- 1College of Agro-Grassland Sciences, Nanjing Agricultural University, Nanjing, China
- 2Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Grassland Science, Beijing Forestry University, Beijing, China
Related to ABSCISIC ACID INSENSITIVE3 (ABI3)/VIVIPAROUS1(VP1)(RAV) transcription factors, which encode a B3 domain and an APETALA2(AP2) domain, belong to the APETALA2/ethylene-responsive element binding factor(AP2/ERF) or B3 superfamily and play an important role in regulating plant growth and development and responding to abiotic stress. Although there have been many functional studies on RAV, the functional differences between RAVs are not clear. Therefore, in this study, the functional differences of RAVs of Medicago truncatula were analyzed. Based on sequence data from the plant transcription factor database and the M. truncatula genome database, we cloned three RAV genes from M. truncatula, named MtRAV1, MtRAV2, and MtRAV3. The cis-acting elements of these genes promoters were predicted, and the expression patterns of MtRAVs under exogenous conditions (4°C, NaCl, Polyethylene Glycol, Abscisic acid) were analyzed. MtRAVs transgenic Arabidopsis thaliana were obtained and subjected to adversity treatment. Subcellular localization results indicated that MtRAVs were located in the nucleus. A much lower expression level was observed for MtRAV3 than the levels of MtRAV1 and MtRAV2 in M. truncatula for growth in normal conditions, but under 4°C or PEG and NaCl treatment, the expression level of MtRAV3 was significantly increased. Only the MtRAV3 overexpression transgenic plants showed strong cold resistance, but the overexpressed MtRAV1 and MtRAV2 transgenic plants showed no difference from wild type plants. MtRAV transgenic plants exhibited similar response to exogenous mannitol, NaCl, and ABA, and the expression of some adverse-related marker genes were up-regulated, such as COLD REGULATED 414 THYLAKOID MEMBRANE 1 (COR414-TM1), Arabidopsis thaliana drought-induced 21 (AtDI21), and Arabidopsis thaliana phosphatidylinositol-specific phospholipase C (ATPLC). MtRAVs transgenic Arabidopsis thaliana exhibited increasing of branch number. These results indicated that there was some function redundancy during MtRAVs proteins of M. truncatula, and MtRAV3 has increased function compared to the other two genes. The results of this study should provide the foundation for future application of MtRAVs in legumes.
Introduction
RAV (Related to ABI3/VP1) transcription factors belong to the AP2 or B3 superfamilies. These proteins encompass an AP2 domain in the N-terminal region, and a B3 domain in the C-terminal region, having two unrelated DNA binding domains, a property that is unique among transcription factors (Swaminathan et al., 2008; Mizoi et al., 2012). RAVs and some TFs, such as GRAS and NAC, are plant-specific transcription factors, and much less is known about these plant-specific proteins compared to TF families that have orthologs in animals and bacteria (Riechmann et al., 2000). AP2/ERF proteins are classified into four subfamilies: AP2, DREB (for dehydration response element-binding protein), ERF, and RAV (Mizoi et al., 2012). The B3 superfamily also encompasses four distinct protein families, including ARF (auxin response factor), LAV (leafy cotyledon2 [LEC2]–abscisic acid insensitive three [ABI3]–val), REM (reproductive meristem) and RAV families (Swaminathan et al., 2008). AP2 and B3 proteins were shown to be involved in the regulation of biotic and abiotic stress responses, plant growth and development (Park et al., 2001; Zhang et al., 2005; Swaminathan et al., 2008). Transcription factors, such as AP2, DREB (CBF), ERF and LAV (ABI) families in plants, regulate the expression of defense genes in responses to biotic and abiotic stresses, seed maturation and seedling growth (Swaminathan et al., 2008; Dietz et al., 2010; Mizoi et al., 2012).
Compared with other subfamilies of the AP2/ERF superfamily, the plant RAV family is a relatively small transcription factor family, with orthologs found only in higher plant species. There are seven members in Arabidopsis thaliana, five in Glycine max, and three in M. truncatula. Some genes of the RAV family have been studied in plants such as Arabidopsis thaliana, Zea mays, Oryza sativa, Solanum lycopersicum, and Capsicum annuum (Hu et al., 2004; Lee et al., 2010; Li et al., 2011; Min et al., 2014; Zhu et al., 2018), with functions related to growth and development, hormone regulation, and response to adverse situation (Endres et al., 2010; Matías-Hernández et al., 2014).
Previous research revealed that the expression levels of GhRAV1, ZmRAV1 and GmRAV-03 genes were up-regulated by salt, polyethylene glycol (PEG), or dehydration, respectively, in Gossypium hirsutum, Zea mays, and Glycine max (Min et al., 2014; Li et al., 2015; Zhao et al., 2017). AtRAV1 or AtRAV2, GmRAV-03, ZmRAV1, CaRAV1, and SlRAV2 transgenic plants exhibited increased resistance to high salt and drought compared to wild-type, by activation of resistance-related genes in the pathway of response to abiotic stress (Sohn et al., 2006; Li et al., 2011; Min et al., 2014; Mittal et al., 2014; Zhao et al., 2017).
Cold stress is an important factor that affects plant growth and distribution. RAV play a regulatory role in the CBF-RAV-ERF-PR signaling cascade, part of the defense mechanism related to biotic stress response (Li et al., 2011). RAV1 functions as a cold-responsive transcription factor that may also be involved in a regulatory pathway independent of DREB/CBF (Fowler and Thomashow, 2002). The BnaRAV-1-HY15 gene was induced by cold treatment (Zhuang et al., 2011), and a separate report determined that cold stress induced nearly half of the RAV subfamily transcripts (Du et al., 2016). The overexpression of SlRAV2 in Solanum lycopersicum resulted in stronger tolerance to cold stress (Li et al., 2011). When Zea mays seedlings were immersed in Hoagland solution supplemented with 25 mM LiCl, the expression of ZmRAV1 was induced initially and remained at high level for 24 h (Min et al., 2014). These results suggested that RAV proteins play important roles in the salt, drought and cold response.
RAV has been reported to be involved in ABA signal pathways. For example, RAV was induced rapidly by ABA in some plants such as Glycine max, Zea mays, Gossypium hirsutum, and Capsicum annuum (Sohn et al., 2006; Min et al., 2014; Li et al., 2015; Zhao et al., 2017). However, GmRAV-03 transgenic plants were insensitive to exogenous ABA treatment, although ABA dramatically induced the expression of all GmRAV genes (Zhao et al., 2017). RAV also was shown to be involved in signaling crosstalk of other plant hormones, such as GA, BR, and ethylene (Zhu et al., 2018).
AtRAV1 and its orthologs in other species have been reported to participate in various biological processes such as plant growth and development. In Arabidopsis thaliana, overexpression of TEM (belong to RAV subfamily) genes resulted in retarded-growth phenotypes of transgenic plants, through repressing the expression of GA4 biosynthetic genes (Osnato et al., 2012). Arabidopsis thaliana AtRAV1 was reported to act as a positive regulator to cause leaf senescence (Woo et al., 2010). Transgenic tobacco overexpressing GmRAV exhibited slower plant growth rate, delayed flowering time, decreased root elongation, down-regulated photosynthetic rate, and decreased chlorophyll contents of leaves (Zhao et al., 2008; Lu et al., 2014). In addition, both ZmRAVL1 RNAi and knockout lines exhibited smaller leaf angle in lower, middle, and upper leaves compared with wild-type plants (Tian et al., 2019). These results indicate the involvement of RAV proteins in the complex regulation network of plant growth and development.
However, there is a certain difference or even the opposite function during RAV TFs of different species or the same species. AtRAV1-overexpressing transgenic Arabidopsis thaliana displayed strong growth inhibition accompanied by early senescence, while the AtRAV1L or AtRAV2-overexpressing transgenic plants did not show any alterations in the growth and development compared to wild type (Fu et al., 2014). Gossypium hirsutum GhRAV1 transgenic Arabidopsis thaliana displayed weaker resistance than wild type to the treatment of NaCl or drought stress (Li et al., 2015). While Zea mays ZmRAV1 transgenic Arabidopsis thaliana enhanced salt and osmotic stress tolerance compared to the wild type, through increasing survival rate, longer primary roots and lower relative electrolyte leakages (Min et al., 2014).
RAVs are important genes affecting plant growth and development, hormone regulation, and response to adverse situation. Although there have been many reports on the functions of RAV family proteins, new functions have been discovered continuously in recent years, suggesting a strong exploration potential. There have been few systematic studies on the relationships between RAVs, and it is not clear to what extent there is functional redundancy between genes. Since M. truncatula is a model plant, its genome information is relatively complete, so used M. truncatula to conduct a systematic study on the RAV family. The purpose of this study was: (I) to investigate the functions of RAV genes in M. truncatula and (II) to further explore the regulatory network involved in RAV gene effects on stress tolerance, particularly by analysis of the expression characteristics of downstream genes. The findings of this study on M. truncatula RAVs can be extended to other legumes such as alfalfa, laying a foundation for research into the basic functions of RAVs for improved utilization of genetic resources.
Materials and Methods
Identification and Isolation of M. truncatula RAVs
Transcription factor prediction of PlantTFDB and the M. truncatula genome information provided by NCBI indicated that there are three RAV genes in M. truncatula, MtRAV1, MtRAV2 and MtRAV3. None of the genes contain introns. M. truncatula genomic DNA was used as the template, and MtRAV1P-F/R, MtRAV2P-F/R, and MtRAV3P-F/R (Supplementary Table S1) were used as the primers to amplify the CDS regions of MtRAV1, MtRAV2, and MtRAV3.
Bioinformatics Analysis
The presence of cis-acting elements in the promoters of the three MtRAV genes was analyzed in Plant CARE. We performed a multiple alignment analysis with ClustalX (2.0) and constructed a phylogenetic tree with the neighbor-joining (NJ) method. The phylogenetic tree diagram was constructed using MEGA7.0 Bootstrap.
Subcellular Localization Analysis in M. truncatula
The obtained sequences of the CDS regions of MtRAV1, MtRAV2, and MtRAV3 were PCR-amplified with SAT-RAV-F and SAT-RAV-R primers (Supplementary Table S1). The resulting DNA fragments were fused to plasmid pSAT6-GFP cut with NCO I in the presence of seamless ligase presence to create 35S:MtRAVs-GFP fusions driven by the CaMV35S promoter. After confirmation of the fusion plasmids by DNA sequencing, the recombinant plasmids were transformed into protoplasts of M. truncatula mesophyll cells, with the empty pSAT6-GFP vector as a control. Mesophyll cell protoplasts were transformed as previously described (Yoo et al., 2007). After 18 h of darkness, the transformed protoplasts were monitored by confocal laser scanning microscope.
