
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 June 2020
Sec. Plant Nutrition
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00588
This article is part of the Research Topic Nutrient Use-Efficiency in Plants: An Integrative Approach View all 16 articles
 Tian Fan1,2†
Tian Fan1,2† Wu Yang2†
Wu Yang2† Xuan Zeng2
Xuan Zeng2 Xinlan Xu2
Xinlan Xu2 Yanling Xu3
Yanling Xu3 Xiaorong Fan3
Xiaorong Fan3 Ming Luo2
Ming Luo2 Changen Tian1
Changen Tian1 Kuaifei Xia2,4
Kuaifei Xia2,4 Mingyong Zhang2,4*
Mingyong Zhang2,4*Enhancing nitrogen (N) use efficiency is a potential way to reduce excessive nitrogen application and increase yield. Autophagy is a conserved degradation system in the evolution of eukaryotic cells and plays an important role in plant development and stress response. Autophagic cores have two conjugation pathways that attach the product of autophagy-related gene 8 (ATG8) to phosphatidylethanolamine (PE) and ATG5 to ATG12, respectively, which then help with vesicle elongation and enclosure. Rice has six ATG8 genes, which have not been functionally confirmed so far. We identified the rice gene OsATG8b and characterized its role in N remobilization to affect grain quality by generating transgenic plants with its over-expression and knockdown. Our study confirmed the autophagy activity of OsATG8b through the complementation of the yeast autophagy-defective mutant scatg8 and by observation of autophagosome formation in rice. The autophagy activity is higher in OsATG8b-OE lines and lower in OsATG8b-RNAi than that in wild type (ZH11). 15N pulse-chase analysis revealed that OsATG8b-OE plants conferred higher N recycling efficiency to grains, while OsATG8b-RNAi transgenic plants exhibited lower N recycling efficiency and poorer grain quality. The autophagic role of OsATG8b was experimentally confirmed, and it was concluded that OsATG8b-mediated autophagy is involved in N recycling to grains and contributes to the grain quality, indicating that OsATG8b may be a potential gene for molecular breeding and cultivation of rice.
Nitrogen (N) is one of the most limiting nutrients for crop yield. Increasing N utilization efficiency (NUE) is not only important for increasing yield and reducing production cost but also for avoiding environmental pollution and keeping agriculture sustainable (Good et al., 2004; Masclaux-Daubresse et al., 2010). Therefore, it is very important to find effective genes to improve NUE and yield. Plant N utilization involves complex mechanisms of absorption, translocation, assimilation, and remobilization. Of those steps, N remobilization plays an important role during seed filling (Masclaux-Daubresse et al., 2008, 2010). At the vegetative stages, most N uptake is directed to leaves, in which most proteins are synthesized. During the reproductive stage, leaf proteins degrade rapidly to amino acids and small peptides, which are transported to seeds (Masclaux-Daubresse et al., 2008). N remobilization of cereals in senescent leaves has been shown to account for 50–90% of the grain N content (Kichey et al., 2007). The 26S proteasome/ubiquitin system and autophagy are the two main pathways of protein degradation (Wada et al., 2009; Roberts et al., 2012). Autophagy can degrade proteins, bulk organelles and cytosolic macromolecules with low selectivity and high throughput (Suzuki and Ohsumi, 2007).
Autophagy is a conserved degradation system in the evolution of eukaryotic cells. In the process of autophagy, the cytoplasm and organelles are separated by bilayer vesicles called autophagosomes and transported to vacuoles of yeast and plant cells or lysosomes of animal cells for degradation and recycling (Nakatogawa et al., 2009; Li and Vierstra, 2012; Yoshimoto, 2012). More than 30 autophagy-related genes (ATGs) have been identified in yeast, and 17 of them are necessary for autophagosome formation (Xie and Klionsky, 2007; Yoshimoto, 2012). Recently, orthologs of most yeast core ATG genes have been found in Arabidopsis and rice (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Bassham et al., 2006; Xia et al., 2011). ATG8 is one of the core proteins for forming autophagosome. It covalently binds to membrane lipid phosphatidylethanolamine (PE) through a ubiquitin-related binding system (Xie and Klionsky, 2007). ATG8 is a scaffold for membrane expansion and elongation during autophagosome formation (Nakatogawa et al., 2007; Xie et al., 2008). Yeast ATG8 also participates in the cytoplasm-to-vacuole targeting (CVT) pathway. Vacuole hydrolases, such as the precursor of aminopeptidase 1 (APE1), are selectively transported into the vacuole to produce mature APE1 (Yamaguchi et al., 2010). Unlike yeast, which has a single copy of the ATG8 gene, plants usually have an ATG8 family, comprising nine genes in Arabidopsis (Yoshimoto et al., 2004), five in maize (Chung et al., 2009), and six in rice (Xia et al., 2011). The different expression patterns of Arabidopsis ATG8s suggest that some ATG8s possess functional diversity besides possible redundancy (Slavikova et al., 2005).
Like yeast and animals, plant autophagy plays an important role in nutrient recycling under N- and C-starvation conditions (Li and Vierstra, 2012; Ohsumi, 2014). Currently, research on autophagy often focuses on the remobilization of N (Guiboileau et al., 2012, 2013; Xia et al., 2012; Li et al., 2015). Most Arabidopsis ATG genes are up-regulated by N-starvation and during leaf senescence (Doelling et al., 2002; Rose et al., 2006). Loss of function of Arabidopsis autophagy (atg5, atg7, atg10, and atg13a atg13b) caused hypersensitivity to N-limiting conditions in Arabidopsis and accelerated senescence even under N-rich conditions (Hanaoka et al., 2002; Phillips et al., 2008; Suttangkakul et al., 2011). Overexpression of AtATG8f and GmATG8c made Arabidopsis more tolerant to both N- and C-starvation (Slavikova et al., 2008; Xia et al., 2012). Autophagy mutants of Arabidopsis and maize (atg5 and atg7 in Arabidopsis and atg12 in maize) showed reduced seed yield, seed N content, and N remobilization efficiency (NRE) (Guiboileau et al., 2012, 2013; Li et al., 2015). About 50% of the remobilized N of Arabidopsis is proven to come from autophagy (Guiboileau et al., 2012). These studies showed that autophagy plays a central role in N remobilization.
Since evidence for the contribution of autophagy to plant physiology largely comes from the study of Arabidopsis, little is known about crop autophagy except for maize. Rice is an important cereal crop for the world population, especially in Asia. Currently, little is known about the contribution of autophagy to rice seed quality. Rice OsATG7 plays a role in NUE at the vegetative stage (Wada et al., 2015), and overexpression of rice gene osatg8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency in Arabidopsis (Zhen et al., 2019). However, the male sterility of osatg7 limits research on autophagy-mediated N recycling to grains in rice.
In our study, we functionally analyzed OsATG8b in rice. Complementation of a yeast atg mutant and subcellular localization analysis demonstrated the role of OsATG8b in the autophagy process. In addition, we characterized the role of OsATG8b in N remobilization and seed quality by generating transgenic plants with over-expression and knockdown of OsATG8b. The phenotypic and 15N-partitioning analysis showed that OsATG8b plays a role in N remobilization and grain quality. This result may provide strategic guidance for N application in molecular breeding and production of rice.
From spring to autumn, the japonica rice cultivar Zhonghua11 (ZH11) and transgenic plants were grown in a controlled paddy with normal planting. In winter, they were grown in a greenhouse at 28°C with 14-h light and 10-h dark per day. For hydroponic experiments, we used the modified rice nutrient solution of the International Rice Research Institute (IRRI, 1.43 mM NH4NO3, 0.32 mM NaH2PO4, 0.51 mM K2SO4, 1 mM CaCl2, 1.65 mM MgSO4, 8.9 mM MnSO4, 0.5 mM Na2MoO4, 18.4 mM H3BO3, 0.14 mM ZnSO4, 0.16 mM CuSO4, 40 mM FeSO4) in a growth room with a 30°C, 14 h light/10 h dark photoperiod (Yoshida et al., 1976). The solution was refreshed every 3-day. For nitrogen treatments, after the plants were germinated in water, they were grown on the IRRI solution for 7 days, and then plants were grown in the IRRI solution supplemented with high nitrogen (HN, 5 mM NH4NO3) and low nitrogen (LN, 0.2 mM NH4NO3) for different times.
Total RNA isolation, cDNA synthesis, and qRT-PCR of the rice were performed as previously described (Xia et al., 2011). Relative gene expression was normalized to the expression level of e-EF-1a with triplicate repeat. All primers are listed in Supplementary Table S1. qRT-PCR was repeated with three biological replicates, and each sample was assayed in triplicate by PCR.
OsATG8b ORF was cloned downstream of promoter GAL1 of the yeast vector pYES260. Wild-type yeast KVY55 and the scatg8 mutant KVY5 (MATa leu2 ura3 trp1 lys2 his3 suc2-△9△atg8::HIS3) were gifts from Dr. Yoshinori Ohsumi (Tokyo Institute of Technology, Japan). The vector was transformed into scatg8 according to the LiAc/SS-DNA/PEG TRAFO protocol (Clontech). Yeast were cultured and shaken at 30°C in SC medium supplemented with 0.67% (w/v) YNB (yeast N base without NH4SO4 and amino acids), 2% (w/v) galactose, 0.5% (w/v) NH4SO4, and Ura DO Supplement. When the yeast grew to the logarithmic metaphase of growth (OD600 = 1), yeast cells were collected by centrifugation, washed, and incubated for another 5 h in 0.67% YNB medium without amino acids, galactose and NH4SO4 for nutrient deprivation to induce autophagy. The collected cells were used for immunoblotting with anti-APE1 antibody (Santa Cruz); the immunoblot analysis process used was as previously described (Hamasaki et al., 2005).
Rice seeds were used for scanning electron microscopy (SEM) analysis. Samples were fixed overnight with 3% glutaraldehyde-sodium phosphate buffer (0.1M) at room temperature and rinsed three times with 0.1M sodium phosphate buffer. The samples were dehydrated through an ethanol series and infiltrated with an isoamyl acetate series. Seeds were then sputter-coated with gold/palladium in six different 30 s pulses (Hitachi JEE-420) and analyzed by scanning electron microscope (Hitachi S-3000N).
GFP-OsATG8b and sGFP-OsATG8b were constructed to analyze the subcellular localization of OsATG8b in yeast and rice, respectively. For yeast subcellular localization, the fused construct was inserted downstream of promoter GAL1 in pYES260 vector. For rice subcellular localization, the fused construct was inserted downstream of 35S promoter (Okano et al., 2008). For root imaging, 7-day seedlings were treated with 1 μM concanamycin A for 6 h at 28°C in darkness, and 5 mm of root from the root tip was cut off for observation. The green fluorescent protein (GFP) fusion protein was analyzed by confocal laser scanning microscope (ZEISS-710 Meta) with a 488-nm exciting wavelength. The thickness of the optical sections (pinhole) was 2.1 μm. The images presented are average projections of 8–20 optical sections.
