- College of Grassland Science and Technology, China Agricultural University, Beijing, China
Melatonin (N-acetyl-5-methoxytryptamine) is a pleiotropic signaling molecule that plays important roles in plant growth, development and stress responses. Alfalfa (Medicago sativa L.) is an important and widely cultivated leguminous forage crop with high biomass yield and rich nutritional value. The effects of exogenous melatonin content regulation on alfalfa stress tolerance have been investigated in recent years. Here, we isolated and introduced the MsASMT1 (N-acetylserotonin methyltransferase) gene into alfalfa, which significantly improved the endogenous melatonin content. Compared with wild-type (WT) plants, MsASMT1 overexpression (OE-MsASMT1) plants exhibited a series of phenotypic changes, including vigorous growth, increased plant height, enlarged leaves and robust stems with increased cell sizes, cell numbers and vascular bundles, as well as more branches. We also found that the flavonoid content and lignin composition of syringyl to guaiacyl ratio (S/G) were decreased and the cellulose content was increased in OE-MsASMT1 transgenic alfalfa. Further transcriptomic and metabolomic exploration revealed that a large group of genes in phenylalanine pathway related to flavonoids and lignin biosynthesis were significantly altered, accompanied by significantly reduced concentrations of the glycosides of quercetin, kaempferol, formononetin and biochanin in OE-MsASMT1 transgenic alfalfa. Our study first uncovers the effects of endogenous melatonin on alfalfa growth and metabolism. This report provides insights into the regulation effects of melatonin on plant growth and phenylalanine metabolism, especially flavonoids and lignin biosynthesis.
Introduction
Melatonin (N-acetyl-5-methoxytryptamine) is a ubiquitous molecule in higher plants and is broadly known as a biological regulator of circadian rhythm and plant growth, development, senescence and stress responses (Reiter et al., 2015; Wang et al., 2018). Melatonin synthesis occurs in chloroplasts and mitochondria (Tan et al., 2013), which has been determined in various species; however, melatonin levels vary dramatically among different plant species and organs (Reiter et al., 2007).
The biosynthesis of melatonin in plants derives from the essential amino acid tryptophan, which is catalyzed successively by tryptophan decarboxylase (TDC), tryptamine 5-hydroxylase (T5H), serotonin N-acetyltransferase (SNAT) and N-acetylserotonin methyltransferase (ASMT)/caffeic acid O-methyltransferase (COMT). ASMT catalyzes the ultimate melatonin biosynthetic step and plays a vital role in melatonin biosynthesis in plants (Byeon et al., 2014; Tan et al., 2016).
With the sessile nature of plants, they are constantly challenged by various biotic and abiotic stresses during growth and development. Plants have evolved a series of defense mechanisms to protect them from adverse conditions. Melatonin is well known as a free radical scavenger and potent antioxidant. The endogenous or exogenous regulation of melatonin significantly alleviates oxidative damage in plants under drought (Antoniou et al., 2017), salinity (Liang et al., 2015), cold (Fu et al., 2017), heat (Shi et al., 2015; Xu et al., 2016), high light (Lee and Back, 2018a), and pathogen/pest stresses (Mandal et al., 2018; Wei et al., 2018). Melatonin is also essential for plant growth and development, such as seed germination (Posmyk et al., 2009; Zhang et al., 2014), lateral root formation (Park and Back, 2012), flowering (Byeon and Back, 2014; Zhang et al., 2018) and senescence (Lee and Back, 2017). The functional diversification of melatonin in plants is accompanied by crosstalk with various plant hormones, such as auxin, ethylene (ET), abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), brassinosteroid (BR), and strigolactone (SL) (Arnao and Hernández-Ruiz, 2018a, 2019; Lee and Back, 2018b; Zhang Z. et al., 2019).
Melatonin also functions in metabolic processes, including primary metabolism (Zhao et al., 2015; Kobyliñska et al., 2018) and secondary metabolism (Yuan et al., 2016; Zhang et al., 2016; Ai and Zhu, 2018). Pretreatment with melatonin improved the accumulation of anthocyanin by upregulating the transcript levels of anthocyanin biosynthetic genes in cabbage (Zhang et al., 2016). However, melatonin antagonized JA in anthocyanin biosynthesis in Arabidopsis (Ai and Zhu, 2018). Our previous research found that genes related to flavonoid biosynthesis and phenylalanine metabolism pathways were upregulated in ovine HIOMT overexpression switchgrass plants (Yuan et al., 2016), which suggests that melatonin may participate in the biological process of flavonoid biosynthesis. Flavonoids are widely distributed throughout the plant kingdom, functioning in plant growth and development and in responses to various biotic and abiotic stresses, including UV protection and defense responses to pathogens/insects (Kumar and Pandey, 2013; Mitsunami et al., 2014). Lee et al. (2018) found that flavonoids acted as potent inhibitors of melatonin synthesis. The exogenous application of flavonoids (morin and myricetin) or the overexpression of a putative flavonol synthase gene (FLS) resulted in the increased production of flavonoids but reduced accumulation of melatonin in rice. However, it is still unclear whether high levels of melatonin affect flavonoids biosynthesis, and which flavonoid are mainly regulated.
In human beings, melatonin is widely known as a dietary supplement that improves the quality of sleep (Cornu et al., 2010), has anti-aging properties (Tresguerres et al., 2014) and alleviates the effects of jet lag (Waterhouse et al., 1997). Numerous pharmacological studies confirmed that melatonin could also be associated with the prevention of several diseases, including cardiovascular and neurodegenerative diseases or cancer (Claustrat et al., 2005; Arnao and Hernández-Ruiz, 2018b). Alfalfa (Medicago sativa L.) is an important and widely cultivated leguminous forage crop worldwide and is known as the “King of Forages,” mainly due to its high biomass yield, high protein content and rich nutritional value (Aung et al., 2015; Fu et al., 2015). Alfalfa has been successfully used as a raw material for health products, and its dietary market is developing rapidly. Improving the melatonin content in alfalfa has profound implications for improving forage quality and nutritional value. To date, there are only two reports related to the function of melatonin in alfalfa on improving drought (Antoniou et al., 2017) and waterlogging resistance (Zhang Q. et al., 2019) with exogenous melatonin pretreatment, while the function of endogenous melatonin in alfalfa has not been reported. Alfalfa is rich in various flavonoids, including quercetin, luteolin, formononetin, and so on. It is of great significance to explore the effect of melatonin on flavonoids biosynthesis in alfalfa.
Here, we report that the overexpression of MsASMT1 in alfalfa led to melatonin accumulation but exhibited a contrasting effect on flavonoids accumulation. In this report, we first cloned the N-acetylserotonin methyltransferase (ASMT) gene from alfalfa and produced OE-MsASMT1 transgenic alfalfa by Agrobacterium-mediated transformation. The growth and development phenotype of OE-MsASMT1 transgenic alfalfa were evaluated compared to WT plants. OE-MsASMT1 transgenic alfalfa had a higher melatonin content and lower flavonoid content. Further transcriptomic and metabolomic analyses found a large group of genes in phenylalanine pathway were downregulated and the concentration of quercetin, kaempferol, formononetin, and biochanin were decreased. Our study first reported the effects of endogenous melatonin on alfalfa plant growth and indicated that melatonin biosynthesis inhibited flavonoids accumulation in alfalfa plants.
Materials and Methods
Gene Isolation, Bioinformatics Analysis and Vector Construction
The apple ASMT and rice ASMT1 genes were obtained for BLAST queries against Medicago truncatula resources at https://phytozome.jgi.doe.gov/, and MtASMT (Medtr5g074600.1) was found to share high identity (60.20%) with apple ASMT and relatively lower identity (38.33%) with rice ASMT1. Due to homology and phylogenetic analysis between alfalfa and M. truncatula, primers were designed based on the M. truncatula ASMT sequence to amplify the coding sequence (CDS) in the alfalfa genome.
Total RNA was extracted from the leaves, stems and roots of alfalfa using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), and intact RNA was immediately reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the manufacturer’s instructions. cDNAs were used as templates to amplify the CDS of MsASMT1 (GenBank accession number: MN092350); primers are listed in Supplementary Table S1.
PCR products were verified by sequencing. Nucleotide sequence and amino acid sequence alignments were performed with DNAMAN. The conserved domain1 and physicochemical properties2 of MsASMT1 were analyzed online.
Amino acid sequences of alfalfa (M. sativa), M. truncatula and other selected species (Oryza sativa OsASMT1 (AK072740), OsASMT2 (AK069308), OsASMT3 (AAL34945.1), Malus zumi MzASMT (KJ123721), Phaseolus vulgaris PvASMT (XP_007142077), Setaria italica SiASMT (XP_004973630.1), Triticum urartu TuASMT (EMS45259.1), Brachypodium distachyon BdASMT (XP_003571634.3), Cicer arietinum CaASMT (XM_004490639.2), Cucumis sativus CsASMT (XP_004151735), Glycine max GmASMT (XP_003536188), Hordeum vulgare HvASMT (BAK00281), and Panicum hallii PhASMT (XP_025822442.1)] were aligned using Clustal X 1.83, and a neighbor-joining phylogenetic tree was constructed based on the amino acid sequence using MEGA 7 software (Park et al., 2013; Zuo et al., 2014; Byeon et al., 2015).
To construct the overexpression vector, the MsASMT1 complementary DNA fragment was amplified using unique primers containing restriction sites for BamHI/KpnI and cloned into the destination vector pZh01. The pZh01-MsASMT1 vector was introduced into Agrobacterium strain EHA105.
Subcellular Localization of MsASMT1
For subcellular location, MsASMT1 was fused to the C terminus of GFP in pBWA(V)HS-GLosgfp by EcoRV. One-month-old Nicotiana benthamiana leaves were used for infiltration with the GV3101 strain carrying pBWA(V)HS-MsASMT1-GLosgfp. The subcellular localization of MsASMT1 was determined using a Nikon C2-ER laser confocal microscope after two days.
Circadian Rhythm Analysis of MsASMT1 Gene and Melatonin Content
For circadian rhythm analysis of MsASMT1 gene, total RNA was extracted from the leaves of wild type (WT) plants at different time points (3:00, 6:00, 9:00, 12:00, 15:00, 18:00, 21:00, and 24:00) during one day. cDNAs were generated by reverse transcription enzyme, and the relative expression level of MsASMT1 gene were analyzed by quantitative real-time PCR (qRT-PCR) with the specific primer (MsASMT1-F, MsASMT1-R).
For circadian rhythm analysis of melatonin, the melatonin contents of WT plant leaves at the same time points with MsASMT1 gene expression analysis were extracted and quantified using a plant melatonin enzyme-linked immunosorbent assay (ELISA) kit (Suzhou Comin Biotechnology Co., Ltd., Suzhou, China) according to the manufacturer’s protocol.
Agrobacterium-Mediated Transformation
Agrobacterium strain EHA105 harboring pZh01-MsASMT1 was transformed into alfalfa (Medicago sativa L.) cultivar ‘Zhongmu No. 1.’ The T-DNA region of pZh01-MsASMT1 is illustrated in Supplementary Figure S1A. A modified Agrobacterium-mediated transformation protocol was used in this paper (Zhang and Wang, 2015). Cotyledons and hypocotyls from the seven-day-old seedlings were used as explants for transformation. After Agrobacterium inoculation, explants were subjected to cocultivation for 3 days and then transferred to callus induction medium for one month with 5 mg/L hygromycin B (hyg). The induced calluses were transferred to differential medium for somatic embryo induction, and the medium was changed every 2 weeks until torpedo-shaped embryos emerged. Then, the green buds were transferred to MS medium for further germination and plantlet formation. The resistant plantlets were transplanted to flowerpots filled with a mixture of vermiculite and humus, and the plants were maintained in a greenhouse under a 16 h/8 h (light/dark) photoperiod with 200 μmol m–2s–2 light intensity at 25 ± 2°C.
Transgenic Plants Verification
Total genomic DNA from every transgenic plant was extracted using the CTAB method. PCR was performed to confirm the positive transformants using specific primers of the selectable marker gene hpt for MsASMT1 gene were derived from alfalfa plants and which could be detected in WT plants.
Total RNA was extracted from leaves of different OE-MsASMT1 transformants. cDNAs were generated by reverse transcription enzyme. Positive transgenic plants were selected randomly for RT-PCR and qRT-PCR tests, which were performed with a pair of primers (MsASMT1-F, MsASMT1-R) specific to MsASMT1 gene. An actin gene of alfalfa was used as the internal control for RNA normalization (Castonguay et al., 2015).
The relative expression levels of MsTDC, MsT5H, MsSNAT, and MsCOMT in positive transgenic plants were analyzed by quantitative real-time PCR with the specific primers listed in Supplementary Table S1.
