
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 06 May 2020
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00452
This article is part of the Research Topic Chromatin Stability and Dynamics: Targeting and Recruitment of Chromatin Modifiers View all 10 articles
 Linhao Xu
Linhao Xu Hua Jiang*
Hua Jiang*In eukaryotes, histone H3 lysine 9 methylation (H3K9me) mediates the silencing of invasive and repetitive sequences by preventing the expression of aberrant gene products and the activation of transposition. In Arabidopsis, while it is well known that dimethylation of histone H3 at lysine 9 (H3K9me2) is maintained through a feedback loop between H3K9me2 and DNA methylation, the details of the H3K9me2-dependent silencing pathway have not been fully elucidated. Recently, the regulation and the function of H3K9 methylation have been extensively characterized. In this review, we summarize work from the recent studies regarding the regulation of H3K9me2, emphasizing the process of deposition and reading and the biological significance of H3K9me2 in Arabidopsis.
In eukaryotic cells, chromatin is divided into two major types of compartments: heterochromatin and euchromatin, reflecting the repressive and permissive potential for transcription in these regions, respectively (Ding et al., 2007). Chromatin is rich in repetitive sequences and transposable elements inside and near centromeres, posing a risk for genome instability through their potential for transposition and meiotic recombination. Thus, during the whole life cycle, it is necessary to keep these regions inaccessible, condensed, and transcriptionally silent. Such regions are classified as constitutive heterochromatin (Saksouk et al., 2015). In contrast, facultative heterochromatin refers to regions whose compaction and silencing are dynamic in the life cycle or under stress stimuli, mainly distributed in chromosomal arms (Trojer and Reinberg, 2007).
Chromatin states are modulated by modifications at the N-terminal tails of histones, DNA methylation, and different histone variants (Jenuwein and Allis, 2001). Histone H3K9 methylation is a critical marker for transcriptional silencing and heterochromatin formation, mostly constitutive heterochromatin formation. Methylation states at H3K9 can be mono- (H3K9me1), di- (H3K9me2), or tri- (H3K9me3) methylation. In mammals, H3K9me3 is the most abundant marker in constitutive heterochromatin (Peters et al., 2003; Rice et al., 2003). However, in plants, the modification of H3K9me1 and H3K9me2 is rich in constitutive heterochromatin and only slightly present in facultative chromatin, whereas H3K9me3 is distributed with a high concentration in euchromatin and at expressed genes (Naumann et al., 2005). In Arabidopsis, H3K9me3 methylation broadly marks 40% of all genes (Roudier et al., 2009), but only a low level of H3K9me3 can be detected in regions with transposons and pseudogenes (Charron et al., 2009). Thus, the function of H3K9me3 has been altered in Arabidopsis compared to H3K9me3 in yeast and mammals.
H3K9me2 is mainly catalyzed by the histone methyltransferases KRYPTONITE (KYP), SUVH5, and SUVH6 in Arabidopsis and is maintained through the feedback loop between H3K9me2 and non-CG methylation (Du et al., 2015). Several studies have shown more details of H3K9me2 deposition with the structural analysis of KYP/SUVH5/SUVH6 and their role in H3K9me2 deposition (Du et al., 2014; Li et al., 2018) and other H3K9 methyltransferases (Caro et al., 2012) and cofactors (Yu et al., 2017). The downstream part of H3K9me2-dependent silencing has also been investigated by identifying a novel H3K9 reader (Zhang C. et al., 2018; Zhao et al., 2019). In this article, we review the writing, reading, and biological roles of H3K9 methylation in Arabidopsis.
Histone lysine methyltransferases usually contain a catalytic SET domain, which is named after three Drosophila melanogaster genes, Su(var)3-9, E(z), and Trx (Jenuwein et al., 1998). In fission yeast, there is only one H3K9 methyltransferase, Clr4/KMT1, which is responsible for all three states of H3K9 methylation (Nakayama et al., 2001; Figure 1). In mammals, there are multiple H3K9 methyltransferases with different catalytic activities and target genes (Sims et al., 2003; Dodge et al., 2004; Shinkai and Tachibana, 2011; Figure 1). SUV39H1 and SUVH39H2 mono- and dimethylase catalyze di-and trimethylation in constitutive heterochromatic regions, SETDB1 monomethylates at the pericentromeric region, and the heterodimer of G9a and G9a-like protein (GLP) catalyzes di- and trimethylation in euchromatic regions.
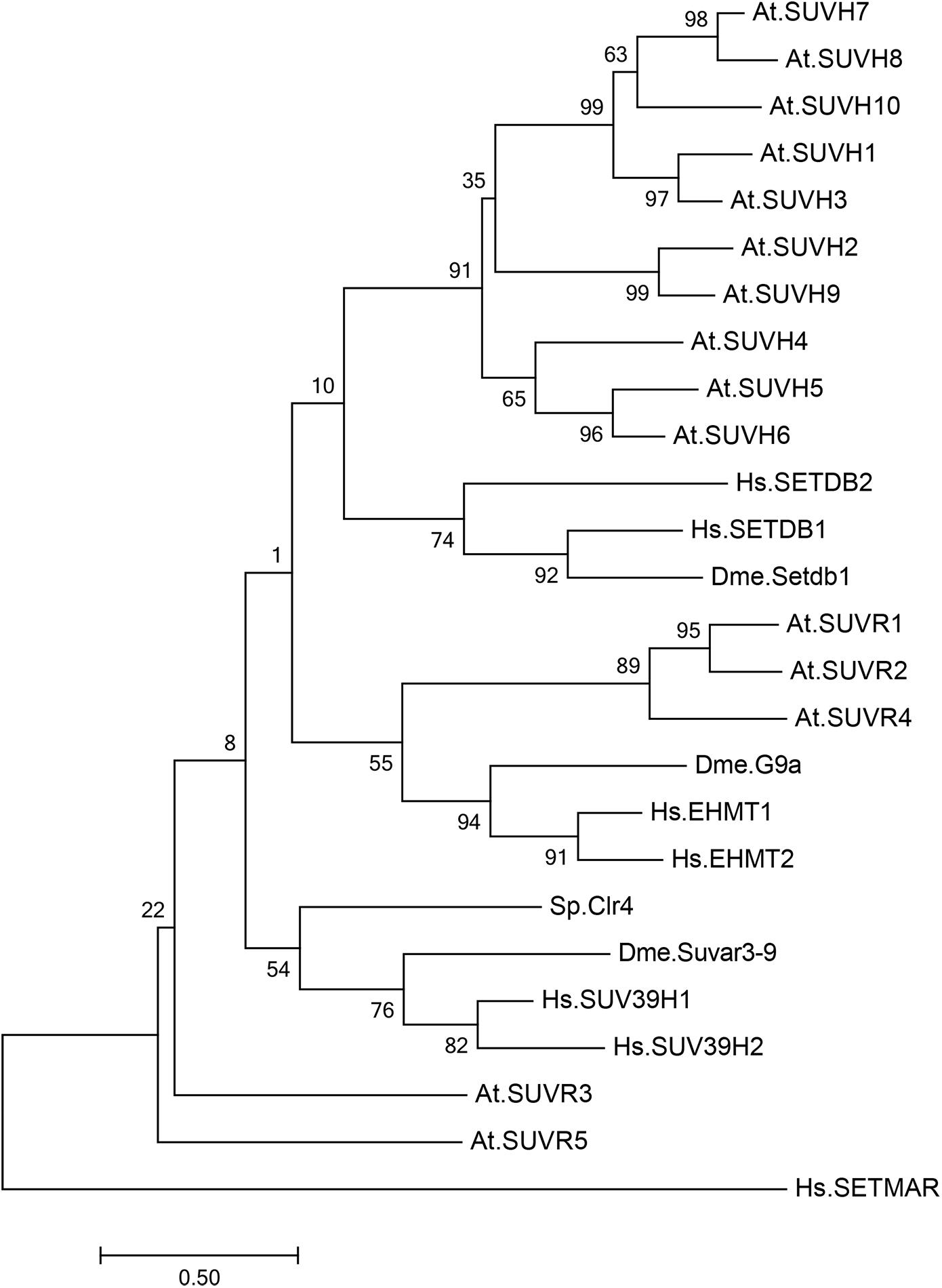
Figure 1. Phylogenetic analysis of SU(VAR)3–9 homologous proteins in Arabidopsis thaliana, Drosophila melanogaster, Schizosaccharomyces pombe, and Homo sapiens. Phylogenetic analysis of 15 SU(VAR)3–9 homologous protein sequences from Arabidopsis thaliana (At), three SU(VAR)3–9 homologous protein sequences from Drosophila melanogaster (Dme), one SU(VAR)3–9 homologous protein sequence from Schizosaccharomyces pombe (Sp), and five SU(VAR)3–9 homologous protein sequences from Homo sapiens (Hs). The evolutionary history was inferred by using the maximum likelihood method based on the Poisson correction model. Phylogenetic analysis was performed using MEGA 7.0.
