
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 May 2020
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00623
This article is part of the Research Topic Regulatory Mechanisms of Leaf Senescence under Environmental Stresses View all 11 articles
 Irfan Ullah Khan1†
Irfan Ullah Khan1† Akhtar Ali1†
Akhtar Ali1† Haris Ali Khan1
Haris Ali Khan1 Dongwon Baek2
Dongwon Baek2 Junghoon Park1
Junghoon Park1 Chae Jin Lim1
Chae Jin Lim1 Shah Zareen1
Shah Zareen1 Masood Jan1
Masood Jan1 Sang Yeol Lee2
Sang Yeol Lee2 Jose M. Pardo3
Jose M. Pardo3 Woe Yeon Kim2
Woe Yeon Kim2 Dae-Jin Yun1*
Dae-Jin Yun1*Drought stress adversely affects plant growth and development and significantly reduces crop productivity and yields. The phytohormone abscisic acid (ABA) rapidly accumulates in response to drought stress and mediates the expression of stress-responsive genes that help the plant to survive dehydration. The protein Powerdress (PWR), which interacts with Histone Deacetylase 9 (HDA9), has been identified as a critical component regulating plant growth and development, flowering time, floral determinacy, and leaf senescence. However, the role and function of PWR and HDA9 in abiotic stress response had remained elusive. Here we report that a complex of PWR and HDA9 interacts with ABI4 and epigenetically regulates drought signaling in plants. T-DNA insertion mutants of PWR and HDA9 are insensitive to ABA and hypersensitive to dehydration. Furthermore, the expression of ABA-responsive genes (RD29A, RD29B, and COR15A) is also downregulated in pwr and hda9 mutants. Yeast two-hybrid assays showed that PWR and HDA9 interact with ABI4. Transcript levels of genes that are normally repressed by ABI4, such as CYP707A1, AOX1a and ACS4, are increased in pwr. More importantly, during dehydration stress, PWR and HDA9 regulate the acetylation status of the CYP707A1, which encodes a major enzyme of ABA catabolism. Taken together, our results indicate that PWR, in association with HDA9 and ABI4, regulates the chromatin modification of genes responsible for regulation of both the ABA-signaling and ABA-catabolism pathways in response to ABA and drought stress.
During their life cycles, plants are continuously exposed to environmental challenges including light, heat, cold, flooding, high salinity, and drought stress. Among them, drought stress results in considerable damage to plant growth, and more than 40% of crop production is lost to drought (Guy et al., 1985; May et al., 1998; Hasegawa et al., 2000). Upon exposure to drought stress, plants initiate the expression of resistance genes and subsequent activation of signaling pathways. Plants have developed complex molecular and signaling mechanisms to adapt to water deficit condition. They respond to drought stress either through osmotic adjustment and regulation of ion homeostasis or by controlling the damage repair system and the detoxification and removal of reactive oxygen species (ROS) (Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2002). The phytohormone abscisic acid (ABA) plays a crucial role in plant physiological processes, regulating many aspects of plant growth and development including seed dormancy, seed maturation, and seedling growth. ABA is also required for drought stress tolerance, which regulates stomatal movement during drought stress and helps plants tolerate extreme water-deficient conditions (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001). However, the molecular and biochemical mechanism of these signaling pathways are not yet fully understood.
The mechanisms of chromatin modification and the constitution of chromatin complexes regulating gene expression are highly conserved in plants, mammals and yeast (Henderson and Jacobsen, 2007). Chromatin structure and its modification form the basic mechanism of genetic and epigenetic regulation of stress-related gene expression (Horn and Peterson, 2002). Histone acetylation is one of the most important features of chromatin remodeling, which removes positive charges by adding an acetyl group to the lysine residues of histone proteins, thereby reducing the histone–DNA affinity and resulting in chromatin decondensation and active transcription (Bannister and Kouzarides, 2011). Histone acetylation levels are dynamically regulated through the combined actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDACs are enzymes conserved in the eukaryotes that function in biological processes including transcription, genome stability, development and in biotic and abiotic stress responses (Haberland et al., 2009; Seto and Yoshida, 2014; Bosch-Presegué and Vaquero, 2015).
Human NCOR1 is a homolog of the Arabidopsis protein powerdress (PWR), which is involved in the floral determinacy network (Yumul et al., 2013). The gene encoding PWR was named Powerdress because of the appearance of the single mutant, which has bulged carpel tips reminiscent of excessively padded suit or dress shoulders. PWR has two conserved SWI3/DAD2/N-CoR/TFIII-B (SANT) domains that together function as a histone-interaction module that couples histone tail binding to enzyme catalysis for the remodeling of nucleosomes. PWR interacts with REDUCED Potassium Dependency Protein 3 (RPD3), a class-1-type Histone Deacetylase 9 (HDA9) and mediates Histone 3 (H3) deacetylation. The complex of PWR–HDA9 also regulates flowering time in Arabidopsis by repressing Agamous-Like 19 (AGL19) transcription (Kim et al., 2016).
HDA9 requires PWR for its nuclear transport and binding to the promoter region of key negative regulator genes involved in leaf senescence (Yumul et al., 2013; Chen et al., 2016). HDA9 interacts with PWR and WRKY53 to regulate leaf aging, and hda9 and pwr loss-of-function mutants exhibit late senescence phenotype (Chen et al., 2016). Furthermore, ABA promotes leaf senescence and loss of function of its receptors PYL8 and PYL9, resulting in delayed leaf senescence (Zhao et al., 2015). To date, approximately 18 Arabidopsis histone deacetylases (HDACs) have been identified. These are divided into three main types. Twelve belong to the reduced Potassium Dependency Protein 3/HDA1 Histone Deacetylase 1 (RPD3) superfamily and are named as HDAs; two are in the histone deacetylase 2 (HD2) family and are named HDTs; and two belong to the silent information regulator protein 2 (SIR2) family and are named SRTs (Pandey et al., 2002; Hollender and Liu, 2008). RPD3-type class 1 HDA6 and HDA19 are involved in the regulation of seed germination, ABA response, salt stress and other abiotic stresses. Unlike hda6 and hda19 mutants, mutants of HDA9 (hda9-1 and hda9-2) are insensitive to ABA and to salt stress during seed germination and root growth (van Zanten et al., 2014; Kang et al., 2015).
Plant endogenous ABA concentration is determined by the rate of ABA metabolism (i.e., biosynthesis, and catabolism). The molecular mechanisms of ABA signaling, biosynthesis, and catabolism have been characterized using genetic and biochemical approaches (Nambara, 2005). Arabidopsis contains several NCED family genes, including AtNCED3, which plays the central role in ABA biosynthesis in response to drought stress (Iuchi et al., 2001). The transcript level of AtNCED3 rapidly increases in response to drought stress, while a nced3 mutant carrying a T-DNA insertion is defective in accumulation of endogenous ABA under drought stress and impaired in drought stress tolerance. For ABA catabolism, at least two crucial pathways have been characterized: the oxidative pathway and the sugar-conjugation pathway (Nambara, 2005). The oxidative pathway is stimulated by CYP707A-induced hydroxylation of ABA C-80 to phaseic acid (Kushiro et al., 2004; Saito et al., 2004). The four Arabidopsis CYP707A-family genes encoding ABA 8′-hydroxylases are induced by exogenous ABA, as well as by dehydration and other abiotic stresses (Kushiro et al., 2004; Saito et al., 2004). By contrast, CYP707A2 transcripts predominantly accumulate in dry seeds, and the gene is immediately upregulated after seed imbibition. The cyp707a2 mutant maintains a high level of ABA and exhibits enhanced seed dormancy as compared to the wild type (WT) (Kushiro et al., 2004). These reports indicate that CYP707A2 is a component in ABA catabolism during seed germination and regulation of seed dormancy. However, the physiological role of other CYP707A genes remained unclear.
The interaction and binding of ABA with PYL/PYR1/RCAR receptors results in the deactivation of protein phosphatase type-2C (PP2C) proteins (ABI1, ABI2, HAB1 and HAB2), thereby releasing SNF1-related protein kinase (SnRK2) kinases. The ABA-mediated disassociation of PP2C from SnRK2s leads to autophosphorylation and subsequently to transphosphorylation, activation of the downstream targets (such as ABI3, ABI4, and ABI5) and ultimately regulation of downstream signaling pathways (Fujii et al., 2009; Umezawa et al., 2009; Antoni et al., 2012). ABI4 is an important transcription factor that was initially identified as a member of the AP2/ERF family and that binds to ABA-responsive cis-regulatory elements (CREs), ABRE and regulates the expression of genes in response to abiotic stresses (Mizoi et al., 2012). ABI4 is also a versatile activator and a repressor of several genes. Beside inducing the expression of genes involved in seed dormancy, ABA signaling, salt stress and floral transition (Söderman et al., 2000; Nakabayashi et al., 2005; Koussevitzky et al., 2007; Bossi et al., 2009; Giraud et al., 2009; Reeves et al., 2011; Shu et al., 2018), ABI4 also represses the expression of genes involved in ABA catabolism (CYP707A genes), ethylene biosynthesis (ACS genes) and retrograde signaling (AOX1a), genes encoding Arabidopsis response regulators (ARRs) (Huang et al., 2017), as well as genes involved in fatty acid biosynthesis, photosynthesis, pigment and wax metabolic processes (Shu et al., 2013; Dong et al., 2016), by directly binding to their promoters.