To overexpress OsATG8b, the full-length CDS of OsATG8b was amplified by PCR and was inserted into the intermediate vector pUC18-sGFP. The whole cassette was finally PCR-amplified and inserted into the binary vector pCAMBIA1301 to replace the GUS via Nco I and BstE II. For the construction of the RNAi vector, a 230-bp fragment of the non-conserved 5′ end of OsATG8b was amplified with primers OsATG8b-Ri-F and OsATG8b-Ri-R and inserted in vector pTCK303 by BamH I and Kpn I for the sense strand and by Spe I and Sac I for the antisense strand (Wang et al., 2004). These vectors were transformed into A. tumefaciens EHA105 and then transformed into ZH11 with the Agrobacterium-mediated transformation method (Hiei et al., 1997).
Antibodies of OsATG8b were made with 6 × His-OsATG8b proteins as the antigen; these were purified using a Ni column (Novagen) and injected directly into rabbits by Beijing ComWin Biotech Co., Ltd.
Two-week old seedlings were used for total cell extracts and were ground in liquid N. The powders were extracted with the lysis buffer (25 mM Tris-HCl pH7.5, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, and Complete Protease Inhibitor Cocktail from Roche). The solution was then centrifuged at 13,000g for 20 min at 4°C, and the supernatant was used as total protein. The supernatant was run by SDS-PAGE with or without 6M urea and then transferred to nitrocellulose filter membranes for immunoblot analysis. The membranes were blocked and then incubated with mouse GFP antibodies (Santa Cruz) at a dilution of 1:1,000, while rabbit serum of OsATG8b was diluted by 1:500. All results came from three independent plant materials.
Rice plants were grown in IRRI solution in a greenhouse with 16-h light/8-h dark cycling. At 40 days after germination (DAG), plants were labeled with 15N for 5-day by adding 10 atom% excess Na15NO3 to the IRRI solution. The plants were then washed thoroughly with distilled H2O and transferred in the field for further growth. For 15N uptake measurements, thirteen plants of each genotype were harvested 2-day after 15N labeling. The 15N-labeled plants were further grown in the field to maturity, and grains and remains were separated for N recycling assessment. A dry weight (DW) of each sample was assayed for 15N and total N content using an isotope ratio mass spectrometer coupled with an N elemental analyzer (IsoPrime100, Elemental Scientific, United States). The 15N content of each sample was calculated as a % of total N, which was calculated as atom% or A%sample = 100 × (15N)/(15N + 14N) (Li et al., 2015).
Factors of calculation for NUE and NRE were estimated through the procedure provided by Guiboileau et al. (2012) and Li et al. (2015). The HI (harvest index) for yield evaluation was defined as the DWgrain/(DWremain + DWgrain). The N harvest index (NHI) for assessing grain filling with N was calculated as N%grain × DWgrain/(N%remain × DWremain + N%grain × DWgrain). NUE was then calculated as the NHI/HI ratio, and NUE values of different genotypes were compared. The efficiency of N recycling to grains was shown by 15NHI (15N harvest index), which was calculated by (A%grains × N%grains × DWgrains)/[(A%remain × N%remain × DWremain) + (A%grains × N%grains × DWgrains)]. The 15NHI:HI ratio was used to compare the NRE of different transgenic plants. 15N-labeling data were compiled from three biological replicates involving five plants for each genotype.
Total protein concentration and starch content were determined as described previously (Masclaux-Daubresse et al., 2002; Carlsson et al., 2011). Quantification data were compiled from three biological replicates involving 40 seeds from five plants for each genotype.
Six OsATG8s have been identified in the rice genome (Xia et al., 2011). The ATG8 phylogenetic tree generated from amino acid sequences showed that plant ATG8s are clustered into two main subgroups. Subgroup I covers most of the plant ATG8 family members, comprising OsATG8a, b, and c. Subgroup II covers 1–3 plant ATG8 family members from each species, containing OsATG8d, e, and f (Supplementary Figure S1). The existence of two subgroups may imply specific functions to each, besides possible redundancy. To explore the relationship between N remobilization derived by autophagy and rice grain quality, we analyzed the expression of OsATG8s in developing endosperm by searching the Rice Expression Profile Database (RiceXpro)1 and found that only OsATG8b expression increased with endosperm development compared with OsATG8a and OsATG8c (Supplementary Figure S8). These data indicated that the OsATG8b may be a potential rice ATG8 gene in grain filling, and it was chosen for further analyses. OsATG8b is encoded by a single gene (Os04g0642400) in rice. It is a soluble protein of 119 amino acids, with a predicted molecular mass of 13.7 kD and pI of 8.78. OsATG8b shares 81.8% amino acid identity with yeast ScATG8, 71.4% identity with human HsGABARAP, and 86.9% identity with Arabidopsis AtATG8a (Supplementary Figure S2A). Like other ATG8 proteins, OsATG8b has a conserved Gly residue at the C-terminus for PE conjugation (Supplementary Figure S2A). The result of 3D model prediction revealed that OsATG8b protein contains an N-terminal helical domain, two hydrophobic pockets named the W-site and the L-site, and a C-terminal ubiquitin-like domain, similar to yeast ScATG8 (Supplementary Figures S2A,B) (Noda et al., 2010). This implies that OsATG8b may have the autophagic function, similar to yeast ScATG8.
To verify the autophagic function of OsATG8b, we investigated whether OsATG8b rescues defects of ATG8-deficient (scatg8) yeast KVY5 (Kirisako et al., 1999). OsATG8b cDNA containing the entire ORF was driven by the yeast GAL1 promoter in a plasmid (pYES260) and expressed in scatg8 yeast. OsATG8b can rescue the abnormal cell morphology of the scatg8 yeast under N starvation (Figure 1A). In yeast, the precursor amino-peptidase1 (prAPE1) was delivered to the vacuole for processing into mature APE1 (mAPE1) through the Cvt/autophagy pathway (Yamaguchi et al., 2010). Thus, we monitored the protein levels of both prAPE1 and mAPE1 after 5 h of starvation. Both wild-type yeast and scatg8 cells complemented with OsATG8b accumulated mAPE1. In contrast, mAPE1 was detected in neither scatg8 cells nor the scatg8 cells transformed with the empty vector (Figure 1B). This suggests that prAPE1 was delivered to the vacuole and processed to mAPE1 in scatg8b yeast when OsATG8b was expressed in these cells. These results confirmed the autophagic activity of OsATG8b and showed that OsATG8b is a functional homolog of yeast ScATG8.
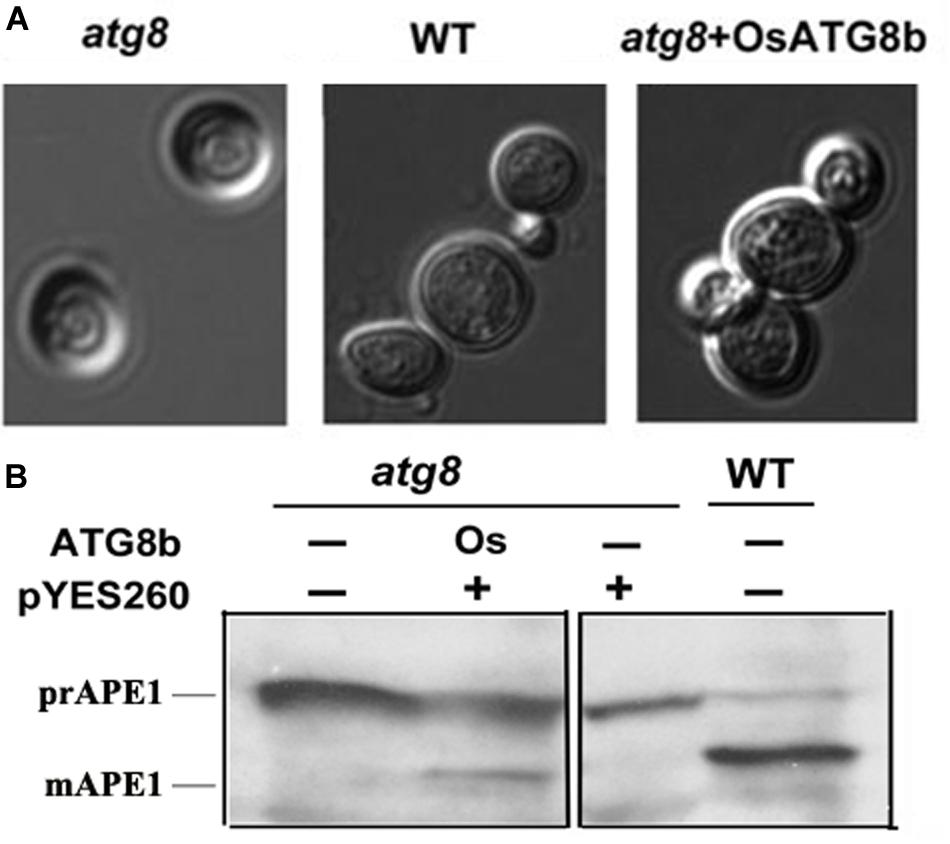
Figure 1. Functional complementation of yeast atg8 mutant by OsATG8b. OsATG8b cDNA was cloned into the plasmid pYES260 and expressed in yeast atg8 mutant KVY5 strain. (A) OsATG8b restores normal cell morphology of the yeast scatg8 mutant under N starvation. Abnormal morphology of scatg8 yeast cell (left), compared with normal cell of wild-type (WT) yeast (middle), and scatg8 yeast transformed with OsATG8b (right). (B) API protein identification by Western blot. Yeast cells were grown to mid-log phase to be collected and washed, incubated in the presence of 2% galactose to induce OsATG8b expression, and then incubated for another 5 h in nutrient deprivation medium and harvested for protein extraction. Proteins were then resolved by SDS-PAGE followed by immunoblotting with anti-APE1 antibody. prAPE1, precursor of aminopeptidase 1; mAPE1, mature aminopeptidase 1.