In Arabidopsis, there are 15 SET-domain proteins that are related to SU(VAR)3-9 (Baumbusch et al., 2001; Lei et al., 2012; Zhang and Ma, 2012; Figure 1). Ten of these proteins are classified as SU(VAR)3-9 HOMOLOGS (SUVH1-SUVH9), and the remaining five are classified as SU(VAR)3-9-RELATED proteins (SUVR1-SUVR5) (Table 1). Among the nine SUVHs, KYP/SUVH4, SUVH5, and SUVH6 have been well identified as H3K9 methyltransferases responsible for maintaining H3K9 methylation. KYP mediates the majority of H3K9me2 methylation in both constitutive and facultative heterochromatin in Arabidopsis, while SUVH5 and SUVH6 only play minor roles in H3K9me2 methylation (Jackson et al., 2002, 2004; Stroud et al., 2014; Li et al., 2018). Crystal structures of KYP, SUVH5, and SUVH6 reveal that the post-SET domain is critical for enzymatic activity (Li et al., 2018); thus, SUVH2 and SUVH9, which lack the post-SET domain, are enzymatically inactive (Johnson et al., 2014). The remaining SUVH1, SUVH3, SUVH7, and SUVH8 were recently reported to function in transcriptional activation but not silencing, expanding the roles of SUVHs in transcriptional regulation (Harris et al., 2018; Xiao et al., 2019). Nevertheless, SUVH7 and SUVH8 are both primarily expressed and imprinted in the endosperm (Gehring et al., 2011; Wolff et al., 2011), indicating an endosperm-specific targeting mechanism favoring a relatively specific chromatin environment. Indeed, SUVH7 has already been shown to play a role in establishing postzygotic hybridization barriers established by H3K9me2 (Wolff et al., 2015; Jiang et al., 2017). Interestingly, computational characterization predicts that SUVH7 and SUVH8 are capable of catalyzing H3K9me1 and H3K9me2 methylation. Two critical residues in the catalytic pocket, Tyr1124 and Phe1209, determine the product specificity in GLP, a G9a-related methyltransferase (Wu et al., 2010). Meanwhile, H3K9me1 or H3K9me2 is correlated with the presence of Tyr in one of the positions and non-Tyr in the other, indicating that the two SUVHs are capable of catalyzing H3K9me1 or H3K9me2. Thus, SUVH7 and SUVH8 may function as methyltransferases for endosperm-specific H3K9me2 deposition. Taken together, KYP, SUVH5, and SUVH6 are the general H3K9 methyltransferases in Arabidopsis, and it is possible that SUVH7 and SUVH8 act as endosperm-specific methyltransferases.
Among the five SUVRs, SUVR1, and SUVR2 have shown no HMTase activity in an in vitro enzymatic assay, but SUVR4 has HMTase activity to convert H3K9me1 to H3K9me2 (ubiquitin) and H3K9me3 (without ubiquitin) in vitro (Thorstensen et al., 2006; Veiseth et al., 2011). The level of H3K9me3 is correlated with the amount of SUVR4-GFP in Arabidopsis nuclei, but the effect of genome-wide H3K9mer3 has not been determined (Veiseth et al., 2011). SUVR5 is capable of establishing H3K9me2 in a DNA methylation–independent manner and is involved in the response to environmental or developmental cues (Caro et al., 2012).
DNA methylation is tightly connected with H3K9 methylation. In Neurospora crassa, the H3K9 methyltransferase DIM5 establishes H3K9me3, and then heterochromatin protein 1 (HP1) recognizes H3K9me3 to facilitate the targeting of the DNA methyltransferase DIM2 (Tamaru and Selker, 2003). In mammals, knockout of either G9a or Suv39 H1, Suv39 H2 results in reduced DNA methylation in mice (Ikegami et al., 2007). Moreover, H3K9 methylation is dependent on DNA methylation in human cancer cells (Espada et al., 2004). Likewise, in Arabidopsis, KYP, SUVH5, and SUVH6 are primarily recruited to the targets through SET and RING-associated (SRA) domain binding to DNA that is methylated in the CHG context (H stands for any base except G). H3K9me2 is known to recruit the DNA methyltransferases CMT2 and CMT3, which mediate CHH and CHG DNA methylation, respectively, in a feedback loop with H3K9me2 (Figure 2; Johnson et al., 2007; Bernatavichute et al., 2008; Du et al., 2012; Zemach et al., 2013; Stroud et al., 2014). Considering the targeting of H3K9 methyltransferases to CHG-methylated DNA, KYP, SUVH5, and SUVH6 have distinct DNA binding preferences. KYP, which is responsible for the majority of H3K9me2, has high affinity to the CWG (W stands for A or T) context but has low affinity to the CCG context (Li et al., 2018). The differential binding affinity is consistent with the phenotype that DNA methylation at CWG is strongly lost in kyp, but loss of CCG methylation is very low in kyp but high in suvh5 and suvh6. Consistent with the in vivo consequence of DNA methylation, SUVH5 has a preference for the CCG context, and SUVH6 can bind to both the CWG and CCG contexts, which act as a backup of KYP to ensure H3K9me2 in all CHG contexts (Figure 2).
