
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 May 2020
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00576
This article mentions parts of:
The Role of EjSOC1s in Flower Initiation in Eriobotrya japonica
 Yuanyuan Jiang1,2†
Yuanyuan Jiang1,2† Yunmei Zhu2†
Yunmei Zhu2† Ling Zhang2
Ling Zhang2 Wenbing Su2
Wenbing Su2 Jiangrong Peng2
Jiangrong Peng2 Xianghui Yang2
Xianghui Yang2 Huwei Song3
Huwei Song3 Yongshun Gao2,4*
Yongshun Gao2,4* Shunquan Lin2*
Shunquan Lin2*TERMINAL FLOWER1 (TFL1), a key factor belonging to the phosphatidyl ethanolamine-binding protein (PEBP) family, controls flowering time and inflorescence architecture in some plants. However, the role of TFL1 in loquat remains unknown. In this study, we cloned two TFL1-like genes (EjTFL1-1 and EjTFL1-2) with conserved deduced amino acid sequences from cultivated loquat (Eriobotrya japonica Lindl.). First, we determined that flower bud differentiation occurs at the end of June and early July, and then comprehensively analyzed the temporal and spatial expression patterns of these EjTFL1s during loquat growth and development. We observed the contrasting expression trends for EjTFL1s and EjAP1s (APETALA 1) in shoot apices, and EjTFL1s were mainly expressed in young tissues. In addition, short-day and exogenous GA3 treatments promoted the expression of EjTFL1s, and no flower bud differentiation was observed after these treatments in loquat. Moreover, EjTFL1s were localized to the cytoplasm and nucleus, and both interacted with another flowering transcription factor, EjFD, in the nucleus, and EjTFL1s-EjFD complex significantly repressed the promoter activity of EjAP1-1. The two EjTFL1s were overexpressed in wild-type Arabidopsis thaliana Col-0, which delayed flowering time, promoted stem elongation, increased the number of branches, and also affected flower and silique phenotypes. In conclusion, our results suggested that EjTFL1-1 and EjTFL1-2 do not show the same pattern of expression whereas both are able of inhibiting flower bud differentiation and promoting vegetative growth in loquat by integrating GA3 and photoperiod signals. These findings provide useful clues for analyzing the flowering regulatory network of loquat and provide meaningful references for flowering regulation research of other woody fruit trees.
Flowering in angiosperm plants marks the change from the vegetative to the reproductive phase, and the time of floral initiation is regulated by environmental (photoperiod and temperature) and endogenous (age, autonomous and GA) pathways (Wellmer and Riechmann, 2010; Andres and Coupland, 2012). In Arabidopsis, these signaling pathways converge on the floral integrator gene FLOWERING LOCUS T (FT), which belongs to PEBP gene family that also includes TERMINAL FLOWER 1 (TFL1), MOTHER OF FT AND TFL1-like (MFT), BROTHER OF FT(BFT), TWIN SISTER OF FT (TSF), and Arabidopsis thaliana RELATIVE OF CENTRORADIALIS (ATC) (Karlgren et al., 2011; Wickland and Hanzawa, 2015). FT and TSF are floral activators (Yamaguchi et al., 2005; Jang et al., 2009) whereas TFL1, BFT and ATC act as floral inhibitors (Bradley et al., 1997; Yoo et al., 2010; Huang et al., 2012). FT is expressed in leaves and then transported to the shoot apices meristems to positively regulate downstream flowering genes and to induce floral differentiation and flowering (Kobayashi and Weigel, 2007; Liu et al., 2012).
Although TFL1 and FT genes belong to the same family and are highly homologous, differences in key amino acids lead to dissimilar functions (Hanzawa et al., 2005; Ahn et al., 2006; Ho and Weigel, 2014). His88/Asp144 in TFL1 and corresponding Tyr85/Gln140 in FT are the most critical residues for functional divergence between TFL1 and FT.
In the Arabidopsis, both FT and TFL1 bind to FD but play different roles. Binding of FT to the transcription factor FD produces a strong activator that promotes expression of AP1 and flowering, and the complex of FD and TFL1 serves as a strong inhibitor of downstream flower meristem identity genes such as AP1 and LFY, which delay flowering (Ratcliffe et al., 1998, 1999; Abe et al., 2005; Ahn et al., 2006; Hanano and Goto, 2011). The cloning and functional analysis of the TFL1 gene in other plants, such as apple (Kotoda and Wada, 2005; Mimida et al., 2009) pear (Freiman et al., 2012; Wang and Pijut, 2013) strawberry (Iwata et al., 2012; Koskela et al., 2012, 2016, 2017; Nakano et al., 2015), poplar (Igasaki et al., 2008; Ruonala et al., 2008; Mohamed et al., 2010) and rose (Iwata et al., 2012; Randoux et al., 2014), has been conducted. Silencing of MdTFL1 significantly accelerates flowering in transformed apple plants (Flachowsky et al., 2012; Yamagishi et al., 2016; Charrier et al., 2019) and overexpression of the apple TFL1 gene in Arabidopsis delays flowering (Kotoda and Wada, 2005). Similarly, silencing of pear TFL1 caused early flowering (Freiman et al., 2012; Yamagishi et al., 2016; Charrier et al., 2019). These studies indicated that the function of TFL1 is relatively conserved among different species, mainly as a flowering inhibitory factor. However, there are no reports on the functional verification of TFL1 homologous genes in loquat.
Loquat (Eriobotrya japonica Lindl.), which belongs to the Maloideae subfamily of the Rosaceae family, is a tropical and subtropical evergreen fruit tree. In Rosaceae fruit trees such as apples, pears, and plums, the time of flower bud differentiation and flowering do not occur in the same year (Kurokura et al., 2013). After flower bud differentiation, a period of dormancy is required, and flowering begins in the next year (Kurokura et al., 2013). However, the flower bud differentiation and flowering time of loquat do occur in the same year (Lin, 2007) as confirmed by Jiang et al. (2019): flower bud differentiation of loquat occurred at the end of June, and the flower buds continued to develop until flowering in November-December. This unique flower development pattern is very important and interesting and may provide different perspectives for exploring the flower development pathway of Rosaceae; nonetheless, there are few reports on flower development in loquat. EjLFY, EjAP1, EjFT, and EjSOC1 have been cloned from cultivated loquat (Esumi et al., 2005; Liu et al., 2013; Reig et al., 2017; Jiang et al., 2019); and EdGI, EdCO, EdFT, and EdFD have been cloned from wild loquat Eriobotrya deflexa Nakai forma koshunensis (Zhang L. et al., 2016; Zhang et al., 2019).
Although two TFL1 homologues have been cloned from cultivated loquat (Esumi et al., 2005) their expression patterns and roles in the regulation of flowering have not yet been elucidated. In this study, two TFL1 homologues were isolated from loquat, namely EjTFL1-1 and EjTFL1-2. To explore their functions during the growth and development of loquat, we comprehensively analyzed their temporal and spatial expression patterns and tested their ability to interact with EjFD using BiFC and the yeast two-hybrid assay. In addition, we examined the effects of gibberellin and photoperiod signaling on the expression patterns of EjTFL1 and overexpressed both EjTFL1s in Arabidopsis for functional analysis.
Tissue samples were collected from ‘Jiefangzhong’ loquat (Eriobotrya japonica Lindl.) field-gown in the loquat germplasm resource preservation garden, South China Agricultural University (Guangzhou, China N23°09′N,113°20′E). The trees used in the experiment were 12-year-old trees, and showed normal flowering. The 5th-6th leaves with the same maturity at the upper end of the stem in the same period (the leaves turned into dark green as the standard), and the young leaves, buds (terminal buds), flowers and fruits were collected in the same period and the phenotype was consistent (tissues were taken at 16:00). All samples for quantitative analysis were frozen with liquid nitrogen immediately after collection and then stored at −80°C until use. Three independent experiments were conducted from three independent trees. Year-round follow-up observations, and sampling were conducted every 2 weeks, and paraffin sections of the shoot apices meristems were observed using fluorescence microscope (Observer. D1, Zeiss, Germany) in a bright-field channel and photographed color images. Arabidopsis thaliana wild-type Col-0 was used in this study. Arabidopsis and Nicotiana benthamiana were grown under long-day conditions (16 h light/8 h dark cycle) at 22°C.
Total RNA from flowers, fruits, stems, leaves and buds of ‘Jiefanzhong’ was extracted using EASY Spin Plus Plant RNA Extraction Kit (Aidlab, China), and total RNA from whole Arabidopsis plants was extracted using HiPure Plant RNA Mini Kit (Magen, China). First-strand cDNA was synthesized using PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Japan). Genomic DNA was extracted from loquat buds using HiPure Plant DNA Mini Kit (Magen, China).
EjTFL1-1, EjTFL1-2, and EjFD cDNA and the promoter regions of EjTFL1-1, EjTFL1-2, EjAP1-1, and EjAP1-2 were amplified using Prime STAR® Max DNA Polymerase (TaKaRa, Japan) with the primer sequences listed in Supplementary Table S1; the fragments were inserted into the pGEM-T easy vector (Promega, United States). Sequence information was obtained from the de novo genome sequencing project of loquat by our research group, which has not yet been published. Amino acid sequences were aligned using Clustal X and Gene Doc. A phylogenetic tree was generated using MEGA 6 by the neighbor-joining (N-J) method with 1000 bootstrap replications. Predictive analysis of cis-acting elements of the promoter fragments was performed online at PlantCARE.1
iTaqTM universal SYBR Green Super mix kit (Bio-Rad, United States) was used for qRT-PCR (quantitative real time polymerase chain reaction) in the LightCyclerR 480 system (Roche, United States). qRT-PCR experiments and analyses were carried out as described by Zhang L. et al. (2016). Gene expression levels were normalized using Ejβ-Actin for loquat (Shan et al., 2008) and AtTUB2(AT5G62690) for Arabidopsis (Yan et al., 2014). The primer sequences used in real-time PCR are listed in Supplementary Tables S2, S3. Data were collected from three biological replicates.
For short-day treatments, the shading was set for 8 h (10:00–18:00) of natural light and 16 h of full darkness (18:00–10:00 [second day]) each day; no shading was performed for control plants. For GA3 treatments, plants were sprayed once every 2 weeks with a 300 mg L–1 aqueous solution of GA3 (DingGuo, China); 0.1% (v/v) phosphoric acid and 0.025% (v/v) Triton X-100 were added as surfactants; the control plants were sprayed with 0.1% (v/v) phosphoric acid and 0.025% (v/v) Triton X-100 aqueous solution. The two experimental treatment times were from 18th May to 10th August. Three independent experiments were conducted.
We used the Matchmaker Gold Yeast two-hybrid system for Y2H assay (Clontech). AD-EjTFL1 (full-length EjTFL1-1 and EjTFL1-2 fused to pGADT7) and BD-EjFD (full-length EjFD fused to pGBKT7) plasmids were constructed for the Y2H assay. The experimental procedure was carried out in strict accordance with the manufacturer’s instructions. The primer sequences used in plasmid construction are listed in Supplementary Table S4.
EjTFL1-1, EjTFL1-2, and EjFD were amplified by PCR and cloned into pGreen-35S-GFP (Lee et al., 2012) for subcellular localization analysis. For the BiFC assay, PCR-amplified EjTFL1-1, EjTFL1-2, and EjFD were cloned into pGreen-35S-cYFP/nYFP vectors (Hou et al., 2014). Primer sequences used in the plasmid construction are listed in Supplementary Table S4. The 35S-YFP-C-EjFD + 35S-EjTFL1s-YFP-N, 35S-YFP-C-EjFD + 35S-YFP-N, 35S-YFP-C + 35S-EjTFL1s-YFP-N, 35S-EjTFL1s-GFP, and 35S-EjFD-GFP fusion constructs were transiently transformed into N. benthamiana leaves using an Agrobacterium tumefaciens GV3101::pSoup-mediated technique, as previously described (Sparkes et al., 2006). Two days later, fluorescent signals were observed by fluorescence microscopy (Observer. D1, Zeiss, Germany).
