
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 May 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00471
This article is part of the Research Topic Developmental Modification under Biotic Interactions in Plants View all 14 articles
 Tomoko Hirano1
Tomoko Hirano1 Seisuke Kimura2
Seisuke Kimura2 Tomoaki Sakamoto2
Tomoaki Sakamoto2 Ayaka Okamoto1
Ayaka Okamoto1 Takumi Nakayama1
Takumi Nakayama1 Takakazu Matsuura3
Takakazu Matsuura3 Yoko Ikeda3
Yoko Ikeda3 Seiji Takeda4
Seiji Takeda4 Yoshihito Suzuki5
Yoshihito Suzuki5 Issei Ohshima6
Issei Ohshima6 Masa H. Sato1*
Masa H. Sato1*Insect galls are unique organs that provide shelter and nutrients to the gall-inducing insects. Although insect galls are fascinating structures for their unique shapes and functions, the process by which gall-inducing insects induce such complex structures is not well understood. Here, we performed RNA-sequencing-based comparative transcriptomic analysis of the early developmental stage of horned gall to elucidate the early gall-inducing process carried out by the aphid, Schlechtendalia chinensis, in the Chinese sumac, Rhus javanica. There was no clear similarity in the global gene expression profiles between the gall tissue and other tissues, and the expression profiles of various biological categories such as phytohormone metabolism and signaling, stress-response pathways, secondary metabolic pathways, photosynthetic reaction, and floral organ development were dramatically altered. Particularly, master transcription factors that regulate meristem, flower, and fruit development, and biotic and abiotic stress-responsive genes were highly upregulated, whereas the expression of genes related to photosynthesis strongly decreased in the early stage of the gall development. In addition, we found that the expression of class-1 KNOX genes, whose ectopic overexpression is known to lead to the formation of de novo meristematic structures in leaf, was increased in the early development stage of gall tissue. These results strengthen the hypothesis that gall-inducing insects convert source tissues into fruit-like sink tissues by regulating the gene expression of host plants and demonstrate that such manipulation begins from the initial process of gall induction.
Galls are plant tissues or organs formed by hyperplasia (increased cell number) and/or hypertrophy (increased cell size) induced by parasitic or pathogenic organisms including viruses, fungi, bacteria, nematodes, mites, and insects (Mani, 1964). Among galls formed by various organisms, insect galls are extraordinarily complex and highly organized structures comprised of several specialized tissue types (Stone and Schönrogge, 2003; Giron et al., 2016). Insect galls range in complexity from relatively simple mine-galls (Guiguet et al., 2018), open- or folded-type galls such as pit galls, blister galls, and roll galls to complex structures in which the gall-inducing insects are entirely enclosed by plant tissues to form covering galls or mark galls in leaf, stem, and bud (Dreger-Jauffret and Shorthouse, 1992; Guiguet et al., 2019).
The most complex gall structures are generated by gall wasps, gall midges, and gall-inducing aphids in which the galls have extra-floral nectarines, and a coating of hair, spines, and sticky resins (Price et al., 1987; Stone and Schönrogge, 2003; Wool, 2004). The complex insect galls consist of various tissues such as nutritive and protective tissues. The nutritive tissues consist of callus cells and vascular cells, which transport nutrients to the callus; the tissues are ingested by gall-inducing insects. The protective tissues (sclerenchyma) are composed of lignified cells arranged as a layer on the outside of the nutritive tissues and function as a physical shelter against natural enemies and outside environment.
Several lines of evidence indicate that many gall-inducing insects have the potential to precisely secrete effectors into plant tissues using their mouthparts or ovipositors, and such effectors are likely to play a central role in gall induction (Sopow et al., 2003; Matsukura et al., 2009; Stuart et al., 2012; Giron et al., 2016). Thus, gall-inducing insects are believed to manipulate plant developmental programs to generate complex gall structures by secretion of certain chemical compounds in plants (Miles, 1968), and this idea has been supported by histological observations and physiological analyses of insect galls (for a review Giron et al., 2016). The most important characteristic of insect galls is their function as a sink for insect nutrition (Rohfritsch and Shorthouse, 1982). The existence of insect galls near the source organs redirects the flow of plant resources such as carbohydrates, lipids, proteins from the original sink organs to the induced galls. Thus, gall formation results in development of a stronger sink of nutrients for gall-inducing insects than the original sink organs such as buds, flowers, fruits, and storage roots (Weis and Kapelinski, 1984; McCrea et al., 1985; Burstein et al., 1994; Larson and Whitham, 1997).
Darwin (1868) pointed out that the shapes of some complex insect galls resemble flowers or fruits. Indeed, many remarkable flower- and fruit-like traits are observed in insect galls, in particular those that are induced by gall midges and cynipids on various plant species (Rohfritsch and Shorthouse, 1982), suggesting that the formation of gall tissues is similar to the development of flowers or fruits (Kurosu and Aoki, 1990; Ferreira and Isaias, 2014). Recently, Schultz et al. (2018) reported that the gene expression pattern during Phylloxera leaf-gall development is similar to that during carpel development. These results indicate that the parasite may, at least partly, hijack the processes of flower development during gall formation (Schultz et al., 2018), supporting the hypothesis that flower- or fruit-like galls are generated by manipulation of flower development, although the initial process of induction of gall tissues in vegetative tissues is still largely unknown.
Aphids are small phloem sap-feeding insects belonging to the super family Aphidoidea, which embraces approximately 5,000 species in nature (Blackman and Eastop, 2007). Of these, no more than 10% of the aphid species can induce apparent galls on their host plants (Wool, 2004). Like other aphids, the gall-inducing aphids have complicated life cycles, in which a fundatrix or stem mother emerges from a fertilized egg in spring, and initiates the induction of a gall on the primary host plant. Then, the fundatrix parthenogenetically produces offspring inside the gall, and this parthenogenetic production is continued over several generations in particular aphid taxa. In summer or early autumn, winged adults appear and exit from the gall for migrating to the secondary host plant, where they spend several generations in autumn and winter (Wool, 2004; Aoki and Kurosu, 2010). A gall-inducing aphid, Schlechtendalia chinensis, induces large, single-chamber galls called horned galls on the leaf wings of several Rhus species (Anacardiaceae) in China, Korea, Taiwan, Malaysia, and Japan (Blackman and Eastop, 2007). Galls are first induced when the fundatrix of S. chinensis feeds on the adaxial side of the leaf wings. After the fundatrix is enclosed in the gall, the gall is enlarged quickly to form large horned galls with forked structures. During gall development, drastic morphological rearrangement occurs in the leaf wing tissues, in which the palisade tissues of the galled leaf wings are reorganized and replaced by parenchyma cells, and galled zones connect to non-galled zones by newly formed vascular bundles (Liu et al., 2014). Such complexity both in the developmental process and in the structure of S. chinensis galls implies that modified but well-organized host-plant gene networks could be incorporated in the process of gall development. However, the underlying molecular mechanisms contributing to the gall formation are largely unknown.
In this study, we performed RNA-sequencing-based comparative transcriptomics of a host plant, R. javanica, to understand the molecular characteristics of the early phase of gall development induced by S. chinensis. We found that there was no clear similarity in the global gene expression profiles between the gall tissue and other tissues. The genes involved in the phytohormone metabolic and signaling pathways, abiotic and biotic stress responses, and organ development were significantly upregulated, whereas photosynthetic genes were dramatically downregulated. These results imply that the gall-inducing aphid manipulates the plant reproductive programs to convert source tissues into fruit-like sink tissues during the initial process of gall induction.
The phase 4 of developmental stage of galls (Liu et al., 2014) (collected in May 22, 2017, Figure 1a), young leaves (collected in May 22, 2017), flowers (collected in September 9, 2017), and fruits (collected in September 28, 2017) (Figure 1b) of R. javanica were collected from a natural plantation located in the Kyoto Prefecture of Japan (35°06′00.83″N 135°72′86.94″E).