Quantification of the Melatonin Content
Melatonin was extracted with a modified method according to Zhang et al. (2014). Approximately 2 g of fresh leaves and stems from the top 10 cm portion were ground into powder with liquid nitrogen and then homogenized with 5 mL methanol at −20°C for 30 min. Homogenate was ultrasonicated at 45°C for 40 min. After centrifugation at 12000 rpm at 4°C for 15 min, the supernatants were collected and dried using nitrogen gas. The extracts were dissolved in 5% methanol and purified using a C18 solid phase extraction (SPE) column (Kromasil, 250 mm × 4.6 mm, 5 μm). The cartridge was then washed with 1 mL 5% methanol, and melatonin was finally eluted with 1 mL 80% methanol. The extract was subsequently filtered through a 0.22 μm PTFE syringe filter before UHPLC-ESI-MS/MS analysis. Melatonin determination and quantification was analyzed using a UHPLC-ESI-MS/MS (Rigol L3000, China).
Phenotypic and Histochemical Analyses
The WT plants and OE-MsASMT1 transgenic plants were cultivated in a greenhouse for two months. Five OE-MsASMT1 transgenic plants (OE3, OE12, OE15, OE16, and OE25) were selected for the comparison of leaf size, stem diameter and plant height with several WT plants. The second fully expanded mature leaves from the top were selected for comparing leaf size. The stem diameter was measured 3 cm above the ground with four mature tillers of the same transgenic plants with a Vernier caliper.
The excised stems of the third internode from the top of three-month-old OE-MsASMT1 transgenic alfalfa and WT plants were used for paraffin section analysis. Tissues were fixed in FAA (formalin-aceto-alcohol) solution for 24 h, dehydrated using 85% ethanol, permeated and embedded in wax. Cross sections were sliced for saffron and green staining to observe the cell morphology and vascular bundles.
Quantification of the Lignin and Flavonoid Contents
Stems of the OE-MsASMT1 transgenic plants (OE16, OE25) and WT plants were collected and dried at 65°C for 48 h for the lignin content measurement. The quantification of lignin content was performed according to Li et al. (2011) using a modified Klason method. Three biological replicates were performed.
Fresh samples from the top 10 cm portions of WT plants and OE-MsASMT1 transgenic plants were harvested for total flavonoids content measurement. The extraction and quantification of total flavonoids were performed using a plant flavonoid kit (Suzhou Comin Biotechnology Co., Ltd) following the manufacturer’s instructions with a spectrophotometer (Hitachi UH5300, Japan). Three biological replicates were performed.
Quantification of IAA Content
Approximately 50 mg fresh samples from the top 10 cm portions of three-month-old WT plants and OE-MsASMT1 transgenic plants (OE16, OE25) were ground into powder with liquid nitrogen. The samples were extracted in 0.5 mL extracting solution and 50 μL internal standard fluid at 4°C for 30 min. Then, the homogenate was extracted with 1 mL trichloromethane. After centrifugation at 14000 rpm at 4°C for 10 min, the subnatant was collected and dried using nitrogen gas. Extracts were dissolved in 100 μL methanol, filtered through a 0.1 μm membrane and transferred to sample vials for LC-MS analysis (UPLC I-Class; Q-Exactive MS). Three biological replicates were performed.
RNA-Sequencing
Fresh samples of the top 10 cm portions (leaves and stems) of three-month-old OE-MsASMT1 transgenic plants (OE3, OE12, OE15, OE16, and OE25) and WT plants with five biological replications were used for RNA-seq analysis without a reference genome. The purity, concentration and integrity of the RNA samples were tested to ensure the use of qualified samples for RNA sequencing. A total of 1 μg RNA per sample was used as input material for the RNA sample preparations. The generation of a sequence library and gene functional annotation were performed in Biomarker Technology Co., Ltd. (Beijing, China). The raw data were submitted to the NCBI Sequence Read Archive (SRA) database3, and the BioProject accession number is PRJNA555673.
The RNA-seq results were validated by quantitative real-time PCR, the relative expression levels of 14 genes selected from the RNA-seq results were tested in WT plants and OE-MsASMT1 transgenic plants (OE16 and OE25 with lower and moderate expression level of MsASMT1 gene) with the specific primers listed in Supplementary Table S1.
Widely Targeted Metabolomic Analysis
The plant materials used for metabolomic analysis were the same as those used for RNA-seq analysis (OE3, OE12, OE15, OE16, OE25, and WT plants). Approximately 1.5 g of each fresh sample from OE-MsASMT1 transgenic plants and WT plants were collected and frozen immediately in liquid nitrogen and stored at −80 °C. Then, the samples were delivered to Biomarker Technology Co., Ltd., for widely targeted metabolomic analysis.
Determination of Forage Quality
Shoots of OE-MsASMT1 transgenic plants and WT plants at the squaring stage were harvested and dried at 65°C for 48 h. According to Reddy et al. (2005), dried samples were ground with a mill and passed through a 1 mm sieve. Approximately 50 mg dry powder samples were used for neutral detergent fiber (NDF), acid detergent fiber (ADF) and acid detergent lignin (ADL) analyses by a fiber analyzer (ANKOM 2000). Three biological replicates were performed.
Statistical Analysis
All experiments were performed with at least three biological replicates. The values presented are the means ± SE of three replications. Statistical analyses were conducted using one-way analysis of variance (ANOVA) with SPSS 18.0.
Results
Isolation of MsASMT1 and Genetic Analysis
A full-length coding sequence (CDS) of 1077 bp was isolated from the alfalfa genome and shared up to 88.61% identity with MtASMT (Medtr5g074600.1) (Supplementary Figure S2A). This gene was named MsASMT1, and encodes a protein of 358 amino acids, contains a dimerization domain and is a S-adenosylmethionine-dependent methyltransferase (SAM or AdoMet-MTase superfamily), which is similar to ASMT genes reported in other plant species (Supplementary Figure S2B). The neighbor-joining phylogenetic tree clearly showed that the ASMTs clustered into two groups for monocots and dicots, which demonstrated that the ASMT gene was evolutionarily conserved (Supplementary Figure S3).
To further understand the characteristics of MsASMT1, we performed subcellular localization and determined the expression patterns of MsASMT1 in different tissues by quantitative real-time PCR. The results showed that MsASMT1 was mainly located in cytoplasm, and also existed in nucleus (Figure 1A). As shown in Figure 1B, MsASMT1 was mainly expressed in the stems and roots, followed by young and mature leaves, while the MsASMT1 gene was rarely expressed in flowers, indicating that MsASMT1 might especially function in stem and root growth. Because melatonin plays an important role in plant circadian rhythm, we investigated the melatonin levels and MsASMT1 expression levels in alfalfa throughout one day. As shown in Figure 1C, the peak of MsASMT1 expression occurred at 15:00 pm and 21:00 pm, while the melatonin content peaked at 9:00 am and 15:00 pm and remained at a stable level at night from 21:00 pm to 3:00 am.
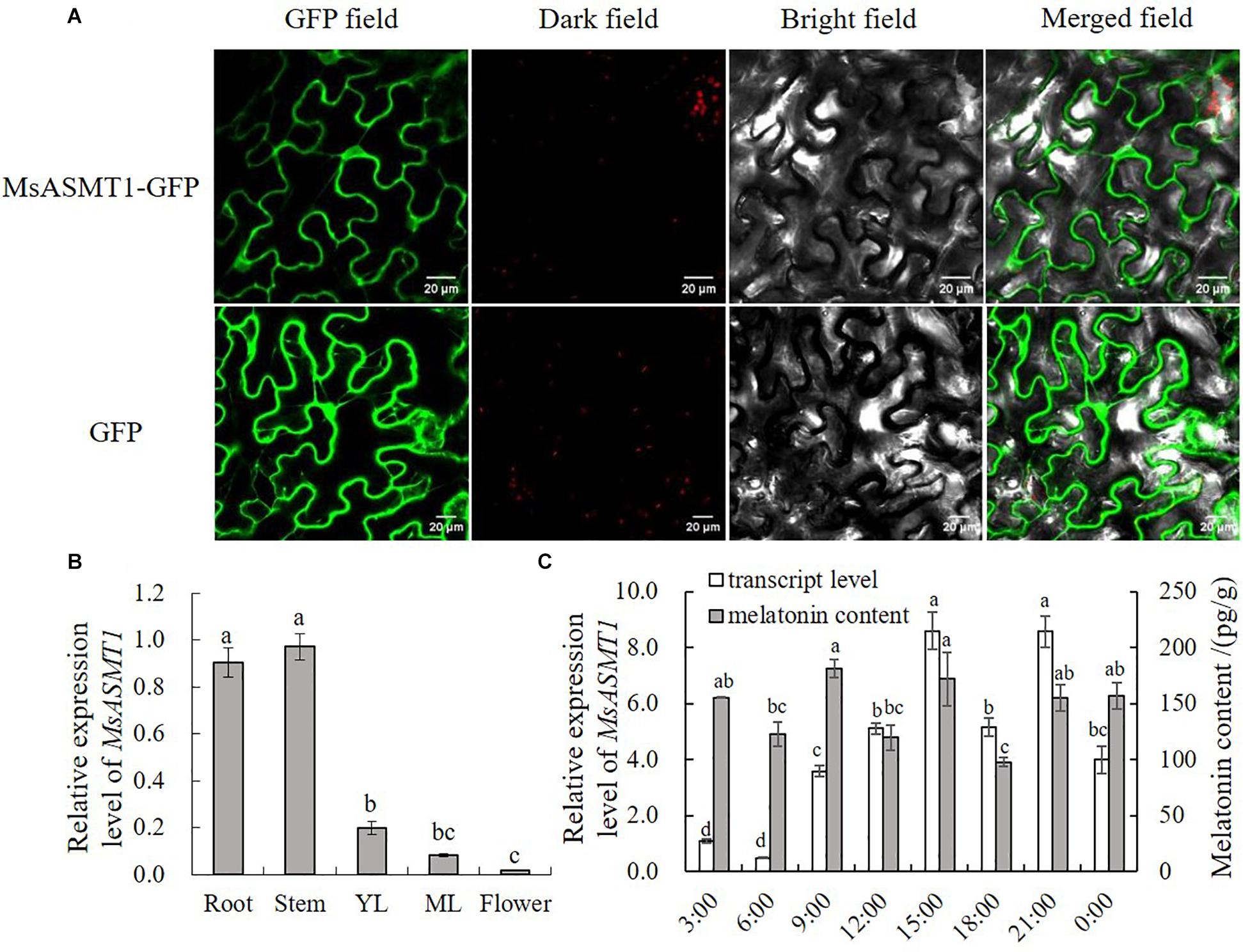
Figure 1. Characterization of MsASMT1. (A) Subcellular location of MsASMT1 in Nicotiana tabacum. From left to right: green fluorescence, dark field, bright field, merged field of MsASMT1-GFP and GFP; (B) Relative expression level of MsASMT1 in different tissues of alfalfa. YL, young leaves, ML, mature leaves. The letters indicate the differences among different tissues of alfalfa according to ANOVA analysis (p < 0.05); (C) Circadian rhythm of MsASMT1 transcript levels and melatonin content in alfalfa throughout one day. Data are presented as means ± SE (n = 3), the letters indicate the differences of MsASMT1 transcript levels and melatonin content respectively among different time points according to ANOVA analysis (p < 0.05).
Overexpression of MsASMT1 Increases Melatonin Biosynthesis in Transgenic Plants
To characterize the function of MsASMT1 in alfalfa, we generated a large number of MsASMT1-overexpressing (OE-MsASMT1) transgenic plants using tissue culture (Supplementary Figures S1B–E). And most of them were PCR-positive, the expected size of the selectable marker gene hpt (741 bp) was detectable in OE-MsASMT1 transgenic plants (Supplementary Figure S4A). To further detect the expression of MsASMT1 in transgenic plants, we performed RT-PCR and qRT-PCR, respectively. The results showed that MsASMT1 gene had expressed in most of the tested transgenic plants (Supplementary Figures S4B,C). And the relative expression levels of MsASMT1 in transgenic plants varied from 5 to 20 times higher than that in WT plants (Figure 2A). Transgenic plants OE3, OE12 with high expression level, OE25 with middle expression level and OE15, OE16 with low expression level were selected for further research.