The EjAP1-1 and EjAP1-2 promoters were cloned into the pGreenII 0800-LUC double-reporter vector as reporters. The reporter constructs were fused to LUC luciferase and REN luciferase driven by CaMV35S; EjTFL1s and EjFD were cloned into the pGreenII62-SK vector as effectors driven by CaMV35S (Hellens et al., 2005). The primer sequences used in vector construction are listed in Supplementary Table S4. The constructed effector and reporter plasmids with different combinations were co-transformed into tobacco leaves using the A. tumefaciens strain GV3101::pSoup method (Hellens et al., 2005). After 48–72 h, LUC and REN luciferases were quantified using the Dual Luciferase Assay kit (Promega, United States) with a Luminoskan Ascent Microplate Luminometer (Thermo, United States). The binding activities of the effectors and reporters were calculated by the ratio of LUC/REN.
EjTFL1-1 and EjTFL1-2 were amplified by PCR and cloned into pGreen-35S (Lee et al., 2012). The constructed vectors were transformed into A. tumefaciens GV3101::pSoup and introduced into A. thaliana wild-type Col-0 using the floral dip method (Zhang et al., 2006). Transformed plants were selected on soil with Basta.
Two TFL1 homologs, EjTFL1-1 and EjTFL1-2, were isolated from loquat buds, and the specific primers were designed using data from the unpublished loquat genome sequence. The length of EjTFL1-1 and EjTFL1-2 CDSs were 519 bp, encoding putative proteins of 172 amino acids that belong to the PEBP family (see Supplementary Sequences). Their sequences were highly similar, with amino acids sequence identity of 90.7%. Amino acid sequence analysis revealed that loquat EjTFL1s and EjFTs are highly similar (Figure 1A). Phylogenetic tree analysis revealed that EjTFL1s is closely related to TFL1 of the sub-family Maloideae, which clustered together into a small clade, and grouped with other plants of Rosaceae family (Figure 1B).
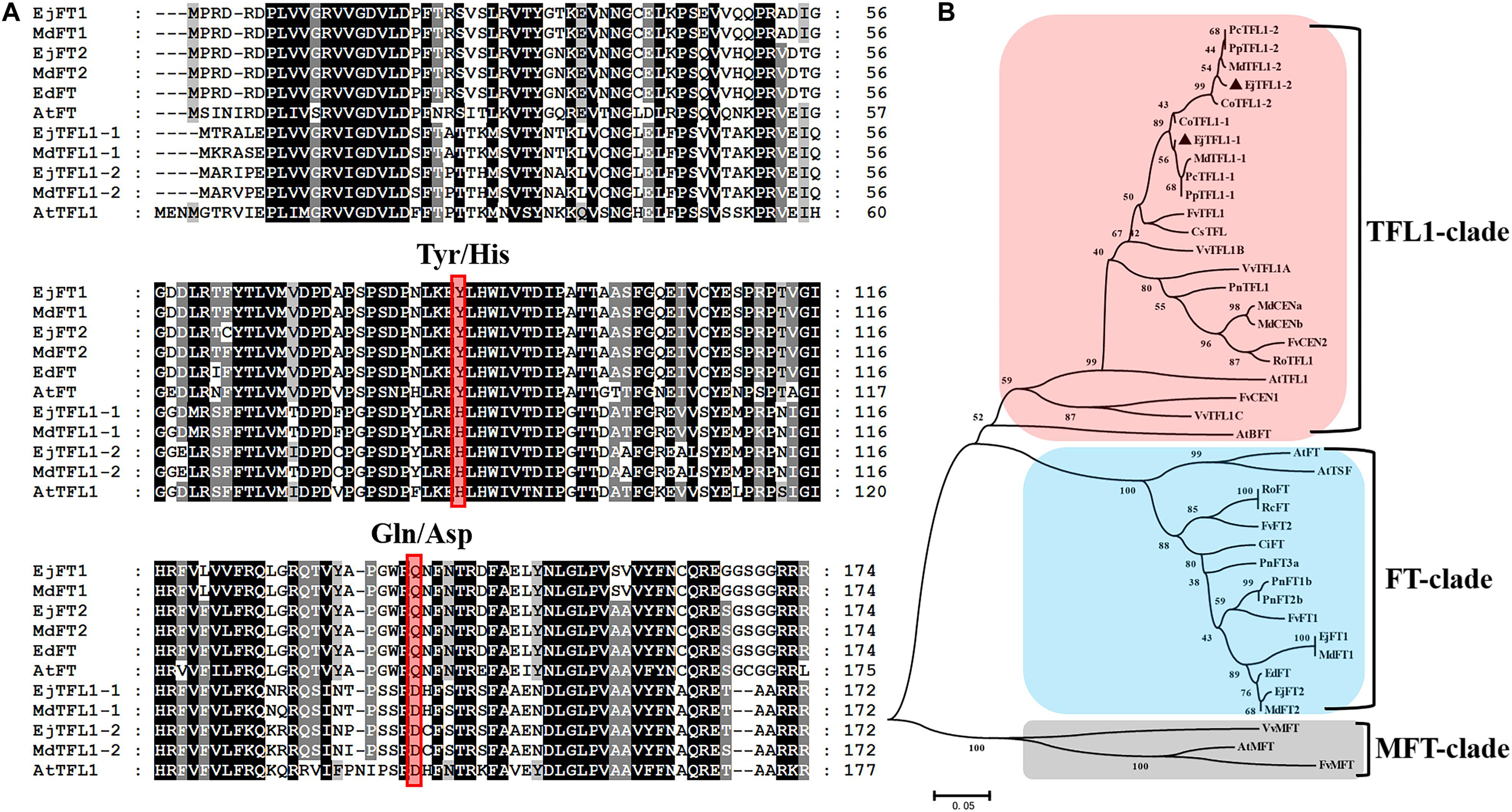
Figure 1. Sequence and phylogenetic analyses of EjTFL1-1 and EjTFL1-2 (At, Arabidopsis thaliana; Ed, Eriobotrya deflexa; Ej, Eriobotrya japonica; Md, Malus domestica; Fv, Fragaria vesca; Ro, Rosa lucieae; Rc, Rosa chinensis; Ci, Citrus unshiu; Pn, Populus nigra; Pc, Pyrus communis; Cs, Citrus sinensis; Pp, Pyrus pyrifolia; Co, Cydonia oblonga; Vv, Vitis vinifera). (A) Amino acid sequence alignment of TFL1 and FT proteins of loquat and other species. The red box indicates the key amino acids Tyr85/His88 and Gln140/Asp144 that differ in function between TFL1 and FT. (B) Phylogenetic analysis of PEBP gene family proteins from loquat and other species. The protein sequences used in this study were retrieved from NCBI. Accession IDs: AtFT (BAA77838), AtTFL1 (AAB41624), AtBFT (Q9FIT4), AtTSF(BAD93590), AtMFT (Q9XFK7), EdFT (AMB72867), EjFT1(ALA56299), EjFT2 (ALA56300), MdFT1 (BAD08340), MdFT2 (BAI77728), MdTFL1-1 (BAD10961), MdTFL1-2 (BAD10967), MdCENa (BAG31957), MdCENb (BAG31958), FvFT1 (AEP23098), FvFT2 (XP_004297273), FvTFL1 (AEP23097), FvCEN1 (XP_004308107), FvCEN2 (XP_004291610), FvMFT (XP_004299541), RoFT (CAX46426), RoTFL1 (CAX46398), RcFT (CBY25182), CiFT (BAA77836), PnFT1b (BAD08338), PnFT2b (BAD08337), PnFT3a (BAD08336), PnTFL1 (BAG12897), PcTFL1-1 (BAD10963), PcTFL1-2 (BAD10969), CsTFL (AAR04683), PpTFL1-1 (BAD10962), PpTFL1-2 (BAD10968), CoTFL1-1 (BAD10964), CoTFL1-2 (BAD10970), VvTFL1A (ABI99466), VvTFL1B (ABI99467), VvTFL1C (ABI99468), VvMFT (ABI99469).
Alignment of the amino acid sequences showed that the EjTFL1s are similar to MdTFL1s (98% identity to MdTFL1-1 for EjTFL1-1 and 98% identity to MdTFL1-2 for EjTFL1-2). EjTFL1-1 and EjTFL1-2 contain His84 and Asp139 residues, which are conserved among other plant species and clearly distinguish FT homologous genes from other species, including loquat (Figure 1A). The EjTFL1 structure suggests a functional conservation. Moreover, phylogenetic tree also revealed that there is a very close genetic evolutionary relationship between loquat, apple and other Rosaceae plants.
To explore the function and regulatory mechanisms of EjTFL1s, we analyzed the relative expression levels of EjTFL1-1 and EjTFL1-2 in different tissues of loquat (Figure 2A). EjTFL1-1 was mainly expressed in roots and leaf buds, but the expression was relatively low in shoots, flower buds and flowers, with no expression in mature leaves and mature fruits (Figure 2B). EjTFL1-2 was mainly expressed in leaf buds, flowers, and fruits and was not expressed in the other tissues (Figure 2C). Thus, the two EjTFL1s exhibit different tissue-specific expressions, which also suggests that they may have functional differences with regard to the growth and development of loquat.
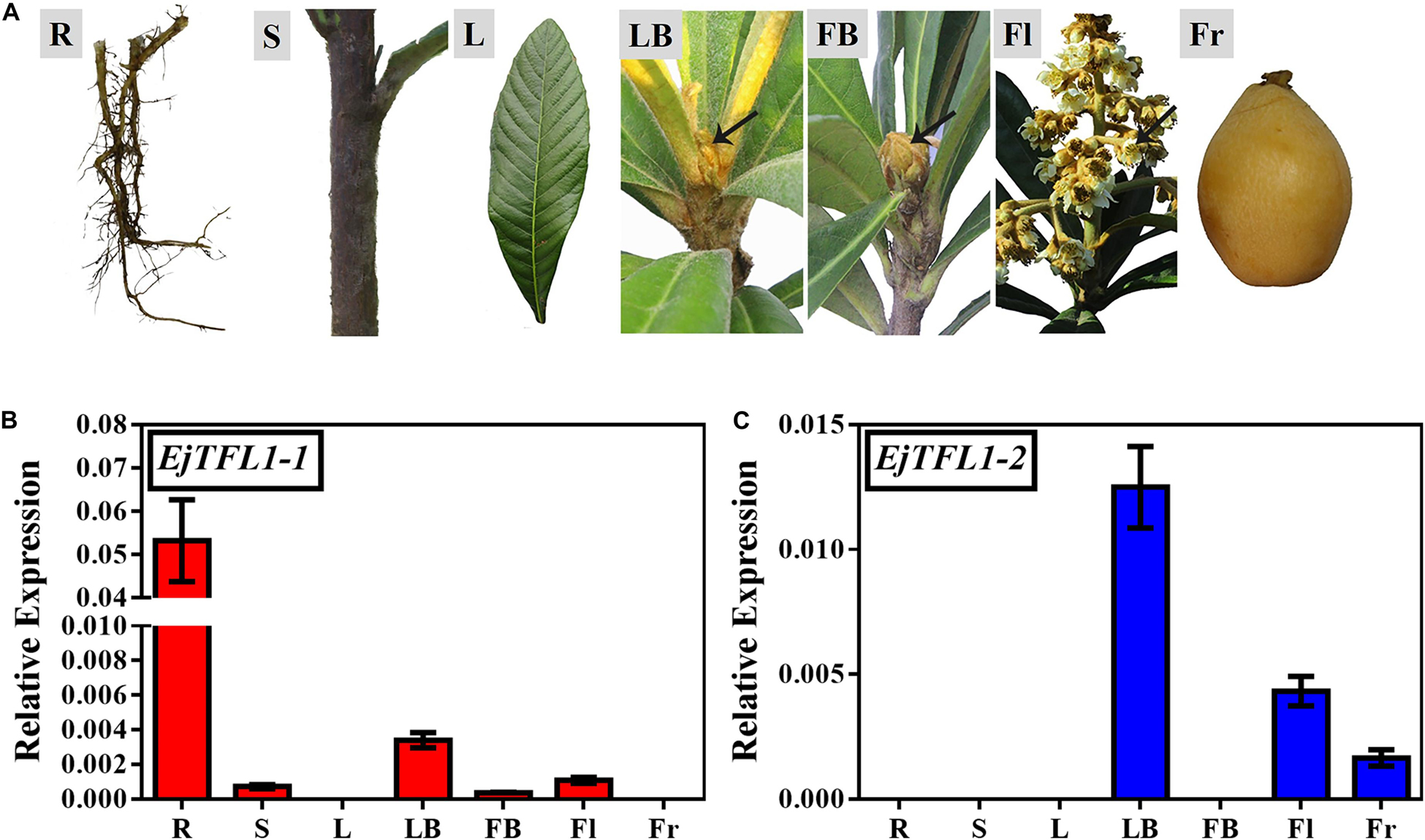
Figure 2. Tissue-specific expression of EjTFL1-1 and EjTFL1-2. (A) Different tissues from loquat (Eriobotrya japonica Lindl) were analyzed. (B,C) Relative expression of EjTFL1-1 and EjTFL1-2 in different tissues shown in (A) (error bars indicate ± SE from three biological replicates). R, root (from rootstock); S, shoot; L, leaf (5.26); LB, leaf bud (5.26); FB, flower bud (8.18); Fl, flower (12.8); Fr, fruit (3.30). Leaf bud, flower bud and flower are phenotypes of the shoot apex at different developmental stages.