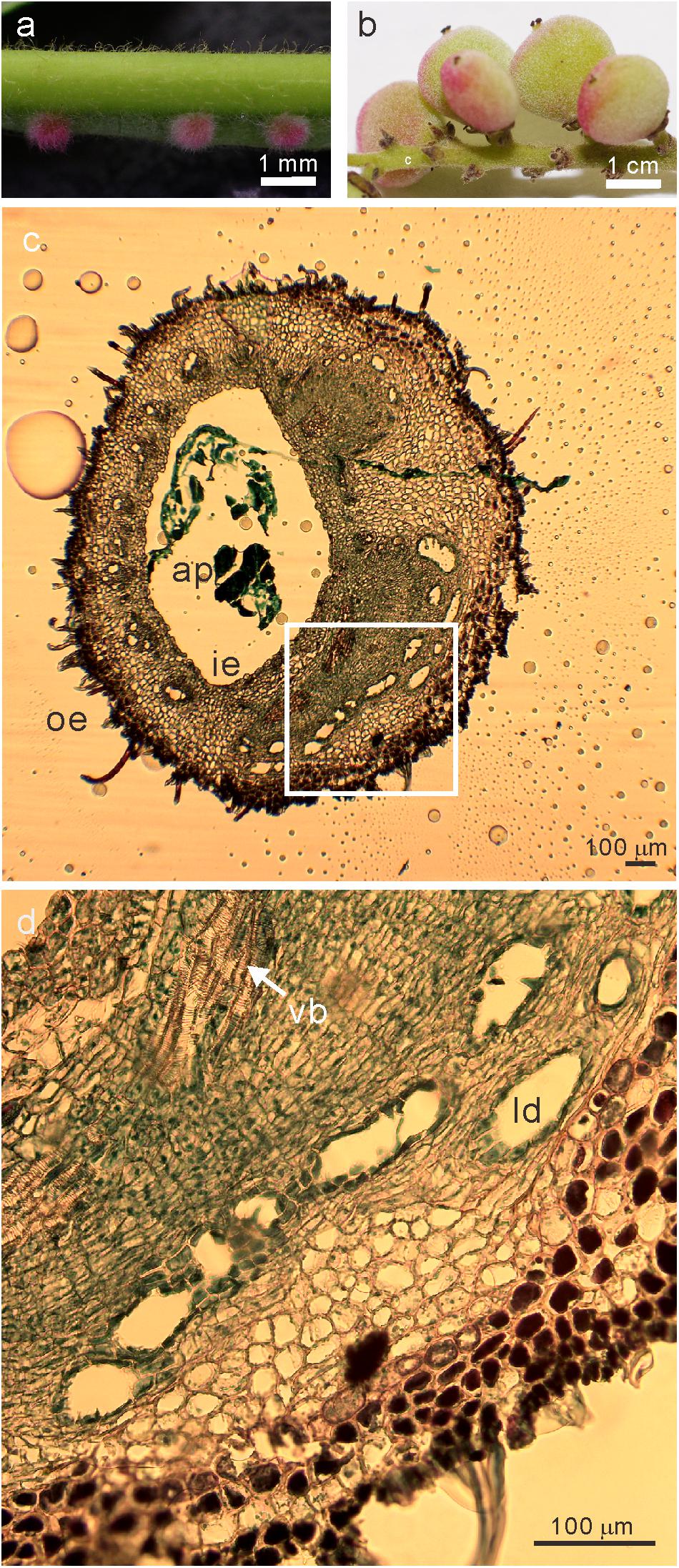
Figure 1. Images of the sampled (a) galls at the early developmental stage of (phase 4) and (b) fruits (c) transverse section of stage 4 galls (d) the magnified image (white square in c) of R. javanica. Abbreviations: ap; aphid; ie, inner epidermis; oe, outer epidermis; ld, latex duct; vb, vascular bundles.
Total RNA was extracted from the young leaves, female flowers, fruits, and gall tissues with an RNeasy Plant Mini Kit (QIAGEN). RNA-seq libraries were prepared using the Illumina TruSeq Stranded RNA LT Kit (Illumina, CA, United States) according to the manufacturer’s instructions. Three independent RNA samples for each tissue were used for the analysis. The qualities of the prepared libraries were checked using the QuantiFluor dsDNA System and Agilent High Sensitivity DNA Assay (Agilent, CA, United States). The pooled libraries were sequenced on the NextSeq500 sequencing platform (Illumina, CA, United States) and paired-end reads were obtained. Then, the obtained reads were assembled into transcriptome contigs using Trinity with the default settings. Blastx searches of the obtained contigs against non-redundant protein sequences from GenPept, SwissProt, PIR, PDF, PDB, and NCBI RefSeq (nr) databases using the DIAMOND software (Buchfink et al., 2015) were conducted to find similar protein sequences. Each contig was classified into a taxon group based on the top hits of the blastx results and NCBI taxonomy lineage data. Finally, the contigs classified into the Virdiplantae (plant) kingdom were extracted as R. javanica reference transcript contigs to exclude contigs from aphids or other contaminants. RNA-seq analysis of R. javanica tissues was biologically repeated at least three times per each tissue sample (Supplementary Table S1).
The obtained reads were mapped to the R. javanica reference transcript contigs using the Burrows-Wheeler alignment tool (BWA)1. The count data were subjected to the trimmed mean of M-values normalization in EdgeR. Multi-dimensional scaling was performed by calculating the log-fold changes between the accessions and by using differentially expressed genes (DEGs) to compute distances in EdgeR with the plotMDS function. Transcript expression profiles and DEGs were defined with the EdgeR general linear models approach (Robinson et al., 2010). The threshold for DEGs was a log-fold change of >2 and a false discovery rate of <0.01.
The gall, young leaf, flower, and fruit samples were frozen in liquid nitrogen. The total RNA was isolated using the NucleoSpin RNA Plant and Fungi Kit (Takara), and the cDNA library construction was performed using the ReverTra Ace qPCR RT Master Mix (TOYOBO) as per the manufacturer’s instructions. The same amount of cDNA was used as a template for the qPCR, which was performed with the THUNDERBIRD SYBR qPCR Mix (TOYOBO) and gene-specific primers. UBQ10 was used as an internal control for normalization. The primers used in this study are listed in Supplementary Table S2.
The endogenous levels of the indole-3-acetic acid (IAA) and cytokinins (CKs) in the whole S. chinensis bodies were quantitatively analyzed according to Tanaka et al. (2013). Briefly, the endogenous levels of IAA and cytokinins in the aphids were analyzed by extracting the samples that were spiked with stable isotope-labeled internal standards ([2H5]tZ, [2H5]tZR, [2H6]iP, [2H6]iPR, and [13C6]IAA), pre-purifying them with solid-phase extractions, and quantifying them by liquid chromatography/tandem mass spectrometry (3200 QTrap, AB Sciex).
tZ, iP, IAA contents in R. javanica leaves and galls were determined according to Yamane et al. (2019) with minor modifications. Briefly, leaf and gall samples (approximately 100 -200 mg per sample) were collected and frozen in liquid nitrogen, and the weight of each tissue was measured. Then the samples were ground and subjected to extraction in 80% acetonitrile and 1% acetic acid containing stable isotope-labeled compounds for internal standards [D5-tZ, D6-iP, 13C6-IAA] (OlChemim, Czech Republic). After sample purification by HLB and MCX columns (Waters), Phytohormones were analyzed with a 6410 Triple Quad LC/MS System (Agilent Technologies Inc., United States) equipped with a ZORBAX Eclipse XDB-C18 column and an XDB-C8 Guard column (Agilent Technologies Inc.), and peak areas were determined using MassHunter Workstation software (vB.04.00; Agilent Technologies). Four independent samples were analyzed for calculation of averages and standard deviations.
Young gall tissues were fixed in the fixative solution consisting of 4% (w/v) paraformaldehyde in 1X PBS under vacuum condition until the samples were drawn to the bottom of the tube. After fixation, the sample were dehydrated through a graded ethanol series, and then followed by a D-Limonene series, and embedded in Paraplast Plus (Sherwood Medical). Microtome sections (4 μm) were deparaffinized in D-Limonene, and rehydrated through a graded ethanol series. In the case of histological observation, the sections were stained with Hematoxylin-Safranin-Fast Green FCF.
Full-length cDNA of RjKNAT6 (KNAT6 of Rhus javanica) was cloned into the pENTR vector, and then was subcloned into pGEM11 vector by the Gateway system. Labeled RNA probes were synthesized using in vitro transcription in the presence of Digoxigenin-11-UTP by RNA polymerases T7 or SP6 (DIG RNA labeling Mix, Roche). Samples were washed twice in PBST (1X PBS plus 0.1% (v/v) Tween-20) for 10 min and then incubated with 1 μg ml–1 proteinase K (Roche) for 15 min. Digestion was stopped by incubating the samples in 1X PBS plus 0.2% glycine for 5 min and then washing them twice in PBST for 10 min. Samples were washed twice in PBST for 10 min and once in the hybridization solution 50% (v/v) formamide in 2X SSC (20X SSC: 3 M NaCl, 300 mM sodium citrate, pH 7.0 with 1 M HCl) for 10 min, and then preincubated in the same solution for 1 h at 50°C. The hybridization step was performed overnight at 42°C by incubating samples in supplemented hybridization solution containing a cocktail of denatured (80°C for 2 min) labeled RNA probes (20–100 ng per ml of the hybridization solution). Samples were washed: three times (10, 60, and 20 min) in a solution of 50% (v/v) formamide. Thereafter samples were incubated with a mixture of the selected primary antibodies (Chicken anti-digoxigenin, Immunology Consultant Laboratory) diluted (1:100) in (PBST + BSA), for 2 h at RT under gentle shaking. Subsequently, samples were washed three times for 10 min in PBST, once for 30 min in PBST plus BSA and then incubated with a mixture of the secondary antibodies (Alexa Fluor dyes 555 Goat Anti-Chicken, INVITROGEN) diluted (1:100) in PBST plus BSA overnight at RT in the dark. After incubation samples were washed twice for 15 min in PBST under gentle shaking in the dark. Fluorescence and differential interference contrast (DIC) images were obtained using Leica TCS SP8 laser scanning confocal microscope. The captured images were processed using Leica LAS X.