PWR also interacts with HOS15 (HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15), a homolog of human transducin-β-like protein 1 (TBL1). HOS15 contains a LisH and a WD40-repeat domain and is involved in histone modification and deacetylation during abiotic stresses. Furthermore, HOS15 also interacts with HISTONE DEACETYLASE 9 (HDA9), as determined by affinity purification of HOS15-interacting proteins (Park et al., 2018). Loss-of-function hos15-2 mutant plants are hypersensitive to ABA during germination and extremely tolerant to drought stress, indicating the importance of HOS15 as a negative regulator (Ali et al., 2019). On the other hand, the function of PWR in abiotic stresses is largely unknown.
Here we report that T-DNA insertion mutants of PWR (pwr-2 and pwr-3) are ABA insensitive and display drought-sensitive phenotypes. Using yeast two-hybrid screening, we observed that both PWR and HDA9 interact with ABI4 along with ABI3. The expression of ABA-responsive genes is downregulated in pwr and hda9 mutants. Transcript levels of genes that are normally repressed by ABI4, such as CYP707A genes, AOX1a and ACS4, are upregulated in the pwr and hda9 mutants. Moreover, in response to drought stress, PWR and HDA9 regulate acetylation at the promoter of CYP707A1, which encodes the major enzyme of ABA catabolism. Taking these results together, we conclude that PWR in association with HDA9 and ABI4 regulates the chromatin modification of genes responsible for ABA catabolism in response to drought stress.
Plants from the Arabidopsis thaliana ecotype Columbia-0 (Col-0) background were used in this study. Seeds of the WT and mutants were surface sterilized in a solution containing 3% sodium hypochlorite solution (Yakuri Pure Chemicals, Kyoto, Japan) for 5 min and then rinsed five times with sterilized water. After stratification for 3 day at 4°C in the dark, the plants were grown on half-strength Murashige and Skoog (1/2 MS) medium or soil at 23°C under a 16-h light/8-h dark condition. The T-DNA insertion mutant pwr-2 (SALK_0718811C) seeds were obtained from ABRC stock center and previously described by Yumul et al. (2013) and the mutant pwr-3 (SALK_006823), was also obtained from ABRC stock center. The T-DNA insertions in these plants were confirmed by genotyping PCR. The hda9-1 (Gk_305G03) and hda9-2 (SALK_007123) mutants were obtained from NASC1 and ABRC2, respectively (Alonso et al., 2003; Rosso et al., 2003; Kang et al., 2015). The pwr-2/hda9-1 double-mutant plants were created by crossing.
Total RNAs (2 μg) from plants (harvested at the time points described in the text for each experiment) were extracted with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), treated with DNase I (Sigma, St. Louis, MO, United States) and used to synthesize first-strand cDNA using the ThermoscriptTM RT-PCR System (Invitrogen, Carlsbad, CA, United States). Quantitative RT-PCR (qRT-PCR) was performed using SYBR Green PCR Master Mix kit (Bio-Rad SYBR Green Supermix, Hercules, CA, United States) according to the manufacturer’s instructions with the CFX96 or CFX384 Realtime PCR detection system (Bio-Rad, CA, United States). The relative expression levels were calculated using the comparative cycle threshold (2–ΔΔCT) method. The sequences of the primers used in qRT-PCR are listed in Supplementary Table S1.
For ABA germination assays, seeds were grown on 1/2 MS medium containing 1.5% sucrose and different concentrations of ABA (Sigma, St. Louis, MO, United States). Successful germination in the presence of ABA was determined by germination rate and the presence of green cotyledons at the indicated concentrations. For drought test, 3-week-old plants were subjected to drought stress by withholding water for 14 day while control plants were watered as before. The drought-stressed plants were then re-watered, and their recovery was monitored. Three experimental repeats were carried out, each involving at least 36 plants from each category.
The full-length PWR, HDA9 and ABI4 coding sequences were amplified with the primers listed in Supplementary Table S1 to generate the entry vector [PWR, HDA9, ABI4 with or without stop codons in the pDONRTM/Zeo vector (Invitrogen, Carlsbad, CA, United States)]. pDONRTM/Zeo-HDA9 and pDONRTM/Zeo-ABI4 were fused into the pGWB14 destination vectors by in vitro recombination using Gateway BP and LR reaction kits (Invitrogen, Carlsbad, CA, United States) to generate HDA9-3xHA. ABI4 was cloned into the pK7WGF destination vector to construct GFP-ABI4. The specific primer sequences are provided in Supplementary Table S1.
For yeast two-hybrid experiments, pDONRTM/Zeo-PWR and pDONRTM/Zeo-HDA9 were fused into the yeast two-hybrid destination vector pDEST22 (harboring the activation domain) and pDONRTM/Zeo-ABI4 was fused into the destination vector pDEST32 (harboring the DNA binding domain) to generate the construct vectors pDEST22-PWR, pDEST22-HDA9, and pDEST32-ABI4, respectively. These plasmids were transformed into the Saccharomyces cerevisiae strain PJ-694-A. Individual colonies of transformants were streaked on agar plates containing synthetic complete (SC) medium lacking tryptophan and leucine, and then grown for 48 h. The interaction of PWR, HDA9, and ABI4 was tested on plates containing medium without histidine and further tested in growth medium containing 3-amino-1,2,4-triazole (3-AT). Empty vector was used as a negative control, while the combination of pDEST22-SOS2 and pDEST32-SOS3 was used as positive control.
Nuclear proteins were extracted from 2-week-old seedlings treated with dehydration stress for indicated time point by CELLYTPN1 CelLytic PN Isolation/Extraction Kit (Sigma-Aldrich), crude preparation. Anti-H3 (Abcam) and anti-AcH3 (Millipore) antibodies and antigen proteins were visualized by chemiluminescence using ECL detecting reagent (Bio-Rad).
For co-immunoprecipitation assays, 35S:ABI4-GFP and 35S:HDA9-HA expression cassettes were co-infiltrated into leaves of N. benthamiana, and after 3 day of incubation, total protein was extracted from the leaves, pulled down with α-GFP, and immunoblotted with α-HA. Each immunoblot was incubated with the appropriate primary antibody (α-HA antibody, 1:2000; α-GFP antibody) for 2 h at room temperature or overnight at 4°C. The membranes were developed using peroxidase-conjugated secondary antibody: 1:2000 for α-rabbit antibody (GE, Little Chalfont, Buckinghamshire, United Kingdom) and 1:1000 for α-rat IgG (Sigma, St. Louis, MO, United States).
ChIP assays were carried out following an established method as previously described (Saleh et al., 2008). Two-week-old control and dehydration-treated Arabidopsis plants were treated with 1% formaldehyde for 15 min to fix the chromatin structure and this cross-linking reaction was subsequently stopped by treatment with 0.1 M glycine for 5 min. The DNA-fixed plant tissues were ground with liquid nitrogen and washed with water and then the nuclei were isolated. Nuclear proteins were extracted and sonicated with a Bioruptor (BMS) to fragment the chromosomal DNA. Immunoprecipitation was performed using an antibody to total anti-acetylated H3 (Millipore), with salmon sperm DNA and protein A agarose (upstate Biotechnology).
Leaves of 12-day-old seedlings were floated on stomatal opening buffer (5 mM 2-(N-morpholino) ethanesulfonic acid [MES], 5 mM KCl, 50 mM CaCl2 [pH 5.6]) under light for 3 h. After 5 μM ABA treatment for 2 h, leaves were fragmented in a warning blender. Samples were rinsed with pure water three times for 10 min each. Washed samples were incubated over–night in the secondary fixative solution, 2% OsO4, in the dark at 4C. After fixation, OsO4 was removed by washing the samples three times for 10 min each. The samples were then dehydrated chemically for embedding in a series of EtOH solutions: 20, 50, 70, 90%, and finally 100% EtOH sequentially for 40 min each. Epidermal fragments were quickly mounted for scanning electron microscopy (SEM) (JSM-6380LV; JEOL, Eching, Munchen, Germany) assay. At least 10 stomata from three different plants of each genotype were used to measure the stomatal aperture with three biological repeats. Each experiment was replicated three times.