We conducted year-round follow-up observation and sampling of the shoot apices and found obvious inflorescences from the end of August to the beginning of September (Figures 3A,F). To examine the time of floral bud differentiation, we prepared paraffin sections of the shoot apices in different periods and examined them by microscope. In late June, the growing point of the apical bud was flat (June 27, Figure 3B), indicating the vegetative growth period. Subsequently, the growing point was raised and a distinct floral meristem can be seen (July 11, Figure 3B), and the floral bud began to differentiate, marking the change from the vegetative stage to the reproductive stage. At the same time, EjAP1-1 and EjAP1-2 began to be expressed in the shoot apices in early July (Figure 3C), indicating that the floral bud began to differentiate. Therefore, we determined that the loquat flower bud differentiation of ‘Jiefangzhong’ loquat in Guangzhou occurs at the end of June and early July 2017.
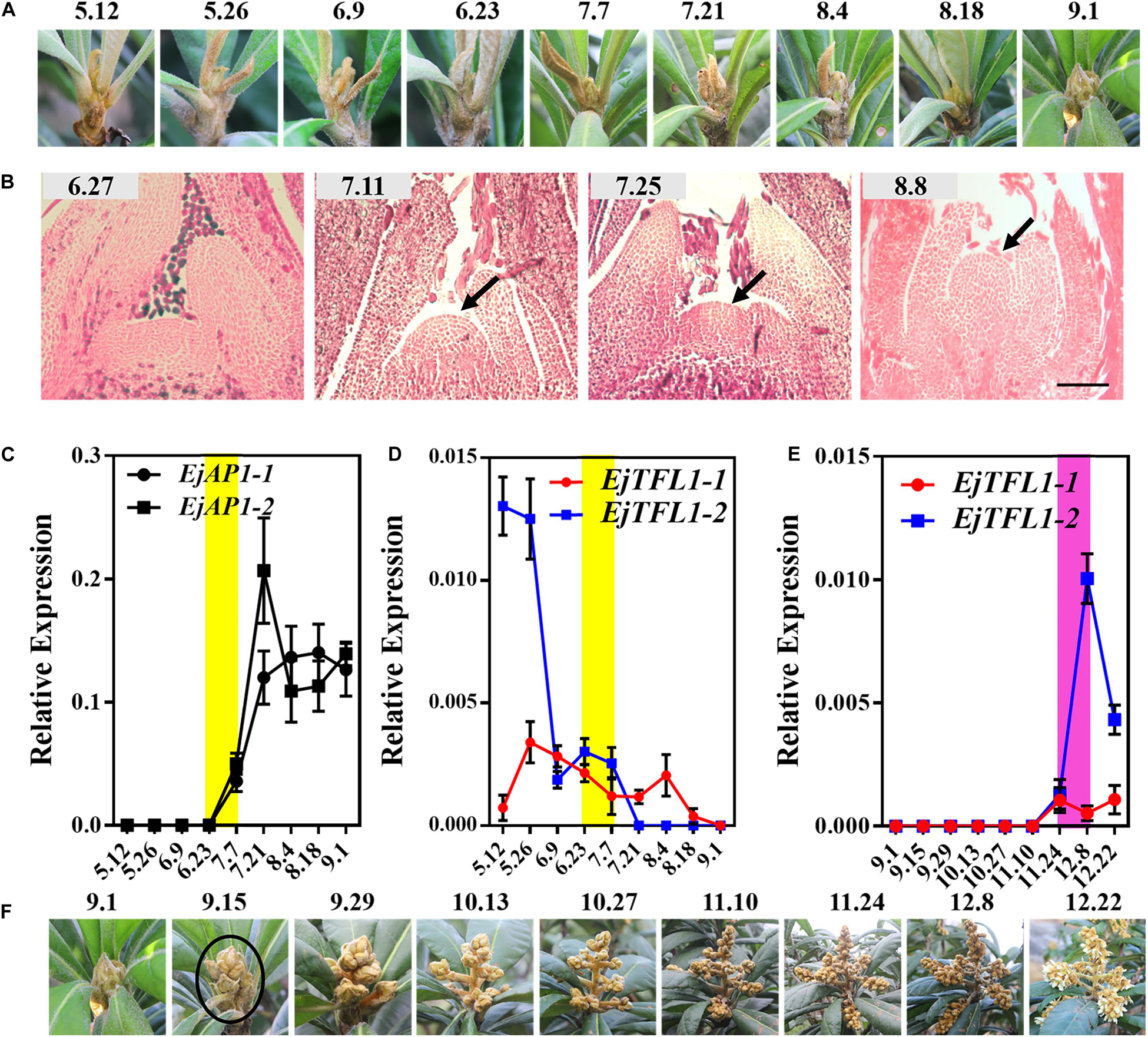
Figure 3. Expression patterns of EjTFL1-1 and EjTFL1-2 in the shoot apices. (A) The phenotype of the shoot apices from May to September. (B) Paraffin sections of the shoot apices from June to August. (C) Relative expression levels of the flower meristem-specific genes EjAP1-1 and EjAP1-2 in the shoot apices from May to September. (D) Relative expression levels of EjTFL1-1 and EjTFL1-2 in the shoot apices from May to September. (E) Relative expression levels of EjTFL1-1 and EjTFL1-2 during inflorescence development. (F) The phenotype of the inflorescence from September to December. The yellow background indicates the initiation period of flower bud differentiation, and the purple background represents the flower-opening period. The black arrow points to the floral meristem. The black circle points to the panicle of loquat. Error bars represent ± SE from three biological replicates.
To identify whether the expression levels of EjTFL1s changed in the shoot apices during different development stages, the qRT-PCR analysis was performed from May to December. The expression level of EjTFL1-1 was the highest in May and then gradually decreased before floral bud differentiation, when almost no expression was observed (July 24th to November 10th) (Figure 3D). In contrast, EjTFL1-2 expression was high in late May and decreased thereafter until reaching a minimum level in early September (Figure 3D). Interestingly, we found that the EjFTL1s started to be expressed again during flower-opening period (Figure 3E). In summary, EjTFL1s showed a relatively high expression level in the vegetative growth stage, and showed a downward trend after entering flower bud differentiation until no expression was found.
To further explore the function of EjTFL1s, we performed qRT-PCR analysis of different tissues of flowers and flowers at different stages of maturity in opening inflorescences (December). EjTFL1-1 expression was extremely low in these tissues and organs or almost undetectable (Figures 4A,B). EjTFL1-2 gradually decreased in Fl1-Fl3 and was almost undetectable in Fl3, Fl4, and Fl5, being expressed only in the stamen (Figures 4A,B).
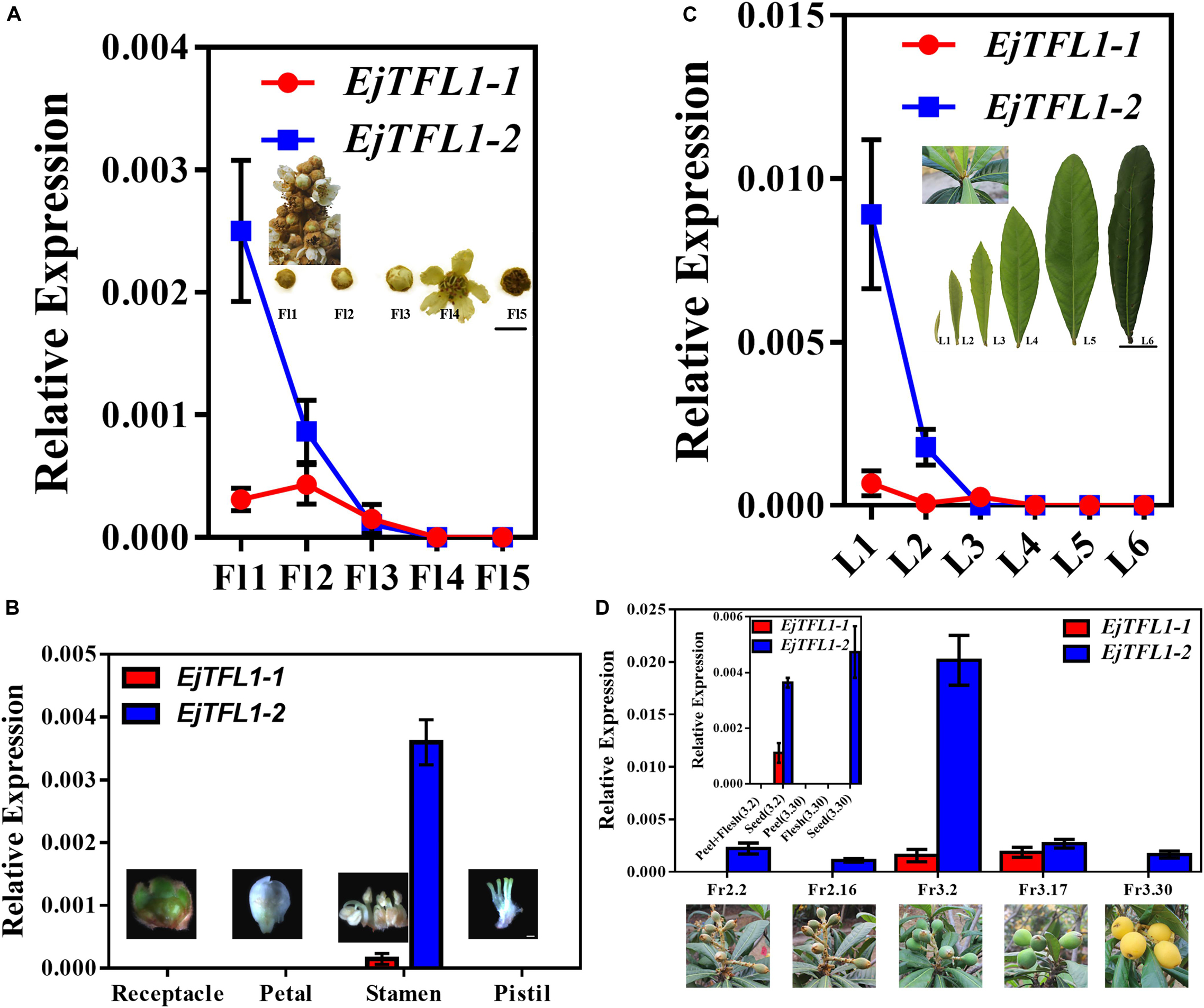
Figure 4. Expression patterns of EjTFL1-1 and EjTFL1-2 in leaves, flowers, and fruits of loquat. (A) Relative expression levels of EjTFL1-1 and EjTFL1-2 in flowers of different maturities in the same period (scale is 1 cm). (B) Relative expression levels of EjTFL1-1 and EjTFL1-2 in different tissues of flower buds (scale 2 mm). (C) Relative expression levels of EjTFL1-1 and EjTFL1-2 in leaves at different levels of maturity in the same period (scale is 5 cm). (D) Relative expression levels of EjTFL1-1 and EjTFL1-2 in fruit development (the small figure shows the relative expression levels of EjTFL1-1 and EjTFL1-2 in different tissues of fruits at different times). ‘Fl1’ indicate the stage 1 of flowers, ‘L1’ indicate the stage 1 of leaves, ‘Fr2.2’ indicates the fruit of February 2, and so on. Error bars represent ± SE from three biological replicates.