Liu et al. (2014) reported the histological analysis of the developmental process of R. chinensis gall; they categorized the developmental process of the gall into six different phases. To investigate the changes in the gene expression profile during the early phase of gall development, we isolated total RNA from various tissues including entire galls in the phase 4 of development (about 1 mm diameter) in which the gall is completely closed (Figure 1a and Supplementary Figure S1a), young leaves (Supplementary Figure S1b), female flowers (Supplementary Figure S1c), and fruits (Figure 1b). The cross-section of the phase 4 gall revealed that the inner and outer epidermis had two to three layers and the parenchyma cells were well developed between the outer and inner epidermal cell layers with vascular bundles and latex ducts (Figures 1c,d). Phase 4 galls enlarged slowly and contained 1–2 aphids inside of a gall, then in phase 5, the size of the gall increased quickly from August to late September, and finally the horn-like or fork shaped galls were formed (Liu et al., 2014). According to the enlarging gall size, the number of aphids inside of galls increases exponentially (Supplementary Figure S2).
Paired-end reads for the gall, young leaf, flower, and fruit tissues were obtained by RNA-seq (Supplementary Table S1). De novo assembly of all the reads yielded 265,145 transcript contigs by Trinity with N50 and average lengths of 1842 and 905.3 nt, respectively. The reference transcript contigs for R. javanica were extracted from the raw assembled contigs based on the blastx results against known protein databases, and their N50 and average lengths were 2267 nucleotides and 1331 nucleotides, respectively. Based on the N50 length, which is an indicator of assembly quality, we confirmed that the quality of the de novo assembly was sufficient for the subsequent analyzes (Supplementary Table S2).
We aligned all single reads to the contigs and compared the number of DEGs in the galls, flowers, and fruits to those in the young leaves. First, we performed a principal component analysis to compare the gene expression profiles of the gall, leaf, flower, and fruit tissues of R. javanica. The eigen values of the two components were greater than 1, and the first component and second component explained 41.4 and 34.6% of the variation, respectively (Supplementary Figure S3a). The factorial map of the principal component analysis showed that the four dots corresponding to each tissue were widely distributed in the graph (Supplementary Figure S2b). These results suggested that there was no clear similarity in the global gene expression profiles between the gall tissue and other tissues.
Compared with the transcripts for young leaves, the transcripts for the gall, flower, and fruit tissues showed upregulation of 1829, 1330, and 2583 DEGs, and downregulation of 1879, 1554, and 4409 DEGs, respectively (log-fold change of >2 and a false discovery rate of <0.01) (Figure 2 and Supplementary Tables S3, S4). As the R. javanica genome has not yet been read and no functional annotation exists, we assigned R. javanica transcripts to Arabidopsis thaliana orthologs using functional annotations from The Arabidopsis Information Resource (TIAR). To assess the similarity between the R. javanica and Arabidopsis orthologs, we cloned and sequenced several R. javanica full-length transcripts (AG, AP1, CLE41/44), SEP2, CYCD4;1, and SEOR1) according to the deduced sequences assembled by Trinity, and then compared them with the full-length sequences of the Arabidopsis counterparts. The cloned transcripts have considerable similarity with the Arabidopsis counterparts (Supplementary Figure S4), implying that most of the R. javanica transcripts could be correctly assigned to the Arabidopsis orthologs. To evaluate the DEGs identified by RNA-seq, we measured the differences in the expression levels of several of the upregulated genes by quantitative real-time PCR and found that SHP1, KNAT6, CLE44, AG, AP1, KNAT1, HEC1, VND7, and CYCD4;1 were considerably upregulated in the gall tissue (Figure 3).
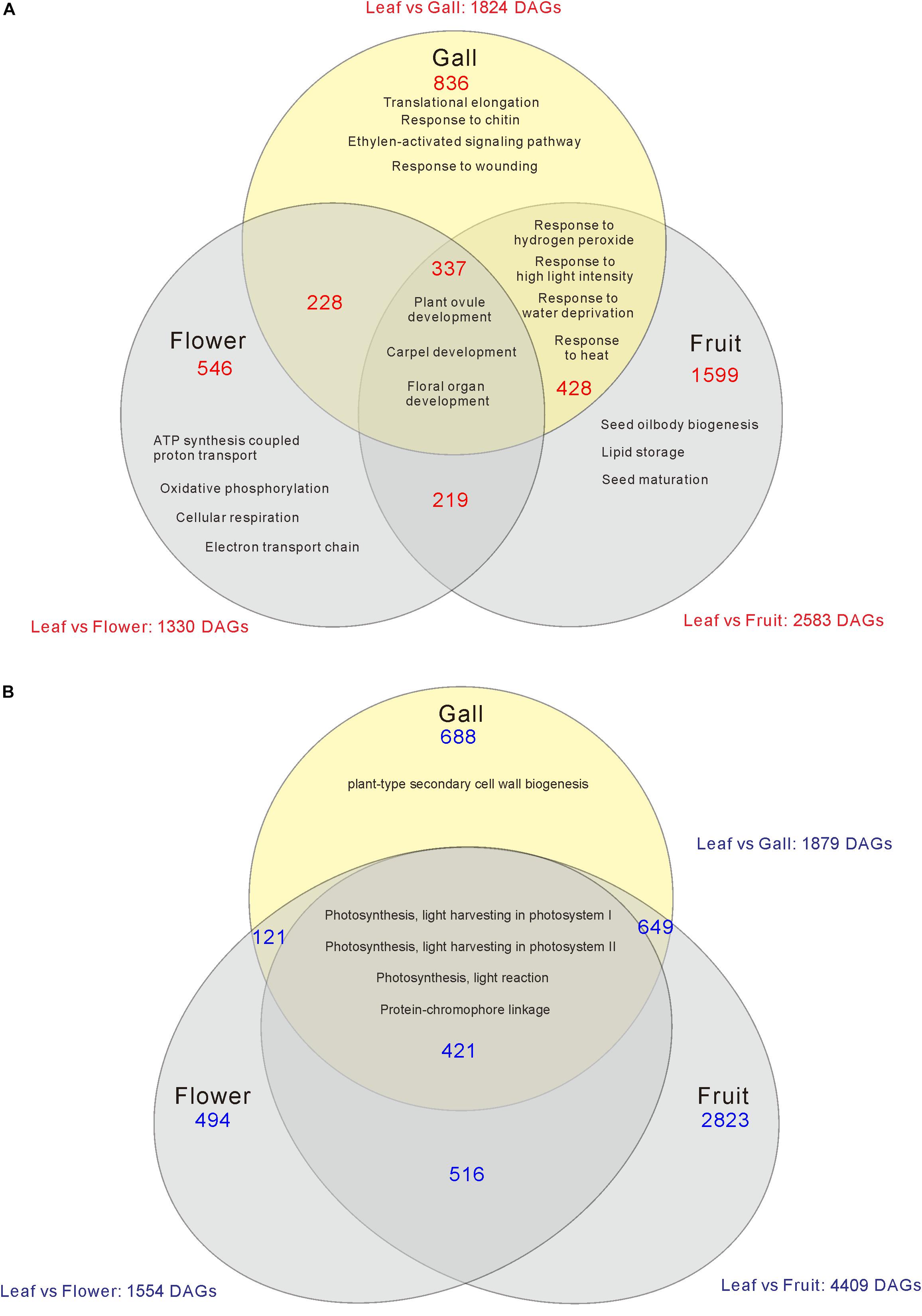
Figure 2. Venn diagram analysis of the number of increased (A) or decreased (B) differentially expressed genes (DEGs) in the gall, flower, and fruit tissue compared with the young leaves. The numbers in each region indicate the DEGs in each tissue. Overlapping regions of the Venn diagram indicates shared DEGs among corresponding groups. Descriptions in each region indicate typically enriched gene ontology categories of the corresponding groups.
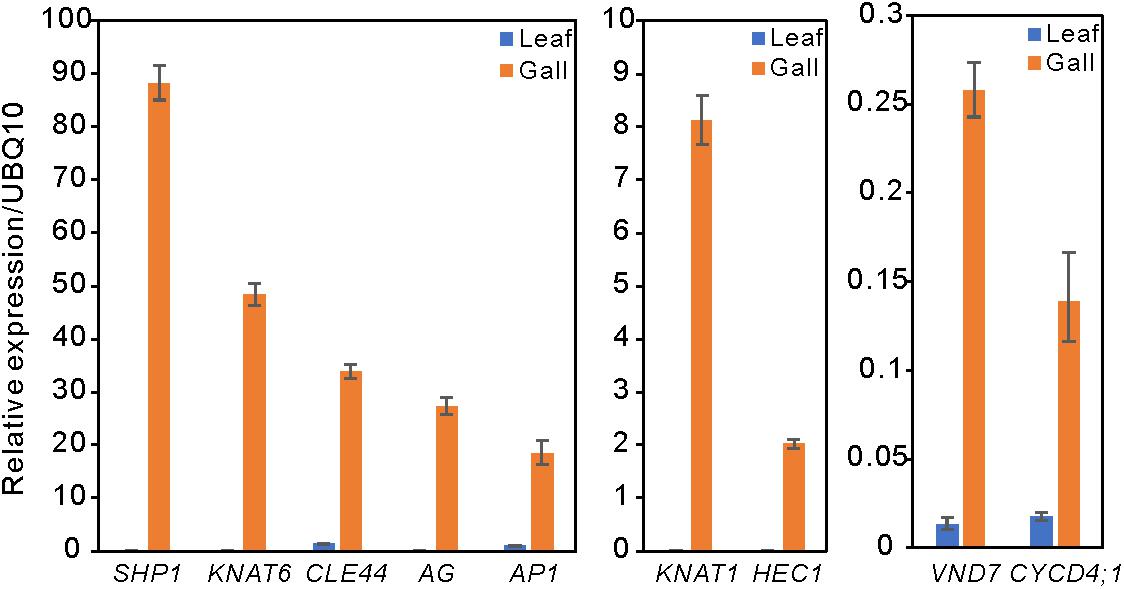
Figure 3. Relative expression levels of several upregulated DEGs in the leaf and gall tissues. The gene expression levels were analyzed by quantitative reverse transcription PCR. UBQ10 served as the internal expression control. Experiments were repeated three times using three biologically distinct samples, and gave similar results. The data is represented as the mean ± standard deviation (Four technical replicates).