Plant Materials and Growth Conditions
The wild type used in this study was Arabidopsis thaliana Columbia (Col-0). Seeds were surface-sterilized with 10% NaClO aqueous solution for 10 min, followed by washing eight times with sterile water. Seeds were germinated on 1/2 MS Phytagar plates (pH = 5.8) supplemented with the appropriate antibiotics, vernalized for 2-day in the dark at 4°C, then plates were transferred to the light at 22°C and incubated under a 16 h light/8 h dark cycle. Upon emergence of the third and fourth leaves of seedlings, seedlings were transferred to sterilized soil to continue to grow or to 1/2 MS Phytagar plates containing other treatment materials.
M. truncatula R108 seeds were provided by our lab. After breaking dormancy, seeds were surface-sterilized with 75% alcohol, and then washed eight times with sterile water. These sterilized seeds were placed on aseptic filter paper to sprout, and then were cultivated in Hoagland nutrient solution for 21 days or transferred to sterilized soil for continued growth under conditions of 16 h light at 26°C/8 h dark at 24°C.
Treatment of Various Abiotic Stress and ABA
M. truncatula R108 seedlings grown in Hoagland nutrient solution for 21 days were sprayed with ABA and then sampled at 0, 2, 4, 8, 12, and 24 h. For treatment with salt and PEG abiotic elicitors, M. truncatula 21-day-old seedlings were transferred to Hoagland nutrient solution containing 200 mM NaCl or 15% PEG and incubated for 24 h. For cold treatment, 21-day-old M. truncatula seedlings grown in soil were placed in a low temperature illumination incubator at 4°C for 24 h.
Real-Time Quantitative PCR Analysis
Plant total RNA was isolated with the EastepTM Super Total RNA Extraction Kit and then cDNA synthesis was performed using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time). Total RNA and cDNA were assayed by electrophoresis and quantified by NanoPhotometer 2000. The cDNA was mixed with TB Green Premix Ex Taq for qRT-PCR using an Applied Biosystems (ABI) 7300 Real Time PCR System. The obtained values of the corresponding gene expression were normalized according to the amounts of Mtactin or Atactin for the comparative ΔΔCt method. The primers used for quantitative realtime PCR are presented in Supplementary Table S1. All quantitative analysis were performed on three independent biological replicates.
Generation of Transgenic Plants
The obtained complete sequences of the CDS regions of MtRAV1, MtRAV2 and MtRAV3 were cloned into the T5 pEASY®-T5 Zero Cloning vector. Subsequently, these target gene fragments were amplified with the primers listed in Supplementary Table S1, and then were ligated into the NcoI-digested pCAMBIA3301 (p3301) vector that harbors the CaMV35S promoter in the presence of seamless ligase. To generate the MtRAVs-overexpressing transgenic plants, the constructs were transformed into Agrobacterium tumefaciens (GV3101), and introduced by plant transformation using the floral dipping method (Clough and Bent, 1998). All T1 generation transgenic Arabidopsis thaliana lines of three MtRAV genes were sampled and examined the mRNA transcription level by qRT-PCR methond (Supplementary Figure S4). All experiments were performed with homozygous lines in the T3 generation.
Tolerance Assays Under Stress Conditions
For the ABA, NaCl, and osmotic stress tolerance assays, the seedlings grown on 1/2 MS medium in the normal condition of 16 h light/8 h dark cycle for 5 days and then transferred to 1/2 MS medium containing 20 μm ABA, 200 mM NaCl, or 500 mM mannitol. Three transgenic lines of each MtRAV gene were used for various treatment, include 35S:MtRAV1-#9, #11 and #4, 35S:MtRAV2-#7, #2 and #5, 35S:MtRAV3-#12, #7 and #5 lines (Supplementary Figure S4). All assays were performed in triplicate.
For the NaCl stress assay, the results were observed after seedlings grew for 3 days on the 1/2 MS medium containing 200 mM NaCl, and the number of surviving seedlings was counted. For the ABA and osmotic stress assays, 12 and 7 days after treatment, root length and fresh weight were measured, respectively.
For the cold tolerance assay, wild-type and transgenic plants were grown on 1/2 MS medium containing 1% agar and grown at 22°C for an additional 18-day. The plates were transferred to a -15°C growth chamber and incubated for 2 h, then were incubated for 24 h at 22°C before counting the number of surviving seedlings. Three transgenic lines of each MtRAV gene were used for various treatment, include 35S:MtRAV1-#9, #11 and #4, 35S:MtRAV2-#7, #2 and #5, 35S:MtRAV3-#12, #7 and #5 lines. All assays were performed in triplicate.
Results
Identification of RAV Genes From M. truncatula
To identify RAV transcription factors in M. truncatula, we performed BLAST searching in the NCBI database by using the publicly known RAV subfamily TF protein sequence AT1G13260 as a query sequence. We identified three homologous genes, MTR_1g093600, MTR_5g053920, and MTR_1g116920, now named as MtRAV1, MtRAV2, and MtRAV3, respectively. These genes contain both AP2 and B3 domains. Phylogenetic tree analysis was performed on these three genes plus 17 previously described RAV genes using neighbor-joining method in MEGA7.0 software (Figure 1A). M. truncatula is a model plant of leguminosae, and the RAV proteins sequences are close to those of other leguminosae (Figure 1B). Glycine max RAV (Gm10g204400) has higher homology with MtRAV1 and MtRAV2, with alignment of 89.8 and 86.6% (Figure 1B), respectively. MtRAV3 and GmRAV (Gm20g247300) share 65.2% homology (Figure 1B). There is 86.1% homology between MtRAV1 and MtRAV2, while MtRAV3 has only 45.5 and 46.5% homology with MtRAV1 and MtRAV2 (Figure 1B), respectively. Thus, there may be functional differences between the three MtRAV genes.
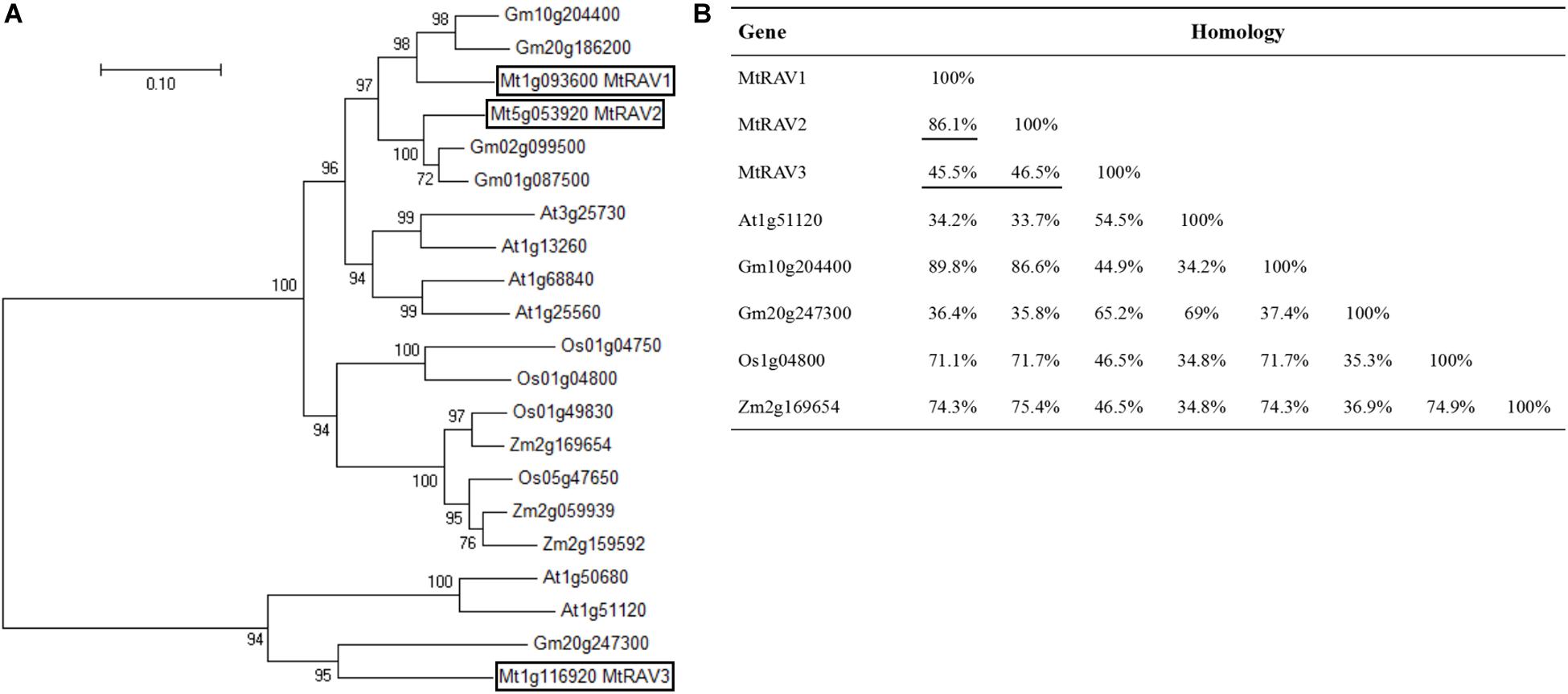
Figure 1. Phylogenetic tree analysis and homologous relation of RAV proteins from different species. (A) Phylogenetic tree analysis of 21 RAV proteins. Sequences were obtained from NCBI. Tree was constructed using neighbor-joining method in MEGA7.0 software. (B) Sequence homology of RAV proteins in the indicated species.
Expression Analysis of RAV Genes in M. truncatula
Based on homology alignment result of the three MtRAV genes, we next examined the expression patterns of the three genes by qRT-PCR in the flower, leaf, stem, and root of M. truncatula. M. truncatula plants grown under conditions of 16 h light at 26°C/8 h dark at 24°C for 52 days were used and RNA samples were extracted from flowers, leaves, stems, and roots. These samples were then used in qRT-PCR analysis. All quantitative analysis were performed on three independent biological replicates. A histogram was constructed to present the expression patterns based on qRT-PCR data (Figure 2). The results showed that MtRAV proteins can be detected in all tissues of M. truncatula during the flowering development period, with different expression levels of the same gene in different tissues. For example, MtRAV1 was expressed at a higher level in leaf, MtRAV2 expression level was higher in root, and MtRAV3 expression level was higher in flower samples. These results suggest that the three MtRAV genes may play different regulatory roles in M. truncatula plants. Next, we examined the relative expression level of the three MtRAV genes in the whole M. truncatula plant by qRT-PCR. RNA of 21-day-old M. truncatula seedlings were extracted and used in qRT-PCR analysis. The results revealed a wide range of expression levels of the three genes. The MtRAV1 transcripts were highest in all tissues, compared to the amounts of MtRAV2 and MTRAV3 transcripts. The expression level of MtRAV1 was 115,076-fold than that of MtRAV3. These results indicated that MtRAV3 may play a particular role in the RAV subfamily in M. truncatula.