Moreover, EjTFL1-1 was slightly expressed in L1 and L3 and not expressed in the other periods in leaves at different stages of maturity in the same period (Figure 4C). EjTFL1-2 was not expressed in L3–L6 but showed relatively high expression in young and tender leaves (L1, L2) (Figure 4C). Previous studies have shown that the L1–L2 period in leaves involves rapid cell proliferation (Gonzalez et al., 2012; Nelissen et al., 2016). Interestingly, flower (stamen) ripening occurs in Fl1-Fl3. These results suggested that EjTFL1-2 may be involved in cell proliferation. Additionally, in the fruit, we found that EjTFL1-1 and EjTFL1-2 were mainly expressed during the fruit-expansion (cell expansion) (see Su et al., 2017) process (Fr3.2-Fr3.17) and were only expressed in seeds during the fruit development process (Figure 4D).
To investigate the effects of the two EjTFL1s in regulating flowering time and inflorescence phenotypes, we overexpressed the EjTFL1s under control of the CaMV 35S promoter in wild-type Col-0 Arabidopsis. Nine transgenic plants were obtained for both groups (35S:EjTFL1-1 and 35S:EjTFL1-2), and two T3 homozygous lines of each genotype were used for further investigation.
Compared with wild-type Col-0 plants, the 35S:EjTFL1-1- and 35S:EjTFL1-2-transgenic plants displayed delayed flowering (Figure 5A). The 35S:EjTFL1-1- and 35S:EjTFL1-2-transgenic plants produced approximately 17-18 rosette leaves, and 35S:EjTFL1-2 line4 (#4) produced approximately 36 rosette leaves (Figure 5B). In contrast, wild-type Col-0 plants produced approximately 12–13 rosette leaves (Figure 5B). qRT-PCR was applied to examine whether the late-flowering phenotype was associated with EjTFL1s in transgenic plants. The expression levels of EjTFL1s were significantly increased in the transgenic plants (Figures 5C,D). In addition, the floral meristem identity genes AtAP1 and AtLFY were clearly downregulated in different transgenic lines (Figures 5E,F). Interestingly, 35S:EjTFL1s transgenic lines exhibited late flowering, and the stem lengths were much higher than those of wild-type Col-0 plants (Figures 5A,G,H).
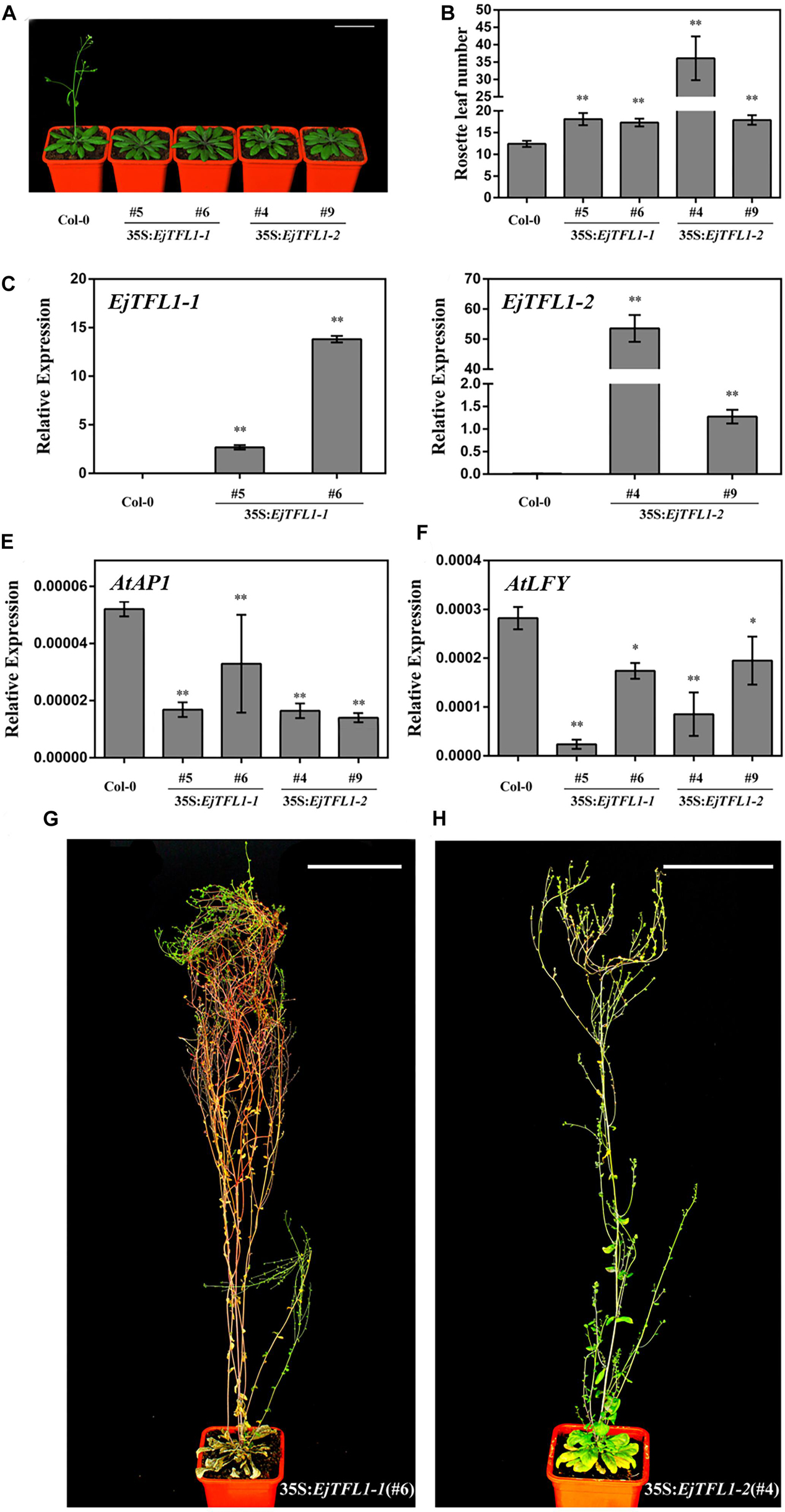
Figure 5. Ectopic expression of EjTFL1s in Arabidopsis. (A) Forty-day-old 35S:EjTFL1s transgenic lines flowered later than did wild-type Col-0 plants. Scale bars = 5 cm. (B) Statistics of 35S:EjTFL1 transgenic lines and wild-type Col-0 rosette leaves (n ≥ 10). (C,D) Expression levels of EjTFLs in 40-day-old transgenic lines and wild-type Col-0 plants. (E,F) Expression levels of AtAP1 and AtLFY in 40-day-old transgenic lines and wild-type Col-0 plants. (G,H) Phenotype of 35S:EjTFL1-transgenic lines, the scale is 10 cm. The whole-plant was used for qRT-PCR. Asterisks indicate significant differences from Col-0, *p < 0.05, **p < 0.01, by Student’s t-test. Error bars indicate ± SE from 3 biological replicates.
In addition, overexpression of EjTFL1s in wild-type Col-0 caused morphological and structural changes. The transgenic plants displayed more inflorescence branches than did wild-type Col-0 (Supplementary Figures S2A–C), with 2–4 inflorescence branches in the latter but as many as 9–17 in the former (Supplementary Figure S3). Compared with wild-type Col-0 plants (Supplementary Figure S2D), the inflorescence phenotypes of some 35S:EjTFL1s-transgenic lines were not significantly different (Supplementary Figures S2E,F). However, there were significant differences in the inflorescence phenotype of other transgenic lines compared to the wild-type Col-0 plants (Supplementary Figures S2G,H). As shown in Supplementary Figure S2G,H, a leaf-like organ appearance, similar to the phenotype described by Ferrandiz et al. (2000), was produced. Moreover, deformed siliques and abnormal cauline leaves were generated and easily distinguished these plants from wild-type Col-0 (Supplementary Figures S2I–L).
Studies in other woody fruit trees, such as apple (Zhang S. et al., 2016) mango (Nakagawa et al., 2012), and citrus (Goldberg-Moeller et al., 2013), have shown that GA3 treatment results in no or less flowering. And that, it is worth noting that loquat flower bud differentiation occurs during the longest period of sunshine in the year, that is, after the summer solstice (June 22), a time when the expression level of EjTFL1s decreased (Figure 3D). Therefore, we speculate that long-day conditions inhibit EjTFL1s expression, thereby promoting flower bud differentiation in loquat. To assess whether EjTFL1s are regulated by GA3 or photoperiod, we designed two experimental protocols (exogenous GA3 and short-day treatments).
Interestingly, after exogenous GA3 and short-day treatments, the loquat plants were unable to undergo flower bud differentiation, though the control and normally grown plants did show flower bud differentiation (Figures 6A,B,K,L). We also found that the flower-specific genes EjAP1-1 and EjAP1-2 were not expressed after GA3 or short-day treatment (Figures 6C,D,G,H). Nonetheless, after the release of treatment (August 10th), the EjAP1s showed a lower level of expression in early September (Figures 6C,G,H). Combining phenotypic observations and qRT-PCR results, we found that GA3 and short-day treatments inhibit flower bud differentiation in loquat.