When comparing the genes upregulated in the galls with those upregulated in the flowers and fruits, we found that expression of 337 genes was increased in the gall, and in the floral and reproductive organs. Gene ontology (GO) term enrichment analysis of these upregulated genes revealed that the genes assigned to the GO categories of the floral organ development (GO: 0048481, 0048440, 0048437, and 0090567) were enriched by over 5-fold (Figure 4). Of these, several genes encoding transcription factors involved in the regulation of floral organ morphogenesis were upregulated in the early development stage of gall tissue. For example, the upregulated genes included a floral integrator (LFY), class-1 KNOX genes (KNAT1/2/6, and STM), and MADS-box-type transcription factors (SEP1, SEP2, SEP3, AP1, AP3 AG, TT16, FUL, and SHP1) (Supplementary Table S5). In situ hybridization analysis revealed that KNAT6 was predominantly expressed in the parenchyma cells of developing gall tissues (Figure 5).
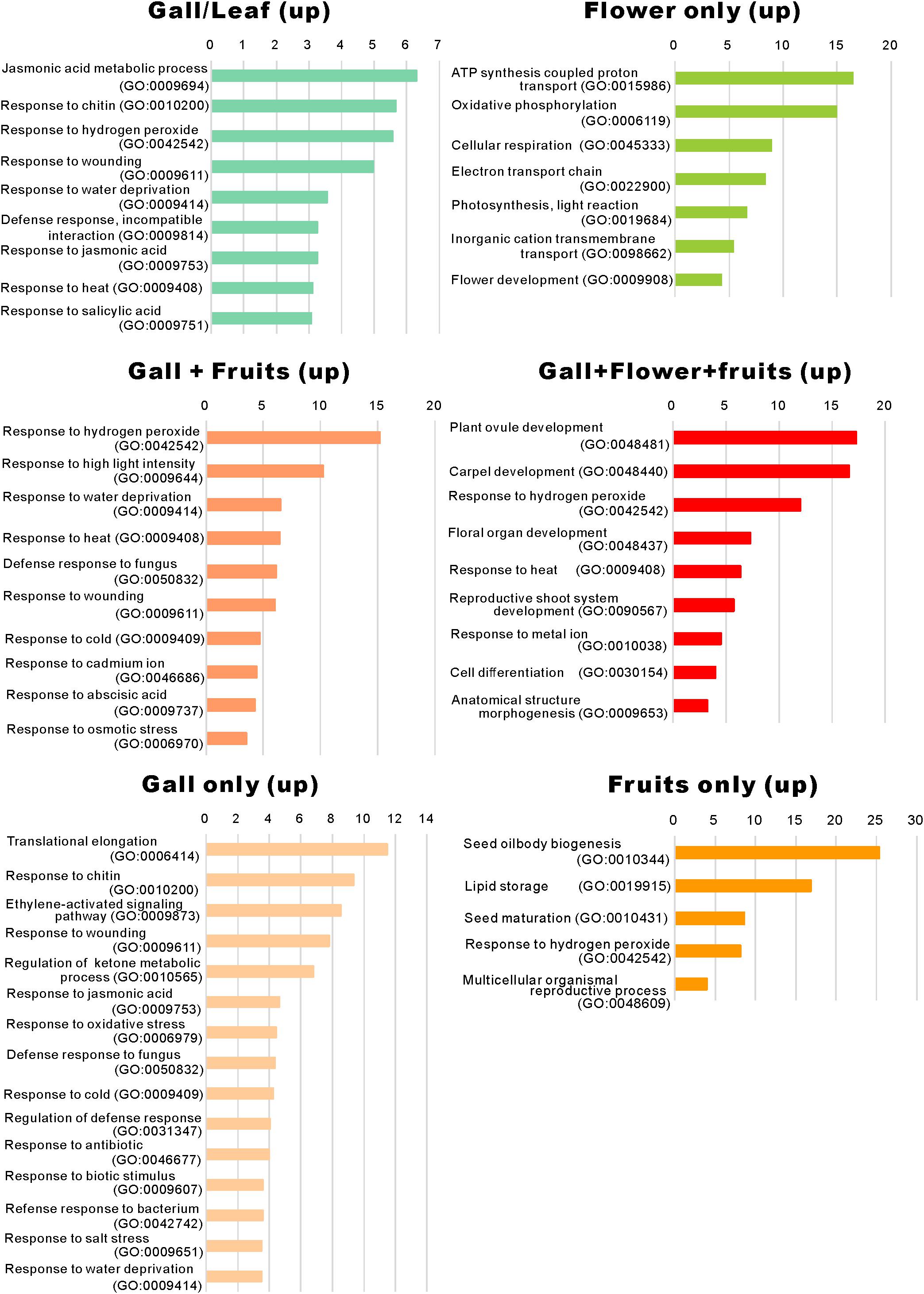
Figure 4. Gene ontology term enrichment of the upregulated genes. The upregulated differentially expressed genes in Figure 2A were subjected to the enrichment analysis (Mi et al., 2019) using the TAIR website (https://www.arabidopsis.org/tools/go_term_enrichment.jsp).
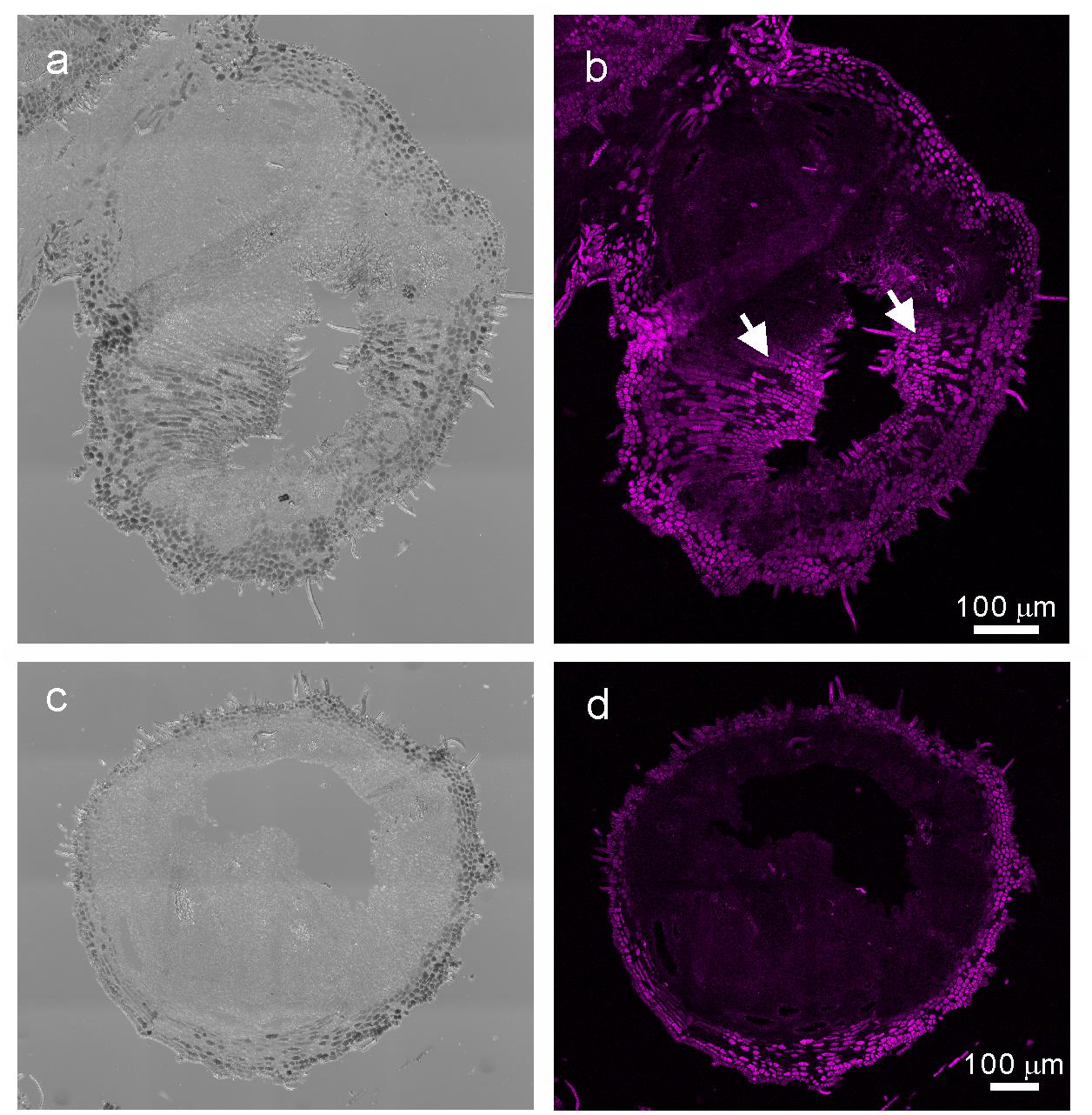
Figure 5. In situ hybridization analysis of R. javanica KNAT6 in the early developing stage of gall. (a) Differential interference contrast (DIC) image of cross-section of the developing gall hybridized with RjKNAT6 anti-sence probe. (b) Fluorescent image of the developing gall hybridized with RjKNAT6 anti-sence probe. (c) Differential interference contrast (DIC) image of cross-section of the developing gall hybridized with RjKNAT6 sence probe. Arrows indicate the positive signals of the anti-sense probe. (d) Fluorescent image of the developing gall hybridized with RjKNAT6 sense probe. The images were taken at least three biologically distinct samples and gave the same results. Scale bars: 200 μm.