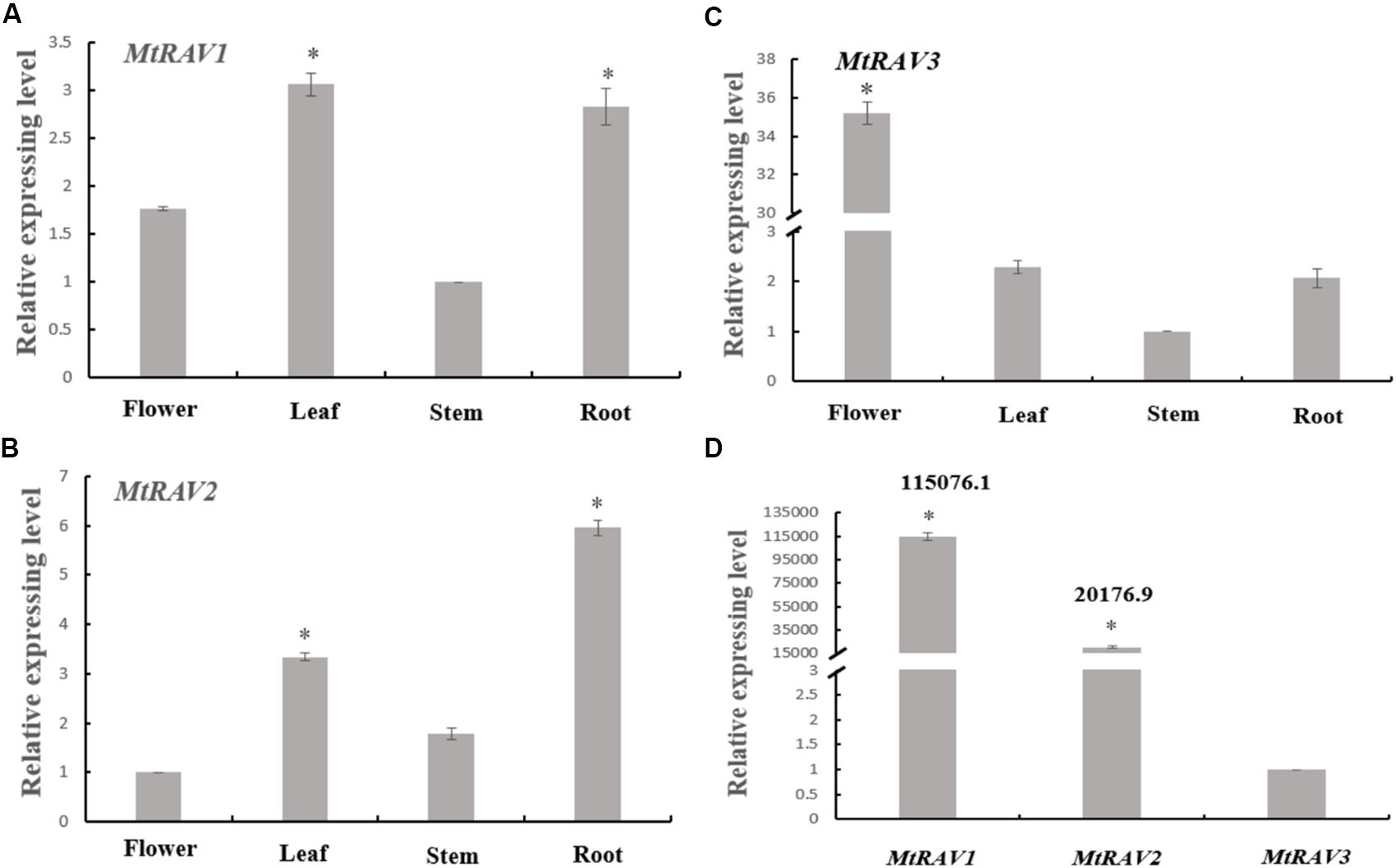
Figure 2. Expression of MtRAVs genes in M. truncatula. (A–C) Relative expression level analysis by qRT-PCR of MtRAV genes in different plant organs of M. truncatula grown for 52 days under conditions of 16 h light at 26°C/8 h dark at 24°C, when the flowers bloom. Flower, leaf, stem, and root samples were analyzed. (A,C) Expression analysis relative to the expression amounts of the MtRAV1 and MtRAV3 gene in the stem sample. (B) Expression analysis relative to the expression amounts of MtRAV2 in flower. (D) The expression level difference analysis of the three MtRAV genes in the whole M. truncatula plant grown for 21 days by qRT-PCR. The results are relative to the expression amounts of MtRAV3. The data are shown as mean values ± SD (n = 3). Independent t-tests demonstrated that there was significant difference (∗P < 0.05).
Subcellular Localization of MtRAVs Genes
Sequence analysis using NCBI showed that MtRAV genes are transcription factors, so to confirm whether MtRAVs localize to the nucleus, we constructed 35S:MtRAVs-GFP fusion proteins. These were delivered into mesophyll protoplast cells of M. truncatula as described in a previous method (Yoo et al., 2007). Next, subcellular localization of GFP expression in M. truncatula protoplasts was observed by confocal microscopy 18 h after transformation of the fused plasmid pSAT6:MtRAV-GFP. The empty 35S:GFP vector was transformed as the control. The results revealed that M. truncatula RAV fusion proteins all localized in the cell nucleus (Figure 3), which suggests that these are nuclear proteins, and may act as transcription factors.

Figure 3. Subcellular localization of the MtRAV proteins. (A,B) The 35S:MtRAV-GFP fusion proteins, were delivered into protoplasmic cells of M. truncatula and transiently expressed, then visualized 18 h after transformation with confocal laser scanning microscopy. Under the green fluorescence excitation channel, MtRAV1-GFP and MtRAV2-GFP obviously localized to the nucleus (green). Scale bars = 8 μm. (C) MtRAV3-GFP also localized to the nucleus (green). Scale bars = 10 μm.
Promoter Prediction and Analysis for MtRAVs
Since localization analysis revealed that all three RAV proteins are located in the nucleus, the promoter sequences were analyzed to predict potential cis-element sequences. We obtained the sequences of the promoters of the three MtRAV genes from NCBI, and then used PlantCARE to identify the cis-elements for the three MtRAV genes. Different cis-elements were identified in the MtRAV gene promoters (Table 1): Only MtRAV1 contained three ABREs, which were absent in the MtRAV2 and MtRAV3 promoter sequences. However, MYC and BOX 4 elements were found in all three MtRAV gene promoters. Additionally, MtRAV2 and MtRAV3 contained MYB repeats, and TC-rich repeats and CGTCA-motif elements were found in MtRAV1 and MtRAV3 promoters. Analyses of these cis-elements in the promoters of the M. truncatula RAV genes provide insight into predicted functions.

Table 1. Distribution of abiotic stress-related cis-acting elements in M. truncatula RAV gene promoters.
Expression Analysis of MtRAVs to Abiotic Stress and ABA
Based on the cis-element sequence analysis of the MtRAV genes promoter, we predicted that RAV proteins might be involved in abiotic stress and ABA signaling regulation pathways. Therefore, we further examined the expression profiles of MtRAV genes by qRT-PCR under ABA and abiotic stress treatment, including cold, NaCl, and PEG. All quantitative analysis were performed on three independent biological replicates. The results showed significant induction of MtRAV3 by cold, salt, and PEG to higher levels than MtRAV1 and MtRAV2 (Figure 4). The expression amounts of MtRAV3 were initially upregulated by low temperature 4°C and by 24 h, increased to 154-fold higher than the control plants grown under normal conditions (Figure 4A). As shown in Figure 4B, in response to exogenous treatment with 200 mM NaCl, the amounts of the three MtRAV transcripts were rapidly upregulated within an hour, and subsequently showed a gradual decline. The results indicated a rapid response of MtRAVs to salt stress. However, under the 15%PEG treatment, the up-regulation degree of MtRAV3 was greater than those of MtRAV1 and MtRAV2 (Figure 4C). For exogenous 100 μM ABA treatment, the expression levels of MtRAV1, MtRAV2, and MtRAV3 were induced, and by 12 h, reached levels that were more than twofold higher than the control plants without ABA treatment. These results suggest that MtRAV3 was more sensitive to abiotic stress than MtRAV1 and MtRAV2.
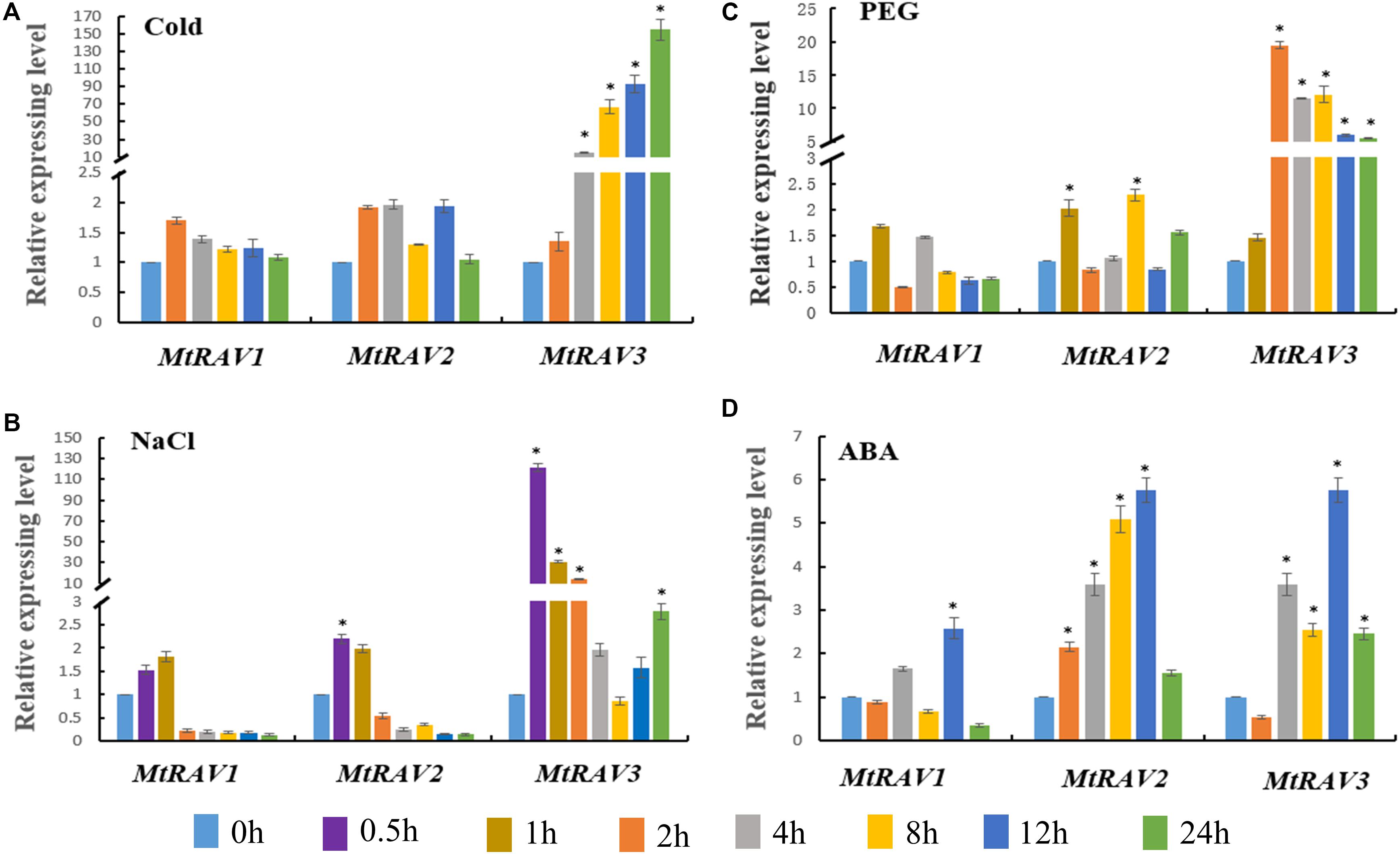
Figure 4. Expression profiles of MtRAV genes in plants treated with Cold, NaCl, PEG, and exogenous application of ABA. MtRAV mRNA expression levels were measured by qRT-PCR analysis. (A) For cold treatment, 21-day-old M. truncatula seedlings grown in soil were placed in a low temperature, illumination incubator at 4°C, and samples were taken at 0, 2, 4, 8, 12, and 24 h. (B) For NaCl treatment, 21-day-old M. truncatula seedling roots were immersed in Hoagland solution supplemented with 200 mM NaCl and samples were removed at 0, 0.5, 1, 2, 4, 8, 12, and 24 h. (C) For osmotic treatment, 21-day-old M. truncatula seedlings were transferred to Hoagland nutrient solution containing 15% PEG and samples were removed at 0, 1, 2, 4, 8, 12, and 24 h. (D) For ABA treatment, 100 μM ABA was sprayed on 21-day-old M. truncatula seedlings grown in Hoagland nutrient solution. Seedlings were sampled at 0, 2, 4, 8, 12, and 24 h. All experiments were replicated three times. Data are shown as the mean values ± SD. Independent t-tests demonstrated that there was significant difference (∗P < 0.05).