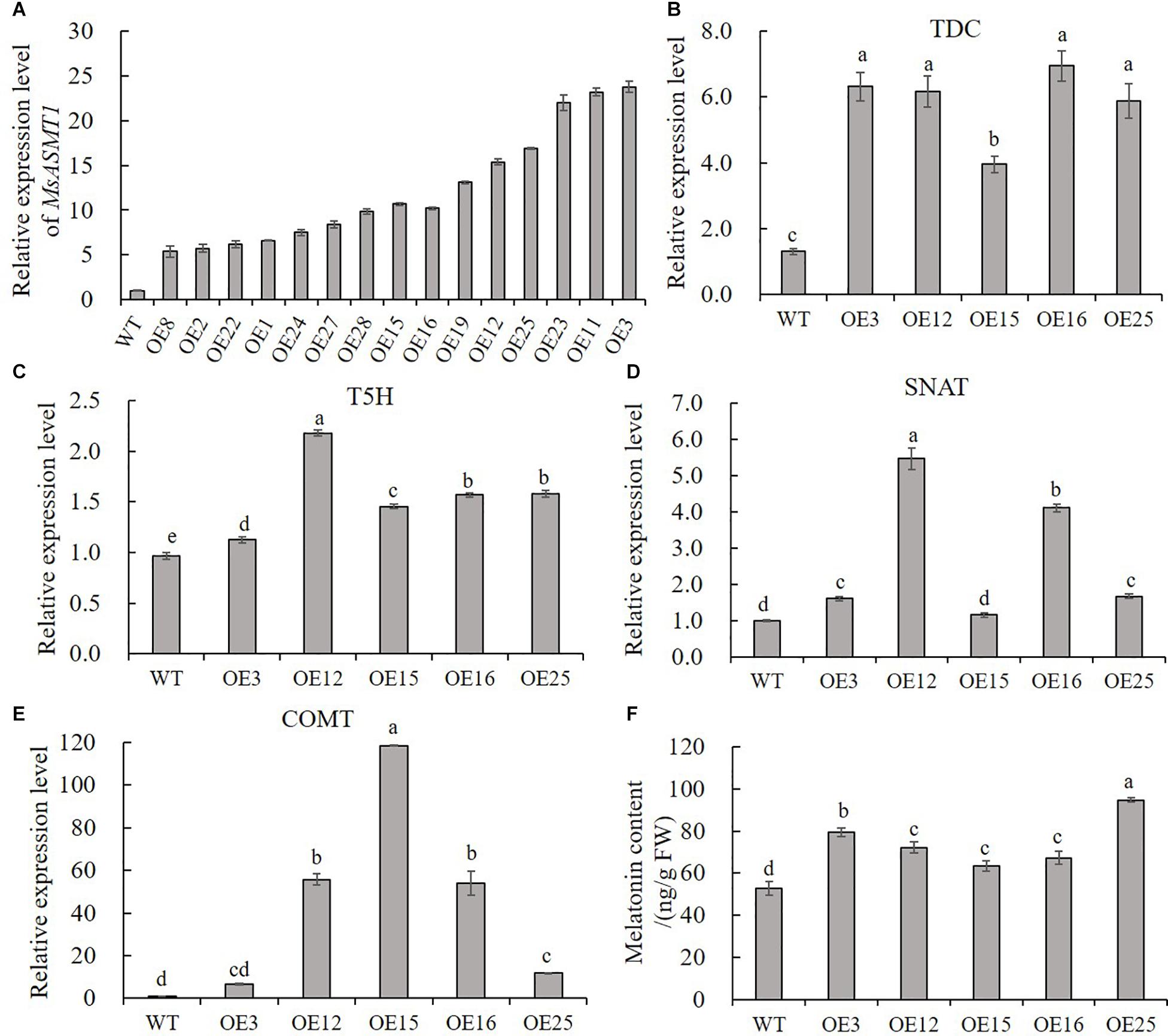
Figure 2. Relative expression level of MsASMT1 and the upstream genes of ASMT in melatonin biosynthesis pathway and the melatonin content in WT plants and OE-MsASMT1 transgenic plants. (A) Relative expression level of MsASMT1 in various transgenic alfalfa plants; (B–E) MsTDC (B) MsT5H (C) MsSNAT (D) and MsCOMT (E) in WT plants and OE-MsASMT1 transgenic plants; (F) Melatonin content in WT plants and OE-MsASMT1 transgenic plants, the correlation between melatonin content and relative expression level of MsASMT1 is analyzed by SPSS with Pearson Correlation Coefficient (p < 0.05). Data are presented as mean ± SE (n = 3). The letters indicate the differences between WT and OE-MsASMT1 transgenic plants according to ANOVA analysis (p < 0.05).
Because ASMT catalyzes the last critical enzymatic step in melatonin biosynthesis, we wondered whether feedback in this pathway occurred once the expression level of MsASMT1 changed in alfalfa. We analyzed the transcripts of MsTDC, MsT5H, MsSNAT, and MsCOMT in these transgenic plants. Quantitative real-time PCR analysis showed that the expression levels of all these genes in OE-MsASMT1 transgenic plants were increased compared to those in WT plants (Figures 2B–E). These results indicated that the overexpression of MsASMT1 directly or indirectly upregulated the expression of upstream genes.
Since ASMT is responsible for melatonin biosynthesis and with the upregulation of other genes in melatonin biosynthesis pathway, we directly measured the melatonin levels in the WT plants and OE-MsASMT1 transgenic plants. The results revealed that the increases of melatonin content in OE-MsASMT1 transgenic plants varied from 21.2% to 80.8% compared to that in WT plants (Figure 2F), and there was a positive correlation between melatonin content and the relative expression level of MsASMT1, the Pearson Correlation Coefficient is 0.782 without significant correlation.
Overexpression of MsASMT1 Increases Leaf and Stem Sizes and Increases Plant Growth in Transgenic Plants
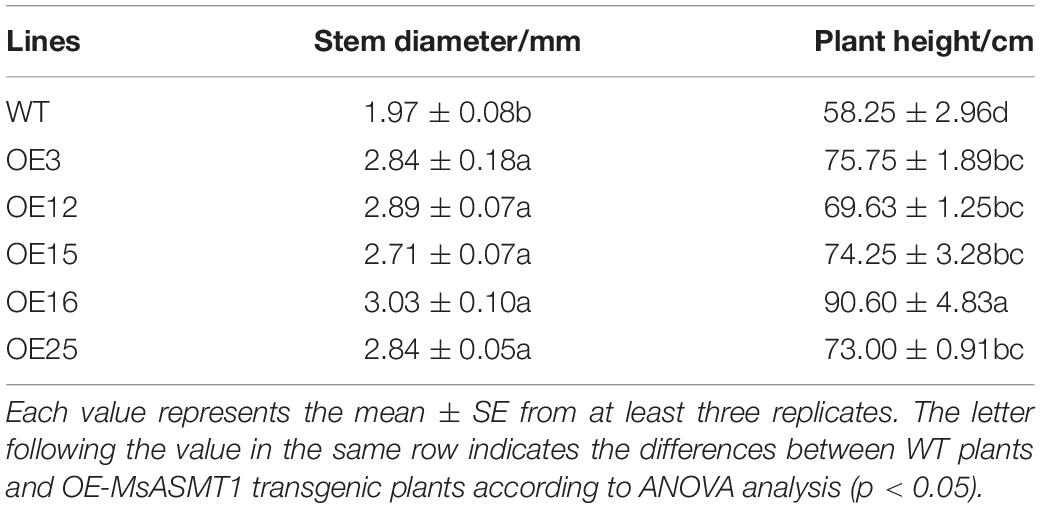
Compared to WT plants, OE-MsASMT1 transgenic plants exhibited vigorous growth with larger leaves and thicker stems (Figures 3A–C). The plant height of OE-MsASMT1 transgenic plants ranged from 69.63 cm to 90.60 cm, which was significantly higher than that of WT plants (58.25 cm). The stem diameter of OE-MsASMT1 transgenic plants ranged from 2.71 mm to 3.03 mm, which was significantly thicker than that of WT plants (1.97 mm; Table 1).
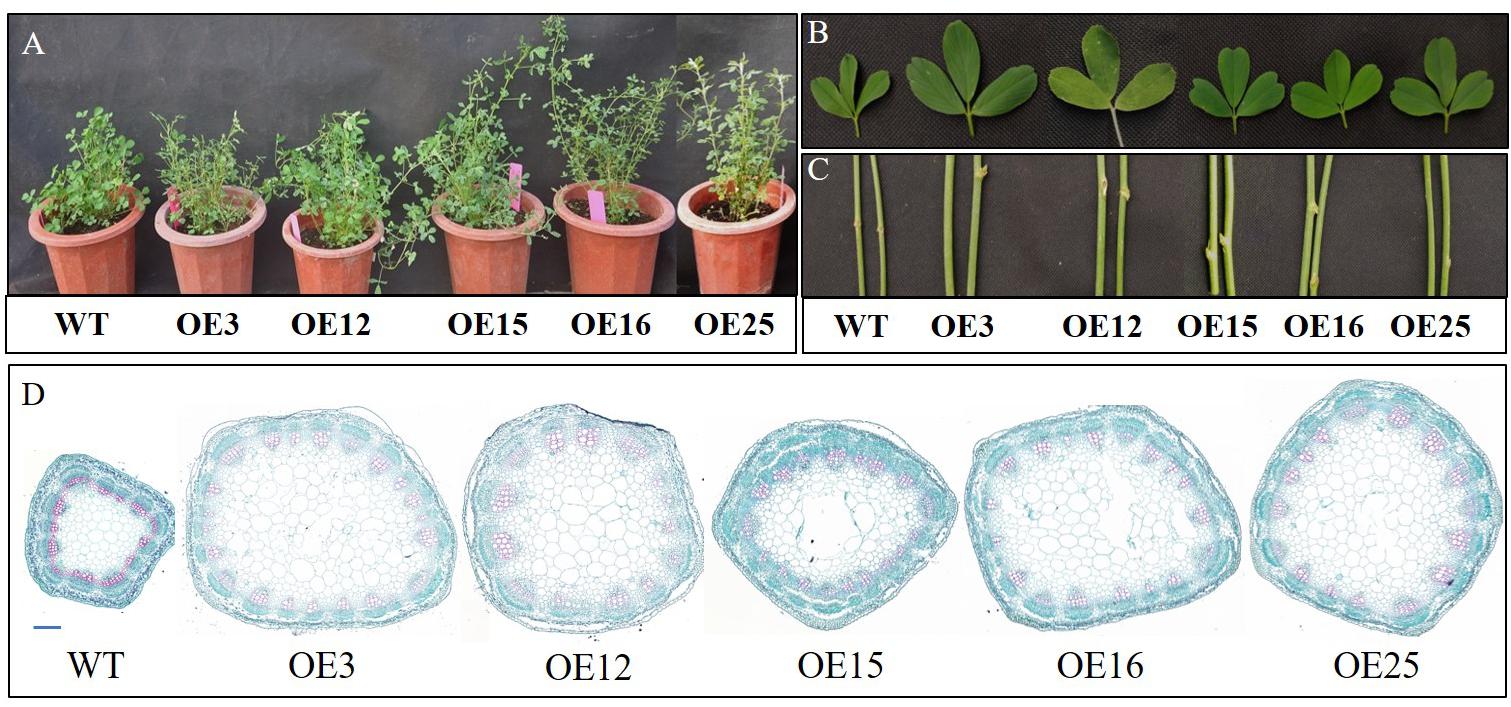
Figure 3. Effects of MsASMT1 overexpression on alfalfa growth and development. (A) Growth situation of WT plants and OE-MsASMT1 transgenic plants after growing in greenhouse for two months; (B,C) the leaf (B) and stem (C) comparison between WT plants and several OE-MsASMT1 transgenic plants, scale bar, 1 cm; (D) Stem cross sections with safranin green staining of WT plants and OE-MsASMT1 transgenic plants, scale bar, 1 mm.
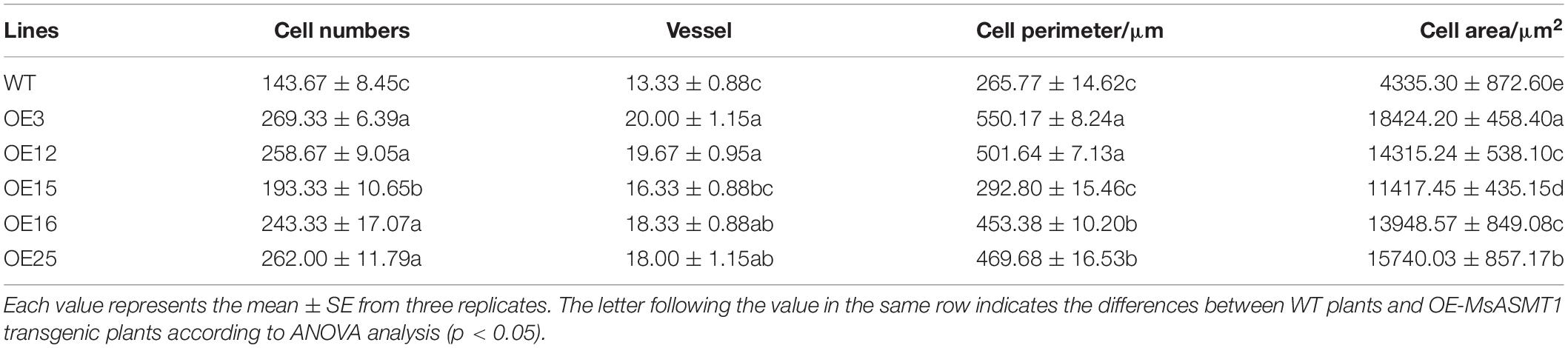
In agreement with the morphological changes, histological analysis showed that the cell size, cell number and vascular bundle in OE-MsASMT1 transgenic plant stems significantly increased compared to those in WT plants, the cells were disordered with irregular shapes and the intercellular spaces became more sparsely arranged (Figure 3D, Table 2).