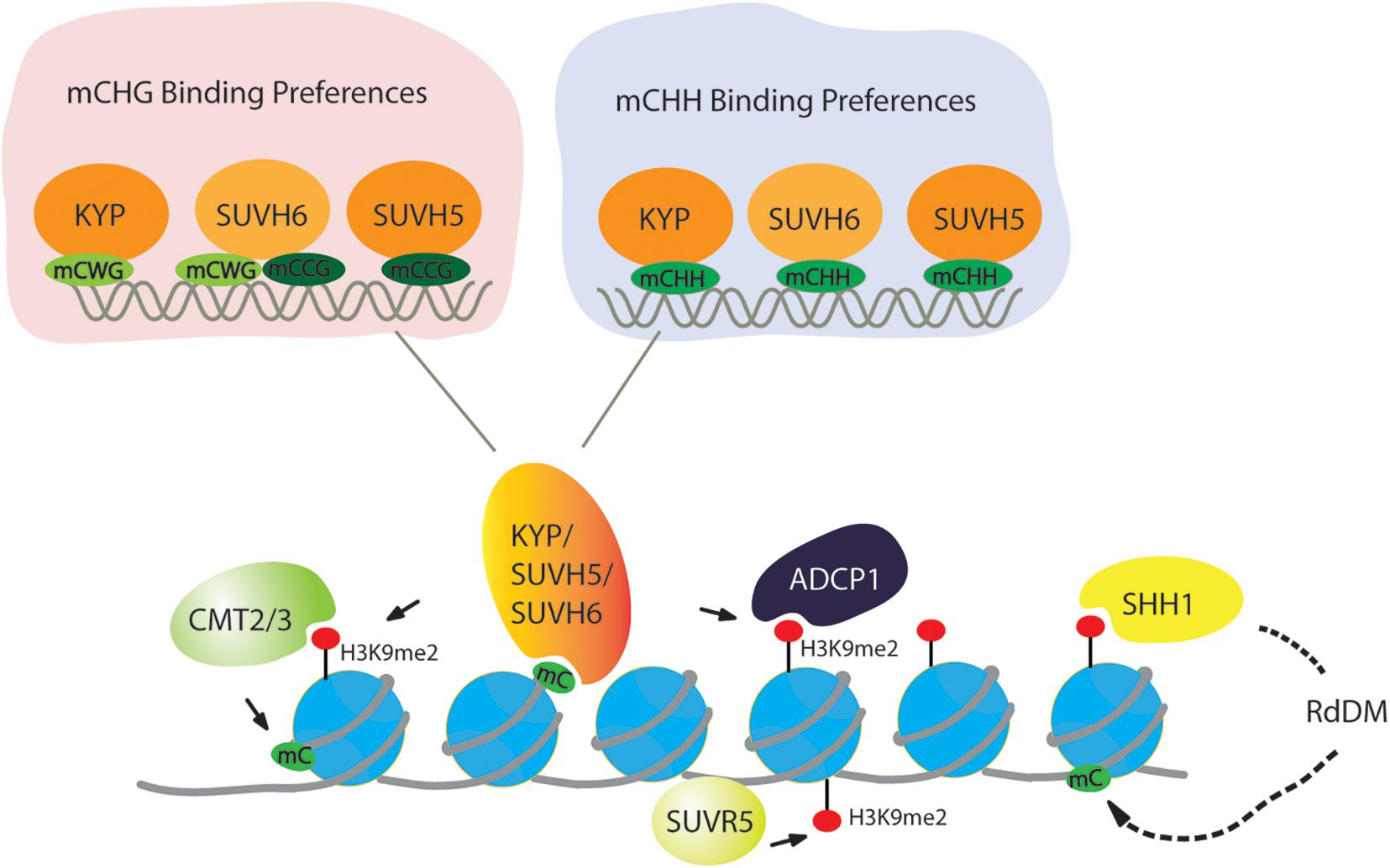
Figure 2. Targeting of H3K9 dimethylation in a DNA methylation-dependent and -independent manner. For DNA methylation, CMT2 and CMT3 recognize the H3K9me2 mark and catalyze DNA methylation in the CHG and CHH context (H = A, T, or C), respectively. For H3K9 methylation, histone methyltransferases KYP/SUVH4, SUVH5, and SUVH6 bind at methylated DNA in the CHG and CHH context to deposit H3K9me2, creating a reinforcing loop between DNA methylation and Histone modification. In the CHG context, KYP has a preference of mCWG (W = A or T), while SUVH5 and SUVH6 have a high affinity to the mCCG. All three SUVHs have similar sequence specificities at mCHH sites. SUVR5 binds to DNA through its zinc finger domain to facilitate H3K9me2 independently of DNA methylation. H3K9me2 is captured by SHH1, then through RdDM pathway to methylate DNA.
While the feedback loop between CHG DNA methylation is frequently discussed, CG and CHH methylation also contribute to H3K9me2 deposition through SRA domains of KYP, SUVH5, and SUVH6 binding to DNA that is methylated in CG or CHH context. In a large-scale comparative epigenome analysis, MET1 was indeed found to be required for the maintenance of CMT2-dependent asymmetric CHH methylation at loci with H3K9me2 (Zhang Y. et al., 2018). Moreover, SUVH5 and SUVH6 can bind to DNA that is methylated in the CG context in vitro (Li et al., 2018), supporting the view that CG methylation also contributes to H3K9me2 deposition. In addition to CG methylation, it has been known for many years that CHH methylation generated by the RNA-directed DNA methylation (RdDM) pathway is also involved in H3K9me2 deposition (Wierzbicki et al., 2008; Zheng et al., 2009; Shin et al., 2013; Liu Z.W. et al., 2014). Recent biochemical evidence indeed supports this hypothesis; all three SUVHs can bind to CHH-methylated DNA (Figure 2), and there is no sequence preference among the three SUVHs in targeting CHH-methylated DNA (Li et al., 2018).
Apart from the specificity of the SRA domain on the sequence context, other factors may also affect KYP, SUVH5, and SUVH6 targeting to methylated DNA. It was reported that SUVH4 and SUVH5 prefer to control transposable elements, but SUVH4 and SUVH6 prefer to target transcribed inverted repeat sources of dsRNA. Thus, in addition to DNA methylation states, chromatin state may also govern SUVH activities (Ebbs and Bender, 2006).
Apart from DNA methylation-dependent H3K9me2 deposition, there are known exceptions. G9a is one of the primary enzymes for H3K9me1 and H3K9me2 and usually interacts with another enzyme, GLP, to form a heteromeric complex that appears to be a functional H3K9 methyltransferase in vivo (Shinkai and Tachibana, 2011). In murine embryonic stem cells (mESCs), H3K9me2 at the newly integrated proviral LTR is reduced in cells with G9a silencing. Since there is no H3K9me2 or DNA methylation at the newly integrated region, G9a is considered to be responsible for de novo H3K9me2 (Leung et al., 2011). In addition to mESCs, G9a-dependent H3K9me2 has also been associated with gene repression in multiple human cell lines (Chen et al., 2009; Liu C. et al., 2014; Yoshida et al., 2015; Kramer, 2016; Scheer and Zaph, 2017).