In the R. javanica gall tissue, genes involved in the auxin- (IAA17, PILS1, GH3.1, GH3.3, WRKY23, and PBP1), ethylene- (ERF017, ERF022, ERF13, ERF72, and ERF109), and abscisic acid- (NHL6, MAPK3, AHG1, CBF4, ABR1, and RDUF2) response pathways were significantly upregulated (Supplementary Tables S5, S7), whereas genes belonging to the GO category “response to cytokinin (GO:0009735)” were downregulated (Supplementary Table S6), suggesting that several phytohormone signaling pathways may be activated by the actions of the gall-inducing aphid. Since active phytohormones such as indole-3-acetic acid (IAA), abscisic acid, and cytokinins (CKs) have been identified in several gall-inducing insects (Mapes et al., 2001a, b; Dorchin et al., 2009; Yamaguchi et al., 2012; Tanaka et al., 2013), we reasoned that S. chinensis would also produce phytohormones for gall induction on R. javanica leaves. Therefore, we measured the contents of IAA and CKs in the whole aphid bodies, and identified a considerable amount of IAA (718.9 ± 269.0 ng/g FW) and CKs, particularly iP (5.106 ± 1.503 ng/g FW), iPA (7.726 ± 1.451 ng/g FW), and tZR (7.726 ± 1.451 ng/g FW) in the whole aphid body (Table 1). In contrast, the concentrations of these phytohormones in gall and leaf tissues were lower than those in the aphid body (Supplementary Figure S5).
During the gall growth process, the latex ducts and vascular elements become denser in the inner gall layer, the outer epidermal cell layer hardens due to the construction of a lignified secondary cell walls, and the palisade tissue of the galled leaf wings is reorganized and replaced by parenchyma cells (Liu et al., 2014). In this study, we observed that the latex ducts and vascular bundles emerged, and the outer epidermal cell layer began to harden by lignification during the early developmental phase of the galls (Figures 1c,d). In this stage, the genes involved in cell wall synthesis (e.g., CSLD2/6, CESA8, XTH4, and CEL2), production of lignin and suberin deposition (e.g., CAD5, PRX25, CYP84A1, PRX72, MYB58, FAR4, and CER9), and vascular tissue morphogenesis (e.g., CLE44, SEOR, VND7, and TDR) were upregulated (Supplementary Table S7). The changes in these genes might reflect the morphological changes in the early stage of gall development.
We identified that a significant number of genes related to “the jasmonic acid metabolic process” (GO: 0009694) and “response to jasmonic acid” (GO: 0009753), “response to chitin” (GO: 0010200), “response to hydrogen peroxide” (GO: 0042542), “response to wounding” (GO: 0009611), and “response to salicylic acid” (GO: 0009751) were considerably enriched only in the developing gall tissues (Figure 5). After a detailed analysis of these genes, we found that the expressions of AOC3/4, JAZ1/2/8/10, CYP94C1, and JOX2 were dramatically upregulated in the developing gall tissues. Also, a significant number of genes involved in plant–pathogen interactions were upregulated in the gall tissues. In particular, transcription factors related to PAMP-triggered immunity (e.g., WRKY33, WRKY40, MYB51, and TGA9) were highly expressed in the gall tissues (Supplementary Table S7). Additionally, the expression of a significant number of abiotic stress-responsive genes, including those that respond to water deprivation (GO: 0009414) and heat (GO: 0009408), was increased in the gall tissues (Figure 4). Most of these genes (e.g., RD17, LTI45, ERD14, HSFB2A, DDF1, LSR3, and ERD7) respond to drought and heat stresses.
We next categorized the 3809 downregulated genes and found that the genes belonged to GO categories related to photosynthesis (GO: 0097868, 0018298, 009769, 0015977, and 0015995) (Supplementary Tables S4, S6). For instance, the components of photosystem I (PsaD, PsaE, PsaF, PsaG, PsaH, PsaK, PsaN, and PsaO), photosystem II (PsbO, PsbP, PsbR, and PsbW), and carbon fixation (GAPA2, RBCS1A, SBPASE, and CFBBP1) were dramatically downregulated (Figures 2, 6 and Supplementary Table S4). In contrast, the transcripts of various transporters including amino acid transporters (UMAMIT14, and AAP3/4), sugar transporters (SUC2, SWEET7), metal transporters (AMT2, ZIP1), and water transporters (TIP1;3, PIP1;2) were increased in the gall tissues (Figure 6 and Supplementary Table S7).
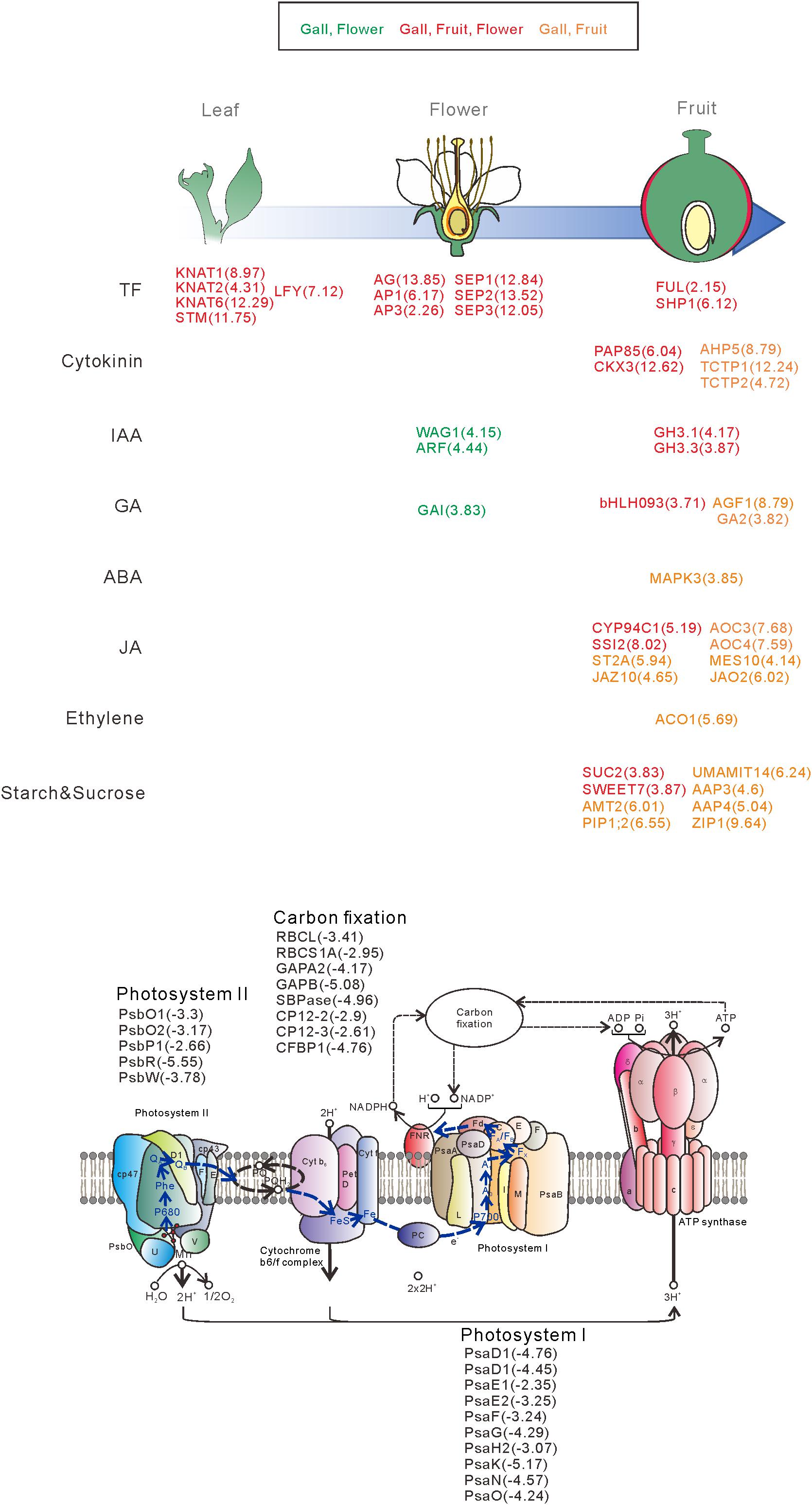
Figure 6. Overview of the representative genes expressed in the galls, flowers, and fruit. The genes are categorized by the developmental stage, the role of the transcription factors, different hormonal signals, and transporters. The numbers in parentheses represent the log-fold change values of the differentially expressed genes compared with the young leaves. Upregulatd genes in gall and flower (Green), gall, fruit, and flower (red), gall and fruit (orange).