Overexpression of MtRAV3 Improved Arabidopsis thaliana Tolerance to Cold
The MtRAVs expression level of all T1-generation transgenic plants were detected by qRT-PCR, and three transgenic strains with the higher MtRAVs expression level were selected for subsequent assays of stress tolerance and phenotypic observation. The results showed that the higher MtRAVs expression level were respectively in the MtRAV1-#4/9/11 lines, MtRAV2-#2/5/7 lines and MtRAV3-#5/7/12 lines.
According to expression pattern analysis of the three MtRAV genes, MtRAV3 was more sensitive to a low temperature of 4°C, with a rapid increase in expression level. Some transcription factors, such as MYB73, RAV1, CRF2, MYB44, ERF6, WRKY33, and CRF3, are considered to be CBF-independent, first-wave transcription factors that act after cold treatment, playing important roles in cold acclimation (Park et al., 2015). We next investigated the induced expression level of cold-related genes in 35S:MtRAV transgenic Arabidopsis thaliana plants (Figure 5). We selected three genes: RD29B (Responsive to desiccation), COR78 (cold-regulated), and COR414-TM1 and analyzed expression of these genes in the MtRAV1, MtRAV2, and MtRAV3 transgenic plants by qRT-PCR (Figure 5A and Supplementary Figure S2). All quantitative analysis were performed on three independent biological replicates. The RD29B expression levels was markedly up-regulated nearly fivefold in MtRAV3-12 transgenic plants, compared to that in WT, and the level was also significantly higher than the expression in MtRAV1-9 and MtRAV2-7. The expression levels of COR78 and COR414-TM1 were also significantly increased, sevenfold and 61-fold in MtRAV3 transgenic plants, respectively. In contrast, the expression of COR414-TM1 was down-regulated in MtRAV1 transgenic plants, compared to that in WT. We further examined the tolerance of three MtRAV-overexpresssion transgenic Arabidopsis thaliana plants to cold treatment. Only the MtRAV3 transgenic plants showed strong cold tolerance ability (Figure 5B), while the MtRAV1 or MtRAV2 transgenic plants did not show significant difference (data not shown) compared with those of the wild type. After cold acclimation, we observed that wild-type Col showed greater freezing sensitivity compared to the lines of MtRAV3. The Col appeared withered and yellow, consistent with freezing that is leading to death. The MtRAV3 showed strikingly stronger tolerance than WT to freezing stress. The survival rates of MtRAV3 transgenic plants were significantly higher than that of WT (Figure 5B). These results clearly showed that overexpression of MtRAV3 enhanced the cold tolerance of transgenic Arabidopsis thaliana and the effects on the expression of stress-responsive genes RD29B, COR78, and COR414-TM1 implied the importance of RAV protein in cold acclimation.
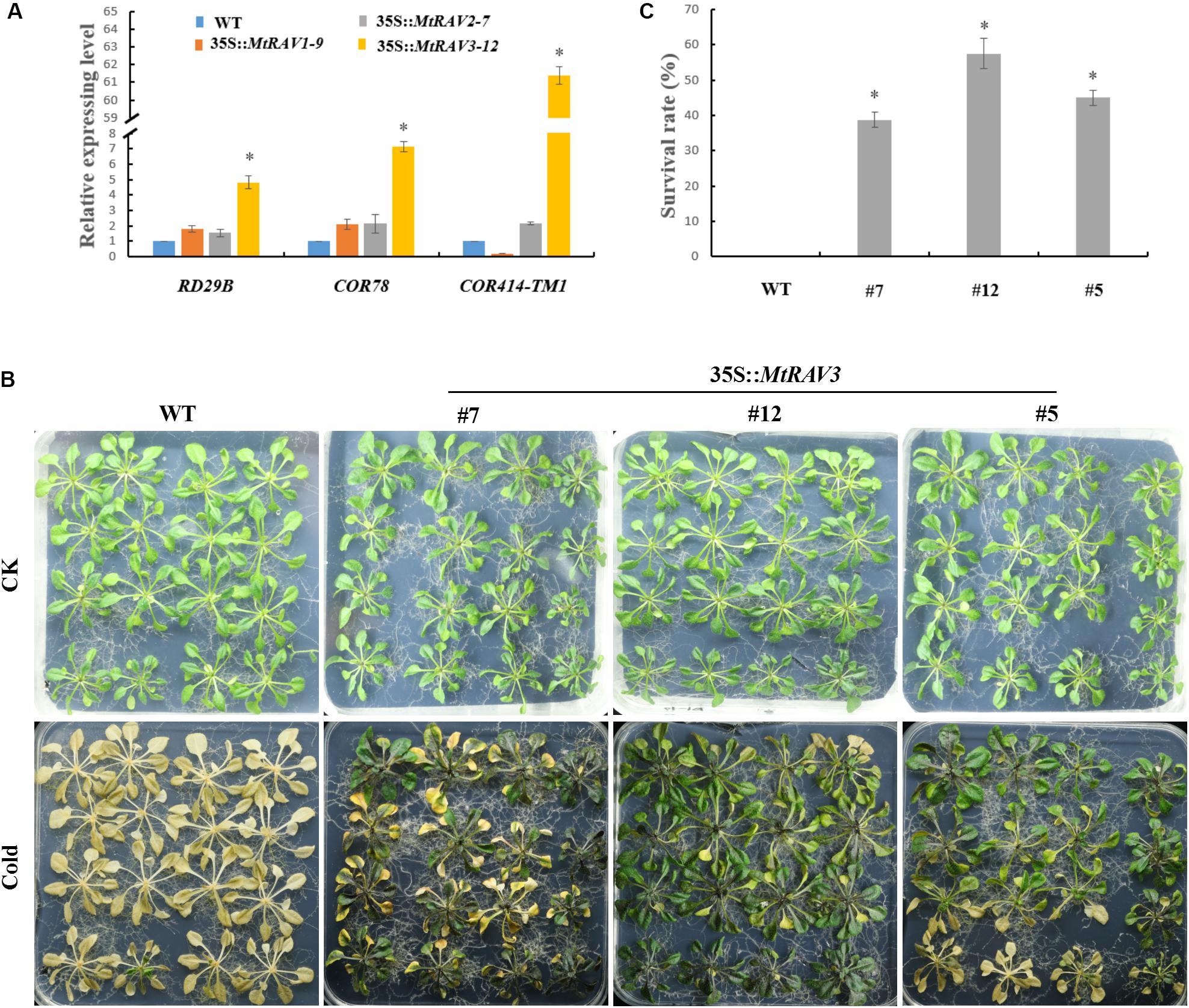
Figure 5. Phenotypic comparison of wild type and 35S:MtRAV transgenic Arabidopsis thaliana under cold treatment. (A) Expression analysis of cold-related marker genes in 35S:MtRAVs transgenic plants by qRT-PCR: RD29B, COR78, and COR414-TM1. (B) Wild-type and 35S:MtRAV3 transgenic Arabidopsis thaliana were grown on 1/2 MS medium containing 1% agar and grown at 22°C for an additional 18-day. The plates were transferred to a -15°C growth chamber and incubated for 2 h, and then returned to normal growth for 24 h at 22°C. (C) After 24 h normal growth after the -15°C cold treatment, we counted the rate of surviving seedlings. This rate was expressed as a percentage (%). Independent t-tests demonstrated that there was significant difference (∗P < 0.05).
Overexpression of MtRAVs Improved Arabidopsis thaliana Tolerance to Osmotic Stress and High Salinity
Since MtRAV transgenic seedlings showed different tolerance to cold and only MtRAV3 transgenic seedlings showed significant cold resistance, we next explored the phenotypes of MtRAV transgenic Arabidopsis thaliana under osmotic stress and salt stress.
All lines showed similar phenotypes in response to osmotic stress when grown on 1/2 MS medium (Figure 6A). However, with exposure to 500 mM mannitol for 7 days (Figure 6), the WT seedlings gradually became yellow and purple. The root length was also shorter than that of transgenic lines (Figures 6B–D). The root lengths of the MtRAV1, MtRAV2, and MtRAV3 transgenic plants were 13.73, 17.37, and 8.53% longer than that of wild-type (Figure 6E), respectively. Meanwhile, there was significant increase of fresh weight in the MtRAV1, MtRAV2, and MtRAV3 transgenic plants, 32.74, 25.76, and 32.87% compared to wild-type plants (Figure 6F). Collectively, MtRAVs transgenic plants were more tolerant than wild-type plants to osmotic stress caused by mannitol at the seedling stage.