In contrast to WT plants, OE-MsASMT1 transgenic plants exhibited bushy branches at the preliminary vegetative stage (Figure 4A). The number of branches per unit length of OE3 exhibited a 2-fold increase, and the internode length of OE3 was 75% shorter than that of WT plants (Figures 4B,C). The leaf/stem ratios of OE3, OE12, and OE25 were also increased compared with that of WT plants (Figure 4D). Identified genes that are known to be involved in branch formation, such as IAA (indoleacetic acid) and CCD8 (carotenoid cleavage dioxygenase 8), were significantly increased in OE16 and OE25, while RAX1 (regulator of axillary meristems 1) and RAX3 exhibited a contrasting trend (Figure 4E) (Foo et al., 2005; Rubio-Moraga et al., 2014; Guo et al., 2015); the IAA content was also significantly increased in the top of OE-MsASMT1 transgenic plants stems (Figure 4F).
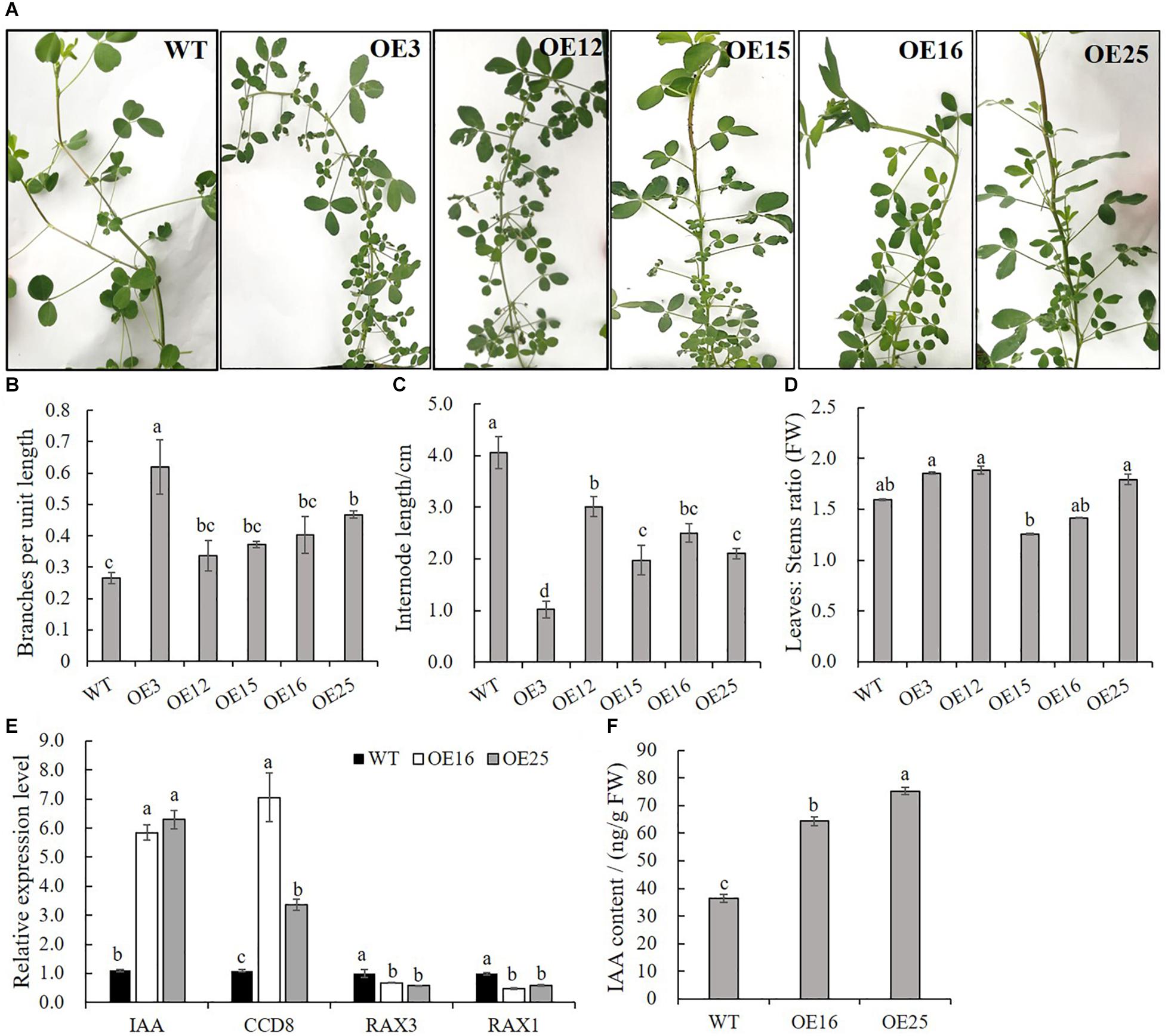
Figure 4. Branching characteristics of WT plants and OE-MsASMT1 transgenic plants. (A) Branching phenotype of three-month-old WT plants and several OE-MsASMT1 transgenic plants; (B–D) The number of lateral branches per unit length on the main branch (B), the internode length between two lateral branches (C) and the ratio of fresh leaves to fresh stems (D) of WT plants and OE-MsASMT1 transgenic plants after grown in greenhouse for three months; (E) Relative expression level of genes related to plant branching in WT plants and OE-MsASMT1 transgenic plants; (F) IAA content of WT plants and OE-MsASMT1 transgenic plants. Data are presented as mean ± SE (n = 3). The letters indicate the differences between WT and OE-MsASMT1 transgenic plants according to ANOVA analysis (p < 0.05).
Overexpression of MsASMT1 Reduces Flavonoids Biosynthesis and Changes Lignin Composition in Transgenic Plants
Due to our previous study about HIOMT gene overexpression in switchgrass regulated flavonoid biosynthesis (Yuan et al., 2016), and to further detect the effects of MsASMT1 overexpression on flavonoids biosynthesis, we measured the total flavonoids contents in WT plants and OE-MsASMT1 transgenic plants directly. The results showed that the flavonoid content in all OE-MsASMT1 transgenic plants was significantly decreased compared to that in WT plants, especially in OE3 and OE25 (Figure 5A). For the stems of OE-MsASMT1 transgenic alfalfa were thickened, and to explore the effects of melatonin on lignin biosynthesis, we measured the lignin content and lignin composition in WT plants and OE16, OE25 transgenic plants. The lignin content and lignin monomers guaiacyl (G) and syringyl (S) contents showed no significant differences between WT plants and OE-MsASMT1 transgenic plants (Figures 5B,C), while the S/G ratio was significantly reduced in OE-MsASMT1 transgenic alfalfa compared to that in WT plants (Figure 5D). The results demonstrated that the increased melatonin content affected flavonoids biosynthesis and lignin monomer composition in phenylalanine metabolism pathway.
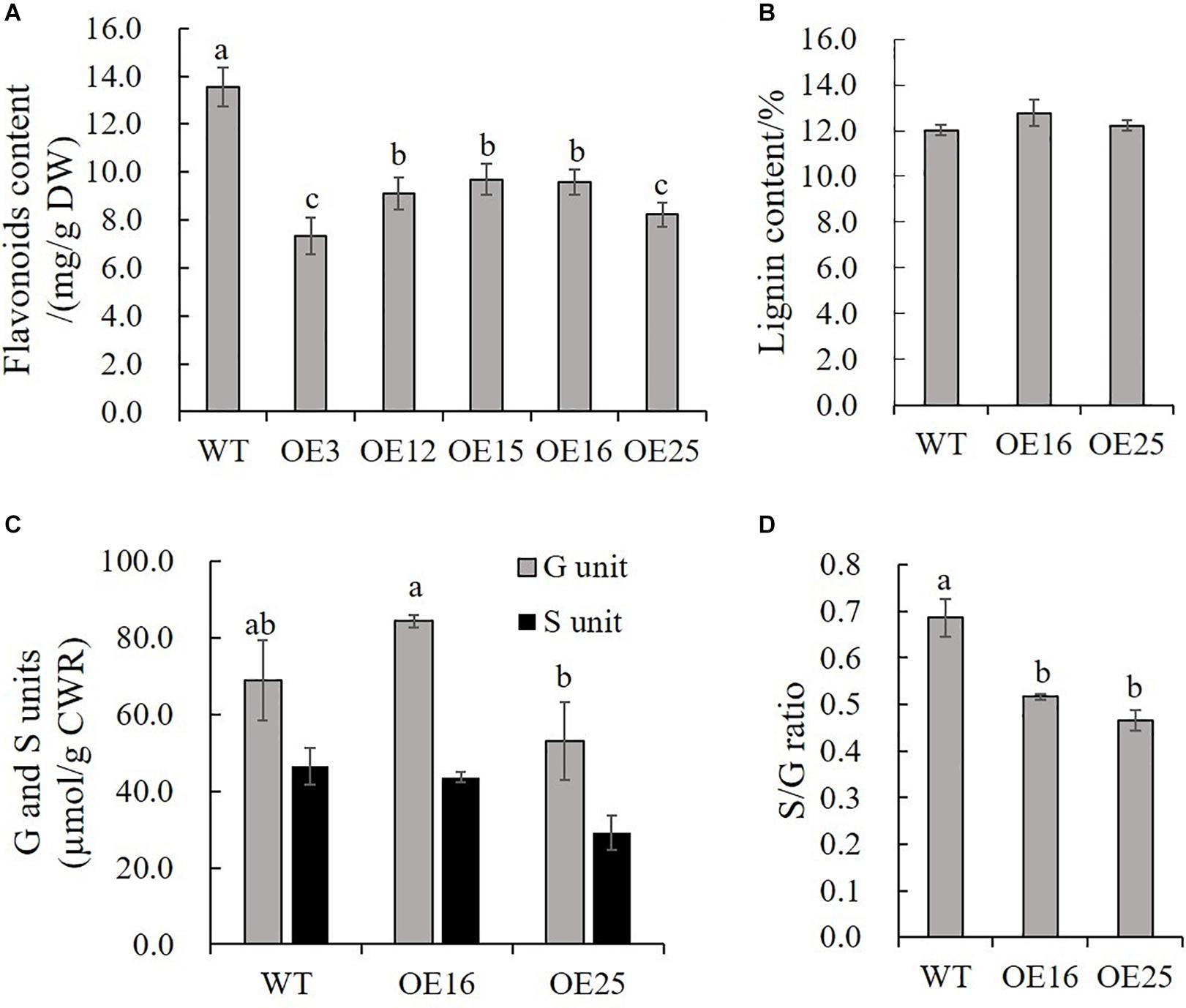
Figure 5. Flavonoids content and lignin content of WT plants and OE-MsASMT1 transgenic plants. (A,B) Total flavonoids content (A) and lignin content (B) in WT plants and OE-MsASMT1 transgenic plants; (C) The lignin monomer G and S unit content in WT plants and OE-MsASMT1 transgenic plants; (D) The S/G ratio in WT plants and OE-MsASMT1 transgenic plants. Data are presented as mean ± SE (n = 3). The letters indicate the differences between WT and OE-MsASMT1 transgenic plants according to ANOVA analysis (p < 0.05).
Transcript Profiling Revealed Genes in Phenylalanine Pathway Associated With Flavonoid and Lignin Metabolism Are Changed in OE-MsASMT1 Transgenic Plants
To gain insight into the underlying molecular mechanism of MsASMT1 overexpression in plant growth and metabolism, we conducted transcript profiling analysis using RNA-seq with WT plants and OE-MsASMT1 transgenic plants and five biological replicates were sequenced for each group. A total of 7712 differentially expressed genes (DEGs) were obtained, of which 3587 genes were upregulated, while 4125 genes were downregulated (Figure 6A). GO enrichment analysis indicated that a wide range of DEGs clustered into metabolic process (1329), cellular process (1374), catalytic activity (1306), single-organism process (822), biological regulation (472), response to stimulus (476), signaling (209), transporter activity (130), etc. (Supplementary Figure S5). In the large group of DEGs in metabolic process, a large number of genes in phenylalanine metabolism pathway were altered and 28 of these genes participated in flavonoids biosynthesis and 13 genes participated in lignin biosynthesis and degradation. Some of these genes were identified as UDP-glucosyltransferase family genes, which are involved in flavonoid glycosylation. The heatmap of DEGs related to flavonoid biosynthesis is shown in Figure 6B. We also observed a large amount of genes response to plant hormones, including ethylene (ET), gibberellin (GA), auxin (IAA), abscisic acid (ABA), jasmonic acid (JA) and salicylic acid (SA) biosynthesis and signaling response genes, were altered in OE-MsASMT1 transgenic alfalfa, which provided evidence of melatonin playing function in plant growth and metabolism by crosstalk with phytohormones. The significant DEGs (|log2FC| > 1.5) related to flavonoid and lignin biosynthesis, hormone signaling and defense responses are listed in Supplementary Table S2.