Similar exceptions also exist in Arabidopsis. It was reported that SUVR5 is able to establish H3K9me2 independently of DNA methylation (Figure 2; Caro et al., 2012). Unlike KYP/SUVH5/SUVH6, SUVR5 does not have the SRA domain which can bind at methylated DNA but relies on a set of three C2H2 zinc fingers in tandem, which can bind at the sequence context of “TACTAGTA” in vitro. This motif also occurs at a minor part of transposable elements (TEs) and surrounds substantial genes losing H3K9me2 in suvr5, further supporting the role of zinc fingers in targeting H3K9me2 deposition. While H3K9 methyltransferases in yeast or mammals do not contain zinc fingers, DNA binding proteins recruiting H3K9 methyltransferases contain zinc fingers (Kim and Huang, 2003; Fog et al., 2012; Bian et al., 2015). In mammalian cells, ZNF644 has eight zinc finger motifs and WIZ contains 12 zinc finger motifs that are the binding partners of the G9a-GLP complex (Bian et al., 2015). The N-terminus of ZNF644 interacts with the transcriptional domain (TAD) of G9a, but the C-terminus of WIZ interacts with the TAD of GLP to facilitate the targeting of the G9a-GLP complex at specific genomic loci with the preference of the promoter region (Bian et al., 2015). Thus, it seems that targeting H3K9me2 by the zinc finger domain is a conserved mechanism in plants and mammals, but in plants, the zinc finger domain has been integrated into H3K9 methyltransferase. Interestingly, the combination of zinc fingers and a C-terminal SET domain can be found in all plant species (Caro et al., 2012). Thus, SUVR5 depositing H3K9me2 independently of DNA methylation might be conserved in plants. Another exception in Arabidopsis is SUVR4, which can bind to ubiquitin through the N-terminal WIYLD domain to facilitate the conversion from H3K9me1 to H3K9me3 in vitro, but it is not clear if the WIYLD domain binds to ubiquitin in vivo and if this domain binds to histone or other proteins with ubiquitination (Thorstensen et al., 2006; Veiseth et al., 2011). Recently, it was reported that the CRL4DCAF8 ubiquitin ligase is capable of targeting H3 for polyubiquitination at K79 in mice, which may further promote H3K9me2 deposition (Li et al., 2017), suggesting a similar connection between histone ubiquitination and H3K9me3. Taken together, the deposition of H3K9me2 is not only DNA methylation dependent but can also be independent.
The distribution of histone acetylation is usually anti-correlated with histone methylation, such as H3K9Ac and H3K9me2 (Zhou et al., 2010), indicating that the removal of H3K9Ac or relevant protein complexes functions in H3K9me2 deposition. Histone deacetylation is processed by histone deacetylases (HDACs), which play important roles in chromatin regulation (Liu X. et al., 2014). In mammals, SUV39H1 can interact with HDAC1 and HDAC2 (Vaute et al., 2002). Moreover, transcriptional repression by SUV39H1 is abolished by treatment with the HDAC inhibitor trichostatin A (TSA), indicating that the function of SUV39H1 is dependent on HDAC activity (Vaute et al., 2002). Likewise, in Arabidopsis, one of the HDACs, HDA6, also physically interacts with H3K9 methyltransferases KYP, SUVH5, and SUVH6, regulating a group of transposable elements and repetitive sequences (Yu et al., 2017). The mutant with compromised HDA6 has reduced H3K9me2 levels compared with the wild type, suggesting that H3K9me2 deposited by KYP, SUVH5, and SUVH6 is partly dependent on HDA6, but it is not clear that H3K9me2 deposition in Arabidopsis is dependent on the level of H3K9Ac at the targets or depends on the interaction between HDA6 and KYP/SUVH5/SUVH6, or perhaps both mechanisms exist in Arabidopsis, which has not been clearly dissected to date. Given the remaining H3K9me2 level in the hda6 mutant, it will be interesting to know if other HDACs are also involved in H3K9me2 deposition in the future.
Matrix attachment regions (MARs) are important for chromatin organization and gene expression (Tetko et al., 2006; Zhao et al., 2014). MARs are stretches of AT-rich sequences that guide the binding of DNA to the nuclear matrix by recruiting MAR-binding proteins. Proteins with AT-hook motifs bind to MARs and play roles in regulating H3K9me2 levels. In Neurospora crassa, CHAP, a protein with AT-hook motifs, was demonstrated to recognize heterochromatic regions through AT-hook motifs and to recruit the H3K9 methyltransferase DIM5 to targets (Honda et al., 2016). In Arabidopsis, overexpression of AT-hook motif nuclear localized 22 (AHL22) causes delayed flowering time by increasing H3K9me2 at MAR located in an intron of the FLOWERING LOCUS T (FT) locus (Xiao et al., 2009; Yun et al., 2012). AHL16 regulates the expression of the floral repressor genes FLOWERING LOCUS C (FLC) and FLOWERING WAGENINGEN (FWA) by adjusting the H3K9me2 level (Xu et al., 2013). Overexpressed AHL10 increases genome-wide H3K9me2 levels in the endosperm of triploid seeds (Jiang et al., 2017). Consistent with the distribution of MARs that are mainly in chromosome arms, TEs that are methylated by H3K9me2 via AHL10 in the endosperm of triploid seeds are usually euchromatic AT-rich TEs (Jiang et al., 2017). To date, it has not been determined how AHLs regulate H3K9me2 levels. While there is no direct physical interaction between AHLs and H3K9 methyltransferase in Arabidopsis, AHLs usually interact with HDAC complexes both in vitro and in vivo, such as AHL22 interacting with HDA1, HDA6, and HDA9 (Xiao et al., 2009) and AHL16 interacting with FVE and MSI5, which are core components of the HDA6 complex (Gu et al., 2011; Xu et al., 2013). Thus, it is possible that AHLs participate in H3K9me2 deposition through interaction with HDACs, especially HDA6. Nevertheless, other chromatin-relevant proteins also occur in AHL complexes, such as SUVH9 in the AHL10 complex. Apart from interacting with the DDR complex and mediating Pol V recruitment in RdDM (Johnson et al., 2014; Liu Z.W. et al., 2014), SUVH9 also interacts with MORC6 and its two close homologs, MORC1 and MORC2, required for heterochromatin condensation and formation of 3D chromatin architecture at SUPPRESSOR OF DRM1 DRM2 CMT3 (SDC) and Solo-LTR loci (Jing et al., 2016). Recently, the mammalian nuclear matrix protein scaffold attachment factor B (SAFB) was found to participate in stabilizing heterochromatin architecture partially through phase separation, which is a phenomenon in which different biological molecules spontaneously separate into two coexisting liquid phases and result in miscellaneous non-membrane-bound cellular compartments. Depletion of SAFB results in more interchromosomal interactions around pericentromeric heterochromatin and a decrease in genomic compartmentalization, which could result from the decondensation of pericentromeric heterochromatin (Huo et al., 2020). Thus, it is also possible that AHLs and MARs participate in H3K9me2 regulation by affecting heterochromatin architecture and phase separation.