To determine the spatial and temporal expression pattern of OsATG8b, we employed qRT-PCR to examine OsATG8b expression. qRT-PCR analysis showed that OsATG8b transcripts accumulated in all studied organs, including roots, stems, leaves, leaf sheaths, and panicles at different growth stages (Figure 2A). The results showed that OsATG8b transcript levels were higher in roots of plants at 45 days after germination (DAG) than in those of plants at other growth stages. At 60 DAG, OsATG8b transcript was relatively abundant in stems, leaf, and panicle (Figure 2A). The expression level of OsATG8b was also examined under N deficiency and darkness treatment for C starvation, respectively (Figures 2B,C). OsATG8b transcript level increased in response to both N deficiency and darkness treatment. When rice seedlings were subjected to the N-free treatment, the expression level of OsATG8b gradually increased, peaking at 10-day after treatment application. Similarly, darkness treatment rapidly induced a roughly three-fold increase in OsATG8b expression within 2-day after treatment. Taken together, these results suggest that OsATG8b may play a crucial role in regulating multiple developmental processes and in response to nutrient stresses.
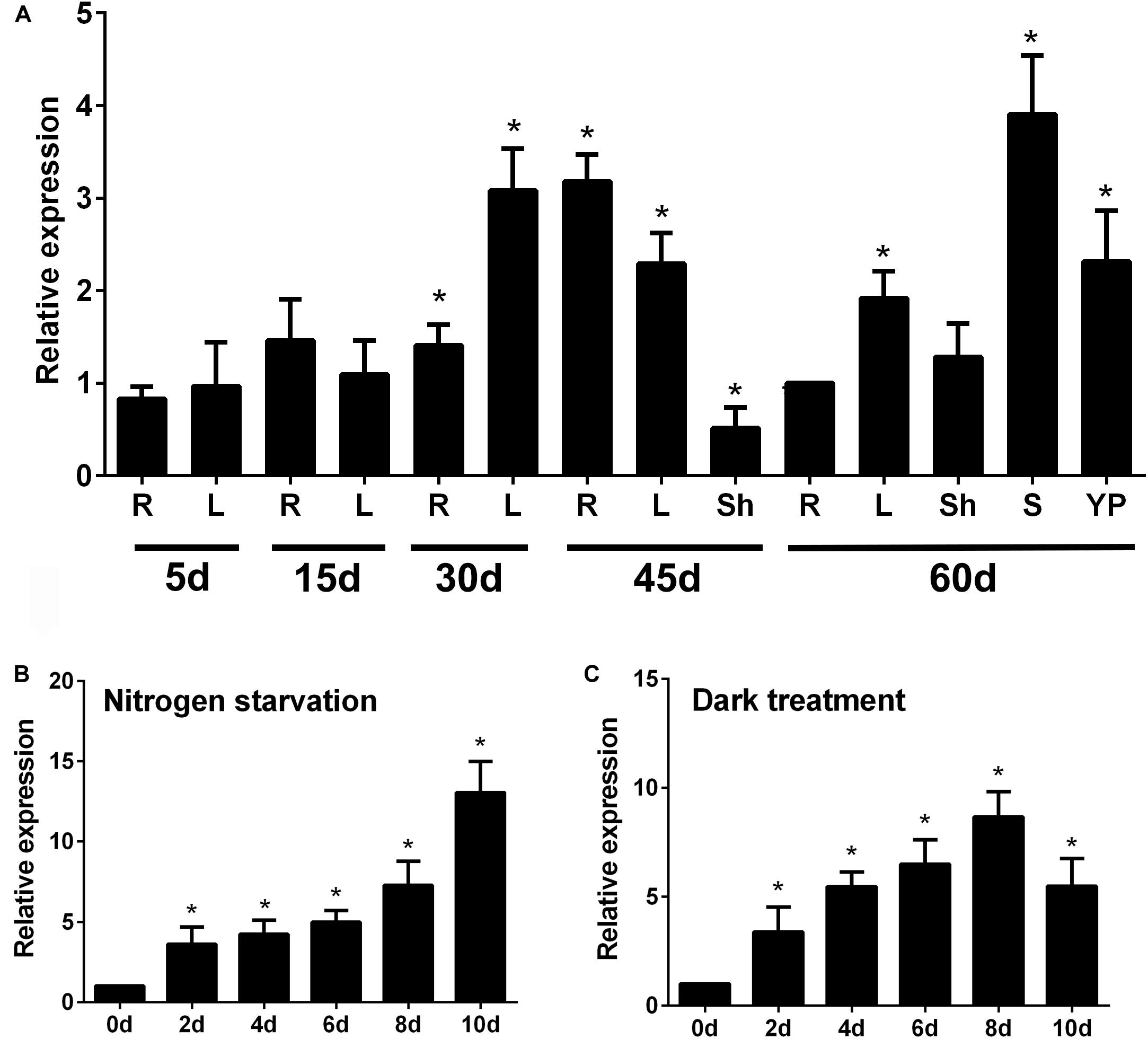
Figure 2. Expression patterns of OsATG8b in developing tissues of rice by qRT-PCR analysis. (A) Expression of OsATG8b in developing tissues. Total RNA was isolated from roots €, sheaths (Sh), young panicles (YP), stems (S), and leaves (L) at different growth stages. OseEF-1a was used as an internal reference. (B,C) Expression profiles of OsATG8b in an N-starvation solution (B) and in darkness (C). Total RNA was isolated from 5-leaf seedlings treated for 2, 4, 6, 8, and 10-day with N-starvation and darkness. Error bars indicate standard deviations of independent biological replicates (n = 3). One asterisk (*p < 0.05, t-test) represents significant difference.
To determine whether OsATG8b is conjugated onto the autophagosome membrane and completed or delivered into the vacuole, GFP was fused to its N-terminus and transformed into scatg8 yeast cells. Under control conditions, GFP-OsATG8b was mainly localized in the cytosol with punctate distribution, whereas after starvation, it accumulated within the vacuole of yeast (Figure 3A). These data suggest that OsATG8b may be localized to the autophagosomes of cytosol under the control conditions and translocate from the cytosol to the vacuole in an autophagy-dependent manner after starvation in yeast. To further verify the above result in rice, sGFP-OsATG8b was also transiently expressed in rice protoplasts, but the data showed that the sGFP-OsATG8b fusion protein was localized to the membrane, cytoplasm, and nucleus (Supplementary Figure S3), similar to the free sGFP control. To further confirm sub-cellular localization, transgenic rice expressing sGFP-OsATG8b were generated under the control of 35S promoter (Figure 3B). The 5 mm of the roots from the tip were cut off and immediately observed by LSCM. In sGFP-OsATG8b, GFP fluorescence was detected in the cytoplasm and nucleus; however, after 6 h of incubation in darkness with concanamycin A (an inhibitor of vacuolar H+-ATPase) to help in the observation of autophagic bodies through increasing vacuolar pH (Ishida et al., 2008; Izumi et al., 2015), many vesicles with a strong GFP signal and the spread of a faint GFP signal were observed (Figure 3B). These results indicate that the sGFP-OsATG8b-labeled puncta located in autophagosomes and the sGFP-OsATG8b can be used to visualize the progression of autophagy in rice, and overexpression of OsATG8b could increase the autophagic activity. Immunoblot analysis using proteins isolated from either ZH11 or transgenic sGFP and sGFP-OsATG8b rice plants showed that the OsATG8b antibodies recognized the endogenous as well as the GFP fusion proteins (Supplementary Figures S4A,B). Meanwhile, we performed an sGFP-ATG8 processing assay by the levels of free GFP moiety in anti-GFP immunoblots. After entering into the vacuole, sGFP-ATG8 is digested and releases free GFP, which is in a soluble and relatively stable form during autophagic body turnover (Suttangkakul et al., 2011). This free vacuolar GFP accumulates to a higher level when autophagy accelerates, so it represents the transport of ATG8 to vacuoles. Since OsATG8b antibody can also recognize OsATG8a and c (Supplementary Figure S9), the endogenous OsATG8(a/b/c) band of ZH11 and G1 (Supplementary Figure S4B) is very weak, but in OsATG8b-OE, the OsATG8(a/b/c) band is very strong, indicating that OsATG8b is overexpressed. These results indicated that the OsATG8 has already conjugated onto the autophagosome membrane and is able to be completed or delivered into the vacuole (Supplementary Figure S4A), which further confirms the autophagic activity of OsATG8b.
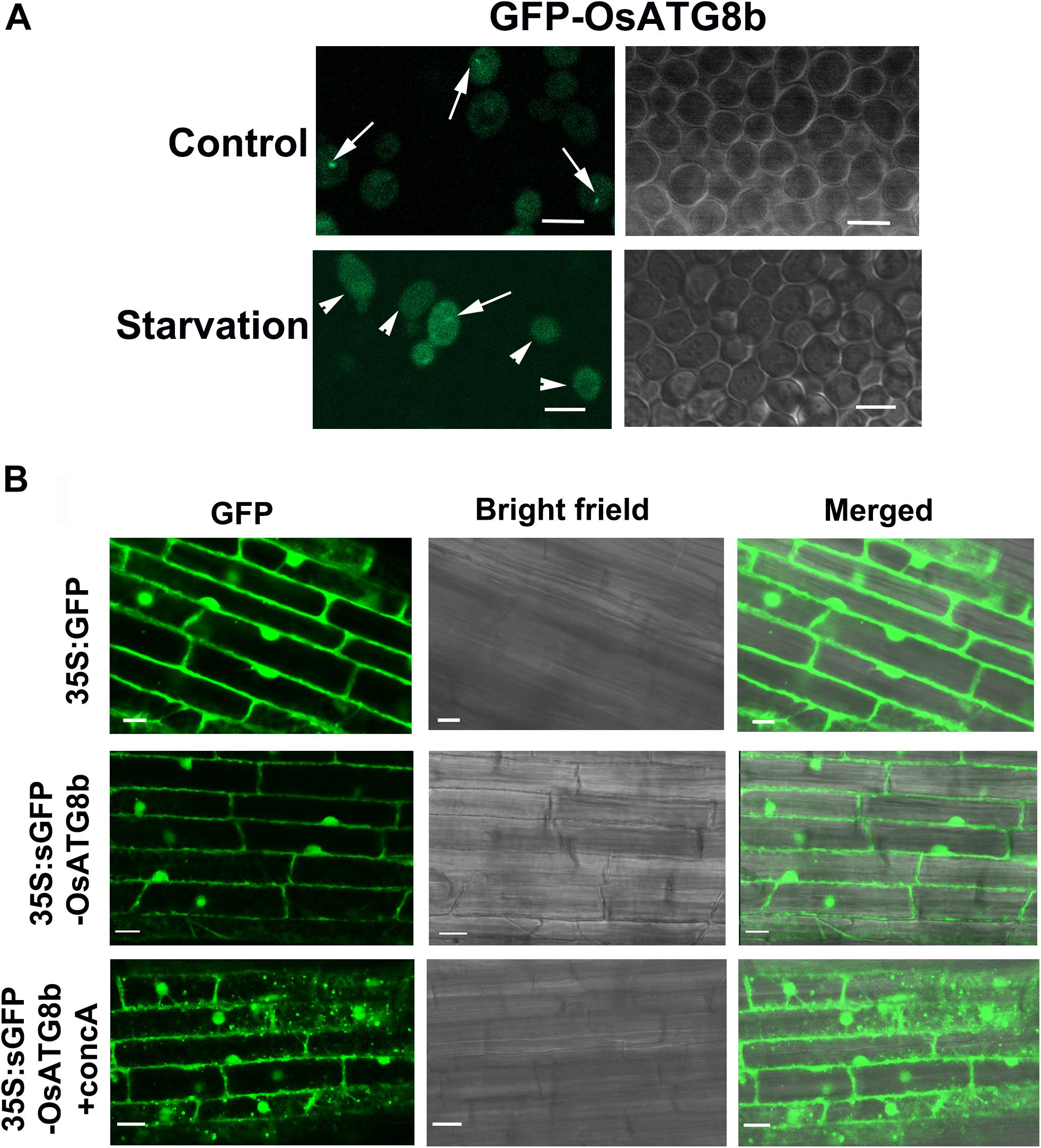
Figure 3. Subcellular localization of OsATG8b. (A) Subcellular localization of GFP-OsATG8b in yeast cells. Yeast cells were grown to mid-exponential phase and then incubated in the presence of 2% galactose to induce GFP-OsATG8b expression. Cells grown in both the control and starvation media were visualized by confocal laser scanning microscope. Arrows indicate the pre-autophagosomal structure (PAS), and arrowheads indicate GFP within vacuoles. Bars = 5 μm. (B) Subcellular localization of sGFP-OsATG8b in roots of transgenic rice. Fresh root samples of transgenic rice plants expressing 35S:sGFP-OsATG8b were excised and observed immediately (0 h) and after 6-h treatment with 1 μM concanamycin A. The region at approximately 5 mm from the root tip was observed in a laser scanning confocal microscope. Simultaneously obtained sGFP fluorescence images and DIC images are shown. Bars = 10 μm.