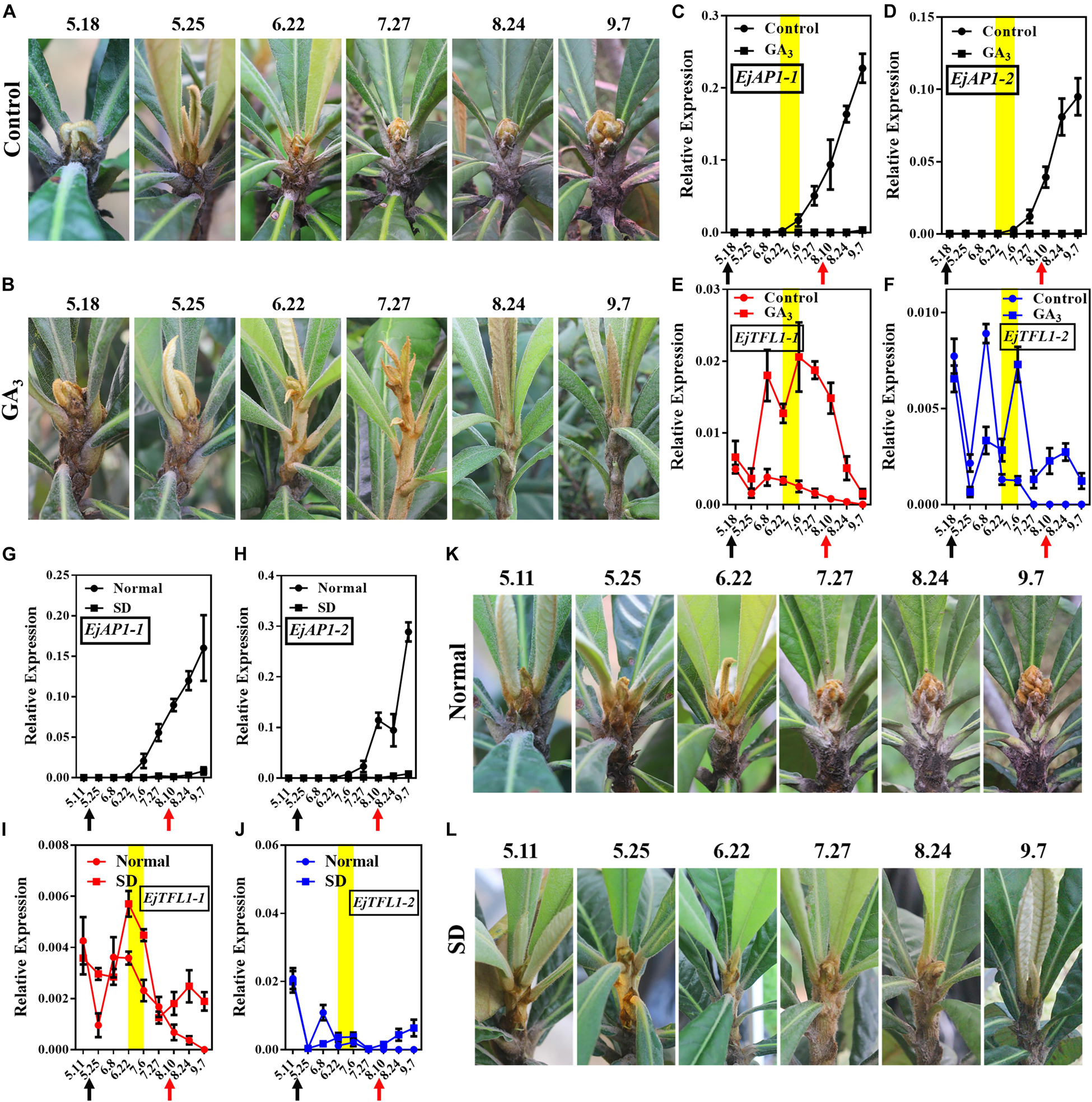
Figure 6. Effects of GA3 treatment and short-day treatment on flowering in loquat. (A) The shoot apices phenotype of the control group from May 18 to September 7. (B) The shoot apices phenotype of the gibberellin-treated group from May 18 to September 7. (C–F) Relative expression levels of EjAP1s and EjTFL1s in the shoot apices of the gibberellin treatment and control groups. (G–J) Relative expression levels of EjAP1s and EjTFL1s in the shoot apices of short-day-treated and normally grown plants. (K) The shoot apices phenotype of normal growth from May 11 to September 7. (L) The shoot apices phenotype of the short-day treatment from May 11 to September 7. The yellow background indicates the initiation period of flower bud differentiation. The two experimental treatment times were from 18th May to 10th August. The black arrow indicates the time at which the treatment started (18th May) and the red arrow indicates the time at which the treatment ended (10th August). Error bars represent ± SE from three biological replicates.
Importantly, EjTFL1-1 and EjTFL1-2 were abundantly expressed after GA3 treatment and maintained high expression level at the end of June, a time when the control group was undergoing flower bud differentiation and EjTFL1-1 and EjTFL1-2 begin to gradually decrease until they are not expressed (Figures 6E,F). In the short-day treatment group, we found that the change in EjTFL1-1 expression after treatment was not as significant as that after GA3 treatment; however, from July 6 to September 7, we still observed a relatively high expression level. The expression level of EjTFL1-2 was not significantly increased at the beginning of the short-day treatment, but a relatively high level of expression began after July 27.
In summary, GA3 clearly promoted EjTFL1-1 expression, while SD treatment may promote EjTFL1-1 expression with a delay, but it does not show clear results on EjTFL1-2 after these treatments.
Furthermore, the stem tip showed strong vegetative growth after GA3 treatment, and we calculated the stem length of the control group and the GA3 treatment group from September 7. The average stem length of the control group was 8.7 cm, while average stem length of the GA3 treatment group was 47.9 cm (Figures 7A,B).
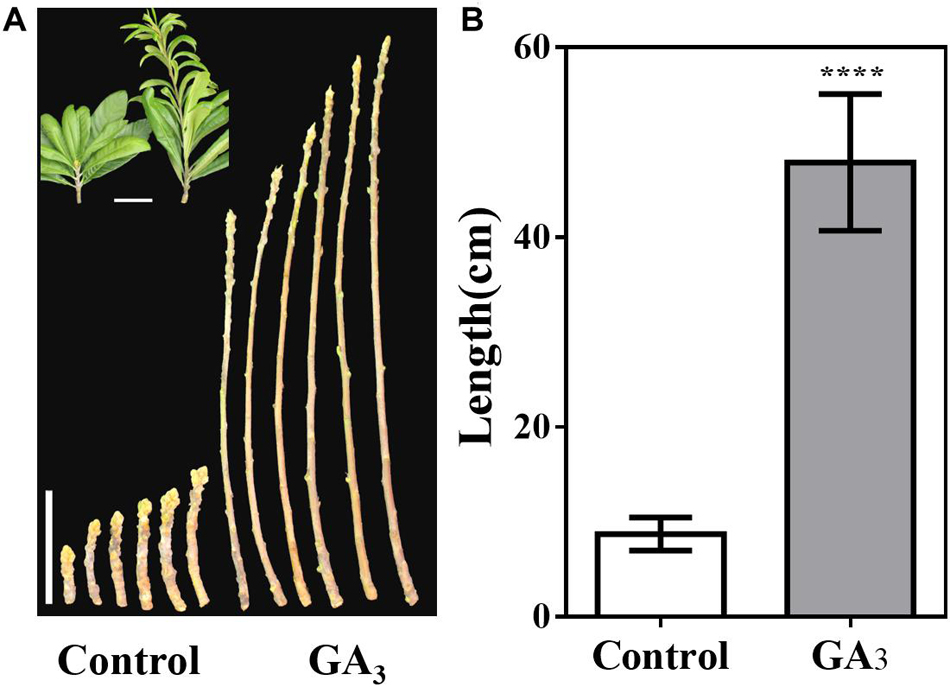
Figure 7. GA3-treatment promotes stem growth. (A) Stem phenotype of GA3-treated and control plants. (B) Stem length statistics of GA3-treated and control plants (n = 30). The GA3-treatment times were from 18th May to 10th August. Asterisks indicate significant differences from Control, ****p < 0.0001, by Student’s t-test. Error bars represent the standard deviation from n = 30.
To investigate the potential reasons leading to the difference in temporal and spatial expression of EjTFL1-1 and EjTFL1-2 during the growth and development of loquat and the difference in response to GA3 and short-day treatment, we searched through the whole genome information of loquat and found that the structural elements of the promoter region of EjTFL1-1 and EjTFL1-2 were significantly different in the 2 kbp upstream regions of the start codon of the EjTFL1 genes. So, we cloned the 2169 bp promoter region upstream of the EjTFL1-1 initiation codon ATG and the 2000 bp promoter region upstream of the EjTFL1-2 initiation codon ATG (see Supplementary Sequences).
Both of these promoter sequences contain a large number of CAAT-boxes associated with transcription initiation and TATA-box promoter core elements of the eukaryotic RNA polymerase II recognition site. Both also contain the following: photoresponsive elements, Box 4, GATA-motif, AE-box and GT1-motif; jasmonic acid (JA) response elements TGACG-motif and CGTCA-motif; and antioxidant response elements (AREs) (Table 1). In addition, the EjTFL1-1 promoter contains the photoresponsive elements TCCC-motif, LAMP-element, MRE and Sp1, as well as the auxin response element TGA-element and salicylic acid (SA) response element TCA-element (Table 1). The EjTFL1-2 promoter contains the photoresponsive element ACE, G-Box, GA-motif, I-box, and TCT-motif, as well as the abscisic acid (ABA) response element (ABRE) and the circadian rhythm regulatory element (Table 1). Importantly, a gibberellin response element, TATC-box, is present in the EjTFL1-1 promoter, and a cis-acting regulatory element, CAT-box, related to meristem expression is present in the EjTFL1-2 promoter (Table 1).
In Arabidopsis, TFL1 forms a complex with FD and inhibits flowering by antagonizing the activity of FT-FD complexes (Abe et al., 2005; Hanano and Goto, 2011). Similar results have been reported in kiwifruit (Varkonyi-Gasic et al., 2013), rose (Randoux et al., 2014), and chrysanthemum (Higuchi and Hisamatsu, 2015). We isolated one FD homologous gene from loquat, EjFD (see Supplementary Sequences), and phylogenetic analysis indicated that EjFD shares high identity with FD in other plants, especially those of the Rosaceae family, such as 98, 74, and 75% identity with EdFD1, EdFD2, and MdFD, respectively (Supplementary Figure S1A). Alignment of deduced amino acid sequences revealed the presence of a conserved bZIP domain (Supplementary Figure S1B).
To determine the subcellular localization of EjTFL1s and EjFD, we constructed plasmids for expressing 35S:EjTFL1-1-GFP, 35S:EjTFL1-2-GFP and 35S:EjFD-GFP fusion proteins and transiently expressed them in N. benthamiana leaves. GFP signals in the nucleus and cytoplasm were found for the two EjTFL1-GFP fusion proteins, whereas the EjFD-GFP fusion protein was only detected in the nucleus (Figure 8). These results showed that both EjTFL1-1 and EjTFL1-2 localized to the cytoplasm and nucleus but that EjFD localized only to the nucleus.
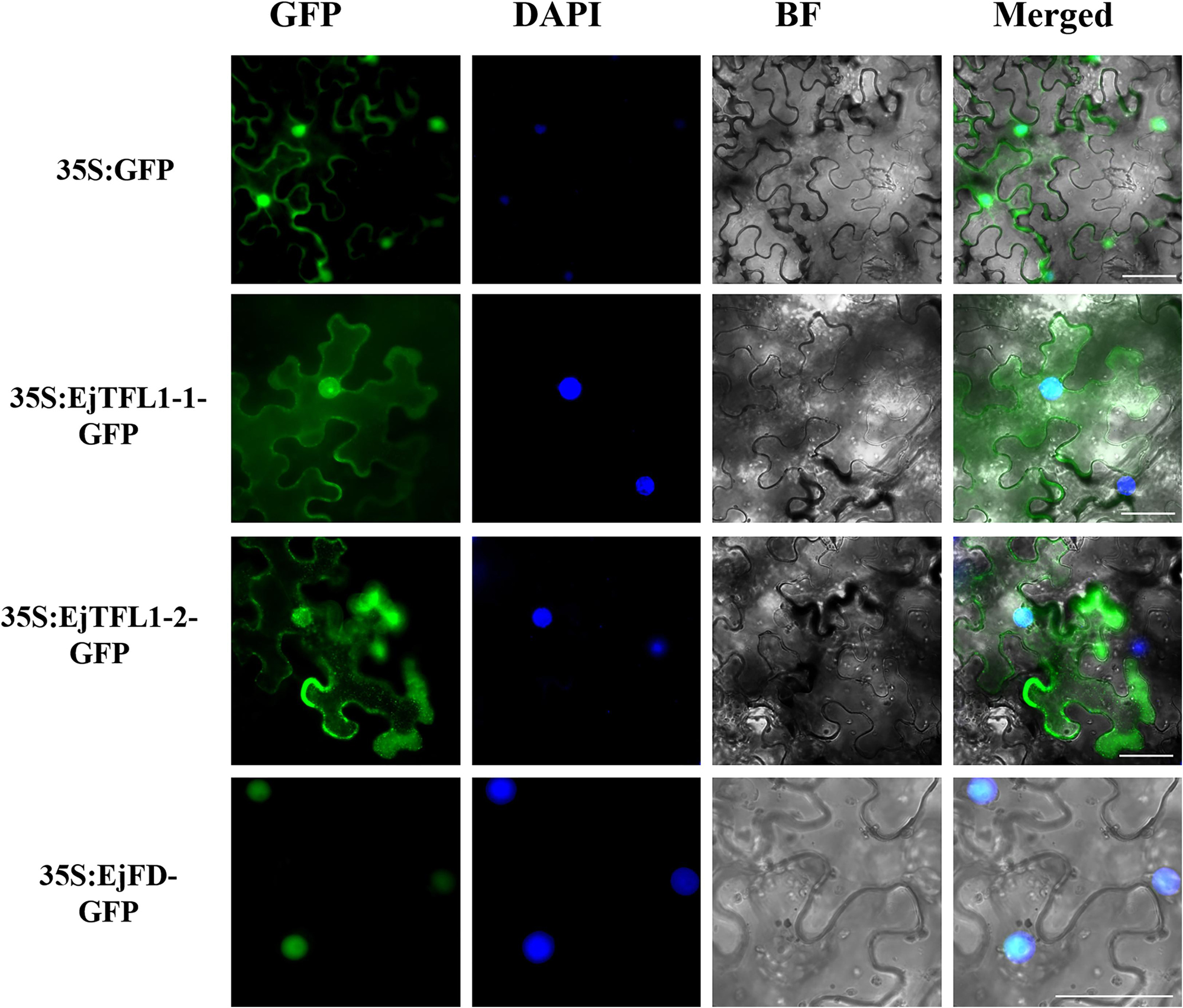
Figure 8. Subcellular localization of EjTFL1s and EjFD. 35S-EjTFL1s-GFP and 35S-EjFD-GFP were transiently transformed into N. benthamiana leaves. GFP, GFP fluorescence; DAPI(4,6-diamidino-2-phenylindole), staining indicates nuclear localization; BF, bright field; Merged, merged image of GFP, BF, and DAPI. Scale bars = 50 μm. 35S: GFP as a control.