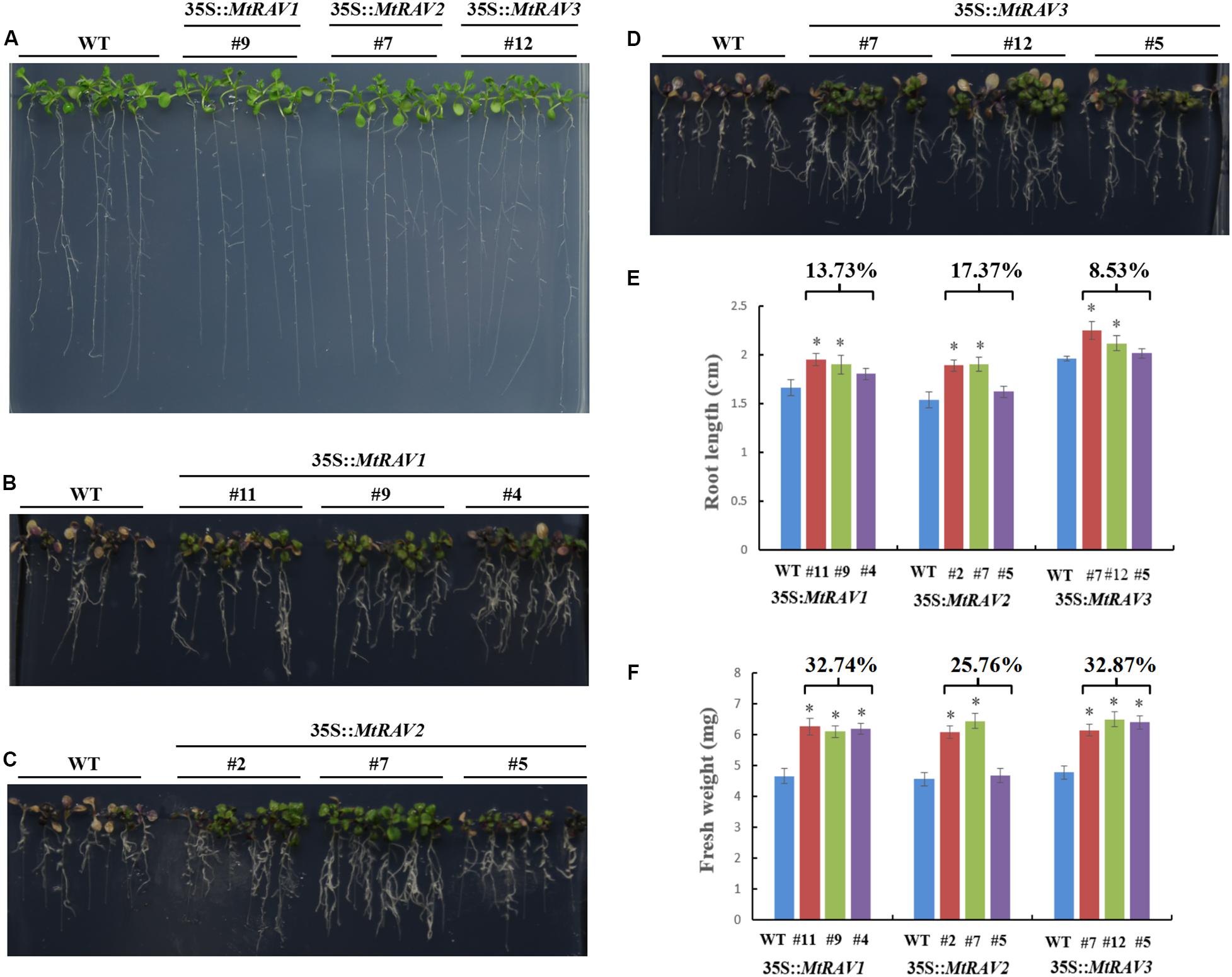
Figure 6. Phenotypic comparison of wild type and 35S:MtRAVs transgenic plants under 500 mM mannitol treatment. (A) 5-day-old seedlings of three transgenic lines and WT Arabidopsis thaliana were planted on 1/2 MS medium without mannitol for 7 days. (B–D) 5-day-old seedlings of three transgenic lines and WT Arabidopsis thaliana were planted on 1/2 MS medium with 500 mM mannitol for 7 days. (E) The root lengths of MtRAVs transgenic lines and WT were measured 7 days after 500 mM mannitol treatment. The percentage represents the average increase of root length in MtRAV transgenic seedlings compared to WT, for 500 mM mannitol treatment. (F) The fresh weight of MtRAV transgenic lines and WT were measured 7 days after 500 mM mannitol treatment. The percentage represents the average increase of fresh weight in MtRAV transgenic seedlings compared to WT, under 500 mM mannitol treatment. Independent t-tests demonstrated that there was significant difference (∗P < 0.05).
In the salt treatment assay, 5-day-old seedlings were placed on 1/2 MS medium containing 200 mM NaCl for 3 days. The leaves of wild-type plants gradually became bleached or died, but the vast majority of leaves of 35S:MtRAVs transgenic Arabidopsis thaliana remained green, indicating they were less affected by NaCl. In the seedling stage, the survival rates were measured as 69.4, 66.7, and 64.8%,for the MtRAV1, MtRAV2, and MtRAV3 transgenic plants compared to wild-type plants as the control (Figure 7E). Collectively, the results suggested that MtRAV proteins may participate in plant response to high salinity, and overexpression of MtRAV genes contributes to the enhanced tolerance to salt stress during the seedling stage.
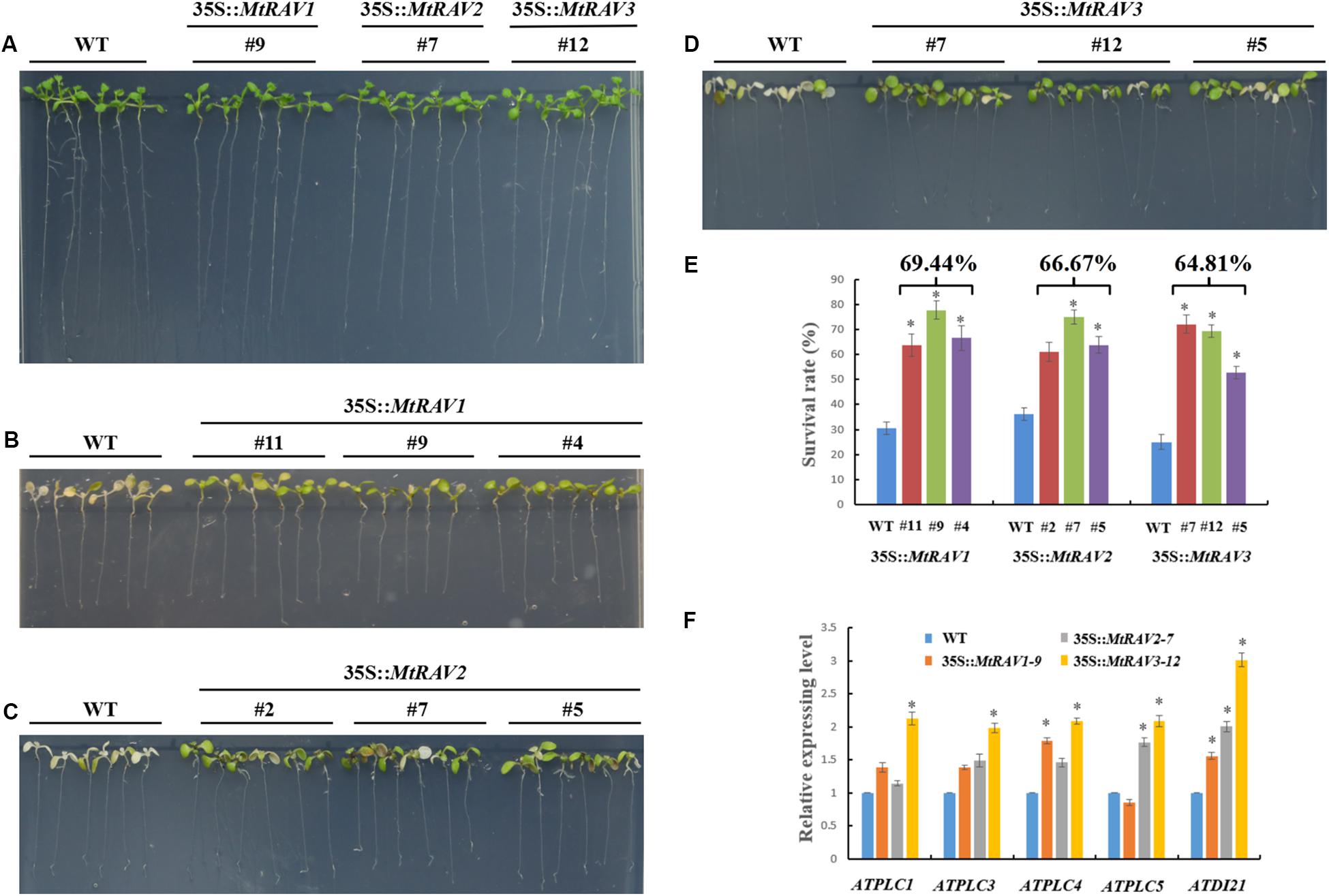
Figure 7. Responses of wild type and 35S:MtRAVs transgenic plants to 200 mM NaCl treatment. (A) 5-day-old seedlings of three transgenic lines and WT Arabidopsis thaliana were planted on 1/2 MS medium without NaCl for 3 days. (B–D) 5-day-old seedlings of three transgenic lines and WT Arabidopsis thaliana were planted on 1/2 MS medium with 200 mM NaCl for 3 days. (E) The survival rate of MtRAVs transgenic lines and WT were measured 3 days after 200 mM NaCl treatment. The percentage represents the average survival rate in MtRAVs transgenic seedlings under 200 mM NaCl treatment. (F) Expression analysis of adversity related marker genes in 35S:MtRAVs transgenic plants compared to WT; levels of ATDI21, ATPLC1, ATPLC3, ATPLC4, and ATPLC5 were determined. Independent t-tests demonstrated that there was significant difference (∗P < 0.05).
To further explore the regulation mechanism of MtRAVs on osmotic stress and salt stress, we next examined the expression of some marker genes related to abiotic stress by qRT-PCR, including ATDI21, AtPLC1, AtPLC3, AtPLC4, and AtPLC5 (Figure 7 and Supplementary Figure S2), induced by abiotic stress. All quantitative analysis were performed on three independent biological replicates. AtPLC3 and AtPLC4 were measured in MtRAV1, MtRAV2, and MtRAV3 transgenic plants and compared to WT. The expression of the AtPLC1 gene was almost unchanged in MtRAV2 transgenic plants, and the expression of AtPLC5 was slightly down-regulated in MtRAV1 transgenic plants. However, the expression levels of AtPLC1, AtPLC3, AtPLC4 and AtPLC5 were all increased in MtRAV3 transgenic Arabidopsis thaliana plants compared to wild type. The previous results revealed that the up-regulated expression of ATDI21 was conducive to drought tolerance in Arabidopsis thaliana (Fang et al., 2015). There was increased expression of AtDI21 in MtRAV1, MtRAV2, and MtRAV3 transgenic plants. There was a greater than twofold effect for AtDI21 in MtRAV3 transgenic plants, with a comparatively lower effect (<2-fold) in MtRAV1 transgenic plants. The results indicate that the overexpression of MtRAV genes induced the expression of AtPLC1, AtPLC3, AtPLC4, AtPLC5, and ATDI21 in Arabidopsis thaliana, with the largest effect observed for the MtRAV3 transgenic plants.