H3K9 methylation recruits downstream effectors containing specific reader domains to further mediate gene silencing. In metazoans, heterochromatin protein 1 (HP1) is known to read the trimethylated lysine 9 residue of histone H3 (H3K9me3) (Bannister et al., 2001; Jacobs et al., 2001), which is a hallmark histone modification for transcriptionally silenced heterochromatin in mammals (Zeng et al., 2010). HP1 contains a conserved chromodomain (CD) at the N-terminus and a chromo shadow domain (CSD) at the C-terminus (Li et al., 2002). CD is able to directly bind to H3K9me3 (Jacobs et al., 2001). Based on sequence similarity and early biochemistry analyses, the homolog of HP1 in Arabidopsis, LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) was first proposed to be the H3K9me reader that mediates H3K9me2-dependent heterochromatic silencing, as it was shown to bind H3K9me2 in vitro (Jackson et al., 2002). Nevertheless, several lines of evidence have indicated that LHP1 is a plant-specific PRC1 H3K27me3 reader subunit. The CHROMO domain of LHP1 specifically binds to H3K27me3 but not H3K9me in Arabidopsis, and the genome-wide distribution of LHP1 displays significant overlap with H3K27me3-enriched sites (Turck et al., 2007; Exner et al., 2009; Lu et al., 2011). While SHH1, CMT2, and CMT3 have the capability to bind to histones with H3K9me2 (Law et al., 2013; Stroud et al., 2014; Figure 1), their function is to maintain or initiate non-CG methylation but not the downstream H3K9me2 reader; therefore, the reader of H3K9me2 in plants had not been determined until two recent studies were conducted in Arabidopsis. Agenet domain (AGD)-containing p1 (AGDP1), also known as ADCP1, appears to be a plant-specific H3K9 reader and functions as an HP1 equivalent protein (Zhang C. et al., 2018; Zhao et al., 2019). The tandem AGDs of AGDP1 can specifically recognize H3K9me2 and unmethylated K4 on the H3 tail (H3K4me0) through two negatively charged surface pockets. In structural studies, AGD12 adopts a tandem Tudor-like conformation, which resembles the human UHRF1 tandem Tudor and Arabidopsis SHH1 SAWADEE domains, both of which function as H3K9me2 readers with similar recognition mechanisms (Arita et al., 2012; Cheng et al., 2013; Law et al., 2013). ADCP1 is responsible for H3K9me2-dependent silencing, and the in vivo binding site of ADCP1 largely overlaps with the regions enriched by H3K9me2, further supporting that ADCP1 is indeed an H3K9me2 reader (Zhao et al., 2019).
While ADCP1 has been successfully identified, how ADCP1 mediates H3K9me2-dependent transcriptional silencing still needs to be discovered. Given that ADCP1 is essential for heterochromatin formation and TE silencing, but ADCP1 itself is only a histone binding protein without any repressor domain (Zhao et al., 2019), other chromatin modeling proteins must be recruited by ADCP1 to heterochromatin. Recently, SMC4, a core subunit of condensins I and II, was identified to act in conjunction with CG methylation, CHG methylation, the chromatin remodeler DDM1 (DECREASE IN DNA METHYLATION 1), and histone modifications, including H3K9me2 and H3K27me1 (Wang et al., 2017). Considering the function of SMC4 in H3K9me2-mediated transcriptional silencing (Wang et al., 2017), it is worth knowing whether SMC4 works together with ADCP1 to mediate heterochromatic silencing. Another possibility is that ADCP1 mediates downstream silencing by driving nucleosome phase separation. It has been demonstrated that human HP1α and Drosophila HP1a may demix from aqueous solution to form phase-separated droplets (Larson et al., 2017; Strom et al., 2017), which rapidly induce compacted chromatin. Similarly, ADCP1 can mediate heterochromatin phase separation together with reconstituted nucleosomes bearing H3K9me3 in vitro (Zhao et al., 2019). Thus, ADCP1 probably has a similar ability to mediate phase separation as the functional analog of mammalian HP1.
The mutant with compromised KYP, SUVH5, and SUVH6 has no obvious abnormality in development; thus, H3K9 was considered to play minor roles in Arabidopsis development. Nevertheless, the role of H3K9me2 in Arabidopsis development has been identified with more careful observations and new approaches.
The main H3K9me2 methyltransferase, KYP, was proven to repress primary seed dormancy by suppressing the expression of dormancy and ABA pathway-related genes, such as DOG1, which is a master regulator in the control of seed dormancy (Bentsink et al., 2006), and ABI3 and ABI4, which are components of ABA signaling (Koornneef et al., 2002; Zheng et al., 2012). However, evidence that H3K9me2 directly regulates the expression of these genes via H3K9me2 levels is not available. Until recently, SUVH5 was revealed to directly repress the expression of genes related to the ABA signaling pathway, DOG1, and its homologs via H3K9me2 in light-mediated seed germination (Gu et al., 2019). Thus, SUVH5-mediated H3K9me2 directly participates in controlling seed germination in Arabidopsis. After seed germination, plants enter the vegetative stage. While the role of H3K9me2 in the vegetative stage is not clear, H3K9me2 is crucial for the transition to flowering. Knockdown of AHL16 leads to obvious late flowering, which results in increased expression of two flowering repressors, FLOWERING LOCUS C (FLC) and FWA. Consistent with the increased expression pattern, the H3K9me2 levels at the intron of FLC and FWA loci were reduced. Interestingly, the phenotype of late flowering in the ahl16 mutant only occurs in Landsberg (Ler) accession but not in Columbia (Col) accession, indicating the ecotype-dependent regulation process (Xu et al., 2013).
During male meiosis, H3K9me2 is crucial for the distribution of meiotic recombination (Underwood et al., 2018). In plants, meiotic recombination is enriched in euchromatic regions, rather than pericentromeric heterochromatin, associated with H3K4me3 and histone variant H2A.Z but inversely correlated with DNA methylation. Suppression of meiotic recombination within the centromeric region is thought to be important for maintaining the fidelity of genome transmission during meiosis (Choi et al., 2018). Loss of DNA methylation in the met1 mutant leads to epigenetic activation of meiotic double-strand breaks (DSBs) in proximity to centromeres (Choi et al., 2018). In addition, non-CG methylation and the H3K9me2 pathway are also responsible for suppressing pericentromeric recombination (Underwood et al., 2018). Epigenetic activation of recombination and crossovers (COs) can be induced via loss of H3K9me2 and non-CG methylation in the kyp, suvh5, suvh6, or cmt3 mutant (Underwood et al., 2018), making it possible to induce COs near centromeres, which are otherwise very low-frequency CO regions in Arabidopsis and crops (Taagen et al., 2020).
In Arabidopsis thaliana and Arabidopsis lyrata seed development, H3K9me2 and CHG methylation are involved in the regulation of genomic imprinting that leads to differential expression of parent-of-origin alleles by maintaining or reinforcing the repression of maternal alleles of imprinted paternally expressed genes (PEGs) (Klosinska et al., 2016; Moreno-Romero et al., 2019). Moreover, the presence of the three repressive epigenetic marks H3K27me3, H3K9me2, and CHG methylation on the maternal alleles in endosperm can be considered a specific epigenetic signature of paternally expressed imprinted genes in the endosperm of Arabidopsis (Moreno-Romero et al., 2019). These marks are able to predict known PEGs at high accuracy and identify several new PEGs that were confirmed by INTACT-based endosperm transcriptomes (Moreno-Romero et al., 2019). In addition to maintaining genomic imprinting in the endosperm of diploid Arabidopsis seeds, H3K9me2 also functions in establishing a hybridization barrier from interploidy cross in the endosperm of triploid seeds (the triploid block) (Jiang et al., 2017). The triploid block acts as an instant reproductive barrier that prevents backcrossing of the newly formed polyploid plants with their progenitors (Schatlowski and Kohler, 2012). Multiple PEGs are enhanced in the endosperm of triploid seeds (Kradolfer et al., 2013; Wolff et al., 2015). Increased H3K9me2 levels in AT-rich TEs derived from overexpressed ADM and AHL10 contribute to enhancing the expression of PEGs, such as PEG2, which is a crucial component in establishing the triploid block. Moreover, H3K9me2 levels in AT-rich TEs are also associated with the different phenotypes of the triploid block in Col and Ler accessions.