To evaluate whether EjTFL1s and EjFD can interact, we performed Y2H and BiFC assay. The yeast cells co-transformed with AD-EjTFL1s and BD-EjFD were able to grow on selective medium (Leu-, Trp-, His-, Ade-) and appeared blue after X-α-Gal treatment (Figure 9A). Thus, EjTFL1s can interact with EjFD in yeast cells. In our BiFC experiment, a yellow fluorescent signal in the nucleus was detected in N. benthamiana leaf epidermal cells co-expressing EjTFL1s and EjFD, with no fluorescent signal observed for the control group (Figure 9B). These results confirmed that EjTFL1s can interact with EjFD in N. benthamiana cells.
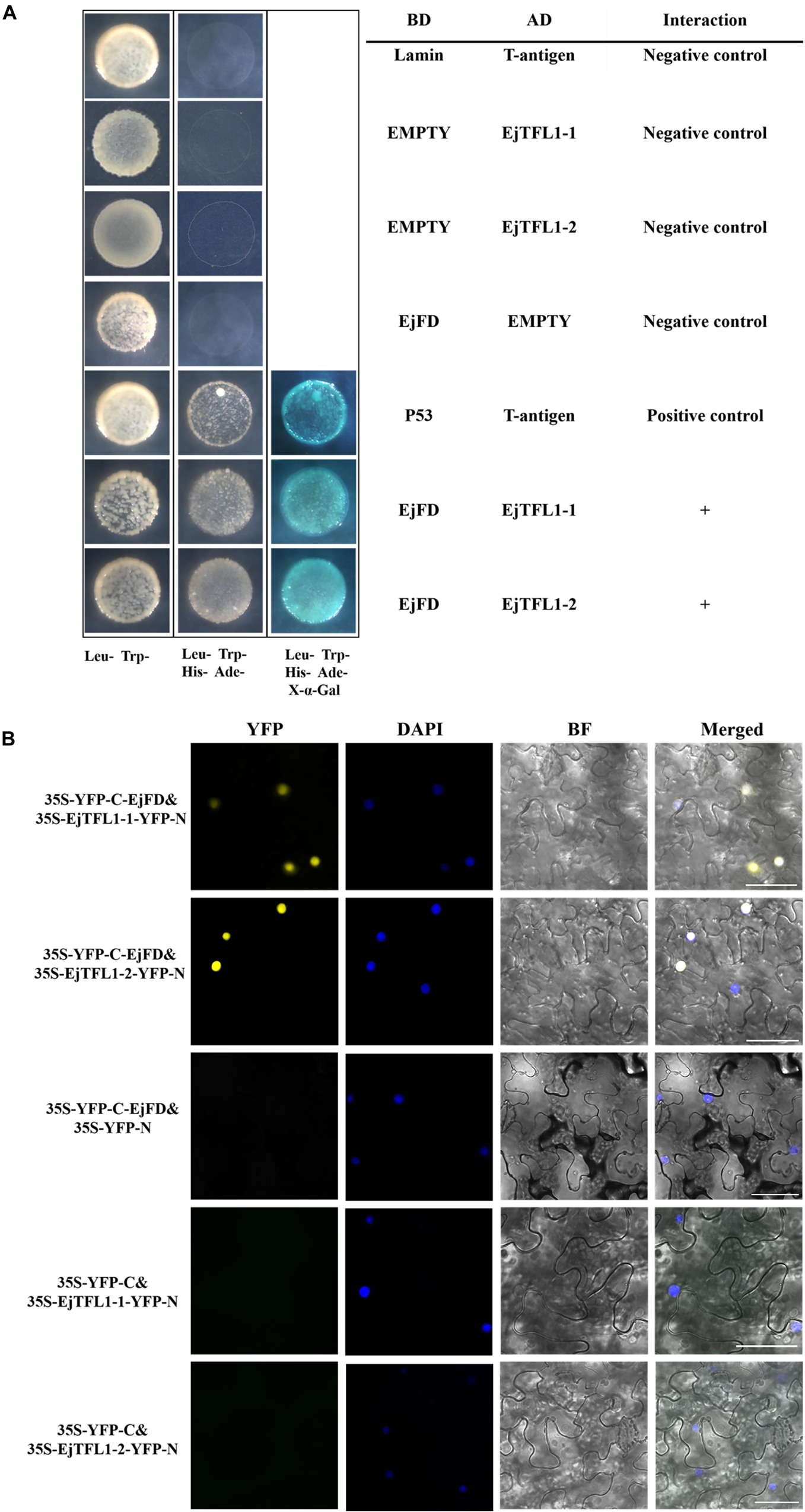
Figure 9. EjTFL1s interacts with EjFD. (A) Y2H analysis of the interaction of EjTFL1s and EjFD in yeast cells. pGBKT7-EjFD is a fusion bait protein, pGADT7-EjTFL1-1 and pGADT7-EjTFL1-2 are fusion prey proteins, pGBKT7-Lam + pGADT7-T, pGBKT7 + pGADT7-EjTFL1s, and pGBKT7-EjFD + pGADT7-T are negative controls, pGBKT7-53 + pGADT7-T are positive controls. The yeast cells co-transfused with the plasmid can grow in the selective medium lacking leucine (Leu-), tryptophan (Trp-), histidine (His-), and adenine (Ade-) but contain 600 μM Aureobasidin A, and turn blue in the presence of X-α-Gal (20 mg/mL), such yeast cells are identified as positive interactions. (B) BiFC system analysis of the interaction of EjTFL1s and EjFD in N. benthamiana leaf epidermal cells. The 35S-YFP-C-EjFD + 35S-EjTFL1s-YFP-N, 35S-YFP-C-EjFD + 35S-YFP-N, 35S-YFP-C + 35S-EjTFL1s-YFP-N were transiently transformed into N. benthamiana leaves. YFP, YFP fluorescence; DAPI (4,6-diamidino-2-phenylindole) staining indicates nuclear localization; BF, bright field; Merged, combined YFP, BF and DAPI images; 35S-YFP-C-EjFD + 35S-YFP-N and 35S-YFP-C + 35S-EjTFL1s-YFP-N as negative control. Scale bar = 50 μm.
In summary, EjTFL1-1 and EjTFL1-2 proteins were able to interact with EjFD proteins, suggesting that EjTFL1s may inhibit floral bud differentiation in loquat by interacting with EjFD, which is the same as the AtTFL1 regulation pattern.
AtAP1 is downregulated in transgenic lines, and the expression trends of EjTFL1s and EjAP1s are opposite during flower bud differentiation. We hypothesized that EjTFL1s may also regulate flower bud differentiation by inhibiting expression of EjAP1s in loquat. Thus, we cloned EjAP1-1 and EjAP1-2 gene promoter regions [1761 bp (EjAP1-1) and 2001 bp (EjAP1-2) upstream of the start codon ATG, see Supplementary Sequences] and tested whether EjTFL1s or EjTFL1-EjFD complex activated or repressed the expression of the EjAP1s using a dual luciferase reporter system (see Figure 10A for the details of vector construction).
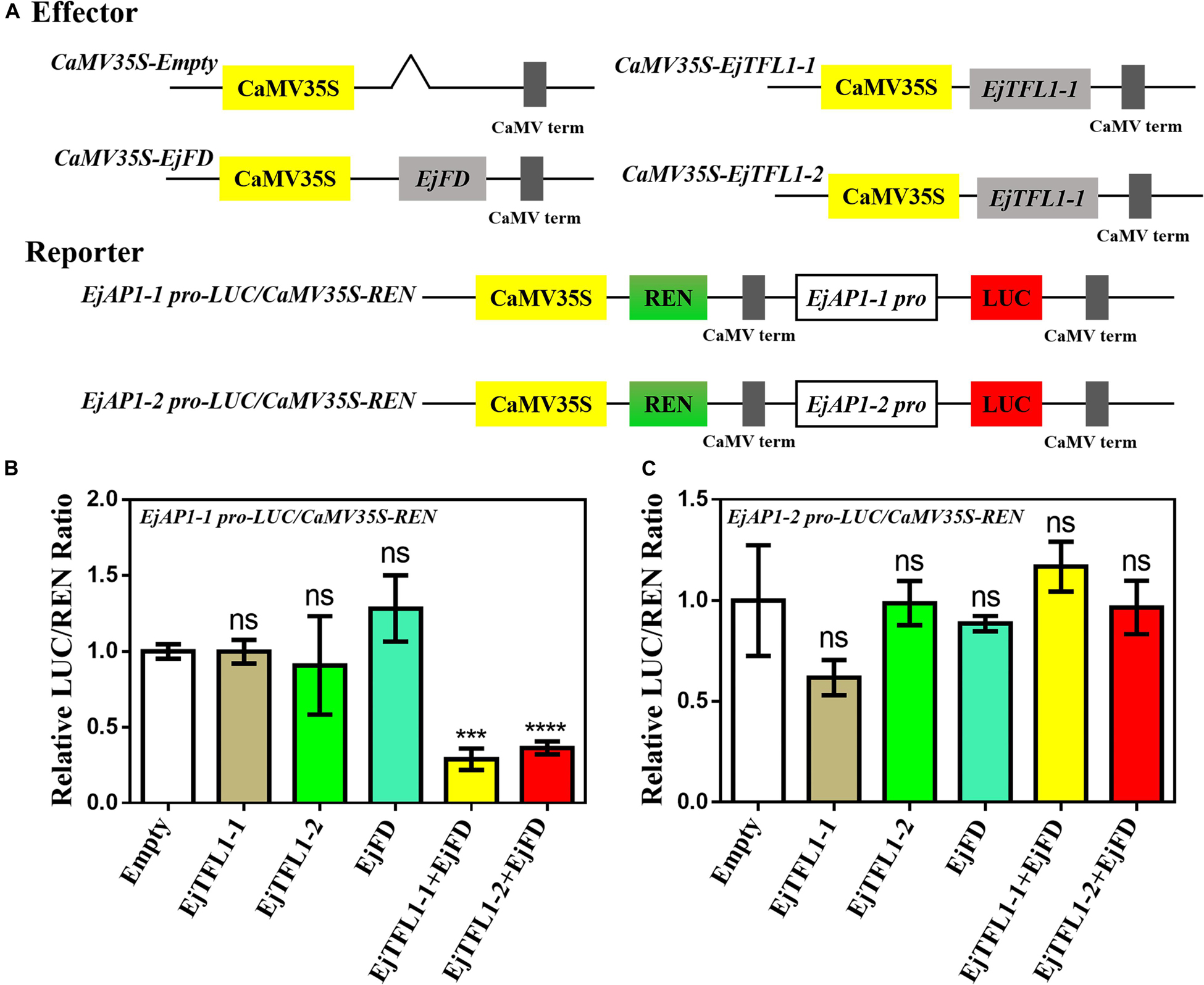
Figure 10. A dual luciferase reporter assay validated the transcriptional activation of EjAP1 promoters by EjTFL1s, EjFD, and EjTFL1s-EjFD. (A) Schematic diagram of effector and reporter plasmid construction. (B) EjTFL1s, EjFD, and EjTFL1s-EjFD act on the EjAP1-1 promoter. (C) EjTFL1s, EjFD, and EjTFL1s-EjFD act on the EjAP1-2 promoter. The activation capabilities of EjTFL1s and EjFD were calculated by the ratio of LUC to REN. The ratio of empty vector LUC/REN was used as a control and set as 1. Error bars represent ± SE from three biological replicates. Asterisks indicate significantly different values compared with those for the empty vector (calculated by Student’s t-test), 0.0001 < *** p < 0.001; ****p < 0.0001; ns, indicates that the difference was not significant.