Powerdress regulates plant growth and developmental processes (Yumul et al., 2013; Chen et al., 2016; Kim et al., 2016); however, we were interested in assessing its involvement in abiotic stresses. Therefore, to investigate PWR possible involvement in ABA signaling, we tested the physiological response of pwr-2 and pwr-3 to exogenously applied ABA. We germinated seeds of WT (Col-0), pwr-2 and pwr-3 lines, as well as the loss-of-function abi2-2 mutant (which is hypersensitive to ABA) as an experimental control, on Murashige and Skoog (MS) medium containing ABA. In the presence of ABA, pwr-2 and pwr-3 seedlings exhibited greater germination than WT seedlings, and abi2-2 seedlings showed even poorer germination and cotyledon greening (Figures 1A–C). After exposure to 0.5 μM ABA for 7 days, the percentages of green cotyledons for pwr-2 and pwr-3 were 62 and 52–55% respectively, compared with 27–30% for WT and 10–15% for abi2-2 while, percentages of green cotyledons for pwr-2 and pwr-3 were 53 and 49% respectively as compared to 25% of WT on 0.8 μM ABA (Figure 1B). Since PWR loss-of-function mutants displayed ABA-insensitive phenotypes, it seemed likely that PWR might play a role in regulating plant response to drought. To test this hypothesis, we exposed 3-week-old WT, abi2-2, pwr-2, and pwr-3 plants to 14 days of drought stress. Plants were re-watered after the drought period and their survival rates recorded 2 days after re-watering. The WT and abi2-2 plants survived the dehydration stress at rates of 80% and 100%, respectively (Supplementary Figure S1A). By contrast, pwr-2 and pwr-3 mutants were unable to tolerate water-deficient condition and survived at rates of only 10 and 15%, respectively (Supplementary Figure S1B). Furthermore, pwr mutants showed impaired stomatal closure in leaf epidermal fragments after treatment with exogenous ABA (Figures 1D,E), indicating that the drought sensitivity of pwr mutants is correlated with reduced stomatal closure. Taken together these results indicating that PWR plays a central role in plant sensitivity to ABA in seed germination and confers tolerance of drought.
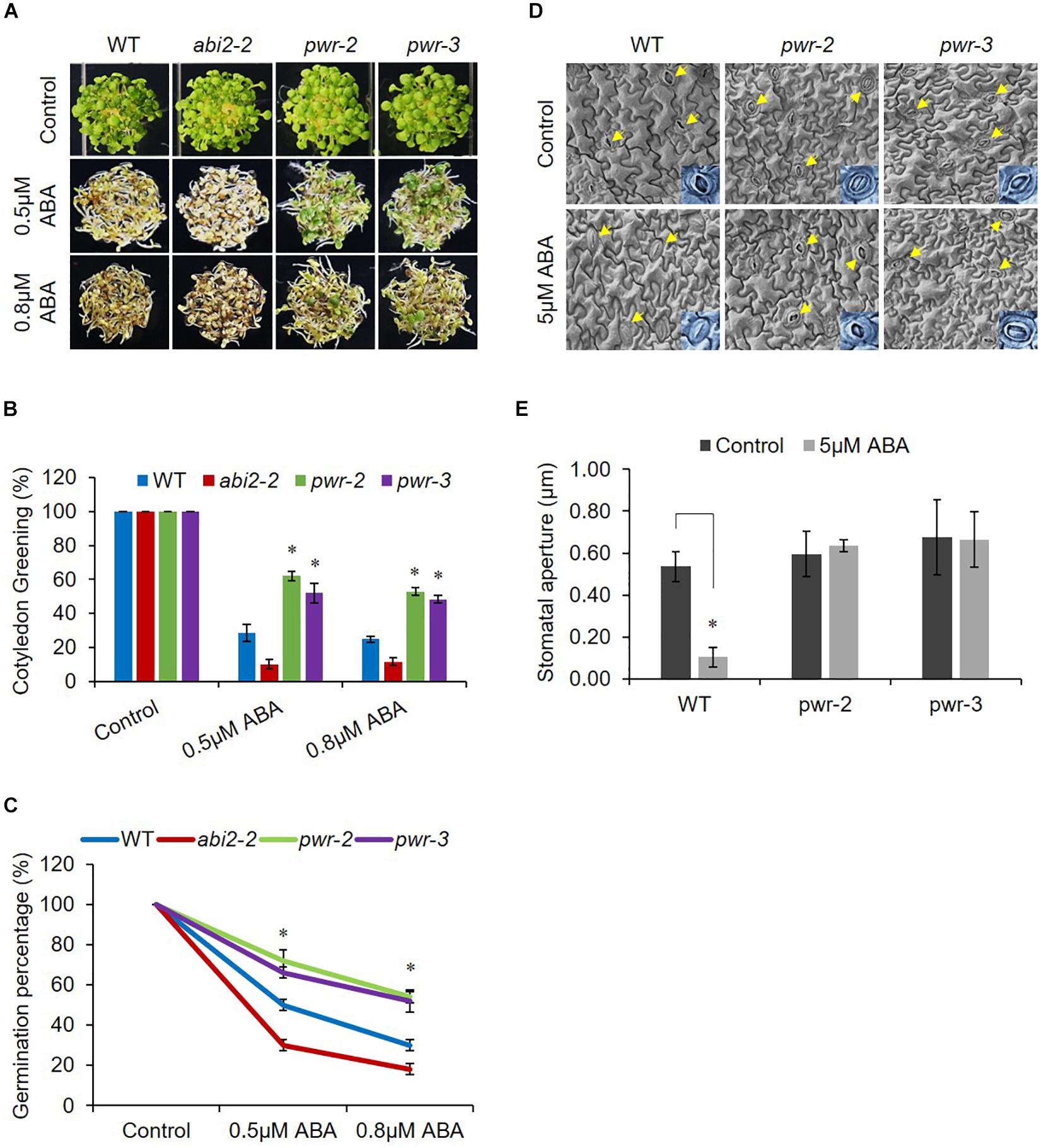
Figure 1. pwr mutants are insensitive to ABA. (A) WT, abi2-2, pwr-2, and pwr-3 Arabidopsis seeds were germinated on 1/2 MS medium supplemented with 0.5 μM and 0.8 μM ABA (or without ABA for controls). Photographs were taken 7 day after germination. (B) The numbers of green cotyledons from each line were counted after 7 day. Mean ±SD values were determined from three replicates (n = 144). Significant difference was determined by Student t-test (∗p < 0.01). (C) Germination rates of indicated genotypes from (A). Mean ±SD values were determined from three replicates (n = 144). Significant difference was determined by Student t-test (∗p < 0.01). (D) pwr mutation impairs ABA-induced stomatal closure. Seedlings of WT (Col-0) and pwr mutants were exposed to 2-h ABA (5 μM) treatment. Epidermal peels from WT, pwr-2, and pwr-3 were measured for stomatal aperture in control condition and in response to ABA (arrows indicate stomata). (E) Quantitative analysis of (D) using Image J 1.47V software. At least 10 stomata from three different plants of each genotype were used to measure stomatal aperture. Error bars represent SE. Significant difference was determined by a Student’s t-test (∗p < 0.01).
As previously described, HDA9 represses the seedling trait and negatively regulates salt and drought stress tolerance; in addition, HDA9 requires PWR for its nuclear transport and promoter association. Furthermore, PWR and HDA9 also regulate flowering through repression of AGL19 (van Zanten et al., 2014; Kang et al., 2015; Kim et al., 2016; Zheng et al., 2016). In this context, we were interested in investigating whether PWR and HDA9 also work together in response to the phytohormone ABA. We therefore tested the physiological response of PWR and HDA9 mutants to exogenous ABA. Compared to WT, pwr, hda9, and pwr/hda9 double-mutant seedlings were relatively insensitive to exogenous ABA (Figure 2A). In the presence of 0.8 and 0.5 μM ABA (in Supplementary Figure S2), the percentages of green cotyledons for pwr-2, pwr-3, hda9-1, hda9-2, and pwr-2/hda9-1 were approximately 83, 85, 82, 84, and 85%, respectively, compared with 55% for WT, 35% for abi2-2 and 98% for abi4-1 (Figure 2B and Supplementary Figure S2). To test root growth phenotypes in the presence of ABA, we transferred 4-days-old seedlings of WT (Col-0), abi2-2, pwr2, pwr-3, hda9-1, hda9-2, and pwr-2/hda9-1 Arabidopsis to MS medium containing 30 μM ABA and allowed them to grow for a further 10 days. Compared to that of the WT, the root growth of single and double mutants of PWR and HDA9 was insensitive to ABA (Figure 2C). The relative reductions in root length were 37.5–43.3% for pwr, 42.8–45.7% for hda9 and 39.4% for the pwr-2/hda9-1 double mutant, as compared to 70 and 85%, respectively, for WT and abi2-2 (Figure 2D). Consistent with phenotypes, the induction of ABA-responsive genes, including RD29A, RD29B, and COR15A, upon ABA was lower in pwr-2, hda9-1 and pwr-2/hda9-1 than in WT plants (Figure 2E). Moreover, we also tested the dehydration responses of PWR and HDA9 mutants by exposing 3-week-old plants to 14-days drought stress, re-watering them and then recording the survival rate 2 days later. As expected, like the pwr-2 and pwr-3 mutants, hda9 mutants were extremely sensitive to drought stress (Figure 3). Taken together, these results suggested that PWR and HDA9 work together and play a critical role in plant ABA response and drought tolerance.