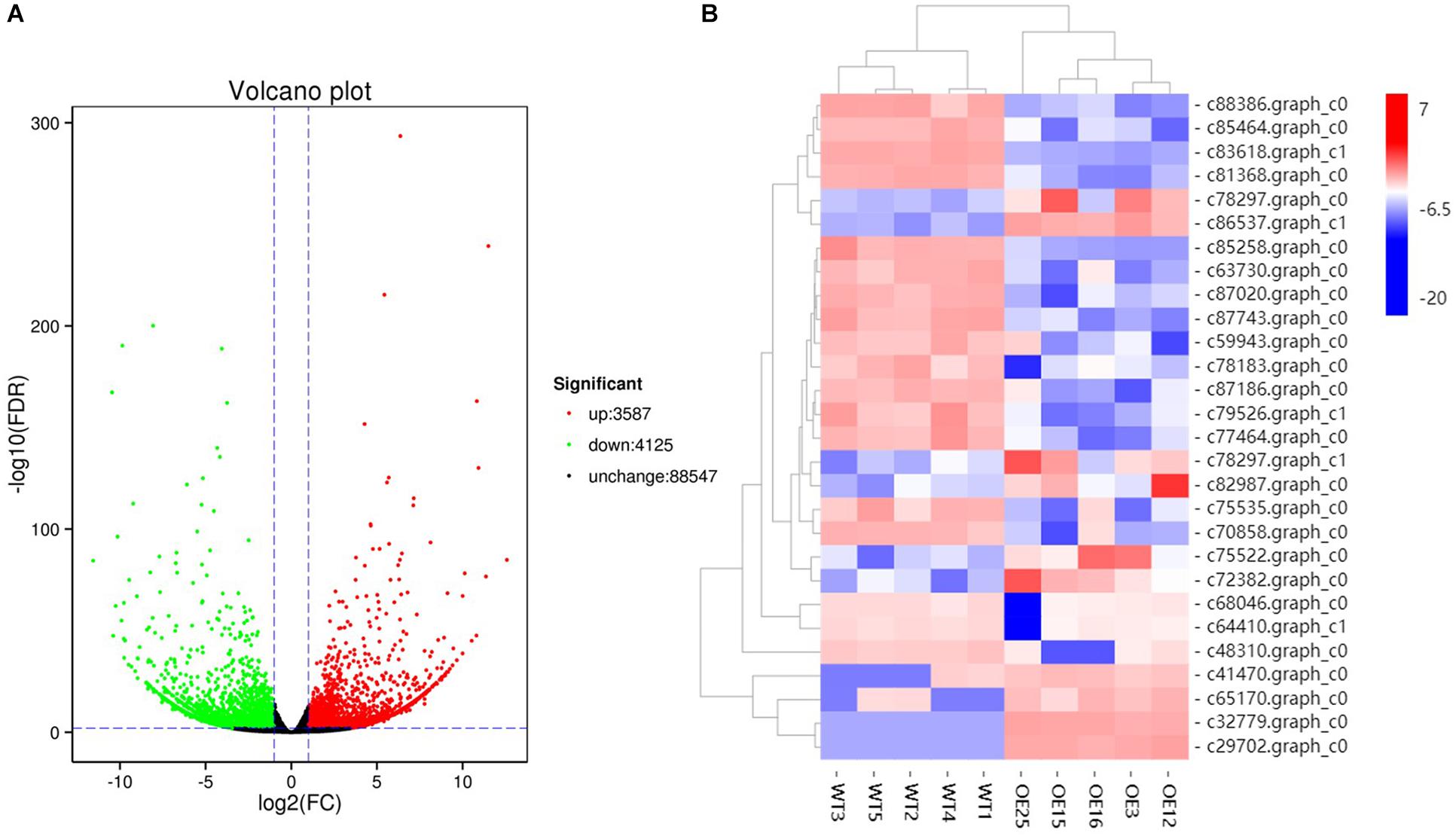
Figure 6. The volcano plot of total differential expression genes (DEGs) (A) and heatmap of DEGs associated with flavonoids biosynthesis (B) between WT plants and OE-MsASMT1 transgenic plants.
To confirm that the overexpression of MsASMT1 affected the expression of flavonoid biosynthesis genes, we selected 14 DEGs for verification in transgenic plants OE16 and OE25 by qRT-PCR. The results showed that the expression levels of UDP-glucosyltransferase UGT74B1 (c87186.graph_c0), UGT85A24 (c83618.graph_c1), and UGT87A1 (c78183.graph_c0) were significantly decreased in OE16 and OE25, which were involved in the flavonoid glucuronidation process with quercetin 3-O-glucosyltransferase activity and quercetin 7-O-glucosyltransferase activity. Transcripts of F3OGT (UDP-glucose flavonoid 3-O-glucosyltransferase; c41470.graph_c0) and I7OMT (isoflavone 7-O-methyltransferase; c32779.graph_c0) were obviously upregulated in OE16 and OE25 plants (Figure 7A). Similarly, the transcript levels of several genes involved in lignin biosynthesis were found to be significantly changed in OE-MsASMT1 transgenic plants. The relative expression levels of 4CL (4-coumarate: CoA ligase; c67220.graph_c0) and CCoAOMT (caffeyl CoA O-methyltransferase; c89170.graph_c0) were downregulated, and the relative expression levels of CAD (coniferyl-aldehyde dehydrogenase; c57796.graph_c0), HCT (hydroxycinnamoyl transferase; c29702.graph_c0), COMT (caffeic acid O-methyltransferase; c71088.graph_c0), and F5H (ferulate 5-hydroxylase; c69879.graoh_c0) were significantly upregulated in OE-MsASMT1 transgenic plants (Figures 7B,C). In addition, the relative expression levels of lignin peroxidase (LIP) and laccase (LAC11, LAC12) genes, which also involved in lignin biosynthesis, were also significantly upregulated in OE-MsASMT1 transgenic plants compared with WT plants (Figure 7D).
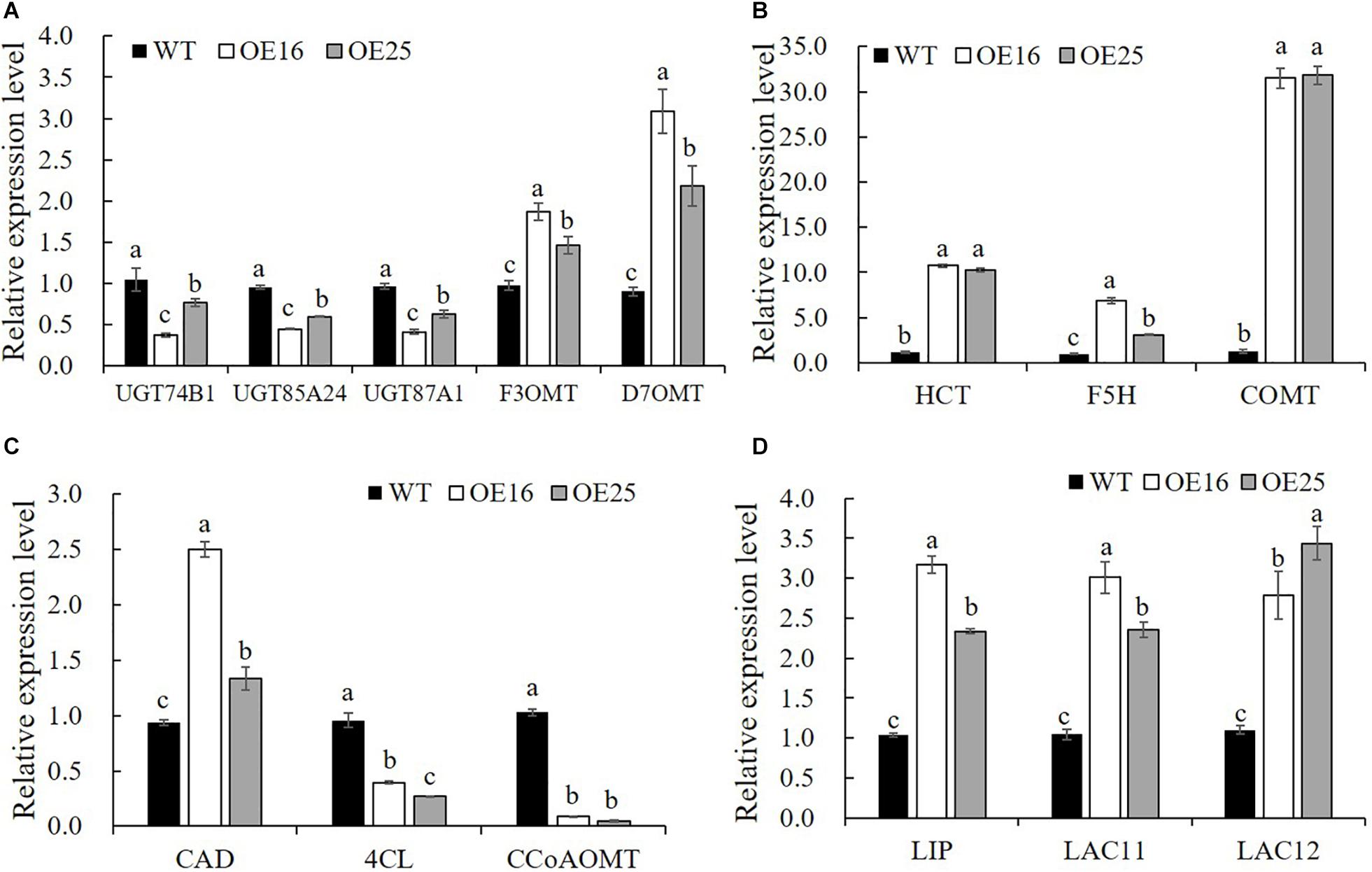
Figure 7. Relative expression levels of genes related to flavonoids biosynthesis (A) and lignin biosynthesis (B–D) in WT plants and OE-MsASMT1 transgenic plants. Data are presented as mean ± SE (n = 3). The letters indicate the differences between WT and OE-MsASMT1 transgenic plants according to ANOVA analysis (p < 0.05).
Metabolic Profiling Revealed Flavonoid Accumulation Changes in OE-MsASMT1 Transgenic Plants
To further explore changes in compounds that accompanied changes in gene regulation, a widely targeted metabolomics assay was performed with WT plants and OE-MsASMT1 transgenic plants. A total of 174 differential secondary metabolites were identified, in which 120 metabolites were decreased and 54 were increased in OE-MsASMT1 transgenic plants. The top 10 upregulated and downregulated metabolites are shown in Figure 8. There were 78 flavonoid metabolites among these 174 identified metabolites, including 37 flavones, 12 flavonols, 6 flavanones, 6 isoflavones, 8 anthocyanins, and 9 flavonoids. The heatmaps of the total differential metabolites (Figure 9A) and the flavonoid metabolites (Figure 9B) compared between WT plants and OE-MsASMT1 transgenic plants are shown in Figure 9. The concentrations of these significantly decreased and increased flavonoid metabolites (|log2FC| > 1.5) in OE-MsASMT1 transgenic plants are listed in Supplementary Tables S3, S4, respectively. Quercetin 7-O-malonylhexosyl-hexoside (-12.9-fold), quercetin O-acetylhexoside (-13.4-fold), quercetin 4’-O-glucoside (-4.01-fold) and quercetin 3-alpha-L-arabinofuranoside (-3.53-fold), all of which were quercetin glycoside derivative, were significantly decreased in OE-MsASMT1 transgenic plants. The concentration of kaempferol 3-O-glucoside, belonging to kaempferol glycoside derivates, in OE-MsASMT1 transgenic plants was 14.4-fold lower than that in WT plants. Furthermore, the concentrations of formononetin (-3.1-fold), formononetin 7-O-glucoside (-4.23-fold) and biochanin 7-O-glucoside (-14.3-fold) were also decreased in OE-MsASMT1 transgenic plants (Supplementary Table S3). However, the concentrations of chrysoeriol derivates including chrysoeriol O-hexosyl-O-hexoside (1.52-fold), chrysoeriol O-glucuronic acid-O-hexoside (1.51-fold), O-methylChrysoeriol 5-O-hexoside (2.94-fold) and O-methylChrysoeriol 7-O-hexoside (2.42-fold) were increased in OE-MsASMT1 transgenic plants (Supplementary Table S4), and the concentrations of naringenin derivates naringenin C-hexoside (1.88-fold) and naringenin 7-O-glucoside (2.81-fold), tricin (2.06-fold) and its derivates tricin 5-O-hexoside (13.7-fold), tricin 7-O-hexoside (1.98-fold), and tricin 5-O-feruloylhexoside (1.73-fold) were increased in OE-MsASMT1 transgenic plants (Supplementary Table S4). In addition, there were also some other metabolites belong to alkaloids, terpene, polyphenol and phenylpropanoids were changed in OE-MsASMT1 transgenic plants, which might be responsible to plants response to environmental stimulus. Limonin, a compound of highly oxidized triterpenoids and a source of bitterness, potentially functions in preventing diseases and decreased almost 16.4-fold in OE-MsASMT1 plants. The concentrations of xanthotoxol (a natural furanocoumarins with various biological activities) and methyl p-coumarate (an esterified derivative of p-coumaric acid) in OE-MsASMT1 transgenic plants were -19.4 and -18.8 folds, respectively, to WT plants (Supplementary Table S5).