In plants, H3K9 methylation, mainly H3K9me2, functions importantly in suppressing TEs and repetitive sequences, protecting plant genomes from TE transposition and genome instability. To enable plants to correctly deposit H3K9me2 in the genome, multiple H3K9 methyltransferases are in charge of H3K9me2 deposition in different sequence contexts via DNA methylation-dependent and -independent activities. Apart from playing a role in genome stability, H3K9me2 also plays roles in plant development and environmental stimuli. Recent studies have enhanced our understanding of the structure and recruitment of H3K9 methyltransferases and the downstream effector of H3K9me2, but open questions remain.
Given that H3K9me2 plays important roles in plant development and environmental stress, how the H3K9 methylation pathway is in response to developmental cues or environmental stimuli will be highly interesting to explore. In addition, our mechanistic understanding of downstream effectors of H3K9 methylation is also limited, while the H3K9me reader has been identified in Arabidopsis. The mechanism by which ADCP1 mediates transcriptional silencing, the existence of other H3K9me downstream effectors, the role of phase separation in chromatin condensation in vivo, and how H3K9me functions in response to developmental cues or environmental stimuli remains to be elucidated. Answering these questions will further broaden our understanding of H3K9 methylation-dependent transcriptional silencing.
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arita, K., Isogai, S., Oda, T., Unoki, M., Sugita, K., Sekiyama, N., et al. (2012). Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. U.S.A. 109, 12950–12955. doi: 10.1073/pnas.1203701109
Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., et al. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. doi: 10.1038/35065138
Baumbusch, L. O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., et al. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29, 4319–4333. doi: 10.1093/nar/29.21.4319
Bentsink, L., Jowett, J., Hanhart, C. J., and Koornneef, M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 17042–17047. doi: 10.1073/pnas.0607877103
Bernatavichute, Y. V., Zhang, X., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2008). Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3:e3156. doi: 10.1371/journal.pone.0003156
Bian, C., Chen, Q., and Yu, X. (2015). The zinc finger proteins ZNF644 and WIZ regulate the G9a/GLP complex for gene repression. eLife 4:e05606. doi: 10.7554/eLife.08168
Cao, X., and Jacobsen, S. E. (2002). Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144. doi: 10.1016/s0960-9822(02)00925-9
Caro, E., Stroud, H., Greenberg, M. V., Bernatavichute, Y. V., Feng, S., Groth, M., et al. (2012). The SET-domain protein SUVR5 mediates H3K9me2 deposition and silencing at stimulus response genes in a DNA methylation-independent manner. PLoS Genet. 8:e1002995. doi: 10.1371/journal.pgen.1002995
Charron, J. B., He, H., Elling, A. A., and Deng, X. W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21, 3732–3748. doi: 10.1105/tpc.109.066845
Chen, X., El Gazzar, M., Yoza, B. K., and Mccall, C. E. (2009). The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J. Biol. Chem. 284, 27857–27865. doi: 10.1074/jbc.M109.000950
Cheng, J., Yang, Y., Fang, J., Xiao, J., Zhu, T., Chen, F., et al. (2013). Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 288, 1329–1339. doi: 10.1074/jbc.M112.415398
Choi, K., Zhao, X., Tock, A. J., Lambing, C., Underwood, C. J., Hardcastle, T. J., et al. (2018). Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28, 532–546. doi: 10.1101/gr.225599.117
Ding, Y., Wang, X., Su, L., Zhai, J., Cao, S., Zhang, D., et al. (2007). SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19, 9–22. doi: 10.1105/tpc.106.048124
Dodge, J. E., Kang, Y. K., Beppu, H., Lei, H., and Li, E. (2004). Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486. doi: 10.1128/mcb.24.6.2478-2486.2004
Du, J., Zhong, X., Bernatavichute, Y. V., Stroud, H., Feng, S., Caro, E., et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180. doi: 10.1016/j.cell.2012.07.034
Du, J. M., Johnson, L. M., Groth, M., Feng, S. H., Hale, C. J., Li, S. S., et al. (2014). Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 55, 495–504. doi: 10.1016/j.molcel.2014.06.009
Du, J. M., Johnson, L. M., Jacobsen, S. E., and Patel, D. J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532. doi: 10.1038/nrm4043
Ebbs, M. L., and Bender, J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18, 1166–1176. doi: 10.1105/tpc.106.041400
Espada, J., Ballestar, E., Fraga, M. F., Garea, A. V., Juarranz, A., Stockert, J. C., et al. (2004). Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 279, 37175–37184. doi: 10.1074/jbc.M404842200
Exner, V., Aichinger, E., Shu, H., Wildhaber, T., Alfarano, P., Caflisch, A., et al. (2009). The Chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is Essential for H3K27me3 binding and function during arabidopsis development. PLoS One 4:e5335. doi: 10.1371/journal.pone.0005335
Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. U.S.A. 93, 8449–8454. doi: 10.1073/pnas.93.16.8449
Fog, C. K., Galli, G. G., and Lund, A. H. (2012). PRDM proteins: important players in differentiation and disease. Bioessays 34, 50–60. doi: 10.1002/bies.201100107
Gehring, M., Missirian, V., and Henikoff, S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6:e23687. doi: 10.1371/journal.pone.0023687
Gu, D. C., Ji, R. J., He, C. M., Peng, T., Zhang, M. Y., Duan, J., et al. (2019). Arabidopsis histone methyltransferase SUVH5 is a positive regulator of light-mediated seed germination. Front. Plant Sci. 10:841. doi: 10.3389/fpls.2019.00841
Gu, X., Jiang, D., Yang, W., Jacob, Y., Michaels, S. D., and He, Y. (2011). Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 7:e1002366. doi: 10.1371/journal.pgen.1002366
Han, Y. F., Dou, K., Ma, Z. Y., Zhang, S. W., Huang, H. W., Li, L., et al. (2014). SUVR2 is involved in transcriptional gene silencing by associating with SNF2-related chromatin-remodeling proteins in Arabidopsis. Cell Res. 24, 1445–1465. doi: 10.1038/cr.2014.156
Harris, C. J., Scheibe, M., Wongpalee, S. P., Liu, W. L., Cornett, E. M., Vaughan, R. M., et al. (2018). A DNA methylation reader complex that enhances gene transcription. Science 362, 1182–1186. doi: 10.1126/science.aar7854
Honda, S., Bicocca, V. T., Gessaman, J. D., Rountree, M. R., Yokoyama, A., Yu, E. Y., et al. (2016). Dual chromatin recognition by the histone deacetylase complex HCHC is required for proper DNA methylation in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 113, E6135–E6144. doi: 10.1073/pnas.1621475114
Huo, X. R., Ji, L. Z., Zhang, Y. W., Lv, P., Cao, X., Wang, Q. F., et al. (2020). The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol. Cell 77, 368–383.e7. doi: 10.1016/j.molcel.2019.10.001
Ikegami, K., Iwatani, M., Suzuki, M., Tachibana, M., Shinkai, Y., Tanaka, S., et al. (2007). Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells 12, 1–11. doi: 10.1111/j.1365-2443.2006.01029.x
Jackson, J. P., Johnson, L., Jasencakova, Z., Zhang, X., Perezburgos, L., Singh, P. B., et al. (2004). Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112, 308–315. doi: 10.1007/s00412-004-0275-7
Jackson, J. P., Lindroth, A. M., Cao, X., and Jacobsen, S. E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560. doi: 10.1038/nature731
Jacobs, S. A., Taverna, S. D., Zhang, Y., Briggs, S. D., Li, J., Eissenberg, J. C., et al. (2001). Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20, 5232–5241. doi: 10.1093/emboj/20.18.5232
Jenuwein, T., Laible, G., Dorn, R., and Reuter, G. (1998). SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 54, 80–93. doi: 10.1007/s000180050127
Jiang, H., Moreno-Romero, J., Santos-Gonzalez, J., De Jaeger, G., Gevaert, K., Van De Slijke, E., et al. (2017). Ectopic application of the repressive histone modification H3K9me2 establishes post-zygotic reproductive isolation in Arabidopsis thaliana. Genes Dev. 31, 1272–1287. doi: 10.1101/gad.299347.117
Jing, Y., Sun, H., Yuan, W., Wang, Y., Li, Q., Liu, Y., et al. (2016). SUVH2 and SUVH9 couple two essential steps for transcriptional gene silencing in Arabidopsis. Mol. Plant 9, 1156–1167. doi: 10.1016/j.molp.2016.05.006
Johnson, L. M., Bostick, M., Zhang, X., Kraft, E., Henderson, I., Callis, J., et al. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17, 379–384. doi: 10.1016/j.cub.2007.01.009
Johnson, L. M., Du, J., Hale, C. J., Bischof, S., Feng, S., Chodavarapu, R. K., et al. (2014). SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128. doi: 10.1038/nature12931
Johnson, L. M., Law, J. A., Khattar, A., Henderson, I. R., and Jacobsen, S. E. (2008). SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 4:e1000280. doi: 10.1371/journal.pgen.1000280
Kim, K. C., and Huang, S. (2003). Histone methyltransferases in tumor suppression. Cancer Biol. Ther. 2, 491–499.