One of important characteristics of galls is their function as sinks for insect nutrients (Rohfritsch and Shorthouse, 1982). The existence of insect galls near the source organs changes the flow of plant resources by partially blocking and redirecting the resources from the original sink organs to the galls. Thus, gall formation results in making a stronger sink than plant sink organs such as buds, flowers, and fruit (Weis and Kapelinski, 1984; McCrea et al., 1985; Burstein et al., 1994; Larson and Whitham, 1997). In R. javanica, galls start to develop on the leaf wing when the fundatrix of S. chinensis feeds on the surface of the leaf wing. In the early stage of gall development, the feeding site on the leaf wing tissue grows abnormally to form hyperplastic tissues, in which the outer epidermal layer of the gall is covered with denser trichomes and is lignified to form a rigid structure. The palisade tissues of the leaf wing are reorganized and replaced by dedifferentiated parenchyma cells, and the latex ducts and vascular elements become denser in the inner gall layer and close to the gall cavity (Liu et al., 2014). Throughout this process, a galling aphid such as S. chinensis creates horned galls as a novel source organ on the leaf wing.
In the case of the grape gall formation by phylloxera, the expression of genes associated with “light harvesting and photosynthetic carbon assimilation” strongly decreased, whereas that of the transcripts associated with “sucrose mobilization” and “glycolysis and fermentation” considerably increased in gall tissues compared to that in ungalled leaf tissues (Nabity et al., 2013). In this study, we revealed that the photosynthesis-related genes involved in the photosystem I (GO: 0009768), photosystem II (GO: 0009769), and carbon fixation pathway (GO: 0015977) were dramatically downregulated during the early gall development in R. chinensis. In contrast, the expression of the genes involved in the translational elongation process (GO: 0006414) increased, suggesting that genes related to de novo protein synthesis necessary for secondary metabolites in gall tissues are activated during gall formation. Through a detailed analysis of the upregulated genes involved in this process, we found that a significant number of molecular chaperons, ribosomal proteins, and various transporter genes such as sugar and amino acid transporters were highly expressed in the gall tissues (Supplementary Tables S5, S7).
Pathogens acquire glucose from their hosts, thereby hijacking host sugar efflux systems (Sutton et al., 1999; Voegele et al., 2001). In particular, SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTERS (SWEETs) sugar transporters have been reported to be utilized by pathogens for the acquisition of sugars. For instance, the rice homologs SWEET11 and SWEET14 are specifically exploited by bacterial symbionts and fungal and bacterial pathogens, indicating that the sugar efflux function of SWEET transporters is probably targeted by pathogens and symbionts for nutritional gain (Chen et al., 2010). Plasmodiophora brassicae is the causal agent of clubroot, a severe disease of Brassica crops. The pathogen lives inside roots, and hijacks nutrient sink in infected roots to trigger active sugar translocation between the sugar producing tissues and the clubbed tissues recruiting the SWEET sucrose transporters within developing galls (Li et al., 2018; Walerowski et al., 2018). In this study, we found that the expression of Arabidopsis SWEET7 homolog is increased in the early development stage of gall tissues, indicating that the expression of SWEET sugar transporter gene of R. javanica is likely activated by the feeding action of S. chinensis.
Collectively, the changes in the expression profile during gall formation imply that the cells of the palisade tissues in the leaf wing were reorganized to be de-differentiated into parenchyma cells, thereby losing their photosynthetic and reconstruction cellular functions, to convert the tissues architecture of the leaf wing from source to sink tissues during the gall development process.
Plants respond to herbivory with the induction of a combination of defense responses such as salicylic acid, jasmonic acid, and ethylene signaling pathways to produce toxins and defensive proteins that target physiological processes in insects (Agrawal et al., 1999; De Vos et al., 2005). Aphid feeding is perceived by plants as pathogenic and herbivory; hence, on sensing the phytopathogens and mechanical damage caused by stylet probing, the plants elicit a defense response that involves both salicylic and jasmonic acid pathways (Kaloshian and Walling, 2005; Thompson and Goggin, 2006; Gao et al., 2007). It has been reported that the other known minor pathways including ethylene, abscisic acid, gibberellic acid, nitric oxide, and auxin are also activated in response to aphid feeding (Smith and Boyko, 2007).
An investigation on the molecular response of gall formation by the gall-inducing aphid S. chinensis on R. chinensis by comparing expression profiles of leaves and 100-day grown galls has demonstrated that the genes involved in the biosynthesis of secondary metabolites, plant–aphid interactions, and plant hormone signal transduction were highly expressed in galls (Wang et al., 2017). In this study, we compared the expression profile of the early developmental galls with that of the early developmental leaves, and found that the salicylic acid-, jasmonic acid-, and ethylene-response pathways were considerably activated in the gall tissues. In particular, genes related to the wounding response and PAMP-triggered immunity (Supplementary Table S7) were highly expressed in the gall tissues. These defense responses were likely induced by sensing the feeding stress of gall-inducing aphid for protecting the plant body from the aphid’s invasion of plant signaling pathways driven by jasmonic acid, salicylic acid, ethylene, abscisic acid, and gibberellic acid. However, the growth and proliferation of S. chinensis inside the gall seemed to be unaffected, which was probably because the aphids adapted to, and overcame the host’s defense systems, suggesting that the gall-growing leaves or plants to become more tolerant against other pathogenic organisms than those are not induced gall.
During the growing process of the horned gall, caused by the gall-inducing aphid S. chinensis, a combination of cell division (hyperplasia) and growth (hypertrophy) occurs in several layers of tissues, inducing the formation of nutritive and protective gall tissues (Liu et al., 2014). It has long been hypothesized that phytohormones produced by gall-inducing insects play a key role in gall formation (Tooker and Helms, 2014). Indeed, active phytohormones such as IAA and CKs have been identified in several gall-inducing insects at various concentrations (e.g., IAA = 60–9000 ng g fw–1, iP = 3–350 ng g fw–1, iPR = 8–190 ng g fw–1, tZ = 2–1300 ng g fw–1, and tZR = 0.4–70 ng g fw–1) (Mapes et al., 2001a, b; Dorchin et al., 2009; Yamaguchi et al., 2012; Tanaka et al., 2013), and the application of exogenous phytohormones led to induction of gall-like structures in plants (Bartlett and Connor, 2014). We found that the concentrations of IAA and CKs in S. chinensis fell within the ranges of those in the gall-inducing insects. The contents of IAA and CKs were considerably higher than those in all and leaf tissues, suggesting that the aphids that may control gall-inducing process on R. javanica leaves by producing phytohormones such as IAA and CKs themselves. Given that IAA and CKs are involved in abnormal cell division, cellular enlargement, and differentiation, our results suggest that the phytohormones secreted from the aphids may be involved in the gall formation reported here, although we could not completely rule out the possibility that the phytophormones in aphid are derived from the host plant tissues.
Some of insect galls are believed to resemble flowers or fruits in their morphology (Darwin, 1868), suggesting that the developmental program of the reproductive organs of plants may be hijacked and exploited by gall-inducing insects. Recently, Schultz et al. (2018) reported that reproductive genes involved in floral organ development were significantly enriched in the developing galls of wild grapevine that were induced by Phylloxera, suggesting that a galling insect utilizes plant reproductive programs during gall development. In the present study, we also found that a significant number of transcription factors involved in floral organ development were upregulated during R. javanica gall development. Of these genes, LFY is an important master regulator for the transition from vegetative to reproductive phase during meristem development (Blázquez et al., 1997). Constitutive expression of LFY under the 35S promoter causes the conversion of indeterminate lateral meristems into flowers and the conversion of the inflorescence meristem into a flower (Weigel and Nilsson, 1995; Siriwardana and Lamb, 2012). AP1, AP2, and AG determine the floral organ identity, and A-, B-, and C-type floral MADS-box genes (Alvarez-Buylla et al., 2010) in combination with the SEP1/2/3 MADS-box subfamily being required for specifying the “floral state” (Alvarez-Buylla et al., 2010). The FUL and SHP1 genes are also members of the MADS-box transcription factors, and are involved in valve development and differentiation of both the lignified layer and separation layer of the valve margin in the developing ovaries, respectively (Gu et al., 1998; Liljegren and Bowman, 2000).