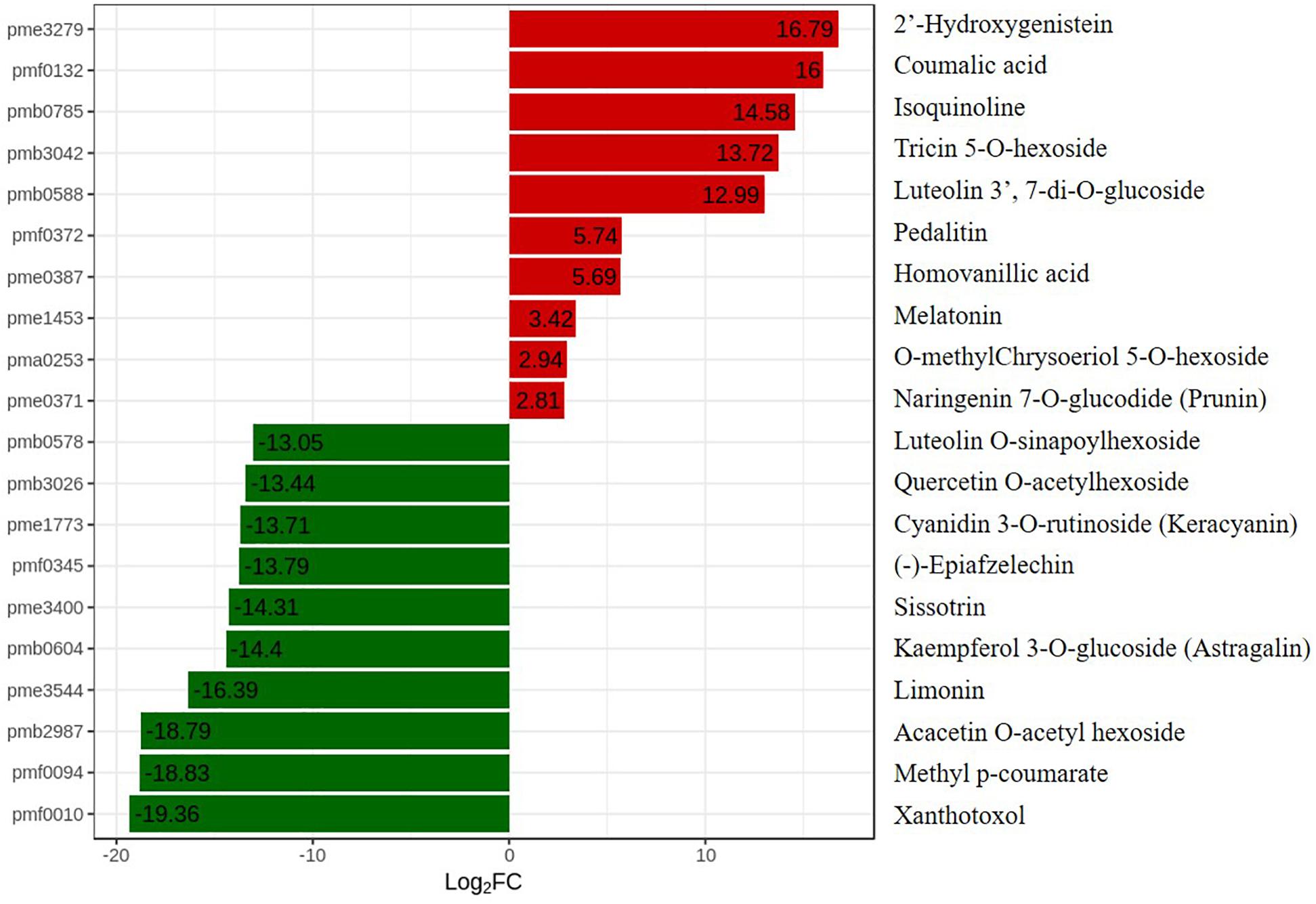
Figure 8. The top ten upregulated and downregulated metabolites between WT plants and OE-MsASMT1 transgenic plants. Red represent the upregulated metabolites in OE-MsASMT1 transgenic plants; green represent the downregulated metabolites in OE-MsASMT1 transgenic plants.
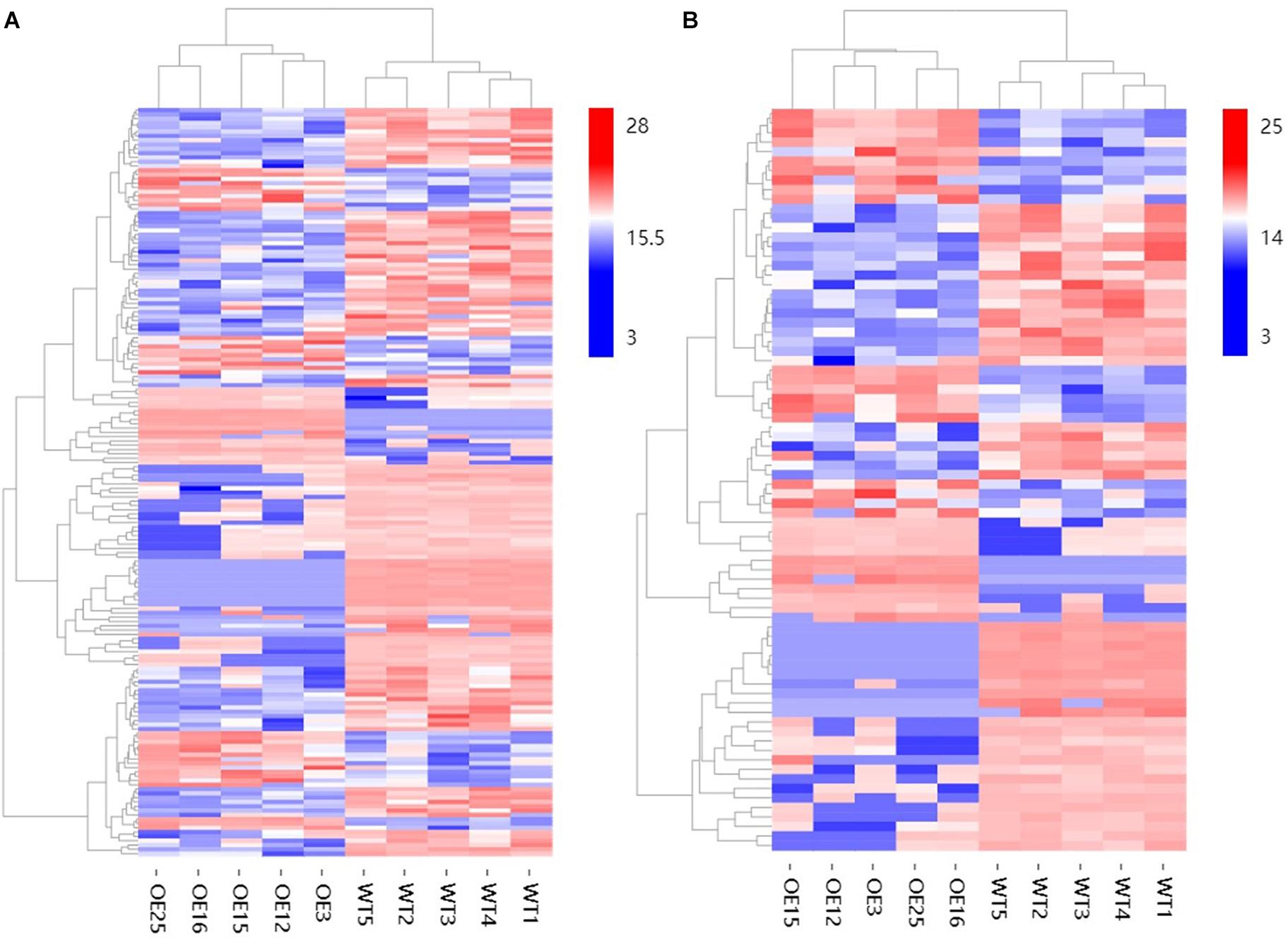
Figure 9. The heatmaps of total differential metabolites (A) and the differential metabolites belong to flavonoids (B) between WT plants and OE-MsASMT1 transgenic plants.
Overexpression of MsASMT1 Affects Forage Quality
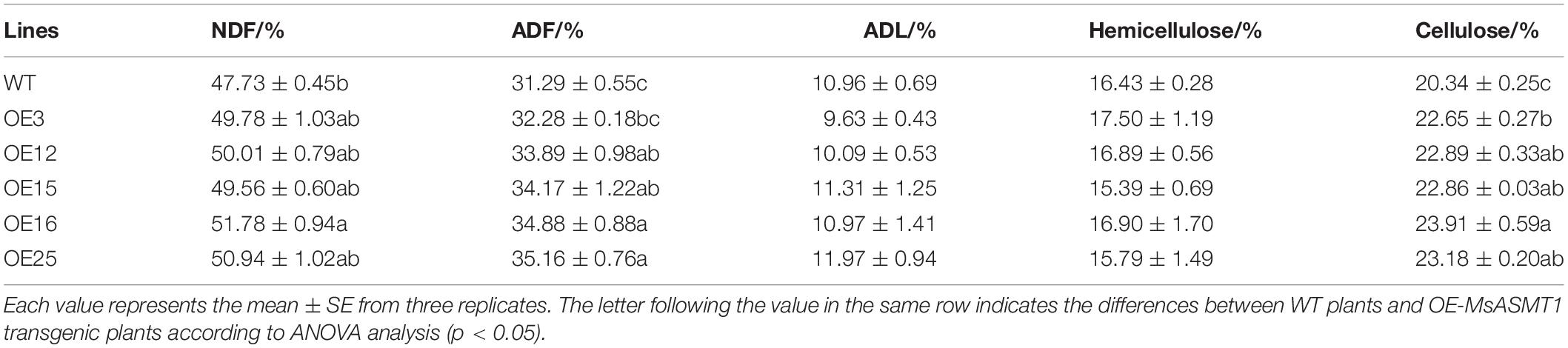
To evaluate the effects of MsASMT1 on forage quality, the shoots of WT plants and OE-MsASMT1 transgenic plants at the early flowering stage were harvested for forage quality evaluation. The overexpression of MsASMT1 led to a significant increase in acid detergent fiber content (ADF) and neutral detergent fiber content (NDF). The increases of NDF and ADF contents in OE-MsASMT1 transgenic plants varied from 1.8% to 4.0% and from 1.0% to 3.9%, respectively, compared to those in WT plants. The cellulose contents in OE-MsASMT1 transgenic plants were also significantly increased (Table 3). Other nutritive quality traits, including ADL and hemicellulose, did not show obvious changes between OE-MsASMT1 transgenic plants and WT plants.
Discussion
Melatonin, a bioactive molecule with multiple functions, is ubiquitously present in various plant species. For the reported melatonin biosynthesis pathway, six genes were implicated in the biosynthesis of melatonin in plants (Back et al., 2016). SNAT and ASMT are associated with the conversion of serotonin to melatonin, which are the rate-limiting steps for melatonin biosynthesis. In our previous study, we ectopically expressed AANAT and HIOMT from Ovis aries in switchgrass, which led to an increase in the melatonin content, promoted plant growth and development, and improved salt tolerance in transgenic plants (Huang et al., 2017). The overexpression of HIOMT regulated the phenylalanine metabolism pathway in transgenic switchgrass, especially flavonoid biosynthesis (Yuan et al., 2016), while the regulation effects and mechanisms have not been studied. The function of melatonin and the genes participating in melatonin biosynthesis were conserved, but there might be some fine distinction among different plant species. Alfalfa is an important forage grass with a rich nutritional value and powerful health-promoting properties. Taking advantage of the health-promoting properties of melatonin, the forage quality improvement of alfalfa has profound implications.