Overexpression of MtRAVs Resulted in the Transgenic Arabidopsis thaliana Hyposensitive to Exogenous ABA
Because exogenous ABA increased MtRAV gene expression and ABA is involved in regulation of stress response pathways, we next investigated whether overexpression of MtRAV genes affects the response of plants to exogenous ABA. We examined response of plant growth to ABA for three homozygous MtRAVs transgenic lines. When germinated on 1/2 MS medium, all lines showed similar growth. However, in the presence of exogenous ABA (MS + ABA), the growth of both WT and MtRAV transgenic seedlings were distinctly inhibited, but the degree of inhibition was greater for the WT than for the MtRAVs transgenic plants (Figure 8). The root lengths of MtRAV transgenic seedlings were significantly longer than those of WT. The transgenic seedlings of the three MtRAVs all displayed an ABA-hyposensitive phenotype. These results suggest that MtRAV genes may be involved in the regulation of abiotic stress in an ABA-independent manner.
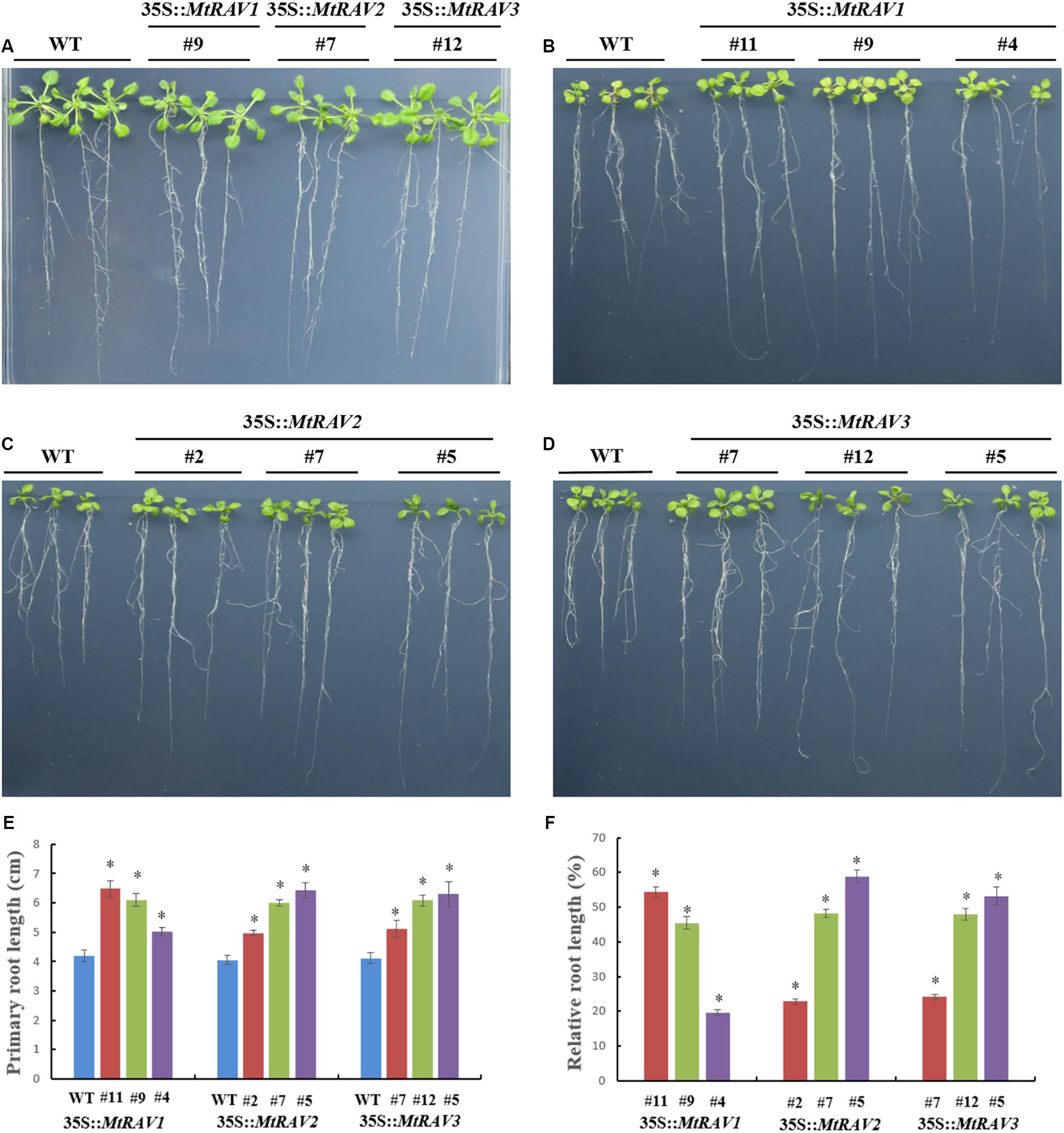
Figure 8. Phenotype assay of 35S:MtRAVs transgenic Arabidopsis thaliana seedlings grown in MS medium, containing ABA. (A) 5-day-old seedlings of the three transgenic lines and WT Arabidopsis thaliana were planted on 1/2 MS medium without ABA for 12 days. (B–D) 5-day-old seedlings of three transgenic lines and WT Arabidopsis thaliana were planted on 1/2 MS medium with 20 μm ABA for 12 days. (E) The primary root length of MtRAVs transgenic lines and WT were measured 12 days after 20 μm ABA treatment. (F) The relative root lengths show an increase of primary root length in MtRAVs transgenic seedlings compared to WT. Independent t-tests demonstrated that there was significant difference (∗P < 0.05).
Overexpression of MtRAVs Increase the Branch Number of Arabidopsis thaliana
In addition to the regulatory abiotic stress described above, MtRAV transgenic Arabidopsis thaliana also exhibit a growth phenotype characterized by increased branch number. Some MtRAV transgenic lines of higher expression level showed increased bolting compared with wild type (Figure 9 and Supplementary Figure S3). We analyzed the statistical results of primary branch, secondary branch, and tertiary branch for these lines (MtRAV1-9, MtRAV2-7 and MtRAV3-12, respectively). The number of primary branches compared to WT exhibited a 0.76∼1.2-fold increase and secondary branches increased 1.4∼1.48-fold in MtRAVs transgenic lines for plants grown in normal conditions. There was not a significant difference in the number of tertiary branch of transgenic plants compared to that of WT. The total number of branches showed a significantly increase in MtRAV1, MtRAV2, and MtRAV3 overexpression lines compared to controls, in a range of 0.87∼1.07-fold. We next analyzed the expression pattern of the AtSUS1 gene in plants with MtRAV overexpression (Figure 9 and Supplementary Figure S2). All quantitative analysis were performed on three independent biological replicates. As previously shown, the SUS gene may be related with shoot branching (Poovaiah et al., 2015). In normal growth conditions, the expression of AtSUS1 in line MtRAV2-7 was obviously up-regulated (2.64-fold), with 2.3- and 1.8-fold increase of AtSUS1 gene expression in lines MtRAV1-9 and MtRAV3-12, respectively. These results suggest that MtRAV1, MtRAV2, and MtRAV3 positively regulate the expression of AtSUS1 gene. MtRAV proteins may be involved in the regulation of shoot architecture process.
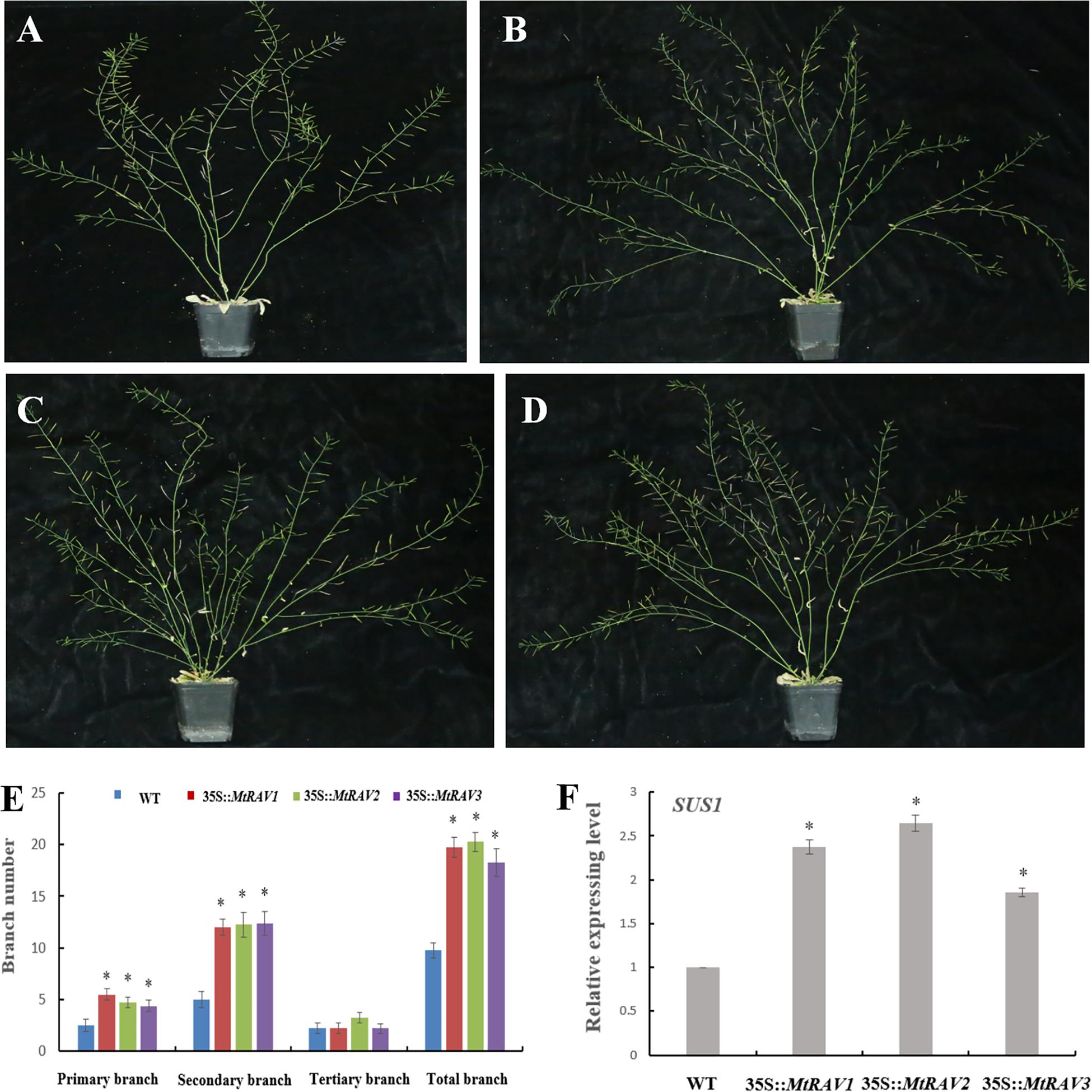
Figure 9. Phenotype of MtRAVs transgenic plants and statistical analysis of branch number compared to wild type Arabidopsis thaliana. (A) Wild type Arabidopsis thaliana grown under normal conditions. (B) 35S:MtRAV1-9 transgenic Arabidopsis thaliana grown under normal conditions. (C) 35S:MtRAV2-7 transgenic Arabidopsis thaliana grown under normal conditions. (D) 35S:MtRAV3-12 transgenic Arabidopsis thaliana grown under normal conditions. (E) The number of branches for WT and 35S:MtRAVs transgenic plants. (F) The expression level of AtSUS1 in MtRAVs transgenic Arabidopsis thaliana. Independent t-tests demonstrated significant difference (∗P < 0.05).