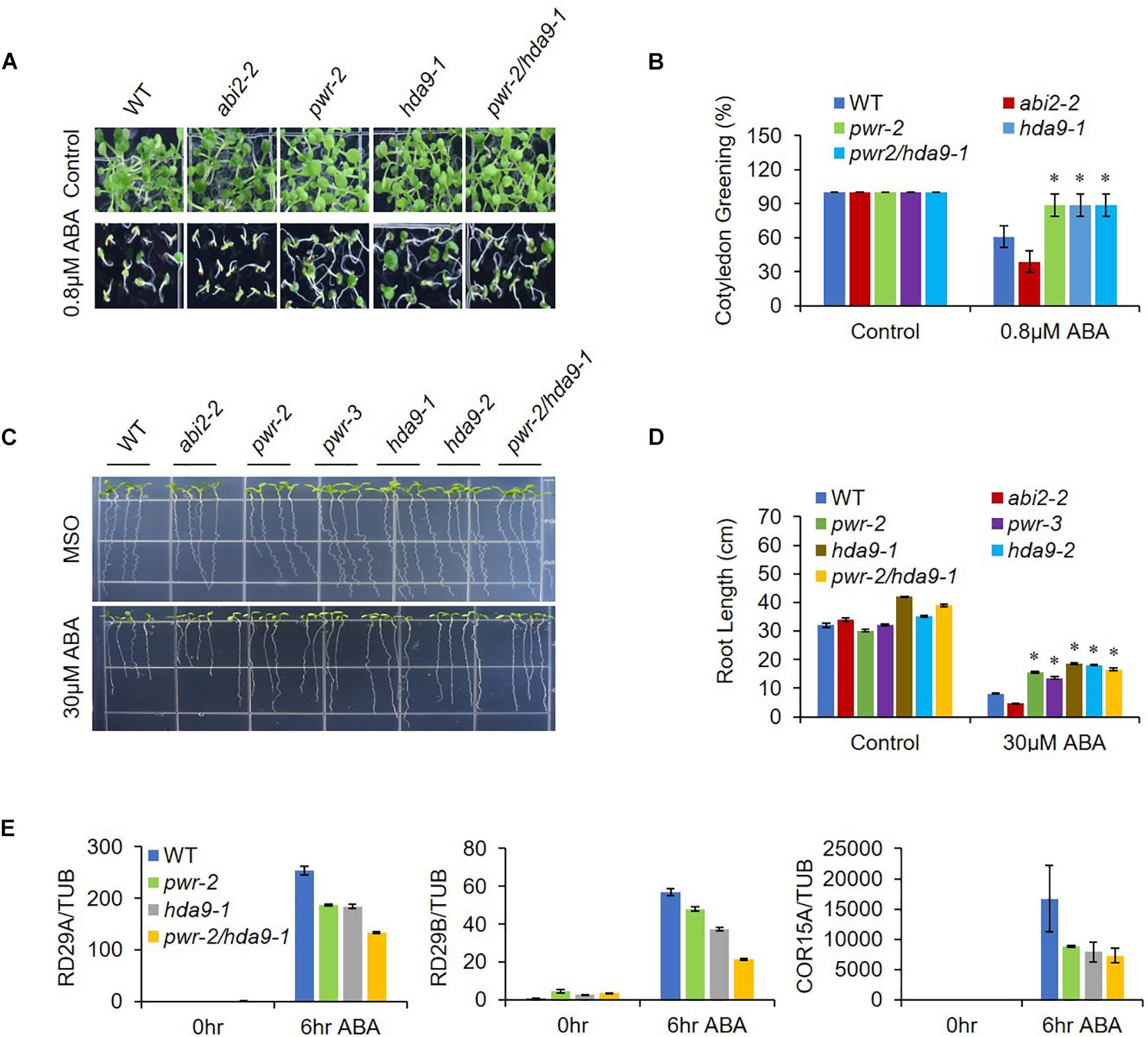
Figure 2. pwr and hda9 mutants are insensitive to ABA. (A) Seeds WT, abi2-2, pwr, hda9, and pwr/hda9 double mutant were germinated on 1/2 MS medium with 0.8 μM ABA. Photographs were taken 7 day after germination. (B) Numbers of green cotyledons were counted after 7 day. Mean ±SD values were determined from three replicates (n = 45). Significant difference was determined by Student t-test (∗p < 0.01). (C) Root growth phenotypes of WT, abi2-2, pwr-2, pwr-3, had9-1, hda9-2, and pwr-2/hda9-1. Photographs were taken 10 day after transfer of 4-day-old seedlings to MS medium with or without 30 μM ABA. (D) Quantification of primary root lengths of the seedling depicted in C. Error bars represent means ± SE of three replicates (n = 20 seedlings per replicate). Significant difference was determined by Student t-test (∗p < 0.01). (E) Expression of three ABA-responsive stress-related genes (RD29A, RD29B, and COR15A) in WT, pwr-2, hda9-1, and pwr-2/hda9-1 Arabidopsis during ABA stress was analyzed by qRT-PCR. The imbibed seeds were germinated and grown on MS medium for 10 days, and then the seedlings were harvested and treated with 50 μM ABA to obtain cDNA for qRT-PCR. Samples were collected at the indicated time points: 0 h (no treatment) and 6 h. Total RNAs were extracted from those seedlings, and RT-qPCR analysis was performed. TUB4 was used as internal control. Error bars show SD.
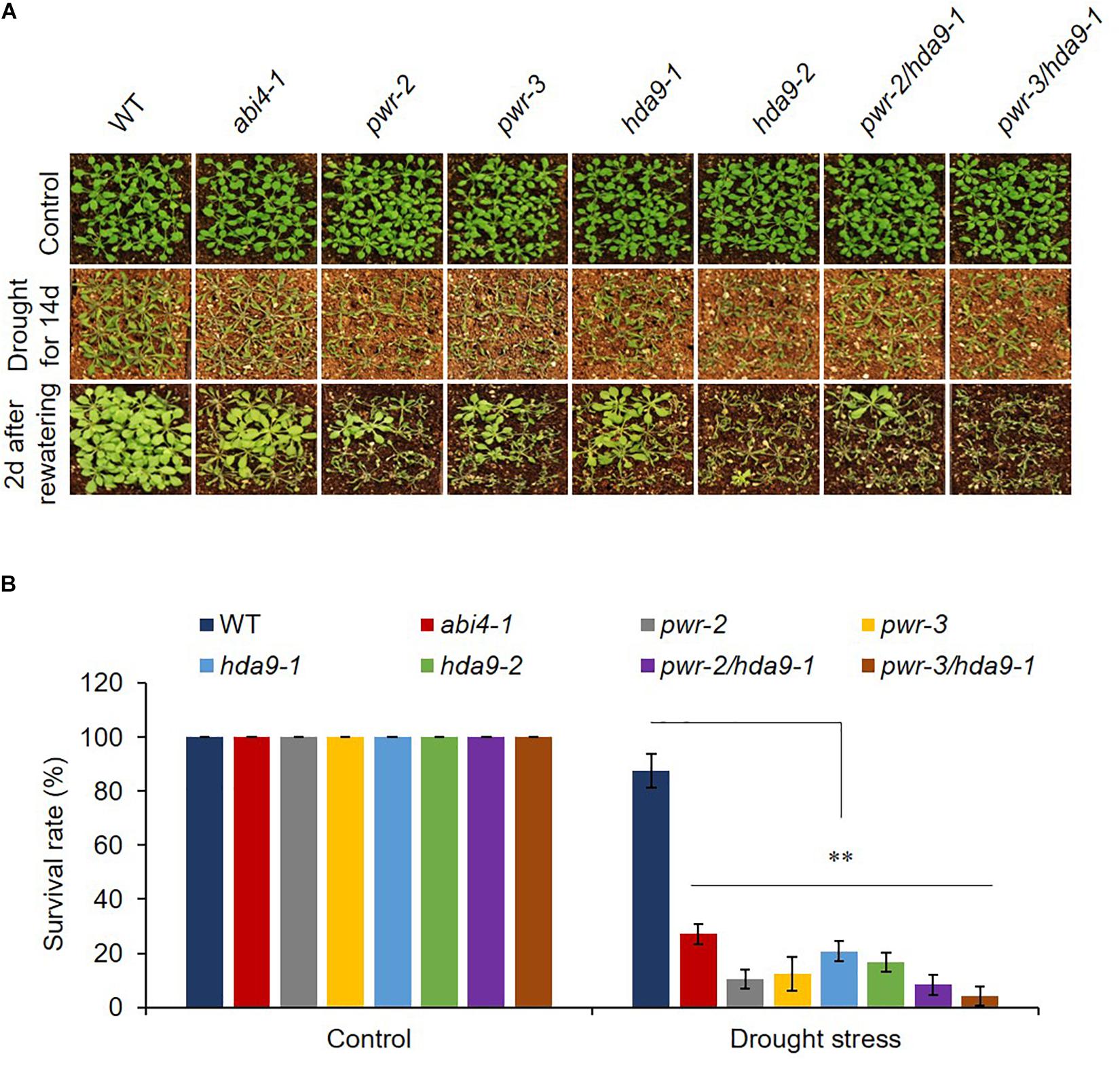
Figure 3. pwr and hda9 mutant plants are drought sensitive. (A) 3-week-old plants of genotypes WT, abi4-1, pwr, hda9 single, and pwr/hda9 double mutants exposed to drought assay by withholding water for 14 days and subsequently rewatered the plants after drought period. Photographs were taken 2 days after rewatering. (B) Survival rate (percentage) of genotypes shown in (A). Results are means of three independent biological repeats. Significant difference was determined by Student t-test (∗∗p < 0.001).