In addition to these genes, in the present study, we found that the class-1 KNOTTED1-like homeobox (KNOX) genes (KNAT1/2/6, and STM) were highly expressed in the developing gall tissues. The KNOX genes are found in all higher plant species and encode homeodomain transcription factors similar to those that regulate development in animals (Scofield and Murray, 2006; Hay and Tsiantis, 2010). The class-1 KNOX genes in Arabidopsis are expressed in the shoot apical meristem (SAM) but not in the lateral organs (Lincoln et al., 1994; Pautot et al., 2001; Lenhard et al., 2002; Dean et al., 2004). It has been reported that the ectopic overexpression of STM or KNAT1 leads to the formation of ectopic knot-like meristematic structures, which results in formation of lobed leaves on the adaxial surface of the leaf (Chuck et al., 1996; Brand et al., 2002; Lenhard et al., 2002). When gall formation begins, the development of ectopic meristematic structures could be made from leaf wing tissues. Similarities in organ structure between the lobed leaves caused by STM or KNAT1 overexpression and the initiation stage of the gall structure imply that the ectopic overexpression of the class-1 KNOT genes induced by gall-inducing insects may initiate de-differentiation of the leaf wing cells to generate a meristematic region followed by the formation of an initial gall structure on the leaf wing.
From these results, we propose the following molecular mechanisms of the early stage of gall formation in R. javanica: (i) an ectopic meristematic structure is generated by the overexpression of class-1 KNOX genes, (ii) the ectopic meristem is converted to floral-like meristem by the expression of LFY, (iii) the floral-like meristem develops to form fruit-like gall structures induced by expression of floral regulatory genes, and (iv) during the transformation from leaf to gall tissues, many photosynthetic genes are downregulated, while transporter and secondary metabolic genes are upregulated to change the tissue functions (Figure 6).
The datasets generated for this study can be found in the NCBI using the following link: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJDB8441.
TH, MS, and IO conceived and designed the study. TH, ST, and AO collected samples and extracted RNA. TH and TN performed qRT-PCR analysis. YS performed a quantitative analysis of indole-3-acetic acid and cytokinins in S. chinensis. TM and YI measured the content of various phytohormones in R. javanica tissues. TS and SK constructed library, and performed RNA-sequencing. TS, SK, TH, ST, and MS analyzed the data.
This work was supported by the Japan Society for the Promotion of Science; a-Grant-in-Aid for Scientific Research (A) (Grant No. 19H00933) to MS, a-Grant-in-Aid for challenging Exploratory Research (Grant No. 17H06260) to IO, the strategic prioritized research in KPU to IO and MS, and a Grant-in-Aid for Scientific Research on Innovative Areas (Grant Nos. 18H04787 and 18H04844) to SK, and by a grant from the MEXT Supported Program for the Strategic Research Foundation at Private Universities of the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant No. S1511023) to SK. This work was supported by the Joint Usage/Research Center, Institute of Plant Science and Resources, Okayama University to YI.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ms. Kaori Kaminoyama (Kyoto Sangyo University) for technical help for library construction and RNA–sequencing. We also thank N. Ohtsubo (Kyoto Prefectural University) for fruitful discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00471/full#supplementary-material
FIGURE S1 | Images of the sampled (a) R. javanica leaf attaching several phase 4 stage galls at the wing region, (b) young leaf (c) female flowers.
FIGURE S2 | Sizes and numbers of aphids inside of various stages of galls. (a) The number of aphids inside of the developmental galls. (b–e) Images of the developmental galls corresponding to the plots in (a).
FIGURE S3 | Principal component analysis of the gene expression profile. (a) Eigenvalues and the cumulative contribution ratio (8%). Bars and open circles represent the eigenvalues and cumulative contribution ratio, respectively. (b) The global expression profile of each transcript as principal components 1 and 2.
FIGURE S4 | Amino acid sequence alignment of several orthologous genes of A. thaliana and R. javanica. AP1, APETALA1 (a); SEP2, SEPALLATA2; CLE41, CLAVATA3/ESR-RELATED 41 (b); CYCD4;1, CYCLIND4; 1 (c).
FIGURE S5 | Phytohormone contents in R. javanica gall and leaf tissues. The results are given as the mean ± standard deviation from five replicates. Abbreviations: tZ, trans-zeatin iP, isopentenyladenine; IAA, indole-3-acetic acid. Experiments were repeated at least three times using three biologically distinct samples.
TABLE S1 | Results of RNA-seq analyzes of various R. javanica tissues.
TABLE S2 | List of primers used for qRT-PCR.
TABLE S3 | Transcriptome sequencing and summary statics of de novo assembly.
TABLE S4 | List of upregulated genes in gall, flower, fruit of R. javanica.
TABLE S5 | List of downregulated genes in gall, flower, fruit of R. javanica.
TABLE S6 | Result of GO enrichment analysis of up-regulated gens in gall of R. javanica.
TABLE S7 | Result of GO enrichment analysis of down-regulated gens in gall of R. javanica.
TABLE S8 | List of upregulated genes categorized by the biological processes.
Agrawal, A. A., Tuzun, S., and Bent, E. (1999). Induced Plant Defenses Against Pathogens and Herbivores. Biochemistry, Ecology, and Agriculture. St Paul, MN: APS Press. doi: 10.2307/177119
Alvarez-Buylla, E. R., Benítez, M., Corvera-Poiré, A., Chaos Cador, Á., de Folter, S., Gamboa de Buen, A., et al. (2010). Flower development. Arabidopsis Book 8:e0127. doi: 10.1199/tab.0127
Aoki, S., and Kurosu, U. (2010). A review of the biology of Cerataphidini (Hemiptera, Aphididae, Hormaphidinae), focusing mainly on their life cycles, gall formation, and soldiers. Psyche 2010:380351. doi: 10.1155/2010/380351
Bartlett, L., and Connor, E. F. (2014). Exogenous phytohormones and the induction of plant galls by insects. Arthropod Plant Interact. 8, 339–348.
Blackman, R. L., and Eastop, V. F. (2007). Aphids on the World’ s Herbaceous. Chichester: John Wiley & Sons Ltd.
Blázquez, M. A., Soowal, L. N., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844.
Brand, U., Gru, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. doi: 10.1104/pp.001867.1
Buchfink, B., Xxie, C., and Hunson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Burstein, M., Wool, D., and Eshel, A. (1994). Sink strength and clone size of sympatric, gall-forming aphids. Eur. J. Entomol. 91, 57–61.
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. doi: 10.1105/tpc.8.8.1277
Darwin, C. (1868). The Variation of Animals and Plants Under Domestication, Vol. 2. London: John Murray.
De Vos, M., Van Oosten, V. R., Van Poecke, R. M., Van Pelt, J. A., Pozo, M. J., Mueller, M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18, 923–937. doi: 10.1094/mpmi-18-0923
Dean, G., Casson, S., and Lindsey, K. (2004). KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol. Biol. 54, 71–84. doi: 10.1023/B:PLAN.0000028772.22892.2d
Dorchin, N., Hoffmann, J. H., Stirk, W. A., Novák, O., Strnad, M., and van Staden, J. (2009). Sexually dimorphic gall structures correspond to differential phytohormone contents in male and female wasp larvae. Physiol. Entomol. 34, 359–369. doi: 10.1111/j.1365-3032.2009.00702.x
Dreger-Jauffret, F., and Shorthouse, J. D. (1992). “Diversity of gall-inducing insects and their galls,” in Biology of Insect-Induced Galls, eds J. D. Shorthouse and O. Rohfritsch (New York, NY: Oxford University Press), 8–33.
Ferreira, B. G., and Isaias, R. M. S. (2014). Floral-like destiny induced by a galling Cecidomyiidae on the axillary buds of Marcetia taxifolia (Melastomataceae). Flora 209, 391–400. doi: 10.1016/j.flora.2014.06.004
Gao, L., Anderson, J. P., Klingler, J. P., Nair, R. M., Edwards, O. R., and Singh, K. B. (2007). Involvement of the octadecanoid pathway in bluegreen aphid resistance in Medicago truncatula. Mol. Plant Microbe Interact. 20, 82–93. doi: 10.1094/mpmi-20-0082
Giron, D., Huguet, E., Stone, G. N., and Body, M. (2016). Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J. Insect Physiol. 84, 70–89. doi: 10.1016/j.jinsphys.2015.12.009
Gu, Q., Ferrandiz, C., Yanofsky, M. F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. doi: 10.1105/tpc.1.1.37
Guiguet, A., Hamatani, A., Amano, T., Takeda, S., Lopez-vaamonde, C., Giron, D., et al. (2018). Inside the horn of plenty: leaf-mining micromoth manipulates its host plant to obtain unending food provisioning. PLoS One 13:e0209485. doi: 10.1371/journal.pone.0209485
Guiguet, A., Ohshima, I., Takeda, S., Laurans, F., Lopez-Vaamonde, C., and Giron, D. (2019). Origin of gall-inducing from leaf-mining in a micromoth showing a mixed feeding strategy. Sci. Rep. 9:6794.