Discussion
RAV transcription factors belonging to the AP2/EREBP or B3 superfamily, which exists only in plants. Previous studies were mostly functional explorations of individual RAV genes in different species. Herein, we identified three MtRAV genes in M. truncatula, and named these three genes MtRAV1, MtRAV2, and MtRAV3. MtRAV1, MtRAV2, and MtRAV3 genes encode proteins of 384, 378, and 298 amino acids (Supplementary Figure S1), respectively. There is 86.1% homology between MtRAV1 and MtRAV2, and MtRAV3 has only 45.5 and 46.5% homology with MtRAV1 and MtRAV2 (Figure 1B), respectively. The MtRAV3 protein is smaller in length than MtRAV1 and MtRAV2, and MtRAV3 also exhibits lower homology, compared to MtRAV1 and MtRAV2. The expression level of MtRAV3 was lower than the expression levels of MtRAV1 and MtRAV2 in M. truncatula plants grown in normal conditions. There may be functional differences in the MtRAV genes, and MtRAV3 may differ from the others. Similarly, a difference was reported for Glycine max GmRAV genes (Zhao et al., 2017).
Previous studies indicated that Glycine max RAVs, Capsicum annuum RAV1, Gossypium hirsutum RAV1, Zea mays RAV1, and Arabidopsis thaliana RAV1 proteins are localized in the cell nucleus (Lee et al., 2010; Woo et al., 2010; Min et al., 2014; Li et al., 2015; Zhao et al., 2017). Green fluorescence of the three pSAT6:MtRAVs-GFP fusion proteins were observed in the cell nuclei, consistent with the localization result of RAV transcription factors in other plants.
The cis-element sequences of a gene promoter are related to the regulatory functions of the gene in plant growth. Our analysis revealed different cis-elements in the promoters of the three MtRAV genes, although they all contained AP2 and B3 domains. This suggested that MtRAV1, MtRAV2, and MtRAV3 proteins may have different functions in plant growth and development, even in the regulation of the stress signaling pathway. The promoters of MtRAV genes mainly contained MYC, MYB, ABRE, TC-rich repeats, BOX4, and CGTCA-motif elements. The MYC element is involved in signaling pathways related to cold regulation (Chinnusamy et al., 2003). The MYB element is involved in the responses of drought and salt stress (Dubos et al., 2010). And ABRE element is involved in plant responses to drought and ABA through their interaction with ABRE binding proteins (Sakuraba et al., 2014; Hwang et al., 2019). BOX4 elements are photosynthetic response elements, TC-rich repeats elements are involved in defense and stress responsiveness, and CGTCA-motifs are related to MeJA-responsiveness (Ciolkowski et al., 2008). Overall, some transcription factors may affect expression of MtRAVs to regulate plant growth, development, and interaction with the environment.
According to the cis-element analysis of MtRAVs gene promoters, we further investigate the expression variation of MtRAVs by qRT-PCR in the response to various abiotic stress treatments. As predicted, our results revealed increased expression of MtRAV3 from the start of 4°C treatment, reaching a level 154-fold higher than the un-treated control at 24 h. The amounts of the three MtRAVs transcripts were rapidly induced within 1 h of NaCl treatment, and the expression of MtRAV3 also markedly increased under PEG treatment. The expression profile of ZmRAV1 in Zea mays in response to 4°C cold treatment immediately increased after treatment (Min et al., 2014), however, CARAV1 to cold exhibited a gradual increased after 6 h of cold (4°C) treatment in Capsicum annuum (Lee et al., 2010). Zea mays ZmRAV1, Gossypium hirsutum GhRAV1, Capsicum annuum CaRAV1, and Brassica napus BnaRAV-1-HY15 were also induced by NaCl, dehydration, and PEG treatments in different stages of growth and development (Lee et al., 2010; Zhuang et al., 2011; Min et al., 2014; Li et al., 2015). However, a separate report showed transcriptional down-regulation of two Arabidopsis thaliana RAV genes (AtRAV1L and AtRAV2) by drought and salt stress (Fu et al., 2014). Overall, these results suggest that MtRAVs are involved in the regulatory pathways of abiotic stress.
We next examined the tolerance of MtRAV transgenic Arabidopsis thaliana to abiotic stress. To do this, we first examined the expression level of cold-related marker genes, including RD29B, COR78, and COR414-TM1, which were markedly up-regulated in MtRAV3 transgenic plants under normal growth conditions compared to the levels in MtRAV1, MtRAV2, and wild type plants. We next observed that only MtRAV3 transgenic Arabidopsis thaliana plants displayed significant cold to lerance ability, compared to MtRAV1 and MtRAV2 transgenic Arabidopsis thaliana, as well as WT. Overexpression of SlRAV2 in Solanum lycopersicum resulted in stronger tolerance ability to cold stress (Li et al., 2011). The ICE–CBF–COR signaling pathway is considered important for cold resistance of plants (Liu et al., 2019). CBFs activate downstream COR genes (Chinnusamy et al., 2003; Park et al., 2015; Zhao C. et al., 2016; Kidokoro et al., 2017). And other transcription factors which are induced during cold acclimation in Arabidopsis thaliana, such as HSFC1, ZAT12, and CZF1, are also regulate the expression of COR genes (Zhao C. et al., 2016; Shi et al., 2017). Overexpression of these genes could induce the expression of COR genes even in normal growth conditions, implying that they could directly regulate COR genes (Park et al., 2015). And AtCBF1 is involved in the regulation of RAV family and ERF family (Li et al., 2011). Thus, the MtRAVs may directly regulate cold-related genes, such as RD29B, COR78, and COR414-TM1. MtRAV3 may have stronger regulation ability than MtRAV1 and MtRAV2 in the cold regulation pathway, but this should be investigated further.
Our results revealed that all three MtRAVs transgenic Arabidopsis thaliana lines increased resistance to salt and osmotic stress to different degrees. Similarly, overexpression Arabidopsis thaliana plants of CaRAV1, GmRAV-03, and ZmRAV1 showed good tolerance to high salinity and osmotic stress compared to wild type plants (Lee et al., 2010; Min et al., 2014; Zhao et al., 2017). Additionally, the overexpression of ZmRAV1 increased the survival rate and primary root length of Arabidopsis thaliana, and decreased relative electrolyte leakage under stress condition (Min et al., 2014). Transgenic Gossypium hirsutum expressing AtRAV1/2 exhibited resistance to drought stress under field and greenhouse conditions, by scavenging reactive oxygen species and osmotic adjustment (Mittal et al., 2014). However, Gossypium hirsutum GhRAV1 transgenic Arabidopsis thaliana exhibited greater sensitivity than wild type plants for NaCl and PEG stress (Li et al., 2015). Thus, there may be species-specific differences in the functions of RAV genes. We next examined the expression of some marker genes related to abiotic stress by qRT-PCR, including ATDI21, AtPLC1, AtPLC3, AtPLC4, and AtPLC5, induced by abiotic stresses (Hirayama et al., 1995; Lee et al., 2005; Tasma et al., 2008; Fang et al., 2015). The results showed that MtRAVs induced the expression of these genes to different degrees. Increased expression levels of ATDI21, AtPLC1, AtPLC3, AtPLC4, and AtPLC5 were observed in MtRAV3 transgenic Arabidopsis thaliana plants compared to wild type. Overall, the results suggested different regulatory functions of MtRAVs in response to different abiotic stresses.
As suggested by the previous cis-element analysis of MtRAV promoters, MtRAVs may be involved in ABA signaling pathways. All MtRAV transgenic Arabidopsis thaliana were hyposensitive to ABA, but the three MtRAV genes were affected to different degrees by ABA, compared to control plants without ABA treatment. Previous studies reported that some stress-responsive genes enhance tolerance to stresses through the ABA-signaling pathway, using both ABA-dependent and ABA-independent methods (Dietz et al., 2010; Fu et al., 2014; Sato et al., 2018). For example, the ABA-PYL-PP2Cs-SnRK2s-AtRAV1-NAC-SAGs signal pathway increased tolerance of stress by causing water to preferentially flow to developing tissues (Zhao Y. et al., 2016). Other RAV genes also showed increased expression in response to ABA, including GhRAV1 and GmRAV-03, but their transgenic Arabidopsis thaliana were insensitive to exogenous ABA (Finkelstein and Lynch, 2000; Li et al., 2015; Zhao et al., 2017). MtRAVs may be involved in ABA signaling pathways through ABA-independent methods.
Previous RAV studies suggested that MtRAVs may act in plant growth and development. The T3 MtRAV transgenic Arabidopsis thaliana revealed a significant increase in the total number of branches in the normal growth condition compared to WT. However, this phenotype is different for other RAV transgenic Arabidopsis thaliana, with reduced lateral roots and rosette leaf reported in AtRAV1-overexpression plants (Hu et al., 2004), strong growth inhibition in AtRAV1-overexpressing transgenic plants (Fu et al., 2014), and late flowering of TEM overexpressing transgenic plants (Castillejo and Pelaz, 2008). Interestingly, Castanea sativa CsRAV1 induced the early formation of sylleptic branches in hybrid poplar Populus tremula × P. alba plantlets. We found 1.8∼2.6-fold increased expression of the sucrose synthase (SUS) gene in MtRAV transgenic Arabidopsis thaliana plants compared to the level in WT. Sucrose synthase (SUS) converts sucrose and uridine di-phosphate (UDP) into UDP-glucose and fructose. Panicum virgatum PvSUS1 transgenic switchgrass showed increased plant tiller number by up to 79% compared to control plants (Poovaiah et al., 2015). These results suggests that a subset of MtRAVs positively regulate the expression of AtSUS1 and possibly regulate axillary bud differentiation. This should be a focus of future work.