Arabidopsis HDA9 regulates several aspects of biological processes such as seed dormancy and maturation, flowering time, and stress responses. Moreover, previous reports have shown that developmental and stress-related genes are hyperacetylated and upregulated in hda9 mutant. The direct interaction between PWR and HDA9 and the similar type of molecular and morphological defects in pwr and hda9 mutants strongly suggest that PWR and HDA9 are working together in same complex (Kim et al., 2013; van Zanten et al., 2014; Kang et al., 2015; Lee et al., 2016; Zheng et al., 2016). To investigate the interaction of proteins with PWR and HDA9 that are involve in seed germination and response to drought stress, we carried out yeast two-hybrid screening of ABA- and drought-stress-responsive transcription factors, and we observed that PWR and HDA9 specifically interact with ABI3 and ABI4 (Figure 4A and Supplementary Figure S3). To validate the interaction between ABI4 and the HDA9-PWR complex, we performed a co-immunoprecipitation assay using Nicotiana benthamiana leaves transiently expressing 35S:HDA9-HA and 35S:ABI4-GFP. ABI4-GFP successfully pulled down HDA9-HA, indicating that ABI4 forms a complex with HDA9 and PWR (Figure 4B).
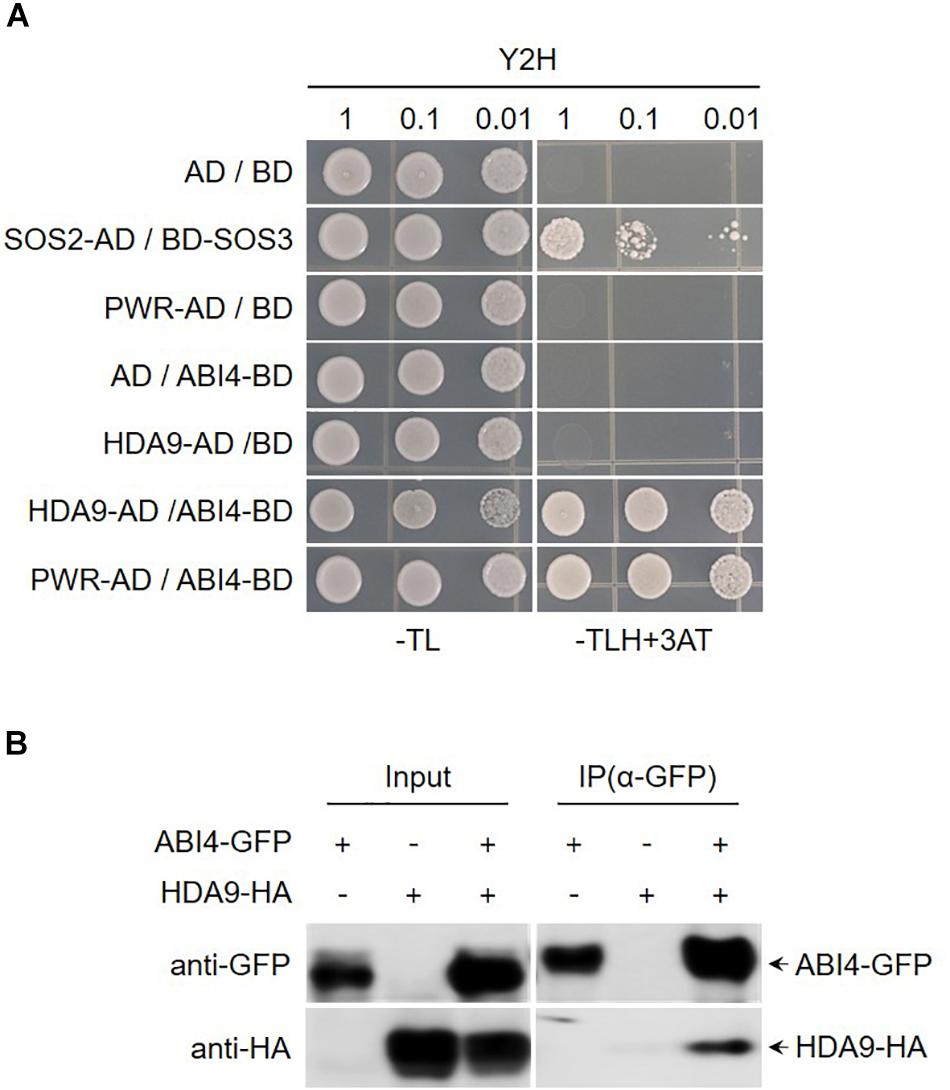
Figure 4. PWR and HDA9 physically interact with ABI4. (A) Screening of PWR and HDA9 with ABA transcription factors using yeast two-hybrid assay. BD, pDEST32 (bait plasmid); AD, pDEST22 (prey plasmid). The co-transformed yeast strains were plated on the control −TL medium and selective −TLH + 3-AT medium. The combinations with empty plasmid were used as negative controls and SOS2-AD/SOS3-BD was used as positive control. (B) Co-immunoprecipitation assay between HDA9 and ABI4. Protein extracts obtained from N. benthamiana leaves infiltrated with Agrobacterium tumefaciens harboring 35S:HDA9-HA and 35S:ABI4-GFP were analyzed using anti-GFP and anti-HA antibodies. Protein extracts (input) were immunoprecipitated (IP) with anti-GFP. Immunoblots were analyzed with anti-GFP and anti-HA to detect interaction between HDA9 and ABI4.
ABI4 is identified as a member of the AP2/ERF superfamily that binds specifically to ABRE elements and regulates abiotic-stress-related gene expression (Mizoi et al., 2012). ABI4 plays dual function in regulating gene expression, serving both as an activator and as a repressor (Söderman et al., 2000; Nakabayashi et al., 2005; Koussevitzky et al., 2007; Bossi et al., 2009; Giraud et al., 2009; Reeves et al., 2011; Shu et al., 2018). To identify the transcriptional regulatory function of PWR in drought stress response, we monitored the expression of genes repressed by ABI4 in pwr mutants treated with different time point of dehydration stress. Surprisingly, the expression of ABA hydroxylase gene CYCP707A1 was upregulated in pwr-2 and pwr-3 mutants as compared with WT (Figure 5), although the expression of other CYCP707A genes were unchanged. Similarly, the expression of other ABI4 target genes such AOX1a (a retrograde signaling gene) and ACS4 (an ethylene biosynthesis genes) in pwr-2 and pwr-3 in response to dehydration stress was also high (Figure 5). These results indicated that ABI4 alone is not enough to suppress these genes and that ABI4 requires the PWR and HDA9 repressor complex to target different genes in regulating plant stress tolerance.
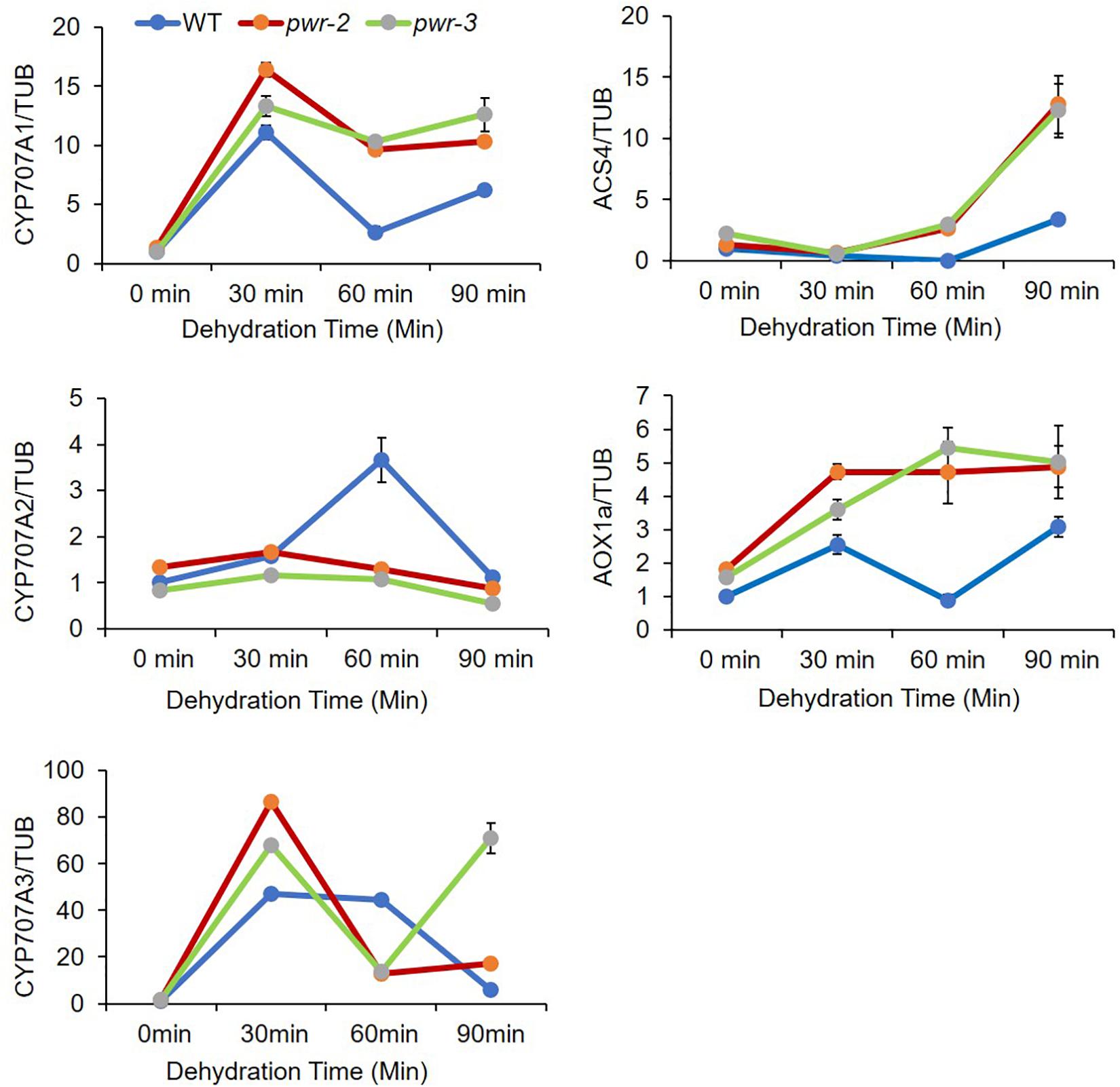
Figure 5. PWR negatively regulates ABI4-responsible genes. The relative expression of ABA hydroxylase genes analyzed by qRT-PCR. 10-day-old seedlings of WT, pwr-2, and pwr-3 were dehydrated at room temperature for indicated time point. Total RNAs were extracted from those seedlings, and RT-qPCR was performed to analyze the abundance of CYP707A1, CYP707A2, CYP707A3, ACS4, and AOX1a expression. Expression levels were normalized to that of TUB4. Normalized expression is shown as mean ± standard deviation (SD). The experiment was repeated three times.