The results showed that EjTFL1s and EjFD had no obvious inhibitory effect on the EjAP1 promoters and that the EjTFL1-EjFD complex had no significant inhibitory effect on the EjAP1-2 promoter (Figures 10B,C). Importantly, the EjTFL1-1-EjFD complex and EjTFL1-2-EjFD complex significantly repressed the promoter activity of EjAP1-1, and the LUC/REN ratio was 0.29-fold and 0.36-fold that of the empty vector, respectively (Figure 10B). These results suggest that the interaction between EjTFL1s and EjFD inhibits flower bud differentiation and maintains vegetative growth in loquat by inhibiting expression of EjAP1-1.
Different from Rosaceae relatives such as apple or pear (flower bud differentiation in summer or autumn, flowering in the next spring), cultivated loquat flower bud differentiation occur in summer, followed by blossoming from late fall into winter and fruits ripening from late spring into summer (Lin, 2007). As there are few other fresh fruits in late spring and early summer, the loquat just complements the fruit requirement in the market at this time. Moreover, various Eriobotrya species, such as Eriobotrya deflexa Nakai, blossom in spring as do apple and pear (Gu and Spongberg, 2003). The changes in the flowering period of Eriobotrya species suggest that these plants have undergone some unknown changes during evolution. Therefore, it is meaningful to explore the flower-regulating network of loquat, which may also provide some references for the study of flowering regulation in other fruit trees in Rosaceae.
In Arabidopsis, TFL1 act as floral inhibitor (Bradley et al., 1997; Yoo et al., 2010; Hanano and Goto, 2011; Huang et al., 2012). Similarly, in this study, our results indicated that the structure and functions of flowering regulation of EjTFL1s are relatively conserved. EjTFL1s might be mainly involved in vegetative growth and inhibition of in flower bud differentiation. However, EjTFL1-1 and EjTFL1-2 have some different functions during the growth and development of loquat. For example, their expression patterns are different in roots and fruits. (Figures 2, 4D). Furthermore, the expression levels in the same tissue of the same period were much different (Figures 2–4). Interestingly, PopCEN1 and PopCEN2, two TFL1 homologs in populus, also have different tissue-type expression patterns and exhibited relatively conserved functions in flowering (Mohamed et al., 2010). Similarly, EjTFL1-1 and EjTFL1-2 may also have functional differences or functional redundancy, ensuring the normal growth and development of loquat. Changes in the function of homologous genes in plants may play a positive role in the response to sophisticated exogenous and endogenous environments during growth and development.
In the study of Eriobotrya deflexa Nakai forma koshunensis, it was found that EdFT can interact with EdFDs, which may promote flower bud differentiation in loquat (Zhang L. et al., 2016). In Arabidopsis, TFL1 forms a complex with FD and inhibits flowering by antagonizing the activity of FT-FD complexes, and thereby inhibit the expression of downstream AP1 (Abe et al., 2005). AP1 was reported to bind to at least two sites in the 3′ region of TFL1 to inhibit its expression (Kaufmann et al., 2010). In this study, EjTFL1s was able to interact with EjFD, and the expression trends of EjTFL1s and EjAP1s were completely opposite (Figures 3C,D); In addition, the EjTFL1-EjFD complex significantly repressed the promoter activity of EjAP1-1 (Figure 10).
We speculated that the regulatory network of EjTFL1s in the loquat should be as follow: EjTFL1s show relatively high expression in the vegetative growth stage (Figure 3D), and EjTFL1s and EjFD form a complex (Figure 9) to inhibit expression of EjAP1-1, thereby suppressing flower bud differentiation and promoting vegetative growth. Additionally, EjTFL1s integrate photoperiod signal. (Figures 3D, 6I,J), and EjAP1s begin to be abundantly expressed in long-days (Figure 3D). EjAP1s regulates expression of EjTFL1s through negative feedback, promoting a rapid decrease in EjTFL1s expression and at the same time relieving inhibition of EjAP1-1 by EjTFL1s and accelerating flower bud differentiation. This type of feedback mechanism promotes flowering transformation and allows plants to reproduce with high efficiency.
It has been found that TFL1 homologues in different plants are relatively conserved in function, which promotes vegetative growth and delays flowering, such as in Arabidopsis (Hanano and Goto, 2011), rice (Nakagawa et al., 2002), chrysanthemum (Higuchi and Hisamatsu, 2015), dogwood (Liu et al., 2016), apple (Flachowsky et al., 2012), black cherry (Wang and Pijut, 2013), and pomegranate (Patil et al., 2018). Furthermore, ectopic expression of EjTFL1s in Arabidopsis has multiple effects, including modified plant height, branching, and inflorescence, silique and cauline leaf morphology (Figures 5G,H and Supplementary Figure S3). For the TFL1 homolog SP (SELF-PRUNING) in tomato, sp. mutations induce sympodial prolongation and decreased internode numbers (Pnueli et al., 1998). Overexpression of RCN1 or RCN2 and rice TFL1/CEN homologues produced more branches and denser panicle morphology (Nakagawa et al., 2002), and the Cornus TFL1 homolog controls inflorescence development (Liu et al., 2016). These findings suggest that TFL1-like genes control inflorescence architecture in different species.
The differences in the cis-acting elements of the EjTFL1-1 and EjTFL1-2 promoters may be responsible for their different patterns of their response to GA3. In our results, EjTFL1-1 responded to the GA3 signal more rapidly and strongly than did EjTFL1-2 (Figures 6E,F), and the EjTFL1-1 promoter contains a gibberellin signal element, suggesting that EjTFL1-1 might directly respond to the GA3 signal and EjTFL1-2 may respond to the GA3 signal through another pathway. Studies have shown that GA regulates cell expansion and division primarily by eliminating inhibition by DELLA repressors (Yang et al., 1996; Cheng et al., 2004; Ubeda-Tomas et al., 2008; Lee et al., 2012). In this study, the transcription level of EjTFL1s increased significantly after GA3 treatment (Figures 6E,F), and loquat plants showed vigorous vegetative growth (Figures 6B, 7A). The expression trend illustrated in Figure 4 indicates that EjTFL1s are mainly expressed in young tissues. Additionally, Arabidopsis 35S:EjTFL1-transgenic lines displayed significantly increased stem length and delayed flowering. All this evidence indicates that EjTFL1-1 and EjTFL1-2 may play a very important role in the vegetative growth process. Both the previous qRT-PCR analysis and the results of promoter analysis suggest that GA3 regulates the vegetative growth of loquat by modulating the expression of EjTFL1s. The results of this study provide some references for analyses of other woody fruit trees and also provide a new idea for the selection of fast-growing tree species.
Moreover, the EjTFL1-1 and EjTFL1-2 promoters contain a large number of photoresponsive elements, and their expression is affected by SD-treatment (Figures 6I,J), which may be regulated by photoperiod. Interestingly, in short-day Fragaria vesca, short days suppressed FvTFL1 expression and long-day conditions activated it (Koskela et al., 2012). However, in our study, qRT-PCR results showed that EjTFL1s were inhibited under long-day conditions and increased under short-day conditions (Figures 3D, 6I,J). This difference provides an important clue for us to further explore the unique flowering habits of loquat. The EjTFL1-2 promoter contains the CAT-box, a regulatory component involved in meristem expression, and this result suggesting that EjTFL1-2 may be involved in cell proliferation. Importantly, Koskela et al. (2017) found that a deletion located 2547 bp upstream of the transcription start site of FvTFL1 have consequences on expression and function of FvTFL1. This suggested that further away from the transcription start site of the EjTFL1s may also contain more structural elements that affect its function and expression.
EjTFL1-1 and EjTFL1-2 possess conserved amino acids and are mainly expressed in young tissues. Furthermore, the EjTFL1s-EjFD complex significantly repressed the promoter activity of EjAP1-1. Ectopic overexpression of the two EjTFL1s in wild-type Arabidopsis Col-0 delayed flowering time. Our findings suggest that EjTFL1-1 and EjTFL1-2 inhibit flower bud differentiation and promote vegetative growth in loquat by integrating GA3 and photoperiod signals. These findings contribute to our understanding of the molecular mechanism of loquat flowering time regulation and may have wider implications for future manipulation of the time of flowering or bearing fruit.
All datasets generated for this study are included in the article/Supplementary Material.
YJ, YZ, YG, and SL conceived and designed the research. YJ and YZ mainly performed the research and wrote the manuscript. LZ, WS, and JP finished some parts of the experiments. HS and XY supervised the research. YG and SL revised and approved the manuscript.
This study was supported by Key Realm R&D Program of GuangDong Provide (2018B020202011), Natural Science Foundation of Guangdong Province (2017A030313129).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful for the experimental instruments provided by the Key Laboratory of Innovation and Utilization of Horticultural Crop Resources in South China (Ministry of Agriculture). We wish to acknowledge Dr. Xingliang Hou for providing pGreen-35S-GFP, and pGreen-35S-cYFP/nYFP plasmids and Arabidopsis seeds, and thank Dr. Muhammad Qasim Shahid for his help in editing this manuscript, and thank American Journal Experts for their language editing services for this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00576/full#supplementary-material
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Lee, J. H., Yoo, S. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614. doi: 10.1038/sj.emboj.7600950
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. doi: 10.1126/science.275.5296.80
Charrier, A., Vergne, E., Dousset, N., Richer, A., Petiteau, A., and Chevreau, E. (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10:40. doi: 10.3389/fpls.2019.00040
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Esumi, T., Tao, R., and Yonemori, K. (2005). Isolation of LEAFY and TERMINAL FLOWER 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sex. Plant Reproduct. 17, 277–287. doi: 10.1007/s00497-004-0239-3
Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734.
Flachowsky, H., Szankowski, I., Waidmann, S., Peil, A., Trankner, C., and Hanke, M. V. (2012). The MdTFL1 gene of apple (Malus x domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 32, 1288–1301. doi: 10.1093/treephys/tps080
Freiman, A., Shlizerman, L., Golobovitch, S., Yablovitz, Z., Korchinsky, R., Cohen, Y., et al. (2012). Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta. 235, 1239–1251. doi: 10.1007/s00425-011-1571-0
Goldberg-Moeller, R., Shalom, L., Shlizerman, L., Samuels, S., Zur, N., Ophir, R., et al. (2013). Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in Citrus buds. Plant Sci. 198, 46–57. doi: 10.1016/j.plantsci.2012.09.012
Gonzalez, N., Vanhaeren, H., and Inze, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Gu, C., and Spongberg, S. A. (2003). “ERIOBOTRYA Lindley,” in Flora of China, eds W. Zhengyi, P. H. Raven, and D. Y. Hong, (Jefferson, MO: Missouri Botanical Garden Press).