As shown above, MtRAVs are involved in regulation of abiotic stress and plant growth and development. The functions of MtRAVs may be diverse, and there may be some gene-specific functions, especially for MtRAV3. Further work is required to explore the functions of MtRAVs.
Conclusion
In summary, three MtRAV genes were cloned from M. truncatula based on transcriptional factor prediction by PlantTFDB and the M. truncatula genome sequence. Constructs encoding these genes were transformed into Arabidopsis thaliana. Expression analysis showed a much lower expression level of MtRAV3 in wild-type M. truncatula plants growing in normal conditions than that of MtRAV1 and MtRAV2, but the expression level of MtRAV3 was significantly increased by application of exogenous low temperature, PEG, and NaCl. The results showed that only MtRAV3-overexpressed transgenic plants showed strong cold resistance. MtRAV transgenic Arabidopsis thaliana plants were hyposensitive to ABA, but more tolerant to high salt stress and osmotic stress. Additionally, increased branch number was observed for MtRAV transgenic plants. According to expression analysis and the observations of transgenic plants, the MtRAVs from M. truncatula are partly redundant, and the function of MtRAV3 is more comprehensive. This study lays the foundation for application of MtRAV genes in legumes.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
YS and SW conceived and designed the experiments. SW and TG performed the experiments. SW and RL analyzed the data. QY and JK contributed reagents, materials, and analysis tools. SW wrote the manuscript. ZW revised the manuscript. All authors read and approved the final manuscript.
Funding
The research was supported by the Agricultural Science and Technology Innovation Program (ASTIP-IAS14), the Fundamental Research Funds for Central Non-profit Scientific Institution (2017ywf-2d-3), and National Natural Science Foundation of China (31772663).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00611/full#supplementary-material
Abbreviations
ABA, abscisic acid; COR, cold-regulated; M. truncatula, Medicago truncatula;; PEG, polyethylene glycol; PLC, Phospholipase C; qRT-PCR, quantitative real-time polymerase chain reaction; RAV, Related to ABI3/VP1.
References
Castillejo, C., and Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. doi: 10.1016/j.cub.2008.07.075
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., and Agarwal, M. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dietz, K., Vogel, M. O., and Viehhauser, A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14. doi: 10.1007/s00709-010-0142-8
Du, C., Hu, K., Xian, S., Liu, C., Fan, J., Tu, J., et al. (2016). Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus l.). Mol. Genet. Genomics 291, 1053–1067. doi: 10.1007/s00438-015-1161-0
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Endres, M. W., Gregory, B. D., Gao, Z., Foreman, A. W., Mlotshwa, S., Ge, X., et al. (2010). Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 6:e1000729. doi: 10.1371/journal.ppat.1000729
Fang, Z., Zhang, X., Gao, J., Wang, P., Xu, X., Zhixiong, L., et al. (2015). A buckwheat (Fagopyrum esculentum) DRE-binding transcription factor gene, FeDREB1, enhances freezing and drought tolerance of transgenic Arabidopsis. Plant Mol. Biol. Rep. 33, 1510–1525. doi: 10.1007/s11105-015-0851-4
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Fu, M., Kang, H. K., Son, S., Kim, S., and Nam, K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55, 1892–1904. doi: 10.1093/pcp/pcu118
Hirayama, T., Ohto, C., Mizoguchi, T., and Shinozaki, K. (1995). A gene encoding a phosphatidylinositol-specific phospholipase c is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 92, 3903–3907. doi: 10.1073/pnas.92.9.3903
Hu, Y. X., Wang, Y. X., Liu, X. F., and Li, J. Y. (2004). Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 14, 8–15. doi: 10.1038/sj.cr.7290197
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., and Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505. doi: 10.1016/j.molp.2019.01.002
Kidokoro, S., Yoneda, K., Takasaki, H., Takahashi, F., Shinozaki, K., Yamaguchi-Shinozaki, K., et al. (2017). Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29, 760–774. doi: 10.1105/tpc.16.00669
Lee, B., Henderson, D. A., and Zhu, J. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175. doi: 10.1105/tpc.105.035568
Lee, S. C., Choi, D. S., Hwang, I. S., and Hwang, B. K. (2010). The pepper oxidoreductase CaOXR1 interacts with the transcription factor CaRAV1 and is required for salt and osmotic stress tolerance. Plant Mol. Biol. 73, 409–424. doi: 10.1007/s11103-010-9629-0
Li, C. W., Su, R. C., Cheng, C. P., Sanjaya, You, S. J., Hsieh, T. H., et al. (2011). Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 156, 213–227. doi: 10.1104/pp.111.174268
Li, X., Li, M., Zhou, Y., Hu, S., Hu, R., Chen, Y., et al. (2015). Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One 10:e118056. doi: 10.1371/journal.pone.0118056
Liu, Y., Dang, P., Liu, L., and He, C. (2019). Cold acclimation by the CBF–COR pathway in a changing climate: lessons from Arabidopsis thaliana. Plant Cell Rep. 38, 511–519. doi: 10.1007/s00299-019-02376-3
Lu, Q., Zhao, L., Li, D., Hao, D., Zhan, Y., Rousselle, T., et al. (2014). A GmRAV ortholog is involved in photoperiod and sucrose control of flowering time in soybean. PLoS One 9:e89145. doi: 10.1371/journal.pone.0089145
Matías-Hernández, L., Aguilar-Jaramillo, A. E., Marín-González, E., Suárez-López, P., and Pelaz, S. (2014). RAV genes: regulation of floral induction and beyond. Ann. Bot. 114, 1459–1470. doi: 10.1093/aob/mcu069
Min, H., Zheng, J., and Wang, J. (2014). Maize ZmRAV1 contributes to salt and osmotic stress tolerance in transgenic Arabidopsis. J. Plant Biol. 57, 28–42. doi: 10.1007/s12374-013-0284-2
Mittal, A., Gampala, S. S. L., Ritchie, G. L., Payton, P., Burke, J. J., Rock, C. D., et al. (2014). Related to ABA-Insensitive3 (ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol. J. 12, 578–589. doi: 10.1111/pbi.12162
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Osnato, M., Castillejo, C., Matías-Hernández, L., and Pelaz, S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3:808. doi: 10.1038/ncomms1810
Park, J. M., Park, C., Lee, S., Ham, B., Shin, R., and Paek, K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Park, S., Lee, C., Doherty, C. J., and Gilmour, S. J. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Poovaiah, C. R., Mazarei, M., Decker, S. R., Turner, G. B., Sykes, R. W., Davis, M. F., et al. (2015). Transgenic switchgrass (Panicum virgatum l.) Biomass is increased by overexpression of switchgrass sucrose synthase (PvSUS1). Biotechnol. J. 10, 552–563. doi: 10.1002/biot.201400499
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Sakuraba, Y., Jeong, J., Kang, M., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Sato, H., Takasaki, H., Takahashi, F., Suzuki, T., Iuchi, S., Mitsuda, N., et al. (2018). Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. U.S.A. 115, E11178–E11187. doi: 10.1073/pnas.1811491115
Shi, Y., Huang, J., Sun, T., Wang, X., Zhu, C., and Ai, Y. (2017). The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integ. Plant Biol. 59, 118–133. doi: 10.1111/jipb.12515
Sohn, K. H., Lee, S. C., Jung, H. W., Hong, J. K., and Hwang, B. K. (2006). Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 61, 897–915. doi: 10.1007/s11103-006-0057-0
Swaminathan, K., Peterson, K., and Jack, T. (2008). The plant B3 superfamily. Trends Plant Sci. 13, 647–655. doi: 10.1016/j.tplants.2008.09.006
Tasma, I. M., Brendel, V., Whitham, S. A., and Bhattacharyya, M. K. (2008). Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 46, 627–637. doi: 10.1016/j.plaphy.2008.04.015
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., and Wu, W. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 6454, 658–664. doi: 10.1126/science.aax5482
Woo, H. R., Kim, J. H., Kim, J., and Kim, J. (2010). The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61, 3947–3957. doi: 10.1093/jxb/erq206
Yoo, S., Cho, Y., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, X., Zhang, Z., Chen, J., and Chen, Q. (2005). Expressing TERF1 in tobacco enhances drought tolerance and abscisic acid sensitivity during seedling development. Planta 222, 494–501. doi: 10.1007/s00425-005-1564-y
Zhao, C., Zhang, Z., Xie, S., Si, T., Li, Y., Zhu, J. K., et al. (2016). Mutational evidence for the critical role of CBF genes in cold acclimation in Arabidopsis. Plant Physiol. 171, 2744–2759. doi: 10.1104/pp.16.00533
Zhao, Y., Chan, Z., Gao, J., Xing, L., Cao, M., Yu, C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113, 1949–1954. doi: 10.1073/pnas.1522840113
Zhao, L., Luo, Q., Yang, C., Han, Y., and Li, W. (2008). A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 227, 1389–1399. doi: 10.1007/s00425-008-0711-7
Zhao, S., Xu, Z., Zheng, W., Zhao, W., Wang, Y., Yu, T. F., et al. (2017). Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front. Plant Sci. 8:905. doi: 10.3389/fpls.2017.00905
Zhu, X. F., Yuan, D. P., Zhang, C., Li, T. Y., and Xuan, Y. H. (2018). RAVL1, an upstream component of brassinosteroid signalling and biosynthesis, regulates ethylene signalling via activation of EIL1 in Oryza sativa. Plant Biotechnol. J. 16, 1399–1401. doi: 10.1111/pbi.12925
Keywords: RAV, Medicago truncatula, abiotic stress response, ABA, cold tolerance, branch number increasement
Citation: Wang S, Guo T, Wang Z, Kang J, Yang Q, Shen Y and Long R (2020) Expression of Three Related to ABI3/VP1 Genes in Medicago truncatula Caused Increased Stress Resistance and Branch Increase in Arabidopsis thaliana. Front. Plant Sci. 11:611. doi: 10.3389/fpls.2020.00611
Received: 30 January 2020; Accepted: 21 April 2020;
Published: 25 May 2020.
Edited by:
Eric Ruelland, UMR 7618 Institut d’Écologie et des Sciences de l’Environnement de Paris (IEES), FranceReviewed by:
Mohsin Tanveer, University of Tasmania, AustraliaSeverine Planchais, Université Pierre et Marie Curie, France
Copyright © 2020 Wang, Guo, Wang, Kang, Yang, Shen and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yixin Shen, eXhzaGVuQG5qYXUuZWR1LmNu; Ruicai Long, ZHJhZ29uZ29kc2dvZEAxNjMuY29t
†These authors have contributed equally to this work
 Shumin Wang
Shumin Wang Tao Guo
Tao Guo Zhen Wang
Zhen Wang Junmei Kang2
Junmei Kang2 Qingchuan Yang
Qingchuan Yang Ruicai Long
Ruicai Long