Klosinska, M., Picard, C. L., and Gehring, M. (2016). Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat. Plants 2:16145. doi: 10.1038/nplants.2016.145
Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36.
Kradolfer, D., Wolff, P., Jiang, H., Siretskiy, A., and Kohler, C. (2013). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26, 525–535. doi: 10.1016/j.devcel.2013.08.006
Kramer, J. M. (2016). Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP. Biochem. Cell Biol. 94, 26–32. doi: 10.1139/bcb-2015-0017
Larson, A. G., Elnatan, D., Keenen, M. M., Trnka, M. J., Johnston, J. B., Burlingame, A. L., et al. (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240. doi: 10.1038/nature22822
Law, J. A., Du, J. M., Hale, C. J., Feng, S. H., Krajewski, K., Palanca, A. M. S., et al. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389. doi: 10.1038/nature12178
Lei, L., Zhou, S. L., Ma, H., and Zhang, L. S. (2012). Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 12:51. doi: 10.1186/1471-2148-12-51
Leung, D. C., Dong, K. B., Maksakova, I. A., Goyal, P., Appanah, R., Lee, S., et al. (2011). Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl. Acad. Sci. U.S.A. 108, 5718–5723. doi: 10.1073/pnas.1014660108
Li, G., Ji, T., Chen, J., Fu, Y., Hou, L., Feng, Y., et al. (2017). CRL4(DCAF8) ubiquitin ligase targets histone H3K79 and promotes H3K9 methylation in the liver. Cell Rep. 18, 1499–1511. doi: 10.1016/j.celrep.2017.01.039
Li, X., Harris, C. J., Zhong, Z., Chen, W., Liu, R., Jia, B., et al. (2018). Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 115, E8793–E8802. doi: 10.1073/pnas.1809841115
Li, Y. H., Kirschmann, D. A., and Wallrath, L. L. (2002). Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. U.S.A. 99, 16462–16469. doi: 10.1073/pnas.162371699
Lindroth, A. M., Cao, X., Jackson, J. P., Zilberman, D., Mccallum, C. M., Henikoff, S., et al. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080. doi: 10.1126/science.1059745
Liu, C., Yu, Y., Liu, F., Wei, X., Wrobel, J. A., Gunawardena, H. P., et al. (2014). A chromatin activity-based chemoproteomic approach reveals a transcriptional repressome for gene-specific silencing. Nat. Commun. 5:5733. doi: 10.1038/ncomms6733
Liu, X., Yang, S., Zhao, M., Luo, M., Yu, C. W., Chen, C. Y., et al. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7, 764–772. doi: 10.1093/mp/ssu033
Liu, Z. W., Shao, C. R., Zhang, C. J., Zhou, J. X., Zhang, S. W., Li, L., et al. (2014). The SET domain proteins SUVH2 and SUVH9 are required for Pol V Occupancy at RNA-Directed DNA methylation loci. PLoS Genet. 10:e1003948. doi: 10.1371/journal.pgen.1003948
Lu, F. L., Cui, X., Zhang, S. B., Jenuwein, T., and Cao, X. F. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Moreno-Romero, J., Del Toro-De Leon, G., Yadav, V. K., Santos-Gonzalez, J., and Kohler, C. (2019). Epigenetic signatures associated with imprinted paternally expressed genes in the Arabidopsis endosperm. Genome Biol. 20:41. doi: 10.1186/s13059-019-1652-0
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D., and Grewal, S. I. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113. doi: 10.1126/science.1060118
Naumann, K., Fischer, A., Hofmann, I., Krauss, V., Phalke, S., Irmler, K., et al. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24, 1418–1429. doi: 10.1038/sj.emboj.7600604
Peters, A. H., Kubicek, S., Mechtler, K., O’sullivan, R. J., Derijck, A. A., Perez-Burgos, L., et al. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589. doi: 10.1016/s1097-2765(03)00477-5
Rice, J. C., Briggs, S. D., Ueberheide, B., Barber, C. M., Shabanowitz, J., Hunt, D. F., et al. (2003). Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598. doi: 10.1016/s1097-2765(03)00479-9
Roudier, F., Teixeira, F. K., and Colot, V. (2009). Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet. 25, 511–517. doi: 10.1016/j.tig.2009.09.013
Saksouk, N., Simboeck, E., and Dejardin, J. (2015). Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 8:3. doi: 10.1186/1756-8935-8-3
Schatlowski, N., and Kohler, C. (2012). Tearing down barriers: understanding the molecular mechanisms of interploidy hybridizations. J. Exp. Bot. 63, 6059–6067. doi: 10.1093/jxb/ers288
Scheer, S., and Zaph, C. (2017). The lysine methyltransferase G9a in immune cell differentiation and function. Front. Immunol. 8:429. doi: 10.3389/fimmu.2017.00429
Shin, J. H., Wang, H. L., Lee, J., Dinwiddie, B. L., Belostotsky, D. A., and Chekanova, J. A. (2013). The role of the Arabidopsis Exosome in siRNA-independent silencing of heterochromatic loci. PLoS Genet. 9:e1003411. doi: 10.1371/journal.pgen.1003411
Shinkai, Y., and Tachibana, M. (2011). H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 25, 781–788. doi: 10.1101/gad.2027411
Sims, R. J. III, Nishioka, K., and Reinberg, D. (2003). Histone lysine methylation: a signature for chromatin function. Trends Genet. 19, 629–639. doi: 10.1016/j.tig.2003.09.007
Strom, A. R., Emelyanov, A. V., Mir, M., Fyodorov, D. V., Darzacq, X., and Karpen, G. H. (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245. doi: 10.1038/nature22989
Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., et al. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72. doi: 10.1038/nsmb.2735
Taagen, E., Bogdanove, A. J., and Sorrells, M. E. (2020). Counting on crossovers: controlled recombination for plant breeding. Trends Plant Sci. doi: 10.1016/j.tplants.2019.12.017 [Epub ahead of print]
Tamaru, H., and Selker, E. U. (2003). Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol. Cell Biol. 23, 2379–2394. doi: 10.1128/mcb.23.7.2379-2394.2003
Tetko, I. V., Haberer, G., Rudd, S., Meyers, B., Mewes, H. W., and Mayer, K. F. (2006). Spatiotemporal expression control correlates with intragenic scaffold matrix attachment regions (S/MARs) in Arabidopsis thaliana. PLoS Comput. Biol. 2:e21. doi: 10.1371/journal.pcbi.0020021