To investigate the function of OsATG8b, OsATG8b over-expression (OsATG8b-OE), and RNA-interference (OsATG8b-RNAi), transgenic lines were generated. RT-PCR analysis showed that OsATG8b expression increased in flag leaves of OsATG8b-OE lines and decreased in flag leaves of OsATG8b-RNAi lines (Figures 4A,B). The OsATG8b-RNAi construct was targeted specifically to the non-conserved 5′ end (Supplementary Figure S5A) of OsATG8b outside the ubiquitin domain to avoid interference with other OsATG8 proteins. Three of the OsATG8b-RNAi lines (Ri20, Ri24, and Ri25) and three of the OsATG8-OE lines (OE3, OE4, and OE6) were selected for subsequent analysis. In order to observe the effect of altered OsATG8b expression on OsATG8a and OsATG8c, we detected the expression of OsATG8a and OsATG8c in the shoots and roots of the transgenic rice seedling at four-leaf stage (Supplementary Figures S5B,C). The results showed that there is no significant difference in OsATG8a or OsATG8c transcript level among ZH11, the OsATG8-OE lines, and the OsATG8b-RNAi lines.
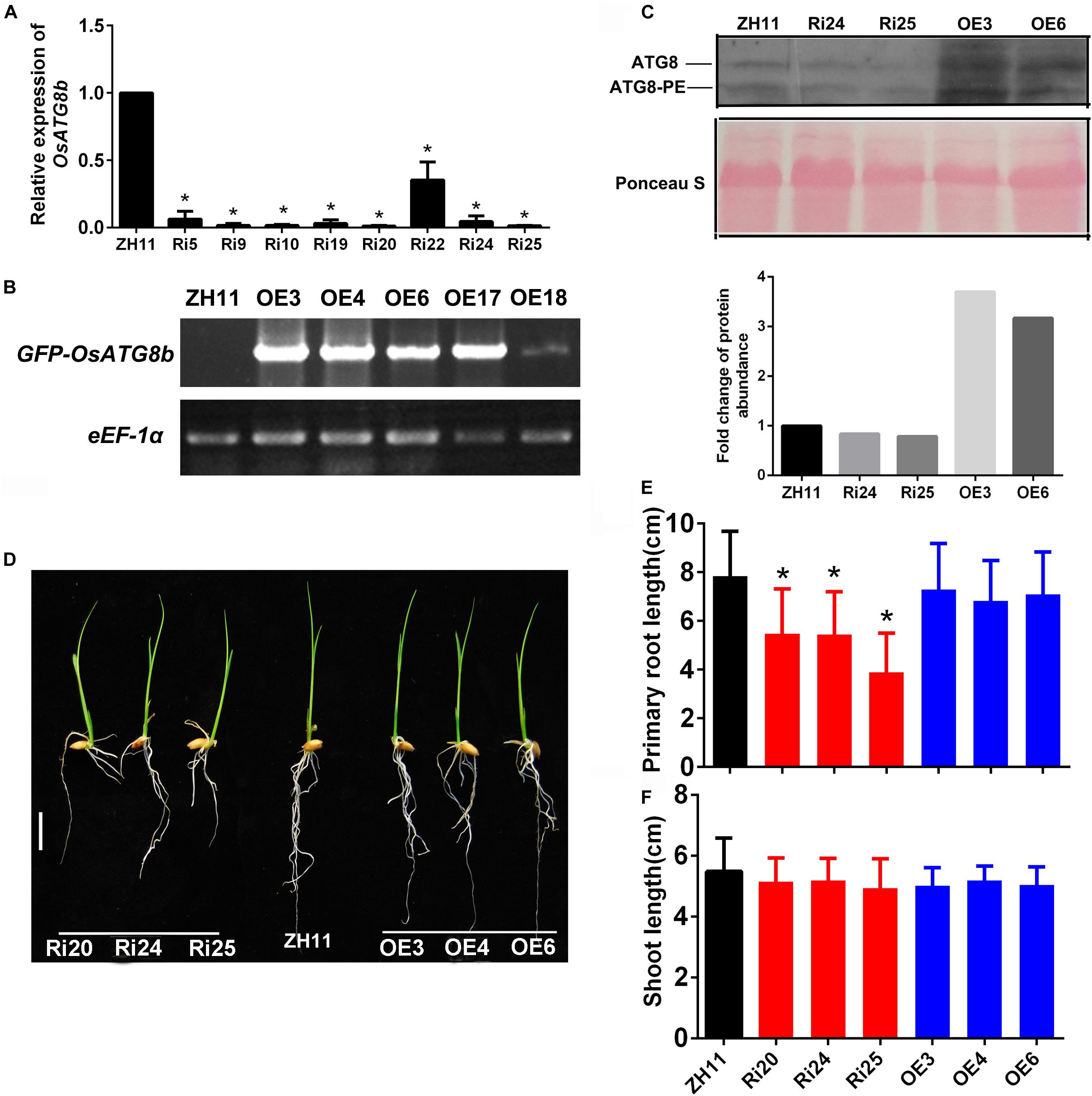
Figure 4. OsATG8b affects root growth at seed germination stage. (A,B) Expression levels of OsATG8b in flag leaves of OsATG8b-RNAi (Ri) (A) and OsATG8b-overexpression (OE) lines (B), measured by qRT-PCR and semi-qRT-PCR analyses, respectively. OseEF-1a was used as an internal reference. Error bars indicate standard deviations of independent biological replicates (n = 3). One asterisk (*p < 0.05, t-test) represents significant difference. (C) Immunoblot detection of the OsATG8a/b/c and OsATG8a/b/c lipidation level in 14 days-after-germination shoots of ZH11, OsATG8b-RNAi, and OsATG8b-OE lines. Total protein extracts were subjected to SDS-PAGE in the presence of 6M urea and immunoblotted with OsATG8b antibody. The fold change of ATG8a/b/c-PE was quantified using Image J software v1.45. (D–F) Seedlings (D) and statistical analysis of root (E) and shoot length (F) of OsATG8b-RNAi and OsATG8b-OE lines and ZH11 at 7-day after germination in water. Bar = 1 cm. Three biological replicates, each containing 80 plants, were used for data analysis. Results are the mean ± SD from 80 plants. *p < 0.05 (t-test): significantly different from ZH11. No asterisks mean no significant difference.
To confirm whether autophagic activities are altered in the OsATG8b-RNAi and OsATG8b-OE lines, we examined the ATG8a/b/c autophagic activities in 14 days-after-germination shoots of OsATG8b-RNAi, OsATG8b-OE, and ZH11 lines using OsATG8b antibodies (Figure 4C). The bands of ATG8 and ATG8-PE respectively represent the sum of OsATG8a/b/c or OsATG8a/b/c-PE (Figure 4C), since OsATG8b antibody can also recognize OsATG8a and OsATG8c (Supplementary Figure S9). The immunoblot analysis showed that the levels of OsATG8a/b/c-PE (representing the forming or completed autophagosomes) and cytosolic OsATG8a/b/c form were remarkably increased in OsATG8b-OE lines compared with ZH11 lines, and the quantified results showed that there is a slight decrease (about 17–20%) in them in OsATG8b-RNAi compared with in ZH11. Because OsATG8a/c expression does not change in these transgenic rice (Supplementary Figures S5B,C), these changes to the immunoblot bands should represent the changes of OsATG8b and OsATG8b-PE in OsATG8b-OE and OsATG8b-RNAi (Figure 4C). These results indicated that the autophagic activity is higher in OsATG8b-OE lines and may be a little lower in OsATG8b-RNAi than that in ZH11.
When the role of OsATG8b in growth at the vegetative stage was analyzed, we observed that the roots of 7-day-old OsATG8b-RNAi seedlings were much shorter than those of ZH11 and OsATG8b-OE lines (Figures 4D,E) when germinated in water. To reveal how the N level affects autophagy in rice, growth of OsATG8b-RNAi and OsATG8b-OE lines was measured under low nitrogen (LN, 0.2 mM NH4NO3) and high nitrogen (HN, 5 mM NH4NO3) for 30- or 60-day. Under LN and HN levels, the OsATG8b-RNAi and OsATG8b-OE lines exhibited a relatively normal phenotype and a similar growth rate when compared with ZH11 at 30 (Supplementary Figures S6A–D) or 60 DAG (Supplementary Figures S6E–H). Neither root nor shoot length showed any significant difference among these lines (Supplementary Figures S6C,D,G,H). These data may indicate that knocking down OsATG8b affects root growth at the stage of seed germination.
The phenotypes of OsATG8b-RNAi and OsATG8b-OE rice at the reproductive stage were investigated in the paddy field under normal N conditions. Previous studies have shown that the autophagy-defective rice mutant osatg7 displayed complete sporophytic male sterility. However, OsATG8b-RNAi and OsATG8b-OE plants produced healthy pollen grains and could be fertilized normally. The statistical results showed that grain number and grain yield per plant increased in OsATG8b-OE plants but decreased in OsATG8b-RNAi ones compared with ZH11 plants (Figure 5). These data indicate that OsATG8b may be involved in grain development and yield.