PWR and HDA9 together regulate the acetylation status of numerous genes, specifically at H3K9, H3K14, and H3K27. SANTb-domain proteins such as PWR preferentially bind to acetylated histone H3 but not H4. Moreover, it has been proposed that the SANTb domain of PWR defines the protein’s substrate specificity for binding to HDA9 (Kim et al., 2016). Therefore, we were interested to test the acetylation status of histone H3 in response to dehydration stress in pwr and hda9 mutants. Compare to that in WT, the total AcH3 levels in the pwr and hda9 single mutants as well as the pwr/hda9 double mutant were increased by drought stress (Figure 6A). Since the total acetylation level of AcH3 was increased in pwr and hda9 mutant, we assumed that the hyper-induction of CYP707A1 gene expression in the pwr mutant (Figure 5) was largely due to hyperacetylation of the CYP707A1 promoter. The transcript levels of all four CYP707A genes increase in response to ABA and to abiotic stresses, including dehydration (Kushiro et al., 2004; Saito et al., 2004). To determine whether the increase in CYP707A1 gene expression in pwr and hda9 upon dehydration is due to chromatin remodeling, we carried out chromatin immunoprecipitation (ChIP) assays to assess the AcH3 level of CYP707A1. The level of AcH3 at one region of the CYP707A1 promoter, P1 (Figure 6B), was higher in pwr mutant plants than in WT after 90-min dehydration stress, whereas we saw no differences at the P2 and P3 promoter regions (Supplementary Figures S4A,B). Interestingly, the level of AcH3 at the P1 region was also high in the hda9 mutant after dehydration (Figure 6B). However, we detected no difference in AcH3 levels in the CYP707A2 promoter during dehydration stress in the pwr mutant compared with the WT (Supplementary Figure S4C). Taken together, these results indicated that during drought stress, PWR and HDA9 repress the expression of CYP707A1 through histone deacetylation to allow ABA accumulation, and that the P1 region is important for the activation or repression of CYP707A1 during drought stress.
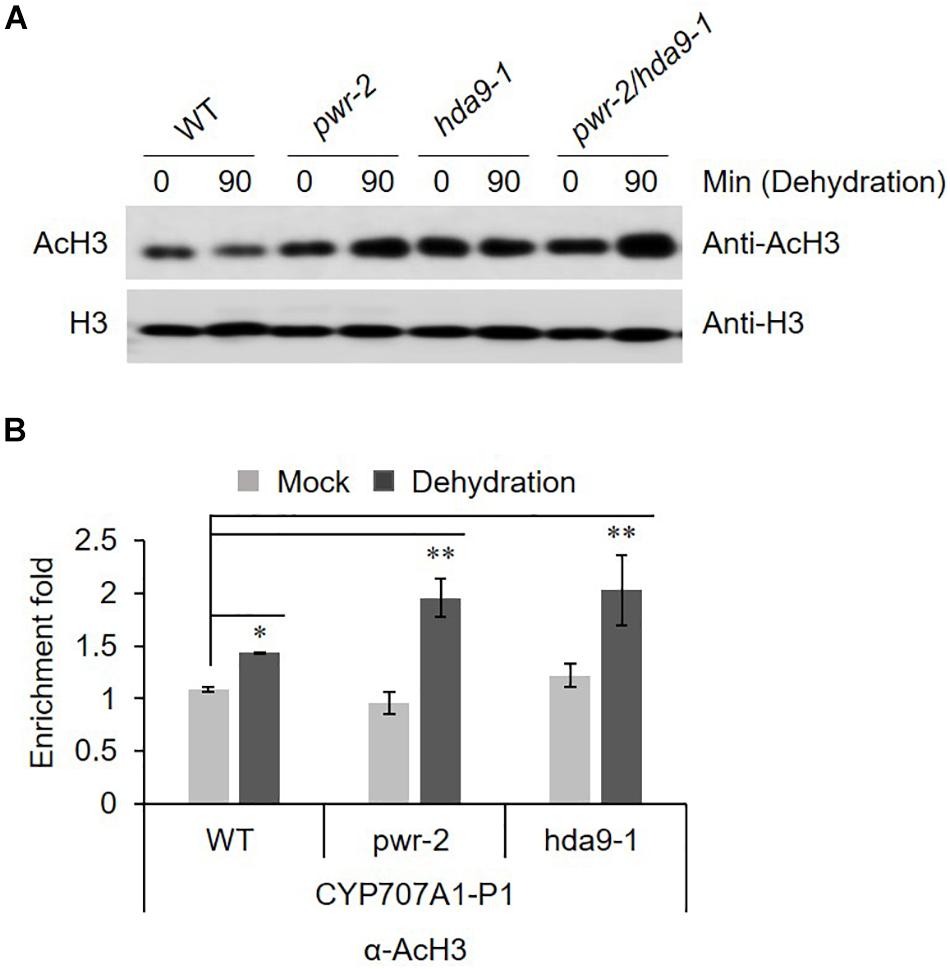
Figure 6. (A) AcH3 level is increased during drought stress. Nuclear protein was isolated from 2 weeks-old Arabidopsis WT (Col-0), pwr-2, hda9-1, and pwr-2/hda9-1 seedlings after drought stress treatment for 0 or 90 min and then separated by SDS-PAGE. The anti-H3 band from a duplicate gel shows that similar amounts of proteins were loaded. (B) ChIP-qPCR assays of the CYP707A1 promoter using antibody to acetylated histone 3 (AcH3). Chromatin from 2-week-old WT Col-0, pwr-2 and hda9-1 mutant plants subjected to drought treatment for 0 or 90 min was immunoprecipitated with anti-AcH3, and the amount of DNA in the immune complex was determined by qRT-PCR. Error bars representing SE (n = 3 independent experiments). Significant difference was determined by Student t-test (∗p < 0.05, ∗∗p < 0.01).
Posttranslational protein/histone modifications such as acetylation, methylation, phosphorylation, and ubiquitination play pivotal roles in plant growth and development, genome integrity, and stress responses. Histone acetylation and deacetylation, mediated by histone acetyltransferases (HATs) or histone deacetylases (HDACs), are reversible processes that promote or repress gene expression (Struhl, 1998). The RPD3/HDA1-type class 1 HISTONE DEACETYLASE 9 (HDA9), among the 18 histone deacetylases (HDACs) identified in Arabidopsis, interacts directly with PWR, a homolog of the human protein NCOR1 (Pandey et al., 2002; Kim et al., 2016). PWR, which was initially identified as being involved in regulating floral determinacy network, contains two important SANT domains known as SANTa and SANTb, which are required for its interaction with HDA9 and mediation of HISTONE (H3) deacetylation. PWR and HDA9 regulate several processes in Arabidopsis, including regulating flowering time by repressing AGAMOUS-LIKE 19 (AGL19). PWR is known for its role in chromatin modification and regulation of developmental processes. Here, we have identified a previously unknown function of PWR regulating abiotic stresses.