Thorstensen, T., Fischer, A., Sandvik, S. V., Johnsen, S. S., Grini, P. E., Reuter, G., et al. (2006). The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res. 34, 5461–5470. doi: 10.1093/nar/gkl687
Trojer, P., and Reinberg, D. (2007). Facultative heterochromatin: Is there a distinctive molecular signature? Mol. Cell 28, 1–13. doi: 10.1016/j.molcel.2007.09.011
Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M. L., Guillaume, E., Buisine, N., et al. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3:e86. doi: 10.1371/journal.pgen.0030086
Underwood, C. J., Choi, K., Lambing, C., Zhao, X., Serra, H., Borges, F., et al. (2018). Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28, 519–531. doi: 10.1101/gr.227116.117
Vaute, O., Nicolas, E., Vandel, L., and Trouche, D. (2002). Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 30, 475–481. doi: 10.1093/nar/30.2.475
Veiseth, S. V., Rahman, M. A., Yap, K. L., Fischer, A., Egge-Jacobsen, W., Reuter, G., et al. (2011). The SUVR4 histone lysine methyltransferase binds ubiquitin and converts H3K9me1 to H3K9me3 on transposon chromatin in Arabidopsis. PLoS Genet. 7:e1001325. doi: 10.1371/journal.pgen.1001325
Wang, J., Blevins, T., Podicheti, R., Haag, J. R., Tan, E. H., Wang, F., et al. (2017). Mutation of Arabidopsis SMC4 identifies condensin as a corepressor of pericentromeric transposons and conditionally expressed genes. Genes Dev. 31, 1601–1614. doi: 10.1101/gad.301499.117
Wierzbicki, A. T., Haag, J. R., and Pikaard, C. S. (2008). Noncoding transcription by RNA Polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. doi: 10.1016/j.cell.2008.09.035
Wolff, P., Jiang, H., Wang, G., Santos-Gonzalez, J., and Kohler, C. (2015). Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. eLife 4:e10074. doi: 10.7554/eLife.10074
Wolff, P., Weinhofer, I., Seguin, J., Roszak, P., Beisel, C., Donoghue, M. T., et al. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis Endosperm. PLoS Genet. 7:e1002126. doi: 10.1371/journal.pgen.1002126
Wu, H., Min, J., Lunin, V. V., Antoshenko, T., Dombrovski, L., Zeng, H., et al. (2010). Structural biology of human H3K9 methyltransferases. PLoS One 5:e8570. doi: 10.1371/journal.pone.0008570
Xiao, C., Chen, F., Yu, X., Lin, C., and Fu, Y. F. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50. doi: 10.1007/s11103-009-9507-9
Xiao, X., Zhang, J., Li, T., Fu, X., Satheesh, V., Niu, Q., et al. (2019). A group of SUVH methyl-DNA binding proteins regulate expression of the DNA demethylase ROS1 in Arabidopsis. J. Integr. Plant Biol. 61, 110–119. doi: 10.1111/jipb.12768
Xu, Y. F., Wang, Y. Z., Stroud, H., Gu, X. F., Sun, B., Gan, E. S., et al. (2013). A matrix protein silences transposons and repeats through interaction with retinoblastoma-associated proteins. Curr. Biol. 23, 345–350. doi: 10.1016/j.cub.2013.01.030
Yoshida, K., Maekawa, T., Zhu, Y., Renard-Guillet, C., Chatton, B., Inoue, K., et al. (2015). The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat. Immunol. 16, 1034–1043. doi: 10.1038/ni.3257
Yu, C. W., Tai, R., Wang, S. C., Yang, P., Luo, M., Yang, S., et al. (2017). HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 29, 1970–1983. doi: 10.1105/tpc.16.00570
Yun, J., Kim, Y. S., Jung, J. H., Seo, P. J., and Park, C. M. (2012). The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 287, 15307–15316. doi: 10.1074/jbc.M111.318477
Zemach, A., Kim, M. Y., Hsieh, P. H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., et al. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205. doi: 10.1016/j.cell.2013.02.033
Zeng, W., Ball, A. R. Jr., and Yokomori, K. (2010). HP1: heterochromatin binding proteins working the genome. Epigenetics 5, 287–292. doi: 10.4161/epi.5.4.11683
Zhang, C., Du, X., Tang, K., Yang, Z., Pan, L., Zhu, P., et al. (2018). Arabidopsis AGDP1 links H3K9me2 to DNA methylation in heterochromatin. Nat. Commun. 9:4547. doi: 10.1038/s41467-018-06965-w
Zhang, L. S., and Ma, H. (2012). Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 195, 248–263. doi: 10.1111/j.1469-8137.2012.04143.x
Zhang, Y., Harris, C. J., Liu, Q., Liu, W., Ausin, I., Long, Y., et al. (2018). Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E1069–E1074. doi: 10.1073/pnas.1716300115
Zhao, J. F., Favero, D. S., Qiu, J. W., Roalson, E. H., and Neff, M. M. (2014). Insights into the evolution and diversification of the AT-hook Motif Nuclear Localized gene family in land plants. BMC Plant Biol. 14:266. doi: 10.1186/s12870-014-0266-7
Zhao, S., Cheng, L., Gao, Y., Zhang, B., Zheng, X., Wang, L., et al. (2019). Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation. Cell Res. 29, 54–66. doi: 10.1038/s41422-018-0104-9
Zheng, B., Wang, Z., Li, S., Yu, B., Liu, J. Y., and Chen, X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23, 2850–2860. doi: 10.1101/gad.1868009
Zheng, J., Chen, F., Wang, Z., Cao, H., Li, X., Deng, X., et al. (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 193, 605–616. doi: 10.1111/j.1469-8137.2011.03969.x
Zhou, J., Wang, X., He, K., Charron, J. B., Elling, A. A., and Deng, X. W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72, 585–595. doi: 10.1007/s11103-009-9594-7
Keywords: epigenetics, histone, heterochromatin, H3K9 methylation, transcriptional silencing
Citation: Xu L and Jiang H (2020) Writing and Reading Histone H3 Lysine 9 Methylation in Arabidopsis. Front. Plant Sci. 11:452. doi: 10.3389/fpls.2020.00452
Received: 22 November 2019; Accepted: 27 March 2020;
Published: 06 May 2020.
Edited by:
Iva Mozgova, Centre for Biology, Academy of Sciences of the Czech Republic, CzechiaReviewed by:
Paul Fransz, University of Amsterdam, NetherlandsCopyright © 2020 Xu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Jiang, amlhbmdoQGlway1nYXRlcnNsZWJlbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.