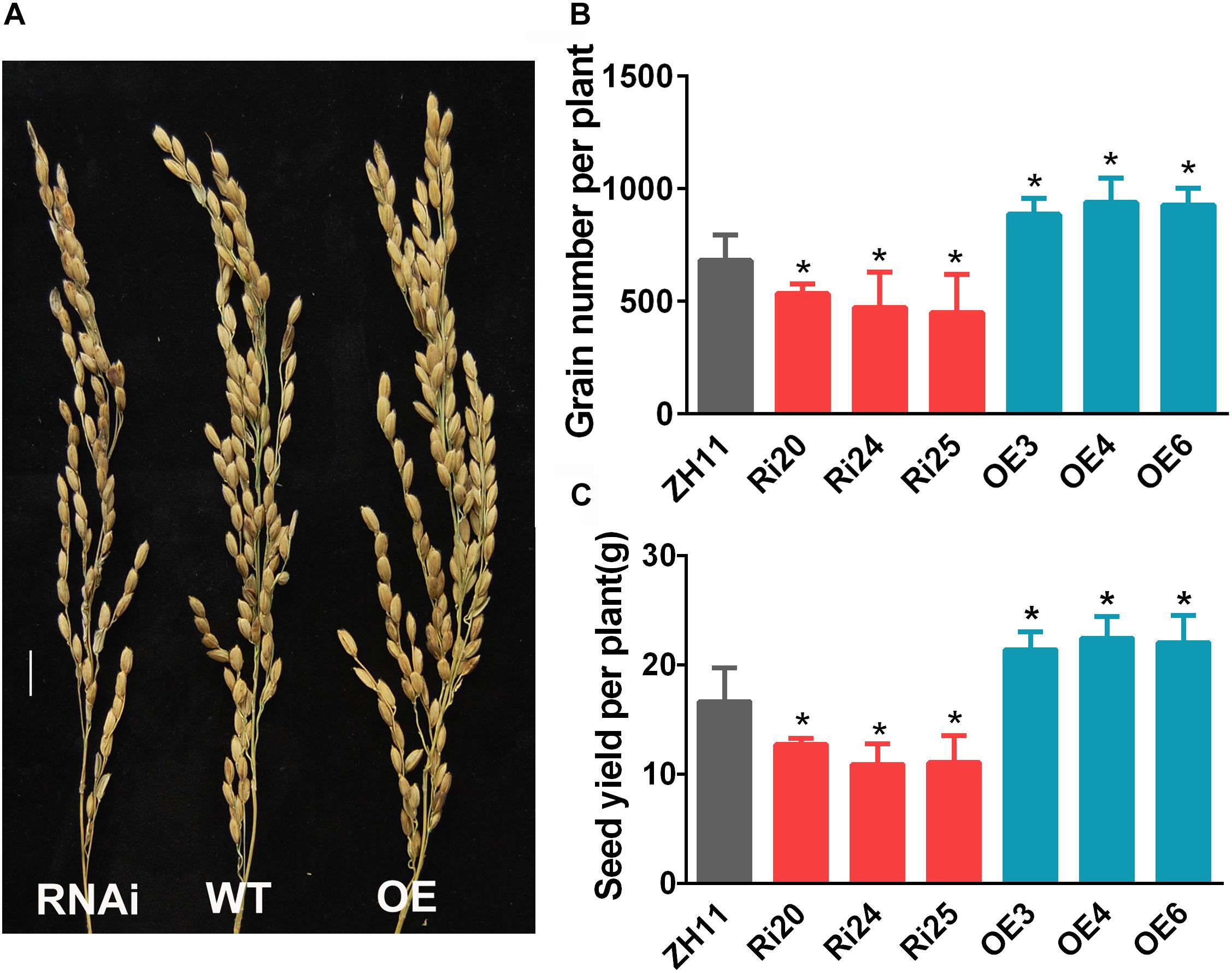
Figure 5. OsATG8b affects rice grain yield. OsATG8b-RNAi (Ri) and OsATG8b-OE (OE) lines and ZH11 were grown in the paddy field with normal fertilizer. (A–C) Panicle architecture (A), grain number (B), and grain yield (C) per plant of OsATG8b-RNAi and OsATG8b-OE lines and ZH11. Bar = 1 cm. Three biological replicates, each containing 80 plants, were used for data analysis. Results are the mean ± SD from 80 plants. *p < 0.05 (t-test): significantly different from ZH11.
The grains of OsATG8b-RNAi have a brown-spotted hull and contain chalky endosperm (Figures 6A,B). This showed that it produced poor quality seeds. The percentage of hulled rice with chalkiness was higher in OsATG8b-RNAi lines than in ZH11 (Figure 6C). SEM revealed that there are many loosely packed and small starch granules in the endosperm of OsATG8b-RNAi, which differed from the large and tightly packed starch granules in ZH11 (Figure 6D). Conversely, endosperm starch granules of OsATG8b-OE and ZH11 grains seemed larger and tighter (Figure 6D). Compared with ZH11, soluble protein content in OsATG8b-RNAi lines was lower, while that in OsATG8b-OE lines was higher (Figure 6E). However, starch content showed no significant difference among those lines (Figure 6F).
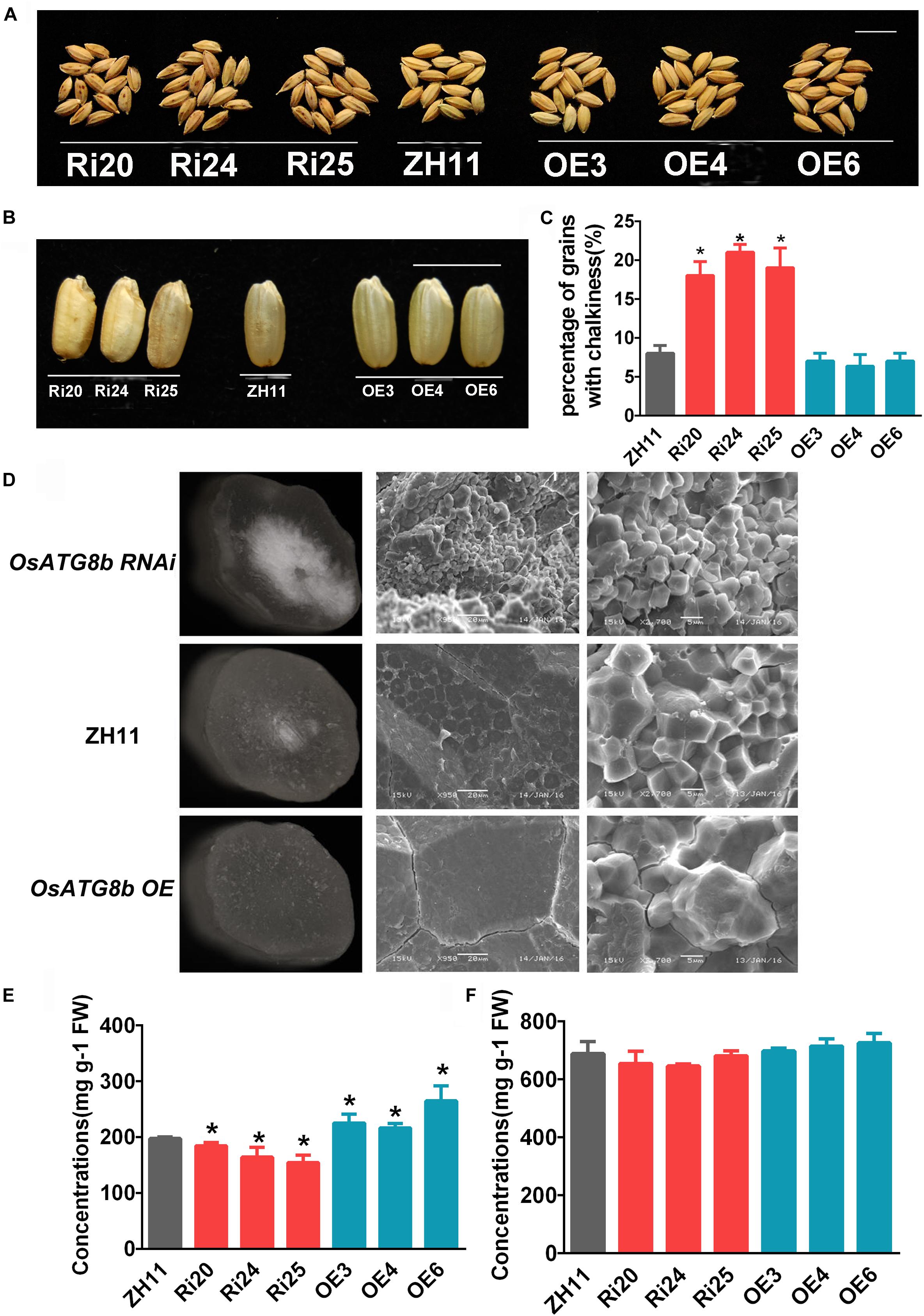
Figure 6. OsATG8b-mediated autophagy affects grain quality in rice. OsATG8b-RNAi (Ri) and OsATG8b-OE (OE) lines and ZH11 were grown in a paddy field under normal growth conditions. (A) Seed grains. OsATG8b-RNAi lines produced grains with brown spotted hulls. Bar = 1 cm. (B) Hulled OsATG8b-RNAi rice lines showed a chalky endosperm phenotype. Bar = 1 cm. (C) Percentage of hulled rice with chalkiness. (D) Scanning electron micrographs of cracked mature caryopses of rice grains under different magnifications. Endosperms of OsATG8b-RNAi lines had small, loosely packed starch granules, which differed markedly from the large, tightly packed starch granules of ZH11 and OsATG8b-OE lines. (E) Soluble protein concentration in grains. (F) Starch concentration in grains. Three biological replicates, each containing 40 seeds from five plants, were used for data analysis. Results are the mean ± SD from five plants. *p < 0.05 (t-test): significantly different from ZH11 (C,E,F).
To investigate whether OsATG8b plays a role in N recycling to grains in rice, we performed a pulse-chase assay with 15NO3–, as previously conducted with Arabidopsis (Masclaux-Daubresse and Chardon, 2011; Guiboileau et al., 2012). 15N and the 14N/15N ratio were measured (Figure 7A). Plant dry weight (DW) was higher in OsATG8b-OE lines and lower in OsATG8b-RNAi lines than in ZH11 (Figure 7B). This is similar to what was observed in Arabidopsis mutants (atg5, atg7) (Doelling et al., 2002; Guiboileau et al., 2012). HI, an important productivity indicator (Yang and Zhang, 2010), was lower in OsATG8b-RNAi lines but higher in OsATG8b-OE lines than in ZH11 (Figure 7C), which shows that autophagy plays an important role at the grain-filling stage.