Hanano, S., and Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23, 3172–3184. doi: 10.1105/tpc.111.088641
Hanzawa, Y., Money, T., and Bradley, D. (2005). A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. U.S.A. 102, 7748–7753. doi: 10.1073/pnas.0500932102
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 1:13. doi: 10.1186/1746-4811-1-13
Higuchi, Y., and Hisamatsu, T. (2015). CsTFL1, a constitutive local repressor of flowering, modulates floral initiation by antagonising florigen complex activity in chrysanthemum. Plant Sci. 237, 1–7. doi: 10.1016/j.plantsci.2015.04.011
Ho, W. W., and Weigel, D. (2014). Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26, 552–564. doi: 10.1105/tpc.113.115220
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. doi: 10.1038/ncomms5601
Huang, N. C., Jane, W. N., Chen, J., and Yu, T. S. (2012). Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J. 72, 175–184. doi: 10.1111/j.1365-313X.2012.05076.x
Igasaki, T., Watanabe, Y., Nishiguchi, M., and Kotoda, N. (2008). The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in lombardy poplar. Plant Cell Physiol. 49, 291–300. doi: 10.1093/pcp/pcn010
Iwata, H., Gaston, A., Remay, A., Thouroude, T., Jeauffre, J., Kawamura, K., et al. (2012). The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 69, 116–125. doi: 10.1111/j.1365-313X.2011.04776.x
Jang, S., Torti, S., and Coupland, G. (2009). Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60, 614–625. doi: 10.1111/j.1365-313X.2009.03986.x
Jiang, Y., Peng, J., Zhu, Y., Su, W., Zhang, L., Jing, Y., et al. (2019). The role of EjSOC1s in flower initiation in Eriobotrya japonica. Front. Plant Sci. 10:253. doi: 10.3389/fpls.2019.00253
Karlgren, A., Gyllenstrand, N., Kallman, T., Sundstrom, J. F., Moore, D., Lascoux, M., et al. (2011). Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol. 156, 1967–1977. doi: 10.1104/pp.111.176206
Kaufmann, K., Wellmer, F., Muino, J. M., Ferrier, T., Wuest, S. E., Kumar, V., et al. (2010). Orchestration of floral initiation by APETALA1. Science 328, 85–89. doi: 10.1126/science.1185244
Kobayashi, Y., and Weigel, D. (2007). Move on up, it’s time for change–mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21, 2371–2384. doi: 10.1101/gad.1589007
Koskela, E. A., Kurokura, T., Toivainen, T., Sonsteby, A., Heide, O. M., Sargent, D. J., et al. (2017). Altered regulation of TERMINAL FLOWER 1 causes the unique vernalisation response in an arctic woodland strawberry accession. New Phytol. 216, 841–853. doi: 10.1111/nph.14734
Koskela, E. A., Mouhu, K., Albani, M. C., Kurokura, T., Rantanen, M., Sargent, D. J., et al. (2012). Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 159, 1043–1054. doi: 10.1104/pp.112.196659
Koskela, E. A., Sonsteby, A., Flachowsky, H., Heide, O. M., Hanke, M. V., Elomaa, P., et al. (2016). TERMINAL FLOWER1 is a breeding target for a novel everbearing trait and tailored flowering responses in cultivated strawberry (Fragaria x ananassa Duch.). Plant Biotechnol. J. 14, 1852–1861. doi: 10.1111/pbi.12545
Kotoda, N., and Wada, M. (2005). MdTFL1, a TFL1-like gene of apple, retards the transition from the vegetative to reproductive phase in transgenic Arabidopsis. Plant Sci. 168, 95–104. doi: 10.1016/j.plantsci.2004.07.024
Kurokura, T., Mimida, N., Battey, N. H., and Hytonen, T. (2013). The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 64, 4131–4141. doi: 10.1093/jxb/ert233
Lee, L. Y., Hou, X., Fang, L., Fan, S., Kumar, P. P., and Yu, H. (2012). STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development. 139, 1568–1576. doi: 10.1242/dev.079426
Lin, S. Q. (2007). “World loquat production and research with special reference to China,” in Proceedings of the Second International Symposium on Loquat (Leuven: International Society for Horticultural Science), 37–43. doi: 10.17660/ActaHortic.2007.750.2
Liu, L., Liu, C., Hou, X., Xi, W., Shen, L., Tao, Z., et al. (2012). FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 10:e1001313. doi: 10.1371/journal.pbio.1001313
Liu, X., Zhang, J., Abuahmad, A., Franks, R. G., Xie, D. Y., and Xiang, Q. Y. (2016). Analysis of two TFL1 homologs of dogwood species (Cornus L.) indicates functional conservation in control of transition to flowering. Planta 243, 1129–1141. doi: 10.1007/s00425-016-2466-x
Liu, Y., Song, H., Liu, Z., Hu, G., and Lin, S. (2013). Molecular characterization of loquat EjAP1 gene in relation to flowering. Plant Growth Regul. 70, 287–296. doi: 10.1007/s10725-013-9800-0
Mimida, N., Kotoda, N., Ueda, T., Igarashi, M., Hatsuyama, Y., Iwanami, H., et al. (2009). Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus x domestica Borkh.). Plant Cell Physiol. 50, 394–412. doi: 10.1093/pcp/pcp001
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62, 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Nakagawa, M., Honsho, C., Kanzaki, S., Shimizu, K., and Utsunomiya, N. (2012). Isolation and expression analysis of FLOWERING LOCUS T-like and gibberellin metabolism genes in biennial-bearing mango trees. Sci. Horticult. 139, 108–117. doi: 10.1016/j.scienta.2012.03.005
Nakagawa, M., Shimamoto, K., and Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. doi: 10.1046/j.1365-313x.2002.01255.x
Nakano, Y., Higuchi, Y., Yoshida, Y., and Hisamatsu, T. (2015). Environmental responses of the FT/TFL1 gene family and their involvement in flower induction in Fragaria x ananassa. J. Plant Physiol. 177, 60–66. doi: 10.1016/j.jplph.2015.01.007
Nelissen, H., Gonzalez, N., and Inze, D. (2016). Leaf growth in dicots and monocots: so different yet so alike. Curr. Opin. Plant Biol 33, 72–76. doi: 10.1016/j.pbi.2016.06.009
Patil, H. B., Chaurasia, A. K., Azeez, A., Krishna, B., Subramaniam, V. R., Sane, A. P., et al. (2018). Characterization of two TERMINAL FLOWER1 homologs PgTFL1 and PgCENa from pomegranate (Punica granatum L.). Tree Physiol. 38, 772–784. doi: 10.1093/treephys/tpx154
Pnueli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., et al. (1998). The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989.
Randoux, M., Daviere, J. M., Jeauffre, J., Thouroude, T., Pierre, S., Toualbia, Y., et al. (2014). RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol. 202, 161–173. doi: 10.1111/nph.12625
Ratcliffe, O. J., Amaya, I., Vincent, C. A., Rothstein, S., Carpenter, R., Coen, E. S., et al. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125, 1609–1615.
Ratcliffe, O. J., Bradley, D. J., and Coen, E. S. (1999). Separation of shoot and floral identity in Arabidopsis. Development 126, 1109–1120.
Reig, C., Gil-Munoz, F., Vera-Sirera, F., Garcia-Lorca, A., Martinez-Fuentes, A., Mesejo, C., et al. (2017). Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Planta 246, 915–925. doi: 10.1007/s00425-017-2740-6
Ruonala, R., Rinne, P. L., Kangasjarvi, J., and van der Schoot, C. (2008). CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell 20, 59–74. doi: 10.1105/tpc.107.056721
Shan, L. L., Li, X., Wang, P., Cai, C., Zhang, B., Sun, C. D., et al. (2008). Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. doi: 10.1007/s00425-008-0696-2
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Su, W., Zhu, Y., Zhang, L., Yang, X., Gao, Y., and Lin, S. (2017). The cellular physiology of loquat (Eriobotrya japonica Lindl.) fruit with a focus on how cell division and cell expansion processes contribute to pome morphogenesis. Sci. Horticult. 224, 142–149. doi: 10.1016/j.scienta.2017.06.012
Ubeda-Tomas, S., Swarup, R., Coates, J., Swarup, K., Laplaze, L., Beemster, G. T., et al. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10, 625–628. doi: 10.1038/ncb1726
Varkonyi-Gasic, E., Moss, S. M. A., Voogd, C., Wang, T., Putterill, J., and Hellens, R. P. (2013). Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytol. 198, 732–746. doi: 10.1111/nph.12162
Wang, Y., and Pijut, P. M. (2013). Isolation and characterization of a TERMINAL FLOWER 1 homolog from Prunus serotina Ehrh. Tree Physiol. 33, 855–865. doi: 10.1093/treephys/tpt051
Wellmer, F., and Riechmann, J. L. (2010). Gene networks controlling the initiation of flower development. Trends Genet. 26, 519–527. doi: 10.1016/j.tig.2010.09.001
Wickland, D. P., and Hanzawa, Y. (2015). The flowering locus t/terminal flower 1 gene family: functional evolution and molecular mechanisms. Mol. Plant. 8, 983–997. doi: 10.1016/j.molp.2015.01.007
Yamagishi, N., Li, C. J., and Yoshikawa, N. (2016). Promotion of flowering by apple latent spherical virus vector and virus elimination at high temperature allow accelerated breeding of apple and pear. Front. Plant Sci. 7:171. doi: 10.3389/fpls.2016.00171
Yamaguchi, A., Kobayashi, Y., Goto, K., Abe, M., and Araki, T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46, 1175–1189. doi: 10.1093/pcp/pci151
Yan, Y., Shen, L., Chen, Y., Bao, S., Thong, Z., and Yu, H. (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 30, 437–448. doi: 10.1016/j.devcel.2014.07.004
Yang, T., Davies, P. J., and Reid, J. B. (1996). Genetic dissection of the relative roles of auxin and gibberellin in the regulation of stem elongation in intact light-grown peas. Plant Physiol. 110, 1029–1034. doi: 10.1104/pp.110.3.1029
Yoo, S. J., Chung, K. S., Jung, S. H., Yoo, S. Y., Lee, J. S., and Ahn, J. H. (2010). Brother of ft and tfl1 (BFT) has tfl1-like activity and functions redundantly with tfl1 in inflorescence meristem development in Arabidopsis. Plant J. 63, 241–253. doi: 10.1111/j.1365-313X.2010.04234.x
Zhang, L., Jiang, Y., Zhu, Y., Su, W., Long, T., Huang, T., et al. (2019). Functional characterization of GI and CO homologs from eriobotrya deflexa nakai forma koshunensis. Plant Cell Rep. 38, 533–543. doi: 10.1007/s00299-019-02384-3
Zhang, L., Yu, H., Lin, S., and Gao, Y. (2016). Molecular characterization of FT and FD homologs from Eriobotrya deflexa Nakai forma koshunensis. Front. Plant Sci. 7:8. doi: 10.3389/fpls.2016.00008
Zhang, S., Zhang, D., Fan, S., Du, L., Shen, Y., Xing, L., et al. (2016). Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus domestica Borkh.). Plant Physiol Biochem. 107, 178–186. doi: 10.1016/j.plaphy.2016.06.005
Keywords: loquat, EjTFL1, flowering time, GA3, TERMINAL FLOWER1
Citation: Jiang Y, Zhu Y, Zhang L, Su W, Peng J, Yang X, Song H, Gao Y and Lin S (2020) EjTFL1 Genes Promote Growth but Inhibit Flower Bud Differentiation in Loquat. Front. Plant Sci. 11:576. doi: 10.3389/fpls.2020.00576
Received: 21 September 2019; Accepted: 17 April 2020;
Published: 15 May 2020.
Edited by:
Patrick Laufs, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Francisco Madueño, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2020 Jiang, Zhu, Zhang, Su, Peng, Yang, Song, Gao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongshun Gao, eW9uZ3NodW5nYW9AMTYzLmNvbQ==; Shunquan Lin, bG9xdWF0QHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.