PWR regulates plant growth and development, flowering time and the floral determinacy network. Moreover, PWR interacts with HDA9 and regulates flowering time in Arabidopsis by repressing Agamous-Like 19 (AGL19) (Yumul et al., 2013; Kim et al., 2016). While HDA9 requires PWR for its nuclear transport and promoter association, and the HDA9-PWR-WRKY53 complex integrates and regulates multiple signaling pathways to mediate global gene expression responsible for leaf senescence (Chen et al., 2016). To explore the relationship between PWR and HDA9 during abiotic stress, we investigated the phenotypes of loss-of-function mutants of PWR (pwr-2 and pwr-3) in the presence of ABA. Since HDA9 positively regulates plant response to ABA and dehydration (van Zanten et al., 2014; Kang et al., 2015), we expected that PWR might also play a positive role in ABA signaling and dehydration stress. We observed that pwr-2 and pwr-3 mutants were less sensitive than WT (Col-0) to ABA and impaired stomatal closure in the presence of exogenous ABA (Figure 1). Loss of PWR resulted in ABA-associated phenotypes such as ABA insensitivity in seed germination and post-germinative growth (Figure 2 and Supplementary Figure S2) and enhanced water loss, ultimately leading to drought sensitivity (Figure 3). Taken together, these data suggest that unlike HDA9, PWR is a positive regulator of ABA signaling. As previously reported, the phytohormone ABA quickly accumulates in response to stresses and plays a pivotal role in plant survival (Xiong et al., 2002, Xiong and Zhu, 2003). ABA is also a key regulator in stomatal movement that regulates water loss (Luan, 2002; Zhu, 2002). Indeed, ABA-related genes were suppressed in both pwr and hda9 mutants (Figure 2E), indicating that PWR is a central regulator of ABA response. Since PWR and HDA9 physically interact and work together in the same complex (Kim et al., 2016), we tested the genetic interaction of PWR and HDA9. As expected, pwr, hda9, and pwr/hda9 double mutants displayed the same insensitivity toward exogenously applied ABA and hypersensitivity to drought stress (Figures 2, 3). HDA9 binds to the active genes and may either prevent promiscuous cryptic gene expression or compete with other HDACs for binding to the same site. Transcriptome and ChIP-seq analyses have shown that HDA9 also binds the PYL gene promoter (Chen et al., 2016). These results indicated that PWR and HDA9 might work with specific transcription factors to repress their target genes during seed germination and drought stress.
HDA9 directly interacts with PWR, and the two proteins work together in the same repressor complex to regulate morphological and developmental processes in plants (Pandey et al., 2002; Kim et al., 2016). Given that the pwr and hda9 mutants were insensitive to ABA and sensitive to drought stress (Figures 2, 3), we assessed the interactions of PWR and HDA9 with the components of the ABA signaling and drought stress pathways. PWR and HDA9 interacted with ABI4 along with ABI3 (Figure 4 and Supplementary Figure S3). ABI4 was initially identified on the basis of the insensitivity of abi4 mutants to ABA, and later to salt and mannitol (Finkelstein, 1994; Quesada et al., 2000). ABI4 binds specifically to ABRE elements and regulates the expression of several genes in response to abiotic stresses (Mizoi et al., 2012). Notably, ABI4 is a versatile activator and a repressor of gene expression (Söderman et al., 2000; Nakabayashi et al., 2005; Koussevitzky et al., 2007; Bossi et al., 2009; Giraud et al., 2009; Reeves et al., 2011; Shu et al., 2018). ABI4 induces the expression of genes involved in seed dormancy and ABA signaling (Reeves et al., 2011), while repressing genes involved in ABA catabolism (CYP707A genes), ethylene biosynthesis (ACS genes), retrograde signaling (AOX1a), ARR genes, which are involved in cytokinin-induced degradation of ABI5 (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001; Huang et al., 2017) and genes involved in fatty acid biosynthesis, photosynthesis, pigment and wax metabolic processes (Shu et al., 2013; Dong et al., 2016). These lines of evidence indicate that ABI4 both induces and represses target genes. We therefore tested the genes suppressed by ABI4 in loss-of-function mutants of PWR (pwr-2 and pwr-3). In response to dehydration, the transcript levels of CYP707A1, ACS4 and AOX1a in pwr mutants were significantly higher than those in WT (Figure 5), whereas there were no significant differences in the expression of CYP707A2 and CYP707A3 in pwr mutants as compared to WT. As PWR and HDA9 regulate the acetylation status of numerous genes specifically at H3K9, H3K14, and H3K27, we measured the total acetyl histone H3 (AcH3) levels in pwr and hda9 mutants (Figure 6A). The AcH3 level was significantly increased in pwr and hda9 mutants upon dehydration stress, suggesting that the total AcH3 increases in response to dehydration stress in the mutants. However, effects of the acetylation status at the specific promoter regions of target genes responsible for drought stress responses could not be excluded. Therefore, we analyzed the AcH3 level at the CYP707A1 promoter and observed that after dehydration, the association of AcH3 was higher at the P1 region, but not the P2 or P3 regions, of the CYP707A1 promoter in the pwr-2 mutant compared to WT (Figure 6B and Supplementary Figure S4). Surprisingly, AcH3 association at the CYP707A1 P1 region was also increased in the hda9 mutant upon dehydration (Figure 6B). However, there was no difference in the CYP707A2 promoter AcH3 level during dehydration stress in the pwr mutant compared to the WT (Supplementary Figure S4), suggesting that PWR and HDA9 are both required for repression of CYP707A1 in response to drought stress.
Besides its role in regulating plant morphological and developmental processes, little is known about how PWR regulate signal transduction and chromatin modification in response to abiotic stresses. Based on our results, we propose a model for how PWR, together with HDA9 and ABI4, mediates biological functions by negatively regulating the expression of genes through chromatin modification during ABA-dependent drought stress (Figure 7). On one hand, PWR and HDA9 mutants displayed the same phenotype with and without exogenous ABA and interact specifically with ABI4, indicating that PWR, HDA9, and ABI4 might regulate the same set of genes in ABA pathway. On the other hand, in the absence of drought stress, ABA-catabolism-related genes such as CYP707A1 are activated by histone acetylation; the CYP707A1 gene product, (+)-abscisic acid 8′-hydroxylase, then converts active ABA to the inactive form (8′-hydroxy-ABA) inside guard cells, resulting in loss of turgor pressure and stomatal opening (Xiong et al., 2002, Xiong and Zhu, 2003). ABI4 was previously reported to suppress the CYP707A genes by directly binding to their promoters (Shu et al., 2013), and here we report that ABI4 together with PWR and HDA9 may associate with the CYP707A1 promoter. This possible association with the PWR-HDA9-ABI4 repressor complex represses CYP707A1 expression through histone deacetylation and results in drought tolerance. In addition to repressing genes, ABI4 is also involved in the activation of several genes. Since HDA9 and PWR regulate a wide range of genes involved in several key physiological processes including autophagy, pathogenesis and senescence, there might be some other unidentified genes through which HDA9 and PWR regulate ABA signaling and drought stress. Further study is required to uncover whether the PWR-HDA9-ABI4 complex is also involved in activating genes responsible for ABA signaling and drought tolerance.
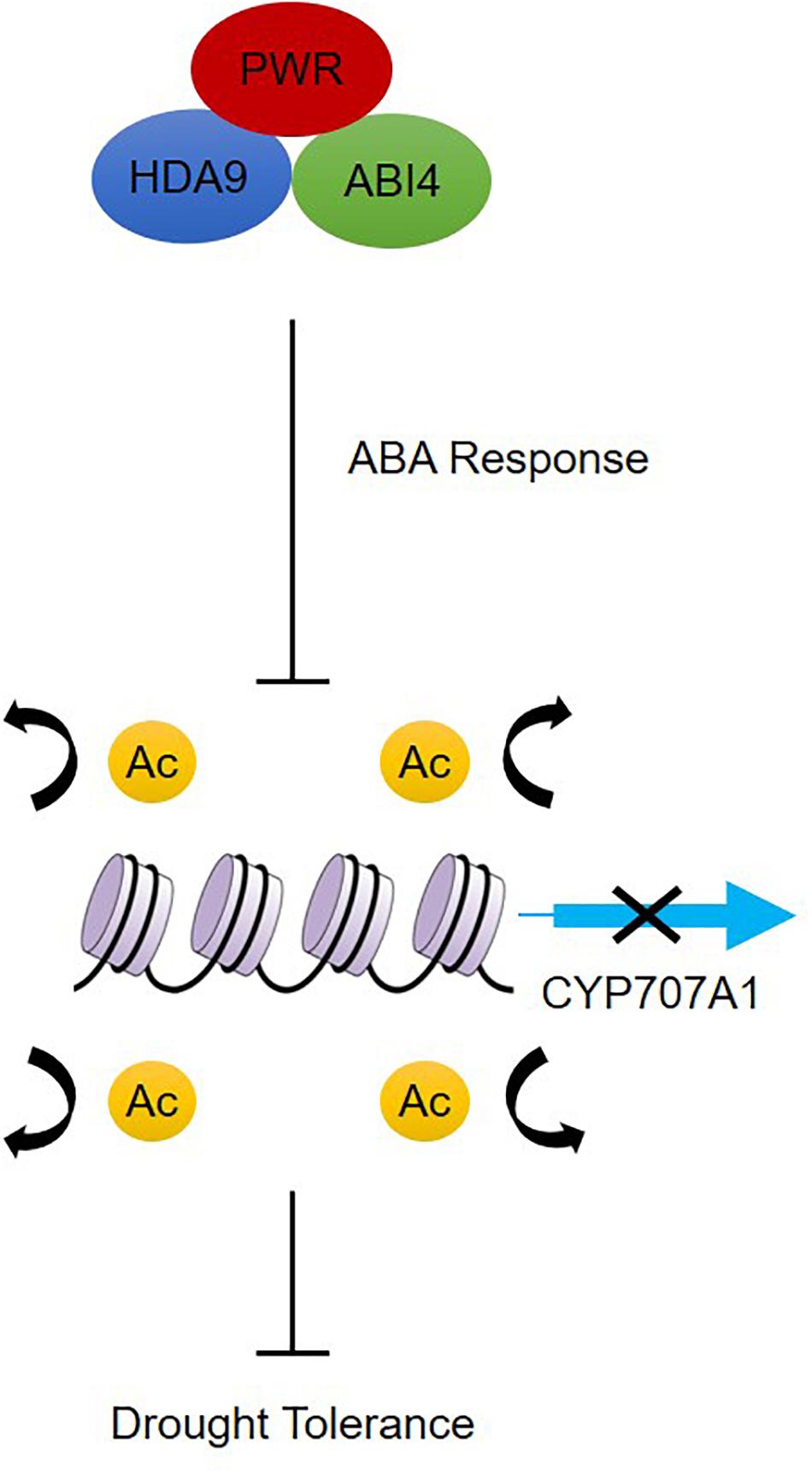
Figure 7. Proposed working model of PWR function in dehydration response. PWR together with HDA9 and ABI4 positively regulate ABA response either by acting at ABA receptors or by repressing negative regulators of the ABA pathway, which results in downstream expression of ABA-responsive genes. In the absence of dehydration stress, CYP707A1 is expressed and regulates ABA hydroxylation, but dehydration stress triggers PWR-HDA9-ABI4-mediated repression of CYP707A1 expression through the process of deacetylation and activates drought response.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
IK, AA, and D-JY conceived and designed the experiments. IK, HK, SZ, MJ, DB, JP, and CL performed the experiments. IK, AA, JP, WK and D-JY analyzed the data and wrote the manuscript. All authors reviewed and approved the final manuscript.