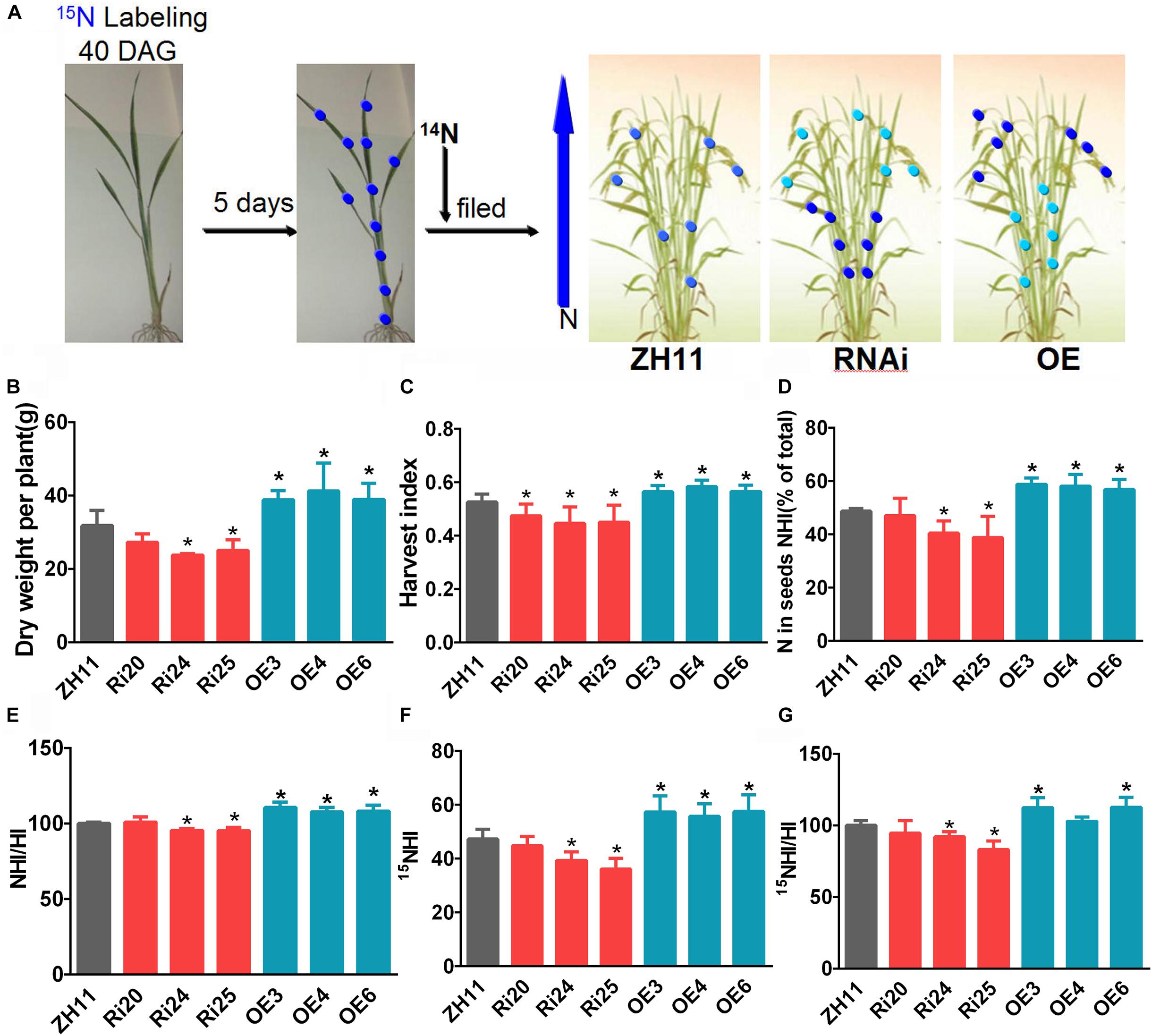
Figure 7. OsATG8b-mediated autophagy significantly affects N recycling efficiency (NRE). OsATG8b-RNAi (Ri) and OsATG8b-OE (OE) lines and ZH11 were grown in IRRI solution with 14NO3– supplementation until 40-day after germination and then labeled by a 5-day pulse of 15NO3– and subsequently grown in normal conditions to mature stage. After grain filling, 15N and the 14N/15N ratio were analyzed by isotopic ratio mass spectrometry. (A) Overview of 15N labeling and subsequent N partitioning. The deeper the blue of the dot, the higher the N content. (B) Biomass accumulation as measured by dry weight (DW) per plant. (C) Harvest index (HI) as measured by ratio of DW of grains to DW of the aboveground plant parts. (D) Nitrogen harvest index (NHI) as measured by the partitioning of total plant N in grains. (E) NHI:HI ratio as an estimate of N use efficiency. (F) Partitioning of total 15N in grains. (G) 15NHI:HI ratio as an indicator of N recycling efficiency. Three biological replicates, each containing five plants, were used for data analysis. Results are the mean ± SD from five plants. *p < 0.05 (t-test): significantly different from ZH11.
NHI is the main index of the efficiency of N distribution to grains and N grain filling (Guiboileau et al., 2012). The NHI of OsATG8b-RNAi was lower than that of ZH11, while that in OsATG8b-OE was higher (Figure 7D). As the NHI/HI ratio is considered a good indicator of NUE in plants (Masclaux-Daubresse and Chardon, 2011), we then measured the NHI/HI ratio of OsATG8b-RNAi, OsATG8b-OE, and ZH11. The results showed that the NHI/HI ratio increased dramatically in OsATG8b-OE lines but decreased in OsATG8b-RNAi lines when compared to ZH11 (Figure 7E). These data indicate that OsATG8b-mediated autophagy plays a role in grain NUE.
On the seventh day after 15NO3– labeling, the 15N contents of OsATG8b-RNAi, OsATG8b-OE, and ZH11 showed no significant differences. This is consistent with the normal growth of the OsATG8b-RNAi and OsATG8b-OE lines under N-rich conditions (Supplementary Figure S7). The abundances of 15N in grains and remains were determined using isotopic ratio mass spectrometry, which enabled us to calculate the partitioning of 15N in grains (15NHI) by combining these values with DW and N% data. 15NHI and the 15NHI:HI ratio, an indicator for NRE, were lower in OsATG8b-RNAi lines and higher in OsATG8b-OE lines than in ZH11 (Figures 7F,G). Taken together, these 15N partitioning results show that OsATG8b-mediated autophagy significantly affects NRE during the grain-filling stage.
Plant autophagy plays important roles in growth and development, grain filling, response to pathogen infection and to abiotic and biotic stresses, and N recycling (Wada et al., 2009; Yoshimoto et al., 2009; Guiboileau et al., 2012). All of these functions have major agricultural relevance, and most ATG orthologs in crops have been identified in maize and rice (Chung et al., 2009; Xia et al., 2011). Here, we report that rice OsATG8b is involved in N recycling and thus affects rice yield and seed quality.
Evolutionarily, autophagy is a highly conserved intracellular mechanism of degradation of cellular components in eukaryotic cells (Michaeli et al., 2016). At the elongation and final enclosure stages of the autophagosome, the linkage of ATG8 to PE anchors the former to both inner and outer membranes of the phagophore (Zientara-Rytter and Sirko, 2016). Therefore, the ATG8 protein is a useful molecular marker of autophagosomes, allowing for their distinction from other cellular vesicles and intracellular membranes (Yoshimoto et al., 2004; Zientara-Rytter and Sirko, 2016). Unlike yeast, which has a single ATG8, higher eukaryotes usually have an ATG8 family. Rice has six ATG8s (Xia et al., 2011), and five of their proteins have the conservative glycine in the C-terminal for PE conjugation except for OsATG8f. OsATG8a, b, and c belong to subgroup I of the plant ATG8 phylogenetic tree (Supplementary Figure S1), as all three proteins have extra amino acids behind the conserved Gly residue and need cleavage by ATG4 to expose the Gly residue (Supplementary Figure S2). On the other hand, OsATG8d and e belong to subgroup II (Supplementary Figure S1), as both have an innate C-terminal-exposed Gly residue, which makes OsATG8 quickly proceed conjugate with PE without ATG4 processing (Supplementary Figure S2). Expression of nine AtATG8 genes showed different patterns (Slavikova et al., 2005), which indicates that different ATG8 members may have multiple non-redundant functions and that individual ATG8s may have specific functions.
Plant ATG8s can functionally complement yeast atg8 mutants, such as those in Arabidopsis (Slavikova et al., 2005), soybean (Xia et al., 2012), and wheat (Pei et al., 2014). In our study, OsATG8b expression restored autophagy defects in the corresponding yeast atg8 mutant (Figure 1). This indicated that OsATG8b has an autophagic function similar to yeast ATG8. At present, observation of GFP-ATG8 puncta has been shown to be the best and most convenient detection method for autophagic activity (Kliosnky, 2016). However, it is showed that GFP-ATG8 signal foci in cytoplasm might not be the true autophagosomes in the cytoplasm of atg4a-1atg4b-1 double mutants (Yoshimoto et al., 2004) and atg7-2 mutants (Suttangkakul et al., 2011) since the foci may be GFP-ATG8 aggregates (Kuma et al., 2007; Kim et al., 2012). However, in the presence of concanamycin A, the mutants (atg7-2, atg5, atg10, and atg4a-1atg4b-1 in Arabidopsis and atg7 in rice) always lack GFP-ATG8 labeled autophagic foci in the vacuole (Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008; Izumi et al., 2015). This indicates that vacuolar GFP-ATG8 spots should be utilized as an autophagy indicator instead of GFP-ATG8 dots (Chung, 2011). The sGFP-OsATG8b puncta in vacuoles of rice root cells in the presence of concanamycin A were observed (Figure 3B); therefore, sGFP-OsATG8b is considered to be a marker for measuring the autophagic activity of rice cells. We also detected autophagosomes in vacuoles of sGFP-ATG8b transgenic rice (Figure 3B). Free GFP released from fused sGFP-ATG8b also supports this transfer and accumulates in vacuoles (Supplementary Figure S4B). Therefore, the sGFP-ATG8b test is a biochemical way to monitor the autophagic flux of rice cells.