To obtain alfalfa with a high melatonin level, we successfully isolated an ASMT gene from the alfalfa genome according to the reported gene sequences in other plant species. The melatonin contents in OE-MsASMT1 transformants were significantly increased and were positively correlated with the transcript levels of MsASMT1. The expression of other genes related to melatonin biosynthesis, including MsTDC, MsT5H, MsSNAT, and MsCOMT, was upregulated in MsASMT1 overexpression lines, leading to increased melatonin contents in OE-MsASMT1 transgenic plants. In our study, the circadian rhythm analysis results showed that the peaks of melatonin levels and MsASMT1 expression levels in alfalfa throughout one day weren’t at the same time. MsASMT1 expression and the melatonin content all have two peaks during one day, while there were no regular relationships between MsASMT1 expression level and melatonin content, which might result from the endogenous melatonin content is regulated by all genes in the melatonin biosynthesis pathway, and the biosynthesis of metabolites is always later than the transcripts of genes.
The exogenous or endogenous regulation of melatonin promoted plant growth and development (Byeon and Back, 2014), which also occurred in alfalfa. OE-MsASMT1 transgenic alfalfa exhibited a consistent phenotype, they were more vigorous growth, possessed larger leaves, thicker stems and grew faster than WT plants, all of which was accompanied by bushy branches at the early vegetative stage. The vigorous stems of OE-MsASMT1 transgenic plants might arise from the highly specific expression pattern of MsASMT1 in alfalfa. Presumably related to the quantification of auxin, OE-MsASMT1 stems grew faster, and the internode length also increased rapidly. Due to the consistent phenotype, we only chose OE16 and OE25 with low and moderate expression level of MsASMT1 gene respectively to measure the IAA content and the relative expression level of genes related to branch formation to reflect the difference between OE-MsASMT1 transgenic plants and WT plants. Compared to cells of stems in WT plants, cells of stems in OE-MsASMT1 transgenic alfalfa were irregularly arranged, and the intercellular spaces were larger, with several hollow areas existing in the cell population, which might result from a strong mechanical strength between cells of stems for rapid growth of thick stems.
Flavonoids are important secondary metabolites in plants, which have versatile physiological functions in plant growth and development. Previous studies reported that flavonoids inhibited the biosynthesis of melatonin (Lee et al., 2018). In our study, melatonin-rich transgenic alfalfa produced less flavonoids compared to WT plants, which suggested that melatonin in turn inhibited the biosynthesis of flavonoids, especially quercetin, kaempferol, formononetin, and biochanin. For flavonoids playing an important role in defending against pathogen/insect attacks, the reduced flavonoid content in OE-MsASMT1 transgenic alfalfa might lead transgenic plants sensitive to pathogen/insect attacks. However, many studies have shown that the exogenous application or endogenous improvement of melatonin enhances various abiotic or biotic stress tolerances (Shi et al., 2015; Antoniou et al., 2017; Mandal et al., 2018; Liu et al., 2019). To test whether the elevated melatonin or the depressed flavonoids had a greater effect on stress resistance of OE-MsASMT1 transgenic alfalfa, we will select several abiotic or biotic stresses to detect the adverse defense ability of OE-MsASMT1 transgenic alfalfa in the future. Furthermore, in our RNA-seq data, we found a large cluster of genes related to plants defense responses and disease resistance were changed in OE-MsASMT1 transgenic alfalfa, which also portended that the defense responses might alter in OE-MsASMT1 transgenic plants.
Flavonoids and lignin are major metabolites in phenylalanine pathway. According to our transcriptomic and metabolomic data, a large number of genes clustered into flavonoids and lignin biosynthesis and many metabolites belong to the precursors of flavonoids and lignin were changed (Figure 10). Among these genes in flavonoids biosynthesis, a large subgroup of genes are UDP-glucosyltransferase family genes, which is involved in flavonoid glycosylation, the major modification in flavonoid biosynthesis (Jones and Vogt, 2001; Yin et al., 2017). And many genes clustered into the MYB transcription factor family. R2R3-MYB transcription factors are widely involved in the phenylalanine metabolic pathway and play an important role in regulating flavonoid biosynthesis and stress responses in plants (Liu et al., 2015; Yan et al., 2015). There are 36 MYB transcription factors were changed in OE-MsASMT1 transgenic alfalfa, and 16 of them were downregulated, such as MYB4 and MYB14. The significantly changed (| log2FC| > 1.5) MYB transcription factors between OE-MsASMT1 transgenic plants and WT plants were listed in Supplementary Table S2. The significant downregulation of these genes might also be involved in the decreased flavonoids content in OE-MsASMT1 transgenic alfalfa. Lignin is a major component of plant secondary cell wall and is essential for mechanical support, water transport and defense responses (Boerjan et al., 2003; Rao et al., 2019). In our report, the transcripts of many genes involved in lignin biosynthesis were altered. And the contents of two lignin alcohol monomers coniferyl alcohol and p-coumaryl alcohol in the metabolomics results were significantly decreased in OE-MsASMT1 transgenic plants. Then, due to the complexity of lignin measurement and the same principle with IAA detection, we only chose OE16 and OE25 plants to quantify the lignin content and lignin monomer composition, and found that the lignin content and lignin monomers (G and S) contents showed no significant differences between WT plants and OE16, OE25 transgenic plants, while the S/G ratio was decreased in OE-MsASMT1 transgenic plants which might influence the saccharification efficiency in transgenic plants. The results indicated that melatonin plays a role in regulating flavonoids and lignin biosynthesis and metabolism. To further explain the function of MsASMT1 in alfalfa, transgenic plants with suppressed expression of the endogenous ASMT gene through RNAi or CRISPR-Cas9 would be generated to further explore the function of melatonin in alfalfa growth and secondary metabolism.
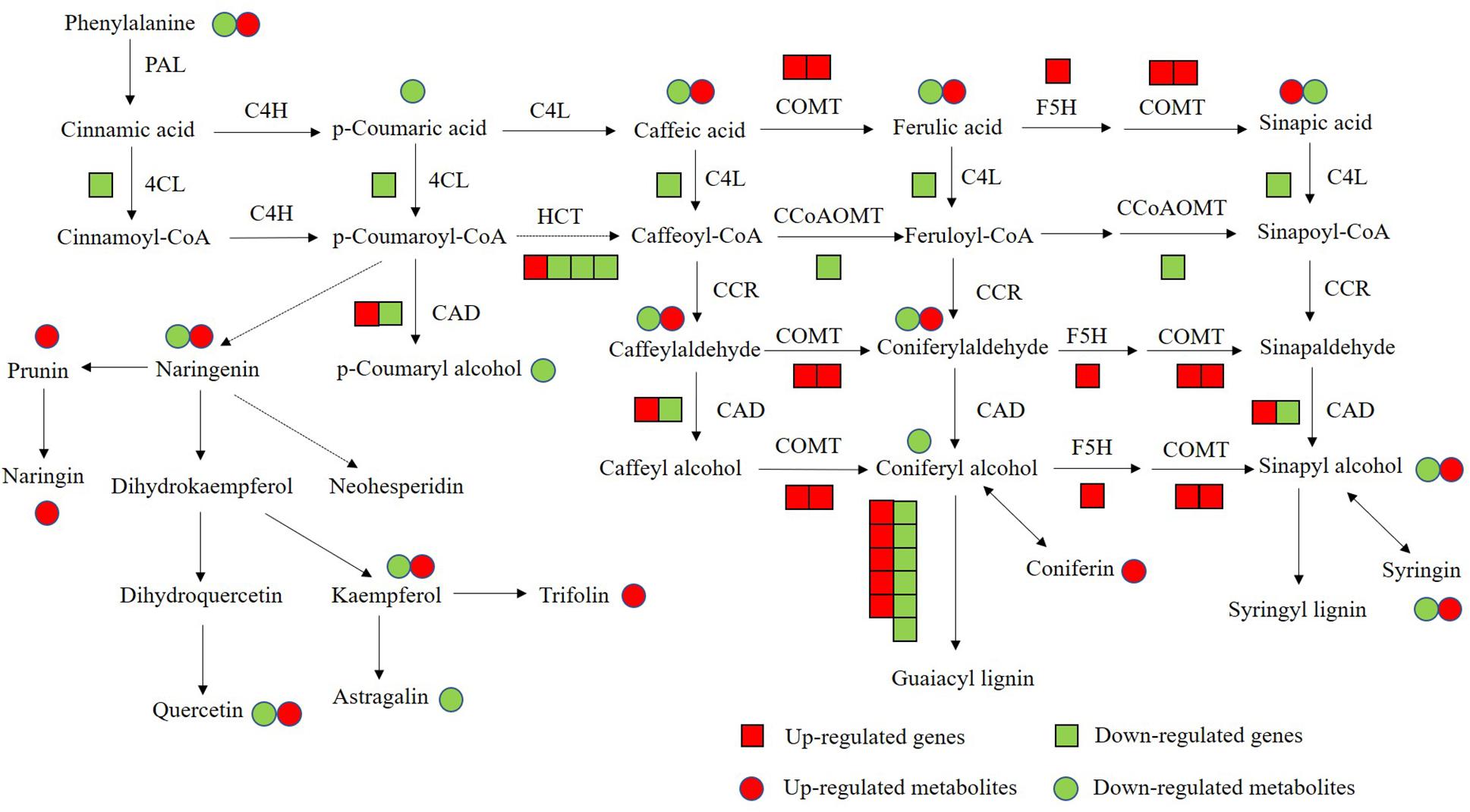
Figure 10. The conjoint analysis of genes and metabolites changed in phenylalanine pathway between WT plants and OE-MsASMT1 transgenic plants. The square represents changed genes, the circle represents changed metabolites. And red color represents the up-regulated genes or metabolites, green represents the down-regulated genes or metabolites.
Conclusion
In summary, the overexpression of MsASMT1 improved endogenous melatonin levels in transgenic alfalfa. OE-MsASMT1 transgenic alfalfa grew faster, had larger leaves and robust stems with increased cell sizes, cell numbers and vascular bundles. Nevertheless, OE-MsASMT1 transgenic alfalfa with rich melatonin accumulated less flavonoids, mainly quercetin, kaempferol, formononetin, and biochanin on account of the downregulation of genes associated with flavonoid biosynthesis. In this report, we found that melatonin plays a role in regulating flavonoids and lignin biosynthesis and inhibits flavonoids biosynthesis in alfalfa. We first reported the effects of endogenous melatonin on alfalfa plant growth and metabolism. This report provides insight into the function of melatonin in plant secondary metabolism and broadens the biological function of endogenous melatonin in plants.
Data Availability Statement
The datasets generated for this study can be found in the GenBank accession number: MN092350, NCBI Sequence Read Archive (SRA) database accession number: PRJNA555673.
Author Contributions
HC and YZ contributed the conception and design of the study. HC, TW, and HL performed the experiments. DT, XL, and XC organized the database. CG, HZ, and ML performed the statistical analysis. HC wrote the first draft of the manuscript. HW and YZ revised the manuscript. All authors read and approved the submitted manuscript.
Funding
This work was supported by National Nature Science Foundation of China (31672478) and Natural Science Foundation of Beijing, China (6162016) to YZ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00489/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ https://web.expasy.org/protparam/
- ^ https://submit.ncbi.nlm.nih.gov/subs/sra/
References
Ai, Y., and Zhu, Z. (2018). Melatonin antagonizes jasmonate-triggered anthocyanin biosynthesis in Arabidopsis. J. Agr. Food Chem. 66, 5392–5400. doi: 10.1021/acs.jafc.8b01795
Antoniou, C., Chatzimichail, G., Xenofontos, R., Pavlou, J. J., Panajiotou, E., Christou, A., et al. (2017). Melatonin systemically ameliorates drought stress-induced damage in Medicago sativa plants by modulating nitro-oxidative homeostasis and proline metabolism. J. Pineal Res. 62:e12401. doi: 10.1111/jpi.12401
Arnao, M. B., and Hernández-Ruiz, J. (2018a). Melatonin and its relationship to plant hormones. Ann. Bot. 121, 963–974.
Arnao, M. B., and Hernández-Ruiz, J. (2018b). The potential of phytomelatonin as a nutraceutical. Molecules 23:238. doi: 10.3390/molecules23010238
Arnao, M. B., and Hernández-Ruiz, J. (2019). Melatonin: a new plant hormone and/or a plant master regulator? Trends Plant Sci. 24, 38–48. doi: 10.1016/j.tplants.2018.10.010
Aung, B., Gruber, M. Y., Amyot, L., Omari, K., Bertrand, A., and Hannoufa, A. (2015). MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. doi: 10.1111/pbi.12308
Back, K., Tan, D. X., and Reiter, R. J. (2016). Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 61, 426–437. doi: 10.1111/jpi.12364
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546.