This work was supported by the Next Generation Bio-Green21 Program, Rural Development Administration (RDA), Republic of Korea (SSAC, PJ01318201 to D-JY and PJ01318205 to JP); the National Research Foundation of Korea (NRF) funded by the Korean Government (2019R1A2C2084096 to D-JY) and Global Research Lab (2017K1A1A2013146 to D-JY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00623/full#supplementary-material
Ali, A., Kim, J. K., Jan, M., Khan, H. A., Khan, I. U., Shen, M., et al. (2019). Rheostatic control of ABA signaling through HOS15-mediated OST1 degradation. Mol. Plant 12, 1447–1462. doi: 10.1016/j.molp.2019.08.005
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome- wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657.
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A., and Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158, 970–980. doi: 10.1104/pp.111.188623
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Bosch-Presegué, L., and Vaquero, A. (2015). Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 282, 1745–1767. doi: 10.1111/febs.13053
Bossi, F., Cordoba, E., Dupré, P., Mendoza, M. S., Román, C. S., and León, P. (2009). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59, 359–374. doi: 10.1111/j.1365-313X.2009.03877.x
Chen, X., Lu, L., Mayer, K. S., Scalf, M., Qian, S., Lomax, A., et al. (2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5:e17214. doi: 10.7554/eLife.17214
Dong, Z., Yu, Y., Li, S., Wang, J., Tang, S., and Huang, R. (2016). Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol. Plant 9, 126–135. doi: 10.1016/j.molp.2015.09.007
Finkelstein, R. R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771.
Finkelstein, R. R., and Lynch, T. J. (2000). Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol. 122, 1179–1186. doi: 10.1104/pp.122.4.1179
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S.-Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Giraud, E., Van Aken, O., Ho, L. H., and Whelan, J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296. doi: 10.1104/pp.109.139782
Guy, C. L., Niemi, K. J., and Brambl, R. (1985). Altered gene expression during cold acclimation of spinach. Proc. Natl. Acad. Sci. U.S.A. 82, 3673–3677. doi: 10.1073/pnas.82.11.3673
Haberland, M., Montgomery, R. L., and Olson, E. N. (2009). The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42. doi: 10.1038/nrg2485
Hasegawa, P. M., Bressan, A. B., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Henderson, I. R., and Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature 447, 418–424.
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Horn, P. J., and Peterson, C. L. (2002). Molecular biology: chromatin higher order folding–wrapping up transcription. Science 297, 1824–1827. doi: 10.1126/science.1074200
Huang, X., Zhang, X., Gong, Z., Yang, S., and Shi, Y. (2017). ABI4 represses the expression of type- ARRs to inhibit seed germination in Arabidopsis. Plant J. 89, 354–365. doi: 10.1111/tpj.13389
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Kang, M. J., Jin, H. S., Noh, Y. S., and Noh, B. (2015). Repression of flowering under a noninductive photoperiod by the HDA9- AGL19-FT module in Arabidopsis. New Phytol. 206, 281–294. doi: 10.1111/nph.13161
Kim, W., Latrasse, D., Servet, C., and Zhou, D. X. (2013). Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432, 394–398. doi: 10.1016/j.bbrc.2012.11.102
Kim, Y. J., Wang, R., Gao, L., Li, D., Xu, C., Mang, H., et al. (2016). POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 14858–14863. doi: 10.1073/pnas.1618618114
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719.
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8#-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Lee, K., Park, O. S., Jung, S. J., and Seo, P. J. (2016). Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J. Plant Physiol. 191, 95–100. doi: 10.1016/j.jplph.2015.12.006
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Luan, S. (2002). Signaling drought in guard cells. Plant Cell Environ. 25, 229–237. doi: 10.1046/j.1365-3040.2002.00758.x
May, M. J., Vernoux, T., Leaver, C., Montagu, M. V., and Inzé, D. (1998). Glutathione homeostasis in plants: implications for environmental sensing and plant development. J. Exp. Bot. 49, 649–667.
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., and Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709. doi: 10.1111/j.1365-313X.2005.02337.x
Nambara, E. (2005). Marion-poll A: abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185.
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Park, J., Lim, C. J., Khan, I. U., Jan, M., Khan, H. A., Park, H. J., et al. (2018). Identification and molecular characterization of HOS15 interacting proteins in Arabidopsis thaliana. J. Plant Biol. 61, 336–345. doi: 10.1186/s12870-015-0461-1
Quesada, V., Ponce, M. R., and Micol, J. L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436.
Reeves, W. M., Lynch, T. J., Mobin, R., and Finkelstein, R. R. (2011). Direct targets of the transcription factors ABA-Insensitive (ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 75, 347–363. doi: 10.1007/s11103-011-9733-9
Rosso, M. G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449. doi: 10.1104/pp.103.037614
Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Seto, E., and Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6:a018713. doi: 10.1101/cshperspect.a018713
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223.
Shu, K., Chen, F., Zhou, W., Luo, X., Dai, Y., and Shuai, H. (2018). ABI4 regulates the floral transition independently of ABI5 and ABI3. Mol. Biol. Rep. 45, 2727–2731. doi: 10.1007/s11033-018-4290-9
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Söderman, E. M., Brocard, I. M., Lynch, T. J., and Finkelstein, R. R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124, 1752–1765. doi: 10.1104/pp.124.4.1752
Struhl, K. (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606. doi: 10.1101/gad.12.5.599
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
van Zanten, M., Zoll, C., Wang, Z., Philipp, C., Carles, A., Li, Y., et al. (2014). HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 80, 475–488. doi: 10.1111/tpj.12646
Xiong, L., Lee, H., Ishitani, M., and Zhu, J. K. (2002). Regulation of osmotic stress responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277, 8588–8596. doi: 10.1074/jbc.M109275200
Xiong, L., and Zhu, J. K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36.
Yumul, R. E., Kim, Y. J., Liu, X., Wang, R., Ding, J., Xiao, L., et al. (2013). POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLoS Genet. 9:e1003218. doi: 10.1371/journal.pgen.1003218
Zhao, T., Rui, L., Li, J., Nishimura, M. T., Vogel, J. P., Liu, N., et al. (2015). A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 11:e1004945. doi: 10.1371/journal.pgen.1004945
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Keywords: Arabidopsis thaliana, epigenetic regulation, powerdress (PWR), HDA9, ABI4, chromatin remodeling, deacetylation, drought stress
Citation: Khan IU, Ali A, Khan HA, Baek D, Park J, Lim CJ, Zareen S, Jan M, Lee SY, Pardo JM, Kim WY and Yun D-J (2020) PWR/HDA9/ABI4 Complex Epigenetically Regulates ABA Dependent Drought Stress Tolerance in Arabidopsis. Front. Plant Sci. 11:623. doi: 10.3389/fpls.2020.00623
Received: 19 December 2019; Accepted: 22 April 2020;
Published: 26 May 2020.
Edited by:
Nam-Chon Paek, Seoul National University, South KoreaReviewed by:
Gyeong Mee Yoon, Purdue University, United StatesCopyright © 2020 Khan, Ali, Khan, Baek, Park, Lim, Zareen, Jan, Lee, Pardo, Kim and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dae-Jin Yun, ZGp5dW5Aa29ua3VrLmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.