Arabidopsis and maize atg mutants are sensitive to nutrient-limiting conditions (Hanaoka et al., 2002; Slavikova et al., 2005; Li et al., 2015). However, the OsATG8b-RNAi and OsATG8b-OE lines showed a relatively normal phenotype. In rice, there are six ATG8s, of which OsATG8a, OsATG8b, and OsATG8c have high homology. Data from RiceXpro (Supplementary Figure S8) showed that these three genes have similar expression patterns at vegetative stage. However, there are some different patterns during grain development; in particular, only OsATG8b expression increases with endosperm development (Supplementary Figure S8B). These data indicate that OsATG8s function redundantly in response to nutrient stress at the vegetative stages, but individual ATG8s may have specific functions in grain development. Indeed, in our study, OsATG8b-RNAi lines showed a chalky endosperm phenotype and carried small, loosely packed starch granules (Figures 6B,D), while in OsATG8b-OE lines endosperm, starch granules seemed larger and tighter (Figure 6D). Many genes and environmental factors control the grain endosperm chalkiness of rice (Siebenmorgen et al., 2013). Starch is the main storage material in rice grains, accounting for nearly 90% of the total dry weight, while protein accounts for about 8% of the endosperm weight of rice, filling the area between starch grains (Lin et al., 2014). Previous studies have shown that incomplete accumulation of starch and inadequate accumulation of proteins cannot fully fill the gap between starch granules, which may lead to the formation of chalk (Delrosario et al., 1968; Lin et al., 2014).
Starch and protein of rice grain are products of C and N, which are transported from source organs to produce starch and protein in precise quantities and proportions (Duan and Sun, 2005). C and N statuses are affected in Arabidopsis atg (atg5 and atg7) mutants (Guiboileau et al., 2013; Masclaux-Daubresse, 2016). We showed that soluble protein content decreased in OsATG8b-RNAi lines and increased in OsATG8b-OE lines, while starch content showed no difference between these lines (Figures 6E,F). In OsATG8b-RNAi lines, autophagic activity was slightly inhibited, and grain yield and quality were reduced (Figures 4A,C). The root shortening phenotype in 7-day-old OsATG8b-RNAi seedling (Figures 4D,E) may be caused by this impaired grain, since there are no obvious morphological differences between OsATG8b-RNAi and ZH11 at other vegetative stages (Supplementary Figure S6). We deduced that knocking-down OsATG8b in grains may cause decreased degradation of stored proteins in the germinating grains and then attenuate the growth rate of roots at the grain germination stage. These results suggest that OsATG8b-RNAi lines produced chalky endosperm mainly by breaking the balance between C and N in rice grains.
During early reproductive stage, panicle primordia and spikelets differentiate and develop in the shoot apical meristems, and the top four leaves and their respective internodes are developed on the mature dwarf stem and leaves. All of these events are mainly maintained by the N storage in the epiphylls of the dwarf stem and supply of new N from soil (Yoneyama et al., 2016). Therefore, spikelet number is determined by the N obtained from both recycling from leaves and root uptake. Our data showed that grain number per plant in OsATG8b-OE lines increased while that in OsATG8b-RNAi lines decreased compared with that in ZH11 in the field, indicating that OsATG8b-mediated autophagy affects grain number mainly by influencing N recycling from the dwarf stem-attached leaves to spikelet development.
Grain yield is affected by both soil N and remobilized N during reproductive stage (Kichey et al., 2007). To increase the NUE and crop yield, traditional methods focus on the operation of basic genes for N uptake and assimilation, such as NRT, NR, etc. (Pathak et al., 2008). In the grain-filling process, leaf organic N supply is more important because it contributes to plant N economy and limits the demand for exogenous N after flowering (Hanaoka et al., 2002). That is to say, the available N of grain was obtained from existing organic storage through recycling rather than from soil sources. Recently, studies on Arabidopsis and maize have shown that autophagy is the main factor affecting N recycling from senescent leaves to seeds (Guiboileau et al., 2012; Li et al., 2015). N recycling in senescent leaves was suppressed in osatg7 at the vegetative stage, but the male sterility of osatg7 limited evaluation of autophagy on both N economy and grain yield (Kurusu et al., 2014). Thus, we inferred that N recycling contributed by autophagy from the plant remains to grains in rice by over-expression and RNA interference of OsATG8b. Immunoblotting analysis results showed that autophagy activity is higher in OsATG8b-OE lines and a little lower in OsATG8b-RNAi than that in ZH11. Previous studies showed that OsATG8b antibody can also recognize OsATG8a and OsATG8c (Supplementary Figure S9). In OsATG8b RNAi lines, the band recognized by OsATG8b antibody represents the total OsATG8s, including OsATG8a, OsATG8b, and OsATG8c, so it is difficult to observe obvious differences in OsATG8b protein level with this method. Therefore, in our study, OsATG8b-RNAi lines showed slightly inhibited autophagic activity, which leads to reduced NRE from vegetative tissues to developing grains and finally results in reduced grain yield and quality. Meanwhile, reduced grain quality may cause decreased degradation of stored proteins in the germinating grains and then slow down the root growth at the grain germination stage. Conversely, OsATG8b-OE plants have higher yield and increased NRE (Figures 6, 7), and higher autophagic activity (Figure 4C). Thus, higher autophagic activity causes increased NRE, which leads to better grain yield. These results confirm that autophagy plays a crucial role in the N recycling process in rice. Therefore, improving N recycling by operating autophagy may be a useful strategy for increasing rice yield.
Data of this study are included in this article and its additional files. The material that supports the findings of this study is available from the corresponding author on request.
MZ and TF designed the research. TF, WY, XZ, XX, YX, and ML performed experiments. TF, MZ, XF, KX, and CT analyzed data. TF and MZ wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Key Research and Development Program of China (2017YFD0100100), the National Natural Science Foundation of China (31772384/31601817), STS of the Chinese Academy of Sciences (KFJ-STS-QYZX-032) and Outstanding youth of Jiangsu Province (BK20160030), the Strategic Priority Research Program, and the Guangdong Provincial Key Laboratory of Applied Botany of the Chinese Academy of Sciences (AB2018007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Yoshinori Ohsumi of Tokyo Institute of Technology for providing the atg8 yeast strain.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00588/full#supplementary-material
FIGURE S1 | Phylogenetic tree of ATG8s by amino sequence alignment of different species. Glycine max (Gm), Arabidopsis thaliana (At), Saccharomyces cerevisiae (Sc), Selaginella moellendorffii (Sm), Oryza sativa (Os), Homo sapiens (Hs), Solanum lycopersicum (Sly), Triticum aestivum (Ta), and Brachypodium distachyon (Bd). Deduced amino acid sequences were aligned by CLUSTALX; the phylogenetic tree was generated by the neighbor-joining method and constructed using MEGA4.
FIGURE S2 | Alignment of ATG8 amino acid sequence and 3D model of OsATG8b. (A) Alignment of ATG8 amino acid sequences from rice, Arabidopsis, human, and yeast. Arrows indicate the C-terminal glycine residue, which is processed by ATG4 cysteine protease. Residues constituting W- and L-sites are colored red and green, respectively. Sc, S. cerevisiae; Hs, Homo sapiens; At, Arabidopsis thaliana; Os, Oryza sativa. (B) 3D models of OsATG8b. Two hydrophobic pockets responsible for the recognition of Trp and Leu are labeled W-site and L-site, respectively, and circled.
FIGURE S3 | Subcellular localization of sGFP-OsATG8b in rice protoplasts. Bars = 1 μm.
FIGURE S4 | Immunoblot detection of the vacuolar delivery of GFP in GFP-OsATG8b lines and immunoblot analysis with OsATG8b antibodies. Total proteins extracted from shoots of 14-day-old-seedlings in GFP-OsATG8b (OE) and GFP (G1) transgenic lines and ZH11. (A) Total proteins were subjected to immunoblot analysis with GFP antibody. (B) OsATG8b antibodies recognize the endogenous proteins OsATG8(a/b/c) as well as the GFP fusion proteins in ZH11 and GFP-OsATG8b transgenic lines.
FIGURE S5 | The expression of OsATG8a and OsATG8c in ZH11, OsATG8b-OE, and OsATG8b-RNAi lines. (A) Sequence comparison with other homologous genes for construction of OsATG8b RNAi. The RNAi fragment is demarcated by the box. (B,C) qRT-PCR analysis of OsATG8a and OsATG8c expression. The seedlings of ZH11, OsATG8b-OE, and OsATG8b-RNAi at four-leaf stage were divided into the shoots and roots. OseEF-1a was used as an internal reference. Error bars indicate standard deviations of independent biological replicates (n = 3). No asterisks mean no significant difference (t-test).
FIGURE S6 | OsATG8b-RNAi (Ri) and OsATG8b-OE (OE) lines exhibited a relatively normal phenotype and a similar growth rate when compared with ZH11 at 30 and 60-day after germination (DAG). (A,B) Phenotype of OsATG8b-RNAi and OsATG8b-OE plants grown under low (LN, 0.2 mM NH4NO3) (A) and high N contents (HN, 5 mM NH4NO3) (B) at 30 DAG. (C,D) Statistical analysis of root (C) and shoot (D) length of OsATG8b-RNAi and OsATG8b-OE plants grown under both LN and HN conditions at 30 DAG. (E,F) Phenotype of OsATG8b-RNAi and OsATG8b-OE plants grown under LN (E) and HN (F) conditions at 60 DAG. (G,H) Statistical analysis of root (G) and shoot (H) length of OsATG8b-RNAi and OsATG8b-OE plants grown under both LN and HN conditions, at 60 DAG. Three biological replicates, each containing thirty plants, were used for data analysis. Error bars indicate standard deviations of independent biological replicates. No asterisks mean no significant difference (t-test).
FIGURE S7 | 15N-labeling efficiency of ZH11, OsATG8b-RNAi (Ri), and OsATG8b-OE (OE) lines. Plants at 40-day after germination were labeled for 5-day with 15NO3–, harvested 7-day later, and then assayed for 15N content in seedlings. Results are the mean ± SD from three plants. Three biological replicates, each containing three plants, were used for data analysis.
FIGURE S8 | Spatio-temporal expression of OsATG8a (A), OsATG8b (B), and OsATG8c (C) in various tissues/organs throughout the entire plant growth in the field. Data were obtained from RiceXpro (http://ricexpro.dna.affrc.go.jp/).
FIGURE S9 | OsATG8b antibody cannot distinguish OsATG8a, OsATG8b, and OsATG8c. The proteins of OsATG8a, OsATG8b, and OsATG8c were expressed in E. coli, and detected by the anti-OsATG8b antibody.
TABLE S1 | All primers used in this study.