Byeon, Y., and Back, K. (2014). An increase in melatonin in transgenic rice causes pleiotropic phenotypes, including enhanced seedling growth, delayed flowering, and low grain yield. J. Pineal Res. 56, 408–414. doi: 10.1111/jpi.12129
Byeon, Y., Lee, H. J., Lee, H. Y., and Back, K. (2015). Cloning and functional characterization of the Arabidopsis N-acetylserotonin O-methyltransferase responsible for melatonin synthesis. J. Pineal Res. 60, 65–73. doi: 10.1111/jpi.12289
Byeon, Y., Lee, H. Y., Lee, K., Park, S., and Back, K. (2014). Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 56, 107–117.
Castonguay, Y., Michaud, J., and Dubé, M. P. (2015). Reference genes for RT-qPCR analysis of environmentally and developmentally regulated gene expression in alfalfa. Amer. J. Plant Sci. 6, 132–143. doi: 10.4236/ajps.2015.61015
Claustrat, B., Brun, J., and Chazot, G. (2005). The basic physiology and pathophysiology of melatonin. Sleep Med. Rev. 9, 11–24. doi: 10.1016/j.smrv.2004.08.001
Cornu, C., Remontet, L., Noel-Baron, F., Nicolas, A., Feugier-Favier, N., Roy, P., et al. (2010). A dietary supplement to improve the quality of sleep: a randomized placebo controlled trial. BMC Complem. Altern. 10:29. doi: 10.1186/1472-6882-10-29
Foo, E., Bullier, E., Goussot, M., Foucher, F., Rameau, C., and Beveridge, C. A. (2005). The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17, 464–474. doi: 10.1105/tpc.104.026716
Fu, C., Hernandez, T., Zhou, C., and Wang, Z. Y. (2015). Alfalfa (Medicago sativa L.). Methods Mol. Biol. 1223, 213–221.
Fu, J., Wu, Y., Miao, Y., Xu, Y., Zhao, E., Wang, J., et al. (2017). Improved cold tolerance in Elymus nutans by exogenous application of melatonin may involve ABA-dependent and ABA-independent pathways. Sci. Rep. 7: 39865.
Guo, D., Zhang, J., Wang, X., Han, X., Wei, B., Wang, J., et al. (2015). The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27, 3112–3127. doi: 10.1105/tpc.15.00829
Huang, Y. H., Liu, S. J., Yuan, S., Guan, C., Tian, D. Y., Cui, X., et al. (2017). Overexpression of ovine AANAT and HIOMT genes in switchgrass improved growth performance and salt-tolerance. Sci. Rep. 7:12212.
Jones, P., and Vogt, T. (2001). Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213, 164–174. doi: 10.1007/s004250000492
Kobyliñska, A., Borek, S., and Posmyk, M. M. (2018). Melatonin redirects carbohydrates metabolism during sugar starvation in plant cells. J. Pineal Res. 64:e12466. doi: 10.1111/jpi.12466
Kumar, S., and Pandey, A. K. (2013). Chemistry and biological activities of flavonoids: an overview. Sci. World J. 2013:162750.
Lee, H., and Back, K. (2018a). Melatonin induction and its role in high light stress tolerance in Arabidopsis thaliana. J. Pineal Res. 65:e12504. doi: 10.1111/jpi.12504
Lee, K., and Back, K. (2017). Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 62:e12392. doi: 10.1111/jpi.12392
Lee, K., and Back, K. (2018b). Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 66:e12537. doi: 10.1111/jpi.12537
Lee, K., Hwang, O. J., Reiter, R. J., and Back, K. (2018). Flavonoids inhibit both rice and sheep serotonin -acetyltransferases and reduce melatonin levels in plants. J. Pineal Res. 65:e12512. doi: 10.1111/jpi.12512
Li, Q., Min, D., Wang, J. P. Y., Peszlen, I., Horvath, L., Horvath, B., et al. (2011). Down-regulation of glycosyltransferase 8D genes in Populus trichocarpa caused reduced mechanical strength and xylan content in wood. Tree Physiol. 31, 226–236. doi: 10.1093/treephys/tpr008
Liang, C., Zheng, G., Li, W., Wang, Y., Hu, B., Wang, H., et al. (2015). Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 59, 91–101. doi: 10.1111/jpi.12243
Liu, C., Chen, L., Zhao, R., Li, R., Zhang, S., Yu, W., et al. (2019). Melatonin induces disease resistance to botrytis cinereal in tomato fruit by activating jasmonic acid signaling pathway. J. Agr. Food Chem. 67, 6116–6124. doi: 10.1021/acs.jafc.9b00058
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Mandal, M. K., Suren, H., Ward, B., Boroujerdi, A., and Kousik, C. (2018). Differential roles of melatonin in plant-host resistance and pathogen suppression in cucurbits. J. Pineal Res. 65:e12505. doi: 10.1111/jpi.12505
Mitsunami, T., Nishihara, M., Galis, I., Alamgir, K. M., Hojo, Y., Fujita, K., et al. (2014). Overexpression of the PAP1 transcription factor reveals a complex regulation of flavonoid and phenylpropanoid metabolism in Nicotiana tabacum plants attacked by Spodoptera litura. PLoS One 9:e108849. doi: 10.1371/journal.pone.0108849
Park, S., and Back, K. (2012). Melatonin promotes seminal root elongation and root growth in transgenic rice after germination. J. Pineal Res. 53, 385–389. doi: 10.1111/j.1600-079x.2012.01008.x
Park, S., Byeon, Y., and Back, K. (2013). Functional analyses of three ASMT gene family member in rice plants. J. Pineal Res. 55, 409–415.
Posmyk, M. M., Wieczorek, B. M., and Janas, E. S. L. M. (2009). Melatonin applied to cucumber (Cucumis sativus L.) seeds improves germination during chilling stress. J. Pineal Res. 46, 214–223. doi: 10.1111/j.1600-079x.2008.00652.x
Rao, X., Chen, X., Shen, H., Ma, Q., Li, G., Tang, Y., et al. (2019). Gene regulatory networks for lignin biosynthesis in switchgrass (Panicum virgatum). Plant Biotechnol. J. 17, 580–593. doi: 10.1111/pbi.13000
Reddy, M. S., Chen, F., Shadle, G., Jackson, L., Aljoe, H., and Dixon, R. A. (2005). Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. U.S.A. 102, 16573–16578. doi: 10.1073/pnas.0505749102
Reiter, R. J., Tan, D. X., Manchester, L. C., Simopoulos, A. P., Maldonado, M. D., Flores, L. J., et al. (2007). Melatonin in edible plants (phytomelatonin): identification, concentrations, bioavailability and proposed functions. World Rev. Nutr. Diet. 97, 211–230. doi: 10.1159/000097917
Reiter, R. J., Tan, D. X., Zhou, Z., Cruz, M. H. C., Fuentes-Broto, L., and Galano, A. (2015). Phytomelatonin: assisting plants to survive and thrive. Molecules 20, 7396–7437. doi: 10.3390/molecules20047396
Rubio-Moraga, A., Ahrazem, O., Pérez-Clemente, R. M., Gómez-Cadenas, A., Yoneyama, K., López-Ráez, J. A., et al. (2014). Apical dominance in saffron and the involvement of the branching enzymes CCD7 and CCD8 in the control of bud sprouting. BMC Plant Biol. 14:171. doi: 10.1186/1472-6882-10-171
Shi, H., Tan, D. X., Reiter, R. J., Ye, T., Yang, F., and Chan, Z. (2015). Melatonin induces class A1 heat-shock factors (HSFA1s) and their possible involvement of thermotolerance in Arabidopsis. J. Pineal Res. 58, 335–342. doi: 10.1111/jpi.12219
Tan, D. X., Hardeland, R., Back, K., Manchester, L. C., Alatorre-Jimenez, M. A., and Reiter, R. J. (2016). On the significance of an alternate pathway of melatonin synthesis via 5-methoxyltryptamine: comparisons across species. J. Pineal Res. 61, 426–437.
Tan, D. X., Manchester, L. C., Liu, X., Rosales-Corral, S. A., Acuna-Castroviejo, D., and Reiter, R. J. (2013). Mitochondria and chloroplasts as the original sites of melatonin synthesis: a hypothesis related to melatonin’s primary function and evolution in eukaryotes. J. Pineal Res. 54, 127–138. doi: 10.1111/jpi.12026
Tresguerres, I. F., Tamimi, F., Eimar, H., Barralet, J. E., Prieto, S., Torres, J., et al. (2014). Melatonin dietary supplement as an anti-aging therapy for age-related bone loss. Rejuvenation Res. 17, 341–346. doi: 10.1089/rej.2013.1542
Wang, Y., Reiter, R. J., and Chan, Z. L. (2018). Phytomelatonin: a universal abiotic stress regulator. J. Exp. Bot. 69, 963–974. doi: 10.1093/jxb/erx473
Wei, Y., Chang, Y., Zeng, H., Liu, G., He, C., and Shi, H. (2018). RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. J. Pineal Res. 64, e12454. doi: 10.1111/jpi.12454
Xu, W., Cai, S. Y., Zhang, Y., Wang, Y., Ahammed, G. J., Xia, X. J., et al. (2016). Melatonin enhances thermotolerance by promoting cellular protein protection in tomato plants. J. Pineal Res. 61, 457–469. doi: 10.1111/jpi.12359
Yan, J., Wang, B., Zhong, Y., Yao, L., Cheng, L., and Wu, T. (2015). The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 89, 35–48. doi: 10.1007/s11103-015-0349-3
Yin, Q., Shen, G., Di, S., Fan, C., Chang, Z., and Pang, Y. (2017). Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in glycine max. Plant Cell Physiol. 58, 1558–1572. doi: 10.1093/pcp/pcx081
Yuan, S., Guan, C., Liu, S., Huang, Y., Tian, D., Cui, X., et al. (2016). Comparative transcriptomic analyses of differentially expressed genes in transgenic melatonin biosynthesis ovine HIOMT gene in switchgrass. Front. Plant Sci. 7:1613. doi: 10.3389/fpls.2016.01613
Zhang, H., Wang, L., Shi, K., Shan, D., Zhu, Y., Wang, C., et al. (2018). Apple tree flowering is mediated by low level of melatonin under the regulation of seasonal light signal. J. Pineal Res. 66:e12551. doi: 10.1111/jpi.12551
Zhang, H. J., Zhang, N., Yang, R. C., Wang, L., Sun, Q. Q., Li, D. B., et al. (2014). Melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA4 interaction in cucumber (Cucumis sativus L.). J. Pineal Res. 57, 269–279. doi: 10.1111/jpi.12167
Zhang, N., Sun, Q., Li, H., Li, X., Cao, Y., Zhang, H., et al. (2016). Melatonin improved anthocyanin accumulation by regulating gene expressions and resulted in high reactive oxygen species scavenging capacity in cabbage. Front. Plant Sci. 7:197. doi: 10.3389/fpls.2016.01197
Zhang, Q., Liu, X., Zhang, Z., Liu, N., Li, D., and Hu, L. (2019). Melatonin improved waterlogging tolerance in alfalfa (Medicago sativa) by reprogramming polyamine and ethylene metabolism. Front. Plant Sci. 10:44. doi: 10.3389/fpls.2016.044
Zhang, Z., Hu, Q., Liu, Y., Cheng, P., Cheng, H., Liu, W., et al. (2019). Strigolactone represses the synthesis of melatonin, thereby inducing floral transition in Arabidopsis thaliana in an FLC-dependent manner. J. Pineal Res. 67:e12582.
Zhang, W., and Wang, T. (2015). Enhanced salt tolerance of alfalfa (Medicago sativa) by rstB gene transformation. Plant Sci. 234, 110–118. doi: 10.1016/j.plantsci.2014.11.016
Zhao, H., Xu, L., Su, T., Jiang, Y., Hu, L., and Ma, F. (2015). Melatonin regulates carbohydrate metabolism and defenses against Pseudomonas syringae pv. Tomato DC3000 infection in Arabidopsis thaliana. J. Pineal Res. 59, 109–119.
Keywords: transgenic, MsASMT1, melatonin, flavonoids, lignin, alfalfa
Citation: Cen H, Wang T, Liu H, Wang H, Tian D, Li X, Cui X, Guan C, Zang H, Li M and Zhang Y (2020) Overexpression of MsASMT1 Promotes Plant Growth and Decreases Flavonoids Biosynthesis in Transgenic Alfalfa (Medicago sativa L.). Front. Plant Sci. 11:489. doi: 10.3389/fpls.2020.00489
Received: 17 December 2019; Accepted: 01 April 2020;
Published: 28 April 2020.
Edited by:
Zeng-Yu Wang, Qingdao Agricultural University, ChinaReviewed by:
Huanzhong Wang, University of Connecticut, United StatesXiang Gao, Northeast Normal University, China
Copyright © 2020 Cen, Wang, Liu, Wang, Tian, Li, Cui, Guan, Zang, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunwei Zhang, enl3ZWlAY2F1LmVkdS5jbg==
 Huifang Cen
Huifang Cen Tingting Wang
Tingting Wang Xin Cui
Xin Cui Cong Guan
Cong Guan Hui Zang
Hui Zang