306
Zhang, J., Li, Q. F., Huang, W. W., Xu, X. Y., Zhang, X. L., Hui, M. X., et al. (2013). A vacuolar processing enzyme RsVPE1 gene of radish is involved in floral bud abortion under heat stress. Int. J. Mol. Sci. 14, 13346–13359. doi: 10.3390/ijms140713346
Keywords: Genome-wide analysis, VPE gene family, diverse expression, salt stress, apple (Malus × domestic)
Citation: Song J, Yang F, Xun M, Xu L, Tian X, Zhang W and Yang H (2020) Genome-Wide Identification and Characterization of Vacuolar Processing Enzyme Gene Family and Diverse Expression Under Stress in Apple (Malus × Domestic). Front. Plant Sci. 11:626. doi: 10.3389/fpls.2020.00626
Received: 24 December 2019; Accepted: 22 April 2020;
Published: 26 May 2020.
Edited by:
Rosa M. Rivero, Spanish National Research Council, SpainReviewed by:
Mingjun Li, Northwest A&F University, ChinaTian Zhong Li, China Agricultural University, China
Copyright © 2020 Song, Yang, Xun, Xu, Tian, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Zhang, emhhbmd3d0BzZGF1LmVkdS5jbg==; Hongqiang Yang, aHF5YW5nQHNkYXUuZWR1LmNu
 Jianfei Song
Jianfei Song Fei Yang
Fei Yang Mi Xun
Mi Xun Longxiao Xu
Longxiao Xu Xiaozhi Tian
Xiaozhi Tian Weiwei Zhang
Weiwei Zhang Hongqiang Yang
Hongqiang Yang