Hay, A., and Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049
Kaloshian, I., and Walling, L. L. (2005). Hemipterans as plant pathogens. Annu. Rev. Phytopathol. 43, 491–521. doi: 10.1146/annurev.phyto.43.040204.135944
Kurosu, U., and Aoki, S. (1990). Formation of a “Cat’s-paw” gall by aphid Ceratovacuna nekoashi (Homoptera). Jpn. J. Entomol. 58, 155–166.
Larson, K. C., and Whitham, T. G. (1997). Competition between gall aphids and natural plant sinks: plant architecture affects resistance to galling. Oecologia 109, 575–582. doi: 10.1007/s004420050119
Lenhard, M., Jürgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206.
Li, H., Li, X., Xuan, Y., Jiang, J., Wei, Y., and Piao, Z. (2018). Genome wide identification and expression profiling of SWEET genes family reveals its role during Plasmodiophora brassicae-induced formation of clubroot in Brassica rapa. Front. Plant Sci. 9:207. doi: 10.3389/fpls.2018.00207
Liljegren, S., and Bowman, J. L. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hakeaibi, S. (1994). A knottedl-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. doi: 10.1105/tpc.6.12.1859
Liu, A. P., Yang, Z. X., Chen, X. M., Foottit, R. G., and Liu, P. (2014). The effect of the gall-forming aphid Schlechtendalia chinensis (Hemiptera: Aphididae) on leaf wing ontogenesis in Rhus chinensis (Sapindales: Anacardiaceae). Ann. Entomol. Soc. Am. 107, 242–250. doi: 10.1603/AN13118
Mapes, C. C., Davies, P. J., and Mapes, C. C. (2001a). Cytokinins in the ball gall of Solidago altissima and in the gall forming larvae of Eurosta solidaginis. New Phytol. 151, 203–212. doi: 10.1046/j.1469-8137.2001.00158.x
Mapes, C. C., Davies, P. J., and Mapes, C. C. (2001b). Indole-3-acetic acid and ball gall development on Solidago altissima. Development 151, 195–202. doi: 10.1046/j.1469-8137.2001.00161.x
Matsukura, K., Matsumura, M., and Tokuda, M. (2009). Host manipulation by the orange leafhopper Cicadulina bipunctata: gall induction on distant leaves by dose-dependent stimulation. Naturwissenschaften 96, 1059–1066. doi: 10.1007/s00114-009-0566-1
McCrea, K. D., Abrahamson, W. G., and Weis, A. E. (1985). Goldenrod ball gall effects on Solidago altissima 14C translocation and growth. Ecology 66, 1902–1907. doi: 10.2307/2937386
Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mils, C., Guo, X., et al. (2019) Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protocols 14, 703–721. doi: 10.1038/s41596-019-0128-8
Miles, P. W. (1968). Insect secretions in plants. Annu. Rev. Phytopathol. 6, 137–164. doi: 10.1146/annurev.py.06.090168.001033
Nabity, P. D., Haus, M. J., Berenbaum, M. R., and Delucia, E. H. (2013). Leaf-galling phylloxera on grapes reprograms host metabolism and morphology. Proc. Natl. Acad. Sci. U.S.A. 110, 16663–16668. doi: 10.1073/pnas.1220219110
Pautot, V., Dockx, J., Hamant, O., Kronenberger, J., Grandjean, O., Jublot, D., et al. (2001). KNAT2: evidence for a link between knotted-like genes and carpel development. Plant Cell 13, 1719–1734. doi: 10.1105/tpc.010184
Price, P. W., Fernandes, G. W., and Waring, G. L. (1987). Adaptive nature of insect galls. Environ. Entomol. 16, 15–24. doi: 10.1093/ee/16.1.15
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinfomatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rohfritsch, O., and Shorthouse, J. D. (1982). “Insect galls,” in Molecular Biology of Plant Tumors, eds G. Kahl and J. Schell (New York, NY: Academic Press), 121–152.
Schultz, J. C., Edger, P. P., Body, M. J. A., and Appel, H. M. (2018). A galling insect activates plant reproductive programs during gall development. Sci. Rep. 9:1833.
Scofield, S., and Murray, J. A. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60, 929–946. doi: 10.1007/s11103-005-4478-y
Siriwardana, N. S., and Lamb, R. S. (2012). The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation. Int. J. Dev. Biol. 56, 207–221. doi: 10.1387/ijdb.113450ns
Smith, C. M., and Boyko, E. V. (2007). The molecular bases of plant resistance and defenses to aphid feeding: current status. Entomol. Exp. Appl. 122, 1–16. doi: 10.1111/j.1570-7458.2006.00503.x
Sopow, S., Shorthouse, J. D., Strong, W., and Quiring, D. T. (2003). Evidence for long-distance, chemical gall induction by an insect. Ecol. Lett. 6, 102–105. doi: 10.1046/j.1461-0248.2003.00410.x
Stone, G. N., and Schönrogge, K. (2003). The adaptive significance of insect gall morphology. J. Trends Ecol. Evol. 18, 512–522. doi: 10.1016/S0169-5347(03)00247-7
Stuart, J. J., Chen, M.-S., Shukle, R., and Harris, M. O. (2012). Gall midges (Hessian flies) as plant pathogens. Annu. Rev. Phytopathol. 50, 339–357. doi: 10.1146/annurev-phyto-072910-095255
Sutton, P. N., Henry, M. J., and Hall, J. L. (1999). Glucose, and not sucrose, is transported from wheat to wheat powdery mildew. Planta 208, 426–430. doi: 10.1007/s004250050578
Tanaka, Y., Okada, K., Asami, T., and Suzuki, Y. (2013). Phytohormones in Japanese mugwort gall induction by a gall-inducing gall midge phytohormones in Japanese mugwort gall induction. Biosci. Biotechnol. Biochem. 77, 1942–1948. doi: 10.1271/bbb.130406
Thompson, G. A., and Goggin, F. L. (2006). Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 57, 755–766. doi: 10.1093/jxb/erj135
Tooker, J. F., and Helms, A. M. (2014). Phytohormone dynamics associated with gall insects, and their potential role in the evolution of the gall-inducing habit. J. Chem. Ecol. 40, 742–753. doi: 10.1007/s10886-014-0457-6
Voegele, R. T., Struck, C., Hahn, M., and Mendgen, K. (2001). The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc. Natl. Acad. Sci. U.S.A. 98, 8133–8138. doi: 10.1073/pnas.131186798
Walerowski, P., Gündel, A., Yahaya, N., Truman, W., Sobczak, M., Olszak, M., et al. (2018). Clubroot disease stimulates early steps of phloem differentiation and recruits SWEET sucrose transporters within developing galls. Plant Cell 30, 3058–3073. doi: 10.1105/tpc.18.00886
Wang, H., Cui, K., Shao, S., Liu, J., Chen, H., Wang, C., et al. (2017). Molecular response of gall induction by aphid Schlechtendalia chinensis (Bell) attack on Rhus chinensis Mill. J. Plant Interact. 12, 465–479. doi: 10.1080/17429145.2017.1392627
Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500. doi: 10.1038/377495a0
Weis, A. E., and Kapelinski, A. (1984). Manipulation of host plant development by the gall-midge Rhabdophaga strobiloides. Ecol. Entomol. 9, 457–465. doi: 10.1111/j.1365-2311.1984.tb00844.x
Wool, D. (2004). Galling aphids: specialization, biological complexity, and variation. Annu. Rev. Entomol. 49, 175–192. doi: 10.1146/annurev.ento.49.061802.123236
Yamaguchi, H., Tanaka, H., Hasegawa, M., Tokuda, M., Asami, T., and Suzuki, Y. (2012). Phytohormones and willow gall induction by a gall-inducing sawfly. New Phytol. 196, 586–595. doi: 10.1111/j.1469-8137.2012.04264.x
Keywords: Rhus javanica, floral organ development, gall formation, Schlechtendalia chinensis, RNA-seq analysis
Citation: Hirano T, Kimura S, Sakamoto T, Okamoto A, Nakayama T, Matsuura T, Ikeda Y, Takeda S, Suzuki Y, Ohshima I and Sato MH (2020) Reprogramming of the Developmental Program of Rhus javanica During Initial Stage of Gall Induction by Schlechtendalia chinensis. Front. Plant Sci. 11:471. doi: 10.3389/fpls.2020.00471
Received: 24 August 2019; Accepted: 30 March 2020;
Published: 15 May 2020.
Edited by:
Essaid Ait Barka, Université de Reims Champagne-Ardenne, FranceReviewed by:
Marion O. Harris, North Dakota State University, United StatesCopyright © 2020 Hirano, Kimura, Sakamoto, Okamoto, Nakayama, Matsuura, Ikeda, Takeda, Suzuki, Ohshima and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masa H. Sato, bWhzYXRvQGtwdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.