APE1, aminopeptidase 1; CVT, cytoplasm-to-vacuole targeting; DAG, days after germination; DW, dry weight; HI, harvest index; IRRI, International Rice Research Institute; LSCM, laser-scanning confocal microscopy; mAPE1, mature APE1; N, nitrogen; NHI, nitrogen harvest index; NUE, nitrogen use efficiency; NRE, nitrogen recycling efficiency; OE, over-expression; ORF, open reading frame; OsATG8b, Oryza sativa autophagic-related gene 8b; PE, phosphatidylethanolamine; qRT-PCR, quantitative real-time PCR; RNAi, RNA interference.
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., et al. (2006). Autophagy in development and stress responses of plants. Autophagy 2, 2–11. doi: 10.4161/auto.2092
Carlsson, N., Borde, A., Wolfel, S., Akerman, B., and Larsson, A. (2011). Quantification of protein concentration by the Bradford method in the presence of pharmaceutical polymers. Anal. Biochem. 411, 116–121. doi: 10.1016/j.ab.2010.12.026
Chung, T. (2011). See how I eat my greens-autophagy in plant cells. J. Plant Biol. 54, 339–350. doi: 10.1007/s12374-011-9176-5
Chung, T., Suttangkakul, A., and Vierstra, R. D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-Lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. doi: 10.1104/pp.108.126714
Delrosario, A. R., Briones, V. P., Vidal, A. J., and Juliano, B. O. (1968). Composition and endosperm structure of developing and mature rice kernel. Cereal Chem. 45, 225–235.
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Duan, M. J., and Sun, S. S. M. (2005). Profiling the expression of genes controlling rice grain quality. Plant Mol. Biol. 59, 165–178. doi: 10.1007/s11103-004-7507-3
Good, A. G., Shrawat, A. K., and Muench, D. G. (2004). Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9, 597–605. doi: 10.1016/j.tplants.2004.10.008
Guiboileau, A., Avila-Ospina, L., Yoshimoto, K., Soulay, F., Azzopardi, M., Marmagne, A., et al. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199, 683–694. doi: 10.1111/nph.12307
Guiboileau, A., Yoshimoto, K., Soulay, F., Bataille, M. P., Avice, J. C., and Masclaux-Daubresse, C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
Hamasaki, M., Noda, T., Baba, M., and Ohsumi, Y. (2005). Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65. doi: 10.1111/j.1600-0854.2004.00245.x
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Hiei, Y., Komari, T., and Kubo, T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35, 205–218.
Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A., et al. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. doi: 10.1104/pp.108.122770
Izumi, M., Hidema, J., Wada, S., Kondo, E., Kurusu, T., Kuchitsu, K., et al. (2015). Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 167, 1307–1316. doi: 10.1104/pp.114.254078
Kichey, T., Hirel, B., Heumez, E., Dubois, F., and Le Gouis, J. (2007). In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crop. Res. 102, 22–32. doi: 10.1016/j.fcr.2007.01.002
Kim, S. H., Kwon, C., Lee, J. H., and Chung, T. (2012). Genes for plant autophagy: functions and interactions. Mol. Cells 34, 413–423. doi: 10.1007/s10059-012-0098-y
Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., et al. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. doi: 10.1083/jcb.147.2.435
Kliosnky, D. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Kuma, A., Matsui, M., and Mizushima, N. (2007). LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy. Autophagy 3, 323–328. doi: 10.4161/auto.4012
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Li, F. Q., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, F. Q., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Lin, Z. M., Zhang, X. C., Yang, X. Y., Li, G. H., Tang, S., Wang, S. H., et al. (2014). Proteomic analysis of proteins related to rice grain chalkiness using iTRAQ and a novel comparison system based on a notched-belly mutant with white-belly. BMC Plant Biol. 14:163. doi: 10.1186/1471-2229-14-163
Masclaux-Daubresse, C. (2016). Autophagy controls carbon, nitrogen, and redox homeostasis in plants. Autophagy 12, 896–897. doi: 10.4161/auto.36261
Masclaux-Daubresse, C., and Chardon, F. (2011). Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J. Exp. Bot. 62, 2131–2142. doi: 10.1093/jxb/erq405
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., and Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. Lond. 105, 1141–1157. doi: 10.1093/aob/mcq028
Masclaux-Daubresse, C., Reisdorf-Cren, M., and Orsel, M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. 10, 23–36. doi: 10.1111/j.1438-8677.2008.00097.x
Masclaux-Daubresse, C., Valadier, M. H., Carrayol, E., Reisdorf-Cren, M., and Hirel, B. (2002). Diurnal changes in the expression of glutamate dehydrogenase andnitrate reductase are involved in the C/N balance of tobacco source leaves. Plant Cell Environ. 25, 1451–1462. doi: 10.1046/j.1365-3040.2002.00925.x
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R., and Avin-Wittenberg, T. (2016). Autophagy in plants – What’s new on the menu? Trends Plant Sci. 21, 134–144. doi: 10.1016/j.tplants.2015.10.008
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Nakatogawa, H., Suzuki, K., Kamada, Y., and Ohsumi, Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467. doi: 10.1038/nrm2708
Noda, N. N., Ohsumi, Y., and Inagaki, F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385. doi: 10.1016/j.febslet.2010.01.018
Ohsumi, Y. (2014). Historical landmarks of autophagy research. Cell Res. 24, 9–23. doi: 10.1038/cr.2013.169
Okano, Y., Miki, D., and Shimamoto, K. (2008). Small interfering RNA (siRNA) targeting of endogenous promoters induces DNA methylation, but not necessarily gene silencing, in rice. Plant J. 53, 65–77. doi: 10.1111/j.1365-313X.2007.03313.x
Pathak, R. R., Ahmad, A., Lochab, S., and Raghuram, N. (2008). Molecular physiology of plant nitrogen use efficiency and biotechnological options for its enhancement. Curr. Sci. India 94, 1394–1403.
Pei, D., Zhang, W., Sun, H., Wei, X. J., Yue, J. Y., and Wang, H. Z. (2014). Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 33, 1697–1710. doi: 10.1007/s00299-014-1648-x
Phillips, A. R., Suttangkakul, A., and Vierstra, R. D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Roberts, I. N., Caputo, C., Criado, M. V., and Funk, C. (2012). Senescence-associated proteases in plants. Physiol. Plantarum. 145, 130–139. doi: 10.1111/j.1399-3054.2012.01574.x
Rose, T. L., Bonneau, L., Der, C., Marty-Mazars, D., and Marty, F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell. 98, 53–67. doi: 10.1042/BC20040516
Siebenmorgen, T. J., Grigg, B. C., and Lanning, S. B. (2013). Impacts of preharvest factors during kernel development on rice quality and functionality. Annu. Rev. Food Sci. T 4, 101–115. doi: 10.1146/annurev-food-030212-182644
Slavikova, S., Shy, G., Yao, Y. L., Giozman, R., Levanony, H., Pietrokovski, S., et al. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56, 2839–2849. doi: 10.1093/jxb/eri276
Slavikova, S., Ufaz, S., Avin-Wittenberg, T., Levanony, H., and Galili, G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043. doi: 10.1093/jxb/ern244
Suttangkakul, A., Li, F. Q., Chung, T., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 23, 3761–3779. doi: 10.1105/tpc.111.090993
Suzuki, K., and Ohsumi, Y. (2007). Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581, 2156–2161. doi: 10.1016/j.febslet.2007.01.096
Thompson, A. R., Doelling, J. H., Suttangkakul, A., and Vierstra, R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. doi: 10.1104/pp.105.060673
Wada, S., Hayashida, Y., Izumi, M., Kurusu, T., Hanamata, S., Kanno, K., et al. (2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73. doi: 10.1104/pp.15.00242
Wada, S., Ishida, H., Izumi, M., Yoshimoto, K., Ohsumi, Y., Mae, T., et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. doi: 10.1104/pp.108.130013
Wang, M., Chen, C., Xu, Y. Y., Jiang, R. X., Han, Y., Xu, Z. H., et al. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22, 409–417. doi: 10.1007/bf02772683
Xia, K. F., Liu, T., Ouyang, J., Wang, R., Fan, T., and Zhang, M. Y. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. doi: 10.1093/dnares/dsr024
Xia, T. M., Xiao, D., Liu, D., Chai, W. T., Gong, Q. Q., and Wang, N. N. (2012). Heterologous expression of ATG8c from Soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS One 7:e37217. doi: 10.1371/journal.pone.0037217
Xie, Z. P., and Klionsky, D. J. (2007). Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109. doi: 10.1038/ncb1007-1102
Xie, Z. P., Nair, U., and Klionsky, D. J. (2008). Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 19, 3290–3298. doi: 10.1091/mbc.e07-12-1292
Yamaguchi, M., Noda, N. N., Nakatogawa, H., Kumeta, H., Ohsumi, Y., and Inagaki, F. (2010). Autophagy-related protein 8 (Atg8) family inte racting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607. doi: 10.1074/jbc.m110.113670
Yang, J. C., and Zhang, J. H. (2010). Crop management techniques to enhance harvest index in rice. J. Exp. Bot. 61, 3177–3189. doi: 10.1093/jxb/erq112
Yoneyama, T., Tanno, F., Tatsumi, J., and Mae, T. (2016). Whole-plant dynamic system of nitrogen use for vegetative growth and grain filling in rice plants (Oryza sativa L.) as revealed through the production of 350 grains from a germinated seed over 150 days: a review and synthesis. Front. Plant Sci. 7:1151. doi: 10.3389/fpls.2016.01151
Yoshida, S., Forno, D. A., Cook, J. H., and Gomez, K. A. (eds) (1976). “Routine procedures for growing rice plants in culture solution”, in Laboratory Manual for Physiological Studies of Rice (Los Banos, Philippines: International Rice Research Institute) 61–66.
Yoshimoto, K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 53, 1355–1365. doi: 10.1093/pcp/pcs099
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 21, 2914–2927. doi: 10.1105/tpc.109.068635
Zhen, X., Xu, F., Zhang, W., and Li, X. (2019). Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS One 14:e0223011. doi: 10.1371/journal.pone.0223011
Keywords: autophagy, OsATG8b, nitrogen recycling, rice, seed quality
Citation: Fan T, Yang W, Zeng X, Xu X, Xu Y, Fan X, Luo M, Tian C, Xia K and Zhang M (2020) A Rice Autophagy Gene OsATG8b Is Involved in Nitrogen Remobilization and Control of Grain Quality. Front. Plant Sci. 11:588. doi: 10.3389/fpls.2020.00588
Received: 10 December 2019; Accepted: 20 April 2020;
Published: 04 June 2020.
Edited by:
Guillermo Esteban Santa María, National University of General San Martín, ArgentinaReviewed by:
Caiji Gao, South China Normal University, ChinaCopyright © 2020 Fan, Yang, Zeng, Xu, Xu, Fan, Luo, Tian, Xia and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingyong Zhang, emhhbmdteUBzY2JnLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.